Abstract
Bifidobacterium is a diverse genus of anaerobic, saccharolytic bacteria that colonize many animals, notably humans and other mammals. The presence of these bacteria in the gastrointestinal tract represents a potential coevolution between the gut microbiome and its mammalian host mediated by diet. To study the relationship between bifidobacterial gut symbionts and host nutrition, we analyzed the genome of two bifidobacteria strains isolated from the feces of a common marmoset (Callithrix jacchus), a primate species studied for its ability to subsist on host-indigestible carbohydrates. Whole genome sequencing identified these isolates as unique strains of Bifidobacterium callitrichos. All three strains, including these isolates and the previously described type strain, contain genes that may enable utilization of marmoset dietary substrates. These include genes predicted to contribute to galactose, arabinose, and trehalose metabolic pathways. In addition, significant genomic differences between strains suggest that bifidobacteria possess distinct roles in carbohydrate metabolism within the same host. Thus, bifidobacteria utilize dietary components specific to their host, both humans and non-human primates alike. Comparative genomics suggests conservation of possible coevolutionary relationships within the primate clade.
Keywords: bifidobacteria, comparative genomics, commensalism, gut microbiota, non-human primate
Data Summary
1. Genome sequence data from isolates UMA51804 and UMA51805 are archived in the DDBJ/ENA/GenBank database (http://www.ncbi.nlm.nih.gov/genbank/) under accession numbers NWTW00000000 and NWTX00000000.
2. The genome sequence data from B. callitrichos JCM 17296T used in this study are available in the GenBank database (http://www.ncbi.nlm.nih.gov/genbank/) under accession number GCA_000741175.1.
Impact Statement.
Microbial commensals colonize mammalian gastrointestinal tracts to assemble into communities known as microbiomes. Bacterial commensals contribute to host physiological and homeostatic operations. It has been postulated that microbial communities coevolved with their hosts, with various degrees of scientific evidence in support of this. Accordingly, bifidobacteria colonize nursing infants early in life due in part to their ability to subsist on human milk oligosaccharides. Bifidobacteria are also members of adult gut microbiomes, albeit to a lesser extent. Herein we describe a comparative genomic analysis of three Bifidobacterium callitrichos strains isolated from common marmoset feces. This was performed to investigate the coevolution of gut commensals with their host mediated by dietary components. In this instance, the common marmoset subsists on oligosaccharide-rich tree gums, and thus the genomic analysis focused on carbohydrate metabolism. Moreover, we tested genomics-derived hypotheses by verifying carbohydrate-related phenotypes. This included evaluating the capacity to utilize milk oligosaccharides as a sole carbohydrate source. This is significant as this trait is shared by select bifidobacterial species that colonize human infants. Interestingly, all three B. callitrichos strains exhibited the milk oligosaccharide utilization phenotype. This investigation provides a foundation for studies of the evolutionary relationship between mother’s milk, the infant, and its gut microbiome.
Introduction
The genus Bifidobacterium contains anaerobic, non-spore-forming, rod-shaped bacteria that present a bifurcated, termed ‘bifid’, morphology under certain growth conditions in some species [1]. Bifidobacteria are commonly found in animal gastrointestinal tracts including mammals, birds, and insects [2–5]. These bacteria have been extensively characterized regarding their ability to metabolize host-indigestible carbohydrates, which often enables their persistence within the gut environment [6–8]. Previous genomic analyses identified genomic signatures underlying oligosaccharide utilization by bifidobacteria [9–11]. It is noteworthy that bifidobacteria associated with different human stages of development have divergent metabolic capabilities. This includes the infant-colonizing Bifidobacterium longum subsp. infantis that metabolizes indigestible oligosaccharides secreted in human milk. This rare phenotype has been linked to a 40 kb gene cluster dedicated to this function [12–14].
The gut microbiome is postulated to have coevolved simultaneously with its host gastrointestinal environment modulated by intrinsic and extrinsic factors such as diet [15–17]. Thus, the study of extant primates provides an opportunity to analyze potential coevolutionary relationships between hosts and microbes. In recent years, there has been an increase in the isolation and characterization of novel bifidobacterial species from non-human primate feces. Since 2012, at least 17 novel species of bifidobacteria have been described from non-human primates, with five from the common marmoset (Callithrix jacchus) feces [18–27]. Marmosets provide an interesting model to study the evolution of diet with gut microbiota, as they are one of the few mammals that subsist on indigestible oligosaccharides found in tree gums or hardened saps [28–32]. This subsistence strategy is relatively rare among mammals, with a very small fraction of mammalian species able to consume plant gums to some extent (n=94), but common among primates (n=78) [33]. The common marmoset is one of 27 mammalian species considered to be obligate exudivores [33]. The exudatory diet of common marmosets provides a rich source of plant β-linked polysaccharides, consisting largely of galactose, arabinose, and rhamnose [34–37].
Bifidobacterium callitrichos JCM 17296T is a facultative anaerobic gastrointestinal symbiont of the common marmoset first isolated in 2012 [19]. A phylogenetic analysis of the family Bifidobacteriaceae using 404 clusters of orthologous groups (COGs) of proteins inferred that B. callitrichos JCM 17296T lies within a clade that includes human-associated Bifidobacterium breve LMG 13208, Bifidobacterium angulatum LMG 11039, Bifidobacterium longum subsp. longum LMG 13197, Bifidobacterium longum subsp. infantis ATCC 15697 and Bifidobacterium longum subsp. suis LMG 21814, and several non-human primate-associated species [22]. Furthermore, a comparative genomic survey of the genus Bifidobacterium noted that the genome of B. callitrichos JCM 17296T is larger than the expected average and may employ metabolic pathways lost from other bifidobacteria. The present study provides a comparative genomic and phenotypic analysis of B. callitrichos JCM 17296T and two B. callitrichos strains recently isolated from the feces of a captive common marmoset.
Methods
Identification of microbial community members within source fecal sample
Total DNA from the marmoset fecal sample was isolated using the MO BIO PowerSoil DNA Isolation Kit as per the manufacturer’s instructions (Qiagen). The sequencing library was prepared with the extracted DNA using the Illumina 16S rRNA Metagenomic Sequencing Library Preparation protocol. Sequencing was performed on an Illumina MiSeq device using the 600-cycle MiSeq V3 reagent kit. Reads underwent quality filtering and were analyzed using the QIIME 1 pipeline [38–42].
Bacterial isolation from fecal specimens
A fecal sample was collected from a 6-year-old female marmoset housed at the University of Massachusetts Amherst in Amherst, MA, USA. The animals were cared for in accordance with the guidelines published in the Guide for the Care and Use of Laboratory Animals, 8th edition. The study was approved by the UMass Institutional Animal Care and Use Committee. The fresh fecal sample was mixed with 5 ml of sterile peptone water and spread onto bifidobacteria-specific medium (BSM) agar, which consists of de Mann Rogosa Sharpe (MRS) agar (Difco), 0.05 % (w/v) l-cysteine (Sigma Aldrich) and 0.05 % (w/v) mupirocin (AppliChem Panreac) [43]. Agar plates were incubated at 37 °C for 24 h in a Coy anaerobic chamber maintained with a gas mix of 7 % H2, 10 % CO2, and N2 to balance (Coy Laboratory Products). Individual colonies were selected and grown in BSM broth for 12 h under the same conditions. Liquid cultures were preserved as freezer stocks at −80 °C in a 25 % (v/v) glycerol solution. Isolates were initially screened via colony PCR with bifidobacteria-specific primers (Bif164f: GGGTGGTAATGCCGGATG, Bif662r: CCACCGTTACACCGGGAA) amplifying a 550 bp fragment of the 16S rRNA [44–46]. PCR products were purified using the QIAquick PCR Purification Kit (Qiagen) and sequenced by Genewiz using Sanger DNA Sequencing (Genewiz). The resulting sequences were used to infer phylogenetic relationships in mega7 using the EzBioCloud 16S rRNA gene sequence database to provide an initial screen [47, 48]. Bifidobacterial isolates were further confirmed using the fructose 6-phosphate phosphoketolase assay. This colorimetric assay determines the presence of a unique phosphoketolase involved in the bifidobacterial-specific fermentative pathway, or ‘bifid shunt’ [49–51].
Whole genome sequencing of bacterial isolates
Genomic DNA was extracted using the MasterPure Gram Positive DNA Purification Kit [Epicentre (an Illumina Company)] and then further processed using the Genomic DNA Clean and Concentrator (Zymo Research) according to the manufacturer’s instructions. DNA quality and quantity were determined using a NanoDrop 2000 Spectrophotometer and a Qubit 2.0 Fluorometer (Thermo Fisher Scientific), respectively. Sequencing libraries were prepared with the Nextera XT 150 bp paired-end library preparation kit according to the manufacturer’s instructions (Illumina). Subsequently, whole genome sequencing was performed on an Illumina NextSeq using the NextSeq V2 reagent kit. Reads were assembled de novo using the SPAdes Genome Assembler 3.9.1 with default stringency parameters [52]. Assemblies were performed on the Massachusetts Green High Performance Computing Cluster (www.mghpcc.org).
Bacterial phylogeny inference
Following whole genome sequencing, phylogenetic analysis was performed using bcgTree, which infers a maximum-likelihood phylogeny using the concatenated sequences of 107 single-copy core genes with 1000 bootstraps [53]. The hmmer v3.1b2 hmmsearch tool was used to locate the single-copy genes amino acid sequences in each genome, muscle 3.8.31 to create a multiple sequence alignment based on the resulting presence/absence table, Gblocks 0.91b to refine the alignment, and RaxML 8.2.4 to build the maximum-likelihood phylogenetic tree [53–58]. Reference Bifidobacterium genomes were obtained from the NCBI RefSeq database under the following accession numbers: NZ_MWWV00000000.1 (B. tissieri JCM 30798T), NZ_AZMV00000000.1 (B. moukalabense JCM 18751T), NZ_MWWY00000000.1 (B. hapali JCM 30799T), NZ_JDUU00000000.1 (B. biavatii JCM 17299T), NZ_BDIS00000000.1 (B. lemurum JCM 30168T), NZ_MWWZ00000000.1 (B. eulemuris JCM 30801T), NZ_MWWW00000000.1 (B. myosotis JCM 30796T), NZ_JGZK00000000.1 (B. reuteri JCM 17295T), NZ_JGZN00000000.1 (B. saguini JCM 17297T), NZ_BCFK00000000 (B. aesculapii JCM 18761T), NZ_JGZP00000000 (B. stellenboschense JCM 17298T), and NZ_JGYS00000000 (B. callitrichos JCM 17296T). [59]. Final phylogenetic trees were visualized and formatted using FigTree v1.4.3 and Phylo.io [60, 61].
Sequencing data analysis
Initial gene model predictions and annotations were performed using the Rapid Annotation using Subsystem Technology (RAST) annotation server [62–64]. Genes were sorted into functional categories using the SEED database through the RAST annotation server, and the percentage of genes belonging to each functional category was calculated relative to the total number of genes in each genome. The genomes of isolates UMA51804 and UMA51805 were submitted for auto-annotation with the Department of Energy’s Joint Genome Institute (JGI) Integrated Microbial Genomes (IMG) Microbial Genome and Metagenome Expert Review Data Submission platform [65]. RAST protein-coding gene predictions were assigned Kyoto Encyclopedia of Genes and Genomes (KEGG) orthology identifications using BlastKOALA [66]. KEGG Orthology (KO) identifiers were converted to Enzyme Commission (EC) numbers using the Carbohydrate Metabolism KEGG reference pathways [67–69]. In addition, genes associated with carbohydrate metabolism were identified using the Carbohydrate-Active Enzymes (CAZy) database (http://www.cazy.org/) via the Database for Automated Carbohydrate-Active Enzyme Annotation (dbCAN) web tool [70, 71]. The presence and absence of genes within each genome was determined using Venny 2.1.0. [72]. Amino acid sequence analysis to determine potential extracellular protein localization was performed with PSORTb version 3.0.2 [73].
Metabolic phenotyping of fermentative substrates
Bacterial strains were tested for their ability to grow on specific carbohydrates as a sole carbon source. Briefly, a 1 % (v/v) overnight culture of each isolate, including biological and technical triplicates, was grown on modified MRS (mMRS) media containing 2 % (w/v) of each carbon substrate: arabinose, cellobiose, fructose, galactose, glucose, acacia gum, arabinoxylan, cranberry xyloglucans (provided by Ocean Spray), lactose, maltose, mannose, N-acetylglucosamine (NAG), rhamnose, sorbitol, tamarind gum, trehalose, xylan and xylose. Biomass production was estimated by measuring the optical density at 600 nm (OD600). To determine final OD600, mMRS was inoculated with an overnight culture at a concentration of 1 % and then incubated for 72 h at 37 °C under anaerobic conditions. Negative controls consisted of inoculated media in the absence of a carbon substrate. Two-way analysis of variance was performed using GraphPad Prism version 6 (GraphPad Software, www.graphpad.com). In addition, the ability of each isolate to grow on 2′-fucosyllactose and pooled human milk oligosaccharides (HMOs) was assayed using a PowerWave HT Microplate Spectrophotometer (BioTek). Overnight cultures grown in MRS broth were used to inoculate mMRS at a concentration of 1 %. Isolates then grew anaerobically for 40 h at 37 °C with shaking and OD600 measurements were taken every 15 min. OD600 values were plotted over time to create a logarithmic growth curve using GraphPad Prism.
Results
Microbial diversity in the marmoset gut is dominated by Proteobacteria and Firmicutes
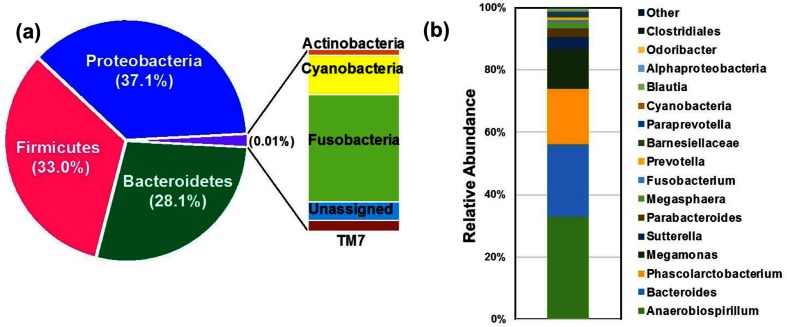
The microbial community structure in the marmoset gut was determined from the source fecal sample using 16S rRNA amplicon sequencing. This sample was dominated by three bacterial phyla: Proteobacteria (37.1 %), Firmicutes (33.0 %), and Bacteroidetes (28.1 %). Other phyla at low abundance (<0.01 %) were members of Actinobacteria, Cyanobacteria, Fusobacteria, TM7, and unassigned classes of bacteria (Fig. 1a). A detailed analysis at the operational taxonomic unit (OTU) level identified the top 16 abundant bacterial genera in this sample, followed by other less abundant (<0.2 %) genera (Fig. 1b). The most abundant genera identified were Anaerobiospirillum (32.5 %), Bacteroides (22.8 %), Phascolarctobacterium (17.8 %) and Megamonas (12.4 %). The genus Anaerobiospirillum consists of Gram-negative, anaerobic, spiral-shaped rods which are known to be the part of the normal gastrointestinal microbiota of dogs and cats [74, 75]. Bacteroides species are important members of the human and animal gut microbiota identified from feces (representing approximately 30 % of the cultured species) [76, 77]. Phascolarctobacterium are Gram-negative, non-spore-forming, saccharolytic Firmicutes, which could produce short-chain fatty acids and have been identified in human and koala feces [78]. Members of the genus Megamonas have been associated with obesity and glucose tolerance in the human gut microbiome, but their role in the marmoset gut is not clearly understood [79, 80].
Fig. 1.
Relative taxon abundance in the marmoset gut. Shown are pie and bar chart representations of the relative abundance values at the (a) phylum and (b) genus level of microbial diversity in the marmoset gut using 16S rRNA gene sequencing. Each color represents a phylum (a) and the top 16 genera (b) identified (at >0.2 % abundance) in the marmoset gut.
Interestingly, the genus Bifidobacterium was detected at low abundance (0.04 %) in the feces of this particular marmoset adult. However, low abundance does not necessarily reflect the functional importance of this genus within the marmoset microbiome. Based on the increased carbohydrate metabolism of bifidobacteria and the high carbohydrate diet of the common marmoset, we chose to isolate and analyze bifidobacterial strains present.
General genome characteristics
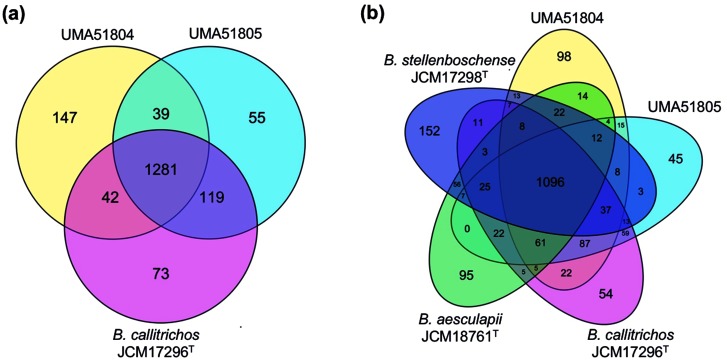
The median genome size of B. callitrichos JCM 17296T is 2.88 Mb with a G+C content of 63.6 mol% and 2230 protein-coding genes in 33 scaffolds [19, 22, 81, 82]. Isolates UMA51804 and UMA51805 have genome sizes of 3.04 and 2.78 Mb with G+C contents of 64.5 and 63.6 mol%, respectively (Table 1). Sequence similarity between UMA51804 and UMA51805 is depicted visually via a dot plot in Fig. S1 (available in the online version of this article). Interestingly, UMA51804 encodes a higher number of protein-coding genes (2528) than UMA51805 (2228) and B. callitrichos JCM 17296T (2230), which is consistent with its larger genome size. The RAST genome annotation for UMA51804 also contains a larger percentage of hypothetical genes (31.59 %) compared with UMA51805 (23.62 %) and JCM 17296T (26.25 %). Genes unique to UMA51804 are not dominated by any particular functional category and include CRISPR- and phage-associated proteins. Average genomic nucleotide identity between UMA51804 and JCM 17296T was 91.41 %, which suggests that UMA51804 may not belong to the same species sensu stricto. An alternative explanation is that this isolate may represent a different subspecies from UMA51805 and B. callitrichos JCM 17296T [83]. Utilization of the alignment fraction method for inter-species determination produced a 40 % probability that UMA51804 represents a separate species from B. callitrichos [84]. UMA51804 and UMA51805 share 39 (2.2 %) genes not found in JCM 17296T, and the core genome of all three strains consists of 1281 genes (Fig. 2a). The closely related strains Bifidobacterium aesculapii JCM 18761T and Bifidobacterium stellenboschense JCM 17298T have 95 and 152 genes, respectively, not identified in the analyzed B. callitrichos genomes by the RAST SEED database (Fig. 2b).
Table 1. General genome characteristics of B. callitrichos strains and closely related species.
| Characteristic |
B. callitrichos UMA51804 |
B. callitrichos UMA51805 |
B. callitrichos JCM 17296T |
B. aesculapii JCM 18761T |
B. stellenboschense JCM 17298T |
B. angulatum JCM 1252T |
|---|---|---|---|---|---|---|
| Isolation source | Callithrix jacchus feces | Callithrix jacchus feces | Callithrix jacchus feces | Callithrix jacchus feces | Saguinus midas feces | Adult human feces |
| Status | Draft | Draft | Draft | Draft | Draft | Complete |
| Number of scaffolds | 92 | 88 | 40 | 93 | 40 | 1 |
| Median genome size (Mb) | 3.02 | 2.76 | 2.87 | 2.69 | 2.81 | 2.01 |
| Median G+C content (mol%) | 64.5 | 63.6 | 63.5 | 64.8 | 65.3 | 59.4 |
| Median protein count | 2465 | 2200 | 2230 | 1989 | 2102 | 1520 |
| GenBank assembly accession no. |
GCA_003024955.1 | GCA_003024945.1 | GCA_000741175.1 | GCA_001417815.1 | GCA_000741785.1 | GCA_001025155.1 |
Fig. 2.
Genomic diversity of B. callitrichos. Venn diagrams showing the number of genes shared and unique between (a) B. callitrichos JCM 17296T, UMA51804, and UMA51805, and (b) genes shared between B. callitrichos strains (UMA51804, UMA51805) and closely related species B. aesculapii JCM 18761T and B. stellenboschense JCM 17298T. The B. callitrichos strains UMA51804 and UMA51805 shared a higher number of genes with B. callitrichos JCM 17296T than with each other and other type strains included in the analysis.
Phylogenetic relatedness within the genus Bifidobacterium
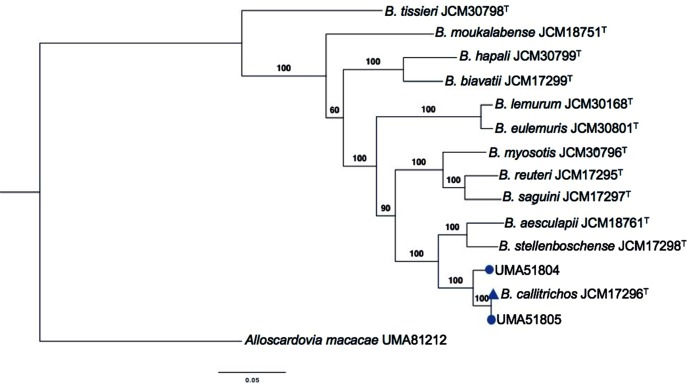
Initial identification of the isolates as members of B. callitrichos used 16S rRNA gene sequences to create a phylogenetic model (Fig. S2). The phylogenetic tree placed UMA51804 and UMA51805 as closely related to B. callitrichos JCM 17296T. The phylogenetic relationships inferred from the whole genome sequences of B. callitrichos JCM 17296T, UMA51804, UMA51805, and other bifidobacteria hosted within non-human primates are depicted in Fig. 3. UMA51804 clusters separately from UMA51805 and B. callitrichos JCM 17296T within an individual clade supported by a bootstrap value of 100, which supports the initial 16S rRNA phylogenetic inference (Fig. S2). The UMA51804, UMA51805, and JCM 17296T clade shares a node with B. aesculapii JCM 18761T and B. stellenboschense JCM 17298T, which were isolated from feces of an infant common marmoset and a red-handed tamarin, respectively [19, 21]. B aesculapii JCM 18761T and B. stellenboschense JCM 17298T, along with many other non-human primate bifidobacteria, have not been studied extensively. Overall, additional strains from these primates will provide greater phylogenetic resolution of evolutionary relatedness between isolates from the same host.
Fig. 3.
Maximum-likelihood phylogenetic tree of members of the genus Bifidobacterium originating from non-human primates. The tree was created using the bcgTree pipeline with 107 essential single-copy core genes, found in a majority of bacteria, using hidden Markov models based on a partitioned maximum-likelihood analysis. Bootstrap confidence values were obtained with 1000 resamplings and are provided at branch points. The scale bar represents the number of amino acid substitutions per site. Microbial isolates identified in this study are shown as blue circles closely clustered with B. callitrichos JCM 17296T (blue triangle). Alloscardovia macacae (UMA81212) was used as the outgroup species within the family Bifidobacterium.
The core genome of B. callitrichos contains carbohydrate utilization genes associated with host diet
Table 2 lists the names and corresponding EC numbers of predicted genes found in the top three highest represented KEGG carbohydrate metabolic pathways. All three B. callitrichos strains exhibit genes coding for ATP-binding cassette (ABC)-type transporters predicted to import exogenous carbohydrate molecules. Specifically, the core genome contains transporters of the multiple sugar metabolism system. These are ABC transporters including ATP-binding proteins, permeases, and substrate binding proteins (see Table S1 for locus tags). This fits with previous studies of bifidobacteria demonstrating the importance of ABC transporters for the import of exogenous carbohydrates into the cell [85, 86]. In addition, several phospho-transferase systems (PTSs) are represented within the genomes. For instance, the core genome contains subunits IIA, IIB, and IIC of the PTS transporter system for the import of lactose and cellobiose, and also has the components of the PTSs specific to trehalose, NAG, and β-glucoside (see Table S2 for locus tags). Bifidobacterial carbohydrate metabolism could involve the secretion of extracellular enzymes which bind or alter carbohydrate structures prior to uptake [87, 88]. Using the CAZy database and the PSORTb subcellular localization tool, three major categories of carbohydrate-active enzymes, glycosyl hydrolase families 25 and 51, carbohydrate esterase family 4, and carbohydrate-binding molecule family 22, were predicted as extracellularly targeted in all three genomes (Tables S3 and S4). The representation of all carbohydrate-active enzyme families is similar among the three strains (Fig. S3 and Table S3).
Table 2. KEGG carbohydrate metabolic pathway genes shared by B. callitrichos JCM 17296T and isolates UMA51804 and UMA51805 identified in this study.
Genes shown are those for which a locus tag in each genome could be identified. Pathways are highlighted based on the different metabolic categories.
| KEGG carbohydrate metabolic pathway | Gene name | EC | UMA51804 locus tag | UMA51805 locus tag | JCM 17296T locus tag |
|---|---|---|---|---|---|
| Galactose metabolism | UTP—glucose 1-phosphate uridylyltransferase | 2.7.7.9 | COO72_RS10475 | CPA40_RS09980 | BCAL_RS08755 |
| Aldose 1-epimerase | 5.1.3.3 | COO72_RS03125 | CPA40_RS00475 | BCAL_RS03445 | |
| Galactokinase | 2.7.1.6 | COO72_RS01650 | CPA40_RS06710 | BCAL_RS05755 | |
| UDP-glucose 4-epimerase | 5.1.3.2 | COO72_RS04485 | CPA40_RS02855 | BCAL_RS02635 | |
| l-Ribulose-5-phosphate 4-epimerase | 5.1.3.4 | COO72_RS03920 | CPA40_RS04240 | BCAL_RS05325 | |
| N-Acylglucosamine 2-epimerase | 5.1.3.8 | COO72_RS09925 | CPA40_RS00610 | BCAL_RS02285 | |
| Starch and sucrose metabolism | α,α-Phosphotrehalase | 3.2.1.93 | COO72_RS08005 | CPA40_RS06370 | BCAL_RS06195 |
| Protein-Nπ-phosphohistidine—trehalose phosphotransferase | 2.7.1.201 | COO72_RS08010 | CPA40_RS06365 | BCAL_RS06200 | |
| β-Fructofuranosidase | 3.2.1.26 | COO72_RS02000 | CPA40_RS06035 | BCAL_RS10725 | |
| α-Glucosidase | 3.2.1.20 | COO72_RS09510 | CPA40_RS01270 | BCAL_RS0830 | |
| Amylosucrase | 2.4.1.4 | COO72_RS05900 | CPA40_RS04855 | BCAL_RS04290 | |
| Sucrose phosphorylase | 2.4.1.7 | COO72_RS10000 | CPA40_RS07310 | BCAL_RS06670 | |
| UTP—glucose-1-phosphate uridylyltransferase | 2.7.7.9 | COO72_RS10475 | CPA40_RS09980 | BCAL_RS08755 | |
| α-Amylase | 3.2.1.1 | COO72_RS11340 | CPA40_RS02640 | BCAL_RS02400 | |
| β-Glucosidase | 3.2.1.21 | COO72_RS09740 | CPA40_RS01270 | BCAL_RS01445 | |
| Isoamylase | 3.2.1.68 | COO72_RS04240 | CPA40_RS05215 | BCAL_RS04710 | |
| Glucose-1-phosphate adenylyltransferase | 2.7.7.27 | COO72_RS01125 | CPA40_RS05455 | BCAL_RS09705 | |
| 1,4-α-Glucan branching enzyme | 2.4.1.18 | COO72_RS03710 | CPA40_RS0112 | BCAL_RS04065 | |
| 4-α-Glucanotransferase | 2.4.1.25 | COO72_RS08655 | CPA40_RS05200 | BCAL_RS04695 | |
| Glycogen phosphorylase | 2.4.1.1 | COO72_RS08360 | CPA40_RS09325 | BCAL_RS09090 | |
| Starch synthase (maltosyl-transferring) | 2.4.99.16 | COO72_RS06885 | CPA40_RS03955 | BCAL_RS11605 | |
| Phosphoglucomutase (α-d-glucose-1,6-bisphosphate-dependent) | 5.4.2.2 | COO72_RS10095 | CPA40_RS03335 | BCAL_RS04990 | |
| Protein-Nπ-phosphohistidine—cellobiose phosphotransferase | 2.7.1.205 | COO72_RS05245 | CPA40_RS08235 | BCAL_RS07995 | |
| Amino sugar and nucleotide sugar metabolism | UDP-N-acetylmuramate dehydrogenase | 1.3.1.98 | COO72_RS09690 | CPA40_RS03405 | BCAL_RS04925 |
| UDP-N-acetylglucosamine 1-carboxyvinyltransferase | 2.5.1.7 | COO72_RS11030 | CPA40_RS10465 | BCAL_RS06425 | |
| UDP-N-acetylglucosamine diphosphorylase | 2.7.7.23 | COO72_RS04345 | CPA40_RS08380 | BCAL_RS00645 | |
| N-Acetylglucosamine-6-phosphate deacetylase | 3.5.1.25 | COO72_RS10300 | CPA40_RS06275 | BCAL_RS02715 | |
| Glucosamine-1-phosphate N-acetyltransferase | 2.3.1.157 | COO72_RS04345 | CPA40_RS08380 | BCAL_RS00645 | |
| Phosphoglucosamine mutase | 5.4.2.10 | COO72_RS05530 | CPA40_RS04580 | BCAL_RS02085 | |
| Glucosamine-6-phosphate deaminase | 3.5.99.6 | COO72_RS08105 | CPA40_RS05730 | BCAL_RS02710 | |
| Glutamine—fructose-6-phosphate transaminase (isomerizing) | 2.6.1.16 | COO72_RS05020 | CPA40_RS04675 | BCAL_RS02180 | |
| Non-reducing end α-l-arabinofuranosidase | 3.2.1.55 | COO72_RS05470 | CPA40_RS04135 | BCAL_RS02145 | |
| Glucose-6-phosphate isomerase | 5.3.1.9 | COO72_RS12210 | CPA40_RS08670 | BCAL_RS05265 | |
| UDP-glucose 6-dehydrogenase | 1.1.1.22 | COO72_RS07800 | CPA40_RS09840 | BCAL_RS07900 | |
| Phosphoglucomutase (α-d-glucose-1,6-bisphosphate-dependent) | 5.4.2.2 | COO72_RS10095 | CPA40_RS03335 | BCAL_RS04990 | |
| UTP—glucose-1-phosphate uridylyltransferase | 2.7.7.9 | COO72_RS10475 | CPA40_RS09980 | BCAL_RS08755 | |
| UDP-glucose 4-epimerase | 5.1.3.2 | COO72_RS04485 | CPA40_RS02855 | BCAL_RS02635 | |
| Galactokinase | 2.7.1.6 | COO72_RS01650 | CPA40_RS06710 | BCAL_RS05755 | |
| UDP-glucose—hexose-1-phosphate uridylyltransferase | 2.7.7.12 | COO72_RS11900 | CPA40_RS05725 | BCAL_RS05750 | |
| UDP-galactopyranose mutase | 5.4.99.9 | COO72_RS07765 | CPA40_RS11110 | BCAL_RS07940 | |
| Phosphoglucomutase (α-d-glucose-1,6-bisphosphate-dependent) | 5.4.2.2 | COO72_RS10095 | CPA40_RS03335 | BCAL_RS04990 | |
| Glucose-1-phosphate adenylyltransferase | 2.7.7.27 | COO72_RS01125 | CPA40_RS05455 | BCAL_RS09705 |
The B. callitrichos core genome is predicted to encode several genes in diverse carbohydrate metabolic pathways. Genomic locus tags are defined with the following convention: the UMA51804 locus tag prefix is COO72_, the UMA51805 locus tag prefix is CPA40_, and the JCM 17296T locus tag prefix is BCAL_. All three genomes contain predicted l-arabinose isomerase genes (COO72_RS01355, CPA40_RS10585, BCAL_RS05330), which could convert arabinose to ribulose and galactose to tagatose. The presence of genes associated with galactose and arabinose metabolism is consistent with the major carbon constituents of tree gums found in common marmoset habitats [34]. α,α-phosphotrehalase (COO72_RS08005, CPA40_RS06370, BCAL_RS06195), which converts trehalose 6-phosphate to d-glucose 6-phosphate, may reflect the marmoset's utilization of alternative nutritive sources in addition to gums. This includes insects that contain trehalose in their hemolymph [89]. Furthermore, several dietary carbohydrates are predicted to be utilized by B. callitrichos through hydrolysis or interconversions to d-glucose including maltose and sucrose via α-glucosidase (COO72_RS09510, COO72_RS09740; CPA40_RS00310, CPA40_RS00625; BCAL_RS03280, BCAL_RS03615), cellobiose via β-glucosidase (COO72_RS03545, COO72_RS09740, COO72_RS05215; CPA40_RS01270, CPA40_RS06850, CPA40_RS08995, CPA40_RS02570; BCAL_RS04215, BCAL_RS10930, BCAL_RS01445), melibiose via α-galactosidase (COO72_RS10210, COO72_RS10225, COO72_RS00655, COO72_RS12035; CPA40_RS10210, CPA40_RS10230, CPA40_RS10525; BCAL_RS06730, BCAL_RS08175, BCAL_RS08225), and lactose via β-galactosidase (COO72_RS01410, COO72_RS05460, COO72_RS07355, COO72_RS08450, COO72_RS08505; CPA40_RS04650, CPA40_RS05890, CPA40_RS09155, CPA40_RS09460 CPA40_RS09665, CPA40_RS09775, CPA40_RS02920; BCAL_RS02155, BCAL_RS02705, BCAL_RS05420, BCAL_RS07405, BCAL_RS10570).
Recently, a genomic analysis of B. breve revealed ~15 kb mannitol/sorbitol utilization cluster in several strains [90]. The genome of B. callitrichos JCM 17296T has a similar cluster of ~12 kb which contains sorbitol dehydrogenase (BCAL_RS04735), alcohol dehydrogenase (BCAL_RS04755), lactoylglutathione lyase (BCAL_RS00215), an araC family transcriptional regulator (BCAL_RS04730), an ROK family transcriptional regulator (BCAL_RS04750), and an MFS superfamily sugar alcohol transporter (BCAL_RS04740). All of these components are clustered in UMA51804 with minor divergence to adjacent genes, and on two separate contigs in UMA51805. In addition, all three genomes contain α-galactosidases which could also catalyse the metabolism of galactinol, d-myo-inositol, sorbitol, melibitol, and glycerol.
Variation in gene representation between isolates may contribute to carbohydrate metabolic diversity
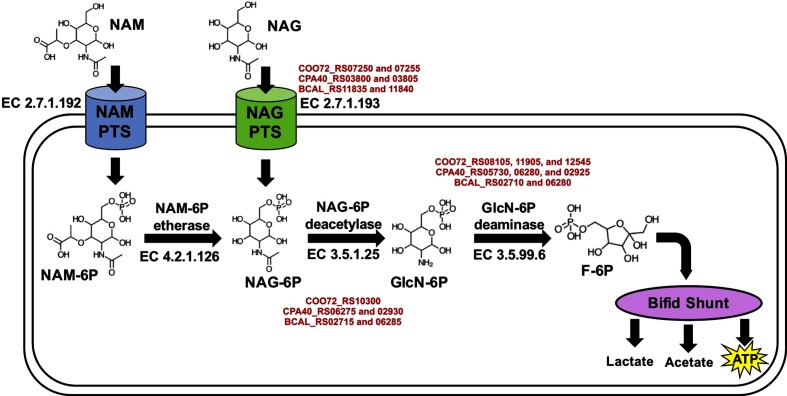
N-Acetylglucosamine (i.e. NAG or GlcNAc) is a moiety incorporated into peptidoglycan, which is a major constituent of the bacterial cell wall [91]. In addition, NAG is incorporated within milk oligosaccharides, which could be utilized as a fermentative substrate by B. longum subsp. infantis and other bifidobacterial species [92]. Extracellular NAG, regardless of source, could be transported into the cell by NAG PTS transporter systems and enter the bifid shunt after several modifications. All three genomes include a NAG-specific PTS intracellular transporter system (Fig. 4, Table S2). Interestingly, a KEGG pathway analysis identified a PTS transporter for a related molecule, N-acetylmuramic acid (NAM), in UMA51804 and UMA51805. NAM has the same structure as NAG but with an ester group bound to the oxygen at the C3 position. B. callitrichos metabolism of NAM is unclear, but it is possible that NAM is further catabolized to fructose 6-phosphate to enter the bifid shunt. NAM 6-phosphate (-6P) and NAG-6P are typically interconverted in the cytoplasm via NAM-6P etherase (EC 4.2.1.126), although this gene sequence was not detected in any of the B. callitrichos genomes. It is possible that NAM-6P is converted to NAG-6P via an as yet uncharacterized mechanism.
Fig. 4.
Proposed mechanism for the utilization of N-acetylmuramic acid (NAM) and N-acetylglucosamine (NAG). NAM and NAG serve as the precursors for fructose 6-phosphate, which feeds into the bifid shunt pathway. The proposed mechanism is depicted to show the structure and EC number for the enzymes involved in the pathway. Text in red represents the locus tags for UMA51804, UMA51805, and JCM 17296T, in that order.
The B. callitrichos UMA51804 genome is significantly larger than that of UMA51805 and JCM 17296T as it contains more protein-coding loci. This has not translated to an increased percentage of carbohydrate-related genes in UMA51804 (Fig. S4). However, UMA51804 does have unique carbohydrate-related genes, for example subunits IIA (COO72_RS02120), IIB (COO72_RS02125), and IIC (COO72_RS02130) of the fructose-specific PTS. In addition, UMA51804 has several genes predicted to be specific to HMO utilization. Of note, the presence of lacto-N-biose phosphorylase (COO72_RS10625), a key enzyme in the lacto-N-biose pathway, suggests that UMA51804 may have the ability to metabolize marmoset milk oligosaccharides [12, 93, 94]. This enzyme phosphorylates the disaccharides lacto-N-biose and galacto-N-biose, carbohydrate residues that comprise milk and mucin glycans, which permits further hydrolysis and catabolism of the substrate [95, 96]. The UMA51804 genome encodes predicted galacto-N-biose/lacto-N-biose ABC transporter components (substrate-binding protein: COO72_RS10640, permeases: COO72_RS10635 and COO72_RS10630) and l-fuconolactone hydrolase (COO72_RS01745), which are active on milk glycans, suggesting a utility early in development of the marmoset. UMA51804 also possesses a predicted N-acetylneuraminate lyase (COO72_RS01755), which could catalyze hydrolysis of negatively charged milk oligosaccharides (i.e. sialylated glycans) [97, 98]. The potential for UMA51804 to utilize marmoset milk oligosaccharides provides a compelling hypothesis related to the role of UMA51804 within the developing marmoset gut and later life stages.
B. callitrichos strains ferment several dietary carbohydrates available to the marmoset
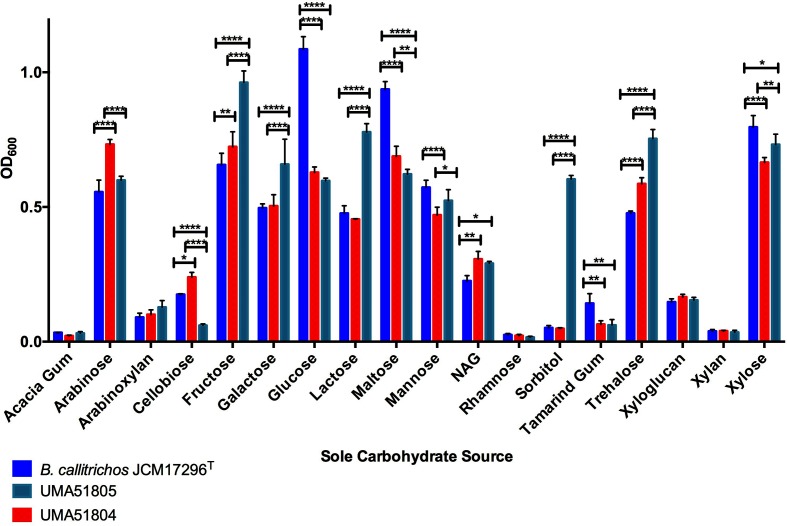
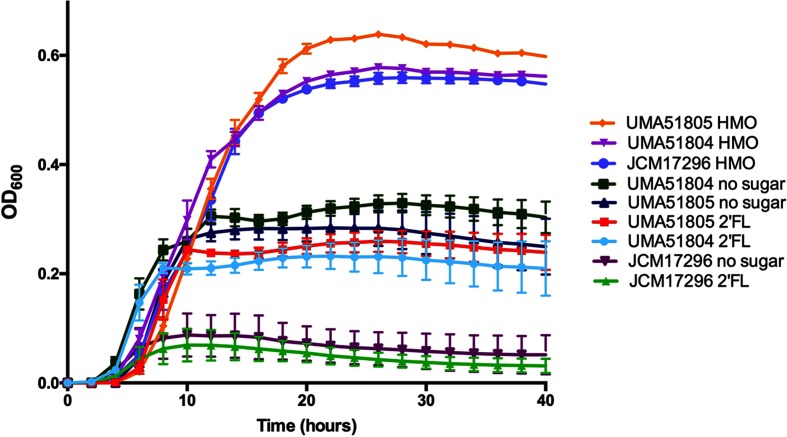
To match genomic predictions with phenotype, the three B. callitrichos strains (UMA51804, UMA51805, and JCM 17296T) were subjected to growth on carbohydrates as the sole carbohydrate source. The results of these analyses are exhibited in Fig. 5 as OD600 values at stationary phase. The carbohydrates to test were selected based on their relevance to bifidobacterial metabolism, the marmoset diet, and the carbohydrate utilization genes predicted within the B. callitrichos genome. All three strains grew on glucose, arabinose, galactose, mannose, xylose, trehalose, lactose and fructose. This is consistent with the findings of genes that are predicted to encode activities that feed these carbohydrates into central metabolism. However, there was significant variation among the three strains depending on the carbohydrate source. Accordingly, B. callitrichos JCM 17296T grew significantly more on glucose (OD600avg=1.086; P<0.0001) than both UMA51804 (OD600avg=0.628) and UMA51805 (OD600avg=0.597). The same trend was observed with growth on maltose and tamarind gum. Conversely, UMA51804 metabolized arabinose to achieve a higher OD600 (OD600avg=0.732; P<0.0001) than UMA51805 (OD600avg=0.599) and JCM 17296T (OD600avg=0.556). Galactose metabolism resulted in OD600 values that were significantly higher (OD600avg=0.659; P<0.0001) for UMA51805 than for UMA51804 (OD600avg=0.504) and JCM 17296T (OD600avg=0.496). UMA51805 achieved greater cell biomass (P<0.0001) while subsisting on trehalose (OD600avg=0.754), lactose (OD600avg=0.778), and fructose (OD600avg=0.962) than UMA51804 or JCM 17296T. Interestingly, UMA51804 grew to a lesser extent than UMA51805 and B. callitrichos JCM 17296T on mannose (OD600avg=0.470; P<0.001) and xylose (OD600avg=0.665; P<0.0001). UMA51804 grew more efficiently than B. callitrichos JCM 17296T on cellobiose (UMA51804 OD600avg=0.239; JCM 17296T OD600avg=0.175; P≤0.05), but both achieved higher final OD600 values than UMA51805 (OD600avg=0.060; P<0.0001). All three strains did not grow appreciably on rhamnose (total OD600avg=0.0214), purified acacia gum (total OD600avg=0.0289), and xylan (total OD600avg=0.0378) as the sole carbohydrate source. UMA51805 grew significantly better on sorbitol (OD600avg=0.603; P<0.0001) than UMA51804 and JCM 17296T. JCM 17296T, UMA51804 and UMA51805 all grew to an similar extent on arabinoxylan (total OD600avg=0.1064) and cranberry xyloglucans (total OD600avg=0.1557). Finally, and significantly, all three strains grew on pooled HMOs suggesting that they may metabolize similar carbohydrates in marmoset milk (Fig. 6). Interestingly, none of the three B. callitrichos strains grew on the typical HMO species 2′-fucosyllactose. It is not currently known to what extent 2′-fucosyllactose appears in marmoset milk. One study found that the molar concentration of fucosylated oligosaccharides in marmoset milk is under 10 % [99]. Somewhat surprising is that the two recent isolates grew moderately under negative control conditions (i.e. no carbohydrate). The assay is validated by the type strain not growing under these conditions, as well as other bifidobacterial strains (data not shown). This is intriguing as it suggests an alternative energy-generating pathway that is potentially independent of carbohydrate fermentation.
Fig. 5.
Growth of B. callitrichos JCM 17296T, UMA51804, and UMA51805 on various sole carbohydrate sources. Shown are growth profiles on acacia gum, arabinose, cellobiose, fructose, galactose, glucose, lactose, maltose, mannose, N-acetylglucosamine (NAG), rhamnose, sorbitol, tamarind gum, trehalose, cranberry xyloglucan, xylan, and xylose as a sole carbohydrate source. Bars represent the average final OD600 of biological triplicates, and error bars show the standard deviation. Sole carbohydrate sources and OD600 values are shown on the x- and y-axis, respectively. Significant differences among the growth profiles of strains on each carbohydrate source are computed using two-way ANOVA with significance at *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Fig. 6.
Growth of B. callitrichos JCM 17296T, UMA51804, and UMA51805 on 2′-fucosyllactose and pooled HMOs. Shown are growth profiles on 2-fucosyllactose and pooled HMOs as sole carbohydrate sources. Curves represent the average OD600 of biological triplicates with error bars showing the standard deviation. Growth times and OD600 values are shown on the x- and y-axis, respectively.
Discussion
Bifidobacteria utilize a broad range of host-indigestible dietary carbohydrates that promote reciprocally beneficial host–microbial interactions within the gut. These traits are postulated to be a product of host–microbial coevolution, and the continued study of bifidobacteria in non-human primate species will further explicate this coevolutionary relationship. Mammalian milk maintains host-indigestible oligosaccharide utilization in infant-associated bifidobacteria, and potentially in non-human primates. A detailed analysis of primate milk oligosaccharides identified 100 oligosaccharide structures in marmoset milk, which was most similar to that of chimpanzees and humans [99]. Power et al. determined that the total gross energy content of marmoset milk is similar to that of several other primates, but that individual marmosets with a lower total gross energy had a higher proportion of energy from sugars [100]. These studies suggest that carbohydrates play an important role in marmoset milk. Lactating marmoset mothers may be under increased energetic stress because they often give birth to twins and immediate postpartum fertility often leads to two births per year [101, 102]. Common marmoset nursing peaks at 2 weeks and solid foods are introduced beginning at 5 weeks. Following weaning, bifidobacteria may continue to benefit their host through metabolism of other dietary carbohydrates in the adult marmoset diet. The exudatory nutritive strategy in marmosets may provide an example of evolutionary pressure to maintain gut microbial fermentation, although more research is required to examine this concept further.
Maintaining an exuditory diet presents somewhat of a nutritional challenge. Plant gums are high in carbohydrates but lower in protein. A recent analysis of marmoset feeding habits determined the carbohydrate and protein content of gums from Anadenanthera peregrina, a food source for marmosets in Brazil, to be 38.2 and 19.0 %, respectively [103]. This may drive marmosets to supplement their protein intake with insects [33]. The relatively low nutritional content of gum exudates increases the need of marmosets to perform highly efficient extraction of substrates for energy and biomass, to which the bifidobacteria and other members of the gut microbiome contribute. Despite these challenges, the common marmoset appears to be physiologically adapted for this purpose [32]. Small body sizes and low nutrient requirements enable common marmosets to subsist on a limited diet, and the relatively slow digestive transit time of gums allows for more complete microbial fermentation [28, 33]. In addition, the marmoset mouth and jaw architecture is hypothesized to have evolved for better access to gums within trees to ultimately provide the gut microbiota with substrates that are not fully digested by their host [104].
The composition of the marmoset gut microbiome and the influence of diet on its function are not thoroughly understood. Ley et al. included two marmoset species, Callimico goeldii and Callithrix geoffroyi, in a larger study of mammalian gut microbiomes and found correlations between these species and the microbiomes of other primates [15]. Another study included C. goeldii in an analysis of dietary strategies and found that this primate grouped with omnivores in an OTU network diagram [16]. Other studies of microbiota in marmoset feces are limited to a few culture-dependent approaches. Accordingly, high concentrations of Gram-negative bacteria (~60 %) have been isolated from the common marmoset large intestine, although this may not be a true representation of the community due to biases in culture-based approaches [105, 106]. Although gastrointestinal pathogens such as Helicobacter spp., enteropathogenic Escherichia coli, and Clostridium difficile have been identified in marmosets, studies of gut microbial symbionts are limited [107–109]. The previous isolation of Lactobacillus casei and Bifidobacterium catenulatum from marmoset feces suggests that gut bacteria with beneficial properties may also be active in maintaining marmoset gastrointestinal health [110]. Microbial diversity analysis (i.e. 16S amplicon sequencing) reveals that community diversity patterns at the genus level are consistent with previous studies of the human gut microbiome. This suggests that the marmoset may serve as a useful model to study the microbial diversity of the primate gut, providing a species-specific diversity signature which can be correlated with infant development and dietary changes early in life to investigate host–microbial interactions.
The microbiome of two other captive primate species, the red-shanked douc (Pygathrix nemaeus) and the mantled howler monkey (Alouatta palliata), had significantly lower alpha diversity compared to those living in the wild [111]. The similarity between the UMA51805 and JCM 17296T genomes suggests that marmoset-hosted B. callitrichos may not vary considerably according to geographical location. However, that UMA51804 was isolated from the same marmoset host adds a measure of uncertainty to this hypothesis. The unexpectedly large variation in the UMA51804 genome may reflect adaptation to B. callitrichos in a captive host. An alternative explanation may be that multi-strain colonization of marmosets occurs frequently and reflects niche partitioning within non-human primates. Future studies will need to determine the extent of genomic and phenotypic plasticity in non-human primate commensals and their microbial community assemblages.
Data bibliography
Sela, D. A., GenBank, NWTW00000000 (2017).
Sela, D. A., GenBank, NWTX00000000 (2017).
University of Parma, Genbank, GCA_000741175.1 (2014).
Funding information
This research was funded in part by grants from USDA-NIFA (2016-67017-24425), Center for Produce Safety (SCB14056), and the University of Massachusetts Amherst Graduate School Dissertation Grant Program.
Acknowledgements
We thank Agnès Lacreuse (UMass Amherst) for providing marmoset feces from which bacteria were isolated, Cynthia (Cindy) Kane and Xiaomeng You for technical assistance, Ezgi Özcan for reviewing the draft manuscript, David Mills (UC Davis) for donating 2′-fucosyllactose and pooled HMOs. Cranberry xyloglucans were donated by Ocean Spray Inc. Illumina is acknowledged for donating reagents used in whole genome sequencing.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Supplementary Data
Footnotes
Abbreviations: ABC, ATP-binding cassette; BSM, Bifidobacteria-specific medium; COGs, clusters of orthologous groups; MRS, de Mann Rogosa Sharpe; JGI, Department of Energy’s Joint Genome Institute; EC, Enzyme Commission; HMO, human milk oligosaccharide; IMG, Integrated Microbial Genomes; KO, KEGG Orthology; KEGG, Kyoto Encyclopedia of Genes and Genomes; MFS, Major Facilitator Superfamily; mMRS, modified MRS; NAG, N-acetylglucosamine; NAM, N-acetylmuramic acid; NCBI, National Center for Biotechnology Information; OTU, operational taxonomic unit; OD600, optical density at 600 nm; RAST, Rapid Annotation using Subsystem Technology; ROK, Rho-associated Protein Kinase; PTS, phosphotransferase system.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Four supplementary tables and four supplementary figures are available with the online version of this article.
References
- 1.Dhanashree SR, Rajashekharan S, Krishnaswamy B, Kammara R. Bifid shape is intrinsic to Bifidobacterium adolescentis. Front Microbiol. 2017;8:478. doi: 10.3389/fmicb.2017.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lamendella R, Santo Domingo JW, Kelty C, Oerther DB. Bifidobacteria in feces and environmental waters. Appl Environ Microbiol. 2008;74:575–584. doi: 10.1128/AEM.01221-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turroni F, van Sinderen D, Ventura M. Genomics and ecological overview of the genus Bifidobacterium. Int J Food Microbiol. 2011;149:37–44. doi: 10.1016/j.ijfoodmicro.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 4.Killer J, Kopecný J, Mrázek J, Rada V, Benada O, et al. Bifidobacterium bombi sp. nov., from the bumblebee digestive tract. Int J Syst Evol Microbiol. 2009;59:2020–2024. doi: 10.1099/ijs.0.002915-0. [DOI] [PubMed] [Google Scholar]
- 5.Killer J, Kopečný J, Mrázek J, Koppová I, Havlík J, et al. Bifidobacterium actinocoloniiforme sp. nov. and Bifidobacterium bohemicum sp. nov., from the bumblebee digestive tract. Int J Syst Evol Microbiol. 2011;61:1315–1321. doi: 10.1099/ijs.0.022525-0. [DOI] [PubMed] [Google Scholar]
- 6.Fushinobu S. Unique sugar metabolic pathways of bifidobacteria. Biosci Biotechnol Biochem. 2010;74:2374–2384. doi: 10.1271/bbb.100494. [DOI] [PubMed] [Google Scholar]
- 7.Turroni F, Milani C, Duranti S, Mahony J, van Sinderen D, et al. Glycan utilization and cross-feeding activities by bifidobacteria. Trends Microbiol. 2018;26:339–350. doi: 10.1016/j.tim.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 8.Milani C, Lugli GA, Duranti S, Turroni F, Mancabelli L, et al. Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut. Sci Rep. 2015;5:15782. doi: 10.1038/srep15782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schnell MA, Karmirantzou M, Snel B, Vilanova D, Berger B, et al. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc Natl Acad Sci. 2002;99:6. doi: 10.1073/pnas.212527599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milani C, Turroni F, Duranti S, Lugli GA, Mancabelli L, et al. Genomics of the genus Bifidobacterium reveals species-specific adaptation to the glycan-rich gut environment. Appl Environ Microbiol. 2016;82:980–991. doi: 10.1128/AEM.03500-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lugli GA, Milani C, Turroni F, Duranti S, Mancabelli L, et al. Comparative genomic and phylogenomic analyses of the Bifidobacteriaceae family. BMC Genomics. 2017;18:568. doi: 10.1186/s12864-017-3955-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci USA. 2008;105:18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sela DA, Mills DA. Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol. 2010;18:298–307. doi: 10.1016/j.tim.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sela DA. Bifidobacterial utilization of human milk oligosaccharides. Int J Food Microbiol. 2011;149:58–64. doi: 10.1016/j.ijfoodmicro.2011.01.025. [DOI] [PubMed] [Google Scholar]
- 15.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moeller AH, Li Y, Mpoudi Ngole E, Ahuka-Mundeke S, Lonsdorf EV, et al. Rapid changes in the gut microbiome during human evolution. Proc Natl Acad Sci USA. 2014;111:16431–16435. doi: 10.1073/pnas.1419136111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michelini S, Modesto M, Oki K, Stenico V, Stefanini I, et al. Isolation and identification of cultivable Bifidobacterium spp. from the faeces of 5 baby common marmosets (Callithrix jacchus L.) Anaerobe. 2015;33:101–104. doi: 10.1016/j.anaerobe.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 19.Endo A, Futagawa-Endo Y, Schumann P, Pukall R, Dicks LMT. Bifidobacterium reuteri sp. nov., Bifidobacterium callitrichos sp. nov., Bifidobacterium saguini sp. nov., Bifidobacterium stellenboschense sp. nov. and Bifidobacterium biavatii sp. nov. isolated from faeces of common marmoset (Callithrix jacchus) and red-handed tamarin (Saguinus midas) Syst Appl Microbiol. 2012;35:92–97. doi: 10.1016/j.syapm.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 20.Michelini S, Oki K, Yanokura E, Shimakawa Y, Modesto M, et al. Bifidobacterium myosotis sp. nov., Bifidobacterium tissieri sp. nov. and Bifidobacterium hapali sp. nov., isolated from faeces of baby common marmosets (Callithrix jacchus L.) Int J Syst Evol Microbiol. 2016;66:255–265. doi: 10.1099/ijsem.0.000708. [DOI] [PubMed] [Google Scholar]
- 21.Modesto M, Michelini S, Stefanini I, Ferrara A, Tacconi S, et al. Bifidobacterium aesculapii sp. nov., from the faeces of the baby common marmoset (Callithrix jacchus) Int J Syst Evol Microbiol. 2014;64:2819–2827. doi: 10.1099/ijs.0.056937-0. [DOI] [PubMed] [Google Scholar]
- 22.Milani C, Lugli GA, Duranti S, Turroni F, Bottacini F, et al. Genomic encyclopedia of type strains of the genus Bifidobacterium. Appl Environ Microbiol. 2014;80:6290–6302. doi: 10.1128/AEM.02308-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Modesto M, Michelini S, Stefanini I, Sandri C, Spiezio C, et al. Bifidobacterium lemurum sp. nov., from faeces of the ring-tailed lemur (Lemur catta) Int J Syst Evol Microbiol. 2015;65:1726–1734. doi: 10.1099/ijs.0.000162. [DOI] [PubMed] [Google Scholar]
- 24.Tsuchida S, Takahashi S, Nguema PPM, Fujita S, Kitahara M, et al. Bifidobacterium moukalabense sp. nov., isolated from the faeces of wild west lowland gorilla (Gorilla gorilla gorilla) Int J Syst Evol Microbiol. 2014;64:449–455. doi: 10.1099/ijs.0.055186-0. [DOI] [PubMed] [Google Scholar]
- 25.Michelini S, Modesto M, Filippini G, Spiezio C, Sandri C, et al. Bifidobacterium aerophilum sp. nov., Bifidobacterium avesanii sp. nov. and Bifidobacterium ramosum sp. nov.: three novel taxa from the faeces of cotton-top tamarin (Saguinus oedipus L.) Syst Appl Microbiol. 2016;39:229–236. doi: 10.1016/j.syapm.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 26.Michelini S, Modesto M, Pisi AM, Filippini G, Sandri C, et al. Bifidobacterium eulemuris sp. nov., isolated from faeces of black lemurs (Eulemur macaco) Int J Syst Evol Microbiol. 2016;66:1567–1576. doi: 10.1099/ijsem.0.000924. [DOI] [PubMed] [Google Scholar]
- 27.Modesto M, Michelini S, Sansosti MC, De Filippo C, Cavalieri D, et al. Bifidobacterium callitrichidarum sp. nov. from the faeces of the emperor tamarin (Saguinus imperator) Int J Syst Evol Microbiol. 2018;68:141–148. doi: 10.1099/ijsem.0.002472. [DOI] [PubMed] [Google Scholar]
- 28.Power ML, Oftedal OT. Differences among captive callitrichids in the digestive responses to dietary gum. Am J Primatol. 1996;40:131–144. doi: 10.1002/(SICI)1098-2345(1996)40:2&lt;131::AID-AJP2&gt;3.0.CO;2-Z. [DOI] [Google Scholar]
- 29.Rylands AB. Exudate-eating and tree-gouging by marmosets (Callitrichidae, Primates) In: Chadwick AC, Sutton SL, editors. Tropical Rain Forest: the Leeds Symposium 1984. Leeds: Leeds Philosophicaland Literary Society; pp. 155–168. (editors) [Google Scholar]
- 30.Garber PA. Vertical clinging, small body size, and the evolution of feeding adaptations in the Callitrichinae. Am J Phys Anthropol. 1992;88:469–482. doi: 10.1002/ajpa.1330880404. [DOI] [PubMed] [Google Scholar]
- 31.Power ML, Myers EW. Digestion in the common marmoset (Callithrix jacchus), a gummivore-frugivore. Am J Primatol. 2009;71:957–963. doi: 10.1002/ajp.20737. [DOI] [PubMed] [Google Scholar]
- 32.Power ML. The Evolution of Exudativory in Primates. New York: Springer; 2010. Nutritional and digestive challenges to being a gum-feeding primate; pp. 25–44. [Google Scholar]
- 33.Cabana F, Dierenfeld ES, Wirdateti, Donati G, Nekaris KAI. Exploiting a readily available but hard to digest resource: a review of exudativorous mammals identified thus far and how they cope in captivity. Integr Zool. 2018;13:94–111. doi: 10.1111/1749-4877.12264. [DOI] [PubMed] [Google Scholar]
- 34.Paula R, Budd PM, Rodrigues JF. Characterization of Anadenanthera macrocarpa exudate polysaccharide. Polym Int. 1997;44:55–60. [Google Scholar]
- 35.Francisco TM, Couto DR, Zanuncio JC, Serrão JE, Silva IO, et al. Vegetable exudates as food for Callithrix spp. (Callitrichidae): exploratory patterns. PLoS One. 2014;9:e112321. doi: 10.1371/journal.pone.0112321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haruna S, Aliyu BS, Bala A. Plant gum exudates (Karau) and mucilages, their biological sources, properties, uses and potential applications: a review. Bayero J Pure Appl Sci. 2016;9:159–165. doi: 10.4314/bajopas.v9i2.30. [DOI] [Google Scholar]
- 37.McWhorter TJ, Karasov WH. Paracellular nutrient absorption in a gum-feeding new world primate, the common marmoset Callithrix jacchus. Am J Primatol. 2007;69:1399–1411. doi: 10.1002/ajp.20443. [DOI] [PubMed] [Google Scholar]
- 38.Caporaso JG, Bittinger K, Bushman FD, Desantis TZ, Andersen GL, et al. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Desantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 42.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turroni F, Foroni E, Pizzetti P, Giubellini V, Ribbera A, et al. Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl Environ Microbiol. 2009;75:1534–1545. doi: 10.1128/AEM.02216-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Satokari RM, Vaughan EE, Akkermans AD, Saarela M, de Vos WM. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2001;67:504–513. doi: 10.1128/AEM.67.2.504-513.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kok RG, De Waal A, Schut F, Welling GW, Weenk G, et al. Specific detection and analysis of a probiotic Bifidobacterium strain in infant feces. Appl Environ Microbiol. 1996;62:3668–3672. doi: 10.1128/aem.62.10.3668-3672.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langendijk PS, Schut F, Jansen GJ, Raangs GC, Kamphuis GR, et al. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl Environ Microbiol. 1995;61:3069–3075. doi: 10.1128/aem.61.8.3069-3075.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, et al. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol. 2017;67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Orban JI, Patterson JA. Modification of the phosphoketolase assay for rapid identification of bifidobacteria. J Microbiol Methods. 2000;40:221–224. doi: 10.1016/S0167-7012(00)00133-0. [DOI] [PubMed] [Google Scholar]
- 50.Yin X, Chambers JR, Barlow K, Park AS, Wheatcroft R. The gene encoding xylulose-5-phosphate/fructose-6-phosphate phosphoketolase (xfp) is conserved among Bifidobacterium species within a more variable region of the genome and both are useful for strain identification. FEMS Microbiol Lett. 2005;246:251–257. doi: 10.1016/j.femsle.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 51.Bogorad IW, Lin T-S, Liao JC. Synthetic non-oxidative glycolysis enables complete carbon conservation. Nature. 2013;502:693–697. doi: 10.1038/nature12575. [DOI] [PubMed] [Google Scholar]
- 52.Nurk S, Bankevich A, Antipov D, Gurevich A, Korobeynikov A, et al. Assembling genomes and Mini-Metagenomes from Highly Chimeric Reads. 2013. pp. 158–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ankenbrand MJ, Keller A. bcgTree: automatized phylogenetic tree building from bacterial core genomes. Genome. 2016;59:783–791. doi: 10.1139/gen-2015-0175. [DOI] [PubMed] [Google Scholar]
- 54.Eddy S. HMMER3: a new generation of sequence homology search software. 2010. http://hmmer.janelia.org
- 55.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 57.Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 58.Dupont CL, Rusch DB, Yooseph S, Lombardo MJ, Richter RA, et al. Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. Isme J. 2012;6:1186–1199. doi: 10.1038/ismej.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ncbi RC. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2017;45:D12. doi: 10.1093/nar/gkw1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rambaut A. FigTree v1. 4. Molecular Evolution, Phylogenetics and Epidemiology. Edinburgh, UK: University of Edinburgh, Institute of Evolutionary Biology; 2012. [Google Scholar]
- 61.Robinson O, Dylus D, Dessimoz C. Phylo.io: interactive viewing and comparison of large phylogenetic trees on the web. Mol Biol Evol. 2016;33:2163–2166. doi: 10.1093/molbev/msw080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aziz RK, Bartels D, Best AA, Dejongh M, Disz T, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, et al. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST) Nucleic Acids Res. 2014;42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, et al. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep. 2015;5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen IA, Markowitz VM, Chu K, Palaniappan K, Szeto E, et al. IMG/M: integrated genome and metagenome comparative data analysis system. Nucleic Acids Res. 2017;45:D507–D516. doi: 10.1093/nar/gkw929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol. 2016;428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 67.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yin Y, Mao X, Yang J, Chen X, Mao F, et al. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012;40:W445–W451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oliveros JC. VENNY. An interactive tool for comparing lists with venn diagrams. 2007.
- 73.Ny Y, Wagner JR, Laird MR, Melli G, Rey S, et al. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schaumburg F, Dieckmann R, Schmidt-Bräkling T, Becker K, Idelevich EA. First description of an Anaerobiospirillum succiniciproducens prosthetic joint infection. New Microbes New Infect. 2017;18:1–2. doi: 10.1016/j.nmni.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hariharan H, Hariharan SH. Zoonotic bacteria associated with cats. Vet Med Open J. 2017;2:68–75. doi: 10.17140/VMOJ-2-118. [DOI] [Google Scholar]
- 76.Moraes SR, Gonçalves RB, Mouton C, Seldin L, Ferreira MC, et al. Use of rep-PCR to define genetic relatedness among Bacteroides fragilis strains. J Med Microbiol. 2000;49:279–284. doi: 10.1099/0022-1317-49-3-279. [DOI] [PubMed] [Google Scholar]
- 77.Almeida FS, Nakano V, Avila-Campos MJ. Occurrence of enterotoxigenic and nonenterotoxigenic Bacteroides fragilis in calves and evaluation of their antimicrobial susceptibility. FEMS Microbiol Lett. 2007;272:15–21. doi: 10.1111/j.1574-6968.2007.00732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu F, Guo X, Zhang J, Zhang M, Ou Z, et al. Phascolarctobacterium faecium abundant colonization in human gastrointestinal tract. Exp Ther Med. 2017;14:3122–3126. doi: 10.3892/etm.2017.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kuang YS, Lu JH, Li SH, Li JH, Yuan MY, et al. Connections between the human gut microbiome and gestational diabetes mellitus. Gigascience. 2017;6:1–12. doi: 10.1093/gigascience/gix058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chiu CM, Huang WC, Weng SL, Tseng HC, Liang C, et al. Systematic analysis of the association between gut flora and obesity through high-throughput sequencing and bioinformatics approaches. Biomed Res Int. 2014;2014:1–10. doi: 10.1155/2014/906168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lugli GA, Milani C, Turroni F, Duranti S, Ferrario C, et al. Investigation of the evolutionary development of the genus Bifidobacterium by comparative genomics. Appl Environ Microbiol. 2014;80:6383–6394. doi: 10.1128/AEM.02004-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sun Z, Zhang W, Guo C, Yang X, Liu W, et al. Comparative genomic analysis of 45 type strains of the genus Bifidobacterium: a snapshot of its genetic diversity and evolution. PLoS One. 2015;10:e0117912. doi: 10.1371/journal.pone.0117912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim M, Oh HS, Park SC, Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 84.Varghese NJ, Mukherjee S, Ivanova N, Konstantinidis KT, Mavrommatis K, et al. Microbial species delineation using whole genome sequences. Nucleic Acids Res. 2015;43:6761–6771. doi: 10.1093/nar/gkv657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Turroni F, Strati F, Foroni E, Serafini F, Duranti S, et al. Analysis of predicted carbohydrate transport systems encoded by Bifidobacterium bifidum PRL2010. Appl Environ Microbiol. 2012;78:5002–5012. doi: 10.1128/AEM.00629-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Khoroshkin MS, Leyn SA, van Sinderen D, Rodionov DA. Transcriptional Regulation of Carbohydrate Utilization Pathways in the Bifidobacterium Genus. Front Microbiol. 2016;7:120. doi: 10.3389/fmicb.2016.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gilad O, Svensson B, Viborg AH, Stuer-Lauridsen B, Jacobsen S. The extracellular proteome of Bifidobacterium animalis subsp. lactis BB-12 reveals proteins with putative roles in probiotic effects. Proteomics. 2011;11:2503–2514. doi: 10.1002/pmic.201000716. [DOI] [PubMed] [Google Scholar]
- 88.Turroni F, Milani C, van Sinderen D, Ventura M. Genetic strategies for mucin metabolism in Bifidobacterium bifidum PRL2010: an example of possible human-microbe co-evolution. Gut Microbes. 2011;2:183–189. doi: 10.4161/gmic.2.3.16105. [DOI] [PubMed] [Google Scholar]
- 89.Arrese EL, Soulages JL. Insect fat body: energy, metabolism, and regulation. Annu Rev Entomol. 2010;55:207–225. doi: 10.1146/annurev-ento-112408-085356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bottacini F, O'Connell Motherway M, Kuczynski J, O'Connell KJ, Serafini F, et al. Comparative genomics of the Bifidobacterium breve taxon. BMC Genomics. 2014;15:170. doi: 10.1186/1471-2164-15-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vollmer W, Blanot D, De Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008;32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 92.Garrido D, Ruiz-Moyano S, Mills DA. Release and utilization of N-acetyl-D-glucosamine from human milk oligosaccharides by Bifidobacterium longum subsp. infantis. Anaerobe. 2012;18:430–435. doi: 10.1016/j.anaerobe.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nishimoto M, Kitaoka M. The complete lacto-N-biose I/galacto-N-biose metabolic pathway in Bifidobacterium longum: identification of N-acetylhexosamine 1-kinase. Appl Environ Microbiol. 2007;73:6444–6449. doi: 10.1128/AEM.01425-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.J-z X, Takahashi S, Nishimoto M, Odamaki T, Yaeshima T, et al. Distribution of in vitro fermentation ability of lacto-N-biose I, a major building block of human milk oligosaccharides, in bifidobacterial strains. Appl Environ Microbiol. 2010;76:54–59. doi: 10.1128/AEM.01683-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wada J, Suzuki R, Fushinobu S, Kitaoka M, Wakagi T, et al. Purification, crystallization and preliminary X-ray analysis of the galacto-N-biose-/lacto-N-biose I-binding protein (GL-BP) of the ABC transporter from Bifidobacterium longum JCM1217. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007;63:751–753. doi: 10.1107/S1744309107036263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nishimoto M, Kitaoka M. Identification of N-acetylhexosamine 1-kinase in the complete lacto-N-biose I/galacto-N-biose metabolic pathway in Bifidobacterium longum. Appl Environ Microbiol. 2007;73:6444–6449. doi: 10.1128/AEM.01425-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Egan M, O'Connell Motherway M, Ventura M, van Sinderen D. Metabolism of sialic acid by Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2014;80:4414–4426. doi: 10.1128/AEM.01114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sánchez-Carrón G, García-García MI, López-Rodríguez AB, Jiménez-García S, Sola-Carvajal A, et al. Molecular characterization of a novel N-acetylneuraminate lyase from Lactobacillus plantarum WCFS1. Appl Environ Microbiol. 2011;77:2471–2478. doi: 10.1128/AEM.02927-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tao N, Wu S, Kim J, An HJ, Hinde K, et al. Evolutionary glycomics: characterization of milk oligosaccharides in primates. J Proteome Res. 2011;10:1548–1557. doi: 10.1021/pr1009367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Power ML, Oftedal OT, Tardif SD. Does the milk of callitrichid monkeys differ from that of larger anthropoids? Am J Primatol. 2002;56:117–127. doi: 10.1002/ajp.1068. [DOI] [PubMed] [Google Scholar]
- 101.Ward JM, Buslov AM, Vallender EJ. Twinning and survivorship of captive common marmosets (Callithrix jacchus) and cotton-top tamarins (Saguinus oedipus) J Am Assoc Lab Anim Sci. 2014;53:7–11. [PMC free article] [PubMed] [Google Scholar]
- 102.Ross AC, Porter LM, Power ML, Sodaro V. Maternal care and infant development in Callimico goeldii and Callithrix jacchus. Primates. 2010;51:315–325. doi: 10.1007/s10329-010-0196-4. [DOI] [PubMed] [Google Scholar]
- 103.Francisco TM, Lopes-Mattos KL, Picoli EA, Couto DR, Oliveira JA, et al. Feeding habits of marmosets: a case study of bark anatomy and chemical composition of Anadenanthera peregrina gum. Am J Primatol. 2017;79:1–9. doi: 10.1002/ajp.22615. [DOI] [PubMed] [Google Scholar]
- 104.Vinyard CJ, Wall CE, Williams SH, Mork AL, Armfield BA, et al. The Smallest Anthropoids. USA: Springer; 2009. The evolutionary morphology of tree gouging in marmosets; pp. 395–409. [Google Scholar]
- 105.Amann RI, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bailey MT, Coe CL. Intestinal microbial patterns of the common marmoset and rhesus macaque. Comp Biochem Physiol A Mol Integr Physiol. 2002;133:379–388. doi: 10.1016/S1095-6433(02)00174-5. [DOI] [PubMed] [Google Scholar]
- 107.de Mello MF, Monteiro AB, Fonseca EC, Pissinatti A, Ferreira AM. Identification of Helicobacter sp. in gastric mucosa from captive marmosets (Callithrix sp.; callitrichidae, primates) Am J Primatol. 2005;66:111–118. doi: 10.1002/ajp.20131. [DOI] [PubMed] [Google Scholar]
- 108.Hayashimoto N, Morita H, Inoue T, Yasuda M, Yamamoto M, et al. Draft genome sequence of enteropathogenic Escherichia coli, isolated from the bloody stool sample of a common marmoset (Callithrix jacchus) Genome Announc. 2015;3:e01161-15. doi: 10.1128/genomeA.01161-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yamazaki Y, Kawarai S, Morita H, Kikusui T, Iriki A. Faecal transplantation for the treatment of Clostridium difficile infection in a marmoset. BMC Vet Res. 2017;13:150. doi: 10.1186/s12917-017-1070-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Endo A, Futagawa-Endo Y, Dicks LM. Diversity of Lactobacillus and Bifidobacterium in feces of herbivores, omnivores and carnivores. Anaerobe. 2010;16:590–596. doi: 10.1016/j.anaerobe.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 111.Clayton JB, Vangay P, Huang H, Ward T, Hillmann BM, et al. Captivity humanizes the primate microbiome. Proc Natl Acad Sci USA. 2016;113:10376–10381. doi: 10.1073/pnas.1521835113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.