Abstract
In recent years, different studies have revealed that adult mammalian cardiomyocytes have the capacity to self-renew under homeostatic conditions and after myocardial injury. Interestingly, data from animal models capable of regeneration, such as the adult zebrafish and neonatal mice, have identified different non-coding RNAs (ncRNAs) as functional RNA molecules driving cardiac regeneration and repair. In this review, we summarize the current knowledge of the roles that a specific subset of ncRNAs, namely microRNAs (miRNA), plays in these animal models. We also emphasize the importance of characterizing and manipulating miRNAs as a novel approach to awaken the dormant regenerative potential of the adult mammalian heart by the administration of miRNA mimics or inhibitors. Overall, the use of these strategies alone or in combination with current cardiac therapies may represent new avenues to pursue for cardiac regeneration.
Keywords: Heart failure, Non-coding RNAs, miRNAs, Animal models, Regeneration
1. Introduction
Cardiovascular diseases (CVDs) represent a leading cause of death worldwide [1], [2]. Among them, coronary artery disease is the most frequent cardiovascular disorder leading to acute myocardial infarction (MI). In the best cases, patients who survive an MI episode face progressive deterioration of their condition over the years, ultimately resulting in heart failure. During the last decades, many efforts have been focused on improving treatments during the acute phase of MI and enhancing the contraction of the surviving myocardium (i.e., β-blockers, angiotensin-converting enzyme inhibitors, and mineralocorticoid receptor blockers, among others). However, none of these approaches are aimed at inducing the formation of new functional cardiac tissue. Since transplantation remains the only therapeutic option for end-stage heart failure, extraordinary efforts have been devoted towards the identification of novel approaches to induce heart regeneration, including: (i) the activation of resident cardiac progenitor cells with proliferative competence and their differentiation into mature cardiomyocytes, (ii) the transplantation of cardiac precursor cells to the damaged myocardium, and (iii) improving the proliferation of pre-existing cardiomyocytes by the administration of compounds [3], [4]. While the two first approaches have not proved successful enough to restore the cardiomyocytes lost after injury, the third approach involving the activation of endogenous cardiac regeneration by manipulating cardiomyocyte proliferation has recently demonstrated promising outcomes.
Certain non-mammalian vertebrates, such as some fish and amphibians are able to regenerate their heart throughout their entire life [5], [6], [7]. For example, adult zebrafish have been shown to elicit a primitive regenerative response upon injuries such as cryoinjury [8], [9], [10], ventricular resection [6], genetic ablation of cardiomyocytes [11] and hypoxia-reoxygenation injury [12]. Recently, lineage tracing approaches in this species have shown that dedifferentiated cardiomyocytes re-enter the cell cycle through increased expression of polo-kinase 1 (plk1) to replenish the lost myocardium [7]. Despite these findings, endogenous mammalian cardiomyocyte dedifferentiation upon injury was unexplored until 2011, when Porrello and colleagues observed a remarkably similar regenerative response in neonatal murine hearts to that seen in the adult zebrafish [13]. In particular, the authors showed that amputating 10%–15% of the ventricular mass in newborn mice elicited a regenerative response during the early days of life (up to 7 days). These newly generated cardiomyocytes restored heart function after approximately 30 days, and arose from pre-existing cardiomyocytes, as demonstrated by Cre/lox genetic lineage-tracing [13]. Supporting this notion, Senyo and colleagues later provided conclusive observations implicating pre-existing cardiomyocytes as the main cellular source of new cardiomyocytes in aging mice and after MI [14]. Altogether, these findings suggest that the mammalian heart possesses all the required elements for regeneration, and that the identification of the molecular pathways sustaining these responses may represent an attracting area to complement existing approaches for repairing the human heart [15].
Currently, regenerative strategies for heart healing rely on developing efficient approaches linked to proliferation, differentiation, and reprogramming. Although these methods have different characteristics and outcomes, they all depend on gene regulatory networks responsible for specialized biological processes during cardiac development and disease [1]. Globally, non-coding RNAs (ncRNAs) refer to those RNAs with no protein-coding potential but control different aspects of gene regulatory network activity, including transcriptional and epigenetic control, post-transcriptional gene regulation, and nuclear genome organization [1]. The different RNAs produced by the noncoding genome are rich and diverse in terms of biogenesis, structure, and function [16]. To date, hundreds of thousands of ncRNAs have been described in humans, however, the precise role of a great majority remains largely unknown. Traditionally, ncRNAs are classified based on their size into two categories: small (<200 nt), which include microRNAs (miRNAs), transfer RNAs, and small nucleolar RNAs; and longer RNAs (>200 nt), which include ribosomal RNAs, natural antisense transcripts and other long ncRNAs (lnRNAs) [17]. Recently, the incorporation of ncRNAs within cardiac gene regulatory networks represents a novel venue for therapeutic intervention in the heart. Mounting evidence highlight the role of lncRNA in cardiac development [18], [19], [20], [21], [22], [23] and cardiovascular diseases [21], [22], [23], [24], [25], [26], [27]. The reader may refer to the following reviews summarizing the effects of lncRNAs on cardiac biology and regeneration [1], [2], [28], [29], and cardiac disease [30]. In this regard, the development of next generation sequencing techniques has allowed the characterization of newly discovered lncRNAs in cardiac homeostasis and disease, which will allow their use in diagnosis, disease progression monitoring and targeted therapies [30].
miRNAs represent the most extensively studied class of small regulatory ncRNAs in the field of cardiac regeneration. These 21–22 nucleotide-long single-stranded ncRNAs guide RNA-inducing silencing complexes to their target messenger RNAs (mRNAs) for degradation or translational repression. Importantly, miRNAs are known to control embryonic development, tissue homeostasis and pathological processes, such as MI [31]. Recent studies highlight that the activation of endogenous cardiac regeneration may be possible by manipulating cardiomyocyte proliferation. Alternatively, another approach to cardiac regeneration that has attracted much attention is the direct reprogramming of fibroblasts into functional cardiomyocytes, raising the possibility of in vivo conversion of cardiac fibroblasts into cardiomyocytes within the injured zone. This review will focus on highlighting the characteristics and biological roles of miRNAs in these processes by paying specific attention to the studies addressing the use of miRNAs that could be targeted for inducing in situ myocardial repair after cardiac injury in a clinical setting.
2. miRNAs controlling cardiomyocyte proliferation after cardiac damage: lessons from animal models
Regardless of the injury model used, cardiac regenerative responses in adult zebrafish are mediated by the proliferation of pre-existing cardiomyocytes, which undergo de-differentiation and re-enter the cell cycle. All these events drive cardiomyocyte migration into the damaged area to restore the ventricular mass lost after injury [7], [32], [33]. Indeed, genetic lineage tracing studies have revealed that existing cardiomyocytes, and not stem cells, are the major source of regenerating cardiac muscle in these species [7], [33], [34]. In this regard, it has been demonstrated that cardiomyocyte de-differentiation and proliferation after damage in the adult zebrafish heart takes place as a major consequence to sarcomere disassembly, which is required for DNA synthesis and cell division to occur. Along this line, a recent study using proteomic analyses has revealed that the zebrafish heart, similar to neonatal mouse hearts, is characterized by an immature myofilament composition, lacking many of the structural proteins present in mature mouse cardiomyocytes [35]. However, such cardiac regeneration capacity found in the adult zebrafish is largely impeded in the adult mammalian heart.
Within the last few years, alternative approaches attempting to recapitulate innate mechanisms of cardiac regeneration as those found in adult zebrafish or neonatal mice have attracted a lot of attention. The inability of the adult mammalian heart to regenerate reflects the postnatal loss of cardiomyocyte proliferative capacity, which occurs during the first few weeks after birth in mice. Indeed, the molecular mechanisms sustaining postnatal cardiomyocyte binucleation and mitotic arrest remain unknown and are currently some of the most challenging questions to answer in cardiac biology. In an attempt to identify miRNAs involved in these processes, Porrello and colleagues performed microarray analyses to profile miRNAs involved in postnatal cardiomyocyte mitotic arrest in 1- and 10- day old mouse hearts. Using this approach, the authors observed that miR-195 expression was upregulated during this period, and that its overexpression in developing cardiomyocytes correlated with premature cell cycle arrest and a predisposition to congenital abnormalities, including ventricular septal defects [36]. On the contrary, postnatal inhibition of the entire miR-15 family (miR-15a, miR-15b, miR-16-1, miR-16-2, miR-195, and miR-497) using locked nucleic acid (LNA)-modified anti-miRs extended the proliferative capacity of neonatal cardiomyocytes beyond the normal window of postnatal cell cycle arrest, resulting in increasing numbers of mitotic cardiomyocytes and the de-repression of checkpoint kinase 1 [36]. Similarly, the same group demonstrated that 1-day old miR-195 transgenic mice failed to regenerate after an infarct, and that cardiac function was severely impaired due to the formation of large fibrotic scars soon after the infarct [37]. Accordingly, inhibition of the miR-15 family in postnatal stages increased cardiomyocyte proliferation and improved left ventricular function after ischemia-reperfusion (I/R) injury in mice [37]. The same group observed similar results after acute inhibition of the miR-15 family in mice and pig models [38]. Overall, these findings suggest that upregulation of the miR-15 family during the neonatal period is an important regulatory mechanism that controls cardiomyocyte cell cycle arrest and highlights its potential use as a therapeutic target for the manipulation of cardiac remodeling and function. Nevertheless, further studies are needed in order to ascertain its effects in cardiac proliferation in adult stages [37].
Of interest, in a work by Yin and colleagues, miR-133, a miRNA that induces defects in cardiac looping and chamber formation in Xenopus [39], which also has known roles in cardiac proliferation [40] and disease [41], was found to diminish its expression during zebrafish heart regeneration [42]. Accordingly, the authors engineered a miR-133 sponge construct encoding an EGFP cDNA followed by triplicate perfect binding sites for miR-133. In this manner, by transgenic miR-133 depletion, authors achieved an enhanced regenerative response seven days after ventricular amputation compared with wild type controls. Remarkably, one of the miR133 targets is Mps1 (monopolar spindle protein 1), a mitotic checkpoint kinase that when mutated hampers zebrafish heart regeneration leading to scar formation after damage [6].
Importantly, a genomic-scale screening performed by Eulalio and colleagues has described several individual miRNAs and miRNA families able to reduce or stimulate cardiomyocyte proliferation when delivered exogenously to neonatal rodent cardiomyocytes [43]. Besides the miR-15 family, another family identified by the authors to largely inhibit cardiac proliferation was the let-7 family, a highly conserved miRNA family that has previously shown to play important roles during development and stem cell differentiation [44]. Notably, recent findings from our laboratory have identified that the miRNA (miR) clusters miR99/Let-7c and miR-100/Let-7a are down regulated during the early stages of zebrafish regeneration, resulting in increased expression of their protein targets: smarca 5 (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily a, member 5) and fntb (beta subunit of farnesyl-transferase) [15]. Interestingly, our results also showed that both human and murine hearts failed to downregulate these miRs after injury, and that the delivery of anti-miRs using adeno-associated virus (AAVs) in a mouse model of MI improved cardiac function and was correlated with increases in the levels of FNTB and SMARCA5 expression. Using this approach we also demonstrated that experimental downregulation of miR-99/100 and/or let-7a/c in vitro (in primary cultures of neonatal cardiomyocytes) and ex vivo (murine heart organotypic slices) resulted in the expression of increased amount of GATA4, a marker associated with dedifferentiated cardiomyocytes, together with markers of proliferation (PCNA, or phosphorylated H3, respectively) [15]. Importantly, it has been shown that both miR99a and Let-7c act as key regulators of cardiomyogenesis during embryonic development [45], and that the manipulation of the let-7-a/c miRNAs by Lin28a allows for the induction of regenerative responses in several adult mouse tissues [46].
Of note, in the above mentioned work by Eulalio and colleagues, at least 40 miRNAs with previously unknown roles in stimulating cardiomyocyte proliferation were also identified, including miR-199a and miR-590, as well as the miR-17/92 and the miR-302/367 clusters [43]. The authors elegantly manipulate miRNA expression in order to evaluate cardiomyocyte proliferation in vivo. Specifically, they made use of high-throughput analyses and high-content microscopy systems to methodically identify proliferation-competent cardiomyocytes using a library of 875 miRNA mimics. The study was focused on the identification of miRNAs that promote expression of proliferative markers (Ki67, H3S10ph) and incorporation of DNA analogs indicative of DNA synthesis. Using this approach, the authors found over 204 miRNAs that enhanced cardiomyocyte proliferation more than 2-fold in both neonatal rats and adult mice. Subsequently, they selected two inducers with the highest potential of cardiomyocyte proliferation, namely miR-199a and miR-590, to be delivered into uninjured and infarcted adult mice using adeno-associated virus serotype 9 (AAV9) transduction. Interestingly, the overexpression of the two miRNAs increased cardiomyocyte proliferation rates, cardiac regeneration and improved cardiac function [43].
As stated above, the miR-17-92 cluster has also been recognized as a critical regulator of cardiac proliferation [43]. Interestingly, this same cluster was previously identified as a human oncogene [47] and a regulator of cardiac function during embryonic heart development [48]. Recently, Chen and colleagues have also demonstrated that transgenic overexpression of the miR-17-92 cluster in the murine heart induces cardiomyocyte proliferation in embryonic, postnatal and adult stages through the inhibition of phosphatase and tensin homolog (PTEN) [49]. Moreover, Chen and colleagues also showed that its overexpression was associated with enhanced cardiac function after MI, highlighting its potential use for the induction of cardiomyocyte proliferation and regeneration in adult stages [49].
In addition to miRNAs that can act by suppressing or enhancing cardiomyocyte proliferation, also several miRNAs have been described to play a role in regulating cardiomyocyte survival. Among them, the miR-34 family is induced after MI and promotes cardiomyocyte cell death [17], [50], [51], [52]. Specifically, miR-34a has been described to be upregulated over time in both endothelial cells and cardiomyocytes [53], [54], after cardiac injury [31], [52], [54], and in patients with heart failure [55]. Moreover, recent findings indicate a cardioprotective effect for miR-34a during cardiac aging [54]. Interestingly, miR-34a targets SIRT1, a deacetylase with cardiac and vasculoprotective functions, which upon de-repression contributes to the antiapoptotic effects of miR-34a inhibition. Moreover, miR-34a represses the protein phosphatase 1 regulator PNUTS (or serine/threonine protein phosphatase 1 regulatory subunit 10 [PPP1R10]), which is involved in mediating DNA damage response and telomere shortening [51], [54]. Similarly, it has been demonstrated that cardiac miR-34a levels are low in the early postnatal period and will rise to adult levels within one week after birth. Accordingly, overexpression of miR-34a in early postnatal mice was found to limit cardiomyocyte proliferation and cardiac regeneration following injury. Conversely, intravenous antagonism of miR-34a using LNA improved cardiac function after MI in adult mice through the modulation of genes previously linked to cellular proliferation and/or survival, such as Bcl2, Cyclin D1 and Sirt1 [54].
Overall, mounting evidence highlights that manipulation of cardiomyocyte proliferation is feasible and represents an effective approach for cardiac regeneration. Nevertheless, the molecular mechanisms promoting cardiomyocyte re-entry into the cell cycle still remain largely unknown. In this regard, it has been recently demonstrated that the miR302/367 cluster stimulates cardiomyocyte proliferation during early heart development by inhibiting the Hippo pathway, and that transient treatment with miR302/367 mimics in mice after MI promotes cardiac regeneration [56]. Interestingly, miR302/367 gain of function led to cardiomegaly in fetal and juvenile hearts, which show a more undifferentiated phenotype, similar to that seen in developing hearts with disrupted Hippo signaling [57]. Conversely, the deletion of Hippo signaling extends the heart's regenerative capacity beyond the first week of postnatal life and sustains cardiomyocyte generation with functional recovery after MI in adult mice [57], [58]. Similarly, deletion of Yap in the embryonic mouse heart causes myocardial hypoplasia, which causes early embryonic lethality [59], [60], whereas in the postnatal heart it induces progressive dilated cardiomyopathy, which is associated with a reduced number of mitotic cardiomyocytes during the neonatal stage [61]. Interestingly, when LAD ligation was performed in 7 day-old transgenic mice overexpressing a constitutively active mutation of Yap (YapS112A) under the control of an αMHC promoter, transgenic mouse hearts could regenerate with almost no fibrosis and showed increased cardiac tissue formation [61]. Importantly, overexpression of the constitutively active YapS112A mutation in cultured cardiomyocytes was associated with the induction of cardiomyocyte proliferation through activation of the insulin growth factor (IGF) signaling pathway and the inactivation of Glycogen synthase kinase 3 beta (GSK3β) [61], highlighting the importance of Hippo and IGF-Wnt/β-catenin crosstalk for cardiomyocyte proliferation during heart development [60], [62].
Research of the Hippo pathway in Drosophila has provided important hints for understanding cell proliferation regulation and organ size in mammals. In the developing mouse heart, many key Hippo pathway components are functionally preserved throughout evolution [63], [64]. In mammals the most downstream Hippo pathway components are the transcriptional coactivators Yap (Yki in Drosophila) and Taz, which promote transcription of pro-proliferative genes. Yap is inactivated by a kinase cascade that includes Mst1/Mst2 (Hippo in Drosophila) and the Lats kinases (Lats1/2), which act together with the Mob1 complex to promote Yap exclusion from the nucleus. Indeed, the work by Tian and colleagues identified that one of the possible mechanisms of action of the miR302/367 cluster is through the repression of Mst1, Mst2 and Mob1b [56]. Recently, Yang and colleagues conducted microarray analyses and observed that miR-206 expression was upregulated by Yap in cardiomyocytes, and that cardiac-specific overexpression of miR-206 in mice induced hypertrophy and protected the heart from I/R injury, whereas its suppression exacerbated I/R injury and prevented pressure overload-induced cardiac hypertrophy. Overall, the authors also identified FoxP1 as a functional target of miR-206, which when overexpressed, attenuated miR-206-induced cardiac hypertrophy and survival [65].
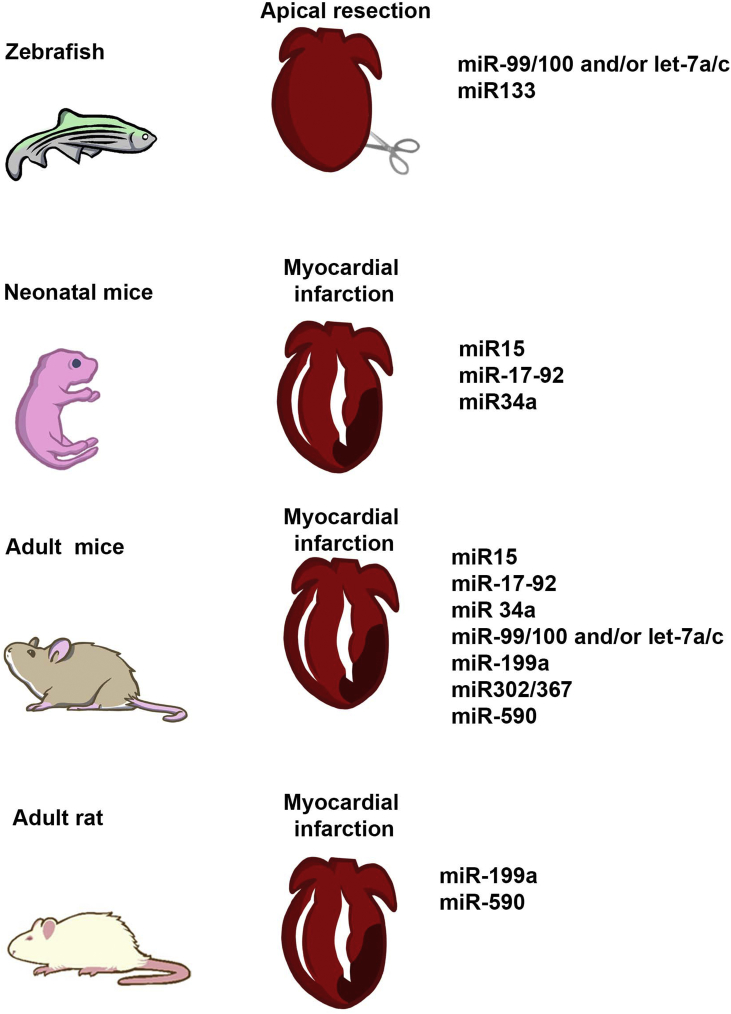
Fig. 1 summarizes the different miRNAs controlling post-natal cardiomyocyte proliferation after injury in different animal models.
Fig. 1.
microRNAs controlling post-natal cardiomyocyte proliferation. The use of different animal models has allowed for the identification of miRNAs driving cardiac endogenous repair by the induction of proliferative responses in cardiomyocytes.
3. miRNAs controlling cardiomyocyte (re)programming
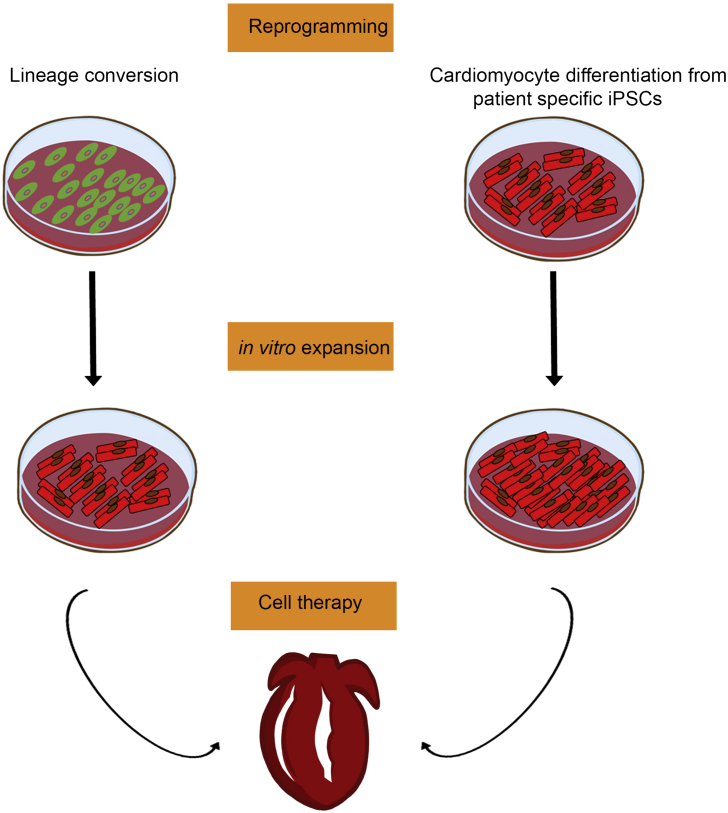
The possibility to isolate and expand cardiac progenitor cells (CPCs) capable of differentiation into cardiomyocytes and vascular cells has encouraged the field of cardiac regeneration with hopes of future uses in regenerative medicine. Several CPC populations have been identified in the developing and adult heart including c-Kit + CPCs [66], [67], cardiosphere-derived cells (CDCs) [68], epicardium derived cells, cardiac side population cells [69], Sca-1+ CPCs [70], Isl-1+ CPCs [71], and PDGRα+ CPCs [72], (further reviewed in Refs. [73], [74]). Despite all the progress, the actual function of these cell populations is still controversial and further studies are needed in order to define their regenerative potential in a clinical setting [4]. Other cell types, such as human embryonic stem cells (hESCs) or induced pluripotent stem cells (iPSCs) have been recently evaluated. However, specification and differentiation towards cardiac cell types must be tightly controlled in order to produce mature cell types and avoid adverse events (i.e., tumor formation after transplantation, among others). In addition, a robust protocol for their derivation into cardiomyocytes has yet to be established, and the generation of mature and functional cardiomyocytes from human pluripotent stem cells still remains a major challenge in the field (Fig. 2).
Fig. 2.
Cellular reprogramming for heart repair. To date reprogramming strategies as lineage conversion and/or guided differentiation from patient-specific iPSCs represent attractive strategies for the treatment of cardiovascular diseases. The possibility to work with unlimited amounts of starting cell populations (either patient fibroblasts or patient-specific iPSCs) would allow for their generation and expansion on demand.
To date several miRNAs have been shown to promote cardiac differentiation from hESCs. Wilson and colleagues reported the first miRNA profiling of cardiomyocytes derived from hESCs and identified that the expression of several miRNAs, including miR-1, miR-133, miR-208 was upregulated during the time course of differentiation. The authors also defined a novel role for miR-499 in cardiac differentiation, since its overexpression caused upregulation of the cardiac transcription factor MEF2C [75]. Interestingly, both miR-499 and miR-208 are also known to affect cardiac function. miR-499 and miR-208 are encoded by an intron of MYH7 and MYH6, respectively, and they share many predicted targets, though miR-208 has been previously shown to play a crucial role in stress adaptation of the adult heart [76]. In recent years, further studies have tried to dissect the functional role of different miRNAs previously shown to be related to cardiac differentiation in stem cells, and identified that whereas miR-499 promotes ventricular specification of hESCs, miR-1 facilitates electrophysiological maturation [77].
In an effort to bypass the use of pluripotent stem cell sources as the initial population for the derivation of cardiac cells, several strategies are being devised to eliminate the risk of teratomas and eventually allow for in vivo reprogramming of resident cardiac cells (Fig. 2). Along this line, a recent report has demonstrated the direct conversion of mouse fibroblasts to a cardiomyocyte-like phenotype using a single transient transfection with a combination of miRNAs with known roles in cardiac development and differentiation (miR-1, miR-133, miR-208, and miR-499) [78]. Interestingly, the reprogrammed cells expressed specific markers for cardiac-like cells and exhibited electrophysiological characteristics related to cardiomyocytes. More importantly, direct administration of these miRNAs into injured myocardia resulted in direct conversion of cardiac fibroblasts to cardiomyocyte-like cells in vivo, as confirmed by genetic tracing using Fsp1Cre-mice [78]. In parallel to these findings, Nam and colleagues found that different human cardiac transcription factors, including GATA binding protein 4, Hand2, T-box5, and myocardin, together with two microRNAs, miR-1 and miR-133, activated cardiac marker expression in neonatal and adult human fibroblasts. Importantly, long-term culture of transduced cells showed that human fibroblasts reprogrammed with these factors exhibited sarcomere-like structures and calcium transients, and that a small subset of reprogrammed cardiac cells showed spontaneous contractility. Interestingly, those changes were in agreement with the acquisition of the expression of cardiac genes and suppression of non-myocyte genes [79].
4. Conclusion
Though the ability to induce organ regeneration has fascinated humanity for centuries, the cellular and molecular events driving the generation of new tissue structures or parts of organs after damage are still unknown. With respect to the heart, early studies in amphibian, axolotls, and newts described the intrinsic capabilities of those organisms to regenerate their hearts after damage [80], [81], [82]. This potential has been believed to be absent in mammals until the recent years, when independent studies highlighted the observation that adult mammalian cardiomyocyte renewal occurs under biological conditions [14], [83], [84]. Identifying the intrinsic repair capacity of the mammalian heart has encouraged the scientific community to develop therapeutic strategies to enhance this residual potential. In this regard, comparative analyses between cardiac repair and regeneration in different animal models and at different stages of development have identified the existence of conserved pathways driving heart regeneration. In this review, we highlight different studies that provide convincing evidence that miRNAs might represent an attractive approach when developing novel strategies for heart healing. Despite these encouraging results, important issues need to be addressed before translating these findings into the clinic. For instance, our understanding of miRNA biology in cardiac tissue is still in its infancy, requiring further studies in order to achieve a full perspective on cardiac regulatory networks under miRNA control. Such information will also benefit the development of effective and safe methods of miRNA-targeting molecules and the avoidance of unwanted off-target effects. Eventually, these advances will result in novel therapeutic approaches targeting cardiac failure, a major unmet need in the clinic.
Acknowledgements
Authors gratefully acknowledge the help of M. Schwarz and P. Schwarz for administrative help and logistic coordination. This project has received founding form the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (StG-2014-640525_REGMAMKID). E.G is supported by StG-2014-640525_REGMAMKID. P.P is partially supported by CardioCel (TerCel, Instituto de Salud Carlos III) and MINECO (SAF2014-59778). J.C.I.B. is supported by the G. Harold and Leila Y. Mathers Charitable Foundation, The Leona M. and Harry B. Helmsley Charitable Trust (2012-PG-MED002), the Moxie Foundation, the Universidad Católica San Antonio de Murcia (UCAM), and Fundación Dr. Pedro Guillén. N.M is supported by CardioCel (TerCel, Instituto de Salud Carlos III), StG-2014-640525_REGMAMKID, MINECO (SAF2014-59778 and RYC-2014-16242) and 2014 SGR 1442.
Contributor Information
Juan Carlos Izpisua Belmonte, Email: belmonte@salk.edu.
Nuria Montserrat, Email: nmontserrat@ibecbarcelona.eu.
References
- 1.Ounzain S., Pedrazzini T. The promise of enhancer-associated long noncoding RNAs in cardiac regeneration. Trends cardiovasc. Med. 2015;25(7):592–602. doi: 10.1016/j.tcm.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 2.Frank S., Aguirre A., Hescheler J., Kurian L. A lncRNA perspective into (Re)Building the heart. Front. Cell Dev. Biol. 2016;4:128. doi: 10.3389/fcell.2016.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguirre A., Sancho-Martinez I., Izpisua Belmonte J.C. Reprogramming toward heart regeneration: stem cells and beyond. Cell Stem Cell. 2013:275–284. doi: 10.1016/j.stem.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Giacca M., Zacchigna S. Harnessing the microRNA pathway for cardiac regeneration. J. Mol. Cell. Cardiol. 2015:68–74. doi: 10.1016/j.yjmcc.2015.09.017. [DOI] [PubMed] [Google Scholar]
- 5.Itou J., Kawakami H., Burgoyne T., Kawakami Y. Life-long preservation of the regenerative capacity in the fin and heart in zebrafish. Biol. Open. 2012;1:739–746. doi: 10.1242/bio.20121057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poss K.D., Wilson L.G., Keating M.T. Heart regeneration in zebrafish. Science (80-. ) 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 7.Jopling C., Sleep E., Raya M., Martí M., Raya A., Izpisúa Belmonte J.C. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chablais F., Veit J., Rainer G., Jaźwińska A. The zebrafish heart regenerates after cryoinjury-induced myocardial infarction. BMC Dev. Biol. 2011;11:21. doi: 10.1186/1471-213X-11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.González-Rosa J.M., Martín V., Peralta M., Torres M., Mercader N. Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish. Development. 2011;138:1663–1674. doi: 10.1242/dev.060897. [DOI] [PubMed] [Google Scholar]
- 10.Schnabel K., Wu C.C., Kurth T., Weidinger G. Regeneration of cryoinjury induced necrotic heart lesions in zebrafish is associated with epicardial activation and cardiomyocyte proliferation. PLoS One. 2011:6. doi: 10.1371/journal.pone.0018503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J., Panáková D., Kikuchi K., Holdway J.E., Gemberling M., Burris J.S. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development. 2011;138:3421–3430. doi: 10.1242/dev.068601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parente V., Balasso S., Pompilio G., Verduci L., Colombo G.I., Milano G. Hypoxia/reoxygenation cardiac injury and regeneration in zebrafish adult heart. PLoS One. 2013;8 doi: 10.1371/journal.pone.0053748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Porrello E.R., Mahmoud A.I., Simpson E., Hill J a, Richardson J a, Olson E.N. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Senyo S.E., Steinhauser M.L., Pizzimenti C.L., Yang V.K., Cai L., Wang M. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2012;493:433–436. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aguirre A., Montserrat N., Zacchigna S., Nivet E., Hishida T., Krause M.N. In vivo activation of a conserved microRNA program induces mammalian heart regeneration. Cell Stem Cell. 2014;15:589–604. doi: 10.1016/j.stem.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mercer T.R., Mattick J.S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 17.Seeger F.H., Zeiher A.M., Dimmeler S. MicroRNAs in stem cell function and regenerative therapy of the heart. Arter. Thromb. Vasc. Biol. 2013;33:1739–1746. doi: 10.1161/ATVBAHA.113.300138. [DOI] [PubMed] [Google Scholar]
- 18.Grote Phillip, Wittler Lars, Währisch Sandra, Hendrix David, Beisaw Arica, Macura K., Bläss Gaby, Kellis Manolis, Werber Martin, BGH The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell. 2014;24:206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klattenhoff C.A., Scheuermann J.C., Surface L.E., Bradley R.K., Fields P.A., Steinhauser M.L. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurian L., Aguirre A., Sancho-Martinez I., Benner C., Hishida T., Nguyen T.B. Identification of novel long noncoding RNAs underlying vertebrate cardiovascular development. Circulation. 2015;131:1278–1290. doi: 10.1161/CIRCULATIONAHA.114.013303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ounzain S., Micheletti R., Arnan C., Plaisance I., Cecchi D., Schroen B. CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis. J. Mol. Cell. Cardiol. 2015;89:98–112. doi: 10.1016/j.yjmcc.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 22.Ounzain S., Micheletti R., Beckmann T., Schroen B., Alexanian M., Pezzuto I. Genome-wide profiling of the cardiac transcriptome after myocardial infarction identifies novel heart-specific long non-coding RNAs. Eur. Heart J. 2015;36:353–368. doi: 10.1093/eurheartj/ehu180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ounzain S., Pezzuto I., Micheletti R., Burdet F., Sheta R., Nemir M. Functional importance of cardiac enhancer-associated noncoding RNAs in heart development and disease. J. Mol. Cell. Cardiol. 2014;76:55–70. doi: 10.1016/j.yjmcc.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han P., Li W., Lin C.-H., Yang J., Shang C., Nurnberg S.T. A long noncoding RNA protects the heart from pathological hypertrophy. Nature. 2014;514:102–106. doi: 10.1038/nature13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michalik K.M., You X., Manavski Y., Doddaballapur A., Zörnig M., Braun T. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ. Res. 2014;114:1389–1397. doi: 10.1161/CIRCRESAHA.114.303265. [DOI] [PubMed] [Google Scholar]
- 26.Yap K.L., Li S., Muñoz-Cabello A.M., Raguz S., Zeng L., Mujtaba S. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang K., Long B., Zhou L.-Y., Liu F., Zhou Q.-Y., Liu C.-Y. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat. Commun. 2014;5:3596. doi: 10.1038/ncomms4596. [DOI] [PubMed] [Google Scholar]
- 28.Devaux Y., Zangrando J., Schroen B., Creemers E.E., Pedrazzini T., Chang C.-P. Long noncoding RNAs in cardiac development and ageing. Nat. Rev. Cardiol. 2015;12:415–425. doi: 10.1038/nrcardio.2015.55. [DOI] [PubMed] [Google Scholar]
- 29.Ounzain S., Crippa S., Pedrazzini T. Small and long non-coding RNAs in cardiac homeostasis and regeneration. Biochim. Biophys. Acta - Mol. Cell Res. 2013:923–933. doi: 10.1016/j.bbamcr.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 30.Uchida S., Dimmeler S. Long noncoding RNAs in cardiovascular diseases. Circ. Res. 2015:737–750. doi: 10.1161/CIRCRESAHA.116.302521. [DOI] [PubMed] [Google Scholar]
- 31.Van Rooij E., Sutherland L.B., Thatcher J.E., DiMaio J.M., Naseem R.H., Marshall W.S. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc. Natl. Acad. Sci. U. S. A. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kikuchi K. Dedifferentiation, transdifferentiation, and proliferation: mechanisms underlying cardiac muscle regeneration in zebrafish. Curr. Pathobiol. Rep. 2015;3:81–88. doi: 10.1007/s40139-015-0063-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kikuchi K., Holdway J.E., Werdich A.A., Anderson R.M., Fang Y., Egnaczyk G.F. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kikuchi K., Gupta V., Wang J., Holdway J.E., Wills A a, Fang Y. Tcf21+ epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration. Development. 2011;138:2895–2902. doi: 10.1242/dev.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gomes R.S.M., Skroblin P., Munster A.B., Tomlins H., Langley S.R., Zampetaki A. “Young at heart”: regenerative potential linked to immature cardiac phenotypes. J. Mol. Cell. Cardiol. Authors. 2016;92:105–108. doi: 10.1016/j.yjmcc.2016.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Porrello E.R., Johnson B.A., Aurora A.B., Simpson E., Nam Y.-J., Matkovich S.J. MiR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circ. Res. 2011;109:670–679. doi: 10.1161/CIRCRESAHA.111.248880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Porrello E.R., Mahmoud A.I., Simpson E., Johnson B.A., Grinsfelder D., Canseco D. Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proc. Natl. Acad. Sci. U. S. A. 2013;110:187–192. doi: 10.1073/pnas.1208863110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hullinger T.G., Montgomery R.L., Seto A.G., Dickinson B.A., Semus H.M., Lynch J.M. Inhibition of miR-15 protects against cardiac ischemic injury. Circ. Res. 2012;110:71–81. doi: 10.1161/CIRCRESAHA.111.244442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen J.-F., Mandel E.M., Thomson J.M., Wu Q., Callis T.E., Hammond S.M. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu N., Bezprozvannaya S., Williams A.H., Qi X., Richardson J.A., Bassel-Duby R. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carè A., Catalucci D., Felicetti F., Bonci D., Addario A., Gallo P. microRNA-133 controls cardiac hypertrophy. Nat. Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 42.Yin V.P., Lepilina A., Smith A., Poss K.D. Regulation of zebrafish heart regeneration by miR-133. Dev. Biol. 2012;365:319–327. doi: 10.1016/j.ydbio.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eulalio A., Mano M., Dal Ferro M., Zentilin L., Sinagra G., Zacchigna S. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492:376–381. doi: 10.1038/nature11739. [DOI] [PubMed] [Google Scholar]
- 44.Roush S., Slack F.J. The let-7 family of microRNAs. Trends Cell Biol. 2008:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 45.Coppola A., Romito A., Borel C., Gehrig C., Gagnebin M., Falconnet E. Cardiomyogenesis is controlled by the miR-99a/let-7c cluster and epigenetic modifications. Stem Cell Res. Authors. 2014;12:323–337. doi: 10.1016/j.scr.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 46.Shyh-Chang N., Zhu H., Yvanka De Soysa T., Shinoda G., Seligson M.T., Tsanov K.M. XLin28 enhances tissue repair by reprogramming cellular metabolism. Cell. 2013:155. doi: 10.1016/j.cell.2013.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ventura A., Young A.G., Winslow M.M., Lintault L., Meissner A., Erkeland S.J. Targeted deletion reveals essential and overlapping functions of the miR-17-92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang J., Greene S.B., Bonilla-Claudio M., Tao Y., Zhang J., Bai Y. Bmp signaling regulates myocardial differentiation from cardiac progenitors through a MicroRNA-mediated mechanism. Dev. Cell. 2010;19:903–912. doi: 10.1016/j.devcel.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen J., Huang Z.P., Seok H.Y., Ding J., Kataoka M., Zhang Z. Mir-17-92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circ. Res. 2013;112:1557–1566. doi: 10.1161/CIRCRESAHA.112.300658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iekushi K., Seeger F., Assmus B., Zeiher A.M., Dimmeler S. Regulation of cardiac MicroRNAs by bone marrow mononuclear cell therapy in myocardial infarction. Circulation. 2012;125:1765–1773. doi: 10.1161/CIRCULATIONAHA.111.079699. [DOI] [PubMed] [Google Scholar]
- 51.Yamakuchi M., Ferlito M., Lowenstein C.J. miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl. Acad. Sci. U. S. A. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bernardo B.C., Gao X.-M., Winbanks C.E., Boey E.J.H., Tham Y.K., Kiriazis H. Therapeutic inhibition of the miR-34 family attenuates pathological cardiac remodeling and improves heart function. Proc. Natl. Acad. Sci. U. S. A. 2012;109:17615–17620. doi: 10.1073/pnas.1206432109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ito T., Yagi S., Yamakuchi M. microRNA-34a regulation of endothelial senescence. Biochem. Biophys. Res. Commun. 2010;398:735–740. doi: 10.1016/j.bbrc.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 54.Boon R a, Iekushi K., Lechner S., Seeger T., Fischer A., Heydt S. MicroRNA-34a regulates cardiac ageing and function. Nature. 2013;495:107–110. doi: 10.1038/nature11919. [DOI] [PubMed] [Google Scholar]
- 55.Matsumoto S., Sakata Y., Suna S., Nakatani D., Usami M., Hara M. Circulating p53-responsive MicroRNAs are predictive indicators of heart failure after acute myocardial infarction. Circ. Res. 2013;113:322–326. doi: 10.1161/CIRCRESAHA.113.301209. [DOI] [PubMed] [Google Scholar]
- 56.Tian Y., Liu Y., Wang T., Zhou N., Kong J., Chen L. A microRNA-Hippo pathway that promotes cardiomyocyte proliferation and cardiac regeneration in mice. Sci. Transl. Med. 2015;7 doi: 10.1126/scitranslmed.3010841. 279ra38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heallen T., Morikawa Y., Leach J., Tao G., Willerson J.T., Johnson R.L. Hippo signaling impedes adult heart regeneration. Development. 2013;140:4683–4690. doi: 10.1242/dev.102798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wackerhage H., Del Re D.P., Judson R.N., Sudol M., Sadoshima J. The Hippo signal transduction network in skeletal and cardiac muscle. Sci. Signal. 2014;7 doi: 10.1126/scisignal.2005096. re4–re4. [DOI] [PubMed] [Google Scholar]
- 59.Von Gise A., Lin Z., Schlegelmilch K., Honor L.B., Pan G.M., Buck J.N. YAP1, the nuclear target of Hippo signaling, stimulates heart growth through cardiomyocyte proliferation but not hypertrophy. Proc. Natl. Acad. Sci. U. S. A. 2012;109:2394–2399. doi: 10.1073/pnas.1116136109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xin M., Kim Y., Sutherland L.B., Qi X., McAnally J., Schwartz R.J. Regulation of insulin-like growth factor signaling by Yap governs cardiomyocyte proliferation and embryonic heart size. Sci. Signal. 2011;4:ra70. doi: 10.1126/scisignal.2002278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xin M., Kim Y., Sutherland L.B., Murakami M., Qi X., McAnally J. Hippo pathway effector Yap promotes cardiac regeneration. Proc. Natl. Acad. Sci. 2013;110:13839–13844. doi: 10.1073/pnas.1313192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heallen T., Zhang M., Wang J., Bonilla-Claudio M., Klysik E., Johnson R.L. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science (80-. ) 2011;332:458–461. doi: 10.1126/science.1199010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pan D. The hippo signaling pathway in development and cancer. Dev. Cell. 2010:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tao G., Wang J., Martin J.F. Small RNA: from development to regeneration. Sci. Transl. Med. 2015;7 doi: 10.1126/scitranslmed.aaa7538. 279fs12. [DOI] [PubMed] [Google Scholar]
- 65.Yang Y., Del Re D.P., Nakano N., Sciarretta S., Zhai P., Park J. MIR-206 mediates YAP-induced cardiac hypertrophy and survival. Circ. Res. 2015;117:891–904. doi: 10.1161/CIRCRESAHA.115.306624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Beltrami A.P., Barlucchi L., Torella D., Baker M., Limana F., Chimenti S. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763–776. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- 67.Bearzi C., Rota M., Hosoda T., Tillmanns J., Nascimbene A., De Angelis A. Human cardiac stem cells. Proc. Natl. Acad. Sci. U. S. A. 2007;104:14068–14073. doi: 10.1073/pnas.0706760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Messina E., De Angelis L., Frati G., Morrone S., Chimenti S., Fiordaliso F. Isolation and expansion of adult cardiac stem cells from human and murine heart. Circ. Res. 2004;95:911–921. doi: 10.1161/01.RES.0000147315.71699.51. [DOI] [PubMed] [Google Scholar]
- 69.Martin C.M., Meeson A.P., Robertson S.M., Hawke T.J., Richardson J.A., Bates S. Persistent expression of the ATP-binding cassette transporter, Abcg2, identifies cardiac SP cells in the developing and adult heart. Dev. Biol. 2004;265:262–275. doi: 10.1016/j.ydbio.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 70.Oh H., Bradfute S.B., Gallardo T.D., Nakamura T., Gaussin V., Mishina Y. Cardiac progenitor cells from adult myocardium: homing, differentiation, and fusion after infarction. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12313–12318. doi: 10.1073/pnas.2132126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Laugwitz K.-L., Moretti A., Lam J., Gruber P., Chen Y., Woodard S. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 2005;433:647–653. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chong J.J.H., Reinecke H., Iwata M., Torok-Storb B., Stempien-Otero A., Murry C.E. Progenitor cells identified by PDGFR-alpha expression in the developing and diseased human heart. Stem Cells Dev. 2013;22:1932–1943. doi: 10.1089/scd.2012.0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Santini M.P., Forte E., Harvey R.P., Kovacic J.C. Developmental origin and lineage plasticity of endogenous cardiac stem cells. Development. 2016;143:1242–1258. doi: 10.1242/dev.111591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Keith M.C.B.R. “String theory” of c-kit(pos) cardiac cells: a new paradigm regarding the nature of these cells that may reconcile apparently discrepant results. Circ. Res. 2015;116:1216–1230. doi: 10.1161/CIRCRESAHA.116.305557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wilson K.D., Hu S., Venkatasubrahmanyam S., Fu J.-D., Sun N., Abilez O.J. Dynamic microRNA expression programs during cardiac differentiation of human embryonic stem cells: role for miR-499. Circ. Cardiovasc. Genet. 2010;3:426–435. doi: 10.1161/CIRCGENETICS.109.934281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Van Rooij E., Sutherland L.B., Qi X., Richardson J a, Hill J., Olson E.N. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science (80-. ) 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 77.Fu J.-D., Rushing S.N., Lieu D.K., Chan C.W., Kong C.-W., Geng L. Distinct roles of microRNA-1 and -499 in ventricular specification and functional maturation of human embryonic stem cell-derived cardiomyocytes. PLoS One. 2011;6:e27417. doi: 10.1371/journal.pone.0027417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jayawardena T.M., Egemnazarov B., Finch E.A., Zhang L., Alan Payne J., Pandya K. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ. Res. 2012;110:1465–1473. doi: 10.1161/CIRCRESAHA.112.269035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nam Y.-J., Song K., Luo X., Daniel E., Lambeth K., West K. Reprogramming of human fibroblasts toward a cardiac fate. Proc. Natl. Acad. Sci. U. S. A. 2013;110:5588–5593. doi: 10.1073/pnas.1301019110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rumyantsev Autoradiographic study on the synthesis of DNA, RNA, and proteins in normal cardiac muscle cells and those changed by experimental injury. Folia Histochen Cytochem. 1966;4:397–424. [PubMed] [Google Scholar]
- 81.Sulima On the regeneration of the myocardium in various injuries to the cardiac wall of reptiles. Arkh Anat. Gistol. Embriol. 1968;55:56–63. [PubMed] [Google Scholar]
- 82.Rumyantsev Post-injury DNA synthesis, mitosis and ultrastructural reorganization of adult frog cardiac myocytes. An electron microscopic-autoradiographic study. Z Zellforsch Mikrosk Anat. 1973;139:431–450. doi: 10.1007/BF00306596. [DOI] [PubMed] [Google Scholar]
- 83.Bergmann O., Bhardwaj R.D., Bernard S., Zdunek S., Barnabé-Heider F., Walsh S. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Beltrami A.P., Urbanek K., Kajstura J., Yan S.-M., Finato N., Bussani R. Evidence that human cardiac myocytes divide after myocardial infarction. N. Engl. J. Med. 2001;344:1750–1757. doi: 10.1056/NEJM200106073442303. [DOI] [PubMed] [Google Scholar]