Abstract
Cancer invasion involves a series of fundamental heterogeneous steps, with each step being distinct in its type regarding its dependence on various oncogenic pathways. Over the past few years, researchers have been focusing on targeted therapies to treat malignancies relying not only on a single oncogenic pathway, but on multiple pathways. Scientists have recently identified potential targets in the human genome considered earlier as non-functional but the discovery of their potential role in gene regulation has put new insights to cancer diagnosis, prognosis and therapeutics. Non coding RNAs (ncRNAs) have been identified as the key gene expression regulators. Long non-coding RNA (lncRNAs) reveal diverse gene expression profiles in benign and metastatic tumours. Improved clinical research may lead to better knowledge of their biogenesis and mechanism and eventually be used as diagnostic biomarkers and therapeutic agents. Small non coding RNAs or micro RNA (miRNA) are capable of reprogramming multiple oncogenic cascades and, thus, can be used as target agents. This review is aimed to give a perspective of non coding transcription in cancer metastasis with an eye on rising clinical relevance of non coding RNAs and their mechanism of action focusing on potential therapeutics for cancer pathogenesis.
Keywords: Metastasis, Oncogene, microRNA, Oncomirs, Transcription, Genetic expression
1. Introduction
Uncontrolled cellular proliferation and inhibition of apoptosis in cells results in the development of tumorigenesis which ultimately leads to the formation of cancer. The hallmark of cancer is the metastasis [1]. Major hindrance to the treatment of metastasis is that cancer cells show biological heterogeneity due to which it is most probably diagnosed at the later stages. Several regulatory factors called oncogenes regulate the proliferation and apoptotic mechanisms by working as tumour promoter genes. The emerging focus of the scientists is to identify certain biomarkers of the cancer related protein coding genes [2]. Only 2% of the entire human genome represents protein coding genes containing 20,000 genes and the vast portion of the genome is comprised of non coding RNA. These non coding RNAs (ncRNAs) are not meant to transcribe into proteins but they have a significant role in the transcription of protein coding transcripts into RNA and translation into functional proteins [3]. The classification of ncRNAs is based upon the targets and functions of the ncRNA molecules i.e. small non coding RNAs called micro RNAs (miRNAs) and long non coding RNAs (lncRNAs). The last version 23 of the Ensemble human genome annotation from March 2015 i.e. GRch38 identifies that there are 27,817 lncRNAs transcripts emerging from 15,931 genes. The number lncRNAs in the human genome is significantly greater than the protein coding genes which exhibit a structure similar to mRNA consisting of poly A tail [4]. Further lncRNAs is classified into five categories i.e Intergenic lnc RNAs (present between two protein coding genes), Intronic lncRNAs (introns of protein coding genes transcribe them), Overlapping lncRNAs (a coding gene is located on the intron), Antisense lncRNAs (the opposite strand of protein coding gene transcribe them) and processed lnc RNAs (lacks an open reading frame ORF) [5]. The functions of lncRNAs are quite intricate. Those lncRNAs which lack an ORF regulate epigenetics, transcription, and post transcriptional modification of RNA genetic expressions in response to vast regulating cues. Therefore, they are sometimes misexpressed in leukemias and primary solid tumours. The expression of many HOX lncRNAs is investigated to be different in solid breast tumours and isolated metastasis regions. The DNA damage in response to carcinogenesis involves p-53 dependent lncRNAs. Many transcriptional variations are under the control of lncRNAs which implies that there is a difference of lncRNAs profiling between normal and carcinogenic cells and lncRNAs are strictly linked with cancer metastasis and progression [6]. Therefore, the differential profiling of lncRNAs expression in normal and cancer cells provides a good biomarker of cancer diagnosis and developing a reasonable therapeutic approach.
Recent studies indicate that other small non coding protein RNAs called micro RNAs (mi RNAs) also aid in tumour suppression and are recognised as one of the largest gene regulators. They comprise approximately 1–4% of the total human genome and are almost 300 in number. The location of existence of miRNAs is on the introns of the non-coding mRNA genes and on the exons of the non-coding mRNA transcripts or either are present at 3' UTR of mRNA transcripts. miRNAs are involved to target multiple genes. However, the identification of these multiple target genes is somewhat difficult because they show partial complementarity with miRNA due to which mismatched and G-U base pairing happens. A simple bioinformatics tool, such as BLAST, is unable to identify the miRNA gene targets [7]. Due to this fact, it is acknowledged that miRNA 5' end depicts the greatest homologous relationship in the whole family. Recent research have also disclosed the important fact of 5' end of mRNA which aids in the loading and stability of miRNA into the miRISC complex system which is of biological importance. This is the reason why most bioinformatics tools use the initial 2–8 bases of miRNA sequence to find sequence complementarity in the 3'end of UTR for all the target genes. All this research has revealed that a single strand of miRNA can target approximately 200 gene sequences, irrespective of the diversity in target genes' functions i.e. they might be receptors, transcription factors or secreted molecules. Therefore, it can be inferred that mi RNA has enormous potential to regulate the expression of about one-third of human mRNA [8].
The biological role of miRNAs is to regulate the processes associated with cancer growth, such as tissue growth and differentiation. Such miRs playing crucial role in cancer development are also termed as oncomirs. These oncomirs encoded by lin-4and let-7 control the cellular proliferation and differentiation in C.elegans. Certain mutations, when appearing in these miRNA genes, result in the development of some abnormalities in the cell cycle arrest which is the hallmark of cancer. Interestingly, a similar fact became known when the function of the lin-4 and let-7 encoded oncomirs was studied that they are strictly related to distinct types of cancers. Several studies have revealed that miRNAs are identified in the serum, peripheral blood circulation, mononuclear cells and whole blood of cancer patients, such as breast cancer (miRNA-195 and let-7a), lung carcinoma (miR-21, miR-210 and miR-486-5p) and prostate cancer (miR-141) [9]. Hence, these miRNA signatures are regarded as diagnostic biomarkers of miRNA detection to differentiate cancer patients from normal individuals. A new evolving concept of circulating miRNA detection in exosomes links miRNA to cell to cell communication. Exosomes are relatively small vesicles containing apoptotic bodies and endosomes including macrophages, platelets and tumour cells. Recently, it was studied that exosomes also consist of miRNA, mRNA, proteins and lipids which renders them to be multivariate molecules [10].
Therapeutics of cancer based miRNA is studied on two levels i.e. either over expression of miRNA tumour suppressor or silencing oncogenic miRNA, both in vivo or in vitro approaches. Recent investigations disclosed the role of transgenic and knock out modelling in the mechanism of a specific miRNA. For instance, in a murine model driven by Kras, the over expression of miR-21 (oncomirs) up regulates the tumerogenesis, whereas its targeted deletion decreased the formation of tumours in the lungs. The most successful application of miRNA targeted therapeutics updated is the treatment of chronic hepatocellular carcinoma by delivering liver specific DNA-LNA (Locked Nucleic acids) miR-122 anti miRNA (SPC3649) as theranostic tool in chronic HCV infected model, but some issues of target toxicity and organ specificity may also persist. The other method of miRNA targeting is through delivery of cholesterol bound 2-O-methyl antagomiRs, which maintain the delivery and stability by decreasing degradation. On the other hand, miRNA targeting through Lock Nucleic Acids (LNAs) is more potent functionally, which locks ribose active sites by methylene bridges. The simplest way of miRNA targeted delivery is through Anti-miRNA oligonucleotides (AMOs) by direct complementarity, thus, inhibiting the target miRNA [11].
2. Molecular mechanism and genetic modification by lncRNAs in cancer
2.1. Deregulation of oncogene HOX-transcript antisense RNA (HOTAIR)
The molecular mechanism of long non coding RNA is strictly based upon controlling the gene expression by direct recruitment of histone modification enzymes to cell chromatin. DNA methylation and the resulting chromatin modification signatures the post translational modification which renders the protein product to be functional. The functional turmoil of these epigenetic changes is the key to the development of carcinogenesis. However, the rate at which DNA mutations occur in somatic cells is much lesser than these epigenetic changes in the cell. That is why these alterations are much more stable and can be inherited through generations. Functional alterations in lncRNAs provide the suitable environment for cells to proliferate undifferentially, leading the path towards tumerogenesis. Here we are explaining this mechanism with the help of an example: An oncogenic long non coding RNA called HOX transcript antisense RNA (HOTAIR) reprograms the state of chromatin promoting cancer metastasis highly expressed in predisposed breast cancer metastasis [12]. Molecular mechanism of this metastasis by HOTAIR is that when it is highly expressed a polycomb repressive complex 2 (PRC2) consisting of methylase EZH2, SUZ12 and EED bring about genetic expression changes via H3K27 methylation which grades the upregulation of cancer metastasis in vivo. Therefore, disappearance or disturbance in the gene expression of HOTAIR or associated PRC2 complex components inhibits cancer metastasis.It is concluded that lncRNAs are potentially involved in the progression of cancer invasiveness and its therapeutics can be designed by deregulation or disturbance of gene expression of HOTAIR to stop cancer invasiveness. HOTAIR is known to be the first identified lncRNAs functional in trans form. However, other antisense lncRNAs are identified to be the gene silencer in cis form. For instance, the transcription of tumor suppressor genes CDKN2B and CDKN2A produce antisense ANRIL which brings interaction with a subunit of PRC1 (CBX7). This results in gene silencing and the production of heterochromatin in cancer cells. Thus antisense HOTAIR works as an oncogene silencer by carrying about epigenetic changes in chromatin and DNA methylation. LNCRNAs also regulate the silencing of genes by controlling chromatin-protein interactions as molecular scaffolds. According to recent findings, PCR2 complex and LSD1 H3K4 demethylase complex are bridged together with the help of HOTAIR and the interaction of these complexes target the specific oncogenes and carry out alterations in histone modifications, resulting in gene silencing.
Another example of gene silencing by lncRNAs is such that an ncRNA (CCND1) can allosterically modify the TLS protein through the transcription of 5'CCDN1 regions when the stimulus of DNA damage is received [13]. This gene specific TLS-CBP/p300 interaction induced by conformational alterations of the TLS protein stops the transcription process of CCDN1. An interesting lncRNA called MALAT-1, known to be extensively expressed in different types of predisposed tumours, regulate the functional SR splicing factors, alternatively splicing to nuclear speckles. An important fact about lncRNAs is that they can control the genetic changes in response to intracellular signalling of transcription factors. One foremost example is the induction of lncRNAs-p21 repressing distinct genes through the recruitment of hnRNP-K protein. Numerous lncRNAs are genetically expressed in different types of cancers with different clinical invasiveness. A two-way ANOVA with pathological risk factors and clinical course suggested that HOTAIR oncogene RNA is a significant and eventual biomarker of cancer metastasis and resulting death. This is because the mRNA profiling requires hundreds of RNA species stratification but the use of a single HOTAIR RNA (lncRNA) provides a novel diagnostic and therapeutic tool for prior prognosis and management of cancer [14].
3. Proteomic and genetic databases of lncRNAs
The identification of a variety of long non coding RNAs requires the establishment of genetic databases which consists of both proteomic and genetic information about lncRNAs, including their molecular functions or functions at the cellular level along with their associations with other RNAs. Important databases with their proteomic functions are listed in Table 1.
Table 1.
Databases for long non coding RNA.
| Database | Functions/Information | Website |
|---|---|---|
| LNCipedia | Annotated human database lncRNAs transcript sequences and structural patterns | www.lncipedia.org |
| DIANA-LncBase | Predicted miRNA recognition elements (MREs) on human and mouse lncRNAs experimentally verified and computationally | http://diana.imis.athena.innovation.gr/DianaTool |
| CHIPBase | An open decoding transcriptional database for regulatory networks of non coding RNAs and protein coding genes from CHIP-seq data | Deepbase.sysu.edu.cn/chipbase |
| Noncode v.3.0 | Annotation of long non coding RNAs integratively excluding t-RNAs and rRNAs | www.ebi.ac.uk/miriam/main/collections/MIR:00000248 |
| lncRNAdb | The reference database for functional comprehensive annotations of eukaryotic long non coding RNA (lncRNAs) | www.lncrnadb.org |
| lnRNome | Preliminary built compared with human genome program. | Genome.igib.res.in/lncRNome |
| The functional lncRNAs database | A database of mammalian long non protein coding repository transcripts to search a specific lncRNAs. Currently, the database contains lncRNAs from Human, Mouse and Rat | www.valadkhanlab.org |
4. Long non coding RNA (lncRNA) decoy function
The lncRNAs decoy function refers to the pseudo gene class of lncRNAs functionally plagiaristic to mutations in non coding genes. A small number of mismatches occur despite originality of sequences with the parental gene. This similarity in structure mimics the structure of mRNA with lncRNAs, influencing the cellular function at the genetic level. This lncRNAs decoy function is of immense importance in cancer metastasis [15]. For instance:
4.1. PTEN
The mutation in PTEN causes the formation of the PTENP1 form by mismatching the sequence at 18 sites. The untranslated region of PTENP1 3'-UTR offers a number of regions for the miRNA to bind in spite of the shorter structure of PTENP1 from PTEN by 1 kb. Therefore, a number of miRNAs mimic the structure of PTENP1 to fight with PTEN [16].
4.2. KRAS
KRAS competes with its pseudo gene KRAS1P to accomplish its decoy function. In numerous tumours, the expression of KRAS1P is enhanced with the increased expression of KRAS by binding to 3'-UTR of KRAS, functioning as a sponge, the miRNA shows positive correlation [17].
4.3. GAS
The decoy function is also applicable to DNA which is depicted by the Growth Arrest Specific 5 (GAS 5) function which mimics the structure of the glucocorticoid response element (GRE) by binding with the GR transcription factor, interrupting the normal function of the glucocorticoid receptor (GR). Normally, the GR is activated when a ligand binds to it and stimulates the GRE to transcribe the genetic expression of downstream genetic transcripts. The genetic sequence of Gas 5 forms 6 hairpin confirmations. The structure of hairpin 5 mimics the actual confirmation of the GRE and, as a result, the GR interacts with Gas 5 in spite of the GRE, consequently, halting the transcription activity of the GR mediated GRE activity [18].
5. Diagnosis and therapeutic considerations using lncRNAs
The association of certain types of cancer with lncRNAs can be explored by determining the comparison of lncRNAs levels in cancer cells verses normal cells which must be confirmed through reverse transcription –quantitative polymerase chain reaction (RT-qPCR). The role of lncRNAs in tumour invasion, metastasis, migration and progression must also be investigated by in vitro and in vivo tissue culture techniques. Statistical correlation analysis also confirms the strong relationship of lncRNAs with progression and invasion of cancer and also the survival rate of these patients. For the last few years, the circulating lncRNAs biomarkers have been given more importance due to cancer diagnosis, prognosis and designing of fruitful cancer therapeutics [19].
5.1. Prostate cancer (PCa)
The diagnosis of PCa is currently based upon androgen regulated-serine protease PSA, highly specific to prostate cancer. The routine testing of PSA, unfortunately (?), addresses many problems due to its high specification for prostatic hyperplasia and prostatitis. The diagnosis and therapy of PCa using lncRNAs, such as DD3, also known as Prostate cancer Antigen 3 (PCA3), is increasingly expressed in such patients, whereas they show no expression in normal individuals. PCA3 is now identified as the most common gene specific to PCa which can be estimated in urine samples [20]. The first ever established molecular test for PCa is the PROGENSA PCA3 test approved by the Food and Drug administration. Multivariate analysis confirmed the validity of PCA3 analysis in urine with a sensitivity of 62% and specificity of 75%. Another one of the most investigated lncRNAs in case of PCa is Metastasis Associated Lung Adenocarcinoma Transcript 1 (MALAT-1) which can be detected in the plasma of the prostate cancer patient in a highly expressed form and has been approved as a biomarker of PCa. The main advantage of using these lncRNAs as diagnostic biomarkers is that it helps to prevent 30.2–46.5% biopsies in patients having serum PSA level of 4–10 ng/ml. Researchers are also working on another lncRNA called PCAT-18 (Prostate Cancer Associated Noncoding RNA Transcript 18) which is overexpressed in prostate cancer, progressing from the localized to the metastatic phase and can be detected highly in the plasma of patients [21].
5.2. Cervical cancer
HOX Transcript Antisense RNA (HOTAIR) is an lncRNA which is specific to tumour invasion in cervical cancer. Polycomb Repressive Complex 2 (PCR2) associates with HOTAIR and undergoes tranmethylation of H3K27. The HOTAIR expression is increased in the serum of cervical cancer patients which is also indicative of the metastatic tumour phase, adenocarcinoma, lymphatic node metastasis and tumour recurrence [22].
5.3. Colorectal cancer (CRC)
CRC is commonly diagnosed by colonoscopy, stool DNA testing and faecal occult blood testing which have been used for many years. The more advanced technique of CLC diagnosis is by using a microarray of lncRNAs, such as XLOC_006844, LOC152578 and XLOC_000303, which are upregulated in the plasma of the CRC patient. These lncRNAs are specific to CRC which signatures the strong AUC value of 0.919–0.975 validation level. Furthermore, this panel of lncRNAs is strongly stable in the plasma of patients and is considered as a valid CRC biomarker [23].
5.4. Breast cancer
The conventional diagnostic biomarkers used for breast cancer are the determination of CEA, CA125, CA153 and AFP, but with the passage of time, more advanced microarray lncRNA analysis has proven to be more useful with confirmation of RT-qPCR. lncRNAs RP11-445H22.4 are specific to breast cancer tissues and can be detected in the serum of patients with test sensitivity of 92% and specificity of 74%, which is significantly better than conventional biomarkers. While it is over expressed in breast cancer tissue samples, this therapy is still under clinical trial and researchers are aiming for its application at the commercial level [24] Table 2.
Table 2.
lncRNAs expression in various cancer types and their genomic location.
| lncRNAs | Cancer type | Expression | Gene loci | Reference |
|---|---|---|---|---|
| HOTAIR | Liver, colorectal, Breast, Cervical and Bladder cancer | Upregulated | 12q13.3 | [25] |
| MALAT-1 | Lung carcinoma | Upregulated | 11 | [26] |
| HULC | Liver cancer | Downregulated | 13q12.2 | [27] |
| SPRY4-IT1 | Ovarian cancer | Upregulated | Multiple loci | [28] |
| PCA3/DD3 | Prostate cancer | Upregulated | 13q14 | [29] |
| H19 | Gastric cancer | Upregulated | 17q23.2 | [30] |
| UCA1 | Bladder cancer | Upregulated | Multiple loci | [31] |
| XLOC_006844 XLOC152578 XLOC_000303 | Colorectal cancer | Upregulated | Multiple loci | [32] |
| ANRIL SPRY4-IT1 NEAT- 1 | Non small cell lung carcinoma | Upregulated | 5q32-33 | [33] |
| POU3F3 HNF1 A-AS1 SPRY 4-IT 1 | Esophageal squamous cell carcinoma | Upregulated | 17q22 | [34] |
| RP11-445H22-4 | Breast cancer | Upregulated | 21q21 | [35] |
| BANCR | Thyroid cancer | Upregulated | 19q6 | [36] |
| BCO40587 | Osteosarcoma | Downregulated | 21q21.2 | [37] |
| CASC2 | Endometerial carcinoma | Downregulated | 22q17.4 | [38] |
| GAS 5 | Cervical cancer | Downregulated | 14 | [39] |
| NEAT1 | Multiple myeloma | Downregulated | 13q31-32 | [40] |
6. Mechanism of action of deregulation of miRNA in cancer
The miRNA processing is a complex process accompanied with several enzymatic reactions which may at some point link to tumour initiation and progression due to deficiencies in miRNA components, impairing the production of miRNA. The cancer cell lines harbouring mutated Exportin 5 gene (XPO5) interferes with the production and function of miRNA. The role of miRNA deregulation in Chronic Lymphocytic Leukemia (CLL) was first elucidated in 2002. The miRNA processing is more prone to epigenetic alterations. That is why a number of miRNAs i.e. miR-29, miR-126, miR-127 and miR-145 are susceptible to methylation [24], [25]. Until now 122 epigenetically modified miRNAs are determined in which 55 miRNAs are specific to malignancies and 67 miRNAs are susceptible to cancer. These epigenetically modified miRNAs are located on chromosome no 1, 7, 11, 14 and 19. Not only are the epigenetic changes or chromosomal defects responsible for miRNA processing, several other factors contributing to miRNA deregulation, such as nucleotide polymorphisms in miRNAs, are also important. As an example, a pri-miR-196a2 holding single nucleotide polymorphisms causes mutation leading to the development of chronic lymphoid leukemia (CLL), lung, breast and gastric solid tumours in vivo. The miRNA quantification is also controlled by deregulation of small nucleolar RNAs (snoRNAs) in cancers. In ribosome biogenesis, snoRNAs (RNU43, RNU44, RNU48) causes 2’-o’- ribose methylation and interacts with growth arrest specific 5 gene (GAS 5) in breast cancer. Therefore, it can be inferred that deregulation of snoRNAs is endogenously controlled to inhibit cancer growth [41].
As described earlier, miRNA functions both as tumour suppressor and oncogene. A multivariate analysis predicted that 50% of the diseased patients' miRNA lies in the fragile sites of cancer which clearly demonstrates the crucial role of miRNA in cancer initiation and progression. Mir-125b-1 (homologue of C.elegans lin-4) is present on chromosomal locus 11q24 which is absent or show deleted mutations in patients of chronic breast, lung, liver, cervical and ovarian cancers [42]. This mir-125b-1gene is also present in B-cell acute lymphoblastic leukemia patients with an insertion mutation of pr-miRNA in the heavy chain of immunoglobulin. These studies render this mir-125-b-1 gene as an oncomirs because of its role in the modulation of tumour cells' functions. Many investigations also provide evidence of the miRNA's function as a tumour suppressor gene. A report was given in which 145 patients were diagnosed with B cell chronic lymphocytic leukemia (CLL) when their molecular analysis was carried out, it was observed that all those patients of CLL were shown to have deletions with different percentages or suppression of miRNA genes i.e. mir-15-a or mir-16-1.chromosomal locus 13q14 show 65% deletion in CLL cases, 16–40% in myeloid leukemia cases, 60% in prostate cancer cases and 50% in lymphomas. These findings support the fact that this miRNA (tumour suppressor gene) is well present in the 30 kb region. Practitioners have revealed that those CLL cases which have deletions at chromosomal locus of 13q14 are more easily prognosed as compared to the patients with deletions at 11q23 or 17p13 chromosomal locus [43].
6.1. miR-15a and miR-16-1 negativley regulate BCL2
BCL2 is an oncogene or antiapoptotic gene that is upregulated in various cancers such as breast cancer, leukemia, prostate cancer and lymphomas. The researchers thought that certain mutations (deletion/down regulation) in mir-16-1 and mir-15a causes the over expression of BCL2, favouring the development of leukemia and lymphomas. This fact is further supported by another report that a germline C to T mutation is present in CLL patients who showed consequently decreased expression of mir-16-1 gene leading to lymphogenesis. Thus, this mir-16-1 also has a role in modulating the immune system and ultimately B-cell differentiation [44].
6.2. miRNA profiling in cancer diagnosis
The tissue specific miRNA genes in humans are well determined through miRNA microassays and the northern blot technique which signatures the miRNA expression profiles in stem cell studies and the differentiation destination of undifferentiated cell types during the development process. Currently, the researchers are conducting trials to diagnose different classes of cancers by using miRNA expression profiles and design miRNA biomarkers that aid in cancer prognosis and diagnosis [45]. Approximately miRNA profiles of about 200 genes are enough to classify different human cancers. The expression of miRNA in cancerous and normal tissues can be studied through advanced novel techniques of bead-based flow cytometry. Through this method, tumours of different tissue origins are grouped in the same class on the basis of their embryonic lineage and miRNA expression. This results in the development of accurate miRNA expression profiles of both normal and cancerous tissues. For instance, the cancers of colon, liver, stomach and pancreas all possess the same endothelial origin and when they are clustered together in the same profile, they can show a single miRNA expression profile [46]. Similarly, tumours of haematopoietic origin are clustered to make one miRNA profile. This method avoids coherent clustering of the same tumour dataset. Therefore, the hepatocellular carcinomas are not grouped in the same class. Hence, miRNA expression profiles reflect the origin, developmental history of both normal and cancerous tissues and metastasis profile of tumours.
The diagnostic path of cancers through miRNA profiling is advanced by comparing the miRNA expression profiles of tumours with those of normal cells. A research reported a miRNA profile of 217 genes was prepared in which 129 miRNAs showed no or low expression irrespective of their tissue origin. Therefore, globally, it is taken into consideration that miRNA expression profiles of tumours signatures the differentiation stage of the cells when compared with normal cells regardless of their tissue origin [47]. These conclusions consider miRNAs simply as ‘oncomirs’ which describe that low or de differentiation in cells causes abnormalities progressing them towards metastasis. That is why proper miRNA signatures libraries or miRNA expression profiles established which help in accurate prognosis, diagnosis and treatment of each type of cancer. The establishment of the miRNA library requires the simple isolation miRNA from a paraffin fixed formalin embedded tissue which can predict the patient's whole prognosis data. Therapeutics of cancer based on miRNA profiling proves to be a powerful tool for researchers to define a proper treatment regimen for every type of tumour, even ones that are poorly differentiated or histologically non diagnostic [48].
7. Small non coding RNAs in cancer therapeutics
The main concept of cancer therapeutics is based upon the identification of molecular targets that are specified to cancer. The specific biological importance and differential expression of every type of non coding RNA for specific cancers may prove to be potential targets. Small non coding RNAs, also known as miRNAs, are potentially targeted by introducing complementary RNA sequences exogenously and by blocking miRNA hybridization through its 3'UTR sequences endogenously [49].
7.1. Negative regulation of ‘Ras’ by ‘Let-7’ family
The first group of oncomirs encoded by let-7 family specifically regulate the expression of an oncogene called Ras genes. Ras proteins are the membrane signalling proteins associated with GTPase specific for the regulation of cell growth and differentiation. It has been investigated that about 15–30% of mutations are associated with mutations in Ras proteins. Therefore, the miRNA that regulates the expression of this Ras protein controls the cellular proliferation or tumour growth in check. That is why, it is indicated that let-7 could be used as a good diagnostic marker for non small cell lung cancer patients because they show poor expression of let-7 and short operative survival. In vitro studies indicate that tissue culture of the human lung adenoma cell line displayed transient cellular proliferation inhibition in the presence of let-7 which suggests that let-7 is a tumour suppressor gene and that let-7 can be used as a therapeutic agent in the treatment of lung cancer caused by stimulation of mutation by Ras genes [50].
7.2. Association of miRNAs with MYC oncogene
Most human cancers often show an increased expression of MYC oncogene which is a potent regulator of growth and is responsible for both cellular apoptosis and proliferation. miRNA is closely related to over expression of MYC, resulting in the formation of B-cell malignancies. Here is an example: when the MYC gene is translocated [t(8;17)] to the place of mir-142 locus under the control of miRNA promoter, it results in the development of B-cell leukemia. This happens because of the disruption of miRNA processing owing to the over expression of MYC and formation of B cell cancer. The miR-155 is also linked to the transformation of B cells [51]. For many years, scientists were trying to explore how RNA that is non coding for protein and possesses a conserved sequence between human, mouse and avian genomes could lead to the formation of lymphomas which might become responsible for the increased expression of MYC. The expression of miR-155 was 100 times increased in Burkitt lymphoma, Hodgkin lymphoma and B cell lymphomas. This means that miR-155 functions as an oncogene by interaction with MYC, however, in normal circumstances it targets those genes that works antagonistically with the MYC pathway. mir-155 expression is also seen to work in other carcinomas, such as breast carcinoma, where its expression is up regulated which is meant for its distinct functions other than haematopoietic cells [52].
7.3. Therapeutic restoration of miRNA with micromolecules
The reason of downregulated expression of miRNA functioning as tumour suppressors is miRNA loci deletion or epigenetic silencing through CpG island hypermethylation through the promoter of miRNA genes. Improving the biogenesis of miRNA and restoring the miRNA through the direct intake of miRNA formulations can solve the problem. In this regard, hypomethylation agents such as decitabine or 5-azacitydine are useful to reserve the miRNA epigenetic silencing [50]. These are the approved treatments for myelodysplastic syndromes by downregulating the expression of several non coding RNAs and mRNAs. Another therapeutic approach is the utilization of a small biomolecule called enoxacin which is a derived fluoroquinolone compound applied usually as an antibacterial agent is a powerful up regulator of miRNA production by interacting with TAR RNA-binding protein 2 (TARBP2). Colon cancer cells have been successfully treated with enoxacin in vitro. Another study also predicted that enoxacin enhanced the expression of about 24 miRNAs in mice models, retarding the tumor growth in xenografts in cancer albino mice models. These studies clearly demonstrate that the anticancer activity is through the regulation of the miRNA mechanism by enoxacin. This illustration highlights the restoration of disrupted miRNA spectrum downregulated in cancer cells [37] Table 3.
Table 3.
Summary of functions of non coding RNAs [53].
| NCRNAs as | |
|---|---|
| Oncogenes or tumour suppressors | Changes in miRNA expression such as miR-155 and miR-21 forces the cell to become neoplastic, develops high grade lymphoma. On the other hand, miR-15 and miR-16 suppress tumour functionally delete lymphocytic leukemia. |
| Cancer metastasis | miR-10b promote distant metastasis in breast cancer to lung by downregulation HOX D10 and downregulating RHOC and miR-31. |
| As diagnostic tool | miRNA, T-UCRs and lincRNAs profiling can be used for diagnosis and prognosis and allows accurate differentiation between normal and cancerous tissue types. |
| Regulatory network | Interplay of miRNA, lncRNAs and protein coding genes display a complex network of interactions in normal cells an d when deregulated causes cancer |
| Genetic variations | Genetic variations in miRNA genes affect miRNA profiling and affect cancer susceptibility |
| Epigenetic regulation | Epigenetic machinery is controlled by a type of miRNAs called epi-miRNAs e.g miR-29 target DNA methyl transferases |
8. Conclusion
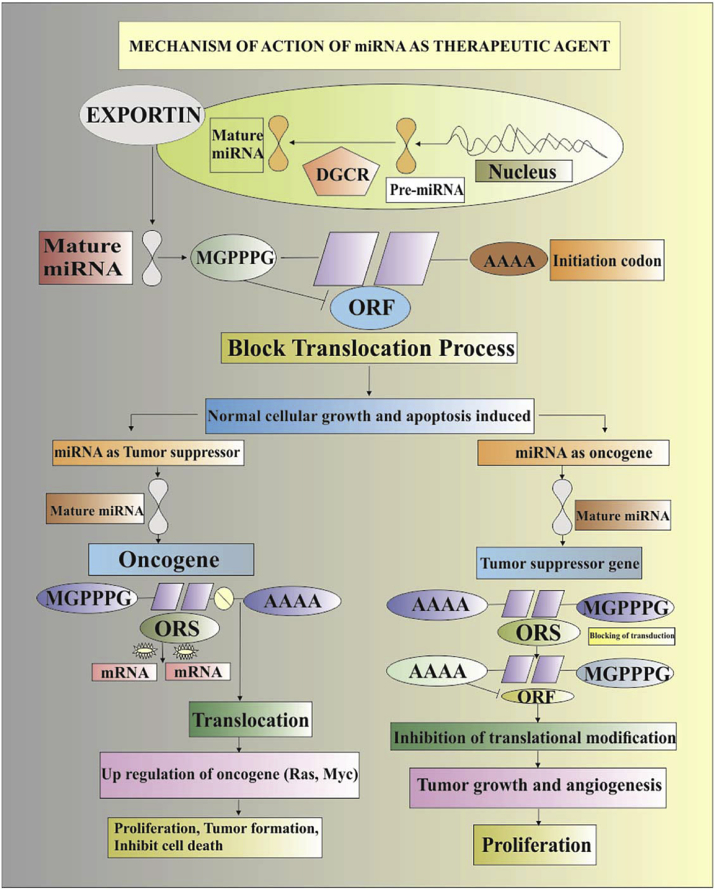
Non coding RNAs involving lncRNAs and miRNAs are potential therapeutic targets as they possess biological significance to be used as strong biomarkers for the diagnosis and prognosis of a number of cancers. The elucidation of the mechanism of non coding RNAs requires systemic study. However, the establishment of RNA profiling using bioinformatics tools and development of NCRNA databases is also very important. Future perspectives which are based upon non coding RNA localization, their genetic profiles and their mechanism elucidate their key roles in gene expression regulation. The utilization of antagomiRs, combined with cholesterol, could play a fundamental role to inhibit oncomirs activity in human cancers. On the other hand, miRNAs used as tumour suppressors, such as those of the let-7 family, could be over expressed for the treatment of specific types of cancers. In the future, the use of synthetic antisense oligonucleotides encoding complementary sequences of mature oncomirs, also known as anti-miRNA oligonucleotides (AMO), might be useful to inhibit the function of oncomirs and halt tumour growth. Therefore, the non coding protein RNA (NCRNAs) might become the magic bullet that provides insight into the treatment of all types of cancers Fig. 1.
Fig. 1.
Therapeutics of cancer through miRNA is based upon either blocking the activity of miRNA as oncogenes or by expressing the activity of miRNA functioning as tumour suppressor agents. In normal cases, complementary binding of miRNA to the target genes results in the induction of apoptosis, cellular growth and ultimately cell death. However, in the case of miRNA functioning as tumour suppressors, the defects of miRNA biogenesis is integrated to reduce in mature miRNA and unfortunate miRNA target oncoprotein expression profile, ultimately consequences cellular proliferation, angiogenesis, halts apoptosis and tumorigenesis. In the second case, where miRNA is functioning as an oncogene, the overexpression of this oncogene due to defects in differentiation tissues at any stage of growth suppresses the expression of miRNA target tumour suppressor genes leading to the progression of metastasis and cancer growth.
Conflict of interest
Declared None.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Mendell J.T., Olson E.N. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 3.Spizzo R., Almeida M.I., Colombatti A. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31:4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pennisi E. Genomics. ENCODE project writes eulogy for junk DNA. Science. 2012;337:1159–1161. doi: 10.1126/science.337.6099.1159. [DOI] [PubMed] [Google Scholar]
- 5.Dunham I. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Djebali S., Davis C.A., Merkel A. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fabian M.R., Sonenberg N., Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 8.Garzon R., Marcucci G., Croce C.M. Targeting microRNAs in cancer: rationale, within cell-cycle promoters. Nat. Genet. 2011;43:621–629. [Google Scholar]
- 9.Thorsen S.B., Obad S., Jensenet N.F. The therapeutic potential of microRNAs in cancer. Cancer J. 2012;18:275–284. doi: 10.1097/PPO.0b013e318258b5d6. [DOI] [PubMed] [Google Scholar]
- 10.Van Rooij E., Olson E.N. MicroRNA therapeutics for cardiovascular disease: opportunities and obstacles. Nat. Rev. Drug Discov. 2012;11:860–872. doi: 10.1038/nrd3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wahlestedt C. Targeting long non-coding RNA to therapeutically upregulate gene strategies and challenges. Nat. Rev. Drug Discov. 2012;9:775–789. doi: 10.1038/nrd4018. [DOI] [PubMed] [Google Scholar]
- 12.Fabbri M. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vickers K.C., Palmisano B.T., Shoucri B.M. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kasinski A.L., Slack F.J. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat. Rev. Cancer. 2011;11:849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pencheva N., Tavazoie S.F. Control of metastatic progression by microRNA regulatory networks. Nat. Cell Biol. 2013;15:546–554. doi: 10.1038/ncb2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Png K.J., Halberg N., Yoshida M., M microRNA regular that mediates endothelial recruitment and metastasis by cancer cells. Nature. 2012;481:190–194. doi: 10.1038/nature10661. [DOI] [PubMed] [Google Scholar]
- 17.Song S.J. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell. 2013;154:311–324. doi: 10.1016/j.cell.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uhlmann S. Global microRNA level regulation of EGFR-driven cell-cycle protein network in breast cancer. Mol. Syst. Biol. 2012;8:570. doi: 10.1038/msb.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Almeida M.I. Strand-specific miR-28-5p and miR-28-3p have distinct effects in colorectal cancer cells. Gastroenterology. 2012;142 doi: 10.1053/j.gastro.2011.12.047. 886–896e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melo S. Small molecule enoxacin is a cancer-specific growth inhibitor that acts by enhancing TAR RNA-binding protein 2-mediated microRNA processing. Proc. Natl. Acad. Sci. U. S. A. 2011;108:4394–4399. doi: 10.1073/pnas.1014720108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bader A.G., Brown D., Stoudemire J. Developing therapeutic microRNAs for cancer. Gene Ther. 2011;18:1121–1126. doi: 10.1038/gt.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tivnan A. Inhibition of neuroblastoma tumor growth by targeted delivery of microRNA-34a using anti-disialoganglioside GD2 coated nanoparticle. PLoS ONE. 2012;7:e38129. doi: 10.1371/journal.pone.0038129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trang P. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol. Ther. 2011;19:1116–1122. doi: 10.1038/mt.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen L. Tumor-specific expression of microRNA-26a suppresses human hepatocellular carcinoma growth via cyclin-dependent and -independent pathways. Mol. Ther. 2011;19:1521–1528. doi: 10.1038/mt.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bader A.G. miR-34-a microRNA replacement therapy is headed to the clinic Front. Genet. 2012;3:120. doi: 10.3389/fgene.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janssen H.L. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y. LNA-mediated anti-miR-155 silencing in low-grade B-cell lymphomas. Blood. 2012;120:1678–1686. doi: 10.1182/blood-2012-02-410647. (km) [DOI] [PubMed] [Google Scholar]
- 28.Kluiver J. Rapid generation of microRNA sponges for microRNA inhibition. PLoS ONE. 2012;7:e29275. doi: 10.1371/journal.pone.0029275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dvinge H. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature. 2013;497:378–382. doi: 10.1038/nature12108. [DOI] [PubMed] [Google Scholar]
- 30.Lehmann S.M. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat. Neurosci. 2012;15:827–835. doi: 10.1038/nn.3113. [DOI] [PubMed] [Google Scholar]
- 31.Jiang K. Biotech comes to its ‘antisenses’ after hard-won drug approval. Nat. Med. 2013;19:252. doi: 10.1038/nm0313-252. [DOI] [PubMed] [Google Scholar]
- 32.Ding L., Hu X.M., Wu H. Combined transfection of Bcl-2 siRNA and miR-15a oligonucleotides enhanced methotrexate-induced apoptosis in Raji cells. Can. Bio Med. 2013;10:16–21. doi: 10.7497/j.issn.2095-3941.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu X., Jung E.J., Calin G.A. The effect of Bcl-2 siRNA combined with miR-15a oligonucleotides on the growth of Raji cells. Med. Oncol. 2013;30:430. doi: 10.1007/s12032-012-0430-6. [DOI] [PubMed] [Google Scholar]
- 34.Bockhorn J., Stoudemire J., Lammers P. MicroRNA-30c inhibits human breast tumour chemotherapy resistance by regulating TWF1 and IL-11. Nat. Commun. 2013;4:1393. doi: 10.1038/ncomms2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Memczal S., Poliseno L., Tay Y. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 36.Ling H., Jung E.J., Calin G.A. 2013. CCAT2, a Novel Non-coding RNA Mapping to 8q24. [Google Scholar]
- 37.Salmena L., Poliseno L., Tay Y. A ceRNA hypothesis: the miR-15a oligonucleotides enhanced methotrexate-induced apoptosis in Raji cells. Cancer. 2013;154:311–324. [Google Scholar]
- 38.Poliseno L., Pencheva N., Tavazoie S.F. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hung T., Li C.H., Chen Y. Extensive and coordinated transcription of noncoding RNAs expression. Nat. Rev. Drug Discov. 2013;12:433–446. [Google Scholar]
- 40.Wheeler T.M., Kino T., Hurt D.E. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488:111–115. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gutschner T. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kino T., Hurt D.E., Ichijo T. Noncoding RNA Gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010;3:8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rusk N. AntagoNATs boost gene expression. Nat. Methods. 2012;9:437. doi: 10.1038/nmeth.2007. [DOI] [PubMed] [Google Scholar]
- 44.Modarresi F. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotech. 2012;30:453–459. doi: 10.1038/nbt.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flemming A. Regulatory watch: pioneering gene therapy on brink of approval. Nat. Rev. Drug Discov. 2012;11:664. doi: 10.1038/nrd3835. [DOI] [PubMed] [Google Scholar]
- 46.Gruber K. Europe gives gene therapy the green light. Lancet. 2012;380:e10. doi: 10.1016/s0140-6736(12)61992-8. [DOI] [PubMed] [Google Scholar]
- 47.Hansen T.B. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 48.Guttman M. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lai F. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Redis R.S., Calin S., Yang Y. Cell-to-cell miRNA transfer: from body homeostasis to therapy. Pharmacol. Ther. 2012;136:169–174. doi: 10.1016/j.pharmthera.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang M. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol. Cancer. 2011;10:117. doi: 10.1186/1476-4598-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang S. MicroRNA-34 suppresses breast cancer invasion and metastasis by directly targeting Fra-1. Oncogene. 2012;32:4294–4303. doi: 10.1038/onc.2012.432. [DOI] [PubMed] [Google Scholar]
- 53.Crawford E.D. Diagnostic performance of PCA3 to detect prostate cancer in men with increased prostate specific antigen: a prospective study of 1,962 cases. J. Urol. 2012;188:1726–1731. doi: 10.1016/j.juro.2012.07.023. [DOI] [PubMed] [Google Scholar]