Gene therapy has now entered a phase of development in which a growing number of recessive single gene defects can be successfully treated by vector-mediated introduction of a wild-type copy of the gene into the appropriate tissue. However, many disease conditions, such as neurodegeneration, cancer, and inflammatory processes, are more complex, requiring either multiple gene corrections or provision of coordinated gene activities to achieve a therapeutic outcome. Although herpes simplex virus (HSV) vectors have the capacity to meet this need, the challenge has been to genetically engineer the HSV genome in a manner to prevent expression of any viral genes while retaining the ability to express multiple therapeutic transgenes under independent transcriptional control. Here, we show that non-HSV insulator elements can be applied to retain at least transient transgene activity from multiple viral loci, thereby opening the door for more complex gene therapy applications in the future.
KEYWORDS: CTCF, HSV-1, ICP0, UCOE, cHS4, chromatin remodeling, gene therapy, insulator, transgene expression, viral vector
ABSTRACT
Inactivation of all herpes simplex virus (HSV) immediate early (IE) genes to eliminate vector cytotoxicity results in rapid silencing of the viral genome, similar to the establishment of HSV latency. We recently reported that silencing of a nonviral reporter cassette could be overcome in nonneuronal cells by positioning the cassette in the viral latency (LAT) locus between resident chromatin boundary elements. Here, we tested the abilities of the chicken hypersensitive site 4 insulator and the human ubiquitous chromatin opening element A2UCOE to promote transgene expression from an IE-gene-inactivated HSV vector. We found that A2UCOE was particularly active in nonneuronal cells and reduced reporter promoter occupancy by a repressive histone mark. We determined whether multiple transgenes could be expressed under the control of different promoters from different loci of the same virus. The results showed abundant coexpression of LAT-embedded and A2UCOE-flanked genes in nonneuronal cells. In addition, a third reporter gene without known protective elements was active in cultured rat sensory neurons. These findings indicate that cellular antisilencing sequences can contribute to the expression of multiple genes from separate promoters in fully IE gene-disabled HSV vectors, providing an opportunity for therapeutic applications requiring mutually independent expression of different gene products from a single vector.
IMPORTANCE Gene therapy has now entered a phase of development in which a growing number of recessive single gene defects can be successfully treated by vector-mediated introduction of a wild-type copy of the gene into the appropriate tissue. However, many disease conditions, such as neurodegeneration, cancer, and inflammatory processes, are more complex, requiring either multiple gene corrections or provision of coordinated gene activities to achieve a therapeutic outcome. Although herpes simplex virus (HSV) vectors have the capacity to meet this need, the challenge has been to genetically engineer the HSV genome in a manner to prevent expression of any viral genes while retaining the ability to express multiple therapeutic transgenes under independent transcriptional control. Here, we show that non-HSV insulator elements can be applied to retain at least transient transgene activity from multiple viral loci, thereby opening the door for more complex gene therapy applications in the future.
INTRODUCTION
Studies over many years have extensively documented that progression of the herpes simplex virus (HSV) lytic life cycle relies on the coordinated expression of three classes of genes, the immediate early (IE), early (E), and late (L) genes (1). Key aspects of this regulation are, first, activation of unique enhancers associated with the promoters of the 5 IE genes by binding of the viral tegument protein VP16 and, second, absolute dependence of E and L viral gene expression on the products of the ICP4 and ICP27 IE genes. The remaining IE gene products have more specialized functions that include overcoming host innate immunity and preventing genome silencing (2). Studies of the expression of members of the viral cascade have employed deletion mutants of the IE genes, and it has been shown that inactivation of all 5 IE genes completely silences the viral genome in nonneuronal cells (3, 4). Such highly defective recombinant HSV vectors are without cytotoxic activity but also fail to express transgenes from heterologous promoters (3, 4). In latently infected neurons, all IE genes are switched off, yet a unique HSV gene referred to as the latency-associated transcript (LAT) gene remains transcriptionally active. The molecular mechanism(s) underlying transition from latency to active infection has remained elusive, although breaking the silencing of the IE genes is undoubtedly an essential component (5, 6).
HSV is an attractive vehicle for transmitting genes to neurons due to its ability to persist in these cells in a nonintegrated state and deliver expansive foreign sequences. However, the challenge of eliminating all IE gene-associated cytotoxicity without losing transgene expression potential has long been a confounding obstacle. We recently showed that a foreign promoter introduced into the LAT locus was protected from genome-wide silencing in the absence of all IE gene products and that this protection was mediated in part by an upstream CTCF-binding repeat element referred to as CTRL1 (7, 8). Here, we have sought to extend these observations by evaluating the ability of cellular antisilencing elements to keep nonviral promoters active in an otherwise silent genome.
In our previous work, we described the replication-defective vector JΔNI5, which has all the IE genes functionally deleted, including the internal repeat (joint) region separating the two unique segments of the viral genome. JΔNI5 is transcriptionally silent in nonneuronal cells and devoid of cytotoxicity (7). We observed that a transgene expression cassette inserted into the JΔNI5 genome adjacent to a reported enhancer-type element (LATP2 or LAP2) (9, 10) in the LAT locus between CTRL1 and a second CTCF motif repeat element, CTRL2, remained robustly active for at least 4 weeks in contact-inhibited cells. While this vector can express multiple transgenes from a single promoter using “self-cleaving” 2A peptides (59; Y. Miyagawa, J. B. Cohen, and J. C. Glorioso, unpublished results), here, we explored the use of ectopic antisilencing elements to create additional active loci that would allow separately controlled expression of multiple transgenes from a single vector. Unlike duplication of the LAT protective elements to serve at more than one location, an ectopic element would not be expected to cause vector instability as a consequence of internal recombination events.
In order to protect chromosomally integrated transgenes against heterochromatin formation, a number of genetic regulatory elements have been characterized and tested in gene transfer vectors. Some of these regulatory elements, including insulators and scaffold/matrix attachment regions (S/MARs), show border functions that shield the transgene promoter from the chromosomal environment to reduce position effects (11, 12). Other elements, including the locus control regions (LCRs) and ubiquitous chromatin opening elements (UCOEs), exhibit a dominant chromatin-remodeling and transcription-activating function (13).
Chicken hypersensitive site 4 (cHS4) is among the most extensively characterized vertebrate insulators (11). It is located in the β-globin locus control region between an ∼16-kb region of condensed chromatin and the β-globin gene cluster. It was found to have both enhancer-blocking and barrier activities. The activity of cHS4 was mapped to a 250-bp core element (14). While some studies have suggested that two copies of the core element on each side of a transgene cassette are sufficient to provide essentially full barrier activity (15, 16), others have used an extended (0.4-kb) core fragment (17, 18) or the 1.2-kb full-length insulator in retroviral (19) and lentiviral (20) vectors to achieve sustained, integration site-independent transgene expression.
UCOEs have also shown antisilencing effects. A prototypical UCOE, referred to as A2UCOE, is located in the human HNRPA2B1-CBX3 locus. This locus contains a pair of divergently oriented promoters that are associated with active histone modification marks (21) within a methylation-free CpG island (22, 23). Based on these features, this region has been proposed to serve a dominant chromatin-remodeling function. Its core activity has been variously mapped, depending on the assay system, to 4.1-, 2.2-, 1.5-, and 1.2-kb subfragments roughly centered on the dual-promoter region (23–27). In a comparison among UCOE, MAR, stabilizing antirepressor (STAR), and cHS4 elements, the 1.5-kb A2UCOE was the most effective in creating cell pools that produced stable, high yields of a transgenic antibody (28). While other studies have shown that the CBX3 component is essential or even sufficient for A2UCOE function (29), it has been reported that a 0.6-kb A2UCOE lacking CBX3 sequences was able to stabilize transgene expression (30).
In this study, we explored the ability of cHS4 and the 4.1-kb A2UCOE encompassing the central CpG island and including unmethylated flanking CBX3 and HNRPA2B1 sequences (21) to increase and prolong transgene expression from a JΔNI5-derived vector in human dermal fibroblasts. We observed that the full-length versions of both elements were able to protect a cellular promoter positioned within a viral intergenic region from global silencing of the HSV-1 genome. The 4.1-kb A2UCOE was superior to cHS4 in terms of transgene expression level and duration and comparable in these regards to the full complement of LAT-protective elements. The antisilencing capacities of cHS4 and A2UCOE were also functional in virus-infected primary human muscle precursor cells and human epidermal keratinocytes. As observed in our previous study (7), the ubiquitin C (UbC) promoter driving mCherry expression in the ICP4 locus of JΔNI5 and derivatives was active in rat sensory neurons, but not in nonneuronal cells, suggesting that expression permissiveness at this site is neuronal cell dependent.
We took advantage of these observations to engineer a novel vector containing three different transgenes (enhanced green fluorescent protein [EGFP], firefly luciferase [fLuc], and mCherry genes) under the control of different promoters at different loci. All three transgenes were simultaneously expressed in individual rat dorsal root ganglion neurons in culture, and two of the three were coexpressed in rat hippocampus following intracranial vector delivery. This vector design has potential for gene therapy of peripheral and central nervous system (CNS) conditions that may benefit from independently regulated expression of more than a single external gene product.
RESULTS
cHS4 and A2UCOE improve transgene expression from a highly defective HSV vector.
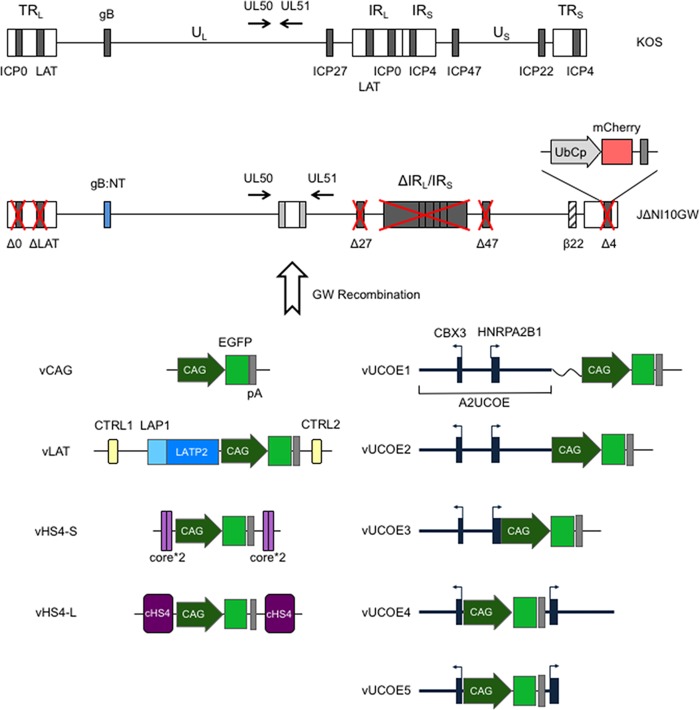
We created a series of HSV-1 recombinant viruses in which an EGFP expression cassette controlled by the cytomegalovirus (CMV) enhancer/β-actin promoter fusion (CAG) was flanked in different configurations by all or components of the 1.2-kb cHS4 insulator or the 4.1-kb A2UCOE fragment. We assembled these extended cassettes in Gateway (GW) entry plasmids and recombined each with a GW destination cassette located in the intergenic region between HSV-1 UL50 and UL51 genes in a previously described HSV-bacterial artificial chromosome (BAC) construct, JΔNI10GW, containing a highly defective HSV-1 genome (7) (Fig. 1). Viruses were produced by transfection of recombinant BAC DNAs into U2OS-ICP4/ICP27 or U2OS-ICP4/ICP27-Cre cells that complement the deleted ICP0, ICP4, and ICP27 genes; the latter cells also excise the BAC region located between loxP sites (7). Viruses were named according to their GW inserts, as indicated in Fig. 1. vCAG and vLAT are JΔNI10GFP and JΔNI10LAT-GFP, respectively, as described by Miyagawa et al. (7).
FIG 1.
JΔNI10 genome engineering. (Top) Compared to the wild-type HSV-1 KOS genome represented at the top, JΔNI10GW (7) has the entire internal repeat (joint) region (IRL and IRS); the remaining ICP0 (Δ0), LAT (ΔLAT), and ICP4 (Δ4) loci; and the single-copy ICP27 (Δ27) gene deleted (red crosses). The promoter and translation initiation codon of the ICP47 (Δ47) gene are also deleted, and the ICP22 promoter acts as an early (β22) promoter due to deletion of its regulatory sequences. The glycoprotein B gene contains a pair of entry-accelerating mutations (gB:NT) (58), and a GW destination cassette is located between the UL50 and UL51 genes, whose directions of transcription are indicated by arrows. A ubiquitin C promoter (UbCp)-mCherry gene cassette is located at the position of the deleted ICP4 gene (Δ4). UL and US, unique long and short segments; TRL and TRS, terminal repeats flanking UL and US, respectively; IRL and IRS, the cognate internal repeats of UL and US, respectively. (Bottom) CAG promoter-EGFP cassettes flanked by antisilencing elements in different configurations were inserted into JΔNI10GW through GW recombination. vCAG contains a CAG promoter-controlled EGFP cassette and the rabbit β-globin polyadenylation (pA) region. vLAT contains the EGFP expression cassette embedded in sequences from the LAT locus between the LATP2 enhancer-type element and CTRL2, with CTRL1 included upstream of the LAT promoter (LAP1). vHS4-S contains two tandem copies of the cHS4 core (core*2) at both sides of the expression cassette, while vHS4-L contains the cassette flanked on each side by a single copy of the full-length (1.2-kb) cHS4. vUCOE1 contains the complete 4.1-kb A2UCOE (black boxes; exons) and a 218-bp cloning sequence (wavy line) upstream of CAG. In vUCOE2, the linker sequence was deleted. vUCOE3 lacks both the linker and the first intron of HNRPA2B1. In vUCOE4 and vUCOE5, the EGFP expression cassette is located in the divergent promoter region separating the CBX3 and HNRPA2B1 transcription start sites, and in vUCOE5, the first intron of HNRPA2B1 is deleted. Black boxes, UCOE exons; thick horizontal lines, UCOE introns and dual-promoter region; arrows, transcription start sites and directions.
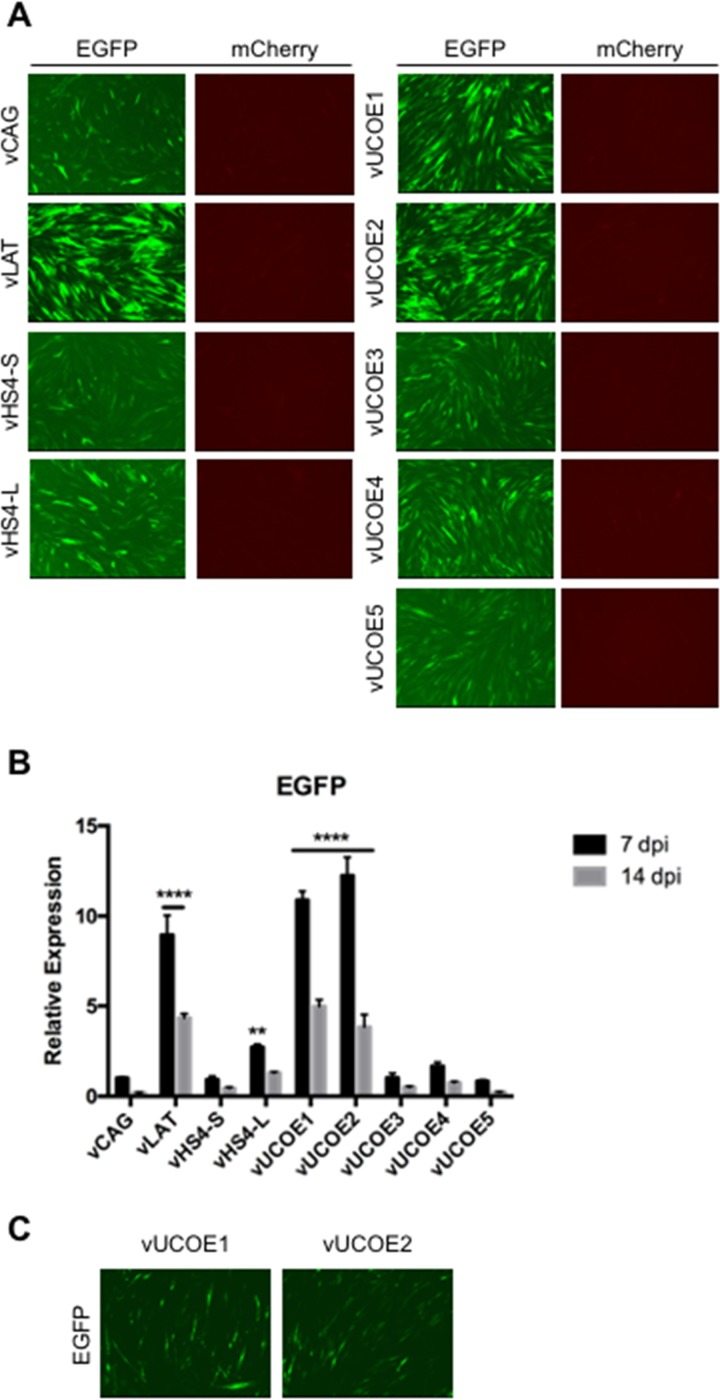
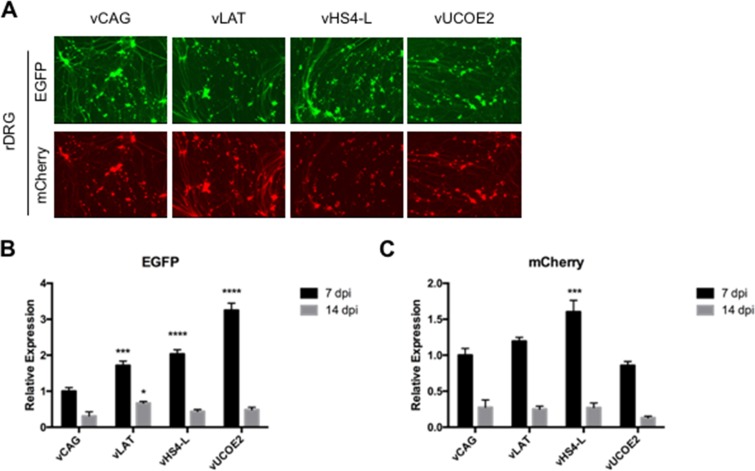
We examined EGFP and mCherry reporter gene expression from the different viruses in human dermal fibroblasts (HDFs) infected with equal numbers of virus particles based on genome copy (gc) titers listed in Table 1. At 7 days postinfection (dpi), none of the viruses showed detectable mCherry expression from the deleted ICP4 locus, while all the viruses produced detectable EGFP fluorescence, although at different levels (Fig. 2A). In agreement with our previous study (7), transplanted sequences from the LAT locus (vLAT) enabled prominent EGFP expression, whereas the CAG-EGFP cassette without additional elements (vCAG) produced a comparatively low level of EGFP fluorescence. Among the viruses, vUCOE2 generated the strongest EGFP fluorescence, while vHS4-S and vUCOE5 produced low levels, similar to vCAG. These results were confirmed and extended by quantitative reverse transcription-PCR (qRT-PCR) measurement of EGFP mRNA levels at 7 and 14 dpi (Fig. 2B). While mRNA levels decreased by an average of approximately 50% between the two time points, EGFP expression in vLAT-, vUCOE2-, and vUCOE1-infected cells was clearly enhanced compared to the vCAG baseline and other infected cells, whereas vHS4-L produced only a modest increase in EGFP RNA levels relative to vCAG (Fig. 2B). These data indicated that the 4.1-kb A2UCOE is capable of protecting a linked transgene expression cassette from the rapid global silencing of the HSV-1 genome that occurs in the absence of functional IE genes and that 2 copies of full-length cHS4 could provide some protection. Notably, omission of the HNRPA2B1 intron abolished the antisilencing activity of A2UCOE (compare vUCOE2 and -3), but preservation of the intron in vUCOE4 did not rescue expression, indicating that disruption of the bidirectional promoter region of A2UCOE was detrimental. Although EGFP fluorescence waned over time, it remained detectable at 28 dpi in vUCOE1- and vUCOE2-infected HDFs (Fig. 2C). Together, these data showed that the uninterrupted 4.1-kb A2UCOE, and to a lesser extent the full-length cHS4, was capable of enhancing transgene expression from an extrachromosomal viral vector in the absence of viral trans-acting, antisilencing gene products. Table 2 summarizes the results of this analysis and our observations in additional cell types presented below.
TABLE 1.
Virus titers
| Virus name | No. of gc/ml | No. of PFU/ml | No. of gc/PFU |
|---|---|---|---|
| vCAG | 1.86E12 | 1.03E9 | 1.80E3 |
| vLAT | 1.62E12 | 4.33E8 | 3.74E3 |
| vHS4-S | 1.75E12 | 4.33E8 | 4.05E3 |
| vHS4-L | 1.45E12 | 3.33E8 | 4.35E3 |
| vUCOE1 | 1.97E12 | 1.13E9 | 1.74E3 |
| vUCOE2 | 2.65E12 | 1.67E9 | 1.59E3 |
| vUCOE3 | 2.85E12 | 1.77E9 | 1.61E3 |
| vUCOE4 | 1.86E12 | 1.37E9 | 1.36E3 |
| vUCOE5 | 1.84E12 | 9.33E8 | 1.97E3 |
| vLuc | 2.18E12 | 1.60E9 | 1.36E3 |
| vU-Luc | 1.52E12 | 1.33E9 | 1.14E3 |
FIG 2.
Transgene expression in infected HDFs at 7, 14, and 28 dpi. The cells were infected with the indicated viruses at 25,000 gc per cell. (A) EGFP and mCherry fluorescence in infected HDFs at 7 dpi. All the images of each marker were collected at a fixed exposure time and automatically adjusted for optimal contrast and brightness (autoscale setting; Meta Imaging Series 7.7 MetaMorph software; Molecular Devices). (B) Relative EGFP mRNA levels in infected HDFs as determined by qRT-PCR analysis. Expression levels were normalized to gc in the same samples and are presented as means and SD relative to the level in vCAG-infected cells at 7 dpi (see “Statistics” in Materials and Methods for details). ****, P ≤ 0.0001; **, P ≤ 0.01 compared to vCAG at the same time point (2-way ANOVA). (C) EGFP fluorescence in vUCOE1- and vUCOE2-infected HDFs at 28 dpi.
TABLE 2.
Summary of EGFP expression in UL50/UL51
| Element | Virus | Feature | Position relative to CAG-EGFP | Fold changea |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HDF |
hMDSC |
hEK |
rDRG |
||||||||
| 7 dpi | 14 dpi | 14 dpi | 28 dpi | 3 dpi | 7 dpi | 7 dpi | 14 dpi | ||||
| cHS4 | vHS4-S | Core*2 (2 × 250 bp) | 5′ + 3′ | 0.92 | 0.44 | ||||||
| vHS4-L | Full length (1.2 kb) | 5′ + 3′ | 2.72 | 1.29 | 2.89 | 1.68 | 2.87 | 0.51 | 2.03 | 0.44 | |
| UCOE | vUCOE1 | 4.1-kb A2UCOE + 218-bp spacer | 5′ | 10.88 | 4.98 | ||||||
| vUCOE2 | 4.1-kb A2UCOE | 5′ | 12.24 | 3.84 | 9.61 | 7.92 | 5.76 | 1.09 | 3.25 | 0.49 | |
| vUCOE3 | A2UCOE without HNRPA2B1 intron | 5′ | 1.02 | 0.51 | |||||||
| vUCOE4 | Split 4.1-kb A2UCOE | CBX3 5′, HNRPA2B1 3′ | 1.66 | 0.75 | |||||||
| vUCOE5 | Split A2UCOE, no HNRPA2B1 intron | CBX3 5′, HNRPA2B1 3′ | 0.84 | 0.23 | |||||||
| LAT | vLAT | Split within 2-kb intron | CTRL1/LAP 5′, CTRL2 3′ | 8.90 | 4.30 | 12.20 | 8.40 | 8.74 | 0.70 | 1.72 | 0.67 |
Fold change in mRNA relative to vCAG at the earlier time point per cell type.
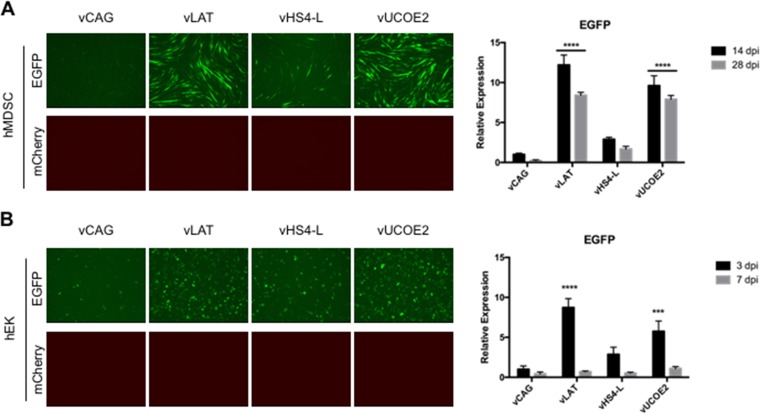
To extend these findings, we used a subset of the viruses to examine EGFP expression in other cell types. In human muscle-derived stem cells (hMDSCs) infected with vLAT or vUCOE2, EGFP fluorescence was elevated compared to vCAG-infected cells through at least 14 dpi, and elevated transcript levels persisted at 28 dpi. In contrast, EGFP expression in vHS4-L-infected cells was only slightly higher than in vCAG-infected cells (Fig. 3A). A similar pattern was observed in human epidermal keratinocytes (hEKs), although expression was more transient than in hMDSCs (Fig. 3B), likely a result of ongoing cell division reducing vector numbers per cell while hMDSCs underwent terminal differentiation. As in HDFs, mCherry fluorescence was not detected in any of the infected cultures (Fig. 3A and B). These results indicated that the antisilencing activity of A2UCOE in a highly defective HSV-1 vector is functional in different nonneuronal cell types.
FIG 3.
Transgene expression in infected hMDSC (50,000 gc/cell) (A) and hEK (25,000 gc/cell) (B) cell cultures. (A) EGFP and mCherry fluorescence at 14 dpi (left) and relative EGFP mRNA levels at 14 and 28 dpi (right). (B) Fluorescence at 3 dpi and relative EGFP mRNA levels at 3 and 7 dpi. Images were collected as for Fig. 2A. mRNA data were obtained by qRT-PCR with normalization to viral gc in the same samples and are presented as expression relative to that in vCAG-infected cells at 14 dpi (A) or 3 dpi (B). The data are means and SD; ***, P ≤ 0.001, and ****, P ≤ 0.0001 compared to vCAG at the same time point (2-way ANOVA).
Transcriptional activity of flanking genes.
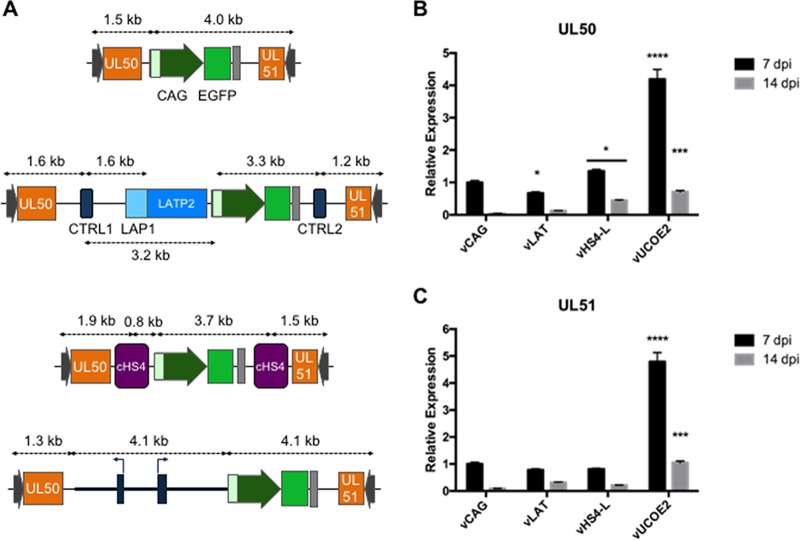
We asked whether cHS4 and A2UCOE affected the transcript levels of neighboring genes. The proximal UL50 and UL51 genes are transcribed in opposite directions toward the CAG-EGFP cassette (Fig. 1 and 4A). Surrounding CAG-EGFP with sequences from the LAT locus did not noticeably alter the expression of either UL50 or UL51 in infected HDFs at 7 and 14 dpi (Fig. 4B and C), indicating that the different elements of the LAT region (CTRLs and LATP2) neither activated neighboring promoters nor protected the promoters from putative stimulation by the CMV enhancer component of the CAG promoter. The cHS4 elements of vHS4-L caused a slight increase in UL50 expression but did not alter the expression of UL51 compared to vCAG, providing no evidence for either a boundary effect shielding these genes from silencing or a CAG enhancer-blocking activity. In contrast, A2UCOE had a stimulatory effect on the expression of both UL50 and UL51, albeit less pronounced than its stimulation of EGFP expression (see Fig. 2B). These results indicated that A2UCOE has an ability to enhance gene expression bidirectionally, although the magnitude may vary with distance (Fig. 4A) and possibly the nature of the promoter (CAG versus non-IE HSV-1 promoters).
FIG 4.
Structure of the UL50-UL51 locus in vCAG, vLAT, vHS4-L, and vUCOE2 (A) and relative UL50 (B) and UL51 (C) mRNA levels in infected HDFs. The data are means and SD; *, P ≤ 0.05; ***, P ≤ 0.001; ****, P ≤ 0.0001 compared to vCAG at the same time point (2-way ANOVA).
Histone modification contributes to the activity of A2UCOE in HDFs.
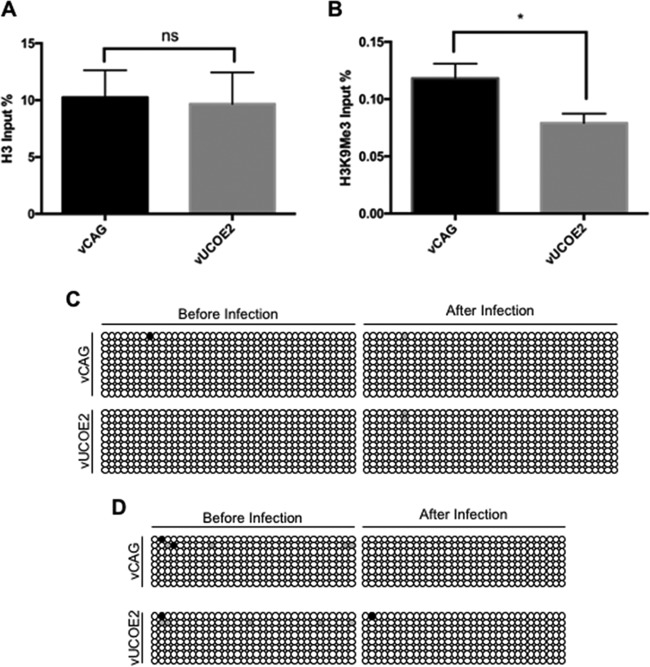
To investigate the mechanism underlying the antisilencing effect of A2UCOE on genes in our highly defective HSV vector, we measured CAG promoter occupancy by total histone H3 and lysine 9-trimethylated histone H3 (H3K9Me3), a repressive histone mark, in vCAG- and vUCOE2-infected HDFs at 7 dpi. As shown in Fig. 5A, anti-H3 antibody precipitated similar fractions of CAG input DNA from vCAG- and vUCOE2-infected cells (∼10%). However, CAG-associated H3K9Me3 was significantly less abundant in vUCOE2-infected cells (0.08%) than in vCAG-infected cells (0.12%) (Fig. 5B), indicating that A2UCOE provides protection against the accumulation of repressive histone marks on the CAG promoter in a quiescent HSV-1 genome.
FIG 5.
(A and B) Total H3 (A) and transcriptionally repressive H3K9Me3 (B) associated with the CAG promoters of vCAG and vUCOE2 7 days after infection of HDFs. qPCR was performed to determine the amount of CAG DNA precipitated by each antibody. The graphs show the percentages of CAG DNA precipitated with anti-histone H3 or K9Me3-specific anti-H3 as a percentage of total CAG DNA prior to precipitation. The error bars represent SD in the results of 4 experiments. ns, not significant; *, P < 0.05; Mann-Whitney test. (C and D) DNA methylation status of two different segments of the CAG promoter in vCAG and vUCOE2 before infection and at 7 days post-HDF infection. Each row of circles represents an individual cloned PCR product from the chicken β-actin component of the CAG promoter after bisulfite conversion. Each circle represents a CpG site. The white circles represent unmethylated CpG sites, and the black circles represent methylated CpG sites; the gray circles represent sites that could not be determined. (C) Primer set M2. (D) Primer set M4.
Since A2UCOE encompasses methylation-free CpG islands, we also explored whether protection against DNA methylation played a role in the stimulatory effect of A2UCOE on transgene expression. Viral DNA extracted prior to infection and at 7 days post-HDF infection was treated with bisulfite to convert unmethylated cytosines to uracil, a portion of the β-actin component of the CAG promoter was amplified by PCR, and a number of cloned fragments were sequenced. As illustrated in Fig. 5C and D, the CAG promoter was substantially free of methylation both in input viral DNA and in viral DNA from infected HDFs. Thus, methylation was not responsible for the repressed activity of the CAG promoter in vCAG-infected cells, and the A2UCOE insert in vUCOE2 did not act by preventing promoter methylation. Together, these results pointed to the involvement of histone modification, but not blockage of DNA methylation, in the mechanism of gene activation by A2UCOE in episomal HSV-1 DNA.
A2UCOE activity in neuronal cells.
In our previous study (7), we observed differences between primary human nonneuronal cells, including HDFs, hMDSCs, and hEKs, and rat dorsal root ganglion (rDRG) cultures in the expression of reporter genes from IE gene-inactive HSV-1 vectors. While the mCherry gene in the ICP4 locus was essentially silent in the human nonneuronal cells, it showed robust activity in rDRGs at 3 dpi. Moreover, while CAG-EGFP inserted between the viral UL3 and UL4 genes was inactive in the human cells, it too showed robust expression in rDRGs, matched in human cells only when the expression cassette was located in the LAT locus (7). Among different explanations, these results raised the unanticipated possibility that the IE gene-disabled genome backbone used in these studies was relatively derepressed in rDRGs compared to nonneuronal cells. Thus, it was of interest to determine whether CAG-EGFP located in the UL50-UL51 intergenic region was detectably expressed in rDRGs and whether its expression was enhanced by regulatory LAT, cHS4, or A2UCOE elements, as was seen in nonneuronal human cells. Infection with vCAG, vLAT, vHS4-L and vUCOE2 produced abundant expression of both EGFP and mCherry in rDRGs at 7 dpi (Fig. 6A) at a virus dose substantially lower than that used in the human cell infections shown in Fig. 2 and 3. Quantitative analysis by qRT-PCR showed small differences in mCherry transcript levels among the 4 viruses but increases in EGFP expression of ∼1.5 to 3-fold per viral genome relative to vCAG (Fig. 6B). These results showed that the LAT, cHS4, and A2UCOE elements had a smaller impact in rDRGs than in the human cells examined earlier, consistent with the interpretation that the IE gene-deficient viral genome exists in DRGs in a less transcriptionally repressed state than in other cells. At 14 dpi, expression of EGFP and mCherry was reduced and was similar among the 4 viruses, indicating that the modest insulator effects observed at 7 dpi were transient in rDRG cultures.
FIG 6.
Fluorescence at 7 dpi (A) and relative EGFP (B) and mCherry (C) mRNA levels at 7 and 14 dpi in cultured rDRGs (3,000 gc/cell). The mRNA data were obtained by qRT-PCR with normalization to viral gc in the same samples and are presented as expression relative to that in vCAG-infected cells at 7 dpi. The data are means and SD; *, P < 0.05; ***, P ≤ 0.001; ****, P ≤ 0.0001 compared to vCAG at the same time point (2-way ANOVA).
Simultaneous expression of 3 distinct transgene cassettes from a single vector.
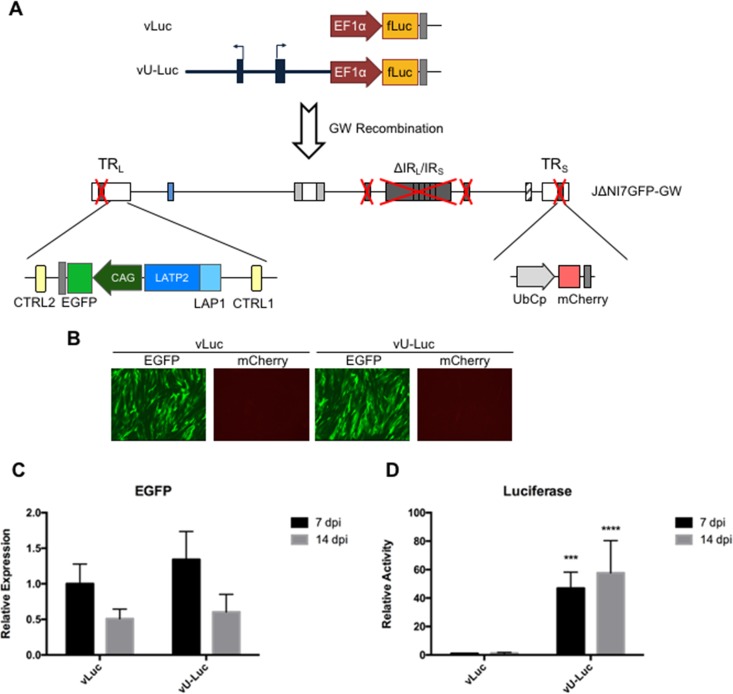
Our observations suggested that A2UCOE would allow the expression in nonneuronal cells of 2 transgenes from different loci in the IE gene-inactive HSV-1 genome, raising the possibility of mutually independent regulation. Furthermore, they suggested that the same virus backbone would be able to express transgenes from 3 different loci in DRGs and perhaps other neuronal cells, at least short term. To test these suggestions, we introduced the fLuc gene under the control of the human EF1α promoter, with or without A2UCOE, into the UL50-UL51 intergenic region of a dual-reporter, IE gene-deficient vector, JΔNI7GFP (7), via a GW intermediate denoted JΔNI7GFP-GW (Fig. 7A); JΔNI7GFP-GW differs from JΔNI10GW (Fig. 1, top) in that it contains CAG-EGFP embedded within the native LAT locus. The resulting viruses, vLuc and vU-Luc (Fig. 7A), produced comparable levels of EGFP fluorescence (Fig. 7B) and mRNA (Fig. 7C, 7 and 14 dpi) in infected HDFs, and neither virus produced detectable mCherry fluorescence. In contrast, vU-Luc-infected cells contained substantially more luciferase enzymatic activity at 7 dpi than vLuc-infected cells, and this level remained relatively unchanged at 14 dpi (Fig. 7D). These results demonstrated directly that A2UCOE provides a means to express 2 transgenes in nonneuronal cells from different promoters in the vector genome. Furthermore, they indicated that A2UCOE, while capable of transiently stimulating the expression of nearby viral genes (Fig. 4), does not significantly affect the activity of distal genes (EGFP and mCherry).
FIG 7.
vLuc and vU-Luc genome structures and transgene expression in HDFs. (A) Schematic drawing of the vLuc and vU-Luc genomes. A CAG-EGFP cassette was located in the LAT loci of both vectors. A GW cassette positioned in the intergenic region between UL50 and UL51 was used to introduce an EF1α promoter-luciferase (fLuc) expression cassette with or without upstream A2UCOE. (B) EGFP and mCherry fluorescence in vLuc- and vU-Luc-infected HDFs at 7 dpi. (C) EGFP mRNA levels of vLuc- or vU-Luc-infected HDFs at 7 and 14 dpi expressed relative to vLuc at 7 dpi (qRT-PCR data were normalized to viral gc in the same samples). (D) Luciferase activity in vLuc- or vU-Luc-infected HDFs at 7 and 14 dpi relative to vLuc at 7 dpi. The data are means and SD; ***, P ≤ 0.001, and ****, P ≤ 0.0001 compared to vLuc at the same time point (2-way ANOVA).
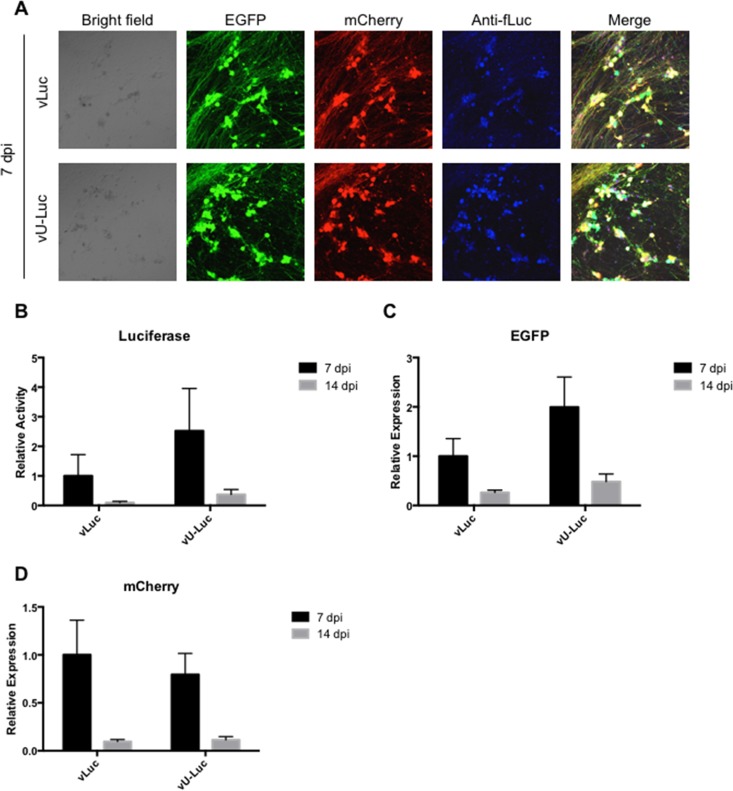
We then sought to confirm that all three reporter genes of vLuc and vU-Luc could be expressed simultaneously in rDRGs. We used immunofluorescence to visualize fLuc protein in the same fields as EGFP and mCherry fluorescence and observed extensive overlap between the three types of signals from either virus (Fig. 8A). Although not statistically significant, luciferase activity appeared moderately higher in vU-Luc- than in vLuc-infected cells (Fig. 8B), consistent with the results shown in Fig. 6B. Likewise, EGFP transcript levels appeared somewhat elevated (Fig. 8C), but mCherry expression was essentially the same from the 2 viruses (Fig. 8D). It should be noted that in the episomal configuration adapted by these viruses, the EGFP and mCherry genes are located near one another while the fLuc locus is distal, arguing that any differential effect of A2UCOE on EGFP and mCherry expression is likely not a function of linear distance.
FIG 8.
Expression of the three vLuc and vU-Luc transgenes in infected rDRG cultures (3,000 gc/cell). (A) EGFP and mCherry fluorescence and luciferase immunofluorescence (Anti-fLuc) at 7 dpi. Merge, overlay of the three fluorescence images. (B) Luciferase activities at 7 and 14 dpi relative to vLuc at 7 dpi. (C and D) EGFP (C) and mCherry (D) mRNA levels at 7 and 14 dpi relative to vLuc-infected cells at 7 dpi. The mRNA data were obtained by qRT-PCR with normalization to viral gc in the same samples. The data are means and SD; P > 0.05 (not significant) for all vU-Luc compared to vLuc at the same time point (2-way ANOVA).
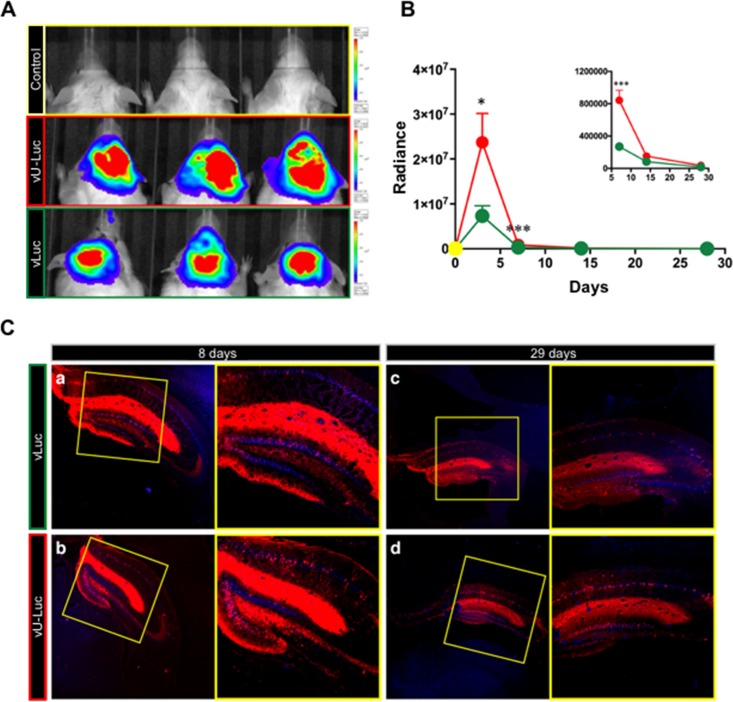
Lastly, we examined vLuc and vU-Luc transgene expression in mouse brain following stereotactic delivery of the vectors to the hippocampus. IVIS monitoring (see below) of luciferin-dependent bioluminescence (Fig. 9A) showed peak luciferase activity at 3 days post-vector inoculation, declining to undetectable at 28 days (Fig. 9B). Notably, at both 3 days and 7 days, the signal in vU-Luc-injected mice was stronger than in vLuc-injected mice (Fig. 9B). Hippocampal sections from animals sacrificed at 8, 15, and 29 days were tested by indirect immunofluorescence for EGFP and mCherry expression. Robust mCherry expression was observed at 8 days and persisted, albeit at reduced levels, at 29 days (Fig. 9C). In contrast, very little, if any, EGFP signal was detected at any time point. These results indicated that A2UCOE can enhance transient, independent expression of a second transgene in the central nervous system to complement the previously described durable expression of a first transgene positioned in the deleted ICP4 locus (31, 32). It remains to be determined what mechanisms can account for the rapid or immediate silencing of the LAT-based EGFP cassette of these vectors in mouse hippocampus.
FIG 9.
Transgene expression in the brains of mice injected with vLuc, vU-Luc, or vehicle (PBS). (A) Luciferase activity in 3 representative mice per group at 3 dpi. The colors indicate activity levels. (B) Quantification of bioluminescence signals (photons per second per square centimeter). Green, vLuc (n = 12 at 3 days and 7 days; n = 8 at 14 days; n = 4 at 28 days); red, vU-Luc (n = 12 at 3 days and 7 days; n = 8 at 14 days; n = 4 at 28 days); yellow, PBS (n = 3 at 3 days and 7 days; n = 2 at 14 days; n = 1 at 28 days). The inset shows differences at the 7-, 14-, and 28-day time points in a lower bioluminescence range. The data are means and standard errors of the mean (SEM) per group. *, P < 0.05; ***, P < 0.001; Mann-Whitney U test. (C) Representative images of coronal sections from animals injected with vLuc (top) or vU-Luc (bottom) sacrificed on the indicated days post-vector delivery. Blue, nuclei of neurons stained with Neurotrace; red, mCherry-positive cells. Higher magnifications of the boxed areas are shown on the right.
DISCUSSION
Replication-defective HSV holds promise as a vector for gene therapy because of its large payload capacity and the establishment of its genome as an extrachromosomal episome without integration into cellular DNA. We recently developed an innocuous HSV vector that is devoid of functional IE genes and is transcriptionally silent, with the exception of transgene expression cassettes inserted into the LAT locus, mimicking neuronal HSV latency in nonneuronal cells (7). We further showed that the privileged status of the LAT locus as a transcriptionally active site in an otherwise silent HSV episome is due in part to the presence of viral insulator-type elements in this region. To expand the utility of this vector platform, here, we explored the question of whether additional transcriptionally active sites can be created by insertion of cellular genetic elements that possess an ability to protect linked genes from epigenetic silencing. We found that the naturally hypomethylated cellular A2UCOE element enables adjacent reporter gene transcription from cellular promoters inserted into the defective HSV backbone, thereby providing a system for mutually independent expression of multiple transgenes from a single vector.
A2UCOE was first described in 2003 (22) and has since been tested in a variety of systems for its ability to support persistent transgene expression from the innate HNRPA2B1 promoter or external promoters. Where comparisons have been performed, A2UCOE has been found to be more effective in blocking transgene silencing than cHS4 (28, 33), consistent with our results. The majority of UCOE studies concern the protection of transgene transcription from position effects at chromosomal integration sites, showing that UCOEs provide uniformity in expression that directly correlates with the transgene copy number and is stable through numerous cell divisions. Nonreplicating, large episomal vectors like the defective HSV vectors used in this study (∼138 to 150 kb) represent a different situation. Most importantly, the transgene position and genetic context are predetermined and unique, excluding variable position effects. The advantage of this situation is that it facilitates the study of regulatory elements in a defined chromatin environment that in some instances can be manipulated. For example, the HSV IE protein ICP0 blocks global heterochromatinization of the viral genome (34), and its deletion, as in our study, allows an assessment of the heterochromatin-blocking activity of cis-acting sequences by comparison of transgene expression in populations of infected cells. HSV DNA, including that of replication-defective virions produced in complementing cells, is largely free of CpG methylation during replication (35). On infection of noncomplementing cells, transgenes are initially expressed from replication-defective vectors lacking ICP0, a cytotoxic protein, but are then rapidly silenced, similar to virus entry into latency in neurons. However, in latency, the viral genome remains unmethylated, and it has been shown that HSV gene expression is regulated by posttranscriptional histone modification (36). In dividing cells, such as preconfluent HDFs, partitioning of replication-defective viral genomes into daughter cells, in addition, reduces viral gene expression per cell by a nonepigenetic mechanism. In nondividing cells (e.g., DRG neurons), the viral genome copy number per cell remains essentially constant.
Previous studies have shown that a 4.0-kb A2UCOE fragment similar to the 4.1-kb region used in our work provided robust, stable transgene expression from the CMV promoter in transfected cells through numerous passages; a 1.5-kb A2UCOE fragment extending from CBX3 intron 1 to HNRPA2B1 intron 1 mediated moderately lower but equally stable expression (23). Likewise, Zhang and colleagues (27) showed that the 1.5-kb fragment and a 1.2-kb subfragment extending to the end of HNRPA2B1 exon 1 enabled stable lentivirus transgene expression from the spleen focus-forming virus (SFFV) long terminal repeat (LTR), whereas the transgene was rapidly silenced in the absence of the UCOE. These authors, in addition, showed that the antisilencing activities of both fragments correlated with orientation-dependent blockage of SSFV promoter methylation. In our study, the CAG promoter was largely unmethylated even in the absence of A2UCOE sequences, and thus, UCOE-mediated promoter protection from methylation was not responsible for the increased transgene expression we observed from vUCOE1 and vUCOE2 compared to vCAG. By the same argument, it is unlikely that the loss of transgene expression we observed on deletion of the HNRPA2B1 intron was due to promoter methylation. Instead, our H3K9Me3 analysis is consistent with the suggestion that A2UCOE prevents the deposition of repressive histone marks on proximal promoters, as previously proposed (37).
We have not excluded the formation of HNRPA2B1-transgene fusion transcripts resulting from transcription read-through from the HNRPA2B1 promoter, as previously observed from the CBX3 promoter (25, 38), and subsequent HNRPA2B1 exon 1 splicing to splice acceptors at the ends of our CAG and EF1α constructs just upstream of the transgene. The intron 1 deletion (vUCOE3) would prevent these splicing events, as it includes the HNRPA2B1 splice donor. Moreover, when we exchanged the positions of the HNRPA2B1 promoter/transcribed region and the CAGp-EGFP cassette by inserting CAGp-EGFP into the A2UCOE dual-divergent promoter region (vUCOE4), transgene expression in HDFs was barely higher than in the absence of A2UCOE (Fig. 2B). Thus, in both cases, reduced green fluorescent protein (GFP) expression was correlated with the predicted absence of a productive read-through/splicing product. In vU-Luc-infected cells, the putative HNRPA2B1-Luc transcription/splicing event would create a short open reading frame (ORF) upstream of the luciferase ORF that might not interfere with luciferase translation. In vUCOE2-infected cells, however, translation from the HNRPA2B1 start codon in exon 1 would read out of frame into the EGFP ORF and prevent the formation of a fluorescent product, arguing against a contribution of read-through transcription to the EGFP signal. As the effect of A2UCOE on expression of the linked reporter gene was larger in vU-Luc-infected than in vUCOE2-infected HDFs, it remains possible that transcripts initiating at the HNRPA2B1 promoter contributed to the vU-Luc luciferase signal in these cells but not to the vUCOE2 EGFP signal and not in neurons, where the difference was minimal.
We found that the 4.1-kb A2UCOE had a greater enhancing effect on transgene expression in HDFs and hMDSCs than in rDRGs. It is possible that this is related to differences between rDRG neurons and other host cells in the activity of the CBX3 promoter, since it has been proposed that the cell-type-dependent strength of the promoter pointing away from the transgene is a key determinant of the chromatin-remodeling and antisilencing activities of A2UCOE (13, 27). However, the additional expression of mCherry in rDRGs is consistent with the alternative suggestion that our highly defective HSV vectors exist in a less transcriptionally repressed state in DRG neurons than in other cultured cells. While our images of vLuc/vU-Luc-infected rDRGs (Fig. 8A) suggest that most of the cells express all three transgenes, mCherry in particular is notably absent in some. This is reminiscent of observations in trigeminal ganglion (TG) cultures infected with the IE gene-disabled d109 vector (39) and likely reflects differences between different neuronal subtypes present in both DRG and TG cultures.
The suggestion that rDRGs in culture are relatively supportive of gene expression from highly defective HSV backbones agrees with our previous observation that an IE gene-inactive vector containing CAG-EGFP in the UL3-UL4 intergenic region produced substantially more EGFP-positive cells per input gc in rDRGs at 3 dpi than in nonneuronal cells (7). In contrast, we did not detect EGFP expression in rat hippocampus from the UL3-UL4 locus in previous work (31). Likewise, while CAG-EGFP in the LAT locus was active in cultured DRG neurons (reference 7 and this study), hippocampal expression was not observed (but see below). These results are suggestive of significant differences between DRG and CNS neurons, although both are predominantly capable of transgene expression from the terminal ICP4 locus, unlike the different nonneuronal cell types we tested here and elsewhere (7).
The absence of detectable EGFP expression from the vLuc and vU-Luc vectors in mouse brain is unexplained. Lilley and coworkers introduced a strong promoter (CMV)-reporter gene cassette downstream of the LAP1-LATP2 components of the LAT gene in an arrangement very similar to our CAG promoter-EGFP insertion in a vector backbone that had the essential ICP4 and ICP27 genes and the IE gene-transactivating activity of VP16 deleted. At 7 days post-vector injection into rat striatum, strong reporter expression was observed in both the striatum and the substantia nigra, increasing from a moderate level at 3 days, and expression remained readily detectable at 1 month (40). While the authors acknowledged the possibility of an apparently nontoxic, undetected residual level of ICP0 expression from their VP16 mutant vector, we note that their vector also differed from ours in other respects, including the strain background (17syn+ versus KOS), a complete deletion of the ICP34.5 neurovirulence gene, and the retention of CTCF-binding sites deleted in our vectors. Guided by these observations, we anticipate that further engineering may provide our vector with the ability to express LAT-based transgene cassettes in the CNS.
The HSV genome contains multiple CTCF-binding regions, in addition to CTRL1 and -2 (8). All of these regions are located within or near the long inverted repeats of the viral genome, and each of the 5 IE genes, in addition to the LAT gene, is flanked by at least one of these sites (8, 36). CTCF has insulator activity, defining chromatin boundaries that allow differential regulation of adjacent transcription domains (41–43). Moreover, in concert with cohesin, CTCF mediates DNA looping to distal CTCF-binding sites (44, 45), suggesting the potential for various types of regulatory interactions among the CTCF-binding regions of HSV (46, 47). Several laboratories have begun to unravel the role of CTCF and its partners in the regulation of HSV latency and reactivation, painting a complex picture of short- and potential long-range effects that may be influenced by the products of some of the regulated genes, LAT and ICP0 in particular (47–53). The extensive deletions in our vectors, which include all but 4 of the CTCF-binding sites in most of the vectors shown in Fig. 1, the ICP0 promoter and coding region, and large portions of the LAT gene, including key splicing signals for the formation of the stable LAT introns (Fig. 7A) or the promoter, enhancer, intron, including CTRL2, and upstream region, including CTRL1 (Fig. 1), can be predicted to profoundly alter the normal CTCF-mediated regulation of viral gene expression in latency compared to the latent-like state assumed by our vectors. We hypothesize that the unexpected long-term reporter gene expression from the terminal ICP4 locus of our defective vectors in central nervous system neurons in vivo (Fig. 9C) (31, 32) and the simultaneous lack of reporter expression from the LAT locus reported here are consequences of the disruption of CTCF interactions and the loss of LAT and ICP0 functions. However, it is uncertain whether A2UCOE activity is influenced by the same mechanisms. While we suspect that the effects of A2UCOE can be duplicated in other locations distant from known CTCF-binding sites, this remains to be shown. Likewise, while we may expect that our vectors will act in human neurons much like they do in rodent neurons, this comparison has yet to be performed.
In conclusion, our study shows that the 4.1-kb A2UCOE can be a valuable component of highly defective HSV gene therapy vectors, particularly in nonneuronal cells. The stimulatory effect of A2UCOE on transgene expression in neuronal cells is short term but may be useful to increase the transient level of a regulator that induces an expression cascade. This work holds promise for the development of increasingly complex vectors for tightly controlled treatment of a variety of pathological conditions.
MATERIALS AND METHODS
Cells.
HDFs, hMDSCs, hEKs, and rDRGs were cultured as described previously (7). For hMDSCs, the medium was changed at 1 dpi to 2% (vol/vol) fetal bovine serum (FBS) with 100 U/ml penicillin and 100 μg/ml streptomycin to induce myogenic differentiation. U2OS-ICP4/ICP27 and U2OS-ICP4/ICP27-Cre cells were as described previously (7).
Plasmids. (i) pHS4-S.
Plasmid pENTR-CAG-EGFP contains a CAG promoter (CMV enhancer/chicken β-actin promoter/chimeric intron)-driven EGFP expression cassette between attL sites (7). To introduce multiple-cloning sites (MCS) both between attL1 and the promoter and between the poly(A) region and attL2, we performed a PCR with primer pair P1 (all the primers mentioned are listed in Table 3) to amplify the plasmid backbone with terminal attL sites and digested the product at unique, attL-external XbaI and StuI sites specified by the respective primers. A SpeI-StuI fragment containing the EGFP expression cassette of pENTR-CAG-EGFP was isolated and ligated to the digested backbone PCR product to generate pCAG-EGFP-MCS1. A SwaI-AscI fragment containing two tandem copies of the 250-bp cHS4 core (54) on both sides of a tdTomato expression cassette was isolated from pBT268 (Addgene plasmid 36880; Liqun Luo laboratory) and cloned between the two MCS loci flanking the CAG-EGFP cassette in pCAG-EGFP-MCS1. The tdTomato cassette of the resulting plasmid, pENTR-cHS4-S, was then replaced with the EGFP expression cassette from pCAG-EGFP-MCS1 to generate pHS4-S containing attL1-(2xcHS4)-[CAG-EGFP-poly(A)]-(2xcHS4)-attL2.
TABLE 3.
Primer sequences
| Primer name | Orientationa | Sequence |
|---|---|---|
| P1 | F | ACTAGTGCGCGCGGCGCGCCACCCAGCTTTCTTGTACAAAGTTGG |
| R | CCAACTTTGTACAAAAAAGCAGGCTATTTAAATGGCCGGCCCATATGGTTAACAGATCTTCTAGAGCTAGCGGGCCC | |
| P2 | F | TTGGGAGCATGCGAATGGAG |
| R | CGCCGGTTCGAAATTCTGGGCACTTATGAATG | |
| P3 | F | GGCCGCTTCGAAGCGTTACATAACTTACGG |
| R | CGCCGCTCTGATTGGCTGCCGCCGCACCTC | |
| P4 | F | CGAATTAGGGACCCTTCGAAGCAAGCTTTATTCCAAGAGGTAGTAACT |
| R | AGCACCTCCGCACGGGACCC | |
| O1 | F | ACCGGTGTATACTGGCCATGGCCGGCCTTAATATC |
| R | GATATTAAGGCCGGCCATGGCCAGTATACACCGGT | |
| Q1 | F | ATCATGGCCGACAAGCAGAAGAAC |
| R | GTACAGCTCGTCCATGCCGAGAGT | |
| Q2 | F | CCTGTCCCCTCAGTTCATGT |
| R | GCTTCAAGTAGTCGGGGATG | |
| Q3 | F | GTGACTACGGCCCTACCG |
| R | GAAACCGGCATCCTCCTCG | |
| Q4 | F | AGCACATCCGCGGACTTGTC |
| R | ACCACCCCGGGTTCGAGG | |
| Q5 | F | CTGACTGACCGCGTTACTCC |
| R | CGTCATTAAACCAAGCGCTAA | |
| M1 | F | GTGAAAGTTTTGAGGGGTTT |
| R | AAAAATACAAAAAAAATTACAACCC | |
| M2 | F | GTTTGTTTTTTTTTTGTGGTTG |
| R | AAAAATACAAAAAAAATTACAACCC | |
| M3 | F | GTAGGGATTTTTTTTGTTTTAAATT |
| R | CATAATTAACAAAAACTCTAAAACC | |
| M4 | F | GTAGTTATTGTTTTTTATGGTAAT |
| R | CATAATTAACAAAAACTCTAAAACC |
F, forward; R, reverse.
(ii) pHS4-L.
A SacI fragment containing the full-length 1.2-kb cHS4 insulator region was isolated from pJC13-1 (54) (kindly provided by G. Felsenfeld, NIH/NIDDK) and cloned into the SacI site of pBluescript to create pBS-cHS4. The SacI fragment was excised, its 3′ overhangs were removed with T4 DNA polymerase, and it was cloned into the filled-in SpeI site upstream of the CAG promoter in pENTR-CAG-EGFP (pENTR-CAG-EGFP-HS4-L-up). The same blunt-ended SacI fragment was then cloned in the same orientation into the StuI site downstream of the poly(A) region in pENTR-CAG-EGFP-HS4-L-up.
(iii) pUCOE1.
A synthetic MCS generated by annealing oligonucleotides O1 was introduced between the SwaI and FseI sites in the upstream MCS of pCAG-EGFP-MCS1 to create pCAG-EGFP-MCS2. A2UCOE (22) was isolated from UCOE Hu-P (Millipore) as an AgeI-MscI fragment and inserted between the same sites in the new MCS of pCAG-EGFP-MCS2.
(iv) pUCOE2.
A 3′ segment of A2UCOE and a 5′ segment of the CAG promoter were separately amplified from pUCOE1 using primer sets P2 and P3, respectively. The two PCR products joined at a BstBI site were cloned into pUCOE1 between an SphI site in the amplified portion of the HNRPA2B1 intron and a SnaBI site in the amplified CAG portion.
(v) pUCOE3.
pUCOE2 was digested with Tth111I and BstBI, cutting at both ends of the HNRPA2B1 intron, and the large fragment was gel purified, its 5′ overhangs were filled in with T4 DNA polymerase, and it was self-ligated.
(vi) pUCOE4.
pUCOE2 was digested with BstBI and HindIII, the 5′ overhangs were filled in with T4 DNA polymerase, and the large fragment was self-ligated to generate pUCOE2 with CAG-EGFP deleted. Adjacent BstBI and HindIII sites were inserted into this plasmid in the CBX3-HNRPA2B1 dual-divergent promoter region by replacing a SanD1 fragment, extending from the center of the region into the HNRPA2B1 exon, with a corresponding PCR fragment generated on pUCOE2 as the template with SanDI-overlapping primers P4-F, specifying the inserted sites, and P4-R. The resulting plasmid was then modified by BstBI-HindIII digestion and ligation to the CAG-EGFP BstBI-HindIII fragment from pUCOE2.
(vii) pUCOE5.
pUCOE2 was digested with Tth111I and HindIII, the 5′ overhangs were filled in with T4 DNA polymerase, and the large fragment was self-ligated to delete CAG-EGFP, along with the HNRPA2B1 intron. Introduction of cloning sites into the CBX3-HNRPA2B1 promoter region, followed by CAG-EGFP insertion, was as described for pUCOE4 above.
(viii) pLuc.
The mir-941 precursor in pEP-mir (Cell Biolabs) was removed by digestion with BamHI and NheI, and the vector piece was blunt ended and self-ligated to generate an EF1α promoter-intron-mCherry/Puro plasmid termed pEP-mCherry. The mCherry/Puro cDNA was replaced by EGFP from pCAG-GFP (7) to create pEP-EGFP. EGFP was subsequently replaced with a blunted BamHI-SacI fragment containing the firefly luciferase gene from pMIR-report (Ambion), producing pEP-luc. The complete EF1α-luc cassette was then isolated from pEP-luc by partial digestion with ClaI and SpeI and used to replace the CAG-EGFP cassette in pENTR-CAG-EGFP.
(ix) pU-Luc.
The EF1α-luc cassette from pEP-luc was isolated as a 5′ ClaI-EcoRI fragment and a 3′ EcoRI-BglII fragment. The two fragments were subcloned into BstBI-BglII-digested pUCOE2, replacing the CAG-EGFP cassette. The PacI-NheI fragment separating A2UCOE from the EF1α-luc cassette was replaced with the PacI-NheI fragment of pLuc to remove differences between pLuc and pU-Luc outside the A2UCOE region.
Viral genome engineering.
Viral genome constructs containing CAG-EGFP in the UL50-UL51 intergenic region were generated by LR clonase (Invitrogen)-mediated recombination between appropriate pENTR-based plasmids and the GW region of JΔNI10GW BAC DNA, essentially as described previously (7). vLuc and vU-Luc were generated by GW recombination between pLuc or pU-Luc, and JΔNI11GW. JΔNI11GW was engineered by Red/ET (Gene Bridges, Heidelberg, Germany)-mediated recombination of a PCR-amplified GW-Zeo cassette (7) into the intergenic region between UL50 and UL51 of JΔNI7GFP BAC (7) in Escherichia coli. All the BAC recombinants were confirmed by PCR analysis, field inversion gel electrophoresis (FIGE) analysis of restriction enzyme digests, and targeted DNA sequencing.
Virus production.
Recombinant viruses were produced by transfection of U2OS-ICP4/ICP27 cells with purified BAC DNA, as described previously (7). Following cytopathic effect (CPE), supernatants were harvested for standard virus amplification in the same cell line. The titers of virus stocks were determined on U2OS-ICP4/ICP27 cells as PFU/ml. Genome copy titers (gc per milliliter) were determined by real-time quantitative PCR (qPCR) for the viral gD gene (7). Deletion of the BAC region, including the associated chloramphenicol resistance and lacZ genes, was essentially as described previously (7). Briefly, viruses were passaged twice through U2OS-ICP4/ICP27-Cre cells, and isolated plaques generated by limiting dilution were screened by PCR across the expected deletion and by staining for β-galactosidase activity. Isolates with the BAC deleted were propagated on U2OS-ICP4/ICP27 cells.
Luciferase assay.
Luciferase assays were performed with a luciferase assay system (Promega) according to the manufacturer's instructions. Cells were lysed with cell culture lysis reagent (CCLR; Promega), and the lysate was used for both luciferase activity measurement in a BioTek Synergy2 microplate reader and PCR determination of viral genome copy numbers for normalization.
Quantitative reverse transcription-PCR.
qRT-PCR was performed as previously described (7) using a Cells-to-cDNA II kit (Ambion) for mRNA extraction and cDNA synthesis. The qPCR primers for GFP (Q1), mCherry (Q2), UL50 (Q3), and UL51 (Q4) are listed in Table 3. The results were normalized to the viral genome copy numbers measured in lysates prior to the DNase step for RNA isolation.
Immunofluorescence.
Transduced rDRGs were fixed in Buffered Formalde-Fresh (Fisher Scientific) at room temperature for 10 min and blocked with 5% normal goat serum–0.2% Triton X-100 in phosphate-buffered saline (PBS) for 1 h at room temperature. After blocking, the cells were incubated with anti-luciferase antibody (Abcam; ab21176; 1:500) at 4°C overnight, and biotin (Sigma; B7389; 1:1,000)-avidin (Invitrogen; A11236; 1:1,000) was added the next day for a 1-h incubation at room temperature. Fluorescence images were collected with an Olympus FV1000 confocal microscope.
ChIP.
Two million HDFs were plated in 10-cm dishes for infection the following day (5,000 gc/cell). At 7 dpi, chromatin immunoprecipitation (ChIP) was carried out as described by Nelson and colleagues (55). Briefly, infected HDFs were cross-linked with 37% formaldehyde for 15 min, and the reaction was quenched with 1 M glycine. Cells were collected by centrifugation, washed in cold PBS, and lysed in immunoprecipitation (IP) buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.5, 5 mM EDTA, 0.5% [vol/vol] NP-40, and 1.0% [vol/vol] Triton X-100). The lysates were sonicated using an S-3000 Misonix sonicator. Immunoprecipitations were carried out overnight at 4°C with normal rabbit IgG (Santa Cruz; sc-2027), rabbit anti-histone H3 antibody (Millipore; 06-755), or rabbit anti-H3K9Me3 (Millipore; 07-442). Dynabeads M-280 sheep anti-rabbit IgG (Life Technologies) was used to precipitate the antibody-bound DNA. A fraction of the sonicated cells was used as the input control. qRT-PCR was carried out using primer set Q5 within the CAG promoter.
Bisulfite conversion and sequencing.
Viral DNA was extracted with a DNeasy blood and tissue kit (Qiagen) prior to (input) and at 7 days after infection of HDFs. Bisulfite conversion was carried out using an Epitech kit (Qiagen) according to the manufacturer's instructions. The converted input DNA was amplified with primer pair M1, and the converted DNA from infected HDFs was amplified with primer pair M2, followed by a nested PCR with primer pair M1 using GoTaq DNA polymerase M300 (Promega) to examine the viral DNA methylation status of a 5′ portion of the β-actin component of the CAG promoter. Likewise, primer pairs M3 and M4 were used for pre- and postinfection viral DNA PCR to determine the methylation status of a 3′ portion of the β-actin segment of the CAG promoter. The PCR products were gel purified and cloned into pCR2.1-TOPO using a TOPO TA Cloning kit (no. 450641; Invitrogen). Inserts of individual clones were sequenced using the M13 reverse primer.
Animals.
Male CD1 mice were purchased from Charles River Italy (Calco, Lecco, Italy). Experiments were performed at the University of Ferrara in compliance with the guidelines for the ethical treatment of experimental animals (authorization from the Italian Ministry for Health D.M. 18/2017-PR).
HSV-1 infusion in vivo.
General anesthesia was induced in 25- to 30-g mice by coadministration of ketamine (45 mg/kg of body weight intraperitoneally [i.p.]) and xylazine (7 mg/kg i.p.) and was maintained with 1.4% isoflurane in air (flow, 1.2 ml/min). The animals were placed in a stereotactic frame on a temperature-controlled heating pad, and a total of 2 × 109 viral genome copies was injected into the right dorsal hippocampus. Viral vectors (vLuc and vU-Luc; n = 12 mice/vector) or PBS (n = 3) was injected by stereotactic placement of a custom-made borosilicate glass needle (56). The needle tips were chamfered by laser microdissector cutting (LMD6500 Laser Microdissection System; Leica Microsystems, Wetzlar, Germany) (angle, 140°; inner and outer diameters at the tip, 80 and 100 μm, respectively). An additional hole (diameter, 10 μm) was opened 100 μm above the tip to favor spreading of the injected virus (56). The needle was bottom filled with the viral solution and linked to a microperfusion pump set to inject a total of 2 μl at a flow rate of 0.3 μl/min. The stereotactic coordinates (57) were 1.6 mm lateral, 1.9 mm posterior to bregma, and 1.9 mm deep from dura. Carprofen (5 mg/kg i.p.) was administered after surgery to prevent pain.
In vivo bioluminescence imaging.
At selected time points (3, 7, 14, and 28 days postsurgery), mice were anesthetized with 2% isoflurane and oxygen, and d-luciferin substrate (Roche) was injected i.p. at a dose of 200 mg/kg. In vivo bioluminescence was detected 8 min after the injection in a total-body luminometer (IVIS Lumina; Caliper-PerkinElmer, Hopkinton, MA, USA). Photon emissions in the region of interest were quantified with Living Image software (Caliper).
Tissue preparation.
Animals were deeply anesthetized with pentobarbital and transcardially perfused with PBS, followed by 4% paraformaldehyde in 0.1 M PBS. Brains were isolated and postfixed in 4% paraformaldehyde for 1 h, cryoprotected in 30% sucrose at 4°C until the tissue sank, and flash-frozen in isopentane prechilled at −80°C.
Tissue immunofluorescence.
Twenty-micrometer coronal sections were cut at −20°C in a cryostat (Leica), rinsed in 0.1 M PBS, permeabilized for 10 min in 0.3% Triton X-100–PBS, and blocked for 30 min in 0.1 M PBS with 10% normal goat serum. The sections were then incubated overnight at 4°C with primary antibodies diluted in 0.1 M PBS: rabbit anti-mCherry (Abcam), 1:500; chicken anti-GFP (Abcam), 1:200. The sections were washed twice in PBS and stained for 2 h at room temperature with 1:500-diluted Alexa Fluor 594 goat anti-rabbit or Alexa Fluor 488 goat anti-chicken antibody (Molecular Probes, Eugene, OR, USA).
NeuroTrace staining.
After rehydration in 0.1 M PBS, sections were permeabilized with 0.1% Triton X-100 in PBS for 10 min, washed twice for 5 min each time in PBS, and stained with NeuroTrace 435/455 Blue Fluorescent Nissl stain (ThermoFisher; dilution, 1:200) for 20 min at room temperature. The sections were then washed twice in PBS and finally left for 2 h at room temperature in PBS before imaging.
Statistics.
For quantitative analyses in cell culture, two plaque-purified isolates per vector construct were tested in parallel in duplicate wells in 48- or 96-well plates. Each experiment was then repeated once on a separate day with one of the two isolates per construct. Data were recorded for each well, and the mean of the duplicate wells was calculated. Viral genome qPCR and mRNA qRT-PCR measurements were performed in duplicate or triplicate per well, and the mean qRT-PCR value was normalized to the mean viral genome qPCR value for each well. The data were analyzed separately for each isolate and repeat experiments by 2-way analysis of variance (ANOVA), with the exception of Fig. 5 (Mann-Whitney test). The representative data shown (means and standard deviations [SD] of duplicate wells) are from one isolate per vector construct, with the exception of Fig. 7 (combined data from duplicate wells for 2 isolates). In vivo bioluminescence results were analyzed by Mann-Whitney U test.
Data availability.
The materials described in this publication are available on request to members of the scientific community for noncommercial purposes.
ACKNOWLEDGMENTS
We are grateful to G. Felsenfeld for the pJC13-1 plasmid, Johnny Huard and Xueqin Gao for hMDSCs, Mingdi Zhang for rat DRG culturing and immunostaining, William Goins for technical advice, and Bonnie Hall for helpful discussions.
This work was supported by grants to J. C. Glorioso from the NIH (NS064988 and DK044935) and the CHDI Foundation (A3777 and A8790) and to M. Simonato from the European Community (FP7-PEOPLE-2011-IAPP project 285827 [EPIXCHANGE]), the Italian Ministry for Education, University and Research (PRIN project 2010N8PBAA [INBDNF]), and the Regione Lombardia (POR FESR 2014-2020 project ID 239047 [NeOn]). F. Han was supported by funding from the China Scholarship Council.
Experiments were designed by F. Han, Y. Miyagawa, G. Verlengia, M. Simonato, J. C. Glorioso, and J. B. Cohen; vector engineering, virus production, and in vitro analyses were performed by F. Han and Y. Miyagawa; animal experiments were carried out by G. Verlengia, S. Ingusci, and M. Soukupova; all the authors conducted data analysis and interpretation; F. Han, Y. Miyagawa, J. C. Glorioso, and J. B. Cohen wrote the manuscript.
G. Verlengia, M. Simonato, and J. C. Glorioso are founders of NuvoVec srl. Y. Miyagawa, J. C. Glorioso and J. B. Cohen are coinventors of intellectual property licensed to Coda Biotherapeutics, Inc., J. C. Glorioso and J. B. Cohen are coinventors of intellectual property licensed to Oncorus, Inc., and J. C. Glorioso is a founder and consultant of Coda Biotherapeutics, Inc., and Oncorus, Inc.
REFERENCES
- 1.Roizman B, Zhou G. 2015. The 3 facets of regulation of herpes simplex virus gene expression: a critical inquiry. Virology 479-480:562–567. doi: 10.1016/j.virol.2015.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knipe DM. 2015. Nuclear sensing of viral DNA, epigenetic regulation of herpes simplex virus infection, and innate immunity. Virology 479-480:153–159. doi: 10.1016/j.virol.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Samaniego LA, Neiderhiser L, DeLuca NA. 1998. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J Virol 72:3307–3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harkness JM, Kader M, DeLuca NA. 2014. Transcription of the herpes simplex virus 1 genome during productive and quiescent infection of neuronal and nonneuronal cells. J Virol 88:6847–6861. doi: 10.1128/JVI.00516-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Du T, Zhou G, Roizman B. 2011. HSV-1 gene expression from reactivated ganglia is disordered and concurrent with suppression of latency-associated transcript and miRNAs. Proc Natl Acad Sci U S A 108:18820–18824. doi: 10.1073/pnas.1117203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raja P, Lee JS, Pan D, Pesola JM, Coen DM, Knipe DM. 2016. A herpesviral lytic protein regulates the structure of latent viral chromatin. mBio 7:e00633-. doi: 10.1128/mBio.00633-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyagawa Y, Marino P, Verlengia G, Uchida H, Goins WF, Yokota S, Geller DA, Yoshida O, Mester J, Cohen JB, Glorioso JC. 2015. Herpes simplex viral-vector design for efficient transduction of nonneuronal cells without cytotoxicity. Proc Natl Acad Sci U S A 112:E1632–E1641. doi: 10.1073/pnas.1423556112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amelio AL, McAnany PK, Bloom DC. 2006. A chromatin insulator-like element in the herpes simplex virus type 1 latency-associated transcript region binds CCCTC-binding factor and displays enhancer-blocking and silencing activities. J Virol 80:2358–2368. doi: 10.1128/JVI.80.5.2358-2368.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palmer JA, Branston RH, Lilley CE, Robinson MJ, Groutsi F, Smith J, Latchman DS, Coffin RS. 2000. Development and optimization of herpes simplex virus vectors for multiple long-term gene delivery to the peripheral nervous system. J Virol 74:5604–5618. doi: 10.1128/JVI.74.12.5604-5618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goins WF, Sternberg LR, Croen KD, Krause PR, Hendricks RL, Fink DJ, Straus SE, Levine M, Glorioso JC. 1994. A novel latency-active promoter is contained within the herpes simplex virus type 1 UL flanking repeats. J Virol 68:2239–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emery DW. 2011. The use of chromatin insulators to improve the expression and safety of integrating gene transfer vectors. Hum Gene Ther 22:761–774. doi: 10.1089/hum.2010.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Argyros O, Wong SP, Harbottle RP. 2011. Non-viral episomal modification of cells using S/MAR elements. Expert Opin Biol Ther 11:1177–1191. doi: 10.1517/14712598.2011.582035. [DOI] [PubMed] [Google Scholar]
- 13.Antoniou MN, Skipper KA, Anakok O. 2013. Optimizing retroviral gene expression for effective therapies. Hum Gene Ther 24:363–374. doi: 10.1089/hum.2013.062. [DOI] [PubMed] [Google Scholar]
- 14.Chung JH, Bell AC, Felsenfeld G. 1997. Characterization of the chicken beta-globin insulator. Proc Natl Acad Sci U S A 94:575–580. doi: 10.1073/pnas.94.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Recillas-Targa F, Pikaart MJ, Burgess-Beusse B, Bell AC, Litt MD, West AG, Gaszner M, Felsenfeld G. 2002. Position-effect protection and enhancer blocking by the chicken beta-globin insulator are separable activities. Proc Natl Acad Sci U S A 99:6883–6888. doi: 10.1073/pnas.102179399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao S. 2003. Retrovirus silencer blocking by the cHS4 insulator is CTCF independent. Nucleic Acids Res 31:5317–5323. doi: 10.1093/nar/gkg742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aker M, Tubb J, Groth AC, Bukovsky AA, Bell AC, Felsenfeld G, Kiem HP, Stamatoyannopoulos G, Emery DW. 2007. Extended core sequences from the cHS4 insulator are necessary for protecting retroviral vectors from silencing position effects. Hum Gene Ther 18:333–343. doi: 10.1089/hum.2007.021. [DOI] [PubMed] [Google Scholar]
- 18.Suttiprapa S, Rinaldi G, Brindley PJ. 2012. Prototypic chromatin insulator cHS4 protects retroviral transgene from silencing in Schistosoma mansoni. Transgenic Res 21:555–566. doi: 10.1007/s11248-011-9556-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emery DW, Yannaki E, Tubb J, Nishino T, Li Q, Stamatoyannopoulos G. 2002. Development of virus vectors for gene therapy of beta chain hemoglobinopathies: flanking with a chromatin insulator reduces gamma-globin gene silencing in vivo. Blood 100:2012–2019. doi: 10.1182/blood-2002-01-0219. [DOI] [PubMed] [Google Scholar]
- 20.Arumugam PI, Scholes J, Perelman N, Xia P, Yee JK, Malik P. 2007. Improved human beta-globin expression from self-inactivating lentiviral vectors carrying the chicken hypersensitive site-4 (cHS4) insulator element. Mol Ther 15:1863–1871. doi: 10.1038/sj.mt.6300259. [DOI] [PubMed] [Google Scholar]
- 21.Lindahl Allen M, Antoniou M. 2007. Correlation of DNA methylation with histone modifications across the HNRPA2B1-CBX3 ubiquitously-acting chromatin open element (UCOE). Epigenetics 2:227–236. doi: 10.4161/epi.2.4.5231. [DOI] [PubMed] [Google Scholar]
- 22.Antoniou M, Harland L, Mustoe T, Williams S, Holdstock J, Yague E, Mulcahy T, Griffiths M, Edwards S, Ioannou PA, Mountain A, Crombie R. 2003. Transgenes encompassing dual-promoter CpG islands from the human TBP and HNRPA2B1 loci are resistant to heterochromatin-mediated silencing. Genomics 82:269–279. doi: 10.1016/S0888-7543(03)00107-1. [DOI] [PubMed] [Google Scholar]
- 23.Williams S, Mustoe T, Mulcahy T, Griffiths M, Simpson D, Antoniou M, Irvine A, Mountain A, Crombie R. 2005. CpG-island fragments from the HNRPA2B1/CBX3 genomic locus reduce silencing and enhance transgene expression from the hCMV promoter/enhancer in mammalian cells. BMC Biotechnol 5:17. doi: 10.1186/1472-6750-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boscolo S, Mion F, Licciulli M, Macor P, De Maso L, Brce M, Antoniou MN, Marzari R, Santoro C, Sblattero D. 2012. Simple scale-up of recombinant antibody production using an UCOE containing vector. N Biotechnol 29:477–484. doi: 10.1016/j.nbt.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 25.Brendel C, Muller-Kuller U, Schultze-Strasser S, Stein S, Chen-Wichmann L, Krattenmacher A, Kunkel H, Dillmann A, Antoniou MN, Grez M. 2012. Physiological regulation of transgene expression by a lentiviral vector containing the A2UCOE linked to a myeloid promoter. Gene Ther 19:1018–1029. doi: 10.1038/gt.2011.167. [DOI] [PubMed] [Google Scholar]
- 26.Zhang F, Thornhill SI, Howe SJ, Ulaganathan M, Schambach A, Sinclair J, Kinnon C, Gaspar HB, Antoniou M, Thrasher AJ. 2007. Lentiviral vectors containing an enhancer-less ubiquitously acting chromatin opening element (UCOE) provide highly reproducible and stable transgene expression in hematopoietic cells. Blood 110:1448–1457. doi: 10.1182/blood-2006-12-060814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang F, Frost AR, Blundell MP, Bales O, Antoniou MN, Thrasher AJ. 2010. A ubiquitous chromatin opening element (UCOE) confers resistance to DNA methylation-mediated silencing of lentiviral vectors. Mol Ther 18:1640–1649. doi: 10.1038/mt.2010.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saunders F, Sweeney B, Antoniou MN, Stephens P, Cain K. 2015. Chromatin function modifying elements in an industrial antibody production platform—comparison of UCOE, MAR, STAR and cHS4 elements. PLoS One 10:e0120096. doi: 10.1371/journal.pone.0120096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muller-Kuller U, Ackermann M, Kolodziej S, Brendel C, Fritsch J, Lachmann N, Kunkel H, Lausen J, Schambach A, Moritz T, Grez M. 2015. A minimal ubiquitous chromatin opening element (UCOE) effectively prevents silencing of juxtaposed heterologous promoters by epigenetic remodeling in multipotent and pluripotent stem cells. Nucleic Acids Res 43:1577–1592. doi: 10.1093/nar/gkv019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uchiyama T, Adriani M, Jagadeesh GJ, Paine A, Candotti F. 2012. Foamy virus vector-mediated gene correction of a mouse model of Wiskott-Aldrich syndrome. Mol Ther 20:1270–1279. doi: 10.1038/mt.2011.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verlengia G, Miyagawa Y, Ingusci S, Cohen JB, Simonato M, Glorioso JC. 2017. Engineered HSV vector achieves safe long-term transgene expression in the central nervous system. Sci Rep 7:1507. doi: 10.1038/s41598-017-01635-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyagawa Y, Verlengia G, Reinhart B, Han F, Uchida H, Zucchini S, Goins WF, Simonato M, Cohen JB, Glorioso JC. 2017. Deletion of the virion host shut-off gene enhances neuronal-selective transgene expression from an HSV vector lacking functional IE genes. Mol Ther Methods Clin Dev 6:79–90. doi: 10.1016/j.omtm.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hagedorn C, Antoniou MN, Lipps HJ. 2013. Genomic cis-acting sequences improve expression and establishment of a nonviral vector. Mol Ther Nucleic Acids 2:e118. doi: 10.1038/mtna.2013.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferenczy MW, DeLuca NA. 2011. Reversal of heterochromatic silencing of quiescent herpes simplex virus type 1 by ICP0. J Virol 85:3424–3435. doi: 10.1128/JVI.02263-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoelzer K, Shackelton LA, Parrish CR. 2008. Presence and role of cytosine methylation in DNA viruses of animals. Nucleic Acids Res 36:2825–2837. doi: 10.1093/nar/gkn121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bloom DC, Giordani NV, Kwiatkowski DL. 2010. Epigenetic regulation of latent HSV-1 gene expression. Biochim Biophys Acta 1799:246–256. doi: 10.1016/j.bbagrm.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Majocchi S, Aritonovska E, Mermod N. 2014. Epigenetic regulatory elements associate with specific histone modifications to prevent silencing of telomeric genes. Nucleic Acids Res 42:193–204. doi: 10.1093/nar/gkt880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Talbot GE, Waddington SN, Bales O, Tchen RC, Antoniou MN. 2010. Desmin-regulated lentiviral vectors for skeletal muscle gene transfer. Mol Ther 18:601–608. doi: 10.1038/mt.2009.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terry-Allison T, Smith CA, DeLuca NA. 2007. Relaxed repression of herpes simplex virus type 1 genomes in murine trigeminal neurons. J Virol 81:12394–12405. doi: 10.1128/JVI.01068-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lilley CE, Groutsi F, Han Z, Palmer JA, Anderson PN, Latchman DS, Coffin RS. 2001. Multiple immediate-early gene-deficient herpes simplex virus vectors allowing efficient gene delivery to neurons in culture and widespread gene delivery to the central nervous system in vivo. J Virol 75:4343–4356. doi: 10.1128/JVI.75.9.4343-4356.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ong CT, Corces VG. 2014. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet 15:234–246. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim S, Yu NK, Kaang BK. 2015. CTCF as a multifunctional protein in genome regulation and gene expression. Exp Mol Med 47:e166. doi: 10.1038/emm.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghirlando R, Felsenfeld G. 2016. CTCF: making the right connections. Genes Dev 30:881–891. doi: 10.1101/gad.277863.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Merkenschlager M, Nora EP. 2016. CTCF and cohesin in genome folding and transcriptional gene regulation. Annu Rev Genomics Hum Genet 17:17–43. doi: 10.1146/annurev-genom-083115-022339. [DOI] [PubMed] [Google Scholar]
- 45.Barrington C, Finn R, Hadjur S. 2017. Cohesin biology meets the loop extrusion model. Chromosome Res 25:51–60. doi: 10.1007/s10577-017-9550-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Busslinger GA, Stocsits RR, van der Lelij P, Axelsson E, Tedeschi A, Galjart N, Peters JM. 2017. Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nature 544:503–507. doi: 10.1038/nature22063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee JS, Raja P, Pan D, Pesola JM, Coen DM, Knipe DM. 2018. CCCTC-binding factor acts as a heterochromatin barrier on herpes simplex viral latent chromatin and contributes to poised latent infection. mBio 9 e02372-. doi: 10.1128/mBio.02372-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang QY, Zhou C, Johnson KE, Colgrove RC, Coen DM, Knipe DM. 2005. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc Natl Acad Sci U S A 102:16055–16059. doi: 10.1073/pnas.0505850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cliffe AR, Knipe DM. 2008. Herpes simplex virus ICP0 promotes both histone removal and acetylation on viral DNA during lytic infection. J Virol 82:12030–12038. doi: 10.1128/JVI.01575-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ferenczy MW, DeLuca NA. 2009. Epigenetic modulation of gene expression from quiescent herpes simplex virus genomes. J Virol 83:8514–8524. doi: 10.1128/JVI.00785-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ertel MK, Cammarata AL, Hron RJ, Neumann DM. 2012. CTCF occupation of the herpes simplex virus 1 genome is disrupted at early times postreactivation in a transcription-dependent manner. J Virol 86:12741–12759. doi: 10.1128/JVI.01655-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lang F, Li X, Vladimirova O, Hu B, Chen G, Xiao Y, Singh V, Lu D, Li L, Han H, Wickramasinghe JM, Smith ST, Zheng C, Li Q, Lieberman PM, Fraser NW, Zhou J. 2017. CTCF interacts with the lytic HSV-1 genome to promote viral transcription. Sci Rep 7:39861. doi: 10.1038/srep39861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Washington SD, Musarrat F, Ertel MK, Backes GL, Neumann DM. 2018. CTCF binding sites in the herpes simplex virus 1 genome display site-specific CTCF occupation, protein recruitment, and insulator function. J Virol 92 e00156-18. doi: 10.1128/JVI.00156-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chung JH, Whiteley M, Felsenfeld G. 1993. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell 74:505–514. doi: 10.1016/0092-8674(93)80052-G. [DOI] [PubMed] [Google Scholar]
- 55.Nelson JD, Denisenko O, Bomsztyk K. 2006. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc 1:179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- 56.Paolone G, Falcicchia C, Verlengia G, Barbieri M, Binaschi A, Paliotto F, Paradiso B, Soukupova M, Zucchini S, Simonato M. 2018. Personalized needles for microinjections in the rodent brain. J Vis Exp doi: 10.1128/JVI.00156-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paxinos GT, Franklin KBJ. 2001. The mouse brain in stereotaxic coordinates, 2nd ed Academic Press, San Diego, CA. [Google Scholar]
- 58.Uchida H, Chan J, Goins WF, Grandi P, Kumagai I, Cohen JB, Glorioso JC. 2010. A double mutation in glycoprotein gB compensates for ineffective gD-dependent initiation of herpes simplex virus type 1 infection. J Virol 84:12200–12209. doi: 10.1128/JVI.01633-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Szymczak AL, Vignali DA. 2005. Development of 2A peptide-based strategies in the design of multicistronic vectors. Expert Opin Biol Ther 5:627–638. doi: 10.1517/14712598.5.5.627. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The materials described in this publication are available on request to members of the scientific community for noncommercial purposes.