Human respiratory syncytial virus (RSV) is an important human pathogen that lacks a licensed vaccine or antiviral drug suitable for routine use. We describe here the evaluation of recombinant murine pneumonia virus (rMPV) as a live-attenuated vector that expresses the RSV F protein, the major RSV neutralization antigen, as an experimental RSV vaccine. The rMPV-RSV-F vectors expressing RSV F from the first, third, or fourth gene position were genetically stable and were not restricted for replication in vitro. In contrast, the vectors exhibited highly attenuated replication in the respiratory tract of rhesus macaques, maintained stable RSV F expression, and induced RSV-neutralizing serum antibodies at high titers similar to those conferred by wild-type RSV. Given the lack of preexisting immunity to MPV in humans and the lack of cross-neutralization and cross-protection between MPV and RSV, an rMPV-vectored RSV vaccine should be immunogenic in both RSV-naive children and RSV-experienced adults.
KEYWORDS: F protein, murine pneumonia virus, RSV F, fusion protein, intranasal vaccine, live-attenuated vaccine, mucosal vaccine, neutralizing antibodies, respiratory syncytial virus, vectored vaccine
ABSTRACT
Human respiratory syncytial virus (RSV) continues to be the leading viral cause of severe acute lower respiratory tract disease in infants and children worldwide. A licensed vaccine or antiviral drug suitable for routine use remains unavailable. Like RSV, Murine pneumonia virus (MPV) is a member of the genus Orthopneumovirus, family Pneumoviridae. Humans are not normally exposed to MPV, and MPV is not cross-protective with RSV. We evaluated MPV as an RSV vaccine vector expressing the RSV fusion (F) glycoprotein. The RSV F open reading frame (ORF) was codon optimized, and the encoded RSV F protein was made identical to an early passage of RSV strain A2. The RSV F ORF was placed under the control of MPV transcription signals and inserted at the first (rMPV-F1), third (rMPV-F3), or fourth (rMPV-F4) gene position of a version of the MPV genome that contained a codon-pair-optimized polymerase (L) gene. The recovered viruses replicated in vitro as efficiently as the empty vector, with stable expression of RSV F protein. Replication and immunogenicity of rMPV-F1 and rMPV-F3 were evaluated in rhesus macaques following intranasal and intratracheal administration. Both viruses replicated at low levels in the upper and lower respiratory tracts, maintained stable RSV F expression, and induced RSV-neutralizing serum antibodies at high levels similar to those induced by wild-type RSV replicating to a 5- to 25-fold-higher titer. In conclusion, this study demonstrated that rMPV provides a highly attenuated yet immunogenic vector for the expression of RSV F protein, with potential application in RSV-naive and RSV-experienced populations.
IMPORTANCE Human respiratory syncytial virus (RSV) is an important human pathogen that lacks a licensed vaccine or antiviral drug suitable for routine use. We describe here the evaluation of recombinant murine pneumonia virus (rMPV) as a live-attenuated vector that expresses the RSV F protein, the major RSV neutralization antigen, as an experimental RSV vaccine. The rMPV-RSV-F vectors expressing RSV F from the first, third, or fourth gene position were genetically stable and were not restricted for replication in vitro. In contrast, the vectors exhibited highly attenuated replication in the respiratory tract of rhesus macaques, maintained stable RSV F expression, and induced RSV-neutralizing serum antibodies at high titers similar to those conferred by wild-type RSV. Given the lack of preexisting immunity to MPV in humans and the lack of cross-neutralization and cross-protection between MPV and RSV, an rMPV-vectored RSV vaccine should be immunogenic in both RSV-naive children and RSV-experienced adults.
INTRODUCTION
Acute respiratory infections during early childhood constitute a major human health burden. Human respiratory syncytial virus (RSV) is the most important viral cause of severe acute pediatric respiratory infections worldwide. Mortality due to RSV in the postneonatal (28 days to 1 year old) population is second only to that due to malaria (1). It is estimated that RSV causes 34 million lower respiratory tract infections, 4 million hospitalizations, and 66,000 to 199,000 deaths every year in children less than 5 years of age (2, 3). Most mortality occurs in the developing world where supportive care is less available (2). Mortality is low in the developed countries, but morbidity is substantial: in the United States alone, RSV is associated with an estimated 132,000 to 172,000 hospitalizations annually in children less than 5 years old (4). There is not yet available a vaccine or an effective antiviral drug suitable for routine use.
RSV is an enveloped, single-stranded negative-sense RNA virus of the genus Orthopneumovirus and the family Pneumoviridae (5) and possesses 10 genes that encode 11 proteins. The fusion (F) and the attachment (G) surface glycoproteins are the two viral neutralization antigens and are the major protective antigens, with F playing the predominant role. RSV F is a type I transmembrane envelope glycoprotein that mediates fusion of the virion envelope with the host cell membrane during entry. RSV F-specific neutralizing antibody titers correlate with protection against RSV infection (6). The clinical use of palivizumab, an F-specific monoclonal antibody, substantially reduces hospitalization for RSV disease in high-risk infants (7). Moreover, the RSV F protein has substantial sequence and antigenic conservation between RSV strains and subgroups, in contrast to the substantial divergence of the G protein. Thus, the F protein is considered to be the most important antigen for an RSV vaccine.
RSV vaccines based on inactivated virus or protein subunits, when administered to RSV-naive infants and children, are associated with enhanced disease (8–10) upon subsequent natural RSV exposure and therefore are contraindicated for this population. In contrast, live-attenuated RSV strains and parainfluenza virus (PIV)-vectored vaccines were not associated with disease enhancement in clinical trials in this population (11–13) and thus are safe for pediatric use when suitably attenuated. A live RSV vaccine would be given by the intranasal route and thus has the additional advantage of inducing local immunity at the site of RSV infection as well as systemic immunity. The phenomenon of enhanced RSV disease has not been observed in RSV-experienced vaccinees, and therefore adults could safely be immunized with either a live or an inactivated vaccine.
Murine pneumonia virus (MPV), previously called pneumonia virus of mice (PVM), is a murine homolog of RSV and belongs to the same genus, Orthopneumovirus, of the family Pneumoviridae (5). MPV has a genome of approximately 15 kb and has essentially the same array of genes encoding the same constellation of proteins as RSV, namely (listed in 3′ to 5′ gene order), nonstructural protein 1 (NS1), nonstructural protein 2 (NS2), nucleoprotein (N), phosphoprotein (P), matrix protein (M), small hydrophobic protein (SH), attachment glycoprotein (G), fusion glycoprotein (F), M2-1 and M2-2 proteins, and polymerase protein (L). The various genes and proteins of MPV share 30 to 62% nucleotide sequence identity and 10 to 60% amino acid sequence identity with RSV (14). Certain characteristics of MPV make it an attractive vector for developing an RSV vaccine: (i) MPV replicates in the superficial epithelial cells of the respiratory tract, similar to RSV, and thus is expected to induce both systemic immunity and local immune responses in the respiratory tract, where RSV infects and replicates; (ii) MPV was shown to have highly attenuated replication in the respiratory tract of African green monkeys (AGMs) and rhesus macaques, presumably due to host range restriction (15), and therefore would be expected to be highly attenuated in the human respiratory tract; (iii) MPV was surprisingly immunogenic in primates despite the high level of restriction, which was the feature that prompted the present study; (iv) we previously reported that humans do not appear to be naturally exposed to MPV; and (v) we also reported a lack of significant antigenic reactivity between human or nonhuman primate RSV-specific serum antibodies and MPV, and a lack of cross-protection in mice between RSV and MPV (15). This lack of cross-reaction and cross-neutralization between RSV and MPV would encourage evaluation of an MPV-vectored RSV vaccine not only in RSV-naive pediatric recipients but also in the RSV-experienced adult population and as a boost following an RSV vaccine in all age groups.
In the present study, we evaluated MPV as a vector to express the RSV F protein as a candidate RSV vaccine for intranasal immunization. Unmodified RSV F protein with a sequence corresponding to that of an early passage of the RSV strain A2 was expressed from the first (pre-NS1), third (between NS2 and N), or fourth (between N and P) gene position of the MPV genome. The vaccine candidates were characterized in vitro for replication, genetic stability, and protein expression and also were evaluated for replication, stability, and immunogenicity against RSV in rhesus macaques.
RESULTS
Recovery of the rMPV-RSV-F vectors expressing the RSV F protein.
We previously showed that recombinant MPV strain 15 (16) is highly attenuated in rhesus macaques and African green monkeys, with only trace levels of shedding on occasional days in only some of the animals (15). Adding a supernumerary gene to a single-stranded negative-sense RNA virus can be further attenuating. In an effort to avoid overattenuation, we used as a vector a version of MPV strain 15 in which most of the L open reading frame (ORF) had been subjected to codon pair optimization (CPO). The construction of this MPV full-length cDNA bearing a CPO L gene (which for simplicity will be called rMPV here) was part of our separate study that will be described elsewhere. CPO of the L ORF was done on the premise that this might increase expression of the L protein, which in turn might increase virus replication. The effects of this modification turned out to be slight, namely, no effect on replication and a modest increase in the pathogenicity in mice (unpublished data), and so the vector is similar to recombinant wild-type (wt) MPV strain 15.
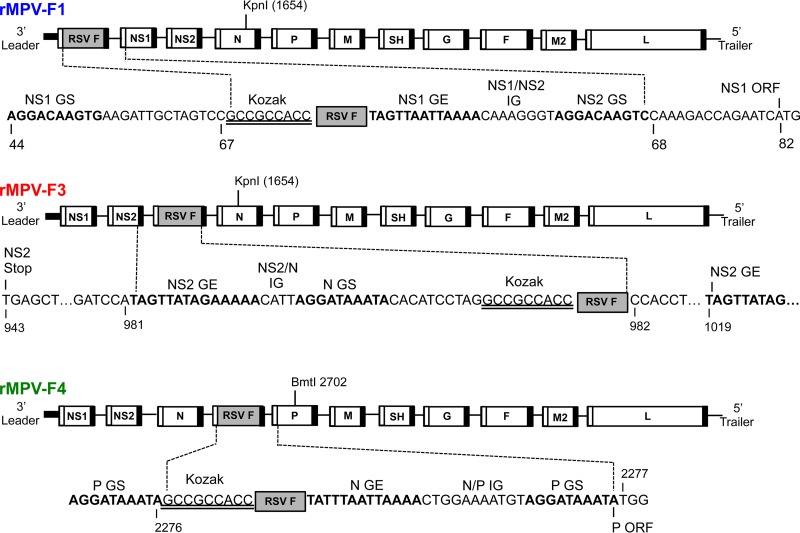
The RSV F ORF (strain A2) was codon optimized for efficient expression in humans, and the encoded protein was modified to contain the two amino acid assignments (K66E and Q101P), called HEK, that make it identical at the amino acid level to that of an early passage of RSV strain A2 (17). This ORF was engineered to be under the control of a set of MPV transcription signals and was inserted at three different gene positions of the rMPV genome (Fig. 1): (i) the first gene position upstream of the NS1 ORF, named rMPV-F1; (ii) the third gene position between the NS2 and N genes, named rMPV-F3; and (iii) the fourth gene position between the N and P genes, named rMPV-F4. RSV F transcription is driven by NS1 gene start-NS1 gene end, N gene start-NS2 gene end, and P gene start-N gene end for rMPV-F1, -F3, and -F4, respectively. All three rMPV-RSV-F viruses were readily recovered by reverse genetics. The viral genomes were sequenced by Sanger sequencing of the uncloned reverse transcription (RT)-PCR products and were found to be free of any adventitious mutations.
FIG 1.
Diagrams of rMPV antigenomes containing the RSV F gene added as a supernumerary gene in the first (F1), third (F3), or fourth (F4) gene position. MPV genes are shown as unfilled rectangles, and the RSV F gene is shown as a shaded rectangle. The MPV gene start (GS) and gene end (GE) transcription signals are indicated by unfilled and filled bars, respectively, flanking each gene, including the supernumerary RSV F gene. The nucleotide sequence flanking the RSV F ORF is shown under each gene map, with the following features identified: RSV F ORF (represented by a shaded box), gene start (GS) and gene end (GE) transcription signals (bold), intergenic regions (IG), and the “Kozak” sequence (double underlined) placed upstream of the RSV F ORF to promote efficient translation.
rMPV-RSV-F vectors maintain expression of RSV F during in vitro replication.
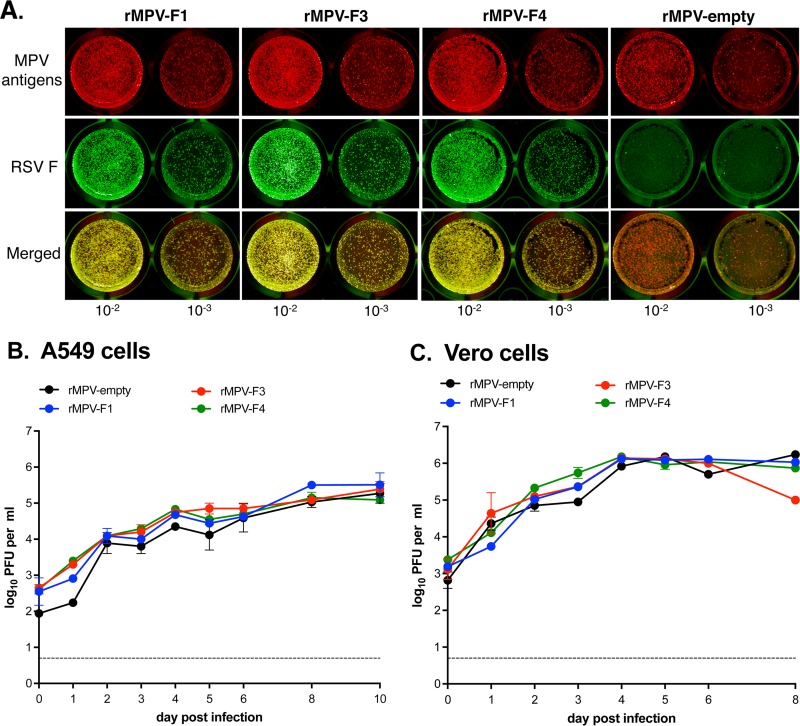
Since the MPV vector retains all of its genes, the supernumerary RSV F gene is not needed for replication and thus has an increased potential for accepting mutations, including ones that might silence expression (18). Therefore, we investigated the stability of expression of RSV F protein following in vitro replication. Specifically, samples from viral stocks (passage 1 [P1]; see Materials and Methods) from four independent recoveries for each of the rMPV-RSV-F vectors were analyzed by a dual-staining plaque assay to detect the coexpression of RSV F and MPV proteins. This showed that, for each of the three MPV-RSV-F vectors, >99% of the viral plaques expressed RSV F protein (Fig. 2A).
FIG 2.
Stability of RSV F expression and kinetics of multicycle replication in vitro. (A) The stability of expression of RSV F by the rMPV vectors was determined by a dual staining plaque assay. Vero cells were inoculated with serial dilutions of P1 virus stocks and incubated for 4 days under a 0.8% methylcellulose overlay. Monolayers were fixed and probed for RSV F and MPV antigens using specific antibodies followed by the corresponding infrared dye-conjugated secondary antibodies. RSV F and MPV antigens appear green and red, respectively, and appear yellow when merged. Displayed are representative images from four independent experiments. (B, C) Multicycle growth kinetics of the rMPV-RSV-F vectors in human A549 lung epithelial cells (B) and African green monkey kidney Vero cells (C). Replicate cultures of A549 and Vero cells were infected with an MOI of 0.1 PFU per cell with the indicated viruses. At 24-h intervals, two cultures per virus in each cell line were harvested by scraping and vortexing, and clarified supernatants were prepared and flash frozen. Viral titers were subsequently determined in duplicate by plaque assay. Data are shown as mean values with the standard errors of the means, although in many cases the error bars are obscured by the symbols, given the small margin of error. The limit of detection was 0.7 log10 PFU per ml (dotted line).
rMPV-RSV-F vectors are not temperature sensitive.
We previously observed with parainfluenza virus type 1 and 3 vectors that the insertion and expression of the RSV F gene could impart temperature sensitivity to the vector (19, 20). To assess possible temperature sensitivity of the MPV vectors, virus stocks were titrated at 42°C, 37°C, and 32°C. In no case was there a 100-fold or greater reduction in titer at 37°C or 42°C compared to the permissive temperature of 32°C, indicating that none of the viruses has a temperature-sensitive phenotype (data not shown).
Multicycle replication kinetics and cytopathology of the rMPV-RSV-F viruses in vitro.
The kinetics of multistep replication of the rMPV-RSV-F vectors in vitro were evaluated in human lung epithelial A549 cells (Fig. 2B) and African green monkey kidney Vero cells (Fig. 2C). The latter cell line does not produce type I interferon (IFN) in response to virus infection (21) and is an approved cell substrate for manufacturing human vaccines. All three rMPV-RSV-F constructs and the empty rMPV vector replicated with similar kinetics and to similar final titers in each cell line. In addition, there was no difference in plaque size for any of the rMPV-RSV-F constructs compared to the empty vector in either cell line (Fig. 2A shows examples of plaques in Vero cells). Thus, the presence of the RSV F insert and the expression of the RSV F protein did not appear to reduce or increase viral replication or spread in vitro. Comparing the two cell lines, the peak titers were obtained much sooner (day 4) in Vero cells than in A549 cells (day 10), likely due to the lack of IFN responsiveness of Vero cells. To evaluate cytopathic effects, Vero cells were infected at a multiplicity of infection (MOI) of 10 PFU per cell with rMPV-F1, -F3, -F4, empty rMPV vector, or wt RSV. Typical syncytium formation was observed in cells infected with wt rRSV at 48 h (data not shown) and at 96 h (Fig. 3), but rMPV-F1, -F3, and -F4 infection did not produce the classical syncytia up to 96 hours postinfection (hpi) and instead formed clumps of fused cells (Fig. 3, top panel), which were absent for the empty vector, suggesting this was mediated by the RSV F protein.
FIG 3.
Cytopathic effects in Vero cells infected with rMPV-RSV-F vectors. Vero cell monolayers were infected with the indicated rMPV-RSV-F vectors, empty vector, or wt rRSV at an MOI of 10 PFU per cell or mock infected. The cultures were incubated for 96 h at 32°C and subjected to light photomicroscopy at a magnification of ×200. The images shown are representative of two independent experiments.
Expression of RSV F and MPV proteins during in vitro infection.
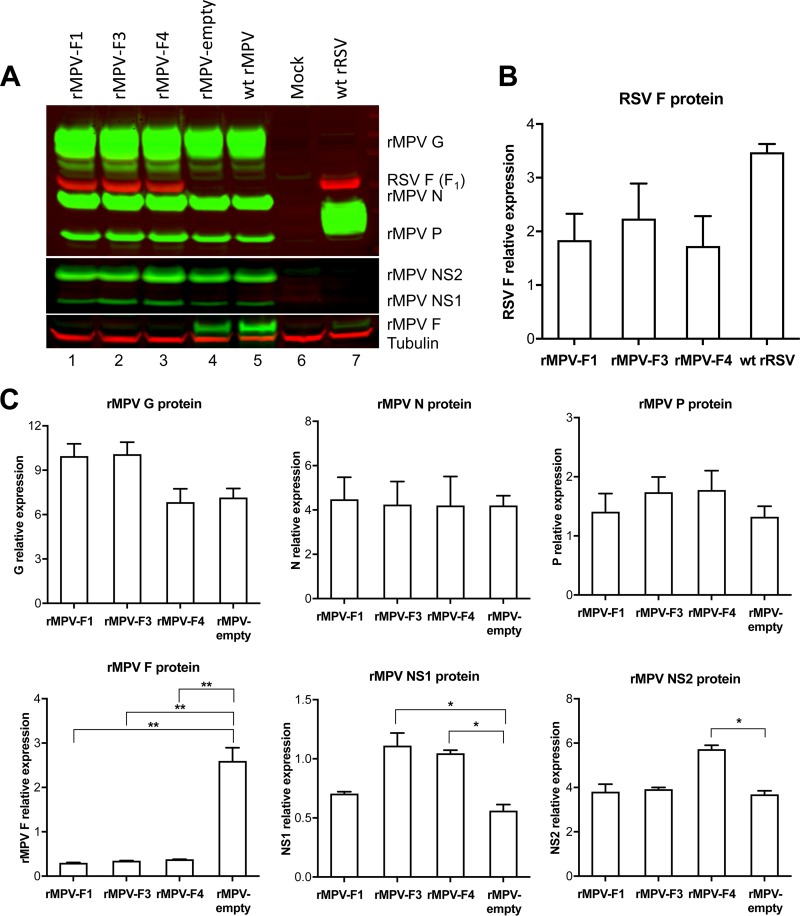
Vero cells were infected with each of the three vectors, or with empty vector, or with wt RSV at an MOI of 10 PFU per cell and incubated for 96 h. Cell lysates were prepared, subjected to SDS-PAGE under reducing and denaturing conditions, and analyzed by Western blotting using antisera and antibodies that detected the RSV F and MPV G, N, P, NS1, NS2, and F proteins (Fig. 4). All three vectors efficiently expressed the RSV F protein, at levels that were somewhat less than that of wt RSV (Fig. 4A and B). There was little or no difference in the level of RSV F protein expression among the three rMPV vectors (Fig. 4A and B, lanes 1 to 3). Furthermore, the presence of the RSV F gene did not decrease the expression of any of the MPV vector proteins that were analyzed (Fig. 4A and C) with the sole exception of the MPV F protein, whose expression was reduced to very low levels in the case of all three rMPV-RSV-F vectors (Fig. 4A and C, lanes 1 to 3 versus lane 4).
FIG 4.
Evaluation of RSV F and rMPV vector protein expression in infected cells. Cell lysates were prepared at 96 hpi using infected cells from the experiment shown in Fig. 3. Denatured and reduced lysates were subjected to Western blot analysis. RSV F was detected with a mouse monoclonal antibody. The MPV G, N, and P proteins were detected with a hyperimmune serum raised against sucrose gradient-purified rMPV virions. The MPV F protein was detected with a rabbit polyclonal antiserum raised against a recombinant vaccinia virus expressing only the MPV F protein. The MPV NS1 and NS2 proteins were detected with individual rabbit hyperimmune sera each raised against a synthetic peptide derived from the respective protein. Tubulin was probed as a loading control and used to normalize each sample. The Western blot images (A) and the quantification plots of protein bands (B and C) are from the same experiment and are representative of three independent experiments. Data shown in panels B and C were analyzed by one-way analysis of variance using Dunnett's multiple-comparison test. The differences in protein expression, when significant, are indicated by asterisks: *, P < 0.05; **, P < 0.01. Standard errors are shown.
rMPV-RSV-F vectors are highly restricted in rhesus macaques.
In the in vitro studies described above, the three rMPV-RSV-F vectors had very similar properties, and rMPV-F1 and rMPV-F3 were chosen for evaluation in rhesus macaques. rMPV-F1 was chosen because its promoter-proximal position might provide increased expression in vivo, even though that was not evident in vitro; and rMPV-F3 was chosen because of its somewhat-greater expression of RSV F in vitro. Empty rMPV vector was not included as a control for viral replication due to an insufficient number of animals. We chose to evaluate two vectors representing two different insertion sites, rather than a single insertion site versus empty vector, because sometimes a given site has disproportionate properties in vivo, such as increased attenuation or instability. Monkeys were confirmed to be seronegative for RSV and MPV by separate RSV- and MPV-specific 60% plaque reduction neutralization tests (PRNT60). The animals were inoculated by the combined intranasal and intratracheal routes with a total of 6.3 log10 PFU of the rMPV-RSV F vectors as described in Materials and Methods. In the interest of using a minimum number of animals, control animals immunized with wt RSV were not included in this study; instead, data from a separate study in which rhesus macaques from the same cohort were inoculated by the same route with a total of 7.3 log10 PFU of wt RSV A2 are shown here for comparison. None of the animals exhibited any signs of clinical illness following inoculation. Replication in the upper and lower respiratory tracts was assessed by collecting, respectively, nasopharyngeal swabs and tracheal lavage samples and determining their virus titers.
In the upper respiratory tract (Table 1), rMPV-F1 and rMPV-F3 had a mean shedding duration of 4.8 days for both vectors. One animal in each of the rMPV-F1 and rMPV-F3 groups did not have detectable replication on any of the sampling days. The mean peak virus titers were 1.6 and 1.4, log10 PFU/ml for rMPV-F1 and rMPV-F3, respectively (Table 1). wt RSV-inoculated animals shed virus for an average of 6.5 days with a peak titer of 2.8 log10 PFU/ml, which was 15- and 25-fold higher than the peak titers for the rMPV-F1 and rMPV-F3 vectors, respectively.
TABLE 1.
Viral titers of nasopharyngeal swab samples from the upper respiratory tract of rhesus macaques inoculated with the indicated rMPV-RSV F vectors or with wt RSVa
| Group | Animal ID(s) | Virus titerb (log10 PFU/ml) on day: |
Peak virus titer | No. of days of sheddingd | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ||||
| rMPV-F1 | A | 2.0 | —c | 2.2 | 1.6 | 2.6 | — | — | — | — | — | 2.6 | 5 |
| B | — | — | 1.8 | 1.5 | 1.5 | 1.0 | 1.7 | — | 1.9 | — | 1.9 | 7 | |
| C | — | — | 1.7 | 1.7 | 1.5 | 1.0 | 1.6 | — | 1.2 | — | 1.7 | 7 | |
| D | — | — | — | — | — | — | — | — | — | — | — | 0 | |
| A to D (mean ± SD) | 1.6 ± 1.1 | 4.8 ± 3.3 | |||||||||||
| rMPV-F3 | E | — | — | — | — | 1.4 | — | — | — | — | — | 1.4 | 1 |
| F | 1.7 | 1.6 | 2.0 | 1.6 | 1.7 | 1.7 | — | 1.6 | 1.3 | — | 2.0 | 9 | |
| G | — | — | — | — | — | — | — | — | — | — | — | 0 | |
| H | 2.3 | 2.0 | 1.7 | 2.0 | 1.6 | 1.4 | — | 1.3 | 1.7 | — | 2.3 | 9 | |
| E to H (mean ± SD) | 1.4 ± 1.0 | 4.8 ± 4.9 | |||||||||||
| wt RSV | I | 0.7 | — | 0.7 | 1.8 | 0.7 | 1.0 | 1.0 | 0.7 | — | — | 1.8 | 8 |
| J | — | 2.4 | 3.6 | 3.3 | 1.9 | 2.2 | 2.2 | — | — | — | 3.6 | 6 | |
| K | — | 2.6 | 2.7 | 3.4 | 1.6 | 1.2 | 1.2 | — | — | — | 3.4 | 6 | |
| L | — | — | 0.7 | 1.9 | 2.0 | 1.0 | 2.2 | 1.3 | — | — | 2.2 | 6 | |
| I to L (mean ± SD) | 2.8 ± 0.9 | 6.5 ± 1.0 | |||||||||||
Rhesus macaques were inoculated by the combined intranasal and intratracheal routes with 6.0 log10 PFU per site with rMPV vectors. In a separate study, animals from the same cohort as those used for rMPV vectors were inoculated with 7.0 log10 PFU per site of recombinant wt RSV. The total dose per animal was 6.3 log10 PFU of rMPV vector or 7.3 log10 PFU of wt RSV. Nasopharyngeal swabs were collected on days 0 to 10, 12, 14, 21, and 28. After day 10, samples had no detectable virus (results not shown).
The lower limit of detection was 0.7 log10 PFU/ml.
—, no detectable virus.
Time period spanning the first day to the last day on which virus was detected, including negative days (if any) in between.
In the lower respiratory tract, rMPV-F1 and rMPV-F3 had mean durations of virus shedding of 3.5 and 1.8 days, respectively, and the mean peak virus titers were 1.8 and 1.7 log10 PFU/ml, respectively (Table 2). All animals had detectable shedding from the lower respiratory tract except for one animal (animal ID G), which was inoculated with rMPV-F3 and also did not have detectable shedding from the upper respiratory tract. Animals inoculated with wt RSV had a mean shedding duration of 4 days, with the mean peak virus titer at 2.5 log10 PFU/ml, which was 5- to 6-fold higher than the peak titers of the vectors.
TABLE 2.
Viral titers of tracheal lavage samples from the lower respiratory tract of rhesus macaques inoculated with the indicated rMPV-RSV F vectors or with wt RSVa
| Group | Animal ID(s) | Virus titer (log10 PFU/ml) on day: |
Peak virus titer | No. of days of sheddingc | |||
|---|---|---|---|---|---|---|---|
| 2 | 4 | 6 | 8 | ||||
| rMPV-F1 | A | 1.7 | —b | 1.7 | — | 1.7 | 5 |
| B | 1.0 | 1.5 | — | — | 1.5 | 3 | |
| C | 1.0 | — | 2.0 | — | 2.0 | 5 | |
| D | 2.1 | — | — | — | 2.1 | 1 | |
| A to D (mean ± SD) | 1.8 ± 0.3 | 3.5 ± 1.9 | |||||
| rMPV-F3 | E | 1.5 | — | 2.0 | — | 2.0 | 5 |
| F | 2.3 | — | — | — | 2.3 | 1 | |
| G | — | — | — | — | — | 0 | |
| H | 2.6 | — | — | — | 2.6 | 1 | |
| E to H (mean ± SD) | 1.7 ± 1.2 | 1.8 ± 2.2 | |||||
| wt RSV | I | 3.2 | 2.5 | — | — | 3.2 | 3 |
| J | 2.2 | 2.1 | 2.7 | — | 2.7 | 5 | |
| K | — | — | — | — | — | 0 | |
| L | 4.1 | 3.0 | 2.6 | 1.5 | 4.1 | 7 | |
| I to L (mean ± SD) | 2.5 ± 1.8 | 4.0 ± 3.6 | |||||
From the experiment described in Table 1, tracheal lavages were performed on days 2, 4, 6, 8, 10, 12, 14, 21, and 28; after day 8, samples had no detectable virus (results not shown). Virus titers were determined by plaque assay as described for Table 1.
—, no detectable virus. The lower limit of detection was 0.7 log10 PFU per ml.
Time period spanning the first day to the last day on which virus was detected, including negative days (if any) in between.
The nasopharyngeal swabs and tracheal lavage samples from rMPV-F1- and rMPV-F3-inoculated rhesus macaques also were analyzed by the fluorescent dual-staining plaque assay to determine the stability of RSV F expression during in vivo replication. The majority of the samples showed 100% of the rMPV plaques expressing RSV F antigen (Table 3), indicating a substantial level of stability of expression for the RSV F protein.
TABLE 3.
Percentage of plaques expressing RSV F in samples collected from the upper and lower respiratory tracts of rhesus macaques inoculated with rMPV vectorsa
| Sample type and virus vector | Monkey ID | % PFU expressing RSV F on day: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ||
| Nasopharyngeal swabs | ||||||||||
| rMPV-F1 | A | — | — | 100 | 50 | 80 | — | — | — | — |
| B | — | — | 100 | 67 | 100 | 100 | 100 | — | 80 | |
| C | — | — | 100 | 100 | 100 | 100 | 100 | — | 50 | |
| D | — | — | — | — | — | — | — | — | — | |
| rMPV-F3 | E | — | — | — | — | 100 | — | — | — | — |
| F | 100 | 83 | 100 | 100 | 100 | 100 | — | 67 | 80 | |
| G | — | — | — | — | — | — | — | — | — | |
| H | 75 | 100 | 100 | 100 | 100 | — | — | 100 | — | |
| Tracheal lavage fluid | ||||||||||
| rMPV-F1 | A | nc | — | nc | — | nc | 75 | nc | — | nc |
| B | nc | — | nc | 100 | nc | — | nc | — | nc | |
| C | nc | 100 | nc | — | nc | — | nc | — | nc | |
| D | nc | 100 | nc | — | nc | — | nc | — | nc | |
| rMPV-F3 | E | nc | 100 | nc | — | nc | 100 | nc | — | nc |
| F | nc | 100 | nc | — | nc | — | nc | — | nc | |
| G | nc | — | nc | — | nc | — | nc | — | nc | |
| H | nc | 94 | nc | — | nc | — | nc | — | nc | |
The nasopharyngeal swabs and tracheal lavage samples collected on the indicated days after intranasal immunization were analyzed by fluorescent dual-staining plaque assay on Vero cells to determine the percentage of viral plaques coexpressing RSV F and MPV antigens during in vivo virus replication. These percent values were determined from approximately 100 plaques per sample. —, no detectable virus; nc, no sample was collected.
Immunization conferred strong virus-neutralizing serum antibody responses against RSV and MPV.
In the rhesus macaque experiment described above, serum samples were collected at 14, 21, and 28 days postimmunization and analyzed by RSV- and MPV-specific PRNT60 assays to determine titers of serum RSV- and MPV-neutralizing antibodies. None of the sera collected on day 0 prior to immunization had detectable virus-neutralizing activity against RSV or MPV (Tables 4 and 5), confirming the animals were seronegative.
TABLE 4.
RSV 60% plaque reduction neutralization titers (PRNT60) of serum samples from rhesus macaques immunized with the rMPV-RSV-F vectors or with wt RSV
| Group | Animal ID(s) | RSV PRNTa (log2 60% titer) on day: |
|||
|---|---|---|---|---|---|
| 0 | 14 | 21 | 28 | ||
| rMPV-F1 | A | <3.3 | 8.2 | 9.8 | 9.6 |
| B | <3.3 | 5.4 | 6.3 | 7.3 | |
| C | <3.3 | 7.1 | 8.4 | 8.6 | |
| D | <3.3 | 6.9 | 7.9 | 7.4 | |
| (A to D) mean ± SD | <3.3 ± 0.0 | 6.9 ± 1.2 | 8.1 ± 1.4 | 8.2 ± 1.1 | |
| rMPV-F3 | E | <3.3 | 5.5 | 6.2 | 7.0 |
| F | <3.3 | 5.0 | 8.3 | 8.2 | |
| G | <3.3 | 7.6 | 8.0 | 7.8 | |
| H | <3.3 | 7.4 | 8.7 | 8.3 | |
| E to H (mean ± SD) | <3.3 ± 0.0 | 6.4 ± 1.3 | 7.8 ± 1.1 | 7.8 ± 0.6 | |
| wt RSV | I | <3.3 | 7.8 | 7.9 | 7.4 |
| J | <3.3 | 7.8 | 8.7 | 8.3 | |
| K | <3.3 | 7.7 | 8.9 | 9.1 | |
| L | <3.3 | 8.1 | 8.7 | 8.8 | |
| I to L (mean ± SD) | <3.3 ± 0.0 | 7.9 ± 0.2 | 8.6 ± 0.4 | 8.4 ± 0.7 | |
The lower limit of detection of the RSV PRNT60 assay was 3.3 log2 60% titer. An RSV serum sample with a log2 PRNT60 value of ≥5.3 was considered positive.
TABLE 5.
MPV 60% plaque reduction neutralization titers (PRNT60) of serum samples from rhesus macaques immunized with the rMPV-RSV F vectorsa
| Group | Animal ID(s) | MPV PRNT (log2 60% titer) on day: |
|||
|---|---|---|---|---|---|
| 0 | 14 | 21 | 28 | ||
| rMPV-F1 | A | <3.3 | 13.7 | 13.8 | 13.7 |
| B | <3.3 | 10.9 | 11.0 | 10.4 | |
| C | <3.3 | 10.7 | 11.7 | 12.5 | |
| D | <3.3 | 8.8 | 8.8 | 8.6 | |
| A to E (mean ± SD) | <3.3 ± 0.0 | 11.0 ± 2.0 | 11.3 ± 2.1 | 11.3 ± 2.3 | |
| rMPV-F3 | E | <3.3 | 11.6 | 11.2 | 11.5 |
| F | <3.3 | 10.4 | 10.2 | 11.8 | |
| G | <3.3 | 9.5 | 9.6 | 10.5 | |
| H | <3.3 | 9.5 | 11.5 | 11.7 | |
| E to H (mean ± SD) | <3.3 ± 0.0 | 10.3 ± 1.0 | 10.6 ± 0.9 | 11.4 ± 0.6 | |
The lower limit of detection of the (PRNT60) assay was 3.3 log2 60% titer. An MPV serum sample with a log2 PRNT60 value of ≥5.3 was considered positive.
At day 14, RSV- and MPV-specific neutralizing activity was detected in sera from monkeys immunized with rMPV-F1 and rMPV-F3. These titers generally did not change significantly over the next 2 weeks, as indicated by analysis of sera from days 21 and 28 (Tables 4 and 5). On days 14, 21, and 28 postimmunization, the mean RSV log2 PRNT60 values were 6.9, 8.1, and 8.2, respectively, for rMPV-F1 and 6.4, 7.8, and 7.8, respectively, for rMPV-F3. The respective mean MPV log2 PRNT60 values on these days were 11.0, 11.3, and 11.3 for rMPV-F1 and 10.3, 10.6, and 11.4 for rMPV-F3. Mean RSV- and MPV-specific log2 PRNT60 values induced by rMPV-F1 and rMPV-F3 were similar to each other on days 14, 21, and 28 postimmunization. Rhesus macaques from the same cohort as those used for rMPV-F1 and -F3 were immunized in a separate study by the same route with a 10-fold-higher dose (7.3 log10 PFU) of wt RSV A2, and the serum RSV PRNT60 values determined side by side with rMPV-F1 and -F3 sera are shown in Table 4. wt RSV induced serum RSV-neutralizing titers at 7.9, 8.6, and 8.4 log2 PRNT60 at days 14, 21, and 28 postimmunization, respectively, which were similar to those conferred by rMPV-F1 and -F3 at those time points.
DISCUSSION
RSV is the most important viral respiratory pathogen of infancy and also has a substantial impact in children and adults, especially the elderly. The high worldwide pediatric morbidity and substantial mortality (particularly in the developing world) associated with RSV emphasize a continuing urgent need for a pediatric RSV vaccine in particular. In addition, the occurrence of substantial RSV disease in older adults with waning RSV immunity indicates the need for a booster RSV vaccine. Thus, a vaccine that could be used in RSV-naive (children) and RSV-experienced (adults) recipients would be highly desirable. A number of different vaccine strategies are presently being pursued, including a live-attenuated RSV and live viral vectors for pediatric use and subunit vaccines for the elderly as well as for pregnant women to increase maternal antibody immunity (reviewed in reference 22), but no candidate has yet been licensed.
In the present study, a codon-optimized RSV F ORF was engineered to be under the control of MPV transcription signals and inserted as an added gene into three different locations in the rMPV genome. The MPV genome that was used in this study contained an L ORF of which the downstream 67% had been subjected to CPO, as part of another study, but this had no effect on replication in mice (our unpublished data). In future work, it would be of interest to compare this CPO version versus wt MPV for replication in rhesus macaques.
The three rMPV-RSV-F vectors and the empty vector were readily recovered by reverse genetics and replicated efficiently in vitro with little evidence of instability of expression of the RSV F protein. Interestingly, insertion of RSV F at any of the three locations yielded essentially the same level of expression of the RSV F insert. This was unexpected because single-stranded negative-sense RNA viruses characteristically exhibit a polar gradient in which gene expression decreases with increasing distance from the single promoter at the 3′ end of the genome (23). For example, in a previous study using a chimeric bovine/human parainfluenza virus type 3 (PIV3) vector, the relative levels of expression of RSV F from gene positions 2 and 3 were 42 to 50% and 9 to 12%, respectively, that of gene position one (19). In another study using two different attenuated HPIV1 vectors, a strong gradient of decreasing expression of RSV F was observed with one attenuated HPIV1 vector but not another (20). In addition to the lack of difference in the level of expression of the RSV F protein, the three rMPV-RSV-F vectors also were very similar with regard to in vitro growth kinetics and final viral yield.
Insertion of the RSV F gene into the rMPV vector did not decrease the expression of the MPV G, N, P, NS1, and NS2 proteins, which is another indication that the vector generally was not inhibited by the presence of the added RSV F gene. However, the expression of one vector protein, namely, MPV F, was reduced to very low levels in all three constructs. The reason why MPV F alone was strongly reduced remains unclear. In previous studies with PIV vectors, the addition of the RSV F insert often reduced the expression of one or more downstream vector genes, particularly the one or two vector genes immediately downstream of the insert (19, 20). A likely explanation is that, due to the polar gradient of transcription, the added RSV F gene decreased the amount of polymerase that reached downstream vector genes. Of the six MPV proteins that were monitored in the present study, three (P, G, and F) were from genes located downstream of the RSV F insert in all three vectors, one additional gene (encoding N) was downstream of the RSV F insert in rMPV-F3, and all six genes were downstream of the RSV F insert in rMPV-F1. Why only the MPV F protein, among all of these downstream genes, was reduced remains unknown. Thus, the factors that affect the expression of a foreign insert and the vector genes in vectors based on single-stranded negative-sense RNA viruses are more complex than the simple model of a polar gradient of transcription.
In previous studies, we investigated a number of strategies to increase the immunogenicity of the RSV F protein expressed from PIV vectors (19, 24–26). Two factors in particular yielded increases in the quantity and quality of RSV-neutralizing serum antibodies, namely, (i) the use of mutations that stabilize RSV F protein in the prefusion (pre-F) conformation and (ii) modification of the RSV F protein to be efficiently packaged in the vector virions. The pre-F conformation is a metastable form in which the newly synthesized RSV F protein appears. The pre-F conformation contains a number of unique, highly effective neutralization epitopes (27–31). When the pre-F protein contacts a target host cell membrane, it is readily triggered to undergo a massive irreversible conformational change that drives fusion between the virion and host cell membranes and also results in the loss of major pre-F-specific epitopes. Pre-F also can readily be triggered prematurely, which also results in the loss of these major epitopes. However, this conformational change can be inhibited by stabilizing this optimal antigenic form through structure-based mutations (30). In addition, RSV F can be packaged efficiently into the vector particle by replacing its transmembrane (TM) and cytoplasmic tail (CT) domains with those of the vector F protein. Both of these strategies are available for evaluation with the rMPV-RSV-F vectors. However, we chose to evaluate unmodified RSV F in the present study because, in previous studies with PIV vectors the optimized forms were very immunogenic and may have approached or reached the limit of the responsiveness of the animals. Therefore, our goal was to first examine whether MPV could be used as a vector and to identify the optimal RSV F insertion site. We subsequently will modify the RSV F insert to increase the quantity and quality of induced RSV-neutralizing antibodies.
As noted, the three rMPV-RSV-F vectors were nearly indistinguishable with regard to in vitro replication and protein expression. The two vectors that were chosen for evaluation in rhesus macaques, rMPV-F1 and rMPV-F3, also were nearly indistinguishable in vivo. Both constructs were very highly restricted for replication but induced high titers of RSV-neutralizing serum antibodies. The titers that were detected in monkeys immunized by either of the two constructs were not greatly different from each other. The responses were strong by day 14 and did not increase significantly by days 21 and 28 postvaccination. One animal did not have detectable shedding in the upper tract (animal D), and another did not have detectable shedding in either the upper or lower tract (animal G). However, there was no evident decrease in RSV-neutralizing serum antibody titers. This suggests that an undetectable low level of replication produced sufficient RSV F protein to induce high titers of RSV-neutralizing serum antibodies. It is also important to note that although the rMPV vectors were administered at a 10-fold-lower dose and replicated 5- to 25-fold less efficiently than wt RSV, they did confer RSV-neutralizing antibodies at high titers similar to those of wt RSV. Thus, we anticipate that due to their desirably attenuated but highly immunogenic phenotype, rMPV-based RSV vaccines may be considered for clinical evaluation.
The high level of attenuation of MPV in primates observed here and in a previous study (15) reflects host range restriction, specifically, inefficient replication due to the incompatibility of the murine virus for the primate system. MPV is a murine homolog of RSV, comparable to bovine PIV3 (BPIV3) as the bovine homolog of HPIV3. BPIV3 has previously been shown to be highly attenuated in nonhuman primates as well as in adult and pediatric volunteers, including young infants, and previously was evaluated as an experimental “Jennerian” HPIV3 vaccine (32). BPIV3 also was used as an attenuated backbone to construct the chimeric B/HPIV3 virus, in which the F and HN genes were replaced by their counterparts from HPIV3, and which has been evaluated as an HPIV3 vaccine (33), as well as a vector to express the RSV F protein (13, 19, 24–26). The percent amino acid sequence identities between the N, P, M, F, and L proteins of BPIV3 versus HPIV3 are 85, 59, 90, 79, and 89%, respectively (34), compared to 60, 33, 42, 43, and 53%, respectively, for the corresponding proteins between MPV and RSV (14). Thus, MPV and RSV are substantially more divergent from each other than are BPIV3 and HPIV3 from each other. Since host range restriction generally is thought to be the cumulative effect of many amino acid (and perhaps nucleotide) sequence differences in multiple genes (35), the high percentage of sequence differences between MPV and RSV suggests that the attenuation phenotype of MPV in humans should be very stable, as appears to be the case for BPIV3-based vaccines. However, this remains to be investigated. It will also be of interest to evaluate MPV vectors in African green monkeys, in which the MPV-neutralizing antibody titers induced by wt MPV are nearly twice those seen in rhesus macaques (15).
A theoretical concern of expressing the RSV F protein from an rMPV vector is that the RSV F protein might enable the murine virus to infect and replicate more efficiently, and in particular to acquire an increased tropism for human cells. However, there was no evidence of this in vitro: there was no increase in replication kinetics or viral yield in human or nonhuman primate cells or in plaque size. Additionally, in vivo the immunized animals were healthy throughout the study and clinical signs were absent. Possible effects of expression of RSV F on replication were not evaluated in rhesus macaques because the empty vector was not included due to an insufficient number of animals. However, the rMPV-RSV-F vectors were highly attenuated, and thus any possible effect would have been modest. In any event, possible effects of RSV F in enhancing replication or changing tropism should not be an issue with vectors expressing stabilized versions of pre-F protein, because these stabilized forms will be largely nonfunctional.
HPIV1 and B/HPIV3 vectors expressing RSV F also are being developed in our laboratory as vectored RSV vaccines (19, 20, 24–26). They have the advantage of conferring bivalent protection against the respective HPIV and RSV. However, they would be anticipated to be strongly restricted in adults by the preexisting immunity acquired by previous natural exposures to the respective HPIV and thus likely could be used only in HPIV-naive infants and children and not in adults. We have previously shown a lack of MPV-neutralizing activity in human sera and also a lack of MPV cross-neutralization by RSV-specific antibodies, as well as a lack of cross-protection between RSV and MPV in mice (15). Therefore, we anticipate that an rMPV vector would not be inhibited by immunity to common human viruses, including RSV, with the caveat that expression of RSV F by this vector might confer some susceptibility to restriction by RSV-specific immunity. Since the rMPV vectors appear to be highly attenuated, it is anticipated that they will be well tolerated as a primary immunization in all age groups. The rMPV-RSV-F vectors also provide the potential for boosting RSV responses in subjects who previously received an RSV vaccine, with the caveat of possible restriction by RSV-specific immunity of the vector expressing RSV F.
In summary, the vaccine candidates developed here show promising immunogenicity, which could be significantly augmented by including the more immunogenic forms of RSV F such as stabilized pre-F (30) and the next generations of single-chain pre-F (36) and by including the packaging signals to enable RSV F incorporation into the MPV virus particles as previously described for PIV-vectored RSV vaccines (24, 25).
MATERIALS AND METHODS
Cells and viruses.
Vero cells (African green monkey kidney cells, CCL-81; ATCC, Manassas, VA) were maintained in Opti-Pro medium supplemented with 4 mM l-glutamine (Thermo Fisher Scientific, Waltham, MA) and 10% fetal bovine serum (FBS; HyClone, Logan, UT). Human lung epithelial A549 (CCL-185; ATCC, Manassas, VA) cells were maintained in F-12K medium (ATCC) supplemented with 4 mM l-glutamine. BHK BSR T7/5 cells are BHK-21 (baby hamster kidney 21) cells (37) that were maintained in Glasgow's minimum essential medium (MEM) (Thermo Fisher Scientific) supplemented with 3% FBS. Geneticin (Thermo Fisher) was included in the medium for every other passage to ensure selection of T7 RNA polymerase-expressing cells.
Recombinant (r) wt RSV strain A2 was used as a control and has been previously described (16, 38). All in vitro tissue culture experiments were done at 37°C unless otherwise noted. rMPV and the rMPV-RSV-F vectors were propagated on Vero cells by infecting at an MOI of 0.1 PFU per cell. Virus stocks were harvested about 2 weeks postinfection, when cytopathic effects disrupted the monolayer. rMPV titers were determined by plaque assay on Vero cells under a 0.8% methylcellulose overlay as described previously (15). Plaques were visualized by immunostaining with rabbit hyperimmune serum raised against sucrose-gradient-purified MPV followed by a horseradish peroxidase-labeled goat anti-rabbit IgG secondary antibody (KPL, Gaithersburg, MD). Bound secondary antibodies were detected by incubation with a peroxidase substrate (KPL). Each sample was tested in duplicate, and the average is reported.
Construction of the rMPV viral vectors expressing RSV F protein from an added gene.
The rMPV-RSV-F vectors were constructed using a previously described reverse genetics system (16). The rMPV vector backbone used for expressing RSV F was derived from MPV (previously called PVM) strain 15 cloned in a pBluescript plasmid vector (pBS) that contained the two introduced restriction sites (AgeI and BstBI) that were absent in the parent strain 15 as described previously (16) and also contained a partially modified L ORF. The downstream 67% of the L ORF was codon pair optimized (CPO) to contain synonymous changes that increased the content of codon pairs associated with efficient expression in humans while preserving both the overall codon usage and the encoded amino acid sequence (39, 40). The RSV F ORF from strain A2 was codon optimized for human codon usage (GenScript, Piscataway, NJ) to obtain greater protein expression, as previously reported (25). RSV F also carried the two previously described HEK amino acid assignments of 66E and 101P (17) that make the encoded F protein amino acid sequence identical to that of an early passage of wt strain A2 and the clinical isolates. We designed three MPV vector constructs in which the RSV F insert was placed in the first gene position, upstream of the NS1 gene (rMPV-F1), or in the third gene position, between the NS2 and N genes (rMPV-F3), or in the fourth gene position, between the N and P genes (rMPV-F4) (Fig. 1). The RSV F inserts were designed so that the RSV F ORF was flanked by MPV gene start (GS) and gene end (GE) transcription signals (16) to enable transcription of RSV F as a separate mRNA. The Kozak consensus sequence GCCGCCACC was placed immediately upstream of the RSV F AUG start codon to provide efficient context for translation initiation as described previously (41). The RSV F inserts were synthesized commercially (Genscript) as long genome segments designed for insertion into the rMPV antigenomic plasmid using the XmaI restriction site in the plasmid (pBS) sequence upstream of the leader region (not shown) and the downstream KpnI site (rMPV-F1 and -F3) or the KpnI and BmtI sites (rMPV-F4) as indicated (Fig. 1). The final construct rMPV-F1 contains 1,768 additional nucleotides, placed immediately after nucleotide position 67 (upstream of NS1); rMPV-F3 contains 1,775 additional nucleotides, placed immediately after nucleotide position 981 (between NS2 and N); and rMPV-F4 contains 1,771 additional nucleotides, placed immediately after nucleotide position 2276 (between N and P) of the MPV genome.
The rMPV vectors were recovered from cDNA as previously described (16) in BHK BSR-T7/5 cells that constitutively express the T7 RNA polymerase (37). The cells were transfected with the MPV antigenome plasmid and support plasmids expressing MPV N, P, M2-1, and L proteins; 24 h later, the cells were scraped and vortexed, and the cell suspension was cocultured with a Vero cell monolayer for approximately 2 weeks to create P1 viral stocks (16). We confirmed that the P1 virus stocks did not contain any adventitious mutations introduced during recovery: specifically, viral RNA was isolated and Sanger sequence analysis of the complete viral genomes was performed using uncloned overlapping RT-PCR fragments. Control RT-PCRs lacking reverse transcriptase did not yield an amplified product, confirming that the PCR products were amplified from the viral RNA and not from cDNA used for virus rescue. Titers of virus stocks were determined by plaque assay and immunostaining as described above.
Fluorescent dual-staining plaque assay quantifying the rMPV plaques coexpressing RSV F and vector antigens.
To evaluate the stability of expression of RSV F protein following in vitro replication, four independent viral recoveries of each rMPV-RSV-F construct were analyzed by a dual-staining plaque assay to determine the stability of RSV F expression. The plaque assay was set up as previously described (15), and cells were fixed using 80% methanol at day 4 postinfection (p.i.). To identify MPV proteins, a rabbit anti-rMPV polyclonal primary antibody described above was used at 1:5,000 and detected by a goat anti-rabbit 680 LT antibody (Li-Cor, Lincoln, NE). RSV F was probed with a mixture of three RSV F-specific mouse monoclonal antibodies (1129, 1243, and 1269) each diluted at 1:200, followed by a goat anti-mouse 800 CW secondary antibody (Li-Cor). Both secondary antibodies were used at a 1:800 dilution. Plates were scanned on an Odyssey infrared imager (Li-Cor), and the images were analyzed to determine the percentage of plaques coexpressing RSV F and rMPV antigens. Plaque images were pseudocolored to appear red and green for MPV and RSV F antigens, respectively. On merging the two channels, rMPV plaques coexpressing RSV F would appear yellow while those with loss of RSV F expression would appear red. To evaluate the stability of expression of RSV F protein following in vivo replication, nasopharyngeal swab and tracheal lavage samples from the rhesus study described below were analyzed by plaque assay followed by fixation and dual-stain plaque assay as described above.
Multicycle growth kinetics.
To evaluate multicycle replication kinetics, replicate monolayer cultures of Vero and A549 cells in T25 flasks were inoculated with each virus at an MOI of 0.1 PFU per cell and incubated at 32°C. The virus inoculum was adsorbed for 3 h, after which monolayers were washed twice and replenished with fresh medium. On days 1 through 6 and days 8 and 10 (A549) or on days 1 through 6 and day 8 (Vero), two cell monolayers per virus and per cell type were scraped into the supernatant, vortexed, clarified by centrifugation, and frozen. Viral titers were determined by plaque assay on Vero cells.
Analysis of expression of RSV F and MPV vector proteins.
Vero cell monolayers in 12-well plates were infected at an MOI of 10 PFU per cell with the rMPV-RSV-F vectors, empty vector, wt rMPV, or wt RSV or were mock infected. At 96 h p.i., images of infected cells were acquired and cell lysates were prepared to examine viral protein expression by Western blotting. Cells were washed twice with 1× phosphate-buffered saline (PBS) and lysed with 200 μl of cell lysis buffer containing 1× NuPAGE LDS sample buffer (Thermo Fisher Scientific) and 1× cOmplete Ultra protease inhibitor (Roche, Basel, Switzerland) in protease-free water. Each lysate was spun through a QIAshredder spin column (Qiagen, Valencia, CA) and flash frozen on dry ice. A 90-μl volume of each lysate was combined with 10 μl of 10× reducing agent (Thermo Fisher Scientific) and denatured and reduced at 70°C for 10 min. Twenty-five microliters was loaded per lane followed by SDS-PAGE and Western blotting. rMPV G, N, and P proteins were detected with hyperimmune serum raised against sucrose-gradient-purified MPV virions as previously described (15). The MPV F protein was detected with a rabbit polyclonal antiserum raised against a recombinant vaccinia virus expressing the MPV F protein (15). NS1 and NS2 were detected with rabbit hyperimmune serum raised individually against the synthetic peptides TNFDRSDLET and SDSEESGDEA derived from the corresponding proteins, respectively. RSV F was probed with monoclonal antibodies as described previously (19). Tubulin was detected as a loading control using mouse monoclonal antitubulin antibody (catalog number A-11126; Thermo Fisher Scientific). Primary antibodies were detected with the corresponding species-specific secondary antibodies conjugated with an infrared dye (Li-Cor). Membranes were scanned on an Odyssey infrared imaging system. Band signal values were obtained and background corrected using ImageStudio Lite, version 5.2.5 (Li-Cor).
Evaluation of selected rMPV-RSV-F viral vectors in rhesus macaques.
The NIH National Institute of Allergy and Infectious Diseases Animal Care and Use Committee approved the nonhuman primate experiment described in this study, which was performed in accordance with the recommendations from the Guide for the Care and Use of Laboratory Animals. Eight young adult rhesus macaques (Macaca mulatta) were pretested to confirm that they were seronegative for both RSV and MPV by separate PRNT60 assays (described in “PRNT60 assays” below). Two groups of four monkeys each were inoculated through the combined intranasal and intratracheal routes with 1.0 ml of inoculum per site containing 106 PFU of rMPV-F1 or -F3 diluted in L-15 medium (Thermo Fisher Scientific). In a separate study, four rhesus macaques from the same cohort as those used for rMPV vectors were inoculated with 7.0 log10 PFU per site of recombinant wt RSV strain A2. All four animals were prescreened to be seronegative for RSV. Clinical observations were made daily. Nasopharyngeal (NP) swabs were collected on days 0 to 10, 12, 14, 21, and 28. Tracheal lavage (TL) samples were collected postinfection on days 2, 4, 6, 8, 10, 12, 14, 21, and 28. The NP and TL samples were analyzed by plaque assay as described above to quantify viral shedding. To assess the immunogenicity, sera were collected on days 0, 14, 21, and 28 postimmunization and were analyzed for RSV- and MPV-neutralizing antibodies by PRNT60.
PRNT60 assays.
Serum samples were analyzed for RSV- and MPV-neutralizing antibody titers by performing PRNT60 assays on Vero cells as previously described (15, 42) using RSV-GFP (where GFP is green fluorescent protein) and MPV-GFP, respectively. Sera from all 12 animals immunized with rMPV-F1, -F3, or wt RSV were analyzed side by side for RSV-neutralizing antibody levels in the same experiment. Briefly, prior to use, serum samples were incubated for 30 min at 56°C to inactivate the serum complement. Serial dilutions of serum were then mixed with an equal volume of diluted RSV-GFP or MPV-GFP and incubated at 37°C for 30 min. The RSV neutralization assay was performed in the presence of 10% guinea pig complement (Lonza, Walkersville, MD) (43). Complement was excluded from the MPV neutralization assay, as it inactivates the virus (15). At day 6 postinfection, images of the RSV-GFP and MPV-GFP plaques were obtained by scanning on a Typhoon imager (GE Healthcare) and PRNT60 was calculated. Each sample was tested in duplicate, and the average values are reported as log2 PRNT60.
ACKNOWLEDGMENTS
This research was supported by the Intramural Research Program of the NIAID, NIH.
We thank Joanna Swerczek from the Experimental Primate Virology Section and the technical staff of the Comparative Medicine Branch, NIAID, NIH, for their technical assistance in the animal experiments and for the care and management of the primates. We also thank Sonja Surman and Lijuan Yang from the Laboratory of Infectious Diseases, NIAID, NIH, for their technical support with the neutralization assays and plaque titrations.
REFERENCES
- 1.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, Alvarado M, Anderson HR, Anderson LM, Andrews KG, Atkinson C, Baddour LM, Barker-Collo S, Bartels DH, Bell ML, Benjamin EJ, Bennett D, Bhalla K, Bikbov B, Bin Abdulhak A, Birbeck G, Blyth F, Bolliger I, Boufous S, Bucello C, Burch M, Burney P, Carapetis J, Chen H, Chou D, Chugh SS, Coffeng LE, Colan SD, Colquhoun S, Colson KE, Condon J, Connor MD, Cooper LT, Corriere M, Cortinovis M, de Vaccaro KC, Couser W, Cowie BC, Criqui MH, Cross M, Dabhadkar KC, Dahodwala N, De Leo D, Degenhardt L, Delossantos A, Denenberg J, Des Jarlais DC, Dharmaratne SD, Dorsey ER, Driscoll T, Duber H, Ebel B, Erwin PJ, Espindola P, Ezzati M, Feigin V, Flaxman AD, Forouzanfar MH, Fowkes FG, Franklin R, Fransen M, Freeman MK, Gabriel SE, Gakidou E, Gaspari F, Gillum RF, Gonzalez-Medina D, Halasa YA, Haring D, Harrison JE, Havmoeller R, Hay RJ, Hoen B, Hotez PJ, Hoy D, Jacobsen KH, James SL, Jasrasaria R, Jayaraman S, Johns N, Karthikeyan G, Kassebaum N, Keren A, Khoo JP, Knowlton LM, Kobusingye O, Koranteng A, Krishnamurthi R, Lipnick M, Lipshultz SE, Ohno SL, Mabweijano J, MacIntyre MF, Mallinger L, March L, Marks GB, Marks R, Matsumori A, Matzopoulos R, Mayosi BM, McAnulty JH, McDermott MM, McGrath J, Mensah GA, Merriman TR, Michaud C, Miller M, Miller TR, Mock C, Mocumbi AO, Mokdad AA, Moran A, Mulholland K, Nair MN, Naldi L, Narayan KM, Nasseri K, Norman P, O'Donnell M, Omer SB, Ortblad K, Osborne R, Ozgediz D, Pahari B, Pandian JD, Rivero AP, Padilla RP, Perez-Ruiz F, Perico N, Phillips D, Pierce K, Pope CA III, Porrini E, Pourmalek F, Raju M, Ranganathan D, Rehm JT, Rein DB, Remuzzi G, Rivara FP, Roberts T, De Leon FR, Rosenfeld LC, Rushton L, Sacco RL, Salomon JA, Sampson U, Sanman E, Schwebel DC, Segui-Gomez M, Shepard DS, Singh D, Singleton J, Sliwa K, Smith E, Steer A, Taylor JA, Thomas B, Tleyjeh IM, Towbin JA, Truelsen T, Undurraga EA, Venketasubramanian N, Vijayakumar L, Vos T, Wagner GR, Wang M, Wang W, Watt K, Weinstock MA, Weintraub R, Wilkinson JD, Woolf AD, Wulf S, Yeh PH, Yip P, Zabetian A, Zheng ZJ, Lopez AD, Murray CJ, AlMazroa MA, Memish ZA. 2012. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, O'Brien KL, Roca A, Wright PF, Bruce N, Chandran A, Theodoratou E, Sutanto A, Sedyaningsih ER, Ngama M, Munywoki PK, Kartasasmita C, Simoes EA, Rudan I, Weber MW, Campbell H. 2010. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet 375:1545–1555. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi T, McAllister DA, O'Brien KL, Simoes EAF, Madhi SA, Gessner BD, Polack FP, Balsells E, Acacio S, Aguayo C, Alassani I, Ali A, Antonio M, Awasthi S, Awori JO, Azziz-Baumgartner E, Baggett HC, Baillie VL, Balmaseda A, Barahona A, Basnet S, Bassat Q, Basualdo W, Bigogo G, Bont L, Breiman RF, Brooks WA, Broor S, Bruce N, Bruden D, Buchy P, Campbell S, Carosone-Link P, Chadha M, Chipeta J, Chou M, Clara W, Cohen C, de Cuellar E, Dang DA, Dash-Yandag B, Deloria-Knoll M, Dherani M, Eap T, Ebruke BE, Echavarria M, de Freitas Lazaro Emediato CC, Fasce RA, Feikin DR, Feng L, Gentile A, Gordon A, Goswami D, Goyet S, Groome M, Halasa N, Hirve S, Homaira N, Howie SRC, Jara J, Jroundi I, Kartasasmita CB, Khuri-Bulos N, Kotloff KL, Krishnan A, Libster R, Lopez O, Lucero MG, Lucion F, Lupisan SP, Marcone DN, McCracken JP, Mejia M, Moisi JC, Montgomery JM, Moore DP, Moraleda C, Moyes J, Munywoki P, Mutyara K, Nicol MP, Nokes DJ, Nymadawa P, da Costa Oliveira MT, Oshitani H, Pandey N, Paranhos-Baccala G, Phillips LN, Picot VS, Rahman M, Rakoto-Andrianarivelo M, Rasmussen ZA, Rath BA, Robinson A, Romero C, Russomando G, Salimi V, Sawatwong P, Scheltema N, Schweiger B, Scott JAG, Seidenberg P, Shen K, Singleton R, Sotomayor V, Strand TA, Sutanto A, Sylla M, Tapia MD, Thamthitiwat S, Thomas ED, Tokarz R, Turner C, Venter M, Waicharoen S, Wang J, Watthanaworawit W, Yoshida LM, Yu H, Zar HJ, Campbell H, Nair H, RSV Global Epidemiology Network. 2017. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet 390:946–958. doi: 10.1016/S0140-6736(17)30938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stockman LJ, Curns AT, Anderson LJ, Fischer-Langley G. 2012. Respiratory syncytial virus-associated hospitalizations among infants and young children in the United States, 1997-2006. Pediatr Infect Dis J 31:5–9. doi: 10.1097/INF.0b013e31822e68e6. [DOI] [PubMed] [Google Scholar]
- 5.Afonso CL, Amarasinghe GK, Banyai K, Bao Y, Basler CF, Bavari S, Bejerman N, Blasdell KR, Briand FX, Briese T, Bukreyev A, Calisher CH, Chandran K, Cheng J, Clawson AN, Collins PL, Dietzgen RG, Dolnik O, Domier LL, Durrwald R, Dye JM, Easton AJ, Ebihara H, Farkas SL, Freitas-Astua J, Formenty P, Fouchier RA, Fu Y, Ghedin E, Goodin MM, Hewson R, Horie M, Hyndman TH, Jiang D, Kitajima EW, Kobinger GP, Kondo H, Kurath G, Lamb RA, Lenardon S, Leroy EM, Li CX, Lin XD, Liu L, Longdon B, Marton S, Maisner A, Muhlberger E, Netesov SV, Nowotny N, Patterson JL, Payne SL, Paweska JT, Randall RE, Rima BK, Rota P, Rubbenstroth D, Schwemmle M, Shi M, Smither SJ, Stenglein MD, Stone DM, Takada A, Terregino C, Tesh RB, Tian JH, Tomonaga K, Tordo N, Towner JS, Vasilakis N, Verbeek M, Volchkov VE, Wahl-Jensen V, Walsh JA, Walker PJ, Wang D, Wang LF, Wetzel T, Whitfield AE, Xie JT, Yuen KY, Zhang YZ, Kuhn JH. 2016. Taxonomy of the order Mononegavirales: update 2016. Arch Virol 161:2351–2360. doi: 10.1007/s00705-016-2880-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prince GA, Horswood RL, Chanock RM. 1985. Quantitative aspects of passive immunity to respiratory syncytial virus infection in infant cotton rats. J Virol 55:517–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson EJ, Carosone-Link P, Yogev R, Yi J, Simoes EAF. 2017. Effectiveness of palivizumab in high-risk infants and children: a propensity score weighted regression analysis. Pediatr Infect Dis J 36:699–704. doi: 10.1097/INF.0000000000001533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kapikian AZ, Mitchell RH, Chanock RM, Shvedoff RA, Stewart CE. 1969. An epidemiologic study of altered clinical reactivity to respiratory syncytial (RS) virus infection in children previously vaccinated with an inactivated RS virus vaccine. Am J Epidemiol 89:405–421. doi: 10.1093/oxfordjournals.aje.a120954. [DOI] [PubMed] [Google Scholar]
- 9.Kim HW, Canchola JG, Brandt CD, Pyles G, Chanock RM, Jensen K, Parrott RH. 1969. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am J Epidemiol 89:422–434. doi: 10.1093/oxfordjournals.aje.a120955. [DOI] [PubMed] [Google Scholar]
- 10.Connors M, Collins PL, Firestone CY, Sotnikov AV, Waitze A, Davis AR, Hung PP, Chanock RM, Murphy BR. 1992. Cotton rats previously immunized with a chimeric RSV FG glycoprotein develop enhanced pulmonary pathology when infected with RSV, a phenomenon not encountered following immunization with vaccinia–RSV recombinants or RSV. Vaccine 10:475–484. doi: 10.1016/0264-410X(92)90397-3. [DOI] [PubMed] [Google Scholar]
- 11.Wright PF, Karron RA, Belshe RB, Shi JR, Randolph VB, Collins PL, O'Shea AF, Gruber WC, Murphy BR. 2007. The absence of enhanced disease with wild type respiratory syncytial virus infection occurring after receipt of live, attenuated, respiratory syncytial virus vaccines. Vaccine 25:7372–7378. doi: 10.1016/j.vaccine.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karron RA, Luongo C, Thumar B, Loehr KM, Englund JA, Collins PL, Buchholz UJ. 2015. A gene deletion that up-regulates viral gene expression yields an attenuated RSV vaccine with improved antibody responses in children. Sci Transl Med 7:312ra175. doi: 10.1126/scitranslmed.aac8463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernstein DI, Malkin E, Abughali N, Falloon J, Yi T, Dubovsky F, MI-CP149 Investigators. 2012. Phase 1 study of the safety and immunogenicity of a live, attenuated respiratory syncytial virus and parainfluenza virus type 3 vaccine in seronegative children. Pediatr Infect Dis J 31:109–114. doi: 10.1097/INF.0b013e31823386f1. [DOI] [PubMed] [Google Scholar]
- 14.Krempl CD, Lamirande EW, Collins PL. 2005. Complete sequence of the RNA genome of pneumonia virus of mice (PVM). Virus Genes 30:237–249. doi: 10.1007/s11262-004-5631-4. [DOI] [PubMed] [Google Scholar]
- 15.Brock LG, Karron RA, Krempl CD, Collins PL, Buchholz UJ. 2012. Evaluation of pneumonia virus of mice as a possible human pathogen. J Virol 86:5829–5843. doi: 10.1128/JVI.00163-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krempl CD, Wnekowicz A, Lamirande EW, Nayebagha G, Collins PL, Buchholz UJ. 2007. Identification of a novel virulence factor in recombinant pneumonia virus of mice. J Virol 81:9490–9501. doi: 10.1128/JVI.00364-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whitehead SS, Juhasz K, Firestone CY, Collins PL, Murphy BR. 1998. Recombinant respiratory syncytial virus (RSV) bearing a set of mutations from cold-passaged RSV is attenuated in chimpanzees. J Virol 72:4467–4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang CF, Wang CK, Malkin E, Schickli JH, Shambaugh C, Zuo F, Galinski MS, Dubovsky F, Study Group, Tang RS. 2013. Implication of respiratory syncytial virus (RSV) F transgene sequence heterogeneity observed in phase 1 evaluation of MEDI-534, a live attenuated parainfluenza type 3 vectored RSV vaccine. Vaccine 31:2822–2827. doi: 10.1016/j.vaccine.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 19.Liang B, Munir S, Amaro-Carambot E, Surman S, Mackow N, Yang L, Buchholz UJ, Collins PL, Schaap-Nutt A. 2014. Chimeric bovine/human parainfluenza virus type 3 expressing respiratory syncytial virus (RSV) F glycoprotein: effect of insert position on expression, replication, immunogenicity, stability, and protection against RSV infection. J Virol 88:4237–4250. doi: 10.1128/JVI.03481-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mackow N, Amaro-Carambot E, Liang B, Surman S, Lingemann M, Yang L, Collins PL, Munir S. 2015. Attenuated human parainfluenza virus type 1 (HPIV1) expressing the fusion glycoprotein of human respiratory syncytial virus (RSV) as a bivalent HPIV1/RSV vaccine. J Virol 89:10319–10332. doi: 10.1128/JVI.01380-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mosca JD, Pitha PM. 1986. Transcriptional and posttranscriptional regulation of exogenous human beta interferon gene in simian cells defective in interferon synthesis. Mol Cell Biol 6:2279–2283. doi: 10.1128/MCB.6.6.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graham BS. 2016. Vaccines against respiratory syncytial virus: the time has finally come. Vaccine 34:3535–3541. doi: 10.1016/j.vaccine.2016.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conzelmann KK. 1998. Nonsegmented negative-strand RNA viruses: genetics and manipulation of viral genomes. Annu Rev Genet 32:123–162. doi: 10.1146/annurev.genet.32.1.123. [DOI] [PubMed] [Google Scholar]
- 24.Liang B, Ngwuta JO, Herbert R, Swerczek J, Dorward DW, Amaro-Carambot E, Mackow N, Kabatova B, Lingemann M, Surman S, Yang L, Chen M, Moin SM, Kumar A, McLellan JS, Kwong PD, Graham BS, Schaap-Nutt A, Collins PL, Munir S. 2016. Packaging and prefusion stabilization separately and additively increase the quantity and quality of respiratory syncytial virus (RSV)-neutralizing antibodies induced by an RSV fusion protein expressed by a parainfluenza virus vector. J Virol 90:10022–10038. doi: 10.1128/JVI.01196-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang B, Ngwuta JO, Surman S, Kabatova B, Liu X, Lingemann M, Liu X, Yang L, Herbert R, Swerczek J, Chen M, Moin SM, Kumar A, McLellan JS, Kwong PD, Graham BS, Collins PL, Munir S. 2017. Improved prefusion stability, optimized codon-usage, and augmented virion packaging enhance the immunogenicity of respiratory syncytial virus (RSV) fusion protein in a vectored vaccine candidate. J Virol doi: 10.1128/JVI.00189-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang B, Surman S, Amaro-Carambot E, Kabatova B, Mackow N, Lingemann M, Yang L, McLellan JS, Graham BS, Kwong PD, Schaap-Nutt A, Collins PL, Munir S. 2015. Enhanced neutralizing antibody response induced by respiratory syncytial virus prefusion F protein expressed by a vaccine candidate. J Virol 89:9499–9510. doi: 10.1128/JVI.01373-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilman MS, Moin SM, Mas V, Chen M, Patel NK, Kramer K, Zhu Q, Kabeche SC, Kumar A, Palomo C, Beaumont T, Baxa U, Ulbrandt ND, Melero JA, Graham BS, McLellan JS. 2015. Characterization of a prefusion-specific antibody that recognizes a quaternary, cleavage-dependent epitope on the RSV fusion glycoprotein. PLoS Pathog 11:e1005035. doi: 10.1371/journal.ppat.1005035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Magro M, Mas V, Chappell K, Vazquez M, Cano O, Luque D, Terron MC, Melero JA, Palomo C. 2012. Neutralizing antibodies against the preactive form of respiratory syncytial virus fusion protein offer unique possibilities for clinical intervention. Proc Natl Acad Sci U S A 109:3089–3094. doi: 10.1073/pnas.1115941109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLellan JS. 2015. Neutralizing epitopes on the respiratory syncytial virus fusion glycoprotein. Curr Opin Virol 11:70–75. doi: 10.1016/j.coviro.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLellan JS, Chen M, Joyce MG, Sastry M, Stewart-Jones GB, Yang Y, Zhang B, Chen L, Srivatsan S, Zheng A, Zhou T, Graepel KW, Kumar A, Moin S, Boyington JC, Chuang GY, Soto C, Baxa U, Bakker AQ, Spits H, Beaumont T, Zheng Z, Xia N, Ko SY, Todd JP, Rao S, Graham BS, Kwong PD. 2013. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science 342:592–598. doi: 10.1126/science.1243283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McLellan JS, Chen M, Leung S, Graepel KW, Du X, Yang Y, Zhou T, Baxa U, Yasuda E, Beaumont T, Kumar A, Modjarrad K, Zheng Z, Zhao M, Xia N, Kwong PD, Graham BS. 2013. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science 340:1113–1117. doi: 10.1126/science.1234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greenberg DP, Walker RE, Lee MS, Reisinger KS, Ward JI, Yogev R, Blatter MM, Yeh SH, Karron RA, Sangli C, Eubank L, Coelingh KL, Cordova JM, August MJ, Mehta HB, Chen W, Mendelman PM. 2005. A bovine parainfluenza virus type 3 vaccine is safe and immunogenic in early infancy. J Infect Dis 191:1116–1122. doi: 10.1086/428092. [DOI] [PubMed] [Google Scholar]
- 33.Karron RA, Thumar B, Schappell E, Surman S, Murphy BR, Collins PL, Schmidt AC. 2012. Evaluation of two chimeric bovine-human parainfluenza virus type 3 vaccines in infants and young children. Vaccine 30:3975–3981. doi: 10.1016/j.vaccine.2011.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bailly JE, McAuliffe JM, Skiadopoulos MH, Collins PL, Murphy BR. 2000. Sequence determination and molecular analysis of two strains of bovine parainfluenza virus type 3 that are attenuated for primates. Virus Genes 20:173–182. doi: 10.1023/A:1008130917204. [DOI] [PubMed] [Google Scholar]
- 35.Skiadopoulos MH, Schmidt AC, Riggs JM, Surman SR, Elkins WR, St Claire M, Collins PL, Murphy BR. 2003. Determinants of the host range restriction of replication of bovine parainfluenza virus type 3 in rhesus monkeys are polygenic. J Virol 77:1141–1148. doi: 10.1128/JVI.77.2.1141-1148.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joyce MG, Zhang B, Ou L, Chen M, Chuang GY, Druz A, Kong WP, Lai YT, Rundlet EJ, Tsybovsky Y, Yang Y, Georgiev IS, Guttman M, Lees CR, Pancera M, Sastry M, Soto C, Stewart-Jones GB, Thomas PV, Van Galen JG, Baxa U, Lee KK, Mascola JR, Graham BS, Kwong PD. 2016. Iterative structure-based improvement of a fusion-glycoprotein vaccine against RSV. Nat Struct Mol Biol 23:811–820. doi: 10.1038/nsmb.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buchholz UJ, Finke S, Conzelmann KK. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol 73:251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collins PL, Hill MG, Camargo E, Grosfeld H, Chanock RM, Murphy BR. 1995. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5′ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc Natl Acad Sci U S A 92:11563–11567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coleman JR, Papamichail D, Skiena S, Futcher B, Wimmer E, Mueller S. 2008. Virus attenuation by genome-scale changes in codon pair bias. Science 320:1784–1787. doi: 10.1126/science.1155761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lauring AS, Acevedo A, Cooper SB, Andino R. 2012. Codon usage determines the mutational robustness, evolutionary capacity, and virulence of an RNA virus. Cell Host Microbe 12:623–632. doi: 10.1016/j.chom.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kozak M. 1987. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res 15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coates HV, Alling DW, Chanock RM. 1966. An antigenic analysis of respiratory syncytial virus isolates by a plaque reduction neutralization test. Am J Epidemiol 83:299–313. doi: 10.1093/oxfordjournals.aje.a120586. [DOI] [PubMed] [Google Scholar]
- 43.Yoder SM, Zhu Y, Ikizler MR, Wright PF. 2004. Role of complement in neutralization of respiratory syncytial virus. J Med Virol 72:688–694. doi: 10.1002/jmv.20046. [DOI] [PubMed] [Google Scholar]