Infection by highly pathogenic avian influenza A virus remains a threat to public health. Our broadly neutralizing antibody, 13D4, is capable of neutralizing all representative H5N1 viruses and protecting mice against lethal challenge. Structural analysis revealed that 13D4 uses heavy-chain complementarity-determining region 3 (HCDR3) to fit the receptor binding site (RBS) via conformational rearrangement. Four conserved residues within the RBS are critical for the broad potency of 13D4. Importantly, polymorphism of Lys193 on the RBS may be associated with the antigenicity shift from H5N1 to other newly emerging viruses, such as H5N6 and H5N8. Our findings may pave the way for highly pathogenic avian influenza virus vaccine development and therapeutic RBS inhibitor design.
KEYWORDS: avian influenza virus, antibody, broad-specific neutralization, receptor binding site
ABSTRACT
Human infection with highly pathogenic avian influenza A viruses causes severe disease and fatalities. We previously identified a potent and broadly neutralizing antibody (bnAb), 13D4, against the H5N1 virus. Here, we report the co-crystal structure of 13D4 in complex with the hemagglutinin (HA) of A/Vietnam/1194/2004 (H5N1). We show that heavy-chain complementarity-determining region 3 (HCDR3) of 13D4 confers broad yet specific neutralization against H5N1, undergoing conformational rearrangement to bind to the receptor binding site (RBS). Further, we show that mutating four critical residues within the RBS—Trp153, Lys156, Lys193, and Leu194—disrupts the binding between 13D4 and HA. Viruses bearing Asn193 instead of Lys/Arg can evade 13D4 neutralization, indicating that Lys193 polymorphism might be, at least in part, involved in the antigenicity of recent H5 genotypes (such as H5N6 and H5N8) as distinguished from H5N1. BnAb 13D4 may offers a template for therapeutic RBS inhibitor design and serve as an indicator of antigenic change for current H5 viruses.
IMPORTANCE Infection by highly pathogenic avian influenza A virus remains a threat to public health. Our broadly neutralizing antibody, 13D4, is capable of neutralizing all representative H5N1 viruses and protecting mice against lethal challenge. Structural analysis revealed that 13D4 uses heavy-chain complementarity-determining region 3 (HCDR3) to fit the receptor binding site (RBS) via conformational rearrangement. Four conserved residues within the RBS are critical for the broad potency of 13D4. Importantly, polymorphism of Lys193 on the RBS may be associated with the antigenicity shift from H5N1 to other newly emerging viruses, such as H5N6 and H5N8. Our findings may pave the way for highly pathogenic avian influenza virus vaccine development and therapeutic RBS inhibitor design.
INTRODUCTION
Highly pathogenic avian influenza viruses (HPAI) are responsible for serious outbreaks in poultry, leading to huge economic loss worldwide, and for occasional human infections since 1997 (1–4). The current H5N1 viruses are considered to be derived from influenza virus A/Goose/Guangdong/1/96, which emerged in 1996 through genetic reassortment (5, 6). It is believed that genetic reassortment of H5N1 has generated H5N6 and H5N8, among others, which are emerging in south China and parts of North America as the most recent development (3, 7–9). Currently, there is a lack of effective antivirals for treatment of human infections with H5N1 or its derivatives (10–12). Vaccines and therapeutics (13–17) that target the H5 subtype are thus urgently needed.
Hemagglutinin (HA) is one of the three membrane proteins on the surface of the influenza virus responsible for receptor recognition and virus entry. The head of the HA1 subunit harbors the major targeting sites of neutralizing antibodies to influenza virus (18, 19). The HA2 subunit is an α-helix-rich “stem” region of the HA trimer that functions to anchor the HA trimer onto the viral envelope membrane (20). We and others have described several broadly neutralizing antibodies (bnAbs) that target either the HA1 receptor binding site (RBS) or the HA2 stem region of H5N1 virus (21–23). Structure analysis of HA complexes with antibodies has provided insight into the molecular basis for the properties of broadly neutralizing antibodies (24–27). Canonically, heavy-chain complementarity-determining region 3 (HCDR3) is most diverse in both length and amino acid sequence and is often the major determinant of the specificity of antibody-antigen recognition (28–30). Upon binding to the RBS of influenza virus, bnAbs commonly use their HCDR loops to plug or fit the structural contour of the narrow pocket of the RBS by mimicking binding of the sialoglycan receptor (31, 32). For example, the monoclonal antibody (MAb) CH65 extends the HCDR3 loop into the pocket and is thus capable of neutralizing 30 representative seasonal H1 viruses (33, 34). In contrast, MAb C05 has an extraordinarily long HCDR3 loop that covers the entire RBS (58), facilitating broad neutralization against H1, H2, H3, H9, and H12. Other MAbs have different actions: HCDR3 of S139/1 recognizes various RBSs of H1, H2, H3, and H13 (59), whereas HCDR3 of F045-092 recapitulates the binding of the α-2,6 of the sialoglycan receptor, neutralizing almost all H3 viruses (60). Recently, a human antibody, AVFluIgG03, was identified, with broad-spectrum activity against most H5N1 clades, except for clades 1, 4, and 8, which were isolated between 1997 and 2008 (22, 35). The crystal structure of AVFluIgG03 in complex with A/Anhui/1/2005 HA highlights its significant neutralization breadth through HCDR3 targeting of the RBS.
Previously, we generated a panel of 14 anti-H5N1 broadly neutralizing MAbs, among which MAb 13D4 presented as the most efficacious in terms of its therapeutic potential, with the widest neutralization capacity. Here, we solve and characterize the crystal structures of the 13D4 Fab and its complex with the A/Vietnam/1194/2004 (VN1194) HA ectodomain. We show that HCDR3 of 13D4 targets the RBS of the HA head region (HAhr) by conformational rearrangement. Using structure-guided mutagenesis, we identify the key binding sites that are specifically conserved among the H5N1 isolates. Furthermore, we show that 13D4 can neutralize the most recent H5N1 strains. To our knowledge, 13D4 might represent one of the most broad-spectrum H5N1 neutralizing antibodies. Intriguingly, the antibody loses its neutralization activity against other H5N1 genetic derivatives, such as H5N6 and H5N8, which emerged in recent years, presumably under evolutionary pressure caused by massive application of poultry vaccine. We used structural comparisons among the HA crystal structures bearing polymorphisms in key 13D4 binding sites to identify the determinants of antigenic shift from H5N1 to other H5 genotypes. Collectively, our findings offer molecular insight into the broad yet specific antibody-mediated neutralization of H5N1, which might be partially related to the antigenic shift from H5N1 to other genotypes, and provide a strategy for the design and optimization of antiviral therapeutics and broad-spectrum vaccines against H5N1 infection.
RESULTS
Overall structure of 13D4:HAhr.
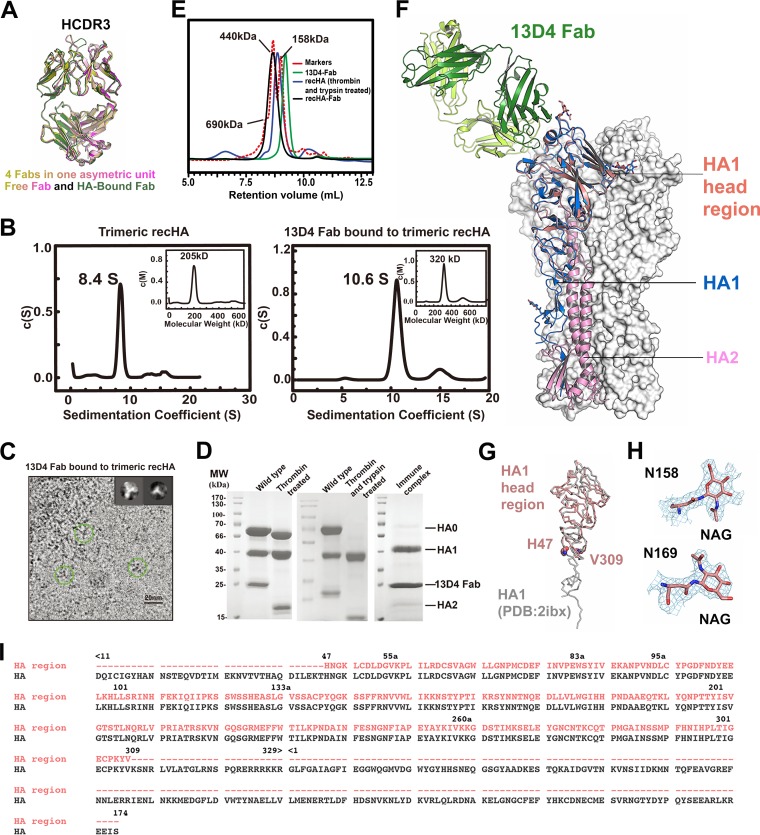
To elucidate how the 13D4 MAb neutralizes all of the representative H5N1 isolates, we solved the crystal structures of the 13D4 Fab alone and in complex with the VN1194 HA head region (13D4:HAhr) to 2.30-Å and 2.33-Å resolution, respectively, and refined to Rwork/Rfree values of 20.5% and 24.3%, and 17.3% and 21.9%, respectively (Table 1). Four 13D4 Fab molecules are found in the asymmetric unit and display consistent conformation of the CDR loops, albeit HCDR3 is involved in crystal packing (Fig. 1A). For crystallization of the immune complex, HA was generated in baculovirus-infected insect cells, as described by Stevens et al. (36). One trimeric HA can bind three 13D4 Fabs in solution, as shown in the continuous molar mass distribution [c(M)] profiles of the analytical ultracentrifugation (AUC) assay (Fig. 1B) and two-dimensional (2D) average figures of cryo-electron microscopy (EM) imaging (Fig. 1C). Purified HA0 was digested with thrombin and trypsin and mixed with the 13D4 Fab for crystallization (Fig. 1D and E). However, in the asymmetric unit of the HA:13D4 Fab complex, one 13D4 Fab binds with only the head region of HA1 (amino acids [aa] 47 to 309; H3 numbering), denoted HAhr (Fig. 1F, G, and I). HA might have degraded due to the length (∼3 months) of the crystallization process. However, we found that the structure of the HA1 head region is congruent with the corresponding moiety of the full-length Viet04 HA trimer structure (Protein Data Bank [PDB] accession no. 2IBX) (superimposition with a root mean square deviation [RMSD] of 0.37 Å for all HA1 C-α atoms) (Fig. 1G). The glycan chains linked to Asn158 and Asn169 in our HA1 structure are well resolved in the electron density map (Fig. 1F and H) and superimpose with HA1 of trimeric HA (Fig. 1F). Overall, the 13D4 interaction interface in HA1 is located on the RBS; this is consistent with our previous result for the escape mutation on 13D4 neutralization. 13D4 Fab in the complex has a well-defined electron density map and shares similar overall structure with the free 13D4 Fab (RMSD, 0.85 Å for all of the C-α atoms), except for a significant change in HCDR3 (Fig. 1A).
TABLE 1.
Data collection and refinement statistics for 13D4:HA complex and 13D4 Fab alone
| Statistic | Value |
|
|---|---|---|
| 13D4:HA | 13D4 Fab | |
| Data collection | ||
| Cell parameters (Å, °) | a = 72.2, b = 72.7, c = 76.7 | a = 81.3, b = 109.0, c = 108.2 |
| α = 90, β = 93.4, γ = 90 | α = 90, β = 90.1, γ = 90 | |
| Space group | P21 | P21 |
| Resolution range (Å) | 50.0–2.33 (2.37–2.33) | 50.0–2.30 (2.34–2.30) |
| Wavelength (Å) | 0.97945 | 0.97946 |
| No. of observed hkl (I > σ) | 127,187 | 611,301 |
| No. of unique hkl | 33,998 | 83,892 |
| Redundancya | 3.7 (3.6) | 7.3 (7.1) |
| Completeness (%)a | 99.8 (99.8) | 99.7 (99.3) |
| Overall (I/σI)a | 19.2 (3.6) | 18.7 (2.2) |
| Rsymb (%)a | 8.7 (48.3) | 11.9 (99.9) |
| Rpimc (%)a | 5.2 (29.4) | 5.1 (47.8) |
| CC1/2 | 0.807 | 0.712 |
| Refinement | ||
| Resolution range (Å) | 34.7–2.3 | 48.5–2.3 |
| No. of reflections | 33,983 | 83,864 |
| Rfactord | 17.3 | 20.5 |
| Rfreee | 21.9 | 24.3 |
| RMSD bond lengths (Å) | 0.004 | 0.003 |
| RMSD bond angles (°) | 0.61 | 0.69 |
| No. of atoms | 5,799 | 13,751 |
| Protein | 5,472 | 13,360 |
| Glycan | 28 | |
| Water | 299 | 391 |
| Wilson B factor (Å2) | 35.4 | 43.9 |
| Average B factors (Å2) | 48.9 | 55.8 |
| Protein | 49.2 | 56.0 |
| Glycan | 65.3 | - |
| Water | 42.4 | 46.3 |
| Ramachandran plot | ||
| Favored and allowed regions (%) | 95.1 | 96.0 |
| Generously allowed regions (%) | 4.6 | 3.6 |
| Disallowed regions (%) | 0.3 | 0.4 |
The numbers in parentheses refer to the highest resolution shell.
Rsym = Σh Σi|I1(h)→I(h)|/Σh Σi I1(h).
Rpim = ΣhSQRT[1/(n − 1)] Σi|I(1h)→I(h)|/Σh Σi I (1h).
Rfactor = Σhkl||Fobs| − k|Fcalc||/Σhkl|Fobs|.
Rfree is calculated using the same equation as for Rfactor, but 5.0% of the reflections were chosen randomly and omitted from the refinement.
FIG 1.
13D4 binds to the RBS on the HAhr. (A) Overlay of four 13D4 Fab molecules in the asymmetric unit and the HA-bound 13D4 Fab. The structure superimposition shows that 13D4 Fabs in the crystal have a consistent loop conformation for CDR loops in the crystal packing. The HCDR3 loop undergoes a substantial conformational change. (B) Sedimentation coefficient measurements of HA alone and in complex with 13D4 Fab. (Left) HA exists as single species in c(s) profile, with an 8.4S sedimentation coefficient and 205-kDa apparent molecular mass; this indicates that HA mainly exists as a trimer in solution. (Right) The HA:13D4 complex presents major species in c(s) profile, 10.6S and 320 kDa, corresponding to one HA trimer binding with 3 Fabs. recHA, recombinant HA. (C) Cryo-EM micrograph of unstained, vitrified HA:13D4 complex. Discernible particles were manually boxed within 20-nm diameter and subjected to 2D classification with Relion software. Two representative classes (morphology) are shown in the insets, corresponding to a top view and a side view of three Fabs binding to the HA trimer. The brightness of the original image was enhanced by 50% to make the trimer shape clearer. (D) Protease treatment of HA. Before protease treatment, HA resolved as a full-length HA0 and separate HA1 and HA2 in SDS-PAGE. The foldon facilitating HA trimerization was removed with thrombin, and the thrombin-treated sample was further catalyzed with trypsin, demonstrating that HA0 was completely cleaved to HA1 and HA2. The purified immune complex sample was further analyzed by SDS-PAGE. (E) HPLC profiles of protease-treated HA and its complex with 13D4 Fab. (F) Overall structure of the 13D4:HAhr complex. The HAhr of the 13D4:HAhr complex overlaps one protomer of the HA trimer (PDB accession no. 2IBX). The HA1 head region of the 13D4:HAhr complex is in salmon; the heavy chain and light chain of 13D4 Fab are in forest green and lime green, respectively; and HA1 and HA2 components of the HAhr-superimposed protomer are in dark blue and pink, respectively. (G) Superimposition of the HA head region on the HA1 monomer in the reported crystal structure (PDB accession no. 2IBX). (H) Sample electron density (2Fo-Fc [Fo, true complex structure factors of a crystal structure; Fc, calculated complex structure factors of any model structure]) maps contoured at 1 σ above the mean are shown for N-linked glycans bound to Asn158 and Asn169 of HA. NAG, N-acetylglucosamine. (I) Sequence alignment of the HA head region (aa 47 to 309; H3 numbering) resolved in the 13D4:HAhr crystal and the HA construct (consisting of HA1 [aa 11 to 329] and HA2 [aa 1 to 174]). The dashes denote the corresponding residues that are missing in the crystal structure.
Interactions between HA1 and 13D4 Fab.
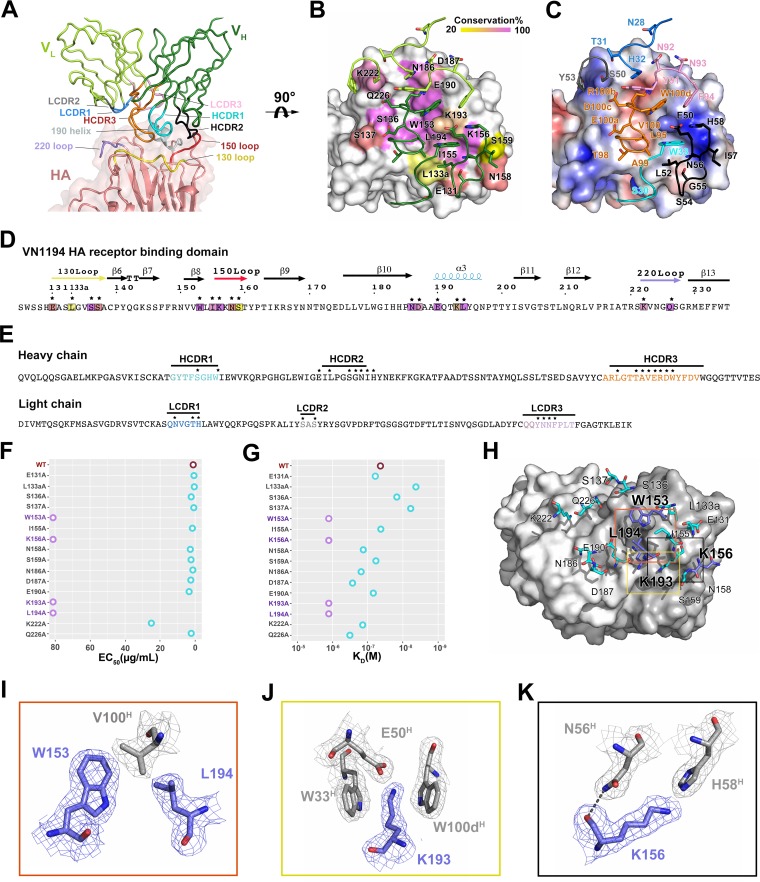
13D4 Fab binds to the membrane-distal region of the globular head of HA at nearly 45° to the 3-fold axis of the HA trimer (Fig. 1F). The HAhr-13D4 interaction buries 857 Å2 of surface area on HA1, as calculated by PISA (37) (Table 2). The heavy chain mediates most of the HAhr-13D4 contacts (75.6% of the total buried surface area of HA1, i.e., 648 Å2 out of 857 Å2). All three loops of the CDR on the 13D4 heavy chain (HCDR) bind the RBS, which is framed by four structural elements: the 130 loop, the 150 loop, the 190 helix, and the 220 loop (Fig. 2A). We found that HCDR1 binds to the ridge between the 130 loop and the 150 loop, HCDR2 binds to the 150 loop, and HCDR3 binds deeply into the negatively charged RBS core region. In contrast, the light-chain CDR (LCDR) loops mostly make contact with the residues outside the RBS, i.e., LCDR1 to -3 bind to the regions near the 190 helix and the 220 loop (Fig. 2A to C). Together, these CDR recognitions define a conformational epitope comprising four stretches of amino acids: Glu131 to Ser137, Trp153 to Ser159, Asn186 to Leu194, and Lys222 to Gln226. These amino acid stretches are highly conserved (83%) among all H5 isolates (n = 5,693; data collected before 2016 from the NCBI Influenza Virus Resource) (Fig. 2B and Table 3). Our observations are also consistent with our previous data on the neutralization breadth and potency of MAb 13D4. In this complex structure, extensive interactions are established, with 16 hydrogen-bonding contacts and 6 salt-bridging linkages (Table 2) formed between the side chains of the epitope residues (Fig. 2D) and the residues of 13D4 Fab (Fig. 2E).
TABLE 2.
Interface identification and interaction analysis of 13D4:HAhr by PISA Program
| HA | 13D4 | BSA (Å2)a | % buried areab | Typec | 13D4 | Distance (Å) |
|---|---|---|---|---|---|---|
| Interface | ||||||
| A:GLU131 | H chain | 19.34 | |||| | |||
| A:LEU133a | H chain | 91.94 | |||||||| | |||
| A:GLY134 | H chain | 1.81 | || | |||
| A:VAL135 | H chain | 29.28 | |||| | |||
| A:SER136 | H chain | 11.94 | |||||||||| | |||
| A:SER137 | H chain | 14.12 | || | |||
| A:SER145 | H chain | 14 | ||| | |||
| A:TRP153 | H chain | 18.69 | |||||||| | |||
| A:ILE155 | H chain | 15.75 | |||||||| | |||
| A:LYS156 | H chain | 40.75 | |||||||||| | |||
| A:LYS157 | H chain | 11.39 | || | |||
| A:ASN158 | H chain | 34.09 | ||| | |||
| A:SER159 | H chain | 47.19 | |||||| | |||
| A:ASN186 | H chain | 4 | || | |||
| A:ASP187 | H chain | 19.52 | |||| | |||
| A:ALA189 | H chain | 22.76 | |||| | |||
| A:GLU190 | H chain | 35.92 | |||||||| | |||
| A:TRP192 | H chain | 9.03 | || | |||
| A:LYS193 | H chain | 132.46 | |||||||||| | |||
| A:LEU194 | H chain | 28.12 | ||||||||| | |||
| A:GLN196 | H chain | 5.88 | | | |||
| A:LYS222 | H chain | 16.98 | || | |||
| A:GLN226 | H chain | 13.48 | |||| | |||
| A:SER227 | H chain | 9.97 | |||| | |||
| A:ASN186 | L chain | 16.54 | ||||||| | |||
| A:ASP187 | L chain | 25.96 | |||||| | |||
| A:ALA188 | L chain | 2.95 | | | |||
| A:ALA189 | L chain | 50.32 | ||||||| | |||
| A:THR192 | L chain | 15.79 | ||| | |||
| A:LYS193 | L chain | 19.90 | || | |||
| A:THR219 | L chain | 69.97 | || | |||
| A:LYS222 | L chain | 59.27 | |||| | |||
| A:SER227 | L chain | 6.15 | ||| | |||
| Interaction | ||||||
| A:LYS156[O] | H | H:ASN56[ND2] | 3.05 | |||
| A:SER159[OG] | H | H:ILE57[N] | 2.79 | |||
| A:ASP187 [OD1] | H | H:ARG100b[NE] | 2.75 | |||
| A:ASP187[OD2] | H | H:ARG100b[NH2] | 2.84 | |||
| A:LYS193[NZ] | H | H:GLU50[OE2] | 2.77 | |||
| A:ASN158[N] | H | H:SER54[O] | 2.91 | |||
| A:SER159[N] | H | H:GLY55[O] | 3.32 | |||
| A:SER159[OG] | H | H:GLY55[O] | 3.10 | |||
| A:ASN158[N] | H | H:ASN56[OD1] | 3.11 | |||
| A:SER159[N] | H | H:ASN56[OD1] | 2.76 | |||
| A:SER159[OG] | H | H:ILE57[O] | 3.43 | |||
| A:SER136[OG] | H | H:GLU100a[OE1] | 3.71 | |||
| A:SER136[OG] | H | H:GLU100a[OE2] | 3.53 | |||
| A:SER137[N] | H | H:GLU100a[OE2] | 2.83 | |||
| A:SER137[OG] | H | H:GLU100a[OE2] | 3.27 | |||
| A:LYS222[NZ] | H | L:SER50[OG] | 3.24 | |||
| A:ASP187[OD2] | S | H:ARG100b[NE] | 3.25 | |||
| A:ASP187[OD1] | S | H:ARG100b[NE] | 2.75 | |||
| A:ASP187[OD2] | S | H:ARG100b[NE2] | 2.84 | |||
| A:ASP187[OD1] | S | H:ARG100b[NE2] | 3.23 | |||
| A:LYS193[NZ] | S | H:GLU50[OE1] | 3.54 | |||
| A:LYS193[NZ] | S | H:GLU50[OE2] | 2.77 |
BSA, buried surface area.
|, 10%.
H, hydrogen bond; S, salt bridge.
FIG 2.
Interaction analysis of the 13D4:HAhr immune complex. (A) Enlarged view of the interface of the 13D4:HAhr complex. (B) Sequence conservation of the 13D4 epitope. HAhr is shown in surface representation with epitope residues colored according to their conservation values. (C) The electrostatic potential surface of HA is depicted in gradient color: red, negative, −4 kT; blue, positive, +4 kT; white, neutral. Contacting residues of the 13D4 CDR loops are in stick form for side chains and cartoonform for main chains. Panels B and C are shown horizontally rotated 90° from panel A. (D and E) Sequences of the RBS domain and 13D4 CDR, respectively. The residues in the interface are marked with asterisks and are depicted with the same color scheme as in panels B and C. (F) Reactivity profiles of HA and its mutants against 13D4 antibody measured through sandwich ELISA. The EC50 values were calculated by sigmoidal fitting as shown in Fig. 3C. The EC50 is plotted as circles along the horizontal axis for each protein. Purple circles, EC50 of >80 μg/ml; cyan, <30 μg/ml; brown, wild-type HA. (G) Binding affinity measurements of HA and its mutants against 13D4 by SPR. The kinetic constants between bnAb 13D4 and W153A, K156A, K193A, and L194A mutants were not determinable and are reported as higher than 1,000 nM. The calculated affinity constants and fitting results are depicted in Fig. 3D. The KD values are plotted as circles along the horizontal axis for each protein. Purple circles, KD of >1,000 nM; cyan, <500 nM; brown, wild-type HA. (H) The 13D4 epitope sites for HA are depicted in stick form on the molecular surface of 13D4 Fab. The epitope residues were identified by the PISA server (Table 2). (I to K) Enlarged views of the interactions for W153 and L194 (red box), K193 (yellow box), and K156 (black box) from the corresponding boxes in panel H. Sample electron density (2Fo-Fc) maps contoured at 1 σ above the mean are shown for all the residues. Hydrogen bonds are shown as black dashed lines.
TABLE 3.
Sequence conservation of 13D4 epitope residues in all H5 isolatesa
| Position | Amino acid type (%) |
|
|---|---|---|
| 13D4 epitope residue | Other | |
| 131 | E (71.0) | D (25.0), Y/N/T/G (4.0) |
| 133a | L (33.8) | S (58.5), M/G/K/A/V/P (7.7) |
| 136 | S (99.4) | T/R/G (0.6) |
| 137 | S (75.1) | A (24.6), P/T/L/V (0.3) |
| 153 | W (100) | |
| 155 | I (86.7) | T (11.7), V/L/N (1.6) |
| 156 | K (99.3) | E/Q/R/N/T/I/G (0.7) |
| 158 | N (75.5) | D (22.8), S/G/Q/E/Y/I/K (1.7) |
| 159 | S (19.0) | N (64.0), D (16.1), T/A/H/G (0.9) |
| 186 | N (99.0) | S/K/T/A/D/I (1.0) |
| 187 | D (87.9) | N (11.1), H/G/S/Y (1.0) |
| 190 | E (99.7) | K/H/A/D/V (0.3) |
| 193 | K (50.0) | R (38.4), N (6.9), M/D/T/G/QE/Q/S (4.7) |
| 194 | L (95.5) | I (4.3), V/F/S (0.2) |
| 222 | K (89.3) | Q (9.0), R/E/P/N (1.7) |
| 226 | Q (99.7) | L/R/K/P (0.3) |
| Avg | 82.8 | |
The cumulative number deposited in the NCBI Influenza Database by 2016 was 5,693.
Structure-based mutational analysis of the 13D4 neutralization sites.
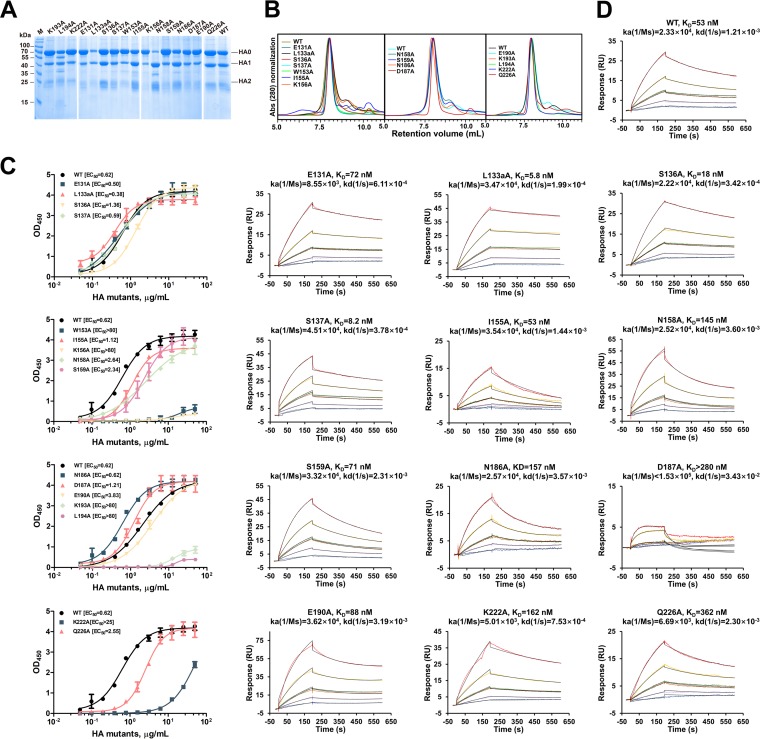
In the HAhr:13D4 complex structure, 16 HA residues at the interface interact with the 13D4 Fab through their side chains (Table 2). To identify the residues critical for 13D4 binding, alanine-scanning mutagenesis was carried out on all 16 amino acids. All of the unprocessed HA mutants were detected in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 3A) and resolved as trimers in solution, similar to wild-type (WT) HA (Fig. 3B). We used 50% effective concentration (EC50) (enzyme-linked immunosorbent assay [ELISA]) and affinity (surface plasma resonance [SPR]) measurements to confirm the importance of each mutation. Notably, 4 mutations (W153A, K156A, K193A, and L194A) dramatically decreased the binding of 13D4 with HA, with the EC50 more than 20 times higher for these mutants than for WT HA (Fig. 2F and G and 3C and D). The association/disassociation SPR responses of the four mutants to MAb 13D4 were undetectable. The other mutants showed affinities similar to that of WT HA, and two mutants (L133aA and S137A) showed significantly enhanced binding affinities to 13D4, with affinity constants (KD) of ∼53 nM to 5 to 8 nM (Fig. 3D).
FIG 3.
Characterization of HA mutants with structure-based alanine substitutions in 13D4 binding sites. (A) SDS-PAGE analysis of the purified HA mutants. Notably, most of the N158A mutant proteins were cleaved into HA1 and HA2. (B) HPLC profiles of the purified HA mutants. The mutants were resolved as major components in HPLC curves with the same retention volume as WT HA, indicating the proteins exist mainly as trimers in solution. Abs, absorbance. (C) The interaction of HA and its mutants against MAb 13D4 was tested by double-antibody sandwich ELISA, and EC50s were calculated by sigmoid trend fitting and plotted as in Fig. 2F. OD450, OD at 450 nm. The error bars represent standard deviations of two repeats. (D) Binding curves of HA, HA mutants, and MAb 13D4 in SPR. A total of 5 concentrations (8.93, 17.9, 35.7, 71.4, and 143 nM) of HA and its mutants (serial dilutions) were injected onto a MAb 13D4-bound chip. The kinetic constants between MAb 13D4 and HA/HA mutants are reported.
The functions of these four strategic residues are structural: Trp153 and Leu194 simultaneously interact with Val100H, where the superscript H denotes heavy chain, making hydrophobic interactions and van der Waals attractions (Fig. 2H and I); Lys193 makes contact with Glu50H through hydrogen bonding and salt bridging, and the stem of its hydrophobic side chain is stabilized by two flanking benzpyrole rings derived from Trp33H and Trp100dH (61) residues (Fig. 2H and J); and Lys156 binds to Asn56H via one hydrogen bond (Fig. 2H and K). Together, Trp153 and Lys156, located on the 150 loop, and Lys193 and Leu194 on the 190 helix are critical for 13D4 neutralization. These residues are highly conserved (Trp153, 100%; Lys156, 99.3%; Lys/Arg193, 88.4%; Leu194, 95.5%) among all H5 isolates, and the RBS localization (Fig. 2B and Table 3) determines the broad neutralization specificity of the 13D4 epitope. These results are also consistent with our previous 13D4 neutralization experiment on virus escape, where we replaced Lys156 and Lys193 with Glu and Met, respectively. In addition, in the present study, we found alanine replacement of the other residues, such as Asn158, Asn186, Asp187, Lys222, and Gln226, could also perturb 13D4 binding to HA to some extent, with 5-fold lower affinity with respect to WT HA (Fig. 3D). These residues, which line the outside the RBS (Fig. 2B), might assist in antibody binding but do not appear to be critical for binding.
13D4 HCDR3 adaptive fitting to the RBS.
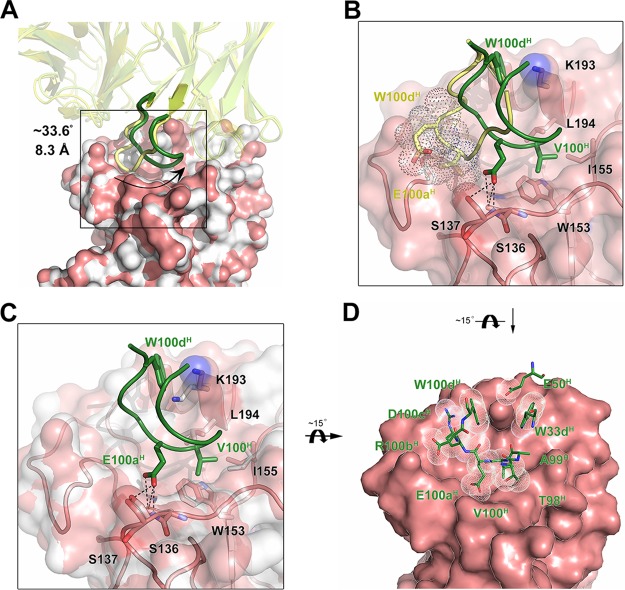
Similar to other reported RBS-directed bnAbs, the HCDR3 loop of 13D4 binds deeply within the RBS groove. Notably, structural superimposition of HA-bound and unbound 13D4 Fabs packing in the crystals revealed substantial conformational changes at the HCDR3 loop (Fig. 1A). Amino acids 93 to 101 of the HCDR3 loop of the free Fab would clash with the 220 loop of receptor binding domain (RBD) if 13D4 underwent direct binding to HA (Fig. 4A and B). To avoid this collision, a subloop, aa 95 to 101, swings from the 220 loop toward the 190 helix at about 33.6°, with Arg94H and Ala99H acting as mechanical fulcrums (Fig. 4A; see Movie S1 in the supplemental material). There, the C-α atoms of Val100H and Glu100aH move 8.3 Å and 7.6 Å, respectively, away to create hydrogen bonding and hydrophobic interactions with Ser136, Ser137, Trp153, Leu194, and Ile155 (Fig. 4B). The conformational fitting over the RBS may determine the broad yet specific nature of MAb 13D4, and this mode of binding is rarely observed in other bnAb-mediated binding. Indeed, in the literature, the HCDR3 loop of MAb H5.3 has been reported to rotate 90° in situ between its unliganded and liganded positions (38), which is distinct to the conformational change of MAb 13D4.
FIG 4.
Conformational change in 13D4 HCDR3 and HA, with HCDR3 adapting to fit into the RBS. (A) The free 13D4 and unbound HA monomer (from PDB accession no. 2IBX) were superimposed on corresponding components of the 13D4:HAhr complex. Fabs are depicted in cartoon mode, with free Fab in yellow and bound Fab in forest green. HA molecules are shown in surface presentation, with unbound HA in white and bound HA in salmon. (B) Enlarged view of the HCDR3 rearrangement (boxed in panel A). For clarity, other parts of the interaction from panel A have been omitted. Hydrogen bonds are shown as black dashed lines. W100dH and E100aH in the free Fab are shown in transparent sphere dot representation, demonstrating clashing with the 220 loop. (C) Conformational changes to HA while binding with 13D4. For clarity, other parts of the interaction from panel A are omitted. (D) Eventual fine fitting of HCDR3 to the RBS. The non-H atoms of HCDR3 up to 4.5 Å from HA are shown in dot sphere representation.
Minor conformational changes were also observed in the RBS during HCDR3 binding; most notably, the side chain of Lys193 rotates to accommodate the gap created by the benzpyrole rings of Trp100dH and Trp33H and donates a hydrogen bond to Glu50H. There is also subtle perturbation to the side chain of Ile155, which constitutes a hydrophobic cavity, together with Trp153 and Leu194, for the binding of Val100H of HCDR3 (Fig. 4C). Compared to the free Fab structure, the fitted conformation of the HCDR3 loop against the RBS might be induced energetically by the spatial restriction of the RBS structural contour (Fig. 4D; see Movie S1 in the supplemental material). These interactive conformational changes manifest an induced fit during HA-13D4 binding, and, given that W153A and L194A mutations abrogate this HA-13D4 binding, it is likely that these residues are critical for the HA-13D4 interaction (Fig. 1F and 2A).
Comparison with receptor analogue and other H5 head region antibodies.
We next sought to compare the 13D4:HAhr structure with the structure of an avian receptor analogue (LSTa [3′-sialyl-N-acetyllactosamine]) bound to HA (PDB accession no. 3ZP0) (39) and with other immune complexes that contain H5 head region antibodies. Overall, the tip residues E100aH and V100H of the 13D4 HCDR3 loop recapitulate the receptor analogue binding scenario (Fig. 5A to D).
FIG 5.
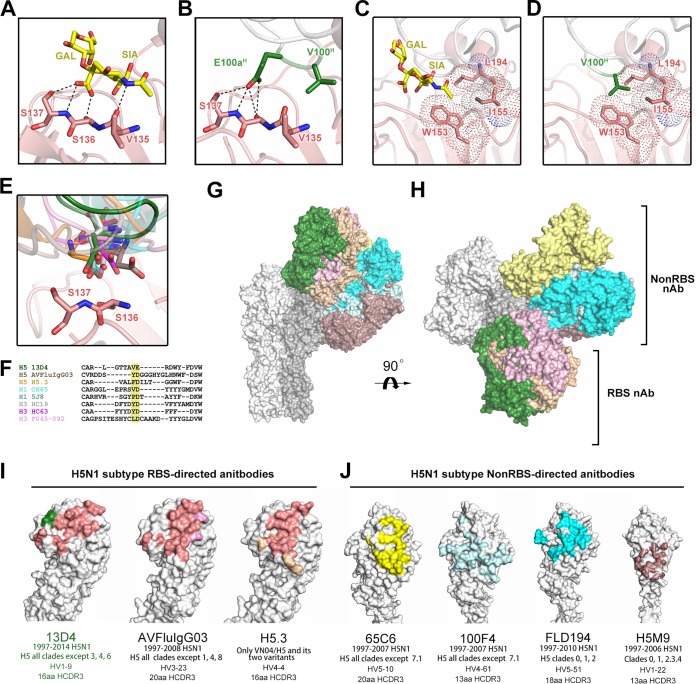
Receptor mimicking of 13D4 and footprint comparison of 13D4 and other H5 neutralizing antibodies. (A) Hydrogen bonding between receptor analogue LSTa and the RBS as shown in the co-crystal structure (PDB no. 3ZP0). (B) Hydrogen bonding between the 13D4 HCDR3 tip and the RBS. (C) The carbohydrate group of the LSTa SIA terminus stretches into a hydrophobic cavity enfolded by side chains of Trp153, Leu194, and Ile155. (D) V100H of HCDR3 mimics the hydrophobic interaction with the RBS, resembling the carboxylate group of the LSTa SIA terminus. (E) Overlay of the 13D4 HCDR tip with representative RBS-directed neutralizing antibodies. The Glu at the tip of 13D4 HCDR3 (green) is different from the Asps of other antibodies; the longer side chain of Glu forms four hydrogen bonds with S136 to S137, more than the hydrogen bond numbers formed by Asp at the HCDR3 tips of other antibodies: H5.3 (PDB accession no. 4XNM; orange), CH65 (3SM5; cyan), 5J8 (4M5Z; teal), HC19 (2VIR; white), HC63 (1KEN; purple), Fo45-092 (4O5I; brown), and AVFluIgG03 (5DUP; pink). (F) Sequence alignment of HCDR3 RBS-directed antibodies highlighting the Glu at the tip of 13D4 HCDR3, which is distinct from those of other antibodies. The same color scheme as in panel E was used. (G) Comparison of the binding sites of 13D4 and other H5-specific antibodies that recognize the HA1 globular head domain. The immune complex structures are overlaid in surface mode: 13D4 (green) in AVFluIg03 (PDB accession no. 5DUP; pink), H5.3 (4XNM; orange), 65C6 (5DUM; yellow), 100F4 (5DUR; pale cyan), FLD194 (5A3I; cyan), and H5M9 (4MHH; brown). (H) The view is horizontally rotated 90° with respect to panel G. (I) Footprints of three H5N1 RBS-targeting antibodies. The HA head region is shown in surface representation with the overlapping region in salmon and the unique binding residues in green, pink, and orange for 13D4, AVFluIgG03, and H5.3, respectively. (J) Footprints of four H5N1 non-RBS-directed antibodies. Table 4 details the neutralization breadths and evolving germ lines for these antibodies.
There are two major interactions between LSTa and the RBS, one through hydrogen bonding and one through hydrophobic interactions. In the first, triple hydrogen bonding occurs between the sialic acid carboxylate and Ser136 and Ser137 inside the RBS (Fig. 5A). The carboxylate of E100aH creates four hydrogen-bonding connections with Ser16 and Ser137, and the hydroxyl group of the side chain of Ser137 receives one more hydrogen bond than LSTa (Fig. 5B). Although other receptor-mimetic antibodies against H1 and H3 show similar hydrogen-bonding scenarios (PDB accession no. 4XNM, 3SM5, 4M5Z, 2VIR, 1KEN, and 4O58) (38, 40, 41), 13D4 uses Glu instead of Asp for its hydrogen-bonding network (Fig. 5A and F). This hydrogen-bonding modality differs from those of other H5 RBS-directed MAbs: H5.3 exclusively uses the main chain of Asp for H bonding, whereas AVFluIgG03 does not form hydrogen bonds via its Asp residue (Fig. 5E). Thus, the use of Glu to recapitulate hydrogen bonding instead of Asp in 13D4 is unique (Fig. 5F). No similar hydrogen bonding between 13D4 Fab and the RBS was observed, as the LSTa acetamide accepts a hydrogen bond from the main chain of Val135 (Fig. 5A and B).
For the second major interaction, the sialic acid of LSTa binds to a hydrophobic region created by the highly conserved Trp153, Ile155, and Leu194 in almost all H5 isolates (Table 3). The tip residue, Val100H, of the 13D4 HCDR3 loop binds to this region via its hydrophobic side chain, which is also observed in many other antibodies, such as CH65 and 2G1 (33, 42).
All of the reported structures for HA-H5 bnAbs show targeting of the HA head region, but they can be further classified into two groups: non-RBS-directed binding (MAbs 65C6, 100F4, FLD194, and H5M9 [43, 44]), with footprints shifted toward the distal membrane and twisting counterclockwise about 60° along the HA 3-fold axis with respect to the RBS (Fig. 5G, H, and J), versus RBS-directed binding, where 13D4, AVFLuIG03, and H5.3 all share similar binding orientations against HA and define an overlapping region (Fig. 5G to I). However, the key residues mediating binding of 13D4, AVFLuIG03, and H5.3 to HA are different, and this may account for the differences noted in the breadths of neutralization. 13D4 corresponds to sites more conserved than those of the other two MAbs (Fig. 5I and Table 4).
TABLE 4.
Summary of H5N1 neutralizing antibodies targeting the HA head region
| MAb parameter | Value |
||||||
|---|---|---|---|---|---|---|---|
| 13D4 | AVFluIgG03 | H5.3 | 65C6 | 100F4 | FLD194 | H5M9 | |
| Neutralization breadth | 1997–2014 H5N1 | 1997–2008H5N1 | Only VN04/05 and its two variants | 1997–2007 H5N1 | 1997–2007 H5N1 | 1997–2007 H5N1 | 1997–2010 H5N1 |
| H5, all clades except 3, 4, 6 | H5, all clades except 1, 4, 8 | H5, all clades except 7.1 | H5, all clades except 7.1 | Clades 0, 1, 2. 3 .4 | H5, all clades except 0, 1, 2 | ||
| Source | Mouse | Human | Human | Human | Human | Human | Mouse |
| PDB accession no. for immune complex structure | 6A0Z | 5DUP | 4XNM | 5DUM | 5DUR | 5A3I | 4MHH |
| HA for cocrystallization | VN1194 HA | Anhui05 HA | VN1203 | Anhui05 HA | Anhui05 HA | VN1203 | VN1203 HA |
| Resolution (Å) | 2.33 | 3.05 | 2.51 | 3.00 | 2.80 | 2.89 | 3.60 |
| Antibody footprint on HA | E131, L133a, S136, S137, W153A, I155A, K156, N158, S159, N186, D187, E190, K193, L194, K222, Q226 | E131, S133, S133a, V135, S136, S137, T144, P145, W153, I155, K156, K157, N158, N159, K193, L194, K222, Q226 | Y98, L133a, V135, S136, S137, Q142, G143, K144, S145, W153, I155, K156, N158, S159, E190, K193, L194, G225, Q226 | I121, Q122, I123, P125, S125b, D129, P162, T163, K165, R166, S167, Y168, N169, T171, N172, N244, E246 | D77, E78, I80, N81, H117, E119, K120, I121, Q122, K125a, Y141, Q142, R149, T171, N172, Q173, E174, Y256, K259, V260a, K261 | I80, N81, K120, I121, Q122, I123, I124, S125b, S126, S128, Y141, Q142, K144, N150, R166, S167, Y168, N169, T171, E255, Y256, K259 | D53, D55, G55a, V56, K57, R62, E78, N81, P83, E83a, H117, E119, E273, Y274, N276, N278 |
| Conservation of critical residues for antibody binding (%) | W153 (100), K156 (99.3), K/R193 (50/38.4), L194 (95.5) | E131 (71.0), S133 (97.4), K156 (99.3, N159 (64.0) | K/R144 (12.3/48.5), K193 (50) | – | D77 (99.4), E119 (98.9) | Q122 (84.7) | D53 (62.4), E83a (99.6), Y274 (99.7), N276 (91.5) |
| Method for key amino acid identification | SPR and ELISA on alanine scanning mutants | Pseudovirus-based neutralization assay | Sequence alignment of multiple reactive strains | Pseudovirus-based neutralization assay | Pseudovirus-based neutralization assay | Neutralization activity using isolated virus | Yeast cell display |
| HCDR3 sequence | ARLGTTAVERDWYFDV | VRDDSYDGGGHYGLHNWFDS | ARVALFDILTGGWFDP | AYHRRGHFYGSGSAWDWFES | ARALLTTVTTFEY | ARHASCSARSCYWGPVDY | VRNYGSSYGYFDV |
| HCDR3 length (aa) | 16 | 20 | 16 | 20 | 13 | 18 | 13 |
| VH germ line | 1-9 | 3-23 | 4-4 | 5-10 | 4-61 | 5-51 | 1-22 |
A key 13D4 binding site associated with antigenicity shift from H5N1 to other H5 viruses.
In our previous study, we showed that MAb 13D4 conferred broad neutralization against all representative H5 isolates identified between the years 1997 and 2009 and was verified as a potential therapeutic candidate for curing H5-infected animals from lethal infection following H5 virus challenge. To confirm the neutralization breadth since our previous study, we tested the emergent H5 subtype viruses isolated from 2012 to 2014 using HA inhibition (Fig. 6A). The data showed that our 13D4 MAb can inhibit hemagglutination of H5N1, but not H5N6 and H5N8, two viral subtypes that first emerged in 2013 and 2006, respectively (Fig. 6A, table). We then analyzed the antigenic shifts of the emergent H5N6 and H5N8 from their ancestral H5N1 (62) using escape neutralization of 13D4. Sequence alignment of the H5 HAs revealed that only 2 residues, D187N and K/R193N, of the 15 13D4 epitope residues are mutated between H5N1 and H5N6/H5N8, both located on the 190 loop (Fig. 6A). In our structure and mutagenesis analysis, Asp187 was not involved in 13D4 binding, whereas the side chain of Lys193 was shown to play a strategic role in the interaction between 13D4 and HA (Fig. 2F, G, and J). Moreover, 13D4 maintains strong neutralization against H5N1 viruses that retain Arg193 in HA, such as HK/5923/2012 and Zhejiang/98/2014 isolates, and also those viruses bearing the Lys193 residue (Fig. 6A). Superimposition of the HA structures with either Lys193 or Arg193 showed similar interaction with Trp33H, Glu50H, and Trp100dH of 13D4 (Fig. 6B). However, Asn193 of H5N6/H5N8 has a shorter side chain than Lys/Arg and thus abrogates 13D4 recognition, possibly because it fails to establish the critical interaction for 13D4 binding (Fig. 6C). Polymorphic analysis of residue 193 among 5,693 H5 isolates revealed high conservation of the Lys/Arg residue at this position, with 88.4% identity (Table 5). Lys/Arg193 is completely conserved in all 13D4-reactive H5 isolates (Table 3).
FIG 6.
Molecular determinants for antigenic shift from H5N1 to other genotypes. (A) RBD sequence alignment of six H5 subtype viruses and HI titer assay of 13D4 or ferret antiserum. The epitope residues of 13D4 are boxed in cyan. Disparate residues between H5N1 and H5N6/H5N8 are indicated by arrowheads (below). Four key sites of 13D4 binding are designated by black stars. The overlapping subset of mutated and key binding residues is designated by the red asterisk. Dots denote that the residue is identical with that of the first VNM sequence. (B) Arg (R) 193 shares a similar side chain rotamer and putative 13D4 interaction mode with Lys (K) 193. Free HA structures of A/duck/Egypt/10185SS/2010 (PDB accession no. 5E30) and A/Vietnam/1194/2004 (PDB accession no. 2IBX), bearing R193 and K193, respectively, are superimposed with the 13D4:HAhr complex. (C) N193 differs from K193 and fails to interact with 13D4. Free HA (A/Sichuan/26221/2014; PDB accession no. 5HU8) containing N193 was overlaid with the free and bound HAs containing K193, as shown in panel B. Dotted ovals highlight the interaction in 193 position. The dashed lines are hydrogen bonds.
TABLE 5.
Polymorphisms of residue 193 in H5 isolates that emerged 2004 to 2016
| Yr | Conservation (%)a |
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H5N1 |
H5N2 |
H5N3 |
H5N5 |
H5N6 |
H5N8 |
Total H5 |
|||||||||||||||
| Lys/Arg | Asn | n | Lys/Arg | Asn | n | Lys/Arg | Asn | n | Lys/Arg | Asn | n | Lys/Arg | Asn | n | Lys/Arg | Asn | n | Lys/Arg | Asn | n | |
| 2004 | 96.3 | 0.1 | 640 | 93.4 | 0.7 | 137 | 100 | 0 | 29 | 100 | 0 | 1 | 100 | 0 | 4 | 100 | 0 | 4 | 93.8 | 0.8 | 1,051 |
| 2006b | 100 | 0 | 4 | 83.3 | 16.7 | 6 | |||||||||||||||
| 2008 | 81.2 | 0.3 | 2,564 | 95.7 | 2.3 | 427 | 98.5 | 65 | 100 | 0 | 5 | 100 | 0 | 4 | 85.7 | 14.3 | 7 | 94.0 | 1.0 | 3,433 | |
| 2012 | 92.6 | 1.6 | 3,517 | 94.1 | 2.8 | 556 | 95.5 | 90 | 100 | 0 | 12 | 100 | 0 | 4 | 47.4 | 52.6 | 19 | 91.5 | 1.2 | 4,665 | |
| 2013c | 5.8 | 9.3 | 18 | ||||||||||||||||||
| 2014 | 82.0 | 0.3 | 3,971 | 9.8 | 86.7 | 68 | 14.0 | 86.0 | 71 | 85.9 | 4.2 | 5,381 | |||||||||
| 2016 | 86.6 | 0.6 | 4,162 | 85.6 | 12 | 667 | 95.9 | 97 | 97.4 | 0 | 39 | 5.9 | 80.5 | 118 | 11.6 | 88.4 | 86 | 88.4 | 6.9 | 5,693 | |
Conservation of the amino acid(s) in all isolate sequences. The statistical result was based on the number of isolate sequences deposited in the NCBI Influenza Database by the specified year.
First year when N193 emerged as H5N8.
First year when N193 emerged as H5N6.
DISCUSSION
Although H5 subtype influenza viruses still exclusively circulate in avian hosts, cross-species infection in sporadic human cases led to severe disease, with a >50% mortality rate. No effective antiviral is available for the treatment of human infection with H5N1 or other highly pathogenic H5 subtype viruses. Like other influenza viruses, H5 subtype virus evolves rapidly in the field. Development of broadly neutralizing antibodies has been considered one of the therapeutic means to respond to the potential threat of highly pathogenic avian influenza viruses. We previously characterized an H5-specific broadly neutralizing antibody, 13D4, and showed potent therapeutic activity against H5N1 infection in mice (21). Here, we report the structure and function of this potent neutralizing antibody against H5N1 isolates. We show that HCDR3 adaptively fits into the shallow groove of the HA RBS and highlight the key residues involved in this binding.
Comparing sequence conservation and genetic variation, there are three highly conserved regions on HA that are associated with the bnAbs, including the RBD, the stem region, and the vestigial esterase region (45). Most isolated bnAbs target the RBD, such as 5J8, 1F1 (63), and CH65, with single-subtype-specific breadth, and C05, S139/1, and F045-092, with cross-subtype activity. The globular head domain bears the conserved RBS, which mediates host specificity and is responsible for the antigenic shift and antigenic drift in the major antigen determinants, which leads to escape from viral neutralization. Notably, bnAbs usually demonstrate receptor mimicry through various binding strategies using the HCDR3 loop. In the case of the H5 subtype, two known bnAbs, AVFluIgG03 and H5.3, are RBS-directed antibodies, and HCDR3 inserts directly into the binding groove. These RBS-directed antibodies neutralize all clades of H5 (except 1, 4, and 8), as well as VN/1203 and two H5 rdt variants only, respectively (Fig. 5I and J). Our 13D4 can neutralize all major H5N1 clades isolated between 1997 and 2014. Structure-guided mutagenesis on 13D4 binding sites has identified four highly conserved amino acids in its targets, which may account for the wide breadth of coverage. 13D4 mimics sialic acid-containing receptor binding, similar to other bnAbs, such as 5J8, CH65, and F045-092. However, we showed that it identifies a unique Val-Glu motif instead of the known Val-Asp to recapitulate the hydrogen-bonding network of receptor binding. Glu harbors a C atom side chain that is longer than that of Asp, which allows it to preferentially donate hydrogen bonds to Ser136 and Ser137 (Fig. 5E). The structural mimicry of the receptor analogue might explain the broad neutralization achieved by 13D4, in concert with conservation of the key sites.
Structural comparison of the unbound and bound 13D4 Fab highlights the significant conformational changes that occur in HCDR3 (Fig. 4A and B), and this rearrangement simultaneously induces subtle side chain rotamer changes on Lys193 and Ile155 to accommodate its fitting (Fig. 4C). Unlike changes invoked by other bnAbs, 13D4 HCDR3 in its original unbound state would bump into the 220 loop, with the other loops also contacting HA (Fig. 4B). We reason that this physical obstruction would cause the HCDR3 loop to “flap away” from the 220 loop and then adaptively fit into the RBS groove. Eventually, the side chains of Ala99H and W100dH would almost fill the space of the RBS rather than the receptor analogue. It is worth noting that fitting is further optimized when the side chain of Asp100cH inserts into the gap instead of extending outside the RBS (Fig. 4D). Intriguingly, replacement of Ser136 or Ser137 with Ala does not decrease but exceptionally enhances the binding affinity of 13D4 for HA (Fig. 2G), as shown in the smoothed dissociation curve of the SPR trace and the decreased dissociation rate (Fig. 3D); this indicates that the initial stage of conformational rearrangement might be related to the final stable binding conformation. We synthesized several oligopeptides according to the 13D4 HCDR3 sequence but found no activity against HA in SPR or isothermal titration calorimetry (ITC) (data not shown), implying that the HCDR3 loop alone is insufficient to undergo the proper conformational changes required for the action of the 13D4 antibody. Thus, a more rational strategy should be devised, such as the use of pharmacophore-based designs or sequential molecular-dynamic simulation (46). Nevertheless, our structures of the two-state HCDR3 loops, the key conserved neutralization sites, and the conformational rearrangement will aid in the design of efficacious polypeptides or therapeutic drugs against the H5 subtype virus.
The threat of HPAI H5 subtype virus spread to humans is still present, given the recent emergence of new H5N1 variant subclades (2.3.4.4) (47, 48) and other reassortants, including H5N2, H5N3, H5N5, H5N6, and H5N8, in poultry in Asia, Europe, and North America. Human infections with the H5N6 strain have been observed (49). The H5N6 virus has undergone more than 17 site mutations (50) in the HA1 primary sequence compared with the original H5N1 in infected humans (Fig. 6A). Most of these mutated sites do not reside in the 13D4 binding sites, with only position 193 related to 13D4 neutralization activity (Fig. 6A). Although HA with Lys193 or Arg193 is reactive to 13D4, 13D4 lost neutralizing activity against H5N6 and H5N8 viruses, which bear an Asn193. As shown in the unbound and bound 13D4:HAhr complex structures, Lys193 or Arg193 may change its side chain rotamer to create elaborate hydrophobic and salt bridge connections with the two benzopyrrole rings of Trp and one negatively charged Glu; mutation of Lys193 or Arg193 to Asn193 abrogates the interaction and allows the virus to escape 13D4 neutralization. Thus, acquisition of polymorphisms at aa 193 might be associated with the evasion of antibody immunity of H5N6, which was considered to have been built up through consistent massive vaccine use against H5N1 virus since 2005 in Asia. It is worthy of note that residue 193 is located at an acknowledged classic antigenic site (the Sb site for H1 [51, 52] and site B for H3 [53, 54]) on the HA RBD; however, it is quite distinct in polymorphism among H1, H3, and H5 subtypes, i.e., the amino acid types with the highest frequencies are Ser (∼78%) and Thr (∼12%) for H1; Phe (∼56.5%), Asn (∼17%), and Ser (∼22%) for H3; and Lys (∼50%) and Arg (38.4%) for H5. Other key residues involved in 13D4 interaction are also attributed to major antigenic sites of other genotypes, i.e., residues 156 and 194 residing in the Sa/Sb site for H1 or site B for H3, whereas residue 153 is located in the RBS of all genotypes. The structural basis characterized from bnAb 13D4 and the HA complex may provide a molecular basis for vaccine strain selection and design of therapeutic reagents for pan-H5 protection.
MATERIALS AND METHODS
Cloning, expression, and purification of HA.
The HA gene of A/Vietnam/1194/2004 (Viet04; accession no. EF541402), encoding HA1 aa 11 to 329 (H3 sequence numbering) and HA2 aa 1 to 174, was cloned into the baculovirus transfer vector pAcGP67B (BD Biosciences) using the Gibson assembly cloning method for baculovirus preparation. The construct contains an N-terminal gp67 secretion signal peptide, a thrombin cleavage site, a T4 fibritin “foldon” domain to assist in trimerization of the product, and a C-terminal 6-His tag for purification, as previously described. Site-directed HA mutants were constructed using a Mut Express II fast mutagenesis kit V2 (Vazyme). For the expression of recombinant HA proteins, Hi5 suspension cells (Invitrogen) were infected with recombinant baculovirus at a multiplicity of infection (MOI) of 5 to 8 at 28°C for 72 h. The supernatant was then dialyzed against phosphate-buffered saline (PBS), pH 7.4, and purified with Ni-nitrilotriacetic acid (NTA) resin (GE Healthcare) by the elution of 250 mM imidazole. HA proteins for crystallization were treated with thrombin to remove the C-terminal fusion elements and further digested with 200 mU/mg HA TPCK (tosylsulfonyl phenylalanyl chloromethyl ketone)-treated trypsin (Sigma-Aldrich) to cleave HA0 into HA1 and HA2. Trypsin was quenched with 2 mM phenylmethylsulfonyl fluoride (PMSF) (Beyotime). The purified HA proteins were mixed with loading buffer (50 mM Tris, pH 6.8, 2% SDS, 5% 2-mercaptoethanol, 0.01% bromophenol blue, 8% glycerol), boiled for 10 min, and subjected to SDS-PAGE.
Purification, crystallization, and structure determination.
The 13D4 Fab was prepared by papain digestion of MAb 13D4 (64) and purified with DEAE-5PW (Tosoh Bioscience, Tokyo, Japan), as described previously (55). HA was mixed with 13D4 Fab in a molar ratio of 1:3.2 and incubated at 37°C for 2 h. The immune complex was purified by gel filtration on a Superdex 200 column (GE Healthcare) in 10 mM Tris, pH 8.0, with 50 mM NaCl. The free 13D4 Fab and its complex with HA were concentrated to ∼20 and ∼8.5 mg/ml, respectively, for crystallization. The crystallization was implemented by sitting-drop vapor diffusion in the screening stage and hanging drop in microseeding optimization at 20°C. Crystals of 13D4 Fab were grown in 12.5% (wt/vol) polyethylene glycol (PEG) 3350, 150 mM potassium acetate, whereas the complex crystals were grown in 0.1 M Bis-Tris-propane, pH 6.5, 0.2 M sodium acetate, and 18% (wt/vol) PEG 3350. Crystal growth of the immune complex took about 3 months for final data collection. Crystals were cryoprotected in reservoir solution supplemented with 30% glycerol at 100 K. Diffraction data were collected at Shanghai Synchrotron Radiation Facility (SSRF) beamline BL17U using a Quantum-315r charge-coupled-device (CCD) area detector. Data sets were processed using the HKL-2000 program package (HKL Research, Inc.). According to the results from multiple input trials with proposed molecular mass, only a protein of 78 kDa (corresponding to one Fab [47 kDa] plus one HA head region [31 kDa], spanning aa 47 to 309 of HA1 in the final model) in one asymmetry unit gives a Matthews coefficient of 2.58 Å3/Da, related to 52.3% solvent content, with probabilities of 1.00 both for high resolution and overall, which indicates a protein(s) of about 78 kDa is harbored in the crystal (56). Then, we used this permutated complex of partial HA and one Fab for phase searching in molecular replacement (MR), which was implemented with the PHASER suite (65), using Phenix to find the initial phases. The search models for the light chain and heavy chain of 13D4 Fab were PDB accession no. 3CLE and 4KI5, respectively. The final 13D4 Fab model and head region of HA (PDB accession no. 2IBX) served for the MR search. The resulting models were manually built in COOT (66), refined with PHENIX (67), and analyzed with MolProbity (57). In brief, one round of rigid-body refinement was performed after MR. The refined models were manually modified in COOT; coordinates and individual B factors were refined in reciprocal space. Translation/libration/screw (TLS) refinement was performed in the later stages with autosearched TLS groups in PHENIX, which are listed in the REMARK 3 sections in the deposited PDB files. The data collection and structure refinement statistics are summarized in Table 1.
Cryo-EM of the HA:13D4 complex.
A 3-μl sample of HA:13D4 immune complex (4 mg/ml) was deposited onto a glow-discharged holey carbon Cu grid (Quantifoil). After blotting for 6 s, the grids were flash frozen in liquid ethane in an FEI Vitrobot. The grids were examined in an FEI G2 Tecnai F30 transmission electron microscope at 300 keV. The data were recorded on a Falcon II direct electron detector, with an electron dose of 30e−/Å2 and a nominal magnification of ×93,000. The defocus ranged from 2 to 3.7 μm, and the pixel size was 1.128. Because the sample dispersed poorly, the particles were manually boxed from the raw micrographs using Relion/1.4. CTF fitting. Reference-free 2D class averages were calculated using Relion/1.4.
BIAcore biosensor analysis.
The anti-mouse IgG Fc antibody was amine coupled to a CM5 sensor chip on a BIAcore 3000 (GE Healthcare). MAb 13D4 was then captured on the sensor surface at a flow rate of 30 μl/min in HBS buffer (10 mM HEPES, 150 mM NaCl, 3 mM EDTA, 0.005% Tween 20, pH 7.4). Injection of the HBS buffer served as a negative control. The kinetics of MAb 13D4 binding with HA and its mutants were measured at a flow rate of 30 μl/min in HBS buffer at 2-fold serially diluted concentrations (8.93, 17.9, 35.7, 71.4, and 143 nM). The flow durations were 200 s for the association stage and 10 min for dissociation. Association rates (ka), dissociation rates (kd), and affinity constants (KD) were calculated using BIAcore evaluation software. All experiments were repeated three times. The mean values of the ka, kd, and KD were reported.
Enzyme-linked immunosorbent assay.
The binding profiles of HA and its mutants against MAb 13D4 were evaluated by double-antibody sandwich ELISA. The wells of a 96-well plate were coated with 200 ng of 13D4 MAb prepared in PBS, pH 7.4, and incubated overnight at 4°C. HA proteins in half-serially diluted concentrations (starting from 2 μg/ml; diluted in PBS) were added to each well and incubated at 37°C for 30 min. This was followed by the addition of horseradish peroxidase (HRP)-conjugated MAb 8G9 (21) and a further 30-min incubation. The color was developed using tetramethylbenzidine substrate. The optical density (OD) was measured using a microplate reader (Sunrise; Tecan) at 450 nm with a reference wavelength of 620 nm. The EC50 was calculated at the half-point of the curve by polynomial fitting.
Gel filtration chromatography.
TSK Gel PW5000xl 7.8- by 300-mm columns (Tosoh Corporation) were equilibrated in PBS using a Waters high-performance liquid chromatography (HPLC) system. Samples of HA, HA mutants, 13D4 Fab, and the immune complex were dialyzed against PBS and diluted to 1 mg/ml before loading. The flow rate was maintained at 0.5 ml/min, and the absorbance at 280 nm was recorded.
Analytical ultracentrifugation.
Sedimentation velocity (SV) experiments were carried out at 20°C in a Beckman XL-A analytical ultracentrifuge equipped with absorbance optics and an An60-Ti rotor. HA and its 13D4 immune complex samples were diluted in PBS to an OD of ∼1.0 at 280 nm in a 1.2-cm light path. The rotor speed was set to 30,000 rpm according to the molecular weights of the control proteins. The sedimentation coefficient was obtained by the continuous sedimentation coefficient distrubution [c(s)] method using Sedfit software, kindly provided by P. Schuck (National Institutes of Health, Bethesda, MD, USA).
Hemagglutinin inhibition assay.
Half-serially diluted MAb 13D4 and anti-SZ ferret serum were incubated with 4 HA units of various live viruses in the wells of a 96-well plate. Red blood cells were added to a final concentration of 0.5%, and the plate was incubated at room temperature for 30 to 60 min. Button formation was scored as evidence of HA inhibition. The maximum dilution titers that produced a hemagglutinin inhibition (HI) phenomenon were reported.
Accession numbers.
The coordinates and structure factors for the HA:13D4 complex and 13D4 Fab have been deposited in the Protein Data Bank (accession no. 6A0Z and 6A0X).
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the use of beamline 17U of the SSRF for data collection and also thank J. Sivaraman and Xuhua Tang (National University of Singapore) for assistance with X-ray structure determination.
N.X., S.L., and Y.C. acknowledge funding support from the Chinese government: National Natural Science Foundation (grants 31670934 and 30901077), Fujian Provincial Science Fund for Distinguished Young Scholars and for Innovative Platform (grants 2017J07005 and 2014Y2004), Xiamen Haicang Science Foundation for Innovative Development (grant 350205Z20154007), and the Scientific Research Foundation of State Key Laboratory of Molecular Vaccinology and Molecular Diagnostics (grant 2016ZY005). H.C. thanks the Research Grants Council of the Hong Kong SAR (grants 7629/13M, 17103214, and 17154516).
S.L. and N.X. designed the study. Q.L., T.L., S.-Y.L., M.W., Y.Z., Z.Z., Q.Y., J.L., Z.L., and D.W. performed experiments. Q.L., T.L., Y.C., Q.Z., H.Y. Y.G., J.Z, H.C., S.L., and N.X. analyzed data. Q.L., T.L., and S.L. wrote the manuscript. Q.L., T.L., Y.C., Q.Z., H.Y., Y.G., J.Z., H.C., S.L., and N.X. participated in discussions and interpretation of the results. We all contributed to experiment design.
We declare that no competing interests exist.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00547-18.
REFERENCES
- 1.Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. 2006. Avian flu: influenza virus receptors in the human airway. Nature 440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 2.Yamada S, Suzuki Y, Suzuki T, Le MQ, Nidom CA, Sakai-Tagawa Y, Muramoto Y, Ito M, Kiso M, Horimoto T, Shinya K, Sawada T, Kiso M, Usui T, Murata T, Lin Y, Hay A, Haire LF, Stevens DJ, Russell RJ, Gamblin SJ, Skehel JJ, Kawaoka Y. 2006. Haemagglutinin mutations responsible for the binding of H5N1 influenza A viruses to human-type receptors. Nature 444:378–382. doi: 10.1038/nature05264. [DOI] [PubMed] [Google Scholar]
- 3.de Vries E, Guo H, Dai M, Rottier PJ, van Kuppeveld FJ, de Haan CA. 2015. Rapid emergence of highly pathogenic avian influenza subtypes from a subtype H5N1 hemagglutinin variant. Emerg Infect Dis 21:842–846. doi: 10.3201/eid2105.141927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park AW, Glass K. 2007. Dynamic patterns of avian and human influenza in east and southeast Asia. Lancet Infect Dis 7:543–548. doi: 10.1016/S1473-3099(07)70186-X. [DOI] [PubMed] [Google Scholar]
- 5.Xu X, Subbarao Cox NJ, Guo Y. 1999. Genetic characterization of the pathogenic influenza A/Goose/Guangdong/1/96 (H5N1) virus: similarity of its hemagglutinin gene to those of H5N1 viruses from the 1997 outbreaks in Hong Kong. Virology 261:15–19. doi: 10.1006/viro.1999.9820. [DOI] [PubMed] [Google Scholar]
- 6.Wan X-F, Dong L, Lan Y, Long L-P, Xu C, Zou S, Li Z, Wen L, Cai Z, Wang W, Li X, Yuan F, Sui H, Zhang Y, Dong J, Sun S, Gao Y, Wang M, Bai T, Yang L, Li D, Yang W, Yu H, Wang S, Feng Z, Wang Y, Guo Y, Webby RJ, Shu Y. 2011. Indications that live poultry markets are a major source of human H5N1 influenza virus infection in China. J Virol 85:13432–13438. doi: 10.1128/JVI.05266-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verhagen JH, Herfst S, Fouchier RA. 2015. Infectious disease. How a virus travels the world. Science 347:616–617. doi: 10.1126/science.aaa6724. [DOI] [PubMed] [Google Scholar]
- 8.Bi Y, Chen Q, Wang Q, Chen J, Jin T, Wong G, Quan C, Liu J, Wu J, Yin R, Zhao L, Li M, Ding Z, Zou R, Xu W, Li H, Wang H, Tian K, Fu G, Huang Y, Shestopalov A, Li S, Xu B, Yu H, Luo T, Lu L, Xu X, Luo Y, Liu Y, Shi W, Liu D, Gao GF. 2016. Genesis, evolution and prevalence of H5N6 avian influenza viruses in China. Cell Host Microbe 20:810–821. doi: 10.1016/j.chom.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 9.Yang L, Zhu W, Li X, Bo H, Zhang Y, Zou S, Gao R, Dong J, Zhao X, Chen W, Dong L, Zou X, Xing Y, Wang D, Shu Y. 2017. Genesis and dissemination of highly pathogenic H5N6 avian influenza viruses. J Virol 91:e02199–16. doi: 10.1128/JVI.02199-16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Vries E, Martin S, Pieter F, Charles B, Albert O. 2013. Influenza virus resistance to antiviral therapy. Adv Pharmacol 67:217–246. doi: 10.1016/B978-0-12-405880-4.00006-8. [DOI] [PubMed] [Google Scholar]
- 11.Kilbourne ED. 2006. Influenza pandemics of the 20th century. Emerg Infect Dis 12:9–14. doi: 10.3201/eid1201.051254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Webster RG, Govorkova EA. 2006. H5N1 influenza: continuing evolution and spread. N Engl J Med 355:2174–2177. doi: 10.1056/NEJMp068205. [DOI] [PubMed] [Google Scholar]
- 13.Stöhr K. 2005. Avian influenza and pandemics: research needs and opportunities. N Engl J Med 352:405–407. doi: 10.1056/NEJMe048344. [DOI] [PubMed] [Google Scholar]
- 14.Ungchusak K, Auewarakul P, Dowell SF, Kitphati R, Auwanit W, Puthavathana P, Uiprasertkul M, Boonnak K, Pittayawonganon C, Cox NJ, Zaki SR, Thawatsupha P, Chittaganpitch M, Khontong R, Simmerman JM, Chunsutthiwat S. 2005. Probable person-to-person transmission of avian influenza A (H5N1). N Engl J Med 352:333–340. doi: 10.1056/NEJMoa044021. [DOI] [PubMed] [Google Scholar]
- 15.Krammer F, Palese P, Steel J. 2015. Advances in universal influenza virus vaccine design and antibody mediated therapies based on conserved regions of the hemagglutinin. Curr Top Microbiol Immunol 386:301–321. doi: 10.1007/82_2014_408. [DOI] [PubMed] [Google Scholar]
- 16.Tan Y, He F, Kwang J. 2015. How can we develop universal H5N1 vaccines. Immunotherapy 7:713–716. doi: 10.2217/imt.15.42. [DOI] [PubMed] [Google Scholar]
- 17.Prabakaran M, Kolpe AB, He F, Kwang J. 2013. Cross-protective efficacy of bivalent recombinant baculoviral vaccine against heterologous influenza H5N1 challenge. Vaccine 31:1385–1392. doi: 10.1016/j.vaccine.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 18.Skehel JJ, Wiley DC. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem 69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 19.Weis W, Brown JH, Cusack S, Paulson JC, Skehel JJ, Wiley DC. 1988. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature 333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 20.Barbey-Martin C, Gigant B, Bizebard T, Calder LJ, Wharton SA, Skehel JJ, Knossow M. 2002. An antibody that prevents the hemagglutinin low pH fusogenic transition. Virology 294:70–74. doi: 10.1006/viro.2001.1320. [DOI] [PubMed] [Google Scholar]
- 21.Chen Y, Qin K, Wu WL, Li G, Zhang J, Du H, Ng MH, Shih JW, Peiris JS, Guan Y, Chen H, Xia N. 2009. Broad cross-protection against H5N1 avian influenza virus infection by means of monoclonal antibodies that map to conserved viral epitopes. J Infect Dis 199:49–58. doi: 10.1086/594374. [DOI] [PubMed] [Google Scholar]
- 22.Zuo T, Sun J, Wang G, Jiang L, Zuo Y, Li D, Shi X, Liu X, Fan S, Ren H, Hu H, Sun L, Zhou B, Liang M, Zhou P, Wang X, Zhang L. 2015. Comprehensive analysis of antibody recognition in convalescent humans from highly pathogenic avian influenza H5N1 infection. Nat Commun 6:8855. doi: 10.1038/ncomms9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen L-M, Santelli E, Stec B, Cadwell G, Ali M, Wan H, Murakami A, Yammanuru A, Han T, Cox NJ, Bankston LA, Donis RO, Liddington RC, Marasco WA. 2009. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol 16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee PS, Wilson IA. 2015. Structural characterization of viral epitopes recognized by broadly cross-reactive antibodies. Curr Top Microbiol Immunol 386:323–341. doi: 10.1007/82_2014_413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Velkov T, Ong C, Baker MA, Kim H, Li J, Nation RL, Huang JX, Cooper MA, Rockman S. 2013. The antigenic architecture of the hemagglutinin of influenza H5N1 viruses. Mol Immunol 56:705–719. doi: 10.1016/j.molimm.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 26.Air GM. 2015. Influenza virus antigenicity and broadly neutralizing epitopes. Curr Opin Virol 11:113–121. doi: 10.1016/j.coviro.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corti D, Lanzavecchia A. 2013. Broadly neutralizing antiviral antibodies. Annu Rev Immunol 31:705–742. doi: 10.1146/annurev-immunol-032712-095916. [DOI] [PubMed] [Google Scholar]
- 28.Tonegawa S. 1983. Somatic generation of antibody diversity. Nature 302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 29.Finn JA, Crowe JE Jr. 2013. Impact of new sequencing technologies on studies of the human B cell repertoire. Curr Opin Immunol 25:613–618. doi: 10.1016/j.coi.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finn JA, Koehler Leman J, Willis JR, Cisneros A, Crowe JE, Meiler J. 2016. Improving loop modeling of the antibody complementarity-determining region 3 using knowledge-based restraints. PLoS One 11:e0154811. doi: 10.1371/journal.pone.0154811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu NC, Wilson IA. 2017. A perspective on the structural and functional constraints for immune evasion: insights from influenza virus. J Mol Biol 429:2694–2709. doi: 10.1016/j.jmb.2017.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corti D, Cameroni E, Guarino B, Kallewaard NL, Zhu Q, Lanzavecchia A. 2017. Tackling influenza with broadly neutralizing antibodies. Curr Opin Virol 24:60–69. doi: 10.1016/j.coviro.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whittle JRR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, Kepler TB, Liao H-X, Harrison SC. 2011. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci U S A 108:14216–14221. doi: 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt AG, Xu H, Khan AR, O'Donnell T, Khurana S, King LR, Manischewitz J, Golding H, Suphaphiphat P, Carfi A, Settembre EC, Dormitzer PR, Kepler TB, Zhang R, Moody MA, Haynes BF, Liao H-X, Shaw DE, Harrison SC. 2013. Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc Natl Acad Sci U S A 110:264–269. doi: 10.1073/pnas.1218256109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun L, Lu X, Li C, Wang M, Liu Q, Li Z, Hu X, Li J, Liu F, Li Q, Belser JA, Hancock K, Shu Y, Katz JM, Liang M, Li D. 2009. Generation, characterization and epitope mapping of two neutralizing and protective human recombinant antibodies against influenza A H5N1 viruses. PLoS One 4:e5476. doi: 10.1371/journal.pone.0005476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stevens J, Corper AL, Basler CF, Taubenberger JK, Palese P, Wilson IA. 2004. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science 303:1866–1870. doi: 10.1126/science.1093373. [DOI] [PubMed] [Google Scholar]
- 37.Krissinel HE. 2007. Inference of macromolecular assemblies from crystalline state. J Mol Biol 372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 38.Winarski KL, Thornburg NJ, Yu Y, Sapparapu G, Crowe JE, Spiller BW. 2015. Vaccine-elicited antibody that neutralizes H5N1 influenza and variants binds the receptor site and polymorphic sites. Proc Natl Acad Sci U S A 112:9346–9351. doi: 10.1073/pnas.1502762112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crusat M, Liu J, Palma AS, Childs RA, Liu Y, Wharton SA, Lin YP, Coombs PJ, Martin SR, Matrosovich M, Chen Z, Stevens DJ, Hien VM, Thanh TT, Nhu LN, Te NT, Nguyet LA, Ha DQ, van Doorn HR, Hien TT, Conradt HS, Kiso M, Gamblin SJ, Chai W, Skehel JJ, Hay AJ, Farrar J, de Jong MD, Feizi T. 2013. Changes in the hemagglutinin of H5N1 viruses during human infection—influence on receptor binding. Virology 447:326–337. doi: 10.1016/j.virol.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hong M, Lee PS, Hoffman RM, Zhu X, Krause JC, Laursen NS, Yoon S-I, Song L, Tussey L, Crowe JE, Ward AB, Wilson IA. 2013. Antibody recognition of the pandemic H1N1 influenza virus hemagglutinin receptor binding site. J Virol 87:12471–12480. doi: 10.1128/JVI.01388-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fleury D, Wharton SA, Skehel JJ, Knossow M, Bizebard T. 1998. Antigen distortion allows influenza virus to escape neutralization. Nat Struct Biol 5:119. doi: 10.1038/nsb0298-119. [DOI] [PubMed] [Google Scholar]
- 42.Xu R, Krause JC, McBride R, Paulson JCJE Jr, Wilson IA. 2013. A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin. Nat Struct Mol Biol 20:363–370. doi: 10.1038/nsmb.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiong X, Corti D, Liu J, Pinna D, Foglierini M, Calder LJ, Martin SR, Lin YP, Walker PA, Collins PJ, Monne I, Suguitan AL, Santos C, Temperton NJ, Subbarao K, Lanzavecchia A, Gamblin SJ, Skehel JJ. 2015. Structures of complexes formed by H5 influenza hemagglutinin with a potent broadly neutralizing human monoclonal antibody. Proc Natl Acad Sci U S A 112:9430–9435. doi: 10.1073/pnas.1510816112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu X, Guo Y-H, Jiang T, Wang Y-D, Chan K-H, Li X-F, Yu W, McBride R, Paulson JC, Yuen K-Y, Qin C-F, Che X-Y, Wilson IA. 2013. A unique and conserved neutralization epitope in H5N1 influenza viruses identified by an antibody against the A/Goose/Guangdong/1/96 hemagglutinin. J Virol 87:12619–12635. doi: 10.1128/JVI.01577-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brandenburg B, Koudstaal W, Goudsmit J, Klaren V, Tang C, Bujny MV, Korse HJ, Kwaks T, Otterstrom JJ, Juraszek J, van Oijen AM, Vogels R, Friesen RH. 2013. Mechanisms of hemagglutinin targeted influenza virus neutralization. PLoS One 8:e80034. doi: 10.1371/journal.pone.0080034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strauch E-MM, Bernard SM, La D, Bohn AJ, Lee PS, Anderson CE, Nieusma T, Holstein CA, Garcia NK, Hooper KA, Ravichandran R, Nelson JW, Sheffler W, Bloom JD, Lee KK, Ward AB, Yager P, Fuller DH, Wilson IA, Baker D. 2017. Computational design of trimeric influenza-neutralizing proteins targeting the hemagglutinin receptor binding site. Nat Biotechnol 35:667–671. doi: 10.1038/nbt.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu T, Song J, Zhang W, Zhao H, Duan B, Liu Q, Zeng W, Qiu W, Chen G, Zhang Y, Fan Q, Zhang F. 2015. Emergence of novel clade 2.3.4 influenza A (H5N1) virus subgroups in Yunnan Province, China. Infect Genet Evol 33:95–100. doi: 10.1016/j.meegid.2015.04.016. [DOI] [PubMed] [Google Scholar]
- 48.Okamatsu M, Ozawa M, Soda K, Takakuwa H, Haga A, Hiono T, Matsuu A, Uchida Y, Iwata R, Matsuno K, Kuwahara M, Yabuta T, Usui T, Ito H, Onuma M, Sakoda Y, Saito T, Otsuki K, Ito T, Kida H. 2017. Characterization of highly pathogenic avian influenza virus A(H5N6), Japan, November 2016. Emerg Infect Dis 23:691–695. doi: 10.3201/eid2304.161957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang H, Wu P, Uyeki TM, He J, Deng Z, Xu W, Lv Q, Zhang J, Wu Y, Tsang TK, Kang M, Zheng J, Wang L, Yang B, Qin Y, Feng L, Fang VJ, Gao GF, Leung GM, Yu H, Cowling BJ. 2017. Preliminary epidemiologic assessment of human infections with highly pathogenic avian influenza A(H5N6) virus, China. Clin Infect Dis 65:383–388. doi: 10.1093/cid/cix334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ren H, Wang G, Wang S, Chen H, Chen Z, Hu H, Cheng G, Zhou P. 2016. Cross-protection of newly emerging HPAI H5 viruses by neutralizing human monoclonal antibodies: a viable alternative to oseltamivir. MAbs 8:1156–1166. doi: 10.1080/19420862.2016.1183083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gerhard W, Yewdell J, Frankel ME, Webster R. 1981. Antigenic structure of influenza virus haemagglutinin defined by hybridoma antibodies. Nature 290:713–717. doi: 10.1038/290713a0. [DOI] [PubMed] [Google Scholar]
- 52.Caton AJ, Brownlee GG, Yewdell JW, Gerhard W. 1982. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 31:417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- 53.Skehel JJ, Stevens DJ, Daniels RS, Douglas AR, Knossow M, Wilson IA, Wiley DC. 1984. A carbohydrate side chain on hemagglutinins of Hong Kong influenza viruses inhibits recognition by a monoclonal antibody. Proc Natl Acad Sci U S A 81:1779–1783. doi: 10.1073/pnas.81.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wiley DC, Wilson IA, Skehel JJ. 1981. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 289:373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- 55.Gu Y, Tang X, Zhang X, Song C, Zheng M, Wang K, Zhang J, Ng M-H, Hew C-L, Li S, Xia N, Sivaraman J. 2015. Structural basis for the neutralization of hepatitis E virus by a cross-genotype antibody. Cell Res 25:604–620. doi: 10.1038/cr.2015.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rupp B. 2010. Biomolecular crystallography: principles, practice, and application to structural biology. Garland Science, New York, NY. [Google Scholar]
- 57.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ekiert DC, Kashyap AK, Steel J, Rubrum A, Bhabha G, Khayat R, Lee JH, Dillon MA, O'Neil RE, Faynboym AM, Horowitz M, Horowitz L, Ward AB, Palese P, Webby R, Lerner RA, Bhatt RR, Wilson IA. 2012. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 489:526–532. doi: 10.1038/nature11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee PS, Yoshida R, Ekiert DC, Sakai N, Suzuki Y, Takada A, Wilson IA. 2012. Heterosubtypic antibody recognition of the influenza virus hemagglutinin receptor binding site enhanced by avidity. Proc Natl Acad Sci U S A 109:17040–17045. doi: 10.1073/pnas.1212371109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee PS, Ohshima N, Stanfield RL, Yu W, Iba Y, Okuno Y, Kurosawa Y, Wilson IA. 2014. Receptor mimicry by antibody F045-092 facilitates universal binding to the H3 subtype of influenza virus. Nat Commun 5:3614. doi: 10.1038/ncomms4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abhinandan KR, Martin ACR. 2008. Analysis and improvements to Kabat and structurally correct numbering of antibody. Mol Immunol. 45:3832–3839. doi: 10.1016/j.molimm.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Z, Li R, Jiang L, Xiong C, Chen Y, Zhao G, Jiang Q. 2016. The complexity of human infected AIV H5N6 isolated from China. BMC Infect Dis 16:600. doi: 10.1186/s12879-016-1932-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsibane T, Ekiert DC, Krause JC, Martinez O, Crowe JE, Wilson IA, Basler CF. 2012. Influenza human monoclonal antibody 1F1 interacts with three major antigenic sites and residues mediating human receptor specificity in H1N1 viruses. PLoS Pathog 8:e1003067. doi: 10.1371/journal.ppat.1003067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu WL, Chen Y, Wang P, Song W, Lau S-Y, Rayner JM, Smith GJ, Webster RG, Peiris JS, Lin T, Xia N, Guan Y, Chen H. 2008. Antigenic profile of avian H5N1 viruses in Asia from 2002 to 2007. J Virol 82:1798–1807. doi: 10.1128/JVI.02256-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 67.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.