Abstract
Background and Aim: MicroRNAs, dysregulated in the circulation of esophageal squamous cell carcinoma (ESCC) patient, have been assumed to be with great potential in the diagnosis and prognosis of esophageal cancer. We aimed to review previous articles on ESCC.
Methods: A search of electronic databases was performed before Nov 12, 2017. We summarized the identification of microRNA imbalance in the blood of ESCC compared with the healthy controls, with the objective to evaluate the efficiency of microRNAs in diagnosing and forecasting ESCC.
Results: A total of 35 studies investigating plasma or serum microRNAs were included in the meta-analysis. Based on the consequences of the quality assessment of each study, the articles involved were appropriate for quantitative synthesis. For diagnostic meta-analysis. The overall pooled sensitivity, specificity, and area under the curve of circulating microRNA is 0.794 (95% CI: 0.765 - 0.820), 0.779 (95%CI: 0.746 - 0.808), 0.86 (95%CI: 0.82 - 0.88). The diagnostic value of each microRNA was calculated respectively. For prognostic meta-analysis, the overall pooled hazard ratios of higher microRNA expression in circulation was 1.34 (95% CI: 1.14-1.58), which could significantly predict poorer survival in ESCC.
Conclusions: Circulating microRNAs distinguish patients with ESCC from healthy controls with high sensitivity and specificity, compared to other invasive currently used screening methods. Simultaneously, there was prognostic value for the prognosis of ESCC.
Keywords: Esophageal neoplasms, microRNA, diagnosis, prognosis, systematic review, meta-analysis
Introduction
Cancer is currently the secondary lethal cause in the world, only inferior to cardiovascular disease 1,2. Esophageal squamous cell carcinoma (ESCC), with the tenth highest cancer morbidity and the sixth highest mortality rate, gradually gained worldwide attention 3. Although with effective and positive treatment, there are still serious challenges waiting to be resolved about the diagnosis and prognosis of esophageal cancer 4. Nowadays, endoscopy examination and pathological biopsy are still the golden standard methods for detecting ESCC, while imaging examination lacks a certain timeliness due to its insensitivity to small lesions 5,6. Nevertheless, patients tend to be reluctant to carry out endoscopic examination because of its intrusiveness and discomfortableness. Conventional biomarkers, such as carcinoembryonic antigen (CEA), carbohydrate antigen 19-9 (CA19-9) and squamous cell carcinoma antigen (SCC), are short of sensitivity and specificity to accelerate the early detection of cancer 7-9.
There aren't highly effective prognostic molecular markers that can forecast the clinical outcome and hereafter furnish guidance for treatment. An aberrant exaltation of the serum SCC antigen level is an effective predictor of advanced esophageal cancer correlative to poor survival after esophagostomy. Serum CEA levels are assumed to be significant in predicting clinically unapparent distant metastasis 7. Hence, there is an enormous requirement to probe fresh and efficient means for ESCC prognosis.
MicroRNAs, non-protein-coding RNA molecules, play an important role in cell differentiation, cell-cycle progression, apoptosis, and tumorigenesis 10,11. Substantial researches have been performed on the appliance of microRNA expression to distinguish between ESCC patients and healthy controls, suggesting the great capacity of microRNA as a novel biomarker in screening ESCC. In the meantime, based on considerable evidences, microRNAs are deemed to be an effective predictor of the clinical outcome owing to its expression level is significantly related to the prognosis of ESCC patients. Therefore, it is essential to summarize the diagnostic efficiencies of these microRNAs via a systematic review.
However, one of the meta-analyses investigated the value of diagnosis and prognosis of single microRNA 12. Moreover, several researchers combined various microRNAs to get conclusions about the value of all microRNAs in ESCC, but overlooked the heterogeneity in diverse microRNAs from inconsistent sample sources 13-16. Taking into consideration the drawbacks of previous publications, a more integrative meta-analysis of microRNA for ESCC, on the basis of all relevant prior studies, was conducted to acquire a better understanding of the diagnostic and prognostic efficiency of microRNA in ESCC.
Methods
Search strategy
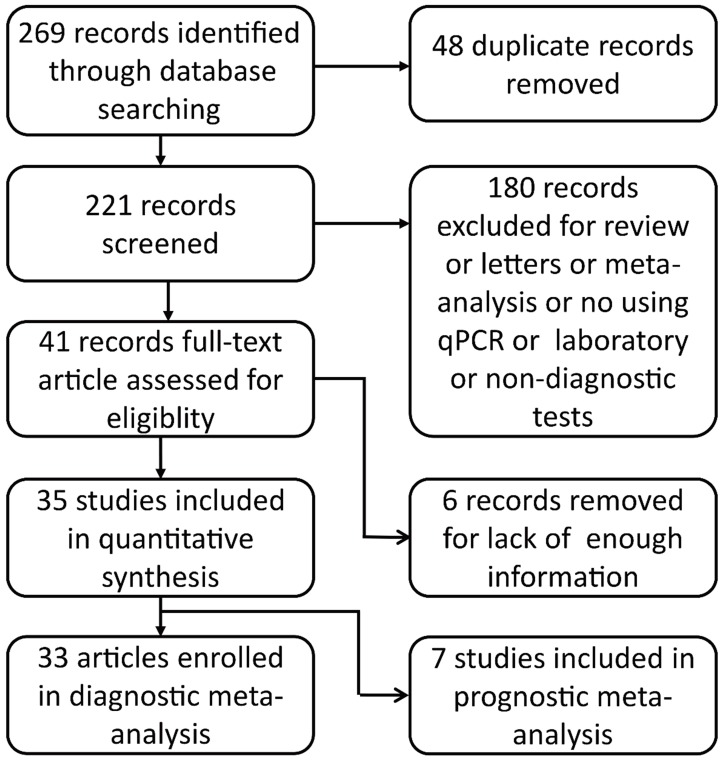
An electronic search of PubMed, Embase and the Chinese Biomedical Literature Database (CBM) was performed for relevant articles published until Nov 12, 2017. The search strategy was (miRNA OR microRNA OR miR) AND ("esophageal neoplasms"[Mesh] OR "esophageal squamous cell carcinoma" OR "esophageal carcinoma" OR "esophageal adenocarcinoma") AND (blood OR serum OR plasma OR circulating) AND (diagnosis OR diagnostic OR diagnose OR prognosis OR prognoses OR prognose OR predict OR prognostic). No language restrictions were set. Duplicates were removed. By screening the title and abstract, eligible manuscripts were obtained for full-text review. The flow-process diagram for the literature is presented in Fig. 1.
Fig 1.
Flow-process diagram.
Inclusion criteria and exclusion criteria
For eligible studies to be enrolled, the following criteria had to be fulfilled 17: (1) studies were conducted comparing ESCC patients versus healthy controls; (2) samples were restricted to serum or plasma; and (3) methods had to include quantitative real-time PCR techniques. Articles were excluded based on the following criteria: (1) review articles, letters or meta-analysis, (2) studies with duplicate data reported in other studies, (3) laboratory studies.
Quality assessment
Quality of all studies included in meta-analysis are systematically assessed based on the criteria as proposed by the Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) 18. For prognostic meta-analysis, information and data were analyzed as previously described.
Data extraction
Each study for diagnosis and prognosis was retrieved and assessed independently by two investigators (CY and HNL). Any discrepancies were resolved by consensus. The extracted data and information included as following: the first author, year of publication, country of origin, ethnicity, characteristics of cases, characteristics of controls, type of blood-based fluid (serum or plasma), reference microRNA and clinical outcomes.
Statistical methods
Diagnostic meta-analysis was performed to determined sensitivity, specificity and area under the curve (AUC) of the summary receiver operating curve (SROC) of all microRNAs identified by previous diagnostic tests 19. For prognostic meta-analysis, hazard ratios (HRs) and 95% confidence intervals (CIs) were calculated. Forest plots were used to estimate the effect of microRNA expression on overall survival (OS) and progression free survival (PFS). The percentage of Higgins I-squared statistic (I2) was used to quantify the heterogeneity. A random effect model would be used if I2 > 50%, otherwise the fixed effect model was applied. Publication bias of diagnostic tests was considered by Deeks' funnel plot asymmetry test 20, while that of prognostic tests was estimated by Egger's test 21 and Begg's test 22, respectively. Level of significance was set at p < 0.05. All data analyses were computed with the Stata 12.0 (StataCorp LP, College Station, TX, USA) and Revman 5.2 (The Nordic Cochrane Centre, Copenhagen, Denmark).
Results
Literature search
269 records from the database search were initially identified. We removed 48 duplicates, 180 irrelevant studies and six articles that failed to provide necessary statistical data. 35 published studies were finally included in this systematic review and meta-analysis 12,23-56. In these studies, 33 articles provided diagnostic information and seven were included in prognostic meta-analysis. 3156 ESCC patients and 2304 healthy controls were included in diagnostic meta-analysis, whereas there were 984 patients in prognostic meta-analysis. The main characteristics of the included articles were listed in Table 1.
Table 1.
Characteristics of the included studies.
| Article ID | First Author | Published Year | Country | Ethnicity | ESCC group | Control group | Specimen | MicroRNA | Reference RNA | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample size | Mean age | Gender | Sample size | Mean age | Gender | ||||||||
| 1 | Zhang C | 2010 | China | Asian | 149 | 61.3 | 116/33 | 100 | 60.0 | 74/26 | Serum | 10a, 22, 100, 127-3p, 133a, 148b, 223 | U6 |
| 2 | Xu H | 2015 | China | Asian | 50 | 60.0 | 27/23 | 50 | 60.0 | 30/20 | Serum | 10b, 29c, 205 | U6 |
| 3 | Xie ZJ | 2013 | China | Asian | 29 | 61.3 | 25/4 | 16 | 57.5 | 13/3 | Serum | 10b | miR-16 |
| 4 | Li JL | 2017 | China | Asian | 106 | 62.3 | 70/36 | 106 | 61.8 | 68/38 | Serum | 15a | U6 |
| 5 | Li BX | 2015 | China | Asian | 38 | - | - | 19 | - | - | Plasma | 16, 21, 185, 375 | miR-1228 |
| 6 | Zhou X | 2017 | China | Asian | 137 | - | - | 155 | - | - | Plasma | 18a, 20b, 106a, 223-3p, 486-5p, 584 | miR-1228 |
| 7 | Hirajima S | 2013 | Japan | Asian | 106 | - | 87/19 | 54 | - | - | Serum | 18a | U6 |
| 8 | Bai YY | 2017 | China | Asian | 89 | 58.0 | 69/20 | 125 | - | - | Plasma | 19a | miR-39 |
| 9 | He FC | 2015 | China | Asian | 70 | 60.5 | 46/24 | 40 | 61.7 | 26/14 | Plasma | 20a | let-7a |
| 10 | Lv HB | 2016 | China | Asian | 126 | 59.3 | 60/66 | 80 | 58.6 | 44/36 | Serum | 21,375 | U6 |
| 11 | Ye MH | 2014 | China | Asian | 100 | - | - | 50 | - | - | Serum | 21 | miR-16 |
| 12 | Komatsu S | 2011 | Japan | Asian | 50 | - | - | 20 | - | - | Serum | 21,375 | U6 |
| 13 | Li W | 2015 | China | Asian | 112 | 54.2 | 65/47 | 100 | 54.2 | 52/48 | Serum | 21 | miR-16 |
| 14 | Guo WT | 2016 | China | Asian | 60 | 52.6 | 37/23 | 60 | 53.1 | 36/24 | Serum | 21 | U6 |
| 15 | Dong W | 2015 | China | Asian | 105 | - | 69/36 | 30 | - | - | Serum | 24 | cel-miR-39 |
| 16 | Komatsu S | 2014 | Japan | Asian | 20 | - | - | 50 | - | - | Plasma | 25 | U6 |
| 17 | Wu CY | 2014 | China | Asian | 28 | - | - | 28 | - | - | Serum | 25, 100, 193a-3p, 194, 223, 337-5p, 483-5p | U6 |
| 18 | Wu CH | 2014 | China | Asian | 194 | 61.4 | 79/115 | 98 | - | - | Serum | 25,223,375 | U6, miR-16 |
| 19 | Zhang TF | 2011 | China | Asian | 120 | 56.3 | 79/41 | 121 | 58.3 | 76/45 | Serum | 31 | miR-16 |
| 20 | She XY | 2016 | China | Asian | 40 | 56.8 | 24/16 | 50 | 55.0 | 28/22 | Serum | 100 | U6 |
| 21 | Zheng SY | 2017 | China | Asian | 128 | 60.0 | 76/52 | 40 | - | - | Serum | 138 | U6 |
| 22 | Wang C | 2015 | China | Asian | 154 | - | - | 154 | - | - | Serum | 146a | miR-16 |
| 23 | Jing RR | 2015 | China | Asian | 28 | - | - | 35 | - | - | Plasma | 185 | U6 |
| 24 | Tanaka K | 2013 | Japan | Asian | 144 | - | - | - | - | - | Serum | 200c | cel-miR-39 |
| 25 | Dong SL | 2016 | China | Asian | 120 | - | 79/41 | 51 | - | - | Plasma | 216a, 216b | miR-16 |
| 26 | Jiang ZJ | 2015 | China | Asian | 106 | 62.0 | 69/37 | 60 | - | - | Serum | 218 | miR-16 |
| 27 | Hui BN | 2015 | China | Asian | 78 | 58.6 | 57/21 | 23 | - | - | Serum | 365 | miR-1228 |
| 28 | Sun JT | 2016 | China | Asian | 35 | 61.5 | 29/6 | - | - | - | Serum | 367 | U6 |
| 29 | Yang Y | 2017 | China | Asian | 50 | 68.0 | 30/20 | 20 | 66.0 | - | Serum | 451 | miR-2911 |
| 30 | Li SP | 2016 | China | Asian | 110 | 59.2 | 55/45 | 40 | - | - | Plasma | 506 | U6 |
| 31 | Guan SH | 2015 | China | Asian | 75 | 65.0 | 43/32 | 75 | - | - | Serum | 613 | U6 |
| 32 | Sun L | 2016 | China | Asian | 120 | 63.0 | 79/41 | 51 | - | - | Plasma | 718 | miR-16 |
| 33 | Takeshita N | 2013 | Japan | Asian | 101 | 65.0 | 89/12 | 46 | - | - | Serum | 1246 | miR-16 |
| 34 | Wang C | 2013 | China | Asian | 156 | - | 81/75 | 156 | - | - | Serum | 1297 | U6, miR-16 |
| 35 | Zhang TF | 2012 | China | Asian | 201 | 58.2 | 128/73 | 201 | 55.7 | 118/83 | Serum | 1322 | U6 |
“-” represents unclear.
Abbreviations: ESCC, esophageal squamous cell carcinoma.
Quality assessment
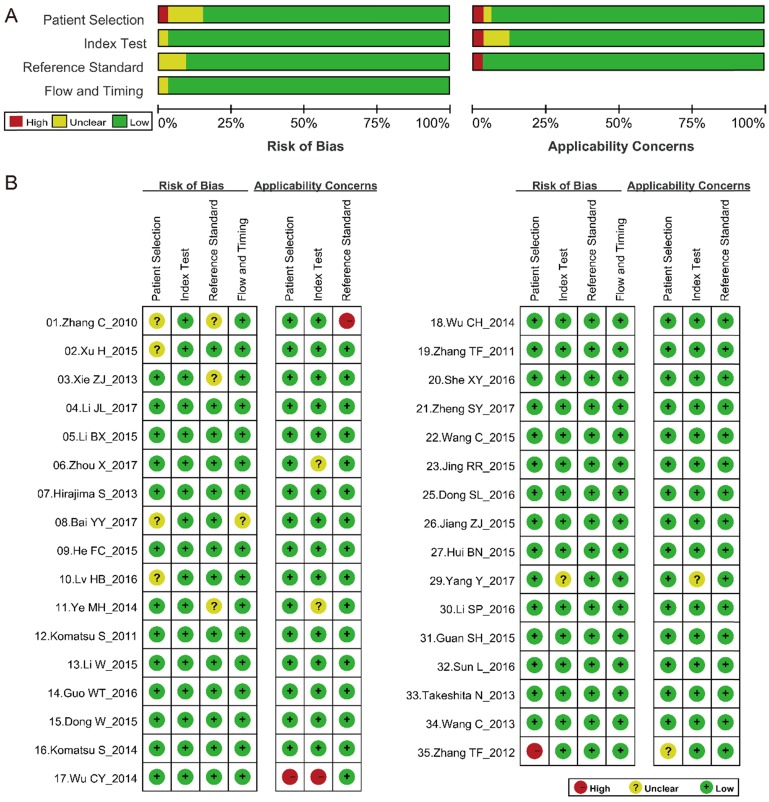
We assessed the quality of the 33 studies according to the QUADAS-2 assessment tool (Fig. 2). Of the articles included, three were considered to be deficient in patient selection and reference standards 48,55,56, while other articles were considered to have higher quality.
Fig 2.
QUADAS-2 quality assessment. Investigators' assessment regarding each domain for included studies. Risk of bias and applicability concerns' (A) graph and (B) summary. The article ID is same as that in Table 1.
Diagnostic accuracy of circulating microRNAs
Studies that reported dysregulated microRNAs in ESCC cases compared with healthy controls, together with the corresponding AUC and sensitivity and specificity for the very microRNA, were enrolled in the meta-analysis. With these criteria, 43 individual microRNAs were included in 33 studies. The diagnostic accuracies for each microRNA are presented in Table 2.
Table 2.
Diagnostic accuracies of the microRNAs mentioned in the literature.
| MicroRNA | Expression | ESCC sample size | Control sample size | Sensitivity (%) | Specificity (%) | AUC | Number of included articles |
|---|---|---|---|---|---|---|---|
| miR-10a | U | 149 | 100 | 81.2 | 80.0 | 0.886 | 1 |
| miR-10b | U & D | 79 | 66 | 79.1 | 91.8 | 0.860 | 2 |
| miR-15a | D | 106 | 106 | 86.4 | 100.0 | 0.951 | 1 |
| miR-16 | U | 38 | 19 | 94.5 | 55.8 | 0.643 | 1 |
| miR-18a | U | 243 | 209 | 87.2 | 97.4 | 0.870 | 2 |
| miR-19a | U | 89 | 125 | 66.3 | 66.4 | 0.712 | 1 |
| miR-20a | U | 70 | 40 | 64.3 | 75.0 | 0.767 | 1 |
| miR-20b | U | 137 | 155 | 56.6 | 67.5 | 0.627 | 1 |
| miR-21 | U | 586 | 379 | 88.5 | 72.0 | 0.840 | 7 |
| miR-22 | U | 149 | 100 | 88.6 | 86.0 | 0.949 | 1 |
| miR-24 | U | 105 | 30 | 81.9 | 83.3 | 0.866 | 1 |
| miR-25 | U | 262 | 176 | 72.4 | 77.0 | 0.800 | 3 |
| miR-29c | D | 50 | 50 | 68.0 | 68.0 | 0.720 | 1 |
| miR-31 | U | 201 | 202 | 86.6 | 82.2 | 0.910 | 2 |
| miR-100 | U | 217 | 178 | 65.5 | 82.5 | 0.700 | 3 |
| miR-106a | U | 137 | 155 | 77.0 | 47.4 | 0.639 | 1 |
| miR-127-3p | U | 149 | 100 | 78.5 | 87.0 | 0.899 | 1 |
| miR-129 | U | 78 | 23 | 78.8 | 73.3 | 0.792 | 1 |
| miR-133a | U | 149 | 100 | 65.1 | 83.0 | 0.830 | 1 |
| miR-138 | D | 128 | 40 | 87.5 | 69.5 | 0.871 | 1 |
| miR-146a | D | 154 | 154 | 83.9 | 76.7 | 0.870 | 2 |
| miR-148b | U | 149 | 100 | 66.4 | 87.0 | 0.855 | 1 |
| miR-185 | U & D | 66 | 54 | 96.9 | 57.5 | 0.590 | 2 |
| miR-193a-3p | U | 28 | 28 | 75.1 | 87.1 | 0.851 | 1 |
| miR-194 | U | 28 | 28 | 65.3 | 90.1 | 0.809 | 1 |
| miR-205 | D | 50 | 50 | 70.0 | 64.0 | 0.720 | 1 |
| miR-216a | D | 120 | 51 | 80.0 | 90.2 | 0.877 | 1 |
| miR-216b | D | 120 | 51 | 55.8 | 90.2 | 0.756 | 1 |
| miR-218 | D | 106 | 60 | 71.7 | 76.7 | 0.833 | 1 |
| miR-223 | U & D | 508 | 381 | 73.1 | 77.1 | 0.810 | 4 |
| miR-337-5p | U | 28 | 28 | 79.1 | 84.0 | 0.848 | 1 |
| miR-365 | U | 78 | 23 | 80.6 | 86.7 | 0.831 | 1 |
| miR-375 | U & D | 408 | 2017 | 85.1 | 65.3 | 0.840 | 4 |
| miR-451 | U | 128 | 43 | 84.4 | 81.4 | 0.900 | 2 |
| miR-483-5p | U | 28 | 28 | 70.8 | 74.5 | 0.739 | 1 |
| miR-486-5p | U | 137 | 155 | 55.3 | 75.8 | 0.688 | 1 |
| miR-506 | U | 110 | 40 | 81.2 | 87.3 | 0.835 | 1 |
| miR-584 | U | 137 | 155 | 65.4 | 63.0 | 0.659 | 1 |
| miR-613 | D | 75 | 75 | 81.3 | 62.7 | 0.728 | 1 |
| miR-718 | D | 120 | 51 | 69.2 | 66.7 | 0.715 | 1 |
| miR-1246 | U | 101 | 46 | 71.3 | 73.9 | 0.754 | 1 |
| miR-1297 | D | 156 | 156 | 82.7 | 84.0 | 0.900 | 2 |
| miR-1322 | U | 201 | 201 | 82.1 | 80.6 | 0.880 | 2 |
Abbreviations: ESCC, esophageal squamous cell carcinoma; AUC, area under the curve; U, up-regulated expression; D, down-regulated expression.
The overall pooled sensitivity, specificity and AUC of all the 43 microRNAs in the diagnosis of ESCC were 0.794 (95%CI: 0.765 - 0.820), 0.779 (95%CI: 0.746 - 0.808) and 0.86 (95%CI: 0.82 - 0.88), respectively. These outcomes indicate excellent distinguishing ability for microRNAs as biomarkers to detect ESCC. The diagnostic odds ratios (DOR) value was 13.518 (95%CI: 10.772 - 16.964). Likelihood ratios are considered with higher clinically value than sensitivity and specificity. The overall positive likelihood ratio (PLR) and negative likelihood ratio (NLR) were 3.584 (95%CI: 3.122 - 4.113) and 0.265 (95%CI: 0.232 - 0.303).
Prognostic accuracy of circulating microRNAs
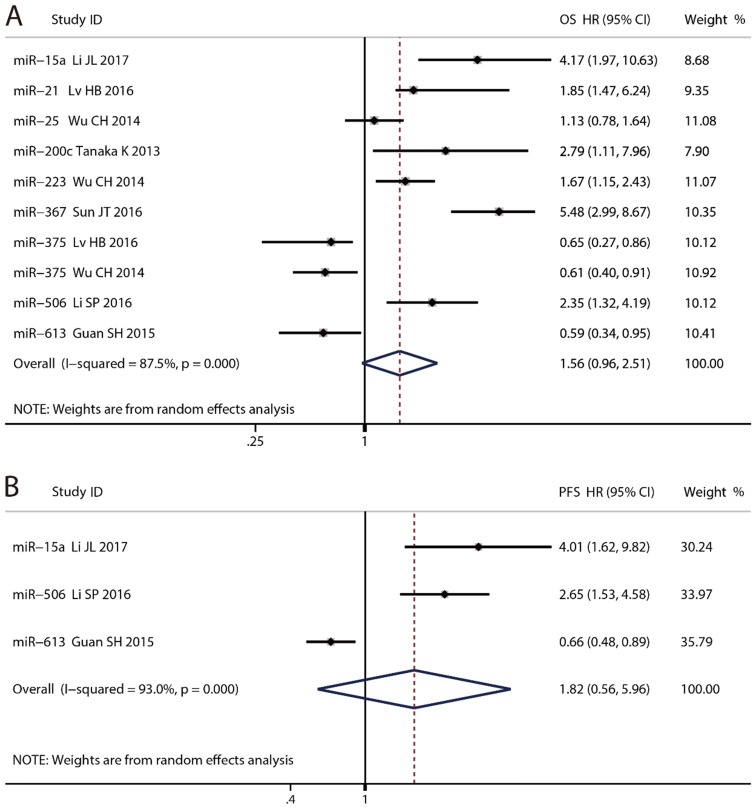
Nine microRNAs were enrolled in OS and three were in PFS. The prognostic accuracy of each microRNA was shown in Fig. 3. The pooled HR of OS was 1.56 (95% CI: 0.96 - 2.51, P = 0.070) with 87.5% of the I², whereas that of PFS was 1.82 (95% CI: 0.56 - 5.96, P = 0.321) with 93.0% of the I². Too high heterogeneity indicated synthesis of the data was inappropriate. Two studies reported the OS of miR-375, and the pooled HR was 0.62 (95% CI: 0.44 - 0.87, P = 0.005) with 0.0% of the I².
Fig 3.
Forest plots for the prognostic meta-analysis. (A) Hazard ratio (HR) for Overall survival (OS). (B) HR for progression free survival (PFS).
Publication bias
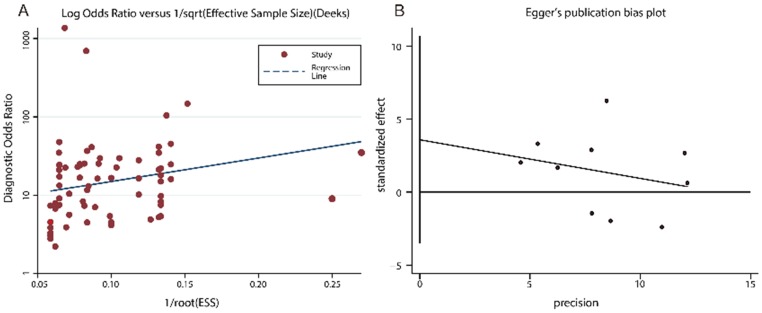
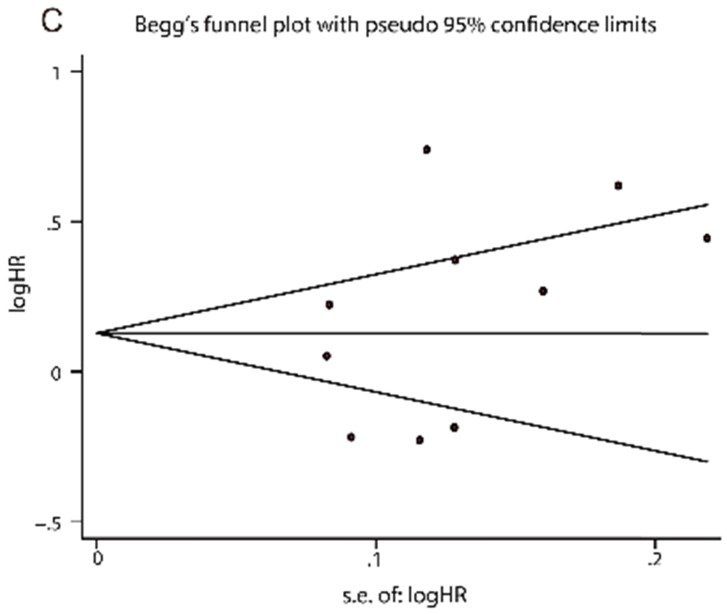
Publication bias for the diagnostic and prognostic meta-analysis was displayed in Fig. 4. The P value of the Deeks', Egger's and Begger's test were 0.120, 0.210 and 0.276, respectively, demonstrating that the meta-analysis was without publication bias.
Fig 4.
Funnel plots of publication bias. (A) Deeks' funnel plot for diagnostic tests. (B) Egger's funnel plot for prognostic tests. (C) Begg's funnel plot for prognostic tests.
Discussion
Circulation biomarkers are often used to diagnose early stages of ESCC. Invasive tests, by contrast, are inconvenience to monitor the progress of the ESCC, while are often rejected by patients because of their discomfort. The development of suitable biomarkers is crucial for diagnosing cancer and predicting the outcome of patients. Traditional biomarkers lack the sensitivity and specificity of early minimal lesions 7-9. Currently, microRNAs are emerging as promising biomarkers to fill the gaps in early diagnosis and prognosis 57.
In circulation, microRNAs are stable by virtue of being bound to Argonaute proteins, and avoid the degradation by RNases 58. As were reported in previous researches, various microRNAs were found to be abnormally expressed in malignant tumors of esophagus, stomach, large intestine, liver, pancreas, lung and ovary 59,60. This suggests that these microRNAs can be used as biomarkers for cancers. Thousands of microRNAs have been discovered, some of which have significant diagnostic and prognostic value in ESCC 16. To comprehensively analyze the value of microRNAs as biomarkers, we performed a meta-analysis of each microRNA, while more literatures than previous meta-analyses were included.
Our meta-analysis reviewed 35 articles and included over five thousand people. There were 28 up-regulated microRNAs and 11 down-regulated microRNAs, while four microRNAs were reported up-regulated and down-regulated in different studies. It is generally believed that the AUC value is greater than 0.9 with high diagnostic value, and the value is between 0.7 and 0.9 with moderate value. If it is less than 0.7, diagnostic value is usually low. In this study, five microRNAs, which were miR-15a, miR-22, miR-31, miR-451 and miR-1297, had high diagnostic value. 32 microRNAs had moderate diagnostic value, and six had low value. Circulation microRNAs achieved a pooled sensitivity of 0.794, specificity of 0.779, AUC of 0.86, and the DOR value was 13.518. The likelihood ratio (PLR and NLR), which are not affected by the prevalence rate, is more clinically meaningful for our diagnostic accuracy. A PLR value of 3.584 and an NLR value of only 0.265 implied that circulation microRNAs were clearly able to discriminate ESCC patients from healthy people.
For prognostic biomarkers, the heterogeneity was too high to merge the HR of all of the microRNAs. It was more appropriate to concentrate the prognostic efficiency of each single microRNA. Higher expression of six microRNAs meant a significantly poorer OS, and that of three microRNAs meant a significantly better OS. PFS was reported in only three microRNAs. The patients with high expression of miR-15a and miR-506 tended to have poorer PFS significantly, while miR-613 was just the opposite.
MicroRNAs have its superiority and weaknesses. Compared to endoscopic biopsies, microRNAs are more readily accepted by patients because of the invasiveness. MicroRNAs are more sensitive and specific than traditional serum markers 7. In addition, microRNA is still stable after the treatment including boiling, freeze-thaw cycles, acid and alkali treatment 61. The combination of microRNAs would make further efforts to increase the diagnostic efficiencies 24,28,31,38,48. Novel and high-performance diagnostic panels of varies combinations of microRNAs should be developed in follow-up studies. However, the cost of microRNA detection is higher than the traditional means, which may affect the wide range of applications of this measure. There were several noteworthy limitations in the present research that should not be overlooked. The study population came from different sources, which suggests that inclusion criteria may be different. Additionally, the ethnicities of the study were restricted to Asians, which may affect the representativeness of the results. The incidence rate in East Asia is the highest, which may be attributed to genetic factors partially 62.
Up-regulation or down-regulation of microRNAs may be related to the different sources of microRNAs in different tumors. As has been reported, blood-based microRNAs are not only synthesized from blood cells but also from the tissues cell, subject to a variety of internal environment and mechanisms such as the appearance and progression of the tumor 61,63, implying that the dysregulated microRNA plasma levels in ESCC patients could be secreted actively or passively by tumor cells. This may explain why different microRNAs are up-regulated or down-regulated. In our study, four microRNAs were reported up-regulated and down-regulated in different studies simultaneously, which could be caused by the choice of reference RNA, the dosage of reagents, the operating process without standardization and the heterogeneity of patients 19.
Now the relationships between circulating microRNAs and ESCC seemingly have been clearly understood. The source of microRNAs is derived from blood cells or tissue cells with different possible mechanisms, whatever the generating mechanism of the microRNAs might be, their circulating levels can be monitored and used for diagnostic and prognostic indicators. In the future, microRNAs may be used as relatively non-invasive circulating biomarkers of ESCC.
Acknowledgments
The authors would like to thank the members of Prof. Xi-Zhong Shen's laboratory for helpful discussions and critical reading of the manuscript. This study was supported by the National Natural Science Foundation of China (No. 81000968; No. 81101540; No. 81101637; No. 81172273; No. 81272388; No. 81301820; No. 81472673), the Doctoral Fund of Ministry of Education of China (20120071110058), and the National Clinical Key Special Subject of China.
Abbreviations
- ESCC
esophageal squamous cell carcinoma
- CBM
Chinese Biomedical Literature Database
- QUADAS-2
Quality Assessment of Diagnostic Accuracy Studies 2
- AUC
area under the curve
- SROC
summary receiver operating curve
- HR
hazard ratio
- CI
confidence interval
- OS
overall survival
- PFS
progression free survival
- DOR
diagnostic odds ratio
- PLR
positive likelihood ratio
- NLR
negative likelihood ratio.
References
- 1.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–52. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 2.Tanday S. Global cancer cases on the rise. Lancet Oncol. 2015;16:e317. doi: 10.1016/S1470-2045(15)00022-4. [DOI] [PubMed] [Google Scholar]
- 3.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 4.Kim T, Grobmyer SR, Smith R, Ben-David K, Ang D, Vogel SB. et al. Esophageal cancer-the five year survivors. J Surg Oncol. 2011;103:179–83. doi: 10.1002/jso.21784. [DOI] [PubMed] [Google Scholar]
- 5.Barber TW, Duong CP, Leong T, Bressel M, Drummond EG, Hicks RJ. 18F-FDG PET/CT has a high impact on patient management and provides powerful prognostic stratification in the primary staging of esophageal cancer: a prospective study with mature survival data. J Nucl Med. 2012;53:864–71. doi: 10.2967/jnumed.111.101568. [DOI] [PubMed] [Google Scholar]
- 6.Tachimori Y, Kanamori N, Uemura N, Hokamura N, Igaki H, Kato H. Salvage esophagectomy after high-dose chemoradiotherapy for esophageal squamous cell carcinoma. J Thorac Cardiovasc Surg. 2009;137:49–54. doi: 10.1016/j.jtcvs.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 7.Kosugi S, Nishimaki T, Kanda T, Nakagawa S, Ohashi M, Hatakeyama K. Clinical significance of serum carcinoembryonic antigen, carbohydrate antigen 19-9, and squamous cell carcinoma antigen levels in esophageal cancer patients. World J Surg. 2004;28:680–5. doi: 10.1007/s00268-004-6865-y. [DOI] [PubMed] [Google Scholar]
- 8.Yilmaz O, Eroglu A, Dag E, Karaoglanoglu N, Yilmaz A. Serum levels of IGF-I and IGFBP-III and their relation with carcinoembryonic antigen and carbohydrate antigen 19-9 in cases of esophageal cancer. Int J Clin Pract. 2006;60:1604–8. doi: 10.1111/j.1742-1241.2006.00854.x. [DOI] [PubMed] [Google Scholar]
- 9.Parenti A, Porzionato A, Pizzi S, Guzzardo V, Fassina G, Macchi V. et al. Expression pattern of squamous cell carcinoma antigen in oesophageal dysplasia and squamous cell carcinoma. Histol Histopathol. 2007;22:989–95. doi: 10.14670/HH-22.989. [DOI] [PubMed] [Google Scholar]
- 10.Aslam MI, Taylor K, Pringle JH, Jameson JS. MicroRNAs are novel biomarkers of colorectal cancer. Br J Surg. 2009;96:702–10. doi: 10.1002/bjs.6628. [DOI] [PubMed] [Google Scholar]
- 11.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 12.Komatsu S, Ichikawa D, Takeshita H, Tsujiura M, Morimura R, Nagata H. et al. Circulating microRNAs in plasma of patients with oesophageal squamous cell carcinoma. Br J Cancer. 2011;105:104–11. doi: 10.1038/bjc.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wan J, Wu W, Che Y, Kang N, Zhang R. Insights into the potential use of microRNAs as a novel class of biomarkers in esophageal cancer. Dis Esophagus. 2016;29:412–20. doi: 10.1111/dote.12338. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Wang Q, Zhang N, Ma H, Gu Y, Tang H. et al. Identification of microRNAs as novel biomarkers for detecting esophageal squamous cell carcinoma in Asians: a meta-analysis. Tumour Biol. 2014;35:11595–604. doi: 10.1007/s13277-014-2350-x. [DOI] [PubMed] [Google Scholar]
- 15.Li M, Wu F, Ji Y, Yang L, Li F. Meta-analysis of microRNAs as potential biomarkers for detecting esophageal carcinoma in Asian populations. Int J Biol Markers. 2017;32:e375–83. doi: 10.5301/ijbm.5000296. [DOI] [PubMed] [Google Scholar]
- 16.Liu F, Tian T, Xia LL, Ding Y, Cormier RT, He Y. Circulating miRNAs as novel potential biomarkers for esophageal squamous cell carcinoma diagnosis: a meta-analysis update. Dis Esophagus. 2017;30:1–9. doi: 10.1111/dote.12489. [DOI] [PubMed] [Google Scholar]
- 17.Liu HN, Wu H, Chen YZ, Chen YJ, Shen XZ, Liu TT. Altered molecular signature of intestinal microbiota in irritable bowel syndrome patients compared with healthy controls: A systematic review and meta-analysis. Dig Liver Dis. 2017;49:331–7. doi: 10.1016/j.dld.2017.01.142. [DOI] [PubMed] [Google Scholar]
- 18.Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB. et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529–36. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 19.Liu HN, Wu H, Chen YJ, Tseng YJ, Bilegsaikhan E, Dong L. et al. Serum microRNA signatures and metabolomics have high diagnostic value in hepatocellular carcinoma. Oncotarget. 2017;8:108810–24. doi: 10.18632/oncotarget.22224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deeks JJ. Systematic reviews in health care: Systematic reviews of evaluations of diagnostic and screening tests. BMJ. 2001;323:157–62. doi: 10.1136/bmj.323.7305.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Egger M, Davey SG, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–101. [PubMed] [Google Scholar]
- 23.Bai Y, Lin H, Fang Z, Luo Q, Fang Y, Su Y. et al. Plasma microRNA-19a as a potential biomarker for esophageal squamous cell carcinoma diagnosis and prognosis. Biomark Med. 2017;11:431–41. doi: 10.2217/bmm-2016-0286. [DOI] [PubMed] [Google Scholar]
- 24.Zhou X, Wen W, Zhu J, Huang Z, Zhang L, Zhang H. et al. A six-microRNA signature in plasma was identified as a potential biomarker in diagnosis of esophageal squamous cell carcinoma. Oncotarget. 2017;8:34468–80. doi: 10.18632/oncotarget.16519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, Li M, Gao F, Ge X. Serum microRNA-15a level acts as a potential diagnostic and prognostic biomarker for human esophageal squamous cell carcinoma. Cancer Biomark. 2017;18:11–7. doi: 10.3233/CBM-160667. [DOI] [PubMed] [Google Scholar]
- 26.Zheng S, Zhang X, Wang X, Li J. Downregulation of miR-138 predicts poor prognosis in patients with esophageal squamous cell carcinoma. Cancer Biomark. 2017;20:49–54. doi: 10.3233/CBM-170079. [DOI] [PubMed] [Google Scholar]
- 27.Yang Y, Du YX, Zhang C, Wei SZ, Li QW. The expression level of microRNA-451 and the role of curative effect evaluation in serum of patients with esophageal squamous cell carcinoma. Journal of Xinjiang Medical University. 2017;40:779–82. [Google Scholar]
- 28.Lv H, He Z, Wang H, Du T, Pang Z. Differential expression of miR-21 and miR-75 in esophageal carcinoma patients and its clinical implication. Am J Transl Res. 2016;8:3288–98. [PMC free article] [PubMed] [Google Scholar]
- 29.Li SP, Su HX, Zhao D, Guan QL. Plasma miRNA-506 as a Prognostic Biomarker for Esophageal Squamous Cell Carcinoma. Med Sci Monit. 2016;22:2195–201. doi: 10.12659/MSM.899377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang C, Li Q, Liu F, Chen X, Nesa EU, Guan S. et al. Serum miR-1297: a promising diagnostic biomarker in esophageal squamous cell carcinoma. Biomarkers. 2016;21:517–22. doi: 10.3109/1354750X.2016.1160291. [DOI] [PubMed] [Google Scholar]
- 31.Dong S, Yin H, Dong C, Sun K, Lv P, Meng W. et al. Predictive Value of Plasma MicroRNA-216a/b in the Diagnosis of Esophageal Squamous Cell Carcinoma. Dis Markers. 2016;2016:1857067. doi: 10.1155/2016/1857067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang C, Guan S, Liu F, Chen X, Han L, Wang D. et al. Prognostic and diagnostic potential of miR-146a in oesophageal squamous cell carcinoma. Br J Cancer. 2016;114:290–7. doi: 10.1038/bjc.2015.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun L, Dong S, Dong C, Sun K, Meng W, Lv P. et al. Predictive value of plasma miRNA-718 for esophageal squamous cell carcinoma. Cancer Biomark. 2016;16:265–73. doi: 10.3233/CBM-150564. [DOI] [PubMed] [Google Scholar]
- 34.Guan S, Wang C, Chen X, Liu B, Tan B, Liu F. et al. MiR-613: a novel diagnostic and prognostic biomarker for patients with esophageal squamous cell carcinoma. Tumour Biol. 2016;37:4383–91. doi: 10.1007/s13277-015-4271-8. [DOI] [PubMed] [Google Scholar]
- 35.Shi XY, Wang Q, Jiang XY, Xu L, Wu J, Zhang C. et al. Value of real-time fluorescent quantitative polymerase chain reaction in detecting expression of miR-100 in patients with esophageal cancer. Int J Lab Med. 2016;37:738–9. [Google Scholar]
- 36.Guo WT, Chang LL, Zhao Q, Zhang QW, Zhu LW, Li YG. et al. Values of magnetic nanoparticles separation-based real-time fluorescent RT-PCR assay for detection of serum;miR-21 in patients with esophageal squamous cell carcinoma. Guangdong Medical Journal. 2016;37:837–40. [Google Scholar]
- 37.Sun J, Song K, Feng X, Gao S. MicroRNA-367 is a potential diagnostic biomarker for patients with esophageal squamous cell carcinoma. Biochem Bioph Res Commun. 2016;473:363–9. doi: 10.1016/j.bbrc.2016.01.042. [DOI] [PubMed] [Google Scholar]
- 38.Hui B, Chen X, Hui L, Xi R, Zhang X. Serum miRNA expression in patients with esophageal squamous cell carcinoma. Oncol Lett. 2015;10:3008–12. doi: 10.3892/ol.2015.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu H, Yao Y, Meng F, Qian X, Jiang X, Li X. et al. Predictive Value of Serum miR-10b, miR-29c, and miR-205 as Promising Biomarkers in Esophageal Squamous Cell Carcinoma Screening. Medicine (Baltimore) 2015;94:e1558. doi: 10.1097/MD.0000000000001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li BX, Yu Q, Shi ZL, Li P, Fu S. Circulating microRNAs in esophageal squamous cell carcinoma: association with locoregional staging and survival. Int J Clin Exp Med. 2015;8:7241–50. [PMC free article] [PubMed] [Google Scholar]
- 41.He FC, Meng WW, Qu YH, Zhou MX, He J, Lv P. et al. Expression of circulating microRNA-20a and let-7a in esophageal squamous cell carcinoma. World J Gastroenterol. 2015;21:4660–5. doi: 10.3748/wjg.v21.i15.4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang Z, Song Q, Yang S, Zeng R, Li X, Jiang C. et al. Serum microRNA-218 is a potential biomarker for esophageal cancer. Cancer Biomark. 2015;15:381–9. doi: 10.3233/CBM-150480. [DOI] [PubMed] [Google Scholar]
- 43.Dong W, Li B, Wang Z, Zhang Z, Wang J. Clinical significance of microRNA-24 expression in esophageal squamous cell carcinoma. Neoplasma. 2015;62:250–8. doi: 10.4149/neo_2015_030. [DOI] [PubMed] [Google Scholar]
- 44.Jing R, Chen W, Wang H, Ju S, Cong H, Sun B. et al. Plasma miR-185 is decreased in patients with esophageal squamous cell carcinoma and might suppress tumor migration and invasion by targeting RAGE. Am J Physiol Gastrointest Liver Physiol. 2015;309:G719–29. doi: 10.1152/ajpgi.00078.2015. [DOI] [PubMed] [Google Scholar]
- 45.Li W, Yan CL, Tan XG. Diagnostic value of application of salivary and plasma microRNA-21 in early esophageal cancer. Chongqing Med; 2015. pp. 1894–6. [Google Scholar]
- 46.Komatsu S, Ichikawa D, Hirajima S, Kawaguchi T, Miyamae M, Okajima W. et al. Plasma microRNA profiles: identification of miR-25 as a novel diagnostic and monitoring biomarker in oesophageal squamous cell carcinoma. Br J Cancer. 2014;111:1614–24. doi: 10.1038/bjc.2014.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ye M, Ye P, Zhang W, Rao J, Xie Z. Diagnostic values of salivary versus and plasma microRNA-21 for early esophageal cancer. J Southern Med Univ. 2014;34:885–9. [PubMed] [Google Scholar]
- 48.Wu C, Wang C, Guan X, Liu Y, Li D, Zhou X. et al. Diagnostic and prognostic implications of a serum miRNA panel in oesophageal squamous cell carcinoma. Plos One. 2014;9:e92292. doi: 10.1371/journal.pone.0092292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu C, Li M, Hu C, Duan H. Clinical significance of serum miR-223, miR-25 and miR-375 in patients with esophageal squamous cell carcinoma. Mol Biol Rep. 2014;41:1257–66. doi: 10.1007/s11033-013-2970-z. [DOI] [PubMed] [Google Scholar]
- 50.Tanaka K, Miyata H, Yamasaki M, Sugimura K, Takahashi T, Kurokawa Y. et al. Circulating miR-200c levels significantly predict response to chemotherapy and prognosis of patients undergoing neoadjuvant chemotherapy for esophageal cancer. Ann Surg Oncol. 2013;20(Suppl 3):S607–15. doi: 10.1245/s10434-013-3093-4. [DOI] [PubMed] [Google Scholar]
- 51.Zhang T, Zhao D, Wang Q, Yu X, Cui Y, Guo L. et al. MicroRNA-1322 regulates ECRG2 allele specifically and acts as a potential biomarker in patients with esophageal squamous cell carcinoma. Mol Carcinog. 2013;52:581–90. doi: 10.1002/mc.21880. [DOI] [PubMed] [Google Scholar]
- 52.Hirajima S, Komatsu S, Ichikawa D, Takeshita H, Konishi H, Shiozaki A. et al. Clinical impact of circulating miR-18a in plasma of patients with oesophageal squamous cell carcinoma. Br J Cancer. 2013;108:1822–9. doi: 10.1038/bjc.2013.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takeshita N, Hoshino I, Mori M, Akutsu Y, Hanari N, Yoneyama Y. et al. Serum microRNA expression profile: miR-1246 as a novel diagnostic and prognostic biomarker for oesophageal squamous cell carcinoma. Br J Cancer. 2013;108:644–52. doi: 10.1038/bjc.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xie ZJ, Chen G, Huang J, Li ZJ. The diagnostic significance of plasma miR-10b for esophageal cancer. Guangdong Medical Journal. 2013;34:2465–8. [Google Scholar]
- 55.Zhang T, Wang Q, Zhao D, Cui Y, Cao B, Guo L. et al. The oncogenetic role of microRNA-31 as a potential biomarker in oesophageal squamous cell carcinoma. Clin Sci (Lond) 2011;121:437–47. doi: 10.1042/CS20110207. [DOI] [PubMed] [Google Scholar]
- 56.Zhang C, Wang C, Chen X, Yang C, Li K, Wang J. et al. Expression profile of microRNAs in serum: a fingerprint for esophageal squamous cell carcinoma. Clin Chem. 2010;56:1871–9. doi: 10.1373/clinchem.2010.147553. [DOI] [PubMed] [Google Scholar]
- 57.Yazbeck R, Jaenisch SE, Watson DI. From blood to breath: New horizons for esophageal cancer biomarkers. World J Gastroenterol. 2016;22:10077–83. doi: 10.3748/wjg.v22.i46.10077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meister G. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet. 2013;14:447–59. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- 59.Liu HN, Liu TT, Wu H, Chen YJ, Tseng YJ, Yao C. et al. Serum microRNA signatures and metabolomics have high diagnostic value in colorectal cancer using two novel methods. Cancer Sci. 2018;109:1185–94. doi: 10.1111/cas.13514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu HN, Wu H, Tseng YJ, Chen YJ, Zhang DY, Zhu L. et al. Serum microRNA signatures and metabolomics have high diagnostic value in gastric cancer. BMC Cancer. 2018;18:415. doi: 10.1186/s12885-018-4343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K. et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Y. Epidemiology of esophageal cancer. World J Gastroenterol. 2013;19:5598–606. doi: 10.3748/wjg.v19.i34.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL. et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]