Herpes simplex virus (HSV) causes infection of the mouth, skin, eyes, and genitals and establishes lifelong latency in humans. gB is conserved among all herpesviruses. HSV gB undergoes reversible conformational changes following exposure to acidic pH which are thought to mediate fusion and entry into epithelial cells. Here, we identified cotranslational folding and oligomerization of newly synthesized gB. A panel of antibodies to gB blocked both low-pH and pH-neutral entry of HSV, suggesting conserved conformational changes in gB regardless of cell entry route. Changes in HSV gB conformation were not triggered by increased temperature alone, in contrast to results with EBV gB. Acid pH-induced changes in the oligomeric conformation of gB are related but distinct from pH-triggered changes in gB antigenic conformation. These results highlight critical aspects of the class III fusion protein, gB, and inform strategies to block HSV infection at the level of fusion and entry.
KEYWORDS: conformational changes, glycoprotein B, herpes simplex virus, herpesviruses, membrane fusion, viral entry
ABSTRACT
Herpes simplex virus (HSV) is an important human pathogen with a high worldwide seroprevalence. HSV enters epithelial cells, the primary site of infection, by a low-pH pathway. HSV glycoprotein B (gB) undergoes low pH-induced conformational changes, which are thought to drive membrane fusion. When neutralized back to physiological pH, these changes become reversible. Here, HSV-infected cells were subjected to short pulses of radiolabeling, followed by immunoprecipitation with a panel of gB monoclonal antibodies (MAbs), demonstrating that gB folds and oligomerizes rapidly and cotranslationally in the endoplasmic reticulum. Full-length gB from transfected cells underwent low-pH-triggered changes in oligomeric conformation in the absence of other viral proteins. MAbs to gB neutralized HSV entry into cells regardless of the pH dependence of the entry pathway, suggesting a conservation of gB function in distinct fusion mechanisms. The combination of heat and acidic pH triggered irreversible changes in the antigenic conformation of the gB fusion domain, while changes in the gB oligomer remained reversible. An elevated temperature alone was not sufficient to induce gB conformational change. Together, these results shed light on the conformation and function of the HSV-1 gB oligomer, which serves as part of the core fusion machinery during viral entry.
IMPORTANCE Herpes simplex virus (HSV) causes infection of the mouth, skin, eyes, and genitals and establishes lifelong latency in humans. gB is conserved among all herpesviruses. HSV gB undergoes reversible conformational changes following exposure to acidic pH which are thought to mediate fusion and entry into epithelial cells. Here, we identified cotranslational folding and oligomerization of newly synthesized gB. A panel of antibodies to gB blocked both low-pH and pH-neutral entry of HSV, suggesting conserved conformational changes in gB regardless of cell entry route. Changes in HSV gB conformation were not triggered by increased temperature alone, in contrast to results with EBV gB. Acid pH-induced changes in the oligomeric conformation of gB are related but distinct from pH-triggered changes in gB antigenic conformation. These results highlight critical aspects of the class III fusion protein, gB, and inform strategies to block HSV infection at the level of fusion and entry.
INTRODUCTION
Herpesviruses cause lifelong, latent infection and result in high morbidity. The human herpes simplex virus (HSV) is responsible for oral and genital lesions, keratitis, and in rare cases, encephalitis. HSV-1 and HSV-2 have seroprevalences of 67% (1) and 11% (2), respectively. As an enveloped virus, HSV must fuse viral and host membranes to initiate entry and infection. Entry of HSV can occur through multiple pathways. Entry into epithelial cells, the primary site of infection, proceeds via low-pH-dependent endocytosis (3–5). Entry into neurons, the site of HSV latency, is via direct penetration at the plasma membrane (6).
HSV has at least 12 virus-encoded glycoproteins in its lipid envelope (7). Glycoprotein B (gB), gD, and the heterodimer gH/gL are required for entry into all pathways, while gE, gG, gI, gJ, gM, pUL45, or pUS9 are considered dispensable for entry (8–19). Glycoprotein D is a key receptor binding protein. Expression of a host cell gD receptor (e.g., nectin-1) in a nonpermissive cell, renders the cell susceptible to HSV-1 infection (20, 21). The heterodimer gH/gL is essential for HSV entry, although its specific role is still incompletely defined. Glycoprotein B is the core fusogen of HSV and is highly conserved among all herpesviruses.
Entry of HSV into epithelial cells is blocked by lysosomotropic agents, which contributes to the notion that the virus fusion machinery is activated by exposure to acidic endosomal pH (3). Acid pretreatment of HSV in the absence of a target membrane results in inactivation of viral entry (3, 8, 9, 22). The target of low-pH inactivation is likely fusion and not attachment to cells or binding to a gD receptor (23, 24).
Herpesviral gB is a member of the class III fusion protein family (25–28), along with rhabdoviral G (29, 30), baculovirus GP64 (31), and thogotovirus Gp (32). HSV gB (22–24, 33), G (34, 35), and GP64 (36) undergo hallmark, reversible conformational changes following exposure to acidic pH. We recently reported that following prolonged acid exposure, the change in a specific epitope of the gB fusion domain becomes irreversible and correlates with an ∼2-log reduction in infectivity (24).
HSV gB also undergoes reversible changes to its oligomeric conformation following exposure to acidic pH. Upon acidification, the virion gB oligomer shifts to a lower density, based on three independent experimental approaches (33). gB, like other viral membrane glycoproteins, must fold and oligomerize prior to becoming functional. These foundational processes occur in the endoplasmic reticulum (ER) (37–42). Whether gB oligomerization or the formation of critical conformation-dependent epitopes occur cotranslationally or posttranslationally is not known.
Our results here suggest that gB oligomerizes rapidly and concurrently with translation. Full-length gB in transfected cells in the absence of other viral glycoproteins undergoes pH-triggered conformational changes. Neutralizing monoclonal antibodies (MAbs) to gB blocked HSV-1 entry regardless of whether the cell supported low-pH or pH-neutral entry. Lastly, an elevated temperature alone does not trigger gB change, but when combined with acid exposure, triggers irreversible antigenic change in gB, while changes detected in the oligomer remained reversible.
RESULTS
HSV gB folding and oligomerization occurs cotranslationally in living cells.
Viral fusion proteins function as oligomers. The active forms of class I and at least some class III fusion proteins are trimers. Polypeptide folding in living cells can begin cotranslationally, i.e., while the nascent chain is still being synthesized by the ribosome (43). Folding and oligomerization of viral glycoproteins typically occurs in the ER and can occur cotranslationally or require termination of monomer translation and release from the ribosome (posttranslational). HSV-1 gB multimers are present in virion and infected cell membranes (39, 44–46). When the formation of important conformation-dependent gB epitopes (folding) and oligomerization occur relative to biosynthesis on the ribosome is not known. We utilized pulse-chase analysis to monitor gB folding and trimerization in the ER of Vero cells. HSV-infected cells were pulsed for 100 s with 35S-labeled cysteine and methionine. Since the pulse (1.7 min) was shorter than the 5-min average time of gB synthesis, the majority of the radiolabel was expected to be in nascent chains (47, 48).
Thus, the detection of radiolabeled full-length gB immediately following the pulse or at early chase times with conformation-dependent or oligomer-specific antibodies reflects cotranslational folding and oligomerization. Following each time point, gB was immunoprecipitated with the designated antibody and, following denaturing SDS-PAGE, detected by autoradiography (Fig. 1).
FIG 1.
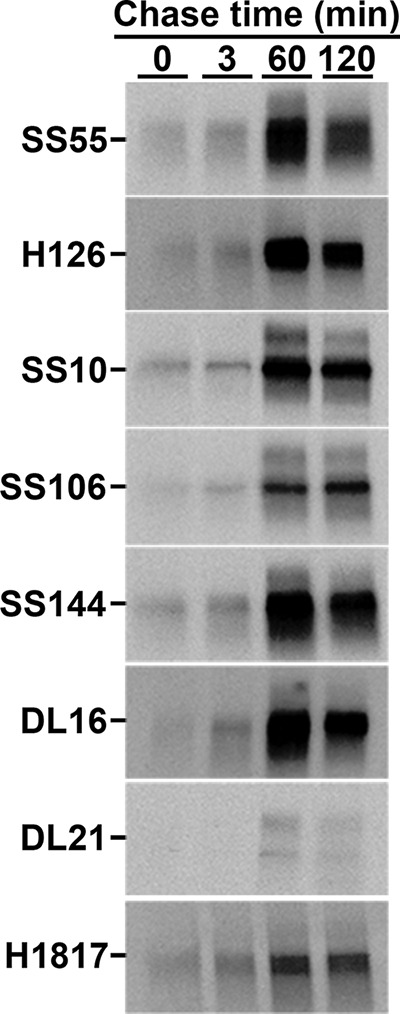
Pulse-chase analysis of gB in HSV-infected cells. After infection of Vero cells with HSV-1 KOS, cells were pulsed with [35S]cysteine and [35S]methionine for 100 s. Detergent lysates were immunoprecipitated with the MAb to gB indicated to the left of each panel, followed by denaturing SDS-PAGE and autoradiography. The bands correspond to monomeric gB.
MAbs DL16, SS10, SS55, and SS144, which detect conformation-dependent epitopes (Table 1), pulled down radiolabeled-gB immediately following the pulse or by 3 min of chase or earlier, suggesting that folding occurs while the nascent gB polypeptide is still being synthesized on the ribosome (i.e., cotranslationally). MAbs against linear epitopes H126, H1817, and SS106 (Table 1), similarly detected gB by 3 min of chase or earlier. The conformation-independent MAbs H126, H1817, and SS106 recognize continuous epitopes on the surface of the folded, native gB structure (25, 33, 49) and can be used along with conformation-dependent MAbs to detect proper folding of gB. Each of the eight antibodies to distinct gB epitopes pulled down labeled gB by 60 min chase. gB MAb DL21 binds to a gB ectodomain epitope distinct from the other MAbs used in this study (50) and detected little to no gB prior to 60 min of chase. Oligomer-specific gB MAb DL16 detected labeled gB immediately following the pulse or by 3 min of chase, suggesting that the gB trimer forms rapidly and cotranslationally. By 60 min of chase, each antibody precipitated an increased amount of labeled gB, which suggests that gB continues to fold after 3 min of chase. While gB clearly begins to fold cotranslationally, there is likely significant posttranslational folding as well. An increase in immunoprecipitation of radiolabeled gB following prolonged chase times has been reported previously (38–40, 45). Together, these results suggest that important gB folded epitopes and gB oligomers begin to form cotranslationally on the ribosome.
TABLE 1.
Summary of monoclonal antibodies used in this study
| Antibody | gB domainb | Binds conformation-dependent epitopec | Neutralizingd | Reduced reactivity with low-pH-treated gBe |
|---|---|---|---|---|
| H126 | I | N | + | + |
| SS55 | I | Y | + | + |
| H1781 | II | N | + | ND |
| H1838 | II | N | + | ND |
| H1359 | III | N | – | – |
| SS10 | IV | Y* | + | – |
| DL16a | V | Y | – | + |
| SS106 | V | N | + | + |
| SS144 | V | Y* | + | + |
| H1817 | VI | N | + | – |
| DL21 | U | Y | – | ND |
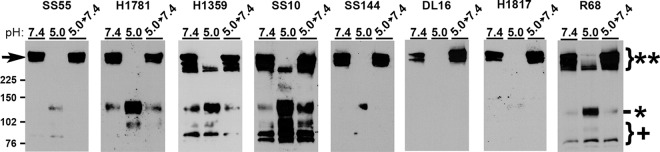
gB MAbs differentially detect low-pH induced oligomeric change in the SDS stability assay.
The HSV gB oligomer undergoes reversible alterations in response to mildly acidic pH as measured by multiple, independent assays (23, 33). In one approach, low-pH pretreatment renders the gB oligomer susceptible to disruption by 1% SDS. This assay has yielded valuable insights into gB conformational change (22–24, 33), e.g., the hydrophobic fusion loops themselves are not required for pH-triggered oligomeric changes, and the threshold of oligomeric change is similar to that of antigenic change. HSV-1 gB oligomers are stable in the presence of 1% SDS and stable when treated with low pH alone. However, following low-pH treatment, a subset of gB oligomeric species becomes susceptible to destabilization by 1% SDS. Notably, the slowest-migrating high-molecular-weight (high-MW) gB species becomes undetectable by this approach (indicated by arrowhead, Fig. 2). These gB-reactive bands correspond to oligomers because they are >225 kDa. The gB monomer is ∼116 kDa. The loss of slower-migrating gB is indicative of a change in oligomeric conformation. Virions that are treated with low pH and then neutralized back to pH 7.4 are resistant to 1% SDS destabilization, indicating this oligomeric change is reversible. The character of these high-MW forms of gB is not known. Previous investigations utilized gB MAb H1359, which detects a linear epitope, or rabbit polyclonal antibody to gB (23, 24, 33).
FIG 2.
Antigenic characterization of gB detected in the oligomeric change assay. HSV-1 KOS was treated at pH 5.0 or 7.4 for 10 min and then neutralized to pH 7.4 for 10 min if testing reversibility. Virions were then treated with 1% SDS and analyzed under “native” SDS-PAGE analysis, followed by Western blotting with the antibody indicated above each panel. **, Oligomeric gB; *, monomeric gB; +, fragmented gB. MW markers in kilodaltons are shown at the left.
To further characterize the pH-induced gB oligomeric change we employed a panel of antibodies to gB to define the antigenic reactivity of the high-MW forms of gB that are differentially susceptible to SDS and pH treatment. As expected, DL16 was specific for oligomeric gB (MW > 225 kDa [**], Fig. 2). The remaining antibodies primarily recognized oligomeric gB (MW > 225 kDa [**], Fig. 2) but also monomeric gB (MW ∼ 116 [*], Fig. 2), particularly following pH 5.0 treatment. Notably, H1359, SS10, and R68 each recognized the faster migrating oligomeric gB species that had been pH 5.0 treated. This suggests that gB oligomers that are resistant to detergent disruption in this assay are reactive with H1359 and SS10 but none of the other MAbs tested. MAbs SS55, SS144, and DL16 all have reduced reactivity with low-pH-treated gB as determined by dot blot analysis (8, 23) and here only detected high-MW gB at a neutral pH. The exact composition of the high-MW gBs in these experiments is incompletely defined. These gB oligomers do not comigrate with gC, gD, gH, or gL (33). Whether they represent gB dimers, trimers, hexamers, etc., remains to be elucidated. Another possibility is that the forms may all be the same order of oligomer, e.g., trimers, but exhibit altered migration due to differences in conformation. MAbs H1359, H1781, and SS10 recognized gB fragments smaller than 102 kDa (+, Fig. 2). These bands were not detected by SS144 or H1817. The fragments are likely cleavage products of gB monomer that result from virus propagation in Vero cells (51). Upon neutralization to pH 7.4 in this experimental approach (5.0 → 7.4), the changes were completely reversible. Together, each of the antibodies detected a reversible oligomer rearrangement of glycoprotein B following acidic pH exposure and 1% SDS treatment.
Full-length HSV-1 gB in transfected cells undergoes pH-triggered oligomeric change in the absence of other viral glycoproteins.
Native, prefusion gB in the virion envelope and gB from virus-infected cells both undergo oligomeric changes following exposure to acid pH (23, 24, 33). In these cases, many other viral proteins are present. Soluble forms of purified gB alone (23) undergo similar changes, indicating that the pH-triggered changes detected do not require that gB is membrane associated nor that any other viral components be present. The minimum essential requirements for cell to cell fusion are HSV-1 gB, gH/gL, gD, and a gD receptor expressed by the target cell (11, 52–54). We tested the susceptibility to pH of cell membrane-associated gB containing the full sequence, both alone and in combination with the other essential viral fusion proteins. This allowed gB conformational changes to be measured under conditions where the complex of fusion proteins are functionally active.
High-MW gB (>180 kDa) corresponding to oligomers was detected in cells transfected with gB subjected to 1% SDS treatment alone (Fig. 3a). After exposure to pH 5.0, the gB high-MW oligomers were not detected, suggesting oligomeric forms of gB became entirely sensitive to 1% SDS treatment. Notably, when virion gB is subjected to similar analysis, only the highest-MW gB species disappear (Fig. 2, H1359 or R68) (23, 24, 33), which may reflect a cell type difference in gB expression. In the presence of gD or gH/gL alone or in combination, oligomers of full-length, transfected cell-associated gB also underwent pH-triggered conformational change (Fig. 3), as indicated by the disappearance of oligomeric species. Less monomeric gB was detected after pH 5.0 treatment, when gB was paired with gH/gL (Fig. 3c and d), for reasons that are not clear. In contrast to virions produced in Vero cells (+, Fig. 2), no fragmented gB was detected in transfected Chinese hamster ovary (CHO) cells regardless of which essential glycoproteins were cotransfected (Fig. 3). These results suggest that full-length wild-type gB with transmembrane and cytoplasmic tail domains intact can undergo low-pH-triggered oligomeric change independent of other HSV proteins.
FIG 3.
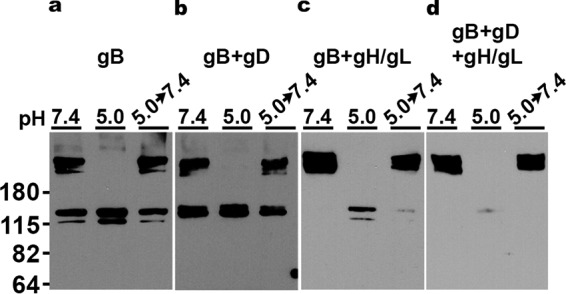
Low-pH-triggered oligomeric change of gB from transfected cells. CHO-K1 cells were transfected with plasmids encoding gB (a), gB+gD (b), gB+gH/gL (c), or gB+gD+gH/gL (d). Cell lysates were treated at pH 7.4 or 5.0 for 10 min. To test reversibility, pH 5.0-treated samples were neutralized back to pH 7.4 for 10 min at 37°C. The samples were then treated with 1% SDS and resolved under “native” PAGE conditions, followed by Western blotting with MAb H1359 to gB. MW markers in kilodaltons are shown at the left.
Glycoprotein B-specific MAbs neutralize HSV entry regardless of the pathway supported by the cell.
CHO cells and mouse melanoma B78 cells are naturally resistant to HSV infection (55, 56). Stable expression of gD receptors, such as nectin-1, allow HSV entry (20, 21). Nectin-1-expressing CHO and B78 cells permit HSV entry via distinct pathways. CHO-nectin-1 cells support low-pH endocytosis entry of HSV (3), while B78-nectin-1 cells support pH-independent endocytosis (57). Little is known about the HSV-1 determinants of low-pH entry versus pH-independent entry. To probe epitopes on gB that are important for one pathway or the other, HSV-1 gB MAbs were assayed for their ability to block entry via these distinct endocytic pathways. gB MAbs H126, H1838, SS10, and SS144 each neutralized HSV entry into both cell types at a similar titer of antibody (Table 2). Antibodies against gB domains I (H126) and V (SS144) have been previously reported to have reduced reactivity with low-pH-treated gB (33), but here they neutralized entry similarly via both low-pH and pH-independent pathways. At 8 μg/ml, MAbs H1359 and H1817 did not neutralize HSV entry into either pathway. H1817 is reportedly neutralizing (58), but 8 μg/ml did not reduce HSV-1 infectivity by >50% (Table 2). These results suggest that the epitopes in gB that are critical for viral entry are conserved regardless of entry pathway. This is consistent with gB conformational changes that are common to low-pH and pH-independent pathways.
TABLE 2.
gB MAb neutralization of HSV-1
| MAb to gB | Domain | Neutralization of HSV-1 (μg/ml)a |
|
|---|---|---|---|
| CHO-nectin-1 | B78-nectin-1 | ||
| H126 | I | 2 | 2 |
| H1838 | II | 1:4,000* | 1:4,000* |
| H1359 | III | >8 | >8 |
| SS10 | IV | 1 | 1 |
| SS144 | V | 0.25 | 0.25 |
| H1817 | VI | >8 | >8 |
The minimum concentration of IgG or (*) the dilution of ascites required to reduce HSV-1 KOS infectivity by >50% as determined by a β-galactosidase reporter assay.
Elevated temperature triggers irreversible low-pH changes in the antigenic conformation of gB but oligomeric change remains reversible.
HSV gB undergoes reversible conformational changes following exposure to acidic pH. Many viruses utilize cellular acidic pH or a host cell receptor to induce conformational changes in viral fusion proteins. For several class I fusion proteins, such as influenza hemagglutinin (59) and parainfluenza virus 5 F protein (60), these cellular triggers can be experimentally substituted by increased temperature. Elevated temperatures trigger relevant conformational changes in the class III fusion protein Epstein-Barr virus gB (61), but not vesicular stomatitis virus G (62). Heat, low pH, and receptor binding are all physicochemical forces that are thought to drive fusion proteins to a fusion active conformation. We determined the impact of elevated temperature in the presence and absence of acidic pH on gB conformational changes and their reversibility.
HSV-1 virions exposed to 37, 45, 50, or 55°C and blotted directly onto nitrocellulose membranes retained reactivity with MAbs H126 and SS10, with only a slight reduction at 55°C (pH 7.4, Fig. 4a), suggesting that temperature alone was not sufficient to induce a conformational change in gB. Treating HSV virions with the mildly acidic pH of 5.0 resulted in a reduction of reactivity with MAb H126 but not SS10 (37°C row, Fig. 4a), as shown previously (23, 24, 33). The reversibility of the low-pH-triggered conformational change in the H126 epitope of gB was confirmed as expected. When virions were treated with pH 5.0 at elevated temperatures, there was also reduced reactivity of H126. The conformational change induced at 45°C was partly reversible, while treatment with 50 or 55°C unexpectedly triggered an irreversible change in the H126 epitope. SS10 had similar reactivity at all pHs tested from 37 to 50°C, suggesting there was no change in this epitope. At 55°C and pH 5.0, there was a loss of SS10 reactivity, which returned upon neutralization to pH 7.4, indicating a reversible conformational change of the SS10 epitope under these conditions.
FIG 4.
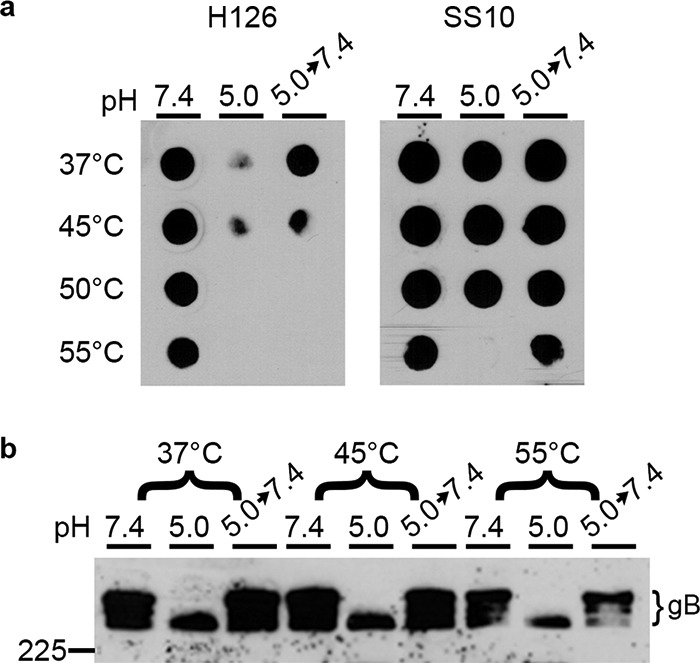
Effect of elevated temperature on acid-induced conformational changes of HSV-1 gB. HSV-1 virions were treated at pH 7.4 or 5.0 at the indicated temperature for 10 min. To test reversibility, pH 5.0-treated samples were neutralized back to pH 7.4 for 10 min at 37°C. Samples were blotted immediately to nitrocellulose membrane and probed at neutral pH with gB-specific MAb H126 or SS10. (b) 1% SDS was added, and samples were resolved under “native” PAGE conditions, followed by Western blotting with MAb H1359 to gB. MW markers in kilodaltons are shown on the left.
We also assessed the impact of elevated temperature and acidic pH on the gB oligomeric changes and their reversibility. As expected, when virions were treated at 37°C with acid and then 1% SDS, there was a loss of slower-migrating oligomeric gB as resolved by “native” SDS-PAGE (Fig. 4b). Upon neutralization to pH 7.4, the slower-migrating gB returned, indicating that this change was reversible. At increased temperatures up to 55°C, this change occurred and remained reversible, unlike the irreversible change in gB antigenic conformation detected by dot blot analysis (Fig. 4a). These results support the notion that changes in gB antigenic conformation do not necessarily correlate with changes in its oligomer conformation (24). Altogether, these results suggest temperature alone is not sufficient to induce antigenic or oligomeric changes in gB.
DISCUSSION
The acidic pH of the host cell endosome is a common trigger of viral membrane fusion (5, 63–65). Intracellular low pH facilitates entry of HSV into epithelial cells, the target of primary and recurrent infection in humans. HSV gB undergoes conformational and oligomeric changes in response to mildly acidic pH exposure both in vitro and during viral entry into cells. These changes are reversible as they are for other class III fusion proteins. Here, we show that gB folds and trimerizes rapidly and cotranslationally while nascent chains are still attached to the ribosome. The low-pH-induced oligomeric change in gB was characterized with a panel of MAbs. Native gB from transfected cells undergoes low-pH alterations in the absence of other viral proteins. gB MAbs neutralize HSV entry similarly into cells that support either the low-pH or the pH-independent pathway, supporting a conserved entry mechanism. Finally, elevated temperatures are not sufficient to induce gB conformational changes. When virions are treated with heat and mildly acidic pH, there is an irreversible change in the H126 epitope in the gB fusion domain, but oligomeric change remained reversible.
Folding and pH-triggered conformational change of oligomeric gB.
The functionally active form of HSV-1 gB is thought to be a trimer. gB trimerizes in the ER concurrently with translation (Fig. 1). Conformation-dependent, viral neutralizing antibodies detect nascent chains of gB on the ribosome, indicating important epitopes of gB fold rapidly and cotranslationally. This contrasts with HIV gp160, which is synthesized in minutes but takes hours to fold (66). Acid-triggered changes in gB oligomeric conformation can be detected with a modified PAGE assay. The high-MW oligomeric species of gB that is susceptible to detergent disruption following low-pH treatment was antigenically distinct, reacting specifically with MAbs SS55, DL16, and H1817. SS55 and DL16 have reduced reactivity with low pH-treated gB, and SS55 is neutralizing, which suggests that the high-MW gB that disappears may be a functionally active form of the oligomer. Oligomeric gB that was resistant to disruption retained reactivity with MAbs H1359 and SS10 (Fig. 2). Fragmented gB smaller than monomeric gB < 115 kDa was detected in virions (Fig. 2), but not in transfected cells (Fig. 3) for reasons that are unclear. Fragmented gB has previously been identified in HSV-infected Vero cells, but not in HEp-2 cells, suggesting that they may result from growing virions in Vero cells (51). gB from transfected cells is shown here for the first time to undergo low-pH-induced oligomeric change (Fig. 3). Transfected cell approaches may now be used to identify gB residues important for conformational change.
Treatment of HSV-1 gB with 45°C or greater did not trigger conformational changes in gB (Fig. 4). However, treatment of virions with 40°C rescued a slow-entry phenotype of HSV (67), suggesting that elevated temperature can help gB transition to a fusion-active form. Interestingly, treating EBV gB with 45°C alone triggers conformational change as detected by proteolytic digestion (61). Low pH triggers reversible conformational change in the H126 epitope of the gB fusion domain (22–24, 33). When treated with both an acidic pH and an elevated temperature, gB undergoes an irreversible change in the H126 epitope (Fig. 4a). Treating HSV-1 for prolonged periods resulted in a similar irreversible change in gB (24).
Conservation of requirements for HSV entry via distinct pathways.
The HSV determinants responsible for directing HSV to a low-pH versus a pH-independent pathway are not clear. Two different strains of HSV-1 enter the same cell type by two distinct entry pathways, suggesting a role for a viral determinant(s) in the selection of entry pathway (68). MAbs to distinct gB domains neutralized HSV-1 entry regardless of the reliance on low pH for entry (Table 2) (49). This supports the notion that conformational changes in the H126, H1838, SS10, and SS144 epitopes are important for fusion and are conserved during entry regardless of the entry pathway. Similarly, acid pretreatment of HSV abolishes virion entry activity regardless of cell type (3). It is clear that gB undergoes changes following exposure to acidic pH (22–24, 33, 69, 70), and changes in gB are also important for pH-independent fusion (71). A cellular trigger(s) for pH-independent changes in gB has not been identified. The gB receptor, paired immunoglobulin-like type 2 receptor α (PILRα), is one candidate, since expression of PILRα in CHO-cells results in pH-independent entry of HSV-1 (72). A key unresolved issue is how HSV mediates entry by both low pH and pH neutral pathways.
MATERIALS AND METHODS
Cells and viruses.
B78 murine melanoma cells expressing nectin-1 (B78-nectin-1 cells) (56) (a gift from G. Cohen and R. Eisenberg, University of Pennsylvania) and Vero cells (American Type Culture Collection, Rockville, MD) were propagated in Dulbecco modified Eagle medium (DMEM; Thermo Fisher Scientific, Waltham, MA) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals. Atlanta, GA) and penicillin, streptomycin, and glutamine (Thermo Fisher Scientific). Chinese hamster ovary cells expressing nectin-1 (CHO-nectin-1 cells [21]; a gift from G. Cohen and R. Eisenberg, University of Pennsylvania) and CHO-K1 cells (a gift from H. Aguilar, Cornell University) were propagated in Ham's F-12 medium supplemented with 10% FBS and penicillin, streptomycin, and glutamine (Thermo Fisher Scientific). B78-nectin-1 and CHO-nectin-1 cells were further supplemented with 250 μg of Geneticin (Sigma, St. Louis, MO) per ml and 6 μg (B78-nectin-1) or 150 μg (CHO-nectin-1) of puromycin (Sigma) per ml. Both B78-nectin-1 and CHO-nectin-1 cells stably express human nectin-1 and contain the Escherichia coli lacZ gene under the control of the HSV-1 ICP4 promoter. Cells were subcultured in nonselective medium prior to use in all experiments. HSV-1 strain KOS (a gift from Priscilla Schaffer, Harvard University) was propagated, and titers were determined on Vero cells.
Preparation of HSV-1.
Subconfluent Vero cells were infected with HSV-1 (multiplicity of infection [MOI] of 0.05) in 175-cm2 flasks. At 72 h postinfection, cells were pelleted at 300 × g at 4°C for 10 min. Pelleted cells were discarded, and cell-free supernatant containing extracellular virions was centrifuged at 27,000 × g for 45 min through a 5% sucrose/phosphate-buffered saline (PBS) cushion. Pellets were resuspended overnight at 4°C in 20 mM HEPES (Thermo Fisher Scientific)-buffered DMEM supplemented with 10% FBS. Concentrated virions were sonicated (model XL2020; Misonix, Inc., Farmingdale, NY) and stored at −80°C.
Antibodies to gB.
Characteristics of anti-HSV-1 gB mouse MAbs are summarized in Table 1. H126, H1359, and H1817 were purchased from Virusys, Taneytown, MD. Ascites fluid of MAbs H1781 and H1838 were provided by L. Pereira, University of California, San Francisco. Anti-gB MAbs DL16, DL21, SS10, SS55, SS144, and SS106, as well as rabbit polyclonal antibody R68 (73), were provided by G. Cohen and R. Eisenberg, University of Pennsylvania.
Pulse-chase analysis of gB folding and oligomerization.
Vero cells in 60-mm culture dishes were infected with HSV-1 KOS (MOI of 5). At 6 h postinfection, cells were washed with PBS and cultured in serum-free DMEM lacking both cysteine and methionine (Invitrogen, Carlsbad, CA) for 20 min. Cells were then pulsed with DMEM containing [35S]cysteine and [35S]methionine (MP Biomedicals, Santa Ana, CA) for 100 s. The medium was replaced with DMEM containing 20 mM HEPES, 5 mM l-cysteine, 5 mM l-methionine, and 1 mM cycloheximide. At the indicated chase time, the reaction was stopped by two washes with ice-cold PBS containing 20 mM N-ethylmaleimide (NEM). The cells were lysed with ice cold 2% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} in HEPES-buffered saline containing NEM and protease inhibitors (Roche, Indianapolis, IN). NEM is an alkylating agent that prevents postlysis oxidation (41). Samples were immunoprecipitated overnight at 4°C with the indicated gB MAb. Immunoprecipitations were subjected to denaturing SDS–6% PAGE and visualized utilizing autoradiography.
Analysis of the gB oligomer by PAGE.
HSV-1 KOS virions were diluted in fusion medium: serum-free, sodium bicarbonate-free DMEM, containing 5 mM HEPES, 5 mM 2-(N-morpholino)ethanesulfonic acid (MES; Sigma), 5 mM sodium succinate (Sigma), and 0.2% BSA. Samples were adjusted to pH 7.4 or pH 5.0 for 10 min at 37°C. To test reversibility, samples were neutralized to the indicated pH for 10 min. After exposure to the designated pH, 1% SDS was added to samples for 2 min at 37°C. Laemmli buffer modified to contain 0.2% SDS, and no reducing agent was added to the samples. Unheated samples were resolved via 8% PAGE and transferred to nitrocellulose (74). Membranes were blocked and probed with anti-gB antibodies. Following incubation with horseradish peroxidase-conjugated secondary antibody, SuperSignal West Dura extended duration substrate (Thermo Fisher Scientific) was added and membranes were exposed to X-ray film (Genesee Scientific, San Diego, CA).
Analysis of transfected cell gB via PAGE.
CHO-K1 cells (∼80% confluent) were transfected with combinations of plasmids pPEP98, pPEP99, pPEP100, or pPEP101 (a gift from P. Spear, Northwestern University, Chicago, IL [54]) encoding HSV-1 gB, gD, gH, or gL, respectively, using Lipofectamine 2000 (Thermo Fisher Scientific). Following a 6 h incubation, complete F-12 medium was added, and cells were incubated for a total of 22 to 24 h. Cells were then subjected to pH treatment and analyzed as described in the previous section.
HSV-1 neutralization by anti-gB antibodies.
HSV-1 KOS virions (105 PFU) were incubated with serial dilutions of gB MAbs for 1 h at 37°C. Virions were then added to B78-nectin-1 cells (MOI of 0.2) or CHO-nectin-1 cells (MOI of 0.1) for 6 to 8 h at 37°C. Cells were lysed with 0.5% IGEPAL (Sigma). The β-galactosidase substrate, chlorophenol red-d-galactopyranoside (Roche, Indianapolis, IN), was added, and the β-galactosidase activity was read at 595 nm with a microtiter plate reader (Biotek, Winooski, VT). The minimum MAb concentration resulting in a reduction of HSV-1 infectivity by >50% was reported.
Heat treatment of HSV virions.
HSV-1 KOS virions were treated at the indicated pH and temperature for 10 min in pH-adjusted medium as described above. If testing reversibility of conformational change, virions were then neutralized to pH 7.4 for 10 min at 37°C. For antigenic analysis, samples were blotted immediately to nitrocellulose membrane and probed with the indicated MAb at neutral pH. For testing oligomeric change, following pH and temperature treatments, 1% SDS was added to samples. SDS-PAGE and Western blotting was then performed as described above.
ACKNOWLEDGMENTS
We thank Sue Pritchard for critical readings of the manuscript. We thank H. Aguilar, G. Cohen, R. Eisenberg, L. Pereira, P. Schaffer, and P. Spear for gifts of reagents.
This study was supported by Public Health Service grants AI119159 (A.V.N.) and GM008336 (D.J.W. and T.K.S.) and National Science Foundation Graduate Research Fellowship DGE 1347973 (D.J.W.).
REFERENCES
- 1.Looker KJ, Magaret AS, May MT, Turner KM, Vickerman P, Gottlieb SL, Newman LM. 2015. Global and regional estimates of prevalent and incident herpes simplex virus type 1 infections in 2012. PLoS One 10:e0140765. doi: 10.1371/journal.pone.0140765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Looker KJ, Magaret AS, Turner KM, Vickerman P, Gottlieb SL, Newman LM. 2015. Global estimates of prevalent and incident herpes simplex virus type 2 infections in 2012. PLoS One 10:e114989. doi: 10.1371/journal.pone.0114989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nicola AV, McEvoy AM, Straus SE. 2003. Roles for endocytosis and low pH in herpes simplex virus entry into HeLa and Chinese hamster ovary cells. J Virol 77:5324–5332. doi: 10.1128/JVI.77.9.5324-5332.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nicola AV, Hou J, Major EO, Straus SE. 2005. Herpes simplex virus type 1 enters human epidermal keratinocytes, but not neurons, via a pH-dependent endocytic pathway. J Virol 79:7609–7616. doi: 10.1128/JVI.79.12.7609-7616.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicola AV. 2016. Herpesvirus entry into host cells mediated by endosomal low pH. Traffic 17:965–975. doi: 10.1111/tra.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lycke E, Hamark B, Johansson M, Krotochwil A, Lycke J, Svennerholm B. 1988. Herpes simplex virus infection of the human sensory neuron. Arch Virol 101:87–104. doi: 10.1007/BF01314654. [DOI] [PubMed] [Google Scholar]
- 7.Loret S, Guay G, Lippé R. 2008. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J Virol 82:8605–8618. doi: 10.1128/JVI.00904-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dollery SJ, Lane KD, Delboy MG, Roller DG, Nicola AV. 2010. Role of the UL45 protein in herpes simplex virus entry via low pH-dependent endocytosis and its relationship to the conformation and function of glycoprotein B. Virus Res 149:115–118. doi: 10.1016/j.virusres.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Komala Sari T, Pritchard SM, Cunha CW, Wudiri GA, Laws EI, Aguilar HC, Taus NS, Nicola AV. 2013. Contributions of herpes simplex virus 1 envelope proteins to entry by endocytosis. J Virol 87:13922–13926. doi: 10.1128/JVI.02500-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicola AV, Straus SE. 2004. Cellular and viral requirements for rapid endocytic entry of herpes simplex virus. J Virol 78:7508–7517. doi: 10.1128/JVI.78.14.7508-7517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weed DJ, Nicola AV. 2017. Herpes simplex virus membrane fusion, p 29–47. In Cell biology of herpes viruses. Springer, New York, NY. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai W, Person S, DebRoy C, Gu B. 1988. Functional regions and structural features of the gB glycoprotein of herpes simplex virus type 1: an analysis of linker insertion mutants. J Mol Biol 201:575–588. doi: 10.1016/0022-2836(88)90639-0. [DOI] [PubMed] [Google Scholar]
- 13.Forrester A, Farrell H, Wilkinson G, Kaye J, Davis-Poynter N, Minson T. 1992. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J Virol 66:341–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ligas MW, Johnson DC. 1988. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by beta-galactosidase sequences binds to but is unable to penetrate into cells. J Virol 62:1486–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roop C, Hutchinson L, Johnson DC.. 1993. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells, and its particles lack glycoprotein H J Virol 67:2285–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balan P, Davis-Poynter N, Bell S, Atkinson H, Browne H, Minson T. 1994. An analysis of the in vitro and in vivo phenotypes of mutants of herpes simplex virus type 1 lacking glycoproteins gG, gE, gI or the putative gJ. J Gen Virol 75:1245–1258. doi: 10.1099/0022-1317-75-6-1245. [DOI] [PubMed] [Google Scholar]
- 17.Longnecker R, Chatterjee S, Whitley RJ, Roizman B. 1987. Identification of a herpes simplex virus 1 glycoprotein gene within a gene cluster dispensable for growth in cell culture. Proc Natl Acad Sci U S A 84:4303–4307. doi: 10.1073/pnas.84.12.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Longnecker R, Roizman B. 1987. Clustering of genes dispensable for growth in culture in the S component of the HSV-1 genome. Science 236:573–576. doi: 10.1126/science.3033823. [DOI] [PubMed] [Google Scholar]
- 19.Weber PC, Levine M, Glorioso JC. 1987. Rapid identification of nonessential genes of herpes simplex virus type 1 by Tn5 mutagenesis. Science 236:576–579. doi: 10.1126/science.3033824. [DOI] [PubMed] [Google Scholar]
- 20.Montgomery RI, Warner MS, Lum BJ, Spear PG. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427–436. doi: 10.1016/S0092-8674(00)81363-X. [DOI] [PubMed] [Google Scholar]
- 21.Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ, Spear PG. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 22.Siekavizza-Robles CR, Dollery SJ, Nicola AV. 2010. Reversible conformational change in herpes simplex virus glycoprotein B with fusion-from-without activity is triggered by mildly acidic pH. Virology J 7:352–352. doi: 10.1186/1743-422X-7-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dollery SJ, Wright CC, Johnson DC, Nicola AV. 2011. Low-pH-dependent changes in the conformation and oligomeric state of the prefusion form of herpes simplex virus glycoprotein B are separable from fusion activity. J Virol 85:9964–9973. doi: 10.1128/JVI.05291-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weed DJ, Pritchard SM, Gonzalez F, Aguilar HC, Nicola AV. 2017. Mildly acidic pH triggers an irreversible conformational change in the fusion domain of herpes simplex virus 1 glycoprotein B and inactivation of viral entry. J Virol 91:e02123-16. doi: 10.1128/JVI.02123-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heldwein EE, Lou H, Bender FC, Cohen GH, Eisenberg RJ, Harrison SC. 2006. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 313:217–220. doi: 10.1126/science.1126548. [DOI] [PubMed] [Google Scholar]
- 26.Backovic M, Longnecker R, Jardetzky TS. 2009. Structure of a trimeric variant of the Epstein-Barr virus glycoprotein B. Proc Natl Acad Sci U S A 106:2880–2885. doi: 10.1073/pnas.0810530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chandramouli S, Ciferri C, Nikitin PA, Caló S, Gerrein R, Balabanis K, Monroe J, Hebner C, Lilja AE, Settembre EC. 2015. Structure of HCMV glycoprotein B in the postfusion conformation bound to a neutralizing human antibody. Nat Commun 6:8176. doi: 10.1038/ncomms9176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vallbracht M, Brun D, Tassinari M, Vaney M-C, Pehau-Arnaudet G, Guardado-Calvo P, Haouz A, Klupp BG, Mettenleiter TC, Rey FA. 2018. Structure-function dissection of pseudorabies virus glycoprotein B fusion loops. J Virol 92:e01203-17. doi: 10.1128/JVI.00376-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roche S, Bressanelli S, Rey FA, Gaudin Y. 2006. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science 313:187–191. doi: 10.1126/science.1127683. [DOI] [PubMed] [Google Scholar]
- 30.Roche S, Rey FA, Gaudin Y, Bressanelli S. 2007. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science 315:843–848. doi: 10.1126/science.1135710. [DOI] [PubMed] [Google Scholar]
- 31.Kadlec J, Loureiro S, Abrescia NG, Stuart DI, Jones IM. 2008. The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines. Nat Struct Mol Bio 15:1024. doi: 10.1038/nsmb.1484. [DOI] [PubMed] [Google Scholar]
- 32.Peng R, Zhang S, Cui Y, Shi Y, Gao GF, Qi J. 2017. Structures of human-infecting Thogotovirus fusogens support a common ancestor with insect baculovirus. Proc Natl Acad Sci 114:E8905–E8912. doi: 10.1073/pnas.1706125114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dollery SJ, Delboy MG, Nicola AV. 2010. Low pH-induced conformational change in herpes simplex virus glycoprotein B. J Virol 84:3759–3766. doi: 10.1128/JVI.02573-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaudin Y. 2002. Reversibility in fusion protein conformational changes the intriguing case of rhabdovirus-induced membrane fusion, p 379–408. In Fusion of biological membranes and related problems. Springer, New York, NY. [DOI] [PubMed] [Google Scholar]
- 35.Gaudin Y, Tuffereau C, Segretain D, Knossow M, Flamand A. 1991. Reversible conformational changes and fusion activity of rabies virus glycoprotein. J Virol 65:4853–4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou J, Blissard GW. 2006. Mapping the conformational epitope of a neutralizing antibody (AcV1) directed against the AcMNPV GP64 protein. Virology 352:427–437. doi: 10.1016/j.virol.2006.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mijnes J, Van der Horst L, Van Anken E, Horzinek M, Rottier P, De Groot R. 1996. Biosynthesis of glycoproteins E and I of feline herpesvirus: gE-gI interaction is required for intracellular transport. J Virol 70:5466–5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamashita Y, Yamada M, Daikoku T, Yamada H, Tadauchi A, Tsurumi T, Nishiyama Y. 1996. Calnexin associates with the precursors of glycoproteins B, C, and D of herpes simplex virus type 1. Virology 225:216–222. doi: 10.1006/viro.1996.0590. [DOI] [PubMed] [Google Scholar]
- 39.Claesson-Welsh L, Spear PG. 1986. Oligomerization of herpes simplex virus glycoprotein B. J Virol 60:803–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laquerre S, Anderson DB, Argnani R, Glorioso JC. 1998. Herpes simplex virus type 1 glycoprotein B requires a cysteine residue at position 633 for folding, processing, and incorporation into mature infectious virus particles. J Virol 72:4940–4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Braakman I, Hoover-Litty H, Wagner KR, Helenius A. 1991. Folding of influenza hemagglutinin in the endoplasmic reticulum. J Cell Biol 114:401–411. doi: 10.1083/jcb.114.3.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doms RW, Lamb RA, Rose JK, Helenius A. 1993. Folding and assembly of viral membrane proteins. Virology 193:545–562. doi: 10.1006/viro.1993.1164. [DOI] [PubMed] [Google Scholar]
- 43.Fedorov AN, Baldwin TO. 1997. Cotranslational protein folding. J Biol Chem 272:32715–32718. doi: 10.1074/jbc.272.52.32715. [DOI] [PubMed] [Google Scholar]
- 44.Laquerre S, Person S, Glorioso JC. 1996. Glycoprotein B of herpes simplex virus type 1 oligomerizes through the intermolecular interaction of a 28-amino-acid domain. J Virol 70:1640–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Highlander SL, Goins WF, Person S, Holland TC, Levine M, Glorioso JC. 1991. Oligomer formation of the gB glycoprotein of herpes simplex virus type 1. J Virol 65:4275–4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu Q, Courtney RJ. 1988. Chemical crosslinking of glycoproteins on the envelope of herpes simplex virus. Virology 167:377–384. doi: 10.1016/S0042-6822(88)90098-0. [DOI] [PubMed] [Google Scholar]
- 47.Chen W, Helenius J, Braakman I, Helenius A. 1995. Cotranslational folding and calnexin binding during glycoprotein synthesis. Proc Natl Acad Sci U S A 92:6229–6233. doi: 10.1073/pnas.92.14.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nicola AV, Chen W, Helenius A. 1999. Cotranslational folding of an alphavirus capsid protein in the cytosol of living cells. Nat Cell Biol 1:341–345. doi: 10.1038/14032. [DOI] [PubMed] [Google Scholar]
- 49.Roller DG, Dollery SJ, Doyle JL, Nicola AV. 2008. Structure–function analysis of herpes simplex virus glycoprotein B with fusion-from-without activity. Virology 382:207–216. doi: 10.1016/j.virol.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 50.Bender FC, Samanta M, Heldwein EE, de Leon MP, Bilman E, Lou H, Whitbeck JC, Eisenberg RJ, Cohen GH. 2007. Antigenic and mutational analyses of herpes simplex virus glycoprotein B reveal four functional regions. J Virol 81:3827–3841. doi: 10.1128/JVI.02710-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pereira L, Dondero D, Norrild B, Roizman B. 1981. Differential immunologic reactivity and processing of glycoproteins gA and gB of herpes simplex virus types 1 and 2 made in Vero and HEp-2 cells. Proc Natl Acad Sci U S A 78:5202–5206. doi: 10.1073/pnas.78.8.5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turner A, Bruun B, Minson T, Browne H. 1998. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J Virol 72:873–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Browne H, Bruun B, Minson T. 2001. Plasma membrane requirements for cell fusion induced by herpes simplex virus type 1 glycoproteins gB, gD, gH, and gL. J Gen Virol 82:1419–1422. doi: 10.1099/0022-1317-82-6-1419. [DOI] [PubMed] [Google Scholar]
- 54.Pertel PE, Fridberg A, Parish ML, Spear PG. 2001. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology 279:313–324. doi: 10.1006/viro.2000.0713. [DOI] [PubMed] [Google Scholar]
- 55.Shieh M-T, WuDunn D, Montgomery RI, Esko JD, Spear PG. 1992. Cell surface receptors for herpes simplex virus are heparan sulfate proteoglycans. J Cell Biol 116:1273–1281. doi: 10.1083/jcb.116.5.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller CG, Krummenacher C, Eisenberg RJ, Cohen GH, Fraser NW. 2001. Development of a syngenic murine B16 cell line-derived melanoma susceptible to destruction by neuroattenuated HSV-1. Mol Ther 3:160–168. doi: 10.1006/mthe.2000.0240. [DOI] [PubMed] [Google Scholar]
- 57.Milne RS, Nicola AV, Whitbeck JC, Eisenberg RJ, Cohen GH. 2005. Glycoprotein D receptor-dependent, low-pH-independent endocytic entry of herpes simplex virus type 1. J Virol 79:6655–6663. doi: 10.1128/JVI.79.11.6655-6663.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pereira L, Ali M, Kousoulas K, Huo B, Banks T. 1989. Domain structure of herpes simplex virus 1 glycoprotein B: neutralizing epitopes map in regions of continuous and discontinuous residues. Virology 172:11–24. doi: 10.1016/0042-6822(89)90102-5. [DOI] [PubMed] [Google Scholar]
- 59.Ruigrok R, Martin S, Wharton S, Skehel J, Bayley P, Wiley D. 1986. Conformational changes in the hemagglutinin of influenza virus which accompany heat-induced fusion of virus with liposomes. Virology 155:484–497. doi: 10.1016/0042-6822(86)90210-2. [DOI] [PubMed] [Google Scholar]
- 60.Connolly SA, Leser GP, Yin H-S, Jardetzky TS, Lamb RA. 2006. Refolding of a paramyxovirus F protein from prefusion to postfusion conformations observed by liposome binding and electron microscopy. Proc Natl Acad Sci U S A 103:17903–17908. doi: 10.1073/pnas.0608678103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chesnokova LS, Ahuja MK, Hutt-Fletcher LM. 2014. Epstein-Barr virus glycoprotein gB and gHgL can mediate fusion and entry in trans, and heat can act as a partial surrogate for gHgL and trigger a conformational change in gB. J Virol 88:12193–12201. doi: 10.1128/JVI.01597-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yao Y, Ghosh K, Epand RF, Epand RM, Ghosh HP. 2003. Membrane fusion activity of vesicular stomatitis virus glycoprotein G is induced by low pH but not by heat or denaturant. Virology 310:319–332. doi: 10.1016/S0042-6822(03)00146-6. [DOI] [PubMed] [Google Scholar]
- 63.Barrow E, Nicola AV, Liu J. 2013. Multiscale perspectives of virus entry via endocytosis. Virol J 10:177. doi: 10.1186/1743-422X-10-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nicola AV, Aguilar HC, Mercer J, Ryckman B, Wiethoff CM. 2013. Virus entry by endocytosis. Adv Virol 2013:469538. doi: 10.1155/2013/469538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marsh M, Helenius A. 1989. Virus entry into animal cells. Adv Virus Res 36:107–151. doi: 10.1016/S0065-3527(08)60583-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Land A, Zonneveld D, Braakman I. 2003. Folding of HIV-1 envelope glycoprotein involves extensive isomerization of disulfide bonds and conformation-dependent leader peptide cleavage. FASEB J 17:1058–1067. doi: 10.1096/fj.02-0811com. [DOI] [PubMed] [Google Scholar]
- 67.Fan Q, Kopp SJ, Connolly SA, Longnecker R. 2017. Structure-based mutations in the herpes simplex virus 1 glycoprotein B ectodomain arm impart a slow-entry phenotype. mBio 8:e00614-17. doi: 10.1128/mBio.00614-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Delboy MG, Patterson JL, Hollander AM, Nicola AV. 2006. Nectin-2-mediated entry of a syncytial strain of herpes simplex virus via pH-independent fusion with the plasma membrane of Chinese hamster ovary cells. Virol J 3:1. doi: 10.1186/1743-422X-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Muggeridge MI. 2012. Glycoprotein B of herpes simplex virus 2 has more than one intracellular conformation and is altered by low pH. J Virol 86:6444–6456. doi: 10.1128/JVI.06668-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stampfer SD, Lou H, Cohen GH, Eisenberg RJ, Heldwein EE. 2010. Structural basis of local, pH-dependent conformational changes in glycoprotein B from herpes simplex virus type 1. J Virol 84:12924–12933. doi: 10.1128/JVI.01750-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gallagher JR, Atanasiu D, Saw WT, Paradisgarten MJ, Whitbeck JC, Eisenberg RJ, Cohen GH. 2014. Functional fluorescent protein insertions in herpes simplex virus gB report on gB conformation before and after execution of membrane fusion. PLoS Pathol 10:e1004373. doi: 10.1371/journal.ppat.1004373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Arii J, Uema M, Morimoto T, Sagara H, Akashi H, Ono E, Arase H, Kawaguchi Y. 2009. Entry of herpes simplex virus 1 and other alphaherpesviruses via the paired immunoglobulin-like type 2 receptor α. J Virol 83:4520–4527. doi: 10.1128/JVI.02601-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Eisenberg RJ, de Leon MP, Friedman HM, Fries LF, Frank MM, Hastings JC, Cohen GH. 1987. Complement component C3b binds directly to purified glycoprotein C of herpes simplex virus types 1 and 2. Microb Pathog 3:423–435. doi: 10.1016/0882-4010(87)90012-X. [DOI] [PubMed] [Google Scholar]
- 74.Cohen G, Isola V, Kuhns J, Berman P, Eisenberg R. 1986. Localization of discontinuous epitopes of herpes simplex virus glycoprotein D: use of a nondenaturing (“native” gel) system of polyacrylamide gel electrophoresis coupled with Western blotting. J Virol 60:157–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cairns TM, Fontana J, Huang Z-Y, Whitbeck JC, Atanasiu D, Rao S, Shelly SS, Lou H, de Leon MP, Steven AC. 2014. Mechanism of neutralization of herpes simplex virus by antibodies directed at the fusion domain of glycoprotein B. J Virol 88:2677–2689. doi: 10.1128/JVI.03200-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kousoulas KG, Pellett PE, Pereira L, Roizman B. 1984. Mutations affecting conformation or sequence of neutralizing epitopes identified by reactivity of viable plaques segregate from syn and ts domains of HSV-1 (F) gB gene. Virology 135:379–394. doi: 10.1016/0042-6822(84)90194-6. [DOI] [PubMed] [Google Scholar]