AUTOPHAGY7, a key autophagy protein that regulates the autophagic process, is a target of pathogen effectors that functions in the ongoing arms race of plant defense and viral counterdefense.
Abstract
Autophagy is a conserved defense strategy against viral infection. However, little is known about the counterdefense strategies of plant viruses involving interference with autophagy. Here, we show that γb protein from Barley stripe mosaic virus (BSMV), a positive single-stranded RNA virus, directly interacts with AUTOPHAGY PROTEIN7 (ATG7). BSMV infection suppresses autophagy, and overexpression of γb protein is sufficient to inhibit autophagy. Furthermore, silencing of autophagy-related gene ATG5 and ATG7 in Nicotiana benthamiana plants enhanced BSMV accumulation and viral symptoms, indicating that autophagy plays an antiviral role in BSMV infection. Molecular analyses indicated that γb interferes with the interaction of ATG7 with ATG8 in a competitive manner, whereas a single point mutation in γb, Tyr29Ala (Y29A), made this protein deficient in the interaction with ATG7, which was correlated with the abolishment of autophagy inhibition. Consistently, the mutant BSMVY29A virus showed reduced symptom severity and viral accumulation. Taken together, our findings reveal that BSMV γb protein subverts autophagy-mediated antiviral defense by disrupting the ATG7-ATG8 interaction to promote plant RNA virus infection, and they provide evidence that ATG7 is a target of pathogen effectors that functions in the ongoing arms race of plant defense and viral counterdefense.
INTRODUCTION
Autophagy is a conserved eukaryotic mechanism that removes damaged or unwanted cellular materials. This process is initiated in the cytoplasm with the formation of double-membraned autophagosomes, followed by the delivery of the cargo to lysosomes or the vacuole for degradation (Xie and Klionsky, 2007). The formation of autophagosomes relies on diverse membrane factors and is mediated by a set of AUTOPHAGY (ATG) proteins (Zhuang et al., 2017). Among these, ATG7 is a key autophagy protein that regulates the autophagic process. In animals, overexpression of ATG7 is sufficient to induce autophagy (Pattison et al., 2011). ATG7 is required for the activity of dual ubiquitin-like conjugation pathways, including the conjugation of ATG12 to ATG5 and ATG8 to the autophagosome membrane (Xie and Klionsky, 2007; Taherbhoy et al., 2011). The N-terminal domain of ATG7 interacts with both the autophagic E2-like enzymes ATG3 and ATG10, whereas the C terminus of ATG7 interacts with ATG8 (Hong et al., 2011).
Autophagy is induced by various abiotic and biotic stresses including pathogen infection (Hayward and Dinesh-Kumar, 2011). Autophagy plays a role in various aspects of immunity against pathogens including viruses (Dagdas et al., 2016; Clavel et al., 2017; Hafrén et al., 2017, 2018; Haxim et al., 2017). Many animal viruses are known to evade or hijack the autophagy pathway to promote their own infection or transmission (Dong and Levine, 2013; Chen et al., 2017). In plants, autophagy regulates immunity-related programmed cell death (Liu et al., 2005; Hofius et al., 2009; Yoshimoto et al., 2009; Clavel et al., 2017). The influence of autophagy on cell death or cell survival may be required to balance plant responses such as reactive oxygen species bursts, salicylic acid signaling, endoplasmic reticulum stress, and the accumulation of ubiquitinated proteins during pathogen infection (Jheng et al., 2014; Munch et al., 2014). Although the role of autophagy in defense against bacterial and viral pathogens in animals has been intensively studied, how autophagy functions in plant-virus interaction remains to be fully understood. In plants, Cucumber mosaic virus 2b and potyviruses HC-Pro are thought to be targeted for autophagic degradation via the calmodulin-like protein rgs-CaM (Nakahara et al., 2012; Jeon et al., 2017). Tomato yellow leaf curl virus proteins are also thought to be partially degraded by autophagy (Gorovits et al., 2016). In addition, some plant viral proteins mediate autophagic degradation of the components of the cellular RNAi-based antiviral defense machinery (Derrien et al., 2012; Cheng and Wang, 2016; Li et al., 2017). Autophagy was recently shown to play an antiviral role during plant infection of some DNA viruses and Turnip mosaic virus (TuMV) (Hafrén et al., 2017, 2018; Haxim et al., 2017). However, the roles of autophagy in compatible plant-virus interactions are largely unknown.
Autophagy targets the geminiviral βC1 protein through its interaction with ATG8 (Haxim et al., 2017). NBR1-mediated selective autophagy limits Cauliflower mosaic virus infection by targeting viral capsid proteins and/or particles (Hafrén et al., 2017) and TuMV infection by targeting the viral RNA silencing suppressor HC-Pro (Hafrén et al., 2018). Furthermore, TuMV VPg and 6K2 proteins block NBR1 and HC-Pro degradation (Hafrén et al., 2018). These findings suggest that autophagy might function in plant responses to viral infection. However, whether and how plant viruses evade or interfere with autophagy are still unknown.
In this study, we show that autophagy plays an antiviral role in plant responses to the RNA virus Barley stripe mosaic virus (BSMV). BSMV is a positive-strand RNA virus consisting of three RNA segments (α, β, and γ) encoding seven major proteins (Jackson et al., 2009). RNAα and RNAγ encode replicase protein αa and γa, respectively. RNAβ encodes coat protein (CP) and triple gene block (TGB) proteins. RNAγ encodes γb protein, which functions as a viral suppressor of RNA silencing (VSR), a helicase enhancer, and a modulator of host defenses (Jackson et al., 2009; Zhang et al., 2017). Our findings indicate that BSMV γb protein subverts autophagy-mediated antiviral defense responses by disrupting the ATG7-ATG8 interaction to facilitate BSMV infection. In addition, we demonstrate that autophagy plays an antiviral role during BSMV infection.
RESULTS
BSMV γb Interacts with ATG7 in Vivo and in Vitro
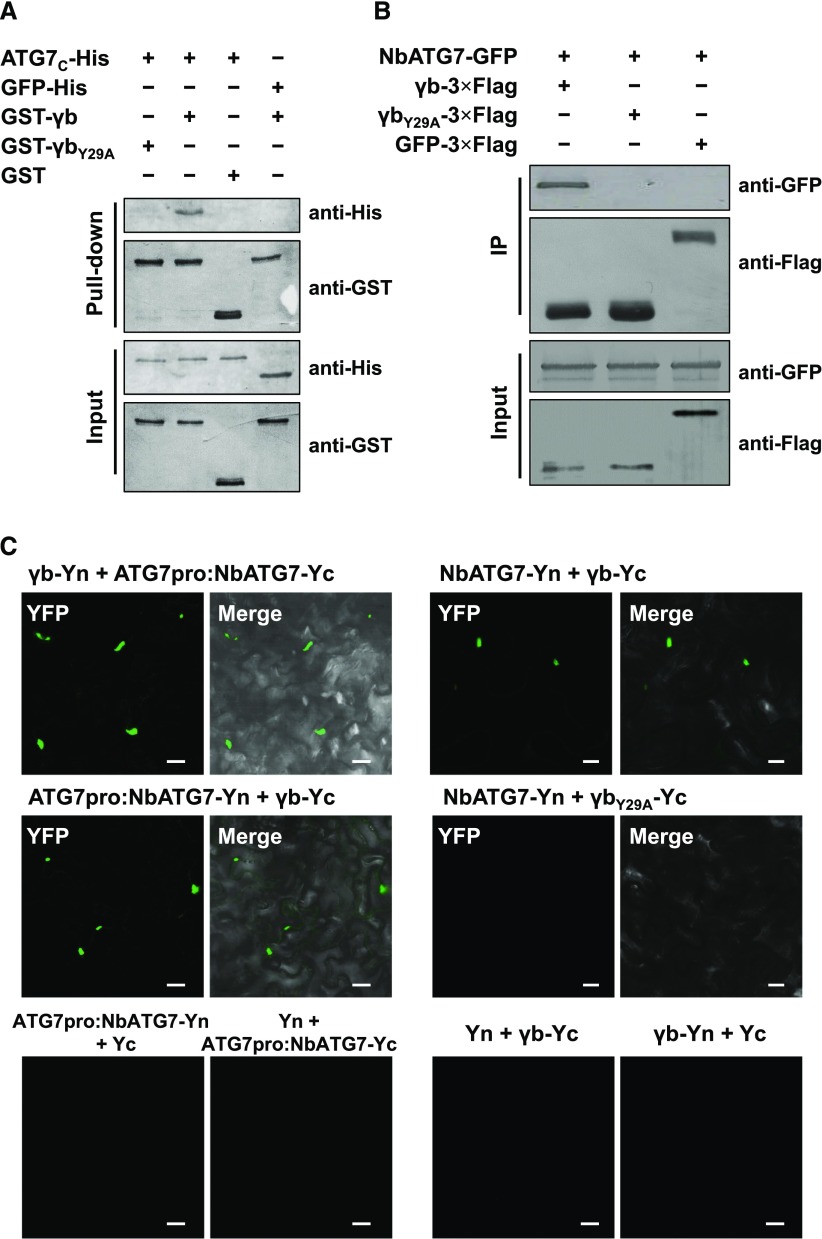
BSMV γb plays multiple roles in viral infection (Jackson et al., 2009; Zhang et al., 2017). To identify the host factors that interact with γb protein, we used γb as a bait to screen a wheat (Triticum aestivum) yeast cDNA library. The C-terminal region of wheat ATG7 (TaATG7) was identified from the yeast two-hybrid screening. To further confirm the interaction between ATG7 and γb protein, we fused the full-length sequence of TaATG7 to the activation domain and tested its interaction with γb fused to the LexA DNA binding domain. Yeast two-hybrid analysis confirmed that full-length TaATG7 interacted with γb (Supplemental Figure 1A).
Since BSMV infects Nicotiana benthamiana, an excellent model plant to study plant-virus interaction (Goodin et al., 2008), we cloned and expressed NbATG7 and tested its interaction with γb. To examine whether NbATG7 directly interacts with γb, we performed a GST pull-down assay. Since full-length NbATG7 is unstable, the C-terminal fragment of NbATG7 (ATG7C, 474–712 amino acids), which was identified as the interaction region of NbATG7 with γb protein (Supplemental Figure 2), was fused with a 6×His tag (ATG7C-His) and purified from Escherichia coli. The GST pull-down assay showed that GST-γb directly interacted with ATG7C-His in vitro, but not with GST alone (control) (Figure 1A). We further mapped the region within γb that is responsible for its interaction with NbATG7 and found that the 19- to 47-amino acid region of γb is sufficient for its interaction with NbATG7 (Supplemental Figure 3). Since the Y62A mutation in ATG8 abolishes the formation of ATG7-ATG8 conjugation in yeast (Saccharomyces cerevisiae) (Hong et al., 2011), and a similar motif exists in γb (amino acids 28–41) and ATG8s (amino acids 61–74) (Supplemental Figure 4), we generated a Y29A mutant form of γb. Interestingly, γbY29A failed to pull down ATG7C-His (Figure 1A). These results indicate that BSMV γb directly interacts with NbATG7 in vitro.
Figure 1.
BSMV γb Directly Interacts with ATG7 in Vivo and in Vitro.
(A) GST pull-down assay showing the interaction between ATG7 and γb in vitro. Purified GST-γb, GST-γbY29A, or GST was incubated with ATG7C-His. After being immunoprecipitated with glutathione-Sepharose beads, the proteins were detected by protein gel blot analysis with anti-His or anti-GST antibodies.
(B) Co-IP analysis of the interaction between γb and ATG7. N. benthamiana leaf tissues agroinfiltrated with various constructs were harvested at 48 hpi. Total proteins were immunoprecipitated with anti-Flag beads. Input and immunoprecipitated protein (IP) were analyzed by protein gel blot analysis with anti-GFP and anti-Flag antibodies.
(C) BiFC analysis of the interaction between ATG7 and γb. ATG7-Yc or ATG7-Yn under the control of the ATG7 native promoter (ATG7pro, left panel) was coexpressed with γb-Yn, γb-Yc, Yn, or Yc. ATG7 under the control of the 35S promoter (right panel) was coexpressed with γb-Yc, or γbY29A-Yc. YFP signals were visualized by confocal microscopy. Bars = 20 μm.
To determine if γb interacts with ATG7 in vivo, we performed a coimmunoprecipitation (co-IP) assay. Flag-tagged γb protein (γb-3×Flag) was transiently coexpressed with GFP-tagged NbATG7 (NbATG7-GFP) or with GFP alone in N. benthamiana by agroinfiltration. Total protein extracts were precipitated by anti-Flag affinity beads at 48 h postinfiltration (hpi) and analyzed by protein gel blot analysis. Co-IP analysis showed that NbATG7-GFP coprecipitated with γb-3×Flag but not with γbY29A-3×Flag or with GFP-3×Flag (Figure 1B).
To further confirm the interaction between γb and NbATG7, we performed a biomolecular fluorescence complementation (BiFC) assay (Walter et al., 2004). To avoid the effects of 35S promoter-driven gene overexpression, NbATG7 fused to C/N terminus of YFP was expressed under the control of its native promoter (ATG7pro:NbATG7-Yc/Yn). Coexpression of ATG7pro:NbATG7-Yc with γb fused to the N terminus of YFP (γb-Yn) or ATG7pro:NbATG7-Yn with γb-Yc resulted in the reconstitution of YFP signals (Figure 1C, top and middle left panels). However, leaves expressing NbATG7-Yn and γbY29A-Yc or other control combinations failed to show YFP signals (Figure 1C, middle right panel and bottom panel), although all BiFC constructs were successfully expressed (Supplemental Figures 1B and 1C).
To further investigate the relationship between ATG7 and γb, we performed a colocalization assay. Agrobacterium tumefaciens cells expressing ATG7-GFP and γb-mCherry or the negative controls (ATG7-GFP/mCherry and GFP/γb-mCherry) were coinfiltrated into N. benthamiana leaves, respectively, followed by confocal microscope observation of their localizations at 3 d postinfiltration (dpi). Many aggregates were observed in plant cells coexpressing ATG7-GFP and γb-mCherry, while puncta were rarely observed in cells expressing the negative controls (Supplemental Figure 5A). Next, we looked for the colocalization of γb-mCherry and CFP-ATG8f. However, no aggregates were observed, and both fusion proteins kept their original localizations (Supplemental Figure 5B).
Together, our results indicate that BSMV γb interacts with ATG7 in vitro and in vivo. Furthermore, the Y29 amino acid of γb N-terminal region crucial for its interaction with ATG7.
BSMV Infection Suppresses Autophagy
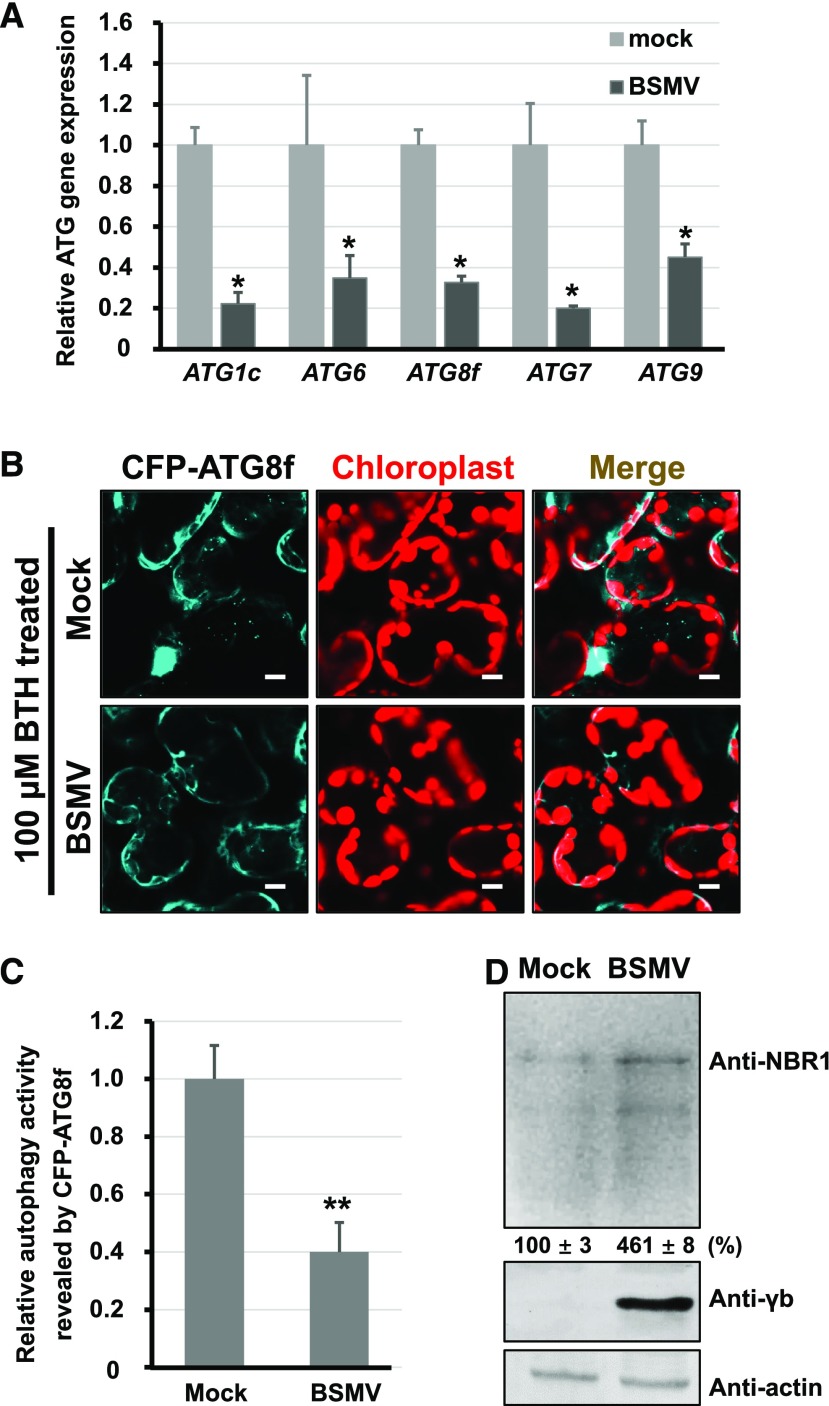
Since BSMV γb interacted with ATG7, we investigated whether BSMV infection influenced autophagy. qRT-PCR indicated that mRNA levels of NbATG1c, NbATG6, NbATG8f, NbATG7, and NbATG9 were reduced at 3 dpi in N. benthamiana leaf tissues containing Agrobacterium strains harboring the pCaBS-α, -β, and -γ plasmids (Figure 2A). To determine the effect of BSMV infection on autophagy, we used CFP-tagged NbATG8f (CFP-ATG8f); fluorescent protein fused to the N terminus of ATG8 is a commonly used autophagosome marker (Han et al., 2015). We found no obvious difference in autophagic activity between the plants infected by BSMV and the control (Supplemental Figure 6), which may be due to low levels of basal autophagic activity. To easily observe autophagic structures, we used autophagy activator benzo-(1,2,3)-thiadiaole-7-carbothioic acid (BTH) (Yoshimoto et al., 2009) to stimulate autophagosome formation in leaf tissues. The number of autophagosomes in BSMV-infected plants was approximate 2.5 times lower than that of empty vector (EV)-infiltrated plants (Figures 2B and 2C). Furthermore, the expression level of NBR1 protein, a selective autophagy substrate that is negatively related to autophagic activity (Kirkin et al., 2009; Klionsky et al., 2016; Hafrén et al., 2017; Haxim et al., 2017; Xu et al., 2017), was 3 times higher in BSMV-infected plants compared with control plants (Figure 2D). These results suggest that BSMV infection downregulates autophagy in N. benthamiana plants.
Figure 2.
BSMV Infection Inhibits Autophagy.
(A) qRT-PCR analysis of the transcript levels of ATG1c, ATG6, ATG8f, ATG7, and ATG9 in mock- and BSMV-infected N. benthamiana. The housekeeping gene PP2A served as the internal control. The data are presented as the relative ratio of gene expression compared with the control (mock), which was set to 1.0. Error bars represent se of the mean (n = 3). The asterisks indicate significant difference by Student’s t test (*P < 0.05) compared with the control.
(B) Relative autophagic activity revealed by CFP-ATG8f in mock- or BSMV-infected leaves treated with 100 μM BTH. Numerous CFP-ATG8f-labeled positive autophagic bodies in vacuoles were observed in mock-inoculated plants, but not in BSMV-infected leaves. Autofluorescence of chloroplasts is pseudocolored in red. Bars = 10 μm.
(C) Relative autophagy activity revealed by the autophagy marker CFP-ATG8f in BSMV-infected leaves was normalized to that of mock-inoculated leaves. Quantification of CFP-ATG8f-labeled autophagic puncta per cell was performed by counting the autophagic bodies. More than 200 cells per treatment were used for quantification. Error bars indicate se from three individual experiments. The asterisks indicate significant differences by Student’s t test (**P < 0.01) compared with mock inoculation.
(D) Protein gel blot analysis of autophagy flux probed with anti-NBR1 antibodies in mock- and BSMV-infected N. benthamiana leaves. Error bars indicate sd of the mean.
BSMV γb Protein Inhibits Autophagy
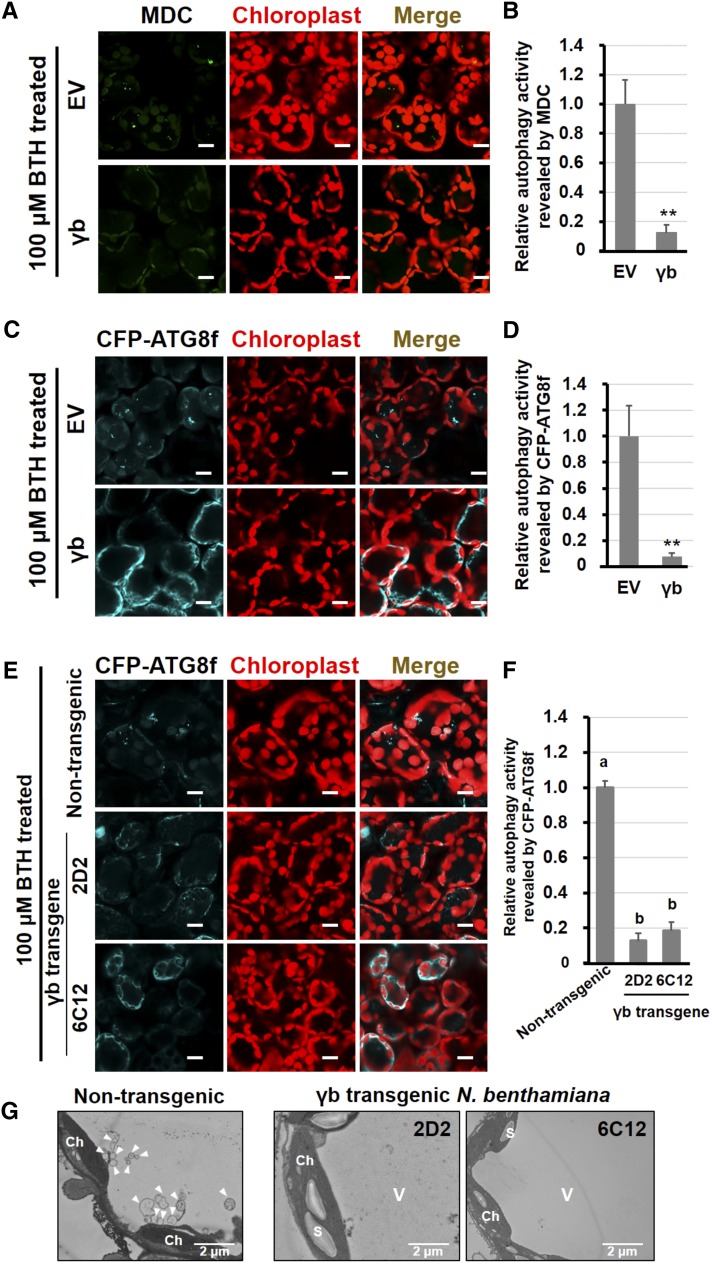
To investigate whether γb plays a direct role in suppressing autophagy, we sprayed 100 μM BTH onto γb- and EV control-infiltrated N. benthamiana leaves and autophagy was monitored by monodansylcadaverine (MDC) staining (Wang et al., 2013) and CFP-ATG8f. We observed 10 times lower levels of autophagy activity in γb-expressing leaves compared with the EV control by MDC staining (Figures 3A and 3B). Similarly, the relative autophagy level (monitored by CFP-ATG8f fluorescence) in γb-expressing leaf tissues was also ∼12 times lower compared with the EV control (Figures 3C and 3D).
Figure 3.
BSMV γb Protein Suppresses Autophagy.
(A) and (C) Representative images of autophagic activity revealed by MDC-staining (A) and the autophagy marker CFP-ATG8f (C) in cells transiently expressing γb (lower panel) or agroinfiltrated with EV (upper panel) after 100 μM BTH treatment. CFP-ATG8f-labeled autophagic bodies and autophagosomes are shown in cyan, and MDC-stained structures and autofluorescence from chloroplasts are pseudocolored in green and red, respectively. Bars = 20 μm.
(B) and (D) Quantification of the autophagy activity shown in (A) and (C) in cells. The autophagy activity in EV-agroinfiltrated leaves was set to 1.0. More than 200 mesophyll cells were analyzed per treatment. Error bars indicate se from three independent experiments. Statistical significance was determined by Student’s t test (**P < 0.01).
(E) Confocal analysis of autophagic activity revealed by the autophagic marker CFP-ATG8f in γb-transgenic (lower panel) and nontransgenic N. benthamiana (upper panel) after 100 μM BTH treatment. Autophagosomes and autophagic bodies are shown by CFP fluorescence, and chloroplasts are shown in red. Bars = 20 μm.
(F) Quantification of the autophagy activity shown in (E). The autophagy activity in nontransgenic plants was set to 1.0. More than 200 mesophyll cells were analyzed per treatment. Error bars indicate se. Statistical significance was determined by Student’s t test (P < 0.05).
(G) TEM analysis of γb or nontransgenic N. benthamiana after 100 μM BTH treatment.
To further confirm these results, we monitored the autophagy activity in γb-expressing transgenic N. benthamiana plants (Zhang et al., 2017; Yang et al., 2018). We treated γb and nontransgenic N. benthamiana plants with BTH and used CFP-ATG8f to observe the autophagosomes. Approximately 8 times fewer autophagosomes were observed in γb transgenic plants compared with nontransgenic plants (Figures 3E and 3F). A similar level of reduction was observed using transmission electron microscopy (TEM) (Figure 3G). Since autophagy contributes to the degradation of starch (Wang et al., 2013), we measured starch accumulation in γb transgenic N. benthamiana leaves by iodine staining. Leaf starch accumulated at the end of the day but not at the end of the night in nontransgenic plants (Supplemental Figure 7A, left panel). However, leaf starch levels remained higher at all times in γb transgenic plants, as revealed by iodine staining (Supplemental Figure 7A, middle and right panels). Furthermore, we quantified starch content in nontransgenic and γb transgenic plant leaves. At both the end of the day and at night, the quality of starch was always higher in γb transgenic plants compared with nontransgenic plants (Supplemental Figure 7B).
Overexpression of ATG7 activates autophagy in animal T cells (Wei et al., 2016). Similarly, we found that the overexpression of NbATG7 also induced autophagy (Figures 4A and 4C, row 2) compared with the control (Figures 4A and 4C, row 1) in plants. NbATG7-overexpressing leaf tissues accumulated significantly higher numbers of autophagic bodies compared with the EV control (Figures 4B and 4D). The number of NbATG7-induced autophagic bodies was significantly reduced in plants coexpressing NbATG7 and γb (Figures 4A and 4C, row 3, 4B, and 4D). By contrast, γbY29A, which failed to interact with ATG7, had no effect on ATG7-induced autophagy (Figures 4A and 4C, row 4). The constructs used for confocal observation were all successfully expressed, as revealed by protein gel blot analysis (Supplemental Figure 8). Similar results were obtained by TEM analysis: ATG7-induced autophagy was inhibited by γb, and the relative autophagy activity in ATG7-overexpressing plants was ∼2.5 times higher than that in plants coexpressing ATG7 and γb (Supplemental Figure 9).
Figure 4.
BSMV γb Inhibits ATG7 Overexpression-Induced Autophagy.
(A) and (C) Representative confocal images of autophagic activity revealed by MDC-staining (A) and the autophagy marker CFP-ATG8f (C) in cells coexpressing ATG7 with γb or γbY29A. EV-agroinfiltrated leaves served as the negative control. MDC-stained autophagic bodies are pseudocolored in green, autophagosomes indicated by CFP-ATG8f are shown in cyan, and the autofluorescence of chloroplasts is shown in red. Bars = 10 μm.
(B) and (D) Quantification of the autophagy activity shown in (A) and (C). The autophagy activities in cells expressing ATG7 alone, coexpressing ATG7 and γb, or ATG7 and γbY29A were normalized to that of the EV control, which was set to 1.0. More than 200 mesophyll cells were analyzed per treatment. Error bars indicate se. a, b, and c indicate statistically significant differences among different groups (ANOVA, P < 0.05).
Together, these results indicate that BSMV γb plays a key role in antagonizing plant autophagy during BSMV infection.
Suppression of Autophagy by γb Protein Is Functionally Distinct from Its VSR Activity
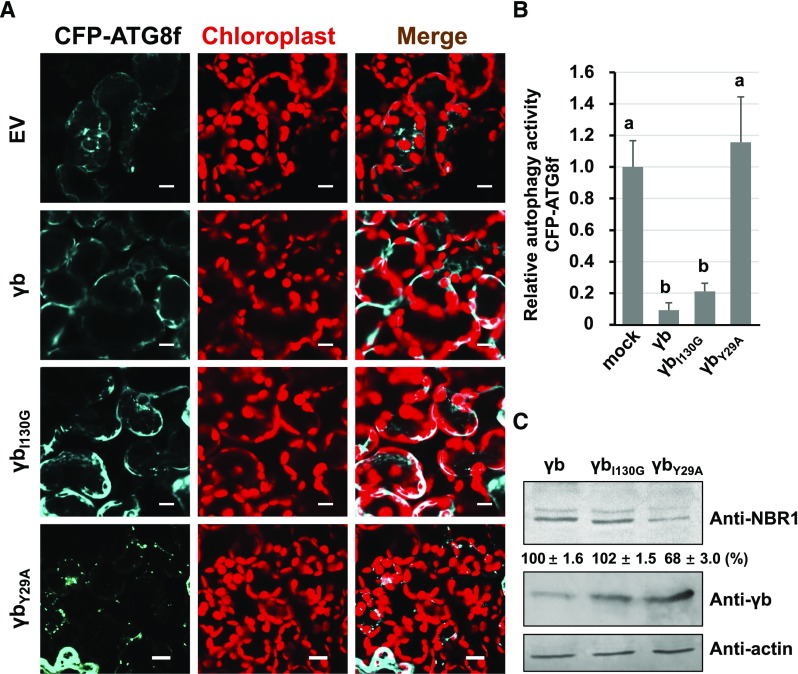
To investigate whether the VSR activity of γb is associated with its autophagy-inhibiting function, we investigated VSR activity in two γb mutant proteins, γbI130G (Zhang et al., 2017) and γbY29A, by coexpressing them with GFP (Bragg and Jackson, 2004; Zhang et al., 2017). Leaf tissues expressing both γb mutant proteins showed much less GFP fluorescence than that expressing γb, indicating that VSR is deficient in these two mutant proteins (Supplemental Figure 10). It should be noted that γbI130G still interacts with ATG7 (Supplemental Figure 11). However, assays of autophagy activity using the autophagic marker CFP-ATG8f showed that either γb or γbI130G, but not γbY29A (which is unable to interact with ATG7), exhibited suppressed autophagy (Figures 5A and 5B). Protein gel blot assays showed that the levels of the autophagy flux indicator NBR1 were ∼2 times higher in γb- or γbI130G-expressing plants than in γbY29A-expressing plants (Figure 5C). These results indicate that the VSR activity of γb is not involved in its autophagy-suppressing function.
Figure 5.
Suppression of Autophagy by γb Is Functionally Distinct from Its VSR Activity.
(A) Representative confocal images of autophagic activity revealed by autophagy marker CFP-ATG8f in N. benthamiana leaves agroinfiltrated with γb, γbI130G, γbY29A, or EV. The plants were observed after 60-h expression followed by 100 μM BTH treatment. Autophagosomes indicated by CFP-ATG8f are shown in cyan, and chloroplasts are shown in red. Bars = 20 μm.
(B) Quantification of the autophagy activity shown in (A). The autophagy activity of γb, γbI130G, and γbY29A was normalized to that of the EV control, which was set to 1. More than 200 mesophyll cells were analyzed per treatment. Error bars indicate se. a and b indicate statistically significant differences among different groups (ANOVA, P < 0.05).
(C) Protein gel blot analysis of autophagy flux probed with anti-NBR1 antibodies in γb, γbI130G, and γbY29A-expressing plant tissues.
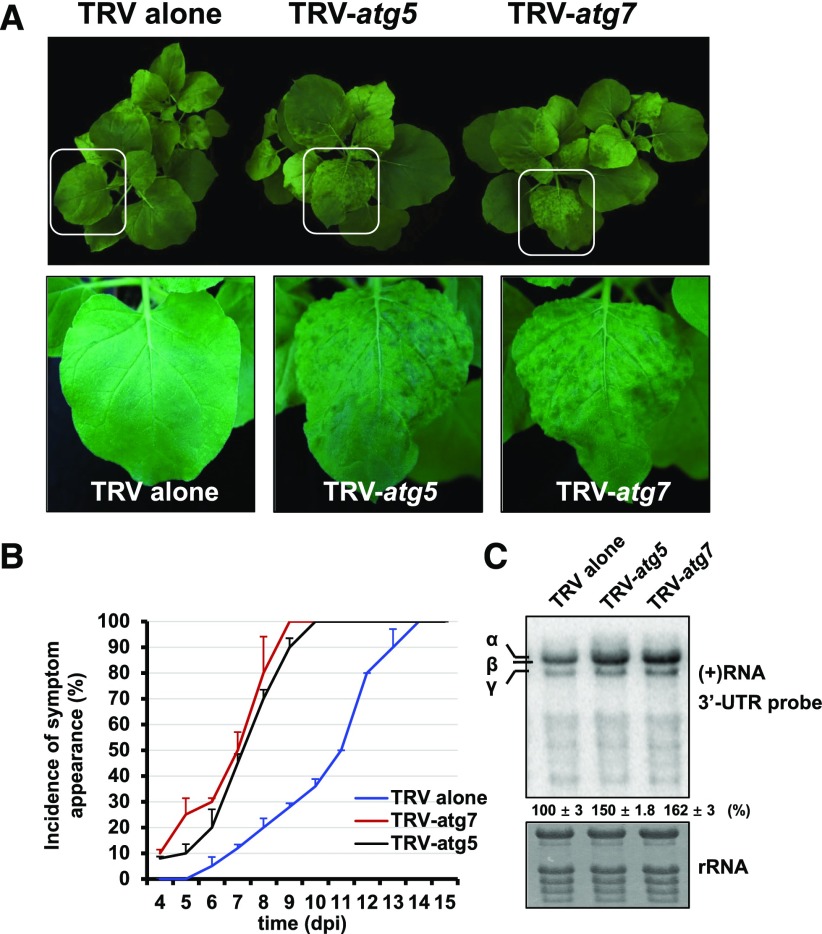
Autophagy Plays an Antiviral Role during BSMV RNA Virus Infection
Autophagy was recently shown to play an important role in defense against plant DNA viruses (Hafrén et al., 2017; Haxim et al., 2017), and NBR1-dependent selective autophagy also plays an antiviral role in TuMV infection (Hafrén et al., 2018). To determine whether autophagy alters BSMV infection, NbATG7 and NbATG5 were silenced by TRV-induced gene silencing, which was previously used to knockdown autophagy (Liu et al., 2002; Wang et al., 2013; Haxim et al., 2017). In NbATG7- and NbATG5-silenced N. benthamiana plants, BSMV spread more rapidly and the plants exhibited enhanced viral symptoms compared with nonsilenced plants (Figures 6A and 6B). Furthermore, BSMV accumulated to 1.5 times higher levels in NbATG7- and NbATG5-silenced plants versus the control (Figure 6C). To confirm the finding that autophagy plays an antiviral role during BSMV infection, we transiently expressed ATG7-GFP or the GFP control into N. benthamiana leaf tissues and coinfiltrated BSMV RNA into the same leaf tissues 24 h later. ATG7-GFP fusion proteins retained the ability to induce autophagy (Supplemental Figure 12A), suggesting that ATG7-GFP was functional. The samples were harvested and examined by protein gel blot analysis at 60 hpi (Supplemental Figure 13). We found that BSMV infection was inhibited in ATG7-expressing plants compared with the GFP control. The protein levels of viral CP in ATG7-overexpressing plants were 5 times lower than that in the control. These results indicate that autophagy functions as an antiviral defense response against BSMV RNA virus.
Figure 6.
Autophagy Plays an Antiviral Role during BSMV Infection.
(A) The symptoms of BSMV in non-silenced control, ATG7-silenced, or ATG5-silenced N. benthamiana via TRV-induced gene silencing. Magnified images of leaves (in white boxes) are shown in the lower panels.
(B) Statistical analysis of symptom appearance after inoculation with BSMV. Error bars indicate se of the mean (n = 3).
(C) RNA gel blot analysis of BSMV RNA accumulation in the nonsilenced control, ATG7-silenced, and ATG5-silenced N. benthamiana. Error bars indicate sd of the mean.
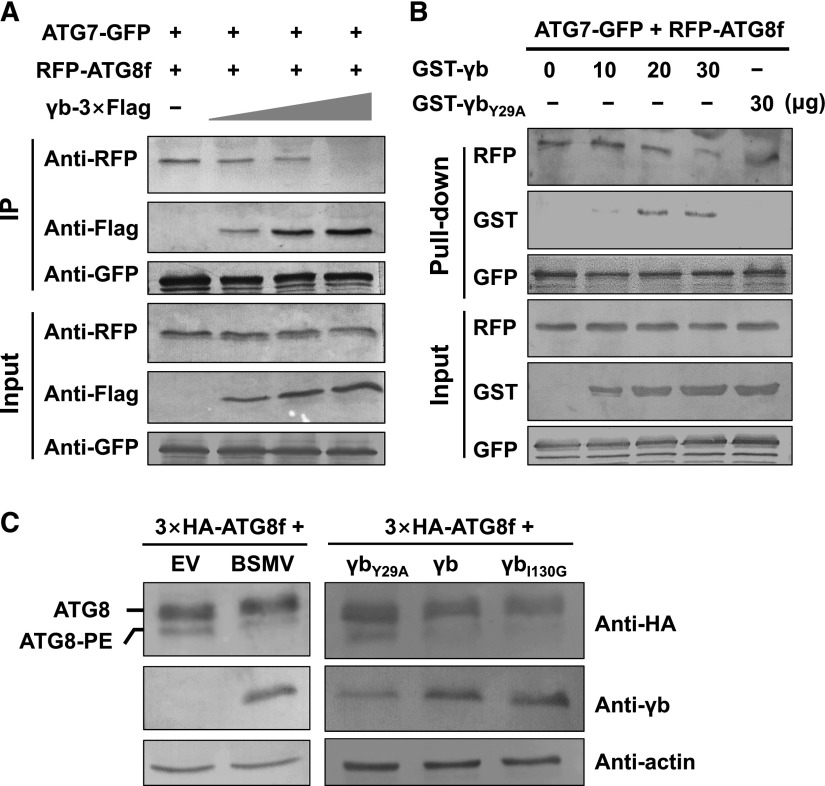
BSMV γb Interferes with the Interaction between ATG7 and ATG8 to Promote Viral Infection
To understand the molecular basis by which γb suppresses autophagy to promote viral infection, we investigated whether γb competitively interferes with the interaction of ATG7 with ATG8. This idea is based on the premise that γb interacts with the C terminus of ATG7 directly (Figure 1A; Supplemental Figure 2) and that the same region is also required for the interaction of ATG7 with ATG8 (Hong et al., 2011). We performed competitive co-IP assays among ATG7-GFP, RFP-ATG8f, and BSMV γb-3×Flag. ATG7-GFP and γb-3×Flag fusions retained the ability to induce or inhibit autophagy (Supplemental Figures 12A and 12 B), suggesting that they were functional. The co-IP samples were harvested after 60 h of expression, total proteins were incubated with anti-GFP antibody and Protein G agarose, and the elutes were examined by protein gel blot analysis. The results showed that the association between RFP-ATG8f and ATG7-GFP decreased with increasing concentration of γb (Figure 7A).
Figure 7.
γb Interferes with the Interaction between ATG7 and ATG8.
(A) Co-IP assays showing the effect of γb on the interaction between ATG7 and ATG8. ATG7-GFP and RFP-ATG8f with increasing concentrations of γb-3×Flag were coinfiltrated into N. benthamiana leaves. After 60 hpi, total proteins were extracted and incubated with anti-GFP agarose. Input and IP proteins were analyzed by protein gel blot analysis with anti-GFP, RFP, or Flag antibodies.
(B) Competitive GST pull-down assays in vitro. GST-γb and GST-γbY29A were purified from E. coli. ATG7-GFP and RFP-ATG8f were coexpressed in N. benthamiana leaves and enriched by incubation with anti-GFP antibody and Protein A/G Plus-Agarose. GFP-trap agarose with ATG7-GFP and RFP-ATG8f proteins was incubated with increasing concentrations of GST-γb protein (10, 20, and 30 μg) or GST-γbY29A (30 μg). Input and IP proteins were analyzed by protein gel blot analysis with anti-GST, RFP, or GFP antibodies.
(C) The two types of ATG8 were detected in different groups. Left panel: 3×HA-ATG8f was coinfiltrated with BSMV RNA strands or EV as a control. Right panel: 3×HA-ATG8f was coinfiltrated with γbI130G, γb, or γbY29A. The plants were treated with E-64d at 48 hpi. At 60 hpi, the samples were examined by protein gel blot analysis with anti-HA, anti-γb, and anti-actin antibodies.
To further confirm these results and the determined whether there is a stronger interaction between ATG7 and γb protein, we performed a pull-down assay in vitro. ATG7-GFP and RFP-ATG8f were coinfiltrated into the leaves, and 60 hpi, total proteins were extracted and immunoprecipitated using anti-GFP antibody. The precipitates were then incubated with increasing amounts of GST-γb (10, 20, and 30 μg) or GST-γbY29A (30 μg). Protein gel blot analysis revealed that less RFP-ATG8f was detected in the immunoprecipitates with increasing amount of GST-γb (Figure 7B). However, γbY29A mutant protein, which failed to interact with ATG7, had no effect on the interaction between ATG7 and ATG8f (Figure 7B). Together, these results indicate that BSMV γb interferes with the interaction of ATG7 with ATG8f competitively by binding to ATG7.
Since ATG8 is an autophagy-specific marker, it is selectively enclosed in autophagosomes, and the conversion of ATG8 to ATG8-phosphatidylethanolamine (PE) is a reliable indicator of autophagy (Ohsumi, 2001). To test whether γb affects the formation of the ATG8-PE complex, 3×HA-ATG8f was coinfiltrated with EV or BSMV. Proteins were harvested at 60 hpi and detected by protein gel blot analysis (Figure 7C). We detected much less ATG8-PE in BSMV-infected leaves compared with EV-infiltrated leaves (Figure 7C, left panel). Furthermore, 3×HA-ATG8f was coinfiltrated into leaves with γbI130G, γb, or γbY29A. Compared with the control, ATG8-PE levels were also much lower in plant tissues expressing γb or γbI130G- (which can interact with ATG7 and inhibit autophagy), while in γbY29A-expressing plant tissues, ATG8-PE levels were much higher (Figure 7C, right panel). These results further support the notion that γb inhibits autophagy by disrupting the conversion of ATG8 to ATG8-PE by competitively binding to ATG7.
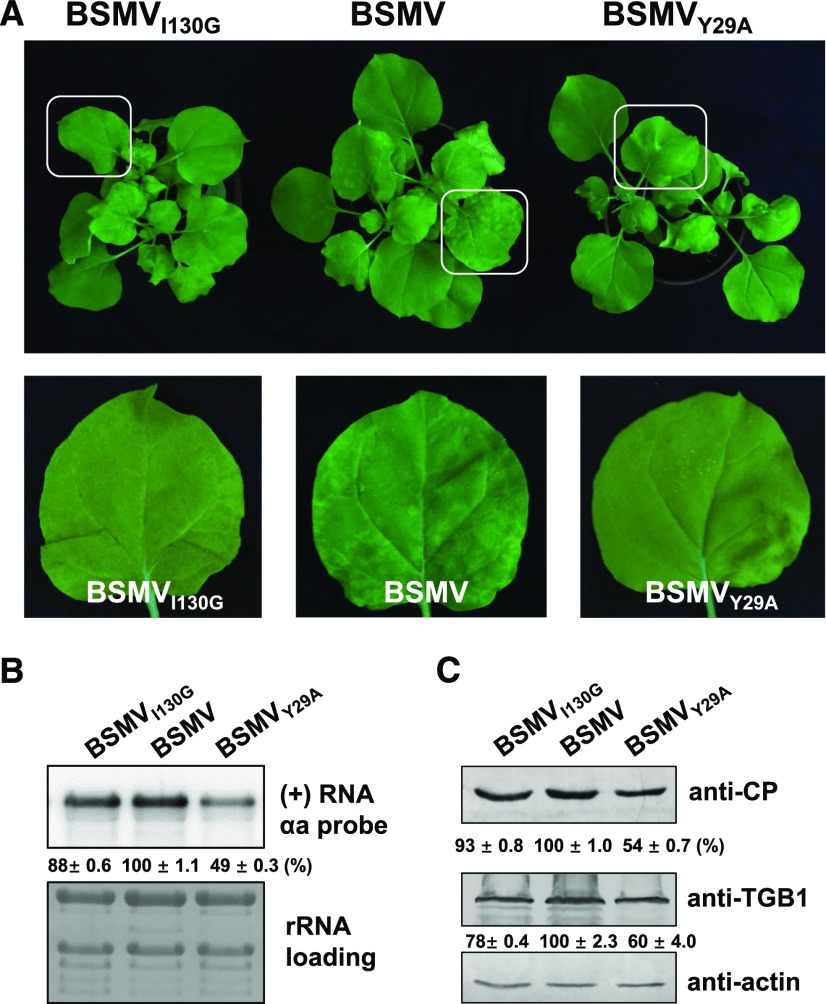
To further explore the biological significance of the γb-ATG7 interaction on autophagy-mediated defense against BSMV infection, we generated BSMVY29A and BSMVI130G mutant viruses and used them to inoculate N. benthamiana plants. It should be mentioned that γbY29A lost the ability to suppress both RNA silencing and autophagy, while γbI130G lost the ability to function as a VSR but could still suppress autophagy (Figure 5; Supplemental Figure 10). Viral symptoms were much milder in BSMVY29A-inoculated plants compared with plants inoculated with wild-type BSMV and BSMVI130G (Figure 8A). Furthermore, lower levels of viral RNA and BSMV proteins were detected in plants inoculated with BSMVY29A compared with plants inoculated with wild-type BSMV and BSMVI130G (Figures 8B and 8C). These results indicate that the interaction of γb with ATG7 is essential for suppressing autophagy and promoting viral infection.
Figure 8.
Plants Infected with BSMVY29A Show Much Milder Symptoms Than Plants Infected with Wild-Type BSMV.
(A) Viral symptoms in BSMV-, BSMVI130G-, or BSMVY29A-infected N. benthamiana. Magnified images of leaves (in white boxes) are shown in the lower panels.
(B) RNA gel blot analysis of BSMV RNA accumulation in BSMV-, BSMVI130G-, or BSMVY29A-infected N. benthamiana. Error bars indicate sd of the mean.
(C) Protein gel blot analysis to detect viral proteins (CP and TGB1) in BSMV-, BSMVI130G-, or BSMVY29A-infected N. benthamiana.
DISCUSSION
In this study, we showed that BSMV γb protein suppresses autophagy by disrupting the interaction between ATG7 and ATG8 through its direct binding to ATG7. Furthermore, we demonstrated that autophagy can function as an antiviral mechanism during compatible plant-RNA virus interactions and provided evidence for how a plant virus subverts autophagy-mediated antiviral defense responses to promote viral infection.
Autophagy plays an active role in host immunity against various invading intracellular pathogens in mammals (Boyle and Randow, 2013; Randow and Youle, 2014; Paul and Münz, 2016). Autophagy was also recently found to function as an antiviral defense response against some DNA viruses in plants (Hafrén et al., 2017; Haxim et al., 2017). Autophagy limits the infection of plants with geminiviruses, a group of single-stranded DNA viruses, by targeting viral proteins for autophagic degradation (Haxim et al., 2017). Cauliflower mosaic virus, a plant double-stranded DNA virus, is targeted by the selective autophagy-related protein NBR1 to suppress viral infection (Han et al., 2011; Hafrén et al., 2017). In this study, silencing of either ATG5 or ATG7 enhanced BSMV accumulation and led to more severe viral symptoms in N. benthamiana plants. Furthermore, the accumulation of mutant virus BSMVY29A, in which γbY29A is incapable of suppressing autophagy, was significantly reduced and failed to spread systemically. These findings suggest that autophagy also plays an active antiviral role in BSMV infection, and they provide evidence that autophagy contributes to antiviral defense responses against RNA viruses in plants.
Despite the antiviral role of autophagy, BSMV is able to successfully infect plants, suggesting that BSMV has a counterdefense mechanism to promote its infection. Consistent with this notion, many pathogens have evolved various mechanisms to evade or suppress host defense responses to promote disease development. In mammals, not only does autophagy eliminate intracellular pathogens, but autophagosomes can also serve as sources of intracellular membranes for the building of viral replication compartments and viral movement (Kirkegaard et al., 2004; Levine, 2005; Levine and Deretic, 2007; Schmid and Münz, 2007). Furthermore, some viruses can target ATG proteins to inhibit immune responses, including human immunodeficiency virus and human cytomegalovirus (de Figueiredo and Dickman, 2016). However, how pathogens disrupt the autophagy pathway in plants remains largely unknown. In this study, we used BSMV, a positive single-stranded RNA virus, as a model system and showed that BSMV γb protein suppresses autophagy to facilitate viral infection by competing with ATG8 for binding to ATG7. Our results show that BSMV γb inhibits autophagy by interfering with the ATG7-ATG8 interaction.
In this study, we found that BSMV γb protein interacts with plant ATG7. ATG7 is not essential for normal growth and development in Arabidopsis thaliana, while the atg7 mutant is hypersensitive to nutrient shortage and displays an early senescence phenotype (Doelling et al., 2002). Autophagy also plays a role in nutrient recycling, as evidenced by plant and mammalian atg mutants (Doelling et al., 2002; Hanaoka et al., 2002; Mizushima et al., 2004; Thompson et al., 2005; Xiong et al., 2005). In agreement with these reports, we detected increased starch accumulation in γb transgenic N. benthamiana (Supplemental Figure 7). In mice, ATG7 deficiency leads to multiple cellular abnormalities, and starved mouse embryonic fibroblasts lacking ATG7 fail to undergo cell cycle arrest (Komatsu et al., 2005; Lee et al., 2012). In human, overexpression of ATG7 is sufficient to induce autophagy in heart muscle cells (Pattison et al., 2011). In this study, we also found that overexpressing ATG7 induced autophagy in plants. ATG7 interacts with several ATG proteins, including ATG3, ATG10, and ATG8, but only ATG8 was shown to interact with the C terminus of ATG7 (Hong et al., 2011), a target of γb (Figure 1C; Supplemental Figure 1B). We found that BSMV γb competes with ATG8 to bind to ATG7 (Figure 7) to inhibit autophagy. These findings suggest that BSMV disrupts autophagy-mediated antiviral defense responses because its encoded protein, γb, interferes with the ATG7-ATG8 interaction to suppress plant autophagy.
METHODS
Plasmid Construction
Genes for BSMV γb and the γb mutants were individually cloned into the pGEX-KG vector (Guan and Dixon, 1991) to express GST-tagged fusion proteins in Escherichia coli. Full-length NbATG7 cDNA was amplified and cloned into pSPYNE/CE-35S vectors. The C terminus of ATG7 (amino acids 475–712) was cloned into pET30a(+) to express His-tagged fusion proteins in E. coli. The same fragment was cloned into pGADT7 for the yeast two-hybrid assay. Full-length NbATG7 was recombined into pSITE2NB (Chakrabarty et al., 2007) with GFP fused at its C terminus under the control of a duplicated 35S promoter. The upstream 2026-bp promoter region of NbATG7 was cloned into the pSPYNE/CE-ATG7 plasmids instead of the 35S promoter. Full-length NbATG8f cDNA was amplified and cloned into pGDR (Goodin et al., 2002). The primers used for these vectors were shown in Supplemental Table 1.
Plant Growth Conditions
Nicotiana benthamiana plants were grown in a growth chamber at 23°C with a 14/10-h light/dark photoperiod with an average light intensity of ∼75 mmol m−2 s−1 provided by three cool daylight tubes (Philips TLD 36W/865) and 60% humidity as described previously (Yuan et al., 2011).
Co-IP
The co-IP assays were performed as described previously (Zhang et al., 2017). Various expression vectors were coinfiltrated in N. benthamiana leaves. The infiltrated leaves were harvested at 3 dpi, and total protein from 3 g of N. benthamiana leaf tissue was agroinfiltrated with pGWB14-γb-3×Flag (Zhang et al., 2017) and pSITE2NB-ATG7 or the controls were ground in a liquid nitrogen-cooled mortar in extraction buffer (10% [v/v] glycerol, 25 mM Tris-HCl, pH 7.5, 1 mM EDTA, 150 mM NaCl, 1% Tween, and protease inhibitor cocktail). The supernatant was incubated with Flag beads (Sigma-Aldrich) for 4 h at 4°C. The precipitates were washed three times with IP buffer at 4°C and analyzed by immunoblotting using anti-Flag antibody (at 1:5000 dilution; Sigma-Aldrich, catalog no. A2220) or anti-GFP antibody at 1:1000 dilution.
GST Pull-Down Assays
The GST pull-down assays were performed as described previously (Zhang et al., 2017). Approximately 10 μg of purified GST fusion proteins and GST were incubated with ATG7C-His in 500 μL incubated buffer (50 mM Tris-HCl, pH 6.8, 300 mM NaCl, 1.5% glycerol, 0.6% Triton X-100, and 0.1% Tween) for 3 h at 4°C. The beads were washed five times with incubation buffer. The washed beads were boiled in 2× SDS loading buffer, and proteins were separated by SDS-PAGE for protein gel blot analysis with anti-GST (at 1:5000 dilution; GenScript, catalog no. A00866) and anti-His (at 1:5000 dilution; Abmart, catalog no. M30111) antibodies.
Starch Quantification and Staining
Iodine staining (for starch observation) was performed as described previously (Wang et al., 2013). N. benthamiana seeds were planted in soil. After 3 week, whole plants were stained with iodine. An EnzyChrom Starch Assay Kit (BioAssay Systems) was used for starch quantification analysis according to the manufacturer’s protocol.
GFP Imaging
VSR detection was performed as described previously (Johansen and Carrington, 2001) using recently reported minor modifications (Zhang et al., 2017). Equal volumes of Agrobacterium tumefaciens cultures (OD600 = 0.3) harboring plasmids expressing positive sense-GFP (Bragg and Jackson, 2004) and plasmids expressing γb, γbY29A, and γbI130G were used for infiltration. The agroinfiltrated leaves were illuminated under a long-wavelength UV lamp (UVP), and photographs were taken under a yellow filter at 3 dpi.
Co-IP and Pull-Down Analysis of the Interaction among Three Proteins
Leaves were agroinfiltrated with pSITE2NB-ATG7, pGDR-ATG8f, and increasing concentrations of pGWB14-γb-3×Flag (OD600 0.2–0.6). After 60 h of expression, total protein from 3 g N. benthamiana leaf tissue was extracted in extraction buffer (10% [v/v] glycerol, 25 mM Tris, pH 7.5, 1 mM EDTA, 150 mM NaCl, 1% Tween, and protease inhibitor cocktail). The leaves were ground in a liquid nitrogen-cooled mortar, and the supernatant was immunoprecipitated with anti-GFP antibody in protein G beads (Santa Cruz Biotechnology) for 4 h at 4°C. The precipitations were washed three times with IP buffer at 4°C and analyzed by immunoblotting using anti-RFP, anti-Flag, and anti-GFP antibodies.
For the pull-down assay, the leaves were infiltrated with pSITE2NB-ATG7 and pGDR-ATG8f, and after 60 h, total protein from 3 g N. benthamiana leaf tissue was extracted in extraction buffer. The leaves were ground in a liquid nitrogen-cooled mortar, and the supernatant was immunoprecipitated with anti-GFP antibody in Protein G beads for 4 h at 4°C. The precipitates were washed three times with IP buffer at 4°C, and various amounts of purified GST-γb (10, 20, and 30 μg) or GST-γbY29A (30 μg) were added to the precipitates. After a 4-h incubation, the pull-down products were analyzed by protein gel blot analysis with anti-RFP, GST, and GFP antibodies.
Detection of the Two Types of ATG8 by Protein Gel Blot Analysis
3×HA-ATG8f and EV, BSMV, γb, γbI130G, or γbY29A were coinfiltrated into N. benthamiana leaves. At 2 dpi, the leaves were infiltrated with 100 μM E-64d (an autophagy inhibitor) for 8 h, and after dark treatment, the samples were examined by protein gel blot analysis with anti-HA antibody (Sigma-Aldrich; H6908) at 1:1000 dilution (Bao et al., 2017).
BiFC Assays
BiFC assays were performed as described (Walter et al., 2004). The BiFC assay was performed using an Olympus FV1000 confocal microscope; YFP was excited at 546 nm. Autophagosomes were observed in N. benthamiana plant tissues as described (Han et al., 2015).
RNA Gel Blot and qRT-PCR
RNA gel blot analyses were performed as described previously (Zhang et al., 2017). Briefly, total RNA was extracted and quantified using a NanoDrop ND-1000 (Thermo Fisher Scientific). To detect the accumulation of viral RNA or RNAα, 3 μg of total RNA was separated on 1.2% agarose/1.1% formaldehyde gels. The RNA was transferred onto Hybond-N+ nylon membranes (GE Healthcare), followed by UV cross-linking and staining with methylene blue solution (0.04% methylene blue and 500 mM NaOAc). The nylon membranes were hybridized using a [γ-32P]UTP-labeled RNAα-specific probe or a [γ-32P]CTP-labeled 3′-untranslated region-specific probe. The resulting data were digitized with a Typhoon 9400 phosphor screen (GE Healthcare).
For qRT-PCR analysis, cDNA was synthesized from 4 μg total RNA (DNase-treated) using an oligo(dT) and M-MLV reverse transcriptase (Promega). The gene fragments were amplified using 2× SsoFast EvaGreen Supermix (Bio-Rad) and the primers shown in Supplemental Table 1. PP2A was used as the internal control (Liu et al., 2012), and the data were analyzed using CFX Manger (Bio-Rad).
Statistical Analysis
Data shown in this study are the means of three independent experiments. The intensity of the protein bands detected by protein gel blot analysis and RNA bands detected by RNA gel blot analysis were quantified using Quantity One software (Bio-Rad). The numbers of autophagosomes recorded were subjected to statistical analysis using SPSS software. The data were compared using one-way ANOVA (Supplemental Tables 2 and 3). When there were only two groups to compare, the numbers of autophagosomes were analyzed by Student’s t test.
Confocal Microscopy and TEM
Confocal microscopy was performed using a Zeiss LSM710 confocal microscope (Carl Zeiss). GFP, YFP, mCherry, chlorophyll autofluorescence, and CFP were visualized at 488, 514, 543, 633, and 440 nm, respectively. Detection bands were optimized for each fluorophore group to avoid emission bleeding.
TEM observation was performed as described previously (Cao et al., 2015). The leaves of γb and nontransgenic plants were pretreated with 100 μM E-64d for 8 h. For electron microscopy, leaves were cut into small fragment (∼1–2 mm2) and vacuum infiltrated in 0.05 M phosphate buffer containing 2.5% glutaraldehyde. The samples were postfixed in 2% osmium tetroxide (OsO4). After dehydration in ethanol and acetone, the leaf samples were embedded in Spurr’s resin (SPI Supplies). The sections were cut with a diamond knife on an ultramicrotome (EM UC7; Leica) and collected on copper grids. The sections were double stained with uranyl acetate and lead citrate before examination.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: TaATG7 (KF294806.1), NbATG7 (KX369398.1), NbATG8f (KU561372.1), NbATG1c (NM_001325605.1), NbATG5 (KX369397), NbATG6 (AY701316.1), NbATG9 (XM_009590092.2), Arabidopsis thaliana ATG8a (NM_001084955.1), and Saccharomyces cerevisiae ATG8 (CP020192.1).
Supplemental Data
Supplemental Figure 1. Interaction between ATG7 and γb in the Y2H assay.
Supplemental Figure 2. BiFC assay showing the interaction region of ATG7 with γb protein.
Supplemental Figure 3. Identification of the region within γb that is responsible for its interaction with ATG7 by GST pull down.
Supplemental Figure 4. Sequence alignment between BSMV γb and ATG8 proteins from Nicotiana benthamiana, Saccharomyces cerevisiae, and Arabidopsis thaliana.
Supplemental Figure 5. The localization of γb and ATG proteins.
Supplemental Figure 6. Autophagosomes are barely observed in BSMV-infected and mock-treated plant cells without autophagy activator treatment.
Supplemental Figure 7. Starch accumulates in γb transgenic N. benthamiana.
Supplemental Figure 8. Protein gel blot analysis to detect the expression of the proteins used for confocal observation shown in Figure 4.
Supplemental Figure 9. γb inhibits ATG7-overexpression induced autophagy, as determined by electron microscopy.
Supplemental Figure 10. RNA silencing suppressor activities of different γb mutant proteins.
Supplemental Figure 11. γbI130G also interacts with ATG7 in a GST pull-down assay.
Supplemental Figure 12. Both ATG7-GFP and γb-3×Flag are functional proteins.
Supplemental Figure 13. BSMV infection is inhibited in ATG7-overexpressing plants.
Supplemental Table 1. List of primers used in this study.
Supplemental Table 2. ANOVA tables for Figures 4B and 4D.
Supplemental Table 3. ANOVA tables for Figure 5B.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Savithramma P. Dinesh-Kumar (University of California at Davis) for constructive criticism and helpful editing and members of the Li lab for useful and crucial discussions. This work was supported by the National Natural Science Foundation of China (31570143) and the National Key R&D Program of China (2016YFD0100502) to D.L., the National Basic Research Program of China (2017YFA0503401) to Y.L., and Fundamental Research Funds for the Central Universities (2017SY003) to Y.Z.
AUTHOR CONTRIBUTIONS
D.L., M.Y., and Y.L. conceived the project. M.Y., Y.Z., X.X., N.Y., J.L., X.-B.W., C.H., Y.L., and D.L. designed the experiments, which were mainly performed by M.Y. All authors analyzed the data. M.Y., Y.L., D.L., J.Y., and Y.Z. wrote the article.
References
- Bao Y., Mugume Y., Bassham D.C. (2017). Biochemical methods to monitor autophagic responses in plants. Methods Enzymol. 588: 497–513. [DOI] [PubMed] [Google Scholar]
- Boyle K.B., Randow F. (2013). The role of ‘eat-me’ signals and autophagy cargo receptors in innate immunity. Curr. Opin. Microbiol. 16: 339–348. [DOI] [PubMed] [Google Scholar]
- Bragg J.N., Jackson A.O. (2004). The C-terminal region of the Barley stripe mosaic virus gammab protein participates in homologous interactions and is required for suppression of RNA silencing. Mol. Plant Pathol. 5: 465–481. [DOI] [PubMed] [Google Scholar]
- Cao X., Jin X., Zhang X., Li Y., Wang C., Wang X., Hong J., Wang X., Li D., Zhang Y. (2015). Morphogenesis of endoplasmic reticulum membrane-invaginated vesicles during Beet black scorch virus infection: Role of auxiliary replication protein and newimplications of three-dimensional architecture. J. Virol. 89: 6184–6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty R., Banerjee R., Chung S.M., Farman M., Citovsky V., Hogenhout S.A., Tzfira T., Goodin M. (2007). PSITE vectors for stable integration or transient expression of autofluorescent protein fusions in plants: probing Nicotiana benthamiana-virus interactions. Mol. Plant Microbe Interact. 20: 740–750. [DOI] [PubMed] [Google Scholar]
- Chen Y., Chen Q., Li M., Mao Q., Chen H., Wu W., Jia D., Wei T. (2017). Autophagy pathway induced by a plant virus facilitates viral spread and transmission by its insect vector. PLoS Pathog. 13: e1006727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X., Wang A. (2016). The potyvirus silencing suppressor protein VPg mediates degradation of SGS3 via ubiquitination and autophagy pathways. J. Virol. 91: e01478–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavel M., Michaeli S., Genschik P. (2017). Autophagy: A double-edged sword to fight plant viruses. Trends Plant Sci. 22: 646–648. [DOI] [PubMed] [Google Scholar]
- Dagdas Y.F., et al. (2016). An effector of the Irish potato famine pathogen antagonizes a host autophagy cargo receptor. eLife 5: e10856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Figueiredo P., Dickman M. (2016). Autophagy under attack. eLife 5: e14447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien B., Baumberger N., Schepetilnikov M., Viotti C., De Cillia J., Ziegler-Graff V., Isono E., Schumacher K., Genschik P. (2012). Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc. Natl. Acad. Sci. USA 109: 15942–15946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling J.H., Walker J.M., Friedman E.M., Thompson A.R., Vierstra R.D. (2002). The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 277: 33105–33114. [DOI] [PubMed] [Google Scholar]
- Dong X., Levine B. (2013). Autophagy and viruses: adversaries or allies? J. Innate Immun. 5: 480–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodin M.M., Dietzgen R.G., Schichnes D., Ruzin S., Jackson A.O. (2002). pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J. 31: 375–383. [DOI] [PubMed] [Google Scholar]
- Goodin M.M., Zaitlin D., Naidu R.A., Lommel S.A. (2008). Nicotiana benthamiana: its history and future as a model for plant-pathogen interactions. Mol. Plant Microbe Interact. 21: 1015–1026. [DOI] [PubMed] [Google Scholar]
- Gorovits R., Fridman L., Kolot M., Rotem O., Ghanim M., Shriki O., Czosnek H. (2016). Tomato yellow leaf curl virus confronts host degradation by sheltering in small/midsized protein aggregates. Virus Res. 213: 304–313. [DOI] [PubMed] [Google Scholar]
- Guan K.L., Dixon J.E. (1991). Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192: 262–267. [DOI] [PubMed] [Google Scholar]
- Hafrén A., Macia J.L., Love A.J., Milner J.J., Drucker M., Hofius D. (2017). Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc. Natl. Acad. Sci. USA 114: E2026–E2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafrén A., Üstün S., Hochmuth A., Svenning S., Johansen T., Hofius D. (2018). Turnip mosaic virus counteracts selective autophagy of the viral silencing suppressor HCpro. Plant Physiol. 176: 649–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Yu B., Wang Y., Liu Y. (2011). Role of plant autophagy in stress response. Protein Cell 2: 784–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Wang Y., Zheng X., Jia Q., Zhao J., Bai F., Hong Y., Liu Y. (2015). Cytoplastic glyceraldehyde-3-phosphate dehydrogenases interact with ATG3 to negatively regulate autophagy and immunity in Nicotiana benthamiana. Plant Cell 27: 1316–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka H., Noda T., Shirano Y., Kato T., Hayashi H., Shibata D., Tabata S., Ohsumi Y. (2002). Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 129: 1181–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haxim Y., et al. (2017). Autophagy functions as an antiviral mechanism against geminiviruses in plants. eLife 6: e23897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward A.P., Dinesh-Kumar S.P. (2011). What can plant autophagy do for an innate immune response? Annu. Rev. Phytopathol. 49: 557–576. [DOI] [PubMed] [Google Scholar]
- Hofius D., Schultz-Larsen T., Joensen J., Tsitsigiannis D.I., Petersen N.H., Mattsson O., Jørgensen L.B., Jones J.D., Mundy J., Petersen M. (2009). Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 137: 773–783. [DOI] [PubMed] [Google Scholar]
- Hong S.B., Kim B.W., Lee K.E., Kim S.W., Jeon H., Kim J., Song H.K. (2011). Insights into noncanonical E1 enzyme activation from the structure of autophagic E1 Atg7 with Atg8. Nat. Struct. Mol. Biol. 18: 1323–1330. [DOI] [PubMed] [Google Scholar]
- Jackson A.O., Lim H.S., Bragg J., Ganesan U., Lee M.Y. (2009). Hordeivirus replication, movement, and pathogenesis. Annu. Rev. Phytopathol. 47: 385–422. [DOI] [PubMed] [Google Scholar]
- Jeon E.J., Tadamura K., Murakami T., Inaba J.I., Kim B.M., Sato M., Atsumi G., Kuchitsu K., Masuta C., Nakahara K.S. (2017). rgs-CaM detects and counteracts viral RNA silencing suppressors in plant immune priming. J. Virol. 91: e00761–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jheng J.R., Ho J.Y., Horng J.T. (2014). ER stress, autophagy, and RNA viruses. Front. Microbiol. 5: 388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen L.K., Carrington J.C. (2001). Silencing on the spot. Induction and suppression of RNA silencing in the Agrobacterium-mediated transient expression system. Plant Physiol. 126: 930–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard K., Taylor M.P., Jackson W.T. (2004). Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat. Rev. Microbiol. 2: 301–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V., et al. (2009). A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 33: 505–516. [DOI] [PubMed] [Google Scholar]
- Klionsky D.J., et al. (2016). Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12: 1–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., et al. (2005). Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169: 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I.H., Kawai Y., Fergusson M.M., Rovira I.I., Bishop A.J., Motoyama N., Cao L., Finkel T. (2012). Atg7 modulates p53 activity to regulate cell cycle and survival during metabolic stress. Science 336: 225–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B. (2005). Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell 120: 159–162. [DOI] [PubMed] [Google Scholar]
- Levine B., Deretic V. (2007). Unveiling the roles of autophagy in innate and adaptive immunity. Nat. Rev. Immunol. 7: 767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Zhao N., Li Z., Xu X., Wang Y., Yang X., Liu S.S., Wang A., Zhou X. (2017). A calmodulin-like protein suppresses RNA silencing and promotes geminivirus infection by degrading SGS3 via the autophagy pathway in Nicotiana benthamiana. PLoS Pathog. 13: e1006213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Shi L., Han C., Yu J., Li D., Zhang Y. (2012). Validation of reference genes for gene expression studies in virus-infected Nicotiana benthamiana using quantitative real-time PCR. PLoS One 7: e46451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Dinesh-Kumar S.P. (2002). Virus-induced gene silencing in tomato. Plant J. 31: 777–786. [DOI] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Czymmek K., Tallóczy Z., Levine B., Dinesh-Kumar S.P. (2005). Autophagy regulates programmed cell death during the plant innate immune response. Cell 121: 567–577. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y. (2004). In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell 15: 1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munch D., Rodriguez E., Bressendorff S., Park O.K., Hofius D., Petersen M. (2014). Autophagy deficiency leads to accumulation of ubiquitinated proteins, ER stress, and cell death in Arabidopsis. Autophagy 10: 1579–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara K.S., et al. (2012). Tobacco calmodulin-like protein provides secondary defense by binding to and directing degradation of virus RNA silencing suppressors. Proc. Natl. Acad. Sci. USA 109: 10113–10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi Y. (2001). Molecular dissection of autophagy: two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2: 211–216. [DOI] [PubMed] [Google Scholar]
- Pattison J.S., Osinska H., Robbins J. (2011). Atg7 induces basal autophagy and rescues autophagic deficiency in CryABR120G cardiomyocytes. Circ. Res. 109: 151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul P., Münz C. (2016). Autophagy and mammalian viruses: Roles in immune response, viral replication, and beyond. Adv. Virus Res. 95: 149–195. [DOI] [PubMed] [Google Scholar]
- Randow F., Youle R.J. (2014). Self and nonself: how autophagy targets mitochondria and bacteria. Cell Host Microbe 15: 403–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid D., Münz C. (2007). Innate and adaptive immunity through autophagy. Immunity 27: 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taherbhoy A.M., Tait S.W., Kaiser S.E., Williams A.H., Deng A., Nourse A., Hammel M., Kurinov I., Rock C.O., Green D.R., Schulman B.A. (2011). Atg8 transfer from Atg7 to Atg3: a distinctive E1-E2 architecture and mechanism in the autophagy pathway. Mol. Cell 44: 451–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A.R., Doelling J.H., Suttangkakul A., Vierstra R.D. (2005). Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138: 2097–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438. [DOI] [PubMed] [Google Scholar]
- Wang Y., Yu B., Zhao J., Guo J., Li Y., Han S., Huang L., Du Y., Hong Y., Tang D., Liu Y. (2013). Autophagy contributes to leaf starch degradation. Plant Cell 25: 1383–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J., Long L., Yang K., Guy C., Shrestha S., Chen Z., Wu C., Vogel P., Neale G., Green D.R., Chi H. (2016). Autophagy enforces functional integrity of regulatory T cells by coupling environmental cues and metabolic homeostasis. Nat. Immunol. 17: 277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Klionsky D.J. (2007). Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 9: 1102–1109. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Contento A.L., Bassham D.C. (2005). AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 42: 535–546. [DOI] [PubMed] [Google Scholar]
- Xu G., Wang S., Han S., Xie K., Wang Y., Li J., Liu Y. (2017). Plant Bax Inhibitor-1 interacts with ATG6 to regulate autophagy and programmed cell death. Autophagy 13: 1161–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Li Z., Zhang K., Zhang X., Zhang Y., Wang X., Han C., Yu J., Xu K., Li D. (2018). Barley stripe mosaic virus γb interacts with glycolate oxidase and inhibits peroxisomal ROS production to facilitate virus infection. Mol. Plant 11: 338–341. [DOI] [PubMed] [Google Scholar]
- Yoshimoto K., Jikumaru Y., Kamiya Y., Kusano M., Consonni C., Panstruga R., Ohsumi Y., Shirasu K. (2009). Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21: 2914–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan C., Li C., Yan L., Jackson A.O., Liu Z., Han C., Yu J., Li D. (2011). A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLoS One 6: e26468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Zhang Y., Yang M., Liu S., Li Z., Wang X., Han C., Yu J., Li D. (2017). The Barley stripe mosaic virus γb protein promotes chloroplast-targeted replication by enhancing unwinding of RNA duplexes. PLoS Pathog. 13: e1006319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X., Chung K.P., Cui Y., Lin W., Gao C., Kang B.H., Jiang L. (2017). ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis. Proc. Natl. Acad. Sci. USA 114: E426–E435. [DOI] [PMC free article] [PubMed] [Google Scholar]