Electron tomography, immunoblot, immunolabeling, and RNA-seq analyses of thylakoid assembly in germinating Arabidopsis revealed five structural assembly stages and the sequential incorporation of photosystem II, cytochrome b6f, ATP synthase, light-harvesting complex II, photosystem I, and CURT1A subunits within 84 h after imbibition.
Abstract
Biogenesis of the complex 3D architecture of plant thylakoids remains an unsolved problem. Here, we analyzed this process in chloroplasts of germinating Arabidopsis thaliana cotyledons using 3D electron microscopy and gene expression analyses of chloroplast proteins. Our study identified a linear developmental sequence with five assembly stages: tubulo-vesicular prothylakoids (24 h after imbibition [HAI]), sheet-like pregranal thylakoids that develop from the prothylakoids (36 HAI), proliferation of pro-grana stacks with wide tubular connections to the originating pregrana thylakoids (60 HAI), structural differentiation of pro-grana stacks and expanded stroma thylakoids (84 HAI), and conversion of the pro-grana stacks into mature grana stacks (120 HAI). Development of the planar pregranal thylakoids and the pro-grana membrane stacks coincides with the appearance of thylakoid-bound polysomes and photosystem II complex subunits at 36 HAI. ATP synthase, cytochrome b6f, and light-harvesting complex II proteins are detected at 60 HAI, while PSI proteins and the curvature-inducing CURT1A protein appear at 84 HAI. If stromal ribosome biogenesis is delayed, prothylakoids accumulate until stromal ribosomes are produced, and grana-forming thylakoids develop after polysomes bind to the thylakoid membranes. In fzo-like (fzl) mutants, in which thylakoid organization is perturbed, pro-grana stacks in cotyledons form discrete, spiral membrane compartments instead of organelle-wide membrane networks, suggesting that FZL is involved in fusing membrane compartments together. Our data demonstrate that the assembly of thylakoid protein complexes, CURT1 proteins, and FZL proteins mediate distinct and critical steps in thylakoid biogenesis.
INTRODUCTION
The plastid family of organelles develops primarily from proplastids in meristem cells (Waters and Langdale, 2009; Charuvi et al., 2012). Proplastids give rise to three subclasses of plastids: the pregranal plastids and etioplasts that serve as precursors forms of chloroplasts; the amyloplasts, elaioplasts, and leucoplasts that develop directly from proplastids; and the chromoplasts that can develop from either proplastids or chloroplasts (Jarvis and López-Juez, 2013; Staehelin, 2015). Each plastid type has unique structural and functional attributes with the chloroplasts being the most prominent due to their distinctive green color and their role in capturing sunlight to produce carbohydrates and molecular oxygen, upon which essentially all life depends.
Each chloroplast is delineated by two envelope membranes that surround an aqueous matrix, the stroma, and a complex 3D network of membranes termed thylakoids. The thylakoids form a continuous chamber, the thylakoid lumen, and are organized into two structural domains: the disc-shaped, stacked grana thylakoids and the interconnecting, nonstacked stroma thylakoids (Austin and Staehelin, 2011). They harbor the functional complexes that trap and convert the sun’s energy into high-energy electrons and transmembrane proton gradients, which generate NADPH, ATP, and O2. The protein complexes that perform these functions are photosystems I and II (PSI and PSII), light-harvesting complex II (LHCII), cytochrome b6f (cyt b6f), and ATP synthase (Nevo et al., 2012; Garab, 2014).
The structural differentiation of the thylakoids results from a nonuniform distribution of these complexes between grana and stroma thylakoids. Thus, most PSII and LHCII complexes are located in grana thylakoids and most PSI and all ATP synthases in stroma thylakoids. In contrast, the cyt b6f complexes are found in similar amounts in grana and stroma thylakoids (Staehelin, 2003; Pribil et al., 2014). The physical parameters of the complexes that lead to the formation of stacked grana membranes and to the confinement of other complexes to stroma membranes have been the subject of many studies. They also play a major role in defining the process of chloroplast biogenesis and development from nonphotosynthetic plastids, the focus of this study (Daum and Kühlbrandt, 2011; Kouřil et al., 2011; Pogson et al., 2015; Rast et al., 2015).
Virtually all recent advances in our understanding of thylakoid architecture and the structural basis of thylakoid dynamics have come from studies of cryofixed cells. The advantages of cryofixation methods such as high-pressure freezing over chemical fixation methods have been known since the 1980s (Gilkey and Staehelin, 1986). Indeed, the use of cryofixation methods in membrane studies has been shown to be of critical importance because the quality of the initial specimen preservation determines the quality of the final electron micrographic and electron tomography data. For example, cryofixation immobilizes all cellular molecules, including proteins, lipids, salts, and water, simultaneously within ∼1 ms compared with the partial stabilization of only certain types of cellular molecules by chemical cross-linking agents that takes minutes and can trigger artifactual membrane vesiculation. In cells subsequently processed via freeze substitution, both dehydration-related structural changes and the extraction of lipids and solutes are greatly reduced (Shimoni et al., 2005; Austin et al., 2006; Staehelin and Kang, 2008; Charuvi et al., 2012). More recently developed cryo techniques involve direct visualization of the frozen samples in the electron microscope (Daum et al., 2010; Engel et al., 2015). Most notably, the images produced by Engel et al. (2015) using cryo-electron tomography imaging of cryo-focused ion beam milled, vitreous Chlamydomonas reinhardtii cells are trendsetting in the clarity with which the structural details of both chloroplast envelope and thylakoid membranes are seen.
Studies of the 3D architecture of thylakoids by means of serial section electron microscopy of chemically fixed cells in the 1960s culminated in the formulation of the “helical model” of thylakoid architecture as formulated by Paolillo (1970). In the helical model, the individual grana stacks are connected through evenly spaced stroma thylakoids that wind around the stacks as right-handed helices and connect to the individual grana cisternae via narrow, tubular membrane junctions (Mustárdy and Garab, 2003). This model has been validated by means of electron tomography analysis of thylakoids preserved by cryofixation methods (Austin and Staehelin, 2011; Daum and Kühlbrandt, 2011).
An important structural feature of thylakoids uncovered in specimens preserved by cryofixation methods includes the demonstration that plastoglobules contain biosynthetic enzymes and are attached to thylakoids through a half-bilayer that encompasses the globule contents and is continuous with the thylakoid membrane via blister-like outgrowths of the stroma-side leaflet of the membrane bilayer (Austin et al., 2006). Cryofixed specimens have also shown that exposure to light changes the volume of the thylakoid lumen (Kirchhoff et al., 2011). Most recently, studies of cryofixed cells have provided clear evidence for distinct contact sites between thylakoid margins and the inner chloroplast envelope membrane both in Arabidopsis thaliana shoot apical meristem and in Chlamydomonas cells (Charuvi et al., 2012; Engel et al., 2015).
Although the above-mentioned studies have contributed much to our understanding of thylakoid architecture, they have provided few insights into how the elaborate architecture of thylakoids arises during the transformation of proplastids into chloroplasts. Charuvi et al. (2012) examined plastid-to-chloroplast conversion in the shoot apical meristem with electron tomography to determine thylakoid structures of young leaf cells and to uncover that thylakoids can either elaborate or degrade in differentiating leaf cells depending the origin of the cells. When a seed germinates underground, its proplastids first develop into etioplasts that contain prolamellar bodies (PLBs) and prothylakoid membranes that are continuous with and extend from marginal PLB tubules into the stroma (Weier et al., 1970). PLBs are paracrystalline bodies of chloroplast lipids (mostly mono- and digalactolipids), protochlorophyllide, and the light-dependent enzyme complex protochlorophyllide oxidoreductase (Ryberg and Sundqvist, 1982; Selstam et al., 2007; Solymosi and Schoefs, 2010). Upon exposure to light and the resulting conversion of protochlorophyllide to chlorophyll, the prothylakoids become photosynthetically active thylakoids that grow and differentiate into grana and stroma thylakoids. Simultaneously, the PLBs shrink in size due to the transfer of lipids into the expanding thylakoid membranes (Henningsen and Boynton, 1974; Adam et al., 2011). The physical continuity between prolamellar body tubules and forming thylakoid membranes and the direct (without vesicle intermediate) transformation of PLB tubules into fenestrated thylakoids were documented with amazing clarity by early electron microscopists (Gunning, 1965). This physical relationship has been recently confirmed in an electron tomographic study of chemically fixed, greening bean chloroplasts (Kowalewska et al., 2016).
How grana stacks are formed remains somewhat of a mystery. As shown by Brangeon and Mustardy (1979), grana stack formation originates with the formation of tongue-like membrane flaps that originate in the margins of membrane fenestrae and grow over the adjacent flat membrane surface to create stacked membrane domains. However, what triggers the formation of membrane flaps and how the initial ministacks become fully developed grana stacks that are interconnected to each other by means of stroma thylakoids that form right-handed helices around the stacks and form bridges between the grana stacks remains to be determined (Austin and Staehelin, 2011). The recently discovered family of membrane curvature-inducing CURT1 proteins (Armbruster et al., 2013) demonstrates that thylakoid architecture is also dependent on membrane-shaping structural proteins.
The goal of this study was to use electron tomography analysis to reexamine the process of thylakoid assembly in proplastids of high-pressure frozen cotyledon cells of germinating Arabidopsis seeds and to correlate these changes with the expression of the major protein complexes at the assembly stages of thylakoids. We also compare the assembly of thylakoids between wild-type chloroplasts, a mutant with impaired ribosome production, and a mutant deficient in the dynamin-like Fzo-like (FZL) membrane fusion protein. Thylakoid-inner envelope membrane contact sites are a typical feature of developing chloroplasts.
RESULTS
Chloroplast Development in Cotyledon Cells of Germinating Arabidopsis Seedlings Takes 120 h from the Onset of Illumination
To monitor the development of chloroplasts from proplastids in germinating Arabidopsis seeds, we examined cotyledon cells at 24, 36, 60, 84, and 120 h after imbibition (HAI) (Figures 1A to 1E) using confocal laser scanning microscopy (Figures 1F to 1J) and transmission electron microscopy (Figures 1K to 1O). The seedlings were grown under continuous light so that light-induced plastid-to-chloroplast conversion was not interrupted by periods of darkness. Autofluorescence from chlorophyll was rarely detected in cotyledon cells of seedlings at 24 HAI, but unambiguously observed in 36 HAI cotyledon cells (Figures 1F and 1G). Chloroplasts enlarged and their autofluorescence intensified gradually from 36 to 120 HAI (Figures 1G to 1J). In the 84 and 120 HAI cotyledon cells, individual chloroplasts could be identified in bright-field micrographs owing to their green coloration (Figures 1I’ and 1J’).
Figure 1.
Arabidopsis Seedling Development.
(A) to (E) Stereomicroscope images of Arabidopsis seedlings at 24 (A), 36 (B), 60 (C), 84 (D), and 120 (E) HAI. Bars = 0.5 mm.
(F) to (J) Confocal micrographs of cotyledon cells of seedlings at 24 (F), 36 (G), 60 (H), 84 (I), and 120 (J) HAI, showing chlorophyll autofluorescence. Bars = 10 μm. (I’) and (J’) are bright-field micrographs of the same are shown in (I) and (J), respectively. Chloroplasts (dark spheroids) are readily identified in (I’) and (J’).
(K) to (O) Transmission electron micrographs of cotyledon cells at 24 (K), 36 (L), 60 (M), 84 (N), and 120 (O) HAI. Plastids/chloroplasts are marked with arrowheads. Bars = 5 μm.
(P) Chloroplast lengths estimated from chloroplast autofluorescence. Confocal micrographs were captured from wild-type, rh3-4 mutant, and fzl mutant cotyledon cells (48 chloroplasts from three seedlings of the three genotypes were measured). Plastids at 24 HAI and rh3-4 plastids at 36 HAI were not examined because their chlorophyll autofluorescence was too weak for reliable measurements.
(Q) The thickness of prothylakoids and pregranal thylakoids in 36 HAI plastids. Forty-one prothylakoids and 21 pregranal thylakoids were measured from tomographic slices of three plastids at 24 and 36 HAI, respectively. Error bars indicate sd in (P) and (O).
Transmission electron microscopy imaging of 24 HAI cotyledon cells demonstrated that the bulk of their cytoplasm was filled with reserve organelles, protein storage vacuoles, and lipid bodies (Figure 1K). The large, darkly stained storage vacuoles together with the nucleus, mitochondria, and plastids occupied the central cytoplasm, whereas the tightly packed lipid bodies were located in the cell periphery (Supplemental Figure 1). In cryofixed/freeze-substituted cells, the darkly staining material in the storage vacuoles corresponds to the storage proteins and the bright spots to holes in the sections left behind by the shattered phytin globoids (Zheng and Staehelin, 2011).
Between 24 and 36 HAI, the first significant changes in the architecture of the pregranal chloroplasts were seen, including a substantial increase in volume and expansion of the nonstacked thylakoids (Figures 1K, 1L, and 2). The cytoplasm of the 36 HAI cotyledon cells still contained large numbers of lipid bodies, and plastoglobules were present in the pregranal chloroplasts. The chloroplasts in the 60 HAI cotyledon cells possessed thylakoids that extended across their entire length and contained small prograna stacks embedded in a darkly stained stroma (Figures 1M and 3). At this stage, the number of cytoplasmic lipid bodies was decreased, and most storage vacuoles had merged into one large, central vacuole (Figure 1M; Supplemental Figures 1C to 1E).
Figure 2.
Arabidopsis Plastids at 24 and 36 HAI.
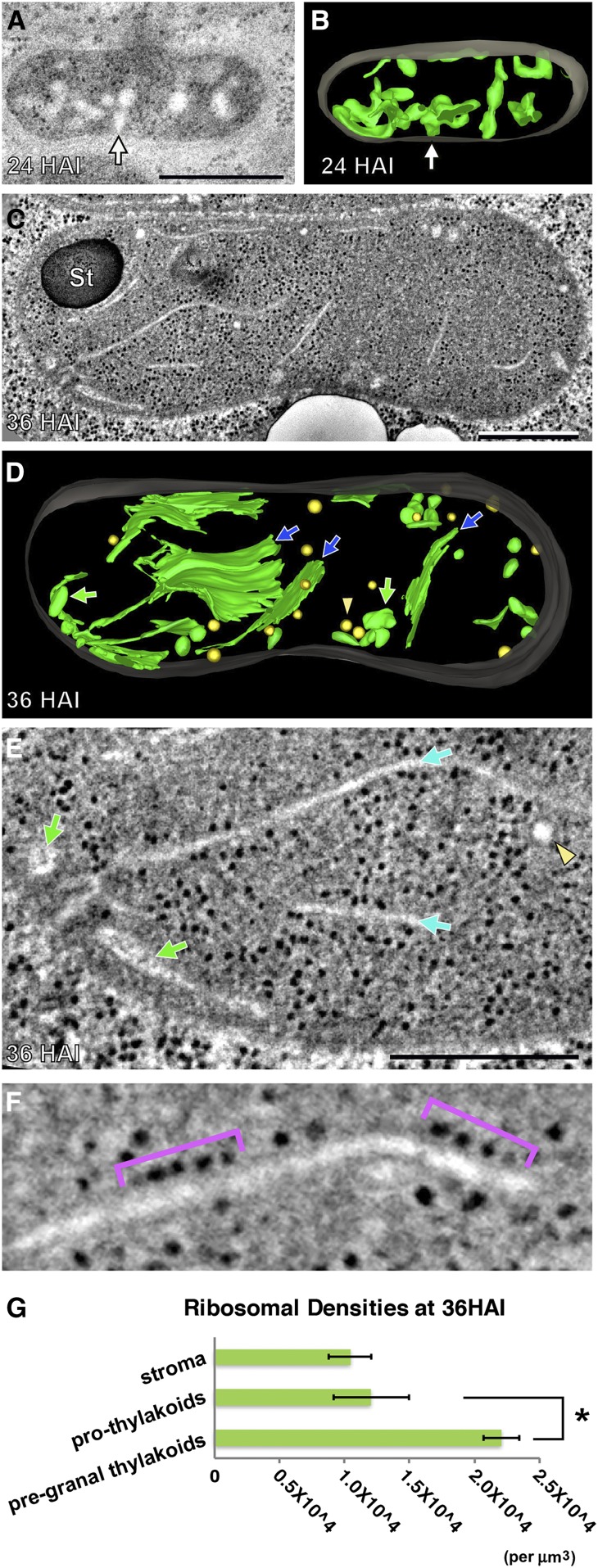
(A) and (B) Electron tomographic slice of a plastid in an Arabidopsis cotyledon cell at 24 HAI (A) and its 3D tomographic model (B). A contact site where the prothylakoid is continuous with the inner envelope is marked with an arrow in (B).
(C) to (F) Electron tomographic slice ([C], [E], and [F]) and 3D model (D) of an Arabidopsis plastid at 36 HAI. Tubulo-vesicular prothylakoids and plastoglobule-like compartments are colored in green and yellow, respectively. In (D) and (E), blue arrows, green arrows, and yellow arrowheads mark rough prothylakoids, thick prothylakoids, and plastoglobule-like compartments, respectively. Membrane-bound ribosomes of a rough prothylakoid are indicated with purple brackets in (F).
(G) Ribosome densities of bulk stroma (stroma), stroma within 20 nm from prothylakoid membranes (prothylakoids), and from pregranal thylakoid membranes (pregranal thylakoids). *P < 0.01 (unpaired Student’s t test).
Bars in (A), (C), and (E) = 500 nm.
Figure 3.
Arabidopsis Plastids at 60 HAI.
(A) and (B) Electron tomographic slice of a plastid in an Arabidopsis cotyledon cell at 60 HAI (A) and its 3D model (B).
(C) Three tomographic slices showing a bud on a rough thylakoid.
(D) 3D model of the bud (light blue) and the rough thylakoid (green) based on the tomogram in (C). Arrows in (C) and (D) point to the same position.
(E) Side view of the model in (D).
(F) Same 3D model in (E) but ribosomes (gold spheres) within 10 nm of the thylakoid membrane are shown.
(G) Three tomographic slice images of a tongue-like thylakoid lying over a larger thylakoid.
(H) 3D model of the thylakoids in (G). Arrows in (G) and (H) point to the same position as the arrowhead in (G).
(I) Same 3D model in (H) but ribosomes (gold spheres) within 10 nm from the thylakoid membrane are shown.
(J) View of the 3D model in (H) after rotating 90°. The membrane contact zone of a double-layered thylakoid is traced with a magenta dashed curve.
(K) to (N) Thylakoid stacks consisting of three layers (red brackets). (N) and (M) show 3D models of the stacks in (K) and (L), respectively. The stack in (K) and (L) has folds on one side while the stack in (M) and (N) has folds on opposite sides constituting an S-shaped stack.
(O) and (P) Tomographic slice images of a thylakoid stack and its 3D model. In (P), cross sections of the stack along the blue and yellow lines are shown ([P’] and [P’’]).
(Q) Ribosome densities of bulk stroma (stroma), stroma within 20 nm from unstacked thylakoid membranes (unstacked thylakoids), and from grana-forming thylakoid membranes (grana-forming thylakoids). *P < 0.01 (unpaired Student’s t test).
Three tomographic slice images from tomograms are shown in (C), (G), (K), (L), and (O). Their slice numbers are indicated in the panels (z). Bars = 500 nm in (A) and 100 nm in (C), (G), (K), (L), and (O).
In the 84 HAI cotyledons, the swollen cells exhibited a rounded shape and were surrounded by large air spaces. Large vacuoles occupied the central regions of the cells and displaced all the other organelles, including the prominent chloroplasts, to the cell periphery (Figure 1N). The thylakoids displayed an increasingly elaborate architecture and alignment with the equatorial plane of the spheroidal chloroplasts (Figures 1N and 4). Cell enlargement continued up to 120 HAI primarily due to an expansion of the central vacuole (Figures 1O and 1P; Supplemental Data Set 1). The chloroplasts in these cells appeared fully developed and possessed extensive grana and stroma thylakoid membrane systems with the margins of the end thylakoids anchored to the chloroplast envelope via contact sites (Figures 1 and 5).
Figure 4.
Arabidopsis Plastids at 84 HAI.
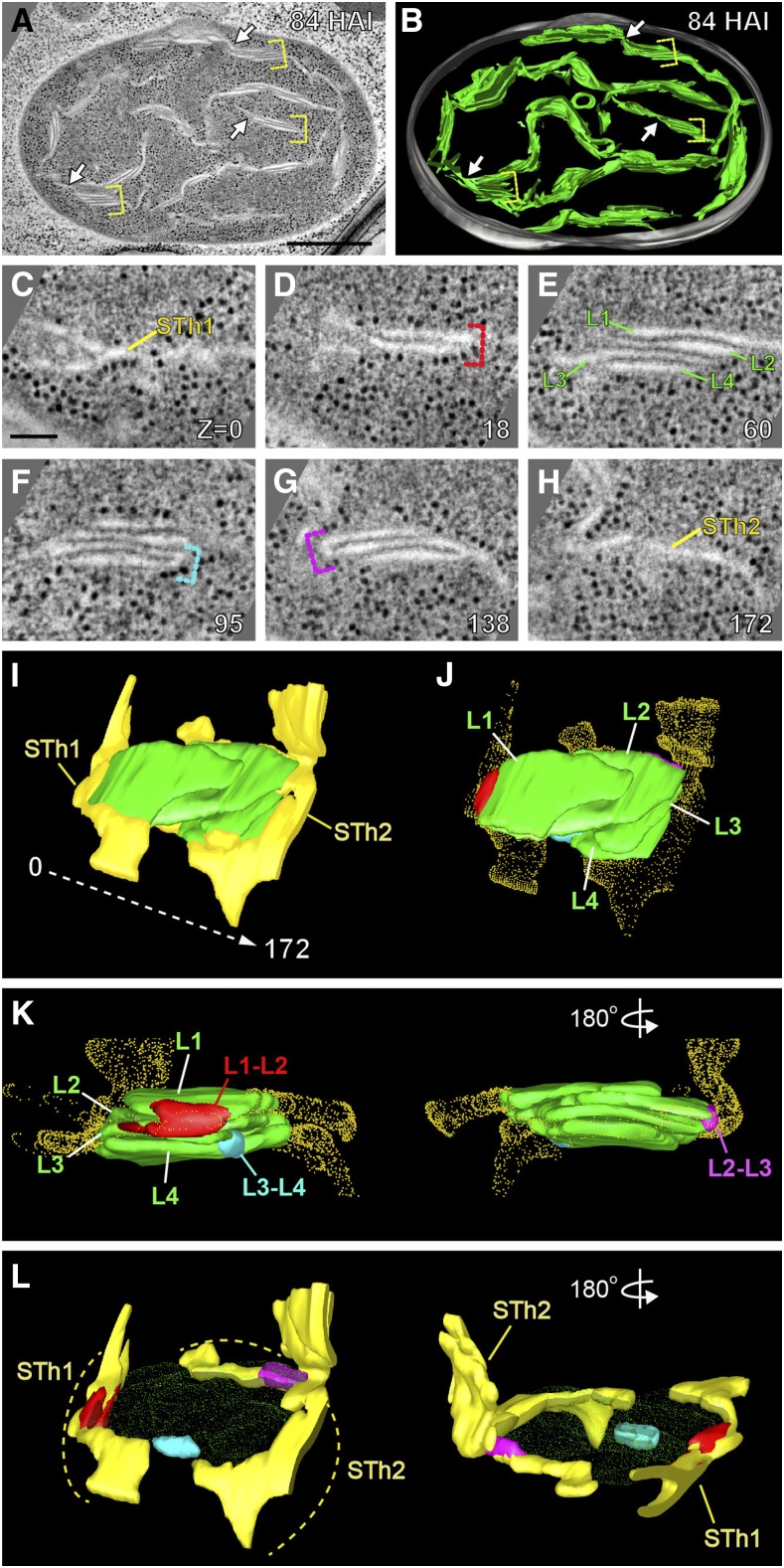
(A) and (B) Electron tomographic slice of a plastid in an Arabidopsis cotyledon cell at 84 HAI (A) and its 3D model (B). Three stacks made of more than three layers and one of their unstacked stroma thylakoids are marked with yellow brackets and arrows, respectively, in (A) and (B).
(C) to (H) Six tomographic slices from a grana stack consisting of four layers (L1–L4 in E) and two stroma thylakoids (STh1 in C and STh2 in [H]) surrounding the stack. Bends connecting adjacent layers are marked with brackets (red in [D], cyan in [F], magenta and dark in [G]). Slice numbers are indicated in the lower right corner of each panel.
(I) 3D model of the grana stack in (C) to (H) showing the four layers (L1–L4).
(J) The grana stack in (I) and its bent membranes between adjacent layers (highlighted according to the colors used in [C] to [H]).
(K) Side views of the 3D model in (J) revealing the three links between adjacent layers.
(L) The two stroma thylakoids around the grana stack (stippled) in (I).
Bars 500 nm in (A) and 100 nm in (C).
Figure 5.
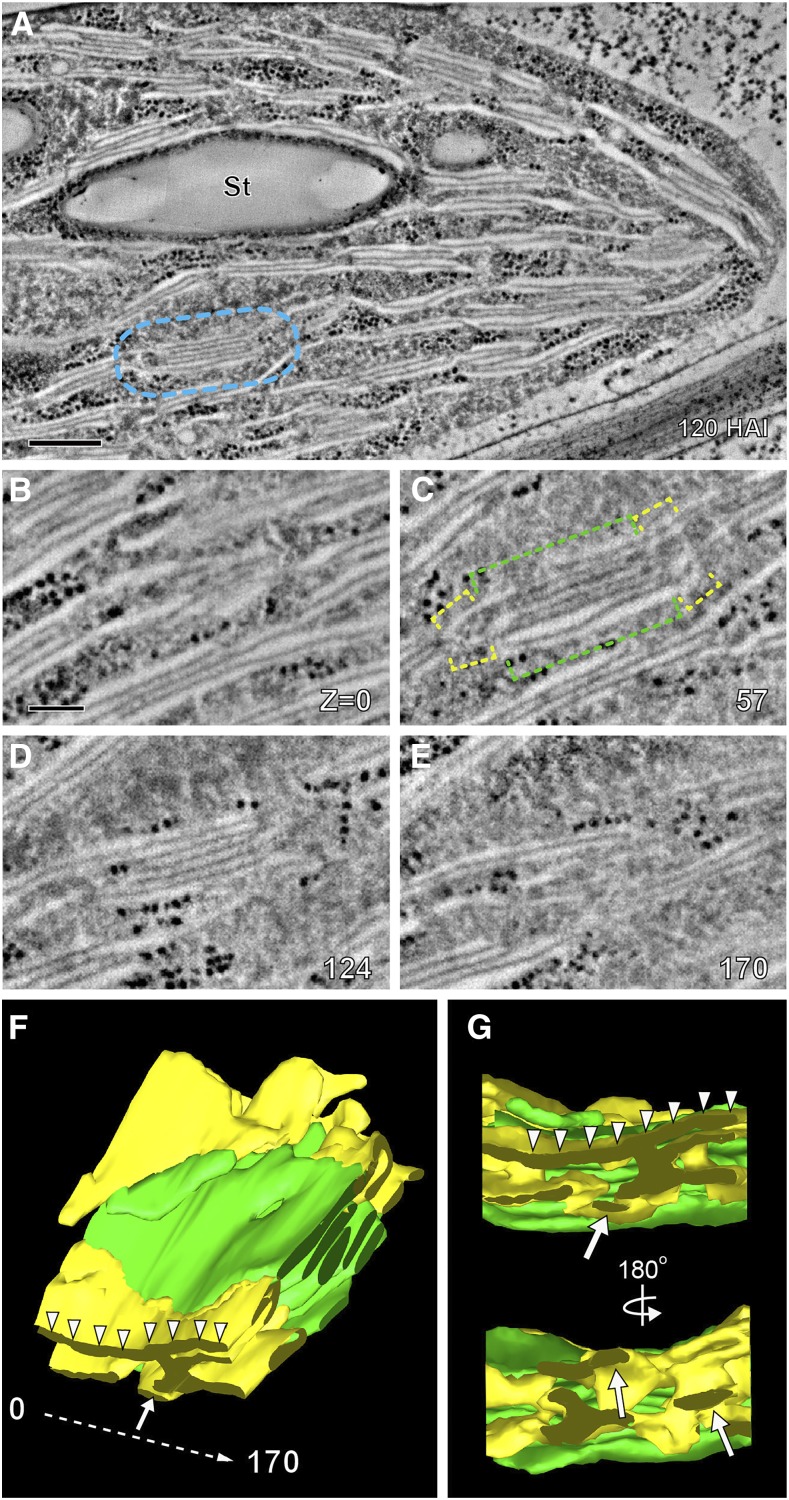
Arabidopsis Chloroplast at 120 HAI.
(A) Electron tomographic slice of a chloroplast in an Arabidopsis cotyledon cell at 120 HAI (A). The grana stack in (B) to (G) is enclosed in the light-blue rectangle. Bars = 500 nm.
(B) to (E) Four tomographic slices of a granum consisting of seven layers (green brackets in [C]) and stroma thylakoids surrounding the stack (yellow brackets in [C]). Slice numbers are indicated in the lower right corner.
(F) 3D model of the grana stack (green) and its stroma thylakoids (yellow) in (B) to (E).
(G) Side views of the 3D model in (G). The convoluted stroma thylakoids consist of sheet-like (arrows in [F] and [G]) and tubular (arrowhead in [F] and [G]) components.
The Assembly of Polysomes on the Thylakoid Membranes Correlates with the Transformation of the Tubulo-Vesicular Prothylakoids into Sheet-Like, Pregranal Thylakoids with Collapsed Lumina
Plastids in 24 HAI cotyledon cells had an ellipsoid shape with an average length of 0.488 μm (sd = 0.224 μm, n = 20) in electron micrographs. Their dense, opaque stroma contained tubulo-vesicular prothylakoids with wide lumina that were attached to the inner envelope membrane by means of membrane contact sites (Figures 2A and 2B, arrows). Some plastoglobules and inner envelope membrane infoldings were also seen (Supplemental Figure 1B).
Between 24 and 36 HAI, the average length of the plastids increased significantly from 0.488 to 1.73 μm (sd = 0. 274 μm, n = 25). During this phase of development, the tubulo-vesicular prothylakoids were gradually converted into larger, nonstacked, and sheet-like pregranal thylakoids that displayed a narrow lumen (Figure 1Q; Supplemental Data Set 2) and extended toward the central regions of the plastids (Figures 2C and 2D). This striking transformation of the thylakoid membranes was always accompanied by the binding of polysomes to the pregranal thylakoids (Figures 2E and 2F), which also possessed contact sites with the inner envelope membrane. We compared ribosome densities (ribosomes per µm3) in the bulk stroma and adjacent to the surfaces of prothylakoids and pregranal thylakoids in electron tomograms (Figure 2G; Supplemental Figure 2 and Supplemental Data Set 3). The ribosome densities in the vicinity of prothylakoids (12,047.2 per μm3, sd: 2888.1) were similar to those in the bulk stroma (10,438.0 per μm3, sd: 1623.9). By contrast, planar pregranal thylakoids had significantly more closely associated ribosomes (22,051.9 per μm3, sd: 1360.1) compared with bulk stroma or tubular prothylakoids. These observations suggest that the flattening of the membranes and their increase in size are related to the insertion of plastid-encoded proteins into the thylakoid membranes and signal the beginning of the assembly of photosynthetic complexes.
Grana-Forming Thylakoid Buds Arise Adjacent to Thylakoid-Bound Polysomes and Become Coated with Additional Polysomes as They Produce More Stacked Membrane Regions
In the 60 HAI plastids, all of the thylakoids formed a single, coherent 3D network that extended along the entire length of the chloroplasts (Figures 3A and 3B). At this time point, the first small prograna stacks, which we define here as grana stacks with wide, slit-like connections between the grana and stroma membrane regions, began to form, giving rise to a variety of membrane shapes in cross-sectional views (Figures 3C to 3P). To understand how these membrane configurations are related to the de novo assembly of grana stacks, we examined the 3D organization of developing prograna in electron tomographic slices and models.
The planar 60 HAI thylakoid domains were morphologically similar to the flat, polysome-bearing thylakoids of the 36 HAI chloroplasts. However, they also possessed prograna-forming, dome-shaped buds coated with polysomes (Figures 3C to 3F; Supplemental Movie 1). As these tongue-like membrane folds expanded over the membrane surfaces adjacent to the outgrowth-forming sites, they developed contact zones that became the first stacked membrane domains (Figures 3G to 3J). The fact that the grana-forming thylakoid buds arose in association with membrane-bound polysomes is consistent with the hypothesis that localized cotranslational protein insertion is an integral part in the grana-forming, membrane outgrowth process (Figures 3F and 3I).
The first polysome-associated stacked membrane domains subsequently seemed to serve as nucleation sites for additional grana-forming membrane folds. Formation of these folds, however, did not seem to follow a precise, 3D developmental program such as a helical pattern program. Instead, membrane sites closely associated with the newly formed adhering membranes appeared to develop buds that grew over the existing grana-forming membrane regions from different directions, thereby creating small membrane stacks that gave rise to a variety of different membrane configurations (Figures 3K to 3P; Supplemental Movies 2 and 3). These prograna stacks consisted of somewhat randomly interconnected membrane folds that had slanted or other noncanonical membrane configurations. Furthermore, unlike in mature chloroplasts where the grana thylakoids can be linked to the stroma thylakoids either through narrow membrane tubules or through wider, sheet-like bridges (Austin and Staehelin, 2011), all of the initial membrane folds were connected to the adjacent membranes through wide, sheet-like membrane structures. The thylakoid-inner envelope membrane contact sites were dispersed around the envelope margins.
Ribosome density in the bulk stroma was higher at 60 HAI (23,078.8 per μm3, sd: 6043.8) than at 36 HAI, indicating that ribosomes accumulated in the stroma during the 24-h period (compare Figures 2G and 3Q). As thylakoids expanded, ribosomes were often concentrated to space between growing thylakoids and these clusters of ribosomes in the stroma contributed to the increase of stromal ribosome density (Supplemental Figures 2E and 2F). On the thylakoid membrane surfaces, ribosomes were more enriched at membrane buds and accessible membranes of thylakoid stacks (24,492.9 per μm3, sd: 3707.8, Supplemental Data Set 3) than at unstacked interconnecting thylakoids (11,722.6 per μm3, sd: 4654.6).
Between 60 and 84 HAI the Prograna Stacks Develop More Uniformly Organized Grana Stacks and Stroma Thylakoids, but Retain Wide, Grana-Stroma Membrane Connections
Defining structural features of mature thylakoids include grana stacks interconnected by stroma thylakoids and grana thylakoids exhibiting uniform margins with small radii of curvature. All of these architectural elements began to emerge in the 84 HAI plastids as evidenced by the development of more regularly shaped grana stacks with larger diameters and more stacked membrane regions, and interconnected by increasing numbers of stroma thylakoids (Figures 4A and 4B). However, the grana stacks were still classified as prograna because the grana-stroma membrane connections were wide and slit-like as in the 60 HAI plastids (Figures 3 and 4L). Hints of a helical arrangement of the connecting stroma membranes around the grana were also seen (Figure 4L, yellow membrane elements encircling the stack and having connections to disks in the stack). Contact sites between thylakoids and inner envelope membranes were observed regularly (arrowheads in Figure 4A).
The grana stack of the 84 HAI plastid depicted in the tomographic slice images Figures 4C to 4H contained four stacked thylakoids connected to two stroma thylakoids that surrounded the stack (Figures 4I to 4K; Supplemental Movie 4). In addition, the adjacent thylakoids in the stack were connected to each other through membrane folds that appeared as U-shaped structures in the cross-sectional views (Figures 4D, 4F, and 4G). These wide connections suggest that the new pro-grana thylakoids continued to be formed as in the 60 HAI plastids (Figure 3).
Narrowing of the Grana to Stroma Membrane Connections in the 120 HAI Plastids Signals the Onset of the Final Phase of Chloroplast Development
The most notable structural difference between the 84 and 120 HAI chloroplasts was the huge increase in thylakoid membranes, which become the dominant structural feature of the latter chloroplasts (Figure 5A). In addition, the grana stacks possessed a greater width, an increased number of membranes per stack (Figures 5B to 5E; Supplemental Movie 5), and most importantly, the transformation of the wide, slit-like grana-to-stroma membrane connections into narrow tubules (compare Figures 4I and 5G), a characteristic feature of mature chloroplasts (Austin and Staehelin, 2011). The sheet-like stroma thylakoids were also seen around the stack (Figure 5F and 5G). Prominent starch granules accumulated between the thylakoids (Figure 5A).
Transcriptomic Analysis Indicates That Transcripts Encoding Subunits for PSII Are Enriched When Polysomes Are Recruited to Pregranal and Grana-Forming Thylakoids
To determine what photosynthetic complexes assembled in the intermediates of thylakoid biogenesis that we observed with electron tomography, we performed transcriptomic analysis of RNA samples isolated from seedlings at the five stages. Total RNA samples were analyzed after degrading rRNA molecules to quantify transcripts from both plastid and nuclear genes encoding constituents of PSI, PSII, LHCs, cyt b6f, and ATP synthase complexes. We repeated the experiments twice, and in each independent experiment, we obtained 12 to 30 million high-quality reads that mapped to the Arabidopsis plastid genome or the five chromosomes (Supplemental Data Set 4). The heat maps (Figure 6; Supplemental Figure 3) illustrate transcriptional profiles of subunits that consist of the major protein complexes in thylakoid membranes during light-induced chloroplast development.
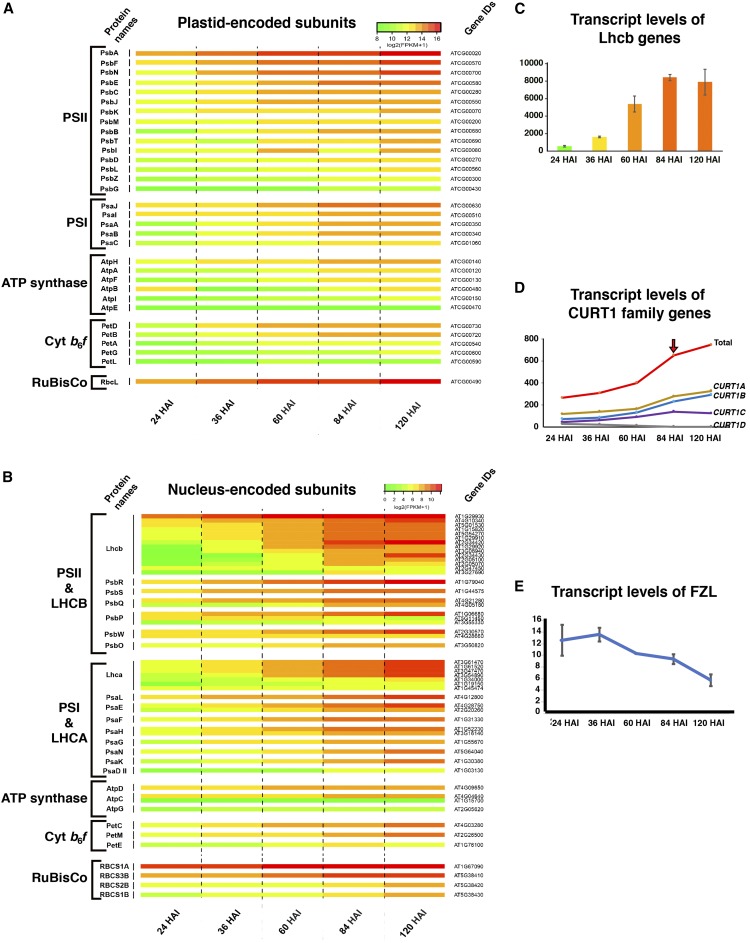
Figure 6.
Transcriptomic Analysis of Genes Encoding Subunits of PSII, PSI, cyt b6f, and ATP Synthase Complexes in the Thylakoid Membrane during Chloroplast Biogenesis.
(A) and (B) Heat maps illustrating changes in the transcript levels of the plastid and nuclear genes encoding complex subunits and Rubisco are shown in (A) and (B), respectively. Color-coding scales for the two heat maps are not equal.
(C) Histogram showing total FPKM values of all Lhcb gene. The amount of Lhcb transcripts is increased steeply from 36 to 84 HAI.
(D) and (E) Transcript levels of CURT1 family genes are depicted in (D) and of FZL in (E).
Names of the subunits constituting the complexes in (A) and (B) followed the nomenclature by Race et al. (1999), and their gene IDs were obtained from www.arabidopsis.org. The charts in (C) to (F) were prepared with average FPKM values and their standard deviations of genes from the two sequencing experiments.
PsbA (ATCG00020) was by far the most highly transcribed gene in the plastid at 24 HAI (fragments per kilobase of transcript per million mapped reads [FPKM]: 10,325–13,840), and its FPKM values surpassed 20,000 at 36 HAI (Figure 6A; Supplemental Data Set 5). FPKM values of some plastid genes encoding PSII proteins reached 10,000 at 36 HAI, while comparable upregulation was observed for plastid genes encoding PSI, cyt b6f, and ATP synthase subunits only after 60 and 84 HAI (Figures 6A; Supplemental Figure 3A). Transcripts for constructing PSII complexes were more abundant in the plastid at the early time points when polysomes concentrated to thylakoid membranes (36 and 60 HAI) than those for the other three complexes. These results suggest that transcripts of PSII subunits encoded by the plastid genome are translated by the polysomes associated with the flattening pregranal thylakoids at 36 HAI (Figure 2) and the grana-forming thylakoids at 60 HAI (Figure 3).
The nuclear genes encoding proteins of the four thylakoid complexes exhibited similar expression patterns as the plastid genes (Figure 6B). PsbR, PsbS, PsbQ, and PsbP became activated at 36 HAI, as some of their partner genes in the plastid did. Nuclear genes encoding PSI, cyt b6f, and ATP synthase subunits were upregulated around 60 HAI. The early transcriptional activation of PSII genes followed by genes for other complexes occurred in both plastid and nucleus. The correlative expression profiles of plastid and nuclear genes encoding components of each complex reinforced the interorganelle communication. Transcript levels of the light-harvesting complex for PSII (Lhcb) increased significantly at 60 and 84 HAI (Figure 6C) when grana-forming thylakoids arose and layers in the prograna stacks increased (Figures 3 and 4).
CURT1A is a thylakoid curvature-inducing membrane protein that localizes to grana margins, and the Arabidopsis genome encodes four CURT1 family proteins (Armbruster et al., 2013). Among the four members, transcripts from CURT1A (AT4G01150) and CURT1B (AT2G46820) accumulated more than CURT1C (AT1G52220) (Figure 6E). FPKM values of CURT1D (AT4G38100) were almost negligible at all the time points (1.3-29.0). CURT1 transcript levels increased sharply at 84 HAI (arrow, Figure 6D) when the distinction between grana stacks and interconnecting thylakoids became more apparent (Figure 4) and CURT1A was readily detected by immunoblot analysis (Figures 7C and 7D).
Figure 7.
Expression Analyses of Genes Encoding PSII, PSI, cyt b6f, and ATP Synthase Subunits in Thylakoid Membranes during Chloroplast Biogenesis.
(A) and (B) ddPCR quantification of transcript levels of genes selected from the RNA-seq data in Figure 6. Heat maps were drawn to illustrate transcript concentrations at the five stages. Average copies per picogram (pg) of RNA sample (for plastid-encoded genes in [A]) and average copies per nanogram (ng) of RNA sample (for nucleus-encoded genes in [B]) from three replicate ddPCR experiments are given. Supplemental Data Set 3 has results from the three ddPCR experiments.
(C) and (D) Immunoblot analysis showing time points when subunits of the four complexes are detected. Amounts of polypeptides were visualized on films (C) or quantified by their chemiluminescence (D). Proteins translated in the stroma are in green letters. For each complex, subunits encoded by the nuclear genome and the plastid genome are shown separately. CURT1A is a thylakoid membrane protein localizing to the grana margin. PBA, a subunit of the Arabidopsis proteasome complex b, was used as the loading control.
For the histograms in (B), readouts from each polypeptide were normalized with those of PBA at the five time points and relative values were calculated after the largest normalized intensity was set to be 1.0. The asterisk in each histogram of (D) indicates the time point when polypeptide readout significantly increased from readout of the previous time point (Student’s t test, one-tailed unpaired test, *P < 0.025).
To substantiate the RNA-seq results, we selected genes exhibiting varying expression levels from the heat maps in Figure 6 and quantified their transcripts using droplet digital PCR (ddPCR). This method is more reliable than quantitative RT-PCR because it does not require external reference genes for measuring amounts of target DNA templates in reaction mixes and has been shown to be more accurate and reproducible (Hindson et al., 2013; Taylor et al., 2017). We chose three PSII subunit genes, two PSI subunit genes, one ATP synthase subunit gene, and two cyt b6f subunit genes, one from the plastid genome and another from the nuclear genome (Figures 7A and 7B; Supplemental Data Set 6). ddPCR analysis indicated that PsbA was most actively transcribed among the three plastid PSII subunit genes, followed by PsbB and PsbZ during the chloroplast biogenesis (Figure 7A). The relative transcript abundances of the three genes agreed with transcriptional activities of the genes measured by RNA-seq in Figure 6A. We were able to observe similar matches between ddPCR and RNA-seq quantifications for plastid-encoded PSI subunit genes (PsaJ and PsaA), nucleus-encoded PSII subunit genes (PsbS, Lhcb-AT1G29930, and Lhcb-AT2G47450), and nucleus-encoded PSI subunit genes (PsaK and Lhca-AT3G54890).
PsbA and PsbS were highly expressed genes of PSII components (Figures 6A and 6B) and their upregulation at 36 and 60 HAI was also observed in ddPCR readouts (Figures 7A and 7B). Transcript concentrations of PsbA (8.6 copies/pg) and PsbS (869 copies/ng) were higher than those of other complex subunits. Amounts of PsaJ, PsaK, and Lhca (AT3G54890) sharply increased 60 to 84 HAI, suggesting that PSI is primarily produced after 60 HAI. These ddPCR results confirm the conclusion from the RNA-seq analyses that transcripts for PSII assembly are more enriched at 36 and 60 HAI than those for other complexes (Figure 6).
PSII, PSI, LHCII, and CURT1 Proteins Appear at Different Times of Thylakoid Development, in Parallel with Changes in Their Transcript Levels
We performed immunoblot analyses for the major protein complexes of thylakoid membranes and CURT1 proteins to test whether accumulation of the transcripts at different times leads to their translation. To evaluate translation in the stroma and cytosol, we employed antibodies against subunits encoded by the nuclear genome and subunits encoded in the plastid genome for each complex. PBA, a subunit of the Arabidopsis proteasome complex β, was used as the loading control (Lee et al., 2013). The immunoblot data were confirmed in three separate experiments and quantified via polypeptide chemiluminescence as measured on film (Figure 7C) or with a digital imaging system (Figure 7D; Supplemental Data Set 7).
PSII proteins were detected earlier in the immunoblots compared with other complex proteins during light-induced chloroplast development, in agreement with the RNA-seq analysis. PsbA, the stroma-translated D1 protein, was seen in the 24 HAI membranes and its amount increased in the 36 HAI thylakoids. PsbN, a chloroplast-encoded, low molecular weight, transmembrane protein that localizes to stroma thylakoids and is needed for the efficient assembly of PSII reaction centers (Torabi et al., 2014), was first detected at 36 HAI and became more abundant at 60 HAI. PsbO, the cytosol-translated 33-kD oxygen-evolving protein, was also noticed in the 36 HAI samples and signals from the polypeptide intensified in later time points. Lhcb, the PSII light-harvesting and membrane adhesion-promoting protein, became enriched from 60 HAI when their transcript levels increased (Figure 6C).
Subunits of the PSI complex were detected only after 84 HAI by immunoblot analysis and ∼24 h after the cyt b6f and the ATP synthase complexes were discerned (Figures 7C and 7D). Both the stroma-translated PsaA (P700 core protein) and the cytosol-translated PsaD (ferredoxin-docking protein) subunits together with the PSI light harvesting antenna protein Lhca became significantly detectable in the 84 HAI samples. The late detection of PSI subunits in the immunoblot is consistent with their transcription profiles (Figures 6A and 6B).
The stroma-translated proteins PetB and AtpB (CF1β) that are associated with the cyt b6f and the ATP synthase complexes, respectively, belonged to the second set of photosynthetic electron transport chain complexes, which appeared ∼24 h after the PSII complex subunits in the greening thylakoids, i.e., at 60 HAI. The membrane curvature-inducing protein CURT1A was on the verge of detection in the 60 HAI membranes and became more abundant in the 84 HAI membranes at the time when discrete stacks were distinguished from stroma thylakoids and the PSI complexes appeared in the immunoblot.
Ribosome Biogenesis in the Stroma Is Required for Grana Formation
In wild-type plastids, the assembly of polysomes on the surface of the 36 HAI thylakoids correlated with the conversion of the tubulo-vesicular prothylakoids into sheet-like pregranal thylakoids (Figures 2C to 2G). Membrane-bound polysomes also assembled close to the thylakoid surface sites where membrane buds gave rise to stacked prograna thylakoids (Figures 3C to 3J). These correlations suggest that protein synthesis by thylakoid-bound polysomes plays a critical role both in the formation of planar thylakoids and in the generation of stacked grana thylakoids.
To test the hypothesis that protein synthesis by chloroplast ribosomes is critical for the assembly of planar thylakoids and of prograna stacks, we examined an Arabidopsis mutant in which ribosome biogenesis in plastids is delayed during germination. RH3 is a plastid-targeted RNA helicase that is required for chloroplast ribosome production. rh3-4 is a weak mutant allele of RH3 in which the density of plastid ribosomes is ∼80% lower than in wild-type plastids until 72 HAI but slowly recovers to the normal level by 2 weeks after germination (Lee et al., 2013). This property has enabled us to examine the effects of reduced and delayed plastid protein synthesis on thylakoid development.
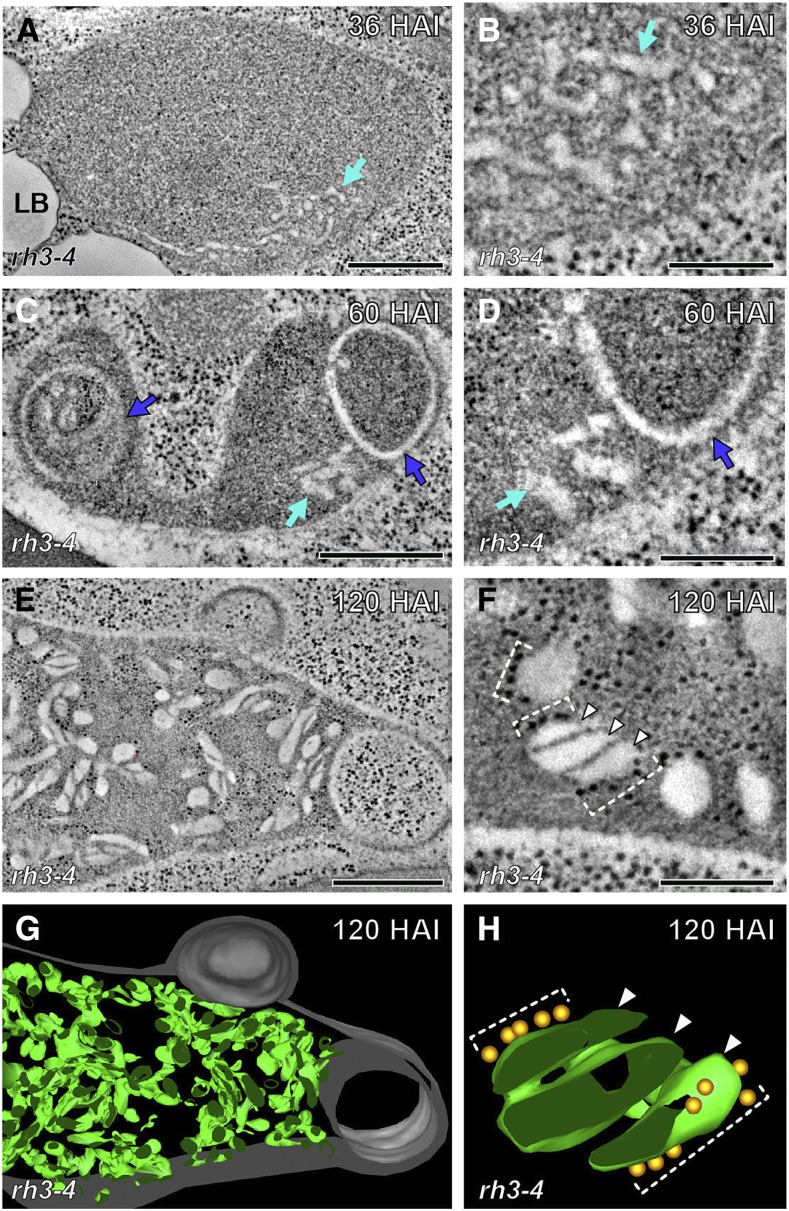
The rh3-4 mutant plastids were significantly smaller than the wild-type plastids at the same HAI times (Figure 1P). The 36 HAI plastids of the rh3-4 mutant cotyledons had tubulo-vesicular prothylakoid membrane compartments (Figures 8A and 8B) similar to those in the 24 HAI plastids of the wild-type cotyledons (Figures 2A and 2B). The 36 HAI rh3-4 mutant plastids also lacked the planar, pregranal-type thylakoids that were a characteristic feature of the 36 HAI wild-type plastids (Figures 2E and 2F). By 60 HAI tubulo-vesicular prothylakoids were no longer seen in the wild-type plastids, whereas in the 60 HAI mutant plastids, many prothylakoids were still present adjacent to rounded, lamellar membrane compartments (Figures 8C and 8D). These membrane lamellae, however, lacked the small prograna stacks seen in the 60 HAI wild-type plastids (Figure 3). Thylakoids coated with polysomes were seen in the rh3-4 plastids at 120 HAI, 84 h later than in the wild-type plastids (Figures 8E to 8H). The appearance of these thylakoid-associated polysomes correlated with thylakoid flattening and stack formation, similar to the events in the 36 and 60 HAI wild-type plastids. Together, these results indicate that thylakoid assembly requires recruitment of polysomes to the thylakoid surface and suggests that the cotranslational insertion of chloroplast DNA-encoded thylakoid membrane proteins is required for the formation of flat thylakoids and prograna stacks.
Figure 8.
Plastids in rh3-4 Cotyledon Cells.
(A) to (F) Electron tomographic slice of plastids in rh3-4 cotyledon cells at 36 ([A] and [B]), 60 ([C] and [D]), and 120 HAI ([E] and [F]). High-magnification images of the mutant plastids at the three time points are shown in (B), (D), and (F), respectively. Convoluted tubular thylakoids of 36 and 60 HAI are marked with light-blue arrows ([A] to [D]). The round thylakoids of 60 HAI are indicated with dark blue ellipses in (C) and (D). Rough thylakoids (bracket in [F]) and grana stacks (arrowheads in [F]) are observed in 120 HAI plastids.
(G) 3D model of the mutant plastid in (E).
(H) 3D model of the primitive stack marked with three arrowheads in (F). The gold spheres represent ribosomes on the thylakoid membrane (brackets).
Bars = 500 nm in (A), (C), and (E) and 150 nm in (B), (D), and (F).
FZL, a Chloroplast-Targeted, Dynamin-Like Protein, Is Required for Connecting the Individual Grana Stacks into a Coherent Thylakoid Membrane System
In 36 HAI wild-type cotyledons, the initially formed pregranal thylakoids were dispersed throughout the stroma (Figure 2), but by 60 HAI, all thylakoids had fused into large, plastid-spanning membrane sheets that were interconnected (Figure 3A). Thylakoid connectivity continued to increase as thylakoid maturation progressed (Figures 4 and 5), suggesting that membrane fusion may be involved in later stages of thylakoid development too.
To investigate this possibility, we examined the role of a protein hypothesized to promote membrane fusion in thylakoid development. Fzo1 is a dynamin-related protein required for fusion of mitochondrial outer membranes (Wang et al., 2016). Arabidopsis has a gene encoding an Fzo1 homolog, termed Fzo-like protein (FZL), that localizes to chloroplasts. In fzl mutant plants, the chloroplasts produce aberrant thylakoids (Gao et al., 2006).
In 36 HAI fzl cotyledons, the plastids contained many vesicle-like compartments and short tubules (Figures 9A and 9B). They also displayed a few small flat thylakoids with ribosomes attached to their surfaces that appeared to be progranal thylakoids. By 60 HAI, most of these vesicles had disappeared and numerous individual, sheet-like pregranal thylakoids and small grana stacks were seen throughout the stroma (Figures 9C and 9D). The thylakoids of the 120 HAI fzl mutant plastids consisted of elongated spirals of large numbers of small diameter, prograna thylakoids (Figures 9E to 9G). However, these elongated, column-like pro-grana stacks were not interconnected by stroma thylakoids. Despite the perturbed thylakoid architecture of their thylakoids, the fzl plastids were as big as the wild-type plastids of the same age (Figure 1P). In agreement with the defects in 36 and 60 HAI mutant chloroplasts, FZL transcript levels were higher at the early time points than at 84 and 120 HAI (Figure 6F).
Figure 9.
Plastids in fzl Cotyledon Cells.
(A) to (F) Electron tomographic slice of fzl plastids at 36, 60, and 120 HAI ([A], [C], and [E]) and their 3D models ([B], [D], and [F]). Extended planar thylakoids and spiral thylakoids are marked with yellow and red brackets, respectively, in (E) and (F).
(G) and (H) A spiral thylakoid in an fzl mutant chloroplast at 120 HAI. The long slanted stack consists of planar layers folded over each other.
(I) to (K) Immunogold labeling of sections from cotyledon cells of fzl mutant seedlings expressing the FZL-GFP fusion protein with a GFP antibody ([I] and [J]) or with an LHCB (light harvesting complex II) antibody (K). The LHCB-specific gold particles are associated with grana stacks where no GFP-specific gold particles are detected. The immunogold labeling experiments were performed with 120 HAI old cotyledon samples.
Bars = 500 nm in (A), (C), (E), and (I) and 200 nm in (J) and (K).
To determine where the FZL proteins function in developing chloroplasts, we performed immunogold labeling of fzl mutant plants that expressed FZL-GFP. Thylakoids in the transgenic plants appeared to be normal when compared with thylakoids of fzl mutant plants, indicating that the GFP fusion protein was functional (Supplemental Figure 4). In such plants, the immunogold particles were detected at the junctions between stroma thylakoids and grana stack margins (Figures 9I and 9J). Thus, the presence of elongated, free-standing and disorganized prograna stacks in the 120 HAI fzl plastids is consistent with the hypothesis that FZL is needed to connect the newly formed grana pro-stacks to each other via fusion of their marginal domains with stroma thylakoids.
DISCUSSION
The process of light-induced thylakoid biogenesis has attracted the interest of plant biologists for over 50 years. However, understanding how the intricate 3D architecture of thylakoid membranes is generated, and how the assembly of the different complexes relates to the structural changes has remained an elusive goal. We investigated this process in Arabidopsis cotyledon chloroplasts using a combination of electron tomography of cryofixed cells, immunolabeling, correlative immunoblot, and transcriptomic analyses. The stages of thylakoid biogenesis in germinating cotyledon cells and chloroplast proteins involved in the assembly activity at each stage are summarized in Figure 10.
Figure 10.
Schematic Diagram of Thylakoid Assembly in the Proplastid of Germinating Arabidopsis.
(A) Structural features and protein complexes associated with the thylakoid membrane noted at the five time points by electron tomography and immunoblot analysis. Black dots on the thylakoid surface represent ribosomes and small arrows point to thylakoid-inner envelope contact sites that become concentrated close to the plane defined by the major axis of the spheroidal chloroplasts during later stages of development. In addition to serving membrane lipid transport functions, these contact sites may help align the thylakoids within the spheroidal chloroplasts.
(B) Cotranslational insertion of PSII by thylakoid-membrane bound polysomes enriches PSII-LHCII at 60 HAI and initiates stack formation. Because of their bulky stromal domains, PSI and ATP syntheses are excluded from the stacked region. Recruitment of CURT1 family proteins define the grana margins and grana become interconnected more extensively by FZL-mediated thylakoid membrane fusion at 84 HAI.
Analysis of the Light-Induced Proplastid Developmental System in Cotyledons Has Uncovered a Remarkably Linear Developmental Program Starting with the Formation of Tubulo-Vesicular Prothylakoids
Proplastids, the colorless precursors of chloroplasts, were discovered in 1927 in a light microscopy study of maize (Zea mays), pondweed (Elodea canadensis), and common toadflax (Lunularia vulgaris) seedlings (Zirkle, 1927). They were subsequently rediscovered by electron microscopists in the 1960s, who characterized them as small, undifferentiated plastids capable of differentiating into all types of plastids during plant development (reviewed in Gunning and Steer, 1975). Despite this long history, it is remarkable how little we know about proplastid structure, function, and differentiation.
Typically, the prothylakoid membranes of proplastids are described as vesicles lacking chlorophyll and photosynthetic complexes (Rudowska et al., 2012; Pogson et al., 2015). As documented in the electron tomography-based 3D reconstructions shown in Figures 2A and 2B, the vesicles seen in thin sections correspond to cross-sectional views of tubulo-vesicular prothylakoids. These membranes lack chlorophyll (Figure 1F) and have defined contact sites with the inner envelope membrane.
The paucity of information about the light-induced greening of proplastids is most likely due to the absence of well-defined structural intermediates in electron micrographs of chemically fixed cells. Furthermore, the analysis of proplastid differentiation in shoot apical meristems is complicated by the fact that their developmental state varies with the specific region and/or layer of the shoot apical meristem in which they are formed (Charuvi et al., 2012). This contrasts with the sizeable literature on the process of photomorphogenesis of etioplasts, the chloroplast precursor-type of plastids of dark-grown seedlings that develop characteristic paracrystalline prolamellar bodies and radiating lamellar prothylakoids (Blomqvist et al., 2008; Solymosi and Schoefs, 2010; Adam et al., 2011). The lipidic prolamellar bodies contain protochlorophyllide molecules that upon exposure to light are rapidly converted to chlorophyll. This induces the transfer of the prolamellar lipids to the prothylakoids that are subsequently converted to pregranal thylakoids (Rudowska et al., 2012).
The downside of the etioplast greening systems is their variability, the result of different experimental growth conditions. This affects prolamellar body formation, growth, and composition. However, prolamellar bodies enable the initial thylakoid assembly events to occur very rapidly and many processes to develop in parallel. This multitude of activities makes it difficult to decipher how each parameter contributes to thylakoid assembly and to compare the results of different studies.
By contrast, the cotyledon proplastid developmental system we have investigated displays a remarkably linear and sequential developmental program that is spread out over a 5-d period. This temporal separation of the different assembly steps and the analysis of chloroplast development mutants has enabled us to untangle the process of thylakoid assembly in ways not previously possible.
The developmental variations in the thylakoids of the shoot apical meristem resemble the sequence of thylakoid assembly events in the cotyledon (Charuvi et al., 2012). Proplastids in the L1 and L3 cell layers have extended thylakoids with simple stacks similar to the thylakoids in 60 HAI chloroplasts (Figure 3). Proplastids in the L2 layer contain discrete thylakoid membranes that are devoid of stacks as in the cotyledon proplastids at 36 HAI (Figure 2). In the leaf primordia, however, chloroplasts consist of grana stacks and interconnecting stroma thylakoids that are easily differentiated. In cotyledons, we observed such partitioning in samples older than 84 HAI (Figure 4). This correlation suggests that cotyledon cells and shoot apical meristem cells operate similar gene expression programs for thylakoid biogenesis.
Thylakoid Biogenesis Starts between 24 and 36 HAI, as Evidenced by Thylakoid Expansion and Flattening, Polysome Binding, and PSII Assembly
The most striking structural change between the 24 and 36 HAI thylakoids is the transformation of the tubulo-vesicular prothylakoids into expanded, sheet-like pregranal thylakoids (Figures 2C to 2F). Additional characteristic features of the pregranal thylakoids include a collapsed lumen, bound polysomes, and the accumulation of PSII subunits, but not other types of thylakoid proteins (Figures 6 and 7). The 36 HAI thylakoids also possess contact sites with the inner envelope membrane and many plastoglobules are seen (Figure 2D). As discussed below, these different structural and biochemical features provide new insights into how pregranal thylakoids are assembled during light-induced plastid development.
Two activities coincide with the thylakoid flattening process: the binding of the polysomes that insert the chloroplast-encoded proteins into the membranes and the forces that drive the collapse of the thylakoid lumen. In our chloroplast greening samples, membrane-bound polysomes were always present on the flattened thylakoids (Figures 2E and 2F), in agreement with the hypothesis that polysome binding to thylakoids is an essential factor in the flattening response. This hypothesis is supported by the fact that a slowdown in chloroplast ribosome assembly in the rh3-4 mutant causes a delay in thylakoid development and flattening (Figure 8). The reversible transformation of endoplasmic reticulum (ER) membrane tubules into membrane sheets in mammalian cells has also been shown to involve polysome binding to translocon proteins, the insertion of coiled-coil membrane proteins, and a reduction of the concentration of membrane curvature-inducing proteins (Shibata et al., 2010). Together, these findings are consistent with our proposal that polysome binding, probably for inserting integral membrane proteins, contributes to the conversion of the prothylakoids into sheet-like pregranal thylakoids.
Assembly of PSII complexes involves both chloroplast-encoded and nuclear-encoded proteins (Nevo et al., 2012). We monitored synthesis of the proteins from the two sources in the early stages of thylakoid development by following the appearance of transcription and translation of PsbA, the stroma-translated D1 core protein, and PsbO and PsbP, the cytosol-translated peripheral 33- and 23-kD oxygen-evolving proteins (Figures 6 and 7). Both transcripts and proteins from the three genes are detected in the 36 HAI thylakoids, but the same samples contain significantly lower amounts of transcripts from genes encoding PSI, LHCII, cyt b6f, and ATP synthase subunits (Figures 6 and 7). Polypeptides from some of their subunit genes were undetectable or barely detectable in the immunoblots (Figures 7C and 7D). Thus, PSII complexes appear to be inserted into the thylakoid membrane when polysomes attach to the flattening thylakoids at 36 HAI. PSII complex assembly starts with the formation of D1-D2 complexes to which other proteins are bound in a precise sequential order (Heinz et al., 2016). Incorporation of the 33- and 23-kD OEC1 and OEC2 proteins is a late event, since their binding to newly formed core complexes occurs only after assembly of the Mn cluster and release of the Psb27 protein (Liu et al., 2011; Heinz et al., 2016). In agreement with these studies, PSII-encoding genes are upregulated at 36 HAI, earlier than activation of the other complexes of the thylakoid (Figures 6 and 7). Together, these results suggest that the first phase of thylakoid biogenesis includes the assembly of PSII complexes. The large diameter and planar geometry of the intrinsic domain of these PSII complexes (Nevo et al., 2012) would be expected to facilitate the thylakoid-flattening process.
Thylakoid Expansion in the 36 HAI Chloroplasts Is Driven in Part by the Incorporation of Membrane Lipids
The thylakoids in the 36 HAI plastids exhibit a greatly expanded surface area and many plastoglobules (Figures 2C and 2D). Both of these changes reflect increased rates of lipid transfer to the thylakoids. Chloroplast membrane lipids are assembled primarily by enzymes in the envelope membranes (Wang and Benning, 2012). Two mechanisms have been proposed to explain how these lipids are transferred to the thylakoids: vesicular transport (Karim and Aronsson, 2014) and nonvesicular lipid transport (Lev, 2012). The vesicular transport hypothesis is supported by the identification of several chloroplast homologs of COPII proteins in bioinformatics studies and by the immunolocalization of CPSAR1 proteins in vesicular structures in chloroplasts of low-temperature-incubated Arabidopsis leaves. However, such vesicles are not typically seen in chloroplasts of room temperature-grown plants preserved by cryofixation methods, and it is often difficult to distinguish plastoglobules from vesicles (Austin et al., 2006; Charuvi et al., 2012). In our opinion, vesicular transport in chloroplasts is at best a minor pathway for transferring chloroplasts lipids and imported chloroplast proteins to developing thylakoids.
In contrast to the limited evidence supporting a vesicular transport mechanism, the evidence for nonvesicular transport mechanisms is substantial. For example, in seed plants the principal proteins involved in intraplastid protein trafficking are components of the Toc-Tic complex, whose functional properties have been characterized in considerable detail (Celedon and Cline, 2013). Similarly, the importance of nonvesicular lipid transport mechanisms for transferring selected lipids between different membrane systems has attracted considerable interest in recent years. Seven types of contact sites (10–30 nm spacing) between ER membranes and other organelle membranes have been identified in plant cells (Staehelin, 2015), and several of those sites have been shown to be involved in lipid transfer activities. Indeed, nonvesicular lipid transport between membranes is most efficient when the membranes are closely linked by tethering molecules (Lebiedzinska et al., 2009; Lev, 2012). A well-characterized nonvesicular lipid transport system of plant cells involves the two-way lipid transport between closely associated chloroplast and ER membranes (Wang and Benning, 2012). In yeast, the Nvj2 protein has been shown to serve as a tethering protein between ER and trans-Golgi membranes and to become activated for lipid transport when ceramides have to be transferred from the ER to the Golgi (Liu et al., 2017).
Contact sites between the inner chloroplast envelope membrane and thylakoids can be visualized in well preserved chemically fixed chloroplasts (see Figure 2 in Staehelin, 2003) but are more reliably seen in cryofixed samples (Figures 2C, 2D, 4A, and 4B; Supplemental Figures 1C to 1F; Charuvi et al., 2012). These contact sites are less frequent but more concentrated in mature chloroplasts than developing ones, suggesting that they may play a role in aligning the thylakoid membranes within the chloroplasts, in addition to lipid transfer (Figure 10).
Bound Polysomes Define the Sites of Grana Stack Assembly, and LHCII, cyt b6f, and ATP Synthase Create the Grana and Stroma Membrane Domains
The two defining structural changes of the 60 HAI plastids are the formation of grana stacks and the lateral differentiation of grana and stroma membrane domains. According to the classical description of grana formation by Brangeon and Mustardy (1979), grana membrane buds originate at the margins of fenestrae of lamellar thylakoids, and as these tongue-like structures expand, they overgrow the existing membranes and create the nascent grana stacks. New overgrowths subsequently expand the size of the stacks. Yet to be determined are the factors that define where a grana stack is formed and which protein complexes participate in this process.
In the cotyledon chloroplast biogenesis system, we investigated, the grana-forming membrane sites are defined by membrane-bound polysomes (Figures 3C to 3J). However, unlike the system described by Brangeon and Mustardy (1979), the membrane bud-forming sites of developing cotyledon chloroplasts originate on the surface of planar membrane domains and not on the margins of fenestrae. Polysomes are also seen attached to the later forming grana thylakoids. All of the newly formed grana folds possess wide, slit-like connections to the originating membranes, a characteristic feature of immature prograna stacks (Figures 3K and 3O), but repeats of unit structures in grana discs (Arvidsson and Sundby, 1999; Shimoni et al., 2005) were not observed.
Our RNA-seq and immunoblot analyses demonstrated that the formation of the grana stacks coincides with the appearance of membrane adhesion-promoting LHCII complexes (McDonnel and Staehelin, 1980). The incorporation of PSII complexes into the stacked membrane regions is likely driven by the tendency of PSII and LHCII to assemble into heteromeric supercomplexes (Dekker and Boekema, 2005). By contrast, the appearance of ATP synthase complexes that are confined to the nonstacked membrane domains (Miller and Staehelin, 1976) most likely helps stabilize the adjacent stroma thylakoid regions. Finally, the addition of cyt b6f complexes to the PSII and ATP synthase complexes may allow for the generation of ATP by the incomplete electron transport chain.
Thylakoid Biosynthesis Appears to Conclude with the Assembly of PSI Complexes, the Incorporation of CURT1A Proteins, and the Narrowing of the Grana-Stroma Membrane Connecting Regions
Genes for PSI construction are upregulated later than those for PSII during thylakoid biogenesis (Figures 6 and 7). The simultaneous appearance of both stroma-translated PsaA and cytosol-translated PsaD subunits (Figures 7C and 7D) is likely indicative of the assembly of functional PSI complexes. The addition of PSI complexes to the developing thylakoids together with the lateral fusion of the grana stacks, possibly mediated by the FZL protein, would be expected to expand the stroma membrane regions, thereby necessitating the addition of more CURT1A proteins to stabilize the boundary regions of the grana stacks.
The final step of thylakoid biogenesis at 120 HAI involves the conversion of the wide grana-stroma membrane-connecting slits into much smaller and well-defined tubular connections. As documented previously (Austin and Staehelin, 2011), the size of these latter connections is highly variable, suggesting that control of their size might be part of a mechanism to regulate grana-to-stroma ion and protein movements. We postulate that this type of membrane structure regulation might be important for optimizing the functional activities of mature chloroplasts.
METHODS
Materials and Plant Growth Conditions
All the chemicals were from Sigma-Aldrich (http://www.sigmaaldrich.com) or Thermo-Fisher (https://www.thermofisher.com/) unless specified. Seeds from wild type (Col-0), homozygous rh3-4 mutant (Lee et al., 2013), homozygous fzl mutant (Gao et al., 2006), and fzl mutant complemented by the FZL-GFP construct (Gao et al., 2006) plants were surface-sterilized and incubated in 4°C overnight. The seeds were germinated and grown on 0.75% phytoagar plates supplemented with half-strength Murashige-Skoog salt and MES buffer (0.5 g/L, pH 5.7) under continuous light (120 μE m−2 s−1, white fluorescent light bulb) in a growth chamber (22°C; Panasonic; catalog no. MLR-352H-PB) before their cotyledons were dissected for high-pressure freezing, immunoblot analyses, and RNA-seq experiments. Some seedlings grew faster or slower than most of the seedlings in the same plates. Seedlings for which developmental stages deviated from the seedlings shown in Figures 1A to 1E were removed before sampling.
High-Pressure Freezing, Sample Processing, and Microscopy
High-pressure freezing, freeze substitution, resin embedding, and ultramicrotomy were performed as described (Kang, 2010). In brief, cotyledon tissues were dissected and rapidly frozen with an HPM100 high-pressure freezer (Leica Microsystems). Before freezing, the seedlings were imaged with a hand-held digital microscope (Dino-Lite; catalog no. AM4113TL), and their cotyledons were examined with a confocal laser scanning microscope (Zeiss) to assess their chlorophyll autofluorescence with the conditions described by Park et al. (2015). The samples were freeze-substituted at −80°C for 72 h, and excess OsO4 was removed by rinsing with precooled acetone. After being slowly warmed up to room temperature over 48 h, tissue samples were separated from planchettes and embedded in Embed-812 resin (Electron Microscopy Sciences; catalog no. 14120). Thin sections (80 nm thick) prepared from sample blocks of each time point were examined with a Hitachi 7400 TEM (Hitachi-High Technologies) operated at 80 kV.
Dual-Axis Electron Tomography, Tomogram Reconstruction, Modeling, and Measuring Morphometric Parameters
Semithick sections (250 nm) were collected on formvar-coated copper slot grids (Electron Microscopy Sciences; catalog no. GS2010-Cu) and stained with 2% uranyl acetate in 70% methanol followed by Reynold’s lead citrate. Tilt series were collected from 60° to −60° (1.5° intervals) with a 200-kV Tecnai F20 intermediate voltage electron microscopy (FEI). For dual-axis tomography analysis, tilt series around two orthogonal axes were acquired from each section using the tomography data acquisition program installed in the microscope by the manufacturer. Tomograms were reconstructed as described (Toyooka and Kang, 2014).
To generate models of complicated thylakoid membranes, we used the autocontour command (bio3d.colorado.edu/imod/doc/3dmodHelp/autox.html) of the 3dmod software package. Noise levels of tomographic slices were reduced by increasing their image thicknesses, and thylakoid membranes in the denoised slice images were traced with the autocontour under the high contrast mode with the threshold values between 140 and 160. The resulting contours were smoothened, and their excess points were deleted with the drawing tool plug-in (http://www.andrewnoske.com/student/downloads/imod/drawingtools.html). This semiautomatic modeling method is explained by Mai and Kang (2017). Thylakoid membranes were outlined in every 4th slice, and obvious errors in the outlines were corrected before the contours were meshed to generate 3D models with the imodmesh command. To compare luminal widths of 24 HAI prothylakoids and 36 HAI pregranal thylakoids, open contours spanning thylakoid lumens were drawn in the tomographic slices after loading in 3dmod and lengths of the contours were extracted with the imodinfo command (bio3d.colorado.edu/imod/doc/3dmodHelp/autox.html). Thirty-one sites in prothylakoids (three plastids) and 20 sites in pregranal thylakoids (three plastids) were measured.
Ribosome Density Measurements
Outlines of stroma space over thylakoid membranes were drawn with the sculpt tool of the Drawing tools (brush size = 18 nm) and the outlines were meshed to calculate the volume inside the mesh. Ribosomes within the meshed volume were identified, counted, and modeled as individual spheres (Supplemental Figures 2A and 2B). Ribosome densities were calculated as the values of ribosome numbers divided by volumes of the 18-nm thickness layer (Supplemental Figures 2C and 2D). Ribosome densities and their standard deviations were measured from three randomly chosen sites in the bulk stroma (36 and 60 HAI), prothylakoids (36 HAI), pregranal thylakoids (36 HAI), unstacked thylakoids (60 HAI), and grana-forming thylakoids (60 HAI). The bar charts and Student’s t tests for significant differences in ribosome densities (two tailed and unpaired tests) in Figures 2G and 3Q were prepared with Microsoft Excel for Mac (version 16.11.1). At each site, 31 to 50 ribosomes were counted.
Immunoblot Analysis and Immunogold Labeling
For immunoblot analysis, cotyledon tissues from seedlings at 24, 36, 60, 84, and 120 HAI were dissected and homogenized in 100 µL of 2× SDS-PAGE sample buffer (Sigma-Aldrich; catalog no. S3401-10VL) and incubated at 95°C for 5 min. The samples were cleared of insoluble debris by centrifugation at 16,100g for 5 min at room temperature. For each lane, 5 to 6 mg of cotyledon protein extract was resolved on a 15% (w/v) SDS-polyacrylamide minigel and analyzed by immunoblotting as described by Kang et al. (2001). After incubating with primary antibodies and horseradish peroxidase-conjugated secondary antibodies (Agrisera; catalog no. AS09 602), chemiluminescence from the blots was generated using the Clarity Max Western ECL Substrates (Bio-Rad, catalog no. 170-5060), and the blots were imaged with ChemiDoc Touch System (Bio-Rad; catalog no. 17001401). Immunoblot analyses were repeated three times with total protein extracts from three independent sets of cotyledon samples. We performed Student’s t test (one-tailed and unpaired test) with the triple replicate immunoblot data to determine when polypeptide quantities increase significantly using Microsoft Excel (version 16.11.1). The immunogold labeling experiments were performed according to the protocol explained by Kang (2010). Antibodies for PsbA (AS10 704), PsbN (AS14 2786), PsaA (AS06 172), PsaD (AS09 461), Lhca (AS01 005), AtpB (AS05 085), 11) AtpC (AS08 312), PetB (AS03 034), 13) PetC (AS08 330), and CURT1A (AS08 316) were purchased from Agrisera. Anti-PBA1 antibody (ab98861) and anti-GFP antibody (ab6556) were purchased from Abcam. Antibodies against PsaA and PsbO were provided by Michael Seibert (National Renewable Energy Laboratory). Antibodies for PsbP (Henry et al., 1997) and Lhcb (Payan and Cline, 1991) were donated by Kenneth Cline (University of Florida)
Transcriptomic Analyses
Total RNA samples were isolated from seedlings at each time point using Qiagen Plant RNA extraction kit (Qiagen; catalog no. 74904) and rRNA in the samples was depleted with the Invitrogen Ribominus Plant Kit for RNA-seq (Thermo Fisher; catalog no. A1083808). cDNA libraries for Illumina sequencing were prepared according to the protocol for Illumina TruSeq RNA-seq library as described previously (https://www.ncbi.nlm.nih.gov/pubmed/21807852). The libraries were sequenced on the Illumina Hi-Seq X10 platform using the PE150 run mode (Cloudhealth). After removing reads mapped to rRNA sequences from the RNA-seq data, we analyzed the remaining reads using TopHat and cufflinks as described previously (Trapnell et al., 2012; https://www.nature.com/nprot/journal/v7/n3/full/nprot.2012.016.html). The reads were mapped to the Arabidopsis TAIR10 reference genome. FPKM values for genes encoding proteins of the thylakoid membrane complexes, CURT1 family genes, and FZL were calculated to estimate their expression levels. The heat maps and the line charts were generated with R Studio (version 1.1.383) and Microsoft Excel (version 15.34), respectively. Two independent RNA-seq experiments were performed (from seedling harvest to preparation of cDNA library for Illumina sequencing). Heat maps and line charts in Figure 6 and Supplemental Figure 3 show results from the two RNA-seq analyses. FPKMs of CURT1 family genes and of FZL at the five time points illustrated in Figures 6E and 6F are their average values from the two data sets.
Gene Expression Analyses by Droplet Digital PCR
We selected several genes encoding subunits of thylakoid membrane complexes in Figure 6 and determined their transcript levels via ddPCR. Total RNA samples from Arabidopsis seedlings at 24, 36, 60, 84, and 120 HAI were isolated with the RNeasy Plant Mini Kit (Qiagen; catalog no. 74903), and equal amounts of RNA samples from each stage (600 ng) were reverse transcribed with a QuantiNova reverse transcription (RT) kit (Qiagen; catalog no. 205411). Three replicates of RT mixes were generated for each stage. We had to dilute the RT mixes before ddPCR because cDNA concentrations of the genes in the mixes were too high and their ddPCR amplitudes surpassed dynamic ranges. To estimate amounts of cDNA templates of the selected genes, qRT-PCR assays were performed (denaturation at 98°C, annealing/extension at 60°C, 40 cycles) using a CFX96 real-time PCR detection system (Bio-Rad). qRT-PCR reaction mixes (20 µL) contained 4.5 ng of RT mix, 10 μL iTaq Universal SYBR Green Supermix (Bio-Rad; catalog no. 1725120), and 14 pmol of 5ʹ and 3ʹ primers. After diluting RT mixes according to qRT-PCR results, ddPCR assay was performed with a QX200 Droplet Digital PCR System (Bio-Rad). ddPCR reaction mixes (20 µL) contained diluted RT mixes, 2 pmol of 5ʹ primer, 2 pmol of 3ʹ primer, and 10 μL EvaGreen Digital PCR Supermix (Bio-Rad; catalog no. 1864034). After generating droplets (Bio-Rad; QX200 Droplet Generation Oil for EvaGreen; catalog no. 1864005), the reaction mixes were transferred to a 96-well plate and the plate was sealed with a PX1 PCR Plate Sealer (Bio-Rad). Fluorescent droplets were quantified with a QX200 Droplet Reader (Bio-Rad; catalog no. 1864003), and the results were analyzed using the QuantaSoft Analysis Pro software package (Bio-Rad; bulletin no. 6827). Absolute amounts of DNA (copy numbers per picogram of cDNA sample for plastid-encoded genes and copy numbers for nanogram of cDNA sample for nucleus-encoded gens) were calculated with Microsoft Excel for Mac (version 16.11.1). Primers for ddPCR are listed in Supplemental Data Set 6. The same primer pairs were used for ddPCR and qRT-PCR.
Accession Numbers
The RNA-seq data have been deposited in NCBI Sequence Read Archive under accession number PRJNA418881.
Supplemental Data
Supplemental Figure 1. Electron micrographs of proplastids and vacuoles in developing cotyledon cells.
Supplemental Figure 2. Tomographic slice and ribosome models for calculating densities of ribosomes associated with thylakoid membranes.
Supplemental Figure 3. Heat maps and line chart showing transcript levels of the four electron transport complex subunits of the thylakoid and Rubisco from the duplicate RNA-seq experiment.
Supplemental Figure 4. Electron micrographs of 120 HAI chloroplasts.
Supplemental Data Set 1. Measurements of chlorophyll autofluorescence sizes.
Supplemental Data Set 2. Measurements of thylakoid thickness.
Supplemental Data Set 3. Ribosome density measurements.
Supplemental Data Set 4. Statistics of the two rounds of RNA-seq experiments.
Supplemental Data Set 5. Transcript levels of genes encoding subunits of the electron transport chain complexes in the thylakoid membrane from the RNA-seq experiment used to prepare the heat maps and line charts in Figure 6 and Supplemental Figure 4.
Supplemental Data Set 6. ddPCR quantification (three replicates) and primers for the experiment
Supplemental Data Set 7. Quantification of polypeptide chemiluminescence with a digital imaging system.
Supplemental Movie 1. Tomographic volume containing the progranal thylakoid with the dome-shaped membrane outgrowth in Figure 3C.
Supplemental Movie 2. Tomographic volume containing the progranal stack in Figure 3K.
Supplemental Movie 3. Tomographic volume containing the progranal stack in Figure 3L.
Supplemental Movie 4. Tomographic volume containing the grana stack in Figures 4C to 4H (inside the dashed red oval).
Supplemental Movie 5. Tomographic volume containing the grana stack in Figures 5B to 5E (inside the dashed red oval).
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Seibert (National Renewable Energy Laboratory, Golden, CO) and Kenneth Cline (University of Florida) for sharing their antibodies. We also thank Anton Titov, Mark McNeely, Dallas Khamiss, Robert Lloyd, and Kwanghee Lee (University of Florida) for their help in preparing preliminary thylakoid models and immunoblots not included here. The ddPCR experiments were assisted by Hong-Ming Lam and Yee-Shan Ku (Chinese University of Hong Kong). This work was supported by the Research Grants Council of Hong Kong (GRF14126116, AoE/M-05/12, C4011-14R, and GRF14119814), by a USDA NIFA Award AFRI grant (2011-67013-30119), and by the Rural Development Administration, Republic of Korea, Cooperative Research Program for Agriculture Science and Technology Development Project PJ010953092018.
AUTHOR CONTRIBUTIONS
B.-H.K., L.A.S., and K.W.O. designed the research. B.-H.K. performed electron microscopy sample preparation, tomography data collection, and tomogram reconstruction. Z.L. and K.K.M. generated 3D models of thylakoids. Z.L. did immunoblot analysis, ECL quantification, immunogold labeling, and transmission electron microscopy imaging. N.Z. carried out RNA-seq experiments. K.K.M. calculated ribosome densities. Z.Y.L. performed ddPCR measurements of transcript concentrations. D.T. and S.Z. analyzed RNA-seq data. B.-H.K., L.A.S., and K.W.O. wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Adam Z., Charuvi D., Tsabari O., Knopf R.R., Reich Z. (2011). Biogenesis of thylakoid networks in angiosperms: knowns and unknowns. Plant Mol. Biol. 76: 221–234. [DOI] [PubMed] [Google Scholar]
- Armbruster U., et al. (2013). Arabidopsis CURVATURE THYLAKOID1 proteins modify thylakoid architecture by inducing membrane curvature. Plant Cell 25: 2661–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvidsson P.-O., Sundby C. (1999). A model for the topology of the chloroplast thylakoid membrane. Funct. Plant Biol. 26: 687–694. [Google Scholar]
- Austin J.R. II, Staehelin L.A. (2011). Three-dimensional architecture of grana and stroma thylakoids of higher plants as determined by electron tomography. Plant Physiol. 155: 1601–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin J.R. II, Frost E., Vidi P.-A., Kessler F., Staehelin L.A. (2006). Plastoglobules are lipoprotein subcompartments of the chloroplast that are permanently coupled to thylakoid membranes and contain biosynthetic enzymes. Plant Cell 18: 1693–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomqvist L.A., Ryberg M., Sundqvist C. (2008). Proteomic analysis of highly purified prolamellar bodies reveals their significance in chloroplast development. Photosynth. Res. 96: 37–50. [DOI] [PubMed] [Google Scholar]
- Brangeon A., Mustardy L.A. (1979). The ontogenetic assembly of intra-chloroplastic lamellae viewed in 3-dimension. Biol. Cell. 36: 71–80. [Google Scholar]
- Celedon J.M., Cline K. (2013). Intra-plastid protein trafficking: how plant cells adapted prokaryotic mechanisms to the eukaryotic condition. Biochim. Biophys. Acta 1833: 341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charuvi D., Kiss V., Nevo R., Shimoni E., Adam Z., Reich Z. (2012). Gain and loss of photosynthetic membranes during plastid differentiation in the shoot apex of Arabidopsis. Plant Cell 24: 1143–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum B., Kühlbrandt W. (2011). Electron tomography of plant thylakoid membranes. J. Exp. Bot. 62: 2393–2402. [DOI] [PubMed] [Google Scholar]
- Daum B., Nicastro D., Austin J. II, McIntosh J.R., Kühlbrandt W. (2010). Arrangement of photosystem II and ATP synthase in chloroplast membranes of spinach and pea. Plant Cell 22: 1299–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J.P., Boekema E.J. (2005). Supramolecular organization of thylakoid membrane proteins in green plants. Biochim. Biophys. Acta 1706: 12–39. [DOI] [PubMed] [Google Scholar]
- Engel B.D., Schaffer M., Kuhn Cuellar L., Villa E., Plitzko J.M., Baumeister W. (2015). Native architecture of the Chlamydomonas chloroplast revealed by in situ cryo-electron tomography. eLife 4: 3583–3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Sage T.L., Osteryoung K.W. (2006). FZL, an FZO-like protein in plants, is a determinant of thylakoid and chloroplast morphology. Proc. Natl. Acad. Sci. USA 103: 6759–6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garab G. (2014). Hierarchical organization and structural flexibility of thylakoid membranes. Biochim. Biophys. Acta 1837: 481–494. [DOI] [PubMed] [Google Scholar]
- Gilkey J.C., Staehelin L.A. (1986). Advances in ultrarapid freezing for the preservation of cellular ultrastructure. J. Electron Microsc. Tech. 3: 177–210. [Google Scholar]
- Gunning B.E.S. (1965). The greening process in plastids. Protoplasma 60: 111–130. [Google Scholar]
- Gunning B., Steer M.W. (1975). Plant Cell Biology. An Ultrastructural Approach (Ultrastructure and Biology of Plant Cells). (New York: Jones & Bartlett Learning; ). [Google Scholar]
- Heinz S., Liauw P., Nickelsen J., Nowaczyk M. (2016). Analysis of photosystem II biogenesis in cyanobacteria. Biochim. Biophys. Acta 1857: 274–287. [DOI] [PubMed] [Google Scholar]
- Henningsen K.W., Boynton J.E. (1974). Macromolecular physiology of plastids. IX. Development of plastid membranes during greening of dark-grown barley seedlings. J. Cell Sci. 15: 31–55. [DOI] [PubMed] [Google Scholar]
- Henry R., Carrigan M., McCaffrey M., Ma X., Cline K. (1997). Targeting determinants and proposed evolutionary basis for the Sec and the Delta pH protein transport systems in chloroplast thylakoid membranes. J. Cell Biol. 136: 823–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindson C.M., Chevillet J.R., Briggs H.A., Gallichotte E.N., Ruf I.K., Hindson B.J., Vessella R.L., Tewari M. (2013). Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods 10: 1003–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P., López-Juez E. (2013). Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 14: 787–802. [DOI] [PubMed] [Google Scholar]
- Kang B.-H. (2010). Electron microscopy and high-pressure freezing of Arabidopsis. Methods Cell Biol. 96: 259–283. [DOI] [PubMed] [Google Scholar]
- Kang B.H., Busse J.S., Dickey C., Rancour D.M., Bednarek S.Y. (2001). The arabidopsis cell plate-associated dynamin-like protein, ADL1Ap, is required for multiple stages of plant growth and development. Plant Physiol. 126: 47–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim S., Aronsson H. (2014). The puzzle of chloroplast vesicle transport - involvement of GTPases. Front. Plant Sci. 5: 472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff H., Hall C., Wood M., Herbstová M., Tsabari O., Nevo R., Charuvi D., Shimoni E., Reich Z. (2011). Dynamic control of protein diffusion within the granal thylakoid lumen. Proc. Natl. Acad. Sci. USA 108: 20248–20253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouřil R., Oostergetel G.T., Boekema E.J. (2011). Fine structure of granal thylakoid membrane organization using cryo electron tomography. Biochim. Biophys. Acta 1807: 368–374. [DOI] [PubMed] [Google Scholar]
- Kowalewska Ł.M., Mazur R., Suski S., Garstka M., Mostowska A. (2016). Three-dimensional visualization of the internal plastid membrane network during runner bean chloroplast biogenesis. Plant Cell 28: 875–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebiedzinska M., Szabadkai G., Jones A.W.E., Duszynski J., Wieckowski M.R. (2009). Interactions between the endoplasmic reticulum, mitochondria, plasma membrane and other subcellular organelles. Int. J. Biochem. Cell Biol. 41: 1805–1816. [DOI] [PubMed] [Google Scholar]
- Lee K.-H., Park J., Williams D.S., Xiong Y., Hwang I., Kang B.-H. (2013). Defective chloroplast development inhibits maintenance of normal levels of abscisic acid in a mutant of the Arabidopsis RH3 DEAD-box protein during early post-germination growth. Plant J. 73: 720–732. [DOI] [PubMed] [Google Scholar]
- Lev S. (2012). Nonvesicular lipid transfer from the endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 4: a013300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Roose J.L., Cameron J.C., Pakrasi H.B. (2011). A genetically tagged Psb27 protein allows purification of two consecutive photosystem II (PSII) assembly intermediates in Synechocystis 6803, a cyanobacterium. J. Biol. Chem. 286: 24865–24871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.-K., Choudhary V., Toulmay A., Prinz W.A. (2017). An inducible ER-Golgi tether facilitates ceramide transport to alleviate lipotoxicity. J. Cell Biol. 216: 131–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai K.K.K., Kang B.-H. (2017). Semiautomatic segmentation of plant Golgi stacks in electron tomograms using 3dmod. In Plant Cell Morphogenesis, Methods in Molecular Biology, Jiang L., ed (New York: Springer; ), pp. 97–104. [DOI] [PubMed] [Google Scholar]
- McDonnel A., Staehelin L.A. (1980). Adhesion between liposomes mediated by the chlorophyll a/b light-harvesting complex isolated from chloroplast membranes. J. Cell Biol. 84: 40–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K.R., Staehelin L.A. (1976). Analysis of the thylakoid outer surface. Coupling factor is limited to unstacked membrane regions. J. Cell Biol. 68: 30–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustárdy L., Garab G. (2003). Granum revisited. A three-dimensional model--where things fall into place. Trends Plant Sci. 8: 117–122. [DOI] [PubMed] [Google Scholar]
- Nevo R., Charuvi D., Tsabari O., Reich Z. (2012). Composition, architecture and dynamics of the photosynthetic apparatus in higher plants. Plant J. 70: 157–176. [DOI] [PubMed] [Google Scholar]
- Paolillo D.J., Jr (1970). The three-dimensional arrangement of intergranal lamellae in chloroplasts. J. Cell Sci. 6: 243–255. [DOI] [PubMed] [Google Scholar]
- Park J., Cui Y., Kang B.-H. (2015). AtPGL3 is an Arabidopsis BURP domain protein that is localized to the cell wall and promotes cell enlargement. Front. Plant Sci. 6: 412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payan L.A., Cline K. (1991). A stromal protein factor maintains the solubility and insertion competence of an imported thylakoid membrane protein. J. Cell Biol. 112: 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson B.J., Ganguly D., Albrecht-Borth V. (2015). Insights into chloroplast biogenesis and development. Biochim. Biophy. Acta 1847: 1017–1024. [DOI] [PubMed] [Google Scholar]
- Pribil M., Labs M., Leister D. (2014). Structure and dynamics of thylakoids in land plants. J. Exp. Bot. 65: 1955–1972. [DOI] [PubMed] [Google Scholar]
- Race H.L., Herrmann R.G., Martin W. (1999). Why have organelles retained genomes?. Trends Genet. 15: 364–370. [DOI] [PubMed] [Google Scholar]
- Rast A., Heinz S., Nickelsen J. (2015). Biogenesis of thylakoid membranes. Biochim. Biophy. Acta 1847: 821–830. [DOI] [PubMed] [Google Scholar]
- Rudowska L., Gieczewska K., Mazur R., Garstka M., Mostowska A. (2012). Chloroplast biogenesis - correlation between structure and function. Biochim. Biophys. Acta 1817: 1380–1387. [DOI] [PubMed] [Google Scholar]
- Ryberg M., Sundqvist C. (1982). Characterization of prolamellar bodies and prothylakoids fractionated from wheat etioplasts. Physiol. Plant. 56: 125–132. [Google Scholar]
- Selstam E., Schelin J., Williams W.P., Brain A.P.R. (2007). Structural organisation of prolamellar bodies (PLB) isolated from Zea mays. Parallel TEM, SAXS and absorption spectra measurements on samples subjected to freeze-thaw, reduced pH and high-salt perturbation. Biochim. Biophys. Acta 1768: 2235–2245. [DOI] [PubMed] [Google Scholar]
- Shibata Y., Shemesh T., Prinz W.A., Palazzo A.F., Kozlov M.M., Rapoport T.A. (2010). Mechanisms determining the morphology of the peripheral ER. Cell 143: 774–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoni E., Rav-Hon O., Ohad I., Brumfeld V., Reich Z. (2005). Three-dimensional organization of higher-plant chloroplast thylakoid membranes revealed by electron tomography. Plant Cell 17: 2580–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solymosi K., Schoefs B. (2010). Etioplast and etio-chloroplast formation under natural conditions: the dark side of chlorophyll biosynthesis in angiosperms. Photosynth. Res. 105: 143–166. [DOI] [PubMed] [Google Scholar]
- Staehelin L.A. (2003). Chloroplast structure: from chlorophyll granules to supra-molecular architecture of thylakoid membranes. Photosynth. Res. 76: 185–196. [DOI] [PubMed] [Google Scholar]
- Staehelin L.A. (2015). Membrane Structure and Membranous Organelles. In Biochemistry and Molecular Biology of Plants, Buchanan B., Gruissem W., Jones R., eds (West Sussex, UK: Wiley; ), pp. 2–44. [Google Scholar]
- Staehelin L.A., Kang B.-H. (2008). Nanoscale architecture of endoplasmic reticulum export sites and of Golgi membranes as determined by electron tomography. Plant Physiol. 147: 1454–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S.C., Laperriere G., Germain H. (2017). Droplet Digital PCR versus qPCR for gene expression analysis with low abundant targets: from variable nonsense to publication quality data. Sci. Rep. 7: 2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torabi S., Umate P., Manavski N., Plöchinger M., Kleinknecht L., Bogireddi H., Herrmann R.G., Wanner G., Schröder W.P., Meurer J. (2014). PsbN is required for assembly of the photosystem II reaction center in Nicotiana tabacum. Plant Cell 26: 1183–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka K., Kang B.-H. (2014). Reconstructing plant cells in 3D by serial section electron tomography. Methods Mol. Biol. 1080: 159–170. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Benning C. (2012). Chloroplast lipid synthesis and lipid trafficking through ER-plastid membrane contact sites. Biochem. Soc. Trans. 40: 457–463. [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhang Y., Chen L., Liang Q., Yin X.-M., Miao L., Kang B.-H., Xue D. (2016). Kinetics and specificity of paternal mitochondrial elimination in Caenorhabditis elegans. Nat. Commun. 7: 12569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M.T., Langdale J.A. (2009). The making of a chloroplast. EMBO J. 28: 2861–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weier T.E., Sjoland R.D., Brown D.L. (1970). Changes induced by low light intensities on the prolamellar body of 8-day, dark-grown seedlings. Am. J. Bot. 57: 276. [Google Scholar]
- Zheng H., Staehelin L.A. (2011). Protein storage vacuoles are transformed into lytic vacuoles in root meristematic cells of germinating seedlings by multiple, cell type-specific mechanisms. Plant Physiol. 155: 2023–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirkle C. (1927). The growth and development of plastids in Lunularia vulgaris, Elodea canadensis, and Zea mays. Am. J. Bot. 14: 429. [Google Scholar]