The KCH kinesin is both a retrograde nuclear transporter and a cytoskeleton cross-linking factor in the moss Physcomitrella patens.
Abstract
Long-distance transport along microtubules (MTs) is critical for intracellular organization. In animals, antagonistic motor proteins kinesin (plus end directed) and dynein (minus end directed) drive cargo transport. In land plants, however, the identity of motors responsible for transport is poorly understood, as genes encoding cytoplasmic dynein are absent in plant genomes. How other functions of dynein are brought about in plants also remains unknown. Here, we show that a subclass of the kinesin-14 family, KCH (kinesin with calponin homology domain), which can also bind actin, drives MT minus end-directed nuclear transport in the moss Physcomitrella patens. When all four KCH genes were deleted, the nucleus was not maintained in the cell center but was translocated to the apical end of protonemal cells. In the knockout (KO) line, apical cell tip growth was also severely suppressed. KCH was localized to MTs, including at the MT focal point near the tip of protonemal cells, where MT plus ends coalesced with actin filaments. MT focus was not stably maintained in KCH KO lines, whereas actin destabilization also disrupted the MT focus in wild-type lines despite KCH remaining on unfocused MTs. KCH had distinct functions in nuclear transport and tip growth, as a truncated KCH construct restored nuclear transport activity, but not tip growth retardation of the KO line. Thus, our study identified KCH as a long-distance retrograde transporter as well as a MT cross-linker, reminiscent of the versatile animal dynein.
INTRODUCTION
Intracellular transport is a critical cellular mechanism for cell organization in eukaryotic cells. Many cellular components, including organelles, proteins, and RNA, are transported to their appropriate positions where they specifically function in response to internal and external signals. Although actin and myosin were long since believed to be the main transporters of cellular components, recent studies have uncovered the prevalence of microtubule (MT)-dependent transport as well (Kong et al., 2015; Miki et al., 2015; Nakaoka et al., 2015; Zhu et al., 2015; Yamada et al., 2017). However, a unique feature of plant motor systems is that the genes encoding cytoplasmic dynein, the sole MT minus end-directed transporter in animals, have been lost during plant evolution. Moreover, dynein function is not limited to cargo transport, as a variety of fundamental cellular processes require dynein, such as MT-based force generation at the cortex (Grill and Hyman, 2005; Gönczy, 2008; McNally, 2013), MT-MT cross-linking (Ferenz et al., 2009; Tanenbaum et al., 2013), and MT-actin cross-linking (Grabham et al., 2007; Perlson et al., 2013; Coles and Bradke, 2015). However, how plants execute these functions without dynein remains unanswered.
The moss Physcomitrella patens is an emerging model plant of cell and developmental biology, in part due to the applicability of homologous recombination and high-resolution live imaging (Cove, 2005; Cove et al., 2006; Vidali and Bezanilla, 2012). The protonemal apical cell of P. patens is an excellent system to study MT-based transport. MTs are predominantly aligned along the cell longitudinal axis with a characteristic overall polarity depending on cell cycle stage. The nucleus, chloroplasts, and newly formed MTs have been identified as cargo that is transported on MT tracks in protonemal cells, wherein nuclear movement was shown to be independent of actin (Miki et al., 2015; Nakaoka et al., 2015; Yamada et al., 2017). Using this model system, kinesin-ARK (armadillo repeat-containing kinesin) was first identified as a plus end-directed nuclear transporter; upon RNAi knockdown of this plant-specific, plus end-directed motor protein, the nucleus migrated toward the cell center after cell division as normal but then moved back to the cell plate, i.e., the nucleus showed an abnormal minus end-directed motility (Miki et al., 2015). It was also revealed that the nonprocessive, minus end-directed KCBP (kinesin-like calmodulin binding protein), a member of the kinesin-14 protein family, is required for minus end-directed nuclear transport. In the absence of KCBP, the nucleus could not move to the cell center immediately after cell division, i.e., minus end-directed motility was inhibited (Yamada et al., 2017). Although a single dimeric KCBP cannot take multiple steps along the MT (nonprocessive), clustered motors exhibit processive motility in vitro and in vivo; thus, multiple KCBP motors associated with the nuclear surface can transport the nucleus toward MT minus ends (Jonsson et al., 2015; Yamada et al., 2017). However, the nuclear transport function of KCBP is limited during the latest stage of cell division, as KCBP is no longer necessary for maintaining the central positioning of the nucleus during interphase. It is plausible that an additional minus end-directed motor protein that antagonizes kinesin-ARK and possibly other plus end-directed kinesins is expressed in moss cells.
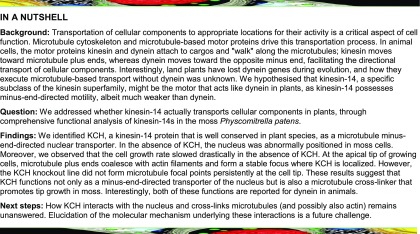
Minus end-directed kinesin-14 is duplicated uniquely in the land plant lineage and constitutes six subfamilies; in Arabidopsis thaliana, it is the most expanded family among the kinesin superfamily (Shen et al., 2012; Zhu and Dixit, 2012). KCBP belongs to class VI of kinesin-14 and transports not only the nucleus but also chloroplasts in moss (Yamada et al., 2017). In Arabidopsis, the cytoskeletal organization of the trichome cell is defective in kcbp mutants, suggesting an additional function to nuclear/chloroplast transport (Tian et al., 2015). The class I kinesin-14 ATK (Arabidopsis thaliana kinesin) conserved in animals is essential for mitotic spindle coalescence and drives minus end-directed transport of newly formed MTs along other MTs in the moss cytoplasm (Ambrose et al., 2005; Yamada et al., 2017). KAC (kinesin-like protein for actin-based chloroplast movement) is a class V kinesin-14 that no longer possesses MT affinity but has acquired an actin binding activity and regulates actin-dependent chloroplast photo-relocation movement and anchorage to the plasma membrane (Suetsugu et al., 2010, 2012). Class III kinesin-14 is localized to the spindle in moss but appears to have lost MT-based motor activity (Miki et al., 2014; Jonsson et al., 2015). The class IV kinesin-14 TBK (tobacco BY-2 kinesin-like polypeptide) has a weak MT motor activity and localizes to cortical MTs, yet its cellular function is unknown (Goto and Asada, 2007; Jonsson et al., 2015). Class II kinesin-14 genes form a large clade in the plant kinesin family, where 9 out of 61 Arabidopsis kinesin genes are classified into this clade (Figure 1A) and multiple activities and cellular functions have been reported. Kinesin14-II possesses the calponin homology (CH) domain in its N-terminal region followed by dimerization and motor domains (hereafter called KCH; Figure 1B). Adjacent to the motor domain, there exists an uncharacterized C-terminal extension in this subfamily that is not found in ATK or KCBP (Preuss et al., 2004; Frey et al., 2009; Shen et al., 2012). Mutant analyses have uncovered divergent functions of KCH, such as cell size regulation (OsKCH1; Frey et al., 2010), mitochondrial respiration (Arabidopsis KP1; Yang et al., 2011), and cell-to-cell movement of a transcription factor (Arabidopsis KinG; Spiegelman et al., 2018). However, the complete picture of KCH function has not been elucidated, since loss-of-function analysis using complete null mutants has not been conducted for this highly duplicated gene subfamily in flowering plants. By contrast, P. patens possesses only four KCH proteins that have high levels of amino acid identity (Figure 1A, boxed in red; Supplemental Data Set 1), suggesting that they redundantly exhibit basal functions of this kinesin subfamily.
Figure 1.
KCH Is Required for Protonema Growth, Gametophore Leaf Morphology, and Rhizoid Elongation.
(A) Phylogenetic tree of plant KCH subgroup members and other members of the kinesin family. At, Arabidopsis thaliana; Os, Oryza sativa; Pp, Physcomitrella patens; Gh, Gossypium hirsutum; Nt, Nicotiana tabacum; Dm, Drosophila melanogaster; Hs, Homo sapiens. P. patens genes are highlighted with a red box. Horizontal branch length is proportional to the estimated evolutionary distance. Bar = 0.1 amino acid substitutions per site.
(B) Schematic diagram of domain organization and coiled-coil (CC) prediction of P. patens KCHa (1–1396 amino acids). CH domain (106–199 amino acids) and kinesin motor domain (642–975 amino acids) were predicted by an NCBI domain search. Predicted coiled-coil domains with a probability >0.7 and 21 residue windows (Coiled-Coils Prediction; PRABI Lyon Gerland) are displayed (454–567 amino acids and 978–1012 amino acids).
(C) MT localization of Citrine-KCHa, -KCHb, and -KCHc at the caulonemal cell tip. Image contrast was individually adjusted for each sample. Bar = 5 µm.
(D) and (E) Colony size comparison between control (parental line expressing GFP-tubulin/histoneH2B-mRFP), KCHabcd KO (KCH KO), and KCH KO/Cerulean-KCHa. Colonies were cultured for 3 weeks from the stage of protoplasts. Bar in (D) = 5 mm. Bars and error bars in (E) represent the mean and se, respectively. Control, n = 20; KCH KO, n = 20; KCH KO/Cerulean-KCHa, n = 20. *P < 0.0001; ns (not significant), P > 0.7 (unpaired t test with equal sd, two-tailed). Experiments were performed three times and the data analyzed twice. The data of one experiment are displayed.
(F) Gametophores and rhizoids cultured for 4 weeks on BCDAT medium. Bars = 1 mm (top) and 0.5 mm (bottom).
In this study, we generated a P. patens plant with a complete deletion of the KCH gene and provide evidence that KCH drives minus end-directed nuclear transport. Furthermore, KCH contributes to cell tip growth likely via cross-linking MTs at the apical tip. These two functions are distinct, as a KCH fragment that lacks the unusual C-terminal extension fulfils the function of nuclear transport but not tip growth. By contrast, the CH domain, which has been assumed to be the cargo (i.e., actin) binding site, was not required for either function. We propose that plant KCH is a versatile cargo transporter that also fulfils other MT-based functions, analogous to the dynein motor in animals.
RESULTS
Complete KCH Deletion Affects Moss Growth and Morphology
The gene expression database (The Bio-Analytic Resource for Plant Biology, Physcomitrella eFP Browser; http://bar.utoronto.ca/efp_physcomitrella/cgi-bin/efpWeb.cgi) (Ortiz-Ramírez et al., 2016) and our previous localization analysis of KCH-Citrine fusion protein (Citrine is a YFP variant) (Miki et al., 2014) suggested that KCHa, b, and c, but not KHCd, are expressed in protonemal cells. However, since C-terminal tagging might perturb the function of the kinesin-14 subfamily, the motor domain of which is generally located closer to the C terminus, we inserted the Citrine gene in front of endogenous KCHa, b, and c (elements other than the Citrine ORF were not integrated) (Supplemental Figures 1A and 1B). With the newly selected Citrine-KCH lines, we confirmed that KCHa, b, and c are indeed expressed in protonemal cells and, furthermore, exhibit MT localization at the cell tip (Figure 1C).
To test the contribution of KCH to nuclear positioning and other intracellular processes, we sequentially deleted four KCH genes by means of homologous recombination in the moss lines expressing GFP-tubulin and histoneH2B-mRFP (Supplemental Figures 1C and 1D). The KCHacd triple knockout (KO) line grew in an indistinguishable manner to wild-type moss. However, when all four KCH genes were deleted, moss colony growth was severely retarded (this line is hereafter called the KCH KO line; Figures 1D and 1E). In addition, the gametophore leaf was curly and the rhizoid was much shorter than in the control line (Figure 1F). These phenotypes were suppressed when Cerulean-tagged KCHa was expressed by a constitutively active promoter in the KCH KO line (Figures 1D to 1F). These results indicate that KCH is a critical motor in moss development, albeit not essential for moss viability.
KCH Is Required for Minus End-Directed Nuclear Transport along MTs during Interphase
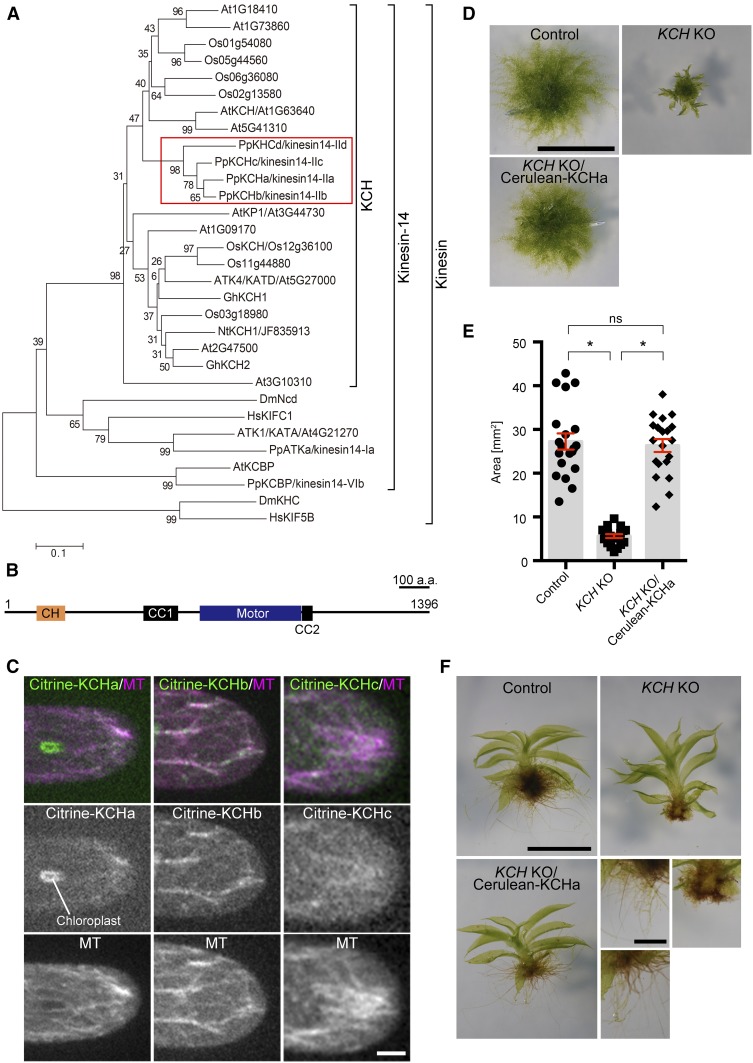
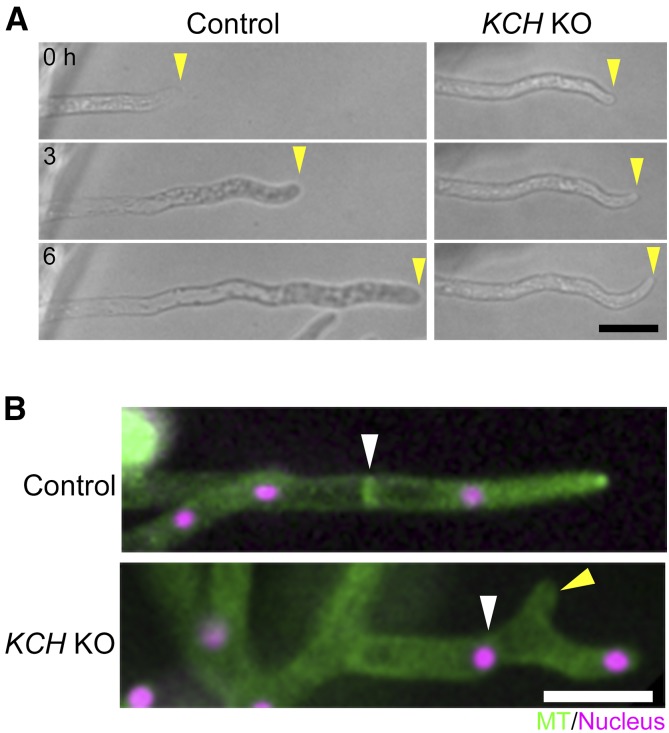
We performed live imaging of the protonemal apical cells of the quadruple KO line. Unlike the KCBP KO line, sister nuclei moved toward the cell center after chromosome segregation, indicating that minus end-directed motility during telophase was not impaired in the absence of KCH (Figure 2B; Supplemental Movie 1). However, unlike the control cells that maintained cell center positioning of the nucleus during tip growth, the nucleus did not stop moving at the cell center but migrated further toward the cell tip (apical cell) or moved back toward the cell plate (subapical cell). Consequently, the nucleus was positioned near the apical cell wall of the apical cell in the KCH KO line; this phenotype was rescued by ectopic Cerulean-KCHa expression (Figures 2C and 2D). During subsequent mitosis of apical cells, spindle assembly took place 10% more apically compared with the control line, resulting in an apical shift of the cell division site (P < 0.05, unpaired t test with equal sd, n = 13 [KO] and 7 [control]); this suggests a physiological role for nuclear positioning. Given the known MT polarity during interphase of apical cells (plus ends predominantly face the apex; Hiwatashi et al., 2014; Yamada et al., 2017; Figure 2A) and the appearance of the nuclear migration defects contrary to kinesin-ARK depletion (the nucleus moves back to the cell plate in apical cells; Miki et al., 2015), it was suggested that KCH drives MT minus end-directed transport of the nucleus during interphase.
Figure 2.
Nuclear Positioning Defects Observed in KCH KO Cells.
(A) Schematic representation of MT polarity and nuclear positioning in apical cells based on previous studies (Hiwatashi et al., 2014; Miki et al., 2015; Yamada et al., 2017).
(B) Nuclear migration defects were observed in the KCH KO line. Snapshot (top) and kymograph (bottom) of a control and KCH KO cell are displayed. White lines in snapshots indicate the position where the kymograph was created. Kymograph starts from the mitotic metaphase. White broken lines indicate the position of the cell wall. Horizontal bar = 50 µm; vertical bar = 2 h.
(C) Nuclei were apically localized in the absence of KCH, and the defect was rescued by Cerulean-KCHa expression. White bars indicate positions of the cell wall, nucleus, and apical tip. Bar = 50 µm.
(D) Relative position of the nucleus within the apical cell was quantified. The “0” corresponds to the cell wall, whereas “100” indicates the cell tip. Bars and error bars represent the mean and se, respectively. Control, n = 21, KCH KO, n = 16; KCH KO/Cerulean-KCHa, n = 18; PRFa RNAi, n = 19. *P < 0.0001 (unpaired t test with equal sd, two-tailed). Experiments and data analyses were performed twice, with data from one experiment displayed.
(E) Distribution of nuclei, chloroplasts, mitochondria, and vacuoles (GFP-tubulin-excluded area). Yellow arrowhead and “N” indicate the position of the cell wall and nucleus, respectively. Mitochondrial images were acquired with six z-sections (separated by 4 μm) and are displayed after maximum projection. Bars = 25 µm.
(F) Nuclear deformation in the apical cell. Triple KO line (KCHacd KO) was used as control. Bar = 5 µm.
To determine whether the distribution of other organelles is perturbed in the absence of KCH, we assessed the intracellular distribution of chloroplasts (by autofluorescence), mitochondria (the N-terminal 78 amino acids of the γ-subunit of the Arabidopsis mitochondrial F1 ATPase) (Uchida et al., 2011; Nakaoka et al., 2015), and vacuoles (the GFP-tubulin-excluded areas). Unlike the nucleus, we did not observe any abnormal distribution or morphology of these organelles, suggesting that the effect of KCH deletion is specific to the nucleus (Figure 2E; Supplemental Figure 2).
In Arabidopsis root and mesophyll cells, the nucleus is transported along actin filaments by the myosin XI-i motor, and mutants of this motor exhibited not only defects in nuclear dynamics but also nuclear deformation (the nucleus becomes rounder) (Tamura et al., 2013). Interestingly, when we observed the KCH KO line with spinning-disc confocal microscopy, we detected stretching and invagination of the nucleus in 55% (n = 29) of apical cells (Figure 2F). The discrepancy in nuclear shape in the Arabidopsis myoXI-i mutant and moss KCH KO is possibly because of the additional force applied on the nuclear surface of moss, for example, by kinesin-ARK. Nevertheless, this observation further supports the idea that KCH is responsible for nuclear motility.
Processive Motility of KCH in Vivo
Cytoskeletal motor proteins that drive cargo transport are generally processive, where a single dimeric motor that attaches to a cytoskeletal filament (MT or actin) takes multiple steps toward one direction before dissociation. Some nonprocessive motors can also be transporters when multiple dimers participate in cargo transport. KCBP represents the latter example; multiple KCBP molecules bind to MTs via the motor domain and to vesicular cargo via the tail domain and execute long-distance transport (Yamada et al., 2017). The purified rice (Oryza sativa) KCH1 motor was also shown to be nonprocessive, but its cohort action drives actin motility along MTs (Walter et al., 2015). On the other hand, there have been contradictory reports as to whether full-length KCH shows processive motility in vivo. When tobacco (Nicotiana tabacum) GFP-NtKCH was expressed in tobacco BY-2 cells, motility of GFP signals (i.e., clustered GFP signals) along MTs was detected (Klotz and Nick, 2012). By contrast, GFP-AtKinG was observed only as static punctae on MTs in Nicotiana benthamiana leaf epidermal cells (Spiegelman et al., 2018). In these studies, however, GFP-tagged constructs were ectopically overexpressed and might not represent native KCH dynamics.
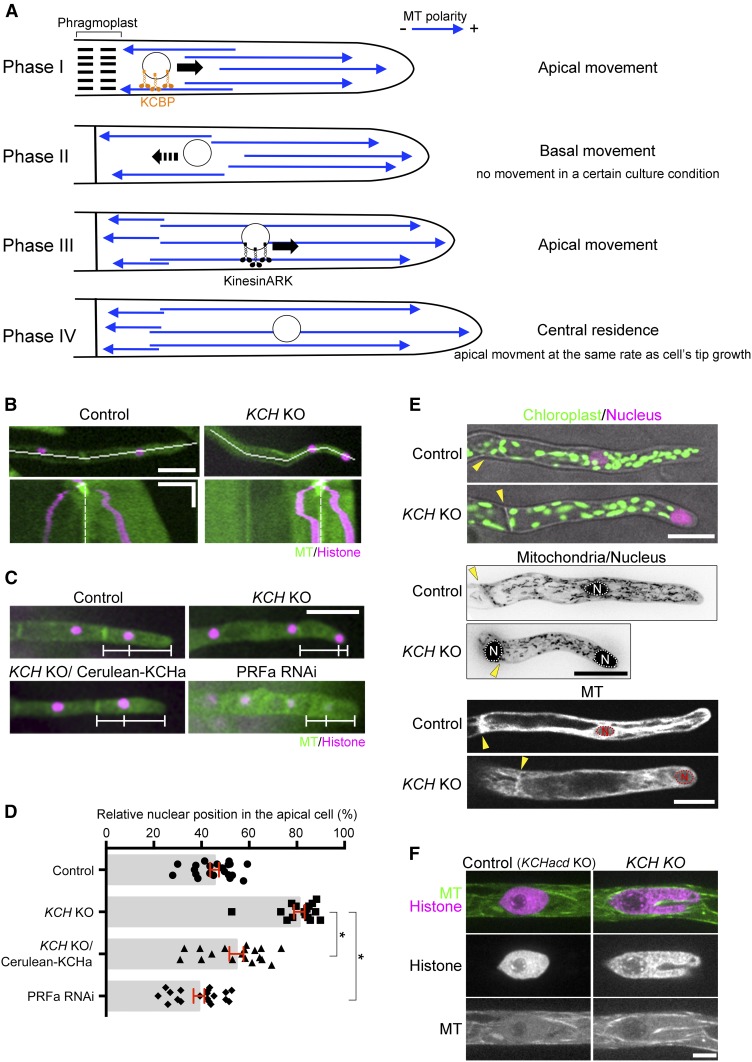
To test if endogenous P. patens KCH exhibits processive motility in moss cells, we acquired time-lapse images of Citrine-KCHa, which was expressed by the native promoter at the endogenous locus, using oblique illumination fluorescence microscopy. This microscopy allows for visualization of the cortex-proximal region, which is largely devoid of autofluorescence derived from chloroplasts. We observed punctate Citrine signals on MTs, and interestingly, the signals moved toward the minus ends at a velocity of 441 ± 226 nm/s (±sd, n = 90) for 1.6 ± 1.5 µm (±sd, n = 74; Figure 3; Supplemental Movie 2); this velocity was 8-fold faster than previously reported for NtKCH (Klotz and Nick, 2012). Since the microscopy technique employed is not sensitive enough to detect individual Citrine molecules (Jonsson et al., 2015), the diffraction-limited spots represent clustered Citrine-KCHa. This observation is consistent with the notion that KCH functions as a minus end-directed transporter of the nucleus.
Figure 3.
Processive, Minus End-Directed Movement of KCH Clusters along MTs in Protonemal Cells.
(A) Localization of Citrine-KCHa on endoplasmic MTs. Scale bar, 5 µm.
(A’) Kymograph showing Citrine-KCHa signals moving toward the MT minus-end. Arrow indicates MT plus-end growth. Horizontal bar = 2 µm; vertical bar = 5 s.
(B) Processive movement of Citrine-KCHa signals (arrowheads) along an endoplasmic MT. Bar = 2 µm.
(C) and (D) Velocity and run length of moving Citrine-KCHa signals. Note that mean run length might be somewhat underestimated as some signals are photobleached during image acquisition.
KCH Promotes Polarized Tip Growth
In addition to defects in nuclear transport, we observed severe tip growth retardation in the complete KCH KO line (Figure 4A; Supplemental Movie 3). Moreover, abnormally branched tip cells were occasionally observed in the KO line, reflecting improperly polarized tip growth (Figure 4B). These defects were suppressed by Cerulean-KCHa expression, confirming that tip growth defects in the KO line were due to the loss of KCH proteins.
Figure 4.
Protonemal Tip Growth Is Suppressed in KCH KO Lines.
(A) Tip growth was retarded in the absence of KCH. Yellow arrowheads indicate apical cell tips.
(B) Branched cell observed in the KCH KO line. White and yellow arrowheads indicate the cell wall and abnormally branched tip, respectively. Bars = 50 µm.
The slow tip growth phenotype raised a possibility that nuclear mislocalization may be a secondary effect of growth retardation in the KCH KO line. However, this was unlikely as the nucleus was mispositioned in the subapical cell, which hardly grows even in wild-type cells (Figures 2B and 2C). Nevertheless, to exclude this possibility, we examined nuclear positioning under another slow-growth condition generated by PRFa (profilin; an actin regulator) RNAi (Vidali et al., 2007; Nakaoka et al., 2012). We confirmed that the nucleus was more centrally localized when tip growth was suppressed following RNAi of PRFa (Figures 2C and 2D), suggesting that nuclear mistranslocation was not a secondary effect of the growth defect associated with KCH KO. Note that induction of PRFa RNAi led to codepletion of histoneH2B-mRFP, resulting in reduced histone signals (Nakaoka et al., 2012).
MT Focus Formation at the Tip Requires Actin and KCH
There are several tip-growing cells in plants, such as moss protonemata, root hairs, and pollen tubes. Actin filaments are essential for tip growth in these cells (Rounds and Bezanilla, 2013), whereas the direction of growth is defined by MTs in some cases. In moss protonemata, MT disruption by inhibitors or depletion of key MT regulators leads to skewed or branched tip growth (Doonan et al., 1988; Hiwatashi et al., 2014). Since KCH binds to both MT and actin in vitro (Frey et al., 2009; Xu et al., 2009; Umezu et al., 2011; Walter et al., 2015; Tseng et al., 2018), we hypothesized that P. patens KCH might regulate cytoskeletal organization at the cell tip.
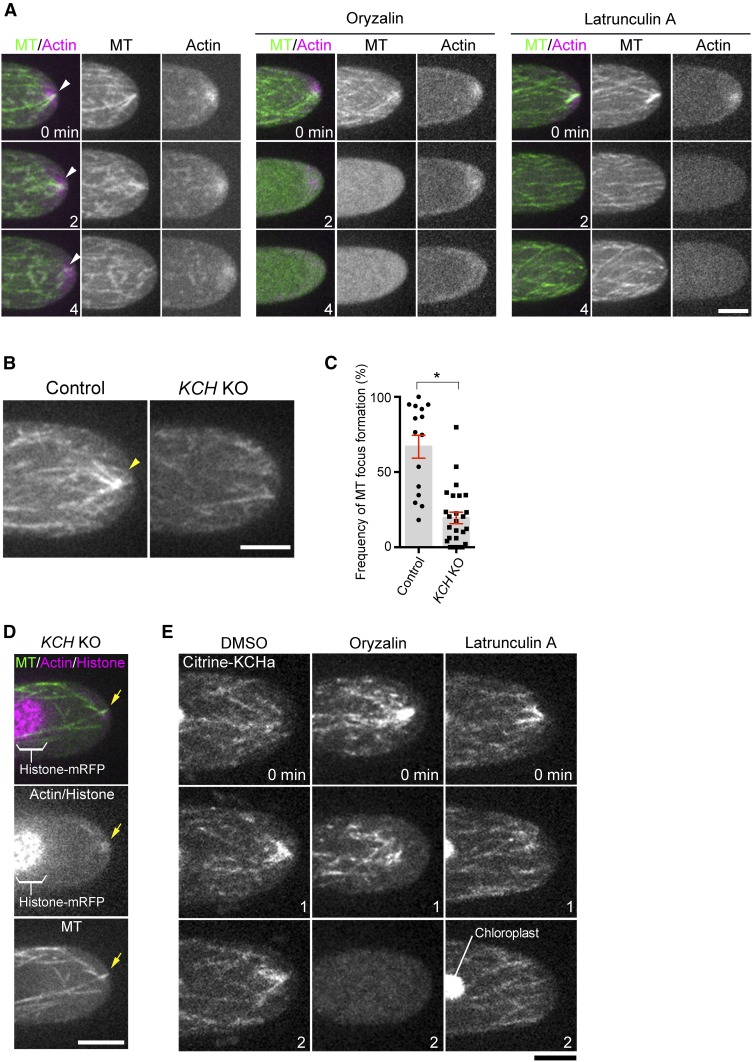
We first performed live imaging of GFP-tubulin and lifeact-mCherry (actin marker) in control apical cells. As reported previously, MT and actin focal points were detected at the tip (Vidali et al., 2009; Hiwatashi et al., 2014); we also observed that they were largely—though not completely—colocalized with each other (Figure 5A; Supplemental Movie 4). These focal points were not maintained when either MT or actin was disrupted by specific drugs (Figure 5A; Supplemental Movie 4); thus, MT and actin focal points at the tip are mutually dependent. Interestingly, in the KCH KO line, the MT focus was smaller and much less persistent (Figures 5B and 5C; Supplemental Movie 5). Furthermore, actin still accumulated at the transiently formed MT focal point, indicating the presence of other factor(s) that crosslink MTs and actin at the tip (Figure 5D).
Figure 5.
MT Focusing at the Apical Cell Tip Requires Actin Filaments and KCH.
(A) Formation of MT and actin foci (arrowheads) at the tip is mutually dependent. The tip of a caulonemal cell expressing GFP-tubulin (MT) and lifeact-mCherry (actin) was observed after oryzalin or latrunculin A treatment. Actin focal points were not maintained at the apical cell tip after oryzalin addition, whereas MT focal points were rarely observed after latrunculin A treatment.
(B) Persistent formation of the MT focus (arrowheads) at the apical cell tip is dependent on KCH.
(C) Frequency of MT focus formation. Images were acquired and analyzed every 3 s for 5 min. Bars and error bars represent the mean and se, respectively. Control, n = 15; KCH KO, n = 26. *P < 0.0001 (unpaired t test with equal sd, two-tailed). Observations were performed independently three times, and the data analyzed twice. The combined data are presented.
(D) Actin (lifeact-mCherry) and the transiently-formed MT focus (GFP-tubulin) in the absence of KCH (arrows).
(E) Actin-independent localization of Citrine-KCHa on MTs. Drugs were added at time 0, and images acquired at 0, 1, and 2 min are displayed. Note that signals derived from chloroplast autofluorescence are visible in some panels. Bars = 5 µm.
KCHa localization at the tip disappeared following MT destabilization by oryzalin treatment, whereas actin destabilization by latrunculin A did not affect KCHa MT association (Figure 5E); thus, KCH and MT colocalize independently of actin, but focal points cannot be formed without actin.
The C-Terminal Region of KCH Is Required for Tip Growth but Not for Nuclear Transport, Whereas the CH Domain Is Dispensable for Either Activity
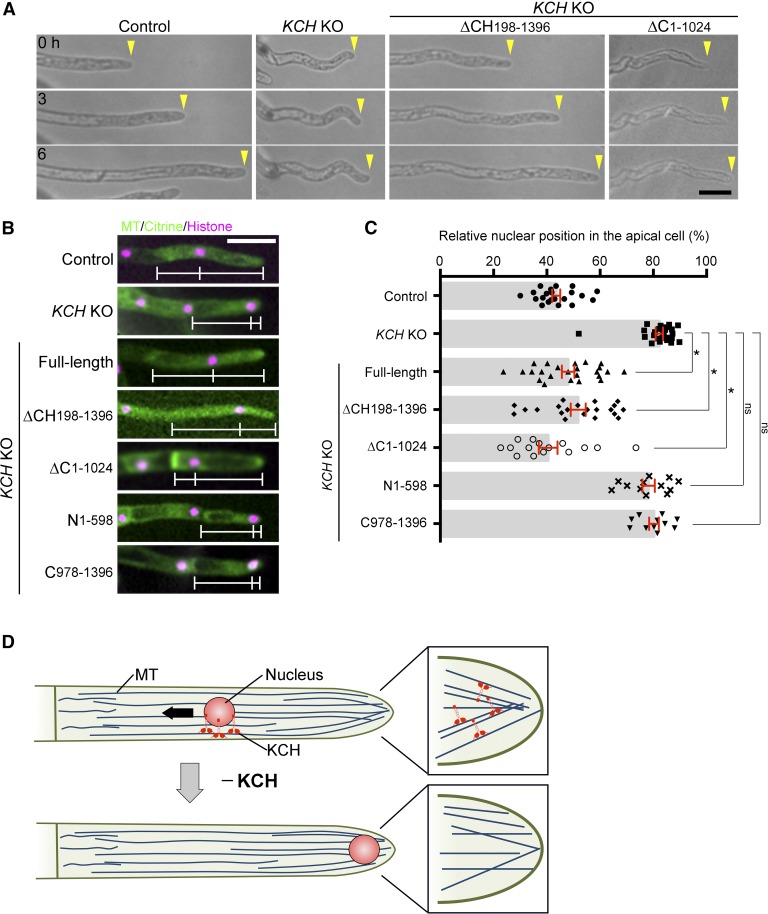
To functionally dissect the KCH protein, we constructed several truncation constructs tagged with Cerulean, transformed them into the KCH KO line, selected for transgenic lines, and assessed if the phenotypes were rescued (Figure 6A; Supplemental Figure 1E). Surprisingly, the truncated KCHa lacking the canonical CH domain, which has been assumed to be the actin binding site, restored protonemal colony growth and rhizoid development (Figures 6B and 6C). The gametophore leaves also showed considerable recovery of morphology (Figure 6B). By contrast, we did not obtain a transgenic line that showed normal protonemal growth or gametophore/rhizoid development after transformation of the KCH fragment with a deleted C-terminal extension (Figures 6B and 6C).
Figure 6.
Mapping of KCH Functional Domains for Moss Development.
(A) Schematic representation of four truncation constructs and their ability to rescue defects in nuclear transport or protonemal growth. Citrine was attached to the N terminus of each fragment.
(B) Protonemal growth, gametophore leaf morphology, and rhizoid development after expression of truncated constructs in the KCH KO line. Bars = 2 mm (top two rows) or 0.5 mm (bottom two rows).
(C) Colony size comparison between control (parental line expressing GFP-tubulin/histoneH2B-mRFP), KCH KO, and KCH KO/Citrine-KCHa truncations. Bars and error bars represent the mean and se, respectively. Control, n = 22; KCH KO, n = 21; KCH KO/Citrine-FL (1–1396), n = 22; KCH KO/Citrine-∆CH (198–1396), n = 22; KCH KO/Citrine-∆C (1–1024), n = 22; KCH KO/Citrine-N (1–598), n = 22; KCH KO/Citrine-C (978–1396), n = 22. **P < 0.0001; ns (not significant), P > 0.06 (unpaired t test with equal sd, two-tailed). †Although colony size was slightly but significantly larger than that of the KO line in this experiment, it was slightly smaller and not significantly larger than the KO line in another experiment, indicating that these two short fragments did not rescue colony growth. Experiments were performed three times and the data analyzed twice. The data of one experiment are displayed.
At the cellular and intracellular levels in the protonemata, tip growth (Figure 7A; Supplemental Movie 3), persistent MT focus formation (Supplemental Movie 6), and nuclear positioning (Figures 7B and 7C) were restored by ∆CH expression. By contrast, we observed recovery of nuclear positioning, but not tip growth or MT focus formation, in the ∆C lines, indicating that the C-terminal extension is dispensable for nuclear transport function but is essential for tip growth.
Figure 7.
C-Terminal Extension of KCH Is Critical for Tip Growth, but Not for Nuclear Transport.
(A) Tip growth was assessed after expression of the truncated KCH in the KO line. Arrowheads indicate the cell tips. Bar = 50 µm.
(B) Nuclear positioning after truncated protein expression in the KCH KO line. White bars indicate the positions of the cell wall, nucleus, and apical tip. Bar = 50 µm. See Supplemental Movie 6 for high-resolution MT images in some transgenic lines.
(C) Relative position of the nucleus within the apical cell was quantified for each line. The “0” corresponds to the cell wall, whereas “100” indicates the cell tip. Bars and error bars represent the mean and se, respectively. Control, n = 22; KCH KO, n = 26; KCH KO/Citrine-FL (1–1396), n = 24; KCH KO/Citrine-∆CH (198–1396), n = 21; KCH KO/Citrine-∆C (1–1024), n = 15; KCH KO/Citrine-N (1–598), n = 12; KCH KO/Citrine-C (978–1396), n = 11. *P < 0.0001; ns (not significant), P > 0.1 (unpaired t test with equal sd, two-tailed). Observations were performed independently two or more times, and the data were analyzed twice. The data of one experiment are displayed.
(D) Role of KCH kinesin in tip cell growth and nuclear transport in P. patens. KCH is a minus end-directed kinesin required for retrograde transport of the nucleus (arrow). KCH is also critical for tip growth as it stably focuses MTs at the apical tip. Actin is enriched around the MT focus (not displayed in this diagram).
DISCUSSION
We generated a plant completely lacking KCH proteins, which exhibited several noticeable phenotypes. We focused our study on two prominent phenotypes associated with protonemal apical cells, nuclear mispositioning, and tip growth retardation. This study also reveals the intracellular dynamics of function-verified KCH protein expressed from the native locus. Our data elucidated the function of KCH as a long-distance retrograde transporter and its role in cytoskeletal coalescence, during which an unanticipated molecular mechanism might be involved (Figure 7D).
KCH Is a Potent Retrograde Transporter
The dimeric, truncated form of rice KCH1 or moss KCHa was shown to be nonprocessive in in vitro motility assays (Jonsson et al., 2015; Walter et al., 2015), whereas a recent report showed processive motility of rice KCH2 as a dimer; OsKCH2 possesses unique sequences adjacent to the C terminus of the motor domain that ensure processivity (Tseng et al., 2018). However, a cohort action of rice KCH1 can transport actin filaments along MTs over long distances, suggesting that KCH family proteins can generally function as long-distance cargo transporters (Walter et al., 2015). Our observation of endogenous KCHa dynamics in the cytoplasm indicated that this kinesin forms a cluster and indeed moves processively toward the minus ends of MTs. Its run velocity (∼440 nm/s) was ∼3-fold faster than that of ATK (kinesin-14-I) and comparable to KCBP (kinesin-14-VI, 413 nm/s)—two other kinesin-14 family proteins for which processive motility in clusters and cargo transport function have been identified (Jonsson et al., 2015; Yamada et al., 2017). These results suggest that KCH proteins are potent minus end-directed transporters.
Paradoxically, a minus end-directed motor is enriched at the MT plus end at the cell tip (Figures 1C and 5E). This might be achieved by interacting with other MT plus end-tracking proteins or actins. However, the results obtained using oblique illumination fluorescence microscopy indicate that many KCH molecules associate only transiently with the MT and then dissociate before moving along the MT (Supplemental Movie 2). Therefore, it is possible that both nonmotile and motile proteins can be visualized using confocal microscopy at the MT-rich cell tip. Consistent with this idea, the mCherry-tubulin signals increased at the tip similar to Citrine-KCH (Figure 1C), and the latter localized uniformly to the MT when the MT focus was disrupted by latrunculin A treatment (Figure 5E, right). Whether minus end-directed motility of KCH is required for cytoskeletal organization at the tip remains to be determined.
KCH Drives Nuclear Transport
Our study identified a defect in nuclear positioning in the absence of KCH. Nuclear translocation during the cell cycle of protonemal apical cells can be divided into four phases (Figure 2A), each driven by MT-based transport. KCBP is a minus end-directed kinesin required for nuclear transport specifically during phase I, the postmitotic phase (Yamada et al., 2017). By contrast, kinesin-ARK is responsible for plus end-directed transport during phase III and possibly also phase IV, which corresponds with the majority of interphase (Miki et al., 2015). KCH is the third kinesin identified required for nuclear positioning; its KO phenotype indicated that KCH also functions during IV. The following three pieces of data strongly suggest that KCH actually transports the nucleus, rather than indirectly affecting nuclear positioning by, for example, altering overall MT polarity: (1) KCH showed minus end-directed, processive motility in cells (Figure 3); (2) the nucleus moved all the way to the cell tip in the KCH KO line (Figure 2B), where the plus ends of MTs are detected regardless of KCH presence (Figure 5B); and (3) in the absence of KCH, abnormal localization was detected for the nucleus, but not for the other three cytoplasmic organelles (Figure 2E). Thus, we envisage that kinesin-ARK and KCH execute bidirectional transport of the nucleus, reminiscent of animal cells where antagonistic cytoplasmic dynein and kinesin-1 (or kinesin-3) motors are involved (Tanenbaum et al., 2010; Tsai et al., 2010). Consistent with this model, the nucleus unidirectionally moved toward the cell plate or cell tip in the absence of kinesin-ARK or KCH, respectively. We could not detect enrichment of KCH or kinesin-ARK (Miki et al., 2015) on the nuclear surface during microscopy; however, failure in detection does not necessarily preclude the possibility that KCH acts as a transporter, as the fluorescence signals might not be distinguishable from cargo-free motors in the cytoplasm or cytoplasmic background signals. This is particularly a likely scenario in moss protonemata, since autofluorescence derived from chloroplasts is dominant in the cytoplasm. Using oblique illumination microscopy, we observed Citrine-KCHa signals for each cortex-proximal MT, suggesting that KCH also associates with nucleus-proximal MTs (note that the nucleus cannot be located in the focal plane using this microscopy technique). Our truncation/rescue experiments suggest that the region downstream of the CH domain and upstream of the motor domain is required for nuclear attachment. It would be interesting to search for the nuclear envelope-associated adaptor of KCH in future studies.
KCH and Actin Ensure MT Focus Formation for Tip Growth
Apical tip growth was severely suppressed in the KCH KO line. Based on previous studies, this phenotype could be attributed to defects in the actin cytoskeleton, MT cytoskeleton, and/or lipid biogenesis at the tip (van Gisbergen et al., 2012; Vidali and Bezanilla, 2012; Hiwatashi et al., 2014; Saavedra et al., 2015). Defects in general housekeeping processes such as protein translation would also perturb cell growth. Although involvement of KCH in lipid production or other general cellular processes was not excluded, our localization and phenotypic data more strongly suggest that KCH regulates MT and possibly also actin cytoskeletons for tip growth. Most compellingly, the characteristic focus composed of MTs—which we observed to largely coincide with the actin focal point—did not persistently form in KCH KO cells. However, unlike for actin or MT destabilization, tip growth was not completely inhibited in KO cells. Phenotypically, this was consistent with the observation that smaller and transient MT/actin foci were still detectable in the KO line. Residual MT bundling activity may be mediated by other proteins such as plant-specific plus end-directed kinesins KINIDa and b, whose double deletion resulted in curved growth accompanied with a lack of MT focus persistence (Hiwatashi et al., 2014). In addition, the mislocalized nucleus also dampened tip growth, as this large organelle occasionally occupied the apical space where MT and actin normally form clusters (e.g., Figure 2B). Thus, another significant role of nuclear positioning in plants—other than division site determination—may be ensuring proper organization of the cytoskeletal network for cell function. Nevertheless, nuclear mislocalization is not the sole factor contributing to tip growth suppression, since MT foci were not stably maintained in KO cells whose nuclei resided fairly distant from the tip (e.g., Figure 5B). Furthermore, the C-terminal deletion construct restored nuclear positioning but not growth defects. Thus, cytoskeletal disorganization independent of nuclear positioning is likely the major factor responsible for growth retardation in KCH KO cells.
The mechanism via which KCH promotes the formation of a large and persistent ensemble of the two cytoskeletal filaments remains unclear. MT-actin coalescence in the KCH KO line indicates the presence of other protein(s) that bridge the two filaments. KCH might support the coalescence via MT cross-linking. Alternatively, the model that KCH, independent of the CH domain, coalesces MTs with actin by directly binding to both filaments remains viable; intriguingly, an in vitro study indicates that actin interacts also with the motor domain of rice KCH-O12 (Umezu et al., 2011). Furthermore, the C-terminal extension of KCH was critical for MT coalescence. Long (>150 amino acids) C-terminal extension from the motor domain is unique to the plant kinesin14-II-V families, andno sequences have been identified in this region from which protein function can be deduced. Our study showed that this unusual extension is a critical element of KCH, yet its exact function remains unclear; it may be required for KCH accumulation at the tip or it may constitute an additional MT or actin interaction interface.
Conservation of KCH Function
In flowering plants, KCH family proteins show more divergence in amino acid sequences than in moss (Figure 1A), and each member appears to play a distinct role. Interestingly, several reported phenotypes associated with KCH depletion and overexpression in flowering plants appear to be consistent with our findings for moss. For example, coleoptile cells were shorter in the rice kch1 mutant, whereas tobacco BY-2 cells were elongated upon heterologous expression of rice KCH (Frey et al., 2010). Tobacco GFP-KCH, when ectopically expressed, decorates cortical MT arrays but is also abundantly present around the nucleus (Klotz and Nick, 2012). Furthermore, the heterologous expression of rice KCH in the tobacco BY-2 cell line induced nuclear positioning defects (Frey et al., 2010). Although these studies did not elucidate the basis of the phenotypes, our study suggests that KCH may transport the nucleus and promote MT-actin interaction in flowering plants as well. However, KCH functions might not be limited to nuclear transport and MT-actin interactions during moss development, as many KCH proteins not associated with the nucleus were observed running along MTs (Figure 3). The complete KO line generated in this study is a potentially valuable resource for uncovering the list of KCH functions throughout plant development.
METHODS
Moss Culture and Transformation
Moss lines used in this study are listed in Supplemental Table 1; all lines originated from the Physcomitrella patens Gransden2004 strain. GFP-tubulin/histoneH2B-mRFP and mCherry-tubulin strains were used as mother strains for gene disruption and Citrine tagging, respectively (Nakaoka et al., 2012; Kosetsu et al., 2013). Methodologies of moss culture, transformation, and transgenic line selection were previously described (Yamada et al., 2016). Briefly, cells were cultured on BCD agar medium for imaging. Transformation was performed by the standard PEG-mediated method and stable lines were selected with antibiotics (blasticidin S, nourseothricin, and hygromycin). Gene knockouts were obtained by replacing endogenous KCH genes with a drug-resistant marker flanked by lox-P sequences. After deleting KCHa and b, the two markers were removed by transient expression of Cre recombinase, followed by replacement of KCHd and b with the same markers. The Citrine gene was inserted into the N terminus of KCHa via homologous recombination. Drug-resistant genes were not integrated into the genome, as nonlinearized, unstable plasmids containing a drug resistant marker were cotransformed with the linearized plasmid containing Citrine tag constructs for transient drug selection. Gene disruption and Citrine tag insertion were confirmed by PCR.
Plasmid Construction
Plasmids (and primers for plasmid construction) used for gene disruption, protein expression, and Citrine tagging are listed in Supplemental Table 2. Gene knockout constructs were designed to replace endogenous KCH genes with a drug-resistant marker flanked by lox-P sequences. One kilobase of genomic DNA sequences upstream/downstream of start/stop codons was amplified and cloned into the vectors containing lox-P sequences and drug-resistant markers. To generate a plasmid for Citrine tagging, 1 kb of genomic DNA sequences upstream/downstream of the KCH gene start codon was cloned into the pKK138 vector (Kosetsu et al., 2013; Yamada et al., 2016). To generate truncation/rescue plasmids, KCHa sequences were amplified from a cDNA library and ligated into the pENTR/D-TOPO vector containing Cerulean or Citrine sequences, followed by a Gateway LR reaction into a pTM153 vector that contains the EF1α promoter, blasticidin-resistance cassette, and PTA1 sequences designated for homologous recombination-based integration (Miki et al., 2016). Lifeact-mCherry was expressed by the actin promoter.
Immunoblotting
Cell extracts were prepared by grinding protonema colonies (Yamada et al., 2016). Immunoblotting of Citrine-tagged proteins was performed with home-made anti-GFP antibody (rabbit “Nishi,” final bleed, 1: 500).
In Vivo Microscopy
Methods for epifluorescence and spinning-disc confocal microscopy were previously described (Yamada et al., 2016). Briefly, protonemal cells were plated onto glass-bottom plates coated with BCD agar medium and cultured for 4 to 5 d. The KO line was precultured on BCD medium covered with cellophane for 2 to 3 d before being placed onto glass-bottom plates. Long-term imaging by a wide-field microscope (low magnification lens) was performed with a Nikon Ti (10× 0.45 NA lens and EMCCD camera Evolve [Roper]). High-resolution imaging was performed with a spinning-disc confocal microscope (Nikon TE2000 or Ti; 100× 1.45 NA lens, CSU-X1 [Yokogawa], and EMCCD camera ImagEM [Hamamatsu]). Oblique illumination microscopy was performed as previously described (Jonsson et al., 2015; Nakaoka et al., 2015); cells were cultured in BCDAT medium covered with cellophane and imaging was performed with a Nikon Ti microscope with a TIRF unit, a 100× 1.49 NA lens, GEMINI split view (Hamamatsu), and EMCCD camera Evolve (Roper). All imaging was performed at 24 to 25°C in the dark, except for the protonema growth assay that utilizes light.
Drug Assay
Mosses were plated on agar gel in 35-mm dishes following standard procedure and incubated for 4 to 5 d (Yamada et al., 2016). Prior to drug treatment, gels were excised from the dish except the central ∼1 cm2 area on which moss colonies grew. Then, 1.5 mL of water was added to the dish and incubated for 30 min (i.e., equilibrated), followed by addition of 1.5 mL drug solution. Cells were treated with 10 µM oryzalin (AccuStandard), 25 µM latrunculin A (Wako), or 0.5 to 1% DMSO as control.
Colony Growth Assay
Protoplasts prepared by the standard driselase treatment were washed three times with 8% mannitol solution, followed by overnight incubation in protoplast liquid medium in the dark (Yamada et al., 2016). Protoplast solution was mixed with PRM-T and plated onto a cellophane-covered PRM plate (the cellophane was a gift from Futamura Chemical) and incubated for 4 d. The cellophane was transferred to a fresh BCDAT plate and incubated for 5 d, and then colonies were picked and used to inoculate a new BCDAT plate. After incubation for a further 10 to 11 d, colonies were imaged with a commercially available digital camera (Olympus C-765) or stereoscopic microscope (Nikon SMZ800N and Sony ILCE-QX1α).
Phylogenetic Tree Construction
Following a previously described method (Miki et al., 2014, 2015), kinesin sequences were aligned with MAFFT and the gaps were removed from the alignment using MacClade. The phylogenetic tree was constructed using the neighbor-joining method using Molecular Evolutionary Genetics Analysis (MEGA) software. Statistical support for internal branches by bootstrap analyses was calculated using 1000 replications.
Data Analysis
To quantify the relative position of the nucleus in the apical cell, microscope images that showed the cell wall, nucleus, and cell tip were analyzed manually with ImageJ. Nonmitotic cells were randomly selected. The velocity and run length of Citrine-KCHa motility along endoplasmic MTs were quantified based on kymographs, which were generated from the images acquired with oblique illumination fluorescence microscopy. To measure the area of moss colonies, cultured moss colony images were acquired by a digital camera (Olympus C-765). All acquired images were aligned side-by-side and the generated single image was analyzed with ImageJ; specifically, the image was processed by “Make binary” and the colonies in the processed image were automatically detected and measured by “Analyze particles” (size, 1.0-Infinity [mm2]; circularity, 0.00–1.00). To quantify the duration of MT focus formation at the tip, GFP-tubulin was imaged every 3 s, and the presence or absence of MT foci was manually judged at each time frame; the focus was recognized when two or more MT ends converged.
Accession Numbers
Sequence data used in this article can be found in the Phytozome database under the following accession numbers: KCHa, Pp3c14_19550; KCHb, Pp3c2_9150; KCHc, Pp3c17_21780; and KCHd, Pp3c24_19380.
Supplemental Data
Supplemental Figure 1. KCH gene tagging and disruption via homologous recombination.
Supplemental Figure 2. Organelle distribution in the KCH KO line.
Supplemental Table 1. Moss lines used in this study.
Supplemental Table 2. List of PCR primers and plasmids used in this study.
Supplemental Data Set 1. Alignments used to generate the phylogeny presented in Figure 1A.
Supplemental Movie 1. Nuclear migration defects in the KCH KO line.
Supplemental Movie 2. Processive motility of clustered Citrine-KCH-a.
Supplemental Movie 3. Tip growth retardation in the KCH KO line.
Supplemental Movie 4. MT and actin focal points at the cell tip.
Supplemental Movie 5. KCH-dependent formation of the MT focus at the apical cell tip.
Supplemental Movie 6. MT focus at the apical cell tip after expression of truncated KCH.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Mitsuyasu Hasebe for providing the plasmids, Momoko Nishina, Tomohiro Miki, and Rie Inaba for technical assistance, as well as Tomomi Kiyomitsu, Elena Kozgunova, Peishan Yi, and Tomohiro Miki for helpful comments regarding the manuscript. This work was funded by the TORAY Science Foundation (14-5503) and JSPS KAKENHI 15K14540 and 17H06471 (to G.G.). M.Y. is a recipient of a JSPS predoctoral fellowship (16J02796).
AUTHOR CONTRIBUTIONS
M.Y. and G.G. conceived and designed the research project. M.Y. performed the experiments and analyzed the data. M.Y. and G.G. wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Ambrose J.C., Li W., Marcus A., Ma H., Cyr R. (2005). A minus-end-directed kinesin with plus-end tracking protein activity is involved in spindle morphogenesis. Mol. Biol. Cell 16: 1584–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles C.H., Bradke F. (2015). Coordinating neuronal actin-microtubule dynamics. Curr. Biol. 25: R677–R691. [DOI] [PubMed] [Google Scholar]
- Cove D. (2005). The moss Physcomitrella patens. Annu. Rev. Genet. 39: 339–358. [DOI] [PubMed] [Google Scholar]
- Cove D., Bezanilla M., Harries P., Quatrano R. (2006). Mosses as model systems for the study of metabolism and development. Annu. Rev. Plant Biol. 57: 497–520. [DOI] [PubMed] [Google Scholar]
- Doonan J.H., Cove D.J., Lloyd C.W. (1988). Microtubules and microfilaments in tip growth: evidence that microtubules impose polarity on protonemal growth in Physcomitrella patens. J. Cell Sci. 89: 533–540. [Google Scholar]
- Ferenz N.P., Paul R., Fagerstrom C., Mogilner A., Wadsworth P. (2009). Dynein antagonizes eg5 by crosslinking and sliding antiparallel microtubules. Curr. Biol. 19: 1833–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey N., Klotz J., Nick P. (2009). Dynamic bridges--a calponin-domain kinesin from rice links actin filaments and microtubules in both cycling and non-cycling cells. Plant Cell Physiol. 50: 1493–1506. [DOI] [PubMed] [Google Scholar]
- Frey N., Klotz J., Nick P. (2010). A kinesin with calponin-homology domain is involved in premitotic nuclear migration. J. Exp. Bot. 61: 3423–3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P. (2008). Mechanisms of asymmetric cell division: flies and worms pave the way. Nat. Rev. Mol. Cell Biol. 9: 355–366. [DOI] [PubMed] [Google Scholar]
- Goto Y., Asada T. (2007). Excessive expression of the plant kinesin TBK5 converts cortical and perinuclear microtubules into a radial array emanating from a single focus. Plant Cell Physiol. 48: 753–761. [DOI] [PubMed] [Google Scholar]
- Grabham P.W., Seale G.E., Bennecib M., Goldberg D.J., Vallee R.B. (2007). Cytoplasmic dynein and LIS1 are required for microtubule advance during growth cone remodeling and fast axonal outgrowth. J. Neurosci. 27: 5823–5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill S.W., Hyman A.A. (2005). Spindle positioning by cortical pulling forces. Dev. Cell 8: 461–465. [DOI] [PubMed] [Google Scholar]
- Hiwatashi Y., Sato Y., Doonan J.H. (2014). Kinesins have a dual function in organizing microtubules during both tip growth and cytokinesis in Physcomitrella patens. Plant Cell 26: 1256–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson E., Yamada M., Vale R.D., Goshima G. (2015). Clustering of a kinesin-14 motor enables processive retrograde microtubule-based transport in plants. Nat. Plants pii: 15087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz J., Nick P. (2012). A novel actin-microtubule cross-linking kinesin, NtKCH, functions in cell expansion and division. New Phytol. 193: 576–589. [DOI] [PubMed] [Google Scholar]
- Kong Z., Ioki M., Braybrook S., Li S., Ye Z.H., Julie Lee Y.R., Hotta T., Chang A., Tian J., Wang G., Liu B. (2015). Kinesin-4 functions in vesicular transport on cortical microtubules and regulates cell wall mechanics during cell elongation in plants. Mol. Plant 8: 1011–1023. [DOI] [PubMed] [Google Scholar]
- Kosetsu K., de Keijzer J., Janson M.E., Goshima G. (2013). MICROTUBULE-ASSOCIATED PROTEIN65 is essential for maintenance of phragmoplast bipolarity and formation of the cell plate in Physcomitrella patens. Plant Cell 25: 4479–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally F.J. (2013). Mechanisms of spindle positioning. J. Cell Biol. 200: 131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T., Naito H., Nishina M., Goshima G. (2014). Endogenous localizome identifies 43 mitotic kinesins in a plant cell. Proc. Natl. Acad. Sci. USA 111: E1053–E1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T., Nishina M., Goshima G. (2015). RNAi screening identifies the armadillo repeat-containing kinesins responsible for microtubule-dependent nuclear positioning in Physcomitrella patens. Plant Cell Physiol. 56: 737–749. [DOI] [PubMed] [Google Scholar]
- Miki T., Nakaoka Y., Goshima G. (2016). Live cell microscopy-based RNAi screening in the moss Physcomitrella patens. Methods Mol. Biol. 1470: 225–246. [DOI] [PubMed] [Google Scholar]
- Nakaoka Y., Miki T., Fujioka R., Uehara R., Tomioka A., Obuse C., Kubo M., Hiwatashi Y., Goshima G. (2012). An inducible RNA interference system in Physcomitrella patens reveals a dominant role of augmin in phragmoplast microtubule generation. Plant Cell 24: 1478–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaoka Y., Kimura A., Tani T., Goshima G. (2015). Cytoplasmic nucleation and atypical branching nucleation generate endoplasmic microtubules in Physcomitrella patens. Plant Cell 27: 228–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Ramírez C., Hernandez-Coronado M., Thamm A., Catarino B., Wang M., Dolan L., Feijó J.A., Becker J.D. (2016). A transcriptome atlas of Physcomitrella patens provides insights into the evolution and development of land plants. Mol. Plant 9: 205–220. [DOI] [PubMed] [Google Scholar]
- Perlson E., Hendricks A.G., Lazarus J.E., Ben-Yaakov K., Gradus T., Tokito M., Holzbaur E.L. (2013). Dynein interacts with the neural cell adhesion molecule (NCAM180) to tether dynamic microtubules and maintain synaptic density in cortical neurons. J. Biol. Chem. 288: 27812–27824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss M.L., Kovar D.R., Lee Y.R., Staiger C.J., Delmer D.P., Liu B. (2004). A plant-specific kinesin binds to actin microfilaments and interacts with cortical microtubules in cotton fibers. Plant Physiol. 136: 3945–3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounds C.M., Bezanilla M. (2013). Growth mechanisms in tip-growing plant cells. Annu. Rev. Plant Biol. 64: 243–265. [DOI] [PubMed] [Google Scholar]
- Saavedra L., Catarino R., Heinz T., Heilmann I., Bezanilla M., Malhó R. (2015). Phosphatase and tensin homolog is a growth repressor of both rhizoid and gametophore development in the moss Physcomitrella patens. Plant Physiol. 169: 2572–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Collatos A.R., Bibeau J.P., Furt F., Vidali L. (2012). Phylogenetic analysis of the Kinesin superfamily from Physcomitrella. Front. Plant Sci. 3: 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelman Z., Lee C.M., Gallagher K.L. (2018). KinG is a plant-specific kinesin that regulates both intra- and intercellular movement of SHORT-ROOT. Plant Physiol. 176: 392–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu N., Yamada N., Kagawa T., Yonekura H., Uyeda T.Q., Kadota A., Wada M. (2010). Two kinesin-like proteins mediate actin-based chloroplast movement in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107: 8860–8865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu N., Sato Y., Tsuboi H., Kasahara M., Imaizumi T., Kagawa T., Hiwatashi Y., Hasebe M., Wada M. (2012). The KAC family of kinesin-like proteins is essential for the association of chloroplasts with the plasma membrane in land plants. Plant Cell Physiol. 53: 1854–1865. [DOI] [PubMed] [Google Scholar]
- Tamura K., Iwabuchi K., Fukao Y., Kondo M., Okamoto K., Ueda H., Nishimura M., Hara-Nishimura I. (2013). Myosin XI-i links the nuclear membrane to the cytoskeleton to control nuclear movement and shape in Arabidopsis. Curr. Biol. 23: 1776–1781. [DOI] [PubMed] [Google Scholar]
- Tanenbaum M.E., Akhmanova A., Medema R.H. (2010). Dynein at the nuclear envelope. EMBO Rep. 11: 649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum M.E., Vale R.D., McKenney R.J. (2013). Cytoplasmic dynein crosslinks and slides anti-parallel microtubules using its two motor domains. eLife 2: e00943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J., Han L., Feng Z., Wang G., Liu W., Ma Y., Yu Y., Kong Z. (2015). Orchestration of microtubules and the actin cytoskeleton in trichome cell shape determination by a plant-unique kinesin. eLife 4: 10.7554/eLife.09351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J.W., Lian W.N., Kemal S., Kriegstein A.R., Vallee R.B. (2010). Kinesin 3 and cytoplasmic dynein mediate interkinetic nuclear migration in neural stem cells. Nat. Neurosci. 13: 1463–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng K.F., Wang P., Lee Y.J., Bowen J., Gicking A.M., Guo L., Liu B., Qiu W. (2018). The preprophase band-associated kinesin-14 OsKCH2 is a processive minus-end-directed microtubule motor. Nat. Commun. 9: 1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida M., Ohtani S., Ichinose M., Sugita C., Sugita M. (2011). The PPR-DYW proteins are required for RNA editing of rps14, cox1 and nad5 transcripts in Physcomitrella patens mitochondria. FEBS Lett. 585: 2367–2371. [DOI] [PubMed] [Google Scholar]
- Umezu N., Umeki N., Mitsui T., Kondo K., Maruta S. (2011). Characterization of a novel rice kinesin O12 with a calponin homology domain. J. Biochem. 149: 91–101. [DOI] [PubMed] [Google Scholar]
- van Gisbergen P.A., Li M., Wu S.Z., Bezanilla M. (2012). Class II formin targeting to the cell cortex by binding PI(3,5)P(2) is essential for polarized growth. J. Cell Biol. 198: 235–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L., Bezanilla M. (2012). Physcomitrella patens: a model for tip cell growth and differentiation. Curr. Opin. Plant Biol. 15: 625–631. [DOI] [PubMed] [Google Scholar]
- Vidali L., Augustine R.C., Kleinman K.P., Bezanilla M. (2007). Profilin is essential for tip growth in the moss Physcomitrella patens. Plant Cell 19: 3705–3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L., Rounds C.M., Hepler P.K., Bezanilla M. (2009). Lifeact-mEGFP reveals a dynamic apical F-actin network in tip growing plant cells. PLoS One 4: e5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter W.J., Machens I., Rafieian F., Diez S. (2015). The non-processive rice kinesin-14 OsKCH1 transports actin filaments along microtubules with two distinct velocities. Nat. Plants 1: 15111. [DOI] [PubMed] [Google Scholar]
- Xu T., Qu Z., Yang X., Qin X., Xiong J., Wang Y., Ren D., Liu G. (2009). A cotton kinesin GhKCH2 interacts with both microtubules and microfilaments. Biochem. J. 421: 171–180. [DOI] [PubMed] [Google Scholar]
- Yamada M., Miki T., Goshima G. (2016). Imaging mitosis in the moss Physcomitrella patens. Methods Mol. Biol. 1413: 263–282. [DOI] [PubMed] [Google Scholar]
- Yamada M., Tanaka-Takiguchi Y., Hayashi M., Nishina M., Goshima G. (2017). Multiple kinesin-14 family members drive microtubule minus end-directed transport in plant cells. J. Cell Biol. 216: 1705–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.Y., Chen Z.W., Xu T., Qu Z., Pan X.D., Qin X.H., Ren D.T., Liu G.Q. (2011). Arabidopsis kinesin KP1 specifically interacts with VDAC3, a mitochondrial protein, and regulates respiration during seed germination at low temperature. Plant Cell 23: 1093–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Dixit R. (2012). Functions of the Arabidopsis kinesin superfamily of microtubule-based motor proteins. Protoplasma 249: 887–899. [DOI] [PubMed] [Google Scholar]
- Zhu C., Ganguly A., Baskin T.I., McClosky D.D., Anderson C.T., Foster C., Meunier K.A., Okamoto R., Berg H., Dixit R. (2015). The fragile Fiber1 kinesin contributes to cortical microtubule-mediated trafficking of cell wall components. Plant Physiol. 167: 780–792. [DOI] [PMC free article] [PubMed] [Google Scholar]