The mosquito Aedes aegypti transmits a number of arthropod-borne viruses (arboviruses), such as dengue virus and Zika virus. Mosquitoes also harbor insect-specific viruses that may affect replication of pathogenic arboviruses in their body. Currently, however, there are only a few insect-specific viruses described from A. aegypti in the literature. Here, we characterize a novel negative-strand virus, AeAV. Meta-analysis of A. aegypti samples showed that it is present in A. aegypti mosquitoes worldwide and is vertically transmitted. Wolbachia-transinfected mosquitoes are currently being used in biocontrol, as they effectively block transmission of several positive-sense RNA viruses in mosquitoes. Our results demonstrate that Wolbachia enhances the replication of AeAV and modestly reduces dengue virus replication in a cell line model. This study expands our understanding of the virome in A. aegypti as well as providing insight into the complexity of the Wolbachia virus restriction phenotype.
KEYWORDS: Aedes aegypti, anphevirus, Mononegavirales, Wolbachia, insect viruses, mosquito, virome
ABSTRACT
Insect-specific viruses (ISVs) of the yellow fever mosquito Aedes aegypti have been demonstrated to modulate transmission of arboviruses such as dengue virus (DENV) and West Nile virus by the mosquito. The diversity and composition of the virome of A. aegypti, however, remains poorly understood. In this study, we characterized Aedes anphevirus (AeAV), a negative-sense RNA virus from the order Mononegavirales. AeAV identified from Aedes cell lines was infectious to both A. aegypti and Aedes albopictus cells but not to three mammalian cell lines. To understand the incidence and genetic diversity of AeAV, we assembled 17 coding-complete and two partial genomes of AeAV from available transcriptome sequencing (RNA-Seq) data. AeAV appears to transmit vertically and be present in laboratory colonies, wild-caught mosquitoes, and cell lines worldwide. Phylogenetic analysis of AeAV strains indicates that as the A. aegypti mosquito has expanded into the Americas and Asia-Pacific, AeAV has evolved into monophyletic African, American, and Asia-Pacific lineages. The endosymbiotic bacterium Wolbachia pipientis restricts positive-sense RNA viruses in A. aegypti. Reanalysis of a small RNA library of A. aegypti cells coinfected with AeAV and Wolbachia produces an abundant RNA interference (RNAi) response consistent with persistent virus replication. We found Wolbachia enhances replication of AeAV compared to a tetracycline-cleared cell line, and AeAV modestly reduces DENV replication in vitro. The results from our study improve understanding of the diversity and evolution of the virome of A. aegypti and adds to previous evidence that shows Wolbachia does not restrict a range of negative-strand RNA viruses.
IMPORTANCE The mosquito Aedes aegypti transmits a number of arthropod-borne viruses (arboviruses), such as dengue virus and Zika virus. Mosquitoes also harbor insect-specific viruses that may affect replication of pathogenic arboviruses in their body. Currently, however, there are only a few insect-specific viruses described from A. aegypti in the literature. Here, we characterize a novel negative-strand virus, AeAV. Meta-analysis of A. aegypti samples showed that it is present in A. aegypti mosquitoes worldwide and is vertically transmitted. Wolbachia-transinfected mosquitoes are currently being used in biocontrol, as they effectively block transmission of several positive-sense RNA viruses in mosquitoes. Our results demonstrate that Wolbachia enhances the replication of AeAV and modestly reduces dengue virus replication in a cell line model. This study expands our understanding of the virome in A. aegypti as well as providing insight into the complexity of the Wolbachia virus restriction phenotype.
INTRODUCTION
The yellow fever mosquito Aedes aegypti is a vector of medically important viruses with worldwide distribution within the tropical and subtropical zones (1). A. aegypti is the principal vector of both dengue virus (DENV) and Zika virus (ZIKV) (family Flaviviridae), with estimates suggesting up to 390 million incidences of DENV infections a year (2) and approximately 400,000 cases of ZIKV during the 2015–2016 Latin American ZIKV outbreak (3).
The ability of mosquitoes to transmit viruses is determined by a complex suite of genetic and extrinsic host factors (4–6). One developing area is the contribution of insect-specific viruses (ISVs), demonstrated not to replicate in mammalian cells, in the vector competence of individual mosquitoes (7, 8). ISVs can suppress the antiviral RNA interference (RNAi) response, as shown in Culex-Y virus (CYV) of the Birnaviridae family (9), or enhance the transcription of host factors. Cell-fusing agent virus (CFAV) (family Flaviviridae) infection of A. aegypti Aa20 cells upregulates the V-ATPase-associated factor RNASEK, allowing more favorable replication of DENV (10). ISVs have also been shown to suppress or exclude replication of arboviruses; prior infection of Aedes albopictus C6/36 cells and A. aegypti mosquitoes with Palm Creek virus (PCV) (family Flaviviridae) has been shown to suppress replication of the zoonotic West Nile virus (WNV) and Murray Valley encephalitis virus (family Flaviviridae) (11, 12). Also, it has recently been demonstrated in Aedes cell lines that dual infection with Phasi Charoen-like virus (PCLV; family Bunyaviridae) and CFAV restricts the cells' permissivity to both DENV and ZIKV infection (13).
Metagenomic and biosurveillance strategies have proved invaluable in describing the virome diversity of wild-caught Culicinae mosquitoes (14, 15). To date, six ISVs have been identified and characterized from wild-caught and laboratory A. aegypti; CFAV (16, 17), PCLV (18), Dezidougou virus from the negevirus taxon (19), Aedes densoviruses (family Parvoviridae) (20), and the unclassified Humaita-Tubiacanga virus (HTV) (21). Recently, transcriptomic analysis of wild-caught A. aegypti mosquitoes from Bangkok, Thailand, and Cairns, Australia, suggested infection of the mosquitoes with up to 27 insect-specific viruses, the majority of which are currently uncharacterized (22). This represents a narrow understanding of the diversity of the circulating virome harbored by A. aegypti mosquitoes.
In this study, we identified and characterized a novel negative-sense RNA Anphevirus, putatively named Aedes anphevirus (AeAV), from the order Mononegavirales in A. aegypti mosquitoes. According to the most recent International Committee on Taxonomy of Viruses (ICTV) report (23), Xīnchéng mosquito virus (XcMV), assembled as part of a metagenomic analysis of Anopheles sinensis mosquitoes in Xīnchéng, China, is the only member of the Anphevirus genus and is closely related to members of Bornaviridae and Nyamiviridae (24). Originally thought to only carry four open reading frames (ORFs), the presence of a number of viruses closely related to XcMV from West African Anopheles gambiae mosquitoes (15) and West Australian Culex mosquitoes (25) suggests that members of this taxon carry six ORFs with a genome size of approximately 12 kb.
The endosymbiotic bacterium Wolbachia pipientis was first shown to restrict RNA viruses in Drosophila melanogaster (26, 27). Transinfection of Wolbachia into A. aegypti was also shown to restrict DENV and Chikungunya virus (family Togaviridae) replication in the host (28). In A. aegypti Aag2 cells stably transinfected with a proliferative strain of Wolbachia (wMelPop-CLA), the endosymbiont restricts CFAV (29) but has no effect on the negative-sense Phasi Charoen-like virus (30). In addition to characterizing AeAV, we also studied the effect of Wolbachia on AeAV replication and coinfection of AeAV and DENV in A. aegypti cells.
(This article was submitted to an online preprint archive [31].)
RESULTS
Identification and assembly of the full AeAV genome from Wolbachia-infected Aedes cells.
During replication of RNA viruses in A. aegypti mosquitoes, the RNA interference (RNAi) pathway cleaves viral double-stranded RNA (dsRNA) intermediates into 21-nucleotide (nt) short interfering RNAs (vsiRNAs) (32, 33). Using the 20- to 32-nt fraction of reads from RNA sequencing (RNA-Seq) data, it is possible to de novo assemble virus genomes (21, 34).
The previously sequenced small RNA fraction of embryonic A. aegypti Aag2 cells and Aag2 cells stably infected with Wolbachia (wMelPop-CLA strain) (35) was trimmed of adapters, filtered for 21-nucleotide (nt) reads, and de novo assembled using CLC Genomics Workbench with a minimum contig length of 100 nt. The resulting contigs were then queried using BLASTX against a local virus protein database downloaded from the National Centre for Biotechnology Information (NCBI). In the Aag2.wMelPop-CLA assembly, four contigs between 396 and 1,162 nt were found to have amino acid similarity (E value of 9.46E−51) to proteins from two closely related Mononegavirales viruses, Culex mononega-like virus 1 (CMLV-1) and Xīnchéng mosquito virus (XcMV), the type species for the Anphevirus genus. No contigs from the Wolbachia-negative A. aegypti Aag2 data set showed any similarity to CMLV-1 or XnMV.
Subsequent reverse transcription-PCR (RT-PCR) analysis between RNA samples from Aag2 and Aag2.wMelPop-CLA cell lines indicated that this tentative virus was exclusive to the Aag2.wMelPop-CLA cell line (Fig. 1A). We hypothesized that the presence of any putative virus was the result of contamination during Wolbachia transinfection. The Wolbachia wMelPop-CLA strain was isolated from the A. albopictus cell line RML-12 and transinfected into Aag2 (36) and the A. albopictus C6/36 (C6/36.wMelPop-CLA) cell lines (37). RT-PCR analysis of RNA extracted from RML-12 and C6/36 cells, as well as the A. aegypti cell line Aa20, showed that the putative virus was present only in RML-12 cells (Fig. 1A).
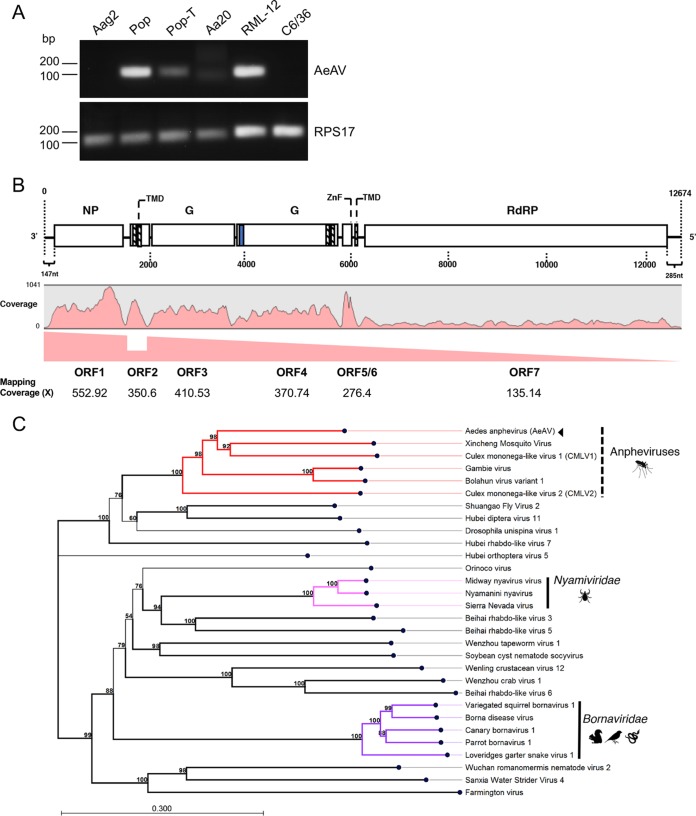
FIG 1.
Presence of AeAV in insect cell lines and genome organization and phylogeny of the virus. (A) RT-PCR analysis of Aedes cell lines Aag2, Aag2.wMelPop-CLA (Pop), wMelPop-CLA.Tet (Pop-T), Aa20, RML-12, and C6/36 for the presence of AeAV. RPS17 was used as a control. (B) Genome organization of the Cali, Colombia, AeAV strain and subgenomic gene transcription profile. Transmembrane domains (TMD) are depicted as boxes with dashed lines, and the signal peptide is depicted as a blue box. NP, nucleoprotein; G, glycoprotein; ZnF, zinc-like finger; RdRP, RNA-dependent RNA polymerase. (C) AeAV is a member of the Anphevirus genus (red), related to members of the Nyamiviridae (pink) and Bornaviridae (purple) in an unassigned family within the order Mononegavirales. A multiple-sequence alignment of the RNA-dependent RNA polymerase and the mRNA capping domain was used to create a maximum likelihood phylogeny. The phylogeny is arbitrarily rooted. One thousand bootstraps were performed, and branches with bootstrap values greater than 85% are highlighted. Branch lengths represent expected numbers of substitutions per amino acid site. Viruses used (GenBank protein accession numbers) are the following: Bolahun virus variant 1 (AOR51366.1), Culex mononega-like virus 1 (CMLV1) (ASA47369.1), Culex mononega-like virus 2 (CMLV2) (ASA47322.1), Gambie virus (AOR51378.1), Xincheng mosquito virus (XcMV) (YP_009302387.1), Borna disease virus (YP_009269418.1), canary bornavirus 1 (YP_009268910.1), Loveridges garter snake virus 1 (YP_009055063.1), parrot bornavirus 1 (AEG78314.1), variegated squirrel bornavirus 1 (SBT82903.1), Midway nyavirus (YP_002905331.1), Nyamanini nyavirus (YP_002905337.1), Sierra Nevada virus (YP_009044201.1), soybean cyst nematode socyvirus (YP_009052467.1), Farmington virus (YP_009091823.1), Beihai rhabdo-like virus 3 (APG78650.1), Beihai rhabdo-like virus 5 (YP_009333422.1), Beihai rhabdo-like virus 6 (YP_009333413.1), Drosophila unispina virus 1 (AMK09260.1), Hubei diptera virus 11 (YP_009337182.1), Hubei orthoptera virus 5 (YP_009336728.1), Hubei rhabdo-like virus 7 (YP_009337121.1), Orinoco virus (ANQ45640.1), Sanxia water strider virus 4 (YP_009288955.1), Shuangao fly virus 2 (AJG39135.1), Wenling crustacean virus 12 (YP_009336618.1), Wenzhou crab virus 1 (YP_009304558.1), Wenzhou tapeworm virus 1 (YP_009342311.1), and Wuchan romanomermis nematode virus 2 (YP_009342285.1).
To recover the remainder of the virus genome, transcriptome RNA-Seq data from RML-12.wMelPop-CLA and C6/36.wMelPop-CLA cells were downloaded (38, 39) and de novo assembled with automatic bubble and word sizes using CLC Genomics Workbench. BLASTN analysis of assembled contigs indicated that a complete 12,940-nt contig from the C6/36.wMelPop-CLA cells and two contigs (9,624 and 3,487 nt) from RML-12.wMelPop-CLA were almost identical (99 to 100% pairwise nucleotide identity) to the virus-like contigs assembled from Aag2.wMelPop-CLA. We were then able to use this reference to recover the full genomes from Aag2.wMelPop-CLA and RML-12.wMelPop-CLA consensus mapping to this reference. To revalidate that AeAV was only present in Aag2.wMelPop-CLA cells, reads from the Wolbachia-negative Aag2 cells were mapped to the representative genome, and only four reads were identified in the data. The results here and from the RT-PCR analysis described above (Fig. 1A) also confirm that the virus found in the Wolbachia-transinfected cells originate from RML-12 cells in which wMelPop-CLA was originally transinfected and subsequently transferred to other cell lines.
Characterization of AeAV.
AeAV genomes assembled in this study were between 12,455 and 13,011 nucleotides in length, with a %GC content of 46.8%, and carry 7 nonoverlapping ORFs (Fig. 1B). Phylogenetic analysis of the RNA-dependent RNA polymerase (RdRp) protein places AeAV within a well-supported clade of the unassigned Anphevirus genus, which are from the order Mononegavirales and closely related to members of Bornaviridae and Nyamiviridae (Fig. 1C).
All members of Mononegavirales have a negative-stranded RNA genome encapsidated within the capsid and the RNA polymerase complex (40). The RNA genome is used as the template by the RNA polymerase complex to sequentially transcribe discrete mRNAs from subgenomic genes. mRNA from each gene is capped and polyadenylated. To analyze the transcriptional activity of AeAV, we used the poly(A)-enriched RNA-Seq libraries prepared from the Cali, Colombia, laboratory strain (41). Read mapping and coverage analysis of the AeAV genome showed that AeAV follows the trend of reduced transcriptional activity seen in other Mononegavirales species (42), with approximately 50% reduction between ORF1 and ORF2 but increased transcription between ORF2 and ORF3 (Fig. 1B). The reduction in transcriptional activity of AeAV genes is conserved for each sequential ORF, with the least transcriptional activity for ORF7/L protein, which is conserved in all AeAV strains in poly(A)-enriched RNA-Seq libraries (see Fig. S1 in the supplemental material).
ORF1 of AeAV encodes a predicted 49-kDa nucleoprotein with no transmembrane domains and closest pairwise amino acid identity (26%) to the nucleoprotein gene from CMLV-1 from Culex mosquitoes in Western Australia (25). Protein homology analysis using HHPred showed that ORF1 was a likely homolog of the p40 nucleoprotein of the Borna disease virus (probability, 98.66%; E value, 7.1e−10). ORF2 encodes an 11-kDa protein with two transmembrane domains in the N terminus of the protein with no similarity to any proteins within the nonredundant protein database or homologs, as predicted by HHPred. ORF3 and ORF4 encode 64-kDa and 72-kDa putative glycoproteins, respectively. ORF3 has no pairwise amino acid similarity to any virus protein or homologs per HHPred analysis. ORF4 was predicted to have a signal peptide in the N terminus followed by a heavily O- and N-linked glycosylated outside region as well as two transmembrane domains in the C terminus of the protein. ORF4 is most closely related to the glycoprotein from the Gambie virus identified from West African A. gambiae mosquitoes, with 45% pairwise amino acid identity (15). Protein homology analysis predicted ORF4 to be a homolog of the human herpesvirus 1 envelope glycoprotein B (probability, 99.88%; E value, 2.2e−22).
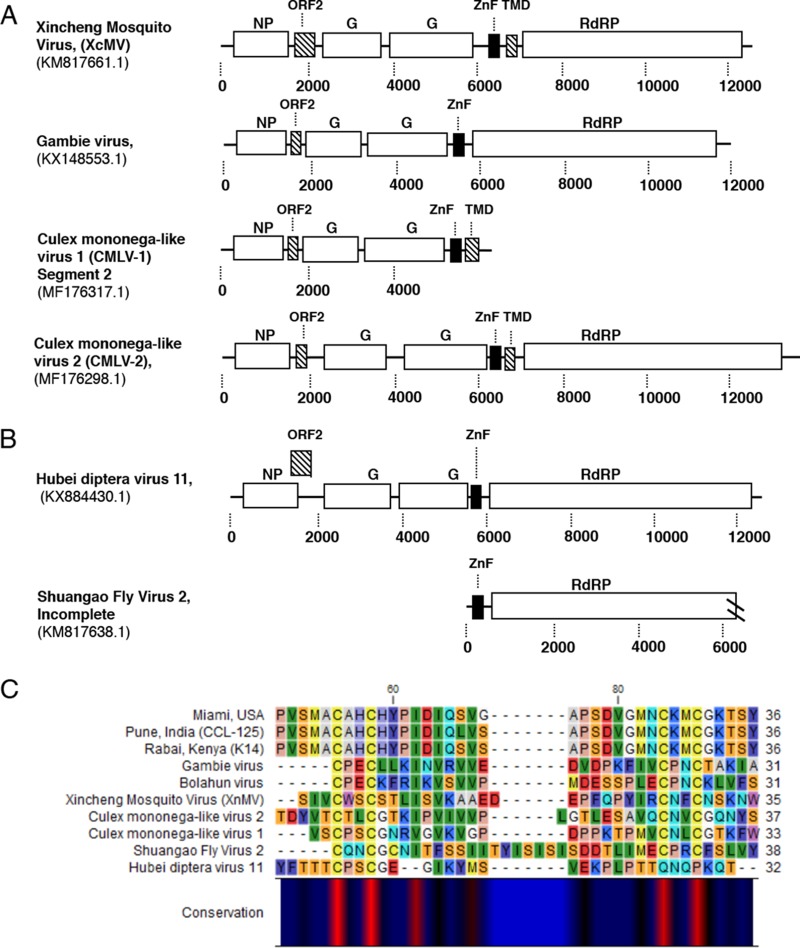
The presence of a Zinc-like finger (ZnF) domain in a small ORF proximal to the L protein previously reported in closely related viruses (15) (Fig. 2A and B) was identified in AeAV based on sequence alignment (Fig. 1B). Reanalysis of putative ORFs from CMLV-1 and CMLV-2 (25) showed the presence of this GATA-like ZnF domain in both of these viruses and the genus type species XcMV identified from Anopheles sinensis (24) (Fig. 2C).
FIG 2.
Conservation of GATA-like zinc finger (ZnF) domain and small transmembrane domain-containing protein between tentative members of the Anphevirus taxon. (A) Genome orientation of previously discovered viruses within the Anphevirus taxon. (B) Two viruses within a closely related clade. Predicted ORF encoding the ZnF domain is indicated by a black square. Predicted ORFs containing transmembrane domains are indicated by dashed lines. GenBank accession numbers are shown below virus names. NP, nucleoprotein; G, glycoprotein; ZnF, zinc-like finger; RdRp, RNA-dependent RNA polymerase. (C) Alignment of predicted GATA-like ZnF protein sequence [C-X(2)-C-X (17–20)-C-X(2)-C] between three representative strains of AeAV (Miami, Florida, USA; Pune, India; Rabai, Kenya) and predicted ZnF domain proteins from panels A and B.
ORF6 encodes a small 4-kDa protein that has a single transmembrane domain in the C terminus. This protein was almost missed in the prediction of ORFs due to having only 37 amino acids; however, it has a strong transcriptional coverage in poly(A) data sets and exists in all assembled strains (Fig. 1B and Fig. S1). It was predicted to share no structural homology or amino acid identity with any previously reported peptide. In addition to this, we were able to identify small transmembrane domain-containing proteins proximal to or overlapping the ZnF protein in CMLV-1, CMLV-2, and XcMV (Fig. 2A), suggesting that this protein is a conserved feature of anpheviruses.
ORF7 encodes the 226-kDa L protein and has 41% pairwise amino acid identity with the RNA-dependent RNA polymerase from CMLV-1. Protein domain analysis of the L protein showed the highly conserved Mononegavirales RNA-dependent RNA polymerase, mRNA capping domain, and an mRNA (guanine-7-)methyltransferase (G-7-MTase) domain conserved in all L proteins in Mononegavirales (43).
AeAV cis-regulatory elements.
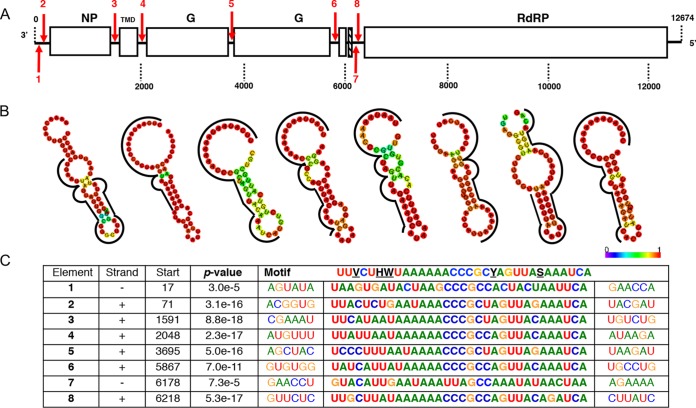
For identification of cis-regulatory elements in the AeAV genome, we used MEME (Multiple Em for Motif Elicitation) to search for overrepresented 5- to 50-nt motifs (44). Using a 0-order Markov model, one 32-nt motif, 3′-UUVCUHWUAAAAAACCCGCYAGUUASAAAUCA-5′, was considered statistically significant (E value of 4.2e−010). Importantly, the motif was proximal to each predicted virus gene ORF, suggesting it is a promoter (Fig. 3A and C). No motif was found between ORF 5 and 6 in AeAV, suggesting that these two genes are under the control of a single cis-regulatory element. Interestingly, the complement of this motif appeared twice on the antigenome, suggesting that it can be used in an antigenome virus intermediate. We noticed that these motifs localized to partial palindromic repeats and predicted that they form stable secondary RNA structures. Using RNAfold, we were able to visualize and predict the MFE structure 20 nt upstream and downstream of the motif (45). All predicted cis-regulatory motifs formed partial or complete stable secondary stem-loops and hairpins with high base-pair probabilities (Fig. 3B). The exception was predicted element 3, which is proximal to the second ORF; as this second gene is transcribed less than ORF3, irrespective of similarity to the motif, the lack of a stable stem-loop structure may be a novel transcriptional regulatory mechanism. The presence of two conserved homopolymeric triplets in the overrepresented motif is very similar to “slippery” −1 ribosome frame shifting (RFS) sites XXX YYY Z (X = A, G, U; Y = A, U; Z = A, C, U) (46). It has been previously demonstrated that similar slippery sequence motifs followed by a predicted stem-loop structure is a feature of rhabdovirus gene overlap regions (47). In AeAV, this feature appears in the intergenic space and is unlikely to represent ribosomal frame shifting events and subsequent extension of a protein. We also searched for additional slippery motifs in the AeAV genome. The genomic context for each predicted “slippery motif” did not extend or produce additional ORFs.
FIG 3.
AeAV cis-regulatory elements. (A) Location and orientation of predicted cis-regulatory element in AeAV indicated by numbered red arrows, with down indicating genome and up indicating antigenome. (B) Predicted minimum free energy (MFE) RNA structure of the region surrounding the motif for each element using the RNAfold web server. Color indicates probability of base pairing, and motif is indicated by the black line. (C) Sequence of the conserved motif as predicted by MEME as well as location and the statistical confidence of the motif. Sequences are written 3′ to 5′, and antigenome motif sequences 1 and 7 are depicted as reverse complement for visual clarity.
AeAV infection is widespread in A. aegypti laboratory colonies, wild-caught mosquitoes, and cell lines.
Taking advantage of the currently published RNA-Seq data, we performed a meta-analysis of global incidence and genetic diversity of this virus. We were able to show that AeAV is ubiquitous in laboratory colonies, cell lines, and wild-caught A. aegypti mosquitoes. During preparation of the manuscript, a partial AeAV genome of 5,313 nt (GenBank accession number MG012486.1) was deposited into the NCBI nucleotide database from a study characterizing the evolution of Piwi-interacting RNA (piRNA) pathways across arthropods (48). Using our AeAV genome reference, we were able to complete the coding sequences (CDS) portion of the genome and also 16 additional strains of AeAV with two additional incomplete genomes (Fig. 4) (Table S1).
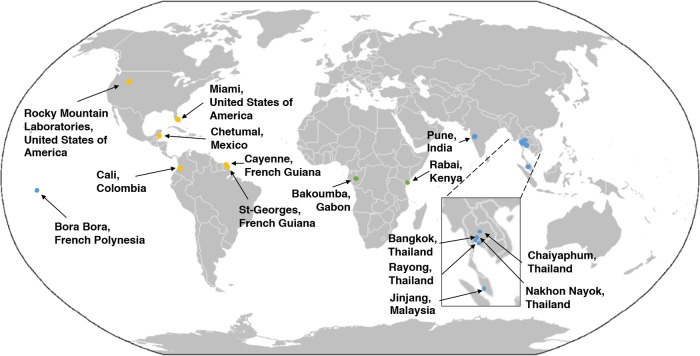
FIG 4.
AeAV has worldwide distribution in A. aegypti laboratory colonies, cell lines, and wild-caught mosquitoes. Locations of mosquito collection from RNA-Seq data that were positive for AeAV are shown (see Table S1). Points refer to collection sites from American (orange), Asia-Pacific (blue), and African (green) locations.
AeAV was present in colonies of A. aegypti established from eggs collected in Bakoumba, Gabon (49), and also from Rabai, Kenya (designated K2 and K14), as well as four mated hybrid strains (50). In colonies wild caught from locations in the Americas, full genomes of AeAV were assembled from Miami, Florida, USA, Cali, Colombia (41), and Chetumal, Mexico (51, 52), laboratory strains (48). Partial genomes of AeAV were assembled from Cayenne and St-Georges, French Guiana (53). AeAV was identified in colonies established from eggs collected in Chaiyaphum and Rayong, Thailand (50, 54), as well as Jinjang, Malaysia (55). AeAV was also identified from the widely used Bora-Bora reference strain from French Polynesia (56). AeAV was also present in eight pools of wild-caught A. aegypti mosquitoes used for ZIKA biosurveillance in Miami, Florida, USA (57), as well as Nakhon Nayok and Bangkok, Thailand (18, 22).
In Aedes cell lines, AeAV was assembled from RNA-Seq data from the larval A. aegypti line CCL-125 origniating from Pune, India (58), and sequenced by the Arthropod Cell Line RNA-Seq Initiative, Broad Institute (broadinstitute.org). With the exception of RNA-Seq data from the three Aedes cell lines stably infected with Wolbachia (RML-12.wMelPop-CLA, C6/36.wMelPop-CLA, and Aag2.wMelPopCLA), AeAV was not identified in any other available C6/36 or Aag2 RNA-Seq libraries.
Genetic variation and evolution of AeAV strains.
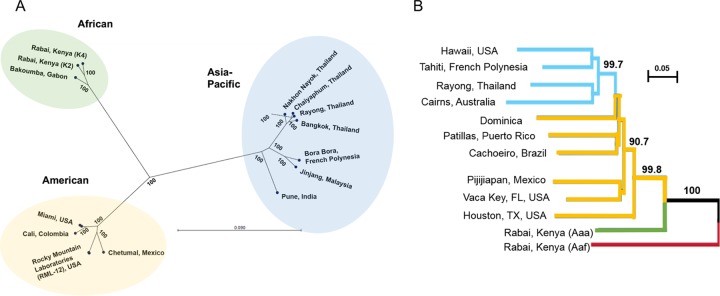
To assess relatedness and evolution between AeAV strains, a maximum likelihood phylogeny (PhyML) was undertaken of the CDS region of all strains with complete genomes (Fig. 5A). The unrooted radial phylogenetic tree indicated three strongly supported monophyletic lineages associated with the geographic origin of the sample. We have designated these lineages of AeAV as African, American, and Asia-Pacific (Fig. 5B).
FIG 5.
AeAV strains have evolved into African, Asia-Pacific, and American lineages. (A) Maximum likelihood phylogeny (PhyML) between AeAV strains using a GTR + G + T model with 1,000 bootstraps. Branch lengths represent expected numbers of substitutions per nucleotide site. For visual clarity, the RML-12 clade and Miami clades were collapsed and single examples are shown. (B) Evolutionary history of worldwide sampling of A. aegypti, adapted from references 60 and 61, from 1,504 SNP species. Bootstrapped neighbor-joining network based on population pairwise chord distances with node support over 90% is shown on relevant branches. New World (American) populations are in yellow, and Asia-Pacific populations are shown in light blue. We have truncated the tree and rooted it to A. aegypti formosus (Aef), shown as a red branch.
In the American lineage of AeAV, all strains that are associated with Wolbachia-infected Aedes cell lines (RML-12.wMelPop-CLA, C6/36.wMelPop-CLA, and Aag2.wMelPopCLA) are almost identical (99.55 to 99.86% identity), supporting the hypothesis that contamination of C6/36 and Aag2 cell lines infected with Wolbachia is likely from the original RML-12 cell line. AeAV from the eight wild-caught pools of A. aegypti mosquitoes from Florida (57) and the laboratory colony established from wild-collected samples from Florida (48) were almost identical (99.86% pairwise identity), with only 17 nt differences over the CDS region. The three African lineage strains of AeAV were slightly closer in pairwise nucleotide identity to the American strains (92.65 to 93.15%) than the Asia-Pacific strains (91.63% to 91.74%). All samples that originated from Thailand form a monophyletic group and are closely related to other Thai strains (99.23 to 99.62%).
We hypothesized that AeAV was harbored as part of the virome of A. aegypti mosquitoes, as A. aegypti expanded from its sub-Saharan African location into the Americas and Asia-Pacific (59). Phylogenetic studies of the A. aegypti genome support the origin of A. aegypti from Africa into the New World (Americas) and a subsequent secondary invasion of A. aegypti from the New World to the Asia-Pacific region (60, 61). Comparing the evolution of the A. aegypti nuclear genome with the evolution of AeAV indicates that the Asia-Pacific strains of AeAV have not evolved from the currently circulating American strain lineage. This indicates that the virus was established independently in both the New-World Americas and also in the Asia-Pacific (Fig. 5B).
Anphevirus-like insertions into the A. aegypti genome.
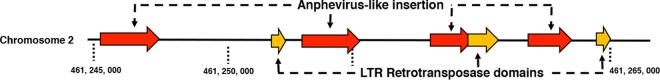
The A. aegypti genome has a large repertoire of virus genes and partial viral genomes, termed endogenous viral elements (EVEs) (49, 62). To explore the possibility of anphevirus-like insertions within the A. aegypti genome, we queried the most recent Liverpool genome (Aaegl5) with the Aag2.wMelPop-CLA AeAV reference strain using the VectorBase BLASTN suite (https://www.vectorbase.org/blast). There were numerous hits of nucleotide similarity (67 to 70%) of 500- to 1,704-nt regions dispersed throughout the A. aegypti genome. EVEs are acquired through recombination with long terminal repeat (LTR) retrotransposons (62). We present one ∼20-kb portion of chromosome 2 of the A. aegypti genome (Fig. 6) with four anphevirus-like insertions and close proximity to LTR retrotransposable fragments in unidirectional orientation. This suggests insertion of viral elements through LTR retrotransposases and a long evolutionary history of challenge with anphevirus-like species in A. aegypti.
FIG 6.
Genomic context for anphevirus-like insertions into the A. aegypti genome. A 21,242-nt portion of chromosome 2 depicting anphevirus insertions (red) with predicted ORFs that encode LTR retrotransposase elements (yellow).
AeAV replicates in Aedes cell lines but does not replicate in three mammalian cell lines.
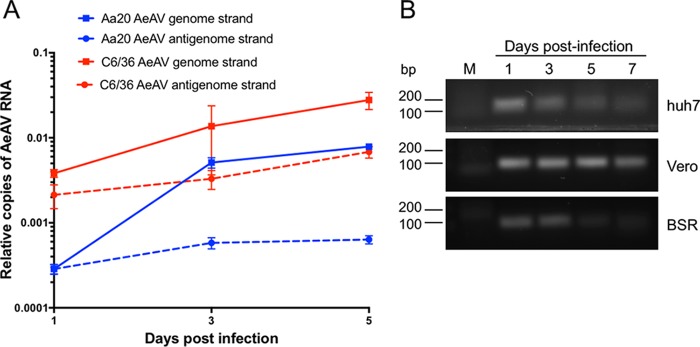
Supernatant of Aag2.wMelPop-CLA cells was infectious to both A. aegypti cells (Aa20) and A. albopictus C6/36 cells over a 5-day time course through RT-quantitative PCR (qPCR) analysis (Fig. 7A). Generally, there were significantly more relative AeAV genome copies detected in C6/36 cells at 1 and 5 days postinfection (dpi) than in Aa20 cells. There were also significantly more antigenome copies of AeAV in C6/36 cells over the 5-day time course. The higher replication of AeAV in C6/36 cells than in Aa20 cells is not unexpected, since C6/36 cells are RNAi deficient and generally RNA viruses replicate more efficiently in these cells (63–65).
FIG 7.
AeAV is infectious to Aedes cell lines but does not replicate in Huh-7, Vero, and BSR vertebrate cell lines. (A) RT-qPCR of AeAV genome and anti-genome in a 5-day time course in A. aegypti Aa20 cells and A. albopictus C6/36 cells. Error bars represent the standard errors of the means (SEM) from three biological replicates. (B) RT-PCR of AeAV genome in a 7-day time course in human hepatocellular carcinoma cells (Huh-7), African green monkey cells (Vero), and baby hamster kidney (BSR). M, mock-infected cells.
We assumed that AeAV is an insect-specific virus based on its phylogenetic position; however, to test if AeAV can replicate in mammalian cells, we inoculated human hepatocellular carcinoma cells (Huh-7), African green monkey cells (Vero), and baby hamster kidney (BSR) cells with medium from AeAV-infected cells and performed RT-PCR for AeAV RNA genome abundance over a 7-day time course. While AeAV RNA (most likely from the inoculum) could be detected by RT-PCR at days 1 and 3 after inoculation, it did not increase over time and was visibly reduced in the mammalian cells by 5 and 7 dpi (Fig. 7B). AeAV was also not detected in the A. gambiae cell line MOS-55 transinfected with wMelPop-CLA from RML-12-wMelPop-CLA (37), sequenced by the Arthropod Cell Line RNA-Seq Initiative, Broad Institute (broadinstitute.org). Taken together, the results suggest that AeAV infection is restricted within the subfamily Culicinae or even the Aedes genus and is insect specific.
Wolbachia pipientis infection in A. aegypti cells enhances AeAV replication.
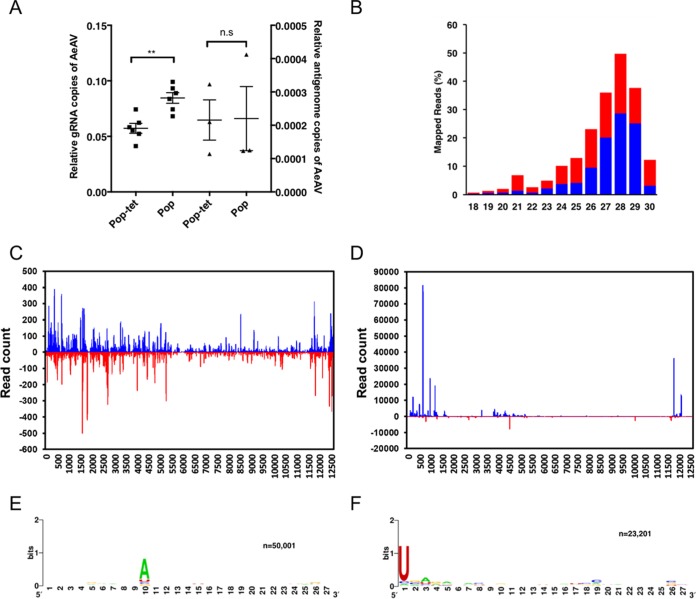
As Wolbachia is being deployed in the field to reduce dengue transmission, we were interested to find out if it has any effect on replication of AeAV. We extracted RNA from Aag2.wMelPop-CLA cells and a previously tetracycline-cured Aag2.wMelPop-CLA cell line (66) and tested the effect of Wolbachia infection on AeAV genome and antigenome copies. AeAV genomic RNA copies were significantly greater in Wolbachia-infected (Aag2.wMelPop-CLA) cells than in tetracycline-cleared (Aag2.wMelPop-CLA.Tet) A. aegypti cells; however, there was no statistically significant difference between the relative antigenome copies of the two cell lines (Fig. 8A).
FIG 8.
AeAV genome replication is enhanced by Wolbachia infection in A. aegypti cells and produces abundant vsiRNAs and vpiRNAs. (A) RT-qPCR of the AeAV genomic (gRNA) and antigenomic RNA in tetracycline-cured Aag2.wMelPop-CLA cells (Pop-tet) and Aag2.wMelPop-CLA cells (Pop) relative to RPS17. Error bars represent the SEM from six (genome) and three (antigenome) biological replicates. n.s, not significant; **, P < 0.01. (B) Mapping profile of pooled small RNA fraction in Aag2.wMelPop-CLA cells. (C) Alignment of the 21-nt sRNA reads (representing siRNAs) and (D) the 26- to 31-nt reads (representing piRNAs) mapped to the AeAV antigenome (blue) and genome (red) in Aag2.wMelPop-CLA cells. Relative nucleotide frequency and conservation of the 28-nt small RNA reads that mapped to the genome (E) and the antigenome (F) of AeAV in Aag2.wMelPop-CLA cells.
To explore the host small RNA response to AeAV, clean reads from previously prepared sRNA libraries from Aag2.wMelPop-CLA and Aag2 (35) were mapped to the AeAV genome. In the cytoplasmic fraction of the Aag2.wMelPop-CLA sample, 870,012 of 4,686,954 reads (18.56%) mapped to AeAV. In the nuclear fraction, 420,215 of 11,406,324 reads (3.68%) mapped to the genome. In the combined Aag2 sRNA library, of 8,600,821 clean reads only four reads mapped to the AeAV genome. The mapping profile of 18- to 31-nt reads mapped from the Aag2.wMelPop-CLA library to AeAV indicates a higher proportion of 27- to 31-nt virus-derived PIWI RNAs (vpiRNAs) than the 21-nt vsiRNAs (Fig. 8B).
Analysis of the profile of mapped AeAV vsiRNAs fairly ubiquitously targeted the AeAV genome and antigenome (Fig. 8C). Hotspots in the vpiRNA mapping profile appeared to target the 3′-untranslated region (UTR) and ORF1 and the 5′UTR of the AeAV antigenome (Fig. 8D). Biogenesis of vpiRNAs are independent of Dicer-2, with a bias for adenosine at position 10 in the sense position and a uracil in the first nucleotide of antisense polarity (67). This ping-pong characteristic signature was apparent in the vpiRNA reads from the cell line (Fig. 8E and F).
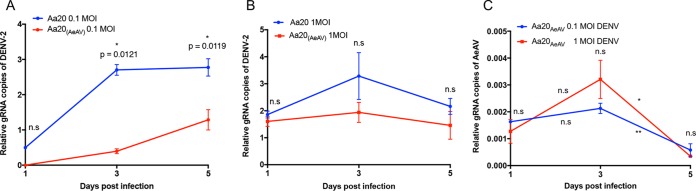
Persistent infection of AeAV in Aa20 cells modestly reduces replication of DENV-2 genomic RNA.
Recently, it has been demonstrated that in Aedes cell lines experimentally infected with two ISVs, replication of DENV and ZIKV was reduced (13). To test if there was any interaction between AeAV and the subsequent challenge of cells with DENV, we generated an Aa20 cell line inoculated with medium from RML-12 and maintained it for three passages. Aa20 cells persistently infected with AeAV were challenged with DENV-2 ET-100 strain at multiplicities of infection (MOIs) of 0.1 and 1. RT-qPCR analysis of DENV-2 genomic RNA showed that accumulation was, on average, less in AeAV-infected Aa20 cells than in the control (Fig. 9A and B). This reduction in DENV-2 genome copies was statistically significant at an MOI of 0.1 at both 3 and 5 days postinfection. No AeAV-related RT-qPCR product was detected in mock-infected Aa20 cells (data not shown). We also examined the effect of DENV infection on AeAV RNA levels in the AeAV persistently infected cells. RT-qPCR analysis showed no significant effect on AeAV levels between 1 and 3 days post-DENV infection; however, AeAV genomic RNA levels significantly declined at 5 days post-DENV infection (Fig. 9C). There was no significant difference in the results between a DENV MOI of 0.1 and 1.
FIG 9.
AeAV reduces dengue virus replication in Aa20 cells. Aa20 cells persistently infected with AeAV were infected with DENV-2 at MOIs of 0.1 (A) and 1 (B). Total RNA was extracted at 0, 1, 3, and 5 days following DENV-2 inoculation and analyzed by RT-qPCR. (C) RT-qPCR analysis of AeAV persistently infected Aa20 cells infected with DENV-2 at MOIs of 0.1 and 1 using primers specific to the AeAV genome. Error bars represent the SEM from three biological replicates. n.s, not significant; *, P < 0.05; **, P < 0.01.
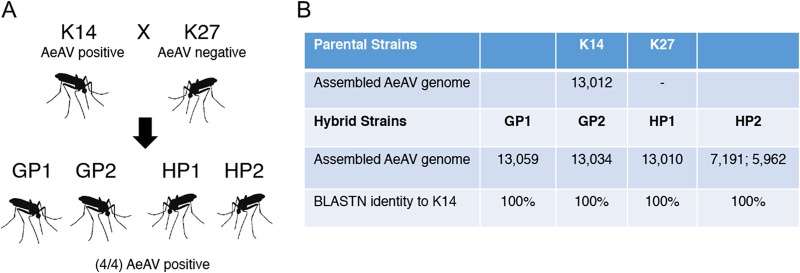
Evidence for vertical transmission of AeAV.
We were fortunate to explore the potential vertical transmission of AeAV by using RNA-Seq data of uninfected and infected mated individuals from a study characterizing the genetic basis of olfactory preference in A. aegypti (50). Briefly, McBride and colleagues used eggs from a number of A. aegypti species in Rabai, Kenya, to establish laboratory colonies for RNA-Seq analysis. We identified AeAV in the domestic K2 and K14 colonies, which was seemingly absent from the other Rabai (K18, K19, and K27) colonies. The K27 colony was interbred with strain K14, which we found to be AeAV positive. In all four resultant hybrid colonies, which were subjected to RNA-Seq analysis, we were able to de novo assemble identical K14 AeAV strain genomes (Fig. 10).
FIG 10.
AeAV is potentially vertically transmitted. (A) Diagram showing the parental (K14 and K27) and hybrid strains (GP1, GP2, HP1, and HP2) from reference 50. (B) Table showing assembly statistics and BLASTN similarity of AeAV genomes assembled from K14 and K27 hybrid strains.
The possibility of vertical transmission also is supported by the presence of AeAV in RNA-Seq data from both the sperm of adult male mosquitoes (55) and the female reproductive tract (54).
DISCUSSION
The ability of AeAV to propagate in A. aegypti and A. albopictus cell lines but not in the three mammalian cell lines suggests that AeAV is an ISV, although this needs to be further confirmed using cell lines from other species. To the best of our knowledge, this is the first comprehensive characterization of any Anphevirus species within Mononegavirales and the first Mononegavirales virus species within A. aegypti; however, as the manuscript was under revision, the complete genome sequence of AeAV and its phylogenetic relationship with other ISVs was published in a short communication (68). While we have demonstrated that AeAV is spread worldwide in A. aegypti mosquitoes, we have limited understanding as to how prevalent AeAV is in individual mosquitoes in wild populations, its tissue tropism, or potential impacts on the host. Although it is likely that AeAV is maintained in wild populations of A. aegypti mosquitoes through vertical transmission, it is possible that AeAV could be maintained through venereal transmission, as we were able to identify the whole AeAV genome from a data set prepared from the sperm of adult male A. aegypti mosquitoes (55).
To our knowledge, the oldest continually maintained colony of laboratory mosquitoes with AeAV present comes from Jinjang, Malaysia, which was established from wild-collected samples from as early as 1975 (55). This suggests vertical transmission rates of AeAV are high or that there is a high incidence of AeAV within the colony. In comparison, in both French Guiana colonies, we were unable to recover the complete genomes from these strains. It is unlikely that this is due to insertion of AeAV into the nuclear A. aegypti genome, as numerous reads mapped to the ORF7/L region of the reference strain; however, there was not enough coverage to reach consensus on the full genome. As these libraries were prepared from homogenates of mosquitoes, it seems likely that the incidence of infection within these colonies will be lower; however, further analysis would have to be conducted.
In our analysis, AeAV was not detected in any of the widely used Liverpool (LVP) and Rockefeller/UGAL, as well as derived “white eye mutant,” strains of A. aegypti. Analysis of published noncoding RNA-Seq data from Australian Townsville and Cairns colonies of wild-caught Australian mosquitoes (69, 70) suggests that there is no RNAi response or presence of AeAV in these mosquitoes. Evidence from this study and others suggests that widely used laboratory strains of A. aegypti harbor a diverse and heterogeneous virome composition and contribute to the variable vector competence between these colonies.
Also in our analysis, the geographic origin of the RNA-Seq samples matched the resulting phylogenetic relationship of each strain. The presence of AeAV in the A. albopictus cell line RML-12, presumably the origin of the AeAV contamination in other Aedes cell lines transinfected with an adapted wMelPop strain, was the only A. albopictus sample in our analysis. During our analysis, we queried all of the 266 currently available A. albopictus RNA-Seq data sets (taxonomy ID 7160) uploaded to the Sequence Read Archive (SRA), none of which indicated the presence of AeAV. We hypothesize that AeAV from RML-12 is due to contamination, as the cell line is often mischaracterized as originating from A. aegypti (71, 72). In many laboratories that study arbovirus interactions, more than one Aedes cell line is maintained. As the RML-12 AeAV strain is genetically placed within the American lineage and the namesake of the cell line, Rocky Mountain Laboratories, (71) is located in Montana, contamination from domestic A. aegypti mosquito samples is possible. While no other A. albopictus RNA-Seq data were positive for AeAV in this study, we cannot rule out the possibility that AeAV exists in American populations of A. albopictus. A. aegypti and A. albopictus coexist in North America and compete for larval habitats of discarded tires and other artificial containers (73).
RNAi response is commonly observed in mosquitoes against RNA viruses. This response includes miRNA, siRNA, and piRNA pathways (74, 75). Similarly, we found a large number of 21-nt vsiRNAs produced against AeAV in infected cells that were evenly mapped to both sense and antisense strands, indicating that dsRNA intermediates produced during replication must be the target of the host cell RNAi response. In addition, a large number of vpiRNAs were found mapped to the 5′UTR, ORF1, and 3′UTR of the AeAV genome. These vpiRNAs had the typical ping-pong signature (U1-A10) of secondary piRNAs. This signature has also been found in vpiRNAs produced during alphavirus (76) and bunyavirus (63, 77) infections but not in vpiRNA-like small RNAs in most flaviviruses, such as DENV (78), ZIKV (79, 80), and an insect-specific flavivirus (81). We found that a higher proportion of small RNAs from Aag2-wMelPop-CLA cells that mapped to AeAV are vpiRNAs (about 50%), and less than 10% are vsiRNAs. Literature suggests that when the siRNA pathway is compromised more vpiRNAs are produced. This has been shown in RNAi-deficient C6/36 cells infected with Sindbis virus, Rift Valley fever virus (RVFV), or La Crosse virus (63–65). The RNA-Seq data from C6/36.wMelPop-CLA cells were for long transcripts rather than small RNAs; therefore, we were not able to confirm if in those cells there are higher proportions of vpiRNAs than vsiRNAs. The overrepresentation of vpiRNAs with respect to vsiRNAs has also been demonstrated in negative-sense Bunyavirales members PCLV and RVFV (30, 65) in Aag2 cells for all segments of the genome. It remains to be seen, however, if the higher vpiRNA-to-vsiRNA ratio in Aag2.wMelPop-CLA is due to suppression of the siRNA pathway by AeAV or, alternatively, if Wolbachia has an effect on the siRNA pathway. However, it seems more likely that as negative-sense RNA viruses produce fewer dsRNA replicative intermediates, these could be simply less targeted by the siRNA pathway and are unable to be resolved by sRNA profiling. These possibilities require further investigations, and the role of the vpiRNAs in AeAV replication or host antiviral response remains to be determined.
The effects of Wolbachia on virus restriction are variable and depend on Wolbachia strain, virus family, and transinfected host (82). Wolbachia was shown to enhance AeAV replication in A. aegypti cells in this study. Recent studies have demonstrated that Wolbachia has no effect on multisegmented negative-sense RNA viruses; for example, PCLV (family Bunyaviridae) in the wMelPop-CLA strain-infected Aag2 cells (30), La Crosse virus (family Bunyaviridae) and vesicular stomatitis virus (family Rhabdoviridae), which is in the same order as AeAV, in wStri strain in the A. albopictus C710 cell line (83), and RVFV (family Phenuiviridae) in Culex tarsalis mosquitoes transinfected with a somatic Wolbachia (strain wAlbB) had no effect on RVFV infection or dissemination (84). RVFV and PCLV belong to the Bunyavirales order and AeAV belongs to Mononegavirales, distinct orders of negative-strand RNA viruses. All three viruses have conserved features that may provide insight into how they might be protected from restriction by Wolbachia. The genome of all negative-sense ssRNA viruses is encapsidated within the nucleoprotein (85, 86) and is attached to RdRp within the virion (87). The RNA-dependent polymerase complex carries out both transcription of virus genes and replication of the genome.
Wolbachia has been shown to restrict a number of positive-sense RNA virus species from the Togaviridae and Flaviviridae families (82). After fusion and entry into the host cell, the genomes of Togaviridae and Flaviviridae species are released into the cytoplasm and translated directly into polypeptide protein(s). These polypeptide proteins are processed by viral and cellular proteases to generate the mature structural and nonstructural proteins, which are then used to replicate the genome (88). While the exact mechanism for RNA virus restriction in Wolbachia-infected insects has remained elusive, it has been shown that restriction of RNA viruses by Wolbachia happens early in infection (89, 90). In the A. albopictus cell line C710 stably infected with wStri, the polypeptide of ZIKV is not produced, as determined by immunoblotting 1 day postinfection (90). Additionally, Wolbachia exploits host innate immunity to establish a symbiotic relationship with A. aegypti (91). Perhaps the combination of protection of the RNA nucleocapsid genome or genome segments when released in the cytoplasm or activity of the RdRp aids in evasion of host immune response enhanced by Wolbachia or Wolbachia effector molecules (92). However, a recent study suggested increases in infection of A. aegypti mosquitoes by insect-specific flaviviruses when they harbor the Wolbachia wMel strain (93).
The ability of AeAV to modestly reduce DENV-2 genomic RNA in a persistently infected cell line was unexpected. While it has previously been shown that members of the same virus family can provide superexclusion of additional viruses (12, 94), little work has been undertaken to look at cross-viral family exclusion effects. Our results showed that less DENV replication occurred in the presence of AeAV, with the difference being particularly significant at lower MOI. If this suppressive effect also occurs in mosquitoes, enhancement of AeAV in A. aegypti mosquitoes infected with Wolbachia may be beneficial in terms of DENV suppression.
As A. aegypti is perhaps the most important vector of arboviruses worldwide, further work should be undertaken to understand and characterize the virome of this mosquito and effects on mosquito life-history traits. Our findings provide new insights into the evolution and genetic diversity of AeAV across a wide geographic range as well as into the virus features and families restricted by Wolbachia in mosquito hosts and its effects on arboviruses they transmit.
MATERIALS AND METHODS
Cell line maintenance and experimental infection with AeAV.
An A. aegypti cell line (Aag2) stably infected with a Wolbachia strain (denoted Aag2.wMelPop-CLA) as previously described for C6/36.wMelPop-CLA (36), with its previously generated tetracycline-treated line (66), and both A. albopictus C6/36 (58) and RML-12 cell lines were maintained in 1:1 Mitsuhashi-Maramorosch and Schneider's insect medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS; Bovogen Biologicals). Aa20 cells established from A. aegypti larvae (95) were maintained in Leibovitz's L15 medium supplemented with 10% FBS (Biowest, France) and 10% tryptose phosphate broth at 27°C. African green monkey cells (Vero) were maintained in Opti-MEM I reduced-serum medium supplemented with 2% FBS and 10 ml/liter penicillin-streptomycin (Sigma-Aldrich). Human hepatocellular carcinoma cells (Huh-7) were maintained in Opti-MEM I reduced-serum medium supplemented with 5% FBS and 10 ml/liter penicillin-streptomycin (Sigma-Aldrich). BSR cells (a clone of baby hamster kidney-21) were maintained in Dulbecco's modified Eagle medium (Gibco), 2% FBS, and 10 ml/liter penicillin-streptomycin (Sigma-Aldrich). All mammalian cells were kept at 37°C with 5% CO2.
For experimental infection of cells, 106 cells of Aedes or mammalian cells were seeded in a 12-well plate. Subsequently, supernatant from Aag2.wMelPop-CLA cells was collected, centrifuged at 2,150 × g for 5 min to remove cells and debris, and used as an AeAV inoculation source. One Aa20 cell line was experimentally inoculated with RML-12 cell supernatant and kept as a persistently infected AeAV cell line. Cells were collected at 1, 3, and 5 days postinoculation for Aedes cell lines for RT-qPCR analysis and 1, 3, 5, and 7 days for mammalian cell lines for RT-PCR analysis.
AeAV and dengue virus (DENV-2) interaction assay.
The third passage of Aa20 cells persistently infected with AeAV (denoted Aa20AeAV) and mock-treated Aa20 cells were seeded at a density of 3 × 105 cells in 12-well plates overnight. Cells were then infected with the East Timor (ET-100) DENV-2 strain at MOIs of 0.1 and 1, cells were rocked for an hour at room temperature, and supernatant was discarded and replaced with fresh medium. Cells were collected at 0, 1, 3, and 5 dpi, with mock-infected cells collected at 5 dpi. Cells were subjected to RNA extraction to quantify the DENV-2 genomic RNA levels by RT-qPCR as described below.
Assembly and identification of AeAV strains from RNA-Seq data.
For detection of AeAV in previously published RNA-Seq data, we used the assembled RML-12 AeAV genome as a BLASTN query for all available A. aegypti (taxonomy ID 7159) RNA-Seq data within the Sequence Read Archive (SRA) on NCBI. SRA run files with positive hits of 90 to 100% identity and an E value of <2E−30 were downloaded and converted to fastq using the NCBI SRA toolkit at https://www.ncbi.nlm.nih.gov/sra/docs/toolkitsoft/ for further analysis. FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used for quality checking of fastq files and adapter identification. Fastq files were then imported into CLC Genomics Workbench (10.1.1) and were adapter and quality trimmed (<0.02; equivalent Phred quality score of 17; ambiguous nucleotides, 2).
Two strategies were used to assemble strains of AeAV: fastq files from the same source of A. aegypti were pooled and de novo assembled using the CLC Genomics Workbench assembly program with automatic bubble and word sizes. This was sufficient to assemble the full coding sequences (CDS) of most strains of AeAV. Table S1 in the supplemental material contains de novo assembly statistics from each data set used.
If de novo assembly did not produce the complete AeAV genome, to complete further sections of the AeAV genome clean reads were mapped to the C6/36.wMelPop-CLA strain of AeAV with stringent alignment criteria (match score, 1; mismatch cost, 2; length fraction, 0.89; similarity fraction, 0.89) to exclude false-positive mapping that derives from endogenous viral elements (EVEs). To confirm accuracy of assembly, the largest contigs of consensus mapping were extracted and then used as a reference for remapping and manually checked. Final sequences of the virus genomes were obtained through the majority consensus of the mapping assembly and were given coding complete (CC) or standard draft (SD) genome quality ratings (96).
RNA isolation, strand-specific cDNA synthesis, and RT-qPCR.
Total RNA was extracted from mosquito cells using QIAzol lysis reagent (Qiagen) and treated with Turbo DNase (Thermo Fisher Scientific) per the manufacturers' instructions. RNA quality and quantity were evaluated using a BioTek Epoch microspot plate spectrophotometer. For the production of AeAV genome and antigenome cDNA, two cDNA reactions were generated using 600 ng of RNA and SuperScript III reverse transcriptase (Thermo Fisher Scientific). The genome cDNA strand was synthesized using a forward primer to AeAV (AeAVGenome-RT, 5′-AGACTTCTAAGCCTGCCCACA-3′), and the AeAV anti-genome cDNA strand was synthesized using a reverse orientation primer (AeAVAntiGenome-RT, 5′-ACACTTGCCATGTGCTCAG-3′). Aedes ribosomal protein subunit 17 (RPS17) primers (A. aegypti, AeRPS17-qR, 5′-GGACACTTCGGGCACGTAGT-3′; A. albopictus, AalRPS17-qR, 5′-ACGTAGTTGTCTCTGCGCTC-3′) were used for reference gene cDNA synthesis. Following RT, qPCRs with AeAV primers (AeAV-qF, 5′-GACAATCGCATTGGCTGCAT-3′; AeAV-qR, 5′-CCCGAGACAATCGGCTTCTT-3′) as well as primer pairs for the RPS17 genes (A. aegypti, AeRPS17-qF, 5′-CACTCCGAGGTCCGTGGTAT-3′; A. albopictus, AalRPS17-qF, 5′-CGCTGGTTTCGTGACACATC-3′) were undertaken. RPS17 was used for normalizing data as described previously in A. aegypti cells (97).
For quantitation of DENV-2 genome copies in Aa20 cells, two SuperScript III reverse transcription (Thermo Fisher Scientific) reaction mixtures with 1,000 ng of RNA were prepared. For the DENV-2 genome copy reaction, the reverse primer (DENV2-qR 5′-CAAGGCTAACGCATCAGTCA-3′) was used, and in a separate cDNA synthesis reaction the A. aegypti RPS17 primers described above were used. Subsequently, qPCR for DENV-2 was carried out using a DENV-2 primer pair (DENV2-qF, 5′-GGTATGGTGGGCGCTACTA-3′, and DENV2-qR), and RPS17 was used as a normalizing control as described above.
Each time point for experimental infection was run with three biological replicates and two technical replicates. Platinum SYBR green mix (Invitrogen) was used for qPCR with 20 ng of RT products in a Rotor-Gene thermal cycler (Qiagen) as described above. The relative abundance of AeAV RNA and DENV-2 to the host reference gene was determined by qGENE software using the ΔΔCT method and analyzed using GraphPad Prism.
To test for the replication of AeAV in mammalian cells, 1,000 ng of total RNA extracted from the cells was extracted and used for first-strand synthesis with SuperScript III reverse transcriptase with the AeAVGenome-RT primer. qPCR was subsequently carried out using the AeAVGenome-RT primer and the qPCR primer AeAVqR2 (5′-ATGAAAGTATGGATACACACCGCG-3′). Products were then visualized on a 1% agarose gel.
Virus genome annotation.
Potential ORFs of AeAV were analyzed using the NCBI Open Reading Frame Finder (https://www.ncbi.nlm.nih.gov/orffinder/) with a minimal ORF length of 150. ORFs were cross referenced with mapping from poly(A)-enriched transcriptomes (Fig. S1) to reduce false-positive identification of ORFs. For determination of putative domains in AeAV, ORFs were translated and searched against the Conserved Domain Search Service (CD Search) (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). For protein homology detection, we used the HHPred webserver on translated AeAV ORFs (98).
To best discriminate N-terminal transmembrane domains from signal peptides, we used the consensus TOPCONS web server (99). Glycosylation sites were predicted by the NetNGlyc 1.0/NetOGlyc 4.0 server (http://www.cbs.dtu.dk/services/).
Phylogenetic analysis.
For placement of AeAV within the order Mononegavirales, ClustalW was used on CLC Genomics Workbench to align amino acid sequences of 30 L proteins of the most closely related Mononegavirales species, as determined by BLASTp of the NCBI nonredundant database. A maximum likelihood phylogeny (PhyML) was constructed using the Whelan and Goldman (WAG) amino acid substitution model with 1,000 bootstraps.
To determine relatedness between different strains of AeAV, genomes that were coding complete and had greater than 30× coverage were trimmed of 3′UTR and 5′UTR and aligned using the ClustalW algorithm on CLC Genomics Workbench. A maximum likelihood phylogeny (PhyML) was constructed. A hierarchical likelihood ratio test (hLRT) with a confidence level of 0.01 suggested that the general time-reversible (GTR) + G (rate variation, 4 categories) and + T (topology variation) nucleotide substitution model was the most appropriate. A total of 1,000 bootstrap replicates were performed with 95% bootstrap branching support cutoff.
Statistical analysis.
Unpaired t test was used to compare differences between two individual groups, while one-way analysis of variance with Tukey's post hoc test was carried out to compare differences between more than two groups.
Accession number(s).
All of the complete and incomplete virus genome sequences generated in this study have been deposited in GenBank under the accession numbers MH037149 and MH430648 to MH430666.
Supplementary Material
ACKNOWLEDGMENTS
This project was funded by the Australian Research Council grant (ARC; DP150101782) to S.A. and University of Queensland scholarship to R.P.
For the supply of the DENV-2 (ET-100) strain, we thank Daniel Watterson and Paul Young from the University of Queensland. We thank Daniel Watterson and Jody Hobson-Peters for providing Huh-7 and BSR cells, respectively. We thank the technical assistance of Sultan Asad, Kayvan Etebari and Hugo Perdomo Contreras, as well as current and former members of the Asgari laboratory for their fruitful discussions.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00224-18.
REFERENCES
- 1.Shriram AN, Sivan A, Sugunan AP. 2017. Spatial distribution of Aedes aegypti and Aedes albopictus in relation to geo-ecological features in South Andaman, Andaman and Nicobar Islands, India. Bull Entomol Res 3:166–174. [DOI] [PubMed] [Google Scholar]
- 2.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. 2013. The global distribution and burden of dengue. Nature 496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.European Centre for Disease Prevention and Control. 2015. Rapid risk assessment: Zika virus epidemic in the Americas: potential association with microcephaly and Guillain-Barré syndrome. European Centre for Disease Prevention and Control, Solna, Sweden: https://ecdc.europa.eu/sites/portal/files/media/en/publications/Publications/zika-virus-americas-association-with-microcephaly-rapid-risk-assessment.pdf. [Google Scholar]
- 4.Tabachnick WJ. 2013. Nature, nurture and evolution of intra-species variation in mosquito arbovirus transmission competence. Int J Environ Res Public Health 10:249–277. doi: 10.3390/ijerph10010249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weiss B, Aksoy S. 2011. Microbiome influences on insect host vector competence. Trends Parasitol 27:514–522. doi: 10.1016/j.pt.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beerntsen BT, James AA, Christensen BM. 2000. Genetics of mosquito vector competence. Microbiol Mol Biol Rev 64:115–137. doi: 10.1128/MMBR.64.1.115-137.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall RA, Bielefeldt-Ohmann H, McLean BJ, O'Brien CA, Colmant AM, Piyasena TB, Harrison JJ, Newton ND, Barnard RT, Prow NA, Deerain JM, Mah MG, Hobson-Peters J. 2016. Commensal viruses, transmission, and interaction with arboviral pathogens. Evol Bioinform Online 12:35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blitvich BJ, Firth A. 2015. Insect-specific flaviviruses: a systematic review of their discovery, host range, mode of transmission, superinfection exclusion potential and genomic organization. Viruses 7:1927–1959. doi: 10.3390/v7041927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Cleef KW, van Mierlo JT, Miesen P, Overheul GJ, Fros JJ, Schuster S, Marklewitz M, Pijlman GP, Junglen S, van Rij RP. 2014. Mosquito and Drosophila entomobirnaviruses suppress dsRNA- and siRNA-induced RNAi. Nucleic Acids Res 42:8732–8744. doi: 10.1093/nar/gku528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang G, Asad S, Khromykh AA, Asgari S. 2017. Cell fusing agent virus and dengue virus mutually interact in Aedes aegypti cell lines. Sci Rep 7:6935. doi: 10.1038/s41598-017-07279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hobson-Peters J, Yam AW, Lu JW, Setoh YX, May FJ, Kurucz N, Walsh S, Prow NA, Davis SS, Weir R, Melville L, Hunt N, Webb RI, Blitvich BJ, Whelan P, Hall RA. 2013. A new insect-specific flavivirus from northern Australia suppresses replication of West Nile virus and Murray Valley encephalitis virus in co-infected mosquito cells. PLoS One 8:e56534. doi: 10.1371/journal.pone.0056534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall-Mendelin S, McLean BJ, Bielefeldt-Ohmann H, Hobson-Peters J, Hall RA, van den Hurk AF. 2016. The insect-specific Palm Creek virus modulates West Nile virus infection in and transmission by Australian mosquitoes. Parasit Vectors 9:414. doi: 10.1186/s13071-016-1683-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schultz MJ, Frydman HM, Connor JH. 2018. Dual insect specific virus infection limits Arbovirus replication in Aedes mosquito cells. Virology 518:406–413. doi: 10.1016/j.virol.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandler JA, Liu RM, Bennett SN. 2015. RNA shotgun metagenomic sequencing of northern California (U S A) mosquitoes uncovers viruses, bacteria, and fungi. Front Microbiol 6:185. doi: 10.3389/fmicb.2015.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fauver JR, Grubaugh ND, Krajacich BJ, Weger-Lucarelli J, Lakin SM, Fakoli LS III, Bolay FK, Diclaro JW II, Dabire KR, Foy BD, Brackney DE, Ebel GD, Stenglein MD. 2016. West African Anopheles gambiae mosquitoes harbor a taxonomically diverse virome including new insect-specific flaviviruses, mononegaviruses, and totiviruses. Virology 498:288–299. doi: 10.1016/j.virol.2016.07.031. [DOI] [PubMed] [Google Scholar]
- 16.Bolling BG, Vasilakis N, Guzman H, Widen SG, Wood TG, Popov VL, Thangamani S, Tesh RB. 2015. Insect-specific viruses detected in laboratory mosquito colonies and their potential implications for experiments evaluating arbovirus vector competence. Am J Trop Med Hyg 92:422–428. doi: 10.4269/ajtmh.14-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cook S, Bennett SN, Holmes EC, De Chesse R, Moureau G, de Lamballerie X. 2006. Isolation of a new strain of the flavivirus cell fusing agent virus in a natural mosquito population from Puerto Rico. J Gen Virol 87:735–748. doi: 10.1099/vir.0.81475-0. [DOI] [PubMed] [Google Scholar]
- 18.Chandler JA, Thongsripong P, Green A, Kittayapong P, Wilcox BA, Schroth GP, Kapan DD, Bennett SN. 2014. Metagenomic shotgun sequencing of a Bunyavirus in wild-caught Aedes aegypti from Thailand informs the evolutionary and genomic history of the phleboviruses. Virology 464-465:312–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vasilakis N, Forrester NL, Palacios G, Nasar F, Savji N, Rossi SL, Guzman H, Wood TG, Popov V, Gorchakov R, Gonzalez AV, Haddow AD, Watts DM, da Rosa AP, Weaver SC, Lipkin WI, Tesh RB. 2013. Negevirus: a proposed new taxon of insect-specific viruses with wide geographic distribution. J Virol 87:2475–2488. doi: 10.1128/JVI.00776-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kittayapong P, Baisley KJ, O'Neill SL. 1999. A mosquito densovirus infecting Aedes aegypti and Aedes albopictus from Thailand. Am J Trop Med Hyg 61:612–617. doi: 10.4269/ajtmh.1999.61.612. [DOI] [PubMed] [Google Scholar]
- 21.Aguiar ER, Olmo RP, Paro S, Ferreira FV, de Faria IJ, Todjro YM, Lobo FP, Kroon EG, Meignin C, Gatherer D, Imler JL, Marques JT. 2015. Sequence-independent characterization of viruses based on the pattern of viral small RNAs produced by the host. Nucleic Acids Res 43:6191–6206. doi: 10.1093/nar/gkv587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zakrzewski M, Rasic G, Darbro J, Krause L, Poo YS, Filipovic I, Parry R, Asgari S, Devine G, Suhrbier A. 2018. Mapping the virome in wild-caught Aedes aegypti from Cairns and Bangkok. Sci Rep 16:4690. doi: 10.1038/s41598-018-22945-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amarasinghe GK, Bao Y, Basler CF, Bavari S, Beer M, Bejerman N, Blasdell KR, Bochnowski A, Briese T, Bukreyev A, Calisher CH, Chandran K, Collins PL, Dietzgen RG, Dolnik O, Durrwald R, Dye JM, Easton AJ, Ebihara H, Fang Q, Formenty P, Fouchier RAM, Ghedin E, Harding RM, Hewson R, Higgins CM, Hong J, Horie M, James AP, Jiang D, Kobinger GP, Kondo H, Kurath G, Lamb RA, Lee B, Leroy EM, Li M, Maisner A, Muhlberger E, Netesov SV, Nowotny N, Patterson JL, Payne SL, Paweska JT, Pearson MN, Randall RE, Revill PA, Rima BK, Rota P, Rubbenstroth D, Schwemmle M, Smither SJ, Song Q, Stone DM, Takada A, Terregino C, Tesh RB, Tomonaga K, Tordo N, Towner JS, Vasilakis N, Volchkov VE, Wahl-Jensen V, Walker PJ, Wang B, Wang D, Wang F, Wang LF, Werren JH, Whitfield AE, Yan Z, Ye G, Kuhn JH. 2017. Taxonomy of the order Mononegavirales: update 2017. Arch Virol 162:2493–2504. doi: 10.1007/s00705-017-3311-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li CX, Shi M, Tian JH, Lin XD, Kang YJ, Chen LJ, Qin XC, Xu J, Holmes EC, Zhang YZ. 2015. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. Elife 4:e05378. doi: 10.7554/eLife.05378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi M, Neville P, Nicholson J, Eden JS, Imrie A, Holmes EC. 2017. High-resolution metatranscriptomics reveals the ecological dynamics of mosquito-associated RNA viruses in Western Australia. J Virol 91:e00680-. doi: 10.1128/JVI.00680-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hedges LM, Brownlie JC, O'Neill SL, Johnson KN. 2008. Wolbachia and virus protection in insects. Science 322:702. doi: 10.1126/science.1162418. [DOI] [PubMed] [Google Scholar]
- 27.Teixeira L, Ferreira A, Ashburner M. 2008. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol 6:e2. doi: 10.1371/journal.pbio.0060002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moreira LA, Iturbe-Ormaetxe I, Jeffery JA, Lu G, Pyke AT, Hedges LM, Rocha BC, Hall-Mendelin S, Day A, Riegler M, Hugo LE, Johnson KN, Kay BH, McGraw EA, van den Hurk AF, Ryan PA, O'Neill SL. 2009. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell 139:1268–1278. doi: 10.1016/j.cell.2009.11.042. [DOI] [PubMed] [Google Scholar]
- 29.Zhang G, Etebari K, Asgari S. 2016. Wolbachia suppresses cell fusing agent virus in mosquito cells. J Gen Virol 97:3427–3432. doi: 10.1099/jgv.0.000653. [DOI] [PubMed] [Google Scholar]
- 30.Schnettler E, Sreenu VB, Mottram T, McFarlane M. 2016. Wolbachia restricts insect-specific flavivirus infection in Aedes aegypti cells. J Gen Virol 97:3024–3029. doi: 10.1099/jgv.0.000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parry R, Asgari S. 2018. Aedes anphevirus: an insect-specific virus distributed worldwide in Aedes aegypti mosquitoes that has complex interplays with Wolbachia and dengue virus infection in cells. bioRxiv doi: 10.1101/335067. [DOI] [PMC free article] [PubMed]
- 32.Sabin LR, Zheng Q, Thekkat P, Yang J, Hannon GJ, Gregory BD, Tudor M, Cherry S. 2013. Dicer-2 processes diverse viral RNA species. PLoS One 8:e55458. doi: 10.1371/journal.pone.0055458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanchez-Vargas I, Scott JC, Poole-Smith BK, Franz AW, Barbosa-Solomieu V, Wilusz J, Olson KE, Blair CD. 2009. Dengue virus type 2 infections of Aedes aegypti are modulated by the mosquito's RNA interference pathway. PLoS Pathog 5:e1000299. doi: 10.1371/journal.ppat.1000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Q, Luo Y, Lu R, Lau N, Lai EC, Li WX, Ding SW. 2010. Virus discovery by deep sequencing and assembly of virus-derived small silencing RNAs. Proc Natl Acad Sci U S A 107:1606–1611. doi: 10.1073/pnas.0911353107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mayoral JG, Etebari K, Hussain M, Khromykh AA, Asgari S. 2014. Wolbachia infection modifies the profile, shuttling and structure of microRNAs in a mosquito cell line. PLoS One 9:e96107. doi: 10.1371/journal.pone.0096107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frentiu FD, Robinson J, Young PR, McGraw EA, O'Neill SA. 2010. Wolbachia-mediated resistance to Dengue virus infection and death at the cellular level. PLoS One 5:e13398. doi: 10.1371/journal.pone.0013398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McMeniman CJ, Lane AM, Fong AW, Voronin DA, Iturbe-Ormaetxe I, Yamada R, McGraw EA, O'Neill SL. 2008. Host adaptation of a Wolbachia strain after long-term serial passage in mosquito cell lines. Appl Environ Microbiol 74:6963–6969. doi: 10.1128/AEM.01038-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Darby AC, Gill AC, Armstrong SD, Hartley CS, Xia D, Wastling JM, Makepeace BL. 2014. Integrated transcriptomic and proteomic analysis of the global response of Wolbachia to doxycycline-induced stress. ISME J 8:925–937. doi: 10.1038/ismej.2013.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woolfit M, Algama M, Keith JM, McGraw EA, Popovici J. 2015. Discovery of putative small non-coding RNAs from the obligate intracellular bacterium Wolbachia pipientis. PLoS One 10:e0118595. doi: 10.1371/journal.pone.0118595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Green TJ, Cox R, Tsao J, Rowse M, Qiu S, Luo M. 2014. Common mechanism for RNA encapsidation by negative-strand RNA viruses. J Virol 88:3766–3775. doi: 10.1128/JVI.03483-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baron OL, Ursic-Bedoya RJ, Lowenberger CA, Ocampo CB. 2010. Differential gene expression from midguts of refractory and susceptible lines of the mosquito, Aedes aegypti, infected with Dengue-2 virus. J Insect Sci 10:41. doi: 10.1673/031.010.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iverson LE, Rose JK. 1981. Localized attenuation and discontinuous synthesis during vesicular stomatitis virus transcription. Cell 23:477–484. doi: 10.1016/0092-8674(81)90143-4. [DOI] [PubMed] [Google Scholar]
- 43.Ferron F, Longhi S, Henrissat B, Canard B. 2002. Viral RNA-polymerases–a predicted 2′-O-ribose methyltransferase domain shared by all Mononegavirales. Trends Biochem Sci 27:222–224. doi: 10.1016/S0968-0004(02)02091-1. [DOI] [PubMed] [Google Scholar]
- 44.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lorenz R, Bernhart SH, Honer Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. 2011. ViennaRNA package 2.0. Algorithms Mol Biol 6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller WA, Giedroc D. 2010. Ribosomal frameshifting in decoding plant viral RNAs, p 193–220. In Atkins JF, Gesteland RF (ed), Recoding: expansion of decoding rules enriches gene expression. Springer, New York, NY. [Google Scholar]
- 47.Walker PJ, Firth C, Widen SG, Blasdell KR, Guzman H, Wood TG, Paradkar PN, Holmes EC, Tesh RB, Vasilakis N. 2015. Evolution of genome size and complexity in the RhabdoviridA. PLoS Pathog 11:e1004664. doi: 10.1371/journal.ppat.1004664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lewis SH, Quarles KA, Yang Y, Tanguy M, Frezal L, Smith SA, Sharma PP, Cordaux R, Gilbert C, Giraud I, Collins DH, Zamore PD, Miska EA, Sarkies P, Jiggins FM. 2018. Pan-arthropod analysis reveals somatic piRNAs as an ancestral defence against transposable elements. Nat Ecol Evol 2:174–181. doi: 10.1038/s41559-017-0403-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suzuki Y, Frangeul L, Dickson LB, Blanc H, Verdier Y, Vinh J, Lambrechts L, Saleh MC. 2017. Uncovering the repertoire of endogenous flaviviral elements in Aedes mosquito genomes. J Virol 91:e00571-. doi: 10.1128/JVI.00571-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McBride CS, Baier F, Omondi AB, Spitzer SA, Lutomiah J, Sang R, Ignell R, Vosshall LB. 2014. Evolution of mosquito preference for humans linked to an odorant receptor. Nature 515:222–227. doi: 10.1038/nature13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bonizzoni M, Dunn WA, Campbell CL, Olson KE, Marinotti O, James AA. 2012. Complex modulation of the Aedes aegypti transcriptome in response to dengue virus infection. PLoS One 7:e50512. doi: 10.1371/journal.pone.0050512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bonizzoni M, Dunn WA, Campbell CL, Olson KE, Marinotti O, James AA. 2012. Strain variation in the transcriptome of the dengue fever vector, Aedes aegypti. G3 2:103–114. doi: 10.1534/g3.111.001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Faucon F, Gaude T, Dusfour I, Navratil V, Corbel V, Juntarajumnong W, Girod R, Poupardin R, Boyer F, Reynaud S, David JP. 2017. In the hunt for genomic markers of metabolic resistance to pyrethroids in the mosquito Aedes aegypti: an integrated next-generation sequencing approach. PLoS Negl Trop Dis 11:e0005526. doi: 10.1371/journal.pntd.0005526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alfonso-Parra C, Ahmed-Braimah YH, Degner EC, Avila FW, Villarreal SM, Pleiss JA, Wolfner MF, Harrington LC. 2016. Mating-induced transcriptome changes in the reproductive tract of female Aedes aegypti. PLoS Negl Trop Dis 10:e0004451. doi: 10.1371/journal.pntd.0004451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sutton ER, Yu Y, Shimeld SM, White-Cooper H, Alphey AL. 2016. Identification of genes for engineering the male germline of Aedes aegypti and Ceratitis capitata. BMC Genomics 17:948. doi: 10.1186/s12864-016-3280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.David JP, Faucon F, Chandor-Proust A, Poupardin R, Riaz MA, Bonin A, Navratil V, Reynaud S. 2014. Comparative analysis of response to selection with three insecticides in the dengue mosquito Aedes aegypti using mRNA sequencing. BMC Genomics 15:174. doi: 10.1186/1471-2164-15-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Metsky HC, Matranga CB, Wohl S, Schaffner SF, Freije CA, Winnicki SM, West K, Qu J, Baniecki ML, Gladden-Young A, Lin AE, Tomkins-Tinch CH, Ye SH, Park DJ, Luo CY, Barnes KG, Shah RR, Chak B, Barbosa-Lima G, Delatorre E, Vieira YR, Paul LM, Tan AL, Barcellona CM, Porcelli MC, Vasquez C, Cannons AC, Cone MR, Hogan KN, Kopp EW, Anzinger JJ, Garcia KF, Parham LA, Ramirez RMG, Montoya MCM, Rojas DP, Brown CM, Hennigan S, Sabina B, Scotland S, Gangavarapu K, Grubaugh ND, Oliveira G, Robles-Sikisaka R, Rambaut A, Gehrke L, Smole S, Halloran ME, Villar L, Mattar S, Lorenzana I, Cerbino-Neto J, Valim C, Degrave W, Bozza PT, Gnirke A, Andersen KG, Isern S, Michael SF, Bozza FA, Souza TML, Bosch I, Yozwiak NL, MacInnis BL, Sabeti PC. 2017. Zika virus evolution and spread in the Americas. Nature 546:411–415. doi: 10.1038/nature22402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh K. 1967. Cell cultures derived from larvae of Aedes albopictus (Skuse) and Aedes aegypti (L.). Curr Sci 36:506–508. [Google Scholar]
- 59.Powell JR, Tabachnick WJ. 2013. History of domestication and spread of Aedes aegypti–a review. Mem Inst Oswaldo Cruz 108(Suppl 1):S11–S17. doi: 10.1590/0074-0276130395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brown JE, Evans BR, Zheng W, Obas V, Barrera-Martinez L, Egizi A, Zhao H, Caccone A, Powell JR. 2014. Human impacts have shaped historical and recent evolution in Aedes aegypti, the dengue and yellow fever mosquito. Evolution 68:514–525. doi: 10.1111/evo.12281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gloria-Soria A, Ayala D, Bheecarry A, Calderon-Arguedas O, Chadee DD, Chiappero M, Coetzee M, Elahee KB, Fernandez-Salas I, Kamal HA, Kamgang B, Khater EI, Kramer LD, Kramer V, Lopez-Solis A, Lutomiah J, Martins A Jr, Micieli MV, Paupy C, Ponlawat A, Rahola N, Rasheed SB, Richardson JB, Saleh AA, Sanchez-Casas RM, Seixas G, Sousa CA, Tabachnick WJ, Troyo A, Powell JR. 2016. Global genetic diversity of Aedes aegypti. Mol Ecol 25:5377–5395. doi: 10.1111/mec.13866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Whitfield ZJ, Dolan PT, Kunitomi M, Tassetto M, Seetin MG, Oh S, Heiner C, Paxinos E, Andino R. 2017. The diversity, structure, and function of heritable adaptive immunity sequences in the Aedes aegypti genome. Curr Biol 27:3511–3519. doi: 10.1016/j.cub.2017.09.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vodovar N, Bronkhorst A, van Cleef K, Miesen P, Blanc H, van Rij RP, Saleh MC. 2012. Arbovirus-derived piRNAs exhibit a ping-pong signature in mosquito cells. PLoS One 7:e30861. doi: 10.1371/journal.pone.0030861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brackney DE, Scott JC, Sagawa F, Woodward JE, Miller NA, Schilkey FD, Mudge J, Wilusz J, Olson KE, Blair CD, Ebel GD. 2010. C6/36 Aedes albopictus cells have a dysfunctional antiviral RNA interference response. PLoS Neglect Trop Dis 4:e856. doi: 10.1371/journal.pntd.0000856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Léger P, Lara E, Jagla B, Sismeiro O, Mansuroglu Z, Coppée J, Bonnefoy E, Bouloy M. 2013. Dicer-2- and Piwi-mediated RNA interference in Rift Valley fever virus-infected mosquito cells. J Virol 87:1631–1648. doi: 10.1128/JVI.02795-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Asad S, Parry R, Asgari S. 2018. Upregulation of Aedes aegypti Vago1 by Wolbachia and its effect on dengue virus replication. Insect Biochem Mol Biol 92:45–52. doi: 10.1016/j.ibmb.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 67.Miesen P, Joosten J, van Rij RP. 2016. PIWIs go viral: arbovirus-derived piRNAs in vector mosquitoes. PLoS Pathog 12:e1006017. doi: 10.1371/journal.ppat.1006017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Di Giallonardo F, Audsley MD, Shi M, Young PR, McGraw EA, Holmes EC. 2018. Complete genome of Aedes aegypti anphevirus in the Aag2 mosquito cell line. J Gen Virol 99:832–836. doi: 10.1099/jgv.0.001079. [DOI] [PubMed] [Google Scholar]
- 69.Lee M, Etebari K, Hall-Mendelin S, van den Hurk AF, Hobson-Peters J, Vatipally S, Schnettler E, Hall R, Asgari S. 2017. Understanding the role of microRNAs in the interaction of Aedes aegypti mosquitoes with an insect-specific flavivirus. J Gen Virol 98:1892–1903. doi: 10.1099/jgv.0.000832. [DOI] [PubMed] [Google Scholar]
- 70.Etebari K, Osei-Amo S, Blomberg SP, Asgari S. 2015. Dengue virus infection alters post-transcriptional modification of microRNAs in the mosquito vector Aedes aegypti. Sci Rep 5:15968. doi: 10.1038/srep15968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kuno G. 1983. Cultivation of mosquito cell lines in serum-free media and their effects on dengue virus replication. In Vitro 19:707–713. doi: 10.1007/BF02628962. [DOI] [PubMed] [Google Scholar]
- 72.O'Neill SL, Kittayapong P, Braig HR, Andreadis TG, Gonzalez JP, Tesh RB. 1995. Insect densoviruses may be widespread in mosquito cell lines. J Gen Virol 76(Part 8):2067–2074. doi: 10.1099/0022-1317-76-8-2067. [DOI] [PubMed] [Google Scholar]
- 73.Juliano SA, Lounibos LP, O'Meara GF. 2004. A field test for competitive effects of Aedes albopictus on A. aegypti in south Florida: differences between sites of coexistence and exclusion? Oecologia 139:583–593. doi: 10.1007/s00442-004-1532-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Blair C, Olson K. 2015. The role of RNA interference (RNAi) in arbovirus-vector interactions. Viruses 7:820–843. doi: 10.3390/v7020820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hussain M, Etebari K, Asgari S. 2016. Functions of small RNAs in mosquitoes. Adv Insect Physiol 51:189–222. doi: 10.1016/bs.aiip.2016.04.001. [DOI] [Google Scholar]
- 76.Schnettler E, Donald CL, Human S, Watson M, Siu RW, McFarlane M, Fazakerley JK, Kohl A, Fragkoudis R. 2013. Knockdown of piRNA pathway proteins results in enhanced Semliki Forest virus production in mosquito cells. J Gen Virol 94:1680–1689. doi: 10.1099/vir.0.053850-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dietrich I, Shi X, McFarlane M, Watson M, Blomstrom AL, Skelton JK, Kohl A, Elliott RM, Schnettler E. 2017. The antiviral RNAi response in vector and non-vector cells against orthobunyaviruses. PLoS Negl Trop Dis 11:e0005272. doi: 10.1371/journal.pntd.0005272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hess AM, Prasad AN, Ptitsyn A, Ebel GD, Olson EN, Barbacioru C, Monighetti C, Campbell CL. 2011. Small RNA profiling of Dengue virus-mosquito interactions implicates the PIWI RNA pathway in anti-viral defense. BMC Microbiol 11:45. doi: 10.1186/1471-2180-11-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Varjak M, Donald CL, Mottram TJ, Sreenu VB, Merits A, Maringer K, Schnettler E, Kohl A. 2017. Characterization of the Zika virus induced small RNA response in Aedes aegypti cells. PLoS Negl Trop Dis 11:e0006010. doi: 10.1371/journal.pntd.0006010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Saldaña MA, Etebari K, Hart CE, Widen SG, Wood TG, Thangamani S, Asgari S, Hughes GL. 2017. Zika virus alters the microRNA expression profile and elicits an RNAi response in Aedes aegypti mosquitoes. PLoS Neg Trop Dis 11:e0005760. doi: 10.1371/journal.pntd.0005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee M, Etebari K, Hall-Mendelin S, van den Hurk AF, Hobson-Peters J, Vatipally S, Schnettler E, Hall R, Asgari S. 2017. Understanding the role of microRNAs in the interaction of Aedes aegypti mosquitoes with an insect-specific flavivirus. J Gen Virol 98:892–1903. doi: 10.1099/jgv.0.000812. [DOI] [PubMed] [Google Scholar]
- 82.Johnson KN. 2015. The impact of Wolbachia on virus infection in mosquitoes. Viruses 7:5705–5717. doi: 10.3390/v7112903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schultz MJ, Tan AL, Gray CN, Isern S, Michael SF, Frydman HM, Connor JH. 2018. Wolbachia wStri blocks Zika virus growth at two independent stages of viral replication. mBio 9:e00738-. doi: 10.1128/mBio.00738-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dodson BL, Andrews ES, Turell MJ, Rasgon JL. 2017. Wolbachia effects on Rift Valley fever virus infection in Culex tarsalis mosquitoes. PLoS Negl Trop Dis 11:e0006050. doi: 10.1371/journal.pntd.0006050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hornak KE, Lanchy JM, Lodmell JS. 2016. RNA encapsidation and packaging in the phleboviruses. Viruses 8:E194. doi: 10.3390/v8070194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Raymond DD, Piper ME, Gerrard SR, Skiniotis G, Smith JL. 2012. Phleboviruses encapsidate their genomes by sequestering RNA bases. Proc Natl Acad Sci U S A 109:19208–19213. doi: 10.1073/pnas.1213553109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Choi KH. 2012. Viral polymerases. Adv Exp Med Biol 726:267–304. doi: 10.1007/978-1-4614-0980-9_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Harak C, Lohmann V. 2015. Ultrastructure of the replication sites of positive-strand RNA viruses. Virology 479–480:418–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rainey SM, Martinez J, McFarlane M, Juneja P, Sarkies P, Lulla A, Schnettler E, Varjak M, Merits A, Miska EA, Jiggins FM, Kohl A. 2016. Wolbachia blocks viral genome replication early in infection without a transcriptional response by the endosymbiont or host small RNA pathways. PLoS Pathog 12:e1005536. doi: 10.1371/journal.ppat.1005536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schultz MJ, Isern S, Michael SF, Corley RB, Connor JH, Frydman HM. 2017. Variable inhibition of Zika virus replication by different Wolbachia strains in mosquito cell cultures. J Virol 91:e00339-. doi: 10.1128/JVI.00339-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pan X, Pike A, Joshi D, Bian G, McFadden MJ, Lu P, Liang X, Zhang F, Raikhel AS, Xi Z. 2018. The bacterium Wolbachia exploits host innate immunity to establish a symbiotic relationship with the dengue vector mosquito Aedes aegypti. ISME J 12:277–288. doi: 10.1038/ismej.2017.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rice DW, Sheehan KB, Newton ILG. 2017. Large-scale identification of Wolbachia pipientis effectors. Genome Biol Evol 9:1925–1937. doi: 10.1093/gbe/evx139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Amuzu HE, Tsyganov K, Koh C, Herbert RI, Powell DR, McGraw EA. 2018. Wolbachia enhances insect-specific flavivirus infection in Aedes aegypti mosquitoes. Ecol Evol 8:5441–5454. doi: 10.1002/ece3.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Karpf AR, Lenches E, Strauss EG, Strauss JH, Brown DT. 1997. Superinfection exclusion of alphaviruses in three mosquito cell lines persistently infected with Sindbis virus. J Virol 71:7119–7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pudney M, Varma M, Leake C. 1979. Establishment of cell lines from larvae of culicine (Aedes species) and anopheline mosquitoes. TCA Man 5:997–1002. doi: 10.1007/BF00919719. [DOI] [Google Scholar]
- 96.Ladner JT, Beitzel B, Chain PS, Davenport MG, Donaldson EF, Frieman M, Kugelman JR, Kuhn JH, O'Rear J, Sabeti PC, Wentworth DE, Wiley MR, Yu GY, Threat Characterization Consortium, Sozhamannan S, Bradburne C, Palacios G. 2014. Standards for sequencing viral genomes in the era of high-throughput sequencing. mBio 5:e01360-14. doi: 10.1128/mBio.01360-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hussain M, Frentiu FD, Moreira LA, O'Neill SL, Asgari S. 2011. Wolbachia uses host microRNAs to manipulate host gene expression and facilitate colonization of the dengue vector Aedes aegypti. Proc Natl Acad Sci U S A 108:9250–9255. doi: 10.1073/pnas.1105469108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zimmermann L, Stephens A, Nam SZ, Rau D, Kubler J, Lozajic M, Gabler F, Soding J, Lupas AN, Alva V. 2017. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J Mol Biol 430:2237–2243. doi: 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 99.Tsirigos KD, Peters C, Shu N, Kall L, Elofsson A. 2015. The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res 43:W401–W407. doi: 10.1093/nar/gkv485. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.