The production of siRNAs and the CHH methylation mediated by the RdDM pathway are enhanced in heterochromatin when DDM1 is nonfunctional, at the expense of silencing mechanisms occurring in euchromatin.
Abstract
In plants, cytosine methylation, an epigenetic mark critical for transposon silencing, is maintained over generations by key enzymes that directly methylate DNA and is facilitated by chromatin remodelers, like DECREASE IN DNA METHYLATION1 (DDM1). Short-interfering RNAs (siRNAs) also mediate transposon DNA methylation through a process called RNA-directed DNA methylation (RdDM). In tomato (Solanum lycopersicum), siRNAs are primarily mapped to gene-rich chromosome arms, and not to pericentromeric regions as in Arabidopsis thaliana. Tomato encodes two DDM1 genes. To better understand their functions and interaction with the RdDM pathway, we targeted the corresponding genes via the CRISPR/Cas9 technology, resulting in the isolation of Slddm1a and Slddm1b knockout mutants. Unlike the single mutants, Slddm1a Slddm1b double mutant plants display pleiotropic vegetative and reproductive phenotypes, associated with severe hypomethylation of the heterochromatic transposons in both the CG and CHG methylation contexts. The methylation in the CHH context increased for some heterochromatic transposons and conversely decreased for others localized in euchromatin. We found that the number of heterochromatin-associated siRNAs, including RdDM-specific small RNAs, increased significantly, likely limiting the transcriptional reactivation of transposons in Slddm1a Slddm1b. Taken together, we propose that the global production of siRNAs and the CHH methylation mediated by the RdDM pathway are restricted to chromosome arms in tomato. Our data suggest that both pathways are greatly enhanced in heterochromatin when DDM1 functions are lost, at the expense of silencing mechanisms normally occurring in euchromatin.
INTRODUCTION
In plants, DNA methylation occurs in all cytosine contexts, mainly to silence repeats and transposable elements (TEs) found in heterochromatic regions. Methylation is maintained over generations by specific proteins, like DNA METHYLTRANSFERASE1 for CG sites or CHROMOMETHYLASEs (CMT2 and CMT3) for CHG and CHH sites (where H is any nucleotide except G). In addition to these well-characterized maintenance pathways, cytosines can be methylated de novo, in all contexts, by an RNA-directed DNA methylation (RdDM) mechanism (Matzke et al., 2015). The RdDM pathway occurs through two sequential steps involving the production of small interfering RNAs (siRNAs) and noncoding transcripts generated by the plant-specific DNA-dependent RNA polymerase V (Pol V). In the canonical form of RdDM, 24-nucleotide siRNAs are first produced by the successive actions of another plant specific polymerase, the Pol IV, coupled to RNA-DEPENDENT RNA POLYMERASE2 (RDR2) and DICER-LIKE3 (DCL3) (Herr et al., 2005; Onodera et al., 2005; Kasschau et al., 2007; Zhang et al., 2007; Jia et al., 2009; Law et al., 2011; Haag et al., 2012; Blevins et al., 2015; Li et al., 2015b; Zhai et al., 2015). The biosynthesis of siRNAs also results from alternative RdDM pathways. For instance, 21/22-nucleotide siRNAs are produced by Pol II, RDR6, DCL4, and ARGONAUTE1 (AGO1), in particular when TEs are transcriptionally reactivated (McCue et al., 2012, 2015; Nuthikattu et al., 2013). The siRNAs then guide either AGO4 or AGO6, by base-pairing association, toward Pol V nascent scaffold transcripts (Wierzbicki et al., 2008, 2009). Finally, the complex formed by AGOs and siRNAs recruits the DNA methyltransferase DOMAIN REARRANGED METHYLTRANSFERASE2 to de novo methylate the genomic region that remained associated with the Pol V transcript (Cao and Jacobsen, 2002; Zhong et al., 2014). Additional proteins, such as DECREASE IN DNA METHYLATION1 (DDM1), a chromatin remodeling protein that belongs to the SWI2/SNF2 family, are involved in preserving heterochromatic features. Indeed, DDM1 was shown to shift nucleosomes in vitro (Brzeski and Jerzmanowski, 2003), assisting enzymes maintaining epigenetic marks on DNA or histones to access condensed heterochromatin (Zemach et al., 2013; Lyons and Zilberman, 2017). In this context, recent data suggest that the function of DDM1 and RdDM are antagonistic (Zemach et al., 2013).
In Arabidopsis thaliana, DDM1 is essential to sustain global levels of DNA methylation and ddm1 mutants are extensively hypomethylated in all cytosine contexts (Vongs et al., 1993; Kakutani et al., 1995, 1996; Lippman et al., 2004; Zemach et al., 2013). Disrupting the mouse LYMPHOID SPECIFIC HELICASE gene, which is the mammalian gene most related to DDM1, also leads to demethylation of the genome (Dennis et al., 2001), suggesting an ancient and widespread role for DDM1 in maintaining methylation. DDM1 preferentially controls the silencing of TEs (Lippman et al., 2004), particularly long TEs located in the heterochromatin (Zemach et al., 2013), preventing their reactivation and transposition. Consequently, Arabidopsis self-pollinating ddm1 lines undergo a burst of uncontrolled retrotransposition events associated with developmental abnormalities gradually acquired over generations (Miura et al., 2001; Singer et al., 2001; Tsukahara et al., 2009). By contrast, some of the phenotypes revealed in a ddm1 background are not alterations in the structure of the genome, but are rather associated with epigenetic modifications that influence gene expression and generate stable epialleles (Kakutani, 1997; Saze and Kakutani, 2007). Accordingly, the epigenetic recombinant inbred lines derived from a ddm1 mutant show heritable phenotypic variation (Cortijo et al., 2014). Aside from Arabidopsis, ddm1 mutants have been isolated in maize (Zea mays; Li et al., 2014) and rice (Oryza sativa; Tan et al., 2016), both species containing two DDM1 homologs. In rice, the T-DNA insertion loss-of-function single mutants Osddm1a and Osddm1b had no distinct phenotype but severe growth defects were observed at the first generation of the double mutant, presenting a major reduction of methylation in all contexts. In maize, two single T-DNA insertion loss-of-function ddm1 mutants showed a significant reduction of methylation in non-CG contexts. Nonetheless, a double ddm1 mutant could not be isolated by crossing the two single mutants.
Tomato (Solanum lycopersicum) is one of the major crops cultivated worldwide, and the regulation of DNA methylation is crucial for fruit ripening in this species (Zhong et al., 2013; Liu et al., 2015; Gallusci et al., 2016; Lang et al., 2017). Tomato pericentromeric regions largely extend beyond centromeres and repeats cover ∼65% of the genome (Tomato Genome Consortium, 2012; Zhong et al., 2013; Jouffroy et al., 2016). Understanding how tomato transposons are controlled and silenced is therefore both of fundamental and agronomic interest. Still, few mutants corresponding to epigenetic pathways have been reported in this model plant (Kravchik et al., 2014a; Gouil and Baulcombe, 2016; Lang et al., 2017), and in particular, ddm1 mutants are yet to be obtained.
In this study, we characterized the methylome, transcriptome, and small RNA content of tomato plants deficient for DDM1. DDM1 is encoded in tomato by two genes for which we generated loss-of-function alleles using the CRISPR/Cas9 technology. We found that the Slddm1a Slddm1b mutant had drastic hypomethylation particularly in TEs of heterochromatic regions in both CG and CHG contexts. As a counterbalancing mechanism, the distribution of both 24-nucleotide siRNAs and CHH methylated sites was strongly modified in this mutant. RNA-seq analyses revealed that the transcriptional reactivation of TEs remained limited in the mutant despite the major changes occurring between heterochromatin and euchromatin.
RESULTS
Generation of Slddm1 Knockout Mutants
To identify the tomato DDM1 genes, the BLASTP program was used to search the tomato protein databases (ITAG 2.40 release) using the Arabidopsis DDM1 protein sequence. We found two proteins showing 81.5% identity to each other and 73% (SlDDM1a, Solyc02g062780) or 70% (SlDDM1b, Solyc02g085390) identity to AtDDM1, suggesting that the tomato genome contains two DDM1 genes (Supplemental Figure 1A). The SlDDM1a and SlDDM1b proteins also display domain architectures similar to their respective Arabidopsis and rice orthologs (Supplemental Figure 1B). Analysis of published RNA-seq data (Tomato Genome Consortium, 2012) reveals that the genes are expressed at relatively low levels at anthesis and are upregulated following fruit set, reaching maximum levels at immature young fruit (Supplemental Figure 1C). The expression of SlDDM1a and SlDDM1b gradually decreases during fruit maturation, and, upon ripening, the expression of both genes declines (Supplemental Figure 1C).
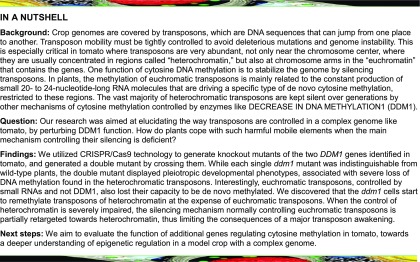
To functionally characterize SlDDM1, we edited the corresponding genes, using the CRISPR/Cas9 technology, to produce loss-of-function mutants. Single-guide RNAs (sgRNAs) were designed to target the second exon of the SlDDM1a gene and the fourth exon of SlDDM1b (Figure 1A). These sgRNAs, together with the plant codon-optimized version of Cas9 (Li et al., 2013), were transgenically expressed in tomato. Genotyping of regenerated T0 transgenic plants identified individual plants that showed Cas9 activity in SlDDM1 genes and nontransgenic homozygous SlDDM1 mutants were successfully isolated among their progenies. Sequencing the crispr-slddm1a-5 and the crispr-slddm1b-16 mutant alleles (hereafter called Slddm1a and Slddm1b, respectively) revealed a large deletion of 131 bp in the 2nd exon of SlDDM1a (nucleotides 1109–1239; Figure 1B) and a 1-bp deletion in the 4th exon of SlDDM1b (nucleotide 1605; Figure 1B), both causing frameshift mutations near the N-terminal parts of the corresponding proteins (Figure 1C). Phenotype analysis of Slddm1a and Slddm1b mutants did not reveal any differences with the wild type (Figure 1D). A Slddm1a Slddm1b double mutant was obtained by crossing Slddm1a and Slddm1b homozygotes. Genotyping of the resulting F2 progeny revealed that while all heterozygous and single homozygous genotypes were indistinguishable from wild-type plants and segregated in a Mendelian manner, the frequencies of the Slddm1b mutant were lower (4%, n = 24/581) than the expected theoretical frequency (6%, χ2 test P = 0.035). The Slddm1a Slddm1b plants exhibited pleiotropic vegetative and reproductive phenotypes. Vegetative phenotypes included variegated cotyledons and leaves and overall smaller size likely due to growth retardation (Figures 1D and 2A). Reproductive phenotypes included smaller floral buds, most of which senesced prematurely except few that produced small flowers displaying partially opened petals and normal anthers and pistils (Figure 2B). Compared with the wild type, mutant anthers produced much less pollen with significantly reduced viability as indicated by Alexander staining (Supplemental Figure 2). Occasionally, mutant flowers could set a small parthenocarpic fruit (Figure 2C) that never produced viable offspring. Altogether, our results indicate that DDM1 is essential for normal vegetative and reproductive tomato development.
Figure 1.
CRISPR/Cas9 Knockout of SlDDM1a and SlDDM1b Genes.
(A) Schematic illustrations of SlDDM1a and SlDDM1b gene regions targeted by the guide RNA. Black bars indicate exons and lines introns. The sgRNA-target sequences are shown, protospacer adjacent motif sequences are colored in red, black triangles mark the predicted Cas9 cut site, restriction enzyme recognition sites are underlined, and black half arrows indicate the location of primers used for mutant genotyping by PCR.
(B) Sequences of isolated mutant Slddm1a and Slddm1b alleles aligned to their respective wild types. The guide RNA target sequence is highlighted in red. PAM, protospacer adjacent motif.
(C) Scheme of SlDDM1 proteins and predicted mutant protein sequences aligned to their respective wild types below. The DNA binding domain (green), SNF2 family N-terminal domain (blue), DEAD-like helicases superfamily domain (yellow), and helicase superfamily C-terminal domain (orange) are indicated. Arrowheads indicate the site of mutation.
(D) Representative 25-d postgermination seedlings of indicated genotypes, all segregated from the same double heterozygote parent plant. Bar = 2 cm.
Figure 2.
Phenotypes of Slddm1a Slddm1b Mutant Plants.
(A) Representative vegetative organs of wild type and Slddm1a Slddm1b. Left to right: adaxial and abaxial sides of cotyledons (bar = 2 mm), mature expanded leaf (bar = 5 cm), terminal leaflet (bar = 4 mm), and a whole plant (bar = 20 cm).
(B) Representative floral organs of wild-type and Slddm1a Slddm1b flowers. Left to right: mature bud (bar = 2 cm), an inflorescence (bar = 1 cm), flower and ovary at anthesis (bars = 2 mm), and a manual cross section of an ovary (bars = 0.4 mm).
(C) Representative wild-type and Slddm1a Slddm1b red fruits and their respective cross sections (bars = 1 cm).
The ddm1 Mutations Have a Limited Effect on Global CHH Methylation Levels in Tomato
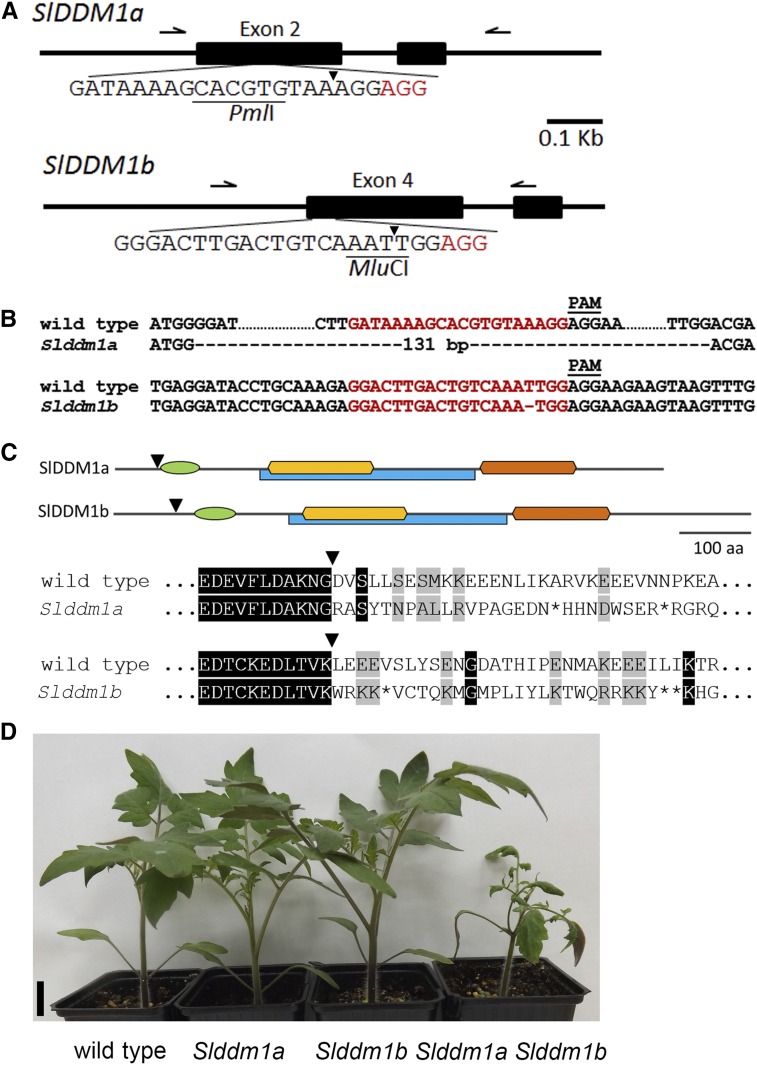
We determined the methylation patterns of Slddm1 single and double mutants by sequencing their genomes after bisulfite conversion. Genomic DNAs were extracted from leaves of two biological replicates per genotype. The levels of methylation per cytosine confirmed that the biological replicates were closely correlated (pairwise Pearson correlation values between biological replicates >0.87 for CGs and CHG and 0.79 for CHH; Supplemental Figure 3) and that the bisulfite conversion rates were >99% because the chloroplast sequences remained unmethylated (Supplemental Table 1). In the Slddm1a Slddm1b mutant, the total number of methylated cytosines was decreased by 49, 64, and 24% in the CG, CHG, and CHH contexts, respectively, compared with the wild type (Supplemental Table 1). We observed similar changes when the average levels of methylation were calculated in 1-kb tiles partitioning the genome (Figures 3A and 3B). Consistent with results obtained in Arabidopsis (Vongs et al., 1993; Kakutani et al., 1995, 1996; Lippman et al., 2004; Zemach et al., 2013) and rice (Tan et al., 2016), the complete disruption of DDM1 genes in tomato led to a drastic hypomethylation of the genome, mainly in the CG and CHG contexts.
Figure 3.
Patterns of Methylation in Tomato Slddm1 Mutants.
(A) Box plots showing mean methylation content of the Slddm1 mutants and the corresponding wild type (WT). The tomato genome (SL2.5 release) was partitioned in 1-kb tiles, and methylation levels correspond to the ratios of methylated cytosines over the total number of cytosines. Only cytosines covered by at least five reads were considered. The average methylation levels were determined by combining the two biological replicates for each genotype.
(B) Pairwise comparison of methylation in the wild type and the Slddm1a Slddm1b mutant. Each dot represents a 1-kb window and their methylation levels were determined as in (A). The color scale measures the density of points (red being very dense). The Pearson correlation coefficients between the samples are 0.29 for mCG, 0.55 for mCHG, and 0.7 for mCHH.
(C) Patterns of methylation in genes and TEs of the Slddm1 mutants. The average methylation levels of genes (upper panels) and TEs (lower panels) were determined by dividing the corresponding annotated regions (ITAG 2.40 release) into 100-bp bins. Regions located 1 kb upstream and 1 kb downstream of the gene bodies and TEs are also presented in these metaplot analyses. TE genes (see Methods) were removed to perform the analysis.
Monitoring the global methylation of genes and TEs revealed several interesting features. On average, the CG, CHG, and CHH methylation levels of the Slddm1a Slddm1b mutant were reduced by 12, 42, and 21% in gene bodies, respectively (Figure 3C). CG methylation is frequently found in genes of plants and is independent of DDM1 (Stroud et al., 2013). The presence of non-CG methylation in tomato genes is more unusual but could possibly be explained by the large number of TEs associated with gene-enriched regions (Jouffroy et al., 2016). When TEs were inspected globally, we found that both CG and CHG methylation levels were drastically reduced (by 41 and 51%, respectively) in the Slddm1a Slddm1b mutant, in agreement with the role played by DDM1 to maintain heterochromatin. However, the CHH methylation level of TEs was decreased by only 7% (Figure 3C), in sharp contrast with the strong global loss of CHH methylation (40%) observed in Arabidopsis or rice ddm1 TEs (Zemach et al., 2013; Ito et al., 2015; Panda et al., 2016; Tan et al., 2016). These data suggest that both CG and CHG methylations of TEs are severely altered in Slddm1a Slddm1b, unlike CHH methylation, and this was confirmed by measuring the methylation levels for different families of TEs and repeats (Supplemental Figure 4).
CHH Methylation Changes between Euchromatic and Heterochromatic Slddm1 TEs
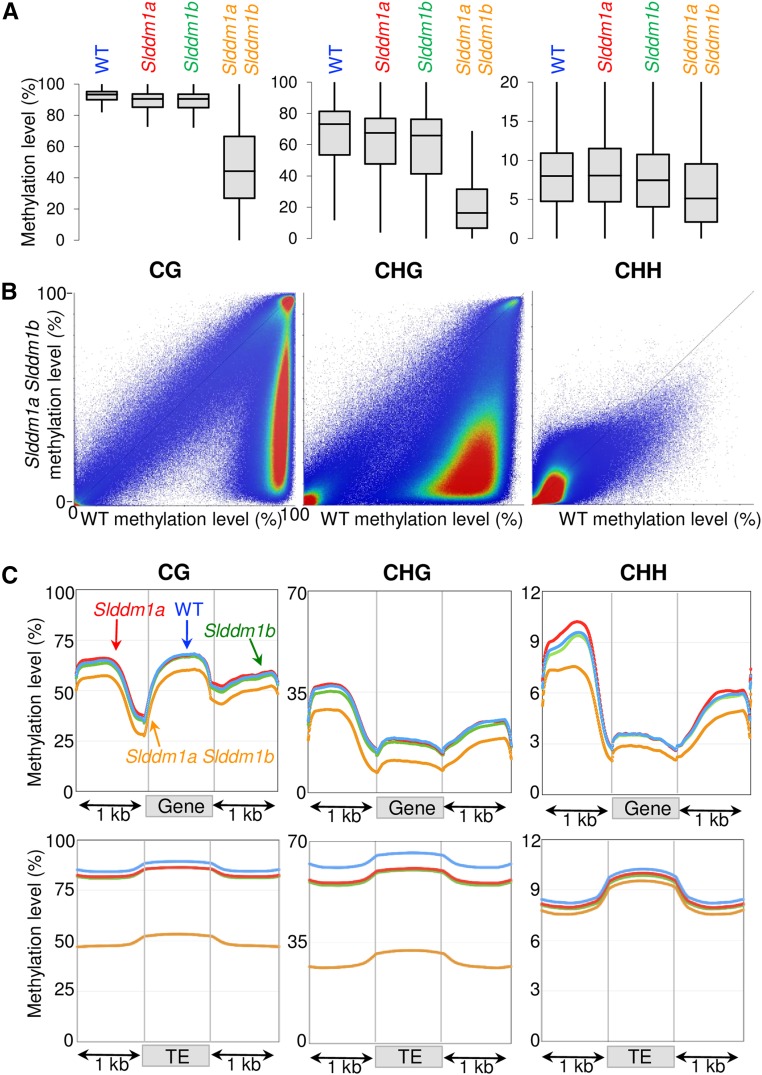
The regions that were significantly differentially methylated (DMRs) between mutants and the wild type were identified. The Slddm1a Slddm1b mutant contained a very high number of DMRs hypomethylated (hypoDMRs) in the CG and CHG contexts (190,026 and 249,593, respectively; Figure 4A; Supplemental Data Set 1) predominantly related to heterochromatic regions (Figure 4B; Supplemental Figure 5, regions in green) and overlapping with TEs (82% for the CG hypoDMRs, 87% for the CHG hypoDMRs; Figure 4C) that were heavily methylated in the wild type (Figure 4D). In parallel, a more limited number of hypermethylated DMRs (hyperDMRs) was identified for CGs and CHGs (719 and 1816 respectively; Figure 4A; Supplemental Data Set 2). Seventy-one percent (507) of the CG hyperDMRs and 64% (1164) of the CHG hyperDMRs were included in repeat-poor regions (Supplemental Data Set 3) and were therefore mostly localized in euchromatic regions (Figure 4B; Supplemental Figure 5, regions in black). They overlap with genes or genes containing TEs (30% of the CG hyperDMRs and 40% of the CHG hyperDMRs), or with TEs alone (16% of the CG hyperDMRs and 23% of the CHG hyperDMRs) (Figure 4C). CG hyperDMRs corresponded to regions unmethylated in the wild type that become methylated at both CGs and CHGs in Slddm1a Slddm1b (Supplemental Figure 6). CHG hyperDMRs were methylated in all contexts in the wild type and gained additional mCHG, and to a lesser extent mCHH (Supplemental Figure 6). Therefore, TEs of the heterochromatin were vastly depleted of CG and CHG methylated sites in Slddm1a Slddm1b; additionally, certain euchromatic regions become methylated in these contexts.
Figure 4.
Number, Localization, and Nature of the DMRs Identified in the Tomato Slddm1 Mutants.
(A) Total number of DMRs found in the three methylation contexts (CG, CHG, and CHH). Hypo- and hyperDMRs are shown. DMRs associated with Chr00 were excluded from the count.
(B) Densities of DMRs between the Slddm1a Slddm1b mutant and the wild type, for chromosomes 3 (SL2.5 genome release). Chromosome 6 is shown in Supplemental Figure 5A, and all chromosomes are presented in Supplemental Figure 5B. The density of hyperDMRs is represented in black, and the density of hypoDMRs is represented in green. The numbers of TEs (Gypsy elements) and genes contained within bins of 100 kb are plotted on histograms. The scale is 0 to 100 elements for TEs and 0 to 30 for genes. Regions were considered to be differentially methylated when the absolute differences of methylation were at least 10% for mCHH, 20% for mCHG, or 30% for mCG, between the mutant and the wild type.
(C) Nature of the DMRs identified in the Slddm1 mutants. CDS+TE corresponds to DMRs overlapping with both genes and TEs, CDS corresponds to DMRs overlapping with genes, and TE corresponds to DMRs overlapping with TEs. All other remaining DMRs are classified as Intergenic. DMRs associated with chr00 were excluded from the count.
(D) Methylation levels of CG and CHG hypoDMRs. The average methylation levels of the DMRs were determined by dividing the corresponding regions into 100-bp bins. Regions located 1 kb upstream and 1 kb downstream of the DMRs are shown.
The opposite situation was observed for regions differentially methylated in the CHH context. The density of CHH hypoDMRs (8518 were identified; Figure 4A; Supplemental Data Set 1) was higher at chromosome arms (Figure 4B; Supplemental Figure 5, regions in green) in regions enriched for genes or localized at the frontiers between euchromatin and heterochromatin (i.e., in repeat-intermediate regions). Indeed, 30% (2582) of the CHH hypoDMRs were localized in repeat-poor regions, 33% (2802) in repeat-intermediate regions, and 39% (3313) in repeat-rich regions, a distribution that differed (χ2 test, P < 10−300) from the one expected if those DMRs were equally distributed in all regions (26, 19, and 55%, respectively). Our results were consistent with previous analyses showing a reduction of mCHH in euchromatin of the rice ddm1 mutants (Tan et al., 2016). The CHH hypoDMRs mostly overlapped with TEs (74% of the CHH hypoDMRs; Figure 4C). A total of 10,297 CHH hyperDMRs (Figure 4A; Supplemental Data Set 2) were detected between Slddm1a Slddm1b and the wild type, but this time, mostly localized in heterochromatic regions (Figure 4B; Supplemental Figure 5, CHH regions in black). They correspond to TEs (85% of CHH hyperDMRs overlap with TEs; Figure 4C) in which mCHH levels increase by almost 3 times in Slddm1a Slddm1b (Figure 5; CHH hyperDMRs). Three (0.1%) of the hypoCHH DMRs found in repeat-poor regions were close to (within 500 bp) a CG hyperDMR and 0.4% (11) to a CHG hyperDMR. Therefore, in repeat-poor regions, CHH hypomethylation and CG/CHG hypermethylation occurred at different locations. By contrast, 64% (4841) of the CHH hyperDMRs found in repeat-rich areas were close to (within 500 bp) a CG hypoDMR and 60% (4478) to a CHG hypoDMR. Therefore, some heterochromatic TEs were actively remethylated at CHH sites, while others, localized within gene-enriched regions, became hypomethylated in this context, altogether resulting in limited quantifiable changes in overall CHH methylation of TEs (Figure 3C).
Figure 5.
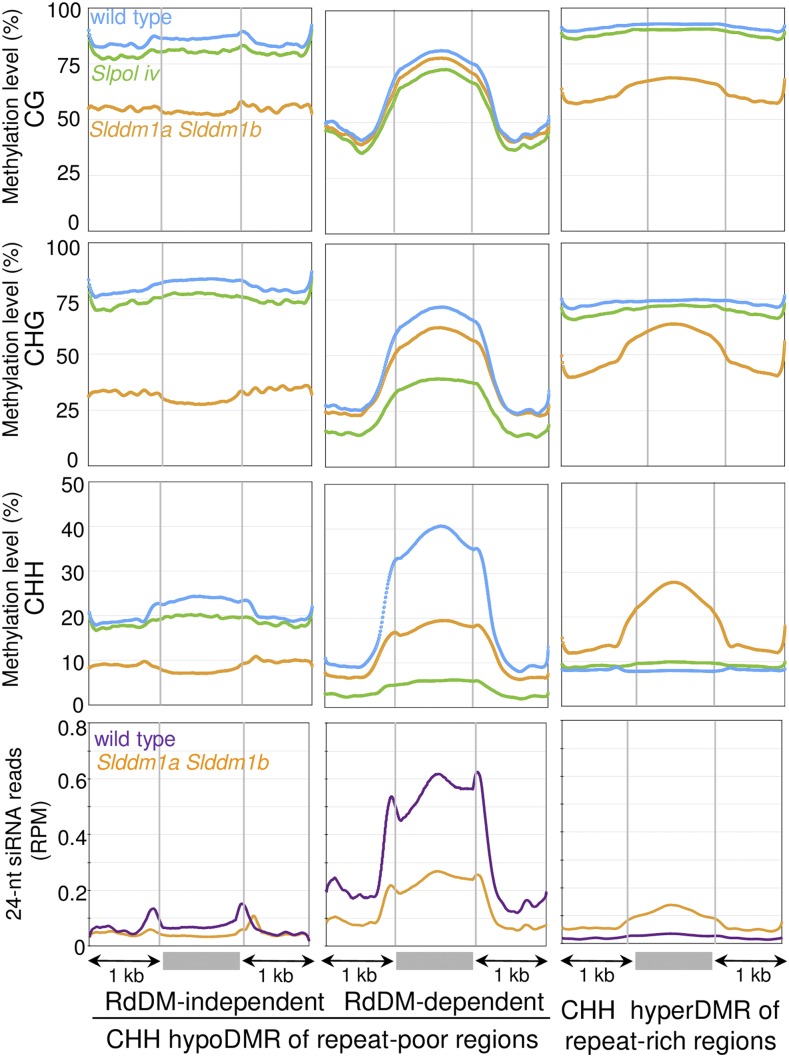
Methylation Levels and siRNA Contents of CHH DMRs.
The average methylation levels and 24-nucleotide siRNA contents of the DMRs were determined by dividing the corresponding regions into 100-bp bins. Regions located 1 kb upstream and 1 kb downstream of the DMRs are shown. CHH hypoDMRs localized within repeat-poor regions (Supplemental Data Set 3) were divided in two groups: one that depends on RdDM (i.e., corresponding to Slddm1a Slddm1b, Slpol iv, and Slpol v overlapping hypoCHH DMRs), and the other one that is RdDM independent (i.e., corresponding to Slddm1a Slddm1b not overlapping with Slpol iv or Slpol v hypoCHH DMRs). The results correspond to the mean values obtained for the two biological repeats.
In Arabidopsis, CHH methylation is both maintained by CMT2 (Zemach et al., 2013; Stroud et al., 2014) or the RdDM pathway that depends on Pol IV and Pol V. To examine whether the RdDM pathway is altered in Slddm1a Slddm1b, thus compromising the methylation of CHH sites in TEs, we retrieved the methylome sequences of both Slpol iv and Slpol v tomato crispr mutants (Gouil and Baulcombe, 2016). A total of 51,482 CHH hypoDMRs were identified between Slpol iv and the corresponding wild type and 41,016 CHH hypoDMRs for Slpol v. Sixty percent of these CHH hypoDMRs were localized in regions enriched for genes and 20% were in repeat-rich regions, confirming that the RdDM is mostly active in euchromatin. Interestingly, we found that 60% (1546) of the CHH hypoDMRs (Supplemental Data Set 1) identified in the euchromatin of Slddm1a Slddm1b overlapped with CHH hypoDMRs of Slpol iv and 50% (1299) overlapped with those of Slpol v. This indicates that euchromatic regions hypomethylated in the CHH context in Slddm1a Slddm1b are mainly TEs targeted by the RdDM. We confirmed this result by dividing the Slddm1a Slddm1b CHH hypoDMRs of repeat-poor regions in two groups: the first one (1198 DMRs) depended on RdDM and corresponded to CHH hypoDMRs overlapping between Slddm1a Slddm1b, pol iv, and pol v. The second group (943 DMRs) corresponded to RdDM-independent CHH hypoDMRs not overlapping with pol iv or pol v DMRs. Seventy-six percent (917) of RdDM CHH hypoDMRs overlapped with short euchromatic TEs (mean length: 238 bp) that had lost almost 13% of mCHG and 52% of mCHH in Slddm1a Slddm1b (Figure 5; Supplemental Figure 7A). Seventy-seven percent (726) of the hypoCHH non-RdDM DMRs corresponded to long heterochromatic TEs (mean length: 751 bp) not targeted by the RdDM in the wild type and losing 39% of mCG, 66% of mCHG, and 69% of mCHH in Slddm1a Slddm1b (Figure 5; Supplemental Figure 7B). Thus, tomato euchromatin contains two types of TEs differently controlled by DDM1 and the RdDM.
The Production of siRNAs Increases in Heterochromatin and Decreases in Euchromatin of Slddm1a Slddm1b
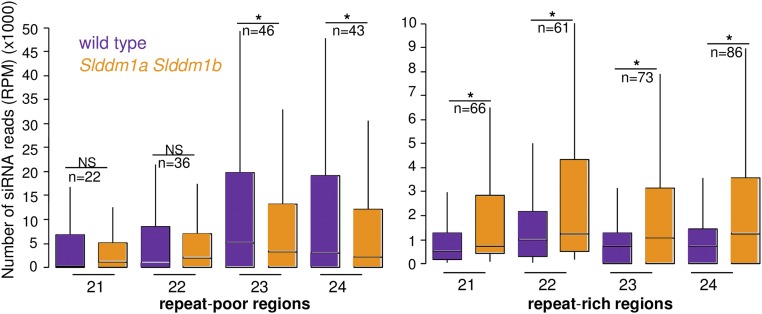
The RdDM pathway depends on the production of small RNAs, in particular 24-nucleotide siRNAs; hence, we sequenced the small RNAs of Slddm1a Slddm1b and compared their distribution along the genome to that in wild-type tomato. Reproducibility between biological replicates was confirmed by performing a principal component analysis to visualize the differences (Supplemental Figure 8A). Mapping the reads revealed that the 24-nucleotide siRNAs of wild-type tomato follow the general patterns of small RNAs along the chromosomes (Tomato Genome Consortium, 2012), being almost excluded from the large pericentromeric regions and accumulating in chromosome arms (Figure 6A). Indeed, repeat-poor regions of the wild-type plants contain 7-fold more 24-nucleotide reads, compared with regions highly enriched in repeats (Figure 6B). While the production of these 24-nucleotide siRNAs, depending on both SlPol IV and SlDCL3 (Figures 6A and 6B), was similar to that in Arabidopsis, their genomic distribution differed sharply as Arabidopsis siRNAs are highly prevalent at pericentromeres (Zhang et al., 2007; Mosher et al., 2008; Law et al., 2013; Liu et al., 2014). The distribution of 24-nucleotide siRNAs in the Slddm1a Slddm1b mutant was drastically modified compared with the wild type. Their levels decreased by almost 2-fold in repeat-poor regions and increased by the same proportion in repeat-rich regions when reads were normalized against total mapped reads (Figure 6B). Similar results were obtained when reads were normalized against miRNA reads (Supplemental Figure 9). Then, 106926 siRNA clusters were defined (see Methods) and we compared their profiles of expression between the wild type and the Slddm1a Slddm1b mutant (Figure 6C). We found that 6913 heterochromatic siRNA clusters showed increased (log2FC(Slddm1aSlddm1b/WT) > 2) expression of 24-nucleotide siRNAs compared with the wild type (DESeq2 significance cutoff of 0.01). In repeat-poor regions, the levels of 24-nucleotide siRNAs were decreased for 967 siRNAs clusters and increased for 680 clusters. Furthermore, we determined the levels of 21-, 22-, and 23-nucleotide siRNAs in repeat-poor and repeat-rich regions containing a significant (DESeq2 significance cutoff of 0.01) number of reads (Figure 7; Supplemental Figure 10). The production of 23/24-nucleotide siRNAs was inhibited and enhanced in gene-rich and gene-poor regions, respectively, and the production of 21/22-nucleotide siRNAs was increased in repeat-rich regions of Slddm1a Slddm1b, likely because the RDR6-RdDM pathway was activated. Less than 3% of CG or CHG DMRs overlapped with siRNAs clusters deregulated (log2FC(Slddm1a Slddm1b/WT) > 2 or < −2) in Slddm1a Slddm1b. The same results were obtained for CHH hypoDMRs found in repeat-poor areas. By contrast, 14% of the CHH hyperDMRs localized in regions enriched for repeats overlapped with 23/24-nucleotide siRNA clusters upregulated (log2FC(Slddm1aSlddm1b/WT) > 2) in Slddm1a Slddm1b and 6% with 21/22-nucleotide siRNA clusters. CHH hyperDMRs corresponded to heterochromatic TEs that were heavily methylated in the wild type but were not targeted by 24-nucleotide siRNAs and the canonical RdDM pathway (Figure 5; CHH hyperDMRs). In Slddm1a Slddm1b, these TEs gained mCHH and became the targets of 24-nucleotide siRNAs, although their levels remained modest (Figure 5; CHH hyperDMRs). Further analyses will help to determine the functional role of these small RNA populations in the Slddm1a Slddm1b mutant.
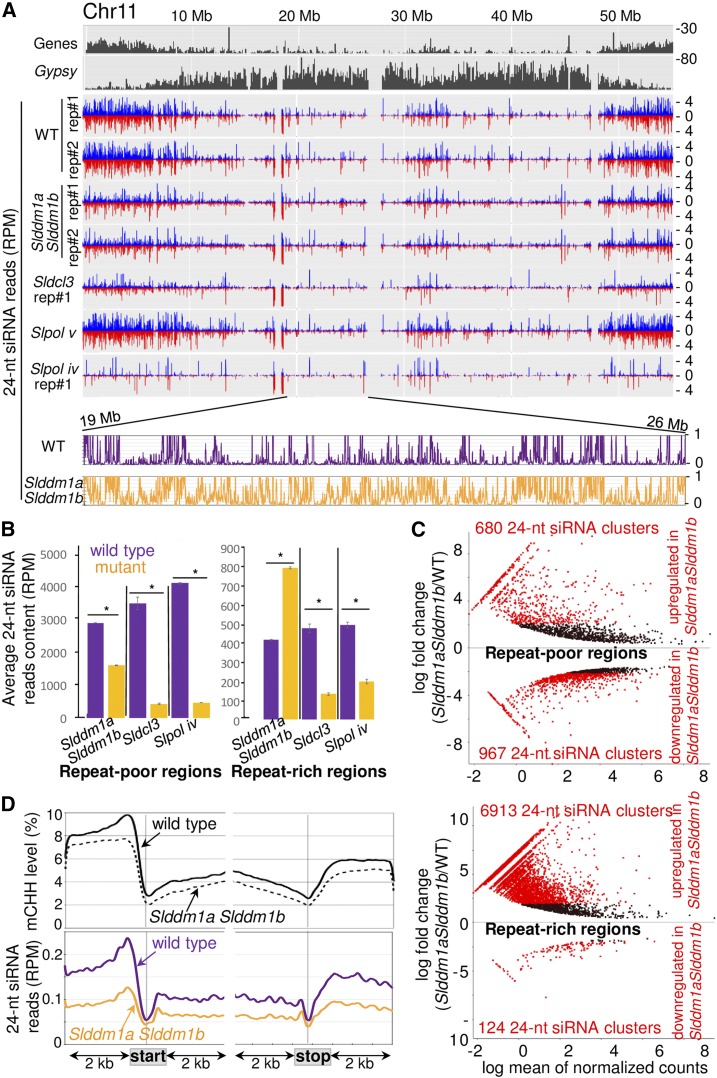
Figure 6.
The 24-Nucleotide siRNA Patterns of Slddm1a Slddm1b Mutants.
(A) Distribution of 24-nucleotide reads along chromosome 11 (chosen as a representative example of all chromosomes). Both sense (blue) and antisense (red) normalized (RPM) reads were plotted. The numbers of TEs (Gypsy elements) and genes contained within bins of 100 kb are plotted on histograms. Sldcl3, Slpol iv, and Slpol v sRNA-seq data were obtained from previous reports (Kravchik et al., 2014a; Gouil and Baulcombe, 2016). The 24-nucleotide read content of one genomic region comprising between 19 and 26 Mb and corresponding to a repeat-rich region are shown below (reads from both strands were collapsed and the values correspond to 5-kb bins).
(B) Average number of 24-nucleotide reads contained within the 106 repeat-rich and 68 repeat-poor regions defined in this study (Supplemental Data Set 3). The numbers of reads were normalized to the total number of mapped reads. Error bars indicate sd (n = 2 biological repeats). Asterisks indicate significant differences (Mann-Whitney U-test, P < 0.01).
(C) MA plots for 24-nucleotide siRNA clusters. Each dot represents a siRNA cluster that was identified as significantly differentially expressed at a 1% FDR using the DESeq2 R package. The clusters differentially expressed by 2-fold [log2FC(Slddm1a Slddm1b/WT) > 2 or < −2] are in red and their numbers are indicated within each panel. Above the center horizontal line are 24-nucleotide clusters that are more expressed in Slddm1a Slddm1b compared with the wild type and below are the ones that are less expressed.
(D) Patterns of CHH methylation and 24-nucleotide siRNAs for regions surrounding the transcription start and stop of genes in the wild type and Slddm1a Slddm1b. The average mCHH levels and normalized 24-nucleotide siRNA contents were determined by dividing the corresponding regions, into 100 bp bins. Regions located 1 kb upstream and 1 kb downstream of the gene bodies and TEs are shown. TE-Genes (see Methods) were removed to perform the analysis. The results correspond to the mean values obtained for the two biological repeats.
Figure 7.
siRNA Content in Repeat-Rich and Repeat-Poor Regions of the Slddm1a Slddm1b Mutant and the Wild Type.
Box plots showing average numbers of 21- to 24-nucleotide reads contained within repeat-rich and repeat-poor regions (n) having a significant number of reads (DESeq2 cutoff of 0.01). The numbers of reads were normalized to the total number of mapped reads. Asterisks indicate significant differences and NS indicates nonsignificant differences (Mann-Whitney U-test, P < 0.01).
Finally, we examined the distribution of siRNAs around genes. In the wild type, the metaprofile of 24-nucleotide siRNAs revealed a peak at ∼500 bp upstream of the transcription start and to a lesser extent downstream of the transcription stop site that corresponded to similar changes in the levels of mCHH (Figure 6D). Interestingly, both peaks were markedly reduced in Slddm1a Slddm1b, further indicating that the RdDM pathway is compromised in euchromatin of the mutant.
The Number of TEs Reactivated in the First Generation of Slddm1a Slddm1b Mutants Is Limited
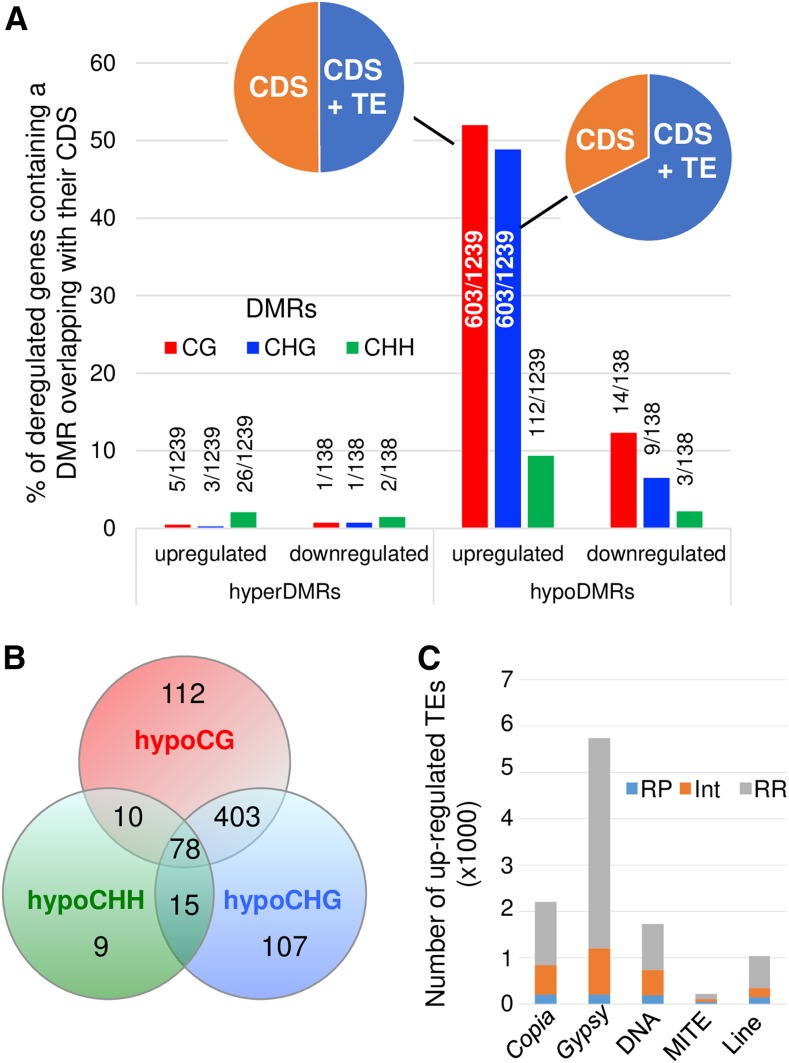
To better understand whether disrupting the RdDM in tomato affects the expression of genes and TEs, we performed RNA-seq analyses using three biological replicates for the wild type and four for the Slddm1a Slddm1b mutant, including those already used for the sRNA-seq (Supplemental Table 2). We verified that all biological replicates were grouped together (Supplemental Figure 8B). We found a total of 138 genes that were significantly (false discover rate [FDR] threshold ≤ 0.01) downregulated [log2FC(Slddm1a Slddm1b/WT) < −1.5] in Slddm1a Slddm1b, compared with the wild type, and 1239 upregulated [log2FC(Slddm1a Slddm1b/WT) > 1.5] genes (Supplemental Data Set 4) that include 390 TE genes, corresponding to TEs incorrectly annotated as genes (Jouffroy et al., 2016). Almost 50% of the upregulated genes were overlapping with hypoDMRs in the CG or CHG contexts and 10% with CHH hypoDMRs (Figure 8A). Moreover, 71% of these different types of hypoDMRs were overlapping between them (Figure 8B), and 50 to 70% overlapped with TEs (Figure 8A). Yet, very few deregulated genes overlapped with hyperDMRs (Figure 8A). Therefore, our RNA-seq data show that TEs localized near or within genes were derepressed.
Figure 8.
Methylation Patterns and Gene Expression Are Correlated in Slddm1a Slddm1b.
(A) Number of genes that are significantly (FDR ≤ 0.01) downregulated [log2FC(Slddm1a Slddm1b/WT) < −1.5] or upregulated [log2FC(Slddm1a Slddm1b/WT) > 1.5] and overlapping a DMR in either one of the three contexts (CG, CHG, and CHH) are indicated. For the upregulated genes corresponding to CG or CHG hypoDMRs, the proportion of DMRs overlapping genes (CDS) or both genes and TEs (CDS+TE) is shown.
(B) Overlap between hypoDMR localized within the CDS of upregulated genes.
(C) Localization of derepressed TEs. RP, repeat-poor regions; INT, regions containing an intermediate number of repeats; RR, repeat-rich regions.
We determined more precisely whether the TEs were transcriptionally reactivated in the Slddm1a Slddm1b mutant using the annotations obtained for each family (see Methods). To monitor their expression, we used both a multiple-mapping strategy, where reads mapping to different locations with a high score were assigned to all these locations and a unique-mapping strategy (see Methods). The results showed that the transcriptional reactivation of TEs was limited to a small fraction of the annotated TEs in Slddm1a Slddm1b mutants. Indeed, in a total of 536643 TE annotations, we found that 2% were upregulated when reads were mapped at unique locations and 3% when they were mapped multiple times (Table 1). On average, 65% of the derepressed TEs were localized in repeat-rich regions, while 12% and 23% were localized in repeat-intermediate and repeat-poor regions, respectively (Figure 8C). Only a fraction of the TEs annotated could potentially be both reactivated transcriptionally and detected by our RNA-seq analysis. Although their exact number was difficult to establish, we hypothesized that longer elements (>2 kb) were the most conserved, containing functional internal genes that could be transcribed. We found that 6.6% of the Gypsy elements longer than 2 kb and 9.4% for the Copia were derepressed in Slddm1a Slddm1b. From these data, we conclude that the fraction of TEs transcriptionally reactivated in the first generation upon loss-of-SlDDM1 function was restricted. The reactivation of additional TEs in Slddm1 might take more generations, or alternatively, the RdDM pathway of tomato might be particularly efficient to rapidly resilence TEs localized in the heterochromatin.
Table 1. TEs Are Derepressed in the Slddm1a Slddm1b Mutant.
| TE/Repeat Type Family | No. of Bases Covered | Fraction Covered (%) | Mean Element Size (bp) | No. of Annotated TEs | Unique-Mapping Strategy | Multiple-Mapping Strategy | ||
|---|---|---|---|---|---|---|---|---|
| TE Down | TE Up | TE Down | TE Up | |||||
| Gypsy | 247,495,673 | 46.2 | 1,394 | 177,553 | 40 (0.02%) | 4,688 (2.6%) | 2,186 (1.2%) | 6,047 (3.4%) |
| Copia | 75,592,636 | 14.1 | 1,175 | 64,329 | 33 (0.05%) | 1,472 (2.3%) | 333 (0.5%) | 2,229 (3.5%) |
| Line | 25,233,169 | 4.7 | 656 | 38,480 | 6 (0.02%) | 401 (1%) | 114 (0.3%) | 1,074 (2.8%) |
| DNA | 20,558,297 | 3.8 | 777 | 26,454 | 15 (0.06%) | 1,162 (4.4%) | 277 (1%) | 1,744 (6.6%) |
| MITE | 6,758,477 | 1.3 | 458 | 14,744 | 4 (0.03%) | 73 (0.5%) | 54 (0.4%) | 223 (1.5%) |
| Others | 159,808,192 | 29.8 | 918 | 215,083 | 53 (0.02%) | 2,506 (1.1%) | 624 (0.3%) | 4,857 (2.3%) |
| Total | 535,446,444 | 100 | 536,643 | 151 (0.03%) | 10,302 (1.9%) | 3,588 (0.7%) | 16,174 (3%) | |
The number of bases covered genome wide, by a specific TE family, is indicated, as well as the corresponding proportions compared to all TEs. The average size and the numbers of TEs annotated with REPET (Flutre et al., 2011) are indicated. The results of the two mapping strategies used are shown (see Methods). The number (and %) of TEs that were significantly (FDR< 0.01) downregulated [log2FC(ddm1/WT) < −1.5] in the Slddm1a Slddm1b mutant, compared to the wild type, and upregulated [log2FC(ddm1/WT) > 1.5] are shown. “Others” corresponds to TEs that were predicted by REPET, but not classified within the Gypsy, Copia, DNA elements, MITE, or Line families.
DISCUSSION
In this study, we used the CRISPR/Cas9 technology to generate loss-of-function Slddm1 alleles and used them to investigate SlDDM1 functions in tomato, a model crop with a complex genome rich in TEs. We isolated single Slddm1a and Slddm1b mutants, as well as the corresponding Slddm1a Slddm1b mutant plants that show severe developmental defects. In plants deficient for SlDDM1, heterochromatic TEs are depleted of mCGs and mCHGs that normally silence them. We also found that some euchromatic TEs have lost DNA methylation, but in the CHH context. Similarly, the production of siRNAs, including 24-nucleotide siRNAs, is enhanced in heterochromatin.
The ddm1 alleles can be propagated in the homozygous state for several generations in Arabidopsis, but not in other plant species. In maize, two DDM1 orthologs were identified. The corresponding single mutants are viable, but the double mutant could not be recovered despite the screening of a substantial amount of offspring (Li et al., 2014). Rice also contains two DDM1 genes and the double mutant exhibits severe developmental abnormalities and sterility (Tan et al., 2016). In tomato, we report that the vegetative and reproductive development of Slddm1a Slddm1b is drastically altered (Figure 2) and that the plants are completely sterile. Although Arabidopsis seems to be particularly tolerant to genome-wide modifications of methylation patterns, other plant species including crops are much more sensitive. One explanation could be that the number of TEs remobilized in a ddm1 background remains limited in Arabidopsis in contrast to these sensitive species. In addition, the chromatin is structured and organized very differently in plants enriched in TEs. For instance, the genome of rice is partitioned in thousands of topologically associated domains that greatly differ from the chromatin packing patterns of Arabidopsis (Liu et al., 2017).
DDM1 is essential for global maintenance of DNA methylation; consequently, the corresponding tomato mutants are extensively hypomethylated, in particular in heterochromatic regions, as observed in Arabidopsis (Vongs et al., 1993; Kakutani et al., 1995, 1996; Lippman et al., 2004; Zemach et al., 2013) and rice (Tan et al., 2016). We also reveal that the Slddm1a Slddm1b mutant is drastically hypomethylated in both the CG and CHG contexts (Figures 3A and 3B) in regions corresponding to heterochromatic TEs (Figure 4). Additionally, our results show that hypoDMRs are already detectable in both Slddm1a and Slddm1b single mutants (Figure 4A), indicating that the absence of one of the two DDM1 proteins is not fully compensated for by the other one. Nevertheless, the two single mutants and the wild type develop similarly (Figure 1D), at least in the first generations. Future work will help to determine whether the heterochromatic hypomethylation observed in the single mutants is stable over generations, whether TEs are derepressed in these backgrounds, and finally if the two tomato DDM1 proteins have specific functions.
In Arabidopsis, the methylomes of first-generation homozygous ddm1 plants diverge from those of the progenies obtained after eight rounds of self-propagation (Ito et al., 2015). Whereas the CG methylation continuously decreases over generations, genes tend to gain ectopic methylation at non-CG sites. A possible explanation is that factors normally targeting heterochromatin are released in a ddm1 background, inducing the spreading of methylation into euchromatic regions in later generations. We observe an ectopic gain of CG and CHG methylation in the euchromatin of the Slddm1a Slddm1b mutant (Figure 4B; Supplemental Figure 6) occurring by two different ways. First, some regions that are not methylated in the wild type, or targeted by siRNAs, are de novo methylated at both CGs and CHGs sites in Slddm1a Slddm1b, by a mechanism independent of RdDM and siRNA targeting (Supplemental Figure 6A; CG hyperDMR). More data are needed to determine whether these regions carry specific features that attract chromatin modifying factors or methyltransferases. Second, the CHG and more slightly the CHH methylations increase in regions that are already methylated and targeted by 23/24-nucleotide siRNAs in the wild type (Supplemental Figure 6A; CHG hyperDMR). These euchromatic regions are likely TEs remethylated by both CMT3 and CMT2 in Slddm1a Slddm1b, reinforcing their silencing (an example is given in Supplemental Figure 6B). The CHG hypermethylation changes, detected after multiple generations in Arabidopsis, occur in only one generation for tomato, suggesting that disrupting SlDDM1 activities has more immediate consequences. However, we found very few genes deregulated in the Slddm1a Slddm1b mutant and associated with hypermethylated CG or CHG-DMRs (Figure 8A) despite the presence of these hyperDMRs in numerous genes (Figure 4C). Thus, the euchromatic CG and CHG methylation gained in one generation in tomato does not significantly alter the transcription of the associated genes. By contrast, we found that almost 50% of the upregulated genes in Slddm1a Slddm1b are associated with regions hypomethylated (Figure 8A) in both the CG and CHG contexts (Figure 8B). The majority of these genes are overlapping TEs (Figure 8A; CDS+TE) that are likely derepressed. Further studies will be required to determine if TE activation within tomato genes controls gene expression.
Two parallel pathways controlling the methylation at CHH sites of TEs were discovered in Arabidopsis. The first pathway involves CMT2, depends on DDM1, and targets the long TEs predominantly found in the constitutive heterochromatin, like the Gypsy elements (Zemach et al., 2013; Stroud et al., 2014). Although CMT2 seems to be absent from maize (Zemach et al., 2013), BLAST analyses reveal the presence of one homolog in the genome of tomato (Gallusci et al., 2016). Direct evidence to confirm the function of this gene is yet to be provided, but indirect evidence exists, such as the methylation profiles of tomato RdDM mutants (Gouil and Baulcombe, 2016). Indeed, the methylation of CHH sites decreases in gene-enriched regions of both Slpol iv and Slpol v crispr mutants, toward chromosome arms, and remains unchanged in heterochromatin where mCHH most likely depends on SlCMT2 (Gouil and Baulcombe, 2016). Second, the RdDM (Matzke et al., 2015) is active at short TEs and edges of long TEs (Zemach et al., 2013; Stroud et al., 2014), is required to maintain the CHH methylation of TEs located within genic regions, relies on Pol IV-dependent 24-nucleotide siRNAs, and is independent of DDM1. In the Slddm1a Slddm1b mutant, the CHH methylation decreases in certain TEs that are found in euchromatic regions (Figures 4B, 4C, and 5; Supplemental Figure 7) and the global content of 24-nucleotide siRNA also decreases in these regions (Figure 6), indicating that the corresponding pathways are compromised in Slddm1a Slddm1b plants. At the same time, the situation is inverted for certain other heterochromatic TEs of Slddm1a Slddm1b that gain CHH methylation (Figures 4B, 4C, and 5) and become the targets of 24-nucleotide siRNAs (Figure 5). In addition, the heterochromatin of Slddm1a Slddm1b is enriched in siRNAs (Figures 6 and 7), indicating that their production is enhanced in regions densely populated by TEs. Altogether, we propose that the homeostasis of the pathways controlling the production of siRNAs and CHH methylation driven by the RdDM is severely compromised in Slddm1a Slddm1b, leading to their partial redistribution toward heterochromatin. In euchromatin, this has different consequences on long or short TEs that are independent or dependent of RdDM, respectively, and both are differently affected by the lack of DDM1 enzymes. In Slddm1a Slddm1b, the canonical RdDM targets short TEs, but much less efficiently compared with the wild type. Long TEs of euchromatin are severely hypomethylated in all contexts in the mutant, implying that SlDDM1 controls their silencing. Few TEs that were targeted by RdDM become targeted by other pathways, likely involving CMTs. In addition, we observed that the distribution of 24-nucleotide siRNAs and mCHH correlates near genes, reaching a peak at ∼500 bp upstream of the transcription start site, that decreases in the Slddm1a Slddm1b mutant (Figure 6D). Thus, RdDM seems to be particularly active at gene boundaries of wild-type tomato and disrupting the SlDDM1 genes strongly impairs this control. Whether some of these regions are similar to CHH islands found in maize (Gent et al., 2013; Li et al., 2015a) remains to be determined.
In tomato, the production of 24-nucleotide siRNAs that depends on both SlPol IV (Gouil and Baulcombe, 2016) and SlDCL3 (Kravchik et al., 2014a) is restricted to gene-enriched regions (Figures 6A and 6B) and follows the pattern previously reported for all categories of small RNAs (Tomato Genome Consortium, 2012). Likewise, a very similar distribution was observed in other Solanaceae such as pepper (Capsicum) (Qin et al., 2014) or potato (Tomato Genome Consortium, 2012). By contrast, Pol IV-dependent siRNAs of Arabidopsis are produced throughout the whole genome, including pericentromeres (Zhang et al., 2007; Mosher et al., 2008; Law et al., 2013; Liu et al., 2014). Therefore, contrary to Arabidopsis, SlPol IV is not efficiently transcribing the heterochromatic regions of tomato, or alternatively, Pol IV transcripts derived from heterochromatin are not efficiently processed, leading to the restriction of Pol IV-siRNA production to chromosome arms. Interestingly, we found that 23/24-nucleotide siRNA production, and therefore the Pol IV activity, can be greatly enhanced in the pericentromeric regions of the Slddm1a Slddm1b mutant, making tomato plants, and this particular mutant, singular model systems in which to study Pol IV recruitment target sites. We observed a similar increase for 21/22-nucleotide siRNAs (Figure 7; Supplemental Figure 10), that are likely produced by the RDR6-RdDM pathway following the transcriptional reactivation of TEs (McCue et al., 2012, 2015; Nuthikattu et al., 2013). Future studies will determine whether tomato miRNAs control the production of epigenetically activated siRNAs like in Arabidopsis (Creasey et al., 2014; Borges et al., 2018). Altogether, this indicates that the heterochromatic regions of wild-type tomato are probably much less accessible than those of Arabidopsis to enzymes involved in the production of small RNAs.
We provide further evidence that DDM1 genes are essential in plants by using the CRISPR/Cas9 technology to obtain the corresponding tomato mutants. In plants deficient for DDM1, heterochromatic TEs are depleted of the mCGs and mCHGs that normally keep them inactive. By contrast, some of them gain CHH methylation, by a counterbalancing mechanism that probably limits the number of reactivated TEs. The small RNAs, including the 24-nucleotide siRNAs, and the methylated CHH sites, are both partially redistributed from euchromatic regions toward heterochromatic regions in Slddm1a Slddm1b, suggesting that both pathways that are under a tight homeostatic control are compromised in the mutant. Additionally, this also strongly suggests that the global production of siRNAs is restricted to chromosome arms in the wild type because the compacted heterochromatin is inaccessible to the enzymes responsible for their synthesis.
METHODS
Plant Material and Growth Conditions
Tomato (Solanum lycopersicum) cv M82 plants were grown in a greenhouse, with temperatures ranging from 15 to 30°C, in 4-liter pots filled with tuff-peat mix with nutrients. For in vitro culture, seeds were surface-sterilized by 3 min treatment with 70% ethanol, followed by 20 min with 2% hypochlorite solution containing 0.1% Tween. After a thorough rinse with sterile distilled water, seeds were sown on sterile solidified medium based on MS (Murashige and Skoog) medium including vitamins (Duchefa), with pH adjusted to 5.7 using KOH. Plant agar (Duchefa) was added to a final concentration of 0.8%. Germinated seedlings were grown in a growth chamber at 22°C under a 16/8-h light/dark regime (photosynthetic photon flux density: 50–70 µmol m−2 s−1; six Osram basic T8 cool daylight lamps model L18w/765).
Generation of ddm1 Tomato Mutants Using the CRISPR/Cas9 Technology
To knockout the SlDDM1a and SlDDM1b genes by CRISPR/Cas9, sgRNAs containing 20-bp target sequences specific to the 5ʹ coding region of the corresponding genes followed by the NGG protospacer adjacent motif were designed. To facilitate mutation detection, target sequences were designed to include a restriction enzyme site (PmlI for SlDDM1a and MluCI for SlDDM1b) overlapping the predicted cut site of the Cas9 nuclease. Then, corresponding sgRNAs were amplified using specific primers (Supplemental Table 3), digested and cloned into the pRCS binary vector SalI-HindIII sites under the control of the synthetic Arabidopsis thaliana U6 promoter (Waibel and Filipowicz, 1990) alongside the plant codon-optimized version of Cas9 (Li et al., 2013) expressed under the constitutive CaMV 35S promoter.
The binary constructs pRCS:Cas9-sgRNA-SlDDM1a and pRCS:Cas9-sgRNA-SlDDM1b were transformed into tomato by cocultivation of cotyledons derived from 14-d-old seedlings using Agrobacterium tumefaciens-mediated transformation (strain GV3101) followed by regeneration on selective kanamycin-containing media as described previously (Kravchik et al., 2014b). Further validation was performed by PCR of genomic DNA with the primer pair Cas9-Fwd and Cas9-Rev to detect the 35S:Cas9 transgene.
To identify mutant plants, genomic DNA was extracted (Phire; Thermo) from each transgenic T0 plant and used for PCR with specific primers flanking the sgRNA target sequences (primer sequences are given in Supplemental Table 3). The resulting amplicons were resolved by agarose gel electrophoresis to detect large indels or digested with PmlI for SlDDM1a and MluCI for SlDDM1b and resolved by agarose gel electrophoresis to detect loss of the corresponding restriction enzyme sites. Then, the progeny of positive T0 plants were screened as above to detect Mendelian segregation of the mutation to confirm its heritability and the absence of the 35S:Cas9 transgene. The amplicons from identified nontransgenic Slddm1 homozygous mutants were sequenced to determine the nature of the mutation.
Pollen Viability and Germination Assay
Freshly harvested anthers from 16 anthesis flowers were sliced and incubated for 3 h in germination medium [10% sucrose, 100 mg/L H3BO3, 300 mg/L CA(NO3)2, 200 mg/L MgSO4, and 100 mg/L KNO3] at room temperature, followed by 1 h incubation in Alexander dye (Alexander, 1969). Pollen grains were counted from 18 arbitrarily selected microscopic fields; they were considered viable when active cytoplasm was evident and considered germinated if the tube length was equal or greater than the grain diameter.
Whole-Genome Bisulfite Sequencing and DMR Analyses
Both Slpol iv and Slpol v BS-seq (Gouil and Baulcombe, 2016) are available from the SRA database (accession number SRP081115). For Slddm1 mutants, genomic DNA was extracted using Genomic DNA extraction kit (Macherey-Nagel) from wild-type (cv M82), Slddm1a mutant (crispr-Slddm1a-5), Slddm1b mutant (crispr-Slddm1b-16), and Slddm1a Slddm1b mutant leaves (leaves 3 and 4). For the wild type and the Slddm1a and the Slddm1b single mutants, we combined the DNAs from 20 plants for each genotype. The DNAs of 12 plants were combined for Slddm1a Slddm1b. We did two biological replicates per genotype. To sequence the methylomes of Slddm1 mutants, bisulfite treatments, library preparations, and whole-genome sequencings (depth of ±19× final; Supplemental Table 1) were performed at BGI (China) using HiSeq technology (Illumina), producing 150-bp paired-end reads (Supplemental Table 1). Reads were trimmed and cleaned (short reads <20 bp removed; paired-end validation) with Trim Galore! version 0.4.2 (Babraham Bioinformatics) and Cutadapt version 1.8.3 (Martin, 2011). Clean reads were aligned to the wild-type reference genomes (version SL2.50; Tomato Genome Consortium, 2012) with Bismark version 0.14.5 (Krueger and Andrews, 2011) and standard options (Bowtie2; 1 nonbisulfite mismatch allowed per read). Reads that were not matching at unique locations were discarded. Identical pairs were collapsed using the script provided with Bismark. Subsequent analyses were done using the following R (version 3.3.3) packages: bsseq version 1.10 (Hansen et al., 2012) and DSS (dispersion shrinkage for sequencing data) version 2.14 (Wu et al., 2015) to call DMRs based on a Wald test procedure and accounting for both biological variations among replicates and sequencing depths. First, differential methylation statistical tests were performed at each C locus by calling the DSS DMLtest function with the parameter smoothing = TRUE. Then, differentially methylated loci were retained when the difference in mean methylation levels was >0.1 for CGs or CHGs and >0.07 for CHH with a posterior probability > 0.9999. DMRs were identified by using the DSS callDMR function with standard parameters (DMR length > 50 bp, number of differentially methylated loci > 3, more than 50% of C sites with P value <0.0001). DMRs closer than 50 bp were merged into longer ones. To define hypo- or hyperDMRs, we applied an additional cutoff to keep DMRs with at least a 10% change in methylation ratio for CHHs, 20% for CHGs, and 30% for CGs. We used the MethylKit software (Akalin et al., 2012) to monitor the levels of methylation per cytosine. To determine the bisulfite conversion rates, reads were aligned to the tomato chloroplast sequence (NCBI reference sequence NC_007898.3) with Bismark (Krueger and Andrews, 2011). Whole-genome bisulfite sequencing statistics are provided in Supplemental Table 1.
Annotation of TEs
Transposable elements were annotated with REPET (Flutre et al., 2011), and the repeat-rich, intermediate, and poor regions were defined as previously described (Jouffroy et al., 2016), using the SL2.50 version of the tomato genome assembly (Tomato Genome Consortium, 2012). The gff file is available on the Sol Genomics Network. Putative TE genes (2246) for which over half of the CDS (coding sequence) fraction is covered by high confidence TEs were annotated previously (Jouffroy et al., 2016).
Analysis of Expression by RNA-Seq
Libraries preparation were done using the INCPM-RNA-seq. Briefly, poly(A) fraction (mRNA) was purified from 500 ng of total RNA extracted from leaves, following by fragmentation and generation of Slddm1a Slddm1b stranded cDNAs. Then, end repair, A base addition, adapter ligation, and PCR amplification steps were performed. Libraries were evaluated by Qubit and TapeStation. Sequencing libraries were constructed with barcodes to allow multiplexing of the samples in one lane. On average, 27 million single-end 60-bp reads were sequenced per sample on Illumina HiSeq 2500 V4 instrument. Raw reads were filtered and cleaned using Trimmomatic (Bolger et al., 2014) to remove adapters and the FASTX-Toolkit version 0.0.13.2 for (1) trimming read-end nucleotides with quality scores <30 using fastq_quality_trimmer and (2) removing reads with <70% base pairs with quality score ≤30 using fastq_quality_filter. Reads were mapped with bowtie2 (Langmead and Salzberg, 2012) against the tomato coding sequences (SL2.5 release) with the parameters: --no-mixed --no-discordant --gbar 1000 --end-to-end -k 20. The transcript quantification was performed using the Expectation-Maximization method (RSEM) (Li and Dewey, 2011) based on the script align_and_estimate_abundance.pl from Trinity software (Haas et al., 2013; https://github.com/trinityrnaseq/trinityrnaseq/wiki). Genes were annotated using the annotation provided by the International Tomato Annotation Group (ITAG, release 2.40).
To quantitate the expression of TEs, we used both a unique and a multiple mapping strategy. TopHat2 (Kim et al., 2013) was used to map the clean reads onto the tomato genome reference (SL2.5 release) with the option -g 1 for unique mapping and -g 200 that allows for up to 200 reported alignments with the best alignment score, for multiple mapping. The Bedtools version 2.25 multicov program including the -D option (that includes duplicate reads) was used for calculation count table for each TE elements based on the annotations generated by REPET (Flutre et al., 2011). Differential expression analyses were done with DESeq2 version 1.16.1 (Love et al., 2014) in the R environment. To define transcripts differentially expressed, we used a significance cutoff of 0.01 and a 1.5-fold change relative to the wild type. RNA-seq statistics are listed in Supplemental Table 2.
Small RNA Analysis
35S:amiR-SlDCL3 (Kravchik et al., 2014a), Slpol iv, and Slpol v (Gouil and Baulcombe, 2016) sRNA-seq data are available from the SRA database (accession SRP032929 for 35S:amiR-SlDCL3 and SRP081115 for both Slpol iv and Slpol v). All mutants are in the same tomato genetic background (cv M82) as the Slddm1 mutants described in this study. In addition, small RNAs were all extracted from leaf samples.
Slddm1 sRNA-seq libraries were prepared from two biological replicates per genotype, using the same RNA as those used for the RNA-seq. Sequences were trimmed and filtered with Trim Galore!, and small RNA reads were mapped to the tomato genome (SL2.50 release) using ShortStack version 3.8.3 (Johnson et al., 2016) with the option --mmap f (placement guided by all mapped reads). To define siRNA clusters, we used ShortStack (distance minimum between alignments, 75 bp; minimum coverage per cluster, 0.5 rpm) with the option --mmap n (placement guided by uniquely mapping reads) to map the reads of 20 to 24 nucleotides corresponding to sRNA-seq data merged from all biological replicates (two Slddm1a Slddm1b mutants and two wild type). siRNA counts were normalized to the total number of mapped reads and analyzed with DESeq2 version 1.16.1 (Love et al., 2014). To define clusters differentially expressed, we used a significance cutoff of 0.01 and a 2-fold change relative to the wild type.
Accession Numbers
RNA-seq, BS-seq, and sRNA-seq data are available from the ENA database under the accession number PRJEB23761. 35S:amiR-SlDCL3, Slpol iv, and Slpol v sRNA-seq data are available from the SRA database (accession number SRP032929 for 35S:amiR-SlDCL3 and SRP081115 for both Slpol iv and Slpol v). Sequence data from this article can be found in the Sol Genomics Network (https://solgenomics.net/) under the following accession numbers: Solyc02g062780, SlDDM1a; Solyc02g085390, SlDDM1b; Solyc08g080210, SlPol IV; and Solyc01g096390, SlPol V. The TE gff file is available on the Sol Genomics Network.
Supplemental Data
Supplemental Figure 1. The tomato DDM1 genes.
Supplemental Figure 2. Effect of SlDDM1 knockout on pollen quality.
Supplemental Figure 3. Correlation between BS-sequencing biological replicates.
Supplemental Figure 4. Patterns of methylation in TEs of the Slddm1 mutants.
Supplemental Figure 5. DMRs identified between the Slddm1a Slddm1b mutant and the wild type.
Supplemental Figure 6. Methylation and siRNA profiles of CG and CHG hyperDMRs localized in repeat-poor regions.
Supplemental Figure 7. Methylation profiles of euchromatic TEs.
Supplemental Figure 8. Principal component analysis plots of small RNA and RNA-seq data.
Supplemental Figure 9. The 24-nucleotide siRNA patterns of Slddm1a Slddm1b mutants normalized against miRNAs.
Supplemental Figure 10. siRNA patterns of Slddm1a Slddm1b mutants along a tomato chromosome.
Supplemental Table 1. Whole-genome bisulfite sequencing statistics.
Supplemental Table 2. RNA-seq statistics.
Supplemental Table 3. Primers used in this study.
Supplemental Data Set 1. HypoDMRs between the wild type and the Slddm1a Slddm1b mutant.
Supplemental Data Set 2. HyperDMRs between the wild type and the Slddm1a Slddm1b mutant.
Supplemental Data Set 3. Regions corresponding to the compartmentalization of the tomato genome in three major regions.
Supplemental Data Set 4. Genes deregulated in the Slddm1a Slddm1b mutant.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This work was supported by a grant from the Chief Scientist of The Israel Ministry of Agriculture and Rural Development (no. 20-10-0039 to T.A.) and by project MemoCROP France-Israel (joint grant 33583WA to N.B. and T.A.). We thank Michal Liberman Lazarovich and Assaf Zemach for critical reading of the manuscript and Filipe Borges and Christine Mézard for the helpful discussions. The Institut Jean-Pierre Bourgin benefits from the support of the LabEx Saclay Plant Sciences-SPS (Project 10-LABX-0040-SPS).
AUTHOR CONTRIBUTIONS
S.C., T.A., and N.B. designed and performed the experiments and wrote the article. S.C., A.D.-F., O.J., F.M., T.A., and N.B. analyzed the data.
References
- Akalin A., Kormaksson M., Li S., Garrett-Bakelman F.E., Figueroa M.E., Melnick A., Mason C.E. (2012). methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 13: R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander M.P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44: 117–122. [DOI] [PubMed] [Google Scholar]
- Blevins T., Podicheti R., Mishra V., Marasco M., Wang J., Rusch D., Tang H., Pikaard C.S. (2015). Identification of Pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in Arabidopsis. eLife 4: e09591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F., Parent J.S., van Ex F., Wolff P., Martínez G., Köhler C., Martienssen R.A. (2018). Transposon-derived small RNAs triggered by miR845 mediate genome dosage response in Arabidopsis. Nat. Genet. 50: 186–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzeski J., Jerzmanowski A. (2003). Deficient in DNA methylation 1 (DDM1) defines a novel family of chromatin-remodeling factors. J. Biol. Chem. 278: 823–828. [DOI] [PubMed] [Google Scholar]
- Cao X., Jacobsen S.E. (2002). Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 12: 1138–1144. [DOI] [PubMed] [Google Scholar]
- Cortijo S., et al. (2014). Mapping the epigenetic basis of complex traits. Science 343: 1145–1148. [DOI] [PubMed] [Google Scholar]
- Creasey K.M., Zhai J., Borges F., Van Ex F., Regulski M., Meyers B.C., Martienssen R.A. (2014). miRNAs trigger widespread epigenetically activated siRNAs from transposons in Arabidopsis. Nature 508: 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis K., Fan T., Geiman T., Yan Q., Muegge K. (2001). Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes Dev. 15: 2940–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flutre T., Duprat E., Feuillet C., Quesneville H. (2011). Considering transposable element diversification in de novo annotation approaches. PLoS One 6: e16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallusci P., Hodgman C., Teyssier E., Seymour G.B. (2016). DNA methylation and chromatin regulation during fleshy fruit development and ripening. Front. Plant Sci. 7: 807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent J.I., Ellis N.A., Guo L., Harkess A.E., Yao Y., Zhang X., Dawe R.K. (2013). CHH islands: de novo DNA methylation in near-gene chromatin regulation in maize. Genome Res. 23: 628–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouil Q., Baulcombe D.C. (2016). DNA methylation signatures of the plant chromomethyltransferases. PLoS Genet. 12: e1006526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag J.R., Ream T.S., Marasco M., Nicora C.D., Norbeck A.D., Pasa-Tolic L., Pikaard C.S. (2012). In vitro transcription activities of Pol IV, Pol V, and RDR2 reveal coupling of Pol IV and RDR2 for dsRNA synthesis in plant RNA silencing. Mol. Cell 48: 811–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B.J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8: 1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen K.D., Langmead B., Irizarry R.A. (2012). BSmooth: from whole genome bisulfite sequencing reads to differentially methylated regions. Genome Biol. 13: R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr A.J., Jensen M.B., Dalmay T., Baulcombe D.C. (2005). RNA polymerase IV directs silencing of endogenous DNA. Science 308: 118–120. [DOI] [PubMed] [Google Scholar]
- Ito T., Tarutani Y., To T.K., Kassam M., Duvernois-Berthet E., Cortijo S., Takashima K., Saze H., Toyoda A., Fujiyama A., Colot V., Kakutani T. (2015). Genome-wide negative feedback drives transgenerational DNA methylation dynamics in Arabidopsis. PLoS Genet. 11: e1005154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y., Lisch D.R., Ohtsu K., Scanlon M.J., Nettleton D., Schnable P.S. (2009). Loss of RNA-dependent RNA polymerase 2 (RDR2) function causes widespread and unexpected changes in the expression of transposons, genes, and 24-nt small RNAs. PLoS Genet. 5: e1000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson N.R., Yeoh J.M., Coruh C., Axtell M.J. (2016). Improved placement of multi-mapping small RNAs. G3 (Bethesda) 6: 2103–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouffroy O., Saha S., Mueller L., Quesneville H., Maumus F. (2016). Comprehensive repeatome annotation reveals strong potential impact of repetitive elements on tomato ripening. BMC Genomics 17: 624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakutani T. (1997). Genetic characterization of late-flowering traits induced by DNA hypomethylation mutation in Arabidopsis thaliana. Plant J. 12: 1447–1451. [DOI] [PubMed] [Google Scholar]
- Kakutani T., Jeddeloh J.A., Richards E.J. (1995). Characterization of an Arabidopsis thaliana DNA hypomethylation mutant. Nucleic Acids Res. 23: 130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakutani T., Jeddeloh J.A., Flowers S.K., Munakata K., Richards E.J. (1996). Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc. Natl. Acad. Sci. USA 93: 12406–12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau K.D., Fahlgren N., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Carrington J.C. (2007). Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 5: e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravchik M., Damodharan S., Stav R., Arazi T. (2014a). Generation and characterization of a tomato DCL3-silencing mutant. Plant Sci. 221-222: 81–89. [DOI] [PubMed] [Google Scholar]
- Kravchik M., Sunkar R., Damodharan S., Stav R., Zohar M., Isaacson T., Arazi T. (2014b). Global and local perturbation of the tomato microRNA pathway by a trans-activated DICER-LIKE 1 mutant. J. Exp. Bot. 65: 725–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F., Andrews S.R. (2011). Bismark: a flexible aligner and methylation caller for bisulfite-seq applications. Bioinformatics 27: 1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang Z., Wang Y., Tang K., Tang D., Datsenka T., Cheng J., Zhang Y., Handa A.K., Zhu J.K. (2017). Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc. Natl. Acad. Sci. USA 114: E4511–E4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J.A., Vashisht A.A., Wohlschlegel J.A., Jacobsen S.E. (2011). SHH1, a homeodomain protein required for DNA methylation, as well as RDR2, RDM4, and chromatin remodeling factors, associate with RNA polymerase IV. PLoS Genet. 7: e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J.A., Du J., Hale C.J., Feng S., Krajewski K., Palanca A.M., Strahl B.D., Patel D.J., Jacobsen S.E. (2013). Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature 498: 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., et al. (2014). Genetic perturbation of the maize methylome. Plant Cell 26: 4602–4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., et al. (2015a). RNA-directed DNA methylation enforces boundaries between heterochromatin and euchromatin in the maize genome. Proc. Natl. Acad. Sci. USA 112: 14728–14733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C.N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.F., Norville J.E., Aach J., McCormack M., Zhang D., Bush J., Church G.M., Sheen J. (2013). Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31: 688–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Vandivier L.E., Tu B., Gao L., Won S.Y., Li S., Zheng B., Gregory B.D., Chen X. (2015b). Detection of Pol IV/RDR2-dependent transcripts at the genomic scale in Arabidopsis reveals features and regulation of siRNA biogenesis. Genome Res. 25: 235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z., et al. (2004). Role of transposable elements in heterochromatin and epigenetic control. Nature 430: 471–476. [DOI] [PubMed] [Google Scholar]
- Liu R., et al. (2015). A DEMETER-like DNA demethylase governs tomato fruit ripening. Proc. Natl. Acad. Sci. USA 112: 10804–10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Cheng Y.J., Wang J.W., Weigel D. (2017). Prominent topologically associated domains differentiate global chromatin packing in rice from Arabidopsis. Nat. Plants 3: 742–748. [DOI] [PubMed] [Google Scholar]
- Liu Z.W., Shao C.R., Zhang C.J., Zhou J.X., Zhang S.W., Li L., Chen S., Huang H.W., Cai T., He X.J. (2014). The SET domain proteins SUVH2 and SUVH9 are required for Pol V occupancy at RNA-directed DNA methylation loci. PLoS Genet. 10: e1003948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons D.B., Zilberman D. (2017). DDM1 and Lsh remodelers allow methylation of DNA wrapped in nucleosomes. eLife 6: e30674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10–12. [Google Scholar]
- Matzke M.A., Kanno T., Matzke A.J. (2015). RNA-directed DNA methylation: the evolution of a complex epigenetic pathway in flowering plants. Annu. Rev. Plant Biol. 66: 243–267. [DOI] [PubMed] [Google Scholar]
- McCue A.D., Nuthikattu S., Reeder S.H., Slotkin R.K. (2012). Gene expression and stress response mediated by the epigenetic regulation of a transposable element small RNA. PLoS Genet. 8: e1002474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCue A.D., Panda K., Nuthikattu S., Choudury S.G., Thomas E.N., Slotkin R.K. (2015). ARGONAUTE 6 bridges transposable element mRNA-derived siRNAs to the establishment of DNA methylation. EMBO J. 34: 20–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura A., Yonebayashi S., Watanabe K., Toyama T., Shimada H., Kakutani T. (2001). Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 411: 212–214. [DOI] [PubMed] [Google Scholar]
- Mosher R.A., Schwach F., Studholme D., Baulcombe D.C. (2008). PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc. Natl. Acad. Sci. USA 105: 3145–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuthikattu S., McCue A.D., Panda K., Fultz D., DeFraia C., Thomas E.N., Slotkin R.K. (2013). The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21-22 nucleotide small interfering RNAs. Plant Physiol. 162: 116–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y., Haag J.R., Ream T., Costa Nunes P., Pontes O., Pikaard C.S. (2005). Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120: 613–622. [DOI] [PubMed] [Google Scholar]
- Panda K., Ji L., Neumann D.A., Daron J., Schmitz R.J., Slotkin R.K. (2016). Full-length autonomous transposable elements are preferentially targeted by expression-dependent forms of RNA-directed DNA methylation. Genome Biol. 17: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C., et al. (2014). Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. USA 111: 5135–5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze H., Kakutani T. (2007). Heritable epigenetic mutation of a transposon-flanked Arabidopsis gene due to lack of the chromatin-remodeling factor DDM1. EMBO J. 26: 3641–3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer T., Yordan C., Martienssen R.A. (2001). Robertson’s mutator transposons in A. thaliana are regulated by the chromatin-remodeling gene Decrease in DNA Methylation (DDM1). Genes Dev. 15: 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H., Greenberg M.V., Feng S., Bernatavichute Y.V., Jacobsen S.E. (2013). Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152: 352–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H., Do T., Du J., Zhong X., Feng S., Johnson L., Patel D.J., Jacobsen S.E. (2014). Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 21: 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan F., Zhou C., Zhou Q., Zhou S., Yang W., Zhao Y., Li G., Zhou D.X. (2016). Analysis of chromatin regulators reveals specific features of rice DNA methylation pathways. Plant Physiol. 171: 2041–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukahara S., Kobayashi A., Kawabe A., Mathieu O., Miura A., Kakutani T. (2009). Bursts of retrotransposition reproduced in Arabidopsis. Nature 461: 423–426. [DOI] [PubMed] [Google Scholar]
- Vongs A., Kakutani T., Martienssen R.A., Richards E.J. (1993). Arabidopsis thaliana DNA methylation mutants. Science 260: 1926–1928. [DOI] [PubMed] [Google Scholar]
- Waibel F., Filipowicz W. (1990). U6 snRNA genes of Arabidopsis are transcribed by RNA polymerase III but contain the same two upstream promoter elements as RNA polymerase II-transcribed U-snRNA genes. Nucleic Acids Res. 18: 3451–3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki A.T., Haag J.R., Pikaard C.S. (2008). Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135: 635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki A.T., Ream T.S., Haag J.R., Pikaard C.S. (2009). RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat. Genet. 41: 630–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Xu T., Feng H., Chen L., Li B., Yao B., Qin Z., Jin P., Conneely K.N. (2015). Detection of differentially methylated regions from whole-genome bisulfite sequencing data without replicates. Nucleic Acids Res. 43: e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A., Kim M.Y., Hsieh P.H., Coleman-Derr D., Eshed-Williams L., Thao K., Harmer S.L., Zilberman D. (2013). The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153: 193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J., et al. (2015). A one precursor one siRNA model for Pol IV-dependent siRNA biogenesis. Cell 163: 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Henderson I.R., Lu C., Green P.J., Jacobsen S.E. (2007). Role of RNA polymerase IV in plant small RNA metabolism. Proc. Natl. Acad. Sci. USA 104: 4536–4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Fei Z., Chen Y.R., Zheng Y., Huang M., Vrebalov J., McQuinn R., Gapper N., Liu B., Xiang J., Shao Y., Giovannoni J.J. (2013). Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 31: 154–159. [DOI] [PubMed] [Google Scholar]
- Zhong X., Du J., Hale C.J., Gallego-Bartolome J., Feng S., Vashisht A.A., Chory J., Wohlschlegel J.A., Patel D.J., Jacobsen S.E. (2014). Molecular mechanism of action of plant DRM de novo DNA methyltransferases. Cell 157: 1050–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]