Hepatitis C virus infection is a global health issue, and there is still no available vaccine, which makes it urgent to reveal the underlying mechanisms of HCV and host factors. Although RIG-I has been recognized as the leading cytoplasmic sensor against HCV for a long time, recent findings that MDA5 regulates the IFN response to HCV have emerged. Our work validates the significant role of MDA5 in IFN signaling and HCV infection and proposes the first lncRNA inhibiting HCV replication by promoting the activation of MDA5 and mediating the association between MDA5 and HCV RNA, the study of which may shed light on the MDA5 function and treatment for hepatitis C patients. Our suggested model of how lncITPRIP-1 orchestrates signal transduction for IFN production illustrates the essential role of lncRNAs in virus elimination.
KEYWORDS: lncRNA, lncITPRIP-1, virus, MDA5, interferon, oligomerization, RNA recognition, ISG
ABSTRACT
Emerging evidence indicates that long noncoding RNAs (lncRNAs) regulate various biological processes, especially innate and adaptive immunity. However, the relationship between lncRNAs and the interferon (IFN) pathway remains largely unknown. Here, we report that lncRNA ITPRIP-1 (lncITPRIP-1) is involved in viral infection and plays a crucial role in the virus-triggered IFN signaling pathway through the targeting of melanoma differentiation-associated gene 5 (MDA5). LncITPRIP-1 can be induced by viral infection, which is not entirely dependent on the IFN signal. Besides, there is no coding potential found in the lncITPRIP-1 transcript. LncITPRIP-1 binds to the C terminus of MDA5, and it possesses the ability to boost the oligomerization of both the full length and the 2 caspase activation and recruitment domains of MDA5 in a K63-linked polyubiquitination-independent manner. Amazingly, we also found that MDA5 can suppress hepatitis C virus (HCV) replication independently of IFN signaling through its C-terminal-deficient domain bound to viral RNA, in which lncITPRIP-1 plays a role as an assistant. In addition, the expression of lncITPRIP-1 is highly consistent with MDA5 expression, indicating that lncITPRIP-1 may function as a cofactor of MDA5. All the data suggest that lncITPRIP-1 enhances the innate immune response to viral infection through the promotion of oligomerization and activation of MDA5. Our study discovers the first lncRNA ITPRIP-1 involved in MDA5 activation.
IMPORTANCE Hepatitis C virus infection is a global health issue, and there is still no available vaccine, which makes it urgent to reveal the underlying mechanisms of HCV and host factors. Although RIG-I has been recognized as the leading cytoplasmic sensor against HCV for a long time, recent findings that MDA5 regulates the IFN response to HCV have emerged. Our work validates the significant role of MDA5 in IFN signaling and HCV infection and proposes the first lncRNA inhibiting HCV replication by promoting the activation of MDA5 and mediating the association between MDA5 and HCV RNA, the study of which may shed light on the MDA5 function and treatment for hepatitis C patients. Our suggested model of how lncITPRIP-1 orchestrates signal transduction for IFN production illustrates the essential role of lncRNAs in virus elimination.
INTRODUCTION
More than 98% of eukaryotic transcriptomes are composed of noncoding RNAs with no specific protein-coding capacity, and these can be classified into a long group (with >200 nucleotides [nt]) and a short group. By some estimates, there may be more than twice as many long noncoding RNA (lncRNA) genes than protein-coding genes in humans (1). It is now recognized that lncRNAs are exquisitely regulated in specific cell types and have frequent evolutionarily conserved regions of microhomology, secondary structure, and function (2). Recent discoveries demonstrate that the “dark genome” significantly contributes to immune responses. Thus, a comprehensive understanding of the roles of lncRNAs would help to better elucidate the regulatory mechanisms of human health and disease.
In the past decade, tens of thousands of lncRNAs have been discovered in the human genome. According to the evidence, lncRNAs can be classified into cytoplasmic and nuclear lncRNAs, which may function through distinct mechanisms. For cytoplasmic lncRNAs, indispensable roles in multiple cellular activities are mainly adopted by modulating mRNA stability, regulating mRNA translation, competing against endogenous RNAs, functioning as precursors of mRNA, and mediating protein modification (3). The physiological roles of lncRNAs in the nucleus are still mostly elusive. Some studies reported that nuclear lncRNAs interact with splice factors in the nuclear speckles, regulating chromatin status, enhancing the initiation of polymerase II of their parent genes, or being involved in centromere/kinetochore formation (4). Although the lncRNA function largely depends on its subcellular localization, the underlying mechanisms are still poorly understood.
Upon pathogen-associated molecular pattern (PAMP) recognition, pattern recognition receptors (PRRs) initiate a series of signaling responses, executing the first line of host defensive responses against infectious microbes (5). Almost at the same time, activation of PRR signaling induces the maturation of dendritic cells (DCs), which is responsible for providing an alert for the induction of the second line of host defense, also known as adaptive immunity (5). At present, three types of PRR sensors that mainly contribute to viral detection have been discovered: Toll-like receptors (TLRs), retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), and cGAS. With the longest research history among PRRs, TLRs only partially explain virus detection, as cell types expressing TLRs are limited (5). Therefore, RLRs and cGAS have emerged as vital sensors that detect intracellular RNA viruses and DNA viruses, respectively.
RIG-I, melanoma differentiation-associated gene 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2) are members of the RLR family (6). The RLRs are expressed within the cytoplasm of nearly every mammalian cell (7). Although they have similar structures, RIG-I and MDA5 function through distinct mechanisms. RIG-I recognizes the triphosphate and diphosphate at the end of a double-stranded RNA (dsRNA) stem and triggers the IFN pathway, while MDA5 recognizes the internal duplex structure of dsRNA (8). LGP2 lacks the caspase activation and recruitment domains (CARDs) required for signaling but shares homology at its DExD/H RNA helicase domain and C-terminal domain (CTD) with RIG-I and MDA5 (9). Although information about LGP2 is limited, LGP2 appears to be a cofactor for the virus RNA sensor and is most likely involved in making the viral RNA more accessible to RIG-I and MDA5 (9).
Hepatitis C virus (HCV) was discovered in 1989 as a non-A, non-B hepatitis virus, infection with which is limited to humans and chimpanzees (10). Approximately 170 million individuals worldwide are infected with HCV, and 700,000 patients die of HCV-related disease each year (10, 11). Over the past few years, many antivirals with activity against HCV have been developed, from the initially administered interferon (IFN) to the recent viral RNA-dependent RNA polymerase (NS5B) inhibitor sofosbuvir, which have helped to combat the disease to an appreciable extent (11, 12). Nonetheless, hepatitis C and its chronic complications are still a major public health issue, as there are intricate challenges, including both clinical challenges and challenges related to the virus itself. Clinical challenges involve the cost of hepatitis C treatment and problems with access to treatment and the lack of infrastructure (13). Challenges related to HCV itself include its high mutation rate, high replication rate, and various shrewd immune evasion mechanisms that grant it the ability to establish chronic persistent infection (13, 14).
With hundreds of years of evolution and variation, viruses have figured out several strategies to dampen the host IFN response, such as hiding the viral genome; inhibiting interactions with key host inducers of the IFN response; and regulating phosphorylation events, ubiquitination and related pathways, cleavage, degradation, and transcriptional shutoff (15). Meanwhile, large amounts of cytokines are produced and play a role in the virus and host interaction. Besides, viral infection and the IFN response induce strong modifications to the cell transcriptome. Among modifications to RNAs, altered expression of lncRNAs is obvious (16). Infected cells may express viral lncRNAs, cellular lncRNAs, and chimeric lncRNAs formed by viral and cellular sequences (16), which suggests that these lncRNAs are likely involved in viral infection and host responses. Here, we identify ITPRIP-1 lncRNA (lncITPRIP-1), which is involved in viral infection and which can enhance innate immunity through the targeting of MDA5. On the one hand, lncITPRIP-1 boosts MDA5 oligomerization to promote signal transduction for IFN production. On the other, lncITPRIP-1 enhances the capacity of MDA5 to recognize HCV RNA and gradually suppresses HCV replication. We confirm that lncITPRIP-1 is an IFN-inducible lncRNA. We find that lnclTPRIP-1 expression is upregulated upon viral infection and is coincident with increased MDA5 expression. Our data indicate that lncITPRIP-1 may function as a cofactor of MDA5 against viral infection.
RESULTS
LncITPRIP-1 is identified as an lncRNA involved in viral infection.
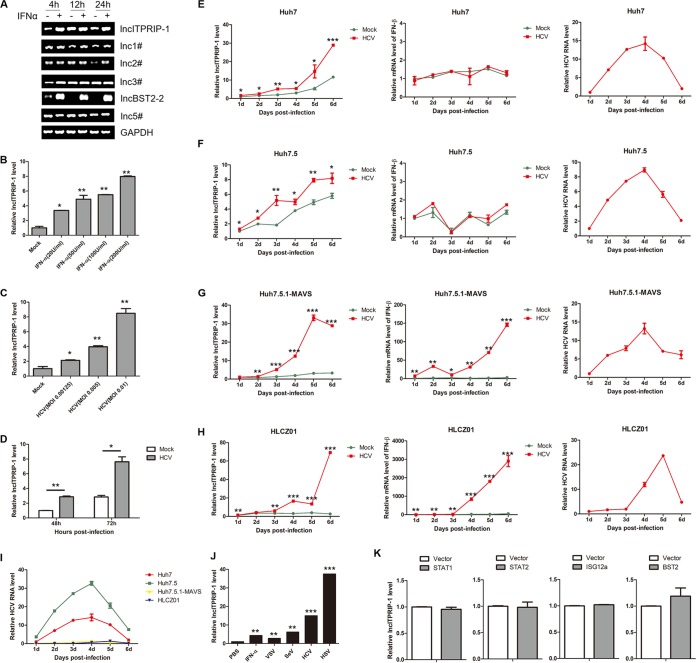
In this study, our preliminary work was to evaluate the expression levels of several candidate lncRNAs in human hepatocytes with or without alpha interferon (IFN-α) stimulation by semi-reverse transcription (RT)-PCR. Six candidate lncRNAs were selected from the 200 upregulated lncRNAs shown in the chip data of Valadkhan and colleagues (17). All candidate lncRNAs exhibited different expression patterns upon IFN stimulation to different degrees (Fig. 1A). RT-PCR data demonstrated that the levels of lncRNA BST2-2 (lncBST2-2) and lncRNA ITPRIP-1 were remarkably upregulated after IFN-α stimulation in HLCZ01 cells (Fig. 1A). One of the selected candidate lncRNAs, lncRNA BST2-2, also known as BISPR, was reported to share a bidirectional promoter with BST2 (18), which was known for its antiviral activity against DNA viruses and RNA viruses (19, 20). Hence, we focused on lncRNA ITPRIP-1 for further study. Further experiments indicated that lncITPRIP-1 is upregulated by IFN-α in a dose-dependent manner (Fig. 1B).
FIG 1.
LncITPRIP-1 is identified as an lncRNA involved in viral infection. (A) HLCZ01 cells were treated with IFN-α (200 U/ml) for the indicated times before semi-RT-PCR. (B) HLCZ01 cells were treated with IFN-α at a dose of 20, 50, 100, or 200 (U/ml) for 6 h. LncITPRIP-1was analyzed by qRT-PCR. (C) Huh7.5 cells were infected with HCV at the indicated MOI for 12 h. qRT-PCR was performed to determine the lncITPRIP-1 level. (D) Huh7.5 cells were infected with HCV (MOI, 0.001) for the indicated times (in hours). LncITPRIP-1 was analyzed by qRT-PCR. (E to H) Huh7 cells (E), Huh7.5 cells (F), Huh7.5.1-MAVS (G) cells, and HLCZ01 cells (H) were infected with HCV at the indicated time points (at an MOI of 0.001 for Huh7 and Huh7.5 cells, an MOI of 3 for Huh7.5.1-MAVS cells, and an MOI of 1 for HLCZ01 cells). LncITPRIP-1 mRNA (left) IFN-β mRNA (middle), and HCV RNA (right) levels were determined by qRT-PCR. (I) Comparison of HCV RNA levels in Huh7, Huh7.5, Huh7.5.1-MAVS, and HLCZ01 cells. (J) HLCZ01 cells were treated with IFN-α (200 U/ml) for 6 h or challenged with VSV (MOI, 1) for 16 h, SeV (MOI, 1) for 12 h, HCV (MOI, 1) for 84 h, and HSV (MOI, 3) for 36 h. LncITPRIP-1 was analyzed by qRT-PCR. (K) HLCZ01 cells were transfected with plasmids expressing STAT1, STAT2, ISG12a, BST2, or the corresponding vector for 48 h, followed by qRT-PCR. Shown are representative RT-PCR and qRT-PCR results from three independent experiments. d, days. *, P < 0.05 versus the control; **, P < 0.01 versus the control; ***, P < 0.001 versus the control.
Type I and type III IFNs are major components in antiviral immune responses (21). To investigate the association between lncITPRIP-1 and viral infection, we analyzed the lncITPRIP-1 level in HCV-infected Huh7.5 cells. LncITPRIP-1 was upregulated in a virus dose- and time-dependent manner (Fig. 1C and D). Then we compared lncITPRIP-1 levels in Huh7, Huh7.5, Huh7.5.1-MAVS, and HLCZ01 cells upon HCV infection. Quantitative RT-PCR (qRT-PCR) data showed that the upregulation of lncITPRIP-1 in Huh7.5.1-MAVS and HLCZ01 cells was evidently stronger than that in Huh7 and Huh7.5 cells upon HCV infection (Fig. 1E to H). Importantly, HCV infection caused robust IFN production in Huh7.5.1-MAVS and HLCZ01 cells but not in Huh7 and Huh7.5 cells (22) (Fig. 1E to H), and HCV replication levels were significantly higher in Huh7 and Huh7.5 cells than in Huh7.5.1-MAVS and HLCZ01 cells (Fig. 1I).
The human lncRNA gene ITPRIP-1 (ENSG00000231233.1; chromosome 10 from positions 106111349 to 106113333 [1.9 kbp]; LNCipedia transcript identifier, lnc-ITPRIP-1:1; 795 nucleotides) is located on chromosome 10 and is adjacent to the coding gene DANGER (also named ITPRIP). To investigate whether lncITPRIP-1 is involved in other viral infections, we examined lncITPRIP-1 levels in HLCZ01 cells infected with vesicular stomatitis virus (VSV), Sendai virus (SeV), HCV, or herpes simplex virus (HSV). Surprisingly, lncITPRIP-1 could be induced by RNA virus (VSV, SeV, and HCV) and DNA virus (HSV) infection (Fig. 1J). IFNs signal through the JAK/STAT pathway, which induces the sequential activation of Janus family kinases and STATs, resulting in the transcriptional activation of interferon-stimulated genes (ISGs), which are mainly involved in host antiviral responses (23, 24). Therefore, we sought to explore whether the upregulation of lncITPRIP-1 was associated with downstream key factors. However, we did not observe a change in the level of lncITPRIP-1 expression after STAT (STAT1 and STAT2) or ISG (ISG12a and BST2) plasmid transfection in HLCZ01 cells (Fig. 1K). Taken together, these data demonstrate that lncITPRIP-1 can be induced by IFN and is involved in viral infection.
LncITPRIP-1 inhibits HCV replication in human hepatocytes.
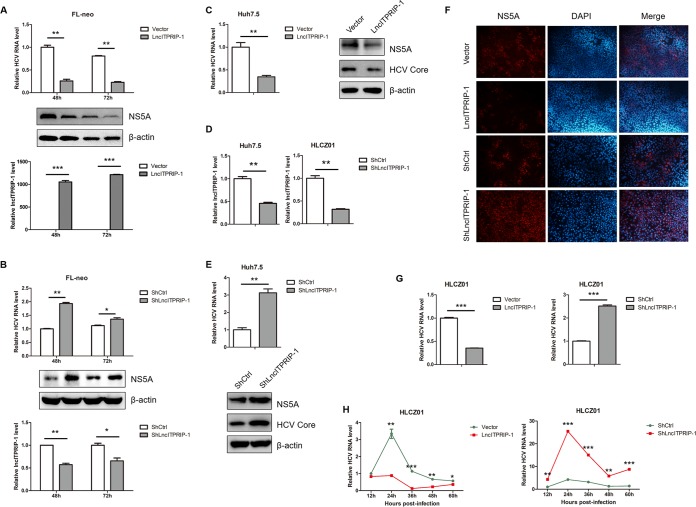
HCV is a hepatotrophic virus that belongs to the Hepacivirus genus of the family Flaviviridae (10, 25). The genome of HCV is a single-stranded positive-sense RNA approximately 9.6 kb in length (10). Upon virus infection, host cells produce large amounts of cytokines to enable elimination of the pathogen (14, 21, 26). We supposed that lncITPRIP-1 might be involved in antiviral responses to HCV. To unveil the role of lncITPRIP-1 in HCV infection, FL-neo cells (containing an HCV genotype 1b full-length replicon) were transfected with plasmids carrying lncITPRIP-1 or short hairpin RNA (shRNA) targeting lncITPRIP-1. qRT-PCR and Western blotting data showed that ectopic expression of lncITPRIP-1 significantly impaired viral replication (Fig. 2A), while lncITPRIP-1 knockdown mildly enhanced viral replication in FL-neo cells (Fig. 2B). Then we investigated the antiviral activity of lncITPRIP-1 in HCV-infected Huh7.5 cells. Ectopic expression of lncITPRIP-1 attenuated virus replication in Huh7.5 cells (Fig. 2C). We noticed that the silencing effects of lncITPRIP-1 in FL-neo cells were not satisfactory. To achieve better silencing effects, we constructed stable lncITPRIP-1-silenced cells of the Huh7.5 and HLCZ01 cell lines (Fig. 2D). LncITPRIP-1-silenced Huh7.5 cells showed higher levels of HCV RNA and viral protein than control cells (Fig. 2E). Furthermore, immunofluorescence results confirmed the antiviral activity of lncITPRIP-1 (Fig. 2F). A newly developed hepatoma cell line, HLCZ01, supporting the entire life cycle of HCV preserves a relatively complete innate immune response to virus infection (27). To have a better understanding of the function of lncITPRIP-1 in HCV infection and the IFN signaling pathway, we concentrated on HLCZ01 cells in the following study. Similarly, we observed that lncITPRIP-1 prominently suppressed virus replication in HCV-infected HLCZ01 cells (Fig. 2G), and the antiviral effects were relatively evident at late time points postinfection (Fig. 2H). The HCV RNA level in lncITPRIP-1-silenced cells was also remarkably higher than that in control HLCZ01 cells regardless of the infection time (Fig. 2G and H). These data demonstrate that lncITPRIP-1 inhibits HCV infection.
FIG 2.
LncITPRIP-1 inhibits HCV replication in human hepatocytes. (A) FL-neo cells were transfected with a plasmid expressing lncITPRIP-1 or the empty vector for the indicated times (in hours), followed by RNA or protein extraction. The replication of HCV was examined by qRT-PCR (top) and Western blotting (middle). The overexpression efficiency of lncITPRIP-1 was confirmed by RT-PCR (bottom). (B) FL-neo cells were transfected with shRNA targeting lncITPRIP-1 for the indicated times (in hours). HCV RNA (top) and the silencing efficiency (bottom) were examined by qRT-PCR, and viral proteins were tested by Western blotting (middle). (C) Huh7.5 cells were infected with HCV (MOI, 0.01) for 6 h before lncITPRIP-1 plasmid transfection for 48 h. HCV RNA was examined by qRT-PCR (left), and viral proteins were measured by Western blotting (right). (D) Silencing effects of lncITPRIP-1 in Huh7.5 (left) or HLCZ01 (right) cells stably expressing shRNA targeting lncITPRIP-1. (E) Huh7.5 cells stably expressing a plasmid carrying shRNA targeting lncITPRIP-1 were infected with HCV (MOI, 0.01) for 48 h before RNA extraction (top) and Western blotting (lower). (F) The HCV NS5A protein was analyzed by immunofluorescence staining. (G) (Left) HCV-infected (MOI, 0.1) HLCZ01 cells were transfected with lncITPRIP-1 or the empty vector plasmid for 48 h. (Right) HLCZ01 cells stably expressing shRNA targeting lncITPRIP-1 were infected with HCV (MOI, 0.1) for 48 h. HCV RNA was tested by qRT-PCR. (H) (Left) HLCZ01 cells were infected with HCV (MOI, 0.1) for 12 h, followed by lncITPRIP-1 plasmid transfection for the indicated times. (Right) HLCZ01 cells stably expressing shRNAs targeting lncITPRIP-1 were infected with HCV (MOI, 0.1) for the indicated times. HCV RNA was examined by qRT-PCR. ShlncITPRIP-1, shRNA targeting lncITPRIP-1; ShCtrl, control shRNA. *, P < 0.05 versus the control; **, P < 0.01 versus the control; ***, P < 0.001 versus the control.
LncITPIRP-1 enhances the HCV-induced antiviral response.
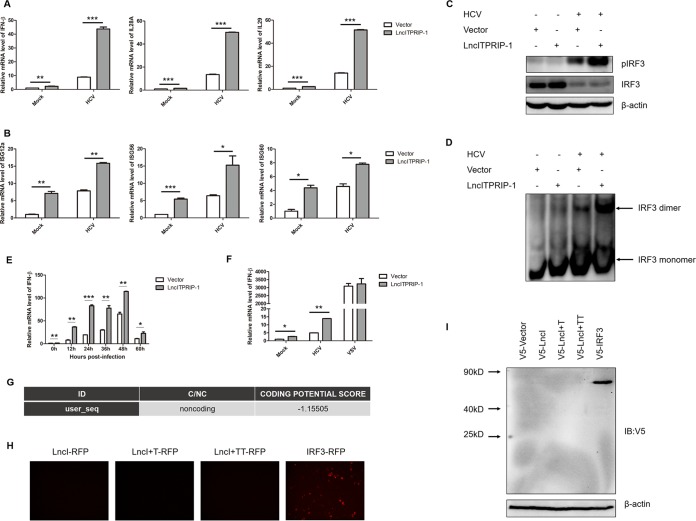
As an IFN-induced lncRNA, there is a strong possibility that lncITPRIP-1 functions through the IFN signaling pathway. Upon HCV infection, RIG-I, MDA5, and protein kinase R (PKR) activate the interferon-responsive genes to set up type I and type III IFN responses to viral infection (10). We speculated that lncITPRIP-1 might play an important role in regulating the HCV-triggered innate immune response. To this end, we investigated the effects of lncITPRIP-1 on HCV-induced cytokines in HLCZ01 cells. As expected, ectopic expression of lncITPRIP-1 enhanced HCV-triggered IFN-β, interleukin-28A (IL-28A), and IL-29 production in HLCZ01 cells (Fig. 3A). Moreover, representative ISGs, including ISG12a, ISG56, and ISG60, were also significantly upregulated in lncITPRIP-1-overexpressing cells (Fig. 3B).
FIG 3.
LncITPRIP-1 enhances the HCV-triggered IFN pathway. (A, B) HLCZ01 cells were infected with HCV (MOI, 0.01) for 24 h before they were transfected with lncITPRIP-1 and the vector plasmid for 48 h. IFN (A) and ISG (B) mRNA was detected by qRT-PCR. (C) HLCZ01 cells were infected with HCV (MOI, 2) for 24 h before transfection of the lncITPRIP-1 plasmid for 48 h. The phosphorylation status of IRF3 was examined by Western blotting. (D) HLCZ01 cells were treated as described in the legend to panel C before running of a native gel for the detection of IRF3 dimerization. (E) HLCZ01 cells were treated as described in the legend to Fig. 2H. IFN-β mRNA was examined by qRT-PCR. (F) HLCZ01 cells were infected with HCV (MOI, 0.01) for 12 h before transfection of the indicated plasmids for 48 h or transfected with the lncITPRIP-1 expression plasmid for 48 h followed by VSV infection (MOI, 1) for 9 h. IFN-β mRNA was examined by qRT-PCR. (G) The coding potential score of lncITPRIP-1 was obtained from the coding potential calculator. ID, identifier; C/NC, coding/noncoding. (H) HEK293T cells were transfected with lncITPRIP-1-RFP, lncITPRIP-1+T-RFP, lncITPRIP-1+TT-RFP, or IRF3-RFP plasmids for 48 h. The RFP-IRF3 plasmid served as a positive control. Images were taken under a fluorescence microscope. (I) HEK293T cells were transfected with the pcDNA3.1a vector (V5-Vector), pcDNA3.1a-lncITPRIP-1 (V5-LncI), pcDNA3.1a-lncITPRIP-1+T (V5-LncI+T), pcDNA3.1a-lncITPRIP-1+TT (V5-LncI+TT), or pcDNA3.1a-IRF3 (V5-IRF3) for 48 h, followed by Western blotting. The pcDNA3.1a-IRF3 plasmid served as a positive control. IB, immunoblotting. *, P < 0.05 versus the control; **, P < 0.01 versus the control; ***, P < 0.001 versus the control.
Upon activation, IFN-regulatory factor 3 (IRF3), which plays a critical role in the IFN signaling pathway, undergoes C-terminal phosphorylation, dimerization, and subsequent nuclear translocation (28, 29). In an attempt to explore whether lncITPRIP-1 has regulatory effects on IRF3 activation upon HCV infection, we determined the status of IRF3 phosphorylation and dimerization by Western blotting and native PAGE, respectively. The activation of IRF3 (i.e., phosphorylation and dimerization of IRF3) by HCV was remarkably augmented in lncITPRIP-1-overexpressing HLCZ01 cells (Fig. 3C and D). Furthermore, lncITPRIP-1 enhanced the IFN induction by HCV at different time points postinfection (Fig. 3E). To figure out whether lncITPRIP-1 had similar effects on the VSV-induced RIG-I-dependent IFN signaling pathway, we tested the induction of IFN-β by VSV in lncITPRIP-1-overexpressing cells. Surprisingly, forced expression of lncITPRIP-1 did not affect the IFN production induced by VSV (Fig. 3F), suggesting that lncITPRIP-1 is not involved in the RIG-I-dependent IFN pathway.
Although lncRNAs are generally non-protein-coding transcripts, recent studies have shown that a fraction of the putative small open reading frames within lncRNAs is translated (30). To investigate whether there was an lncRNA ITPRIP-1-coding peptide involved in the regulation of innate immunity, we got a coding potential score of −1.15505 by running the coding potential calculator (31) (Fig. 3G). To confirm the noncoding ability of lncITPRIP-1, we designed two more reverse primers by adding bases (+T, +TT) and cloned the constructs into pcDNA3.1a vector plasmids and three pairs of corresponding primers and cloned the constructs into pRed-max-N1 vector plasmids (31). For red fluorescent protein (RFP)-labeled lncITPRIP-1 plasmid construction, the lncITPRIP-1 gene was inserted before the initiation signal of the RFP gene. Then, three lncITPRIP-1-RFP plasmids, IRF3-RFP plasmids (as an RFP-tagged positive control), three pcDNA3.1a lncITPRIP-1 plasmids, pcDNA3.1a vector plasmids (as a pcDNA3.1a-tagged negative control), and pcDNA3.1a IRF3 plasmids (as a pcDNA3.1a-tagged positive control) were transfected into HEK293T cells. Consistent with the expectation, no red fluorescence was observed in HEK293T cells transfected with the lncITPRIP-1-RFP, lncITPRIP-1+T-RFP, or lncITPRIP-1+TT-RFP plasmids and no band was shown in Western blot images (Fig. 3H and I), which proved the noncoding capacity of lncITPRIP-1. These data suggest that lncITPRIP-1 is a noncoding transcript and is capable of promoting HCV-triggered IRF3 activation and subsequent IFN production.
LncITPRIP-1 promotes HMW poly(I·C)-triggered IFN production.
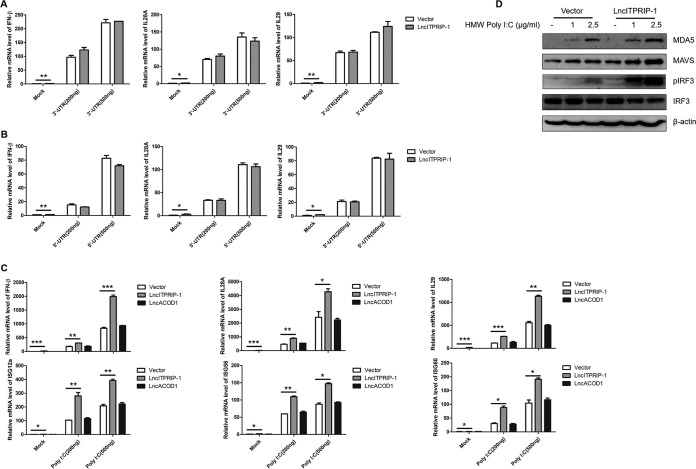
RIG-I, PKR, and MDA5 are three main functional PRRs in the host IFN response to HCV infection (10, 14, 32). To define the mechanism by which lncITPRIP-1 promotes HCV-triggered innate immunity, we used the HCV 3′ untranslated region (UTR), HCV 5′ UTR, and high-molecular-weight (HMW) poly(I·C) as mimics for activating the RIG-I-, PKR-, and MDA5-mediated IFN signaling pathway, respectively. The expressions of IFN-β, IL-28A, and IL-29 triggered by the HCV 3′ UTR and the HCV 5′ UTR were not affected when we overexpressed lncITPRIP-1 in HLCZ01 cells (Fig. 4A and B). However, ectopic expression of lncITPRIP-1 remarkably augmented the production of IFN-β, IL-28A, and IL-29 triggered by poly(I·C) (Fig. 4C). Representative ISGs were also significantly upregulated in lncITPRIP-1-overexpressing cells (Fig. 4C). Based on these data, we speculated that lncITPRIP-1 might impair HCV infection through the promotion of the MDA5-dependent IFN pathway. Western blotting confirmed its function in the MDA5-dependent IFN pathway upon poly(I·C) stimulation. Overexpression of lncITPRIP-1 increased the protein level of MDA5 and MAVS triggered by poly(I·C) (Fig. 4D). Importantly, ectopic expression of lncITPRIP-1 promoted poly(I·C)-induced IRF3 activation (Fig. 4D). In brief, lncITPRIP-1 promotes poly(I·C)-triggered IRF3 activation and IFN production but has no regulatory effects on the HCV 3′ UTR- or 5′ UTR-induced IFN signaling pathway.
FIG 4.
LncITPRIP-1 promotes the HMW poly(I·C)-triggered IFN pathway. (A to C) HLCZ01 cells were transfected with the pcDNA3.1a vector and pcDNA3.1a-tagged lncITPRIP-1 plasmids for 40 h and then transfected with the indicated doses of the HCV 3′ UTR (A), the HCV 5′ UTR (B), or HMW poly(I·C) (C) for 8 h. IFNs or ISGs were measured by qRT-PCR. LncRNA ACOD1 (LncACOD1) served as a negative control. (D) HLCZ01 cells were transfected with lncITPRIP-1 and vector plasmids for 40 h and then transfected with the indicated doses of poly(I·C) for 9 h. MDA5, IRF3, and the phosphorylation status of IRF3 were analyzed by Western blotting. *, P < 0.05 versus the control; **, P < 0.01 versus the control; ***, P < 0.001 versus the control.
LncITPRIP-1 targets and interacts with MDA5.
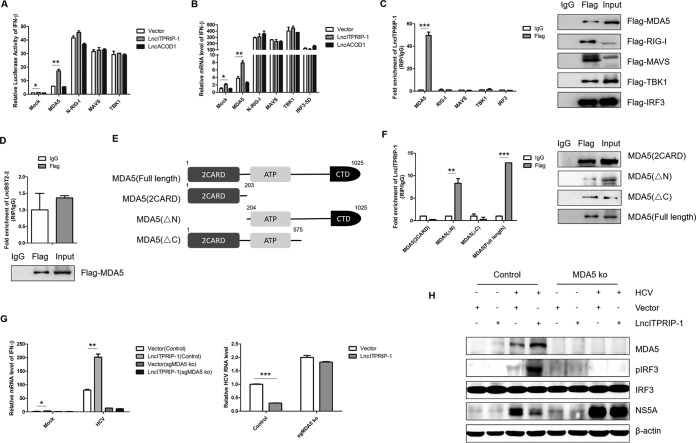
MDA5 is a cytosolic viral RNA sensor that belongs to the RLR family (9). Upon stimulation, MDA5 undergoes a conformational change and recruits MAVS, leading to the sequential activation of downstream TBK1 and IRF3 (21, 22). To explore which component(s) of the MDA5-mediated IFN signaling pathway could be targeted by lncITPRIP-1, we contransfected plasmids carrying lncITPRIP-1 and various candidates in a reporter assay system. Interestingly, lncITPRIP-1 augmented the activation of the IFN-β promoter by MDA5, while no differences in N-RIG-I (N-terminal, 2CARD domains of full-length RIG-I)-, MAVS-, or TBK1-induced IFN-β promoter activation were observed (Fig. 5A), suggesting that lncITPRIP-1 might regulate MDA5 to enhance IFN-β production. We further analyzed the effects of lncITPRIP-1 on IFN-β induced by MDA5, N-RIG-I, MAVS, TBK1, and IRF3-5D (a mutant in which the serine-threonine cluster at aa 396 to 405 was mutated to phosphomimetic Asp [5D]) in HLCZ01 cells. Consistently, lncITPRIP-1 increased the production of IFN-β triggered by MDA5 (Fig. 5B). These data suggest that lncITPRIP-1-mediated IFN production might depend on MDA5.
FIG 5.
LncITPRIP-1 targets and interacts with MDA5. (A) IFN-β luciferase assay in HEK293T cells. HEK293T cells were cotransfected with pRL-CMV, IFN-β luciferase, lncITPRIP-1 plasmids, and MDA5, N-RIG-I, MAVS, and TBK1 plasmids for 24 h. An IFN-β luciferase assay was then performed. LncRNA ACOD1 served as a negative control. (B) HLCZ01 cells were cotransfected with the lncITPRIP-1 plasmid and MDA5, N-RIG-I, MAVS, TBK1, and IRF3-5D plasmids for 48 h. IFN-β mRNA was measured by qRT-PCR. LncRNA ACOD1 served as a negative control. (C) HEK293T cells were cotransfected with the lncITPRIP-1 plasmid and MDA5, RIG-I, MAVS, TBK1, and IRF3 plasmids for 48 h. RIP analysis of the interaction between lncITPRIP-1 and its potential target was then performed. (D) RIP analysis of HEK293T cells cotransfected with lncBST2-2 and MDA5 plasmids for 48 h. (E) Schematic illustration of MDA5 truncations. (F) HEK293T cells were cotransfected with the lncITPRIP-1 plasmid and truncations of MDA5 before RIP. (G, H) Huh7-MAVSR control and Huh7-MAVSR-sgMDA5 knockout (ko) cells were infected with HCV (MOI, 2) for 6 h before they were transfected with the indicated plasmids for 48 h, followed by qRT-PCR (G) and Western blotting (H). *, P < 0.05 versus the control; **, P < 0.01 versus the control; ***, P < 0.001 versus the control.
Based on the observations of the regulatory functions of lncITPRIP-1 described above, we sought to explore the association between lncRNA ITPRIP-1 and MDA5. To verify the possibility of an association, we cotransfected plasmids carrying lncITPRIP-1 and MDA5, RIG-I, MAVS, TBK1, or IRF3, followed by RNA immunoprecipitation (RIP) analysis of the interactions between them. Consistent with our assumption, the RIP assay suggested that there is an association between MDA5 and lncITPRIP-1 (Fig. 5C). Most lncRNAs form a double-strand structure in their native state. As MDA5 is endowed with the ability to recognize dsRNA in the cell, we further investigated whether the association between MDA5 and lncRNA was universal. However, lncBST2-2, a random lncRNA selected from our candidate lncRNAs, did not interact with MDA5 (Fig. 5D), validating that the association between MDA5 and lncITPRIP-1 was a result of specific recognition.
To better understand the molecular mechanisms by which lncITPRIP-1 mediates MDA5-dependent IFN signaling, we examined the interaction between lncITPRIP-1 and truncations of MDA5. MDA5 contains two N-terminal caspase activation and recruitment domains (2CARD) that act as signaling domains, a central DExD/H RNA helicase domain that facilitates ATP hydrolysis and RNA binding, and a C-terminal domain (CTD) that aids in RNA ligand recognition and binding specificity (9). So, we mapped physical truncations of MDA5 and designed three truncated constructs: 2CARD of MDA5 [MDA5(2CARD); aa 1 to 203], MDA5 lacking the N-terminal region [MDA5(ΔN); aa 204 to 1025], and MDA5 lacking the C-terminal region [MDA5(ΔC); aa 1 to 575] (Fig. 5E). Subsequent RIP experiments demonstrated that the C terminus of MDA5 plays a key role in the association between MDA5 and lncITPRIP-1 (Fig. 5F), but the fold enrichment of lncITPRIP-1 associated with MDA5(ΔN) was lower than that of lncITPRIP-1 associated with full-length MDA5 (Fig. 5F), suggesting that the integral structure of MDA5 is essential for the association with lncITPRIP-1.
To verify whether lncITPRIP-1 inhibits HCV replication via MDA5, we performed experiments in MDA5 knockout (ko) Huh7-MAVSR cells. In control cells, forced expression of lncITPRIP-1 enhanced HCV-induced IRF3 activation, MDA5 expression, and IFN production and lowered the HCV replication level (Fig. 5G and H). HCV infection could not cause robust IFN production after MDA5 knockout, as Huh7-MAVSR-sgMDA5 (silencing MDA5 gene) ko cells showed no apparent change in IRF3 phosphorylation and a much higher NS5A level than control cells. As expected, knockout of MDA5 almost reversed the inhibition of HCV replication by lncITPRIP-1 (Fig. 5G and H). These results reveal that lncITPRIP-1 may be a positive regulator of MDA5 to promote IFN production, thereby suppressing HCV infection.
LncITPRIP-1-mediated MDA5 upregulation is IFN dependent.
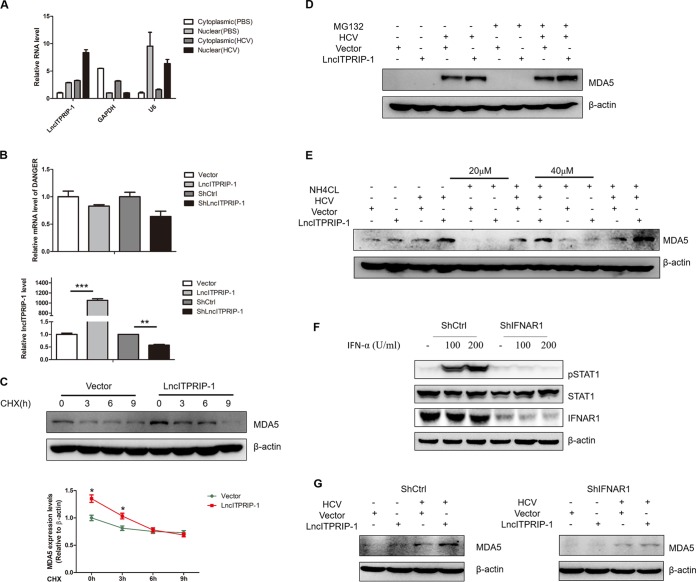
Then, we sought to investigate the mechanisms of how lncITPRIP-1 regulates MDA5. To this end, we determined the cellular localization of lncITPRIP-1. LncITPRIP-1 was localized both in the cytoplasm and in the nucleus, and more lncITPRIP-1 was distributed in the nucleus of HLCZ01 cells (Fig. 6A). Upon HCV infection, the levels of both cytosolic and nuclear lncITPRIP-1 were upregulated (Fig. 6A). The human gene DANGER is a neighbor gene to lncITPRIP-1. To investigate whether lncITPRIP-1 regulates the expression of DANGER, we measured the mRNA level of DANGER after altering lncITPRIP-1 expression. The level of DANGER mRNA was not affected when lncITPRIP-1 was silenced or overexpressed in HLCZ01 cells (Fig. 6B), ruling out the possibility that lncITPRIP-1 functions through cis effects on DANGER.
FIG 6.
LncITPRIP-1-mediated MDA5 upregulation is IFN dependent. (A) The RNA levels of lncITPRIP-1, the cytoplasmic control (GAPDH mRNA), and the nuclear control (U6 RNA) in the cytoplasmic and nuclear fractions from HLCZ01 cells were assessed by qRT-PCR. HLCZ01 cells were infected with HCV for 48 h. (B) HLCZ01 cells were transfected with an lncITPRIP-1 overexpression plasmid or a plasmid carrying shRNA targeting lncITPRIP-1 for 48 h. DANGER (top) or lncITPRIP-1 (bottom) was analyzed by qRT-PCR. (C to E) HLCZ01 cells were infected with HCV for 12 h before transfection of the indicated plasmids and then treated with CHX (C), MG132 (D), or NH4Cl (E). The MDA5 protein was examined by Western blotting. (F) Silencing efficiency of IFNAR1 in HLCZ01 cells expressing control shRNA (ShCtrl) and HLCZ01 cells expressing shRNA targeting IFNAR1 (ShIFNAR1). (G) HLCZ01 cells expressing control shRNA and shRNA targeting IFNAR1 were infected with HCV for 12 h before transfection of the indicated plasmids for 48 h. The MDA5 protein was examined by Western blotting. *, P < 0.05 versus the control; **, P < 0.01 versus the control; ***, P < 0.001 versus the control.
In view of the observation that lncITPRIP-1 increased the amount of the MDA5 protein upon HCV infection (Fig. 5G), we speculated that lncITPRIP-1 might enhance virus-induced IFN production through the regulation of MDA5 expression. To test this hypothesis, HLCZ01 cells infected with HCV were treated with cycloheximide (CHX), MG132, and NH4Cl. CHX almost abolished the upregulation of MDA5 by lncITPRIP-1 at 6-h and 9-h stimulations (Fig. 6C), whereas neither MG132 nor NH4Cl treatment had any effects on MDA5 upregulation upon HCV infection when we delivered lncITPRIP-1 into HLCZ01 cells (Fig. 6D and E). MG132 treatment increased the level of MDA5 protein triggered by HCV (Fig. 6D), while NH4Cl treatment did not have obvious effects on the level of MDA5 upon HCV infection (Fig. 6E), which was consistent with the data from a study of trim40-related MDA5 (33), suggesting that the degradation of MDA5 is mainly through an ubiquitin-proteasome pathway. Then, we performed experiments in HLCZ01 cells stably expressed shRNA targeting IFNAR1 (Fig. 6F). The results showed that silencing of IFNAR1 abrogated the effects of lncITPRIP-1 on the level of MDA5 protein upon HCV infection (Fig. 6G), suggesting that lncITPRIP-1-mediated MDA5 upregulation upon HCV infection is dependent on IFN and that lncITPRIP-1 has no direct regulatory effects on MDA5 expression.
LncITPRIP-1 promotes MDA5 oligomerization.
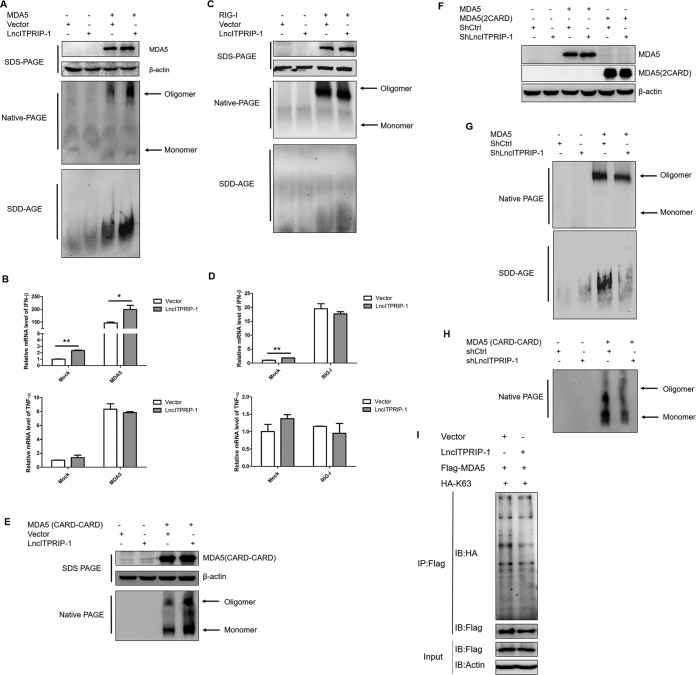
Upon recognizing the internal duplex structure of dsRNA, the 2CARD domains of MDA5 oligomerize into an elongated structure (8). Although lncITPRIP-1 is distributed in both the cytoplasm and the nucleus, MDA5 is mainly located in the cytoplasm. Hence, we sought to explore the protein modification effects of lncITPRIP-1 on MDA5. We focused on MDA5 oligomerization, which is reported to be essential for MDA5-dependent antiviral signal transduction (8, 34). To investigate whether lncITPRIP-1 is involved in the formation of MDA5 oligomerization, we attempted to determine the effects of lncITPRIP-1 on MDA5 oligomerization by native PAGE and semidenaturating detergent agarose gel electrophoresis (SDD-AGE) (35, 36). The data showed that ectopic expression of lncITPRIP-1 significantly promoted exogenous MDA5 oligomerization in HEK293T cells (Fig. 7A). However, the MDA5 monomer was relatively faint in HEK293T cells, and lncITPRIP-1 did not have obvious effects on it. Interestingly, lncITPRIP-1 promoted IFN production triggered by MDA5 but did not have effects on MDA5-induced tumor necrosis factor alpha (TNF-α) expression (Fig. 7B), suggesting that lncITPRIP-1 affected only MDA5-dependent IFN production and that there might be multiple roles of lncITPRIP-1 in mediating innate immunity. Although they have similar activation mechanisms, the oligomerization of exogenous RIG-I and RIG-I-triggered IFN production were not affected when we delivered lncITPRIP-1 into HEK293T cells (Fig. 7C and D). Surprisingly, ectopic expression of lncITPRIP-1 boosted MDA5(2CARD) oligomerization (Fig. 7E), yet no association between the 2CARD of MDA5 and lncITPRIP-1 was found (Fig. 5F), suggesting that lncITPRIP-1 may regulate 2CARD oligomerization through other factors and that the interaction is not completely critical in the regulation. Silencing of lncITPRIP-1 remarkably attenuated the oligomerization of full-length MDA5 and the 2CARD of MDA5 (Fig. 7F to H). These data demonstrate that lncITPRIP-1 plays an essential role in MDA5 oligomerization.
FIG 7.
LncITPRIP-1 promotes MDA5 oligomerization. (A, B) HEK293T cells were cotransfected with V5-tagged lncITPRIP-1 and Flag-tagged MDA5 plasmids for 48 h before Western blotting (A), native PAGE (A), SDD-AGE (A), and qRT-PCR (B). Exogenous MDA5 oligomerization was determined by immunoblotting with antibodies against Flag. (C, D) HEK293T cells were cotransfected with V5-tagged lncITPRIP-1 and Flag-tagged RIG-I plasmids for 48 h before Western blotting (C), native PAGE (C), SDD-AGE (C), and qRT-PCR (D). Exogenous RIG-I oligomerization was determined by immunoblotting with antibodies against Flag. (E) HEK293T cells were cotransfected with V5-tagged lncITPRIP-1 and Flag-tagged MDA5(2CARD) plasmids for 48 h, followed by native PAGE. MDA5(2CARD) was immunoblotted by antibodies against Flag. (F to H) HEK293T cells were cotransfected with shRNA targeting lncITPRIP-1 and Flag-tagged MDA5 or MDA5(2CARD) plasmids for 48 h, followed by Western blotting (F), native PAGE (G, H), and SDD-AGE (G). Oligomerization was tested by detection of antibodies against Flag. (I) Ubiquitination assay in HEK293T cells cotransfected with MDA5, ubiquitin, and a vector or lncITPRIP-1 plasmid for 48 h. An IP assay was performed. *, P < 0.05 versus the control; **, P < 0.01 versus the control.
It is widely known that ubiquitin plays an essential role in protein modification, provoking signal transduction (34, 37, 38). However, the mechanisms of ubiquitination in MDA5 are poorly understood. Earlier study shows that K63-linked polyubiquitination is essential for MDA5 activation (34, 36), while Wu et al. (8) reported that MDA5 can be directly activated by a filamentous structure formed by MDA5 oligomerization around dsRNA in an ubiquitin-independent manner (8). However, we found that lncITPRIP-1 did not have conclusive effects on the K63-linked ubiquitination of MDA5 (Fig. 7I). Briefly, lncITPRIP-1 promotes MDA5 oligomerization and mediates MDA5-triggered IFN production in a K63-linked, ubiquitination-independent manner.
MDA5 is associated with HCV RNA, and lncITPRIP-1 strengthens the association.
Without stimulation, MDA5 maintains a low base level in the majority of cells (7). Upon IFN activation and direct virus signal stimulation, there is a great increase in MDA5 expression (7). We did notice the low expression level of MDA5 in HLCZ01 cells and found that HCV infection induced MDA5 expression (Fig. 5G and 6C to F). To investigate the expression level of MDA5 upon HCV infection in human hepatocytes, we measured the mRNA level of MDA5 in Huh7, Huh7.5, Huh7.5.1-MAVS, and HLCZ01 cells upon HCV infection (Fig. 8A). We found that HCV infection initiated robust MDA5 expression in Huh7.5.1-MAVS and HLCZ01 cells, which was consistent with the IFN level. Surprisingly, MDA5 expression was also upregulated in Huh7.5 cells, and there was an inconspicuous increase in MDA5 expression in Huh7 cells at 6 days postinfection (Fig. 8A).
FIG 8.
MDA5 is associated with HCV RNA, and lncITPRIP-1 strengthens the association. (A) Total cellular RNA was from the assay whose results are presented in Fig. 1E to H. MDA5 mRNA was examined by qRT-PCR. (B) Huh7.5 cells were infected with HCV (MOI, 0.01) for 6 h before transfection of the indicated plasmids for the indicated times. HCV RNA was analyzed by qRT-PCR. (C, D) Huh7.5 cells were treated as described in the legend to panel B. (C) HCV proteins were measured by Western blotting. (D) IFN and ISG were detected by qRT-PCR. (E) RIP assay analysis of HCV-infected Huh7.5 cells transfected with MDA5 or HMGB1 expression plasmids. HMGB1 served as a positive control for RNA binding. (F) Huh7.5 cells were infected with HCV (MOI, 0.01) for 6 h before transfection of MDA5 truncation plasmids for 48 h. HCV RNA was analyzed by qRT-PCR (top), and HCV proteins were tested by Western blotting (bottom). (G) RIP assay analysis of the association between MDA5 truncations and HCV RNA in HCV-infected Huh7.5 cells. (H) LncITPRIP-1-silenced Huh7.5 cells and control cells were infected with HCV for 6 h before they were transfected with the MDA5 expression plasmid for 48 h. The RIP assay was performed to analyze the change in association. (I) Huh7.5 cells were infected with HCV for 6 h before they were transfected with MDA5 and lncITPRIP-1 or vector plasmids for 48 h. The RIP assay was performed to analyze the change in association. (J) RIP assay analysis in HLCZ01 cells infected with HCV for 48 h. (K) HLCZ01 cells were infected with HCV for 12 h before transfection of the indicated plasmids for 48 h. The RIP assay was performed to analyze the change in the interaction. (L, M) HLCZ01 cells expressing control shRNA and shRNA targeting IFNAR1 were infected with HCV (MOI, 1) for 12 h and then transfected with the indicated plasmids. The association between MDA5 and HCV RNA was analyzed by the RIP assay (L), and the HCV RNA level was tested by qRT-PCR (M). *, P < 0.05 versus the control; **, P < 0.01 versus the control; ***, P < 0.001 versus the control.
Accordingly, we speculated that MDA5 might be involved in the innate immune response against HCV in a defective immune system. To characterize the IFN-independent role of MDA5 in HCV replication, we performed experiments in Huh7.5 cells. Interestingly, forced expression of MDA5 suppressed HCV replication in Huh7.5 cells (Fig. 8B and C). MDA5 downregulated the level of viral RNA, NS5A, and core protein but had no effects on the IFN or ISG level (Fig. 8B to D), suggesting that MDA5 possesses antiviral activity against HCV independent of the IFN pathway. MDA5 is reported to be capable of functioning as an effector protein by binding to viral RNA to suppress virus replication (39). Therefore, we attempted to explore the association between MDA5 and HCV RNA. An RIP assay showed that there was an association between MDA5 and HCV RNA in Huh7.5 cells (40), indicating that MDA5 might inhibit HCV replication apart from IFN assistance by directly binding to viral RNA (Fig. 8E). In an attempt to define the mechanism of MDA5 affecting HCV replication independent of IFN, we sought to investigate the antiviral effects of the MDA5 truncations on HCV replication in Huh7.5 cells. qRT-PCR and Western blotting data showed that MDA5(ΔC) inhibited viral replication, while MDA5(2CARD) and MDA5(ΔN) had no obvious effects on it (Fig. 8F). Importantly, the RIP assay indicated that HCV RNA was associated with MDA5(ΔC) but not MDA5(2CARD) or MDA5(ΔN) (Fig. 8G). These data suggest that MDA5 is able to weaken HCV replication via binding to viral RNA.
Next, we determined the functionality of lncITPRIP-1 in the MDA5-HCV RNA association in Huh7.5 cells. Surprisingly, lncITPRIP-1 was able to reinforce the association between MDA5 and HCV RNA. LncITPRIP-1-silenced Huh7.5 cells exhibited a weaker interaction between MDA5 and HCV RNA, and forced expression of lncITPRIP-1 facilitated the recognition of HCV by MDA5 (Fig. 8H and I). A further RIP assay validated the association between MDA5 and HCV RNA (Fig. 8J). Ectopic expression of lncITPRIP-1 strengthened their interaction in HLCZ01 cells (Fig. 8K).
To confirm the role of type I interferon in the association, we investigated the effects of lncITPRIP-1 on the MDA5-HCV RNA interaction in IFNAR1-silenced HLCZ01 cells. RIP assay data indicated that IFNAR1 knockdown had no obvious effects on MDA5-mediated HCV recognition (Fig. 8L and M). Significantly, lncITPRIP-1 still exhibited a positive regulatory role in the MDA5-HCV RNA association and antiviral activity against HCV in IFNAR1-silenced HLCZ01 cells (Fig. 8L and M). These data indicate that MDA5 is able to suppress HCV replication independently of IFN by binding to viral RNA and lncITPRIP-1 is likely to function as a cofactor in the process.
LncITPRIP-1 promotes the innate immune response to viral infection through MDA5.
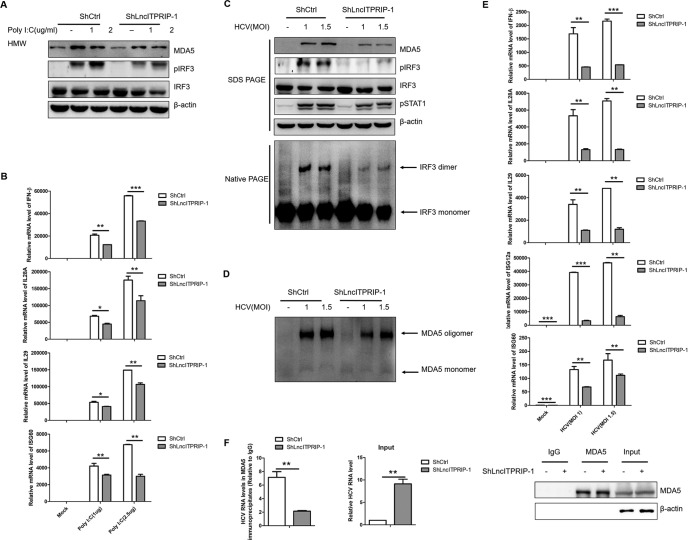
Parallel to MDA5, lncITPRIP-1 remains at a low base level in hepatocytes. Nonetheless, we attempted to investigate the effects of lncITPRIP-1 knockdown on the MDA5-mediated IFN immune response. Silencing of lncITPRIP-1 downregulated the level of MDA5 and the phosphorylation of IRF3 triggered by poly(I·C) (Fig. 9A). Accordingly, impaired expressions of IFN-β, IL-28A, IL-29, and ISGs induced by poly(I·C) were observed in lncITPRIP-1-silenced HLCZ01 cells (Fig. 9B). To further test the role of lncITPRIP-1 in innate immunity against viral infection, stable lncITPRIP-1-silenced HLCZ01 cells were infected by HCV at the multiplicities of infection (MOIs) indicated in Fig. 9. The data showed that silencing of lncITPRIP-1 decreased MDA5 expression and suppressed the phosphorylation of both IRF3 and STAT1 in HCV-infected HLCZ01 cells (Fig. 9C). Importantly, native PAGE data indicated that the knockdown of lncITPRIP-1 remarkably attenuated the formation of the IRF3 dimer and MDA5 oligomerization (Fig. 9C and D). Consistent with this finding, silencing of lncITPRIP-1 significantly impaired the expressions of IFN-β, IL-28A, IL-29, and ISGs triggered by HCV (Fig. 9E).
FIG 9.
Inhibition of lncITPRIP-1 expression significantly attenuated the MDA5-mediated IFN signaling pathway and virus recognition. (A, B) HLCZ01 cells stably expressing shRNA targeting lncITPRIP-1 were transfected with the indicated doses of poly(I·C) for 8 h. MDA5, IRF3, and pIRF3 were detected by Western blotting (A). IFNs and ISG60 were analyzed by qRT-PCR (B). (C to E) HLCZ01 cells stably expressing shRNA targeting lncITPRIP-1 were infected with HCV at the indicated MOIs for 48 h, followed by Western blotting (C), native PAGE (C, D), and qRT-PCR (E). (F) RIP assay analysis of MDA5 and HCV RNA in HLCZ01 cells stably expressing shRNA targeting lncITPRIP-1. The HCV RNA in the input was measured by qRT-PCR. *, P < 0.05 versus the control; **, P < 0.01 versus the control; ***, P < 0.001 versus the control.
Then we sought to explore the effects of lncITPRIP-1 knockdown on MDA5-mediated HCV recognition in HLCZ01 cells, which we considered to be done in an IFN-independent manner. LncITPRIP-1 knockdown remarkably weakened the association between MDA5 and HCV RNA (Fig. 9F), thereby partially explaining the antiviral activity against HCV. All these data reveal that lncITPRIP-1 enhances the MDA5-mediated innate immune response to HCV infection via promotion of the recognition of viral RNA by MDA5 and MDA5-mediated signal transduction for IFN production.
DISCUSSION
There are distinct changes in particular lncRNAs in response to viral protein or virus infection. The lncRNA NRON is reported to be differentially regulated by early and late viral proteins of HIV (41). Tumor suppressor lncRNA DREH is downregulated upon exposure to the X protein (HBx) of hepatitis B virus (HBV) (42). Altered expressions of NEAT1 and NRAV are observed during viral infection (24, 43). However, the latent mechanisms between lncRNAs and HCV infection remain unsettled. Here, we report on an lncRNA, ITPRIP-1, which is reported to be induced by IFN-α and can be induced by HCV, SeV, VSV, and HSV. Interestingly, our data show that the induction of lncITPRIP-1 is not entirely dependent on IFN, as lncITPRIP-1 upregulation is observed in HCV-infected Huh7 and Huh7.5 cells which are defective in IFN production. Although HCV, SeV, VSV, and HSV infection leads to the activation of the JAK/STAT pathway and induces ISG production, neither STAT (STAT1 and STAT2) nor ISG (ISG12a and BST2) overexpression could induce lncITPRIP-1 expression. We suppose that lncITPRIP-1 may be derived from a host antiviral factor downstream of the JAK/STAT pathway and respond to virus infection to enhance innate immunity. MDA5 expression is increased upon HCV infection, which is highly consistent with the expression of lncITPRIP-1. Hence, we hypothesize that lncITPRIP-1 may function as a coworker with MDA5 in innate immunity.
Importantly, our data reveal that lncITPRIP-1 functions as an antiviral factor against HCV infection. LncITPRIP-1 significantly suppresses HCV replication in human hepatocytes (FL-neo, Huh7.5, and HLCZ01 cells). By comparison, we found that the antiviral activity of lncITPRIP-1 is much more obvious in HLCZ01 cells. HLCZ01 cells mount a strong innate immune response to viral infection, so we focused on HLCZ01 cells to explore the role of lncITPRIP-1 in innate immunity.
Over the past decade, studies have disclosed the tricks played by HCV to sustain itself in the host without elimination by the immune system (32). Furthermore, this crafty cytoplasmic virus has all the cellular organelles at its disposal to successfully replicate, from ribosomes and intracellular membranous structures to the nucleus (10). Still, a number of clinical scenarios remain challenging and there is no vaccine available for HCV yet, which urge us to dig for the underlying mechanisms between HCV and host responses. Although much emphasis has been focused on investigating the roles of host coding genes in the activation of the innate immune response to HCV infection, little is known about lncRNAs in these biological processes. LncRNA EGOT favors HCV replication through negatively regulating the immune response (44), and lncRNA GAS5 binds to the NS3 protein to inhibit HCV replication (45). Despite this progress, the specific regulatory roles of these lncRNAs in the host defense response against HCV remain incompletely characterized. IFN responses play crucial parts in innate immunity against pathogens. Then we investigated whether this IFN-induced and virus infection-involved lncRNA plays a role in the immune response to function as an antiviral factor against HCV. Our data show that forced expression of lncITPRIP-1 remarkably enhances the expression of IFNs (IFN-β, IL-28A, and IL-29) and ISGs (ISG12a, ISG56, and ISG60) triggered by HCV. Interestingly, we demonstrate that lncITPRIP-1 possesses the ability to promote IRF3 phosphorylation and dimerization upon HCV infection. However, lncITPRIP-1 does not have effects on VSV-induced IFN production (the traditional RIG-I-mediated IFN pathway). Hence, we speculate that lncITPRIP-1 may play an important role in IFN production independent of the RIG-I-mediated IFN signaling pathway.
HCV presents a myriad of mechanisms to dodge the innate immune system. Notably, upon entry into the host, viral RNA is sensed by RIG-I, PKR, and MDA5. These three PRRs get activated, recruit MAVS, and induce the expression of IFNs through IRF3. However, viral NS3/4A protease cleaves MAVS and TRIF, which deprives them of their function in activating IRF3 (10). The viral NS5A protein blocks the activity of PKR (10). HCV core protein inhibits the dimerization of STATs and, hence, prevents downstream ISG production (10). Nevertheless, lncITPRIP-1 promotes only the poly(I·C)-triggered IFN pathway, which implies that lncITPRIP-1-mediated IFN production is MDA5 dependent. Subsequent IFN-β luciferase assay and cotransfection results confirm that lncITPRIP-1 promotes IFN production through MDA5.
Like RIG-I and LGP2, MDA5 is broadly expressed in most tissues (7). Nonetheless, MDA5 expression is typically maintained at a low level in a variety of cell types other than myeloid cells, epithelial cells, central nervous system cells, and plasmacytoid dendritic cells (7). We did observe a low base level of MDA5 in hepatocytes and a great increase upon exposure to IFN and virus infection. As a cytosolic viral dsRNA receptor, MDA5 shares sequence similarity and signaling pathways with RIG-I, yet it plays essential roles in antiviral signaling by distinct mechanisms. MDA5 recognizes the internal duplex structure of dsRNA, stacks onto dsRNA in a head-to-tail arrangement by direct protein-protein contacts, and decorates the outside the core filament with its tandem CARD, which oligomerize into an elongated structure that activates MAVS (8). During signal transduction, MDA5 oligomerization is indispensable for its activation. Interestingly, lncITPRIP-1 does not have direct regulatory effects on MDA5 expression but exerts a strong influence on the oligomerization of both endogenous and exogenous MDA5. We also demonstrate that the C-terminal domain (CTD) of MDA5 is essential for its association with lncITPRIP-1, and the integral structure of MDA5 makes a difference in the interaction. Hel2i provides a docking site for the MDA5 CTD to be positioned near the target RNA (8). LncITPRIP-1 may work as a cofactor to make MDA5 more accessible to viral RNA. Although there is no association found between lncITPRIP-1 and 2CARD of MDA5, lncITPRIP-1 promotes the oligomerization of exogenous 2CARD of MDA5.
For a long time, plentiful studies have explained the molecular mechanisms regarding the IFN-dependent antiviral activity of MDA5, but study of the hidden IFN-independent role of MDA5 in viral infection has almost been set aside. A previous study revealed that MDA5 suppresses the replication of several dsRNA viruses in an ATP-dependent manner independent of IFN signaling. HCV belongs to single-stranded RNA viruses, yet our results demonstrate that MDA5 is capable of inhibiting HCV replication through its C-terminal-deficient domain of MDA5 (ΔC domain) by directly binding to viral RNA, which has been identified to be an IFN-independent process. Unfortunately, we did not disclose whether there was a bridge participating in their association. Furthermore, lncITPRIP-1 plays a crucial role in MDA5-mediated HCV recognition, facilitating the association between MDA5 and HCV RNA, which may strengthen the IFN-independent antiviral activity of MDA5 against HCV. The effects of lncITPRIP-1 on HCV recognition may partially contribute to its stronger antiviral activity in HLCZ01 cells.
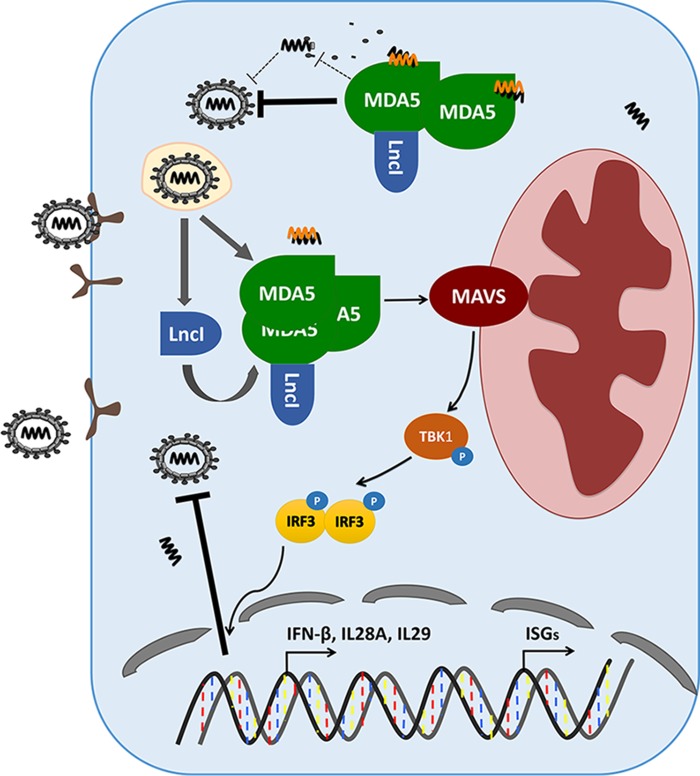
Based on these data, we propose a model of the function of lncITPRIP-1 in regulating HCV-induced innate immunity (Fig. 10). In their original state, lncITPRIP-1 and MDA5 are maintained at a low base level. Upon virus entry into hepatocytes, the level of lncITPRIP-1 along with that of MDA5 is increased. LncITPRIP-1 boosts MDA5 oligomerization and promotes the IFN signaling pathway to facilitate pathogen elimination. In addition, lncITPRIP-1 promotes the association between MDA5 and HCV RNA to suppress virus replication.
FIG 10.
Working model of lncITPRIP-1 in the regulation of the innate immune response to HCV infection. LncITPRIP-1 and MDA5 are induced by viral infection. LncITPRIP-1 binds to MDA5 and facilitates the oligomerization of MDA5, provoking downstream MAVS to promote signal transduction for IFN production, which gradually inhibits viral infection. In addition, lncITPRIP-1 enhances the association between MDA5 and viral RNA to accelerate pathogen elimination.
The binding of unanchored lysine-63 (K63) polyubiquitin chains is essential for the activation of RIG-I, in which TRIM25 plays a crucial part (34, 36). However, whether ubiquitination is vital for MDA5 activation remains unclear. A study has proposed that noncovalent ubiquitin chain binding is necessary for MDA5 activation (34), while another study reported that 2CARD of MDA5 oligomerization could be an ubiquitin-independent process (8). Our study demonstrates that lncITPRIP-1 does not have an effect on the K63-linked polyubiquitination of MDA5. The binding of lncITPRIP-1 may facilitate the conformation change of MDA5 for activating downstream MAVS, or lncITPRIP-1 itself provides a danger signal similar to that provided by specific viral RNA. Further experiments are needed to uncover the explicit mechanisms by which lncITPRIP-1 regulates MDA5 oligomerization.
Although MDA5 is crucial for the induction of the antiviral innate immune response, uncontrolled expression and the excessive activation of MDA5 contribute to the initiation of autoinflammatory and autoimmune diseases (33, 46). Besides, Yao et al. reported that MDA5 functions as an effector protein by displacing viral proteins bound to RNA during ATP hydrolysis independently of the IFN pathway (39). In MDA5-dependent IFN signaling regulation, PP1 and TRIM65 play essential roles. PP1α and PP1β are primary phosphatases responsible for MDA5 dephosphorylation, which leads to its subsequent activation (47), while TRIM65 promotes the activation and oligomerization of MDA5 through K63-linked polyubiquitination (36).
As a whole, our study identifies the first lncRNA, ITPRIP-1, involved in the antiviral activity of MDA5 against HCV infection in both an IFN-dependent and an IFN-independent manner. In IFN signaling regulation, we demonstrate that lncITPRIP-1 enhances HCV-triggered IFN production by targeting MDA5 but not RIG-I. We also confirm that lncITPRIP-1 is a noncoding RNA induced by IFN and virus infection. Although the C terminus of MDA5 is critical for the association between MDA5 and lncITPRIP-1, lncITPRIP-1 promotes the oligomerization of 2CARD of MDA5 independently of a direct association. Importantly, lncITPRIP-1 and MDA5 expression increased coordinately, suggesting that lncITPRIP-1 may function as a cofactor for MDA5 and may have potential relevance to clinical diseases involving MDA5 dysfunction. At the same time, we reveal the significant role of lncITPRIP-1 in the IFN-independent function of MDA5 against HCV infection. Our work validates the essential regulatory effects of MDA5 in HCV elimination and demonstrates that lncITPRIP-1 is involved in MDA5 HCV recognition. Hence, it is a meaningful job to determine the roles of lncRNAs in innate immunity, especially when most biological processes linked to “genome noise” remain ambiguous.
MATERIALS AND METHODS
Cells.
HLCZ01 cells were established in our lab at Hunan University (27). Huh7 cells were purchased from the American Type Culture Collection. Huh7.5 cells and FL-neo cells (an Huh7-based HCV 1b full-length replicon cell line) were kindly provided by Charles M. Rice (Rockefeller University, New York, NY). The Huh7.5.1-MAVS wild-type (WT) cell line, the Huh7-MAVSR control cell line, and the Huh7-MAVSR-sgMDA5 ko cell line were gifts from Jin Zhong (Institute Pasteur of Shanghai, China). HEK293T cells were purchased from Boster. HLCZ01 cells were cultured in collagen-coated tissue culture plates and cultured with Dulbecco's modified Eagle medium (DMEM)–Ham's F-12 medium supplemented with 10% (vol/vol) fetal bovine serum (FBS; Gibco), 40 ng/ml of dexamethasone (Sigma), insulin-transferrin-selenium (ITS; Lonza), penicillin, and streptomycin. Huh7, Huh7.5, Huh7.5.1-MAVS, FL-neo, and HEK293T cells were propagated in DMEM supplemented with 10% FBS, l-glutamine, nonessential amino acids, penicillin, and streptomycin. All cell lines were maintained at 37°C in a humidified 5% CO2 incubator.
Viruses.
The pJFH1 and pJFH1/GND plasmids were gifts from Takaji Wakita (National Institute of Infectious Diseases, Tokyo, Japan). The linearized DNAs from the pJFH1 and pJFH1/GND plasmids were purified and used as the templates for in vitro transcription by use of a MEGAscript kit (Ambion, Austin, TX). In vitro-transcribed genomic JFH1 or JFH1/GND RNA was delivered into Huh7.5 cells by electroporation. The transfected cells were transferred to complete DMEM and cultured for the periods of time indicated above. The medium for Huh7.5 cells was replaced every 3 to 4 days, and the corresponding supernatants were collected and filtered through a 0.45-μm-pore-size filter device. The viral titers are presented as the number of focus-forming units (FFU) per milliliter, determined from the average number of NS5A-positive foci detected in Huh7.5 cells. SeV, VSV, and HSV were kindly provided by Xuetao Cao (Second Military Medical University, Shanghai, China).
Antibodies.
The following antibodies were used in our experiments. Monoclonal antibodies anti-β-actin, anti-Flag tag, and antihemagglutinin (anti-HA) tag were purchased from Sigma. Monoclonal antibody anti-V5 tag was obtained from Invitrogen. Monoclonal antibodies anti-MDA5, anti-STAT1, anti-phosphorylated STAT1 (anti-pSTAT1), anti-IRF3, and anti-phosphorylated IRF3 (anti-pIRF3) were from Cell Signaling Technology (CST). Monoclonal antibody anti-MAVS was purchased from Santa Cruz Biotechnology. Goat anti-mouse IgG-horseradish peroxidase secondary antibody was from Millipore. Mouse monoclonal anti-NS5A and anti-HCV core antibodies were kindly provided by Chen Liu (Rutgers University, NJ). Monoclonal antibody anti-IFNAR1 was purchased from Bioss.
Plasmids.
The IFN-β-luciferase and pRL-CMV plasmids were purchased from InvivoGen. Flag-MDA5 plasmids were kindly provided by Jianguo Wu (Wuhan University, Wuhan, China). MAVS was synthesized from total cellular RNA isolated from HLCZ01 cells by standard RT-PCR and was subsequently cloned into the p3×FLAG-CMV vector. The Flag-TBK1 and pHA-Ub (K63) plasmids were kindly provided by Zhengfan Jiang (Peking University, Beijing, China). The Flag-IRF3-5D plasmid was a gift from Deyin Guo (Wuhan University, Wuhan, China). LncITPRIP-1 was synthesized from total cellular RNA isolated from HLCZ01 cells by standard RT-PCR and was subsequently cloned into the pcDNA3.1a vector and the pRed-max-N1 vector. LncBST2-2 was synthesized from total RNA isolated from HLCZ01 cells and cloned into the pcDNA3.1a vector. Multiple truncation domains of MDA5 were amplified from their full-length templates and were cloned into the p3×FLAG-CMV vector. shRNAs targeting lncTIPRIP-1 were cloned into the pSilencer-neo plasmid (Ambion). Primers for the constructed plasmids are listed in Table 1.
TABLE 1.
Primers for constructed plasmids
| Construct | Primer sequence (orientation) |
|---|---|
| pcDNA3.1a-LncITPRIP-1 | GGGGTACCGGGCAGATGGAGACACCCA (forward) |
| CGGGATCCTGGGTTGTTGGAAAATTTGCTCAC (reverse) | |
| pcDNA3.1a-LncITPRIP-1+T | GGGGTACCGGGCAGATGGAGACACCCA (forward) |
| CGGGATCCATGGGTTGTTGGAAAATTTGCTCAC (reverse) | |
| pcDNA3.1a-LncITPRIP-1+TT | GGGGTACCGGGCAGATGGAGACACCCA (forward) |
| CGGGATCCAATGGGTTGTTGGAAAATTTGCTCAC (reverse) | |
| pcDNA3.1a-LncBST2-2 | GGGGTACCGCTGCTAGTCTAGAAAAAAGCTCGG (forward) |
| CGGGATCCAACCTGATTGCCTTTTCCG (reverse) | |
| pRed-Max-N1-LncITPRIP-1 | GGGGTACCGGGCAGATGGAGACACCCA (forward) |
| CGGGATCCTGGGTTGTTGGAAAATTTGCTCAC (reverse) | |
| pRed-Max-N1-LncITPRIP-1+T | GGGGTACCGGGCAGATGGAGACACCCA (forward) |
| CGGGATCCATGGGTTGTTGGAAAATTTGCTCAC (reverse) | |
| pRed-Max-N1-LncITPRIP-1+TT | GGGGTACCGGGCAGATGGAGACACCCA (forward) |
| CGGGATCCAATGGGTTGTTGGAAAATTTGCTCAC (reverse) | |
| pRed-Max-N1-IRF3 | GGGGTACCATGGGAACCCCAAAGCCACG (forward) |
| GCTCTAGAGCTCTCCCCAGGGCCCTGGA (reverse) | |
| LncBST2-2 | GGGGTACCCCAGGATGGTCTTGATCTCCTGACC (forward) |
| CGGGATCCCCAGCCTGGGTGACAAGGG (reverse) | |
| pSilencer-neo-H1-LncITPRIP-1 | GATCCCCACAGACGCTGTCTTCAATTCAAGAGATTGAAGACAGCGTCTGTGGTTTTTTGGAAA (forward) |
| AGCTTTTCCAAAAAACCACAGACGCTGTCTTCAATCTCTTGAATTGAAGACAGCGTCTGTGGG (reverse) | |
| p3×Flag-MDA5(2CARD) | GGGGTACCATGTCGAATGGGTATTCCAC (forward) |
| GCTCTAGATGAGCAATCAGAGCCTGTTA (reverse) | |
| p3×Flag-MDA5(ΔN) | GGGGTACCATGGAAAGCAATGCAGAGAT (forward) |
| CGGGATCCATCCTCATCACTAAATAAAC (reverse) | |
| p3×Flag-MDA5(ΔC) | GGGGTACCATGTCGAATGGGTATTCCAC (forward) |
| CGGGATCCAGTTCCAAAATCTGACATTG (reverse) |
Quantitative PCR.
Total cellular RNA was extracted in the TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. The obtained RNA was reverse transcribed by use of a PrimeScript RT reagent kit (gDNAEraser; Perfect Real Time; TaKaRa). A SYBR RT-PCR kit (Roche) was used for real-time qRT-PCR analysis. Gene expression was normalized to that of the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene. Semi-RT-PCR was run with a quantitative PCR-RT kit (Toyobo). The primers used for semi-RT-PCR are listed in Table 2.
TABLE 2.
Primers for semi-RT-PCR
| Construct | Primer sequence (orientation) |
|---|---|
| LncITPRIP-1 | GGGGTACCGGGCAGATGGAGACACCCA (forward) |
| CGGGATCCTGGGTTGTTGGAAAATTTGCTCAC (reverse) | |
| Lnc1# | GGGGTACCCAGTCCGTGGCACCGCAAT (forward) |
| CGGGATCCTAAATAGTACGTGAATTCTTTCGGG (reverse) | |
| Lnc2# | GGGGTACCCCCCTCCCCGCCCTGCCCC (forward) |
| CGGGATCCCATCTGTGGCGTGGTGGCTG (reverse) | |
| Lnc3# | GGGGTACCGACGGCTGCAGTAGCGTGG (forward) |
| CGGGATCCGAACACCTCTTTACAAGGAAATGAG (reverse) | |
| LncBST2-2 | GGGGTACCCCAGGATGGTCTTGATCTCCTGACC (forward) |
| CGGGATCCCCAGCCTGGGTGACAAGGG (reverse) | |
| Lnc5# | GGGGTACCGCTGCTAGTCTAGAAAAAAGCTCGG (forward) |
| CGGGATCCAACCTGATTGCCTTTTCCG (reverse) |
Western blotting.
Cells were washed with ice-cold phosphate-buffered saline (PBS) and lysed in radioimmunoprecipitation assay (RIPA) lysis and extraction buffer (Thermo Fisher) supplemented with a protease inhibitor cocktail. Forty micrograms of protein was resolved by SDS-PAGE, transferred to a polyvinylidene difluoride (PVDF) membrane at a constant voltage of 100 V for 2 h, and probed with the appropriate primary and secondary antibodies. The bound antibodies were detected by use of the SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, IL).
Immunofluorescence staining.
Huh7.5 cells were seeded on glass coverslips and infected with HCV for 6 h before transfection of the indicated plasmids. After 48 h of transfection, Huh7.5 cells were fixed with ice-cold acetone for 8 min at −20°C. Cells were blocked with goat serum (diluted in PBS to 1:50) for 30 min and then incubated with mouse monoclonal anti-NS5A (diluted in PBS to 1:100) for 1 h. Cells were then washed three times with PBS and stained with fluorescence-labeled secondary antibodies (diluted in PBS to 1:200; Invitrogen) for 50 min. Finally, the coverslips were washed with PBS (three times, 5 min each time) and counterstained with DAPI (4′,6-diamidino-2-phenylindole; Vector Laboratories, Inc.). Fluorescent images were obtained with a fluorescence microscope (Olympus).
Luciferase reporter assay.
Luciferase reporter assays were performed with a luciferase assay kit (Promega) according to the manufacturer's instructions. HEK293T cells were plated in 12-well plates and transfected with plasmids carrying the IFN-β luciferase reporter (200 ng) and pRL-CMV (10 ng) together with 300 ng Flag-N-RIG-I, Flag-MDA5, Flag-MAVS, or Flag-TBK1 and 500 ng the lncITPRIP-1 expression plasmids. An empty pcDNA3.1a vector was used to maintain equal amounts of DNA among the wells. Cells were collected at 24 h after transfection, and luciferase activity was measured according to the manufacturer's protocol.
Immunoprecipitation and RNA immunoprecipitation.
Cells were washed three times with ice-cold PBS and lysed in lysis buffer supplemented with protease inhibitor cocktail. The cell lysates were incubated at 4°C for 30 min and centrifuged at 12,000 × g at 4°C for 15 min. The lysates were diluted to a concentration of 2 μg/μl total cell protein with PBS before immunoprecipitation (IP). Two hundred micrograms of lysates was immunoprecipitated with the indicated antibodies. The immunocomplex was captured by adding a protein G agarose bead slurry. The protein binding to the beads was boiled in 2× Laemmli sample buffer and then subjected to 10% SDS-PAGE. The protocol for immunoblotting was described above.
For RNA immunoprecipitation, the amounts of MDA5-, RIG-I-, MAVS-, TBK1-, IRF3-, and truncations of MDA5-bound lncITPRIP-1 or lncBST2-2 RNA in the immunoprecipitates obtained from formaldehyde-cross-linked cells were quantified relative to the amount of IgG. The cells were trypsinized to detach them, the trypsin was inactivated, the cells were pelleted, and the supernatant was discarded. The cells were washed by gently resuspending them in 1 ml PBS and pelleted by centrifugation at 3,000 × g for 1 min. Then, the cells were resuspended in 1 ml 1% formaldehyde (diluted in PBS) and allowed to stand for 10 min at room temperature. The cells were pelleted by centrifugation at 3,000 × g for 1 min, resuspended in 1 ml 0.25 M glycine solution (diluted in PBS), and again allowed to stand for 10 min at room temperature. The cells were pelleted and washed with 500 μl PBS, and then the cells were lysed with RIPA buffer supplemented with a protease inhibitor cocktail and an RNase inhibitor (Roche) on ice for 30 min. The subsequent immunoprecipitate was processed as described above. Protein-RNA complexes binding to beads were eluted in PBS at 70°C for 45 min. The eluted material was lysed in ice-cold TRIzol reagent for RT-PCR.
Native PAGE for IRF3 dimerization assay and MDA5 oligomerization assay.
HEK293T and HLCZ01 cells were washed three times with ice-cold PBS and lysed in native lysis buffer (25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100) supplemented with a protease inhibitor cocktail. The cell lysates were incubated on ice for 30 min and centrifuged at 12,000 × g at 4°C for 15 min. The protein was dissolved in 4× native sample buffer (0.25 M Tris-HCl, pH 6.8, 40% glycerol, 4% sodium deoxycholate, 0.005% bromophenol blue) and then subjected to 10% native PAGE (without SDS). After electrophoresis in the running buffer (3 g Tris base, 14.4 g glycine), to which double-distilled H2O was added to 1 liter, the protein was stored at 4°C. Immediately before use, 1% deoxycholate was added to the upper chamber buffer, and electrophoresis was performed for 60 min with a constant voltage of 25 mA at 4°C (the gel was prerun at 40 mA for 30 min). Western immunoblotting was then performed. For the MDA5 and RIG-I oligomerization assay, we made a modification to a 5% gel. For the 2CARD of MDA5 oligomerization assay, we made a modification to a 10% gel.
SDD-AGE.
HEK293T cells were washed three times with ice-cold PBS and lysed in native lysis buffer (25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100) supplemented with a protease inhibitor cocktail. The cell lysates were incubated at 4°C for 30 min and centrifuged at 12,000 × g at 4°C for 15 min. The protein was resuspended in 1× sample buffer (0.5× Tris-borate-EDTA [TBE], 10% glycerol, 2% SDS, 0.0025% bromophenol blue) and loaded onto a vertical 1.5% agarose gel. After electrophoresis in the running buffer (1× TBE, 0.1% SDS) for 60 min with a constant voltage of 80 V at 4°C, Western immunoblotting was performed.
Cytoplasmic and nuclear RNA extraction.
Cytoplasmic and nuclear RNAs were extracted from HLCZ01 cells by use of a Paris kit (Thermo Fisher) according to the manufacturer's instructions.
Statistical analysis.
All results are presented as means and standard deviations. Comparisons between two groups were performed using Student's t test, and P values are indicated in the appropriate figures.
ACKNOWLEDGMENTS
We thank Charles M. Rice for the FL-neo and Huh7.5 cell lines, Jin Zhong for Huh7.5.1-MAVS WT, Huh7-MAVSR, and Huh7-MAVSR-sgMDA5 ko cells, Takaji Wakita for pJFH1 and pJFH1/GND, and Xuetao Cao for SeV, VSV, and HSV. We also appreciate Jianguo Wu, Zhengfan Jiang, and Deyin Guo for kindly sharing research materials.
This work was supported by the National Natural Science Foundation of China (81730064, 81571985, and 31571368), the National Science and Technology Major Project (2017ZX10202201), and the Hunan Natural Science Foundation of Youth Fund (2018JJ3713).
Q.X., S.C., R.T., X.H., B.X., Y.Q., Y.X., J.W., M.G., and J.C. performed the experiments. S.T., Q.X., and S.C. analyzed the data. Q.X., S.C., R.D., and G.L. performed the computational prediction. Q.X. and S.C. wrote the manuscript. Q.X., S.C., and H.Z. designed experiments and revised the manuscript. All authors contributed to this study.
We have no competing financial interests in relation to the work described.
Hunan Provincial Tumor Hospital is an Affiliated Tumor Hospital of the Xiangya Medical School of Central South University.
REFERENCES
- 1.Aune TM, Spurlock CF III. 2016. Long non-coding RNAs in innate and adaptive immunity. Virus Res 212:146–160. doi: 10.1016/j.virusres.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmitt AM, Chang HY. 2016. Long noncoding RNAs in cancer pathways. Cancer Cell 29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rashid F, Shah A, Shan G. 2016. Long non-coding RNAs in the cytoplasm. Genomics Proteomics Bioinformatics 14:73–80. doi: 10.1016/j.gpb.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu B, Shan G. 2016. Functions of long noncoding RNAs in the nucleus. Nucleus 7:155–166. doi: 10.1080/19491034.2016.1179408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawai T, Akira S. 2011. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 6.Komuro A, Bamming D, Horvath CM. 2008. Negative regulation of cytoplasmic RNA-mediated antiviral signaling. Cytokine 43:350–358. doi: 10.1016/j.cyto.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loo YM, Gale M Jr. 2011. Immune signaling by RIG-I-like receptors. Immunity 34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu B, Peisley A, Richards C, Yao H, Zeng X, Lin C, Chu F, Walz T, Hur S. 2013. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell 152:276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- 9.Quicke KM, Diamond MS, Suthar MS. 2017. Negative regulators of the RIG-I-like receptor signaling pathway. Eur J Immunol 47:615–628. doi: 10.1002/eji.201646484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma G, Raheja H, Das S. 2018. Hepatitis C virus: enslavement of host factors. IUBMB Life 70:41–49. doi: 10.1002/iub.1702. [DOI] [PubMed] [Google Scholar]
- 11.Sofia MJ. 2016. Enter sofosbuvir: the path to curing HCV. Cell 167:25–29. doi: 10.1016/j.cell.2016.08.044. [DOI] [PubMed] [Google Scholar]
- 12.Klenerman P, Fitzmaurice K. 2015. An update on hepatitis C virus. Clin Med (Lond) 15(Suppl 6):s33–s36. doi: 10.7861/clinmedicine.15-6-s33. [DOI] [PubMed] [Google Scholar]
- 13.Anonymous. 2016. Bringing the hepatitis C virus to life. Cell 167:39–42. doi: 10.1016/j.cell.2016.08.046. [DOI] [PubMed] [Google Scholar]
- 14.Park SH, Rehermann B. 2014. Immune responses to HCV and other hepatitis viruses. Immunity 40:13–24. doi: 10.1016/j.immuni.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Sastre A. 2017. Ten strategies of interferon evasion by viruses. Cell Host Microbe 22:176–184. doi: 10.1016/j.chom.2017.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fortes P, Morris KV. 2016. Long noncoding RNAs in viral infections. Virus Res 212:1–11. doi: 10.1016/j.virusres.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kambara H, Niazi F, Kostadinova L, Moonka DK, Siegel CT, Post AB, Carnero E, Barriocanal M, Fortes P, Anthony DD, Valadkhan S. 2014. Negative regulation of the interferon response by an interferon-induced long non-coding RNA. Nucleic Acids Res 42:10668–10680. doi: 10.1093/nar/gku713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kambara H, Gunawardane L, Zebrowski E, Kostadinova L, Jobava R, Krokowski D, Hatzoglou M, Anthony DD, Valadkhan S. 2015. Regulation of interferon-stimulated gene BST2 by a lncRNA transcribed from a shared bidirectional promoter. Front Immunol 5:676. doi: 10.3389/fimmu.2014.00676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lv M, Zhang B, Shi Y, Han Z, Zhang Y, Zhou Y, Zhang W, Niu J, Yu XF. 2015. Identification of BST-2/tetherin-induced hepatitis B virus restriction and hepatocyte-specific BST-2 inactivation. Sci Rep 5:11736. doi: 10.1038/srep11736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dafa-Berger A, Kuzmina A, Fassler M, Yitzhak-Asraf H, Shemer-Avni Y, Taube R. 2012. Modulation of hepatitis C virus release by the interferon-induced protein BST-2/tetherin. Virology 428:98–111. doi: 10.1016/j.virol.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 21.Shin EC, Sung PS, Park SH. 2016. Immune responses and immunopathology in acute and chronic viral hepatitis. Nat Rev Immunol 16:509–523. doi: 10.1038/nri.2016.69. [DOI] [PubMed] [Google Scholar]
- 22.Cao X, Ding Q, Lu J, Tao W, Huang B, Zhao Y, Niu J, Liu YJ, Zhong J. 2015. MDA5 plays a critical role in interferon response during hepatitis C virus infection. J Hepatol 62:771–778. doi: 10.1016/j.jhep.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 23.Chow KT, Gale M Jr. 2015. SnapShot: interferon signaling. Cell 163:1808–1808.e1. doi: 10.1016/j.cell.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 24.Ouyang J, Zhu X, Chen Y, Wei H, Chen Q, Chi X, Qi B, Zhang L, Zhao Y, Gao GF, Wang G, Chen JL. 2014. NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon-stimulated gene transcription. Cell Host Microbe 16:616–626. doi: 10.1016/j.chom.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong CM, Tsang FH, Ng IO. 2018. Non-coding RNAs in hepatocellular carcinoma: molecular functions and pathological implications. Nat Rev Gastroenterol Hepatol 15:137–151. doi: 10.1038/nrgastro.2017.169. [DOI] [PubMed] [Google Scholar]
- 26.Ma Z, Damania B. 2016. The cGAS-STING defense pathway and its counteraction by viruses. Cell Host Microbe 19:150–158. doi: 10.1016/j.chom.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang D, Zuo C, Wang X, Meng X, Xue B, Liu N, Yu R, Qin Y, Gao Y, Wang Q, Hu J, Wang L, Zhou Z, Liu B, Tan D, Guan Y, Zhu H. 2014. Complete replication of hepatitis B virus and hepatitis C virus in a newly developed hepatoma cell line. Proc Natl Acad Sci U S A 111:E1264–E1273. doi: 10.1073/pnas.1320071111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumthip K, Yang D, Li NL, Zhang Y, Fan M, Sethuraman A, Li K. 2017. Pivotal role for the ESCRT-II complex subunit EAP30/SNF8 in IRF3-dependent innate antiviral defense. PLoS Pathog 13:e1006713. doi: 10.1371/journal.ppat.1006713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, Du F, Ren J, Wu YT, Grishin NV, Chen ZJ. 2015. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347:aaa2630. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- 30.Matsumoto A, Pasut A, Matsumoto M, Yamashita R, Fung J, Monteleone E, Saghatelian A, Nakayama KI, Clohessy JG, Pandolfi PP. 2017. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature 541:228–232. doi: 10.1038/nature21034. [DOI] [PubMed] [Google Scholar]
- 31.Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, Jiang Z, Xu J, Liu Q, Cao X. 2014. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 344:310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 32.Esteban M. 2009. Hepatitis C and evasion of the interferon system: a PKR paradigm. Cell Host Microbe 6:495–497. doi: 10.1016/j.chom.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 33.Zhao C, Jia M, Song H, Yu Z, Wang W, Li Q, Zhang L, Zhao W, Cao X. 2017. The E3 ubiquitin ligase TRIM40 attenuates antiviral immune responses by targeting MDA5 and RIG-I. Cell Rep 21:1613–1623. doi: 10.1016/j.celrep.2017.10.020. [DOI] [PubMed] [Google Scholar]
- 34.Jiang X, Kinch LN, Brautigam CA, Chen X, Du F, Grishin NV, Chen ZJ. 2012. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity 36:959–973. doi: 10.1016/j.immuni.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lui PY, Wong LR, Ho TH, Au SWN, Chan CP, Kok KH, Jin DY. 2017. PACT facilitates RNA-induced activation of MDA5 by promoting MDA5 oligomerization. J Immunol 199:1846–1855. doi: 10.4049/jimmunol.1601493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lang X, Tang T, Jin T, Ding C, Zhou R, Jiang W. 2017. TRIM65-catalized ubiquitination is essential for MDA5-mediated antiviral innate immunity. J Exp Med 214:459–473. doi: 10.1084/jem.20160592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akutsu M, Dikic I, Bremm A. 2016. Ubiquitin chain diversity at a glance. J Cell Sci 129:875–880. doi: 10.1242/jcs.183954. [DOI] [PubMed] [Google Scholar]
- 38.Hu H, Sun SC. 2016. Ubiquitin signaling in immune responses. Cell Res 26:457–483. doi: 10.1038/cr.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yao H, Dittmann M, Peisley A, Hoffmann HH, Gilmore RH, Schmidt T, Schmidt-Burgk J, Hornung V, Rice CM, Hur S. 2015. ATP-dependent effector-like functions of RIG-I-like receptors. Mol Cell 58:541–548. doi: 10.1016/j.molcel.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu R, Yang D, Lei S, Wang X, Meng X, Xue B, Zhu H. 2015. HMGB1 promotes hepatitis C virus replication by interaction with stem-loop 4 in the viral 5′ untranslated region. J Virol 90:2332–2344. doi: 10.1128/JVI.02795-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Imam H, Bano AS, Patel P, Holla P, Jameel S. 2015. The lncRNA NRON modulates HIV-1 replication in a NFAT-dependent manner and is differentially regulated by early and late viral proteins. Sci Rep 5:8639. doi: 10.1038/srep08639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang JF, Guo YJ, Zhao CX, Yuan SX, Wang Y, Tang GN, Zhou WP, Sun SH. 2013. Hepatitis B virus X protein (HBx)-related long noncoding RNA (lncRNA) down-regulated expression by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by targeting the intermediate filament protein vimentin. Hepatology 57:1882–1892. doi: 10.1002/hep.26195. [DOI] [PubMed] [Google Scholar]
- 43.Ma H, Han P, Ye W, Chen H, Zheng X, Cheng L, Zhang L, Yu L, Wu X, Xu Z, Lei Y, Zhang F. 2017. The long noncoding RNA NEAT1 exerts antihantaviral effects by acting as positive feedback for RIG-I signaling. J Virol 91:e02250-16. doi: 10.1128/JVI.02250-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carnero E, Barriocanal M, Prior C, Unfried JP, Segura V, Guruceaga E, Enguita M, Smerdou C, Gastaminza P, Fortes P. 2016. Long noncoding RNA EGOT negatively affects the antiviral response and favors HCV replication. EMBO Rep 17:1013–1028. doi: 10.15252/embr.201541763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qian X, Xu C, Zhao P, Qi Z. 2016. Long non-coding RNA GAS5 inhibited hepatitis C virus replication by binding viral NS3 protein. Virology 492:155–165. doi: 10.1016/j.virol.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 46.Ablasser A, Hertrich C, Wassermann R, Hornung V. 2013. Nucleic acid driven sterile inflammation. Clin Immunol 147:207–215. doi: 10.1016/j.clim.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 47.Wies E, Wang MK, Maharaj NP, Chen K, Zhou S, Finberg RW, Gack MU. 2013. Dephosphorylation of the RNA sensors RIG-I and MDA5 by the phosphatase PP1 is essential for innate immune signaling. Immunity 38:437–449. doi: 10.1016/j.immuni.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]