Profiling wound-responsive gene transcriptomes in wild Solanum pennellii and domesticated S. lycopersicum sheds light on the contribution of cis-regulatory variation to stress-responsive gene expression divergence during species domestication.
Abstract
The evolution of transcriptional regulatory mechanisms is central to how stress response and tolerance differ between species. However, it remains largely unknown how divergence in cis-regulatory sites and, subsequently, transcription factor (TF) binding specificity contribute to stress-responsive expression divergence, particularly between wild and domesticated species. By profiling wound-responsive gene transcriptomes in wild Solanum pennellii and domesticated S. lycopersicum, we found extensive wound response divergence and identified 493 S. lycopersicum and 278 S. pennellii putative cis-regulatory elements (pCREs) that were predictive of wound-responsive gene expression. Only 24-52% of these wound response pCREs (depending on wound response patterns) were consistently enriched in the putative promoter regions of wound-responsive genes across species. In addition, between these two species, their differences in pCRE site sequences were significantly and positively correlated with differences in wound-responsive gene expression. Furthermore, ∼11-39% of pCREs were specific to only one of the species and likely bound by TFs from different families. These findings indicate substantial regulatory divergence in these two plant species that diverged ∼3-7 million years ago. Our study provides insights into the mechanistic basis of how the transcriptional response to wounding is regulated and, importantly, the contribution of cis-regulatory components to variation in wound-responsive gene expression between a wild and a domesticated plant species.
INTRODUCTION
Natural or artificial selection on diverse phenotypes leads to adaptation and domestication (Andersson, 2001; Doebley et al., 2006). Studies of the regulatory mechanisms underlying phenotypic diversity suggest that the variation in gene expression at the transcriptional level is one of the major contributing factors (Carroll, 2008; Romero et al., 2012). The divergent phenotypes between domesticated and wild plant species are the result of the domestication process in response to human selection (Doebley et al., 2006; Bauchet, 2012; Meyer and Purugganan, 2013; Chen et al., 2015). Comparisons of transcriptome profiles between domesticated and wild maize (Zea mays), carrot (Daucus carota), cotton (Gossypium hirsutum), and tomato (Solanum lycopersicum) species have revealed that the extensive changes of gene expression are associated with phenotypic differences between closely related wild-domesticated species pairs (Swanson-Wagner et al., 2012; Koenig et al., 2013; Ichihashi et al., 2014; Rong et al., 2014). However, it remains unclear to what extent regulatory mechanisms have diverged between domesticated and wild species.
Two of the major components of the transcription regulatory program are trans-acting factors such as DNA binding transcription factors (TFs) and cis-regulatory sites recognized by TFs (Kaufmann et al., 2010; Wittkopp and Kalay, 2011; Spitz and Furlong, 2012). The cis-regulatory sites are typically ∼6 to 15 bp in length and located in close proximity to their target genes. A TF generally recognizes multiple, slightly different cis-regulatory sites that are collectively referred to as a cis-regulatory element (CRE), representing the binding specificity of TFs (Wittkopp and Kalay, 2011). Thus, variation in gene expression may result from the differences in the cis-regulatory sites and/or the TFs that regulate the genes in question. In cross-species studies, CREs have been shown to evolve much slower than individual cis-regulatory sites that have undergone extensive divergence (Doebley and Lukens, 1998; Wray et al., 2003; Carroll, 2008; Romero et al., 2012). For example, CREs among the orthologous TFs from fruit fly, mouse, and human are highly conserved (Nitta et al., 2015). Similarly, by identifying sequence motifs resembling CREs from mouse and human based on DNase I footprints, >94% of the motifs are conserved (Stergachis et al., 2014). Because CREs are distinct TF binding motifs, these findings of CRE conservation indicate a high degree of conservation in trans-regulatory mechanisms. Meanwhile, only ∼20% of mouse DNase I footprints were colocalized with human footprints (Stergachis et al., 2014), suggesting extensive cis-regulatory site divergence. Since mouse and human were diverged ∼100 million years ago, the mammalian regulatory mechanism has significantly diverged cis-regulatory sites but highly conserved CREs and, thus, trans-acting components (Stergachis et al., 2014).
In plants, studies have shown that the divergence of cis-regulatory sites affects the transcript levels of key developmental regulators of multiple domestication traits (Doebley et al., 2006; Ichihashi et al., 2014; Swinnen et al., 2016). In addition, because artificial selection for these domestication traits created bottleneck, genes relevant to biotic/abiotic tolerance could be eliminated in domesticated species (Rosenthal and Dirzo, 1997; Chaudhary, 2013; Chen et al., 2015), contributing to significant divergence in stress response. As a result, the wild species preserves much of the genetic variation and presumably regulatory mechanisms underlying stress tolerance mechanisms (Hajjar and Hodgkin, 2007; Bauchet, 2012; Koenig et al., 2013; Bolger et al., 2014a). To understand how regulatory divergence contributes to stress tolerance traits, the response to wounding in domesticated and wild tomato species serves as a good model because of (1) their significant differences in stress tolerance (Bauchet, 2012; Koenig et al., 2013; Bolger et al., 2014a), (2) their divergence in transcriptional response to stress (Koenig et al., 2013; Bolger et al., 2014a), (3) the available information about the molecular underpinnings of responses to wounding in tomato (Howe and Jander, 2008; Howe and Schaller, 2008), and (4) knowledge of TFs and their corresponding cis-regulatory sites involved in regulating wound-responsive gene expression (Stanković et al., 2000; Boter et al., 2004). Nonetheless, the identities of most CREs and their corresponding cis-regulatory sites underlying stress tolerance regulation in tomato and most other plant species have not been comprehensively examined. It also remains unclear how wound-induced patterns of gene expression differ between domesticated and wild tomato species such as Solanum pennellii and how regulatory divergence contributes to divergence in wound transcriptional response between these species.
To assess the role of regulatory variation in gene expression divergence, one approach is to infer cis- and trans-regulatory divergence indirectly by comparing the differential gene expression of alleles between two parental lines and their F1 hybrid (Wittkopp et al., 2004; Emerson and Li, 2010). However, this strategy does not allow the exact CREs and the critical polymorphisms on binding sites to be evaluated. For this reason, we examined the regulatory mechanisms directly by identifying the CREs and CRE sites across species (Borneman et al., 2007; Sullivan et al., 2014; Nitta et al., 2015). To elucidate the regulatory mechanism divergence across species, we explored (1) to what extent the wound-responsive gene expression has diverged between S. lycopersicum and S. pennellii, (2) what CREs regulate differentially expressed genes between wound-treated and control samples from each species and between species, (3) to what degree CREs are relevant to wound-responsive gene expression conserved across species, and (4) to what extent differences in wound-induced transcriptional responses in these two tomato species are attributed to divergence in cis-regulatory sites.
RESULTS AND DISCUSSION
Temporal and Spatial Expression Profiles of Wound-Responsive Genes in Two Solanum Species
To globally examine how the effects of wounding on gene expression differ in S. pennellii and S. lycopersicum, leaves were wounded mechanically to trigger the response in damaged (local) and undamaged (systemic) tissues and at 0.5 and 2 h time points after wounding, each condition with three biological replicates. Control leaf tissue was collected from unwounded plants. The data reproducibility was high among replicates of all conditions (Supplemental Figure 1A). To evaluate the robustness of the wound-responsive gene expression profile revealed by RNA-seq (see Methods), the expression levels of several known wound-responsive genes were further examined by RT-qPCR (Supplemental Figure 1B). We found that the RNA-seq and RT-qPCR results were generally consistent, suggesting the robust and reliable expression profiles. Thus, in subsequent analysis, we included all conditions that provide replicates and use RNA-seq analyses to identify wound-responsive genes.
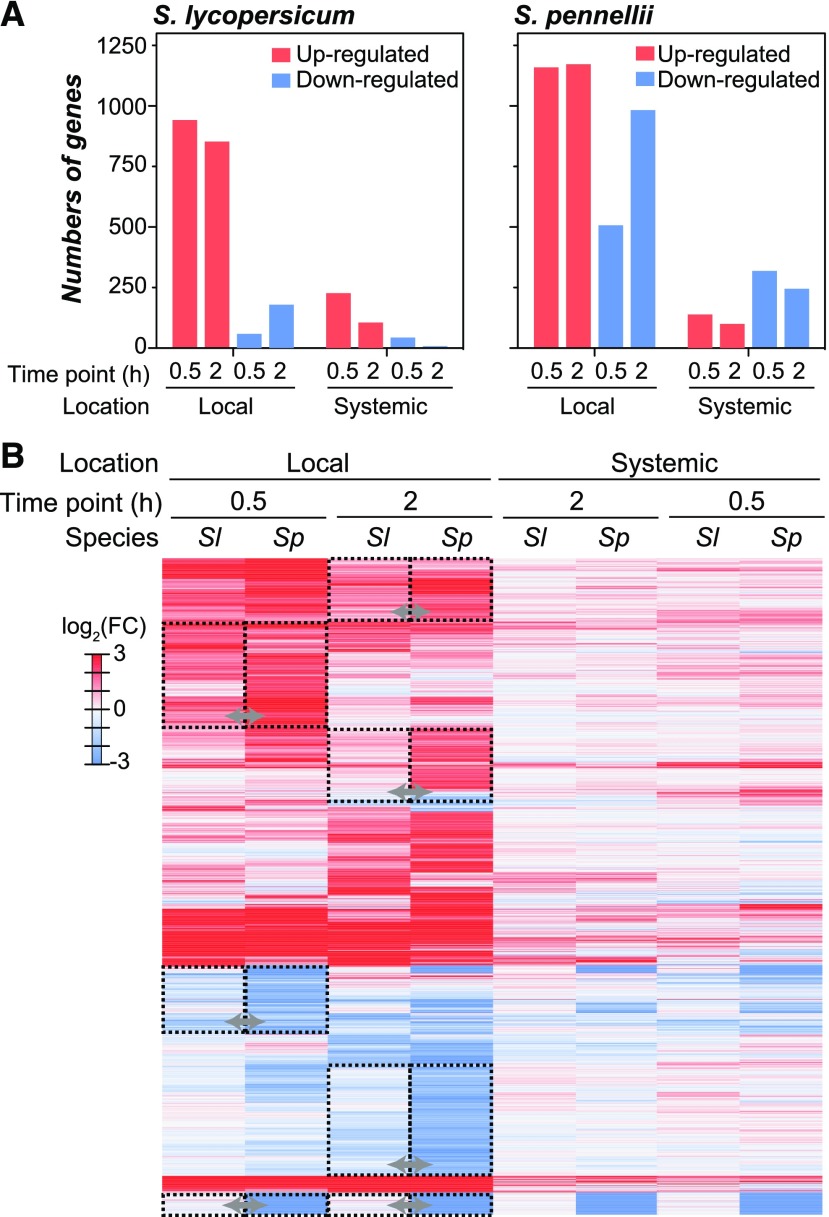
A gene is defined as wound responsive if it is either significantly up- or downregulated [multiple-testing adjusted P < 0.05, |log2(FC)| > 2; fold change (FC)] in a wounded sample compared with the unwounded control. To increase the stringency of our analysis, we chose a FC threshold of 4-fold instead of the conventional twofold to emphasize robust changes in gene expression. This is also because we found that cis-element finding was more fruitful with more robustly differentially expressed genes. In both species, ∼1000 genes were significantly upregulated by wounding (wound-induced) in local leaves during both time points (Figure 1A). Interestingly, the pattern is very different for downregulated genes where, at 0.5 h in the local tissue, there were only 59 downregulated S. lycopersicum genes compared with 507 in S. pennellii (Figure 1A). Similarly, at 2 h in the local tissue, 179 S. lycopersicum genes were downregulated (Figure 1A, left panel) compared with 983 in S. pennellii (Figure 1A, right panel). Similar patterns were also observed for wound-responsive genes identified with the more conventional 2-fold change (Supplemental Figure 2A). In the systemic tissue, far fewer genes were differentially expressed in both species, with S. pennellii having more systemically responsive genes than the cultivated species (353 in S. lycopersicum and 555 in S. pennellii) (Figure 1A). Approximately 52% and 81% of these systemic wound-responsive genes in S. lycopersicum and S. pennellii, respectively, were a subset of the local wound-responsive genes, similar to previous microarray studies (Scranton et al., 2013), indicating similar wound responses between the local and systemic leaf. Taken together, these findings show that in response to wounding, both species have extensive changes in gene expression programs, but the extent of gene expression repression is more prominent in S. pennellii.
Figure 1.
Similarities and Differences in Wound-Responsive Gene Expression between Tomato Species.
(A) Number of significantly differentially regulated genes (|log2(FC)| > 2) upon mechanical wounding in local and systemic leaves of S. lycopersicum (Sl) and S. pennellii (Sp) for the indicated time points [hour(s)] after wounding.
(B) Differential gene expression values of orthologous genes (rows) in different location/species/time points (columns). Only orthologous genes significantly up- or downregulated in ≥1 sample were included (n = 2199). Dashed boxes and arrows indicate clusters of orthologous genes with inconsistent regulatory patterns across species in local tissues.
To assess in more detail how S. lycopersicum and S. pennellii differ in their wound response, orthologous genes that are wound responsive (n = 2199) in any time point or tissue (i.e., local or systemic) in ≥1 species were compared. Hierarchical clustering of the overall expression patterns showed that the samples were clustered first based on the treatment location (local or systemic) and then by time points (0.5 or 2 h) and species (Figure 1B), indicating that the spatial response has higher impact over the species origins or the duration of treatment on wound-responsive gene expression. Nonetheless, although the overall patterns of up- and downregulation are similar between species, there are important differences. In the local leaves at both time points, S. pennellii genes had higher amplitude of differential expression (higher absolute FC values) compared with their S. lycopersicum orthologs (Figure 1B, dashed boxes). Thus, S. pennellii apparently responds to wounding earlier and stronger than S. lycopersicum, which is similar to the heightened tolerance to drought and salt in S. pennellii compared with S. lycopersicum (Tal and Shannon, 1983; Gong et al., 2010; Koenig et al., 2013; Bolger et al., 2014a).
Coexpression Clustering and Functions of Wound-Responsive Genes
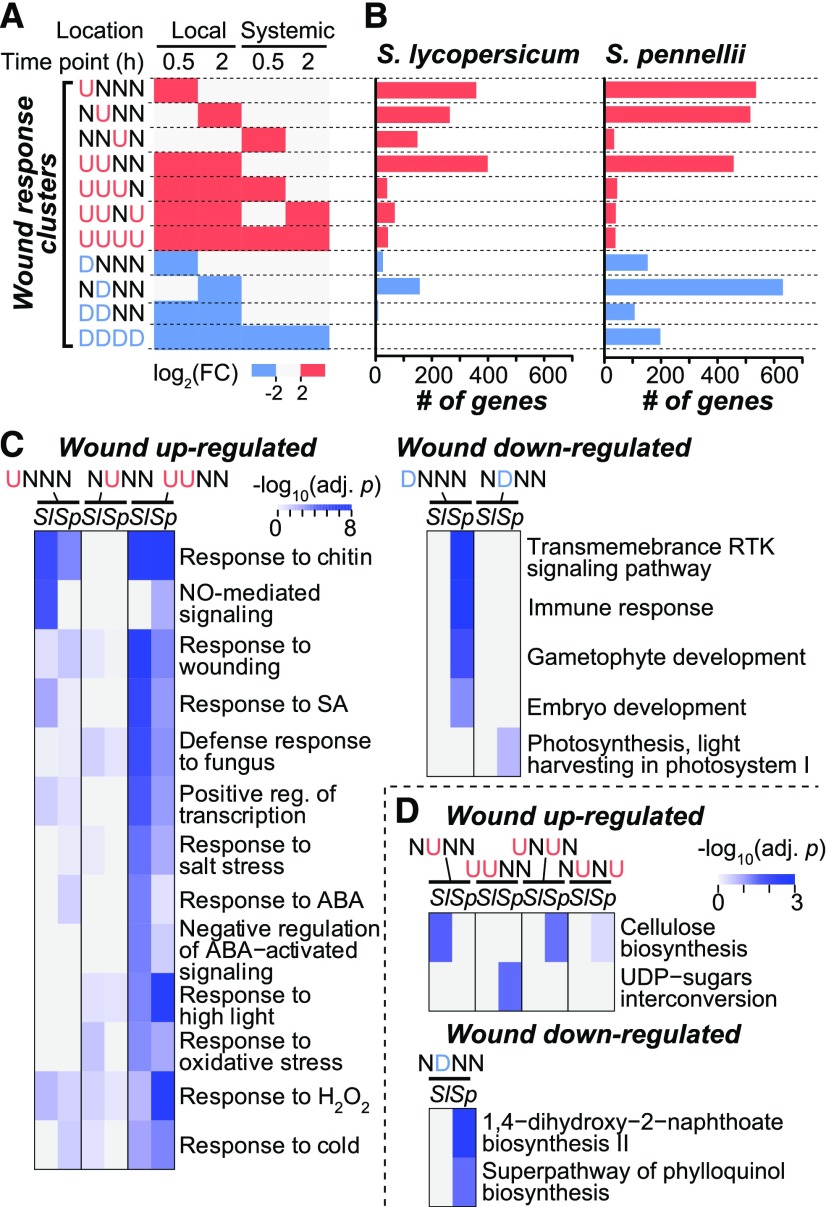
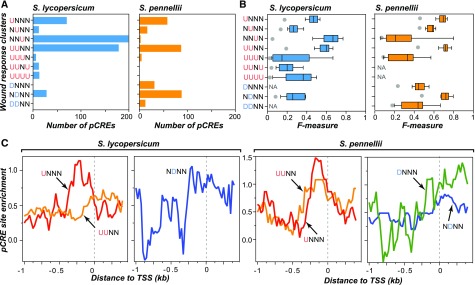
The overall transcript profile showed that wound-responsive genes differed significantly between species and could be classified into categories according to the time of treatment and spatial location of the response (Figure 1). To further investigate how the wound response may have functionally diverged between species, we first categorized a wound-responsive gene from a species into one of 81 “wound response clusters” based on whether the gene in question is upregulated (U), nonregulated (N), and downregulated (D) in response to wounding at a given time/location (major clusters shown in Figure 2A; all clusters comprising <2% of wound-responsive genes in Supplemental Data Set 1). For example, a gene is categorized in the UUDN cluster if it is upregulated at both 0.5 and 2 h in the local wounded leaf, downregulated at the 0.5 h time point in the systemic undamaged leaf, and not changed significantly in the 2 h systemic response. Among the major wound-induced clusters (Figure 2A, red), the UNNN, NUNN, and UUNN clusters were the largest with >250 genes in both species (Figure 2B; Supplemental Data Set 1). The number of upregulated genes in these three major clusters was greater in S. pennellii than in S. lycopersicum. Similarly, the number of genes in the four major wound-repressed clusters (Figure 2A, blue) was greater in S. pennellii (Figure 2B). The same tendency was also observed when differential expression was defined as |log2(FC)| > 1 (Supplemental Figures 2B and 2C). Taken together, these findings suggest that S. pennellii has a more dynamic wound response, particularly in the case of downregulated genes.
Figure 2.
Numbers of Genes and Functional Category Enrichments in Wound Response Clusters.
(A) Definitions of wound response clusters. U (red), upregulation (log2FC) > 2; N (gray), no significant change, 2 > (log2FC) > −2; D (blue), downregulation (log2FC) < −2. Only clusters with >40 genes in ≥1 species were shown.
(B) Numbers of wound-responsive genes in the clusters shown in (A) for S. lycopersicum (left) and S. pennellii (right). Red and blue, up- and downregulated clusters.
(C) GO biological process categories significantly enriched in wound upregulated (adjusted P values < 1e-03) and downregulated (adjusted P values < 1e-02) cluster genes from S. lycopersicum (Sl) or S. pennellii (Sp).
(D) Metabolic pathways significantly enriched (adjusted P values < 5e-02) in S. lycopersicum and S. pennellii genes from wound up- and downregulated clusters. Deeper shades of blue indicate higher −log10(adjusted P value).
Considering the differences in wound-responsive gene expression between S. lycopersicum and S. pennellii (Figures 1 and 2), we assessed the function of wound-responsive genes in each wound response cluster with Gene Ontology (GO) and metabolic pathway annotations (see Methods). Wounding activates broad-spectrum defense responses in tomato (Green and Ryan, 1972; Howe and Jander, 2008; Howe and Schaller, 2008). Consistent with previous findings (Howe and Schaller, 2008; Scranton et al., 2013), the wound upregulated genes in local leaves, especially those in the UNNN and UUNN clusters, were significantly enriched in genes responsive to multiple biotic and abiotic stresses, including those mediated by the stress hormones salicylic acid and abscisic acid [Figure 2C; also true for genes with log2(FC) > 1; Supplemental Figure 2D]. Notably, most biological processes were more significantly enriched in S. lycopersicum than in S. pennellii for the genes with log2(FC) > 2 (Figure 2C), but not in genes with log2(FC) > 1 (Supplemental Figure 2D). This result suggests that, while the defense-related genes were wound-induced both in domesticated and wild species, wound stress results in higher degrees of gene induction and/or a proportionally higher number of defense-related genes in the domesticated tomato than that in the wild species.
Although there was a large number of wound downregulated genes (Figure 1), only two clusters (DNNN and NDNN) containing S. pennellii genes were significantly enriched in plant growth-related GO categories, including photosynthesis (Figure 2C). This is consistent with previous studies showing the trade-offs between growth and stress tolerance in wild species (Huot et al., 2014). The metabolic pathway analyses further showed that genes in the NDNN clusters in S. pennellii were significantly enriched in phylloquinone biosynthesis (Figure 2D). Phylloquinone is an integral part of the photosynthetic electron transport chain (Nowicka and Kruk, 2010). The reduction in the expression levels of genes associated with photosynthetic efficiency suggests an antagonistic relationship between defense response and plant growth in S. pennellii (Figure 2C). In addition, photosynthesis-related functional categories were enriched in wound-repressed genes with log2(FC) < −1 in S pennellii (NDNN cluster in Supplemental Figure 2E), further supporting the trade-offs between growth and stress tolerance in wild tomato, a pattern that was not apparent in the domesticated species.
Taken together, our findings show that wound response genes can be categorized into a few dominant clusters (Figures 2A and 2B). Because some orthologs have differing responses to wounding (Figure 1B), the identity and the enrichment test statistics of some GO categories and metabolic pathways also differ (Figures 2C and 2D). Nonetheless, the number of GO categories and metabolic pathways enriched in genes up- or downregulated in either species was small. This was particularly true for S. pennellii downregulated genes. Since only the orthologs were included in the gene set enrichment analyses (see Methods), the small numbers of GO categories recovered may be due to the lower gene number in a cluster, which consequently decreases statistical power.
Divergence of Wound Responses among Orthologous Genes
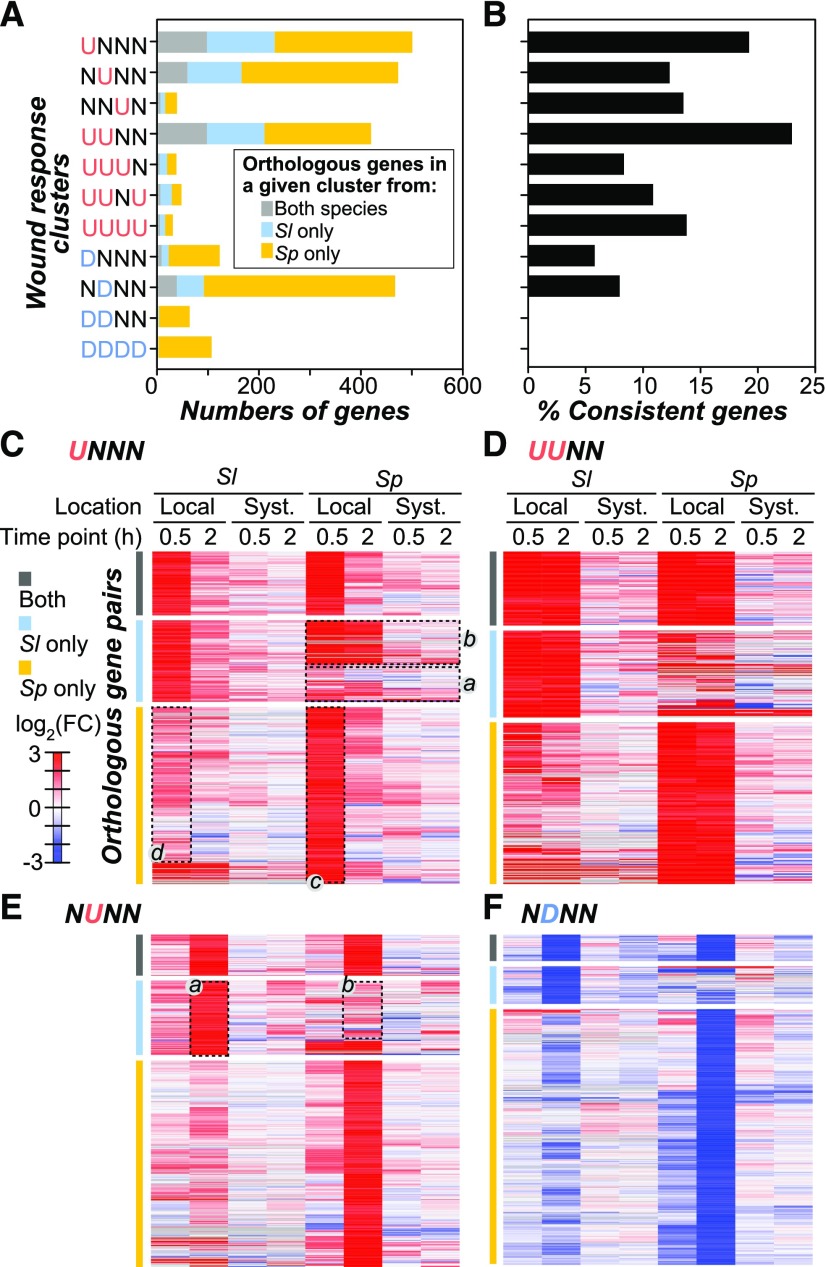
Previous work in maize and tomato has suggested that the domestication process or the adaptation to extreme environments may result in extensive changes in the transcriptional regulation of genes controlling relevant morphological and physiological traits (Swanson-Wagner et al., 2012; Koenig et al., 2013). Our findings showed that there were substantial differences in the wound-responsive expression of S. lycopersicum and S. pennellii genes, as well as differences in the biological processes represented by these genes (Figure 2). One immediate question is to what extent the orthologous genes in these two species differ in their wound response. To address this question, we first assessed which putative orthologous genes (see Methods) have consistent wound response patterns (i.e., both orthologs are in the same wound response cluster; Figure 3A). These genes are referred to as “consistent genes.” Interestingly, depending on the cluster (Figure 3A), only 0 to 24% orthologs were considered consistent (Figure 3B). These results showed that 76 to 100% of the wound-responsive orthologous genes were in different clusters and thus differentially regulated between species. Upon examination of the orthologous gene expression patterns side-by-side between species, some orthologous pairs had substantially different responses (Figures 3C to 3F, cyan and orange bar). For example, in the UNNN cluster (Figure 3C), in 47% of cases the S. pennellii orthologous genes were either in the NNNN cluster (Figure 3C, dotted rectangle a) or in the UUNN cluster (Figure 3C, dotted b). The pattern of low consistency (<25%) in ortholog expression was also observed when genes with |log2(FC)| > 1 were used (Supplemental Figures 2F and 2G). These results suggest that wound responses have diverged among the majority of orthologs in the past 3 to 7 million years (Nesbitt and Tanksley, 2002; Kamenetzky et al., 2010).
Figure 3.
Divergence of Wound Responses among Orthologous Genes.
(A) Number of orthologous genes with |log2(FC)| > 2 in the wound response clusters as defined in Figure 2A. Gray, orthologous genes from both species were in the same cluster; cyan, the S. lycopersicum (Sl) ortholog is in the indicated cluster but not the S. pennellii one; orange, the S. pennellii (Sp) ortholog is in the indicated cluster but not the S. lycopersicum one.
(B) Percentage of the orthologous genes that are considered to have consistent regulatory patterns (in the same cluster) in each cluster.
(C) to (F) Heat maps showing the differential expression levels [log2(FC)] of orthologous genes in UNNN (C), UUNN (D), NUNN (E), and NDNN (F) clusters. The bars on the left of each heat map are colored the same way as in (A). The dotted rectangles highlight differential expression patterns discussed in the main text.
To assess the extent to which the wound response differed between orthologous genes, we compared the wound-induced gene expression levels of “inconsistent orthologs,” defined as orthologous gene pairs not in the same wound response cluster, over the tested durations/tissues in the four largest clusters. In most cases, although the S. lycopersicum and the S. pennellii genes in inconsistent ortholog pairs belonged to different clusters, both orthologs were responsive but at different levels. For example, in the UNNN cluster in which only the S. pennellii genes were significantly upregulated (above threshold) at 0.5 h in the local leaves (Figure 3C, dotted rectangle c), the corresponding S. lycopersicum orthologs were also upregulated but at levels below the threshold (Figure 3C, dotted rectangle d). Similarly, in the NUNN cluster where only the S. lycopersicum orthologs were significantly upregulated (Figure 3E, dotted rectangle a), the expression of most corresponding S. pennellii orthologs was also induced but at levels below threshold (Figure 3E, dotted rectangle b). This pattern was also true for downregulated genes (Figure 3F, cyan and orange bars). Given that most orthologs were wound responsive but at different levels, the ancestral genes of these orthologs were likely wound responsive as well. Thus, when the wound response of orthologous genes diverges, the divergence is not typically due to complete loss or gain of response but more likely due to diverging levels of responsiveness.
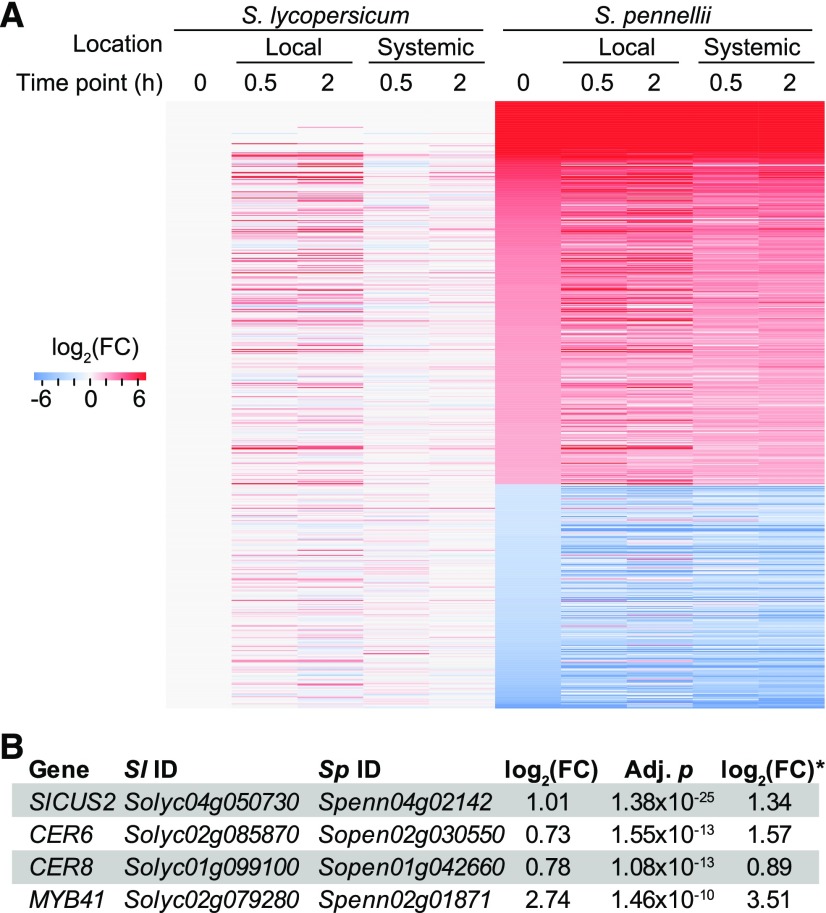
To this point, our analysis focused on differential expression by comparing wounded leaves to unwounded, control leaves. Although induced gene expression is important for kickstarting defense systems in unfavorable environments (Green and Ryan, 1972; Howe and Jander, 2008; Howe and Schaller, 2008), constitutive defenses also contribute to plant resilience to environmental stress (Wittstock and Gershenzon, 2002). Using the S. lycopersicum gene expression level as a reference, we identified 374 and 219 S. pennellii genes that were expressed at significantly higher and lower levels, respectively, than their cultivated tomato orthologs (Figure 4A). This finding indicates that significant differences in gene expression already exist between the two species prior to wounding, contributing to divergence in constitutive defense. For example, cuticular wax and cutin biosynthesis genes CER6, CER8, MYB41, and SICUS2 (Hooker et al., 2002; Cominelli et al., 2008; Lü et al., 2009) were expressed at higher levels in S. pennellii (Figure 4B), consistent with findings of earlier studies (Bolger et al., 2014a). Given that expression levels are already different between the control samples, it is possible that a gene contributing to constitutive defense will have a consistently high expression level before and after wounding. To assess this, we also compared the gene expressions in wound-treated samples in both species against the S. lycopersicum unwounded control. A surprising pattern was that, if a S. pennellii gene had a significantly different (either higher or lower) expression level in unwounded control compared with that of its S. lycopersicum ortholog under control condition, the S. pennellii gene in question tended to remain significantly different in a consistent fashion after wounding in both time points and in both local and systemic tissues (Figure 4A). This finding supports the hypothesis that the basal level of defense response is stronger in S. pennellii (Koenig et al., 2013; Bolger et al., 2014a).
Figure 4.
Genes Differentially Expressed between Species prior to Wounding.
(A) Heat map showing differential expression where FC values of all samples were calculated using the S. lycopersicum unwounded control (time point 0) expression values as the denominator. Only genes in the unwounded control in S. pennellii with significant FC values in comparison to the unwounded control in S. lycopersicum were shown [n = 593, |log2(FC)| > 2].
(B) Differential expression values and test statistics contrasting S. pennellii and S. lycopersicum unwounded controls between orthologous gene pairs from both species involved in biosynthesis of cuticular wax and cutin in this and an earlier study (indicated by an asterisk; Bolger et al., 2014a).
Putative cis-Regulatory Sequences Controlling Wound-Responsive Gene Regulation
The expression patterns in control and wounded tissue between S. lycopersicum and S. pennellii orthologous genes have diverged substantially, suggesting divergence of regulatory mechanisms central to controlling wound-responsive gene expression. Substitutions in cis-regulatory sites may lead to expression divergence due to the inability of orthologous TFs to bind to the site with substitutions. Alternatively, expression divergence may be due to substantial changes in cis-regulatory sites such that the orthologous gene is now bound by a different TF. To assess these two mechanisms, we first need to know what the CREs (representing the TF binding specificity) are and where they are located in the genome. We identified globally the CREs likely controlling wound-responsive gene expression for cross-species comparison with an enriched k-mer approach (an oligomer with the length k ≥ 5 bp; see Methods).
Since the sites of CREs may be located in both the promoter and 5ʹ untranslated regions (UTRs) of a gene (Sullivan et al., 2014), we queried whether an enriched k-mer sequence is located near the transcriptional start sites (TSSs; see Methods) of member genes in each cluster. Zero to hundreds of k-mers were found to have significantly enriched numbers of sites among genes in wound response clusters relative to nonresponsive genes (Figure 5A). These enriched k-mers are referred to as putative CREs (pCREs). The pCREs identified include ones that resemble known CREs relevant to the wound response, including abscisic acid response element, W-box, and G-box (Rushton and Somssich, 1998; Hobo et al., 1999; Sibéril et al., 2001; Boter et al., 2004; Adie et al., 2007), as well as those that do not resemble known CREs (Supplemental Figure 3). To further assess how well these pCREs can jointly explain the wound response in each cluster, we applied a machine-learning algorithm, support vector machine (SVM; see Methods), to predict wound-responsive expression of genes in each wound response cluster based on identified pCREs. Among the 10 clusters with pCREs in S. lycopersicum and/or S. pennellii (Figure 5A), the wound response prediction models based on pCREs performed significantly better than randomly expected (box plots versus gray spot, Wilcoxon signed rank test, all P < 0.01; Figure 5B; Supplemental Figure 4A). In addition, our k-mer approaches led to a differential expression prediction model that outperformed the model built with motifs from the commonly used Multiple EM for Motif Elicitation (MEME; Bailey et al., 2009) (Supplemental Figure 3G; see Methods). These results showed that our approach could efficiently identify short sequences resembling CREs because they are predictive of wound response in multiple clusters. In addition, the pCREs from clusters involving wound-induced expression (e.g., UNNN [red] and UUNN [orange]; Figure 5C) tend to be located within 500 bp upstream of the TSS, consistent with the finding that plant TFs tend to bind preferentially in the upstream region close to TSSs (Franco-Zorrilla et al., 2014; Heyndrickx et al., 2014).
Figure 5.
Evidence Indicating Biological Relevance of Putative CREs.
(A) Number of pCREs identified through the k-mer pipeline (see Methods) for each wound response cluster in S. lycopersicum (blue) or S. pennellii (orange). Only the clusters with pCREs in ≥1 species are shown.
(B) Box plot showing the wound response prediction performance (F-measure) based on a model using pCREs identified from genes in a wound response cluster. F-measure: the harmonic mean of precision (proportion predicted correctly) and recall (proportion true positives predicted). The maximum F-measure is 1, indicating a perfect model. For each wound response cluster, 10 F-measures were calculated from 10-fold cross-validation and are shown as a box plot. Gray dot: the average F-measure of 10,000 random predictions indicating the performance of a meaningless model. NA, not applicable since no pCRE was found in the cluster.
(C) Enrichment of sites of pCREs identified from four different clusters. For each pCRE, the degree of enrichment of its sites around TSSs was represented as the log2 ratio between pCRE site frequencies of genes in a cluster and frequencies of the same pCREs in genes not responsive to wounding. This log ratio was generated for each pCRE in the region from 1 kb upstream to 0.5 kb downstream of TSSs with a sliding window of 100 bp and a step size of 25 bp. For each cluster, the median log ratios of all pCREs identified from the cluster in question was shown.
In contrast to pCREs involved in upregulation, the pCREs identified in wound downregulated clusters (NDNN [blue] and DNNN [green]; Figure 5C) tend to be located downstream of TSSs, including 5ʹUTRs. This is similar to the 5ʹUTR of excision repair cross complementation group-1 gene in human that contains binding sites for a transcription repressor (Yu et al., 2001). Similarly, the cyclin D1 inhibitory element within the 5ʹUTRs represses the expression of the human cyclin D1 gene in an age-dependent manner (Berardi et al., 2003). Nonetheless, we discovered no pCRE from the DDDD cluster, suggesting the potential role of posttranscriptional regulation such as transcript turnover (Narsai et al., 2007) in repression control of these genes. Taken together, the pCREs identified are predictive of wound-responsive gene expression in most clusters and have a position bias resembling the known TF binding sites, suggesting that they are authentic cis-elements in regulating gene expression.
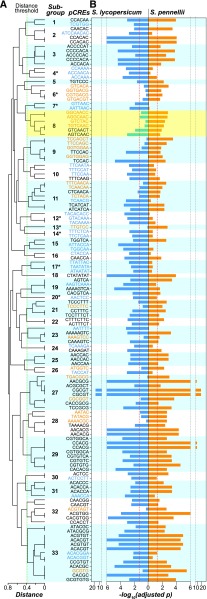
Divergence of Putative CREs between Tomato Species
To assess the degree of regulatory divergence across tomato species, we first examined if similar pCREs are regulating S. lycopersicum and S. pennellii genes with similar wound response patterns. This was accomplished by asking whether a pCRE is consistently enriched in a wound response cluster in both species. If a pCRE is consistently enriched, the pCRE in question is likely a component of a conserved wound response regulatory program. We found that 24 to 52% of pCREs in UNNN, UUNN, and NDNN clusters were consistently enriched between species (Supplemental Data Set 2 for the UNNN cluster, pCREs in black; Figure 6A), suggesting their conserved role in wound response regulatory programs. This result also showed that the remaining 48 to 76% of pCREs, depending on the wound response cluster, were species-specifically enriched (pCREs in blue or orange, Figures 6A and 6B), indicating substantial divergence in regulatory programs. The presence of species-specific pCREs raises the question of whether they are (1) bound by the same sets of orthologous TFs that bind cis-regulatory sites with subtle differences between species (Zhang et al., 2006) or (2) bound by nonorthologous TFs between species. To assess the above possibilities, we first defined two sets of species-specific pCREs as those that were enriched only in S. lycopersicum and only in S. pennellii genes within a cluster, respectively. Next, we asked whether these two sets of species-specific pCREs could be bound by TFs from the same family. We adopted this conservative approach to ensure that we could provide a lower-bound estimate of the proportion of species-specific pCREs that are bound by distinct TFs across-species. We should also emphasize that the pCREs, including the species-specific ones, were identified first based on their enrichments in the putative promoters of genes in wound response clusters relative to nonresponsive genes. Thus, these species-specific pCREs are likely relevant to species-specific wound response regulation, a point supported based on modeling results in the next section.
Figure 6.
Differential Enrichment of pCREs in UNNN Cluster Genes from Two Tomato Species.
(A) Dendrogram showing the distances between the pCREs identified from UNNN cluster genes and enriched in S. lycopersicum only (blue), S. pennellii only (orange), or both species (black). The dotted line indicates the threshold distance defined based on the 95th percentile distances between binding motifs of TFs from distinct families and defines multiple pCRE subgroups (numbered) where each subgroup contains pCREs likely bound by TFs of the same family (distance threshold = 0.39). Single-species subgroups with pCREs from only one species are labeled with asterisks. Note that some pCRE duplicates were due to their identification from both S. lycopersicum and S. pennellii.
(B) Degrees of pCRE site enrichment in S. lycopersicum (blue) and S. pennellii (orange) UNNN genes. Adjusted P value: multiple testing corrected P value. Dashed line, adjusted P < 0.05. Yellow box: pCREs similar to the W-box element.
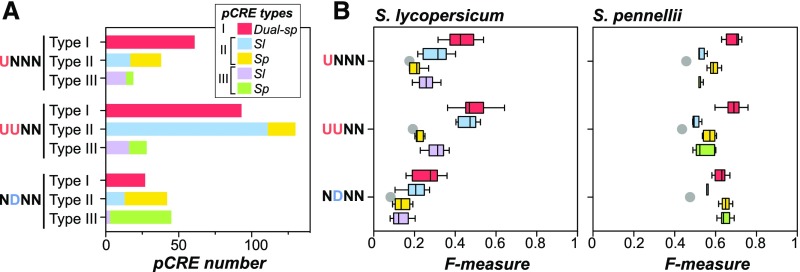
Using in vitro TF binding data (see Methods), we divide pCREs into subgroups where pCREs in a subgroup are likely bound by TFs of the same family (Figure 6A; Supplemental Figure 5). For example, pCREs that were enriched in UNNN wound-responsive genes from ≥1 species could be divided into 33 pCRE subgroups (Figure 6A). A subgroup was defined as “dual-species” if it contained pCREs from both species. By contrast, if all pCREs in a subgroup came from only one species, this subgroup was then designated as “single-species” (Figure 6A, asterisk; Supplemental Figure 5). Together with whether a pCRE was enriched in the putative promoter regions of wound-responsive genes in one or both species, we classified pCREs into three types (Figure 7A): (1) Type I, a pCRE is enriched in both species and belongs to a dual-species subgroup; (2) Type II, a pCRE is enriched only in one species but belongs to a dual-species subgroup; and (3) Type III, a pCRE is enriched only in one species and belong to a single-species subgroup. We should emphasize that Type I, II, and III pCREs are bound by TFs with increasingly divergent binding specificities. We have shown that 24 to 52% pCREs were Type I enriched in both species (Supplemental Data Set 2). Type II pCREs were found in 32, 52, and 37% of subgroups in UNNN, UUNN, and NDNN clusters, respectively (Figure 6; Supplemental Figure 5). In the UNNN cluster, for example, the consensus sequence of six pCREs in the 8th subgroup is GTTGACT (Figure 6, yellow box) similar to the W-box (TTGAC[C/T]) recognized by WRKY TFs that mediate biotic and abiotic stress responses (van Verk et al., 2008; Banerjee and Roychoudhury, 2015). Among these six pCREs, AGTCAAC and GTCAACT were enriched in both species, whereas the remaining pCREs were enriched specifically in S. pennellii. This indicates the conserved role of the same TF family across species in triggering wound responses but also implies the regulatory divergence at the level of individual TF binding cis-regulatory elements.
Figure 7.
Performance of the Type I, II, and III pCREs in Predicting Wound Response.
(A) Numbers of pCREs that were consistently enriched in both species and belong to a dual-species subgroup (red, Type I), belong to “dual-species” subgroup but were specifically enriched in S. lycopersicum (blue, Type II from Sl) or in S. pennellii (orange, Type II from Sp), and belong to “single-species” subgroup and were specifically enriched in S. lycopersicum (purple, Type III from Sl) or in S. pennellii (green, Type III from Sp) in three example wound response clusters.
(B) Box plot showing the wound response prediction performance (F-measure) based on a model using the pCRE sets in (A). For each wound response cluster, 10 F-measures were calculated from 10-fold cross validation and shown as a box plot. Gray dot: the average F-measure of 10,000 random predictions indicating the performance of a meaningless model.
Compared with Types I and II, there are relatively fewer Type III pCREs. Among the largest clusters, 16, 11, and 39% pCREs in UNNN (Figure 6A), UUNN (Supplemental Figure 5A), and NDNN (Supplemental Figure 5C) clusters were Type III (Supplemental Data Set 2). In the UNNN cluster, for example, 14 Type III pCREs were enriched only in S. lycopersicum wound-responsive genes and belonged to a subgroup with only S. lycopersicum pCREs (Figure 6A, blue and asterisk; Supplemental Data Set 2), suggesting these pCREs are specific to S. lycopersicum and likely bound by specific S. lycopersicum TFs where their S. pennellii orthologs are either absent or do not bind. Note that the subgroups were defined to ensure pCREs bound by TFs from the same family can be correctly identified but errs on the side of not calling pCREs truly regulated by distinct TFs. Thus, the 16% represents the lower bound in terms of the degree of regulatory divergence involving pCREs bound by nonorthologous TFs in regulating the UNNN wound response cluster between these two species.
To further assess the regulatory divergence of pCREs on wound response, we examined the enrichment of the species-specific pCREs (Types II and III) among inconsistent orthologs (Figure 3). We found that the species-specific pCREs enriched within a wound response cluster in a particular species were significantly enriched among inconsistent orthologous genes from the species in question but not in the other species (Supplemental Figures 6B and 6C). This finding further supports the species-specific nature of these pCREs and their positive correlation with expression divergence. Taken together, while S. lycopersicum and S. pennellii may have similar pCREs to control wound-induced gene expression, there are distinct preferences of pCREs for wound response across species, supporting the presence of both regulatory conservation and divergence.
Relationship between pCRE Conservation and Gene Regulation across Species
We show that wound response pCREs differ in their enrichment in genes between species and in whether they can be recognized by TFs from the same family (Figure 6; Supplemental Figure 5). Based on their enrichment and subgroup memberships, they can be classified into three types (Figure 7A; Supplemental Data Set 2). To assess which types of pCREs contribute more significantly to wound transcriptional response, we used the Type I, II, and III pCREs to build machine learning models (see Methods) for predicting wound-responsive expression of genes in a wound response cluster.
We found that models built with Type I pCREs were in most cases the best at predicting wound response in both species (Figure 7B, red; Supplemental Figure 4B), suggesting that these pCREs are components of conserved regulatory mechanisms across species. Type II pCREs predicted wound response well within species but not across species (compare blue and yellow, Figure 7B; Supplemental Figure 4B), supporting their roles in species-specific regulatory function. We should note that, except the NDNN clusters in S. pennellii, the prediction performance of Type II and III pCREs was not as accurate as the Type I pCREs (Figure 7B). This suggests that the conserved cis-regulatory elements play a more central role in wound-responsive transcription in both tomato species and that species-specific pCREs, to a lesser extent, contribute to differential gene expression species specifically.
Turnover of Putative CRE Sites between Orthologous Genes and Their Association with Gene Regulation
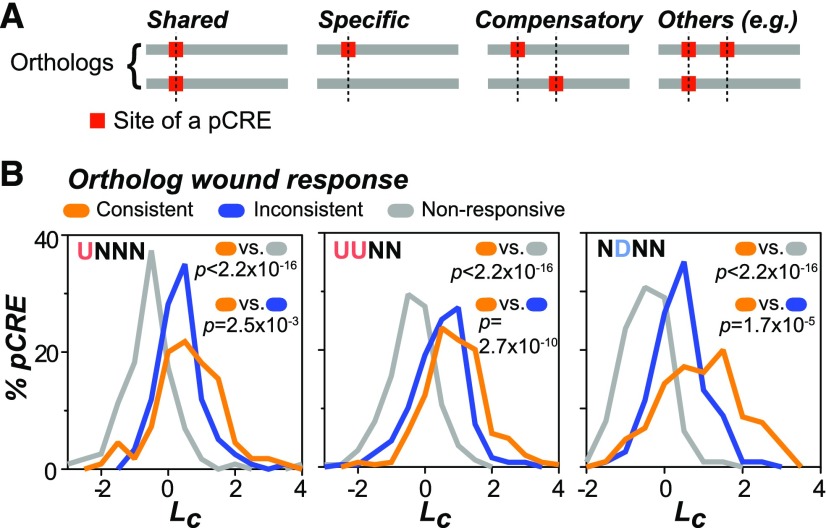
Our findings so far indicate substantial conservation of CREs between domesticated and wild tomato species and their association in predicting wound response (Figures 6 and 7). In addition, we found extensive variation of wound-responsive gene expression among orthologous genes (Figure 3). These differences may result from minor changes in CRE sequences, leading to differences in TF binding specificity (Figure 6). Alternatively, the wound response divergence between orthologs may be the consequence of differential turnover (i.e., the gain and loss) of the cis-regulatory sites within orthologous regions (Carroll, 2008; Wittkopp and Kalay, 2011). To assess these possibilities, we next determined the extent to which these cis-regulatory sites were conserved or turned over across species and their association with gene expression divergence. Based on the relative position of the sites located in regulatory regions of orthologous gene pairs, the sites of a given pCRE were categorized into “shared,” “specific,” “compensatory,” and “other” types (Figure 8A; see Methods). Since the “compensatory” and “other” types accounted for small portions of the pCRE sites (Supplemental Figure 6A), we focused on the “shared” and “specific” pCRE types.
Figure 8.
Relationships of pCRE Site Turnover and Wound-Responsive Gene Expression between Orthologs.
(A) Types of pCRE sites. Shared: The sites of a pCRE are present in both orthologs and located at the same position. Specific: The site of a pCRE is present only in one ortholog but not the other. Compensatory: The sites are present in both species but in different locations. Others: Any situation that does not belong to the previous three types. Gray line: The defined regulatory regions from the orthologous gene pairs (see Methods).
(B) The conservation likelihood (Lc) of a pCRE in the UNNN (left panel), the UUNN (middle panel), and NDNN (right panel) clusters. For a pCRE, its Lc is defined as the log ratio between the proportions of sites that shared and those that are specific (see Methods). The Lc for each pCRE was evaluated using orthologous gene pairs with consistent (belong to the same wound response cluster, orange) and inconsistent (belong to different clusters, blue) wound responses, as well as orthologous genes that are not responsive to wounding (nonresponsive, gray). P values: Testing whether the likelihood scores generating based the blue or gray data sets differ from the orange one (one-sided Mann-Whitney U test).
To summarize the degree of conservation of the sites of each pCRE identified from various wound response clusters (Figure 5A), a conservation likelihood (Lc) for each pCRE was computed by calculating the log2 ratio between the proportion of sites that are shared and the proportion of sites that are specific (see Methods). Thus, a higher Lc indicates a higher degree of enrichment of shared sites relative to that of specific sites. A pCRE with a higher Lc was considered more conserved than that with a lower Lc. First, to assess if the conservation of pCRE sites was correlated with the consistency of the wound response between orthologs, we compared the Lc values for the orthologs with consistent wound response and for those with inconsistent patterns. Using the UNNN cluster as an example (Figure 8B, left panel), we found that the sites of pCREs in orthologous gene pairs with consistent wound response patterns (median Lc = 0.61) had significantly higher Lc values than sites in orthologous pairs with inconsistent patterns (median Lc = 0.28, Mann-Whitney U test, P = 2.5 × 10−3). The same was true when comparing pCRE sites in genes with consistent patterns against sites found in the nonresponsive orthologous genes (median Lc = −0.56; P < 2.2 × 10−16) (Figure 8B, left panel). Similar results were also observed for the pCREs in the UUNN and NDNN cluster (Figure 8B, middle and right panels). Taken together, these results imply that in UNNN, UUNN, and NDNN clusters, the orthologs with consistent gene regulation tend to have more conserved pCRE sites, indicating that, as expected, conservation of pCREs sites contribute to a conserved wound upregulated response across species.
Taken together, these findings suggest a positive correlation between the degrees of pCRE site conservation and the conservation of wound-regulated gene expression between wild and domesticated species.
Conclusion
In this study, we investigated the patterns and mechanisms of transcriptional divergence of environmental stress response in a wild and a domesticated tomato species. Specifically, our analyses focus on wound-responsive gene expression and the cis-regulatory components regulating wound responses. Despite the relatively recent divergence (∼3–7 million years ago) between the wild S. pennellii and domesticated S. lycopersicum species (Nesbitt and Tanksley, 2002; Kamenetzky et al., 2010), the wound-responsive expression patterns of the orthologous genes have diverged significantly, which may be partly attributable to the combined action of natural and artificial selection. In addition, we characterized the pCREs significantly associated with wound response regulation. pCREs identified in S. lycopersicum and S. pennellii were predictive of gene expression. In addition, Type I pCREs (enriched in both species) could better explain gene regulation between species than Type II and III pCREs (species-specifically enriched). This is in line with the conclusion in metazoan studies that the TF binding specificity evolves slowly and is highly conserved among fruit fly, mouse, and human (Stergachis et al., 2014; Nitta et al., 2015). Intriguingly, the Type II and III pCREs partially explain wound response within species, indicating the involvement of divergent TFs after speciation contributing to regulatory divergence. Our results based on the approaches of the differential enrichment of pCREs and whether they may be recognized by TFs from the same family suggest diverging binding preference of some TFs relevant to wound response regulation across species. Further protein-DNA binding studies such as protein binding array and DNA affinity purification sequencing (Weirauch et al., 2014; O’Malley et al., 2016) should be useful to test the regulatory divergence hypothesized here.
Our finding of correlation between the turnover of the pCRE site and the expression divergence of orthologous genes further supports the evolutionary conservation of CREs for wound response in tomato. We should emphasize that, although the correlation is apparent, it is far from perfect. Specifically, some pCRE sites enriched among wound-responsive genes displayed high degrees of conservation between orthologous pairs with inconsistent wound response patterns. One possibility is that these conserved pCREs in orthologs with inconsistent patterns are still regulating weaker wound responses. This is because the wound response clusters were defined based on threshold differential expression; weaker wound responses may not pass the defined threshold. As a result, some orthologous genes were classified into different clusters despite a similar but significantly weaker response (Figure 3). We should also point out that the conservation likelihood (Lc) distribution of some pCREs on orthologs with consistent wound responses may also be low (Figure 8B), indicating that consistent expression patterns cannot be easily attributed to the pCREs analyzed. This highlights the complexity of the transcriptional regulatory systems and the need for studies to further ascertain the mechanistic basis of stress response conservation and divergence. Lastly, among these sites located in the regulatory regions, it is possible that only part of them are the in vivo cis-regulatory sites which can be further narrowed down based on chromatin state, GC content, or DNA structural properties on the surrounding regions (Raveh-Sadka et al., 2012; White et al., 2013; Tsai et al., 2015). Future studies aimed at reducing false-positive identification of pCRE sites based on additional features and at identifying the combinatorial relationship between CREs will be helpful for further understanding the cis-regulatory codes and their evolution.
Our study provides global comparative analyses connecting the divergence of pCREs and turnover of cis-regulatory sites to gene expression divergence between species and orthologous genes. The comparison of pCREs predictive of the wound response revealed both cis-regulatory conservation and divergence. The correlation between the turnover of the cis-regulatory sites and the differential expression of orthologs uncovered cis-regulatory divergence underlying the gene expression variation. Collectively, these findings advance our understanding of the mechanistic basis underlying the stress-responsive gene expression divergence across a wild and a domesticated species.
METHODS
Plant Materials and Growth Conditions
Solanum lycopersicum cv Castlemart was used as the domesticated species. Seeds for the wild species, Solanum pennellii (LA0716), were obtained from Tomato Genetic Resource Center (UC Davis) and grown on Jiffy-7 peat pot (Hummert International) in a growth chamber under a 16-h-light (6:00–22:00, 200 μmol m−2 s−1)/8-h-dark cycle at 28°C. Three- to four-week-old plants with three to four expanded true leaves were used for wound treatment as previously described (Li et al., 2004). For wound elicitation, the lower (older) two leaves were crushed with a hemostat across the midrib of all leaflets. All wounding was performed in the morning (8:00–9:00), 2 h after the start of the light cycle. At the indicated time points, leaflets from multiple tomato seedlings were excised with a razor blade, pooled together, and immediately frozen in liquid nitrogen. Damaged leaflets (local, older leaves) from the first and second leaves and undamaged leaflets (systemic, younger leaves) from the third and fourth leaves were collected separately. Control leaves were harvested from a set of unwounded plants grown side-by-side with the set of wounded plants. Three biological replicates (i.e., three separate sets of plants sampled on different days) were harvested for each treatment and time point. Total RNA was isolated from frozen leaf tissue using an RNeasy Plant Mini Kit (Qiagen). Except the locally 2-h wound-treated samples in S. pennellii, RNA sequencing (100-bp paired-end reads) of the samples was performed with the Illumina HiSeq 2500 platform in the Michigan State University Research Technology Support Facility. RNA sequencing of the locally 2-h wound-treated samples in S. pennellii was performed with HiSeq 4000 (150-bp paired-end reads).
Sequencing Data Processing
To map the RNA-seq reads and determine the gene expression level, the reference genome sequences and gene annotation of S. lycopersicum (ITAG2.4) and S. pennellii (Spenn_v2.0) were retrieved from Sol Genomics Network (https://solgenomics.net). The S. pennellii gene annotation was further reannotated through Maker-P module (Campbell et al., 2014). The cumulative distribution plot of AED (annotation edit distance), which provides the measure of how well the annotations are supported by the EST, protein, and RNA-seq evidence (Campbell et al., 2014), showed that the MAKER-mediated version performed better compared with the Spenn_v2.0 version (red versus black lines, Supplemental Figure 7). To further evaluate MAKER-P performance, we focused on genes annotated in both data sets (n = 22,292) (Supplemental Data Set 3). Among the genes with one-to-one relationship (i.e., overlapped genic regions) between Spenn_v2.0 and MAKER-P annotated version, the gene models annotated by MAKER-P had higher AED values than in Spenn_v2.0 version (green versus gray lines, Supplemental Figure 7). These results showed that MAKER-P improved the gene annotations of S. pennellii; thus, the MAKER-mediated version was adopted in this study.
The paired-end RNA reads were trimmed with Trimmomatic (default setting except leading = 20, trailing = 20, and minlen = 20) (Bolger et al., 2014b) and mapped to the genome with TopHat2 (version 2.0.8) (Kim et al., 2013). Transcript levels of annotated genes were calculated with Cufflinks (version 2.1.1) (Trapnell et al., 2010) and shown as FPKM (fragments per kilobase per million fragments mapped). The numbers of raw, quality-filtered, and mapped reads and the sequencing coverage are reported in Supplemental Data Set 4. To evaluate the reproducibility of gene expressions among replicates and the similarity of gene expression profiles among treatments, the Spearman’s rank correlation coefficient was determined by pairwise comparison of gene expression between samples. The distance (1 − Spearman’s rank correlation coefficient) was used to generate the dendrogram through hierarchical clustering function with “complete” method (Supplemental Figure 1A). The three replicates of a given treatment in one species were clustered together, showing the gene expression profiles were similar and reproducible among replicates (Supplemental Figure 1A).
To identify the significantly wound-responsive genes, only protein-coding genes with the value of FPKM ≥1 in all replicates of any time point and tissue were considered (n = 17,945 in S. lycopersicum and 16,868 in S. pennellii). The transcript abundances of control and wound-treated samples were compared with EdgeR (Robinson et al., 2010). Genes with false discovery rate adjusted P < 0.05 (Benjamini, 1995) and with 4-fold difference in RNA level between wound and control (unwounded) samples was considered to be wound responsive and included for the analyses. Note that the replicates of the local 2-h S. pennellii sample were sequenced in a different Illumina platform that result in significantly higher number of reads (n = 62–153 million) compared with those of the other samples (n = 13–23 million) (Supplemental Data Set 4). Given between sample normalization was part of the modeling process in EdgeR (Robinson et al., 2010), we expected that the DE gene call will not be significantly influenced. Consistent with this, by downsampling reads from the high coverage replicate, we found that the difference of the input size of raw read numbers among samples did not impact the identification of DE genes (Figure 1A; Supplemental Figure 1C).
Identification of Putative cis-Elements and Prediction for Wound Response
Wound-responsive genes are categorized into the different regulatory clusters depending on the levels of differential gene expression in the indicated points as defined in Figure 2A. Genes are regarded as nonresponsive genes if their FC values in all comparisons between wound treatments and controls are between 1.2 and 0.8 (n = 3548 in S. lycopersicum and 1058 in S. pennellii). Note that replicates from a treatment were jointly compared with control replicates in determining FC using EdgeR. The FC values were used for pCRE identification (Figure 5A) and pCRE site turnover analyses (Figure 8). To identify the pCREs associated with wound response (Figure 5A), a k-mer (oligomer with the length of k) pipeline was established by examining the frequency enrichment of a k-mer sequence in the regulatory region among the genes of a given wound response cluster compared with the nonresponsive genes and determining the adjusted P values through Fisher’s exact test and multiple testing (Benjamini-Hochberg method) (Benjamini, 1995). Here, the regulatory region is defined as the region ranging from upstream 1 kb to downstream 0.5 kb of TSS.
Since the cis-regulatory elements range from 5 to ∼30 nucleotides (Stewart et al., 2012), this k-mer pipeline includes several steps to discover the pCREs with various sequence lengths. Step 1: A set of all possible 5-mer oligomers was evaluated for their enrichment among genes in each wound response cluster compared with nonresponsive genes. Only the 5-mers with significant enrichment (adjusted P values < 0.05) were retained for the next step. Step 2: The sequence of each significantly enriched 5-mer from step 1 was extended with 1 nucleotide in either direction, the resulting extended k-mer was examined for enrichment, and the significantly enriched ones (adjusted P values < 0.05) were retained. The step was repeated until no extended k-mer sequence was found to be significantly enriched among the regulated genes. Noted that if two k-mers were both significantly enriched and one k-mer sequence exactly matched the other one, only the one with lower adjusted P value was retained. Step 3: As described in step 1, but starting with a set of all possible 6-mers. The significantly enriched 6-mers were combined with the set of the k-mers identified from step 2. Step 4: As described in step 2, but starting with the set of k-mers from step 3. Finally, the set of k-mers significantly enriched in the indicated wound response cluster was determined and considered as pCREs (Figure 5A). To compare the performance of our k-mer pipeline to the typical motif-finding approaches, we used MEME (Bailey et al., 2009) to identify pCREs in UNNN clusters in both species. The prediction model employing MEME-derived pCREs performed significantly worse than that employing identified k-mers (Supplemental Figure 3G), suggesting that our approach could more efficiently identify short sequences resembling CREs.
The SVM method that allows predicting of wound response of a gene based on a set of pCREs was performed using the LIBSVM implementation of the SVM method through the Weka wrapper with the parameters described previously (Liu et al., 2015). The pCREs were used as attributes whereas the binary status of genes with/without wound regulation was the class we wanted to predict. For training the predictive models for each regulatory pattern, the genes of the given clusters are positive examples whereas the nonresponsive genes are negative examples.
Sequence Similarity of Putative CREs between Species and to the Known TF Binding Motifs
To identify the pCREs whose sequences are more significantly similar than expected between TF families [thus, the pCRE in question are likely bound by a TF(s) from a family that pCREs are similar to], the pairwise distances of known TF binding motifs (TFBMs) across 30 TF families (Weirauch et al., 2014) were calculated and the 5th percentile of distance, 0.39 (with a P value = 0.05) was set as a threshold (Liu et al., 2015).
To determine what pCREs identified in S. lycopersicum and S. pennellii for a given cluster are likely bound by TF(s) of the same family, the pairwise PCC (Pearson’s correlation coefficient) distance of the pCREs was generated with TAMO package (Gordon et al., 2005) and used to construct the average linkage tree using UPGMA method in “cluster” package in R (Maechler et al., 2016). The threshold of 0.39 value that corresponds to the distance of the motifs among TF families was applied such that any pCREs within a branch length <0.39 are considered to be in the same subgroup and likely bound by TFs from the same family (Figure 6A; Supplemental Figure 5 and Supplemental Data Set 2). The pCREs located in a given subgroup were merged through STAMP with default settings (Mahony and Benos, 2007) to summarize the sequence information of these pCREs since the pCREs within a subgroup are likely bound by TFs from the same family but may have subtle nucleotide difference and various lengths (Supplemental Figure 3). Note that the presence of pCRE duplicates in Figure 6 and Supplemental Figure 5 is because some pCREs were identified from both S. lycopersicum and S. pennellii. In these cases, one copy was removed before merging.
The known TFBM data set consists of 256 and 510 CREs from protein binding microarray (Weirauch et al., 2014) and DNA affinity purification sequencing approaches (O’Malley et al., 2016). The similarity between the merged pCREs and known TFBMs were determined with the threshold of PCC distance (P < 0.05) as described previously (Liu et al., 2015) (Supplemental Figure 3).
Identification of Orthologous Genes
Using the longest protein sequences for genes, an all versus all comparison of protein sequences was run on a combined set of genes in S. lycopersicum and S. pennellii using BLAST. Custom python scripts were then used to extract reciprocal best matches between species. The set of the reciprocal best matches was divided into those that were the best overall match (the “overall” set) and those where one of the two proteins had a better match within species (the “reciprocal-only” set). Initially, there were 19,657 overall and 1198 reciprocal-only best matches. For reciprocal-only best matches, the sequences of the better within species matches were obtained, creating a group of three or more protein sequences (i.e., the best match between species gene pairs and any genes that are better matches within species) for each reciprocal-only best match.
For each pair of overall best matches and group of reciprocal-only best matches, protein sequences were aligned using MAFFT. Protein alignments were then back-aligned to the longest coding sequences for genes in each species using custom python scripts. The resulting aligned nucleotide sequences were used to determine the Ks of best matches using PAML. The “yn00” algorithm was used on sequence pairs from overall best matches and the “codeml” algorithm was used for sequence groups from reciprocal-only best matches. Next, we visualized the distribution of Ks values for the “overall” set because they have a clear 1:1 relationship between S. lycopersicum and S. pennellii. Given the recent speciation event, we expected the Ks distribution to follow a normal distribution. We observed a roughly normal distribution with a long right tail. We theorize that the extremely large Ks value in the tail can be attributed to ancient duplication events that experienced reciprocal loss in both species. Therefore, to enrich the set of reciprocal-best matches for orthologs of the recent speciation event, a normal distribution was fit to set of Ks values for the “overall” set in R using nonlinear minimization. The 99th percentile of the fit distribution was determined and applied as a cutoff to both the “overall” and “reciprocal-only” best matches. This resulted in a final set of 16,222 orthologous genes between S. lycopersicum and S. pennellii.
GO and Metabolic Pathway Analyses
The data sets of GO annotation and metabolic pathways of genes in S. lycopersicum were retrieved from the Sol Genomics Network (https://solgenomics.net) and Plant Metabolic Network (http://www.plantcyc.org). To have comparable annotation set of GO and metabolic pathways of genes across species, the annotations of genes from S. lycopersicum were inferred to the orthologous ones in S. pennellii. In the end, 10,091 and 2006 orthologous genes with biological process and metabolic pathway were retrieved for the downstream analyses. The list of orthologous gene pairs between S. lycopersicum and S. pennellii was generated as mentioned above.
The enrichments of GO terms and metabolic pathways in the clusters and differentially regulated gene sets, compared with the total orthologous genes, were determined though Fisher’s Exact test. A P value obtained for each GO term and pathway comparison and was multiple-testing corrected (Bass et al., 2015).
Conservation and Divergence of pCRE Sites in Orthologous Gene Pairs
The region of the 1 kb upstream and 500 bp downstream of TSSs in the orthologous gene pairs was defined to be regulatory regions and aligned with MUSCLE package (Edgar, 2004) (Figure 8A). Based on the positions of the pCRE sites on the aligned sequences, these sites for each pCRE were assigned into four types: (1) shared (i.e., the site from each species was located on the same positions), (2) specific (i.e., the site was present only in one species), (3) compensatory (i.e., the site was present in both species but located in different location), and (4) others (i.e., any cases of pCRE sites were not assigned to the three types mentioned above). A likelihood score representing the conservation degree of pCRE sites for each pCRE was determined by taking the ratio of the pCRE site types (%) between the shared and specific ones. The orthologous gene pairs with consistent patter means the pairs are assigned to the same regulatory cluster as defined in Figure 2A; otherwise, the orthologous gene pairs are considered to be with inconsistent patterns. Nonresponsive orthologous genes are the orthologous genes if their fold-change values in all wound treatment conditions, compared with the control one, are between 1.2 and 0.8 in both S. lycopersicum and S. pennellii (n = 452).
RT-qPCR Analyses
Total RNA from three independent samples was reverse-transcribed with the High Capacity cDNA Reverse Transcription Kit (Life Technologies) according to the manufacturer’s instructions. The resulting cDNA was subsequently used for quantification of transcripts with Power SYBR Green PCR Master Mix (Life Technologies) and analysis of products on an ABI 7500 Fast real-time PCR system (Life Technologies). The relative transcript abundances were calculated using the ΔCt (threshold cycle) method. The ACTIN gene was used as an internal control. Primers were designed to target the conserved regions of genes between S. lycopersicum and S. pennellii and listed in Supplemental Table 1.
Accession Numbers
The RNA-seq data from this study have been submitted to the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE93556. The names and accession numbers of genes described in this study can be found in Supplemental Table 1.
Supplemental Data
Supplemental Figure 1. Similarity of gene expression profiles among replicates of each condition and between RT-qPCR and RNA-seq.
Supplemental Figure 2. Comparison of wound response clusters for genes with |log2(FC)| > 1 and their functional category enrichments.
Supplemental Figure 3. Properties of the putative cis-elements regulating wound response.
Supplemental Figure 4. Model performance employing pCREs in predicting wound response.
Supplemental Figure 5. Differential enrichment of pCREs in UUNN and NDNN cluster genes between tomato species.
Supplemental Figure 6. The types of pCRE sites on the orthologous gene pairs and the differential enrichments of species-specific pCREs in inconsistent orthologs.
Supplemental Figure 7. MAKER-P-mediated improvement on S. pennellii gene annotation.
Supplemental Table 1. Primers used in this study.
Supplemental Data Set 1. Numbers of genes in different wound-responsive clusters in S. lycopersicum and S. pennellii.
Supplemental Data Set 2. The species enrichment, types, and the subgroups of pCREs identified in S. lycopersicum and S. pennellii as indicated in Figure 6 and Supplemental Figure 5.
Supplemental Data Set 3. Genes with one to one relationship (i.e., overlapped genic regions) between Spenn_v2.0 and MAKER-P annotated versions and their corresponding gene name (ID) in each data set.
Supplemental Data Set 4. Numbers of sequencing reads in each read-mapping step.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This work was supported in part by a grant from the Rackham Foundation and Michigan AgBioResearch Project (MICL02278) to G.A.H.; by the National Science Foundation (IOS-1546617 and DEB-1655386) and the U.S. Department of Energy (DE-SC0018409) to S.-H.S.; by a Michigan State University Discretionary Funding Initiative grant to S.-H.S.; by theBiotechnology Center in Southern Taiwan, Academia Sinica, and Ministry of Science and Technology, Taiwan (MOST106-2311-B-001-02), to M.-J.L.; and by the Japan Society for Promotion of Science, Research Fellowship for Young Scientists (24.841) to K.S.
AUTHOR CONTRIBUTIONS
M.-J.L., K.S., M.Y., G.A.H., and S.-H.S. designed the research. K.S. performed the experiments. M.-J.L., S.U., N.P., and M.S.C. analyzed the data. M.-J.L., G.A.H., and S.-H.S. wrote the manuscript with contributions by all authors.
Footnotes
Articles can be viewed without a subscription.
References
- Adie B.A.T., Pérez-Pérez J., Pérez-Pérez M.M., Godoy M., Sánchez-Serrano J.J., Schmelz E.A., Solano R. (2007). ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19: 1665–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson L. (2001). Genetic dissection of phenotypic diversity in farm animals. Nat. Rev. Genet. 2: 130–138. [DOI] [PubMed] [Google Scholar]
- Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37: W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., Roychoudhury A. (2015). WRKY proteins: signaling and regulation of expression during abiotic stress responses. Sci. World J. 2015: 807560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass A.J., Dabney A., Robinson D. (2015). qvalue: Q-value estimation for false discovery rate control. R package, version 2.6.0. https://github.com/StoreyLab/qvalue.
- Bauchet G.C.M. (2012). Genetic diversity in tomato (Solanum lycopersicum) and its wild relatives. In Environmental Sciences, Çaliskan M., ed (Rijeka, Croatia: InTechOpen; ), pp. 133–162. [Google Scholar]
- Benjamini Y.Y.H. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. A Stat. Soc. 57: 289–300. [Google Scholar]
- Berardi P., Meyyappan M., Riabowol K.T. (2003). A novel transcriptional inhibitory element differentially regulates the cyclin D1 gene in senescent cells. J. Biol. Chem. 278: 7510–7519. [DOI] [PubMed] [Google Scholar]
- Bolger A., et al. (2014a). The genome of the stress-tolerant wild tomato species Solanum pennellii. Nat. Genet. 46: 1034–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. (2014b). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman A.R., Gianoulis T.A., Zhang Z.D., Yu H., Rozowsky J., Seringhaus M.R., Wang L.Y., Gerstein M., Snyder M. (2007). Divergence of transcription factor binding sites across related yeast species. Science 317: 815–819. [DOI] [PubMed] [Google Scholar]
- Boter M., Ruíz-Rivero O., Abdeen A., Prat S. (2004). Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 18: 1577–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell M.S., et al. (2014). MAKER-P: a tool kit for the rapid creation, management, and quality control of plant genome annotations. Plant Physiol. 164: 513–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S.B. (2008). Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134: 25–36. [DOI] [PubMed] [Google Scholar]
- Chaudhary B. (2013). Plant domestication and resistance to herbivory. Int. J. Plant Genomics 2013: 572784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.H., Gols R., Benrey B. (2015). Crop domestication and its impact on naturally selected trophic interactions. Annu. Rev. Entomol. 60: 35–58. [DOI] [PubMed] [Google Scholar]
- Cominelli E., Sala T., Calvi D., Gusmaroli G., Tonelli C. (2008). Over-expression of the Arabidopsis AtMYB41 gene alters cell expansion and leaf surface permeability. Plant J. 53: 53–64. [DOI] [PubMed] [Google Scholar]
- Doebley J., Lukens L. (1998). Transcriptional regulators and the evolution of plant form. Plant Cell 10: 1075–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J.F., Gaut B.S., Smith B.D. (2006). The molecular genetics of crop domestication. Cell 127: 1309–1321. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson J.J., Li W.H. (2010). The genetic basis of evolutionary change in gene expression levels. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 2581–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla J.M., López-Vidriero I., Carrasco J.L., Godoy M., Vera P., Solano R. (2014). DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. USA 111: 2367–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong P., Zhang J., Li H., Yang C., Zhang C., Zhang X., Khurram Z., Zhang Y., Wang T., Fei Z., Ye Z. (2010). Transcriptional profiles of drought-responsive genes in modulating transcription signal transduction, and biochemical pathways in tomato. J. Exp. Bot. 61: 3563–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.B., Nekludova L., McCallum S., Fraenkel E. (2005). TAMO: a flexible, object-oriented framework for analyzing transcriptional regulation using DNA-sequence motifs. Bioinformatics 21: 3164–3165. [DOI] [PubMed] [Google Scholar]
- Green T.R., Ryan C.A. (1972). Wound-induced proteinase inhibitor in plant leaves: A possible defense mechanism against insects. Science 175: 776–777. [DOI] [PubMed] [Google Scholar]
- Hajjar R., Hodgkin T. (2007). The use of wild relatives in crop improvement: A survey of developments over the last 20 years. Euphytica 156: 1–13. [Google Scholar]
- Heyndrickx K.S., Van de Velde J., Wang C., Weigel D., Vandepoele K. (2014). A functional and evolutionary perspective on transcription factor binding in Arabidopsis thaliana. Plant Cell 26: 3894–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobo T., Asada M., Kowyama Y., Hattori T. (1999). ACGT-containing abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent. Plant J. 19: 679–689. [DOI] [PubMed] [Google Scholar]
- Hooker T.S., Millar A.A., Kunst L. (2002). Significance of the expression of the CER6 condensing enzyme for cuticular wax production in Arabidopsis. Plant Physiol. 129: 1568–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe G.A., Jander G. (2008). Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59: 41–66. [DOI] [PubMed] [Google Scholar]
- Howe G.A., Schaller A. (2008). Direct Defenses in Plants and Their Induction by Wounding and Insect Herbivores. (Dordrecht, The Netherlands: Springer; ). [Google Scholar]
- Huot B., Yao J., Montgomery B.L., He S.Y. (2014). Growth-defense tradeoffs in plants: a balancing act to optimize fitness. Mol. Plant 7: 1267–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihashi Y., Aguilar-Martínez J.A., Farhi M., Chitwood D.H., Kumar R., Millon L.V., Peng J., Maloof J.N., Sinha N.R. (2014). Evolutionary developmental transcriptomics reveals a gene network module regulating interspecific diversity in plant leaf shape. Proc. Natl. Acad. Sci. USA 111: E2616–E2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamenetzky L., Asís R., Bassi S., de Godoy F., Bermúdez L., Fernie A.R., Van Sluys M.A., Vrebalov J., Giovannoni J.J., Rossi M., Carrari F. (2010). Genomic analysis of wild tomato introgressions determining metabolism- and yield-associated traits. Plant Physiol. 152: 1772–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K., Pajoro A., Angenent G.C. (2010). Regulation of transcription in plants: mechanisms controlling developmental switches. Nat. Rev. Genet. 11: 830–842. [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig D., et al. (2013). Comparative transcriptomics reveals patterns of selection in domesticated and wild tomato. Proc. Natl. Acad. Sci. USA 110: E2655–E2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zhao Y., McCaig B.C., Wingerd B.A., Wang J., Whalon M.E., Pichersky E., Howe G.A. (2004). The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16: 126–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M.J., Seddon A.E., Tsai Z.T., Major I.T., Floer M., Howe G.A., Shiu S.H. (2015). Determinants of nucleosome positioning and their influence on plant gene expression. Genome Res. 25: 1182–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lü S., Song T., Kosma D.K., Parsons E.P., Rowland O., Jenks M.A. (2009). Arabidopsis CER8 encodes LONG-CHAIN ACYL-COA SYNTHETASE 1 (LACS1) that has overlapping functions with LACS2 in plant wax and cutin synthesis. Plant J. 59: 553–564. [DOI] [PubMed] [Google Scholar]
- Maechler M., Rousseeuw P., Struyf A., Hubert M., Hornik K. (2016). Cluster: Cluster Analysis Basics and Extensions. R package, version 2.0.4.
- Mahony S., Benos P.V. (2007). STAMP: a web tool for exploring DNA-binding motif similarities. Nucleic Acids Res. 35: W253–W258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R.S., Purugganan M.D. (2013). Evolution of crop species: genetics of domestication and diversification. Nat. Rev. Genet. 14: 840–852. [DOI] [PubMed] [Google Scholar]
- Narsai R., Howell K.A., Millar A.H., O’Toole N., Small I., Whelan J. (2007). Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell 19: 3418–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbitt T.C., Tanksley S.D. (2002). Comparative sequencing in the genus Lycopersicon. Implications for the evolution of fruit size in the domestication of cultivated tomatoes. Genetics 162: 365–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta K.R., Jolma A., Yin Y., Morgunova E., Kivioja T., Akhtar J., Hens K., Toivonen J., Deplancke B., Furlong E.E., Taipale J. (2015). Conservation of transcription factor binding specificities across 600 million years of bilateria evolution. eLife 4: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowicka B., Kruk J. (2010). Occurrence, biosynthesis and function of isoprenoid quinones. Biochim. Biophys. Acta 1797: 1587–1605. [DOI] [PubMed] [Google Scholar]
- O’Malley R.C., Huang S.C., Song L., Lewsey M.G., Bartlett A., Nery J.R., Galli M., Gallavotti A., Ecker J.R. (2016). Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 166: 1598. [DOI] [PubMed] [Google Scholar]
- Raveh-Sadka T., Levo M., Shabi U., Shany B., Keren L., Lotan-Pompan M., Zeevi D., Sharon E., Weinberger A., Segal E. (2012). Manipulating nucleosome disfavoring sequences allows fine-tune regulation of gene expression in yeast. Nat. Genet. 44: 743–750. [DOI] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero I.G., Ruvinsky I., Gilad Y. (2012). Comparative studies of gene expression and the evolution of gene regulation. Nat. Rev. Genet. 13: 505–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong J., Lammers Y., Strasburg J.L., Schidlo N.S., Ariyurek Y., de Jong T.J., Klinkhamer P.G., Smulders M.J., Vrieling K. (2014). New insights into domestication of carrot from root transcriptome analyses. BMC Genomics 15: 895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal J.P., Dirzo R. (1997). Effects of life history, domestication and agronomic selection on plant defence against insects: Evidence from maizes and wild relatives. Evol. Ecol. 11: 337–355. [Google Scholar]
- Rushton P.J., Somssich I.E. (1998). Transcriptional control of plant genes responsive to pathogens. Curr. Opin. Plant Biol. 1: 311–315. [DOI] [PubMed] [Google Scholar]
- Scranton M.A., Fowler J.H., Girke T., Walling L.L. (2013). Microarray analysis of tomato’s early and late wound response reveals new regulatory targets for Leucine aminopeptidase A. PLoS One 8: e77889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibéril Y., Doireau P., Gantet P. (2001). Plant bZIP G-box binding factors. Modular structure and activation mechanisms. Eur. J. Biochem. 268: 5655–5666. [DOI] [PubMed] [Google Scholar]
- Spitz F., Furlong E.E. (2012). Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 13: 613–626. [DOI] [PubMed] [Google Scholar]
- Stanković B., Vian A., Henry-Vian C., Davies E. (2000). Molecular cloning and characterization of a tomato cDNA encoding a systemically wound-inducible bZIP DNA-binding protein. Planta 212: 60–66. [DOI] [PubMed] [Google Scholar]
- Stergachis A.B., et al. (2014). Conservation of trans-acting circuitry during mammalian regulatory evolution. Nature 515: 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart A.J., Hannenhalli S., Plotkin J.B. (2012). Why transcription factor binding sites are ten nucleotides long. Genetics 192: 973–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan A.M., et al. (2014). Mapping and dynamics of regulatory DNA and transcription factor networks in A. thaliana. Cell Reports 8: 2015–2030. [DOI] [PubMed] [Google Scholar]
- Swanson-Wagner R., Briskine R., Schaefer R., Hufford M.B., Ross-Ibarra J., Myers C.L., Tiffin P., Springer N.M. (2012). Reshaping of the maize transcriptome by domestication. Proc. Natl. Acad. Sci. USA 109: 11878–11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinnen G., Goossens A., Pauwels L. (2016). Lessons from domestication: Targeting cis-regulatory elements for crop improvement. Trends Plant Sci. 21: 506–515. [DOI] [PubMed] [Google Scholar]
- Tal M., Shannon M.C. (1983). Salt tolerance in the wild relatives of the cultivated tomato: Responses of Lycopersicon esculentum, Lycopersicon cheesmanii, Lycopersicon peruvianum, Solanum pennellii and F1 hybrids to high salinity. Aust. J. Plant Physiol. 10: 109–117. [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. (2010). Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Z.T., Shiu S.H., Tsai H.K. (2015). Contribution of sequence motif, chromatin state, and DNA structure features to predictive models of transcription factor binding in yeast. PLOS Comput. Biol. 11: e1004418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Verk M.C., Pappaioannou D., Neeleman L., Bol J.F., Linthorst H.J. (2008). A novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiol. 146: 1983–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch M.T., et al. (2014). Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158: 1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M.A., Myers C.A., Corbo J.C., Cohen B.A. (2013). Massively parallel in vivo enhancer assay reveals that highly local features determine the cis-regulatory function of ChIP-seq peaks. Proc. Natl. Acad. Sci. USA 110: 11952–11957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp P.J., Kalay G. (2011). Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 13: 59–69. [DOI] [PubMed] [Google Scholar]
- Wittkopp P.J., Haerum B.K., Clark A.G. (2004). Evolutionary changes in cis and trans gene regulation. Nature 430: 85–88. [DOI] [PubMed] [Google Scholar]
- Wittstock U., Gershenzon J. (2002). Constitutive plant toxins and their role in defense against herbivores and pathogens. Curr. Opin. Plant Biol. 5: 300–307. [DOI] [PubMed] [Google Scholar]
- Wray G.A., Hahn M.W., Abouheif E., Balhoff J.P., Pizer M., Rockman M.V., Romano L.A. (2003). The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 20: 1377–1419. [DOI] [PubMed] [Google Scholar]
- Yu J.J., Thornton K., Guo Y., Kotz H., Reed E. (2001). An ERCC1 splicing variant involving the 5′-UTR of the mRNA may have a transcriptional modulatory function. Oncogene 20: 7694–7698. [DOI] [PubMed] [Google Scholar]
- Zhang C., Xuan Z., Otto S., Hover J.R., McCorkle S.R., Mandel G., Zhang M.Q. (2006). A clustering property of highly-degenerate transcription factor binding sites in the mammalian genome. Nucleic Acids Res. 34: 2238–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]