A multilayer molecular protection system functioning in Arabidopsis and human cells suppresses the formation of N6-methyl-ATP, possibly to prevent random insertion of N6-methylated adenine into RNA.
Abstract
N6-methylated adenine (m6A) is the most frequent posttranscriptional modification in eukaryotic mRNA. Turnover of RNA generates N6-methylated AMP (N6-mAMP), which has an unclear metabolic fate. We show that Arabidopsis thaliana and human cells require an N6-mAMP deaminase (ADAL, renamed MAPDA) to catabolize N6-mAMP to inosine monophosphate in vivo by hydrolytically removing the aminomethyl group. A phylogenetic, structural, and biochemical analysis revealed that many fungi partially or fully lack MAPDA, which coincides with a minor role of N6A-RNA methylation in these organisms. MAPDA likely protects RNA from m6A misincorporation. This is required because eukaryotic RNA polymerase can use N6-mATP as a substrate. Upon abrogation of MAPDA, root growth is slightly reduced, and the N6-methyladenosine, N6-mAMP, and N6-mATP concentrations are increased in Arabidopsis. Although this will potentially lead to m6A misincorporation into RNA, we show that the frequency is too low to be reliably detected in vivo. Since N6-mAMP was severalfold more abundant than N6-mATP in MAPDA mutants, we speculate that additional molecular filters suppress the generation of N6-mATP. Enzyme kinetic data indicate that adenylate kinases represent such filters being highly selective for AMP versus N6-mAMP phosphorylation. We conclude that a multilayer molecular protection system is in place preventing N6-mAMP accumulation and salvage.
INTRODUCTION
Over 100 chemical modifications, mostly methylations, have been identified in eukaryotic RNA. N6-methylated adenine (m6A) is the most prevalent modification in mRNA but also occurs in other RNA species. In the nucleus, m6A is installed at specific sites in unspliced mRNA by the RNA methyltransferase complex comprising the active methyltransferase METTL3 as well as other proteins (Zhong et al., 2008; Liu et al., 2014; Ping et al., 2014; Meyer and Jaffrey, 2017; Růžička et al., 2017). The methylation pattern may be altered in the nucleus through partial demethylation by the RNA demethylase ALKBH5 (Zheng et al., 2013), although this process affects only a few sites and its purpose is still unclear (Meyer and Jaffrey, 2017). Interestingly, FTO, another demethylase involved in m6A demethylation (Jia et al., 2011; Su et al., 2018), was recently shown to also demethylate cap-structure-associated N6-2’-O-dimethyladenosine (Mauer et al., 2017). In the nucleus, some m6A marks are decoded by the nuclear m6A reader YTHDC1, which regulates mRNA splicing (Xiao et al., 2016). Mature mRNA, containing the bulk of m6A sites (Ke et al., 2017), is exported to the cytoplasm and the methylated sites are recognized by YTHDF1 (Wang et al., 2015), YTHDF2 (Wang et al., 2014), YTHDF3 (Li et al., 2017; Shi et al., 2017), and the eukaryotic initiation factor eIF3 (Meyer et al., 2015), thereby influencing various biological processes, such as mRNA translation efficiency and stability. Quantitatively little methylation or demethylation on mRNA occurs in the cytosol (Ke et al., 2017). Therefore, upon turnover of the mRNA, N6-methyl-AMP (N6-mAMP) will be released together with other mononucleotides. What is the metabolic fate of N6-mAMP, and how does the cell prevent its reuse as an RNA building block and random insertion into newly synthesized RNA species? This report describes how N6-mAMP is catabolized by a conserved mechanism in plants and mammals using Arabidopsis thaliana and the human cervical cancer cell line HeLa as model systems. An enzyme (ADAL, renamed MAPDA) that catabolizes N6-mAMP to inosine monophosphate (IMP) was identified, and its phylogenetic spread and possible specificity determinants for N6-mAMP were analyzed. We also investigated if MAPDA protects RNA from random misincorporation of N6-methylated A during RNA biosynthesis and which other molecular filters might be involved in this task.
RESULTS
Identification of N6-mAMP Deaminase
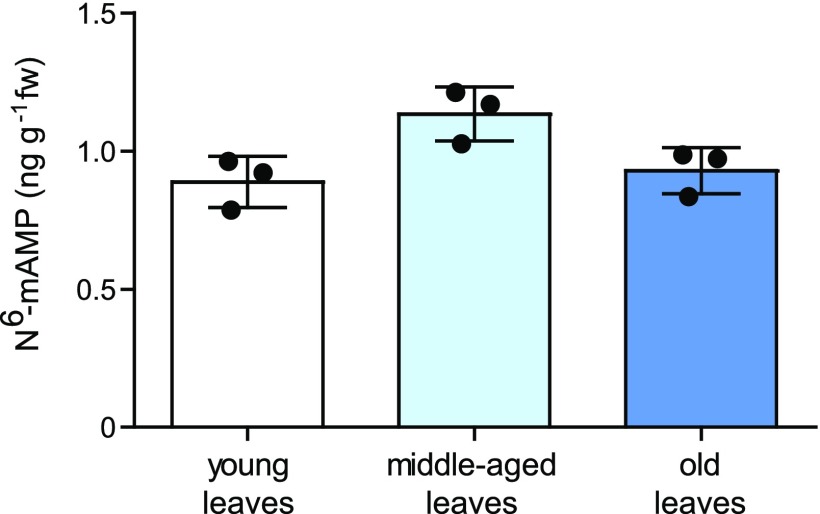
N6-mAMP might accumulate in aging tissue if it is not catabolized. However, we did not observe marked N6-mAMP accumulation with increasing leaf age in Arabidopsis (Figure 1; Supplemental Table 3), indicating that the plant does not simply accumulate N6-mAMP, but it might instead be able to degrade it.
Figure 1.
Absolute Quantification of N6-mAMP in Leaves of Different Ages.
Young leaves (top 9 rosette leaves), middle-aged leaves (leaf position 20 to 22), and old leaves (leaf position 26 to 30) from 33-d-old plants were used. Statistical evaluation with ANOVA indicated a difference at P = 0.0338, but there is no tendency toward increased N6-mAMP content with leaf age. Error bars are sd (n = 3 independent plants).
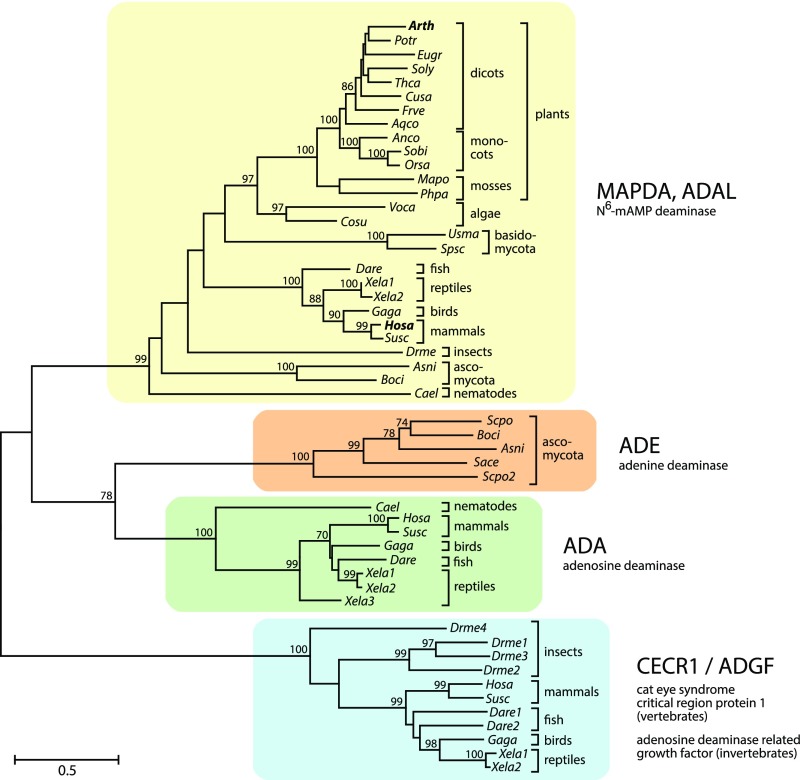
Arabidopsis and many other eukaryotes possess an enzyme, which is annotated as Adenosine/AMP Deaminase Like (ADAL) protein (UniProt Q8LPL7, locus At4g04880 for Arabidopsis). ADAL is structurally most similar to eukaryotic adenosine deaminases (ADAs) but is phylogenetically clearly distinct from these proteins (Figure 2; Maier et al., 2005). Radiotracer experiments have shown that plants do not possess ADA (Stasolla et al., 2003), and mutants of the only known Arabidopsis AMP deaminase (EMBRYONIC FACTOR1 [FAC1]) are embryo lethal (Xu et al., 2005), indicating that ADAL is neither an adenosine deaminase nor an AMP deaminase able to compensate for the lack of FAC1. In the context of screening for enzymes that structurally alter modified nucleosides or nucleotides used as prodrugs in cancer therapy, human ADAL (Uniprot Q6DHV7-1) was found to catalyze the hydrolysis of many N6- or O6-alkylated purine mononucleotides in vitro, including N6-mAMP (Schinkmanová et al., 2006; Murakami et al., 2011), but the physiological substrate and the physiological roles of this enzyme have not been investigated. We hypothesized that ADAL might catalyze the hydrolysis of N6-mAMP to IMP and methylamine in vivo, thereby converting a rare methylated nucleotide into a canonical non-methylated nucleotide (Figure 3A). This notion is additionally supported by the cytosolic subcellular location of ADAL (Figure 4), since its putative substrate, N6-mAMP, is generated in the cytosol.
Figure 2.
Phylogenetic Analysis of the Eukaryotic Adenyl Deaminase Family Including MAPDA (ADAL), ADE, ADA, and the Adenosine Deaminase-Related Growth Factor.
An unrooted maximum likelihood tree was constructed with MEGA 7 focusing on major model species representing a wide taxonomic range. The tree with the best log score is displayed, with branch lengths indicating the number of substitutions per site (legend). Numbers at branches indicate the percentage of trees in which associated taxa clustered together during bootstrap analysis (1000 bootstraps). Values below 70 are not shown. See Methods for details. CECR1/ADGF, adenosine deaminase-related growth factor.
Figure 3.
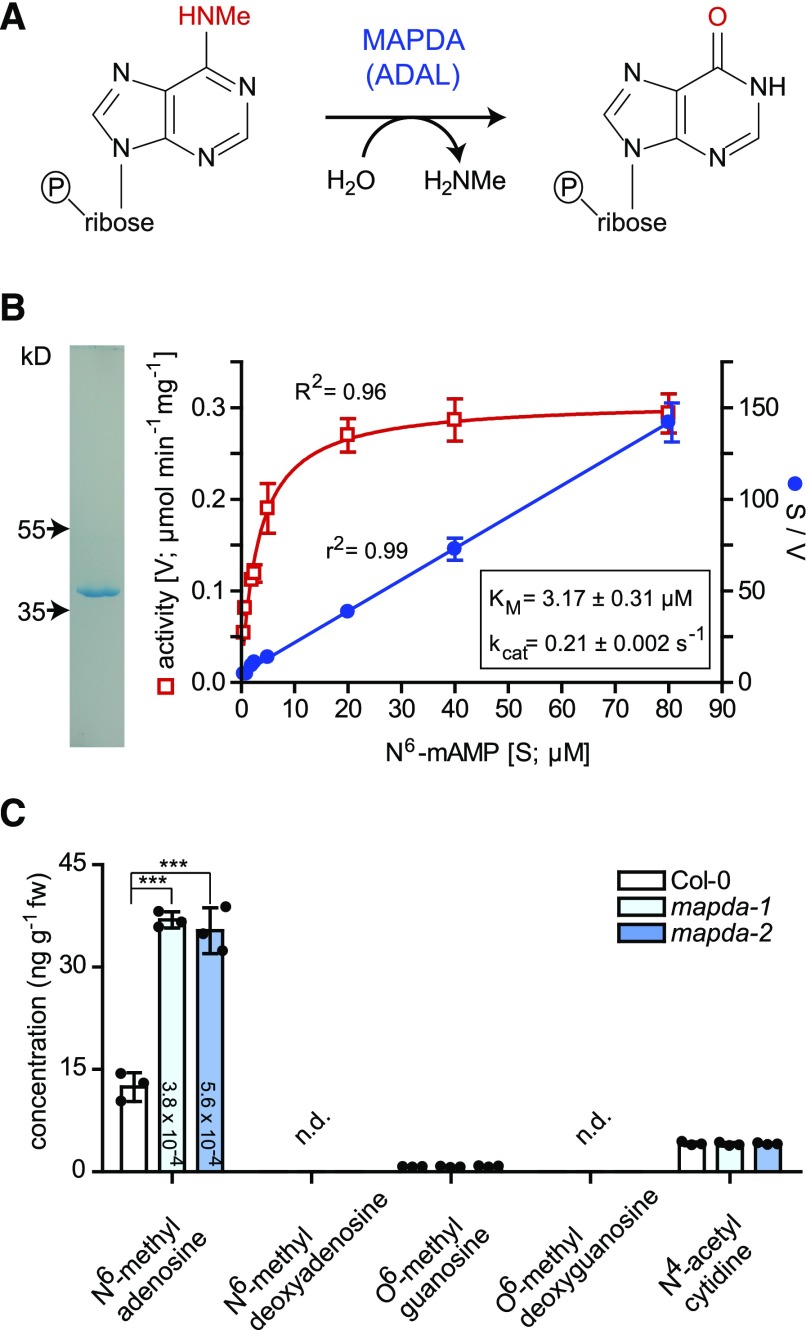
Enzymatic Reaction of AtMAPDA (AtADAL).
(A) Reaction scheme.
(B) Purity after affinity purification of AtMAPDA-Strep shown on a Coomassie-stained gel (left panel) and determination of kinetic constants using this enzyme preparation (right panel). The kinetic data were fitted with the Michaelis-Menten equation (red) or by linear regression for the Hanes plot (S/V over S; blue). Error bars are sd (n = 3 independent reactions).
(C) Concentrations of potential AtMAPDA substrates (after dephosphorylation) in leaf extracts of 7-d-old plants of the wild type and two independent AtMAPDA mutants. Detection limit of 0.0067 ng g−1 for N6-methyl deoxyadenosine and 0.56 ng g−1 for O6-methyl deoxyguanosine. Statistical evaluation with ANOVA followed by Tukey’s post test (relevant P values for the pairwise comparisons between Col-0 and the MAPDA mutants are shown on the columns; ***P < 0.001). Error bars are sd (n = 3 biological replicates, which are three pools containing several seedlings derived from seeds of three independent parental plants per genotype).
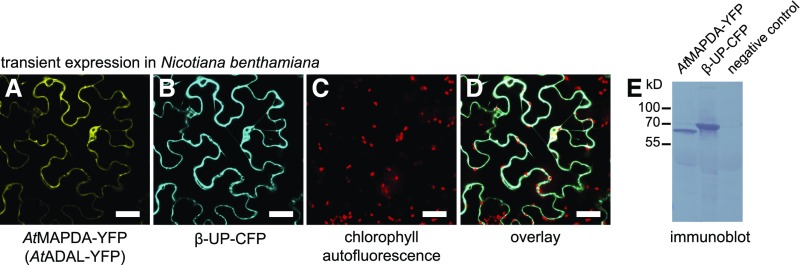
Figure 4.
Subcellular Localization of AtMAPDA (AtADAL).
(A) to (D) Confocal fluorescence microscopy images of cells at the lower leaf epidermis of N. benthamiana transiently coexpressing AtMAPDA C-terminally tagged with YFP (AtMAPDA-YFP) and cytosolic β-ureidopropionase C-terminally tagged with CFP (β-UP-CFP). YFP (A), CFP (B), autofluorescence of chloroplasts (C), and YFP, CFP, and autofluorescence detection (D). Bars = 40 µm.
(E) Stability test of the AtMAPDA-YFP and β-UP-CFP fusion proteins analyzed by immunoblot developed with a GFP-specific antibody. The negative control was generated from leaves infiltrated only with the helper Agrobacterium tumefaciens carrying the silencing inhibitor construct.
AtADAL, which was affinity purified after transient expression in Nicotiana benthamiana, indeed catalyzed N6-mAMP hydrolysis, yielding IMP with a catalytic efficiency (kcat/Km) of 66 mM−1 s−1 (Figure 3B, Table 1; Supplemental Figure 1), a value slightly lower than the reported catalytic efficiency of 126 mM−1 s−1 of the human ADAL homolog (HsADAL) for N6-mAMP (Murakami et al., 2011). Neither adenosine and AMP nor N6-methyladenosine and N6-mATP are substrates of AtADAL. Therefore, we decided to rename ADAL as N6-mAMP deaminase (MAPDA).
Table 1. Kinetic Constants of MAPDA from Arabidopsis, A. nidulans, and Homo sapiens and of the Cytosolic Adenylate Kinases AMPK3 and AMPK4 from Arabidopsis.
| N6-mAMP | AMP | |||||
|---|---|---|---|---|---|---|
| Km (µM) | kcat (s−1) | kcat/Km (mM−1 s−1) | Km (µM) | kcat (s−1) | kcat/Km (mM−1 s−1) | |
| AtMAPDA | 3.17 ± 0.31a | 0.21 ± 0.002 | 66 | n.d.b | n.d. | n.d. |
| AtMAPDA V57F | 3.77 ± 0.79 | 0.012 ± 0.0014 | 3.2 | n.d. | n.d. | n.d. |
| AnMAPDA | 2.70 ± 0.54 | 0.021 ± 0.0025 | 7.8 | n.d. | n.d. | n.d. |
| HsMAPDAc | 12.5 ± 1.7 | 1.58 ± 0.12 | 126 | 32.8 ± 7.5 | 0.055 ± 0.004 | 1.7 |
| AMPK3 | 3220 ± 600 | 0.0088 ± 0.0004 | 0.27 × 10−2 | 99 ± 27 | 3.66 ± 0.91 | 37 |
| AMPK4 | 7080 ± 1620 | 0.0081 ± 0.0017 | 0.11 × 10−2 | 620 ± 110 | 3.65 ± 1.11 | 5.9 |
Errors are sd (n = 3 independent enzymatic reactions).
n.d., not detectable.
Kinetic constants determined by Murakami et al. (2011).
MAPDA Is Required for N6-mAMP Catabolism in Vivo
To investigate the in vivo role of AtMAPDA, we quantified N6- or O6-methylated purine nucleosides by liquid chromatography coupled to quadrupole mass spectrometry (LC-MS/MS) in phosphatase-treated leaf extracts of two independent T-DNA insertion mutants (mapda-1 and mapda-2; Supplemental Figure 2A) and the wild type. Both mutant extracts contained elevated amounts of N6-methyladenosine but not O6-methylguanosine (Figure 3C). The corresponding deoxynucleosides were not detectable, indicating that they did not accumulate significantly. The levels of N4-acetylcytidine, a N-modified cytidine conserved in tRNA (Johansson and Byström, 2004; Ikeuchi et al., 2008), were also unchanged between the wild type and the mutants. Because MAPDA has no activity for N6-methyladenosine, the results indicate that N6-mAMP is the main in vivo substrate of AtMAPDA.
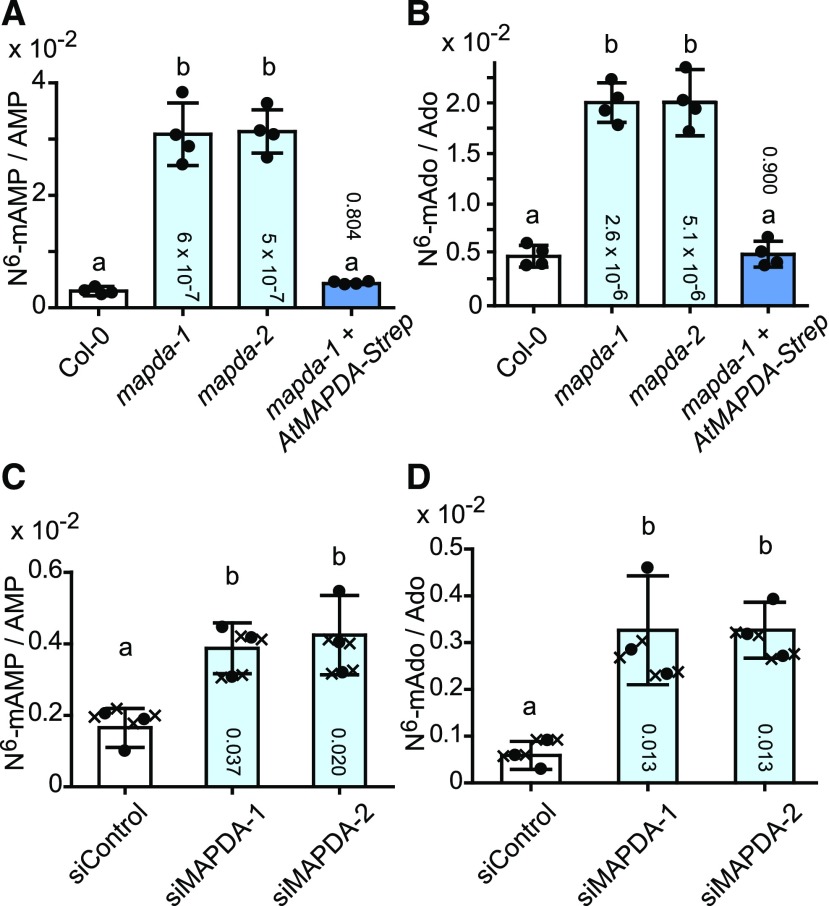
To confirm this finding, we quantified N6-mAMP and N6-methyladenosine directly (without dephosphorylation) in leaf extracts of the wild type, the two mutants, and a complementation line expressing an AtMAPDA-strep cDNA in the mapda-1 mutant background (Supplemental Figure 2B). AMP and adenosine were used as internal references. The N6-mAMP/AMP and N6-methyladenosine/adenosine ratios were ∼10 and 4 times higher, respectively, in 7-d-old seedlings of the AtMAPDA mutants compared with the wild type. In the complementation line, both ratios were similar to those of the wild type (Figures 5A and 5B). Comparable results were obtained later in development with 33-d-old plants for leaves of different ages (Supplemental Figure 3). These results demonstrate that AtMAPDA is required for N6-mAMP catabolism in vivo and that N6-mAMP is not completely stable but is in part dephosphorylated to N6-methyladenosine, especially in the absence of MAPDA.
Figure 5.
Relative N6-mAMP and N6-Methyladenosine Accumulation in Arabidopsis Leaves and Human HeLa Cells Varying in MAPDA (ADAL) Expression.
(A) N6-mAMP/AMP ratios of 7-d-old plants of Arabidopsis wild type (Col-0; white), the two AtMAPDA mutants (light blue), and a complementation line (dark blue). Error bars are sd (n = 4 biological replicates as defined in Figure 3C).
(B) As in (A) but showing N6-methyladenosine/adenosine ratios (N6-mAdo/Ado).
(C) N6-mAMP/AMP ratios in HeLa cells 3 d after transfection with either a control siRNA (siControl; white) or with two anti-HsMAPDA siRNAs (siMAPDA-1 and siMAPDA-2; light blue). Error bars are sd (n = 3 biological replicates from three independent transfections; “x” in contact with a data point of a biological replicate (circle) indicates technical replicates for this data point).
(D) As in (C) but showing N6-methyladenosine/adenosine ratios.
Statistical evaluation with ANOVA followed by Tukey’s post test. Probability values for pairwise comparisons to Col-0 or the siControl, respectively, are shown at the respective bars. Same letters label data that are not significantly different (P > 0.05).
To assess the phylogenetic conservation of MAPDA function, we extended our analysis to HeLa cells, in which the human homolog HsMAPDA (Figure 2) was downregulated by RNA interference. Transient transfection of two independent small interfering RNAs (siRNAs) directed against HsMAPDA (siMAPDA-1 and siMAPDA-2) resulted in a close to 90% reduction in mRNA levels compared with cells transfected with a nonsilencing control siRNA (Supplemental Figure 4). Although HsMAPDA depletion was only short term (3 d), a significant increase of ∼1.6-fold in the N6-mAMP/AMP ratio and of 5.5-fold in the N6-methyladenosine/adenosine ratio was observed in the knockdown cells compared with the control cells (Figures 5C and 5D). As in Arabidopsis, N6-methyladenosine accumulated in addition to N6-mAMP, although the former is not a substrate of HsMAPDA (Murakami et al., 2011), indicating that N6-mAMP is readily dephosphorylated to the nucleoside after the release from RNA in the absence of MAPDA.
Does MAPDA Help Protect RNA from Random N6-A Methylation?
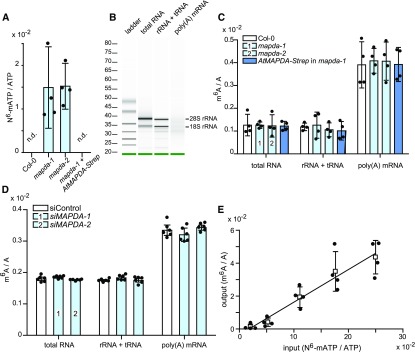
MAPDA function is conserved between Arabidopsis and human, and MAPDA enzymes are widespread in the eukaryotes, forming a clade distinct from related eukaryotic enzymes such as ADAs and adenine deaminases (ADEs) (Figure 2). This suggests that many eukaryotes, including fungi, insects, fish, reptiles, birds, mammals, as well as algae and plants, require MAPDA function. We speculated that MAPDA might protect organisms against the undesired salvage of N6-mAMP to N6-mATP, which could result in random incorporation of m6A into RNA, leading to a higher m6A content. Indeed, N6-mATP concentrations were significantly elevated in Arabidopsis seedlings of the two AtMAPDA mutants compared with the wild type and the complementation line (Figure 6A). However, the m6A/A ratios in the tRNA + rRNA and in the highly purified mRNA pools (Figure 6B) of the respective genotypes were not significantly different (Figure 6C). In HeLa cells, the m6A/A ratio of RNA was not altered by MAPDA knockdown (Figure 6D), but one needs to bear in mind that in this experimental setup, residual MAPDA expression cannot be ruled out. These results demonstrate that MAPDA contributes to the molecular filter preventing the generation of N6-mATP from RNA breakdown products, but MAPDA loss does not result in marked changes in the m6A content of RNA.
Figure 6.
Relative N6-mATP Accumulation and m6A Frequency in Different RNA Species in Arabidopsis Leaves and HeLa Cells Varying in MAPDA (ADAL) Expression and Selectivity of RNA Polymerase II for ATP versus N6-mATP.
(A) N6-mATP/ATP ratio of 7-d-old seedlings of Arabidopsis wild type (Col-0), the two AtMAPDA mutants (light blue), and a complementation line. Error bars are sd (n = 4 biological replicates as defined in Figure 3C).
(B) The integrity of the purified RNA samples and the efficient removal of rRNA from mRNA was analyzed using a Bioanalyzer 2100. The level of rRNA contamination in the mRNA was estimated to be 1.3%.
(C) m6A frequency relative to A in total RNA, nonpolyadenylated RNA (rRNA and tRNA), and mRNA of 7-d-old seedlings of wild type (Col-0; white), the two AtMAPDA mutants (light blue), and the complementation line (dark blue). Error bars are sd (n = 4; two independent RNA extractions and RNA isolations, each RNA digested twice). Statistical evaluation with ANOVA showed no significant (P < 0.05) differences between the respective RNA species from the distinct genotypes.
(D) m6A frequency relative to A in total RNA, nonpolyadenylated RNA (rRNA and tRNA), and mRNA from HeLa cells transfected either with control siRNA (white) or two independent siRNAs directed against HsMAPDA (light blue). Error bars are sd (n = 6 independent digestions from one RNA isolation). Statistical evaluation with ANOVA showed no significant (P < 0.05) differences between the genotypes.
(E) Relative incorporation of m6A per total A into mRNA (output) synthesized in vitro by RNA polymerase II using a commercial HeLa cell transcription assay at varying input ratios of N6-mATP/ATP. Error bars are sd (n = 4 independent transcription reactions).
Using an in vitro transcription assay employing HeLa cell extracts, we investigated the selectivity of a eukaryotic RNA polymerase II for ATP versus N6-mATP. Although the RNA polymerase preferred ATP as a substrate, N6-mATP was used as well (Figure 6E). At an N6-mATP/ATP ratio of 2%, which is close to the ratio occurring in the AtMAPDA mutants (1.49% ± 0.93% for madpa-1; 1.52% ± 0.93% for mapda-2) (Figure 6A), the increase in RNA methylation in vitro was 0.12% ± 0.17% (Figure 6E), which is too little and too variable to be reliably detected above the wild-type background of RNA methylation of 0.39% ± 0.10% in vivo (Figure 6C). This could explain why a possible effect of the MAPDA mutation on the m6A/A ratios in the mRNA is very difficult to demonstrate directly. In any case, the effect would be small. Indirectly, however, our data suggest that the MAPDA mutation will result in some random incorporation of m6A into newly synthesized RNA because the N6-mATP concentrations are elevated in this genetic background, and N6-mATP is a substrate of RNA polymerase II.
Adenylate Kinases Are Strong Selectivity Filters against N6-mAMP Phosphorylation
Seedlings of the MAPDA mutants contain more than 15 times more N6-mAMP than N6-mATP (Supplemental Table 1), indicating that an efficient conversion into the trinucleotide does not occur. Adenylate kinases are prime candidates for molecular filters preventing N6-mAMP phosphorylation, as they catalyze the conversion of AMP and ATP to two molecules of ADP. To test whether adenylate kinases efficiently prevent N6-mAMP phosphorylation, we cloned the two cytosolic adenylate kinase genes (AMPK3, At5g50370; AMPK4, At5g63400) (Lange et al., 2008) of Arabidopsis, expressed them in planta, and affinity purified the proteins. Indeed, AMPK3 and AMPK4 had over 13,700- and 5360-fold higher catalytic efficiencies (kcat/Km), respectively, for AMP compared with N6-mAMP (Table 1; Supplemental Figure 5). Considering that the AMP concentration in Arabidopsis leaf cells exceeds the N6-mAMP concentration by ∼300-fold in seedlings (Figure 5A; Supplemental Table 1) and over 3000-fold in fully grown rosettes (Supplemental Figure 3), it is clear that the AMPKs represent effective molecular filters preventing N6-mAMP phosphorylation. However, without MAPDA, this AMPK filter does not completely abolish the formation of N6-mATP (Figure 6A). Unfortunately, unlike MAPDA, the adenylate kinase filter cannot be removed to study the effect of a compromised filter on m6A-RNA methylation in vivo because these enzymes are central for ATP production in all organisms.
Elucidating Specificity Determinants of MAPDA Suggests That Fungal MAPDAs Might Have Acquired a Different Function
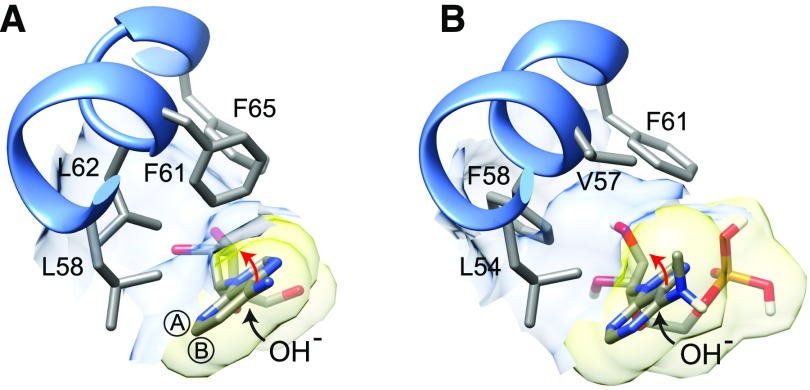
Finally, we investigated the molecular basis for the selectivity of AtMAPDA for N6-methylated AMP. We aligned conserved regions of MAPDA, ADA, and ADE from a selection of phylogenetically distant eukaryotic organisms, highlighting amino acids characteristically conserved in the respective enzyme classes (Supplemental Figure 6). Additionally, we constructed a homology model of AtMAPDA using HsADA as a template and docked N6-mAMP to the putative active site (Figure 7; Supplemental Figure 7). These analyses suggested that Val-57 and Phe-58 of AtMAPDA create a hydrophobic pocket to accommodate the aminomethyl group of the tetrahedral reaction intermediate generated after the addition of an OH group to carbon 6 of N6-mAMP during catalysis (Wilson and Quiocho, 1993). This size of this pocket is reduced in HsADA because the enzyme possesses a bulky phenylalanine (Phe-61) at the position equivalent to Val-57 in AtMAPDA (Figure 7; Supplemental Figure 6). Curiously, a phenylalanine at this position is also found in the putative MAPDA from the filamentous fungus Aspergillus nidulans (Supplemental Figure 6) and is widely conserved in many fungi. Based on this in silico analysis, we hypothesized that the enzyme from A. nidulans might not efficiently catalyze N6-mAMP deamination. This was confirmed experimentally by showing that the catalytic efficiency of AnMAPDA was 8.5- and 16.2-fold lower than that of the Arabidopsis and human enzyme, respectively (Table 1; Supplemental Figure 8). A mutant variant of MAPDA from Arabidopsis in which Val-57 was exchanged for Phe also showed a drastic reduction in catalytic efficiency (20.6-fold lower than the wild type). These diminished efficiencies of MAPDA-F variants were due to lower turnover numbers (kcat), which was predicted by the model (Figure 7) because phenylalanine at this position likely reduces the stabilization of the reaction intermediate by partially filling the cavity for the aminomethyl leaving group. Why is this phenylalanine conserved in fungal enzymes if it is unfavorable for MAPDA activity? A possible explanation is that these fungal MAPDA-F variants have an additional unknown substrate that requires this phenylalanine.
Figure 7.
Models of Substrate Binding to ADA and MAPDA.
(A) Binding of the A-face side of the 1-deaza-adenine ring of 1-deaza-adenosine to the hydrophobic residues between Leu-58 and Phe-65 of human ADA (PDB accession 1ADD). Encircled A and B indicate the A- and B-faces of the purine ring, respectively.
(B) Hypothetical binding of the A-face of the N6-methyladenine ring of N6-mAMP to the hydrophobic residues between Leu-54 and Phe-61 of the Arabidopsis MAPDA protein model. The black arrows in (A) and (B) indicate that during catalysis, the addition of water occurs from the B-face side of the purine ring. This leads to a tetrahydral configuration on C6 moving the amino or methyl-amino group above the A-face side of the ring (red arrow) in the reaction intermediate. Note that in the MAPDA model, a putative hydrophobic pocket is formed by Val-57 and Phe-58, which can potentially accommodate the methyl-amino group.
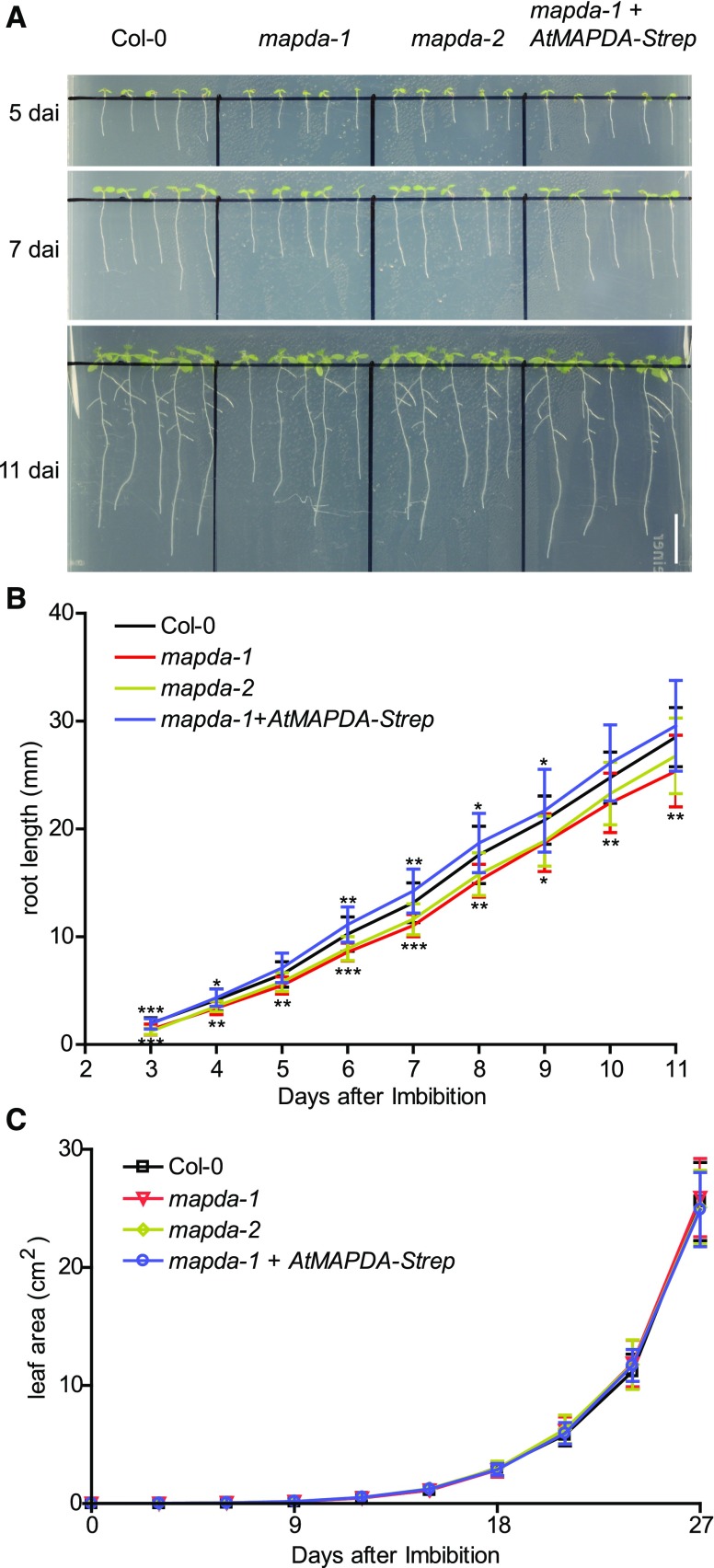
MAPDA Mutation Causes a Slight Reduction in Root Growth
The root growth of mapda-1 andmapda-2 showed a slight but significant reduction compared with the wild type and the complementation line (Figures 8A and 8B), particularly at early growth stages. Rosette growth was not compromised by MAPDA mutation (Figure 8C; Supplemental Figure 9). No further mutant phenotypes were observed.
Figure 8.
Comparison of the Root Length and Rosette Leaf Area of the Wild Type, the MAPDA Mutants, and a Complementation Line.
(A) Representative images of the root development observed for the respective genotypes grown on half-strength Murashige and Skoog plates under long-day conditions (16 h light). Bar = 10 mm. d.a.i., days after imbibition.
(B) Root length analysis from plants grown as shown in (A). Statistical evaluation with ANOVA followed by Tukey’s post test. Significance levels of P < 0.05, P < 0.01, and P < 0.001 is indicated in the figure by single, double, and triple asterisks, respectively. Error bars are sd (n = 25 independent plants per genotype grown on five plates).
(C) Quantification of rosette leave area for plants grown in soil under long-day conditions (16 h light). According to statistical evaluation with ANOVA, there are no significant (P < 0.05) differences between the genotypes. Error bar are sd (n = 10 independent plants per genotype grown as shown in Supplemental Figure 9).
DISCUSSION
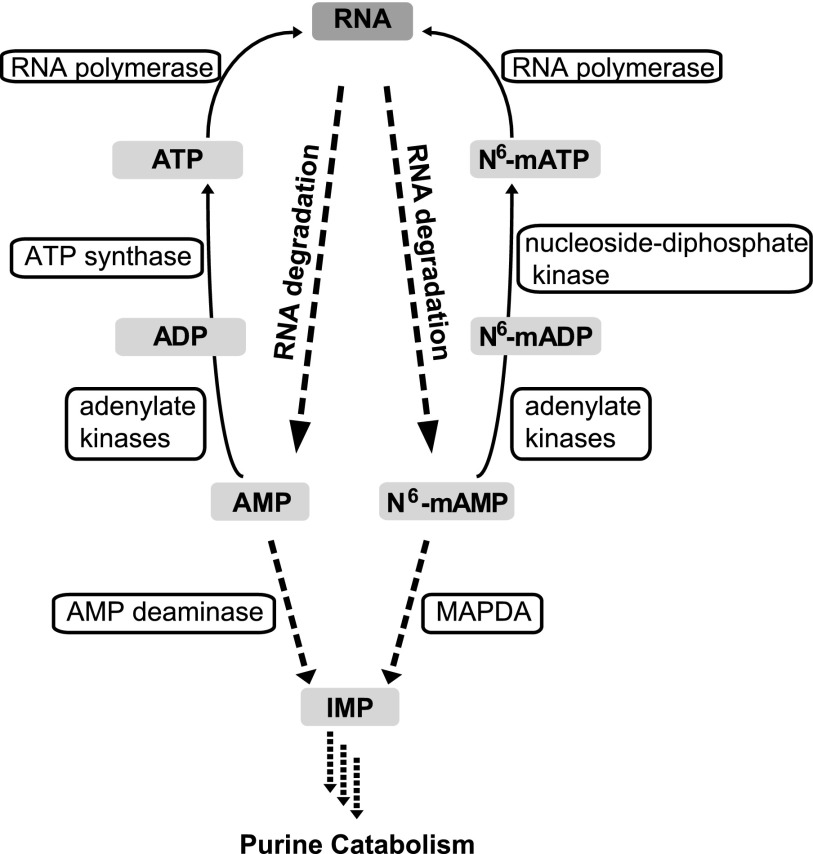
Our work highlights the notion that the events downstream of RNA degradation might be important for the fidelity of the RNA modification pattern, although we cannot show this directly due to experimental constraints. Random reentry of noncanonical nucleotides stemming from degraded RNA into newly synthesized RNA is likely prevented by the catabolism of such nucleotides by specialized enzymes such as MAPDA and by concerted enzymatic selectivity for the canonical nucleotides, as we have shown for the cytosolic adenylate kinases and for RNA polymerase II (Figure 9).
Figure 9.
A Model of Adenine Nucleotide Conversions.
The phosphorylation of ADP to ATP is catalyzed by ATP synthase. By contrast, the phosphorylation of N6-mADP to N6-mATP is probably catalyzed by nucleoside diphosphate kinases, which use ATP to phosphorylate a broad range of purine and pyrimidine dinucleotides and deoxydinucleotides.
Although the MAPDA mutation leads to an increase in N6-mATP concentration in vivo, a possible aberrant m6A introduction into RNA in this mutant background is too small to be measured reliably (Figures 6C and 6D). Therefore, it is possible that the selective pressure that has led to the conservation of MAPDA in most eukaryotes (Figure 2) did not only arise due to the requirement for proper RNA modification. MAPDA might also be needed to protect other cellular processes from excess N6-mAMP. For example, AMP deaminase, which is required to maintain the proper energy charge, is competitively inhibited by N6-mAMP (Merkler and Schramm, 1993). However, there is a considerable excess of AMP versus N6-mAMP in vivo (Supplemental Table 1), which will likely prevent drastic inhibition of AMP deaminase. Interestingly, we found that ATP concentrations in MAPDA mutants appear to be lower than those in the wild type or in our complementation line (without reaching statistical significance at the P < 0.05 level; Supplemental Table 1). Phenotypically the MAPDA mutants are very similar to the wild type under laboratory growth conditions, except for a slight reduction in root growth (Figure 8). By which molecular mechanism the excess N6-mAMP and N6-mATP accumulation in the mapda background causes a root growth defect remains to be elucidated.
In many fungi, such as A. nidulans, a MAPDA variant (MAPDA-F) might have acquired an additional or an alternative function to N6-mAMP deamination. Interestingly, some fungi, for example the yeast Saccharomyces cerevisiae, do not even possess a MAPDA gene. In S. cerevisiae, m6A methylation of mRNA occurs only during meiosis, and its abrogation is not lethal as in other eukaryotes (Yue et al., 2015). This also indicates that in yeast, the turnover of N6-mAMP is of minor importance. Additionally, ADE, which is widespread in fungi but absent in other eukaryotes (Ribard et al., 2003) (Figure 2), can hydrolyze N6 substituted adenines to some extent (Pospísilová et al., 2008), possibly reducing the requirement for MAPDA in fungi that have ADE. In agreement with this hypothesis, the Basidiomycete Ustilago maydis, which lacks ADE, possesses a consensus MAPDA, whereas many fungi, e.g., A. nidulans, contain ADE and MAPDA-F.
MAPDA is a eukaryotic enzyme that is required to remove modified RNA catabolites. It will be interesting to investigate whether other modified nucleotides derived from RNA breakdown, such as pseudouridine monophosphate and 5-methylcytosine monophosphate, also require specialized enzymes for their removal or whether they might be allowed to accumulate during plant development and perhaps be stored in the vacuole.
METHODS
Plant Materials and Cultivation
T-DNA insertion mutants of Arabidopsis thaliana from the SALK collection (SALK_144851, mapda-1; SALK_010573, mapda-2) (Alonso et al., 2003) were ordered from the Nottingham Arabidopsis Stock Centre. Wild type (Col-0) plants were obtained from the offspring of the segregating mutants.
Arabidopsis and Nicotiana benthamiana plants were cultivated under the same conditions described before (Witte et al., 2005). Agar plates were prepared with half-strength Murashige and Skoog nutrients (Duchefa Biochemie). The plates were incubated in a controlled growth chamber (Binder KBFW 720 with Osram Lumilux lights; 16 h light of 55 µmol m−2 s−1, 22°C day, 20°C night). For metabolic profiling, 7-d-old plants were grown on half-strength Murashige and Skoog agar plates, and older plants were grown in soil with full nutrients. Rosette leaf area was quantified from photographs using ImageJ 1.47 software.
HeLa Cell Culture and Transfection
HeLa cells were cultured in Roswell Park Memorial Institute 1640 medium (Life Technologies) supplemented with 10% fetal bovine serum (PAA Laboratories). The cells were incubated in a humidified atmosphere of 5% CO2 at 37°C and transfected with siRNAs at a final concentration of 5 nM, using RNAiMax (Life Technologies) according to the manufacturer’s instructions. The Silencer Select siRNAs targeting HsMAPDA (#1, s46296; #2, s46298) and the negative control (catalog no. 4390843) were obtained from Life Technologies.
RNA Extraction, Cloning, and Quantitative RT-PCR Analysis
For Arabidopsis, total RNA was isolated and cDNA was prepared according to a previous study (Chen et al., 2016). The following primers were employed for PCR: for AtMAPDA, 1901 and 1902; for AMPK3, P780 and P781; and for AMPK4, P782 and P783. All primers are listed in Supplemental Table 2. The resulting PCR products and the vector V69 (Myrach et al., 2017) were digested with EcoRI and XmaI and ligated. To express the MAPDA candidate from Aspergillus nidulans (CBF89034.1), a codon-optimized version of the gene for plant expression including an EcoRI site and a triple A sequence upstream of the start codon as well as an XmaI site replacing the stop codon was designed. The gene fragment was synthesized (Integrated DNA Technologies) and an EcoRI/XmaI fragment was cloned as described for AtMAPDA. Site-directed mutagenesis of AtMAPDA (V57F) was performed using P959 and P961 and a Q5 Site-Directed Mutagenesis Kit (NEB) following the manufacturer’s instructions.
Arabidopsis homozygous mutant lines were screened from a segregating population by PCR using the primers N59 and N60 as well as N59 and N61 for mapda-1 and the primers N59 and N60 as well as N61 and N60 for mapda-2. The PCR products from the mutants were cloned and sequenced to map the exact positions of the insertions. To determine the amount of gene-specific mRNA in the mutants, cDNA from seedlings was prepared. The PCR employed the primers N59 and N60, giving rise to a product of 773 bp from the wild-type allele. In both mutant lines, the T-DNA insertions are flanked by these primers. To amplify ACTIN2 as a control, the primers 2531 and 2532 were used.
For HeLa cells, total RNA was isolated from 5 × 105 to 106 cells using an RNeasy Plus Mini kit (Qiagen) according to the manufacturer’s protocol. Primers used for HsMAPDA detection were S1 and S2. QuantiTect Primer assays (Qiagen) were used to amplify the reference genes Peptidylprolyl Isomerase (PPIA; Hs_PPIA_4_SG) and Beta Actin (ACTB; Hs_ACTB_1_SG). qRT-PCR was performed with a Cfx96 device (Bio-Rad) using a Power SYBR Green RNA-to-Ct 1-Step kit (Applied Biosystems) according to the manufacturer’s protocol. Changes in the relative expression levels were determined using the 2–ΔΔCt method (Bio-Rad CFX Manager software 3.1.).
MRNA and nonpolyadenylated RNA were isolated from total RNA for digestion using the PolyATract mRNA Isolation Systems III (Promega). For mRNA isolation, two rounds of purification were used. The RNA was quantified using a NanoDrop photometer, and the integrity and purity were assessed by applying 100 ng of RNA per sample on a 2100 Bioanalyzer (Agilent).
Protein Purification, Enzymatic Assays, and Immunoblots
StrepII-tagged AtMAPDA, AtAMPK3, and AtAMPK4 were affinity purified after 3 d of transient expression in N. benthamiana as described before (Werner et al., 2008). Purified protein was quantified using the Bradford reagent from Serva using BSA as a standard.
For the deamination assay, deamination activity was assessed by quantifying the conversion of N6-mAMP to IMP using a UV-VIS spectrophotometer (UV-2700; Shimadzu; 1-cm path length; set at 240 nm). IMP dissolved in reaction buffer (0.5, 1, 2.5, 5, and 10 µM) was used as a standard. The enzymatic reaction was conducted at 30°C in a 0.3 mL reaction mixture containing 50 mM Bis-Tris propane buffer, pH 6.8, 2 mM DTT, 100 mM NaCl, and varying concentrations of the substrates. The reaction was started by adding 10 μL (0.58 µg) of purified enzyme. The kinetic constants were determined at 0.5, 0.75, 2, 2.5, 5, 20, 40, and 80 µM N6-mAMP. Kinetic curves were recorded in three replicates, and kinetic constants were determined by fitting the data to the Michaelis-Menten equation using Graph Pad Prism software.
AMP kinase activity was assessed by quantifying the conversion of ATP to ADP by HPLC analysis according to a previous study (Schroeder et al., 2018). The kinetic constants for the AMPKs were determined using the substrate AMP at concentrations of 0.01, 0.04, 0.1, 0.4, 1, and 4 mM and the substrate N6-mAMP at concentrations of 0.1, 0.25, 0.5, 1, 5, 10, and 18 mM. Each measurement was repeated three times. To start the reaction toward AMP as the substrate, 20 μL (0.18 µg) of purified enzyme was added to 80 μL reaction buffer containing 8 mM Tris-HCl, pH 7.5, 0.4 mM MgCl2, 0.1 mM KCl, 0.2 mM ATP, and varying amounts of AMP. The reaction was incubated at 30°C for 0, 3, and 6 min. For the reaction toward N6-mAMP as the substrate, 20 μL (5.52 µg) of purified enzyme was added to the above reaction buffer and incubated at 30°C for 0, 15, and 30 min. The reaction (90 µL) was stopped by adding 45 μL HClO4 (1.2 M) and subsequently 18.5 μL alkaline solution (5 M KOH and 2 M K2CO3). After centrifuging at 20,000g for 15 min, 20 μL supernatant was injected into the HPLC. The kinetic constants were determined as described above.
Monoclonal anti-GFP antibody from mouse (Roche; clones 7.1 and 13.1; 1:5000 diluted) were used together with anti-mouse IgG conjugated to alkaline phosphatase (Sigma-Aldrich A3562; 1:20,000 diluted) to detect YFP and CFP on immunoblots.
Subcellular Localization
AtMAPDA, C-terminally fused to eYFP, was transiently coexpressed with the cytosolic marker protein β-ureidopropionase fused to CFP (β-UP-CFP) in N. benthamiana (Dahncke and Witte, 2013). The samples were analyzed using a Leica True Confocal Scanner SP8 microscope equipped with an HC PL APO CS2 40× 1.10 water immersion objective (Leica). Acquired images were processed using Leica Application Suite Advanced Fluorescence software (Leica Microsystems).
Metabolite Analyses
Metabolites were quantified using an Agilent 1200 HPLC system coupled to an Agilent 6460 triple-quadrupole mass spectrometer (LC-MS/MS) according to the methods described before (Schroeder et al., 2018). For extraction, the plant material was frozen in liquid nitrogen and ground in a mixer mill (Mixer Mill MM400; Retsch) for 4 min at a frequency of 30 Hz. Homogenized plant material (100 mg) was extracted with 1 mL methanol:acetonitrile:water at a ratio of 2:2:1 containing freshly added internal standards (ISTDs; 1000 ng mL-1 15N5 13C10-AMP and 500 ng mL-1 13C5-adenosine). HeLa cells (2 × 10−6) were harvested and washed three times with 10 mM HEPES buffer containing 150 mM NaCl. Frozen cell pellets were resuspended in 0.6 mL extraction buffer as described above. The suspension was slowly forced through a 0.4-mm gauge syringe needle for 10 strokes to ensure complete cell lysis. The extracts were vortexed, incubated on ice for 10 min, and centrifuged at 15,000g at 4°C for 15 min. The supernatant (0.5 mL) was concentrated in a freeze dryer (Alpha 1-2 LDplus) and resuspended in 0.1 mL 5% acetonitrile and 95% water. For bulk dephosphorylation, clarified leaf extracts (88 µL) were mixed with 10 μL 10-fold-concentrated CutSmart buffer (NEB) and 2 μL Shrimp Alkaline Phosphatase (1 U µL−1; NEB) for 2 h. Bulk dephosphorylation was employed because (1) nucleosides could be resolved and detected with higher reproducibility than nucleotides on our LC-MS/MS setup and (2) commercial heavy atom-labeled internal standards were available for most nucleosides but generally not for nucleotides. After dephosphorylation, the solution was filtered in ultrafiltration tubes (3-kD molecular mass cutoff; Pall). To quantify purine nucleoside triphosphates, the samples were extracted as described above, and the extracts were directly applied to the LC-MS/MS after centrifuging at 15,000g at 4°C for 15 min. A sample volume of 20 μL was used for analysis. Nucleotides were separated on a Hypercarb column (50 × 4.6 mm, particle size 5 µm; Thermo Fisher Scientific), and nucleosides were separated on a Polaris 5 C18A column (50 × 4.6 mm, particle size 5 µm; Agilent) at a flow rate of 0.65 mL min−1. Solvent A was 10 mM ammonium acetate (pH 9.5), and solvent B was acetonitrile for the Hypercarb column and methanol for the C18A column. The gradient on the Hypercarb column was 0 min, 10% B; 9 min, 100% B; 10.4 min, 100% B; 10.6 min, 10% B; and 21 min, 10% B. The gradient on the C18A column was 0 min, 5% B; 2 min, 5% B; 5.5 min, 15% B; 9.5 min, 85% B; 11 min, 85% B; 11.1 min, 5% B; and 20 min, 5% B. The following mass transitions were monitored: m/z 348.07 to 136 (AMP), m/z 363.07 to 146 (15N5 13C10-AMP), m/z 362.09 to 150 (N6-mAMP), m/z 348.9 to 137 (IMP), m/z 268.1 to 136 (adenosine), m/z 273.1 to 136 (13C5-adenosine), m/z 282.12 to 150 (N6-methyladenosine), m/z 266.13 to 150 (N6-methyl deoxyadenosine), m/z 298.12 to 166 (O6-methylguanosine), m/z 282.12 to 166 (O6-methyl deoxyguanosine), and m/z 286.2 to 154.1 (N4-acetylcytidine). AMP and adenosine were quantified against their ISTDs. The other compounds were quantified using external standards (ESTDs) added to Col-0 extracts from the corresponding growth conditions as the matrix. N6-methyladenosine (0, 0.5, 2.5, 5, 25, and 50 ng mL−1), O6-methylguanosine (0, 1, 5, 10, 50, and 100 ng mL−1), N4-acetylcytidine (0, 1, 5, 10, 50, and 100 ng mL−1), and N6-mAMP (0, 0.5, 2.5, 5, 25, and 50 ng mL−1) were employed as ESTDs. To test whether N6-mAMP might be dephosphorylated during the extraction, 150 ng mL−1 external N6-mAMP was added while preparing a Col-0 leaf extract. An increase in N6-methyladenosine was not observed, showing that N6-mAMP was stable during sample preparation.
Quantitative Analysis of the m6A/A Ratio in RNA by LC-MS/MS
RNA samples were digested into single nucleosides according to a previous study (Su et al., 2014) with minor modifications. One hundred nanograms of mRNA or 1 µg of total RNA or nonpolyadenylated RNA was digested by benzonase (1 U; Sigma-Aldrich) and phosphodiesterase I (4 × 10−3 U; Sigma-Aldrich) and dephosphorylated by shrimp alkaline phosphatase (1 U; NEB) in 50 μL of buffer containing 10 mM Tris-HCl (pH 8.0), 1 mM MgCl2, and 0.1 mg mL−1 BSA. After an incubation at 37°C for 6 h, the samples were filtered in ultrafiltration tubes (3-kD cutoff; Pall), and 10-μL aliquots were analyzed by LC-MS/MS. The nucleosides were separated and identified as described above. Standard solutions of adenosine (5, 25, 50, 250, 500, and 1000 ng µL−1) and N6-methyladenosine (0.5, 2.5, 5, 25, 50, and 100 ng µL−1) were used for the quantification. The ratio of N6-methyladenosine to adenosine was calculated based on the calibrated concentrations.
The ability of eukaryotic RNA polymerase II to use N6-mATP as substrate was evaluated using the HeLa Scribe nuclear extract in vitro transcription system (Promega). The experiment was performed according to the manufacturer’s instructions with minor modifications: N6-mATP and ATP were used in different ratios (2:98, 5:95, 10:90, 15:85, and 20:80). Additionally ratios of 0:100 and 100:0 were used to assess the background of the system for m6A and A (from preformed RNA), respectively. After incubation of 1 h, RNA was isolated, digested, and the m6A/A ratio determined by LC-MS/MS as described above.
Phylogenetic Analysis
Eukaryotic members of the adenyl deaminase family were identified by BLASTp analysis at the Phytozome V12.1 website for plants and algae (phytozome.jgi.doe.gov/pz/portal.html) and at the National Center for Biotechnology Information (NCBI) for all nonplant organisms, focusing on major model species representing a wide taxonomic range (blast.ncbi.nlm.nih.gov/Blast.cgi). MAPDA from Arabidopsis (At4g04880.1) and human (NP_001311295.1) were used at NCBI as queries for the refseq protein data set employing the substitution matrix BLOSUM45 and recovering all hits with E-values < E-04 (<0.004 for Xenopus laevis). Redundant sequences were removed. A multiple alignment was generated using MUSCLE at the European Bioinformatics Institute (www.ebi.ac.uk/Tools/msa/muscle/). A maximum likelihood tree was constructed with MEGA7 (Kumar et al., 2016). All positions with less than 95% site coverage were eliminated, resulting in 316 positions for analysis. Initial trees were generated by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was employed to model evolutionary rate differences among sites (5 categories (+G, parameter = 2.8565)), while some sites were treated as evolutionarily invariable ([+I], 1.44% sites). Numbers at branches (indicating the percentage of trees in which associated taxa clustered together) were calculated by bootstrap analysis (1000 bootstraps). The sequences used in this analysis are listed in the Accession Numbers section. The following sequences were not included because they contained significant nonconsensus insertions and deletions: Xela_CECR1C, XP_018109923.1; Xela_ADA4, OCT60113.1; Asni_ADAL2, XP_658266.1; and Soly_ADE, Solyc00g031040.1.1 (adenine deaminase gene with nonconsensus sequence alteration from horizontal gene transfer into tomato).
Protein Modeling and Docking
A protein model of AtMAPDA was created using SWISS-MODEL with human ADA as template (PDB: 3AIR). N6-mAMP was docked into the model using AutoDock Vina (Trott and Olson, 2010). Residues involved in Zn binding and in the catalytic process are conserved between MAPDA and ADA (Supplemental Figure 6) (Wilson et al., 1991). Therefore, the only docking position in which the purine ring overlaid strongly with the ring of the substrate analog 1-deaza adenosine in the structure of human ADA (PDB: 1ADD) (Wilson and Quiocho, 1993) was selected because we assumed that the catalytic mechanism is identical in both enzymes, which constrains the position of the purine ring and the methyl-amino leaving group. USCF Chimera (http://www.rbvi.ucsf.edu/chimera/) (Pettersen et al., 2004) was used for structural inspection, manual adjustments of the model, and analyses.
Quantification and Statistical Analysis
ANOVA followed by Tukey’s honestly significant difference test were performed in R for statistical evaluation. Different letters represent differences at the P < 0.05 significance level.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: for Arabidopsis, AtMAPDA (At4g04880), AtAMPK3 (At5g50370), AtAMPK4 (At5g63400), ACTIN2 (At3g18780); MAPDA from A. nidulans (CBF89034.1), and MAPDA from Homo sapiens (NP_001311295.1). The following sequence was used in phylogenetic analysis (Phytozome V12.1 locus identifiers for plant and algae sequences and NCBI locus identifiers for nonplant sequences are given): Arth_ADAL (Arabidopsis, At4g04880.1), Cusa_ADAL (Cucumis sativus, Cucsa.045330.1), Frve_ADAL (Fragaria vesca, mrna19578.1-v1.0-hybrid), Thca_ADAL (Theobroma cacao, Thecc1EG030190t1), Potr_ADAL (Populus trichocarpa, Potri.011G000100.1), Eugr_ADAL (Eucalyptus grandis, Eucgr.D01304.1), Soly_ADAL (Solanum lycopersicum, Solyc10g005310.2.1), Aqco_ADAL (Aquilegia coerulea, Aqcoe4G236300.1), Sobi_ADAL (Sorghum bicolor, Sobic.002G405200.1), Orsa_ADAL (Oryza sativa, Os07g46630.1), Anco_ADAL (Ananas comosus, Aco002363.1), Mapo_ADAL (Marchantia polymorpha, Mapoly0023s0135.1), Phpa_ADAL (Physcomitrella patens, Pp3c11_7970V3.1), Voca_ADAL (Volvox cateri, Vocar.0005s0203.1), Cosu_ADAL (Coccomyxa subellipsoidea C-169, 22,155), Hosa_ADAL (H. sapiens, NP_001311295.1), Hosa_ADA (H. sapiens, NP_000013.2), Hosa_ADGF (H. sapiens, NP_001269154.1), Susc_ADAL (Sus scrofa, XP_005659746.1), Susc_ADA (Sus scrofa, XP_020933637.1), Susc_ADGF (S. scrofa, NP_999181.1), Gaga_ADAL (Gallus gallus, XP_015134377.1), Gaga_ADA (G. gallus, NP_001006290.1), Gaga_ADGF (G. gallus, XP_015141316.1), Dare_ADAL (Danio rerio, NP_001028916.1), Dare_ADA (D. rerio, NP_001002646.1), Dare_ADGF1 (D. rerio, XP_687719.2), Dare_ADGF2 (D. rerio, NP_001028889.2), Cael_ADAL (Caenorhabditis elegans, NP_871955.2), Cael_ADA (C. elegans, NP_872091.1), Xela_ADAL1 (X. laevis, NP_001085299.1), Xela_ADAL2 (X. laevis, XP_018109785.1), Xela_ADA1 (X. laevis, NP_001085740.1), Xela_ADA2 (X. laevis, OCT62325.1), Xela_ADA3 (X. laevis, NP_001087740.1), Xela_ADGF1 (X. laevis, NP_001090531.1), Xela_ADGF2 (X. laevis, NP_001089165.1), Drme_ADAL (Drosophila melanogaster, NP_649866.1), Drme_ADGF1 (Drosophila, NP_649006.2), Drme_ADGF2 (Drosophila, NP_524130.1), Drme_ADGF3 (Drosophila, NP_610977.1), Drme_ADGF4 (Drosophila, NP_650308.3), Sace_ADE (Saccharomyces cerevisiae [strain ATCC 204508/S288c], NP_014258.1), Scpo_ADE (Schizosaccharomyces pombe 972h, NP_595071.1), Scpo_ADE2 (S. pombe 972h, NP_595058.1), Asni_ADAL (A. nidulans, CBF89034.1), Asni_ADE (A. nidulans, XP_663682.1), Boci_ADAL (Botrytis cinerea B05.10, XP_001553511.1), Boci_ADE (B. cinerea B05.10, XP_001559454.1), Usma_ADAL (Ustilago maydis [strain 521/FGSC 9021], XP_011386395.1), and Spsc_ADAL (Sporisorium scitamineum, CDU23211.1).
Supplemental Data
Supplemental Figure 1. Detailed analysis of the AtMAPDA (AtADAL) reaction products.
Supplemental Figure 2. Characterization of AtMAPDA knockout and complementation lines.
Supplemental Figure 3. Relative N6-mAMP and N6-methyladenosine accumulation in the leaves of 33-d-old Arabidopsis plants.
Supplemental Figure 4. Transient HsMAPDA knockdown in HeLa cells.
Supplemental Figure 5. Purification and determination of the kinetic constants of the cytosolic Arabidopsis adenylate kinases AMPK3 and AMPK4.
Supplemental Figure 6. Comparative sequence analysis of N6-mAMP deaminase, adenosine deaminase, and adenine deaminase.
Supplemental Figure 7. 2D models of substrate binding to ADA and MAPDA.
Supplemental Figure 8. Purification and determination of kinetic constants of MAPDA from Aspergillus nidulans and AtMAPDA mutV57F.
Supplemental Figure 9. Experimental setup and representative images for the phenotypic comparison of rosettes of Col-0, mapda-1, mapda-2, and the complementation line before bolting.
Supplemental Table 1. Metabolites quantified in 7-d-old Arabidopsis seedlings.
Supplemental Table 2. Primers used in this study.
Supplemental Table 3. ANOVA analyses.
Supplemental File 1. Alignment used to produce the phylogenetic tree shown in Figure 2.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank André Specht, Hildegard Thölke, and Philipp Rüter for technical support, Xiaoye Liu for help with the statistical analyses, Anting Zhu for assistance with mass spectrometry, and Markus Niehaus for his help with root phenotype analysis. We also thank Rico M. Hartmann, Marian Uhe, and Helge Küster for support with the bioanalyzer. This work was financially supported by the Deutsche Forschungsgemeinschaft (Grants WI3411/2-1, WI3411/4-1, and HE 5949/3-1) and the Leibniz University Hannover (Wege in die Forschung II to M.C.).
AUTHOR CONTRIBUTIONS
C.-P.W. conceived the research and performed the bioinformatics analyses. M.C. designed and performed most experiments. M.J.U. carried out cloning and MAPDA kinetics. I.S.-G. prepared the human cell samples. M.A.O. designed the experiments for the human cell lines. M.H. analyzed the MAPDA-like from A. nidulans. M.C., M.A.O., and C.-P.W. analyzed the data. M.C. and C.-P.W. wrote the article.
References
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Chen M., Herde M., Witte C.-P. (2016). Of the nine cytidine deaminase-like genes in Arabidopsis, eight are pseudogenes and only one is required to maintain pyrimidine homeostasis in vivo. Plant Physiol. 171: 799–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahncke K., Witte C.-P. (2013). Plant purine nucleoside catabolism employs a guanosine deaminase required for the generation of xanthosine in Arabidopsis. Plant Cell 25: 4101–4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi Y., Kitahara K., Suzuki T. (2008). The RNA acetyltransferase driven by ATP hydrolysis synthesizes N4-acetylcytidine of tRNA anticodon. EMBO J. 27: 2194–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y.-G., He C. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7: 885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M.J.O., Byström A.S. (2004). The Saccharomyces cerevisiae TAN1 gene is required for N4-acetylcytidine formation in tRNA. RNA 10: 712–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S., Pandya-Jones A., Saito Y., Fak J.J., Vågbø C.B., Geula S., Hanna J.H., Black D.L., Darnell J.E. Jr., Darnell R.B. (2017). m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 31: 990–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange P.R., Geserick C., Tischendorf G., Zrenner R. (2008). Functions of chloroplastic adenylate kinases in Arabidopsis. Plant Physiol. 146: 492–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A., et al. (2017). Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res. 27: 444–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10: 93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier S.A., Galellis J.R., McDermid H.E. (2005). Phylogenetic analysis reveals a novel protein family closely related to adenosine deaminase. J. Mol. Evol. 61: 776–794. [DOI] [PubMed] [Google Scholar]
- Mauer J., et al. (2017). Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature 541: 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkler D.J., Schramm V.L. (1993). Catalytic mechanism of yeast adenosine 5′-monophosphate deaminase. Zinc content, substrate specificity, pH studies, and solvent isotope effects. Biochemistry 32: 5792–5799. [DOI] [PubMed] [Google Scholar]
- Meyer K.D., Jaffrey S.R. (2017). Rethinking m6A readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 33: 319–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K.D., Patil D.P., Zhou J., Zinoviev A., Skabkin M.A., Elemento O., Pestova T.V., Qian S.-B., Jaffrey S.R. (2015). 5′ UTR m6A promotes cap-independent translation. Cell 163: 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami E., Bao H., Mosley R.T., Du J., Sofia M.J., Furman P.A. (2011). Adenosine deaminase-like protein 1 (ADAL1): characterization and substrate specificity in the hydrolysis of N(6)- or O(6)-substituted purine or 2-aminopurine nucleoside monophosphates. J. Med. Chem. 54: 5902–5914. [DOI] [PubMed] [Google Scholar]
- Myrach T., Zhu A., Witte C.-P. (2017). The assembly of the plant urease activation complex and the essential role of the urease accessory protein G (UreG) in delivery of nickel to urease. J. Biol. Chem. 292: 14556–14565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25: 1605–1612. [DOI] [PubMed] [Google Scholar]
- Ping X.-L., et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24: 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospísilová H., Sebela M., Novák O., Frébort I. (2008). Hydrolytic cleavage of N6-substituted adenine derivatives by eukaryotic adenine and adenosine deaminases. Biosci. Rep. 28: 335–347. [DOI] [PubMed] [Google Scholar]
- Ribard C., Rochet M., Labedan B., Daignan-Fornier B., Alzari P., Scazzocchio C., Oestreicher N. (2003). Sub-families of alpha/beta barrel enzymes: a new adenine deaminase family. J. Mol. Biol. 334: 1117–1131. [DOI] [PubMed] [Google Scholar]
- Růžička K., et al. (2017). Identification of factors required for m6 A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 215: 157–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinkmanová M., Votruba I., Holý A. (2006). N6-methyl-AMP aminohydrolase activates N6-substituted purine acyclic nucleoside phosphonates. Biochem. Pharmacol. 71: 1370–1376. [DOI] [PubMed] [Google Scholar]
- Schroeder R.Y., Zhu A., Eubel H., Dahncke K., Witte C.-P. (2018). The ribokinases of Arabidopsis thaliana and Saccharomyces cerevisiae are required for ribose recycling from nucleotide catabolism, which in plants is not essential to survive prolonged dark stress. New Phytol. 217: 233–244. [DOI] [PubMed] [Google Scholar]
- Shi H., Wang X., Lu Z., Zhao B.S., Ma H., Hsu P.J., Liu C., He C. (2017). YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 27: 315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasolla C., Katahira R., Thorpe T.A., Ashihara H. (2003). Purine and pyrimidine nucleotide metabolism in higher plants. J. Plant Physiol. 160: 1271–1295. [DOI] [PubMed] [Google Scholar]
- Su R., et al. (2018). R-2HG exhibits anti-tumor activity by targeting FTO/m6A/MYC/CEBPA signaling. Cell 172: 90–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su D., Chan C.T.Y., Gu C., Lim K.S., Chionh Y.H., McBee M.E., Russell B.S., Babu I.R., Begley T.J., Dedon P.C. (2014). Quantitative analysis of ribonucleoside modifications in tRNA by HPLC-coupled mass spectrometry. Nat. Protoc. 9: 828–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., Olson A.J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31: 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505: 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhao B.S., Roundtree I.A., Lu Z., Han D., Ma H., Weng X., Chen K., Shi H., He C. (2015). N6-methyladenosine modulates messenger RNA translation efficiency. Cell 161: 1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner A.K., Sparkes I.A., Romeis T., Witte C.-P. (2008). Identification, biochemical characterization, and subcellular localization of allantoate amidohydrolases from Arabidopsis and soybean. Plant Physiol. 146: 418–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D.K., Quiocho F.A. (1993). A pre-transition-state mimic of an enzyme: X-ray structure of adenosine deaminase with bound 1-deazaadenosine and zinc-activated water. Biochemistry 32: 1689–1694. [DOI] [PubMed] [Google Scholar]
- Wilson D.K., Rudolph F.B., Quiocho F.A. (1991). Atomic structure of adenosine deaminase complexed with a transition-state analog: understanding catalysis and immunodeficiency mutations. Science 252: 1278–1284. [DOI] [PubMed] [Google Scholar]
- Witte C.P., Rosso M.G., Romeis T. (2005). Identification of three urease accessory proteins that are required for urease activation in Arabidopsis. Plant Physiol. 139: 1155–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W., et al. (2016). Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61: 507–519. [DOI] [PubMed] [Google Scholar]
- Xu J., Zhang H.-Y., Xie C.-H., Xue H.-W., Dijkhuis P., Liu C.-M. (2005). EMBRYONIC FACTOR 1 encodes an AMP deaminase and is essential for the zygote to embryo transition in Arabidopsis. Plant J. 42: 743–756. [DOI] [PubMed] [Google Scholar]
- Yue Y., Liu J., He C. (2015). RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 29: 1343–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G., et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49: 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Li H., Bodi Z., Button J., Vespa L., Herzog M., Fray R.G. (2008). MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 20: 1278–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]