Abstract
Purpose
Third-generation epidermal growth factor receptor (EGFR) inhibitors like nazartinib are active against EGFR mutation–positive lung cancers with T790M-mediated acquired resistance to initial anti-EGFR treatment, but some patients have mixed responses.
Methods
Multiple serial tumor and liquid biopsies were obtained from two patients before, during, and after treatment with nazartinib. Next-generation sequencing and droplet digital polymerase chain reaction were performed to assess heterogeneity and clonal dynamics.
Results
We observed the simultaneous emergence of T790M-dependent and -independent clones in both patients. Serial plasma droplet digital polymerase chain reaction illustrated shifts in relative clonal abundance in response to various systemic therapies, confirming a molecular basis for the clinical mixed radiographic responses observed.
Conclusion
Heterogeneous responses to treatment targeting a solitary resistance mechanism can be explained by coexistent tumor subclones harboring distinct genetic signatures. Serial liquid biopsies offer an opportunity to monitor clonal dynamics and the emergence of resistance and may represent a useful tool to guide therapeutic strategies.
INTRODUCTION
Epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) are effective therapies against advanced EGFR-mutant lung cancer, but outcomes are limited by acquired resistance.1 EGFR T790M accounts for 50% to 60% of resistance to erlotinib, gefitinib, and afatinib, whereas other mechanisms, including bypass pathway activation (eg, MET or ERRB2 amplification, mutations in BRAF and PIK3CA) and lineage shifts (eg, small-cell transformation, epithelial-to-mesenchymal transition), are less commonly observed.2,3 Third-generation EGFR inhibitors yield responses in 40% to 60% of T790M-positive tumors, and osimertinib is US Food and Drug Administration approved in this setting.4 Other third-generation TKIs, including nazartinib (EGF816; Novartis, Cambridge, MA),5 are in development. However, tumors can harbor extensive molecular heterogeneity, potentially affecting response to subsequent therapies.6
We present two cases of EGFR-mutant lung cancer treated with nazartinib after developing T790M. Serial tissue and liquid biopsies were collected during and after nazartinib treatment. These cases illustrate how T790M and T790 wild-type (WT) subclones can coexist in individual patients and how serial liquid biopsies might provide insight into tumor heterogeneity and clonal dynamics during therapy.
METHODS
Patients
Patients were treated with nazartinib in a phase I/II trial (ClinicalTrials.gov identifier: NCT02108964) at Massachusetts General Hospital (MGH; patient 1: 200 mg daily, patient 2: 150 mg daily). Other therapies were administered at standard doses. Patients provided informed consent under an institutional review board–approved protocol allowing next-generation sequencing (NGS) and exploratory research on their biopsies.
Clinical Genotype Assessments
Tissue biopsies were genotyped at MGH with three Clinical Laboratory Improvement Amendments (CLIA)-certified assays: SNaPshot version 4, a multiplex polymerase chain reaction (PCR) allele-specific assay for somatic mutations in 23 cancer-related genes (Appendix Table A1; SNaPshot; Applied Biosystems7); a PCR sizing assay for in-frame EGFR insertions or deletions; and SNaPshot NGS, an anchored multiplex PCR assessing 39 (V1) or 91 (V2) genes (Appendix Table A1). Foundation Medicine’s FoundationOne NGS (Cambridge, MA) assesses the entire coding sequence of 315 genes and select introns of 28 genes.8
Tissue MET amplification was assessed at MGH with a CLIA-certified dual-color fluorescence in situ hybridization (FISH) assay using the 07Q001B550 C-MET probe (chromosome 7q31 MET, Leica Biosystems, Wetzlar, Germany) and a copy number probe (CEP7; Abbott-Vysis, Lake Bluff, IL). Signal quantitation generates a MET/CEP7 ratio, and 2.2 to 5.0 is considered low-level amplification, whereas > 5.0 or clustered MET signals too numerous to count are considered highly amplified.
Where indicated, plasma was analyzed by the CLIA-certified Guardant360 targeted NGS circulating tumor DNA (ctDNA) panel (coverage as described previously9). The remaining plasma samples were analyzed by droplet digital PCR (ddPCR).
Plasma ctDNA Isolation and ddPCR
ctDNA was extracted from plasma using the QIAamp Circulating Nucleic Acid Kit (QIAGEN, Hilden, Germany) per manufacturer’s instructions. A total of 3 μL circulating free DNA (cfDNA) was used in each reaction. All samples were analyzed in duplicate. PCR reactions were performed using 10 μL final volume containing 5 μL LightCycler 480 SYBR Green I qPCR Master Mix, 2X (Roche, Basel, Switzerland) and LINE-1 (12.5 μmol) forward and reverse primers. DNA at known concentrations was used to build the standard curve. Isolated ctDNA was amplified using ddPCR Supermix for Probes (Bio-Rad, Hercules, CA) with EGFR p.E746_A750del, EGFR p.T790M, BRAF p.V600E, TP53 p.R273C (PrimePCR ddPCR Mutation Assay; Bio-Rad), EGFR p. C797S (T>A; custom designed) ddPCR assays for mutation detection, and MET, EGFR, and EIF2C1 (PrimePCR ddPCR Copy Number Assay; Bio-Rad) ddPCR assays for gene copy number variation.
An 8-µL DNA template was added to 10 µL ddPCR Supermix for Probes (Bio-Rad) and 2 µL primer/probe mixture. This reaction mix and 60 µL Droplet Generation Oil for Probes (Bio-Rad) were added to a DG8 cartridge to generate droplets, then transferred to a 96-well plate (Eppendorf, Hamburg, Germany) and thermal cycled as follows: 5 minutes at 95°C, 40 cycles of 94°C for 30 seconds, 55°C (56.3°C for EGFR p.C797S and 54°C for TP53 p.R273C), for 1 minute followed by 98°C for 10 minutes (Ramp Rate 2°C/s). Droplets were analyzed with the QX200 Droplet Reader (Bio-Rad) for fluorescent measurement of FAM and HEX probes.
QuantaSoft software (Bio-Rad) was used to obtain fractional abundance and copy number variations of mutant DNA alleles in wild-type/normal background. Quantification of target molecules was presented as total copy number (mutant + WT) per sample in each reaction. Fractional abundance was calculated as (Nmut/[Nmut + Nwt]) × 100, where Nmut is number of mutant events and Nwt is number of WT events per reaction. Positive and negative droplet counts were used to calculate the concentration of target and reference DNA sequences as previously described.10 Normal control DNA (human genomic DNA [Promega, Madison, WI]), and no DNA template controls were included. Samples with insufficient positive events were independently repeated at least twice to validate results. Custom probe and primer sequences are available on request.
RESULTS
Patient 1
A 68-year-old never-smoking woman presented with progressive neurologic symptoms and was found to have a lung mass with multiple brain, pulmonary, lymph node, and hepatic metastases. A trans-bronchial biopsy confirmed lung adenocarcinoma and multiplexed, allele-specific PCR (SNaPShot Version 4) identified an EGFR exon 19 deletion (del19). A symptomatic brain metastasis was resected, and erlotinib 150 mg daily was initiated with good response in all sites of disease.
The cancer progressed after 9 months of erlotinib, and biopsy of a growing liver metastasis (Fig 1B; day −35) was analyzed via targeted NGS (SNaPshot NGS-V1), revealing both the original EGFR del19 and acquired EGFR T790M. Hence, she began a clinical trial of nazartinib (ClinicalTrials.gov identifier: NCT02108964; Fig 1A, days 1 to 392) and achieved a partial response.11 After 11 months on nazartinib, she developed a solitary progressing liver metastasis in a location distinct from her prior lesion (Fig 1B, day 332). Biopsy was performed on the oligoprogressing site (confirming adenocarcinoma), and it was treated with percutaneous microwave ablation (Fig 1C, day 351). NGS (SNaPshot NGS-V1) again identified her EGFR del19, this time without T790M, suggesting outgrowth of a T790 WT subclone. In addition, a BRAF V600E mutation, not present on the pre- or post-erlotinib biopsy samples, was detected as the likely nazartinib-resistant mechanism within this lesion. Shortly after the ablation, further progression was noted within the thorax (day 388).
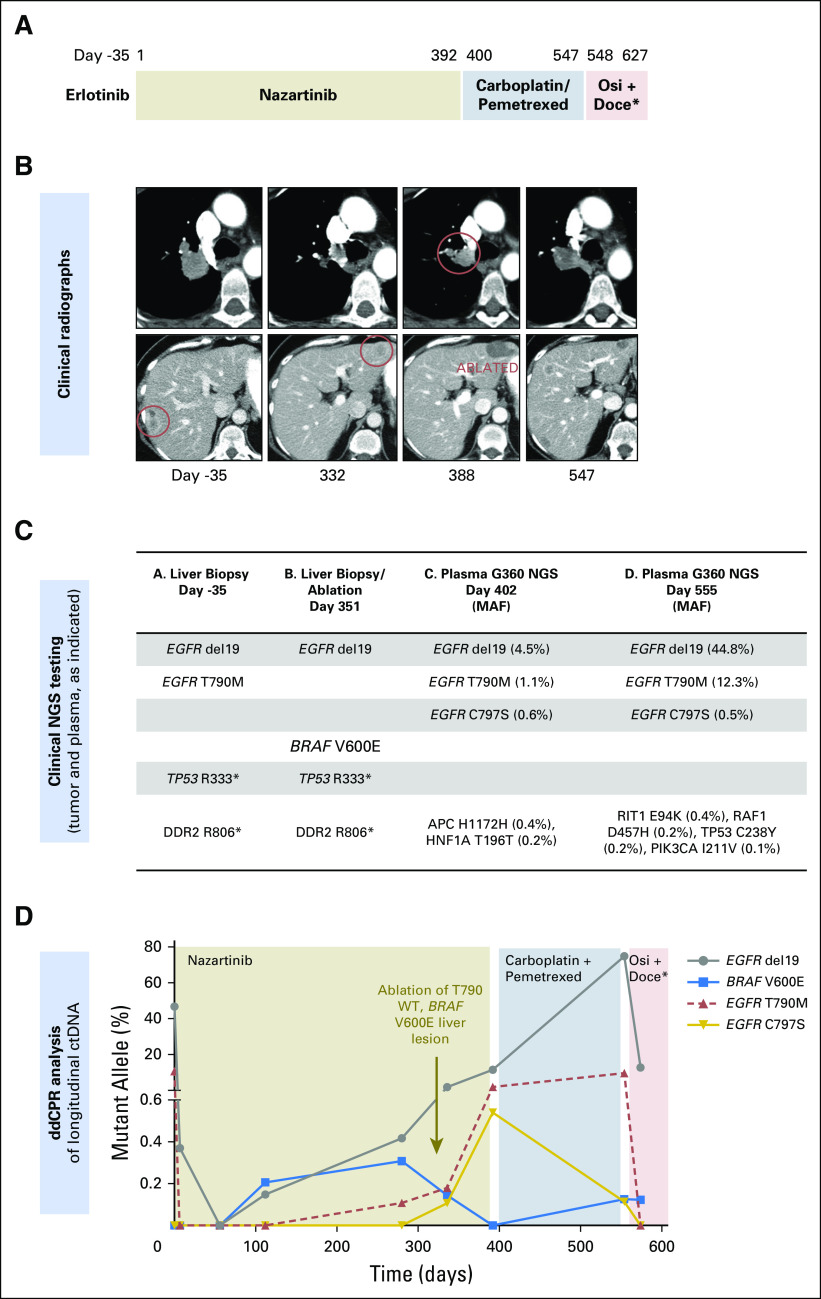
Fig 1.
(A) Treatment history of patient 1. (B) Computed tomography images of the patient’s lung nodule (top) and liver metastases (bottom) during treatment. (C) Results of clinical genotyping on days −35, 351 (tissue biopsies tested by SNaPshot next-generation sequencing [NGS] panel), and days 402 and 555 (liquid biopsies, Guardant360). For plasma results, the mutant allele fraction (MAF) for each mutation is shown in parentheses. (D) Serial circulating tumor DNA (ctDNA) droplet digital polymerase chain reaction (ddPCR) results during treatment. doce, docetaxel; osi, osimertinib; WT, wild type.
ctDNA analysis provides a noninvasive means of monitoring the genomic evolution of cancer during treatment. In this patient, multiple plasma samples were taken for ctDNA analyses, including NGS (Guardant360; Guardant Health, Redwood City, CA) and ddPCR (Fig 1D). On nazartinib initiation, ddPCR revealed a sharp decline in both EGFR del19 (Fig 1D, days 0 to 100, gray line), and T790M (red line), suggesting effective inhibition of the dominant EGFR del19/T790M subclone. By day 100, the mutant allele fraction (MAF) of del19 began to increase again, this time without rebound of T790M, suggesting outgrowth of a distinct T790WT subclone. The increase in EGFR del19 was coincident with emergence of a previously undetected BRAF V600E mutation (ddPCR; blue line), consistent with the EGFR del19/T790WT/BRAF V600E subclone detected in the contemporaneous biopsy of the progressing liver metastasis. Interestingly, ablation of the liver metastasis (Fig 1C, day 351) caused BRAF V600E levels to drop below detection in ctDNA, corresponding with radiographic regression, and suggesting this subclone was likely unique to the ablated metastasis.
Meanwhile, EGFR T790M (Fig 1D, red line) reappeared around day 200, accompanying a continued increase in total del19, emergence of EGFR C797S (ddPCR; gold line), and clinical progression of disease in the thorax. C797S drives resistance to third-generation EGFR TKIs by disrupting the key cysteine residue at the drug-binding site.12-15 Although tissue biopsy was not obtained, we hypothesize based on increasing plasma levels of T790M and C797S that an EGFR del19/T790M/C797S clone may have been driving disease progression in the thorax. Consistent with this, Guardant360 NGS ctDNA analysis on day 402 revealed EGFR del19 (MAF 4.5%), T790M (1.1%), and C797S (0.6%) in cis configuration without detectable BRAF V600E (Fig 1C, day 402; Appendix Figs A1A and A1B).
Although several novel treatment strategies for EGFR C797S are being studied,16,17 there are currently no targeted therapies against T790M/C797S available in the clinic. Thus, the patient discontinued nazartinib and started carboplatin/pemetrexed chemotherapy (Fig 1A, days 400 to 547) with an initial brief response, followed by disease progression in the liver and thorax 5 months later (Fig 1B, day 547). A repeat plasma sample (Guardant360 NGS) demonstrated increasing MAF of EGFR del19 (44.8%) and T790M (12.3%), whereas EGFR C797S (0.5%) remained stable (Fig 1C, day 555; Appendix Fig A1C). EGFR amplification was also detected in the plasma at this time. The pattern of ddPCR-detected ctDNA clones during carboplatin/pemetrexed chemotherapy (Fig 1D, days 400 to 547) suggests there are (at least) two subclones present. We hypothesize that the EGFR del19/T790M/C797S clone decreases after day 400, possibly because of its relative chemosensitivity to or withdrawal of the selective pressure from nazartinib, a scenario observed in other targeted therapy resistance paradigms.18 Another EGFR del19/T790M clone (without C797S) may be more chemoresistant, as evidenced by increasing del19 and steady T790M levels during chemotherapy. It is also possible a third clone existed with more significant chemoresistance and no other detectable EGFR mutations besides del19. The decline in C797S abundance relative to overall del19 suggested a potential clinical opportunity to rechallenge with a third-generation EGFR TKI. On day 548, treatment was switched to docetaxel plus osimertinib (Fig 1A), which yielded a response in the thorax but progression in other sites, including the liver. Interestingly, the high T790M levels quantified through ddPCR before reintroduction of a T790M-specific inhibitor (this time, osimertinib) may provide a molecular predictor of the retreatment effect sometimes observed clinically, as in this patient.19
In summary, this case highlights the complexity and heterogeneity of resistance in EGFR-mutant lung cancer and provides insight into clonal evolution via complementary longitudinal ctDNA and tumor analyses. Here, nazartinib resistance was attributed to two genetically distinct subclones, one BRAF V600E/EGFR T790WT, the second EGFR T790M/C797S, each of which followed a distinct trajectory in response to systemic and local therapies.
Patient 2
A 48-year-old never-smoking woman presented with flank pain and was found to have a lung mass with metastases to mediastinal lymph nodes, ovary, liver, adrenal gland, and bones, and several small brain metastases. A liver biopsy confirmed lung adenocarcinoma. The patient initiated cisplatin/pemetrexed chemotherapy before genotyping results but failed to respond. NGS (SNaPshot NGS-V1) revealed EGFR del19 and TP53 R273C. MET FISH was negative for amplification (MET:CEP7 reference probe copy number ratio, 1.0). The patient began receiving afatinib 40 mg daily with initial response in all disease sites.
Progression occurred at 10 months, and a growing liver lesion underwent biopsy. NGS (FoundationOne) showed EGFR del19, TP53 R273C, and the emergence of T790M (Fig 2C, day −29, core 1), prompting treatment with nazartinib. Interestingly, parallel NGS (SNaPshot NGS-V1) of a second core from the same liver biopsy demonstrated EGFR del19 without T790M, but MET amplification was detected by FISH (abnormal signal clusters, unquantifiable; Fig 2C, day −29, core 2).
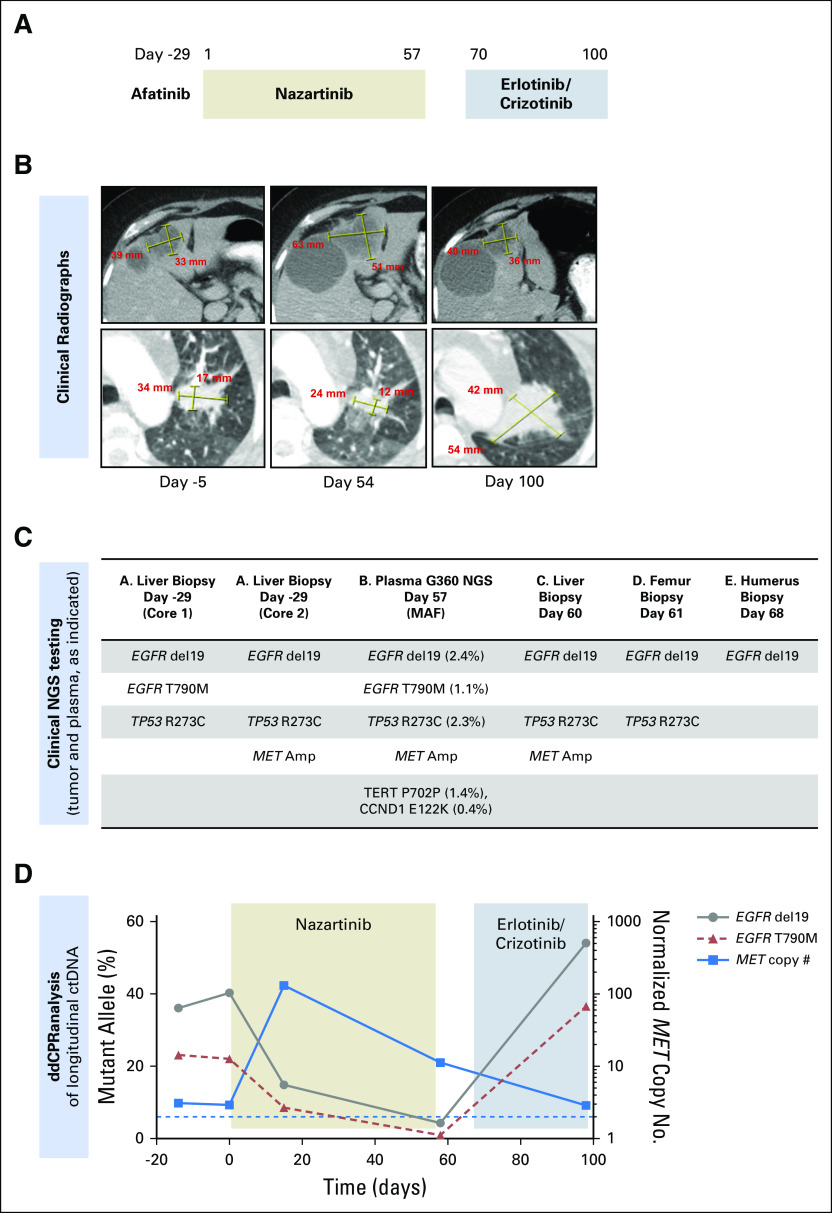
Fig 2.
(A) Treatment history of patient 2. (B) Computed tomography images of the patient’s liver metastases (top) and lung mass (bottom) during treatment. (C) Results of tissue genotyping with SNaPshot next-generation sequencing (NGS) panel on days −29, 60, 61, and 68 and Guardant360 plasma test (mutant allele frequencies [MAFs] in parentheses) on day 57. (D) Serial circulating tumor DNA (ctDNA) droplet digital polymerase chain reaction (ddPCR) results during treatment.
After 2 months receiving nazartinib (Fig 2A, days 1 to 57), a mixed response was observed, with progression of multiple liver metastases but reduction in mediastinal lymph nodes and lung mass (Fig 2B, day 54). Biopsy of the progressive hepatic metastasis on day 60 (Fig 2C) analyzed by NGS (SNaPshot NGS-V1) revealed EGFR del19 without T790M, TP53 R273C, and high-level MET amplification by FISH (abnormal signal clusters, unquantifiable). Concurrent ctDNA NGS (Guardant360; Fig 2C, day 57) demonstrated EGFR del19 (MAF, 2.4%), T790M (1.1%), TP53 R273C (2.3%), and MET amplification (plasma copy number, 2.2; Appendix Fig A2).
Shortly thereafter, the patient suffered pathologic fractures requiring surgical intervention on the femur (Fig 2C, day 61) and humerus (Fig 2C, day 68.) Molecular testing (SNaPshot NGS-V2) of both bone lesions demonstrated EGFR del19 without T790M; MET FISH was also negative. On the basis of MET amplification observed within the liver and ctDNA, nazartinib was discontinued and combination therapy initiated (Fig 2A, days 70 to 100) with erlotinib (100 mg daily) and crizotinib (250 mg daily), a regimen effective in another patient with EGFR-mutant, MET-amplified cancer.20 Scans 1 month later showed reduction in the MET-amplified liver metastasis that previously underwent biopsy but marked disease progression elsewhere (Fig 2B, day 100). The patient transitioned to hospice care.
In this case, ddPCR of longitudinal ctDNA confirmed the mixed molecular response to nazartinib. As expected, EGFR del19 and T790M declined on nazartinib initiation (Fig 2D, days 1 to 25, gray and red lines), but we observed a coincident increase in normalized MET copy number (ddPCR; Fig 2D, blue line). Taken together with the biopsy and radiographic findings, the increasing MET signal likely corresponds to a clone residing within the hepatic metastases, whereas the decline in T790M may arise from a nazartinib-sensitive clone within the thorax. On withdrawal of the T790M-targeted nazartinib and initiation of erlotinib/crizotinib (a combination expected to inhibit MET-driven subclones but not capable of overcoming T790M), the MET copy number declined while T790M increased rapidly. Clinically, we observed growth of the lung nodule but shrinkage of the MET-amplified liver lesion that previously underwent biopsy, highlighting the differential responses of genetically distinct resistant subclones.
DISCUSSION
These two EGFR-mutant lung cancer cases with serial tissue and plasma biopsies present a unique opportunity to study clonal evolution in depth during diverse molecularly targeted therapies. Our findings illustrate the critical role heterogeneity plays in resistance to targeted therapies and the clinical challenge posed by genetically distinct subpopulations that respond differentially to treatment.
Heterogeneity in the context of EGFR resistance has been documented in both autopsy and ctDNA-based studies.14,21,22 We previously described an afatinib-resistant pleural effusion, which was T790M positive on clinical testing, but single-cell cloning revealed both T790M-mutant and T790WT subclones. In a cohort of such patients, either subpopulation could spawn the dominant driver of subsequent resistance to third-generation EGFR TKIs.6 Recently, Blakely et al23 demonstrated increasing heterogeneity of an EGFR-mutant cancer across multiple lines of therapy through whole-exome sequencing of serial biopsies and autopsy specimens. The two cases reported here build on these observations and reveal that one can clinically document coexistent, genetically distinct subpopulations differentially responding as predicted by their genotype in real time to therapies. In patient 2, EGFR T790M and MET amplification were both readily detected in tumor and plasma at the time of afatinib resistance. The presence of two resistant subclones may explain the overall lack of clinical benefit from nazartinib, a drug targeting T790M but not MET amplification. Indeed, Chabon et al14 also demonstrated that coexistence of T790M and MET amplification within ctDNA predicted poorer outcomes than T790M alone on a third-generation TKI.14 In contrast, patient 1 had only T790M detected on erlotinib progression and enjoyed a durable response to nazartinib. Her ultimate nazartinib resistance was driven initially by a T790WT/BRAF-mutant clone localized to a solitary liver metastasis that was transiently eradicated from ctDNA by directed ablation and, subsequently, by a T790M/C797S subclone. In each case, the coexistent clones followed divergent trajectories, which were apparent within plasma, highlighting the complementary role of ctDNA to tumor biopsies and radiographs to assess subclone-specific response to treatment.
Patient 1 also adds to a growing understanding of third-generation EGFR TKI resistance. Although EGFR C797S has previously been described in osimertinib, olmutinib, and rociletinib resistance,12-15 to our knowledge this is the first report of C797S emergent after nazartinib. Bypass pathway activation via BRAF V600E has also been described in resistance to both first- and third-generation EGFR TKIs.24,25 The observation of these bona fide resistance mechanisms in nazartinib resistance confirms the drug’s potency and effective target inhibition.5 However, despite broad molecular testing capable of detecting all described genomic mechanisms of EGFR TKI resistance, we have been unable to identify the etiology of progression within patient 2’s bone metastases. Previous studies of EGFR TKI resistance have focused primarily on genetic resistance mechanisms and histologic transformations but have been limited in characterizing epigenetic and other nongenomic contributors. Broader efforts may be required moving forward to more fully characterize resistance.
Locally ablative therapies (LAT) to treat sites of oligoprogression are often used to prolong disease control among patients with EGFR- and ALK-positive disease.26 In patient 1, microwave ablation of an oligoprogressive EGFR del19/BRAF V600E-mutant liver metastasis caused the corresponding ctDNA signal to fall below detectable levels, suggesting local therapy can effectively eradicate a focally restricted resistant clone and lending biologic rationale to the use of LAT. In this patient, ablation did not provide sufficient overall disease control, likely because of the EGFR del19/T790M/C797S subclone already detectable in plasma at the time of ablation, which emerged clinically shortly thereafter. ctDNA analysis before LAT may distinguish patients with a singular resistance mechanism from those with polyclonal resistance and steer patient selection, although additional prospective study is required to validate such an approach.
Tumor biopsies have formed the cornerstone of research on EGR TKI resistance and have several advantages: they allow for histologic and immunohistochemical analyses, greater depth of molecular characterization including RNA and copy number analyses, and establishment of cell line models for functional studies. However, a single tumor biopsy is spatially limited to a minute fragment of a patient’s overall cancer burden. Conversely, ctDNA is shed from multiple disease sites and thus may illustrate coexistent subclonal genetic alterations. Sometimes, as in patient 1, plasma identifies resistance mechanisms not detected in tissue (here, EGFR C797S), although its utility is limited in patients with low levels of ctDNA. Many plasma assays, on the basis of both ddPCR and NGS, are now clinically available, and additional studies to define optimal strategies are needed. Here, we used a broad NGS-based ctDNA panel at key points of disease progression and ddPCR, which is generally more sensitive and cost-effective than serial NGS, to retrospectively quantify known genetic variants in plasma samples banked during treatment. Although this strategy was well suited to our longitudinal analysis, in practice providers must select the most appropriate test on the basis of gene coverage, turnaround time, and cost.
Ultimately, our cases illustrate the critical role of cancer heterogeneity in targeted therapy–acquired resistance and provide a glimpse of a future where real-time information gleaned from tracking resistant clones during treatment could be clinically meaningful. In each case, ctDNA revealed both T790M-positive and T790WT clones, providing a molecular rationale for nazartinib failure. Currently, T790M status is used to determine whether a patient should be treated with second-line osimertinib,1 but our data suggest that looking beyond T790M might have relevance; incorporating broader tissue and plasma panels into routine testing may identify other clones influencing the success of subsequent therapies. More heterogeneous tumors may have less robust responses to singularly focused therapeutics. This paradigm also translates to other analogous situations, such as acquired resistance to first-line osimertinib and selection of sequential ALK TKIs in patients with ALK rearrangements. Future clinical trials should incorporate longitudinal ctDNA- and tissue-based assessments during therapy to develop a more complete understanding of dynamic evolution under the pressure of treatment and which clones survive and drive resistance. Ultimately, we expect a more sophisticated understanding of resistance will better inform innovative strategies to reduce heterogeneity, eradicate the seeds of eventual resistance, and induce more durable remissions.
Appendix
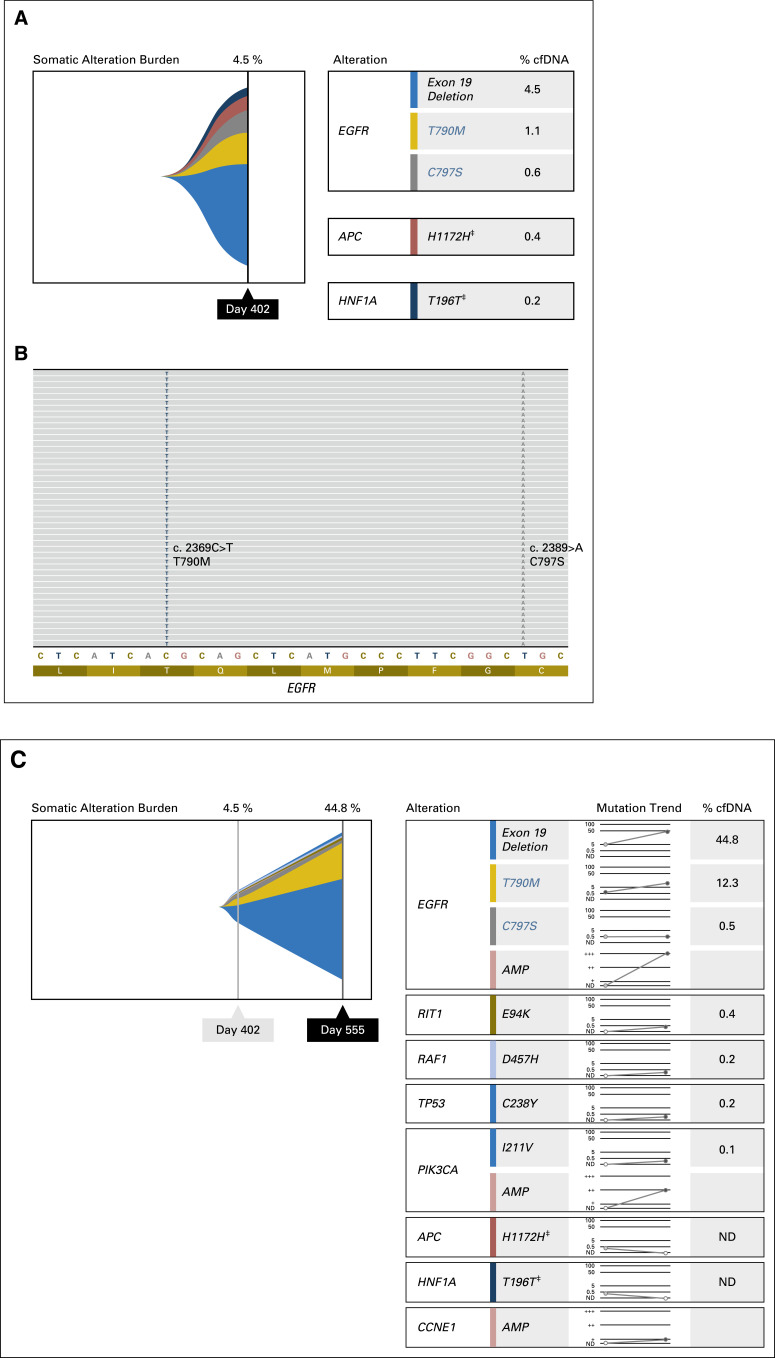
Fig A1.
(A) Results of next-generation sequencing of circulating tumor DNA (ctDNA) by Guardant360 for patient 1, day 402 (progression on nazartinib). The mutant allele fractions (percent cfDNA) are shown for each mutation detected. (B) Integrated Genomics Viewer figure demonstrating that T790M and C797S are coexistent on the same allele (cis configuration). (C) Results of subsequent ctDNA analysis on day 555 (progression on carboplatin/pemetrexed) with an overall increase in the mutant allele fraction of EGFR del19 and T790, whereas EGFR C797S declined slightly. (‡) Synonymous variant.
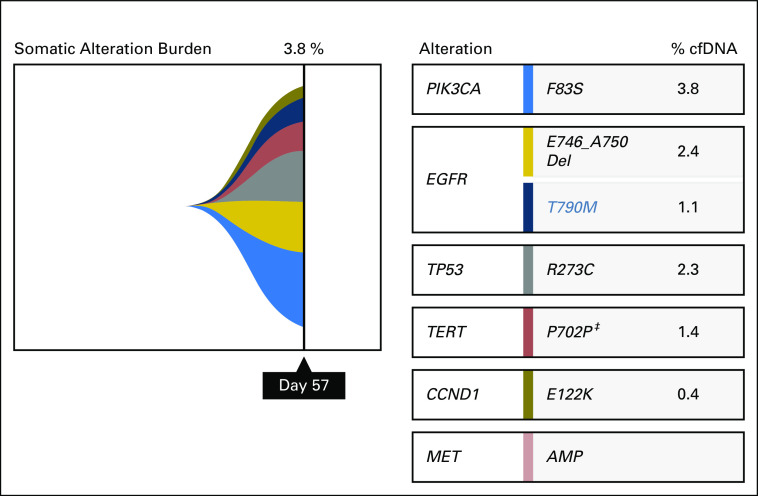
Fig A2.
Results of Guardant360 ctDNA analysis obtained for patient 2 on day 57 (progression on nazartinib) are shown. EGFR del109, T790M, and MET amplification are all detected. (‡) Synonymous variant
Table A1.
Genes Covered by SNaPshot NGS Testing, by Version
Footnotes
Supported by National Institutes of Health Grant No. 2R01CA137008 (L.V.S.), LungStrong (L.V.S.), Targeting a Cure for Lung Cancer (L.V.S.), the Susanne E. Coyne Memorial Fund (L.V.S.), Uniting Against Lung Cancer (Z.P.), the LUNGevity Foundation (Z.P.), a Damon Runyon Clinical Investigator Award (R.B.C.), and National Institutes of Health/National Cancer Institute Grants No. 1K08CA166510 and 1R01CA208437 (R.B.C.).
AUTHOR CONTRIBUTIONS
Conception and design: Zofia Piotrowska, Mehlika Hazar-Rethinam, Jeffrey A. Engelman, Ryan B. Corcoran, Lecia V. Sequist
Financial support: Ryan B. Corcoran
Administrative support: Anthony J. Iafrate, Nicholas A. Jessop, Ryan B. Corcoran
Provision of study material or patients: Zofia Piotrowska, Anthony J. Iafrate, Ryan B. Corcoran, Lecia V. Sequist
Collection and assembly of data: Zofia Piotrowska, Mehlika Hazar-Rethinam, Coleen Rizzo, Brandon Nadres, Emily E. Van Seventer, Heather A. Shahzade, Inga T. Lennes, Anthony J. Iafrate, Nicholas A. Jessop, Haichuan Hu, Subba R. Digumarthy, Rebecca J. Nagy, Matthew J. Niederst, Ryan B. Corcoran
Data analysis and interpretation: Zofia Piotrowska, Mehlika Hazar-Rethinam, Anthony J. Iafrate, Dora Dias-Santagata, Ignaty Leshchiner, Subba R. Digumarthy, Richard B. Lanman, Susan Moody, Jeffrey A. Engelman, Aaron N. Hata, Ryan B. Corcoran
Manuscript writing: All authors
Final approval: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Zofia Piotrowska
Consulting or Advisory Role: Boehringer Ingelheim, AstraZeneca, ARIAD Pharmaceuticals/Takeda, Superdimension, Guardant Health, Novartis, Abbvie
Research Funding: Novartis (Inst), ARIAD Pharmaceuticals/Takeda (Inst), Guardant Health (Inst)
Mehlika Hazar-Rethinam
No relationship to disclose
Coleen Rizzo
No relationship to disclose
Brandon Nadres
No relationship to disclose
Emily E. Van Seventer
No relationship to disclose
Heather A. Shahzade
No relationship to disclose
Inga T. Lennes
Honoraria: Blue Cross and Blue Shield of Massachusetts
Consulting or Advisory Role: Kyruus
Anthony J. Iafrate
Stock and Other Ownership Interests: Archer Biosciences
Consulting or Advisory Role: Debiopharm Group, Chugai Pharmaceutical, Roche
Research Funding: Blueprint Medicines
Patents, Royalties, Other Intellectual Property: ArcherDx exclusive license to Anchored MultiplexPCR technology
Dora Dias-Santagata
No relationship to disclose
Ignaty Leshchiner
No relationship to disclose
Nicholas A. Jessop
No relationship to disclose
Haichuan Hu
No relationship to disclose
Subba R. Digumarthy
No relationship to disclose
Rebecca J. Nagy
Employment: Guardant Health
Stock and Other Ownership Interests: Guardant Health
Richard B. Lanman
Employment: Guardant Health, Veracyte
Leadership: Guardant Health, Biolase
Stock and Other Ownership Interests: Guardant Health, Biolase
Research Funding: Guardant Health
Susan Moody
Employment: Novartis Institutes for BioMedical Research
Stock and Other Ownership Interests: Novartis
Consulting or Advisory Role: Dominion Diagnostics (I)
Travel, Accommodations, Expenses: Dominion Diagnostics (I)
Other Relationship: Acadia Healthcare (I)
Matt J. Niederst
Employment: Novartis
Stock and Other Ownership Interests: Novartis
Jeffrey A. Engelman
Employment: Novartis
Stock and Other Ownership Interests: Loxo Oncology, Agios Pharmaceuticals, Kura Oncology, Gatekeeper Pharmaceuticals, Novartis
Consulting or Advisory Role: Novartis, Agios Pharmaceuticals, Loxo Oncology, Clovis Oncology, Ventana Medical Systems, G1 Therapeutics, Sanofi, Chugai Pharmaceutical, Warp Drive Bio, Asana Biosciences, EMD Serono, Synta, Allostery, Genentech, Aveo/Biodesix, Merck, Cell Signaling Technology, Endo Pharmaceuticals, AstraZeneca, GlaxoSmithKline, Amgen, Bristol-Myers Squibb, Cancer Progress
Research Funding: Novartis, Jounce Therapeutics, Sanofi, Araxes Pharma, AstraZeneca, Amgen (Inst)
Patents, Royalties, Other Intellectual Property: I am a co-inventor on a patent application that has been licensed to Ventana/Roche
Other Relationship: Third Rock Ventures
Aaron N. Hata
Research Funding: Amgen, Novartis, Relay Therapeutics
Ryan B. Corcoran
Consulting or Advisory Role: Avidity Nanomedicines, Taiho Pharmaceutical, Merrimack, Genentech, N-of-One, Astex Pharmaceuticals, Amgen, Bristol-Myers Squibb, Roche, Shire, FOG Pharma, Loxo Oncology, Roivant Sciences, Warp Drive Bio
Research Funding: AstraZeneca, Sanofi
Lecia V. Sequist
Honoraria: AstraZeneca
Consulting or Advisory Role: AstraZeneca, Genentech, Bristol-Myers Squibb, Pfizer
Research Funding: Boehringer Ingelheim (Inst), Clovis Oncology (Inst), Genentech (Inst), Merrimack (Inst), Novartis (Inst), AstraZeneca (Inst), Johnson & Johnson (Inst), Merck (Inst), Pfizer (Inst), Guardant Health (Inst), Incyte (Inst)
REFERENCES
- 1.National Comprehensive Cancer Network Non-small cell lung cancer: (Version 7.2017) https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf
- 2.Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013;19:2240–2247. doi: 10.1158/1078-0432.CCR-12-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jänne PA, Yang JC, Kim DW, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med. 2015;372:1689–1699. doi: 10.1056/NEJMoa1411817. [DOI] [PubMed] [Google Scholar]
- 5.Tan DS-W, Yang JC-H, Leighl NB, et al. Updated results of a phase 1 study of EGF816, a third-generation, mutant-selective EGFR tyrosine kinase inhibitor (TKI), in advanced non-small cell lung cancer (NSCLC) harboring T790M. J Clin Oncol. 2016;34 suppl; abstr 9044. [Google Scholar]
- 6.Piotrowska Z, Niederst MJ, Karlovich CA, et al. Heterogeneity underlies the emergence of EGFRT790 wild-type clones following treatment of T790M-positive cancers with a third-generation EGFR inhibitor. Cancer Discov. 2015;5:713–722. doi: 10.1158/2159-8290.CD-15-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dias-Santagata D, Akhavanfard S, David SS, et al. Rapid targeted mutational analysis of human tumours: A clinical platform to guide personalized cancer medicine. EMBO Mol Med. 2010;2:146–158. doi: 10.1002/emmm.201000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lanman RB, Mortimer SA, Zill OA, et al. Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS One. 2015;10:e0140712. doi: 10.1371/journal.pone.0140712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayden RT, Gu Z, Sam SS, et al. Comparative performance of reagents and platforms for quantitation of cytomegalovirus DNA by digital PCR. J Clin Microbiol. 2016;54:2602–2608. doi: 10.1128/JCM.01474-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 12.Yu HA, Tian SK, Drilon AE, et al. Acquired resistance of EGFR-mutant lung cancer to a T790M-specific EGFR inhibitor: Emergence of a third mutation (C797S) in the EGFR tyrosine kinase domain. JAMA Oncol. 2015;1:982–984. doi: 10.1001/jamaoncol.2015.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thress KS, Paweletz CP, Felip E, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med. 2015;21:560–562. doi: 10.1038/nm.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chabon JJ, Simmons AD, Lovejoy AF, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun. 2016;7:11815. doi: 10.1038/ncomms11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song HN, Jung KS, Yoo KH, et al. Acquired C797S mutation upon treatment with a T790M-specific third-generation EGFR inhibitor (HM61713) in non-small cell lung cancer. J Thorac Oncol. 2016;11:e45–e47. doi: 10.1016/j.jtho.2015.12.093. [DOI] [PubMed] [Google Scholar]
- 16.Uchibori K, Inase N, Araki M, et al. Brigatinib combined with anti-EGFR antibody overcomes osimertinib resistance in EGFR-mutated non-small-cell lung cancer. Nat Commun. 2017;8:14768. doi: 10.1038/ncomms14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jia Y, Yun CH, Park E, et al. Overcoming EGFR(T790M) and EGFR(C797S) resistance with mutant-selective allosteric inhibitors. Nature. 2016;534:129–132. doi: 10.1038/nature17960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siravegna G, Mussolin B, Buscarino M, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med. 2015;21:827. doi: 10.1038/nm0715-827b. [DOI] [PubMed] [Google Scholar]
- 19.Chang GC, Tseng CH, Hsu KH, et al. Predictive factors for EGFR-tyrosine kinase inhibitor retreatment in patients with EGFR-mutated non-small-cell lung cancer–A multicenter retrospective SEQUENCE study. Lung Cancer. 2017;104:58–64. doi: 10.1016/j.lungcan.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 20.Gainor JF, Niederst MJ, Lennerz JK, et al. Dramatic response to combination erlotinib and crizotinib in a patient with advanced, EGFR-mutant lung cancer harboring de novo MET amplification. J Thorac Oncol. 2016;11:e83–e85. doi: 10.1016/j.jtho.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 21.Suda K, Murakami I, Katayama T, et al. Reciprocal and complementary role of MET amplification and EGFR T790M mutation in acquired resistance to kinase inhibitors in lung cancer. Clin Cancer Res. 2010;16:5489–5498. doi: 10.1158/1078-0432.CCR-10-1371. [DOI] [PubMed] [Google Scholar]
- 22.Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 23.Blakely CM, Watkins TBK, Wu W, et al. Evolution and clinical impact of co-occurring genetic alterations in advanced-stage EGFR-mutant lung cancers. Nat Genet. 2017;49:1693–1704. doi: 10.1038/ng.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho CC, Liao WY, Lin CA, et al. Acquired BRAF V600E mutation as resistant mechanism after treatment with osimertinib. J Thorac Oncol. 2017;12:567–572. doi: 10.1016/j.jtho.2016.11.2231. [DOI] [PubMed] [Google Scholar]
- 25.Ohashi K, Sequist LV, Arcila ME, et al. Lung cancers with acquired resistance to EGFR inhibitors occasionally harbor BRAF gene mutations but lack mutations in KRAS, NRAS, or MEK1. Proc Natl Acad Sci USA. 2012;109:E2127–E2133. doi: 10.1073/pnas.1203530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weickhardt AJ, Scheier B, Burke JM, et al. Local ablative therapy of oligoprogressive disease prolongs disease control by tyrosine kinase inhibitors in oncogene-addicted non-small-cell lung cancer. J Thorac Oncol. 2012;7:1807–1814. doi: 10.1097/JTO.0b013e3182745948. [DOI] [PMC free article] [PubMed] [Google Scholar]