Tembusu virus, similar to other mosquito-borne flaviviruses such as WNV, JEV, and BAGV, can be transmitted without the presence of mosquito vectors. We demonstrate that the envelope protein of TMUV and its amino acid (S) at position 156 is responsible for tissue tropism and transmission in ducks. The mutation S156P results in disruption of N-linked glycosylation at amino acid 154 of the E protein and changes the conformation of “150 loop” of the E protein, which induces limited virus replication in lungs and abrogates transmission between ducks. Our findings provide new knowledge about TMUV transmission among ducks.
KEYWORDS: Tembusu virus, flavivirus, envelope protein, mutation, replication, transmission
ABSTRACT
Duck Tembusu virus (TMUV), like other mosquito-borne flaviviruses, such as Japanese encephalitis virus, West Nile virus, and Bagaza virus, is able to transmit vector-independently. To date, why these flaviviruses can be transmitted without mosquito vectors remains poorly understood. To explore the key molecular basis of flavivirus transmissibility, we compared virus replication and transmissibility of an early and a recent TMUV in ducks. The recent TMUV strain FX2010 replicated systemically and transmitted efficiently in ducks, while the replication of early strain MM1775 was limited and did not transmit among ducks. The TMUV envelope protein and its domain I were responsible for tissue tropism and transmissibility. The mutation S156P in the domain I resulted in disruption of N-linked glycosylation at amino acid 154 of the E protein and changed the conformation of “150 loop” of the E protein, which reduced virus replication in lungs and abrogated transmission in ducks. These data indicate that the 156S in the envelope protein is critical for TMUV tissue tropism and transmissibility in ducks in the absence of mosquitos. Our findings provide novel insights on understanding TMUV transmission among ducks.
IMPORTANCE Tembusu virus, similar to other mosquito-borne flaviviruses such as WNV, JEV, and BAGV, can be transmitted without the presence of mosquito vectors. We demonstrate that the envelope protein of TMUV and its amino acid (S) at position 156 is responsible for tissue tropism and transmission in ducks. The mutation S156P results in disruption of N-linked glycosylation at amino acid 154 of the E protein and changes the conformation of “150 loop” of the E protein, which induces limited virus replication in lungs and abrogates transmission between ducks. Our findings provide new knowledge about TMUV transmission among ducks.
INTRODUCTION
Tembusu virus (TMUV) is a member of the Ntaya virus group within the genus Flavivirus, family Flaviviridae (1). TMUV was first isolated in mosquitos in Malaysia in 1955 (2), and since then several mosquito isolates have been reported in Malaysia and Thailand (3, 4). In 2000, an infectious disease caused by TMUV emerged in a broiler farm in Sitiawan district of Perak state, Malaysia (5). This disease was characterized by encephalitis and retarded growth in broiler chicks (5). Ten years later, TMUV caused outbreaks in ducks characterized by a severe drop in egg production and growth retardation in almost all duck farms in China in 2010 (6). TMUV continues to result in annual losses of millions of dollars in China and has spread to duck farms in Southeast Asia (7–10).
As a flavivirus, TMUV was thought to be transmitted by mosquitos at the beginning of duck outbreaks (2, 6). Although arthropod-borne transmission of flaviviruses is still the major route of transmission (11–13), nonvector transmission has occurred between birds and between pigs (14–18). TMUV causes outbreaks in birds in winter, when mosquitos are inactive, suggesting that nonvector transmission routes play a key role in TMUV spread (6). In vivo studies indicate that TMUV can be transmitted efficiently among ducks by both direct contact and aerosol transmission (19). However, limited knowledge is available about the molecular basis determining the transmission of flaviviruses without the presence of vectors.
TMUV contains a positive-sense RNA genome with ∼10,991 nucleotides that encodes a single polyprotein that is processed by viral and host proteases to generate three structural (capsid, C; pre-membrane, prM; and envelope, E) and seven nonstructural (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) proteins. The structural proteins of flavivirus are involved in virion formation, attachment, and entry into host cells (1, 20), whereas the nonstructural proteins participate in genome replication, virion assembly, and evasion of host antiviral responses (21–23). Comparative genomic and phylogenetic analyses showed multiple nucleotide substitutions in different genes of TMUV that may contribute to the outbreaks in ducks (24, 25). Variation analyses of amino acid loci in the TMUV E protein revealed two mutated amino acid loci in strains isolated from Malaysia, Thailand, and mainland China compared to the prototypical strain of the virus (MM1775) isolated from mosquitos (26). Furthermore, TMUV isolates from the Chinese mainland have six common variations in the E protein that differ from the Southeast Asian strains (26).
Like other flaviviruses, the TMUV E protein contains three different structural domains (β-barrel shaped domain I [DI], finger-like domain II [DII], and Ig-like domain III [DIII]) and a transmembrane helix domain (27–30). The central DI acts as a bridge between DII and DIII, is folded into an eight-stranded β-barrel, and contains about 130 residues in segments: residues 1 to 50, 133 to 197, and 279 to 300. DII is formed by two segments, comprising residues 51 to 132 and 197 to 278. The tip of DII contains the fusion loop, which interacts with the host membrane during membrane fusion (25, 31). In most flaviviruses, DIII contains the putative receptor-binding site and has an important role in virus binding and cell entry (27, 32).
In the present study, we showed that the MM1775, an early TMUV strain isolated from mosquitos, which was recovered from cDNA by using reverse genetics system (33), was able to replicate efficiently in the spleens of ducks inoculated intramuscularly (i.m.) or intranasally (i.n.) but was not transmitted among ducks. In contrast, the recent TMUV duck isolates were able to transmit efficiently among ducks by both direct contact and by aerosol transmission (19). Here, we investigated the underlying molecular mechanisms of TMUV efficient transmission without the presence of mosquito vectors. Our results demonstrate that the envelope protein and its amino acid at position 156 are critical for TMUV transmission in ducks.
RESULTS
Amino acid sequence comparison between TMUVs MM1775 and FX2010.
The genome sequences of TMUV strain MM1775 (GenBank accession no. MH414569), isolated from mosquitos in Malaysia in 1955, and TMUV strain FX2010 (GenBank accession no. MH414568), isolated from sick ducks in China in 2010, were analyzed using DNASTAR software. A total of 101 amino acid differences among all 10 proteins were detected between 2 viruses depicted in Table 1. There were 25 amino acid differences located at three structural proteins and 76 other differences distributed among 7 nonstructural proteins (Table 1). There were 4 and 5 amino acid differences in the structural PrM and C proteins, respectively, whereas 16 amino acids were different in the E protein. These included 9 amino acids in DI, 3 amino acids in DII, 2 amino acids in DIII, and 2 amino acids in the transmembrane helix domain which acts as a stem-anchor and is important to the surface architecture of the tick-borne flaviviruses (34). Eighteen amino acids were different in the NS5 protein and 21 amino acid differences were found in the NS1 protein (Table 1). Both NS1 and NS5 are important for flavivirus replication (22, 23, 35).
TABLE 1.
Amino acid sequence comparison of TMUVs MM1775 and FX2010
| Protein | Positionsa |
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amino acids (MM1775/FX2010) | |||||||||||||||||||||
| C | 42 | 61 | 76 | 109 | 111 | ||||||||||||||||
| T/A | K/R | S/N | P/S | V/I | |||||||||||||||||
| PrM | 22 | 24 | 29 | 150 | |||||||||||||||||
| M/L | I/V | T/A | G/S | ||||||||||||||||||
| E | 2 | 38 | 72 | 89 | 148 | 149 | 156 | 157 | 180 | 181 | 185 | 236 | 312 | 332 | 467 | 483 | |||||
| N/S | R/K | P/S | D/E | A/E | V/A | P/S | V/A | L/M | G/E | A/T | R/K | A/V | S/T | V/A | I/M | ||||||
| NS1 | 2 | 9 | 21 | 41 | 57 | 69 | 83 | 99 | 105 | 108 | 112 | 113 | 121 | 139 | 147 | 173 | 174 | 205 | 274 | 339 | 351 |
| M/T | T/A | I/V | K/R | V/I | A/V | S/A | R/K | S/P | E/D | T/M | F/Y | N/S | R/K | K/R | S/T | G/E | H/K | K/V | V/A | V/M | |
| NS2A | 4 | 13 | 15 | 23 | 31 | 35 | 36 | 49 | 63 | 79 | 98 | 147 | 150 | 157 | 165 | 187 | |||||
| D/G | I/M | I/V | V/I | S/P | P/S | S/A | V/I | V/A | I/M | R/K | A/V | L/F | S/L | I/V | N/S | ||||||
| NS2B | 86 | 95 | 96 | 114 | |||||||||||||||||
| K/R | F/L | S/G | L/F | ||||||||||||||||||
| NS3 | 15 | 107 | 210 | 258 | 324 | 358 | 591 | 610 | |||||||||||||
| K/R | M/I | R/K | V/I | D/E | R/K | T/I | A/T | ||||||||||||||
| NS4A | 5 | 89 | 96 | 110 | |||||||||||||||||
| V/I | A/T | V/I | I/V | ||||||||||||||||||
| NS4B | 38 | 46 | 53 | 83 | 203 | ||||||||||||||||
| T/A | N/S | V/M | V/I | T/I | |||||||||||||||||
| NS5 | 4 | 76 | 150 | 188 | 197 | 233 | 246 | 258 | 277 | 288 | 385 | 422 | 435 | 506 | 562 | 729 | 828 | 841 | |||
| A/T | R/K | S/A | T/M | K/R | V/I | R/K | A/V | E/G | K/R | D/N | T/A | N/S | R/K | L/M | V/I | Y/H | I/V | ||||
For each protein, the position numbers are indicated in the first row, and the amino acids are specified in the second row. MM1775 is a TMUV strain isolated from mosquitos in Malaysia in 1955; FX2010 is a TMUV strain isolated from sick ducks in China in 2010.
TMUV stain FX2010 causes systemic infection and efficient transmission in ducks compared to an early strain MM1775.
To explore why TMUV causes outbreaks in ducks, we compared virus replication and transmission of two related strains, FX2010 and MM1775. At 3 day postinoculation (dpi), all ducks inoculated with 103.5 50% tissue culture infective dose(s) (TCID50) of FX2010 through either the i.m. or the i.n. route or inoculated with 103.5 TCID50 of MM1775 through the i.m. route showed swollen spleens and severe ovarian follicle distortion, while only two of three ducks inoculated with the same dose of MM1775 through the i.n. route had these clinical symptoms. FX2010 was detected in all tested tissues (including the liver, spleen, lung, kidney, brain, ovary, pancreas, and trachea) of all infected ducks through i.m. inoculation and in most tissues except for the brain through i.n. inoculation (Table 2). In contrast, MM1775 was only detected in the livers and spleens of most of ducks that were inoculated either i.m. or i.n.; in addition, virus was also detected in the ovaries of two of three birds inoculated through the i.m. route (Table 2). Although the range of tropism tissues were different, the viral titer of two viruses reached to a comparable level in spleens in inoculated ducks, suggesting their replication efficiency in this tissue was similar.
TABLE 2.
Virus titers in different samples of ducks inoculated with TMUV FX2010 or MM1775
| Virus | Inoculation route | Test time (dpia) | Virus titer (log10 TCID50/0.1 g)b |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Liver | Spleen | Lung | Kidney | Brain | Ovary | Pancreas | Trachea | Serum | |||
| FX2010 | i.m. | 3 | 1.17 ± 0.78 | 3.39 ± 0.34 | 2.72 ± 0.25 | 2.33 ± 0.58 | 1.00 ± 0.33 | 3.95 ± 0.47 | 2.32 ± 0.40 | 2.57 ± 0.35 | 2.51 ± 0.10 |
| i.n. | 3 | –/–/1.00 | 3.78 ± 0.63 | 1.72 ± 0.86 | 0.72 ± 0.54 | –/–/– | 0.50/2.33/– | 0.50 ± 0.3 | 1.65 ± 0.42 | 2.10 ± 0.50 | |
| MM1775 | i.m. | 3 | 1.56 ± 0.10 | 3.94 ± 0.42 | –/–/– | –/–/– | –/–/– | 1.67/2.00/– | –/–/– | –/–/– | –/–/– |
| i.n. | 3 | 1.67/2.33/– | 2.50/3.50/– | –/–/– | –/–/– | –/–/– | –/–/– | –/–/– | –/–/– | –/–/– | |
dpi, days postinoculation.
Where virus was detected in all three ducks sampled, the mean TCID50 ± the standard deviation is shown; otherwise, individual titers are shown for each animal. –, No virus was detected.
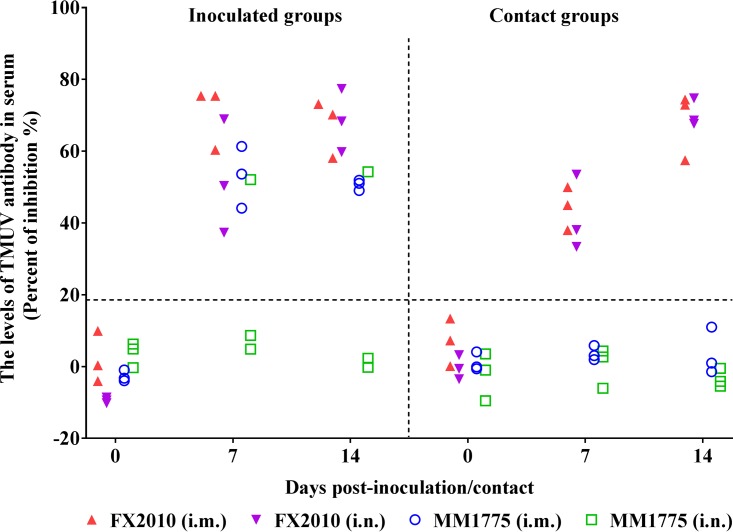
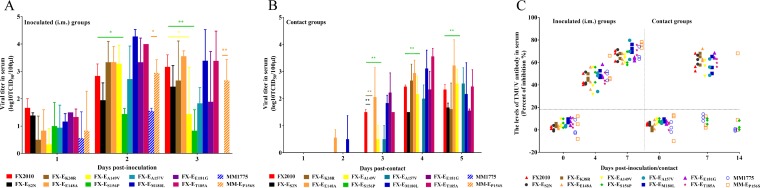
The serum samples of the remaining inoculated and contact ducks were tested using ELISAs. All ducks inoculated with FX2010 via either route, as well as all three birds inoculated with MM1775 through the i.m. route were seropositive at both 7 and 14 dpi, while only one of three ducks inoculated with MM1775 through the i.n. route seroconverted (Fig. 1). Seroconversion was detected in all contact ducks in groups of inoculated FX2010 at both 7 and 14 days postcontact (dpc). In contrast, no seroconversion was detected in contact ducks in the MM1775 groups at 14 dpc (Fig. 1). These results indicate that the FX2010 virus, but not the MM1775 virus, transmits efficiently between ducks in the absence of mosquitos.
FIG 1.
Antibodies against TMUV in FX2010- or MM1775-inoculated and contact ducks. Ducks were inoculated i.m. and i.n. with 103.5 TCID50 of FX2010 or MM1775. One day later, naive ducks were introduced into the isolators. Data show the relative levels of TMUV-specific antibodies detected in serum samples by an ELISA. Serum was considered positive conversion when the PI value was ≥18.4%.
E protein dictates TMUV tissue tropism and transmissibility in ducks.
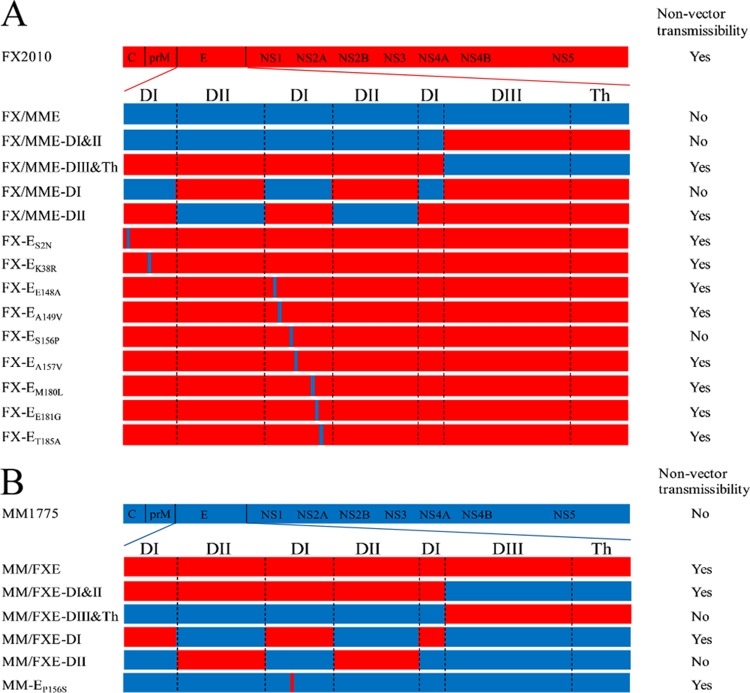
To identify which protein(s) was/were critical for tissue tropism and transmissibility of TMUVs, the E gene was exchanged between FX2010 and MM1775 viruses. Two chimeric viruses, FX/MME containing the MM1775 E gene in the FX2010 background and MM/FXE containing the FX2010 E gene in the MM1775 background, were rescued by PCR-based reverse genetic method (Fig. 2). Their replication and transmissibility were compared to their respective parental viruses. The growth curves showed that E gene substitution viruses had the similar replication capacities on DF-1 cells infected with each virus at a multiplicity of infection (MOI) of 0.0001 compared to their respective parental viruses (Fig. 3A). The chimeric virus FX/MME was detected in tested spleens, kidneys, and ovaries, but not in the lungs of inoculated ducks at 3 dpi (Table 3). Furthermore, FX/MME was not isolated from tested tissues of contact birds at 6 dpc compared to its parental FX2010 virus (Table 3). On the contrary, the MM/FXE with FX2010 E gene obtained the ability to replicate in the lungs, ovaries, and kidneys of inoculated ducks in contrast to its parental MM1775 virus. Noticeably, MM/FXE was detected in multiple organs, including the spleens, lungs, and ovaries of contact ducks, compared to the parental virus (Table 3).
FIG 2.
Generation of chimeric or mutated TMUVs for duck transmissibility studies. The colored bars indicate the origin of the viral protein (red, FX2010; blue, MM1775). The viruses were rescued in a background of FX2010 (A) or MM1775 (B). Whether virus transmission occurred was determined based on the seroconversion of specific TMUV antibody in contact ducks that were cohoused with originally inoculated ducks at 14 days postcontact. The transmissibility of each virus is specified on the right of the figure.
FIG 3.
Growth curves of parental and E gene substitution TMUVs on DF-1 cells and antibodies detected in contact and inoculated ducks with these viruses. (A) Replication of parental and E gene substitution TMUVs. DF-1 cells were infected by the parental and E gene substitution TMUVs at an MOI of 0.0001. Virus samples from the supernatant were collected at different time points and titrated on DF-1 cells. The data for virus titers indicate the means of the results of three repeats, and the error bars indicate standard errors of the means (*, P < 0.05; **, P < 0.01). (B) Relative levels of TMUV-specific antibodies detected in serum samples as a function of time after inoculation with the indicated virus. Serum was considered positive when the PI value was ≥18.4%.
TABLE 3.
Substitution of the E gene changes the replication capacity of Tembusu viruses in ducks
| Virus | Infection route | Test timea | Virus titer (log10 TCID50/0.1 g)b |
|||
|---|---|---|---|---|---|---|
| Spleen | Lung | Kidney | Ovary | |||
| FX2010 | i.m. | 3 dpi | 2.89 ± 0.78c | 1.33 ± 0.87 | 2.17 ± 1.25 | 3.27 ± 0.27 |
| FX/MME | i.m. | 3 dpi | 4.28 ± 0.53 | –/–/– | –/0.67/2.00 | 2.78 ± 0.19 |
| MM1775 | i.m. | 3 dpi | 3.72 ± 0.75 | –/–/– | –/–/– | –/–/0.67 |
| MM/FXE | i.m. | 3 dpi | 3.17 ± 0.76 | 2.67/2.33/– | –/2.33/– | 2.50 ± 0.33 |
| FX2010 | Contact | 6 dpc | 2.72 ± 0.25 | 2.33 ± 0.87 | 2.00 ± 0.44 | 3.00 ± 0.19 |
| FX/MME | Contact | 6 dpc | –/–/– | –/–/– | –/–/– | –/–/– |
| MM1775 | Contact | 6 dpc | –/–/– | –/–/– | –/–/– | –/–/– |
| MM/FXE | Contact | 6 dpc | 4.83 ± 0.44 | 2.17 ± 0.58 | –/0.50/– | 2.57 ± 1.39 |
dpi, days postinoculation; dpc, days postcontact.
Where virus was detected in all three ducks sampled, the mean TCID50 ± the standard deviation is shown; otherwise, individual titers are shown for each animal. –, No virus was detected.
To further confirm virus replication and transmissibility of chimeric viruses, we performed an enzyme-linked immunosorbent assay (ELISA) to determine seroconversion of originally inoculated and contact ducks. All inoculated ducks, except for two birds inoculated with FX/MME, seroconverted at 4 dpi; however, all inoculated birds, including two seronegative ducks at 4 dpi, were seropositive to TMUV at 7 dpi (Fig. 3B). Seroconversion was only found in contact ducks of either FX2010 or MM/FXE groups at the tested dates; none of the contact birds in either MM1775 or FX/MME groups seroconverted, even at 14 dpc (Fig. 3B). These results indicate that the E gene plays a key role in TMUV replication and transmissibility.
Domains I and II of the E protein are crucial for the replication and transmissibility of TMUVs in ducks.
To determine which part of TMUV E protein was critical for efficient replication and transmissibility of TMUVs in ducks, the E protein was divided into two parts: domains I and II (DI&II) and domain III and transmembrane helix (DIII&Th) based on its proposed structure (Fig. 2). Chimeric FX/MME-DI&II, FX/MME-DIII&Th, MM/FXE-DI&II, and MM/FXE-DIII&Th were generated as depicted in Fig. 2 in the background of either FX2010 or MM1775, respectively, and characterized in ducks. All chimeric viruses, as well as their parental viruses, were able to replicate in the spleens of inoculated ducks, but only the viruses containing the E-DI&II of FX2010 were detected in the lungs and kidneys of inoculated ducks at 3 dpi (Table 4). In contrast, the viruses containing the E-DI&II of MM1775 were not detected in other tissues of inoculated birds at 3 dpi (Table 4). Substitutions of E-DIII&Th did not significantly impact virus replication and tissue tropisms of TMUVs compared to the parental viruses (Table 4).
TABLE 4.
Domain I of the E gene determines the replication capacity of Tembusu virus in the i.m.-inoculated ducks
| Virus | Test time (dpia) | Virus titer (log10 TCID50/0.1 g)b |
|||
|---|---|---|---|---|---|
| Spleen | Lung | Kidney | Ovary | ||
| FX2010 | 3 | 4.83 ± 0.44 | 2.89 ± 0.19 | 2.50 ± 0.50 | 3.23 ± 0.39 |
| FX/MME-DI&II | 3 | 3.67 ± 0.67 | –/–/– | –/–/– | –/–/– |
| FX/MME-DIII&Th | 3 | 3.45 ± 0.39 | 1.39 ± 0.10 | –/1.00/– | 3.05 ± 0.48 |
| FX/MME-DI | 3 | 4.39 ± 0.92 | –/–/– | –/0.67/– | –/–/– |
| FX/MME-DII | 3 | 4.22 ± 0.63 | –/2.00/2.67 | –/–/0.67 | 2.67 ± 1.53 |
| MM1775 | 3 | 2.72 ± 0.86 | –/–/– | –/–/– | –/–/– |
| MM/FXE-DI&II | 3 | 3.56 ± 0.84 | 2.33/–/– | –/0.67/0.67 | –/–/– |
| MM/FXE-DIII&Th | 3 | 3.89 ± 0.54 | –/–/– | –/–/– | –/–/– |
| MM/FXE-DI | 3 | 4.39 ± 0.35 | 2.33/–/– | –/–/– | –/–/– |
| MM/FXE-DII | 3 | 2.72 ± 0.75 | –/–/– | –/–/– | –/–/– |
dpi, days postinoculation.
Where virus was detected in all three ducks sampled, the mean TCID50 ± the standard deviation is shown; otherwise, individual titers are shown for each animal. –, No virus was detected.
ELISA results revealed that seroconversion was found in all inoculated ducks with each parental and chimeric virus at 7 dpi (Fig. 4). Seroconversion was only detected in contact ducks in the viruses that contain the E-DI&II of FX2010 groups. Moreover, no seroconversion was detected in contact ducks in the viruses that contain the E-DI&II of MM1775 groups (Fig. 4). The viruses substituted with E-DIII&Th showed the same transmissibility as their parental viruses, consistent with virus replication patterns (Fig. 4 and Table 4). These data indicate that DI&II, and not DIII&Th, of the E protein are crucial for replication and transmissibility of TMUVs in ducks.
FIG 4.
Antibodies detected in contact and inoculated ducks with parental and E domain substitution TMUVs. The data show the relative levels of TMUV-specific antibodies detected in serum samples as a function of time after inoculation with the indicated virus. Serum was considered positive when the PI value was ≥18.4%.
Domain I of E protein is responsible for the replication and transmissibility of TMUVs in ducks.
To identify which domain of E protein determined replication and transmissibility of TMUVs in ducks, the chimeric viruses FX/MME-DI and FX/MME-DII or MM/FXE-DI and MM/FXE-DII, which contain heterogeneous DI or DII in the E protein of FX2010 or MM1775 were rescued as depicted in Fig. 2 and further characterized in ducks. All chimeric viruses substituted with a single domain and parental viruses were able to replicate in the spleens of inoculated ducks (Table 4). Only the chimeric viruses containing the E-DI of FX2010 were detected in the lungs of some inoculated ducks at 3 dpi, while the chimeric viruses containing the E-DI of MM1775 were not detected in the lungs of inoculated ducks at the same date. The FX/MME-DII retained systemic replication in ducks, which is similar to FX2010, while the replication of MM/FXE-DII was still limited in spleens, similar to MM1775 (Table 4).
All ducks inoculated with the chimeric viruses with single E-DI or E-DII showed seropositive at 4 dpi (Fig. 4). One of three contact ducks seroconverted in the MM/FXE-DI infection group, indicating that the parental MM1775 obtained transmissibility in ducks after having the E-DI of FX2010, while the FX2010 lost transmissibility after substitution of the E-DI of MM1775, as none of contact birds seroconverted at 14 dpc (Fig. 4). In addition, substitution of the single E-DII did not impact transmissibility of both FX2010 and MM1775 (Fig. 4). These data indicate that the DI of the E protein is responsible for tissue tropisms and efficient transmissibility of TMUVs in ducks.
The ES156P mutation abolishes the transmissibility of TMUV in ducks.
To determine which amino acids contribute to efficient replication and transmission of TMUV in ducks, nine mutant viruses (FX-ES2N, FX-EK38R, FX-EE148A, FX-EA149V, FX-ES156P, FX-EA157V, FX-EM180L, FX-EE181G, and FX-ET185A) with a single amino acid change located at DI of the E protein (9 different amino acids in DI of the E protein between FX2010 and MM1775) in the background of FX2010 were generated (Fig. 2) and further characterized in ducks. Viremia was detectible in all ducks inoculated i.m. with each recombinant virus with single mutated amino acid from 1 to 3 dpi. Two mutated viruses (FX-EA149V and FX-ES156P) showed significantly lower titers in serum samples of infected ducks than the parental FX2010 at different tested time points (Fig. 5A). Virus titers in the sera of FX-ES156P-inoculated ducks were significantly lower than FX2010 at both 2 (P < 0.05) and 3 dpi (P < 0.01) (Fig. 5A). Viremia was also detected in contact ducks of each group, except for the contact birds in the group of FX-ES156P (Fig. 5B). Virus was only detected in the serum samples of contact birds of the FX-EE148A and FX-EM180L groups as early 2 dpc, whereas contact ducks in the FX-ES2N and FX-EK38R groups showed a delayed viremia starting at 4 dpc in contrast contact birds in other groups. No virus was detected in the sera of any contact ducks in the FX-ES156P group at all tested time points, indicating that ES156P mutation is critical for virus replication and transmission.
FIG 5.
Virus titers and antibody levels in the sera of contact and inoculated ducks with parental and single amino acid mutant TMUVs. (A) Virus titers in sera of infected ducks were determined on DF-1 cells at 1, 2, and 3 dpi. (B) Virus titers in sera of contact ducks were determined on DF-1 cells at 1, 2, 3, 4, and 5 dpc. (C) Serum antibody responses against TMUVs were detected at 0, 4, and 7 dpi for inoculated ducks and at 0, 7, and 14 dpc for contact ducks. Serum was considered positive when the PI value was ≥18.4%. The data for the virus titers (panels A and B) indicate the means of the results of three ducks, and the error bars indicate standard errors of the means (*, P < 0.05; **, P < 0.01).
The ducks inoculated with either the parental FX2010 or its single amino acid mutants showed seroconversion at 4 dpi (Fig. 5C). All single amino acid mutants except for FX-ES156P were transmitted to contact ducks, resulting in seroconversion at 7 dpc. The results further demonstrate that ES156P mutation abolishes the transmissibility of TMUV FX2010 in ducks.
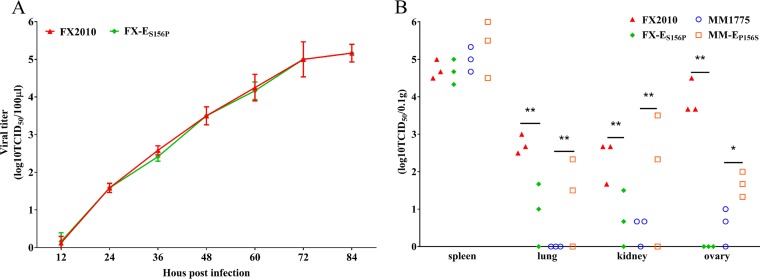
The ES156P mutation restricts tissue tropism of TMUV in ducks.
We further compared virus replication of FX-ES156P with the parental FX2010 in vitro and in ducks. FX-ES156P had the similar replication capacities as FX2010 on DF-1 cells infected with each virus at an MOI of 0.0001 (Fig. 6A). The ducks were inoculated i.m. with 103.5 TCID50 viruses per bird, and at 3 dpi, the virus replication in different tissues was tested. A similar virus titer was detected in spleens of ducks inoculated with either FX-ES156P or FX2010 (Fig. 6B). However, a significantly lower titer was found in both lungs and kidneys of ducks infected with FX-ES156P compared to that detected in FX2010-inoculated ducks. No virus was detected in ovary tissues of any ducks inoculated with FX-ES156P, while virus was found in ovary tissues of all FX2010-inoculated ducks with a high titer (Fig. 6B). These results indicate that the ES156P mutation also restricts the tissue tropism of TMUV in ducks.
FIG 6.
Replication comparison of TMUVs and their E156 mutants. (A) Replication of FX2010 or FX-ES156P on DF-1 cells infected at an MOI of 0.0001. Virus samples from the supernatant were collected at different time points and titrated on DF-1 cells. (B) To compare the replication of TMUVs and E156 mutants, ducks were inoculated i.m. with 103.5 TCID50 of FX2010, FX-ES156P, MM1775, and MM-EP156S, respectively. Three ducks of each group were euthanized at 3 dpi, and the spleens, lungs, kidneys, and ovaries were collected for virus titration. Virus titers were determined on DF-1 cells. The data for virus titers indicate the means of the results of three repeats, and the error bars indicate standard errors of the means (*, P < 0.05; **, P < 0.01).
The EP156S mutation recovers the transmissibility of TMUV in ducks.
To test further the influence of EP156S mutation on tissue tropism and transmissibility of TMUV in ducks, MM-EP156S with a P156S mutation at position 156 of the E protein in the background of MM1775 was rescued and characterized in ducks. Each of six ducks were inoculated i.m. with 103.5 TCID50/bird of MM-EP156S and its parental virus MM1775. The virus replication in different tissues of three inoculated ducks were tested at 3 dpi. A comparable virus titer was detected in the spleens of ducks inoculated with either MM-EP156S or MM1775 (Fig. 6B). However, the virus titers in the kidneys and ovaries of MM-EP156S-inoculated ducks were significantly higher than in those of MM1775-inoculated ducks (Fig. 6B). MM-EP156S was detectable in the lungs of some inoculated ducks, while MM1775 was not detectable in any lungs of inoculated ducks (Fig. 6B). Viremia was detectable in some ducks inoculated both with MM-EP156S and MM1775 at 1 dpi (Fig. 5A). Virus titers in the sera of MM-EP156S-inoculated ducks were higher than those of MM1775-inoculated ducks at 2 dpi (P < 0.05) (Fig. 5A). The virus titers in sera of MM-EP156S-inoculated ducks remained high at 3 dpi, while no virus was detected in the sera of MM1775-inoculated ducks at that time point (Fig. 5A). These results indicate that the EP156S mutation enhances TMUV replication in different organs in ducks. Viremia was not detected in contact ducks in either the MM1775 group or the MM-EP156S group from 1 to 5 dpc (Fig. 5B).
All ducks inoculated i.m. with the MM-EP156S or parental MM1775 were seropositive at 4 and 7 dpi (Fig. 5C). One of three contact ducks was seropositive at 14 dpc in the MM-EP156S group, while all contact ducks in MM1775 group were seronegative at 14 dpc (Fig. 5C). These results indicate that EP156S mutation of TMUV enhances transmissibility in ducks.
ES156P mutation demolishes the N-linked glycosylation in “150 loop” of E protein.
Sequence analysis showed that the FX2010 and its single amino acid mutants, except for FX-ES156P, encoded only one potential N-linked glycosylation site in the “150 loop” of E protein; this glycosylation site could be amino acid ASN at position 154 (154N/155Y/156S) based on the consensus sequence (N-X-S/T) of N-glycosylation (11, 12). To determine whether the specific site is glycosylated, the molecular weight of E protein of the parental FX2010 and its single amino acid mutants was analyzed by Western blotting using frozen viral stocks. The E protein of FX-ES156P migrated faster than those of the parental FX2010 and its other single-site mutants, indicating that its molecular weight is smaller than others. This result suggests that the S156P mutation ablates the N154 glycosylation site in the E protein (Fig. 7A).
FIG 7.
Glycosylation status of FX2010 E protein mutants. (A) The apparent molecular weight of E protein mutants was analyzed by Western blotting against frozen viral stocks. (B) Viruses in supernatants and DF-1 cell lysates from cells infected with FX2010 and FX-ES156P were collected separately at 48 h postinfection and then treated, respectively, with glycosidases Endo Hf, PNGase F, or buffer only (−). Western blots were probed with a monoclonal antibody (1F5) that recognizes TMUV E proteins.
To further confirm the glycosylation at E154N/155Y/156S, DF-1 cells were infected with either FX2010 or FX-ES156P. Viruses in supernatants and cell lysates were collected and prepared separately, and the E protein was analyzed by Western blotting. The virus was digested with or without either endoglycosidase Hf (Endo Hf, removes only immature carbohydrate structures) or peptide N-glycosidase F (PNGase F, removes all N-linked carbohydrate structures). The E protein of FX2010 migrated more quickly in cell lysates following treatment with either Endo Hf or PNGase F, indicating that the E protein of FX2010 was predominantly in high mannose form in DF-1 cells (Fig. 7B). In contrast to the cell-associated forms, the E protein of FX2010 in the supernatants was Endo Hf resistant but not PNGase F resistant (Fig. 7B). This result is consistent with carbohydrate side chain modification as the envelope proteins were transported through the Golgi apparatus. However, the E protein of FX-ES156P in both cell lysates and supernatants was completely Endo Hf and PNGase F resistant, suggesting that S156P mutation abolished the single glycosylation on the E protein of FX2010.
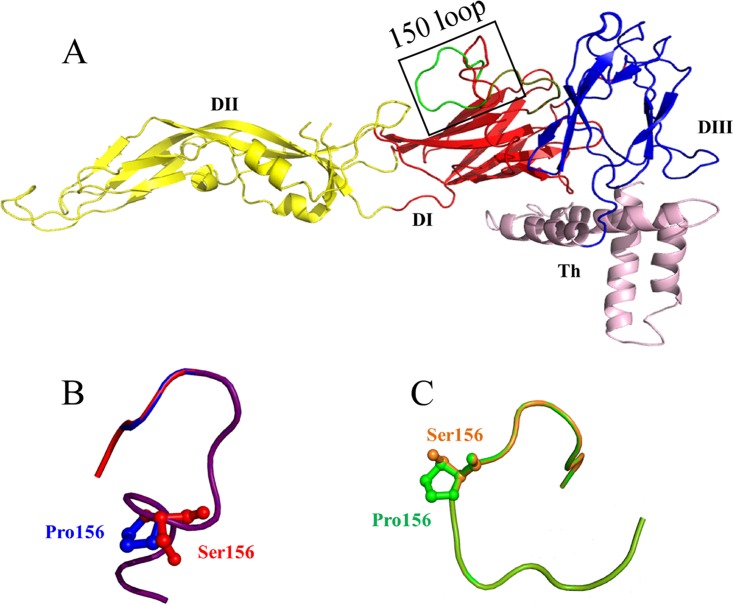
To locate the amino acid at position 156, the images of E proteins of FX2010 and MM1775 were visualized and analyzed by using PyMOL referring to the JEV E protein. The amino acid at position 156 was presented and exposed on the surface and located in the “150 loop” of E proteins (Fig. 8A). The amino acid mutations of S156P and P156S changed the loop conformations slightly on the predicted structures (Fig. 8B and C).
FIG 8.
Constructed homology models of E proteins of TMUVs. To localize amino acid at position156 of the E protein, the images of the E proteins of FX2010 and MM1775 were created with the program Phyre2 by using the JEV E protein structure (PDB accession number 5WSN). Homology models of the E protein of FX2010 (red) and MM1775 (green) (A), the “150 loop” of FX2010 (red) and FX-ES156P (blue) (B), and the “150 loop” of MM1775 (green) and MM-EP156S (orange) (C) are shown.
DISCUSSION
TMUVs, which belong to the Flaviviridae family and have caused significant impacts in the duck industry, have emerged in China since 2010, resulting in millions of dollars worth of losses annually (6, 8, 9, 19). Other flaviviruses such as dengue virus (DENV), West Nile virus (WNV), and Japanese encephalitis virus (JEV), as well as recently the emerged Zika virus (ZIKV), pose a significant threat to public health (36–40). The diseases caused by JEV and WNV not only occur in seasons depending on mosquito vectors but can also be spread through vector-free transmission (14, 16, 41). Bagaza virus can be transmitted by direct contact in experimental partridges (15). In our previous study, TMUV transmitted efficiently among ducks without mosquitos (19). To date, the molecular mechanisms on these pathogens' transmission independent of the mosquito vectors are poorly understood. To determine the genetic determinants that impact the transmissibility of TMUVs in ducks, we used two TMUV viruses: one is a prototypic MM1775 strain, first isolated from Culex mosquitos in 1955 (2), and the other is a recently emerged FX2010, isolated from sick ducks in China in 2010, to address this important question. Our results demonstrate that TMUV transmissibility in ducks is largely attributable to the E protein.
Flavivirus pathogenicity is associated with genetic changes in multiple genes and even noncoding regions (30, 34, 42–56). Amino acid changes in E proteins can result in attenuation of several flaviviruses due to the important role of the protein in host-specific adaption, cell tropism, virus attachment, and membrane fusion with target cells (28, 34, 45, 47, 48, 57–59). Like other flaviviruses, the TMUV E protein also has three different structural domains (28). In this study, we show that the DI of E protein is the key domain that crucial for vector-free transmissibility of TMUVs in ducks because the DI directly impacts virus replication in duck lungs, thereby influencing virus shedding and nonvector transmissibility. Finally, we determined that the amino acid Ser at position 156 in the E protein is responsible for virus tropism and transmission in ducks. The amino acid Ser at position 156 located in a “150 loop” region protrudes from the E protein surface. Similar to other flaviviruses, such as WNV and ZIKV (56, 60), the recent TMUV FX2010 has a single glycosylation site (N154) located in the “150 loop,” while the early TMUV MM1775 lacks this glycosylation due to the presence of amino acid proline at position 156. We further demonstrate that the S156P mutation in the E protein of the TMUV FX2010 results in the loss of N-linked glycosylation, leading to the abrogation of vector-free transmission of TMUV in ducks.
Glycosylation is important for the viral protein functions such as entry into host cells, proteolytic processing and protein trafficking (61). The glycosylation of E protein of TMUV FX2010 is Endo Hf resistant, suggesting that the E protein has been folded in the ER and processed through the Golgi network, before being released into the supernatant (24). In most flaviviruses, their E proteins contain one or more potential N-linked glycosylation motifs, but some flaviviruses lack this N-linked glycosylation site (60, 62–67). Among them, a highly conserved glycosylation motif is located at position 153 or 154 of the E protein (11, 12, 60, 68–70). The N153 or N154 glycosylation site is located at the “150 loop” of DI of the E protein, which could be an attachment region and influence virus transmission (25). The isolation of TMUV MM1775 from mosquito, which lacks this N-linked glycosylation site, suggests that N-linked glycosylation of the E protein is not necessary for early TMUV replication in mosquitos. Similar results have been found in DENV2 (57). However, ablation of the N-linked glycosylation site has been reported to reduce infectivity of DENV in mosquito cells (69). In addition, glycosylation of WNV E proteins can affect the efficiency of virus release and infection in a cell type-specific manner (56); further studies indicate that it is required for virus infection, but not spread within mosquito vectors (12). Recent studies have reported that the single N-linked glycosylation at N154 of ZIKV E protein influences E secretion from mammalian cells (60). Similar results have been also found in both WNV and tick-borne encephalitis virus (TBEV) (59, 66). Moreover, this glycosylation is critical for ZIKV infection of mammalian and mosquito hosts (71, 72) and antagonizing to the vector immune defense (73). In the present study, we demonstrate that nonglycosylation on the E protein of TMUVs results in limited virus replication (not systemic infection in ducks) and abrogation of virus transmission.
Analyses based on available E protein sequences show that N-linked glycosylation at N153 or N154 of E protein is conserved and predominant in most flaviviruses such as ZIKV, JEV, WNV, Murray Valley encephalitis virus, St. Louis encephalitis virus, TBEV, and DENV. This suggests that this glycosylation site is important for the survival of flaviviruses during evolution. Most flaviviruses, such as WNV, DENV, and ZIKV, that pose serious threats to public health contain N153 or N154 glycosylation (36–40). Viral E protein glycosylation is a molecular determinant of the neuroinvasiveness of WNV (74) and ZIKV (20). The presence of the single N-linked oligosaccharide side chain on the E protein and its trimming by glucosidases is necessary for secretion of recombinant subviral particles and truncated E dimers (75). Furthermore, the production of WNV (67), DENV (61), and TBEV (75) was inhibited after E glycan structures were disrupted. The TMUV E protein 156S site is located at the “150 loop” which is involved in the dimer contacts of flavivirus E proteins (30). This “150 loop” region varies among flaviviruses, which suggests that differences in this region influence virus transmission and disease (31). We found that the S156P mutation not only disrupted N154 glycosylation of the E protein but might also change the E protein loop conformation. Whether the predicted change in the structure of “150 loop” contributes to the abolishment of the transmission of TMUV in ducks needs to examined. However, TMUV replicates in the spleens of infected ducks independently of the S156P mutation, suggesting that host factors in different tissues most likely impacts viral replication.
Taken together, we demonstrate that a single amino acid (S) at position 156 of TMUV E protein is critical for virus replication and transmissibility in ducks, and we further reveal that its mutation (S156P) abolishes N-linked glycosylation at N154 and changes the conformation of the “150 loop” of the E protein, thereby influencing virus transmissibility among birds independent of mosquito vectors. Whether this finding also fits other flaviviruses should be explored in future studies.
MATERIALS AND METHODS
Ethics statement.
All animal experiments were carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Ministry of Science and Technology of the People's Republic of China. The protocol (Shvri-po-2015040380) used in the study was approved by the Animal Care Committee of the Shanghai Veterinary Research Institute.
Cells and viruses.
DF-1 cells were obtained from the American Type Culture Collection (ATCC) and maintained in Dulbecco modified Eagle medium (DMEM; HyClone, Logan, UT), supplemented with 5% fetal bovine serum (FBS; Biowest, South America), and 100 U/ml of penicillin and 100 μg/ml of streptomycin (Invitrogen, Carlsbad, CA) at 37°C in a 5% CO2 humidified incubator.
TMUV strain FX2010 was rescued by a PCR-based reverse genetic system using the viral RNAs of FX2010, which was isolated from sick ducks and purified three times in specific-pathogen-free chicken embryonated eggs by a limiting dilution method. TMUV strain MM1775 was rescued by a PCR-based reverse genetic system using the viral RNAs of MM1775 (ATCC VR-1266), which was first isolated from Culex tritaeniorhynchus in Malaya in 1955 and propagated in the brain of a suckling mouse. All of the rescued viruses were propagated once on DF-1 cells, aliquoted, and stored at −80°C.
Sequence analysis.
The viral RNAs were extracted using RNAiso Plus (TaKaRa Biotechnology, Dalian, China), and the first-strand cDNAs were synthesized with specific reverse transcription primers (see Table S1 in the supplemental material) and SuperScript III transcriptase (Invitrogen, USA) (33). The overlapping segments of PCR products covering the whole genome amplified with primers reported previously (24) were sequenced with ABI 3730 automated sequencers (Applied Biosystems). The sequences were analyzed using DNASTAR software.
Plasmid construction.
To prepare the templates for PCR-based virus rescue, the first-strand cDNAs of FX2010 and MM1775 were used to amplify the overlapping segments covering the whole genome by a PCR method using High Fidelity DNA polymerase Pfx (Invitrogen). Four plasmids, pFXT7-1-956, pFX942-2459, pFX2433-3831, and pFX3656-10991, containing nucleotides 1 to 956 with the addition of a T7 promoter at the 5′ terminus, nucleotides 942 to 2459, nucleotides 2433 to 3831, and nucleotides 3656 to 11001 of FX2010 were generated to provide the template for FX2010 genome amplification. For MM1775, the primers were modified to improve the amplification efficiency and, accordingly, the four plasmids pMMT7-1-955, pMM941-2458, pMM2433-3827, and pMM3549-11001, containing nucleotides 1 to 955 with the addition of a T7 promoter at the 5′ terminus, nucleotides 941 to 2458, nucleotides 2433 to 3827, and nucleotides 3549 to 11001 of MM1775, respectively, were generated. Plasmids containing a domain substitution or single-site mutation on E protein were generated through two-step PCR methods or using a site-directed mutagenesis kit (TransGene, Inc., Strasbourg, France) based on the background of the plasmids pFX942-2459 and pMM941-2458, which contained the E genes of each virus.
PCR-based Tembusu virus rescue.
The full-length cDNA with T7 promoter was generated by two rounds of PCR using High Fidelity DNA polymerase pfx (Invitrogen) as reported previously (33). First-round PCR was performed to produce four overlapped fragments, using each of four plasmids (pFXT7-1-956, pFX942-2459, pFX2433-3831, and pFX3656-10991; or pMMT7-1-955, pMM941-2458, pMM2433-3827, and pMM3549-11001) as the templates. For the rescue of the viruses with domain substitution or a single-site mutation, one of four plasmids pFX942-2459 or pMM941-2458 was substituted with the accordingly modified plasmid. The full-length cDNA was then produced by fusion PCR, using the PCR fragments as the templates.
The transcription of infectious viral RNA from full-length cDNA was performed in vitro using an mMESSAGE mMACHINE T7 kit (Ambion, USA). The transcripts were purified by lithium chloride precipitation and used to transfect DF-1 cells on 6-well plate at an amount of 5 μg using Lipofectamine LTX and Plus Reagent (Invitrogen). The cell culture medium was changed to the DMEM containing 2% FBS at 6 h posttransfection. When cytopathic effects appeared in the transfected cells, the supernatants were collected, and rescued viruses were amplified on DF-1 cells, aliquoted, and stored at −80°C. The rescued viruses were confirmed by sequencing.
Growth of TMUVs in DF-1 cells.
To test the viral replication in vitro, DF-1 cells grown in T-25 flask were infected with TMUVs at an MOI of 0.0001. The cells were incubated with DMEM containing 2% FBS at 37°C. Virus samples from the supernatant were harvested every 12 h postinfection and subjected to virus titration on DF-1 cells to determine the virus titer.
Virus titration.
To determine the virus titer, organs were weighed and homogenized in PBS to yield 1:1 (ml/g) tissue homogenates. Tissue homogenates or cell cultures were clarified by centrifugation, and undiluted and 10-fold serially diluted supernatants were titrated for virus infectivity on DF-1 cells in 96-well plates. The lower limit of virus detection was 0.5 log10 TCID50 per 0.1 g of tissue. The virus titer was calculated by the method of Reed and Muench.
Blocking ELISA.
To test for specific antibodies against TMUV, a blocking ELISA method was used as described previously (76). Briefly, each well of ELISA plates was coated with ∼0.1 μg of purified FX2010 in 0.1 M carbonate-bicarbonate buffer (pH 9.6), followed by incubation overnight at 4°C. Antigen-coated plates were washed with PBS (pH 7.4) containing 0.05% Tween 20 (PBST), and the nonspecific binding sites were blocked with 100 μl of blocking buffer (PBS containing 5% skim milk) for 1 h at 37°C. After a 10-fold dilution in PBS, serum samples were added to the wells, followed by incubation for 1 h at 37°C. The wells were then washed three times with PBST and incubated with monoclonal antibody (MAb) 1F5 (1:20) for 1 h at 37°C. After the wells were washed three times with PBST, goat anti-mouse IgG (1:2,000; Sigma, USA) conjugated to horseradish peroxidase (HRP) was added, and the mixture was incubated at room temperature for 1 h. After the wells were rinsed with PBST three times, 100 μl of 3,3′,5,5′-tetramethyl benzidine was added, and the mixture was incubated at room temperature for 5 min. The reaction was then stopped by adding 0.1 N sulfuric acid. The optical density (OD) was measured at 450 nm, and the percent inhibition (PI) was determined using the following formula: PI (%) = [1 − (OD450 of test serum/OD450 of negative-control serum)] × 100. The serum was considered positive for TMUV reactivity when the PI value was ≥18.4%.
Duck experiments.
To test the pathogenicity and transmissibility of TMUV strain FX2010, TMUV strain MM1775, or the modified viruses in ducks, 12- to 15-week-old Shelducks (a local outbred strain) were inoculated i.n. or i.m. with 103.5 TCID50 of each virus at a volume of 0.2 ml, respectively. One day later, naive ducks were introduced into the isolators where the inoculated ducks were housed. The ducks were monitored daily for clinical signs. The specific method is described as follows.
To compare the replication and transmission of TMUV FX2010 and MM1775, groups of six ducks were inoculated i.m. and i.n., respectively, and three naive ducks were introduced 1 day later. On day 3 postinoculation, three inoculated birds in each group were euthanized by CO2 inhalation, and tissues from the spleen, lung, kidney, brain, ovary, pancreas, and trachea, as well as serum samples, were collected to measure viral replication. The serum samples were collected from the remaining inoculated ducks at 0, 4, and 7 dpi and from the contact ducks at 0, 7, and 14 dpc for antibody detection.
To identify the proteins related to the replication and transmissibility of TMUVs, the chimeric viruses with E gene substitution and their parental viruses were inoculated i.m. into six ducks, respectively, and six naive ducks were introduced 1 day later. In each group, three inoculated ducks were euthanized at 3 dpi, and three contact ducks were euthanized at 6 dpc by CO2 inhalation, and tissue samples from the spleens, lungs, kidneys, and ovaries were collected for viral titration. The serum samples were collected from the remaining inoculated ducks at 0, 4, and 7 dpi and from the remaining contact ducks at 0, 7, and 14 dpc for antibody detection.
To determine further which domain of TMUV E protein influenced the replication and transmissibility of TMUVs in ducks, the E domain substitution recombinant viruses were inoculated i.m. into six ducks, and three naive ducks were introduced 1 day later. Three inoculated ducks were euthanized by CO2 inhalation at 3 dpi, and tissue samples from the spleens, lungs, kidneys, and ovaries were collected for viral titration. The serum samples were collected from the remaining inoculated ducks at 0, 4, and 7 dpi and from the contact ducks at 0, 7, and 14 dpc for antibody detection.
To determine the key amino acids attributed to the replication and transmission of TMUV FX2010 in ducks, viruses with single amino acid mutations located at E-DI were inoculated i.m. into three ducks, following by the introduction of three naive ducks 1 day late. Serum samples were collected from the inoculated ducks at 0, 1, 2, 3, 4, and 7 dpi or from the contact ducks at 0, 1, 2, 3, 4, 5, 7, and 14 dpc for virus titration or antibody detection.
To compare the virus replication of FX-ES156P with the parental FX2010 in ducks, each virus was inoculated i.m. into three ducks. The inoculated ducks were euthanized by CO2 inhalation at 3 dpi, and tissue samples from the spleens, lungs, kidneys, and ovaries were collected for viral titration.
To test the influence of EP156S mutation in the replication and transmission, each of six ducks was inoculated i.m. with MM-EP156S and MM1775, respectively, and three naive ducks were introduced into each of isolator 1 day later. Three inoculated ducks were euthanized by CO2 inhalation at 3 dpi, and tissue samples from the spleens, lungs, kidneys, and ovaries were collected for viral titration. The serum samples were collected from remaining inoculated ducks and the contact ducks at different dpi or dpc points for virus titration or antibody detection.
Western blotting.
To analyze the influence of single amino acid mutation on the posttranslational modification of E proteins, Western blot assays were performed. Briefly, samples were denatured in sample preparation buffer (2% sodium dodecyl sulfate[SDS], 50 mM Tris [pH 6.8], 10% glycerol) at 95°C for 10 min. The samples were then analyzed under denaturing conditions by SDS-PAGE (10%) and then transferred to nitrocellulose membrane (PALL, USA) by wet transfer process. The membrane was blocked with 5% (wt/vol) slim milk powder in PBS at room temperature for at least 1 h before probing the protein bands with E MAb 1F5 (1:500) (76) overnight at 4°C.
After three washes with PBST, the membranes were incubated with HRP-linked anti-mouse IgG goat antibody (1:2,000; Sigma) for 2 h at room temperature. Blots were imaged using SuperSignal West Femto maximum sensitivity substrate (Thermo) on a Tanon 5200 automatic chemiluminescence analyzer (Tanon, Inc., Shanghai, China).
Glycosylation assays.
DF-1 cells were plated in a six-well plate at 106 cells/well and infected with each indicated virus at an MOI of 0.0001. At 48 h postinfection, both supernatants and cells were collected. The cells were solubilized in lysis buffer (50 mM Tris, 150 mM NaCl, 2 mM EDTA, and 1% Triton X-100 [pH 7.5] with protease inhibitors [Roche Molecular Biochemicals, Indianapolis, IN]). Lysates were centrifuged at 10,000 × g for 10 min to clear cellular debris. Both supernatants and lysates were denatured in sample preparation buffer (2% SDS, 50 mM Tris [pH 6.8], 10% glycerol) at 55°C for 20 min. Aliquots from each sample were then treated with 50 U of PNGase F or Endo Hf according to the manufacturer's instructions (New England BioLabs, Inc., Beverly, MA) or with PNGase F buffer without enzyme as a control as described previously (56). Samples were then analyzed under denaturing conditions by SDS-PAGE (7.5% or a 10 to 20% gradient) and Western blotting.
Homology modeling.
To analyze the E protein structure, images of the E proteins of FX2010 and MM1775 were created with the program Phyre2 (77). The E protein structure was referred to JEV E protein (Protein Data Bank [PDB] accession number 5WSN) and visualized and analyzed with PyMOL (Schrodinger, 2015).
Statistical analysis.
Virus titers between groups were evaluated using analysis of variance with GraphPad Prism software (GraphPad Software, Inc.). A P value of ≤0.05 was considered significant.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National Key Research and Development Program of China (2016YFD0500106), the National Natural Science Foundation of China (31472205 and 31702237), and the Foundation of Shanghai Key Laboratory of Veterinary Biotechnology (klab201706). This study was a collaborative research by the Shanghai Veterinary Research Institute, CAAS-Kansas State University Research Laboratory Collaboration for Animal influenza and Poultry Disease.
Z.L. designed the study. D.Y., Y.S., H.W., G.L., X.L., B.W., X.S., J.W., Q.T., J.Y., H.C., and Q.L. performed the experiments. D.Y., W.M., and Z.L. analyzed the data and wrote the paper.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00427-18.
REFERENCES
- 1.Lindenbach BDM, Thiel CLRJ, Rice CM. 2013. Flaviviridae, p 712–746. In Knipe DM, Howley PM (ed), Fields virology, 6th ed Lippincott/Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Platt GS, Way HJ, Bowen ET, Simpson DI, Hill MN, Kamath S, Bendell PJ, Heathcote OH. 1975. Arbovirus infections in Sarawak, October 1968–February 1970 Tembusu and Sindbis virus isolations from mosquitoes. Ann Trop Med Parasitol 69:65–71. doi: 10.1080/00034983.1975.11686984. [DOI] [PubMed] [Google Scholar]
- 3.Leake CJ, Ussery MA, Nisalak A, Hoke CH, Andre RG, Burke DS. 1986. Virus isolations from mosquitoes collected during the 1982 Japanese encephalitis epidemic in northern Thailand. Trans R Soc Trop Med Hyg 80:831–837. doi: 10.1016/0035-9203(86)90397-4. [DOI] [PubMed] [Google Scholar]
- 4.Pandey BD, Karabatsos N, Cropp B, Tagaki M, Tsuda Y, Ichinose A, Igarashi A. 1999. Identification of a flavivirus isolated from mosquitos in Chiang Mai Thailand. Southeast Asian J Trop Med Public Health 30:161–165. [PubMed] [Google Scholar]
- 5.Kono Y, Tsukamoto K, Abd Hamid M, Darus A, Lian TC, Sam LS, Yok CN, Di KB, Lim KT, Yamaguchi S, Narita M. 2000. Encephalitis and retarded growth of chicks caused by Sitiawan virus, a new isolate belonging to the genus Flavivirus. Am J Trop Med Hyg 63:94–101. doi: 10.4269/ajtmh.2000.63.94. [DOI] [PubMed] [Google Scholar]
- 6.Yan P, Zhao Y, Zhang X, Xu D, Dai X, Teng Q, Yan L, Zhou J, Ji X, Zhang S. 2011. An infectious disease of ducks caused by a newly emerged Tembusu virus strain in mainland China. Virology 417:1–8. doi: 10.1016/j.virol.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Thontiravong A, Ninvilai P, Tunterak W, Nonthabenjawan N, Chaiyavong S, Angkabkingkaew K, Mungkundar C, Phuengpho W, Oraveerakul K, Amonsin A. 2015. Tembusu-related flavivirus in ducks, Thailand. Emerg Infect Dis 21:2164–2167. doi: 10.3201/eid2112.150600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao Z, Zhang C, Liu Y, Ye W, Han J, Ma G, Zhang D, Xu F, Gao X, Tang Y. 2011. Tembusu virus in ducks, China. Emerg Infect Dis 17:1873. doi: 10.3201/eid1710.101890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su J, Li S, Hu X, Yu X, Wang Y, Liu P, Lu X, Zhang G, Hu X, Liu D. 2011. Duck egg-drop syndrome caused by BYD virus, a new Tembusu-related flavivirus. PLoS One 6:e18106. doi: 10.1371/journal.pone.0018106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang Y, Diao Y, Yu C, Gao X, Ju X, Xue C, Liu X, Ge P, Qu J, Zhang D. 2013. Characterization of a Tembusu virus isolated from naturally infected house sparrows (Passer domesticus) in Northern China. Transboundary Emerg Dis 60:152–158. doi: 10.1111/j.1865-1682.2012.01328.x. [DOI] [PubMed] [Google Scholar]
- 11.Moudy RM, Zhang B, Shi PY, Kramer LD. 2009. West Nile virus envelope protein glycosylation is required for efficient viral transmission by Culex vectors. Virology 387:222–228. doi: 10.1016/j.virol.2009.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moudy RM, Payne AF, Dodson BL, Kramer LD. 2011. Requirement of glycosylation of West Nile virus envelope protein for infection of, but not spread within, Culex quinquefasciatus mosquito vectors. Am J Trop Med Hyg 85:374–378. doi: 10.4269/ajtmh.2011.10-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhuanzhuan L, Tengfei Z, Zetian L, Zhenhong Z, Zhirong J, Guofa Z, Tricia W, Jiabao X, Jinbao G, Xiaohong Z, Lifeng L, Guiyun Y, Xiao-Guang C. 2017. Competence of Aedes aegypti, Ae. albopictus, and Culex quinquefasciatus mosquitoes as Zika virus vectors, China. Emerg Infect Dis J 23:1085–1091. doi: 10.3201/eid2307.161528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banet-Noach C, Simanov L, Malkinson M. 2003. Direct (non-vector) transmission of West Nile virus in geese. Avian Pathol 32:489–494. doi: 10.1080/0307945031000154080. [DOI] [PubMed] [Google Scholar]
- 15.Llorente F, Perez-Ramirez E, Fernandez-Pinero J, Elizalde M, Figuerola J, Soriguer RC, Jimenez-Clavero MA. 2015. Bagaza virus is pathogenic and transmitted by direct contact in experimentally infected partridges, but is not infectious in house sparrows and adult mice. Vet Res 46:93. doi: 10.1186/s13567-015-0233-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ricklin ME, Garcia-Nicolas O, Brechbuhl D, Python S, Zumkehr B, Nougairede A, Charrel RN, Posthaus H, Oevermann A, Summerfield A. 2016. Vector-free transmission and persistence of Japanese encephalitis virus in pigs. Nat Commun 7:10832. doi: 10.1038/ncomms10832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ip HS, Van Wettere AJ, McFarlane L, Shearn-Bochsler V, Dickson SL, Baker J, Hatch G, Cavender K, Long R, Bodenstein B. 2014. West nile virus transmission in winter: the 2013 great salt lake bald eagle and eared grebes mortality event. PLoS Curr doi: 10.1371/currents.outbreaks.b0f031fc8db2a827d9da0f30f0766871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinton MG, Reisen WK, Wheeler SS, Townsend AK. 2015. West Nile virus activity in a winter roost of American crows (Corvus brachyrhynchos): is bird-to-bird transmission important in persistence and amplification? J Med Entomol 52:683–692. doi: 10.1093/jme/tjv040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X, Shi Y, Liu Q, Wang Y, Li G, Teng Q, Zhang Y, Liu S, Li Z. 2015. Airborne transmission of a novel Tembusu virus in ducks. J Clin Microbiol 53:2734–2736. doi: 10.1128/JCM.00770-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Annamalai AS, Pattnaik A, Sahoo BR, Muthukrishnan E, Natarajan SK, Steffen D, Vu HLX, Delhon G, Osorio FA, Petro TM, Xiang SH, Pattnaik AK. 2017. Zika virus encoding non-glycosylated envelope protein is attenuated and defective in neuroinvasion. J Virol doi: 10.1128/JVI.01348-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diamond MS, Pierson TC. 2015. Molecular insight into dengue virus pathogenesis and its implications for disease control. Cell 162:488–492. doi: 10.1016/j.cell.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grant A, Ponia SS, Tripathi S, Balasubramaniam V, Miorin L, Sourisseau M, Schwarz MC, Sanchez-Seco MP, Evans MJ, Best SM, Garcia-Sastre A. 2016. Zika virus targets human STAT2 to inhibit type i interferon signaling. Cell Host Microbe 19:882–890. doi: 10.1016/j.chom.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Lei CQ, Ji Y, Zhou H, Ren Y, Peng Q, Zeng Y, Jia Y, Ge J, Zhong B, Li Y, Wei J, Shu HB, Zhu Q. 2016. Duck Tembusu virus nonstructural protein 1 antagonizes IFN-β signaling pathways by targeting VISA. J Immunol 197:4704–4713. doi: 10.4049/jimmunol.1502317. [DOI] [PubMed] [Google Scholar]
- 24.Li G, Gao X, Xiao Y, Liu S, Peng S, Li X, Shi Y, Zhang Y, Yu L, Wu X, Yan P, Yan L, Teng Q, Tong G, Li Z. 2014. Development of a live attenuated vaccine candidate against duck Tembusu viral disease. Virology 450-451:233–242. doi: 10.1016/j.virol.2013.12.028. [DOI] [PubMed] [Google Scholar]
- 25.Liu P, Lu H, Li S, Moureau G, Deng YQ, Wang Y, Zhang L, Jiang T, de Lamballerie X, Qin CF, Gould EA, Su J, Gao GF. 2012. Genomic and antigenic characterization of the newly emerging Chinese duck egg-drop syndrome flavivirus: genomic comparison with Tembusu and Sitiawan viruses. J Gen Virol 93:2158–2170. doi: 10.1099/vir.0.043554-0. [DOI] [PubMed] [Google Scholar]
- 26.Lei W, Guo X, Fu S, Feng Y, Tao X, Gao X, Song J, Yang Z, Zhou H, Liang G. 2017. The genetic characteristics and evolution of Tembusu virus. Vet Microbiol 201:32–41. doi: 10.1016/j.vetmic.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 27.Bressanelli S, Stiasny K, Allison SL, Stura EA, Duquerroy S, Lescar J, Heinz FX, Rey FA. 2004. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J 23:728–738. doi: 10.1038/sj.emboj.7600064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu K, Sheng ZZ, Huang B, Ma X, Li Y, Yuan X, Qin Z, Wang D, Chakravarty S, Li F, Song M, Sun H. 2013. Structural, antigenic, and evolutionary characterizations of the envelope protein of newly emerging duck Tembusu virus. PLoS One 8:e71319. doi: 10.1371/journal.pone.0071319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao D, Huang X, Liu Y, Han K, Zhang J, Yang J, Xie X, Li Y. 2015. Domain I and II from newly emerging goose Tembusu virus envelope protein functions as a dominant-negative inhibitor of virus infectivity. Res Vet Sci 98:121–126. doi: 10.1016/j.rvsc.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Zhang W, Ogata S, Clements D, Strauss JH, Baker TS, Kuhn RJ, Rossmann MG. 2004. Conformational changes of the flavivirus E glycoprotein. Structure 12:1607–1618. doi: 10.1016/j.str.2004.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi Y, Gao GF. 2017. Structural biology of the Zika virus. Trends Biochem Sci 42:443–456. doi: 10.1016/j.tibs.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 32.Modis Y, Ogata S, Clements D, Harrison SC. 2004. Structure of the dengue virus envelope protein after membrane fusion. Nature 427:313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 33.Wu X, Shi Y, Yan D, Li X, Yan P, Gao X, Zhang Y, Yu L, Ren C, Li G, Yan L, Teng Q, Li Z. 2016. Development of a PCR-based reverse genetics system for an attenuated duck Tembusu virus strain. PLoS One 11:e0156579. doi: 10.1371/journal.pone.0156579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holbrook MR, Ni H, Shope RE, Barrett AD. 2001. Amino acid substitution(s) in the stem-anchor region of Langat virus envelope protein attenuates mouse neurovirulence. Virology 286:54–61. doi: 10.1006/viro.2001.0959. [DOI] [PubMed] [Google Scholar]
- 35.Ashour J, Laurent-Rolle M, Shi PY, Garcia-Sastre A. 2009. NS5 of dengue virus mediates STAT2 binding and degradation. J Virol 83:5408–5418. doi: 10.1128/JVI.02188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campbell GL, Hills SL, Fischer M, Jacobson JA, Hoke CH, Hombach JM, Marfin AA, Solomon T, Tsai TF, Tsu VD, Ginsburg AS. 2011. Estimated global incidence of Japanese encephalitis: a systematic review. Bull World Health Organ 89:766–774, 774A–774E. doi: 10.2471/BLT.10.085233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohen J. 2016. Infectious disease: the race for a Zika vaccine is on. Science 351:543–544. doi: 10.1126/science.351.6273.543. [DOI] [PubMed] [Google Scholar]
- 38.Gray TJ, Webb CE. 2014. A review of the epidemiological and clinical aspects of West Nile virus. Int J Gen Med 7:193–203. doi: 10.2147/IJGM.S59902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murrell S, Wu SC, Butler M. 2011. Review of dengue virus and the development of a vaccine. Biotechnol Adv 29:239–247. doi: 10.1016/j.biotechadv.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 40.Vogel G. 2016. Infectious disease: evidence grows for Zika virus as pregnancy danger. Science 351:1123–1124. doi: 10.1126/science.351.6278.1123. [DOI] [PubMed] [Google Scholar]
- 41.Centers for Disease Control and Prevention. 2003. West Nile virus infection among turkey breeder farm workers–Wisconsin, 2002. MMWR Morb Mortal Wkly Rep 52:1017–1019. [PubMed] [Google Scholar]
- 42.Yu L, Robert Putnak J, Pletnev AG, Markoff L. 2008. Attenuated West Nile viruses bearing 3′ SL and envelope gene substitution mutations. Vaccine 26:5981–5988. doi: 10.1016/j.vaccine.2008.08.064. [DOI] [PubMed] [Google Scholar]
- 43.Butrapet S, Huang CY-H, Pierro DJ, Bhamarapravati N, Gubler DJ, Kinney RM. 2000. Attenuation markers of a candidate dengue type 2 vaccine virus, strain 16681 (PDK-53), are defined by mutations in the 5′ noncoding region and nonstructural proteins 1 and 3. J Virol 74:3011–3019. doi: 10.1128/JVI.74.7.3011-3019.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blaney JE Jr, Sathe NS, Goddard L, Hanson CT, Romero TA, Hanley KA, Murphy BR, Whitehead SS. 2008. Dengue virus type 3 vaccine candidates generated by introduction of deletions in the 3′ untranslated region (3′UTR) or by exchange of the DENV-3 3′UTR with that of DENV-4. Vaccine 26:817–828. doi: 10.1016/j.vaccine.2007.11.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang S, Li L, Woodson SE, Huang CY-H, Kinney RM, Barrett AD, Beasley DW. 2006. A mutation in the envelope protein fusion loop attenuates mouse neuroinvasiveness of the NY99 strain of West Nile virus. Virology 353:35–40. doi: 10.1016/j.virol.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 46.Butrapet S, Childers T, Moss KJ, Erb SM, Luy BE, Calvert AE, Blair CD, Roehrig JT, Huang CY-H. 2011. Amino acid changes within the E protein hinge region that affect dengue virus type 2 infectivity and fusion. Virology 413:118–127. doi: 10.1016/j.virol.2011.01.030. [DOI] [PubMed] [Google Scholar]
- 47.Lee E, Hall RA, Lobigs M. 2004. Common E protein determinants for attenuation of glycosaminoglycan-binding variants of Japanese encephalitis and West Nile viruses. J Virol 78:8271–8280. doi: 10.1128/JVI.78.15.8271-8280.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whiteman MC, Li L, Wicker JA, Kinney RM, Huang C, Beasley DW, Chung KM, Diamond MS, Solomon T, Barrett AD. 2010. Development and characterization of non-glycosylated E and NS1 mutant viruses as a potential candidate vaccine for West Nile virus. Vaccine 28:1075–1083. doi: 10.1016/j.vaccine.2009.10.112. [DOI] [PubMed] [Google Scholar]
- 49.Zhang W, Chipman PR, Corver J, Johnson PR, Zhang Y, Mukhopadhyay S, Baker TS, Strauss JH, Rossmann MG, Kuhn RJ. 2003. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat Struct Mol Biol 10:907–912. doi: 10.1038/nsb990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mitzel DN, Best SM, Masnick MF, Porcella SF, Wolfinbarger JB, Bloom ME. 2008. Identification of genetic determinants of a tick-borne flavivirus associated with host-specific adaptation and pathogenicity. Virology 381:268–276. doi: 10.1016/j.virol.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whiteman MC, Wicker JA, Kinney RM, Huang CY-H, Solomon T, Barrett AD. 2011. Multiple amino acid changes at the first glycosylation motif in NS1 protein of West Nile virus are necessary for complete attenuation for mouse neuroinvasiveness. Vaccine 29:9702–9710. doi: 10.1016/j.vaccine.2011.09.036. [DOI] [PubMed] [Google Scholar]
- 52.Audsley M, Edmonds J, Liu W, Mokhonov V, Mokhonova E, Melian EB, Prow N, Hall RA, Khromykh AA. 2011. Virulence determinants between New York 99 and Kunjin strains of West Nile virus. Virology 414:63–73. doi: 10.1016/j.virol.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rùžek D, Gritsun TS, Forrester NL, Gould EA, Kopecký J, Golovchenko M, Rudenko N, Grubhoffer L. 2008. Mutations in the NS2B and NS3 genes affect mouse neuroinvasiveness of a Western European field strain of tick-borne encephalitis virus. Virology 374:249–255. doi: 10.1016/j.virol.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 54.Puig-Basagoiti F, Tilgner M, Bennett CJ, Zhou Y, Muñoz-Jordán JL, García-Sastre A, Bernard KA, Shi P-Y. 2007. A mouse cell-adapted NS4B mutation attenuates West Nile virus RNA synthesis. Virology 361:229–241. doi: 10.1016/j.virol.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wicker JA, Whiteman MC, Beasley DW, Davis CT, McGee CE, Lee JC, Higgs S, Kinney RM, Huang CY-H, Barrett AD. 2012. Mutational analysis of the West Nile virus NS4B protein. Virology 426:22–33. doi: 10.1016/j.virol.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hanna SL, Pierson TC, Sanchez MD, Ahmed AA, Murtadha MM, Doms RW. 2005. N-linked glycosylation of West Nile virus envelope proteins influences particle assembly and infectivity. J Virol 79:13262–13274. doi: 10.1128/JVI.79.21.13262-13274.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bryant JE, Calvert AE, Mesesan K, Crabtree MB, Volpe KE, Silengo S, Kinney RM, Huang CY, Miller BR, Roehrig JT. 2007. Glycosylation of the dengue 2 virus E protein at N67 is critical for virus growth in vitro but not for growth in intrathoracically inoculated Aedes aegypti mosquitoes. Virology 366:415–423. doi: 10.1016/j.virol.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 58.Lee E, Lobigs M. 2008. E protein domain III determinants of yellow fever virus 17D vaccine strain enhance binding to glycosaminoglycans, impede virus spread, and attenuate virulence. J Virol 82:6024–6033. doi: 10.1128/JVI.02509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mukhopadhyay S, Kuhn RJ, Rossmann MG. 2005. A structural perspective of the flavivirus life cycle. Nat Rev Microbiol 3:13–22. doi: 10.1038/nrmicro1067. [DOI] [PubMed] [Google Scholar]
- 60.Mossenta M, Marchese S, Poggianella M, Slon Campos JL, Burrone OR. 2017. Role of N-glycosylation on Zika virus E protein secretion, viral assembly, and infectivity. Biochem Biophys Res Commun 492:579–586. doi: 10.1016/j.bbrc.2017.01.022. [DOI] [PubMed] [Google Scholar]
- 61.Vigerust DJ, Shepherd VL. 2007. Virus glycosylation: role in virulence and immune interactions. Trends Microbiol 15:211–218. doi: 10.1016/j.tim.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adams SC, Broom AK, Sammels LM, Hartnett AC, Howard MJ, Coelen RJ, Mackenzie JS, Hall RA. 1995. Glycosylation and antigenic variation among Kunjin virus isolates. Virology 206:49–56. doi: 10.1016/S0042-6822(95)80018-2. [DOI] [PubMed] [Google Scholar]
- 63.Ishak H, Takegami T, Kamimura K, Funada H. 2001. Comparative sequences of two type 1 dengue virus strains possessing different growth characteristics in vitro. Microbiol Immunol 45:327–331. doi: 10.1111/j.1348-0421.2001.tb02627.x. [DOI] [PubMed] [Google Scholar]
- 64.Johnson AJ, Guirakhoo F, Roehrig JT. 1994. The envelope glycoproteins of dengue 1 and dengue 2 viruses grown in mosquito cells differ in their utilization of potential glycosylation sites. Virology 203:241–249. doi: 10.1006/viro.1994.1481. [DOI] [PubMed] [Google Scholar]
- 65.Vorndam V, Mathews JH, Barrett AD, Roehrig JT, Trent DW. 1993. Molecular and biological characterization of a non-glycosylated isolate of St Louis encephalitis virus. J Gen Virol 74(Pt 12):2653–2660. doi: 10.1099/0022-1317-74-12-2653. [DOI] [PubMed] [Google Scholar]
- 66.Winkler G, Heinz FX, Kunz C. 1987. Studies on the glycosylation of flavivirus E proteins and the role of carbohydrate in antigenic structure. Virology 159:237–243. doi: 10.1016/0042-6822(87)90460-0. [DOI] [PubMed] [Google Scholar]
- 67.Scherret JH, Mackenzie JS, Khromykh AA, Hall RA. 2001. Biological significance of glycosylation of the envelope protein of Kunjin virus. Ann N Y Acad Sci 951:361–363. doi: 10.1111/j.1749-6632.2001.tb02719.x. [DOI] [PubMed] [Google Scholar]
- 68.McMinn PC, Weir RC, Dalgarno L. 1996. A mouse-attenuated envelope protein variant of Murray Valley encephalitis virus with altered fusion activity. J Gen Virol 77(Pt 9):2085–2088. doi: 10.1099/0022-1317-77-9-2085. [DOI] [PubMed] [Google Scholar]
- 69.Mondotte JA, Lozach PY, Amara A, Gamarnik AV. 2007. Essential role of dengue virus envelope protein N glycosylation at asparagine-67 during viral propagation. J Virol 81:7136–7148. doi: 10.1128/JVI.00116-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shirato K, Miyoshi H, Goto A, Ako Y, Ueki T, Kariwa H, Takashima I. 2004. Viral envelope protein glycosylation is a molecular determinant of the neuroinvasiveness of the New York strain of West Nile virus. J Gen Virol 85:3637–3645. doi: 10.1099/vir.0.80247-0. [DOI] [PubMed] [Google Scholar]
- 71.Fontes-Garfias CR, Shan C, Luo H, Muruato AE, Medeiros DBA, Mays E, Xie X, Zou J, Roundy CM, Wakamiya M, Rossi SL, Wang T, Weaver SC, Shi PY. 2017. Functional analysis of glycosylation of Zika virus envelope protein. Cell Rep 21:1180–1190. doi: 10.1016/j.celrep.2017.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gong D, Zhang T-H, Zhao D, Du Y, Chapa TJ, Shi Y, Wang L, Contreras D, Zeng G, Shi P-Y, Wu T-T, Arumugaswami V, Sun R. 2018. High-throughput fitness profiling of Zika virus E protein reveals different roles for glycosylation during infection of mammalian and mosquito cells. iScience 1:97–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wen D, Li S, Dong F, Zhang Y, Lin Y, Wang J, Zou Z, Zheng A. 2018. N-glycosylation of viral E protein is the determinant for vector midgut invasion by flaviviruses. mBio 9:e00046-. doi: 10.1128/mBio.00046-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Beasley DW, Whiteman MC, Zhang S, Huang CY, Schneider BS, Smith DR, Gromowski GD, Higgs S, Kinney RM, Barrett AD. 2005. Envelope protein glycosylation status influences mouse neuroinvasion phenotype of genetic lineage 1 West Nile virus strains. J Virol 79:8339–8347. doi: 10.1128/JVI.79.13.8339-8347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lorenz IC, Kartenbeck J, Mezzacasa A, Allison SL, Heinz FX, Helenius A. 2003. Intracellular assembly and secretion of recombinant subviral particles from tick-borne encephalitis virus. J Virol 77:4370–4382. doi: 10.1128/JVI.77.7.4370-4382.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li X, Li G, Teng Q, Yu L, Wu X, Li Z. 2012. Development of a blocking ELISA for detection of serum neutralizing antibodies against newly emerged duck Tembusu virus. PLoS One 7:e53026. doi: 10.1371/journal.pone.0053026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction, and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.