ICU11 and CP2 are Arabidopsis epigenetic machinery components that contain a prolyl 4-hydroxylase domain and are required for histone chemical modification, but not for DNA methylation.
Abstract
All critical developmental and physiological events in a plant’s life cycle depend on the proper activation and repression of specific gene sets, and this often involves epigenetic mechanisms. Some Arabidopsis thaliana mutants with disorders of the epigenetic machinery exhibit pleiotropic defects, including incurved leaves and early flowering, due to the ectopic and heterochronic derepression of developmental regulators. Here, we studied one such mutant class, the incurvata11 (icu11) loss-of-function mutants. We have identified ICU11 as the founding member of a small gene family that we have named CUPULIFORMIS (CP). This family is part of the 2-oxoglutarate/Fe(II)-dependent dioxygenase superfamily. ICU11 and its closest paralog, CP2, have unequally redundant functions: although cp2 mutants are phenotypically wild type, icu11 cp2 double mutants skip vegetative development and flower upon germination. This phenotype is reminiscent of loss-of-function mutants of the Polycomb-group genes EMBRYONIC FLOWER1 (EMF1) and EMF2. Double mutants harboring icu11 alleles and loss-of-function alleles of genes encoding components of the epigenetic machinery exhibit synergistic, severe phenotypes, and some are similar to those of emf mutants. Hundreds of genes are misexpressed in icu11 plants, including SEPALLATA3 (SEP3), and derepression of SEP3 causes the leaf phenotype of icu11. ICU11 and CP2 are nucleoplasmic proteins that act as epigenetic repressors through an unknown mechanism involving histone modification, but not DNA methylation.
INTRODUCTION
The phenotypic plasticity of plants depends on a large arsenal of epigenetic regulators, including noncoding RNAs and proteins involved in DNA methylation, histone chemical modification, and nucleosome positioning. This repertoire of epigenetic machinery components is produced by gene families that often include sets of paralogous genes with partially redundant functions. In plants such as Arabidopsis thaliana, null alleles of genes encoding proteins with epigenetic functions are typically viable and can be studied, whereas the corresponding mutations in animals are frequently lethal (Pikaard and Mittelsten Scheid, 2014; Provart et al., 2016).
Genetic screens, particularly in Arabidopsis and maize (Zea mays) have contributed to the discovery of components of the plant epigenetic circuitry. A good example of this is the Polycomb group (PcG) of genes, which encode some of the best studied components of the epigenetic machinery and form different Polycomb Repressive Complexes (PRCs) (Mozgova and Hennig, 2015). The founding PcG member, Polycomb, was first identified by mutation in Drosophila melanogaster (Lewis, 1947). The first PcG gene identified in Arabidopsis was CURLY LEAF (CLF), a homolog of Enhancer of zeste, a PcG gene in Drosophila (Goodrich et al., 1997). PcG proteins exert a type of dynamic transcriptional repression that not only is stable and heritable through mitosis but also responds to developmental and environmental factors (Mozgova and Hennig, 2015). PRC2-induced repression of target genes occurs via the trimethylation of Lys-27 on histone 3 (H3K27me3), and PRC1-induced repression involves its H2A E3 ubiquitin ligase activity. In animals, H3K27me3 deposition by PRC2 allows PRC1 to monoubiquitinate H2A (Wang et al., 2004). In plants, PRC1 recruitment to a specific target may occur via both H3K27me3-dependent and -independent mechanisms (Blackledge et al., 2015). In some cases, PRC1 also recruits PRC2 (Yang et al., 2013; Blackledge et al., 2014; Cooper et al., 2014; Kalb et al., 2014). Moreover, in most plant genes, PRC2-mediated H3K27 trimethylation does not take place before H2A monoubiquitination (Zhou et al., 2017).
The roles of PcG proteins in sporophytic and gametophytic development appear to be conserved from mosses to vascular plants (Mosquna et al., 2009; Okano et al., 2009). In Arabidopsis, the PcG protein complex known as the FERTILIZATION-INDEPENDENT SEED complex (Wang et al., 2006) is required for gametogenesis and early seed formation (Luo et al., 1999; Chanvivattana et al., 2004), and EMBRYONIC FLOWER1 (EMF1) and EMF2 participate in sporophyte development (Sung et al., 1992; Chen et al., 1997). The PcG protein EMF2 is a component of PRC2, and EMF1 is a plant-specific protein required for H3K27me3 deposition (Kim et al., 2012; Yang et al., 2013; Merini and Calonje, 2015). Loss-of-function, recessive alleles of EMF1 and EMF2 cause ectopic and heterochronic expression of many genes, including key flower development genes. This misexpression, in turn, results in the loss of vegetative identity. The strongest homozygous emf mutants are postembryonic lethal and skip the vegetative phase. Instead of developing leaves, these mutants form reproductive structures such as floral buds, carpels, and ovules upon germination (Sung et al., 1992; Chen et al., 1997).
The superfamily of 2-oxoglutarate/Fe(II)-dependent dioxygenases (2OGDs; also known as 2-ODDs) is present in bacteria, fungi, and metazoans (Aravind and Koonin, 2001) and is the second largest family in plant proteomes (Kawai et al., 2014). In Arabidopsis, these non-heme iron-containing soluble proteins localize to the cytosol, nucleus, and plastids (Mielecki et al., 2012). Experimental evidence is available for the activity of only some plant 2OGDs, in a few plant species (Kawai et al., 2014). The 2OGDs require 2-oxoglutarate (α-ketoglutarate) and molecular oxygen as cosubstrates and Fe2+ as a cofactor to catalyze substrate oxidation. The proven and presumptive oxidative reactions catalyzed by 2OGDs include hydroxylation, demethylation, demethylenation, desaturation, ring closure, ring cleavage, epimerization, rearrangement, and halogenation (Farrow and Facchini, 2014).
Analysis of the sequences of 479 2OGD-domain proteins from six plant species, ranging from green algae to angiosperms, grouped these proteins into three classes: DOXA, DOXB, and DOXC (Kawai et al., 2014). DOXA 2OGDs are homologous to Escherichia coli alpha-ketoglutarate-dependent dioxygenase B, a DNA repair protein that is induced in response to alkylating agents (Kataoka et al., 1983) and functions in the oxidative demethylation of nucleic acids and histones (Falnes et al., 2002; Trewick et al., 2002; Korvald et al., 2011; Mielecki et al., 2012; Duan et al., 2017; Martínez-Pérez et al., 2017). The DOXB group comprises proteins with a subtype of the 2OGD domain, the prolyl 4-hydroxylase domain (P4Hc), which participates in the posttranslational modification of proline residues in cell wall proteins and plant peptide hormones (Hieta and Myllyharju, 2002; Matsubayashi, 2011; Velasquez et al., 2015). DOXC proteins are involved in the metabolism of gibberellins, flavonoids, ethylene, and auxin (Shimizu, 2014; Porco et al., 2016). Although not taken into account by Kawai et al. (2014), the Jumonji-containing (JmjC) domain proteins are also considered to be 2OGDs by many authors (Cloos et al., 2008; Dong et al., 2014). These proteins, which are conserved from bacteria to humans, function as histone demethylases in organisms ranging from yeast to humans (Accari and Fisher, 2015). In Arabidopsis, several JmjCs participate in histone demethylation (Chen et al., 2011b). For example, JMJ25/IBM1 and JMJ12/REF6 control flower development and flowering time via H3K9 and H3K27 demethylation, respectively (Miura et al., 2009; Lu et al., 2011).
Kawai et al. (2014) identified 14 DOXA, 14 DOXB, 100 DOXC, and two unclassifiable 2OGDs in Arabidopsis. Here, we found that one of the unclassified 2OGDs is the product of the INCURVATA11 (ICU11) gene, which encodes the founding member of a small protein family that we have named CUPULIFORMIS (CP). At least two proteins of the CP family, ICU11 and CP2, act as epigenetic repressors in an unequally redundant manner through an unknown mechanism involving histone modification, but not DNA methylation.
RESULTS
The icu11-1 Mutation Perturbs Leaf Morphology and Flowering Time
To better understand leaf development, we performed forward (Berná et al., 1999) and reverse (Wilson-Sánchez et al., 2014) genetic screens for Arabidopsis mutants exhibiting defects in leaf shape, size, or pigmentation. We also studied similar leaf mutants isolated by previous authors, including the N242 (cupuliformis [cp]) mutant, which had been isolated after N-nitroso-N-methylurea (NMU) mutagenesis of wild-type S96 Arabidopsis seeds (Relichová, 1976; Serrano-Cartagena et al., 1999). To facilitate our genetic analyses, several phenotypic classes were defined based on the most conspicuous morphological trait of each mutant (Berná et al., 1999). One of these classes was dubbed Incurvata because it contained mutants exhibiting inward curvature of vegetative leaves; the Incurvata class included the N242 mutant, which we renamed icu11 (Serrano-Cartagena et al., 1999) and refer to here as icu11-1.
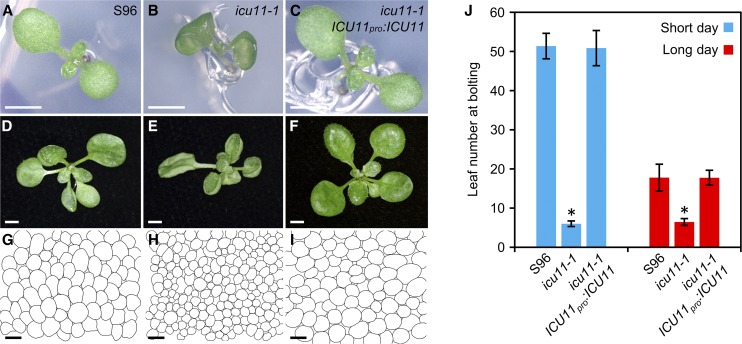
Cotyledons of icu11-1 are epinastic (curved downward) 7 d after stratification (DAS; Figures 1A and 1B). First- and second-node leaves of icu11-1 have leaf margins more involute (curved up, hyponastic; Figure 1E) than the S96 wild type (Figure 1D), a trait that was also observed to various extents in all other vegetative leaves. Analysis of paradermal sections of cleared first-node leaves revealed a significant reduction in icu11-1 palisade mesophyll cell size compared with S96 (Figures 1G and 1H). However, no apparent defect was observed in adaxial or abaxial epidermal cells (Supplemental Figures 1A to 1D and 1G).
Figure 1.
Morphological, Histological, and Flowering-Time Phenotypes of the icu11-1 Mutant.
(A) to (I) Seedlings with fully expanded cotyledons, rosettes, and diagrams of the palisade mesophyll subepidermal layer from the first-node leaves of the S96 wild type ([A], [D], and [G]), the icu11-1 mutant ([B), [E], and [H]), and a mutant and transgenic icu11-1 ICU11pro:ICU11 plant ([C], [F], and [I]).
(J) Flowering time in S96, icu11-1, and icu11-1 ICU11pro:ICU11 plants grown under short- and long-day photoperiods. Error bars indicate sd. Asterisks indicate values significantly different from S96 in a Mann-Whitney U test (*P < 0.001).
Nine plants grown in individual pots were analyzed per genotype. Photographs were taken 7 ([A] to [C]), 16 ([D] to [F]), and 21 ([G] to [I]) DAS. Bars = 2 mm in (A) to (F) and 50 µm in (G) to (I).
Under our standard culture conditions (continuous light), the icu11-1 plants flowered earlier and produced fewer vegetative leaves than S96 plants. The icu11-1 plants bolted at 11.7 ± 1.1 DAS and had 7.9 ± 0.8 rosette leaves at bolting. The wild-type S96 plants bolted at 28.9 ± 1.5 DAS and had 13.3 ± 1.6 rosette leaves at bolting (n = 120; P < 0.001). To determine the basis of the early flowering phenotype, we counted the number of rosette leaves at bolting under both short and long day conditions (Figure 1J). The early flowering of icu11-1 was photoperiod independent. We also dissected siliques from 10 different S96 and icu11-1 plants at 45 DAS and found more unfertilized ovules per half silique in icu11-1 than in S96 (19.1 ± 4.8 and 2.9 ± 1.6, respectively; n = 20; P < 0.001; Supplemental Figures 2A, 2C, and 2L).
The ICU11 Gene Is At1g22950
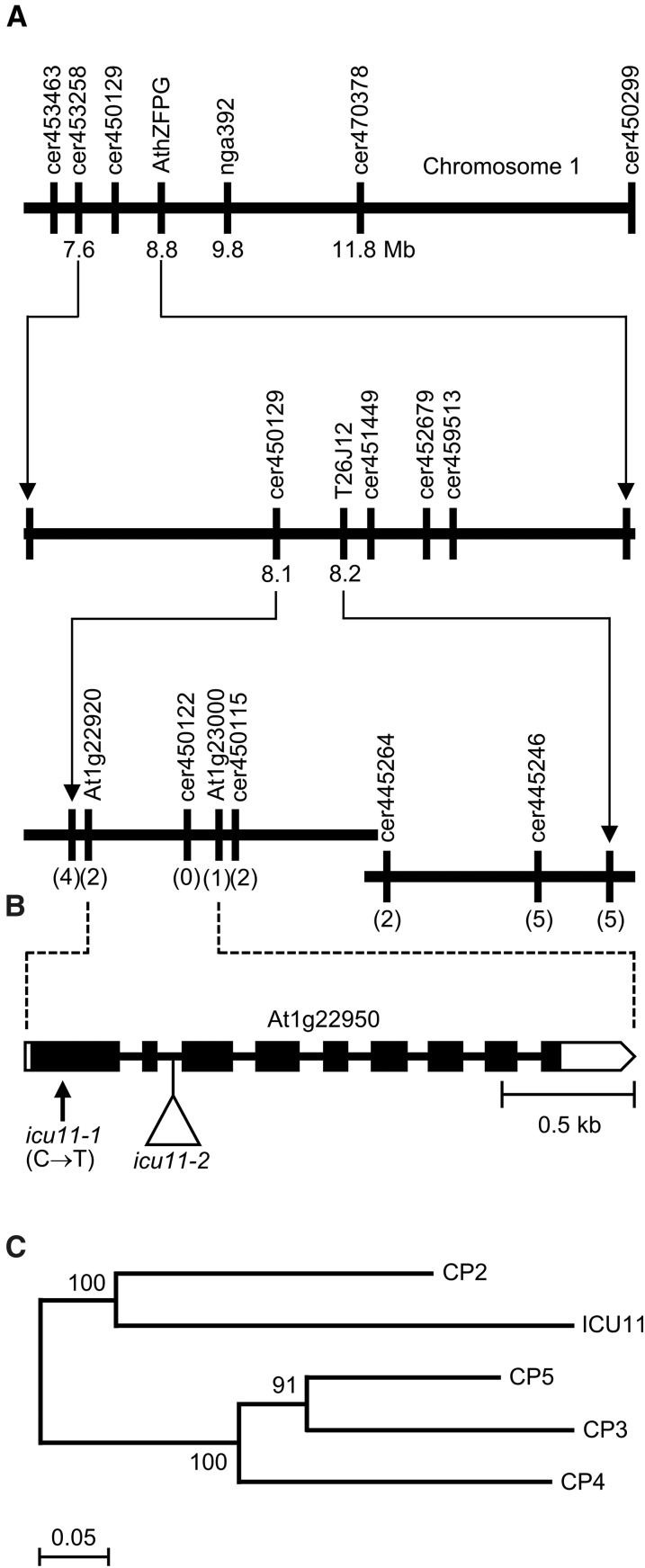
The icu11-1 (cp) mutation was mapped to the short arm of chromosome 1 (Repková et al., 2005). Using linkage analysis, we delimited a 32.6-kb candidate interval for ICU11 on chromosome 1 (Figure 2A). We then used next-generation sequencing to identify NMU-type mutations within the candidate interval (see Methods) (Hartwig et al., 2012; Candela et al., 2015) and found a C→T transition mutation in the first exon of At1g22950. This mutation, which was confirmed by Sanger sequencing, is predicted to cause a Gln22→Stop substitution in ICU11 (Figure 2B).
Figure 2.
Fine Mapping and Structure of ICU11, and Phylogenetic Analysis of CP Proteins.
(A) The use of a mapping population of 455 F2 plants derived from an icu11-1 × Col-0 cross allowed us to delimit a candidate interval of 32.6 kb on chromosome 1. The names and physical map positions of the molecular markers (Supplemental Table 1) used for linkage analysis are indicated. The number of recombinant chromosomes is shown in parentheses. All values not in parentheses indicate Mb.
(B) Structure of the ICU11 gene with the nature and positions of the icu11 mutations indicated. Boxes and lines represent exons and introns, respectively. Open and black boxes represent untranslated and translated regions, respectively. The triangle indicates the T-DNA insertion in icu11-2, and the vertical arrow indicates the point mutation in icu11-1.
(C) Neighbor-joining tree of proteins of the CP family. The tree was constructed with MEGA6 (see Methods) and inferred from 1000 replicates. Numbers indicate bootstrap percentages. The scale bar indicates 5% amino acid sequence changes according to the Poisson correction method (Nei and Kumar, 2000).
To confirm that At1g22950 is ICU11, we transformed icu11-1 plants with a transgene containing a 3.18-kb genomic segment encompassing At1g22950 driven by its own promoter (ICU11pro:ICU11). Five independent transformants carrying ICU11pro:ICU11 were obtained, all of which had normal morphology, histology, flowering time, and fertility (Figures 1C, 1F, 1I, and 1J; Supplemental Figures 2E and 2L). We found additional lines annotated as carrying T-DNA insertions within the At1g22950 transcription unit, including SALK_051945, SALK_054985, and FLAG_402G04, but only the FLAG_402G04 line contained the annotated insertion; we named this line icu11-2. The T-DNA insertion in icu11-2 is located in the second intron of ICU11 (Figure 2B). Plants homozygous for icu11-2 (in a Ws-2 genetic background) exhibited leaf incurvature, early flowering, and reduced palisade mesophyll cell size (Supplemental Figures 3D, 3E, 3G, and 3H) as well as unfertilized ovules (Supplemental Figures 2B, 2D, and 2L), similar to the phenotypes of icu11-1. Heterozygous icu11-1/icu11-2 plants were similar to icu11-1/icu11-1 and icu11-2/icu11-2 homozygotes, confirming that icu11-1 and icu11-2 are allelic (Supplemental Figure 3C). Moreover, the phenotype of icu11-2 was fully restored to the wild type via transformation with the ICU11pro:ICU11 transgene in 29 out of 31 independent transformants obtained (Supplemental Figures 2F, 2L, and 3F). Wild-type ICU11 transcripts were nearly absent in icu11-2 plants, and wild-type levels were restored in icu11-2 ICU11pro:ICU11, as determined by qRT-PCR (Supplemental Figure 3I). These results indicate that At1g22950 is indeed ICU11 and that its two mutant alleles cause similar phenotypes in different wild-type genetic backgrounds.
ICU11 Is the Founding Member of the CUPULIFORMIS Family of 2OGD Proteins
According to TAIR10 annotation, At1g22950 encodes a protein of unknown function that belongs to the 2OGD superfamily and contains an oxoglutarate/iron-dependent oxygenase domain (InterPro:IPR005123) of the prolyl 4-hydroxylase, alpha subunit subtype (P4Hc; InterPro:IPR006620). However, Kawai et al. (2014) did not include the protein encoded by At1g22950 in the DOXB group of 2OGDs, which are characterized by the presence of a P4Hc domain because it exceeded the BLASTP threshold set by these authors (P < 10−7).
BLASTP searches allowed us to identify four close paralogs of ICU11 in the Arabidopsis genome. These five genes define the gene family we have named CP, including CP2 (At3g18210), CP3 (At5g43660), CP4 (At1g48740), and CP5 (At1g48700) (Supplemental Table 2). Phylogenetic analysis indicated that the CP proteins fall into two clades: one contains ICU11 and CP2 and the other contains CP3, CP4, and CP5 (Figure 2C; Supplemental Figure 4).
Kawai et al. (2014) did not describe CP2, CP3, CP4, or CP5 as 2OGDs, even though they are annotated as 2OGDs of unknown function. We found little or no information about these genes and ICU11 in transcriptome and gene coexpression network databases, except for CP2, which is expressed ubiquitously according to the eFP Browser (Winter et al., 2007). In addition, although Kawai et al. (2014) did not include ICU11 in the DOXB group of 2OGDs, we found that all CP family members contain a P4Hc domain.
To determine whether ICU11 orthologs are conserved in plants, we aligned putative ICU11 orthologs from several angiosperms, the moss Physcomitrella patens, and the liverwort Marchantia polymorpha. Multiple sequence alignment (Supplemental Figure 5) revealed that these proteins are evolutionarily conserved, particularly in the P4Hc domain. The full-length sequences share identities with ICU11 ranging from 41.70% (M. polymorpha) to 50.99% (maize). According to the HomoloGene database, ICU11 also has metazoan orthologs, with which it only shares the P4Hc domain, so we included some of these metazoan orthologs in the alignment (Supplemental Figure 6).
The ICU11-CP2 Paralogous Gene Pair Is Essential and Exhibits Unequal Functional Redundancy
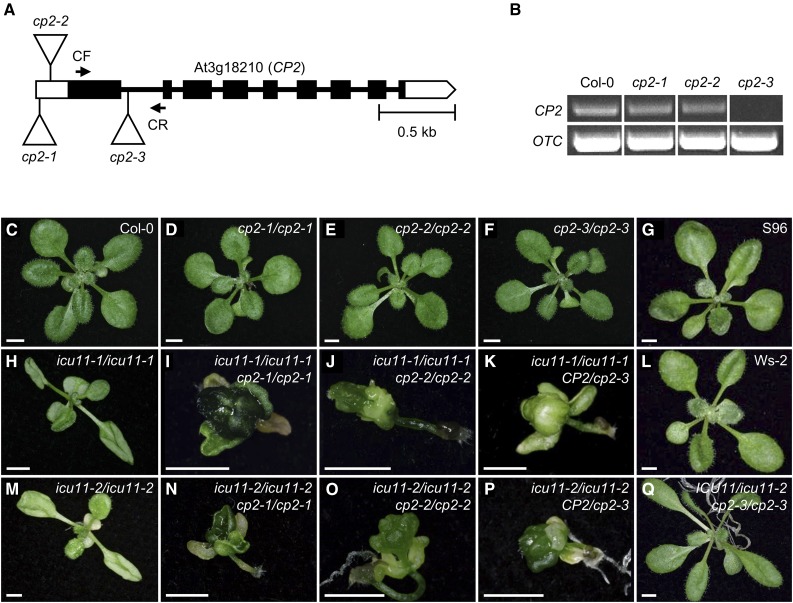
Based on their sequence similarity (Supplemental Figure 4 and Supplemental Table 2), we reasoned that ICU11 and CP2 might have redundant functions. We obtained T-DNA alleles of CP2 harboring an insertion in different parts of the 5′ untranslated region (SAIL_1215_B02 and SAIL_658_E12, which we named cp2-1 and cp2-2, respectively) or in the first intron (SAIL_621_G08, cp2-3) of the gene (Figure 3A). RT-PCR analysis revealed that CP2 mRNA levels were reduced in cp2-1 and cp2-2 compared with the wild type and were undetectable in cp2-3. Therefore, cp2-3 is a likely null allele (Figure 3B).
Figure 3.
Unequal Functional Redundancy between ICU11 and CP2.
(A) Structure of CP2 and the positions of insertional cp2 mutations. Horizontal arrows represent the oligonucleotides q_CP2_F (CF) and qCP2_R (CR) (not drawn to scale) used as PCR primers (Supplemental Table 1) to determine the relative expression of CP2 in the cp2 mutants. Boxes, lines, and triangles are defined in the Figure 2B legend.
(B) RT-PCR analysis of CP2 expression in the cp2 mutants and Col-0. The bands were obtained after 35 cycles of PCR amplification following reverse transcription using RNA extracted from rosette leaves collected 10 DAS as a template. OTC was used as an internal control.
(C) to (Q) Genetic interactions between loss-of-function alleles of ICU11 and CP2. Rosettes of the Col-0 (C), S96 (G), and Ws-2 (L) wild-type lines, the cp2-1/cp2-1 (D), cp2-2/cp2-2 (E), cp2-3/cp2-3 (F), icu11-1/icu11-1 (H), and icu11-2/icu11-2 (M) single mutants, the icu11-1/icu11-1 CP2/cp2-3 (K), icu11-2/icu11-2 CP2/cp2-3 (P), and icu11-2/ICU11 cp2-3/cp2-3 (Q) sesquimutants, and the icu11-1/icu11-1 cp2-1/cp2-1 (I), icu11-1/icu11-1 cp2-2/cp2-2 (J), icu11-2/icu11-2 cp2-1/cp2-1 (N), and icu11-2/icu11-2 cp2-2/cp2-2 (O) double mutants. Photographs were taken 14 ([C] to [P]) and 21 (Q) DAS. Bars = 2 mm.
Although the amino acid sequences of CP2 and ICU11 are almost 50% identical (Supplemental Table 2), no morphological or flowering time mutant phenotypes were observed in plants homozygous for cp2-1, cp2-2, or cp2-3 (Figures 3C to 3F; Supplemental Figure 7). However, we found more unfertilized ovules in cp2-3 siliques than in Col-0 (7.50 ± 4.54 and 0.93 ± 0.91, respectively; P < 0.001; n = 20), but not in cp2-1 or cp2-2 (Supplemental Figures 2G to 2J and 2L). Nonetheless, severe synergistic phenotypes were observed in the icu11 cp2 double mutants: Seeds of the icu11-1 cp2-1 and icu11-1 cp2-2 double mutants germinated, but the seedlings lacked rosette leaves (Figures 3C and 3G to 3J), skipped vegetative growth, and flowered upon germination, generating small, sterile flowers. Cotyledons of these double mutants were usually involute (Supplemental Figure 8A), directly connecting a seemingly normal root system with aberrant floral tissues comprising a mixture of stamens, carpels, and stigma (Supplemental Figure 8B) as well as ovules (Supplemental Figures 8C and 8D).
Similar defects were observed in icu11-1/icu11-1 CP2/cp2-3 sesquimutant (homozygous for one mutation and heterozygous for another) seedlings (Figure 3K; Supplemental Figures 8C, 8E, and 8F) than in the icu11-1/icu11-1 cp2-1/cp2-1 and icu11-1/icu11-1 cp2-2/cp2-2 double homozygotes. Similar results were obtained for genetic combinations involving icu11-2 (Figures 3C and 3L to 3P). Since no icu11-1/icu11-1 cp2-3/cp2-3 or icu11-2/icu11-2 cp2-3/cp2-3 seedlings were found, we dissected siliques from ICU11/icu11-2 cp2-3/cp2-3 sesquimutant plants, finding that 74.9% of the ovules were unfertilized (Supplemental Figure 2K). The remaining ovules produced both seemingly normal seeds (16.3%) and shrunken, embryo-arrested seeds (8.8%) (Supplemental Figures 2K and 2L). These observations confirm that the loss of CP2 function is stronger in cp2-3 than in cp2-1 and cp2-2. Taken together, the relatively mild phenotype of icu11 alleles, the completely wild-type phenotype of cp2 alleles, and the severe, lethal phenotypes of their double mutant or sesquimutant combinations indicate that ICU11 and CP2 are a pair of redundant genes with an essential function. Their functional redundancy is unequal, as shown by comparing the phenotypically wild-type ICU11/icu11-2 cp2-3/cp2-3 and the aberrant and lethal icu11-2/icu11-2 CP2/cp2-3 sesquimutants (Figures 3P and 3Q). In conclusion, the lack of CP2 function can be complemented by a single copy of the wild-type allele of ICU11, but CP2 cannot substitute for ICU11.
ICU11 and CP2 Are Nuclear Proteins Expressed During Vegetative and Reproductive Development
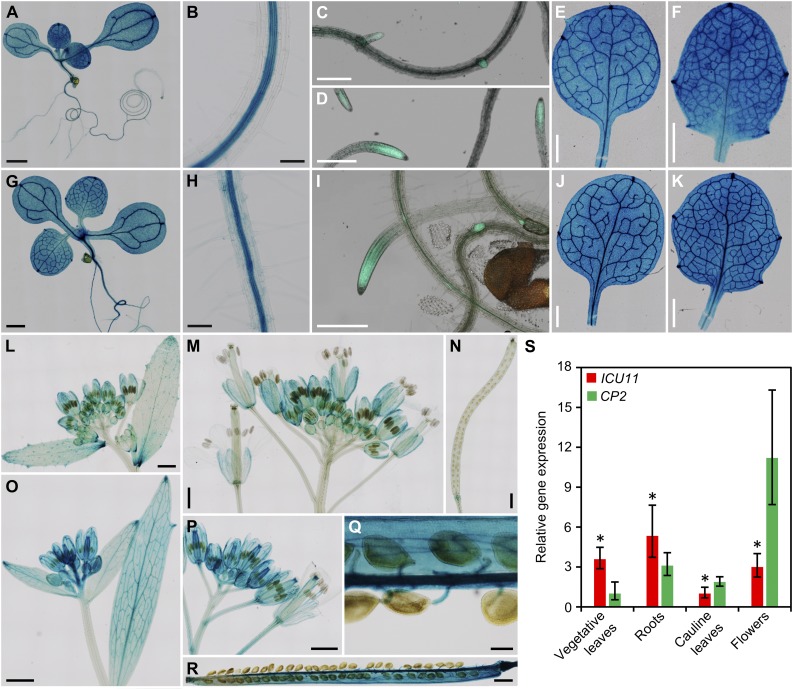
To analyze the expression patterns of ICU11 and CP2, we constructed ICU11pro:GUS and CP2pro:GUS reporter transgenes and transferred them into wild-type S96 and Col-0 plants, respectively. We obtained eight independent lines expressing each transgene and showing reproducible staining patterns. Strong GUS signals were detected in all vegetative tissues of ICU11pro:GUS and CP2pro:GUS transgenic plants collected 8 DAS (Figures 4A and 4G). Weak signals were observed in roots, mainly in the vasculature (Figures 4B and 4H). In plants collected at 14 DAS, GUS activity was still strong in all vegetative tissues (Figures 4E, 4F, 4J, and 4K). At the reproductive stage, the GUS activity was stronger in CP2pro:GUS than in ICU11pro:GUS plants in cauline leaves (Figures 4L and 4O), inflorescences (Figures 4M and 4P), siliques (Figures 4N and 4R), and funiculi (Figure 4Q). We also performed qRT-PCR to analyze the expression of ICU11 and CP2 in various tissues from S96 plants (Figure 4S). At the vegetative stage, ICU11 was strongly expressed in leaves and roots, but at the reproductive stage, CP2 was more strongly expressed in cauline leaves and flowers.
Figure 4.
Expression Patterns of ICU11 and CP2.
(A), (B), (E) to (H), and (J) to (R) GUS staining of seedlings ([A] and [G]), primary roots ([B] and [H]), first- ([E] and [J]) and third-node ([F] and [K]) leaves, cauline leaves ([L] and [O]), inflorescences ([M] and [P]), siliques ([N] and [R]), and funiculi (Q) of ICU11pro:GUS ([A], [B], [E], [F], and [L] to [N]), and CP2pro:GUS transgenic plants ([G], [H], [J], [K], and [O] to [R]).
(C), (D), and (I) GFP signals detected at the root tips and incipient lateral roots in ICU11pro:ICU11:GFP ([C] and [D]) and CP2pro:CP2:GFP (I) transgenic plants.
(S) qRT-PCR analysis of the relative expression of ICU11 and CP2 in various S96 tissues. CP2 expression levels in vegetative leaves were used as the reference value. RNA was extracted from leaves, roots, cauline leaves, and flowers collected 14, 14, 40, and 45 DAS, respectively. Error bars indicate the interval delimited by 2−(ΔΔCT ±sd), where sd is the sd of the ΔΔCT values. Asterisks indicate values significantly different between ICU11 and CP2 in a Mann-Whitney U test (*P < 0.001). Three different biological replicates were analyzed in triplicate.
Photographs were taken 8 ([A] to [D] and [G] to [I]), 14 ([E], [F], [J], and [K]), and 45 ([L] to [R]) DAS. Bars = 1 mm in (A), (E) to (G), (J) to (P) and (R), 100 µm in (B) to (D), (H) and (I), and 200 µm in (Q).
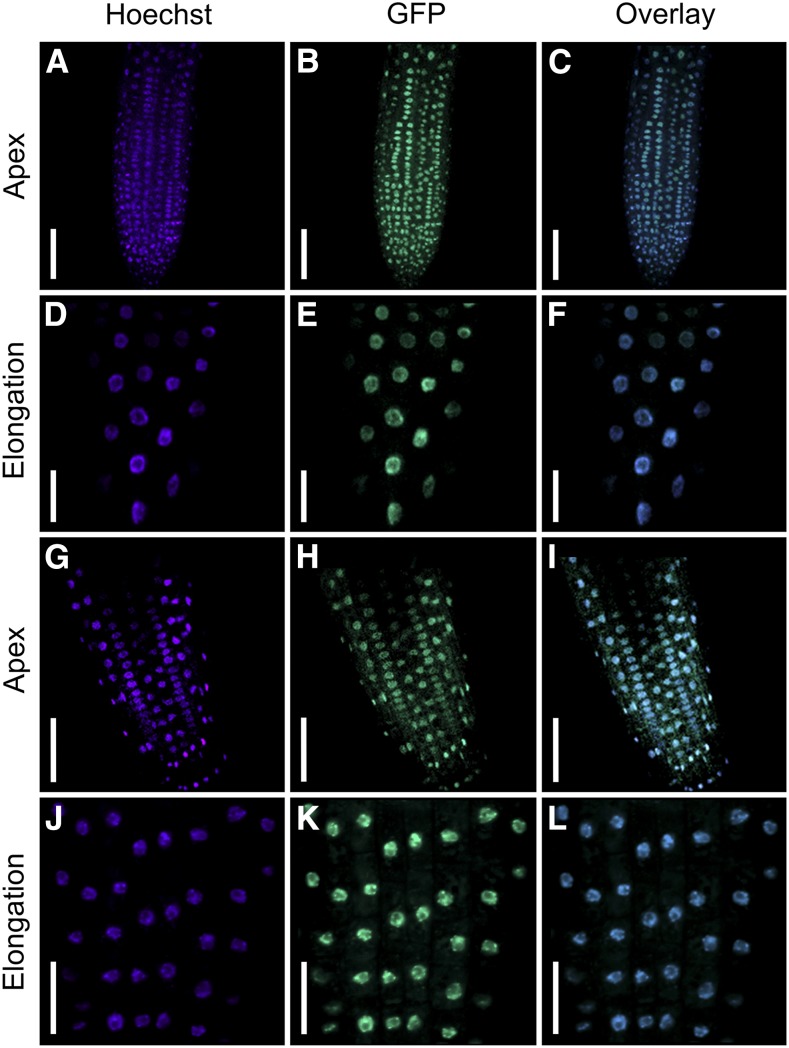
All computational tools (see Methods) that we used consistently predicted that both ICU11 and CP2 localize to the nucleus. To further characterize these proteins, we generated C-terminal GFP fusion proteins; the corresponding in-frame translational fusions were driven by either the ICU11 or CP2 endogenous promoters or the constitutive Cauliflower mosaic virus 35S (hereafter, 35S) promoter. We examined transgenic plants expressing 35Spro:ICU11:GFP (nine independent transformants) and ICU11pro:ICU11:GFP (seven transformants) in the icu11-1 background, and 35Spro:CP2:GFP (seven transformants) and CP2pro:CP2:GFP (ten transformants) in the Col-0 background. The ICU11:GFP fusion protein rescued the mutant phenotype of icu11-1 in all cases (Supplemental Figure 9). In accordance with bioinformatic predictions, both GFP signals were specifically detected in the nucleoplasm, where they colocalized with Hoechst 33342 nuclear dye in root apex cells (Figures 5A to 5C and 5G to 5I). GFP fluorescence was also detected in cells of the root elongation zone in transgenic plants carrying the transgenes driven by the 35S promoter (Figures 5D to 5F and 5J to 5L).
Figure 5.
Subcellular Localization of ICU11 and CP2.
Confocal laser scanning micrographs of icu11-1 ICU11pro:ICU11:GFP ([A] to [C]), icu11-1 35Spro:ICU11:GFP ([D] to [F]), CP2pro:CP2:GFP ([G]to [I]), and 35Spro:CP2:GFP ([J] to [L]) plants. Different parts of the root are shown: apex ([A] to [C] and [G] to [I]) and elongation zone ([D] to [F] and [J] to [L]). Fluorescent signals correspond to the Hoechst 33342 nuclear dye ([A], [D], [G] and [J]), GFP ([B], [E], [H], and [K]), and their overlay ([C], [F], [I], and [L]). Bars = 50 µm in (A) to (C) and (G) to (I) and 20 µm in (D) to (F) and (J) to (L).
As expected based on their functional redundancy, both ICU11 and CP2 proteins appear to function in the same subcellular compartment, the nucleus. We also examined the expression patterns of GFP fusion proteins produced by transgenes driven by the endogenous ICU11 and CP2 promoters in roots, and we found that both were expressed in the root tip and incipient lateral roots (Figures 4C, 4D, and 4I).
The Promoters, but Not the Protein Products of ICU11 and CP2, Are Functionally Equivalent
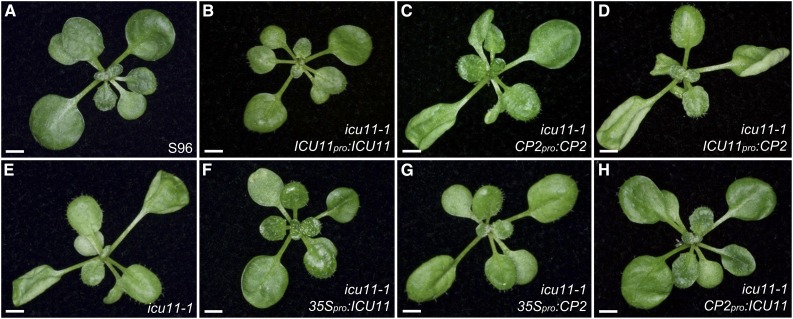
Because the expression patterns of ICU11 and CP2 overlap, we tested the interchangeability of their promoters and protein products in an attempt to explain their unequal functional redundancy. Hence, we generated transgenes with swapped ICU11 and CP2 promoters and transgenes with a double 35S promoter overexpressing either ICU11 or CP2.
Plants homozygous for icu11-1 and the CP2pro:CP2 or ICU11pro:CP2 transgenes were phenotypically mutant, as shown in five and eight independent lines, respectively (Figures 6A and 6C to 6E), showing that CP2 cannot substitute for ICU11. By contrast, in five out of six independent lines, icu11-1 CP2pro:ICU11 plants were phenotypically wild type (Figures 6A, 6E, and 6H), confirming the overlapping expression patterns of ICU11 and CP2. In contrast to our previous results, the icu11-1 35Spro:CP2 plants also exhibited full phenotypic complementation in five out of eight independent lines (Figures 6A, 6E, and 6G). To explain this unexpected observation, we used qRT-PCR to evaluate the CP2 transcript levels in three of the above icu11-1 35Spro:CP2 lines. These plants exhibited increases in CP2 transcript levels of 50- to 112-fold compared with S96, and this was sufficient to complement the lack of ICU11 function (Supplemental Figure 10). We also examined three CP2pro:CP2 and ICU11pro:CP2 lines and found that their CP2 transcript levels were 1.4- to 4.1-fold (Supplemental Figure 10) higher than those of the wild type, but not enough to complement the mutant phenotype of icu11-1. In addition, icu11-1 plants showed a 2.5-fold increase in CP2 transcript levels, suggesting transcriptional feedback between these two paralogs. These results suggest that only high levels of CP2 can substitute for ICU11.
Figure 6.
Effects of Interchanging the ICU11 and CP2 Promoters on the Phenotype of icu11-1.
(A) and (E) Rosettes of S96 (A) and icu11-1 (E).
(B) to (D) and (F) to (H) Transgenic plants in the icu11-1 background expressing ICU11 ([B], [F], and [H]) under the control of its endogenous promoter (B), the constitutive 35S promoter (F), and the CP2 promoter (H), and CP2 ([C], [D], and [G]) under the control of its endogenous promoter (C), the ICU11 promoter (D), and the 35S promoter (G). Photographs were taken 16 DAS. Bars = 2 mm.
ICU11 and CP2 Participate in an Epigenetic Pathway Independent of DNA Methylation
Genes required for flower development are repressed by PRC2 and PRC1 in Arabidopsis. For example, loss-of-function alleles of EMF2 (Yoshida et al., 2001) and EMF1 (Calonje et al., 2008), which encode PRC components, exhibit derepression of genes required for flower development. Upon germination, these mutants activate flowering, rather than initiating vegetative growth (Sung et al., 1992). These defects were also observed in icu11 cp2 double mutants, which had carpel- and anther-like structures instead of rosette leaves (Figures 3I to 3K and 3N to 3P; Supplemental Figure 8). The similar phenotypes of icu11 cp2 double mutant and emf single mutant seedlings suggested that ICU11 and CP2 participate in epigenetic repression of flowering genes.
Some members of the 2OGD superfamily function as demethylases. In Arabidopsis, several JUMONJI proteins have been shown to function as histone demethylases (Chen et al., 2011b), and two members of the ALKBH (DOXA) group have been recently shown to demethylate viral RNA (ALKBH9B) (Martínez-Pérez et al., 2017) or endogenous mRNA (ALKBH10B) (Duan et al., 2017). To determine whether ICU11 plays a role in DNA methylation, we performed whole-genome bisulfite sequencing of S96 and icu11-1 seedlings. We first determined the methylation patterns across the five Arabidopsis chromosomes, distinguishing the CG, CHG, and CHH methylation contexts (Supplemental Figure 11). We also determined the average methylation ratio in protein-coding genes and transposable elements (Supplemental Figure 12). However, no differences were found between icu11-1 and S96, indicating that ICU11 does not affect gene expression through DNA methylation.
ICU11 Synergistically Interacts with Genes Encoding Components of the Epigenetic Machinery
Studies of mutants with involute leaves and early flowering, such as clf and icu2-1, led to the identification of Arabidopsis genes involved in epigenetic regulation (Goodrich et al., 1997; Barrero et al., 2007). Evidence that CLF and ICU2 participate in epigenetic regulation is the synergistic genetic interaction of their alleles with each other and with alleles of other genes involved in epigenetic phenomena. We obtained double mutant combinations of either icu11-1 or icu11-2 with loss-of-function alleles of the following genes: ICU2 and GIGANTEA SUPPRESSOR5 (GIS5), encoding the catalytic subunits of DNA polymerase α and δ, respectively, which are involved in the deposition of epigenetic marks (Barrero et al., 2007; Hyun et al., 2013; Iglesias et al., 2015); CLF, encoding a PRC2 methyltransferase that deposits the H3K27me3 repressive mark (Goodrich et al., 1997); TERMINAL FLOWER2 (TFL2; also known as LIKE HETEROCHROMATIN PROTEIN1), whose product is an epigenetic repressor that recognizes H3K27me3 and physically associates with PRC2 components (Kotake et al., 2003; Turck et al., 2007; Zhang et al., 2007; Derkacheva et al., 2013; Liang et al., 2015); FASCIATA1 (FAS1), encoding a subunit of the heterotrimeric complex chromatin assembly factor-1 (CAF-1) that is involved in the assembly of acetylated histones onto newly synthesized DNA during replication (Kaya et al., 2001); and EARLY BOLTING IN SHORT DAYS (EBS), encoding a chromatin remodeling factor that regulates flowering time (Piñeiro et al., 2003). All of the double mutant combinations of icu11-1 or icu11-2 with loss-of-function alleles of ICU2, GIS5, CLF, TFL2, FAS1, or EBS exhibited synergistic phenotypes, with a strong reduction in body size; double mutant combinations of icu11 alleles with icu2-1, clf-2, or gis-5 showed extremely involute leaves (Figures 7H to 7J, 7O, and 7P). Also, development of the icu11 ebs-1 and icu11 fas1-1 double mutants was arrested at early vegetative stages (Figures 7L, 7M, 7R, and 7S), and the icu11 tfl2-2 double mutants displayed phenotypes similar to those of the emf single mutants and the icu11 cp2 double mutants (Figures 7K and 7Q). Synergistic double mutant phenotypes mainly result from mutations in functionally related genes (Pérez-Pérez et al., 2009a). Therefore, all these synergistic interactions provide further genetic evidence of the functional relationship between ICU11 and the epigenetic machinery.
Figure 7.
Genetic Interactions between icu11 Alleles and Loss-of-Function Alleles of Genes Encoding Components of the Epigenetic Machinery.
(A) to (G) and (N) Single mutants icu11-1 (A), gis-5 (B), icu2-1 (C), clf-2 (D), tfl2-2 (E), ebs-1 (F), fas1-1 (G), and icu11-2 (N).
(H) to (M) and (O) to (S) Double mutants icu11-1 gis-5 (H), icu11-1 icu2-1 (I), icu11-1 clf-2 (J), icu11-1 tfl2-2 (K), icu11-1 ebs-1 (L), icu11-1 fas1-1 (M), icu11-2 icu2-1 (O), icu11-2 clf-2 (P), icu11-2 tfl2-2 (Q), icu11-2 ebs-1 (R), and icu11-2 fas1-1 (S).
Photographs were taken 16 DAS. Bars = 2 mm.
Genes Related to Flowering and Floral Development Are Derepressed in icu11-1 Plants
To obtain molecular evidence for the effects of ICU11 on the transcription of genes known to be epigenetically regulated, we performed RNA-seq analysis of S96 and icu11-1 seedling aerial tissues collected at 10 DAS. We thus identified 840 genes with altered expression patterns in the mutant, including 403 upregulated and 437 downregulated genes (Supplemental Data Set 1). Among the upregulated genes, the most significantly enriched Gene Ontology categories were related to reproductive structures and flower development (Supplemental Data Set 2A). The most highly enriched domains among proteins encoded by the upregulated genes were the keratin-like (K-box) and MADS-box domains (Supplemental Data Set 3A), which are characteristic of the type II subfamily of MADS-box genes, including AGAMOUS (AG) (Yanofsky et al., 1990), AGAMOUS-LIKE42 (AGL42) (Chen et al., 2011a), SHATTERPROOF2 (SHP2; also known as AGL5) (Savidge et al., 1995), SEEDSTICK (STK; also known as AGL11) (Rounsley et al., 1995), APETALA3 (AP3) (Jack et al., 1992), SEPALLATA1 (SEP1), SEP2, and SEP3 (Pelaz et al., 2000), and MADS AFFECTING FLOWERING5 (MAF5) (Kim and Sung, 2010). FLOWERING LOCUS T (FT) (Kobayashi et al., 1999), which does not belong to the MADS-box family, was also upregulated in icu11-1. Among downregulated genes, the most significantly enriched categories were related to stress, stimulus, and defense response genes (Supplemental Data Sets 2B and 3B), as was observed in the icu2-1 mutant (Micol-Ponce et al., 2015).
To validate our RNA-seq results and to further quantify the observed derepression, we analyzed the expression of the 10 genes mentioned above by qRT-PCR in icu11-1 plants. These genes were strongly upregulated (6.5- to 224-fold) in the mutant compared with the wild type (Supplemental Figure 13), demonstrating ectopic and heterochronic activation of flower development programs during the postgerminative phase in icu11-1. To account for local effects on DNA methylation, we determined the methylation levels of these genes, which were found to have almost no methylation, suggesting that their upregulation is not dependent on their methylation pattern (Supplemental Figure 14). In addition, the DNA methylation patterns of downregulated and upregulated genes in icu11-1 plants were indistinguishable from those of S96 (Supplemental Table 3). Taken together, the transcriptomic results, the embryonic flower phenotypes exhibited by icu11 cp2 double mutants and sesquimutants, and the synergistic genetic interactions of icu11 alleles with alleles of genes encoding epigenetic machinery components indicate that the ICU11 and CP2 proteins participate in the epigenetic regulation of Arabidopsis gene expression.
ICU11 and CP2 Are Required for Modulation of Histone Modifications at the Chromatin of Several MADS-Box Genes, Including SEP3
The most severely misregulated genes found in our RNA-seq analysis included the three SEP genes (Supplemental Figure 13). SEP3 is repressed during vegetative development through the deposition of repressive epigenetic marks and the removal of activating epigenetic marks. SHORT VEGETATIVE PHASE (SVP) recruits TFL2 to modulate H3K27me3 marks along the first intron of SEP3 (Liu et al., 2009). SUPPRESSOR OF THE OVEREXPRESSION OF CONSTANS1 (SOC1) and AGL24 physically interact with SIN3 ASSOCIATED POLYPEPTIDE P18 (SAP18) to prevent histone acetylation at SEP3, probably by recruiting the histone deacetylase complex (HDAC) (Liu et al., 2009).
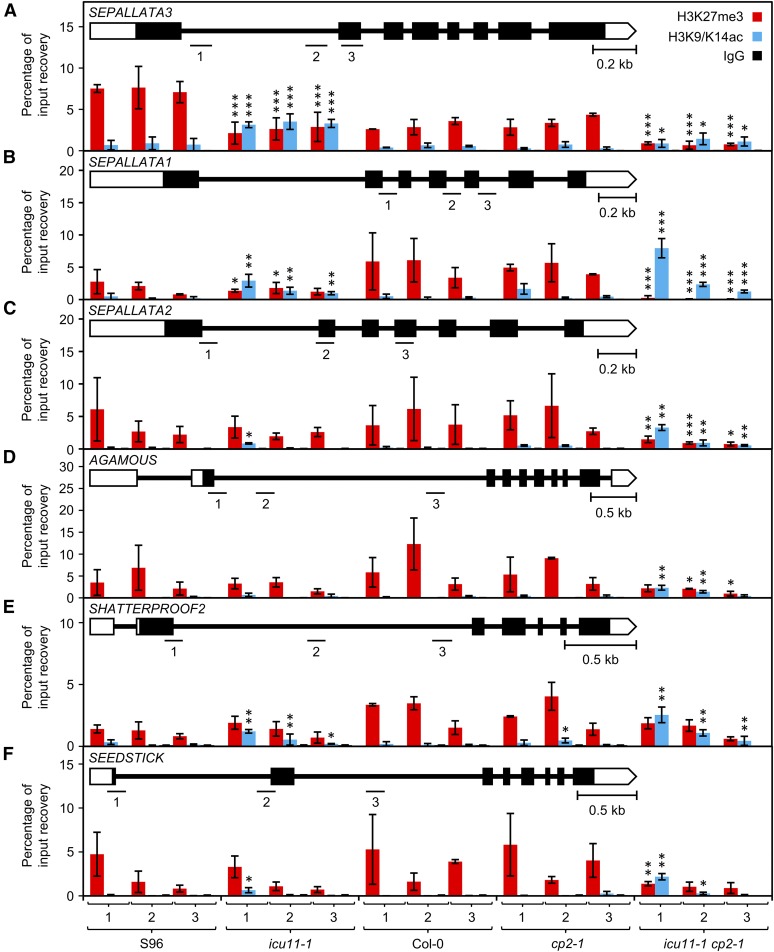
To investigate whether the misregulation of SEP3 observed in icu11-1 plants is caused by the alteration in chromatin modifications, we performed chromatin immunoprecipitations (ChIP) to quantify H3K27me3 and H3K9/K14ac enrichment at the first intron of SEP3 in S96, icu11-1, Col-0, cp2-1, and icu11-1 cp2-1 plants. The level of coimmunoprecipitated DNA in the cp2-1 mutant was indistinguishable from that of wild-type Col-0 in both ChIP analyses (Figure 8A). A significant reduction in H3K27me3 levels was found in icu11-1 compared with wild-type S96 plants, which might explain not only the strong upregulation of SEP3 but also its leaf phenotype (Figure 8A). Consistent with their strong morphological phenotype, icu11-1 cp2-1 seedlings showed the largest reduction in H3K27me3 levels (Figure 8A), demonstrating that both ICU11 and CP2 are required for the deposition of this repressive mark in SEP3 during vegetative development.
Figure 8.
ChIP Analysis of H3K27me3 and H3K9/K14ac Levels at the SEP3, SEP1, SEP2, AG, SHP2, and STK Genes.
Structure and ChIP analysis of the SEP3 (A), SEP1 (B), SEP2 (C), AG (D), SHP2 (E), and STK (F) genes. The numbered horizontal lines indicate regions that were analyzed by qPCR after ChIP. Bars indicate the mean of the percentage of input recovery in ChIP experiments using seedlings collected 10 DAS and antibodies against H3K27me3 (red), H3K9/14ac (blue), or IgG (black). Error bars indicate sd. Asterisks indicate values significantly different from the corresponding wild type in a Mann-Whitney U test (*P < 0.05, **P < 0.01, and ***P < 0.001). Three biological replicates were analyzed in triplicate.
We further analyzed H3K27me3 levels in other genes that we found to be severely deregulated in icu11-1 plants, namely SEP1, SEP2, AG, SHP2, and STK. Small differences between S96 and icu11-1 were only found in SEP1 (Figure 8B), and a consistent pattern was found in the icu11-1 cp2-1 double mutant, which showed a strong decrease of H3K27me3 levels in SEP1, SEP2 (Figures 8B and 8C), and AG (Figure 8D) and lesser decreases in STK (Figure 8F). No differences were found among the genotypes analyzed for H3K27me3 enrichment in SHP2 (Figure 8E). The differential behavior of icu11-1 and cp2-1 in terms of the deposition of epigenetic marks is in agreement with both the absence of a visible leaf mutant phenotype in cp2-1 (Figures 3C and 3D) and the unequal functional redundancy between ICU11 and CP2 (Figures 3P and 3Q).
Patterns of H3K9/K14ac were, in most cases, complementary to those of H3K27me3, with a significant increase in the levels of this activating mark in the SEP3, SEP1, SEP2, SHP2, and STK genes of the icu11-1 mutant compared with S96 (Figure 8). The H3K9/K14ac levels were indistinguishable in Col-0 and cp2-1 in all loci analyzed, with SHP2 being the only exception. Similar to H3K27me3, levels of H3K9/K14ac were more altered in the icu11-1 cp2-1 double mutant compared with the wild type in SEP1, SEP2, AG, SHP2, and STK (Figures 8B to 8F). Although slightly different from S96, the H3K9/K14ac levels of SEP3 in icu11-1 cp2-1 appeared to result from the addition of these levels in icu11-1 and cp2-1 (Figure 8A).
Our ChIP results suggest that ICU11 and CP2 work redundantly to repress several members of the MADS-box transcription factors family, during vegetative development via histone modification. The different H3K27me3 levels of S96 and Col-0 might be an example of natural variation among Arabidopsis accessions (Moghaddam et al., 2011).
The Leaf Phenotype of icu11 Mutants Is Caused by Ectopic Derepression of SEP3
Derepression of SEP3 due to constitutive expression (Castillejo et al., 2005) or mutations in chromatin remodelers accelerates flowering and generates involute leaves (Honma and Goto, 2001; López-Vernaza et al., 2012; Fernandez et al., 2014; Iglesias et al., 2015). Moreover, our ChIP analysis revealed epigenetic alterations at the SEP3 locus in icu11-1 plants (Figure 8A), resulting in increased levels of SEP3 transcript (Supplemental Figure 13).
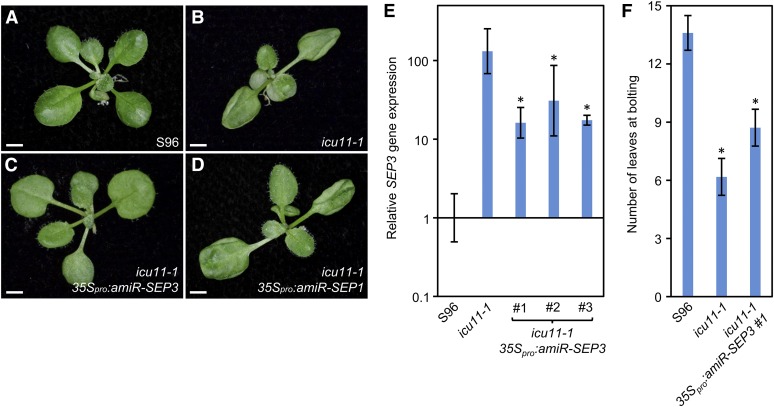
To ascertain whether SEP3 upregulation causes some of the phenotypes of the icu11-1 mutant, we wanted to obtain a sep3 icu11-1 double mutant. However, the ICU11 and SEP3 genes are linked and no insertional alleles of SEP3 are available in the S96 genetic background. Therefore, instead of obtaining the sep3 icu11-1 double mutant, we transformed icu11-1 plants (in the S96 genetic background) with a transgene designed to produce an artificial microRNA (amiRNA) targeting SEP3 transcripts (Iglesias et al., 2015); this transgene should reduce SEP3 expression. Seven transformants carrying the 35Spro:amiR-SEP3 transgene were obtained, all of which showed a clear abolition of leaf hyponasty (Figures 9A to 9C). We also transformed icu11-1 plants with the 35Spro:amiR-SEP1 transgene, obtaining 12 transformants, and this had no effect on the leaf phenotype of the mutant (Figures 9A, 9B, and 9D). We also used qRT-PCR to measure SEP3 transcript levels in seedlings of three independent icu11-1 35Spro:amiR-SEP3 lines (Figure 9E), finding that they were reduced, but not to wild-type levels (from more than 130-fold to 15-fold more than the wild type). This result suggests that a threshold exists for the morphological effects of SEP3 overexpression in leaves. In addition, the early flowering of icu11-1 was not suppressed by 35Spro:amiR-SEP3 (Figure 9F), indicating that this phenotypic trait has one or more other causal genes. These results suggest that the vegetative leaf phenotype of icu11-1 is due to ectopic and heterochronic derepression of SEP3, which is consistent with the previous findings mentioned above (Honma and Goto, 2001; Castillejo et al., 2005; López-Vernaza et al., 2012; Fernandez et al., 2014; Iglesias et al., 2015). However, the early flowering of this mutant does not appear to be caused by SEP3 derepression. Since the icu2-1 ag-1 double mutant exhibits suppression of the leaf phenotype of icu2-1 (Barrero et al., 2007), we obtained the icu11-1 ag-1 double mutant, finding that its leaves were similar to those of icu11-1 (Supplemental Figure 15).
Figure 9.
Effects of Posttranscriptional Silencing of SEP3 on the Phenotype of icu11-1.
(A) to (D) Rosettes of S96 (A), icu11-1 (B), and icu11-1 plants expressing an amiRNA targeting SEP3 (C) and SEP1 (D). Photographs were taken 14 DAS. Bars = 2 mm.
(E) qRT-PCR analysis of the relative expression of SEP3. RNA was extracted from rosette leaves collected 10 DAS. Bars indicate relative expression of SEP3 in S96 (only the error bar is visible), icu11-1, and three independent transgenic lines of icu11-1 35Spro:amiR-SEP3 seedlings. Error bars indicate the interval delimited by 2–ΔΔCT±sd, where sd is the sd of the ΔΔCT values. Asterisks indicate values significantly different from icu11-1 in a Mann-Whitney U test (*P < 0.001). Three biological replicates were analyzed in triplicate.
(F) Flowering time in S96, icu11-1, and icu11-1 35Spro:amiR-SEP3 plants, determined under continuous light. Error bars indicate sd. Asterisks indicate values significantly different from S96 in a Mann-Whitney U test (*P < 0.001). Nine plants grown in individual pots (see Methods) were analyzed per genotype.
DISCUSSION
SEP3 Derepression Is the Major Contributor to Leaf Hyponasty in icu11 Mutants
We conducted genetic screens for Arabidopsis mutants with abnormally shaped or sized leaves (Berná et al., 1999; Serrano-Cartagena et al., 1999; Wilson-Sánchez et al., 2014). In our screens, gene morphology relationships among mutants were reproducible and in some cases predictable: Mutations classified together based on morphological phenotype affected genes involved in a single pathway or molecular mechanism (Nelissen et al., 2005; Horiguchi et al., 2011; Rubio-Díaz et al., 2012). One of the most highly represented phenotypic class in our screens was the icu mutants, most of which carried alleles of genes that fell into three functional classes: genes involved in auxin homeostasis, miRNA metabolism, or epigenetic pathways (Serrano-Cartagena et al., 2000; Pérez-Pérez et al., 2009b).
Some mutations causing leaf hyponasty directly or indirectly affect intrinsic leaf organogenesis mechanisms. For example, mutations in HYPONASTIC LEAVES1, which participates in miRNA biogenesis, alter miRNA160 and miRNA319 levels. These miRNAs regulate genes encoding transcription factors that pattern abaxial and adaxial cell fate, such as FILAMENTOUS FLOWER and PHABULOSA, respectively (Liu et al., 2011). However, in other mutants with hyponastic leaves, the causal genes of their phenotypes are involved in epigenetic processes (Serrano-Cartagena et al., 2000); leaf hyponasty in these mutants is not caused by the failure of intrinsic mechanisms determining leaf flatness but by the ectopic derepression of floral development-related genes in leaves.
Eight icu1 mutants (Serrano-Cartagena et al., 2000) were found to carry alleles of the PcG gene CLF (Goodrich et al., 1997). We also found that icu2-1 is an allele of ICU2, which encodes the catalytic subunit of DNA polymerase α, an essential protein involved in DNA replication as well as in the deposition of histone epigenetic marks (Serrano-Cartagena et al., 2000; Barrero et al., 2007; Hyun et al., 2013; Micol-Ponce et al., 2015). In these icu1 (clf) and icu2 mutants, flower developmental programs are globally derepressed, but only a few genes contribute to altered leaf flatness. In an early study, overexpression of AG caused leaf hyponasty in 35Spro:AG transgenic plants (Mizukami and Ma, 1992). AG derepression was subsequently detected in the clf and icu2 single mutants, whose hyponastic leaves were restored to flatness in the clf ag and icu2 ag double mutants (Goodrich et al., 1997; Barrero et al., 2007). Leaf flatness was also restored in clf sep3 double mutants (López-Vernaza et al., 2012). Moreover, like transgenic plants harboring 35Spro:AG, 35Spro:SEP3 transgenic plants also showed involute leaves. As described for clf and icu2, the icu11 mutants exhibited leaf hyponasty, and this was associated with the generalized derepression of flower development genes in leaves. The depletion of AG transcripts restored leaf flatness in clf and icu2, but not in the icu11 mutants. The depletion of SEP3 transcripts suppressed leaf hyponasty in the icu11 mutants, as observed in icu11-1 35Spro:amiR-SEP3 plants. We conclude that the leaf phenotype of the icu11 mutants is caused by ectopic and heterochronic derepression of SEP3.
ICU11 Is a Component of the Epigenetic Machinery
In addition to leaf hyponasty, the icu2-1 and clf mutants share other morphological and molecular phenotypes, including early flowering and ectopic and heterochronic derepression of many genes, including MADS-box transcription factor genes and those of other transcription factor families. The icu2-1 and clf mutations also render very similar synergistic genetic interactions in double mutant combinations with loss-of-function alleles of genes such as TFL2 and FAS1, which encode components of the epigenetic machinery.
The icu11-1 and icu11-2 mutants exhibited similar morphological and molecular phenotypes and similar genetic interactions to those of the icu2-1 and clf mutants. Further evidence for an epigenetic role for ICU11 was provided by the reduced deposition of the H3K27me3 repressive mark at the SEP3 and SEP1 genes in the icu11 mutants, as well as at the SEP2, AG, and STK genes on the icu11-1 cp2-1 double mutant. Additionally, increased levels of the H3K9/K14ac mark were detected in SEP1, SEP2, SEP3, SHP2, and STK, suggesting a role for ICU11 beyond H3K27me3. We also found that ICU11 is a nucleoplasmic protein and that DNA methylation seems normal in the icu11-1 mutant. In conclusion, genetic and molecular evidence indicates that ICU11 participates in an epigenetic, DNA methylation-independent pathway that is required for histone modification. ICU11 likely performs an ancient function, as suggested by the presence of an ortholog in M. polymorpha and two in P. patens.
ICU11 and CP2 Are 2OGD Proteins That Act as Epigenetic Repressors through an Unknown Mechanism
To date, more than 130 plant genes have been identified that encode epigenetic modifiers in a broad sense, and these can be grouped into five classes: regulators of DNA modification (e.g., cytosine methylation), histone-modifying enzymes (including methylating, acetylating, phosphorylating, and ubiquitinating enzymes), PcG proteins and their interactors, nucleosome-organizing proteins (also known as chromatin remodelers), which control the accessibility of transcription factors to DNA, and protein and RNA molecules that participate in RNA-directed DNA methylation (Chen and Dent, 2014; Pikaard and Mittelsten Scheid, 2014).
The 2OGD superfamily includes some 150 annotated members in Arabidopsis, 128 of which were assigned to three classes by Kawai et al. (2014). ICU11 is one of the two putative 2OGDs that remained unclassified in that study. Four paralogs of ICU11 are also annotated as 2OGDs in the TAIR10 and Araport11 databases (Cheng et al., 2017), but they were not described by Kawai et al. (2014). We have named the family formed by ICU11, CP2, CP3, CP4, and CP5 the CUPULIFORMIS (CP) family. Little information is currently available about these five genes in public databases. The CP genes clustered into two clades, one of which includes ICU11 and CP2.
Many functionally redundant paralogous gene pairs have been described, most of which encode components of signal transduction, metabolic, and developmental pathways, as well as ribosomal proteins (Kafri et al., 2009). Functionally redundant gene pairs originated by ancient gene duplication; these duplicated genes do not diverge but instead remain redundant. Some of these gene pairs appear to be evolutionarily stable, for up to 100 million years in some cases, suggesting that they contribute positively to the fitness of the organism by reducing the phenotypic cost of mutations (Dean et al., 2008). The first example of unequal redundancy is the Arabidopsis APETALA1 (AP1) and CAULIFLOWER (CAL) genes: cal mutants are phenotypically wild type, and ap1 mutants exhibit flower organ homeotic transformations that are strongly enhanced in cal ap1 double mutants (Kempin et al., 1995; Briggs et al., 2006). We found that ICU11 and its closest paralog, CP2, also exhibit unequal genetic redundancy. Indeed, although the cp2 null mutants show mildly reduced fertility but are otherwise indistinguishable from the wild type, the icu11 cp2 double mutants suffer postgerminative lethality.
Several members of the 2OGD superfamily are known components of the epigenetic machinery. Some JmjC proteins function in H3K4, H3K27, and H3K36 demethylation in Arabidopsis (Xiao et al., 2016). DNA demethylase activity has also been demonstrated for the human 2OGD protein TET1 (similar to the Arabidopsis DOXA proteins) in cultured human cells (Tahiliani et al., 2009). The Arabidopsis DOXA protein ALKBH2 participates in DNA demethylation in response to damage by alkylating agents (Meza et al., 2012), and ALKBH10B is an mRNA N6-methyladenosine demethylase involved in the floral transition (Duan et al., 2017). In contrast to other described 2OGDs, ICU11 and CP2 appear to function as epigenetic repressors. No other 2OGD has thus far been shown to be required for either H3K27 methylation or vegetative development. The redundant protein pair ICU11-CP2 is the only example of 2OGDs required for histone methylation, and they are the only P4Hc domain-containing proteins known to be related to the epigenetic machinery.
The icu11 cp2 Double Mutant Synergistic Phenotype Suggests That the ICU11-CP2 Gene Pair Is Functionally Related to PRC1 or PRC2
Few Arabidopsis mutants described to date lack a vegetative phase, a phenotype shown by some emf1 and emf2 single mutants and the icu11 cp2 double mutants and sesquimutants described in this study. These phenotypes have also been found in mutants or transgenic plants with depleted PRC1 or PRC2 activity.
SWINGER (SWN), the closest paralog of CLF, is a PRC2 component. Unequal functional redundancy exists between CLF and SWN: The lack of function of SWN does not cause a mutant phenotype (Chanvivattana et al., 2004), but swn clf double mutants exhibit severe phenotypes similar to those of emf mutants (Chanvivattana et al., 2004). Some PRC1 subunits are encoded by paralogous genes. For example, the function of PRC1 is severely compromised in ring1a ring1b double mutants (Chen et al., 2010) and in bmi1a bmi1b bmi1c triple mutants (Yang et al., 2013), which also yield seedlings exhibiting emf-like structures and skip the vegetative phase. Unlike PRC2, which has histone methylase activity, histone demethylases excise methyl groups to fine tune gene expression. RELATIVE EARLY FLOWERING6 (REF6; also called JMJ12) encodes a member of the JmjC subfamily of 2OGDs that has H3K27me2/me3 demethylase activity (Lu et al., 2011). Mutations in REF6 cause a generalized increase in H3K27me3 methylation, affecting several hundred genes. Conversely, overexpression of REF6 reduces H3K27me3 levels, generating phenotypes reminiscent of those of emf single mutants and icu11 cp2 double mutants (Lu et al., 2011).
Although many epigenetic machinery components are known in Arabidopsis, only a few mutants or transgenic plants have been reported to skip the vegetative phase, as observed in icu11 cp2 double mutants. In addition, all reported cases are directly or indirectly related to the dysfunction of PRC1 or PRC2, suggesting that these complexes and the ICU11-CP2 redundant gene pair share a functional relationship.
ICU11 and CP2 Broaden the Spectrum of Plant Epigenetic Factors
Plants undergo several developmental transitions in their life cycle: the embryogenesis, seed maturation, juvenile, adult vegetative, adult reproductive, and gametophytic stages. The success of all these critical phase transitions relies on the proper integration of endogenous and environmental cues by sets of genes that regulate developmental programs (Huijser and Schmid, 2011). The contribution of transcription factors, posttranscriptional activities of noncoding RNAs, and the epigenetic machinery to plant development as a whole and to phase transitions in particular is a matter of intense interest.
In animals, the establishment and maintenance of cell fates is thought to be controlled (to a large extent) by the epigenetic machinery, and differentiation is considered to be essentially an epigenetic process. In mammals, key developmental embryonic transitions, such as those that occur within blastocysts and during gastrulation, as well as adult cell fate decisions, are regulated by epigenetic factors. Our current understanding of the developmental roles of the components of the metazoan epigenetic machinery was in part obtained from studies of mammalian pluripotent stem cells, which became the workhorse behind investigations of normal embryonic development and of human diseases caused by erroneous repression or activation of regulatory pathways (Lee and Young, 2013; Chen and Dent, 2014; Feinberg et al., 2016).
Several facets of plant physiology and development are known to be modulated by the epigenetic machinery, including flowering time, male and female gametogenesis, stress responses, light signaling, and morphological plasticity. The mitotically heritable processes known to be under epigenetic control in plants include imprinting, vernalization, acclimation, and systemic acquired resistance (Pikaard and Mittelsten Scheid, 2014). The small number of epigenetically regulated processes in plants suggests that epigenetics plays a minor role in plants compared with animals, or, perhaps, that much remains to be learned about plant epigenetics. Further examination of the interactions of CP proteins with PcG proteins will help us to determine the function of CP proteins, and future analyses of the role of the ICU11-CP2 gene pair will shed light on the molecular nature of their epigenetic activity. Together, these should provide insight into the number and diversity of processes regulated by the epigenetic machinery in plants.
METHODS
Plant Material, Culture Conditions, and Crosses
Unless otherwise stated, all Arabidopsis thaliana plants studied in this work were homozygous for the mutations indicated. The Nottingham Arabidopsis Stock Center provided seeds for the wild-type accessions Landsberg erecta (Ler, NW20), Col-0 (N1092), S96 (N914), and Wassilewskija-2 (Ws-2, N1601), as well as seeds for icu11-1 (N242, in the S96 background), SALK_051945 (N551945, Col-0), SALK_054985 (N554985, Col-0), clf-2 (N8853, Ler; Goodrich et al., 1997), tfl2-2 (N3797, Col-0; Kotake et al., 2003), icu2-1 (N329, En-2; Barrero et al., 2007), fas1-1 (N265, En-2; Reinholz, 1966), cp2-1 (N861581, Col-0), cp2-2 (N828642, Col-0), and cp2-3 (N826626, Col-0). Seeds for ebs-1 (Piñeiro et al., 2003) were provided by Manuel Piñeiro, and gis-5 (Iglesias et al., 2015) seeds were provided by Pablo D. Cerdán. Seeds of the icu11-2 line (FLAG_402G04; EGZ248; in the Ws-2 background) were provided by the Versailles Arabidopsis Stock Center (Brunaud et al., 2002). The presence and positions of all T-DNA insertions were confirmed by PCR amplification using gene-specific primers, and the LbB1.3, LB1, and LB4 primers for the SALK, SAIL, and FLAG T-DNA insertions, respectively (Supplemental Table 1).
Unless otherwise stated, plants were grown under sterile conditions on 150-mm Petri dishes containing 100 mL half-strength Murashige and Skoog agar medium with 1% sucrose at 20°C ± 1°C, 60 to 70% relative humidity, and continuous illumination at 75 µmol m−2 s−1, as previously described (Ponce et al., 1998). Crosses were performed as previously described (Quesada et al., 2000).
Flowering Time Analysis
Flowering time was determined based on the total vegetative leaf number at bolting (leaves were counted when internode elongation was visible), as previously described (Andrés and Coupland, 2012). To determine flowering time, plants were grown in Petri dishes for 5 d and transferred to individual pots. All flowering time determinations were performed in the same Conviron TC30 growth chamber using different photoperiods: short day (8 h light/16 h dark), long day (16 h light/8 h dark), and continuous light.
Statistical Analysis
For hypothesis testing, we used the parametric Student’s t test when the number of observations (n) was 10 or more, under the assumption that most, if not all the data obtained were in the center of a normal distribution. Student’s t values were calculated at https://www.graphpad.com/quickcalcs/ttest1.cfm. The normality assumption, however, cannot be made for samples with n < 10, for which we used the nonparametric Mann-Whitney U test, which does not require any assumption on the type of distribution under study. Mann-Whitney U tests for two samples of unpaired data (also known as Wilcoxon signed rank tests) were run by calculating U values at http://www.socscistatistics.com/tests/mannwhitney/.
Gene Constructs
To construct the ICU11pro:ICU11 and CP2pro:CP2 transgenes (including the 3′ regions of ICU11 and CP2, respectively), 3.18- and 4.76-kb segments of chromosome 1 and chromosome 3 were PCR amplified from Col-0 genomic DNA using Phusion High Fidelity DNA Polymerase (Finnzymes, Thermo Fisher Scientific), as recommended by the manufacturer using oligonucleotides that contained attB sites at their 5′ ends. These constructs included genomic regions starting from nucleotide −889 of At1g22950 and −453 of At3g18210; they also included 382 nucleotides and 2154 nucleotides downstream of the respective stop codons. The PCR products obtained were purified using a 30% polyethylene glycol/30 mM MgCl2 solution and cloned into the pGEM-T Easy221 vector using a BP Clonase II Kit (Life Technologies). Chemically competent Escherichia coli DH5α cells were transformed by the heat-shock method (Dagert and Ehrlich, 1979), and the structural integrity of the inserts carried by the transformants was verified by Sanger sequencing in an ABI Prism 3130xl Genetic Analyzer (Applied Biosystems). The inserts in pGEM-T Easy221 were subcloned into the pMDC107 Gateway-compatible destination vector (Curtis and Grossniklaus, 2003) via an LR Clonase II (Life Technologies) reaction.
For the ICU11pro:ICU11:GFP and CP2pro:CP2:GFP transgenes, 2.79- and 2.58-kb segments of ICU11 and CP2, respectively, were amplified from Col-0 genomic DNA, cloned into pGEM-T Easy221, and subcloned into pMDC107 as described above. For the 35Spro:ICU11:GFP and 35Spro:CP2:GFP transgenes, the corresponding transcription units were amplified from Col-0 genomic DNA, cloned into pGEM-T Easy221, and subcloned into pMDC83 as described above. Primers were designed so that ICU11 and CP2 were fused in-frame with the GFP6 gene harbored by pMDC107 and pMDC83. For the ICU11pro:GUS and CP2pro:GUS transgenes, 2.66- and 2.50-kb segments from the upstream regions of the corresponding start codons of ICU11 and CP2, respectively, were amplified from Col-0 genomic DNA, cloned into pGEM-T Easy221, and subcloned into pMDC164, which harbors a gusA gene (Curtis and Grossniklaus, 2003). For the 35Spro:ICU11 and 35Spro:CP2 transgenes, 1.88- and 2.13-kb segments (from the start to stop codons) of ICU11 and CP2, respectively, were amplified from Col-0 genomic DNA, cloned into pGEM-T Easy221, and subcloned into pMDC32, which harbors a double 35S promoter (Curtis and Grossniklaus, 2003).
For promoter interchange analyses, promoters and transcription units were separately amplified from Col-0 genomic DNA and fused in a second amplification. All primers used in these PCR amplifications are described in Supplemental Table 1. For example, for the CP2pro:ICU11 transgene, the CP2 promoter was amplified using the CP2pro_F and CP2pro_R primers, and ICU11 was amplified using ICU11_F and ICU11_R; both PCR products were fused using CP2pro_F and ICU11_R. The final PCR products were purified using an Illustra GFX PCR DNA and Gel Band Purification Kit (GE Healthcare Biosciences), cloned into pGEM-T Easy221, and subcloned into pMDC107.
All constructs were mobilized into Agrobacterium tumefaciens GV3101 (C58C1 RifR) cells, which were used to transform S96, icu11, and Col-0 plants by the floral dip method (Clough and Bent, 1998). T1 transgenic plants were selected on plates supplemented with 15 mg L−1 hygromycin B (Invitrogen). The 35Spro:amiRNA-SEP3 and 35Spro:amiRNA-SEP1 transgenes were previously generated (Iglesias et al., 2015). T1 transgenic plants carrying the latter two transgenes were selected on plates supplemented with 50 mg L−1 kanamycin as previously described (Harrison et al., 2006).
Morphological and Histological Analyses
Morphometric analysis of palisade mesophyll and epidermal cells was performed as previously described (Pérez-Pérez et al., 2011). In brief, 10 first-node leaves were excised, cleared with chloral hydrate, and mounted on glass slides. Micrographs were taken of leaves halfway along the primary vein and leaf margin and were then transformed into diagrams by drawing cell outlines on the screen of a Cintiq 18SX Interactive Pen Display (Wacom) using Adobe Photoshop CS3. Individual cell areas were scored using the NIS Elements AR2.30 image analysis package (Nikon).
Confocal laser scanning microscopy images were obtained and processed using the operator software EZ-C1 for the Nikon C1 confocal microscope (Nikon Instruments). GFP was excited at 488 nm with an argon ion laser and its emission analyzed between 515 and 530 nm. To confirm the nuclear localization of ICU11:GFP and CP2:GFP, seedlings were stained with a 5 µg mL−1 solution of 2,5′-Bi-1H-benzimidazole, 2′-(4-ethoxyphenyl)-5-(4-methyl-1-piperazinyl) (Hoechst 33342; Thermo Fisher Scientific) for 5 min, washed extensively with water, and excited at 408 nm with a modulated diode laser.
Bioinformatic Analysis
To identify ICU11 paralogs, BLASTP searches (Altschul et al., 1997) were performed at NCBI using default parameters. Sequences were selected based on a BLAST E-value cutoff of 1 × 10−17 and a query coverage higher than 50%, aligned using ClustalW (Thompson et al., 1994), and shaded with BOXSHADE3.21 (https://embnet.vital-it.ch/software/BOX_form.html). A phylogenetic tree was constructed using the neighbor-joining clustering method inferred from 1000 replicates with MEGA6 (Tamura et al., 2013) using default parameters (model: Poisson; rates among sites: uniform rates; gaps/missing data treatment: complete deletion). Identity and similarity comparisons were performed using the “Iden and Sim” tool of the Sequence Manipulation Suite (Stothard, 2000; http://www.bioinformatics.org/sms2/ident_sim.html) from a Muscle alignment (Edgar, 2004). The computational tools used to predict the subcellular localization of CP proteins included SubLoc (Hua and Sun, 2001), WoLF PSORT (Horton et al., 2007), and YLoc (Briesemeister et al., 2010).
Mapping-by-Sequencing
ICU11 was cloned by combining traditional iterative linkage analysis (map-based cloning) with massively parallel sequencing. First, the icu11-1 mutation was mapped to a 32.6-kb candidate interval on chromosome 1 (Figure 2A) as previously described (Ponce et al., 1999, 2006). The icu11-1 mutant was then backcrossed to S96 and two F2 sibling populations were collected, comprising 150 phenotypically mutant and 150 phenotypically wild-type plants. DNA was extracted using a DNeasy Plant Mini Kit (Qiagen) and bulked into two samples. Both samples were subjected to massive parallel sequencing at the Beijing Genomics Institute (BGI) using the Illumina HiSeq 2000 platform. The resulting reads were mapped to TAIR10 using Bowtie 2 (Langmead and Salzberg, 2012), and mutations were listed usingSHORE (http://sourceforge.net/projects/shore/files; Schneeberger et al., 2009). Only one NMU-type mutation was found within the otherwise relatively short candidate interval, which was present in all sequencing reads from the phenotypically mutant population and in one-third of the reads from the phenotypically wild-type population.
RNA Isolation, cDNA Synthesis, and qRT-PCR
For qRT-PCR, total RNA was extracted using TRI reagent (Sigma-Aldrich). Contaminating DNA was removed using a TURBO DNA-free Kit (Invitrogen). First-strand cDNA was synthesized using random hexamers and the Maxima Reverse Transcriptase system (Fermentas). OTC was used as an internal control for relative expression analysis (Quesada et al., 1999). Three different biological replicates of the aerial tissues of five seedlings were used, except for Figure 4S, where three biological replicates of five first-node vegetative leaves, five whole root systems, five cauline leaves, or 15 open flowers were used. Biological replicates were analyzed in triplicate. PCR was performed in a 20-μL volume containing 7.5 μL of Maxima SYBR Green/ROX qPCR Master Mix (Fermentas), 5 μL of the corresponding primer pair (1.5 µM each), and 1 μL of cDNA template. Relative quantification of gene expression data was performed using the comparative CT method (2−ΔΔCT) on a Step One Plus System (Applied Biosystems). The results obtained were analyzed with the Expression Suite Software v1.1 (Thermo Fisher Scientific) and Relative Expression Software Tool v2.0.13 (Pfaffl et al., 2002). The primer sets used are listed in Supplemental Table 1.
RNA-Seq Analysis
Total RNA was isolated from 100 mg of aerial tissues collected from S96 and icu11-1 seedlings at 10 DAS using TRIzol (Thermo Fisher Scientific). RNA quality was checked in a 2100 Bioanalyzer (Agilent Genomics) and its RNA integrity number (based on the height of different rRNA peaks in a microcapillary electrophoresis and obtained using the algorithm described in Schroeder et al., 2006) values were always found to be higher than 7. More than 60 µg of RNA per sample were sent to the BGI for massive parallel sequencing in an Illumina HiSeq 2000. Reads were mapped to TAIR10 using the 0.1.6-beta version of HISAT (Kim et al., 2015). Differentially expressed genes were detected based on a Poisson distribution with a fold change ≥ 2 and a false discovery rate ≤ 0.001. Biological process categorization of the differentially expressed genes was performed using the web-based tool DAVID (Huang et al., 2009a, 2009b). For Gene Ontology, terms within GOTERM_BP_ALL, GOTERM_CC_ALL, and GOTERM_MF_ALL were used. For protein domains, terms from the INTERPRO database were used.
Whole-Genome Bisulfite Sequencing
Genomic DNA was isolated from S96 and icu11-1 seedlings collected 10 DAS using a DNeasy Plant Maxi Kit (Qiagen). More than 6 µg of DNA per sample was sent to the BGI, where they were treated with a mixture of sodium bisulfite and sodium metabisulfite and massively sequenced in triplicate. Clean reads were mapped to TAIR10 using BSMAP (Xi and Li, 2009). Methylation levels were determined by dividing the number of reads covering each methylcytosine by the total number of reads covering that cytosine with the Python script methratio.py (Xiang et al., 2010).
ChIP
Chromatin was cross-linked by formaldehyde fixation of 0.5 g tissue from S96, icu11-1, Col-0, cp2-1, and icu11-1 cp2-1 seedlings collected 10 DAS. Chromatin was isolated as previously described (Lázaro et al., 2008) and sonicated in a Bioruptor Pico (Diagenode) three times for 5 min (30 s ON/OFF) intervals at 4°C. The H3K27me3 and H3K9/K14ac fractions were immunoprecipitated using 1 µg of the specific ChIP-seq grade polyclonal antibodies pAb-069-050 (lot number: A1821D) and pAb-005-050 (lot number: A381-004) (Diagenode) and an Auto Plant ChIP-seq Kit (Diagenode) in the IP-STAR automated system (Diagenode). Incubation times were 4, 11, and 0.5 h for antibody coating, immunoprecipitation reaction, and washes, respectively. The immunoprecipitated DNA was purified using an Auto IPure Kit v2 (Diagenode) and qPCR analyzed in a Step One Plus System (Applied Biosystems, now Thermo Fisher Scientific). qPCR amplifications were performed in a 20-μL volume including 10 μL of Maxima SYBR Green/ROX qPCR Master Mix (Fermentas), 1 μL of the corresponding primer pair (5 µM each), and 5 μL of a 1:10 dilution of immunoprecipitated DNA. The primers used are listed in Supplemental Table 1. The results were analyzed with the Expression Suite v1.1 (Thermo Fisher Scientific).
Accession Numbers
Sequence data from this article can be found at The Arabidopsis Information Resource (http://www.arabidopsis.org) under the following accession numbers: AG (At4g18960), SHP2 (At2g42830), STK (At4g09960), AGL42 (At5g62165), AP3 (At3g54340), CLF (At2g23380), CP2 (At3g18210), CP3 (At5g43660), CP4 (At1g48740), CP5 (At1g48700), EBS (At4g22140), FAS1 (At1g65470), FT (At1g65480), GIS5 (At5g63960), ICU2 (At5g67100), ICU11 (At1g22950), MAF5 (At5g65080), OTC (At1g75330), SEP1 (At5g15800), SEP2 (At3g02310), SEP3 (At1g24260), and TFL2 (At5g17690). The massive sequencing raw data from mapping-by-sequencing, RNA-seq, and bisulfite-seq experiments were deposited in the Short Read Archive sequence database (NCBI; https://www.ncbi.nlm.nih.gov/sra) under accession numbers SRP076217, SRP076215, and SRP076378, respectively.
Supplemental Data
Supplemental Figure 1. Leaf Cell Phenotypes of S96, icu11-1, and icu11-1 ICU11pro:ICU11 Plants.
Supplemental Figure 2. Fertility Traits Observed in icu11 and cp2 Mutants.
Supplemental Figure 3. Some Morphological, Histological, and Molecular Phenotypes of icu11-2.
Supplemental Figure 4. Alignment of the Amino Acid Sequences of Arabidopsis CP Proteins.
Supplemental Figure 5. Alignment of the Amino Acid Sequences of Arabidopsis ICU11 and its Orthologs in other Plant Species.
Supplemental Figure 6. Alignment of the Amino Acid Sequences of ICU11 Orthologs in Metazoa.
Supplemental Figure 7. Flowering Time in the cp2 Mutants.
Supplemental Figure 8. Aberrant Floral Organ Structures Observed in icu11 cp2 Double Mutants and Sesquimutants.
Supplemental Figure 9. Transgene-Mediated Complementation of the Mutant Phenotype of icu11-1 Using an ICU11:GFP Translational Fusion.
Supplemental Figure 10. Quantitative RT-PCR Analysis of CP2 Gene Expression.
Supplemental Figure 11. DNA Methylation along Each icu11-1 Chromosome.
Supplemental Figure 12. DNA Methylation Patterns across All icu11-1 Genes and Transposable Elements.
Supplemental Figure 13. Quantitative RT-PCR Analysis of the Expression of Several MADS-Box Genes and FT in icu11-1 Leaves.
Supplemental Figure 14. DNA Methylation Patterns of Genes Found Upregulated in icu11-1 Seedlings.
Supplemental Figure 15. Phenotypic Traits of the icu11-1 ag-1 Double Mutant.
Supplemental Table 1. Primer Sets Used in This Work.
Supplemental Table 2. Identity and Similarity among CP Protein Sequences.
Supplemental Table 3. DNA Methylation across Misregulated Genes in icu11-1.
Supplemental Data Set 1. Genes Found Deregulated in an RNA-Seq Analysis of icu11-1 Plants.
Supplemental Data Set 2. Gene Ontology Term Enrichment among Genes Deregulated in icu11-1 Plants.
Supplemental Data Set 3. Protein Domain Enrichment among Genes Deregulated in icu11-1 Plants.
Supplemental File 1. Text File of the Alignment Used for the Phylogenetic Analysis Shown in Figure 2C.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank P.D. Cerdán and M. Piñeiro for providing seeds and constructs, and J.M. Serrano and T. Trujillo for their excellent technical assistance. Research in the laboratory of J.L.M. was supported by grants from the Ministerio de Economía, Industria y Competitividad of Spain (BIO2014-53063-P) and the Generalitat Valenciana (PROMETEOII/2014/006). E.M.-B. and L.J.-V. held predoctoral fellowships from the Ministerio de Educación, Cultura y Deporte of Spain (FPU13/00371 and FPU16/03772) and D.E.-B. and R.N. from the Generalitat Valenciana (BFPI/2009/015 and GRISOLIAP/2016/131, respectively).
AUTHOR CONTRIBUTIONS
J.L.M. conceived and supervised the study, provided resources, and obtained funding. J.L.M., M.R.P., J.M.P.-P., D.E.-B., and E.M.-B. designed the methodology. F.M.L., H.C., E.M.-B, D.E.-B., L.J.-V., and R.N. performed the research. J.L.M., E.M.-B., and L.J.-V. wrote the original draft. All authors reviewed and edited the manuscript.
References
- Accari S.L., Fisher P.R. (2015). Emerging roles of JmjC domain-containing proteins. Int. Rev. Cell Mol. Biol. 319: 165–220. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés F., Coupland G. (2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13: 627–639. [DOI] [PubMed] [Google Scholar]
- Aravind L., Koonin E.V. (2001). The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol. 2: research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero J.M., González-Bayón R., del Pozo J.C., Ponce M.R., Micol J.L. (2007). INCURVATA2 encodes the catalytic subunit of DNA Polymerase alpha and interacts with genes involved in chromatin-mediated cellular memory in Arabidopsis thaliana. Plant Cell 19: 2822–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berná G., Robles P., Micol J.L. (1999). A mutational analysis of leaf morphogenesis in Arabidopsis thaliana. Genetics 152: 729–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge N.P., et al. (2014). Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 157: 1445–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge N.P., Rose N.R., Klose R.J. (2015). Targeting Polycomb systems to regulate gene expression: modifications to a complex story. Nat. Rev. Mol. Cell Biol. 16: 643–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briesemeister S., Rahnenführer J., Köhlbacher O. (2010). YLoc--an interpretable web server for predicting subcellular localization. Nucleic Acids Res. 38: W497–W502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs G.C., Osmont K.S., Shindo C., Sibout R., Hardtke C.S. (2006). Unequal genetic redundancies in Arabidopsis--a neglected phenomenon? Trends Plant Sci. 11: 492–498. [DOI] [PubMed] [Google Scholar]
- Brunaud V., et al. (2002). T-DNA integration into the Arabidopsis genome depends on sequences of pre-insertion sites. EMBO Rep. 3: 1152–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calonje M., Sánchez R., Chen L., Sung Z.R. (2008). EMBRYONIC FLOWER1 participates in polycomb group-mediated AG gene silencing in Arabidopsis. Plant Cell 20: 277–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candela H., Casanova-Sáez R., Micol J.L. (2015). Getting started in mapping-by-sequencing. J. Integr. Plant Biol. 57: 606–612. [DOI] [PubMed] [Google Scholar]
- Castillejo C., Romera-Branchat M., Pelaz S. (2005). A new role of the Arabidopsis SEPALLATA3 gene revealed by its constitutive expression. Plant J. 43: 586–596. [DOI] [PubMed] [Google Scholar]
- Chanvivattana Y., Bishopp A., Schubert D., Stock C., Moon Y.H., Sung Z.R., Goodrich J. (2004). Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131: 5263–5276. [DOI] [PubMed] [Google Scholar]
- Chen T., Dent S.Y. (2014). Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat. Rev. Genet. 15: 93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Molitor A., Liu C., Shen W.H. (2010). The Arabidopsis PRC1-like ring-finger proteins are necessary for repression of embryonic traits during vegetative growth. Cell Res. 20: 1332–1344. [DOI] [PubMed] [Google Scholar]
- Chen L., Cheng J.C., Castle L., Sung Z.R. (1997). EMF genes regulate Arabidopsis inflorescence development. Plant Cell 9: 2011–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.K., Hsu W.H., Lee P.F., Thiruvengadam M., Chen H.I., Yang C.H. (2011a). The MADS box gene, FOREVER YOUNG FLOWER, acts as a repressor controlling floral organ senescence and abscission in Arabidopsis. Plant J. 68: 168–185. [DOI] [PubMed] [Google Scholar]
- Chen X., Hu Y., Zhou D.X. (2011b). Epigenetic gene regulation by plant Jumonji group of histone demethylase. Biochim. Biophys. Acta 1809: 421–426. [DOI] [PubMed] [Google Scholar]
- Cheng C.Y., Krishnakumar V., Chan A.P., Thibaud-Nissen F., Schobel S., Town C.D. (2017). Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 89: 789–804. [DOI] [PubMed] [Google Scholar]
- Cloos P.A., Christensen J., Agger K., Helin K. (2008). Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 22: 1115–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cooper S., Dienstbier M., Hassan R., Schermelleh L., Sharif J., Blackledge N.P., De Marco V., Elderkin S., Koseki H., Klose R., Heger A., Brockdorff N. (2014). Targeting polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment. Cell Reports 7: 1456–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagert M., Ehrlich S.D. (1979). Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene 6: 23–28. [DOI] [PubMed] [Google Scholar]
- Dean E.J., Davis J.C., Davis R.W., Petrov D.A. (2008). Pervasive and persistent redundancy among duplicated genes in yeast. PLoS Genet. 4: e1000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkacheva M., Steinbach Y., Wildhaber T., Mozgová I., Mahrez W., Nanni P., Bischof S., Gruissem W., Hennig L. (2013). Arabidopsis MSI1 connects LHP1 to PRC2 complexes. EMBO J. 32: 2073–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C., Zhang H., Xu C., Arrowsmith C.H., Min J. (2014). Structure and function of dioxygenases in histone demethylation and DNA/RNA demethylation. IUCrJ 1: 540–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H.C., Wei L.H., Zhang C., Wang Y., Chen L., Lu Z., Chen P.R., He C., Jia G. (2017). ALKBH10B is an RNA N(6)-Methyladenosine demethylase affecting Arabidopsis floral transition. Plant Cell 29: 2995–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falnes P.O., Johansen R.F., Seeberg E. (2002). AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature 419: 178–182. [DOI] [PubMed] [Google Scholar]
- Farrow S.C., Facchini P.J. (2014). Functional diversity of 2-oxoglutarate/Fe(II)-dependent dioxygenases in plant metabolism. Front. Plant Sci. 5: 524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A.P., Koldobskiy M.A., Göndör A. (2016). Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat. Rev. Genet. 17: 284–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez D.E., Wang C.T., Zheng Y., Adamczyk B.J., Singhal R., Hall P.K., Perry S.E. (2014). The MADS-Domain Factors AGAMOUS-LIKE15 and AGAMOUS-LIKE18, along with SHORT VEGETATIVE PHASE and AGAMOUS-LIKE24, are necessary to block floral gene expression during the vegetative phase. Plant Physiol. 165: 1591–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J., Puangsomlee P., Martin M., Long D., Meyerowitz E.M., Coupland G. (1997). A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386: 44–51. [DOI] [PubMed] [Google Scholar]
- Harrison S.J., Mott E.K., Parsley K., Aspinall S., Gray J.C., Cottage A. (2006). A rapid and robust method of identifying transformed Arabidopsis thaliana seedlings following floral dip transformation. Plant Methods 2: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig B., James G.V., Konrad K., Schneeberger K., Turck F. (2012). Fast isogenic mapping-by-sequencing of ethyl methanesulfonate-induced mutant bulks. Plant Physiol. 160: 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieta R., Myllyharju J. (2002). Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from Arabidopsis thaliana. Effective hydroxylation of proline-rich, collagen-like, and hypoxia-inducible transcription factor alpha-like peptides. J. Biol. Chem. 277: 23965–23971. [DOI] [PubMed] [Google Scholar]
- Honma T., Goto K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409: 525–529. [DOI] [PubMed] [Google Scholar]
- Horiguchi G., Mollá-Morales A., Pérez-Pérez J.M., Kojima K., Robles P., Ponce M.R., Micol J.L., Tsukaya H. (2011). Differential contributions of ribosomal protein genes to Arabidopsis thaliana leaf development. Plant J. 65: 724–736. [DOI] [PubMed] [Google Scholar]
- Horton P., Park K.J., Obayashi T., Fujita N., Harada H., Adams-Collier C.J., Nakai K. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35: W585–W587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua S., Sun Z. (2001). Support vector machine approach for protein subcellular localization prediction. Bioinformatics 17: 721–728. [DOI] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., Lempicki R.A. (2009a). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., Lempicki R.A. (2009b). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser P., Schmid M. (2011). The control of developmental phase transitions in plants. Development 138: 4117–4129. [DOI] [PubMed] [Google Scholar]
- Hyun Y., Yun H., Park K., Ohr H., Lee O., Kim D.H., Sung S., Choi Y. (2013). The catalytic subunit of Arabidopsis DNA polymerase α ensures stable maintenance of histone modification. Development 140: 156–166. [DOI] [PubMed] [Google Scholar]
- Iglesias F.M., Bruera N.A., Dergan-Dylon S., Marino-Buslje C., Lorenzi H., Mateos J.L., Turck F., Coupland G., Cerdán P.D. (2015). The arabidopsis DNA polymerase δ has a role in the deposition of transcriptionally active epigenetic marks, development and flowering. PLoS Genet. 11: e1004975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack T., Brockman L.L., Meyerowitz E.M. (1992). The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68: 683–697. [DOI] [PubMed] [Google Scholar]
- Kafri R., Springer M., Pilpel Y. (2009). Genetic redundancy: new tricks for old genes. Cell 136: 389–392. [DOI] [PubMed] [Google Scholar]
- Kalb R., Latwiel S., Baymaz H.I., Jansen P.W., Müller C.W., Vermeulen M., Müller J. (2014). Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat. Struct. Mol. Biol. 21: 569–571. [DOI] [PubMed] [Google Scholar]
- Kataoka H., Yamamoto Y., Sekiguchi M. (1983). A new gene (alkB) of Escherichia coli that controls sensitivity to methyl methane sulfonate. J. Bacteriol. 153: 1301–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai Y., Ono E., Mizutani M. (2014). Evolution and diversity of the 2-oxoglutarate-dependent dioxygenase superfamily in plants. Plant J. 78: 328–343. [DOI] [PubMed] [Google Scholar]
- Kaya H., Shibahara K.I., Taoka K.I., Iwabuchi M., Stillman B., Araki T. (2001). FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell 104: 131–142. [DOI] [PubMed] [Google Scholar]
- Kempin S.A., Savidge B., Yanofsky M.F. (1995). Molecular basis of the cauliflower phenotype in Arabidopsis. Science 267: 522–525. [DOI] [PubMed] [Google Scholar]
- Kim D.H., Sung S. (2010). The Plant Homeo Domain finger protein, VIN3-LIKE 2, is necessary for photoperiod-mediated epigenetic regulation of the floral repressor, MAF5. Proc. Natl. Acad. Sci. USA 107: 17029–17034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S.L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.Y., Lee J., Eshed-Williams L., Zilberman D., Sung Z.R. (2012). EMF1 and PRC2 cooperate to repress key regulators of Arabidopsis development. PLoS Genet. 8: e1002512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y., Kaya H., Goto K., Iwabuchi M., Araki T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962. [DOI] [PubMed] [Google Scholar]
- Korvald H., Mølstad Moe A.M., Cederkvist F.H., Thiede B., Laerdahl J.K., Bjørås M., Alseth I. (2011). Schizosaccharomyces pombe Ofd2 is a nuclear 2-oxoglutarate and iron dependent dioxygenase interacting with histones. PLoS One 6: e25188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake T., Takada S., Nakahigashi K., Ohto M., Goto K. (2003). Arabidopsis TERMINAL FLOWER 2 gene encodes a heterochromatin protein 1 homolog and represses both FLOWERING LOCUS T to regulate flowering time and several floral homeotic genes. Plant Cell Physiol. 44: 555–564. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lázaro A., Gómez-Zambrano A., López-González L., Piñeiro M., Jarillo J.A. (2008). Mutations in the Arabidopsis SWC6 gene, encoding a component of the SWR1 chromatin remodelling complex, accelerate flowering time and alter leaf and flower development. J. Exp. Bot. 59: 653–666. [DOI] [PubMed] [Google Scholar]
- Lee T.I., Young R.A. (2013). Transcriptional regulation and its misregulation in disease. Cell 152: 1237–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis P.H. (1947). Pc: Polycomb. Drosoph. Inf. Serv. 21: 69. [Google Scholar]
- Liang S.C., et al. (2015). Kicking against the PRCs - a domesticated transposase antagonises silencing mediated by polycomb group proteins and is an accessory component of Polycomb Repressive Complex 2. PLoS Genet. 11: e1005660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Xi W., Shen L., Tan C., Yu H. (2009). Regulation of floral patterning by flowering time genes. Dev. Cell 16: 711–722. [DOI] [PubMed] [Google Scholar]
- Liu Z., Jia L., Wang H., He Y. (2011). HYL1 regulates the balance between adaxial and abaxial identity for leaf flattening via miRNA-mediated pathways. J. Exp. Bot. 62: 4367–4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Vernaza M., Yang S., Müller R., Thorpe F., de Leau E., Goodrich J. (2012). Antagonistic roles of SEPALLATA3, FT and FLC genes as targets of the polycomb group gene CURLY LEAF. PLoS One 7: e30715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F., Cui X., Zhang S., Jenuwein T., Cao X. (2011). Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat. Genet. 43: 715–719. [DOI] [PubMed] [Google Scholar]
- Luo M., Bilodeau P., Koltunow A., Dennis E.S., Peacock W.J., Chaudhury A.M. (1999). Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96: 296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Pérez M., Aparicio F., López-Gresa M.P., Bellés J.M., Sánchez-Navarro J.A., Pallás V. (2017). Arabidopsis m6A demethylase activity modulates viral infection of a plant virus and the m6A abundance in its genomic RNAs. Proc. Natl. Acad. Sci. USA 114: 10755–10760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y. (2011). Post-translational modifications in secreted peptide hormones in plants. Plant Cell Physiol. 52: 5–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merini W., Calonje M. (2015). PRC1 is taking the lead in PcG repression. Plant J. 83: 110–120. [DOI] [PubMed] [Google Scholar]
- Meza T.J., Moen M.N., Vågbø C.B., Krokan H.E., Klungland A., Grini P.E., Falnes P.O. (2012). The DNA dioxygenase ALKBH2 protects Arabidopsis thaliana against methylation damage. Nucleic Acids Res. 40: 6620–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micol-Ponce R., Sánchez-García A.B., Xu Q., Barrero J.M., Micol J.L., Ponce M.R. (2015). Arabidopsis INCURVATA2 regulates salicylic acid and abscisic acid signaling, and oxidative stress responses. Plant Cell Physiol. 56: 2207–2219. [DOI] [PubMed] [Google Scholar]
- Mielecki D., Zugaj D.L., Muszewska A., Piwowarski J., Chojnacka A., Mielecki M., Nieminuszczy J., Grynberg M., Grzesiuk E. (2012). Novel AlkB dioxygenases--alternative models for in silico and in vivo studies. PLoS One 7: e30588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura A., Nakamura M., Inagaki S., Kobayashi A., Saze H., Kakutani T. (2009). An Arabidopsis jmjC domain protein protects transcribed genes from DNA methylation at CHG sites. EMBO J. 28: 1078–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami Y., Ma H. (1992). Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell 71: 119–131. [DOI] [PubMed] [Google Scholar]
- Moghaddam A.M., Roudier F., Seifert M., Bérard C., Magniette M.L., Ashtiyani R.K., Houben A., Colot V., Mette M.F. (2011). Additive inheritance of histone modifications in Arabidopsis thaliana intra-specific hybrids. Plant J. 67: 691–700. [DOI] [PubMed] [Google Scholar]
- Mosquna A., Katz A., Decker E.L., Rensing S.A., Reski R., Ohad N. (2009). Regulation of stem cell maintenance by the Polycomb protein FIE has been conserved during land plant evolution. Development 136: 2433–2444. [DOI] [PubMed] [Google Scholar]
- Mozgova I., Hennig L. (2015). The polycomb group protein regulatory network. Annu. Rev. Plant Biol. 66: 269–296. [DOI] [PubMed] [Google Scholar]
- Nei M., Kumar S. (2000). Molecular Evolution and Phylogenetics. (New York: Oxford University Press; ). [Google Scholar]
- Nelissen H., Fleury D., Bruno L., Robles P., De Veylder L., Traas J., Micol J.L., Van Montagu M., Inzé D., Van Lijsebettens M. (2005). The elongata mutants identify a functional Elongator complex in plants with a role in cell proliferation during organ growth. Proc. Natl. Acad. Sci. USA 102: 7754–7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano Y., Aono N., Hiwatashi Y., Murata T., Nishiyama T., Ishikawa T., Kubo M., Hasebe M. (2009). A polycomb repressive complex 2 gene regulates apogamy and gives evolutionary insights into early land plant evolution. Proc. Natl. Acad. Sci. USA 106: 16321–16326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz S., Ditta G.S., Baumann E., Wisman E., Yanofsky M.F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405: 200–203. [DOI] [PubMed] [Google Scholar]
- Pérez-Pérez J.M., Candela H., Micol J.L. (2009a). Understanding synergy in genetic interactions. Trends Genet. 25: 368–376. [DOI] [PubMed] [Google Scholar]
- Pérez-Pérez J.M., Candela H., Robles P., Quesada V., Ponce M.R., Micol J.L. (2009b). Lessons from a search for leaf mutants in Arabidopsis thaliana. Int. J. Dev. Biol. 53: 1623–1634. [DOI] [PubMed] [Google Scholar]
- Pérez-Pérez J.M., Rubio-Díaz S., Dhondt S., Hernández-Romero D., Sánchez-Soriano J., Beemster G.T., Ponce M.R., Micol J.L. (2011). Whole organ, venation and epidermal cell morphological variations are correlated in the leaves of Arabidopsis mutants. Plant Cell Environ. 34: 2200–2211. [DOI] [PubMed] [Google Scholar]
- Pfaffl M.W., Horgan G.W., Dempfle L. (2002). Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikaard C.S., Mittelsten Scheid O. (2014). Epigenetic regulation in plants. Cold Spring Harb. Perspect. Biol. 6: a019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñeiro M., Gómez-Mena C., Schaffer R., Martínez-Zapater J.M., Coupland G. (2003). EARLY BOLTING IN SHORT DAYS is related to chromatin remodeling factors and regulates flowering in Arabidopsis by repressing FT. Plant Cell 15: 1552–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponce M.R., Quesada V., Micol J.L. (1998). Rapid discrimination of sequences flanking and within T-DNA insertions in the Arabidopsis genome. Plant J. 14: 497–501. [DOI] [PubMed] [Google Scholar]
- Ponce M.R., Robles P., Micol J.L. (1999). High-throughput genetic mapping in Arabidopsis thaliana. Mol. Gen. Genet. 261: 408–415. [DOI] [PubMed] [Google Scholar]
- Ponce M.R., Robles P., Lozano F.M., Brotóns M.A., Micol J.L. (2006). Low-resolution mapping of untagged mutations. Methods Mol. Biol. 323: 105–113. [DOI] [PubMed] [Google Scholar]
- Porco S., et al. (2016). Dioxygenase-encoding AtDAO1 gene controls IAA oxidation and homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. USA 113: 11016–11021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provart N.J., et al. (2016). 50 years of Arabidopsis research: highlights and future directions. New Phytol. 209: 921–944. [DOI] [PubMed] [Google Scholar]
- Quesada V., Ponce M.R., Micol J.L. (1999). OTC and AUL1, two convergent and overlapping genes in the nuclear genome of Arabidopsis thaliana. FEBS Lett. 461: 101–106. [DOI] [PubMed] [Google Scholar]
- Quesada V., Ponce M.R., Micol J.L. (2000). Genetic analysis of salt-tolerant mutants in Arabidopsis thaliana. Genetics 154: 421–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinholz E. (1966). Radiation induced mutants showing changed inflorescence characteristics. Arab. Inf. Serv. 3: 19–20. [Google Scholar]
- Relichová J. (1976). Some new mutants. Arab. Inf. Serv. 13: 25–28. [Google Scholar]
- Repková J., Hlavacová S., Lízal P., Kyjovská Z., Relichová J. (2005). Molecular mapping of some Arabidopsis thaliana genes determining leaf shape and chlorophyll defects. Biologia 60: 443–449. [Google Scholar]
- Rounsley S.D., Ditta G.S., Yanofsky M.F. (1995). Diverse roles for MADS box genes in Arabidopsis development. Plant Cell 7: 1259–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Díaz S., Pérez-Pérez J.M., González-Bayón R., Muñoz-Viana R., Borrega N., Mouille G., Hernández-Romero D., Robles P., Höfte H., Ponce M.R., Micol J.L. (2012). Cell expansion-mediated organ growth is affected by mutations in three EXIGUA genes. PLoS One 7: e36500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savidge B., Rounsley S.D., Yanofsky M.F. (1995). Temporal relationship between the transcription of two Arabidopsis MADS box genes and the floral organ identity genes. Plant Cell 7: 721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger K., Ossowski S., Lanz C., Juul T., Petersen A.H., Nielsen K.L., Jørgensen J.E., Weigel D., Andersen S.U. (2009). SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat. Methods 6: 550–551. [DOI] [PubMed] [Google Scholar]
- Schroeder A., Mueller O., Stocker S., Salowsky R., Leiber M., Gassmann M., Lightfoot S., Menzel W., Granzow M., Ragg T. (2006). The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 7: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Cartagena J., Robles P., Ponce M.R., Micol J.L. (1999). Genetic analysis of leaf form mutants from the Arabidopsis Information Service collection. Mol. Gen. Genet. 261: 725–739. [DOI] [PubMed] [Google Scholar]
- Serrano-Cartagena J., Candela H., Robles P., Ponce M.R., Pérez-Pérez J.M., Piqueras P., Micol J.L. (2000). Genetic analysis of incurvata mutants reveals three independent genetic operations at work in Arabidopsis leaf morphogenesis. Genetics 156: 1363–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu B. (2014). 2-Oxoglutarate-dependent dioxygenases in the biosynthesis of simple coumarins. Front. Plant Sci. 5: 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stothard P. (2000). The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 28: 1102–1104. [DOI] [PubMed] [Google Scholar]
- Sung Z.R., Belachew A., Shunong B., Bertrand-Garcia R. (1992). EMF, an Arabidopsis gene required for vegetative shoot development. Science 258: 1645–1647. [DOI] [PubMed] [Google Scholar]
- Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L., Rao A. (2009). Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewick S.C., Henshaw T.F., Hausinger R.P., Lindahl T., Sedgwick B. (2002). Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature 419: 174–178. [DOI] [PubMed] [Google Scholar]
- Turck F., Roudier F., Farrona S., Martin-Magniette M.L., Guillaume E., Buisine N., Gagnot S., Martienssen R.A., Coupland G., Colot V. (2007). Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 3: e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasquez S.M., et al. (2015). Complex regulation of prolyl-4-hydroxylases impacts root hair expansion. Mol. Plant 8: 734–746. [DOI] [PubMed] [Google Scholar]
- Wang D., Tyson M.D., Jackson S.S., Yadegari R. (2006). Partially redundant functions of two SET-domain polycomb-group proteins in controlling initiation of seed development in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 13244–13249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R.S., Zhang Y. (2004). Role of histone H2A ubiquitination in Polycomb silencing. Nature 431: 873–878. [DOI] [PubMed] [Google Scholar]
- Wilson-Sánchez D., Rubio-Díaz S., Muñoz-Viana R., Pérez-Pérez J.M., Jover-Gil S., Ponce M.R., Micol J.L. (2014). Leaf phenomics: a systematic reverse genetic screen for Arabidopsis leaf mutants. Plant J. 79: 878–891. [DOI] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y., Li W. (2009). BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics 10: 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang H., et al. (2010). Single base-resolution methylome of the silkworm reveals a sparse epigenomic map. Nat. Biotechnol. 28: 516–520. [DOI] [PubMed] [Google Scholar]
- Xiao J., Lee U.S., Wagner D. (2016). Tug of war: adding and removing histone lysine methylation in Arabidopsis. Curr. Opin. Plant Biol. 34: 41–53. [DOI] [PubMed] [Google Scholar]
- Yang C., Bratzel F., Hohmann N., Koch M., Turck F., Calonje M. (2013). VAL- and AtBMI1-mediated H2Aub initiate the switch from embryonic to postgerminative growth in Arabidopsis. Curr. Biol. 23: 1324–1329. [DOI] [PubMed] [Google Scholar]
- Yanofsky M.F., Ma H., Bowman J.L., Drews G.N., Feldmann K.A., Meyerowitz E.M. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346: 35–39. [DOI] [PubMed] [Google Scholar]
- Yoshida N., Yanai Y., Chen L., Kato Y., Hiratsuka J., Miwa T., Sung Z.R., Takahashi S. (2001). EMBRYONIC FLOWER2, a novel polycomb group protein homolog, mediates shoot development and flowering in Arabidopsis. Plant Cell 13: 2471–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Germann S., Blus B.J., Khorasanizadeh S., Gaudin V., Jacobsen S.E. (2007). The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat. Struct. Mol. Biol. 14: 869–871. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Romero-Campero F.J., Gómez-Zambrano Á., Turck F., Calonje M. (2017). H2A monoubiquitination in Arabidopsis thaliana is generally independent of LHP1 and PRC2 activity. Genome Biol. 18: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]