Abstract
Mutation of the gene encoding dystrophin leads to Duchenne and Becker muscular dystrophy (DMD and BMD). Currently, dystrophin is thought to function primarily as a structural protein, connecting the muscle cell actin cytoskeleton to the extra-cellular matrix. In addition to this structural role, dystrophin also plays an important role as a scaffold that organizes an array of signaling proteins including sodium, potassium, and calcium channels, kinases, and nitric oxide synthase (nNOS). Many of these signaling proteins are linked to dystrophin via syntrophin, an adapter protein that is known to bind directly to two sites in the carboxyl terminal region of dystrophin. A search of the dystrophin sequence revealed three additional potential syntrophin binding sites (SBSs) within the spectrin-like repeat (SLR) region of dystrophin. Binding assays revealed that the site at SLR 17 bound specifically to the α isoform of syntrophin while the site at SLR 22 bound specifically to the β-syntrophins. The SLR 17 α-SBS contained the core sequence known to be required for nNOS–dystrophin interaction. In vitro and in vivo assays indicate that α-syntrophin facilitates the nNOS–dystrophin interaction at this site rather than nNOS binding directly to dystrophin as previously reported. The identification of multiple SBSs within the SLR region of dystrophin demonstrates that this region functions as a signaling scaffold. The signaling role of the SLR region of dystrophin will need to be considered for effective gene replacement or exon skipping based DMD/BMD therapies.
Introduction
Duchenne and Becker muscular dystrophies are caused by the absence of a full-length form of dystrophin and the subsequent loss of its associated proteins. A main function of dystrophin in normal muscle is to organize and localize a complex of proteins at specific sites of the sarcolemma. Many proteins bind directly to dystrophin including cytoskeletal proteins (actin, microtubules, intermediate filament proteins, ankyrin B, plectin), at least one transmembrane protein (β-dystroglycan) and several intracellular proteins (dystrobrevins, syntrophins, Par-1B) [reviewed in (1)]. Dystrophin organizes these proteins into a dynamic functional complex that prevents muscle degeneration and preserves muscle health.
Traditionally, dystrophin has been divided into four domains based on structure and binding properties (2). The N-terminal domain binds actin, the cysteine rich domain contains the β-dystroglycan binding region and the C-terminal region binds dystrobrevin and syntrophin. The bulk of the protein is made of 24 spectrin-like repeats (SLRs) that originally were thought to function as a spring or shock absorber. However, recently this region has also been implicated in the binding of lipids, cholesterol and proteins. Specifically, the proteins binding the SLRs include actin filaments (3), microtubules (4), intermediate filaments (5,6), Par-1B (7) and neuronal nitric oxide synthase (nNOS) (8).
Proper localization of nNOS to the sarcolemma requires both α-syntrophin and the region of dystrophin forming SLRs 16/17. nNOS binds directly to the PDZ domain of α-syntrophin (9). Biochemical and crystallography studies show that the β-finger adjacent to the PDZ domain of nNOS binds with high affinity to the ligand-binding groove in the syntrophin PDZ domain. α-Syntrophin is required for sarcolemmal localization of nNOS as demonstrated in Snta−/− mice using both immunofluorescence and western blots of membrane enriched fractions (10,11). Furthermore, the Snta−/− mice fail to attenuate adrenergic stimulated vasoconstriction, indicating that nNOS function is also lost in the α-syntrophin−/− mice (12). The nNOS requirement for full-length dystrophin was demonstrated by similar experiments using mdx mice (a mouse model lacking dystrophin). In mdx mouse muscle, nNOS is not localized to the sarcolemma (13) and adrenergic stimulated vasoconstriction is not attenuated (14). The requirement that a portion of the rod domain of dystrophin was needed for nNOS localization was originally observed in DMD and Becker patients carrying genomic deletions that removed rod domain portions spanning SLRs 16 and 17. Muscle from these patients had sarcolemmal α-syntrophin but nNOS was absent from the sarcolemma (15). The region in dystrophin required for nNOS localization was further narrowed to SLRs 16/17 by experiments that restored this region of dystrophin in mdx mice (8).
Following the initial identification of syntrophin as a dystrophin-binding protein, three independent labs identified a region in the C-terminal domain of dystrophin as the syntrophin binding site (SBS) (16–18). The SBS is also present in the dystrophin homologs, utrophin, DRP2 and dystrobrevin. Newey et al. (19), showed that the SBS was actually comprised of two adjacent sites, allowing two syntrophins to bind dystrophin (or utrophin, DRP2, dystrobrevin). However, none of these studies searching for the SBS used full-length dystrophin, only the C-terminal region. Also, Crawford et al. showed that otherwise full-length dystrophins lacking the C-terminal domain still co-localized with syntrophin in skeletal muscles (20). We therefore investigated if additional SBSs were present in other regions of dystrophin (the N-terminal and SLR regions). Here we identify and characterize additional SBSs in dystrophin.
The results presented in this paper allow three major conclusions. First, it is α-syntrophin that links nNOS to the SLR 16/17 region of dystrophin, not nNOS binding directly to dystrophin as reported by Lai et al. (8). Second, although α-syntrophin can bind to either or both of the dystrophin C-terminal SBSs, only the α-syntrophin bound to SLR 16/17 binds nNOS. Finally, we have discovered at least one more SBS in SLR 22 of dystrophin that specifically binds β-syntrophins. These results need to be considered when designing the most effective gene therapy construct and for correcting dystrophin mutations by exon skipping or gene replacement.
Results
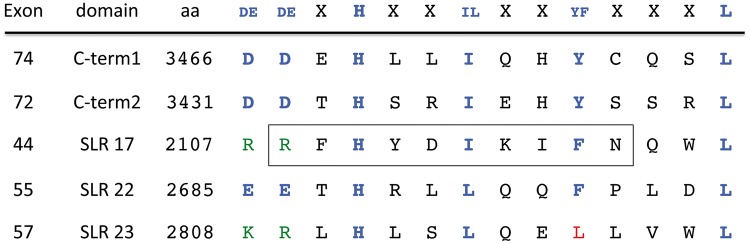
We searched for potential SBSs within dystrophin by using the Newey study as a guide (19). A similar approach had been used previously to identify a third SBS in dystrobrevin present in an alternatively spliced exon (21). We compared the two known SBSs in the dystrophin C-terminal region with those in dystrobrevin, utrophin and DRP2. We used the sequence of these proteins from multiple species (including mammals, birds, fish and insects) to develop a consensus sequence for the dystrophin family SBS (Fig. 1). The consensus sequence shows an absolutely conserved histidine residue, designated Position 0. The consensus SBS occurs in a predicted amphipathic α-helix with hydrophobic residues at Positions 3, 6 and 10. The hydrophobic residue in Position 6 is almost always a tyrosine or phenylalanine. The −2 and −3 positions are primarily charged amino acids with the majority being negatively charged aspartic and glutamic acid residues. Using this consensus sequence, we scanned full-length human dystrophin and identified a total of 5 potential SBSs (Fig. 1). The first two sites listed in Figure 1 are the previously characterized sites located in the C-terminal region (Exons 74 and 72). The remaining three potential SBSs are located in SLRs 17, 22 and 23. Each occurs in a similar position in the A-helix of the SLR. Interestingly, the SBS in repeat 17 corresponds to the minimal ‘nNOS binding’ site reported by Lai et al. (22) (boxed sequence in Fig. 1).
Figure 1.
Dystrophin contains five SBS consensus sequences. In addition to the two known SBSs in the C-terminal region, dystrophin contains three SBSs located in the spectrin-like repeats (SLRs). The SBS located in SLR-17 encompasses the minimal sequence required for nNOS association with dystrophin (22) (boxed region).
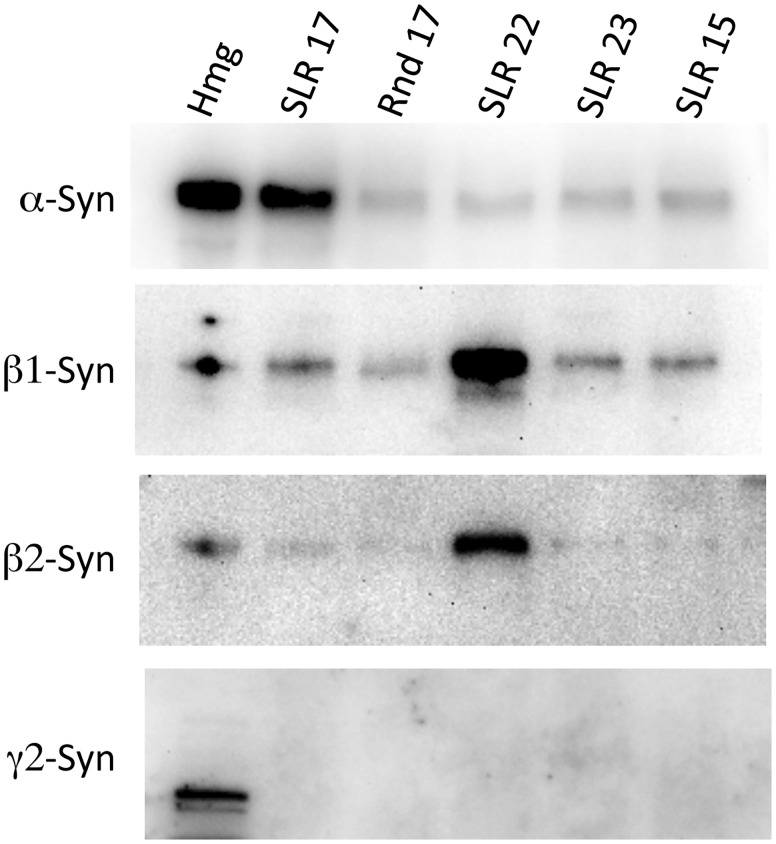
To confirm that these putative SBSs are, in fact, capable of binding syntrophin we synthesized peptides corresponding to each of the new SBSs and two control peptides (randomized SLR 17 sequence, and the corresponding A-helix sequence of repeat 15). The synthesized peptides were biotinylated at the N-terminus and used to ‘pull down’ binding partners from mouse muscle homogenates. Western blot analysis of the proteins pulled down using each peptide is shown in Figure 2. The SBS in repeat 17 was the only SLR SBS to bind specifically to α-syntrophin. The SBS in repeat 22 bound to both β1 and β2 syntrophin but did not bind α-syntrophin. The isoform binding specificity provides additional evidence that the pulldown experiments are not artifactual. No syntrophin binding was observed with the potential SBS from repeat 23 under the conditions used.
Figure 2.
α- and β-Syntrophins bind specifically to different repeats. Peptides corresponding to the SBSs in SLRs were used to pull down syntrophins from mouse muscle homogenates. The associated syntrophins were identified by western blot. Hmg—muscle homogenate before pull down assay, Rnd 17—peptide from SLR-17 with amino acids in random order, SLR 15—negative control peptide with SLR 15 sequence that does not contain a SBS.
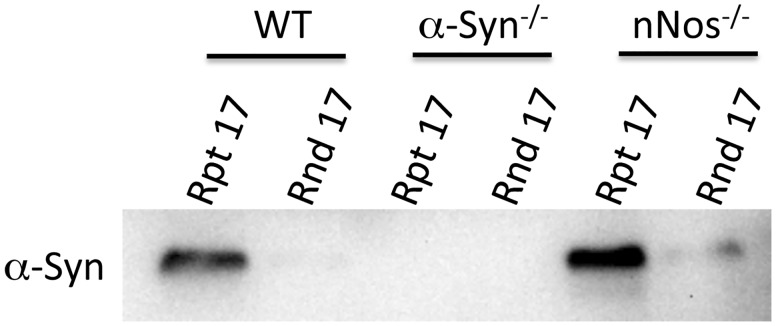
After establishing that repeat 17 SBS can bind to α-syntrophin in wild-type mouse muscle homogenates, we tested if this binding was dependent on the presence of nNOS. This is necessary since it is possible that α-syntrophin only binds to the 17 SBS in the presence of nNOS. We repeated the pull down experiments using mouse muscle homogenates from wild-type (positive control), α-syntrophin KO (negative control) and nNOS KO mice (Fig. 3). Even in the absence of nNOS, α-syntrophin binds to repeat 17 SBS.
Figure 3.
α-Syntrophin binding to SLR 17 does not require nNOS. The SLR-17 SBS peptide was used in mouse muscle homogenate pull down assays and α-syntrophin was detected by western blot. Homogenates were generated from C57bl6 (WT), α-syntrophin knockout (Snta−/−) and nNOS knockout (Nos1−/−) mice.
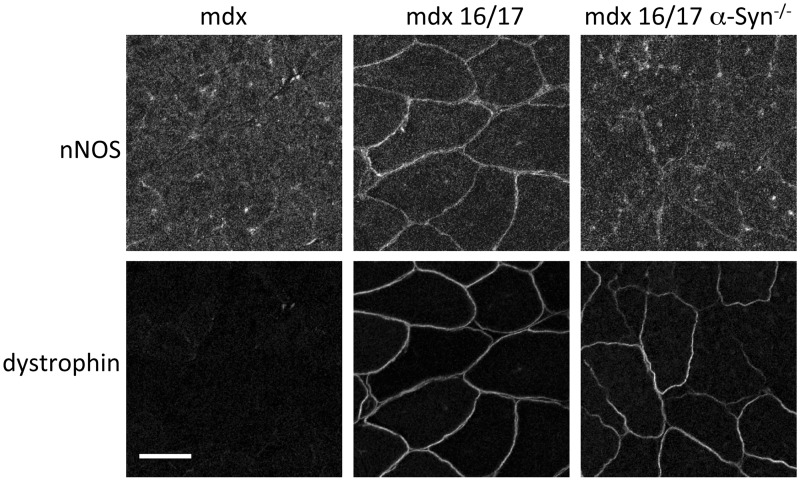
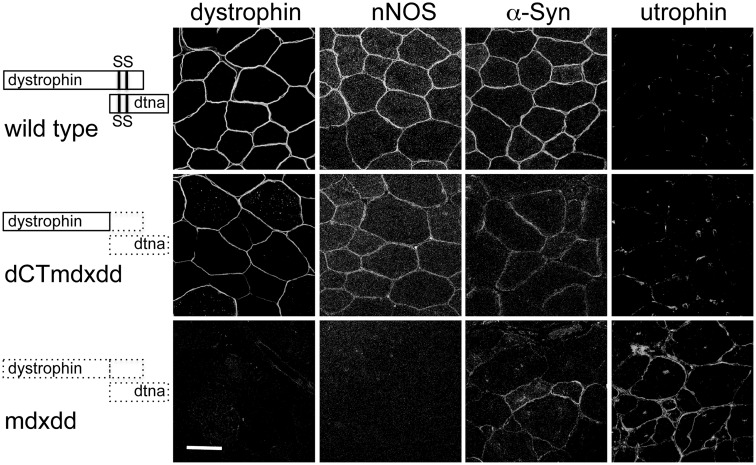
To test these in vitro experiments in a more in vivo setting, we used several mdx mice lines expressing microdystrophins. Previous studies showed that nNOS was restored to the sarcolemma of mdx mice by AAV mediated expression of a microdystrophin construct that included repeats 16 and 17 (22,23). We investigated if nNOS restoration by this microdystrophin construct also required α-syntrophin. We expressed the same microdystrophin construct containing repeats 16/17 in both mdx and mdx/Snta−/− mice (mice lacking dystrophin or dystrophin and α-syntrophin, respectively). Immunofluorescence of muscle sections showed that restoration of nNOS at the mdx sarcolemma required α-syntrophin (Fig. 4).
Figure 4.
Restoration of nNOS to mdx sarcolemma by AAV-microdystrophin requires α-syntrophin. AAV-microdystrophin containing SLRs 16 and 17 was injected into the TA muscle of mdx and mdx/Snta−/− mice. Immunofluorescence of cryosectioned muscle showed that both treated strains expressed the microdystrophin but nNOS was only restored to the sarcolemma in the mdx strain that expressed α-syntrophin. Scale bar = 50 μm.
Investigation of the binding of α-syntrophin to the SBS in SLR-17 in vivo presented several challenges. Endogenous α-SBSs on both dystrophin and on other dystrophin family member proteins (dystrobrevins, utrophin) must be considered. Previously, we generated a transgenic mouse line that expresses a modified dystrophin that is lacking the deleted C-terminal domain (dCT) on the mdx background (20). Both the previously known SBSs and the dystrobrevin binding site are missing in these mice. Surprisingly, at the time, characterization of this mouse showed that it did retain some α-syntrophin on the sarcolemma. However, it could not be concluded that this α-syntrophin was binding to the SLR region since this mouse also retained some α-dystrobrevin at the sarcolemma that could potentially recruit the α-syntrophin. To address this issue, we bred this mouse onto the α-dystrobrevin KO background producing a dCT:mdx:dtn−/− mouse (see Fig. 5). Immunofluorescence microscopy of the TA muscle of these mice and control mdx:dtn−/− mice show that the dCT transgene restores both nNOS and α-syntrophin to the sarcolemma (Fig. 5). This association of α-syntrophin with dCT dystrophin in vivo is consistent with α-syntrophin binding to SLR-17. We did observe α-syntrophin sarcolemmal labeling in the mdx:dtn−/− control. In the absence of dystrophin (or in this case absence of dCT dystrophin), utrophin is upregulated. The α-syntrophin observed in the control most likely is binding to the C-terminal region of utrophin. While utrophin lacks the SBS consensus sequence analogous to that in dystrophin SLR 17 and does not recruit nNOS to the sarcolemma (15), the C-terminal SBS can still bind α-syntrophin.
Figure 5.
Both α-syntrophin and nNOS are restored to the mdx sarcolemma by a dystrophin transgene lacking the C-terminal SBSs and α-dystrobrevin. Immunofluorescence of gastrocnemious muscle cross sections shows that α-syntrophin and nNOS are restored to the sarcolemma of mdx mice expressing dystrophin lacking the C-terminal region (dCT) and lacking α-dystrobrevin (dd). Scale bar = 100 μm.
The in vitro and in vivo experiments described above validate that the SBS consensus sequences in the SLR repeat region of dystrophin bind specifically to syntrophins. We searched for SBSs in other proteins. Table 1 lists 17 proteins that contain SBS that are conserved across species. The first three proteins listed (KIAA 0556, VPS13C and ATP11C) were previously identified as potentially linked to syntrophin. Lyssand et al. (24) used a TAP Tag strategy to isolate syntrophins expressed in HEK cells and identified potential interacting proteins by mass spectroscopy. These three proteins were identified in that study. The second three proteins (ATR, SULT1C3 and DST) have also been linked to syntrophin in a previous study. Steen (25) performed gene chip analysis comparing wild-type and α-syntrophin−/− muscle RNA expression. The levels of RNA encoding each of these three proteins showed significant differences in expression in the α-syntrophin−/− muscle. The remaining potential syntrophin binding proteins are listed in the table because their SBS consensus sequence is conserved across species. The authenticity and significance of syntrophin binding to these proteins remain to be determined.
Table 1.
Potential SBSs in proteins other than members of the dystrophin family
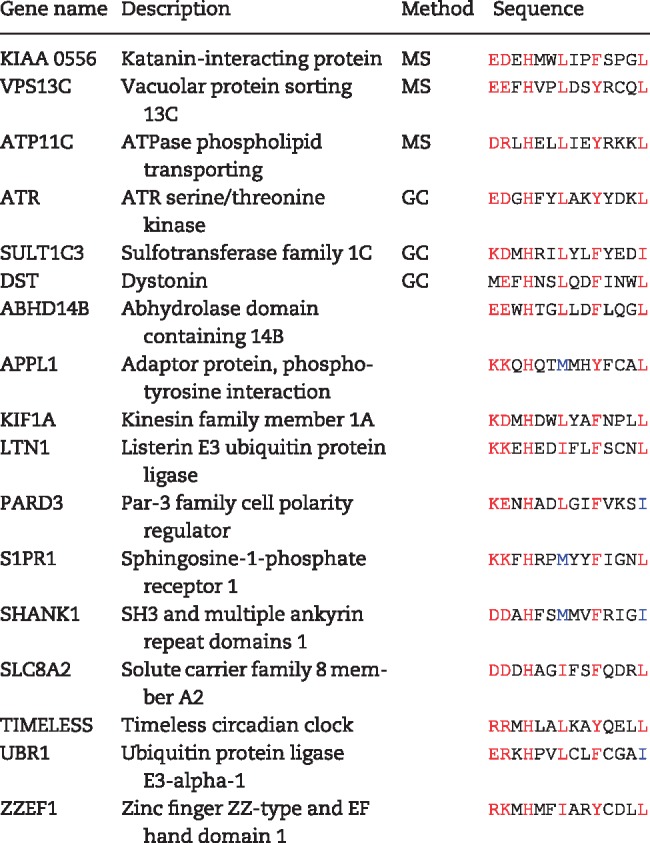 |
MS, mass spectroscopy; GC, gene chip.
Discussion
The findings described above lead us to three main conclusions. First, α-syntrophin specifically binds to SLR 17 in dystrophin and mediates the binding of nNOS in skeletal muscle. Second, the α-syntrophin bound to SLR 17 is the only dystrophin-associated syntrophin that binds nNOS: the syntrophins attached to the other SBSs do not bind nNOS. Third, the SLR region of dystrophin contains two additional SBS consensus sequences, one of which (SLR 22) specifically binds β-syntrophins. These findings strongly support the concept that the repeats are not simply ‘springs’ or ‘shock absorbers’, but are also capable of assembling adapters and signaling proteins that coordinate signaling pathways in time and space.
We have previously used pull down assays to determine which syntrophins bind to the two C-terminal SBSs in dystrophin family members (dystrophin, utrophin, dystrobrevin and DRP2) (26). Both sites in dystrophin bound α, β1 and β2 syntrophin with no indication of isoform specificity. In contrast, the current study shows specific syntrophin interactions among the SBSs located in the SLRs. SLR 22 bound only to the beta syntrophins (both β1 and β2) while SLR 17 bound only α-syntrophin. The potential SBS in SLR 23 did not show any binding to syntrophins in our pull down assays. This site (which contains a non-ideal leucine in Position 6) may not bind to syntrophins or do so only under specific (as yet unidentified) conditions.
The specific syntrophin binding to sites in SLR 17 and 22 may lead to the localization of unique PDZ ligands to these locations. The specific ligand binding to the beta syntrophins at SLR 22 is not known but several PDZ ligands are known to preferentially bind to the beta syntrophins, including ABCA1 (27) and ICA512 (28). The most obvious PDZ ligand binding to α-syntrophin at SLR 17 is nNOS. The sarcolemmal localization of nNOS depends on the presence of α-syntrophin as demonstrated in α-syntrophin null mice (10,11). Sarcolemmal nNOS is also dependent on the presence of the SLR 17 as demonstrated in both Becker patients (15) and mouse studies (8,22). In fact, the core amino acid sequence responsible for nNOS association with dystrophin (22) is embedded within the SLR-17 SBS (Fig. 1). The N-terminal region of nNOS and the α-syntrophin PDZ domain have been co-crystalized and the crystal structure revealed that the α-syntrophin PDZ domain binds directly to a β-finger region of nNOS (9). Together, these data demonstrate that α-syntrophin bound to SLR 17 binds nNOS and localizes it to the sarcolemma.
Additional data indicate that only the α-syntrophin bound to the SLR 17 SBS localizes nNOS. α-Syntrophin bound to either of the C-terminal two binding sites does not appear to bind nNOS. Transgenic mdx mice expressing the dystrophin C-terminal SBSs restore α-syntrophin to the sarcolemma but fail to restore nNOS (29). Similarly, AAV constructs that express C-terminal SBSs but not the SLR 17 SBS fail to restore nNOS but do express α-syntrophin on the sarcolemma (30). Lai et al. (22) showed that removal of the C-terminal dystrophin region (including both SBSs) does not appreciably affect the levels of nNOS associated with the membrane. This explains why some Becker patients have syntrophin on the sarcolemma but are missing nNOS (15).
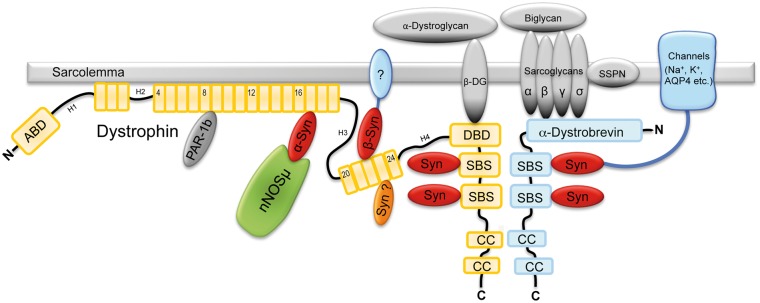
The α-syntrophin bound to SLR 17 binds nNOS while α-syntrophin bound to the C-terminal SBSs does not bind nNOS (Fig. 6). How does the location of the SBS affect the PDZ ligand affinity? While we cannot fully answer this question here, we do have several clues as to how this may happen. First, one must remember that the interaction of the internal nNOS β-finger with the α-syntrophin PDZ domain is unique and different than the normal C-terminal PDZ ligand utilized by other syntrophin PDZ domain interactors (sodium channels, potassium channels, aquaporin-4, etc.). The nNOS β-finger is a loop and requires more space than a C-terminal ligand therefore the α-syntrophin PDZ domain must be in a slightly different conformation to accommodate the nNOS β-finger. Second, the SBS consensus sequence shows that the −2 and −3 positions are normally negatively charged amino acids (D, E) but in the SLR 17 SBS they are both positively charged arginine. This may alter the α-syntrophin confirmation to facilitate binding of the nNOS β-finger. Finally, the N-terminal region of α-syntrophin (PH1a) appears to be involved in the nNOS binding. In a previous study we showed that transgenic mice that express PH1a-deleted α-syntrophin fail to restore sarcolemmal nNOS in Snta−/− mice (31). This is despite the transgene localizing to the sarcolemma and expressing a functional PDZ domain. Functionality of the PDZ domain was demonstrated by its ability to restore sarcolemmal aquaporin-4 which is normally absent in Snta−/− mice.
Figure 6.
Model of syntrophin/dystrophin interactions. α-Syntrophin can bind to SLR-17 and to either C-terminal SBSs but only the α-syntrophin at SLR-17 binds nNOS. β1/β2-Syntrophins can bind to SLR-22 and to either C-terminal SBSs.
Based largely on the observation that α-syntrophin at the C-terminal SBSs does not bind nNOS and on experiments with microdystrophin containing SLR16/17, the idea that nNOS binds directly to dystrophin at this region was proposed. The direct binding of nNOS to dystrophin has been generally accepted despite the inability of this model to address the fundamental question of why α-syntrophin would be required for sarcolemmal localization if nNOS binds directly to dystrophin. The only evidence for a direct interaction comes from yeast two hybrid experiments that showed that repeats 16/17 gave a positive Y2H interaction while repeat 16 or 17 alone did not (8). The more appropriate control would have been another repeat dimer (e.g. 14/15) with a similar coiled-coil structure. The repeats are coiled coils which are notorious for providing false positive Y2H results (32). In light of our discovery of an SBS in dystrophin SLR 17, it is now apparent that the nNOS/dystrophin interaction is mediated via α-syntrophin at the SLR 17 site.
Discovery of at least two SBSs embedded within the SLR domain unleashes new possibilities for potential protein interactions with dystrophin. While the SLRs are known to interact with Par-1b (SLR 8/9) (7) and with cytoskeletal proteins (actin, intermediate filaments and microtubules) this dystrophin region has been largely viewed as a structural ‘spring’. Our data suggest that the SLR region may also serve as a scaffold for protein interactions that may be crucial for the function of dystrophin in muscle. Identifying and characterizing proteins/pathways interaction with the SLRs of dystrophin will be important for optimizing DMD gene therapy constructs and other treatments of this disease. AAV constructs with both the SLR 16/17 and SLR 22 SBSs may show greater efficacy than current constructs.
Finally, the refinement and validation of the consensus syntrophin binding sequence has allowed us to search for SBS in proteins outside of the dystrophin protein family. Based on our identification of six proteins previously linked to syntrophin, it is likely that many proteins other than the dystrophins utilize syntrophin as an adapter protein to increase functionality.
Materials and Methods
Mice
The control strain C57Bl/6, nNOS−/− mice and mdx mice were obtained from Jackson Labs (Bar Harbor, ME). Dtna−/− mice (33) were a gift from Drs Josh Sanes and Mark Grady. We have previously generated the Snta−/− mice (34) and the transgenic mice expressing dystrophin with the C-terminal region deleted (designated dCT) (20). All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Washington.
Antibodies
We have previously characterized our rabbit antibodies to α-syntrophin, β1-syntrophin, β2-syntrophin (35), γ2-syntrophin (26) and utrophin (36). The dystrophin antibody used was MANEX1011b (1C7) (U. of Iowa, Developmental Studies Hybridoma Bank, deposited by Morris, G.E.) conjugated with Alexa Flour-488 (DyLight, Thermo). The nNOS rabbit antibody was purchased from ThermoFisher Scientific (Waltham, MA, USA).
Peptides
Biotinylated peptides were synthesized (Anaspec, Fremont, CA, USA) with sequence corresponding to the SBS regions of human dystrophin SLRs 17, 22 and 23. Additionally two control peptides were synthesized, one corresponding to the N-terminal region of SLR 15 [The A-helix (37) part of the SLR three-helix bundle that contains the SBS sequence in SLR17], and a peptide with the same amino acid composition as the SLR 17 peptide but in random order. The peptide sequences are:
SLR 17—Biotin-GSVEKWRRFHYDIKIFNQWLTEAE
SLR 22—Biotin-GREAALEETHRLLQQFPLDLEKFL
SLR 23—Biotin-GSSDQWKRLHLSLQELLVWLQLKD
SLR 15—Biotin-GISHQWYQYKRQADDLLKCLDDIE
SLR 17 randomized—Biotin-GHFIRSKNWFWEQERIETDYVLAK
Pull down assays
Streptavidin magnetic beads (ThermoFisher Scientific) were incubated 12–16 h at 4°C with mouse muscle membrane enriched homogenates. Homogenates were generated by homogenizing mouse skeletal muscle and bone in 10 volumes of buffer 1 [phosphate buffered saline (PBS), pH 7.4, 1 mM EDTA, with protease inhibitors (ThermoFisher Scientific)], centrifuging for 15 min at 8000×g, removing the supernatant and resuspending the membrane enriched pellet in buffer 1 plus 0.5% Tween 20 and re-centrifuging for 25 min at 37 000×g. Beads were washed six times with 1 ml of buffer 1 and the sample eluted in 70 μl of sample prep buffer for western blot analysis performed as described previously (38).
Immunofluorescence
Mouse tibialis anterior (TA) and gastrocnemius muscle was snap frozen in liquid nitrogen cooled isopentane. Cryosections (10 μm) were treated as described (34) and incubated with the designated primary antibody and detected with a secondary antibody (goat anti-rabbit) labeled with either Alexa 555 or Alexa 488 fluorophores (ThermoFisher Scientific). Imaging was performed using a Leica SL confocal microscope (W.M. Keck Microscopy Center, University of Washington).
AAV
The expression construct, adapted from a second generation microdystrophin (39,40) (ΔR2–R15/ΔR18–R23/ΔCT) was driven by the immediate early promoter and enhancer of cytomegalovirus (CMV), and contained the N-terminal domain, SLRs 1, 16, 17 and 24 followed by the CR domain and the rabbit β-globin poly-adenylation signal. Thirty units of AAV (1× 1011 vector genomes) were injected intramuscularly in the TA and gastrocnemius muscles of the right leg of 8-week-old mdx or mdx/Snta−/− mice. The left (uninjected) muscle served as a negative control. Four weeks following injection, the hindlimb muscles were analyzed by immunofluorescence.
Funding
This work was supported by grants from Parent Project Muscular Dystrophy; The Raymond and Beverly Sackler Foundation; and National Institutes of Health [NS14871 to S.F., HL122332 to J.C.]. Viral vectors were developed and produced in the University of Washington Senator Paul D. Wellstone Muscular Dystrophy Cooperative Research Center [U54 AR065139 to J.C.].
Conflict of Interest statement. None declared.
References
- 1. Allen D.G., Whitehead N.P., Froehner S.C. (2016) Absence of dystrophin disrupts skeletal muscle signaling: roles of Ca2+, reactive oxygen species, and nitric oxide in the development of muscular dystrophy. Physiol. Rev., 96, 253–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Koenig M., Monaco A.P., Kunkel L.M. (1988) The complete sequence of dystrophin predicts a rod-shaped cytoskeletal protein. Cell, 53, 219–228. [DOI] [PubMed] [Google Scholar]
- 3. Rybakova I.N., Amann K.J., Ervasti J.M. (1996) A new model for the interaction of dystrophin with F-actin. J. Cell Biol., 135, 661–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Belanto J.J., Olthoff J.T., Mader T.L., Chamberlain C.M., Nelson D.M., McCourt P.M., Talsness D.M., Gundersen G.G., Lowe D.A., Ervasti J.M. (2016) Independent variability of microtubule perturbations associated with dystrophinopathy. Hum. Mol. Genet., 25, 4951–4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rezniczek G.A., Konieczny P., Nikolic B., Reipert S., Schneller D., Abrahamsberg C., Davies K.E., Winder S.J., Wiche G. (2007) Plectin 1f scaffolding at the sarcolemma of dystrophic (mdx) muscle fibers through multiple interactions with beta-dystroglycan. J. Cell Biol., 176, 965–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bhosle R.C., Michele D.E., Campbell K.P., Li Z., Robson R.M. (2006) Interactions of intermediate filament protein synemin with dystrophin and utrophin. Biochem. Biophys. Res. Commun., 346, 768–777. [DOI] [PubMed] [Google Scholar]
- 7. Yamashita K., Suzuki A., Satoh Y., Ide M., Amano Y., Masuda-Hirata M., Hayashi Y.K., Hamada K., Ogata K., Ohno S. (2010) The 8th and 9th tandem spectrin-like repeats of utrophin cooperatively form a functional unit to interact with polarity-regulating kinase PAR-1b. Biochem. Biophys. Res. Commun., 391, 812–817. [DOI] [PubMed] [Google Scholar]
- 8. Lai Y., Thomas G.D., Yue Y., Yang H.T., Li D., Long C., Judge L., Bostick B., Chamberlain J.S., Terjung R.L., Duan D. (2009) Dystrophins carrying spectrin-like repeats 16 and 17 anchor nNOS to the sarcolemma and enhance exercise performance in a mouse model of muscular dystrophy. J. Clin. Invest., 119, 624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hillier B.J., Christopherson K.S., Prehoda K.E., Bredt D.S., Lim W.A. (1999) Unexpected modes of PDZ scaffolding revealed by structure of nNOS-syntrophin complex. Science, 284, 812–815. [PubMed] [Google Scholar]
- 10. Kameya S., Miyagoe Y., Nonaka I., Ikemoto T., Endo M., Hanaoka K., Nabeshima Y., Takeda S. (1999) Alpha-syntrophin gene disruption results in the absence of neuronal-type nitric oxide synthase at the sarcolemma but does not induce muscle degeneration. J. Biol. Chem., 274, 2193–2200. [DOI] [PubMed] [Google Scholar]
- 11. Adams M.E., Kramarcy N., Krall S.P., Rossi S.G., Rotundo R.L., Sealock R., Froehner S.C. (2000) Absence of alpha-syntrophin leads to structurally aberrant neuromuscular synapses deficient in utrophin. J. Cell Biol., 150, 1385–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thomas G.D., Shaul P.W., Yuhanna I.S., Froehner S.C., Adams M.E. (2003) Vasomodulation by skeletal muscle-derived nitric oxide requires alpha-syntrophin-mediated sarcolemmal localization of neuronal Nitric oxide synthase. Circ. Res., 92, 554–560. [DOI] [PubMed] [Google Scholar]
- 13. Brenman J.E., Chao D.S., Xia H., Aldape K., Bredt D.S. (1995) Nitric oxide synthase complexed with dystrophin and absent from skeletal muscle sarcolemma in Duchenne muscular dystrophy. Cell, 82, 743–752. [DOI] [PubMed] [Google Scholar]
- 14. Thomas G.D., Sander M., Lau K.S., Huang P.L., Stull J.T., Victor R.G. (1998) Impaired metabolic modulation of alpha-adrenergic vasoconstriction in dystrophin-deficient skeletal muscle. Proc. Natl. Acad. Sci. USA., 95, 15090–15095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chao D.S., Gorospe J.R., Brenman J.E., Rafael J.A., Peters M.F., Froehner S.C., Hoffman E.P., Chamberlain J.S., Bredt D.S. (1996) Selective loss of sarcolemmal nitric oxide synthase in Becker muscular dystrophy. J. Exp. Med., 184, 609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Suzuki A., Yoshida M., Ozawa E. (1995) Mammalian alpha 1- and beta 1-syntrophin bind to the alternative splice-prone region of the dystrophin COOH terminus. J. Cell Biol., 128, 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ahn A.H., Kunkel L.M. (1995) Syntrophin binds to an alternatively spliced exon of dystrophin. J. Cell Biol., 128, 363–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang B., Jung D., Rafael J.A., Chamberlain J.S., Campbell K.P. (1995) Identification of alpha-syntrophin binding to syntrophin triplet, dystrophin, and utrophin. J. Biol. Chem., 270, 4975–4978. [DOI] [PubMed] [Google Scholar]
- 19. Newey S.E., Benson M.A., Ponting C.P., Davies K.E., Blake D.J. (2000) Alternative splicing of dystrobrevin regulates the stoichiometry of syntrophin binding to the dystrophin protein complex. Curr. Biol., 10, 1295–1298. [DOI] [PubMed] [Google Scholar]
- 20. Crawford G.E., Faulkner J.A., Crosbie R.H., Campbell K.P., Froehner S.C., Chamberlain J.S. (2000) Assembly of the dystrophin-associated protein complex does not require the dystrophin COOH-terminal domain. J. Cell Biol., 150, 1399–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bohm S.V., Constantinou P., Tan S., Jin H., Roberts R.G. (2009) Profound human/mouse differences in alpha-dystrobrevin isoforms: a novel syntrophin-binding site and promoter missing in mouse and rat. BMC Biol., 7, 85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lai Y., Zhao J., Yue Y., Duan D. (2013) alpha2 and alpha3 helices of dystrophin R16 and R17 frame a microdomain in the alpha1 helix of dystrophin R17 for neuronal NOS binding. Proc. Natl. Acad. Sci. USA., 110, 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hakim C.H., Wasala N.B., Pan X., Kodippili K., Yue Y., Zhang K., Yao G., Haffner B., Duan S.X., Ramos J.. et al. (2017) A five-repeat micro-dystrophin gene ameliorated dystrophic phenotype in the severe DBA/2J-mdx model of Duchenne muscular dystrophy. Mol. Ther. Methods Clin. Dev., 6, 216–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lyssand J.S., Lee K.S., DeFino M., Adams M.E., Hague C. (2011) Syntrophin isoforms play specific functional roles in the alpha1D-adrenergic receptor/DAPC signalosome. Biochem. Biophys. Res. Commun., 412, 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Steen M.S. (2008) In Physiology and Biophysics. Doctor of Philosophy, University of Washington, pp. 28–85. [Google Scholar]
- 26. Alessi A., Bragg A.D., Percival J.M., Yoo J., Albrecht D.E., Froehner S.C., Adams M.E. (2006) Gamma-syntrophin scaffolding is spatially and functionally distinct from that of the alpha/beta syntrophins. Exp. Cell Res., 312, 3084–3095. [DOI] [PubMed] [Google Scholar]
- 27. Buechler C., Boettcher A., Bared S.M., Probst M.C., Schmitz G. (2002) The carboxyterminus of the ATP-binding cassette transporter A1 interacts with a beta2-syntrophin/utrophin complex. Biochem. Biophys. Res. Commun., 293, 759–765. [DOI] [PubMed] [Google Scholar]
- 28. Ort T., Maksimova E., Dirkx R., Kachinsky A.M., Berghs S., Froehner S.C., Solimena M. (2000) The receptor tyrosine phosphatase-like protein ICA512 binds the PDZ domains of beta2-syntrophin and nNOS in pancreatic beta-cells. Eur. J. Cell Biol., 79, 621–630. [DOI] [PubMed] [Google Scholar]
- 29. Judge L.M., Haraguchiln M., Chamberlain J.S. (2006) Dissecting the signaling and mechanical functions of the dystrophin-glycoprotein complex. J. Cell Sci., 119, 1537–1546. [DOI] [PubMed] [Google Scholar]
- 30. Yue Y., Liu M., Duan D. (2006) C-terminal-truncated microdystrophin recruits dystrobrevin and syntrophin to the dystrophin-associated glycoprotein complex and reduces muscular dystrophy in symptomatic utrophin/dystrophin double-knockout mice. Mol. Ther., 14, 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Adams M.E., Anderson K.N., Froehner S.C. (2010) The alpha-syntrophin PH and PDZ domains scaffold acetylcholine receptors, utrophin, and neuronal nitric oxide synthase at the neuromuscular junction. J. Neurosci., 30, 11004–11010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Serebriiskii I.G., Golemis E.A. (2001) Two-hybrid system and false positives. In MacDonald P.N. (ed.), Methods in Molecular Biology. Humana Press, Inc, Totowa, NJ, Vol. 177, pp. 123–134. [DOI] [PubMed] [Google Scholar]
- 33. Grady R.M., Grange R.W., Lau K.S., Maimone M.M., Nichol M.C., Stull J.T., Sanes J.R. (1999) Role for alpha-dystrobrevin in the pathogenesis of dystrophin-dependent muscular dystrophies. Nat. Cell Biol., 1, 215–220. [DOI] [PubMed] [Google Scholar]
- 34. Adams M.E., Kramarcy N., Fukuda T., Engel A.G., Sealock R., Froehner S.C. (2004) Structural abnormalities at neuromuscular synapses lacking multiple syntrophin isoforms. J. Neurosci., 24, 10302–10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Peters M.F., Adams M.E., Froehner S.C. (1997) Differential association of syntrophin pairs with the dystrophin complex. J. Cell Biol., 138, 81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kramarcy N.R., Vidal A., Froehner S.C., Sealock R. (1994) Association of utrophin and multiple dystrophin short forms with the mammalian Mr 58, 000 dystrophin-associated protein (syntrophin). J. Biol. Chem., 269, 2870–2876. [PubMed] [Google Scholar]
- 37. Muthu M., Richardson K.A., Sutherland-Smith A.J. (2012) The crystal structures of dystrophin and utrophin spectrin repeats: implications for domain boundaries. PLoS One, 7, e40066.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rebolledo D.L., Kim M.J., Whitehead N.P., Adams M.E., Froehner S.C. (2015) Sarcolemmal targeting of nNOSmu improves contractile function of mdx muscle. Hum. Mol. Genet., 25, S290–S166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Banks G.B., Judge L.M., Allen J.M., Chamberlain J.S. (2010) The polyproline site in hinge 2 influences the functional capacity of truncated dystrophins. PLoS Genet., 6, e1000958.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Park J., Wicki J., Knoblaugh S.E., Chamberlain J.S., Lee D. (2015) Multi-parametric MRI at 14T for muscular dystrophy mice treated with AAV vector-mediated gene therapy. PLoS One, 10, e0124914.. [DOI] [PMC free article] [PubMed] [Google Scholar]