Abstract
Hyper-activation of extracellular signal-regulated kinase (ERK) 1/2 contributes to heart dysfunction in cardiomyopathy caused by mutations in the lamin A/C gene (LMNA cardiomyopathy). The mechanism of how this affects cardiac function is unknown. We show that active phosphorylated ERK1/2 directly binds to and catalyzes the phosphorylation of the actin depolymerizing factor cofilin-1 on Thr25. Cofilin-1 becomes active and disassembles actin filaments in a large array of cellular and animal models of LMNA cardiomyopathy. In vivo expression of cofilin-1, phosphorylated on Thr25 by endogenous ERK1/2 signaling, leads to alterations in left ventricular function and cardiac actin. These results demonstrate a novel role for cofilin-1 on actin dynamics in cardiac muscle and provide a rationale on how increased ERK1/2 signaling leads to LMNA cardiomyopathy.
Introduction
Mutations in the lamin A/C gene (LMNA) cause an autosomal dominant inherited form of dilated cardiomyopathy (hereafter referred to as LMNA cardiomyopathy), often with concurrent muscular dystrophy (1,2). LMNA encodes the A-type nuclear lamins, which arise from alternative RNA splicing (3–5) and along with B-type lamins are the main constituents of nuclear lamina (6). Much of the current research on A-type lamins is focused on how mutations leading to alterations in these proteins cause dilated cardiomyopathy and other inherited diseases. We previously demonstrated that extracellular signal-regulated kinase (ERK) 1/2 is hyper-activated in the heart in LMNA cardiomyopathy (7). However, insights into the molecular mechanisms bridging ERK1/2 activation and depressed cardiac function are lacking.
Alterations in cardiomyocyte (CM) mechanotransduction likely underlie molecular mechanisms of dilated cardiomyopathy and progression to heart failure (8,9). Actin is one of the major cytoskeletal proteins in eukaryotic cells that play an essential role in several cellular processes, including mechano-resistance and contractile force generation. Actin filaments within sarcomeres, the contractile units of CMs, are uniform in length and precisely oriented with their barbed-ends (+) facing the Z-disc, which are capped by CapZ (10) and their pointed-ends (−) directed toward the M-band, which are associated with tropomodulin. Actin filaments are additionally decorated along their length by tropomyosin and a large number of actin-binding proteins, which contribute to maintaining sarcomere structure and organization (11–16). A number of actin-binding proteins enhance their turnover, promoting polymerization, depolymerization or filament severing (17–19). Defective regulation of the length or the organization of actin filaments in sarcomeres, owing to genetic mutations or de-regulated expression of cytoskeletal proteins, is a hallmark of many heart and skeletal muscle disorders (20). Among the regulators of actin, cofilins, which are actin-depolymerizing factors, play an essential role in the dynamics of filaments. Cofilins enhance actin filament turnover by severing and promoting dissociation of actin monomers from the pointed-ends (−) (21). We now show in a large array of unique in vitro and in vivo disease models that phosphorylated ERK1/2 (pERK1/2) binds to and activates cofilin-1 in LMNA cardiomyopathy. The disassembly of actin occurs in CMs from the mouse model, leading to left ventricular dysfunction.
Results
pERK1/2 alters F-actin dynamics in LMNA cardiomyopathy
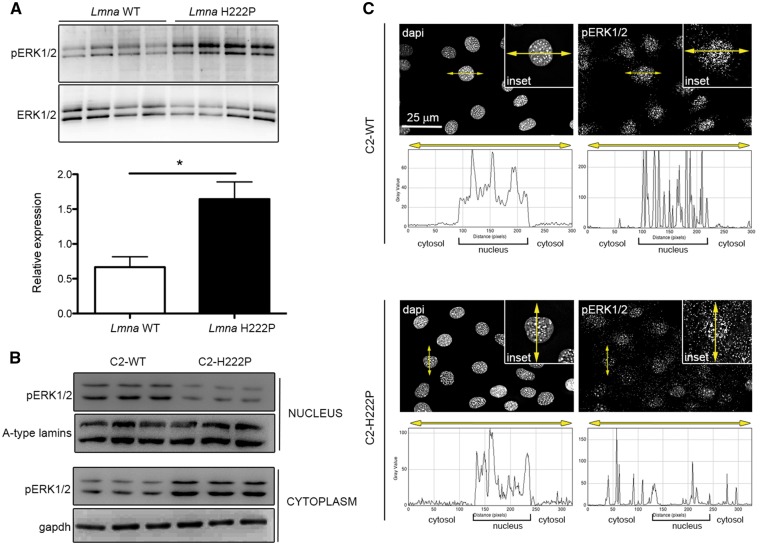
We set out to unravel the consequences of abnormal ERK1/2 signaling in the heart of LmnaH222P/H222P mice, a model for dilated cardiomyopathy caused by mutation in LMNA (22). As in previous studies (7,23), we demonstrated an increase in pERK1/2 in hearts of LmnaH222P/H222P mice compared with wild-type mice (Fig. 1A). We also previously demonstrated increased pERK1/2 primarily in the nucleus of transiently transfected C2C12 cells over-expressing the lamin A H222P variant (7). When we examined protein extracts from stably transfected C2C12 cells expressing H222P lamin A (C2-H222P) at lower levels (24,25), we observed an increase in cytoplasmic relative to nuclear pERK1/2 compared with cells expressing wild-type lamin A (C2-WT) (Fig. 1B). Total cellular pERK1/2 was not changed (data not shown), which is consistent with previous results showing that it is only increased after subjecting these cells to stress (24). The increased cytoplasmic pERK1/2 in C2-H222P cells was further demonstrated by immunofluorescence microscopy (Fig. 1C).
Figure 1.
Increased ERK1/2 activation in hearts of LmnaH222P/H222P mice and in the cytoplasm of C2-H222P cells. (A) Immunoblots showing pERK1/2 and total ERK1/2 in hearts from LmnaH222P/H222P mice (H222P) and wild-type mice (WT). Data in bar graph below are represented as means±SEM (n=4; *P <0.05) from three independent repeats. (B) Immunoblots showing pERK1/2 and total ERK1/2 in extracts of nucleus and cytoplasm from C2-WT (n=3) and C2-H222P (n=3) cells. A-type lamins and gapdh are shown as loading controls. (C) Representative immuofluorescence micrographs of pERK1/2 staining of C2-WT and C2-H222P cells. Nuclei counter-stained with dapi are also shown. Insets show a higher magnification. Scan line graphs represent the intensity of pERK1/2 and dapi staining along the yellow arrow lines.
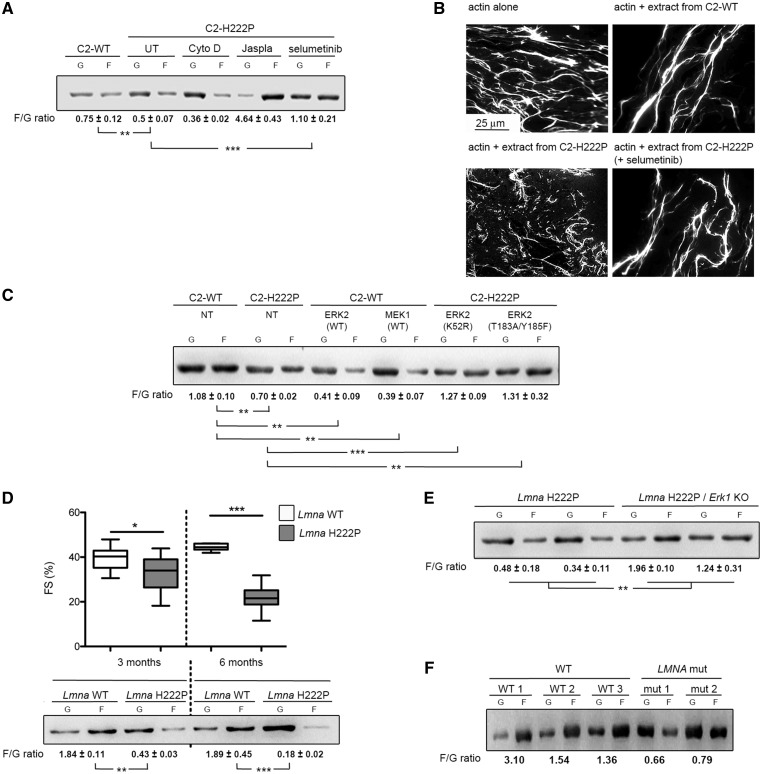
Although most ERK1/2 substrates are localized in the nucleus, several cytoskeletal proteins are targets (26). We hypothesized that cytoplasmic pERK1/2 in cells expressing the lamin A H222P variant catalyzes the phosphorylation of cytosolic proteins. Given that A-type lamins modulate cytosolic actin polymerization (27), we focused on actin dynamics. When examined by immunoblotting, the ratio of filamentous (F) to globular (G) actin was significantly lower in C2-H222P cells compared with C2-WT cells (Fig. 2A). Treating C2-H222P cells with cytochalasin D, an inhibitor of actin polymerization, lowered the ratio while treating with jasplakinolide, which promotes actin polymerization and stabilization, increased the ratio (Fig. 2A). Treating C2-H222P cells with selumetinib, an inhibitor of MEK1/2, the kinases that specifically phosphorylate ERK1/2, led to F-actin polymerization (Fig. 2A). These results were confirmed by immunofluorescence microscopic analysis of F- and G-actin (Supplementary Material, Fig. S1A). Similar to selumetinib, other inhibitors of MEK1/2 gave the same results (Supplementary Material, Fig. S1B and C). When selumetinib was washed out from the media in which C2-H222P cells were cultured, the F/G actin ratio analyzed by immunoblotting progressively returned to a value similar to that in untreated cells (Supplementary Material, Fig. S1D). This was also observed by immunofluorescence microscopy (Supplementary Material, Fig. S1E). Treating C2-WT and C2-H222P cells with cytochalasin D or latrunculin B induced depolymerization of the actin network and washing out these drugs led to progressive re-polymerization of F-actin (Supplementary Material, Fig. S1E). However, actin re-polymerization was delayed in C2-H222P cells compared with C2-WT cells and this delay was diminished when we added selumetinib (Supplementary Material, Fig. S1E).
Figure 2.
Altered F-actin dynamics in LMNA cardiomyopathy. (A) Immunoblot from one representative experiment showing the effect of lamin A H222P on the amounts of G-actin and F-actin and the calculated F/G actin ratios. C2-H222P cells were either untreated (UT), or treated with cytochalasin D, jasplakinolide or selumetinib. Data are represented as means±SEM (n=3; **P <0.005, ***P <0.0005) from three independent repeats. (B) Microscopic analyses of F-actin depolymerization and/or severing. Micrographs show in vitro F-actin filaments alone or incubated in the presence of protein extracts from C2-WT or C2-H222P cells with or without selumetinib treatment. (C) Immunoblot from one representative experiment illustrating the effects of transfection with ERK2 and MEK1 constructs on the amount of G-actin and F-actin and the calculated F/G actin ratio. NT indicates not transfected. Data are represented as means±SEM (n=3; **P<0.005, ***P <0.0005) from three independent repeats. (D) Graph showing left ventricular fractional shortening (FS) in 3- and 6-month-old male LmnaH222P/H222P mice (Lmna H222P) (n=10) compared with Lmna+/+ (Lmna WT) (n=19) mice. The central line represents the median, the box limits the interquartile range and the whiskers the minimum and maximum (***P <0.0005). Immunoblot illustrating the effect of lamin A H222P on the amount of G-actin and F-actin and the calculated F/G actin ratio protein extracts of hearts from mice at 3 and 6 months of age. Data are represented as means±SEM (n=3; **P<0.005, ***P <0.0005) from three independent repeats. (E) Immunoblot of two representative experiments showing G-actin and F-actin and the calculated F/G actin ratio in protein extracts of hearts from LmnaH222P/H222P; Erk1+/+ mice (Lmna H222P) and LmnaH222P/H222P; Erk1-/- mice (Lmna H222P/Erk1 KO). Data are represented as means ± SEM (n=3; **P<0.005) from three independent repeats. (F) Immunoblot illustrating the amount of G-actin and F-actin and the calculated F/G actin ratio in hearts from unaffected subjects (WT) and from two patients with cardiomyopathy caused by LMNA mutation (LMNA mut).
We next compared the effect of protein extracts from C2-WT or C2-H222P cells on the length of F-actin in vitro by microscopic analysis of fluorescently labeled actin. When actin was polymerized in the presence of extracts from C2-H222P cells, the length of F-actin was shorter than in the presence of extracts from C2-WT cells (Fig. 2B). This effect on F-actin dynamics was blunted when an extract of C2-H222P cells treated with selumetinib was used (Fig. 2B). To test if ERK1/2 contributed directly to F-actin dynamics, we transiently transfected C2-WT cells with wild-type ERK2 or MEK1 constructs. This led to a decrease in the F/G actin ratio compared with non-transfected cells (Fig. 2C). Conversely, C2-H222P cells transfected with plasmids encoding ERK2-K52R (kinase dead) or ERK2-T183A/Y185F (dominant negative), both of which competitively inhibit activation of endogenous ERK2, had an increased F/G actin ratio compared with non-transfected C2-H222P cells with the quantity of F-actin in these transfected C2-H222P cells similar to that in C2-WT cells (Fig. 2C). These data suggest that ERK1/2 triggers depolymerization of actin in C2-H222P cells expressing a lamin A variant that causes dilated cardiomyopathy.
To determine whether other lamin A variants have the same effect on actin dynamics as lamin A H222P, we transiently transfected C2C12 cells with plasmids encoding lamin A E358K, L271P and N456I. These mutations have been previously shown to cause cardiomyopathy (28–30). Similar to lamin A H222P, C2C12 cells expressing these pathogenic lamin A variants showed increased pERK1/2 (Supplementary Material, Fig. S2A) and altered ratios of F-actin to G-actin (Supplementary Material, Fig. S2B). These findings further suggest that altered F-actin dynamics arises from LMNA mutations as a result of abnormal ERK1/2 activation.
Owing to limitations of using stably transfected C2C12 cells expressing H222P lamin A as a cellular model to study LMNA cardiomyopathy, we next assessed altered F-actin dynamics in a mouse model of the disease. Male LmnaH222P/H222P mice have enhanced activation of ERK1/2 signaling in the heart starting at 4 weeks of age (7,31). We therefore investigated whether this was correlated with alterations in the actin cytoskeleton in vivo. Compared with Lmna+/+ mice, the F/G actin ratio was decreased in hearts from LmnaH222P/H222P mice at 3 months of age with slight decreased left ventricular fractional shortening, and further decreased at 6 months of age, when the mice had impaired left ventricular systolic function (Fig. 2D). This altered F/G actin ratio was reversed in vivo when LmnaH222P/H222P mice were crossed with Erk1-/- mice (Fig. 2E). We have previously shown that cardiac function is partially restored in these mice (32). The F/G actin ratio was also lower in heart tissue from humans carrying cardiomyopathy-causing LMNA mutations compared with controls (Fig. 2F). These data imply a central role of pERK1/2 in the regulation of F-actin dynamics in LMNA cardiomyopathy.
pERK1/2 catalyzes the phosphorylation of cofilin-1 on threonine 25 and alters its activity
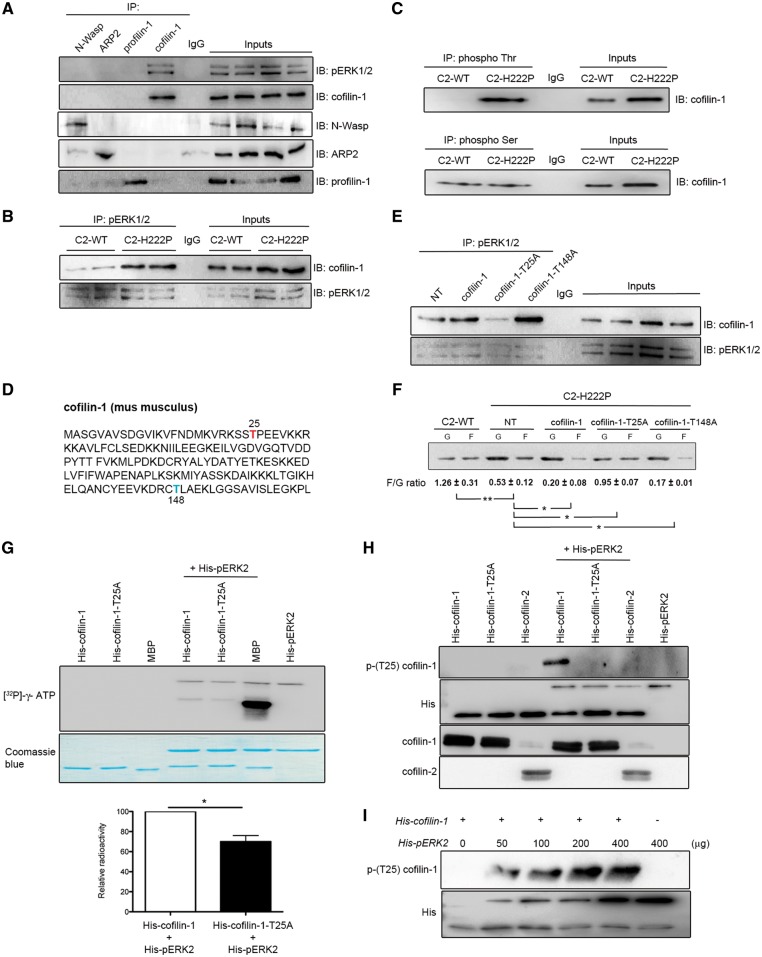
We hypothesized that pERK1/2 binds to a nucleation promoting factor or an actin depolymerizing factor to alter F-actin cycling that leads to cardiac muscle dysfunction. To test our hypothesis, we performed immunoprecipitations (IPs) using extracts of C2-H222P cells and antibodies against cofilin-1, N-Wasp, ARP2 or profilin-1. We found that pERK1/2 interacted only with cofilin-1 (Fig. 3A). The interaction between pERK1/2 and cofilin-1 was enhanced in C2-H222P cells compared with C2-WT cells (Fig. 3B). This confirmed that the higher level of pERK1/2 in the cytoplasm of C2-H222P cells promoted greater interaction with cofilin-1. Un-phosphorylated (inactive) ERK1/2 did not interact with cofilin-1 (Supplementary Material, Fig. S3A). A similar interaction between pERK1/2 and cofilin-1 was found in hearts of 3- and 6-month-old LmnaH222P/H222P mice but was not detected in hearts of Lmna+/+ mice (Supplementary Material, Fig. S3B). Therefore, these results showed that pERK1/2 bound to cofilin-1 in cells expressing lamin A H222P (and also lamin C H222P in the mouse hearts). Cofilin-1 was highly phosphorylated on threonine residues in C2-H222P cells compared with C2-WT cells, while phosphorylation on serine residues was similar in both cell types (Fig. 3C). This was consistent with the known ability of ERK1/2 to catalyze the phosphorylation of serine or threonine residues (33).
Figure 3.
Interaction between cofilin-1 and pERK1/2. (A) Immunoblot showing interaction of cofilin-1 and pERK12 in IP experiments. Proteins extracted from C2C12 cells were subjected to IP using antibodies against cofilin-1, N-Wasp, ARP2 or profilin-1. Proteins in immunoprecipitates were separated by SDS-PAGE and IB using antibodies against pERK1/2, cofilin-1, N-Wasp, ARP2 or profilin-1. Immunoglobulin G (IgG) was used as a negative control. Representative from three independent repeats. (B) Immunoblot showing interaction of cofilin-1 and pERK12 in IP experiments from C2-WT and C2-H222P cells. Proteins extracted from C2-WT and C2-H222P cells were subjected to IP using antibodies against pERK1/2. Proteins in immunoprecipitates were separated by SDS-PAGE and IB using antibodies against pERK1/2 and cofilin-1. IgG was used as a negative control. Representative from three independent repeats. (C) Proteins extracted from C2-WT and C2-H222P cells were subjected to IP using antibodies specific to phospho Thr or phospho Ser. The immunoprecipitates were separated by SDS-PAGE and IB using antibody against cofilin-1. IgG was used as a negative control. Representative from three independent repeats. (D) Amino acid sequence of murine cofilin-1 with highlighted threonine 25 (red) and threonine 148 (blue). (E) C2-H222P cells transfected or not transfected (NT) with plasmids encoding cofilin-1, cofilin-1-T25A or cofilin-1-T148A were subjected to IP using antibodies against pERK1/2. Proteins in immunoprecipitates were separated by SDS-PAGE and IB using antibodies against cofilin-1 or pERK1/2. IgG was used as a negative control. Representative from three independent repeats. (F) Immunoblot illustrating the effect of transfection with different cofilin-1 constructs on the amount of G-actin and F-actin and the calculated F/G actin ratio. Data are represented as means±SEM (n=3; *P <0.05, **P <0.005) from three independent repeats. (G) Representative immunoblot of in vitro kinase assay using recombinant histidine-tagged wild-type cofilin-1 (His-cofilin-1-WT) and cofilin-1-T25A (His-cofilin-T25A), without or with the addition of recombinant pERK2. MBP was used as a positive control. [32P]-γ-ATP indicates recombinant protein phosphorylation. Corresponding Coomassie blue-stained gel is shown below immunoblot. Data in bar graph below are means±SEM (n=3; *P<0.05) from three independent repeats. (H) Representative immunoblot of in vitro kinase assay using recombinant His-cofilin-1, His-cofilin-1-T25A and His-cofilin-2 without or with the addition of recombinant pERK2. Phosphorylated (Thr25) cofilin-1 was detected using a specific antibody. Representative from three independent repeats. (I) Representative immunoblot of in vitro kinase assay using recombinant cofilin-1 without or with the addition of recombinant pERK2 at increasing doses. Phosphorylated (Thr25) cofilin-1 was detected using the specific antibody. Representative from three independent repeats.
Threonine phosphorylation on cofilin-1 was blunted when C2-H222P cells were treated with selumetinib (Supplementary Material, Fig. S3C). Threonine phosphorylation of cofilin-1 occurred in C2-WT cells transfected with a plasmid encoding wild-type ERK2 (Supplementary Material, Fig. S3C). Pro-Xaa-Ser/Thr-Pro is the most common consensus sequence for substrate recognition by pERK1/2, although Ser/Thr-Pro can also serve as a substrate (34). Cofilin-1 contains eight threonine residues but only Thr25 is N-terminal to a proline, therefore suggesting that it is a potential target residue for pERK1/2-catalyzed phosphorylation (Fig. 3D). We therefore created plasmids encoding cofilin-1 variants T25A and T148A. The interaction between pERK1/2 and cofilin-1-T25A was attenuated, while the interaction with cofilin-1-T148A was not altered (Fig. 3E). This suggested that Thr25 was involved in the interaction with pERK1/2. Cofilin-2, another actin depolymerizing factor expressed mainly in striated muscles, does not have the Thr-Pro consensus site for phosphorylation catalyzed by ERK1/2 and therefore did not bind to it (Supplementary Material, Fig. S3D). We further asked whether cofilin-1-T25A could alter actin dynamics. C2-H222P cells transfected with plasmids that expressed wild-type cofilin-1 or cofilin-1-T148A had a decreased F/G actin ratio compared with non-transfected C2-H222P cells (Fig. 3F). Expression of cofilin-1-T25A partially rescued the F/G actin ratio in C2-H222P cells (Fig. 3F). Cofilin-1-T25D, a phosphomimetic variant, did not reverse actin depolymerization when expressed in C2-H222P cells (Supplementary Material, Fig. S3E). Similar results were observed when C2-WT cells were transfected with plasmids that expressed wild-type cofilin-1 or cofilin-1-T25D (Supplementary Material, Fig. S3F). Expressing cofilin-1-T25A did not alter the F/G actin ratio in C2-WT cells (Supplementary Material, Fig. S3F).
We next generated an antibody recognizing the region of phosphorylated (Thr25) cofilin-1 (Supplementary Material, Fig. S4A). To ensure that the identified phosphorylated (Thr25) cofilin-1 was increased in C2-H222P cells, immunoblots of protein lysates were probed with these antibodies. The level of phosphorylated (Thr25) cofilin-1 was significantly increased in C2-H222P cells compared with C2-WT cells and reduced when treated with selumetinib (Supplementary Material, Fig. S4B). Like ERK1/2, phosphorylated (Thr25) cofilin-1 was present in the cytoplasm and nucleus of C2-H222P cells, while it is mostly in the nucleus of C2-WT cells and essentially absent from C2-H222P cells treated with selumetinib (Supplementary Material, Fig. S4C). To determine whether other lamin A variants have the same effect on the phosphorylation of cofilin-1 on Thr25 as lamin A H222P, we transiently transfected C2C12 cells with plasmids encoding other pathogenic lamin A variant associated with cardiomyopathy. Similar to lamin A H222P, C2C12 cells expressing these lamin A variants showed increased level of phosphorylated (Thr25) cofilin-1 (Supplementary Material, Fig. S4D). Expression of the phosphorylated (Thr25) cofilin-1 was increased in heart tissue from LmnaH222P/H222P mice compared with wild-type mice (Supplementary Material, Fig. S4F). The increased expression of phosphorylated cofilin-1-T25A occurs as early as 2 months of age, when the cardiac function is not altered (22). We then examined the phosphorylated (Thr25) cofilin-1 in human heart samples. We saw an increase of phosphorylated (Thr25) cofilin-1 as well as pERK1/2 in heart tissue from humans carrying cardiomyopathy-causing LMNA mutations compared with controls (Supplementary Material, Fig. S4F).
We next demonstrated that pERK1/2 catalyzed the phosphorylation of cofilin-1 on Thr25. In an in vitro kinase assay, incubation of histidine-tagged pERK2 (His-pERK2) with histidine-tagged cofilin-1 (His-cofilin-1) and [32P]-γ-ATP resulted in the phosphorylation of cofilin-1 (Fig. 3G). This in vitro assay also showed that incubation of His-pERK2 with [32P]- γ-ATP resulted in 32P incorporation into ERK2, which confirmed the previously reported autophosphorylation of ERK2 (35,36). The pERK2-catalyzed phosphorylation of cofilin-1 was significantly reduced in the presence of His-cofilin-1-T25A (Fig. 3G). This suggested that pERK2 catalyzed the phosphorylation of cofilin-1 on Thr25. We further incubated pERK2 with His-cofilin-1, His-cofilin-1-T25A or His-cofilin-2. pERK2 catalyzed the phosphorylation of His-cofilin-1 on Thr25, but not His-cofilin-1-T25A or His-cofilin-2 (Fig. 3H). The phosphorylation of cofilin-1 correlated with amount of His-pERK2 (Fig. 3I), indicating that its phosphorylation was catalyzed by ERK2. This phosphorylation of cofilin-1 catalyzed by pERK2 was blunted in the presence of a His-cofilin-1 variant with Thr25 replaced by an alanine (Fig. 3H). Overall, these results demonstrated that pERK1/2 catalyzed the phosphorylation of cofilin-1 on Thr25 in cells expressing lamin A H222P, which correlated with altered F-actin dynamics.
To detect endogenous cofilin-1 phosphorylated on Thr25, we analyzed protein extracts from C2-WT, C2-H222P, and C2-H222P cells treated with selumetinib using 2-dimensional gel electrophoresis (Supplementary Material, Fig. S5A). Immunoblotting of the separated proteins with antibodies against cofilin-1 identified a specific pattern of expression of cofilin-1 in C2-H222P cells compared with C2-WT cells and C2-H222P cells treated with selumetinib (Supplementary Material, Fig. S5B). Given that these specific patterns of expression of cofilin-1 could be owing to difference in phosphorylation status, each protein isoform revealed by the antibody against cofilin-1 was excised and subjected to in-gel digestion and mass spectrometry analysis (Supplementary Material, Fig. S5C). This analysis identified a total of 12 phosphorylation sites distributed in 8 polypeptides corresponding to cofilin-1 (Supplementary Material, Table S1). Phosphopeptide D (Supplementary Material, Fig. S5D) contained the Thr25 phosphorylation site (SSTPEEVKK) and was observed only in extracted proteins from C2-H222P cells (Table 1; Supplementary Material, Table S2). This phosphopeptide was predominantly observed in C2-H222P cells when immunoblotted (IB) with a specific antibody against phosphorylated (Thr25) cofilin-1 (Supplementary Material, Fig. S5D). Phosphopeptides B and C (Supplementary Material, Fig. S5C) showed increased intensity in C2-H222P cells (Supplementary Material, Fig. S5B). Similar to phosphopeptide D, the increase in phosphopeptide B was abrogated by selumetinib, suggesting an ERK1/2 dependent phosphorylation of cofilin-1. The intensity of phosphopeptide C was not affected by selumetinib (Supplementary Material, Fig. S5B), indicating that cofilin-1 could be phosphorylated by other kinases.
Table 1.
Cofilin-1 phospho-residues identified by mass spectrometry analysis from C2-WT, C2-H222P and C2-H222P cells treated with selumetinib
| Serine | S3 |
S23 |
S41 |
S94 |
S156 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | C2-WT | C2-H222P | C2-H222P (selumetinib) | C2-WT | C2-H222P | C2-H222P (selumetinib) | C2-WT | C2-H222P | C2-H222P (selumetinib) | C2-WT | C2-H222P | C2-H222P (selumetinib) | C2-WT | C2-H222P | C2-H222P (selumetinib) |
| A | × | × | × | × | × | × | × | ||||||||
| B | × | × | × | ||||||||||||
| C | × | × | × | ||||||||||||
| D | × | × | × | × | × | × | × | × | × | ||||||
| Threonine |
T25 |
T63 |
T88 |
||||||||||||
| Genotype | C2-WT | C2-H222P | C2-H222P (selumetinib) | C2-WT | C2-H222P | C2-H222P (selumetinib) | C2-WT | C2-H222P | C2-H222P (selumetinib) | ||||||
| A | × | × | × | ||||||||||||
| B | |||||||||||||||
| C | |||||||||||||||
| D | x | × | × | × | × | × | |||||||||
| Tyrosine |
Y82 |
Y85 |
Y89 |
Y140 |
|||||||||||
| Genotype | C2-WT | C2-H222P | C2-H222P (selumetinib) | C2-WT | C2-H222P | C2-H222P (selumetinib) | C2-WT | C2-H222P | C2-H222P (selumetinib) | C2-WT | C2-H222P | C2-H222P (selumetinib) | |||
| A | × | × | × | × | × | × | |||||||||
| B | × | × | × | × | |||||||||||
| C | × | × | × | × | |||||||||||
| D | × | × | × | × | × | × | × | × | |||||||
RhoA and Rho kinase (ROCK) regulate cofilin-1-mediated F-actin disassembly through LIMK-catalyzed phosphorylation of the protein (37). LIMK catalyzes the phosphorylation of cofilin-1 on Ser3, and inactivates its actin-severing activity (38). Phosphorylated (Ser3) cofilin-1 was detected in the protein extracts analyzed by mass spectrometry (Table 1; Supplementary Material, Table S2). However, the level of phosphorylated (Ser3) cofilin-1 was not different between C2-WT and C2-H222P cells (Supplementary Material, Fig. S6). Treatment with Y27632, a selective inhibitor of ROCK, blocks the phosphorylation of LIMK and in turn the LIMK-catalyzed phosphorylation of cofilin-1, which leads to its activation (39). Treating C2-WT and C2-H222P cells with Y27632 decreased phosphorylation of cofilin-1 on Ser3 without affecting the phosphorylation of Thr25 (Supplementary Material, Fig. S6). These results suggested that phosphorylation of Thr25 on cofilin-1 was independent of the ROCK pathway.
Phosphorylated (Thr25) cofilin-1 stimulates cardiac dysfunction
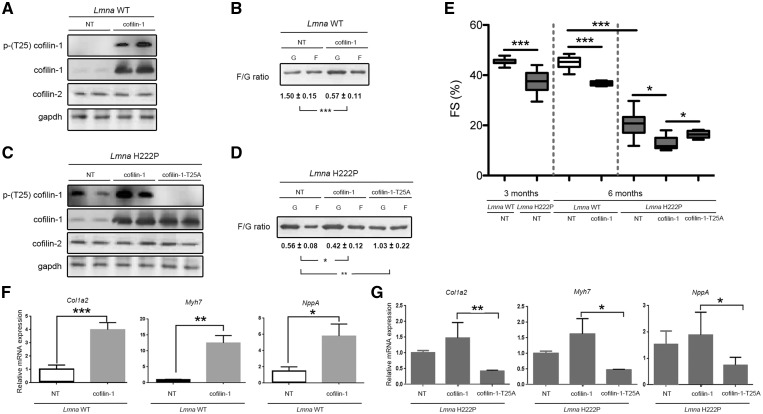
We next tested the hypothesis that phosphorylated (Thr25) cofilin-1 influences left ventricular function in vivo. We injected adeno-associated virus (AAV) vectors expressing cofilin-1 into 3-month-old Lmna+/+ mice. Three months after injection they had increased phosphorylated (Thr25) cofilin-1 expression in the heart (Fig. 4A). Expression of cofilin-2 in the heart was unchanged by over-expression of cofilin-1 (Fig. 4A). There was depolymerization of F-actin in Lmna+/+ mice expressing viral vector-encoded cofilin-1 (Fig. 4B). Compared with non-transduced mice, left ventricular end-systolic diameter was significantly increased and fractional shortening significantly decreased in Lmna+/+ mice that expressed viral-encoded cofilin-1 (Fig. 4E;Table 2). Lmna+/+ mice that received the viral vector encoding cofilin-1 had significantly increased expression of Col1a2, which encodes a collagen, Myh7, which encodes myosin heavy chain, and Nppa, which encodes atrial natriuretic precursor (Fig. 4F). These genes have been previously shown to be up-regulated in LMNA cardiomyopathy (23,40–43).
Figure 4.
Phosphorylated (Thr25) cofilin-1 induces cardiac dysfunction. (A) Immunoblots showing cofilin-1, phosphorylated (Thr25) cofilin-1, cofilin-2 and gapdh in protein extracts from hearts of Lmna+/+ (Lmna WT) transduced with AAV vectors encoding cofilin-1; NT indicates not transduced. Representative from three independent repeats. (B) Immunoblot illustrating the effect of AAV expressing cofilin-1 construct on the amount of G-actin and F-actin and the calculated F/G actin ratio in hearts from mice; NT indicates not transduced. Data are represented as means±SEM (n=3; ***P<0.0005) from three independent repeats. (C) Immunoblots showing cofilin-1, phosphorylated (Thr25) cofilin-1, cofilin-2 and gapdh in protein extracts from hearts of LmnaH222P/H222P (Lmna H222P) transduced with AAV vectors encoding the indicated proteins; NT indicates not transduced. Representative from three independent repeats. (D) Immunoblot illustrating the effect of AAV expressing cofilin-1 constructs on the amount of G-actin and F-actin and the calculated F/G actin ratio in hearts from mice transduced with AAV vectors encoding the indicated proteins; NT indicates not transduced. Data are represented as means±SEM (n=3; *P<0.05, **P<0.005) from three independent repeats. (E) Graph showing left ventricular fraction shortening (FS) in 3-month-old (prior injection of AAV) and 6-month-old male transduced Lmna WT mice and Lmna H222P mice (see Table 2 for details). The central line represents the median, the box limits the interquartile range and the whiskers the minimum and maximum (*P<0.05, ***P<0.0005). (F) Expression of Col1a2, Myh7 and NppA mRNAs in hearts from 6-month-old male Lmna+/+ mice (Lmna WT) transduced with AAV expressing cofilin-1 or not transduced (NT). Data are represented as means±SEM (n=4; *P<0.05, **P <0.005, ***P <0.0005). (G) Expression of Col1a2, Myh7 and NppA mRNAs in hearts from 6-month-old male LmnaH222P/H222P mice (Lmna H222P) transduced with AAV expressing cofilin-1 or AAV expressing cofilin-1-T25A. Data from Lmna H222P mice not transduced (NT) are showed for comparison. Data are represented as means±SEM (n=4; *P<0.05, **P <0.005).
Table 2.
Echocardiographic data for Lmna+/+ (Lmna WT) transduced or not transduced with AAV-cofilin-1 and LmnaH222P/H222P (Lmna H222P) mice transduced or not transduced with AAV-cofilin-1 or AAV-cofilin-1-T25A
| Treatment | n | Age (months) | Heart rate (beats/min) | LVEDD (mm) | LVESD (mm) | FS (%) |
|---|---|---|---|---|---|---|
| Lmna WT | ||||||
| NT | 27 | 3 | 362.8±19.6 | 3.5±0.2 | 1.8±0.1 | 47.3±1.9 |
| NT | 21 | 6 | 374.8±17.6 | 3.8±0.2 | 2.1±0.2 | 45.1±2.5 |
| AAV-cofilin-1 | 5 | 6 | 338.6±41.7 | 3.8±0.3 | 2.4±0.1* | 36.8±0.9*** |
| Lmna H222P | ||||||
| NT | 16 | 3 | 306.0±43.1 | 3.9±0.3# | 2.6±0.5### | 34.4±6.6### |
| NT | 6 | 6 | 255.3±48.9 | 4.8±0.6 | 3.8±0.7 | 20.5±5.7 |
| AAV-cofilin-1 | 7 | 6 | 226.4±28.5 | 4.8±0.4 | 4.2±0.5† | 12.7±2.8†† |
| AAV-cofilin-1-T25A | 6 | 6 | 234.2±36.9 | 4.6±0.4 | 3.9±0.5 | 16.4±1.5§ |
P < 0.05 between not transduced Lmna WT mice and Lmna H222P mice (3 months of age).
P < 0.0005 between not transduced Lmna WT mice and Lmna H222P mice (3 months of age).
P < 0.005 between not transduced and AAV-cofilin-1-transduced Lmna WT mice (6 months of age).
P < 0.0005 between not transduced and AAV-cofilin-1-transduced Lmna WT mice (6 months of age).
P < 0.005 between not transduced and AAV-cofilin-1-transduced Lmna H222P mice (6 months of age).
P < 0.05 between AAV-cofilin-1-transduced and AAV-cofilin-1-T25A transduced Lmna H222P mice (6 months of age).
LVEDD, left ventricular end diastolic diameter; LVESD, left ventricular end systolic diameter; FS, fractional shortening. Data are represented as means±SEM.
We next tested the effect of cofilin-1 variants T25A on left ventricular function in vivo. Expressing cofilin-1-T25A in 3-month-old LmnaH222P/H222P mice lead to a decrease of the relative expression of phosphorylated (Thr25) to total cofilin-1 in the heart (Fig. 4C). Expression of cofilin-2 in the heart was unchanged by overexpression of cofilin-1 or cofilin-1-T25A (Fig. 4C). Expression of cofilin-1-T25A rescued the cardiac F/G actin ratio of LmnaH222P/H222P mice (Fig. 4D). Compared with mice that expressed virally encoded cofilin-1, left ventricular fractional shortening significantly improved in LmnaH222P/H222P mice that expressed viral encoded cofilin-1-T25A (Fig. 4E;Table 2). LmnaH222P/H222P mice that received the viral vector encoding cofilin-1-T25A had decreased expression of Col1a2, Myh7 and Nppa (Fig. 4G), compared with mice that expressed virally encoded cofilin-1.
Phosphorylated (Thr25) cofilin-1 alters sarcomere organization
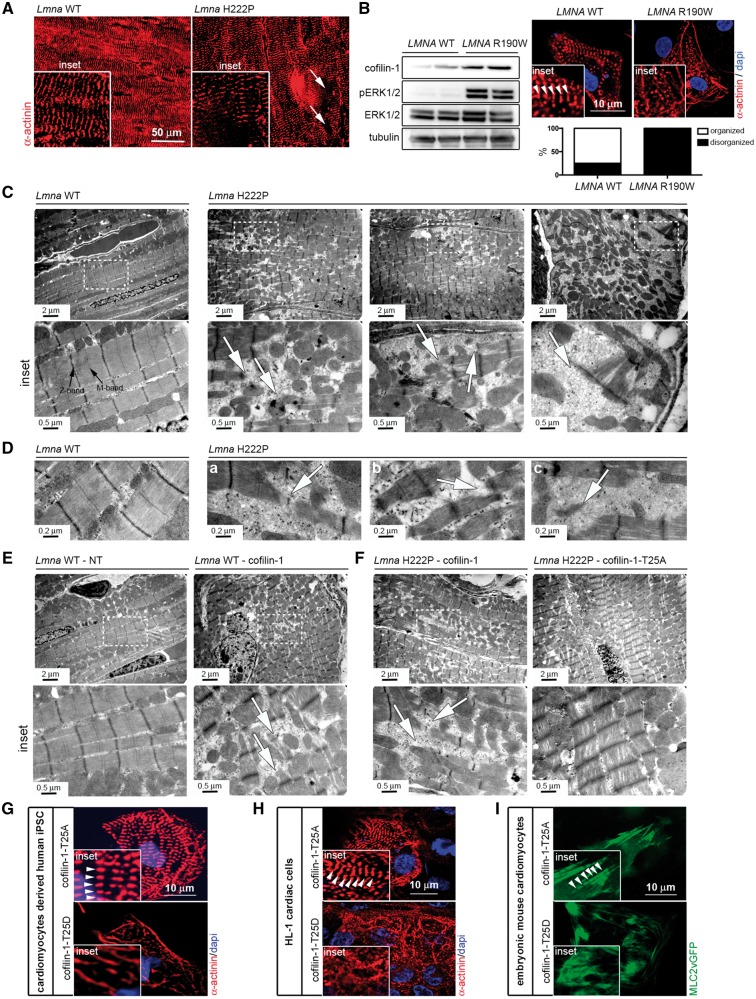
We hypothesized that phosphorylation of Thr25 on cofilin-1 has detrimental effects on cardiac muscle cells. Given that sarcomeres are composed of myosin and actin, we hypothesized that phosphorylated (Thr25) cofilin-1 may affect the dynamics of sarcomeric actin. Immunostaining using an α-actinin antibody showed disruption of sarcomere organization (Fig. 5A). Similar myofibrillars alterations were observed in CMs derived from patient-specific human induced pluripotent stem cells (iPSCs) carrying LMNA p.R190W mutation, for which ERK1/2 signaling was abnormally activated (Fig. 5B). Transmission electron microscopy further showed that left ventricular tissue from LmnaH222P/H222P mice exhibited severe disruption of myofibrillar structure including areas of sarcomere disorganization alongside of normal-looking fibers (Fig. 5C and D). Left ventricular tissue from Lmna+/+ mice that received the vector encoding cofilin-1 exhibited severe disruption of myofibrillars, similar to the ones observed in LmnaH222P/H222P mice (Fig. 5E). The sarcomeric organization is improved in left ventricular tissue from LmnaH222P/H222P mice that received the vector encoding cofilin-1-T25A, compared with LmnaH222P/H222P mice that received the vector encoding cofilin-1 (Fig. 5F). We further showed that phosphorylated (Thr25) cofilin-1 influences sarcomeric organization in vitro. Transfection with a plasmid encoding the cofilin-1 T25D variant in CMs derived from human iPSCs (Fig. 5G), embryonic CMs (Fig. 5H), and HL-1 cardiac muscle cells (Fig. 5I) led to altered myofibrillar organization compared with transfection with a plasmid encoding the cofilin-1 T25A variant.
Figure 5.
Phosphorylated (Thr25) cofilin-1 alters sarcomeric organization. (A) Micrographs showing α-actinin labeling of cross-sections of hearts from 6-month-old male Lmna+/+ (Lmna WT) mice and LmnaH222P/H222P (Lmna H222P) mice. Arrows indicate areas of disorganized sarcomeres (inset). (B) Immunoblots showing cofilin-1, pERK1/2, ERK1/2 and tubulin in protein extracts from CMs derived from patient-specific human iPSCs carrying the LMNA p.R190W mutation (LMNA R190W) and control (LMNA WT). Micrographs showing α-actinin labeling of CMs derived from patient-specific human iPSCs carrying LMNA p.R190W mutation (LMNA R190W) and control (LMNA WT). Dapi counter-staining (blue) of nuclei is also shown. Inset indicates altered myofibrillar organization. Arrows indicate Z-bands of sarcomeres. Means of organized and (black bar) and disorganized (while bar) sarcomeres are shown in each bar for each condition. (C) Electron micrographs showing disruption of sarcomeric organization in hearts from 6-month-old male LmnaH222P/H222P (Lmna H222P) mice compared with Lmna+/+ (Lmna WT) mice. Insets show a higher magnification. Arrows indicate disorganized sarcomeres. (D) Electron micrographs showing disorganized myofibrillar apparatus (a), areas without rods with normal sarcomere appearance (b) and sparse sarcomere structures (c) indicated by arrows in hearts from 6-month-old male LmnaH222P/H222P (Lmna H222P) mice. (E) Electron micrographs showing sarcomeric organization in hearts from 6-month-old Lmna+/+ (Lmna WT) mice transduced with AAV expressing cofilin-1 construct. NT indicates not transduced. Insets show a higher magnification. Arrows indicate disorganized sarcomeres. (F) Electron micrographs showing sarcomeric organization in hearts from 6-month-old LmnaH222P/H222P (Lmna H222P) mice transduced with AAV expressing cofilin-1 or cofilin-1-T25A constructs. Insets show a higher magnification. Arrows indicate disorganized sarcomeres. (G) Micrographs showing α-actinin labeling of CMs derived from human iPSCs transfected with plasmids encoding cofilin-1-T25A and cofilin-1-T25D. Insets highlight altered myofibrillar organization. Arrows indicate Z-bands of sarcomeres (inset). Dapi counter-staining (blue) of nuclei is also shown. (H) Micrographs showing α-actinin labeling of mouse HL-1 cardiac cells transfected with plasmids encoding cofilin-1-T25A and cofilin-1-T25D. Insets highlight altered myofibrillar organization. Arrows indicate Z-bands of sarcomeres (inset). Dapi counter-staining (blue) of nuclei is also shown. (I) Micrographs showing sarcomeric organization (MLC2vGFP) of embryonic mouse CMs transfected with plasmids encoding cofilin-1-T25A and cofilin-1-T25D. Insets highlight altered myofibrillar organization. Arrows indicate Z-bands of sarcomeres (inset).
Discussion
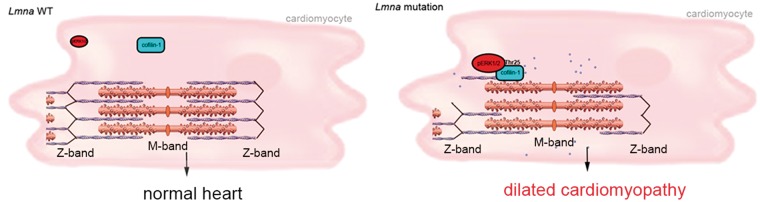
We have shown that ERK1/2-catalyzed phosphorylation of cofilin-1 in cells expressing a cardiomyopathy-causing lamin A variant leads to depolymerization of F-actin. In a mouse model of LMNA cardiomyopathy, this participates in the development of cardiac dysfunction. These results suggest a novel model for A-type lamins in the regulation of actin dynamics and pathophysiology (Fig. 6). In this model, active pERK1/2 binds to cofilin-1, an F-actin binding partner, and catalyzes its phosphorylation on Thr25. This phosphorylation event activates the F-actin depolymerizing function of cofilin-1 in CMs. Inhibition of ERK1/2 suppresses the phosphorylation of cofilin-1 events on Thr25 and improves left ventricular function. The mechanism by which LMNA mutation leads to ERK1/2 activation remains to be elucidated but inference can be made from the literature. ERK1/2 signaling activation occurred prior to significant cardiomyopathy in LmnaH222P/H222P mice (7,31) and in LmnaH222P/+ mice, which do not develop clinical heart disease until 2 years of age (7). This is consistent with the hypothesis that ERK1/2 signaling activation underlies the development of LMNA cardiomyopathy instead of occurring as a consequence of the cardiac disease. Accordingly, lamin A/C-deficient cells subjected to cyclic strain respond with altered expression of the mechanosensitive genes, which are downstream targets of the ERK1/2 signaling (44). While it remains unclear how A-type lamins with amino acid substitutions activate ERK1/2 signaling, our results show that they do so when expressed in transfected cells. An intriguing question is how certain alterations in A-type lamins, which are expressed in virtually all differentiated somatic cells, activate ERK1/2 signaling specifically in cardiac muscle (7). Given their role in maintaining normal cellular mechanics, a hypothesis is that A-type lamins variants make contractile cells more susceptible to stress-induced damage, which activates ERK1/2 signaling. Our results demonstrate downstream alterations by which ERK1/2 promotes cardiac dysfunction caused by such mutations. Although cofilin-1 has been known to function in F-actin depolymerization (45), its role in sarcomeric organization and the development of cardiomyopathy was unclear.
Figure 6.
A schematic representation of the mechanism of cofilin-1 phosphorylation by ERK1/2 in the heart from Lmna H222P mice with consequences on sarcomeric actin depolymerization. Blue circle indicates G-actin.
We previously showed that hearts from LmnaH222P/H222P mice at 6 months of age have an increase in mostly nuclear pERK1/2 (7). Relatively increased cytoplasmic pERK1/2 may therefore reflect an early-stage in the development of LMNA cardiomyopathy. The effect of metabolic stress in C2-H222P cells (24) or age in mouse hearts (7) might trigger nuclear import of pERK1/2 when total cellular pERK1/2 significantly increases. Nonetheless, there is still a pool of cytoplasmic pERK1/2 that can catalyze the phosphorylation of resident proteins such as cofilin-1. ERK1/2 can be present in both the nucleus and cytoplasm and catalyze the phosphorylation of proteins in either subcellular compartment (46). Gene expression changes induced by nuclear pERK1/2 may also contribute to the pathology of LMNA cardiomyopathy along with the effects on cytoplasmic cofilin-1 we have delineated in this study.
In striated muscle cells, actin and several scaffolding and regulatory proteins are arranged into contractile sarcomeres (47). Sarcomeric actin is decorated along its length by tropomyosin (48) and other actin-binding proteins, which contribute to controlling sarcomere structure and organization. Mutations in human genes encoding regulators of actin induce cardiomyopathy (49–55). These demonstrate the functional significance of sarcomeric actin dynamics on normal heart function. Abnormalities of actin dynamics will hamper sarcomeric organization and may be a pathway common to left ventricular dysfunction.
Cofilin-1 and -2, both expressed in cardiac muscle (56), contribute to the dynamic turnover of F-actin in contractile cells (57). Monomeric G-actin exchange occurs primarily at the pointed-ends (−) of sarcomeric actin, near the M-band (58), where cofilin-2 functions with tropomodulin in actin disassembly (48). Some results have suggested that cofilin-2 is more effective than cofilin-1 in sarcomeric actin binding (59). This implicates cofilin-2 as having a more significant role in sarcomeric actin depolymerization (57,60,61). We have shown that cofilin-2 is not activated by ERK1/2-catalyzed phosphorylation in LMNA cardiomyopathy, as it lacks the consensus Ser/Thr-Pro amino acid sequence. Our results demonstrate that only cofilin-1, under certain condition, disassembles F-actin in CMs and participates to the pathogenesis of left ventricular dysfunction.
Cofilin-1 maintains the pool of G-actin monomers and thus remodels actin filaments by enhancing assembly/disassembly dynamics. Cofilin-1 regulation of F-actin dynamics is controlled by reversible phosphorylation. Several mechanisms regulate the activation of cofilin-1, including phosphorylation at Ser3 catalyzed by LIMK1/2 or testicular protein kinase 1 (62,63). The LIMK-catalyzed phosphorylation of cofilin-1 on Ser3 inactivates its F-actin depolymerization, leading to accumulation of polymerized F-actin filaments (64). Phosphorylation of cofilin-1 at Tyr68 by v-Src, which does not change its activity, induces its ubiquitination and degradation through the proteasome pathway (65). Aurora A kinase also catalyzes the phosphorylation of cofilin-1 on Ser3, Ser8 and Thr25 during mitosis (66). Our results now identify Thr25 as a phosphorylated residue in cofilin-1 that specifically activates its F-actin depolymerizing function, participating to impaired left ventricular contractility. It would be interesting in the future to further mechanistically assess how active cofilin-1 phosphorylated on Thr25 stimulates the F-actin depolymerization.
We observed that the presence of active cofilin-1 phosphorylated on Thr25 in hearts from LmnaH222P/H222P mice impedes the organization of contractile apparatus. These data suggest that cofilin-1 phosphorylated on Thr25 participates to the disassembly of sarcomeric actin and loss of integrity of sarcomeres. The actin length in sarcomeres is a process coordinated by several actin-binding proteins that regulate pointed-ends dynamics (67). A role of cofilin for the sarcomeric actin organization in striated muscle has been previously proposed (57,68). We cannot exclude that cofilin-1 phosphorylated on Thr25 impedes the emergence of sarcomeric structures from the actin clusters coalescence (69) or from ARP2/3-dependent network actin-integrin connection known to strengthen newly formed myofibril structures (70). In addition, the disassembly of cytoskeletal F-actin could directly control the nuclear activity of transcriptional cofactors of the myocardin protein family. The cytosolic G-actin buffers the myocardin-related transcription factors, which in turn, control the activity of serum response factor (SRF), a nuclear transcription factor (71). There is thus a requirement for a relation between actin dynamics and transcription. SRF is playing a critical role in the normal cardiac muscle development and in the sarcomerogenesis (72–74). Cardiac SRF-null mice display severe defects in the cardiac contractile apparatus (74), similar to the ones we observed in LmnaH222P/H222P mice. Given that SRF controls the expression of sarcomeric genes (74,75), it has been suggested that SRF is essential for the maintenance of normal cardiac sarcomere organization. It would be interesting to assess the role played by SRF in the alteration on sarcomeric structure in hearts from LmnaH222P/H222P mice, where actin dynamics in impaired.
Our results indicate that cofilin-1-mediated modulation of actin dynamics is a cellular consequence of ERK1/2 activation in cardiac cells. We cannot exclude form our study that a protein, other that cofilin-1, is directly implicated in F-actin disassembly. Indeed, alteration of actin dynamics was also observed in cells lacking A-type lamins (76), which could result from alteration of the linker of the nucleoskeleton and cytoskeleton (LINC complex) (77,78) or emerin (27,79,80). These data indicate that nuclear envelope is an important actor for actin cohesion. More work is needed to delineate the role played by the nuclear envelope on cytoskeleton stability in diseases. Given that both actin cytoskeleton and A-type lamins are necessary for the regulation of mechanosensing (81–84), and that cofilin-1 has recently been involved in nuclear integrity (78), we plan future studies to assess the role played by this novel phosphorylated (Thr25) cofilin-1 on mechanosensitivity in cardiac cells carrying LMNA mutations. We showed that cofilin-1 phosphorylation catalyzed by ERK1/2 is observed as early as 2 months of age in hearts from LmnaH222P/H222P mice. We further showed that phosphorylated (Thr25) cofilin-1 influences sarcomeric organization in vitro. These data suggest that cofilin-1 phosphorylation catalyzed by ERK1/2 is a pathological event in LMNA cardiomyopathy. Enhanced phosphorylation of ERK1/2 and cofilin-1 on Thr25 in hearts from a mouse model of LMNA cardiomyopathy and from patients with this disease supports our conclusion that this mechanism contributes to the pathology. Our work encourages further approaches to mechanistically assess the role played by this novel phosphorylated (Thr25) cofilin-1 on actin dynamics. These findings suggest that drugs that could correct impaired actin dynamics would ameliorate left ventricular dysfunction in LMNA cardiomyopathy. Similar pathogenic mechanism of cofilin-1 mediated modulation of F-actin dynamics may play a role in left ventricular dysfunction in other forms of cardiomyopathy, in which there appears to be abnormal activation of ERK1/2 signaling (85–88).
Materials and Methods
Cell culture and reagents
Generation of C2-WT and C2-H222P cells has been described previously (24). Cells were transiently transfected with plasmids using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Plasmids encoding GFP-ERK2, RFP-MEK1, GFP-ERK2 K52R and GFP-ERK2 T183A/Y185F were kindly provided by P. Stork (Oregon Health and Science University). Plasmids encoding GFP-lamin A, GFP-lamin A E358K, GFP-Lamin A L271P and GFP-LaminA N456I have been previously described (89). Plasmid encoding MLC2vGFP has been previously described (90). Working concentrations of 100 nM cytochalasin D, 100 nM latrunculin B, 200 nM jasplakinolide, 10 μM Y27632, 50 μM PD0325901, 10 μM U0126 and 50 μM selumetinib were prepared from stocks diluted in DMSO. Cells were incubated with cytochalasin D for 45 min, jasplakinolide for 40 min and selumetinib for 15 h.
To generate mouse embryonic CMs, hearts were dissected out from E9.5 mouse embryos; ventricles were cut and myocytes dissociated using 1 mg/ml collagenase and 0.3 m/ml pancreatin (15 min, 37°C), cells were spun down and replated on gelatin coated wells (DMEM supplemented with NEAA, glutamine and 10% FCS). HL-1 cardiac muscle cells were plated according to the manufacturer’s instructions (Merck Millipore). Cells were transiently transfected using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions.
Mice
Lmna H222P/H222P mice (22) were fed chow and housed in a disease-free barrier facility at 12 h/12 h light/dark cycles. All animal experiments were approved by the French Ministry of Health at the Center for Research in Myology for the Care and Use of Experimental Animals. The animal experiments were performed according to the guidelines from Directive 2010/63/EU of the European Parliament on the protection of animals used for scientific purposes.
Human heart tissue
Sections of explanted hearts from human subjects with LMNA mutations were obtained without identifiers from Myobank-AFM de l’lnstitut de Myologie. Myobank-AFM received approval from the French Ministry of Health and from the Committee for Protection of Patients to share tissues and cells of human origin for scientific purposes, ensuring the donors’ anonymity, respect of their volition and consent according to the legislation. The subjects were a 23-year-old man with cardiomyopathy associated with muscular dystrophy and LMNA delK261 mutation, a 53-year-old man with cardiomyopathy and LMNA E33D mutation and a 47-year-old woman with cardiomyopathy and LMNA R60G mutation. Control human heart samples were obtained from the National Disease Research Interchange; information regarding donor confidentiality and consent can be found at http://www.ndriresource.org. Control human heart samples were obtained from a 57-year-old man with an intracranial bleed, a 15-year-old woman who died of a drug overdose and a 46-year-old man who died from end-stage liver disease.
iPSCs generation and differentiation into CMs
iPSCs have been generated from peripheral blood mononuclear cells (PBMNCs) obtained from a patient carrying the LMNA p.R190W mutation, after signed informed consent. Reprogramming has been induced using the Cytotune iPS-2.0 Sendai Reprogramming kit (ThermoScientific) on T-lymphocytes activated from PBMNCs using CD3 and CD28 ligands, as described (91). Reprogrammed clones have been selected and characterized as previously (92) and two fully characterized lines been used for the experiments. As controls, already generated wild-type lines have been employed (93). Differentiation into CMs has been achieved using a chemically defined serum free induction protocol, which is based on the modulation of the Wnt pathway, as previously reported (94,95). In brief, iPSCs were induced with CHIR99021 for 24 h, which mediates Wnt activation, and subsequently exposed to IWR-1 (Wnt inhibitor) for 3 days in RPMI-B27 medium. At Day 10 of induction, medium was supplemented with insulin. Spontaneous contracting activity usually appears around Days 8–10 and CMs were used for experiments around 25–30 days after spontaneous contraction has started. For the immunofluorescence staining, CMs were plated onto glass coverslips coated with fibronectin and laminin, fixed in 2% paraformaldehyde.
Measurement of F-actin/G-actin ratio
The ratio of F-actin to G-actin was determined using the G-actin/F-actin in vivo assay kit (Cytoskeleton) according to the manufacturer's instructions. Briefly, 2 mg of protein from cells or frozen heart tissues were homogenized in Lysis and F-actin Stabilization Buffer and centrifuged at 2000 rpm for 5 min to remove unbroken cells. F-actin was separated from G-actin by centrifugation at 100 000g for 60 min at 37°C. The F-actin-containing pellet was resuspended in F-actin Depolymerizing Buffer at a volume equivalent to the G-actin-containing supernatant volume. The resuspended F-actin pellet was kept on ice for 60 min and was gently mixed every 15 min to dissociate F-actin. Proteins in equivalent volumes (10 μl) of supernatant and pellet were separated by SDS-PAGE and subjected to immunoblot analysis using an anti-pan actin antibody supplied in the kit. F/G actin ratio was quantified using ImageJ software.
RNA isolation and real-time PCR
Total RNA was extracted from cells or mice hearts using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. One microgram of total RNA was subjected to cDNA synthesis using the First-Strand cDNA Synthesis Kit (Life Technologies). Mouse primer sequences used for transcriptional analyses were as follows: mCofilin-1 5′-CGCAAGTCTTCAACACCAGA-3′, 5′-TGAACACCAGGTCCTCCTTC-3′; Myh7 5′-ACTGTCAACACTAAGAGGGTCA-3′, 5′-TTGGATGATTTGATCTTCCAGGG-3′; Col1a2 5′-CCGTGCTTCTCAGAACATC-3′, 5′-GAGCAGCCATCGACTAGGAC-3′; Nppa 5′-GCTTCCAGGCCATATTGGAG-3′, 5′-GGGGGCATGACCTCATCTT-3′. Real-time quantitative PCR reactions were performed on a LightCycler 480 (Roche) using the SYBR Green PCR Master Mix (Applied Biosystems). The PCR products were subjected to melting curve analysis to exclude the synthesis of non-specific products. Cycle threshold (Ct) values were quantified using a standard curve for the specific gene and relatively quantified using RplR0 as an internal reference control. The Ct values were then normalized to the average expression levels of samples, calculated according to the △△Ct method (96) and are presented as fold change over wild-type controls. All experiments were performed in triplicates.
Protein extraction and immunoblotting
Total proteins were isolated by resuspending mouse heart tissue or cultured cells in extraction buffer (Cell Signaling) with the addition of protease inhibitors (25 mg/ml aprotinin, 10 mg/ml leupeptin, 1 mM 4-[2-aminoethyl]-benzene sulfonylfluoride hydrochloride and 2 mM Na3VO4). The lysates were sonicated (3 pulses of 10 s at 30% amplitude) to allow dissociation of protein from chromatin and solubilization. Cytosolic and nuclear fractions were prepared using the NE-PER Nuclear and Cytosolic Extraction Reagents (ThermoFisher Scientific) according to the manufacturer’s instructions. Sample protein content was determined by the BiCinchoninic Acid Assay protein assay (ThermoFisher Scientific). Extracts were analyzed by SDS-PAGE using a 10% gel and transferred onto nitrocellulose membranes (Invitrogen). Subsequent to being washed with Tris–buffered saline containing 1% Tween 20 (TBS-T), the membranes were blocked in 5% bovine serum albumin (BSA) in TBS-T for 1 h at room temperature, then incubated with the appropriate antibody overnight at 4°C. Subsequent to being washed with TBS-T, the membranes were incubated with horseradish peroxidase-conjugated anti-rabbit or anti-mouse antibodies for 1h at room temperature. After washing with TBS-T, the signal was revealed using Immobilon Western Chemiluminescent HorseRadish Peroxidase (HRP) Substrate (Millipore) on a G-Box system with GeneSnap software (Ozyme).
Antibodies
Primary antibodies used were: anti-cofilin1 (Cell Signaling), anti-phosphorylated (Ser3) cofilin-1 (Cell Signaling), anti-cofilin-2 (ThermoFisher Scientific), anti-profilin1 (Cell Signaling), anti-N-wasp (Cell Signaling), anti-Arp2 (Cell Signaling), anti-pERK1/2 antibody (Cell Signaling), anti-ERK1/2 (Santa Cruz Biotechnology), anti-lamin A/C (Santa Cruz Biotechnology), anti-gapdh (Santa Cruz Biotechnology) and anti-α-actinin (Abcam). Secondary antibodies for immunofluorescence were Alexa Fluor-488 conjugated goat anti-rabbit IgG, Alexa Fluor 568-conjugated goat anti-mouse IgG and Alexa-Fluor-488-conjugated donkey anti-goat IgG (Life Technologies). Secondary antibodies for immunoblotting were HRP-conjugated rabbit anti-mouse and goat-anti rabbit IgG (Jackson ImmunoResearch). We generated custom-made anti-phosphorylated (Thr25) cofilin-1 (GenScript). Briefly, we used cofilin-1 phosphopeptide (RKSS{phosphoT}PEEVKKRKKA) conjugated with keyhole limpet hemocyanin (KLH) for rabbit immunization. Two animals were immunized with 200 μg of purified cofilin-1 phosphopeptide. The animals received three booster injections at 2 weeks intervals. The antibody was tested by ELISA on the crude blood serum.
Immunofluorescence microscopy
For immunofluorescence microscopy, frozen tissues were cut into 8-μm-thick sections. Cryosections were fixed [15 min, 4% paraformaldehyde in phosphate-buffered saline (PBS) at room temperature], permeabilized (10 min, 0.5% Triton X-100 in PBS) and blocked (1 h, PBS with 0.3% Triton X-100, 5% BSA). Sections were incubated with primary antibodies (overnight, 4°C, in PBS with 0.1% Triton X-100 and 1% BSA) and washed in PBS. The sections were then incubated for 1 h with secondary antibodies. Sections were washed with PBS and slides were mounted in Vectashield mounting medium with dapi (Vector Laboratories).
C2C12 cells were grown on coverslips, washed with PBS and fixed with 4% paraformaldehyde in PBS for 10 min. Cells were permeabilized with 0.2% Triton X-100 diluted in PBS for 7 min and non-specific signals were blocked by incubation in 0.2% Triton X-100, 5% BSA for 30 min. The samples were then incubated with primary antibody for 1 h in PBS with 0.1% Triton X-100 and 1% BSA at room temperature. Cells were washed with PBS and incubated for 1 h with secondary antibodies. F-actin was stained with Alexa Fluor 568-phalloidin and G-actin with Alexa Fluor 488-deoxyribonuclease I for 1 h at room temperature. Cells and slides were then mounted in Vectashield mounting medium with dapi (Vector Laboratories). Immunofluorescence microscopy was performed using an Axiophot microscope (Carl Zeiss). All the images were digitally deconvolved using Autodeblur v9.1 (Autoquant) deconvolution software and were processed using Adobe Photoshop 6.0 (Adobe Systems).
Electron microscopy
Freshly harvested left ventricle apex was cut into small pieces and immediately fixed by immersion in 2.5% glutaraldehyde diluted in PBS for 1 h at room temperature. After washing in PBS, samples were post-fixed with 1% OsO4, dehydrated in a graded series of acetone and embedded in an epoxy resin. Ultrathin sections were cut at 90 nm and stained with uranyl acetate and lead citrate, examined using a transmission electron microscope (JEOL 1011) and photographed with a digital Erlangshen 1000 camera (GATAN), using Digital Micrograph software.
Immunoprecipitation
Cells and cardiac tissues were lysed in 0.5 ml of lysis buffer [50 mM Tris–HCl (pH 7.5), 0.15 M NaCl, 1 mM EDTA, 1% NP-40]. Mouse cardiac muscle tissue lysate was diluted to 2 mg/ml in lysis buffer, pre-cleared with 20 µl washed protein G-Sepharose 4 fast-flow (GE Healthcare) and incubated with 20 µg of specific antibody overnight at 4°C. Next, 30 µl of washed beads was added and incubated for 2 h at 4°C. Pelleted beads were collected in sample buffer [0.25 M Tris–HCl (pH 7.5), 8% SDS, 40% glycerol, 20% β-mercaptoethanol] and subjected to SDS-PAGE and immunoblotting. For control reactions, we used rabbit immunoglobulin G1.
Construction of plasmids encoding wild-type and cofilin-1 variants
Mutagenesis was carried out using QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies) according to the manufacturer's instructions. Cofilin-pmCherryC1 was a gift from Christien Merrifield (Addgene plasmid # 27687) (97). Cofilin-1-pmCherryC1 was used as a template for substituting threonine for alanine at position 25 and 148. Primers used for the introduction of single mutations are: mCofilin-1-T25A 5′-CTTCACTTCTTCTGGTGCTGAAGACTTGCGAACCT-3′, 5′-AGGTTCGCAAGTCTTCAGCACCAGAAGAAGTGAAG-3′; mCofilin-1-T148A 5′-TTCTCTGCCAGGGCGCAGCGGTCCTTG-3′, 5′-CAAGGACCGCTGCGCCCTGGCAGAGAA-3′, mCofilin-1-T25D 5′-CGTTTCTTCACTTCTTCTGGATCTGAAGACTTGCGAACCTTCATGTCA-3′, 5′-TGACATGAAGGTTCGCAAGTCTTCAGATCCAGAAGAAGTGAAGAAACG-3′. The final plasmids were transformed in XL1-Blue super competent cells. Mutations were verified by DNA sequencing using an appropriate set of oligonucleotides to cover the full length of the cofilin1 cDNA.
Protein expression and purification
Cofilin-1, cofilin-1-T25A and cofilin-2 cDNAs cloned in pNIC-CTHF vector expressing a cleavable C-terminal His6 and FLAG tag were obtained from Nicola Burgess-Brown (Structural Genomics Consortium, Oxford). The plasmid encoding constitutively MEK1 and His-ERK2 kinase, pET-His6-ERK2-MEK1 R4F co-expression, was a gift from Melanie Cobb (Addgene plasmid # 39212) (98). His-cofilin-1 and His-cofilin-2 recombinant proteins were expressed in Rosetta (DE3) E. coli and His-MEK1-ERK2 was expressed in BL21 (DE3) E. coli. Bacteria were grown in 2× YT medium until they reached an optical density (OD600) of 0.8 and recombinant protein expression was induced by the addition of 0.5 mM isopropyl-thiogalactopyranoside for 16–24 h at 30°C for BL21 (DE3) or 18°C for Rosetta (DE3) before harvesting. Cells were sonicated in lysis buffer [50 mM HEPES (pH 7.5), 500 mM NaCl, 5% glycerol, 5 mM imidazole 0.5 mM TCEP with complete EDTA-free Protease Inhibitor Cocktail, Roche]. After centrifugation at 40 000g for 30 min at 4°C, the soluble fraction was filtered through a 0.8 µm filter and subjected to immobilized metal affinity Ni2+-nitrilotriacetic resin chromatography. Recombinant His6-tagged proteins eluted with elution buffer [50 mM HEPES (pH 7.5), 300 mM NaCl, 5% glycerol and 250 mM imidazole]. His-tagged cofilin proteins were purified by size exclusion chromatography on a HiLoad 16/60 Superdex 75 preparative-grade column. His-tagged active ERK2 was purified as described previously (98). Briefly, following affinity purification, eluted His-pERK2 was dialyzed overnight in 50 mM HEPES (pH 7.5), 50 mM NaCl, 1 mM dithiotreitol, 20% glycerol, diluted 1:1 in the same buffer without glycerol then applied to a MonoQ GL 5/50 column and eluted on a gradient from 50 mM to 1 M NaCl.
In vitro kinase assay
Recombinant His-tagged cofilin-1, cofilin-1-T25A and cofilin-2 (5 μg) were incubated alone or with 0.5 μg recombinant active His-ERK2, in kinase buffer [50 mM HEPES (pH 7.5), 150 M NaCl, 5 mM MgCl2, 0.5 mM TCEP] in a total volume of 100 µl containing 1 mM ATP. Reaction mixtures were incubated at 37°C for 1 h and were terminated by the addition of 100 µl of 2× SDS sample buffer. Proteins were resolved by SDS-PAGE. Blots were incubated with anti-His (Sigma-Aldrich H1029), cofilin-1 and phosphorylated (Thr25) cofilin-1 antibodies.
Radioactive in vitro kinase assay
Cofilin-1, cofilin-1-T25A or dephosphorylated myelin basic protein (MBP) (Merck-Millipore) were incubated alone or in the presence of purified pERK2 in buffer comprising 50 mM HEPES (pH 7.5), 150 mM NaCl, 5 mM MgCl2, 0.5 mM TCEP and 0.5 mM ATP. A 10 pmol of protein was used and 0.5 µCi [32P]-γ-ATP was added per sample (total volume 25 µl). The samples were incubated for 30 min at 37°C and the reaction terminated by the addition of 5 µl 6× SDS-PAGE sample buffer. Proteins were then separated by SDS-PAGE and blotted onto nitrocellulose. The nitrocellulose membrane was then used to expose a phosphor screen (typically for 24 h), which was then imaged using a Typhoon FLA 7000 system.
Construction and injection of AAV encoding cofilin-1
AAV vectors of serotype rh10, carrying wild-type or mutant cofilin-1 under the control of the cytomegalovirus immediate/early promoter were prepared by the triple transfection method in HEK293T cells as previously described (99). Briefly, cells were transfected with (i) the adenovirus helper plasmid, (ii) the AAV packaging plasmid encoding the rep2 and cap-rh10 genes and (iii) the AAV2 plasmid expressing cytomegalovirus promoter cofilin-1 cDNA. Seventy-two hours after transfection, cells were harvested and AAV vectors were purified by ultracentrifugation through an iodixanol gradient and concentrated with Ultra–Ultra cell 100 K filter units (Amicon) in 0.1 M PBS, 1 mM MgCl2 and 2.5 mM KCl. Aliquots were stored at −80°C until use. Vector titers were determined by real-time PCR. Three-month-old mice were injected with scAAVrh10-cofilin-1 into the retro-orbital vein (5×1013 viral genomes/kg in 100 μl) with an insulin syringe.
Two-dimensional gel electrophoresis
Proteins were extracted from cells in a buffer composed of 7 M urea, 2 M thiourea, 1% CHAPS, 10% isopropanol, 10% isobutanol, 0.5% Triton X-100 and 0.5% SB3-10. Proteins were precipitated using the Perfect Focus kit (G-Biosciences) and pellets were resuspended in the same buffer supplemented with 25 mM Tris−HCl (pH 8.8). The protein content was assessed by the Quick StartTM Bradford protein assay (BioRad) using BSA as standard. For each sample, 50 µg of proteins were labeled with 400 pmol of Cy2 (Fluoprobes), incubated 30 min on ice and then quenched with 0.35 mM lysine for 10 min. Then, 700 µg of unlabeled proteins were added to 50 µg of Cy2-labeled proteins.
Proteins were separated using 24 cm gels with a pH range 3–10 using commercial strips (GE Healthcare). Strips were passively rehydrated overnight directly with the samples supplemented with 40 mM dithiothreitol and 0.5% ampholites. Isoelectric focusing migration was set as follows: 3 h at 50 V, 3 h at 200 V, 2 h gradient from 200 to 1000 V, 2 h at 1000 V, 2 h gradient from 1000 to 10 000 V, 8 h at 10 000 V. A second step of isoelectric focusing was performed using the following parameters: 30 min at 200 V, 1 h gradient from 200 to 10 000 V, 4 h at 10 000 V. The area of the strips corresponding to the pH range from 5.5 to 9 was excised and frozen at −20°C until use for the second dimension. Strips were incubated for 15 min in equilibration buffer [6 M urea, 75 mM Tris–HCl (pH 8.8), 26 and 2% SDS] supplemented with 65 mM dithiothreitol and then for 20 min in equilibration buffer supplemented with 135 mM iodoacetamide. The second dimension was run in 12% acrylamide gels at 25 V for the first hour then 150 V and 12 mA per gel in a Tris–glycine buffer. Gel images were acquired on Ettan DIGE Imager (GE Healthcare). Each gel was run two times using the same conditions: first gel was used for immunoblotting and the second for protein identification.
In-gel digestion and mass spectrometry
Gels were stained with silver nitrate for 1 min sensitizing in 0.02% sodium thiosulfate, rinsed two times in ultra-pure water, incubated for 30 min in 0.212% silver nitrate, rinsed two times in ultra-pure water and developed in 3% sodium carbonate, 0.00125% sodium thiosulfate and 0.03% formalin. Staining was stopped by soaking the gels in 5% acetic acid. Stained protein spots of interest were manually excised, sliced into 1 mm cubes, destained in 50 mM sodium thiosulfate, 15 mM potassium ferricyanure and washed several times alternatively in water and acetonitrile. Tryptic digestion was performed overnight at 37°C, using 200 ng of mass spectrometry grade trypsin (G-Biosciences) in 50 mM ammonium bicarbonate, 5% acetonitrile. Supernatants were collected and gel pieces washed two times for 15 min in an ultrasonic bath with 50 µl of 0.1% trifluoroacetic acid and 60% acetonitrile. Peptides solutions were dried using a vacuum centrifuge and resuspended in 12 µl of 2.5% acetonitrile and 0.1% formic acid.
Identification of phosphopeptides by mass spectrometry
Three microliters of each sample were analyzed in LC–MS–MS using an Ultimate 3000 Rapid Separation liquid chromatographic system coupled to an Orbitrap Fusion mass spectrometer (ThermoFisher Scientific). Peptides were loaded on a C18 reverse phase resin (2 µm particle size, 100 Å pore size, 75 µm i.d., 15 cm length) with a 50 min ‘effective’ gradient from 99% A (0.1% formic acid and 100% H2O) to 40% B (80% acetonitrile, 0.085% formic acid and 20% H2O). The Orbitrap Fusion mass spectrometer acquired data throughout the elution process and operated in a data-dependent scheme with full MS scans acquired with the Orbitrap, followed by stepped HCD MS/MS (top speed in 3 s maximum) on the most abundant ions detected in the MS scan. Mass spectrometer settings were: full MS (AGC: 4E5, resolution: 1.2E5, m/z range 350–1500, maximum ion injection time: 60 ms); MS/MS (Normalized Collision Energy: 30, resolution: 17 500, intensity threshold: 1E4, isolation window: 1.6 m/z, dynamic exclusion time setting: 15 s, AGC Target: 1E4 and maximum injection time: 100 ms). The fragmentation was permitted for precursors with a charge state of 2–4. Proteome discoverer 1.4 software was used to generate .mpg files. Data analysis was performed with X! tandem (win-15-04-01) and X! tandem pipeline v3.3.4 (http://pappso.inra.fr/bioinfo/xtandempipeline/) using the SwissProt M. musculus database (16 762 entries, 2 January 2016). The following parameters were used: 1 missed cleavage, 5 ppm MS error tolerance, 0.6 Da MS2 error tolerance, peptide charge max 4+ and Cys-CAM as a fixed modification and methionine oxidation as a variable modification. N-terminal acetylation and phosphorylation of Y, S and T were set as variable modifications for a refine search and the phosphopeptide option of X! tandem pipeline was run to determine phosphorylated residues localization. Peptides were validated if e value <0.05 and proteins when e value <0.001.
Direct observation of F-actin severing
The F-actin-severing assay was performed as previously described (100). Purified muscle actin (Cytoskeleton) monomers were polymerized in 500 mM KCL, 10 nM ATP at room temperature for 1 h. Actin filaments were allowed to adhere on glass coverslips overnight at 4°C, washed with buffer [5 mM Tris–HCl (pH 8.0), 0.2 mM CaCl2, 0.2 mM ATP and 0.5 mM dithiothreitol] and incubated with protein extracts (10 μg/μl) for 30 min at 4°C. Actin filaments were thereafter incubated with phalloidin Alexa Fluor 555 (ThermoFisher Scientific) and mounted with Vectashield mounting media (Vector Laboratories). Labeled F-actin filaments were viewed using an Axiovert 200 fluorescent microscope (Zeiss).
Echocardiography
Lmna H222P/H222P and wild-type mice were anesthetized with 0.75% isoflurane in O2 and placed on a heating pad (25°C). Echocardiography was performed using an ACUSON 128XP/10 ultrasound with an 11 MHz transducer applied to the chest wall. Cardiac ventricular dimensions and fractional shortening were measured in 2D mode and M-mode three times for the number of animals indicated. A ‘blinded’ echocardiographer, unaware of the genotype and the treatment, performed the examinations.
Statistics
Statistical analyses were performed using GraphPad Prism software. Statistical significance between groups of mice analyzed by echocardiography was analyzed with a corrected parametric test (Welch’s t test), with a value of P < 0.05 being considered significant. To validate results of echocardiographic analyses, we performed a non-parametric test (Wilcoxon–Mann–Whitney test). For all other experiments, a two-tailed Student’s t test was used with a value of P < 0.05 considered significant. Values are represented as means±standard errors of mean (SEM). Sample sizes are indicated in the figure and table legends.
Supplementary Material
Acknowledgements
We thank 3P5 proteomic facility (Institut Cochin, Paris) for LC–MS/MS data acquisition. We thank Christien Merrifield, Melanie Cobb, Nicola Burgess-Brown and Philip Stork for essential reagents.
Conflict of Interest statement. H.J.W. and A.M. are members of the scientific advisory board of AlloMek Therapeutics, LLC, a privately held pharmaceutical company developing small molecules to inhibit ERK1/2 signaling. All other authors have declared no conflicts of interest.
Funding
This work was supported by the Association Française contre les Myopathies, Cure CMD and Fundación Andrés Marcio to A.M., and a collaborative grant (Agence Nationale de la Recherche) between W.H.G., H.J.W. and A.M. H.J.W. was supported by NIH/NIAMS grant AR048997.
References
- 1. Bonne G., Di Barletta M.R., Varnous S., Bécane H.M., Hammouda E.H., Merlini L., Muntoni F., Greenberg C.R., Gary F., Urtizberea J.A.. et al. (1999) Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat. Genet., 21, 285–288. [DOI] [PubMed] [Google Scholar]
- 2. Fatkin D., MacRae C., Sasaki T., Wolff M.R., Porcu M., Frenneaux M., Atherton J., Vidaillet H.J., Spudich S., De Girolami U.. et al. (1999) Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N. Engl. J. Med., 341, 1715–1724. [DOI] [PubMed] [Google Scholar]
- 3. Fisher D.Z., Chaudhary N., Blobel G. (1986) cDNA sequencing of nuclear lamins A and C reveals primary and secondary structural homology to intermediate filament proteins (lamin sequence identity/divergent carboxyl terminal/a-helical domains/coiled coils/nuclear localization sequence). Cell Biol., 83, 6450–6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McKeon F.D., Kirschner M.W., Caput D. (1986) Homologies in both primary and secondary structure between nuclear envelope and intermediate filament proteins. Nature, 319, 463–468. [DOI] [PubMed] [Google Scholar]
- 5. Lin F., Worman H.J. (1993) Structural organization of the human gene encoding nuclear lamin A and nuclear lamin C. J. Biol. Chem., 268, 16321–16326. [PubMed] [Google Scholar]
- 6. Gerace L., Blobel G. (1980) The nuclear envelope lamina is reversibly depolymerized during mitosis. Cell, 19, 277–287. 1980). [DOI] [PubMed] [Google Scholar]
- 7. Muchir A., Pavlidis P., Decostre V., Herron A.J., Arimura T., Bonne G., Worman H.J. (2007) Activation of MAPK pathways links LMNA mutations to cardiomyopathy in Emery–Dreifuss muscular dystrophy. J. Clin. Invest., 117, 1282–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sheehy S.P., Grosberg A., Parker K.K. (2012) The contribution of cellular mechanotransduction to cardiomyocyte form and function. Biomech. Model. Mechanobiol., 11, 1227–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lyon R.C., Zanella F.J., Omens H., Sheikh F. (2015) Mechanotransduction in cardiac hypertrophy and failure. Circ. Res., 116, 1462–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Casella J.F., Craig S.W., Maack D.J., Brown A.E. (1987) Cap Z(36/32), a barbed end actin-capping protein, is a component of the Z-line of skeletal muscle. J. Cell Biol., 105, 371–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Labeit S., Kolmerer B. (1995) Titins: giant proteins in charge of muscle ultrastructure and elasticity. Science, 270, 293–296. [DOI] [PubMed] [Google Scholar]
- 12. Agarkova I., Perriard J.-C. (2005) The M-band: an elastic web that crosslinks thick filaments in the center of the sarcomere. Trends Cell Biol., 15, 477–485. [DOI] [PubMed] [Google Scholar]
- 13. Chereau D., Boczkowska M., Skwarek-Maruszewska A., Fujiwara I., Hayes D.B., Rebowski G., Lappalainen P., Pollard T.D., Dominguez R. (2008) Leiomodin is an actin filament nucleator in muscle cells. Science, 320, 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lange S., Ehler E., Gautel M. (2006) From A to Z and back? Multicompartment proteins in the sarcomere. Trends Cell Biol., 16, 11–18. [DOI] [PubMed] [Google Scholar]
- 15. Pappas C.T., Krieg P.A., Gregorio C.C. (2010) Nebulin regulates actin filament lengths by a stabilization mechanism. J. Cell Biol., 189, 859–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sanger J.M., Sanger J.W. (2008) The dynamic Z bands of striated muscle cells. Sci. Signal, 1, pe37.. [DOI] [PubMed] [Google Scholar]
- 17. Paavilainen V.O., Bertling E., Falck S., Lappalainen P. (2004) Regulation of cytoskeletal dynamics by actin-monomer-binding proteins. Trends Cell Biol., 14, 386–394. [DOI] [PubMed] [Google Scholar]
- 18. Nicholson-Dykstra S., Higgs H.N., Harris E.S. (2005) Actin dynamics: growth from dendritic branches. Curr. Biol., 15, R346–3R357. [DOI] [PubMed] [Google Scholar]
- 19. Ono S. (2007) Mechanism of depolymerization and severing of actin filaments and its significance in cytoskeletal dynamics. Int. Rev. Cytol., 258, 1–82. [DOI] [PubMed] [Google Scholar]
- 20. Kho A.L., Perera S., Alexandrovich A., Gautel M. (2012) The sarcomeric cytoskeleton as a target for pharmacological intervention. Curr. Opin. Pharmacol., 12, 347–354. [DOI] [PubMed] [Google Scholar]
- 21. Van Troys M., Huyck L., Leyman S., Dhaese S., Vandekerkhove J., Ampe C. (2008) Ins and outs of ADF/cofilin activity and regulation. Eur. J. Cell Biol., 87, 649–667. [DOI] [PubMed] [Google Scholar]
- 22. Arimura T., Helbling-Leclerc A., Massart C., Varnous S., Niel F., Lacène E., Fromes Y., Toussaint M., Mura A.M., Kelle D.I.. et al. (2005) Mouse model carrying H222P-Lmna mutation develops muscular dystrophy and dilated cardiomyopathy similar to human striated muscle laminopathies. Hum. Mol. Genet., 14, 155–169. [DOI] [PubMed] [Google Scholar]
- 23. Muchir A., Reilly S.A., Wu W., Iwata S., Homma S., Bonne G., Worman H.J. (2012) Treatment with selumetinib preserves cardiac function and improves survival in cardiomyopathy caused by mutation in the lamin A/C gene. Cardiovasc. Res., 93, 311–319. [DOI] [PubMed] [Google Scholar]
- 24. Choi J.C., Wu W., Muchir A., Iwata S., Homma S., Worman H.J. (2012) Dual specificity phosphatase 4 mediates cardiomyopathy caused by lamin A/C (LMNA) gene mutation. J. Biol. Chem., 287, 40513–40524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Muchir A., Kim Y.J., Reilly S.A., Wu W., Choi J.C., Worman H.J. (2013) Inhibition of extracellular signal-regulated kinase 1/2 signaling has beneficial effects on skeletal muscle in a mouse model of Emery-Dreifuss muscular dystrophy caused by lamin A/C gene mutation. Skelet. Muscle, 3, 17–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chatzifrangkeskou M., Muchir A. (2015) Extracellular signal-regulated kinases 1/2 and their role in cardiac diseases. Sci. Proc., 2, e457. [Google Scholar]
- 27. Ho C.Y., Jaalouk D.E., Vartiainen M.K., Lammerding J. (2013) Lamin A/C and emerin regulate MKL1-SRF activity by modulating actin dynamics. Nature, 497, 507–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Quijano-Roy S., Mbieleu B., Bonnemann C.G., Jeannet P.Y., Colomer J., Clarke N.F., Cuisset J.M., Roper H., De Meirleir L., D’Amico A.. et al. (2008) De novo LMNA mutations cause a new form of congenital muscular dystrophy. Ann. Neurol., 64, 177–186. [DOI] [PubMed] [Google Scholar]
- 29. Kichuk Chrisant M.R., Drummond-Webb J., Hallowell S., Friedman N.R. (2004) Cardiac transplantation in twins with autosomal dominant Emery–Dreifuss muscular dystrophy. J. Hear. Lung Transpl., 23, 496–417. [DOI] [PubMed] [Google Scholar]
- 30. Brown C.A., Lanning R.W., McKinney K.Q., Salvino A.R., Cherniske E., Crowe C.A., Darras B.T., Gominak S., Greenberg C.R., Grosmann C.. et al. (2001) Novel and recurrent mutations in lamin A/C in patients with Emery–Dreifuss muscular dystrophy. Am. J. Med. Genet., 102, 359–367. [DOI] [PubMed] [Google Scholar]
- 31. Choi J.C., Muchir A., Wu W., Iwata S., Homma S., Morrow J.P., Worman H.J. (2012) Temsirolimus activates autophagy and ameliorates cardiomyopathy caused by lamin A/C gene mutation. Sci. Transl. Med., 4, 144ra102.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wu W., Iwata S., Homma S., Worman H.J., Muchir A. (2014) Depletion of extracellular signal-regulated kinase 1 in mice with cardiomyopathy caused by lamin A/C gene mutation partially prevents pathology before isoenzyme activation. Hum. Mol. Genet., 23, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wortzel I., Seger R. (2011) The ERK Cascade: distinct functions within various subcellular organelles. Genes Cancer, 2, 195–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gonzalez F.A., Raden D.L., Davis R.J. (1991) Identification of substrate recognition determinants for human ERK1 and ERK2 protein kinases. J. Biol. Chem., 266, 22159–22163. [PubMed] [Google Scholar]
- 35. Lorenz K., Schmitt J.P., Schmitteckert E.M., Lohse M.J. (2009) A new type of ERK1/2 autophosphorylation causes cardiac hypertrophy. Nat. Med., 15, 75–83. [DOI] [PubMed] [Google Scholar]
- 36. Rossomando A.J., Wu J., Michel H., Shabanowitz J., Hunt D.F., Weber M.J., Sturgill T.W. (1992) Identification of Tyr-185 as the site of tyrosine autophosphorylation of recombinant mitogen-activated protein kinase p42mapk. Proc. Natl. Acad. Sci. USA., 89, 5779–5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bamburg J.R., McGough A., Ono S. (1999) Putting a new twist on actin: aDF/cofilins modulate actin dynamics. Trends Cell Biol., 9, 364–370. [DOI] [PubMed] [Google Scholar]
- 38. Agnew B.J., Minamide L.S., Bamburg J.R. (1995) Reactivation of phosphorylated actin depolymerizing factor and identification of the regulatory site. J. Biol. Chem., 270, 17582–17587. [DOI] [PubMed] [Google Scholar]
- 39. Maekawa M., Ishizaki T., Boku S., Watanabe N., Fujita A., Iwamatsu A., Obinata T., Ohashi K., Mizuno K., Narumiya S. (1999) Signaling from Rho to the actin cytoskeleton through protein kinases ROCK and LIM-kinase. Science, 285, 895–898. [DOI] [PubMed] [Google Scholar]
- 40. Wu W., Muchir A., Shan J., Bonne G., Worman H.J. (2011) Mitogen-activated protein kinase inhibitors improve heart function and prevent fibrosis in cardiomyopathy caused by mutation in lamin A/C gene. Hum. Mol. Genet., 123, 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Muchir A., Wu W., Choi J.C., Iwata S., Morrow J., Homma S., Worman H.J. (2012) Abnormal p38a mitogen-activated protein kinase signaling in dilated cardiomyopathy caused by lamin A/C gene mutation. Hum. Mol. Genet., 21, 4325–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wu W., Chordia M.D., Hart B.P., Kumarasinghe E.S., Ji M.K., Bhargava A., Lawlor M.W., Shin J.Y., Sera F., Homma S.. et al. (2017) Macrocyclic MEK1/2 inhibitor with efficacy in a mouse model of cardiomyopathy caused by lamin A/C gene mutation. Bioorg. Med. Chem., 25, 1004–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chatzifrangkeskou M., Le Dour C., Wu W., Morrow J.P., Joseph L.C., Beuvin M., Sera F., Homma S., Vignier N., Mougenot N.. et al. (2016) ERK1/2 directly acts on CTGT/CCN2 expression to mediate myocardial fibrosis in cardiomyopathyt caused by mutations in the lamin A/C gene. Hum. Mol. Genet., 25, 2220–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lammerding J., Schulze P.C., Takahashi T., Kozlov S., Sullivan T., Kamm R.D., Stewart C.L., Lee R.T. (2004) Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J. Clin. Invest., 113, 370–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mohri K., Takano-Ohmuro H., Nakashima K., Hayakawa K., Endo T., Hanaoka K., Obinata T. (2000) Expression of cofilin isoforms during development of mouse striated muscles. J. Muscle Res. Cell Motil., 21, 49–57. [DOI] [PubMed] [Google Scholar]
- 46. Chuderland D., Konson A., Seger R. (2008) Identification and characterization of a general nuclear translocation signal in signaling proteins. Mol. Cell, 31, 850–861. [DOI] [PubMed] [Google Scholar]
- 47. Ono S. (2010) Dynamic regulation of sarcomeric actin filaments in striated muscle. Cytoskeleton, 67, 677–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yamashiro S., Gokhin D.S., Kimura S., Nowak R.B., Fowler V.M. (2012) Tropomodulins: pointed-end capping proteins that regulate actin filament architecture in diverse cell types. Cytoskeleton, 69, 337–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gatayama R., Ueno K., Nakamura H., Yanagi S., Ueda H., Yamagishi H., Yasui S. (2013) Nemaline myopathy with dilated cardiomyopathy in childhood. Pediatrics, 131, e1986–e1990. [DOI] [PubMed] [Google Scholar]
- 50. Mir A., Lemler M., Ramaciotti C., Blalock S., Ikemba C. (2012) Hypertrophic cardiomyopathy in a neonate associated with nemaline myopathy. Congenit. Heart Dis., 7, E37–E41. [DOI] [PubMed] [Google Scholar]
- 51. D’Amico A., Graziano C., Pacileo G., Petrini S., Nowak K.J., Boldrini R., Jacques A., Feng J.J., Porfirio B., Sewry C.A.. et al. (2006) Fatal hypertrophic cardiomyopathy and nemaline myopathy associated with ACTA1 K336E mutation. Neuromuscul. Disord., 16, 548–552. [DOI] [PubMed] [Google Scholar]
- 52. Skyllouriotis M.L., Marx M., Skyllouriotis P., Bittner R., Wimmer M. (1999) Nemaline myopathy and cardiomyopathy. Pediatr. Neurol., 20, 319–321. [DOI] [PubMed] [Google Scholar]
- 53. Lynn Van Antwerpen C., Gospe S.M., Dentinger M.P. (1988) Nemaline myopathy associated with hypertrophic cardiomyopathy. Pediatr. Neurol., 4, 306–308. [DOI] [PubMed] [Google Scholar]
- 54. Arimura T., Takeya R., Ishikawa T., Yamano T., Matsuo A., Tatsumi T., Nomura T., Sumimoto H., Kimura A. (2013) Dilated cardiomyopathy-associated FHOD3 variant impairs the ability to induce activation of transcription factor serum response factor. Circ. J., 77, 2990–2996. [DOI] [PubMed] [Google Scholar]
- 55. Rosenson R.S., Mudge G.H. Jr, Sutton M.G. (1986) Nemaline cardiomyopathy. Am. J. Cardiol., 58, 175–177. [DOI] [PubMed] [Google Scholar]
- 56. Skwarek-Maruszewska A., Hotulainen P., Mattila P.K., Lappalainen P. (2009) Contractility-dependent actin dynamics in cardiomyocyte sarcomeres. J. Cell Sci., 122, 2119–2126. [DOI] [PubMed] [Google Scholar]
- 57. Kremneva E., Makkonen M.H., Skwarek-Maruszewska A., Gateva G., Michelot A., Dominguez R., Lappalainen P. (2014) Cofilin-2 controls actin filament length in muscle sarcomeres. Dev. Cell, 31, 215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Littlefield R., Almenar-Queralt A., Fowler V.M. (2001) Actin dynamics at pointed ends regulates thin filament length in striated muscle. Nat. Cell Biol., 3, 544–551. [DOI] [PubMed] [Google Scholar]
- 59. Nakashima K., Sato N., Nakagaki T., Abe H., Ono S., Obinata T. (2005) Two mouse cofilin isoforms, muscle-type (MCF) and non-muscle type (NMCF), interact with F-actin with different efficiencies. J. Biochem., 138, 519–526. [DOI] [PubMed] [Google Scholar]
- 60. Subramanian K., Gianni D., Balla C., Assenza G.E., Joshi M., Semigran M.J., Macgillivray T.E., Van Eyk J.E., Agnetti G., Paolocci N.. et al. (2015) Cofilin-2 phosphorylation and sequestration in myocardial aggregates: novel pathogenetic mechanisms for idiopathic dilated cardiomyopathy. J. Am. Coll. Cardiol., 65, 1199–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Agrawal P.B., Joshi M., Savic T., Chen Z., Beggs A.H. (2012) Normal myofibrillar development followed by progressive sarcomeric disruption with actin accumulations in a mouse Cfl2 knockout demonstrates requirement of cofilin-2 for muscle maintenance. Hum. Mol. Genet., 21, 2341–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Arber S., Barbayannis F.A., Hanser H., Schneider C., Stanyon C.A., Bernard O., Caroni P. (1998) Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature, 393, 805–809. [DOI] [PubMed] [Google Scholar]
- 63. Yang N., Higuchi O., Ohashi K., Nagata K., Wada A., Kangawa K., Nishida E., Mizuno K. (1998) Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature, 393, 809–812. [DOI] [PubMed] [Google Scholar]
- 64. Sumi T., Matsumoto K., Takai Y., Nakamura T. (1999) Cofilin phosphorylation and actin cytoskeletal dynamics regulated by Rho- and Cdc42-activated LIM-kinase 2. J. Cell Biol., 147, 1519–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yoo Y., Ho H.J., Wang C., Guan J.-L. (2010) Tyrosine phosphorylation of cofilin at Y68 by v-Src leads to its degradation through ubiquitin-proteasome pathway. Oncogene, 29, 263–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ritchey L., Chakrabarti R. (2014) Aurora A kinase modulates actin cytoskeleton through phosphorylation of Cofilin: implication in the mitotic process. Biochim. Biophys. Acta, 1843, 2719–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Littlefield R., Fowler V.M. (1998) Defining actin filament length in striated muscle: rulers and caps or dynamic stability? Annu. Rev. Cell Dev. Biol., 14, 487–525. [DOI] [PubMed] [Google Scholar]
- 68. Yamashiro S., Cox E.A., Baillie D.L., Hardin J.D., Ono S. (2008) Sarcomeric actin organization is synergistically promoted by tropomodulin, ADF/cofilin, AIP1 and profilin in C. elegans. J. Cell Sci., 121, 3867–3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Friedrich B.M., Fischer-Friedrich E., Gov N.S., Safran S.A. (2012) Sarcomeric pattern formation by actin cluster coalescence. PLoS Comput. Biol., 8, e1002544.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rui Y., Bai J., Perrimon N. (2010) Sarcomere formation occurs by the assembly of multiple latent protein complexes. PLoS Genet., 6, e1001208.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Olson E.N., Nordheim A. (2010) Linking actin dynamics and gene transcription to drive cellular motile functions. Nat. Rev. Mol. Cell Biol., 11, 353–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Niu Z., Iyer D., Conway S.J., Martin J.F., Ivey K., Srivastava D., Nordheim A., Schwartz R.J. (2008) Serum response factor orchestrates nascent sarcomerogenesis and silences the biomineralization gene program in the heart. Proc. Natl. Acad. Sci. USA., 105, 17824–17829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Miano J.M., Ramanan N., Georger M.A., de Mesy Bentley K.L., Emerson R.L., Balza R.O., Xiao Q., Weiler H., Ginty D.D., Misra R.P. (2004) Restricted inactivation of serum response factor to the cardiovascular system. Proc. Natl. Acad. Sci. USA., 101, 17132–17137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Balza R.O., Misra R.P. (2006) Role of the serum response factor in regulating contractile apparatus gene expression and sarcomeric integrity in cardiomyocytes. J. Biol. Chem, 281, 6498–6510. [DOI] [PubMed] [Google Scholar]
- 75. Sun Q., Chen G., Streb J.W., Long X., Yang Y., Stoeckert C.J., Miano J.M. (2006) Defining the mammalian CArGome. Genome Res., 16, 197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Corne T.D.J., Sieprath T., Vandenbussche J., Mohammed D., Te Lindert M., Gevaert K., Gabriele S., Wolf K., De Vos W.H. (2017) Deregulation of focal adhesion formation and cytoskeletal tension due to loss of A-type lamins. Cell Adh. Migr., 11, 447–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hale C.M., Shrestha A.L., Khatau S.B., Stewart-Hutchinson P.J., Hernandez L., Stewart C.L., Hodzic D., Wirtz D. (2008) Dysfunctional connections between the nucleus and the actin and microtubule networks in laminopathic models. Biophys. J., 95, 5462–5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kanellos G., Zhou J., Patel H., Ridgway R.A., Huels D., Gurniak C.B., Sandilands E., Carragher N.O., Sansom O.J., Witke W.. et al. (2015) ADF and cofilin1 cotrol actin stress fiber, nuclear integrity, and cell survival. Cell Rep., 13, 1949–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Chang W., Folker E.S., Worman H.J., Gundersen G.G. (2013) Emerin organizes actin flow for nuclear movement and centrosome orientation in migrating fibroblasts. Mol. Biol. Cell, 24, 3869–3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Holaska J.M., Kowalski A.K., Wilson K.L. (2004) Emerin caps the pointes end of actin filaments: evidence for an actin cortical network at the nucelar inner membrane. PLoS Biol., 2, E231.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Gardel M.L., Nakamura F., Hartwig J.H., Crocker J.C., Stossel T.P., Weitz D.A. (2006) Prestressed F-actin networks cross-linked by hinged filamins replicate mechanical properties of cells. Proc. Natl. Acad. Sci. USA., 103, 1762–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lange J.R., Steinwachs J., Kolb T., Lautscham L.A., Harder I., Whyte G., Fabry B. (2015) Micro-constriction arrays for high-throughput quantitative measurements of cell mechanical properties. Biophys. J., 109, 26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lange J.R., Metzner C., Richter S., Schneider W., Spermann M., Kolb T., Whyte G., Fabry B. (2017) Unbiased high-precision cell mechanical measurements with micro-constrictions. Biophys. J., 112, 1472–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Schwartz C., Fischer M., Mamchaoui K., Bigot A., Lok T., Verdier C., Duperray A., Michel R., Holt I., Voit T.. et al. (2017) Lamins and nesprin-1 mediate inside-out mechanical coupling in muscle cell precursors through FHOD1. Sci. Rep., 7, 1253.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Huang H., Petkova S.B., Pestell R.G., Bouzahzah B., Chan J., Magazine H., Weiss L.M., Christ G.J., Lisanti M.P., Douglas S.A.. et al. (2000) Trypanosoma cruzi infection (Chagas’ disease) of mice causes activation of the mitogen-activated protein kinase cascade and expression of endothelin-1 in the myocardium. J. Cardiovasc. Pharmacol., 36, S148–S150. [DOI] [PubMed] [Google Scholar]
- 86. Wu X., Simpson J., Hong J.H., Kim K.H., Thavarajah N.K., Backx P.H., Neel B.G., Araki T. (2011) MEK-ERK pathway modulation ameliorates disease phenotypes in a mouse model of Noonan syndrome associated with the Raf1L613V mutation. J. Clin. Invest., 121, 1009–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Haq S., Choukroun G., Lim H., Tymitz K.M., del Monte F., Gwathmey J., Grazette L., Michael A., Hajjar R., Force T.. et al. (2001) Differential activation of signal transduction pathways in human hearts with hypertrophy versus advanced heart failure. Circulation, 103, 670–677. [DOI] [PubMed] [Google Scholar]
- 88. Chen M.H., Kerkelä R., Force T. (2008) Mechanisms of cardiac dysfunction associated with tyrosine kinase inhibitor cancer therapeutics. Circulation, 118, 84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Östlund C., Bonne G., Schwartz K., Worman H.J. (2001) Properties of lamin A mutants found in Emery–Dreifuss muscular dystrophy, cardiomyopathy and Dunnigan-type partial lipodystrophy. J. Cell Sci., 114, 4435–4445. [DOI] [PubMed] [Google Scholar]
- 90. Grey C., Méry A., Pucéat M. (2005) Fine-tuning in Ca2+ homeostasis underlies progression of cardiomyopathy in myocytes derived from genetically modified embryonic stem cells. Hum. Mol. Genet., 14, 1367–1377. [DOI] [PubMed] [Google Scholar]
- 91. Seki T., Yuasa S., Fukuda K. (2012) Generation of indiced pluripotent ste cells from a small amount of human peripheral blood using a combination of activated T cells and Sensai virus. Nat. Protoc., 7, 718–728. [DOI] [PubMed] [Google Scholar]
- 92. Lodola F., Morone D., Denegri M., Bongianino R., Nakahama H., Rutigliano L., Gosetti R., Rizzo G., Vollero A., Buonocore M.. et al. (2016) Adeno-associated virus-mediated CASQ2 delivery rescues phenotypic alterations in a patient-specific model of recessive catecholaminergic polymorphic ventricular tachycardia. Cell Death Dis., 7, e2393.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Di Pasquale E., Lodola F., Miragoli M., Denegri M., Avelino-Cruz J.E., Buonocore M., Nakahama H., Portararo P., Bloise R., Napolitano C.. et al. (2013) CaMKII inhibition rectifies arrhythmic phenotype in a pateint-specific model of catecholaminergic polymorphic ventricular tachycardia. Cell Death Dis., 4, e843.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Lian X., Selekman J., Bao X., Hsiao C., Zhu K., Palecek S.P. (2013) A small molecule inhibitor of SRC family kimnases promotes simple epithelial differentiation of human pluripotent stem cells. PLoS One, 8, e60016.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Nakahama H., Di Pasquale E. (2016) Generation of cardiomyocytes from pluripotent stem cells. Methods Mol Biol., 1353, 181–190. [DOI] [PubMed] [Google Scholar]
- 96. Ponchel F., Toomes C., Bransfield K., Leong F.T., Douglas S.H., Field S.L., Bell S.M., Combaret V., Puisieux A., Mighell A.J.. et al. (2003) Real-time PCR based on SYBR-Green I fluorescence: an alternative to the TaqMan assay for a relative quantification of gene rearrangements, gene amplifications and micro gene deletions. BMC Biotechnol., 3, 18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Taylor M.J., Perrais D., Merrifield C.J. (2011) A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol., 9, e1000604.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Khokhlatchev A., Xu S., Wu P., Schaefer E., Cobb M.H., English J. (1997) Reconstitution of mitogen-activated protein kinase phosphorylation cascades in bacteria. J. Biol. Chem., 272, 11057–11057. [DOI] [PubMed] [Google Scholar]
- 99. Dominguez E., Marais T., Chatauret N., Benkhelifa-Ziyyat S., Duque S., Ravassard P., Carcenac R., Astord S., de Moura A.P., Voit T.. et al. (2011) Intravenous scAAV9 delivery of a codon-optimized SMN1 sequence rescues SMA mice. Hum. Mol. Genet., 20, 681–693. [DOI] [PubMed] [Google Scholar]
- 100. Ono S., Mohri K., Ono K. (2004) Microscopic evidence that actin-interacting protein 1 actively disassembles actin-depolymerizing factor/cofilin-bound actin filaments. J. Biol. Chem., 279, 14207–14212. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.