SUMMARY
Cells have evolved quality control pathways to prevent the accumulation of improperly localized proteins, which are often toxic. One of these pathways, regulation of aberrant protein production (RAPP), recognizes aberrant secretory proteins during translation and degrades the associated mRNA. Here, we demonstrate endogenous RAPP substrates. Haploinsufficiency of the secretory protein progranulin (GRN) is associated with the neurodegenerative disease frontotemporal lobar degeneration (FTLD). Our results show FTLD-associated GRN mutations W7R and A9D disrupt co-translational interaction with a targeting factor, signal recognition particle (SRP). This triggers RAPP and initiates specific mRNA degradation. Conversely, wild-type GRN and the naturally occurring polymorphism V5L GRN are efficiently expressed and secreted. Thus, RAPP plays a role in the molecular pathology of A9D GRN and W7R GRN.
In Brief
Progranulin mutations, which reduce its secretion, cause the disease FTLD (frontotemporal lobar degeneration). Here, Pinarbasi et al. show that one such mutation, A9D, prevents recruitment of the trafficking factor SRP (signal recognition particle). This triggers a quality control response, which results in degradation of A9D mRNA.
Graphical Abstract
INTRODUCTION
A typical secretory protein contains an N-terminal signal sequence, which is recognized by the targeting factor SRP (signal recognition particle) and directed to the endoplasmic reticulum (ER) membrane for translocation (von Heijne, 1985; Walter et al., 1981). SRP is recruited during translation, once the signal sequence emerges from the ribosomal tunnel, and is the critical first step for secretion (Walter et al., 1981).
SRP recruitment is also critical for stability of the mRNA being translated; if the integrity of a signal sequence is compromised and the secretory protein fails to recruit SRP during translation, the associated mRNA is selectively degraded. This quality control process, termed RAPP (regulation of aberrant protein production), thus protects the cell from accumulating aberrant protein products, which might aggregate or misfold (Karamyshev et al., 2014; Popp and Maquat, 2014). The molecular mechanism of RAPP is largely unknown, but data supported a model that mutant secretory proteins that fail to recruit SRP come into close proximity of Ago2, which is required for efficient mutant mRNA degradation (Karamyshev et al., 2014).
However, to date, endogenous RAPP substrates have not been reported. Here, we explore disease-linked signal sequence mutations in the secretory protein progranulin (GRN) as potential RAPP substrates.
A single loss-of-function GRN mutation results in GRN haploinsufficiency and the neurodegenerative disease FTLD (frontotemporal lobar degeneration) (Baker et al., 2006; Gass et al., 2006). The function of GRN is unclear; however, GRN −/− patients develop the lysosomal storage disease NCL (neuronal ceroid lipofuscinosis), suggesting GRN promotes lysosomal function (Almeida et al., 2016; Smith et al., 2012).
Of the ~100 unique GRN polymorphisms reported in FTLD patients, three are within the signal sequence: V5L; W7R; and A9D (Horaitis et al., 2007). Of these, only A9D GRN has been established as a cause of FTLD. GRN haploinsufficiency in patients carrying the A9D GRN mutation has been demonstrated (Finchet al., 2009); moreover, A9D GRN is found in families and segregates with disease (Mukherjee et al., 2006). In contrast, the impact of V5L and W7R on GRN secretion and function is unclear, and genetic linkage to FTLD has not been established (Le Ber et al., 2008; López de Munain et al., 2008).
We hypothesized that the A9D mutation, which disrupts GRN secretion, prevents SRP recruitment and triggers RAPP. We further hypothesized that the effect of the V5L and W7R mutations on GRN secretion predicts whether RAPP is triggered. The data presented here demonstrate the far-reaching implications of the recently discovered protein quality control pathway, RAPP, on human disease.
RESULTS
GRN Variants W7R and A9D Alter Interaction with the Targeting Factor SRP
While signal sequences are highly variable, all contain a central, hydrophobic “H” domain (von Heijne, 1981, 1985). Both W7R GRN and A9D GRN introduce a polar amino acid into the “H” domain, dramatically altering local hydrophobicity (Figure 1A). Thus, these substitutions would be predicted to disrupt SRP recruitment whereas the conservative GRN V5L substitution would not.
Figure 1. W7R and A9D Mutations Disrupt SRP Recruitment, Leading to Inefficient Expression and Secretion.
(A) Kyte-Doolittle hydrophobicity plot of GRN signal sequence: WT (black); W7R (blue); and A9D (red).
(B) Experimental schema of in vitro translation and site-specific photocrosslinking.
(C) Site-specific photocrosslinking of WT, A9D, W7R, and V5L GRN nascent chains: 86 codons, photocrosslinking probe in position 13. Autoradiograph of the SDS-PAGE is shown. Arrow denotes position of SRP54 photoadducts.
(D) Western blot of media and whole-cell lysate (WCL) of HeLa cells transfected with either WT, V5L, W7R, or A9D GRN. GRN, loading control actin, and transfection control GFP are shown.
(E) GRN immunofluorescence of HeLa cells transfected with either WT, V5L, W7R, or A9D GRN. GFP, expressed from the same plasmid, is shown as a transfection control. Nuclei were detected with Hoechst stain. The scale bars represent 10 μm.
Images in (D) and (E) are representative of 3 independent experiments.
To test this prediction, we used site-specific photocrosslinking in vitro (Etchells et al., 2005; Karamyshev et al., 2014; Krieg et al., 1986; Nilsson et al., 2015). With this method, truncated mRNAs are translated in vitro to create a pool of ribosomes with a set length of exposed nascent chain (Figure 1B). Nascent chains contain a photocrosslinking probe, which after UV irradiation forms a covalent bond to proteins in close proximity. Photocrosslinking adducts are detected by the shift in electrophoretic mobility of the radiolabeled nascent chain (schema, Figure 1B). For further details, see Experimental Procedures.
In vitro translation reactions containing 66-codon, 86-codon, and 132-codon wild-type (WT) GRN mRNAs produced nascent chains of the expected length; all three nascent chains formed an adduct with a ~50-kDa protein consistent with SRP54, the SRP subunit that binds signal sequences (Figures 1C and S1A; Karamyshev et al., 2014; Krieg et al., 1986; Nilsson et al., 2015; Walter and Blobel, 1983).
To confirm the identity of the band, we compared the photo-crosslinking pattern of 86 WT GRN (86-codon WT GRN) to that of 86 WT PPL (86-codon WT preprolactin). We have previously shown that 86 WT PPL efficiently crosslinks to SRP54 when the reaction is supplemented with purified SRP (Karamyshev et al., 2014). The SRP54 adduct in the 86 WT PPL reaction migrates identically to the ~50-kDa adduct in the 86 WT GRN reaction (Figure S1A).
Notably, substitutions A9D or W7R in the GRN nascent chain inhibited photo-crosslinking to SRP54, demonstrating that these mutant proteins did not recruit SRP (Figure 1C). However, the V5L GRN photocrosslinking pattern was indistinguishable from WT GRN, demonstrating that V5L GRN does not disrupt SRP recruitment.
The two other strong bands seen on the gel are unlikely to be true adducts (~100 kDa and ~46 kDa), as their migration pattern is unchanged in the 66-, 86-, and 132-codon WT GRN reactions (Figure S1A). These bands likely originate from the translational lysate or the [35S]methionine labeling, as has been previously observed (Jackson and Hunt, 1983).
Secretion of W7R and A9D GRN Is Impaired
We next assessed GRN secretion and localization using western blotting and immunofluorescence of HeLa cells transfected with either WT GRN, V5L GRN, W7R GRN, or A9D GRN. WT GRN and V5L GRN, which efficiently recruit SRP, were detected both in whole-cell lysate and in media of transfected cells (Figure 1D). Immunofluorescence of transfected cells demonstrated intracellular WT and V5L GRN; WT GRN was shown to colocalize with ER and Golgi (Figures 1E, S1B, and S1C). In contrast, we did not detect W7R and A9D GRN in transfected cells with either western blotting or immunofluorescence, despite adequate expression of the transfection control GFP (Figures 1D, 1E, S1B, and S1C).
However, several other groups have reported intracellular A9D GRN (Li et al., 2018; Mukherjee et al., 2008). To improve detection, we generated flag-tagged WT and A9D GRN, which were detected using a Flag antibody. The flag tag did not disrupt WT GRN localization (Figure S1D). However, unlike untagged A9D GRN, Flag A9D GRN generated a punctate signal (Figure S1D). These puncta looked grossly similar, regardless of whether the Flag tag was within the protein sequence (replacing one of the 7.5 repeated GRN domains; Bateman and Bennett, 1998) or at the C terminus (data not shown), suggesting the puncta represent fully translated GRN.
W7R GRN and A9D GRN mRNA Levels Are Decreased Relative to WT GRN and V5L GRN
We hypothesized that disrupting SRP recruitment triggers GRN mRNA degradation. To test this, we compared steady-state mRNA levels of WT and V5L GRN, which recruit SRP, and W7R and A9D GRN, which do not. We performed GRN qPCR in HeLa cells transfected with four GRN overexpression constructs, normalizing to the transfection control GFP (Figure 2A). Despite comparable transfection efficiency, GRN mRNA levels segregated into two groups. mRNA levels of efficiently secreted GRN variants, WT GRN and V5L GRN, were high, whereas mRNA levels of variants that cannot efficiently recruit SRP, W7R GRN and A9D GRN, were ~70% lower (Figure 2A).
Figure 2. W7R and A9D mRNA Are Subject to Specific, Transferable Degradation.
(A) GRN qPCR of HeLa cells transfected with WT (black), V5L (dark gray), W7R (light gray), or A9D GRN (white). Mean and SD (error bars) of 3 independent experiments are shown. One-way ANOVA analysis, with pairwise comparison using Holm-Sidak method, is shown: WT versus V5L no significant difference (n.s.d.); W7R versus A9D n.s.d.; all other pairwise comparisons: p < 0.001. *p < 0.001.
(B) GRN qPCR in HeLa Tet-OFF cells transfected with WT (black) or A9D (white) GRN. mRNA levels (mean and SD [error bars] for 3 independent experiments) on natural log scale as a function of time after doxycycline treatment in hours are shown. Lines of best fit (using mean squares method) for WT and A9D are also shown. Each mRNA was normalized to the initial quantity of that mRNA species, set at 1. Inset: relative mRNA levels of WT and A9D GRN at time zero are shown. t test analysis: WT versus A9D at 0 hr (p = 0.009, inset) and WT versus A9D at 4 hr (p = 0.018) and at 8 hr (p = 0.029). Asterisk indicates p <0.05.
(C) WT and A9D GRN qPCR (using variant-specific primers) in HeLa Tet-OFF cells transfected with WT alone (black), A9D alone (white), or WT and A9D GRN (striped). A9D GRN levels were normalized using a factor of 0.709 (see Figure S2 for derivation of normalization factor). Mean and SD (error bars) for 3 independent experiments are shown. One-way ANOVA with pairwise comparison using Holm-Sidak method is shown: WT GRN in single versus co-transfected cells: n.s.d.; A9D GRN in single versus co-transfected cells: n.s.d.; and WT GRN versus A9D GRN in co-transfected cells: p = 0.011.
(D) qPCR analysis of chimeric mRNAs in transfected HeLa cells: WT or A9D GRN signal sequence fused to PPL mature protein sequence: WT GRN-PPL, black bar and A9D GRN-PPL, white bar. Hybrid mRNAs were detected with PPL primers. Mean and SD (error bars) for 3 independent experiments are shown. t test analysis: WT GRN-PPL versus A9D GRN-PPL: p < 0.001. * p <0.001.
A9D GRN mRNA Degradation Is Specific and Transferable to Chimeric mRNAs
To distinguish between decreased transcription of mutant mRNA and increased degradation of mutant mRNA, we assessed mRNA turnover. HeLa Tet-OFF cells were transfected with constructs containing WT or A9D GRN behind a tetracycline responsive promoter. 20 hr after transfection, cells were treated with doxycycline to inhibit GRN transcription, and GRN qPCR was performed at 0, 1, 4, and 8 hr. As expected, GRN A9D mRNA levels were reduced compared to WT at time 0 (Figure 2B, inset). GRN A9D mRNA was degraded faster than WT, with significantly reduced mRNA levels at 4 and 8 hr (Figure 2B). To rule out nonspecific degradation, GRN mRNA turnover was assessed in cells co-expressing WT GRN and A9D GRN. The percent of GRN WT mRNA remaining after 8 hr of transcriptional inhibition was unchanged in the presence of GRN A9D mRNA and vice versa (Figure 2C), consistent with mutant-specific mRNA degradation. Sequence-specific primers did not cross-amplify (Figure S2A). However, A9D GRN amplification was less efficient than WT GRN amplification, leading to underestimation of A9D GRN in a mixture. To correct for this, A9D GRN mRNA levels were normalized using the experimentally derived 0.709 (Figure S2B).
To determine whether the A9D signal sequence is sufficient to trigger mRNA degradation, two fusion proteins were constructed: the signal sequence of PPL was replaced by either the WT or A9D GRN signal sequence (referred to as WT GRN-PPL or A9D GRN-PPL). Chimeric proteins were expressed in cultured cells, and GRN-PPL mRNA was assessed using qPCR with PPL-specific primers. A9D GRN-PPL mRNA levels were significantly reduced compared to those of WT GRNPPL, suggesting that a mutant signal sequence is sufficient to trigger mRNA degradation (Figure 2D).
SRP Protects WT GRN mRNA from Degradation
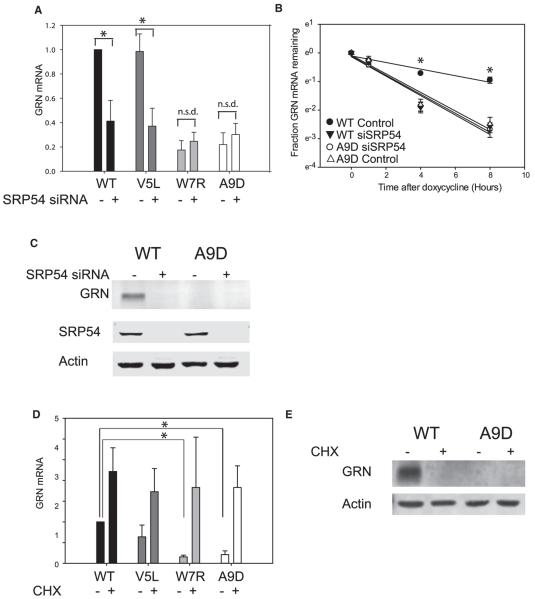
The mRNA levels of four GRN variants correlated with the ability of the translating nascent chain to recruit SRP. To determine whether SRP was required for differential mRNA stability, mRNA levels were assessed in control or SRP54 small interfering RNA (siRNA)-treated cells. In control cells, W7R and A9D GRN mRNA levels were reduced compared to WT. However, in SRP54 siRNA-treated cells, W7R and A9D GRN mRNA levels were unchanged whereas WT and V5L GRN mRNA levels were significantly depleted (Figure 3A). As expected, WT GRN protein levels were also reduced in SRP54 siRNA-treated cells (Figure 3C).
Figure 3. Differential Stability of WT GRN mRNA Is SRP and Translation Dependent.
(A) GRN qPCR analysis in HeLa cells pretreated with either control siRNA or SRP54 siRNA, transfected with WT (black), V5L (dark gray), W7R (light gray), or A9D GRN (white). Mean and SD (error bars) for 4 independent experiments are shown. One-way ANOVA pairwise comparison: *p < 0.001.
(B) GRN qPCR analysis in HeLa Tet-OFF cells pretreated with either control siRNA (circles) or SRP54 siRNA (triangles), transfected with WT GRN (black) or A9D GRN (white). mRNA levels (mean and SD [error bars] for 3 independent experiments; 2 biological replicates/experiment) are shown on natural log scale as a function of time after doxycycline treatment. Each mRNA is normalized to the initial quantity of that mRNA species, which is set at 1. One-way ANOVA pairwise comparison: *p < 0.001. All other comparisons: no significant difference.
(C) Western blot of whole-cell lysates from HeLa cells treated with control siRNA or SRP54 siRNA, transfected with either WT GRN or A9D GRN. Lysates were immunoblotted for GRN, SRP54, and the loading control actin. Images represent 3 independent experiments.
(D) GRN qPCR in HeLa cells transfected with WT GRN (black), V5L GRN (dark gray), W7R GRN (light gray), or A9D GRN (white), +/− cycloheximide (CHX) (50 μg/mL for 24 hr). Mean and SD (error bars) for 3 independent experiments are shown. One-way ANOVA pairwise comparison: *p < 0.05. All other comparisons: no significant difference.
(E) Western blot of whole-cell lysates from HeLa cells transfected with WT GRN or A9D GRN, +/− CHX (50 μg/mL for 24 hr). Lysates were immunoblotted for GRN and the loading control actin. Images are representative of 3 independent experiments.
Next, GRN WT and GRN A9D mRNA turnover were assessed in control or SRP54 siRNA-treated cells. Consistent with earlier results, A9D GRN mRNA levels were significantly reduced compared to WT GRN at 4 and 8 hr in control siRNA-treated cells (Figure 3B). However, there was no significant difference at these time points in SRP54 siRNA-treated cells (Figure 3B). Together, these results suggest SRP recruitment is critical for WT GRN mRNA stability.
Mutant mRNA Degradation Is Translation Dependent
To assess the role of translation in mutant mRNA degradation, transfected HeLa cells were treated with the elongation inhibitor cycloheximide (CHX). Unlike vehicle-treated cells, in CHX-treated cells, W7R GRN and A9D GRN mRNA levels were restored to WT GRN mRNA levels (Figure 3D). Effective translation inhibition was confirmed by western blotting for GRN (Figure 3E).
Based on these results, we conclude that W7R GRN and A9D GRN are RAPP substrates, whereas V5L GRN is not. Because the signal sequence is generally cleaved in the ER (von Heijne, 1983), secreted V5L GRN is likely identical to secreted WT GRN. Thus, we predict that V5L GRN is a benign polymorphism, whereas W7R GRN, like A9D GRN, disrupts GRN secretion and causes FTLD.
Ago2 Does Not Play a Role in A9D GRN mRNA Degradation
Original studies of RAPP uncovered a role for Ago2 in mutant mRNA degradation (Karamyshev et al., 2014). To determine whether this role is preserved, GRN mRNA turnover was assessed in HeLa Tet-OFF cells treated with either control or Ago2 siRNA. As seen previously, A9D GRN mRNA is rapidly degraded compared to WT GRN mRNA; A9D GRN mRNA levels are significantly reduced at both 4 and 8 hr (Figure S3A). However, there was no significant difference in WT GRN mRNA turnover between cells pretreated with control versus Ago2 siRNA (Figure S3A). Similarly, A9D GRN mRNA turnover was unchanged in Ago2 siRNA-treated cells (Figure S3A).
Additionally, we used Myc-Ago2 constructs to assess the effect of Ago2 overexpression on GRN mRNA turnover. Again, A9D GRN mRNA turnover was rapid compared with WT GRN mRNA turnover, with significantly less A9D GRN mRNA remaining at both the 4- and 8-hr time points (Figure S3B). However, WT GRN mRNA turnover was not significantly different in cells expressing Myc-Ago2 (Figure S3B). A9D GRN mRNA turnover was similarly unchanged with Myc-Ago2 overexpression (Figure S3B). Western blotting confirmed both Ago2 knockdown and Myc-Ago2 overexpression (Figure S3C).
A9D GRN Triggers RAPP in Patient-Derived Fibroblasts
To assess RAPP in a disease-relevant system, we obtained GRN +/A9D fibroblasts from an FTLD patient (genotype confirmed by sequencing; Figure 4A).
Figure 4. A9D mRNA Is Specifically Degraded in Patient-Derived Cells, and mRNA Degradation Is Translation Dependent.
(A) Sequence traces of PCR-amplified GRN from primary human fibroblasts. Top: PCR amplified genomic DNA is shown; bottom: PCR-amplified cDNA (reverse-transcribed mRNA) is shown.
(B) WT (black) and A9D (white) GRN qPCR (using variant-specific primers) in patient fibroblasts, using genotype specific primers. Mean and SD (error bars) for 8 independent experiments are shown. Mann-Whitney Rank sum test: WT GRN versus A9D GRN; p < 0.001. *p < 0.001.
(C) WT (closed circle) and A9D (open circle) GRN qPCR in patient fibroblasts as a function of time after CHX treatment in hours (CHX: 10 μg/mL). Mean and SD (error bars) for 3 independent experiments plotted on a log 10 scale are shown. t test WT versus A9D at t = 0: p < 0.001. t test WT versus A9D at t = 48 hr: n.s.d. t test A9D t = 0 versus A9D t = 48 hr: p = 0.046. *p < 0.001.
(D) WT (black) and A9D (white) GRN qPCR in patient fibroblasts treated with either control virus or SRP54 shRNA virus. Mean and SD (error bars) for 3 independent experiments are shown. t test: *p < 0.001. For (B)–(D), A9D GRN mRNA levels normalized using factor 0.709 (see Figure S2B for derivation of normalization factor).
First, we compared steady-state levels of WT GRN with A9D GRN mRNA. We used peak height on sequencing tracings as a semiquantitative measure of DNA. In the genomic DNA tracing, WT and A9D peaks were of similar height; however, in the cDNA tracing (prepared from mRNA), the WT peak was far larger than the A9D peak, consistent with the previous report (Gass et al., 2006). We also performed qPCR on untreated fibroblasts with WT and A9D-specific primers (validated in Figure S2). By this method, after normalization, only ~6% of the GRN mRNA in the patient fibroblasts is A9D (Figure 4B). Together, these results demonstrate that A9D GRN mRNA is significantly underrepresented in patient fibroblasts.
Next, GRN qPCR was performed on fibroblasts treated with either vehicle or CHX. Whereas WT GRN mRNA levels showed little response to CHX treatment, A9D GRN mRNA levels increased ~8-fold over 48 hr (Figure 4C). After 48 hr of CHX, the ratio of A9D GRN:WT GRN had also changed significantly, from ~0.10 to ~0.68.
Finally, we assessed GRN mRNA in fibroblasts infected with either virus containing SRP54 short hairpin RNA (shRNA) or control virus containing non-targeting shRNA. WT GRN mRNA levels were dramatically reduced in cells infected with SRP54 shRNA, to ~45% of those infected with control virus (Figure 4D). However, levels of A9D mRNA did not differ significantly between the two conditions (Figure 4D). Because most of the GRN mRNA is WT, we also expected reduced total GRN mRNA in SRP54 shRNA-treated cells. Indeed, total GRN mRNA levels were similarly reduced in SRP54 shRNA-treated cells (~47% of level in control cells; Figure S4A). SRP54 knockdown was confirmed with SRP54 qPCR, which demonstrated a significant reduction in cells treated with SRP54 shRNA (~22% of SRP54 mRNA in control cells; Figure S4B).
Thus, our major findings in the overexpression system were conserved in patient fibroblasts: underrepresentation of A9D GRN mRNA is dependent on translation and SRP54.
DISCUSSION
Here, we provide evidence that RAPP plays a role in the pathophysiology of human diseases caused by signal sequence mutations. However, some results have forced us to re-examine our model of RAPP. We originally conceived of secretory mRNA stability as dependent on the balance between two nascent chain interactors: SRP and Ago2 (Karamyshev et al., 2014). However, we found that whereas SRP depletion affected GRN mRNA turnover, Ago2 knockdown and overexpression did not. Perhaps Ago2 only plays a role in a subset of RAPP substrates that has not yet been defined. Additionally, our model does not explain the increase in WT GRN mRNA in CHX-treated cells. This may reflect that WT GRN fails to interact with SRP at a high rate. Alternatively, this may represent the influence of a unique translation-dependent regulatory pathway. Finally, we originally described that mutant secretory proteins that triggered RAPP were not detectable (Karamyshev et al., 2014). However, through the use of a flag tag, we detected a punctate A9D GRN signal, which other groups have observed colocalizes with stress markers (Liet al., 2018; Mukherjee et al., 2008). We concluded that the use of Flag-tagged A9D GRN improved detection both due to an absence of background and higher affinity antibody. However, we cannot exclude the possibility this punctate signal is an artifact of the flag tag. Moreover, the punctate signal may be an artifact of overexpressing A9D GRN; RAPP machinery may be overwhelmed by mutant secretory mRNAs, which can escape degradation but are then sequestered into stress granules. However, it is tantalizing to speculate that the punctate signal represents a true RAPP intermediate, especially given the characteristic protein aggregates seen in FTLD.
The extent of the role RAPP plays in the molecular pathogenesis of the A9D GRN mutation is unclear. Does the continuous generation of aberrant protein products use up RAPP factors, reducing the capacity of the cell to handle additional proteotoxic stress? If this were true, one might expect A9D GRN to have a more severe phenotype than other loss-of-function GRN mutations. However, there is only modest evidence to support this. In a study limited by small sample size, A9D GRN carriers were found to have a lower age of disease onset and a lower age of death compared with patients carrying other GRN mutations (Chen-Plotkin et al., 2011).
Our demonstration of an endogenous RAPP substrate, A9D GRN, confirms the physiological relevance of this quality control pathway. We anticipate that RAPP plays a role in many disparate genetic diseases. Additional signal sequence mutations predicted to disrupt SRP recruitment include the L15R mutation in UGT1A1, which causes Crigler-Najjar type II (Seppen et al., 1996), and the L25R mutation in COL5A1, which causes Ehlers-Danlos (Symoens et al., 2009); these both substitute a polar amino acid for the WT hydrophobic residue. This study of A9D GRN reveals a mechanism of molecular pathology that likely underlies multiple genetic diseases.
EXPERIMENTAL PROCEDURES
Cell Lines and Drug Concentrations
HeLa Tet-ON (Clontech), HeLa Tet-OFF (Clontech), and A9D GRN patient fibroblasts (ND40082; obtained from NINDS Human Genetics Resource Center DNA and Cell Line Repository; https://www.coriell.org/1/ninds) were used.
For several experiments, HeLa Tet-ON cells were used for continuity, but doxycycline induction was not used. In these cases, we refer to these cells as “HeLa” in the manuscript to avoid confusion. Cell lines have been further specified in Supplemental Experimental Procedures.
Cycloheximide 50 μg/mL was used for 24 hr in HeLa Tet-ON and 10 μg/mL for indicated time in A9D GRN fibroblasts.
Doxycycline 1 μg/mL was used for indicated time in HeLa Tet-OFF.
Plasmids, Lentivirus, and siRNA
GRN cDNA was obtained from DNASU (HscD00022612) and subcloned into the following vectors: pBI-CMV2 AcGFP (Clontech Laboratories) and pTRE2-hyg (Clontech).
Lentiviral particles were purchased from Sigma, both control (SHC002V) and SRP54 shRNA (TRCN0000278368).
Control siRNA was purchased from Ambion. SRP54 and Ago2 siRNAs were synthesized by Dharmacon. For siRNA sequences, plasmid details, and details of shRNA and siRNA experiments, see Supplemental Experimental Procedures.
In Vitro Translation and Site-Specific Photocrosslinking
DNA fragments for in vitro transcription were generated as before by standard PCR techniques, then transcribed in vitro using SP6 RNA polymerase, and then purified using QIAGEN RNeasy Mini kit (Flanagan et al., 2003).
In vitro translation and site-specific photocrosslinking were conducted in rabbit reticulocyte system (Etchells et al., 2005; Karamyshev et al., 2014).
EANB-[14C]Lys-tRNAamb for photocrosslinking experiments was prepared as described earlier (Flanagan et al., 2003) or was purchased from tRNA Probes. Samples were analyzed by electrophoresis in the gradient 4%–15% SDS-PAGE and autoradiography.
Western Blots, Immunofluorescence, and Antibodies
WT, V5L, W7R, and A9D GRN expression and secretion were assessed by western blotting and immunofluorescence of transiently transfected HeLa Tet-ON cells. Immunofluorescence images were obtained on a Nikon Eclipse TE2000-U with NIS Elements software, using 60× objective.
Subcellular localization of GRN was assessed with immunofluorescence of HeLa Tet-OFF cells transiently expressing GRN variants (pTRE hyg2) and an ER marker, mcherry-KDEL (Chang et al., 2013) or Golgi marker, CFP-Golgi (Clontech). Images were captured with LSM 880 laser scanning confocal microscope (Zeiss). Plan Apochromat 63×/1.4 numerical aperture (NA) oil objective was used.
For sample preparation and antibodies, please see Supplemental Experimental Methods.
Statistical Analysis
All statistical analyses were performed using SigmaPlot 12.0 software. Statistical method, n, and p value are indicated within the figure legends.
Supplementary Material
Highlights.
Mutations in progranulin that prevent its secretion cause FTLD
FTLD-linked A9D progranulin cannot recruit the trafficking factor SRP
Inability to recruit SRP prevents secretion and triggers A9D mRNA degradation
ACKNOWLEDGMENTS
Authors thank Jerry Shay for help culturing patient fibroblasts. This study used samples from the NINDS Cell Line Repository: sample number ND40082. This research was supported by a grant from the National Institute of Diabetes and Digestive and Kidney Diseases (DK049835) to P.J.T. and by the Center of Excellence for Translational Neuroscience and Therapeutics (CTNT) grant PN-CTNT 2017-05 AKHRJDHW and the National Institute of Neurological Disorders and Stroke of the NIH under award number R03NS102645 to A.L.K. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures and four figures and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.05.003.
AUTHOR CONTRIBUTIONS
E.S.P. designed, conceived, and performed the majority of the experiments; designed all figures; and wrote and edited the manuscript. A.L.K. designed and supervised in vitro translation and photocrosslinking assays and conducted some photocrosslinking experiments. E.B.T. prepared constructs for in vitro translations, conducted photocrosslinking experiments, and edited the manuscript. P.J.T. and A.L.K. provided intellectual expertise and guidance and edited the manuscript. I.-H.W. performed progranulin immunofluorescence co-staining with ER and Golgi. H.H. suggested the chimeric protein experiments and edited the manuscript. P.J.T. coordinated the study.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Almeida MR, Macario MC, Ramos L, Baldeiras I, Ribeiro MH, and Santana I (2016). Portuguese family with the co-occurrence of frontotemporal lobar degeneration and neuronal ceroid lipofuscinosis phenotypes due to progranulin gene mutation. Neurobiol. Aging 41, 200.e1–200.e5. [DOI] [PubMed] [Google Scholar]
- Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, Snowden J, Adamson J, Sadovnick AD, Rollinson S, et al. (2006). Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 442, 916–919. [DOI] [PubMed] [Google Scholar]
- Bateman A, and Bennett HP (1998). Granulins: the structure and function of an emerging family of growth factors. J. Endocrinol 158, 145–151. [DOI] [PubMed] [Google Scholar]
- Chang CL, Hsieh TS, Yang TT, Rothberg KG, Azizoglu DB, Volk E, Liao JC, and Liou J (2013). Feedback regulation of receptor-induced Ca2+ signaling mediated by E-Syt1 and Nir2 at endoplasmic reticulum-plasma membrane junctions. Cell Rep 5, 813–825. [DOI] [PubMed] [Google Scholar]
- Chen-Plotkin AS, Martinez-Lage M, Sleiman PM, Hu W, Greene R, Wood EM, Bing S, Grossman M, Schellenberg GD, Hatanpaa KJ, et al. (2011). Genetic and clinical features of progranulin-associated frontotemporal lobar degeneration. Arch. Neurol 68, 488–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchells SA, Meyer AS, Yam AY, Roobol A, Miao Y, Shao Y, Carden MJ, Skach WR, Frydman J, and Johnson AE (2005). The cotranslational contacts between ribosome-bound nascent polypeptides and the subunits of the hetero-oligomeric chaperonin TRiC probed by photocross-linking. J. Biol. Chem 280, 28118–28126. [DOI] [PubMed] [Google Scholar]
- Finch N, Baker M, Crook R, Swanson K, Kuntz K, Surtees R, Bisceglio G, Rovelet-Lecrux A, Boeve B, Petersen RC, et al. (2009). Plasma progranulin levels predict progranulin mutation status in frontotemporal dementia patients and asymptomatic family members. Brain 132, 583–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan JJ, Chen JC, Miao Y, Shao Y, Lin J, Bock PE, and Johnson AE (2003). Signal recognition particle binds to ribosome-bound signal sequences with fluorescence-detected subnanomolar affinity that does not diminish as the nascent chain lengthens. J. Biol. Chem 278, 18628–18637. [DOI] [PubMed] [Google Scholar]
- Gass J, Cannon A, Mackenzie IR, Boeve B, Baker M, Adamson J, Crook R, Melquist S, Kuntz K, Petersen R, et al. (2006). Mutations in progranulin are a major cause of ubiquitin-positive frontotemporal lobar degeneration. Hum. Mol. Genet 15, 2988–3001. [DOI] [PubMed] [Google Scholar]
- Horaitis O, Talbot CC Jr., Phommarinh M, Phillips KM, and Cotton RG (2007). A database of locus-specific databases. Nat. Genet 39, 425. [DOI] [PubMed] [Google Scholar]
- Jackson RJ, and Hunt T (1983). Preparation and use of nuclease-treated rabbit reticulocyte lysates for the translation of eukaryotic messenger RNA. Methods Enzymol 96, 50–74. [DOI] [PubMed] [Google Scholar]
- Karamyshev AL, Patrick AE, Karamysheva ZN, Griesemer DS, Hudson H, Tjon-Kon-Sang S, Nilsson I, Otto H, Liu Q, Rospert S, et al. (2014). Inefficient SRP interaction with a nascent chain triggers a mRNA quality control pathway. Cell 156, 146–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg UC, Walter P, and Johnson AE (1986). Photocrosslinking of the signal sequence of nascent preprolactin to the 54-kilodalton polypeptide of the signal recognition particle. Proc. Natl. Acad. Sci. USA 83, 8604–8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Ber I, Camuzat A, Hannequin D, Pasquier F, Guedj E, Rovelet-Lecrux A, Hahn-Barma V, van der Zee J, Clot F, Bakchine S, et al. ; French research network on FTD/FTD-MND (2008). Phenotype variability in progranulin mutation carriers: a clinical, neuropsychological, imaging and genetic study. Brain 131, 732–746. [DOI] [PubMed] [Google Scholar]
- Li S, Chen Y, Sun D, Bai R, Gao X, Yang Y, Sheng J, and Xu Z (2018). Angiogenin Prevents Progranulin A9D Mutation-Induced Neuronal-Like Cell Apoptosis Through Cleaving tRNAs into tiRNAs. Mol. Neurobiol 55, 1338–1351. [DOI] [PubMed] [Google Scholar]
- López de Munain A, Alzualde A, Gorostidi A, Otaegui D, Ruiz-Martínez J, Indakoetxea B, Ferrer I, Pérez-Tur J, Saenz A, Bergareche A, et al. (2008). Mutations in progranulin gene: clinical, pathological, and ribonucleic acid expression findings. Biol. Psychiatry 63, 946–952. [DOI] [PubMed] [Google Scholar]
- Mukherjee O, Pastor P, Cairns NJ, Chakraverty S, Kauwe JS, Shears S, Behrens MI, Budde J, Hinrichs AL, Norton J, et al. (2006). HDDD2 is a familial frontotemporal lobar degeneration with ubiquitin-positive, tau-negative inclusions caused by a missense mutation in the signal peptide of progranulin. Ann. Neurol 60, 314–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee O, Wang J, Gitcho M, Chakraverty S, Taylor-Reinwald L, Shears S, Kauwe JS, Norton J, Levitch D, Bigio EH, et al. (2008). Molecular characterization of novel progranulin (GRN) mutations in frontotemporal dementia. Hum. Mutat 29, 512–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson I, Lara P, Hessa T, Johnson AE, von Heijne G, and Karamyshev AL (2015). The code for directing proteins for translocation across ER membrane: SRP cotranslationally recognizes specific features of a signal sequence. J. Mol. Biol 427 (6 Pt A), 1191–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp MW, and Maquat LE (2014). Defective secretory-protein mRNAs take the RAPP. Trends Biochem. Sci 39, 154–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppen J, Steenken E, Lindhout D, Bosma PJ, and Elferink RP (1996). A mutation which disrupts the hydrophobic core of the signal peptide of bilirubin UDP-glucuronosyltransferase, an endoplasmic reticulum membrane protein, causes Crigler-Najjar type II. FEBS Lett 390, 294–298. [DOI] [PubMed] [Google Scholar]
- Smith KR, Damiano J, Franceschetti S, Carpenter S, Canafoglia L, Morbin M, Rossi G, Pareyson D, Mole SE, Staropoli JF, et al. (2012). Strikingly different clinicopathological phenotypes determined by progranulin-mutation dosage. Am. J. Hum. Genet 90, 1102–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symoens S, Malfait F, Renard M, André J, Hausser I, Loeys B, Coucke P, and De Paepe A (2009). COL5A1 signal peptide mutations interfere with protein secretion and cause classic Ehlers-Danlos syndrome. Hum. Mutat 30, E395–E403. [DOI] [PubMed] [Google Scholar]
- von Heijne G (1981). On the hydrophobic nature of signal sequences. Eur. J. Biochem 116, 419–422. [DOI] [PubMed] [Google Scholar]
- von Heijne G (1983). Patterns of amino acids near signal-sequence cleavage sites. Eur. J. Biochem 133, 17–21. [DOI] [PubMed] [Google Scholar]
- von Heijne G (1985). Signal sequences. The limits of variation. J. Mol. Biol 184, 99–105. [DOI] [PubMed] [Google Scholar]
- Walter P, and Blobel G (1983). Signal recognition particle: a ribonucleoprotein required for cotranslational translocation of proteins, isolation and properties. Methods Enzymol 96, 682–691. [DOI] [PubMed] [Google Scholar]
- Walter P, Ibrahimi I, and Blobel G (1981). Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to in-vitro-assembled polysomes synthesizing secretory protein. J. Cell Biol 91, 545–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.