Abstract
Following injury, skeletal muscles can regenerate from muscle specific stem cells, called satellite cells. Quiescent in uninjured muscles, satellite cells become activated, proliferate and differentiate into myotubes. Muscle regeneration occurs following distinct main overlapping phases, including inflammation, regeneration and maturation of the regenerated myofibers. Each step of muscle regeneration is orchestrated through complex signaling networks and gene regulatory networks, leading to the expression of specific set of genes in each concerned cell type. Apart from the well-established transcriptional mechanisms involving the myogenic regulatory factors of the MyoD family, increasing data indicate that each step of muscle regeneration is controlled by a wide range of non-coding RNAs. In this review, we discuss the role of two classes of non-coding RNAs (microRNAs and long non-coding RNAs) in the inflammatory, regeneration and maturation steps of muscle regeneration.
Keywords: Muscle regeneration, lncRNA, miRNA, MyoD, Differentiation, Satellite cells
1. Introduction
Skeletal muscles are able to regenerate following a range of muscle injuries, including trauma linked to physical exercise and chronic damage in degenerative diseases, as muscle dystrophies [1]. Located between the basal lamina and the plasma membrane of muscle fibers, the muscle stem cells or satellite cells (SC) are the myogenic source that is absolutely necessary for muscle regeneration following an acute injury [2]. Immobile and quiescent in uninjured muscles, these Pax7 expressing cells become activated and migrate along the persistent basal lamina of the degenerated myofiber following muscle injury [3]. Strong in vitro evidence based on cultured fibers and myoblasts and a recent in vivo study in zebrafish suggest that some of the first satellite cell divisions are asymetric generating a self-renewing stem cell and a myogenic precursor cell (mpc) prone to proliferate and differentiate [4], [5]. Mpc fuse to each other to form new myofibers or fuse with existing neighbor myofibers [5], [6].
Each step of myogenesis is regulated and controlled by complex networks of signaling pathways. Historically, the identified networks were based on proteins and the messenger RNAs encoding them. However, protein-coding sequences represent less than 2% of the human genome [7], while the vast majority of the genome (about 62%) encodes for non-coding RNAs [8], [9], which possess intrinsic abilities to modulate gene expression via transcriptional, post-transcriptional, epigenetic mechanisms and nuclear genome organization [10], [11], [12]. Non-coding RNAs have been divided into two classes depending on their size. Small ncRNAs (<200 nt) include microRNAs (miRs), small nucleolar RNAs (snRNAs) and piwi-interacting RNAs (piRNAs). Long ncRNAs (>200 nt) include circular non coding RNAs (circRNAs) and other non coding RNAs (lncRNAs) [10]. Transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs) belong to the class of structural RNAs. Among the ncRNAs, two of them received lately extensive attention: miRNAs and lncRNAs. Several review the role of miRNAs and/or lncRNAs in embryonic or adult skeletal myogenesis and muscle diseases [13], [14], [15], [16], [17]. In this review, we present an update of the implication of both miRNAs and long ncRNAs in the main events of muscle regeneration in healthy mammals (Tables 1 and 2).
Table 1.
Role of major miRNAs in skeletal muscle regeneration.
| miRNA | Function in muscle regeneration | Experimental design | Ref |
|---|---|---|---|
| miR-1 | Promotes satellite cell differentiation | In vitro gain and loss of function experiments | [121] |
| Promotes myogenic differentiation |
In vitro gain and loss of function experiments; in vivo gain of function experiments |
[157] | |
| miR-17-92 | Promotes myoblast proliferation | In vitro and in vivo gain of function experiments | [129] |
| miR-20a/b | Repress myoblast proliferation | In vitro gain and loss of functions experiments | [130] |
| miR-23a | Protection from myofiber atrophy |
In vitro gain and loss of function experiments; transgenic mice |
[168] |
| Specification of muscle fibers | In vitro gain and loss of function experiments | [167] | |
| miR-27b | Promotes satellite cell differentiation |
In vitro loss of function experiments; In vivo gain and loss of function experiments |
[96] |
| Inhibition of adipogenic differentiation of FAPs | In vitro loss of function experiments | [90] | |
| miR-29a | Promotes satellite cell proliferation | In vitro and in vivo loss of function experiments | [128] |
| miR-31 | Maintenance of satellite cell quiescence |
In vitro gain of function experiments; In vivo loss of function experiments |
[102] |
| miR-34c | Represses myoblast proliferation | In vitro gain and loss of function experiments | [147] |
| miR-125b | Inhibits myoblast differentiation | In vitro and in vivo gain and loss of function experiments | [150] |
| miR-133 | Promotes myoblast proliferation |
In vitro gain and loss of function experiments; in vivo gain of function experiments |
[157] |
| miR-155 | Activation of macrophages | Knock out mice | [87] |
| miR-195/497 | Maintenance of satellite cell quiescence |
In vitro gain of function experiments; in vivo gain and loss of function experiments |
[105] |
| miR-206 | Promotes satellite cell differentiation | In vitro gain and loss of function experiments | [118] |
| miR-208b | Specification of muscle fibers | In vitro gain of function experiments; Knock out mice | [165] |
| miR-351 | Promotes myoblast proliferation | In vitro gain and loss of function experiments | [84] |
| miR-378 | Promotes myogenic differentiation | In vitro gain and loss of function experiments | [143] |
| miR-410 | Promotes myogenic differentiation | In vitro gain and loss of function experiments | [162] |
| miR-431 | Promotes satellite cell differentiation | In vitro gain of function experiments; Transgenic mice | [117] |
| miR-433 | Promotes myogenic differentiation | In vitro gain and loss of function experiments | [162] |
| miR-486 | Promotes satellite cell differentiation | In vitro gain and loss of function experiments | [118] |
| miR-489 | Maintenance of satellite cell quiescence | In vitro and in vivo gain and loss of function experiments | [104] |
| miR-499 | Specification of muscle fibers |
In vitro gain of function experiments; Knock out and transgenic mice |
[165] |
| miR-501 | Promotes myogenic differentiation |
In vitro gain and loss of function experiments; in vivo loss of function experiments |
[164] |
| miR-675 | Promotes myogenic differentiation | In vitro and in vivo gain and loss of function experiments | [109] |
| miR-715 | Inhibits myogenic differentiation | In vitro gain and loss of function experiments | [148] |
Table 2.
Role of major lncRNAs in skeletal muscle regeneration.
| lncRNA | Function in muscle regeneration | Experimental design | Ref |
|---|---|---|---|
| CEeRNA | Promotes myogenic differentiation | In vitro loss of function experiments | [136] |
| Dum | Promotes satellite cell differentiation | In vitro and in vivo loss of function experiments | [126] |
| Linc-MD1 | Promotes myogenic differentiation | In vitro gain and loss of function experiments | [160] |
| Linc-MYH | Specification of muscle fibers | In vivo gain and loss of function experiments | [166] |
| Linc-RAM | Promotes myogenic differentiation |
In vitro loss and gain of function experiments; Knock out mice |
[141] |
| Linc-YY1 | Promotes myogenic differentiation |
In vitro gain and loss of function experiments; in vivo loss of function experiments |
[144] |
| Lnc-31 | Promotes myoblast proliferation | In vitro loss of function experiments | [135] |
| LncMyoD | Promotes myoblast differentiation | In vitro loss of function experiments | [138] |
| Malat1 | Promotes myoblasts differentiation | In vitro loss of function experiments | [154] |
| MUNC | Promotes myogenic differentiation |
In vitro gain and loss of function experiments; in vivo loss of function experiments |
[137] |
| Sirt1 AS | Promotes myoblast proliferation | In vitro and in vivo gain of function experiments | [132] |
| SRA | Promotes myogenic differentiation | In vitro gain and loss of function experiments | [139] |
| Yam-1 | Inhibits myogenic differentiation | In vitro and in vivo loss of function experiments | [148] |
2. miRNAs
The miRNA class of small ncRNAs is a group of RNAs transcribed by type II or type III RNA polymerase [18]. The primary transcript is a long primary miRNA (pri-miRNA) with a cap and a poly-A tail, and contains a double stranded stem of about 30 base pairs, a terminal loop and two flanking single stranded tails. Within the nucleus, the RNase III enzyme Drosha associated to a double stranded-RNA binding protein, Di George syndrome critical region 8 gene (DGCR8), cleaves the pri-miRNA into short 70 nucleotides precursor miRNAs (pre-miRNAs) [18]. The precursor miRNA is then exported into the cytoplasm via exportin-5 [19], [20]. Once in the cytoplasm, pre-miRNAs are cleaved by the RNase III enzyme Dicer, producing miRNA duplexes (ds-miRNAs) of around 22 nt with its 2 nt overhang at its 3′ ends [21], [22], [23]. The mature ds-miRNA, called miRNA/miRNA* duplex, binds with a member of the Argonaute (Ago) family proteins into the RNA-induced silencing complex (RISC) [24]. One strand of the miRNA duplex, the “passenger” strand, is recognized by the TRBP protein (trans-activation response RNA-binding protein), which loads the ds-miRNA in the correct orientation on Ago proteins. Ago proteins eject the passenger strand from the RISC complex and retain the other miRNA strand, the “guide” strand [20], [25].
Most of the miRNAs are processed following this canonical pathway. However, alternative (non canonical) pathways exist, bypassing several biogenesis steps described above. Mirtrons are a family of microRNAs, which are processed independently from Drosha. Mirtrons are localized inside introns of host genes. The pre-miRNA is generated by splicing and debranching of the host intron and later cleaved by Dicer according to the miRNA canonical maturation [26], [27]. A new class of miRNA-like small non-coding RNAs has been recently identified, the agotrons [28]. They are localized in short introns and their mature form resembles to pre-miRNA (80–100 nt). Their biogenesis is independent from Drosha and Dicer, and they associate with Ago proteins as an unprocessed, full-length intron to regulate gene expression in a miRNA-like manner.
miRNAs regulate gene expression at cytoplasmic and nuclear levels by complex mechanisms, recently reviewed [11]. The minimal RISC (a particular Ago protein combined with a miRNA molecule) is associated with proteins from the Ago and TNRC6 families (the mammalian homologs of the Drosophila protein GW182) together with many other accessory proteins. The fate of the target mRNA is defined by the composition of the RISC complex, combined with the degree of complementarity between the miRNA seed sequence (sequence spanning from position 2 to 8 at the 5′ end) and sequences usually located on the 3′UTR of the target mRNA. A perfect match between the miRNA seed sequence with the mRNA sequence could induce a direct degradation of the latter mRNA by Ago2, the only Ago isoform containing an endonucleolytic activity [29]. However, this mRNA degradation is very rare in mammals. Increasing works demonstrate that microRNAs first repress translation through competing with the cap binding protein elF4E and/or interfering with ribosome scanning, before inducing mRNA decay by recruiting deadenylating/decapping enzymes (for details see Ref. [30]). Increasing studies show the presence of RISC proteins and microRNAs in mammalian nuclei and suggest its implication in the regulation of transcription and splicing (for review, see Ref. [31]). Complementary sequences of miRNA seed sequences exist on gene promoters where miRNA can activate or silence gene transcription by multiple mechanisms, sharing similarities to classical protein mechanisms of transcription and splicing [31].
3. lncRNAs
Despite their poor conservation at the sequence level across species, lncRNAs represent the vast majority of ncRNAs [32]. Their promoter regions have been shown to be generally more conserved than promoters of protein-coding genes [8]. Based on their genomic localization in relation to protein-coding genes, most of the lncRNAs can be classified in four classes [33]. 1/Antisense lncRNAs are transcripts intersecting any coding-protein exon on the opposite strand or have been shown to be an antisense regulation of a coding gene. 2/Long intergenic ncRNAs (lincRNAs) are transcribed between coding gene loci. 3/Sense overlapping lncRNAs contain a coding gene within one of its introns. 4/Sense intronic ncRNAs are transcribed within an intron of a protein coding gene.
Most of these lncRNAs share mRNAs-like features, as they are transcribed by RNA polymerase II [34], they appear to be capped, spliced and polyadenylated at their 3′ end. However, they tend to be expressed at lower levels than protein-coding genes, and specific to tissues [33]. They are devoid of any typical open reading frame (ORF) as found in mRNAs [8]. However, most of them are associated to ribosomes, which could suggest that they might be a source of peptides [35]. In 2006, Frith et al. analyzed for the first time the size and the nature of the mammalian peptidome [36]. They identified novel translated proteins, shorter than 100 amino acids, raising the question of functional short (<100 nt) open reading frames within the genome (sORF). Many sORF have been described within the genome. 7.3% of them found in intergenic regions likely encodes novel peptides, half of them are conserved in humans [36]. Most of these intergenic sORF were found on expressed RNAs, indicating a putative expression. In most lncRNAs, such sORF were identified [37]. In rare and very recent cases in mammals, some functional peptides have indeed been described to be translated from lncRNAs [38], [39], [40]. These recent works support that many transcripts annotated as “lncRNAs” might encode for small peptides with important biological functions, even if they are named “non-coding RNAs” (see below, section 4.6).
Despite their possibility to code for peptides, lncRNAs are believed to be bifunctional as they are also involved, as transcripts, in diverse biological functions, as positive or negative regulators of chromatin remodeling, transcription, splicing and translation in development, differentiation and human diseases [13], [41], [42]. LncRNAs are found in the nucleus and in the cytoplasm of cells, giving them the possibility to modulate a vast number of cellular processes. How lncRNAs exert their functions is poorly understood. In a limited number of cases, a function has been defined. In the nucleus, lncRNAs are proposed to act in cis (acting on nearby genes) and/or trans (acting on genes in a different locus) to alter transcription by diverse mechanisms. LncRNAs transcribed from the promoter region have a strong influence on the neighboring protein-coding genes. In mammals, promoters with CpG islands tend to have a bidirectional promoter activity. Antisense lncRNAs derived from such promoters (pancRNAs) participate to the activation of their partner genes through sequence-specific DNA methylation [43], [44], [45]. Some lncRNAs regulate gene expression through direct base pairing between RNA and DNA sequences forming a triple helix in the promoter of the target gene, interfering with its transcription [46]. Furthermore, transcription is also activated by distal regulatory enhancer elements, which are brought close to their target promoters by chromatin looping. This mechanism involves the recruitment of transcription factors [47] and a class of lncRNA, called enhancer RNA (eRNA) [48], [49]. LncRNAs can also modulate chromatin remodeling. Indeed, some lncRNAs interact with the chromatin remodeling complex SWI/SNF either to inhibit binding of this complex to chromatin [50], [51], to promote its binding on promoters of target genes [52], or to inhibit the chromatin remodeling activity [53]. Furthermore, genome-wide RNA immunoprecipitation sequencing revealed that many lncRNAs associate with the polycomb repressive complex PRC2, which mediates methylation of lysine 27 on histone 3 (H3K27me) [54], to overall promote PRC occupancy at genomic target sites and therefore repressing gene transcription [55]. Transcription is also regulated by lncRNAs, through a direct interaction with RNA polymerase II to repress transcription [56], [57]. Moreover, several lncRNAs act directly on specific transcription factors, such as SMAD2/3 [58], STAT3 [59], transcription factors of the NF-κB pathway [60] to positively or negatively regulate transcription of target genes. Besides their role in transcription regulation, some lncRNAs have been shown to interfere with splicing events, globally by interacting with splicing factors [61], [62], [63], [64], and in the nuclear organization, particularly in the formation of speckles, paraspeckles and polycomb bodies (for review, see Ref. [65]).
In the cytoplasm, many lncRNAs could regulate translation through different mechanisms. SINEUPs are a new class of antisense lncRNAs, which carry an inverted SINEB2 (short interspersed repetitive element B2) sequence to up-regulate translation in a gene-specific manner [66]. Their activity depends on two RNA regions: a SINEUP binding domain, that overlaps in an antisense orientation to the sense target protein-coding mRNA; and an effector domain, containing the inverted SINEB2 sequence, which is required to activate translation [66]. Translation is also down-regulated by miRNAs, which could be targeted by competing endogenous RNAs (ceRNAs). These RNAs are sponges for miRNAs, leading to a reduction in the level of free miRNAs, thus reducing the availability of these miRNAs to bind their target mRNAs and thereby protecting their target RNAs from repression [67]. On the other hand, lncRNAs can also be a source of miRNAs [68]. Some non-coding RNAs from another type of non-coding RNAs, circular RNAs (circRNAs), have been shown to potentially serve as miRNA sponges or competing endogenous RNAs [69]. Due to their circular structure, these ncRNAs are resistant to degradation by exonucleases and are highly stable in cells, making them excellent candidates for competing endogenous RNA activity [70]. However, the ceRNA function remains controversial, since no explanation has been found so far to understand how the change in expression of one single ceRNA gene, which corresponds to a very small fraction of the miRNA targets, could significantly influence the activity of this miRNA across all of its target sites, in physiological conditions [71], [72], [73].
Non-coding RNAs are important players with many different biological functions. In this review, we will explore the current literature on the role of ncRNAs in regulating skeletal muscle regeneration, in non-pathological conditions (Tables 1 and 2).
4. Non-coding RNA in skeletal muscle regeneration
The muscle regeneration process is triggered by myofiber necrosis, after acute injury or in degenerating diseased muscles [74]. Following necrosis, three distinct overlapping phases are commonly distinguished: 1/an inflammatory reaction, mainly characterized by the invasion of macrophages on the injured area, to phagocyte necrotic debris and promote the myogenic response; 2/the activation and differentiation of satellite cells; 3/the maturation of newly formed myofibers and remodeling of regenerated muscle [1].
4.1. Necrosis and inflammatory response
Very shortly after acute muscle injury (15 min later), likely during the acute muscle degeneration phase, specific miRNAs (miR-1, miR-133a and miR-206) are released into the circulation [75]. These extracellular miRNAs appear to be highly stable in a nuclease-rich extracellular environment as serum. They are known to be released in vesicle-free protein [76] or lipoprotein [77] complexes, or in lipid membrane bound vesicles, as microvesicles, exosomes [78] or apoptotic bodies [79]. In the case of muscle regeneration after experimentally induced muscle injury or in dystrophic mice, miRNAs released in the serum are protected from RNase-mediated degradation by association with proteins as Ago2 rather than encapsulation in extracellular vesicles [75]. The physiological function of these circulating miRNAs is currently not understood, but we can suggest that they might participate to intercellular communication, what could improve muscle regeneration. This process is indeed influenced by different cell types. MiRNAs-containing exosomes released from non muscle cells are indeed able to promote skeletal muscle regeneration. In vitro myogenic differentiation as well as in vivo skeletal muscle regeneration are promoted by exosomes purified from mesenchymal stem cells [80]. Altogether, these data suggest that muscle regeneration might be regulated by many different cell types and extracellular miRNAs could be one of the communication tools between these cells.
Inflammatory cells are among the non muscle cells which actively participate to muscle regeneration. A large number of them infiltrate shortly the injured area [81]. This inflammatory phase plays an important role in the first steps of regeneration and involves the recruitment of specific myeloid cells. This is characterized by a transient wave of neutrophils, followed by an infiltration of macrophages mainly derived from circulating monocytes. Two populations of macrophages were described in regenerating muscles: M1 and M2 macrophages [82]. The proinflammatory M1 macrophages appear soon after injury and they stimulate satellite cell proliferation. In contrast, M2 macrophages play an anti-inflammatory role and they promote myoblast fusion and myotube hypertrophy [83]. A genome wide analysis has revealed that a number of miRNAs are up-regulated immediately after injury [84]. Most of these miRNAs are probably involved in the early steps of inflammation. Indeed, two of them, miR-147 and miR-223, can be associated to inflammation, since they are induced in multiple immune cells, such as neutrophils and macrophages [85], [86]. However, very few miRNAs were further analyzed in a context of muscle regeneration. MiR-155 has been shown to be enriched in myeloid cells and its expression is increased upon muscle injury [87]. This miRNA promotes muscle regeneration by regulating the balance between M1 and M2 macrophage activation, and targeting SOCS1 (suppressors of cytokine signaling 1), a negative regulator of the JAK/STAT signaling pathway. In absence of miR-155, skeletal muscle regeneration is delayed.
Other inflammatory cell types appear to be involved in muscle regeneration. Eosinophils release interleukin-4, which stimulates fibro-adipocyte progenitors (FAPs), a muscle resident mesenchymal stem cell population [88], [89]. In turn, FAPs inhibit their adipogenic differentiation, proliferate as fibroblasts and phagocyte necrotic debris. MiR-27b is highly expressed in proliferating FAPs and its expression is reduced when FAPs undergo adipocyte differentiation [90]. One of the target mRNAs of miR-27b codes for the pro-adipogenic protein PPARγ (peroxysome proliferator-activated receptor γ) [91]. Therefore, miR-27b could participate to the inhibition of adipogenic differentiation of FAPs by down-regulating pro-adipogenic genes, such as PPARγ.
Very recently, a new type of inflammatory cells has been shown to accumulate in injured skeletal muscles [92], [93]. The regulatory T cells (Treg) would promote the M1 to M2 switch in macrophages and the activation of satellite cells [92]. The gene expression profile of Treg cells isolated from injured muscles is different from Treg cells from lymphoid organs [92]. Since Treg cells from lymphoid organs have been shown to express many different miRNAs [94], it could be very interesting to extend the analysis of non-coding gene expression in the muscle Treg cells.
Inflammatory and immune cells have a central role in the regeneration process. Very few studies focused their analyses on the role of non-coding RNAs in regulating these inflammatory cells in a context of muscle injury. This inflammatory response is a coordinated process which should be finely regulated to efficiently regenerate the muscle tissue. The identification of ncRNAs involved in this process would give valuable information to better understand how each inflammatory cell participates to this process.
4.2. Satellite cell quiescence and activation
In resting adult muscles, satellite cells are quiescent (in a non-dividing state) and express high levels of the transcription factor, Pax7, although a subset of satellite cells also expresses Pax3 in specific adult muscles [95], [96]. The expression of Pax7 is essential for the establishment and maintenance of adult satellite cells [95], [97], [98]. The myogenic determination factors MyoD and Myf5 are direct targets of Pax3/7 [99], [100]. However, these mRNA are not translated in quiescent satellite cells, until they are activated to regenerate injured myofibers [101]. It has been shown that Myf5 mRNAs are sequestered in mRNA-containing ribonucleoprotein (mRNP) granules as long as satellite cells remain quiescent [101], [102]. miR-31 is sequestered with Myf5 transcripts in mRNP granules of quiescent cells, where they target Myf5 transcripts preventing the accumulation of Myf5 proteins [102]. As satellite cells are activated, mRNP granules dissociate, Myf5 transcripts are released leading to the translation and the accumulation of Myf5 proteins, and promoting myogenesis. MiR-31 is therefore a key element to maintain satellite cells in a quiescent state.
MyoD translation is repressed through a different mechanism [103]. The mRNA decay factor Tristetrapolin is expressed in quiescent SCs, binds the 3′UTR of myod transcripts promoting its decay. Once SC activates, Tristetrapolin is inhibited by p38α/β MAPK signaling and myod mRNAs are stabilized and translated. These studies indicate that the quiescent state of SCs is protected by several critical post-transcriptional regulatory mechanisms. SC-specific conditional deletion of Dicer in mice shows that many miRNAs might protect SCs from activation [104]. Indeed, SCs spontaneously exit quiescence in absence of Dicer to enter into the cell cycle. 351 miRNAs are differentially regulated in activated SCs compared to quiescent ones. Among them, 22 are highly expressed in quiescent SCs and their level decreases as SCs are activated [104]. MiR-489 is one of them. In this study, the authors showed that miR-489 targets mRNAs coding for the oncogene Dek in quiescent cells, blocking therefore the proliferative expansion of myogenic progenitors. Cell cycle arrest of quiescent satellite cells is also controlled by two other miRNAs, miR-195 and miR-497, members of the miR-15 family with the same seed sequence [105]. Highly expressed in quiescent SCs, miR-195/497 target transcripts of genes coding for cell cycle activators of the cyclin family (Ccnd2) and cell division cycle families (Cdc25a and Cdc25b), protecting quiescent SCs from cell cycle entry.
The imprinted gene encoding the H19 lncRNA is highly expressed in satellite cells [106]. Mice carrying a targeted deletion of the H19 gene present a 50% loss of SCs in adult muscles. This analysis suggests that the H19 lncRNA is probably essential to maintain the quiescent SC pool. Conditional deletion of the maternal H19-DMR (an epigenetic regulator that controls expression of H19) has previously been shown to reduce adult hematopoietic stem cell quiescence, by activating the Igf2-Igf1r (Insulin-like Growth factor 2–Igf-1 receptor) pathway [107]. In the placenta, H19 has been shown to also be a precursor of miR-675, which in turn targets Igf1r mRNAs [108], [109]. Therefore, the authors showed in hematopoietic stem cells that miR-675 is reduced in H19 deficient cells, and the Igf2-Igf1r pathway activated, leading to activation of stem cells [107]. It remains to determine whether the H19 lncRNA protects SCs from activation through a similar mechanism. In the adult tibialis muscle, Igf2 transcript levels were increased in H19 deficient muscles compared to control muscles but Igf1r was not modulated [106]. It would be very interesting to analyze the activation of the Igf2-Igf1r signaling pathway in purified quiescent vs activated satellite cells to better characterize the molecular mechanism by which H19 participates to the maintain of satellite cells in a quiescent state.
4.3. Satellite cell activation and early differentiation
Upon injury, satellite cells are activated, which is mediated in part by the expression of the muscle specific transcription factors, MyoD and Myf5 [110], [111], [112], [113], [114]. A genome wide analysis comparing proliferating vs. quiescent human satellite cells in culture showed a global up-regulation of miRNAs in proliferating SCs [115], suggesting that miRNAs might be largely implicated in the activation of SCs and their subsequent proliferative step (Table 1). The involvement of non-coding RNAs in these steps of muscle regeneration is indeed well documented.
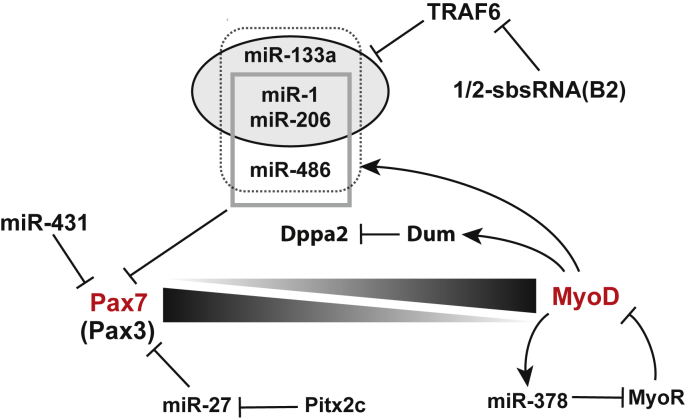
Once activated, satellite cell progeny follow a fate, which depends on MyoD expression. If SCs downregulate MyoD, they self-renew, and replenish the satellite cell pool. In the case SC progeny starts expressing MyoD proteins, Pax7 will be down-regulated, leading to the rapid expansion of muscle progenitor cells (myoblasts) to repair the damaged muscle fibers. In adult muscles, two subpopulations of SCs have been identified, depending on their level of Pax7 expression [116]. Pax7Lo cells are primed to differentiate in myogenic cells, whereas Pax7Hi cells better self-renew. Controlling Pax7 expression would therefore establish and maintain the two SC subpopulations. In miR-431 overexpressing mice, the Pax7Lo population is highly enriched showing the involvement of miR-431 in the control of the ratio between Pax7Hi and Pax7Lo SCs in adult muscles [117]. miR-431 is probably not involved in the self-renewal of these cells, since the satellite cell pool was not depleted after two rounds of injury in miR-431 transgenic mice. However, miR-431 promotes myogenic differentiation by targeting Pax7 transcripts in Pax7Lo SCs (Fig. 1). Muscle regeneration is indeed accelerated in miR-431 transgenic mice compared to control. Pax7 transcripts are targeted by several other miRNAs. Once MyoD is expressed, it directly activates the expression of the miR-486 host gene, Ank1.5, leading to the expression of miR-486 [118]. MyoD also activates expression of miR-206 [119], miR-1 and miR-133a [120], all muscle enriched miRNAs that are part of the myomiR family of short ncRNAs. miR-486, miR-206 and miR-1 target Pax7 transcripts and facilitate SC differentiation (Fig. 1) [118], [121]. miR-1, miR-133a and miR-206 expression are inhibited by an adaptor protein, which also functions as an E3 ubiquitin ligase, the Tumor necrosis factor (TNF) receptor-associated factor 6 (TRAF6) [122]. Absence of TRAF6 expression in SCs induces increased levels of these myomiRs leading to a precocious differentiation of these satellite cells. Although the mechanism by which TRAF6 inhibits miR1, miR-206 and miR-133a remains unknown, data suggest that the c-Jun/AP-1 transcription factor is probably involved in this regulation. By inhibiting miR-1, miR-206 and miR-133a expression, TRAF6 favors Pax7 expression in satellite cells. Traf6 mRNAs harbor a Staufen-binding site in their 3′ UTR [123]. These types of mRNAs are prone to Staufen 1 (Stau1)-mediated mRNA decay (SMD), which involves a base-pairing interaction between the mRNA 3′UTR containing short interspersed element (SINE) and SINE-containing lncRNAs. The duplex is stabilized by Stau1 and mRNA degraded. Traf6 mRNAs are targeted by the ½-sbsRNA(B2) lncRNA, which reduces the half-life of Traf6 mRNAs and therefore the levels of Traf6 proteins in cultured C2C12 cells. Whether this lncRNA is involved in activation of satellite cells remains to be determined.
Fig. 1.
Role of ncRNAs in the early differentiation of satellite cells. Upon muscle injury, Pax7 positive satellite cells are activated and differentiate into MyoD positive myogenic cells. Several non-coding RNAs have been described in the literature as regulating Pax7, Pax3 or MyoD expression in the early differentiation process of satellite cells.
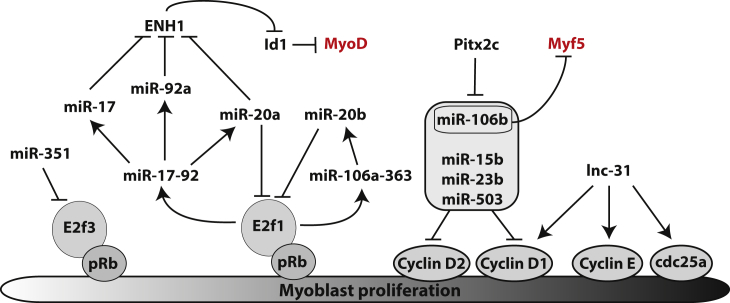
In addition, miR-27 expression is turned on in activated SCs, inducing in vitro and in vivo a repression of Pax3 by targeting the 3′UTR of Pax3 transcripts [96]. miR-27 is down-regulated by the homeobox transcription factor Pitx2c in muscle progenitors, maintaining cells in a pre-differentiated state [124] (Fig. 1). Independently from Pax3 and miR-27 regulation, Pitx2c also promotes myoblast proliferation, by down-regulating the expression of a miRNA set, comprising miR-15b, miR-23b, miR-106b, and miR-503 [125]. All four miRNAs directly target mRNAs coding for cyclin D1 and cyclin D2. Furthermore, miR-106b directly targets Myf5 transcripts in satellite cells [125] (Fig. 2). In total, Pitx2 promotes satellite cells to get a myogenic fate and to proliferate.
Fig. 2.
Control of the proliferative step of myoblasts by non-coding RNAs. During muscle regeneration, MyoD positive cells proliferate before differentiating into myotubes. This proliferative step is regulated by many non-coding RNAs targeting key cell cycle proteins and repressing the myogenic differentiation gene program.
The lncRNA, Dum (developmental pluripotency-associated 2 (Dppa2) Upstream binding Muscle lncRNA) was found to be induced during the early regeneration stage, when satellite cells become activated, proliferate and start to differentiate [126]. Its expression is induced by MyoD upon muscle differentiation. Dum transcripts silence in cis transcription of its far upstream neighbor gene, Dppa2, coding for a pluripotency regulator, promoting then early differentiation (Fig. 1). Dum inhibits Dppa2 expression by interacting and recruiting DNA methyltransferases (Dnmts) to Dppa2 promoter, leading to CpG site hypermethylation and gene silencing. Dum silencing in injured muscles decreased Pax7 and MyoD positive cells suggesting that Dum could be involved in SC activation, proliferation or self-renewal capacity.
4.4. Myoblast proliferation
A subset of satellite cells expresses MyoD and rapidly proliferates before differentiating into mature myofibers. Both proliferation and differentiation programs represent critical points in muscle regeneration. Several non-coding RNAs have been identified as regulator of the proliferative step of myoblasts (Tables 1 and 2).
Even if MyoD is expressed in proliferating myoblasts, its level remains relatively low compared to cells undergoing terminal differentiation. MyoD expression is indeed limited through Transforming Growth Factor - beta signals (TGFβ) [127]. TGFβ reduces MyoD expression in differentiating C2C12 cells by suppressing levels of miR-29 and miR-206. Both miRNAs target HDAC4 (histone deacetylase 4) transcripts (Fig. 3). An increase in HDAC4 expression has negative effects on muscle specific genes, limiting therefore MyoD expression.
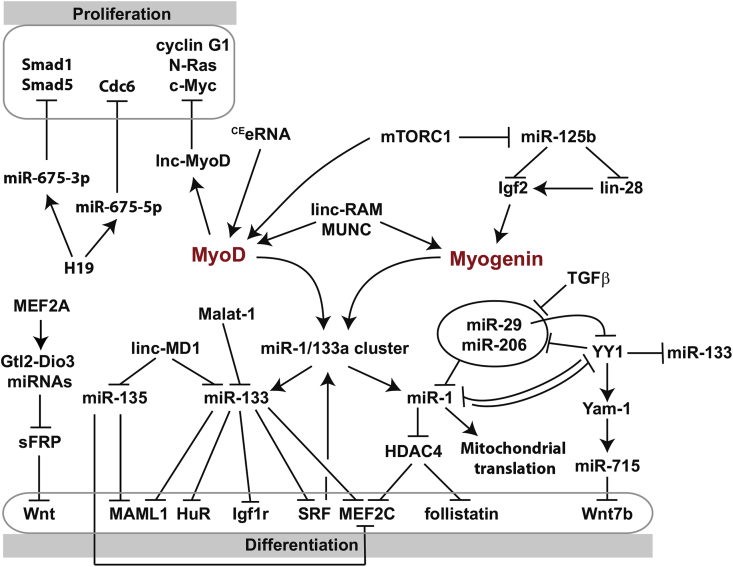
Fig. 3.
Role of ncRNAs in skeletal muscle differentiation. The committed myoblasts exit cell cycle to elongate and fuse into myotubes. Cell cycle withdrawal and myogenic differentiation are orchestrated by a network of non-coding RNAs controlling the muscle specific transcription factors, MyoD and myogenin, and different signaling pathways promoting myogenic differentiation.
Myoblast proliferation is promoted by many growth factors, released in the extracellular matrix, including Fibroblast Growth Factor 2 (FGF2). During muscle regeneration, FGF2 induces the expression of miR-29a, which leads to reduction of key elements of the basement membrane, as fibrillin-1 (Fbn1), sparc, collagen-IV (Col4a1), laminin gamma-1 (lamc1), nidogen-2 (Nid2) and heparane sulfate proteoglycan-2 (Hspg2) [128]. This could promote the dismantling of the basement membrane, promoting the release of growth factors from the basement membrane and facilitating their access to their receptors.
MiR-351 is transiently expressed in vitro by proliferating myoblasts and in vivo during muscle regeneration [84]. In vitro, miR-351 is required for myoblast proliferation and would protect them from apoptosis as myoblasts start differentiating. The cell cycle is controlled by a transcriptional network involving the retinoblastoma protein (pRb). miR-351 would control the cell cycle of myoblasts by targeting transcripts coding for the E2F3 transcription factor, which directly interacts with pRb [84] (Fig. 2). Another member of the E2F family, E2F1, activates the expression of two miRNA clusters, miR-17-92 and miR-106a-363, in proliferating myoblasts [129], [130]. The three miRNAs produced by miR-17-92 (miR-17, miR-20a and miR-92a) target the 3′UTR of transcripts coding for enigma homolog 1 (ENH1). This scaffold protein modulates the subcellular localization of a natural inhibitor of myogenic factors, Id1. When ENH1 is silenced, Id1 is accumulated in the nucleus, where it can repress myogenic differentiation gene expression [129], [131]. By this mechanism, E2F1 promotes myoblast proliferation (Fig. 2). However, a negative feedback loop exists in which miR-20a and miR-20b, produced respectively by the E2F1 transcriptional targets pri-miR-17-92 and pri-miR-106a-363, directly target E2F1 mRNAs [130] (Fig. 2).
Myoblast proliferation is also promoted by an antisense lncRNA, Sirt1 AS [132]. Sirt1 AS lncRNA is transcribed from the antisense strand of the gene coding for Sirt1, a NAD-dependent class III protein deacetylase, which promotes cell proliferation by inhibiting cell cycle inhibitor expression [133], [134]. When Sirt1 AS is overexpressed, cyclin B, D and E are overexpressed and C2C12 cell differentiation inhibited. Sirt1 AS lncRNA interacts with the 3′UTR of Sirt1 mRNA forming an RNA duplex to protect Sirt1 transcripts from degradation mediated by miR-34a. As Sirt1 AS, lnc-31 sustains myoblast proliferation and inhibit their differentiation, although the mechanism remains unknown [135]. Lnc-31 harbors precursor sequences for miR-31 and can be converted into miR-31 or into mature lnc-31. Lnc-31 is only expressed in proliferating myoblasts and is required to maintain the expression levels of cyclin D1 (Ccnd1), cyclin E (Ccne1) and Cdc25a cell cycle genes, independently from miR-31 (Fig. 2).
4.5. Muscle differentiation
Among the MyoD target genes activated during differentiation, myogenin expression is a key step in the commitment of myogenic progenitors to differentiate. Both MyoD and myogenin have been shown to be regulated by eRNAs, transcribed from two developmentally important MyoD enhancers, the core enhancer (CE) and the distal regulatory regions (DRR), located upstream of the MyoD coding sequence [136]. CEeRNA stimulates MyoD expression by facilitating in cis chromatin access to RNA polymerase II on the MyoD locus (Fig. 3). In contrast to CEeRNA, DRReRNA (also called MyoD upstream noncoding or MUNC) acts in trans to enhance MyoD binding to certain MyoD target sites, activating MyoD, myogenin, and Myh3 (myosin heavy chain 3) transcription [136], [137] (Fig. 3). Even if some MUNC dependent genes are not recognized as MyoD target genes, MUNC stimulates overall similar pro-myogenic genes as MyoD, to promote in vitro and in vivo muscle differentiation. MyoD directly activates the expression of another lncRNA, lncMyoD, encoded 30 kb upstream the MyoD1 gene [138]. As myoblast differentiate, lncMyoD is accumulated and binds Igf2 mRNA-binding proteins (IMPs). Its binding with IMP2 particularly competes out other mRNAs including transcripts coding for proliferation genes, such as c-Myc, Nras, and Ccng1 (coding for Cyclin G1) [138] (Fig. 3). Translation of these mRNAs is then inhibited and myoblasts are able to exit the cell cycle and therefore to differentiate. Other pro-proliferative genes, such as the DNA replication initiation factor, Cdc6 or the proliferation promoting Smad transcription factors are repressed by two miRNAs, miR-675-5p and miR-675-3p respectively [109]. These two miRNAs are produced by the first exon of the H19 gene and their expression is required for proper muscle differentiation in vitro and muscle regeneration in vivo (Fig. 3).
During muscle differentiation, MyoD protein levels and activity increase. The SRA (steroid receptor RNA activator) ncRNA is a co-activator of MyoD, which accumulates during differentiation of primary human muscle cells [139]. SRA serves as a scaffold for MyoD and two members of the DEAD-box family of RNA helicases, p68 and p72, to mediate transcriptional activation of MyoD-regulated genes [140]. p68/p72 and SRA are required for undifferentiated myoblasts to properly mature into myotubes. Alternative spliced variants of SRA transcripts encode for an SRA protein (SRAP), which is more abundant in myoblasts than in myotubes and prevent the SRA RNA-activation of the MyoD activity [139]. linc-RAM (linc-RNA Activator of Myogenesis) is a lncRNA, whose expression is regulated by MyoD during muscle differentiation [141]. Once expressed, this lncRNA directly interacts with MyoD and enhances MyoD transcriptional activity by promoting the assembly of the epigenetic complex MyoD-Baf60c-Brg1 on regulatory elements of target genes, independently from the linc-RAM encoded micropeptide, myoregulin (see below, section 4.6) [141].
MyoD activity is also modulated by its natural inhibitor, MyoR, a bHLH protein, which binds E-proteins directly on MyoD target DNA sequences [142]. A feed forward loop has been identified where MyoD down-regulates its own inhibitor by activating the expression of miR-378, which targets MyoR during myoblast differentiation [143], leading to a progressive up-regulation of MyoD-responsive target genes. Among the MyoD target genes, the lncRNA, linc-YY1, is generated from the opposite strand of the transcription factor Yin Yang 1 (YY1) gene and transcribed from YY1 promoter [144]. Linc-YY1 binds YY1 to promote myogenic differentiation and muscle regeneration. In proliferating myoblasts, YY1 is activated by NF-κB and recruits the PRC2 histone methyltransferase Ezh2, to silence transcription of its target genes, such as Myosin Heavy chain (MyHC), but also miR-29, miR-1, miR-133, and miR-206. miR-29 and miR-1 are able, in turn, to target their repressor YY1 [145], [146] (Fig. 3). As myoblasts undergo differentiation, YY1 is inhibited by miR-34c [147] and MyoD activates linc-YY1 expression, which could destabilize the YY1/PRC2 complexes, leading to the activation of these pro-differentiation genes [144]. In addition to its function as a gene repressor, YY1 also activates many genes among which 63 potential lincRNAs, named Yam (YY1 associated muscle lincRNAs) [148]. Yam-1 is activated by YY1 in proliferating C2C12 cells, as well as in the first steps of muscle regeneration. Yam-1 activates in cis miR-715 expression, which in turn directly targets Wnt7b transcripts to inhibit myogenic differentiation (Fig. 3). Other Yams regulate myogenic differentiation, but their specific mechanism should be determined. As Yam-1, Yam-3 promotes the expansion of proliferating myoblasts, whereas Yam-2 and Yam-4 enhance early myogenic differentiation [148].
The Igf signaling, and particularly Igf2, is essential for efficient muscle differentiation, in part by up-regulating myogenin expression [149]. During myoblast differentiation, the mammalian target of rapamycin (mTOR) signaling pathway is also activated, that induces a down-regulation of miR-125b [150]. Consequently, miR-125b targets, as Igf2 and Lin-28 transcripts, are released [150], [151] (Fig. 3). Lin-28 is a small RNA binding protein, which increases efficiency of target mRNAs translation, as Igf2 [152]. Therefore, Igf2 transcripts are more abundant and their translation is more efficient, thus increasing Igf2 protein synthesis and probably reinforcing myogenin expression (Fig. 3). Myogenin up-regulation activates in parallel a negative feedback loop, by up-regulating miR-133 expression [149]. MiR-133 directly represses Igf1r expression, modulating then the Igf signaling pathway.
During myoblast differentiation, the competing endogenous lncRNA Malat1 (Metastasis-associated lung adenocarcinoma transcript 1) is up-regulated [153] and acts as a sponge for miR-133, preventing miR-133 from inhibiting its target mRNAs, such as SRF (serum response factor) [154] (Fig. 3). SRF is therefore expressed and able to promote terminal differentiation of the muscle progenitor cells. miR-133a is derived from the same miRNA polycistron as miR-1. Expression of this polycistron is controlled by SRF [155], but also by the mTOR complex 1 (mTORC1) signaling through MyoD [156] (Fig. 3). In contrast to miR-133, miR-1 targets the 3′UTR of mRNA coding for HDAC4. HDAC4 interacts with the transcription factor MEF2C (myocyte-specific enhancer factor 2C) to repress MEF2C target gene expression [157], [158]. Therefore miR-1 promotes myogenic differentiation, by releasing MEF2C from HDAC4 (Fig. 3). Repression of HDAC4 by miR-1 also leads to the expression of follistatin, which promotes the fusion of myocytes in vitro and in vivo [156] (Fig. 3). If miR-1 represses its target transcripts in cytoplasm of differentiating myoblasts, Zhang et al. described an unexpected mechanism, in which miR-1 enters the mitochondria to stimulate mitochondrial translation of multiple mtDNA-encoded transcripts during muscle differentiation [159] (Fig. 3). By this mechanism, miR-1 enhances protein synthesis and ATP production in the mitochondria of differentiated muscle cells, without any increase in mtDNA copy number or transcription.
miR-133 and miR-135 are both targeted by a competing endogenous lncRNA, linc-MD1 (long intergenic noncoding RNA-muscle differentiation 1) [160]. The pro-myogenic transcription factors MEF2C and MAML1 were identified as two natural targets of miR-133 and miR-135. Linc-MD1 would therefore activate the expression of these transcription factors, by sponging miR-133 and miR-135 (Fig. 3). As an alternative to its “sponge” function, linc-MD1 transcripts could be a source of miR-133b, since it contains a pri-miRNA-133b sequence, which could be cleaved by Drosha [160]. The balance between generating or sponging miR-133b is controlled by the RNA binding HuR protein (Human antigen R), whose transcripts are targeted by miR-133 [161]. HuR proteins associate with linc-MD1 and protect the lncRNA from Drosha cleavage and then repress miR-133b synthesis from linc-MD1. On the other hand, HuR facilitates the interaction between linc-MD1 and miR-133/135, enhancing the sponge activity of linc-MD1 [161].
Another member of the MEF2 family of transcription factors, MEF2A, directly activates the expression of a cluster of miRNAs, called Gtl2-Dio3 [162]. More than 40 miRNAs are expressed from this cluster. Many of them target mRNAs coding for secreted Frizzled-related proteins (sFRPs) 1, 2 and 4 members of inhibitors of the canonical Wnt signaling pathway. Two miRNAs within the Gtl2-Dio3 cluster, miR-410 and miR-433, directly inhibit Sfrp2 expression [162]. MEF2A therefore activates the Wnt signaling pathway during muscle regeneration by repressing Wnt inhibitors via a series of miRNAs (Fig. 3).
4.6. Maturation of regenerating myofibers
The regenerative process is complete when injured myofibers rescue their functional performance and contractile apparatus, which is efficient when myofibers are effectively innervated. The role of the nerve in controlling maturation of myofibers can be monitored by the expression of MyHC isoforms. Nascent regenerating myotubes initially express developmental MyHC forms, as embryonic or neonatal MyHC, whereas maturating myofibers express adult fast and slow MyHC forms [163]. Expression of embryonic MyHC (coded by the MYH3 gene) is protected in newly regenerating myofibers by a miRNA, miR-501, expressed from an intron of isoform-2 of the Clcn5 (chloride channel 5) gene [164]. Inhibition of miR-501 in regenerating skeletal muscle inhibits appearance of embryonic MyHC and to a lesser extent adult myosin isoforms. MiR-501 directly represses mRNAs coding for gigaxonin, an E3 ligase adaptor, targeting proteins, such as MyHC-emb, for degradation by the proteasome [164]. A family of miRNA, including miR-208a/b and miR-499, are all intronic miRNAs located and co-transcribed with different MYH genes. MiR-208a is processed from the intronic RNA of the cardiac specific MYH6 gene, whereas miR-208b and miR-499 are processed from introns in MYH7 and MYH7b genes, respectively and are both expressed in slow-type skeletal muscle fibers [165]. Loss and gain of function analyses revealed that miR-208b and miR-499 redundantly reinforce the slow-muscle gene expression program by directly repressing translation of the transcriptional repressors Sox6, Purβ, Sp3 and HP-1β. On the other hand, slow genes are silenced in fast myofibers by a lncRNA identified in the cluster of fast MYH genes [166]. Linc-MYH is transcribed from the same enhancer as fast MYH genes and is required for full expression of fast genes [166]. The mechanism by which linc-MYH regulates expression of these fiber-type specific genes remains to be determined. mRNAs coding for all three adult fast MyHC (MyHC IIa, IIx and IIb) have been shown to be directly repressed by miR-23a [167]. Since miR-23a is highly expressed in the slow-twitch soleus muscle, compared to the fast-twitch plantaris muscle [168], we could argue that miR-23a could protect slow fibers from expression of fast fiber specific genes. Despite its role in repressing expression of fast MyHC, miR-23a also regulates fiber growth, by protecting cells from atrophy. One of the hallmarks of muscle atrophy is protein degradation through activation of the ubiquitin-proteasome pathway. Two E3 ubiquitin ligases appear to be essential for accelerated muscle protein loss in muscle atrophy: MAFbx/atrogin-1 (muscle atrophy F-box/muscle-specific ubiquitin E3-ligases atrophy gene-1) and MuRF1 (muscle ring-finger protein 1). The 3′UTR of mRNAs coding for these two E3 ubiquitin ligases are targeted by miR-23a, resulting in resistance to muscle atrophy when overexpressed in myotubes and myofibers [168].
Very recent papers demonstrate that a subset of transcripts annotated as lncRNAs may encode 'hidden' polypeptides. This is the case of LINC00961 which encodes a lysosomal type I transmembrane polypeptide, SPAR (Small regulatory Polypeptide of Amino acids Response) [169]. This peptide is localized to the late endosome/lysosome to negatively regulate mTORC1 activation. Upon acute muscle injury, LINC00961 is down-regulated in skeletal muscles, enabling efficient activation of mTORC1 and promoting muscle regeneration.
Muscle contraction depends on release of Ca2+ from the sarco/endoplasmic reticulum (S/ER), the main source of intracellular calcium, and reuptake by the S/ER Ca2+ Adenosine triphosphatase (SERCA) pump. Four functional polypeptides encoded by lncRNAs have been reported to modulate calcium handling into muscle cells. Myoregulin (MLN) is a micropeptide encoded by an annotated lncRNA, recently called linc-RAM (LINC00948 in humans and 2310015B20Rik in mice) [39], [141]. As phospholamban or sarcolipin proteins, MLN interacts with SERCA to inhibit its activity. In contrast to phospholamban and sarcolipin which are only expressed in slow muscles, MLN inhibits the SERCA pump in all skeletal muscles. In only slow muscles, the inhibitory effect of MLN, phospholamban and sarcolipin peptides on the SERCA pump activity is counteracted by another micropeptide, DWORF (Dwarf Open Reading Frame), encoded by a “lncRNA” (NONMMUG026737 in mice and LOC100507537 in humans) [38]. Very recently, two new micropeptides encoded by so called lncRNAs have been identified: endoregulin (ELN), encoded by the lncRNA 1110017F19Rik/SMIM6 and another-regulin (ALN), encoded by the 1810037I17Rik lncRNA [40]. As MLN, they both bind and inhibit the SERCA pump activity, but their contribution in muscle cell contraction is lower than MLN since they are weakly expressed in adult muscles. The involvement of these described SERCA-inhibitory micropeptides remains to be determined in a context of muscle regeneration. Even if these lncRNAs code for micropeptides, we cannot exclude that they could play additional functions as ncRNAs.
5. Conclusions
Increasing evidence indicate that non-coding RNAs participate to the regulation of multiple steps of skeletal muscle regeneration, from the inflammatory response to the maturation of the regenerating myofibers (Tables 1 and 2). However, the understanding of the different mechanisms by which these ncRNAs modulate genome expression is in its infancy. If the cytoplasmic function of miRNAs in regulating gene expression seems to be well characterized, a recent paper described an unconventional function, where miR-1 stimulates translation of its target transcripts in mitochondria [159]. In addition, growing data indicate that miRNAs also have specific nuclear functions, among them a miRNA-guided transcriptional control of gene expression [31]. In contrast to miRNAs, the role of lncRNAs in myogenesis has just begun to emerge and the molecular mechanisms through which most of the lncRNAs modulate gene expression and cellular functions is still poorly understood. Future investigations will likely uncover new molecular mechanisms for ncRNAs and characterize new ncRNAs involved in the different steps of skeletal muscle regeneration. Muscle regeneration is compromised in many muscular dystrophies. Increasing data describe an up-or down-regulation of ncRNAs expression in pathological tissues, as dystrophic tissues [170]. Future studies will likely uncover to what extent dysregulation of ncRNAs would impact muscle waste and regeneration in muscle dystrophies. Examining their interaction with other ncRNAs and proteins may provide very interesting mechanistic insights into their mode of action in healthy and pathological myogenesis.
Acknowledgements
We would like to thank Suzie Lefebvre (Inserm UMRS 1124) for helpful discussions and Mario Pende (Inserm U1151) for critical reading of the manuscript. Work in the lab has been supported by Université Paris Descartes, Inserm (France) and grants from the Association Française contre les Myopathies [AFM18802].
References
- 1.Ciciliot S., Schiaffino S. Regeneration of mammalian skeletal muscle Basic mechanisms and clinical implications. Curr. Pharm. Des. 2010;16:906–914. doi: 10.2174/138161210790883453. [DOI] [PubMed] [Google Scholar]
- 2.Lepper C., Partridge T.A., Fan C.-M. An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development. 2011;138:3639–3646. doi: 10.1242/dev.067595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Webster M.T., Manor U., Lippincott-Schwartz J., Fan C.-M. Intravital imaging reveals ghost fibers as architectural units guiding myogenic progenitors during regeneration. Cell Stem Cell. 2016;18:243–252. doi: 10.1016/j.stem.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dumont N.A., Wang Y.X., Rudnicki M.A. Intrinsic and extrinsic mechanisms regulating satellite cell function. Development. 2015;142:1572–1581. doi: 10.1242/dev.114223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gurevich D.B., Nguyen P.D., Siegel A.L., Ehrlich O.V., Sonntag C., Phan J.M.N., Berger S., Ratnayake D., Hersey L., Berger J., Verkade H., Hall T.E., Currie P.D. Asymmetric division of clonal muscle stem cells coordinates muscle regeneration in vivo. Science. 2016;353:aad9969. doi: 10.1126/science.aad9969. [DOI] [PubMed] [Google Scholar]
- 6.Gayraud-Morel B., Chrétien F., Tajbakhsh S. Skeletal muscle as a paradigm for regenerative biology and medicine. Regen. Med. 2009;4:293–319. doi: 10.2217/17460751.4.2.293. [DOI] [PubMed] [Google Scholar]
- 7.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Funke R., Gage D., Harris K., Heaford A., Howland J., Kann L., Lehoczky J., LeVine R., McEwan P., McKernan K. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 8.Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Kodzius R., Shimokawa K., Bajic V.B., Brenner S.E., Batalov S., Forrest A.R.R., Zavolan M., Davis M.J., Wilming L.G., Aidinis V. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 9.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quan M., Chen J., Zhang D. Exploring the secrets of long noncoding RNAs. Int. J. Mol. Sci. 2015;16:5467–5496. doi: 10.3390/ijms16035467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Catalanotto C., Cogoni C., Zardo G. MicroRNA in control of gene expression: an overview of nuclear functions. Int. J. Mol. Sci. 2016;17 doi: 10.3390/ijms17101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai L., Chang H., Fang Y., Li G. A comprehensive characterization of the function of LincRNAs in transcriptional regulation through long-range chromatin interactions. Sci. Rep. 2016;6:36572. doi: 10.1038/srep36572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sohi G., Dilworth F.J. Noncoding RNAs as epigenetic mediators of skeletal muscle regeneration. FEBS J. 2015;282:1630–1646. doi: 10.1111/febs.13170. [DOI] [PubMed] [Google Scholar]
- 14.Horak M., Novak J., Bienertova-Vasku J. Muscle-specific microRNAs in skeletal muscle development. Dev. Biol. 2016;410:1–13. doi: 10.1016/j.ydbio.2015.12.013. [DOI] [PubMed] [Google Scholar]
- 15.Simionescu-Bankston A., Kumar A. Noncoding RNAs in the regulation of skeletal muscle biology in health and disease. J. Mol. Med. 2016;94:853–866. doi: 10.1007/s00109-016-1443-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kusakabe R., Inoue K. Developmental regulation and evolution of muscle-specific microRNAs, Semin. Cell Dev. Biol. 2015;47–48:9–16. doi: 10.1016/j.semcdb.2015.10.020. [DOI] [PubMed] [Google Scholar]
- 17.Liu N., Bassel-Duby R. Regulation of skeletal muscle development and disease by microRNAs. In: Brand-Saberi B., editor. Vertebrate Myogenesis. Springer; Berlin Heidelberg: 2015. pp. 165–190. [DOI] [PubMed] [Google Scholar]
- 18.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 19.Yi R., Qin Y., Macara I.G., Cullen B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lund E., Güttinger S., Calado A., Dahlberg J.E., Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 21.Knight S.W., Bass B.L. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grishok A., Pasquinelli A.E., Conte D., Li N., Parrish S., Ha I., Baillie D.L., Fire A., Ruvkun G., Mello C.C. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 23.Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi H., Tomari Y. RISC assembly: coordination between small RNAs and Argonaute proteins. Biochim. Biophys. Acta. 2016;1859:71–81. doi: 10.1016/j.bbagrm.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 25.Chendrimada T.P., Gregory R.I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berezikov E., Chung W.-J., Willis J., Cuppen E., Lai E.C. Mammalian mirtron genes. Mol. Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okamura K., Hagen J.W., Duan H., Tyler D.M., Lai E.C. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hansen T.B., Venø M.T., Jensen T.I., Schaefer A., Damgaard C.K., Kjems J. Argonaute-associated short introns are a novel class of gene regulators. Nat. Commun. 2016;7:11538. doi: 10.1038/ncomms11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meister G., Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 30.Wilczynska A., Bushell M. The complexity of miRNA-mediated repression. Cell Death Differ. 2015;22:22–33. doi: 10.1038/cdd.2014.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalantari R., Chiang C.-M., Corey D.R. Regulation of mammalian transcription and splicing by Nuclear RNAi. Nucleic Acids Res. 2016;44:524–537. doi: 10.1093/nar/gkv1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guttman M., Amit I., Garber M., French C., Lin M.F., Feldser D., Huarte M., Zuk O., Carey B.W., Cassady J.P., Cabili M.N., Jaenisch R., Mikkelsen T.S., Jacks T., Hacohen N., Bernstein B.E., Kellis M., Regev A., Rinn J.L., Lander E.S. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G., Lagarde J., Veeravalli L., Ruan X., Ruan Y., Lassmann T., Carninci P., Brown J.B., Lipovich L., Gonzalez J.M., Thomas M. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bunch H., Lawney B.P., Burkholder A., Ma D., Zheng X., Motola S., Fargo D.C., Levine S.S., Wang Y.E., Hu G. RNA polymerase II promoter-proximal pausing in mammalian long non-coding genes. Genomics. 2016;108:64–77. doi: 10.1016/j.ygeno.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruiz-Orera J., Messeguer X., Subirana J.A., Alba M.M. Long non-coding RNAs as a source of new peptides. Elife. 2014;3:e03523. doi: 10.7554/eLife.03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frith M.C., Forrest A.R., Nourbakhsh E., Pang K.C., Kai C., Kawai J., Carninci P., Hayashizaki Y., Bailey T.L., Grimmond S.M. The abundance of short proteins in the mammalian proteome. PLoS Genet. 2006;2:e52. doi: 10.1371/journal.pgen.0020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andrews S.J., Rothnagel J.A. Emerging evidence for functional peptides encoded by short open reading frames. Nat. Rev. Genet. 2014;15:193–204. doi: 10.1038/nrg3520. [DOI] [PubMed] [Google Scholar]
- 38.Nelson B.R., Makarewich C.A., Anderson D.M., Winders B.R., Troupes C.D., Wu F., Reese A.L., McAnally J.R., Chen X., Kavalali E.T., Cannon S.C., Houser S.R., Bassel-Duby R., Olson E.N. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science. 2016;351:271–275. doi: 10.1126/science.aad4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson D.M., Anderson K.M., Chang C.-L., Makarewich C.A., Nelson B.R., McAnally J.R., Kasaragod P., Shelton J.M., Liou J., Bassel-Duby R., Olson E.N. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson D.M., Makarewich C.A., Anderson K.M., Shelton J.M., Bezprozvannaya S., Bassel-Duby R., Olson E.N. Widespread control of calcium signaling by a family of SERCA-inhibiting micropeptides. Sci. Signal. 2016;9:ra119. doi: 10.1126/scisignal.aaj1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.S.A. Bhat, S.M. Ahmad, P.T. Mumtaz, A.A. Malik, M.A. Dar, U. Urwat, R.A. Shah, N.A. Ganai, Long non-coding RNAs: mechanism of action and functional utility, Non-Coding RNA Res., (n.d.). [DOI] [PMC free article] [PubMed]
- 42.Schmitz S.U., Grote P., Herrmann B.G. Mechanisms of long noncoding RNA function in development and disease. Cell. Mol. Life Sci. 2016;73:2491–2509. doi: 10.1007/s00018-016-2174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uesaka M., Nishimura O., Go Y., Nakashima K., Agata K., Imamura T. Bidirectional promoters are the major source of gene activation-associated non-coding RNAs in mammals. BMC Genomics. 2014;15:35. doi: 10.1186/1471-2164-15-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tomikawa J., Shimokawa H., Uesaka M., Yamamoto N., Mori Y., Tsukamura H., Maeda K., Imamura T. Single-stranded noncoding RNAs mediate local epigenetic alterations at gene promoters in rat cell lines. J. Biol. Chem. 2011;286:34788–34799. doi: 10.1074/jbc.M111.275750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Imamura T., Yamamoto S., Ohgane J., Hattori N., Tanaka S., Shiota K. Non-coding RNA directed DNA demethylation of Sphk1 CpG island. Biochem. Biophys. Res. Commun. 2004;322:593–600. doi: 10.1016/j.bbrc.2004.07.159. [DOI] [PubMed] [Google Scholar]
- 46.Martianov I., Ramadass A., Serra Barros A., Chow N., Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 47.Heintzman N.D., Ren B. Finding distal regulatory elements in the human genome. Curr. Opin. Genet. Dev. 2009;19:541–549. doi: 10.1016/j.gde.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lai F., Orom U.A., Cesaroni M., Beringer M., Taatjes D.J., Blobel G.A., Shiekhattar R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ørom U.A., Derrien T., Beringer M., Gumireddy K., Gardini A., Bussotti G., Lai F., Zytnicki M., Notredame C., Huang Q., Guigo R., Shiekhattar R. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prensner J.R., Iyer M.K., Sahu A., Asangani I.A., Cao Q., Patel L., Vergara I.A., Davicioni E., Erho N., Ghadessi M., Jenkins R.B., Triche T.J., Malik R., Bedenis R., McGregor N., Ma T., Chen W., Han S., Jing X., Cao X. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat. Genet. 2013;45:1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han P., Li W., Lin C.-H., Yang J., Shang C., Nurnberg S.T., Jin K.K., Xu W., Lin C.-Y., Lin C.-J., Xiong Y., Chien H.-C., Zhou B., Ashley E., Bernstein D., Chen P.-S., Chen H.-S.V., Quertermous T., Chang C.-P. A long noncoding RNA protects the heart from pathological hypertrophy. Nature. 2014;514:102–106. doi: 10.1038/nature13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Y., He L., Du Y., Zhu P., Huang G., Luo J., Yan X., Ye B., Li C., Xia P., Zhang G., Tian Y., Chen R., Fan Z. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 2015;16:413–425. doi: 10.1016/j.stem.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 53.Cajigas I., Leib D.E., Cochrane J., Luo H., Swyter K.R., Chen S., Clark B.S., Thompson J., Yates J.R., Kingston R.E., Kohtz J.D. Evf2 lncRNA/BRG1/DLX1 interactions reveal RNA-dependent inhibition of chromatin remodeling. Development. 2015;142:2641–2652. doi: 10.1242/dev.126318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao J., Ohsumi T.K., Kung J.T., Ogawa Y., Grau D.J., Sarma K., Song J.J., Kingston R.E., Borowsky M., Lee J.T. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li L., Liu B., Wapinski O.L., Tsai M.-C., Qu K., Zhang J., Carlson J.C., Lin M., Fang F., Gupta R.A., Helms J.A., Chang H.Y. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep. 2013;5:3–12. doi: 10.1016/j.celrep.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mariner P.D., Walters R.D., Espinoza C.A., Drullinger L.F., Wagner S.D., Kugel J.F., Goodrich J.A. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol. Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 57.Allen T.A., Von Kaenel S., Goodrich J.A., Kugel J.F. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat. Struct. Mol. Biol. 2004;11:816–821. doi: 10.1038/nsmb813. [DOI] [PubMed] [Google Scholar]
- 58.Jiang W., Liu Y., Liu R., Zhang K., Zhang Y. The lncRNA DEANR1 facilitates human endoderm differentiation by activating FOXA2 expression. Cell Rep. 2015;11:137–148. doi: 10.1016/j.celrep.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang P., Xue Y., Han Y., Lin L., Wu C., Xu S., Jiang Z., Xu J., Liu Q., Cao X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344:310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 60.Rapicavoli N.A., Qu K., Zhang J., Mikhail M., Laberge R.-M., Chang H.Y. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife. 2013;2:e00762. doi: 10.7554/eLife.00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramos A.D., Andersen R.E., Liu S.J., Nowakowski T.J., Hong S.J., Gertz C.C., Salinas R.D., Zarabi H., Kriegstein A.R., Lim D.A. The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell. 2015;16:439–447. doi: 10.1016/j.stem.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tripathi V., Shen Z., Chakraborty A., Giri S., Freier S.M., Wu X., Zhang Y., Gorospe M., Prasanth S.G., Lal A., Prasanth K.V. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013;9:e1003368. doi: 10.1371/journal.pgen.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tripathi V., Ellis J.D., Shen Z., Song D.Y., Pan Q., Watt A.T., Freier S.M., Bennett C.F., Sharma A., Bubulya P.A., Blencowe B.J., Prasanth S.G., Prasanth K.V. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ishizuka A., Hasegawa Y., Ishida K., Yanaka K., Nakagawa S. Formation of nuclear bodies by the lncRNA Gomafu-associating proteins Celf3 and SF1. Genes cells. 2014;19:704–721. doi: 10.1111/gtc.12169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Quinodoz S., Guttman M. Long noncoding RNAs: an emerging link between gene regulation and nuclear organization. Trends Cell Biol. 2014;24:651–663. doi: 10.1016/j.tcb.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zucchelli S., Fasolo F., Russo R., Cimatti L., Patrucco L., Takahashi H., Jones M.H., Santoro C., Sblattero D., Cotella D., Persichetti F., Carninci P., Gustincich S. SINEUPs are modular antisense long non-coding RNAs that increase synthesis of target proteins in cells. Front. Cell Neurosci. 2015;9:174. doi: 10.3389/fncel.2015.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Röther S., Meister G. Small RNAs derived from longer non-coding RNAs. Biochimie. 2011;93:1905–1915. doi: 10.1016/j.biochi.2011.07.032. [DOI] [PubMed] [Google Scholar]
- 69.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 70.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Denzler R., Agarwal V., Stefano J., Bartel D.P., Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol. Cell. 2014;54:766–776. doi: 10.1016/j.molcel.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Denzler R., McGeary S.E., Title A.C., Agarwal V., Bartel D.P., Stoffel M. Impact of MicroRNA levels, target-site complementarity, and cooperativity on competing endogenous RNA-regulated gene expression. Mol. Cell. 2016;64:565–579. doi: 10.1016/j.molcel.2016.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bosson A.D., Zamudio J.R., Sharp P.A. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Mol. Cell. 2014;56:347–359. doi: 10.1016/j.molcel.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lefaucheur J.P., Sébille A. The cellular events of injured muscle regeneration depend on the nature of the injury. Neuromuscul. Disord. 1995;5:501–509. doi: 10.1016/0960-8966(95)00012-c. [DOI] [PubMed] [Google Scholar]
- 75.Roberts T.C., Godfrey C., McClorey G., Vader P., Briggs D., Gardiner C., Aoki Y., Sargent I., Morgan J.E., Wood M.J.A. Extracellular microRNAs are dynamic non-vesicular biomarkers of muscle turnover. Nucleic Acids Res. 2013;41:9500–9513. doi: 10.1093/nar/gkt724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang K., Zhang S., Weber J., Baxter D., Galas D.J. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38:7248–7259. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vickers K.C., Palmisano B.T., Shoucri B.M., Shamburek R.D., Remaley A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J.J., Lötvall J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 79.Zernecke A., Bidzhekov K., Noels H., Shagdarsuren E., Gan L., Denecke B., Hristov M., Köppel T., Jahantigh M.N., Lutgens E., Wang S., Olson E.N., Schober A., Weber C. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci. Signal. 2009;2:ra81. doi: 10.1126/scisignal.2000610. [DOI] [PubMed] [Google Scholar]
- 80.Nakamura Y., Miyaki S., Ishitobi H., Matsuyama S., Nakasa T., Kamei N., Akimoto T., Higashi Y., Ochi M. Mesenchymal-stem-cell-derived exosomes accelerate skeletal muscle regeneration. FEBS Lett. 2015;589:1257–1265. doi: 10.1016/j.febslet.2015.03.031. [DOI] [PubMed] [Google Scholar]
- 81.Tidball J.G., Villalta S.A. Regulatory interactions between muscle and the immune system during muscle regeneration. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2010;298:R1173–R1187. doi: 10.1152/ajpregu.00735.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mills C.D., Kincaid K., Alt J.M., Heilman M.J., Hill A.M. M-1/M-2 macrophages and the Th1/Th2 paradigm. J. Immunol. 2000;164:6166–6173. doi: 10.4049/jimmunol.164.12.6166. [DOI] [PubMed] [Google Scholar]
- 83.Arnold L., Henry A., Poron F., Baba-Amer Y., van Rooijen N., Plonquet A., Gherardi R.K., Chazaud B. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J. Exp. Med. 2007;204:1057–1069. doi: 10.1084/jem.20070075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen Y., Melton D.W., Gelfond J.A.L., McManus L.M., Shireman P.K. MiR-351 transiently increases during muscle regeneration and promotes progenitor cell proliferation and survival upon differentiation. Physiol. Genomics. 2012;44:1042–1051. doi: 10.1152/physiolgenomics.00052.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu G., Friggeri A., Yang Y., Park Y.-J., Tsuruta Y., Abraham E. miR-147, a microRNA that is induced upon Toll-like receptor stimulation, regulates murine macrophage inflammatory responses. Proc. Natl. Acad. Sci. U. S. A. 2009;106:15819–15824. doi: 10.1073/pnas.0901216106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Johnnidis J.B., Harris M.H., Wheeler R.T., Stehling-Sun S., Lam M.H., Kirak O., Brummelkamp T.R., Fleming M.D., Camargo F.D. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 87.Nie M., Liu J., Yang Q., Seok H.Y., Hu X., Deng Z.-L., Wang D.-Z. MicroRNA-155 facilitates skeletal muscle regeneration by balancing pro- and anti-inflammatory macrophages. Cell Death Dis. 2016;7:e2261. doi: 10.1038/cddis.2016.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Uezumi A., Fukada S., Yamamoto N., Takeda S., Tsuchida K. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat. Cell Biol. 2010;12:143–152. doi: 10.1038/ncb2014. [DOI] [PubMed] [Google Scholar]
- 89.Heredia J.E., Mukundan L., Chen F.M., Mueller A.A., Deo R.C., Locksley R.M., Rando T.A., Chawla A. Type 2 innate signals stimulate fibro/adipogenic progenitors to facilitate muscle regeneration. Cell. 2013;153:376–388. doi: 10.1016/j.cell.2013.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cordani N., Pisa V., Pozzi L., Sciorati C., Clementi E. Nitric oxide controls fat deposition in dystrophic skeletal muscle by regulating fibro-adipogenic precursor differentiation. Stem Cells. 2014;32:874–885. doi: 10.1002/stem.1587. [DOI] [PubMed] [Google Scholar]
- 91.Karbiener M., Fischer C., Nowitsch S., Opriessnig P., Papak C., Ailhaud G., Dani C., Amri E.-Z., Scheideler M. microRNA miR-27b impairs human adipocyte differentiation and targets PPARgamma. Biochem. Biophys. Res. Commun. 2009;390:247–251. doi: 10.1016/j.bbrc.2009.09.098. [DOI] [PubMed] [Google Scholar]
- 92.Burzyn D., Kuswanto W., Kolodin D., Shadrach J.L., Cerletti M., Jang Y., Sefik E., Tan T.G., Wagers A.J., Benoist C., Mathis D. A special population of regulatory T cells potentiates muscle repair. Cell. 2013;155:1282–1295. doi: 10.1016/j.cell.2013.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schiaffino S., Pereira M.G., Ciciliot S., Rovere-Querini P. Regulatory T cells and skeletal muscle regeneration. FEBS J. 2016;284:517–524. doi: 10.1111/febs.13827. [DOI] [PubMed] [Google Scholar]
- 94.Cobb B.S., Hertweck A., Smith J., O'Connor E., Graf D., Cook T., Smale S.T., Sakaguchi S., Livesey F.J., Fisher A.G., Merkenschlager M. A role for Dicer in immune regulation. J. Exp. Med. 2006;203:2519–2527. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Seale P., Sabourin L.A., Girgis-Gabardo A., Mansouri A., Gruss P., Rudnicki M.A. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- 96.Crist C.G., Montarras D., Pallafacchina G., Rocancourt D., Cumano A., Conway S.J., Buckingham M. Muscle stem cell behavior is modified by microRNA-27 regulation of Pax3 expression. Proc. Natl. Acad. Sci. U. S. A. 2009;106:13383–13387. doi: 10.1073/pnas.0900210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Günther S., Kim J., Kostin S., Lepper C., Fan C.-M., Braun T. Myf5-positive satellite cells contribute to Pax7-dependent long-term maintenance of adult muscle stem cells. Cell Stem Cell. 2013;13:590–601. doi: 10.1016/j.stem.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.von Maltzahn J., Jones A.E., Parks R.J., Rudnicki M.A. Pax7 is critical for the normal function of satellite cells in adult skeletal muscle. Proc. Natl. Acad. Sci. U. S. A. 2013;110:16474–16479. doi: 10.1073/pnas.1307680110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.McKinnell I.W., Ishibashi J., Le Grand F., Punch V.G.J., Addicks G.C., Greenblatt J.F., Dilworth F.J., Rudnicki M.A. Pax7 activates myogenic genes by recruitment of a histone methyltransferase complex. Nat. Cell Biol. 2008;10:77–84. doi: 10.1038/ncb1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bajard L., Relaix F., Lagha M., Rocancourt D., Daubas P., Buckingham M.E. A novel genetic hierarchy functions during hypaxial myogenesis: Pax3 directly activates Myf5 in muscle progenitor cells in the limb. Genes Dev. 2006;20:2450–2464. doi: 10.1101/gad.382806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liu L., Cheung T.H., Charville G.W., Hurgo B.M.C., Leavitt T., Shih J., Brunet A., Rando T.A. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep. 2013;4:189–204. doi: 10.1016/j.celrep.2013.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Crist C.G., Montarras D., Buckingham M. Muscle satellite cells are primed for myogenesis but maintain quiescence with sequestration of Myf5 mRNA targeted by microRNA-31 in mRNP granules. Cell Stem Cell. 2012;11:118–126. doi: 10.1016/j.stem.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 103.Hausburg M.A., Doles J.D., Clement S.L., Cadwallader A.B., Hall M.N., Blackshear P.J., Lykke-Andersen J., Olwin B.B. Post-transcriptional regulation of satellite cell quiescence by TTP-mediated mRNA decay. Elife. 2015;4:e03390. doi: 10.7554/eLife.03390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cheung T.H., Quach N.L., Charville G.W., Liu L., Park L., Edalati A., Yoo B., Hoang P., Rando T.A. Maintenance of muscle stem-cell quiescence by microRNA-489. Nature. 2012;482:524–528. doi: 10.1038/nature10834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sato T., Yamamoto T., Sehara-Fujisawa A. miR-195/497 induce postnatal quiescence of skeletal muscle stem cells. Nat. Commun. 2014;5:4597. doi: 10.1038/ncomms5597. [DOI] [PubMed] [Google Scholar]
- 106.Martinet C., Monnier P., Louault Y., Benard M., Gabory A., Dandolo L. H19 controls reactivation of the imprinted gene network during muscle regeneration. Development. 2016;143:962–971. doi: 10.1242/dev.131771. [DOI] [PubMed] [Google Scholar]
- 107.Venkatraman A., He X.C., Thorvaldsen J.L., Sugimura R., Perry J.M., Tao F., Zhao M., Christenson M.K., Sanchez R., Yu J.Y., Peng L., Haug J.S., Paulson A., Li H., Zhong X., Clemens T.L., Bartolomei M.S., Li L. Maternal imprinting at the H19-Igf2 locus maintains adult haematopoietic stem cell quiescence. Nature. 2013;500:345–349. doi: 10.1038/nature12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Keniry A., Oxley D., Monnier P., Kyba M., Dandolo L., Smits G., Reik W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat. Cell Biol. 2012;14:659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dey B.K., Pfeifer K., Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 2014;28:491–501. doi: 10.1101/gad.234419.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yablonka-Reuveni Z., Rudnicki M.A., Rivera A.J., Primig M., Anderson J.E., Natanson P. The transition from proliferation to differentiation is delayed in satellite cells from mice lacking MyoD. Dev. Biol. 1999;210:440–455. doi: 10.1006/dbio.1999.9284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Megeney L.A., Kablar B., Garrett K., Anderson J.E., Rudnicki M.A. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 1996;10:1173–1183. doi: 10.1101/gad.10.10.1173. [DOI] [PubMed] [Google Scholar]
- 112.Gayraud-Morel B., Chrétien F., Flamant P., Gomès D., Zammit P.S., Tajbakhsh S. A role for the myogenic determination gene Myf5 in adult regenerative myogenesis. Dev. Biol. 2007;312:13–28. doi: 10.1016/j.ydbio.2007.08.059. [DOI] [PubMed] [Google Scholar]
- 113.Cooper R.N., Tajbakhsh S., Mouly V., Cossu G., Buckingham M., Butler-Browne G.S. In vivo satellite cell activation via Myf5 and MyoD in regenerating mouse skeletal muscle. J. Cell. Sci. 1999;112(Pt 17):2895–2901. doi: 10.1242/jcs.112.17.2895. [DOI] [PubMed] [Google Scholar]
- 114.Ustanina S., Carvajal J., Rigby P., Braun T. The myogenic factor Myf5 supports efficient skeletal muscle regeneration by enabling transient myoblast amplification. Stem Cells. 2007;25:2006–2016. doi: 10.1634/stemcells.2006-0736. [DOI] [PubMed] [Google Scholar]
- 115.Koning M., Werker P.M.N., van Luyn M.J.A., Krenning G., Harmsen M.C. A global downregulation of microRNAs occurs in human quiescent satellite cells during myogenesis. Differentiation. 2012;84:314–321. doi: 10.1016/j.diff.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 116.Rocheteau P., Gayraud-Morel B., Siegl-Cachedenier I., Blasco M.A., Tajbakhsh S. A subpopulation of adult skeletal muscle stem cells retains all template DNA strands after cell division. Cell. 2012;148:112–125. doi: 10.1016/j.cell.2011.11.049. [DOI] [PubMed] [Google Scholar]
- 117.Wu R., Li H., Zhai L., Zou X., Meng J., Zhong R., Li C., Wang H., Zhang Y., Zhu D. MicroRNA-431 accelerates muscle regeneration and ameliorates muscular dystrophy by targeting Pax7 in mice. Nat. Commun. 2015;6:7713. doi: 10.1038/ncomms8713. [DOI] [PubMed] [Google Scholar]
- 118.Dey B.K., Gagan J., Dutta A. miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Mol. Cell. Biol. 2011;31:203–214. doi: 10.1128/MCB.01009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rosenberg M.I., Georges S.A., Asawachaicharn A., Analau E., Tapscott S.J. MyoD inhibits Fstl1 and Utrn expression by inducing transcription of miR-206. J. Cell Biol. 2006;175:77–85. doi: 10.1083/jcb.200603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rao P.K., Kumar R.M., Farkhondeh M., Baskerville S., Lodish H.F. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc. Natl. Acad. Sci. U. S. A. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen J.-F., Tao Y., Li J., Deng Z., Yan Z., Xiao X., Wang D.-Z. microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J. Cell Biol. 2010;190:867–879. doi: 10.1083/jcb.200911036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hindi S.M., Kumar A. TRAF6 regulates satellite stem cell self-renewal and function during regenerative myogenesis. J. Clin. Invest. 2016;126:151–168. doi: 10.1172/JCI81655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang J., Gong C., Maquat L.E. Control of myogenesis by rodent SINE-containing lncRNAs. Genes Dev. 2013;27:793–804. doi: 10.1101/gad.212639.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lozano-Velasco E., Contreras A., Crist C., Hernández-Torres F., Franco D., Aránega A.E. Pitx2c modulates Pax3+/Pax7+ cell populations and regulates Pax3 expression by repressing miR27 expression during myogenesis. Dev. Biol. 2011;357:165–178. doi: 10.1016/j.ydbio.2011.06.039. [DOI] [PubMed] [Google Scholar]
- 125.Lozano-Velasco E., Vallejo D., Esteban F.J., Doherty C., Hernández-Torres F., Franco D., Aránega A.E. A pitx2-MicroRNA pathway modulates cell proliferation in myoblasts and skeletal-muscle satellite cells and promotes their commitment to a myogenic cell fate. Mol. Cell. Biol. 2015;35:2892–2909. doi: 10.1128/MCB.00536-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang L., Zhao Y., Bao X., Zhu X., Kwok Y.K.-Y., Sun K., Chen X., Huang Y., Jauch R., Esteban M.A., Sun H., Wang H. LncRNA Dum interacts with Dnmts to regulate Dppa2 expression during myogenic differentiation and muscle regeneration. Cell Res. 2015;25:335–350. doi: 10.1038/cr.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Winbanks C.E., Wang B., Beyer C., Koh P., White L., Kantharidis P., Gregorevic P. TGF-beta regulates miR-206 and miR-29 to control myogenic differentiation through regulation of HDAC4. J. Biol. Chem. 2011;286:13805–13814. doi: 10.1074/jbc.M110.192625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Galimov A., Merry T.L., Luca E., Rushing E.J., Mizbani A., Turcekova K., Hartung A., Croce C.M., Ristow M., Krützfeldt J. MicroRNA-29a in adult muscle stem cells controls skeletal muscle regeneration during injury and exercise downstream of fibroblast growth Factor-2. Stem Cells. 2016;34:768–780. doi: 10.1002/stem.2281. [DOI] [PubMed] [Google Scholar]
- 129.Qiu H., Liu N., Luo L., Zhong J., Tang Z., Kang K., Qu J., Peng W., Liu L., Li L., Gou D. MicroRNA-17-92 regulates myoblast proliferation and differentiation by targeting the ENH1/Id1 signaling axis. Cell Death Differ. 2016;23:1658–1669. doi: 10.1038/cdd.2016.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Luo W., Li G., Yi Z., Nie Q., Zhang X. E2F1-miR-20a-5p/20b-5p auto-regulatory feedback loop involved in myoblast proliferation and differentiation. Sci. Rep. 2016;6:27904. doi: 10.1038/srep27904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Langlands K., Yin X., Anand G., Prochownik E.V. Differential interactions of Id proteins with basic-helix-loop-helix transcription factors. J. Biol. Chem. 1997;272:19785–19793. doi: 10.1074/jbc.272.32.19785. [DOI] [PubMed] [Google Scholar]
- 132.Wang G., Wang Y., Xiong Y., Chen X.-C., Ma M., Cai R., Gao Y., Sun Y., Yang G.-S., Pang W.-J. Sirt1 AS lncRNA interacts with its mRNA to inhibit muscle formation by attenuating function of miR-34a. Sci. Rep. 2016;6:21865. doi: 10.1038/srep21865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang Y., Pang W.-J., Wei N., Xiong Y., Wu W.-J., Zhao C.-Z., Shen Q.-W., Yang G.-S. Identification, stability and expression of Sirt1 antisense long non-coding RNA. Gene. 2014;539:117–124. doi: 10.1016/j.gene.2014.01.037. [DOI] [PubMed] [Google Scholar]
- 134.Rathbone C.R., Booth F.W., Lees S.J. Sirt1 increases skeletal muscle precursor cell proliferation. Eur. J. Cell Biol. 2009;88:35–44. doi: 10.1016/j.ejcb.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ballarino M., Cazzella V., D'Andrea D., Grassi L., Bisceglie L., Cipriano A., Santini T., Pinnarò C., Morlando M., Tramontano A., Bozzoni I. Novel long noncoding RNAs (lncRNAs) in myogenesis: a miR-31 overlapping lncRNA transcript controls myoblast differentiation. Mol. Cell. Biol. 2015;35:728–736. doi: 10.1128/MCB.01394-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mousavi K., Zare H., Dell’orso S., Grontved L., Gutierrez-Cruz G., Derfoul A., Hager G.L., Sartorelli V. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol. Cell. 2013;51:606–617. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mueller A.C., Cichewicz M.A., Dey B.K., Layer R., Reon B.J., Gagan J.R., Dutta A. MUNC, a long noncoding RNA that facilitates the function of MyoD in skeletal myogenesis. Mol. Cell. Biol. 2015;35:498–513. doi: 10.1128/MCB.01079-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gong C., Li Z., Ramanujan K., Clay I., Zhang Y., Lemire-Brachat S., Glass D.J. A long non-coding RNA, LncMyoD, regulates skeletal muscle differentiation by blocking IMP2-mediated mRNA translation. Dev. Cell. 2015;34:181–191. doi: 10.1016/j.devcel.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 139.Hubé F., Velasco G., Rollin J., Furling D., Francastel C. Steroid receptor RNA activator protein binds to and counteracts SRA RNA-mediated activation of MyoD and muscle differentiation. Nucleic Acids Res. 2011;39:513–525. doi: 10.1093/nar/gkq833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Caretti G., Schiltz R.L., Dilworth F.J., Di Padova M., Zhao P., Ogryzko V., Fuller-Pace F.V., Hoffman E.P., Tapscott S.J., Sartorelli V. The RNA helicases p68/p72 and the noncoding RNA SRA are coregulators of MyoD and skeletal muscle differentiation. Dev. Cell. 2006;11:547–560. doi: 10.1016/j.devcel.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 141.Yu X., Zhang Y., Li T., Ma Z., Jia H., Chen Q., Zhao Y., Zhai L., Zhong R., Li C., Zou X., Meng J., Chen A.K., Puri P.L., Chen M., Zhu D. Long non-coding RNA Linc-RAM enhances myogenic differentiation by interacting with MyoD. Nat. Commun. 2017;8:14016. doi: 10.1038/ncomms14016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lu J., Webb R., Richardson J.A., Olson E.N. MyoR: a muscle-restricted basic helix-loop-helix transcription factor that antagonizes the actions of MyoD. Proc. Natl. Acad. Sci. U.S.A. 1999;96:552–557. doi: 10.1073/pnas.96.2.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gagan J., Dey B.K., Layer R., Yan Z., Dutta A. MicroRNA-378 targets the myogenic repressor MyoR during myoblast differentiation. J. Biol. Chem. 2011;286:19431–19438. doi: 10.1074/jbc.M111.219006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhou L., Sun K., Zhao Y., Zhang S., Wang X., Li Y., Lu L., Chen X., Chen F., Bao X., Zhu X., Wang L., Tang L.-Y., Esteban M.A., Wang C.-C., Jauch R., Sun H., Wang H. Linc-YY1 promotes myogenic differentiation and muscle regeneration through an interaction with the transcription factor YY1. Nat. Commun. 2015;6:10026. doi: 10.1038/ncomms10026. [DOI] [PubMed] [Google Scholar]
- 145.Lu L., Zhou L., Chen E.Z., Sun K., Jiang P., Wang L., Su X., Sun H., Wang H. A Novel YY1-miR-1 regulatory circuit in skeletal myogenesis revealed by genome-wide prediction of YY1-miRNA network. PLoS ONE. 2012;7:e27596. doi: 10.1371/journal.pone.0027596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhou L., Wang L., Lu L., Jiang P., Sun H., Wang H. A novel target of microRNA-29, Ring1 and YY1-binding protein (Rybp), negatively regulates skeletal myogenesis. J. Biol. Chem. 2012;287:25255–25265. doi: 10.1074/jbc.M112.357053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wang M., Liu C., Su Y., Zhang K., Zhang Y., Chen M., Ge M., Gu L., Lu T., Li N., Yu Z., Meng Q. miRNA-34c inhibits myoblasts proliferation by targeting YY1. Cell Cycle. 2017:0. doi: 10.1080/15384101.2017.1281479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lu L., Sun K., Chen X., Zhao Y., Wang L., Zhou L., Sun H., Wang H. Genome-wide survey by ChIP-seq reveals YY1 regulation of lincRNAs in skeletal myogenesis. EMBO J. 2013;32:2575–2588. doi: 10.1038/emboj.2013.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Huang M.-B., Xu H., Xie S.-J., Zhou H., Qu L.-H. Insulin-like growth factor-1 receptor is regulated by microRNA-133 during skeletal myogenesis. PLoS ONE. 2011;6:e29173. doi: 10.1371/journal.pone.0029173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ge Y., Sun Y., Chen J. IGF-II is regulated by microRNA-125b in skeletal myogenesis. J. Cell Biol. 2011;192:69–81. doi: 10.1083/jcb.201007165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang J., Cao N., Yuan M., Cui H., Tang Y., Qin L., Huang X., Shen N., Yang H.-T. MicroRNA-125b/Lin28 pathway contributes to the mesendodermal fate decision of embryonic stem cells. Stem Cells Dev. 2012;21:1524–1537. doi: 10.1089/scd.2011.0350. [DOI] [PubMed] [Google Scholar]
- 152.Polesskaya A., Cuvellier S., Naguibneva I., Duquet A., Moss E.G., Harel-Bellan A. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev. 2007;21:1125–1138. doi: 10.1101/gad.415007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Watts R., Johnsen V.L., Shearer J., Hittel D.S. Myostatin-induced inhibition of the long noncoding RNA Malat1 is associated with decreased myogenesis. Am. J. Physiol. Cell Physiol. 2013;304:C995–C1001. doi: 10.1152/ajpcell.00392.2012. [DOI] [PubMed] [Google Scholar]
- 154.Han X., Yang F., Cao H., Liang Z. Malat1 regulates serum response factor through miR-133 as a competing endogenous RNA in myogenesis. FASEB J. 2015;29:3054–3064. doi: 10.1096/fj.14-259952. [DOI] [PubMed] [Google Scholar]
- 155.Zhao Y., Samal E., Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 156.Sun Y., Ge Y., Drnevich J., Zhao Y., Band M., Chen J. Mammalian target of rapamycin regulates miRNA-1 and follistatin in skeletal myogenesis. J. Cell Biol. 2010;189:1157–1169. doi: 10.1083/jcb.200912093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chen J.-F., Mandel E.M., Thomson J.M., Wu Q., Callis T.E., Hammond S.M., Conlon F.L., Wang D.-Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lu J., McKinsey T.A., Zhang C.L., Olson E.N. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol. Cell. 2000;6:233–244. doi: 10.1016/s1097-2765(00)00025-3. [DOI] [PubMed] [Google Scholar]
- 159.Zhang X., Zuo X., Yang B., Li Z., Xue Y., Zhou Y., Huang J., Zhao X., Zhou J., Yan Y., Zhang H., Guo P., Sun H., Guo L., Zhang Y., Fu X.-D. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell. 2014;158:607–619. doi: 10.1016/j.cell.2014.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M., Tramontano A., Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Legnini I., Morlando M., Mangiavacchi A., Fatica A., Bozzoni I. A feedforward regulatory loop between HuR and the long noncoding RNA linc-MD1 controls early phases of myogenesis. Mol. Cell. 2014;53:506–514. doi: 10.1016/j.molcel.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Snyder C.M., Rice A.L., Estrella N.L., Held A., Kandarian S.C., Naya F.J. MEF2A regulates the Gtl2-Dio3 microRNA mega-cluster to modulate WNT signaling in skeletal muscle regeneration. Development. 2013;140:31–42. doi: 10.1242/dev.081851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Schiaffino S., Reggiani C. Fiber types in mammalian skeletal muscles. Physiol. Rev. 2011;91:1447–1531. doi: 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- 164.Mizbani A., Luca E., Rushing E.J., Krützfeldt J. MicroRNA deep sequencing in two adult stem cell populations identifies miR-501 as a novel regulator of myosin heavy chain during muscle regeneration. Development. 2016;143:4137–4148. doi: 10.1242/dev.136051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.van Rooij E., Quiat D., Johnson B.A., Sutherland L.B., Qi X., Richardson J.A., Kelm R.J., Jr., Olson E.N. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev. Cell. 2009;17:662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sakakibara I., Santolini M., Ferry A., Hakim V., Maire P. Six homeoproteins and a Iinc-RNA at the fast MYH locus lock fast myofiber terminal phenotype. PLoS Genet. 2014;10:e1004386. doi: 10.1371/journal.pgen.1004386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wang L., Chen X., Zheng Y., Li F., Lu Z., Chen C., Liu J., Wang Y., Peng Y., Shen Z., Gao J., Zhu M., Chen H. MiR-23a inhibits myogenic differentiation through down regulation of fast myosin heavy chain isoforms. Exp. Cell Res. 2012;318:2324–2334. doi: 10.1016/j.yexcr.2012.06.018. [DOI] [PubMed] [Google Scholar]
- 168.Wada S., Kato Y., Okutsu M., Miyaki S., Suzuki K., Yan Z., Schiaffino S., Asahara H., Ushida T., Akimoto T. Translational suppression of atrophic regulators by microRNA-23a integrates resistance to skeletal muscle atrophy. J. Biol. Chem. 2011;286:38456–38465. doi: 10.1074/jbc.M111.271270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Matsumoto A., Pasut A., Matsumoto M., Yamashita R., Fung J., Monteleone E., Saghatelian A., Nakayama K.I., Clohessy J.G., Pandolfi P.P. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature. 2016;541:228–232. doi: 10.1038/nature21034. [DOI] [PubMed] [Google Scholar]
- 170.Falcone G., Perfetti A., Cardinali B., Martelli F. Noncoding RNAs: emerging players in muscular dystrophies. Biomed. Res. Int. 2014;2014:503634. doi: 10.1155/2014/503634. [DOI] [PMC free article] [PubMed] [Google Scholar]