Adaptation of viruses to their hosts is critical for viral transmission between different species. Previous studies had identified changes in a protein from the APOBEC3 family that influenced the species specificity of simian immunodeficiency viruses (SIVs) in African green monkeys. We studied the evolution of a related protein in the same system, APOBEC3H, which has experienced a loss of function in humans. This evolutionary approach revealed that recurrent loss of APOBEC3H activity has taken place during primate evolution, suggesting that APOBEC3H places a fitness cost on hosts. The variability of APOBEC3H activity between different primates highlights the differential selective pressures on the APOBEC3 gene family.
KEYWORDS: APOBEC3H, African green monkey, evolution, human immunodeficiency virus, innate immunity, lentiviruses
ABSTRACT
Genes in the APOBEC3 family encode cytidine deaminases that provide a barrier against viral infection and retrotransposition. Of all the APOBEC3 genes in humans, APOBEC3H (A3H) is the most polymorphic: some genes encode stable and active A3H proteins, while others are unstable and poorly antiviral. Such variation in human A3H affects interactions with the lentiviral antagonist Vif, which counteracts A3H via proteasomal degradation. In order to broaden our understanding of A3H-Vif interactions, as well as its evolution in Old World monkeys, we characterized A3H variation within four African green monkey (AGM) subspecies. We found that A3H is highly polymorphic in AGMs and has lost antiviral activity in multiple Old World monkeys. This loss of function was partially related to protein expression levels but was also influenced by amino acid mutations in the N terminus. Moreover, we demonstrate that the evolution of A3H in the primate lineages leading to AGMs was not driven by Vif. Our work suggests that the activity of A3H is evolutionarily dynamic and may have a negative effect on host fitness, resulting in its recurrent loss in primates.
IMPORTANCE Adaptation of viruses to their hosts is critical for viral transmission between different species. Previous studies had identified changes in a protein from the APOBEC3 family that influenced the species specificity of simian immunodeficiency viruses (SIVs) in African green monkeys. We studied the evolution of a related protein in the same system, APOBEC3H, which has experienced a loss of function in humans. This evolutionary approach revealed that recurrent loss of APOBEC3H activity has taken place during primate evolution, suggesting that APOBEC3H places a fitness cost on hosts. The variability of APOBEC3H activity between different primates highlights the differential selective pressures on the APOBEC3 gene family.
INTRODUCTION
The seven members of the APOBEC3 (A3) gene family in primates encode cytidine deaminases involved in innate immune defense against retroviruses and retroelements (1–3). Four A3 enzymes are known to potently restrict the replication of lentiviruses, like simian immunodeficiency virus (SIV) and human immunodeficiency virus (HIV): A3D, A3F, A3G, and A3H (4). A3 proteins are packaged into budding virions and cause G-to-A hypermutation of viral DNA, although deamination-independent modes of restriction have also been characterized (5, 6). Hypermutation of viral DNA results in detrimental mutations that render the virus inactive and thus protects new cells from infection. However, lentiviruses have evolved a mechanism to evade A3 restriction by encoding a viral antagonist, Vif, that binds and targets A3 proteins for proteasomal degradation via a cellular E3 ubiquitin ligase complex (7). Given that A3-Vif interactions often act in a species-specific manner, adaptation of Vif to host A3 proteins is important for successful adaptation of lentiviruses to their hosts (8–12).
Various A3 proteins differ in their abilities to restrict viral infection. For instance, A3G is the most potent A3-mediated inhibitor of HIV-1, while A3A and A3B have limited antiviral potential (4, 13). In humans, the A3H protein is especially remarkable because multiple polymorphisms drastically impact its antiviral activity (14–17). Two independent mutations have occurred in human evolution that destabilized A3H (14); genes that encode R105G or a deletion of amino acid 15 make unstable proteins (haplotypes I, III, IV, and VI) that have lost antiviral activity, while those without these changes make stable proteins (haplotypes II, V, and VII) that potently restrict HIV-1 (14–16, 18). Stability and antiviral activity of human A3H have been further linked to subcellular localization: unstable/poorly antiviral proteins are more nuclear, while stable/antiviral proteins remain cytoplasmic (19). A3H haplotype I has been associated with breast and lung cancer (20), as has another nuclear A3, A3B (21), suggesting that in some cases A3 activity may be detrimental to the host. While such events have occurred in humans, examples of gains or losses of A3 activity over evolutionary time in other primates have been less explored.
The A3H genetic polymorphisms present in human populations also impact the interactions between human A3H and HIV-1 Vif (18, 22–24). Stable A3H proteins are only partially susceptible to degradation by Vif from the LAI isolate of HIV-1 and not at all by HIV-1 NL4-3 Vif (22, 24). However, studies using human cohorts carrying different haplotypes of A3H have shown that Vif proteins from some primary virus strains are able to antagonize stable A3H proteins (18, 23, 25, 26). These studies suggest that there is selection in vivo for Vif strains that counteract the stably expressed forms of A3H in infected people (18, 23, 25, 26). Furthermore, the cross-species transmissions that led to adaptation of SIV from monkeys to chimpanzees to humans, giving rise to HIV-1, involved adaptation of Vif to antagonize the A3 proteins found in each host (8, 9, 11, 12), including SIVcpz Vif to antagonize human A3H (12).
The study of the evolution of lentivirus-host interactions within Old World monkeys has provided insights into the longer-term dynamics of the evolutionary arms race between host antiviral proteins and their lentiviral targets (8, 10, 27). African green monkeys (AGMs), in particular, provide a unique opportunity to assess the evolutionary forces governing interactions between lentiviruses and their hosts, since the genus Chlorocebus encompasses four geographically distinct subspecies (vervets, sabaeus, tantalus, and grivets), each of which is infected with a species-specific subtype of SIVagm (28–30). These SIV subtypes are named for the subspecies they infect: SIVagm.ver, SIVagm.sab, SIVagm.tan, and SIVagm.gri (30). Furthermore, this system is particularly powerful because, although these subspecies are closely genetically related, enough divergence exists to have allowed species-specific lentiviral infections to occur.
Previous studies have demonstrated that many genes involved in antiviral immunity are polymorphic in AGMs, and some changes at the protein level are critical for interactions with viral antagonists (9, 31, 32). A3G in particular was discovered to encode species-specific polymorphisms (9). Amino acid changes in the Vif binding domain of AGM A3G (sites 128 and 130) confer protection against SIVagm strains from other subspecies. For example, K128E, found in grivet monkeys, is resistant to all Vifs except SIVagm.gri, while D130H, found in sabaeus monkeys, is resistant to Vifs from SIVagm.ver and SIVagm.tan (9). This demonstrates that Vif continues to drive the evolution of A3G and contributes to the species specificity of lentiviruses in AGM populations. It is unknown whether A3H has a similar role in these primates.
In this study, we asked whether we could identify a host-virus “arms race” between A3H in AGMs and Vif proteins encoded by the SIVs that infect these species. Surprisingly, we found that although A3H is highly polymorphic in AGMs, the antiviral activity of A3H has been largely lost. The reduced antiviral activity is in part caused by lower protein expression levels, although some amino acid changes also lower antiviral activity independently of protein expression levels. By reconstructing ancestral A3H proteins spanning evolution in AGMs and in other Old World monkeys, we found that there has been recurrent loss of A3H activity in some, but not all, Old World monkeys. We also identified amino acids that affect A3H restriction which map to regions implicated in RNA binding (33–35). Thus, our data support a model where A3H antiviral activity has been repeatedly lost throughout evolution. This argues that there is a longer-scale dynamic between the cost and benefit for A3H function in primates that is not necessarily driven by interactions with its antagonist.
(This article was submitted to an online preprint archive [36].)
RESULTS
A3H is highly polymorphic in AGM subspecies, but all tested alleles have low antiviral activity.
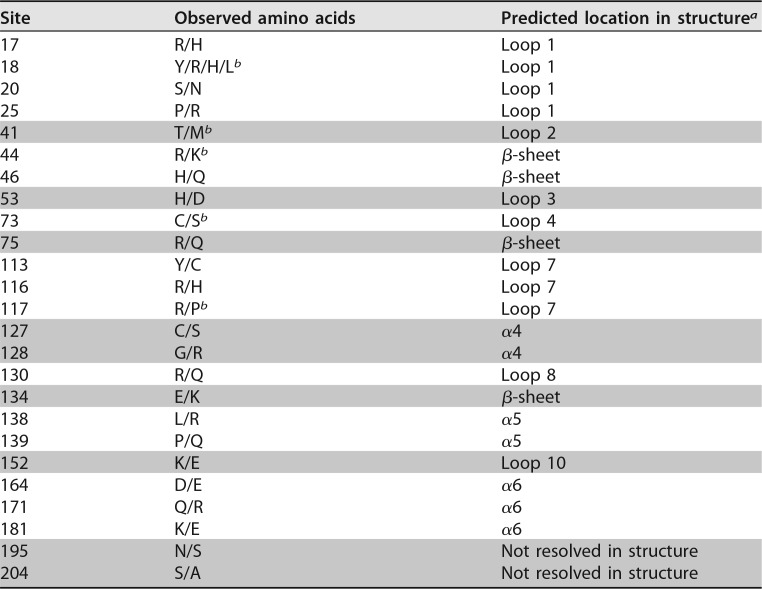
Polymorphisms in human A3H are known to affect its antiviral activity, as well as its interactions with Vif (14, 16, 18, 23, 24). Similarly, polymorphisms in AGM A3G impact interactions with Vif, suggesting an ongoing and ancient genetic conflict between A3G and SIVagm Vif (9). These observations prompted us to explore the genetic landscape of A3H in AGMs and other Old World monkeys to determine if A3H evolution has been driven by Vif over a broad evolutionary scale. First, we sequenced A3H in 50 AGM samples collected from all four subspecies infected with a species-specific SIV: vervet, sabaeus, tantalus, and grivet monkeys. The mitochondrial DNA of these animals had also been previously sequenced and was confirmed to cluster by AGM subspecies (9). Sequence analysis of 80 independent A3H genes revealed 34 single-nucleotide polymorphisms (SNPs), 6 of which are synonymous changes at a frequency higher than 5%. Twenty-eight SNPs are nonsynonymous, and 23 are found in more than one individual (Table 1). Most polymorphisms are represented in all four subspecies (Fig. 1A and B), except for low-frequency variants found in only one AGM and two nonsynonymous mutations found in two vervet sequences at amino acid residues 113 and 116. Phylogenetic analysis showed that A3H sequences from all AGM subspecies are paraphyletic (Fig. 1A), similar to a previous study examining A3G in these species (9).
TABLE 1.
Polymorphisms identified in AGM A3H sequences
FIG 1.
Sequence and phylogenetic analyses of A3H in African green monkeys. (A) The evolutionary relationship among 80 full-length AGM A3H genes was inferred by Bayesian MCMC phylogenetic reconstruction. The red asterisks mark nodes that have a posterior probability of >0.5. The colored boxes indicate the subspecies of origin (red, vervet; yellow, sabaeus; blue, tantalus; and green, grivet). The blue asterisks mark cloned haplotypes, numbered 1 to 11 on the right. The arrow marks divergence of the phylogenetic tree between two clades. (B) Numbers of AGM individuals carrying genes encoding haplotypes 1 to 11, color coded by subspecies, similar to A3H phylogeny.
Nonsynonymous SNPs are spread throughout the protein (Table 1), although one group is clustered between amino acids 127 and 139. The changes are tightly linked and result in divergence of the phylogenetic tree between two clades (Fig. 1A, arrow). Haplotypes with a CGRELP motif comprise one clade on the tree, while the other has an SRQKRQ motif. Residues 127 and 128 are located on the α4 helix, which has been implicated in A3H-Vif binding (37, 38). Furthermore, the regions of A3H with the greatest numbers of nonsynonymous mutations are in the predicted loops 1 and 7 and the α6 helix (Table 1), which were recently shown by structural studies to be involved in an interaction between A3H and a cocrystalized RNA (34, 35). These results indicate that, similar to A3G, SAMHD1, and other antiviral genes in AGMs (8, 31, 32), there is extensive polymorphism in AGM A3H.
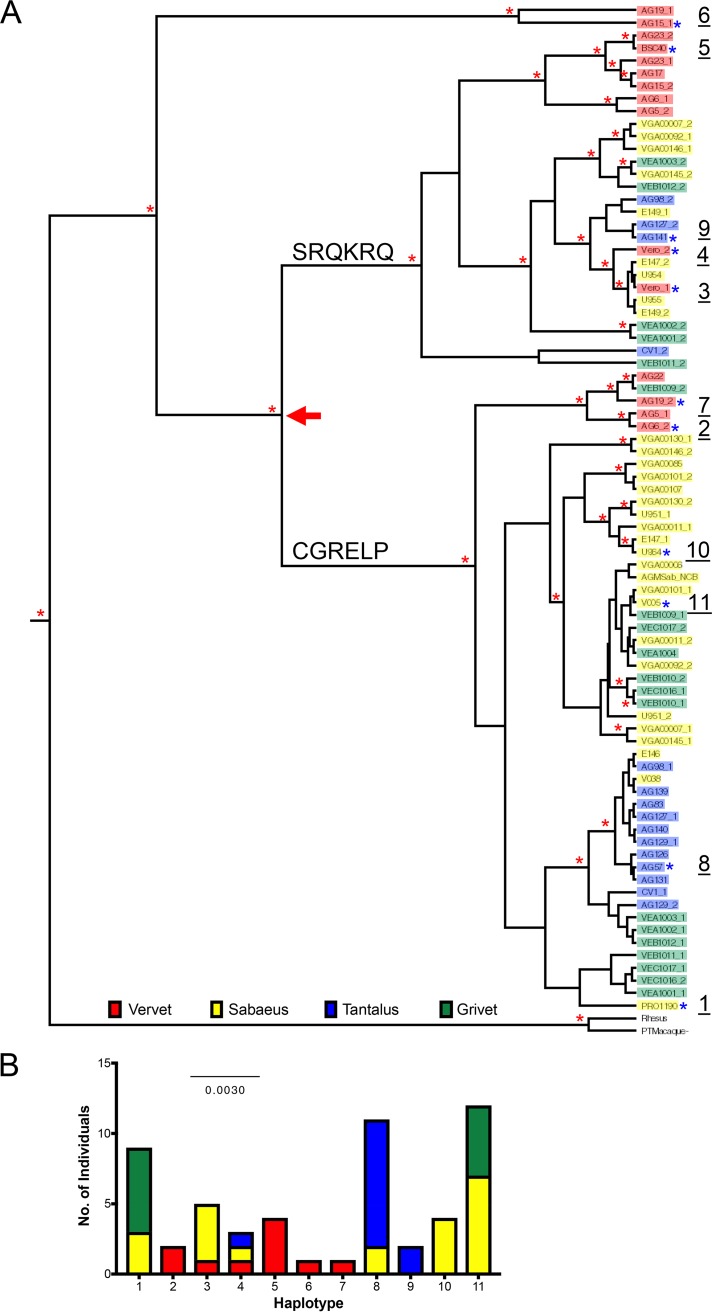
The presence of numerous polymorphisms suggests there may be functional consequences for either antiviral activity or Vif antagonism. To determine whether nonsynonymous polymorphisms in A3H impact antiviral activity, we tested the abilities of A3H protein variants to restrict lentiviruses. We cloned haplotypes, numbered 1 to 11, that are representative of each clade in the phylogenetic tree into a mammalian expression vector for functional analysis (Fig. 1B). These haplotypes also represent all four subspecies of AGMs. Out of the 28 total nonsynonymous SNPs found in AGMs (Table 1), 11 unique protein sequences were tested and 20 polymorphic sites were characterized (Table 2).
TABLE 2.
Polymorphic sites in tested A3H haplotypes
a Haplotype number of A3H allele.
b Amino acid positions with nonsynonymous mutations across the tested haplotypes. Shading indicates a different amino acid residue than in AGM A3H haplotype 1.
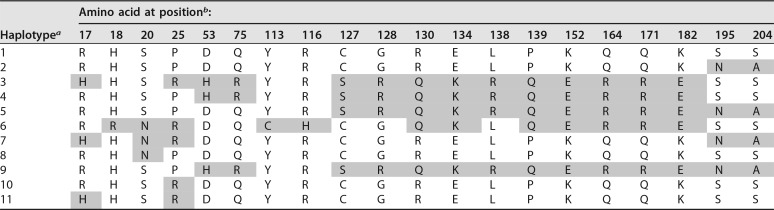
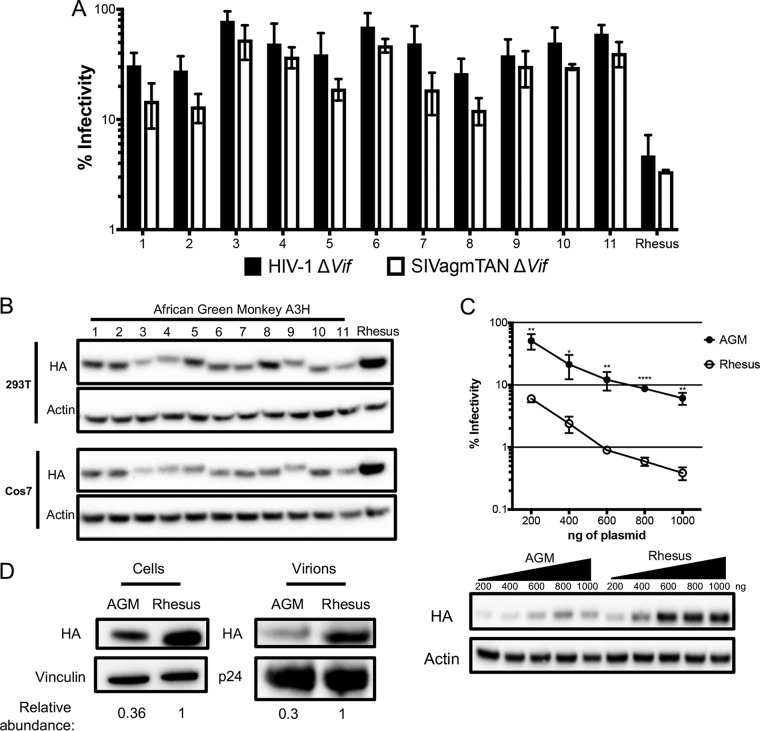
A3H-expressing plasmids were cotransfected into HEK293T cells for single-round infectivity experiments with HIVΔenvΔvif or SIVagm.TANΔenvΔvif proviruses and a VSV-G expression plasmid for pseudotyping to measure antiviral activity. Viral supernatants were collected, normalized for reverse transcriptase (RT) activity, and used to infect SupT1 cells, and luciferase activity from a gene carried in the proviral plasmid was measured relative to a no-A3 control (see Materials and Methods). We found that none of the AGM A3H variants restricted lentiviruses as potently as the A3H from rhesus macaque (Fig. 2A), an Old World monkey that has been previously characterized for its A3H activity (4, 14, 15). Rhesus A3H restricted viral infection with HIVΔvif by approximately 21-fold, while AGM A3H variants restricted viral infection by no more than 3-fold, and some not at all (Fig. 2A). Although activity against HIV-1Δvif correlates with activity against other lentiviruses, we also validated this result using SIVagm.TANΔvif, a strain originally isolated from tantalus monkeys. We found that the AGM A3H variants were also poorly restrictive against SIVagm.TANΔvif compared to the activity of rhesus macaque A3H (Fig. 2A). SIVagm.TANΔvif was observed to be more sensitive to restriction by all A3H proteins tested, similar to a previous study from our laboratory using A3G (9), although the difference in viral inhibition for specificity between inhibition of HIV-1 or SIVagm.TAN within haplotypes was not statistically significant (unpaired t test; P > 0.05). This indicates that A3H variants encoded by AGMs have poor antiviral activity against at least two separate lentiviruses and that this is not due to species specificity (Fig. 2B).
FIG 2.
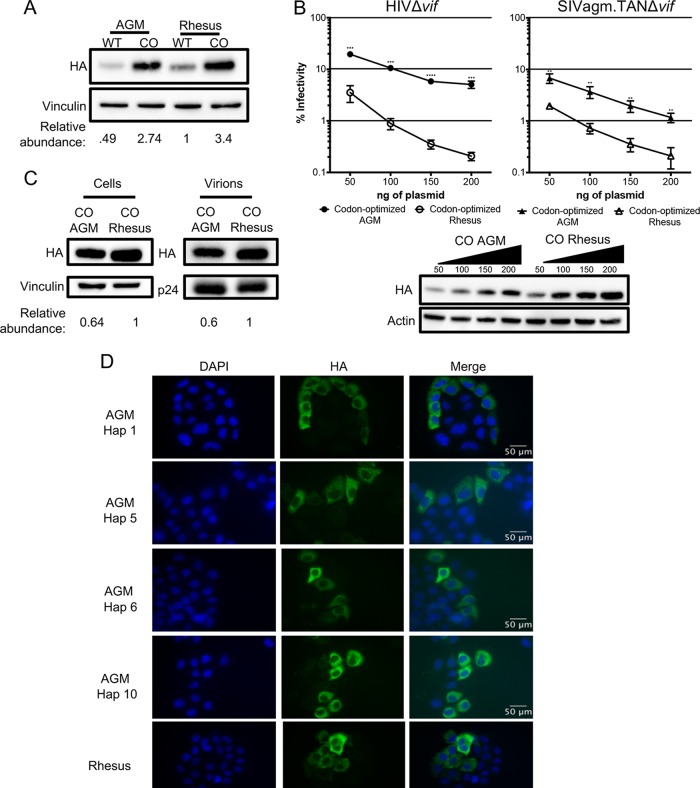
Antiviral activity of A3H is lower in AGMs than in another Old World monkey. (A) Single-cycle infectivity assays were performed in the presence or absence of A3 proteins against HIVΔvif and SIVagmΔvif. Rhesus macaque was included as a positive control. Relative infection was normalized to viral infectivity in the absence of A3 proteins. Averages of three replicates, each with triplicate infections (±SEM), are shown. All the samples were statistically significantly different than rhesus macaque A3H, except AGM haplotype 7 against SIVagm.TAN (P = 0.0502), as measured by unpaired t tests. Furthermore, no significant differences in restriction between HIV and SIVagm.TAN within individual haplotypes were found. (B) Western blot analysis of HA-tagged AGM A3H protein expression in human (HEK293T) and AGM (Cos7) cell lines. The different-size bands for different AGM A3H haplotypes were reproducible. β-Actin is shown as a loading control. (C) (Top) Single-cycle infectivity assay of HIVΔvif in the presence of increasing amounts of A3H-expressing plasmids. AGM A3H haplotype 1 (black circles) and rhesus macaque A3H (open circles) were compared. Relative infection was normalized to viral infectivity in the absence of A3 proteins. Averages of three replicates, each with triplicate infections (±SEM), are shown. Statistical differences were determined by unpaired t tests: *, P ≤ 0.05; **, P ≤ 0.01; ****, P ≤ 0.0001. (Bottom) Western blot analysis of protein expression levels with the same amounts of plasmid as in the graph above. β-Actin is shown as a loading control. (D) Packaging of A3H proteins into virions analyzed by Western blotting. Relative abundances in cellular expression (left) and virion incorporation (right) were determined compared to rhesus macaque A3H.
The differences in restriction could be explained by changes in expression, since we found that no variant of AGM A3H is as strongly expressed as rhesus macaque A3H in HEK293T cells (Fig. 2B, top). This lower expression of AGM A3H proteins relative to rhesus macaque A3H was not due to the species of origin of the cells used for transfection, since when we transfected AGM-derived Cos7 cells, the protein expression levels of AGM A3H were similarly poor relative to the expression of the rhesus A3H (Fig. 2B, bottom). These data indicate that lower protein expression levels are correlated with less potent antiviral activity, similar to unstable haplotypes of human A3H.
To investigate whether lower antiviral activity of AGM A3H proteins was due to low expression levels alone, we selected one of the most potent AGM A3H proteins, haplotype 1 (Fig. 2A), and increased the amount of transfected plasmid from 200 ng to 1,000 ng in parallel with rhesus macaque A3H. At the highest plasmid concentration, haplotype 1 was able to restrict HIVΔvif 16-fold, while rhesus A3H restricted HIVΔvif 19-fold at the lowest concentration used (Fig. 2C, top). This corresponds to approximately equal levels of protein expression at these amounts of plasmid transfected (Fig. 2C, bottom; compare the band for AGM A3H at the highest level of plasmid transfected with the level of rhesus A3H at the lowest level of plasmid transfected). At all levels, rhesus macaque A3H was significantly better than AGM A3H (unpaired t test; P < 0.05). This demonstrates that AGM A3H has not inherently lost its function, since as the protein concentration increased, the antiviral activity correspondingly became more potent.
One explanation for the difference in antiviral activity is that AGM A3H is not packaged as efficiently into virions as rhesus macaque A3H. To explore this possibility, we measured the amounts of A3H in virions and compared the relative abundances in AGM A3H and rhesus A3H. In virions, there was 3.3-fold less AGM A3H (Fig. 2D, right), which is reflective of 3-fold less protein observed in cells (Fig. 2D, left). Therefore, AGM A3H and rhesus A3H are packaged at similar efficiencies. However, since there is a more than 10-fold difference in viral inhibition, there must be an additional defect in antiviral activity for AGM A3H that is independent of packaging.
In order to more fully explore the relationship between expression levels and antiviral activity of AGM A3H, we codon optimized both AGM A3H haplotype 1 and rhesus macaque A3H sequences to remove rare codons that might negatively affect protein translation efficiency. Based on codon usage statistics in primates (see Materials and Methods), 100 out of 211 codons were replaced with more frequent codons in the codon-optimized AGM haplotype 1 A3H and 106 out of 211 were replaced in the codon-optimized rhesus macaque A3H. We found that codon optimization of both AGM A3H and rhesus macaque A3H increased their expression levels relative to the native codons in each gene (Fig. 3A, compare wild type [WT] to CO [codon optimized]). Moreover, after codon optimization, AGM A3H haplotype 1 and rhesus macaque A3H are expressed at similar levels (Fig. 3A and B, bottom). However, while codon optimization increased the antiviral activity of both AGM and rhesus macaque A3H over that of the wild type (compare Fig. 3B to 2C), codon-optimized rhesus macaque A3H still restricted viral infection 10-fold better than codon-optimized AGM A3H against HIVΔvif and 5-fold better against SIVagm.TANΔvif (Fig. 3B, top). We also tested the virion-packaging efficiency of AGM versus rhesus A3H by measuring the relative abundance of each codon-optimized construct in the virion. We found that codon-optimized AGM A3H is packaged with efficiency similar to that of rhesus macaque A3H (Fig. 3C), This shows that while increasing the expression level of the protein is sufficient to improve its antiviral activity, other factors after viral incorporation impact the poor antiviral activity of AGM A3H.
FIG 3.
Codon optimization increases protein expression and antiviral activity. (A) Western blot analysis for the expression of AGM A3H haplotype 1, codon-optimized haplotype 1 A3H, rhesus macaque A3H, and codon-optimized rhesus macaque A3H. Vinculin was used as a protein-loading control. Quantification was done relative to rhesus macaque A3H (normalized to 1). (B) (Top) Single-cycle infectivity assay of HIVΔvif and SIVagm.TANΔvif in the presence of increasing amounts of A3H plasmid comparing codon-optimized AGM haplotype 1 A3H (black) and codon-optimized rhesus macaque A3H (open). Relative infection was normalized to viral infectivity in the absence of A3 proteins. Averages of three replicates, each with triplicate infections (±SEM), are shown. Statistical differences were determined by unpaired t tests: **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. (Bottom) Western blot analysis of protein expression levels with amounts of plasmid added as for the graph above. β-Actin is shown as a loading control. (C) Packaging of A3H proteins into virions analyzed by Western blotting. Relative abundances in cellular expression (left) and virion incorporation (right) were determined compared to codon-optimized rhesus macaque A3H. (D) Subcellular localization of wild-type rhesus macaque and wild-type AGM A3H haplotypes (Hap) 1, 5, 6, and 10 in HeLa cells. A3H proteins were detected with an anti-HA antibody (green), and DAPI staining was used to detect the nucleus (blue). The images are representative of 135 total images over 3 replicates.
Studies in humans have shown that less active A3H proteins also localize to the nucleus (14, 19). We therefore asked whether wild-type AGM A3H proteins were expressed at lower levels with low antiviral activity due to a change in localization. Wild-type AGM A3H haplotypes 1, 5, 6, and 10 and rhesus macaque A3H were transfected into HeLa cells and visualized using immunofluorescence microscopy. However, all AGM A3H variants and rhesus macaque A3H were mainly present in the cytoplasm (Fig. 3D), demonstrating that a drastic change in localization was not responsible for the lack of potent antiviral activity of AGM A3H.
Our results suggest that the extensive diversity observed in AGM A3H results in lower antiviral activity linked both to protein levels and to other functional differences after packaging due to amino acid divergence between species.
Reconstruction of ancestral A3H proteins demonstrates the loss of activity in more recent evolution of AGMs and other primates.
We previously explored the evolutionary dynamic of hominoid A3H by reconstructing the ancestor of human/chimpanzee A3H and found that the predicted A3H protein of the human/chimpanzee ancestor had higher antiviral activity than either the extant chimpanzee or human protein (14). This suggests that there has been a loss of some activity in both lineages over their evolution. Due to the finding that all tested AGM A3H proteins are poorly active relative to the rhesus macaque A3H, we wanted to reconstruct the ancestral history of A3H leading to the modern AGM lineage. Thus, in order to gain statistical power in the ancestral sequence predictions at each node, we determined the A3H sequence from a broader panel of Old World monkeys in sister clades, including De Brazza's monkey, Allen's swamp monkey, Wolf's guenon, mustached guenon, talapoin, and patas monkey (Fig. 4A).
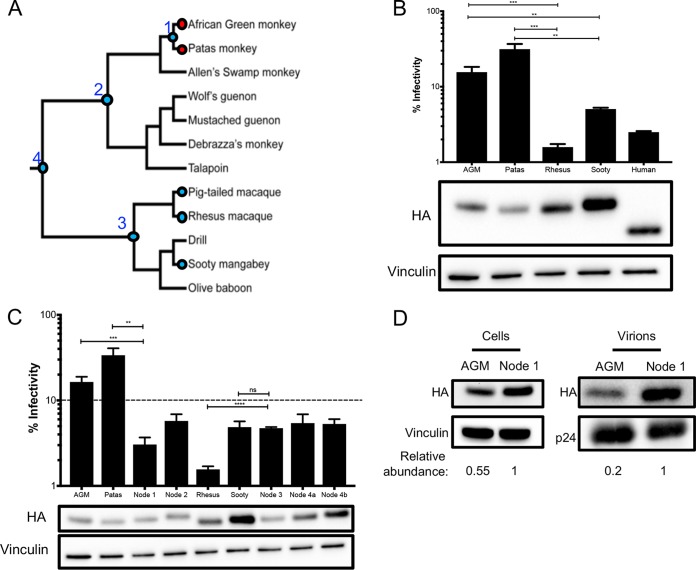
FIG 4.
AGM ancestors produce potent antiviral proteins. (A) Phylogeny, depicted as a cladogram, based on the accepted species tree of all sequenced Old World primates included in the study (28). The blue circles denote active antiviral proteins; the red circles denote less active antiviral proteins. Ancestral nodes are labeled with numbers (1 to 4). (B) (Top) Single-cycle infectivity assay for HIVΔvif against extant primate A3H proteins. Relative infection was normalized to viral infectivity in the absence of A3 proteins. Averages of three replicates, each with triplicate infections (+SEM), are shown. Statistical differences were determined by unpaired t tests. (Bottom) Western blot analysis of the protein expression level. Vinculin was used as a loading control. Human A3H is missing 28 C-terminal amino acids due to a natural premature stop codon (15) and thus ran lower on the blot. (C) (Top) Single-cycle infectivity assay for HIVΔvif against ancestral A3H proteins and their extant descendants. Relative infection was normalized to viral infectivity in the absence of A3 proteins. Averages of three replicates, each with triplicate infections (+SEM), are shown. The dashed line at 10% is an arbitrary reference point. (Bottom) Western blot analysis of the protein expression level. Vinculin was used as a loading control. (D) Packaging of A3H proteins into virions analyzed by Western blotting. Relative abundance in cellular expression (left) and virion incorporation (right) were determined compared to the node 1 A3H ancestor. **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001; ns, not significant.
We tested the A3H activity of the closest sister species to the AGMs, patas monkeys, and the A3H activity of a sister species to the rhesus, the sooty mangabey (Fig. 4A). Upon transfection into HEK293T cells, the protein expression level of patas monkey A3H was lower than that of rhesus macaque A3H, similar to AGM A3H (Fig. 4B, bottom). Patas monkey A3H, correspondingly, had low antiviral activity when tested against HIVΔvif; that is, while AGM and rhesus macaque A3H proteins restricted viral infection 6-fold and 63-fold, respectively, patas monkey A3H restricted HIVΔvif infection only 3-fold (Fig. 4B, top). However, active A3H proteins from sooty mangabey and the human A3H haplotype II restrict viral infection 20-fold and 40-fold, respectively. These data show that the antiviral activity is low in a species closely related to AGMs but a relative of the rhesus macaque and humans produce more active A3H proteins. This finding further suggests that changes in A3H activity may have evolutionary origins deeper in primate evolution.
In order to determine whether A3H antiviral activity was gained in the rhesus macaque/sooty mangabey lineage or was lost in the AGM/patas monkey lineage, we reconstructed the A3H ancestors at various nodes in the Old World monkey phylogeny (Fig. 4A). These included the common ancestor of AGMs and patas monkeys (node 1) and AGMs, patas monkeys, and their sister clade (node 2), as well as the common ancestor of rhesus macaque, pig-tailed macaque, and sooty mangabey (node 3) and the common ancestor of both groups (node 4). Each ancestor was constructed using maximum likelihood with FastML (39) based on the primate species phylogeny. Although the majority of codons were reconstructed with a probability of >99%, site 207 was ambiguous in the common ancestor of all tested Old World monkeys, and two codons were possible, encoding either an isoleucine (node 4a) or a threonine (node 4b) at position 207. In this case, both possible ancestors were generated using point mutagenesis.
We then tested the predicted A3H protein at the reconstructed ancestral nodes of the Old World monkey phylogenetic tree for antiviral activity and protein expression level. All A3H ancestors inhibited viral infection between 17- and 33-fold (Fig. 4C). Thus, this result suggests that activity was lost within the AGM/patas monkey clade. Similarly, while the A3H node 3 ancestor restricted viral infection 21-fold, rhesus macaque A3H inhibited HIVΔvif by 64-fold, indicating that some activity may have been gained along that lineage. Moreover, the node 1 ancestor representing the common ancestor of AGM and patas monkeys restricted viral infection 33-fold, which was more potent than its descendants, AGMs (6-fold) and patas monkeys (3-fold), despite having similar protein expression levels (Fig. 4C). Therefore, loss of activity in A3H in AGMs and patas monkeys that occurred after the common ancestor at node 1, which diverged at least 4 million years ago (28), included mutations that both decreased protein expression levels and led to the losses of antiviral activity that are independent of protein expression.
To determine whether differences in packaging of A3H into virions was responsible for the difference in antiviral activity between the node 1 ancestor and an extant AGM A3H, we performed a packaging assay to compare protein amounts in virions. AGM A3H is packaged 2.4-fold less efficiently than the node 1 ancestor (Fig. 4D). Moreover, the node 1 ancestor is present at 5-fold higher levels than AGM A3H haplotype 1 in the virion (Fig. 4D), which corresponds to the 5-fold better viral inhibition of the ancestor (Fig. 4C). This suggests that in the ancestral node directly leading to the AGM lineage, a decrease in packaging of A3H led to a further loss of antiviral activity.
Multiple amino acid mutations are responsible for loss of A3H antiviral activity.
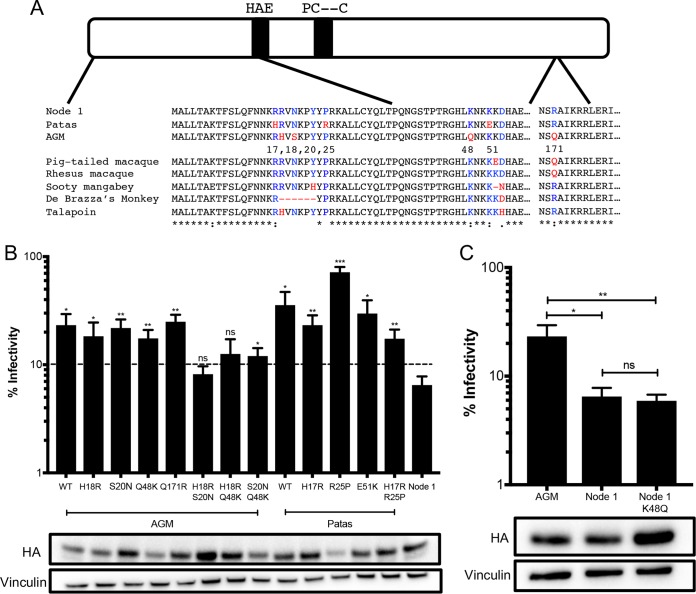
The ancestral A3H protein at node 1 representing the common ancestor of AGMs and patas monkeys (Fig. 4A) has stronger antiviral activity than its extant descendants (Fig. 4C). Therefore, we wanted to trace the amino acid mutations that resulted in the subsequent loss along the branches leading from node 1 to AGMs and patas monkeys. The sequence alignment of the node 1 ancestor with AGM haplotype 1 and patas monkey A3H revealed 4 (sites 18, 20, 48, and 171) and 3 (sites 17, 25, and 51) amino acid differences, respectively (Fig. 5A). One site in AGMs, S20N, created a protein sequence identical to that of AGM A3H haplotype 8. We introduced the other residues found in the node 1 ancestor into AGM and patas monkey A3H backgrounds to test the expression level and antiviral activity of each mutant.
FIG 5.
Multiple amino acid mutations required for an increase in antiviral activity. (A) Schematic of the A3H protein. The black bars outline the A3H catalytic site. Numbered amino acid residues that are different between AGM A3H, patas A3H, and the node 1 ancestor are in red in the protein sequence alignment, and ancestral residues are in blue. Amino acids that are different in other primates are similarly colored. (B) (Top) Single-cycle infectivity assay for HIVΔvif against extant mutants. Relative infection was normalized to viral infectivity in the absence of A3 proteins. Averages of three replicates, each with triplicate infections (+SEM), are shown. Statistical differences were determined by unpaired t tests: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ns, not significant. Statistically significant differences from the node 1 ancestor are depicted. (Bottom) Western blot analysis of protein expression levels of HA-tagged extant mutants made in the AGM and patas A3H backgrounds. Vinculin was used as a loading control. (C) (Top) Single-cycle infectivity assay for HIVΔvif against AGM haplotype 1, node 1 ancestor, and node 1 mutant A3H. Relative infection was normalized to viral infectivity in the absence of A3 proteins. Averages of three replicates, each with triplicate infections (+SEM), are shown. Statistical differences were determined by unpaired t tests: *, P ≤ 0.05; **, P ≤ 0.01; ns, not significant. (Bottom) Western blot analysis of protein expression levels of HA-tagged proteins made in the AGM and patas A3H backgrounds. Vinculin was used as a loading control.
Notably, no single amino acid mutation in AGM A3H increased the antiviral activity compared to wild-type AGM A3H, and all single mutants inhibited viral infection around 5-fold relative to the no-A3 control (Fig. 5B). In contrast, for the double mutants H18R/S20N, H18R/Q48K, and S20N/Q48K, antiviral activity increased to around 13-fold (Fig. 5B). Double mutants with the H18R mutation were not statistically significantly different from the node 1 ancestor, indicating that this amino acid may be most important to rescue antiviral activity. Similarly, patas monkey A3H with single mutations at sites 25 (R25P) and 51 (E51K) did not improve restriction, while an H17R mutation slightly increased the antiviral activity compared to the wild type (3-fold to 5-fold) (Fig. 5B). However, this single mutation at position 17 did not make patas monkey A3H comparable to the node 1 ancestor. Inserting both H17R and R25P mutations in patas monkey A3H similarly did not further improve restriction (Fig. 5B). Taken together, the inability of single point mutations to restore antiviral activity to its ancestral state emphasizes that changes at multiple sites have functional consequences in AGM and patas monkey A3H proteins.
Of interest, site 48 is the only residue that is fixed in all AGMs sequenced for this study. This demonstrates that it occurred first, whereas the other sites are polymorphic and have not yet become fixed in the species (Table 1). To determine whether site 48 alone is sufficient to decrease antiviral activity, we added a K48Q mutation in the node 1 background and tested its antiviral activity. However, node 1 K48Q has similar antiviral activity and expression (Fig. 5C, bottom), showing that epistasis between multiple amino acids may play an essential role for viral restriction by A3H.
Overall, these data establish that amino acids in the N-terminal portion of A3H are important for antiviral activity. Two out of 3 residues responsible for the loss of activity in patas monkey A3H, sites 17 and 25, are also polymorphic in AGMs (Table 1), suggesting that the loss began at a shared common ancestor not sampled by our analysis. Significantly, many Old World monkeys carry additional mutations near amino acid 15, whose loss in human A3H is known to affect protein stability (14). For example, residues 18 to 23 have been deleted in De Brazza's monkey (Fig. 5A). Such a large deletion in this region would likely impact the ability of A3H to inhibit viral infection in the species. Similarly, talapoin A3H encodes a histidine at position 18 instead of an arginine, similar to AGM A3H, suggesting that its activity may also be lost (Fig. 5A). In contrast, rhesus macaque and sooty mangabey A3H genes encode active ancestral amino acids at such residues, such as two arginines at positions 17 and 18 (Fig. 5A), which may result in the potent antiviral activity of these modern proteins. This implies that A3H function has been lost independently at various points of evolutionary history in both hominoids and Old World monkeys.
Lack of evidence that evolution in A3H leading to AGMs has been driven by Vif.
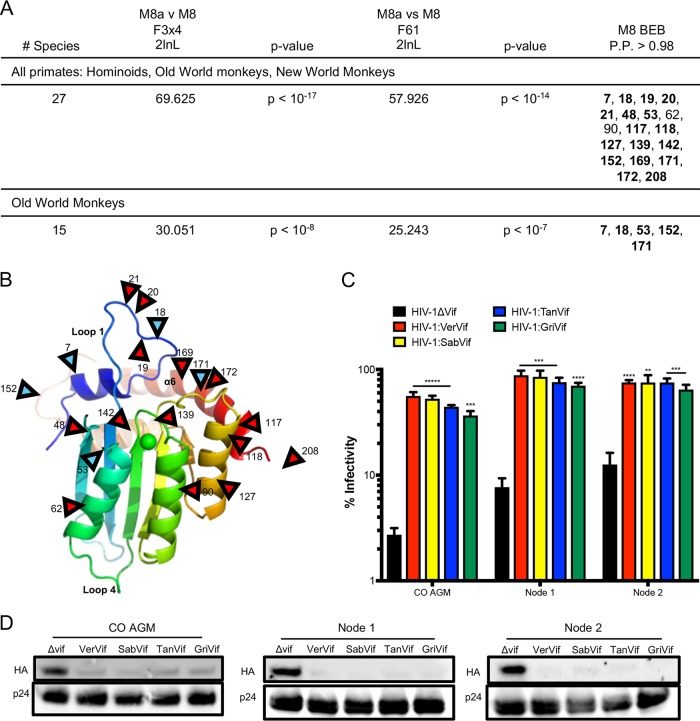
The variability in A3H sequence throughout Old World monkey evolution could be driven by escape from viral antagonists such as the Vif protein of SIVs or by other outside selective pressures. Antiviral restriction factors are often rapidly evolving and undergo positive selection (40), which is defined by an excess of nonsynonymous mutations compared to synonymous mutations. Evolutionary conflicts between host restriction factors and viral proteins to either maintain or escape interactions result in an accumulation of nonsynonymous mutations at binding interfaces. A previous study found that A3H is under positive selection (15). However, the study used fewer primate sequences, which can bias the analysis. Therefore, we wanted to retest positive selection in primate A3H using additional sequences we obtained from Old World monkeys. Using the PAML (phylogenetic analysis by maximum likelihood) program (41), we calculated the number of nonsynonymous mutations (dN) over the number of synonymous mutations (dS) for the entire A3H gene, as well as individual codons. In agreement with previous data, we found that A3H is under positive selection, with a dN/dS ratio of 1.3 in all primates. Additionally, models that allow codons to evolve under positive selection fit the data significantly better than models that do not for all primate clades (Fig. 6A). We evaluated individual sites within the gene and found that in all primates, a total of 19 residues are under positive selection with a posterior probability of >0.98. One site, position 90, has been implicated in Vif binding interactions but had a posterior probability of >0.98 in only one codon model (F3x4) (Fig. 6A). Since selection driven by primate lentiviruses would be expected to be concentrated in the Old World monkey clade, we also condensed our analysis to Old World monkeys alone and found that 5 sites remain under positive selection (Fig. 6B, blue arrowheads), all of which are polymorphic in AGMs (Table 1). No positively selected residues in Old World monkey A3H proteins are in the putative Vif binding region (37, 38). Thus, we conclude that multiple sites are under diversifying selection in primates but are likely not driven by lentiviral Vif.
FIG 6.
Evolution of A3H is not driven by Vif. (A) Results of positive-selection analyses of primate A3H. The far-right column lists sites under positive selection with dN/dS values of >1 with a posterior probability of >0.98 under M8 Bayes empirical Bayes (BEB) implemented in PAML model 8. The sites are relative to African green monkey A3H. Sites that had a posterior probability of >0.98 in both codon models (F3x4 and F61) are in boldface. (B) A3H from individual PR01190 was modeled onto the pig-tailed macaque A3H structure (Protein Data Bank [PDB] accession no. 5W3V). Locations of positively selected sites are indicated by red arrowheads. The blue arrowheads indicate sites also under positive selection in Old World primates. Site 208 is part of a region missing from the crystallized protein and is therefore not resolved in the model. (C) Single-cycle infectivity assay done with HIVΔvif and HIV-1 expressing SIVagm Vif in the presence of codon-optimized AGM A3H haplotype 1 (CO AGM), the node 1 ancestor, and the node 2 ancestor. Relative infection was normalized to viral infectivity in the absence of A3 proteins. Averages of three replicates, each with triplicate infections (+SEM), are shown. Statistical differences were determined by unpaired t tests: **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. Statistically significant differences from the restriction of HIVΔvif are depicted. (D) Western blot analyses demonstrating virion incorporation of A3H proteins in the absence and presence of Vif. p24 was used as a loading control.
In order to test the hypothesis that the changes in A3H in the lineages leading to the AGMs were not driven by the Vif proteins of the lentiviruses that infect AGMs, we tested the restriction capabilities of the node 1 and node 2 ancestors against HIV-1 proviruses expressing Vifs from four SIVagm strains. We also included the codon-optimized AGM A3H, since, as it is isolated from an AGM, it should be susceptible to degradation by all Vifs encoded by SIVagm. Of interest, the node 2 ancestor encodes an aspartic acid (D) at amino acid 100, which has previously been important for differences in Vif binding for SIVcpz and HIV-1, while the node 1 ancestor and all AGM A3Hs have an asparagine (N) (37, 38). The antiviral activities of all the ancestors, as well as codon-optimized AGM A3H, were counteracted by each Vif, resulting in the rescue of viral infection (Fig. 6C). Moreover, antagonism of A3H by each Vif protein was accompanied by a loss of A3H packaging into virions (Fig. 6D). Because there were no differences in the ability of Vif to rescue viral infection, these data suggest that Vif is not the primary force in A3H evolution in AGMs, which is instead driven by different selective pressures.
DISCUSSION
We have shown evidence for the recurrent functional loss of APOBEC3H in primates. We found that a decrease in protein expression levels, as well as amino acid mutation in the N-terminal region, resulted in lower antiviral activity. Using molecular reconstruction of ancestral A3H sequences, we found that the most recent common ancestor of AGMs and patas monkeys likely possessed an active A3H, similar to other common ancestors throughout evolutionary history. This suggests that the recurrent loss is a more recent event in primate evolution. Selective pressure by Vif does not appear to be a primary force behind the evolution of A3H in the AGM clade, but as loss of A3H function has occurred both in humans and in Old World monkeys, there may be a fitness cost to encoding this mutator protein over long evolutionary periods.
Molecular evolution of A3H protein impacts expression levels and antiviral activity.
While increasing the amount of A3H present in cells did increase its capability to inhibit viral infection, greater antiviral activity does not perfectly correlate with higher protein expression. Although codon optimization increased both the expression level and antiviral activity of A3H, codon-optimized AGM A3H haplotype 1 was still not as potent as codon-optimized rhesus macaque A3H. Thus, both protein expression levels and other amino acid differences that affect the function of A3H have led to the loss of antiviral A3H activity in the AGM lineage. Moreover, although packaging of A3H was equivalent between AGM and rhesus A3H proteins, the packaging efficiency of A3H did decrease after AGM A3H diverged from the node 1 ancestor. These results and the results of testing each mutation along the AGM lineage show that multiple mutations that affected the antiviral functions of the extant AGM A3H protein were acquired. Position 48 is fixed in AGMs, indicating that this change likely occurred first, while subsequent amino acid changes concurrently altered the antiviral activity of the protein. Other AGM A3H haplotypes have also accumulated additional nonsynonymous mutations (Fig. 2A), indicating that additional genetic drift may be further mutating A3H to become less antiviral in AGMs.
Why has primate A3H maintained partial activity rather than a complete loss?
Why has the antiviral activity of A3H not been lost completely in AGMs? It is possible that A3H has been coopted for a nonlentiviral function in Old World monkeys, and thus, it has been preserved. Alternatively, A3H could be retained through linkage to protective A3G haplotypes. A3G and A3H are located close together on chromosome 19 in AGMs, meaning A3H haplotypes that have ultimately lost most of their function may still be passed on to offspring, particularly if the A3G allele encodes a protein providing a selective advantage. In support of this idea, we noticed poorly active A3H proteins in a monkey producing protective A3G, as characterized in a separate study (9). This individual, V005, possesses A3H haplotype 11, as well as an A3G haplotype that cannot be antagonized by Vif proteins from SIVagm.ver or SIVagm.tan. Since A3G is a more potent antiviral and the protein protects individuals from two strains of SIVagm, the protection it supplies may supersede any detrimental effects incurred due to the presence of a less antiviral A3H haplotype.
Loss of activity of A3H due to recurrent mutations in a putative RNA binding domain.
Two mutations in human A3H gave rise to less active protein variants: R105G and a deletion of amino acid 15. Of interest, amino acid 15 is positioned within loop 1 of the A3H protein. Recent work has revealed that loops 1, 7, and α6 are important for binding to an RNA duplex (33–35). We similarly identified that amino acids 18 and 20, found within loop 1, are important for increasing the antiviral activity of AGM A3H (Fig. 5B). The analogous locations of such residues may suggest that these changes impact the antiviral activity of A3H by affecting its capability to bind to viral RNA. Dual amino acid changes at residues 18 and 20 in AGMs increase the antiviral activity of A3H close to the levels of a recent common ancestor. An additional mutation close to the catalytic site, Q48K, together with either H18R or S20N in AGMs, further increased antiviral activity. Furthermore, we found that amino acid 17 similarly increased the antiviral activity of patas monkey A3H. This residue is polymorphic in AGMs, suggesting that the change may have occurred in a common ancestor not tested in this study. We also found that other Old World monkeys have changes within loop 1, such as a 6-amino-acid deletion of residues 18 to 23 in De Brazza's monkey and an R18H mutation in talapoin (Fig. 5A). The diversity within loop 1 of Old World monkey A3H is indicative that A3H activity was possibly lost multiple times independently. RNA binding has been shown to play an important role in the antiviral activity of A3H (33, 34), and thus, loss of RNA binding may result in functional defects.
One caveat to our studies is that we have not measured A3H protein levels in primary AGM cells or in animals. Nonetheless, analysis of activities and protein levels of cloned human A3H genes did mimic their endogenous protein levels and antiviral activities (18).
Vif does not appear to play a role in the evolution of A3H along lineages leading to AGMs.
AGMs are highly polymorphic in a number of antiviral genes that are specific to infection with lentiviruses in both AGMs specifically and other primates (32), further highlighting the adaptation to SIV within this particular species. Indeed, we found that multiple sites in A3H are under positive selection in primates, which is suggestive of an evolutionary arms race between A3H and another protein. However, we found that Vif did not play a role in A3H's evolution in AGMs, since ancestral proteins were comparably susceptible to antagonism by Vif. Loss of A3H function may facilitate cross-species transmission of SIVagm strains between AGM subspecies (42) due a diminished A3 repertoire. Although many A3 proteins in the family may be redundant, encoding a diverse range of A3s is likely important to achieve maximum protection against lentiviruses.
Since lentiviruses have infected simian primates for millions of years (8), it is unlikely that the lack of residues altering A3H-Vif interactions in AGMs stems from recent infection of Old World monkeys. It is possible that the changes are the result of genetic drift or are driven by a different viral pathogen in these primates. Conversely, positively selected residues may indicate evolutionary toggling to preserve or eliminate protein function. It is also possible that the relative importance of different A3 proteins may change depending on the evolutionary history of a species, driven by the redundancy of the protein family. In the Old World monkeys studied here, less active A3H proteins may impart an increased risk for host genome mutations; thus, its function was lost due to the balance between viral protection and host fitness.
Our data imply that A3H function was lost prior to the divergence of different SIVagm strains due to its inactivity in all AGMs. Thus, Vif proteins from the ancestral virus may not have required adaptation to escape the antiviral activity of A3H and instead evolved in response to pressure from the more potent A3G. Expansion of the primate A3 locus provides flexibility of this antiviral protein family to take different trajectories throughout evolution. In addition to previous work in humans, the A3H homolog in felines, APOBEC3Z3, was recently shown to have a similar functional loss (43), demonstrating that loss of A3H and its homologs are frequent throughout a variety of animal species. Such widespread loss of function is suggestive of a potential fitness cost to hosts, although the presence of modern and active A3H proteins exemplifies the importance of a diverse A3 locus in primates.
MATERIALS AND METHODS
APOBEC3H cDNA amplification and sequencing.
APOBEC3H cDNAs were cloned by nested RT-PCR (Qiagen One-step RT PCR kit) or PCR (Accuprime Pfx) from RNA or genomic DNA (gDNA) isolated from AGM peripheral blood mononuclear cells (PBMC) or cell lines. Sample origins and extractions have been previously described (9, 31). For each sample, PCR products were amplified and sequenced using primers designed to amplify African green monkey A3H (forward, CACGAATTCGCCACCATGTATCCATACGATGTTCCAGATTACGCTGCTCTGCTAACAGCCAAA; reverse, CACGAGCTCATCTTGAGTTGAGTGT). The primers for gDNA amplification were designed to target intronic regions. Heterozygous sequences were cloned using the pGEM T-Easy vector system (Promega) and a TOPO TA cloning kit (Invitrogen) to analyze individual clones. Additional primate sequences were obtained from gDNA isolated from immortalized cell lines using a Qiagen DNeasy blood and tissue kit. The following cell lines from Coriell Cell Repositories (Camden, NJ) were used: patas monkey (Erythrocebus patas; ID no. 6254), De Brazza's monkey (Cercopithecus neglectus; PR01144), Wolf's guenon (Cercopithecus wolfi; PR01241), mustached guenon (Cercopithecus cephus; PR00527), Allen's swamp monkey (Allenopithecus nigroviridis; PR00198), and Francois' leaf monkey (Trachypithecus francoisi; PR01099). Talapoin (Miopithecus talapoin; OR755) cells were obtained from Frozen Zoo (San Diego, CA).
Expression constructs and plasmids.
Primate A3H genes were cloned from cDNA. A 5′ hemagglutinin (HA) tag was added via PCR (forward, GTGGTGGAATTCATGTATCCATACGATGTTCCAGATTACGCTGCTCTGCT; reverse, CTAGACTCGAGTCATCTTGAGTT). The products were digested using EcoRI/XhoI restriction enzymes and ligated into a mammalian pcDNA 3.1 vector (Invitrogen; V79020). Site-directed mutagenesis was completed with the QuikChange II site-directed mutagenesis kit (Agilent; 200524) to construct all ancestral genes and mutants. The A3H gene from patas monkeys was generated by gene synthesis (IDT) and cloned into the pcDNA 3.1 backbone. A3H genes from AGM and rhesus macaque were codon optimized based on usage frequencies in primates (human and rhesus macaque) to remove rare codons within the gene in Geneious (Biomatters Ltd.). Codon-optimized sequences were generated by gene synthesis and cloned into pcDNA 3.1.
Cell lines, transfections, and Western blot analysis.
HEK293T, HeLa, and Cos7 cell lines (ATCC) were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (Corning; 35-015-CV) and 100 μg/ml penicillin-streptomycin (Gibco; 15140-122) at 37°C. SupT1 cells (ATCC) were maintained in RPMI 1640 with 10% fetal bovine serum and 100 μg/ml penicillin-streptomycin under the same conditions. Transfections were done in serum-free DMEM with TransIT-LT1 transfection reagent (Mirus Bio; MIR 2305) at a reagent/plasmid DNA ratio of 3:1. For Western blot analysis, cells were lysed in ice-cold NP-40 buffer (0.5% NP-40, 20 mM NaCl, 50 mM Tris, pH 7.5) with protease inhibitors (Roche Complete Mini, EDTA-free tablets; 11836170001). Lysates were quantified using a Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Scientific; 23225), and 10 μg of protein was resolved by SDS-PAGE, transferred to a polyvinylidene difluoride (PVDF) membrane, and probed with anti-HA (BioLegend; 901503) and anti-actin (Sigma; A2066) or anti-vinculin (Proteintech; 66305-1) antibodies at a 1:2,000 dilution. Samples from viral supernatants were probed with a p24 antibody (NIH-ARP; 3537) at a 1:1,000 dilution. Anti-mouse or anti-rabbit secondary antibody was used at a 1:5,000 dilution (Santa Cruz Biotechnology; sc-2005 and sc-2004).
Immunofluorescence.
HeLa cells were seeded onto 18-mm (VWR; 48380 046) coverslips at 4 × 104 cells/ml and transfected with 500 ng of A3H-expressing plasmids the next day. Forty-eight hours after transfection, the coverslips were fixed in 2% paraformaldehyde, permeabilized in 0.5% PBS–Triton-X, and blocked in PBS-bovine growth serum. HA-tagged proteins were detected using the same HA antibody used for Western blots at a 1:1,000 dilution, followed by an anti-mouse AF488 antibody at 1:400 (Invitrogen; A11001). Nuclei were stained in SlowFade Gold antifade reagent with DAPI (4′,6-diamidino-2-phenylindole) mounting medium (Life Technologies; S36939). Images were taken on a Nikon E800 microscope.
Phylogenetic analysis.
AGM A3H sequences were analyzed phylogenetically using a Bayesian Monte Carlo Markov chain (MCMC) approach implemented in BEAST v1.7.1. Sequence alignments were constructed using the MAFFT align function in Geneious (Biomatters Ltd.) and underwent 10,000,000 MCMC generations using a HKY85 substitution model, a gamma site heterogeneity model, estimated base frequencies, and a constant population size coalescent as the tree prior.
Positive-selection analysis.
The alignment of 27 primate A3H sequences was analyzed using HyPhy GARD analysis to ensure there was no recombination in the gene (44). The species phylogeny (28) was input into the CODEML site model of PAML (41), along with the nucleotide alignment, to detect positive selection at individual sites. The P value was calculated as twice the difference in log likelihood between models M7 and M8, as well as M8 and M8a, with two degrees of freedom. Analysis was conducted with both the F3x4 and F61 codon frequency models, with omega values of 0.4 and 1.5. Data from the F3x4 and F61 models are shown in Table 1. Positively selected sites were categorized as those with an M8 Bayes empirical Bayes posterior probability greater than 98%.
Ancestral reconstruction.
The ancestors at specific nodes within the Old World monkey clade were reconstructed using the FASTML Web server (fastml.tau.ac.il; last accessed March 2018) (39). The alignment of 27 primate A3H sequences was used in conjunction with the species tree to generate marginal reconstructions of codon sequences.
Single-cycle infectivity assays.
HEK293T cells were plated in 1 ml in 12-well plates at 1.25 × 105 cells/ml. After the cells reached between 50 and 70% confluence, they were cotransfected with 250 ng of A3H or empty expression plasmid, 600 ng of a luciferase-encoding proviral plasmid with a frameshift mutation in env and deletion of vif, and 100 ng of L-VSV-G (vesicular stomatitis virus glycoprotein) for pseudotyping in 100 μl serum-free medium with TransIT-LT1 transfection reagent (Mirus Bio). Supernatants containing virus were harvested after 48 h and clarified through 0.2-μm filters. Viral titers were determined by measuring RT activity by quantitative PCR (qPCR) as described previously (45). In short, viral supernatants were lysed in 2× lysis buffer (0.25% Triton X-100, 50 mM KCl, 100 mM Tris-HCl, 40% glycerol) in the presence of 4 U RNase inhibitor (Fermentas; EO0382). qRT-PCRs were set up with an MS2 RNA template using the Takyon Rox SYBR MasterMix dTTP Blue kit (Eurogentec; UF-RSMT-B0101) alongside a standard curve made with a stock virus of previously determined titers. The primers used to amplify duplicate reactions were TCCTGCTCAACTTCCTGTCGAG (forward) and CACAGGTCAAACCTCCTAGGAATG (reverse). qRT-PCR was performed on an ABI QuantStudio5 real-time PCR machine; 2,000 mU/ml was used to infect SupT1 cells plated at 2 × 104 cells/well in a 96-well plate in medium supplemented with 20 μg/ml DEAE-dextran. Infections were done in triplicate for 48 to 64 h. Luciferase activity was measured with Bright-Glo Luciferase Assay Reagent (Promega; E2620) on a Lumistar Omega luminometer.
Packaging assay.
Packaging of A3 proteins into virions was measured by cotransfection of 600 ng of A3 plasmid with 1,000 ng of the proviral plasmid described for single-cycle assays into a 6-well plate seeded with 2 × 105 cells/ml. Two days after transfection, cell lysates were collected, and 1.5 ml of supernatant was clarified through 0.2-μm filters. Viral supernatants were concentrated by pelleting by centrifugation at 13,000 rpm for 60 min. Medium was aspirated from the virion pellet until ∼10 μl remained, and virions were lysed with 4× NuPAGE LDS sample buffer (Invitrogen; NP0007). Virion and cell lysates were analyzed by Western blotting.
Statistical analysis.
Data analyses were done using GraphPad Prism 7.0 software. All the data are shown as means and standard errors of the mean (SEM). Statistical analysis was performed using unpaired two-tailed t tests with 95% confidence. P values of less than 0.05 were considered statistically significant.
Accession number(s).
The GenBank accession numbers for new Old World monkey A3H sequences, including those of De Brazza's monkey, Wolf's guenon, mustached guenon, patas monkey, talapoin, Francois' leaf monkey, Allen's swamp monkey, and AGM haplotype 1, reported here are MH231602 to MH231609.
ACKNOWLEDGMENTS
We thank the following investigators for providing samples used in this study: M. Muller-Trutwin (AGM PBMC), C. Apetrei (AGM PBMC and SIVagm.Sab92018), and N. Freimer and A. Jasinska (AGM DNA). We additionally thank the National Institutes of Health (NIH) Nonhuman Primate Research Resource for the V038 and AG23 AGM cells, as well as SIVagm.Gri+ plasma. We thank the Fred Hutchinson Cancer Research Center Shared Resources Genomics Core for sequencing. We thank Ferdinand Roesch, Nicholas Chesarino, Mollie McDonnell, and Emily Hsieh for constructive comments on the manuscript. We also thank Harmit Malik, Rick McLaughlin, and Rossana Colon-Thillet for helpful discussions.
African sabaeus samples used in this study were collected as a part of the Systems Biology Sample Repository funded by NIH grants R01RR016300 and R0100010980, with the particular sabaeus samples used in this study obtained through the Department of Parks and Management Ministry of Forestry and the Environment and Medical Research Council Unit, The Gambia.
This work was supported by NIH grant R01 AI30927 (M.E.) and a Viral Pathogenesis Training Grant T32AI083203 (E.I.G.).
REFERENCES
- 1.Harris RS, Dudley JP. 2015. APOBECs and virus restriction. Virology 479-480: 131–145. doi: 10.1016/j.virol.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harris RS, Liddament MT. 2004. Retroviral restriction by APOBEC proteins. Nat Rev Immunol 4:868–877. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- 3.Stavrou S, Ross SR. 2015. APOBEC3 proteins in viral immunity. J Immunol 195:4565–4570. doi: 10.4049/jimmunol.1501504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hultquist JF, Lengyel JA, Refsland EW, LaRue RS, Lackey L, Brown WL, Harris RS. 2011. Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1. J Virol 85:11220–11234. doi: 10.1128/JVI.05238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belanger K, Savoie M, Rosales Gerpe MC, Couture JF, Langlois MA. 2013. Binding of RNA by APOBEC3G controls deamination-independent restriction of retroviruses. Nucleic Acids Res 41:7438–7452. doi: 10.1093/nar/gkt527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pollpeter D, Parsons M, Sobala AE, Coxhead S, Lang RD, Bruns AM, Papaioannou S, McDonnell JM, Apolonia L, Chowdhury JA, Horvath CM, Malim MH. 2018. Deep sequencing of HIV-1 reverse transcripts reveals the multifaceted antiviral functions of APOBEC3G. Nat Microbiol 3:220–233. doi: 10.1038/s41564-017-0063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simon V, Bloch N, Landau NR. 2015. Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nat Immunol 16:546–553. doi: 10.1038/ni.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Compton AA, Emerman M. 2013. Convergence and divergence in the evolution of the APOBEC3G-Vif interaction reveal ancient origins of simian immunodeficiency viruses. PLoS Pathog 9:e1003135. doi: 10.1371/journal.ppat.1003135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Compton AA, Hirsch VM, Emerman M. 2012. The host restriction factor APOBEC3G and retroviral Vif protein coevolve due to ongoing genetic conflict. Cell Host Microbe 11:91–98. doi: 10.1016/j.chom.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCarthy KR, Kirmaier A, Autissier P, Johnson WE. 2015. Evolutionary and FUNCTIONAL Analysis of Old World primate TRIM5 reveals the ancient emergence of primate lentiviruses and convergent evolution targeting a conserved capsid interface. PLoS Pathog 11:e1005085. doi: 10.1371/journal.ppat.1005085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Etienne L, Bibollet-Ruche F, Sudmant PH, Wu LI, Hahn BH, Emerman M. 2015. The role of the antiviral APOBEC3 gene family in protecting chimpanzees against lentiviruses from monkeys. PLoS Pathog 11:e1005149. doi: 10.1371/journal.ppat.1005149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Z, Gu Q, de Manuel Montero M, Bravo IG, Marques-Bonet T, Haussinger D, Munk C. 2017. Stably expressed APOBEC3H forms a barrier for cross-species transmission of simian immunodeficiency virus of chimpanzee to humans. PLoS Pathog 13:e1006746. doi: 10.1371/journal.ppat.1006746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol 14:1392–1396. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 14.OhAinle M, Kerns JA, Li MM, Malik HS, Emerman M. 2008. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe 4:249–259. doi: 10.1016/j.chom.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.OhAinle M, Kerns JA, Malik HS, Emerman M. 2006. Adaptive evolution and antiviral activity of the conserved mammalian cytidine deaminase APOBEC3H. J Virol 80:3853–3862. doi: 10.1128/JVI.80.8.3853-3862.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Abudu A, Son S, Dang Y, Venta PJ, Zheng YH. 2011. Analysis of human APOBEC3H haplotypes and anti-human immunodeficiency virus type 1 activity. J Virol 85:3142–3152. doi: 10.1128/JVI.02049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harari A, Ooms M, Mulder LC, Simon V. 2009. Polymorphisms and splice variants influence the antiretroviral activity of human APOBEC3H. J Virol 83:295–303. doi: 10.1128/JVI.01665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Refsland EW, Hultquist JF, Luengas EM, Ikeda T, Shaban NM, Law EK, Brown WL, Reilly C, Emerman M, Harris RS. 2014. Natural polymorphisms in human APOBEC3H and HIV-1 Vif combine in primary T lymphocytes to affect viral G-to-A mutation levels and infectivity. PLoS Genet 10:e1004761. doi: 10.1371/journal.pgen.1004761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li MM, Emerman M. 2011. Polymorphism in human APOBEC3H affects a phenotype dominant for subcellular localization and antiviral activity. J Virol 85:8197–8207. doi: 10.1128/JVI.00624-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Starrett GJ, Luengas EM, McCann JL, Ebrahimi D, Temiz NA, Love RP, Feng Y, Adolph MB, Chelico L, Law EK, Carpenter MA, Harris RS. 2016. The DNA cytosine deaminase APOBEC3H haplotype I likely contributes to breast and lung cancer mutagenesis. Nat Commun 7:12918. doi: 10.1038/ncomms12918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burns MB, Temiz NA, Harris RS. 2013. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet 45:977–983. doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li MM, Wu LI, Emerman M. 2010. The range of human APOBEC3H sensitivity to lentiviral Vif proteins. J Virol 84:88–95. doi: 10.1128/JVI.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ooms M, Brayton B, Letko M, Maio SM, Pilcher CD, Hecht FM, Barbour JD, Simon V. 2013. HIV-1 Vif adaptation to human APOBEC3H haplotypes. Cell Host Microbe 14:411–421. doi: 10.1016/j.chom.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Ooms M, Letko M, Binka M, Simon V. 2013. The resistance of human APOBEC3H to HIV-1 NL4-3 molecular clone is determined by a single amino acid in Vif. PLoS One 8:e57744. doi: 10.1371/journal.pone.0057744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakurai D, Iwatani Y, Ohtani H, Naruse TK, Terunuma H, Sugiura W, Kimura A. 2015. APOBEC3H polymorphisms associated with the susceptibility to HIV-1 infection and AIDS progression in Japanese. Immunogenetics 67:253–257. doi: 10.1007/s00251-015-0829-2. [DOI] [PubMed] [Google Scholar]
- 26.Naruse TK, Sakurai D, Ohtani H, Sharma G, Sharma SK, Vajpayee M, Mehra NK, Kaur G, Kimura A. 2016. APOBEC3H polymorphisms and susceptibility to HIV-1 infection in an Indian population. J Hum Genet 61:263–265. doi: 10.1038/jhg.2015.136. [DOI] [PubMed] [Google Scholar]
- 27.Fregoso OI, Ahn J, Wang C, Mehrens J, Skowronski J, Emerman M. 2013. Evolutionary toggling of Vpx/Vpr specificity results in divergent recognition of the restriction factor SAMHD1. PLoS Pathog 9:e1003496. doi: 10.1371/journal.ppat.1003496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perelman P, Johnson WE, Roos C, Seuanez HN, Horvath JE, Moreira MA, Kessing B, Pontius J, Roelke M, Rumpler Y, Schneider MP, Silva A, O'Brien SJ, Pecon-Slattery J. 2011. A molecular phylogeny of living primates. PLoS Genet 7:e1001342. doi: 10.1371/journal.pgen.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wertheim JO, Worobey M. 2007. A challenge to the ancient origin of SIVagm based on African green monkey mitochondrial genomes. PLoS Pathog 3:e95. doi: 10.1371/journal.ppat.0030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Locatelli S, Peeters M. 2012. Cross-species transmission of simian retroviruses: how and why they could lead to the emergence of new diseases in the human population. AIDS 26:659–673. doi: 10.1097/QAD.0b013e328350fb68. [DOI] [PubMed] [Google Scholar]
- 31.Spragg CJ, Emerman M. 2013. Antagonism of SAMHD1 is actively maintained in natural infections of simian immunodeficiency virus. Proc Natl Acad Sci U S A 110:21136–21141. doi: 10.1073/pnas.1316839110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Svardal H, Jasinska AJ, Apetrei C, Coppola G, Huang Y, Schmitt CA, Jacquelin B, Ramensky V, Muller-Trutwin M, Antonio M, Weinstock G, Grobler JP, Dewar K, Wilson RK, Turner TR, Warren WC, Freimer NB, Nordborg M. 2017. Ancient hybridization and strong adaptation to viruses across African vervet monkey populations. Nat Genet 49:1705–1713. doi: 10.1038/ng.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ito F, Yang H, Xiao X, Li SX, Wolfe A, Zirkle B, Arutiunian V, Chen XS. 2018. Understanding the structure, multimerization, subcellular localization and mC selectivity of a genomic mutator and anti-HIV factor APOBEC3H. Sci Rep 8:3763. doi: 10.1038/s41598-018-21955-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaban NM, Shi K, Lauer KV, Carpenter MA, Richards CM, Salamango D, Wang J, Lopresti MW, Banerjee S, Levin-Klein R, Brown WL, Aihara H, Harris RS. 2018. The antiviral and cancer genomic DNA deaminase APOBEC3H is regulated by an RNA-mediated dimerization mechanism. Mol Cell 69:75–86. e79. doi: 10.1016/j.molcel.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bohn JA, Thummar K, York A, Raymond A, Brown WC, Bieniasz PD, Hatziioannou T, Smith JL. 2017. APOBEC3H structure reveals an unusual mechanism of interaction with duplex RNA. Nat Commun 8:1021. doi: 10.1038/s41467-017-01309-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia EI, Emerman M. 2018. Recurrent loss of APOBEC3H activity during primate evolution. bioRxiv 10.1101/311878. [DOI] [PMC free article] [PubMed]
- 37.Nakashima M, Tsuzuki S, Awazu H, Hamano A, Okada A, Ode H, Maejima M, Hachiya A, Yokomaku Y, Watanabe N, Akari H, Iwatani Y. 2017. Mapping region of human restriction factor APOBEC3H critical for interaction with HIV-1 Vif. J Mol Biol 429:1262–1276. doi: 10.1016/j.jmb.2017.03.019. [DOI] [PubMed] [Google Scholar]
- 38.Ooms M, Letko M, Simon V. 2017. The structural interface between HIV-1 Vif and human APOBEC3H. J Virol 91:e02289-16. doi: 10.1128/JVI.02289-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ashkenazy H, Penn O, Doron-Faigenboim A, Cohen O, Cannarozzi G, Zomer O, Pupko T. 2012. FastML: a web server for probabilistic reconstruction of ancestral sequences. Nucleic Acids Res 40:W580–W584. doi: 10.1093/nar/gks498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daugherty MD, Malik HS. 2012. Rules of engagement: molecular insights from host-virus arms races. Annu Rev Genet 46:677–700. doi: 10.1146/annurev-genet-110711-155522. [DOI] [PubMed] [Google Scholar]
- 41.Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 42.Bell SM, Bedford T. 2017. Modern-day SIV viral diversity generated by extensive recombination and cross-species transmission. PLoS Pathog 13:e1006466. doi: 10.1371/journal.ppat.1006466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Konno Y, Nagaoka S, Kimura I, Takahashi Ueda M, Kumata R, Ito J, Nakagawa S, Kobayashi T, Koyanagi Y, Sato K. 3 April 2018. A naturally occurring feline APOBEC3 variant that loses anti-lentiviral activity by lacking two amino acid residues. J Gen Virol. doi: 10.1099/jgv.0.001046. [DOI] [PubMed] [Google Scholar]
- 44.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. 2006. GARD: a genetic algorithm for recombination detection. Bioinformatics 22:3096–3098. doi: 10.1093/bioinformatics/btl474. [DOI] [PubMed] [Google Scholar]
- 45.Vermeire J, Naessens E, Vanderstraeten H, Landi A, Iannucci V, Van Nuffel A, Taghon T, Pizzato M, Verhasselt B. 2012. Quantification of reverse transcriptase activity by real-time PCR as a fast and accurate method for titration of HIV, lenti- and retroviral vectors. PLoS One 7:e50859. doi: 10.1371/journal.pone.0050859. [DOI] [PMC free article] [PubMed] [Google Scholar]