Abstract
Extensive genome-wide transcriptome study mediated by high throughput sequencing technique has revolutionized the study of genetics and epigenetic at unprecedented resolution. The research has revealed that besides protein-coding RNAs, large proportions of mammalian transcriptome includes a heap of regulatory non protein-coding RNAs, the number encoded within human genome is enigmatic. Many taboos developed in the past categorized these non-coding RNAs as ‘‘dark matter” and “junks”. Breaking the myth, RNA-seq-- a recently developed experimental technique is widely being used for studying non-coding RNAs which has acquired the limelight due to their physiological and pathological significance. The longest member of the ncRNA family-- long non-coding RNAs, acts as stable and functional part of a genome, guiding towards the important clues about the varied biological events like cellular-, structural- processes governing the complexity of an organism. Here, we review the most recent and influential computational approach developed to identify and quantify the long non-coding RNAs serving as an assistant for the users to choose appropriate tools for their specific research.
Keywords: Transcriptome, High throughput sequencing, Genetic and epigenetic, Long non-coding RNA, RNA-sequencing, RNA-seq
1. Introduction
All the disciplines of biological research perhaps were more edged towards identifying the coding genetic materials in human genome. The molecular biology beliefs circumference around Crick's central dogma ethos-- that genes are generally protein-coding [1]. This belief was accepted for several decades, until the major breakthrough triumphed to understand the fact that proportion of protein-coding region fails to explain the complexity of higher multi-cellular organisms, such as mammals [2]. This postulation has led to several reasonable assumptions, the most obvious one designating that the complexity of a genome is structured with the pillars of infrastructural RNAs essential for protein-coding as well as non-translated regulatory RNAs, the mild expressed ones. Measuring the current conceptions of regulatory RNAs the information needs to be re-examined. The newly developed techniques should be brought into measures to understand the true functioning of weird complex processes of gene imprinting, chromosome inactivation, translational repression, messenger RNA (mRNA) degradation, and so on, which can serve the basis of individual and species diversity [3].
The emergence of Sanger chain termination method in 1977 motivated the scientific community to sequence DNA in a definitive and consistent manner [4]. The trivial sequencing method encouraged plethora of research practices which ended up at the longest running project in the history of mankind (Human Genome Project, NIH). The natural events like the formations of DNA secondary structures which alter sequencing fidelity, non-specific primer binding, limited number of samples usage, posed problem to Sanger chain termination method [5]. Sooner, the method was replaced by a high throughput sequencing technique, Microarray. The coming of microarray empowered the researchers to measure the expression levels of vast number of genes concurrently which paved the ways to understand the underlying principles of genetic causes of abnormality, answerable for improper functioning of human body [6]. However, few shortcomings led to the obliteration of the existing technology more rapidly replacing it with Next Generation Sequencing (NGS) technique. The low dynamic range of microarray technology negatively affected the accuracy of the results reporting low sensitivity and specificity [7]. Above all, microarrays restrict the expression profiling data to specific annotations and contents. Usage of the digital expression profiling, NGS showed the potential to minimize or completely eliminate these flaws having the potential to sequence numerous DNA templates in a single run [8], [9]. A comparison between microarray and NGS showing the advantages and disadvantages of the technologies is given in Table 1.
Table 1.
Comparison between Next Generation Sequencing technique and Microarray technique.
| Next generation sequencing | Microarray |
|---|---|
| Advantages | |
| Species- or transcript-specific probes are not required in the case of NGS technology. | Specific probes are required in the case of microarray technologies. |
| NGS technology computes the sequencing read counts, analyzing the result for studying gene expression. | Gene expression measurement based on array hybridization technology is restricted by background and signal saturation noise. |
| NGS shows increased specificity and sensitivity for wide range of applications. | Specificity and sensitivity is low as compared to NGS for identifying differentially expressed genes. |
| Sequencing coverage depth is high in NGS technology facilitating the detection of rare or single transcripts per cell as well as in identifying weakly expressed genes. | Rare and low-abundance transcripts cannot be easily detected and are lost using microarray technology. |
| NGS technology is able to detect multiple splice sites and novel isoforms. | Microarray technologies cannot detect multiple splice sites and novel isoforms. |
| NGS technology is able to do de novo analysis of sample without reference genome. | Reference genome is required for the analysis of sample. |
| Disadvantages | |
| NGS based techniques are very expensive. | Microarrays are cheaper in comparison to NGS. |
| Accuracy and longevity of this approach remains questionable. | Microarray is more reliable methods in long run. |
| Low yield of high-quality sequences are obtained using NGS techniques. | Comparatively high yield of high-quality sequences is obtained using microarray technologies. |
| NGS technologies have a drawback of generating shorter sequences with more noise. | Microarray offers lesser errors and is more accurate. |
| NGS assembly algorithms show poor performance in presence of identical repeats. | Homologous repeats are identified using microarray technologies. |
| Annotation is challenging when considering complex genomes with higher repeat and duplication content. | Microarray technologies are more successful when considering complex genomes with higher repeat and duplication content. |
Of the past 30 years NGS is among the most convincing technology happened to the biological world. Concurrent sequencing of several genomes in a single instrument run can be completed with the help of NGS sequencers [10]. These sequences in the form of reads/fragments are taken as input by the varied number of computational tools and packages, which in a culture-free environment provides valuable outlook to analyse the data, to study its compositions, predicts variants, detects expression-- taking the research to a new and brighter level. A higher genome output can be obtained with targeted DNA enhancement approach at a much lower cost per sample [11].
NGS techniques provided highly attractive platform for sequencing genomes as compared to other sequencing modalities and has been widely implemented for various applications such as DNA sequencing, de novo genome sequencing, epigenomics- and transcriptomics- profiling. In clinical areas, NGS has been implemented in identification of genetic variants, somatic or inherited mutations, epigenetic changes, etc weaving around infected genes (or epi-genes) enabling the analysis of an individual's whole genome/transcriptome or disease specific targeted genome where a comprehensive match of variants (such as SNP) can be easily detected [12]. A large amount of genetic information is communicated by sequencing the entire genome, or transcriptome in which a notable amount will either be of unknown clinical importance (or novel) and very few of them can be interpreted and actionable. It holds multifarious capability in identifying the uncharacterized non-coding RNAs and confronting the fact that most of them are of biological significance [13], [14].
2. Non-coding RNA: the missing link
In the mid-20th century with the discovery of DNA as the genetic material, the information flow from double helical nucleic acid macromolecule into the beaded stretch of amino acid sequence embedded in proteins was traced. The extracted information incompletely governed the diversified cellular functions belonging to complex organisms [15]. This dilemma was explicitly answered following comprehensive experiments and collective insights about the role of another important macromolecule messenger ribonucleic acid (mRNA), which stores genetic information collected from the nucleotide sequence bases along a nucleic acid chain, thus passing it from one generation to another generation with high flexibility and high fidelity. mRNA functions to transfer information from DNA to the ribosome during the process of ‘translation’, knitting and evidencing the concept of “central dogma” (DNA→ RNA→ Protein) [16].
Our knowledge regarding the complex genetic makeup of higher organisms is inadequate. The sole reason behind the concept is that the major part of the genome of higher eukaryotes comprises of genetically inactive material collectively termed as non-coding RNAs (ncRNAs) [17]. However, the earlier research was only focussed towards the identification of the protein-coding genes only. Perhaps the lime-light has shifted towards the ∼98% of genomic content in humans, i.e. ncRNAs, made up of introns and other structures that do not encode proteins. Therefore it can be hypothesized that either the genomes of higher organisms are abounded with useless transcription, or there is some mysterious functioning of ncRNAs yet to be uncovered [18]. If the puzzle gets decoded, the suspicious account of ncRNAs in complex organisms would be able to explain the significance of genetic programming particularly related to regulatory information.
These RNAs which played crucial role in central dogma can further be characterized by series of contrasting features like their activation, modification, transportation, and digression profiles [19]. RNA that does not encode for a protein are collectively grouped under the class of ncRNAs: the housekeeping RNAs (tRNA, transfer RNA and rRNA, ribosomal RNA) and other regulatory ncRNAs (snRNA, small nuclear RNA; snoRNA, small nucleolar RNA; snoRNP, small nucleolar ribonucleoprotein RNA; gRNA, guide RNA; siRNA, small silencing RNAs; miRNA, micro RNA; piRNA, piwi-interacting RNA and circRNA, circular RNA) [20]. If it is strongly evidenced that the regulatory RNAs are the backbone of unexplained cascaded events taking place in cells of different species then it is a matter of reconsideration that why the phenomenon has gone unnoticed for so many years. This could be explained for the likelihood of protein-coding RNAs as a key player in most of the regulatory activities shrinking the knowledge network to a confined class of housekeeping genes where any inherited changes becomes biochemically visible. Therefore, the best way to understand the regulatory mechanisms is by fusing the molecular genetics practices with the comparative genomics studies.
RNA splicing and degradation ultimately makes it non-functional but equally if the vice versa is effective it can be said that introns are genetically efficient and transfer the functionality into meaningful regulatory events occurring inside the cell [21]. It came as a surprising that the biologists till date had completely ignored the possibility of the presence of these functional intronic sequences in a genome.
3. LncRNAs: unraveling the dark matter
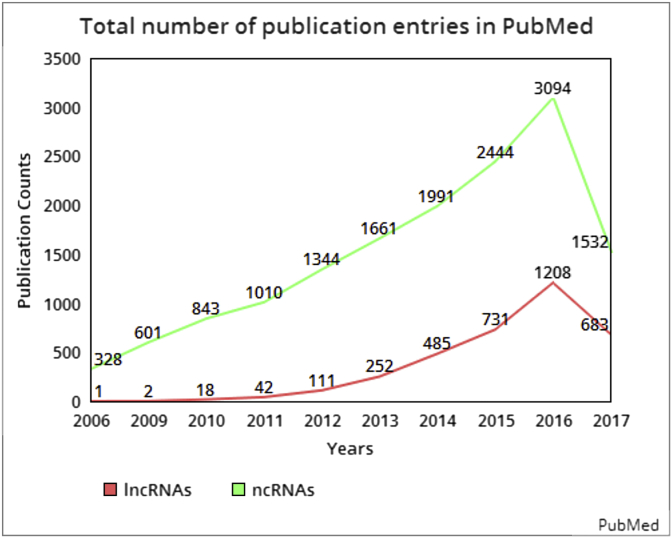
Molecular biologists have intense beliefs and explanations for the flow of information from genes to proteins via ribonucleic acid. The protein outcome in the form of crypted information occupies no more than ∼2% of the genome sequence. The dark matters longest component, lncRNA clearly demonstrates that they can perform different functions more than being a mere messenger. Clear insights can end up the research at the “RNA-level” extracting information from the “hidden-jewel” ignoring and blocking the movement towards the next-protein level. It has also recently been reported that in past few years the attention from short ncRNAs (sncRNAs) has shifted towards the long ncRNAs (lncRNAs) [22] the highly ignored section in the past. Fig. 1 show how the research trend has moved from other ncRNAs towards the lncRNAs.
Fig. 1.
The progressive and substantial research on long non-coding RNAs is rising. Cumulative plot of the total number of publication entries in PubMed related to non-coding RNAs is represented in green line and of entries related to long non-coding RNAs is represented in red line and axis.
So far it has been understood that lncRNAs are endogenous cellular RNAs which lack significant positive strand of open reading frame (ORF), i.e. they lack protein coding potential. These are of more than 200 nucleotides in length that make it distinct from any known functional RNA classes. This group of ncRNA does not constitute the homogeneous class of functionally related molecules [20]. LncRNAs are less expressed as compared to the protein coding genes as well less conserved, structurally residing in nucleus (e.g., MIAT) and cytoplasm (e.g., GAS5). LncRNAs are present in both the kingdoms of life, i.e. Plantae and Animalia but are more diverse in the higher group of organisms [23]. The GENCODE 25 release catalog is much larger and expansive than previously expected accounting for approximately 15,787 lncRNA, as recorded in human genome (https://www.gencodegenes.org/).
4. Computational analysis & application in decoding non-coding RNAs
As previously said, NGS has miraculously revolutionised the profiling of transcriptomic data. NGS monitors the sequential addition of nucleotides to immobilized and spatially arrayed DNA templates. The technique is being widely applied for transcriptome sequencing, for the characterization of the long non-coding transcriptomes. Millions of samples are being analyzed in a given limited time for detecting these lncRNAs as never before [24]. The efficiency of NGS platforms can be measured as per the yield and experiment run performed during each cycle. The advance sequencing technique has already proved its efficiency in identifying the protein-coding genes and is pacing towards the identification and characterization of the regulatory lncRNAs [25].
Before taking into account the in-depth workflow of the sequencing technique step by step, it is worth discussing about the NGS platforms which can successfully provide the clues for undertaking the bench studies towards the functional analysis. At present, the sequencing platforms used massively for NGS are Illumina/Solexa Genome Analyzer, Applied Biosystems SOLiD TM System, Roche/454 FLX technology, Pacific Biosciences SMRT and Helicos Heliscope. An intricate interaction of chemistry, high-resolution optics, enzymology, software and hardware engineering is shown by all these platforms. DNA sequencing samples are prepared efficiently with the minimum associated equipment requirement. Roche 454 FLX Pyrosequencer was the first instrument to be commercially introduced in 2004 functioning on the principle of pyrosequencing. In the process of pyrosequencing, DNA polymerase incorporates nucleotides resulting in the release of pyrophosphate. This incorporation of pyrophosphate causes the initiation of a number of downstream reactions that conclusively produce light using luciferase enzyme. The number of nucleotides incorporated can be detected by the amount of light produced. The agarose beads carrying oligonucleotides on the surfaces are mixed with the library fragments. These beads are specifically paired to the adapter sequences on the library of fragments which allows each bead to associate with only a single fragment. The linked fragment-bead complied along with PCR reactants, produces multiple copies of each fragment using PCR [24], [26], [27]. Illumina Genome Analyzer works on the principal of bridge amplification, the method produces definitive copies of DNA molecule. The flow cell is a micro-fabricated device which is 8-channel sealed allowing, on its surface bridge amplification of fragments. Multiple copies of DNA are produced in cluster consisting of molecules in it generated by means of amplification. Each eight may contain either a distinct library, or utilization of the same library may occur [28]. Applied Biosystems SOLiD TM Sequencer works on an adapter-ligated fragment library comparable with the libraries used in other sequencing platforms amplified by means of emulsion PCR technique [20]. User defined read lengths (∼25–400 bp), and the yield length (∼2–16 Gb) of DNA sequence data is obtained from the mentioned sequencing platform. Once the low quality reads are removed, reads of quality values are then base called. The sequencing steps involve library preparation, cluster generation, amplification, and read generation. The broad categories of targeted enrichment methods are PCR-amplicon which is a PCR based approach. The approach has a more uniform coverage than comparative hybridisation and is highly specific, provided that the PCR products concentrations are normalised before sequencing and pooling. Hybridisation capture approaches which is used for capturing of exons and larger target regions from hundreds of genes. Hybridisation enrichment offers the advantage of easy capture of large regions within a single tube assay [29], [30], [31].
5. LncRNA profiling using RNA-seq analysis tools
Genome-wide searches and screening of ncRNAs are performed in a variety of species analyzing full-length cDNA libraries, or transcriptional sequence data from other sources, with the intent to identify non-coding transcripts using varied experimental and computational approaches. Experimental method includes identification of lncRNAs through cDNA libraries, i.e. these methods are based on the fact that the expression of most of these ncRNA is lower than other protein coding transcripts [32]. However, the reported trivial experimental approaches have certain limitations, the ncRNA species, which exceeds in size range, cannot be directly analyzed, they are cleaved into smaller pieces prior to the analysis [20]. As well as the rule based approaches remains computationally challenging, they have certain limitations such as— not sensitive enough to detect RNA transcripts with low-expression level (microarray), or more expensive (SAGE). Learning based methods based on ORF length strategy, sequence and secondary structure conservation strategy and machine learning strategies have led to the development of classification tools to characterize these lncRNAs [33]. However, these approaches have limitations; they lack common conserved secondary structures specific for lncRNAs and use of these structural features are not sufficiently statistically robust enough to get detected. This is because a random RNA with low GC content can also fold into a low-energy structure. Contrastingly, computational methods using stable sequence and less densely structured features have successfully identified highly conserved and low expressed lncRNAs [34].
Reliable identification of lncRNAs interfaces are critical for understanding the structural bases, functional implications and for developing effective computational methods that offers a fast, feasible as well as cost-effective way to recognize putative lncRNAs. Compared to traditional technologies, RNA-seq experiment has many advantages in studying gene expression that can be highly specific for cell and tissue types. The technique is more sensitive in detecting less-abundant transcripts, and identifying novel alternative splicing isoforms and novel interacting ncRNA transcripts [35], [36], [37]. Existing computational tools have tried to predict some ncRNA features by testing against the available experimentally validated high-throughput generated datasets including physical interactions, genetic interactions, and phylogenetic profiles [38].
RNA-seq has bloomed as a powerful experimental technique with wide scope of applications determining the lncRNA expression levels more precisely as a quantitative approach [39]. In the following section we will review method and tools available for the analysis of these lncRNAs precisely considering the higher eukaryotic transcriptomes designed for the reads generated using Illumina platform. But the recommendation is not limited to the specific organism and platform and can be equally applied on different systems by including slight alterations. In depth knowledge regarding the comparison of NGS platforms and strategies can be found in Ref. [40].
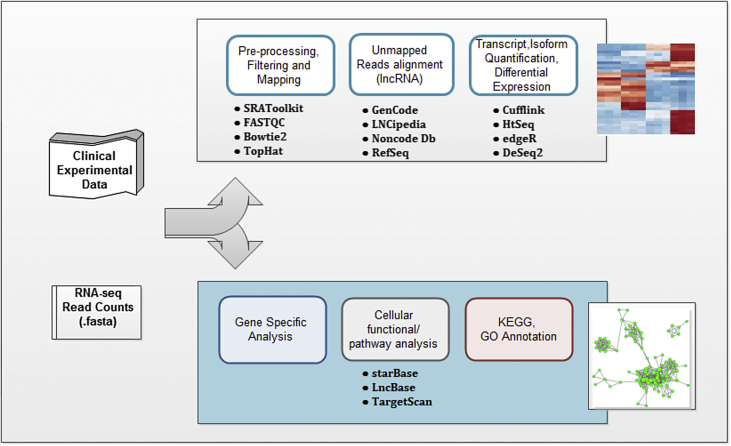
In the initial step, numerous fragmented sequences (‘reads’) are generated from the sequencers optimizing different protocols (as discussed in the above section). Sequencers try to mimic the sequencing process closet to the real world by taking into consideration all the steps that could particularly influence the characteristics of the reads. The length of the read is platform dependent varying from ∼75 bp to ∼400 bp generated by Illumina, IonTorrent, respectively. As well as sequencing depth of 50–100 million reads can be achieved using the machines covering most part of the genome. Paired-end (PE) sequencing is much preferred over the single-end (SE) sequencing, which improves the detection of lncRNAs enhancing their characterization [41], [42], [43]. The experiment requires large number of tools and analytical steps for the processing of sequences as shown in the flowgram (Fig. 2).
Fig. 2.
The RNA sequencing (RNA-seq) process commences with the input of sequences (in fasta format) generated using sequencers. Further the process requires pre-processing events involving the filtering and mapping of the input sequences (reads) followed by gene quantification and topological analysis.
The raw reads are stored in different formats (FASTQ, FASTA or SAM/BAM) in repositories (GEO database or ENA database) and later on filtered to remove the low quality reads developed due to smudges or debris attached to the flow cell, PCR artefacts, base-call errors, etc. Filtering and trimming is done using quality control packages viz. FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), filteR (http://scbb.ihbt.res.in/SCBB_dept/filter.php), NGS QC Toolkit (http://www.nipgr.res.in/ngsqctoolkit.html), FASTQsim (https://sourceforge.net/projects/fastqsim/), SimSeq (https://github.com/jstjohn/SimSeq), Trimmomatic (http://www.usadellab.org/cms/?page=trimmomatic) depending upon the individual's requirement [44], [45]. The pre-processing steps also necessitate the removal of other cellular RNAs (such as rRNA, tRNA, mRNA) which is achieved by using Sortmerna package (http://bioinfo.lifl.fr/RNA/sortmerna/). FastQC is the most preferred choice among the users since it applies different parameters to carry out the pre-processing step as well as support the output with the images of the reads generated before and after the filtering process [44].
After checking and cleaning the reads, next step in the pipeline is mapping or alignment of the filtered reads to the targeted genome [46]. The reference genome for the human is available in GENCODE (https://www.gencodegenes.org/releases/current.html), RefSeq (https://www.ncbi.nlm.nih.gov/refseq/), UCSC (https://genome.ucsc.edu/) and ENSEMBL (http://www.ensembl.org/index.html) annotation databases containing information for both protein coding genes as well as non-coding genes. Apart from these databases and genome browsers specific annotation files confined for the lncRNAs annotation is also available in LNCipedia (https://lncipedia.org/), NONCODE (http://www.noncode.org/), lncRNAdb (http://www.lncrnadb.org/) databases. A large number of mapping tools have been developed working on different mapping algorithms, including Bowtie (http://bowtie-bio.sourceforge.net/index.shtml), Bowtie2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml), BWA (http://bio-bwa.sourceforge.net/), TopHat (https://ccb.jhu.edu/software/tophat/), SeqMonk (https://www.bioinformatics.babraham.ac.uk/projects/seqmonk/), STAR (https://github.com/alexdobin/STAR), and MapSplice (http://www.netlab.uky.edu/p/bioinfo/MapSplice2). For the analysis of lncRNAs, short sequences are aligned using efficient read mapping algorithm, such as splice aware aligner (TopHat, STAR, MapSplice, GSNAP). For the genome mapping where the reference is unavailable, de novo assemblers are widely used (TRINITY, SOAPdenovo) [47], [48], [49]. Since, lncRNAs are less expressed as compared to protein coding genes and the choice of de novo alignment may eliminate the lowly expressed transcripts from the datasets therefore reference based alignment should be the preferred choice [50]. The mappers take sequencing reads in the form of SAM/BAM input files which can be generated using SAMtools [51]. The aligned reads can be visualized using different genome viewers, such as Integrated Genome Viewer, IGV (http://software.broadinstitute.org/software/igv/) and GiTools (http://www.gitools.org/) in order to check the position of split reads against the splice-junctions. Visualization will also ensure the expression changes, if any, in the reads mapped across the genome or transcriptome.
Mapping is followed by isoform, transcript and/or gene quantification which forms the base of differential expression analysis followed by annotation of genes and novel transcript discoveries. The quantification can be obtained using a python based package, HTseq (http://www-huber.embl.de/HTSeq/doc/count.html) which generates count by estimating the abundance of read mapping to a particular segment of the genome. Transcript level expression quantification is achieved using RSEM (https://deweylab.github.io/RSEM/), Sailfish (http://sailfish.readthedocs.io/en/master/sailfish.html), Salmon (http://salmon.readthedocs.io/en/latest/salmon.html) tools. Other differential expression analysis tools such as edgeR (https://bioconductor.org/packages/release/bioc/html/edgeR.html), Cufflink (http://cole-trapnell-lab.github.io/cufflinks/), DESeq (http://bioconductor.org/packages/release/bioc/html/DESeq.html) functioning is divided into two main processes: (i) how to identify isoforms and (ii) how to estimate their abundance? In an RNA-seq experiment the fragments length- and depth- coverage is used to generate the significant number of differentially expressed genes/transcripts by calculating RPKM/FPKM/TPM values [52]. Moreover, it is worth to point out that the accurate analysis of isoforms is highly dependent on the availability of replicates extracted from multiple samples which is an initial step in differential expression analysis at the level of isoforms. Statistically the estimation of isoforms from multiple RNA-seq samples is effective in precisely identifying ncRNAs [53], [54].
If the experiment carried out turns to be a success there is a good percentage of chances to find a handful of novel, uncharacterized lncRNAs. Further, the protein-coding potential check ensures the accurate identification of the novel transcripts achieved using the rigorous process of RNA-seq approach. As, it has been proved that bulk of lncRNAs lack ORF, the initial step in characterizing is doing ORF analysis using NCBI Open Reading Frame finder (https://www.ncbi.nlm.nih.gov/orffinder/), ORF identifier (http://bioportal.bioontology.org/ontologies/EDAM?p=classes&conceptid=data_2795) or similar tools. Different tools are developed to find the coding potential of the sequences based on different parameters- likewise based on the phylogenetic codon substitution frequency (PhyloCSF- https://github.com/mlin/PhyloCSF/wiki); based on the robustness of ORFs and protein-coding features (CONC [55] and CPC- http://cpc.cbi.pku.edu.cn); k-mer frequencies using DNN, SVM algorithm to distinguish lncRNAs and protein-coding RNAs (DeepLNC- http://bioserver.iiita.ac.in/deeplnc/; CNCI- https://github.com/www-bioinfo-org/CNCI and PLEK- http://www.ibiomedical.net/plek/); and by the process of identifying specific ORF size and coverage (CPAT- https://omictools.com/coding-potential-assessment-tool-tool). An intense search of the sequences against the protein databases and domains (Pfam- http://pfam.xfam.org/; PRIDE- http://www.ebi.ac.uk/pride); UniProt- http://www.uniprot.org/; SwissProt- http://web.expasy.org/docs/swiss-prot_guideline.html) can help to separate the non-coding from the coding ones. The sponge- activity of the lncRNAs can be detected using online available tools (TargetScan- http://www.targetscan.org/vert_71/; LncBase- http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2; StarBase- http://starbase.sysu.edu.cn/) based on ChIP sequencing technique [56], [57].
A comprehensive study provides a landscape of lncRNA mediated expression among the diverse species indicating their dynamic regulations governing several intrinsic and extrinsic activities. The promising intricacy involved will definitely lead to significant insights on the developmental role which may either act as boon if turns out to be regulatory or curse if is de-regulatory [58]. The identified differentially expressed lncRNAs can also be executed to demarcate the critical functioning of a system to further reveal the complex ncRNA-ncRNA regulatory relationships (acting as Sponge-pairs). Biological experiments need validation for warranting the potential regulatory role of the ncRNAs and their participation in cellular processes can be performed using simple and widely used technique, RT-PCR [59], [60], [61].
5.1. RNA sequencing (RNA-seq): boon for biologists
RNA-seq - the technology compiles experimental and computational methods in order to extract the information about RNA abundance in biological sample content. RNA-seq offers several advantages over all the conventional approaches. Firstly, the former technique is capable of identifying new genes as the identification range of the approach is not restricted to set of fixed probes and non-specific hybridization as compared to the microarray technology. The updated technique can detect expression at the transcript, exon, gene and coding DNA sequence (CDS) levels [62]. One of the most important characteristics of RNA-seq experiment is that it can identify structural variants, novel transcripts, raised by the activity of gene fusion and alternative splicing. The process of alternative splicing, reshuffles the exonic and intronic regions of the pre-mRNA transcripts engendering multiple isoforms from a single gene [63]. The diversity at genomic level is directly or indirectly carried to the proteomic level significantly affecting varied processes of tissue specificity, cellular and molecular functioning, developmental and differentiation patterning. The shuffling-error may lead to disorders and diseases; therefore the accurate gene quantification is needed, which may be accompanied by proper sequencing methodology. Hence, in the run RNA-seq is becoming an attractive technology as it provides additional dynamic genomic information at a lower price and instance as depicted in recent research practices [64], [65]. For eg., Wang et al. investigated the expression profiles of endometrial tissue samples of pig by using RNA sequencing which showed that differentially expressed lncRNAs were involved in different biological functions and signaling pathways during pre-implantation phases. The lncRNAs TCONS_01729386 and TCONS_01325501 were found to play a vital role in embryo pre-implantation [66]. Tsoi et al. evidenced the involvement of lncRNAs in immune-pathogenesis of psoriasis and other autoimmune diseases using RNA-seq data from lesional psoriatic, uninvolved psoriatic and normal skin biopsies, respectively. They also concluded that most of the differentially expressed lncRNAs are found to be co-expressed with genes involved in immune related functions [67]. In another experiment performed by Verma et al. 2632 novel lncRNAs were identified in DLBCL lymphoma transcriptome and their potential roles in lymphomagenesis and/or tumour maintenance [68]. Tripathi et al. in their paper thoroughly described the integrated analysis of dysregulated lncRNAs, lnc-PCP4, and lnc-FAM in breast cancer expression using RNA-seq study [69]. In a parallel study done by a group of researchers from Scripps Research Institute, RNA seq data was analyzed to demonstrate region specific enrichment of populations of lncRNAs and mRNAs in the mouse hippocampus and pre-frontal cortex (PFC) which were found to be involved in memory storage and neuropsychiatric disorders [70]. Several studies have showed the role of lncRNAs in different biological processes using the current RNA sequencing technique. The rapid rise in the discovery of novel set of classified lncRNAs in different species can be considered as valuable resource for future genomic experimental studies in these organisms. Therefore, discussing about all the discoveries and novel lncRNAs are beyond the scope of this review.
5.2. RNA-seq: targeted method to perk up the long non-coding RNA's profile
The proper characterization of these regulatory RNAs is somewhere lagging behind due to lack of intense knowledge of the specificity. Therefore, the thorough research can only withstand and explain the complexities, challenges involved in the emerging part of the “dark” genome. Discovery of epigenetically modified pathways and involved ncRNAs in different biological system can show new behaviour to allow researchers to embellish the applicative path of these regulatory RNAs [70], [71]. RNA-seq will help in detecting ncRNAs as well as underlying protocols for their identification will broaden the applicative areas within a biological system, their origin, biological-, evolutionary-, and regulatory functions.
The major objective of RNA-seq is to identify the sequence, structure and related abundance of the RNA molecules in the complete genome/transcriptome and enable the quantification of expressed transcripts, as well as to detect the novel genes and isoform composition [72], [73]. Since changes at the genetic and epigenetic govern the phenotypic differences between two individuals, a researcher's main aim should be to understand the regulatory mechanisms underlying the changes. The biological significance of sequences in terms of differential expression during diagnostic evolution along with their involvement in structural and functional maintenance is broadly studied in regulatory network pathways. Cellular expression patterns of these ncRNAs should reflect their functional relevance in the contexts of genomic and cellular distribution [22]. Furthermore, through cellular level transcriptome analysis, wide range of sense/antisense lncRNAs are implicated in functions like epigenetic regulation during lineage specification, reported to express in mammalian tissues and their deregulated expression can be linked to diseases and abnormalities [74]. The expression of protein coding mRNAs is comparatively higher than the lncRNAs, suggesting their involvement more in regulatory function rather than cellular structuring. These ncRNAs participate in gene expression regulation by the process of chromatin modification, transcriptional-, post- transcriptional processing and translational repression [75]. LncRNAs (such as H19, XIST, MALAT1, HOTAIR, etc) showing differential expression patterns involved with different tumour entities have already established their role as new source of biomarkers. LncRNAs have significant role in cancer, regulating cell proliferation, apoptosis, metastasis, invasion, etc and controlling varying expression levels in cells relative to normal tissue. Regulating the tumour suppressor genes (TSGs) or proto-oncogenes (PGs), the loss or gain in expression of lncRNAs has diagnostic and prognostic significance displaying additional traits in malignant tumours [76]. This class of ncRNAs are the recent discovered attractive therapeutic targets as they snatch away the normal functioning of mRNAs by their direct degradation or indirectly targeting them via smaller ncRNAs such as miRNAs. Since, lncRNAs are composed of a very heterogeneous group of RNA molecules varying in molecular and cellular functions, observations conclude that they show tissue-specific expression and are deregulated in several human diseases such as cancers, Alzheimer's, growth abnormality, etc [77]. H19 was the first lncRNA to be discovered in murine foetal liver cells. A significant positive link has been established between H19 and breast cancer. In breast adenocarcinoma, the expression of the discussed lncRNA is said to be increased compared to the healthy tissues. The increased expression rate has its effect on the size of the tumour and hormonal receptor expression [78]. H19 is said to repress the transcription with the help of epigenetic modification as well as regulate the translation. H19 has also been linked with other cancer such as bladder, ovarian, lung, oesophageal, colorectal and liver cancer. H19 is said to be up-regulated in differentiated embryonic stem cell as well as in hypoxic conditions. It is said to be involved in genetic imprinting, exclusively expressed in maternal chromosomes. Some studies have reported that H19 also have tumour suppressive role to play, for example children suffering from Beckwith-Wiedemann Syndrome show silencing of H19 genes. The contrasting activity of H19 is surprising and widely can be explained with the environment and cell type of investigations [60], [61]. HOTAIR a 2.2 kb long transcript arising from the HOXC locus, is one of the most common lncRNA to be known. It functions in transcriptional silencing by the process of chromatin modification. HOTAIR is said to be over expressed in several tumours which is an indicator in diagnosis of cancers [79]. The overexpression indicates metastasis and invasion in tumorous conditions and indicates poor survival of the cells. The de-regulated expression is reported in the breast and liver carcinoma. Its oncogenic property is fuelled by chromatin remodelling which alters the H3K27 methylation. Cancer epigenome gets re-juvenated by the altering activity of TSGs which ultimately causes the progression of cancerous cells [80]. XIST is highly characterized lncRNAs located on chrXq13.2 region. The reported length is about 17 kb. It has been widely studies in developmental biology due to its involvement in dosage compensation in human's genetic chromosomes [81]. XIST is implicated in female cancers like breast, ovarian and cervical cancers by the loss of expression. XIST has been linked up with the TSG, BRCA1, where the expression of the lncRNA is highly inflated in breast cancer cell lines. However, significantly deregulated expression in different types of cancer makes it a potential biomarker to study disease progression [82], [83]. MALAT1 (previously known as NEAT2), 8 kb long lncRNA located on 11q13 chromosome and is found to metastasize in NSCLC (non-small cell lung cancer) and are reported to be abundantly transcribed in the oncogenic tissues. MALAT1 participates in metastasis by promoting the motility. This can be observed by examining the expression change in motility related genes. MALAT1 shows deregulation in several human cancers like cervical, lung, breast, liver and breast cancer. MALAT1 promotes cell motility in cancer cells through transcriptional and post-transcriptional regulation of motility-related genes. MALAT1 shows overexpression in the reported cancers suggesting its use as potential biomarker [84], [85], [86].
Non-coding sequences are relevant and contain significant information in the form of ncRNAs of functional importance. Some of these characterized lncRNAs are highly expressed, at various levels of functioning such as chromatin modification, transcription, and post-transcriptional processing [87]. These abruptly expressed lncRNAs serve as an extensive source of new biomarkers shedding light on novel insights mechanisms underlying in cancer pathogenesis and tumour development which may serve as new targets for future biomarker development correlated with cancer therapy [88]. The dysregulated lncRNA expression in cancer characterizes the entire range of disease and the abnormal functioning drives cancer by disrupting normal cell processes, by facilitating epigenetic repression of downstream target genes [89], [90].
6. Conclusion
In this review, we discussed the NGS technique- RNA-seq to study and detect novel transcripts and isoforms related to lncRNAs from medical genomics, phylogenetic, epigenomics and environmental barcoding. RNA-seq has a very promising future in identifying new isoforms recognizing the structural changes involved in disease genomics. RNA-seq technique has succeeded in building a valuable conceptual platform by evaluating high-throughput data, exposing their advantages and drawbacks under different circumstances. Although, in past years more focus was towards short ncRNAs, but now the attention has shifted towards the lncRNAs. By learning about the conventional interaction between two ncRNAs one can unfold the hidden ways of malignancy in a cell. Revelation of the interactive network can not only solve the complex cellular mysteries, but can even take the medical science to a new level where one can completely win over the disease. Identifying these changes and analysing it in a proper way is the current demand of research practices which is purely inclined towards the bioinformatics approach to proceed with the uncharacterized genetic data. Outspokenly, the boom in the field will solve the riddle of transcriptome complexity as well as will scratch the layer of veiled tumorigenic events with more robustness. Along with the improvements at each step of RNA-sequencing, some more new improved tools and software packages based on practical considerations are required which can help the researchers to peep through an insightful window towards the transcriptomics data.
Contributor Information
Rashmi Tripathi, Email: rashmi.tripathi12@gmail.com.
Pavan Chakraborty, Email: pavan@iiita.ac.in.
Pritish Kumar Varadwaj, Email: pritish@iiita.ac.in.
References
- 1.Crick Francis. Central dogma of molecular biology. Nature. 1970;227.5258:561–563. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- 2.Kuznetsov Vladimir A. Springer; US: 2003. “Statistics of the Numbers of Transcripts and Protein Sequences Encoded in the genome.” Computational and Statistical Approaches to Genomics; pp. 125–171. [Google Scholar]
- 3.Cimatti Laura. 2012. Long non-coding RNA antisense to Uchl1 increases its protein translation and identifies a new class of protein translation activators. [Google Scholar]
- 4.Heather J.M., Chain B. The sequence of sequencers: the history of sequencing DNA. Genomics. 2016;107(1):1–8. doi: 10.1016/j.ygeno.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morozova O., Marra M.A. Applications of next-generation sequencing technologies in functional genomics. Genomics. 2008;92:255–264. doi: 10.1016/j.ygeno.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Slonim D.K., Yanai I. Getting started in gene expression microarray analysis. PLoS Comput. Biol. 2009;5 doi: 10.1371/journal.pcbi.1000543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao S., Fung-Leung W.P., Bittner A., Ngo K., Liu X. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS One. 2014;9(1) doi: 10.1371/journal.pone.0078644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mardis E.R. Next-generation DNA sequencing methods. Annu. Rev. genomics Hum. Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- 9.Meldrum C., Ma Doyle, Tothill R.W. Next-generation sequencing for cancer diagnostics: a practical perspective. Clin. Biochem. Rev. 2011;32:177–195. [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Yi. An integrative variant analysis pipeline for accurate genotype/haplotype inference in population NGS data. Genome Res. 2013;23.5:833–842. doi: 10.1101/gr.146084.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagalakshmi Ugrappa, Waern Karl, Snyder Michael. RNA-Seq: a method for comprehensive transcriptome analysis. Curr. Protoc. Mol. Biol. 2010:4–11. doi: 10.1002/0471142727.mb0411s89. [DOI] [PubMed] [Google Scholar]
- 12.Nowrousian Minou. Next-generation sequencing techniques for eukaryotic microorganisms: sequencing-based solutions to biological problems. Eukaryot. Cell. 2010;9.9:1300–1310. doi: 10.1128/EC.00123-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Metzker M.L. Sequencing technologies - the next generation. Nat. Rev. Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 14.Marguerat S., Wilhelm B.T., Bähler J. Next-generation sequencing: applications beyond genomes. Biochem. Soc. Trans. 2008;36:1091–1096. doi: 10.1042/BST0361091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie Hongbo. Functional anthology of intrinsic disorder. 1. Biological processes and functions of proteins with long disordered regions. J. Proteome Res. 2007;6.5:1882–1898. doi: 10.1021/pr060392u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perkins Diana O., Jeffries Clark, Sullivan P. 2005. Expanding the ‘Central Dogma’: the Regulatory Role of Nonprotein Coding Genes and Implications for the Genetic Liability to Schizophrenia; pp. 69–78. [DOI] [PubMed] [Google Scholar]
- 17.Irma Lozada-Chávez, Stadler Peter F., Prohaska Sonja J. “Hypothesis for the modern RNA world”: a pervasive non-coding RNA-based genetic regulation is a prerequisite for the emergence of multicellular complexity. Orig. Life Evol. Biospheres. 2011;41.6:587–607. doi: 10.1007/s11084-011-9262-1. [DOI] [PubMed] [Google Scholar]
- 18.Costa Fabrício F. Non-coding RNAs, epigenetics and complexity. Gene. 2008;410.1:9–17. doi: 10.1016/j.gene.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 19.Ravasi Timothy. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006;16.1:11–19. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tripathi Rashmi. DeepLNC, a long non-coding RNA prediction tool using deep neural network. Netw. Model. Anal. Health Inf. Bioinforma. 2016;5.1:1–14. [Google Scholar]
- 21.Zhang Jianzhi. 2010. “Evolutionary Genetics: Progress and challenges.” Evolution since Darwin: the First 150 Years; pp. 87–118. [Google Scholar]
- 22.Fatica Alessandro, Bozzoni Irene. Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 2014;15.1:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 23.Lennox Kim A., Behlke Mark A. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2015;44.2:863–877. doi: 10.1093/nar/gkv1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vitali Patrice. Long nuclear-retained non-coding RNAs and allele-specific higher-order chromatin organization at imprinted snoRNA gene arrays. J. Cell Sci. 2010;123.1:70–83. doi: 10.1242/jcs.054957. [DOI] [PubMed] [Google Scholar]
- 25.Xuan Jiekun. Next-generation sequencing in the clinic: promises and challenges. Cancer Lett. 2013;340.2:284–295. doi: 10.1016/j.canlet.2012.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Von Bubnoff Andreas. Next-generation sequencing: the race is on. Cell. 2008;132.5:721–723. doi: 10.1016/j.cell.2008.02.028. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki Shingo. Comparison of sequence reads obtained from three next-generation sequencing platforms. PLoS One. 2011;6.5:e19534. doi: 10.1371/journal.pone.0019534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer M., Stenzel U., Myles S., Prüfer K., Hofreiter M. Targeted high-throughput sequencing of tagged nucleic acid samples. Nucleic Acids Res. 2007;35(15) doi: 10.1093/nar/gkm566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo C., Tsementzi D., Kyrpides N., Read T., Konstantinidis K.T. Direct comparisons of Illumina vs. Roche 454 sequencing technologies on the same microbial community DNA sample. PLoS One. 2012;7(2) doi: 10.1371/journal.pone.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caporaso J.G., Lauber C.L., Wa Walters, Berg-Lyons D., Huntley J., Fierer N., Owens S.M., Betley J., Fraser L., Bauer M., Gormley N., Gilbert J., Smith G., Knight R. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yegnasubramanian S. Preparation of fragment libraries for next-generation sequencing on the applied biosystems SOLiD platform. Methods Enzym. 2013;529:185–200. doi: 10.1016/B978-0-12-418687-3.00015-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kristensen Vessela N. Principles and methods of integrative genomic analyses in cancer. Nat. Rev. Cancer. 2014;14.5:299–313. doi: 10.1038/nrc3721. [DOI] [PubMed] [Google Scholar]
- 33.Sønderkær Mads. 2012. Bioinformatic tools for next generation DNA sequencing. [Google Scholar]
- 34.Yotsukura Sohiya. Computational recognition for long non-coding RNA (lncRNA): software and databases. Briefings Bioinforma. 2017;18.1:9–27. doi: 10.1093/bib/bbv114. [DOI] [PubMed] [Google Scholar]
- 35.Xie Chaoyong. NONCODEv4: exploring the world of long non-coding RNA genes. Nucleic Acids Res. 2013;42.D1:D98–D103. doi: 10.1093/nar/gkt1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oshlack Alicia, Robinson Mark D., Young Matthew D. From RNA-seq reads to differential expression results. Genome Biol. 2010;11.12:220. doi: 10.1186/gb-2010-11-12-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prensner John R., Chinnaiyan Arul M. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1.5:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wan Yue. Understanding the transcriptome through RNA structure. Nat. Rev. Genet. 2011;12.9:641–655. doi: 10.1038/nrg3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilhelm B.T., Landry J.R. RNA-Seq-quantitative measurement of expression through massively parallel RNA-sequencing. Methods. 2009;48:249–257. doi: 10.1016/j.ymeth.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 40.Tripathi R., Sharma P., Chakraborty P., Varadwaj P.K. Next-generation sequencing revolution through big data analytics. Front. Life Sci. 2016 [Google Scholar]
- 41.Ilott Nicholas E., Ponting Chris P. Predicting long non-coding RNAs using RNA sequencing. Methods. 2013;63.1:50–59. doi: 10.1016/j.ymeth.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 42.Feng Huijuan, Qin Zhiyi, Zhang Xuegong. Opportunities and methods for studying alternative splicing in cancer with RNA-Seq. Cancer Lett. 2013;340.2:179–191. doi: 10.1016/j.canlet.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 43.Korbel Jan O. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318.5849:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andrews S. 2010. FastQC: a Quality Control Tool for High Throughput Sequence Data. doi: citeulike-article-id:11583827. [Google Scholar]
- 45.Patel R.K., Jain M. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 2012;7(2) doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langmead B. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinforma. 2010 doi: 10.1002/0471250953.bi1107s32. no. SUPP.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., Zehua Chen, Mauceli E., Hacohen N., Gnirke A., Rhind N., Palma F di, Birren B.W., Nusbaum C., Lindblad-Toh K., Friedman N., Regev A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Bo. Evaluation of de novo transcriptome assemblies from RNA-Seq data. Genome Biol. 2014;15.12:553. doi: 10.1186/s13059-014-0553-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sims David. Sequencing depth and coverage: key considerations in genomic analyses. Nat. Rev. Genet. 2014;15.2:121–132. doi: 10.1038/nrg3642. [DOI] [PubMed] [Google Scholar]
- 51.Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25.16:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi Yang. 2016. Statistical and Computational Methods for Differential Expression Analysis in High-throughput Gene Expression Data. [Google Scholar]
- 53.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinforma. Oxf. Engl. 2010;2:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trapnell C., Williams B., Pertea G., Mortazavi A., Kwan G., Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Jinfeng, Gough Julian, Rost Burkhard. Distinguishing protein-coding from non-coding RNAs through support vector machines. PLoS Genet. 2006;2.4:e29. doi: 10.1371/journal.pgen.0020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paci Paola, Colombo Teresa, Farina Lorenzo. Computational analysis identifies a sponge interaction network between long non-coding RNAs and messenger RNAs in human breast cancer. BMC Syst. Biol. 2014;8.1:83. doi: 10.1186/1752-0509-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karreth Florian A., Pandolfi Pier Paolo. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3.10:1113–1121. doi: 10.1158/2159-8290.CD-13-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liz Julia, Esteller Manel. lncRNAs and microRNAs with a role in cancer development. Biochimica Biophysica Acta (BBA)-Gene Regul. Mech. 2016;1859.1:169–176. doi: 10.1016/j.bbagrm.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 59.Jia Hui. Genome-wide computational identification and manual annotation of human long noncoding RNA genes. Rna. 2010;16.8:1478–1487. doi: 10.1261/rna.1951310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kartha Reena V., Subramanian Subbaya. Competing endogenous RNAs (ceRNAs): new entrants to the intricacies of gene regulation. Front. Genet. 2014;5 doi: 10.3389/fgene.2014.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pasquinelli Amy E. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat. Rev. Genet. 2012;13.4:271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 62.Wilhelm Brian T., Landry Josette-Renée. RNA-Seq—quantitative measurement of expression through massively parallel RNA-sequencing. Methods. 2009;48.3:249–257. doi: 10.1016/j.ymeth.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 63.Pickrell Joseph K. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464.7289:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.F Milos PM. RNA sequencing: advances, challenges and opportunities. Nat. Rev. Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kukurba K.R., Montgomery S.B. RNA sequencing and analysis. Cold Spring Harb. Protoc. 2015. 2015:951–969. doi: 10.1101/pdb.top084970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Yueying. Analyses of long non-coding RNA and mRNA profiling using RNA sequencing during the pre-implantation phases in pig endometrium. Sci. Rep. 2016;6 doi: 10.1038/srep20238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsoi Lam C. Analysis of long non-coding RNAs highlights tissue-specific expression patterns and epigenetic profiles in normal and psoriatic skin. Genome Biol. 2015;16.1:24. doi: 10.1186/s13059-014-0570-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Verma Akanksha. Transcriptome sequencing reveals thousands of novel long non-coding RNAs in B cell lymphoma. Genome Med. 2015;7.1:110. doi: 10.1186/s13073-015-0230-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tripathi Rashmi, Soni Apoorva, Varadwaj Pritish Kumar. Integrated analysis of dysregulated lncRNA expression in breast cancer cell identified by RNA-seq study. Non-coding RNA Res. 2016;1.1:35–42. doi: 10.1016/j.ncrna.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kadakkuzha Beena M. Transcriptome analyses of adult mouse brain reveal enrichment of lncRNAs in specific brain regions and neuronal populations. Front. Cell. Neurosci. 2015;9:63. doi: 10.3389/fncel.2015.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dennis Carina. Gene regulation: the brave new world of RNA. Nature. 2002;418.6894:122–124. doi: 10.1038/418122a. [DOI] [PubMed] [Google Scholar]
- 72.Pertea Mihaela. The human transcriptome: an unfinished story. Genes. 2012;3.3:344–360. doi: 10.3390/genes3030344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pandey, Ashutosh K., and Robert W. Williams. “Genetics of gene expression in CNS.” Int. Rev. Neurobiol.. [DOI] [PMC free article] [PubMed]
- 74.Clark Brian S., Blackshaw Seth. Long non-coding RNA-dependent transcriptional regulation in neuronal development and disease. 2014;116(2014):195. doi: 10.3389/fgene.2014.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guttman M., Rinn J.L. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fabbri M. vol 2014. Springer; New York: 2014. pp. 1–284. (Non-coding RNAs and Cancer). [Google Scholar]
- 77.Morris K.V. Long antisense non-coding RNAs function to direct epigenetic complexes that regulate transcription in human cells. Epigenetics Off. J. DNA Methylation Soc. 2009;4:296–301. doi: 10.4161/epi.4.5.9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gibb E.A., Vucic E.A., Enfield K.S.S., Stewart G.L., Lonergan K.M., Kennett J.Y., Becker-Santos D.D., MacAulay C.E., Lam S., Brown C.J., Lam W.L. Human cancer long non-coding RNA transcriptomes. PLoS One. 2011:6. doi: 10.1371/journal.pone.0025915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang J., Zhang P., Wang L., Piao H.L., Ma L. Long non-coding RNA HOTAIR in carcinogenesis and metastasis. Acta Biochimica Biophysica Sinica. 2014;46:1–5. doi: 10.1093/abbs/gmt117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bhan A., Mandal S.S. LncRNA HOTAIR: a master regulator of chromatin dynamics and cancer. Biochimica Biophysica Acta - Rev. Cancer. 2015;1856:151–164. doi: 10.1016/j.bbcan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brown C.J., Hendrich B.D., Rupert J.L., Lafrenière R.G., Xing Y., Lawrence J., Willard H.F. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 82.Lee J.T., Davidow L.S., Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat. Genet. 1999;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- 83.Sirchia S.M., Tabano S., Monti L., Recalcati M.P., Gariboldi M., Grati F.R., Porta G., Finelli P., Radice P., Miozzo M. Misbehaviour of XIST RNA in breast cancer cells. PLoS One. 2009;4(5) doi: 10.1371/journal.pone.0005559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gutschner T., Hämmerle M., Diederichs S. MALAT1-A paradigm for long noncoding RNA function in cancer. J. Mol. Med. 2013;91:791–801. doi: 10.1007/s00109-013-1028-y. [DOI] [PubMed] [Google Scholar]
- 85.Martens-Uzunova Elena S., Böttcher R., Croce C.M., Jenster G., Visakorpi T., Calin G.A. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur. Urol. 2014;65.6:1140–1151. doi: 10.1016/j.eururo.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 86.Gutschner T., Hmmerle M., Eimann M., Hsu J., Kim Y., Hung G., Revenko A., Arun G., Stentrup M., Gro M., Zrnig M., MacLeod A.R., Spector D.L., Diederichs S. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu J.H., Chen G., Dang Y.W., Li J., Luo D.Z. Expression and prognostic significance of lncRNA MALAT1 in pancreatic cancer tissues. Asian Pac. J. Cancer Prev. APJCP. 2014;15:2971–2977. doi: 10.7314/apjcp.2014.15.7.2971. [DOI] [PubMed] [Google Scholar]
- 88.Morris K.V., Vogt P.K. Long antisense non-coding RNAs and their role in transcription and oncogenesis. Cell Cycle. 2010;9:2544–2547. doi: 10.4161/cc.9.13.12145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dey B.K., Mueller A.C., Dutta A. Long non-coding RNAs as emerging regulators of differentiation, development, and disease. Transcription. 2014;5(4):e944014. doi: 10.4161/21541272.2014.944014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gibb E.A., Vucic E.A., Enfield K.S.S., Stewart G.L., Lonergan K.M., Kennett J.Y., Becker-Santos D.D., MacAulay C.E., Lam S., Brown C.J., Lam W.L. Human cancer long non-coding RNA transcriptomes. PLoS One. 2011:6. doi: 10.1371/journal.pone.0025915. [DOI] [PMC free article] [PubMed] [Google Scholar]