Double mutants of the two genes encoding chromomethylases or the two genes encoding DDM1-type nucleosome remodelers lose RNA-directed DNA methylation in maize.
Abstract
Plants make use of distinct types of DNA methylation characterized by their DNA methyltransferases and modes of regulation. One type, RNA-directed DNA methylation (RdDM), is guided by small interfering RNAs (siRNAs) to the edges of transposons that are close to genes, areas called mCHH islands in maize (Zea mays). Another type, chromomethylation, is guided by histone H3 lysine 9 methylation to heterochromatin across the genome. We examined DNA methylation and small RNA expression in plant tissues that were mutant for both copies of the genes encoding chromomethylases as well as mutants for both copies of the genes encoding DECREASED DNA METHYLATION1 (DDM1)-type nucleosome remodelers, which facilitate chromomethylation. Both sets of double mutants were nonviable but produced embryos and endosperm. RdDM was severely compromised in the double mutant embryos, both in terms of DNA methylation and siRNAs. Loss of 24-nucleotide siRNA from mCHH islands was coupled with a gain of 21-, 22-, and 24-nucleotide siRNAs in heterochromatin. These results reveal a requirement for both chromomethylation and DDM1-type nucleosome remodeling for RdDM in mCHH islands, which we hypothesize is due to dilution of RdDM components across the genome when heterochromatin is compromised.
INTRODUCTION
Chromatin modification directed by RNA interference and related processes is essential to genome defense in most eukaryotes. The molecular mechanisms vary even within the same cell, but key features are small interfering RNAs (siRNAs), which guide argonaute proteins and induce methylation of histone H3 lysine 9 (H3K9me) (Holoch and Moazed, 2015). RNA-directed DNA methylation (RdDM) is a well-characterized example in plants (Cuerda-Gil and Slotkin, 2016). RdDM leads to H3K9me2 (Jackel et al., 2016; Fultz and Slotkin, 2017), but as its name suggests, is better known for methylating DNA. At least two Arabidopsis thaliana proteins that function in RdDM physically interact with a DNA methyltransferase, indicating a direct connection between RdDM and DNA methylation (Gao et al., 2010; Zhong et al., 2014). RdDM represses repetitive and foreign DNA but can also influence gene expression, at gene regulatory elements for example (Rowley et al., 2017), and is involved in epigenetic phenomena such as paramutation (Hollick, 2017) and genomic imprinting (Satyaki and Gehring, 2017).
DNA methylation occurs at cytosines in all sequence contexts and is catalyzed by three distinct types of methyltransferases in plants (Du et al., 2015). The first, related to Arabidopsis METHYLTRANSFERASE1 (MET1), which is homologous to mammalian DNA Methyltransferase 1 (DNMT1), is responsible for replication-coupled CG methylation. The second, related to Arabidopsis CHROMOMETHYLASE1 (CMT1), CMT2, and CMT3, methylates CHGs and CHHs, where H is A, T, or C. The chromodomains of these methyltransferases guide their activity to regions of H3K9me1 or H3K9me2. The third, related to Arabidopsis DOMAINS REARRANGED METHYLTRANSFERASE1 (DRM1) and DRM2, methylates cytosines in all sequence contexts through RdDM. Depending on the species, methylation in the CHH context (mCHH) can be used as an indicator of RdDM, as mCHH produced by CMT is of lesser magnitude than that of RdDM (Niederhuth et al., 2016). Division of methylation contexts into mCG, mCHG, and mCHH is a helpful simplification, but methyltransferases have additional nucleotide preferences (Gouil and Baulcombe, 2016).
Although RdDM was discovered as a mechanism that initiates methylation at previously unmethylated DNA (referred to as de novo methylation; Wassenegger et al., 1994), discoveries since then have revealed that most RdDM occurs at already-methylated and repressed loci (Cuerda-Gil and Slotkin, 2016). This maintenance form of RdDM, called canonical RdDM, depends on the activity of the RNA polymerase II variants Pol IV and Pol V and is responsible for the majority of 24-nucleotide siRNAs. Canonical RdDM in Arabidopsis relies on histone H3 lysine 9 methylation (H3K9me) and lysine 4 demethylation (Johnson et al., 2008, 2014; Kuhlmann and Mette, 2012; Greenberg et al., 2013; Law et al., 2013; Zhang et al., 2013; Blevins et al., 2014; Stroud et al., 2014; Li et al., 2015b). Canonical RdDM also requires DNA methylation, as drm and cmt mutants have reduced levels of 24-nucleotide siRNAs (Law et al., 2013; Stroud et al., 2014; Li et al., 2015b). DNA methylation may act indirectly by impacting the levels of H3K9me2, as histone methyltransferase mutants also lose 24-nucleotide siRNAs (Stroud et al., 2014). H3K9me2 recruits Pol IV (Law et al., 2013; Zhang et al., 2013), but DNA methylation also promotes RdDM through Pol V interacting factors (Johnson et al., 2014; Liu et al., 2014).
The heterochromatic middle regions of long transposons are depleted of RdDM relative to their euchromatin-flanking ends (Lee et al., 2012; Zhong et al., 2012; Stroud et al., 2013, 2014; Zemach et al., 2013; Li et al., 2015b). Enrichment for RdDM at heterochromatin-euchromatin boundaries is especially clear in maize (Zea mays) because its heterochromatin and euchromatin are highly interspersed (Gent et al., 2013, 2014; Li et al., 2015a; Niederhuth et al., 2016). That heterochromatin inhibits RdDM is also suggested by activation of RdDM in normally heterochromatic regions in plants that lack the SNF2 family nucleosome remodeling protein DECREASED DNA METHYLATION1 (DDM1) (Nuthikattu et al., 2013; Stroud et al., 2013; Creasey et al., 2014; McCue et al., 2015; Panda et al., 2016), known as LYMPHOID SPECIFIC HELICASE in mammals (Dennis et al., 2001). DDM1 is required for access of MET1 and CMT-type methyltransferases to nontranscribed, nucleosome-bound DNA in Arabidopsis (Lyons and Zilberman, 2017). In its absence, mCG, mCHG, and H3K9me2 are reduced and chromocenters partially decondensed (Gendrel et al., 2002; Soppe et al., 2002).
The vegetative cell of pollen in Arabidopsis provides another line of evidence that heterochromatin inhibits RdDM because this cell type undergoes a dramatic decondensation of heterochromatin coupled with activation of RdDM (Schoft et al., 2009; Slotkin et al., 2009; Mérai et al., 2014). DDM1 may be reduced or absent from vegetative cells, as transgene-driven expression of a DDM1 fusion protein from a ddm1 promoter produced signal in sperm but not in vegetative cells (Slotkin et al., 2009). Vegetative cells have increased mCHH (Calarco et al., 2012; Ibarra et al., 2012) primarily driven by CMT2, but also by DRM2 activity (Hsieh et al., 2016). The apparent contradiction between chromatin modifications associated with heterochromatin promoting RdDM, yet heterochromatin itself inhibiting RdDM may be explained by the relative abundance of chromatin modifications (e.g., by the ratio of H3K9me1 to H3K9me2; Stroud et al., 2014), and by higher-order chromatin structure that affects chromatin accessibility to RNA polymerases.
Its quick life cycle, small genome, and resilience to loss of DNA methylation have made Arabidopsis the plant model of choice for research on DNA methylation and chromatin. The discoveries made with Arabidopsis have been tremendously helpful in understanding similar phenomena plants that are more difficult to work with, such as maize, with its large, repetitive genome and nonviable methylation mutants (Li et al., 2015a). However, the differences between maize and Arabidopsis also limit the extent to which results can be projected from Arabidopsis to maize. Maize lacks a CMT2-type chromomethylase (Zemach et al., 2013; Bewick et al., 2017), has multiple copies of the major subunits of Pol IV and Pol V complexes with potential for specialized functions (Haag et al., 2014), has abundant meiotic phased siRNAs (Johnson et al., 2009; Zhai et al., 2015), and has abundant 22-nucleotide siRNAs (Nobuta et al., 2008; Wang et al., 2009).
The maize genome encodes two chromomethylases, named ZMET2 and ZMET5 (also known as DMT102 and DMT105) (Li et al., 2014). Both are functionally more similar to CMT3 than to CMT1 or CMT2 (Bewick et al., 2017). The maize genome also encodes two DDM1-like nucleosome remodelers, CHR101 and CHR106 (Li et al., 2014). The effects of single mutants of all four genes on whole genome methylation have been investigated previously (Gent et al., 2014; Li et al., 2014; Gouil and Baulcombe, 2016). Single mutants of zmet2 and zmet5 have reduced mCHG in both leaves and developing ears and reduced mCHH in leaves but not in developing ears. Single mutants of chr101 have reduced mCHH in leaves, and single mutants of chr106 have reduced mCHH and mCHG in leaves (Li et al., 2014). Neither chr101 nor chr106 has been tested in developing ears. The effects of single mutants on siRNAs have not been reported, and it is not clear whether reduced RdDM explains their reduced mCHH.
Here, we investigated the relationships between RdDM and chromomethylases and DDM1-type nucleosome remodelers using mutants in maize. We found that double mutants of chr101 and chr106 and double mutants of zmet2 and zmet5, while nonviable (Li et al., 2014), can produce embryos and endosperm. Analysis of DNA methylation and siRNAs in both tissues revealed that RdDM was severely compromised in developing embryos, with near complete loss of both 24-nucleotide siRNAs and mCHH from mCHH islands. The loss of 24-nucleotide siRNAs from mCHH islands was accompanied by dramatic gains of 21- and 22-nucleotide siRNAs at heterochromatic loci in the genome, but these siRNAs did not direct DNA methylation.
RESULTS
Generation of ddm1 double and cmt Double Mutant Embryo and Endosperm
To make plants that lacked chromomethylases or DDM1-type nucleosome remodelers, we obtained UniformMu stocks with Mu insertions in exons of Zmet2 (mu1013094) (Gent et al., 2014) and Zmet5 (mu1017456), and in the DDM1-encoding genes Chr101 (mu1044815) and Chr106 (mu1021319) (Supplemental Table 1). Here, we will refer to zmet2 zmet5 homozygous double mutants as cmt, and the chr101 and chr106 double mutants as ddm1. Mutants that carried a single wild-type copy of either Zmet2 or Zmet5 were viable and fertile, and crosses between such mutants produced kernels with sectors of pigmented aleurone (Supplemental Figure 1). This phenotype is characteristic of Mutator transposon activity in UniformMu stocks, where excision of a Mu from the bz1-mum9 allele of the Bz1 gene can restore pigmentation in small sectors during development (McCarty et al., 2005). Mu activation has been observed in a maize mutant lacking the RNA-directed RNA polymerase MEDIATOR OF PARAMUTATION1 (MOP1), which synthesizes the antisense strand of Pol IV transcripts (Woodhouse et al., 2006; Haag et al., 2014). We found a tight correlation between pigmented sectors in the aleurone and zmet2 zmet5 (cmt) double mutant genotype (Supplemental Table 2). The cmt kernels contained both endosperm and embryos but were usually incapable of more than a couple centimeters of root development upon germination. A second pair of zmet2 and zmet5 alleles (zmet2-m1 and zmet5-m1) that was introgressed into the B73 genetic background produced homozygous double mutant kernels at the expected ratios and were also nonviable (Supplemental Table 3). Failure to produce double mutant plants with these alleles was previously reported (Li et al., 2014). These kernels lacked the Mu insertion in the Bz1 gene and so were incapable of pigmentation sectoring. Mutants that carried a single wild-type copy of either Chr101 or Chr106 were viable and fertile but produced homozygous double mutant (ddm1) kernels that were nonviable and had small embryos (Supplemental Table 2 and Supplemental Figure 1). Although in the bz1-mum9 background, these ddm1 kernels did not exhibit the sectoring phenotype of cmt mutants. They also did not germinate, not even to produce a root tip. A second set of chr101 and chr106 alleles (chr101-m3 and chr106-m1) that was introgressed into B73 did not produce any homozygous double mutant kernels (Supplemental Table 3), consistent with the prior study (Li et al., 2014).
Loss of DNA Methylation Flanking Genes in cmt and ddm1 Embryo and Endosperm
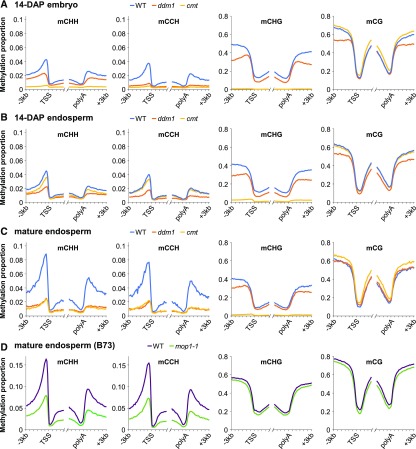
To determine the effects of the mutations on DNA methylation, we performed whole-genome bisulfite sequencing (WGBS) using the methylC-seq method (Urich et al., 2015) in three tissues: mature endosperm, developing endosperm 14 d after pollination (DAP), and 14-DAP embryos. In cmt mutants, mCHG was reduced to near background levels in all three tissues, with little or no effect on mCG (Figure 1). In ddm1, both mCHG and mCG were mildly reduced (40% reduction in mCG; 50% in mCHG in 14-DAP embryos). These effects on mCHG (nearly absent in cmt) and mCHG and mCG (reduced in ddm1) are consistent with the expected roles of chromomethylases and DDM1-like nucleosome remodelers in transcriptionally silent heterochromatin. Unexpectedly, we found that mCHH was nearly absent in cmt 14-DAP embryos, strongly reduced in mature endosperm, and slightly reduced in 14-DAP endosperm (Figure 1). The effect on mCHH was not due to a linked background mutation in the UniformMu-derived cmt mutant, as 14-DAP sibling embryos with a single wild-type copy of either Zmet2 or Zmet5 had near wild-type levels of mCHH (Supplemental Figure 2). mCHH was reduced in ddm1 embryos and endosperm, particularly in mature endosperm (Figure 1). The effect of ddm1 on RdDM is more evident when considered in the light of the fact that ZMET2 and ZMET5 can methylate CHH, particularly in the CAA and CTA contexts (Gouil and Baulcombe, 2016). The mCCH subset of mCHH is more specific to RdDM. The ddm1 mutant had a strong effect on mCCH and less of an effect on mCHH in 14-DAP embryos (Figure 1). A single wild-type copy of either Chr101 or Chr106 largely restored mCHH in mature endosperm, indicating that the loss of RdDM in ddm1 was not due to a background mutation linked to either gene (Supplemental Figure 3).
Figure 1.
DNA Methylation Profiles Near Genes in Double Mutants.
(A) Methylation in 14-DAP embryo. All genes were defined by their annotated transcription start sites (TSS) and polyadenylation sites (polyA) and split into nonoverlapping 100-bp intervals. Methylation for each sample was calculated as the proportion of methylated C over total C in each sequence context (CHH, CCH, CHG, and CG) averaged for each 100-bp interval.
(B) Methylation in 14-DAP endosperm, as in (A).
(C) Methylation in mature endosperm, as in (A).
(D) Methylation in mature B73 endosperm, as in (A). The wild type was derived from Mop1 homozygous siblings of the mop1-1 mutants.
In developing ear, mCHH is greatly reduced in mop1 mutants lacking the RNA-directed RNA polymerase MOP1, which is coupled with Pol IV in synthesis of 24-nucleotide siRNAs (Gent et al., 2014; Haag et al., 2014; Li et al., 2015a). Mop1 is highly expressed in reproductive tissue, in embryo, and in endosperm (Sekhon et al., 2011). We performed WGBS on mop1-1 mature endosperm and found ∼50% reduction in mCHH flanking genes, indicating MOP1 function in endosperm, though probably not as the only source of antisense RNA in RdDM (Figure 1D). This experiment also revealed a high level of variation in mCHH in mature endosperm, as the wild-type Mop1 siblings (B73-related) had methylation levels nearly twice as high as the wild-type mature endosperm in Figure 1C and slightly higher than the ddm1 heterozygote mature endosperm (W22-related) (Supplemental Figure 3).
Loss of 24-Nucleotide siRNAs in cmt and ddm1 Developing Embryo and Endosperm
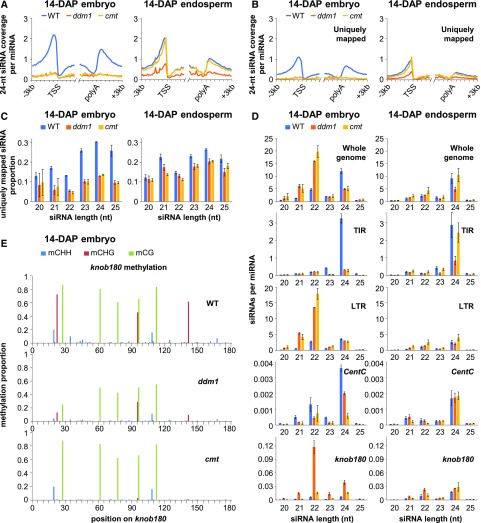
Since 24-nucleotide siRNAs guide argonautes in RdDM, we sequenced small RNA from 14-DAP endosperm and embryo. To quantify siRNA abundance, we normalized siRNA counts by microRNA (miRNA) counts. We included all mappable small RNAs, both uniquely mapping and multimapping. There was a nearly complete loss of 24-nucleotide siRNAs from gene flanks in both cmt and ddm1 14-DAP embryos and a partial loss in 14-DAP endosperm (Figure 2A). Analysis of the uniquely mapping subset of siRNAs showed similar trends and revealed a >50% decrease in the proportion of uniquely mapping siRNAs in ddm1 and in cmt embryos (Figures 2B and 2C). Loss of 24-nucleotide siRNAs was also evident from the distribution of total siRNA lengths: In homozygous wild-type individuals and heterozygous mutants, the dominant siRNA length was 24 nucleotides, but in cmt and ddm1, it shifted to 22 nucleotides and to a lesser extent 21 nucleotides (Figure 2D). DNA transposons with terminal inverted repeats of the Harbinger, Mutator, hAT, and Mariner superfamilies are enriched in mCHH islands (Gent et al., 2013). The 24-nucleotide siRNAs from these terminal inverted repeat (TIR) transposons were reduced ∼8-fold in cmt and in ddm1 (Figure 2D).
Figure 2.
Loss of 24-Nucleotide siRNAs and Gain of 21- and 22-Nucleotide siRNAs in Double Mutants.
(A) The 24-nucleotide siRNA coverage near genes. All genes were defined by their annotated transcription start sites (TSS) and polyadenylation sites (polyA) and split into nonoverlapping 100-bp intervals. The siRNA coverage for each 100-bp interval was summed for the complete set of genes and normalized by miRNAs.
(B) Uniquely mapped 24-nucleotide siRNA coverage near genes, as in (A).
(C) Proportion of uniquely mapped siRNAs for each length.
(D) Length distribution of various classes of siRNAs. “Whole genome” includes all mapped siRNAs. “TIR” includes all siRNAs that overlapped by at least half their lengths to Mutator, hAT, Harbinger, or Mariner TIR transposons. “LTR” includes all siRNAs that overlapped by at least half their lengths with Gypsy or Copia LTR retrotransposons. CentC includes all siRNAs that aligned to a CentC consensus sequence and knob180 includes all siRNAs that aligned to a knob180 consensus sequence. siRNA counts were normalized per miRNA. Error bars are standard errors of the means for the biological replicates of each genotype.
(E) Single-base-pair DNA methylation in knob180 repeats. WGBS reads were mapped to the knob180 consensus sequence and methylation calculated as the proportion of methylated C over total C in each sequence context.
Gain of siRNAs in Heterochromatin in cmt and ddm1
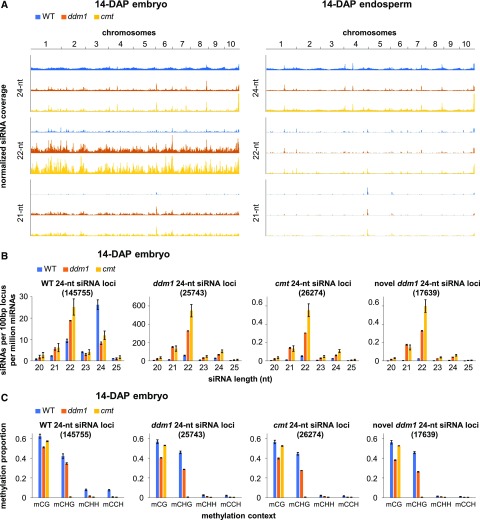
Retrotransposons with long terminal repeats (LTRs), which are relatively depleted in mCHH islands but have high copy numbers elsewhere in maize heterochromatin, gained both 21- and 22-nucleotide siRNAs in the mutants (8.8-fold gain of 21-nucleotide siRNAs in ddm1, 7.0-fold gain of 21-nucleotide siRNAs in cmt- 4.6-fold gain of 22-nucleotide siRNAs in ddm1, and 6.2-fold gain of 22-nucleotide siRNAs in cmt; Figure 2D). We also examined siRNAs at two types of high-copy tandem repeats: centromeric CentC and noncentromeric knob180. Both types are depleted of mCHH and siRNAs in wild-type plants (Gent et al., 2012, 2014), but in the ddm1 14-DAP embryos, knob180 produced 8-fold more 21-nucleotide siRNAs, 18-fold more 22-nucleotide siRNAs, and 6-fold more 24-nucleotide siRNAs than in the wild type (Figure 2D). This increase in knob180 siRNAs was not accompanied by an increase in mCHH (Figure 2E). A chromosome-level view of 24-nucleotide siRNA coverage showed enrichment toward chromosome arms in wild-type 14-DAP endosperm and embryo, corresponding to gene density and mCHH islands, whereas 21- and 22-nucleotide siRNAs had a more uneven distribution with large numbers of siRNAs at discrete loci (Figure 3A). In cmt and ddm1 14-DAP embryos, 24-nucleotide siRNAs showed a distribution similar to wild-type 21- and 22-nucleotide siRNAs, with a strong reduction at the majority of loci across the genome. We split the genome into 100-bp bins and counted the number of 24-nucleotide siRNAs normalized per 500,000 miRNAs that overlapped each bin. We required that at least 14 bp of each siRNA overlap with the bin in order to be counted. Any locus with at least five overlapping siRNAs and with siRNAs spanning at least 50 of its 100 bp was defined as a 24-nucleotide siRNA locus. In wild-type 14-DAP embryos, 176,342 loci met these criteria, while only 26,519 did in ddm1 embryos and 26,546 did in cmt embryos. We also identified the subset of 17,985 novel ddm1 24-nucleotide siRNAs that did not meet the criteria in the wild type. Despite the ddm1 and cmt 24-nucleotide siRNA loci being defined solely by 24-nucleotide siRNAs, they were more strongly enriched for 21- and 22-nucleotide siRNAs than 24-nucleotide siRNAs (Figure 3B). Similar to the knob180 tandem repeat (Figure 2E), the cmt, ddm1, and novel 24-nucleotide siRNA loci were highly methylated in mCG and mCHG and poorly methylated in mCHH in the wild type and did not gain mCHH in either mutant (Figure 3C).
Figure 3.
24-Nucleotide siRNA Loci in Mutants.
(A) Whole-chromosome siRNA patterns. Average siRNA coverage on 5-Mb intervals (per 500,000 miRNAs) is shown for each of the 10 chromosomes for 14-DAP embryo and 14-DAP endosperm. Coverage is on the same scale in every track, with a maximum value of 0.4 overlapping siRNAs per base pair. The peaks are cut off at loci that exceed this value.
(B) siRNA lengths in 24-nucleotide siRNA loci in embryos. siRNA counts were normalized by miRNA count and by the number of loci in each set (shown in parentheses). All siRNAs that overlapped by at least half their lengths with each type of locus were included. Error bars are standard errors of the means for the biological replicates of each genotype.
(C) DNA methylation in 24-nucleotide siRNA loci in embryos. Average methylation for each set of loci was calculated as the proportion of methylated C over total C in each sequence context. Error bars are standard errors of the means for the biological replicates of each genotype.
DISCUSSION
We found that double mutants of the chromomethylases zmet2 and zmet5 (cmt) and double mutants of the DDM1-type nucleosome remodelers chr101 and chr106 (ddm1) were deficient in canonical RdDM, as indicated by loss of DNA methylation and 24-nucleotide siRNAs in mCHH islands. The near complete loss of mCHG in cmt is what would be expected for mu1013094 and mu1017456 being null alleles (Figures 1 and 3). The smaller reductions in mCHG/mCG in ddm1 is consistent with null alleles of ddm1 in Arabidopsis and rice (Oryza sativa; Zemach et al., 2013; Tan et al., 2016; Lyons and Zilberman, 2017), but in the absence of a clear expectation for a ddm1 phenotype in maize, residual DDM1 activity in mu1044815 or mu1021319 is theoretically possible. The complete nonviability of the ddm1 kernels indicates that the mutants are at least severe hypomorphs. What was unexpected for both cmt and ddm1 was the near complete loss of RdDM. In 14-DAP embryos, methylation in the CHH context (mCHH) was more strongly reduced in cmt than in ddm1. The residual mCHH in ddm1 could be explained by continued activity of ZMET2 and ZMET5, as the mCCH subcategory of mCHH, which is more strictly dependent on RdDM (Gouil and Baulcombe, 2016), was reduced to similar levels in both ddm1 and cmt (Figure 1A). In maize, the effect of cmt and ddm1 mutants was weaker in 14-DAP endosperm than in 14-DAP embryo and weaker than in mature endosperm. Transfer of wild-type maternal products directly into developing endosperm might explain these differences.
In agreement with loss of RdDM, 24-nucleotide siRNAs in embryos were reduced to nearly background levels in ddm1 and in cmt in regions that had high levels of RdDM in the wild type (Figures 2 and 3). The corresponding gain of 21-, 22-, and 24-nucleotide siRNAs in heterochromatin, particularly in retrotransposons, is consistent with their increased siRNA and mRNA expression in ddm1 mutants in Arabidopsis (Onodera et al., 2005; Creasey et al., 2014; McCue et al., 2015). The knob180 tandem repeat in maize is associated with an extreme form of heterochromatin (Peacock et al., 1981). knob180 siRNAs increased up to 18-fold in abundance in ddm1 but not in cmt (Figure 2D). Even though 24-nucleotide siRNAs also increased, mCHH decreased, indicating that these siRNAs did not lead to productive RdDM. The fact that production of siRNAs requires transcription indicates transcriptional derepression of knob180 in ddm1 and suggests that DDM1-type nucleosome remodelers can have roles in transcriptional silencing independent of chromomethylation. The total number of 24-nucleotide siRNAs was only decreased to about half of wild-type levels relative to miRNAs (Figure 2B), and they were retained at high levels at discrete loci throughout the genome (Figure 3A). Loci that retained or gained 24-nucleotide siRNAs in ddm1 or cmt embryos tended to have abundant 21- and 22-nucleotide siRNAs, even in the wild type (Figure 3B). In Arabidopsis, the additional siRNAs produced in ddm1 mutants can direct mCHH using alternative forms of RdDM in developing flower buds, but not in leaves (Nuthikattu et al., 2013; McCue et al., 2015; Panda et al., 2016). In maize embryos, the loci that gained or retained 24-nucleotide siRNAs in ddm1 and cmt embryos, and that were rich in 21- and 22-nucleotide siRNAs, had low mCHH in the wild type and even lower in mutants. This was true even for novel ddm1 24-nucleotide siRNA loci that did not qualify as 24-nucleotide siRNA loci in the wild type (Figure 3C).
De novo RdDM can initiate CMT activity (Jackel et al., 2016; Fultz and Slotkin, 2017), yet it appears that in maize CMT is required to maintain canonical RdDM. One explanation for loss of RdDM in the cmt mutant is that CMT produces mCHG, which leads to H3K9me2. In Arabidopsis, the H3K9me2 binding protein SAWADEE HOMEODOMAIN HOMOLOG1 (SHH1) recruits Pol IV (Law et al., 2013; Zhang et al., 2013). Maize SHH1 interacts with both the Pol IV and Pol V protein complexes (Haag et al., 2014). In addition, since chromomethylases can also produce mCHH (Gouil and Baulcombe, 2016), not all mCHH is dependent on RdDM even at loci that are undergoing RdDM. Some of the decrease in mCHH in cmt is a direct consequence of loss of CMT rather than a loss of RdDM. This is evident in 14-DAP embryos, where cmt had a more severe loss of mCHH than ddm1 did (Figure 1A). The similarly strong loss of 24-nucleotide siRNAs in cmt and ddm1 indicates that RdDM was similarly affected in both mutants.
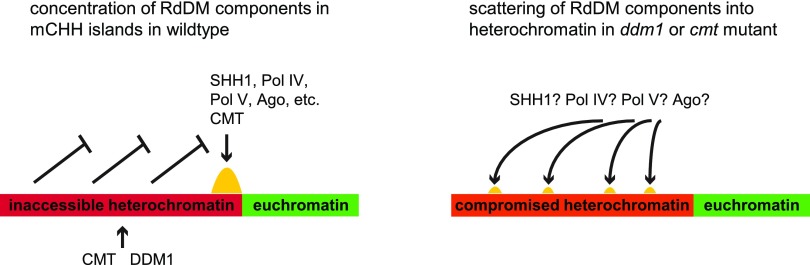
However, the principle reason for loss of RdDM in cmt and ddm1 may be simple dilution. The loss of methylation from heterochromatin could result in the spreading of at least one critical RdDM component from mCHH islands into the newly accessible heterochromatin (Figure 4). Rather than an increase in RdDM at new sites across the genome, we might expect a global decrease because no specific loci would reproducibly recruit the full complement of RdDM components needed to sustain sufficient DRM activity to keep up with the constant loss of methylation during DNA replication. The absence of mCHH in the genome in ddm1 and cmt could be explained if even a single critical RdDM component were present at too low a concentration.
Figure 4.
A Hypothetical Explanation for Loss of RdDM in ddm1 and cmt Mutants.
In wild-type conditions, heterochromatin is maintained in an inaccessible state that excludes RdDM. All the components required for RdDM are then concentrated at a small set of loci in the genome (mCHH islands) where they function in concert to methylate DNA. In the absence of chromomethylases or DDM1-type nucleosome remodelers, heterochromatin no longer excludes RdDM, and RdDM components are scattered over a larger area of repetitive DNA.
Three lines of evidence support the dilution hypothesis. First is the >50% loss of RdDM observed in single mutants of either zmet2 or zmet5 in leaf tissues (Supplemental Figure 4) (Li et al., 2015a). A mild increase in heterochromatin accessibility genome wide could have a large effect in diluting RdDM components from mCHH islands and exacerbate these single mutant phenotypes. The second is the loss of RdDM without the loss of mCHG in mCHH islands in ddm1 mutants (Figure 1). This observation rules out the simple scenario that DDM1 is required for chromomethylation, which is then required for RdDM, and suggests an alternative explanation such as the RdDM dilution model. Finally, there is no evidence for DDM1 functioning directly in RdDM. Instead, nucleosome remodeling in each stage of RdDM is accomplished by distinct SNF2 family nucleosome remodelers: CLSY and DRD1 in Arabidopsis (Kanno et al., 2004; Smith et al., 2007) and CHR167 and RMR1 in maize (Hale et al., 2007; Haag et al., 2014). In contrast, an increase in heterochromatin accessibility to RdDM components genome wide in ddm1 is strongly consistent with its known function and mutant phenotypes (Gendrel et al., 2002; Soppe et al., 2002; Onodera et al., 2005; Zemach et al., 2013; Creasey et al., 2014; McCue et al., 2015; Lyons and Zilberman, 2017).
The severity of the effect of ddm1 and cmt mutants on RdDM may be related to genome size. RdDM in rice, with a genome one-quarter the size of maize, appears to have an intermediate dependence on ddm1: A double mutant of DDM1-type nucleosome remodelers in rice had an ∼50% reduction in mCHH near genes and a slight increase in mCHH in regions not normally undergoing RdDM (Tan et al., 2016). In Arabidopsis, with a genome one-nineteenth the size of maize, there is minor loss of mCHH from RdDM loci in a ddm1 mutant (as indicated by analysis of differentially methylated regions; Stroud et al., 2013) and substantial loss of 24-nucleotide siRNA from many non-LTR transposons (Creasey et al., 2014). It is possible that loss of RdDM in Arabidopsis mutants that lack H3K9me2 might be partially explained by the dilution model rather than simply by direct involvement of H3K9me2 in RdDM (Stroud et al., 2014). If so, Arabidopsis H3K9 methyltransferase mutants or cmt mutants (particularly cmt triple mutants) would also be expected to exhibit increased siRNA production in heterochromatin, as has been demonstrated with ddm1 (Onodera et al., 2005; Creasey et al., 2014; McCue et al., 2015). We look forward to learning whether the dilution model for loss of RdDM in ddm1 and cmt holds true not only in maize, but also as a general feature of plant genomes. What is clear from our data, however, is that both DDM1 and CMT are required for canonical RdDM in maize, and in their absence, siRNAs of multiple lengths are produced in normally heterochromatic regions.
METHODS
Plant Materials and Plant Growth Conditions
PCR primers, alleles, and gene names for all ddm1 and cmt mutants used in this study are listed in Supplemental Table 1, including published zmet alleles (Li et al., 2014). Primers for mop1-1 genotyping are as published previously (Madzima et al., 2014). Wild-type controls for ddm1 and cmt were derivatives of the same UniformMu lineage carrying the ddm1 Mu insertions mu1044815 and mu1021319 and were progeny of homozygous wild-type parents. Wild-type controls for mop1-1 were homozygous Mop1 siblings of the homozygous mop1-1 mutants in the B73 background (derived from Madzima et al. [2014], but further introgressed into B73 another generation).
Developing endosperm and embryos were collected 14 DAP and flash-frozen in liquid nitrogen for later nucleic acid extraction. Prior to freezing, pericarps were removed from kernels and the embryos separated from the endosperm. The parent plants were grown in a winter greenhouse with supplemental lighting from high pressure sodium bulbs for 14 h a day. Each endosperm was cut into two halves, one for DNA extraction and one for RNA extraction. Genotypes of embryos were inferred by genotyping of endosperm. For mature endosperm, dry kernels were soaked in 6% NaOH in water at 57°C for 8 min and the embryos and pericarps removed with forceps. Each mature endosperm was ground to a powder with a mortar and pestle. Frozen 14-DAP endosperms and embryos were ground with micropestles in 2-mL microcentrifuge tubes without thawing. DNA was extracted from all three tissue types, each individual sample separately, with the DNeasy Plant Mini Kit (Qiagen; no. 69104).
WGBS and Small RNA Sequencing
WGBS libraries were prepared using the methylC-seq method (Urich et al., 2015) with no more than seven cycles of PCR amplification for endosperm and no more than 10 for embryo. For the mature endosperm libraries of Figure 1C, DNA from three individuals was combined. For all other libraries, separate libraries were made from each individual embryo or endosperm. All results shown are the average of two to four individuals per genotype, except Zmet2/zmet2 zmet5/zmet5 in Supplemental Figure 2, which is derived from a single embryo.
RNA was extracted from individual 14-DAP embryos and 14-DAP endosperm using mirVana miRNA isolation kits (Thermo Fisher Scientific; no. AM1560) using the total RNA method. For the 14-DAP endosperm, Plant RNA Isolation Aid (Thermo Fisher Scientific; no. AM9690) was added at the lysis step. Small RNA sequencing libraries were prepared from individual embryos and endosperm (two or three for each genotype) using the NEXTflex Small RNA-Seq Kit v3 (Bioo Scientific; no. 5132-05) with 13 cycles of PCR amplification for endosperm and 17 cycles for embryo. The 150-nucleotide single-end Illumina sequencing was performed on an Illumina NextSeq 500 system at the Georgia Genomics Facility, University of Georgia.
BS-seq reads were trimmed and quality filtered using cutadapt (version 1.9.dev1 with Python 2.7.8) (Martin, 2011), command line parameters “-q 20 -a AGATCGGAAGAGC -e .1 -O 1 -m 50.” Trimmed reads were aligned to the B73 RefGen_v4 maize genome (Jiao et al., 2017) using BS-Seeker2 (v2.1.1 with Python 2.7.8 and Bowtie2 2.2.9) with default parameters except –m 1 to allow for a single mismatch. All libraries were aligned to the Zea consensus sequences of the 156-bp tandem repeat CentC and the 180-bp tandem repeat knob180 (Gent et al., 2017) using BS-seeker2 in the same way, except up to four mismatches were allowed per read.
Small RNA-seq reads were trimmed and quality filtered using cutadapt (version 1.14 with Python 2.7.8) (Martin, 2011), command line parameters “-u 4 -q 20 -a TGGAATTCTCGGGTGCCAAGG -e .05 -O 20 --discard-untrimmed -m 24 -M 29,” followed by a second trim with just “-u -4.” In this way adapter sequences and the four random nucleotides at each end of each RNA were trimmed and all reads outside the range of 20 to 25 nucleotides were removed. NCBI BLAST (version 2.2.26) was used to identify reads corresponding to the set of maize mature miRNA sequences from miRBase (version 20; Kozomara and Griffiths-Jones, 2011). The blastall Expectation value was set to 1e-5. Reads corresponding to the tandem repeats CentC and knob180 (Gent et al., 2017) were identified similarly, except the blastall Expectation value was set to 1e-6. The consensus sequence for each was turned into a dimer to allow reads that spanned the junctions between monomers to be identified. After removing all identified miRNAs from the small RNA reads, the remaining 20- to 25-nucleotide reads were mapped to the B73 RefGen_v4 maize genome (Jiao et al., 2017) using BWA-backtrack (version 0.7.15; Li and Durbin, 2009), command line parameters “aln -t 8 -l 10.” All mapping reads were included in the set of siRNAs, including nonuniquely mapping reads, except in the analyses of Figures 2B and 2C, where the subset of uniquely mapping siRNAs were identified based on MAPQ values of at least 20. All results shown are averages from two or three individual embryos or endosperms. Whole-genome coverage was calculated on 5-Mb intervals and visualized using the Integrative Genomics Viewer (Thorvaldsdóttir et al., 2013).
Accession Numbers
All sequencing reads have been deposited in the Sequence Read Archive under accession number SRP127627. Read counts for each experiment and SRA accession numbers are listed in Supplemental Tables 4 and 5. Alleles and gene IDs are listed in Supplemental Table 1.
Supplemental Data
Supplemental Figure 1. Pigment sectoring and small embryo phenotypes of double mutant kernels.
Supplemental Figure 2. DNA methylation in 14-DAP embryos of zmet2 zmet5 heterozygote and homozygote combinations.
Supplemental Figure 3. DNA methylation in mature endosperm of chr101 chr106 heterozygote and homozygote combinations.
Supplemental Figure 4. DNA methylation in leaves and developing ears of single mutants of zmet2-m1 and zmet5-m1.
Supplemental Table 1. Mutant alleles and genotyping primers.
Supplemental Table 2. Kernel genotypes and phenotypes.
Supplemental Table 3. Segregation of zmet2 and zmet5 alleles and chr101 and chr106 alleles.
Supplemental Table 4. Whole-genome bisulfite sequencing libraries.
Supplemental Table 5. Small RNA sequencing libraries.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Sean Trostel for assistance with genotyping, the Maize Genetics Cooperation Stock Center for providing UniformMu stocks, and Nathan Springer for providing other stocks and for comments on the manuscript. This study was supported in part by resources and technical expertise from the Georgia Advanced Computing Resource Center, a partnership between the University of Georgia’s Office of the Vice President for Research and Office of the Vice President for Information Technology. Funding for this study was provided by the National Science Foundation (0922703 and 1118550 to R.K.D. and 1547760 to J.I.G.).
AUTHOR CONTRIBUTIONS
F.-F.F. designed and performed research. R.K.D. designed research and wrote the article. J.I.G. designed and performed research, analyzed data, and wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Bewick A.J., Niederhuth C.E., Ji L., Rohr N.A., Griffin P.T., Leebens-Mack J., Schmitz R.J. (2017). The evolution of CHROMOMETHYLASES and gene body DNA methylation in plants. Genome Biol. 18: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins T., Pontvianne F., Cocklin R., Podicheti R., Chandrasekhara C., Yerneni S., Braun C., Lee B., Rusch D., Mockaitis K., Tang H., Pikaard C.S. (2014). A two-step process for epigenetic inheritance in Arabidopsis. Mol. Cell 54: 30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco J.P., Borges F., Donoghue M.T., Van Ex F., Jullien P.E., Lopes T., Gardner R., Berger F., Feijó J.A., Becker J.D., Martienssen R.A. (2012). Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 151: 194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creasey K.M., Zhai J., Borges F., Van Ex F., Regulski M., Meyers B.C., Martienssen R.A. (2014). miRNAs trigger widespread epigenetically activated siRNAs from transposons in Arabidopsis. Nature 508: 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuerda-Gil D., Slotkin R.K. (2016). Non-canonical RNA-directed DNA methylation. Nat. Plants 2: 16163. [DOI] [PubMed] [Google Scholar]
- Dennis K., Fan T., Geiman T., Yan Q., Muegge K. (2001). Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes Dev. 15: 2940–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., Johnson L.M., Jacobsen S.E., Patel D.J. (2015). DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 16: 519–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fultz D., Slotkin R.K. (2017). Exogenous transposable elements circumvent identity-based silencing, permitting the dissection of expression-dependent silencing. Plant Cell 29: 360–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z., et al. (2010). An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature 465: 106–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel A.V., Lippman Z., Yordan C., Colot V., Martienssen R.A. (2002). Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297: 1871–1873. [DOI] [PubMed] [Google Scholar]
- Gent J.I., Dong Y., Jiang J., Dawe R.K. (2012). Strong epigenetic similarity between maize centromeric and pericentromeric regions at the level of small RNAs, DNA methylation and H3 chromatin modifications. Nucleic Acids Res. 40: 1550–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent J.I., Ellis N.A., Guo L., Harkess A.E., Yao Y., Zhang X., Dawe R.K. (2013). CHH islands: de novo DNA methylation in near-gene chromatin regulation in maize. Genome Res. 23: 628–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent J.I., Madzima T.F., Bader R., Kent M.R., Zhang X., Stam M., McGinnis K.M., Dawe R.K. (2014). Accessible DNA and relative depletion of H3K9me2 at maize loci undergoing RNA-directed DNA methylation. Plant Cell 26: 4903–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent J.I., Wang N., Dawe R.K. (2017). Stable centromere positioning in diverse sequence contexts of complex and satellite centromeres of maize and wild relatives. Genome Biol. 18: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouil Q., Baulcombe D.C. (2016). DNA methylation signatures of the plant chromomethyltransferases. PLoS Genet. 12: e1006526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg M.V., Deleris A., Hale C.J., Liu A., Feng S., Jacobsen S.E. (2013). Interplay between active chromatin marks and RNA-directed DNA methylation in Arabidopsis thaliana. PLoS Genet. 9: e1003946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag J.R., et al. (2014). Functional diversification of maize RNA polymerase IV and V subtypes via alternative catalytic subunits. Cell Reports 9: 378–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale C.J., Stonaker J.L., Gross S.M., Hollick J.B. (2007). A novel Snf2 protein maintains trans-generational regulatory states established by paramutation in maize. PLoS Biol. 5: e275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollick J.B. (2017). Paramutation and related phenomena in diverse species. Nat. Rev. Genet. 18: 5–23. [DOI] [PubMed] [Google Scholar]
- Holoch D., Moazed D. (2015). RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 16: 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh P.H., He S., Buttress T., Gao H., Couchman M., Fischer R.L., Zilberman D., Feng X. (2016). Arabidopsis male sexual lineage exhibits more robust maintenance of CG methylation than somatic tissues. Proc. Natl. Acad. Sci. USA 113: 15132–15137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra C.A., et al. (2012). Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science 337: 1360–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackel J.N., Storer J.M., Coursey T., Bisaro D.M. (2016). Arabidopsis RNA polymerases IV and V are required to establish H3K9 methylation, but not cytosine methylation, on geminivirus chromatin. J. Virol. 90: 7529–7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., et al. (2017). Improved maize reference genome with single-molecule technologies. Nature 546: 524–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C., Kasprzewska A., Tennessen K., Fernandes J., Nan G.L., Walbot V., Sundaresan V., Vance V., Bowman L.H. (2009). Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res. 19: 1429–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L.M., Law J.A., Khattar A., Henderson I.R., Jacobsen S.E. (2008). SRA-domain proteins required for DRM2-mediated de novo DNA methylation. PLoS Genet. 4: e1000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L.M., Du J., Hale C.J., Bischof S., Feng S., Chodavarapu R.K., Zhong X., Marson G., Pellegrini M., Segal D.J., Patel D.J., Jacobsen S.E. (2014). SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature 507: 124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T., Mette M.F., Kreil D.P., Aufsatz W., Matzke M., Matzke A.J. (2004). Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr. Biol. 14: 801–805. [DOI] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S. (2011). miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39: D152–D157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlmann M., Mette M.F. (2012). Developmentally non-redundant SET domain proteins SUVH2 and SUVH9 are required for transcriptional gene silencing in Arabidopsis thaliana. Plant Mol. Biol. 79: 623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J.A., Du J., Hale C.J., Feng S., Krajewski K., Palanca A.M., Strahl B.D., Patel D.J., Jacobsen S.E. (2013). Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature 498: 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.F., Gurazada S.G., Zhai J., Li S., Simon S.A., Matzke M.A., Chen X., Meyers B.C. (2012). RNA polymerase V-dependent small RNAs in Arabidopsis originate from small, intergenic loci including most SINE repeats. Epigenetics 7: 781–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., et al. (2014). Genetic perturbation of the maize methylome. Plant Cell 26: 4602–4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., et al. (2015a). RNA-directed DNA methylation enforces boundaries between heterochromatin and euchromatin in the maize genome. Proc. Natl. Acad. Sci. USA 112: 14728–14733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Vandivier L.E., Tu B., Gao L., Won S.Y., Li S., Zheng B., Gregory B.D., Chen X. (2015b). Detection of Pol IV/RDR2-dependent transcripts at the genomic scale in Arabidopsis reveals features and regulation of siRNA biogenesis. Genome Res. 25: 235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z.W., Shao C.R., Zhang C.J., Zhou J.X., Zhang S.W., Li L., Chen S., Huang H.W., Cai T., He X.J. (2014). The SET domain proteins SUVH2 and SUVH9 are required for Pol V occupancy at RNA-directed DNA methylation loci. PLoS Genet. 10: e1003948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons D.B., Zilberman D. (2017). DDM1 and Lsh remodelers allow methylation of DNA wrapped in nucleosomes. eLife 6: e30674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madzima T.F., Huang J., McGinnis K.M. (2014). Chromatin structure and gene expression changes associated with loss of MOP1 activity in Zea mays. Epigenetics 9: 1047–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17: 3. [Google Scholar]
- McCarty D.R., et al. (2005). Steady-state transposon mutagenesis in inbred maize. Plant J. 44: 52–61. [DOI] [PubMed] [Google Scholar]
- McCue A.D., Panda K., Nuthikattu S., Choudury S.G., Thomas E.N., Slotkin R.K. (2015). ARGONAUTE 6 bridges transposable element mRNA-derived siRNAs to the establishment of DNA methylation. EMBO J. 34: 20–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mérai Z., et al. (2014). The AAA-ATPase molecular chaperone Cdc48/p97 disassembles sumoylated centromeres, decondenses heterochromatin, and activates ribosomal RNA genes. Proc. Natl. Acad. Sci. USA 111: 16166–16171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederhuth C.E., et al. (2016). Widespread natural variation of DNA methylation within angiosperms. Genome Biol. 17: 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobuta K., et al. (2008). Distinct size distribution of endogeneous siRNAs in maize: Evidence from deep sequencing in the mop1-1 mutant. Proc. Natl. Acad. Sci. USA 105: 14958–14963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuthikattu S., McCue A.D., Panda K., Fultz D., DeFraia C., Thomas E.N., Slotkin R.K. (2013). The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21-22 nucleotide small interfering RNAs. Plant Physiol. 162: 116–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y., Haag J.R., Ream T., Costa Nunes P., Pontes O., Pikaard C.S. (2005). Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120: 613–622. [DOI] [PubMed] [Google Scholar]
- Panda K., Ji L., Neumann D.A., Daron J., Schmitz R.J., Slotkin R.K. (2016). Full-length autonomous transposable elements are preferentially targeted by expression-dependent forms of RNA-directed DNA methylation. Genome Biol. 17: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock W.J., Dennis E.S., Rhoades M.M., Pryor A.J. (1981). Highly repeated DNA sequence limited to knob heterochromatin in maize. Proc. Natl. Acad. Sci. USA 78: 4490–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley M.J., Rothi M.H., Böhmdorfer G., Kuciński J., Wierzbicki A.T. (2017). Long-range control of gene expression via RNA-directed DNA methylation. PLoS Genet. 13: e1006749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyaki P.R., Gehring M. (2017). DNA methylation and imprinting in plants: machinery and mechanisms. Crit. Rev. Biochem. Mol. Biol. 52: 163–175. [DOI] [PubMed] [Google Scholar]
- Schoft V.K., Chumak N., Mosiolek M., Slusarz L., Komnenovic V., Brownfield L., Twell D., Kakutani T., Tamaru H. (2009). Induction of RNA-directed DNA methylation upon decondensation of constitutive heterochromatin. EMBO Rep. 10: 1015–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhon R.S., Lin H., Childs K.L., Hansey C.N., Buell C.R., de Leon N., Kaeppler S.M. (2011). Genome-wide atlas of transcription during maize development. Plant J. 66: 553–563. [DOI] [PubMed] [Google Scholar]
- Slotkin R.K., Vaughn M., Borges F., Tanurdzić M., Becker J.D., Feijó J.A., Martienssen R.A. (2009). Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L.M., Pontes O., Searle I., Yelina N., Yousafzai F.K., Herr A.J., Pikaard C.S., Baulcombe D.C. (2007). An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell 19: 1507–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppe W.J., Jasencakova Z., Houben A., Kakutani T., Meister A., Huang M.S., Jacobsen S.E., Schubert I., Fransz P.F. (2002). DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 21: 6549–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H., Greenberg M.V., Feng S., Bernatavichute Y.V., Jacobsen S.E. (2013). Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152: 352–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H., Do T., Du J., Zhong X., Feng S., Johnson L., Patel D.J., Jacobsen S.E. (2014). Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 21: 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan F., Zhou C., Zhou Q., Zhou S., Yang W., Zhao Y., Li G., Zhou D.X. (2016). Analysis of chromatin regulators reveals specific features of rice DNA methylation pathways. Plant Physiol. 171: 2041–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. (2013). Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urich M.A., Nery J.R., Lister R., Schmitz R.J., Ecker J.R. (2015). MethylC-seq library preparation for base-resolution whole-genome bisulfite sequencing. Nat. Protoc. 10: 475–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Elling A.A., Li X., Li N., Peng Z., He G., Sun H., Qi Y., Liu X.S., Deng X.W. (2009). Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell 21: 1053–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger M., Heimes S., Riedel L., Sänger H.L. (1994). RNA-directed de novo methylation of genomic sequences in plants. Cell 76: 567–576. [DOI] [PubMed] [Google Scholar]
- Woodhouse M.R., Freeling M., Lisch D. (2006). The mop1 (mediator of paramutation1) mutant progressively reactivates one of the two genes encoded by the MuDR transposon in maize. Genetics 172: 579–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A., Kim M.Y., Hsieh P.H., Coleman-Derr D., Eshed-Williams L., Thao K., Harmer S.L., Zilberman D. (2013). The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153: 193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J., Zhang H., Arikit S., Huang K., Nan G.L., Walbot V., Meyers B.C. (2015). Spatiotemporally dynamic, cell-type-dependent premeiotic and meiotic phasiRNAs in maize anthers. Proc. Natl. Acad. Sci. USA 112: 3146–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., et al. (2013). DTF1 is a core component of RNA-directed DNA methylation and may assist in the recruitment of Pol IV. Proc. Natl. Acad. Sci. USA 110: 8290–8295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Hale C.J., Law J.A., Johnson L.M., Feng S., Tu A., Jacobsen S.E. (2012). DDR complex facilitates global association of RNA polymerase V to promoters and evolutionarily young transposons. Nat. Struct. Mol. Biol. 19: 870–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Du J., Hale C.J., Gallego-Bartolome J., Feng S., Vashisht A.A., Chory J., Wohlschlegel J.A., Patel D.J., Jacobsen S.E. (2014). Molecular mechanism of action of plant DRM de novo DNA methyltransferases. Cell 157: 1050–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]