Abstract
The long non-coding RNA CRNDE is an oncogene that promotes tumor growth in glioblastoma multiforme (GBM). At least five CRNDE transcript variants with possibly different functional roles have been described in recent studies. Here, we report our preliminary findings on the differential expressions of CRNDE transcript variants in GBM, and their prognostic significance. Our preliminary data suggest that different transcript variants of CRNDE might have different functions in GBM and should be further studied as potential biomarkers for clinical prognostication.
Keywords: Long non-coding RNA, CRNDE, Transcript variant, Glioblastoma
1. Introduction
Glioblastoma multiforme (GBM) is classified as the most malignant type of brain cancer characterized by its highly proliferative and invasive behavior. Patients with GBM have poor clinical outcomes with a median survival of 14.6 months and 30% two-year survival following standard treatment. A host of factors account for its poor prognosis including heterogeneity of cell types, differential responses to therapies as well as the development of chemoresistance [1]. The treatment regimen for GBM is multimodal in nature that includes surgical resection followed by concurrent chemo-irradiation and chemotherapy with temozolomide [2], [3].
Long non-coding RNAs (lncRNAs) have recently emerged as important regulators in normal biological processes and disease pathogenesis. Colorectal neoplasia differentially expressed (CRNDE), one of the most upregulated lncRNAs in GBM [4], has been identified to be an oncogene closely associated with malignant phenotypes in glioma [5], [6], [7]. It is noteworthy that CRNDE has different transcript variants resulting from alternative splicing [8]. These transcript isoforms may differ in expression levels and functions in glioma [6], [9]. Here, we investigate the differential expression of CRNDE transcript isoforms and provide preliminary evidence on how they may be further exploited as prognostic markers in GBM.
2. Materials and methods
2.1. Clinical specimens
A total of 13 GBM specimens (mean age: 53.92 ± 16.05) and normal brain tissues were retrieved from our institutional tissue bank. The diagnosis of GBM was confirmed by a certified pathologist and classified according to the World Health Organization (WHO) brain tumor classification system [10]. The study protocol was approved by the Institutional Review Board of our institution, with signed informed consent obtained from the patients or their legal guardians before sample collection.
2.2. Cell lines
Human GBM cell lines U87-MG and U138-MG were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). U87-MG and U138-MG were cultured in Minimum essential medium (MEMα) (GIBCO, Invitrogen, Carlsbad, CA, USA) and Dulbecco's modified eagle's medium with high glucose (DMEM GlutaMAX) (GIBCO), respectively. Media were supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 IU/mL penicillin, and 100 μg/mL streptomycin (all from GIBCO). Primary normal human astrocytes (NHA; Lonza, Basel, Switzerland) were used as non-transformed controls and cultured in DMEM GlutaMAX (GIBCO). Cells were cultured at 37 °C in 5% CO2 and 90% relative humidity.
2.3. Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted using Trizol reagent (Invitrogen) and reverse transcribed into cDNA using the reverse transcription kit (Takara Bio, Shiga, Japan). cDNAs were then mixed with gene-specific primers (TV-1: NR_034105.3, TV-2: NR_034106.2, TV-3: NR_110453.1, TV-4: NR_110454.1 and TV-5: NM_001308963.1) and SYBR reagent (Takara), and amplified in an ABI Prism 7900HT Fast Real-Time PCR System. GAPDH was used as an internal control for PCR quality. mRNA expression levels were analyzed using the 2-ΔΔ Ct calculation method.
2.4. Survival and statistical analysis
For survival analysis, patients were first stratified into low and high expression groups by using the median expression value of TV-1 or TV-5 as the cut-off. Kaplan Meier estimation was used to examine overall survival, and a log-rank test was used to assess the difference between the two groups. Data analysis was performed using GraphPad Prism 7.0 (La Jolla, CA, USA). Continuous data were presented as mean ± SD. The p values were calculated by One-way ANOVA analysis for comparison of more than 2 groups. P values of equal or less than 0.05 were considered statistically significant.
3. Results and discussion
3.1. Differential expression and clinical significance of CRNDE transcript variants
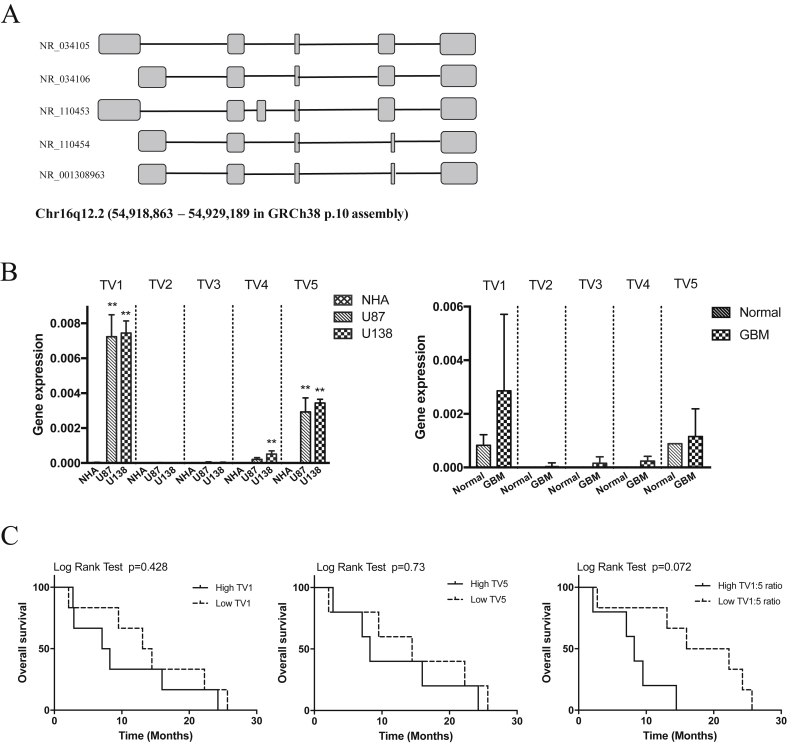
According to National Center for Biotechnology Information (NCBI), five transcript variants (TV1-5) of CRNDE have been reported. These variants are generated through alternative splicing, and consist of different numbers of exons as illustrated in Fig. 1A. In a preliminary survey of GBM cell lines and specimens, the expressions of both TV-1 and TV-5 were highly upregulated as compared to normal astrocytes; TV-2, TV-3 and TV-4 were expressed only in trace amounts both in normal and malignant brain tissues (Fig. 1B). To further evaluate the clinical significance of these transcripts in GBM, we performed Kaplan Meier survival analysis based on TV-1 and TV-5 expression among the GBM patient specimens. Interestingly, while the expression levels of TV-1 and TV-5 alone did not correlate with survival, a trend can be observed between clinical survival and TV-1 to TV-5 ratio (TV-1:TV-5) on Kaplan Meier analysis (Fig. 1C). Though the p value in log rank test did not reach statistical significance (p = 0.072), the findings suggest a potential clinical utility of CRNDE transcripts expression level in predicting patient survival.
Fig. 1.
Differential expression of CRNDE transcript variants (TV) and its clinical significance in glioblastoma multiforme. A) CRNDE is located at Chr 16q12.2 and presented with different TV. Corresponding exons are illustrated in boxes with different length. NCBI reference (TV-1: NR_034105.3, TV-2: NR_034106.2, TV-3: NR_110453.1, TV-4: NR_110454.1, TV-5: NM_001308963.1). B) Differential mRNA expressions of TV1-TV5 were detected by qRT-PCR in GBM cell lines (U87 and U138) vs. normal human astrocytes (NHA) and in GBM specimens vs. normal brain tissues. Graphs show the mean ± SD of three independent experiments. **p < 0.001 C) Kaplan Meier survival curve on TV1 and TV5 expression of GBM specimens. Patients were stratified by median expression of TV1 and TV5 into two groups (High or Low).
This finding also suggests different functional roles among the CRNDE transcripts. In our preliminary results, a low TV-1:TV-5 ratio was associated with a more favorable disease outcome, implying that the oncogenic activities of TV-1 and TV-5 may in fact oppose one another. Indeed, tumor-specific splice variants have been discovered in some other cancers [11], and individual variants may serve as highly specific “molecular signatures” [12]. The characterization of these tumor-specific variants may also allow for high specificity targeting therapies on cancerous tissues with only minimal impact on normal tissues. In that regard, antisense therapies that aimed to modify splicing patterns have been shown to yield promising results [11], [12].
Different functional roles of the two variants of CRNDE have been described. TV-1, also known as CRNDE-h, is the reference variant with full-length sequence. Strikingly, it was found that this variant could be detected in plasma with elevated expressions in colorectal cancer patients, suggesting that TV-1 isoform may be a promising biomarker [8], [13]. An open reading frame (ORF) has recently been identified on TV-5 which is able to encode for a nuclear peptide (CRNDEP). The latter may have functional roles in the formation of stress granule and the regulation of cell proliferation [9].
4. Conclusion
Our study provides preliminary findings that direct future investigations into specific transcript variants of CRNDE. Researches that focus on the identification of tumor-specific variants are likely to become increasingly important for the development of biomarkers and effective therapeutic strategies.
Conflicts of interest
All authors declare no conflicts of interest.
References
- 1.Ostrom Q.T. American brain tumor association adolescent and young adult primary brain and central nervous system tumors diagnosed in the United States in 2008-2012. Neuro Oncol. 2016;18(Suppl 1):i1–i50. doi: 10.1093/neuonc/nov297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stupp R. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Stupp R. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 4.Zhang X. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol. Dis. 2012;48(1):1–8. doi: 10.1016/j.nbd.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Kiang K.M. CRNDE expression positively correlates with EGFR activation and modulates glioma cell growth. Target Oncol. 2017;12(3):353–363. doi: 10.1007/s11523-017-0488-3. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y. CRNDE, a long-noncoding RNA, promotes glioma cell growth and invasion through mTOR signaling. Cancer Lett. 2015;367(2):122–128. doi: 10.1016/j.canlet.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 7.Zheng J. CRNDE affects the malignant biological characteristics of human glioma stem cells by negatively regulating miR-186. Oncotarget. 2015;6(28):25339–25355. doi: 10.18632/oncotarget.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graham L.D. Colorectal neoplasia differentially expressed (CRNDE), a novel gene with elevated expression in colorectal adenomas and adenocarcinomas. Genes Cancer. 2011;2(8):829–840. doi: 10.1177/1947601911431081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szafron L.M. The novel gene CRNDE encodes a nuclear peptide (CRNDEP) which is overexpressed in highly proliferating tissues. PLoS One. 2015;10(5):e0127475. doi: 10.1371/journal.pone.0127475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Louis D.N. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blair C.A., Zi X. Potential molecular targeting of splice variants for cancer treatment. Indian J. Exp. Biol. 2011;49(11):836–839. [PMC free article] [PubMed] [Google Scholar]
- 12.Li H.R. Two-dimensional transcriptome profiling: identification of messenger RNA isoform signatures in prostate cancer from archived paraffin-embedded cancer specimens. Cancer Res. 2006;66(8):4079–4088. doi: 10.1158/0008-5472.CAN-05-4264. [DOI] [PubMed] [Google Scholar]
- 13.Liu T. Increased expression of the long noncoding RNA CRNDE-h indicates a poor prognosis in colorectal cancer, and is positively correlated with IRX5 mRNA expression. Onco Targets Ther. 2016;9:1437–1448. doi: 10.2147/OTT.S98268. [DOI] [PMC free article] [PubMed] [Google Scholar]