Abstract
Bacteraemia caused by Escherichia coli is a growing problem with a significant mortality. The factors that influence the acquisition and outcome of these infections are not clear. Here, we have linked detailed genetic data from the whole-genome sequencing of 162 bacteraemic isolates collected in Scotland, UK, in 2013–2015, with clinical data in order to delineate bacterial and host factors that influence the acquisition in hospital or the community, outcome and antibiotic resistance. We identified four major sequence types (STs) in these isolates: ST131, ST69, ST73 and ST95. Nearly 50 % of the bacteraemic isolates had a urinary origin. ST69 was genetically distinct from the other STs, with significantly less sharing of accessory genes and with a distinct plasmid population. Virulence genes were widespread and diversely distributed between the dominant STs. ST131 was significantly associated with hospital-associated infections (HAIs), and ST69 with those from the community. However, there was no association of ST with outcome, although patients with HAI had a higher immediate mortality compared to those with community-associated infections (CAIs). Genome-wide association studies revealed genes involved in antibiotic persistence as significantly associated with HAIs and those encoding elements of a type VI secretion system with CAIs. Antibiotic resistance was common, and there were networks of correlated resistance genes and phenotypic antibiotic resistance. This study has revealed the complex interactions between the genotype of E. coli and its ability to cause bacteraemia, and some of the determinants influencing hospital or community acquisition. In part, these are shaped by antibiotic usage, but strain-specific factors are also important.
Keywords: bacteraemia, antibiotic resistance, genome sequencing, genome-wide association study
Data Summary
Illumina sequences have been deposited in the European Nucleotide Archive under project PRJEB12513.
Impact Statement.
Blood stream invasion with the bacterium Escherichia coli is increasing across the UK and carries a poor prognosis. About half of these infections are acquired in healthcare settings, with the rest arising from the community. The microbial and host factors that determine the acquisition and outcome of these infections are not known. To uncover some of these factors, we have undertaken whole-genome sequencing of bloodstream isolates of E. coli from patients and linked these to their clinical data. We identified the major genetic families of E. coli causing bloodstream infection in Scotland. Genetic analysis showed that genes conferring an ability to microbes to survive at low growth rates in the presence of antibiotics were important in hospital-associated infections (HAIs). Genes encoding a mechanism to compete with other microbes were important in community-associated infections (CAIs). Outcome was worse in HAI and was associated with urinary catheter use. Genes encoding resistance to antibiotics used extensively in hospitals clustered together in strains from HAIs, suggesting co-inheritance of multiple resistance genes in these strains. These results show how antibiotic use is driving the evolution of strains causing bloodstream infection in hospitals; factors responsible for CAI are less clear.
Introduction
The Gram-negative bacterium Escherichia coli is a common gut commensal in man, as well as an important cause of a range of intestinal and extra-intestinal infections [1, 2]. It is the leading cause of bloodstream infection across Europe and the USA [3, 4], with a high attributable mortality of between 10–30 % that rises with age [5–7]. Rates of E. coli bacteraemia have increased dramatically over the last 10–20 years, rising 25.6 % between 2012/3 and 2016/7 in England and Wales [8], and 46 % between 1992 and 2006 in Denmark [5], with similar increases within the USA [3]. Across Europe, isolates of E. coli from blood collected by the European Antimicrobial Resistance Surveillance System increased by 60 % from 2002 to 2008 [4]. About half these bacteraemias are hospital or healthcare associated, with reported percentages of just over 50 % in Scotland in 2016 [9, 10] and in England in 2012/3 [11]. Thus, community-associated infections (CAIs) are also a significant cause of bacteraemias.
The mechanisms underlying these increases in incidence worldwide remain obscure. Data from Europe show that the increase has been more marked in antibiotic-resistant organisms, many of which are multidrug resistant [4]. A more focussed study in England also found an increasing incidence of antibiotic-resistant strains [12]. Because of the rise in incidence of E. coli bacteraemias, enhanced surveillance programs have been instituted in both England and Scotland. Preliminary data from the surveillance in England has shown about half of all patients had previous healthcare exposure in the month prior to the bacteraemia [11]. The source of bacteria was estimated to be the urogenital tract in about 50 % of cases. The Scottish data also showed a urinary source in about half of the bacteraemic cases [9].
A number of studies have analysed the genetic make-up of E. coli lineages isolated from the urinary tract and/or bacteraemias [2, 13–17]. These have characterized the dominant multilocus sequence types (MLSTs) associated with these infections: sequence types (STs) ST73, ST131, ST95 and ST69 are the most prevalent in both the UK and the USA. ST131 has come to prominence in the last few decades as a widely distributed strain that typically has a multidrug-resistant phenotype [18, 19]. A recent study estimated that ST131 underwent rapid expansion in about 1995 [20]. However, this expansion stabilized quite rapidly, with a new equilibrium established with other E. coli STs. Antibiotic resistance is not the only factor involved in the success of E. coli strains, since ST73 is the most abundant phenotype found in some studies and is typically antibiotic susceptible. Although whole-genome sequencing has been performed on large collections of clinical isolates, these have lacked detailed clinical data on the source patients and, thus, the clinical factors involved in selection of different bacterial genes that might be associated with the observed increase in rates of bacteraemia and their origin remain unknown.
The ability to link full clinical data with whole-genome sequences of causative microbes greatly increases the ability to identify key pathogen factors involved in acquisition and subsequent development of disease. Here, we have taken advantage of the rich clinical data provided by enhanced surveillance of E. coli bacteraemias within Scotland to link this with whole-genome sequencing of the causative strains. Our aim was to explore potential factors associated with the microbe and the host that lead to bacteraemia, and in particular to examine the determinants of CAI versus hospital-associated infection (HAI). A total of 162 bacteraemic E. coli isolates from 2013 and 2015 were fully sequenced and analysed in tandem with the clinical data available on each source patient that was recorded as part of an enhanced surveillance program. The results presented here allowed us to examine in depth the population structure of E. coli strains causing bacteraemia, and to relate this to the clinical risk factors, antibiotic sensitivities and course of infection. We identified genes important in HAIs versus CAIs by genome-wide association study, which revealed that acquisition of antibiotic tolerance is an important factor in bacterial fitness in healthcare environments. We observed differing virulence determinants between different strains, and evidence of co-selection of multiple antibiotic-resistance genes (ABRs).
Methods
Collection of E. coli isolates and whole-genome sequencing
A total of 162 blood culture E. coli isolates were selected retrospectively from the microbiology laboratories of the Queen Elizabeth University Hospital, Glasgow, Scotland, UK, and Dumfries Royal Infirmary, Dumfries, Scotland, UK. For the Queen Elizabeth Hospital, all non-duplicate isolates in the last quarter of 2013 (97) and from a 6 week period in the last quarter of 2015 (41) were taken. From Dumfries, all non-duplicate isolates from the last quarter of 2013 were collected (24). The total numbers of E. coli bacteraemic isolates during these periods were 428 in 2013 and 576 in 2015 from Glasgow, and 95 in Dumfries for 2013. Genomic DNA was extracted at each site using QIAmp (Qiagen) and index-tagged paired-end Illumina sequencing libraries were prepared. Sequencing was performed at the Welcome Trust Sanger Centre (Hinxton, UK) using the 96-plex Illumina HiSeq 2500 platform to generate 2×200 bp paired-end reads. These raw Illumina sequences have been deposited in the European Nucleotide Archive under project PRJEB12513.
Clinical data for linkage analysis
The clinical data for the Scottish isolates had been collected from blood specimens with patient outcome (e.g. died in hospital or discharged) along with date of death/discharge, catheter association during hospital admission, age, sex, infection type (e.g. urosepsis, bone, skin, respiratory tract or gastrointestinal) and mode of acquisition whether it was a HAI or a CAI. Bacteria isolated from samples taken greater than 48 h after admission to hospital were classified as HAI and those within 48 h of admission to hospital were classed as CAI. Bacterial minimum inhibitory concentration (MIC) and antibiotic-sensitivity data for each isolate were also collected. Multilocus sequence typing analysis of all the isolates was carried out with SRST2 [21], using maximum number of mismatches per read for MLST allele calling as 10.
Phylogenetic analysis and variant detection
The raw Illumina reads were mapped to the E. coli reference genome EC958 (strain ST131, accession no. NZ_HG941718.1) using SAMtools mpileup [22] and were called for single nucleotide polymorphisms (SNPs) through VarScan [23] (read depth ≥2×, variant allele frequency ≥0.08 and P value ≥0.005). Mobile genetic elements were masked and recombination filtration was performed using Gubbins [24]. Maximum likelihood (ML) trees were inferred using RAxML [25] with the generalized time-reversible (GTR) model and a gamma distribution to model site-specific rate variation. One hundred bootstraps were conducted for the support of the SNP-based ML phylogenetic tree.
Pangenome analysis
De novo assembly of the Illumina reads and the annotation of the coding sequence regions were conducted using the Sanger pipeline [26]. Core and accessory genes were identified using Roary [27] with blastp identity ≥95 % and percentage of isolates to be in the core genome taken as 99 %. The accessory gene (>5 and <99 % isolates) accumulation curves were generated separately for each of the dominant STs using R. Non-metric multidimensional scaling (NMDS) was performed using the vegan package in R (http://cran.r-project.org/web/packages/vegan/index.html). The variation of core gene and accessory gene content was performed using pairwise distances. For each pair of isolates, we calculated their cophenetic distance from the core gene phylogenetic tree and the Jaccard distance based on presence or absence of accessory genes. These two distances were plotted against each other to compare the diversity of core and accessory genes in Fig. 2(c). Plasmid incompatibility types and subtypes were identified using PlasmidFinder [28].
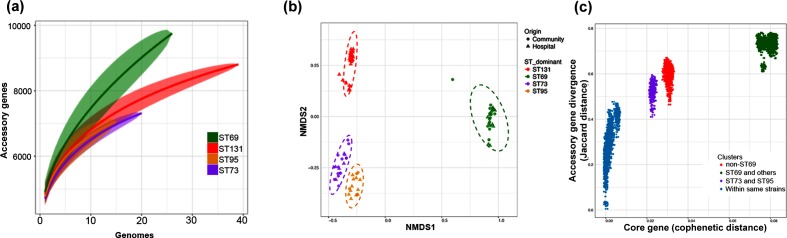
Fig. 2.
(a) Accessory gene accumulation curves for the dominant STs with respect to the number of genomes compared. At each value x of the number of genomes compared, a random sample n of genomes was compared in n!/(n−x)!x! ways. The solid line shows the median value at each value of x; the shaded area contains the interquartile values. (b) NMDS plots of the different dominant STs based on their shared accessory gene content. Community or hospital origin are as indicated. Dotted lines are 95 % confidence limits of values around the centroid for each ST. (c) Comparison of core and accessory genomes of different STs. Each circle is the core genome similarity as cophenetic distance plotted against accessory gene similarity as Jaccard distance for each possible paired comparison within the different groups as indicated in the key.
Pathway analysis
Genes were extracted from all the isolates using an in-house python script and linked to KEGG (Kyoto Encyclopedia of Genes and Genomes) orthologue or KO groups using david [29]. The fold enrichment for each pathway was evaluated with reference to E. coli K-12 genome.
Virulence genes
Virulence genes were identified using the VirulenceFinder database [30] of SRST2 with percentage of coverage cut-off for calling a gene ≥90, keeping default values for all other parameters. We found 437 virulence genes in our 162 isolates. A Kruskal–Wallis non-parametric test (with Bonferroni correction adjusted P values for multiple testing) was performed in R using the stats package.
Survival analysis
Survival was analysed by the Kaplan-Meier method and comparisons between subgroups were examined by the log rank test using the survival package in R. Cox proportional hazards analysis was carried out to identify the predictors for 6 months mortality that were independent (univariate analysis) or dependent of age and gender (multivariate analysis).
Genome-wide association study
Pangenome-wide association study was carried out on pangenome genes extracted from all the isolates. Using Scoary [31], the significant genes were identified that were different between the HAI versus CAI groups of isolates. A Kruskal–Wallis significance test with Bonferroni correction was also conducted to reconfirm the significance. The Manhattan plot has genes along the x-axis in no particular order and the y-axis gives the negative logarithm with base 10 of the significant P values.
ABRs and network analysis
ABRs were detected using ariba [32] using the card database. Altogether 52 ABRs were identified (see the Supplementary Material available in the online version of this article). The correlation networks of ABRs gene presence/absence matrix and clinical susceptibility (R/S) matrices were constructed using Pearson’s correlation and the Hmisc v4.0–2 package in R. The cut-off edge weight value was set at a correlation of >0.5 for the network plot.
Results
Study design
We selected isolates of E. coli from blood cultures taken from patients within the Greater Glasgow and Clyde Health Board over a 2 month period in 2013 and in 2015. We also collected similar isolates from patients within Dumfries and Galloway in 2013. After removal of potential duplicates, samples that failed to grow and those that failed to provide full genomic sequences, we were left with a total of 162 isolates that were fully analysed as below.
Population structure of isolates
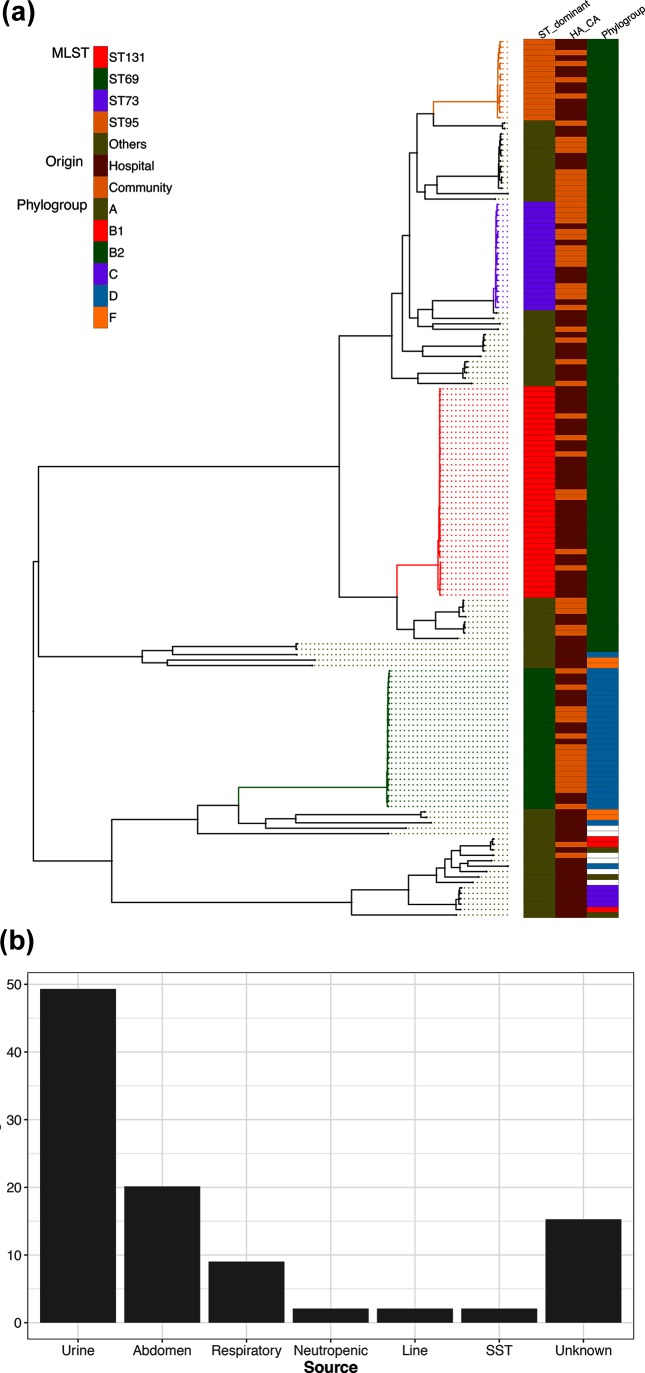
De novo genome assembly produced sequences ranging in length from 4.6 to 5.1 Mbp genomes. A total of 20 855 genes were identified from these assemblies, of which 13 % (2804 genes) were conserved in more than 99 % of genomes. These core genes were used to generate a 2.68 Mbp base pair alignment, which was used to construct a phylogenetic clustering analysis (Fig. 1a). The MLST scheme identified four dominant STs found in more than 10 isolates within our 162 isolates: ST131 (39, 24 %), ST69 (26, 16 %), ST73 (20, 12 %) and ST95 (15, 9 %). Other strains accounted for 38 % of the isolates. The ML phylogenetic analysis, with 100 % bootstrap support, corresponded very well with the MLST identified strain types. The tree was rooted on the longest branch, separating the ST69 strains (phylogroup D) from the other strains ST131, ST73 and ST95, which belong to the phylogroup B2. The source of the bacteraemic isolates was identified by case note review for 144 of the isolates (Fig. 1b). A urinary origin was the commonest source (49 %), followed by abdominal (20 %) and respiratory (9 %) origins.
Fig. 1.
(a) ML SNP core gene phylogeny of the E. coli isolates. The STs were identified using the MLST scheme. STs with fewer than 10 isolates are clustered in the category ‘others’. The key indicates phylogroups and whether the isolates were community associated (CA) or hospital associated (HA). (b) Source of bacteraemia as percentages of total isolates. Line, indwelling vascular device; SST, skin and soft tissue.
Sub-analysis of the ST131 isolates into the clades described by Price et al. [33] and Petty et al. [34] showed that the majority of the ST131 isolates were in clade C (29/39; 74 %), characterized by being of serotype O25H4 and possessing the fimh30 allele (see Fig. S1). Further subdivision of the C clade was made using clade-specific SNPs as described by Ben Zakour et al. [35]. Five isolates were sensitive to ciprofloxacin (C clade). Of the ciprofloxacin-resistant strains, 5 were in clade C1 and 19 in clade C2. Possession of the extended spectrum β-lactamase CTMX-15 has been described as a marker of clade C2. We found 10/19 clade 2 isolates had this gene; in addition, 1 had the gene for CTXM-9. C1 isolates lacked CTXM-15, but one had the CTXM-9 gene. CTXM-15 has, thus, been acquired/lost repeatedly within the C2 clade as noted also by Kallonen et al. [20]. One clade A isolate had also acquired the CTXM-15 gene.
Accessory gene analysis
We hypothesized that the accessory gene content of the isolates would contain genes important in establishing blood stream infection that were common across all the ST types. Within the 162 isolates, 69 % (n=14 328) of the pangenome was identified within the dominant strain types ST131, ST73, ST95 and ST69. We identified 5757 accessory genes that are present in (≥5 and <99 %) our 162 genomes. Most of the accessory genes were rare, and only 22 % have been identified functionally. To measure the size of the accessory genome, we determined the numbers of new genes identified with increasing numbers of genomes sampled for each of the dominant ST types (Fig. 2a). This showed that for all STs, there was a steady rise in the number of new genes identified with increasing sampling. As noted before for E. coli, this did not reach a plateau for any of the STs. The accumulation curves fitted a power law as given by Heaps’ law, as described by Tettelin et al. [36]. The derived constant α was less than 1 for all ST, indicating an open pangenome. However, as Fig. 2(a) shows, the accumulation curve for ST69 rises more rapidly than the other STs. This is reflected in the derived α constants, which is lowest for the ST69 isolates. Comparing ST69 to the largest group, ST131, the difference in the distributions was significant (two sample Kolgomov Smirnoff test, P=0.0071). Overall measures of accessory gene diversity showed all STs were highly diverse, although ST131 and ST69 were marginally more diverse than the other two STs (see Fig. S2).
Next, we assessed whether the accessory gene composition reflected the ST or grouped the isolates in another fashion. Similarity between the accessory gene content of each isolate was analysed using NMDS based on the Jaccard distance of accessory genes between each isolate (Fig. 2b). This showed that isolates grouped according to ST rather than any other parameter. However, there were significant differences in which STs were associated with HAI or CAIs (multinomial goodness of fit test, P<0.05, exact binomial probability). ST131 was highly associated with HAI (82 % of isolates, exact binomial compared to overall rate of HAI of 62 %, P<0.05) and ST69 significantly associated with CAI (62 % of isolates, exact binomial compared to overall rate of CAI of 38 %, P<0.05)
The variation in the accessory gene pool was further assessed by comparing the core genome divergence and accessory gene content in pairwise comparisons (Fig. 2c). Comparison of isolates with other members of the same ST showed little difference in their accessory gene content, clustering towards the origin of the plot (blue dots). Pairwise comparison between all isolates of different ST groups other than ST69 are shown in red, and cluster quite separately from the intra-ST pairwise comparisons. This discontinuity reflects a distinct clonal structure to the population, since higher rates of recombination between STs has been shown to generate a more homogeneous distribution [37]. Comparison of ST69 with all other isolates shows a more extreme clustering in the upper right of the plot (green), highlighting the divergence of ST69 from the other STs, with a significantly different accessory gene content that does not appear to be undergoing exchange from the other STs. This does not reflect any difference between the recombination rates in ST69 compared to the other STs; recombination rates between the different STs were very similar (see Fig. S3). As a comparison, we compared core gene divergence and accessory gene content between ST73 and ST95, which are more closely related in core gene content (purple dots). Accessory gene content was more divergent between these groups than for the intra-strain comparison, but not so extreme as between ST69 and the other STs.
A proportion of accessory genes will be present on different plasmids between the isolates. Although from the short reads provided by Illumina sequencing we were unable to reconstruct plasmids, we identified plasmid profiles within the isolates based on the presence of specific replicon genes that places them in different incompatibility (Inc) groups (Fig. S4). Ten plasmid Inc groups were identified across all the strains. The broad range IncQ plasmids were predominantly found in isolates of ST69 (42 % of the isolates, Fig. S4); the differences between the groups were significant by G test of independence (P<0.001 with Williams correction), with significant increases in the percentage of IncQ plasmids in ST69 compared to ST131 and ST73 (G test false discovery rate corrected P values of 0.002 and 0.01, respectively). Other plasmids without recognized replicon genes may also be present in these strains.
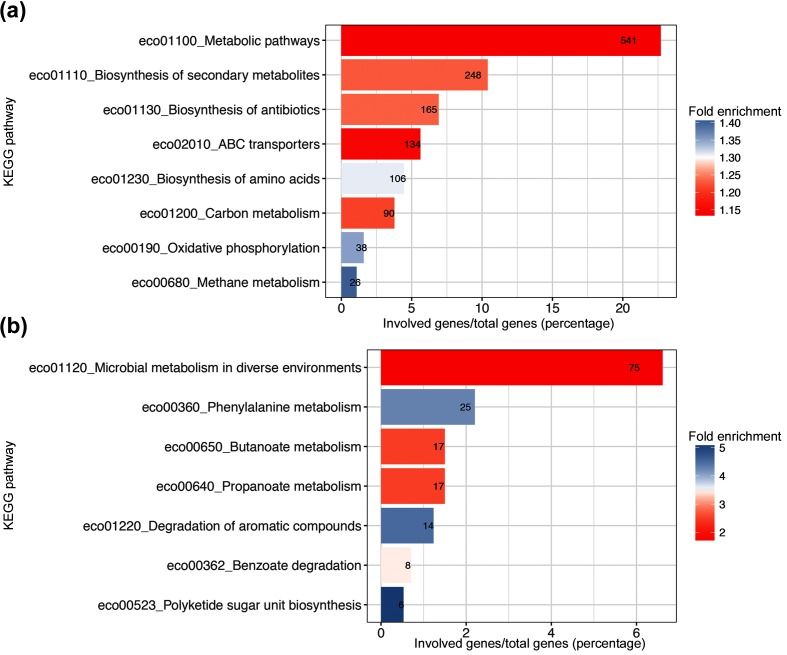
We compared the pathway designations of the core and accessory genomes of the different isolates (Fig. 3). Compared to the E. coli reference strain K12, the core genome showed modest but significant enrichment principally of metabolic pathways (Fig. 3a). The accessory genome had over-representation of different pathways (Fig. 3b). These included phenylalanine metabolism, products of which have been implicated in bacterial pathogenesis [38]. We also determined which genes were common in the accessory genome of the different isolates but not at the 99 % threshold of the core genome. A total of 495 genes were identified as shared between 80 % of the commonest isolates ST131, ST69 and ST73. Pathway analysis of these genes did not reveal any significant common pathways.
Fig. 3.
KEGG pathway analysis of (a) core and (b) accessory genes of the different isolates. Comparison was made to the KEGG assignation of genes with the E. coli reference strain K12. Bars show the genes significantly associated within each KEGG group as a percentage of the total number of the core or accessory genes with a KEGG pathway assignation. Fold enrichment of the different groups is as indicated in the colour key. Superimposed numbers are the number of genes assigned within the indicated KEGG pathway.
Virulence genes
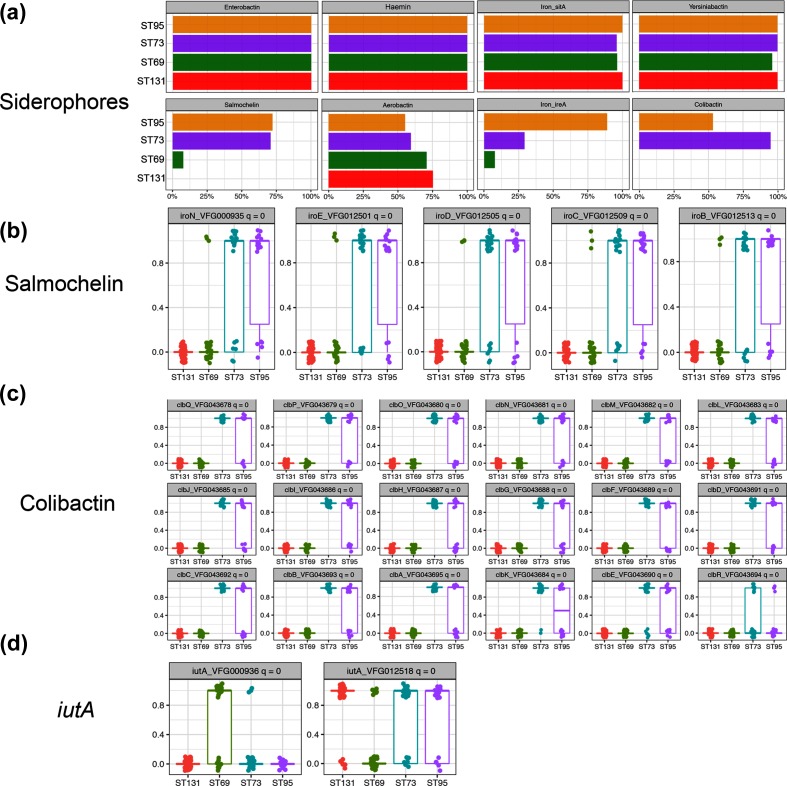
We used the Virulence Factor Database [39] to determine the different virulence genes within the different E. coli strains. A total of 363 different virulence genes were detected. In keeping with the predominantly urinary origin of about 50 % of the isolates, genes previously identified as important in urinary pathogenic E. coli were highly prevalent. Genes encoding siderophores showed considerable variation between the different dominant ST strains (Fig. 4). Genes encoding enterobactin, haemin, yersinobactin and the metal transporter SitA were found in virtually all of the dominant STs. However, genes for salmochelin and colibactin were almost exclusively present in ST95 and ST73 (Fig. 4). Fig. S5 (a–d) shows the distribution of individual siderophore genes between the STs. Genes of the salmochelin operon (iroBCDEN) were present together in the salmochelin-positive strains. Likewise, genes of the colibactin operon were also found within ST95 and ST73. Two different alleles of the iutA gene that is involved in aerobactin synthesis were distributed differentially between the different STs; the allele VFG012518 has been found in plasmids isolated from avian pathogenic E. coli strains, suggesting possible transmission of plasmids or elements of these plasmids between humans and farmed poultry [40].
Fig. 4.
Distribution of siderophores within the major STs. (a) Percentage of each major ST containing the genes for the indicated siderophore systems. (b–d) Individual occurrences of each indicated siderophore gene within the different STs. Genes are scored as present (1) or absent (0), with each circle representing a single isolate; symbols are dodged to aid in viewing. Calculated q values are shown in each graph header; values <10−6 are assigned as 0.
Virulence genes that showed a significant difference between the dominant STs are shown in Figs S5 and S6. The diversity of virulence gene content between the different STs is very clear. Some of these genes were only found within one ST. Numerous adhesins that are known to be important in the pathogenesis of urinary infection were present in these isolates, and some significant differences were detected. Some of the divergence is associated with different alleles of adhesion genes, such as papG, which is the terminal component of P type pili that is key in mediating attachment of bacteria to receptors within the kidney and initiating pyelonephritis. This allele (vfg000890) is a class II papG that is known to be important in attachment of E. coli to cells in the upper urinary tract and in the development of sepsis [41, 42]. However, some of the ST73 isolates were unique in possessing genes encoding components of the S and FIC pili [43], which were not seen in any of the other isolates. There were significant differences in the presence of members of the serine protease autotransporters of Enterobacteriacae (SPATE) family [44]. A previous study of ST131 found the SPATE family gene espC in 99 % of ST131 isolates [20]; however, we only found one of our isolates contained this gene, which was an ST131. ST69 strains contained many genes that are part of a second type III secretion system (ETT2) [45] previously described in strains of enterohaemorrhagic and enteroadhesive E. coli, which we found intact in a complete operon. Finally, members of strains ST73 and ST131 possessed genes of the haemolysin operon, a toxin important in the pathogenesis of urinary E. coli infections [46].
Outcome and risk factors for death
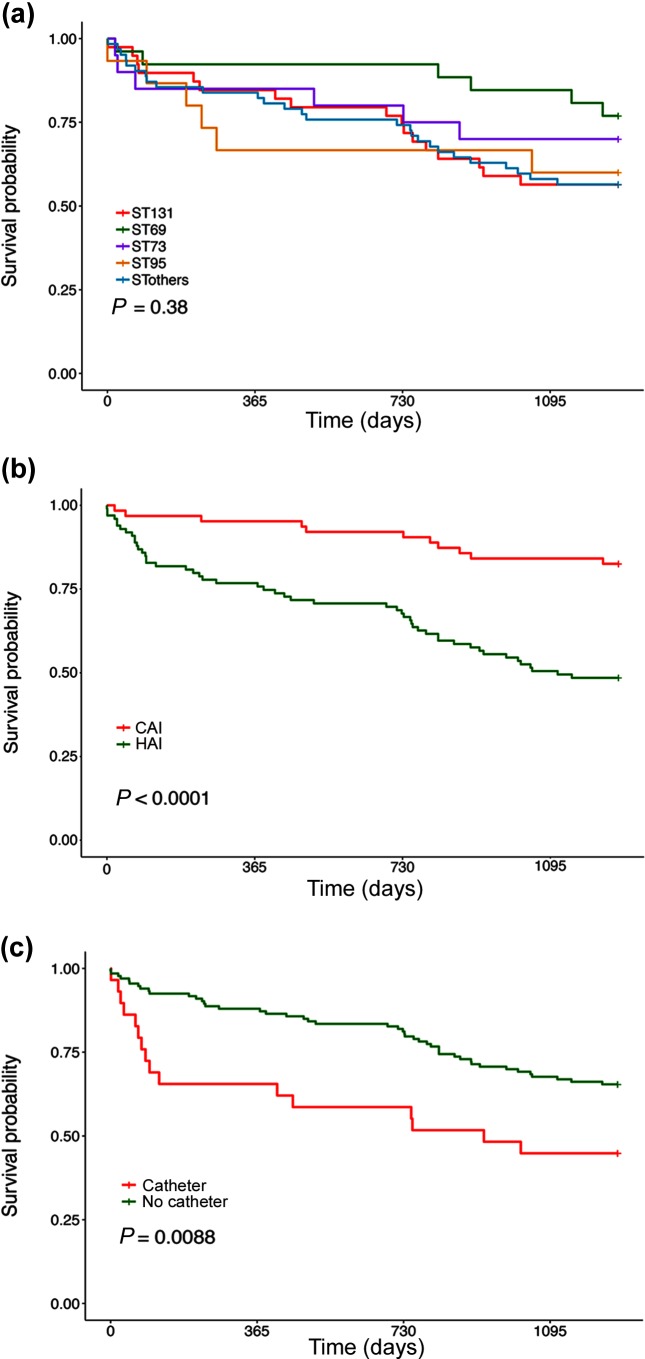
To begin to dissect the contribution of this diverse genomic repertoire to disease, we compared the outcome of patients infected with the different dominant ST types (Fig. 5a). The Kaplan–Meir survival curves show little difference between the different STs in mortality; ST69 shows a slower rate but multiple comparisons did not reveal statistically significant differences (Cox proportional hazards model with correction for multiple testing). Mortality between HAI and CAI showed a significant difference (Fig. 5b). HAIs showed a worse outcome; the curves separated markedly in the first 90 days after the E. coli was isolated and then became parallel. We also tested whether presence of a urinary catheter influenced outcome (Fig. 5c). Early mortality was significantly more marked in patients who had a urinary catheter at some point in their hospital stay. We performed univariate and multivariate analysis of 6 month mortality in this group of bacteraemic patients with a number of clinical factors (Table 1). Age, presence of a urinary catheter and a respiratory source were all significant risk factors for 6 month mortality in a univariate analysis. Presence of a urinary catheter and respiratory source remained significant in a multivariate analysis.
Fig. 5.
Kaplan–Meir survival curves of the patients within the study stratified according to (a) ST, (b) CAI or HAI and (c) presence or absence of urinary catheter. P values show the significance of the differences between the outcomes using the Cox proportional hazards model.
Table 1. Univariate and multivariate analysis for predictors of 6 months mortality calculated on 162 isolates.
P values (P val) and confidence intervals (CI) are shown. P values less than 0.05 are shown in bold.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Variable | Odds ratio | P val | 95 % CI | Odds ratio | P val | 95 % CI |
| Gender – female (ref: male) | 0.91 | 0.8369 | 0.37–2.23 | |||
| Age>65 (years)(ref: ≤65 (years)) | 3.66 | P=0.0384 | 1.07–12.48 | |||
| Hospital (ref: community) | 1.73 | 0.2627 | 0.66–4.49 | 1.62 | 0.3411 | 0.60–4.34 |
| Urinary catheter (ref: no catheter) | 4.83 | P=0.0005 | 1.97–11.83 | 6.22 | P=0.001 | 2.09–18.48 |
| Diagnosis (ref: gastrointestinal) | ||||||
| Urosepsis | 1.83 | 0.4371 | 0.40–8.33 | 1.34 | 0.7121 | 0.27–6.45 |
| Respiratory | 7.03 | P=0.0243 | 1.29–38.43 | 5.55 | P<0.05 | 0.97–31.499 |
| Others | 1.36 | 0.7210 | 0.25–7.44 | 1.91 | 0.4873 | 0.31–11.95 |
| Strain types (ref: 0) | ||||||
| ST131 | 0.78 | 0.6531 | 0.26–2.33 | |||
| ST69 | 0.57 | 0.4451 | 0.13–2.44 | |||
| ST73 | 1.30 | 0.6735 | 0.38–4.44 | |||
| ST95 | 1.10 | 0.8982 | 0.26–4.47 | |||
Determinants of HAI versus CAI
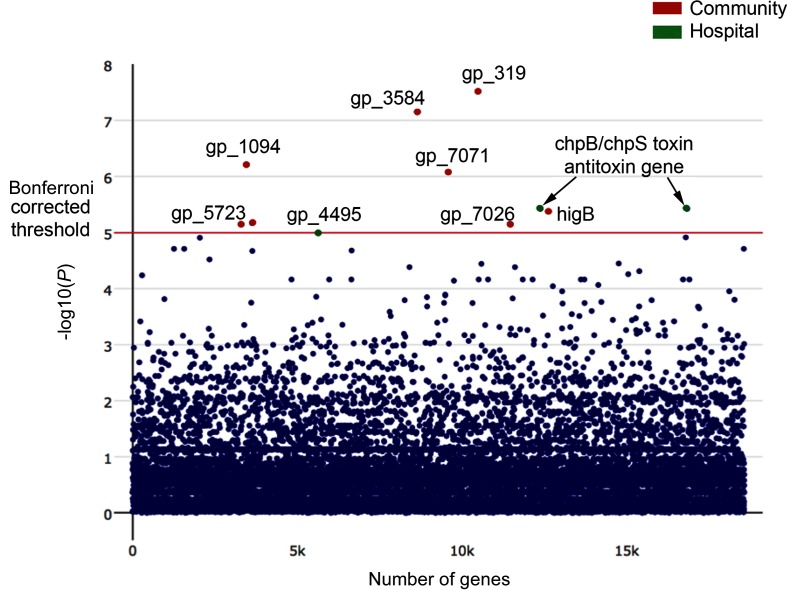
Sixty-two per cent of all the bacteraemic isolates were from HAI. We wished to discover the determinants of HAI versus CAI using the whole-genome sequence data. To analyse the determinants within isolates that favour healthcare-associated or community-acquired bacteraemia, we performed a genome-wide association study to identify those genes preferentially present in the two different groups (Figs 6 and S7). We used the software programme Scoary [31], which scores the pangenome of isolates associated with phenotypic characteristics while accounting for population stratification. This correction is important, as spurious associations of genes with phenotypic characteristics may arise because of the strong association of some genes with different STs in E. coli.
Fig. 6.
Manhattan plot of association of genes with CAI or HAI detected by a genome-wide association study. Association of genes with phenotype was determined by multiple Kruskal–Wallis tests. Uncorrected P values are shown with the Bonferroni corrected P value for significance threshold indicated by the red horizontal line. Genes more highly represented in HAI are shown in green and those more highly represented in CAIs are in red. Individual distributions of Bonferroni corrected significant associations are shown in Fig. S7.
In the healthcare-associated bacteraemic isolates, we did not find any significant association with genes mediating antibiotic resistance, but there was a significant association with the chpB/chpS genes encoding a chromosomal toxin/antitoxin pair. ChpB is a toxin of a type II toxin/antitoxin module that is an endoribonuclease, inhibiting protein translation and inducing bacterial stasis [47, 48]. Such chromosomal toxin/antitoxin modules are increasingly recognized as crucial in the phenomenon of bacterial persistence [49]. These are cells that enter into a dormant non-growing state on exposure to antibiotics or other adverse conditions that allows them to survive until such environmental stress is relieved. Another gene found significantly more frequently in isolates from HAIs is BglH (YieC), which encodes an outer-membrane carbohydrate-specific porin [50]. In isolates from CAIs, a number of genes associated with a type VI secretion system were identified: an Rhs/Vgr-family protein (gp_319), and a PAAR-repeat-containing protein (gp_3584). Additionally, the gene higB-1 was more common in the community-acquired isolates; this encodes a putative mRNA interferase as part of a toxin/antitoxin pair [51].
Resistance to antimicrobials
In the collection, 98 resistance genes were identified; their distribution is shown in Fig. S8. Mutations conferring ciprofloxacin resistance were also present. There were significantly higher numbers of ABRs in ST131 and ST69 compared to the other STs (Fig. 7a). However, there was no difference between the ABR counts between isolates from HAI and CAI (Fig. 7b). We compared phenotypic resistance to five key antibiotics between the four main STs: 3rd generation cephalosporins, ciprofloxacin, co-amoxiclav, gentamicin and trimethoprim (Fig. 7c). This revealed significant differences between the main STs (overall G test, P=1.84×10−11); differences between all possible pairs were also significant (G test pairwise nominal test for independence with false discovery rate correction, P values all <0.05). ST131 was notable for significant antibiotic resistance in all of these classes, as has been noted elsewhere [19]. In particular, some ST131 strains contained extended spectrum β-lactamases of the CTX-M class, which mediate resistance to 3rd generation cephalosporins. The predominantly community-associated ST69 showed high levels of phenotypic resistance to co-amoxiclav (31 % of isolates) as did ST131 (51 % of isolates). Both ST131 and ST69 showed a high percentage of resistance to trimethoprim (69 % of isolates for both STs).
Fig. 7.

ABRs and phenotypes within the isolates. (a) Distribution of ABRs within the different STs. Each circle is the value of single isolate; boxes show the interquartile range and the horizontal line the median. * indicates a significant difference between ST131 and others, with P value as shown. (b) Distribution of ABRs between isolates from community or hospital sources. Symbols as in (a). There was no significant difference between the groups. (c) Percentage of dominant STs having phenotypic resistance to the indicated antibiotics.
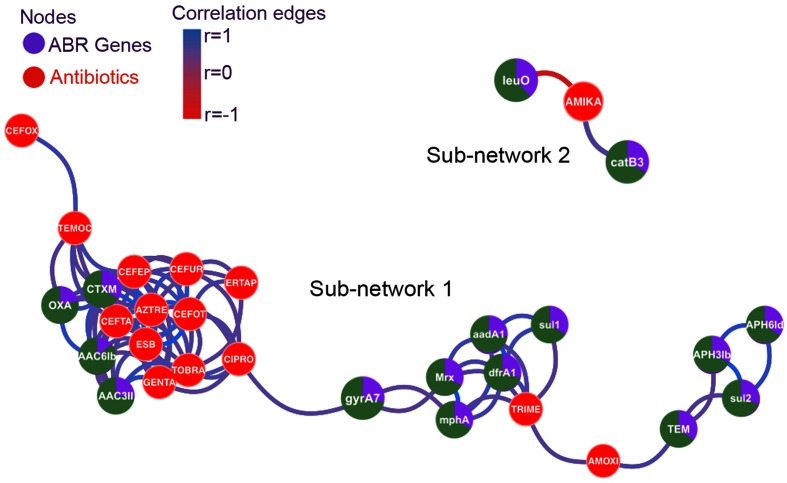
We wished to determine whether there were significant correlations between phenotypic resistance to certain antibiotics and the presence of known ABRs. We hypothesized that increased levels of antibiotic use as found for HAI might result in selection for multiply resistant strains. Moreover, the common organization of ABRs within integrons [52] and within plasmids [53] could result in the co-selection of many different ABRs as a group. We tested this hypothesis by calculating the Pearson correlation coefficient between presence and absence of ABRs within strains and antibiotic susceptibility, coded as sensitive or resistance. The correlation matrices were visualized with a network plot (Fig. 8). Nodes for ABRs are shown in green/purple, and phenotypic resistance in red. Linking edges show the degree of correlation; the higher the degree of correlation, the shorter the edge. Only correlations with a value >0.5 are shown. The plot shows a number of key relationships. Firstly, there is a marked strong correlation between antibiotic resistance to a group of antibiotics whose use is largely confined to hospital use, such as gentamicin, tobramycin, aztreonam, ertapenem and 3rd generation cephalosporins (all clustered closely to the left of the figure). A number of ABRs are associated with these phenotypic resistances. These include extended spectrum β-lactamases of the CTXM class as well as β-lactamases of the OXA class that are associated with resistance to co-amoxiclav [54]. Aminoglycoside-resistance genes are also contained within this group: aac(3′)II encodes an aminoglycoside N-acetyltransferase that mediates resistance to gentamicin and tobramycin, while aac(6)Ib encodes a protein that mediates resistance to gentamicin and other aminoglycosides [55]. On the right of the network is a looser cluster of ABRs and phenotypic resistances. These are centred around amoxicillin and trimethoprim, both antibiotics with extensive community use. Trimethoprim resistance is linked to the dfrA1 gene, a dihydrofolate reductase allele that is not inhibited by trimethoprim. However, there is close correlation also with the streptomycin/spectinomycin resistance gene aadA1, genes within an erythromycin-resistance operon (mrx and mphA), and sul1, a sulphonamide-resistance determinant. This would seem to be an example of co-selection, as these genes are commonly found linked as part of an integron [56]. These alleles were extremely closely linked; of 51 isolates with the dfrA1 trimethoprim-resistance gene, 47 also contained the aadA1 gene. Similar considerations apply to the linkage between the β-lactamase of the TEM class that mediates amoxicillin resistance, and aph3Ib and aph6Id, which mediate kanamycin/streptomycin resistance, and sul2, which mediates sulphonamide resistance. sul2, aph3Ib and aph6Id are commonly genetically linked on DNA fragments found in plasmids, integrative elements and chromosomal islands [55].
Fig. 8.
Network analysis of ABRs and phenotypic resistance. The network is based on Pearson correlation coefficients between the different determinants. Only correlations with absolute value >0.5 are shown, colour coded as indicated. Strength of correlation is also coded by proximity of nodes. Genes are shown in green/purple with the green sector indicating the proportion of these genes found in hospital-acquired isolates. Phenotypic resistance nodes are coloured red.
A separate sub-network links amikacin resistance to the genes encoding transcriptional regulator leuO and catB3, which mediates chloramphenicol resistance. LeuO has been shown to influence expression of a number of operons involved in drug resistance [57]. The linkage between catB3 and amikacin resistance is not clear.
Discussion
We have analysed whole-genome sequences of E. coli isolated from blood cultures of patients in Scotland in 2013 and 2015, and linked them to clinical data on the same patients. This has allowed us to explore the relationships between genes within these isolates and the origins and clinical course of these infections, as well as detailed information on the range of strains producing these infections and their sensitivity to antibiotics. The four most prevalent STs found in our study – ST131, ST69, ST73 and ST95 – are similar to those in reports of isolates from elsewhere in the UK, Ireland and the USA [2, 13–17, 20]. About half of our isolates had a urinary origin and this was reflected in the content of virulence genes, which contained many genes known to be involved in the pathogenesis of urinary infection and pyelonephritis. Most of the isolates were from the known virulent phylogroups B2 and D [58].
A recent study of isolates from the UK showed the appearance of ST131 and ST69 from about 2002 [20]. ST131 was the commonest ST in our study and was more prevalent in hospital-acquired bacteraemias, as found in other studies [19]. ST73 was only the third most prevalent ST (12 %) of our isolates, compared to a recent study of bacteraemic isolates in England from 2001 to 2011 in which it was the most common isolate [20]. ST69 was the second commonest ST in our isolates and shows some significant differences from the other STs causing bacteraemias. ST69 was more common in CAI and diverged the most from the other dominant STs in its core gene content as well as its accessory genome (Fig. 2). The marked divergence of ST69 accessory genome from the accessory genomes of the other dominant STs suggests very little recombination is occurring with these other STs. We also found a higher content of plasmids with IncQ replicons within ST69 compared to the other STs. Taken together, these differences highlight that ST69 is significantly distinct from the other STs and would appear to occupy a rather distinct ecological niche compared to the other STs.
Virulence determinants were widely distributed within the isolates. However, no common pattern of such determinants was found. Rather, there are multiple different combinations of such factors within bacteraemic isolates that segregate between STs rather than any other factor. Although some determinants were found in several of the dominant STs, some were only present in one ST. This suggests that these factors were either acquired subsequent to the divergence of the different STs or lost in those that no longer possess the factor. It is difficult to determine which of these alternatives have given rise to the observed lineages. Establishing the ancestral history of E. coli is problematic because of the high degree of recombination and horizontal gene transfer [59]. Loss of gene function can allow a pathogen to be better adapted to a new niche [60]. ST69 was unusual in possessing an intact second type III secretion system (ETT2) [45] previously described in strains of enterohaemorrhagic and enteroadhesive E. coli, which we found intact in a complete operon. What contribution this might make to the pathogenesis of bacteraemia produced by this ST is not clear. That most other E. coli strains contain degenerate versions of the ETT2 [45] suggests it is ancestral in ST69. Genes that were present in isolates of one or two STs were more likely to have been acquired during evolution of the various strains.
Analysis of the accessory gene content of the different isolates gave a similar result as seen with analysis of the virulence genes. We had hypothesized that there might be a ‘common’ set of accessory genes present in our isolates that would be important in producing bacteraemia. This was not the case; no particular functional class of genes were found in the accessory genome. The accessory gene pool of E. coli is open, essentially unbounded. This strongly suggests that different STs must exist in different ecological niches, as they do not share significant accessory genome content. The origin of these pathogenic strains is most likely from strains carried within the human gut that then colonize the urinary tract and from there invade the bloodstream. The maintenance of distinct accessory genomes between the different STs could be explained by a number of different mechanisms. Colonization of the human gut from another source may be in evolutionary terms a too recent event for significant gene transfer between the different STs to have occurred. Alternatively, these pathogenic strains may be only transiently present in the gut microbiota and are replaced from time to time from another source. Studies of faecal carriage of extended spectrum β-lactamase-producing E. coli in patients who had a urinary infection with these pathogens show that they are cleared from the gut over a period of a few years [61]. Whether the same holds for non-extended spectrum β-lactamase producing STs is not clear. Hospitals might provide an environmental source for reinfection of patients, but the origin of community-acquired E. coli infections is not clear.
We used the whole genome sequences of the bacteraemic isolates to identify genetic determinants that are associated with CAI versus HAI. Chromosomal genes encoding the toxin/antitoxin pair chpB/chpS were strongly associated with HAIs. Toxin/antitoxin pairs were originally described in plasmids, where they act as ‘addiction mechanisms’ for plasmid maintenance. Continued expression of the labile antitoxin is required to ensure the more stable toxin is neutralized. Thus, plasmid loss results in unfettered expression of the more stable toxin and death of the cell, ensuring a strong selection pressure for plasmid maintenance. A number of chromosomal toxin/antitoxin pairs have also now been described. Such toxin/antitoxins are important in mediating the phenomenon of bacterial persistence, survival in the face of adverse environmental conditions such as antibiotic exposure [47, 62]. Stress results in protease destruction of the antitoxin molecule, resulting in unchecked toxin activity, which produces growth inhibition. When the stress is relieved, the bacteria recover and can resume their normal growth. This is not antibiotic resistance, but importantly has been found to be closely associated with the onset of true antibiotic resistance in laboratory experiments [63]. We speculate that the association of genes conferring antibiotic tolerance in HAI reflects the general burden of antibiotic use in this setting. Selection of these genes then will facilitate the subsequent generation and selection of ABRs in these isolates. Community-acquired strains showed an increase in genes encoding components of the type 6 secretory systems present in E. coli. Studies have shown that this system is a potent mediator of inter-species killing that allows the organism to compete more effectively with other bacteria to establish colonization [64]. This may be important to allow strains such as ST69 that are more frequently the cause of CAIs to establish colonization within the gut and/or urinary tract. Together with the data showing the genetic divergence of ST69 from the other STs that cause bacteraemias, this supports the view that ST69 occupies a distinct ecological niche and has evolved adaptations favouring its survival within the community and the ability to cause bacteraemia. We hypothesize that type VI secretion by ST69 may be an important factor allowing this successful adaptation and could potentially be exploited therapeutically.
Antibiotic resistance was highly prevalent in the isolates examined here. Our network analysis of resistance genes and phenotypic resistance of isolates revealed two main clusters. There was a strongly correlated cluster of genes conferring phenotypic resistance to typical ‘hospital’ antibiotics, such as third-generation cephalosporins, aminoglycosides and β-lactamase-resistant penicillins. This is strong evidence that within this setting, use of these antibiotics is generating cross-resistance to other unrelated antibiotics, most likely through selection of plasmids containing multiple antibiotic-resistance determinants. This phenomenon also is seen with the predominantly ‘community’ utilized antibiotics trimethoprim and amoxicillin. However, in these cases, the co-selected genes mediate resistance to aminoglycosides that are not currently used to any degree in Scotland (e.g. streptomycin). However, if, say, an integron mediating trimethoprim resistance were to acquire a resistance gene to a more commonly used antibiotic, then this could be strongly selected by trimethoprim use.
The CTXM-15 extended spectrum β-lactamase was found almost exclusively in ST131 isolates of clade C2, but was present in one isolate ST131 clade A and one isolate of ST405. This gene is widespread in isolates of ST131 and is of high clinical significance given that it confers resistance to the widely used 3rd generation cephalosporins cefotaxime and ceftazidime. CTX-M15 has also been reported in ST405 and other isolates [65]. The CTXM group of genes originated from chromosomal β-lactamases present in species of the microbial genus Kluyvera. The spread within ST131 occurred during the 1990s and it is now present worldwide [66], but its presence of this gene in ST405 and other STs suggests that it has the potential to become even more widespread.
Outcome of the patients studied here after the isolation of the infecting E. coli showed no significant dependence on ST. Patients with HAIs fared worse than those with infections acquired from the community; the excess mortality was in the first 90 days after infection suggesting a causal link. Patients who had a catheter also fared worse in the initial 90 days. However, it proved difficult to establish from patients’ notes exactly when catheters were placed. Since more severely ill patients may be catheterized by preference, the causal link with catheterization will require further study. In multivariate analysis, apart from catheterization, a respiratory source of infection was also a risk factor for death within 6 months. Previous studies have identified non-urinary source as a risk factor for death [67], and a comprehensive study in France showed a number of host factors and portal of entry were more important than bacterial determinants as predictors of death [68]. The association with a respiratory source has not previously been specifically identified.
In conclusion, this study has revealed the complex genetic patterns present in E. coli isolates from bacteraemic patients in the West of Scotland in 2013–5. We identify ST69 as an important contributor to the burden of disease, showing that it is predominantly a CAI with a distinct genetic make-up. Using a genome-wide association study, we have identified genes mediating antibiotic tolerance as important in HAI and elements of a type VI secretion system in CAI. The close correlation of resistance to multiple antibiotics that are largely used in hospitals, show how antibiotic use is driving selection of such resistance determinants, which subsequently can spread to community isolates. It is unlikely given the diverse mechanisms by which these strains of E. coli can cause disease that a single intervention will reduce the incidence of such infections. However, this study has identified a number of factors that could be targeted to reduce the burden of this disease.
Data bibliography
Scottish Healthcare Associated Infection Prevention Institute (SHAIPI). European Nucleotide Archive, PRJEB12513 (2017).
Funding information
The work was funded by the Scottish Executive via the Chief Scientist Office through the provision of a grant to establish the Scottish Healthcare Associated Infection Prevention Institute (SHAIPI). The funders had no role in the study design; the collection, analysis or interpretation of data; nor the writing of the manuscript.
Conflicts of interest
The authors declare there are no conflicts of interest.
Ethical statement
Advice was sought from the Local Research Ethics Committee of Greater Glasgow and Clyde NHS Board. Specific ethical permission was deemed not to be required as the study was viewed as service improvement. Approval for access to clinical patient data was given by the Caldicott Guardian of the relevant health boards, who is the designated regulator of confidential patient information within NHS Scotland.
Supplementary Data
Footnotes
Abbreviations: ABR, antibiotic-resistance gene; CAI, community-associated infection; HAI, hospital-associated infection; KEGG, Kyoto Encyclopedia of Genes and Genomes; ML, maximum likelihood; MLST, multilocus sequence type; NMDS, non-metric multidimensional scaling; SNP, single nucleotide polymorphism; ST, sequence type.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Eight supplementary figures are available with the online version of this article.
References
- 1.Poolman JT, Wacker M. Extraintestinal pathogenic Escherichia coli, a common human pathogen: challenges for vaccine development and progress in the field. J Infect Dis. 2016;213:6–13. doi: 10.1093/infdis/jiv429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dale AP, Woodford N. Extra-intestinal pathogenic Escherichia coli (ExPEC): disease, carriage and clones. J Infect. 2015;71:615–626. doi: 10.1016/j.jinf.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Laupland KB. Incidence of bloodstream infection: a review of population-based studies. Clin Microbiol Infect. 2013;19:492–500. doi: 10.1111/1469-0691.12144. [DOI] [PubMed] [Google Scholar]
- 4.de Kraker ME, Jarlier V, Monen JC, Heuer OE, van de Sande N, et al. The changing epidemiology of bacteraemias in Europe: trends from the European Antimicrobial Resistance Surveillance System. Clin Microbiol Infect. 2013;19:860–868. doi: 10.1111/1469-0691.12028. [DOI] [PubMed] [Google Scholar]
- 5.Søgaard M, Nørgaard M, Dethlefsen C, Schønheyder HC. Temporal changes in the incidence and 30-day mortality associated with bacteremia in hospitalized patients from 1992 through 2006: a population-based cohort study. Clin Infect Dis. 2011;52:61–69. doi: 10.1093/cid/ciq069. [DOI] [PubMed] [Google Scholar]
- 6.Abernethy JK, Johnson AP, Guy R, Hinton N, Sheridan EA, et al. Thirty day all-cause mortality in patients with Escherichia coli bacteraemia in England. Clin Microbiol Infect. 2015;21:251.e1–25251. doi: 10.1016/j.cmi.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Roubaud Baudron C, Panhard X, Clermont O, Mentré F, Fantin B, et al. Escherichia coli bacteraemia in adults: age-related differences in clinical and bacteriological characteristics, and outcome. Epidemiol Infect. 2014;142:2672–2683. doi: 10.1017/S0950268814000211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thelwall S, Nsonwu N, Bhattacharya A, Wasti S, Gerver S, et al. Annual Epidemiological Commentary: Mandatory MRSA, MSSA and E. coli Bacteraemia and C. difficile Infection Data 2016/17. London:: Public Health England; 2017. document no. 2017158. [Google Scholar]
- 9.Health Protection Scotland . Scottish National Point Prevalence Survey of Healthcare Associated Infection and Antimicrobial Prescribing 2016. Glasgow:: Health Protection Scotland; 2017. [Google Scholar]
- 10.Health Protection Scotland . Healthcare Associated Infections 2016. Glasgow:: Health Protection Scotland; 2017. [Google Scholar]
- 11.Abernethy J, Guy R, Sheridan EA, Hopkins S, Kiernan M, et al. Epidemiology of Escherichia coli bacteraemia in England: results of an enhanced sentinel surveillance programme. J Hosp Infect. 2017;95:365–375. doi: 10.1016/j.jhin.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 12.Schlackow I, Stoesser N, Walker AS, Crook DW, Peto TE, et al. Increasing incidence of Escherichia coli bacteraemia is driven by an increase in antibiotic-resistant isolates: electronic database study in Oxfordshire 1999– 2011. J Antimicrob Chemother. 2012;67:1514–1524. doi: 10.1093/jac/dks082. [DOI] [PubMed] [Google Scholar]
- 13.Day MJ, Doumith M, Abernethy J, Hope R, Reynolds R, et al. Population structure of Escherichia coli causing bacteraemia in the UK and Ireland between 2001 and 2010. J Antimicrob Chemother. 2016;71:2139–2142. doi: 10.1093/jac/dkw145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horner C, Fawley W, Morris K, Parnell P, Denton M, et al. Escherichia coli bacteraemia: 2 years of prospective regional surveillance (2010–12) J Antimicrob Chemother. 2014;69:91–100. doi: 10.1093/jac/dkt333. [DOI] [PubMed] [Google Scholar]
- 15.Banerjee R, Johnston B, Lohse C, Chattopadhyay S, Tchesnokova V, et al. The clonal distribution and diversity of extraintestinal Escherichia coli isolates vary according to patient characteristics. Antimicrob Agents Chemother. 2013;57:5912–5917. doi: 10.1128/AAC.01065-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adams-Sapper S, Diep BA, Perdreau-Remington F, Riley LW. Clonal composition and community clustering of drug-susceptible and -resistant Escherichia coli isolates from bloodstream infections. Antimicrob Agents Chemother. 2013;57:490–497. doi: 10.1128/AAC.01025-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibreel TM, Dodgson AR, Cheesbrough J, Fox AJ, Bolton FJ, et al. Population structure, virulence potential and antibiotic susceptibility of uropathogenic Escherichia coli from Northwest England. J Antimicrob Chemother. 2012;67:346–356. doi: 10.1093/jac/dkr451. [DOI] [PubMed] [Google Scholar]
- 18.Riley LW. Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin Microbiol Infect. 2014;20:380–390. doi: 10.1111/1469-0691.12646. [DOI] [PubMed] [Google Scholar]
- 19.Nicolas-Chanoine MH, Bertrand X, Madec JY. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev. 2014;27:543–574. doi: 10.1128/CMR.00125-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kallonen T, Brodrick HJ, Harris SR, Corander J, Brown NM, et al. Systematic longitudinal survey of invasive Escherichia coli in England demonstrates a stable population structure only transiently disturbed by the emergence of ST131. Genome Res. 2017;;27:1437–1449. doi: 10.1101/gr.216606.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inouye M, Dashnow H, Raven LA, Schultz MB, Pope BJ, et al. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014;6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koboldt DC, Larson DE, Wilson RK. Using VarScan 2 for germline variant calling and somatic mutation detection. Curr Protoc Bioinformatics. 2013;44:11–17. doi: 10.1002/0471250953.bi1504s44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stamatakis A. Using RAxML to infer phylogenies. Curr Protoc Bioinformatics. 2015;51:11–14. doi: 10.1002/0471250953.bi0614s51. [DOI] [PubMed] [Google Scholar]
- 26.Page AJ, de Silva N, Hunt M, Quail MA, Parkhill J, et al. Robust high-throughput prokaryote de novo assembly and improvement pipeline for Illumina data. Microb Genom. 2016;2:e000083. doi: 10.1099/mgen.0.000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joensen KG, Scheutz F, Lund O, Hasman H, Kaas RS, et al. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol. 2014;52:1501–1510. doi: 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brynildsrud O, Bohlin J, Scheffer L, Eldholm V. Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol. 2016;17:238. doi: 10.1186/s13059-016-1108-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hunt M, Mather AE, Sánchez-Busó L, Page AJ, Parkhill J, et al. ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb Genom. 2017;3:e000131. doi: 10.1099/mgen.0.000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price LB, Johnson JR, Aziz M, Clabots C, Johnston B, et al. The epidemic of extended-spectrum-β-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. mBio. 2013;4:e00377-13. doi: 10.1128/mBio.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petty NK, Ben Zakour NL, Stanton-Cook M, Skippington E, Totsika M, et al. Global dissemination of a multidrug resistant Escherichia coli clone. Proc Natl Acad Sci USA. 2014;111:5694–5699. doi: 10.1073/pnas.1322678111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ben Zakour NL, Alsheikh-Hussain AS, Ashcroft MM, Khanh Nhu NT, Roberts LW, et al. Sequential acquisition of virulence and fluoroquinolone resistance has shaped the evolution of Escherichia coli ST131. mBio. 2016;7:e00347. doi: 10.1128/mBio.00347-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tettelin H, Riley D, Cattuto C, Medini D. Comparative genomics: the bacterial pan-genome. Curr Opin Microbiol. 2008;11:472–477. doi: 10.1016/j.mib.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 37.Croucher NJ, Coupland PG, Stevenson AE, Callendrello A, Bentley SD, et al. Diversification of bacterial genome content through distinct mechanisms over different timescales. Nat Commun. 2014;5:5471. doi: 10.1038/ncomms6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teufel R, Mascaraque V, Ismail W, Voss M, Perera J, et al. Bacterial phenylalanine and phenylacetate catabolic pathway revealed. Proc Natl Acad Sci USA. 2010;107:14390–14395. doi: 10.1073/pnas.1005399107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen L, Zheng D, Liu B, Yang J, Jin Q. VFDB 2016: hierarchical and refined dataset for big data analysis - 10 years on. Nucleic Acids Res. 2016;44:D694–D697. doi: 10.1093/nar/gkv1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skyberg JA, Johnson TJ, Johnson JR, Clabots C, Logue CM, et al. Acquisition of avian pathogenic Escherichia coli plasmids by a commensal E. coli isolate enhances its abilities to kill chicken embryos, grow in human urine, and colonize the murine kidney. Infect Immun. 2006;74:6287–6292. doi: 10.1128/IAI.00363-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tseng CC, Wu JJ, Liu HL, Sung JM, Huang JJ. Roles of host and bacterial virulence factors in the development of upper urinary tract infection caused by Escherichia coli. Am J Kidney Dis. 2002;39:744–752. doi: 10.1053/ajkd.2002.32992. [DOI] [PubMed] [Google Scholar]
- 42.Spaulding CN, Hultgren SJ. Adhesive pili in UTI pathogenesis and drug development. Pathogens. 2016;5:30. doi: 10.3390/pathogens5010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wiles TJ, Kulesus RR, Mulvey MA. Origins and virulence mechanisms of uropathogenic Escherichia coli. Exp Mol Pathol. 2008;85:11–19. doi: 10.1016/j.yexmp.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dautin N. Serine protease autotransporters of enterobacteriaceae (SPATEs): biogenesis and function. Toxins. 2010;2:1179–1206. doi: 10.3390/toxins2061179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ren CP, Chaudhuri RR, Fivian A, Bailey CM, Antonio M, et al. The ETT2 gene cluster, encoding a second type III secretion system from Escherichia coli, is present in the majority of strains but has undergone widespread mutational attrition. J Bacteriol. 2004;186:3547–3560. doi: 10.1128/JB.186.11.3547-3560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ristow LC, Welch RA. Hemolysin of uropathogenic Escherichia coli: a cloak or a dagger? Biochim Biophys Acta. 2016;1858:538–545. doi: 10.1016/j.bbamem.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 47.Page R, Peti W. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat Chem Biol. 2016;12:208–214. doi: 10.1038/nchembio.2044. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Y, Zhu L, Zhang J, Inouye M. Characterization of ChpBK, an mRNA interferase from Escherichia coli. J Biol Chem. 2005;280:26080–26088. doi: 10.1074/jbc.M502050200. [DOI] [PubMed] [Google Scholar]
- 49.Maisonneuve E, Shakespeare LJ, Jørgensen MG, Gerdes K. Bacterial persistence by RNA endonucleases. Proc Natl Acad Sci USA. 2011;108:13206–13211. doi: 10.1073/pnas.1100186108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Andersen C, Rak B, Benz R. The gene bglH present in the bgl operon of Escherichia coli, responsible for uptake and fermentation of beta-glucosides encodes for a carbohydrate-specific outer membrane porin. Mol Microbiol. 1999;31:499–510. doi: 10.1046/j.1365-2958.1999.01191.x. [DOI] [PubMed] [Google Scholar]
- 51.Christensen-Dalsgaard M, Jørgensen MG, Gerdes K. Three new RelE-homologous mRNA interferases of Escherichia coli differentially induced by environmental stresses. Mol Microbiol. 2010;75:333–348. doi: 10.1111/j.1365-2958.2009.06969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cambray G, Guerout AM, Mazel D. Integrons. Annu Rev Genet. 2010;44:141–166. doi: 10.1146/annurev-genet-102209-163504. [DOI] [PubMed] [Google Scholar]
- 53.Mathers AJ, Peirano G, Pitout JD. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev. 2015;28:565–591. doi: 10.1128/CMR.00116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bajaj P, Singh NS, Virdi JS. Escherichia coli β-lactamases: what really matters. Front Microbiol. 2016;7:417. doi: 10.3389/fmicb.2016.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramirez MS, Tolmasky ME. Aminoglycoside modifying enzymes. Drug Resist Updat. 2010;13:151–171. doi: 10.1016/j.drup.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gestal AM, Stokes HW, Partridge SR, Hall RM. Recombination between the dfrA12-orfF-aadA2 cassette array and an aadA1 gene cassette creates a hybrid cassette, aadA8b. Antimicrob Agents Chemother. 2005;49:4771–4774. doi: 10.1128/AAC.49.11.4771-4774.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shimada T, Yamamoto K, Ishihama A. Involvement of the leucine response transcription factor LeuO in regulation of the genes for sulfa drug efflux. J Bacteriol. 2009;191:4562–4571. doi: 10.1128/JB.00108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Picard B, Garcia JS, Gouriou S, Duriez P, Brahimi N, et al. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect Immun. 1999;67:546–553. doi: 10.1128/iai.67.2.546-553.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wirth T, Falush D, Lan R, Colles F, Mensa P, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 2006;60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maurelli AT. Black holes, antivirulence genes, and gene inactivation in the evolution of bacterial pathogens. FEMS Microbiol Lett. 2007;267:1–8. doi: 10.1111/j.1574-6968.2006.00526.x. [DOI] [PubMed] [Google Scholar]
- 61.Jørgensen SB, Søraas A, Sundsfjord A, Liestøl K, Leegaard TM, et al. Fecal carriage of extended spectrum β-lactamase producing Escherichia coli and Klebsiella pneumoniae after urinary tract infection - a three year prospective cohort study. PLoS One. 2017;12:e0173510. doi: 10.1371/journal.pone.0173510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lewis K. Persister cells. Annu Rev Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 63.Levin-Reisman I, Ronin I, Gefen O, Braniss I, Shoresh N, et al. Antibiotic tolerance facilitates the evolution of resistance. Science. 2017;355:826–830. doi: 10.1126/science.aaj2191. [DOI] [PubMed] [Google Scholar]
- 64.Ho BT, Dong TG, Mekalanos JJ. A view to a kill: the bacterial type VI secretion system. Cell Host Microbe. 2014;15:9–21. doi: 10.1016/j.chom.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Coque TM, Novais A, Carattoli A, Poirel L, Pitout J, et al. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum β-lactamase CTX-M-15. Emerg Infect Dis. 2008;14:195–200. doi: 10.3201/eid1402.070350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cantón R, González-Alba JM, Galán JC. CTX-M enzymes: origin and diffusion. Front Microbiol. 2012;3:110. doi: 10.3389/fmicb.2012.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Laupland KB, Gregson DB, Church DL, Ross T, Pitout JD. Incidence, risk factors and outcomes of Escherichia coli bloodstream infections in a large Canadian region. Clin Microbiol Infect. 2008;14:1041–1047. doi: 10.1111/j.1469-0691.2008.02089.x. [DOI] [PubMed] [Google Scholar]
- 68.Lefort A, Panhard X, Clermont O, Woerther PL, Branger C, et al. Host factors and portal of entry outweigh bacterial determinants to predict the severity of Escherichia coli bacteremia. J Clin Microbiol. 2011;49:777–783. doi: 10.1128/JCM.01902-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.