Abstract
Parkinson’s Disease affects more than 4 million people worldwide, and biomarkers to bolster the therapeutic development pipeline are urgently needed. The recent advent of an “ecosystem” of shared biosample biorepositories and data enables us to consider how to focus PD biomarker activity to best translate efforts into real-world impact.
MAIN TEXT
Parkinson’s Disease (PD) currently affects >4 million people worldwide, with numbers projected to double in the next few decades. We urgently need treatments to slow or stop the progression of this disease. Of >550 open studies for PD listed on ClinicalTrials.gov, only ~10% are aimed at testing disease-modifying or neuroprotective therapies. Of the existing disease-modifying or neuroprotective trials, only 2 are in Phase III. Why is the pipeline meager? Why have previous disease-modifying trials been unsuccessful? Most importantly, what can we do to ameliorate this situation?
Biomarkers may represent an important tool in bolstering that pipeline towards therapeutic discovery. Biomarkers have been defined variously, with a highly inclusive definition emerging from the 2000 NIH Working Group on this subject: “A characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention” (1).
As a body of scientists from academic, industrial, government, and private foundation sectors, we highlight here the types of biomarkers that are likely to have the greatest immediate utility and impact on the development of disease-modifying therapeutic trials. Moreover, we stress the urgency of finding such biomarkers in the near future given the likely entry of several classes of mechanism-based therapies (e.g. therapies targeting the aggregation or propagation of alpha-synuclein or targeting LRRK2 kinase activity) into late pre-clinical and early human trials.
Specifically, we argue that biomarkers that are likely to be useful in a clinical trial context should be (1) of reasonable effect size alone or in combination (e.g. demonstrate areas under the curve >0.8), and (2) robust, by which we mean that they must demonstrate clear reproducibility across cohorts. We note, moreover, that it would be preferable if the biomarker could be verified in neuropathologically-proven cases of PD, given recent studies indicating that the accuracy of PD diagnosis may not be high at early stages (2). Finally, practical considerations, such as cost and complexity of assays, and capability for frequent or serial testing, need to be considered early in biomarker discovery and development.
What should these robust, accurate, reproducible markers tell us? While many goals are worth considering, the most urgent generic goal centers on patient stratification/enrichment, reducing clinical heterogeneity by discriminating individuals likely to have different PD trajectories. For example, objective markers that predict whether a patient may progress faster or slower in cognitive or motor symptoms would be powerful in selecting patients for clinical trials of potential neuroprotective agents.
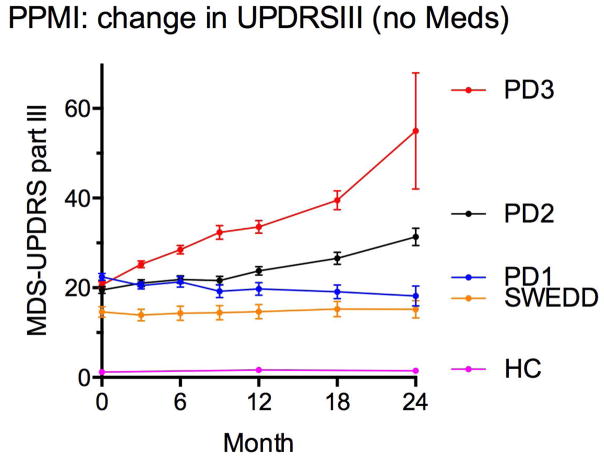
Indeed, within the first two years of longitudinal data collected on 400 PD patients from the international Parkinson’s Progression Marker Initiative (PPMI) (3), one can discern groups with distinct trajectories of motor decline (Figure 1). Should biomarkers be found that could predict at baseline who might follow each trajectory, they could reduce heterogeneity in clinical trial design, increasing the signal-to-noise ratio. Such biomarkers are not unprecedented. Dopamine transporter SPECT (DAT-SPECT) scans and serum urate measurements were used for patient enrichment in the SURE-PD trial (4). Such an approach could be augmented in future trials, should robust biomarkers emerge from our efforts. We note that while we focus on biomarkers derived from patient biofluids, these are by no means the only possible markers. Indeed, as the SURE-PD example demonstrates, other types of markers based on imaging or other modalities may also be useful.
Figure 1.
Means and standard errors of the mean (SEM) for scores on the Movement Disorders Society-Unified Parkinson’s Disease Rating Scale (MDS-UPDRS)-III for PPMI cohort participants. Pink=Healthy controls (HC). Yellow=subjects without evidence of dopaminergic deficit (SWEDD). Parkinson’s Disease patients stratified by tertile of MDS-UPDRS-III score are shown for the lowest tertile (blue), middle tertile (black), and highest tertile (red).
We also acknowledge the utility that markers of target engagement may have in acceleration of clinical trials. These sui generis biomarkers, however, will likely be developed in conjunction with specific therapies and are difficult to discuss without prior knowledge of the therapeutic target, obvious candidates such as alpha-synuclein, glucocerebrosidase, or LRRK2 excepted.
A pipeline for biomarker discovery
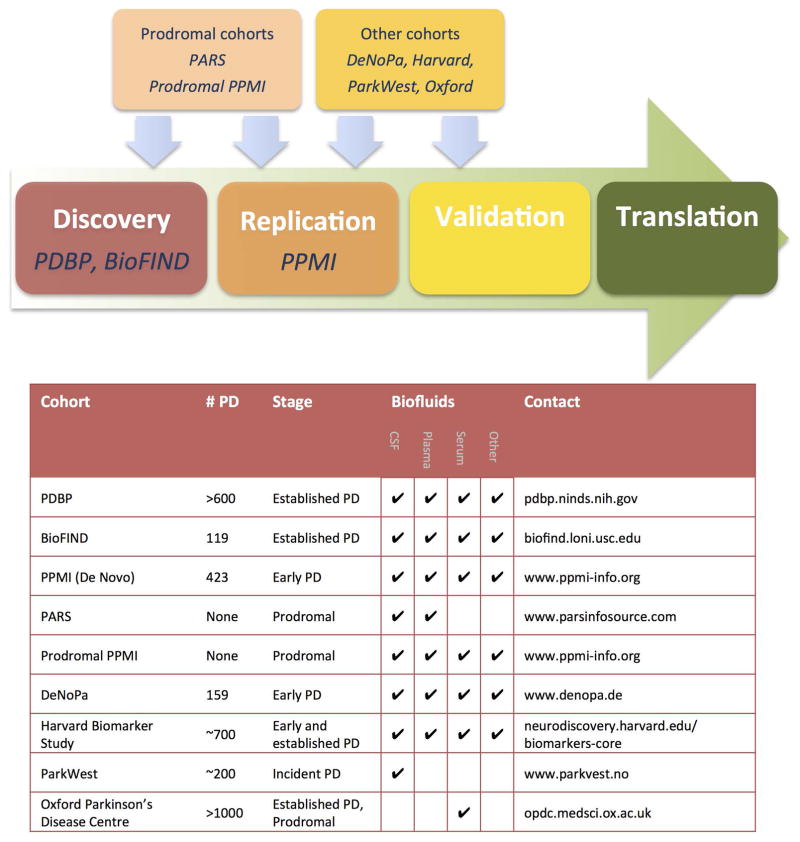
Underlying our preceding statements is the implicit assumption that the creation of tools enabling development of disease-modifying therapies in PD is a reachable, immediate goal, worthy of research investment. This goal is achievable because an “ecosystem” of shared data and specimen biorepositories has emerged, stressing standardized protocols for sample acquisition and storage, data analysis and distribution. Such an infrastructure then serves as a pipeline for de novo discovery, replication of discovery findings in additional cohorts of subjects, and eventual validation of biomarker candidates. Prior to the advent of these shared cohorts and biobanks, investigators depended on their own ability to collect hundreds or thousands of samples for testing, obstructing potential researchers lacking ready access to large clinical populations from entering the biomarker discovery arena. Within the last five years, however, multiple public-private PD efforts have laid the groundwork for investigators from both academic and industrial sectors to access well-annotated clinical samples. We summarize these large PD cohort efforts with biorepositories in the paragraphs below and then place them in a potential pipeline for how they may be used to take biomarkers from concept to reality. While many PD cohorts exist and are summarized in an excellent recent review (5), we focus on large, multicenter cohorts with associated biofluid collections, and clear protocols for requesting samples (Figure 2).
Figure 2.
A pipeline for Parkinson’s Disease biomarker discovery and development.
Discovery cohorts: Parkinson’s Disease Biomarkers Program, BioFIND
The Parkinson’s Disease Biomarkers Program (PDBP, pdbp.ninds.nih.gov) was launched by the National Institute for Neurological Disorders and Stroke (NINDS) in late 2012 as a longitudinal study with biologic sampling every 6 months (6). The PDBP features a dedicated biorepository (BioSEND) comprising DNA, RNA, and biofluid samples from >1000 individuals (>600 with PD) from multiple US centers. Imaging data exists on a subsample of this cohort. An important feature of the PDBP is its associated database, the Data Management Resource (DMR), which can be directly queried by interested researchers, and serves as a portal for biosample and data requests.
Also launched in 2012, the BioFIND study (biofind.loni.usc.edu) is a collaboration between the Michael J. Fox Foundation (MJFF) and the NINDS (7). BioFIND collected clinical data and DNA, RNA, whole blood pellets, as well as multiple biofluids (CSF, saliva, urine, plasma) from 119 PD patients and 96 neurologically healthy controls; samples are requested through the DMR.
The average PD disease duration for both PDBP and BioFIND is >5 years, and many patients are on dopaminergic medications. BioFIND does, however, feature biosample collection dates “on” and “off” dopaminergic medication, with the latter occurring after an overnight washout period. As multicenter PD cohorts with standardized protocols for biosample handling, PDBP and BioFIND are optimized for biomarker discovery.
Replication cohort: Parkinson’s Progression Marker Initiative
Like PDBP, the Parkinson’s Progression Marker Initiative (PPMI, www.ppmi-info.org) is a multicenter, longitudinally followed PD cohort with an associated biorepository (3). However, PPMI was designed to be a replication cohort for PD biomarkers discovered in other cohorts. Moreover, for inclusion in the De Novo cohort of PPMI, PD patients must be within 2 years of diagnosis and naïve to dopaminergic medications at study entry. PPMI De Novo subjects comprise 423 PD patients and 196 normal controls followed clinically, with DNA, RNA and biofluid (plasma, serum, whole blood, urine, saliva) collection; DAT scans are obtained at entry. In addition, PPMI has recently integrated peripheral blood mononuclear cell (PBMC) collection, and a subset of PPMI subjects are participating in an induced pluripotent stem cell (iPSC) generation ancillary study. Imaging measures, such as functional and resting state MRI, are available from a subset of subjects, and a new Pathology Core may offer brain tissue in the future.
Additional cohorts with notable features
A generic pipeline for biomarker development might involve samples from the PDBP and BioFIND as discovery cohorts, with PPMI as the replication cohort. In addition, we highlight five other resources with notable features that aid in PD biomarker discovery efforts.
The National Brain and Tissue Resource for Parkinson’s Disease and Related Disorders (www.ninds.nih.gov/research/parkinsonsweb/brain_banks/U24_BSHRI.htm), housed at the Banner Sun Health Research Institute, comprises fixed and frozen brain tissue from >150 subjects with PD, and >250 elderly control subjects (8). Serum and CSF samples collected postmortem from subjects are also available.
The De Novo Parkinson (DeNoPa) study (www.denopa.de) follows subjects (159 PD, 110 matched healthy controls) from a single center in Germany, with a biorepository housing DNA, RNA, and biofluids (plasma, serum, whole blood, feces, saliva) (9). Importantly, DeNoPa PD subjects are earlier in the course of PD than most PDBP and BioFIND PD patients. Specifically, they are drug-naïve at enrollment, with an average disease duration of <2 years, thus resembling PPMI PD subjects.
The Norwegian ParkWest study (www.parkwest.no) sought to capture and recruit all incident PD cases in Western and Southern Norway in a 22-month period starting in 2004 (10). 265 cases were identified, and follow-up continues. The ParkWest study is mentioned for its unusually comprehensive design and lengthy follow-up; CSF and DNA have been collected.
The Oxford Parkinson’s Disease Centre (OPDC, opdc.medsci.ox.ac.uk) houses a multicenter, prospective, longitudinal biomarker study of PD (11). Over 1000 PD cases have been recruited, with DNA and biofluids collected.
The Harvard Biomarkers Study (HBS, neurodiscovery.harvard.edu/biomarkers-core) has collected and longitudinally phenotyped more than 2,500 individuals since 2007. Notable features of this biorepository include the inclusion of >700 patients with early (drug-naïve and treated) PD and accessibility through the PDBP DMR for plasma, serum, RNA, DNA, and (in a subset) CSF samples.
Prodromal cohorts
Pathogenic processes underlying neurodegenerative diseases such as PD and Alzheimer’s Disease (AD) may be underway years or decades before onset of overt clinical features. Moreover, if the example of AD is informative (12, 13), trials of disease-modifying therapies in PD may enroll pre-symptomatic or high-risk individuals, as well as individuals with overt PD. Thus, the development of biomarkers that can discriminate high-risk individuals who will go on to develop PD vs. those who will not, will be important in the planning of such trials. We next discuss current efforts to enroll and follow asymptomatic individuals at high risk for PD by virtue of genetic (e.g. LRRK2 genetic mutation carriers) or clinical characteristics (e.g. REM behavior disorder (14), or hyposmia (15)). Samples from these cohorts represent an opportunity to develop biomarkers for pre-symptomatic diagnosis or enrichment of prodromal PD clinical trial populations. Moreover, biomarkers emerging from studies in cohorts of individuals with overt PD symptoms (e.g. PDBP, PPMI) may be worth investigating in these prodromal cohorts as well, to better understand the characteristics of these signals in early pathogenesis.
The Parkinson’s Associated Risk Study (PARS, www.parsinfosource.com) screened >10,000 asymptomatic individuals for PD risk factors, identifying 669 hyposmic subjects, 203 of whom underwent DAT imaging and DNA and biofluid collection (CSF in a subset, plasma in all) along with 100 normosmic subjects. At least 50 subjects have putaminal DAT binding <80% of age-expected norms (16), suggesting that many subjects will eventually acquire a PD diagnosis.
Additional cohorts within the PPMI umbrella study include asymptomatic subjects at risk for PD: the prodromal subject study (Prodromal PPMI, or P-PPMI) includes 65 asymptomatic individuals characterized by hyposmia or RBD, while the currently-recruiting genetic cohort study will enroll 600 subjects with or without PD who have a PD-associated genetic mutation in LRRK2, GBA, or SNCA. Protocols for DNA, RNA, and biofluid sample collections mirror those of the De Novo PPMI cohort.
Current efforts to identify prodromal PD subjects rely heavily on Mendelian genetics, suggesting a potential weakness. For example, PARS and PPMI together yield <100 subjects at high risk for non-Mendelian PD, which may be too few to reliably test prodromal biomarkers.
The role of standardization
Attempts to harmonize efforts among some of these biorepositories are aided by a combined Biospecimen Review Access Committee (BRAC, pdbp.ninds.nih.gov/content/application-webform) that reviews requests for samples from PDBP, BioFIND, and HBS.
Harmonization of access to biorepository samples streamlines the process of biomarker discovery. Standardization of other key steps may also accelerate creation of biomarkers that can translate rapidly to clinical trials. Indeed, while replication of a specific association between candidate biomarker and biological process or response of interest is important, many other practical steps are needed to ultimately validate a biomarker for real-world use. For example, the experience of the AD community, which, through the Alzheimer’s Disease Neuroimaging Initiative (ADNI), has translated both biochemical biomarkers (in the form of CSF measures of amyloid-beta and tau) and imaging biomarkers (PET ligands specific for amyloid-beta deposition) into clinical trial use (12), suggests that standardization of sample collection protocols is very important (17). Also recognizing the value that standardization may bring to biomarker studies, the European Union funded a large consortium within its neurodegenerative disease program entitled BIOMARKAPD (biomarkapd.org) starting in 2012, with >48 sites from 21 European countries elaborating standard operating procedures (SOPs) for sample collection, handling and analysis across biomarkers for AD and PD (18).
One key step for standardization entails SOPs for the collection and storage of biofluid samples. BioFIND, PDBP, and PPMI have detailed and harmonized SOPs, readily accessed online (see www.ppmi-info.org/wp-content/uploads/2016/04/PPMI-Biologics-Manual-Complete-03-02-2016.pdf for PPMI protocols). While we recognize that both logistic difficulties and points of scientific disagreement may lead individual investigators and clinical sites to favor more individualized SOPs, we stress the many benefits to collective action in this regard. As the harmonized BioFIND, PDBP and PPMI SOP applies already to the collection of biofluids from 1000+ longitudinally-followed PD patients across >50 clinical sites, we strongly urge that these detailed SOPs serve as “best-practice” guidelines for the collection of biofluids for PD biomarker discovery.
A second key step in which standardization is important is that of assay standardization. The AD experience, especially with CSF tau and amyloid-beta assays (19), again provides relevant precedent. In the early stages of biomarker discovery, the best assay platform may not be clear. The community, however, will need ways to compare results obtained on alternative platforms. To that end, the PDBP developed pools of reference samples (6). These reference samples are simply a large set of identical aliquots obtained by pooling many samples of the biofluids in question. By including reference samples in individual assays, cross-laboratory comparisons of values obtained for a given protein assay, for example, are possible, and normalization across assays may be feasible. For a subset of biomarkers nominated beyond the discovery phase, reference standards are critical. These should be developed and shared across multiple sites.
Finally, we note that the marriage of PD biomarker discovery efforts to downstream use in clinical trials is aided by a Food and Drug Administration (FDA) program designed to facilitate this exact transition. Specifically, the FDA Biomarker Qualification Program (www.fda.gov/Drugs/DevelopmentApprovalProcess/DrugDevelopmentToolsQualificationProgram/BiomarkerQualificationProgram/default.htm) outlines a process by which the Center for Drug Evaluation and Research (CDER) may guide biomarker discovery scientists on practices most likely to integrate a given biomarker into drug development processes.
What types of biomarkers, what types of patients?
With samples collected and available, what markers should we seek? While there are myriad potential uses for biomarker discovery efforts – including the unexpected discovery of potential therapeutic pathways – we focus here on marker types most likely to accelerate the pace of disease-modifying clinical trials in PD. The average clinical trial length in PD is ~2 years, with the majority of disease-modifying trials aimed at early symptomatic PD patients. It is likely that future disease-modifying trials will also enroll the earliest symptomatic PD patients, with possible extension into the prodromal phase. As a consequence, we argue that the following points should be considered in biomarker discovery.
We need to study the right patient population
If the ultimate goal is use in an early PD trial of ~2 years’ length, the replication cohort should be similar, and PPMI is well-suited as a longitudinal cohort of de novo patients. In earlier stages of biomarker discovery and assay development, populations with more “exaggerated” signals might be used – for example, PD subjects with established disease such as those in the PDBP and BioFIND cohorts.
We need large-scale discovery efforts
The current state of biomarkers in PD has been reviewed previously (20–22), so we will confine our remarks to the observation that we do not yet have, despite years of effort, any biomarkers that meet the criteria mentioned previously of acting robustly, with high sensitivities and specificities, in a way that can reduce heterogeneity for clinical trials. Many discovery studies are hampered, moreover, by small numbers. For example, a systematic review of published studies investigating the utility of alpha-synuclein species as a PD biomarker finds 84% of publications with data from sample sizes of 100 PD patients or fewer (23). Such studies risk “losing” discoveries to lack of statistical power; we should be conducting discovery studies in reasonably well-powered cohorts.
We may need to use markers in combination
While most existing biomarker studies evaluated performance characteristics of markers used alone or in very small groups, these approaches may not yield clinically useful receiver operating curve (ROC) characteristics. Of the protein biomarker studies published by PDBP investigators since 2012, only three report distinguishing groups with ROCs of ~0.8. Two of these three studies aimed to differentiate PD from normal controls or from AD (24, 25), while one aimed to differentiate PD patients with and without dementia (26). What all three studies have in common is an approach of incorporating panels of 5–21 biomarkers into classifying algorithms; these algorithms, rather than individual markers, reach high classification accuracies. Such a strategy of constructing an aggregate measure from multiple individual markers has been fruitful in genetic studies of PD risk (27). Indeed, the use of markers in combination may need to span multiple modalities (e.g. genetic, clinical, biochemical, and imaging-based markers) to maximize utility.
We may need to reconsider our overwhelming focus on markers that discriminate PD from controls or from other diseases, and consider markers that separate subgroups within PD
To date, most PD biomarker studies have focused on the differentiation of PD from controls or other diseases. Confirmation of diagnosis is an important goal, given the difficulty posed by non-PD parkinsonian syndromes such as multiple system atrophy (MSA). These “mimic” disorders complicate PD clinical trial enrollment, and the inclusion of disease controls is important in biomarker validation studies. However, confirmation of diagnosis is by no means the only goal. Indeed, a more urgent need may be for markers that differentiate subgroups within PD. Unfortunately, few studies have sought to find and develop markers differentiating PD subgroups.
Two lines of reasoning argue for a shift of focus in PD biomarker investigations towards inclusion of markers of differential progression. First, clinical trials aimed at testing potential neuroprotective therapies are hampered by significant heterogeneity in PD progression. As shown in Figure 1, PPMI participants not receiving any symptomatic medications differed greatly in their rates of motor progression over 24 months. Whereas the fastest third of progressors had substantial increase in UPDRS-III score, the slowest third saw essentially no change in UPDRS-III score over the same time period. Such heterogeneity makes demonstration of disease-modifying effect difficult, requiring the enrollment of large numbers of subjects. If one could establish biomarkers capable of reducing the variability of expected disease course, one might substantially improve the speed and reduce the cost of clinical trial designs.
Second, there is reason to believe biological heterogeneity may underlie the relatively monolithic entity we regard as PD. We already recognize, for example, phenomenological subtypes of PD such as tremor-predominant and postural-instability-gait-disorder subtypes, which may also show differences in rates of motor progression and may be tagged by genetic markers (28). Moreover, we can now identify large subgroups of PD based on single genes – carriers of mutations in the glucocerebrosidase (GBA) or LRRK2 genes. Groups stratified by GBA or LRRK2 mutation status show clinical and biochemical differences (29, 30), and drug development efforts aimed particularly at these molecular targets are underway. Although few in number, biochemical markers separating groups with differential disease progression have also been reported. Indeed, one such marker – serum or plasma urate, lower levels of which associate with increased risk for PD and faster rates of PD motor decline – is both an outcome measure and enrollment criterion for the Phase III trial of inosine as a neuroprotective agent in PD (4).
We must insist on replication and confirmation
Genetics and genomics have taught us that unbiased large-scale discovery efforts may yield promising leads. However, we have also learned that large-scale discovery efforts, and particularly those generating large datasets in limited numbers of people, yield many false positives. As a consequence, we need to insist on replication and confirmation of promising biomarker candidates. This requires collective change, so that we both increase incentive for, and remove roadblocks to, replication and confirmation.
The specific delineation of replication cohorts – such as PPMI – for confirmation of early results is crucial. While study design features may mitigate concerns of over-fitting and false positives (e.g. randomized subsampling of samples into discovery and replication sets), these cannot entirely obviate concerns. Thus, the development of multiple cohorts, where biomarkers can be cross-replicated, is a priority.
The development of easily accessible, appropriate cohorts, with standardized SOPs, serves to remove roadblocks to replication, but the problem of increasing incentive still remains. How might funding structures and publishing venues work to combat a widespread bias for the “novel” over the confirmatory? How might we elicit the will to invest substantial time and effort in studies that definitively tell us which early hits to move forwards vs. eliminate from further consideration? These are not questions with easy answers. However, the growing recognition of widespread problems of reproducibility in the biomedical community (31) argues that we cannot defer asking.
Summary remarks
Biomarkers emerging from such large-scale screening efforts may or may not relate to known molecular pathways involving PD. This begs the question of whether to prioritize markers for which prior knowledge suggests a clear biological link. We argue that a practical method for weighing these considerations may be as follows. Markers with no prior known biological connection to PD (“biologically agnostic markers”) should move forward provided that they are consistently reproducible across cohorts and platforms, an approach that has been taken in genomics with success. For example, we point to measures of plasma ApoA1, for which lower levels have consistently been found to associate with younger age of PD onset and more severe motoric impairment (16, 32). Such an approach allows “room” for the fact that many aspects of PD pathobiology may yet remain unknown, posing a significant challenge for top-down weighting of potential biomarker candidates, as well as potential therapeutic targets. At the same time, markers with a clear connection to PD based on prior knowledge (“biologically attractive markers”) should move forward provided that they are reasonably reproducible, with more attention to modifications of assay design to reduce noisy (and therefore less robust) measures. Here, we point to the many efforts that are aimed at developing CSF alpha-synuclein as a PD biomarker (23).
Regardless of the type of biomarker emerging from discovery efforts, standardization will be an important next step. We will need to make difficult decisions, about assay choice, measurement conditions, in order to make the candidate markers reproducible across cohorts and labs. The use of pools of reference samples early in the biomarker discovery process, mentioned before, may help in our assessments of reproducibility across sites.
Harmonization among biomarker development efforts remains a daunting challenge
Currently, platforms, methods of processing raw data, and analysis of these “usable” data may all differ by investigator or biomarker. Indeed, we fully appreciate the vast leap in complexity around issues of harmonization when one moves from assaying binary, relatively robust genetic markers to assaying biochemical or other phenotypic markers that may be affected by many other factors and yield a continuum of values. There is hope, however! In the early days of RNA expression analysis, standards were developed and have been adopted widely in the field, first in the arena of microarray data. Alternately, in the AD biomarker field, large-scale funding efforts with active industrial partnerships such as the ADNI took a guiding role in the choice of imaging and biochemical biomarkers for which to develop specific assays. Both are viable options for moving forwards, but continuing to work in an un-harmonized fashion beyond the early discovery stage is likely to waste money and effort.
Finally, we would like to end on a forward-looking note – that of how one might envision the first biomarkers crossing over from research into PD disease-modifying trials. Should clearly-reproducible biomarkers enter the PD arena that predict, for example, the presence of PD pathology or motor trajectory, these markers may first be measured in actively enrolling PD clinical trials. While there may not yet be a strong enough body of evidence behind them to influence selection of trial participants, these markers could be ascertained in clinical trial participants and used as enrichment criteria in pre-specified analyses to increase the probability of detecting therapeutic efficacy in sub-populations of PD patients. Moreover, future clinical trials could be aided greatly by making data and biosamples collected during clinical trials available for independent researchers to mine after the fact. One might imagine, for example, analyses of treatment effect or disease progression based on biomarker status, or inclusion of biomarkers as covariates in additional analyses, to control for some of the underlying heterogeneity that currently hampers clinical trials. This is a win-win scenario: data gained in clinical trials could further strengthen our confidence in the biomarker candidates, possibly leading to their use in future clinical trial enrollment, and helping to define specific phenotypes and sub-types of Parkinsonism at the molecular level. Indeed, this type of “biomarker-enriched design” has been proposed for oncology trials (33).
We believe that these goals are eminently achievable. Success depends, though, on close collaboration across multiple sectors of the biomedical and drug development endeavor, including academic groups of many stripes (clinicians, pharmacologists, basic biologists, experts in technical assay development), industry (drug developers, technology experts), government (funding sources such as NIH, standardization bureaus, drug regulatory agencies worldwide), and private non-profit funding agencies. These players have at times acted in distinct spheres. However, with a disease affecting so many and lacking disease-modifying therapies, broad collaboration is essential.
References
- 1.Biomarkers Definitions Working Group. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69:89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- 2.Adler CH, Beach TG, Hentz JG, Shill HA, Caviness JN, Driver-Dunckley E, Sabbagh MN, Sue LI, Jacobson SA, Belden CM, Dugger BN. Low clinical diagnostic accuracy of early vs advanced Parkinson disease: clinicopathologic study. Neurology. 2014;83:406–412. doi: 10.1212/WNL.0000000000000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marek K, Jennings D, Lasch S, Siderowf A, Tanner C, Simuni T, Coffey C, Kieburtz K, Flagg E, Chowdhury S. The parkinson progression marker initiative (PPMI) Prog Neurobiol. 2011;95:629–635. doi: 10.1016/j.pneurobio.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parkinson Study Group SURE-PD Investigators. Inosine to increase serum and cerebrospinal fluid urate in Parkinson disease: a randomized clinical trial. JAMA Neurol. 2014;71:141–150. doi: 10.1001/jamaneurol.2013.5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lerche S, Liepelt-Scarfone I, Alves G, Barone P, Behnke S, Ben-Shlomo Y, Berendse H, Burn D, Dodel R, Grosset D, Heinzel S, Hu M, Kasten M, Kruger R, Maetzler W, Moccia M, Mollenhauer B, Oertel W, Roeben B, Sunkel U, Walter U, Wirdefeldt K, Berg D. Methods in Neuroepidemiology Characterization of European Longitudinal Cohort Studies in Parkinson’s Disease--Report of the JPND Working Group BioLoC-PD. Neuroepidemiology. 2015;45:282–297. doi: 10.1159/000439221. [DOI] [PubMed] [Google Scholar]
- 6.Rosenthal LS, Drake D, Alcalay RN, Babcock D, Bowman FD, Chen-Plotkin A, Dawson TM, Dewey RB, Jr, German DC, Huang X, Landin B, McAuliffe M, Petyuk VA, Scherzer CR, Hillaire-Clarke CS, Sieber BA, Sutherland M, Tarn C, West A, Vaillancourt D, Zhang J, Gwinn K PDBP consortium. The NINDS Parkinson’s disease biomarkers program. Mov Disord. 2016;31:915–923. doi: 10.1002/mds.26438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang UJ, Goldman JG, Alcalay RN, Xie T, Tuite P, Henchcliffe C, Hogarth P, Amara AW, Frank S, Rudolph A, Casaceli C, Andrews H, Gwinn K, Sutherland M, Kopil C, Vincent L, Frasier M. The BioFIND study: Characteristics of a clinically typical Parkinson’s disease biomarker cohort. Mov Disord. 2016;31:924–932. doi: 10.1002/mds.26613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beach TG, Adler CH, Sue LI, Serrano G, Shill HA, Walker DG, Lue L, Roher AE, Dugger BN, Maarouf C, Birdsill AC, Intorcia A, Saxon-Labelle M, Pullen J, Scroggins A, Filon J, Scott S, Hoffman B, Garcia A, Caviness JN, Hentz JG, Driver-Dunckley E, Jacobson SA, Davis KJ, Belden CM, Long KE, Malek-Ahmadi M, Powell JJ, Gale LD, Nicholson LR, Caselli RJ, Woodruff BK, Rapscak SZ, Ahern GL, Shi J, Burke AD, Reiman EM, Sabbagh MN. Arizona Study of Aging and Neurodegenerative Disorders and Brain and Body Donation Program. Neuropathology. 2015;35:354–389. doi: 10.1111/neup.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mollenhauer B, Trautmann E, Sixel-Doring F, Wicke T, Ebentheuer J, Schaumburg M, Lang E, Focke NK, Kumar KR, Lohmann K, Klein C, Schlossmacher MG, Kohnen R, Friede T, Trenkwalder C DeNoPa Study Group. Nonmotor and diagnostic findings in subjects with de novo Parkinson disease of the DeNoPa cohort. Neurology. 2013;81:1226–1234. doi: 10.1212/WNL.0b013e3182a6cbd5. [DOI] [PubMed] [Google Scholar]
- 10.Alves G, Bronnick K, Aarsland D, Blennow K, Zetterberg H, Ballard C, Kurz MW, Andreasson U, Tysnes OB, Larsen JP, Mulugeta E. CSF amyloid-beta and tau proteins, and cognitive performance, in early and untreated Parkinson’s disease: the Norwegian ParkWest study. J Neurol Neurosurg Psychiatry. 2010;81:1080–1086. doi: 10.1136/jnnp.2009.199950. [DOI] [PubMed] [Google Scholar]
- 11.Lawton M, Baig F, Rolinski M, Ruffman C, Nithi K, May MT, Ben-Shlomo Y, Hu MT. Parkinson’s Disease Subtypes in the Oxford Parkinson Disease Centre (OPDC) Discovery Cohort. J Parkinsons Dis. 2015;5:269–279. doi: 10.3233/JPD-140523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sevigny J, Chiao P, Bussiere T, Weinreb PH, Williams L, Maier M, Dunstan R, Salloway S, Chen T, Ling Y, O’Gorman J, Qian F, Arastu M, Li M, Chollate S, Brennan MS, Quintero-Monzon O, Scannevin RH, Arnold HM, Engber T, Rhodes K, Ferrero J, Hang Y, Mikulskis A, Grimm J, Hock C, Nitsch RM, Sandrock A. The antibody aducanumab reduces Abeta plaques in Alzheimer’s disease. Nature. 2016;537:50–56. doi: 10.1038/nature19323. [DOI] [PubMed] [Google Scholar]
- 13.Sperling RA, Rentz DM, Johnson KA, Karlawish J, Donohue M, Salmon DP, Aisen P. The A4 study: stopping AD before symptoms begin? Sci Transl Med. 2014;6:228fs13. doi: 10.1126/scitranslmed.3007941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Postuma RB, Aarsland D, Barone P, Burn DJ, Hawkes CH, Oertel W, Ziemssen T. Identifying prodromal Parkinson’s disease: pre-motor disorders in Parkinson’s disease. Mov Disord. 2012;27:617–626. doi: 10.1002/mds.24996. [DOI] [PubMed] [Google Scholar]
- 15.Siderowf A, Jennings D, Eberly S, Oakes D, Hawkins KA, Ascherio A, Stern MB, Marek K Investigators PARS. Impaired olfaction and other prodromal features in the Parkinson At-Risk Syndrome Study. Mov Disord. 2012;27:406–412. doi: 10.1002/mds.24892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiang JK, Wong YC, Siderowf A, Hurtig HI, Xie SX, Lee VM, Trojanowski JQ, Yearout D, Leverenz J, Montine TJ, Stern M, Mendick S, Jennings D, Zabetian C, Marek K, Chen-Plotkin AS. Plasma apolipoprotein A1 as a biomarker for parkinson’s disease. Ann Neurol. 2013 doi: 10.1002/ana.23872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaw LM, Vanderstichele H, Knapik-Czajka M, Clark CM, Aisen PS, Petersen RC, Blennow K, Soares H, Simon A, Lewczuk P, Dean R, Siemers E, Potter W, Lee VM, Trojanowski JQ Alzheimer’s Disease Neuroimaging Initiative. Cerebrospinal fluid biomarker signature in Alzheimer’s disease neuroimaging initiative subjects. Ann Neurol. 2009;65:403–413. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reijs BL, Teunissen CE, Goncharenko N, Betsou F, Blennow K, Baldeiras I, Brosseron F, Cavedo E, Fladby T, Froelich L, Gabryelewicz T, Gurvit H, Kapaki E, Koson P, Kulic L, Lehmann S, Lewczuk P, Lleo A, Maetzler W, de Mendonca A, Miller AM, Molinuevo JL, Mollenhauer B, Parnetti L, Rot U, Schneider A, Simonsen AH, Tagliavini F, Tsolaki M, Verbeek MM, Verhey FR, Zboch M, Winblad B, Scheltens P, Zetterberg H, Visser PJ. The Central Biobank and Virtual Biobank of BIOMARKAPD: A Resource for Studies on Neurodegenerative Diseases. Front Neurol. 2015;6:216. doi: 10.3389/fneur.2015.00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaw LM, Vanderstichele H, Knapik-Czajka M, Figurski M, Coart E, Blennow K, Soares H, Simon AJ, Lewczuk P, Dean RA, Siemers E, Potter W, Lee VM, Trojanowski JQ Alzheimer’s Disease Neuroimaging Initiative. Qualification of the analytical and clinical performance of CSF biomarker analyses in ADNI. Acta Neuropathol. 2011;121:597–609. doi: 10.1007/s00401-011-0808-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parnetti L, Castrioto A, Chiasserini D, Persichetti E, Tambasco N, El-Agnaf O, Calabresi P. Cerebrospinal fluid biomarkers in Parkinson disease. Nat Rev Neurol. 2013;9:131–140. doi: 10.1038/nrneurol.2013.10. [DOI] [PubMed] [Google Scholar]
- 21.Chen-Plotkin AS. Unbiased approaches to biomarker discovery in neurodegenerative diseases. Neuron. 2014;84:594–607. doi: 10.1016/j.neuron.2014.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mollenhauer B, Rochester L, Chen-Plotkin A, Brooks D. What can biomarkers tell us about cognition in Parkinson’s disease? Mov Disord. 2014;29:622–633. doi: 10.1002/mds.25846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simonsen AH, Kuiperij B, El-Agnaf OM, Engelborghs S, Herukka SK, Parnetti L, Rektorova I, Vanmechelen E, Kapaki E, Verbeek M, Mollenhauer B. The utility of alpha-synuclein as biofluid marker in neurodegenerative diseases: a systematic review of the literature. Biomark Med. 2016;10:19–34. doi: 10.2217/BMM.14.105. [DOI] [PubMed] [Google Scholar]
- 24.Shi M, Movius J, Dator R, Aro P, Zhao Y, Pan C, Lin X, Bammler TK, Stewart T, Zabetian CP, Peskind ER, Hu SC, Quinn JF, Galasko DR, Zhang J. Cerebrospinal fluid peptides as potential Parkinson disease biomarkers: a staged pipeline for discovery and validation. Mol Cell Proteomics. 2015;14:544–555. doi: 10.1074/mcp.M114.040576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Bryant SE, Xiao G, Zhang F, Edwards M, German DC, Yin X, Como T, Reisch J, Huebinger RM, Graff-Radford N, Dickson D, Barber R, Hall J, O’Suilleabhain P, Grammas P. Validation of a serum screen for Alzheimer’s disease across assay platforms, species, and tissues. J Alzheimers Dis. 2014;42:1325–1335. doi: 10.3233/JAD-141041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berlyand Y, Weintraub D, Xie SX, Mellis IA, Doshi J, Rick J, McBride J, Davatzikos C, Shaw LM, Hurtig H, Trojanowski JQ, Chen-Plotkin AS. An Alzheimer’s Disease-Derived Biomarker Signature Identifies Parkinson’s Disease Patients with Dementia. PLoS One. 2016;11:e0147319. doi: 10.1371/journal.pone.0147319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nalls MA, McLean CY, Rick J, Eberly S, Hutten SJ, Gwinn K, Sutherland M, Martinez M, Heutink P, Williams NM, Hardy J, Gasser T, Brice A, Price TR, Nicolas A, Keller MF, Molony C, Gibbs JR, Chen-Plotkin A, Suh E, Letson C, Fiandaca MS, Mapstone M, Federoff HJ, Noyce AJ, Morris H, Van Deerlin VM, Weintraub D, Zabetian C, Hernandez DG, Lesage S, Mullins M, Conley ED, Northover CA, Frasier M, Marek K, Day-Williams AG, Stone DJ, Ioannidis JP, Singleton AB Parkinson’s Disease Biomarkers Program and Parkinson’s Progression Marker Initiative investigators. Diagnosis of Parkinson’s disease on the basis of clinical and genetic classification: a population-based modelling study. Lancet Neurol. 2015;14:1002–1009. doi: 10.1016/S1474-4422(15)00178-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cooper CA, Jain N, Gallagher MD, Weintraub D, Xie SX, Berlyand Y, Espay AJ, Quinn J, Edwards KL, Montine T, Van Deerlin VM, Trojanowski J, Zabetian CP, Chen-Plotkin AS. Common variant rs356182 near SNCA defines a Parkinson’s disease endophenotype. Annals of Clinical and Translational Neurology. 2016 doi: 10.1002/acn3.371. n/a–n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alcalay RN, Levy OA, Waters CC, Fahn S, Ford B, Kuo SH, Mazzoni P, Pauciulo MW, Nichols WC, Gan-Or Z, Rouleau GA, Chung WK, Wolf P, Oliva P, Keutzer J, Marder K, Zhang X. Glucocerebrosidase activity in Parkinson’s disease with and without GBA mutations. Brain. 2015;138:2648–2658. doi: 10.1093/brain/awv179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fraser KB, Moehle MS, Alcalay RN, West AB LRRK2 Cohort Consortium. Urinary LRRK2 phosphorylation predicts parkinsonian phenotypes in G2019S LRRK2 carriers. Neurology. 2016;86:994–999. doi: 10.1212/WNL.0000000000002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ioannidis JP. Why most published research findings are false. PLoS Med. 2005;2:e124. doi: 10.1371/journal.pmed.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swanson CR, Berlyand Y, Xie SX, Alcalay RN, Chahine LM, Chen-Plotkin A. Plasma ApoA1 levels associate with Age at Onset and Disease Progression in Drug-Naïve early Parkinson Disease Patients. Manuscript in submission. [Google Scholar]
- 33.Freidlin B, Korn EL. Biomarker enrichment strategies: matching trial design to biomarker credentials. Nat Rev Clin Oncol. 2014;11:81–90. doi: 10.1038/nrclinonc.2013.218. [DOI] [PubMed] [Google Scholar]