Multiple pattern-recognition receptors activate mitogen-activated protein kinase cascades and trigger disease resistance in plants via direct phosphorylation and positive feedback regulation.
Abstract
Plants deploy numerous cell surface-localized pattern-recognition receptors (PRRs) to perceive host- and microbe-derived molecular patterns that are specifically released during infection and activate defense responses. The activation of the mitogen-activated protein kinases MPK3, MPK4, and MPK6 (MPK3/4/6) is a hallmark of immune system activation by all known PRRs and is crucial for establishing disease resistance. The MAP kinase kinase kinase (MAPKKK) MEKK1 controls MPK4 activation, but the MAPKKKs responsible for MPK3/6 activation downstream of diverse PRRs and how the perception of diverse molecular patterns leads to the activation of MAPKKKs remain elusive. Here, we show that two highly related MAPKKKs, MAPKKK3 and MAPKKK5, mediate MPK3/6 activation by at least four PRRs and confer resistance to bacterial and fungal pathogens in Arabidopsis thaliana. The receptor-like cytoplasmic kinases VII (RLCK VII), which act downstream of PRRs, directly phosphorylate MAPKKK5 Ser-599, which is required for pattern-triggered MPK3/6 activation, defense gene expression, and disease resistance. Surprisingly, MPK6 further phosphorylates MAPKKK5 Ser-682 and Ser-692 to enhance MPK3/6 activation and disease resistance, pointing to a positive feedback mechanism. Finally, MEKK1 Ser-603 is phosphorylated by both RLCK VII and MPK4, which is required for pattern-triggered MPK4 activation. These findings illustrate central mechanisms by which multiple PRRs activate MAPK cascades and disease resistance.
INTRODUCTION
In plants, cell surface-localized pattern-recognition receptors (PRRs) perceive host- and microbe-derived molecular patterns, leading to pattern-triggered immunity (Dodds and Rathjen, 2010; Tang et al., 2017). This serves as the first layer of a plant surveillance system that is essential for plants to ward off diverse pathogens. Well-known PRRs include FLAGELLIN SENSITIVE2 (FLS2), EF-TU RECEPTOR (EFR), LYSIN MOTIF RECEPTOR KINASE5 (LYK5), and PLANT ELICITOR PEPTIDE RECEPTORs (PEPRs). These PRRs perceive a conserved 22-amino acid epitope (flg22) of the N terminus of bacterial flagellin and a conserved N-terminal epitope (elf18) of the bacterial elongation factor Tu, the fungal cell wall component chitin, and plant elicitor peptides, respectively (Chinchilla et al., 2006; Zipfel et al., 2006; Yamaguchi et al., 2010; Cao et al., 2014). Upon ligand binding, FLS2, EFR, and PEPRs immediately heterodimerize with their shared coreceptor BRASSINOSTEROID INSENSITIVE1-ASSOCIATED KINASE1 (BAK1) to form active receptor complexes (Chinchilla et al., 2007; Heese et al., 2007; Sun et al., 2013; Tang et al., 2015). LYK5 forms a complex with its coreceptor CHITIN ELICITOR RECEPTOR KINASE1 (CERK1) in the presence of chitin (Miya et al., 2007; Wan et al., 2008; Cao et al., 2014). Within minutes, pattern recognition triggers a number of downstream responses, including a transient influx of calcium ions from the apoplast, activation of calcium-dependent protein kinases, the production of extracellular reactive oxygen species (ROS), and the activation of mitogen-activated protein kinases (MAPKs) (Nühse et al., 2000; Zhang et al., 2007; Kadota et al., 2014; Li et al., 2014).
Downstream, PRRs and/or coreceptors directly associate with members of receptor-like cytoplasmic kinase subfamily VII (RLCK VII), which are thought to mediate the activation of multiple downstream signaling pathways (Tang et al., 2017). Among these, BOTRYTIS-INDUCED KINASE1 (BIK1) is a central component that integrates signals from multiple PRRs, including FLS2, EFR, PEPRs, and LYK5, and is required for resistance to both fungal and bacterial pathogens (Veronese et al., 2006; Lu et al., 2010; Zhang et al., 2010; Laluk et al., 2011; Liu et al., 2013; Li et al., 2014). Upon pattern recognition, BIK1 directly phosphorylates the NADPH oxidase RBOHD to prime ROS production (Kadota et al., 2014; Li et al., 2014). The importance of RLCK VII is further supported by the finding that its members are attacked by multiple pathogen effectors during pathogenesis (Zhang et al., 2010; Feng et al., 2012; Feng and Zhou, 2012). However, RLCK VII contains 46 members, and obtaining a detailed understanding of their role in defense is hampered by the functional redundancy of various members.
The activation of MAPK cascades is essential for plant immunity (Boller and Felix, 2009; Meng and Zhang, 2013). All PRRs studied to date activate two MAPK cascades. One cascade is composed of the MAP kinase kinase kinase (MAPKKK) MEKK1, the MAP kinase kinases MKK1 and MKK2, and the MAP kinase MPK4 (Suarez-Rodriguez et al., 2007; Gao et al., 2008). The second cascade is composed of two MKKs, MKK4 and MKK5, and two MAPKs, MPK3 and MPK6 (Asai et al., 2002). The latter cascade plays a far more important role in plant immunity by activating defense gene expression, phytoalexin and ethylene biosynthesis, and stomatal immunity (Liu and Zhang, 2004; Menke et al., 2004; Bethke et al., 2009; Li et al., 2012; Lassowskat et al., 2014; Xu et al., 2016; Su et al., 2017). Paradoxically, the MAPKKKs that govern the second MAPK cascade downstream of various PRRs remain elusive. While different PRRs are believed to converge on the same MAPKKKs for MAPK activation, a recent report suggests that MAPKKK5 and PBL27, an RLCK VII subfamily member, positively regulate chitin-triggered MPK3/6 activation but negatively regulate flg22-triggered MAPK activation (Yamada et al., 2016b). Thus, the underlying mechanisms by which different PRRs activate MAPK cascades remain unclear.
Here, we show that MAPKKK3 and MAPKKK5 function redundantly to activate MPK3/6 downstream of at least four PRRs and confer resistance to both bacterial and fungal pathogens in Arabidopsis thaliana. We further show that a clade of RLCK VII (RLCK VII-4) directly links PRRs to MPK3/6 activation by phosphorylating MAPKKK5 Ser-599 and that this phosphorylation positively regulates defenses and disease resistance. In addition, we show that MAPKKK5 Ser-682 and Ser-692 are phosphorylated by MPK6 to further enhance the activation of MPK3/6 and disease resistance, pointing to a positive feedback mechanism. Moreover, RLCK VII-4 and MPK4 also phosphorylate MEKK1 at Ser-603 to positively regulate MPK4 activation. Together, the findings uncover a mechanism by which multiple PRRs activate MPKs during plant immune responses.
RESULTS
MAPKKK3/5 Are Phosphorylated in Response to Multiple Patterns
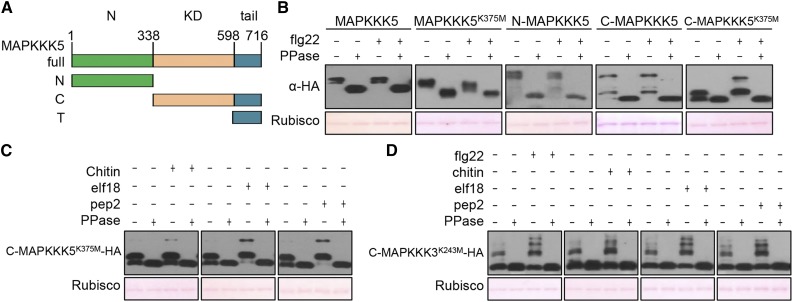
An examination of the public Arabidopsis gene expression database revealed a number of MAPKKKs whose transcription in plants is triggered by various patterns and Pseudomonas syringae bacteria (Hruz et al., 2008) (Supplemental Figure 1A). Among these, MAPKKK5 transcripts displayed the greatest induction upon treatment with various patterns and pathogens. We therefore first focused on MAPKKK5 in our analyses. We reasoned that a MAPKKK involved in pattern-triggered MAPK activation must be phosphorylated upon exposure to patterns. To facilitate detection of phosphorylation in MAPKKK5, we constructed a series of HA-tagged constructs including the full-length MAPKKK5, N-terminal domain, and the C terminus containing a kinase domain followed by a C-terminal tail (Figure 1A). Because autophosphorylation of MAPKKK5 may obscure phosphorylation by upstream kinases, we also introduced a Lys375Met mutation into the kinase domain, which abolishes its kinase activity. We expressed these constructs in Arabidopsis protoplasts, treated the protoplasts with flg22, and subjected the proteins to Phos-tag analysis. The full-length MAPKKK5 and MAPKKK5K375M constructs showed a phosphatase-sensitive mobility-shift indicative of protein phosphorylation (Figure 1B). However, the mobility-shift was insensitive to flg22 treatment. Because large phosphoproteins are often poorly resolved in Phos-tag assays, the results were not informative. The C-MAPKKK5 fragment showed constitutive mobility shift in the absence of flg22 treatment. Interestingly, the C-MAPKKK5K375M showed an flg22-specific mobility shift, and this shift was sensitive to phosphatase treatment, suggesting flg22-triggered phosphorylation. The differential flg22-specific phosphorylation in C-MAPKKK5 and C-MAPKKK5K375M suggested that autophosphorylation of C-MAPKKK5 might have masked the detection of flg22-induced phosphorylation in the Phos-tag assays. The N-terminal fragment displayed phosphorylation independently of flg22 treatment. While it remains unknown whether flg22 induces phosphorylation in the N-terminal fragment not resolved by Phos-tag assays, the aforementioned results suggest that flg22 treatment induces phosphorylation in MAPKKK5. Kinase-dead forms of additional family members representative of various groups of MAPKKKs were similarly examined for flg22- and chitin-triggered phosphorylation. Only MAPKKK3, which is the closest homolog of MAPKKK5, clearly showed flg22- and chitin-triggered phosphorylation (Supplemental Figure 1B). YODA, which is also homologous to MAPKKK3/5, showed minor flg22- or chitin-triggered phosphorylation. All patterns tested, including flg22, chitin, elf18, and Pep2, triggered band shifts of C-MAPKKK3K243M and C-MAPKKK5K375M in the Phos-tag assays (Figures 1C and 1D), suggesting that MAPKKK3/5 are activated by a variety of PRRs.
Figure 1.
Patterns Trigger MAPKKK3/5 Phosphorylation.
(A) Schematic representation of MAPKKK5 domain structure and deletion constructs. Green, N-terminal domain (N); orange, kinase domain (KD); aqua, C-terminal tail (T). The C terminus construct (C) contains both the KD and T.
(B) Flg22-induced phosphorylation of MAPKKK5 in the C terminus. Protoplasts expressing full-length, truncated, or kinase-dead (K375M) variants of MAPKKK5 were treated with flg22. Total protein treated with (+) or without (−) λ protein phosphatase (PPase) was subjected to SDS-PAGE containing Phos-tag acrylamide and detected by immunoblotting with anti-HA antibody. Ponceau staining of Rubisco indicates equal loading.
(C) and (D) Pattern-triggered phosphorylation of MAPKKK3 and MAPKKK5 C termini. Protoplasts expressing C-MAPKKK5K375M-HA (C) and C-MAPKKK3K243M-HA (D) were treated with chitin, elf18, Pep2, or flg22. Total protein was treated with (+) or without (−) λ protein phosphatase (PPase) and subjected to SDS-PAGE containing Phos-tag acrylamide and detected by immunoblotting with anti-HA antibody.
MAPKKK3/5 Are Required for MPK3/6 Activation Triggered by Multiple Patterns
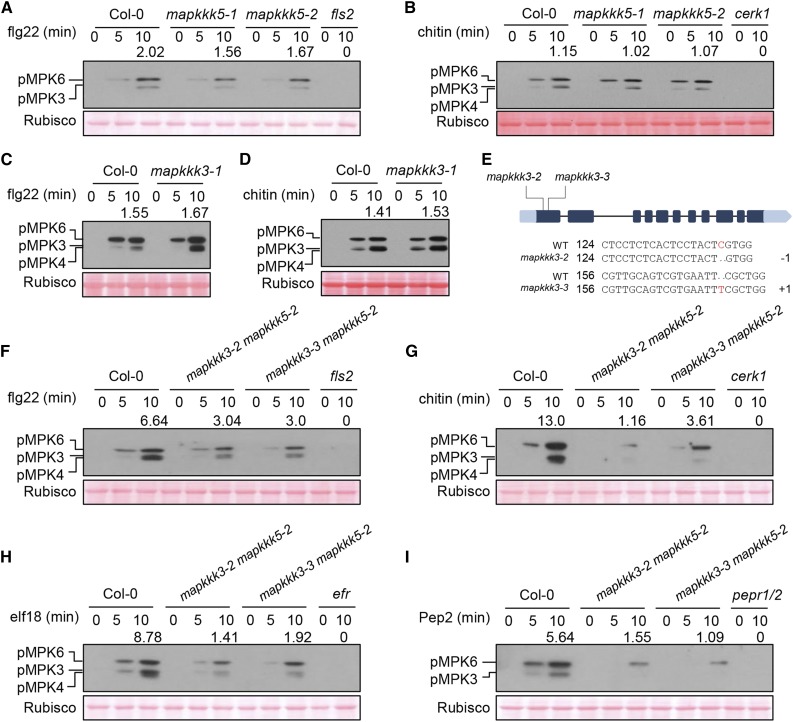
We next tested pattern-triggered MPK3/6 activation in Arabidopsis mapkkk3 (mapkkk3-1) and mapkkk5 (mapkkk5-1 and mapkkk5-2) single mutants. The flg22-triggered MPK3/6 activation was slightly reduced in mapkkk5 mutants compared with wild-type (Col-0) seedlings, whereas the chitin-, elf18-, and Pep2-triggered MPK3/6 activation was completely normal (Figures 2A and 2B; Supplemental Figures 2A and 2B). Furthermore, MPK3/6 activation in mapkkk3 was not affected, regardless of which patterns were tested (Figures 2C and 2D; Supplemental Figures 2C and 2D). A previous report suggested that MAPKKK5 plays a negative role in flg22-triggered MPK3/6 activation but a positive role in chitin-triggered MPK3/6 activation (Yamada et al., 2016b), which was not observed in our study or a recent study (Yan et al., 2018). Given that MAPKKK3/5 are highly homologous and may act redundantly in MAPK activation, the contradictory observations in different laboratories likely reflect experimental variations attributed to the weak phenotypes of single mutants. We therefore introduced two independent mapkkk3 mutations created by CRISPR-CAS9 into the mapkkk5-2 mutant to generate the mapkkk3-2 mapkkk5-2 and mapkkk3-3 mapkkk5-2 double mutants (Figure 2E). Strikingly, the flg22-triggered MPK3/6 activation in mapkkk3 mapkkk5 double mutant seedlings was reduced to ∼45% compared with that in Col-0, whereas chitin-, elf18-, and Pep2-triggered MPK3/6 activation in the mapkkk3 mapkkk5 mutants was reduced to ∼10 to 20% (Figures 2F to 2I). As expected, the PRR mutants fls2, efr, cerk1, and pepr1/2 were completely nonresponsive in terms of MAPK activation upon treatment with their corresponding patterns. These results were reliably obtained in numerous experiments, indicating that MAPKKK3/5 are required for MPK3/6 activation in response to microbe- and plant-derived patterns. The residual MPK3/6 activation in the mapkkk3 mapkkk5 double mutants suggests the presence of additional MAPKKK family members that play a minor role in pattern-triggered MPK3/6 activation. We conclude that all tested PRRs, including FLS2, EFR, CERK1, and PEPRs, activate MPK3/6 through MAPKKK3/5.
Figure 2.
MAPKKK3 and MAPKKK5 Regulate Pattern-Induced MAPK Activation.
(A) to (D) Pattern-triggered MAPK activation is normal in mapkkk5 ([A] and [B]) and mapkkk3 ([C] and [D]) single mutants. Ten-day-old seedlings were sprayed with flg22 or chitin, and samples were harvested at the indicated times for immunoblot analysis with anti-pERK antibody. Note that MPK4 and MPK3 are highly similar in size and were not well-separated in some experiments. Numbers indicate arbitrary densitometry units of phosphorylated MPK3/6 (pMPK3/6) bands normalized to Rubisco 10 min after pattern stimulation. All assays were performed three times, and a representative photograph is shown.
(E) CRISPR-Cas9-mediated mutations in the first exon of MAPKKK3 in the mapkkk3 mapkkk5 (mapkkk3-2 mapkkk5-2 and mapkkk3-3 mapkkk5-2) double mutant lines. The numbers 124 and 156 indicate the nucleotide positions in the MAPKKK3 coding sequence. −1 and +1 indicate frame shifts in the mutant lines.
(F) to (I) Pattern-triggered MPK3/6 activation is diminished in mapkkk3 mapkkk5 double mutants. Seedlings were sprayed with flg22 (F), chitin (G), elf18 (H), or Pep2 (I), and immunoblot analysis with anti-pERK antibody was performed at the indicated times. Note that MPK4 and MPK3 are highly similar in size and were not well separated in some experiments. Numbers indicate relative protein band density of pMPK3/6 normalized to the loading control (Rubisco). All assays were performed at least three times, and a representative photograph is shown. Ponceau staining of Rubisco indicates equal loading.
MAPKKK3/5 Are Required for Pattern-Triggered Defense Gene Expression and Disease Resistance
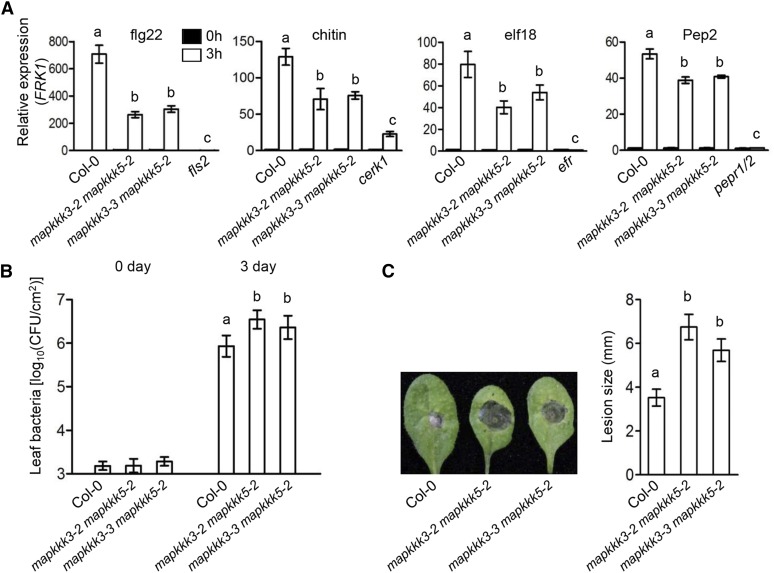
To further investigate the role of MAPKKK3/5 in MAPK signaling, we examined the expression of FLG22-INDUCED RECEPTOR KINASE1 (FRK1), NDR1/HIN1-LIKE10 (NHL10), and CYP81F2, which are known to be regulated by MAPK signaling in response to patterns (Boudsocq et al., 2010). The transcript levels in the mapkkk3 mapkkk5 double mutants were reduced by 27 to 70% compared with that in Col-0 in response to flg22, chitin, elf18, and Pep2, and the differences were statistically significant and reproducible (Figure 3A; Supplemental Figure 3). MPK3/6 play crucial roles in bacterial and fungal resistance (Ren et al., 2008; Galletti et al., 2011; Su et al., 2017). We challenged wild-type and mapkkk3 mapkkk5 mutant plants with P. syringae DC3000 and Botrytis cinerea. The double mutants supported 3- to 4-fold more bacterial growth and developed ∼2-fold larger fungal disease lesions compared with Col-0 plants (Figures 3B and 3C). The enhanced disease susceptibility to both bacterial and fungal pathogens was highly reproducible and statistically significant. These results are consistent with the diminished MAPK activation in response to both bacterial and fungal patterns (Figures 2F to 2H), and they demonstrate that MAPKKK3/5 are required for disease resistance to both bacterial and fungal pathogens.
Figure 3.
MAPKKK3/5 Are Required for Defense Gene Expression and Disease Resistance.
(A) Pattern-induced FRK1 expression is impaired in mapkkk3 mapkkk5 mutants. Ten-day-old seedlings were sprayed with flg22, chitin, elf18, or Pep2, and samples were harvested at 3 h. ACTIN1 was used as the internal standard. Data are presented as mean ± sd. Different letters indicate significant difference at P < 0.05 (n = 3, one-way ANOVA, Tukey post-test, three independent experiments).
(B) mapkkk3 mapkkk5 mutants display increased susceptibility to P. syringae pv tomato (Pto) DC3000. Plants were infiltrated with Pto DC3000, and bacterial population in the leaf was determined 3 d after inoculation. Data are presented as mean ± sd. Different letters indicate significant difference at P < 0.05 (n ≥ 8, one-way ANOVA, Tukey post-test, three independent experiments).
(C) mapkkk3 mapkkk5 mutants display increased susceptibility to B. cinerea. Disease lesions were recorded 3 d after inoculation with B. cinerea. Lesion size represents mean ± sd. Different letters indicate significant difference at P < 0.05 (n ≥ 14, one-way ANOVA, Tukey post-test, three independent experiments).
RLCK VII-4 Subfamily Members Link PRRs to MAPKKK5
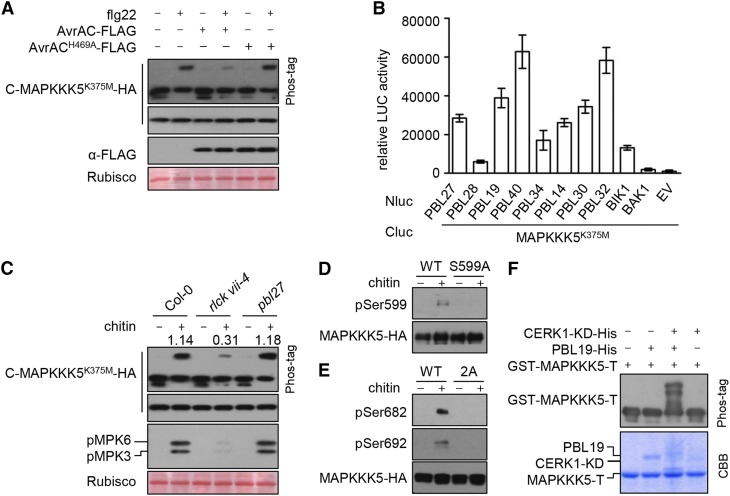
The RLCK VII subfamily contains 46 members, several of which have been shown to play a role in pattern-triggered immunity (Lin et al., 2013). However, individual knockout mutants of BIK1 and other RLCK VII subfamily members are largely normal in terms of pattern-triggered MAPK activation (Feng et al., 2012). We previously showed that the Xanthomonas campestris pv campestris effector AvrAC, a uridylyltransferase that specifically targets multiple RLCK VII subfamily members, can block flg22-triggered MAPK activation (Feng et al., 2012). The results suggest that RLCK VII subfamily members are functionally redundant and collectively play a role in pattern-triggered MAPK activation. We investigated whether RLCK VII subfamily members are required for MAPKKK5 phosphorylation in response to patterns by expressing AvrAC in the protoplasts. The expression of AvrAC-FLAG in protoplasts inhibited flg22-triggered phosphorylation of C-MAPKKKK375M-HA (Figure 4A), suggesting that certain members of the RLCK VII subfamily are required for pattern-triggered MAPKKK5 phosphorylation. Most of the 46 RLCK VII subfamily members belong to nine phylogenetic clades. We tested selected members of each clade for interactions with MAPKKK5 using luciferase complementation assays in Nicotiana benthamiana. Because MAPKKK5 induces cell death in N. benthamiana leaves (Yamada et al., 2016b), we used the kinase-dead mutant MAPKKK5K375M for this test. MAPKKK5K375M interacted with all RLCK VII subfamily members tested, but not BAK1 (Figure 4B), indicating that MAPKKK5 interacts with multiple RLCK VII subfamily members in plants.
Figure 4.
RLCK VII Subfamily Members Link PRRs to MAPKKK5.
(A) AvrAC inhibits flg22-induced phosphorylation of MAPKKK5 C terminus. C-MAPKKK5K375M-HA was coexpressed with AvrAC or AvrACH469A in Col-0 protoplasts, treated with flg22 for 10 min, and total protein was subjected to SDS-PAGE containing Phos-tag acrylamide and detected by immunoblotting with anti-HA antibody.
(B) MAPKKK5K375M interacts with selected members of RLCK VII. The indicated Nluc and Cluc constructs were transiently expressed in N. benthamiana plants for luciferase complementation assays. Error bars indicate sd.
(C) Chitin-triggered phosphorylation of MAPKKK5 C terminus and MPK3/6 is impaired in rlck vii-4 sextuple mutant, but occurs normally in pbl27. Protoplasts expressing C-MAPKKK5K375M-HA were treated with chitin. Total protein was subjected to SDS-PAGE containing Phos-tag acrylamide and detected by immunoblotting with anti-HA antibody for MAPKKK5 phosphorylation and anti-pERK antibody for MPK3/6 phosphorylation. Numbers indicate relative band density of pMPK3/6 normalized to the loading control (Rubisco).
(D) and (E) Chitin-triggered phosphorylation of MAPKKK5 at Ser-599, Ser-682, and Ser-692. Stable transgenic seedlings expressing MAPKKK5-HA (WT), MAPKKK5S599A-HA (S599A), and MAPKKK5S682/692A-HA (2A) were sprayed with chitin. The proteins were affinity purified with anti-HA antibodies and detected by immunoblot analysis with anti-pSer599 (D), anti-pSer682 (E), and anti-pSer692 (E) antibodies to investigate MAPKKK5 phosphorylation.
(F) In the presence of the CERK1 kinase domain (CERK1-KD), PBL19 phosphorylates MAPKKK5 C-terminal tail in vitro. GST-MAPKKK5-T was incubated with PBL19-His and/or CERK1-KD-His proteins, and total protein was subjected to SDS-PAGE containing Phos-tag acrylamide and detected by immunoblot analysis with anti-GST antibodies. The smeared appearance of CERK1-KD and PBL19 in lanes 3 and 4 of the Coomassie Brilliant Blue (CBB)-stained gel likely reflects phosphorylation of these proteins.
We recently constructed higher-order mutants in which multiple members of each RLCK VII clade are mutated (Rao et al., 2018). Systematic analyses of these mutants showed that the six RLCK VII-4 subfamily members play an additive role in chitin-triggered immune responses and are required for MAPK activation (Rao et al., 2018). An rlck vii-4 sextuple mutant displays marked reduction in chitin-triggered MAPK activation but normal MAPK activation in response to flg22 (Rao et al., 2018). Chitin-triggered phosphorylation of C-MAPKKK5K375M was reduced to ∼25% in protoplasts from the rlck vii-4 sextuple mutant compared with Col-0, whereas flg22-triggered C-MAPKKK5K375M phosphorylation was normal (Figure 4C; Supplemental Figure 4), suggesting that RLCK VII-4 specifically mediates chitin-triggered MAPKKK5 phosphorylation. PBL27, an RLCK VII-1 subfamily member, was reported to mediate chitin-triggered MAPK activation (Shinya et al., 2014). However, we were unable to reproduce this result, even when the entire VII-1 clade was mutated (Rao et al., 2018). Phos-tag assays showed normal C-MAPKKK5K375M phosphorylation in the pbl27 mutant (Figure 4C). Together, our results indicate that RLCK VII-4, but not PBL27, is required for chitin-triggered MAPKKK5 phosphorylation and MAPK activation.
The observation that C-MAPKKK5K375M is phosphorylated upon various pattern treatments prompted us to examine phospho-sites in MAPKKK5. We affinity-purified MAPKKK5-FLAG from protoplasts pretreated with flg22 and subjected the protein to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis for phospho-peptides. The identified peptides covered 75% of the MAPKKK5 sequence and contained at least 19 phosphorylated amino acid residues (Supplemental Table 1 and Supplemental Figure 5A). Interestingly, three phospho-sites, Ser-599, Ser-682, and Ser-692, were located in the C-terminal tail spanning the last 199 amino acids, whereas other phospho-sites were located in the N terminus. We focused on these three sites because the Phos-tag assays described above showed that pattern-triggered phospho-sites are located in the C terminus of MAPKKK5. Notably, alignment of the MAPKKK5 C-terminal tail from various flowering plant species indicated that Ser-599 is a highly conserved residue (Supplemental Figure 5B). This residue is also conserved in Arabidopsis MAPKKK3.
To analyze these phospho-sites in plants, we transformed mapkkk3 mapkkk5 double mutant plants with HA-tagged wild-type MAPKKK5, phospho-dead MAPKKK5S599A (S599A) and MAPKKK5S682/692A (2A), and phospho-mimetic MAPKKK5S599D (S599D) and MAPKKK5S682/692E (2E) transgenes under the control of the MAPKKK5 native promoter. We obtained 50 to 80 lines for each construct and identified lines that accumulated similar amounts of MAPKKK5 protein by immunoblotting, as described below. All transgenic lines were morphologically normal compared with Col-0. We also generated a control transgenic line by transforming mapkkk3 mapkkk5 double mutant plants with an empty vector (EV).
To determine whether pattern-induced phosphorylation of MAPKKK5 occurs at Ser-599, Ser-682, and Ser-692, we developed phosphopeptide-specific antibodies that recognize phospho-Ser599 (pSer599), phospho-Ser682 (pSer682), and phospho-Ser692 (pSer692) in MAPKKK5 and used these antibodies for immunoblot analysis. To verify chitin-induced phosphorylation of Ser-599, Ser-682, and Ser-692 in full-length MAPKKK5, we treated transgenic seedlings carrying the MAPKKK5, MAPKKK5S599A, and MAPKKK5S682/692A transgenes with chitin, enriched MAPKKK5 from the seedlings by affinity purification, and performed immunoblot analyses with the aforementioned phosphopeptide-specific antibodies. While no signal was detected in noninduced samples, strong signals indicative of phosphorylation at Ser-599, Ser-682, and Ser-692 in MAPKKK5 were detected in the chitin-treated samples (Figures 4D and 4E). Importantly, these signals were completely absent in the MAPKKK5S599A and MAPKKK5S682/692A mutant proteins, indicating that the antibodies were specific for the designated sites. These results demonstrate that chitin indeed triggers phosphorylation of MAPKKK5 at these three sites.
Because chitin-triggered C-MAPKKK5K375M phosphorylation requires RLCK VII-4, we tested whether one of its members, PBL19, can phosphorylate MAPKKK5 at Ser-599 in the presence or absence of the CERK1 kinase domain (CERK1-KD). We were unable to express the full-length MAPKKK5 or C-MAPKKK5 recombinant protein in Escherichia coli. Therefore, we generated a GST-tagged MAPKKK5 C-terminal tail (GST-MAPKKK5-T) recombinant protein as a substrate and incubated it with PBL19 and CERK1-KD in kinase assay buffer. Phos-tag assays revealed two slowly migrating bands of GST-MAPKKK5-T when it was incubated with both PBL19 and CERK1-KD, but not with PBL19 or CERK1-KD alone (Figure 4F), indicating that GST-MAPKKK5-T was phosphorylated only when both kinases was present. LC-MS/MS analysis of the in vitro phosphorylated GST-MAPKKK5-T product confirmed that Ser-599 is indeed phosphorylated (Supplemental Table 2).
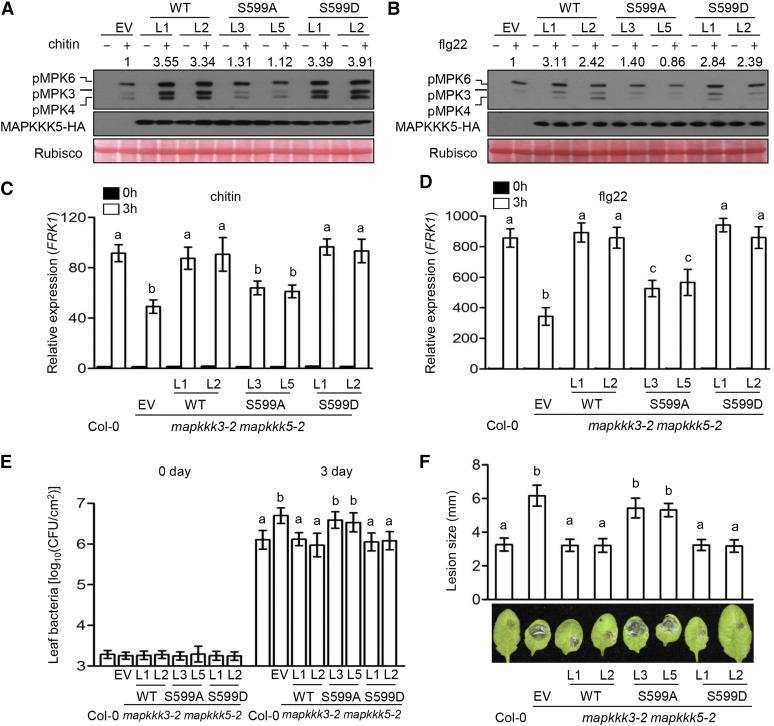
Phosphorylation of MAPKKK5 at Ser-599 Is Required for MAPK Activation and Disease Resistance
We treated seedlings of mapkkk3 mapkkk5 lines carrying EV, MAPKKK5, MAPKKK5S599A, and MAPKKK5S599D transgenes with chitin to determine the role of Ser-599 phosphorylation in the activation of MPKs. Compared with the EV line, which displayed weak, chitin-triggered MPK3/6 activation typical of mapkkk3 mapkkk5 plants, the MAPKKK5 and MAPKKK5S599D lines exhibited ∼3.5-fold levels of MPK3/6 activation (Figure 5A), indicating complementation of the mapkkk3 mapkkk5 mutations. By contrast, the MAPKKK5S599A lines were similar to the EV line in terms of chitin-induced MPK3/6 activation, indicating that the phosphorylation of Ser-599 is essential for the activation of MAPKKK5. It should be noted, however, that the MAPKKK5S599D lines showed no activation of MPK3/6 in the absence of chitin, suggesting that the phosphorylation of Ser-599 is required, but not sufficient, for activating MAPKKK5. Together, these results indicate that RLCK VII-4 subfamily members directly phosphorylate Ser-599 to positively regulate chitin-triggered MAPKKK5 activation. We also examined the role of MAPKKK5 Ser-599 phosphorylation in flg22-triggered MPK3/6 activation. Again, the MAPKKK5S599A lines were similar to the EV line, whereas the MAPKKK5S599D lines were comparable to the MAPKKK5 lines (Figure 5B). These results indicate that Ser-599 phosphorylation is also required for flg22-triggered MPK3/6 activation and suggest that other RLCK VII subfamily members also regulate flg22-triggered MAPKKK5 activation through phosphorylating Ser-599.
Figure 5.
Phosphorylation of MAPKKK5 at Ser-599 Is Required for MPK3/6 Activation and Disease Resistance.
(A) and (B) Phosphorylation of Ser-599 in MAPKKK5 is required for MAPK activation induced by chitin (A) and flg22 (B). Seedlings of mapkkk3-2 mapkkk5-2 mutant T2 transgenic lines complemented with EV, wild-type MAPKKK5 (WT), MAPKKK5S599A (S599A), or MAPKKK5S599D (S599D) were sprayed with chitin (A) or flg22 (B), and total protein was subjected to immunoblot analysis with anti-pERK and anti-HA antibody. Numbers indicate arbitrary densitometry units of pMPK3/6 normalized to EV. All assays were performed at least three times, and a representative photograph is shown.
(C) and (D) Phosphorylation of Ser-599 in MAPKKK5 is required for pattern-induced FRK1 expression. Ten-day-old seedlings were sprayed with chitin (C) and flg22 (D), and the samples were harvested at 3 h after treatment. ACTIN1 was used as the internal standard. Data are presented as mean ± sd. Different letters indicate significant difference at P < 0.05 (n = 3, one-way ANOVA, Tukey post-test, three independent experiments).
(E) Phosphorylation of Ser-599 in MAPKKK5 is required for immunity to P. syringae DC3000. Plants were infiltrated with Pto DC3000, and bacterial population in the leaf was determined 3 d after inoculation. Data are presented as mean ± sd. Different letters indicate significant difference at P < 0.05 (n ≥ 8, one-way ANOVA, Tukey post-test, three independent experiments).
(F) Phosphorylation of Ser-599 in MAPKKK5 is required for immunity to B. cinerea. Plants were inoculated with B. cinerea, and disease lesions were recorded 3 d after inoculation with B. cinerea. Lesion size represents mean ± sd. Different letters indicate significant difference at P < 0.05 (n ≥ 14, one-way ANOVA, Tukey post-test, four independent experiments).
Next, we examined these transgenic lines for flg22- and chitin-induced FRK1 expression. While the MAPKKK5 and MAPKKK5S599D lines showed the highest FRK1 transcript levels, i.e., 2- to 3-fold that of the EV line (Figures 5C and 5D), the MAPKKK5S599A lines expressed FRK1 at levels similar to (chitin treatment) or only mildly higher than (flg22 treatment) the EV line but significantly lower than the MAPKKK5 lines. We challenged these mutants with P. syringae DC3000 and B. cinerea. As expected, the EV line supported greater amounts of bacterial growth and developed much larger lesions compared with Col-0 plants (Figures 5E and 5F). The MAPKKK5 and MAPKKK5S599D lines were indistinguishable compared with Col-0 plants, indicating full restoration of disease resistance in these lines. By contrast, the MAPKKK5S599A lines were significantly more susceptible to both P. syringae DC3000 and B. cinerea compared with Col-0 plants and statistically indistinguishable from the EV line, indicating that MAPKKK5 Ser-599 phosphorylation is essential for disease resistance.
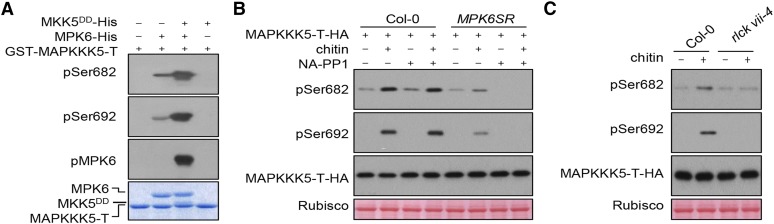
MPK6 Phosphorylates MPKKK5 at Ser-682/692
Unexpectedly, the phospho-sites Ser-682 and Ser-692 are within a predicted MAPK phosphorylation motif (Pitzschke, 2015). We sought to test whether these sites are indeed phosphorylated by MPK6 in vitro. Incubation of GST-MAPKKK5-T with recombinant MPK6 resulted in weak phosphorylation of both Ser-682 and Ser-692 in vitro (Figure 6A). Inclusion of recombinant MKK5DD, a constitutively active form of MKK5, resulted in strong phosphorylation of Ser-682/692, indicating that MPK6 can directly phosphorylate these sites. We next asked whether Ser-682/692 are phosphorylated in vivo in an MPK3/6-dependent manner. Overexpression of the full-length MAPKKK5 induces cell death in Arabidopsis protoplasts, making it difficult to enrich sufficient amounts of MAPKKK5 protein for phosphorylation assays. We therefore examined MAPKKK5-T phosphorylation in protoplasts of a conditional mpk3 mpk6 double mutant, mpk3 mpk6 PMPK6:MPK6YG (MPK6SR) (Xu et al., 2014). This mutant is rescued with MPK6YG, which contains a Tyr144Gly substitution that enlarges the ATP binding pocket and renders MPK6YG sensitive to a bulky ATP analog kinase inhibitor NA-PP1 (4-amino-1-tert-butyl-3-(1'-naphthyl) pyrazolo [3,4-d] pyrimidine). The addition of NA-PP1 specifically inhibited phosphorylation of both Ser-682 and Ser-692 in MPK6SR protoplasts (Figure 6B), supporting the notion that MPK6 mediates pattern-triggered Ser-682/692 phosphorylation in plant cells. To further investigate whether RLCK VII-4 subfamily members are required for chitin-triggered phosphorylation of Ser-682/692, we expressed the MAPKKK5-T-HA fusion protein in Col-0 and rlck vii-4 protoplasts, treated the protoplasts with chitin, enriched MAPKKK5-T-HA protein by affinity purification, and performed immunoblot analysis. In the absence of chitin treatment, we detected background phosphorylation of Ser-682 and no phosphorylation of Ser-692 (Figure 6C). In rlck vii-4 mutant protoplasts, both basal and chitin-induced levels of Ser-682 phosphorylation were reduced, whereas Ser-692 phosphorylation was abolished (Figure 6C). These results indicate that RLCK VII-4 subfamily members are required for chitin-triggered phosphorylation of Ser-682/692 in plant cells, and they are consistent with the notion that MPK6 activation was reduced in rlck vii-4.
Figure 6.
MPK6 Phosphorylated MAPKKK5 at Ser-682 and Ser-692.
(A) MPK6 phosphorylates MAPKKK5 in vitro. GST-MAPKKK5-T was incubated with MPK6-His and/or MKK5DD-His in kinase buffer and detected by immunoblotting with anti-pSer682 and pSer692 antibodies.
(B) MAPKKK5 phosphorylation induced by chitin is blocked in the conditional mpk3 mpk6 double mutant. Protoplasts of Col-0 and the conditional mpk3 mpk6 double mutant were transfected with MAPKKK5-T-HA, incubated in the presence of DMSO (solvent control) or 2 μM NA-PP1 overnight, and treated with chitin. MAPKKK5-T-HA was affinity purified with anti-HA antibodies and detected by immunoblotting with anti-pSer682 and -pSer692 antibodies.
(C) Chitin-triggered phosphorylation of Ser682/692 is impaired in the rlck vii-4 sextuple mutant. Protoplasts expressing MAPKKK5-T-HA were treated with chitin. MAPKKK5-T-HA was affinity purified with anti-HA antibody and detected by immunoblotting with anti-pSer682 and pSer692 antibodies.
Phosphorylation of MAPKKK5 at Ser-682/692 Enhances MPK3/6 Activation and Immunity
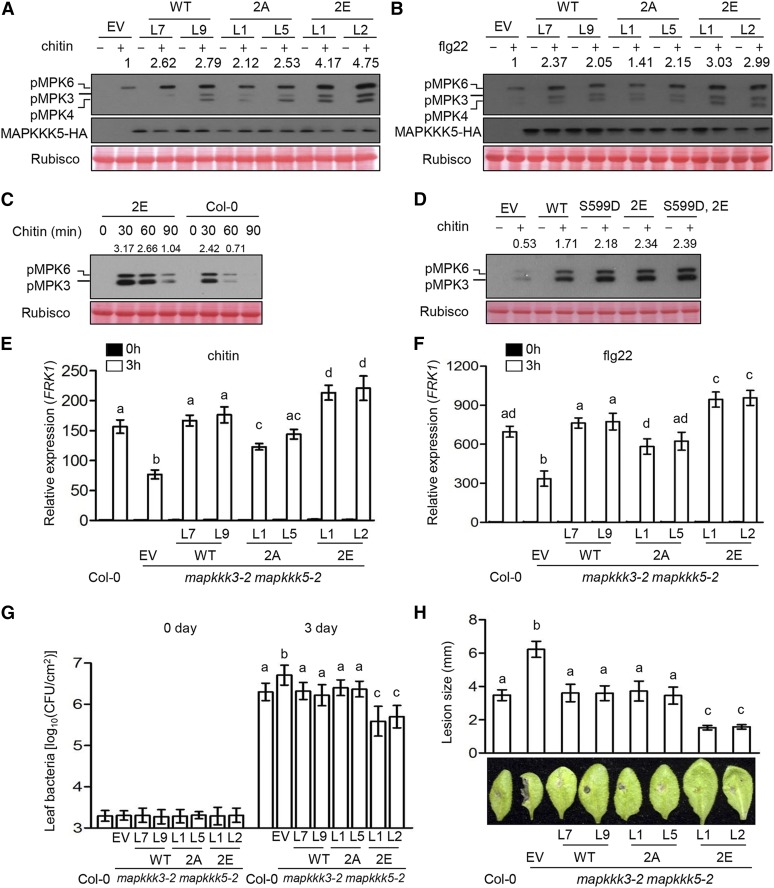
To determine whether Ser-682/692 play a role in pattern-triggered MAPKKK5 activation, we treated mapkkk3 mapkkk5 seedlings carrying MAPKKK5, MAPKKK52A, and MAPKKK52E transgenes with chitin and flg22. The MAPKKK52A lines showed a mild but reproducible reduction of chitin- and flg22-triggered MPK3/6 activation compared with MAPKKK5 lines, while the MAPKKK52E lines displayed elevated MPK3/6 activation compared with MAPKKK5 (Figures 7A and 7B), indicating that Ser-682/692 phosphorylation is required for the full activation of MAPKKK5. Time-course analysis of chitin-triggered MPK3/6 activation showed that MAPKKK52E seedlings displayed extended MPK3/6 activation compared with Col-0 plants (Figure 7C), suggesting that Ser-682/692 phosphorylation positively controls the duration of MPK3/6 activation. We tested whether simultaneous phospho-mimetic mutations of S599/682/692 would further enhance MPK3/6 activation. While transfection of mapkkk3 mapkkk5 mutant protoplasts with MAPKKK52E enhanced chitin-triggered MPK3/6 activation compared with transfection with MAPKKK5, transient expression of MAPKKK5S599D,2E did not further enhance chitin-triggered MPK3/6 activation or lead to constitutive MPK3/6 activation (Figure 7D).
Figure 7.
Phosphorylation of MAPKKK5 Ser-682/692 Enhances MPK3/6 Activation and Immunity.
(A) and (B) Phosphorylation of Ser-682/692 in MAPKKK5 is required for MAPK activation induced by chitin (A) and flg22 (B). Seedlings of mapkkk3-2 mapkkk5-2 mutant T2 transgenic lines complemented with EV, wild-type MAPKKK5 (WT), MAPKKK5S682/S692A (2A), or MAPKKK5S682/692E (2E) were sprayed with chitin (A) or flg22 (B), and total protein was subjected to immunoblot analysis with anti-pERK and anti-HA antibodies. Numbers indicate arbitrary densitometry units of phosphorylated MPK3/6 normalized to EV. All assays were performed at least three times, and a representative photograph is shown.
(C) Phospho-mimetic mutations at Ser682/692 prolong chitin-triggered MPK3/6 activation. Seedlings of mapkkk3-2 mapkkk5-2 transgenic lines complemented with MAPKKK5S682/692E (2E) and Col-0 were treated with chitin for the indicated times, and total protein was subjected to immunoblotting with anti-pERK antibodies. Numbers indicate arbitrary densitometry units of phosphorylated MPK3/6 normalized to Rubisco. All assays were performed at least three times, and a representative photograph is shown.
(D) Phospho-mimetic mutations at Ser-599/682/692 do not further enhance chitin-triggered MPK3/6 activation compared with phospho-mimetic mutations at Ser-682/692. Protoplasts of the mapkkk3-2 mapkkk5-2 double mutant expressing EV, the wild type, phospho-dead (S599D), phospho-mimicking Ser682/692E (2E), and phospho-mimicking S599D, S682/692E (S599D,2E) forms of MAPKKK5 were treated with chitin. Total protein was subjected to immunoblot analysis with anti-pERK antibody. Numbers indicate relative protein band density of phosphorylated MPKs normalized to Rubisco. All assays were performed at least three times, and a representative photograph is shown.
(E) and (F) Phosphorylation of Ser-682/692 in MAPKKK5 is required for pattern-induced FRK1 expression. Ten-day-old seedlings were sprayed with chitin (E) and flg22 (F), and the samples were harvested at 3 h after treatment. ACTIN1 was used as the internal standard. Data are presented as mean ± sd. Different letters indicate significant difference at P < 0.05 (n = 3, one-way ANOVA, Tukey post-test, three independent experiments).
(G) Phosphorylation of Ser-682/692 in MAPKKK5 is required for immunity to P. syringae DC3000 and B. cinerea. Plants were infiltrated with Pto DC3000, and bacterial population size in the leaf and diseased lesions was determined 3 d after inoculation. Data are presented as mean ± sd. Different letters indicate significant difference at P < 0.05 (n ≥ 8, one-way ANOVA, Tukey post-test, three independent experiments).
(H) Phosphorylation of Ser-682/692 in MAPKKK5 is required for immunity to B. cinerea. Plants were inoculated with B. cinerea, and disease lesions were recorded 3 d after inoculation with B. cinerea. Lesion size represents mean ± sd. Different letters indicate significant difference at P < 0.05 (n ≥ 14, one-way ANOVA, Tukey post-test, three independent experiments).
To further investigate the role of Ser-682/692 phosphorylation in defense responses, we examined the expression of FRK1 in response to chitin and flg22. FRK1 expression was partially restored in the MAPKKK52A lines compared with the complete restoration in the MAPKKK5 lines, whereas the MAPKKK52E lines exhibited stronger FRK1 expression compared with the MAPKKK5 lines (Figures 7E and 7F), indicating that Ser-682/692 phosphorylation positively regulates flg22- and chitin-triggered immune responses. We challenged these mutants with P. syringae DC3000 and B. cinerea. The MAPKKK52A lines were indistinguishable from the MAPKKK5 lines, whereas the MAPKKK52E lines showed much stronger resistance against both P. syringae DC3000 and B. cinerea compared with the MAPKKK5 lines (Figures 7G and 7H), indicating that phosphorylation of Ser-682/692 can enhance resistance to both bacterial and fungal pathogens.
The phosphorylation of Ser-599/682/692 may affect pattern-triggered MAPK activation through one or multiple mechanisms. We reasoned that this process might modulate interactions of MAPKKK5 with MKK5 or RLCKs. Coimmunoprecipitation assays showed that MAPKKK5, MAPKKK5S599A, and MAPKKK5S682/692A were indistinguishable in their interactions with MKK5 and PBL19 (Supplemental Figures 6A and 6B), indicating that the phosphorylation of these sites did not affect protein-protein interactions. We then attempted to investigate whether phosphorylation regulates the kinase activity of MAPKKK5. The inability to express full-length MAPKKK5 or C-MAPKKK5 protein E. coli prevented us from testing its kinase activity in vitro. Attempts to isolate the full-length MAPKKK5 protein from plants or protoplasts were similarly hampered by cell death upon overexpression of full-length MAPKKK5. Transgenic plants expressing MAPKKK5-HA under the control of its native promoter did not produce sufficient amounts of protein for the kinase assay. Finally, the phosphorylation of these sites might affect the subcellular localization of MAPKKK5, a possibility that remains to be tested in future studies.
MEKK1 Is Phosphorylated by Both RLCK VII-4 and MPK4
The above results demonstrate that RLCK VII subfamily members directly mediate MAPKKK5 activation downstream of PRRs. We extended the analysis to investigate whether MEKK1 is regulated in a similar manner. We first confirmed MPK4 activation in summ2-8 mekk1 mutant plants treated with various patterns. summ2-8 mekk1 was used in this study because the nucleotide binding leucine-rich repeat immune receptor SUMM2 (SUPPRESSOR OF MKK1 MKK2 2) monitors the MEKK1-MKK1/2-MPK4 pathway and activates immunity when this pathway is attacked by pathogens or genetically impaired, which masks the true function of MPK4 and MEKK1 in signaling (Zhang et al., 2012). In contrast to summ2 plants, which displayed strong MPK4 activation in response to flg22, elf18, and chitin, summ2-8 mekk1 plants failed to show detectable MPK4 phosphorylation (Supplemental Figures 7A to 7C), which is consistent with previous reports that flg22-induced MPK4 phosphorylation is undetectable in mekk1 (Ichimura et al., 2006; Suarez-Rodriguez et al., 2007).
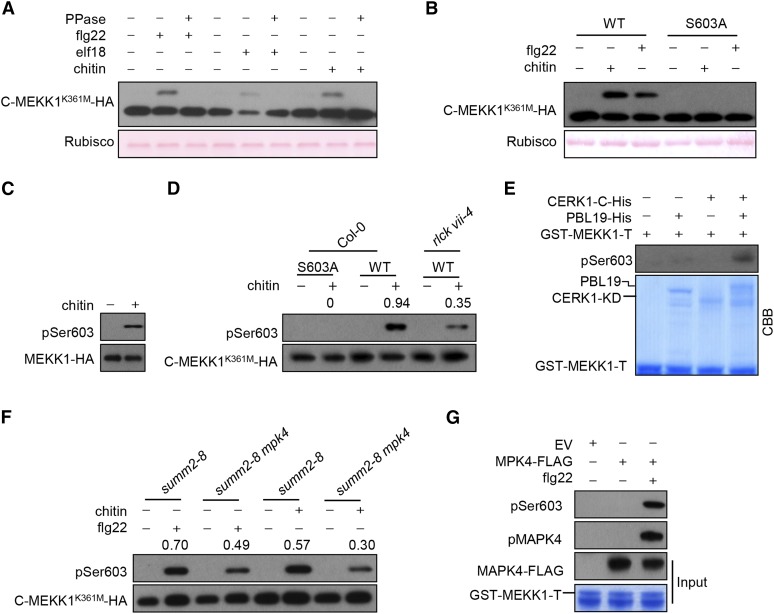
We next tested whether MEKK1 is also phosphorylated upon exposure to various patterns. An examination of a kinase-dead C-terminal MEKK1 fragment (C-MEKK1K361M-HA), which carried a Lys361Met mutation in the ATP binding site, showed a phosphatase-sensitive mobility shift in SDS-PAGE upon treatment of protoplasts with flg22, elf18, and chitin (Figure 8A), indicating pattern-triggered phosphorylation of MEKK1. To identify the phospho-sites in MEKK1 that occurred independently of autophosphorylation, we expressed the full-length MEKK1K361M-FLAG in protoplasts. After flg22 treatment, we affinity-purified MEKK1K361M-FLAG protein and subjected it to LC-MS/MS analysis. Approximately 74% of the MEKK1 peptide sequence was covered, and 21 phospho-peptides corresponding to at least 15 phospho-sites were identified (Supplemental Figure 8A and Supplemental Table 3). Strikingly, amino acids 589 to 607 in the C terminus of MEKK1 were phosphorylated one to three times, as identified in multiple peptides. We examined whether these residues were major phospho-sites by site-directed mutagenesis. Substitution of Ser-603 (but not the four other residues) to Ala resulted in the complete elimination of the chitin- or flg22-induced band shift in C-MEKK1K361M (Figure 8B; Supplemental Figure 8B), indicating that Ser-603 is a major phospho-site during pattern-triggered MEKK1 phosphorylation. To determine whether pattern-induced phosphorylation of full-length MEKK1 occurs at Ser-603, we developed phosphopeptide-specific antibodies that recognize phospho-Ser603 (pSer603) in MEKK1 and used these antibodies for immunoblot analysis. We transformed summ2-8 mekk1 double mutant plants with a FLAG-tagged wild-type MEKK1 transgene under the control of the MEKK1 native promoter. After treatment of the transgenic seedlings with chitin, we enriched the full-length MEKK1-FLAG protein by affinity purification and performed immunoblot analysis with the abovementioned phosphopeptide-specific antibodies. While no signals were detected in the control samples, strong signals indicative of phosphorylation at Ser-603 in MEKK1 were detected in the chitin-treated sample (Figure 8C). These results demonstrate that chitin indeed triggers phosphorylation of MEKK1 at Ser-603. An alignment of the Arabidopsis MEKK1 with homologous sequences from other plants indicated that Ser-603 is highly conserved in vascular plants including Physcomitrella patens (Supplemental Figure 8C) (Bressendorff et al., 2016), implying that this site is functionally important.
Figure 8.
Both RLCK VII-4 and MPK4 Phosphorylate MEKK1 at Ser-603.
(A) Pattern-triggered phosphorylation of MEKK1 at the C terminus. Protoplasts expressing C-MEKK1K361M-HA were treated with flg22, elf18, or chitin. Total protein was fractionated by SDS-PAGE containing Phos-tag acrylamide and detected by immunoblotting with anti-HA antibody.
(B) Ser-603 is the major phospho-site in MEKK1 C terminus MEKK1 upon chitin and flg22 induction. C-MEKK1K361M-HA (WT) or C-MEKK1K361M/S603A-HA (S603A) was expressed in protoplasts and total protein was detected by immunoblotting with anti-HA antibody.
(C) Chitin-triggered phosphorylation of MEKK1 at Ser-603. Stable transgenic seedlings expressing MEKK1-FLAG were sprayed with chitin. MEKK1-FLAG was affinity purified with anti-FLAG antibody and detected by immunoblotting with anti-pSer603 antibodies for MEKK1 phosphorylation.
(D) Chitin-triggered phosphorylation of Ser-603 is impaired in the rlck vii-4 sextuple mutant. Protoplasts of the indicated genotypes transfected with the C-MEKK1 K361M-HA constructs containing a wild-type Ser603 (WT) or a Ser603Ala substitution (S603A) were treated with chitin. The C-MEKK1K361M-HA protein was immunoprecipitated with anti-HA antibodies, and Ser-603 phosphorylation was detected by immunoblotting with anti-pSer603 antibodies. Total C-MEKK1K361M-HA protein was detected by immunoblotting with anti-HA antibody. Numbers indicate arbitrary densitometry units of pSer603 normalized to total C-MEKK1 K361M-HA protein.
(E) CERK1-activated PBL19 phosphorylates the MEKK1 C-terminal tail in vitro. GST-MEKK1-T was incubated with PBL19-His and/or CERK1-KD-His (CERK1-C-His) proteins, and MEKK1 phosphorylation was detected by immunoblotting with anti-pSer603 antibodies.
(F) Flg22- or chitin-triggered Ser-603 phosphorylation of MEKK1 is impaired in the summ2-8 mpk4 mutant. Numbers indicate arbitrary densitometry units of pSer603 normalized to total C-MEKK1 K361M-HA protein.
(G) Activated MPK4 phosphorylates MEKK1 in vitro. Protoplasts expressing MPK4-FLAG were treated with (+) or without (−) flg22. MPK4-FLAG proteins were affinity-purified with anti-FLAG antibodies and incubated with GST-MEKK1-T protein in kinase buffer, and MEKK1 phosphorylation was detected by immunoblotting with anti-pSer603 antibodies.
We next asked whether RLCK VII-4 subfamily members are required for the phosphorylation of MEKK1 at Ser-603 in vivo. The aforementioned phosphopeptide-specific antibodies specifically detected chitin-triggered phosphorylation in C-MEKK1K361M but not C-MEKK1K361M/S603A (Figure 8D). Moreover, chitin-triggered phosphorylation of C-MEKK1K361M was diminished in rlck vii-4 sextuple mutant protoplasts compared with Col-0, indicating that RLCK VII-4 subfamily members are required for pattern-triggered MEKK1 Ser-603 phosphorylation. We then conducted an in vitro kinase assay to determine whether CERK1-activated PBL19 can phosphorylate MEKK1 Ser-603. We detected phosphorylation at Ser-603 only when the GST-tagged MEKK1 C-terminal tail (GST-MEKK1-T) recombinant protein was incubated with both PBL19 and CERK1-KD (CERK1-C), but not with PBL19 or CERK1-C individually (Figure 8E). Together, these results support the notion that RLCK VII-4 subfamily members directly phosphorylate MEKK1 at Ser-603.
The sequence adjacent to Ser-603 resembles a MAPK phosphorylation motif (S/TP) (Supplemental Figure 8C), suggesting that this site is also phosphorylated by MAPKs. We investigated whether MPK4 also contributes to the overall phosphorylation of MEKK1 Ser-603 in vivo by examining MEKK1 phosphorylation in summ2-8 mpk4 double mutant protoplasts (Zhang et al., 2012). Chitin- and flg22-induced MEKK1 phosphorylation in summ2-8 mpk4 was reduced compared with that in summ2-8 (Figure 8F). In addition, MPK4-FLAG immunopurified from protoplasts phosphorylated the GST-MEKK-T recombinant protein at Ser-603 (Figure 8G), indicating that MPK4 directly phosphorylates MEKK1 at Ser-603.
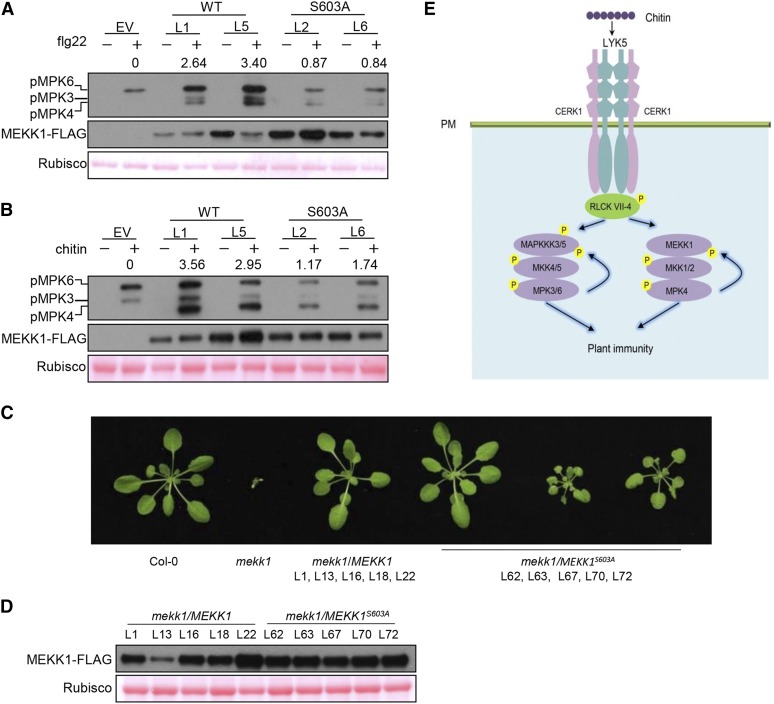
Phosphorylation of MEKK1 at Ser-603 Is Required for MPK4 Activation and the Suppression of Autoimmunity
To determine whether Ser-603 phosphorylation plays a role in MEKK1 activation, we generated stable transgenic lines expressing wild-type MEKK1 and MEKK1S603A (phospho-dead) under the control of the native MEKK1 promoter in the summ2-8 mekk1 double mutant background. As indicated in Figures 9A and 9B, flg22- and chitin-induced MPK4 activation was diminished in the MEKK1S603A lines compared with plants transformed with wild-type MEKK1, indicating that Ser-603 phosphorylation is required for MEKK1 activation. We next asked whether Ser-603 phosphorylation is required for the rescue of SUMM2-mediated autoimmunity in mekk1 plants. We generated five MEKK1 and five MEKK1S603A transgenic lines in the mekk1 background. The dwarf phenotype was fully rescued in all plants transformed with MEKK1 (Figures 9C and 9D). However, among the five lines complemented with MEKK1S603A, each included individual plants with a range of phenotypes, with approximately one-third fully restored to the wild type and two-thirds showing intermediate phenotypes compared with wild-type and mekk1 plants. These results suggest that Ser-603 phosphorylation plays an important role in MEKK1 activation.
Figure 9.
Phosphorylation of MEKK1 at Ser-603 Is Required for the Activation of MPK4 and the Suppression of Autoimmunity.
(A) and (B) Phosphorylation of Ser-603 in MEKK1 is required for flg22- (A) and chitin-triggered (B) MPK4 activation. Seedlings of summ2-8 mekk1 T2 lines complemented with EV, wild-type MEKK1 (WT), or phospho-dead MEKK1S603A (S603A) transgene were sprayed with flg22 (A) and chitin (B) prior to immunoblot analysis with anti-pERK antibody. Numbers indicate arbitrary densitometry units of pMPK4 normalized to Rubisco.
(C) Phosphorylation of Ser-603 in MEKK1 is required for the suppression of autoimmune phenotypes. Wild-type MEKK1-FLAG and MEKK1S603A-FLAG were transformed into heterozygous MEKK1 (+/−) plants. T2 plants homozygous for mekk1 and carrying MEKK1-FLAG and MEKK1S603A-FLAG transgenes were identified by genotyping and immunoblot analysis with anti-FLAG antibody. Plants of the indicated genotypes were scored for phenotypes, and representative photographs are shown. All plants of lines complemented with wild-type MEKK1-FLAG (mekk1/MEKK1) displayed a wild-type phenotype, whereas lines complemented with MEKK1S603A-FLAG (mekk1/MEKK1S603A) displayed a range of phenotypes from wild-type-like (approximately one-third) to partially stunted growth (approximately two-thirds) in individual plants.
(D) Accumulation of MEKK1-FLAG protein in lines shown in (C). Immunoblot analysis with anti-FLAG antibody was performed on mature plants of the indicated lines. Ponceau staining indicates equal loading of protein.
(E) A model for MPK3/6/4 activation triggered by chitin. LYK5 forms a receptor complex with CERK1 for chitin perception, leading to the activation of RLCK VII-4 subfamily members, which subsequently phosphorylate MAPKKK5 Ser-599 and MEKK1 Ser-603. This phosphorylation positively regulates MAPKKK3/5 and MEKK1 activity to activate MKK4/5-MPK3/6 and MKK1/2-MPK4, respectively. The activated MPK3/6 phosphorylate MAPKKK5 Ser-682/692 to further enhance MAPKKK5 activity, and the activated MPK4 phosphorylates MEKK1 to amplify the MAPK cascade.
DISCUSSION
In this study, we demonstrated that MAPKKK3/5 function redundantly to activate MPK3/6 downstream of multiple PRRs and confer resistance to both bacterial and fungal pathogens. We further show that downstream of the chitin coreceptor CERK1, RLCK VII-4 subfamily members directly phosphorylate MAPKKK5 at Ser-599 for MPK3/6 activation and disease resistance, providing an example in which RLCK VII subfamily members directly link a PRR to MAPKKK5 for MPK3/6 activation. We show that MAPKKK5 is additionally phosphorylated by MPK6 at Ser-682/692 to enhance both flg22- and chitin-triggered MPK3/6 activation, pointing to a positive feedback mechanism. RLCK VII-4- and MPK-mediated phosphorylation regulate MEKK1 for MPK4 activation in a similar manner.
A previous study reported that MAPKKK5 positively regulates MPK3/6 activation downstream of CERK1 but negatively regulates MPK3/6 downstream of FLS2 (Yamada et al., 2016b), while a more recent study reported that MAPKKK5 has no role in chitin-triggered MPK3/6 activation but positively regulates flg22-triggered MPK3/6 activation (Yan et al., 2018). These contradictory results are likely due to the mild phenotypes of single mapkkk5 mutants, which are difficult to reproduce. Indeed, we did not observe any significant defects in the mapkkk5 or mapkkk3 single mutants, whereas two mapkkk3 mapkkk5 double mutants reproducibly displayed reduced MPK3/6 activation in response to flg22, elf18, chitin, and Pep2, indicating that the same MAPKKKs operate downstream of at least four distinct PRRs. Furthermore, we showed that MAPKKK3/5 are required for defense gene activation and resistance to both bacterial and fungal pathogens. Thus, MAPKKK3/5 are key components utilized by diverse PRRs for MPK3/6 activation and to mediate disease resistance.
It should be noted, however, that the mapkkk3 mapkkk5 double mutants still exhibited low levels of MPK3/6 activation, suggesting that additional MAPKKKs are involved in this process. ARABIDOPSIS NUCLEUS-AND PHRAGMOPLAST-LOCALIZED KINASE1-RELATED PROTEIN KINASEs (ANPs) including ANP1, ANP2, and ANP3 are required for MAPK activation triggered by oligogalacturonides, but not elf18 (Savatin et al., 2014). Further studies are needed to determine whether these MAPKKKs function redundantly with MAPKKK3/5 in pattern-triggered MPK3/6 activation.
We showed that MAPKKK3/5 and MEKK1 are phosphorylated upon treatment with multiple patterns and identified MAPKKK5 Ser-599 and MEKK1 Ser-603 as phospho-sites in proteins isolated from plant cells. Ser-599 is highly conserved in MAPKKK5 in angiosperms and is also conserved in Arabidopsis MAPKKK3, whereas Ser-603 is highly conserved in MEKK1 homologs in vascular plants including P. patens, the latter containing a functional MPK4 pathway (Bressendorff et al., 2016). MAPKKK5 Ser-599 phosphorylation is essential for MPK3/6 activation by flg22 and chitin, whereas MEKK1 Ser-603 phosphorylation is required for full MPK4 activation by flg22 and chitin. Furthermore, Ser-599 plays a crucial role in pattern-induced defense gene expression and disease resistance to both bacterial and fungal pathogens. A previous study reported that MAPKKK5 is phosphorylated at Ser-617/622/658/660/677/685 and suggested that the phosphorylation of these sites is required for chitin-triggered MPK3/6 activation (Yamada et al., 2016b). Unlike Ser-599, these reported sites are not conserved in angiosperms. Importantly, none of these reported sites were identified in vivo, and it remains unknown whether they are relevant in the context of disease resistance.
We recently showed that RLCK VII-4 subfamily members are the most important RLCKs required for chitin-triggered ROS production, defense gene expression, and MAPK activation (Rao et al., 2018). In this study, we showed that RLCK VII-4 subfamily members are required for chitin-triggered phosphorylation of MAPKKK5 and MEKK1 in vivo (Figures 4C and 8C). The RLCK VII-4 subfamily member PBL19 can phosphorylate MAPKKK5 at Ser-599 and MEKK1 at Ser-603 in the presence of the CERK1 kinase domain, providing strong evidence that RLCK VII-4 directly activates MAPKKK5 and MEKK1 downstream of CERK1. A previous study reported that PBL27, an RLCKVII-1 subfamily member, phosphorylates MAPKKK5 in vitro and is required for chitin-triggered MAPK activation (Yamada et al., 2016b). However, we were unable to reproduce this result (Figure 4C) (Rao et al., 2018). Even the higher-order rlck vii-1 mutant failed to show any defects in chitin-triggered MAPK activation (Rao et al., 2018). We conclude that RLCKVII-4, but not RLCK VII-1 subfamily members, are the primary RLCKs that directly phosphorylate MAPKKK5 and MEKK1 for the activation of two MAPK cascades upon chitin perception.
Although we have not yet pinpointed specific RLCK VII clades that contribute to MAPKKK5 and MEKK1 phosphorylation in response to patterns other than chitin, the RLCK VII-4 subfamily members PCRK1 and PCRK2 have been shown to play a minor role in flg22-triggered MAPK activation (Kong et al., 2016), whereas the RLCK VII-8 subfamily members BIK1 and PBL1 play a minor role in Pep2-triggered MAPK activation (Yamada et al., 2016a). These mutants did not appear to be affected in terms of MAPKKK5 phosphorylation, but this could be attributed to the weak phenotypes of these mutants. It is likely that high levels of redundancy exist in RLCK VII-mediated MAPKKK3/5 activation in response to various patterns, a possibility consistent with the interaction of multiple RLCK VII subfamily members with MAPKKK5 (Figure 4B). Indeed, overexpression of AvrAC, which inhibits multiple RLCKVII subfamily members (Feng et al., 2012), strongly inhibited flg22-triggered MAPK activation and MAPKKK5 phosphorylation (Figure 4A). Furthermore, MAPKKK5 Ser-599 and MEKK1 Ser-603 phosphorylation are required for flg22-triggered MPK3/6 and MPK4 activation, respectively (Figures 5A and 9B). Given the similarity among RLCK VII subfamily members, these sites are likely phosphorylated by RLCKs operating downstream of FLS2 and other PRRs. Taken together, our data support the notion that RLCK VII subfamily members act downstream of multiple PRRs/coreceptors to phosphorylate MAPKKK3/5 and MEKK1 for MAPK activation.
RLCKs may also connect RKs to MAPK activation during plant development. SHORT SUSPENSOR (SSP), an RLCK-XII family member that controls embryonic patterning, is required for the activation of the MAPK cascade by acting upstream of YODA (Bayer et al., 2009; Costa et al., 2014; Yuan et al., 2017), raising the possibility that SSP activates YODA and the MAPK cascade through a phosphorylation relay in a similar manner.
We also identified Ser-682/692 as phospho-sites in MAPKKK5 isolated from plant cells. Ser-682/692 are phosphorylated by MPK6, and the phosphorylation is required for the full activation of MPK3/6 and FRK1 expression in response to patterns. The phospho-mimetic MAPKKK52E lines exhibited elevated and prolonged MPK3/6 activation, FRK1 expression, and resistance to bacterial and fungal pathogens. These results indicate that the phosphorylation of Ser-682/692 can enhance MPK3/6 activation and immunity, and they uncover a positive feedback regulation mechanism. MEKK1 Ser-603 is similarly phosphorylated by MPK4, which is required for the full activation of MPK4 by flg22 and chitin, indicating that positive feedback regulation also functions for MEKK1. Interestingly, MAPK-mediated feedback regulation of MAPKKKs has also been reported in animals. The activation of the MAPKKK7 TAK1 by Tumor necrosis factor-a and interleukin-1a is negatively regulated by the MAPK p38a (Cheung et al., 2003), although the precise mechanism remains unknown.
Ser-599/682/692 are located in the C-terminal tail of MAPKKK5, indicating that the C-terminal tail plays a critical role in the regulation of MAPKKKs activity during pattern-triggered immunity. The C-terminal tail of YODA, a MAPKKK that controls stomatal development and extra-embryonic cell fate (Lukowitz et al., 2004; Wang et al., 2007), is required for its full function (Lukowitz et al., 2004), although it is not known whether this also involves phosphorylation. The phospho-mimetic MAPKKK5S599D, MAPKKK52E, and MAPKKK5S599D,2E mutant proteins did not confer constitutive MPK3/6 activation, indicating that Ser-599/682/692 phosphorylation is essential but not sufficient for MAPKKK5 activation. Perhaps the phosphorylation of the N terminus is involved in the full activation of MAPKKK5. The N terminus negatively regulates MEKK1, YODA, and ANPs, and deletion of the N terminus results in constitutive active forms of these MAPKKKs (Kovtun et al., 2000; Asai et al., 2002; Lukowitz et al., 2004). Our LC-MS/MS analysis identified at least 16 phospho-sites in the MAPKKK5 N terminus and at least 11 phospho-sites in the MEKK1 N terminus (Supplemental Tables 1 and 3), although Phos-tag assays were unable to resolve a pattern-dependent phosphorylation of these sites. Indeed, a recent report showed that Ser-289 in the N terminus of MAPKKK5 is required for MAPKKK5-mediated immunity (Yan et al., 2018). It is possible that the phosphorylation of both the N and C termini coordinate MAPKKK5 and MEKK1 activation.
In summary, our findings show that MAPKKK3/5 are the MAPKKKs required for the activation of the major MAPK cascade downstream of diverse PRRs (Figure 9E). Upon pattern recognition, RLCK VII subfamily members directly phosphorylate MAPKKK3/5, as evidenced by the direct phosphorylation of MAPKKK5 Ser-599 by RLCK VII-4. This phosphorylation positively regulates MAPKKK3/5 activity for MPK3/6 activation. MPK3/6 further phosphorylate Ser-682/692 to enhance MAPKKK5 activity and to amplify MPK3/6 activation. In addition, MEKK1 is regulated in a similar manner to activate MPK4. The findings uncover two common mechanisms, namely, RLCK-mediated phosphorylation and MAPK-mediated feedback regulation on MAPKKKs, that are utilized by multiple PRRs for the activation of two MAPK cascades. MAPK cascades are common signaling modules that act downstream of receptor kinases in plants, but the regulatory mechanisms are poorly understood. The mechanisms described here may serve as a paradigm for diverse plant processes controlled by receptor kinases.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana single mutants used in this study include mapkkk5-1 (SAIL_1219_E11), mapkkk5-2 (SALK_122847), mapkkk3-1 (SALK_203147C), pbl27 (GABI_01C07), mekk1 (SAIL_167-G05), summ2-8, summ2-8 mekk1, and summ2-8 mpk4 (Zhang et al., 2012). The mapkkk3-2 mapkkk5-2 and mapkkk3-3 mapkkk5-2 double mutants were generated by introducing MAPKKK3 mutations into the mapkkk5-2 mutant background using CRISPR-CAS9 (see the Constructs and Transgenic Plants section). The rlck vii-4 sextuple mutant was described elsewhere (Supplemental Table 4) (Rao et al., 2018).
The Nicotiana benthamiana plants used for the luciferase-complementation assay and the Arabidopsis plants used for protoplast preparation and the pathogen infection assays were grown in soil at 23°C and 70% relative humidity with a 10/14-h day/night photoperiod for 4 to 5 weeks. Lighting was provided with white fluorescent bulbs (YZ28RR16; TOPSTAR) at 90 μE m−2 s−1.
Constructs and Transgenic Plants
To generate full-length and truncated constructs for transient expression in protoplasts, the corresponding cDNA fragments for MAPKKK5, MAPKKK3, MEKK1, and the other MAPKKKs were amplified from Col-0 total RNA by RT-PCR and cloned into pUC19-35S-HA-RBS1 vector (Li et al., 2005) using a ClonExpress II One Step Cloning Kit (no. C112; Vazyme Biotech). Mutant plasmids were generated by site-directed mutagenesis. To generate GST- and His-fusion proteins, the coding sequences were PCR amplified and cloned into pGEX-6p-1 and pET28a, respectively. For the MAPKKK5K375M-Cluc and PBLs-Nluc constructs, the cDNAs were amplified and cloned into pCAMBIA1300-Cluc and pCAMBIA1300-Nluc, respectively (Chen et al., 2008).
To generate the mapkkk3 mapkkk5 double mutants, two 20-bp sequences targeting MAPKKK3 were cloned into the BsaI site of AtU6-26-sgRNA-SK. The cassettes were obtained by digestion with SpeI and NheI, inserted into the SpeI position of pCAMBIA1300-pYAO:Cas9 (Yan et al., 2015), and introduced into the mapkkk5-2 mutant by Agrobacterium tumefaciens-mediated transformation (Clough and Bent, 1998).
To generate ProMAPKKK5:MAPKKK5-HA transgenic plants containing mutated phospho-sites, an 1892-bp fragment upstream of the start codon was PCR amplified from Col-0 genomic DNA, fused with the MAPKKK5 coding sequence, and cloned into pCAMBIA1300. Approximately 30 independent transgenic lines were identified by immunoblotting with an anti-HA antibody (CW0092M; CWBIO) at 1:4000 dilution, and lines with similar MAPKKK5-HA protein levels were chosen for further analysis. To generate ProMEKK1:MEKK1-FLAG transgenic plants, a genomic fragment spanning 1732 bp upstream of the start codon and the entire coding region of MEKK1 was PCR amplified and cloned into pCAMBIA1300. Transgenic plants expressing the transgene were identified by with an anti-FLAG immunoblots antibody (F1804; Sigma-Aldrich) at 1:4000 dilution.
Luciferase Complementation Assay
The luciferase complementation assay was performed as previously described (Chen et al., 2008). Leaves of 5-week-old N. benthamiana plants were infiltrated with Agrobacterium cells containing the indicated plasmids, and leaf discs were harvested 2 d after inoculation. The luminescence was recorded with a GLO-MAX 96 microplate luminometer. Each data point consisted of eight leaf discs.
MAPK Activation Assay
Ten-day-old Arabidopsis seedlings grown on 0.5× MS medium were sprayed with 100 nM flg22, 100 nM elf18, 100 nM pep2, or 200 μg/mL chitin (Sigma-Aldrich) containing 0.02% Silwet L-77 and frozen in liquid nitrogen. The protein was extracted using IP buffer (50 mM HEPES-KOH, pH 7.5, 150 mM KCl, 1 mM EDTA, 0.2% Triton X-100, 1 mM DTT, and 1× complete protease inhibitors [04693116001, Roche]). To investigate MAPK activation in protoplasts, the desired plasmids were transfected into protoplasts prepared from mapkkk3-2 mapkkk5-2 plants as described (Yoo et al., 2007), incubated for 12 h, and treated with 200 μg/mL chitin or water for 10 min. For each reaction, 10 μg plasmids were transfected in 4 mL 105 protoplasts. MAPK activation was detected by immunoblotting analysis with an anti-pERK monoclonal antibody (4370; Cell Signaling Technology) at 1:4000 dilution, and the Rubisco bands were stained with Ponceau as the loading control.
Phospho-Site Identification
To identify MAPKKK5 and MEKK1 phospho-sites in vivo, wild-type Arabidopsis protoplasts derived from ∼500 leaves were transfected with 2.4 mg MAPKKK5-FLAG or MEKK1-FLAG plasmid, treated with 100 nM flg22 for 10 min, and protein was extracted with 12 mL IP buffer. Total protein was incubated for 4 h with 70 μL agarose-conjugated anti-FLAG antibodies (A2220; Sigma-Aldrich), washed three times with IP buffer, and eluted in 60 μL 1 μg/μL 3×FLAG peptide. The eluted protein was separated in a 10% NuPAGE gel (Invitrogen) followed by mass spectrometry as previously described (Li et al., 2014).
To identify MAPKKK5 phospho-sites in vitro, the His-CERK1-KD and/or His-PBL19 recombinant proteins were incubated with GST-MAPKKK5-T protein in kinase reaction buffer (25 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM DTT, and 100 mM ATP) at 30°C for 30 min. The sample was separated in a 10% NuPAGE gel, and the GST-MAPKKK5-T band was excised and subjected to mass spectrometry.
Anti-Phospho-Peptide Antibodies and Detection of Site-Specific Phosphorylation in Vivo
MAPKKK5 and MEKK1 phospho-site-specific antibodies were produced by Abmart as previously described (Li et al., 2014). For anti-pSer599, the phosphopeptide was [CAERPTA(pS)MLL], and the control peptide was [CAERPTASMLL]. For anti-pSer682, the phosphopeptide was [CGTVNRL(pS)PRS], and the control peptide was [CGTVNRLSPRS]. For anti-pSer692, the phosphopeptide was [TLEAIP(pS)PCP], and the control peptide was [TLEAIPSPCP]. For anti-pSer603, the phosphopeptide was [CGGSGSA(pS)PLL], and the control peptide was [CGGSGSASPLL].
Approximately 60 10-d-old seedlings grown on 0.5× MS agar plates were treated with 200 μg/mL chitin or water for 10 min, and protein was extracted with 1 mL IP buffer. Total protein was incubated for 4 h with 50 μL protein A agarose (2854209; Merck Millipore) and 4 μg anti-HA antibody, washed five times with IP buffer, and boiled with loading buffer for 5 min. Site-specific phosphorylation was detected by immunoblotting analysis with anti-phospho-peptide antibodies at 1:500 dilution.
In Vitro Phosphorylation Assays
HIS- and GST-tagged recombinant proteins were expressed in Escherichia coli strain BL21 and affinity-purified with 200 μL glutathione-Sepharose and 200 μL Ni-Sepharose, respectively. For PBL19-mediated phosphorylation, 200 ng PBL19-HIS and 200 ng CERK1-KD-HIS were incubated with 2 μg GST-MAPKKK5-T or GST-MEKK1-T in 20 μL reaction buffer (25 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM DTT, and 100 mM ATP) for 30 min at 30°C, and the reaction was stop by adding SDS loading buffer. GST-MAPKKK5-T phosphorylation was detected by Phos-tag assay with an anti-GST monoclonal antibody (AB101-02; Tiangen) at 1:4000 dilution, while GST-MEKK1-T phosphorylation was detected by immunoblotting with anti-pSer603 antibodies at 1:500 dilution. For MPK-mediated phosphorylation, 200 ng MPK6-HIS and 50 ng MKK5DD-HIS were incubated with 200 ng GST-MAPKKK5-T in 20 μL reaction buffer for 30 min at 30°C, and GST-MAPKKK5-T phosphorylation was detected by immunoblotting analysis with anti-pSer682 and anti-pSer692 antibodies at 1:500 dilution.
Phos-Tag Assays
Protoplasts from wild-type and mutant plants were transfected with the indicated plasmids, treated with 100 nM flg22, 200 μg/mL chitin or water for 10 min, and protein was extracted with IP buffer without EDTA. The samples were subsequently analyzed using 10% SDS-polyacrylamide gels containing 100 μM MnCl2 and 50 μM Phos-tag Acrylamide AAL-107 (NARD Institute). After electrophoresis, the gel was soaked for 10 min in transfer buffer (20 mM Tris and 150 mM glycine) containing 1 mM EDTA, followed by 10 min in transfer buffer without EDTA. The proteins were then transferred to PVDF membrane, followed by immunoblot analysis with the desired antibodies.
RNA Isolation and qRT-RCR
Ten-day-old seedlings grown on 0.5× MS were sprayed with 100 nM flg22, 100 nM elf18, 100 nM Pep2, or 200 μg/mL chitin (Sigma-Aldrich) in 0.02% Silwet L-77 and harvested at 0 and 3 h. Total RNA was extracted from a pool of 10 seedlings using an RNeasy Plant Mini Kit (Qiagen). Reverse transcription was performed using a first-strand cDNA synthesis kit (Thermo), and PCR was performed (three technical replicates per gene; e.g., three PCR reactions from the same cDNA sample) using a SYBR Premix Ex Taq Kit (TaKaRa). Relative expression levels were normalized to the ACTIN1 (ACT1) housekeeping gene. The primers used for qRT-PCR analysis are listed in Supplemental Table 4.
Pathogen Infection Assays
Leaves of 4-week-old Arabidopsis plants were infiltrated with Pseudomonas syringae DC3000 at 5 × 105 colony-forming units/mL, and bacterial number in leaves was determined at 0 and 3 d after inoculation (Li et al., 2014). Each data point included eight plants for each genotype, and two leaf discs were collected from each plant as one sample.
For B. cinerea inoculation, the central vein of the leaf of a 4-week-old Arabidopsis plant was punctured with a needle. A droplet of conidia suspension at 5 × 105 conidia/mL in potato dextrose broth was placed on the wound, and lesion size was record at 3 d after inoculation. Each data point included 30 leaves per genotype.
Number of Experiments and Statistical Analyses
All experiments were independently performed at least three times, and the results were analyzed with one-way ANOVA with Tukey’s test (Supplemental Table 5).
Accession Numbers
Sequence data for the genes described in this article can be found in the TAIR database (https://www.arabidopsis.org) under the following accession numbers: At1g53570 for MAPKKK3, At5g66850 for MAPKKK5, At1g63700 for YODA, At4g08500 for MEKK1, At5g47070 for PBL19, and At5g18610 for PBL27.
Supplemental Data
Supplemental Figure 1. MAPKKK gene expression profiles and MAPKKK protein phosphorylation triggered by various patterns.
Supplemental Figure 2. Pattern-triggered MAPK activation is normal in mapkkk3 and mapkkk5 single mutants.
Supplemental Figure 3. CYP81F2 and NHL10 expression in Col-0 and mapkkk3 mapkkk5 mutant plants upon pattern treatment.
Supplemental Figure 4. Flg22-triggered phosphorylation of MAPKKK5 C terminus is normal in the rlck vii-4 mutant.
Supplemental Figure 5. Identification of phospho-sites in MAPKKK5.
Supplemental Figure 6. Phosphorylation of Ser-599 and Ser-682/692 is not required for the interaction of MAPKKK5 with PBL19 and MKK5.
Supplemental Figure 7. MEKK1 is required for pattern-induced MPK4 activation.
Supplemental Figure 8. Identification of phospho-sites in MEKK1.
Supplemental Table 1. List of MAPKKK5 phospho-peptides identified in vivo.
Supplemental Table 2. List of phospho-peptides in the MAPKKK5 C-terminal tail that is phosphorylated in vitro.
Supplemental Table 3. Phospho-peptides of MEKK1 identified in vivo.
Supplemental Table 4. Primers used for genotyping, cloning, and qRT-PCR.
Supplemental Table 5. ANOVA analyses.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Qi Xie for providing pCAMBIA1300-pYAO:Cas9 vector, Tsutomu Kawasaki for providing the mapkkk3-1 mutant, Shuqun Zhang for providing the conditional mpk3 mpk6 double mutant, and Yuelin Zhang for summ2-8, summ2-8 mpk4, and summ2-8 mekk1 mutants. The work was supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant XDB11020200) and the Chinese Ministry of Science and Technology (Grant 2015CB910200) to J.-M.Z.
AUTHOR CONTRIBUTIONS
J.-M.Z. and G.B. coordinated the research and wrote the article. G.B., Z.Z., and W.W. performed majority of the experiments. S.R. and X.Z. contributed to MAPK activation analyses. L.L., F.L.H.M., and S.C. performed mass spectrometry analyses. Y.W. performed bioinformatics analysis.
Footnotes
Articles can be viewed without a subscription.
References
- Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983. [DOI] [PubMed] [Google Scholar]
- Bayer M., Nawy T., Giglione C., Galli M., Meinnel T., Lukowitz W. (2009). Paternal control of embryonic patterning in Arabidopsis thaliana. Science 323: 1485–1488. [DOI] [PubMed] [Google Scholar]
- Bethke G., Unthan T., Uhrig J.F., Pöschl Y., Gust A.A., Scheel D., Lee J. (2009). Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc. Natl. Acad. Sci. USA 106: 8067–8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60: 379–406. [DOI] [PubMed] [Google Scholar]
- Boudsocq M., Willmann M.R., McCormack M., Lee H., Shan L., He P., Bush J., Cheng S.H., Sheen J. (2010). Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464: 418–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressendorff S., Azevedo R., Kenchappa C.S., Ponce de León I., Olsen J.V., Rasmussen M.W., Erbs G., Newman M.A., Petersen M., Mundy J. (2016). An innate immunity pathway in the moss Physcomitrella patens. Plant Cell 28: 1328–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Liang Y., Tanaka K., Nguyen C.T., Jedrzejczak R.P., Joachimiak A., Stacey G. (2014). The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife 3: e03766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.M. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung P.C., Campbell D.G., Nebreda A.R., Cohen P. (2003). Feedback control of the protein kinase TAK1 by SAPK2a/p38alpha. EMBO J. 22: 5793–5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D., Bauer Z., Regenass M., Boller T., Felix G. (2006). The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18: 465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J.D., Felix G., Boller T. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Costa L.M., et al. (2014). Central cell-derived peptides regulate early embryo patterning in flowering plants. Science 344: 168–172. [DOI] [PubMed] [Google Scholar]
- Dodds P.N., Rathjen J.P. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11: 539–548. [DOI] [PubMed] [Google Scholar]
- Feng F., Zhou J.M. (2012). Plant-bacterial pathogen interactions mediated by type III effectors. Curr. Opin. Plant Biol. 15: 469–476. [DOI] [PubMed] [Google Scholar]
- Feng F., Yang F., Rong W., Wu X., Zhang J., Chen S., He C., Zhou J.M. (2012). A Xanthomonas uridine 5′-monophosphate transferase inhibits plant immune kinases. Nature 485: 114–118. [DOI] [PubMed] [Google Scholar]
- Galletti R., Ferrari S., De Lorenzo G. (2011). Arabidopsis MPK3 and MPK6 play different roles in basal and oligogalacturonide- or flagellin-induced resistance against Botrytis cinerea. Plant Physiol. 157: 804–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., Liu J., Bi D., Zhang Z., Cheng F., Chen S., Zhang Y. (2008). MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 18: 1190–1198. [DOI] [PubMed] [Google Scholar]
- Heese A., Hann D.R., Gimenez-Ibanez S., Jones A.M., He K., Li J., Schroeder J.I., Peck S.C., Rathjen J.P. (2007). The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA 104: 12217–12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T., Laule O., Szabo G., Wessendorp F., Bleuler S., Oertle L., Widmayer P., Gruissem W., Zimmermann P. (2008). Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv. Bioinforma. 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura K., Casais C., Peck S.C., Shinozaki K., Shirasu K. (2006). MEKK1 is required for MPK4 activation and regulates tissue-specific and temperature-dependent cell death in Arabidopsis. J. Biol. Chem. 281: 36969–36976. [DOI] [PubMed] [Google Scholar]
- Kadota Y., Sklenar J., Derbyshire P., Stransfeld L., Asai S., Ntoukakis V., Jones J.D., Shirasu K., Menke F., Jones A., Zipfel C. (2014). Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell 54: 43–55. [DOI] [PubMed] [Google Scholar]
- Kong Q., Sun T., Qu N., Ma J., Li M., Cheng Y.T., Zhang Q., Wu D., Zhang Z., Zhang Y. (2016). Two redundant receptor-like cytoplasmic kinases function downstream of pattern recognition receptors to regulate activation of SA biosynthesis. Plant Physiol. 171: 1344–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun Y., Chiu W.L., Tena G., Sheen J. (2000). Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc. Natl. Acad. Sci. USA 97: 2940–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laluk K., Luo H., Chai M., Dhawan R., Lai Z., Mengiste T. (2011). Biochemical and genetic requirements for function of the immune response regulator BOTRYTIS-INDUCED KINASE1 in plant growth, ethylene signaling, and PAMP-triggered immunity in Arabidopsis. Plant Cell 23: 2831–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassowskat I., Böttcher C., Eschen-Lippold L., Scheel D., Lee J. (2014). Sustained mitogen-activated protein kinase activation reprograms defense metabolism and phosphoprotein profile in Arabidopsis thaliana. Front. Plant Sci. 5: 554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Meng X., Wang R., Mao G., Han L., Liu Y., Zhang S. (2012). Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genet. 8: e1002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Li M., Yu L., Zhou Z., Liang X., Liu Z., Cai G., Gao L., Zhang X., Wang Y., Chen S., Zhou J.M. (2014). The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15: 329–338. [DOI] [PubMed] [Google Scholar]
- Li X., Lin H., Zhang W., Zou Y., Zhang J., Tang X., Zhou J.M. (2005). Flagellin induces innate immunity in nonhost interactions that is suppressed by Pseudomonas syringae effectors. Proc. Natl. Acad. Sci. USA 102: 12990–12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., Ma X., Shan L., He P. (2013). Big roles of small kinases: the complex functions of receptor-like cytoplasmic kinases in plant immunity and development. J. Integr. Plant Biol. 55: 1188–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Zhang S. (2004). Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16: 3386–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Wu Y., Yang F., Zhang Y., Chen S., Xie Q., Tian X., Zhou J.M. (2013). BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc. Natl. Acad. Sci. USA 110: 6205–6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Wu S., Gao X., Zhang Y., Shan L., He P. (2010). A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA 107: 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W., Roeder A., Parmenter D., Somerville C. (2004). A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell 116: 109–119. [DOI] [PubMed] [Google Scholar]
- Meng X., Zhang S. (2013). MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 51: 245–266. [DOI] [PubMed] [Google Scholar]
- Menke F.L.H., van Pelt J.A., Pieterse C.M.J., Klessig D.F. (2004). Silencing of the mitogen-activated protein kinase MPK6 compromises disease resistance in Arabidopsis. Plant Cell 16: 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya A., Albert P., Shinya T., Desaki Y., Ichimura K., Shirasu K., Narusaka Y., Kawakami N., Kaku H., Shibuya N. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 19613–19618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nühse T.S., Peck S.C., Hirt H., Boller T. (2000). Microbial elicitors induce activation and dual phosphorylation of the Arabidopsis thaliana MAPK 6. J. Biol. Chem. 275: 7521–7526. [DOI] [PubMed] [Google Scholar]
- Pitzschke A. (2015). Modes of MAPK substrate recognition and control. Trends Plant Sci. 20: 49–55. [DOI] [PubMed] [Google Scholar]
- Rao S., Zhou Z., Miao P., Bi G., Hu M., Wu Y., Feng F., Zhang X., Zhou J.M. (2018). Roles of receptor-like cytoplasmic kinase VII members in pattern-triggered immune signaling. Plant Physiol. 177: 1679–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Liu Y., Yang K.Y., Han L., Mao G., Glazebrook J., Zhang S. (2008). A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 105: 5638–5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savatin D.V., Bisceglia N.G., Marti L., Fabbri C., Cervone F., De Lorenzo G. (2014). The Arabidopsis NUCLEUS- AND PHRAGMOPLAST-LOCALIZED KINASE1-Related Protein Kinases Are Required for Elicitor-Induced Oxidative Burst and Immunity. Plant Physiol. 165: 1188–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinya T., et al. (2014). Selective regulation of the chitin-induced defense response by the Arabidopsis receptor-like cytoplasmic kinase PBL27. Plant J. 79: 56–66. [DOI] [PubMed] [Google Scholar]
- Su J., Zhang M., Zhang L., Sun T., Liu Y., Lukowitz W., Xu J., Zhang S. (2017). Regulation of stomatal immunity by interdependent functions of a pathogen-responsive MPK3/MPK6 cascade and abscisic acid. Plant Cell 29: 526–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Rodriguez M.C., Adams-Phillips L., Liu Y., Wang H., Su S.H., Jester P.J., Zhang S., Bent A.F., Krysan P.J. (2007). MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol. 143: 661–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Li L., Macho A.P., Han Z., Hu Z., Zipfel C., Zhou J.M., Chai J. (2013). Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342: 624–628. [DOI] [PubMed] [Google Scholar]
- Tang D., Wang G., Zhou J.M. (2017). Receptor kinases in plant-pathogen interactions: more than pattern recognition. Plant Cell 29: 618–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Han Z., Sun Y., Zhang H., Gong X., Chai J. (2015). Structural basis for recognition of an endogenous peptide by the plant receptor kinase PEPR1. Cell Res. 25: 110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veronese P., Nakagami H., Bluhm B., Abuqamar S., Chen X., Salmeron J., Dietrich R.A., Hirt H., Mengiste T. (2006). The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 18: 257–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J., Zhang X.C., Neece D., Ramonell K.M., Clough S., Kim S.Y., Stacey M.G., Stacey G. (2008). A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20: 471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Ngwenyama N., Liu Y., Walker J.C., Zhang S. (2007). Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19: 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Xie J., Yan C., Zou X., Ren D., Zhang S. (2014). A chemical genetic approach demonstrates that MPK3/MPK6 activation and NADPH oxidase-mediated oxidative burst are two independent signaling events in plant immunity. Plant J. 77: 222–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Meng J., Meng X., Zhao Y., Liu J., Sun T., Liu Y., Wang Q., Zhang S. (2016). Pathogen-responsive MPK3 and MPK6 reprogram the biosynthesis of indole glucosinolates and their derivatives in Arabidopsis Immunity. Plant Cell 28: 1144–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., et al. (2016b). The Arabidopsis CERK1-associated kinase PBL27 connects chitin perception to MAPK activation. EMBO J. 35: 2468–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Yamashita-Yamada M., Hirase T., Fujiwara T., Tsuda K., Hiruma K., Saijo Y. (2016a). Danger peptide receptor signaling in plants ensures basal immunity upon pathogen-induced depletion of BAK1. EMBO J. 35: 46–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Huffaker A., Bryan A.C., Tax F.E., Ryan C.A. (2010). PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 22: 508–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H., Zhao Y., Shi H., Li J., Wang Y., Tang D. (2018). BRASSINOSTEROID-SIGNALING KINASE1 phosphorylates MAPKKK5 to regulate immunity in Arabidopsis. Plant Physiol. 176: 2991–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Wei S., Wu Y., Hu R., Li H., Yang W., Xie Q. (2015). High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol. Plant 8: 1820–1823. [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yuan G.L., Li H.J., Yang W.C. (2017). The integration of Gβ and MAPK signaling cascade in zygote development. Sci. Rep. 7: 8732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., et al. (2007). A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1: 175–185. [DOI] [PubMed] [Google Scholar]
- Zhang J., et al. (2010). Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7: 290–301. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Wu Y., Gao M., Zhang J., Kong Q., Liu Y., Ba H., Zhou J., Zhang Y. (2012). Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe 11: 253–263. [DOI] [PubMed] [Google Scholar]
- Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J.D., Boller T., Felix G. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760. [DOI] [PubMed] [Google Scholar]