Abstract
The genus Lactobacillus encompasses a diversity of species that occur widely in nature and encode a plethora of metabolic pathways reflecting their adaptation to various ecological niches, including humans, animals, plants and food products. Accordingly, their functional attributes have been exploited industrially and several strains are commonly formulated as probiotics or starter cultures in the food industry. Although divergent evolutionary processes have yielded the acquisition and evolution of specialized functionalities, all Lactobacillus species share a small set of core metabolic properties, including the glycolysis pathway. Thus, the sequences of glycolytic enzymes afford a means to establish phylogenetic groups with the potential to discern species that are too closely related from a 16S rRNA standpoint. Here, we identified and extracted glycolysis enzyme sequences from 52 species, and carried out individual and concatenated phylogenetic analyses. We show that a glycolysis-based phylogenetic tree can robustly segregate lactobacilli into distinct clusters and discern very closely related species. We also compare and contrast evolutionary patterns with genome-wide features and transcriptomic patterns, reflecting genomic drift trends. Overall, results suggest that glycolytic enzymes provide valuable phylogenetic insights and may constitute practical targets for evolutionary studies.
Keywords: Lactobacillus, phylogeny, glycolysis, evolution
Data Summary
RNA sequencing data has been deposited at the National Center for Biotechnology Information, BioProject PRJNA420353.
Impact Statement.
Though 16S rRNA-based phylogeny methods have been broadly used, they have a limited ability to precisely ascribe genus species across the prokaryotic branch of the tree of life. In this study, we have shown that using glycolysis enzyme sequences for phylogenetic analyses can be applied to the diverse genus Lactobacillus, and is able to consistently unravel phylogenetic groups and precisely ascertain relatedness, even between species nearly identical on the classical ribosomal tree. Because of their universal presence and greater diversity compared to 16S rRNA sequences, we posit that these sequences could be valuable markers in future phylogenetic and microbiome studies, specifically by providing connections to the other major branches, and enabling increased resolution. This can also be used to help identify unknown and un-culturable species, as the glycolysis enzymes are widespread, variable and allow for greater discriminatory power. Importantly, variability within some of the hypervariable regions within glycolytic sequences can also provide discrimination within a species. Looking forward, expanding this analysis to other genera and phylogenetic branches could open new avenues for evolutionary studies, and for investigating the phylogeny, composition and diversity of microbial populations in complex microbiomes.
Introduction
Genome adaptation is an important feature for speciation, and evolutionary processes balance various adaptive techniques for optimal growth and survival. At the genome level, adaptation features may include gene synteny conservation, G+C mol% drift, as well as codon bias optimization [1–3]. A working balance of these and other forces enable an organism to become uniquely adapted to its niche, and build up competitive advantages in shifting environmental conditions, or overcome predators and competitors. Such unique adaptations are the basis of phylogenetic studies and allow researchers various degrees of discrimination. At the genus and species levels, additions and deletions of genes can be used to define the pan- and core-genome and genome architecture can be used to evaluate synteny [4]. At the strain level, nucleotide polymorphisms afford the highest resolution opportunities, with the ability to compare and contrast nearly identical isolates and even clonal relatives [5, 6].
For prokaryotic species, various tools and methodologies have been used to compare and contrast genomes, but the challenges are often genus- or species-specific, and approaches can vary depending on the desired resolution and encompassed genetic diversity [7]. In some cases where within genus diversity is extensive, such as in bifidobacteria and lactobacilli, using canonical housekeeping genes or universal markers (i.e. 16S rRNA) has proven difficult or limited [8–11]. Also, there has yet to be defined a consistent set of genes to be utilized for multilocus sequence typing studies. Indeed, while universally conserved 16S rRNA sequences afford opportunities for metagenomic analyses, their shortcomings and biases are increasingly under scrutiny [12–14].
For some genera, it has become obvious that the 16S rRNA resolution limit has been met and a new set of criteria must be established. One such genus is Lactobacillus. Belonging to the lactic acid bacteria (LAB) group, this genus is composed of over 150 Gram-positive, low G+C species [15, 16]. Lactobacilli have been used as starter cultures in the food industry for decades, and by humankind for millennia, and as such have been labelled generally regarded as safe (GRAS) and benefit from the qualified presumption of safety (QPS) [17]. Food-related studies have led to the assertion that some strains in select species are to be considered probiotic (‘live microorganisms which when administered in adequate amounts confer a health benefit on the host’) [18] and, as such, are now predominantly featured in dairy foods and widely formulated in probiotic dietary supplements [19]. Recently, the advent of microbiome studies has revealed that microbial populations are more numerous, diverse and variable than originally thought [20, 21]. With both qualitative and quantitative considerations, associations and sometimes even correlations have been established between members of the microbiome and host health, though the accuracy and precision with which bacteria are identified vary widely and are not universally satisfactory. One such instance concerns the genus Lactobacillus, which has been established as an important colonizer of the human gastrointestinal tract [22]. Additional research is thus needed in this area, as researchers better grasp the role of this genus in health and disease [23–28]. Some lactobacilli are already being exploited, for example, as a tool to deliver vaccines [29]. Arguably, we are far from exhausting all the possible uses of this functional genus. However, in order to be able to fully utilize the numerous functions of Lactobacillus, we must first establish a method that enables us to properly identify and relate the many diverse species within this genus. While 16S rRNA sequencing has gotten us this far, it has a limited ability to distinguish between closely related species and represent overall genomic content and reflect genome-wide trends. These shortcomings are certainly not unique to Lactobacillus, and with the ever-increasing expansion of our understanding of the microbial world [30], there is a need to identify 16S rRNA-independent genomic features that capture diversity on a more granular level. Thus, it is imperative that a standard method be developed that allows the proper identification of species. In order to achieve this, we assessed the potential of the widespread glycolysis pathway enzyme sequences to inform phylogeny.
In this paper, we applied a previously described method of phylogenetic analysis using the classical glycolysis enzymes as phylogenetic markers [31] to a diverse set of Lactobacillus species in order to establish its effect on a complicated genus. Though previous studies had used glycolysis as an expansion of ribosomal trees [32], we determined how a broad glycolysis-based phylogeny compares to the ribosomal tree. Specifically, previous studies have applied glycolysis-based approaches to LAB in order to define an evolutionary pathway. By adding data from the entirety of the glycolysis and pentose phosphate pathways, Salvetti et al. [32] were able to apply phenotypic data to explain the branching of the LAB tree, as well as highlight some areas of misclassification in the 16S rRNA tree [32]. Here, we propose using the entirety of the canonical glycolysis pathway as a replacement phylogenetic marker for the 16S rRNA. Conveniently, the glycolysis pathway, much like the 16S rRNA, is universally present, at least partially, conserved, and constitutes a set of suitable candidates for phylogenetic analyses [33, 34]. Here, we demonstrate that this method can assign phylogenetic relationships consistent with what is known from the 16S rRNA marker, though at a much higher discriminatory power. Specifically, we compared sequence-based alignment trees of a representative set of lactobacilli using 16S rRNA- and glycolysis-based approaches. We also analysed the occurrence and location, expression, and G+C mol% of each glycolysis gene. The location and transcriptional profiles confirm that these genes are conserved and highly transcribed with varying levels of drift.
Methods
Genomes
We selected 52 diverse species and subspecies of Lactobacillus for analysis, sampled across and throughout the 16S rRNA and core- and pan-genome tree (Table 1). We ensured this set was representative of this paraphyletic genus and included species from various niches, as previously established [16]. The genomes were mined using Geneious version 9.0.5 [35] to identify the classical glycolysis genes in each species (Figs S1 and S2, available with the online version of this article). Four reference genomes were used to make a curated database for the glycolysis genes, namely Lactobacillus acidophilus, Lactobacillus gasseri, Lactobacillus reuteri and Lactobacillus rhamnosus. The Annotate from Database feature was used to annotate the other genomes. To validate the glycolysis annotations, especially in the case of multiple hits, a combination of blast, get_homologues and mRNA-Seq (mRNA sequencing) data was used [36, 37]. The 16S rRNA sequences were extracted from the genomes and blast was used to validate any cases where there were multiple hits. Once annotated and curated, the genes were extracted from the genome. The glycolysis genes were then translated and confirmed by ExPASy [38]. For the concatenated tree, the amino acid sequences were joined together in order of their presence in the glycolysis pathway (Fig. S1).
Table 1. Species and genomes list.
This shows the representative set of 52 Lactobacillus species and sub-species used in this study. Accession numbers and naming conventions are included.
| Genus | Species | Subspecies | Strain | Accession no. | Naming convention | Locus tag |
|---|---|---|---|---|---|---|
| Lactobacillus | acidipiscis | KCTC 13900 | NZ_BACS00000000 | L_acidipiscis | GSS | |
| Lactobacillus | acidophilus | NCFM | NC_006814 | L_acidophilus | LBA | |
| Lactobacillus | algidus | DSM 15638 | NZ_AZDI00000000 | L_algidus | FC66 | |
| Lactobacillus | amylolyticus | DSM 11664 | NZ_ADNY00000000 | L_amylolyticus | HMPREF0493 | |
| Lactobacillus | amylovorus | GRL1118 | NC_017470 | L_amylovorus | LAB52 | |
| Lactobacillus | animalis | DSM 20602 | NZ_AEOF00000000 | L_animalis | LACAN | |
| Lactobacillus | aquaticus | DSM 21051 | NZ_AYZD00000000 | L_aquaticus | FC19 | |
| Lactobacillus | brevis | ATCC 367 | NC_008497 | L_brevis | LVIS | |
| Lactobacillus | buchneri | CD034 | NC_018610 | L_buchneri | LBUCD034 | |
| Lactobacillus | cacaonum | DSM 21116 | NZ_AYZE00000000 | L_cacaonum | FC80 | |
| Lactobacillus | casei | DSM 20011 | NZ_AZCO00000000 | L_casei | FC13 | |
| Lactobacillus | coryniformis | torquens | DSM 20004 | NZ_AEOS00000000 | L_coryniformis_t | EWE |
| Lactobacillus | crispatus | ST1 | NC_014106 | L_crispatus | LCRIS | |
| Lactobacillus | curvatus | CRL 705 | NZ_AGBU00000000 | L_curvatus | CRL705 | |
| Lactobacillus | delbrueckii | bulgaricus | ATCC BAA-365 | NC_008529 | L_delbrueckii_b | LBUL |
| Lactobacillus | farciminis | DSM 20184 | NZ_AEOT00000000 | L_farciminis | LACFC | |
| Lactobacillus | fermentum | CECT 5716 | NC_017465 | L_fermentum | LC40 | |
| Lactobacillus | floricola | DSM 23037 | NZ_AYZL00000000 | L_floricola | FC86 | |
| Lactobacillus | gallinarum | DSM 10532 | NZ_BALB00000000 | L_gallinarum | JCM2011 | |
| Lactobacillus | gasseri | ATCC 33323 | NC_008530 | L_gasseri | LGAS | |
| Lactobacillus | helveticus | CNRZ32 | NC_021744 | L_helveticus | LHE | |
| Lactobacillus | hilgardii | DSM 20176 | NZ_ACGP00000000 | L_hilgardii | HMPREF0519 | |
| Lactobacillus | hominis | DSM 23910 | NZ_CAKE00000000 | L_hominis | BN55 | |
| Lactobacillus | iners | DSM 13335 | NZ_ACLN00000000 | L_iners | HMPREF0520 | |
| Lactobacillus | jensenii | DSM 20557 | NZ_AYYU00000000 | L_jensenii | FC45 | |
| Lactobacillus | johnsonii | NCC 533 | NC_005362 | L_johnsonii | LJ | |
| Lactobacillus | kimchicus | JCM_15530 | NZ_AZCX00000000 | L_kimchicus | FC96 | |
| Lactobacillus | lindneri | DSM 20690 | NZ_JQBT00000000 | L_lindneri | IV52 | |
| Lactobacillus | mali | DSM 20444 | NZ_AKKT00000000 | L_mali | LMA | |
| Lactobacillus | mindensis | DSM 14500 | NZ_AZEZ00000000 | L_mindensis | FD29 | |
| Lactobacillus | mucosae | LM1 | NZ_CP011013 | L_mucosae | LBLM1 | |
| Lactobacillus | nasuensis | JCM_17158 | NZ_AZDJ00000000 | L_nasuensis | FD02 | |
| Lactobacillus | oeni | DSM 19972 | NZ_AZEH00000000 | L_oeni | FD46 | |
| Lactobacillus | oris | F0423 | NZ_AFTL00000000 | L_oris | HMPREF9102 | |
| Lactobacillus | otakiensis | DSM 19908 | NZ_BASH00000000 | L_otakiensis | LOT | |
| Lactobacillus | parabuchneri | DSM 5707 | NZ_AZGK00000000 | L_parabuchneri | FC51 | |
| Lactobacillus | paracasei | N1115 | NZ_CP007122 | L_paracasei | AF91 | |
| Lactobacillus | pasteurii | DSM 23907 | NZ_CAKD00000000 | L_pasteurii | BN53 | |
| Lactobacillus | pentosus | DSM 20314 | NZ_AZCU00000000 | L_pentosus | FD24 | |
| Lactobacillus | plantarum | 16 | NC_021514 | L_plantarum | LP16 | |
| Lactobacillus | reuteri | DSM 20016 | NC_009513 | L_reuteri | LREU | |
| Lactobacillus | rhamnosus | GG | NC_013198 | L_rhamnosus | LGG | |
| Lactobacillus | rossiae | DSM 15814 | NZ_AZFF00000000 | L_rossiae | FD35 | |
| Lactobacillus | ruminis | ATCC 27782 | NC_015975 | L_ruminis | LRC | |
| Lactobacillus | sakei | sakei | DSM 20017 | NZ_BALW00000000 | L_sakei_s | JCM1157 |
| Lactobacillus | salivarius | CECT 5713 | NC_017481 | L_salivarius | CECT 5713 | |
| Lactobacillus | sanfranciscensis | TMW 1.1304 | NC_015978 | L_sanfranciscensis | LSA | |
| Lactobacillus | suebicus | DSM 5007 | NZ_BACO00000000 | L_suebicus | GSK | |
| Lactobacillus | sunkii | DSM 19904 | NZ_AZEA00000000 | L_sunkii | FD17 | |
| Lactobacillus | vaginalis | DSM 5837 | NZ_ACGV00000000 | L_vaginalis | HMPREF0549 | |
| Lactobacillus | versmoldensis | DSM 14857 | NZ_BACR00000000 | L_versmoldensis | GSQ | |
| Lactobacillus | zymae | DSM 19395 | NZ_AZDW00000000 | L_zymae | FD38 |
Transcriptional profiles of glycolysis genes
We analysed RNA transcription profiles from mRNA-Seq data for six species (L. acidophilus, Lactobacillus amylovorus, Lactobacillus crispatus, Lactobacillus delbrueckii subsp. bulgaricus, L. gasseri, and Lactobacillus helveticus) with the previously published isolation method, mRNA sequencing and analyses [39]. Briefly, we used mRNA-Seq data generated in our laboratory to determine the boundaries and quantitative amounts of RNA transcripts for glycolysis genes as previously described. Samples were grown to mid-log phase and flash-frozen. Single-read RNA sequencing was performed on the extracted RNA using an Illumina HiSeq 2500. Data was then quality assessed, trimmed, filtered and mapped on the reference genomes. Presumably, levels of constitutive transcription reflect biological relevance in the tested conditions and transcript boundaries inform on co-transcribed functional pairs.
Alignments and trees
Alignments and trees were generated using a previously described methodology [31]. Briefly, once curated sequences were extracted, we aligned the sequences using clustalw (IUB, gap penalty of 15, gap extension of 6.66), muscle (eight iterations), Geneious [global alignment with free end gaps, cost matrix was BLOSUM62 (amino acids) or 65 % similarity (nucleotide)] and mafft [algorithm was auto, scoring matrix was BLOSUM62 and BLOSUM80 (amino acids) or 100PAM and 200PAM (nucleotide), gap penalty of 1.53, offset 0.123], then used trimAl (compareset and automated1) to find a consistent alignment [35, 40–43]. Trees were then generated using RaxML [CAT BLOSUM62 (amino acids) or CAT GTR (nucleotide), Bootstrap using rapid hill climbing with random seed 1, replicates were 100] [44]. A consensus tree was then established using a 50 % threshold level.
R analyses
Statistical analyses were performed using R version 3.2.2. [45]. R was used to create plots, graphs and quantitative data. Statistical tests used included a two-tailed t-test for comparing G+C contents. Default settings were used to preform statistical analyses and assess quantitative distributions.
Results
16S rRNA phylogeny
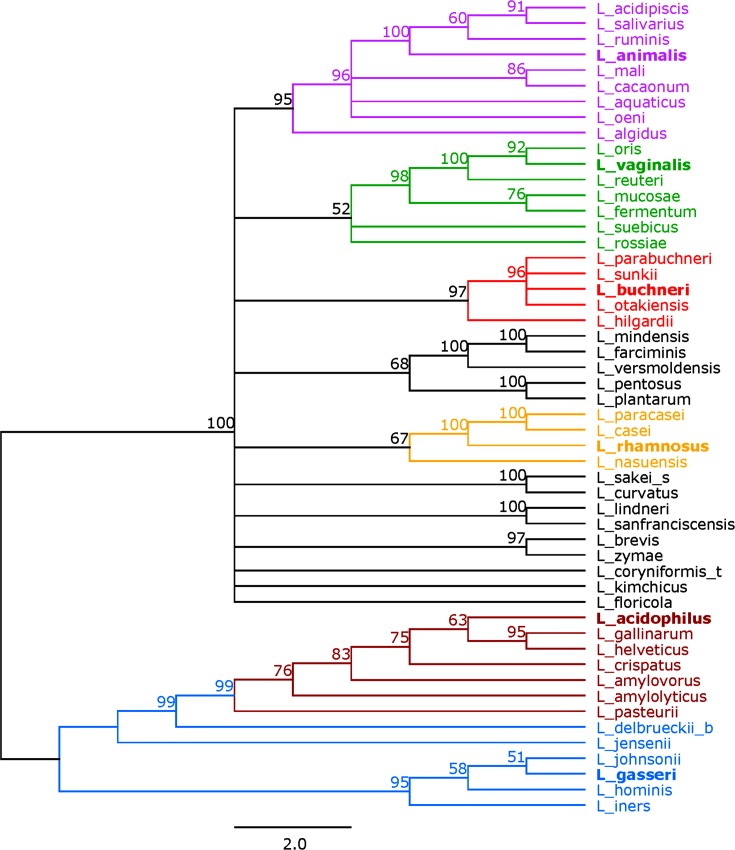
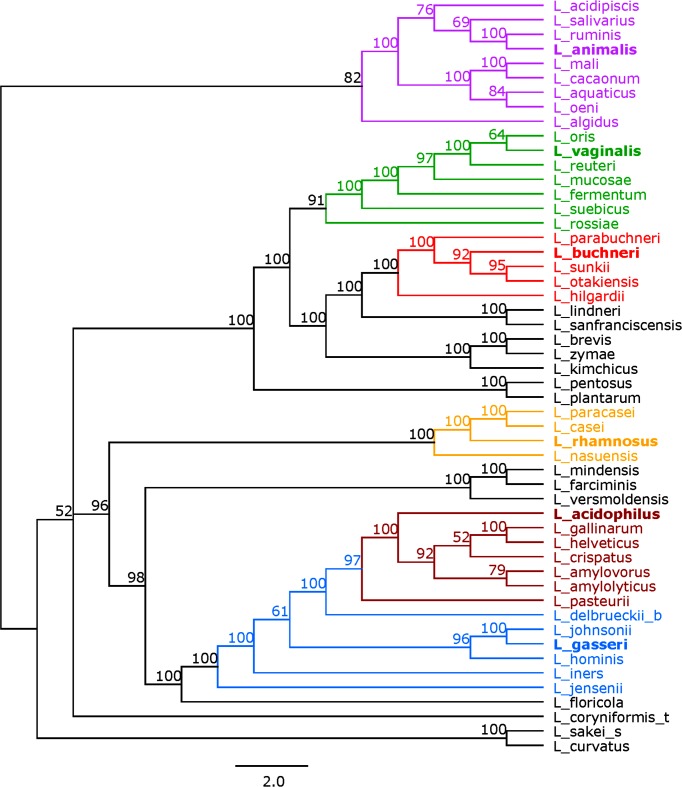
We first generated a 16S rRNA-based tree to use as a reference for our subsequent analyses. A phylogenetic tree based on the alignment of the 16S rRNA sequences from a representative set of 52 species and sub-species of Lactobacillus is depicted in Fig. 1. Six phylogenetic groups were identified based on their branching: the Lactobacillus animalis group, the Lactobacillus vaginalis group, the Lactobacillus buchneri group, the L. rhamnosus group, the L. acidophilus group and the L. gasseri group. These groupings are consistent with historically established relationships, as well as recent core-genome analyses [16, 46]. Some of these groups also encompass species that have been historically associated with distinct niches and points of isolation (i.e. mucosal vs intestinal vs dairy origins) [16]. The groups ranged in size from four to nine genomes with the L. rhamnosus group as the smallest and the L. animalis group as the largest. The bootstrap values for the 16S rRNA tree ranged from 51 to 100. There were 27 nodes that had a bootstrap of 70 or greater (Fig. S3). We used these six phylogenetic groups as references for our subsequent analyses, though some species were not assigned to one of these six groups.
Fig. 1.
16S rRNA tree. Tree based on the alignment of the 16S rRNA sequences using RaxML. Bootstrap values are recorded on the nodes. Groups are coloured as follows: the L. animalis group in purple, the L. vaginalis group in green, the L. buchneri group in red, the L. rhamnosus group in yellow, the L. acidophilus group in maroon, and the L. gasseri group in blue. The representative species in each group is in bold. Species names follow the naming convention shown in Table 1.
Glycolysis gene expression
Before using the glycolysis enzymes as phylogenetic markers, we first explored their genetic properties in Lactobacillus. Of the 52 Lactobacillus species and sub-species selected, 35 species encoded all ten of the classical glycolytic genes. In contrast, 16 species (encompassing the L. vaginalis and L. buchneri groups) presented eight of the canonical genes (missing pfk and fba) (Fig. S2). In such cases, alternative metabolic pathways may be utilized, such as the pentose phosphate pathway (Lactobacillus fermentum) or the phosphoketolase pathway (L. buchneri) [47, 48]. L. reuteri uses a mixture of the Embden–Meyerhof pathway and phosphoketolase pathway and, thus, was the only species with six of the glycolysis genes (Fig. S2) [49].
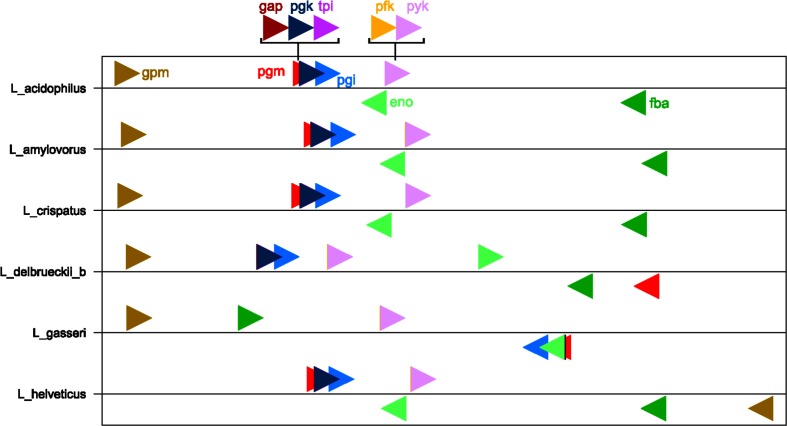
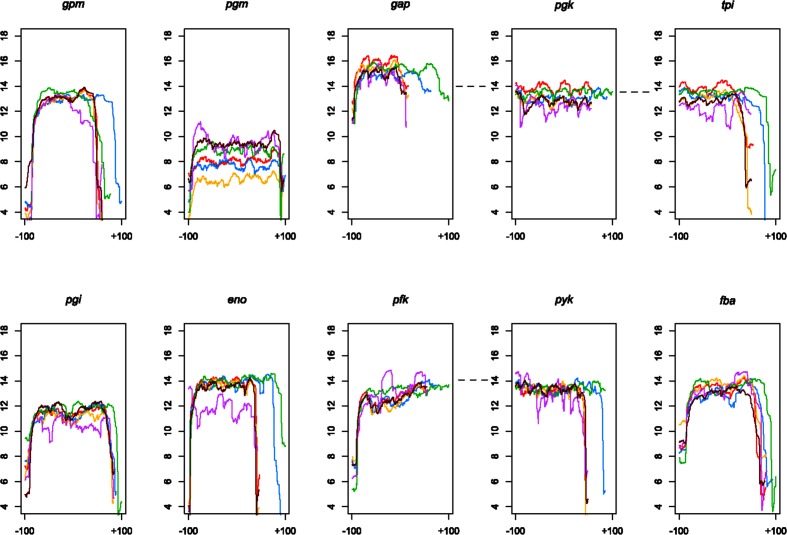
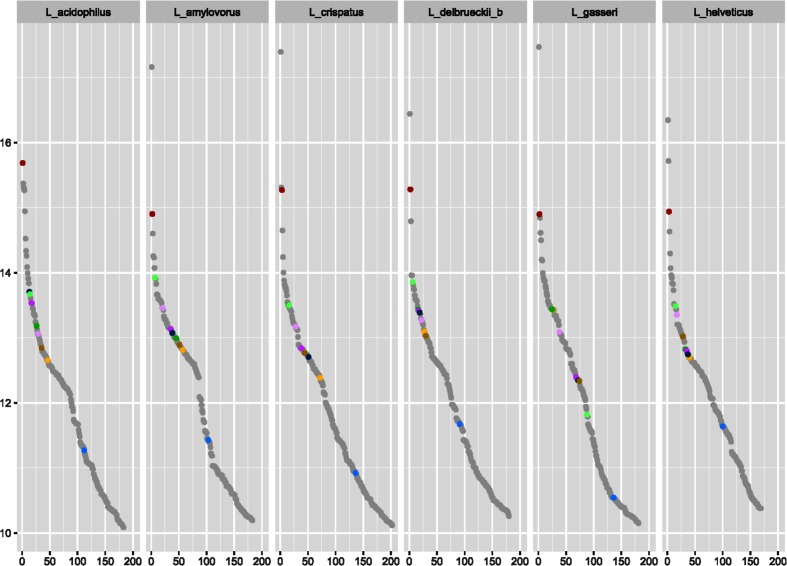
Next, we characterized the transcripts of glycolysis genes in Lactobacillus. Chromosome location and mRNA sequence data were analysed from six species: L. acidophilus, L. amylovorus, L. crispatus, L. delbrueckii subsp. bulgaricus, L. gasseri and L. helveticus. These six species fall into the L. acidophilus and L. gasseri groups, and all six species contain the complete complement of glycolysis genes, allowing for inferences on all of the genes in this study, instead of just a subset. Fig. 2 depicts the location of the glycolysis genes on normalized chromosomes for each of these six species. It is noteworthy that two operons can be visualized: the gap, pgk and tpi operon, as well as the pfk and pyk operon. Furthermore, the operon boundaries are clearly seen in the mRNA coverage data for each of the six species (Fig. 3). The remaining five genes have clear start and stop boundaries. Notably, L. helveticus has a unique arrangement of the glycolysis genes compared to the other five species, possibly due to the large number of IS elements leading to genome decay; however, the operons remain conserved [50]. Next, we compared the expression levels of the glycolysis genes to the whole transcriptome. We found that the glycolysis genes are among the most highly expressed genes. Indeed, considering the top 10 % of the most highly expressed genes in the cell, nine of the ten glycolysis genes are listed (Fig. 4). The only gene absent from the top 10 % is pgm. Strikingly, the gap gene is consistently among the top three most highly expressed genes in all six species. Such a consistently high transcription level indicates that the gap gene is critical to the functionality of the cell and perhaps, as such, less susceptible to changes. This is also reflected by the conserved location of gap in the genome and operon structure amongst the strains studied (Fig. 2), potentially indicating uses for gap in identification. These results demonstrate that glycolysis genes are genomically conserved, organizationally syntenous and transcriptionally important, showcasing their use as potential phylogenetic markers.
Fig. 2.
Genomic location. Normalized glycolysis gene locations in L. acidophilus, L. amylovorus, L. crispatus, L. delbrueckii subsp. bulgaricus, L. gasseri and L. helveticus. Normalization was calculated by dividing the location on the genome by the total genome size. Right arrows indicate forward direction, left reverse direction. The genomes are organized in the 5′ to 3′ direction. Colours are as follows: pgm in red, pgi in blue, pfk in yellow, fba in dark green, tpi in purple, gap in maroon, pgk in navy, gpm in mustard, eno in light green and pyk in lavender.
Fig. 3.
Glycolysis genes transcription. Each plot represents the mRNA-Seq coverage, log2 transformed, for the corresponding glycolysis gene over its length; ±100 represents the number of bases away from the start/end of the gene. The species are plotted as follows: L. acidophilus is red, L. amylovorus in blue, L. crispatus in yellow, L. delbrueckii subsp. bulgaricus in green, L. gasseri in purple and L. helveticus in maroon.
Fig. 4.
Ranked order of mRNA expression. Top 10 % most highly expressed genes in L. acidophilus, L. amylovorus, L. crispatus, L. delbrueckii subsp. bulgaricus, L. gasseri and L. helveticus. Data is represented as a log2 transformed RPKM (Reads Per Kilobase of transcript, per Million mapped reads). Transcripts are ranked from most abundant to least abundant. Glycolysis genes are coloured as follows: pgm in red, pgi in blue, pfk in yellow, fba in dark green, tpi in purple, gap in maroon, pgk in navy, gpm in mustard, eno in light green and pyk in lavender.
Glycolysis-based phylogeny
To create a glycolysis-based phylogeny for the 52 selected Lactobacillus species and subspecies, the concatenated amino acid sequences of the glycolysis enzymes were used (Fig. 5). The enzymes were concatenated in their order of occurrence in the glycolysis pathway (Fig. S1). For organisms with all enzymes present, this meant ten sequences were concatenated together, whereas only six to eight amino acid sequences were concatenated for the other species (Fig. S2). The six phylogenetic groups identified from the 16S rRNA reference tree, namely L. animalis, L. vaginalis, L. buchneri, L. rhamnosus, L. acidophilus and L. gasseri, were also identified in the concatenated tree and follow the same clustering (colouring) scheme. The bootstrap values for the concatenated tree ranged from 52 to 100. Nodes with bootstrap values equal to or greater than 70 numbered 43, a 59 % increase from that of the 16S rRNA tree. Overall, the concatenated tree correctly assigned the phylogenetic groups established from the 16S rRNA tree. In addition, the concatenated tree better discerned how the phylogenetic groups relate to one another, even within groups. This is supported by the higher bootstrap values (Fig. S3). Trees based on the individual glycolysis enzymes can be found in Figs S4–S13. The sum of branch lengths for each tree can be found in Table S1. A detailed comparative analysis of various trees structures revealed that overall there is high congruence in clustering both between and within the six established groups, though with various levels of discrimination across each protein sequence. Repeatedly, glycolysis-based trees provided more discriminatory power than the 16S rRNA tree.
Fig. 5.
Concatenated glycolysis tree. Tree based on the alignment of concatenated amino acid sequences of glycolysis enzymes using RaxML. Bootstrap values are recorded on the nodes. Groups are coloured as follows: the L. animalis group in purple, the L. vaginalis group in green, L. buchneri group in red, the L. rhamnosus group in yellow, the L. acidophilus group in maroon, and the L. gasseri group in blue. The representative species in each group is in bold. Species name follows the naming convention shown in Table 1.
G+C content analyses
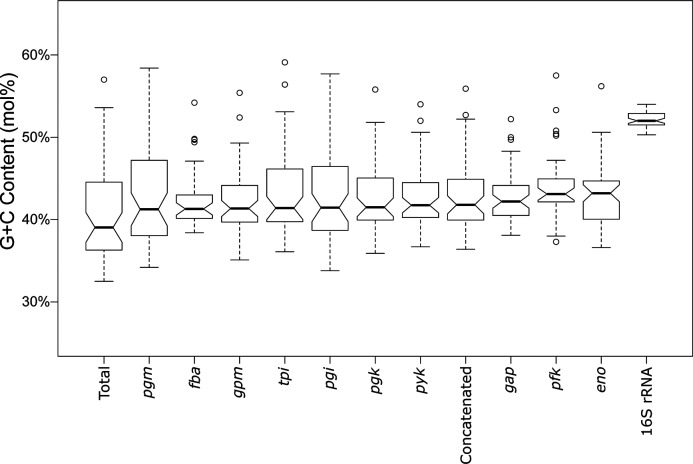
Next, we looked at the G+C mol% and genomic drift of the glycolysis genes across the various species. Fig. 6 shows notched boxplots comparing the G+C mol% of each sequence set (the 16S rRNA sequence, the 10 genes and the concatenated sequences) in this study, compared to the genome-wide G+C mol%, ranked in increasing order. The G+C mol% of the pgm gene is closest to that of the total genome, while the 16S rRNA gene is the farthest. The notches are indicative of strong evidence that the medians differ when the notches do not overlap [51]. The 16S rRNA gene does not overlap with any other gene. In fact, a two-tailed t-test with a P value less than 0.001 (2.2×10−16) revealed that the G+C mol% of the 16S rRNA sequence was statistically distinct from that of the total genome G+C mol%. This indicates that the 16S rRNA gene is not matching the pace of drift of the total genome with regards to G+C mol%. In contrast, all of the glycolysis genes, with the exception of pfk and eno, were not statistically different from the total genome G+C mol% (P value greater than 0.01), indicating that G+C mol% drift for glycolysis genes provide insights into the genome-wide G+C mol% drift. This further supports glycolytic sequences as intriguing candidates for both phylogenetic studies and representatives of genome-wide trends.
Fig. 6.
G+C mol% analysis of Lactobacillus glycolysis genes. Depicted are notched boxplots of G+C mol% for each glycolysis gene, concatenated genes, 16S rRNA and total genome. Genes are placed in order of increasing median. If two notches do not overlap, it is an indication of strong evidence for differing medians.
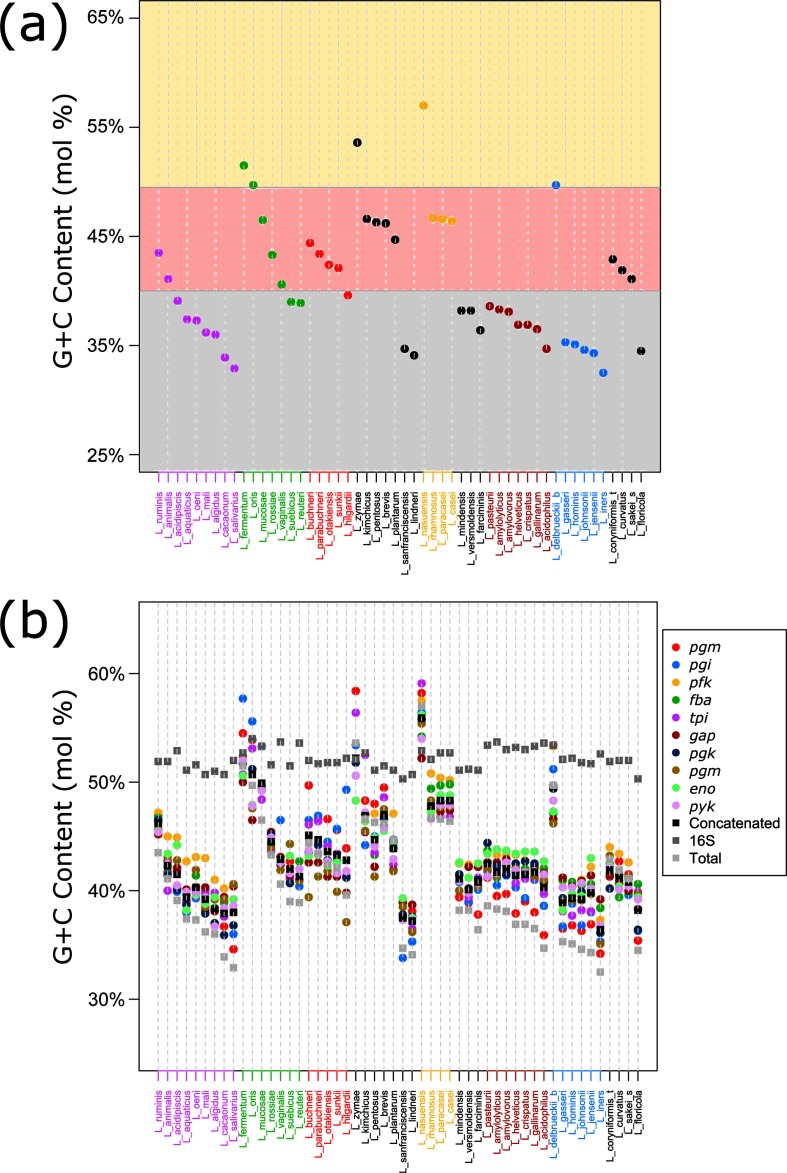
The genome sizes in this study ranged from 1.28 Mb (Lactobacillus iners) to 3.65 Mb (Lactobacillus pentosus), again reflecting the extensive genomic diversity within this genus. The total G+C mol% ranged from 32.50 % (L. iners) to 57.00 % (Lactobacillus nasuensis), which is intriguing given the general assumption that all lactobacilli are low G+C mol% organisms. Nevertheless, the mean G+C mol% was 40.70 %, consistent with Lactobacillus being generally perceived as low G+C mol% organisms. Splitting the species into high, medium and low categories, it becomes apparent that most species are trending towards the lower end of the spectrum, and away from the higher G+C mol% range (Fig. 7a). Some of the phylogenetic groups are closely clustered, such as the L. acidophilus group, L. gasseri group and the L. rhamnosus group, with the exception of L. delbrueckii subsp. bulgaricus (a dairy bacterium) and L. nasuensis (an aforementioned ex in G+C mol%). The L. animalis group and L. buchneri group are similarly clustered, albeit more loosely. These observations hold true when comparing the G+C mol% of all the individual genes in their respective genomes, perhaps reflecting a consistent and genome-wide pace of drift, rather than variable speeds of drift for each gene (Fig. 7b). Again, the 16S rRNA sequence has a much higher G+C mol% than most of the other studied genes, with the outlier L. nasuensis deviating from the consensus. The G+C mol% of the glycolysis genes within clusters are often times very close, as exemplified by the L. acidophilus group.
Fig. 7.
G+C mol% analysis of Lactobacillus genomes. (a) shows the total G+C mol% for each species. Species are coloured according to their phylogenetic group. (b) shows the G+C mol% of the glycolysis genes, the concatenated glycolysis genes, the 16S rRNA and total G+C mol% for each species. Species are named according to Table 1.
Discussion
The genomic and functional attributes of Lactobacillus render it a pervasive genus, both in research and in industry. The benefits and uses of this diverse set of species are well-established and exhaustive, and yet, the list continues to grow. Many Lactobacillus strains are now considered to be health-promoting in the form of probiotics and are often found to be a part of a healthy microbiome [26]. They are also being engineered to promote healthy host–microbe interactions and deliver bioactive compounds such as vaccines [52]. As microbiome studies expand, we anticipate that the interest in Lactobacillus is set to increase, especially given their occurrence in several human-associated microbiomes, encompassing intestinal, vaginal, oral and skin communities [21]. Many studies have been published discussing the role of Lactobacillus in the microbiota, including research into the microbiota changes through disease, enhancing the microbiome as a form of treatment, and how the microbiome reacts to drugs [53–55]. The continuously expanding list of uses and studies just illustrates how important it is to accurately identify Lactobacillus species. While all species of Lactobacillus share some classical features of LAB organisms, notably their ability to produce lactic acid, the similarities between species are relatively few. In fact, even basic characteristics such as niche and isolation source can vary radically. Proper identification is an increasing concern especially when it comes to disease modelling in the human microbiome, as well as the formulation, tracking and efficacy of probiotic strains. Innovative techniques are continuously being developed and often use a combination of 16S rRNA with developing technologies, such as MALDI-TOF [56]. However, these tools are not broadly accessible and still rely partially on the sometimes unsatisfactory 16S rRNA. Here, we provide a practical alternative to the classical use of 16S rRNA sequencing.
In this paper, we applied the previously proposed methodology of using glycolysis sequences to perform phylogenetic studies [31] in the genus Lactobacillus. We demonstrated that this method is a practical and robust approach for Lactobacillus. Compared to the traditional 16S rRNA method, this approach was able to consistently identify phylogenetic groupings, with notably high-resolution between closely related species. While the 16S rRNA-based tree was able to identify the six phylogenetic groups, the concatenated tree was able to add more discrimination both between and within groups, evidenced by the higher bootstrap values in the glycolysis-based tree. Our grouping is consistent with a previous study using glycolysis sequences for phylogenetic analysis of Lactobacillus species [32]. Further analyses based on genomic content revealed clues as to why the glycolysis-based tree was better able to assign species.
First, looking at the organization of the genes in the genomes revealed two conserved operons in Lactobacillus, the gap operon and the pfk operon, with the remaining enzymes showing clear start and stop boundaries. This shared synteny emphasizes the importance of glycolysis gene conservation. Next, we looked at expression level. The glycolysis genes were consistently among the most highly expressed genes in the cell, with the gap gene always in the top three most abundant transcripts. These high expression levels indicate a great use and energy expenditure and, thus, arguably reflect the biological importance of this gene to the cell. Because of this importance, the glycolysis genes are much less likely to be subjected to loss. The operon structures and expression levels of the glycolysis genes are significant because a main criterion for selecting the 16S rRNA as a phylogenetic marker was its high conservation among species [57]. Next, we looked at how the glycolysis genes reflected genomic drift in terms of G+C mol%. First, it would appear that the genus is reaching a stabilizing point in its G+C mol% drift, though some species with high G+C mol% still have margin for extending the trend (L. nasuensis, Lactobacillus zymae, and L. fermentum). Next, we saw that the glycolysis gene G+C mol% was extremely close to that of the genome-wide G+C mol%, while the 16S rRNA was startlingly higher (P<0.001), underscoring the fact that the 16S rRNA is by all accounts much different than that of the total genome, whereas the majority of the glycolysis genes are significantly similar to the total genome G+C mol% (Fig. 6). This provides a possible explanation for the reason why the 16S rRNA analyses have been limited at a high-resolution level in Lactobacillus and why the glycolysis-based tree was able to reach a higher-resolution level. In fact, it has long been noted that 16S rRNA is unable to discriminate between species of lactobacilli due to its high similarity amongst them [58]. The individual glycolysis genes are much more similar to the genome as a whole (Fig. 6). Additionally, individual glycolysis genes are also able to accurately assign species to groups with a high resolution (Figs S4–S13). The gap gene is of particular note, due to its presence in an operon, consistently high expression, G+C mol% and ability to accurately define species groups. Overall, the glycolysis-based approach was able to provide a highe-resolution phylogeny for Lactobacillus, due in part to its conservation, expression and reflection of genomic drift.
Funding information
This study was supported by start-up funds from North Carolina State University (Raleigh, USA). K. B. is a recipient of a National Institute of Environmental Health Sciences (NIEHS) training grant.
Acknowledgements
The authors thank the funding sources for their support and the CRISPR lab for insightful conversations.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Supplementary Data
Footnotes
Abbreviations: eno, enolase-encoding gene; fba, fructose bisphosphate aldolase-encoding gene; gap, glyceraldehyde 3-phosphate dehydrogenase-encoding gene; gpm, phosphoglycerate mutase-encoding gene; LAB, lactic acid bacteria; mRNA-Seq, mRNA sequencing; pfk, 6-phosphofructokinase-encoding gene; pgi, glucose-6-phosphate isomerase-encoding gene; pgk, phosphoglycerate kinase-encoding gene; pgm, phosphoglucomutase-encoding gene; pyk, pyruvate kinase-encoding gene; tpi, triosephosphate isomerase-encoding gene.
All supporting data, code and protocols have been provided within the article or through supplementary data files. One supplementary table and thirteen supplementary figures are available with the online version of this article.
References
- 1.Dandekar T, Snel B, Huynen M, Bork P. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem Sci. 1998;23:324–328. doi: 10.1016/S0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- 2.Karlin S, Campbell AM, Mrázek J. Comparative DNA analysis across diverse genomes. Annu Rev Genet. 1998;32:185–225. doi: 10.1146/annurev.genet.32.1.185. [DOI] [PubMed] [Google Scholar]
- 3.Karlin S, Mrázek J, Campbell A, Kaiser D. Characterizations of highly expressed genes of four fast-growing bacteria. J Bacteriol. 2001;183:5025–5040. doi: 10.1128/JB.183.17.5025-5040.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boekhorst J, Siezen RJ, Zwahlen MC, Vilanova D, Pridmore RD, et al. The complete genomes of Lactobacillus plantarum and Lactobacillus johnsonii reveal extensive differences in chromosome organization and gene content. Microbiology. 2004;150:3601–3611. doi: 10.1099/mic.0.27392-0. [DOI] [PubMed] [Google Scholar]
- 5.Hoffmann M, Zhao S, Pettengill J, Luo Y, Monday SR, et al. Comparative genomic analysis and virulence differences in closely related Salmonella enterica serotype eidelberg isolates from humans, retail meats, and animals. Genome Biol Evol. 2014;6:1046–1068. doi: 10.1093/gbe/evu079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Losada PM, Tümmler B. SNP synteny analysis of Staphylococcus aureus and Pseudomonas aeruginosa population genomics. FEMS Microbiol Lett. 2016;363:fnw229-fnw. doi: 10.1093/femsle/fnw229. [DOI] [PubMed] [Google Scholar]
- 7.Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, et al. Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci USA. 2006;103:15611–15616. doi: 10.1073/pnas.0607117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Claesson MJ, van Sinderen D, O'Toole PW. Lactobacillus phylogenomics-towards a reclassification of the genus. Int J Syst Evol Microbiol. 2008;58:2945–2954. doi: 10.1099/ijs.0.65848-0. [DOI] [PubMed] [Google Scholar]
- 9.Felis GE, Dellaglio F, Mizzi L, Torriani S. Comparative sequence analysis of a recA gene fragment brings new evidence for a change in the taxonomy of the Lactobacillus casei group. Int J Syst Evol Microbiol. 2001;51:2113–2117. doi: 10.1099/00207713-51-6-2113. [DOI] [PubMed] [Google Scholar]
- 10.Milani C, Turroni F, Duranti S, Lugli GA, Mancabelli L, et al. Genomics of the genus Bifidobacterium reveals species-specific adaptation to the glycan-rich gut environment. Appl Environ Microbiol. 2016;82:980–991. doi: 10.1128/AEM.03500-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milani C, Lugli GA, Turroni F, Mancabelli L, Duranti S, et al. Evaluation of bifidobacterial community composition in the human gut by means of a targeted amplicon sequencing (ITS) protocol. FEMS Microbiol Ecol. 2014;90:493–503. doi: 10.1111/1574-6941.12410. [DOI] [PubMed] [Google Scholar]
- 12.Baker GC, Smith JJ, Cowan DA. Review and re-analysis of domain-specific 16S primers. J Microbiol Methods. 2003;55:541–555. doi: 10.1016/j.mimet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 13.Clarridge JE. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev. 2004;17:840–862. doi: 10.1128/CMR.17.4.840-862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de La Cuesta-Zuluaga J, Escobar JS. Considerations for optimizing microbiome analysis using a marker gene. Front Nutr. 2016;3:26. doi: 10.3389/fnut.2016.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salvetti E, Torriani S, Felis GE. The genus Lactobacillus: a taxonomic update. Probiotics Antimicrob Proteins. 2012;4:217–226. doi: 10.1007/s12602-012-9117-8. [DOI] [PubMed] [Google Scholar]
- 16.Sun Z, Harris HM, McCann A, Guo C, Argimón S, et al. Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat Commun. 2015;6:8322. doi: 10.1038/ncomms9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernardeau M, Vernoux JP, Henri-Dubernet S, Guéguen M. Safety assessment of dairy microorganisms: the Lactobacillus genus. Int J Food Microbiol. 2008;126:278–285. doi: 10.1016/j.ijfoodmicro.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 18.Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11:506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 19.Saxelin M. Probiotic formulations and applications, the current probiotics market, and changes in the marketplace: a European perspective. Clin Infect Dis. 2008;46:S76–S79. doi: 10.1086/523337. [DOI] [PubMed] [Google Scholar]
- 20.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conlon M, Bird A. The impact of diet and lifestyle on gut microbiota and human health. Nutrients. 2015;7:17–44. doi: 10.3390/nu7010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, Wang N, Yin B, Fang D, Jiang T, et al. Effects of Lactobacillus plantarum CCFM0236 on hyperglycaemia and insulin resistance in high-fat and streptozotocin-induced type 2 diabetic mice. J Appl Microbiol. 2016;121:1727–1736. doi: 10.1111/jam.13276. [DOI] [PubMed] [Google Scholar]
- 24.Feng XB, Jiang J, Li M, Wang G, You JW, et al. Role of intestinal flora imbalance in pathogenesis of pouchitis. Asian Pac J Trop Med. 2016;9:786–790. doi: 10.1016/j.apjtm.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 25.Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- 26.Gerritsen J, Smidt H, Rijkers GT, de Vos WM. Intestinal microbiota in human health and disease: the impact of probiotics. Genes Nutr. 2011;6:209–240. doi: 10.1007/s12263-011-0229-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shreiner AB, Kao JY, Young VB. The gut microbiome in health and in disease. Curr Opin Gastroenterol. 2015;31:69–75. doi: 10.1097/MOG.0000000000000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okai S, Usui F, Yokota S, Hori-I Y, Hasegawa M, et al. High-affinity monoclonal IgA regulates gut microbiota and prevents colitis in mice. Nat Microbiol. 2016;1:16103. doi: 10.1038/nmicrobiol.2016.103. [DOI] [PubMed] [Google Scholar]
- 29.O'Flaherty S, Klaenhammer TR. Multivalent chromosomal expression of the Clostridium botulinum serotype a neurotoxin heavy-chain antigen and the Bacillus anthracis protective antigen in Lactobacillus acidophilus. Appl Environ Microbiol. 2016;82:6091–6101. doi: 10.1128/AEM.01533-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, et al. A new view of the tree of life. Nat Microbiol. 2016;1:16048. doi: 10.1038/nmicrobiol.2016.48. [DOI] [PubMed] [Google Scholar]
- 31.Brandt K, Barrangou R. Phylogenetic analysis of the Bifidobacterium genus using glycolysis enzyme sequences. Front Microbiol. 2016;7:00657. doi: 10.3389/fmicb.2016.00657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salvetti E, Fondi M, Fani R, Torriani S, Felis GE. Evolution of lactic acid bacteria in the order Lactobacillales as depicted by analysis of glycolysis and pentose phosphate pathways. Syst Appl Microbiol. 2013;36:291–305. doi: 10.1016/j.syapm.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 33.Fothergill-Gilmore LA. The evolution of the glycolytic pathway. Trends Biochem Sci. 1986;11:47–51. doi: 10.1016/0968-0004(86)90233-1. [DOI] [Google Scholar]
- 34.Fothergill-Gilmore LA, Michels PA. Evolution of glycolysis. Prog Biophys Mol Biol. 1993;59:105–235. doi: 10.1016/0079-6107(93)90001-Z. [DOI] [PubMed] [Google Scholar]
- 35.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 37.Contreras-Moreira B, Vinuesa P. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microbiol. 2013;79:7696–7701. doi: 10.1128/AEM.02411-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, et al. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson BR, Hymes J, Sanozky-Dawes R, Henriksen ED, Barrangou R, et al. Conserved S-layer-associated proteins revealed by exoproteomic survey of S-layer-forming lactobacilli. Appl Environ Microbiol. 2016;82:134–145. doi: 10.1128/AEM.01968-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 43.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Core Team R. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2015. [Google Scholar]
- 46.Canchaya C, Claesson MJ, Fitzgerald GF, van Sinderen D, O'Toole PW. Diversity of the genus Lactobacillus revealed by comparative genomics of five species. Microbiology. 2006;152:3185–3196. doi: 10.1099/mic.0.29140-0. [DOI] [PubMed] [Google Scholar]
- 47.Heinl S, Wibberg D, Eikmeyer F, Szczepanowski R, Blom J, et al. Insights into the completely annotated genome of Lactobacillus buchneri CD034, a strain isolated from stable grass silage. J Biotechnol. 2012;161:153–166. doi: 10.1016/j.jbiotec.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 48.Cárdenas N, Laiño JE, Delgado S, Jiménez E, Juárez del Valle M, et al. Relationships between the genome and some phenotypical properties of Lactobacillus fermentum CECT 5716, a probiotic strain isolated from human milk. Appl Microbiol Biotechnol. 2015;99:4343–4353. doi: 10.1007/s00253-015-6429-0. [DOI] [PubMed] [Google Scholar]
- 49.Arsköld E, Lohmeier-Vogel E, Cao R, Roos S, Rådström P, et al. Phosphoketolase pathway dominates in Lactobacillus reuteri ATCC 55730 containing dual pathways for glycolysis. J Bacteriol. 2008;190:206–212. doi: 10.1128/JB.01227-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Broadbent JR, Hughes JE, Welker DL, Tompkins TA, Steele JL. Complete genome sequence for Lactobacillus helveticus CNRZ 32, an industrial cheese starter and cheese flavor adjunct. Genome Announc. 2013;1:e00590-13. doi: 10.1128/genomeA.00590-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chambers JM. GraphicalMethods for Data Analysis. Belmont, CA: Wadsworth International Group; 1983. Notched box plots; pp. 60–63. [Google Scholar]
- 52.Seegers JF. Lactobacilli as live vaccine delivery vectors: progress and prospects. Trends Biotechnol. 2002;20:508–515. doi: 10.1016/S0167-7799(02)02075-9. [DOI] [PubMed] [Google Scholar]
- 53.Bhat M, Arendt BM, Bhat V, Renner EL, Humar A, et al. Implication of the intestinal microbiome in complications of cirrhosis. World J Hepatol. 2016;8:1128–1136. doi: 10.4254/wjh.v8.i27.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bull-Otterson L, Feng W, Kirpich I, Wang Y, Qin X, et al. Metagenomic analyses of alcohol induced pathogenic alterations in the intestinal microbiome and the effect of Lactobacillus rhamnosus GG treatment. PLoS One. 2013;8:e53028. doi: 10.1371/journal.pone.0053028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shin CM, Kim N, Kim YS, Nam RH, Park JH, et al. Impact of Long-term proton pump inhibitor therapy on gut microbiota in F344 rats: pilot study. Gut Liver. 2016;10:896–901. doi: 10.5009/gnl15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Foschi C, Laghi L, Parolin C, Giordani B, Compri M, et al. Novel approaches for the taxonomic and metabolic characterization of lactobacilli: integration of 16S rRNA gene sequencing with MALDI-TOF MS and 1H-NMR. PLoS One. 2017;12:e0172483. doi: 10.1371/journal.pone.0172483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eisen JA. The RecA protein as a model molecule for molecular systematic studies of bacteria: comparison of trees of RecAs and 16S rRNAs from the same species. J Mol Evol. 1995;41:1105–1123. doi: 10.1007/BF00173192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fox GE, Wisotzkey JD, Jurtshuk P. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.