Lacto-N-biose I units in the PDB were investigated. Instances and conformational features of the disaccharide in proteins and enzymes in the PDB from various organisms are reviewed.
Keywords: human milk oligosaccharides, Lewis blood group antigens, glycosphingolipids, LS-tetrasaccharides, type-1 ABO(H) blood group antigens
Abstract
The lacto-N-biose I (Galβ1–3GlcNAc; LNB) disaccharide is present as a core unit of type-1 blood group antigens of animal glycoconjugates and milk oligosaccharides. Type-1 antigens often serve as cell-surface receptors for infection by pathogens. LNB in human milk oligosaccharides functions as a prebiotic for bifidobacteria and plays a key role in the symbiotic relationship of commensal gut microbes in infants. Protein Data Bank (PDB) entries exhibiting the LNB unit were investigated using the GlycoMapsDB web tool. There are currently 159 β-LNB and nine α-LNB moieties represented in ligands in the database. β-LNB and α-LNB moieties occur in 74 and six PDB entries, respectively, as NCS copies. The protein and enzyme structures are from various organisms including humans (galectins), viruses (haemagglutinin and capsid proteins), a pathogenic fungus, a parasitic nematode and protist, pathogenic bacteria (adhesins) and a symbiotic bacterium (a solute-binding protein of an ABC transporter). The conformations of LNB-containing glycans in enzymes vary significantly according to their mechanism of substrate recognition and catalysis. Analysis of glycosidic bond conformations indicated that the binding modes are significantly different in proteins adapted for modified or unmodified glycans.
1. Introduction
The lacto-N-biose I (Galβ1–3GlcNAc, LNB or, to be more precise, β-d-Galpβ1–3-d-GlcpNAc) disaccharide unit, also called ‘type-1’ to distinguish it from the ‘type-2’ N-acetyllactosamine (Galβ1–4GlcNAc) unit, is present in blood group antigens of protein glycans, glycolipids and milk oligosaccharides (Stanley & Cummings, 2015 ▸). LNB is a core unit of fucosylated blood groups such as type-1 ABO(H) and Lewis antigens, which often serve as surface receptors of animal cells for infection by viruses, pathogenic microbes and parasites (Cooling, 2015 ▸). Human milk contains 12–24 g l−1 of a complex array of oligosaccharides (with a degree of polymerization of greater than three), which consist of more than 160 identified molecular species, and they are collectively termed human milk oligosaccharides (HMOs; Urashima et al., 2018 ▸). HMOs are especially rich in type-1 oligosaccharides (Urashima et al., 2011 ▸). Lacto-N-tetraose (Galβ1–3GlcNAcβ1–3Galβ1–4Glc; LNT) consists of an LNB and a lactose moiety connected by a β-1,3-bond and is a major core structure of type-1 HMOs. Three of the four major molecular species in HMOs [LNT, Fucα1–2Galβ1–3GlcNAcβ1–3Galβ1–4Glc (LNFP I) and Fucα1–2Galβ1–3(Fucα1–4)GlcNAcβ1–3Galβ1–4Glc (LNDFH I)] contain the LNT structure. The other is 2′-fucosyllactose (Fucα1–2Galβ1–4Glc), but the milk of about 15% of human donors does not contain this trisaccharide (Kobata et al., 2010 ▸). Recent studies on HMOs and bifidobacteria revealed the important role of LNB in the symbiosis between human infants and commensal gut microbes (Fushinobu, 2010 ▸; Kitaoka, 2012 ▸). Interestingly, the predominance of type-1 oligosaccharides is a specific feature of human milk. All breast-milk oligosaccharides from nonhuman mammals analyzed to date, including those from primates such as chimpanzees, bonobos and orangutans, exclusively or predominantly contain type-2 oligosaccharides (Urashima et al., 2012 ▸). Infant gut-associated bifidobacteria (Bifidobacterium longum, B. longum subsp. infantis, B. bifidum and B. breve) have specific degradation and assimilation pathways for type-1 HMOs and LNB (Pacheco et al., 2015 ▸; Katayama, 2016 ▸). Consequently, LNB has a prebiotic effect promoting the growth of infant gut-associated bifidobacterial strains (Xiao et al., 2010 ▸). In this review, I summarize the incidences of LNB units in protein structures published to date and describe their conformations, focusing on the sugar ring conformations, exocyclic groups and glycosidic bond torsion angles of the disaccharide.
2. LNB units in the Protein Data Bank
LNB units in entries in the current Protein Data Bank (PDB) as of Februay 2018 were surveyed using GlycoMapsDB on the glycosciences.de website (http://www.glycosciences.de/modeling/glycomapsdb/; Frank et al., 2007 ▸). 74 PDB entries contained LNB(s) exhibiting the β-anomer of GlcNAc (β-LNB) and a total of 159 β-LNB units were present including copies in noncrystallographic symmetry (NCS; Table 1 ▸). In contrast, only nine NCS copies of α-LNB were found in six PDB entries. The dominance of β-LNB in the PDB is probably because proteins and enzymes recognizing LNT-containing glycans have mainly been studied. The structures of glycan-binding proteins (for example, lectins, adhesins, haemagglutinins and viral capsid proteins) from various organisms including humans and pathogens have been extensively studied and represent the majority of the LNB-containing structures in the PDB. The pathogens include a fungus (or yeast, Candida glabrata; Diderrich et al., 2015 ▸), a nematode (Toxascaris leonina; Hwang et al., 2016 ▸), a protist (Toxoplasma gondii; Garnett et al., 2009 ▸), bacteria (Helicobacter pylori, Shiga toxin-producing Escherichia coli and Pseudomonas aeruginosa; Moonens et al., 2012 ▸, 2016 ▸; Boukerb et al., 2016 ▸) and viruses (influenza virus, norovirus, rotavirus and polyomavirus; Vachieri et al., 2014 ▸; Choi et al., 2008 ▸; Liu et al., 2017 ▸; Stehle & Harrison, 1997 ▸). Eight PDB entries for enzymes contain the β-LNB unit as part of their substrate (discussed below). Two PDB entries of human proteins (one each for β-LNB and α-LNB) include LNB as a part of a sialylated O-glycan attached to a threonine residue, but they may be incorrectly modelled in place of the galacto-N-biose (Galβ1–3GalNAc) unit of mucin-type core structures (Brockhausen & Stanley, 2015 ▸). The remaining five PDB entries containing α-LNB units consist of a lectin from a mushroom (Agaricus bisporus; Carrizo et al., 2005 ▸), haemagglutinin from influenza virus (Wang et al., 2007 ▸) and a capsid protein from norovirus (Singh et al., 2015 ▸).
Table 1. Number of LNB structures in the PDB surveyed by GlycoMapsDB.
| β-LNB | α-LNB | |
|---|---|---|
| PDB entries† | 74 (159) | 6 (9) |
| Enzymes | 8 | 0 |
| Sugar-binding proteins and lectins | ||
| From human (galectins) | 8 | 0 |
| From fungi | 2 | 1 |
| From other eukaryotes‡ | 4 | 0 |
| From bacteria | 18 | 0 |
| From viruses | 33 | 4 |
| Protein O-glycans§ | 1 | 1 |
Values in parentheses indicate the total number of occurrences including NCS copies.
Lectins from a plant (G. simplicifolia) and a nematode (T. leonina), and an adhesin from a protist (T. gondii).
Apparent incorrectly modelled galacto-N-biose (Galβ1–3GalNAc) core structure.
2.1. Human galectins and viral binding proteins
Galectins bind β-Gal-containing glycans (Cummings et al., 2015 ▸). The PDB contains LNB or LNT structures bound to human galectins (galectin-1, galectin-3, galectin-4, galectin-7 and galectin-8) as a part of a lacto-series of glycosphingolipids. The LNB unit of LNT bound to the N-terminal domain of the galectin-8 structure was clearly visible (PDB entry 5t7t), probably because its nonreducing-end β-Gal residue exhibits extensive hydrogen-bonding interactions with the protein (Bohari et al., 2016 ▸). LNT was modelled in the crystal structure of galectin-3 determined at 1.55 Å resolution (PDB entry 4lbm; Fig. 1 ▸ a). However, galectin-3 interacts exclusively with the lactose unit, and the LNB moiety was much less well defined in the electron density (Collins et al., 2014 ▸).
Figure 1.
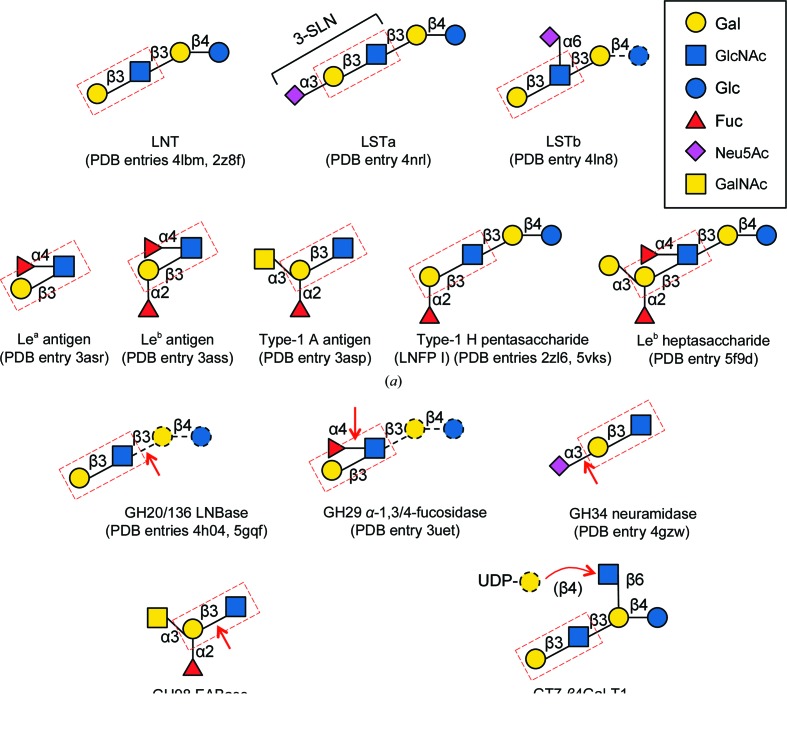
Schematic representation of β-LNB-containing glycan structures in the PDB. (a) Glycan structures found in sugar-binding proteins and lectins. (b) Glycans observed in enzyme structures. The cleavage sites of the glyoside hydrolases and the transferase reaction site of the gycosyltransferase are indicated by red arrows. Disordered (and hence unmodelled) sugar moieties in sugar-binding protein structures, and substrate moieties additional to the crystallographic ligand of the enzymes, are shown with black dashed lines. The LNB unit is boxed with a red dashed line. For α-LNB-containing glycan structures, α-LNB disaccharide, LSTa and Lewis antigens were observed in a lectin from A. bisporus, in haemagglutinin from influenza virus and in VP1 (P domain) in norovirus, respectively.
Haemagglutinins of influenza viruses bind sialic acid-containing glycans, and their interactions with avian and human receptors have been extensively studied. 17 PDB entries currently represent haemagglutinin structures complexed with LNB-containing ligands. In particular, LS-tetrasaccharide a (Neu5Acα2–3Galβ1–3GlcNAcβ1–3Galβ1–4Glc; LSTa) has frequently been used as an avian receptor analogue (Fig. 1 ▸ a). However, the reducing-end lactose moiety of the sialylated pentasaccharide is not recognized by haemagglutinins, and its nonreducing-end trisaccharide part, 3′-sialyl-N-acetyllactosamine (3-SLN), was usually modelled in the crystal structures. The entire pentasaccharide of LSTa was modelled in the F95Y mutant of haemagglutinin from influenza virus B/Lee/40 (PDB entry 4nrl) but only one hydrogen-bond interaction was observed for the lactose moiety (Ni et al., 2014 ▸). The complex structure of an α-2,6-sialylated pentasaccharide, LS-tetrasaccharide b [Galβ1–3(Neu5Acα2–6)GlcNAcβ1-3Galβ1-4Glc; LSTb], with haemagglutinin from the H7N9 influenza virus has also been determined, and the tetrasaccharide excluding the reducing-end Glc was modelled (PDB entry 4ln8; Yang et al., 2013 ▸).
Interactions of histo-blood group antigen-binding proteins from diarrhoea-causing viruses such as norovirus and rotavirus have been also extensively studied. Complex structures of the P domain of a major capsid protein VP1 from norovirus with fucosylated oligosaccharides of Lewis a [Galβ1–3(Fucα1–4)GlcNAcβ-R; Lea], Lewis b [Fucα1–2Galβ1–3(Fucα1–4)GlcNAcβ-R, Leb] and type-1 A antigens have been determined (PDB entries 3asr, 3ass and 3asp; Fig. 1 ▸ a; Kubota et al., 2012 ▸). In addition, the type-1 H antigen pentasaccharide (or LNFP I) was entirely defined in a high-resolution crystal structure of a norovirus P domain with clear electron density (PDB entry 2zl6; Choi et al., 2008 ▸). A complex structure with LNFP I has also been reported for the VP8* domain of the rotavirus spike protein VP4 (PDB entry 5vks; Liu et al., 2017 ▸).
2.2. Binding proteins from other eukaryotes and microbes
Complex crystal structures of plant lectin IV from the African shrub Griffonia simplicifolia with Leb tetrasaccharide (Delbaere et al., 1993 ▸), microneme protein 1 from a protozoan parasite (T. gondii) with 3-SLN (Garnett et al., 2009 ▸), galectin Tl-gal from a canine gastrointestinal nematode parasite (T. leonina) with LNT (Hwang et al., 2016 ▸) and the adhesin Epa6A domain from a fungal pathogen (C. glabrata) with LNB (Diderrich et al., 2015 ▸) have been reported. For bacterial glycan-binding proteins and lectins, several crystal structures, including those of the adhesin BabA from H. pylori, the fimbrial adhesin subunit FedF from Shiga toxin-producing E. coli and the LecB lectin from P. aeruginosa, complexed with Lewis and type-1 ABO(H) blood group antigens have been reported (Moonens et al., 2012 ▸, 2016 ▸; Boukerb et al., 2016 ▸). PDB entry 5f9d for BabA contains the whole Leb heptasaccharide (Fig. 1 ▸ a).
In the LNB-degrading pathway of infant gut-associated bifidobacteria, a set of genes for a three-component ABC transporter and four intracellular enzymes play key roles in HMO utilization (Kitaoka, 2012 ▸) and the solute-binding protein of the transporter (GL-BP) is responsible for the recognition and specific import of LNB into the bifidobacterial cells (Suzuki et al., 2008 ▸). GL-BP from B. longum strongly binds LNB (K d = 0.087 µM), while it also binds LNT with a higher K d value (11 µM). In the crystal structure of GL-BP, electron density for a whole LNT molecule was clearly visible at 1.65 Å resolution (PDB entry 2z8f).
2.3. Enzymes
Currently, five kinds of enzymes, four glycoside hydrolases (GHs) and one glycosyltransferase (GT), have been reported to contain the LNB unit in their crystal structures (Fig. 1 ▸ b). The four major infant gut-associated bifidobacterial species employ different strategies for the utilization of type-1 HMOs (Katayama, 2016 ▸): B. longum subsp. infantis and B. breve probably have transporter(s) for LNT and degrade the tetrasaccharide using intracellular enzymes, while B. bifidum and B. longum employ extracellular enzymes to release and import LNB. The key extracellular enzyme is lacto-N-biosidase (LNBase; EC 3.2.1.140), which recognizes the LNB moiety of LNT (and type-1 HMOs) and hydrolyzes the GlcNAcβ1–3Gal bond. Interestingly, B. bifidum and B. longum have completely different types of LNBases belonging to GH20 and GH136, respectively, and the crystal structures of both enzymes complexed with LNB have been determined at resolutions of higher than 1.82 Å (PDB entries 4h04 and 5gqf; Ito et al., 2013 ▸; Yamada et al., 2017 ▸). The crystal structure of an intracellular GH29 α-1,3/4-fucosidase (EC 3.2.1.111) from B. longum subsp. infantis, which is involved in intracellular HMO degradation, has also been determined in complex with its substrate Lea antigen trisaccharide using an inactive D172A/E217A mutant (PDB entry 3uet; Sakurama et al., 2012 ▸). In this case the LNB moiety represents the aglycone, and is bound to plus subsites in the enzyme, while the LNBs in LNBases are bound to minus subsites.
A GH98 endo-β-galactosidase from Streptococcus pneumoniae SP3-BS71 that removes the whole trisaccharides of type-2 A and B antigens (EABase; EC 3.2.1.102) has been engineered to improve its activity towards type-1 chains (Kwan et al., 2015 ▸). The crystal structure of an inactive E558A mutant of the engineered GH98 EABase was determined in complex with a type-1 A antigen tetrasaccharide at 1.65 Å resolution (PDB entry 4d6h). For GH34 neuraminidase (EC 3.2.1.18) from influenza virus, a complex structure with 3-SLN has been reported (PDB entry 4gzw; Zhu et al., 2012 ▸). However, GlcNAc in the LNB moiety of 3-SLN was poorly defined in the electron-density map because the neuraminidase mostly interacts with the sialic acid group. The sole representative of the LNB moiety in GT family enzyme structures, a pentasaccharide analogue of I-antigens (which are branched mammalian polylactosamines), was observed in the structure of the complex with human GT7 β1–4-galactosyltransferase-I (β4Gal-T1; EC 2.4.1.22/38/90) as a receptor molecule bound to the plus subsites (PDB entry 4ee3; Ramakrishnan et al., 2012 ▸). Electron density for the LNB moiety of the pentasaccharide was relatively ambiguous as it protrudes out of the protein and is not involved in substrate recognition.
3. Conformations of LNB units
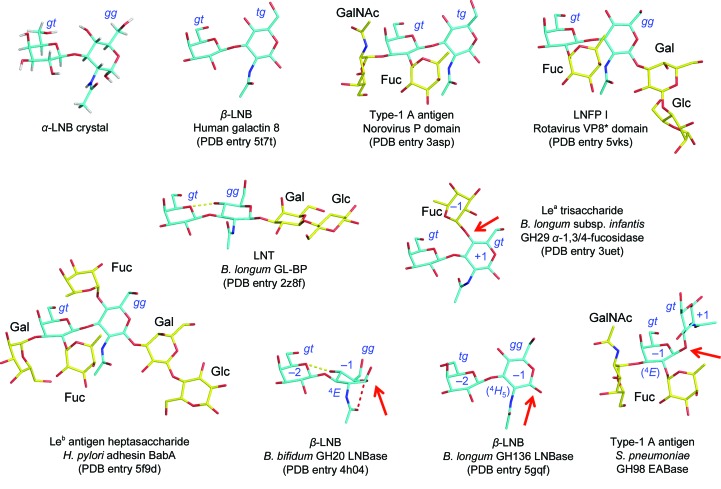
Fig. 2 ▸ shows selected LNB and LNB-containing glycan structures in protein crystal structures for which the LNB moiety was able to be confidently modelled in the electron density, as well as the small-molecule crystal structure of α-LNB (Wada et al., 2009 ▸).
Figure 2.
Crystal structures of α-LNB (small molecule) and a selection of LNB-containing glycans in PDB entries.
3.1. Conformations of sugar rings and exocyclic groups
The sugar rings of Gal and GlcNAc in LNB are in a favoured chair (4 C 1) conformation in most cases. In several cases of poorly recognized groups with ambiguous electron density (for example in the reducing-end region of glycans in haemagglutinin structures), the sugar rings were sometimes modelled as high-energy distorted conformations, as reported by Agirre et al. (2015 ▸). The N-acetyl group of GlcNAc is usually in an extended conformation, with its carbonyl oxygen and methyl groups in ‘up’ and ‘down’ orientations, respectively (Fig. 2 ▸). The β-LNB molecule observed in the GH20 LNBase is an exception. GH20 enzymes utilize a substrate-assisted mechanism in which the 2-acetamido (N-acetyl) group acts as the nucleophile. GlcNAc bound to the GH20 LNBase adopts a distorted 4 E conformation, and the N-acetyl group is oriented so that the carbonyl O atom sits beneath the C1 atom, facilitating in-line nucleophilic attack (Ito et al., 2013 ▸). In the GH136 LNBase, which uses a classical retaining GH mechanism employing an aspartate residue as the nucleophile, GlcNAc is nearly in the chair conformation and its N-acetyl group is in a typical extended conformation. The engineered GH98 EABase is an inverting enzyme that cleaves the internal Galβ1–3 bond in LNB (Kwan et al., 2015 ▸), and its Gal moiety also appears to have a conformation similar to 4 C 1. However, it has been suggested that all glycosidases acting on β-glycosidic bonds in principle require distortion of the sugar ring bound at subsite −1 to move the scissile glycosidic bond into a pseudoaxial orientation, to remove potential steric hindrance from H1 and to facilitate direct inline nucleophilic attack (Davies et al., 2003 ▸). Investigation using the Cremer–Pople parameter (Cremer & Pople, 1975 ▸) indicates that the sugar rings at subsite −1 in these enzymes are slightly distorted towards 4 H 5 in the GH136 LNBase (θ = 17–21°, φ = 267–270°) and 4 E in the GH98 EABase (θ = 27°, φ = 249°).
In addition to the N-acetyl group, the two C5–C6 hydroxymethyl groups are rotatable exocyclic groups in LNB. The C5–C6 group in GlcNAc adopts various rotamers (gg, tg and gt) in protein crystal structures, while that in Gal is mostly in the gt conformer (Fig. 2 ▸). A survey of aldohexopyranoses in carbohydrate crystal structures in 1979 showed that the rotamer populations of gluco-type sugars are preferentially in the gg conformer (60%) over gt (40%), with no observation of tg, while those of galacto-type sugars are in the order gt (58%), tg (34%), gg (8%) (Marchessault & Perez, 1979 ▸). These distributions were explained by steric hindrance with the O4 hydroxyl. The rare tg conformer of GlcNAc in human galectin-8 is stabilized by a hydrogen bond to Glu89 (Bohari et al., 2016 ▸), whereas that in type-1 A antigen bound to the norovirus P domain is not supported by any protein interactions and the electron density is relatively ambiguous in this region (Kubota et al., 2012 ▸). The rare tg conformer of the Gal in β-LNB bound to the GH136 LNBase is stabilized by the shape complementarity of surrounding hydrophobic residues (Yamada et al., 2017 ▸).
3.2. Glycosidic bond conformations
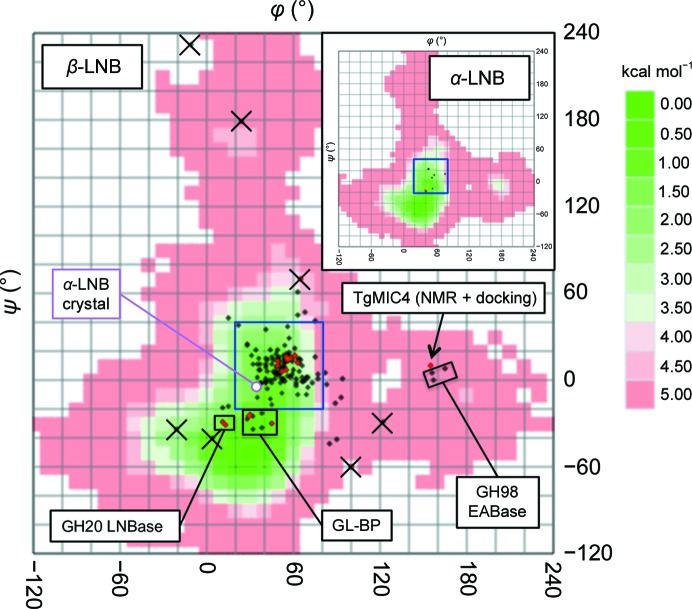
The glycosidic torsion angles of the LNB disaccharide, defined as H1′—C1′—O3—C3 for φ and C1′—O3—C3—H3 for ψ, were investigated. The GlycoMapsDB web tool can map the glycosidic torsions in PDB entries onto conformational energy maps calculated for more than 2500 disaccharide units of oligosaccharides (Frank et al., 2007 ▸). As shown in Fig. 3 ▸ and Table 2 ▸, glycosidic bond conformations of both β-LNBs and α-LNBs are mostly mapped in a limited area (20 < φ < 80° and −20 < ψ < 40°; blue box in Fig. 3 ▸) of the low-energy conformation area in the energy map (shown in green), which extends to ψ ≃ −70°. The glycosidic bond torsion angles of the α-LNB small-molecule structure are φ = 38.7° and ψ = −2.9°, and they are mapped near the area that is frequently observed in protein structures. As shown in Fig. 2 ▸, this frequently observed glycosidic bond conformation places the two sugar rings at an angle of approximately 60° to each other. This conformation is apparently suitable for binding the disaccharide unit even when it is decorated with fucose or other sugars. This observation may reflect the fact that the LNB unit is often modified by fucosylation or sialylation in type-1 chains and is not present in an unmodified form in animal glycans. In contrast, the LNB units in GL-BP and GH20 LNBase have a distinct conformation (Fig. 3 ▸; 0 < φ < 50° and −40 < ψ < −20°). The two sugar rings are almost parallel and the endocyclic O5 atom of Gal and the O4 hydroxyl of GlcNAc form a direct intramolecular hydrogen bond (Fig. 2 ▸). This is probably because these proteins exclusively bind the unmodified LNB unit and form extensive hydrogen bonds to all of the hydroxyls (Suzuki et al., 2008 ▸; Ito et al., 2013 ▸) that are formed after the action of bifidobacterial fucosidases and sialidases (Katayama, 2016 ▸).
Figure 3.
Glycosidic bond torsion angles of the LNB disaccharide (red) and LNB-containing glycans (black) plotted on calculated conformational free-energy maps of β-LNB (GlycoMAP ID 7430) and α-LNB (inset, GlycoMAP ID 7777) (coded green to red for low to high-energy conformations). Unreliable conformations owing to disorder are marked with ‘×’ symbols.
Table 2. Glycosidic bond torsion angles of LNB units.
| PDB code (chain) | Resolution (Å) | φ (°) | ψ (°) | Organism | Protein | Ligand |
|---|---|---|---|---|---|---|
| 4lbm (A) | 1.55 | 44.3† | −12.6† | Human | Galectin-3 | LNT |
| 5t7t (A) | 1.96 | 49.8 | 13.5 | Human | Galectin-8 | LNB |
| 2z8f (A) | 1.65 | 38.2 | −32.7 | B. longum | GL-BP | LNT |
| 4nrl (A) | 2.72 | 48.9 | 4.8 | Influenza virus | Hemagglutinin | LSTa |
| 4ln8 (A) | 2.50 | 37.8 | −1.2 | Influenza virus | Hemagglutinin | LSTb |
| 3asp (A) | 1.60 | 56.6 | 13.7 | Norovirus | VP1 (P domain) | Type-1 A antigen |
| 2zl6 (B) | 1.43 | 42.0 | 14.1 | Norovirus | VP1 (P domain) | LNFP I |
| 5vks (A) | 1.94 | 53.6 | 10.5 | Rotavirus | VP4 (VP8* domain) | LNFP I |
| 5f9d (A) | 2.59 | 59.7 | 13.4 | H. pylori | Adhesin BabA | Leb heptasaccharide |
| 4h04 (A) | 1.80 | 13.6 | −30.5 | B. bifidum | GH20 LNBase | β-LNB |
| 5gqf (A) | 1.82 | 49.6 | 10.3 | B. longum | GH136 LNBase | β-LNB |
| 3uet (A) | 2.10 | 36.8 | 15.5 | B. infantis | GH29 α-1,3/4-fucosidase | Lea trisaccharide |
| 4gzw (B) | 2.45 | −10.9† | 232.0† | Influenza virus | GH34 neuraminidase | 3-SLN |
| 4d6h (A) | 1.65 | 156.4 | 5.8 | S. pneumoniae | GH98 EABase | Type-1 A antigen |
| 4ee3 (A) | 2.30 | 62.6 | −1.7 | Human | GT7 β4Gal-T1 | Pentasaccharide acceptor |
The electron-density map for the LNB unit is partly ambiguous.
High-energy conformers are found in the PDB structures of two proteins, T. gondii microneme protein 4 (TgMIC4) and the S. pneumoniae GH98 EABase (Fig. 3 ▸; 150 < φ < 170° and −10 < ψ < 10°). The TgMIC4 structure was determined by solution NMR, and LNB was modelled by calculation using a molecular docking program (HADDOCK) based on NOE-derived distance restraints (Marchant et al., 2012 ▸). The type-1 A tetrasaccharide in GH98 EABase mutants contains an LNB in which the sugars adopt an L-shaped conformation (Fig. 2 ▸; the Gal and GlcNAc sugars are oriented perpendicularly). This is probably because the aglycon (subsite +1) specificity of the enzyme was engineered from the original type-2 chain specificity, and the β-1,3-linked GlcNAc moiety is bound in an unusual position.
4. Concluding remarks
In the GlycoMapsDB, there are 640 fragments of the type-2 chain core unit (Galβ1–4GlcNAcβ; GlycoMAP ID 7761), indicating that the type-2 chain is approximately four times more prevalent in the PDB compared with the type-1 chain. However, as exemplified in the case of symbiosis between human infants and bifidobacteria, type-1 chains are often involved in the interaction between animals and microbes. In addition to the prebiotic effects on bifidobacteria, HMOs can also function as decoy receptors that inhibit the attachment of pathogens to the colonic mucosa (Urashima et al., 2018 ▸). Further studies will not only shed light on host–pathogen interactions but also on host–symbiont interactions, which play an important role in maintaining human health.
Acknowledgments
I thank Dr Martin Frank and the glycosciences.de team for technical help and management of the excellent web-based tool, and Ms Wendy A. Offen for proofreading the manuscript.
References
- Agirre, J., Davies, G., Wilson, K. & Cowtan, K. (2015). Nature Chem. Biol. 11, 303. [DOI] [PubMed]
- Bohari, M. H., Yu, X., Zick, Y. & Blanchard, H. (2016). Sci. Rep. 6, 39556. [DOI] [PMC free article] [PubMed]
- Boukerb, A. M., Decor, A., Ribun, S., Tabaroni, R., Rousset, A., Commin, L., Buff, S., Doléans-Jordheim, A., Vidal, S., Varrot, A., Imberty, A. & Cournoyer, B. (2016). Front. Microbiol. 7, 811. [DOI] [PMC free article] [PubMed]
- Brockhausen, I. & Stanley, P. (2015). Essentials of Glycobiology, 3rd ed., edited by A. Varki, R. D. Cummings, J. D. Esko, P. Stanley, G. W. Hart, M. Aebi, A. G. Darvill, T. Kinoshita, N. H. Packer, J. H. Prestegard, R. L. Schnaar & P. H. Seeberger, pp. 113–123. New York: Cold Spring Harbor Laboratory Press. [PubMed]
- Carrizo, M. E., Capaldi, S., Perduca, M., Irazoqui, F. J., Nores, G. A. & Monaco, H. L. (2005). J. Biol. Chem. 280, 10614–10623. [DOI] [PubMed]
- Choi, J.-M., Hutson, A. M., Estes, M. K. & Prasad, B. V. (2008). Proc. Natl Acad. Sci. USA, 105, 9175–9180. [DOI] [PMC free article] [PubMed]
- Collins, P. M., Bum-Erdene, K., Yu, X. & Blanchard, H. (2014). J. Mol. Biol. 426, 1439–1451. [DOI] [PubMed]
- Cooling, L. (2015). Clin. Microbiol. Rev. 28, 801–870. [DOI] [PMC free article] [PubMed]
- Cremer, D. & Pople, J. A. (1975). J. Am. Chem. Soc. 97, 1354–1358.
- Cummings, R. D., Liu, F.-T. & Vasta, G. R. (2015). Essentials of Glycobiology, 3rd ed., edited by A. Varki, R. D. Cummings, J. D. Esko, P. Stanley, G. W. Hart, M. Aebi, A. G. Darvill, T. Kinoshita, N. H. Packer, J. H. Prestegard, R. L. Schnaar & P. H. Seeberger, pp. 469–480. New York: Cold Spring Harbor Laboratory Press. [PubMed]
- Davies, G. J., Ducros, V. M., Varrot, A. & Zechel, D. L. (2003). Biochem. Soc. Trans. 31, 523–527. [DOI] [PubMed]
- Delbaere, L. T., Vandonselaar, M., Prasad, L., Quail, J. W., Wilson, K. S. & Dauter, Z. (1993). J. Mol. Biol. 230, 950–965. [DOI] [PubMed]
- Diderrich, R., Kock, M., Maestre-Reyna, M., Keller, P., Steuber, H., Rupp, S., Essen, L.-O. & Mösch, H.-U. (2015). J. Biol. Chem. 290, 19597–19613. [DOI] [PMC free article] [PubMed]
- Frank, M., Lütteke, T. & von der Lieth, C. W. (2007). Nucleic Acids Res. 35, 287–290. [DOI] [PMC free article] [PubMed]
- Fushinobu, S. (2010). Biosci. Biotechnol. Biochem. 74, 2374–2384. [DOI] [PubMed]
- Garnett, J. A., Liu, Y., Leon, E., Allman, S. A., Friedrich, N., Saouros, S., Curry, S., Soldati-Favre, D., Davis, B. G., Feizi, T. & Matthews, S. (2009). Protein Sci. 18, 1935–1947. [DOI] [PMC free article] [PubMed]
- Hwang, E. Y., Jeong, M. S., Park, S. K., Ha, S. C., Yu, H. S. & Jang, S. B. (2016). J. Biol. Chem. 291, 25326–25338. [DOI] [PMC free article] [PubMed]
- Ito, T., Katayama, T., Hattie, M., Sakurama, H., Wada, J., Suzuki, R., Ashida, H., Wakagi, T., Yamamoto, K., Stubbs, K. A. & Fushinobu, S. (2013). J. Biol. Chem. 288, 11795–11806. [DOI] [PMC free article] [PubMed]
- Katayama, T. (2016). Biosci. Biotechnol. Biochem. 80, 621–632. [DOI] [PubMed]
- Kitaoka, M. (2012). Adv. Nutr. 3, 422S–429S. [DOI] [PMC free article] [PubMed]
- Kobata, A. (2010). Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 86, 1–17. [DOI] [PMC free article] [PubMed]
- Kubota, T., Kumagai, A., Ito, H., Furukawa, S., Someya, Y., Takeda, N., Ishii, K., Wakita, T., Narimatsu, H. & Shirato, H. (2012). J. Virol. 86, 11138–11150. [DOI] [PMC free article] [PubMed]
- Kwan, D. H., Constantinescu, I., Chapanian, R., Higgins, M. A., Kötzler, M. P., Samain, E., Boraston, A. B., Kizhakkedathu, J. N. & Withers, S. G. (2015). J. Am. Chem. Soc. 137, 5695–5705. [DOI] [PubMed]
- Liu, Y., Xu, S., Woodruff, A. L., Xia, M., Tan, M., Kennedy, M. A. & Jiang, X. (2017). PLoS Pathog. 13, e1006707. [DOI] [PMC free article] [PubMed]
- Marchant, J. et al. (2012). J. Biol. Chem. 287, 16720–16733. [DOI] [PMC free article] [PubMed]
- Marchessault, R. H. & Perez, S. (1979). Biopolymers, 18, 2369–2374.
- Moonens, K., Bouckaert, J., Coddens, A., Tran, T., Panjikar, S., De Kerpel, M., Cox, E., Remaut, H. & De Greve, H. (2012). Mol. Microbiol. 86, 82–95. [DOI] [PubMed]
- Moonens, K. et al. (2016). Cell Host Microbe, 19, 55–66. [DOI] [PMC free article] [PubMed]
- Ni, F., Nnadi Mbawuike, I., Kondrashkina, E. & Wang, Q. (2014). Virology, 450–451, 71–83. [DOI] [PMC free article] [PubMed]
- Pacheco, A. R., Barile, D., Underwood, M. A. & Mills, D. A. (2015). Annu. Rev. Anim. Biosci. 3, 419–445. [DOI] [PMC free article] [PubMed]
- Ramakrishnan, B., Boeggeman, E. & Qasba, P. K. (2012). J. Biol. Chem. 287, 28666–28674. [DOI] [PMC free article] [PubMed]
- Sakurama, H., Fushinobu, S., Hidaka, M., Yoshida, E., Honda, Y., Ashida, H., Kitaoka, M., Kumagai, H., Yamamoto, K. & Katayama, T. (2012). J. Biol. Chem. 287, 16709–16719. [DOI] [PMC free article] [PubMed]
- Singh, B. K., Leuthold, M. M. & Hansman, G. S. (2015). J. Virol. 89, 2024–2040. [DOI] [PMC free article] [PubMed]
- Stanley, P. & Cummings, R. D. (2015). Essentials of Glycobiology, 3rd ed., edited by A. Varki, R. D. Cummings, J. D. Esko, P. Stanley, G. W. Hart, M. Aebi, A. G. Darvill, T. Kinoshita, N. H. Packer, J. H. Prestegard, R. L. Schnaar & P. H. Seeberger, pp. 161–178. New York: Cold Spring Harbor Laboratory Press. [PubMed]
- Stehle, T. & Harrison, S. C. (1997). EMBO J. 16, 5139–5148. [DOI] [PMC free article] [PubMed]
- Suzuki, R., Wada, J., Katayama, T., Fushinobu, S., Wakagi, T., Shoun, H., Sugimoto, H., Tanaka, A., Kumagai, H., Ashida, H., Kitaoka, M. & Yamamoto, K. (2008). J. Biol. Chem. 283, 13165–13173. [DOI] [PubMed]
- Urashima, T., Asakuma, S., Leo, F., Fukuda, K., Messer, M. & Oftedal, O. T. (2012). Adv. Nutr. 3, 473S–482S. [DOI] [PMC free article] [PubMed]
- Urashima, T., Fukuda, K., Kitaoka, M., Ohnishi, M., Terabayashi, T. & Kobata, A. (2011). Milk Oligosaccharides. New York: Nova Science Publishers.
- Urashima, T., Hirabayashi, J., Sato, S. & Kobata, A. (2018). Trends Glycosci. Glycotechnol. 30, SE31–SE65.
- Vachieri, S. G., Xiong, X., Collins, P. J., Walker, P. A., Martin, S. R., Haire, L. F., Zhang, Y., McCauley, J. W., Gamblin, S. J. & Skehel, J. J. (2014). Nature (London), 511, 475–477. [DOI] [PubMed]
- Wada, M., Kobayashi, K., Nishimoto, M., Kitaoka, M. & Noguchi, K. (2009). Acta Cryst. E65, o1781–o1782. [DOI] [PMC free article] [PubMed]
- Wang, Q., Tian, X., Chen, X. & Ma, J. (2007). Proc. Natl Acad. Sci. USA, 104, 16874–16879. [DOI] [PMC free article] [PubMed]
- Xiao, J.-Z., Takahashi, S., Nishimoto, M., Odamaki, T., Yaeshima, T., Iwatsuki, K. & Kitaoka, M. (2010). Appl. Environ. Microbiol. 76, 54–59. [DOI] [PMC free article] [PubMed]
- Yamada, C., Gotoh, A., Sakanaka, M., Hattie, M., Stubbs, K. A., Katayama-Ikegami, A., Hirose, J., Kurihara, S., Arakawa, T., Kitaoka, M., Okuda, S., Katayama, T. & Fushinobu, S. (2017). Cell. Chem. Biol. 24, 515–524.e5. [DOI] [PubMed]
- Yang, H., Carney, P. J., Chang, J. C., Villanueva, J. M. & Stevens, J. (2013). J. Virol. 87, 12433–12446. [DOI] [PMC free article] [PubMed]
- Zhu, X., McBride, R., Nycholat, C. M., Yu, W., Paulson, J. C. & Wilson, I. A. (2012). J. Virol. 86, 13371–13383. [DOI] [PMC free article] [PubMed]