Human herpesviruses pose a constant threat to human health. Reactivation of persisting herpesvirus infections, particularly in immunocompromised individuals and the elderly, can cause severe diseases, such as zoster, pneumonia, encephalitis, or cancer. The interferon system is relevant for the control of herpesvirus replication as exemplified by fatal disease outcomes in patients with primary immunodeficiencies. Here, we describe the interferon-induced, human MX2 gene product MxB as an efficient restriction factor of alpha-, beta-, and gammaherpesviruses. MxB has previously been described as an inhibitor of HIV-1. Importantly, our mutational analyses of MxB reveal an antiviral mechanism of herpesvirus restriction distinct from that against HIV-1. Thus, the dynamin-like MxB GTPase serves as a broadly acting intracellular restriction factor that controls retrovirus as well as herpesvirus infections.
KEYWORDS: HCMV, HSV-1, herpesviruses, MHV68, human MX2 gene, MxB protein, cytomegalovirus, herpes simplex virus, innate immunity, interferons
ABSTRACT
Herpesvirus infections are highly prevalent in the human population and persist for life. They are often acquired subclinically but potentially progress to life-threatening diseases in immunocompromised individuals. The interferon system is indispensable for the control of herpesviral replication. However, the responsible antiviral effector mechanisms are not well characterized. The type I interferon-induced, human myxovirus resistance 2 (MX2) gene product MxB, a dynamin-like large GTPase, has recently been identified as a potent inhibitor of HIV-1. We now show that MxB also interferes with an early step of herpesvirus replication, affecting alpha-, beta-, and gammaherpesviruses before or at the time of immediate early gene expression. Defined MxB mutants influencing GTP binding and hydrolysis revealed that the effector mechanism against herpesviruses is thoroughly different from that against HIV-1. Overall, our findings demonstrate that MxB serves as a broadly acting intracellular restriction factor that controls the establishment of not only retrovirus but also herpesvirus infection of all three subfamilies.
IMPORTANCE Human herpesviruses pose a constant threat to human health. Reactivation of persisting herpesvirus infections, particularly in immunocompromised individuals and the elderly, can cause severe diseases, such as zoster, pneumonia, encephalitis, or cancer. The interferon system is relevant for the control of herpesvirus replication as exemplified by fatal disease outcomes in patients with primary immunodeficiencies. Here, we describe the interferon-induced, human MX2 gene product MxB as an efficient restriction factor of alpha-, beta-, and gammaherpesviruses. MxB has previously been described as an inhibitor of HIV-1. Importantly, our mutational analyses of MxB reveal an antiviral mechanism of herpesvirus restriction distinct from that against HIV-1. Thus, the dynamin-like MxB GTPase serves as a broadly acting intracellular restriction factor that controls retrovirus as well as herpesvirus infections.
INTRODUCTION
Herpesviruses are highly adapted to their natural hosts, establishing mostly asymptomatic infections that develop into lifelong latency. Moreover, latent genomes can readily reactivate upon exposure to external stimuli or under conditions of waning immunity or immunosuppression, leading to severe disease outcomes such as herpes simplex virus (HSV) encephalitis, cytomegalovirus (CMV) pneumonia, or tumor induction by gammaherpesviruses (1 to 3). Herpesvirus replication is controlled by the interferon (IFN)-induced host defense (4, 5), and defects in the innate immune system can cause severe diseases (3, 6, 7). Recently, Liu et al. identified human MxB as a potential inhibitor of murine gammaherpesvirus (MHV68) in a broad screen for antiviral active IFN-stimulated genes (ISGs) (8).
Myxovirus resistance (Mx) proteins are key players in the innate immune response to viral infections (9). Humans express two paralogous Mx genes, MX1 and MX2, encoding the MxA and MxB proteins, under strict IFN type I and type III control (9, 10). These dynamin-like GTPases contain a G domain responsible for GTP hydrolysis and a stalk region involved in oligomerization (11, 12). MxA has long been known for its broad antiviral activity against many RNA viruses, including influenza A viruses (IAV) and vesicular stomatitis virus (VSV) (9). However, the function of the second human Mx protein, MxB, was not identified for the longest period. MxB has only recently been described to block the replication of MHV68 (8) as well as of human immunodeficiency virus 1 (HIV-1) (13, 14). MxB is expressed in IFN-treated cells in two isoforms, full-length MxB (positions 1 to 715) [MxB(1-715)] and MxB(26-715), the latter of which is lacking the first 25 N-terminal amino acids that function as a nuclear localization signal (NLS) (15). Interestingly, the extended N-terminal region of the full-length MxB(1-715) is crucial for the anti-HIV-1 activity, rendering the truncated MxB(26-715) inactive against retroviruses (13, 14).
A recent analysis of MX2 evolution in primates suggested a broader antiviral specificity of MxB extending its antiretroviral activity (16). In the present study, we therefore analyzed the activity of MxB against members of the three herpesvirus subfamilies and directly compared the effects with those seen against HIV-1. Activities against HIV and activities against herpesviruses both depend on the N-terminal 25 amino acids of MxB(1-715). However, analysis of MxB mutants in the N-terminal extension and the G domain revealed virus-specific mechanistic differences in the control of herpesviruses that were clearly distinct from that against HIV-1. Thus, our study indicates that MxB serves as a broadly acting intracellular restriction factor controlling retrovirus as well as herpesvirus infections.
RESULTS
MxB inhibits herpesvirus replication.
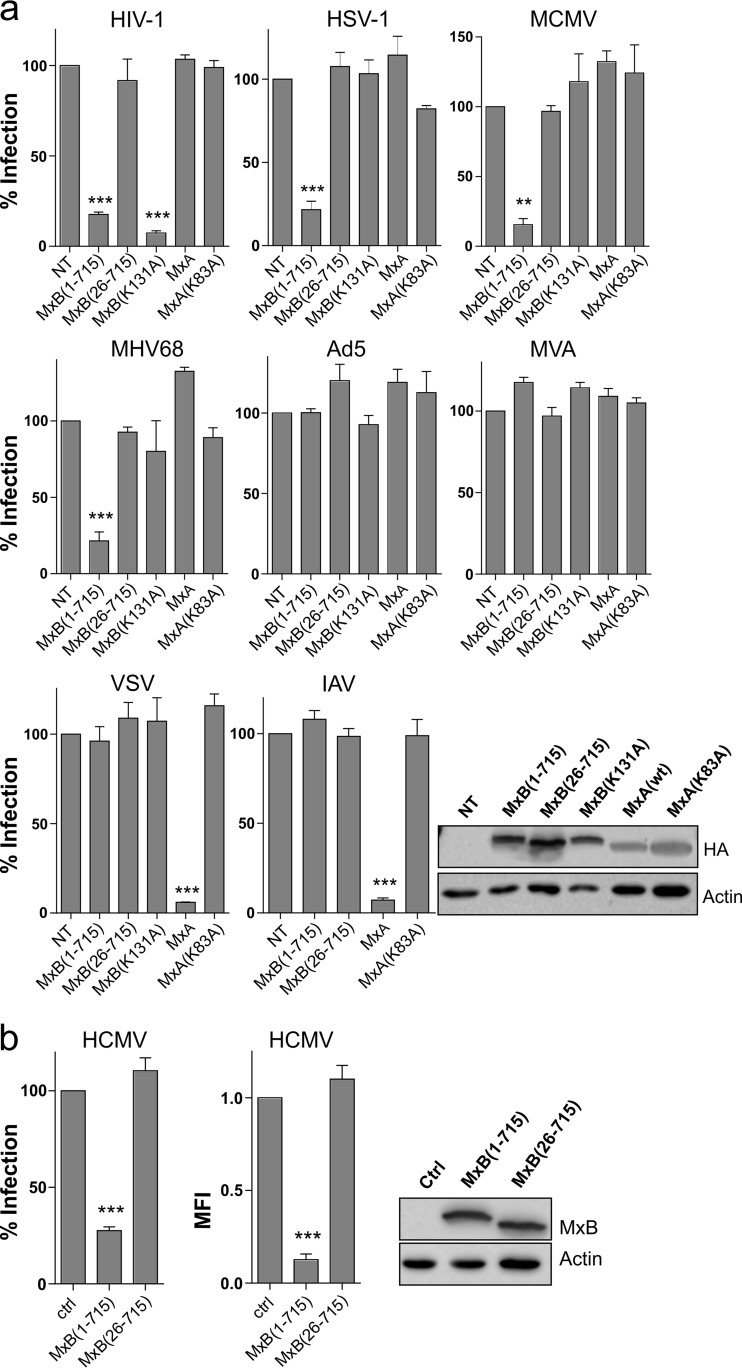
To investigate the antiviral spectrum of human MxB against different herpesviruses, human glioblastoma U87MG cells stably expressing the full-length MxB(1-715) or the MxB(26-715) isoform or a full-length MxB mutant, MxB(K131A), devoid of GTPase activity (17), were generated. Cell lines expressing wild-type MxA or GTPase-defective MxA(K83A) (18) served as controls. We used green fluorescent protein (GFP)-expressing viruses for infection of the different cell lines that allowed quantitative measurement of viral gene expression in single cells via flow cytometry. As a control, we used a vesicular stomatitis virus (VSV) G protein (VSV-G) pseudotyped HIV-1 lentiviral vector. As expected, MxB(1-715) and GTPase-defective MxB(K131A) but not the MxB(26-715) isoform or MxA blocked GFP expression by the HIV-1 lentiviral vector (Fig. 1a). To test activity against herpesviruses, the cell lines were infected with HSV-1, murine cytomegalovirus (MCMV), and MHV68, representing alpha-, beta-, and gammaherpesviruses, respectively. Strikingly, in all cases MxB(1-715) reduced the percentage of infected GFP-positive (GFP+) cells by about 75% at 20 h postinfection (hpi) (Fig. 1a). However, in cells expressing the MxB(26-715) isoform or the GTPase mutant MxB(K131A), rates of infection were comparable to the rates seen with the control. To analyze the specificity of this antiviral effect, we infected the cells with two other DNA viruses, recombinant adenovirus-5 (Ad5) and modified vaccinia virus strain Ankara (MVA), which were both unaffected by MxA or MxB expression (Fig. 1a). Furthermore, two RNA viruses, VSV and IAV, were strongly suppressed by MxA, but not by MxB(1-715), as expected (19). The effect of MxB on human CMV (HCMV), a human betaherpesvirus, was tested in human lung epithelial A549 cells stably expressing the MxB isoforms. Again, MxB(1-715) but not MxB(26-715) restricted infectivity of HCMV strain TB40/E (Fig. 1b).
FIG 1.
Human MxB is a herpesvirus restriction factor. (a) U87MG cells expressing HA-tagged Mx proteins were infected (MOI of 0.5) with GFP-encoding HIV-1 VSV-G pseudotyped lentiviral vector, HSV-1, MCMV, MHV68, Ad5, MVA, VSV, or IAV (H7N7) and subjected to fluorescence-activated cell sorter (FACS) analysis 20 hpi or 6 hpi (only VSV) or 48 hpi (only HIV-1). The percentage of GFP+ cells relative to nontransduced control cells (NT) is shown. For Western blot analysis, Mx was detected by anti-HA antibody. Actin served as a control. (b) A549 cells expressing untagged Mx proteins were infected (MOI of 50) with GFP-encoding HCMV. Data represent percentages of GFP+ cells relative to cells transduced with empty vector (ctrl) and the relative values of the mean fluorescence intensities (MFI). For Western blot analysis, MxB was detected by anti-MxB antibody. Actin served as a control. Error bars represent the SEM of results from three independent experiments. Statistical analysis was performed via one-way ANOVA with a post hoc Tukey's test. ***, <0.001; **, < 0.01; *, < 0.1.
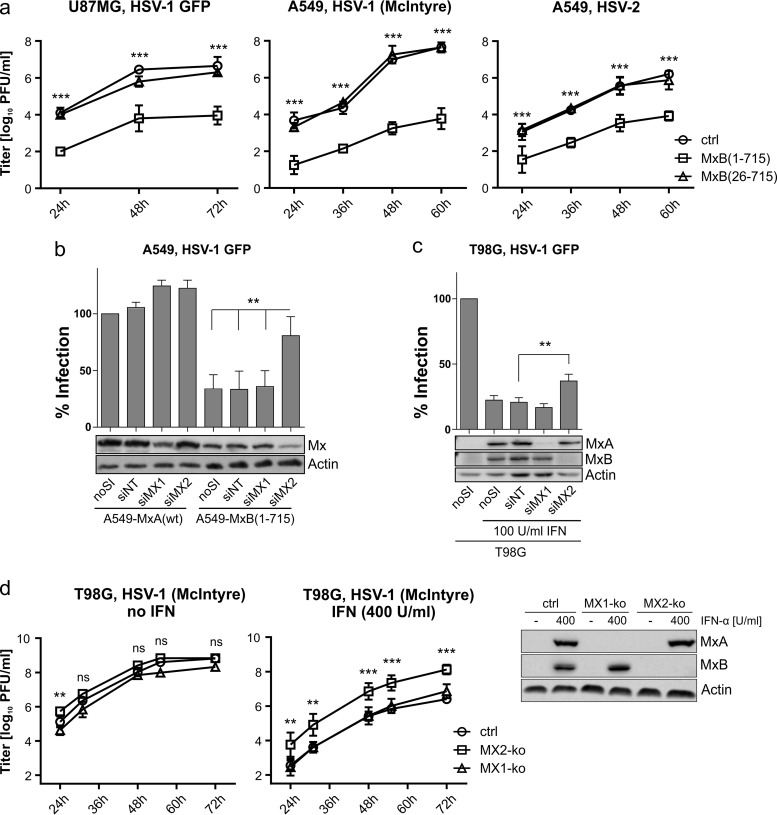
To analyze virus propagation, we infected the U87MG- and A549-based cell lines with HSV-1 and HSV-2. Up to 72 hpi, MxB(1-715) efficiently reduced viral titers compared to control and MxB(26-715)-expressing cells (Fig. 2a). Whereas overexpression of MxB(1-715) reduced the percentage of GFP+ cells upon infection (Fig. 1), RNA interference (RNAi)-mediated knockdown of MX2 but not MX1 increased the susceptibility of the MxB(1-715)-expressing A549 cells to HSV-1 (Fig. 2b), confirming the specificity of our findings. To evaluate the physiological relevance of the antiviral MxB effect for the type I IFN-mediated host response, endogenous MX2 was knocked down by the use of RNAi in human glioblastoma T98G cells, which have previously been shown to express both Mx proteins upon alpha interferon (IFN-α) pretreatment (20). HSV-1 infection was reduced 5-fold by the IFN treatment. However, HSV-1 infectivity could be partially restored (2-fold increase) by treating the cells with the MX2-specific small interfering RNA (siRNA) but not by treating the cells with the MX1-targeting siRNA or a nontargeting control siRNA (Fig. 2c), confirming that MxB represented one of the factors restricting herpesvirus replication in the context of the IFN response. Furthermore, virus propagation on IFN-α-treated T98G cells was enhanced about 100-fold upon MX2-specific gene knockout in these cells compared to the results seen with MX1-specific knockout or control cells (Fig. 2d).
FIG 2.
Herpesvirus infectivity is restricted at multiple time points and can partially be rescued by MxB depletion. (a) Growth kinetics (MOI of 0.0001) of HSV-1 GFP (17+) on U87MG cells and HSV-1 (McIntyre) and HSV-2 (MS) on A549 cells. The titers determined by plaque assay are shown as arithmetic means ± standard deviations (SD) of results from three independent experiments. Statistical analysis was performed on log values of the titers via two-way ANOVA with a post hoc Tukey's test. P values were determined by comparing the mean titers of control (ctrl) and MxB(1-715) cells at the depicted time points. (b) MxB- or MxA-expressing A549 cells were treated with siRNA as indicated and subsequently infected with HSV-1 GFP (MOI of 0.5) for 20 h. Infectivity was analyzed as described for Fig. 1. Error bars represent the SEM of results from three independent experiments. Statistical analysis was performed via one-way ANOVA with a post hoc Tukey's test. (c) Nontreated or IFN-α-pretreated T98G cells (100 U/ml for 16 h) were treated and analyzed as described for panel b. (d) T98G cells with specific MX1 and MX2 gene knockouts were left untreated or pretreated with IFN-α (400 U/ml for 16 h) and then infected with HSV-1 strain McIntyre (MOI of 0.001). IFN-α treatment was continued during the whole experiment. Virus titers in the supernatants of the cultures were determined at the indicated time points by plaque assay. The expression of MX1 and MX2 in the different cell lines was detected by Western blotting using MxA- and MxB-specific antibodies. The titers determined by plaque assay are shown as arithmetic means ± SD of results from three independent experiments. Significance is depicted for the MX2 knockout (MX2-ko) strain relative to the control cells and was determined via two-way ANOVA with a post hoc Tukey's test. ***, <0.001; **, <0.01; ns, nonsignificant.
GTP binding and hydrolysis are necessary for herpesvirus restriction.
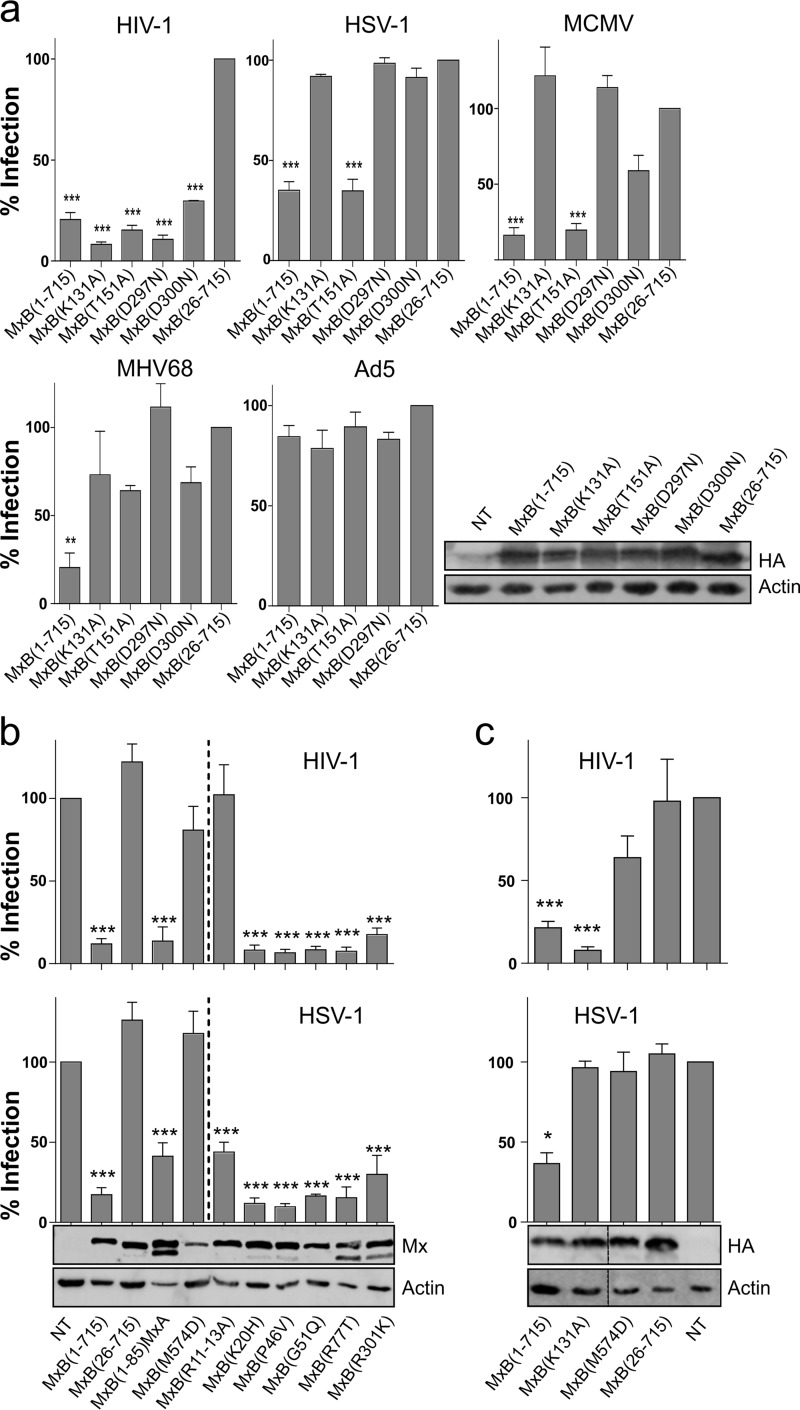
The anti-IAV potential of MxA depends critically on its capacity to bind and hydrolyze GTP (18). In contrast, MxB is active against HIV-1 in the absence of GTPase activity (13, 14). To test the importance for antiherpesvirus activity of MxB, mutations at key positions in the G domain were generated based on sequence homologies to MxA (11, 18). Position K131 is located in the phosphate-binding loop, and position T151 is crucial for stabilizing the transition state during GTP hydrolysis; therefore, the corresponding mutant does not hydrolyze GTP (17). D297 contacts the purine base in cis and prevents GTP binding, whereas D300 contacts the purine base of the opposing G domain and the mutant binds but does not hydrolyze GTP (18). As expected, none of the mutations reduced the capacity of MxB(1-715) to restrict HIV-1 (Fig. 3a). In contrast, the antiherpesvirus activity of MxB was decreased by all but one of the G domain mutations. MxB(T151A) exhibited anti-HSV-1 and anti-MCMV activity in a manner similar to that seen with wild-type MxB(1-715) (Fig. 3a). In contrast, all MHV68-infected cells carrying MxB G domain mutants displayed no restriction or only very low restriction compared to MxB(26-715). Importantly, infection with Ad5 showed no decreased infectivity, excluding the possibility of off-target or toxicity effects of MxB mutants (Fig. 3a).
FIG 3.
GTPase activity, the N terminus, and oligomerization of MxB determine antiviral function. (a) U87MG cells expressing MxB variants were infected (MOI of 0.5) with GFP-encoding HIV-1 pseudotyped particles, HSV-1, MCMV, MHV68, or Ad5. Cells were subjected to FACS analysis and Western blotting as described for Fig. 1a. Depicted is the percentage of GFP-positive cells relative to cells expressing MxB(26-715). (b) A549 cells expressing MxB variants were infected (MOI of 0.5) with VSV-G pseudotyped HIV-1 vector or HSV-1, both encoding GFP, and subjected to FACS analysis and Western blotting as described for Fig. 1a. Shown are data from four independent experiments. Significance is depicted relative to the empty vector control. (c) U87MG cells expressing MxB variants were infected and analyzed as described for panel b. Shown are data from four independent experiments. The vertical line in the Western blot indicates a cut combining two parts of the same blot. Significance is depicted relative to the empty vector control. ***, <0.001; **, <0.01; *, <0.1. The chimeric MxB(1–85)-MxA(38-662) construct was described in reference 22. Single point mutations in MxB were determined according to the method previously described by Mitchell et al. (16).
The N terminus and oligomerization of MxB are required for antiherpesvirus activity.
The unique N-terminal extension of human MxB(1-715) contains an NLS (15, 21) and serves as a major determinant of antiviral activity against HIV-1 (22) and, as shown here, also against herpesviruses (Fig. 1). As seen for HIV-1 (22), a chimeric MxB(1-85)-MxA, with the first 85 N-terminal amino acids of MxB fused to a truncated version of human MxA(35-662), also significantly inhibited HSV-1 (Fig. 3b). In addition, we tested MxB(M574D), a mutant that prevents MxB oligomerization due to inactivation of self-assembly interface 2 in the stalk region of MxB (12, 23). Monomeric MxB(M574D) was inactive against both HIV-1 and HSV-1 in stably transduced A549 as well as U87MG cells (Fig. 3b and c).
A recent evolutionary analysis of primate MxB sequences identified several codons under the control of diversifying selection, mostly located in the N-terminal extension of MxB (16). The selected residues in human MxB(1-715) were changed to the corresponding amino acids found in more distant Old or New World monkey MxBs, generating MxB(K20H), MxB(P46V), MxB(G51Q), MxB(R77T), and MxB(R301K) (16). Introduction of the respective substitutions had no effect on HSV-1 or HIV-1 restriction (Fig. 3b), indicating that these positions are not directly participating in a genetic conflict between MxB and HSV-1, possibly due to the long history of HSV-1 coevolution with its human host (24). Strikingly, however, mutants with a mutation of a triple-arginine motif in MxB(R11–13A) that is required for anti-HIV-1 activity (25) maintained considerable activity against HSV-1 (Fig. 3b).
MxB inhibits herpesvirus replication before or at immediate early gene transcription.
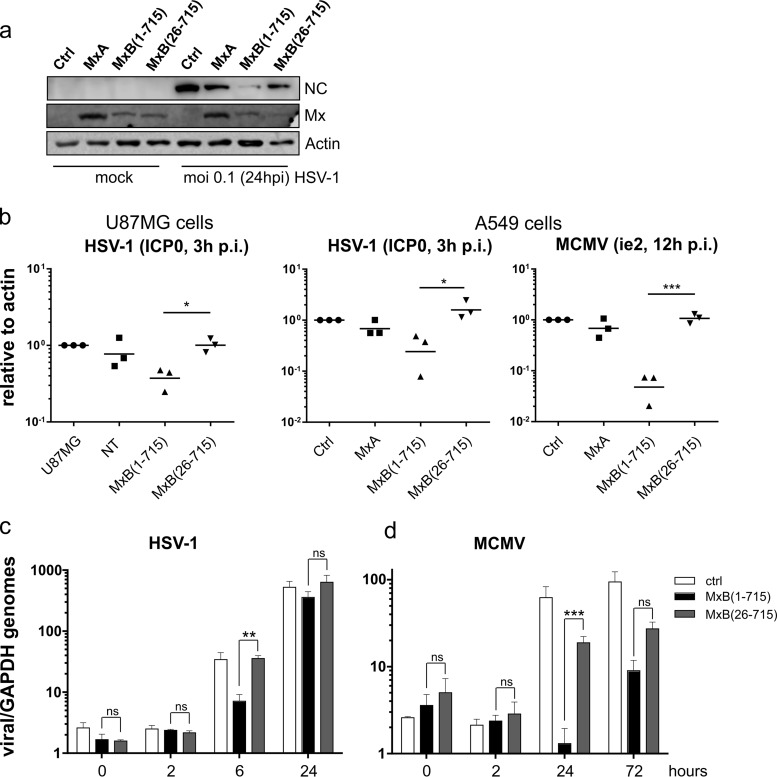
Reduced GFP expression (Fig. 1a) and production of viral progeny (Fig. 2a) in MxB(1-715)-expressing cells indicate that MxB inhibits an early step of the herpesvirus replication cycle. Accordingly, we found diminished HSV-1 capsid protein levels in MxB(1-715) cells (Fig. 4a). Next, we determined the effect of MxB on early gene expression in infected cells by detecting ICP0 transcript levels for HSV-1 and ie2 mRNA levels for MCMV via reverse transcription-quantitative PCR (RT-qPCR). MxB(1-715) had already inhibited this process in infected cells after 3 h of HSV-1 infection and 12 h of MCMV infection (Fig. 4b) as had been shown previously for MHV68 (8). Furthermore, we monitored genome amplification of HSV-1 and MCMV by qPCR at early and late time points of infection. Comparable amounts of attached (0 hpi) and incoming (2 hpi) viral genomes were detected (Fig. 4c and d). At later time points, genome amplification of both HSV-1 (6 hpi) and MCMV (24 hpi) was significantly reduced by MxB(1-715). However, late in infection, the inhibitory effect of MxB(1-715) was reduced, possibly due to the massive increase in viral gene expression and genome amplification, which might overrun the limited inhibitory capacity of MxB at these late time points.
FIG 4.
MxB reduces viral gene expression and amplification of the herpesvirus genome. (a) HSV-1 capsid protein (anti-VP5) expression in A549 cells was analyzed by Western blotting at 24 hpi (MOI of 0.1). Shown is a blot representative of two independent experiments. (b) Expression of immediate early genes of HSV-1 (ICP0) and MCMV (ie2) in infected U87MG or A549 cells was measured by RT-qPCR. CT values were normalized to actin and plotted relative to the ΔCT values determined for the infected control cell line. Error bars represent geometric means of results from three independent experiments with technical duplicates. (c and d) Viral genome equivalents relative to GAPDH as a host-specific probe of HSV-1-infected (c) or MCMV-infected (d) A549 cells (MOI of 1). Error bars represent the SEM of (c) three independent HSV-1 experiments with technical quadruplicates or (d) two independent MCMV experiments with technical duplicates. Statistical analysis was performed using an unpaired t test for each time point. Significance is depicted relative to the control cells. ***, <0.001; **, <0.01; *, <0.1; ns, nonsignificant.
DISCUSSION
An evolutionary analysis of primate MX2 cDNAs suggested a broader antiviral activity of MxB that exceeds its well-established inhibition of lentiviral replication (16). We can now report that MxB actually targets viral replication of all three subfamilies, i.e., alpha-, beta-, and gammaherpesviruses, after testing a panel of both human and murine herpesviruses. Our data confirmed the effect of human MxB on MHV68 as previously shown (8) and demonstrated a generic decrease of early herpesviral gene expression by MxB for HSV-1, HSV-2, and HCMV as well as for nonhuman MCMV. A recent study by Crameri et al. (26) confirmed this broad antiviral effect of MxB against HSV-1, HSV-2, and Kaposi's sarcoma-associated herpesvirus. Remarkably, GTPase-defective MxB variants revealed that both GTP binding and hydrolysis are essential for the antiherpesvirus activity, distinguishing it from the anti-HIV-1 effect but showing its resemblance to MxA in its requirements for antiviral activity. Intriguingly, MxB(T151A), known to be deficient in GTP hydrolysis but not GTP binding (17, 18), was still active against HSV-1 and MCMV. Most likely, MxB(T151A) exhibits an antiviral conformation able to recognize and block the viral target (9). Such an activation induced solely by GTP binding has been demonstrated for the corresponding Dynamin(T65A) variant during clathrin-mediated endocytosis (27).
The N-terminal 25 amino acids of MxB are pivotal for both anti-HIV-1 and antiherpesvirus activity. This N-terminal region of MxB was shown to target MxB to the nuclear envelope and is involved in recognition of the HIV-1 capsid (14, 21, 22). In addition, our analysis identified amino acids in the N-terminal region determining antiviral specificity. For example, inactivation of the triple-arginine motif at positions 11 to 13 in MxB(R11–13A) still restricted HSV-1 but not HIV-1. These differences most likely reflect specific recognition of viral target structures that differ between the two viruses. In the case of HIV-1, the viral capsid is proposed to be targeted by MxB accumulating at the nuclear pore. Thus, distinct capsid protein mutants escape MxB restriction (21, 28). Similarly, herpesviral capsids may be directly targeted by MxB at the nuclear pore and may be hindered from final uncoating. This model was substantiated by a recent study that showed a defect in delivery of viral DNA into the nucleus of HSV-1-infected cells (26).
Interestingly, MxB initally reduced HSV-1 and MCMV genome synthesis during early replication but viral genome accumulation recovered over time. This suggests that initial infection is blocked by MxB but that the viruses are is able to overcome this during the course of replication, possibly indicating the presence of a viral MxB antagonist. In future studies we will address whether MxB directly targets herpesviral nucleocapsids and whether herpesviruses are able to escape the restriction by MxB as described for HIV-1.
In conclusion, our results have identified MxB as a novel interferon-induced restriction factor of herpesviruses preventing lytic infection. The molecular details of MxB antiherpesvirus action remain to be clarified, but its characterization is of great interest in understanding the herpesvirus-specific antiviral host defense, which may provide new therapeutic options for the treatment of life-threatening infections.
MATERIALS AND METHODS
Cell lines, plasmids, and lentiviral transduction.
Human glioblastoma fibroblast U87MG cells (ATCC HTB-14) (13) and T98G cells (ATCC CRL-1690) (20, 29), human lung epithelial A549 cells (ATCC CCL-185), and African green monkey kidney Vero cells (ATCC CCL-81) were cultured at 37°C in Dulbecco's modified Eagle's medium (DMEM; Life Technologies) containing 10% fetal calf serum, 50 U/ml penicillin, and 50 μg/ml streptomycin.
Mx constructs were cloned into pLVX-Puro vector (Clontech) or pCSGW vector (30) by PCR at EcoRI and NotI (pLVX) or BamHI and NotI (pCSxW) sites. MxB (GenBank NM_002463) was either left untagged (pLVX) or subjected to hemagglutinin (HA) tagging at the C terminus (pCSxW). MxA (GenBank accession no. NM_001178046) was subjected to HA tagging at the N terminus.
U87MG cells were transduced using lentiviral particles produced by transfection of 293T cells (ATCC CCL-3216) with HIV-1 Gag-pol-encoding pCMV-ΔR8.91 and VSV-G-encoding pMD.G as well as pCSxW (30). Pools of transduced cells were ∼99% Mx-positive as judged by immunofluorescence and were used without further selection.
A549 cells were transduced using lentiviral particles produced by transfection of the pLVX vector encoding the gene of interest. Thereafter, A549 cells were selected for Mx expression by treatment with 2 μg/ml puromycin.
Viruses, infection, flow cytometry analysis, and growth curves.
The following viruses were used: HSV-1 (McIntyre) (ATCC, VR-539D); HSV-1 (F) (31); HSV-1 GFP (17+) (32); HSV-2 (MS) (ATCC, VR-734); HCMV (TB40-SE-EGFP) (33); MCMV Δ1Δ6 GFP (generated by inserting a GFP expression cassette into a ΔMCMV bacterial artificial chromosome [BAC] via its FLP recombination target [FRT] site [34]); wild-type MCMV rescued from BAC pSM3fr-MCK-2fl clone 3.3 (35); MHV68 GFP (36); Ad5 GFP (37); MVA GFP (38); VSV GFP (39); and IAV GFP (A/Seal/MA/1/80-SC35M, H7N7) (40). Pseudotyped HIV-1 lentiviral particles were produced as described previously (13).
The multiplicity of infection (MOI) for GFP-expressing viruses or lentiviral particles was calculated according to the results of direct titration on the corresponding cell line. The MOI for all non-GFP-encoding viruses was calculated according to the results of titration on VeroCH cells via plaque assay.
Infection of cells was performed either in a 48-well format for flow cytometry analysis or in a 6-well format to determine growth kinetics and gene expression, using centrifugal enhancement (2,000 × g for 30 min).
For analysis of GFP expression by flow cytometry (FACS Fortessa; BD Biosciences), cells were detached from the well by the use of trypsin (Gibco) and fixed with 2% paraformaldehyde at 20 hpi (HSV-1, MCMV, MHV68, Ad5, MVA, and IAV), 6 hpi (VSV), or 48 hpi (pseudotyped HIV-1 lentiviral particles). HCMV-infected cells were detached with Accutase (Sigma) and fixed as described above. For each cell line and condition, 10,000 cells were analyzed and the percentage of GFP+ cells was determined. For data analysis, BD FACSDiva software and FlowJo vX.0.7 were used. To evaluate viral growth, supernatants of infected cells were collected at the indicated time points and titrated by plaque assay.
Western blotting and immunofluorescence analysis.
For Western blot analysis, cells were lysed in tissue protein extraction reagent (T-PER; Thermo Fisher Scientific) and the samples were incubated at 95°C for 5 min. The protein lysates were separated on 10% SDS-PAGE gels and transferred onto a polyvinylidene difluoride (PVDF) membrane (Merck Millipore). As primary antibodies, we used anti-HA (rabbit; Sigma catalog no. H6908), anti-Mx2(N-17) (goat; Santa Cruz Biotechnology catalog no. sc-47197), anti-Mx (mouse; M143 [41]), anti-VP5(SY4563) capsid protein of HSV-1 (rabbit; gift from B. Sodeik, Hannover, Germany [42]), and anti-actin (rabbit; Sigma catalog no. A5060). Detection of the primary antibodies was performed by the use of peroxidase-conjugated (Jackson ImmunoResearch) or fluorescent-labeled (Li-COR) secondary antibodies.
siRNA-mediated knockdown and CRISPR-Cas9-mediated knockout.
T98G cells and A549 cells stably expressing MxA or MxB were transfected with either specific siRNAs (Hs_MX1_11 [targeting sequence, AATGTTCTTCCTGATAGATAA] and Hs_MX2_4 [targeting sequence, GAGCACGATTGAAGACATAAA]; Qiagen) or a nontargeting control siRNA pool (ON-TARGETplus control pool, lot 2027072; Dharmacon) at a final concentration of 30 nM in Opti-MEM (Gibco) using RNAiMax (Invitrogen). At 48 h posttransfection, T98G cells were pretreated with 100 U/ml human IFN-α for 16 h and then infected with HSV-1 GFP (MOI of 0.5). A549 cells were directly infected after the 48-h siRNA treatment. At 20 hpi, cells were harvested for flow cytometry analysis and cell lysates were analyzed by Western blotting for the accumulation of MxA and MxB.
MX gene knockout was performed according to a method previously described by Shalem et al. (43) using lentiviral pXPR vector (Addgene) coding for MX1- or MX2-specific guide RNAs (gRNA) or no gRNA as a control, the Cas9 endonuclease, and a puromycin resistance cassette. The gRNA sequences correspond to positions 381 to 400 of the MX1 open reading frame and positions 162 to 181 of the MX2 open reading frame. T98G cells were transduced with the lentiviral vectors or with empty vector as a control, and positive cell clones were selected by incubation with puromycin. The successful knockdowns of MX1 and MX2 were confirmed by Western blotting.
DNA extraction and qPCR.
For detection of viral DNA replication by qPCR, cells seeded on 12-well plates were infected with HSV-1 and MCMV MOIs of 0.5 and 3, respectively. Cells were lysed at different time points after infection, and total cell-associated DNA was extracted using a NucleoSpin tissue kit (Macherey-Nagel). In duplicate qPCRs performed using a QuantiTect SYBR green PCR kit (Qiagen), 10 ng DNA served as the template. For detection of virus genomes, specific primer pairs were used, amplifying either the UL27 gene of HSV-1 (forward [for], 5′-ATGACCATGTCGGTGACCTTGG-3′; reverse [rev], 5′-TGCAGAGCAACCCCATGAAG-3′) or the M45 gene of MCMV (for, 5′-ATCTCCTCGAAGGGGAATGA-3′; rev, 5′-TCGACAGACAGCCGTTCGT-3′) (44). To normalize the viral genome counts, host cell genomes were quantified using genomic GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene-specific primers (for, 5′-GGACTGAGGCTCCCACCTTT-3′; rev, 5′-GCATGGACTGTGGTCTGCAA-3′). The viral DNA replication was quantified by determination of the increasing ratio between the virus and host genomes using the threshold cycle (ΔΔCT) method.
RNA extraction, reverse transcription, and RT-qPCR.
The U87MG and A549 cell lines were seeded in technical replicates in a 12-well format and infected with MCMV and HSV-1 at an MOI of 1. Cells were lysed at different time points postinfection, and total RNA was extracted using NucleoSpin kit RNA II (Macherey-Nagel) and reverse transcribed using a QuantiTect reverse transcription kit (Qiagen). cDNA (1 μg) was used for the subsequent qPCRs performed with a QuantiTect SYBR green PCR kit (Qiagen). CT values were normalized to actin (ΔCT) and plotted relative to the ΔCT values of the nontransduced control cell line.
For the detection of the ICP0 immediate early gene of HSV-1, we used primer pair P3 (5′-TTCGGTCTCCGCCTCAGAGTC-3′) and P5 (5′-GACCCTCCAGCCGCATACGA-3′) (45). For the detection of the ie2 immediate early gene of MCMV by RT-qPCR, the following primers were designed: for, 5′-CTGACCTGTTGTTACTAATATGG-3′; rev, 5′-AGACCCGCTGCATAAAGATA-3′.
Statistical analyses.
Statistics were performed with GraphPad Prism 6 software using one-way or two-way analysis of variance (ANOVA) with a post hoc Tukey's test and unpaired t test (Fig. 4c and d) (***, <0.001; **, <0.01; *, <0.1). Bars in the figure panels show means and standard errors of the means (SEM).
ACKNOWLEDGMENTS
We thank Heiko Adler, Helmholtz Zentrum Munich, for MHV68-GFP; John Hiscott, Vaccine & Gene Therapy Institute of Florida, for VSV-GFP; Jürgen Hausmann, Bavaria Nordic Martinsried, for MVA-GFP; Martin Schwemmle, Virology Freiburg, for SC35M-GFP; Beate Sodeik, Virology Hannover, for HSV-1-GFP and antibodies; and Christian Sinzger and Kerstin Laib Sampaio, Virology Ulm, for the TB40-SE-EGFP BAC.
This work was supported by grants of the Deutsche Forschungsgemeinschaft to G.K. (Ko 1579/8-2) and to T.S. (SCHA 1950/1-1). The funder had no role in study design, data collection and publishing. T.S. declares competing financial interests, being presently employed with Heidelberg Immunotherapeutics GmbH. The rest of us disclose conflicts of interest and declare no competing financial interests.
REFERENCES
- 1.Griffiths PD. 2012. Burden of disease associated with human cytomegalovirus and prospects for elimination by universal immunisation. Lancet Infect Dis 12:790–798. doi: 10.1016/S1473-3099(12)70197-4. [DOI] [PubMed] [Google Scholar]
- 2.Wen KW, Damania B. 2010. Kaposi sarcoma-associated herpesvirus (KSHV): molecular biology and oncogenesis. Cancer Lett 289:140–150. doi: 10.1016/j.canlet.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herman M, Ciancanelli M, Ou YH, Lorenzo L, Klaudel-Dreszler M, Pauwels E, Sancho-Shimizu V, Perez de Diego R, Abhyankar A, Israelsson E, Guo Y, Cardon A, Rozenberg F, Lebon P, Tardieu M, Heropolitanska-Pliszka E, Chaussabel D, White MA, Abel L, Zhang SY, Casanova JL. 2012. Heterozygous TBK1 mutations impair TLR3 immunity and underlie herpes simplex encephalitis of childhood. J Exp Med 209:1567–1582. doi: 10.1084/jem.20111316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mossman KL, Ashkar AA. 2005. Herpesviruses and the innate immune response. Viral Immunol 18:267–281. doi: 10.1089/vim.2005.18.267. [DOI] [PubMed] [Google Scholar]
- 5.Paludan SR, Bowie AG, Horan KA, Fitzgerald KA. 2011. Recognition of herpesviruses by the innate immune system. Nat Rev Immunol 11:143–154. doi: 10.1038/nri2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dupuis S, Jouanguy E, Al-Hajjar S, Fieschi C, Al-Mohsen IZ, Al-Jumaah S, Yang K, Chapgier A, Eidenschenk C, Eid P, Al Ghonaium A, Tufenkeji H, Frayha H, Al-Gazlan S, Al-Rayes H, Schreiber RD, Gresser I, Casanova JL. 2003. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat Genet 33:388–391. doi: 10.1038/ng1097. [DOI] [PubMed] [Google Scholar]
- 7.Hoyos-Bachiloglu R, Chou J, Sodroski CN, Beano A, Bainter W, Angelova M, Al Idrissi E, Habazi MK, Alghamdi HA, Almanjomi F, Al Shehri M, Elsidig N, Alaa Eldin M, Knipe DM, AlZahrani M, Geha RS. 2017. A digenic human immunodeficiency characterized by IFNAR1 and IFNGR2 mutations. J Clin Invest doi: 10.1172/JCI93486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu SY, Sanchez DJ, Aliyari R, Lu S, Cheng G. 2012. Systematic identification of type I and type II interferon-induced antiviral factors. Proc Natl Acad Sci U S A 109:4239–4244. doi: 10.1073/pnas.1114981109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haller O, Staeheli P, Schwemmle M, Kochs G. 2015. Mx GTPases: dynamin-like antiviral machines of innate immunity. Trends Microbiol 23:154–163. doi: 10.1016/j.tim.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 10.Holzinger D, Jorns C, Stertz S, Boisson-Dupuis S, Thimme R, Weidmann M, Casanova JL, Haller O, Kochs G. 2007. Induction of MxA gene expression by influenza A virus requires type I or type III interferon signaling. J Virol 81:7776–7785. doi: 10.1128/JVI.00546-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao S, von der Malsburg A, Dick A, Faelber K, Schroder GF, Haller O, Kochs G, Daumke O. 2011. Structure of myxovirus resistance protein a reveals intra- and intermolecular domain interactions required for the antiviral function. Immunity 35:514–525. doi: 10.1016/j.immuni.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 12.Fribourgh JL, Nguyen HC, Matreyek KA, Alvarez FJ, Summers BJ, Dewdney TG, Aiken C, Zhang P, Engelman A, Xiong Y. 2014. Structural insight into HIV-1 restriction by MxB. Cell Host Microbe 16:627–638. doi: 10.1016/j.chom.2014.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goujon C, Moncorge O, Bauby H, Doyle T, Ward CC, Schaller T, Hue S, Barclay WS, Schulz R, Malim MH. 2013. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature 502:559–562. doi: 10.1038/nature12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kane M, Yadav SS, Bitzegeio J, Kutluay SB, Zang T, Wilson SJ, Schoggins JW, Rice CM, Yamashita M, Hatziioannou T, Bieniasz PD. 2013. MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature 502:563–566. doi: 10.1038/nature12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melén K, Keskinen P, Ronni T, Sareneva T, Lounatmaa K, Julkunen I. 1996. Human MxB protein, an interferon-alpha-inducible GTPase, contains a nuclear targeting signal and is localized in the heterochromatin region beneath the nuclear envelope. J Biol Chem 271:23478–23486. doi: 10.1074/jbc.271.38.23478. [DOI] [PubMed] [Google Scholar]
- 16.Mitchell PS, Young JM, Emerman M, Malik HS. 2015. Evolutionary analyses suggest a function of MxB immunity proteins beyond lentivirus restriction. PLoS Pathog 11:e1005304. doi: 10.1371/journal.ppat.1005304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.King MC, Raposo G, Lemmon MA. 2004. Inhibition of nuclear import and cell-cycle progression by mutated forms of the dynamin-like GTPase MxB. Proc Natl Acad Sci U S A 101:8957–8962. doi: 10.1073/pnas.0403167101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dick A, Graf L, Olal D, von der Malsburg A, Gao S, Kochs G, Daumke O. 2015. Role of nucleotide binding and GTPase domain dimerization in dynamin-like myxovirus resistance protein A for GTPase activation and antiviral activity. J Biol Chem 290:12779–12792. doi: 10.1074/jbc.M115.650325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pavlovic J, Zurcher T, Haller O, Staeheli P. 1990. Resistance to influenza virus and vesicular stomatitis virus conferred by expression of human MxA protein. J Virol 64:3370–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aebi M, Fah J, Hurt N, Samuel CE, Thomis D, Bazzigher L, Pavlovic J, Haller O, Staeheli P. 1989. cDNA structures and regulation of two interferon-induced human Mx proteins. Mol Cell Biol 9:5062–5072. doi: 10.1128/MCB.9.11.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Busnadiego I, Kane M, Rihn SJ, Preugschas HF, Hughes J, Blanco-Melo D, Strouvelle VP, Zang TM, Willett BJ, Boutell C, Bieniasz PD, Wilson SJ. 2014. Host and viral determinants of Mx2 antiretroviral activity. J Virol 88:7738–7752. doi: 10.1128/JVI.00214-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goujon C, Moncorge O, Bauby H, Doyle T, Barclay WS, Malim MH. 2014. Transfer of the amino-terminal nuclear envelope targeting domain of human MX2 converts MX1 into an HIV-1 resistance factor. J Virol 88:9017–9026. doi: 10.1128/JVI.01269-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao S, von der Malsburg A, Paeschke S, Behlke J, Haller O, Kochs G, Daumke O. 2010. Structural basis of oligomerization in the stalk region of dynamin-like MxA. Nature 465:502–506. doi: 10.1038/nature08972. [DOI] [PubMed] [Google Scholar]
- 24.Sharp PM. 2002. Origins of human virus diversity. Cell 108:305–312. doi: 10.1016/S0092-8674(02)00639-6. [DOI] [PubMed] [Google Scholar]
- 25.Goujon C, Greenbury RA, Papaioannou S, Doyle T, Malim MH. 2015. A triple-arginine motif in the amino-terminal domain and oligomerization are required for HIV-1 inhibition by human MX2. J Virol 89:4676–4680. doi: 10.1128/JVI.00169-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crameri M, Bauer M, Caduff N, Walker R, Steiner F, Franzoso FD, Gujer C, Boucke K, Kucera T, Zbinden A, Munz C, Fraefel C, Greber UF, Pavlovic J. 2018. MxB is an interferon-induced restriction factor of human herpesviruses. Nat Commun 9:1980. doi: 10.1038/s41467-018-04379-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marks B, Stowell MH, Vallis Y, Mills IG, Gibson A, Hopkins CR, McMahon HT. 2001. GTPase activity of dynamin and resulting conformation change are essential for endocytosis. Nature 410:231–235. doi: 10.1038/35065645. [DOI] [PubMed] [Google Scholar]
- 28.Fricke T, White TE, Schulte B, de Souza Aranha Vieira DA, Dharan A, Campbell EM, Brandariz-Nunez A, Diaz-Griffero F. 2014. MxB binds to the HIV-1 core and prevents the uncoating process of HIV-1. Retrovirology 11:68. doi: 10.1186/s12977-014-0068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stein GH. 1979. T98G: an anchorage-independent human tumor cell line that exhibits stationary phase G1 arrest in vitro. J Cell Physiol 99:43–54. doi: 10.1002/jcp.1040990107. [DOI] [PubMed] [Google Scholar]
- 30.Schaller T, Ocwieja KE, Rasaiyaah J, Price AJ, Brady TL, Roth SL, Hue S, Fletcher AJ, Lee K, KewalRamani VN, Noursadeghi M, Jenner RG, James LC, Bushman FD, Towers GJ. 2011. HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS Pathog 7:e1002439. doi: 10.1371/journal.ppat.1002439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ejercito PM, Kieff ED, Roizman B. 1968. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol 2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 32.Snijder B, Sacher R, Ramo P, Liberali P, Mench K, Wolfrum N, Burleigh L, Scott CC, Verheije MH, Mercer J, Moese S, Heger T, Theusner K, Jurgeit A, Lamparter D, Balistreri G, Schelhaas M, De Haan CA, Marjomaki V, Hyypia T, Rottier PJ, Sodeik B, Marsh M, Gruenberg J, Amara A, Greber U, Helenius A, Pelkmans L. 2012. Single-cell analysis of population context advances RNAi screening at multiple levels. Mol Syst Biol 8:579. doi: 10.1038/msb.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sampaio KL, Weyell A, Subramanian N, Wu Z, Sinzger C. 2017. A TB40/E-derived human cytomegalovirus genome with an intact US-gene region and a self-excisable BAC cassette for immunological research. Biotechniques 63:205–214. doi: 10.2144/000114606. [DOI] [PubMed] [Google Scholar]
- 34.Cicin-Sain L, Bubic I, Schnee M, Ruzsics Z, Mohr C, Jonjic S, Koszinowski UH. 2007. Targeted deletion of regions rich in immune-evasive genes from the cytomegalovirus genome as a novel vaccine strategy. J Virol 81:13825–13834. doi: 10.1128/JVI.01911-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jordan S, Krause J, Prager A, Mitrovic M, Jonjic S, Koszinowski UH, Adler B. 2011. Virus progeny of murine cytomegalovirus bacterial artificial chromosome pSM3fr show reduced growth in salivary glands due to a fixed mutation of MCK-2. J Virol 85:10346–10353. doi: 10.1128/JVI.00545-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adler H, Messerle M, Wagner M, Koszinowski UH. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J Virol 74:6964–6974. doi: 10.1128/JVI.74.15.6964-6974.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wodrich H, Henaff D, Jammart B, Segura-Morales C, Seelmeir S, Coux O, Ruzsics Z, Wiethoff CM, Kremer EJ. 2010. A capsid-encoded PPxY-motif facilitates adenovirus entry. PLoS Pathog 6:e1000808. doi: 10.1371/journal.ppat.1000808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chakrabarti S, Sisler JR, Moss B. 1997. Compact, synthetic, vaccinia virus early/late promoter for protein expression. Biotechniques 23:1094–1097. [DOI] [PubMed] [Google Scholar]
- 39.Stojdl DF, Lichty BD, ten Oever BR, Paterson JM, Power AT, Knowles S, Marius R, Reynard J, Poliquin L, Atkins H, Brown EG, Durbin RK, Durbin JE, Hiscott J, Bell JC. 2003. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell 4:263–275. doi: 10.1016/S1535-6108(03)00241-1. [DOI] [PubMed] [Google Scholar]
- 40.Reuther P, Gopfert K, Dudek AH, Heiner M, Herold S, Schwemmle M. 2015. Generation of a variety of stable Influenza A reporter viruses by genetic engineering of the NS gene segment. Sci Rep 5:11346. doi: 10.1038/srep11346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flohr F, Schneider-Schaulies S, Haller O, Kochs G. 1999. The central interactive region of human MxA GTPase is involved in GTPase activation and interaction with viral target structures. FEBS Lett 463:24–28. doi: 10.1016/S0014-5793(99)01598-7. [DOI] [PubMed] [Google Scholar]
- 42.Döhner K, Ramos-Nascimento A, Bialy D, Anderson F, Hickford-Martinez A, Rother F, Koithan T, Rudolph K, Buch A, Prank U, Binz A, Hugel S, Lebbink RJ, Hoeben RC, Hartmann E, Bader M, Bauerfeind R, Sodeik B. 2018. Importin alpha1 is required for nuclear import of herpes simplex virus proteins and capsid assembly in fibroblasts and neurons. PLoS Pathog 14:e1006823. doi: 10.1371/journal.ppat.1006823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Victória JM, Guimarães AL, da Silva LM, Kalapothakis E, Gomez RS. 2005. Polymerase chain reaction for identification of herpes simplex virus (HSV-1), cytomegalovirus (CMV) and human herpes virus-type 6 (HHV-6) in oral swabs. Microbiol Res 160:61–65. doi: 10.1016/j.micres.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 45.Majerciak V, Zheng Z-M. 2013. Detection of viral RNA splicing in diagnostic virology, p 693–748. In Advanced techniques in diagnostic microbiology. Springer, New York, NY. [Google Scholar]