An isoform of the EPSP synthase enzyme possesses a DNA-binding helix-turn-helix motif and has dual roles in shikimate biosynthesis and transcriptional repressor activity.
Abstract
Long-lived perennial plants, with distinctive habits of inter-annual growth, defense, and physiology, are of great economic and ecological importance. However, some biological mechanisms resulting from genome duplication and functional divergence of genes in these systems remain poorly studied. Here, we discovered an association between a poplar (Populus trichocarpa) 5-enolpyruvylshikimate 3-phosphate synthase gene (PtrEPSP) and lignin biosynthesis. Functional characterization of PtrEPSP revealed that this isoform possesses a helix-turn-helix motif in the N terminus and can function as a transcriptional repressor that regulates expression of genes in the phenylpropanoid pathway in addition to performing its canonical biosynthesis function in the shikimate pathway. We demonstrated that this isoform can localize in the nucleus and specifically binds to the promoter and represses the expression of a SLEEPER-like transcriptional regulator, which itself specifically binds to the promoter and represses the expression of PtrMYB021 (known as MYB46 in Arabidopsis thaliana), a master regulator of the phenylpropanoid pathway and lignin biosynthesis. Analyses of overexpression and RNAi lines targeting PtrEPSP confirmed the predicted changes in PtrMYB021 expression patterns. These results demonstrate that PtrEPSP in its regulatory form and PtrhAT form a transcriptional hierarchy regulating phenylpropanoid pathway and lignin biosynthesis in Populus.
INTRODUCTION
Evolution of the phenylpropanoid pathway has been proposed as one of the primary events leading to successful colonization of terrestrial environments by plants (Tohge et al., 2013). Specifically, lignin biosynthesis evolved from the phenylpropanoid pathway to overcome obstacles related to structural support and defense against biotic and abiotic stresses as plants moved from aquatic to terrestrial environments (Weng et al., 2008). Long-lived perennial plants, which cover approximately one-third of the Earth’s terrestrial surface, are of special economic and ecological importance providing woody biomass as renewable feedstock for materials and energy while harboring substantial biodiversity and providing immeasurable environmental services (Ragauskas et al., 2006; Achard, 2009; Hinchee et al., 2009). Unlike herbaceous plants, woody plants exhibit extensive cell division and secondary cell wall thickening to generate biomass from secondary xylem tissue (Iqbal, 1990).
Carbon flow into the phenylpropanoid pathway is vital for terrestrial plants, as it provides precursors for secondary metabolites including monolignols (Barros et al., 2015). Besides monolignols, the phenylpropanoid pathway also provides precursors for various nonstructural, carbon-rich secondary metabolites, such as flavonoids, isoflavonoids, and coumarins, which have important functions in plant defense against pathogens and predators (Vogt, 2010; Fraser and Chapple, 2011). The phenylpropanoid pathway begins at phenylalanine, an end-product of the shikimate pathway (Fraser and Chapple, 2011). After three reactions, carbon precursors from phenylalanine are transferred to 4-coumaroyl CoA, which serves as the precursor of all downstream phenylpropanoids, including lignin and nonstructural metabolites. Consequently, most lignin biosynthetic enzymes also play critical roles in the phenylpropanoid pathway, such as PHENYLALANINE AMMONIA-LYASE (PAL), CINNAMATE 4-HYDROXYLASE (C4H), 4-COUMARATE:COA LIGASE (4CL), HYDROXYCINNAMOYL-COA SHIKIMATE:QUINATE HYDROXY-CINNAMOYL-TRANSFERASE (HCT), P-COUMAROYL SHIKIMATE 3′ HYDROXYLASE (C3H), CAFFEOYL COA 3-O-METHYLTRANSFERASE (CCoAOMT), CINNAMOYL-COA REDUCTASE (CCR), CINNAMYL ALCOHOL DEHYDROGENASE (CAD), FERULATE 5-HYDROXYLASE (F5H), and LACCASE (Vanholme et al., 2010; Vogt, 2010).
The transcription factor MYB DOMAIN PROTEIN46 (MYB46) is one of the master regulators of the phenylpropanoid pathway and lignin biosynthesis in plant species. Genetic and biochemical studies in Arabidopsis thaliana have demonstrated that MYB46 directly targets and activates the expression of multiple lignin biosynthetic genes, including PAL1, C4H, 4CL, HCT, C3H1, F5H1, CCR, CAD6, and CCoAOMT1 (Kim et al., 2014). In addition to lignin biosynthetic genes, MYB46 also activates the expression of MYB58 and MYB63, two master regulators of lignin biosynthesis (Kim et al., 2014). Consistent with transcriptional data, transgenic Arabidopsis plants overexpressing MYB46 displayed ectopic lignin deposition in stem cells (Kim et al., 2014). Besides Arabidopsis, pine (Pinus taeda) and cider gum (Eucalyptus gunnii) MYB46 homologs were found to be functional in the regulation of the phenylpropanoid pathway and lignin biosynthesis (Patzlaff et al., 2003; Goicoechea et al., 2005). Phylogenetic analyses identified four close homologs of MYB46 in Populus trichocarpa: PtrMYB002, PtrMYB003, PtrMYB020, and PtrMYB021 (Wilkins et al., 2009; McCarthy et al., 2010; Zhong et al., 2013). All these four genes are functional because the heterologous expression of each gene in Arabidopsis could induce ectopic lignin deposition, which was observed in plants overexpressing the Arabidopsis MYB46 (McCarthy et al., 2010; Zhong et al., 2013). Transient expression assays in protoplasts demonstrated that the four Populus MYB46 genes were able to activate promoters of Populus lignin biosynthetic genes, such as 4CL1, CCoAOMT1, and CAFFEIC ACID O‐METHYLTRANSFERASE2 (McCarthy et al., 2010; Zhong et al., 2013). Moreover, in transgenic Populus plants overexpressing PtrMYB003 or PtrMYB021, lignin was ectopically deposited in cell walls of stem cells (Zhong et al., 2013).
In addition to lignin biosynthesis, Arabidopsis and Populus MYB46s also regulate the biosynthesis of other major components of the secondary cell wall including cellulose and xylan. The expression of cellulose synthases (CesAs) and xylan synthetic genes (IRREGULAR XYLEMs [IRXs]) could be directly activated by both Arabidopsis and Populus MYB46s (McCarthy et al., 2010; Kim et al., 2013b, 2014; Zhong et al., 2013). Because of the ability to activate the biosynthesis of all three major components of the secondary cell wall, MYB46 has been defined as one master regulator of secondary cell wall biosynthesis and wood formation. On the other hand, MYB46 is also under transcriptional regulation during secondary cell wall formation. NAC transcription factors, including SECONDARY WALL-ASSOCIATED NAC DOMAINPROTEIN1 (SND1), VASCULAR-RELATEDNAC DOMAIN6 (VND6), and VND7, were found to directly activate MYB46 expression in Arabidopsis (Zhong et al., 2007; Ohashi-Ito et al., 2010; Yamaguchi et al., 2011). In Populus, the activation of MYB46 expression by a set of SND1 homologs (PtrWNDs) has been confirmed (Zhong and Ye, 2010). However, since perennial woody species clearly possess unique attributes during cell wall biosynthesis, little is known about their regulatory repertoire outside of those inferred from model systems.
The advent of next-generation sequencing technologies and high-resolution linkage disequilibrium (LD)-based association mapping has created opportunities for non-inference-based discovery in target species such as Populus. We previously demonstrated the ability to use association mapping to characterize the genetic architecture underlying complex traits in Populus with single-gene resolution (McKown et al., 2014; Muchero et al., 2015). Despite these successes, linking the causal DNA variant to the molecular basis of such association remains a challenge. In this study, we employed LD-based association mapping to identify significant associations between lignin content and a P. trichocarpa 5-enolpyruvylshikimate 3-phosphate synthase (EPSP) gene (PtrEPSP). Transgenic Populus plants overexpressing PtrEPSP showed ectopic deposition of lignin, accumulations of phenylpropanoid metabolites, and differential expressions of secondary cell wall biosynthesis genes. At the molecular level, we used subcellular localization, transcriptional activity, and electrophoretic mobility shift assays to demonstrate that PtrEPSP accumulates in the nucleus and acts as a transcriptional repressor by directly binding to the promoter element of its target. Our in vivo and in vitro results further indicate that PtrEPSP directly regulates one hAT transposase family gene (PtrhAT). We also show that PtrhAT is a nuclear protein and has transcriptional repressor activity. Furthermore, we demonstrate that the direct repression target of PtrhAT is PtrMYB021. Through repressing PtrhAT expression, PtrEPSP activates the expression of PtrMYB021 and the phenylpropanoid pathway. Finally, we establish that the transcription of PtrEPSP is under the control of the master regulator PtrWND1B, a homolog of SECONDARY WALL-ASSOCIATED NAC DOMAIN1 (SND1). In total, we uncovered a novel transcriptional regulatory mechanism regulating MYB46 expression and the flow of carbon into the phenylpropanoid pathway and lignin biosynthesis.
RESULTS
SNPs in the Potri.002G146400 Gene Have Significant Associations with Lignin Content in Populus
In a study designed to assess the genetic basis of lignin biosynthesis across multiple environments in Populus, we used the Populus association mapping panel characterized for lignin content in two different environments. Wood samples were taken from 1081 mature P. trichocarpa genotypes in 2008 across the species range as the association mapping panel was assembled (Muchero et al., 2015). Subsequently, the same panel of genotypes was established in a field site in Clatskanie, OR, in 2009 and wood samples were obtained in July 2012. Phenotyping results for these samples were published previously (Muchero et al., 2015). To identify single-nucleotide polymorphism (SNP) markers associated with lignin content, we targeted chromosome II, which was previously shown to harbor major quantitative trait loci for lignin content and syringyl-to-guaiacyl ratio (Yin et al., 2010). We performed genotype-to-phenotype correlations using 2352 SNPs selected for even coverage on chromosome II. This analysis revealed that multiple SNPs within a 4.2-kb interval exhibited associations with lignin content across two environments ranking 4th, 5th, 7th, and 8th in the native environment from where the parent trees were sampled. Three of the same four SNPs ranked 1st, 6th, and 7th out of the 2352 markers in the Clatskanie field site when 4-year-old clones were sampled and assessed for lignin content (Table 1). These SNPs fell within a Populus gene model, POPTR_0002s14740 (Potri.002G146400, v.3.1) annotated as an EPSP synthase (scaffold_2_10944029 [C/T] intron, scaffold_2_10945723 [A/G] intron, scaffold_2_10947571 [A/C] noncoding region, and scaffold_2_10948215 [G/T] noncoding region). The fact that this interval exhibited association with lignin content in decades-old mature trees in their native environments across the species range as well as juvenile 4-year-old trees suggested that this locus might play a key role in lignin biosynthesis in Populus. To rule out the possibility of false associations due to the low frequency of these alleles, we generated Potri.002G146400 overexpression Populus and analyzed phenotypic and molecular changes related with lignin biosynthesis.
Table 1. SNP Markers Ranking in the Top 10 Associations out of 2352 SNPs on Chromosome II of the Populus v2.2 Reference Genome Assembly.
| Native Environment_2008 | Clatskanie_2012 | ||||
|---|---|---|---|---|---|
| SNP Marker | P Value | DF | SNP Marker | P Value | DF |
| scaffold_2_6958732 | 0.0000321 | 682 | scaffold_2_10947571* | 0.000574308 | 382 |
| scaffold_2_782169 | 0.0000581 | 683 | scaffold_2_286493 | 0.000722477 | 365 |
| scaffold_2_6956134 | 0.0000704 | 683 | scaffold_2_11628510 | 0.000765244 | 348 |
| scaffold_2_10945723* | 0.0000752 | 681 | scaffold_2_358872 | 0.001743983 | 363 |
| scaffold_2_10948215* | 0.0000861 | 683 | scaffold_2_11990358 | 0.003210959 | 380 |
| scaffold_2_6976928 | 0.0000905 | 682 | scaffold_2_10948215* | 0.00379822 | 382 |
| scaffold_2_10944029* | 0.0000909 | 683 | scaffold_2_10944029* | 0.004179331 | 382 |
| scaffold_2_10947571* | 0.0000928 | 683 | scaffold_2_23037473 | 0.004660356 | 381 |
| scaffold_2_15761393 | 0.000307 | 679 | scaffold_2_7096789 | 0.00473181 | 382 |
| scaffold_2_15757811 | 0.000562 | 675 | scaffold_2_7109565 | 0.00484312 | 382 |
Bold font and asterisks indicate SNP markers located in the Populus gene model Potri.002G146400 ranking in the top 10 across two test environments. DF, degrees of freedom. Bold SNPs were represented among the top 10 SNPs across both environments.
Overexpression of Potri.002G146400 Alters the Deposition of Lignin, Accumulation of Phenylpropanoid Metabolites, and Expression of Secondary Cell Wall Biosynthesis Genes
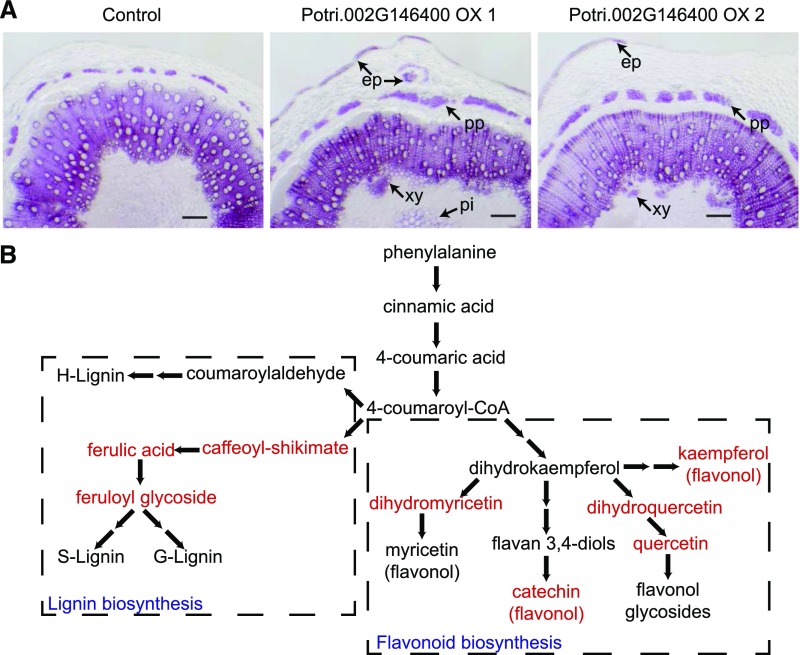
To experimentally link the Potri.002G146400 locus with lignin biosynthesis, we first investigated lignin deposition in stem sections from 1-month-old transgenic Populus overexpressing Potri.002G146400. Lignin depositions of two independent transgenic lines (Potri.002G146400 OX-1 and Potri.002G146400 OX-2) were compared with that of plants transformed with empty vector (control). Phloroglucinol-HCl staining was performed to visualize lignin in cell walls (red-violet). In Potri.002G146400 overexpression plants, we observed ectopic deposition of lignin in epidermis, phloem fiber, and pith cells (Figure 1A), suggesting that Potri.002G146400 affects lignin biosynthesis in Populus.
Figure 1.
Potri.002G146400 Affects the Phenylpropanoid Pathway and Lignin Biosynthesis.
(A) Phloroglucinol-HCl staining of stems from transgenic Populus plants overexpressing Potri.002G146400. Ectopic lignin depositions in different tissues are illustrated by black arrows. ep, epidermis; pp, phloem fibers; xy, secondary xylem; pi, pith cells. Bar = 200 µm.
(B) Changes of phenylpropanoid pathway metabolites in two independent Potri.002G146400 overexpression plants. Up-regulated metabolites in both transgenic lines are in red, based on the metabolism data in Supplemental Table 1.
To provide further evidence supporting the connection between PtrEPSP-TF and lignin biosynthesis, we measured secondary metabolites in Potri.002G146400 overexpression lines using gas chromatography-mass spectrometry. Metabolites in the lignin biosynthesis pathway, including feruloyl glycoside, ferulic acid, and caffeoyl conjugates, exhibited significant increases, between 7% and 87%, in the overexpression plants relative to controls (Figure 1B; Supplemental Table 1). Besides lignin-related metabolites, other products of the phenylpropanoid pathway also showed increased accumulation. As shown in Figure 1B and Supplemental Table 1, levels of quercetins, dihydromyricetin, and catechins exhibited up to 2.8-fold increases in the overexpression lines.
To fully define the molecular effects of Potri.002G146400 in Populus, we extracted total RNAs from stems of transgenic Populus plants and performed RNA-seq analysis. Differential gene expression (DGE) analysis identified a total of 89 genes, which had consistent expression changes between two independent Potri.002G146400 overexpression lines. Based on DGE analysis performed using DESeq2 (v1.2.10) (Love et al., 2014), 71 upregulated genes and 18 downregulated genes were identified (P value < 0.01, false discovery rate < 0.05; Supplemental Table 2). This set was too small for Gene Ontology enrichment. By searching biological functions of individual gene, a number of secondary cell wall biosynthesis genes were identified among upregulated genes: Potri.006G087100 (LACCASE17), Potri.006G087500 (LACCASE17), and Potri.006G096900 (LACCASE4) for lignin biosynthesis; Potri.009G006500 (IRX7) and Potri.011G132600 (IRX8) for xylan biosynthesis; and Potri.011G058400 (SND2; which directly activate CesA8 [cellulose], but not IRX9 [xylan] or 4CL1 [lignin] in Arabidopsis; Zhong et al., 2008) for cellulose biosynthesis (Supplemental Table 2). More importantly, the expression of two master regulators of secondary cell wall biosynthesis, including Potri.009G053900 (PtrMYB021/MYB46) and Potri.011G153300 (NAC SECONDARYWALL THICKENING PROMOTING FACTOR1 [NST1]) (Mitsuda et al., 2007), were upregulated by Potri.002G146400 overexpression (Supplemental Table 2). By contrast, the 18 downregulated genes exhibit little association with secondary cell wall biosynthesis.
Based on these cumulative observations, we hypothesized that Potri.002G146400 may affect lignin biosynthesis and the phenylpropanoid pathway via regulating the expression of master regulators of secondary cell wall biosynthesis, such as MYB46 and NST1.
Potri.002G146400 Encodes an EPSP Synthase Protein with an Additional Motif at Its N Terminus
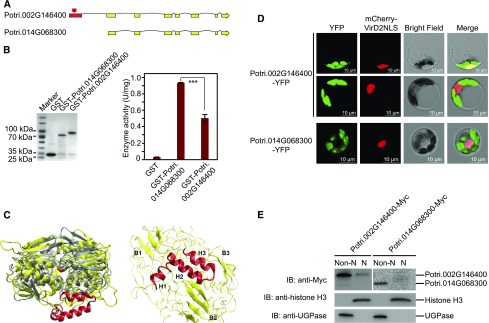
In Populus, Potri.002G146400 has one paralog Potri.014G068300 that, presumably, arose from the Salicoid whole-genome duplication event (Tuskan et al., 2006). Both are annotated as EPSP synthases. Based on RNA-seq coverage (https://phytozome.jgi.doe.gov), Potri.002G146400 has a longer N-terminal than Potri.014G068300. Although the proteins encoded by these two genes share 90.0% sequence similarity, Potri.002G146400, the candidate gene, carries an extra exon in the 5′ region, resulting in a longer cDNA transcript with a total of 1557 nucleotides and encoding a protein with 518 amino acids (∼56 kD), which is larger than the canonical EPSP synthase (∼46 kD) (Figure 2A). By contrast, the transcript for the putative paralog Potri.014G068300 is 1173 nucleotides long and encodes a protein with 390 amino acids, corresponding to the canonical EPSP sequences reported in multiple organisms (Garg et al., 2014). To determine whether the two putative Populus paralogs retain EPSP synthase activity, we expressed GST-tagged proteins in Escherichia coli and purified them (GST-Potri.002G146400 and GST-Potri.014G068300; Figure 2B). A GST tag-only was purified as a negative control. EPSP synthase activity was measured with the presence of 1 mM phosphoenolpyruvate, 1 mM shikimate-3-phosphate, and 100 mM KCl. As shown in Figure 2B, both Potri.002G146400 (0.506 ± 0.041 U/mg) and Potri.014G068300 (0.936 ± 0.003 U/mg) displayed enzymatic activities. The shorter paralog (Potri.014G068300) exhibited stronger EPSP synthase activity than the longer paralog (Potri.002G146400; Figure 2B).
Figure 2.
Potri.002G146400 Encodes an HTH Motif Containing Protein.
(A) Comparison of two putatively paralogous genes, Potri.002G146400 and Potri.014G068300, shows an additional exon (red) encoding an N-terminal HTH motif.
(B) Left panel: Purified GST, GST-Potri.002G146400, and GST-Potri.014G068300 were resolved on an SDS-PAGE gel and stained with Coomassie blue. The protein molecular weights are indicated on the left. GST only was used as a negative control. Right panel: EPSP synthase activities of GST, GST-Potri.002G146400, and GST-Potri.014G068300. For each sample, three reactions were performed in parallel to calculate the mean value and sd (error bar), which were used in Student’s t tests (***P < 0.001, **P < 0.01, *P < 0.05, and ns P > 0.05).
(C) Left: Potri.002G146400 (yellow) superimposed on an EPSP synthase from the Agrobacterium strain CP4 crystallographic structure (gray, PDB entry 2GG6) bound with a shikimate-3-phosphate substrate (blue and red spheres). The HTH domain is shown in red. Right: Detailed view of the HTH domain of Potri.002G146400 comprised of three characteristic α-helices H1 to H3, which are surrounded by β-sheets B1 to B3 in the enzyme. The rest of the enzyme is shown in ribbons.
(D) Subcellular localization of Potri.002G146400-YFP and Potri.014G068300-YFP in Populus protoplasts (green). The nuclear marker mCherry-VirD2NLS is shown in red. Bar = 10 µm.
(E) Immunoblot showing the accumulation of Potri.002G146400-Myc in non-nuclear and nuclear fractions. Potril002G146400-Myc and Potri.014G068300-Myc were blotted using anti-Myc. UGPase and histone H3 were blotted to indicate the purity of non-nuclear and nuclear fractions, respectively. Non-N, non-nuclear fraction; N, nuclear fraction. P value comparison is calculated using two-tailed Student’s t tests (***P < 0.001, **P < 0.01, *P < 0.05, and ns P > 0.05).
Given the sequence variation between EPSP synthase paralogs, we employed atomistic modeling and molecular dynamics simulations to characterize differences between the two Populus paralogs. Characterization of the tertiary structure ascending from the longer N terminus revealed a putative helix-turn-helix (HTH) motif spanning amino acid residues 30 to 70 of Potri.002G146400 (Figure 2C). This motif has the classic three α-helices surrounded by three β-sheets that are characteristically found in nucleic acid binding HTH domains of transcription factors (Aravind et al., 2005). The remainder of the protein, like the paralog Potri.014G068300, shared high similarity with the Agrobacterium tumefaciens-derived cp4 EPSP synthase with an intact substrate binding domain (Figure 2C). Given that EPSP synthase, as the name implies, was only known as a biosynthesis enzyme catalyzing reactions in chloroplasts, the presence of a predicted DNA binding domain in Potri.002G146400 suggested the possibility of an evolved or co-opted novel function.
Protein Encoded by Potri.002G146400 Accumulates in the Nucleus
The presence of a putative HTH motif in the protein encoded by Potri.002G146400 suggested that it may have novel or additional functions divergent from the canonical EPSP synthase in the shikimate pathway. Because the molecular function of a protein is closely linked to its subcellular localization (Lu and Hunter, 2005) and to explore potential functions of Potri.002G146400, we evaluated Potri.002G146400 subcellular localization using Populus protoplasts. We generated the construct of Potri.002G146400 fused to YFP at its C terminus (Potri.002G146400-YFP) and cotransfected it with a nuclear marker fused with mCherry tag (mCherry-VirD2NLS) (Lee et al., 2008) into Populus protoplasts (Figure 2D). Although the majority of Porti.002G146400-YFP signal (shown in green) was detected in chloroplasts, we observed that in ∼10% transfected cell, the fluorescence signal of Potri.002G146400-YFP overlapped with that of mCherry-VirD2NLS. To further determine the accumulation of EPSP synthase encoded by Pori.002G146400 in the nucleus, we expressed C-terminal Myc-tagged Potri.002G146400 (Potri.002G146400-Myc) in protoplasts and performed cell fractionation analysis. Immunoblotting with anti-Myc was used to detect the accumulation of Potri.002G146400-Myc in non-nuclear and nuclear fractions. The cytosolic marker UGPase and nuclear marker histone H3 were blotted simultaneously to indicate the purity of each fraction (Figure 2E). Consistent with subcellular localization results, Potri.002G146400 accumulation was detected in both non-nuclear and nuclear fractions, whereas the homologous construct Potri.014G068300-Myc was only detected in the non-nuclear fraction (Figure 2E). Based on these assays, Potri.002G146400 exhibits dissimilar subcellular localization from canonical EPSP synthase and accumulates in both chloroplasts and the nucleus, leading to the hypothesis that Potri.002G146400-encoded PtrEPSP functions as a transcriptional regulator, as further described below.
Potri.002G146400-Encoded PtrEPSP Is a Transcriptional Repressor
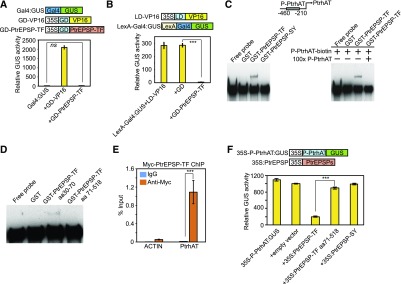
HTH motifs are commonly found in transcription factors (Aravind et al., 2005). To test whether Potri.002G146400 has transcriptional activity, we applied the Populus protoplast transient expression system to evaluate both the transcriptional activator and repressor activity of Potri.002G146400. We generated constructs to overexpress Gal4-DNA binding domain-fused Potri.002G146400 (GD-Potri.002G146400; Figure 3A). Two reporter constructs were generated to analyze activator and repressor activity. To analyze activator activity, the GUS reporter gene was fused downstream of the Gal4 DNA binding site (Gal4:GUS; Figure 3A), which is bound by GD. In this assay, Potri.002G146400 was recruited to the upstream region of the GUS gene coding sequence via the association between GD and the Gal4 DNA binding site. If Potri.002G146400 acts as a transcriptional activator, one would expect the activation of the expression of GUS reporter downstream of the Gal4 DNA binding site when cotransfecting the two constructs into protoplasts. However, little GUS activity was detected in protoplasts cotransfected with GD-Potri.002G146400 and Gal4:GUS (Figure 3A). By contrast, the positive control, a construct in which GD is fused with a transactivator, the Herpes simplex virus VP16 (GD-VP16) (Tiwari et al., 2003), was capable of activating GUS reporter expression to high levels (Figure 3A). These results suggested that Potri.002G146400 has no transcriptional activator activity.
Figure 3.
PtrEPSP-TF Is a Transcriptional Repressor and Directly Targets PtrhAT.
(A) Potril002G146400 has no activator activity. In the reporter construct (Gal4:GUS), the GUS reporter gene was fused downstream of the Gal4 DNA binding site. The construct GD-PtrEPSP-TF was used to express Gal4 binding domain (GD) fused with PtrEPSP-TF. The construct only expressing GD was used as the negative control. Transactivator GD-VP16 was used as the positive control.
(B) Potril002G146400 represses the expression of the GUS reporter activated by the transactivator LD-VP16, in which the transcription activator VP16 is fused with LexA binding domain (LD). In the reporter construct (LexA-Gal4:GUS), the GUS reporter gene was fused downstream of the LexA DNA binding site and Gal4 DNA binding site. The construct only expressing GD was used as the negative control. Luciferase activity was used to normalize and calculate the relative GUS activity and the 35S:Luciferase construct was cotransfected with reporters and effectors.
(C) DNA binding assays of PtrEPSP-TF and PtrEPSP-SY with the promoter region of PtrhAT. GST only was used as a negative control in the DNA binding assays. 100× unlabeled DNAs with the same sequence of biotin-labeled PtrhAT promoter DNA were used for the competition assay.
(D) DNA binding activities of the HTH motif (PtEPSP-TF amino acids 30–70) and truncated PtEPSP-TF without HTH motif (PtEPSP-TF amino acids 71–518).
(E) PtrEPSP-TF binds to the PtrhAT promoter in vivo. µChIP was performed in protoplasts to analyze in vivo targets of PtrEPSP-TF. To quantify DNA enrichment, input DNA was analyzed by quantitative PCR to calculate ChIP signal (% INPUT). Reactions with IgG were used as negative controls. The promoter region of PtACTIN (Potri.019G010400) was amplified to indicate the specificity of ChIP. Means and standard derivations (error bars) of three technical repeats are presented. P value comparison was calculated using two tailed Student’s t tests (***P < 0.001). µChIP was performed at least three times.
(F) Transcriptional repression of EPSP-TF on the PtrhAT promoter. The repression activity of the blank vector was analyzed in parallel as a negative control.
For transcription activity analysis of (A), (B), and (F), all transfection assays were performed in triplicate to calculate the mean value and sd (error bar), which were used in Student’s t tests. P value comparison is calculated using two tailed Student’s t tests (***P < 0.001, **P < 0.01, *P < 0.05, and ns P > 0.05).
To analyze repressor activity, a reporter construct containing the LexA DNA binding site, Gal4 DNA binding site, and GUS reporter gene was used (LexA-Gal4:GUS; Figure 3B). The GUS expression of LexA-Gal4:GUS can be activated by a transactivator construct containing LexA-DNA binding domain-fused VP16 (LD-VP16; Figure 3B). GD-Potri.002G146400 and LD-VP16 have different binding sites on LexA-Gal4:GUS reporter and do not compete for binding sites. When these three constructs were cotransfected into protoplasts, the GUS activity activated by LD-VP16 was abolished (Figure 3B). As a negative control, the effector containing only GD had no effect on the expression of GUS reporter (Figure 3B). These results demonstrated that Potri.002G146400 may function as a transcriptional repressor. As a result of this apparent transcriptional activity, henceforth Potri.002G146400 is referred to as PtrEPSP-transcription factor (PtrEPSP-TF) and Potri.014G068300 as PtrEPSP-synthase (PtrEPSP-SY).
PtrEPSP-TF Directly Binds to the PtrhAT Promoter in Vivo and in Vitro
To investigate the molecular mechanisms linking the transcriptional repressor function of PtrEPSP-TF and its role in lignin biosynthesis or the phenylpropanoid pathway, we sought to identify the direct target genes of PtrEPSP-TF by mining RNA-seq data from PtrEPSP-TF overexpression lines and then validating the candidates via an in vitro electrophoretic mobility shift assay (EMSA) approach. Given PtrEPSP-TF functions as a repressor in the Populus protoplast assays, we targeted the top three genes downregulated by PtrEPSP-TF (Supplemental Table 2). Among the three genes, PtrEPSP-TF, but not PtrEPSP-SY, displayed binding affinity to the promoter of a hAT transposase family gene (Potri.016G026100, designated as PtrhAT) (Figure 3C; Supplemental Figure 1). PtrhAT shares amino acid sequence similarity with the Arabidopsis DAYSLEEPER protein, which is a known global transcriptional regulator (Bundock and Hooykaas, 2005). PtrEPSP-TF does not bind to promoters of tested upregulated genes in PtrEPSP-TF overexpression lines, such as Potri.009G053900 (PtrMYB021) and Potri.011G153300 (NST1; Supplemental Figure 1). As shown in Figure 3C, incubating GST-PtrEPSP-TF and biotin-labeled PtrhAT promoter (P-PtrhAT-biotin, −460 to −210) resulted in a mobility shift above the free probe band. By contrast, neither GST nor GST-PtrEPSP-SY generated the same mobility shift (Figure 3C). Furthermore, the fact that the binding between PtrEPSP-TF and P-PtrhAT-biotin was abolished by competition with 100 times unlabeled P-PtrhAT DNA suggests that binding of PtrEPSP-TF to the PtrhAT promoter is direct and specific under in vitro conditions (Figure 3C). To further determine whether the HTH motif in PtrEPSP-TF is responsible for the DNA binding activity, we measured binding affinities of the HTH motif (PtrEPSP-TF amino acids 30–70) and truncated PtrEPSP-TF without the HTH motif (PtrEPSP-TF amino acids 71–518) using EMSA. As shown in Figure 3D, it is the HTH motif that is responsible for binding to the PtrhAT promoter.
We further performed chromatin immunoprecipitation (ChIP) experiments to examine the in vivo binding of PtrEPSP-TF to the PtrhAT promoter. Because the generation of transgenic Populus is very time-consuming, we combined transient protein expression technique in protoplast and micro-ChIP (µChIP) approaches modified from mammalian studies (Dahl and Collas, 2008; Para et al., 2014). µChIP is capable of discovering protein-DNA binding from limited numbers of cells, such as cell suspensions and protoplasts. The 10xMyc-tagged PtrEPSP-TF (Myc-PtrEPSP-TF) was overexpressed in Populus protoplasts (Supplemental Figure 2) and then subjected to a µChIP assay. The ChIPed DNA was analyzed by quantitative PCR to detect DNA enrichment. As shown in Figure 3E, fragments from the PtrhAT promoter, but not from the PtACTIN promoter (Potri.019G010400) (Li et al., 2014), were enriched in Myc-PtrEPSP-TF precipitates, confirming in vivo association between PtrEPSP-TF and the PtrhAT promoter.
If PtrEPSP-TF directly binds to the PtrhAT promoter, as a repressor, PtrEPSP-TF is expected to suppress the activity of the PtrhAT promoter. To test this, we performed repressor activity analysis using an in vivo protoplast system. We generated a construct to overexpress PtrEPSP-TF without any tag (35S:PtrEPSP-TF; Figure 3F). In the reporter construct, the PtrhAT promoter (−460 to −210 bp from the start codon) region was inserted between the CaMV 35S promoter and GUS reporter gene (35S-P-PtrhAT:GUS; Figure 3F). This reporter construct showed high GUS gene expression in protoplasts (Figure 3F), whereas cotransfection of 35S:PtrEPSP-TF and 35S-P-PtrhAT:GUS showed reduced GUS expression (Figure 3F), suggesting that PtrEPSP-TF directly binds to the PtrhAT promoter and represses its activity. Consistently, neither the truncated PtrEPSP-TF without the HTH motif (PtrEPSP-TF amino acids 71–518) nor PtrEPSP-SY repressed the expression of GUS, which is downstream of the PtrhAT promoter (Figure 3F).
PtrhAT Is a Transcriptional Repressor
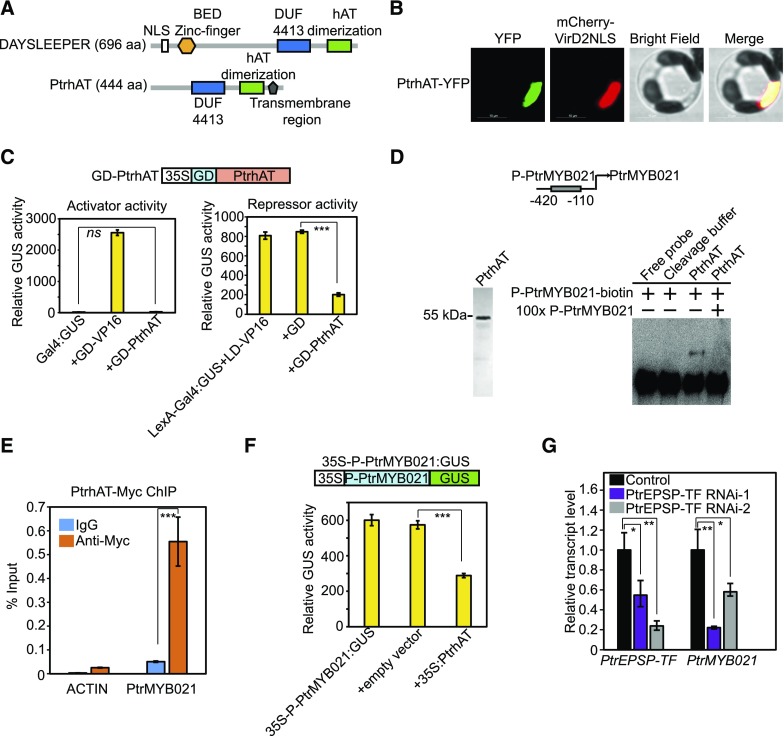
Although the angiosperm-specific SLEEPER genes have been reported to be essential for plant growth and development (Bundock and Hooykaas, 2005), no direct connection has been made between SLEEPER genes and the established cell wall biosynthesis transcriptional hierarchy (Hussey et al., 2013). It should be noted that PtrhAT is much shorter than the DAYSLEEPER in Arabidopsis (444 versus 696 amino acids). Amino acid alignment of PtrhAT and AtDAYSLEEPER indicated that PtrhAT also lacked the K/R rich nuclear localization domain (NLS) adjacent to the BED zinc finger domain near the N-terminal region (Figure 4A). Because CYTOSLEEPER (encoded by AT1G15300), which also lacks the K/R-rich NLS adjacent to the BED zinc finger domain, was shown to be localized in the cytosol, it has been proposed that this K/R-rich NLS domain is necessary for nuclear localization of DAYSLEEPER proteins (Knip et al., 2012). However, phylogenetic analysis of Populus DAYSLEEPER-like genes illustrated that PtrhAT might not belong to either the DAYSLEEPER or CYTOSLEEPER group (Supplemental Figure 3; Supplemental Data Set 1). This prompted us to examine the subcellular localization of PtrhAT. By examining the fluorescence of PtrhAT-YFP fusion protein transiently expressed in the Populus protoplasts, we found that PtrhAT is localized in the nucleus (Figure 4B).
Figure 4.
PtrhAT Directly Targets PtrMYB021.
(A) Comparison of protein domains and motifs between Populus PtrhAT and Arabidopsis DAYSLEEPER.
(B) Nuclear localization of PtrhAT in Populus protoplasts. PtrhAT-YFP (green) was transiently expressed in Populus protoplasts. MCherry-VirD2NLS (red) was cotransfected to indicate nucleus localization. Bar = 10 µm.
(C) PtrhAT has repressor but not activator activity. Transcriptional activity of PtrhAT was analyzed in a similar protoplast transient expression system as the analyses on PtrEPSP-TF. 35S:Luciferase was cotransfected with reporters and effectors and Luciferase activity was used to normalize and calculate the relative GUS activity. All transfection assays were performed in triplicate to calculate the mean value and sd (error bar), which were used in Student’s t tests.
(D) EMSA. Left: Purified PtrhAT protein. GST-PtrhAT was expressed and purified in E. coli. The GST tag was subsequently cleaved off using PreScission Protease. Right: PtrhAT directly binds to the PtrMYB021 promoter in vitro. Cleavage buffer was used as the negative control in EMSA. 100× unlabeled DNAs with the same sequence of biotin-labeled PtrMYB021 promoter region were used for the competition assay.
(E) µChIP shows in vivo association of PtrhAT and the PtrMYB021 promoter. Reactions with IgG were used as negative controls. The promoter region of PtACTIN was amplified to indicate the specificity of ChIP. Means and standard derivations (error bars) of three technical repeats are presented. µChIP was performed at least three times.
(F) PtrhAT represses the activity of the PtrMYB021 promoter. PtrMYB021 promoter (−420 to −110 nucleotides from the start codon) was inserted between the 35S promoter and GUS reporter and then cotransfected with vectors overexpressing PtrhAT. Blank vector was used as negative control. All transfection assays were performed in triplicate to calculate the mean value and sd (error bar), which were used in Student’s t tests.
(G) RT-qPCR analysis of transcript levels of PtrEPSP-TF and PtrMYB021 in two Populus PtrEPSP-TF RNAi lines (PtrEPSP-TF RNAi-1 and PtrEPSP-TF RNAi-2). RT-qPCR analysis was performed in triplicate to calculate the mean value and sd (error bar), which were used in Student’s t tests.
P value comparisons of (C) and (E) to (G) were calculated using two-tailed Student’s t tests (***P < 0.001, **P < 0.01, *P < 0.05, and ns P > 0.05).
As the direct target of PtrEPSP-TF, PtrhAT appears to be an intermediary step in PtrEPSP-TF-triggered transcriptional regulation. Thus, PtrhAT may also have transcriptional activity. To test this possibility, PtrhAT was fused in frame with GD (GD-PtrhAT) for transcriptional activity analyses in protoplasts. Similar to PtrEPSP-TF, GD-PtrhAT reduced GUS expression of the LaxA-Gal4:GUS reporter (activated by LD-VP16) but had no effect on GUS expression of the Gal4:GUS reporter (Figure 4C), suggesting that PtrhAT also functions as a transcriptional repressor.
PtrhAT Directly Binds to the PtrMYB021 Promoter and Represses PtrMYB021 Expression
Results showing that both PtrEPSP-TF and its direct target PtrhAT are repressors prompted us to examine whether PtrhAT directly targets Potri.009G053900 (PtrMYB021) and/or Potri.011G153300 (NST1), which are upregulated in PtrEPSP-TF overexpression lines. If this is the case, by suppressing PtrhAT expression, PtrEPSP-TF would upregulate the expression of master regulators of secondary cell wall biosynthesis, which would be consistent with our RNA-seq results (Supplemental Table 1). To test this possibility, we expressed and purified PtrhAT in vitro (Figure 4D) and used it for in vitro EMSA. As expected, PtrhAT specifically bound to a 310-bp PtrMYB021 promoter (−420 to −110 nucleotides from the start codon; Figure 4D). However, no binding to the NST1 promoter was detected (Supplemental Figure 1). Consistent with the EMSA results, our µChIP results also showed drastic enrichment of the PtrMYB021 promoter in PtrhAT precipitates (Figure 4E), illustrating in vivo binding of PtrhAT to the PtrMYB021 promoter. By inserting the PtrMYB021 promoter (−420 to −110 nucleotides from the start codon) between 35S promoter and the GUS reporter gene (35S-P-PtrMYB021:GUS) in the reporter construct for repressor activity analysis, we examined the effect of PtrhAT on PtrMYB021 promoter activity. As predicted, PtrhAT reduced the activity of the PtrMYB021 promoter (Figure 4F).
Systematically, we conclude that PtrMYB021 is a direct target of PtrhAT. Combined with the functional characterization of PtrEPSP-TF, we propose that PtrEPSP-TF and PtrhAT form a previously undescribed hierarchical transcriptional regulation on PtrMYB021 expression. In support of this model, the knockdown of PtrEPSP-TF via RNAi reduced the transcript level of PtrMYB021 in Populus (Figure 4G).
Transcriptional Regulation of PtrEPSP-TF by PtWND1B
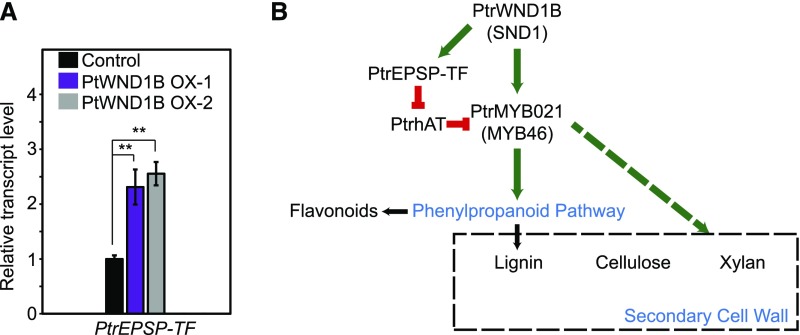
Current Arabidopsis and Populus models of the transcriptional regulatory hierarchy of secondary cell wall biosynthesis is that SND1 (PtWNDs in Populus) activates MYB46 (PtMYB002, PtMYB003, PtMYB020, and PtrMYB021 in Populus) and the expression of downstream genes (Kim et al., 20131b, 2014; Zhong et al., 2007, 2013; McCarthy et al., 2010; Zhong and Ye, 2010). To place the PtrEPSP-TF/PtrhAT mechanism into the current model, we evaluated whether PtrEPSP-TF is downstream of SND1 in the transcriptional regulation of MYB46 in Populus. RT-qPCR analysis showed that PtrEPSP-TF was upregulated in two independent Populus transgenic lines overexpressing PtWND1B (homolog of SND1; Ohtani et al., 2011; Zhao et al., 2014) relative to the empty vector control (Figure 5A), suggesting that the PtrEPSP-TF/PtrhAT mechanism is downstream of SND1.
Figure 5.
PtrEPSP-TF/PtrhAT Mechanism Is a Novel Transcription Regulatory Hierarchy Regulating MYB46 Expression and the Phenylpropanoid Pathway in Populus.
(A) Transcriptional response of PtrEPSP-TF to overexpression of PtWND1B, the homolog of Arabidopsis SND1. RT-qPCR analysis was performed in triplicate to calculate the mean value and sd (error bar), which were used in Student’s t tests. P value comparison was calculated using two-tailed Student’s t tests (**P < 0.01, *P < 0.05, and ns P > 0.05).
(B) A simplified scheme illustrating the transcriptional regulation of the PtrEPSP-TF/PtrhAT mechanism in MYB46 expression and the phenylpropanoid pathway. Solid line indicates processes studied in this work. The dashed line indicates processes not studied in this work. Green indicates transcriptional activation. Red indicates transcriptional repression.
The HTH Motif Is Highly Variable in the Plant Kingdom
Finally, since the HTH motif is not found in homologs of the ancestral prokaryotic progenitor of EPSP synthase, we assessed penetrance of this motif in plants by surveying 57 EPSP synthase isoforms derived from 42 phylogenetically distributed plant genomes. As previously observed (Tohge et al., 2013; Garg et al., 2014), phylogenetic relatedness of sequences reflects the broader classification delineating monocot, dicot, non-vascular and algal clades of the kingdom plantae (Supplemental Figure 4). The most striking observation was that the HTH motif was almost entirely missing in nonvascular, algal, and monocot clades, but interestingly was found in many dicots (Supplemental Figure 5). Sequence alignments suggested that, unlike other plants, dicots share a start codon and a conserved MAQV(A/L/I)S(T) amino acid residue in this additional exon (Supplemental Figure 5). Functional studies of these sequences are outside the scope of this work and will be required to establish penetrance of the transcriptional regulatory function in other plant species.
DISCUSSION
For decades, EPSP synthases were believed to have the single function of catalyzing the conversion from shikimate-3-phosphate to 5-enolpyruvylshikimate-3-phosphate in the shikimate pathway in prokaryotes and eukaryotes (Maeda and Dudareva, 2012; Mir et al., 2015). In plants, the shikimate pathway is upstream of the phenylpropanoid pathway and provides phenylalanine for the biosynthesis of phenylpropanoid compounds (Tohge et al., 2013). Here, we provide evidence for an unrelated function of a Populus isoform, PtrEPSP-TF, that indirectly regulates the expression of MYB46, a master regulator of the phenylpropanoid pathway and secondary cell wall biosynthesis. Although PtrEPSP-TF retains EPSP synthase activity, its enzymatic activity is much weaker than PtrEPSP-SY (Figure 2B). With an additional N-terminal HTH DNA binding motif, PtrEPSP-TF exhibited nuclear accumulation and functioned as a transcriptional repressor (Figures 2 and 3B). Our subcellular localization and cell fraction results demonstrated that PtrEPSP-TF protein, but not its paralog PtrEPSP-SY, accumulates in the nucleus (Figures 2D and 2E). A previous proteomic study on Populus secondary cell wall formation provided clues for the existence of this novel function. The abundance of PtrEPSP-TF, but not PtrEPSP-SY or any other enzymes involved in the shikimate pathway, was found to increase during primary to secondary growth of Populus stem development (Liu et al., 2015). To support this observation, we found that the expression of PtrEPSP-TF was highest in the relatively chloroplast-devoid developing xylem tissue, where Pt-EPSP-SY exhibited extremely low expression in the same tissue (Supplemental Figure 6). These findings were reinforced by an independent proteomic study on nuclear-enriched proteins from Populus developing xylem (Loziuk et al., 2015). In that study, peptides aligning to the N terminus of PtrEPSP-TF, but not PtrEPSP-SY, were detected, again suggesting a nuclear presence for the PtrEPSP-TF protein. Although the direct mechanism underlying the observed nuclear presence remains to be determined, a possible explanation is that other proteins interact with PtrEPSP-TF and block the effect of the N-terminal chloroplast transit peptide, mimicking the effect observed after we tagged the N terminus leading to almost 100% nuclear localization (Supplemental Figure 7). Our ChIP and EMSA results demonstrated that unlike PtrEPSP-SY, PtrEPSP-TF does have DNA binding activity and directly binds to its target (i.e., PtrhAT promoter) under both in vivo and in vitro conditions (Figure 3). Physiologically, the level of lignin biosynthesis during secondary growth is higher than that during primary growth, which requires more monolignols produced via the phenylpropanoid pathway. Considering PtrEPSP-TF is capable of activating the expression of MYB46 and subsequently activating the phenylpropanoid pathway, the accumulation of PtrEPSP-TF proteins from primary to secondary growth in developing xylem is consistent with the regulatory function we described.
Being one close homolog of Arabidopsis MYB46, Populus PtrMYB021 regulates not only the phenylpropanoid pathway and lignin biosynthesis, but also the biosynthesis of cellulose and xylan (Zhong et al., 2013). As an upstream regulator of PtrMYB021, PtrEPSP-TF also displayed tight association with lignin biosynthesis and the phenylpropanoid pathway in Populus. In addition to lignin biosynthesis, multiple genes involved in xylan and cellulose biosynthesis were upregulated in PtrEPSP-TF overexpression Populus (Supplemental Table 2). Whether PtrMYB021 has similar effects as PtrMYB021 on the biosynthesis of xylan and cellulose merits future research. Among the two master regulators with increased expression levels in PtrEPSP-TF overexpression Populus plants, the roles of PtrMYB021 in secondary cell wall formation have been well established. However, the functions of Potri.011G153300 (NST1) remain poorly studied in Populus. Further experiments will be performed to understand how PtrEPSP-TF affects Potri.011G153300 (NST1) expression and whether Potri.011G153300 (NST1) is involved in the regulation of secondary cell wall biosynthesis.
The phenylpropanoid pathway provides key secondary metabolites for secondary cell wall formation and plant immunity. As such, the regulation of MYB46 expression is critical for plants to respond to various developmental and environmental changes. Currently, only the mechanism that SND1 directly activates, MYB46 expression, has been well studied in both Arabidopsis and Populus (Zhong et al., 2007; Ohtani et al., 2011). Such a singular regulatory mechanism seems vulnerable and insufficient to comprehensively regulate MYB46 expression and the phenylpropanoid pathway under diverse and variable developmental and environmental changes experienced by long-lived perennials. The PtrEPSP-TF/PtrhAT mechanism identified in this study broadens our understanding of the regulation of MYB46 expression, and concomitantly the phenylpropanoid pathway, and provides additional targets for engineering the phenylpropanoid pathway to meet the needs of the bioeconomy. Furthermore, the role of SLEEPER hAT transposase family genes in regulating gene expression is poorly studied. Our study illustrates the involvement of a SLEEPER gene in the transcriptional regulation of the phenylpropanoid pathway.
It is noteworthy that exaptation of transposases, such as PtrhAT, and protein moonlighting by biosynthesis enzymes, exemplified by PtrEPSP-TF, which retains chloroplastic localization, has been proposed as adaptive mechanisms yielding novel genetic regulators in eukaryotes, where organismal complexity necessitated a complementary expansion of transcription factors (Wray et al., 2003; Feschotte, 2008). Our results support this proposition. The evolutionary origin of the HTH motif in EPSP-TF, leading to novel repressor activity, could not be exhaustively addressed in this work; however, the presence of secondary cell walls is a key distinguishing feature separating dicots from algae and mosses. It is intriguing that this shikimate pathway derived-EPSP synthase isoform appears to have obtained a regulatory function modulating expression of processes that are ubiquitous in dicots relative to other plants. With this in mind, we hypothesized that domain co-option may have occurred during the course of evolution when early dicotyledonous plants attained complex cell wall structure (Weng et al., 2008; Tohge et al., 2013).
Finally, our characterization of the molecular mechanism linking PtrEPSP-TF function to lignin and subsequently phenylpropanoid biosynthesis in Populus provides a solid foundation for functional studies and confirmation of discoveries from association mapping studies. As it is becoming rapidly clear that model system-based studies have significant limitations in informing the biology of complex organisms, data-driven, non-inference-based methods of linking genes to phenotypes hold tremendous potential in facilitating discovery in target plant species.
METHODS
Populus LD-Based Association Mapping
A genome-wide association mapping was performed using the Populus trichocarpa mapping population that was genotyped using the Populus 34K Illumina Bead Array (Geraldes et al., 2013) and phenotyped for cell wall chemistry as described previously (Muchero et al., 2015). Lignin content was measured in increment cores collected from 683 mature trees in their native environment and 382 4-year-old trees grown in a field site at Clatskanie, OR (46°6′11″N, 123°12′13″W) (Iqbal, 1990). Genotype-phenotype associations were evaluated for 2352 single nucleotide polymorphisms (SNPs) on chromosome II using the mixed linear model analysis with kinship and population structure as covariates (Yu et al., 2006). The analyses were performed in TASSEL software (http://www.maizegenetics.net). Correction for multiple testing was performed using the false discovery rate method (Benjamini and Hochberg, 1995).
PtrEPSP-TF Cloning and Generation of Transgenic Populus
Full-length Potri.002G146400 containing the HTH motif was cloned from a Populus deltoides cDNA library via PCR using primers for sequences from Potri.002G146400: forward primer 5′-CACCCCCGGGAAAGCCATGGCTCAAGTGA-3′ and reverse primer 5′-ACGCGTTTTGAGTGCAACTCAATGCTT-3′. For the PtrEPSP-TF RNAi lines, a 207-bp fragment was cloned using forward primer 5′-CACCCCCGGGGAGGTTCTTGAGAGGTACAC-3′ and reverse primer 5′- TCTAGATTCACATATGACCAGTCTCCA-3′. For PtWND1B overexpression lines, the full-length coding region of PtWND1B (gene model: Potri.001G448400; 1235 bp) was amplified using the forward primer 5′-CACCCCCGGGATGCCTGAGGATATGATGAA-3′ and reverse primer 5′-ACGCGTTTGTTATACCGATAAGTGGCAT-3′. The integrity of fragments was verified by DNA sequencing (ACGT Inc.) after cloning into the Gateway entry vector, pENTR/D-TOPO (Life Technologies). For PtrEPSP-TF overexpression and RNAi lines, the fragment was transferred to a binary Gateway destination plasmid, pAGW560 (Karve et al., 2012) for overexpression and pAGSM552 (GenBank accession number KP259613.2) for knockdown, using LR Clonase II recombination (Life Technologies). The resulting overexpression and knockdown cassettes comprised the Arabidopsis UBIQUITIN3 promoter, the PtrEPSP-TF coding or target sequence, and the nopaline synthase terminator. For PtWND1B overexpression lines, the gene fragment was subcloned into a binary vector under the control of vasculature-specific 4CL promoter. The binary transformation vector was then transformed into Agrobacterium tumefaciens strain EHA105 via electroporation. P. deltoides genotype WV94 was transformed using a modified Agrobacterium-based method (Tsai et al., 1994; Mingozzi et al., 2009). Shoots regenerated from isolated calli were tested using PCR to verify the presence of the transformed construct. Empty vector transformed plants were used as controls. Plants were propagated in a greenhouse maintained at 25°C at 16-h daylength.
Metabolite Profiling
Freeze-dried bark peels were ground with a micro-Wiley mill and ∼50 mg dry weight was subsequently twice extracted with 2.5 mL 80% ethanol overnight and then combined prior to drying a 0.5-mL aliquot in a nitrogen stream. Sorbitol was added before extraction as an internal standard to correct for differences in extraction efficiency, subsequent differences in derivatization efficiency and changes in sample volume during heating. Dried extracts were silylated for 1 h at 70°C to generate trimethylsilyl derivatives, which were analyzed after 2 d with an Agilent Technologies 5975C inert XL gas chromatograph-mass spectrometer as described elsewhere (Li et al., 2012; Tschaplinski et al., 2012). Metabolite peak extraction, identification, and quantification were described previously (Tschaplinski et al., 2012), and unidentified metabolites were denoted by their retention time as well as key m/z ratios. There were 10 replicate plants of upregulated EPSP-TF and empty vector controls from three lines each. Data were pooled across lines and treatment means were tested for statistical significance (P < 0.05) using Student’s t tests.
Structural Modeling and Molecular Dynamics Simulations
PtrEPSP-TF and PtrEPSP-SY models were built using the iterative threading assembly refinement (I-TASSER, v3.0) protein structure modeling toolkit (Roy et al., 2010), which integrates the ab initio (free) modeling and template-based modeling on the basis of the multiple threading alignments for protein structure building (Zhang, 2014). Structure-based functional annotations and ligand/cofactor predictions of the constructed models were performed using COFACTOR (Roy et al., 2012). The structure-based phylogenetic analysis was performed using the MultiSeq (Roberts et al., 2006) bioinformatics toolkit embedded in Visual Molecular Dynamics (Humphrey et al., 1996). A 200-ns molecular dynamics (MD) simulation without any restraint was performed for the best model constructed by I-TASSER. The online program MolProbity (Chen et al., 2010) was applied to validate the rotamers of Asn, Gln, and His and to determine the protonation states of titratable residues of Glu, Asp, Lys, Arg, and His. Missing hydrogen atoms were added using the HBUILD module in CHARMM (Brooks et al., 2009). A water box with at least 15 Å to the edge of the protein was used, and sodium/chloride ions were added to balance the net charge of the whole system. The MD simulations were performed using the software NAMD (Phillips et al., 2005). The CHARMM protein force field (Best et al., 2012) and TIP3P water model (Jorgensen et al., 1983) were adopted in all MD simulations. A time step of 2 fs was applied with the SHAKE algorithm to fix the bonds involving hydrogen atoms. In each MD simulation, after a 50,000-step energy minimization, the temperature of the system was gradually heated to 300K at a rate of 0.001K per time step. The MD simulations were performed under the constant-temperature, constant-pressure (NPT) ensemble with the system pressure of 1 atm and temperature of 300K maintained by the Langevin piston controls. Cutoff of switching between 9 and 11 Å was applied for the nonbonded interactions, and particle mesh Ewald summation with a grid spacing of 1.35 Å was applied for long-range electrostatic interactions, respectively. For each of the 200-ns MD simulations, analyses were performed on the last 100 ns of all MD trajectories.
EPSP Synthase Activity
The enzymatic activities of purified PtrEPSP-TF and PtrEPSP-SY were assayed in 100 μL of 150 mM HEPES-NaOH (pH 7.0), 2 mM DTT, 100 mM KCl, 1 mM phosphoenolpyruvate (Sigma-Aldrich), and 1 mM shikimate-3-phosphate (Sigma-Aldrich). After incubating the samples at room temperature for 10 min, the enzymatic activity was then determined by measuring the amount of inorganic phosphate using malachite green (Carter and Karl, 1982). The enzymatic activity (U/mg) represents µmol product/min of reaction/mg of EPSP protein. For each sample, three reactions were performed in parallel to calculate the mean value and sd (error bar), which were used in Student’s t tests.
Subcellular Localization in Populus Protoplasts
Protoplasts from Populus were isolated and subsequently transfected, as previously described (Guo et al., 2012). For EPSP subcellular localization, 8 µg of YFP fusion PtrEPSP-TF, PtrEPSP-SY, and PtrhAT constructs (CaMV 35S promoter driving) were cotransfected with 2 µg of VirD2NLS-mCherry construct (nuclear marker) into 100 μL of protoplasts, respectively. After 12 h incubation, YFP and mCherry fluorescence were examined and photographed. Images were collected on a Zeiss LSM 710 confocal microscope and images were processed using the Zeiss ZEN software package.
Transcriptional Activity Assay
The protoplast transfection-based transcriptional activity assay was performed according to a previously described method (Tiwari et al., 2003). Ten micrograms of effector, reporter, and/or transactivator plasmids was cotransfected into 100 μL of Populus protoplasts using the PEG-calcium transfection method and incubated under darkness for 18 to 20 h at room temperature. A GUS activity assay was performed as described (Yoo et al., 2007). GUS activity was measured using a Fluoroskan microplate reader. To normalize GUS activity, 100 ng of 35S:Luciferase plasmid was cotransfected for each transfection. Luciferase activity was measured using a Promega Luciferase Assay System according to the manufacturer’s manual. All transfections were performed in triplicate to calculate the mean value and sd, which were used in Student’s t tests.
EMSA
PtrEPSP-TF (full-length and truncated fragments) and PtrEPSP-SY were cloned into the pGEX-6P-1 vector (GE Healthcare) by BamHI and EcoRI for GST fusion constructs. PtrhAT was cloned into the pGEX-6P-1 vector by EcoRI and XhoI. The constructs were transformed into Escherichia coli strain BL21(DE3)pLysS (Invitrogen) for protein expression. GST fusion proteins were extracted and purified as previously described using Glutathione Sepharose 4B beads (GE Healthcare) (Xie et al., 2012). To perform EMSA, GST-PtrEPSP-TF (full-length and truncated fragments) and GST-PtrEPSP-SY were then eluted from beads by incubating with Elution Buffer (50 mM Tris-HCl, pH 8.0, and 10 mM reduced glutathione) at 4°C for 30 min. PtrhAT was cleaved from beads by PreScission Protease (GE Healthcare) in cleavage buffer (50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, and 1 mM DTT, pH 7.0) at 4°C overnight. For DNA probes, DNA fragments inside the region 500 bp upstream from the start codon were amplified by PCR from Populus clone 717-1B4 (female, Populus tremula × alba) genomic DNA, gel purified, and end labeled with biotin using a DNA 3′ End Biotinylation Kit (Thermo Fisher Scientific) according to the manufacturer’s manual. The DNA binding reaction included 0.25 nM biotin-labeled probe, 0.4 µg of purified protein, 10 mM Tris-HCl (pH 7.5), 50 mM KCl, 1 mM DTT, 2.5% glycerol, 5 mM MgCl2, 1 µg poly(dI-dC), and 0.05% Nonidet P-40. Reactions were incubated at room temperature for 20 min. The reaction mixtures were then resolved in a 6% DNA retardation gel (Novex) by electrophoresis at 100 V for 1 to 2 h and electrophoretically transferred to nylon membrane. Signals of biotin were detected using the Chemiluminescent Nucleic Acid Detection Module (Thermo Fisher Scientific) as suggested by the manufacturer.
µChIP in Populus Protoplasts
µChIP was performed as previously described with a few modifications (Dahl and Collas, 2008; Para et al., 2014). Myc fusion PtrEPSP-TF and PtrhAT were transfected and transiently expressed in protoplasts. After 14 h incubation at room temperature, ∼50,000 transfected protoplasts were then used for µChIP. Cells were cross-linked by 1% formaldehyde in W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, and 2 mM MES, pH 5.7) for 8 min at room temperature with a gentle rotation. The formaldehyde was subsequently quenched by adding 1.25 M glycine (Sigma-Aldrich) to the final concentration of 125 mM and incubating the samples for 5 min at room temperature with gentle rotation. After two washes with W5 solution, cells were collected by centrifugation (2000 rpm for 10 min, 4°C) and lysed in 50 μL of lysis buffer (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 10 mM EDTA, pH 8.0, 1% SDS, 1 mM PMSF, and protease inhibitor [Sigma-Aldrich]) with intermittent vortexing for 20 min. The concentration of SDS was then diluted to less than 0.1% by adding 800 μL of ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.0, 167 mM NaCl, 1 mM PMSF, and protease inhibitor [Sigma-Aldrich]). After dilution, cell lysate was sonicated for 150 s with 0.7 s ON and 1.3 s OFF pulses at 20% power amplitude using the Branson 450 Digital sonifier machine to achieve chromatin fragments of 150 to 600 bp (Adli and Bernstein, 2011). The sonicated cell lysate was added to an additional 150 μL of ChIP dilution buffer and centrifuged at 10,000g for 10 min at 4°C to remove cellular debris. After centrifugation, the supernatant was aliquoted into three clean 1.5-mL tubes: 25 μL for input sample, 450 μL for IgG control, and 450 μL for ChIP with antibody. Additional ChIP dilution buffer was then added: 75 μL for input sample, 450 μL for IgG control, and 450 μL for ChIP with antibody. The protein-DNA complexes were captured using anti-Myc antibody (Sigma-Aldrich) at 4°C overnight and then incubated with washed Magnetic Protein-A beads (GE Healthcare) at 4°C for 1 h. After six washes (two washes with low-salt buffer [150 mM NaCl, 0.1% SDS, 20 mM Tris-HCl, pH 8.0, 2 mM EDTA, pH 8.0, and 1% Triton X-100], two LiCl buffer washes [0.25 M LiCl, 1% Na-deoxycholate, 10 mM Tris-HCl, pH 8.0, 1% Nonidet P-40, and 1 mM EDTA, pH 8.0], and two TE buffer washes [10 mM Tris-HCl, pH 8.0, and 1 mM EDTA, pH 8.0]), DNAs were eluted from beads as described (Dahl and Collas, 2008). The ChIPed DNA and input DNA were then cleaned and concentrated using a Qiagen MinElute PCR purification kit (Qiagen). qPCR was then performed to quantify DNA enrichment. Three biological replicates were performed. The following primers were used for qPCR: Promoter∼PtActin (Potri.019G010400) (forward, ACCTACTTCGTTTGGTCATTGTTA; reverse, CAAATACAACATACTAGTTCCTCCAC); Promoter∼PtrhAT (Potri.016G026100) (forward, CCCACAACAATCAACCCATA; reverse, GGGGAAAATAAGGGAAAAAGG); Promoter∼PtrMYB021 (Potri.009G053900) (forward TGAGCAGTAAAACGGTTTGG; reverse, GGAAAAGGACAAGATCATGGA). µChIP analyses were performed in triplicate using independent transfected cells to ensure results were consistent.
RNA-Seq Analysis
For each transgenic line, mature stems (internodes 6 to 8) were collected between 12:00 and 2:00 pm from four individual plants growing in the greenhouse for 6 months after propagation from cuttings. Total RNAs were extracted from the developing xylem (scrapped stem under the bark of the mature stem [internodes 6 to 8]). Stranded RNA-seq libraries were created and quantified by qPCR. Sequencing was performed using an Illumina HiSeq 2500 instrument. Raw fastq file reads were filtered and trimmed using the JGI QC pipeline resulting in the filtered fastq file (*.anqrp.fastq.gz files). Using BBDuk (https://sourceforge.net/projects/bbmap/), raw reads were evaluated for artifact sequence by k-mer matching (k-mer = 25), allowing one mismatch and detected artifact was trimmed from the 3′ end of the reads. RNA spike-in reads, PhiX reads, and reads containing any Ns were removed. Quality trimming was performed using the phred trimming method set at Q6. Finally, following trimming, reads under the length threshold were removed (minimum length 25 bases or one-third of the original read length, whichever is longer). Raw reads from each library were aligned to the reference genome using TopHat (Kim et al., 2013a) with only unique mapping allowed (BAMs/directory). If a read mapped to more than one location, it was ignored. FeatureCounts (Liao et al., 2014) was used to generate the raw gene counts. Raw gene counts were used to evaluate the level of correlation between biological replicates using Pearson’s correlation and to determine which replicates would be used in the DGE analysis. DESeq2 (v1.2.10) (Love et al., 2014) was subsequently used to determine which genes were differentially expressed between pairs of conditions. The parameters used to call a gene DE between conditions were P ≤ 0.05.
qPCR Analyses
For RNA extraction and gene expression, RNA was extracted from stem and shoot tip samples using Plant RNA extraction kit (Sigma-Aldrich) with modifications as described in our previous report (Payyavula et al., 2014). cDNA synthesis was performed using DNase free total RNA (1.5 µg), oligo(dT) primers, and RevertAid Reverse Transcriptase (Thermo Fisher Scientific). RT-qPCR was performed using 3 ng cDNA, 250 nM gene-specific primers, and iTaq Universal SYBR Green Supermix (Bio-Rad). Gene expression was calculated by the delta-delta cT method (Livak and Schmittgen, 2001) using the expression of housekeeping genes (18S rRNA and Ubiquitin-conjugating enzyme E2) for template normalization. The following RT-qPCR primers were used: PtrEPSP-TF (forward, ACCTGAGATCGTTTTGCAACC; reverse, CAACAGTCGTACCCTCAGAGA). Total RNAs were extracted from root, young leaf (1st to 3rd from apex), mature leaf (4th to 6th from apex), senescent leaf (yellow leaf), young stem (internodes 1 to 3), mature stem (internodes 6 to 8), petiole of mature leaf, phloem (bark of mature stem), and developing xylem (scrapped stem under bark of mature stem) to analyze the expression of PtrEPSP-TF in various tissue and organs. RT-qPCR analyses were performed using samples from three individual plants to ensure results were consistent.
Phylogenetic Tree Construction
Hmmer (v3.1) (Eddy, 2011) was used to identify PtrEPSP-TF homologs using the EPSP domain training file obtained from Pfam (Finn et al., 2006) in all species from Phytozome (http://phytozome.jgi.doe.gov/pz/portal.html) except Salix. Suchowensis, which was obtained from the willow genome website (http://115.29.234.170/node/5) (Dai et al., 2014). Multiple alignment analysis of these EPSP genes was performed using Muscle (version 3.8) (Edgar, 2004) at the amino acid level with default settings. Amino acid alignments were translated into nucleotide alignments using ad hoc perl scripts. The phylogenetic tree of the EPSP gene family was constructed by Mrbayes (v3.2.2) (Ronquist and Huelsenbeck, 2003) using the Bayesian inference method with substitution model set as GTR + I + Γ and a Markov Chain Mont Carlo set as 1,000,000 generations.
Populus DAYSLEEPER-like genes were identified by searching for sequence similarity to the Arabidopsis DAYSLEEPER (AT3G42170) amino acid sequence in the Populus V3.1 reference genome assembly. After multiple sequence alignment using Clustal X2.1, the unrooted tree was generated using the neighbor-joining method with 1000 bootstrap replicates with MEGA 5.1 software.
Cell Fractionation and Protein Gel Blots
One milliliter of transfected protoplasts (2 × 105 mL−1) was incubated at room temperature for 14 h for protein expression and then collected by centrifugation at 2000 rpm for 10 min at 4°C. The non-nuclear and nuclear fractions were separated according to a previously published method (Yin et al., 2016). Total proteins were extracted by incubating protoplasts in extraction buffer (20 mM Tris-HCl, pH 7.4, 25% glycerol, 20 mM KCl, 2 mM EDTA, 2.5 mM MgCl2, 250 mM sucrose, 1 mM DTT, and 1 mM PMSF) for 1 h at 4°C. After centrifugation at 1500g for 10 min at 4°C, the clear supernatant was taken and enriched using acetone precipitation as the non-nuclear fraction. The pellet was washed twice with nuclei resuspension Triton buffer (20 mM Tris-HCl, pH 7.4, 25% glycerol, 2.5 mM MgCl2, and 0.2% Triton X-100) and once with nuclei resuspension buffer (20 mM Tris-HCl, pH 7.4, 25% glycerol, and 2.5 mM MgCl2). Non-nuclear and nuclear proteins were separated by SDS-PAGE and transferred to PVDF membrane (Bio-Rad). Anti-Myc (Sigma-Aldrich; C3956), anti-histone H3 (Abcam; ab1791), and anti-UGPase (Agrisera; AS05 086) were used as primary antibodies. Anti-Rabbit IgG peroxidase antibody (Sigma-Aldrich; A9169) were used as secondary antibodies. Chemiluminescent signals were generated using the ECL Immunoblotting Detection Reagents (GE Health) and detected with ChemiDoc XRS+ system (Bio-Rad).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: PtrEPSP-TF, XM_002301243; PtrEPSP-SY, XM_024584967; PtrhAT, XM_024588158; PtrMYB021, KF148678; and PtWND1B, HQ215848.
Supplemental Data
Supplemental Figure 1. Negative EMSA results.
Supplemental Figure 2. The expression of Myc-PtrEPSP-TF and PtrhAT-Myc in protoplasts.
Supplemental Figure 3. Unrooted neighbor-joining phylogenies based on full-length amino acid sequences of 26 Populus DAYSLEEPER like proteins.
Supplemental Figure 4. Phylogenetic tree of EPSP synthase.
Supplemental Figure 5. Protein sequence alignment of EPSP synthases from dicots, monocots, algae, and nonvascular plants.
Supplemental Figure 6. PtrEPSP-TF, but not PtrEPSP-SY, exhibits high expression level in developing xylem tissue.
Supplemental Figure 7. Subcellular localization of N-terminal YFP-tagged PtrEPSP-TF and PtrEPSP-SY in Populus protoplasts.
Supplemental Table 1. Metabolite changes in P. deltoides PtrEPSP-TF overexpression lines.
Supplemental Table 2. Up- and downregulated genes in PtrEPSP-TF overexpression lines.
Supplemental Data Set 1. Alignments for the phylogenetic analysis of 26 Populus DAYSLEEPER like proteins in Supplemental Figure 3.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This research was funded by The BioEnergy Science Center and The Center for Bioenergy Innovation from 2007 to 2017 and from 2017 to 2018, respectively. Both are U.S. Department of Energy Research Centers (Grant DE-AC05-00OR22725) supported by the Office of Biological and Environmental Research in the DOE Office of Science. Oak Ridge National Laboratory is managed by UT-Battelle for the U.S. Department of Energy.
AUTHOR CONTRIBUTIONS
W.M., J.-G.C., G.A.T., and M.X. designed experiments. W.M., M.X., A.C.B., J.-G.C., and G.A.T. wrote the manuscript. W.M., S.P.D., L.M.E., and G.A.T. performed association mapping. M.X., A.C.B., R.S.P., V.R.S., J.B., S.S.J., and L.E.G. performed molecular experiments. K.Y., R.W.S., O.T., and M.R. performed cell wall compositional analysis. K.W. and C.C. generated transgenic plants. H.-B.G. and H.G. performed molecular dynamic simulations. T.J.T. and N.E. performed metabolomic analysis. W.M., J.Z., T.J.T., E.L., R.D., M.D., J.S., and U.K. analyzed data.
Footnotes
Articles can be viewed without a subscription.
References
- Achard F. (2009). Vital Forest Graphics. (Arendal, Norway: UNEP/GRID-Arendal; ). [Google Scholar]
- Adli M., Bernstein B.E. (2011). Whole-genome chromatin profiling from limited numbers of cells using nano-ChIP-seq. Nat. Protoc. 6: 1656–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L., Anantharaman V., Balaji S., Babu M.M., Iyer L.M. (2005). The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 29: 231–262. [DOI] [PubMed] [Google Scholar]
- Barros J., Serk H., Granlund I., Pesquet E. (2015). The cell biology of lignification in higher plants. Ann. Bot. 115: 1053–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Best R.B., Zhu X., Shim J., Lopes P.E., Mittal J., Feig M., Mackerell A.D. Jr (2012). Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 8: 3257–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks B.R., et al. (2009). CHARMM: the biomolecular simulation program. J. Comput. Chem. 30: 1545–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundock P., Hooykaas P. (2005). An Arabidopsis hAT-like transposase is essential for plant development. Nature 436: 282–284. [DOI] [PubMed] [Google Scholar]
- Carter S.G., Karl D.W. (1982). Inorganic phosphate assay with malachite green: an improvement and evaluation. J. Biochem. Biophys. Methods 7: 7–13. [DOI] [PubMed] [Google Scholar]
- Chen V.B., Arendall W.B. III, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66: 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl J.A., Collas P. (2008). A rapid micro chromatin immunoprecipitation assay (microChIP). Nat. Protoc. 3: 1032–1045. [DOI] [PubMed] [Google Scholar]
- Dai X., et al. (2014). The willow genome and divergent evolution from poplar after the common genome duplication. Cell Res. 24: 1274–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S.R. (2011). Accelerated profile HMM searches. PLOS Comput. Biol. 7: e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C. (2008). Transposable elements and the evolution of regulatory networks. Nat. Rev. Genet. 9: 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R.D., et al. (2006). Pfam: clans, web tools and services. Nucleic Acids Res. 34: D247–D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser C.M., Chapple C. (2011). The phenylpropanoid pathway in Arabidopsis. The Arabidopsis Book 9: e0152.doi/10.1199/tab.0152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg B., Vaid N., Tuteja N. (2014). In-silico analysis and expression profiling implicate diverse role of EPSPS family genes in regulating developmental and metabolic processes. BMC Res. Notes 7: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldes A., et al. (2013). A 34K SNP genotyping array for Populus trichocarpa: design, application to the study of natural populations and transferability to other Populus species. Mol. Ecol. Resour. 13: 306–323. [DOI] [PubMed] [Google Scholar]
- Goicoechea M., Lacombe E., Legay S., Mihaljevic S., Rech P., Jauneau A., Lapierre C., Pollet B., Verhaegen D., Chaubet-Gigot N., Grima-Pettenati J. (2005). EgMYB2, a new transcriptional activator from Eucalyptus xylem, regulates secondary cell wall formation and lignin biosynthesis. Plant J. 43: 553–567. [DOI] [PubMed] [Google Scholar]
- Guo J., Morrell-Falvey J.L., Labbé J.L., Muchero W., Kalluri U.C., Tuskan G.A., Chen J.-G. (2012). Highly efficient isolation of Populus mesophyll protoplasts and its application in transient expression assays. PLoS One 7: e44908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchee M., Rottmann W., Mullinax L., Zhang C., Chang S., Cunningham M., Pearson L., Nehra N. (2009). Short-rotation woody crops for bioenergy and biofuels applications. In Vitro Cell. Dev. Biol. 45: 619–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., Schulten K. (1996). VMD: visual molecular dynamics. J. Mol. Graph. 14: 33–38, 27–28. [DOI] [PubMed] [Google Scholar]
- Hussey S.G., Mizrachi E., Creux N.M., Myburg A.A. (2013). Navigating the transcriptional roadmap regulating plant secondary cell wall deposition. Front. Plant Sci. 4: 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal M. (1990). The Vascular Cambium. (Taunton, UK: Research Studies Press; ). [Google Scholar]
- Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. (1983). Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79: 926–935. [Google Scholar]
- Karve A.A., Jawdy S.S., Gunter L.E., Allen S.M., Yang X., Tuskan G.A., Wullschleger S.D., Weston D.J. (2012). Initial characterization of shade avoidance response suggests functional diversity between Populus phytochrome B genes. New Phytol. 196: 726–737. [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. (2013a). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W.C., Ko J.H., Kim J.Y., Kim J., Bae H.J., Han K.H. (2013b). MYB46 directly regulates the gene expression of secondary wall-associated cellulose synthases in Arabidopsis. Plant J. 73: 26–36. [DOI] [PubMed] [Google Scholar]
- Kim W.-C., Kim J.-Y., Ko J.-H., Kang H., Han K.-H. (2014). Identification of direct targets of transcription factor MYB46 provides insights into the transcriptional regulation of secondary wall biosynthesis. Plant Mol. Biol. 85: 589–599. [DOI] [PubMed] [Google Scholar]
- Knip M., de Pater S., Hooykaas P.J. (2012). The SLEEPER genes: a transposase-derived angiosperm-specific gene family. BMC Plant Biol. 12: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L.-Y., Fang M.-J., Kuang L.-Y., Gelvin S.B. (2008). Vectors for multi-color bimolecular fluorescence complementation to investigate protein-protein interactions in living plant cells. Plant Methods 4: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Lin Y.-C., Li Q., Shi R., Lin C.-Y., Chen H., Chuang L., Qu G.-Z., Sederoff R.R., Chiang V.L. (2014). A robust chromatin immunoprecipitation protocol for studying transcription factor-DNA interactions and histone modifications in wood-forming tissue. Nat. Protoc. 9: 2180–2193. [DOI] [PubMed] [Google Scholar]
- Li Y., Tschaplinski T.J., Engle N.L., Hamilton C.Y., Rodriguez M. Jr., Liao J.C., Schadt C.W., Guss A.M., Yang Y., Graham D.E. (2012). Combined inactivation of the Clostridium cellulolyticum lactate and malate dehydrogenase genes substantially increases ethanol yield from cellulose and switchgrass fermentations. Biotechnol. Biofuels 5: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. [DOI] [PubMed] [Google Scholar]
- Liu J., Hai G., Wang C., Cao S., Xu W., Jia Z., Yang C., Wang J.P., Dai S., Cheng Y. (2015). Comparative proteomic analysis of Populus trichocarpa early stem from primary to secondary growth. J. Proteomics 126: 94–108. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loziuk P.L., Parker J., Li W., Lin C.-Y., Wang J.P., Li Q., Sederoff R.R., Chiang V.L., Muddiman D.C. (2015). Elucidation of xylem-specific transcription factors and absolute quantification of enzymes regulating cellulose biosynthesis in Populus trichocarpa. J. Proteome Res. 14: 4158–4168. [DOI] [PubMed] [Google Scholar]
- Lu Z., Hunter L. (2005). GO molecular function terms are predictive of subcellular localization. Pacific Symposium on Biocomputing Pacific Symposium on Biocomputing (NIH Public Access). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H., Dudareva N. (2012). The shikimate pathway and aromatic amino Acid biosynthesis in plants. Annu. Rev. Plant Biol. 63: 73–105. [DOI] [PubMed] [Google Scholar]
- McCarthy R.L., Zhong R., Fowler S., Lyskowski D., Piyasena H., Carleton K., Spicer C., Ye Z.-H. (2010). The poplar MYB transcription factors, PtrMYB3 and PtrMYB20, are involved in the regulation of secondary wall biosynthesis. Plant Cell Physiol. 51: 1084–1090. [DOI] [PubMed] [Google Scholar]
- McKown A.D., et al. (2014). Genome-wide association implicates numerous genes underlying ecological trait variation in natural populations of Populus trichocarpa. New Phytol. 203: 535–553. [DOI] [PubMed] [Google Scholar]
- Mingozzi M., Montello P., Merkle S. (2009). Adventitious shoot regeneration from leaf explants of eastern cottonwood (Populus deltoides) cultured under photoautotrophic conditions. Tree Physiol. 29: 333–343. [DOI] [PubMed] [Google Scholar]
- Mir R., Jallu S., Singh T.P. (2015). The shikimate pathway: review of amino acid sequence, function and three-dimensional structures of the enzymes. Crit. Rev. Microbiol. 41: 172–189. [DOI] [PubMed] [Google Scholar]
- Mitsuda N., Iwase A., Yamamoto H., Yoshida M., Seki M., Shinozaki K., Ohme-Takagi M. (2007). NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19: 270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchero W., et al. (2015). High-resolution genetic mapping of allelic variants associated with cell wall chemistry in Populus. BMC Genomics 16: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi-Ito K., Oda Y., Fukuda H. (2010). Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 22: 3461–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani M., Nishikubo N., Xu B., Yamaguchi M., Mitsuda N., Goué N., Shi F., Ohme‐Takagi M., Demura T. (2011). A NAC domain protein family contributing to the regulation of wood formation in poplar. Plant J. 67: 499–512. [DOI] [PubMed] [Google Scholar]
- Para A., et al. (2014). Hit-and-run transcriptional control by bZIP1 mediates rapid nutrient signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 10371–10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzlaff A., McInnis S., Courtenay A., Surman C., Newman L.J., Smith C., Bevan M.W., Mansfield S., Whetten R.W., Sederoff R.R., Campbell M.M. (2003). Characterisation of a pine MYB that regulates lignification. Plant J. 36: 743–754. [DOI] [PubMed] [Google Scholar]
- Payyavula R.S., Tschaplinski T.J., Jawdy S.S., Sykes R.W., Tuskan G.A., Kalluri U.C. (2014). Metabolic profiling reveals altered sugar and secondary metabolism in response to UGPase overexpression in Populus. BMC Plant Biol. 14: 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kalé L., Schulten K. (2005). Scalable molecular dynamics with NAMD. J. Comput. Chem. 26: 1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragauskas A.J., Williams C.K., Davison B.H., Britovsek G., Cairney J., Eckert C.A., Frederick W.J., Hallett J.P., Leak D.J., Liotta C.L. (2006). The path forward for biofuels and biomaterials. Science 311: 484–489. [DOI] [PubMed] [Google Scholar]
- Roberts E., Eargle J., Wright D., Luthey-Schulten Z. (2006). MultiSeq: unifying sequence and structure data for evolutionary analysis. BMC Bioinformatics 7: 382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J.P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Roy A., Kucukural A., Zhang Y. (2010). I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5: 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A., Yang J., Zhang Y. (2012). COFACTOR: an accurate comparative algorithm for structure-based protein function annotation. Nucleic Acids Res. 40: W471–W477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S.B., Hagen G., Guilfoyle T. (2003). The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15: 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T., Watanabe M., Hoefgen R., Fernie A.R. (2013). Shikimate and phenylalanine biosynthesis in the green lineage. Front. Plant Sci. 4: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C.-J., Podila G.K., Chiang V.L. (1994). Agrobacterium-mediated transformation of quaking aspen (Populus tremuloides) and regeneration of transgenic plants. Plant Cell Rep. 14: 94–97. [DOI] [PubMed] [Google Scholar]
- Tschaplinski T.J., et al. (2012). Down-regulation of the caffeic acid O-methyltransferase gene in switchgrass reveals a novel monolignol analog. Biotechnol. Biofuels 5: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan G.A., Difazio S., Jansson S., Bohlmann J., Grigoriev I., Hellsten U., Putnam N., Ralph S., Rombauts S., Salamov A. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604. [DOI] [PubMed] [Google Scholar]
- Vanholme R., Demedts B., Morreel K., Ralph J., Boerjan W. (2010). Lignin biosynthesis and structure. Plant Physiol. 153: 895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt T. (2010). Phenylpropanoid biosynthesis. Mol. Plant 3: 2–20. [DOI] [PubMed] [Google Scholar]
- Weng J.-K., Li X., Bonawitz N.D., Chapple C. (2008). Emerging strategies of lignin engineering and degradation for cellulosic biofuel production. Curr. Opin. Biotechnol. 19: 166–172. [DOI] [PubMed] [Google Scholar]
- Wilkins O., Nahal H., Foong J., Provart N.J., Campbell M.M. (2009). Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol. 149: 981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray G.A., Hahn M.W., Abouheif E., Balhoff J.P., Pizer M., Rockman M.V., Romano L.A. (2003). The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 20: 1377–1419. [DOI] [PubMed] [Google Scholar]
- Xie M., Ren G., Costa-Nunes P., Pontes O., Yu B. (2012). A subgroup of SGS3-like proteins act redundantly in RNA-directed DNA methylation. Nucleic Acids Res. 40: 4422–4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M., Mitsuda N., Ohtani M., Ohme-Takagi M., Kato K., Demura T. (2011). VASCULAR-RELATED NAC-DOMAIN7 directly regulates the expression of a broad range of genes for xylem vessel formation. Plant J. 66: 579–590. [DOI] [PubMed] [Google Scholar]
- Yin R., Skvortsova M.Y., Loubéry S., Ulm R. (2016). COP1 is required for UV-B-induced nuclear accumulation of the UVR8 photoreceptor. Proc. Natl. Acad. Sci. USA 113: E4412–E4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin T., Zhang X., Gunter L., Priya R., Sykes R., Davis M., Wullschleger S.D., Tuskan G.A. (2010). Differential detection of genetic Loci underlying stem and root lignin content in Populus. PLoS One 5: e14021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.-D., Cho Y.-H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yu J., Pressoir G., Briggs W.H., Vroh Bi I., Yamasaki M., Doebley J.F., McMullen M.D., Gaut B.S., Nielsen D.M., Holland J.B., Kresovich S., Buckler E.S. (2006). A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38: 203–208. [DOI] [PubMed] [Google Scholar]
- Zhang Y. (2014). Interplay of I-TASSER and QUARK for template-based and ab initio protein structure prediction in CASP10. Proteins 82 (suppl. 2): 175–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Sun J., Xu P., Zhang R., Li L. (2014). Intron-mediated alternative splicing of WOOD-ASSOCIATED NAC TRANSCRIPTION FACTOR1B regulates cell wall thickening during fiber development in Populus species. Plant Physiol. 164: 765–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Ye Z.-H. (2010). The poplar PtrWNDs are transcriptional activators of secondary cell wall biosynthesis. Plant Signal. Behav. 5: 469–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Richardson E.A., Ye Z.-H. (2007). The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 19: 2776–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Lee C., Zhou J., McCarthy R.L., Ye Z.-H. (2008). A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20: 2763–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., McCarthy R.L., Haghighat M., Ye Z.-H. (2013). The poplar MYB master switches bind to the SMRE site and activate the secondary wall biosynthetic program during wood formation. PLoS One 8: e69219. [DOI] [PMC free article] [PubMed] [Google Scholar]