Abstract
As part of the superfamily of long noncoding RNAs, circular RNAs (circRNAs) are emerging as a new type of regulatory molecules that partake in gene expression control. Here, we review the current knowledge about circRNAs in cardiovascular disease. CircRNAs are not only associated with different types of cardiovascular disease, but they have also been identified as intracellular effector molecules for pathophysiological changes in cardiovascular tissues, and as cardiovascular biomarkers. This evidence is put in the context of the current understanding of general circRNA biogenesis and of known interactions of circRNAs with DNA, RNA, and proteins.
Keywords: Non-coding RNA, Circular RNA, circRNA, Cardiovascular disease, Splicing, Transcription
1. Introduction
Today, nearly 28000 genes for long noncoding RNAs (lncRNAs) have been mapped. These lncRNA genes can be located in genomes separate from any other protein-coding gene, or overlapping in complex patterns with other genes [1]. LncRNAs have been shown to be expressed in a cell type- and state-specific mode during cell differentiation and during pathophysiological cell state changes including in diseases of the cardiovascular system [2]. Surprisingly, although genes are linear, and mRNAs are linear, and although typical lncRNA transcripts are linear as well, cells also express circular non-protein-coding RNAs, termed “circRNAs”. In a seminal study by Salzman et al. [3], advances in high-throughput sequencing and in annotating splice-junctions have allowed accumulating solid evidence that thousands of eukaryotic genes form circular RNAs as a physiologically normal process (see Ref. [4] for review). These circRNAs are not encoded as separate genetic units, like lncRNAs, but are produced by a variant form of splicing from the pre-mRNA of any transcribed gene in cis, may it be protein-coding or noncoding. To form circRNAs, the spliceosome covalently fuses an RNA's 5′ end to its 3′ end, which results in a circular ribonucleic acid. Thus, circRNAs lack 5′Cap and poly(A) tail. These features are used to biochemically enrich for circRNAs. For example, one can negatively select against polyA-containing mRNA, or digest all linear RNAs in circRNA preparations by adding exonucleases like RNase R that attack open linear ends [5]. Irrespective of origin, and with only a tiny minority of exceptions, circRNAs are not translated into proteins and are thus falling into the class of noncoding RNAs. More specifically, with a median size of around 500 ribonucleotides, circRNAs are part of the larger superfamily of lncRNAs [3,[6], [7], [8], [9]].
Two seminal findings showed that circRNAs do have important regulatory and developmental functions in animals [8,10]. This triggered a number of studies on circRNAs in basic and applied research, including studies relevant to cardiovascular research. Despite the large number of annotated circRNAs, so far only a handful of these has been approached by functional studies. This is due to technical challenges in detecting circRNAs, as well as problems in genetically manipulating circRNAs without affecting linear RNA expression from their host genes. By numbers, so far, 10 circRNAs have been firmly associated with cardiovascular pathophysiology by experimental evidence in cell culture systems that serve to model aspects of cardiovascular disease. Of these, only 3 circRNAs have been studied in vivo, that is in animal CVD disease models or by decisive evidence from genome-wide association studies (GWAS) in human patient cohorts. Another 27 circRNAs have been implicated as biomarkers for CVD but have not been functionally investigated. Given that many, if not most, circRNAs have been suggested to affect the expression of linear mRNA from their host genes, many more circRNAs can be expected to be linked to CVD in the future. Therefore, we will review here all the CVD-linked circRNAs, but start out by highlighting milestones in the investigation of general circRNA biogenesis and molecular function. In these introductory passages (chapters 2–4) we distill general concepts with relevance for cardiovascular disease, as then discussed later in the review of studies on specific CVD-linked circRNAs (chapters 5.1–5.3). Throughout, the main focus is the review of studies profiling circRNA expression in cardiovascular tissues, of studies exploring cellular functions of circRNAs in cardiovascular physiology and disease, and of studies translating these data to human pathophysiology. In the review, one major question will be how to prioritize circRNAs during the investigation of cardiovascular disease pathways, in times when expression profiles of thousands of circRNAs are accumulating through RNA high-throughput sequencing efforts in tissues, cells or blood samples. A second important question will be how to interpret genetic polymorphisms associated with CVD risk from GWAS, for example when risk single nucleotide polymorphisms (SNPs) are found to locate in CVD risk genes. Third, we will discuss how the spliceosome mediates circRNA biogenesis, and how alterations in splicing patterns are linked to the onset of cardiovascular disease.
2. Classes of circular RNAs
Three major classes of circular RNAs can be distinguished based on the type of covalent linkage underlying circularization, based on the molecular machinery circularizing 5′ and 3′ ends and based on the typical subcellular localization of the circularized RNA products. The large majority of circular RNAs in eukaryotes are produced by the spliceosome. Thereby, the major U2-containing spliceosome is the major machinery that produces circular RNAs. 3 groups of circular RNAs are produced by the U2-containing spliceosome: a. 3′-5′-linked circRNAs that only contain exonic sequences, b. 3′-5′-linked EIciRNAs, which contain both exons and introns, and c. ciRNAs, which contain only intronic sequences and have been circularized by a 2′-5′ covalent phosphodiester bond [[11], [12], [13]] (Table 1, Fig. 1). For the biogenesis of all these three groups of circular RNAs, in a reaction termed “backsplicing”, the spliceosome uses existing canonical splice signals [3,13] for cutting out and circularizing internal parts of pre-mRNA sequences in cis. In the following, we describe the classes of circular RNAs in more detail. The accompanying Box 1 contains a short list of vocabulary that can be used as an entry point into the field of circRNA biology.
Table 1.
Spliceosome-dependent circular RNAs. Key features of circular RNAs produced by the spliceosome in eukaryotic cells. n (estimated number of relevant circRNAs expressed from a typical mammalian genome; counted per host gene), ntotal (total number of all predicted circRNA transcript isoforms) as deduced from mapping RNAseq reads during transcriptome profiling of different human organs/tissues/cell types.
| Common features |
|
| Exon-containing (3′-5′)-linked circRNAs |
|
| Exon- and intron-containing (3′-5′)-linked EIciRNAs |
|
| Intron-only (2′-5′)-linked ciRNA |
|
Abbreviations: circbase (curated database for circular RNAs [35]), HeLa (human cervical cancer cell line), hESCs (cultured human embryonic stem cell line).
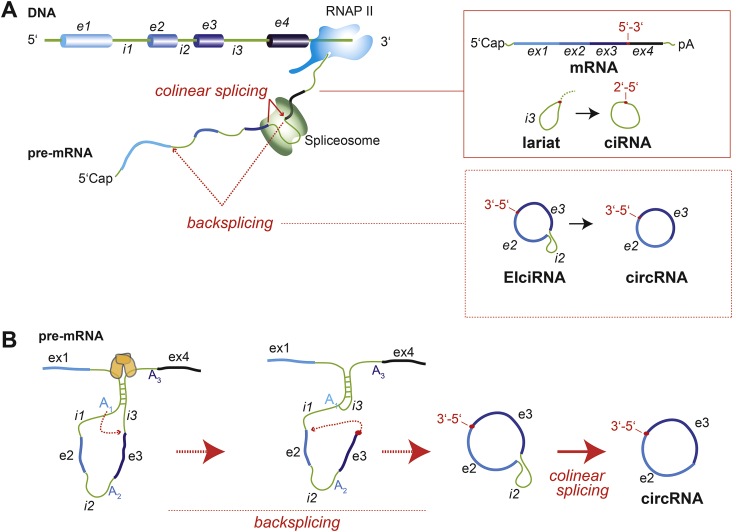
Fig. 1.
Biogenesis of spliceosome-dependent circular RNAs.
Overview of the three classes of spliceosome-dependent circular RNAs treated in this review: ciRNAs, EIciRNAs and circRNAs (A). Shown is the arrangement of exons (e1-4) and introns (i1-3) in 5′-3′ order in a gene in the genome, and the transcription into an exon- and intron-containing pre-mRNA, which is either collinearly spliced (top) or backspliced (bottom). Colinear splicing results in a major product (the exon-containing mRNA, where exons are arranged in the same 5′-3′ order as in encoded in the genome), and the byproducts (introns in the form of 2′-5′ branched lariats, of which only one case is shown). Lariats are usually rapidly degraded in the nucleus, but can become processed and parental molecules for ciRNAs. Backsplicing (bottom) results in a major product (3′-5′-linked EIciRNAs and circRNAs), and the byproduct (2′-5′ branched mRNA, not shown). Details of backsplicing are depicted in detail in (B). (B) Backsplicing resulting in 3′-5′-linked circRNA formation: Intramolecular backfolding between inverted intronic repeats in the pre-mRNA, assisted by dimerization of RNA-binding proteins with binding motifs on the pre-mRNA, brings canonical splice junction in such a three-dimensional context, that a downstream end of an exon (red dot) is spliced to the begin of an upstream exon. The two transesterification reactions are shown with small red dotted arrows (indicating the direction of the underlying nucleophilic attack). The result is a 3′-5′-linked EIciRNA that displays an intervening intron. This intron is further processed by conventional splicing to an exon-only 3′-5′-linked circRNA.
Introns (light green), exons (different shades of blue, indicating more 5′ or more 3′ location in a gene or mRNA), RNA-binding proteins serving as circRNA biogenesis regulators like Quaking or Muscleblind (orange), RNA polymerase III holocomplex (RNAP II), Spliceosome (dark green).
Box 1. List of terminologies from circRNA biology used in this review.
| Term/Reference | Definition |
|---|---|
| Splicing [214] |
|
| Splice sites [215] |
|
| Splicing factors [216] |
|
| Branchpoint [217] |
|
| Lariat [218,219] |
|
| Colinear splicing [220,221] |
|
| Circular RNAs [3] |
|
| Backsplicing [56] |
|
| circRNAs [3] |
|
| 5′Cap [222] |
|
| 3′polyA tail [223] |
|
| EIciRNAs [14] |
|
| ciRNAs [17] |
|
| circRNA host gene [3] |
|
| Backsplice junction [4] |
|
| ORF [114] |
|
| 5′ UTR |
|
| IRES |
|
| RNAP II [224] |
|
| Preinitiation complex [225] |
|
| TFIIH [225] |
|
| P-TEFb [226] |
|
| RNAP II pausing [227] |
|
| R-loop |
|
| Pre-rRNA processing [104] |
|
Alt-text: Box 1
2.1. Spliceosomal, exon-containing (3′-5′)-linked circRNAs
The first and major class of circular RNAs is 3′-5′-linked circRNA produced by backsplicing, a reaction whereby the spliceosome fuses a downstream splice donor to an upstream splice acceptor [6]. In the simplest case by this reaction a 5’→3′-linked circRNA is formed that consist of a single exon, where the end of the respective exon attacks its own start in the second transesterification reaction. circRNAs can, however also contain multiple exons, whereby intervening introns are spliced out in a subsequent step (Fig. 1). Exon-only circRNAs can exhibit a number of roles, ranging from regulating linear splicing, over binding proteins and modulating their activity, to binding and sequestering microRNAs, termed microRNA sponging. Since circRNAs are sequences of fused exons but do not display a 5′Cap and other upstream features necessary for ribosome entry to linear mRNAs, circRNAs are classically not translated. However, the small possibility exists that protein-coding potential is still exhibited, and this is a known function of a tiny subset of circRNAs. All these functions are described in detail in chapter 4.
2.2. Spliceosomal, exon- and intron-containing (3′-5′)-linked EIciRNAs
When one or several intervening intronic sequences are maintained during the biogenesis of a circRNA, these specialized forms are termed circular exon-intron-containing circRNAs (EIciRNAs) [14] (Fig. 1). In a typical cell, approximately hundred EIciRNAs exist. They are, thus, a minority compared to exon-only circRNAs. EIciRNAs are mostly nuclear and have been involved in stimulating RNAP II-dependent initiation, likely via the intronic sequences they contain.
2.3. Spliceosomal, intron-only (2′-5′)-linked ciRNA
The third class of circular RNAs is 2′-5′-linked circular intronic RNA (ciRNA). ciRNAs are products of normal colinear splicing. ciRNAs essentially represent lariats, and are composed exclusively of intronic sequence. In the two transesterification steps of conventional colinear splicing, the adenosine at the intron branchpoint is linked to the 5′ of the excised intron, resulting in a lasso-like circular RNA, the 2’→5′-branched lariat. In contrast to normal lariats, which are rapidly degraded in the nucleus [15,16], ciRNAs do not get degraded [17]. Instead, their signature 3′ single-stranded extensions are nibbled off, and they resist further 2’→5′ debranching because of the presence of a 7 nucleotides (nts) long motif near the 5′ splice site and another 11 nts motif near the branchpoint [17]. As a result, ciRNAs are perfect RNA circle without branches that become stable and exhibit cellular functions (Fig. 1). Two independent experimental approaches basing on high-throughput sequencing have defined two different types of 2′-5′-linked circular intronic RNA: Firstly, several hundred circular intronic RNAs (ciRNAs) were identified by specifically screening for intronic sequences in poly(A)-negative and rRNA-depleted RNA pools of cultured human cell lines [17,18]. CiRNAs were found to be localized in the nucleus of cells. Three ciRNAs have so far been functionally investigated to some extent, and they play a role in stimulating RNAP II-dependent transcriptional initiation at their host gene loci. Secondly, a family of several thousand stabilized intronic sequence RNAs (sisRNAs) were found as part of the nuclear RNA pool in the Xenopus tropicalis oocyte [19] and later also in other organisms and cells. sisRNAs are a more broadly defined class of RNAs compared to ciRNAs, as they contain both linear and circular intronic RNAs. sisRNAs are overall not well understood, their relevance in cardiovascular settings not studied, and they will, thus, not be treated in any detail in this review.
2.4. Non-spliceosomal circular RNAs
A small number of circular RNAs are not produced by the spliceosome. This class of circular RNAs is rather mixed and includes specialized cases of circularization that mostly happen in lower eukaryotes, for example, tRNA ligase-mediated circularization of tRNAs introns [20], or circularization by the ribozyme (self-splicing) activity of group I and group II introns encoded in mitochondrial and chloroplast genomes or in rRNA [[21], [22], [23]]. This class also contains circular RNAs that represent genomes of ssRNA viruses, like the hepatitis delta virus in mammals or virusoids in plants [24]. The relevance of the circular nature of all these types of RNAs is not specifically clear, and it can only be hypothesized that circularity promotes stability against nucleases and, as such, could hypothetically be of potential advantage for propagation on an evolutionary scale. Since these circular RNAs have not been studied in cardiovascular context, they will not further be treated in this review.
Pertaining to the spliceosomal circRNA, EIciRNAs, and ciRNAs, the formation of these circular RNAs seems to be a regulated process, as different cell types have been shown to exhibit differential circRNA expression profiles [9,11,25,26]. This is then reflected in differing circRNA expression profiles when comparing different tissues and organs [9,[26], [27], [28], [29], [30]]. Moreover, a single multi-exon gene can give rise to a number of different isoforms of circular RNA, depending on which splice donors and acceptors have been accessible in the pre-mRNA and how the biogenesis of a circular RNA proceeded [9,31,32] or depending on internal alternative splicing in circRNA-generating sequences [33]. Together, spliceosomal RNA circularization greatly enhances the complexity of transcriptomes.
2.5. Tools and databases to study circular RNAs
Researchers in the circRNA field are beginning to realize that both, the bioinformatics mapping of circRNAs, as well as the experimental validation of circularity and the quantification of circRNAs, are far less trivial than anticipated and prone to variation.
2.5.1. Bioinformatic tools and databases
Studies using different circRNA profiling algorithms come to circRNA datasets that differ by up to 40%. Thus, when exploring existing circRNA databases that become ever more specialized (like circBase [35], CircNet [36], starBase [37] or CSCD [38]), or when choosing from existing circRNA detection tools (circRNA_finder [12], find_circ [8], CIRCexplorer [32], CIRI [39], DCC [40], KNIFE [11], MapSplice [41], NCLScan [42], PTESFinder [43], Segemehl [44], Uroborus [45]) care has to be taken that no gold standards exist yet in assessing the accuracy of bioinformatics algorithms to map circRNAs.
2.5.2. Biochemical circRNA detection and quantification
No gold standard exists, either, for the decision which biochemical approach is taken for RNA preparation during circRNA analysis, or for molecular techniques used to quantify circRNAs. Specifically, no benchmarks for enzymatic procedures have yet been developed that could be followed when enriching circRNAs in RNA preparations. In fact, potential artefacts in the enrichment steps for circRNAs and the depletion steps of linear RNAs in biochemical preparations may account for high rates of false-positive circRNAs (up to 30%). For example, RNase R is employed to deplete non-circular RNAs, but may lead to decay also of some circRNAs [6], and is also not always completely efficient [34]. Excellent recent reviews and comparative studies summarize the current understanding of the underlying problems in biochemical circRNA prediction, detection, and quantification [4,11,[46], [47], [48]]. Consequently, it is advised to use a number of complementary methods instead of a single method, for example for circRNA quantification, or for the experimental validation of circularity of candidate circRNAs (e.g. applying also other enzymes equivalent to RNase R treatment, and quantifying circRNAs not only by RT-PCR, which can cause artefacts due to template switching, but also by Northern blotting or 2D gels [34]).
2.5.3. Functional genetic analysis of circRNAs
Not last, care must be taken when making functional arguments based on genetic activation of inactivation of circRNAs, for example by overexpression or knockdown approaches in cells, or by mutation analysis or genomic knockouts in vivo. In a commonly used cellular approach, ectopic overexpression of circRNAs is accomplished by transfecting into cells mini-gene constructs on plasmids that carry inverted repeats in the introns that flank the exons to be circularized. Such mini-gene constructs have been experimentally shown to promote artificial spliceosomal exon circularization [34], but have also been found to give rise to RNA concatemers, off-target effects [34], and unphysiologically high levels of circRNAs. The latter is especially problematic when dose-dependent functions like microRNA sponging are implicated as prime effector mechanism by such a study [49]. Finally, an in-depth analysis of circRNA function may ultimately involve genome-engineering of the endogenous host locus, but this can be complicated, as it is not often clear how to avoid affecting linear RNA expression from the host locus [50].
3. Molecular mechanism of circRNA biogenesis
Colinear splicing and alternative splicing are regulated processes and under the influence of a number of factors. This includes a range of sequence motifs and contexts, splicing factors, chromatin-dependent processes, and RNAP II-dependent parameters. Similarly, the regulation of circRNA formation is expected to be equally complex, but only part of the determining factors have so far been detected. Conceptually, two different modes of circRNA biogenesis are distinguished: a. cotranscriptional circRNA formation from within the linear pre-mRNA, and b. posttranscriptional backsplicing from within an already excised exon-containing lariat, which is not connected anymore with the linear pre-mRNA molecule [51,52]. This distinction is relevant because co-transcriptional circRNA biogenesis is able to negatively inflict the fate of the linear mRNA molecule directly in cis [53] (Fig. 1B), while posttranscriptional circRNA biogenesis cannot (see Ref. [34] for review). As a consequence, circRNA formation from a gene with disease relevance may impact disease parameters.
3.1. RNA sequence motifs promoting RNA circularization
A number of bioinformatics, biochemical and genetic experiments have explored which factors promoted RNA circularization from pre-mRNA. Bioinformatic approaches involve the analysis of the sequence space in an around circularizing exons, biochemical approaches involve the isolation of circular RNA species, their quantification and the identification of circRNA-binding proteins and factors, and genetic experiments involve the use of mutation screens or reverse genetic overexpression or knockdown of circRNA-regulatory candidates or of circRNAs themselves. Especially genetic screens combining mutation analysis with circRNA biogenesis reporter assays, sometimes also in different model systems such as yeast, Drosophila or mouse, have been very successfully in identifying circRNA-regulating factors [52,54,55]. From these combinatorial approaches, some higher-order parameters for circRNA biogenesis became apparent, such as a sufficient length of the exon(s), an above-average length of flanking introns, the relative positioning of a circularization event in the gene body, and the absence of secondary RNA structures that might disfavor circularization [3,52,56]. However, also RNA sequence content per se appears to be important for circRNA biogenesis. RNA sequence content determines circRNA biogenesis in three different ways:
3.1.1. Splice donor and splice acceptor sequence motifs in RNA
High-throughput sequencing and genetic experiments have determined that the vast majority of circular RNAs in cells were formed by the cellular spliceosome. Three-quarters of all circularization events employ canonical splice donors (GU) and acceptors (AG) and, thus, the same sequence signals that are also used during conventional linear splicing by the U2-containing spliceosome [11,13,53,57]. In the rest of cases, existing cryptic splice sites are used, or the U12-containing minor spliceosome is involved, but these features, as such, do not make the difference to colinear splicing [13,52,58]. Rather, what determines the frequency of RNA circularization is whether upstream splice acceptor sites are still available for backsplicing (and not already used up by colinear splicing). In fact, colinear splicing occurs rapidly, within seconds, after the pre-mRNA is synthesized and exits from the RNAP II complex [59] (Fig. 1A). Splice sites are then used colinearly, one by one, as the pre-mRNA emerges from the exit-channel of the RNAP II and becomes accessible to the spliceosome [53,60]. The spliceosome fuses splice sites with each other, first come first serve. A head-to-tail backsplice reaction is, thus, disfavored compared to linear splicing [51] and for many genes, the amount of circRNA produced is an order of magnitude smaller than mRNA expression [6,7,9]. Less than 0.1% of genes express more circRNA than linear mRNA [9].
3.1.2. Reverse-complementary RNA repeat motifs in introns flanking a circularization event
Backsplicing has been found to be especially favored when the downstream splice site is brought in close 3-dimensional proximity of the upstream splice site because of base-pairing between sequence-complementary inverted repeats in the two introns flanking the circularization event in the pre-mRNA [6,13,32,[60], [61], [62], [63]] (Fig. 1B). A number of different types of repeat motifs have been identified in introns, some of which are oriented in reverse-complementary orientation to each other. Alu repeats are one example. In humans, Alu elements are a major type of repeats associated with circRNA formation and exist in more than 1 million copies in our genomes [60]. Alu elements derived from the 7SL noncoding RNA and are primate-specific retrotransposons that insert in gene-rich genomic regions but are losing their mobility as they accumulate mutations on an evolutionary timescale. As a consequence of their insertion preference, they are prevalent in expressed RNAs, and also in noncoding parts, such that 66% of Alu elements reside in introns [64,65]. It has been suggested that the likelihood and strength of repeat-mediated backfolding of introns contained within pre-mRNA can determine the frequency of backsplicing [32,51,54,63,66]. This applies to both cotranscriptional as well as to posttranscriptional backsplicing. Genome engineering approaches have documented that deleting one such repeat on one side of the circularization event with CRISPR/Cas9was sufficient to abolish circularization, corroborating the repeat-induced backfolding model of biogenesis [51,66].
3.1.3. Binding motifs for RNA-binding proteins and splicing factors in RNA
circRNA formation is suggested to be additionally supported by proteins that bind to introns or intron-exon-junctions and stabilize backfolding. A number of circRNA biogenesis-regulating proteins have been identified in genetic screens using synthetic backsplice reporters. Some of these were known from before to be regulators of normal alternative mRNA splicing. Common to these proteins is their capacity to bind RNA, and some additionally homodimerize to assist RNA backfolding and appropriately position backsplice junctions next to each other (Fig. 1B). Examples are the evolutionarily conserved proteins Quaking (QKI) [29] and Muscleblind (MBNL) [53], which bind to introns flanking a circularization event and promote circRNA biogenesis. These two proteins have, however, also been previously described as regulators of developmentally programmed alternative linear mRNA splicing in mouse and in Drosophila melanogaster, respectively [67,68]. Beyond that they also influence translation, decay, and localization of sets of mRNAs, making the selective analysis of circRNA regulation very complex [69,70]. Another circRNA regulator is Fused-in-sarcoma (FUS) [71], a well-known RNA- and DNA-binding protein that pleiotropically regulates also transcription start and transcript site control, mRNA length control and alternative linear mRNA splicing. Similar to QKI and MBNL, FUS was found to bind to the exon-intron junctions that formed the RNA circle, consistent with stabilizing a microenvironment that favored backsplicing. Dozens of other RNA-binding proteins have since been implicated in RNA circularization and stabilization by measuring circRNA abundance during RNAi-based screens in Drosophila cells [54] or in human cells in culture [55]. Among the hits, serine-arginine-rich SR proteins and hnRNPs turned up, which are well-known mRNA splicing regulators, but also factors with still unclear function. It should also be mentioned that not all of the identified circRNA regulator candidates were stimulators, as some also repressed circularization by still unknown mechanisms [54,55]. Important for the aitiology of cardiovascular diseases, some of these circRNA-regulating proteins have been studied in vivo, and have been clearly implicated in both development of the cardiovascular system, as well as in pathophysiological cellular changes, yet without any evidence whether these functions were due to regulating linear mRNA splicing, or circRNA biogenesis. This will be discussed in chapter 5.3 below.
Most recent models for circRNA biogenesis base on the observation of an inversely correlated link between the speed and efficiency of transcription-coupled mRNA processing, and the efficiency of circRNA formation [72]. CircRNA biogenesis has been found to be favored when levels of splicing factors and core spliceosomal factors dropped and when linear splicing slowed down. This may have to do with the fact that in such conditions, spliceosomal “cross-exon” interactions [73] are only slowly converted to linear splicing-competent “cross-intron” interactions [73], which would then allow the alternative pathways of circRNA biogenesis to prevail [72].
4. Molecular functions of circular RNAs
Similar to lncRNAs, circular RNAs have been proposed to be functional because their biogenesis itself has functional consequences, or because the RNA exerts functions as a molecular entity. In the following chapters 4.1–4.6, we will describe the already known roles of circRNA biogenesis and of circular RNAs themselves (Fig. 2). We will highlight relevant aspects that also play a role in how circRNAs affect cardiovascular pathophysiology.
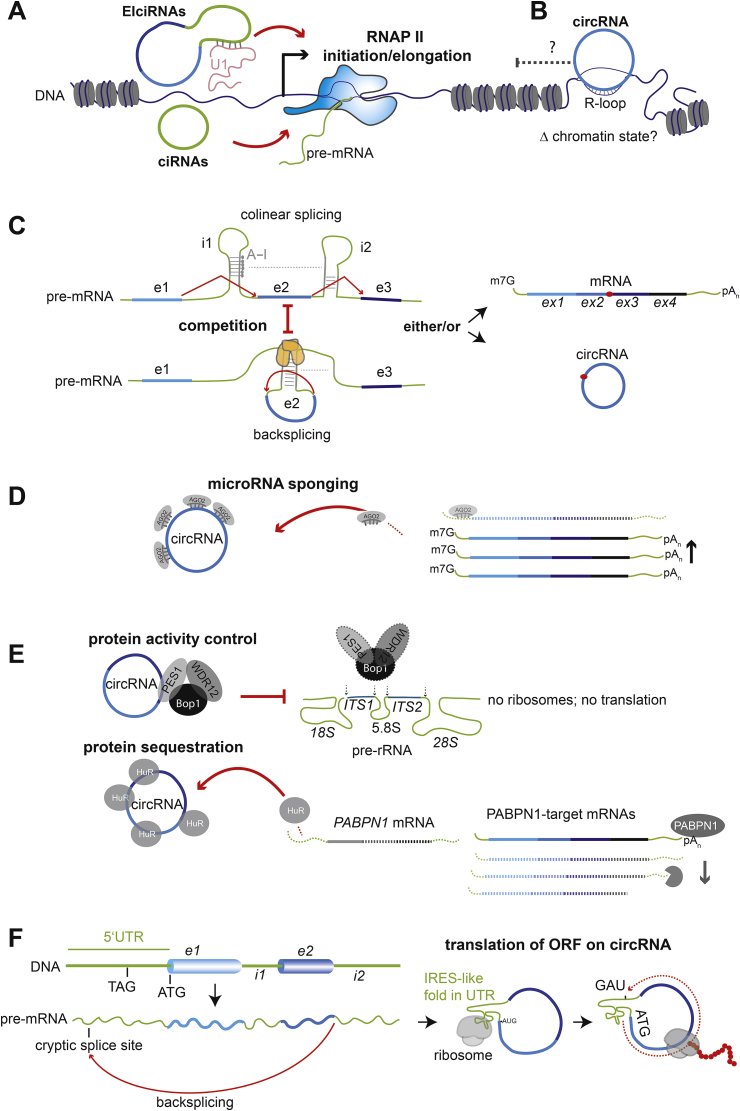
Fig. 2.
General molecular functions of circular RNAs.
Proposed functions of circular RNAs in the cell nucleus (A–C) and the cytoplasm (D–F). Functions are not arranged by the prominence where circular RNAs are expected to be most important but instead are ordered by how gene expression proceeds, starting from transcription and ranging over co-/post-transcriptional processing to cytoplasmic regulation and finally to translation. (A) EiciRNAs and ciRNAs binding to host locus and stimulating RNAP II initiation and/or elongation. (B) circRNA binding to DNA sequence encoding the circRNA-generating exon in the host locus and forming and R-loop at this site. This is proposed to impair RNAP II progression, a function so far not studied as a transcription-regulating process, but rather as a splicing regulating process. (C) Model for competition between colinear splicing (top) and co-transcriptional backsplicing (bottom): competition is due to backfolding between different intronic repeat motifs (grey segments). Selective adenosine deamination to inosine by ADAR enzymes within double-stranded RNA patches in the backfolded regions can promote linear splicing (top). RNA-binding protein like Quaking or Muscleblind (orange) bind and homodimerize, and thereby can promote backsplicing by favoring backfolding of introns flanking the circularization event (bottom). (D) Sequestration of Argonaute 2-bound microRNAs on the circRNA (left) reduces the cellular pool of active microRNAs and, thereby, increases the abundance of mRNAs that would otherwise be degraded by these microRNAs (right). (E) Known examples of circRNA:protein interactions: circANRIL binding PES1 protein of the PeBoW complex. This interaction blocks PeBoW activity in ribosomal RNA (rRNA) maturation (Top). The second example (bottom): Sequestration of HuR proteins by circPABPN reduces the free levels of the PABPN mRNA-stabilizing HuR and leads to reduced levels of PABPN1 protein, leading to destabilization of PABPN1-dependent mRNAs. (F) An exceptional case of protein translation from an open reading frame (ORFs) encoded on a circRNA: Sequences in the genes 5′UTR that serve as unconventional internal ribosome entry site (IRES) for translation beginning with the ATG start codon. Translation proceeds until an in-frame stop codon (UAG) located, for example, in the 5′ UTR.
4.1. Circular RNAs and the regulation of transcription
A number of different circular RNAs have been found to regulate the transcription of their cognate host gene, either by affecting initiation or elongation of RNA polymerase II. These will be reviewed in this paragraph. In general terms, how circular RNAs affect transcription is different from how many linear noncoding RNAs control transcription: One of the best understood and prototypical functions of linear lncRNAs is their ability to tether chromatin-regulatory complexes to specific gene loci, and, concurrently, to scaffold bound complexes. Both functions are used in regulating transcription of genes but have so far not been described in circular RNAs. A second paradigm is that a linear lncRNA can interact with multiple proteins at once. Through their modular buildup, lncRNAs can bind, for example, several different transcription-regulating enzymes (see Ref. [74] for review). Conceptually, circular RNAs, at least in published reports, have so far not been found to function as scaffolds of chromatin regulatory protein complexes, either. This may, in part, have to do with the fact that circular RNAs have rather recently been identified.
4.1.1. 3′-5′-linked circRNAs and the regulation of RNAP II?
Exon-only circRNAs have so far not been directly linked to the regulation of transcriptional initiation at gene promoters. Although the cytoplasmic abundance prevails for circRNAs, as experimentally observed in individual cases as well as biochemically by fractionation [3,6,8,10,63,66,75], some mature circRNAs do also exist in the nucleus. Examples are circ-SEP3 [76], circ-Amotl1 [77], circRNAs from the CAMSAP1, GLIS3, or c-01, c-87 and c-88 [71], or circRNA from the HIPK3 gene, which has cardiovascular relevance [78]. The specific role ofcircHIPK3 in CVD will be summarized in detail in chapter 5.2. For some of these nuclear circRNAs, a nuclear function may exist. The only relevant evidence for a nuclear function of a circRNA comes, however, from a study in Arabidopsis thaliana, a model plant. A circRNA was found that stemmed from a transcription factor gene that regulated floral development. The circRNA was shown to bind to its parental DNA, from which it had been transcribed [76]. In vitro, it could be documented, that during binding, the investigated circRNA locally opened the DNA helix and hybridized to one DNA strand. At this site, the second DNA strand is locally displaced (Fig. 2B). Such a configuration is called an R-loop (see Ref. [79] for review). The investigated plant circRNA formed an R-loop even with slightly higher efficiency than a linearized RNA with same sequence [76]. Functionally, the circRNA:DNA R-loop was shown to impair transcriptional elongation at this site, and possibly as a consequence stimulated skipping of the exon from which the very circRNA derived [76]. Thus, this circRNA established a feed-forward loop in stimulating its own production by hybridizing to its locus of origin (Fig. 2B). How often circRNAs form R-loops on a genome-wide scale is unknown, but if this mechanism was more far-spread the implications could be very broad and be relevant for every cell type of the cardiovascular system. R-loops will be described in more detail in the following chapter on the role of circRNAs in splicing (4.2).
Also in mammalian systems, an indirect link has been established between circRNAs and RNAP II progression: It has been suggested that the process of circRNA biogenesis (backsplicing) was influenced by the speed of RNAP II transcription: A faster progression of RNAP II was suggested to give circRNA-forming splice sites more chance to engage with each other, decreasing the frequency by which linear splicing could use them up [51,53]. Together, at least in some cases, it may be possible that circRNA biogenesis and RNAP II progression mutually impair each other, even though 3′-5′-linked circRNAs are not known to themselves bind RNAP II or transcription factors or affect their activity. Overall, circRNAs exist in rather low copy numbers, which makes the biochemical pulldown and functional analysis of circular RNA-binding complexes more challenging. By numbers, 90% of circRNAs are present with only 1–10 molecules per cell, which is at 10-fold lower levels than their cognate host gene [7]. Only 2% of circRNAs are present at levels >50% of their cognate linear mRNA [7]. Yet, circRNAs would not need to exist in high copy numbers, if the very process of circRNA biogenesis was the essential determinant in affecting the expression of a circRNA host locus.
Specific classes of different circular RNAs have been linked to RNAP II-dependent transcription more directly, namely EIciRNAs and ciRNAs, as described in the following:
4.1.2. EIciRNAs stimulate transcription initiation at gene promoters
EIciRNAs are a separate class of approximately hundred circular RNAs that also stimulate transcription. They have been found to co-immunoprecipitate with RNAP II at their parent locus [14]. The understanding of EIciRNA function is a bit more advanced compared to ciRNAs. EIciRNAs stimulate transcription by interacting with the small nuclear U1 that is otherwise well known for its classical and conserved role in the spliceosome [14] (Fig. 2A). U1 has, however also life on its own at promoters, where it functions independently of the other snRNAs and spliceosomal proteins [80]. There, U1 binds the general transcription factor TFIIH to stimulate transcriptional initiation [81], as well as the transcription elongation factor P-TEFb to stimulate elongation of RNAP II [82]. U1 can also determine the directionality of transcription at promoters and can prevent premature polyadenylation and cleavage of linear mRNAs [83]. Which of these functions EIciRNAs associates with is still unknown, but a number of different RNAP II-dependent processes might be under their influence.
EIciRNAs have not yet been specifically studied in cardiovascular context, but are expressed also in cardiovascular tissues, as determined by RNA-seq analyses, for example of the heart. 5–18% of circular RNAs in hearts do contain introns [[84], [85], [86]], a ratio that fits with the overall initial estimation that up to 20% of circular RNAs expressed from mammalian genomes contain introns [7]. Of those, of course, only a fraction belongs to the class of EIciRNAs/ciRNAs, but this fraction is also predicted to be significantly large (e.g. 30% of all intronic RNAs >200 nts are circular [17]). Also, intron-containing circRNAs have been reported to map to a gene with cardiovascular relevance, such as to MYH7, Slc8a1, Ryr2, or Camk2d [[84], [85], [86]]. Yet, any function of such intron-containing RNAs in cardiovascular cell types remains to be established.
4.1.3. ciRNAs stimulate transcription elongation by RNAP II
Around 100 ciRNAs were found in cultured cell lines [17,18]. Of these, three representatives were functionally tested in more detail. In biochemical purifications ciRNAs from the ANKRD52, MCM5, and SIRT7 genes segregated with the nuclear insoluble fraction, which represents chromatinized DNA and strongly associated molecules. These ciRNAs stimulated the transcriptional elongation of RNAP II on the gene from which they derived. Thereby, the ciRNAs were activating only when expressed from within the locus, and not when provided from plasmids in trans. The actual molecular mechanism remains to be dissected [17]. Interestingly, a number of nuclear ciRNA foci were observed by immunostaining. This indicated that ciRNAs regulate also other gene loci, but direct evidence for trans-regulation is still missing, as well as which sequence determinants would guide ciRNAs to other loci [17]. Overall ciRNAs are expected not to lead to an all or nothing decision in transcription, but to a certain degree of modulation of transcriptional activity. ciRNAs have not yet been specifically studied in cells of the cardiovascular system, but given their generality of function, similar to EIciRNAs, ciRNAs can be expected to be found and to be functional in hearts, vessels and blood cells. For example, if it holds true in vivo that circSIRT7 regulates linear SIRT7 mRNA transcription [17], circSIRT7 may very well be of cardiovascular relevance because linear SIRT7 is known from before to be important for survival and stress resistance in cardiomyocytes [87], especially also in the infarcted heart [88]. This remain to be determined, and the specific list of ciRNAs and or EIciRNAs in cardiovascular cell types to be derived.
4.2. 3′-5′-linked circRNA:DNA interaction and the regulation of splicing
Cotranscriptional RNA circularization has been found to anticorrelate with the abundance of linear mRNA levels produced from the very circRNA-hosting gene (Fig. 2C). Insight from these experiments constitutes a very generalizable model for circRNAs function that affects all classes of genes, and thus also different traits and disease, including cardiovascular disease. The underlying mechanism, how circRNA competes with linear mRNA expression, is not yet fully understood and will be explained in detail in the following:
A number of independent studies have documented that there is competition between circRNA biogenesis and linear mRNA formation. mRNA formation usually is dominant in this competition, but circRNAs formation can have measurable negative effects on linear mRNA abundance of a given locus [51,53]. In other cases, when assessing this competition in higher temporal resolution in single cells, linear and backsplicing were suggested to be completely mutually exclusive [89]. A first possible explanation for competition between mRNA formation and circRNA formation concerns the accessibility of splice sites as a result of competition between different flanking introns to pair with each other in the pre-mRNA [32] (Fig. 2C). A second explanation follows the concept of “kinetic coupling” defined earlier in studies of alternative splicing regulation (see Ref. [90] for a recent review): It has been suggested that the rate of backsplicing was influenced by the speed of RNAP II transcription: A faster progression of RNAP II was suggested to give circRNA-forming splice sites more chance to engage with each other, decreasing the frequency by which linear splicing could use them up [51,53]. A third explanation is that, once backsplicing has indeed occurred co-transcriptionally, the linear mRNA may hypothetically be less stable, because carrying an internal 2′-5′-linked covalent branch [52,56]. In fact, in 55% of cases, the linear mRNAs that have likely undergone circRNA formation (because lacking circRNA-generating exons) cannot be detected anymore in cells [6]. In a fourth possible explanation, backsplicing can also change the downstream splicing pattern and render linear host pre-mRNA less stable after producing a circRNA: in one case, knockdown of a ciRNA led to the abnormal retention of a downstream intron in the host mRNA, and this intron contained a premature stop codon [17,91]. Whether this observation related to nonsense-mediated mRNA decay has, however, not been decisively explored yet.
Together, though the frequency of circRNA formation is on average an order of magnitude smaller than linear mRNA formation, circRNA biogenesis is thought to be potent enough to compete with concurrent mRNA formation.
4.3. circRNA:RNA interaction and the regulation of mRNA stability
A major topic in many of currently published papers on circRNAs is their role in sequestering microRNAs. The ease to predict potential microRNA seed sites on noncoding RNAs has led to a flurry of papers implicating microRNA sponging as effector mechanism, and this is occurring also in papers on circRNAs with cardiovascular relevance. The function of microRNA sponging is, however, seen more and more controversially and will be discussed in some more detail here (Fig. 2D).
4.3.1. CDR1as as bona-fide microRNA sponge
The idea that certain RNA transcripts can serve as decoys or sponges for microRNAs to regulated sequence homologous target mRNAs is not new. It originally emerged from tools of synthetic biology, which used antisense sequences to sequester microRNAs [92]. Later, endogenous microRNA-sponging lncRNAs were found, including those transcribed from noncoding pseudogenes [93,94]. As a consequence of sequestering microRNAs, microRNA-sponging lncRNAs were shown to promote the expression of their cognate protein-coding genes. By coincidence, the first published report on a physiologically relevant circRNA had picked one of the very few circRNAs that do indeed contain multiple binding sites for microRNAs, and which are expressed at substantially high levels, namely the circular antisense to the cerebellar degeneration-related protein 1 transcript (CDR1as) [8,10]. CDR1as displays 74 microRNA seed regions for the miR-7 microRNA. Since CDR1as lacks full sequence complementarity with this microRNA, CDR1as degradation is thought to be avoided [8,10]. Indeed, CDR1as does form stabilized microRNA:AGO2 endonuclease complexes, as would be expected for a microRNA sponge [10]. That sponging was indeed a biological function for CDR1as was concluded from genetic experiments: The depletion of CDR1as caused the upregulation of miR-7 targeted mRNAs, and the overexpression of CDR1as phenocopied a miR-7 loss of function [8]. This did not only apply in cells in vitro but also in situations in vivo. A mouse knockout of the CDR1as locus has recently been reported [95]: Since the CDR1as locus had been thought to produce only a circRNA and no linear RNA, the straight knockout of the exon encompassing the circRNA was used to explore the function of the CDR1as circRNA as microRNA sponge in vivo. The authors showed that CDR1as mutant mice upregulated miR-7 targeted mRNAs in neurons. Neurons are the cell type most strongly expressing this circRNA in the brain, and the deregulation of excitatory synaptic transmission and abnormal sensorimotoric gating in the CDR1as mutant mice could, at least phenotypically and in part, be ascribed to deregulation of miR-7 targets [95]. The picture is likely more complex, because a recent reinvestigation of the CDR1as loci in human and mouse revealed that the CDR1as circRNA (also known as ciRS-7) is not a stand-alone circRNA, but embedded in a previously unannotated linear long noncoding RNA (ASINC/LINC00632), from which it is produced by splicing [50]. In the reported CDR1as circRNA mouse knockout the levels of this linear lncRNA were increase. The lncRNA transcripts from the locus were located in the nucleus and thus were unlikely to participate in the cytoplasmic microRNA sponging typical of the circular CiRS-7. Together, this suggests that, normally, the circRNA impaired the expression or stability of the cognate linear host lncRNA. Consequently the question is raised whether all neurological phenotypes were solely due to circRNA misregulation [50]. On a more general level, the recent study by Salzman et al. also raises the possibility that the transcriptional start sites of other circRNAs in the genome may still be misannotated, which is a problem in studying how circRNAs crosstalk with transcription factor-based gene regulatory mechanisms. Despite the undisputed evidence for a function of transcripts from the CDR1as locus in the central nervous system, CDR1as has also been studied in cardiovascular context, and one report suggested that it promoted myocardial infarction size (see in detail in chapter 5.2).
4.3.2. microRNA sponging is not a common function for circRNAs
MicroRNA sponging is, however, not considered a very common function for many circRNAs on a genome-wide scale for several reasons [9]: Few other circRNAs contain microRNA binding sites in numbers >10, and equally critically, circRNAs are expressed mostly at relatively low levels, certainly overall lower than many linear mRNAs and microRNAs [7]. MicroRNA sponging can also not be a specific function of circRNAs, as circularized exons were found to associate with Ago2 protein complexes not more frequently than exons of the same sequence content inside the linear mRNAs [7,26]. Also, only very few circRNAs and microRNAs are present in cells at such stoichiometric levels – relative for example to the number of their seed-targets in mRNAs - that they possibly can participate in a sponge effect [49,[96], [97], [98], [99], [100]]. For a sponge effect to occur, calculations suggest that competing endogenous RNA abundance has to be close to the target mRNA abundance [101]. While a number of microRNAs have been described as important regulators of tissue-specific gene expression networks underlying cell proliferation, cell-cell communication or cell type specification in development and disease [102], such as in cardiovascular disease (see Ref. [103] for a recent review), microRNA sponging is thought to be a rather exceptional mechanism for circRNAs [9,49].
4.4. circRNA:protein interaction
CircRNAs do not only bind other nucleic acids but can also interact with proteins (Fig. 2E). Three different examples for how circRNAs interact with protein complexes are reviewed in the following, one of which (circANRIL) has direct functional relevance for cardiovascular disease.
4.4.1. circANRIL regulates the PeBoW protein complex
In a first example, circANRIL was found to bind to members of the evolutionarily conserved PeBoW complex (Pes1, Bop1, and WDR12-containing). The PeBoW complex is essential in organisms ranging from yeasts to animals and plants. It is required in the nucleolus for pre- 60 S ribosome maturation and it is thought to stabilize RNases that excise the internal transcribed spacer 1 (ITS1) from pre-rRNAs [104,105]. circANRIL impaired rRNA maturation, which led to impaired cellular translation capacity (Fig. 2E, top panel) [106]. circANRIL stems from the long noncoding RNA ANRIL [106,107]. ANRIL is the acronym for antisense noncoding RNA in the INK4 locus because the ANRIL locus is located in antisense to the INK4 tumor suppressor locus on chromosome 9p21. ANRIL is considered a central effector of cardiovascular risk, as will be described separately in chapters 5.1 and 5.2 on circRNAs in cardiovascular disease.
4.4.2. circPABPN1 as decoy for the protein HuR
In a second example, circPABPN1 was suggested to sequester HuR, a central RNA-binding protein that is known to stabilize specific linear mRNAs (Fig. 2E, bottom panel) [108]. The mRNA encoding polyadenylate-binding nuclear protein 1 (PABPN1) was found as one of the most important targets of HuR [108]. Among other functions, PABPN1 protein defines the site of polyadenylation and polyA tail length in a number of mRNAs and augments their stability and thus translation. The notion emerged that circPABPN1 competed with its parental linear mRNA in binding to HuR, such that the circPABPN1 served as a decoy for HuR, and impaired PABPN1 translation and, secondarily, the translation of PABPN1-dependent mRNAs [108]. Together, the activity of RNA binding proteins may be fine-tuned by the relative abundance of circular and linear RNAs emerging from a locus, but a generalization of this theme remains to be determined.
4.4.3. circRNAs associate with hundreds of different RNA-binding proteins
Thirdly, genetic screens have identified over 100 RNA binding proteins (RBPs) that have the potential to regulate circRNA biogenesis [55]. Of these, some have been tested for their capacity to physically bind to circRNAs. For example, the RBPs NF90/NF110 (NFAR1/2) represent protein isoforms expressed from the interleukin enhancer binding factor 3 (ILF3) gene. These RBPs contain double-strand RNA-binding motifs that were found to interact with backfolded intronic RNA during circRNA biogenesis [55]. Coincidently, for still unknown molecular reasons, NF90/NF110 also interacted with mature circRNAs, and a quantitative preference was observed in circRNA-binding over binding the cognate linear RNA sequences [55]. The functional implications of circRNAs binding to NF90/NF110 are not so clear. NF90/NF110 proteins are usually required for an efficient host immune active in defending against RNA viruses [109,110]. Correlative evidence suggests that circRNA biogenesis decreases during viral infection at a time when NF90/NF110 is released from circRNAs and starts to bind the invading viral genomic RNA instead [55]. Currently, it is thought that structured RNA motifs in some circRNAs are similar to viral RNA motifs and may, thus, be effective in binding NF90/NF110. This circRNA:protein binding is thought to serve as kind of reservoir and to provide a sufficient amount of ready-and-go NF90/NF110 for fast antiviral surveillance [55]. The causality in this hypothetical model remains to be demonstrated. Lastly and as evident from independent co-immunoprecipitation experiments, EIciRNAs and ciRNAs interact somehow with the initiating or elongating forms of RNAP II in the nucleus [14,17]. Details of the nature of the underlying presumptive circular RNA:protein interactions have not been reported, and these circular RNAs may very well also interact with histones or chromatin proteins or DNA or RNA at the locus (Fig. 2A).
Together, it is likely that many other circRNAs functionally interact with proteins, and reported cases will likely increase as the biochemical purification of circRNA-bound complexes becomes routine.
4.5. Exons encoded on circRNAs are not generally translated into proteins
Linear lncRNAs do not contain open reading frames (ORFs) that are longer than 300 nucleotides (encoding for more than 100 amino acids), which is the central definition of the non-coding character of lncRNAs [111,112]. The vast majority of lncRNAs has been experimentally found not to be translated into proteins [113]. In reality, there is, however, a grey zone in distinguishing non-coding from coding transcripts: In fact, small ORFs are present just by chance in any sufficiently long RNA transcript, including the best characterized long lncRNAs [114], and experimental evidence demonstrates that more and more micropeptides are translated from such small ORFS in regions that have been previously annotated as noncoding.
4.5.1. circRNAs do not productively associate with ribosomes
At first glance, the question whether circRNAs are translated or not seems clear: Many circRNAs derive from protein-coding genes and constitute fusions of exonic sequences. Thus, circRNAs do contain open reading frames per definition. The vast majority of endogenous circRNAs is, however, not translated because circRNAs do not contain 5′Caps which could serve as entry points for ribosomes for translation. Several ribosome footprinting (RFP) studies agree on that the vast majority of circRNAs are not associated with active polyribosomes and are thus not translated [6,7,26]. This is likely a consequence of circularization from internal portions of genes and, consequently, the absence of a 5′ Cap, of linear ends, and of the Kozak sequence for ribosome entry and translation initiation in linear 5′capped mRNAs (see Ref. [115] for review). Advanced biochemical conditions in RFP experiments have, however, succeeded to demonstrate that some hundred circRNAs could be potentially translated [58], and a few dozens of those had the potential to encode a polypeptide [58] (Fig. 2F). So far, translation of only two circRNA molecules, circMbl [58] and circZNF609, have been documented with confidence [75]. Paramount for their translation was that RNA circularization led to the inclusion of the endogenous start codon, as well as of specialized 5′ untranslated regions (UTRs) that folded into specific secondary RNA structures with a potential to serve as internal ribosome entry site (IRES) [58,75].
For the cardiovascular field, especially the study on the translation of circZNF609 is of potential relevance because knockdown of circZNF609 revealed a role of this circRNA in myoblast proliferation in vitro [75]. This suggests a role of this circRNA in muscle biology, with implications also for heart function. As explained in chapter 4.6.2 the role of protein derived from circZNF609 is much less clear.
4.5.2. Exceptional translation of protein fragments from selected circRNAs
Whether circRNA-encoded polypeptides from the Mbl or ZNF609 genes exhibited any function is still unknown. Current experiments have succeeded in knocking down the relevant circRNAs by siRNAs targeting the circRNA-specific backsplice sequence and, thus, avoiding to affect the linear host mRNA [75]. However, the knockdown depleted the circRNA, and, thus, it remained unclear whether the observed proliferative defects were due to a function of the circRNA itself, or due to the circRNA-derived polypeptide [75]. In this specific case, the circRNA-derived 251 amino acid long ZNF609* protein is a truncation of the native ZNF609, and as such, may theoretically behave as a dominant-negative version of this transcriptional regulator (Fig. 2F). More experiments are needed to test this hypothesis in muscle cells and in pathophysiologically relevant settings.
4.5.3. Micropeptides translated from circRNAs?
Small ORF-encoded peptides have recently been found to be much more common than previously thought. Specifically, this relates to the observation that there are rather many instances where functional protein translation occurs outside of annotated protein-coding open reading frames [116,117]. Translation has been observed to occur also on RNAs that have been thought to be purely noncoding. Micropeptides from small ORFS are well-known to have especially important functions in cardiac physiology: Several of such micropeptides have been found to be translated from linear long noncoding RNAs and to regulate heart function. This includes the sarcolamban family of ORFs which repress the Ca2+ pump SERCA to terminate muscle contraction [118,119]. Also, another small ORF is important, namely DWORF, which displaces SERCA inhibitors and enhances contraction [120] and shows reduced expression in mouse models of dilated cardiomyopathy and ischemic failing human hearts [120]. At the moment, all known small peptides stem from linear RNAs and not from circRNAs. Also, circRNA formation from the relevant linear lncRNAs has not been specifically addressed, nor whether any of the few circRNAs that are associating with productively translating ribosomes do encode peptides that have the potential to regulate SERCA or function in other aspects of muscle biology. Thus, while the view emerges that translation of small ORFs from circRNAs is not impossible in theory [58,75], current evidence does not yet support the view that many such cases with relevance for cardiovascular physiology exist. More focused analyses are required to address this problem.
5. circRNAs in cardiovascular disease
So far, only a single circRNA knockout has been described in an animal model system, and this knockout affected neurological function [95]. Evidence from loss-of-function or transgenic overexpression of specific circRNAs in cardiovascular development in vivo does not yet exist. Those circRNAs that will be described in this chapter have been associated with cardiovascular disease by three conceptually different approaches:
First, circRNA expression profiling has been performed in cardiovascular tissue as well as in blood. Some of these studies compared circRNAs in control and diseased cardiovascular states. For biomarker studies, RNA preparations of whole blood, or of specific blood cells were analyzed. Such studies ended in speculations whether a specific circRNA had the potential for biomarker development, or could serve as candidate effector of cardiovascular pathophysiology.
Secondly, some studies have functionally implicated circRNAs as effectors of cardiovascular disease. None of these circRNAs have been selected based on measuring differential abundance alone. Evidence was collected from multiple pieces of evidence. For example, such studies analyzed circRNAs together with host mRNA expression investigated the competition between circRNAs and parental RNA, or they explored GWAS datasets with a focus on circRNAs that stemmed from disease-linked loci (e.g circANRIL). Other cases followed the rationale to focus on a highly abundant or enriched circRNAs that carried seed regions for well-known microRNAs (e.g. HRCR).
Thirdly, in yet another type of approach, some studies focused on specific RNA-binding proteins that regulated splicing of mRNAs and that had, separately, been linked to cardiovascular pathophysiology in mouse knockout or transgenic models. Since some of these RNA-binding proteins and splicing regulators (e.g. QKI, MBNL, RBM20) have recently also been found in genetic screens to serve as regulators of circRNA biogenesis, the question arose whether cardiovascular functions were due to circRNA regulation. We will review reports how these three factors related to cardiovascular pathophysiology, although current evidence supports the view that these roles are due to regulation of linear mRNA splicing.
Considering these three major routes, we will summarize the current state of knowledge about circRNAs in cardiovascular pathophysiology in the three chapters 5.1–5.3: a. evidence determined by differential circRNA expression profiling in cardiovascular tissue and blood, b. evidence determined by functional tests of candidate disease effectors, and c. evidence determined by deducing functionality from studies of circRNA-regulating proteins. Evidence from circRNA expression profiling studies will be summarized in Table 2, and in vivo, functional evidence from candidate approaches in Table 3. A summary of the suggested cellular functions of circRNAs and circRNA regulators, as determined from experiments with cells cultured in vitro, is presented in Fig. 3. To enable a quick overview of key relevant papers, we have highlighted the major publications in Box 2.
Table 2.
circRNA as biomarkers for cardiovascular disease.
| Name | Disease | Regulation (in Disease) | Function | human GWAS/Mouse models | Role in vivo | Ref. |
|---|---|---|---|---|---|---|
| circNPPA | Heart defects (multiple) |
n.a. (expressed in diseased tissue) |
n.a. |
Right atrium, vena cava, heart tissues |
n.a. (but produced from disease-linked mRNAs): | [234] |
| circCORIN | NPPA: heart failure [228] | [85] | ||||
| circRYR2 | CORIN: heart failure [229] | [84] | ||||
| circMYH6 | RYR2: atrial fibrillation [230] | [195] |
||||
| circQKI | MYH6: hypertrophic cardiomyopathies [231] | |||||
| circSLC8A1 | QKI: CAD [193] | |||||
| circTitin | SLC8A1/NCX: heart failure and arrhythmia, hypertension [232] | |||||
| (h.s.) | ||||||
|
TITIN: DCM [233]) | ||||||
|
circANRIL (h.s.) |
CAD |
Down |
Protective |
Blood T-cells, PBMCs, carotid endarterectomy tissue |
n.a. (Human GWAS: Associated with CAD risk SNPs (chr9p21); Generally antiproliferative, proapoptotic; Anticorrelation with linear ANRIL isoforms) |
[107] |
| [106] | ||||||
| [148,149,235] | ||||||
| hsa_circ_0124644 | CAD |
Up |
n.a. |
PBMCs |
n.a. |
[137] |
| hsa_circ_0082081 | ||||||
| hsa_circ_0113854 | ||||||
| hsa_circ_0098964 | ||||||
| hsa_circRNA5974-1 | ||||||
| (h.s.) | ||||||
| hsa_circ_0089378 | CAD |
Up |
n.a. |
Blood plasma |
n.a. |
[138] |
| hsa_circ_0083357 | ||||||
| hsa_circ_0082824 | ||||||
| hsa_circ_0068942 | ||||||
| hsa_circ_0057576 | ||||||
| hsa_circ_0054537 | ||||||
| hsa_circ_0051172 | ||||||
| hsa_circ_0032970 | ||||||
| hsa_circ_0006323 | ||||||
| (h.s.) | ||||||
| circHIPK3 | Diabetic retinopathy (TIID) |
Up |
n.a. |
Blood plasma, aqueous humor of eyes |
n.a. |
[139] |
| orthologs: | ||||||
| hsa_circ_0000284 | ||||||
| mmu_circ_0001052 | ||||||
|
(h.s., m.m.) # | ||||||
| Hsa_circ_0054633 | Prediabetic state (TIID) |
Up |
n.a. |
Peripheral whole blood |
n.a. |
[140] |
|
(h.s.) # | ||||||
| hsa-circ-0005870 | Hypertension |
Down |
n.a. |
Blood plasma |
n.a. |
[141] |
| (h.s.)# | ||||||
|
MICRA (h.s.) # |
MI: LVD |
Down |
n.a. |
Peripheral whole blood |
n.a. (predictor for LVD) |
[142] |
| circR-284:mir-221 ratio | Stroke (acute phase) | Up | n.a. | Blood serum | n.a. (circRNA Up/microRNA Down) | [144] |
| (h.s.) |
# (marking circRNAs associated with CAD risk factors), DCM (dilated cardiomyopathy), MI (myocardial infarction), LVD (Left ventricular dysfunction), LAD (permanent ligation of the left anterior descending), PBMCs (blood mononuclear cells), ECs (vascular endothelial cells), h.s. (homo sapiens), m.m. (mus musculus).
Table 3.
circRNAs and circRNA regulators in cardiovascular disease in vivo. List of circRNAs implicated as regulators of cardiovascular disease entities by in vivo evidence. Levels of expression and proposed functions in vivo are indicated. Asterisks (*) mark RNA-binding proteins that have been functionally studied and linked to CVD, and which have independently been found to regulate also circRNA biogenes - It is not known whether their role as circRNA regulators has any impact on their role in CVD.
| Name (Species) | Disease | Regulation (in Disease) | Function | Mouse Models | Role in vivo | Ref. |
|---|---|---|---|---|---|---|
| HRCR (m.m.) | Heart injury | Down | Protective | Cardiomyocytes? | ISO-induced heart injury mouse model: vein-injected HRCR-expressing construct rescued from cardiac hypertrophy | [170] |
| MFACR (m.m.) | Heart injury | Up | Deleterious | Cardiomyocytes? | I/R-induced heart injury mouse model: intracoronary injection of MFCR siRNA rescued from heart dysfunction | [171] |
| Cdr1as (m.m.) | MI | Up | Deleterious | Heart | LAD ligation-induced MI in mouse: intracardially injected CDR1as-expressing construct prior to MI increased infarction size | [143] |
| MBNL* (h.s. m.m. d.r.) | DM | Down | Protective | Cardiac muscle |
|
[236] [237] [238] |
| MBNL* (h.s., m.m.) | MI | Down | Protective | Cardiac myofibroblasts? |
|
[180] |
| QKI* (h.s. m.m.) | CAD | Up | Deleterious | VSMCs and macrophages? |
|
[190] [193] |
| QKI* (h.s. m.m. d.r.) | MI | Down | Protective | Cardiomyocytes? |
|
[191,192] [187] |
| RBM20* (h.s, m.m. d.r., r.n.) | DCM | Down | Protective | Heart |
|
[239] [194] [195] |
Abbreviations: CAD (coronary artery disease), ISO (isoproterenol), I/R (ischemia/reperfusion), T-cell (T-lymphocyte), PBMC (peripheral blood mononuclear cell), VSMC (vascular smooth muscle cell), GWAS (genome-wide association study), MI (myocardial infarction), LAD (left anterior descending artery), DCM (dilated cardiomyopathy), ER (endoplasmatic reticulum), CM (congenital myopathy), DM (myotonic dystrophy), KO (knockout), h.s. (homo sapiens), m.m. (mus musculus), d.r. (danio rerio), r.n. (rattus norvegicus).
Fig. 3.
Cellular roles of circRNAs in cardiovascular cell types in vitro.
Blood cells and cells of the cardiovascular system that are known to contribute to cardiovascular diseases are depicted on the left. Evidence for cellular roles of (A) circRNAs and of (B) RNA-binding proteins that have recently also been identified as circRNA biogenesis regulators. Evidence stems from in vitro experiments in cultured cells. Note that it is unknown whether the cardiovascular roles of the indicated RNA-binding proteins are related in any way to their function as circRNA biogenesis regulators.
Box 2. Reading highlights.
| Historical detection of circRNAs |
|
|
|
| Benchmarking circRNA detection and quantification |
|
| CircRNA biogenesis |
|
|
| Molecular functions of circular RNAs |
|
|
|
| CircRNA expression profiling in cardiovascular tissue |
|
| CircRNAs as biomarkers in the blood |
|
| CircRNAs as effectors of cardiovascular disease |
|
|
|
Alt-text: Box 2
5.1. circRNAs associated with cardiovascular disease by RNA expression profiling
5.1.1. CircRNA expression profiling in cardiovascular tissues
A number of studies have reported on profiling heart-specific sets of circRNAs, either in physiological or pathological conditions. For example, myocardial circRNAs were delineated by profiling hearts samples from different species: samples from healthy mouse and rat were compared to mouse hearts after pressure overload and to human samples, including RNA from hearts of heart failure patients [84]. Likewise, both human and mouse heart-specific circRNA sets were determined in a recent comprehensive approach that aimed to delineate cardiac circRNAs expressed in healthy or in pathological conditions [85]. The pathologies included ischaemic and non-ischaemic dilated cardiomyopathy and hypertrophic cardiomyopathy in humans, and material was derived from left ventricle myocardium in these cases. In mouse, RNA was analyzed from cardiomyocytes that were isolated from hearts after pressure-overload-induced hypertrophy as a consequence of transverse-aortic constriction (TAC) [85]. Independent papers focused on cataloging heart-specific circRNAs, from fetal to adult stages: For example, circRNAs profiles were established in young and maturing adult mouse hearts [121] or mapped based on existing publically available datasets [122]. Yet another circRNA profile was established by profiling expression in mouse ventricular tissue after permanent ligation of the left anterior descending coronary artery [123].
In another set of approach, circRNAs profiles were determined while profiling differentiation of human embryonic stem cell (hESC) to cardiomyocytes over 3 weeks in culture conditions [85]. In this study, among all the identified circRNAs, 1664 were heart-specific as compared to expression in other organs. Some of these stemmed from genes that had been independently functionally studied in the heart (Titin, RYR2 gene, HIPK3). A circRNA from the SLC8A1 gene, which encodes a Na+/Ca2+ exchanger, was the most abundant circRNA in human hearts and the second most abundant in mouse cardiomyocytes. Conceptually similar experiments let to the definition of circRNA species expressed when induced pluripotent stem cell (iPSC)-derived cardiomyocytes matured upon beta-adrenergic stimulation in culture [86]. The latter condition is used to reflect acute sympathetic stimulation of heart contractility or when chronic, hypertrophic proliferation and changes in apoptosis (see Ref. [124] for review). Top candidates from this diverse range of circRNA profiling studies may serve as entry points for future tests whether these circRNAs influenced cardiovascular diseases also in functional terms.
In another very recent approach, a team of researchers sequenced multiple human tissues of a single person in parallel, in order to focus on potentially disease-relevant circRNAs [125]. Among the identified candidates, those tissue-specific circRNAs were identified, for example, that mapped to genes with known physiological roles, as well as to genes where mutations had previously been associated with tissue-specific diseases. Related to cardiovascular disease, an atrial circRNA was identified in the NPPA gene, which encodes the natriuretic peptide and had been associated with diabetes. Other circRNAs identified were from CORIN, which is a peptidase that activates NPPA. With a similar rationale, circRNAs were highlighted in the ryanodine receptor gene RYR2, which is linked to atrial fibrillation, in myosin heavy chain MYH6 which associates with hypertrophic cardiomyopathy, and in the solute carrier SLC8A1 which controls Ca2+ levels and associates with cardiomyocyte dysfunction, heartbeat and regulation of hypertension [125] (Table 2). Follow-up studies on these circRNAs will reveal whether these circRNAs may be biomarkers or even effectors of cardiovascular disease endpoints.
5.1.2. Expression profiling establishes cell-free circRNAs as potential CVD risk biomarkers in the blood
Apart from tissue-centered expression profiling, the minimal-invasive molecular analysis of peripheral blood is gaining diagnostic importance. In the field of cardiometabolic diseases, classically, protein serum markers like cardiac troponins T/I and creatine kinases (CK, CK-MB) are profiled in blood samples. More recently, also microRNA profiles in blood have been shown to be potentially useful. For example, miR-1, miR-133, miR-208, and miR-499, as well as circulating lncRNAs, such as LIPCAR, MYHEART or UCA1, can be informative for monitoring myocardial infarction and heart failure, and a number of other small and long linear noncoding RNAs are emerging as possible biomarkers also of coronary artery disease (see Ref. [126] for review). Studies are currently performed that assess whether also cell-free circRNAs may serve as meaningful molecular targets in the analysis of blood. Exploratory studies have documented that circRNAs are not an exception from all other nucleic acids, and can be found in the cell-free transcriptome of bioliquids like blood [74,[152], [153], [154]] or saliva [127]. Thereby, circRNAs are expected to be particularly interesting during blood profiling because of their increased stability against cellular exonucleases compared to nucleic acids with linear ends [6,35,125,[128], [129], [130], [131], [132], [133]]. In blood, a major degradation pathway is the action of RNase A-like endonucleases and RNase A also degrades circular RNAs very efficiently [155]. Mechanisms exist that protect linear RNAs and circRNAs in blood. For example, RNAs (and other cellular components) are encapsulated in phospholipid-membrane-bound vesicles. These can be exosomes or the slightly larger microvesicles, or other fragments of cells like ER fragments [73,74,153,154,157,158]. RNAs may also be protected by coating with diverse RNA binding proteins, high-density and low-density lipoproteins or Argonaute proteins [[134], [135], [136]], which abolishes recognition by RNase A. Together, circRNA degradation routes are still an open field, but circRNAs stability, whatever the underlying mechanism, may indeed turn out to be an advantage when screening for disease-initiating that occurred in distant tissues or at some time point in the past. Subsequent studies have probed whether circRNAs can serve as biomarkers for cardiovascular disease. For example, specific circANRIL isoforms in whole blood-derived T-cells or in blood mononuclear cells (PBMCs) were found to anti-correlate with atherosclerosis risk genotypes and disease phenotype severity [106,107]. Conversely, levels of linear noncoding ANRIL RNA correlated with atherosclerosis (Table 2). This suggested that measuring both circular and linear ANRIL could be a robust indicator of atherosclerosis. ANRIL may also be an example, where one could think of gaining robust information by combining information from RNA and DNA sequencing of blood cells since specific predisposing atherosclerosis risk SNPs in the ANRIL locus have been suggested to affect the splicing pattern of ANRIL pre-mRNA and the decision between linear and circular RNA formation [107]. This is to our knowledge the only known case where such advanced insight is available and GWAS data have been firmly linked to circRNA biogenesis.
In another case, circRNA profiling in PBMCs from venous blood has delivered several candidate circRNAs, like crc_0124644, circ_0082081, circ_0113854, circ_0098964 and circRNA5974-1, which may also be useful as biomarkers of coronary artery disease (CAD) [137]. Dozens of other circRNAs were found in another study to be differentially expressed in plasma samples from (CAD) patients and control individuals. Based on existing microRNA profiling data, and using predictions of putative sponge effects to find anticorrelating circRNA:microRNA pairs, modest upregulation of hsa_circ_0089378, hsa_circ_0083357, hsa_circ_0082824, hsa_circ_0068942, hsa_circ_0057576, hsa_circ_0054537, hsa_circ_0051172, hsa_circ_0032970, and hsa_circ_0006323 was suggested to be indicative of CAD [138] (Table 2).
A set of separate studies investigated whether circRNAs could serve as biomarkers for cardiometabolic risk factors, such as dyslipidemia, diabetes or hypertension (Table 2): No circRNAs have yet been found to indicate dyslipidemia in blood. With respect to CAD risk factors, circHIPK3 has been shown to be upregulated in diseased retinal vascular tissue in diabetic patients. Compared to controls, circHIPK3 is more abundant also in blood plasma of patients as well as in the aqueous humor (fluid) of the eye [139]. This finding also shows that circRNAs will likely be found also in other bioliquids. Hsa_circ_0054633 was identified in a separate study as a potential biomarker in whole blood for the onset of the pre-diabetic phase in type II diabetes [140]. Another example of a circRNA for a CAD risk factor is hsa-circ-0005870, which was found to be less abundant in blood plasma of individuals with hypertension [141] (Table 2).
Relevant to myocardial infarction, myocardial infarction associated circular RNA (MICRA) in preparations of peripheral blood has been proposed to be useful as a predictor of left ventricular dysfunction [142]. Also, the well-described Cdr1as circRNA was shown to be upregulated in whole blood from acute myocardial infarction patients [143] (Table 2).
Finally, circR-284 was recently shown to be elevated to some extent in blood serum of patients with CAD and suffering from a recent cerebrovascular stroke, as compared to CAD patients having not yet experienced plaque rupture leading to stroke [144].
Summarizing, a few dozen human circRNAs have been suggested to potentially serve as blood biomarkers for state or stage of cardiovascular diseases. Since specific cells of the blood mechanistically contribute to CAD onset and progression, it is not excluded that some of the reported circRNAs in this list may also have functional pathophysiological relevance. For example, specific monocyte and macrophage subtypes, diverse T-lymphocyte subtypes, and diverse antigen-presenting cell types shape, indirectly or in the lesion itself, when and how atherosclerotic plaques form and develop (see Ref. [145] for review). On the other hand, a number of circRNAs have been shown to be differentially regulated in tissues of mouse models of myocardial fibrosis, cardiac hypertrophy, and myocardial infarction or in cultured models of vascular cell types. It is, however, currently unclear whether circRNAs orthologous to these mouse candidates are also differentially abundant in blood samples or in human patients. Also, more work is needed to independently consolidate the initial biomarker candidates and to rigorously assess biomarker performance in larger, independent, and appropriately control-matched cohorts. This relates not only to the normal range of circRNA biomarker levels in healthy individuals and the quantification of false-positive- and false-negative classification rates (see Ref. [146] for review) but also to benchmarking biochemical and bioinformatics approaches used to sequence and identify circRNAs [4].
5.2. Candidate approaches to delineate circRNAs as potential effectors of cardiovascular disease
Only a few circRNAs have been functionally studied so far. Therefore the number of circRNAs that serve as candidate effectors of cardiovascular disease is rather small. Relevant studies concurrently analyzed circRNA regulation during cardiovascular injury in rodent models in vivo mimicked pathological processes in physiological relevant cell types in culture, and eventually correlated circRNA expression with known disease risk genotypes in human association studies. From the existing evidence, it becomes clear that no single overarching effector mechanism is apparent. In the few cases where circRNAs were tested in the context of cardiovascular disease in vivo, such as by classical transgenic expression in mice, or from injection of virus-encoded expression cassettes into the circulation or into the heart, the induced expression was system-wide (not cell-type specific) and constituted ectopic overexpression (not from the endogenous locus). This is important to realize when interpreting the results of these studies. In the following, we will first report on circRNAs that have been studied with connotation to human cardiovascular disease, and then circRNAs that have emerged in animal disease models.
In humans, the strongest known genetic risk determinant for atherosclerosis resides on a locus on chromosome 9p21, where independent studies have discovered tightly-clustering SNPs that associate with myocardial infarction [147], coronary artery disease (CAD) [148,149] and aneurysms [150]. These SNPs are independent of the traditional risk factors like dyslipidemia, hypertension, type II diabetes (TIID), or obesity. While independent from association with TIID, a TIID-linked haplotype block lies adjacent to the CAD and myocardial infarction-linked haplotype blocks on 9p21 [[151], [152], [153]]. Regarding CAD/MI, the relevant SNPs correlate with atherosclerosis plaque development and the risk for MI is a consequence of increased atherosclerosis burden [154]. The major effector of this locus is not a protein-coding gene but a long noncoding RNA, human ANRIL, whose expression levels associate with more severe atherosclerosis phenotypes [149,[155], [156], [157], [158], [159], [160]]. Mechanistically, on the one hand, human linear ANRIL has been implicated in the chromatin-based silencing of the INK4 locus: ANRIL was shown to bind both PRC1 and PRC2 polycomb complexes, which are one of the most well-studied repressive histone modifying complexes in multicellular organisms [161,162]. INK4 silencing by ANRIL in cis, thus, may contribute to cell cycle progression by promoting G0-G1 cell cycle entry. This may hypothetically be relevant for ANRIL's described roles in activating atherosclerosis [163]. On the other hand, ANRIL can also interact with transcriptional coactivators to alter the transcription of target genes in trans and, thereby, affect cell adhesion, apoptosis and inflammation in ways that stimulate atherosclerosis [164,165].
In any case, the human ANRIL locus has recently also been shown to yield a number of circular RNA isoforms [106,107] making the picture more complex. Specifically, circANRIL expression was found to correlate with the occurrence of cardiovascular risk alleles [106,107]: Specifically, atherosclerosis disease severity correlated with reduced circANRIL levels in a number of cell types, such as PBMCs, blood T-lymphocytes and cells of the vascular wall [106,107] (Table 2). This evidence and subsequent results from functional testing suggested that circANRIL protects from CAD. Indeed, when expressed in cultured cells, including vascular cell types, circANRIL was found to reduce cell proliferation and to increase the sensitivity to pro-apoptotic signals, functions that are conceptually able to protect from atherosclerotic progression in the vascular wall in specific contexts [106] (Table 3, Fig. 3). For such a net atheroprotective function, circANRIL was suggested to bind to and inhibit the PES1 protein, a member of the PeBoW complex, which is known to be essential for pre-rRNA processing and consequently for the formation of functional ribosomes. As a consequence of increased circANRIL levels, rRNA maturation was impaired, the capacity of cells to translate proteins dropped, and this results in nucleolar stress, p53 activation in cultured cells and antigrowth conditions [106].
A number of studies investigated the role of the first described circRNA, CDR1as circRNA, in diverse tissues, cells, and contexts. CDR1as circRNA was found to be induced in the blood of acute myocardial infarction patients. Separately, CDR1as circRNA was also described to be upregulated in a mouse model for myocardial infarction, both in infarcted tissue and in circulating blood. The model was put forward that CDR1as circRNA might not only serve as a biomarker for disease, but that CDR1as circRNA also might be a stimulator of infarction size. The authors suggested that for this function CDR1-as circRNA served as a sponge for miR-7a and inhibited the antiapoptotic effects of this microRNA in cardiomyocytes [143]. This latter interpretation has, however, to be met with caution, as the implication of both CDR1as and miR-7, and their genetic interaction, was made based on overexpression of both factors, and functional experiments were not performed in vivo. Moreover, CDR1as and miR-7 are known to be highly expressed in brain tissues, their expression is low or absent in non-neuronal tissues [8] and the mouse knockout shows a specific neurological deficit [95]. Thus, further experiments have to substantiate the proposed model.
Also, the gene encoding the zinc finger ZNF292 expresses circRNAs (cZNF292) [166]. cZNF292 was identified by RNA high-throughput sequencing of cultured human umbilical vein endothelial cells (HUVECs). Hypoxic culture conditions were found to affect the abundance of selected circRNAs, and among those, cZNF292 became one of the most abundant circRNAs [166]. siRNA-mediated inhibition of a skipped exon that was included in cZNF292 allowed to deplete this circRNA in HUVECs. This intervention inhibited the typical proangiogenic phenotypes that can be tested in cultured endothelial cells such as sprouting or tube formation in Matrigel. The mechanism of action of cZNF292 is unknown, but the authors excluded the possibility that a microRNA sponge effect was involved [166] (Fig. 3).
Hsa_circ_0010729 is another human circRNA that was found to be induced by hypoxic conditions in HUVEC cells [167]. It was implicated as an antisurvival factor in HUVEC cells under hypoxic conditions. The effector mechanism for hsa_circ_0010729 is not clear [167]. Hsa_circ_0003575 is yet another circRNA that is differentially regulated in stressed HUVECs. It is upregulated upon stimulation with oxidized LDL, a classical in vitro model for studying effects of cardiovascular pathology [138]. siRNA-mediated knockdown showed that hsa_circ_0003575 was antiproliferative and antiangiogenic in HUVEC cells (Fig. 2).
Another cell type relevant for cardiovascular diseases is the vascular smooth muscle cell (VSMC). For example, several dozens of circRNAs were found to be differentially regulated in human VSMCs cultured in vitro under high glucose administration, a culture system used to mimic diabetic conditions [168]. Among the differentially expressed circRNAS, circWDR77 was found to be induced. Based on siRNA-mediated knockdown, it was concluded that circWDR77 contributed to cell survival, S-phase cell cycle entry and migrative behavior [168]. Since miR-124 bound circWDR77, and since miR-124 knockdown rescued the cellular defects of circWDR77 depletion, this circRNA was suggested to act as a sponge for miR-124 [168]. As a consequence, one possibility is, that FGF2, a well-known promigrative and pro-proliferative factor for VSCMs, and targeted by miR-124, is one of the potential effectors of circWDR77 in this pathway [168]. A similar study explored circRNA profiles in cultured VSMCs under hypoxic conditions as well as in aortic tissue from patients with aortic aneurysms [169]. hsa-circ-000595 was found to be induced in these cases, and siRNA-mediated knockdown experiments suggested that this circRNA slightly contributed to apoptosis under chemically-induced HIF-1 activation (hypoxic-like conditions) in cell culture [169] (Fig. 3).
In yet another mouse model of ischemia/reperfusion, the MFACR circRNA was found to be upregulated [171]. Nuclear-encoded mitochondrial protein MTP18 is known to be important for cellular viability by facilitating mitochondrial fission during the complex homeostatic balance of fusion and fission that is necessary for mitochondrial function. Especially the unbalanced overshooting of mitochondrial fission in many different cell types, including cardiomyocytes, has been previously implicated in cardiometabolic syndrome. In the injury model, mouse circRNA MFACR promoted MTP18 protein levels and mitochondrial fission. In the injury model in vivo, based on systemic manipulation of MFACR levels, the authors showed that this circRNA stimulated MTP18, mitochondrial fission, apoptosis and heart dysfunction [171]. The authors suggested that miR-652-3p was impaired downstream of MFAC function and repressed MTP18 in a deleterious pathway (Fig. 3).
In another set of experiments, some circRNAs were mouse circ_000203, as well as mouse circRNA_010567, were hypothesized to function as potential promoters of cardiac fibrosis by two separate studies that used the same experimental setup [172,173]: In these reports, circ_000203 and the transcript of its linear host gene Myosin 9a, as well as circ_010567 were found to be induced in the myocardium of db/db mutant mice, which serve as a model for type II diabetes. Further, in vivo evidence was not reported, and insight is limited to experiments in cardiomyocytes stimulated with angiotensin II. Under these conditions, overexpression of circ_010567 induced the expression of three fibrosis marker genes that were concurrently repressed by miR-26b [172]. The authors of this study discussed the possibility that circ_000203 served as a sponge for miR-26b. In the second report, circ_010567 knockdown in cultured cardiac fibroblasts repressed the same three fibrosis marker genes under conditions of angiotensin II exposure. Since miR-141 was repressed by angiotensin II, and circ_010567 inhibition partially rescued the downregulation of miR-141, the authors hypothesized that circ_010567 might stimulate fibrosis as an upstream inhibitor of miR-141 transcription [173]. The mechanism underlying the proposed negative regulation is unknown.
Finally, and with respect to CAD risk factors, circHIPK3 was identified as an abundantly expressed circRNA induced in diabetic conditions. circHIPK3 was expressed both, in mice, as well as in diabetic human retinas and in cultured human retinal vascular endothelial cells (HRVECs) under glucose stimulation, which served as a relevant stressor in vitro [139]. circHIPK3 was found to be required for optimal growth of HRVECs in baseline and stressed conditions and for expression of selected miR-30 targets. The possibility was discussed that circHIPK3 served as a sponge for miR-30 targets not only in vitro but also in the retinal vasculature. Systemic depletion of circHIPK3 by expression of shRNAs targeting the backsplice junction from injected adenoviral constructs, the phenotype of retinopathy in the diabetic mouse model was partially ameliorated. Conversely, miR-30 downregulation in a similar experiment increased retinopathy. Since an epistatic relationship was found in double-knockdown experiments for circHIPK3 and miR-30, and since miR-30 was pulled down by circHIPK3 in Ago2 complexes, the possibility exists that circHIPK3 is indeed a physiologically relevant sponge of miR-30 [139]. A similar pro-proliferative cellular function for circHIPK3 was previously also suggested in a separate study, that showed that circHIPK3 more generally stimulated proliferation of a number of different cell types by sponging growth-suppressive microRNAs like miR-124 [66]. Together, circHIPK3 may exert a pro-proliferative role in different tissues, possibly by sponging different growth-restrictive microRNAs (Fig. 3).
5.3. Some circRNA-regulatory proteins have an independently reported function as regulators of cardiovascular physiology
Some upstream regulators of circRNA biogenesis or stability have been studied already before the re-discovery of circRNAs in the context of cardiovascular disease. For example, more than 700 RBPs are known to be encoded in the genome, and more than a hundred of these have been shown to specifically affect circRNA biogenesis [174]. Despite their expected pleiotropic roles as RNA-binding proteins, some tissue- and lineage-specific RBPs and splicing regulators have been successfully connected to rather specific sets of linear target mRNAs (see Refs. [175,176] for review). Still, the large number of targets makes it then difficult to associate a single mRNA or a single splicing event with a phenotype, let alone to associate a cardiovascular function with the so far independent role in circRNA regulation. Therefore it is only a curiosity, that rather specific cardiovascular functions have been reported for three RBPs that have recently (but independently) also been revealed as circRNA biogenesis regulators:
5.3.1. The Muscleblind family of RNA-binding proteins
The first of these RBPs is the Muscleblind protein family (mouse Mbnl1-3, human MBNL1-3). Mbnl/MBNL1-3 proteins are RNA binding proteins, splicing regulators [177,178], alternative polyadenylation regulators [70] and circRNA regulators [53]. Muscleblind proteins are not specifically expressed in cardiovascular tissues, but compared to other organs they are relatively highly expressed in hearts of both mice and man. Mbnl1 knockout studies have shown that Muscleblind it is required for normal heart valve development [179] and for fibroblast-to-myofibroblast differentiation [180] (Fig. 3). Besides a number of other phenotypes, and with relevance to disease, Mbnl1 knockout mice also develop myotonic dystrophy 1 (DM)-related cardiac defects including myocardial fiber death, fibrosis, cardiac hypertrophy and conduction problems [180] (Table 3). These heart problems are the second most frequent cause of death in DM patients [181]. Cell differentiation defects in Mbnl1 mutants have been ascribed to a failure of cells in transitioning from a naïve and undifferentiated splicing pattern to a differentiation-determining splicing pattern [177,178]. In fact, Mbnl regulates mRNA splicing in the maturing heart [182]. The question that has not yet been addressed is whether Mbnl-dependent regulation in the heart is due to circRNA regulation or primarily to regulation of linear splicing. The implication of Mbnl1 in DM is better understood in this regard: Critical to the onset of DM is that specific RNAs acquire CUG repeat extensions, which causes these mutant RNAs to sequester Mbnl and to impair Mbnl-dependent linear mRNA splicing [183,184]. In the cardiovascular system, it might be the mis-splicing of surprisingly few important cardiovascular regulatory mRNAs, such as of SCN5A, that trigger disease. SCN5A mRNA splicing has been found to contribute to a significant part of the cardiac-conduction delay and heart arrhythmia in Mbnl1 mutants [185]. Yet, also in a disease context, any involvement of circRNA biogenesis is speculative. To the contrary, as a test circRNA, circMbnl1 levels were quantified in DM patients and in transgenic CUG-repeat-expressing mice [183] that serve as a mouse model for Mbnl1 -protein inhibition in DM, but no changes were found in the diseased state [186]. This may speak against an implication of circRNAs in DM and be explained by the known upregulation of Mbnl1 transcription upon Mbnl1 protein loss, which would effectively counteract decreases in circMbnl1 by increasing Mbnl1 transcription.
5.3.2. The RNA-binding protein Quaking
The second relevant RBP is the global linear splicing regulator Quaking (QKI). Apart from its major role in nervous system function, it is also required for muscle fibril formation [187], visceral endoderm differentiation, and vessel formation in embryogenesis [188,189]. QKI also partakes in the onset of atherosclerosis, where it regulates neointimal vascular smooth muscle cell (VSMC) proliferation in adult human coronary lesions [190]. QKI may also participate in the resolution of heart injury [191] (Table 3). Others studies showed that QKI is involved in endothelial layer formation. Not last SNPs in QKI have recently been associated with risk for myocardial infarction and coronary heart disease [192] (Table 3). It is still not fully clear in which cell types QKI would function in disease, but Qki is known to promote monocyte adhesion, and to stimulate differentiation of monocytes to inflammatory macrophages and foam cells in atherosclerotic plaques [193] (Fig. 3). Together, QKI has a surprisingly large number of functions in the cardiovascular system, but as for Mbnl, whether any of these is due to circRNA regulation is unknown.
5.3.3. The RNA-binding protein RBM20
A third example is Rbm20, a splicing regulator implicated in hereditary dilated cardiomyopathy, one of the leading causes of heart failure [194] (Table 3). Rbm20 has been found to be responsible for alternative splicing of at least 30 genes, some of which are known to regulate cardiomyocytes, sarcomere organization or biomechanical muscle function, or are otherwise implicated in heart failure or cardiomyopathy [194] (Fig. 3). Among these is Titin, a gene encoding a sarcomeric filament protein that is inherently linked to force generation in muscle cells and hence to myocardial function. The Titin gene is known to be massively alternatively spliced and, thus, became the focus of interest with respect to circRNA biology. Indeed, both mouse and human Titin express up to 80 different circular RNA isoforms in the heart [195]. Moreover, the splicing factor Rbm20 has been found to be involved in the biogenesis of circTitin, because it potentially binds to introns flanking the circularization events. An Rbm20 mutation was generated in the mouse, and it was found that these knockout mice experienced early onset dilated cardiomyopathy [195]. In the absence of Rbm20, the formation of a specific subset of circRNAs from the Titin gene was found to be defective in heart tissue [195]. Together, this raised the possibility that RBM20-dependent changes in circRNA biogenesis of Titin contribute to disease onset [195,196], hypothetically related to the fact that circTitin formation occurs in exons encompassing the region determining linear Titin's biomechanical rigidity [195]. This possibility has, however, so far not been tested and the implication of RBM20-dependent circRNAs in CVD is solely correlative at this point.
Complicating the analysis, in addition to the already described functions in regulating linear mRNA expression, Muscleblind [178,[197], [198], [199]], Quaking [[200], [201], [202]], or also other RBPs like HuR [[203], [204], [205], [206], [207], [208], [209], [210]], have specific pleiotropic and context-dependent functions in cardiovascular cell types. Therefore, whether and to what extent the regulation of circRNAs, as opposed to the regulation of alternative linear splicing, contributes to the known Mbnl- or Qki- or Rbm20-dependent processes in cardiovascular physiology and disease will be difficult to be teased apart.
6. Outlook
During the past years, it has become clear that thousands of genes express circular RNAs and that these have gene regulatory potential. CircRNA formation is a regulated process, and deregulation of circRNA formation can contribute to cellular and organismal defects. CircRNAs have not only been involved in cardiovascular disease but also in neurological functions (see Ref. [211] for review) and in cancer-related processes such as overproliferation, transformation, metastasis, and chemotherapeutic drug resistance (see Ref. [212] for review). So far it has not been reported whether circRNAs can serve as successful targets or therapeutic agents in preclinical settings. Approaches focusing on RNA as a therapeutic molecule, are, however, gaining importance. What makes them interesting candidates is their intracellular stability. So far, approaches centering on RNA transgenics pertain to linear RNAs, such as mRNAs or interfering RNAs: technologies for their delivery into cells of living tissues are improving, whereby chemical modifications are used to increase their stability [213]. In the future, also circRNAs might be administered for therapeutic purposes in vivo, their endogenous stability may be superiors, and additional covalent modification of their backbone may increase their application even further. Also, one could imagine that antisense oligonucleotides could be transfected into tissues in vivo, which bind to splice-regulatory sequences and could be designed to affect backsplicing at a specific locus. Thereby one might be in the position to either increase or decrease the frequency of endogenous circRNA biogenesis from a specific locus. In this context, and what is maybe best shown in the example of ANRIL, it is likely the defined expression levels of both, linear and circular RNA, that determines the function of a gene. Finally, even if not all circRNAs were always functional, circRNAs may still be used as biomarkers for defining cell or disease states.
Disclosures
None. There are no competing interests.
Sources of funding
This work was funded by the University Hospital Munich and by the German Research Foundation as part of the Collaborative Research Center CRC1123 “Atherosclerosis - Mechanisms and Networks of Novel Therapeutic Targets” (project B1). Part of this project was also funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation, # 394237736).
Acknowledgements
None.
References
- 1.Ulitsky I. Evolution to the rescue: using comparative genomics to understand long non-coding RNAs. Nat. Rev. Genet. 2016;17:601–614. doi: 10.1038/nrg.2016.85. [DOI] [PubMed] [Google Scholar]
- 2.Rizki G., Boyer L.A. Lncing epigenetic control of transcription to cardiovascular development and disease. Circ. Res. 2015;117:192–206. doi: 10.1161/CIRCRESAHA.117.304156. [DOI] [PubMed] [Google Scholar]
- 3.Salzman J., Gawad C., Wang P.L., Lacayo N., Brown P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7 doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szabo L., Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat. Rev. Genet. 2016;17:679–692. doi: 10.1038/nrg.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki H., Zuo Y., Wang J., Zhang M.Q., Malhotra A., Mayeda A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006;34:e63. doi: 10.1093/nar/gkl151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo J.U., Agarwal V., Guo H., Bartel D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Memczak S. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 9.Salzman J., Chen R.E., Olsen M.N., Wang P.L., Brown P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 11.Szabo L. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015;16:126. doi: 10.1186/s13059-015-0690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Westholm J.O. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9:1966–1980. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Starke S., Jost I., Rossbach O., Schneider T., Schreiner S., Hung L.H., Bindereif A. Exon circularization requires canonical splice signals. Cell Rep. 2015;10:103–111. doi: 10.1016/j.celrep.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 14.Li Z. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 15.Clement J.Q., Qian L., Kaplinsky N., Wilkinson M.F. The stability and fate of a spliced intron from vertebrate cells. RNA. 1999;5:206–220. doi: 10.1017/s1355838299981190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacquier A., Rosbash M. RNA splicing and intron turnover are greatly diminished by a mutant yeast branch point. Proc. Natl. Acad. Sci. U. S. A. 1986;83:5835–5839. doi: 10.1073/pnas.83.16.5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 18.Yang L., Duff M.O., Graveley B.R., Carmichael G.G., Chen L.L. Genomewide characterization of non-polyadenylated RNAs. Genome Biol. 2011;12:R16. doi: 10.1186/gb-2011-12-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gardner E.J., Nizami Z.F., Talbot C.C., Jr., Gall J.G. Stable intronic sequence RNA (sisRNA), a new class of noncoding RNA from the oocyte nucleus of Xenopus tropicalis. Genes Dev. 2012;26:2550–2559. doi: 10.1101/gad.202184.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu Z., Filonov G.S., Noto J.J., Schmidt C.A., Hatkevich T.L., Wen Y., Jaffrey S.R., Matera A.G. Metazoan tRNA introns generate stable circular RNAs in vivo. RNA. 2015;21:1554–1565. doi: 10.1261/rna.052944.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nielsen H., Fiskaa T., Birgisdottir A.B., Haugen P., Einvik C., Johansen S. The ability to form full-length intron RNA circles is a general property of nuclear group I introns. RNA. 2003;9:1464–1475. doi: 10.1261/rna.5290903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zaug A.J., Grabowski P.J., Cech T.R. Autocatalytic cyclization of an excised intervening sequence RNA is a cleavage-ligation reaction. Nature. 1983;301:578–583. doi: 10.1038/301578a0. [DOI] [PubMed] [Google Scholar]
- 23.Murray H.L., Mikheeva S., Coljee V.W., Turczyk B.M., Donahue W.F., Bar-Shalom A., Jarrell K.A. Excision of group II introns as circles. Mol. Cell. 2001;8:201–211. doi: 10.1016/s1097-2765(01)00300-8. [DOI] [PubMed] [Google Scholar]
- 24.Branch A.D., Robertson H.D. A replication cycle for viroids and other small infectious RNA's. Science. 1984;223:450–455. doi: 10.1126/science.6197756. [DOI] [PubMed] [Google Scholar]
- 25.Rybak-Wolf A. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 26.You X. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 2015;18:603–610. doi: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Broadbent K.M., Broadbent J.C., Ribacke U., Wirth D., Rinn J.L., Sabeti P.C. Strand-specific RNA sequencing in Plasmodium falciparum malaria identifies developmentally regulated long non-coding RNA and circular RNA. BMC Genom. 2015;16:454. doi: 10.1186/s12864-015-1603-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang P.L. Circular RNA is expressed across the eukaryotic tree of life. PLoS One. 2014;9 doi: 10.1371/journal.pone.0090859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conn S.J. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 30.Kelly S., Greenman C., Cook P.R., Papantonis A. Exon skipping is correlated with exon circularization. J. Mol. Biol. 2015;427:2414–2417. doi: 10.1016/j.jmb.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X.O., Dong R., Zhang Y., Zhang J.L., Luo Z., Zhang J., Chen L.L., Yang L. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016;26:1277–1287. doi: 10.1101/gr.202895.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X.O., Wang H.B., Zhang Y., Lu X., Chen L.L., Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 33.Gao Y., Wang J., Zheng Y., Zhang J., Chen S., Zhao F. Comprehensive identification of internal structure and alternative splicing events in circular RNAs. Nat. Commun. 2016;7:12060. doi: 10.1038/ncomms12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barrett S.P., Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143:1838–1847. doi: 10.1242/dev.128074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glazar P., Papavasileiou P., Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y.C. CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res. 2016;44:D209–D215. doi: 10.1093/nar/gkv940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang J.H., Li J.H., Shao P., Zhou H., Chen Y.Q., Qu L.H. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011;39:D202–D209. doi: 10.1093/nar/gkq1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia S. CSCD: a database for cancer-specific circular RNAs. Nucleic Acids Res. 2018;46:D925–D929. doi: 10.1093/nar/gkx863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao Y., Wang J., Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015;16:4. doi: 10.1186/s13059-014-0571-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng J., Metge F., Dieterich C. Specific identification and quantification of circular RNAs from sequencing data. Bioinformatics. 2016;32:1094–1096. doi: 10.1093/bioinformatics/btv656. [DOI] [PubMed] [Google Scholar]
- 41.Wang K. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010;38:e178. doi: 10.1093/nar/gkq622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chuang T.J., Wu C.S., Chen C.Y., Hung L.Y., Chiang T.W., Yang M.Y. NCLscan: accurate identification of non-co-linear transcripts (fusion, trans-splicing and circular RNA) with a good balance between sensitivity and precision. Nucleic Acids Res. 2016;44:e29. doi: 10.1093/nar/gkv1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Izuogu O.G., Alhasan A.A., Alafghani H.M., Santibanez-Koref M., Elliott D.J., Jackson M.S. PTESFinder: a computational method to identify post-transcriptional exon shuffling (PTES) events. BMC Bioinf. 2016;17:31. doi: 10.1186/s12859-016-0881-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoffmann S. A multi-split mapping algorithm for circular RNA, splicing, trans-splicing and fusion detection. Genome Biol. 2014;15:R34. doi: 10.1186/gb-2014-15-2-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song X., Zhang N., Han P., Moon B.S., Lai R.K., Wang K., Lu W. Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res. 2016;44:e87. doi: 10.1093/nar/gkw075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roy C.K., Olson S., Graveley B.R., Zamore P.D., Moore M.J. Assessing long-distance RNA sequence connectivity via RNA-templated DNA-DNA ligation. Elife. 2015;4 doi: 10.7554/eLife.03700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hansen T.B., Veno M.T., Damgaard C.K., Kjems J. Comparison of circular RNA prediction tools. Nucleic Acids Res. 2016;44:e58. doi: 10.1093/nar/gkv1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeng X., Lin W., Guo M., Zou Q. A comprehensive overview and evaluation of circular RNA detection tools. PLoS Comput. Biol. 2017;13 doi: 10.1371/journal.pcbi.1005420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomson D.W., Dinger M.E. Endogenous microRNA sponges: evidence and controversy. Nat. Rev. Genet. 2016;17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 50.Barrett S.P., Parker K.R., Horn C., Mata M., Salzman J. ciRS-7 exonic sequence is embedded in a long non-coding RNA locus. PLoS Genet. 2017;13 doi: 10.1371/journal.pgen.1007114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y., Xue W., Li X., Zhang J., Chen S., Zhang J.L., Yang L., Chen L.L. The biogenesis of nascent circular RNAs. Cell Rep. 2016;15:611–624. doi: 10.1016/j.celrep.2016.03.058. [DOI] [PubMed] [Google Scholar]
- 52.Barrett S.P., Wang P.L., Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife. 2015;4 doi: 10.7554/eLife.07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ashwal-Fluss R. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 54.Kramer M.C., Liang D., Tatomer D.C., Gold B., March Z.M., Cherry S., Wilusz J.E. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015;29:2168–2182. doi: 10.1101/gad.270421.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li X. Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol. Cell. 2017;67(2):214–227.e7. doi: 10.1016/j.molcel.2017.05.023. [DOI] [PubMed] [Google Scholar]
- 56.Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y., Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21:172–179. doi: 10.1261/rna.048272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pamudurti N.R. Translation of CircRNAs. Mol. Cell. 2017;66(1):9–21. doi: 10.1016/j.molcel.2017.02.021. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oesterreich F.C., Herzel L., Straube K., Hujer K., Howard J., Neugebauer K.M. Splicing of nascent RNA coincides with intron exit from RNA polymerase II. Cell. 2016;165:372–381. doi: 10.1016/j.cell.2016.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ivanov A. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170–177. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 61.Pasman Z., Been M.D., Garcia-Blanco M.A. Exon circularization in mammalian nuclear extracts. RNA. 1996;2:603–610. [PMC free article] [PubMed] [Google Scholar]
- 62.Dubin R.A., Kazmi M.A., Ostrer H. Inverted repeats are necessary for circularization of the mouse testis Sry transcript. Gene. 1995;167:245–248. doi: 10.1016/0378-1119(95)00639-7. [DOI] [PubMed] [Google Scholar]
- 63.Liang D., Wilusz J.E. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lander E.S. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 65.Sela N., Mersch B., Gal-Mark N., Lev-Maor G., Hotz-Wagenblatt A., Ast G. Comparative analysis of transposed element insertion within human and mouse genomes reveals Alu's unique role in shaping the human transcriptome. Genome Biol. 2007;8:R127. doi: 10.1186/gb-2007-8-6-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng Q. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ho T.H., Charlet B.N., Poulos M.G., Singh G., Swanson M.S., Cooper T.A. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fujita N., Sato S., Kurihara T., Inuzuka T., Takahashi Y., Miyatake T. Developmentally regulated alternative splicing of brain myelin-associated glycoprotein mRNA is lacking in the quaking mouse. FEBS Lett. 1988;232:323–327. doi: 10.1016/0014-5793(88)80762-2. [DOI] [PubMed] [Google Scholar]
- 69.Fagg W.S., Liu N., Fair J.H., Shiue L., Katzman S., Donohue J.P., Ares M., Jr. Autogenous cross-regulation of Quaking mRNA processing and translation balances Quaking functions in splicing and translation. Genes Dev. 2017;31:1894–1909. doi: 10.1101/gad.302059.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Batra R. Loss of MBNL leads to disruption of developmentally regulated alternative polyadenylation in RNA-mediated disease. Mol. Cell. 2014;56:311–322. doi: 10.1016/j.molcel.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Errichelli L. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat. Commun. 2017;8:14741. doi: 10.1038/ncomms14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liang D., Tatomer D.C., Luo Z., Wu H., Yang L., Chen L.L., Cherry S., Wilusz J.E. The output of protein-coding genes shifts to circular RNAs when the Pre-mRNA processing machinery is limiting. Mol. Cell. 2017;68:940–954. doi: 10.1016/j.molcel.2017.10.034. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schneider M., Will C.L., Anokhina M., Tazi J., Urlaub H., Luhrmann R. Exon definition complexes contain the tri-snRNP and can be directly converted into B-like precatalytic splicing complexes. Mol. Cell. 2010;38:223–235. doi: 10.1016/j.molcel.2010.02.027. [DOI] [PubMed] [Google Scholar]
- 74.Guttman M., Rinn J.L. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Legnini I. Circ-ZNF609 is a circular RNA that can Be translated and functions in myogenesis. Mol. Cell. 2017;482(7385):339–346. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Conn V.M. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nature Plants. 2017;3:17053. doi: 10.1038/nplants.2017.53. [DOI] [PubMed] [Google Scholar]
- 77.Yang Z.G. The circular RNA interacts with STAT3, increasing its nuclear translocation and wound repair by modulating Dnmt3a and miR-17 function. Mol. Ther. 2017;25:2062–2074. doi: 10.1016/j.ymthe.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schneider T. CircRNA-protein complexes: IMP3 protein component defines subfamily of circRNPs. Sci. Rep. 2016;6:31313. doi: 10.1038/srep31313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chedin F. Nascent connections: r-loops and chromatin patterning. Trends Genet. 2016;32:828–838. doi: 10.1016/j.tig.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Engreitz J.M. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell. 2014;159:188–199. doi: 10.1016/j.cell.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kwek K.Y., Murphy S., Furger A., Thomas B., O'Gorman W., Kimura H., Proudfoot N.J., Akoulitchev A. U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nat. Struct. Biol. 2002;9:800–805. doi: 10.1038/nsb862. [DOI] [PubMed] [Google Scholar]
- 82.Fong Y.W., Zhou Q. Stimulatory effect of splicing factors on transcriptional elongation. Nature. 2001;414:929–933. doi: 10.1038/414929a. [DOI] [PubMed] [Google Scholar]
- 83.Kaida D., Berg M.G., Younis I., Kasim M., Singh L.N., Wan L., Dreyfuss G. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468:664–668. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Werfel S., Nothjunge S., Schwarzmayr T., Strom T.M., Meitinger T., Engelhardt S. Characterization of circular RNAs in human, mouse and rat hearts. J. Mol. Cell. Cardiol. 2016;98:103–107. doi: 10.1016/j.yjmcc.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 85.Tan W.L. A landscape of circular RNA expression in the human heart. Cardiovasc. Res. 2017;113:298–309. doi: 10.1093/cvr/cvw250. [DOI] [PubMed] [Google Scholar]
- 86.Siede D. Identification of circular RNAs with host gene-independent expression in human model systems for cardiac differentiation and disease. J. Mol. Cell. Cardiol. 2017;109:48–56. doi: 10.1016/j.yjmcc.2017.06.015. [DOI] [PubMed] [Google Scholar]
- 87.Vakhrusheva O., Smolka C., Gajawada P., Kostin S., Boettger T., Kubin T., Braun T., Bober E. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ. Res. 2008;102:703–710. doi: 10.1161/CIRCRESAHA.107.164558. [DOI] [PubMed] [Google Scholar]
- 88.Araki S. Sirt7 contributes to myocardial tissue repair by maintaining transforming growth factor-beta signaling pathway. Circulation. 2015;132:1081–1093. doi: 10.1161/CIRCULATIONAHA.114.014821. [DOI] [PubMed] [Google Scholar]
- 89.Koh W., Gonzalez V., Natarajan S., Carter R., Brown P.O., Gawad C. Dynamic ASXL1 exon skipping and alternative circular splicing in single human cells. PLoS One. 2016;11 doi: 10.1371/journal.pone.0164085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Saldi T., Cortazar M.A., Sheridan R.M., Bentley D.L. Coupling of RNA polymerase II transcription elongation with Pre-mRNA splicing. J. Mol. Biol. 2016;428:2623–2635. doi: 10.1016/j.jmb.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lewis B.P., Green R.E., Brenner S.E. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl. Acad. Sci. U. S. A. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ebert M.S., Neilson J.R., Sharp P.A. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Br. J. Pharmacol. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Poliseno L., Salmena L., Zhang J., Carver B., Haveman W.J., Pandolfi P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Franco-Zorrilla J.M. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 95.Piwecka M. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357 doi: 10.1126/science.aam8526. [DOI] [PubMed] [Google Scholar]
- 96.Denzler R., Agarwal V., Stefano J., Bartel D.P., Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol. Cell. 2014;54:766–776. doi: 10.1016/j.molcel.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bosson A.D., Zamudio J.R., Sharp P.A. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Mol. Cell. 2014;56:347–359. doi: 10.1016/j.molcel.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Karreth F.A. The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell. 2015;161:319–332. doi: 10.1016/j.cell.2015.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Denzler R., McGeary S.E., Title A.C., Agarwal V., Bartel D.P., Stoffel M. Impact of MicroRNA levels, target-site complementarity, and cooperativity on competing endogenous RNA-regulated gene expression. Mol. Cell. 2016;64:565–579. doi: 10.1016/j.molcel.2016.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mullokandov G. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Br. J. Pharmacol. 2012;9:840–846. doi: 10.1038/nmeth.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yuan Y., Liu B., Xie P., Zhang M.Q., Li Y., Xie Z., Wang X. Model-guided quantitative analysis of microRNA-mediated regulation on competing endogenous RNAs using a synthetic gene circuit. Proc. Natl. Acad. Sci. U. S. A. 2015;112:3158–3163. doi: 10.1073/pnas.1413896112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Alberti C., Cochella L. A framework for understanding the roles of miRNAs in animal development. Development. 2017;144:2548–2559. doi: 10.1242/dev.146613. [DOI] [PubMed] [Google Scholar]
- 103.Small E.M., Olson E.N. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469:336–342. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lapik Y.R., Fernandes C.J., Lau L.F., Pestov D.G. Physical and functional interaction between Pes1 and Bop1 in mammalian ribosome biogenesis. Mol. Cell. 2004;15:17–29. doi: 10.1016/j.molcel.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 105.Rohrmoser M. Interdependence of Pes1, Bop1, and WDR12 controls nucleolar localization and assembly of the PeBoW complex required for maturation of the 60S ribosomal subunit. Mol. Cell Biol. 2007;27:3682–3694. doi: 10.1128/MCB.00172-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Holdt L.M. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016;7:12429. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Burd C.E., Jeck W.R., Liu Y., Sanoff H.K., Wang Z., Sharpless N.E. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Abdelmohsen K. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017:1–9. doi: 10.1080/15476286.2017.1279788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Isken O., Grassmann C.W., Sarisky R.T., Kann M., Zhang S., Grosse F., Kao P.N., Behrens S.E. Members of the NF90/NFAR protein group are involved in the life cycle of a positive-strand RNA virus. EMBO J. 2003;22:5655–5665. doi: 10.1093/emboj/cdg562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Harashima A., Guettouche T., Barber G.N. Phosphorylation of the NFAR proteins by the dsRNA-dependent protein kinase PKR constitutes a novel mechanism of translational regulation and cellular defense. Genes Dev. 2010;24:2640–2653. doi: 10.1101/gad.1965010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Frith M.C. Discrimination of non-protein-coding transcripts from protein-coding mRNA. RNA Biol. 2006;3:40–48. doi: 10.4161/rna.3.1.2789. [DOI] [PubMed] [Google Scholar]
- 112.Clamp M. Distinguishing protein-coding and noncoding genes in the human genome. Proc. Natl. Acad. Sci. U. S. A. 2007;104:19428–19433. doi: 10.1073/pnas.0709013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Guttman M., Russell P., Ingolia N.T., Weissman J.S., Lander E.S. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154:240–251. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dinger M.E., Pang K.C., Mercer T.R., Mattick J.S. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput. Biol. 2008;4 doi: 10.1371/journal.pcbi.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sonenberg N., Hinnebusch A.G. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Andrews S.J., Rothnagel J.A. Emerging evidence for functional peptides encoded by short open reading frames. Nat. Rev. Genet. 2014;15:193–204. doi: 10.1038/nrg3520. [DOI] [PubMed] [Google Scholar]
- 117.Cabrera-Quio L.E., Herberg S., Pauli A. Decoding sORF translation - from small proteins to gene regulation. RNA Biol. 2016;13:1051–1059. doi: 10.1080/15476286.2016.1218589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Magny E.G., Pueyo J.I., Pearl F.M., Cespedes M.A., Niven J.E., Bishop S.A., Couso J.P. Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science. 2013;341:1116–1120. doi: 10.1126/science.1238802. [DOI] [PubMed] [Google Scholar]
- 119.Anderson D.M. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nelson B.R. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science. 2016;351:271–275. doi: 10.1126/science.aad4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jakobi T., Czaja-Hasse L.F., Reinhardt R., Dieterich C. Profiling and validation of the circular RNA repertoire in adult murine hearts. Dev. Reprod. Biol. 2016;14:216–223. doi: 10.1016/j.gpb.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Xu T., Wu J., Han P., Zhao Z., Song X. Circular RNA expression profiles and features in human tissues: a study using RNA-seq data. BMC Genom. 2017;18:680. doi: 10.1186/s12864-017-4029-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wu H.J., Zhang C.Y., Zhang S., Chang M., Wang H.Y. Microarray expression profile of circular RNAs in heart tissue of mice with myocardial infarction-induced heart failure. Cell. Physiol. Biochem. 2016;39:205–216. doi: 10.1159/000445617. [DOI] [PubMed] [Google Scholar]
- 124.Vaniotis G., Allen B.G., Hebert T.E. Nuclear GPCRs in cardiomyocytes: an insider's view of beta-adrenergic receptor signaling. Am. J. Physiol. Heart Circ. Physiol. 2011;301:H1754–H1764. doi: 10.1152/ajpheart.00657.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Maass P.G. A map of human circular RNAs in clinically relevant tissues. J. Mol. Med. (Berl.) 2017;95:1179–1189. doi: 10.1007/s00109-017-1582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Viereck J., Thum T. Circulating noncoding RNAs as biomarkers of cardiovascular disease and injury. Circ. Res. 2017;120:381–399. doi: 10.1161/CIRCRESAHA.116.308434. [DOI] [PubMed] [Google Scholar]
- 127.Bahn J.H., Zhang Q., Li F., Chan T.M., Lin X., Kim Y., Wong D.T., Xiao X. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin. Chem. 2015;61:221–230. doi: 10.1373/clinchem.2014.230433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Memczak S., Papavasileiou P., Peters O., Rajewsky N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One. 2015;10 doi: 10.1371/journal.pone.0141214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet. 2016;32:309–316. doi: 10.1016/j.tig.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Enuka Y., Lauriola M., Feldman M.E., Sas-Chen A., Ulitsky I., Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016;44:1370–1383. doi: 10.1093/nar/gkv1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 132.Schoenberg D.R., Maquat L.E. Regulation of cytoplasmic mRNA decay. Nat. Rev. Genet. 2012;13:246–259. doi: 10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kilchert C., Wittmann S., Vasiljeva L. The regulation and functions of the nuclear RNA exosome complex. Nat. Rev. Mol. Cell Biol. 2016;17:227–239. doi: 10.1038/nrm.2015.15. [DOI] [PubMed] [Google Scholar]
- 134.Arroyo J.D. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. U. S. A. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Vickers K.C., Palmisano B.T., Shoucri B.M., Shamburek R.D., Remaley A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Turchinovich A., Weiz L., Langheinz A., Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhao Z., Li X., Gao C., Jian D., Hao P., Rao L., Li M. Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci. Rep. 2017;7:39918. doi: 10.1038/srep39918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pan R.Y. Circular RNAs promote TRPM3 expression by inhibiting hsa-miR-130a-3p in coronary artery disease patients. OncoTargets. 2017;8:60280–60290. doi: 10.18632/oncotarget.19941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Shan K. Circular non-coding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation. 2017;136(17):1629–1642. doi: 10.1161/CIRCULATIONAHA.117.029004. [DOI] [PubMed] [Google Scholar]
- 140.Zhao Z., Li X., Jian D., Hao P., Rao L., Li M. Hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre-diabetes and type 2 diabetes mellitus. Acta Diabetol. 2017;54:237–245. doi: 10.1007/s00592-016-0943-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wu N., Jin L., Cai J. Profiling and bioinformatics analyses reveal differential circular RNA expression in hypertensive patients. Clin. Exp. Hypertens. 2017;39:454–459. doi: 10.1080/10641963.2016.1273944. [DOI] [PubMed] [Google Scholar]
- 142.Vausort M. Myocardial infarction-associated circular RNA predicting left ventricular dysfunction. J. Am. Coll. Cardiol. 2016;68:1247–1248. doi: 10.1016/j.jacc.2016.06.040. [DOI] [PubMed] [Google Scholar]
- 143.Geng H.H., Li R., Su Y.M., Xiao J., Pan M., Cai X.X., Ji X.P. The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS One. 2016;11 doi: 10.1371/journal.pone.0151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bazan H.A., Hatfield S.A., Brug A., Brooks A.J., Lightell D.J., Jr., Woods T.C. Carotid plaque rupture is accompanied by an increase in the ratio of serum circR-284 to miR-221 levels. Circ. Cardiovasc. Genet. 2017;10 doi: 10.1161/CIRCGENETICS.117.001720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Witztum J.L., Lichtman A.H. The influence of innate and adaptive immune responses on atherosclerosis. Annu. Rev. Pathol. 2014;9:73–102. doi: 10.1146/annurev-pathol-020712-163936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Geyer P.E., Holdt L.M., Teupser D., Mann M. Revisiting biomarker discovery by plasma proteomics. Mol. Syst. Biol. 2017;13:942. doi: 10.15252/msb.20156297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Helgadottir A. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 148.Cooper J.D., Walker N.M., Smyth D.J., Downes K., Healy B.C., Todd J.A., Type I.D.G.C. Follow-up of 1715 SNPs from the wellcome trust case control consortium genome-wide association study in type I diabetes families. Gene Immun. 2009;10(Suppl 1):S85–S94. doi: 10.1038/gene.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.McPherson R. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Helgadottir A. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat. Genet. 2008;40:217–224. doi: 10.1038/ng.72. [DOI] [PubMed] [Google Scholar]
- 151.Diabetes Genetics Initiative of Broad Institute of H. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 152.Scott L.J. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zeggini E. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Dandona S. Gene dosage of the common variant 9p21 predicts severity of coronary artery disease. J. Am. Coll. Cardiol. 2010;56:479–486. doi: 10.1016/j.jacc.2009.10.092. [DOI] [PubMed] [Google Scholar]
- 155.Broadbent H.M. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum. Mol. Genet. 2008;17:806–814. doi: 10.1093/hmg/ddm352. [DOI] [PubMed] [Google Scholar]
- 156.Liu Y. INK4/ARF transcript expression is associated with chromosome 9p21 variants linked to atherosclerosis. PLoS One. 2009;4:e5027. doi: 10.1371/journal.pone.0005027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Jarinova O. Functional analysis of the chromosome 9p21.3 coronary artery disease risk locus. Arterioscler. Thromb. Vasc. Biol. 2009;29:1671–1677. doi: 10.1161/ATVBAHA.109.189522. [DOI] [PubMed] [Google Scholar]
- 158.Holdt L.M. ANRIL expression is associated with atherosclerosis risk at chromosome 9p21. Arterioscler. Thromb. Vasc. Biol. 2010;30:620–627. doi: 10.1161/ATVBAHA.109.196832. [DOI] [PubMed] [Google Scholar]
- 159.Cunnington M.S., Santibanez Koref M., Mayosi B.M., Burn J., Keavney B. Chromosome 9p21 SNPs associated with multiple disease phenotypes correlate with ANRIL expression. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1000899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Holdt L.M., Teupser D. Recent studies of the human chromosome 9p21 locus, which is associated with atherosclerosis in human populations. Arterioscler. Thromb. Vasc. Biol. 2012;32:196–206. doi: 10.1161/ATVBAHA.111.232678. [DOI] [PubMed] [Google Scholar]
- 161.Yap K.L. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kotake Y., Nakagawa T., Kitagawa K., Suzuki S., Liu N., Kitagawa M., Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kuo C.L. Cdkn2a is an atherosclerosis modifier locus that regulates monocyte/macrophage proliferation. Arterioscler. Thromb. Vasc. Biol. 2011;31:2483–2492. doi: 10.1161/ATVBAHA.111.234492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Holdt L.M. Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhou X. Long non-coding RNA ANRIL regulates inflammatory responses as a novel component of NF-kappaB pathway. RNA Biol. 2016;13:98–108. doi: 10.1080/15476286.2015.1122164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Boeckel J.N. Identification and characterization of hypoxia-regulated endothelial circular RNA. Circ. Res. 2015;117:884–890. doi: 10.1161/CIRCRESAHA.115.306319. [DOI] [PubMed] [Google Scholar]
- 167.Dang R.Y., Liu F.L., Li Y. Circular RNA hsa_circ_0010729 regulates vascular endothelial cell proliferation and apoptosis by targeting the miR-186/HIF-1alpha axis. Biochem. Biophys. Res. Commun. 2017;490:104–110. doi: 10.1016/j.bbrc.2017.05.164. [DOI] [PubMed] [Google Scholar]
- 168.Chen J., Cui L., Yuan J., Zhang Y., Sang H. Circular RNA WDR77 target FGF-2 to regulate vascular smooth muscle cells proliferation and migration by sponging miR-124. Biochem. Biophys. Res. Commun. 2017;494(1–2):126–132. doi: 10.1016/j.bbrc.2017.10.068. [DOI] [PubMed] [Google Scholar]
- 169.Zheng C. Cyclic RNA hsacirc000595 regulates apoptosis of aortic smooth muscle cells. Mol. Med. Rep. 2015;12:6656–6662. doi: 10.3892/mmr.2015.4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wang K. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 2016;37:2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 171.Wang K. Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ. 2017;24:1111–1120. doi: 10.1038/cdd.2017.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Tang C.M. CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci. Rep. 2017;7:40342. doi: 10.1038/srep40342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhou B., Yu J.W. A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-beta1. Biochem. Biophys. Res. Commun. 2017;487:769–775. doi: 10.1016/j.bbrc.2017.04.044. [DOI] [PubMed] [Google Scholar]
- 174.Li X. Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol. Cell. 2017;67:214–227. doi: 10.1016/j.molcel.2017.05.023. e7. [DOI] [PubMed] [Google Scholar]
- 175.Calarco J.A., Zhen M., Blencowe B.J. Networking in a global world: establishing functional connections between neural splicing regulators and their target transcripts. RNA. 2011;17:775–791. doi: 10.1261/rna.2603911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Jangi M., Sharp P.A. Building robust transcriptomes with master splicing factors. Cell. 2014;159:487–498. doi: 10.1016/j.cell.2014.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wang E.T. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell. 2012;150:710–724. doi: 10.1016/j.cell.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Han H. MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature. 2013;498:241–245. doi: 10.1038/nature12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Coram R.J., Stillwagon S.J., Guggilam A., Jenkins M.W., Swanson M.S., Ladd A.N. Muscleblind-like 1 is required for normal heart valve development in vivo. BMC Dev. Biol. 2015;15:36. doi: 10.1186/s12861-015-0087-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Davis J., Salomonis N., Ghearing N., Lin S.C., Kwong J.Q., Mohan A., Swanson M.S., Molkentin J.D. MBNL1-mediated regulation of differentiation RNAs promotes myofibroblast transformation and the fibrotic response. Nat. Commun. 2015;6:10084. doi: 10.1038/ncomms10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Groh W.J. Electrocardiographic abnormalities and sudden death in myotonic dystrophy type 1. N. Engl. J. Med. 2008;358:2688–2697. doi: 10.1056/NEJMoa062800. [DOI] [PubMed] [Google Scholar]
- 182.Kalsotra A., Xiao X., Ward A.J., Castle J.C., Johnson J.M., Burge C.B., Cooper T.A. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc. Natl. Acad. Sci. U. S. A. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Mankodi A., Logigian E., Callahan L., McClain C., White R., Henderson D., Krym M., Thornton C.A. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 184.Kanadia R.N. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 185.Freyermuth F. Splicing misregulation of SCN5A contributes to cardiac-conduction delay and heart arrhythmia in myotonic dystrophy. Nat. Commun. 2016;7:11067. doi: 10.1038/ncomms11067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Konieczny P., Stepniak-Konieczna E., Taylor K., Sznajder L.J., Sobczak K. Autoregulation of MBNL1 function by exon 1 exclusion from MBNL1 transcript. Nucleic Acids Res. 2017;45:1760–1775. doi: 10.1093/nar/gkw1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Bonnet A., Lambert G., Ernest S., Dutrieux F.X., Coulpier F., Lemoine S., Lobbardi R., Rosa F.M. Quaking RNA-binding proteins control early myofibril formation by modulating tropomyosin. Dev. Cell. 2017;42:527–541. doi: 10.1016/j.devcel.2017.08.004. e4. [DOI] [PubMed] [Google Scholar]
- 188.Li Z. Defective smooth muscle development in qkI-deficient mice. Dev. Growth Differ. 2003;45:449–462. doi: 10.1111/j.1440-169x.2003.00712.x. [DOI] [PubMed] [Google Scholar]
- 189.Noveroske J.K., Lai L., Gaussin V., Northrop J.L., Nakamura H., Hirschi K.K., Justice M.J. Quaking is essential for blood vessel development. Genesis. 2002;32:218–230. doi: 10.1002/gene.10060. [DOI] [PubMed] [Google Scholar]
- 190.van der Veer E.P. Quaking, an RNA-binding protein, is a critical regulator of vascular smooth muscle cell phenotype. Circ. Res. 2013;113:1065–1075. doi: 10.1161/CIRCRESAHA.113.301302. [DOI] [PubMed] [Google Scholar]
- 191.Guo W., Jiang T., Lian C., Wang H., Zheng Q., Ma H. QKI deficiency promotes FoxO1 mediated nitrosative stress and endoplasmic reticulum stress contributing to increased vulnerability to ischemic injury in diabetic heart. J. Mol. Cell. Cardiol. 2014;75:131–140. doi: 10.1016/j.yjmcc.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 192.Dehghan A. Genome-wide association study for incident myocardial infarction and coronary heart disease in prospective cohort studies: the charge consortium. PLoS One. 2016;11 doi: 10.1371/journal.pone.0144997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.de Bruin R.G. Quaking promotes monocyte differentiation into pro-atherogenic macrophages by controlling pre-mRNA splicing and gene expression. Nat. Commun. 2016;7:10846. doi: 10.1038/ncomms10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Guo W. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat. Med. 2012;18:766–773. doi: 10.1038/nm.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Khan M.A. RBM20 regulates circular RNA production from the titin gene. Circ. Res. 2016;119:996–1003. doi: 10.1161/CIRCRESAHA.116.309568. [DOI] [PubMed] [Google Scholar]
- 196.Schafer S. Titin-truncating variants affect heart function in disease cohorts and the general population. Nat. Genet. 2017;49:46–53. doi: 10.1038/ng.3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Savkur R.S., Philips A.V., Cooper T.A. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- 198.Charlet B.N., Savkur R.S., Singh G., Philips A.V., Grice E.A., Cooper T.A. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol. Cell. 2002;10:45–53. doi: 10.1016/s1097-2765(02)00572-5. [DOI] [PubMed] [Google Scholar]
- 199.Giudice J. Alternative splicing regulates vesicular trafficking genes in cardiomyocytes during postnatal heart development. Nat. Commun. 2014;5:3603. doi: 10.1038/ncomms4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Guo W. RNA binding protein QKI inhibits the ischemia/reperfusion-induced apoptosis in neonatal cardiomyocytes. Cell. Physiol. Biochem. 2011;28:593–602. doi: 10.1159/000335755. [DOI] [PubMed] [Google Scholar]
- 201.Wang F., Yuan Y., Yang P., Li X. Extracellular vesicles-mediated transfer of miR-208a/b exaggerate hypoxia/reoxygenation injury in cardiomyocytes by reducing QKI expression. Mol. Cell. Biochem. 2017;431:187–195. doi: 10.1007/s11010-017-2990-4. [DOI] [PubMed] [Google Scholar]
- 202.Cochrane A. Quaking is a key regulator of endothelial cell differentiation, neovascularization, and angiogenesis. Stem Cell. 2017;35:952–966. doi: 10.1002/stem.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Pullmann R., Jr., Juhaszova M., Lopez de Silanes I., Kawai T., Mazan-Mamczarz K., Halushka M.K., Gorospe M. Enhanced proliferation of cultured human vascular smooth muscle cells linked to increased function of RNA-binding protein HuR. J. Biol. Chem. 2005;280:22819–22826. doi: 10.1074/jbc.M501106200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Aguado A. HuR mediates the synergistic effects of angiotensin II and IL-1beta on vascular COX-2 expression and cell migration. Br. J. Pharmacol. 2015;172:3028–3042. doi: 10.1111/bph.13103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Rhee W.J., Ni C.W., Zheng Z., Chang K., Jo H., Bao G. HuR regulates the expression of stress-sensitive genes and mediates inflammatory response in human umbilical vein endothelial cells. Proc. Natl. Acad. Sci. U. S. A. 2010;107:6858–6863. doi: 10.1073/pnas.1000444107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Stellos K. Adenosine-to-inosine RNA editing controls cathepsin S expression in atherosclerosis by enabling HuR-mediated post-transcriptional regulation. Nat. Med. 2016;22:1140–1150. doi: 10.1038/nm.4172. [DOI] [PubMed] [Google Scholar]
- 207.Levy N.S., Chung S., Furneaux H., Levy A.P. Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J. Biol. Chem. 1998;273:6417–6423. doi: 10.1074/jbc.273.11.6417. [DOI] [PubMed] [Google Scholar]
- 208.Zhang J. Macrophage beta2 integrin-mediated, HuR-dependent stabilization of angiogenic factor-encoding mRNAs in inflammatory angiogenesis. Am. J. Pathol. 2012;180:1751–1760. doi: 10.1016/j.ajpath.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Chang S.H. Antagonistic function of the RNA-binding protein HuR and miR-200b in post-transcriptional regulation of vascular endothelial growth factor-A expression and angiogenesis. J. Biol. Chem. 2013;288:4908–4921. doi: 10.1074/jbc.M112.423871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Paukku K., Backlund M., De Boer R.A., Kalkkinen N., Kontula K.K., Lehtonen J.Y. Regulation of AT1R expression through HuR by insulin. Nucleic Acids Res. 2012;40:5250–5261. doi: 10.1093/nar/gks170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Lu D., Xu A.D. Mini review: circular RNAs as potential clinical biomarkers for disorders in the central nervous system. Front. Genet. 2016;7:53. doi: 10.3389/fgene.2016.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Greene J., Baird A.M., Brady L., Lim M., Gray S.G., McDermott R., Finn S.P. Circular RNAs: biogenesis, function and role in human diseases. Front. Mol. Biol. 2017;4:38. doi: 10.3389/fmolb.2017.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Dowdy S.F. Overcoming cellular barriers for RNA therapeutics. Nat. Biotechnol. 2017;35:222–229. doi: 10.1038/nbt.3802. [DOI] [PubMed] [Google Scholar]
- 214.Hang J., Wan R., Yan C., Shi Y. Structural basis of pre-mRNA splicing. Science. 2015;349:1191–1198. doi: 10.1126/science.aac8159. [DOI] [PubMed] [Google Scholar]
- 215.Aebi M., Hornig H., Padgett R.A., Reiser J., Weissmann C. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell. 1986;47:555–565. doi: 10.1016/0092-8674(86)90620-3. [DOI] [PubMed] [Google Scholar]
- 216.Braunschweig U., Gueroussov S., Plocik A.M., Graveley B.R., Blencowe B.J. Dynamic integration of splicing within gene regulatory pathways. Cell. 2013;152:1252–1269. doi: 10.1016/j.cell.2013.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Rautmann G., Breathnach R. A role for branchpoints in splicing in vivo. Nature. 1985;315:430–432. doi: 10.1038/315430a0. [DOI] [PubMed] [Google Scholar]
- 218.Padgett R.A., Konarska M.M., Grabowski P.J., Hardy S.F., Sharp P.A. Lariat RNA's as intermediates and products in the splicing of messenger RNA precursors. Science. 1984;225:898–903. doi: 10.1126/science.6206566. [DOI] [PubMed] [Google Scholar]
- 219.Ruskin B., Krainer A.R., Maniatis T., Green M.R. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984;38:317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- 220.Puttaraju M., Been M.D. Group I permuted intron-exon (PIE) sequences self-splice to produce circular exons. Nucleic Acids Res. 1992;20:5357–5364. doi: 10.1093/nar/20.20.5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.de la Mata M., Lafaille C., Kornblihtt A.R. First come, first served revisited: factors affecting the same alternative splicing event have different effects on the relative rates of intron removal. RNA. 2010;16:904–912. doi: 10.1261/rna.1993510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Topisirovic I., Svitkin Y.V., Sonenberg N., Shatkin A.J. Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip. Rev. RNA. 2011;2:277–298. doi: 10.1002/wrna.52. [DOI] [PubMed] [Google Scholar]
- 223.Moore M.J., Proudfoot N.J. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 224.Sainsbury S., Bernecky C., Cramer P. Structural basis of transcription initiation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2015;16:129–143. doi: 10.1038/nrm3952. [DOI] [PubMed] [Google Scholar]
- 225.Hantsche M., Cramer P. Conserved RNA polymerase II initiation complex structure. Curr. Opin. Struct. Biol. 2017;47:17–22. doi: 10.1016/j.sbi.2017.03.013. [DOI] [PubMed] [Google Scholar]
- 226.Kwak H., Lis J.T. Control of transcriptional elongation. Annu. Rev. Genet. 2013;47:483–508. doi: 10.1146/annurev-genet-110711-155440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Jonkers I., Lis J.T. Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2015;16:167–177. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Sergeeva I.A., Hooijkaas I.B., Ruijter J.M., van der Made I., de Groot N.E., van de Werken H.J., Creemers E.E., Christoffels V.M. Identification of a regulatory domain controlling the Nppa-Nppb gene cluster during heart development and stress. Development. 2016;143:2135–2146. doi: 10.1242/dev.132019. [DOI] [PubMed] [Google Scholar]
- 229.Dong N., Chen S., Wang W., Zhou Y., Wu Q. Corin in clinical laboratory diagnostics. Clin. Chim. Acta. 2012;413:378–383. doi: 10.1016/j.cca.2011.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Ather S., Respress J.L., Li N., Wehrens X.H. Alterations in ryanodine receptors and related proteins in heart failure. Biochim. Biophys. Acta. 2013;1832:2425–2431. doi: 10.1016/j.bbadis.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Jiang J., Wakimoto H., Seidman J.G., Seidman C.E. Allele-specific silencing of mutant Myh6 transcripts in mice suppresses hypertrophic cardiomyopathy. Science. 2013;342:111–114. doi: 10.1126/science.1236921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Gao Z. Genetic inhibition of Na+-Ca2+ exchanger current disables fight or flight sinoatrial node activity without affecting resting heart rate. Circ. Res. 2013;112:309–317. doi: 10.1161/CIRCRESAHA.111.300193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Gerull B. Mutations of TTN, encoding the giant muscle filament titin, cause familial dilated cardiomyopathy. Nat. Genet. 2002;30:201–204. doi: 10.1038/ng815. [DOI] [PubMed] [Google Scholar]
- 234.Maass P.G. A map of human circular RNAs in clinically relevant tissues. J. Mol. Med. (Berl.) 2017;95(11):1179–1189. doi: 10.1007/s00109-017-1582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Liu Y., Sanoff H.K., Cho H., Burd C.E., Torrice C., Ibrahim J.G., Thomas N.E., Sharpless N.E. Expression of p16(INK4a) in peripheral blood T-cells is a biomarker of human aging. Aging Cell. 2009;8:439–448. doi: 10.1111/j.1474-9726.2009.00489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Miller J.W., Urbinati C.R., Teng-Umnuay P., Stenberg M.G., Byrne B.J., Thornton C.A., Swanson M.S. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Machuca-Tzili L.E., Buxton S., Thorpe A., Timson C.M., Wigmore P., Luther P.K., Brook J.D. Zebrafish deficient for Muscleblind-like 2 exhibit features of myotonic dystrophy. Dis. Model. Mech. 2011;4:381–392. doi: 10.1242/dmm.004150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Lee K.Y. Compound loss of muscleblind-like function in myotonic dystrophy. EMBO Mol. Med. 2013;5:1887–1900. doi: 10.1002/emmm.201303275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Brauch K.M., Karst M.L., Herron K.J., de Andrade M., Pellikka P.A., Rodeheffer R.J., Michels V.V., Olson T.M. Mutations in ribonucleic acid binding protein gene cause familial dilated cardiomyopathy. J. Am. Coll. Cardiol. 2009;54:930–941. doi: 10.1016/j.jacc.2009.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Pasman Z., Garcia-Blanco M.A. Early history of circular RNAs, children of splicing. RNA Biol. 2017;14:975–977. doi: 10.1080/15476286.2016.1227903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Legnini I. Circ-ZNF609 is a circular RNA that can Be translated and functions in myogenesis. Mol. Cell. 2017;66:22–37. doi: 10.1016/j.molcel.2017.02.017. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]