EBV infection contributes to the development of cancers, such as nasopharyngeal carcinoma, Burkitt lymphoma, Hodgkin's disease, and posttransplant lymphomas, in immunocompromised or genetically susceptible individuals. LMP1 is an important viral protein expressed by EBV in these cancers. LMP1 is secreted in extracellular vesicles (EVs), and the transfer of LMP1-modified EVs to uninfected cells can alter their physiology. Understanding the cellular machinery responsible for sorting LMP1 into EVs is limited, despite the importance of LMP1-modified EVs. Here, we illustrate the roles of different regions of LMP1 in EV packaging. Our results show that the N terminus and TM1 are sufficient to drive LMP1 EV trafficking. We further show the existence of potential positive and negative regulatory mechanisms for LMP1 vesicle sorting. These findings provide a better basis for future investigations to identify the mechanisms of LMP1 targeting to EVs, which could have broad implications in understanding EV cargo sorting.
KEYWORDS: exosomes, microvesicles, Epstein-Barr virus, trafficking, extracellular vesicles, herpesvirus, tetraspanin, exosomes, protein trafficking
ABSTRACT
EBV latent membrane protein 1 (LMP1) is released from latently infected tumor cells in small membrane-enclosed extracellular vesicles (EVs). Accumulating evidence suggests that LMP1 is a major driver of EV content and functions. LMP1-modified EVs have been shown to influence recipient cell growth, migration, differentiation, and regulation of immune cell function. Despite the significance of LMP1-modified exosomes, very little is known about how this viral protein enters or manipulates the host EV pathway. In this study, LMP1 deletion mutants were generated to assess protein regions required for EV trafficking. Following transfection of LMP1 or mutant plasmids, EVs were collected by differential centrifugation, and the levels of specific cargo were evaluated by immunoblot analysis. The results demonstrate that, together, the N terminus and transmembrane region 1 of LMP1 are sufficient for efficient sorting into EVs. Consistent with these findings, a mutant lacking the N terminus and transmembrane domains 1 through 4 (TM5-6) failed to be packaged into EVs, and exhibited higher colocalization with endoplasmic reticulum and early endosome markers than the wild-type protein. Surprisingly, TM5-6 maintained the ability to colocalize and form a complex with CD63, an abundant exosome protein that is important for the incorporation of LMP1 into EVs. Other mutations within LMP1 resulted in enhanced levels of secretion, pointing to potential positive and negative regulatory mechanisms for extracellular vesicle sorting of LMP1. These data suggest new functions of the N terminus and transmembrane domains in LMP1 intra- and extracellular trafficking that are likely downstream of an interaction with CD63.
IMPORTANCE EBV infection contributes to the development of cancers, such as nasopharyngeal carcinoma, Burkitt lymphoma, Hodgkin's disease, and posttransplant lymphomas, in immunocompromised or genetically susceptible individuals. LMP1 is an important viral protein expressed by EBV in these cancers. LMP1 is secreted in extracellular vesicles (EVs), and the transfer of LMP1-modified EVs to uninfected cells can alter their physiology. Understanding the cellular machinery responsible for sorting LMP1 into EVs is limited, despite the importance of LMP1-modified EVs. Here, we illustrate the roles of different regions of LMP1 in EV packaging. Our results show that the N terminus and TM1 are sufficient to drive LMP1 EV trafficking. We further show the existence of potential positive and negative regulatory mechanisms for LMP1 vesicle sorting. These findings provide a better basis for future investigations to identify the mechanisms of LMP1 targeting to EVs, which could have broad implications in understanding EV cargo sorting.
INTRODUCTION
Epstein-Barr virus (EBV) is a member of the gammaherpesvirus family that has established a persistent infection in greater than 90% of the world's population (1). In immunocompromised or genetically susceptible individuals, EBV infection can contribute to cancer development and has been linked to nasopharyngeal carcinoma, Burkitt lymphoma, Hodgkin's disease, posttransplant lymphomas, and a subset of gastric carcinomas (2, 3). Latent membrane protein 1 (LMP1) is one of the major viral oncogenes expressed in most EBV-associated cancers (4–6). LMP1 alone is sufficient to transform cells, and recombinant EBV lacking LMP1 is incapable of immortalizing B cells in vitro (4, 7, 8). Additionally, transgenic mice expressing LMP1 behind a B cell-specific promoter develop lymphomas (9).
LMP1 is an integral transmembrane protein consisting of a short N-terminal cytoplasmic domain, six transmembrane domains, and a cytoplasmic C-terminal domain containing the C-terminal activating region (CTAR) domains CTAR1 and CTAR2 (6, 10, 11). The oncogenic properties of LMP1 are due to its ability to mimic CD40 receptor signaling through the recruitment of tumor necrosis factor receptor-associated factors (TRAFs) and other effector molecules to CTARs in its C-terminal cytoplasmic tail (8, 11–15). The molecular events orchestrated by LMP1 result in the activation of a plethora of signaling pathways, including mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK), phosphatidylinositol 3-kinase (PI3K)/AKT, NF-κB, mTOR, and c-Jun N-terminal kinase (JNK). The activation of these pathways results in transcriptional upregulation of multiple genes that are involved with regulation of apoptosis, cell cycle progression, cell proliferation, migration, and invasion (8, 16–26). In addition to its functions within infected cells, LMP1 has been found to be present in extracellular vesicles (EVs) and to induce similar cellular responses in cells that receive LMP1-modified EVs (27–31).
EVs represent a heterogeneous population of membrane-enclosed vesicles released from cells ranging from 40 nm to 1,000 nm in size (32, 33). EVs are classified into at least three major groups based on their size and cellular origin: they include exosomes, microvesicles, and apoptotic bodies (34–37). Apoptotic bodies are released from cells undergoing programmed cell death, while microvesicles bud directly off the plasma membrane into the extracellular space. In contrast, exosomes are small endocytically derived extracellular vesicles that form following budding and fusion events at the limiting membranes of multivesicular bodies (MVBs). These newly formed intraluminal vesicles are released from the cell as exosomes following fusion of the MVB membrane with the plasma membrane (34, 35). Exosomes and other EVs can transfer proteins, mRNAs, microRNAs (miRNAs), and lipids to neighboring or distant cells (38–40). The ability to package and transfer diverse cargoes from cell to cell makes EVs important mediators of intercellular communication events that are involved in normal physiological processes, as well as in pathological conditions, such as cancer (41–43). In malignancy, EVs play a major role in cell growth, invasion, and metastasis (44, 45). In the case of virus-infected cells, EVs can transfer modified protein and RNA cargo to uninfected cells, which can enhance viral pathogenesis (34, 35). Some viruses even package infectious genomes into EVs that can facilitate productive infections and pathogen spread (46–48).
Transfer of the LMP1-containing EVs can activate MAPK/ERK and PI3K/Akt signaling pathways within neighboring uninfected cells (27). More recently, EBV has been shown to dramatically alter the protein content of EVs released from latently infected B cells, with many of the significant changes correlating with LMP1 expression (27, 36, 40). EBV-modified EVs enhance proliferation, migration, invasion, and B cell differentiation toward a plasmablast-like phenotype (29–31). Together, these data suggest that EBV-infected cells can manipulate the tumor microenvironment through the transfer of virus-modified EVs and contribute to cancer development and progression. Based on these findings and others, it is likely that LMP1 is an important viral factor contributing to EV content and function during EBV infection and viral tumorigenesis.
Despite the significance of LMP1-modified EVs, very little is known about how the viral protein actually enters the host EV pathway. In this study, deletion mutants of LMP1 were constructed to assess protein regions of LMP1 important for EV targeting and release from cells. Deletion analyses revealed that, together, the N terminus and transmembrane region 1 are sufficient to guide efficient sorting into the exosome pathway. Consistent with these findings, a mutant (TM5-6) lacking the N terminus and transmembrane domains 1 through 4 that failed to be packaged into EVs exhibited higher colocalization with endoplasmic reticulum (ER) and early endosome markers than the wild-type (WT) protein. Interestingly, some mutations within the transmembrane regions of LMP1 resulted in enhanced EV trafficking, suggesting potential positive and negative regulation mechanisms of LMP1 secretion. Additional data suggest that other transmembrane regions may be able to substitute for transmembrane domain 1 (TM1) when fused to the N terminus. Overall, this study provides a detailed analysis of the roles different LMP1 structural domains play in endosomal sorting and EV packaging.
RESULTS
The C terminus of LMP1 is not required for EV packaging.
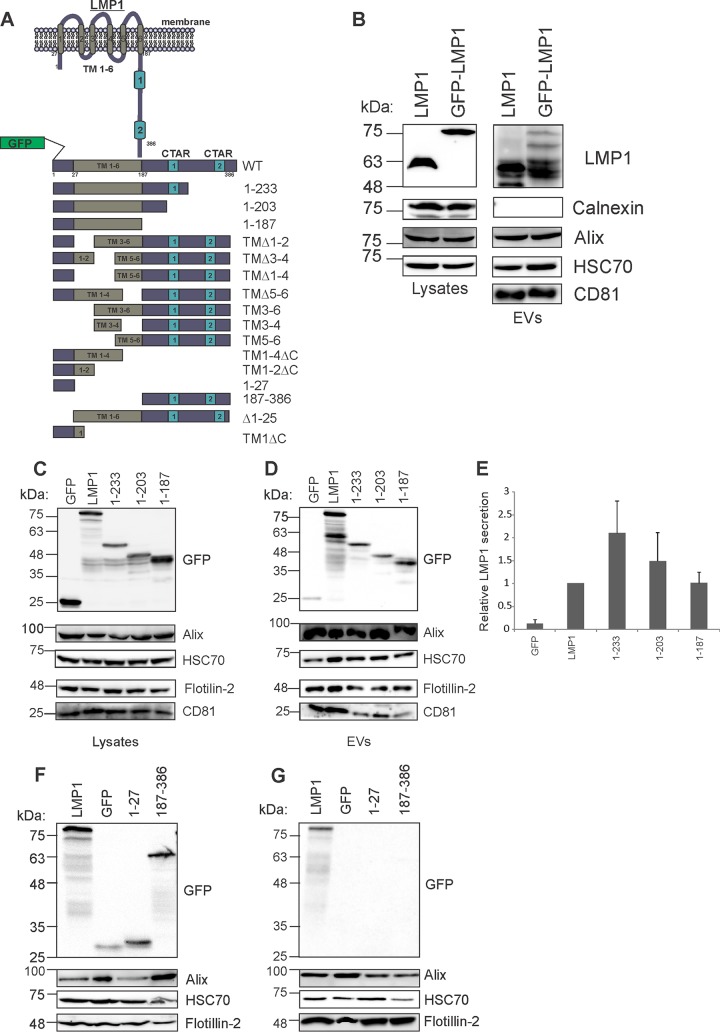
LMP1 is an integral transmembrane protein (386 amino acids [aa]) consisting of a short N-cytoplasmic domain (aa 1 to 24), six transmembrane domains (aa 25 to 186), and the cytoplasmic C-terminal domain (aa 187 to 386), which contains C-terminal activating region 1 (CTAR1) and CTAR2 (16). The C-terminal tail of LMP1 protrudes into the cytoplasm, and the majority of LMP1-mediated signal transductions involve protein interactions with the CTAR1 and CTAR2 domains. Because the C-terminal tail interacts with many cellular proteins, we hypothesized that deletions within the C-terminal tail region would result in reduced EV packaging of LMP1. To assess which LMP1 domains are important for EV packaging, green fluorescent protein (GFP)-tagged LMP1 deletion mutants were generated (Fig. 1A). Comparison of GFP-LMP1 to nontagged LMP1 showed that the two constructs are packaged into EVs at comparable levels, confirming that the addition of GFP to the N terminus of LMP1 does not interfere with EV packaging (28, 49) (Fig. 1B). HEK293 cells were transfected with GFP-LMP1 or mutants lacking regions of the C-terminal tail (aa 1 to 233 or 1 to 203) or the entire tail (aa 1 to 187) (Fig. 1C). Transfections with GFP alone were used as controls in these experiments. The results revealed that LMP1 C-terminal deletion mutants (aa 1 to 233 and 1 to 203) could be packaged into EVs (Fig. 1D and E). Surprisingly, the mutant lacking the entire C-terminal tail (aa 1 to 187) was also incorporated into EVs. These data imply that the C-terminal tail is dispensable for sorting LMP1 into EVs. We further assessed whether the N-terminal (aa 1 to 27) or C-terminal (aa 187 to 386) cytoplasmic tails alone are sufficient for EV targeting. These truncated mutants could not be packaged into EVs (Fig. 1F and G). Taken together, these data suggest that the cytoplasmic tails of LMP1 do not contain molecular information sufficient for EV secretion and further demonstrate that the C-terminal cytoplasmic tail is not required for sorting of LMP1 into EVs.
FIG 1.
The C-terminal cytoplasmic tail of LMP1 is not required for EV packaging. (A) Schematic depiction of WT GFP-tagged LMP1 and deletion mutants. LMP1 is a six-pass transmembrane protein with N- and C-terminal cytoplasmic tails. Various GFP-tagged mutants of the full-length wild-type LMP1 protein were constructed by PCR mutagenesis and used throughout the study. (B) The levels of nontagged LMP1 and GFP-LMP1 were compared in cell lysates and EVs. (C) Lysate immunoblot analysis of cells transiently transfected with LMP1 or mutants containing deletions in the C terminus (1-233, 1-203, and 1-187) (equal protein masses were loaded). (D) EVs (equal volumes were loaded) were harvested by differential centrifugation and analyzed by immunoblot analysis for GFP-tagged proteins and common EV markers (Alix, HSC70, Flotillin-2, and CD81). (E) Semiquantitative immunoblot analysis of the results of 3 independent experiments. The levels of LMP1 mutant secretion are relative to wild-type LMP1 secretion and normalized to cellular expression and EV production [(LMP1EV/HSC70EV)/(LMP1cell/HSC70cell)]. The error bars indicate standard deviations. (F) Cell lysates and (G) EVs harvested from GFP, GFP-LMP1, or GFP-tagged N (1-27) and C (187-386) termini of LMP1. Cells and EVs were harvested by differential centrifugation and analyzed by immunoblotting for expression with anti-GFP antibody and common EV markers (Alix, HSC70, and Flotillin-2). TM, transmembrane domains.
Transmembrane domains mediate LMP1 release from cells into EVs.
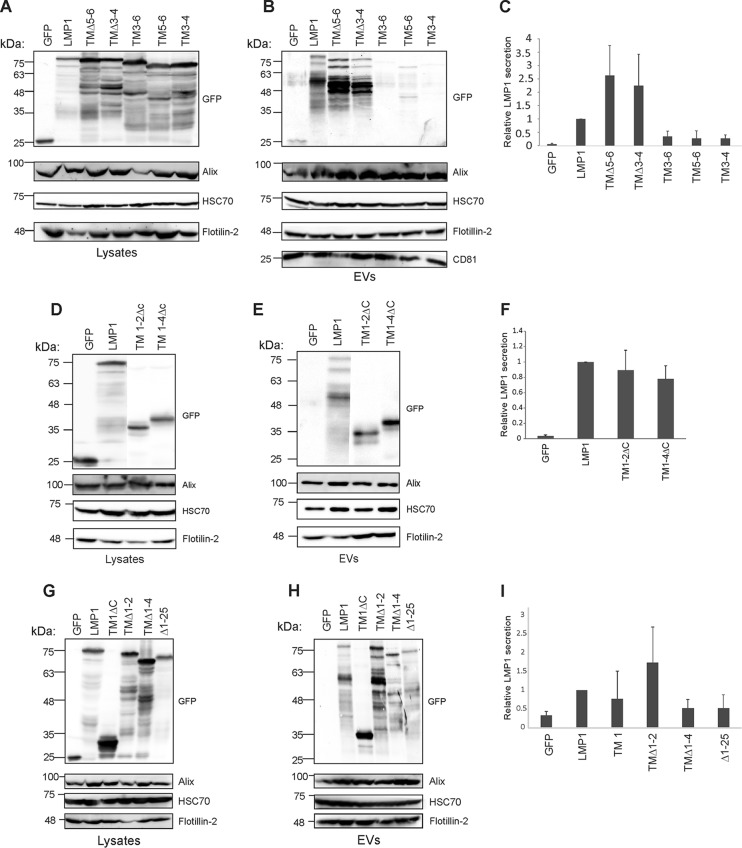
The LMP1 N terminus and transmembrane domains play a major active role in LMP1 oligomerization, signaling, and intracellular trafficking (10). To determine the region within LMP1 transmembrane domains responsible for sorting into EVs, various transmembrane deletion mutants were constructed and tested for their presence in EVs isolated from transfected cells. Deletion of transmembrane segments 5 and 6 (TMΔ5-6) or 3 and 4 (TMΔ3-4) did not abrogate LMP1 sorting into EVs, but instead resulted in higher levels of secretion from cells than wild-type protein (Fig. 2A to C). Conversely, deletion of the N terminus and transmembrane segments 1 and 2 (TM3-6) or 1 to 4 (TM5-6) impaired trafficking of LMP1 into EVs (Fig. 2A to C). Consistent with this result, trafficking of LMP1 to EVs was also impaired when a mutant containing transmembrane segments 3 and 4 fused to the C terminus (TM3-4) was assessed. Quantification of three independent experiments revealed lower relative vesicular secretion of these mutants to a level that was similar to the packaging of GFP, a soluble protein that is not efficiently sorted into EVs (Fig. 2B and C). We further assessed deletion mutants containing only the N terminus and transmembrane segments 1 and 2 (TM1-2ΔC) or 1 to 4 (TM1-4ΔC) but lacking the C-terminal tail. Western blot analysis showed that both mutants (TM1-2ΔC and TM1-4ΔC) could be targeted into EVs (Fig. 2D and E). Moreover, secretion of TM1-2ΔC and TM1-4ΔC into EVs was comparable to that of wild-type LMP1 (Fig. 2F). We next examined whether deletion of the N terminus alone (Δ1-25) or deletion of transmembrane domains 1 and 2 (TMΔ1-2) or 1 to 4 (TMΔ1-4) could prevent sorting of LMP1 into EVs (Fig. 2G and H). Immunoblot analysis determined that Δ1-25, TMΔ1-2, and TMΔ1-4 were detectable in EVs. Likewise, expression of the N terminus and transmembrane domain 1 without the C-terminal tail (TM1ΔC) was capable of targeting to EVs (Fig. 2G and H). Semiquantitative Western blot analysis from three independent experiments revealed that mutants Δ1-25 and TMΔ1-4 were packaged into EVs at lower levels than the WT LMP1 (Fig. 2I). Furthermore, TM1 and TMΔ1-2 quantitative analysis showed higher EV packaging than the WT (Fig. 2I). Taken together, these data suggest the N terminus, in combination with specific domains within the transmembrane regions of LMP1, is required for efficient sorting into EVs. Our data further show that deletion of some transmembrane segments increases the sorting of LMP1 into EVs, supporting the possibility of potential positive and negative regulatory mechanisms of LMP1 secretion.
FIG 2.
Transmembrane domains control the vesicular release of LMP1 from cells. HEK293 cells were transfected with GFP, GFP-LMP1, or transmembrane deletion mutants (TMΔ5-6, TMΔ3-4, TM3-6, TM5-6, and TM3-4). (A) Whole-cell and (B) EV lysates were separated by SDS-PAGE and analyzed by immunoblot analysis for GFP and common EV markers (Alix, HSC70, Flotillin-2, and CD81). (C) Semiquantitative immunoblot analysis of the results of four to six independent experiments examining relative vesicular LMP1 secretion of the transmembrane deletion mutants. Immunoblot analysis of (D) cell and (E) vesicle lysates for deletion mutants (TM1-2ΔC and TM1-4ΔC). (F) Semiquantitative immunoblot analysis from three independent experiments showing the relative EV LMP1 secretion for TM1-2ΔC and TM1-4ΔC deletion mutants. Immunoblot analysis of (G) cell and (H) vesicle lysates for mutants (TM1ΔC, TMΔ1-2, TMΔ1-4, and Δ1-25). (I) Semiquantitative immunoblot analysis from three independent experiments showing the relative EV LMP1 secretion for TM1ΔC, TMΔ1-2, TMΔ1-4, and Δ1-25 deletion mutants. The error bars indicate standard deviations.
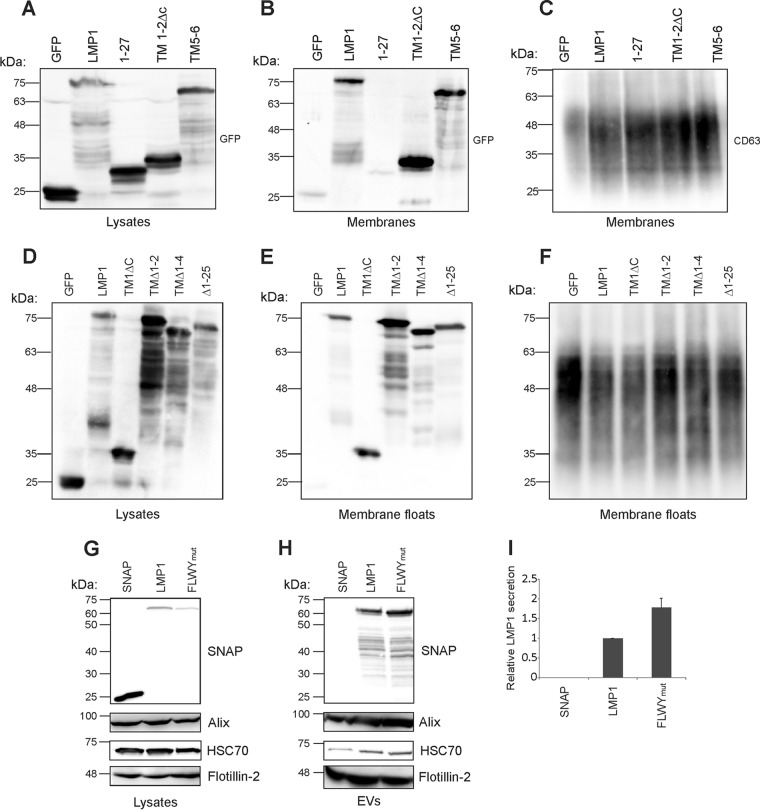
Deletion mutations in LMP1 could render the protein unable to properly associate with membranes, and this could be the reason for the observed decrease in EV packaging. Therefore, to test membrane association of LMP1 mutants, sucrose density flotation assays were performed to isolate membrane fractions. The mutants utilized for this purpose were N terminus (1-27), TM1-2ΔC, TM5-6, TM1ΔC, TMΔ1-2, TMΔ1-4, and Δ1-25. The N terminus (1-27) mutant and GFP alone were not detected in the membrane float fraction despite being readily expressed in cells (Fig. 3A, B, D, and E). However, TM1-2ΔC, TM5-6, TM1ΔC, TMΔ1-2, TMΔ1-4, and Δ1-25 were present in membrane fractions at levels similar to those of wild-type LMP1, suggesting that these mutants are still able to associate with membranes despite their defects in packaging (Fig. 3A, B, D, and E). The tetraspanin protein CD63 was used as a positive control in these experiments as a cellular transmembrane protein that associates with membranes (Fig. 3C and F).
FIG 3.
Mutation of the lipid-raft anchoring domain FWLY increases LMP1 EV release. Immunoblot analysis of the levels of GFP-tagged wild-type or mutant LMP1 in (A and D) cell lysates or (B and E) purified membranes. (C and F) CD63 was detected by immunoblotting in all isolated membranes. (G) Whole-cell or (H) EV lysates were analyzed by immunoblot analysis for SNAP, HSC70, Alix, and Flotillin-2 following transfection with SNAP, SNAP-LMP1, or the LMP1-FWLY mutant. (I) Semiquantitative immunoblot analysis of the results of 3 independent experiments. The levels of LMP1 mutant secretion are relative to wild-type LMP1 secretion and normalized to cellular expression and EV production. The error bars indicate standard deviations.
Mutation of the lipid raft-targeting domain FWLY enhances vesicle release of LMP1.
LMP1 transmembrane domains contain critical residues that are required for LMP1 lipid raft microdomain localization and signaling. Lipid rafts have also been suggested to function in EV biogenesis and cargo sorting (50–52). The persistent ability of TM1-2ΔC to traffic to EVs led us to evaluate whether inhibiting lipid raft targeting would affect vesicle sorting, as mutation of the lipid raft-anchoring domain within TM1 of LMP1 (FWLY38–41 to AAAA) has been shown to inhibit LMP1 lipid raft localization (53). HEK293 cells were transfected with SNAP, SNAP-LMP1, and the SNAP-LMP1-FWLY mutant. Immunoblot analysis demonstrated that the LMP1-FWLY mutant was detectable in EVs at reproducibly higher levels than wild-type LMP1 when normalized for cellular expression of each protein (Fig. 3G and H). Quantitation of these results showed a marked increase in the relative EV sorting of FWLY compared with WT LMP1 (Fig. 3I). These data reveal that lipid raft targeting is not required for LMP1 exosomal packaging and further suggest that redirecting LMP1 away from lipid raft microdomains results in more efficient sorting into the EV pathway.
LMP1 deletion mutants exhibit altered subcellular localization.
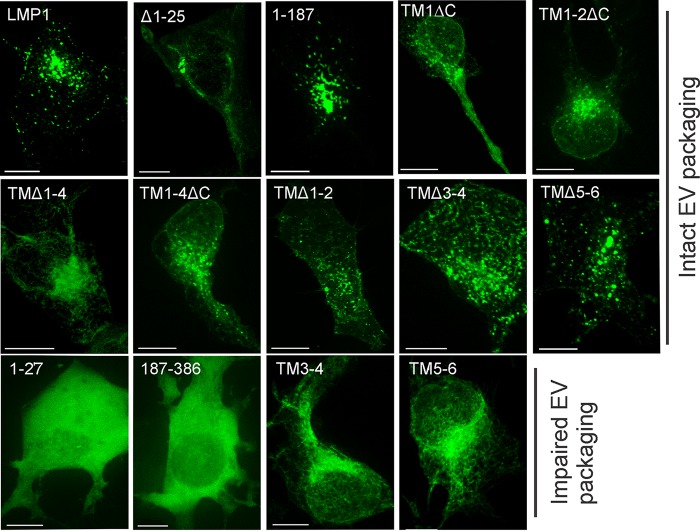
Intact LMP1 has been shown to localize in vesicular structures in the internal perinuclear membranes (23, 54). Since we observed alterations in EV sorting with various transmembrane deletion mutants, we hypothesized that the mutants would similarly exhibit altered subcellular localization compared to WT protein. To verify the localization of LMP1, we examined HEK293 cells transfected with GFP-tagged mutants by live-cell fluorescent confocal microscopy. Wild-type LMP1 localized around the perinuclear region in vesicle-like structures, as previously described (Fig. 4). LMP1 mutants Δ1-25, 1-187, TM1, TM1-2ΔC, TMΔ1-4, and TM1-4ΔC exhibited similar punctate perinuclear localization patterns, with some mutants exhibiting enhanced staining around the nucleus (Fig. 4). Transmembrane deletion mutants TMΔ1-2, TMΔ3-4, and TMΔ5-6 displayed punctate-like structures scattered around the whole cell (Fig. 4). However, transmembrane mutants TM3-4, TM5-6, and TMΔ1-4 demonstrated more diffuse perinuclear localization, lacking distinct punctate structures (Fig. 4). The N terminus (1-27) and C terminus (187-386) mutants revealed diffuse localization throughout the cells (Fig. 4). In conclusion, these data suggest that mutants that fail to be packaged into EVs exhibit altered subcellular localization, lacking the distinct punctate perinuclear patterns observed with WT protein.
FIG 4.
LMP1 mutants that fail to traffic to EVs exhibit altered subcellular localization. HEK293 cells were transfected with the indicated LMP1 constructs and imaged 24 h posttransfection by live-cell confocal microscopy. Scale bars, 10 μm.
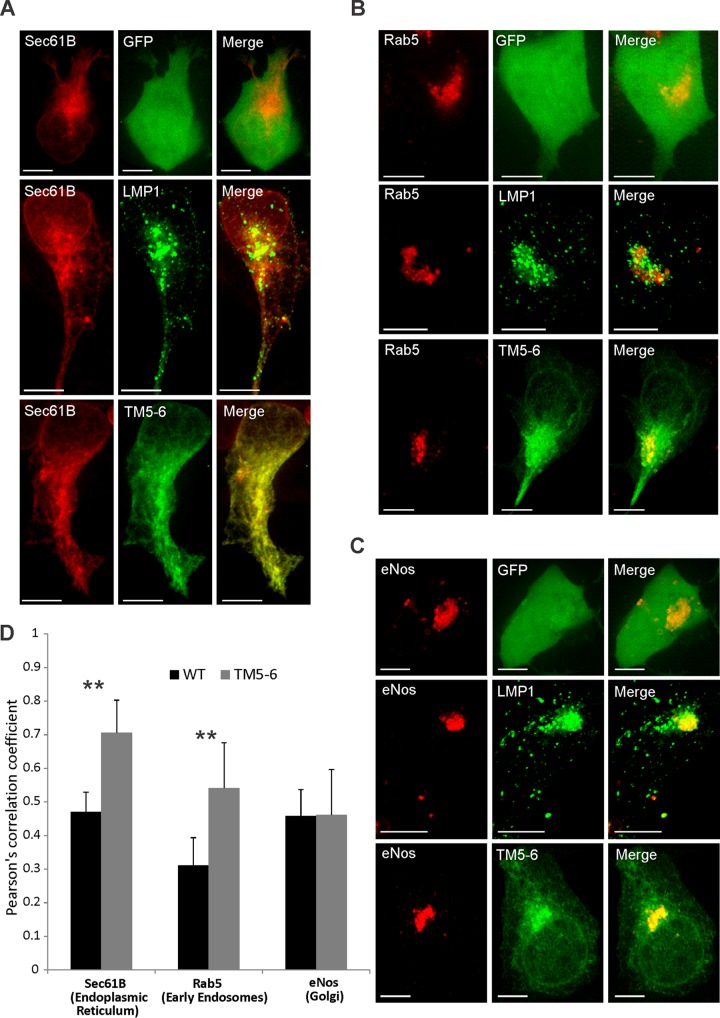
TM5-6 accumulates in ER and early endosome membranes.
We hypothesized that mutants exhibiting reduced EV secretion would similarly display altered localization in subcellular compartments compared to WT protein. For these experiments, TM5-6 was chosen as a representative mutant because it contained the minimal transmembrane regions for proper membrane insertion and orientation, was not packaged into EVs, and lacked the distinct punctate perinuclear staining observed for WT LMP1. To further explore the localization of TM5-6 to specific regions in the cell, fluorescently tagged ER, Golgi, and early endosome markers were cotransfected with either GFP-LMP1 or GFP–TM5-6 and analyzed by confocal microscopy. The LMP1 mutant TM5-6 was found to localize in greater proportion to the endoplasmic reticulum (Pearson correlation coefficient [PCC] = 0.71) than wild-type LMP1 (PCC = 0.47), displaying greater colocalization with the ER marker Sec61B (P < 0.05) (Fig. 5A and D). Similarly, TM5-6 (PCC = 0.54) displayed greater colocalization with an early endosomal marker (Rab5) than LMP1 WT protein (PCC = 0.32; P < 0.05) (Fig. 5C and D). However, LMP1 WT protein and the mutant TM5-6 exhibited similar levels of localization in the Golgi apparatus (endothelial nitric oxide synthase [eNos]) (Fig. 5B and D). In conclusion, confocal imaging indicated that TM5-6 exhibits altered subcellular localization, accumulating to a greater extent in the endoplasmic reticulum and early endosomal compartments than WT protein.
FIG 5.
TM5-6 accumulates in ER and early endosome membranes. HEK293 cells were cotransfected with GFP-tagged LMP1 constructs and RFP-tagged (A) Sec61B, (B) Rab5, or (C) eNos. Live-cell confocal images were acquired 24 h posttransfection. The images were analyzed using Imaris software. (D) Colocalization was quantified using Pearson's correlation coefficient and graphed with standard errors of the mean (n ≥ 12 cells). Representative maximum-projection images are shown. **, P < 0.001. Scale bars, 10 μm.
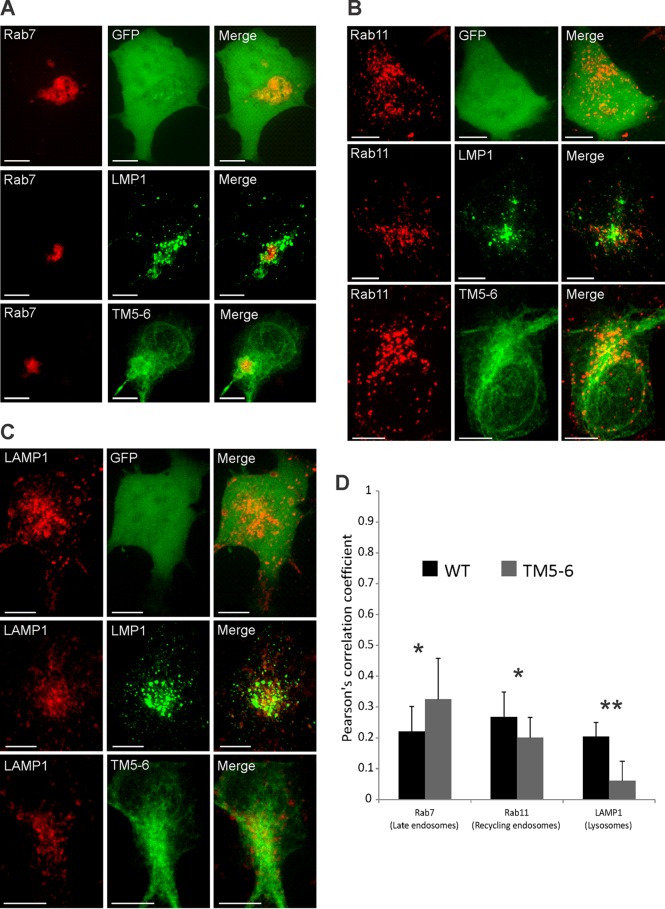
TM5-6 displays altered colocalization with endolysosomal markers.
The sorting of LMP1 into EVs is thought to permit evasion of lysosomal and proteasomal degradation (49). Intracellular LMP1 has been found to localize to late endosomes, lysosomes, and MVBs (49). Since TM5-6 exhibited defective exosomal trafficking, we predicted the mutant would have altered subcellular localization in endolysosomal components compared to the WT protein. To test this, colocalization of wild-type LMP1 or mutant TM5-6 with the subcellular compartment markers Rab7 (late endosomes), Rab11 (recycling endosomes), and Lamp1 (lysosomes) was examined. TM5-6 displayed greater colocalization with Rab7 (PCC = 0.32; P < 0.05) than LMP1 WT (PCC = 0.21) (Fig. 6A and D). However, LMP1 WT displayed greater colocalization with both Rab11 (PCC = 0.26; P < 0.05) and Lamp1 (PCC = 0.21; P < 0.001) (Fig. 6B to D). Taken together, these data suggest that TM5-6 accumulates in the ER, early endosomes, and late endosomes with concomitant reduced localization to recycling endosomes and lysosomes. These results were likely due, at least in part, to the defective sorting of TM5-6 into exosomes.
FIG 6.
TM5-6 exhibits altered colocalization with endolysosomal markers. HEK293 cells were cotransfected with GFP-tagged LMP1 constructs and RFP-tagged (A) Rab7, (B) Rab11, or (C) LAMP1. Live-cell confocal images were acquired 24 h posttransfection. The images were analyzed using Imaris software. (D) Colocalization was quantified using Pearson's correlation coefficient and graphed with standard errors of the mean (n ≥ 12 cells). Representative maximum-projection images are shown. *, P < 0.05; **, P < 0.001. Scale bars, 10 μm.
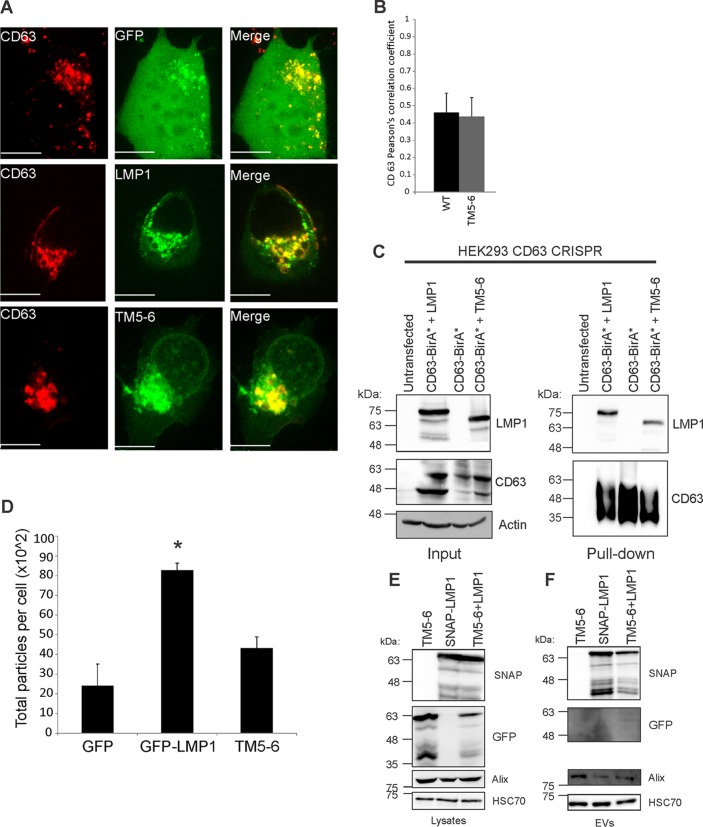
TM5-6 associates and forms a complex with CD63.
LMP1 has been shown to associate with the tetraspanin protein CD63, which is enriched in late endosomes, lysosomes, and MVBs (49). The LMP1-CD63 association has been demonstrated to be important for LMP1 exosomal trafficking and its upregulation of exosome secretion (28). To investigate whether TM5-6 still colocalizes with CD63, coexpression of WT LMP1 or TM5-6 with red fluorescent protein-tagged CD63 (CD63-RFP) was examined by live-cell confocal imaging. The data indicate that TM5-6 colocalizes with CD63 at levels similar to those of WT LMP1 (Fig. 7A and B). We further explored the association of CD63 and TM5-6 by performing a Bio-ID proximity-dependent biotinylation assay (55). As previously observed, CD63 readily biotinylated LMP1 in cells (56). Surprisingly, CD63 similarly biotinylated TM5-6 despite the inability of TM5-6 to be packaged into EVs. These results reveal that TM5-6 maintains the ability to associate with CD63 (Fig. 7C), which is consistent with the colocalization of these proteins observed by confocal microscopy. Since LMP1 has previously been shown to increase EV production, we wondered whether TM5-6 similarly enhances EV levels. Nanoparticle tracking analysis of cells transfected with LMP1 confirmed previous findings of elevated EV production (P < 0.05) (28, 57); however, expression of TM5-6 produced levels of particles comparable to those of control GFP-transfected cells (P = 0.16) (Fig. 7D). Taken together, these data suggest that LMP1 packaging and enhancement of EV production require factors downstream of its interaction with CD63. It is likely that regions in the N terminus of LMP1 are important for recruiting cellular components that mediate these processes.
FIG 7.
TM5-6 colocalizes and associates with CD63. (A) HEK293 cells were cotransfected with GFP-tagged LMP1 constructs and CD63-RFP. Live-cell confocal images were acquired 24 h posttransfection. The images were analyzed using Imaris software. Representative maximum-projection images are shown. Scale bar, 10 μm. (B) Colocalization was quantified using Pearson's correlation coefficient and graphed with standard errors of the mean (n ≥ 12 cells). (C) CD63 knockout cells were cotransfected with CD63-BirA* plus WT LMP1 or CD63-BirA* plus TM5-6 or transfected with CD63-BirA* alone. Biotinylated protein complexes were isolated and separated by SDS-PAGE and analyzed by immunoblotting with LMP1 and CD63-specific antibodies. (D) EVs were harvested from HEK293 cells transiently transfected with GFP, GFP-LMP1, or TM5-6 and quantified by nanoparticle tracking. The data are represented as total numbers of particles harvested per cell. *, P < 0.05. (E and F) HEK293 cells were transfected with GFP-TM5-6, SNAP-LMP1 or cotransfected with GFP-TM5-6 and SNAP-LMP1. (E) Whole-cell and (F) EV lysates were separated by SDS-PAGE and analyzed by immunoblot analysis for GFP, SNAP, and common EV markers (Alix and HSC70).
LMP1 NF-κB activation has previously been shown to be complemented by different intermolecular interactions among transmembrane domains; therefore, we wondered whether TM5-6 EV trafficking could be enhanced by the presence of WT LMP1 (53). Coexpression of TM5-6 and WT LMP1 revealed that mutant TM5-6 EV trafficking could not be rescued by WT LMP1 (Fig. 7E and F). Surprisingly, LMP1 EV packaging was reduced (Fig. 7F) following cotransfection with TM5-6, suggesting the mutant construct may act as a dominant negative for LMP1 EV trafficking. Taken together, intermolecular interactions between transmembrane domains 5 and 6 and 1 to 4 likely do not drive EV incorporation of LMP1. This may not be surprising, since Soni et al. showed that intermolecular interactions between transmembrane domains 1 and 2 and 3 to 6 are required for dimerization and NF-κB activation (53).
DISCUSSION
Despite the potential significance of LMP1-modified EVs for modulating the tumor microenvironment, the mechanisms by which LMP1 hijacks the endolysosomal pathways for its own secretion and the enhanced release of cellular cargo remain unknown. To begin to better understand the trafficking of LMP1 to EVs, we investigated the abilities of deletion mutants of LMP1 to be packaged into EVs. Here, it was found that the N terminus and transmembrane domain 1 are sufficient for targeting LMP1 to EVs. The mutants lacking the N terminus and transmembrane domains failed to be packaged into EVs, pointing to the importance of these regions in EV targeting. However, the N terminus alone could not direct GFP to EVs unless it was combined with other transmembrane regions. Therefore, other transmembrane regions may be able to substitute for the function of TM1 when fused to the N terminus by supporting membrane association and oligomerization.
Prior studies have demonstrated that plasma membrane targeting and higher-order oligomerization are sufficient for protein trafficking to EVs (58–60). Various protein modifications have been shown to be responsible for EV targeting, including myristoylation, prenylation, and palmitoylation (58–60). Furthermore, phosphatidylinositol 4,5-bisphosphate and phosphatidylinositol (3,4,5)-trisphosphate-binding domains can act as membrane anchors for EV incorporation (58, 59). LMP1 contains prenylation, palmitoylation, and other signaling motifs that could mediate LMP1 exosomal trafficking and the efficient sorting of LMP1-interacting proteins into EVs. Indeed, previous findings suggested that a single point mutation at the C78 palmitoylation site within TM3 impairs LMP1 EV secretion (61). However, mutants lacking TM3 in our study were still incorporated into EVs, and TM1 was sufficient to target GFP to EVs. Although we did not directly analyze a C78 mutant, these discrepancies may reflect the different EV isolation methods used in each study. Here, we used slightly longer and higher-speed centrifugation steps than in the previous study, a method that is more likely to concentrate smaller extracellular vesicles. It will be interesting to determine if distinct molecular requirements guide LMP1 sorting into different EV subpopulations. Regardless, it is clear from our data that amino acids 1 to 44 of LMP1 contain EV-targeting information.
We further demonstrated that the N- and C-terminal tails of LMP1 are not sufficient for EV incorporation. The primary function of the N-terminal tail is to ensure proper orientation of the transmembrane domains (62, 63). Absence of the N-terminal domain, or a smaller proline/arginine-rich subdomain, results in altered LMP1 stability, localization, and oligomerization (64). Therefore, the LMP1 mutants used in this study were designed with an arginine-rich N terminus to maintain the tethering of the first transmembrane domain and to ensure proper membrane orientation of LMP1, as previously described (53, 65). The GFP-LMP1 and derivative constructs exhibited varying levels of proteolytic cleavage, which could complicate immunoblot analyses. To account for this, an antibody against GFP was used, and the signal from the entire lane was quantified in determining relative EV packaging. The proteolytic products could also influence the interpretation of the localization studies. However, the minor cleaved products likely have little effect, as the GFP-LMP1 construct exhibited localization patterns similar to those of nontagged LMP1 described in other studies. It is also well established that LMP1 is degraded through proteasomal and lysosomal processes.
The cytoplasmic C-terminal tail of LMP1 contains 200 amino acids, with CTAR1 and CTAR2 mediating the majority of the signal transduction events described to date. Despite the importance of these signaling motifs in interacting with multiple proteins to initiate unique signal transduction events, the motifs were not necessary or sufficient to target LMP1 into EVs. In fact, greater release of LMP1 into EVs was observed when LMP1-TRAF2 association domains were mutated, suggesting that CTAR interaction with TRAFs may limit the incorporation of LMP1 into EVs (61). Together, our data indicate that the LMP1 C-terminal tail is dispensable for packaging and support previous observations that mutants lacking CTAR domains are efficiently sorted into EVs (61).
In this study, TM1ΔC and TM1-2ΔC were found to efficiently traffic to EVs and to exhibit perinuclear intracellular localization similar to that of intact LMP1. These constructs contain minimal transmembrane regions attached to the N terminus. To better understand the molecular requirements for LMP1 EV sorting, we examined the significance of trafficking motifs within the first transmembrane domain. The inhibition of LMP1 lipid raft localization by mutating the FWLY sequence within the first transmembrane region increased sorting of LMP1 into EVs. Mutation of the FWLY domain causes plasma membrane accumulation, which may enhance LMP1 shedding from the plasma membrane in microvesicles or increase the sorting of LMP1 into exosomes through its reuptake into the endocytic pathway (49). Furthermore, the data may reflect the inversely related nature of LMP1 signaling and EV secretion (28, 49, 57). The first transmembrane segments of LMP1 have multiple leucine residues that are similar to a leucine heptad motif (66). The leucine heptad has been found to be important for efficient trafficking of LMP1 to membrane microdomains, which are thought to be platforms for exosome biogenesis and cargo sorting (67, 68). The leucine heptad possesses multiple dileucine motifs that have been reported to be efficient for the endosomal/lysosomal targeting of transmembrane protein 192 (69). These leucine residues mediate movement of the proteins from the trans-Golgi network to endosomes and from there to exosomes or lysosomes. Alternatively, the proteins are directed to the plasma membrane and then endocytosed to be targeted to endosomes. These data suggest that the leucine heptad within TM1 could serve as an endosomal sorting motif to target LMP1 to the endosomal pathway and ultimately toward trafficking into EVs.
Mutants containing deletions of transmembrane segments 3 and 4 (TMΔ3-4) or 5 and 6 (TMΔ5-6) exhibited greater packaging into EVs. LMP1 binds the Rab-associated protein PRA1 through transmembrane domains 3 to 6 (70). PRA1 plays a significant role in LMP1 intracellular trafficking from the ER to the Golgi network and its induction of NF-κB signaling (70). Deleting TM3-4 or TM5-6 of LMP1 impairs its ability to interact with PRA1 and hence results in defects in ER-to-Golgi transport. Similarly, we observed diffuse punctate localization of these mutants throughout the cell (Fig. 4) (70–72). TMΔ1-2, a mutant lacking the lipid raft trafficking anchor, was found to be targeted into EVs at higher levels than WT protein (61, 73). TMΔ1-2 induces NF-κB activation; accumulates in the plasma membrane, where it is endocytosed; and is secreted into EVs to escape lysosomal degradation (61, 73). Greater EV release displayed by TM3-4 and TM5-6 suggests that these constructs too may evade lysosomal degradation to be sorted into exosomes or, alternatively, shed straight from the plasma membrane in microvesicles. Future mechanistic studies on mutants with enhanced and reduced EV secretion will likely provide novel insights into the diverse EV biogenesis and trafficking pathways present within the cell.
The levels of EV packaging that we observed for TM3-4, TM3-6, and TM5-6 were decreased compared to LMP1 WT. Though these mutants contained transmembrane domains harboring important signaling and trafficking motifs, they were not sufficiently packaged to EVs. Transmembrane domains 3 to 6 are important for LMP1 interaction with PRA1, and transmembrane domains 5 and 6 also contain the leucine heptad repeat (66, 70). Yasui et al. demonstrated that TM3-4, TM3-6, and TM5-6 mutants do not activate NF-κB signaling (65), which may be explained by the altered subcellular localization and deficiencies in EV sorting. LMP1 is synthesized in the ER and traffics to the Golgi network and endosomes, where it activates NF-κB. Retention of LMP1 in the ER results in decreased NF-κB activation (70). Our data demonstrate altered intracellular trafficking of TM5-6 with accumulation of the mutant protein in the ER and early endosomes concomitant with the attenuation of extracellular release. Notably, a smaller proportion of TM5-6 than of WT LMP1 colocalized with Lamp1, indicating the mutant is not targeted as efficiently to lysosomes. Surprisingly, TM5-6 maintained its ability to colocalize and form a complex with CD63 despite the inefficient EV targeting. LMP1 has been shown to associate with the tetraspanin protein CD63, and LMP1 sorting into EVs is severely inhibited when CD63 is knocked down or out in cells (28, 49). Taken together, these results suggest new functional roles for the N terminus and LMP1 transmembrane domain 1 in intra- and extracellular trafficking, which are downstream of an interaction with CD63. The observation that a mutant with transmembrane domains 1 to 4 deleted (TMΔ1-4) is still capable of trafficking to EVs was surprising and suggests that the transmembrane regions 5 and 6 may be able to substitute for TM1 when fused to the N terminus. If the secretion of LMP1 from cells is important for regulating intercellular signaling, inhibiting the immune system, and modulating the infected microenvironment, then it may be expected that the virus has built-in redundancy to ensure LMP1 EV sorting (27, 28, 57, 74). In support of this hypothesis, TM5-6 also contains heptad repeats similar to those found in TM1 that may direct EV trafficking.
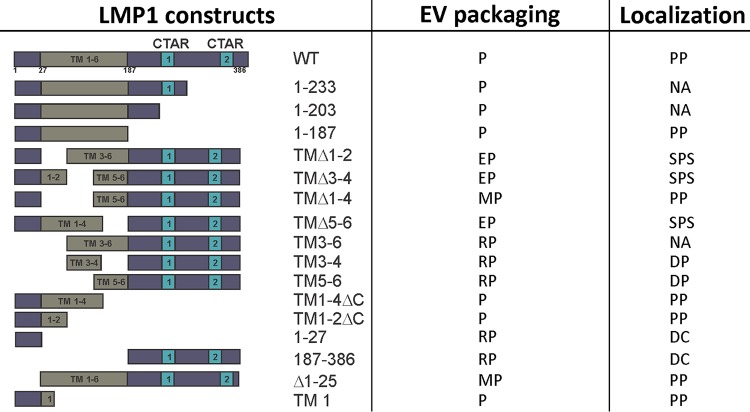
Finally, the deletion of the N terminus only from LMP1 did not impair its sorting into EVs, supporting the importance of TM1 in EV trafficking. Our results further identify the minimal regions of LMP1 required for EV targeting. A summary of the EV packaging and localization of all mutants analyzed in this study is provided in Fig. 8. The presence of trafficking motifs like the leucine heptad in TM1 may play a role in orchestrating LMP1 EV secretion. It is likely that cellular proteins bind to regions in the N terminus and the first transmembrane domain of LMP1 that direct its intra- and extracellular trafficking. Future research is needed to identify LMP1 protein interactors necessary for the incorporation of LMP1 into exosomes and other EVs.
FIG 8.
Summary of LMP1 mutant data. P, packaged; EP, enhanced packaging; MP, moderate packaging; RP, reduced packaging; PP, punctate perinuclear; SPS, scattered punctate structures; DP, diffuse perinuclear; DC, diffuse in cytoplasm; NA, not analyzed.
MATERIALS AND METHODS
Cell culture and transfection.
HEK293 cells (ATTC CLR-1573) were cultured in Dulbecco's modified Eagle medium (Lonza; 12-604Q). The medium was supplemented with 10% fetal bovine serum (FBS) (Seradigm; 1400-500), 2 mM l-glutamine (Corning; 25-005-CI), 100 international units (IU) penicillin-streptomycin (Corning; 30-002-CI), and 100 IU:100 μg/ml:0.25 μg/ml antibiotic/antimycotic (Corning; 30-002-CI). The cells were maintained at 37°C with 5% CO2. HEK293 cells were transfected with plasmids using JetPrime transfection reagent (Polyplus; 114-15) according to the manufacturer's instructions. The HEK293 cells were transduced with CD63 lentiCRISPRv2 plasmid as previously described (28).
Plasmids. (i) LMP1 mutants.
The GFP-LMP1 (LMP1 WT) and mutant (1-231, 1-203, and 1-187) plasmids were constructed by PCR amplification of LMP1 from pBabe-HA-LMP1 (a kind gift from Nancy Raab-Traub) using primers containing HindIII and EcoRI restriction sites. The resulting PCR products were digested with HindIII and EcoRI and cloned in frame into pENTR4-GFP-C1 (Addgene; number 17396; a gift from Eric Campeau). The mutants were recombined into the destination vectors pLenti PGK Neo DEST (Addgene; number 19067; a gift from Eric Campeau and Paul Kaufman) and pQCXP CMV/TO (Addgene; number 17386; a gift from Eric Campeau) (75) by recombination using Gateway LR Clonase II (Invitrogen; number 11791-020).
GFP-LMP1 mutants (1-27, TM1ΔC, TM1-4ΔC, and TM1-2ΔC) were constructed using the GeneArt site-directed mutagenesis system (Invitrogen; number A13282) to introduce premature stop codons using GFP-LMP1 WT as a template.
GFP-LMP1 mutants (187-386, TMΔ5-6, TM3-6, TM5-6, TM3-4, Δ1-25, TMΔ3-4, TMΔ1-2, and TMΔ1-4) were constructed by inverse PCR of WT GFP-LMP1 to introduce internal deletions. The resulting PCR products were T4 ligated with the pENTR4 vector, and clones were verified by sequencing. GFP-LMP1 mutants were constructed by LR recombination (Invitrogen; number 11791-020) between entry vectors and the pGKNeo destination vector.
SNAP-LMP1 was constructed by PCR amplification of LMP1 from the pBabe-HA-LMP1 vector with primers containing BamHI and XbaI restriction sites. The resulting PCR products were digested and cloned in frame into pENTR4-SNAPf (Addgene; number 29652; a gift from Eric Campeau). SNAP-LMP1-QCXP was constructed by LR recombination (Invitrogen; number 11791-020) between entry vectors and pQCXIZ CMV/TO DEST (Addgene; number 17401; a gift from Eric Campeau and Paul Kaufman). SNAP-LMP1-FWLY mutants were constructed by site-directed mutagenesis (Agilent Quick Change; number 200523) of pQCXIZ-SNAP-LMP1. The LMP1-FWLY mutant has been described previously (65).
CD63-BirA was generated by PCR amplification from pCT-CD63-GFP (SBI System Biosciences; number CYTO120-PA-1) with primers containing EcoRI and XmnI restriction sites (57). The resulting PCR product was digested with EcoRI and XmnI and cloned in frame into pENTR1A-BirA (Addgene; number 29645; a gift from Eric Campeau). CD63-BirA pGKNeo vectors were constructed by LR recombination (Invitrogen; number 11791-020) between entry vectors and the pLenti-pGKNeo (Addgene; number 19067) destination vector.
Nontagged LMP1 was generated by digesting pQCXIZ-SNAP-LMP1 with BamH1 and Xbal restriction enzymes to remove the SNAP tag. The resulting product was cloned into pENTR1A no ccDB (Addgene; number 17398; a gift from Eric Campeau and Paul Kaufman) following digestion with the same restriction enzymes. pENTR1A LMP1 was then recombined into the destination vector pLenti PGK Neo DEST (Addgene; number 19067; a gift from Eric Campeau and Paul Kaufman).
(ii) Subcellular compartment markers.
mCh-Sec61 beta (Addgene; number 49155; a gift from Gia Voeltz) (76), Lamp1-RFP (Addgene; number 1817; a gift from Walther Mothes) (77), DsRed-rab11 WT (Addgene; number 12679) (78), DsRed-rab7 WT (Addgene; number 12661) (78), and DsRed-Rab5 WT (Addgene; plasmid number 13050) (79) were gifts from Richard Pagano. Golgi-RFP was constructed by cloning the eNos sequence from Golgi-CFP (Addgene; number 14873; a gift from Alexandra Newton) into the entry vector pENTR dsRed2 N1 (Addgene; number 22523), followed by an LR recombination into the destination vector pQCXP CMV/TO (Addgene; number 17386). The RFP-CD63 plasmid was constructed by PCR amplification of CD63 from pCT-CD63-GFP (System Biosciences) using primers containing EcoRI and BamHI cut sites. The CD63 PCR product was cloned into pENTRDsRedEx2 (a gift from Nathan Lawson; Addgene; number 22451) and then recombined into the pQCXP CMV/TO destination vector by LR recombination using Gateway LR Clonase II (Invitrogen) according to the manufacturer's instructions.
Extracellular vesicle isolation.
EVs were harvested from HEK293 cells following transfection with WT LMP1 or LMP1 mutants. Forty-eight hours posttransfection, the vesicle-containing culture medium was harvested. EVs were isolated from the culture medium by differential centrifugation. The medium was centrifuged at 500 × g for 5 min (twice) and 2,000 × g for 10 min (twice) in an Eppendorf 5804R using an S-4-104 rotor, followed by 10,000 × g centrifugation for 30 min (twice) in an Eppendorf 5804R using the FA-45-630 rotor to remove cells and cellular debris. EVs were pelleted with a 100,000 × g centrifugation for 70 min in a Beckman Optima XL using a SW41 Ti rotor. The EV pellet was washed with 1× phosphate-buffered saline (PBS), centrifuged at 100,000 × g for 70 min in a Beckman MAX-E using the TLA120.2 rotor, and resuspended in 2× Laemmli sample buffer (4% SDS, 100 mM Tris, pH 6.8, 0.4 mg/ml bromophenol blue, 0.2 M dithiothreitol [DTT], 20% glycerol, 2% β-mercaptoethanol [BME]). All centrifugations were performed at 4°C. Nanoparticle tracking analysis (NTA) was performed on a Malvern Nanosight LM10 as previously described (80).
Immunoblot analysis.
HEK293 cells were harvested 48 h posttransfection and centrifuged at 500 × g for 5 min to collect cell pellets. The cells were washed with 1× PBS to remove excess medium and recentrifuged at 500 × g for 5 min. The cell pellets were resuspended in radioimmunoprecipitation assay (RIPA) buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 0.1% SDS, 0.5% deoxycholic acid) plus 1× HALT protease inhibitor (Thermo Scientific; number 78429), Phosphatase Arrest (G-Biosciences; number 786-450), and 1× phenylmethylsulfonyl fluoride (PMSF) (0.5 mM PMSF) and incubated on ice for 30 min. The cell lysates were centrifuged at 22,220 × g for 10 min at 4°C to harvest the solubilized protein. The lysates were mixed with 5× Laemmli sample buffer (10% SDS, 250 mM Tris, pH 6.8, 1 mg/ml bromophenol blue, 0.5 M DTT, 50% glycerol, 5% BME) to a final concentration of 1× and boiled at 95°C for 10 min. An equal amount of protein was loaded into an SDS-10% PAGE gel for electrophoresis and then transferred to a nitrocellulose membrane. The blots were blocked in Tris-buffered saline solution containing 0.1% Tween 20 and 5% nonfat dry milk. The primary antibodies used included Alix (Q-19; Santa Cruz Biotechnology), HSC70 (B-6; Santa Cruz), TSG101 (C-2; Santa Cruz), CD81 (sc-9158; Santa Cruz), Flotillin-2 (H-90; Santa Cruz), CD63 (TS63; Abcam), GFP (600-101-215; Rockland), LMP1 (CS1-4; Dako). The blots were subsequently probed with the following horseradish peroxidase (HRP)-conjugated secondary antibodies: rabbit anti-mouse IgG (Genetex; 26728), rabbit anti-goat IgG (Genetex; 26741), goat anti-rabbit IgG (Fab fragment) (Genetex; 27171), or anti-mouse kappa light chain (H139-52.1; Abcam). The blots were incubated with Pico ECL (Thermo; number 34080). The blots were imaged using an Image Quant LAS4000 (General Electric) and processed with ImageQuant TL v8.1.0.0 software, Adobe Photoshop CS6, and CorelDraw Graphic Suite X5.
Live-cell confocal microscopy.
HEK293 cells were seeded in 35-mm glass bottom plates (Greiner Bio-One; number 627860) and transfected 24 h later. Twenty-four hours posttransfection, nuclei were labeled with Hoechst 33342 (Thermo Scientific; number 62249) to a final concentration of 5 μg/ml according to the manufacturer's instructions. Cells were imaged on the Andor Revolution spinning-disk laser confocal microscope or the Zeiss LSM 880 confocal microscope. Images were analyzed using the Imaris (Andor), Zen 2.1 Black, or ImageJ software package.
Bio-ID pulldown assay.
HEK293 CD63 CRISPR (clustered regularly interspaced short palindromic repeats) cells were seeded in 100-mm dishes and transfected with CD63*-BirA and GFP, CD63*-BirA and GFP-LMP1, or CD63*-BirA and TM5-6. The medium was replaced with fresh medium containing 50 mM biotin 14 h posttransfection, and the cells were allowed to grow for another 24 h. Cells were lysed in 700 μl of 2× Bio-ID lysis buffer (50 mM Tris, pH 7.6, 500 mM NaCl, 0.4% SDS, 5 mM EDTA, 2% Triton X-100, 1 mM DTT) and HALT protease inhibitor (Thermo Scientific; number 78429) and then sonicated using a Diagenode Bioruptor-300 water bath sonicator. Insoluble materials were cleared by centrifuging for 15 min at 20,200 × g at 4°C. An equal volume of 50 mM Tris (pH 7.6) was added to the cell lysate to dilute the high detergent concentration, and then biotinylated proteins were pulled down by overnight incubation with streptavidin-conjugated magnetic beads (Pierce; 88817) at 4°C. The magnetic beads were collected and washed twice for 8 min at room temperature in 1 ml wash buffer 1 (2% SDS in distilled water [dH2O]). This was then repeated with wash buffer 2 (0.1% deoxycholate, 1% Triton X-100, 500 mM NaCl, 1 mM EDTA, and 50 mM HEPES, pH 7.5), once with wash buffer 3 (250 mM LiCl, 0.5% NP-40, 0.5% deoxycholate, 1 mM EDTA, and 10 mM Tris, pH 8.1), and twice with wash buffer 4 (50 mM Tris, pH 7.4, and 50 mM NaCl). The bound proteins were eluted using 1× Laemmli SDS sample buffer and boiled at 98°C for 5 min. The proteins were resolved on SDS-10% PAGE for further immunoblotting. This protocol has been previously described by Roux et al. (55).
Membrane flotation assay.
HEK293 cells were seeded into 150-mm plates and transfected with GFP-LMP1 or selected mutants for 24 h. The cell pellets (equivalent to 300 μl of packed cells) were collected as described above and frozen at −80°C prior to lysis. The cells were lysed in 700 μl of hypotonic buffer (20 mM HEPES, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA) and then homogenized with a tight-fit Dounce homogenizer (30 to 35 strokes). Membranes were isolated from the crude lysates by sucrose gradient purification, as previously described (28).
ACKNOWLEDGMENTS
We thank Ruth Didier at the Florida State University (FSU) College of Medicine Confocal Microscopy Laboratory for assistance with live-cell microscopy and Mark Rider for helpful discussions. We also thank the Molecular Cloning Facility in the Department of Biological Science at FSU for help with cloning some of the mutants described in this study.
This work was supported by the National Institutes of Health under grant RO1CA204621 and grant R15CA188941 awarded to D.G.M.
We report no conflicts of interest.
D.G.M. conceived of and designed the study. D.N., L.A.H., M.R.C., S.N.H., D.C.T., and X.L. designed and performed experiments and interpreted results. The manuscript was written by D.N. and D.G.M. with edits from all authors.
REFERENCES
- 1.Henle G, Henle W, Klein G, Gunven P, Clifford P, Morrow RH, Ziegler JL. 1971. Antibodies to early Epstein-Barr virus-induced antigens in Burkitt's lymphoma. J Natl Cancer Inst 46:861–871. [PubMed] [Google Scholar]
- 2.Hsu JL, Glaser SL. 2000. Epstein-Barr virus-associated malignancies: epidemiologic patterns and etiologic implications. Crit Rev Oncol Hematol 34:27–53. doi: 10.1016/S1040-8428(00)00046-9. [DOI] [PubMed] [Google Scholar]
- 3.Pattle SB, Farrell PJ. 2006. The role of Epstein-Barr virus in cancer. Expert Opin Biol Ther 6:1193–1205. doi: 10.1517/14712598.6.11.1193. [DOI] [PubMed] [Google Scholar]
- 4.Kaye KM, Izumi KM, Kieff E. 1993. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc Natl Acad Sci U S A 90:9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kempkes B, Robertson ES. 2015. Epstein-Barr virus latency: current and future perspectives. Curr Opin Virol 14:138–144. doi: 10.1016/j.coviro.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kieser A, Sterz KR. 2015. The latent membrane protein 1 (LMP1), p 119–149. In Münz C. (ed), Epstein Barr virus, vol 2 One herpes virus: many diseases. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 7.Wang D, Liebowitz D, Kieff E. 1985. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell 43:831–840. doi: 10.1016/0092-8674(85)90256-9. [DOI] [PubMed] [Google Scholar]
- 8.Kilger E, Kieser A, Baumann M, Hammerschmidt W. 1998. Epstein-Barr virus-mediated B-cell proliferation is dependent upon latent membrane protein 1, which simulates an activated CD40 receptor. EMBO J 17:1700–1709. doi: 10.1093/emboj/17.6.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kulwichit W, Edwards RH, Davenport EM, Baskar JF, Godfrey V, Raab-Traub N. 1998. Expression of the Epstein-Barr virus latent membrane protein 1 induces B cell lymphoma in transgenic mice. Proc Natl Acad Sci U S A 95:11963–11968. doi: 10.1073/pnas.95.20.11963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soni V, Cahir-McFarland E, Kieff E. 2007. LMP1 TRAFficking activates growth and survival pathways, p 173–187. In Wu H. (ed), TNF receptor associated factors (TRAFs). Springer, New York, NY. [DOI] [PubMed] [Google Scholar]
- 11.Kuhne MR, Robbins M, Hambor JE, Mackey MF, Kosaka Y, Nishimura T, Gigley JP, Noelle RJ, Calderhead DM. 1997. Assembly and regulation of the CD40 receptor complex in human B cells. J Exp Med 186:337–342. doi: 10.1084/jem.186.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Busch LK, Bishop GA. 1999. The EBV transforming protein, latent membrane protein 1, mimics and cooperates with CD40 signaling in B lymphocytes. J Immunol 162:2555–2561. [PubMed] [Google Scholar]
- 13.Uchida J, Yasui T, Takaoka-Shichijo Y, Muraoka M, Kulwichit W, Raab-Traub N, Kikutani H. 1999. Mimicry of CD40 signals by Epstein-Barr virus LMP1 in B lymphocyte responses. Science 286:300–303. doi: 10.1126/science.286.5438.300. [DOI] [PubMed] [Google Scholar]
- 14.Miller WE, Cheshire JL, Baldwin AS Jr, Raab-Traub N. 1998. The NPC derived C15 LMP1 protein confers enhanced activation of NF-κB and induction of the EGFR in epithelial cells. Oncogene 16:1869. doi: 10.1038/sj.onc.1201696. [DOI] [PubMed] [Google Scholar]
- 15.Mosialos G, Birkenbacht M, Yalamanchill R, Van Arsdale T, Ware C, Kleff E. 1995. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell 80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 16.Morris MA, Dawson CW, Young LS. 2009. Role of the Epstein-Barr virus-encoded latent membrane protein-1, LMP1, in the pathogenesis of nasopharyngeal carcinoma. Future Oncol 5:811–825. doi: 10.2217/fon.09.53. [DOI] [PubMed] [Google Scholar]
- 17.Luftig M, Prinarakis E, Yasui T, Tsichritzis T, Cahir-McFarland E, Inoue J-I, Nakano H, Mak TW, Yeh W-C, Li X, Akira S, Suzuki N, Suzuki S, Mosialos G, Kieff E. 2003. Epstein–Barr virus latent membrane protein 1 activation of NF-κB through IRAK1 and TRAF6. Proc Natl Acad Sci U S A 100:15595–15600. doi: 10.1073/pnas.2136756100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eliopoulos AG, Gallagher NJ, Blake SMS, Dawson CW, Young LS. 1999. Activation of the p38 mitogen-activated protein kinase pathway by Epstein-Barr virus-encoded latent membrane protein 1 coregulates interleukin-6 and interleukin-8 production. J Biol Chem 274:16085–16096. doi: 10.1074/jbc.274.23.16085. [DOI] [PubMed] [Google Scholar]
- 19.Eliopoulos AG, Young LS. 1998. Activation of the cJun N-terminal kinase (JNK) pathway by the Epstein-Barr virus-encoded latent membrane protein 1 (LMP1). Oncogene 16:1731. doi: 10.1038/sj.onc.1201694. [DOI] [PubMed] [Google Scholar]
- 20.Dawson CW, Tramountanis G, Eliopoulos AG, Young LS. 2003. Epstein-Barr virus latent membrane protein 1 (LMP1) activates the phosphatidylinositol 3-kinase/Akt pathway to promote cell survival and induce actin filament remodeling. J Biol Chem 278:3694–3704. doi: 10.1074/jbc.M209840200. [DOI] [PubMed] [Google Scholar]
- 21.Kieser A, Kilger E, Gires O, Ueffing M, Kolch W, Hammerschmidt W. 1997. Epstein-Barr virus latent membrane protein-1 triggers AP-1 activity via the c-Jun N-terminal kinase cascade. EMBO J 16:6478–6485. doi: 10.1093/emboj/16.21.6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saito N, Courtois G, Chiba A, Yamamoto N, Nitta T, Hironaka N, Rowe M, Yamamoto N, Yamaoka S. 2003. Two carboxyl-terminal activation regions of Epstein-Barr virus latent membrane protein 1 activate NF-κB through distinct signaling pathways in fibroblast cell lines. J Biol Chem 278:46565–46575. doi: 10.1074/jbc.M302549200. [DOI] [PubMed] [Google Scholar]
- 23.Meckes DG, Menaker NF, Raab-Traub N. 2013. Epstein-Barr virus LMP1 modulates lipid raft microdomains and the vimentin cytoskeleton for signal transduction and transformation. J Virol 87:1301–1311. doi: 10.1128/JVI.02519-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shair KHY, Schnegg CI, Raab-Traub N. 2008. Epstein-Barr virus latent membrane protein-1 effects on plakoglobin, cell growth and migration. Cancer Res 68:6997–7005. doi: 10.1158/0008-5472.CAN-08-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mainou BA, Everly DN Jr, Raab-Traub N. 2005. Epstein–Barr virus latent membrane protein 1 CTAR1 mediates rodent and human fibroblast transformation through activation of PI3K. Oncogene 24:6917. doi: 10.1038/sj.onc.1208846. [DOI] [PubMed] [Google Scholar]
- 26.Kung CP, Meckes DG, Raab-Traub N. 2011. Epstein-Barr virus LMP1 activates EGFR, STAT3, and ERK through effects on PKCdelta. J Virol 85:4399–4408. doi: 10.1128/JVI.01703-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meckes DG, Shair KH, Marquitz AR, Kung CP, Edwards RH, Raab-Traub N. 2010. Human tumor virus utilizes exosomes for intercellular communication. Proc Natl Acad Sci U S A 107:20370–20375. doi: 10.1073/pnas.1014194107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hurwitz SN, Nkosi D, Conlon MM, York SB, Liu X, Tremblay DC, Meckes DG. 2017. CD63 regulates Epstein-Barr virus LMP1 exosomal packaging, enhancement of vesicle production, and noncanonical NF-κB signaling. J Virol 91:e02251-16. doi: 10.1128/JVI.02251-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gutzeit C, Nagy N, Gentile M, Lyberg K, Gumz J, Vallhov H, Puga I, Klein E, Gabrielsson S, Cerutti A, Scheynius A. 2014. Exosomes derived from Burkitt's lymphoma cell lines induce proliferation, differentiation, and class-switch recombination in B cells. J Immunol 192:5852–5862. doi: 10.4049/jimmunol.1302068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aga M, Bentz GL, Raffa S, Torrisi MR, Kondo S, Wakisaka N, Yoshizaki T, Pagano JS, Shackelford J. 2014. Exosomal HIF1α supports invasive potential of nasopharyngeal carcinoma-associated LMP1-positive exosomes. Oncogene 33:4613–4622. doi: 10.1038/onc.2014.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nanbo A, Kawanishi E, Yoshida R, Yoshiyama H. 2013. Exosomes derived from Epstein-Barr virus-infected cells are internalized via caveola-dependent endocytosis and promote phenotypic modulation in target cells. J Virol 87:10334–10347. doi: 10.1128/JVI.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colombo M, Raposo G, Théry C. 2014. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol 30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 33.Maas SLN, Breakefield XO, Weaver AM. 2017. Extracellular vesicles: unique intercellular delivery vehicles. Trends Cell Biol 27:172–188. doi: 10.1016/j.tcb.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meckes DG, Raab-Traub N. 2011. Microvesicles and viral infection. J Virol 85:12844–12854. doi: 10.1128/JVI.05853-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meckes DG. 2015. Exosomal communication goes viral. J Virol 89:5200–5203. doi: 10.1128/JVI.02470-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meckes DG, Gunawardena HP, Dekroon RM, Heaton PR, Edwards RH, Ozgur S, Griffith JD, Damania B, Raab-Traub N. 2013. Modulation of B-cell exosome proteins by gamma herpesvirus infection. Proc Natl Acad Sci U S A 110:E2925–E2933. doi: 10.1073/pnas.1303906110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nolte-‘t Hoen E, Cremer T, Gallo RC, Margolis LB. 2016. Extracellular vesicles and viruses: are they close relatives? Proc Natl Acad Sci U S A 113:9155–9161. doi: 10.1073/pnas.1605146113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Camussi G, Deregibus MC, Bruno S, Cantaluppi V, Biancone L. 2010. Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney Int 78:838–848. doi: 10.1038/ki.2010.278. [DOI] [PubMed] [Google Scholar]
- 39.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. 2007. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biol 9:654. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 40.Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MAJ, Hopmans ES, Lindenberg JL, de Gruijl TD, Würdinger T, Middeldorp JM. 2010. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U S A 107:6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Théry C. 2011. Exosomes: secreted vesicles and intercellular communications. F1000 Biol Rep 3:15. doi: 10.3410/B3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mathivanan S, Ji H, Simpson RJ. 2010. Exosomes: extracellular organelles important in intercellular communication. J Proteomics 73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 43.Graner MW, Alzate O, Dechkovskaia AM, Keene JD, Sampson JH, Mitchell DA, Bigner DD. 2009. Proteomic and immunologic analyses of brain tumor exosomes. FASEB J 23:1541–1557. doi: 10.1096/fj.08-122184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang N, Li S, Li G, Zhang S, Tang X, Ni S, Jian X, Xu C, Zhu J, Lu M. 2017. The role of extracellular vesicles in mediating progression, metastasis and potential treatment of hepatocellular carcinoma. Oncotarget 8:3683–3695. doi: 10.18632/oncotarget.12465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soung YH, Nguyen T, Cao H, Lee J, Chung J. 2016. Emerging roles of exosomes in cancer invasion and metastasis. BMB Rep 49:18–25. doi: 10.5483/BMBRep.2016.49.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng Z, Hensley L, McKnight KL, Hu F, Madden V, Ping L, Jeong S-H, Walker C, Lanford RE, Lemon SM. 2013. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 496:367–371. doi: 10.1038/nature12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gastaminza P, Dryden KA, Boyd B, Wood MR, Law M, Yeager M, Chisari FV. 2010. Ultrastructural and biophysical characterization of hepatitis C virus particles produced in cell culture. J Virol 84:10999–11009. doi: 10.1128/JVI.00526-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramakrishnaiah V, Thumann C, Fofana I, Habersetzer F, Pan Q, de Ruiter PE, Willemsen R, Demmers JAA, Stalin Raj V, Jenster G, Kwekkeboom J, Tilanus HW, Haagmans BL, Baumert TF, van der Laan LJW. 2013. Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7.5 cells. Proc Natl Acad Sci U S A 110:13109–13113. doi: 10.1073/pnas.1221899110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verweij FJ, Eijndhoven MAJv Hopmans ES, Vendrig T, Wurdinger T, Cahir-McFarland E, Kieff E, Geerts D, Kant Rvd Neefjes J, Middeldorp JM, Pegtel DM. 2011. LMP1 association with CD63 in endosomes and secretion via exosomes limits constitutive NF-κB activation. EMBO J 30:2115–2129. doi: 10.1038/emboj.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Gassart A, Géminard C, Février B, Raposo G, Vidal M. 2003. Lipid raft-associated protein sorting in exosomes. Blood 102:4336. doi: 10.1182/blood-2003-03-0871. [DOI] [PubMed] [Google Scholar]
- 51.Valapala M, Vishwanatha JK. 2011. Lipid raft endocytosis and exosomal transport facilitate extracellular trafficking of annexin A2. J Biol Chem 286:30911–30925. doi: 10.1074/jbc.M111.271155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keerthikumar S, Gangoda L, Liem M, Fonseka P, Atukorala I, Ozcitti C, Mechler A, Adda CG, Ang C-S, Mathivanan S. 2015. Proteogenomic analysis reveals exosomes are more oncogenic than ectosomes. Oncotarget 6:15375–15396. doi: 10.18632/oncotarget.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soni V, Yasui T, Cahir-McFarland E, Kieff E. 2006. LMP1 transmembrane domain 1 and 2 (TM1-2) FWLY mediates intermolecular interactions with TM3-6 ATivate NF-κB. J Virol 80:10787–10793. doi: 10.1128/JVI.01214-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lam N, Sugden B. 2003. LMP1, a viral relative of the TNF receptor family, signals principally from intracellular compartments. EMBO J 22:3027–3038. doi: 10.1093/emboj/cdg284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roux KJ, Kim DI, Raida M, Burke B. 2012. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rider MA, Cheerathodi MR, Hurwitz SN, Nkosi D, Howell LA, Tremblay DC, Liu X, Zhu F, Meckes DG. 2018. The interactome of EBV LMP1 evaluated by proximity-based BioID approach. Virology 516:55–70. doi: 10.1016/j.virol.2017.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hurwitz SN, Cheerathodi MR, Nkosi D, York SB, Meckes DG. 2018. Tetraspanin CD63 bridges autophagic and endosomal processes to regulate exosomal secretion and intracellular signaling of Epstein-Barr virus LMP1. J Virol 92:e01969-17. doi: 10.1128/JVI.01969-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shen B, Wu N, Yang J-M, Gould SJ. 2011. Protein targeting to exosomes/microvesicles by plasma membrane anchors. J Biol Chem 286:14383–14395. doi: 10.1074/jbc.M110.208660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang J-M, Gould Stephen J. 2013. The cis-acting signals that target proteins to exosomes and microvesicles. Biochem Soc Trans 41:277. doi: 10.1042/BST20120275. [DOI] [PubMed] [Google Scholar]
- 60.Fang Y, Wu N, Gan X, Yan W, Morrell JC, Gould SJ. 2007. Higher-order oligomerization targets plasma membrane proteins and HIV Gag to exosomes. PLoS Biol 5:e158. doi: 10.1371/journal.pbio.0050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Verweij FJ, Heus CD, Kroeze S, Cai H, Kieff E, Piersma SR, Jimenez CR, Middeldorp JM, Pegtel DM. 2015. Exosomal sorting of the viral oncoprotein LMP1 is restrained by TRAF2 association at signalling endosomes. J Extracell Vesicles 4:26334. doi: 10.3402/jev.v4.26334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Coffin WF III, Geiger TR, Martin JM. 2003. Transmembrane domains 1 and 2 of the latent membrane protein 1 of Epstein-Barr virus contain a lipid raft targeting signal and play a critical role in cytostasis. J Virol 77:3749–3758. doi: 10.1128/JVI.77.6.3749-3758.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Coffin WF, Erickson KD, Hoedt-Miller M, Martin JM. 2001. The cytoplasmic amino-terminus of the Latent Membrane Protein-1 of Epstein-Barr virus: relationship between transmembrane orientation and effector functions of the carboxy-terminus and transmembrane domain. Oncogene 20:5313–5330. doi: 10.1038/sj.onc.1204689. [DOI] [PubMed] [Google Scholar]
- 64.Izumi KM, Kaye KM, Kieff ED. 1994. Epstein-Barr virus recombinant molecular genetic analysis of the LMP1 amino-terminal cytoplasmic domain reveals a probable structural role, with no component essential for primary B-lymphocyte growth transformation. J Virol 68:4369–4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yasui T, Luftig M, Soni V, Kieff E. 2004. Latent infection membrane protein transmembrane FWLY is critical for intermolecular interaction, raft localization, and signaling. Proc Natl Acad Sci U S A 101:278–283. doi: 10.1073/pnas.2237224100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaykas A, Worringer K, Sugden B. 2002. LMP-1's transmembrane domains encode multiple functions required for LMP-1's efficient signaling. J Virol 76:11551–11560. doi: 10.1128/JVI.76.22.11551-11560.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaykas A, Worringer K, Sugden B. 2001. CD40 and LMP-1 both signal from lipid rafts but LMP-1 assembles a distinct, more efficient signaling complex. EMBO J 20:2641–2654. doi: 10.1093/emboj/20.11.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lingwood D, Simons K. 2010. Lipid rafts as a membrane-organizing principle. Science 327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 69.Behnke J, Eskelinen E-L, Saftig P, Schröder B. 2011. Two dileucine motifs mediate late endosomal/lysosomal targeting of transmembrane protein 192 (TMEM192) and a C-terminal cysteine residue is responsible for disulfide bond formation in TMEM192 homodimers. Biochem J 434:219. doi: 10.1042/BJ20101396. [DOI] [PubMed] [Google Scholar]
- 70.Liu H-P, Wu C-C, Chang Y-S. 2006. PRA1 promotes the intracellular trafficking and NF-κB signaling of EBV latent membrane protein 1. EMBO J 25:4120–4130. doi: 10.1038/sj.emboj.7601282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liebowitz D, Wang D, Kieff E. 1986. Orientation and patching of the latent infection membrane protein encoded by Epstein-Barr virus. J Virol 58:233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liebowitz D, Mannick J, Takada K, Kieff E. 1992. Phenotypes of Epstein-Barr virus LMP1 deletion mutants indicate transmembrane and amino-terminal cytoplasmic domains necessary for effects in B-lymphoma cells. J Virol 66:4612–4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Verweij FJ, Middeldorp JM, Pegtel DM. 2012. Intracellular signaling controlled by the endosomal-exosomal pathway. Commun Integr Biol 5:88–93. doi: 10.4161/cib.18452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dukers DF, Meij P, Vervoort MB, Vos W, Scheper RJ, Meijer CJ, Bloemena E, Middeldorp JM. 2000. Direct immunosuppressive effects of EBV-encoded latent membrane protein 1. J Immunol 165:663–670. doi: 10.4049/jimmunol.165.2.663. [DOI] [PubMed] [Google Scholar]
- 75.Campeau E, Ruhl VE, Rodier F, Smith CL, Rahmberg BL, Fuss JO, Campisi J, Yaswen P, Cooper PK, Kaufman PD. 2009. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One 4:e6529. doi: 10.1371/journal.pone.0006529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zurek N, Sparks L, Voeltz G. 2011. Reticulon short hairpin transmembrane domains are used to shape ER tubules. Traffic 12:28–41. doi: 10.1111/j.1600-0854.2010.01134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sherer NM, Lehmann MJ, Jimenez-Soto LF, Ingmundson A, Horner SM, Cicchetti G, Allen PG, Pypaert M, Cunningham JM, Mothes W. 2003. Visualization of retroviral replication in living cells reveals budding into multivesicular bodies. Traffic 4:785–801. doi: 10.1034/j.1600-0854.2003.00135.x. [DOI] [PubMed] [Google Scholar]
- 78.Choudhury A, Dominguez M, Puri V, Sharma DK, Narita K, Wheatley CL, Marks DL, Pagano RE. 2002. Rab proteins mediate Golgi transport of caveola-internalized glycosphingolipids and correct lipid trafficking in Niemann-Pick C cells. J Clin Invest 109:1541–1550. doi: 10.1172/JCI0215420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sharma DK, Choudhury A, Singh RD, Wheatley CL, Marks DL, Pagano RE. 2003. Glycosphingolipids internalized via caveolar-related endocytosis rapidly merge with the clathrin pathway in early endosomes and form microdomains for recycling. J Biol Chem 278:7564–7572. doi: 10.1074/jbc.M210457200. [DOI] [PubMed] [Google Scholar]
- 80.Hurwitz SN, Conlon MM, Rider MA, Brownstein NC, Meckes DG. 2016. Nanoparticle analysis sheds budding insights into genetic drivers of extracellular vesicle biogenesis. J Extracell Vesicles 5:31295. doi: 10.3402/jev.v5.31295. [DOI] [PMC free article] [PubMed] [Google Scholar]