Abstract
Autism spectrum disorders (ASDs) include a group of syndromes characterized by impaired language, social and communication skills, in addition to restrictive behaviors or stereotypes. However, with a prevalence of 1.5% in developed countries and high comorbidity rates, no clear underlying mechanism that unifies the heterogeneous phenotypes of ASD exists. 5-hydroxymethylcytosine (5hmC) is highly enriched in the brain and recognized as an essential epigenetic mark in developmental and brain disorders. To explore the role of 5hmC in ASD, we used the genomic DNA isolated from the postmortem cerebellum of both ASD patients and age-matched controls to profile genome-wide distribution of 5hmC. We identified 797 age-dependent differentially hydroxymethylated regions (DhMRs) in the young group (age ≤ 18), while no significant DhMR was identified in the groups over 18 years of age. Pathway and disease association analyses demonstrated that the intragenic DhMRs were in the genes involved in cell–cell communication and neurological disorders. Also, we saw significant 5hmC changes in the larger group of psychiatric genes. Interestingly, we found that the predicted cis functions of non-coding intergenic DhMRs strikingly associate with ASD and intellectual disorders. A significant fraction of intergenic DhMRs overlapped with topologically associating domains. These results together suggest that 5hmC alteration is associated with ASD, particularly in the early development stage, and could contribute to the pathogenesis of ASD.
Introduction
Autism spectrum disorders (ASDs) encompass a broad range of neurodevelopmental problems, whose symptoms arise early in development as clinically significant functional deficits in linguistic and social aptitude, impaired communication skills, and restrictive or stereotypic behaviors (1). ASD presents in the population at a prevalence of 1.5% in developed countries, with males accounting for four times more cases than females. Moreover, they occur in various severities and different, specific combinations of behaviors (2). The development of ASD is predominantly attributed to genetic factors as shown in twin and family studies (3,4); while environmental factors, including prenatal chemical exposure, infection, inflammation, and emotional health are also reported to increase ASD risk (5). Despite the progress in identifying genetic risk factors, the heterogeneity of ASD remains a barrier to identifying the specific underlying mechanisms.
Given the most common features of ASD patients are the defects in social interactions, specific regions of cerebral cortex, such as the prefrontal cortex that is highly involved in complex cognitive behavior remain the primary research focus (6–8). However, the cerebellum, which plays fundamental roles in motor control, has non-motor functions associated with psychiatric disorders (9). The number of Purkinje cells, which provide connections to a wide range of other central nervous system structures to control movement and influence many other functions, is remarkably reduced in the brains of autism patients (10). This finding is consistent throughout postmortem studies (11,12). More convincing evidence exists in mouse models, where the Purkinje cell-specific deletion of several essential genes, such as Tsc1/2 (Tuberous sclerosis 1/2) and En2 (engrailed homeobox 2), leads to various autism-like behaviors, including impaired memory, repetitive behaviors and altered vocalizations (13–15).
DNA methylation serves as a critical epigenetic mark, as an interface between genetic and environmental factors, which modifies DNA–protein interactions that influence transcriptional states and cellular identity (16). Current studies suggest that DNA methylation is an essential player in ASD etiology. A vital clue arose from the results of an epigenomic analysis, where significant variations in DNA methylation patterns were found in ASD-discordant monozygotic twins who share the same genotype and identical environmental interactions (17). In ASD patients, previous research reported that DNA methylation was altered in different brain regions during development and involved in multiple dysregulated biological pathways (18–21). Loss of DNA methylation in particular hotspots within the genome could be prone to mutations, which are linked to various disorders, including autism (22). 5-methylcytosine (5mC) is a stable covalent modification to DNA; however, the fact that 5mC can be enzymatically modified to 5-hydroxymethylcytosine (5hmC) by the ten-eleven translocation (Tet) protein family gives a new perspective on the previously observed plasticity in 5mC-dependent regulatory processes (23,24). 5hmC plays a core role in the process of DNA demethylation, where 5hmC can be further oxidized by Tet proteins to produce 5-formylcytosine and 5-carboxylcytosine, which are quickly removed from the genome by thymine-DNA glycosylase (TDG) to initiate base excision repair (25–27). In addition to the role of transient intermediate in DNA demethylation, 5hmC is reported to be enriched and stable in the brain, bound by specific 5hmC-binding proteins and has its biological function in some developmental and neurological diseases (28–31). Few studies, however, have explored the role of 5hmC in ASD patient brains.
In this study, we used the genomic DNA isolated from the postmortem cerebellum of both ASD patients and age-matched controls to profile genome-wide distribution of 5hmC. We found a set of differentially hydroxymethylated regions (DhMRs) that are exclusive to the young group (age ≤ 18), suggesting the dynamic change of 5hmC could play essential roles in early neurodevelopment and ASD pathogenesis. Pathway and disease association analyses showed that the intragenic DhMRs identified in the young group were in the genes involved in cell–cell communication and neurological disorders. Strikingly, we found that the intergenic DhMRs could potentially have cis functions that associate with ASD and intellectual disorders (IDs). About 40% of the intergenic DhMRs overlapped with predicted enhancers. These results together suggest that 5hmC dynamic alteration is associated with ASD, particularly in the early development stage, and contributes to the pathogenesis of ASD.
Results
Identification of age-dependent DhMRs in ASD patient cerebellum
We isolated genomic DNA from the postmortem cerebellum of 17 ASD patients and 19 age-matched controls for profiling the genome-wide 5hmC distribution by employing a previously established chemical labeling and affinity purification method, coupled with high-throughput sequencing technology to explore the role of 5hmC in ASD (30).
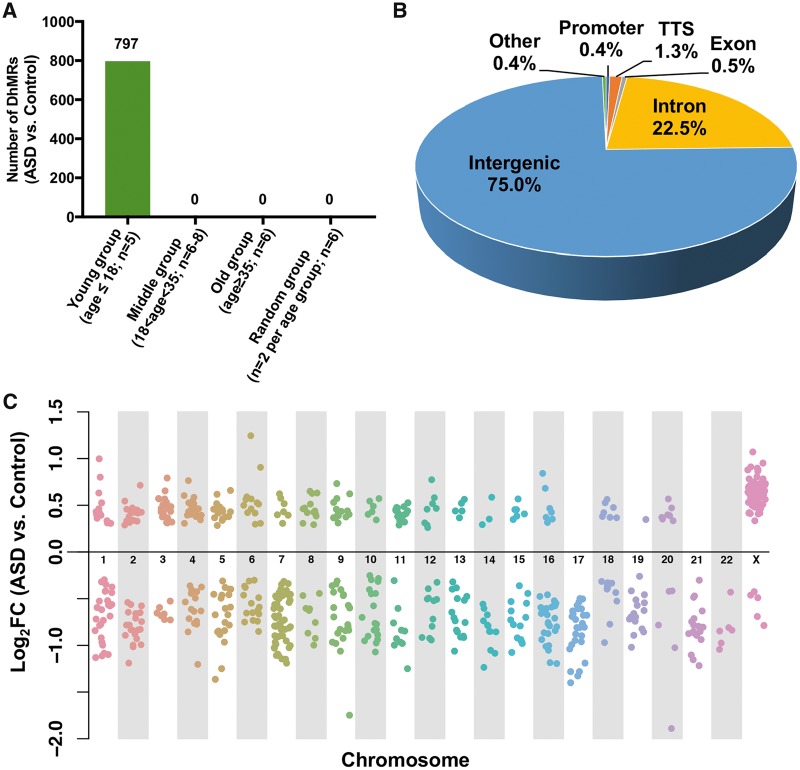
The number of 5hmC reads in each 20 kb bin of human genome (hg19) was statistically analyzed between ASD and control samples to detect DhMRs. We divided our samples into young (age ≤18), middle (18 < age < 35), and old age groups (age ≥ 35) (Supplementary Material, Fig. S1). Interestingly, we identified 797 DhMRs exclusively in the young group, but none from the two older groups or a ‘random’ group, where two samples were randomly chosen from each age group (Fig. 1A). We further examined the 797 regions in the old group and plotted the histogram of the SDs of these 797 regions in the old group, as well as in the young group for comparison (Supplementary Material, Fig. S2A). We found that these 797 regions in the old group (mean of SDs: 16.7) do not have larger variations than the young group (mean of SDs: 27.9), suggesting the pattern of 5hmC modification becomes indistinguishable in the older ASD cohorts and their controls.
Figure 1.
Identification and characterization of age-dependent DhMRs in ASD cerebellum. (A) The number of 5hmC reads in each 20 kb bin of hg19 were analyzed between ASD and control samples to find DhMRs. Only 797 DhMRs (q-value < 0.05, FDR adjusted) were found in the young group (n = 5), while no DhMR was found in the middle (n = 6–8), old (n = 6), or random groups (two samples were randomly chosen from each age group; n = 6). (B) Genomic annotation of 797 DhMRs to show their percentage of each genomic region. (C) Chromosomal distribution of DhMRs. When compared with control samples, 71% of the DhMRs show reduced 5hmC level (Log2FC < 0) in ASD samples.
Genomic annotation of these 797 DhMRs revealed that 75% of DhMRs are found within intergenic regions, while only 25% of DhMRs overlap the gene bodies, including the intron, TSS (transcription start site), promoter, and exon regions (Fig. 1B). These DhMRs, including both gain-of-5hmC and loss-of-5hmC regions, could be found on both autosome and sex chromosomes (Fig. 1C). Numbers of 5hmC reads are higher in autosomes (Mean ± SD = 51.60 ± 67.6) than in sex chromosomes (Mean ± SD = 17.76 ± 26.3) (Supplementary Material, Fig. S2B). When compared with control samples, 71% of DhMRs showed the decreased 5hmC level in ASD samples, suggesting that the maintenance of 5hmC in certain loci may be important to the pathogenesis of ASD, particularly in the early development stage. To investigate whether DhMRs are involved in genome-wide significant risk loci, we obtained ASD risk single nucleotide polymorphisms (SNPs) and their associated regions from a previously published paper (32) and found that 5hmC differentially changed in the associated regions of 3 ASD risk SNPs, including rs4392770, rs7024761 and rs2564899 (Supplementary Material, Fig. S2C). The permutation test indicated that this finding is more than chance (P-value < 0.05). Together these data indicate that, except for the partial association with 5mC, 5hmC may also independently play important roles in ASD.
DhMRs-associated genes significantly involved in ASD and other psychiatric disorders
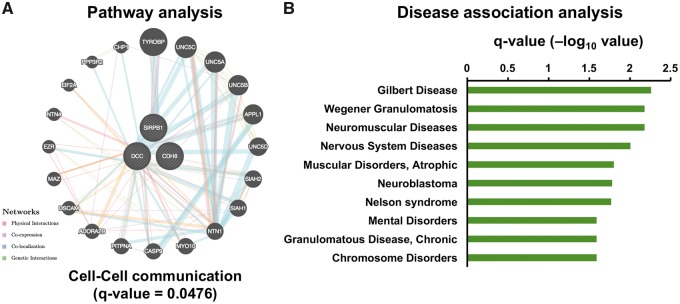
To further explore the biological relevance of these DhMRs found in the young group, we performed gene ontology (GO) analysis on the 181 genes that associate with intragenic DhMRs. Pathway analysis indicated that the intragenic DhMRs were in the genes involved in ‘Cell–Cell communication’ and in ‘Heparan sulfate biosynthesis’ (Fig. 2A andSupplementary Material, Fig. S3A). It is consistent with the fact that many ASD risk genes play roles in regulating the development of the synapse, where brain cells communicate with each other (33). Also, the heparan sulfate deficiency is associated with autism-like socio-communicative deficits and stereotypes (34). Moreover, the disease association analysis revealed a substantive relationship between DhMRs-associated genes and neurological disorders (Fig. 2B).
Figure 2.
DhMRs-associated genes involved in neurological disease. Gene ontology analysis on the pathway (A) and disease association (B) revealed that the genes with intragenic DhMRs were involved in ‘Cell–Cell communication’ pathway and several neurological disorders (q-value < 0.05, FDR adjusted).
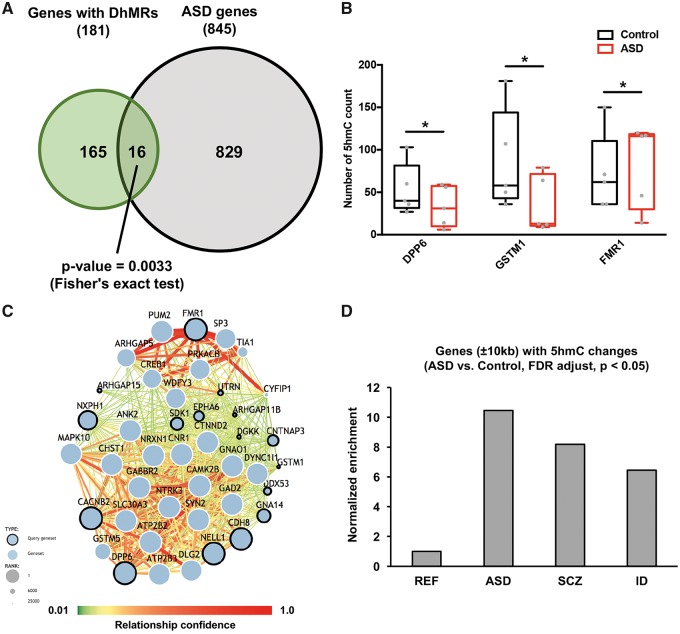
To shed light on the functional relevance of 5hmC dynamics in ASD, we overlapped 181 DhMRs-associated genes with 845 ASD risk genes (obtained from The Simons Foundation Autism Research Initiative (SFARI)). We found a significant overlap between the DhMRs-associated genes and ASD risk genes (P-value = 0.0033, Fisher’s exact test), suggesting that 5hmC may have essential roles in ASD by regulating the genes involved in ASD (Fig. 3A). The 16 overlapped genes, such as FMR1, GSTM1 and DPP6 were associated with significant gain- or loss-of-5hmC [P-value < 0.05, false discovery rate (FDR) adjusted] (Fig. 3B, Supplementary Material, Fig. S3B and C). Given the potential impact of 5hmC in ASD, we generated a predicted ASD–gene network to explore brain-specific interactions between ASD candidate genes, including the 16 DhMRs-associated genes (Fig. 3C). In the predicted network, we found that the dynamic change in 5hmC potentially influences the expression of ASD candidate genes. For instance, PUM2, a member of the Pumilio proteins, has multiple roles in neuronal functions through both translational and post-translational regulation (35–37). Interestingly, Pum2 is a mRNA target of fragile X mental retardation protein (FMRP) in mouse brains as reported in two independent studies (38,39). Therefore, the significantly increased 5hmC in FMR1 gene may influence the expression of FMRP that further affects mRNA targets of FMRP, including PUM2, and contributes to the pathogenesis of ASD.
Figure 3.
Bioinformatics analyses reveal the regulatory roles of intragenic DhMRs in ASD. (A) DhMRs-associated genes significantly overlapped with ASD risk genes (P-value = 0.0033, Fisher’s exact test). (B) 5hmC changes in three ASD risk genes, including DPP6, GSTM1 and FRM1, in ASD and control samples (*P < 0.05, FDR adjusted). (C) 5hmC-mediated regulation in ASD candidate genes interactome. Predicted ASD–gene network exploring brain-specific interactions between ASD candidate genes were generated, including 16 DhMRs-associated genes shown with the dots marked with bold lines. (D) 5hmC changes in the ±10 kb region of psychiatric genes, including ASD, SCZ and IDs risk genes. Compared to reference genes; we found a significantly higher fraction of ASD, SCZ and ID risk genes associated with 5hmC changes (P-value = 2.2e-16, Chi-square test).
We further investigated 5hmC changes in the ±10 kb region of psychiatric genes, including ASD, schizophrenia (SCZ), and ID risk genes (Fig. 3D andSupplementary Material, S3D). When compared with reference genes; we found a significantly higher number of ASD, SCZ and ID risk genes associated with 5hmC changes (P-value = 2.2e-16, Chi-square test), suggesting 5hmC may play significant roles in regulating ASD and other neuropsychiatric genes.
Predicted cis functions of intergenic DhMRs associate with ASD
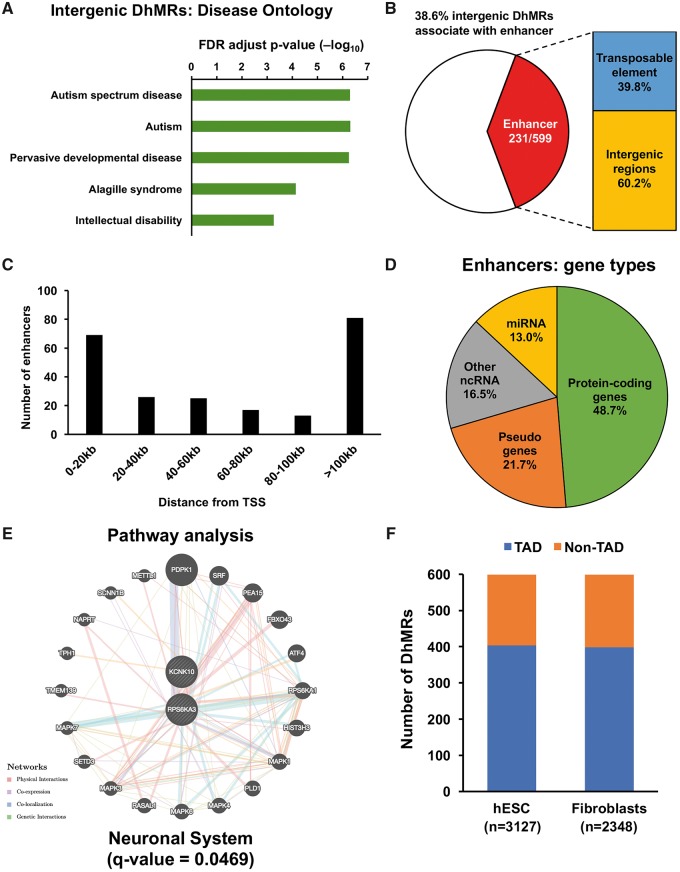
Given the fact that 75% of the DhMRs were in the intergenic regions, we next sought to investigate the cis function of these non-coding intergenic DhMRs. To this end, we used GREAT (Genomic Regions Enrichment of Annotations Tool, version 3.0), which calculates statistics by associating genomic regions with nearby genes and applying the gene annotations to the regions, to predict the cis function of 599 intergenic DhMRs. By incorporating long-range regulatory domains and genomic region-based enrichment test, GREAT analysis can highlight biologically meaningful terms and their associated cis-regulatory regions and genes (40). Strikingly, we found a significant association between the intergenic DhMRs and neurodevelopmental disorders (Fig. 4A). For example, FDR-adjusted P-values for ASD, autism, and pervasive developmental disease are <1e-6, indicating the dynamic change of 5hmC in the pathogenesis of neurodevelopmental disorders, particularly in ASD.
Figure 4.
Predicted cis-regulatory functions of intergenic DhMRs in ASD. (A) Prediction of the cis function of 599 non-coding intergenic DhMRs. A significant association was identified between the intergenic DhMRs and neurodevelopment disorders, particularly ASD and IDs. Intergenic DhMRs overlap with general enhancers in the brain. (B) These identified brain enhancers overlap with 231 out of 599 intergenic DhMRs, 40% of which were located on transposable elements. These DhMRs-associated enhancers were located in different distances from TSS, with the most abundant in 0–20 kb and >100 kb regions (C) and associated with different gene types (D). The pathway and disease association analyses indicated that those protein-coding genes (with enhancers in 100 kb from TSS) were significantly involved in the neuronal system (E). (F) A significant portion of intergenic DhMRs was associated with TADs that were found in hESC (n = 3127) and human IMR90 fibroblasts (n = 2348) cell lines.
To further explore the mechanistic roles of 5hmC in gene regulation, we investigated the overlap between the intergenic DhMRs and published general enhancer regions in the brain (41). We found that 231 intergenic DhMRs, where 40% were located on transposable elements overlapped with the predicted enhancers (Fig. 4B), suggesting 5hmC may be involved in gene regulation through influencing the function of enhancers. These enhancers are found at different distances from TSS. The most abundant were in 0–20 kb and >100 kb regions and associated with different gene types (Fig. 4C and D). The pathway and disease association analyses indicated that those protein-coding genes (with enhancers in 100 kb from TSS) were significantly involved in the neuronal system (Fig. 4E), as well as heart and neurological diseases (Supplementary Material, Fig. S4). Indeed, an unexpected genetic link exists between congenital heart disease and neurodevelopmental disorders (42).
Topologically associating domains (TADs) are cell-type independent and evolutionarily conserved genomic regions that often overlap with regulatory landscapes and their target genes. They can be associated with distinct patterns of epigenetic marks (43–45). By overlapping intergenic DhMRs with the TADs identified in human embryonic stem cells (hESCs) and human IMR90 fibroblasts (43), we found a significant proportion of intergenic DhMRs were associated with TADs (Fig. 4F). TAD-mediated GO analysis was recently proposed to be an alternative way to annotate the genome-wide non-coding regions according to function (46). By using the SNPs located in the intergenic DhMRs, we identified 329 TAD-associated genes by employing ‘TAD_Pathways Software’ (46). Consistent with the predicted cis function of intergenic DhMRs, the GO analysis indicated that these 329 TAD-associated genes were significantly involved in autistic disorders and their signal transduction pathways (Supplementary Material, Fig. S5). These data together suggest that the dynamic change of 5hmC in non-coding regions may play important roles in regulating expression of nearby genes involved in the pathogenesis of ASD.
Discussion
5hmC is critical for normal neurodevelopment. The dysregulation of 5hmC may contribute to many neurodevelopmental disorders (31). So far, however, only a small number of studies explored the role of 5hmC in autism (28,47,48). Our data show for the first time a dynamic change in 5hmC between ASD patients and age-matched controls. The fact that DhMRs are only found in the young group (age ≤ 18) suggests that this change in 5hmC plays essential roles in early neurodevelopment and ASD pathogenesis. GO analyses indicate that the intragenic DhMRs are within genes involved in cell–cell communication and neurological disorders. Interestingly, we have found that the intergenic DhMRs, 40% of which overlapped with predicted enhancers, potentially have cis functions that associate with ASD and IDs. Together, these results suggest that the dynamic change of 5hmC is associated with ASD, particularly in the early development stage, and could contribute to the pathogenesis of ASD.
Epigenetic plasticity in DNA methylation-related regulatory processes influences activity-dependent gene regulation and learning and memory in the central nervous system (49). Therefore, hydroxylation of 5mC to 5hmC presents a particularly intriguing epigenetic regulatory paradigm in the development of the mammalian brain, where its dynamic regulation is critical. Our previous study indicated that DhMRs between the fetus and the adult were highly enriched in the genes that have been implicated in autism, suggesting 5hmC-mediated epigenetic regulation may broadly impact brain development, and its dysregulation could contribute to autism (48). The present results reveal an interesting age-dependent 5hmC alteration, where DhMRs are only identified in the cerebella from the young group (age ≤ 18) of ASD patients. These DhMRs may play important roles in ASD, proving by the GO analyses that the DhMRs-associated genes are involved in neurodevelopment and significantly overlapped with ASD risk genes. Given that 5hmC is markedly increased from the early postnatal stage to adulthood (28), the significant 5hmC changes in specific loci would be critical in the early development stage and associate the pathogenesis of ASD.
Increasing attention is placed on the role of the cerebellum on nonmotor neural circuitry and cognitive development linked to ASD (50). Different from other brain regions; the cerebellum often exhibits a distinct pattern of DNA methylation (51–53). Purkinje neurons, which manage the development of the cerebellum, contain more 5hmC than any other type of neurons (23,28). Therefore, our results could have further implications for the 5hmC-mediated regulation in ASD. Methyl-CpG-binding protein 2 (MECP2) acts as a significant 5hmC binding protein in the cerebellum and can bind 5hmC and 5mC containing DNA with similar high affinities (54). Mutations in the MECP2 gene occur in 90% of patients with Rett syndrome, a severe monogenic developmental disorder with autistic phenotypes (55). Therefore, the DhMRs identified in our study may be preferentially bound by MECP2 and associate with the dysfunction of MECP2-mediated regulation in autistic phenotypes. Fragile X syndrome (FXS) is another well-known monogenic disorder associated with attention deficit, hyperactivity, aggression, and autistic behavior (56–58). Moreover, 5hmC is enriched in the FMR1 gene promoter in primary neurons derived from FXS patients (59). Consistently, our results show an increase of 5hmC in the promoter/TSS region of FMR1 in ASD patient cerebella from the young group (age ≤ 18) (Fig. 3B), suggesting the dysregulation of FMR1 in the early development stage may significantly contribute to the pathogenesis of ASD. Although these monogenic disorders collectively only account for a minority of autism cases (10–15%), a better understanding of the molecular alterations in these disorders could reveal common pathogenic pathways shared by autism.
Protein-coding DNA only accounts for barely 2% of the hg19, while a significant amount of the genomic non-coding DNA, initially recognized as ‘junk DNA,’ is actively transcribed as essential and functional contributors to gene regulation (60,61). In the present study, we show that the majority of DhMRs are in intergenic regions, of which cis functions are highly involved in ASD. Many of these intergenic DhMRs are overlapped with predicted enhancers and TADs, suggesting the dynamic change of 5hmC in non-coding regions may be highly involved in regulating the expression of non-coding DNA sequences and nearby genes. Indeed, there is increasing evidence showing that non-coding regions in the genome can play important roles in ASD. For example, genome-wide characterization of de novo mutations (DNMs) in autism revealed a significant enrichment of predicted damaging DNMs in ASD cases, of which ∼40% were in the non-coding regions, suggesting that DNMs in the non-coding regions of the genome could contribute to the etiology of ASD (62). Also, a genome-wide association study comparing thousands of autism and controls identified an ASD risk SNP (rs4307059) on chromosome 5p14.1 located on an intergenic region between CDH9 and CDH10 (63). The DNA sequence in this locus was later noted to encode an antisense non-coding RNA MSNP1AS (moesin pseudogene 1, antisense) that can bind the MSN transcript, regulate MSN expression, and contribute to ASD risk (64). Therefore, in the future, it would be important to further understand the precise roles of 5hmC in specific non-coding regions in the genome.
In summary, we profiled the genome-wide distribution of 5hmC in the cerebellum of ASD patients and age-matched controls. Our results indicated an age-dependent change of 5hmC in a young group of ASD patients. The DhMRs located within genes associated with neurodevelopment pathways and ASD risk genes. Interestingly, we found that the intergenic DhMRs were strongly associated with ASD. Together, our finding implies that the abnormal alteration of 5hmC may contribute to ASD pathogenesis.
Materials and Methods
Case materials
Frozen brain tissues from 17 ASD and 19 control individuals were acquired from the Autism Tissue Program at the Harvard Brain Tissue Resource Center and NICHD Brain and Tissue Bank for Developmental Disorders. Brain sample and individual level metadata are provided in Supplementary Material, Figure S1. Subjects with ages ≤ 18 were assigned to the Young group, those with ages between 18 and 35 were assigned to the Middle group, and those with age ≥ 35 were assigned to the Old group. To eliminate the sampling bias, we randomly picked two samples from each age group and defined those as a Random group (n = 6).
Genomic DNA preparation
Genomic DNA was isolated from brain samples with standard protocols. Tissues were homogenized on ice and then treated with proteinase K (0.667 µg/µl) in 600 μl digestion buffer (100 mm Tris-HCl, pH 8.5, 5 mm EDTA, 0.2% SDS, 200 mm NaCl) at 55°C for overnight. The second day, 600 μl of Phenol: Chloroform: Isoamyl Alcohol (25:24:1 saturated with 10 mm Tris, pH 8.0, 1 mm EDTA) (P-3803, Sigma) was added to samples, mixed completely, and centrifuged for 10 min at 12 000 rpm. The aqueous layer solution was transferred into a new Eppendorf tube, and the genomic DNA precipitated with 600 μl isopropanol. The pellet was washed with 75% ethanol, air-dried and eluted with Nuclease-Free Water (Ambion).
5hmC-specific chemical labeling, affinity purification and sequencing
5hmC enrichment was performed using a previously described procedure with an improved selective chemical labeling method (28). DNA libraries were generated following the Illumina protocol for ‘Preparing Samples for ChIP Sequencing of DNA’ (Part no. 111257047 Rev. A) using 25–50 ng of input genomic DNA or 5hmC-captured DNA to initiate the protocol. All sequencing libraries were run on Illumina Hi-seq 2000 machines.
Bioinformatics analysis
FASTQ sequence file from each sample was aligned to the Homo sapiens reference genome (hg19) using Bowtie 1.1.0, keeping unique non-duplicate genomic matches with no more than two mismatches within the first 25 bp. All subsequent data analyses were performed using R language and Bioconductor packages. We first cut the whole hg19 into equal sized bins of 20 kb. Numbers of reads overlapped with each bin were obtained to represent the level of 5hmC at the corresponding regions. Only regions with a mean 5hmC level > 2 across all samples were kept for follow-up analysis. Bin counts are compared between ASD samples and controls for DhMRs detection using the Bioconductor package DSS (65), which uses a negative binomial distribution to model the bin counts, and a Wald statistical test for testing differences in two groups. DhMRs are defined as the regions with FDR < 0.05. DSS function estNormFactor() was used to adjust the variations in total reads and alignment rates. Since the age/gender of cases and controls matched well, we did not include age and gender in the analysis. Cell composition is also excluded in the analyses since there are no systematic biases in cell composition (e.g. compositions are very different from ASD and control), thus the effects of cell composition will be removed and have no impact on the results.
Overlapping of DhMRs and different genomic features were obtained using HOMER (Hypergeometric Optimization of Motif EnRichment) software (66). A chromosome distribution plot was generated by the generic plotting function in R. WebGestalt (WEB-based GEne SeT AnaLysis Toolkit), and GeneMANIA was used for GO analysis (67,68). To investigate whether DhMRs are involved in genome-wide significant risk loci, we obtained ASD risk SNPs and their associated regions from previously published results (32). We ran a permutation test to evaluate whether our findings are more than chance. The permutation test procedures are described as follows: (i) Randomly generate 797 regions with the same lengths as our DhMRs; (ii) Overlap the simulated random regions with the ASD SNP list; (iii) Repeat steps (i) and (ii) for 100 times to obtain a null distribution for some overlaps. Predicted ASD-associated gene interaction network was performed by genome-wide predictions of autism-associated genes (http://asd.princeton.edu/; date last accessed May 26, 2018). SCZ-associated genes were obtained from SCZ Gene Resource (http://bioinfo.mc.vanderbilt.edu/SZGR/; date last accessed May 26, 2018) and Gene List Automatically Derived For You (GLAD4U) (69). ASD-associated genes were obtained from SFARI (https://gene.sfari.org/autdb/HG_Home.do; date last accessed May 26, 2018) (70). ID-associated genes were obtained from the Intellectual Disability Project (http://gfuncpathdb.ucdenver.edu/iddrc/home.php; date last accessed May 26, 2018). Human brain enhancers were obtained from the GSE40465 (41). GREAT (version 3.0) was used to predict the cis function non-coding intergenic DhMRs (40). The reported SNPs were obtained from the UCSC genome browser (https://genome.ucsc.edu/cgi-bin/hgTables; date last accessed May 26, 2018) using Common SNPs track from Variation group of hg19. In total, we found 52135 SNPs were located within the 599 intergenic DhMRs. Those SNPs were then used as input for ‘tad_pathyway_pipeline’ (https://github.com/greenelab/tad_pathways_pipeline; date last accessed May 26, 2018) to discover disease candidate genes (46).
Data and Code Availability
The sequencing data have been deposited at Gene Expression Omnibus under accession number GSE107012. The customized R codes were used in this study and are available upon request.
Supplementary Material
Acknowledgements
We would like to acknowledge the support of the Autism Tissue Program (ATP) at the Harvard Brain Tissue Resource Center and NICHD Brain and Tissue Bank for Development.
Conflict of Interest statement. None declared.
Funding
This work was supported in part by NIH grants [NS051630, NS079625, MH102690 and NS097206 to P.J. and MH098114, MH104316, HD087795, HD088007, HD077197 to Y.H.J.] and the Simons Foundation Autism Research Initiative [239320 to P.J.].
References
- 1. Kelleher R.J. 3rd, Bear M.F. (2008) The autistic neuron: troubled translation? Cell, 135, 401–406. [DOI] [PubMed] [Google Scholar]
- 2. Lyall K., Croen L., Daniels J., Fallin M.D., Ladd-Acosta C., Lee B.K., Park B.Y., Snyder N.W., Schendel D., Volk H.. et al. (2017) The changing epidemiology of autism spectrum disorders. Ann. Rev. Public Health, 38, 81–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sandin S., Lichtenstein P., Kuja-Halkola R., Hultman C., Larsson H., Reichenberg A. (2017) The heritability of autism spectrum disorder. JAMA, 318, 1182–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tick B., Bolton P., Happe F., Rutter M., Rijsdijk F. (2016) Heritability of autism spectrum disorders: a meta-analysis of twin studies. J. Child Psychol. Psychiatry, 57, 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ornoy A., Weinstein-Fudim L., Ergaz Z. (2015) Prenatal factors associated with autism spectrum disorder (ASD). Reprod. Toxicol., 56, 155–169. [DOI] [PubMed] [Google Scholar]
- 6. Gilbert S.J., Meuwese J.D.I., Towgood K.J., Frith C.D., Burgess P.W. (2009) Abnormal functional specialization within medial prefrontal cortex in high-functioning autism: a multi-voxel similarity analysis. Brain, 132, 869–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Courchesne E., Mouton P.R., Calhoun M.E., Semendeferi K., Ahrens-Barbeau C., Hallet M.J., Barnes C.C., Pierce K. (2011) Neuron number and size in prefrontal cortex of children with autism. JAMA, 306, 2001–2010. [DOI] [PubMed] [Google Scholar]
- 8. Stoner R., Chow M.L., Boyle M.P., Sunkin S.M., Mouton P.R., Roy S., Wynshaw-Boris A., Colamarino S.A., Lein E.S., Courchesne E. (2014) Patches of disorganization in the neocortex of children with autism. N. Engl. J. Med., 370, 1209–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wolf U., Rapoport M., Schweizer T.A. (2007) Evaluating the affective component of the cerebellar cognitive-affective syndrome. J. Neuropsychiatry. Clin. Nuerosci., 19, 235–235. [DOI] [PubMed] [Google Scholar]
- 10. Bauman M.L., Kemper T.L. (1996) Observations on the purkinje cells in the cerebellar vermis in autism. Int. J. Dev. Neurosci., 55, 613. [Google Scholar]
- 11. Bauman M.L., Kemper T.L. (2005) Neuroanatomic observations of the brain in autism: a review and future directions. Int. J. Dev. Neurosci., 23, 183–187. [DOI] [PubMed] [Google Scholar]
- 12. Palmen S.J., van Engeland H., Hof P.R., Schmitz C. (2004) Neuropathological findings in autism. Brain, 127, 2572–2583. [DOI] [PubMed] [Google Scholar]
- 13. Tsai P.T., Hull C., Chu Y., Greene-Colozzi E., Sadowski A.R., Leech J.M., Steinberg J., Crawley J.N., Regehr W.G., Sahin M. (2012) Autistic-like behaviour and cerebellar dysfunction in Purkinje cell Tsc1 mutant mice. Nature, 488, 647–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rossman I.T., Lin L., Morgan K.M., Digiovine M., Van Buskirk E.K., Kamdar S., Millonig J.H., Dicicco-Bloom E. (2014) Engrailed2 modulates cerebellar granule neuron precursor proliferation, differentiation and insulin-like growth factor 1 signaling during postnatal development. Mol. Autism, 5, 9.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reith R.M., McKenna J., Wu H., Hashmi S.S., Cho S.H., Dash P.K., Gambello M.J. (2013) Loss of Tsc2 in Purkinje cells is associated with autistic-like behavior in a mouse model of tuberous sclerosis complex. Neurobiol. Dis., 51, 93–103. [DOI] [PubMed] [Google Scholar]
- 16. Jaenisch R., Bird A. (2003) Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet., 33, 245–254. [DOI] [PubMed] [Google Scholar]
- 17. Wong C.C., Meaburn E.L., Ronald A., Price T.S., Jeffries A.R., Schalkwyk L.C., Plomin R., Mill J. (2014) Methylomic analysis of monozygotic twins discordant for autism spectrum disorder and related behavioural traits. Mol. Psychiatry, 19, 495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ladd-Acosta C., Hansen K.D., Briem E., Fallin M.D., Kaufmann W.E., Feinberg A.P. (2014) Common DNA methylation alterations in multiple brain regions in autism. Mol. Psychiatry, 19, 862–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nardone S., Sams D.S., Reuveni E., Getselter D., Oron O., Karpuj M., Elliott E. (2014) DNA methylation analysis of the autistic brain reveals multiple dysregulated biological pathways. Transl. Psychiatry, 4, e433.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lister R., Mukamel E.A., Nery J.R., Urich M., Puddifoot C.A., Johnson N.D., Lucero J., Huang Y., Dwork A.J., Schultz M.D.. et al. (2013) Global epigenomic reconfiguration during mammalian brain development. Science, 341, 1237905.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schroeder D.I., Lott P., Korf I., LaSalle J.M. (2011) Large-scale methylation domains mark a functional subset of neuronally expressed genes. Genome Res., 21, 1583–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li J., Harris R.A., Cheung S.W., Coarfa C., Jeong M., Goodell M.A., White L.D., Patel A., Kang S.H., Shaw C.. et al. (2012) Genomic hypomethylation in the human germline associates with selective structural mutability in the human genome. PLoS Genet., 8, e1002692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kriaucionis S., Heintz N. (2009) The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science, 324, 929–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L.. et al. (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science, 324, 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. He Y.F., Li B.Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L.. et al. (2011) Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science, 333, 1303–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ito S., Shen L., Dai Q., Wu S.C., Collins L.B., Swenberg J.A., He C., Zhang Y. (2011) Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science, 333, 1300–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maiti A., Drohat A.C. (2011) Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J. Biol. Chem., 286, 35334–35338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Szulwach K.E., Li X., Li Y., Song C.X., Wu H., Dai Q., Irier H., Upadhyay A.K., Gearing M., Levey A.I.. et al. (2011) 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci., 14, 1607–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Spruijt C.G., Gnerlich F., Smits A.H., Pfaffeneder T., Jansen P.W., Bauer C., Munzel M., Wagner M., Muller M., Khan F.. et al. (2013) Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell, 152, 1146–1159. [DOI] [PubMed] [Google Scholar]
- 30. Song C.X., Szulwach K.E., Fu Y., Dai Q., Yi C., Li X., Li Y., Chen C.H., Zhang W., Jian X.. et al. (2011) Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol., 29, 68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cheng Y., Bernstein A., Chen D., Jin P. (2015) 5-Hydroxymethylcytosine: a new player in brain disorders? Exp. Neurol., 268, 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Anney R.J.L., Ripke S., Anttila V., Grove J., Holmans P., Huang H., Klei L., Lee P.H., Medland S.E., Neale B.. et al. (2017) Meta-analysis of GWAS of over 16, 000 individuals with autism spectrum disorder highlights a novel locus at 10q24.32 and a significant overlap with schizophrenia. Mol. Autism, 8, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ebert D.H., Greenberg M.E. (2013) Activity-dependent neuronal signaling and autism spectrum disorder. Nature, 493, 327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Irie F., Badie-Mahdavi H., Yamaguchi Y. (2012) Autism-like socio-communicative deficits and stereotypies in mice lacking heparan sulfate. Proc. Natl. Acad. Sci. USA., 109, 5052–5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vessey J.P., Vaccani A., Xie Y., Dahm R., Karra D., Kiebler M.A., Macchi P. (2006) Dendritic localization of the translational repressor Pumilio 2 and its contribution to dendritic stress granules. J. Neurosci., 26, 6496–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vessey J.P., Schoderboeck L., Gingl E., Luzi E., Riefler J., Di Leva F., Karra D., Thomas S., Kiebler M.A., Macchi P. (2010) Mammalian Pumilio 2 regulates dendrite morphogenesis and synaptic function. Proc. Natl. Acad. Sci. USA., 107, 3222–3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang M., Chen D., Xia J., Han W., Cui X., Neuenkirchen N., Hermes G., Sestan N., Lin H. (2017) Post-transcriptional regulation of mouse neurogenesis by Pumilio proteins. Genes Dev., 31, 1354–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Brown V., Jin P., Ceman S., Darnell J.C., O'Donnell W.T., Tenenbaum S.A., Jin X., Feng Y., Wilkinson K.D., Keene J.D.. et al. (2001) Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell, 107, 477–487. [DOI] [PubMed] [Google Scholar]
- 39. Darnell J.C., Van Driesche S.J., Zhang C., Hung K.Y., Mele A., Fraser C.E., Stone E.F., Chen C., Fak J.J., Chi S.W.. et al. (2011) FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell, 146, 247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G. (2010) GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol., 28, 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vermunt M.W., Reinink P., Korving J., de Bruijn E., Creyghton P.M., Basak O., Geeven G., Toonen P.W., Lansu N., Meunier C.. et al. (2014) Large-scale identification of coregulated enhancer networks in the adult human brain. Cell Rep., 9, 767–779. [DOI] [PubMed] [Google Scholar]
- 42. Homsy J., Zaidi S., Shen Y., Ware J.S., Samocha K.E., Karczewski K.J., DePalma S.R., McKean D., Wakimoto H., Gorham J.. et al. (2015) De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science, 350, 1262–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B. (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature, 485, 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nora E.P., Lajoie B.R., Schulz E.G., Giorgetti L., Okamoto I., Servant N., Piolot T., van Berkum N.L., Meisig J., Sedat J.. et al. (2012) Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature, 485, 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sexton T., Yaffe E., Kenigsberg E., Bantignies F., Leblanc B., Hoichman M., Parrinello H., Tanay A., Cavalli G. (2012) Three-dimensional folding and functional organization principles of the Drosophila genome. Cell, 148, 458–472. [DOI] [PubMed] [Google Scholar]
- 46. Way G.P., Youngstrom D.W., Hankenson K.D., Greene C.S., Grant S.F.A. (2017) Implicating candidate genes at GWAS signals by leveraging topologically associating domains. Eur. J. Hum. Genet., 25, 1286–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhubi A., Chen Y., Dong E., Cook E.H., Guidotti A., Grayson D.R. (2014) Increased binding of MeCP2 to the GAD1 and RELN promoters may be mediated by an enrichment of 5-hmC in autism spectrum disorder (ASD) cerebellum. Transl. Psychiatry, 4, e349.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang T., Pan Q., Lin L., Szulwach K.E., Song C.X., He C., Wu H., Warren S.T., Jin P., Duan R.. et al. (2012) Genome-wide DNA hydroxymethylation changes are associated with neurodevelopmental genes in the developing human cerebellum. Hum. Mol. Genet., 21, 5500–5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Feng J., Zhou Y., Campbell S.L., Le T., Li E., Sweatt J.D., Silva A.J., Fan G. (2010) Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci., 13, 423–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang S.S., Kloth A.D., Badura A. (2014) The cerebellum, sensitive periods, and autism. Neuron, 83, 518–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ladd-Acosta C., Pevsner J., Sabunciyan S., Yolken R.H., Webster M.J., Dinkins T., Callinan P.A., Fan J.B., Potash J.B., Feinberg A.P. (2007) DNA methylation signatures within the human brain. Am. J. Hum. Genet., 81, 1304–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hernandez D.G., Nalls M.A., Gibbs J.R., Arepalli S., van der Brug M., Chong S., Moore M., Longo D.L., Cookson M.R., Traynor B.J.. et al. (2011) Distinct DNA methylation changes highly correlated with chronological age in the human brain. Hum. Mol. Genet., 20, 1164–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Christensen B.C., Houseman E.A., Marsit C.J., Zheng S., Wrensch M.R., Wiemels J.L., Nelson H.H., Karagas M.R., Padbury J.F., Bueno R.. et al. (2009) Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. Plos Genet., 5, e1000602.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mellen M., Ayata P., Dewell S., Kriaucionis S., Heintz N. (2012) MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell, 151, 1417–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Amir R.E., Van den Veyver I.B., Wan M., Tran C.Q., Francke U., Zoghbi H.Y. (1999) Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet., 23, 185–188. [DOI] [PubMed] [Google Scholar]
- 56. Fisch G.S., Carpenter N.J., Holden J.J., Simensen R., Howard-Peebles P.N., Maddalena A., Pandya A., Nance W. (1999) Longitudinal assessment of adaptive and maladaptive behaviors in fragile X males: growth, development, and profiles. Am. J. Med. Genet., 83, 257–263. [DOI] [PubMed] [Google Scholar]
- 57. O'Donnell W.T., Warren S.T. (2002) A decade of molecular studies of fragile X syndrome. Ann. Rev. Neurosci., 25, 315–338. [DOI] [PubMed] [Google Scholar]
- 58. Bassell G.J., Warren S.T. (2008) Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron, 60, 201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Esanov R., Andrade N.S., Bennison S., Wahlestedt C., Zeier Z. (2016) The FMR1 promoter is selectively hydroxymethylated in primary neurons of fragile X syndrome patients. Hum. Mol. Genet., 25, 4870–4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Birney E., Stamatoyannopoulos J.A., Dutta A., Guigó R., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T., Stamatoyannopoulos J.A.. et al. (2007) Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature, 447, 799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pennisi E. (2012) Genomics. ENCODE project writes eulogy for junk DNA. Science, 337, 1159, 1161–1161. [DOI] [PubMed] [Google Scholar]
- 62. Yuen R.K., Merico D., Cao H., Pellecchia G., Alipanahi B., Thiruvahindrapuram B., Tong X., Sun Y., Cao D., Zhang T.. et al. (2016) Genome-wide characteristics of de novo mutations in autism. NPJ Genom. Med., 1, 160271–1602710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang K., Zhang H.T., Ma D.Q., Bucan M., Glessner J.T., Abrahams B.S., Salyakina D., Imielinski M., Bradfield J.P., Sleiman P.M.A.. et al. (2009) Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature, 459, 528–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kerin T., Ramanathan A., Rivas K., Grepo N., Coetzee G.A., Campbell D.B. (2012) A Noncoding RNA Antisense to Moesin at 5p14.1 in Autism. Sci. Transl. Med., 4, 128ra40.. [DOI] [PubMed] [Google Scholar]
- 65. Wu H., Wang C., Wu Z. (2013) A new shrinkage estimator for dispersion improves differential expression detection in RNA-seq data. Biostatistics, 14, 232–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell, 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang J., Duncan D., Shi Z., Zhang B. (2013) WEB-based GEne SeT analysis toolkit (WebGestalt): update 2013. Nucleic Acids Res., 41, W77–W83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Warde-Farley D., Donaldson S.L., Comes O., Zuberi K., Badrawi R., Chao P., Franz M., Grouios C., Kazi F., Lopes C.T.. et al. (2010) The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res., 38, W214–W220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Jourquin J., Duncan D., Shi Z., Zhang B. (2012) GLAD4U: deriving and prioritizing gene lists from PubMed literature. BMC Genomics, 13(Suppl 8), S20.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Abrahams B.S., Arking D.E., Campbell D.B., Mefford H.C., Morrow E.M., Weiss L.A., Menashe I., Wadkins T., Banerjee-Basu S., Packer A. (2013) SFARI Gene 2.0: a community-driven knowledge base for the autism spectrum disorders (ASDs). Mol. Autism, 4, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data have been deposited at Gene Expression Omnibus under accession number GSE107012. The customized R codes were used in this study and are available upon request.