Abstract
Metastasis and resistance to therapy significantly contribute to cancer-related deaths. Growing body of evidence suggest that altered expression of microRNAs (miRNAs) is one of the root cause of adverse clinical outcome. miRNAs such as let-7 are the new fine tuners of signaling cascade and cellular processes which regulates the genes in post-transcriptional manner. In this review, we described the regulation of let-7 expression and the involvement of molecular factors in this process. We discussed the mechanism by which let-7 alter the expression of genes involved in the process of tumorigenesis. Further, we listed the pathways targeted by let-7 to reduce the burden of the tumor. In addition, we described the role of let-7 in breast cancer metastasis and stemness properties. This article will provide the in-depth insight into the biology of let-7 miRNA and its role in the breast cancer progression.
Keywords: miRNA, Let-7, Breast cancer, Metastasis, Stemness
Abbreviations: miRNAs, MicroRNAs; BC, breast cancer; IL-6, interleukin-6; 3′ UTRs, 3′ untranslated regions; SNPs, single nucleotide polymorphisms; NF, nuclear factor; JAK, Janus protein tyrosine kinase; STAT3, signal transducer and activator of transcription 3; CSC, cancer stem cell
1. Introduction
Breast cancer (BC) is most frequently diagnosed cancer and remains one of leading cause of cancer-related death in women worldwide [1], [2]. Altered signaling pathways [3], [4], [5], [6], [7], mutation in genes [8], activation of oncogenic pathways [9], [10], [11], DNA damage [12], [13] and non-targeted effects of chemotherapeutic agents [14], [15], [16], [17] significantly contributes in cancer progression. Therapeutic strategies including chemotherapy [18], application of toxins obtained from pathogen [19], [20], [21], [22], [23], [24] have shown limited clinical efficacy against cancer. During past one and half decade, enormous growth in the field of microRNAs (miRNAs) biology have been witnessed and it has been suggested that targeting these small molecules holds potential therapeutic efficacy for cancer [25], [26], [27]. miRNAs are evolutionary conserved, single-stranded and contains approximately 22 nucleotides RNA molecules that alter the expression of gene at the post-transcriptional level [28]. In nucleus miRNAs are transcribed by RNA polymerase II as pri-miRNAs and subsequently cleaved by ribonuclease III, Drosha, to form a ∼70 nucleotide long pre-miRNA. Thereafter, the pre-miRNA are transported to the cytoplasm and processed by the RNase III protein, Dicer, to yield 18–25 nucleotide long miRNA duplex. After unwinding, one of the strands incorporated into the RNA-induced silencing complex, that subsequently interacts with complementary sequences in the 3′ untranslated regions (3′ UTRs) of the target mRNA transcripts. A single miRNA is capable of regulating multiple mRNAs of various functions. Further, dysregulation of miRNAs abrogate the normal functioning of the cellular system that promotes several pathological conditions such as cancer [29], [30]. Abnormal expression of miRNAs such as let-7 has been reported in several malignancies including BC. In year 2000 Reinhart et al. demonstrated that let-7 miRNA alter the phenotype of nematode and regulates the development of Caenorhabditis elegans [30]. In human, 10 members of the let-7 family have been identified, including let-7a, let-7b, let-7c, let-7d, let-7e, let-7f, let-7g, let-7i, miR-98 and miR-202. In normal physiological conditions let-7 is primarily involved in gene regulation, cell adhesion and muscle formation. Accumulating evidence suggests that let-7 is downregulated in numerous types of cancer, including gastric tumors [31], colon cancer [32], lung cancer [33], Burkitt's lymphoma [34] and BC [35]. The let-7 family of miRNA is associated with apoptosis, proliferation and invasion of cancer cells. Further, let-7 regulates several signaling pathways that are crucial for the biological characteristics of tumor cells. In this review article we have explored the possible factors associated with let-7 expression and its mechanisms of action. Further, we described the target of let-7 that are important for BC cell growth, aggressiveness and explored the benefits of targeting let-7 to control the BC progression.
2. Regulation of miRNA let-7 expression
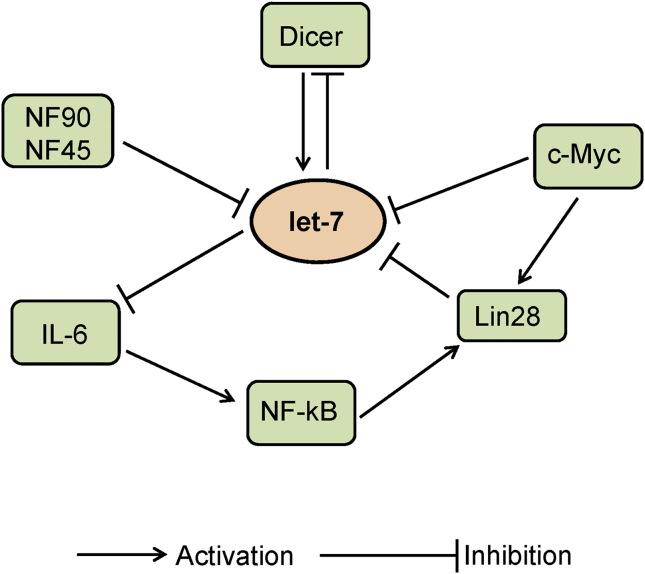
Role of let-7 in cell proliferation and differentiation have been demonstrated in animal and human cell lines [36], [37], [38]. Interestingly let-7 has been implicated in inhibiting the growth of cancer cells [39], [40]. microRNA let-7 expression is important to explore as it involved in the tumor suppression. let-7 expression is controlled at various stages biogenesis which involves numerous factors and signaling molecule (Fig. 1). In this section we described the factors that are known to regulate the expression of let-7 in BC.
Fig. 1.
Signaling pathways involved in miRNA let-7 expression.
2.1. Regulation of miRNA let-7 by Lin28
Lin28 encodes a RNA-binding protein that is known to bind let-7 pre-microRNA. The activity of let-7 was demonstrated to be affected by mutations in Lin28 [30]. Lin28 and its subtype Lin28B have been suggested to bind to hairpin and the stem of pri-let-7 and inhibit the binding of Dicer, thus inhibiting its processing and biogenesis [41], [42]. In addition, binding of Lin28 to the terminal loop region of let-7g has also been demonstrated [43]. Importantly, the zinc-finger and cold-shock domains in Lin28 were determined to be crucial for pre-let-7 binding. Further, upregulation of Lin28 were shown to inhibit the let-7g processing. Ectopic expression of Lin28 abrogates the processing of pri-let-7a suggesting, that Lin28 is important to block the microprocessor-mediated cleavage of pri-let-7 miRNAs [44]. Further, the transfection of Lin28 reduces the endogenous levels of let-7 [44]. Other than Drosha/Dicer inhibition, Lin28/Lin28B is shown to block the let-7 processing by terminal uridylation of pre-let-7 that leads to the irreversibly re-routing pre-let-7 to a degradation pathway [45]. Several enzymes including Zcchc11, a terminal uridylyl transferase 4 (TUT4) have been suggested to be involved in the progress of terminal uridylation. The TUT4 has been found to promote the pre-let-7 uridylation and blockade of let-7 processing in mouse embryonic stem cells [46]. Lin28 recruit TUT4 to pre-let-7 by recognizing tetranucleotide sequence motif (GGAG) in the loop. Later the TUT4 adds an oligouridine tail to pre-let-7 that subsequently blocks Dicer processing [47]. Further, the interaction of PUP-2 with Lin28 controls the stability of Lin28-blockaded let-7 pre-miRNA which suppress the action of Dicer and contribute to the Lin28-stimulated uridylation of let-7 pre-miRNA [48].
2.2. Regulation of miRNA let-7 by nuclear factor 90 and nuclear factor 45
Nuclear factor (NF) 90 and NF45 are the member of Drosha family, which is crucial for the production of pre-miRNA from pri-miRNA. Altered expression of NF90 and NF45 is found to be associated with the level of pri-miRNA. The NF90-NF45 complex is shown to bind with the majority of pri-miRNAs, including pri-let-7a-1 and has higher binding affinity than the DGCR8-Drosha complex, which also binds to pri-miRNAs. Due to elevated binding affinity, NF90-NF45 complex attenuate the processing of pri-miRNA by the DGCR8-Drosha complex. The NF90-NF45 have been shown to have higher binding affinity for pri-let-7a-1 than the other pri-miRNAs [49].
2.3. Regulation of miRNA let-7 by other factors
DNA methylation is considered to be one of the reason that alter miRNA let-7 expression [50], [51], [52]. The human let-7 gene is located on chromosome 22q13.31, which is known to be methylated by the DNA methyltransferases such as DNMT3B and DNMT1. The miRNA let-7a-3 is found to be methylated in lung samples. Interestingly the hypomethylation of let-7a-3 promotes the expression of miRNA and reduce the growth of lung adenocarcinomas cells [52]. Moreover, hypermethylation downregulated the let-7a-3 in epithelial ovarian cancer and associated with unfavorable prognosis [53]. Several factors act at the time of let-7 biogenesis and control the expression of let-7 via regulatory loops. These loops can be either Lin28-dependent or Lin28-independent. The Lin28-dependent regulatory feedback loop involves the NFκB-Lin28-let-7-interleukin (IL)-6-NFκB, and Lin28-let-7-Lin28 loops. The NFκB is shown to activate Lin28 transcription and reduces let-7 levels. Further, let-7 can inhibit IL-6 expression that can activate NFκB, and completing a positive feedback loop [54]. c-Myc, an oncogene is one of the target of let-7. The expression of c-Myc regulated by IMP1 which is believed to be negatively and directly regulated by let-7 [55], [56]. Further, c-Myc was demonstrated to transactivate Lin28B, which inhibit let-7 expression. In addition, activation of Lin28B was found to associate with Myc-mediated let-7 expression [57], [58]. Moreover, let-7 can also affect Lin28 expression as the binding of let-7 to the 3′ UTR of Lin28 transcripts represses Lin28 expression [58]. Lin28 is believed to be a classical direct inhibitor of let-7, which create a double-negative regulatory loop for let-7. Alteration in regulatory circuits affects the expression of let-7 that can promote normal and abnormal responses. A single nucleotide polymorphisms (SNPs) in tumor suppressor miRNA is believed to be responsible for several malignancies [59], [60]. A SNP of the Lin28 gene, rs3811463 is shown to be involved in downregulation of let-7 via the let-7-Lin28 double negative feedback loop. rs3811463 was therefore believed to involved in breast cancer [61].
2.4. Mechanism of miRNA let-7 mediated response
The best explained mechanism of let-7 miRNA action is binding to the 3′ UTR of target mRNAs to alter their expression. Further, let-7 induces its effect when it was targeted to the 3′ or 5′ UTRs of mRNAs, suggesting that let-7 can act via binding to sites other than the 3′ UTR [62]. In addition, let-7 is capable to bind directly to coding regions to target mRNAs to alter its expression [63]. It has been suggested that let-7a can inhibit the translation of target mRNAs by binding and inhibiting the translating polyribosomes [64]. Deadenylation that is removal of adenylate group from protein is another process that can be exploited by let-7 to inhibit or decay the translation of mRNA. However, deadenylation alone seems insufficient to participate in mRNA repression [65].
2.5. Signaling pathways targeted by miRNA let-7
Let-7 is one of the miRNA that targets multiple signaling pathways including Janus protein tyrosine kinase (JAK), signal transducer and activator of transcription 3 (STAT3) and c-Myc. These pathways are crucial for tumor cell growth and aggressiveness. By suppressing these oncogenic pathways let-7 act as a tumor suppressor.
2.6. Regulation of JAK-STAT3 pathway by miRNA let-7
The JAK is a member of the intracellular, non-receptor tyrosine kinases that mediates signals via JAK-STAT3 signaling pathway. Activated JAK promotes STATs activation, which transmits the information from extracellular chemical to the nucleus and induces the expression of genes involved in differentiation, proliferation, apoptosis, oncogenesis and immunity [66]. The JAK-STAT3 pathway is believed to be activated in several types of malignancies [67], [68]. Interestingly, STAT3 was found to be a target of let-7a, which mediates cell proliferation in HepG2 cells [69]. It is speculated that let-7 may regulate the activity of cancer cells by targeting JAK-STAT3 signaling pathway.
2.7. Regulation of Myc oncogene pathway by miRNA let-7
Myc (c-Myc) is a transcription factor that plays an important role in cell cycle progression and apoptosis. This gene is usually activated in tumors [70]. Activation of Myc promotes cell growth, and survival by enhancing the synthesis of its target proteins, which are associated with cell cycle and apoptosis [71]. In addition the mutation in Myc gene have been reported in many cancers which causes this gene to be persistently expressed that leads to the altered expression of several genes in which are involved in growth and aggressiveness of the cancer. Altered expression of Myc has been found in the cancers of colon [72], cervix [73] and breast [74]. Several reports have been suggested that let-7a downregulated Myc mRNA and protein [34], [75]. Further, it has been suggested that miRNA let-7 regulates Myc expression by binding to its 3′ UTR. However, let-7 has been found to regulate the cell cycle by altering the expression of several downstream proteins such as cyclin D1 and cyclin-dependent kinase (CDK) 6 that are the part of Myc oncogene signaling pathway [76], [77].
2.8. miRNA let-7 and breast cancer
BC is one of the leading causes of cancer related death in women. The poor outcome of BC is attributed to the heterogenous nature and metastasis. Another mechanism that helps tumor cell to grow unchecked is the development of cancer stem cell (CSC) phenotype. The let-7 microRNAs that regulate the expression of multiple genes related to the metastasis and stem cell phenotype and attracted scientific community to develop targeted therapies against BC. In this section we briefly discussed the role of let-7 in BC metastasis and stemness.
2.9. Role of miRNA let-7 in breast cancer metastasis
Metastasis is a process in which cancer cell break away from the original site and travels the blood or lymphatic system to the other parts of body and form new tumors. Chemotaxis is believed to be a fundamental cause of metastasis in which external signals orient and attract tumor cells. Overexpression of certain receptors facilitates BC cells to get attracted and move to other site. CXCR7 is one of the receptor that is found to be overexpressed in BC and participate in metastasis [78]. CCR7 is activated by binding chemokines CCL21 and CCL19 [79], [80]. T cells utilize CCL21 to enter lymphoid tissues from circulation. As for as CCL19 is concerned, it is expressed by mature dendritic cells which activates T cells [81]. However, tumor cells exploit the conditions by expressing CCR7 that helps them to localize in lymph node after receiving the chemotactic signals from CCL19 or CCL21. miRNAs have been suggested to suppress the expression of numerous cancer-related genes that subsequently reduces tumorigenesis and metastasis in BC and several other cancers [82], [83], [84]. A study examined the roles of CCR7 and miRNA in breast cancer metastasis suggested that let-7 family binds participate in the process of metastasis [85]. Let-7a was found to influence the CCR7 down expression by targeting 3′UTR of CCR7, thereby downregulating BC cell invasion and migration. Similar results were confirmed in study performed using zebrafish embryo models. Let-7a, a member of family let-7 act as a tumor suppressor by regulating the expression of RAS and HMGA2 oncogenes [86], [87], [88]. Further, decreased levels of let-7a was found to associate with elevated RAS expression in lung squamous carcinoma [86].
2.10. Role of miRNA let-7 in breast cancer stemness
CSCs, a sub-population of tumor cells is believed to be largely responsible for the therapy resistance and unfavorable clinical outcome. miRNAs have been suggested to contribute in tumor initiation by regulating the properties of CSC, including de-differentiation, self-renewal and therapy resistance [89], [90]. Let-7 is emerged as a master regulator of CSC properties including self-renewal and tumor-seeding ability [91]. CSCs demonstrated CD44+/CD24−/low antigen phenotype in mammosphere culture conditions and treatment with anti-cancer reagents and have significant down-regulation of let-7 expression in BC cells. Further, let-7 is shown to inhibit the self-renewal and de-differentiation capacity of BC cells by targeting several genes such as RAS and high mobility group AT-hook 2 (HMGA2) [92]. Further, in BC let-7 is found to suppress tumorigenicity and self-renewal capability by targeting H-RAS and HMGA2 [92]. Delivery of miRNAs in lung cancer using nucleic acid aptamers specific to the receptor tyrosine kinase oncogene Axl, conjugated with let-7g, which target HMGA2, showed effective inhibition of tumor growth [93], [94]. Thus, combining let-7 with specific antibodies or aptamers against breast CSCs, might be a useful approach to improve the treatment of BC patients [95], [96]. Considerable efforts have been made to improve the target specific delivery of miRNA, however, more research is required to improve the therapeutic efficacy and design of vehicles and methods for their delivery in vivo.
3. Conclusion
The research on miRNA have delivered several new aspects of therapeutics and crossed a long way since its discovery. Multiple parameters such as their small size and conserved sequence make them a potential candidate for drug development program. Further, miRNA targeted downstream genes that function in numerous ways including growth, aggressiveness, and therapy resistance, thus targeting single miRNA can have multiple implications. As let-7 have shown multiple connections with metastasis and stemness of BC, the day is not far off when let-7 will be a good therapeutic target for the patients suffering from BC and other malignancies. In addition altered expression of let-7 family members have been shown to influence several other malignancies (Table 1). However, the understanding of precise mechanism is required to gain more insights into the therapeutic efficacy of miRNAs.
Table 1.
miRNA let-7 family target genes in various cancers.
| S. No. | Type of cancer | Target gene | Gene title | References |
|---|---|---|---|---|
| 1 | Thyroid cancer | SLC5A5 | Sodium/iodide cotransporter or solute carrier family 5, member 5 | [97] |
| 2 | Oral Squamous cell Carcinoma | OCT4 | octamer-binding transcription factor 4 | [98] |
| 3 | Ovarian Cancer | KRAS | Kirsten rat sarcoma viral oncogene homolog | [99] |
| 4 | Pancreatic Cancer | STAT3 | Signal transducer and activator of transcription 3 | [100] |
| 5 | Neuroblastoma | MYCN | V-Myc Avian Myelocytomatosis Viral Oncogene Homolog | [101] |
| 6 | Lung Cancer | ITGB3 | Integrin beta-3 | [102] |
| 7 | Liver Cancer | HMGA2 | High-mobility group AT-hook 2 | [103] |
| 8 | Prostate Cancer | IL-6 | Interleukin 6 | [104] |
| 9 | Endometrial Cancer | Aurora-B | Aurora B kinase | [105] |
| 10 | Colorectal Cancer | KRAS | Kirsten rat sarcoma viral oncogene homolog | [106] |
| 11 | Gastric Cancer | CMYC | V-Myc Avian Myelocytomatosis Viral Oncogene a Derived Homolog | [107] |
| 12 | Multiple Myeloma | MYC | V-Myc Avian Myelocytomatosis Viral Oncogene Homolog | [108] |
| 13 | Burkitts Lymphoma | MYC | V-Myc Avian Myelocytomatosis Viral Oncogene Homolog | [34] |
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2015. CA. Cancer J. Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2016. CA. Cancer J. Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Deshmukh S.K., Srivastava S.K., Bhardwaj A., Singh A.P., Tyagi N., Marimuthu S., Dyess D.L., Dal Zotto V., Carter J.E., Singh S. Resistin and interleukin-6 exhibit racially-disparate expression in breast cancer patients, display molecular association and promote growth and aggressiveness of tumor cells through STAT3 activation. Oncotarget. 2015;6:11231–11241. doi: 10.18632/oncotarget.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tyagi N., Bhardwaj A., Srivastava S.K., Arora S., Marimuthu S., Deshmukh S.K., Singh A.P., Carter J.E., Singh S. Development and characterization of a novel in vitro progression model for UVB-induced skin carcinogenesis. Sci. Rep. 2015;5:13894. doi: 10.1038/srep13894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tyagi N., Marimuthu S., Bhardwaj A., Deshmukh S.K., Srivastava S.K., Singh A.P., McClellan S., Carter J.E., Singh S. p-21 activated kinase 4 (PAK4) maintains stem cell-like phenotypes in pancreatic cancer cells through activation of STAT3 signaling. Cancer Lett. 2016;370:260–267. doi: 10.1016/j.canlet.2015.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tripathi K., Mani C., Somasagara R.R., Clark D.W., Ananthapur V., Vinaya K., Palle K. Detection and evaluation of estrogen DNA-adducts and their carcinogenic effects in cultured human cells using biotinylated estradiol. Mol. Carcinog. 2016 doi: 10.1002/mc.22566. [DOI] [PubMed] [Google Scholar]
- 7.Tripathi K., Mani C., Clark D.W., Palle K. Rad18 is required for functional interactions between FANCD2, BRCA2, and Rad51 to repair DNA topoisomerase 1-poisons induced lesions and promote fork recovery. Oncotarget. 2016;7:12537–12553. doi: 10.18632/oncotarget.7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Futreal P.A., Coin L., Marshall M., Down T., Hubbard T., Wooster R., Rahman N., Stratton M.R. A census of human cancer genes. Nat. Rev. Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Croce C.M. Oncogenes and cancer. N. Engl. J. Med. 2008;358:502–511. doi: 10.1056/NEJMra072367. [DOI] [PubMed] [Google Scholar]
- 10.Tripathi K., Mani C., Barnett R., Nalluri S., Bachaboina L., Rocconi R.P., Athar M., Owen L.B., Palle K. Gli1 protein regulates the S-phase checkpoint in tumor cells via Bid protein, and its inhibition sensitizes to DNA topoisomerase 1 inhibitors. J. Biol. Chem. 2014;289:31513–31525. doi: 10.1074/jbc.M114.606483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palle K., Mani C., Tripathi K., Athar M. Aberrant GLI1 activation in DNA damage response, carcinogenesis and chemoresistance. Cancers. 2015;7:2330–2351. doi: 10.3390/cancers7040894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tyagi N., Srivastava S.K., Arora S., Omar Y., Ijaz Z.M., Al-Ghadhban A., Deshmukh S.K., Carter J.E., Singh A.P., Singh S. Comparative analysis of the relative potential of silver, Zinc-oxide and titanium-dioxide nanoparticles against UVB-induced DNA damage for the prevention of skin carcinogenesis. Cancer Lett. 2016;383:53–61. doi: 10.1016/j.canlet.2016.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rakiman I., Mani C., Baraneedharan U., Paul S., Venkatachalam P. γ-H2AX assay: a technique to quantify DNA double strand breaks. Adv. Biotech. 2008:39–41. [Google Scholar]
- 14.Chinnadurai M., Chidambaram S., Ganesan V., Baraneedharan U., Sundaram L., Paul S.F.D., Venkatachalam P. Bleomycin, neocarzinostatin and ionising radiation-induced bystander effects in normal diploid human lung fibroblasts, bone marrow mesenchymal stem cells, lung adenocarcinoma cells and peripheral blood lymphocytes. Int. J. Radiat. Biol. 2011;87:673–682. doi: 10.3109/09553002.2010.549536. [DOI] [PubMed] [Google Scholar]
- 15.Chinnadurai M., Rao B., Deepika R., Paul S., Venkatachalam P. Role of reactive oxygen species and nitric oxide in mediating chemotherapeutic drug induced bystander response in human cancer cells exposed in-vitro. World J. Oncol. 2012;3:64–72. doi: 10.4021/wjon474w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Basheerudeen S.A.S., Mani C., Kulkarni M.A.K., Pillai K., Rajan A., Venkatachalam P. Human brain glioblastoma cells do not induce but do respond to the bleomycin-induced bystander response from lung adenocarcinoma cells. Mutat. Res. 2013;757:114–119. doi: 10.1016/j.mrgentox.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 17.Chinnadurai M., Paul S.F.D., Venkatachalam P. The effect of growth architecture on the induction and decay of bleomycin and X-ray-induced bystander response and genomic instability in lung adenocarcinoma cells and blood lymphocytes. Int. J. Radiat. Biol. 2013;89:69–78. doi: 10.3109/09553002.2012.726397. [DOI] [PubMed] [Google Scholar]
- 18.Akahori T., Sho M., Tanaka T., Kinoshita S., Nagai M., Nishiwada S., Nishiofuku H., Ohbayashi C., Kichikawa K., Nakajima Y. Factors associated with failure to complete adjuvant chemotherapy in pancreatic cancer. Am. J. Surg. 2016;211:787–792. doi: 10.1016/j.amjsurg.2015.10.034. [DOI] [PubMed] [Google Scholar]
- 19.Mathur D.D., Deshmukh S., Kaushik H., Garg L.C. Functional and structural characterization of soluble recombinant epsilon toxin of Clostridium perfringens D, causative agent of enterotoxaemia. Appl. Microbiol. Biotechnol. 2010;88:877–884. doi: 10.1007/s00253-010-2785-y. [DOI] [PubMed] [Google Scholar]
- 20.Chhabra G., Sharma P., Anant A., Deshmukh S., Kaushik H., Gopal K., Srivastava N., Sharma N., Garg L.C. Identification and modeling of a drug target for Clostridium perfringens SM101. Bioinformation. 2010;4:278–289. doi: 10.6026/97320630004278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaushik H., Deshmukh S., Mathur D.D., Tiwari A., Garg L.C. Recombinant expression of in silico identified Bcell epitope of epsilon toxin of Clostridium perfringens in translational fusion with a carrier protein. Bioinformation. 2013;9:617–621. doi: 10.6026/97320630009617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Theys J., Lambin P. Clostridium to treat cancer: dream or reality? Ann. Transl. Med. 2015;3:S21. doi: 10.3978/j.issn.2305-5839.2015.03.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts N.J., Zhang L., Janku F., Collins A., Bai R.-Y., Staedtke V., Rusk A.W., Tung D., Miller M., Roix J., Khanna K.V., Murthy R., Benjamin R.S., Helgason T., Szvalb A.D., Bird J.E., Roy-Chowdhuri S., Zhang H.H., Qiao Y., Karim B., McDaniel J., Elpiner A., Sahora A., Lachowicz J., Phillips B., Turner A., Klein M.K., Post G., Diaz L.A., Riggins G.J., Papadopoulos N., Kinzler K.W., Vogelstein B., Bettegowda C., Huso D.L., Varterasian M., Saha S., Zhou S. Intratumoral injection of Clostridium novyi-NT spores induces antitumor responses. Sci. Transl. Med. 2014;6:249ra111. doi: 10.1126/scitranslmed.3008982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siliprandi N., Di Lisa F., Menabò R. Propionyl-L-carnitine: biochemical significance and possible role in cardiac metabolism. Cardiovasc. Drugs Ther. Spons. Int. Soc. Cardiovasc. Pharmacother. 1991;5(Suppl 1):11–15. doi: 10.1007/BF00128238. [DOI] [PubMed] [Google Scholar]
- 25.Hydbring P., Badalian-Very G. Clinical applications of microRNAs. F1000Research. 2013;2:136. doi: 10.12688/f1000research.2-136.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barger J.F., Nana-Sinkam S.P. MicroRNA as tools and therapeutics in lung cancer. Respir. Med. 2015;109:803–812. doi: 10.1016/j.rmed.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tyagi N., Arora S., Deshmukh S.K., Singh S., Marimuthu S., Singh A.P. Exploiting nanotechnology for the development of MicroRNA-based Cancer therapeutics. J. Biomed. Nanotechnol. 2016;12:28–42. doi: 10.1166/jbn.2016.2172. [DOI] [PubMed] [Google Scholar]
- 28.Nilsen T.W. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. TIG. 2007;23:243–249. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 29.Carthew R.W., Sontheimer E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 31.Zhang H.-H., Wang X.-J., Li G.-X., Yang E., Yang N.-M. Detection of let-7a microRNA by real-time PCR in gastric carcinoma. World J. Gastroenterol. 2007;13:2883–2888. doi: 10.3748/wjg.v13.i20.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akao Y., Nakagawa Y., Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol. Pharm. Bull. 2006;29:903–906. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- 33.Takamizawa J., Konishi H., Yanagisawa K., Tomida S., Osada H., Endoh H., Harano T., Yatabe Y., Nagino M., Nimura Y., Mitsudomi T., Takahashi T. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 34.Sampson V.B., Rong N.H., Han J., Yang Q., Aris V., Soteropoulos P., Petrelli N.J., Dunn S.P., Krueger L.J. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–9770. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 35.Huang X., Yang Y., Guo Y., Cao Z.L., Cui Z.W., Hu T.C., Gao L.B. Association of a let-7 KRAS rs712 polymorphism with the risk of breast cancer. Genet. Mol. Res. GMR. 2015;14:16913–16920. doi: 10.4238/2015.December.14.19. [DOI] [PubMed] [Google Scholar]
- 36.Sokol N.S., Xu P., Jan Y.-N., Ambros V. Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 2008;22:1591–1596. doi: 10.1101/gad.1671708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caygill E.E., Johnston L.A. Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. Curr. Biol. CB. 2008;18:943–950. doi: 10.1016/j.cub.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomson J.M., Parker J., Perou C.M., Hammond S.M. A custom microarray platform for analysis of microRNA gene expression. Nat. Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- 39.Lou Y., Mai W., Jin J. Simultaneous presentation of acute myocardial infarction and acute promyelocytic leukemia. Ann. Hematol. 2006;85:409–410. doi: 10.1007/s00277-006-0106-4. [DOI] [PubMed] [Google Scholar]
- 40.Chang T.-C., Mendell J.T. microRNAs in vertebrate physiology and human disease. Annu. Rev. Genomics Hum. Genet. 2007;8:215–239. doi: 10.1146/annurev.genom.8.080706.092351. [DOI] [PubMed] [Google Scholar]
- 41.Pérez-Llarena F.J., Cartelle M., Mallo S., Beceiro A., Pérez A., Villanueva R., Romero A., Bonnet R., Bou G. Structure-function studies of arginine at position 276 in CTX-M beta-lactamases. J. Antimicrob. Chemother. 2008;61:792–797. doi: 10.1093/jac/dkn031. [DOI] [PubMed] [Google Scholar]
- 42.Newman M.A., Thomson J.M., Hammond S.M. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA N. Y. N. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piskounova E., Viswanathan S.R., Janas M., LaPierre R.J., Daley G.Q., Sliz P., Gregory R.I. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J. Biol. Chem. 2008;283:21310–21314. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- 44.Viswanathan S.R., Daley G.Q., Gregory R.I. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heo I., Joo C., Cho J., Ha M., Han J., Kim V.N. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol. Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 46.Hagan J.P., Piskounova E., Gregory R.I. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heo I., Joo C., Kim Y.-K., Ha M., Yoon M.-J., Cho J., Yeom K.-H., Han J., Kim V.N. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 48.Lehrbach N.J., Armisen J., Lightfoot H.L., Murfitt K.J., Bugaut A., Balasubramanian S., Miska E.A. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 2009;16:1016–1020. doi: 10.1038/nsmb.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sakamoto S., Aoki K., Higuchi T., Todaka H., Morisawa K., Tamaki N., Hatano E., Fukushima A., Taniguchi T., Agata Y. The NF90-NF45 complex functions as a negative regulator in the microRNA processing pathway. Mol. Cell. Biol. 2009;29:3754–3769. doi: 10.1128/MCB.01836-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lujambio A., Ropero S., Ballestar E., Fraga M.F., Cerrato C., Setién F., Casado S., Suarez-Gauthier A., Sanchez-Cespedes M., Git A., Gitt A., Spiteri I., Das P.P., Caldas C., Miska E., Esteller M. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 51.Yoshitomi T., Kawakami K., Enokida H., Chiyomaru T., Kagara I., Tatarano S., Yoshino H., Arimura H., Nishiyama K., Seki N., Nakagawa M. Restoration of miR-517a expression induces cell apoptosis in bladder cancer cell lines. Oncol. Rep. 2011;25:1661–1668. doi: 10.3892/or.2011.1253. [DOI] [PubMed] [Google Scholar]
- 52.Brueckner B., Stresemann C., Kuner R., Mund C., Musch T., Meister M., Sültmann H., Lyko F. The human let-7a-3 locus contains an epigenetically regulated microRNA gene with oncogenic function. Cancer Res. 2007;67:1419–1423. doi: 10.1158/0008-5472.CAN-06-4074. [DOI] [PubMed] [Google Scholar]
- 53.Lu L., Katsaros D., de la Longrais I.A.R., Sochirca O., Yu H. Hypermethylation of let-7a-3 in epithelial ovarian cancer is associated with low insulin-like growth factor-II expression and favorable prognosis. Cancer Res. 2007;67:10117–10122. doi: 10.1158/0008-5472.CAN-07-2544. [DOI] [PubMed] [Google Scholar]
- 54.Iliopoulos D., Hirsch H.A., Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boyerinas B., Park S.-M., Shomron N., Hedegaard M.M., Vinther J., Andersen J.S., Feig C., Xu J., Burge C.B., Peter M.E. Identification of let-7-regulated oncofetal genes. Cancer Res. 2008;68:2587–2591. doi: 10.1158/0008-5472.CAN-08-0264. [DOI] [PubMed] [Google Scholar]
- 56.Ioannidis P., Mahaira L.G., Perez S.A., Gritzapis A.D., Sotiropoulou P.A., Kavalakis G.J., Antsaklis A.I., Baxevanis C.N., Papamichail M. CRD-BP/IMP1 expression characterizes cord blood CD34+ stem cells and affects c-myc and IGF-II expression in MCF-7 cancer cells. J. Biol. Chem. 2005;280:20086–20093. doi: 10.1074/jbc.M410036200. [DOI] [PubMed] [Google Scholar]
- 57.Shaposhnikov I.G., Tabatadze K.G., Zhukova O.V., Kondrat’eva I.E., Filianin A.M., Rychkov I.G. [Wound healing as affected by constitutional immunity factors] Khirurgiia (Sofiia) 1991:23–27. [PubMed] [Google Scholar]
- 58.Dangi-Garimella S., Yun J., Eves E.M., Newman M., Erkeland S.J., Hammond S.M., Minn A.J., Rosner M.R. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J. 2009;28:347–358. doi: 10.1038/emboj.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liang D., Meyer L., Chang D.W., Lin J., Pu X., Ye Y., Gu J., Wu X., Lu K. Genetic variants in MicroRNA biosynthesis pathways and binding sites modify ovarian cancer risk, survival, and treatment response. Cancer Res. 2010;70:9765–9776. doi: 10.1158/0008-5472.CAN-10-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Permuth-Wey J., Kim D., Tsai Y.-Y., Lin H.-Y., Chen Y.A., Barnholtz-Sloan J., Birrer M.J., Bloom G., Chanock S.J., Chen Z., Cramer D.W., Cunningham J.M., Dagne G., Ebbert-Syfrett J., Fenstermacher D., Fridley B.L., Garcia-Closas M., Gayther S.A., Ge W., Gentry-Maharaj A., Gonzalez-Bosquet J., Goode E.L., Iversen E., Jim H., Kong W., McLaughlin J., Menon U., Monteiro A.N.A., Narod S.A., Pharoah P.D.P., Phelan C.M., Qu X., Ramus S.J., Risch H., Schildkraut J.M., Song H., Stockwell H., Sutphen R., Terry K.L., Tyrer J., Vierkant R.A., Wentzensen N., Lancaster J.M., Cheng J.Q., Sellers T.A. Ovarian Cancer Association Consortium, LIN28B polymorphisms influence susceptibility to epithelial ovarian cancer. Cancer Res. 2011;71:3896–3903. doi: 10.1158/0008-5472.CAN-10-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen A.-X., Yu K.-D., Fan L., Li J.-Y., Yang C., Huang A.-J., Shao Z.-M. Germline genetic variants disturbing the Let-7/LIN28 double-negative feedback loop alter breast cancer susceptibility. PLoS Genet. 2011;7:e1002259. doi: 10.1371/journal.pgen.1002259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lytle J.R., Yario T.A., Steitz J.A. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5’ UTR as in the 3’ UTR. Proc. Natl. Acad. Sci. U. S. A. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Forman J.J., Legesse-Miller A., Coller H.A. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc. Natl. Acad. Sci. U. S. A. 2008;105:14879–14884. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nottrott S., Simard M.J., Richter J.D. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat. Struct. Mol. Biol. 2006;13:1108–1114. doi: 10.1038/nsmb1173. [DOI] [PubMed] [Google Scholar]
- 65.Beilharz T.H., Humphreys D.T., Clancy J.L., Thermann R., Martin D.I.K., Hentze M.W., Preiss T. microRNA-mediated messenger RNA deadenylation contributes to translational repression in mammalian cells. PLoS One. 2009;4:e6783. doi: 10.1371/journal.pone.0006783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leeman R.J., Lui V.W.Y., Grandis J.R. STAT3 as a therapeutic target in head and neck cancer. Expert Opin. Biol. Ther. 2006;6:231–241. doi: 10.1517/14712598.6.3.231. [DOI] [PubMed] [Google Scholar]
- 67.Bowman T., Garcia R., Turkson J., Jove R. STATs in oncogenesis. Oncogene. 2000;19:2474–2488. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- 68.Ma X.-T., Wang S., Ye Y.-J., Du R.-Y., Cui Z.-R., Somsouk M. Constitutive activation of Stat3 signaling pathway in human colorectal carcinoma. World J. Gastroenterol. 2004;10:1569–1573. doi: 10.3748/wjg.v10.i11.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Y., Lu Y., Toh S.T., Sung W.-K., Tan P., Chow P., Chung A.Y.F., Jooi L.L.P., Lee C.G.L. Lethal-7 is down-regulated by the hepatitis B virus x protein and targets signal transducer and activator of transcription 3. J. Hepatol. 2010;53:57–66. doi: 10.1016/j.jhep.2009.12.043. [DOI] [PubMed] [Google Scholar]
- 70.Albihn A., Johnsen J.I., Henriksson M.A. MYC in oncogenesis and as a target for cancer therapies. Adv. Cancer Res. 2010;107:163–224. doi: 10.1016/S0065-230X(10)07006-5. [DOI] [PubMed] [Google Scholar]
- 71.Ruggero D. The role of Myc-induced protein synthesis in cancer. Cancer Res. 2009;69:8839–8843. doi: 10.1158/0008-5472.CAN-09-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kugimiya N., Nishimoto A., Hosoyama T., Ueno K., Enoki T., Li T.-S., Hamano K. The c-MYC-ABCB5 axis plays a pivotal role in 5-fluorouracil resistance in human colon cancer cells. J. Cell. Mol. Med. 2015;19:1569–1581. doi: 10.1111/jcmm.12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu D., Jiang X.-H., Jiang Y.-H., Ding W.-C., Zhang C.-L., Shen H., Wang X.-L., Ma D., Hu Z., Wang H. Amplification and overexpression of TP63 and MYC as biomarkers for transition of cervical intraepithelial neoplasia to cervical cancer. Int. J. Gynecol. Cancer Off. J. Int. Gynecol. Cancer Soc. 2014;24:643–648. doi: 10.1097/IGC.0000000000000122. [DOI] [PubMed] [Google Scholar]
- 74.von Eyss B., Jaenicke L.A., Kortlever R.M., Royla N., Wiese K.E., Letschert S., McDuffus L.-A., Sauer M., Rosenwald A., Evan G.I., Kempa S., Eilers M. A myc-driven change in mitochondrial dynamics limits YAP/TAZ function in mammary epithelial cells and breast Cancer. Cancer Cell. 2015;28:743–757. doi: 10.1016/j.ccell.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 75.Barh D., Malhotra R., Ravi B., Sindhurani P. MicroRNA let-7: an emerging next-generation cancer therapeutic. Curr. Oncol. Tor. Ont. 2010;17:70–80. doi: 10.3747/co.v17i1.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun X., Tang S.-C., Xu C., Wang C., Qin S., Du N., Liu J., Zhang Y., Li X., Luo G., Zhou J., Xu F., Ren H. DICER1 regulated let-7 expression levels in p53-induced cancer repression requires cyclin D1. J. Cell. Mol. Med. 2015;19:1357–1365. doi: 10.1111/jcmm.12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guo Y., Yan K., Fang J., Qu Q., Zhou M., Chen F. Let-7b expression determines response to chemotherapy through the regulation of cyclin D1 in glioblastoma. J. Exp. Clin. Cancer Res. CR. 2013;32:41. doi: 10.1186/1756-9966-32-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Müller A., Homey B., Soto H., Ge N., Catron D., Buchanan M.E., McClanahan T., Murphy E., Yuan W., Wagner S.N., Barrera J.L., Mohar A., Verástegui E., Zlotnik A. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 79.Kim C.H., Pelus L.M., Appelbaum E., Johanson K., Anzai N., Broxmeyer H.E. CCR7 ligands, SLC/6Ckine/Exodus2/TCA4 and CKbeta-11/MIP-3beta/ELC, are chemoattractants for CD56(+)CD16(-) NK cells and late stage lymphoid progenitors. Cell. Immunol. 1999;193:226–235. doi: 10.1006/cimm.1999.1483. [DOI] [PubMed] [Google Scholar]
- 80.Yoshida R., Nagira M., Imai T., Baba M., Takagi S., Tabira Y., Akagi J., Nomiyama H., Yoshie O. EBI1-ligand chemokine (ELC) attracts a broad spectrum of lymphocytes: activated T cells strongly up-regulate CCR7 and efficiently migrate toward ELC. Int. Immunol. 1998;10:901–910. doi: 10.1093/intimm/10.7.901. [DOI] [PubMed] [Google Scholar]
- 81.Steinman R.M., Pack M., Inaba K. Dendritic cells in the T-cell areas of lymphoid organs. Immunol. Rev. 1997;156:25–37. doi: 10.1111/j.1600-065x.1997.tb00956.x. [DOI] [PubMed] [Google Scholar]
- 82.McGuire A., Brown J.A.L., Kerin M.J. Metastatic breast cancer: the potential of miRNA for diagnosis and treatment monitoring. Cancer Metastasis Rev. 2015;34:145–155. doi: 10.1007/s10555-015-9551-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marino A.L.F., Evangelista A.F., Vieira R.A.C., Macedo T., Kerr L.M., Abrahão-Machado L.F., Longatto-Filho A., Silveira H.C.S., Marques M.M.C. MicroRNA expression as risk biomarker of breast cancer metastasis: a pilot retrospective case-cohort study. BMC Cancer. 2014;14:739. doi: 10.1186/1471-2407-14-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lin Y., Liu A.Y., Fan C., Zheng H., Li Y., Zhang C., Wu S., Yu D., Huang Z., Liu F., Luo Q., Yang C.J., Ouyang G. MicroRNA-33b inhibits breast Cancer metastasis by targeting HMGA2, SALL4 and Twist1. Sci. Rep. 2015;5:9995. doi: 10.1038/srep09995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cunningham H.D., Shannon L.A., Calloway P.A., Fassold B.C., Dunwiddie I., Vielhauer G., Zhang M., Vines C.M. Expression of the C-C chemokine receptor 7 mediates metastasis of breast cancer to the lymph nodes in mice. Transl. Oncol. 2010;3:354–361. doi: 10.1593/tlo.10178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Johnson S.M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A., Labourier E., Reinert K.L., Brown D., Slack F.J. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 87.Qian Z.R., Asa S.L., Siomi H., Siomi M.C., Yoshimoto K., Yamada S., Wang E.L., Rahman M.M., Inoue H., Itakura M., Kudo E., Sano T. Overexpression of HMGA2 relates to reduction of the let-7 and its relationship to clinicopathological features in pituitary adenomas. Mod. Pathol. Off. J. U. S. Can. Acad. Pathol. Inc. 2009;22:431–441. doi: 10.1038/modpathol.2008.202. [DOI] [PubMed] [Google Scholar]
- 88.Lee Y.S., Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Song S.J., Poliseno L., Song M.S., Ala U., Webster K., Ng C., Beringer G., Brikbak N.J., Yuan X., Cantley L.C., Richardson A.L., Pandolfi P.P. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell. 2013;154:311–324. doi: 10.1016/j.cell.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kang H.-Y. MicroRNA-21 regulates stemness in cancer cells. Stem Cell Res. Ther. 2013;4:110. doi: 10.1186/scrt321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chiu S.-C., Chung H.-Y., Cho D.-Y., Chan T.-M., Liu M.-C., Huang H.-M., Li T.-Y., Lin J.-Y., Chou P.-C., Fu R.-H., Yang W.-K., Harn H.-J., Lin S.-Z. Therapeutic potential of microRNA let-7: tumor suppression or impeding normal stemness. Cell Transpl. 2014;23:459–469. doi: 10.3727/096368914X678418. [DOI] [PubMed] [Google Scholar]
- 92.Yu F., Yao H., Zhu P., Zhang X., Pan Q., Gong C., Huang Y., Hu X., Su F., Lieberman J., Song E. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 93.Esposito C.L., Cerchia L., Catuogno S., De Vita G., Dassie J.P., Santamaria G., Swiderski P., Condorelli G., Giangrande P.H., de Franciscis V. Multifunctional aptamer-miRNA conjugates for targeted cancer therapy. Mol. Ther. J. Am. Soc. Gene Ther. 2014;22:1151–1163. doi: 10.1038/mt.2014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kortylewski M., Nechaev S. How to train your dragon: targeted delivery of microRNA to cancer cells in vivo. Mol. Ther. J. Am. Soc. Gene Ther. 2014;22:1070–1071. doi: 10.1038/mt.2014.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yang T., Rycaj K. Targeted therapy against cancer stem cells. Oncol. Lett. 2015;10:27–33. doi: 10.3892/ol.2015.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Esposito C.L., Catuogno S., de Franciscis V. Aptamer-mediated selective delivery of short RNA therapeutics in cancer cells, J. RNAi Gene Silenc. Int. J. RNA Gene Target. Res. 2014;10:500–506. [PMC free article] [PubMed] [Google Scholar]
- 97.Damanakis A.I., Eckhardt S., Wunderlich A., Roth S., Wissniowski T.T., Bartsch D.K., Di Fazio P. MicroRNAs let7 expression in thyroid cancer: correlation with their deputed targets HMGA2 and SLC5A5. J. Cancer Res. Clin. Oncol. 2016;142:1213–1220. doi: 10.1007/s00432-016-2138-z. [DOI] [PubMed] [Google Scholar]
- 98.Chien C.-S., Wang M.-L., Chu P.-Y., Chang Y.-L., Liu W.-H., Yu C.-C., Lan Y.-T., Huang P.-I., Lee Y.-Y., Chen Y.-W., Lo W.-L., Chiou S.-H. Lin28B/Let-7 regulates expression of Oct4 and Sox2 and reprograms oral squamous cell carcinoma cells to a stem-like state. Cancer Res. 2015;75:2553–2565. doi: 10.1158/0008-5472.CAN-14-2215. [DOI] [PubMed] [Google Scholar]
- 99.Wang X., Cao L., Wang Y., Wang X., Liu N., You Y. Regulation of let-7 and its target oncogenes (Review) Oncol. Lett. 2012;3:955–960. doi: 10.3892/ol.2012.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Patel K., Kollory A., Takashima A., Sarkar S., Faller D.V., Ghosh S.K. MicroRNA let-7 downregulates STAT3 phosphorylation in pancreatic cancer cells by increasing SOCS3 expression. Cancer Lett. 2014;347:54–64. doi: 10.1016/j.canlet.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Buechner J., Tømte E., Haug B.H., Henriksen J.R., Løkke C., Flægstad T., Einvik C. Tumour-suppressor microRNAs let-7 and mir-101 target the proto-oncogene MYCN and inhibit cell proliferation in MYCN-amplified neuroblastoma. Br. J. Cancer. 2011;105:296–303. doi: 10.1038/bjc.2011.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhao B., Han H., Chen J., Zhang Z., Li S., Fang F., Zheng Q., Ma Y., Zhang J., Wu N., Yang Y. MicroRNA let-7c inhibits migration and invasion of human non-small cell lung cancer by targeting ITGB3 and MAP4K3. Cancer Lett. 2014;342:43–51. doi: 10.1016/j.canlet.2013.08.030. [DOI] [PubMed] [Google Scholar]
- 103.Chen K.-J., Hou Y., Wang K., Li J., Xia Y., Yang X.-Y., Lv G., Xing X.-L., Shen F. Reexpression of Let-7g microRNA inhibits the proliferation and migration via K-Ras/HMGA2/snail axis in hepatocellular carcinoma. Biomed. Res. Int. 2014;2014:742417. doi: 10.1155/2014/742417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sung S.-Y., Liao C.-H., Wu H.-P., Hsiao W.-C., Wu I.-H., Lin S.-H., Hsieh C.-L. Loss of let-7 microRNA upregulates IL-6 in bone marrow-derived mesenchymal stem cells triggering a reactive stromal response to prostate cancer. PLoS One. 2013;8:e71637. doi: 10.1371/journal.pone.0071637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu P., Qi M., Ma C., Lao G., Liu Y., Liu Y., Liu Y. Let7a inhibits the growth of endometrial carcinoma cells by targeting Aurora-B. FEBS Lett. 2013;587:2523–2529. doi: 10.1016/j.febslet.2013.05.065. [DOI] [PubMed] [Google Scholar]
- 106.Zhang W., Winder T., Ning Y., Pohl A., Yang D., Kahn M., Lurje G., Labonte M.J., Wilson P.M., Gordon M.A., Hu-Lieskovan S., Mauro D.J., Langer C., Rowinsky E.K., Lenz H.-J. A let-7 microRNA-binding site polymorphism in 3’-untranslated region of KRAS gene predicts response in wild-type KRAS patients with metastatic colorectal cancer treated with cetuximab monotherapy. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2011;22:104–109. doi: 10.1093/annonc/mdq315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yang X., Cai H., Liang Y., Chen L., Wang X., Si R., Qu K., Jiang Z., Ma B., Miao C., Li J., Wang B., Gao P. Inhibition of c-Myc by let-7b mimic reverses mutidrug resistance in gastric cancer cells. Oncol. Rep. 2015;33:1723–1730. doi: 10.3892/or.2015.3757. [DOI] [PubMed] [Google Scholar]
- 108.Manier S., Powers J.T., Sacco A., Glavey S.V., Huynh D., Reagan M.R., Salem K.Z., Moschetta M., Shi J., Mishima Y., Roche-Lestienne C., Leleu X., Roccaro A.M., Daley G.Q., Ghobrial I.M. The LIN28B/let-7 axis is a novel therapeutic pathway in multiple myeloma. Leukemia. 2016 doi: 10.1038/leu.2016.296. [DOI] [PMC free article] [PubMed] [Google Scholar]