Abstract
Myocardial infarction (MI), the globally leading cause of heart failure, morbidity and mortality, involves post-MI ventricular remodeling, a complex process including acute injury healing, scar formation and global changes in the surviving myocardium. The molecular mechanisms involved in adverse post-infarct left ventricular remodeling still remain poorly defined. Recently, microRNAs have been implicated in the development and progression of various cardiac diseases as crucial regulators of gene expression. We previously demonstrated that in a murine model of pressure overload, a model of heart failure secondary to aortic stenosis or chronic high blood pressure, elevated myocardial expression of miR-199b-5p is sufficient to activate calcineurin/NFAT signaling, leading to exaggerated cardiac pathological remodeling and dysfunction. Given the differences in left ventricular remodeling secondary to post-infarct healing and pressure overload, we evaluated miR-199b function in post-MI remodeling. We confirmed that the expression of miR-199b is elevated in the post-infarcted heart. Transgenic animals with cardiomyocyte-restricted overexpression of miR-199b-5p displayed exaggerated pathological remodeling after MI, reflected by severe systolic and diastolic dysfunction and fibrosis deposition. Conversely, therapeutic silencing of miR-199b-5p in MI-induced cardiac remodeling by using an antagomir to specifically inhibit endogenous miR-199b-5p in vivo, resulted in efficient suppression of cardiac miR-199b-5p expression and attenuated cardiac dysfunction and dilation following MI. Mechanistically, miR-199b-5p influenced the expression of three predicted target genes in post-infarcted hearts, dual specificity tyrosine-phosphorylation-regulated kinase 1A (Dyrk1a), the notch1 receptor and its ligand jagged1. In conclusion, here we provide evidence supporting that stress-induced miR-199b-5p participates in post-infarct remodeling by simultaneous regulation of distinct target genes.
1. Introduction
Acute myocardial infarction (MI) is one of the major causes of mortality and morbidity in humans [1]. MI occurs when blood supply to the left ventricular (LV) wall of the heart is hindered due to an occlusion in the coronary arteries [2] and consequently leading to massive cardiomyocyte death. A complex remodeling process is then initiated involving scar formation in the infarct zone, interstitial fibrosis in the border zone between the infarct and non-infarct area, cardiomyocyte hypertrophy and capillary rarefaction in the non-infarct remote zone. Eventually, these changes provoke impaired cardiac contractility and, ultimately, heart failure [3]. Moreover, post-infarction remodeling is associated with a higher incidence of arrhythmia and sudden cardiac death [4]. Hence, therapeutic strategies targeting pathological remodeling following MI could provide a promising strategy for post-MI management and heart failure. In this regard, we and others have identified a variety of stress-induced microRNAs (miRNAs) as regulators of crucial mechanisms in the development of heart failure and acting as promising therapeutic targets [5], [6], [7], [8], [9], [10]. These short (∼22 nucleotide) single stranded RNA molecules modulate gene expression by binding to ‘seed regions’ on protein-coding transcripts and leading to translational inhibition or degradation of mRNAs [11]. Importantly, a single miRNA likely simultaneously targets dozens of mRNAs, while individual mRNAs display several seed regions for different miRNAs, providing and enormous regulatory capacity for post-transcriptional gene regulation.
Previously, stress-induced miR-199b-5p was identified as a potent regulator of pathologic cardiac hypertrophy by functioning as an activator of calcineurin/nuclear factor of activated T-cell (CnA/NFAT) signaling [5]. miR-199b-5p exerts its function by targeting Dual-specificity tyrosine-phosphorylation regulated kinase 1a (Dyrk1a), a nuclear kinase responsible for NFAT rephosphorylation and translocation from the nucleus to the cytoplasm, essentially acting as an inhibitor of CnA/NFAT signaling. Therapeutic inhibition of miR-199b-5p by using a cholesterol-conjugated antagomir provoked inhibition of cardiac CnA/NFAT activity, attenuated pathological remodeling and preserved cardiac contractility after pressure overload, identifying miR-199b-5p as a therapeutic target to treat hypertension-induced forms of heart failure [5]. In the present study, we aimed at exploring the function of miR-199b-5p (referred to as miR-199b), if any, in post-infarct remodeling in the mouse using gain-of-function and therapeutic silencing approaches. We confirmed that the expression of miR-199b is elevated in the post-infarcted heart. Transgenic animals with cardiomyocyte-restricted overexpression of miR-199b displayed exaggerated pathological remodeling after MI, reflected by severe systolic and diastolic dysfunction and fibrosis deposition. Conversely, therapeutic silencing of miR-199b in MI-induced cardiac remodeling by using an antagomir to specifically inhibit endogenous miRNA-199b in vivo, resulted in efficient suppression of cardiac miR-199b expression and attenuated cardiac dysfunction and dilation following MI. Mechanistically, miR-199b influenced the expression of three predicted target genes in post-infarcted hearts, dual specificity tyrosine-phosphorylation-regulated kinase 1A (Dyrk1a), the notch1 receptor and its ligand jagged1. In conclusion, here we provide evidence supporting that stress-induced miR-199b participates in post-infarct remodeling by simultaneous regulation of distinct target genes.
2. Material and methods
2.1. Mouse models
Mice used in this study were male and female B6129S2F1 wild-type mice (Charles River Laboratories) of 2–6 months of age. Other mice used in this study were transgenic mice overexpressing miR-199b-5p in the postnatal myocardium under control of the 5.5 kb murine alpha-myosin heavy chain (MHC) promoter (miR-199b TG) [5]. Sample size was determined by a power calculation based on an echocardiographic effect size. Randomization of subjects to experimental groups was based on a single sequence of random assignments. Animal caretakers and investigators were blinded to group allocation during the experiment and/or when assessing the outcome. All protocols were performed according to institutional guidelines and approved by local Animal Care and Use Committees. All mice were housed on a 12hr:12hr light:dark cycle in a temperature-controlled environment with ad libitum access to water and chow at Innoser Netherlands BV, a commercial mouse breeding company with a quarterly animal health monitoring system that complies with FELASA guidelines and recommendations.
2.2. Myocardial infarction and transthoracic echocardiography
Myocardial infarction (MI) was performed in 2–3 month-old mice by permanent ligation of the left coronary artery as previously described [13]. Sham-operated animals underwent the same procedure without the occlusion of the left coronary artery. For Doppler-echocardiography, mice were shaved, lightly anaesthetized with isoflurane (mean 1% in oxygen) and allowed to breathe spontaneously through a nasal cone. Non-invasive, echocardiographic parameters were measured using a digital cardiac ultrasound platform (Vevo 770, VisualSonics, Toronto, Canada), and employing a single-element mechanical transducer with a center frequency of 30 MHz, (M − and B-mode), on self-breathing mice under anesthesia (2% isoflurane and 98% oxygen) to evaluate left ventricular dimensions and function [5]. In M-mode, the following parameters were obtained: AWthd, anterior wall thickness in diastole; LVIDd, left ventricular internal diameter in diastole; PWthd, posterior wall thickness in diastole; AWths, anterior wall thickness in systole; LVIDs, left ventricular internal diameter in systole; PWths, posterior wall thickness in systole; PWths, posterior wall thickness in systole; LVmass, left ventricular mass and FS, fractional shortening.
2.3. Antagomir administration
Chemically modified antisense oligonucleotides designed to target mmu-miR-199b-5p (5′-GAACAGGUAGUCUAAACACUGGG//3CholTEG-3'; antagomir-199b) with a 3′ cholesterol conjugation and 2 phosphorothioate (PT) bonds at the very first 5′ end and 4 PT bonds between the last 3′ bases [14] was synthesized at Integrated DNA Technologies (IDT, Leuven, Belgium). As control we generated an antagomir against C. elegans miR-39-5p (5′-AAGGCAAGCUGACCCUGAAGUU-3′/3CholTEG-3′), which does not target mammalian sequences (antagomir-ctrl). Female and male B6129S2F1 mice were first subjected to sham or MI surgery and injected intraperitoneally with antagomir-199b (80 mg/kg body weight dissolved in sterile PBS) or antagomir-ctrl for 3 consecutive days.
2.4. Quantitative real-time PCR
Total RNA (1 μg) was applied to either miR-based or mRNA-based reverse transcription. Real-time PCR was performed on a BioRad iCycler (Biorad) using SYBR Green. Transcript quantities were compared using the relative Ct method, where the amount of target normalized to the amount of endogenous control (L7 or U6 for mRNAs or miRNAs, respectively) and relative to the control sample is given by 2–ΔΔCt. Primers used for protein-coding transcripts are provided in Supplementary Table 1. For microRNA real-time PCR, miRNAs were isolated with TRIzol reagent (Invitrogen) and cDNA was generated with the miScript Reverse Transcription Kit (Qiagen). For real-time PCR detection of miRNAs, miScript Primer Assays and the miScript SYBR Green PCR Kit (Qiagen) were used. Primers for miRNA detection included miR-199b-5p, 5′- CCCAGTGTTTAGACTACCTGTTC and Universal reverse 5′- GAATCGAGCACCAGTTACGC.
2.5. Western blotting
SDS PAGE electrophoresis and blotting were performed as previously described [15]. In short, whole tissue or cell lysates were produced in RIPA buffer supplemented with PhosSTOP (Roche) and Protease inhibitor cocktail (Roche). Subsequently samples were boiled in 4× Leammli buffer, including 2% β-mercaptoethanol, for 5 min at 95 °C. SDS-PAGE and Western blotting were performed using the Mini-PROTEAN 3 system (Bio-Rad). Blotted membranes were blocked in 5% BSA/TBS-Tween. Primary antibody labeling was performed overnight at 4 °C. Secondary IgG–horseradish peroxidase (HRP)-conjugated antibodies were applied for 2 h at room temperature. Following incubation with an antibody, blots were washed for 3 × 10 min in TBS-Tween. Images were generated using Supersignal West Dura Extended Duration ECL Substrate (Pierce) and the LAS-3000 documentation system (FujiFilm Life Science). Stripping was performed with Restore Western blot stripping buffer (Pierce). Outputs were normalized for loading and results are expressed as an n-fold increase over the values of the control group in densitometric arbitrary units. Primary antibodies that were used included rabbit polyclonal anti-Dyrk1A (Santa Cruz, 1:500), mouse monoclonal anti-α-tubulin (Sigma, 1:1000). Secondary antibodies included polyclonal rabbit anti-mouse IgG–HRP (DAKO, 1:5000) and polyclonal swine anti-rabbit IgG–HRP (DAKO, 1:5000).
2.6. Histological analysis and (immunofluorescence) microscopy
For histological analysis, hearts were arrested in diastole, perfusion-fixed with 4% paraformaldehyde, embedded in paraffin and cut into 4-μm sections. Paraffin sections were stained with haematoxylin and eosin (H&E) for routine histological analysis; Sirius Red for detection of fibrillar collagen and FITC-labeled wheat-germ-agglutinin (WGA) to visualize and quantify the cell cross-sectional area. Cell surface areas and infarct size were determined using ImageJ software. Sirius Red stained sections were used to determine the infarct size. Epicardial infarct ratio was obtained by dividing the epicardial infarct lengths by the epicardial circumferences from all sections. Endocardial infarct ratio was calculated similarly. Infarct size derived from this approach was calculated as [(epicardial infarct ratio + endocardial infarct ratio)/2] × 100 [16]. Slides were visualized using a Leica DM 2000 for bright field and Leica DM3000 for fluorescence imaging.
2.7. microRNA target prediction
Putative microRNA-199b target genes were identified using the microRNA databases and target prediction tools miRBase (http://microrna.sanger.ac.uk/), PicTar (http://pictar.mdc-berlin.de/) and TargetScan (http://targetscan.org/index.html).
2.8. Statistical analysis
The results are presented as mean ± standard error of the mean. Statistical analyses were performed using Prism software (GraphPad Software Inc.), and consisted of ANOVA followed by Tukey's test when group differences were detected at the 5% significance level, or Student's t-test when comparing two experimental groups. Differences were considered significant when P < 0.05.
3. Results
3.1. Cardiac overexpression of miR-199b exacerbates LV remodeling following MI
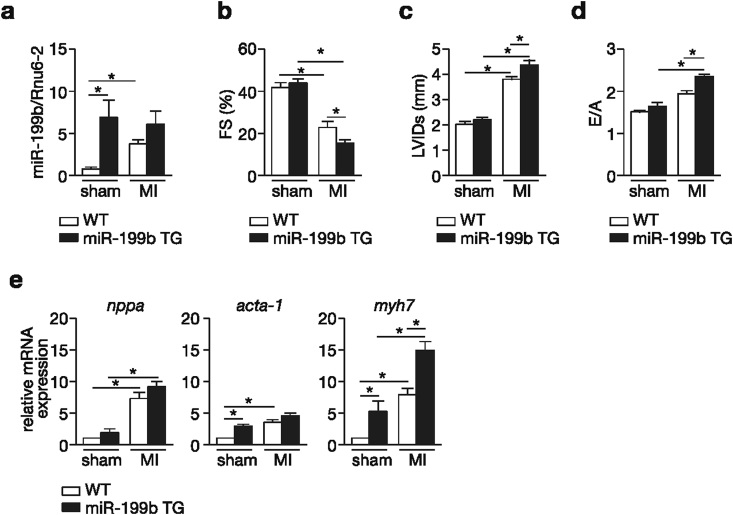
We have previously established the therapeutic potential of targeting miR-199b in a murine model of pressure overload-induced heart failure [5]. In the present follow-up study we aimed at determining the involvement of miR-199b, if any, during pathological cardiac remodeling induced by myocardial infarction (MI). To this end, wild-type (WT) and cardiac-specific miR-199b transgenic mice (miR-199b TG or TG) [5], were subjected to either sham surgery or MI for 4 weeks. WT animals displayed increased cardiac miR-199b expression levels when subjected to MI compared to the sham group (Fig. 1a). As expected, higher miR-199b expression was observed in transgenic hearts compared to WT mice. All animals subjected to 4 weeks of MI developed severe cardiac dysfunction as evidenced by decreased fractional shortening (FS), a parameter of systolic function, and increased left ventricular internal diameters (LVIDd), indicating a profound dilation post-infarction (Fig. 1b and c; Table 1). TG mice developed severe systolic dysfunction compared to WT animals indicating that increased miR-199b expression levels sensitized the heart to MI-induced dysfunction (Fig. 1b). We also observed diastolic dysfunction in all experimental post-MI groups as evidenced by an increased E/A ratio (‘E’ early; ‘A’ late ventricular filling velocity measured by mitral valve Doppler; Fig. 1d; Table 1), but only post-infarcted TG animals displayed an E/A ratio above 2, indicating restrictive filling of the LV. The observed cardiac phenotypes correlated with increased transcript abundance of cardiac stress genes (Fig. 1e) including atrial natriuretic factor (Nppa), α-skeletal actin (Acta1) and β-myosin heavy chain (Myh7). Taken together, these results indicate that cardiac miR-199b overexpression sensitizes the myocardium to geometric post-infarction remodeling, cardiac dysfunction and the induction of stress marker gene expression.
Fig. 1.
Cardiac specific overexpression of miR-199b sensitizes the heart to MI. (a) Real-time PCR analysis of miR-199b transcript abundance in hearts from wild type (WT) or miR-199b transgenic mice (TG) subjected to either sham or MI. Rnu6-2 was used as a reference gene for normalization. (b-d)Assessment of cardiac functional parameters by cardiac echocardiography, in WT and miR-199b transgenic mice (TG) after 4 weeks of either sham or MI surgery: (b) fractional shortening (FS), (c) LV internal diameter at systole (LVIDs) and (d) early to late ventricular filling velocity (E/A). (e) Real-time PCR analysis of transcript abundance for the fetal gene markers natriuretic peptide atrial natriuretic factor (nppa), α-skeletal actin (acta1) and β−myosin heavy chain (myh7) in the hearts from WT or miR-199b transgenic mice (TG), 4 weeks after either sham or MI operation. n = 6–9, *P < 0.05 (mean ± s.e.m.).
Table 1.
Morphometric and echocardiographic characteristics of WT and miRNA-199b TG mice subjected to 4 weeks of sham or MI.
| Sham |
MI |
|||
|---|---|---|---|---|
| WT | miR-199b TG | WT | miR-199b TG | |
| n | 11 | 8 | 7 | 13 |
| HW/BW | 4.61 ± 0.09 | 4.31 ± 0.18 | 6.30 ± 0.59* | 6.10 ± 0.25* |
| HW/TL | 6.82 ± 0.21 | 7.06 ± 0.23 | 7.99 ± 0.19* | 8.83 ± 0.21*# |
| LV mass (mg) | 187.72 ± 13.42 | 208.11 ± 19.52 | 354.71 ± 9.24* | 343.03 ± 19.48* |
| LV mass/BW (mg/g) | 5.89 ± 0.73 | 5.50 ± 0.67 | 12.48 ± 0.40* | 9.50 ± 0.56*# |
| LV mass/TL (mg/mm) | 8.5 ± 0.54 | 8.9 ± 0.81 | 14.78 ± 1.16* | 15.51 ± 0.93* |
| IVSd (mm) | 1.59 ± 0.08 | 1.64 ± 0.10 | 2.09 ± 0.07* | 2.06 ± 0.09* |
| IVSs (mm) | 1.22 ± 0.12 | 1.61 ± 0.08* | 1.87 ± 0.09* | 1.84 ± 0.11* |
| LVIDd (mm) | 3.57 ± 0.18 | 3.83 ± 0.58 | 4.58 ± 0.18* | 4.52 ± 0.22* |
| LVIDs (mm) | 2.08 ± 0.15 | 2.18 ± 0.11 | 3.85 ± 0.13* | 4.30 ± 0.15*# |
| LVPWd (mm) | 1.31 ± 0.02 | 1.40 ± 0.08 | 1.64 ± 0.06* | 1.38 ± 0.06*# |
| LVPWs (mm) | 2.29 ± 0.05 | 2.15 ± 0.11 | 1.48 ± 0.08* | 1.71 ± 0.04*# |
| FS (%) | 41.73 ± 3.25 | 43.08 ± 2.57 | 22.69 ± 2.59* | 16.01 ± 1.32*# |
| E/A (mm/s) | 1.59 ± 0.06 | 1.68 ± 0.06 | 1.79 ± 0.09 | 2.24 ± 0.06*# |
Data are expressed as means ± SEM. HW, heart weight; BW, body weight; TL, tibia length; LV, left ventricular; IVSd, interventricular septal thickness at end-diastole; IVSs, interventricular septal thickness at end-systole; LVIDd, left ventricular internal dimension at end-diastole; LVIDs, left ventricular internal dimension at end-systole; LVPWd, left ventricular posterior wall thickness at end-diastole; LVPWs, left ventricular posterior wall thickness at end-systole; FS, fractional shortening; E/A, Doppler E/A ratio. *, indicates P < 0.05 vs wt-sham group; #, indicates P < 0.05 vs experimental group.
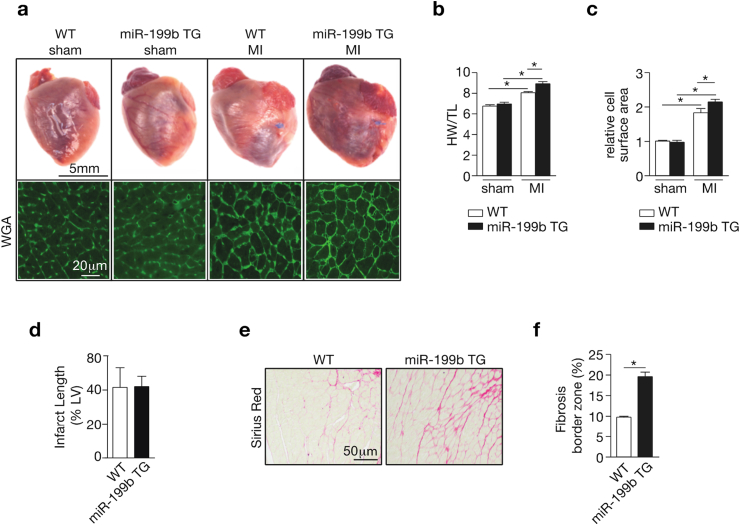
Next, we evaluated the extent of cardiac hypertrophic growth and fibrotic scar formation following MI [3], [17]. Although post-infarcted hearts displayed increased heart weights, this effect was exacerbated in TG animals. Accordingly, cross-sectional cardiomyocytes surface areas were increased after MI with the cells being slightly more hypertrophic in the TG animals (Fig. 2c). This finding of mild increase in cardiac cell hypertrophy in the TG animals after MI, in combination with the observed exaggerated dilation response secondary to MI suggests that miR-199b overexpression promotes a switch from concentric to eccentric hypertrophy response in the viable myocardium after MI. Next, we investigated infarct scar size, a main determinant of outcome after MI [18]. Although the scar size on the left anterior wall distal to the occluded LAD was comparable between WT and TG hearts (Fig. 2d), the amount of fibrosis in the border zone of TG hearts was more extensive compared to WT hearts (Fig. 2e and f). Conclusively, miR-199b overexpression promotes, on the one hand, a mild cardiac concentric hypertrophic phenotype in the viable remote myocardium, and on the other hand, LV dilatation. Furthermore, it does not influence infarct healing but stimulates exaggerated fibrotic deposition in the remote myocardium in post-infarcted hearts.
Fig. 2.
miR-199b overexpression results in exaggerated fibrosis in the border zone during post-myocardial infarct remodeling. (a) Representative images of whole hearts (top panels) or wheat-germ agglutinin (WGA)-labeled (lower panels) histological sections, (b) gravimetric analysis of corrected heart weights and (c) quantification of cardiomyocyte surface area of WT or miR-199b transgenic (TG) mice hearts after either 4 weeks of sham or MI surgery. (d) Quantification of infarct sizes in WT and miR-199b transgenic (TG) animals, 4 weeks after MI. (e) Representative images of Sirius red-stained histological sections and (f) quantification of collagen deposition in the border zone of WT or miR-199b transgenic (TG) hearts after MI. n = 6–10, *P < 0.05 (mean ± s.e.m.).
3.2. In vivo miR-199b silencing attenuates post-infarction remodeling
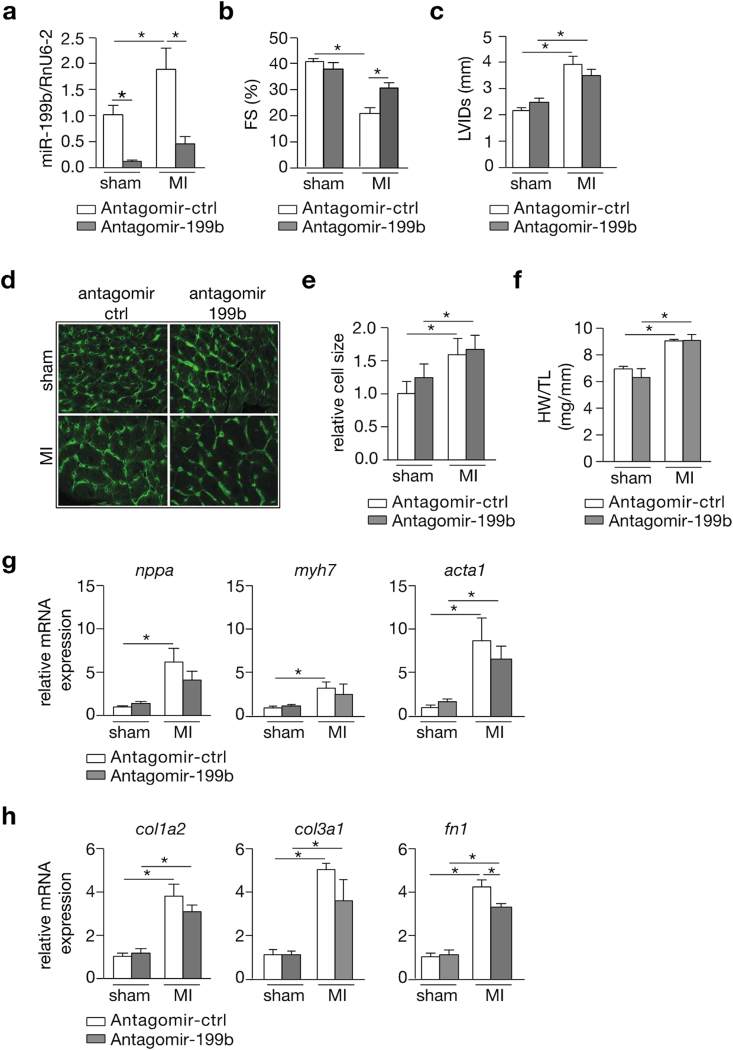
Next, we evaluated post-infarction remodeling following an in vivo silencing approach for miR-199b. To this end, animals were randomized to either receive antagomir-control or antagomir-199b treatment for 3 consecutive days before being subjected to sham or MI surgery. We observed that while miR-199b expression was augmented in the LV after MI in antagomir-control treated mice, miR-199b was effectively silenced in the myocardium after administration of antagomir-199b (Fig. 3a). As expected, antagomir-control treated mice developed severe systolic dysfunction and LV dilation following MI (Fig. 3b and c; Table 2). Interestingly, infarcted mice receiving antagomir-199b demonstrated a clear attenuation of cardiac dysfunction accompanied by a mild suppression of myocardial dilation (Fig. 3b and c; Table 2). Histological analysis demonstrated that cross-sectional surface areas of cardiomyocytes were increased to the same extent in antagomir-control or antagomir-199b-treated mice following MI (Fig. 3d and e), which was also reflected at the whole organ level by measuring heart weight-to-tibia length ratios (Fig. 3f). No major inhibition of stress marker gene induction was observed between infarcted mice receiving either antagomir-control or antagomir-199b (Fig. 3g). As we did not detect major differences regarding scar size and collagen deposition at the histological level (data not shown), we also assessed the expression levels of several fibrosis-related genes such as collagen type I alpha 2 chain (col1a2), collagen type 3 alpha 1 chain (col3a1) and fibronectin-1 (fn1) (Fig. 3h). Whereas for the collagens there was reduced mRNA expression in the mice treated with antagomir-199b but this effect did not reach statistical significance, we did observe significant reduction in fibronectin1 expression in the mice treated with antagomir-199b compared to the control group (Fig. 3h). In conclusion, our in vivo silencing approach for miR-199b shows therapeutic effects on post-infarction remodeling in mice, with pronounced improvement of systolic contractility, reduction of LV dilation and reduced fibrosis-related gene expression, but no major differences in cardiomyocyte hypertrophy or “fetal” gene induction in the remote myocardium.
Fig. 3.
Pharmacological inhibition of miR-199b attenuates cardiac dysfunction following MI. (a) Real-time PCR analysis of miR-199b expression levels in the LV of antagomir-control or antagomir-199b-treated animals after subjection to either 4 weeks of sham or MI. Rnu6-2 was used as a reference gene for normalization. (b,c) Quantification of (b) fractional shortening (FS) and (c) LV internal diameter at systole (LVIDs) by echocardiography in antagomir-control or antagomir-199b-treated mice after subjection to 4 weeks of either sham or MI. (d) Representative images of WGA-labeled cardiac histological sections from mice subjected to sham or MI, and treated either with antagomir-control or antagomir-199b. (e) Quantification of cardiomyocyte surface area in histological sections from mice subjected to sham or MI, and treated either with antagomir-control or antagomir-199b. (f) Gravimetric analysis of corrected hearts weights of animals that underwent sham or MI surgery and after treatment with antagomir-control or antagomir-199b. (g) Real-time PCR analysis of transcript abundance of the fetal gene markers: natriuretic peptide atrial natriuretic factor (nppa), β−myosin heavy chain (myh7) and the sarcomeric proteins α-skeletal actin (acta1) in antagomir-control or antagomir-199b treated mice after subjection to 4 weeks of sham or MI. (h) Real-time PCR analysis of transcript abundance of fibrosis related genes: collagen type I alpha 2 chain (col1a2), collagen type 3 alpha 1 chain (col3a1) and fibronectin-1 (fn1) in antagomir-control or antagomir-199b treated mice after subjection to 4 weeks of sham or MI. n = 8–13 *P < 0.05 (mean ± s.e.m.).
Table 2.
Morphometric and echocardiographic characteristics of mice subjected to 4 weeks of sham or MI and treated either with antagomir-control or antagomir-199b.
| Sham |
MI |
|||
|---|---|---|---|---|
| Antagomir-control | Antagomir-199b | Antagomir-control | Antagomir-199b | |
| n | 13 | 8 | 11 | 11 |
| HW/BW (mg/g) | 7.16 ± 0.43 | 5.65 ± 0.81 | 8.28 ± 1.03 | 8.64 ± 0.53 |
| HW/TL (mg/mm) | 7.07 ± 0.28 | 6.47 ± 0.76 | 9.01 ± 0.43* | 9.06 ± 0.67* |
| LV mass (mg) | 95.01 ± 6.51 | 113.32 ± 6.01 | 151.71 ± 24.18* | 182.29 ± 14.30* |
| LV mass/BW (mg/g) | 4.04 ± 0.22 | 4.42 ± 0.17 | 5.58 ± 0.28* | 6.93 ± 0.46*# |
| LV mass/TL (mg/mm) | 4.32 ± 0.43 | 4.97 ± 0.51 | 6.94 ± 0.23* | 7.99 ± 0.33* |
| IVSd (mm) | 0.99 ± 0.03 | 1.02 ± 0.05 | 0.84 ± 0.08 | 0.87 ± 0.08 |
| IVSs (mm) | 1.46 ± 0.05 | 1.49 ± 0.06 | 1.21 ± 0.10* | 1.19 ± 0.10* |
| LVIDd (mm) | 3.70 ± 0.10 | 3.83 ± 0.12 | 4.97 ± 0.28* | 5.20 ± 0.25* |
| LVIDs (mm) | 2.19 ± 0.12 | 2.38 ± 0.16 | 3.91 ± 0.33* | 3.55 ± 0.28* |
| LVPWd (mm) | 0.79 ± 0.06 | 0.92 ± 0.04 | 0.92 ± 0.07 | 1.04 ± 0.09* |
| LVPWs (mm) | 1.20 ± 0.05 | 1.35 ± 0.05 | 1.23 ± 0.08 | 1.29 ± 0.07 |
| FS (%) | 40.81 ± 2.02 | 37.85 ± 2.36 | 21.32 ± 2.69* | 31.73 ± 1.74*# |
| E/A (mm/s) | 1.40 ± 0.05 | 1.24 ± 0.04 | 1.36 ± 0.07 | 1.33 ± 0.05 |
Data are expressed as means ± SEM. HW, heart weight; BW, body weight; TL, tibia length; LV, left ventricular; IVSd, interventricular septal thickness at end-diastole; IVSs, interventricular septal thickness at end-systole; LVIDd, left ventricular internal dimension at end-diastole; LVIDs, left ventricular internal dimension at end-systole; LVPwd, left ventricular posterior wall thickness at end-diastole; LVPws, left ventricular posterior wall thickness at end-systole; FS, fractional shortening; EF, ejection fraction. *, indicates P < 0.05 vs wt-sham group; #, indicates P < 0.05 vs experimental group.
3.3. Downstream effectors of miR-199b in post-MI remodeling
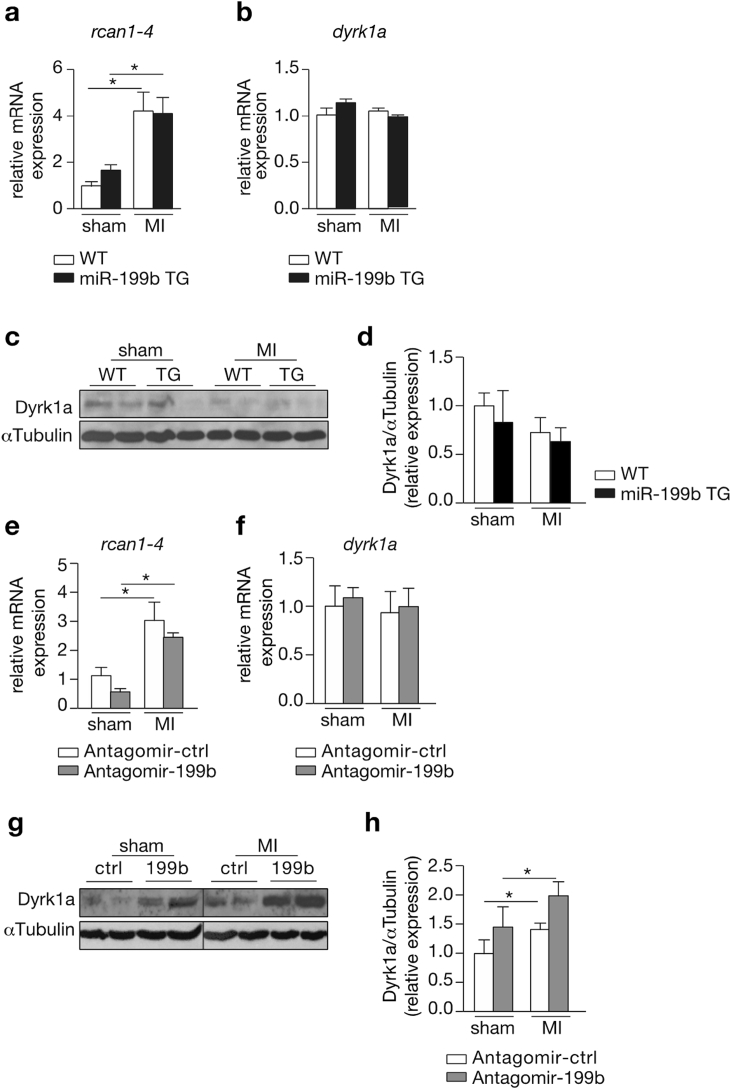
Next, we investigated the mechanisms by which miR-199b may induce an exaggerated cardiac dysfunction and LV remodeling following MI. Our group earlier established that miR-199b exerts its pro-hypertrophic function in the pressure-overloaded heart by regulating signaling strength of the calcineurin/nuclear factor of activated T-cell (CnA/NFAT) pathway. In line, we found Rcan1-4 transcript expression, a sensitive marker of cardiac NFAT activity, upregulated in response to MI, indicative of activation of this signaling pathway following MI. However, no additional activation was discernable in post-infarcted TG mice with miR-199b overexpression (Fig. 4a). We also could not observe differences in transcript or abundance or protein expression of Dual-specificity tyrosine-phosphorylation regulated kinase 1a (Dyrk1a), a previously validated target gene of miR-199b in the myocardium (Fig. 4b–d). Conversely, we also evaluated Rcan1-4 and Dyrk1a transcript abundance in the experimental infarction groups receiving antagomir-control or antagomir-199b. Although the data show no significant differences for both transcripts (Fig. 4e and f), western blot analysis revealed an increase in Dyrk1a expression levels in infarcted hearts after antagomir-199b treatment (Fig. 4g and h). Altogether, these data suggest that miR-199b silencing in the post-infarcted heart may still involve CnA/NFAT signaling by direct regulation of Dyrk1a expression, but to a less extent than what we previously observed in pressure-overloaded hearts [5]. Therefore, we conclude that miR-199b expression promotes adverse remodeling and cardiac dysfunction following MI in a response that must require additional target genes to play a synergistic effect along with specific players of CnA/NFAT signaling.
Fig. 4.
Improved cardiac function after pharmacological inhibition of miRNA-199b involves CnA/NFAT signaling. (a,b) Real-time PCR analysis of transcript abundance of (a) rcan1-4 and (b) dyrk1a in WT or miR-199b transgenic (TG) murine hearts after subjection to 4 weeks of sham or MI. (c) Western blot analysis of endogenous Dyrk1a and αTubulin in WT or miR-199b transgenic (TG) murine hearts after subjection to 4 weeks of sham or MI. (d) Quantification of αTubulin-corrected Dyrk1A western blot signals from (c). (e,f) Real-time PCR analysis of transcript abundance of (e) rcan1-4 and (f) dyrk1a in hearts from mice subjected to sham or MI, and treated either with antagomir-control or antagomir-199b. (g) Western blot analysis of endogenous Dyrk1a and αTubulin in murine hearts subjected to sham or MI, and treated either with antagomir-control or antagomir-199b. (h) Quantification of αTubulin-corrected Dyrk1A western blot signals from (g). n = 5–9 *P < 0.05 (mean ± s.e.m.).
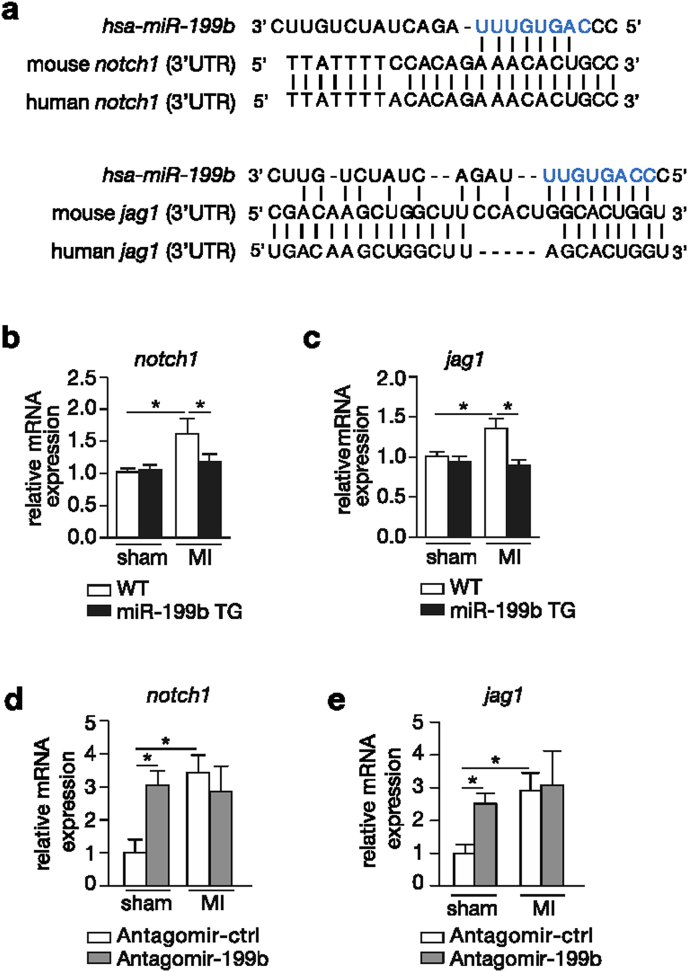
Because miRNAs target multiple transcripts by virtue of the presence of a specific seed region, we evaluated the expression of additional miR-199b target genes in post-infarcted hearts. To this end, we analyzed target prediction databases for miR-199b and identified two members of the Notch signaling pathway as potential targets of miR-199b, Notch1 and Jagged1. The Notch pathway is of particular interest here because increased activity of Notch1 in the adult heart after a cardiac insult has been associated with beneficial effects on cardiac function and remodeling [19], [20], [21], [22], [23], [24], [25]. Moreover, Jagged1 is a validated direct target of miR-199b and an inverse correlation between miR-199b and Notch1 expression in cancer cells was previously demonstrated [26], [27], [28]. The sequence alignments between mature mmu-miR-199b-5p and sequences in the 3′UTR of Jagged1 and Notch1 displayed perfect sequence complementarity between the transcripts and miR-199b (Fig. 5a). In line, we found that transcript abundance for both Jagged1 and Notch1 were enhanced in WT post-infarcted hearts, while in TG animals subjected to MI their expression was at control level (Fig. 5b and c). Vice versa, the hearts of WT mice receiving antagomir-199b displayed a clear derepression of both Jagged1 and Notch1 transcripts (Fig. 5d and e). Conclusively, these results demonstrate that miR-199b acts as a negative regulator of the protective Notch signaling pathway during LV remodeling following MI.
Fig. 5.
miR-199b regulation of post-MI cardiac remodeling involves the Notch signaling pathway. (a) Sequence alignment of human miR-199b seed region and 3′UTRs of human and mouse notch1 and jagged1. (b,c) Real-time PCR analysis of transcript abundance of (b) notch1 and (c) jagged1 in WT or miR-199b transgenic (TG) murine hearts after 4 weeks of sham or MI. (d,e) Real-time PCR analysis of transcript abundance of (b) notch1 and (c) jagged1 in murine hearts subjected to sham or MI, and treated either with antagomir-control or antagomir-199b. n = 4–6 *P<0.05 (mean ± s.e.m.).
4. Discussion
Our study suggests that under volume overload conditions, miR-199b is still able to affect CnA/NFAT signaling by direct regulation of Dyrk1A expression but also interferes with the Notch pathway during pathological remodeling after myocardial infarction (MI), which is associated with elevated levels of fibrosis in the border zone. We detected activation of the CnA/NFAT signaling pathway in the LV of MI-subjected hearts by measuring transcript levels of calcineurin 1 isoform 4 (Rcan1-4), a sensitive target gene activated by CnA/NFAT and often used to evaluate signaling strength of this cascade [29]. We observed higher Rcan1-4 expression levels in hearts from mice receiving myocardial infarction, suggesting that CnA/NFAT signaling is indeed activated in the post-infarcted heart as observed previously by us [13]. In contrast, Dyrk1a transcript levels, a previously validated target of miR-199b, were not dysregulated in LV after MI, which in part may explain why we did not observe a further activation of pathological CnA/NFAT signaling in post-infarcted hearts from transgenic mice [5]. In line, the cardiac hypertrophic response induced by MI was also similar in both miR-199b transgenic and WT animals providing another piece of evidence for a seemingly equal activation of CnA/NFAT signaling regardless of miR-199b overexpression [30], [31], [32]. While miR-199b knockdown does not affect Dyrk1a transcript levels, we observed an increase in protein expression levels in hearts that received miR-199b antagomir treatment after undergoing a myocardial infarct. Nevertheless, antagomir treatment seems to have a milder effect on Dyrk1A expression when compared to our previous observations in pressure overloaded hearts [5]. All in all, our data implies that miR-199b in post-infarct remodeling may exert a slightly different function through regulation of different downstream targets compared to situations where pressure overload ensues maladaptive hypertrophy.
Target prediction databases and literature searches highlighted two additional targets of miR-199b, Notch1 and Jagged1, both previously associated with MI-induced cardiac remodeling. Previously, it was demonstrated that miR-199b is involved in chemotaxis resistance of ovarian cancer via targeting Jagged1 and thereby altering the Notch signaling pathway [26]. In meduloblastoma cells, miR-199b impairs the engrafting potential in the cerebellum of athymic/nude mice by influencing Hes1 expression, a transcription factor of the Notch pathway [27], [28]. Finally, miR-199b is also a key modulator of endothelial cell differentiation derived from induced pluripotent stem cells by targeting the Notch ligand jagged1 [33]. Evolutionarily conserved, the Notch pathway is an essential signaling cascade in the development of metazoans [34]. In mammals, signal transduction is initiated once one of the four Notch receptors (Notch1-4) interacts with membrane tethered ligands such as jagged (jagged 1 and 2) and the delta-like (Delta-like 1, 3 and 4) family [34]. Both receptors and ligands are transmembrane proteins involving the Notch pathway in the communication between two neighboring cells [35]. Upon ligand binding, activation of Notch receptor is followed by proteolytic cleavage by TACE (TNF-α converting enzyme) and the multicomponent γ-secretase complex to release the intercellular domain of Notch (NICD) and allowing translocate to the nucleus. Once in the nucleus, a transcription activator complex is formed with the DNA binding protein family CSL [CBF1/RBPJ-kappa/Su (H)/Lag1] [36], [37] inducing the transcription of Notch downstream target genes such as bHLH (basic helix-loop-helix) repressors of the Hairy/enhancer of split (Hes) family. The Hes family includes Hes-1 and Herp (Hairy-related transcription factors) also known as Hey [38], [39], [40].
While mutations in the Notch signaling pathway have been associated with human congenital heart defects including Alagille syndrome, bicuspid aortic valve disease, calcification of the heart valves, and ventricular septal defects [41], [42], [43], [44], and the essential role of Notch pathway during cardiac development has been experimentally established [45], the function of Notch in the adult heart is somewhat a matter of controversy [46]. Several of the aforementioned studies revealed induced Notch signaling activity in the myocardium either by direct injection of a Notch1-activating antibody in the border zone or by cardiac-specific overexpression of Jagged1, with both approaches leading to improved cardiac function following MI [20], [25]. In our study, we also observed increased Notch1 and Jagged1 expression after antagomir-199b treatment in the sham-operated animals suggesting that these two genes act downstream of miR-199b. Analysis of protein levels and target gene validation experiments are still necessary to establish these two genes as direct target genes of miR-199b involved in pathological cardiac remodeling post-MI.
In summary, we demonstrated that myocardial overexpression of miR-199b results in exaggerated pathological remodeling in a mouse model of MI, while in vivo pharmacological silencing of miR-199b using an antagomir approach had therapeutic functional effects. The observed effects may result from a synergism between the CnA/NFAT and Notch signaling pathways. Given that stress-induced miRNAs target a variety of genes, here we provided evidence that miRNAs may regulate different targets and act on different signaling pathways depending on the context of the stress. Previously, miR-199b was shown to exert its function via activation of the CnA/NFAT pathway in pressure overload-induced cardiac remodeling whereas upon volume overload we observe a milder effect on CnA/NFAT signaling but in addition, miR-199b also exerts its cardio-protective effects by influencing the protective Notch signaling pathway.
Author contributions
B.D., E.M.P., R.J., N.B., S.O. and P.D.C.M performed experiments. B.D., E.M.P., R.J., L.D.W. and P.D.C.M analyzed data. B.D., L.D.W. and P.D.C.M. designed the study. B.D., L.D.W. and P.D.C.M. wrote the manuscript.
Acknowledgements
We gratefully acknowledge members of the laboratory for technical support and helpful discussions. E.D. is supported by a VENI award 916-150-16 from the Netherlands Organisation for Health Research and Development. L.D.W. acknowledges support from the Netherlands CardioVascular Research Initiative: the Dutch Heart Foundation, Dutch Federation of University Medical Centers, ZonMW and the Royal Netherlands Academy of Sciences. L.D.W. was further supported by grant 311549 from the European Research Council and a VICI award 918-156-47 from NWO. P.D.C.M. is supported by a MEERVOUD grant from The Netherlands Organisation for Scientific Research (NWO) and is an Established Investigator of the Dutch Heart Foundation (2015T066).
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.ncrna.2016.12.002.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Mozaffarian D. Heart disease and stroke Statistics–2015 update: a report from the American heart association. Circulation. 2015;131 doi: 10.1161/CIR.0000000000000152. [DOI] [PubMed] [Google Scholar]
- 2.Maseri A., Chierchia S., Davies G. Pathophysiology of coronary occlusion in acute infarction. Circulation. 1986;73:233–239. doi: 10.1161/01.cir.73.2.233. [DOI] [PubMed] [Google Scholar]
- 3.French B.A., Kramer C.M. Mechanisms of postinfarct left ventricular remodeling. Drug Discov. Today Dis. Mech. 2007;4:185–196. doi: 10.1016/j.ddmec.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaudron P. Time course of cardiac structural, functional and electrical changes in asymptomatic patients after myocardial infarction: their inter-relation and prognostic impact. J. Am. Coll. Cardiol. 2001;38:33–40. doi: 10.1016/s0735-1097(01)01319-5. [DOI] [PubMed] [Google Scholar]
- 5.da Costa Martins P.A. MicroRNA-199b targets the nuclear kinase Dyrk1a in an auto-amplification loop promoting calcineurin/NFAT signalling. Nat. Cell Biol. 2010;12:1220–1227. doi: 10.1038/ncb2126. [DOI] [PubMed] [Google Scholar]
- 6.Ren X.P. MicroRNA-320 is involved in the regulation of cardiac ischemia/reperfusion injury by targeting heat-shock protein 20. Circulation. 2009;119:2357–2366. doi: 10.1161/CIRCULATIONAHA.108.814145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hinkel R. Inhibition of microRNA-92a protects against ischemia/reperfusion injury in a large-animal model. Circulation. 2013;128:1066–1075. doi: 10.1161/CIRCULATIONAHA.113.001904. [DOI] [PubMed] [Google Scholar]
- 8.Meloni M. Local inhibition of microRNA-24 improves reparative angiogenesis and left ventricle remodeling and function in mice with myocardial infarction. Mol. Ther. 2013;21:1390–1402. doi: 10.1038/mt.2013.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fiedler J. MicroRNA-24 regulates vascularity after myocardial infarction. Circulation. 2011;124:720–730. doi: 10.1161/CIRCULATIONAHA.111.039008. [DOI] [PubMed] [Google Scholar]
- 10.Hu S. MicroRNA-210 as a novel therapy for treatment of ischemic heart disease. Circulation. 2010;122 doi: 10.1161/CIRCULATIONAHA.109.928424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 13.van Rooij E. MCIP1 overexpression suppresses left ventricular remodeling and sustains cardiac function after myocardial infarction. Circ. Res. 2004;94:e18–26. doi: 10.1161/01.RES.0000118597.54416.00. [DOI] [PubMed] [Google Scholar]
- 14.Krützfeldt J. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 15.Dirkx E. Nfat and miR-25 cooperate to reactivate the transcription factor Hand2 in heart failure. Nat. Cell Biol. 2013;15:1282–1293. doi: 10.1038/ncb2866. [DOI] [PubMed] [Google Scholar]
- 16.Takagawa J. Myocardial infarct size measurement in the mouse chronic infarction model: comparison of area- and length-based approaches. J. Appl. Physiol. 2007;102:2104–2111. doi: 10.1152/japplphysiol.00033.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfeffer M.A., Braunwald E. Ventricular remodeling after myocardial infarction. Experimental observations and clinical implications. Circulation. 1990;81:1161–1172. doi: 10.1161/01.cir.81.4.1161. [DOI] [PubMed] [Google Scholar]
- 18.Ørn S. Effect of left ventricular scar size, location, and transmurality on left ventricular remodeling with healed myocardial infarction. Am. J. Cardiol. 2007;99:1109–1114. doi: 10.1016/j.amjcard.2006.11.059. [DOI] [PubMed] [Google Scholar]
- 19.Croquelois A. Control of the adaptive response of the heart to stress via the Notch1 receptor pathway. J. Exp. Med. 2008;205:3173–3185. doi: 10.1084/jem.20081427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kratsios P. Distinct roles for cell-autonomous notch signaling in cardiomyocytes of the embryonic and adult heart. Circ. Res. 2010;106:559–572. doi: 10.1161/CIRCRESAHA.109.203034. [DOI] [PubMed] [Google Scholar]
- 21.Øie E. Activation of Notch signaling in cardiomyocytes during post-infarction remodeling. Scand. Cardiovasc. J. 2010;44:359–366. doi: 10.3109/14017431.2010.511256. [DOI] [PubMed] [Google Scholar]
- 22.Boopathy A.V. Intramyocardial delivery of notch ligand-containing hydrogels improves cardiac function and angiogenesis following infarction. Tissue Eng. - Part A. 2015;21:2315–2322. doi: 10.1089/ten.tea.2014.0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y., Hiroi Y., Liao J.K. Notch signaling as an important mediator of cardiac repair and regeneration after myocardial infarction. Trends Cardiovasc. Med. 2010;20:228–231. doi: 10.1016/j.tcm.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gude N.A. Activation of Notch-mediated protective signaling in the myocardium. Circ. Res. 2008;102:1025–1035. doi: 10.1161/CIRCRESAHA.107.164749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nemir M. The Notch pathway controls fibrotic and regenerative repair in the adult heart. Eur. Heart J. 2014;35:2174–2185. doi: 10.1093/eurheartj/ehs269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu M.X. Epigenetic silencing of microRNA-199b-5p is associated with acquired chemoresistance via activation of JAG1-Notch1 signaling in ovarian cancer. Oncotarget. 2014;5:944–958. doi: 10.18632/oncotarget.1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garzia L. MicroRNA-199b-5p impairs cancer stem cells through negative regulation of HES1 in medulloblastoma. PLoS One. 2009;4 doi: 10.1371/journal.pone.0004998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andolfo I. The micro-RNA 199b-5p regulatory circuit involves Hes1, CD15, and epigenetic modifications in medulloblastoma. Neuro. Oncol. 2012;14:596–612. doi: 10.1093/neuonc/nos002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J. Independent signals control expression of the calcineurin inhibitory proteins MCIP1 and MCIP2 in striated muscles. Circ. Res. 2000;87:E61–E68. doi: 10.1161/01.res.87.12.e61. [DOI] [PubMed] [Google Scholar]
- 30.Wilkins B.J. Calcineurin/NFAT coupling participates in pathological, but not physiological, cardiac hypertrophy. Circ. Res. 2004;94:110–118. doi: 10.1161/01.RES.0000109415.17511.18. [DOI] [PubMed] [Google Scholar]
- 31.Heineke J., Molkentin J.D. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat. Rev. Mol. Cell Biol. 2006;7:589–600. doi: 10.1038/nrm1983. [DOI] [PubMed] [Google Scholar]
- 32.Molkentin J.D. Parsing good versus bad signaling pathways in the heart role of calcineurin-nuclear factor of activated t-cells. Circulation Res. 2013;113:16–19. doi: 10.1161/CIRCRESAHA.113.301667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen T. MicroRNA-199b modulates vascular cell fate during ips cell differentiation by targeting the notch ligand jagged1 and enhancing VEGF signaling. Stem Cells. 2015;33:1405–1418. doi: 10.1002/stem.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andersson E.R., Sandberg R., Lendahl U. Notch signaling: simplicity in design, versatility in function. Development. 2011;138:3593–3612. doi: 10.1242/dev.063610. [DOI] [PubMed] [Google Scholar]
- 35.Lai E.C. Notch signaling: control of cell communication and cell fate. Development. 2004;131:965–973. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- 36.Blaumueller C.M., Qi H., Zagouras P., Artavanis-Tsakonas S. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell. 1997;90:281–291. doi: 10.1016/s0092-8674(00)80336-0. [DOI] [PubMed] [Google Scholar]
- 37.Lieber T., Kidd S., Young M.W. Kuzbanian-mediated cleavage of Drosophila notch. Genes Dev. 2002;16:209–221. doi: 10.1101/gad.942302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishimura M. Structure, chromosomal locus, and promoter of mouse Hes2 gene, a homologue of Drosophila hairy and Enhancer of split. Genomics. 1998;49:69–75. doi: 10.1006/geno.1998.5213. [DOI] [PubMed] [Google Scholar]
- 39.Maier M.M., Gessler M. Comparative analysis of the human and mouse Hey1 promoter: Hey genes are new Notch target genes. Biochem. Biophys. Res. Commun. 2000;275:652–660. doi: 10.1006/bbrc.2000.3354. [DOI] [PubMed] [Google Scholar]
- 40.Kovall R.a. More complicated than it looks: assembly of Notch pathway transcription complexes. Oncogene. 2008;27:5099–5109. doi: 10.1038/onc.2008.223. [DOI] [PubMed] [Google Scholar]
- 41.Oda T. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat. Genet. 1997;16:235–242. doi: 10.1038/ng0797-235. [DOI] [PubMed] [Google Scholar]
- 42.Kamath B.M. NOTCH2 mutations in Alagille syndrome. J. Med. Genet. 2012;49:138–144. doi: 10.1136/jmedgenet-2011-100544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garg V. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- 44.Donovan J., Kordylewska A., Jan Y.N., Utset M.F. Tetralogy of fallot and other congenital heart defects in Hey2 mutant mice. Curr. Biol. 2002;12:1605–1610. doi: 10.1016/s0960-9822(02)01149-1. [DOI] [PubMed] [Google Scholar]
- 45.Niessen K., Karsan A. Notch signaling in cardiac development. Circulation Res. 2008;102:1169–1181. doi: 10.1161/CIRCRESAHA.108.174318. [DOI] [PubMed] [Google Scholar]
- 46.Felician G. Epigenetic modification at Notch responsive promoters blunts efficacy of inducing Notch pathway reactivation after myocardial infarction. Circ. Res. 2014;115:636–649. doi: 10.1161/CIRCRESAHA.115.304517. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.