Abstract
Background
Mast cells (MCs) are long-lived immune cells of the connective tissue which play a key role in development and amplification of inflammatory process initiated inter alia by allergic reactions or microbial infections. They reside in strategic locations in the body that are notably exposed to deleterious factors disturbing homeostasis, which enables them to become one of the first-line defense strategy. MCs have developed a wide range of various mechanisms to deal with invading intruders and harmful endogenic factors. Those include storage and synthesis with a subsequent release of inflammatory mediators, forming of MC-extracellular traps, and phagocytosis.
Findings
Particularly, important role in microbial sensing is achieved due to the presence of different pattern recognition receptors (PRRs). The best-described receptors are Toll-like receptors activated by different pathogen- and damage-associated molecular patterns. However, MCs express also C-type lectin receptors specialized in antifungal defense, NOD-like receptors detecting bacterial peptidoglycans, and RIG-like receptors relevant in viral sensing.
Conclusion
This review will focus on the current knowledge of PRRs expressed within different types of MCs.
Keywords: Mast cells, PRRs, TLRs, CLRs, NLRs, RLRs.
Mast cells
For a long time, mast cells (MCs) have been recognized as the critical immune system weapon in the allergic reactions [1, 2], as well as during parasitic infections [3]. Indeed, these immune system’s chief sentinels hold a multitude of functions in the host defense against a variety of microorganisms, mainly bacteria and viruses [4–7]. So far, the contribution of MCs to the protection against fungal infections is an almost unexplored area [8]. MCs are long-lived resident cells located preferentially throughout vascularized connective tissues. They are numerous at the junction point of the host and external environment, including the skin, the respiratory system, the gastrointestinal and genitourinary tracts; therefore, they belong to the first immune cells, which encounter intruders [9]. Since MCs are rich source of numerous biologically active mediators, such as granule-associated preformed mediators (e.g., neutral proteases, histamine, proteoglycans, metalloproteinases), de novo generated arachidonic acid metabolites (e.g., leukotrienes, prostaglandins, thromboxanes), as well as many newly synthesized cytokines and chemokines, they can alter the functions of the surrounding cells and tissues. MC-derived proinflammatory mediators, cytokines and chemokines induce the development of inflammation at the site of pathogen entry [10–14]. MCs are highly efficient effector cells that release not only active mediators but also antimicrobial peptides, such as cathelicidins [15]. Moreover, these cells can phagocytose and subsequently kill bacteria, via oxidative and non-oxidative systems [16, 17]. MCs also contribute to host defense by forming extracellular traps (MCETs) composed of DNA, histones, and granule proteins, which can entrap and kill various microbes [18]. Furthermore, following phagocytosis these cells have the capability of processing bacterial antigens for presentation through class I and II MHC molecules, which leads to the development of adaptive antimicrobial immunity [17, 19, 20].
To respond rapidly to infection or cellular damage, MCs express germline-encoded pattern recognition receptors (PRRs) that recognize unique bacterial, viral, fungal or parasitic components known as pathogen-associated molecular patterns (PAMPs) and host-derived molecules, called damage-associated molecular patterns (DAMPs). Hitherto, PRRs have comprised five families: Toll-like receptors (TLRs), C-type lectin-like receptors (CLRs), retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs) (Table 1), and recently designated absent-in-melanoma (AIM)-like receptors (ALRs). While TLRs constitute the best characterized PRRs in MCs, recent findings suggest that other types of recognition receptors act as surveillance molecules in these cells. In this review, we will provide a concise overview of the TLR, CLR, RLR, and NLR family members in MCs.
Table 1.
Members of the TLR, CLR, NLR and RLR families with their alternative names (separated by forward slashes), divided into relevant subgroups or subfamilies
| Type | Family member | ||
|---|---|---|---|
| TLRs |
TLR1-10
TLR11-13 |
||
| CLRs | Group I |
MR/CD206 DEC205/LY75/CD205 |
|
| Group II | DC-SIGN/CD209 langerin/CLEC4K/CD207 MGL/CLEC10A/CD301 CLEC5A/MDL1 |
||
| Dectin 1 subfamily |
Dectin 1/CLEC7A MICL/CLEC12A/DCAL2 CLEC2/CLEC1B DNGR1/CLEC9A CLEC12B |
||
| DCIR subfamily | DCIR/CLEC4A Dectin 2/CLEC6A BDCA2/CLEC4C/CD303 Mincle/CLEC4E |
||
| NLRs | NLRA | CIITA | |
| NLRB | NAIP | ||
| NLRC |
NLRC1/NOD1 NLRC2/NOD2 NLRC3-5 NLRX1 |
||
| NLRP | NLRP1 NLRP2 NLRP3 NLRP4-14 |
||
| RLRs |
RIG-I
MDA5 LGP2 |
||
The underlined receptors are those that have been documented in MCs
Toll-like receptors (TLRs)
TLR family, their ligands and molecular structure
TLRs are the best studied and described molecules among other PRRs. They comprise a large group of transmembrane receptors expressed in various types of cells including hepatocytes [21], monocytes/macrophages [22], dendritic cells (DCs) [22, 23], B lymphocytes [24], T lymphocytes [25], epithelial cells [26], endothelial cells [27], neutrophils [28], and NK cells [29]. Most importantly, TLRs are expressed in MCs [30, 31]. The TLR family include TLR1, TLR2, TLR3, TLR4, TLR5, TLR6, TLR7, TLR8, TLR9, TLR10, TLR11 [32], TLR12 [33] and TLR13 [34]. Hitherto sources strengthen the conviction that certain TLRs are present merely in a particular membrane structure, e.g., cellular membrane and endosome membrane [32]. TLRs recognize a wide range of molecular patterns associated with bacteria, viruses, protozoa or parasites, as well as different endogenous factors arising as a result of any tissue damage [32, 35]. Various members of TLR family may sense the same molecular patterns or entirely different, depending on the type of TLR. Mentioned ligands include lipoproteins, diacyl and triacyl lipoproteins, lipoteichoic acid (LTA), lipopolysaccharide (LPS), single- and double-stranded RNA, unmethylated CpG oligodeoxynucleotide DNA, flagellin, heat shock protein (HSP)70, profilin-like molecule, myeloid differentiation factor 2 (MD2), oxidized phospholipids, fibrinogen, and many others [30–32]. TLRs are composed of the N-terminal ligand recognition domain, transmembrane region and a C-terminal cytoplasmic signaling domain exhibiting homology of a domain present in interleukin (IL) type 1 receptor (IL-1R). Thus, it is called Toll IL-1R (TIR) domain. TIR domains are a part of adaptor proteins and play a crucial role in the early phase of signaling pathways. Transmembrane domains are responsible for the interaction with multispan transmembrane protein Unc-93 homolog B1 (UNC93B1), resulting in trafficking of TLRs within endocytic compartments of the cell. TLRs which transmembrane domains do not bind to UNC93B1 are directed to the surface of the cell. N-terminal ectodomains (ECDs), built of leucine-reach repeat (LRR) motif, constitute glycoproteins that recognize molecular patterns associated with pathogens [36].
TLR signaling pathways
Recognition of any inducing agents by TLRs leads to expression of different genes accordingly to the type of cell and TLR engaged in this process. TLR signaling pathway is usually based on two different mechanisms, which depend on TIR domain-containing adaptors that are recruited to TLRs, namely myeloid differentiation primary response gene 88 (MyD88) and TIR domain-containing adaptor-inducing interferon (IFN)-β [TRIF or Toll-like receptor adaptor molecule 1 (TICAM-1)]. However, there are three other adaptor molecules, that may be required for bridging and activating MyD88 or TRIF pathways: TIR domain containing adaptor protein/MyD88-adapter-like (TIRAP/Mal), TRIF-related adaptor molecule (TRAM) and Sterile-alpha and Armadillo motif-containing protein (SARM). Various TLRs, but not TLR3, may initiate the MyD88 signaling pathway. Moreover, in the case of particular TLRs, e.g., TLR1, TLR2, TLR4 and TLR6, MyD88 signaling is preceded by TIRAP/Mal pathway. TLR3 and TLR4 may initiate the TRIF signaling pathway, however, in case of TLR4, this process is preceded by TRAM signaling. The signaling cascade leading to the activation of nuclear factor kappa B (NF-κB) results in the expression of various cytokines in a cell engaged in the entire process, while activation of IFN regulatory factor (IRF) 3 or IRF7 leads to the expression of type I IFNs [32]. The signal cascade initiated by the activated TLR is complex, which makes it possible to regulate this process in many stages. A recent report has shown that leukotrienes C4 and D4 have a considerable influence on the down-regulation of TLR1–7 and this effect is mediated by activation of the CysLT1 receptor [37]. Karpov et al. have hypothesized that inflammatory GPCR signaling (via CysLTR) interacts with and modifies mast cell TLR expression and activation pathways, ultimately blocking activation of NF-κB.
TLRs in MCs
Indisputably, all the studies concerning the presence of TLRs in MCs carried out to date confirm the expression of various members of the TLR family within different types of MCs to a greater or lesser degree. The expression of TLRs in MCs has been confirmed both at mRNA and protein level in a wide range of MCs including murine MCs, e.g., murine bone marrow-derived MCs (BMMCs), murine fetal skin-derived MCs (FSDMCs), murine peritoneal cell-derived MCs (PCMCs); rat peritoneal MCs (RPMCs); human MCs comprising immature human MCs, e.g., human BMMCs, cord blood-derived MCs (CBMCs), and mature human MCs, e.g., MCs isolated from human lungs, MCs isolated from human skin; human MC lines, e.g., human MC line-1 (HMC-1), laboratory of allergic diseases 2 MCs (LAD2); as well as mouse MC lines, e.g., MC/9, P815. Data indicate that MCs express TLR1–10 and there is no information confirming the presence of TLR11–13. Additionally, our recent data revealed that nearly every TLR might be expressed either in MC plasma membrane as well as inside the cell [38, 39]. More detailed data concerning this issue are already described in other reviews [30, 31].
C-type lectin receptors
CLR family, their ligands and molecular structure
CLRs constitute a group of PRRs that play a pivotal role in antifungal defense strategies and may cooperate with other PRRs, such as TLRs, in various immunological mechanisms [40]. CLRs are expressed in different cell types including endothelial cells [41] and DCs [42, 43]. Additionally, their presence is noticed in myeloid cells, e.g., neutrophils [44], monocytes/macrophages [45], and Langerhans cells [42]. Current knowledge assumes that CLR family is slightly greater and more complex in comparison to TLR members, and consists of two major groups with subgroups of receptors. Group I CLRs includes the Mannose Receptor family with Mannose Receptor (MR or CD206) and DEC205 (LY75 or CD205) as representatives. Group II CLRs comprise asialoglycoprotein receptor family that consists of DC-specific intercellular adhesion molecule-3-grabbing non-integrin (DC-SIGN or CD209), langerin (C-type lectin domain family (CLEC) 4 member K or CD207), macrophage galactose-type lectin (MGL, CLEC10A or CD301) and CLEC5A [myeloid DAP12-associating lectin 1 (MDL1)]. Moreover, group II CLRs includes two subgroups, namely Dectin 1 subfamily and DC immunoreceptor (DCIR) subfamily. The former one consists of Dectin 1 (CLEC7A), myeloid C-type lectin-like receptor [MICL, CLEC12A or DC-associated lectin 2 (DCAL2)], CLEC2 (CLEC1B), DC NK lectin group receptor 1 (DNGR1 or CLEC9A) and CLEC12B, while the latter one comprise DCIR (CLEC4A), Dectin 2 (CLEC6A), blood DC antigen 2 (BDCA2, CLEC4C or CD303) and macrophage inducible Ca2+-dependent lectin (Mincle or CLEC4E) [46]. CLRs are associated with membranes, and they constitute a family of transmembrane signaling proteins [32]. Those receptors are capable of sensing various carbohydrate molecules present in cell walls of fungi. The vast majority of them may bind to mannose and fucose sugars, while some of them can also recognize sulphated sugars, β-1,3-glucan (in the case of Dectin receptors) and spliceosome-associated protein 130 (SAP130, for Mincle receptor) [32, 46, 47]. CLRs possess one or more carbohydrate-recognition domains (CRDs), that act in a calcium-dependent manner, sensing and binding sugar molecular patterns by ligation to Ca2+. CRDs form compact and spherical structures comprising conserved regions, which is responsible for CLRs affinity to specific carbohydrates. However, some CLRs such as Dectin 1 can bind carbohydrates in a Ca2+-independent manner due to the presence of non-canonical sugar binding site [46, 48]. Certain CLRs also possess immunoreceptor tyrosine-based activation motif (ITAM) domains or immunoreceptor tyrosine-based inhibition motif (ITIM) domains or do not clearly express any of this domain within cytoplasmic tail [49].
CLR signaling pathways
CLRs signaling pathway is directly dependent on the presence or absence of ITAM or ITIM domains. Receptors such as DNGR1, Dectin 1, Dectin 2, Mincle and CLEC5A expressing ITAM domain can initiate a signaling cascade through spleen tyrosine kinase (Syk). Moreover, DNGR1 and Dectin 1 activate Syk via direct interaction with ITAM domain, while others recruit Syk by ITAM-associated adaptor molecules, e.g., Fc receptor γ-chain (FcRγ) or DAP12 [32, 49]. Syk activation leads to a formation of caspase recruitment domain-containing protein 9 (CARD9), mucosa-associated lymphoid tissue lymphoma translocation protein 1 (Malt1) and B cell lymphoma 10 (Bcl10) complex, which in turn triggers mitogen-activated protein kinases (MAPK) and NF-κB. In consequence, this signaling pathway results in initiation of pro-inflammatory processes such as synthesis of mediators, chemotaxis, phagocytosis, and activation of inflammasomes. DCIR express ITIM domain is interacting via Src homology region 2 domain-containing phosphatase (SHP)-1 and SHP-2 that is responsible mainly for inhibition of mediator production initiated by different PRR, e.g., TLR8, TLR9. Receptors without ITAM or ITIM domains include MR, DEC205, and DC-SIGN. While in the case of MR and DEC205 there is no clearly defined mode of a signaling pathway, DC-SIGN, as well as Dectin 1, are able to induce pro-inflammatory processes independently from Syk via Raf1 Proto-Oncogene [40, 47, 49].
CLRs in MCs
Data showing the expression of CLRs within MCs, unlike TLRs presence, are still limited. Furthermore, among investigated receptors from the CLR family, the most commonly presented studies concerned Dectin 1, and only a few data revealed the expression of MR, MGL, and Mincle within MCs. Thus, this issue is yet to be explored. Out of various MCs and their cell lines, murine BMMCs were used predominantly to investigate the presence of CLRs within MCs, however murine PCMCs, RPMCs, peripheral blood-derived MCs (PBMCs), and human CBMCs were studied as well. Moreover, human KU812 and rat RBL-2H3 basophils cell lines were the subject of research for those studies, since they share certain characteristics with MCs, e.g., MC IgE-mediated degranulation. mRNA expression of different CLRs within MCs was studied exclusively by one method, namely RT-PCR, sometimes also carried out with real-time analysis, RT-qPCR, and in one case even confirmed with RNA direct sequencing. Unlike mRNA, various CLR protein expression in MCs was investigated using more techniques and tools including Western blot, immunohistochemistry, immunoblotting, fluorescence-activated cell sorting (FACS) and even ELISA in the sense of checking the secretion of MC cytokines after the use of certain CLR inhibitor [50–56].
Dectin-1 is the best known and described receptor from the whole CLR family. It is responsible for the recognition of β-glucan, comprising the main building block for fungal cell wall, and stimulation of tumor necrosis factor (TNF) and reactive oxygen species (ROS) production. Dectin-1 is expressed within various immune cells including neutrophils, monocytes/macrophages, NK cells, DCs, and fibroblasts, but also within murine BMMCs supplemented with mouse plasma. RT-PCR, with a subsequent product visualization by agarose gel electrophoresis, revealed a low mRNA expression of Dectin-1 isoform A and isoform B in murine BMMCs stimulated with zymosan, a β-glucan from yeast cells. FACS analysis with a Dectin-1 specific antibody showed a weak cell surface expression of Dectin-1 on untreated murine BMMCs and a significantly higher expression in cells stimulated with zymosan. However, this functional protein expression was not dependent on the presence of mouse plasma. Interestingly, cell surface expression of Dectin-1 was significantly lower in cells treated with a Dectin-1 inhibitor, namely laminarin. Furthermore, FACS enabled the measurement of ROS production mediated by Dectin-1, since ROS levels were significantly lower in laminarin-treated cells compared to those untreated [56].
Further studies with another β-glucan, specifically curdlan that constitute an agonist for Dectin-1, indicate the induction of murine BMMC degranulation, including a significant increase of histamine release measured in supernatants by ELISA technique. Moreover, murine BMMCs released from their granules β-hexosaminidase, which levels were estimated by an enzymatic colorimetric assay. Entirely different observations were made in the case of de novo synthesized mediators, such as LTC4, IL-6, and CCL2. Analysis of supernatants by ELISA technique revealed that murine BMMCs treated with curdlan did not produce mentioned mediators unless the cells were activated through IgE receptor cross-linking. Interestingly, those mechanisms differ greatly from processes initiated by TLR signaling pathways, where de novo synthesis of lipid mediators and cytokines is triggered independently of degranulation [50].
The presence of Dectin-1 within murine BMMCs and RPMCs was also confirmed through their direct stimulation with Candida albicans yeasts or hyphae, which significantly induced murine BMMC and RPMCs degranulation, assessed by β-hexosaminidase release. Analysis of supernatants by ELISA technique also revealed a considerably high level of TNF, which increased even more with time after murine BMMC fungal stimulation compared to untreated cells. Furthermore, C. albicans induced production of other cytokines or chemokines including IL-1β in the case of yeasts, IL-6, delayed synthesis of IL-10, CCL3, and CCL4, as well as induced ROS production. To confirm Dectin-1 mediated initiation of cytokine synthesis, Nieto-Patlán et al. [52] treated murine BMMCs with anti-Dectin-1 and measured the release of TNF. They observed a significant decrease in TNF production when compared to murine BMMCs incubated with anti-Dectin-1 isotype control.
Dectin-1 receptor expression was assessed as well within other MCs and MC-like cell types including human CBMCs and human KU812 cell line. mRNA expression of Dectin-1 isoform B obtained through RT-PCR was observed in both untreated MC types, which was also confirmed by direct sequencing that revealed a complete sequence identity of the obtained product when compared to known Dectin-1 isoform B sequence. Likewise, the presence of the functional Dectin-1 isoform B protein was observed by Western blot in human KU812 when treated with IL-4 or sodium butyrate, while not stimulated human CBMCs weakly expressed Dectin-1 isoform B as well as isoform A. Moreover, supernatants from human CBMCs incubated with zymosan and peptidoglycan (PGN) were analyzed for certain cytokine and lipid mediator secretion. Although, the levels of IL-1β, granulocyte–macrophage colony-stimulating factor (GM-CSF), LTB4, and LTC4 were significantly higher in human CBMCs treated with zymosan and PGN compared with not stimulated cells, only LTC4 production was found to be Dectin-1 mediated, since pre-treatment with laminarin and other Dectin-1 inhibitor, glucan phosphate, significantly reduced LTC4 release in human CBMCs incubated with zymosan [53].
Ribbing et al. [54] studied the expression of Dectin-1 and, responsible mainly for glucose or mannose sensing, Mincle receptor within human PBMCs isolated from healthy control (HC) and atopic eczema (AE) patients. Low mRNA expression of both receptors obtained through RT-qPCR was observed in untreated PBMCs from HC as well as from AE subjects. A significantly higher expression of those receptors was noticed in PBMCs stimulated via IgE receptor cross-linking together with Malassezia sympodialis in both groups of patients. MCs activation through IgE receptor cross-linking and incubation with M. sympodialis separately, revealed a higher expression of Dectin-1 in HC and a slight increase in mRNA levels in AE patients when compared to not stimulated cells, while increased expression of Mincle was comparable in HC and AE subjects. Western blot analysis confirmed the functional protein expression of both receptors, Mincle and Dectin-1, including Dectin-1 isoform A and isoform B, in PBMCs after IgE receptor cross-linking. Furthermore, biopsy samples taken from lesional HC and AE skin subjects and stained by immunohistochemistry revealed the presence of Dectin-1.
Expression of Mincle mRNA studied by RT-PCR and confirmed by electrophoresis was as well observed in rat MC-like RBL-2H3 cell line. However, due to the lack of commercially available anti-Mincle antibody which would recognize and attach to rat Mincle, any further studies involving assessment of functional Mincle protein presence and levels is not possible, unless the cells are transfected with vectors expressing functional Mincle [51].
Finally, expression of MR, which recognize fucose, mannose or N-acetylglucosamine, and MGL, sensing N-acetylgalactosamine within particular MCs was studied among various CLRs. The expression of functional proteins of both receptors was confirmed in murine BMMCs and murine PCMCs with the use of FACS. Interestingly, ELISA technique revealed a reduced production of TNF and IL-6 in murine BMMCs and murine PCMCs, both treated with anti-MR or anti-MGL antibodies and Bordetella pertussis, and IFN-γ only in case of murine PCMCs, incubated with anti-MR or anti-MGL antibodies and B. pertussis when compared to not stimulated cells [55].
NOD-like receptors (NLRs)
NLR family, their ligands and molecular structure
The cytosolic nucleotide-binding oligomerization domain receptors (also known as CATERPILLERs) are multi-domain proteins with a tripartite architecture. NOD-like receptors are typically composed of four variable N-terminal effector domains, which are used to subdivide NLRs into four groups: the acidic transactivation domain (subfamily NLRA), the baculoviral inhibitory repeat-like domain (subfamily NLRB), the CARD (subfamily NLRC), and the pyrin domain (subfamily NLRP). The N-terminal domain performs effector functions by interacting with other proteins [57, 58]. The central nucleotide-binding domain NACHT (also referred to as NOD domain) shares homology with the AAA + superfamily of ATPases and is involved in dNTPase activity and oligomerization [58–60]. The C-terminal LRRs are involved in ligand binding or activator sensing [58, 61]. NLRs comprise a large family of 23 members in human and constitute a major group of intracellular PRRs [57]. The MHC-II transactivator (CIITA) is the only member of the NLRA subfamily. Likewise, the NLRB subfamily has only one receptor, NAIP. The NLRC subfamily consists of NLRC1 (NOD1), NLRC2 (NOD2), NLRC3-5, and NLRX1 [57, 58, 61]. The best characterized NLRs, NOD1 and NOD2, were the first receptors from this family identified as sensors of microbial patterns [62, 63]. NOD1 and NOD2 detect distinct motifs of PGN, which is a fundamental component of the bacterial cell wall. NOD1 binds the d-γ-glutamyl-meso-DAP dipeptide (iE-DAP), which is found in all Gram-negative and certain Gram-positive bacteria (Listeria sp. and Bacillus sp.) [64]. In turn, NOD2 identify the muramyl dipeptide (MDP) structure found widely in nearly all bacteria [65]. Thus, NOD1 is involved in the identification of a specific subset of bacteria and NOD2 acts as a general sensor of PGN. The last subfamily—NLRP consists of 14 members, NLRP1-14 [57, 58, 61]. Interestingly, NLRP1 and NLRP3 induce the formation of the inflammasome complex that stimulates caspase-1 activation to promote the secretion of proinflammatory cytokines [66].
NLR signaling pathways
The current model assumes that the crucial step in NLR activation is the self-oligomerization of the N-terminal NACHT domain upon recognition of their ligands. This step is followed by the transient recruitment and activation of the CARD-containing kinase RIPK2 (also called RIP2/RICK), a potent activator of NF-κB and MAPKs, through CARD–CARD interaction. Recruitment of RIPK2 leads to NF-κB and the inhibitor of NF-κB-(IκB-) kinase (IKK) complex activation through phosphorylation and ubiquitination of IκBα. It is accomplished via the recruitment of the E3 ubiquitin ligase TRAF6 to RIPK2, followed by TRAF6 autoubiquitination, polyubiquitination of RIPK2, and the ubiquitination-dependent signaling and activation of the TAK1 complex. The activated TAK1 complex promotes the K63-type polyubiquitination of IKK-β, culminating in the degradation of the NF-κB repressor IκB, the translocation of NF-κB to the nucleus, and the transcription of proinflammatory genes. In addition, recruitment of RIPK2 also activates p38, ERK, and JNK, and it seems to be possible that NLRs participate in the activation of the type I IFN pathway. Although, RIPK2 is critical for the induction of NF-κB activation, the molecular basis how the NOD/RIPK2 complex stimulates NF-κB activation is poorly understood [57].
NLRs in MCs
Few sources indicate that NLRs are expressed in RPMCs, mouse MC line P815, and mouse BMMCs. Zabucchi et al. [67] stated by immunogold analysis that in resting RPMCs, NOD1 and NOD2 receptors are associated with the same subcellular compartments such as surface and matrix of secretory granules or with cytoplasmic vesicles. Additionally, gold particles labeling showed that free cytosolic NLRs in RPMCs are rare. Interestingly, after ingestion of opsonized Escherichia coli, the extent of labeling for NOD1 and NOD2 in RPMC granules decreased significantly.
Expression of NOD2 mRNA was observed in P815 cells [68]. Real-time PCR results showed that NOD2 mRNA expression was increased considerably in P815 cells after Staphylococcus aureus infection. Furthermore, IL-6, CXCL8 and histamine secretion was significantly up-regulated in the S. aureus-infected cells pretreated with NOD2 siRNA before infection. Noteworthy, S. aureus infection could trigger NF-κB activation in P815 cells, but NOD2 siRNA treatment significantly reduced the NF-κB activation in response to S. aureus and these results may indicate that the NF-κB activation was closely associated with NOD2.
Ueno et al. [69] established transient induction of endogenous NLRP3 protein in BMMCs after stimulation with stem cell factor (SCF) alone or after stimulation via FcɛRI crosslinking by anti-DNP-IgE and DNP-BSA. Moreover, in BMMCs stimulated with the cytokine triad: SCF + IL-1β + IL-10, there was a severe induction of NLRP3 protein expression that continued to increase over 24 h. Persistent induction of NLRP3 protein was preceded by the transient expression of NLRP3 mRNA, which peaked after 2 h and declined after 6 h as assessed by Northern blotting. Authors also examined the expression of other inflammasome-associated genes such as ASC and RIP2, which are known to form a complex with NLRP3. RT-PCR revealed that the expression of RIP2 was increased (as in the case of NLRP3), after stimulation for 2 h with the cytokine triad. Intriguingly, NLRP3 had a negative regulatory role in the cyclooxygenase (COX)-2 dependent production of prostaglandin (PGD)2 by BMMCs.
The expression of NLRs was also confirmed in human CBMCs, MCs from human colon specimens and HMC-1. Enoksson et al. [70] observed by FACS that CBMCs isolated from healthy donors expressed NOD1 and NOD2 protein. All tested individuals expressed NOD1 protein (NOD1+ CBMCs: 75.4 ± 8.3%), but only one donor expressed NOD2 receptor at low level (NOD2+ CBMCs: 6.6 ± 6.0%). Moreover, NOD1 specific agonist M-TriDAP induced an increase in CXCL8, CCL3, CCL4, and TNF secretion by CBMCs and this effect was mediated through p38 and/or RIP2-dependent pathways. Interestingly, a synergistic effect of the co-stimulation with M-TriDAP in combination with LPS on CXCL8 release by CBMCs was also observed. Analysis of FACS-sorted human CBMCs confirmed the expression of NOD2 and NLRP3, two of the major receptors for MDP [71]. Moreover, the effect of specific NLR agonists on sustained CBMCs activation was also observed. CBMCs were incubated with S. aureus PGN, MDP, and murabutide (a synthetic MDP analogue). PGN induced the production of IL-1β, IL-6, and CXCL8 by MCs. Neither NLR activator MDP nor synthetic analogue alone induced detectable MC response elicited by cytokine release. Interestingly, NLR agonists potentiated the Pam3CSK4-induced MC IL-6 response in a concentration-dependent manner.
Analysis of sequential sections of healthy human colon specimens with anti-tryptase and anti-NOD2 monoclonal antibodies (mAbs) revealed MCs with NOD2 [72]. Moreover, analysis of inflamed colon specimens from Crohn’s disease (CD) and ulcerative colitis (UC) patients revealed that NOD2+ MCs were localized in the lamina propria. Interestingly, the analysis of adjacent sections showed that the densities of NOD2+ MCs were significantly increased in CD specimens in comparison to UC specimens and controls. The same authors stressed that NOD2 mRNA and protein expression was markedly higher in IFN-γ-treated PBMCs and human lung MCs than in IFN-γ-untreated MCs.
The qPCR results showed that NOD2 mRNA was also detected in HMC-1 [73]. To establish the role of NOD2 in PGN-induced MC activation, HMC-1 were preincubated with either anti-NOD2 antibody or transfected with siRNA-NOD2. As a result, histamine release from pretreated or transfected HMC-1 was significantly inhibited as compared with controls. These results strongly suggest that NOD2 plays a critical role in PGN-induced HMC-1 activation.
RIG-like receptors (RLRs)
RLR family, their ligands and molecular structure
RLRs are a family of DExD/H box RNA helicases that function as cytoplasmic sensors of viral RNA. So far, three RLR members have been identified: RIG-I, MDA5 (melanoma differentiation associated factor 5), and LGP2 (laboratory of genetics and physiology 2) [74]. All RLR family members have a central DExD/H box RNA helicase domain with ATPase activity. The C-terminal region consists of a domain termed the RNA recognition domain which is responsible for binding RNA. In addition, the C-terminal domains of RIG-I and LGP2 have been shown to act as repressor domains (RD), ensuring that the receptors remain in an inactive form. Both RIG-I and MDA5 have two N-terminal CARDs, while LGP2 lacks this domain. For this reason, LGP2 is rather considered to be RIG-I and MDA5 signaling regulator than active receptor [74, 75]. Because RLRs detect various types of viral RNA, these molecules are one of the first line of defense against virus infection [76]. RIG-I senses short (< 300 bp) 5′ triphosphorylated dsRNA or ssRNA with double-stranded regions. Additionally, it binds specific dsRNAs lacking a 5′ phosphate or containing a 5′ monophosphate. In turn, MDA5 binds to longer (> 1 kb) dsRNA [77–79].
RLR signaling pathways
In the RIG-I pathway, RNA binding is modulated by polyubiquitination with TRIM25 and Riplet (RNF135) which act as E3 ubiquitin ligases. They mediate the covalent attachment of the K63-linked polyubiquitin chains. In turn, little is known about the molecules required for the MDA5 polyubiquitination. Following this, the CARDs of RIG-I and MDA5 trigger signaling cascades by interacting with the N-terminal CARD-containing adaptor IFN-β-promoter stimulator 1 (IPS-1/MAVS/CARDIF/VIsA) localized on the mitochondrial membrane. Dimerization of IPS-1 activates TRAF-2, TRAF-6, and TRADD, which recruits TRAF-3 and TRAF family member-associated NF-κB activator (TANK) to trigger the activation of tank-binding kinase-1 (TBK1) and I κB kinase epsilon (IKK-ε). Interestingly, NLRX1 may act as an inhibitor of IPS-1 signaling. In addition to the TRAFs and TRADD, translocase of the outer membrane 70 (TOM70) in the mitochondria and stimulator of interferon genes (STING) localizes to the outer membrane of the endoplasmic reticulum may also be involved in facilitating RIG-I signaling [32].
RLRs in MCs
Scarce data indicate that BMMCs and CBMCs express RLRs, and these observations were confirmed upon virus infection. Fukuda et al. [80] observed that resting mouse BMMCs express mRNA for MDA5 and RIG-I, and transcript levels of each receptor increased notably upon vesicular stomatitis virus (VSV) infection. Receptor expression was also established at the protein level by western blotting. Consistent with mRNA results, increased expression of MDA5 and RIG-I protein was noted upon VSV infection. To determine the receptors responsible for the recognition and activation of MCs after VSV infection, BMMCs were treated with siRNA for the particular molecules and examined the antiviral cytokine responses. siRNA inhibition of MDA5 and RIG-I significantly reduced the level of IFN-α, IFN-β, and IL-6 production by MCs in response to VSV. This specific knockdown of each receptor confirmed the contribution of MDA5 and RIG-I in MC anti-VSV defense. Additionally, Sendai virus infection of mature human MCs resulted in a potent up-regulation of RIG-I and MDA-5 mRNA expression that was noticeable after 4 h and was increased further upon 20 h after the onset of infection [81]. Furthermore, stimulation of human MCs by IFN-β strongly up-regulated the expression of MDA-5 and RIG-I. In turn, Brown et al. [82] stated that mRNA RIG-I and MDA5 in human CBMCs was increased following dengue virus and dengue-immune sera treatment.
Concluding remarks
In summary, hitherto sources assume that MCs, as one of the first line of defense, are adapted to fulfill a wide range of functions in defending against various exogenous intruders or endogenous deleterious factors. Notably, MCs have a broad set of pattern recognition molecules, thus can identify and bind bacterial, viral, fungal, and parasitic PAMPs as well as various endogenous molecules generated in response to infection (Fig. 1). Therefore, MCs may be considered as our internal bodyguards. These abilities allow to conclude that MCs contribute to the control of a wide range of infections and orchestrate mobilization of cells involved in innate and adaptive immune responses.
Fig. 1.
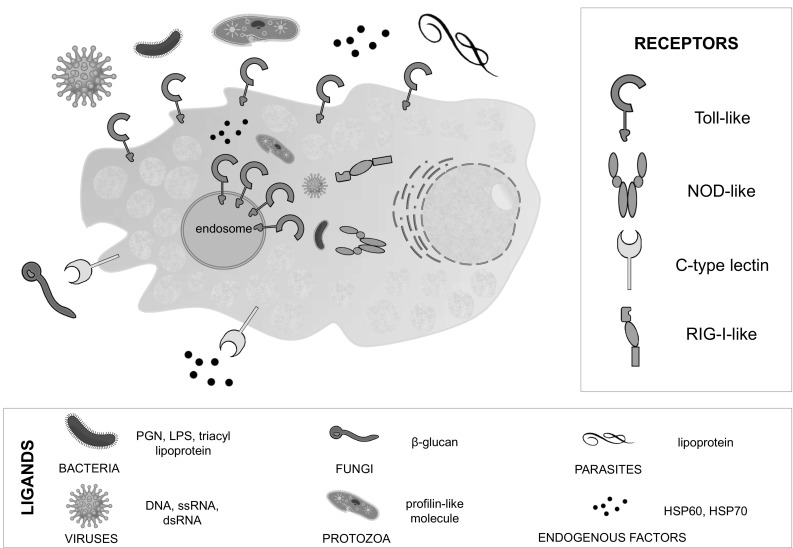
TLR, CLR, NLR and RLR arrangement in MCs: intracellularly in the cytoplasm or in MC membranes, including cell membrane and endosome. Ligand legend contains exemplary ligands derived from a given type of organism/molecule
Acknowledgements
This work was supported by Medical University of Lodz (Grant nos. 503/6-164-01/503-61-001 and 503/6-164-01/503-66-001).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Amin K. The role of mast cells in allergic inflammation. Respir Med. 2012;106:9–14. doi: 10.1016/j.rmed.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 2.Galli SJ, Tsai M. IgE and mast cells in allergic disease. Nat Med. 2012;18:693–704. doi: 10.1038/nm.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu F, Huang S. The roles of mast cells in parasitic protozoan infections. Front Immunol. 2017;8:363. doi: 10.3389/fimmu.2017.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abraham SN, St John AL. Mast cell-orchestrated immunity to pathogens. Nat Rev Immunol. 2010;10:440–452. doi: 10.1038/nri2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conti P, Carinci F, Caraffa A, Ronconi G, Lessiani G, Theoharides TC. Link between mast cells and bacteria: antimicrobial defense, function and regulation by cytokines. Med Hypotheses. 2017;106:10–14. doi: 10.1016/j.mehy.2017.06.018. [DOI] [PubMed] [Google Scholar]
- 6.Marshall JS. Mast-cell responses to pathogens. Nat Rev Immunol. 2004;4:787–799. doi: 10.1038/nri1460. [DOI] [PubMed] [Google Scholar]
- 7.Urb M, Sheppard DC. The role of mast cells in the defence against pathogens. PLoS Pathog. 2012;8:e1002619. doi: 10.1371/journal.ppat.1002619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saluja R, Metz M, Maurer M. Role and relevance of mast cells in fungal infections. Front Immunol. 2012;3:146. doi: 10.3389/fimmu.2012.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collington SJ, Williams TJ, Weller CL. Mechanisms underlying the localisation of mast cells in tissues. Trends Immunol. 2011;32:478–85. doi: 10.1016/j.it.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Krystel-Whittemore M, Dileepan KN, Wood JG. Mast cell: a multi-functional master cell. Front Immunol. 2016;6:620. doi: 10.3389/fimmu.2015.00620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pastwińska J, Agier J, Dastych J, Brzezińska-Błaszczyk E. Mast cells as the strength of the inflammatory process. Pol J Pathol. 2017;68:187–196. doi: 10.5114/pjp.2017.71526. [DOI] [PubMed] [Google Scholar]
- 12.Theoharides TC, Alysandratos KD, Angelidou A, Delivanis DA, Sismanopoulos N, Zhang B, et al. Mast cells and inflammation. Biochim Biophys Acta. 2012;1822:21–33. doi: 10.1016/j.bbadis.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robuffo I, Toniato E, Tettamanti L, Mastrangelo F, Ronconi G, Frydas I, et al. Mast cell in innate immunity mediated by proinflammatory and antiinflammatory IL-1 family members. J Biol Regul Homeost Agents. 2017;31:837–842. [PubMed] [Google Scholar]
- 14.Conti P, Caraffa Al, Kritas SK, Ronconi G, Lessiani G, Toniato E, Theoharides TC. Mast cell, pro-inflammatory and anti-inflammatory: Jekyll and Hyde, the story continues. J Biol Regul Homeost Agents. 2017;31:263–267. [PubMed] [Google Scholar]
- 15.Di Nardo A, Yamasaki K, Dorschner RA, Lai Y, Gallo RL. Mast cell cathelicidin antimicrobial peptide prevents invasive group A Streptococcus infection of the skin. J Immunol. 2008;180:7565–7573. doi: 10.4049/jimmunol.180.11.7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malaviya R, Ross EA, MacGregor JI, Ikeda T, Little JR, Jakschik BA, Abraham SN. Mast cell phagocytosis of FimH-expressing enterobacteria. J Imunol. 1994;152:1907–1914. [PubMed] [Google Scholar]
- 17.Malaviya R, Twesten NJ, Ross EA, Abraham SN, Pfeifer JD. Mast cells process bacterial Ags through a phagocytic route for class I MHC presentation to T cells. J Immunol. 1996;156:1490–1496. [PubMed] [Google Scholar]
- 18.Möllerherm H, von Köckritz-Blickwede M, Branitzki-Heinemann K. Antimicrobial activity of mast cells: role and relevance of extracellular DNA traps. Front Immunol. 2016;7:265. doi: 10.3389/fimmu.2016.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dimitriadou V, Mécheri S, Koutsilieris M, Fraser W, Al-Daccak R, Mourad W. Expression of functional major histocompatibility complex class II molecules on HMC-1 human mast cells. J Leukoc Biol. 1998;64:791–799. doi: 10.1002/jlb.64.6.791. [DOI] [PubMed] [Google Scholar]
- 20.Kambayashi T, Allenspach EJ, Chang JT, Zou T, Shoag JE, Reiner SL, et al. Inducible MHC class II expression by mast cells supports effector and regulatory T cell activation. J Immunol. 2009;182:4686–4695. doi: 10.4049/jimmunol.0803180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Z, Xu MJ, Gao B. Hepatocytes: a key cell type for innate immunity. Cell Mol Immunol. 2016;13:301–315. doi: 10.1038/cmi.2015.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Visintin A, Mazzoni A, Spitzer JH, Wyllie DH, Dower SK, Segal DMJ. Regulation of Toll-like receptors in human monocytes and dendritic cells. J Immunol. 2001;166:249–255. doi: 10.4049/jimmunol.166.1.249. [DOI] [PubMed] [Google Scholar]
- 23.Hemmi H, Akira S. TLR signalling and the function of dendritic cells. Chem Immunol Allergy. 2005;86:120–135. doi: 10.1159/000086657. [DOI] [PubMed] [Google Scholar]
- 24.Buchta CM, Bishop GA. Toll-like receptors and B cells: functions and mechanisms. Immunol Res. 2014;59:12–22. doi: 10.1007/s12026-014-8523-2. [DOI] [PubMed] [Google Scholar]
- 25.Kabelitz D. Expression and function of Toll-like receptors in T lymphocytes. Curr Opin Immunol. 2007;19:39–45. doi: 10.1016/j.coi.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 26.McClure R, Massari P. TLR-dependent human mucosal epithelial cell responses to microbial pathogens. Front Immunol. 2014;5:386. doi: 10.3389/fimmu.2014.00386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fitzner N, Clauberg S, Essmann F, Liebmann J, Kolb-Bachofen V. Human skin endothelial cells can express all 10 TLR genes and respond to respective ligands. Clin Vaccine Immunol. 2008;15:138–146. doi: 10.1128/CVI.00257-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prince LR, Whyte MK, Sabroe I, Parker LC. The role of TLRs in neutrophil activation. Curr Opin Pharmacol. 2011;11:397–403. doi: 10.1016/j.coph.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 29.Souza-Fonseca-Guimaraes F, Parlato M, Philippart F, Misset B, Cavaillon JM, Adib-Conquy M. Toll-like receptors expression and interferon-γ production by NK cells in human sepsis. Crit Care. 2012;16:R206. doi: 10.1186/cc11838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brzezińska-Błaszczyk E, Wierzbicki M. Mast cell Toll-like receptors (TLRs) Postepy Hig Med Dosw. 2010;64:11–21. [PubMed] [Google Scholar]
- 31.Sandig H, Bulfone-Paus S. TLR signaling in mast cells: common and unique features. Front Immunol. 2012;3:185. doi: 10.3389/fimmu.2012.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 33.Koblansky AA, Jankovic D, Oh H, Hieny S, Sungnak W, Mathur R, et al. Recognition of profilin by toll-like receptor 12 is critical for host resistance to Toxoplasma gondii. Immunity. 2013;38:119–130. doi: 10.1016/j.immuni.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi Z, Cai Z, Sanchez A, Zhang T, Wen S, Wang J, et al. A novel Toll-like receptor that recognizes vesicular stomatitis virus. J Biol Chem. 2011;286:4517–4524. doi: 10.1074/jbc.M110.159590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uematsu S, Akira S. Toll-like receptors (TLRs) and their ligands. Handb Exp Pharmacol. 2008;183:1–20. doi: 10.1007/978-3-540-72167-3_1. [DOI] [PubMed] [Google Scholar]
- 36.Botos I, Segal DM, Davies DR. The structural biology of Toll-like receptors. Structure. 2011;19:447–459. doi: 10.1016/j.str.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karpov V, Ilarraza R, Catalli A, Kulka M. Cysteinyl leukotrienes C4 and D4 downregulate human mast cell expression of toll-like receptors 1 through 7. J Biol Regul Homeost Agents. 2018;32:233–239. [PubMed] [Google Scholar]
- 38.Agier J, Żelechowska P, Kozłowska E, Brzezińska-Błaszczyk E. Expression of surface and intracellular Toll-like receptors by mature mast cells. Cent Eur J Immunol. 2016;41:333–338. doi: 10.5114/ceji.2016.65131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agier J, Brzezińska-Błaszczyk E, Żelechowska P, Wiktorska M, Pietrzak J, Różalska S. Cathelicidin LL-37 affects surface and intracellular Toll-like receptor expression in tissue mast cells. J Immunol Res. 2018 doi: 10.1155/2018/7357162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vautier S, MacCallum DM, Brown GD. C-type lectin receptors and cytokines in fungal immunity. Cytokine. 2012;58:89–99. doi: 10.1016/j.cyto.2011.08.031. [DOI] [PubMed] [Google Scholar]
- 41.Thebault P, Lhermite N, Tilly G, Le Texier L, Quillard T, Heslan M, et al. The C-type lectin-like receptor CLEC-1, expressed by myeloid cells and endothelial cells, is up-regulated by immunoregulatory mediators and moderates T cell activation. J Immunol. 2009;183:3099–3108. doi: 10.4049/jimmunol.0803767. [DOI] [PubMed] [Google Scholar]
- 42.Figdor CG, van Kooyk Y, Adema GJ. C-type lectin receptors on dendritic cells and Langerhans cells. Nat Rev Immunol. 2002;2:77–84. doi: 10.1038/nri723. [DOI] [PubMed] [Google Scholar]
- 43.van Kooyk Y. C-type lectins on dendritic cells: key modulators for the induction of immune responses. Biochem Soc Trans. 2008;36:1478–1481. doi: 10.1042/BST0361478. [DOI] [PubMed] [Google Scholar]
- 44.Kennedy AD, Willment JA, Dorward DW, Williams DL, Brown GD, DeLeo FR. Dectin-1 promotes fungicidal activity of human neutrophils. Eur J Immunol. 2007;37:467–478. doi: 10.1002/eji.200636653. [DOI] [PubMed] [Google Scholar]
- 45.Arce I, Martínez-Muñoz L, Roda-Navarro P, Fernández-Ruiz E. The human C-type lectin CLECSF8 is a novel monocyte/macrophage endocytic receptor. Eur J Immunol. 2004;34:210–220. doi: 10.1002/eji.200324230. [DOI] [PubMed] [Google Scholar]
- 46.Geijtenbeek TBH, Gringhuis SI. Signalling through C‑type leptin receptors: shaping immune responses. Nat Rev Immunol. 2009;9:465–479. doi: 10.1038/nri2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hardison SE, Brown GD. C-type lectin receptors orchestrate antifungal immunity. Nat Immunol. 2012;13:817–822. doi: 10.1038/ni.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Drickamer K, Taylor ME. Recent insights into structures and functions of C-type lectins in the immune system. Curr Opin Struct Biol. 2015;34:26–34. doi: 10.1016/j.sbi.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoving JC, Wilson GJ, Brown GD. Signalling C-type lectin receptors, microbial recognition and immunity. Cell Microbiol. 2014;16:185–194. doi: 10.1111/cmi.12249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barbosa-Lorenzi VC, Peyda S, Scheynius A, Nilsson G, Lunderius-Andersson C. Curdlan induces selective mast cell degranulation without concomitant release of LTC4, IL-6 or CCL2. Immunobiology. 2017;222:647–650. doi: 10.1016/j.imbio.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 51.Honjoh C, Chihara K, Yoshiki H, Yamauchi S, Takeuchi K, Kato Y, et al. Association of C-type lectin mincle with FcεRIβγ subunits leads to functional activation of RBL-2H3 cells through Syk. Sci Rep. 2017;7:46064. doi: 10.1038/srep46064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nieto-Patlán A, Campillo-Navarro M, Rodríguez-Cortés O, Muñoz-Cruz S, Wong-Baeza I, Estrada-Parra S, et al. Recognition of Candida albicans by dectin-1 induces mast cell activation. Immunobiology. 2015;220:1093–1100. doi: 10.1016/j.imbio.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 53.Olynych TJ, Jakeman DL, Marshall JS. Fungal zymosan induces leukotriene production by human mast cells through a dectin-1-dependent mechanism. J Allergy Clin Immunol. 2006;118:837–843. doi: 10.1016/j.jaci.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 54.Ribbing C, Engblom C, Lappalainen J, Lindstedt K, Kovanen PT, Karlsson MA, et al. Mast cells generated from patients with atopic eczema have enhanced levels of granule mediators and an impaired dectin-1 expression. Allergy. 2011;66:110–119. doi: 10.1111/j.1398-9995.2010.02437.x. [DOI] [PubMed] [Google Scholar]
- 55.Vukman KV, Ravidá A, Aldridge AM, O’Neill SM. Mannose receptor and macrophage galactose-type lectin are involved in Bordetella pertussis mast cell interaction. J Leukoc Biol. 2017;4:439–448. doi: 10.1189/jlb.0313130. [DOI] [PubMed] [Google Scholar]
- 56.Yang Z, Marshall JS. Zymosan treatment of mouse mast cells enhances dectin-1 expression and induces dectin-1-dependent reactive oxygen species (ROS) generation. Immunobiology. 2009;214:321–330. doi: 10.1016/j.imbio.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 57.Shaw MH, Thornik R, Kim YG, Nuñez G. NOD-like receptors (NLRs): bona fide intracellular microbial sensors. Curr Opin Immunol. 2008;20:377–382. doi: 10.1016/j.coi.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ting JP, Lovering RC, Alnemri ES, Bertin J, Boss JM, Davis BK, et al. The NLR gene family: a standard nomenclature. Immunity. 2008;280:285–287. doi: 10.1016/j.immuni.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koonin EV, Aravind L. The NACHT family—a new group of predicted NTPases implicated in apoptosis and MHC transcription activation. Trends Biochem Sci. 2000;25:223–224. doi: 10.1016/S0968-0004(00)01577-2. [DOI] [PubMed] [Google Scholar]
- 60.MacDonald JA, Wijekoon CP, Liao KC, Muruve DA. Biochemical and structural aspects of the ATP-binding domain in inflammasome-forming human NLRP proteins. IUBMB Life. 2013;65:851–862. doi: 10.1002/iub.1210. [DOI] [PubMed] [Google Scholar]
- 61.Jha S, Ting JP. Inflammasome-associated nucleotide-binding domain, leucine-rich repeat proteins and inflammatory diseases. J Immunol. 2009;183:7623–7629. doi: 10.4049/jimmunol.0902425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, Saab L, et al. An essential role for Nod1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol. 2003;4:702–707. doi: 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- 63.Inohara N, Ogura Y, Fontalba A, Gutierrez O, Pons F, Crespo J, et al. Host recognition of bacterial muramyl dipeptide mediated through Nod2. Implications for Crohn’s disease. J Biol Chem. 2003;278:5509–5512. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 64.Girardin SE, Boneca IG, Carneiro LA, Antignac A, Jéhanno M, Viala J, et al. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science. 2003;300:1584–1587. doi: 10.1126/science.1084677. [DOI] [PubMed] [Google Scholar]
- 65.Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G, et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003;278:8869–8872. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 66.Ogura Y, Sutterwala FS, Flavell RA. The inflammasome: first line of the immune response to cell stress. Cell. 2006;126:659–662. doi: 10.1016/j.cell.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 67.Zabucchi G, Trevisan E, Vita F, Soranzo MR, Borelli V. NOD1 and NOD2 interact with the phagosome cargo in mast cells: a detailed morphological evidence. Inflammation. 2015;38:1113–1125. doi: 10.1007/s10753-014-0077-x. [DOI] [PubMed] [Google Scholar]
- 68.Xie X, Wang L, Gong F, Xia C, Chen J, Song Y, et al. Intracellular Staphylococcus aureus-induced NF-κB activation and proinflammatory responses of P815 cells are mediated by NOD2. J Huazhong Univ Sci Technolog Med Sci. 2012;32:317–323. doi: 10.1007/s11596-012-0055-1. [DOI] [PubMed] [Google Scholar]
- 69.Ueno N, Taketomi Y, Koga K, Atsumi Y, Kikuchi-Yanoshita R, Kudo I, Murakami M. A negative regulator of delayed prostaglandin D2 production in mouse mast cells. Biochim Biophys Acta. 2008;1781:415–421. doi: 10.1016/j.bbalip.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 70.Enoksson M, Ejendal KF, McAlpine S, Nilsson G, Lunderius-Andersson C. Human cord blood-derived mast cells are activated by the Nod1 agonist M-TriDAP to release pro-inflammatory cytokines and chemokines. J Innate Immun. 2011;3:142–149. doi: 10.1159/000321933. [DOI] [PubMed] [Google Scholar]
- 71.Haidl ID, McAlpine SM, Marshall JS. Enhancement of mast cell IL-6 production by combined toll-like and nucleotide-binding oligomerization domain-like receptor activation. Int Arch Allergy Immunol. 2011;154:227–235. doi: 10.1159/000321109. [DOI] [PubMed] [Google Scholar]
- 72.Okumura S, Yuki K, Kobayashi R, Okamura S, Ohmori K, Saito H, et al. Hyperexpression of NOD2 in intestinal mast cells of Crohn’s disease patients: preferential expression of inflammatory cell-recruiting molecules via NOD2 in mast cells. Clin Immunol. 2009;130:175–185. doi: 10.1016/j.clim.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 73.Wu L, Feng B-S, He S-H, Zheng PY, Croitoru K, Yang PC. Bacterial peptidoglycan breaks down intestinal tolerance via mast cell activation: the role of TLR2 and NOD2. Immunol Cell Biol. 2007;85:538–545. doi: 10.1038/sj.icb.7100079. [DOI] [PubMed] [Google Scholar]
- 74.Onoguchi K, Yoneyama M, Fujita T. Retinoic acid-inducible gene-I-like receptors. J Interferon Cytokine Res. 2011;31:27–31. doi: 10.1089/jir.2010.0057. [DOI] [PubMed] [Google Scholar]
- 75.Loo YM. Gale MJr. Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reikine S, Nguyen JB, Modis Y. Pattern recognition and signaling mechanisms of RIG-I and MDA5. Front Immunol. 2014;5:342. doi: 10.3389/fimmu.2014.00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hornung V, Ellegast J, Kim S, Brzózka K, Jung A, Kato H, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 78.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 79.Pichlmair A, Schulz O, Tan CP, Näslund TI, Liljeström P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 80.Fukuda M, Ushio H, Kawasaki J, Niyonsaba F, Takeuchi M, Baba T, et al. Expression and functional characterization of retinoic acid-inducible gene-I-like receptors of mast cells in response to viral infection. J Innate Immun. 2013;5:163–173. doi: 10.1159/000343895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lappalainen J, Rintahaka J, Kovanen PT, Matikainen S, Eklund KK. Intracellular RNA recognition pathway activates strong anti-viral response in human mast cells. Clin Exp Immunol. 2013;172:121–128. doi: 10.1111/cei.12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brown MG, McAlpine SM, Huang YY, Haidl ID, Al-Afif A, Marshall JS, Anderson R. RNA sensors enable human mast cell anti-viral chemokine production and IFN-mediated protection in response to antibody-enhanced dengue virus infection. PLoS One. 2012;7:e34055. doi: 10.1371/journal.pone.0034055. [DOI] [PMC free article] [PubMed] [Google Scholar]


