Coronaviruses cause widespread respiratory, gastrointestinal, and central nervous system diseases in humans and other animals, threatening human health and causing economic loss. Coronavirus nsp9, a member of the replication complex, is an important RNA binding subunit in the RNA-synthesizing machinery of all coronaviruses. However, the mechanisms of the dimerization and nucleic acid binding of nsp9 remain elusive. In this study we determined the nsp9 crystal structures of PDCoV and PEDV. We first found that the N-finger of nsp9 from PDCoV plays a critical role in dimerization. Meanwhile, PEDV nsp9 is distinguished by the presence of a disulfide bond in the dimer interface. This study provides a structural and functional basis for understanding the mechanism of dimerization and shows that the diverse dimerization modes of coronavirus nsp9 proteins enhance their nucleic acid binding affinity. Importantly, these findings may provide a new insight for antiviral drug development.
KEYWORDS: coronavirus nsp9, N-finger, dimerization, disulfide bond, nucleic acid binding
ABSTRACT
Coronaviruses pose serious health threats to humans and other animals. Understanding the mechanisms of their replication has important implications for global health and economic stability. Nonstructural protein 9 (nsp9) is an essential RNA binding protein for coronavirus replication. However, the mechanisms of the dimerization and nucleic acid binding of nsp9 remain elusive. Here, we report four crystal structures, including wild-type porcine delta coronavirus (PDCoV) nsp9, PDCoV nsp9-ΔN7 (N-terminal 7 amino acids deleted), wild-type porcine epidemic diarrhea virus (PEDV) nsp9, and PEDV nsp9-C59A mutant. These structures reveal the diverse dimerization forms of coronavirus nsp9. We first found that the N-finger of nsp9 from PDCoV plays a critical role in dimerization. Meanwhile, PEDV nsp9 is distinguished by the presence of a disulfide bond in the dimer interface. Interestingly, size exclusion chromatography and analytical ultracentrifugation analyses indicate that the PDCoV nsp9-ΔN7 and PEDV nsp9-C59A mutants are monomeric in solution. In addition, electrophoretic mobility shift assays and microscale thermophoresis analysis indicate that the monomeric forms of PDCoV nsp9 and PEDV nsp9 still have nucleic acid binding affinity, although it is lower than that of the wild type. Our results show that the diverse dimerization forms of coronavirus nsp9 proteins enhance their nucleic acid binding affinity.
IMPORTANCE Coronaviruses cause widespread respiratory, gastrointestinal, and central nervous system diseases in humans and other animals, threatening human health and causing economic loss. Coronavirus nsp9, a member of the replication complex, is an important RNA binding subunit in the RNA-synthesizing machinery of all coronaviruses. However, the mechanisms of the dimerization and nucleic acid binding of nsp9 remain elusive. In this study we determined the nsp9 crystal structures of PDCoV and PEDV. We first found that the N-finger of nsp9 from PDCoV plays a critical role in dimerization. Meanwhile, PEDV nsp9 is distinguished by the presence of a disulfide bond in the dimer interface. This study provides a structural and functional basis for understanding the mechanism of dimerization and shows that the diverse dimerization modes of coronavirus nsp9 proteins enhance their nucleic acid binding affinity. Importantly, these findings may provide a new insight for antiviral drug development.
INTRODUCTION
Coronaviruses (CoVs) are enveloped viruses, with single-stranded RNA genomes of positive polarity approximately 30 kb in length, that can be divided into four genera: α-CoV, β-CoV, γ-CoV, and δ-CoV (1, 2). The ideal hosts of α-CoV and β-CoV are mammals, and γ-CoV primarily infects birds, while δ-CoV has been identified in both mammals and birds (3). In the past 15 years, there have been two epidemics of fatal pneumonia in addition to other outbreaks caused by CoVs. The severe acute respiratory syndrome CoV (SARS-CoV) emerged in China in 2002, and the Middle East respiratory syndrome CoV (MERS-CoV) emerged in Saudi Arabia in 2012 (4–6). In addition, the PEDV and the PDCoV, two reemerging and emerging epizootic swine viruses, have caused major economic losses in Asia and the United States (7–9).
CoVs encode the open reading frame (ORF) 1a and ORF 1ab polyproteins, which are then processed into 15 to 16 nonstructural proteins (nsp's) by two virus-encoded proteinases, the papain-like and 3C-like proteinases (10–14). All of these nsp's, except for nsp1 and nsp2, are considered essential for transcription, replication, and translation of the viral RNA (15–17). nsp9 with nsp7, nsp8, and nsp10 localizes within the replication complex and is likely a member of the replication complex (18). The deletion of nsp9 in mouse hepatitis virus (MHV) prevents RNA synthesis and productive virus infection, while fusion of a nsp9-10 oligoprotein results in viability but attenuated growth, indicating that the mature form of the nsp9 protein is critical for viral replication (19). Another study showed that nsp9 is a nucleic acid binding protein that is essential for replication (20). Meanwhile, the dimerization of nsp9 is critical for viral replication (21). For nsp9, there are five crystallographic structures that showed a variety of dimeric interfaces (22–25). However, the dimerization mechanism of all CoV nsp9s, whether from emerging viruses or not, is not so clear yet.
In this study, we elucidated the crystal structures and functions of PDCoV nsp9 and PEDV nsp9. The critical amino acids or motifs for nsp9 dimerization and nucleic acid binding were further investigated.
RESULTS
Overall structure of the nsp9.
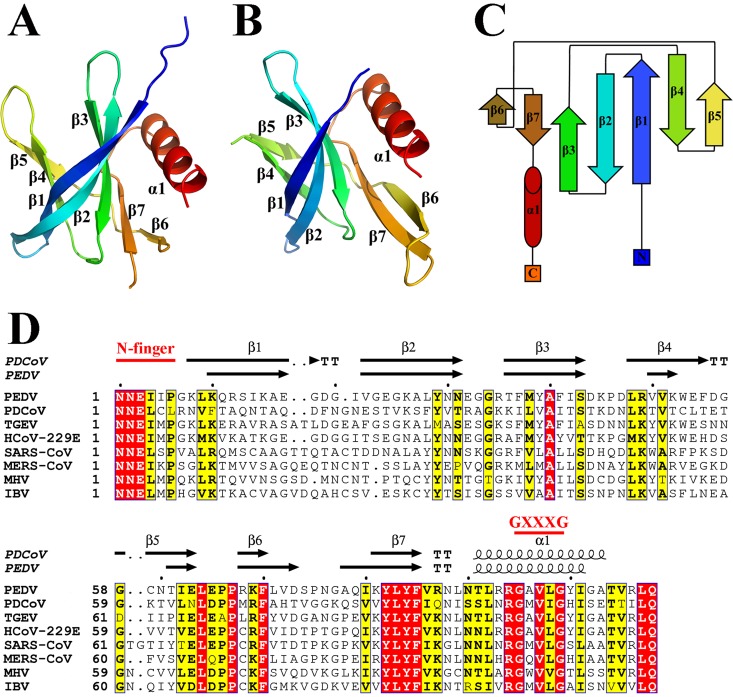
The PDCoV nsp9 and PEDV nsp9 structures are refined to 1.80- and 2.89-Å resolutions, respectively (Table 1). Their space groups are P21 and P64, respectively. With the exception of the regions encompassing amino acids 72 to 79, and 106 to 109 of PDCoV nsp9 and 1 to 6, 33, 55 to 58, and 106 to 108 of PEDV nsp9, all residues of both nsp9 proteins can be built in the final models (Fig. 1A and B). The crystal structures of PDCoV nsp9 and PEDV nsp9 reveal that their monomers contain seven antiparallel β-strands and one α-helix appended at the C terminus of the polypeptide chain (residues 91 to 103), as well as loops connecting the β-strands (Fig. 1A and B). All seven β-strands form a β-sandwich, with sheet β1-3 interleaved between sheets β4-5 and β6-7 (Fig. 1C).
TABLE 1.
Data collection and refinement statistics
| Parameter | Value(s)a |
|||
|---|---|---|---|---|
| Wild-type PDCoV nsp9 | PDCoV nsp9-ΔN7 | Wild-type PEDV nsp9 | PEDV nsp9-C59A | |
| Data collection | ||||
| Space group | P21 | P21 | P64 | P43212 |
| Cell parameters (a, b, c [Å]) | 57.97, 55.19, 69.44 | 31.55, 83.86, 36.34 | 73.87, 73.87, 91.55 | 58.73, 58.73, 193.24 |
| α, β, γ (°) | 90.00, 90.01, 90.00 | 90.00, 106.08, 90.00 | 90.00, 90.00, 120.00 | 90.00, 90.00, 90.00 |
| Wavelength (Å) | 0.97917 | 0.97917 | 0.97917 | 0.97917 |
| Resolution range (Å) | 29.79–1.80 | 34.92–1.99 | 28.75 −2.89 | 38.15–3.00 |
| Completeness (%) | 97.8 (97.0) | 99.7 (100.0) | 99.4 (99.2) | 99.9 (100.0) |
| Rmergeb (%) | 6.4 (46.6) | 7.5 (10.3) | 6.3 (67.7) | 5.9 (84.9) |
| I/σ (last shell) | 15.71 (3.38) | 24.51 (23.27) | 35.33 (3.00) | 29.8 (3.74) |
| Redundancy (last shell) | 3.8 (3.8) | 7.0 (7.2) | 11.1 (11.4) | 7.6 (7.7) |
| Refinement | ||||
| Resolution range (Å) | 29.79–1.80 | 34.92–2.00 | 28.75–2.90 | 38.15–3.00 |
| Rwork/Rfreec (%) | 20.6/24.6 | 21.3/24.8 | 30.7/32.7 | 29.5/33.5 |
| No. reflections | 39,706 | 12,364 | 6,340 | 6,925 |
| No of protein atoms | 2,970 | 1,463 | 1,498 | 2,249 |
| No. of solvent atoms | 207 | 70 | 0 | 0 |
| No. of molecules | 4 | 2 | 2 | 3 |
| RMSD | ||||
| Bond length (Å) | 0.007 | 0.009 | 0.009 | 0.005 |
| Bond angle (°) | 1.018 | 1.070 | 1.577 | 1.348 |
| Avg B factor (Å2) | 33 | 20 | 109 | 103 |
| Ramachandran plot: core, allow, disallow (%) | 97.29, 2.71, 0.00 | 97.81, 2.19, 0.00 | 92.22, 6.67, 1.11 | 95.40, 3.83, 0.77 |
The highest resolution values are given in parentheses.
Rmerge = Σ Σ Ii − <I>/Σ Σ Ii (where Ii is the intensity measurement of reflection h, and <I> is the average intensity from multiple observations).
Rwork = Σ ||Fo| − |Fc||/Σ |Fo|, where Fo and Fc are the observed and calculated structure factors, respectively. Rfree is equivalent to Rwork but where 5% of the measured reflections have been excluded from refinement and set aside for cross-validation.
FIG 1.
Stereoview of the structures of PDCoV nsp9 and PEDV nsp9. (A) Ribbon representation of the PDCoV nsp9 monomer. (B) Ribbon representation of the PEDV nsp9 monomer. (C) Schematic illustration of nsp9 topology. The β strands are drawn as arrows, and α-helices are drawn as cylinders. (D) Sequence alignment of CoV nsp9 homologs to PDCoV nsp9 and PEDV nsp9. Identical residues are highlighted in red, and conserved residues are shown in yellow. The table was produced using ESpript3.0 with the secondary structure elements for PDCoV nsp9 and PEDV nsp9 assigned using DSSP (43). Residues boxed in red are completely conserved.
The N-finger plays an important role in the dimerization of PDCoV nsp9.
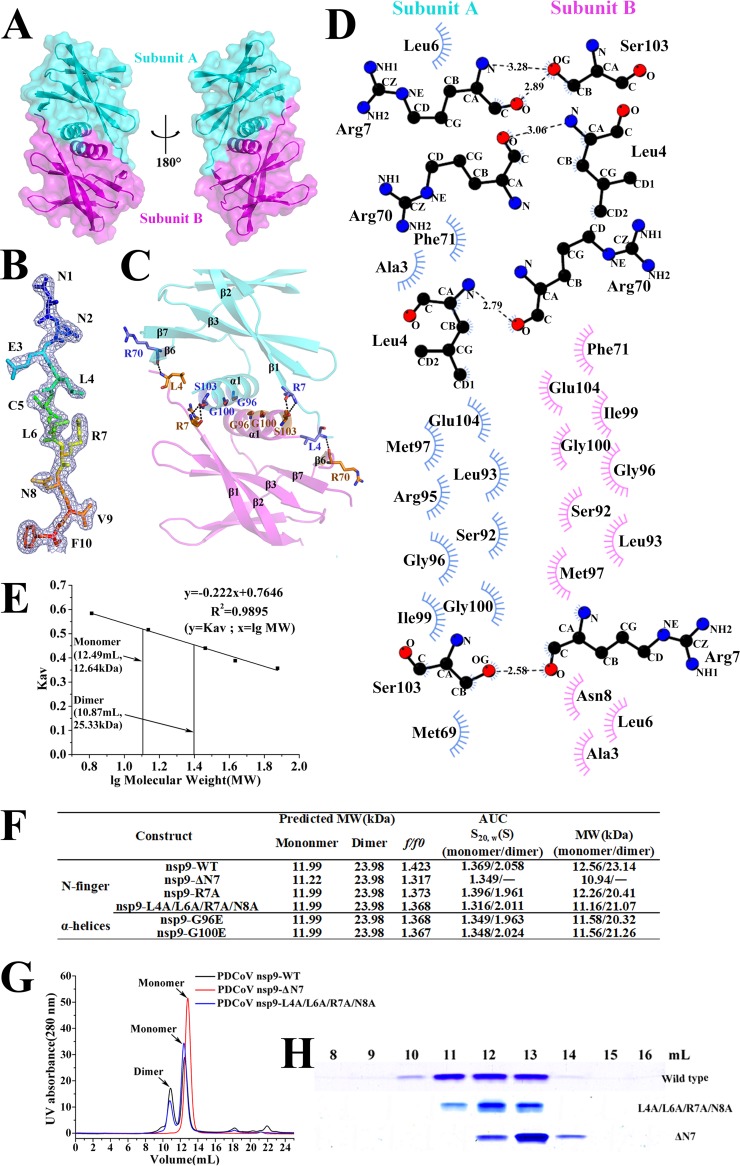
Based on the PDCoV nsp9 crystal structure, the dimer interface is formed by the N-fingers and the parallel association of the C-terminal α-helices. A total surface area of 924.3 Å2 is buried upon dimerization (Fig. 2A). Although the N terminus of nsp9 may be flexible in other coronaviruses, we see clear electron density for this region in the 2Fo–Fc map (Fig. 2B). One hydrogen bond forms between the Leu4 main chain of subunit B and the Arg70 main chain of subunit A, and one hydrogen bond forms between the Arg7 main chain of subunit A and the Ser103 side chain of subunit B (Fig. 2C and D). Leu4 and Arg7, which form part of the N-finger (Fig. 1D), clip onto the edge of the inner β-sheet (Arg70 on strand 6) and the edge of the α-helix (Ser103) from its dimer partner (Fig. 2C and D). Further stabilization is derived from the close packing of the helices from the two monomers because the heart of the dimer interface consists of two glycines (Gly96 and Gly100), which form a hydrophobic interface and stabilize the dimer (Fig. 2C and D).
FIG 2.
The N-finger of the PDCoV nsp9 has a strong effect on dimerization. (A) The two monomers are colored cyan and magenta, respectively, and depicted in cartoon and surface representations. (B) 2Fo–F electron density map of the N-finger at 1.5σ. This region includes 10 residues. (C) Dimerization interface of PDCoV nsp9. The interacting residues (Leu4, Arg7, Arg70, Gly96, Gly100, and Ser103) between two monomers are shown with stick illustrations. (D) All of the interacting residues between subunits A (blue) and B (magenta) were determined using LIGPLOT. (E) Calculated molecular weights of the PDCoV nsp9 protein peaks with the values obtained for known calibration standards (GE Healthcare). The calculated molecular weight of the PDCoV nsp9 peaks was determined by fitting to the calibration curve (Kav = volumes of elution [Ves]/24); volumes of elution of 10.87 ml (approximately 25.33) and 12.49 ml (approximately 12.64) are indicated by arrows. (F) Velocity AUC analysis of the PDCoV nsp9 and mutant proteins. The molecular mass represented by each major peak is indicated, and the biophysical data are shown in the table. (G) Size exclusion experiment with the wild-type and mutants (ΔN7 and L4A/L6A/R7A/N8A) of PDCoV nsp9. (H) SDS-PAGE analysis of wild-type and mutant (L4A/L6A/R7A/N8A and ΔN7) nsp9. The elution volume is described in panel E. Molecular mass markers are shown.
To determine whether the PDCoV nsp9 dimer found in the crystal is also present in solution, we performed size exclusion chromatography (SEC) and analytical ultracentrifugation (AUC) experiments to confirm the oligomeric state of PDCoV nsp9 in solution. Our data show that PDCoV nsp9 eluted in two peaks with calculated molecular masses of approximately 12.64 and 25.33 kDa, corresponding to a monomer and a dimer, respectively (Fig. 2E). Meanwhile, the AUC data, which are consistent with the SEC results, show that the PDCoV nsp9 protein existed in both monomeric and dimeric states in solution (Fig. 2F).
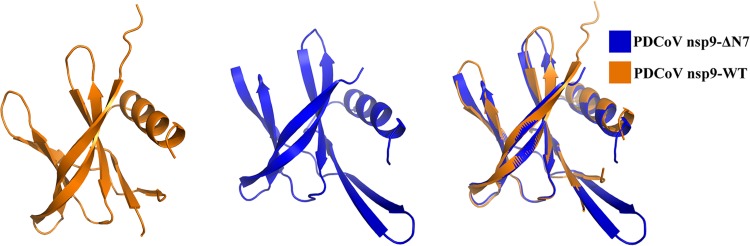
Then, we determined whether the N-finger plays important roles in nsp9 dimer formation. The nsp9-ΔN7 protein was prepared, and SEC and AUC assays were used to analyze the oligomeric states of the two mutants. The SEC data show that the nsp9-ΔN7 mutant eluted in only one peak, which corresponded to a monomer (Fig. 2G and H). Meanwhile, the nsp9-L4A/L6A/R7A/N8A mutant has a monomeric form (Fig. 2G and H). The AUC results reveal a monomeric state of the nsp9-ΔN7 mutant with a calculated molecular weight of 10.94 kDa, a sedimentation coefficient (S20,W) of 1.349, and a frictional ratio (f/f0) of 1.317 (Fig. 2F). In addition, PDCoV nsp9-ΔN7 is crystallized in the space group P21. The structure is determined by molecular replacement using the structure of wild-type PDCoV nsp9 as the search template and is refined at a 1.99-Å resolution (Table 1). The structure of PDCoV nsp9-ΔN7 remains basically consistent with wild-type PDCoV nsp9 except for the N-finger. The structural superposition of the wild-type nsp9 monomer with the nsp9-ΔN7 mutant monomer shows a root mean square deviation (RMSD) of 0.9 Å between the 90 Cα atoms (Fig. 3). According to these results, the N-finger plays an important role in maintaining the dimer stability. Meanwhile, the SEC data show that the nsp9-G96E and nsp9-G100E mutants have a monomeric form (data not shown), indicating that the Gly96 and Gly100 of GXXXG motif can also impact the dimerization.
FIG 3.
Illustration of the superimposition of PDCoV nsp9-WT and PDCoV nsp9-ΔN7 monomer structures. The PDCoV nsp9-WT and PDCoV nsp9-ΔN7 monomers are colored orange and blue, respectively, and depicted in cartoon representation. The structural superposition shows an RMSD of 0.9 Å between the 90 Cα atoms.
The importance of the dimerization-related amino acids in nucleic acid binding of PDCoV nsp9.
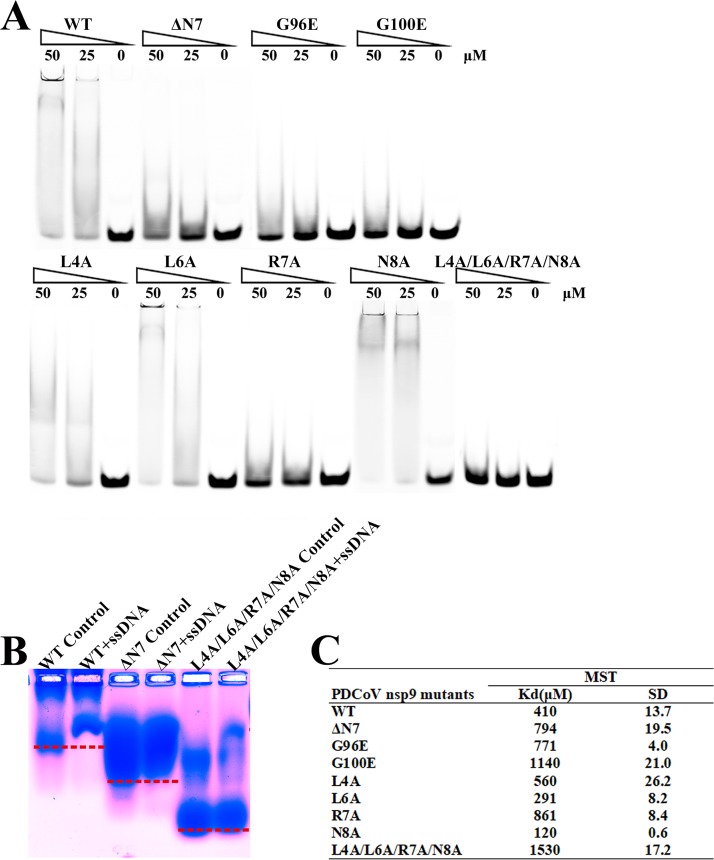
Seven mutants, including nsp9-L4A, nsp9-L6A, nsp9-R7A, nsp9-N8A, nsp9-L4A/L6A/R7A/N8A, nsp9-G96E, and nsp9-G100E, were designed to test whether the N-finger and the glycines in the α-helix of PDCoV nsp9 are related to nucleic acid binding. Then, two electrophoretic mobility shift assays (EMSAs) were performed to examine the nucleic acid binding affinity of each mutant, and microscale thermophoresis (MST) assays were used to determine the quantitative value of the binding affinity. The results reveal that Leu4 and Leu6 are not important amino acids for nucleic acid binding. The wild-type PDCoV nsp9 protein bound to ssDNA with a Kd of 410 μM, and the single-stranded DNA (ssDNA) binding affinities of the nsp9-R7A, nsp9-G96E, and nsp9-G100E mutants are 2.5∼5-fold weaker than that of wild-type PDCoV nsp9. In addition, nsp9-L4A/L6A/R7A/N8A shows almost no binding affinity (Fig. 4). The results of the EMSAs and MST assays indicate that Arg7 in the N-finger and the glycines in the α-helix of PDCoV nsp9 play important roles in nucleic acid binding. Based on the sequence alignment, the N-finger motif and GXXXG motif of the protein are highly conserved (Fig. 1D). To determine whether these motifs are actually critical for the dimerization of other reemerged coronavirus nsp9s, the PEDV nsp9 should be further investigated.
FIG 4.
The N-finger motif can affect the ssDNA binding abilities of PDCoV nsp9. (A) The ssDNA binding abilities of nsp9-WT, nsp9-ΔN7, nsp9-G96E, nsp9-G100E, nsp9-L4A, nsp9-L6A, nsp9-R7A, nsp9-N8A, and nsp9-L4A/L6A/R7A/N8A as determined by EMSA. Each protein was assayed at different concentrations (0, 25, and 50 μM) with 5 μM 5′-Cy5-labeled 36-mer ssDNA. (B) Zone-interference gel electrophoresis illustrating the association of nsp9-WT, nsp9-ΔN7, and nsp9-L4A/L6A/R7A/N8A with 36-mer ssDNA. The red dotted lines represent the position of the bands of nsp9-WT, nsp9-ΔN7 and nsp9-L4A/L6A/R7A/N8A without reaction with ssDNA. (C) The ssDNA binding affinities of nsp9-WT and mutants were measured by MST assays. Each protein was assayed in 2-fold concentration steps with 10 nM 5′-Cy5-labeled 36-mer ssDNA. The measured Kd of each protein is shown in the table.
The disulfide bond is critical for the dimerization of PEDV nsp9.
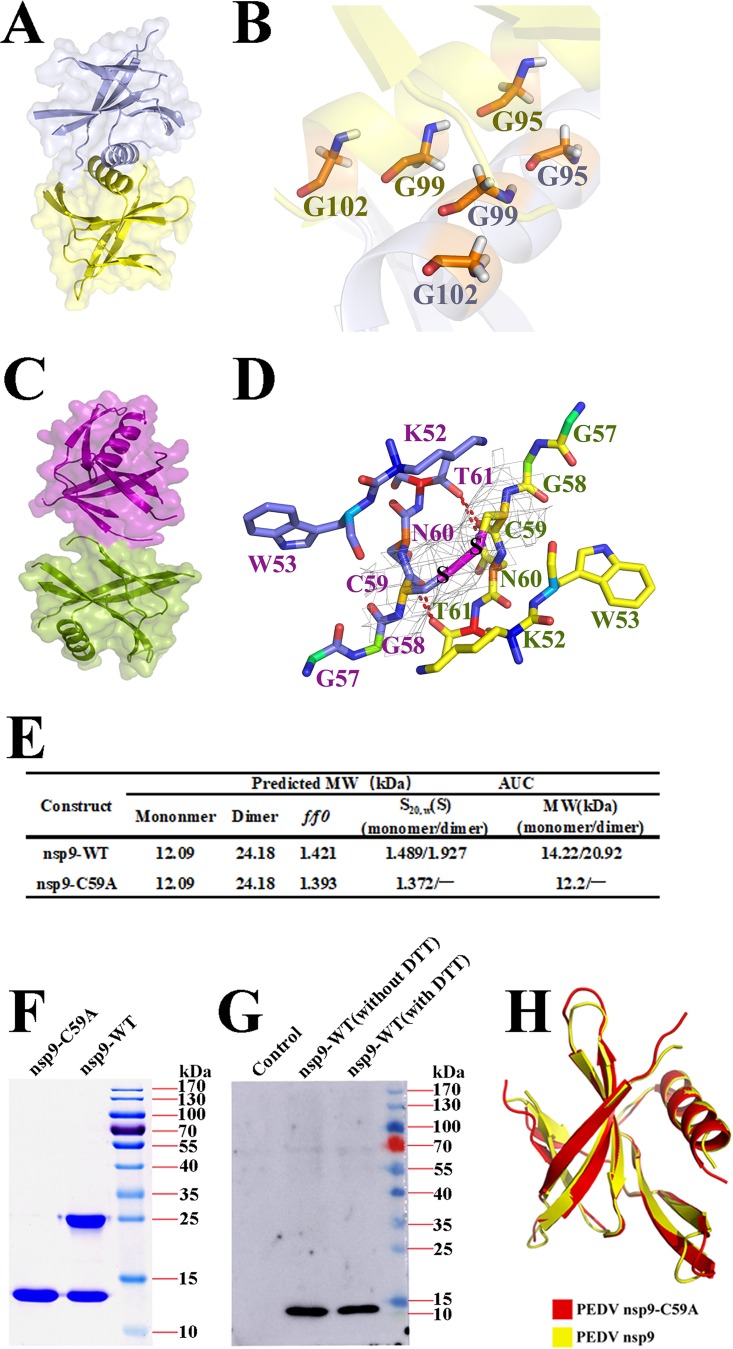
Two potential dimer interfaces are observed in the PEDV nsp9 crystal. One of the dimers observed in the crystals is formed by the parallel association of the C-terminal α-helices (Fig. 5A). The helices from the two monomers pack together closely because the heart of the dimer interface consists of three glycines (Gly95, Gly99, and Gly102), which form a hydrophobic interface and stabilize the dimer. The α-helix atoms of the two Gly95 residues show the closest distance of 3.3 Å, and the distance between the two Gly102 residues is 10.3 Å, with a very weak hydrophobic interaction (Fig. 5B). Compared with PDCoV nsp9, the N-finger of PEDV nsp9 may be flexible and cannot be built in the model.
FIG 5.
Two potential types of PEDV nsp9 dimers and the crucial role of cysteine. (A) Orthogonal views of the helix dimer. The two monomers are colored yellow and light blue and are depicted in cartoon and surface representations. (B) The heart of the dimer interface is formed from three glycines (Gly95, Gly99, and Gly102). (C) Orthogonal views of the disulfide-linked dimer. The two monomers are colored purple and split-pea and are depicted as cartoon and surface representations. (D) The disulfide bond is depicted as a magenta dotted line and the hydrogen bonds as red dotted lines, and residues located in the dimer interface are labeled in green. (E) Velocity AUC analysis of the PEDV nsp9 and nsp9-C59A mutant proteins. The molecular mass represented by each major peak is indicated, and the biophysical data are shown in the table. (F) SDS-PAGE analysis of freshly prepared proteins treated with 5× SDS-PAGE loading buffer without DTT. The differences between nsp9 and the nsp9-C59A mutant proteins were detected on polyacrylamide gels. (G) HEK293T cells were transfected with an empty vector or wild-type PEDV nsp9 plasmids. At 48 h posttransfection, the cell lysates were treated with SDS-PAGE loading buffer with or without DTT and subjected to Western blotting. (H) Illustration of the superimposition of PEDV nsp9 and PEDV nsp9-C59A monomeric structures.
Another dimer of PEDV nsp9 forms a disulfide-linked homodimer (Fig. 5C), in which the two monomers are linked by the disulfide bond formed between the Cys59 residues. Two β-strands, 4 and 5, are also involved in dimerization through the formation of two hydrogen bonds between the Cys59 side chain and the main chain of residue 61, two H-bonds between the Asn60 main chain and the side chain of residue 61, and hydrophobic interactions among residues Lys52, Gly58, Cys59, Asn60, and Thr61 (Fig. 5D).
We applied biochemical techniques to determine the oligomeric state of PEDV nsp9 in solution. We performed AUC and confirmed the monomeric and dimeric states of PEDV nsp9, with calculated molecular weights of 14.22 and 20.92 kDa, respectively. The sedimentation coefficients (S20,W) are 1.489 and 1.927, respectively, and the frictional ratio (f/f0) is 1.421 (Fig. 5E). The AUC assays show that monomers and dimers are the primary forms of PEDV nsp9 protein in solution, coexisting with a small number of higher oligomers.
To determine whether Cys59 plays important roles in nsp9 dimer formation, one mutant, namely, nsp9-C59A, was designed. Then, the AUC assay was used to analyze the oligomeric states of the mutant. The AUC results for the nsp9-C59A mutant reveal a monomeric state, with a calculated molecular weight of 12.2 kDa, a measured sedimentation coefficient (S20,W) of 1.372, and a frictional ratio (f/f0) of 1.393 (Fig. 5E). Then, PEDV nsp9 and the nsp9-C59A mutant were treated with SDS-PAGE loading buffer without DTT, a reagent that can be used to disrupt the intramolecular or intermolecular disulfide bond formed by cysteines, and SDS-PAGE was performed (Fig. 5F). We used freshly prepared PEDV nsp9 protein from Escherichia coli to perform the AUC assay and SDS-PAGE, which have proved the disulfide bond may be formed in E. coli and indicated that the PEDV nsp9 dimer has a disulfide bond and Cys59 is an important residue for PEDV nsp9 dimer formation. In addition, to determine whether the PEDV nsp9 protein has the disulfide bond in mammalian cells, HEK293T cells were transfected with an empty vector or wild-type PEDV nsp9 plasmids. At 48 h posttransfection, cells were treated with radioimmunoprecipitation assay (RIPA) lysis buffer. Then the lysates were treated with SDS-PAGE loading buffer with or without dithiothreitol (DTT) and immunoblotting was performed. The results show that the PEDV nsp9 is monomer in cells and indicate that the disulfide-bonded nsp9 dimer might be an artifact in the infected cells (Fig. 5G).
To further validate the importance of the disulfide bond in dimer formation, we analyzed the structure of the nsp9-C59A mutant. The crystals belong to space group P43212. The structure is determined by molecular replacement and refined to a 3.0-Å resolution (Table 1). The crystal structure of the nsp9-C59A mutant monomer also shows 7 β-strands and an α-helix appended at the C terminus (Fig. 5H). Neither a disulfide bond nor any analogous dimer interface to that formed by the disulfide bond is observed in the structure. The structural superposition of the wild-type nsp9 monomer with the nsp9-C59A mutant monomer yields 94 Cα atoms with an RMSD of 0.2 Å (Fig. 5H).
Taken together, the crystal structure and biochemical assays reveal the crucial role of Cys59 in PEDV nsp9 dimer formation, and the disulfide-linked homodimer is the primary form of the PEDV nsp9 dimer. However, PEDV nsp9 is a monomer in cells, and the disulfide-bonded nsp9 dimer might be an artifact in the infected cells.
The importance of dimerization-related amino acids in nucleic acid binding of PEDV nsp9.
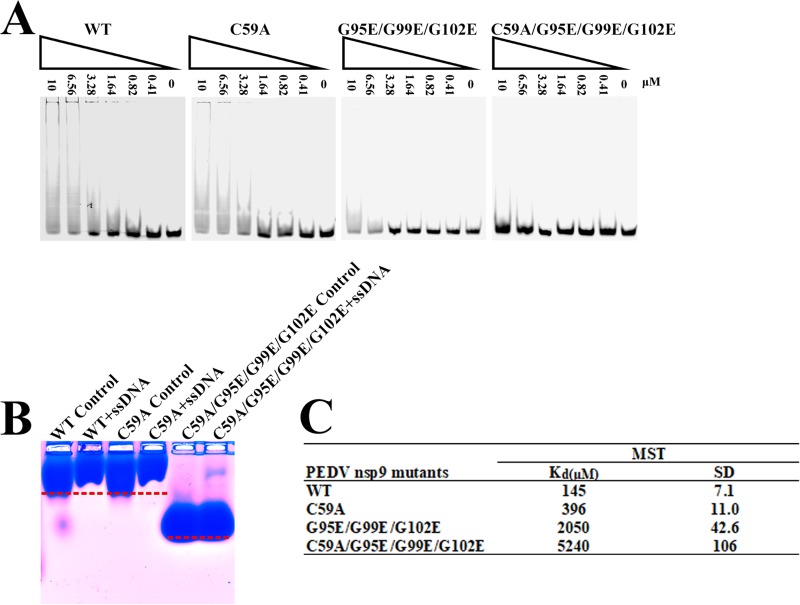
To test whether Cys59 and the glycines in the α-helix (Gly95, Gly99, and Gly102) are related to nucleic acid binding of PEDV nsp9, we designed three mutants: nsp9-C59A, nsp9-G95E/G99E/G102E, and nsp9-C59A/G95E/G99E/G102E. Two EMSAs were performed to examine the nucleic acid binding affinity of each mutant. The ssDNA binding affinity of the nsp9-C59A mutant is somewhat weaker than that of the wild-type nsp9 protein, whereas the nsp9-G95E/G99E/G102E mutant shows very weak binding affinity, and the nsp9-C59A/G95E/G99E/G102E mutant shows almost no binding affinity (Fig. 6A and B). Furthermore, MST assays were performed to obtain quantitative measurements of the binding affinities. The results show that the wild-type PEDV nsp9 protein bound to ssDNA with a Kd of 145 μM, the nsp9-C59A mutant shows a 2.7-fold reduction in ssDNA binding, the nsp9-G95E/G99E/G102E mutant shows a 14-fold reduction, and the nsp9-C59A/G95E/G99E/G102E mutant shows much weaker binding affinity, with a 36-fold reduction (Fig. 6C). The results of the EMSAs and MST assays indicate that Cys59 and the glycines in the α-helix of PEDV nsp9 play important roles in nucleic acid binding.
FIG 6.
The disulfide bond can affect the ssDNA binding abilities of PEDV nsp9. (A) The ssDNA binding abilities of nsp9-WT, nsp9-C59A, nsp9-G95E/G99E/G102E, and nsp9-C59A/G95E/G99E/G102E as determined by EMSA. Each protein was assayed at different concentrations (0, 0.41, 0.82, 1.64, 3.28, 6.56, and 10 μM) with 5 μM 5′-Cy5-labeled 36-mer ssDNA. (B) Zone-interference gel electrophoresis illustrating the association of nsp9-WT, nsp9-C59A, and nsp9- C59A/G95E/G99E/G102E with 36-mer ssDNA. The red dotted lines represent the position of the bands of nsp9-WT, nsp9-C59A, and nsp9-C59A/G95E/G99E/G102E without reaction with ssDNA. (C) The ssDNA binding affinities of nsp9-WT, nsp9-C59A, nsp9-G95E/G99E/G102E, and nsp9-C59A/G95E/G99E/G102E were measured by MST assays. Each protein was assayed in 2-fold concentration steps with 10 nM 5′-Cy5-labeled 36-mer ssDNA. The measured Kd of each protein is shown in the table.
The importance of positively charged amino acids in nucleic acid binding of PEDV nsp9.
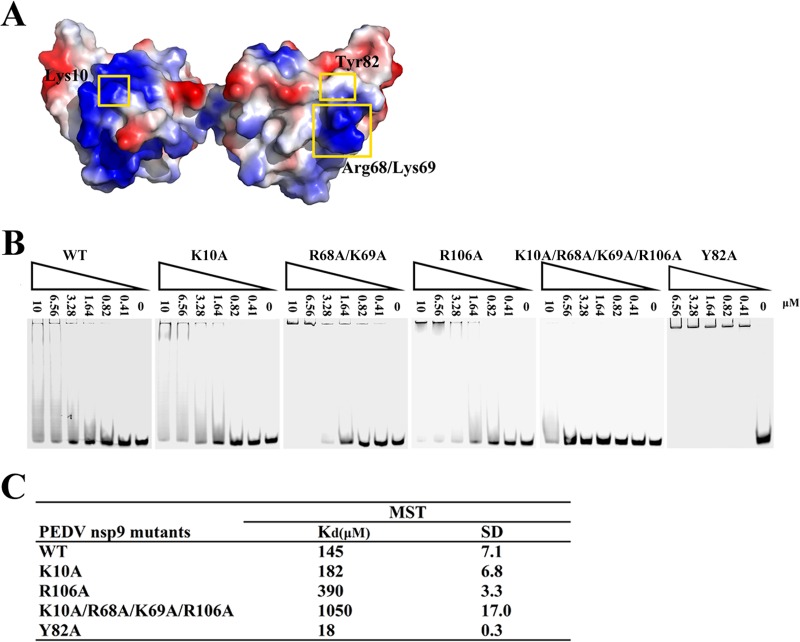
The surface electrostatic potential analysis of PEDV nsp9 reveals a highly positively charged patch suggestive of a potential site for nucleic acid binding (Fig. 7A). To determine whether this surface contributes to the ssDNA binding activity of PEDV nsp9, we introduced individual substitutions of positively charged residues, and their effects on ssDNA binding were assessed by EMSA and MST assays. In this study, residues Lys10, Arg68, Lys69, and Arg106 were mutated to alanine because of their highly positively charged potential and their exposure on the surface of the nsp9 structure. One aromatic amino acid, Tyr82, was also mutated to alanine (Fig. 7A). The EMSA results show that the nsp9-K10A, nsp9-R68A/K69A, and nsp9-R106A mutants show slightly weaker ssDNA binding activity, whereas the nsp9-K10A/R68A/K69A/R106A mutant reveals a significant loss of ssDNA binding affinity, and the nsp9-Y82A mutant shows clearly enhanced ssDNA binding affinity (Fig. 7B). The MST assay indicates that the nsp9-K10A and nsp9-R106A mutants have 1.3- and 2.7-fold reductions in ssDNA binding affinity compared to the wild-type protein, respectively, whereas the nsp9-K10A/R68A/K69A/R106A mutant shows much weaker binding affinity, with a 7.2-fold reduction, and nsp9-Y82A shows much stronger binding affinity, with an 8.0-fold increase (Fig. 7C). The ssDNA binding activity of the nsp9-R68A/K69A mutant could not be measured by the MST assay because of the protein's sedimentation characteristics. The results of the EMSA and MST assay indicate that the positively charged surface of the PEDV nsp9 does indeed contribute to the ssDNA interaction.
FIG 7.
Impact of positively charged amino acid substitutions on ssDNA binding of PEDV nsp9. (A) Depictions of the electrostatic potential surface of the PEDV nsp9 dimer created by APBS tools. The mutated amino acids exposed on the protein surface are labeled. (B) The ssDNA binding abilities of nsp9-WT, nsp9-K10A, nsp9-R68A, K69A, nsp9-R106A, nsp9-K10A/R68A/K69A/R106A, and nsp9-Y82A as determined by EMSA. Each protein was assayed at different concentrations (0, 0.41, 0.82, 1.64, 3.28, 6.56, and 10 μM) with 5 μM 5′-Cy5-labeled 36-mer ssDNA. (C) The ssDNA binding affinities of nsp9-WT, nsp9-K10A, nsp9-R106A, nsp9-K10A/R68A/K69A/R106A, and nsp9-Y82A as measured by MST assays. Each protein was used in 2-fold concentration steps with 10 nM 5′-Cy5-labeled 36-mer ssDNA. The measured Kd of each protein is shown in the table.
DISCUSSION
Diverse dimerization of coronavirus nsp9.
In the emerging CoV PDCoV, we first confirm that the N-finger motif is critical for dimerization. Moreover, the dimerization of PDCoV nsp9 is also supported by the parallel association of the C-terminal α-helix via strong hydrophobic interactions. Although the monomeric structures of nsp9 from PDCoV exhibited significant similarity to those from other CoVs, the dimerization modes showed high diversity. Infectious bronchitis virus (IBV) nsp9 forms a homodimer via interactions across a hydrophobic interface, which consists of two parallel α-helixes near the carboxy terminus of the protein (25). For the SARS-CoV nsp9 protein, the N-terminal extended β chain and the C-terminal α-helices stabilize its interface (24). The N-terminal extended β chain of SARS nsp9 is supported by the extended tag, which may lead to the unclear function of its N-finger motif. Here, SARS-CoV nsp9-ΔN7 mutant is monomeric in solution, which is consistent with wild-type PDCoV nsp9-ΔN7 (data not shown). The N and C termini of the protein are more conserved than the central core region, and the GXXXG motif is strictly conserved (24, 26). In other proteins, mutation of the Gly residues in the motif led to the decreased or the complete loss of dimerization of transmembrane helices (21, 27, 28). The GXXXG motif of nsp9 is important for maintaining the dimer form. PDCoV nsp9 ΔN7 without the N-finger forms the monomeric stage. We conclude that the N-finger motif and the GXXXG motif are both important for dimerization of PDCoV nsp9.
In the dimer interface of PEDV nsp9, similar to HCoV-229E nsp9, there is a disulfide bond. However, the cysteine is located at a different position, resulting in a very different dimer conformation. PEDV nsp9 primary forms a disulfide-linked homodimer, in which the two monomers are linked by the disulfide bond formed between the Cys59 residues. For HCoV-229E nsp9, a disulfide bond is involved in dimerization (23), which is consistent with our results. However, the weak effect of the C59A mutation on nucleic acid binding indicate that dimerization of PEDV nsp9 by disulfide formation may not be a major form in the infected cells. Moreover, the overall milieu is reductive, and disulfide bonds are rare in the cytoplasm of the infected cell (29). The biological function of the disulfide bond should be further investigated by a reverse genetics system.
Nucleic acid binding by nsp9.
The crystal structures of CoV nsp9s share great similarity in the oligosaccharide-binding fold, which is characteristic of proteins that bind to ssDNA or ssRNA (30). Our data show that PDCoV nsp9 and PEDV nsp9 both have binding activity with nucleic acids, which is consistent with other CoV nsp9 proteins (21, 23, 25). In addition, the dimeric state of the PDCoV nsp9 and PEDV nsp9 can increase the binding activity with nucleic acids. Our EMSA and MST experiments also suggest that the N-finger of PDCoV nsp9 can influence the affinity to nucleic acids. In a previous study, the SARS-CoV nsp9 dimer was shown to be required for viral growth (21). The nsp9 dimerization is critical for the function of this replicase protein for the IBV infectious clone system (31). We hypothesize that the dimer of PDCoV nsp9 and PEDV nsp9 may be critical for viral replication. Whether the N-finger of PDCoV nsp9 or the cysteine of PEDV nsp9 affects viral replication should be further studied by using reverse genetic approaches.
Nucleotides interact with proteins primarily via aromatic amino acid side chains, hydrophobic side chains or the aliphatic portions of polar and positively charged groups, such as lysine and arginine (30). A previous study of SARS-CoV nsp9 suggested that the most likely site of RNA binding is on the loops L23, L45, and L7H1, since this face is accessible in the preferred helix-stabilized dimer but largely occluded in the putative β sheet-stabilized dimer (24). Based on the structure of SARS-CoV nsp9, we observe that positively charged amino acids are present on the loops. In our study, the PEDV nsp9-K10A, nsp9-R68A/K69A, and nsp9-R106A mutants mildly affect ssDNA binding activity, whereas the nsp9-K10A/R68A/K69A/R106A mutant reveals a significant reduction in ssDNA binding (Fig. 7B and C), which confirmed the importance of highly positively charged residues exposed on the protein surface in ssDNA binding. Many amino acids in a single protein may participate in ssDNA binding, and it is thus reasonable that a single-residue mutation could have a moderate effect on ssDNA binding activity. We hypothesize that the potential site of RNA binding is on the face of PEDV nsp9 present on strands β1 and β6 (Fig. 1B). Surprisingly, the nsp9-Y82A mutant strongly enhances the ssDNA binding affinity (Fig. 7B and C), and L6A and N8A of PDCoV nsp9 unexpectedly induced an enhancement of the ssDNA binding affinity, which may be caused by decreasing steric hindrance. Whether the nsp9-Y82A mutant affects viral replication should be further investigated. In addition, we have attempted to solve the structure of the nsp9-RNA or DNA complexes. Unfortunately, we were unable to obtain the crystals of the nsp9-RNA or DNA complexes.
Nsp9 is able to bind ssDNA or ssRNA (22, 25), although binding of ssRNA is expected to be the native function. In the infected cell, the coupling/compartmentation of the viral RNA synthesis with the RNA-binding function of nsp9 might render RNA versus DNA specificity unnecessary (22). Moreover, as a member of the replication complex, nsp9 may not have a specific binding sequence but may act in conjunction with other nsp's as a processivity factor (21). According to a model for the initiation of coronavirus negative-strand RNA synthesis, a protein complex (including nsp8 and nsp9) binds to viral RNA initially (32). The potential site of RNA binding may provide a new insight into the binding of the replication/transcription complexes to viral RNA.
CoV nsp9 has diverse dimerization, while its function of enhancement nuclear binding affinity is conservative (22–25). We propose that most nsp9 proteins originated from ancestral coronavirus, which have N-finger motifs and GXXXG motifs that both play critical roles in dimerization. The N-terminal loops and GXXXG motifs of PDCoV, IBV, and SARS-CoV nsp9 are also involved in dimerization, while the extra amino acids (the residues of fusion tag) in the N-finger may block the fully function of the N-finger (22, 24, 25). Interestingly, PEDV and HCoV-229E evolve different disulfide bonds to enhance the dimerization (23). Using diverse dimerization strategies, nsp9 might increase the nucleic acid binding interface and then promote its nucleic acid binding affinity, which might stabilize nascent viral RNAs during replication or transcription, thus providing protection from nucleases (22, 24).
In summary, CoV nsp9s have diverse forms of dimerization that promote their biological function, which may help elucidate the mechanism underlying CoVs replication and contribute to the development of antiviral drugs.
MATERIALS AND METHODS
Protein production and crystallization.
The full-length nsp9 genes of PDCoV and PEDV were amplified by PCR from virus-derived cDNA fragments and cloned separately into pET-42b. Both nsp9s were expressed with a C-terminal His6 tag in E. coli BL21(DE3). High-quality soluble PDCoV nsp9 protein and PEDV nsp9 protein were obtained by adding 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) to Luria broth when the bacteria had grown to an optical density at 600 nm of approximately 0.6 at 37°C, followed by shaking at 37°C for 4 h and at 27°C for 10 h, respectively. For protein purification, the E. coli cells were harvested, resuspended with phosphate-buffered saline (PBS), and lysed via passage through an AH-1500 homogenizer. After centrifugation at 8,500 rpm for 40 min, the supernatant was filtered and loaded onto a HisTrap HP column (GE Healthcare). The PDCoV nsp9-ΔN7, PEDV nsp9-C59A, and other nsp9 mutants were expressed and purified as described above.
Crystallization screening was performed by the sitting drop vapor diffusion method at 20°C. PDCoV nsp9 (5.0 mg/ml) was mixed at a 1:1 ratio with crystallization cocktails. Wild-type protein crystals and Se-Met-labeled nsp9 crystals appeared within 3 days at 20°C in 2.0 M sodium chloride and 0.1 M citric acid (pH 3.5). The PDCoV nsp9-ΔN7 mutant protein crystals appeared within 3 days at 20°C in 0.2 M sodium chloride, 0.1 M Na2HPO4-citric acid (pH 4.2), and 20% (wt/vol) PEG 8000. After optimization, the wild-type crystals or Se-Met-labeled nsp9 crystals were stabilized in 3.0 M sodium chloride and 0.1 M citric acid (pH 3.5), and the nsp9-ΔN7 crystal was stabilized in 0.1 M Na2HPO4-citric acid (pH 4.2) and 22% (wt/vol) PEG 8000. Meanwhile, PEDV nsp9 (6.5 mg/ml) was mixed at a 1:1 ratio with crystallization cocktails. Wild-type protein crystals appeared within 40 days at 20°C in 0.1 M Tris (pH 8.5) and 25% (vol/vol) tert-butanol, and PEDV nsp9-C59A mutant protein crystals appeared within 2 days at 20°C in 0.2 M sodium thiocyanate (pH 6.9) and 12% PEG 3350. After optimization, the wild-type crystals were stabilized in 0.3 M Tris (pH 8.5) and 13% (vol/vol) tert-butanol, and the nsp9-C59A mutant crystals were stabilized in 0.25 M sodium thiocyanate (pH 7.7), 10% PEG 3350, and 2% ethylene glycol. There was no reductant (DTT) in the crystallization setup.
Data collection and structure determination.
X-ray diffraction data were collected at beamline BL17U at the Shanghai Synchrotron Radiation Facility. The structure of PDCoV nsp9 was solved using the single-wavelength anomalous dispersion method and a Se-Met-derivative nsp9 wild-type protein. Both potential selenium atoms in the nsp9 monomer were located, and the initial phases were calculated using the AutoSol program from the PHENIX software suite (33). The molecular replacement program HKL-3000 (34) was used to solve the structure of PEDV nsp9, with a monomer of HCoV-229E nsp9 (PDB 2J97) as the search model. Manual model rebuilding was performed using COOT (35) and then refined in the PHENIX software suite. The figures were created using PyMOL (Schrödinger).
Structural analysis and sequence alignment.
Detailed molecular interactions between the two monomers of nsp9 were determined using LIGPLOT (36), and the other structure figures were generated using PyMOL (Schrödinger). The buried surface areas between the two monomers and the RMSD were analyzed using PDBePISA (http://pdbe.org/pisa/) and PDBeFold (http://pdbe.org/fold/), respectively. In addition, the amino acid sequences of coronavirus nsp9 were aligned using ClustalW2 (37) and visualized with the ESPript 3 server (http://espript.ibcp.fr) (38). The analyzed viruses (definition; NCBI accession numbers) were as follows: PEDV (porcine epidemic diarrhea virus; AHZ94880), PDCoV (porcine delta coronavirus; APG38197), TGEV (transmissible gastroenteritis virus; AML22776), HCoV-229E (human coronavirus 229E; NP_073550), SARS-CoV (SARS coronavirus Shanhgai LY; AAP82976), MERS-CoV (middle east respiratory syndrome-related coronavirus; AHI48749), MHV (murine hepatitis virus strain A59; ACO72881), and IBV (infectious bronchitis virus; ACJ12833).
Size exclusion chromatography.
Oligomerization of wild-type (0.1 mg) and mutant (ΔN7 and L4A/L6A/R7A/N8A) (0.1 mg) PDCoV nsp9 proteins was analyzed using a Superdex 75 10/300GL column (GE Healthcare) with a buffer containing 20 mM Tris-HCl (pH 7.4) and 200 mM NaCl at a flow rate of 0.6 ml/min (4°C). Wild-type and mutant (ΔN7 and L4A/L6A/R7A/N8A) nsp9 proteins eluted in different fractions were analyzed by SDS-PAGE.
Immunoblotting.
To obtain high expression in eukaryotic cells, PEDV nsp9 wild-type, flanked by an C-terminal hemagglutinin (HA) tag, was cloned into pCAGGS vector using the EcoRI and XhoI restriction sites.
HEK293T cells cultured in 6-well plates (Corning, Tewksbury, MA) were transfected with an empty vector or wild-type PEDV nsp9 plasmids (2.5 μg) using Lipofectamine 2000. At 48 h posttransfection, cells were treated with RIPA lysis buffer (Beyotime). The lysates were then treated with SDS-PAGE loading buffer with or without DTT. For the immunoblot analysis, 10 μl of lysate was electrophoresed on 12% SDS-PAGE gels (Bio-Rad). Proteins were transferred to polyvinylidene difluoride membranes (Bio-Rad) according to the manufacturer's recommendations. Membranes were incubated in blocking buffer (5% nonfat dried milk in TBS-Tween [50 mM Tris, pH 7.6; 150 mM NaCl]) for 3 h at room temperature. Anti-HA antibody (Ab; MBL) was diluted 1:10,000 in blocking buffer and incubated with the membrane for 3 h at room temperature. Membranes were washed three times for 15 min at room temperature with TBS-Tween. Goat anti-mouse horseradish peroxidase-conjugated antibodies (BOSTER) were diluted 1:5,000 in blocking buffer, followed by incubation with the membrane for 1 h at room temperature. Membranes were washed three times for 15 min at room temperature with TBS-Tween. The membranes were then visualized using an enhanced chemiluminescence system (Amersham Imager 600; GE Healthcare).
Analytical ultracentrifugation.
Sedimentation velocity was assessed with an XL-I analytical centrifuge (Proteome Lab, Huazhong Agricultural University, Wuhan) at 60,000 rpm and 18°C in 400 μl of double-sector cells for sedimentation analytical ultracentrifugation. The sedimentation boundary was monitored every minute at a wavelength of 280 nm. Samples were at a concentration of 1 mg/ml in 20 mM Tris–200 mM NaCl (pH 7.4) buffer. Data were interpreted with the model-based distribution of Lamm equation solutions c(s) using the software Sedfit (39). Weight-averaged molar masses were calculated using Vector NTI software.
Electrophoretic mobility shift assay.
An electrophoretic mobility shift assay was performed as described previously (40). A 5′-Cy5-labeled 36-mer ssDNA oligonucleotide (TAGTACCGCCACCCTCAGAACCTTTTTTTTTTTTTT; synthesized by Sangon Biotech, Shanghai, China) was used for the gel shift assay (22). Initially, 5 μM ssDNA was incubated with different concentrations of PDCoV and PEDV nsp9s and the mutant proteins in 10 mM Tris (pH 8.0) and 200 mM NaCl for 45 min at room temperature, followed by the addition of 10% glycerol to the mixture. Samples were then run on 6.5% nondenaturing Tris-borate-EDTA polyacrylamide gels for 37 min at a voltage of 180 V, and the results were determined with an FLA-2000 fluorescent image analyzer (Fuji, Stamford, CT).
Zone-interference gel electrophoresis was described in the publication by Ponnusamy et al. (23). The ssDNA was dissolved at a concentration of 2 nmol before loading onto a 1% agarose gel. The gel was run for 50 min with running buffer (20 mM Tris, 50 mM boric acid, 1 mM sodium EDTA [pH 8.3]) at 100 mA and 4°C. With the poles of the electrodes interchanged, 1 nmol of nsp9 protein was loaded and run for another 50 min. After electrophoresis, the gel was fixed in 3.5% (wt/vol) α-sulfosalicylic acid–10% (wt/vol) trichloroacetic acid until the dye turned yellow. For the detection of protein bands, the gel was washed for 15 min in 15% (vol/vol) ethanol–8% (vol/vol) acetic acid and stained for 30 min with 0.25% (wt/vol) Coomassie brilliant blue in the same solution with an additional 10% (vol/vol) methanol. The gel was washed in 15% (vol/vol) ethanol–8% (vol/vol) acetic acid and stored in 10% (vol/vol) acetic acid.
Microscale thermophoresis assay.
Specific binding between nsp9 or the nsp9 mutant proteins and ssDNA was measured by MST assays as previously described (41, 42). Briefly, nsp9 or nsp9 mutant proteins were serially diluted with buffer containing 10 mM Tris (pH 8.0) and 200 mM NaCl, and then 10 nM Cy5-labeled ssDNA diluted with buffer containing 10 mM Tris (pH 8.0)–200 mM NaCl and 0.05% Tween 20 was added. The mixtures were loaded into NT.115 standard capillaries (Nanotemper Technologies), and thermophoresis was carried out at room temperature, 100% light-emitting diode (LED) power, and 20% IR-laser power with a Monolith NT.115 (Nanotemper Technologies). The data were analyzed using Nanotemper Analysis software v.1.2.101.
Accession number(s).
The atomic coordinates of wild-type PDCoV nsp9, PDCoV nsp9-ΔN7, wild-type PEDV nsp9, and the nsp9-C59A mutant have been deposited in the RCSB Protein Data Bank under accession codes 5YM6, 5YM8, 5HIZ, and 5HIY, respectively.
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (grant 31722056), the National Key R&D Plan of China (program no. 2016YFD0500103), and the Huazhong Agricultural University Scientific and Technological Self-Innovation Foundation (program no. 2662015JQ003 and 2662017PY028).
We thank the staff at the SSRF BL17U1 beamline for assistance with X-ray data collection. Moreover, we also thank research associates at the Center for Protein Research (CPR), Huazhong Agricultural University, for technical support.
REFERENCES
- 1.Woo PC, Lau SK, Huang Y, Yuen KY. 2009. Coronavirus diversity, phylogeny and interspecies jumping. Exp Biol Med (Maywood, NJ) 234:1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- 2.Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, Lau JH, Bai R, Teng JL, Tsang CC, Wang M, Zheng BJ, Chan KH, Yuen KY. 2012. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol 86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan JF, To KK, Tse H, Jin DY, Yuen KY. 2013. Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol 21:544–555. doi: 10.1016/j.tim.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drosten C, Gunther S, Preiser W, van der Werf S, Brodt HR, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RA, Berger A, Burguiere AM, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra JC, Muller S, Rickerts V, Sturmer M, Vieth S, Klenk HD, Osterhaus AD, Schmitz H, Doerr HW. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 5.Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling AE, Humphrey CD, Shieh WJ, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang JY, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ, SARS Working Group. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 6.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 7.Mai K, Feng J, Chen G, Li D, Zhou L, Bai Y, Wu Q, Ma J. 2017. The detection and phylogenetic analysis of porcine deltacoronavirus from Guangdong Province in Southern China. Transbound Emerg Dis 65:166–173. doi: 10.1111/tbed.12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saeng-Chuto K, Lorsirigool A, Temeeyasen G, Vui DT, Stott CJ, Madapong A, Tripipat T, Wegner M, Intrakamhaeng M, Chongcharoen W, Tantituvanont A, Kaewprommal P, Piriyapongsa J, Nilubol D. 2017. Different lineage of porcine deltacoronavirus in Thailand, Vietnam, and Lao PDR in 2015. Transbound Emerg Dis 64:3–10. doi: 10.1111/tbed.12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scott A, McCluskey B, Brown-Reid M, Grear D, Pitcher P, Ramos G, Spencer D, Singrey A. 2016. Porcine epidemic diarrhea virus introduction into the United States: root cause investigation. Prev Vet Med 123:192–201. doi: 10.1016/j.prevetmed.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thiel V, Ivanov KA, Putics A, Hertzig T, Schelle B, Bayer S, Weissbrich B, Snijder EJ, Rabenau H, Doerr HW, Gorbalenya AE, Ziebuhr J. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J Gen Virol 84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 11.Liu DX, Tibbles KW, Cavanagh D, Brown TD, Brierley I. 1995. Identification, expression, and processing of an 87-kDa polypeptide encoded by ORF 1a of the coronavirus infectious bronchitis virus. Virology 208:48–57. doi: 10.1006/viro.1995.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim KP, Ng LF, Liu DX. 2000. Identification of a novel cleavage activity of the first papain-like proteinase domain encoded by open reading frame 1a of the coronavirus Avian infectious bronchitis virus and characterization of the cleavage products. J Virol 74:1674–1685. doi: 10.1128/JVI.74.4.1674-1685.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai MM, Cavanagh D. 1997. The molecular biology of coronaviruses. Adv Virus Res 48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herold J, Gorbalenya AE, Thiel V, Schelle B, Siddell SG. 1998. Proteolytic processing at the amino terminus of human coronavirus 229E gene 1-encoded polyproteins: identification of a papain-like proteinase and its substrate. J Virol 72:910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graham RL, Sims AC, Brockway SM, Baric RS, Denison MR. 2005. The nsp2 replicase proteins of murine hepatitis virus and severe acute respiratory syndrome coronavirus are dispensable for viral replication. J Virol 79:13399–13411. doi: 10.1128/JVI.79.21.13399-13411.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hurst-Hess KR, Kuo L, Masters PS. 2015. Dissection of amino-terminal functional domains of murine coronavirus nonstructural protein 3. J Virol 89:6033–6047. doi: 10.1128/JVI.00197-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neuman BW, Chamberlain P, Bowden F, Joseph J. 2014. Atlas of coronavirus replicase structure. Virus Res 194:49–66. doi: 10.1016/j.virusres.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bost AG, Carnahan RH, Lu XT, Denison MR. 2000. Four proteins processed from the replicase gene polyprotein of mouse hepatitis virus colocalize in the cell periphery and adjacent to sites of virion assembly. J Virol 74:3379–3387. doi: 10.1128/JVI.74.7.3379-3387.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deming DJ, Graham RL, Denison MR, Baric RS. 2007. Processing of open reading frame 1a replicase proteins nsp7 to nsp10 in murine hepatitis virus strain A59 replication. J Virol 81:10280–10291. doi: 10.1128/JVI.00017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frieman M, Yount B, Agnihothram S, Page C, Donaldson E, Roberts A, Vogel L, Woodruff B, Scorpio D, Subbarao K, Baric RS. 2012. Molecular determinants of severe acute respiratory syndrome coronavirus pathogenesis and virulence in young and aged mouse models of human disease. J Virol 86:884–897. doi: 10.1128/JVI.05957-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miknis ZJ, Donaldson EF, Umland TC, Rimmer RA, Baric RS, Schultz LW. 2009. Severe acute respiratory syndrome coronavirus nsp9 dimerization is essential for efficient viral growth. J Virol 83:3007–3018. doi: 10.1128/JVI.01505-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Egloff MP, Ferron F, Campanacci V, Longhi S, Rancurel C, Dutartre H, Snijder EJ, Gorbalenya AE, Cambillau C, Canard B. 2004. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc Natl Acad Sci U S A 101:3792–3796. doi: 10.1073/pnas.0307877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ponnusamy R, Moll R, Weimar T, Mesters JR, Hilgenfeld R. 2008. Variable oligomerization modes in coronavirus nonstructural protein 9. J Mol Biol 383:1081–1096. doi: 10.1016/j.jmb.2008.07.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sutton G, Fry E, Carter L, Sainsbury S, Walter T, Nettleship J, Berrow N, Owens R, Gilbert R, Davidson A, Siddell S, Poon LL, Diprose J, Alderton D, Walsh M, Grimes JM, Stuart DI. 2004. The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure 12:341–353. doi: 10.1016/j.str.2004.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu T, Chen C, Li H, Dou Y, Zhou M, Lu D, Zong Q, Li Y, Yang C, Zhong Z, Singh N, Hu H, Zhang R, Yang H, Su D. 2017. Structural basis for dimerization and RNA binding of avian infectious bronchitis virus nsp9. Protein Sci 26:1037–1048. doi: 10.1002/pro.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gimpelev M, Forrest LR, Murray D, Honig B. 2004. Helical packing patterns in membrane and soluble proteins. Biophys J 87:4075–4086. doi: 10.1529/biophysj.104.049288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munter LM, Voigt P, Harmeier A, Kaden D, Gottschalk KE, Weise C, Pipkorn R, Schaefer M, Langosch D, Multhaup G. 2007. GxxxG motifs within the amyloid precursor protein transmembrane sequence are critical for the etiology of Abeta42. EMBO J 26:1702–1712. doi: 10.1038/sj.emboj.7601616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneider D, Engelman DM. 2004. Motifs of two small residues can assist but are not sufficient to mediate transmembrane helix interactions. J Mol Biol 343:799–804. doi: 10.1016/j.jmb.2004.08.083. [DOI] [PubMed] [Google Scholar]
- 29.Bessette PH, Aslund F, Beckwith J, Georgiou G. 1999. Efficient folding of proteins with multiple disulfide bonds in the Escherichia coli cytoplasm. Proc Natl Acad Sci U S A 96:13703–13708. doi: 10.1073/pnas.96.24.13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Theobald DL, Mitton-Fry RM, Wuttke DS. 2003. Nucleic acid recognition by OB-fold proteins. Annu Rev Biophys Biomol Struct 32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen B, Fang S, Tam JP, Liu DX. 2009. Formation of stable homodimer via the C-terminal alpha-helical domain of coronavirus nonstructural protein 9 is critical for its function in viral replication. Virology 383:328–337. doi: 10.1016/j.virol.2008.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sola I, Almazan F, Zuniga S, Enjuanes L. 2015. Continuous and discontinuous RNA synthesis in coronaviruses. Annu Rev Virol 2:265–288. doi: 10.1146/annurev-virology-100114-055218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58:1948–1954. doi: 10.1107/S0907444902016657. [DOI] [PubMed] [Google Scholar]
- 34.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. 2006. HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr 62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 35.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.Laskowski RA, Swindells MB. 2011. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Infect Model 51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 37.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 38.Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schuck P. 2000. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys J 78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Min B, Collins K. 2010. Multiple mechanisms for elongation processivity within the reconstituted tetrahymena telomerase holoenzyme. J Biol Chem 285:16434–16443. doi: 10.1074/jbc.M110.119172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh S, Katzer K, Lambert J, Cerri M, Parniske M. 2014. CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe 15:139–152. doi: 10.1016/j.chom.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 42.Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S. 2010. Protein-binding assays in biological liquids using microscale thermophoresis. Nat Commun 1:100. doi: 10.1038/ncomms1093. [DOI] [PubMed] [Google Scholar]
- 43.Zacharias J, Knapp EW. 2014. Protein secondary structure classification revisited: processing DSSP information with PSSC. J Chem Infect Model 54:2166–2179. doi: 10.1021/ci5000856. [DOI] [PubMed] [Google Scholar]