Abstract
The 94-nt full-length Y4-RNA is thought to have roles in the initiation of DNA replication and RNA quality control. Although its 31/32-nt fragment also exists abundantly in plasma, little is known about its physiological role. Since the 31/32-nt Y4-RNA fragment in sera is reported to be more abundant in patients with coronary artery disease than healthy persons, the fragment may have a potential for a diagnostic and/or prognostic biomarker for some diseases regardless of its functionality. As a step toward further investigation of its potential utility, we examined if the 31/32-nt Y4-RNA fragment also exists in saliva that can be obtained noninvasively, and showed that, in addition to the 31/32-nt fragment, 14- and 11-nt Y4-RNA fragments are present in all saliva RNA samples from four healthy persons. We established a PCR method to accurately quantitate the amount of the 31/32-nt Y4-RNA fragment, and estimated its amount in saliva of healthy persons to be 0.06 ± 0.04 fmol per nanogram of saliva RNA. We also tried to develop an easier quantitation method using a DNA molecular beacon.
Keywords: Y4-RNA fragment, Saliva RNA, Diagnostic/prognostic marker, Next-generation sequencing, RT-PCR, Molecular beacon
1. Introduction
Cellular RNA is generated first as a primary transcript of DNA, which is then processed in various steps such as RNA chain shortenings and nucleotide modifications to produce functional RNA molecules. These functional molecules are finally degraded into small pieces with no function. Homogeneous RNA molecules transcribed from a single gene would be processed to generate various functional molecules with a different length and/or different nucleotide modifications depending on cell types and environments. In some cases, apparently degraded RNA fragments can be functional [1], [2].
One of the founding examples is the fragments from mammalian tRNAs. Mature tRNA molecules that are used for translation are ∼80–90 nucleotides (nt), whereas ∼67-nt 3′-truncated tRNAs and ∼35-nt 5′-half-tRNAs also exist in cells [3], [4], [5], [6], [7], [8]. We have shown that these apparent tRNA degradation products can work as small guide RNAs (sgRNAs) for tRNase ZL to cleave target RNAs [4], [9]. The functionality of 5′-half-tRNAs has been corroborated by the efficacy of tRNase ZL-utilizing efficacious (TRUE) gene silencing with the aid of artificially designed 5′-half-tRNAs [10], [11], [12], [13], [14].
The 31-nt fragment from Y4-RNA could be another example. The 94-nt full-length Y4-RNA is thought to have roles in the initiation of DNA replication and RNA quality control [15], [16]. Although the 31-nt fragment exists abundantly in plasma and little is known about its physiological role, interestingly, it can form a 5′-half-tRNA-like structure and function as an sgRNA for tRNase ZL [7]. And we have shown that two 31-nt Y4-RNA-related fragments with a C-to-A substitution at the 31st or 26th base, which may be generated from Y4-RNA pseudogenes, exist significantly more abundantly in plasma from multiple myeloma patients than in that from healthy persons [7]. Furthermore, one group has reported that the ∼31-nt Y4-RNA fragment in sera is more abundant in coronary artery disease patients than controls [17]. And similarly, another group has demonstrated that the level of the ∼31-nt plasma Y4-RNA fragment correlates with platelet function in patients with acute coronary syndrome and platelet activation markers in a general population [18].
From these observations, the 31-nt Y4-RNA fragment appears to have a potential for a diagnostic and/or prognostic biomarker regardless of its functionality. As a step toward further investigation of its potential utility, in this study, we examined if the 31-nt Y4-RNA fragment also exists in saliva that can be obtained noninvasively, and established a PCR method to accurately quantitate the amount of the 31-nt Y4-RNA fragment in saliva. We also tried to develop an easier quantitation method using a DNA molecular beacon.
2. Materials and methods
2.1. Preparation of saliva RNA
Naturally secreted saliva (5 ml) was obtained from four healthy persons, and filtrated with a sterile syringe filter with a 0.45-μm pore size membrane to remove cells. In a subset of experiments, a meal and aspirin tablets (660 mg) were taken ∼90 min before saliva collection. Total RNA was extracted from the saliva filtrate with RNAiso Blood (Takara Bio, Otsu, Japan) according to manufacturer's instructions. The extracted RNA was quantitated first with a spectrophotometer, NanoDrop 2000c (Thermo Fisher Scientific, Massachusetts, USA) and then with the fluorescent dye RiboGreen and a Qubit 3.0 Fluorometer (Life Technologies, Carlsbad, USA) according to manufacturer's protocol. Written informed consent was obtained from each person, and this study was approved by the ethics committee of Niigata University of Pharmacy and Applied Life Sciences (Permit Number: H28-001).
2.2. cDNA synthesis and deep sequencing
A cDNA library was constructed from each of the four saliva RNA samples using a TruSeq Small RNA Sample Prep Kit-Set A (Illumina, California, USA), T4 RNA Ligase 2, truncated (New England Biolabs, Massachusetts, USA), and SuperScript II Reverse Transcriptase (Invitrogen, Massachusetts, USA) by 14 cycles of PCR according to manufacturers' protocols. A mixture of the four cDNA libraries was separated by polyacrylamide gel electrophoresis, and ∼120–170-base-pair cDNAs, which correspond to ∼5–55-nt original RNAs, were recovered and purified. The purified cDNA sample was subjected to the next-generation sequencer MiSeq (Illumina) using a MiSeq Reagent Kit v3 (150 Cycles) and a PhiX Control Kit v3.
2.3. Synthetic RNA/DNA
The following RNA and DNA were chemically synthesized by Nippon Bioservice (Saitama, Japan): the 31-nt Y4-RNA fragment with a 5′-phosphate without and with full 2′-O-methyl modifications, 5′-pGGCUGGUCCGAUGGUAGUGGGUUAUCAGAAC-3′; Beacon-1, 5′-(6-carboxyfluorescein)-CGACGACTACCATCGGACCACGTCG-(dabclyl)-3′; Beacon-2, 5′-(6-carboxyfluorescein)-CGACGACCATCGGACCAGCCCGTCG-(dabclyl)-3′; Target-1, 5′-TGGTCCGATGGTAGT-3′; Target-2, 5′-GGCTGGTCCGATGGT-3′; Target-3, 5′-AGGTAGTAGGTTGTG-3′; Target-4, 5′-AATGTAGATAAGGGA-3′; Target-5, 5′-GGGGCGGATTAGTAG-3′.
The following DNA primers were obtained from Sigma-Aldrich (Tokyo, Japan): SL-forward_Y4_3, 5′-GGCGGCTGGTCCGATGGT-3′; Y4RNA_R4, 3′-CCCAATAGTCTTGGCG-5′; Y4RNA_R5, 3′-TTTAAACTGACCGGCG-5′.
2.4. Reverse transcription (RT) PCR
An RNA test sample was reverse transcribed using a reverse primer, Y4RNA_R4 or Y4RNA_R5, with a PrimeScriptTM RT reagent Kit with gDNA Eraser (Perfect Real Time) (Takara Bio, Otsu, Japan) according to manufacturer's protocol. Subsequently, a real time PCR for the obtained cDNA was conducted using the primer pair SL-forward_Y4_3/Y4RNA_R4 or SL-forward_Y4_3/Y4RNA_R5 and SYBR Premix Ex Taq II (Tli RNaseH Plus) (Takara Bio, Otsu, Japan) with a Thermal Cycler Dice Real Time System (Takara Bio, Otsu, Japan) under the standard conditions according to manufacturer's protocol. We confirmed that the PCR product obtained with the primer pair SL-forward_Y4_3/Y4RNA_R4 contains the AGT sequence of Y4-RNA that is not overlapped with the primer pair sequences by Sanger sequencing of a further extended PCR product.
The statistical significance of differences between groups was evaluated by the two-tailed Student's t-test. The differences with a P value < 0.05 were considered as statistically significant.
2.5. Fluorometric quantitation with molecular beacons
An RNA/DNA test sample was incubated with a DNA molecular beacon [19] in a 200-μl solution containing 10 mM Tris-HCl (pH 7.5), 100 mM NaCl, and 10 mM MgCl2 at room temperature for 5–30 min, and a fluorescence intensity was measured with the Qubit 3.0 Fluorometer.
3. Results and discussion
3.1. The 31-nt Y4-RNA fragment exists in saliva
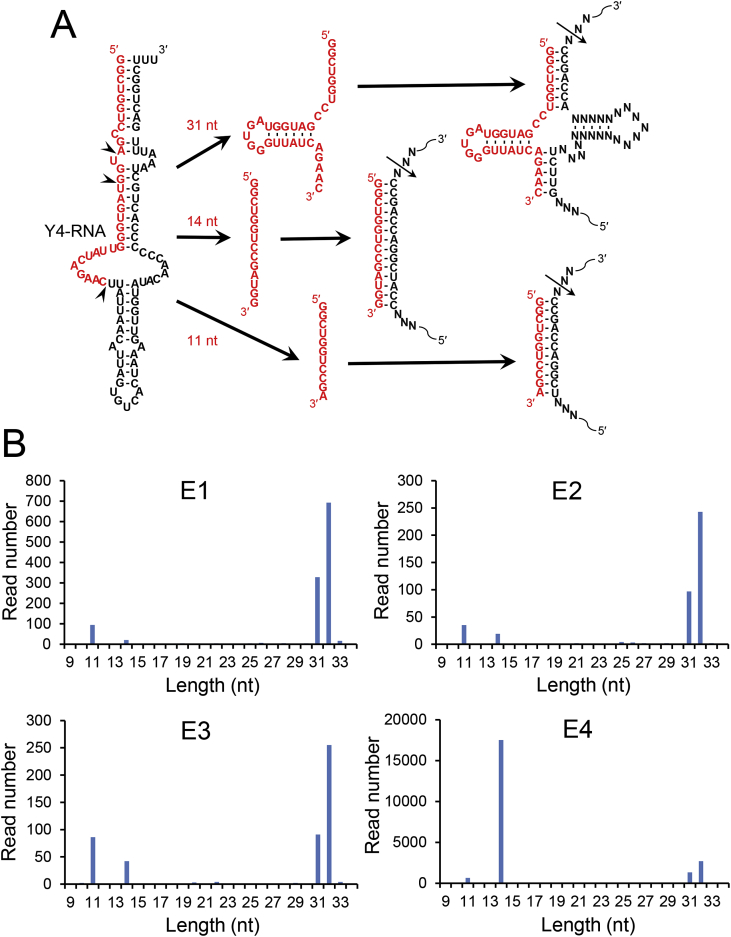
To see if the 31-nt Y4-RNA fragment also exists in saliva, we extracted total RNAs from filtrated saliva of four healthy persons (E1–E4) and analyzed their sequences with a next-generation sequencer. Age and gender of the examinees were as follows: E1, 57 male; E2, 48 male; E3, 26 female; E4, 24 male. Frequencies of 9- to 34-nt Y4-RNA-related sequences are graphed in Fig. 1B. We detected the 31-nt Y4-RNA fragment in every RNA sample and also found that 32-, 14-, and 11-nt Y4-RNA fragments are present in all saliva RNA samples (Fig. 1B).
Fig. 1.
Deep sequencing analysis of saliva RNA for Y4-RNA fragments. (A) Human full-length Y4-RNA and its fragments. Arrowheads on the 94-nt Y4-RNA denote the cleavage sites to generate 31-, 14-, and 11-nt fragments, which can function as 5′-half-tRNA-type, linear-type, and linear-type sgRNAs, respectively, for tRNase ZL. The expected cleavage sites by tRNase ZL are indicated by thin arrows in model target RNAs. (B) Frequencies of Y4-RNA fragments in saliva from four healthy persons, E1–E4. Read numbers of the 9–34-nt fragments are presented.
The 32-nt Y4-RNA fragment was 2–3-fold as abundant as the 31-nt fragment, and the read numbers of the 11-nt fragment were 29–95% of those of the 31-nt fragment. The read numbers of the 14-nt Y4-RNA fragment were 6–46% of those of the 31-nt fragment with the exception that the 14-nt fragment was 13-fold as abundant as the 31-nt fragment in the examinee E4. This exceptionally high abundance of the 14-nt Y4-RNA fragment in E4 is interesting and may imply that the ratio of the 14-nt to 31-nt fragment levels can be a diagnostic and/or prognostic marker for some diseases.
Although we do not know how these Y4-RNA fragments are generated from the primary Y4-RNA transcript, they would be able to work as sgRNA for tRNase ZL to cleave appropriate target RNA: the 31- and 32-nt fragments as 5′-half-tRNA-type sgRNA, and the 11- and 14-nt fragments as linear-type sgRNA (Fig. 1A). And these fragments may be functioning as signaling molecules between cells.
The 32-nt Y4-RNA fragment was more abundant than the 31-nt fragment in this analysis and another analysis for plasma/sera Y4-RNA fragments [20], whereas the 31-nt fragment was much more abundant in our previous study about the plasma Y4-RNA fragments [7]. This difference may be due to the difference in the procedure for cDNA construction. In any case, we call these two fragments collectively the 31/32-nt Y4-RNA fragment.
The difference in the total read number of the Y4-RNA fragments among the four examinees (Fig. 1B) appeared to reflect the difference in the saliva RNA amount used for cDNA construction. This was due to low accuracy of the extracted RNA concentrations, which were estimated by measuring OD260 values with a spectrophotometer. In the following experiments, we used more accurate values of the RNA concentrations re-evaluated using a fluorescent dye, RiboGreen.
3.2. Quantitation of the 31/32-nt Y4-RNA fragment in saliva by RT-PCR
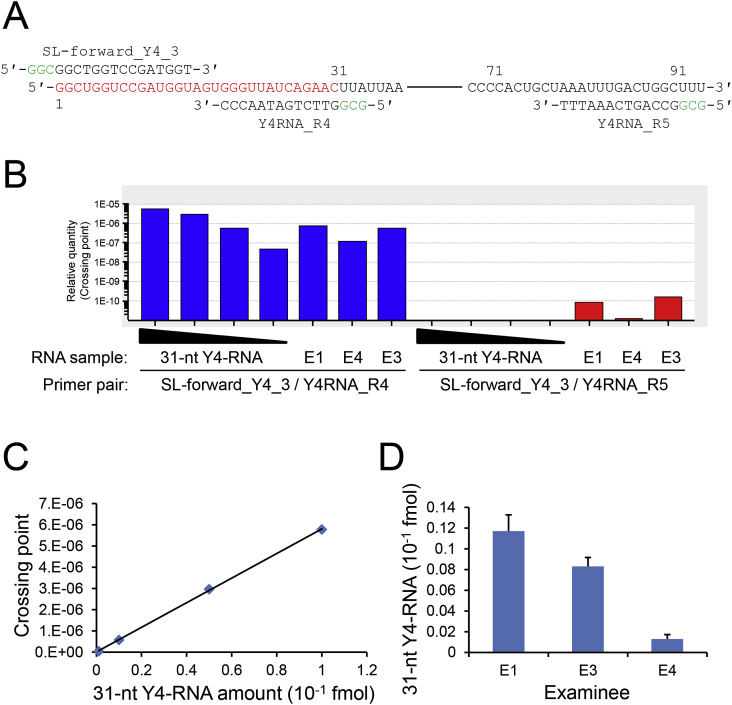
To accurately quantitate the amount of the 31/32-nt Y4-RNA fragment in saliva, we designed a primer pair for the 31/32-nt Y4-RNA fragment (Fig. 2A) and performed RT-PCR. In case that the 94-nt full-lengthY4-RNA co-exists in saliva RNA samples and contributes to overestimation, we also carried out RT-PCR for it. Although the full-lengthY4-RNA was detected in all saliva RNA samples examined, its amount was negligible compared with that of the 31/32-nt Y4-RNA fragment in each examinee (Fig. 2B). The amount of the 31/32-nt Y4-RNA fragment in saliva total RNA of the examinees E1, E3, and E4 was quantitated using the synthetic 31-nt Y4-RNA fragment as a standard (Fig. 2C). Its amount ranged from 0.0013 to 0.012 fmol/ng (Fig. 2D). Although there was another possibility that a 32-nt similar sequence with two base substitutions in the 3′-UTR of the ATP5G3 mRNA (GenBank accession number: NM_001689) might contribute to overestimation, the sequence was not detected in saliva RNA samples by either next-generation sequencing or RT-PCR (data not shown).
Fig. 2.
Quantitative RT-PCR. (A) Primer pairs. The sequences of the primers SL-forward_Y4_3 and Y4RNA_R4 for amplification of the 31/32-nt Y4-RNA fragment and the sequences of the primers SL-forward_Y4_3 and Y4RNA_R5 for amplification of the full-length Y4-RNA are presented together with the sequence of Y4-RNA, a middle part of which is omitted. Three nucleotides are added to the 5′ terminus of each primer to stabilize the primer/template hybrid. (B) A representative RT-PCR data for the synthetic 31-nt Y4-RNA fragment (0.001, 0.01, 0.05, and 0.1 fmol) and saliva RNA (1 ng) from the examinees E1, E3, and E4. The right and left halves represent the data for the full-length Y4-RNA and the 31/32-nt fragment, respectively. (C) A standard curve was drawn using the RT-PCR data for the synthetic 31-nt Y4-RNA fragment. (D) Quantitation of the 31/32-nt Y4-RNA fragment in saliva RNA from the examinees E1, E3, and E4. Error bars denote standard deviations (n = 3).
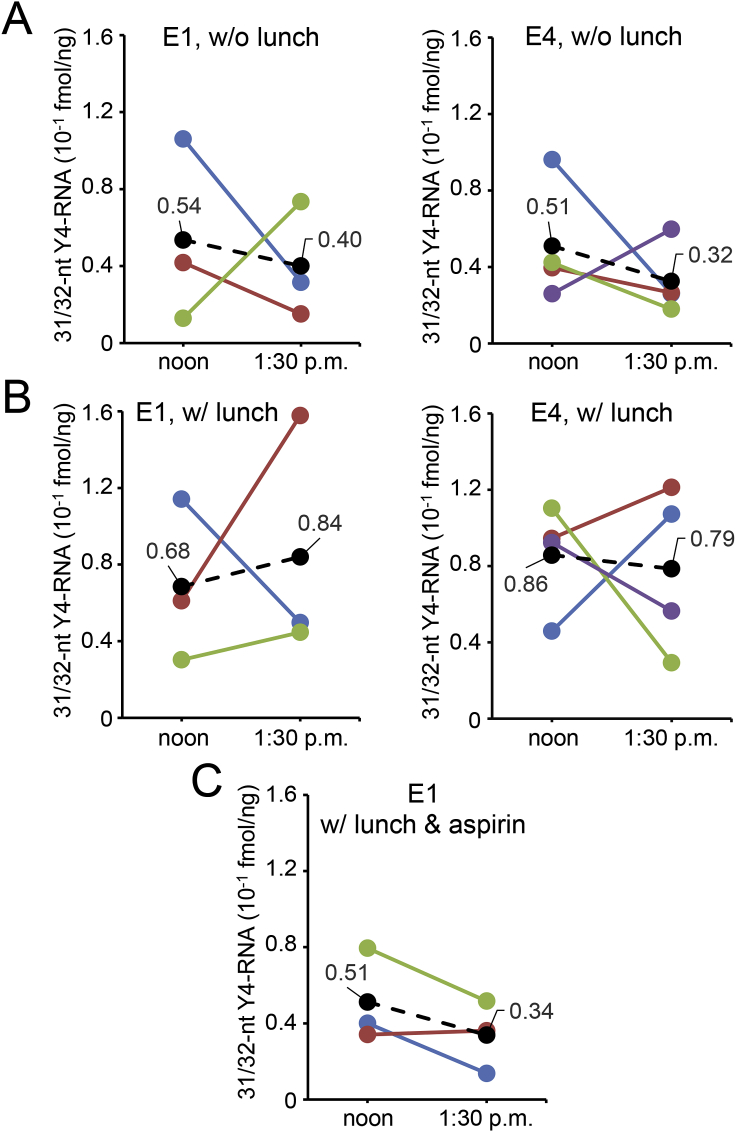
Further, we examined daily variations in the amount of the 31/32-nt Y4-RNA fragment in saliva. Saliva samples were collected from the examinees E1 and E4 on 3 and 4 different days, respectively, at noon and 1:30 p.m. without having lunch. As shown in Fig. 3A, the saliva Y4-RNA fragment level varied daily and within 90 min, and its means and standard deviations were 0.047 ± 0.036 fmol/ng (E1) and 0.042 ± 0.026 fmol/ng (E4). Next, we examined if taking a meal affects the amount of the 31/32-nt Y4-RNA fragment in saliva. Although its mean value increased for E1 and decreased for E4 after lunch, the differences were not statistically significant (Fig. 3B), suggesting that taking a meal does not affect the saliva Y4-RNA fragment level meaningfully. We also examined how aspirin affects the Y4-RNA fragment level in saliva, and found that the level decreased from 0.051 to 0.034 fmol/ng with no statistical significance (Fig. 3C).
Fig. 3.
Quantitation of the 31/32-nt saliva Y4-RNA fragment by RT-PCR. The values connected to a black dot denote mean values, and each of the other dots represents a value of one of the independent saliva RNA samples. (A, B) Saliva was collected at noon and 1:30 p.m. from the examinees E1 and E4 on 3 and 4 different days, respectively, and each saliva RNA (0.1 ng) was subjected to RT-PCR. A meal was not taken between noon and 1:30 p.m. in (A), and a meal was taken soon after saliva collection at noon in (B). (C) Saliva was collected at noon and 1:30 p.m. from E1 on 3 different days, and each saliva RNA (0.1 ng) was subjected to RT-PCR. A meal was taken soon after saliva collection at noon, and aspirin (660 mg) was taken soon after having the meal.
From all the data presented in Fig. 3, the means and standard deviations for E1 and E4 were calculated to be 0.055 ± 0.039 and 0.062 ± 0.036 fmol/ng, respectively, and, thus, the 31/32-nt Y4-RNA fragment level in saliva of healthy persons could be estimated to be 0.06 ± 0.04 fmol/ng. The very low level (0.0013 fmol/ng) of the saliva Y4-RNA fragment shown by the examinee E4 on one day (Fig. 2D) might have reflected some abnormal health condition.
3.3. Easy quantitation of Y4-RNA fragments with a molecular beacon
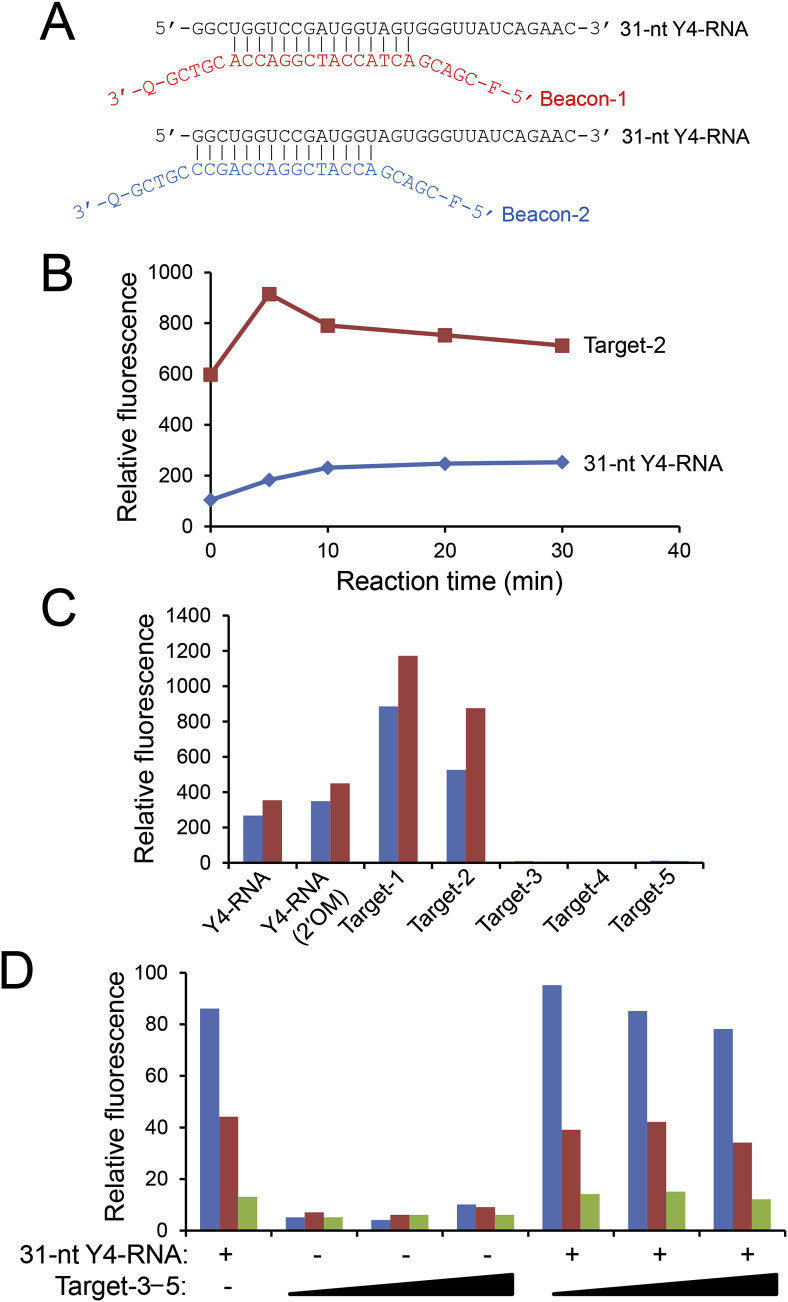
We tried to develop an easier method to quantitate the 31/32-nt Y4-RNA fragment using a molecular beacon [19]. We designed two 25-nt DNA molecular beacons and synthesized them chemically. These molecular beacons, Beacon-1 and Beacon-2, have a 15-nt sequence complementary to a part of the 31-nt Y4-RNA fragment in the middle, 5-nt sequences complementary to each other at both termini, and a fluorescent dye, 6-carboxyfluorescein, and a fluorescent quencher, dabcyl, at the 5′ and 3′ ends, respectively (Fig. 4A). First, to determine an appropriate reaction time, we performed a time course analysis for the interaction between Beacon-2 and the synthetic 31-nt Y4-RNA fragment or Target-2, which is a 15-nt DNA complementary to the middle 15 nt of Beacon-2. Since the fluorescence strength reached almost a plateau in 20–30 min, the reaction time was set 25 min in the following assays (Fig. 4B).
Fig. 4.
Fluorometric quantitation with molecular beacons. (A) DNA molecular beacons for human Y4-RNA fragments. The hybrid of the 31-nt Y4-RNA fragment and Beacon-1 or Beacon-2 is shown. F, 6-carboxyfluorescein; G, dabcyl. (B) Reaction time dependency of molecular beacon fluorescence. Beacon-2 (10 pmol) was incubated with Target-2 (4 pmol) or the synthetic 31-nt Y4-RNA fragment (4 pmol) at room temperature. (C) Sensitivity tests. Ten pmol of Beacon-1 (left bars) or Beacon-2 (right bars) were incubated at room temperature for 25 min with 4 pmol of the synthetic 31-nt Y4-RNA fragment without or with 2′-O-methyl modifications, or Target-1–Target-5. (D) Non-specific target effects. One (left bars), 0.4 (middle bars), or 0.1 (right bars) pmol of Beacon-2 were incubated at room temperature for 25 min with the synthetic 31-nt Y4-RNA fragment (4 pmol) and/or a Target-3–Target-5 mixture (1, 2, or 3 pmol each).
Next, we examined sensitivity and specificity of both molecular beacons using the synthetic 31-nt Y4-RNA fragments with and without 2′-O-methyl modifications, Target-2, Target-1, which is a 15-nt DNA complementary to the middle 15 nt of Beacon-1, and Target-3–Target-5, which are 15-nt DNA molecules with no related sequences to both beacons. In both molecular beacons, the interactions with DNA targets were more stable than that with RNA targets (Fig. 4C). And the 2′-O-methyl modifications appeared to stabilize the interactions, and the interaction with Target-1 was more stable than that with Target-2. The reason that the Beacon-2 interaction with Target-1 was more stable than that with its authentic target, Target-2, may be because Target-2 tends to form a dimer due to partial self-complementarity. Three non-specific targets were not reactive to both beacons. Judging from these results, we decided to use more sensitive Beacon-2 in the following tests.
We examined how non-specific targets affect the fluorescence intensity from the interaction between Beacon-2 and the 31-nt Y4-RNA fragment. Beacon-2 was reacted in three different concentrations with the 31-nt Y4-RNA fragment in the presence of increasing concentrations of a non-specific Target-3–Target-5 mixture, and the fluorescence intensity of the reaction was measured (Fig. 4D). Although on the whole the fluorescence intensity tended to decrease with the increasing concentrations of the non-specific targets, their interference was not so strong.
Considering these characteristics of Beacon-2, we measured the amount of the 31/32-nt Y4-RNA fragment in saliva. And we found that this method overestimates its amount in saliva RNA by ∼1000-fold, probably partly due to the presence of many other RNA molecules in saliva that can interact with Beacon-2 (data not shown). Thus, finding a way to reduce the background noise intensively is needed before this method is put into practical use.
3.4. Y4-RNA fragments as a potential diagnostic and/or prognostic marker
We are currently planning to conduct a relatively large cohort study to assess the 31/32-nt Y4-RNA fragment level in saliva or plasma as a diagnostic and/or prognostic marker for coronary artery disease and various cancers by applying our RT-PCR quantitation method with a correction function for the full-length Y4-RNA level. We will also investigate which cells produce the 31/32-nt fragment, how it is generated from the full-length Y4-RNA, and what its physiological role is, if any, to better evaluate its value as a biomarker. Lastly, the ratio of the 14-nt to 31/32-nt fragment levels in saliva or plasma, which can be determined by next-generation sequencing, may be proved to be an indicator of some pathophysiological conditions, in which the amounts and/or the activities of intra- and/or extra-cellular ribonucleases targeting Y4-RNA may be abnormal.
Conflict of interest
The authors declare no competing interests.
Funding information
This work was supported by Nikkyoko Research Award.
References
- 1.Tuck A.C., Tollervey D. RNA in pieces. Trends Genet. 2011;27:422–432. doi: 10.1016/j.tig.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 2.Jackowiak P., Nowacka M., Strozycki P.M. RNA degradome–its biogenesis and functions. Nucleic Acids Res. 2011;39:7361–7370. doi: 10.1093/nar/gkr450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nashimoto M., Sakai M., Nishi S. Transfer RNA lacking its 3′ terminus is required for spermidinedependent ribonuclease 65 activity in mouse FM3A cell extracts. Biochem. Biophys. Res. Commun. 1991;178:1247–1252. doi: 10.1016/0006-291x(91)91027-a. [DOI] [PubMed] [Google Scholar]
- 4.Elbarbary R.A., Takaku H., Uchiumi N. Modulation of gene expression by human cytosolic tRNase ZL through 5′-half-tRNA. PLoS One. 2009;4:e5908. doi: 10.1371/journal.pone.0005908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thompson D.M., Parker R. Stressing out over tRNA cleavage. Cell. 2009;138:215–219. doi: 10.1016/j.cell.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Ivanov P., Emara M.M., Villen J. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell. 2011;43:613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ninomiya S., Kawano M., Abe T. Potential small guide RNAs for tRNase ZL from human plasma, peripheral blood mononuclear cells, and cultured cell lines. PLoS One. 2015;10:e0118631. doi: 10.1371/journal.pone.0118631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma U., Conine C.C., Shea J.M. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016;351:391–396. doi: 10.1126/science.aad6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nashimoto M. Conversion of mammalian tRNA 3′ processing endoribonuclease to four-base-recognizing RNA cutters. Nucleic Acids Res. 1995;23:3642–3647. doi: 10.1093/nar/23.18.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nashimoto M. Specific cleavage of target RNAs from HIV-1 with 5′ half tRNA by mammalian tRNA 3′ processing endoribonuclease. RNA. 1996;2:2523–2524. [PMC free article] [PubMed] [Google Scholar]
- 11.Habu Y., Miyano-Kurosaki N., Kitano M. Inhibition of HIV-1 gene expression by retroviral vector-mediated small-guide RNAs that direct specific RNA cleavage by tRNase ZL. Nucleic Acids Res. 2005;33:235–243. doi: 10.1093/nar/gki164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakashima A., Takaku H., Shibata H.S. Gene-silencing by the tRNA maturase tRNase ZL under the direction of small guide RNA. Gene Ther. 2007;14:78–85. doi: 10.1038/sj.gt.3302841. [DOI] [PubMed] [Google Scholar]
- 13.Elbarbary R.A., Takaku H., Tamura M. Inhibition of vascular endothelial growth factor expression by TRUE gene silencing. Biochem. Biophys. Res. Commun. 2009;379:924–927. doi: 10.1016/j.bbrc.2008.12.173. [DOI] [PubMed] [Google Scholar]
- 14.Iizuka S., Oridate N., Nashimoto M. Growth inhibition of head and neck squamous cell carcinoma cells by sgRNA targeting the cyclin D1 mRNA based on TRUE gene silencing. PLoS One. 2014;9:e114121. doi: 10.1371/journal.pone.0114121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang A.T., Langley A.R., Christov C.P. Dynamic interaction of Y RNAs with chromatin and initiation proteins during human DNA replication. J. Cell Sci. 2011;124:2058–2069. doi: 10.1242/jcs.086561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sim S., Wolin S.L. Emerging roles for the Ro 60-kDa autoantigen in noncoding RNA metabolism. Wiley Interdiscip. Rev. RNA. 2011;2:686–699. doi: 10.1002/wrna.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Repetto E., Lichtenstein L., Hizir Z. RNY-derived small RNAs as a signature of coronary artery disease. BMC Med. 2015;13:259. doi: 10.1186/s12916-015-0489-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaudewitz D., Skroblin P., Bender L.H. Association of microRNAs and YRNAs with platelet function. Circ. Res. 2016;118:420–432. doi: 10.1161/CIRCRESAHA.114.305663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tyagi S., Kramer F.R. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 20.Dhahbi J.M., Spindler S.R., Atamna H. 5′-YRNA fragments derived by processing of transcripts from specific YRNA genes and pseudogenes are abundant in human serum and plasma. Physiol. Genomics. 2013;45:990–998. doi: 10.1152/physiolgenomics.00129.2013. [DOI] [PubMed] [Google Scholar]