Abstract
Breast cancer is a major cause of cancer mortality amongst women. Chemokine (C-C motif) ligand 4 is encoded by the CCL4 gene; specific CCL4 gene polymorphisms are related to the risks and prognoses of various diseases. In this study, we examined whether CCL4 gene single nucleotide polymorphisms (SNPs) predict the risk and progression of breast cancer. Between 2014 and 2016, we recruited 314 patients diagnosed with breast cancer and a cohort of 209 healthy participants (controls) without a history of cancer. Genotyping of the CCL4 rs1634507, rs10491121 and rs1719153 SNPs revealed no significant between-group differences for these polymorphisms. However, amongst luminal A and luminal B subtypes, compared with patients with the AA genotype, those carrying the AG genotype at SNP rs10491121 were less likely to develop lymph node metastasis. In addition, compared with AA carriers, those carrying the AG + GG genotype at SNP rs10491121 were at lower risk of developing distant metastasis, while the presence of the AT genotype at SNP rs1719153 increased the likelihood of pathologic grade (G3 or G4) disease. Variations in the CCL4 gene may help to predict breast cancer progression and metastasis.
Keywords: single nucleotide polymorphism, breast cancer, chemokine C-C motif ligand 4 (CCL4), genotype
Introduction
Breast cancer is the second leading cause of cancer deaths amongst women worldwide. Nearly million women worldwide are diagnosed with breast cancer annually and more than 500,000 die from this disease 1. Besides age, reproductive and gynecologic factors, other risk factors such as family history and environmental factors including tobacco and alcohol consumption, as well as overall amount of physical activity, can greatly modify the risk of developing breast cancer 2. In addition, gynecologic diseases including polycystic ovarian syndrome and adenomyosis have been found to enhance the risk of breast cancer 3, 4.
Mammography screening and genetic testing have limited sensitivity and specificity for estimating breast cancer risk 2. It is uncertain as to whether single nucleotide polymorphism (SNP) genotyping could more accurately predict breast cancer risk and guide disease management 5, 6. Susceptibility to breast cancer appears to be influenced by certain SNPs, as well as clinicopathologic status 7. BRCA1 and BRCA2 gene mutations increase the risk of breast cancer 8, 9. Fascin-1 (FSCN1) and high-mobility group box protein 1 (HMGB1) genetic polymorphisms have also been identified as predictive biomarkers for breast cancer 10.
Chemokine (C-C motif) ligand 4 (CCL4) is a protein that is encoded by the CCL4 gene and acts as a chemoattractant for natural killer cells, monocytes and various other immune cells in the site of inflamed or damaged tissue. CCL4 polymorphisms influence gene expression, protein function and susceptibility to various diseases, including hepatocellular carcinoma, oral cancer, and psoriasis 11-14. CCL4 belongs to a cluster of genes located in the chromosomal region 17q11-q21. The CCL4 protein acts as the chemokine being secreted under mitogenic signals and antigens and attracting monocytes, dendritic cells, natural killer cells and other effector cells into the site of inflamed or damaged tissue 15, 16. On the other hand, the CCL4 gene polymorphisms has been associated with risk and development in oral cancer and hepatocellular carcinoma 12, 17. Despite the well-known impact of chemokines on cancer progression and the recognition that CCL4 gene SNPs play important roles in a variety of human diseases, little is known about the association between these SNPs and the susceptibility to breast cancer and its progression. In this study, we evaluated the predictive capacity of three CCL4 SNPs as candidate biomarkers for breast cancer risk.
Materials and Methods
Participants
Between 2014 and 2016, we collected 314 blood specimens from patients (cases) diagnosed with breast cancer at Dongyang People's Hospital. A total of 209 healthy, cancer-free individuals served as controls. Written informed consent was obtained from all participants before study entry. The Ethics Committee of Dongyang People's Hospital granted study approval. Pathohistologic diagnosis used the World Health Organization breast tumor classification and tumors were graded using the Scarff-Bloom-Richardson method 18. Breast cancer cases were categorized by estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2) and Ki-67 status and grouped under 1 of 4 subtypes: Luminal A (ER-positive [+] and/or PR+, HER2-negative [-], Ki-67 <14%); Luminal B (ER+ and/or PR+, HER2-, Ki-67 ≥14%; or ER+ and/or PR+, HER2+); HER2-enriched (ER-, PR-, HER2+); or as triple-negative breast cancer (TNBC; ER-, PR-, HER2-) 19-21. A standardized questionnaire at study entry collected sociodemographic data and electronic medical records provided clinicopathologic information.
Selection of CCL4 polymorphisms
The CCL4 SNPs selected for this study were identified from multi-allelic copy number variation (CNV) profiles encompassing the q12 region of chromosome 17 containing CCL4 genes. Nonsynonymous SNPs rs1634507, rs10491121 and rs1719153 were extracted from a search of the National Center for Biotechnology Information (NCBI) dbSNP database.
Genomic DNA extraction
The QIAamp DNA Blood Mini Kit (Qiagen, Inc., Valencia, CA, USA) purified genomic DNA from peripheral blood leukocytes. The DNA was dissolved in TE buffer (10 mM Tris, 1 mM EDTA; pH 7.8), quantified by OD260, then stored at -20℃ for further analysis.
Real-time PCR
The ABI StepOne™ real-time polymerase chain reaction (PCR) system (Applied Biosystems, Foster City, CA, USA) assessed sequencing of allelic discrimination for the CCL4 SNP. The TaqMan assay used Software Design Specification version 3.0 software (Applied Biosystems) to analyze the discrimination data. Primers and probes consisted of rs1634507 “AGTTTTCTTGACCTCATGAATGCTG-[G/T]TGAGGCTTTATCCCTCTCTCAGGAA” (product ID: C_7451708_10), rs10491121 “CCTATCCCCTTCCTGAATTAAGTCC-[A/G]AATATAGTCAGTCTTTGAGTGTGGA” (product ID: C_11626804_10) and rs1719153 “TAGGGACTGTTGCACCGAGTTTCAC-[A/T]GTTAAGGAAACAGAGGCACAGAGAG” (product ID: C_12120537_10). PCRs were performed in a total volume of 10 μL containing Master Mix (5 μL), probes (0.25 μL) and genomic DNA (10 ng). The real-time PCR reaction included an initial denaturation step at 95°C for 10 min, then 40 amplification cycles of 95°C for 15 secs and 60°C for 1 min 19, 22.
Statistical analysis
Between-group differences were considered significant if p-values were less than 0.05. Chi-square analysis tested for Hardy-Weinberg equilibrium in the SNP genotype distributions. The Mann-Whitney U-test and Fisher's exact test were utilized for between-group demographic comparisons. Multiple logistic regression models adjusted for confounding variables estimated adjusted odds ratios (AORs) and 95% confidence intervals (CIs) for associations between genotype frequencies and the risk of breast cancer or clinicopathologic characteristics. Haplotype frequencies were analyzed using Haploview 23. All data were analyzed with the software program Statistical Analytic System version 9.1 and are reported as the sample mean ± the standard deviation (SD).
Results
All study participants were Chinese Han (Table 1). The majority were nonsmokers and did not consume alcohol. There was a significantly higher proportion of younger age participants in the control group compared with the breast cancer cohort (p<0.05). Most patients (77.1%) had stage I/II breast cancer; 22.9% had stage III/IV disease (Table 1). In an analysis of hormone receptor status, tumors were mostly ER- (69.7%), PR- (54.1%), or HER2+ (63.1%) (Table 1).
Table 1.
Demographic and clinicopathologic characteristics among healthy cancer-free controls and patients with breast cancer.
| Variable | Controls N=209 (%) |
Patients N=314 (%) |
p value |
|---|---|---|---|
| Age (years) | Mean ± SD | Mean ± SD | |
| 38.5±16.7 | 53.1±11.4 | *p<0.05 | |
| Tobacco smokers | |||
| No | 202 (96.7) | 313 (99.7) | |
| Yes | 7 (3.3) | 1 (0.3) | *p<0.05 |
| Alcohol consumption | |||
| No | 203 (97.1) | 295 (93.9) | |
| Yes | 6 (2.9) | 19 (6.1) | p>0.05 |
| Clinical stage | |||
| I/II | 242 (77.1) | ||
| III/IV | 72 (22.9) | ||
| Tumor size | |||
| ≤T2 | 298 (94.9) | ||
| >T2 | 16 (5.1) | ||
| Lymph node status | |||
| N0+N1 | 247 (78.7) | ||
| N2+N3 | 67 (21.3) | ||
| Distant metastasis | |||
| M0 | 304 (96.8) | ||
| M1 | 10 (3.2) | ||
| Histological grade | |||
| G1+G2 | 218 (69.4) | ||
| G3+G4 | 96 (30.6) | ||
| ER status | |||
| Positive | 95 (30.3) | ||
| Negative | 219 (69.7) | ||
| PR status | |||
| Positive | 144 (45.9) | ||
| Negative | 170 (54.1) | ||
| HER2 status | |||
| Positive | 198 (63.1) | ||
| Negative | 116 (36.9) | ||
The Mann-Whitney U-test and Fisher's exact test were used to compare values between controls and patients with breast cancer. *p < 0.05 was statistically significant. T2 = tumor >20 mm but ≤50 mm in greatest dimension; N0 = lymph node-negative; N1 = cancer has spread to 1-3 lymph node(s); N2 = 4-9 lymph nodes; N3 = ≥10 positive lymph nodes; M0 = noninvasive cancer; M1 = cancer has metastasized to organs or lymph nodes away from the breast; G1 = well differentiated (low grade); G2 = moderately differentiated (intermediate grade); G3 = poorly differentiated (high grade); G4 = undifferentiated (high grade); ER = estrogen receptor; PR = progesterone receptor; HER2 = human epidermal growth factor receptor 2.
Polymorphism frequencies are shown in Table 2. All genotypes were in Hardy-Weinberg equilibrium (p > 0.05). In both study groups, the most frequent genotypes for SNPs rs10491121, rs1634507 and rs1719153 were homozygous for A/A, homozygous for G/G and homozygous for A/A. Analyses that adjusted for potential confounders found no significant between-group differences for the polymorphism frequencies.
Table 2.
Distribution frequencies of CCL4 genotypes among healthy cancer-free controls and patients with breast cancer.
| Variable | Controls N=209 (%) |
Patients N=314 (%) |
OR (95% CI) |
|---|---|---|---|
| rs10491121 | |||
| AA | 64 (41) | 79 (34.2) | 1.00 (reference) |
| AG | 92 (59) | 152 (65.8) | 1.338 (0.88-2.035) |
| GG | 53 (45.3) | 83 (51.2) | 1.269 (0.787-2.044) |
| AG+GG | 145 (69.4) | 235 (74.8) | 1.313 (0.89-1.938) |
| rs1634507 | |||
| GG | 101 (54.9) | 135 (49.5) | 1.00 (reference) |
| GT | 83 (45.1) | 138 (50.5) | 1.244 (0.855-1.810) |
| TT | 25 (19.8) | 41 (23.3) | 1.227 (0.701-2.148) |
| GT+TT | 108 (51.7) | 179 (57) | 1.240 (0.873-1.762) |
| rs1719153 | |||
| AA | 101 (55.5) | 149 (52.7) | 1.00 (reference) |
| AT | 81 (44.5) | 134 (47.3) | 1.121 (0.771-1.630) |
| TT | 27 (21.1) | 31 (17.2) | 0.778 (0.438-1.382) |
| AT+TT | 108 (51.7) | 165 (52.5) | 1.036 (0.73-1.470) |
The odds ratios (ORs) with their 95% confidence intervals (CIs) were estimated by logistic regression analysis. The adjusted ORs (AORs) with their 95% CIs were estimated by multiple logistic regression analysis that controlled for tobacco smoking, alcohol consumption and age.
A comparison of clinicopathologic characteristics and CCL4 genotypes revealed no significant differences (Table 3). Similarly, an analysis of CCL4 genotypic frequencies amongst breast cancer subtypes failed to identify any significant differences between patients and controls (Table 4). However, among luminal A and luminal B subtypes, patients carrying the AG genotype at SNP rs10491121 were less likely to develop lymph node metastasis compared with AA genotype carriers (AOR, 0.298; 95% CI: 0.1-0.885) (Table 5). In addition, patients with the rs10491121 AG + GG genotype were at lower risk of developing distant metastasis compared with AA genotype carriers (AOR, 0.106; 95% CI: 0.011-1.038). Moreover, the presence of the TT haplotype at the SNP rs1719153 (AOR 3.316; 95% CI: 1.12-9.815) increased the likelihood of developing pathologic grade (G3+G4) disease (Table 5).
Table 3.
Odds ratios and their confidence intervals for clinical status and CCL4 genotypic frequencies in patients with breast cancer.
| Genotype | Patients N=314 (%) |
OR (95% CI) | |
|---|---|---|---|
| Clinical stage | |||
| Stage I/II | Stage III/IV | ||
| rs10491121 | |||
| AA | 55 (25) | 24 (25.5) | 1.00 (reference) |
| AG+GG | 165 (75) | 70 (74.5) | 0.972 (0.558-1.694) |
| rs1634507 | |||
| GG | 98 (44.5) | 37 (39.4) | 1.00 (reference) |
| GT+TT | 122 (55.5) | 57 (60.6) | 1.237 (0.757-2.024) |
| rs1719153 | |||
| AA | 109 (49.5) | 40 (42.6) | 1.00 (reference) |
| AT+TT | 111 (50.5) | 54 (57.4) | 1.326 (0.815-2.157) |
| Tumor size | |||
| ≤T2 | >T2 | ||
| rs10491121 | |||
| AA | 76 (25.5) | 3 (18.8) | 1.00 (reference) |
| AG+GG | 222 (74.5) | 13 (81.2) | 1.483 (0.412-5.347) |
| rs1634507 | |||
| GG | 130 (43.6) | 5 (31.2) | 1.00 (reference) |
| GT+TT | 168 (56.4) | 11 (68.8) | 1.702 (0.577-5.021) |
| rs1719153 | |||
| AA | 144 (48.3) | 5 (31.2) | 1.00 (reference) |
| AT+TT | 154 (51.7) | 11 (68.8) | 2.057 (0.698-6.065) |
| Lymph node status | |||
| N0+N1 | N2+N3 | ||
| rs10491121 | |||
| AA | 68 (86.1) | 11 (13.9) | 1.00 (reference) |
| AG+GG | 215 (91.5) | 20 (8.5) | 0.575 (0.262-1.260) |
| rs1634507 | |||
| GG | 121 (89.6) | 14 (10.4) | 1.00 (reference) |
| GT+TT | 162 (90.5) | 17 (9.5) | 0.907 (0.403-1.911) |
| rs1719153 | |||
| AA | 136 (91.3) | 13 (8.7) | 1.00 (reference) |
| AT+TT | 147 (89.1) | 18 (10.9) | 1.281 (0.605-2.713) |
| Distant metastasis | |||
| M0 | M1 | ||
| rs10491121 | |||
| AA | 74 (93.7) | 5 (6.3) | 1.00 (reference) |
| AG+GG | 230 (97.9) | 5 (2.1) | 0.322 (0.91-1.142) |
| rs1634507 | |||
| GG | 130 (96.3) | 5 (3.7) | 1.00 (reference) |
| GT+TT | 174 (97.2) | 5 (2.8) | 0.747 (0.212-2.635) |
| rs1719153 | |||
| AA | 144 (96.6) | 5 (3.4) | 1.00 (reference) |
| AT+TT | 160 (97) | 5 (3) | 0.9 (0.255-3.172) |
| Histologic grade | |||
| G1+G2 | G3+G4 | ||
| rs10491121 | |||
| AA | 58 (73.4) | 21 (26.6) | 1.00 (reference) |
| AG+GG | 160 (68.1) | 75 (31.9) | 1.295 (0.732-2.288) |
| rs1634507 | |||
| GG | 99 (73.3) | 36 (26.7) | 1.00 (reference) |
| GT+TT | 119 (66.5) | 60 (33.5) | 1.387 (0.848-2.267) |
| rs1719153 | |||
| AA | 109 (73.2) | 40 (26.8) | 1.00 (reference) |
| AT+TT | 109 (66.1) | 56 (33.9) | 1.4 (0.862-2.274) |
The odds ratios (ORs) with their 95% confidence intervals (CIs) were estimated by logistic regression analysis. The adjusted odds ratios (AORs) with their 95% CIs were estimated by multiple logistic regression analysis that controlled for smoking, consumption and age.
T2 = tumor >20 mm but ≤50 mm in greatest dimension; N0 = lymph node-negative; N1 = cancer has spread to 1-3 lymph node(s); N2 = 4-9 lymph nodes; N3 = ≥10 positive lymph nodes; M0 = noninvasive cancer; M1 = cancer has metastasized to organs or lymph nodes away from the breast; G1 = well differentiated (low grade); G2 = moderately differentiated (intermediate grade); G3 = poorly differentiated (high grade); G4 = undifferentiated (high grade).
Table 4.
Distribution frequencies of CCL4 genotypes in breast cancer subtypes.
| Variable | Control N= 209(%) | Patients N= 220(%) Lumina A + Lumina B |
OR (95% CI) | Variable | Control N= 209(%) | Patients N= 94(%) HER2 overexpression + TNBC |
OR (95% CI) |
|---|---|---|---|---|---|---|---|
| rs10491121 | rs10491121 | ||||||
| AA | 64 (53.8) | 55 (46.2) | 1.00 (reference) | AA | 64 (76.2) | 20 (23.8) | 1.00 (reference) |
| AG | 92 (45.8) | 109 (54.2) | 1.379 (0.875-2.173) | AG | 92 (74.2) | 32 (25.8) | 1.113 (0.585-2.118) |
| GG | 53 (48.6) | 56 (51.4) | 1.23 (0.731-2.069) | GG | 53 (72.6) | 20 (27.4) | 1.208 (0.588-2.478) |
| AG+GG | 145 (46.8) | 165 (53.2) | 1.324 (0.867-2.023) | AG+GG | 145 (73.6) | 52 (26.4) | 1.148 (0.634-2.078) |
| rs1634507 | rs1634507 | ||||||
| GG | 101 (50.8) | 98 (49.2) | 1.00 (reference) | GG | 101 (77.7) | 29 (22.3) | 1.00 (reference) |
| GT | 83 (46.6) | 95 (53.4) | 1.18 (0.787-1.768) | GT | 83 (69.7) | 36 (30.3) | 1.511 (0.855-2.668) |
| TT | 25 (48.1) | 27 (49.8) | 1.113 (0.604-2.050) | TT | 25 (78.1) | 7 (21.9) | 0.975 (0.383-2.482) |
| GT+TT | 108 (47) | 122 (53) | 1.164 (0.796-1.702) | GT+TT | 108 (74.4) | 72 (25.6) | 1.387 (0.805-2.388) |
| rs1719153 | rs1719153 | ||||||
| AA | 101 (48.1) | 109 (51.9) | 1.00 (reference) | AA | 101 (75.9) | 32 (24.1) | 1.00 (reference) |
| AT | 81 (46.3) | 94 (53.7) | 1.075 (0.719-1.607) | AT | 81 (69.8) | 35 (30.2) | 1.364 (0.778-2.391) |
| TT | 27 (61.4) | 17 (38.6) | 0.583 (0.3-1.134) | TT | 27 (84.4) | 5 (15.6) | 0.584 (0.208-1.643) |
| AT+TT | 108 (49.3) | 111 (50.7) | 0.952 (0.652-1.391) | AT+TT | 108 (73) | 40 (27) | 1.169 (0.682-2.002) |
The odds ratios (ORs) with their 95% confidence intervals (CIs) were estimated by logistic regression analysis. The adjusted odds ratios (AORs) with their 95% CIs were estimated by multiple logistic regression analysis that controlled for smoking, consumption and age.
HER2 = human epidermal growth factor receptor 2; TNBC = triple-negative breast cancer.
Table 5.
Odds ratios and their confidence intervals for clinical status and CCL4 genotypic frequencies in breast cancer subtypes.
| Variable | Luminal A + Luminal B N=220 (%) |
HER2 overexpression + TNBC N=94 (%) |
|||||
|---|---|---|---|---|---|---|---|
| Clinical Stage | Clinical Stage | ||||||
| Stage I/II | Stage III/IV | OR (95% CI) | Stage I/II | Stage III/IV | OR (95% CI) | ||
| rs10491121 | |||||||
| AA | 40 (72.7) | 15 (27.3) | 1.00 (reference) | 19 (79.2) | 5 (20.8) | 1.00 (reference) | |
| AG | 93 (85.3) | 16 (14.7) | 0.459 (0.207-1.017) | 27 (62.8) | 16 (37.2) | 2.252 (0.704-7.206) | |
| GG | 40 (71.4) | 16 (28.6) | 1.067 (0.465-2.445) | 23 (85.2) | 4 (14.8) | 0.661 (0.155-2.813) | |
| AG+GG | 133 (80.6) | 32 (19.4) | 0.642 (0.316-1.302) | 50 (71.4) | 20 (28.6) | 1.52 (0.499-4.627) | |
| rs1634507 | |||||||
| GG | 77 (78.6) | 21 (21.4) | 1.00 (reference) | 29 (78.4) | 8 (21.6) | 1.00 (reference) | |
| GT | 74 (77.9) | 21 (22.1) | 1.041 (0.525-2.062) | 28 (65.1) | 15 (34.9) | 1.942 (0.712-5.294) | |
| TT | 22 (81.5) | 5 (18.5) | 0.833 (0.282-2.464) | 12 (85.7) | 2 (14.3) | 0.604 (0.112-3.272) | |
| GT+TT | 96 (78.7) | 26 (21.3) | 0.993 (0.519-1.899) | 40 (70.2) | 17 (29.8) | 1.541 (0.586-4.051) | |
| rs1719153 | |||||||
| AA | 85 (78) | 24 (22) | 1.00 (reference) | 32 (80) | 8 (20) | 1.00 (reference) | |
| AT | 74 (78.7) | 20 (21.3) | 0.957 (0.49-1.871) | 25 (62.5) | 15 (37.5) | 2.4 (0.879-6.556) | |
| TT | 14 (82.4) | 3 (17.6) | 0.759 (0.201-2.86) | 12 (85.7) | 2 (14.3) | 0.667 (0.124-3.597) | |
| AT+TT | 88 (79.3) | 23 (20.7) | 0.926 (0.486-1.764) | 37 (68.5) | 17 (31.5) | 1.838 (0.701-4.821) | |
| Tumor size | Tumor size | ||||||
| ≤T2 | >T2 | OR (95% CI) | ≤T2 | >T2 | OR (95% CI) | ||
| rs10491121 | |||||||
| AA | 53 (96.4) | 2 (3.6) | 1.00 (reference) | 23 (95.8) | 1 (4.2) | 1.00 (reference) | |
| AG | 106 (97.2) | 3 (2.8) | 0.75 (0.122-4.626) | 38 (88.4) | 5 (11.6) | 3.026 (0.332-27.548) | |
| GG | 54 (96.4) | 2 (3.6) | 0.981 (0.133-7.225) | 24 (88.9) | 3 (11.1) | 2.875 (0.279-29.677) | |
| AG+GG | 160 (97) | 5 (3) | 0.828 (0.156-4.395) | 62 (88.6) | 8 (11.4) | 2.968 (0.352-25.054) | |
| rs1634507 | |||||||
| GG | 95 (96.9) | 3 (3.1) | 1.00 (reference) | 35 (94.6) | 2 (5.4) | 1.00 (reference) | |
| GT | 92 (96.8) | 3 (3.2) | 1.033 (0.203-5.248) | 37 (86) | 6 (14) | 2.838 (0.537-15.01) | |
| TT | 26 (96.3) | 1 (3.7) | 1.218 (0.122-12.201) | 13 (92.9) | 1 (7.1) | 1.346 (0.112-16.13) | |
| GT+TT | 118 (96.7) | 4 (3.3) | 1.073 (0.235-4.914) | 50 (87.7) | 7 (12.3) | 2.45 (0.48-12.501) | |
| rs1719153 | |||||||
| AA | 106 (97.2) | 3 (2.8) | 1.00 (reference) | 38 (95) | 2 (5) | 1.00 (reference) | |
| AT | 91 (96.8) | 3 (3.2) | 1.165 (0.229-5.913) | 34 (85) | 6 (15) | 3.353 (0.634-17.738) | |
| TT | 16 (94.1) | 1 (5.9) | 2.208 (0.216-22.548) | 13 (92.9) | 1 (7.1) | 1.462 (0.122-17.482) | |
| AT+TT | 107 (96.4) | 4 (3.6) | 1.321 (0.289-6.044) | 47 (87) | 7 (13) | 2.83 (0.555-14.423) | |
| Lymph node status | Lymph node status | ||||||
| N0+N1 | N2+N3 | OR (95% CI) | N0+N1 | N2+N3 | OR (95% CI) | ||
| rs10491121 | |||||||
| AA | 46 (83.6) | 9 (16.4) | 1.00 (reference) | 22 (91.7) | 2 (8.3) | 1.00 (reference) | |
| AG | 103 (94.5) | 6 (5.5) | 0.298 (0.1-0.885)* | 37 (86) | 6 (14) | 1.784 (0.331-9.619) | |
| GG | 48 (85.7) | 8 (14.3) | 0.852 (0.303-2.397) | 27 (100) | 0 (0) | 0.917 (0.813-1.034) | |
| AG+GG | 151 (91.5) | 14 (8.5) | 0.474 (0.193-1.166) | 64 (91.4) | 6 (8.6) | 1.031 (0.194-5.489) | |
| rs1634507 | |||||||
| GG | 87 (88.8) | 11 (11.2) | 1.00 (reference) | 34 (91.9) | 3 (8.1) | 1.00 (reference) | |
| GT | 87 (91.6) | 8 (8.4) | 0.727 (0.279-1.896) | 38 (88.4) | 5 (11.6) | 1.491 (0.331-6.712) | |
| TT | 23 (85.2) | 4 (14.8) | 1.375 (0.401-4.721) | 14 (100) | 0 (0) | 0.919 (0.835-1.011) | |
| GT+TT | 110 (90.2) | 23 (10.5) | 0.863 (0.363-2.049) | 52 (91.2) | 5 (8.8) | 1.09 (0.244-4.861) | |
| rs1719153 | |||||||
| AA | 99 (90.8) | 10 (9.2) | 1.00 (reference) | 37 (92.5) | 3 (7.5) | 1.00 (reference) | |
| AT | 84 (89.4) | 10 (10.6) | 1.179 (0.468-2.968) | 35 (87.5) | 5 (12.5) | 1.762 (0.392-7.929) | |
| TT | 14 (82.4) | 3 (17.6) | 2.121 (0.52-8.658) | 14 (100) | 0 (0) | 0.925 (0.847-1.01) | |
| AT+TT | 98 (88.3) | 13 (11.7) | 1.313 (0.55-3.136) | 49 (90.7) | 5 (9.3) | 1.259 (0.283-5.605) | |
| Distant metastasis | Distant metastasis | ||||||
| M0 | M1 | OR (95% CI) | M0 | M1 | OR (95% CI) | ||
| rs10491121 | |||||||
| AA | 52 (94.5) | 3 (5.5) | 1.00 (reference) | 22 (91.7) | 2 (8.3) | 1.00 (reference) | |
| AG | 109 (100) | 0 (0) | 0.945 (0.887-1.007)* | 40 (93) | 3 (7) | 0.825 (0.128-5.317) | |
| GG | 55 (98.2) | 1 (1.8) | 0.315 (0.032-3.127) | 26 (96.3) | 1 (3.7) | 0.423 (0.036-4.985) | |
| AG+GG | 164 (99.4) | 1 (0.6) | 0.106 (0.011-1.038)* | 66 (94.3) | 4 (5.7) | 0.667 (0.114-3.893) | |
| rs1634507 | |||||||
| GG | 95 (96.9) | 3 (3.1) | 1.00 (reference) | 35 (94.6) | 2 (5.4) | 1.00 (reference) | |
| GT | 95 (100) | 0 (0) | 0.969 (0.936-1.004) | 39 (90.7) | 4 (9.3) | 1.795 (0.31-10.408) | |
| TT | 26 (96.3) | 1 (3.7) | 1.218 (0.122-12.201) | 14 (100) | 0 (0) | 0.946 (0.876-1.022) | |
| GT+TT | 121 (99.2) | 1 (0.8) | 0.262 (0.027-2.556) | 53 (93) | 4 (7) | 1.321 (0.229-7.602) | |
| rs1719153 | |||||||
| AA | 106 (97.2) | 3 (2.8) | 1.00 (reference) | 38 (95) | 2 (5) | 1.00 (reference) | |
| AT | 94 (100) | 0 (0) | 0.972 (0.942-1.004) | 36 (90) | 4 (10) | 2.111 (0.364-12.24) | |
| TT | 16 (94.1) | 1 (5.9) | 2.208 (0.216-22.548) | 14 (100) | 0 (0) | 0.95 (0.885-1.02) | |
| AT+TT | 110 (99.1) | 1 (0.9) | 0.321 (0.033-3.137) | 50 (92.6) | 4 (7.4) | 1.52 (0.264-8.738) | |
| Histological grade | Histological grade | ||||||
| G1+G2 | G3+G4 | OR (95% CI) | G1+G2 | G3+G4 | OR (95% CI) | ||
| rs10491121 | |||||||
| AA | 45 (81.8) | 10 (18.2) | 1.00 (reference) | 13 (54.2) | 11 (45.8) | 1.00 (reference) | |
| AG | 95 (87.2) | 14 (12.8) | 0.663 (0.274-1.608) | 16 (37.2) | 27 (62.8) | 1.994 (0.724-5.495) | |
| GG | 40 (71.4) | 16 (28.6) | 1.8 (0.734-4.417) | 9 (33.3) | 18 (66.7) | 2.364 (0.761-7.343) | |
| AG+GG | 135 (81.8) | 30 (18.2) | 1 (0.453-2.206) | 25 (35.7) | 45 (64.3) | 2.127 (0.831-5.446) | |
| rs1634507 | |||||||
| GG | 81 (82.7) | 17 (17.3) | 1.00 (reference) | 18 (48.6) | 19 (51.4) | 1.00 (reference) | |
| GT | 81 (85.3) | 14 (14.7) | 0.824 (0.381-1.781) | 16 (37.2) | 27 (62.8) | 1.599 (0.654-3.906) | |
| TT | 18 (66.7) | 9 (33.3) | 2.382 (0.916-6.196) | 4 (28.6) | 10 (71.4) | 2.368 (0.628-8.926) | |
| GT+TT | 99 (81.1) | 23 (18.9) | 1.107 (0.554-2.212) | 20 (35.1) | 37 (64.9) | 1.753 (0.754-4.074) | |
| rs1719153 | |||||||
| AA | 90 (82.6) | 19 (17.4) | 1.00 (reference) | 19 (47.5) | 21 (52.5) | 1.00 (reference) | |
| AT | 80 (85.1) | 14 (14.9) | 0.829 (0.39-1.76) | 13 (32.5) | 27 (67.5) | 1.879 (0.759-4.655) | |
| TT | 10 (58.8) | 7 (41.2) | 3.316 (1.12-9.815)* | 6 (42.9) | 8 (57.1) | 1.206 (0.354-4.115) | |
| AT+TT | 90 (81.1) | 21 (18.9) | 1.105 (0.557-2.195) | 19 (35.2) | 35 (64.8) | 1.667 (0.723-3.841) | |
The odds ratios (ORs) with their 95% confidence intervals (CIs) were estimated by logistic regression analysis. The adjusted odds ratios (AORs) with their 95% CIs were estimated by multiple logistic regression analysis that controlled for smoking, consumption and age. * p<0.05.
HER2 = human epidermal growth factor receptor 2; TNBC = triple-negative breast cancer; T2 = tumor >20 mm but ≤50 mm in greatest dimension; N0 = lymph node-negative; N1 = cancer has spread to 1-3 lymph node(s); N2 = 4-9 lymph nodes; N3 = ≥10 positive lymph nodes; M0 = noninvasive cancer; M1 = cancer has metastasized to organs or lymph nodes away from the breast; G1 = well differentiated (low grade); G2 = moderately differentiated (intermediate grade); G3 = poorly differentiated (high grade); G4 = undifferentiated (high grade).
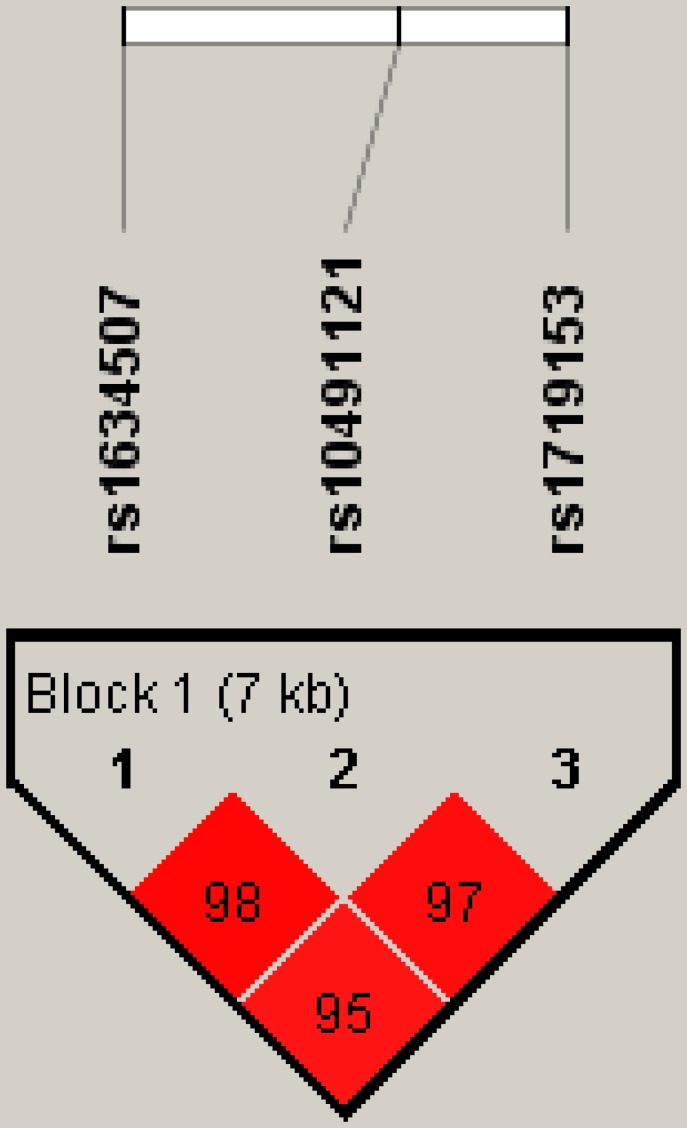
Figure 1 represents the reconstructed linkage disequilibrium plot of the genotyped polymorphisms in our study population. In one haploblock, rs1634507 and rs10491121 displayed 98% linkage disequilibrium. CCL4 SNPs rs1634507 and rs1719153 expressed 95% linkage disequilibrium; rs10491121 and rs1719153 expressed 97% linkage disequilibrium (Fig. 1).
Figure 1.
Linkage disequilibrium patterns of three single nucleotide polymorphisms in the CCL4 gene.
Discussion
CCL4, also known as macrophage inflammatory protein-1β (MIP-1β), belongs to the pro-inflammatory CC subfamily. MIP proteins recruit pro-inflammatory cells and thus play a crucial role in acute and chronic inflammatory responses in various conditions including asthma, granuloma formation, wound healing, arthritis, multiple sclerosis, pneumonia, and psoriasis 16. Accumulating evidences indicated CCL4 expression associated with cancer progression such as oral cancer and hepatocellular carcinoma 12, 17. We have previously suggested that CCL4 gene polymorphisms influence susceptibility to oral cancer and hepatocellular carcinoma and affect their progression 11, 12. We found that CCL4 rs1634507 G/T polymorphism increased a risk in oral-cancer susceptibility, but CCL4 rs10491121 A/G polymorphism decreased a risk in hepatocellular carcinoma. Now, the findings from this study indicate that CCL4 SNPs may serve as candidate biomarkers for susceptibility to breast cancer.
The 5-year relative survival rate for breast cancer has gradually increased since the early 1990s; between 2007 and 2011 it was ~89.2%. As breast cancer prognosis depends upon the disease stage at the time of diagnosis, increasing screening rates and making genetic testing more widely available increase the chances of early diagnosis 24, 25. Our study is the first to examine the expression of SNPs rs1634507, rs10491121 and rs1719153 and their possible association with the development of breast cancer. Our investigation into possible associations between these CCL4 SNPs, clinicopathologic markers, and disease susceptibility failed to find any significant differences between patients and healthy controls. Moreover, CCL4 SNPs did not differ significantly according to breast cancer clinical aspects. Amongst luminal A and luminal B subtypes, patients carrying the AG haplotype at SNP rs10491121 were less likely to develop lymph node metastasis compared with patients with the AA haplotype, while patients carrying the AG + GG haplotype at rs10491121 were less likely to develop distant metastasis. The presence of the AT haplotype at the SNP rs1719153 increased the likelihood of developing pathologic grade (G3+G4) disease.
Linkage disequilibrium is expressed across the human genome. Thus, loci can be used as genetic markers to locate adjacent variants that participate in the detection and treatment of disease. Haplotype analyses clarify genetic contribution to disease susceptibility 26, 27. We observed 98% linkage disequilibrium between rs1634507 and rs10491121, 95% linkage disequilibrium between rs1634507 and rs1719153, and 97% between rs10491121 and rs1719153. These results suggest that these CCL4 haplotypes play an important role in breast cancer development.
This is the first study to demonstrate a correlation between CCL4 polymorphisms and breast cancer risk. CCL4 may prove to be a diagnostic marker and therapeutic target for breast cancer therapy.
Acknowledgments
This work was supported by two grants from China Medical University Hospital (CMU106-S-05) of Taiwan and Medical and Health Science and Technology Project of Zhejiang Province (2012KYB230) of China.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Amir E, Freedman OC, Seruga B, Evans DG. Assessing women at high risk of breast cancer: a review of risk assessment models. J Natl Cancer Inst. 2010;102:680–91. doi: 10.1093/jnci/djq088. [DOI] [PubMed] [Google Scholar]
- 3.Shen CC, Yang AC, Hung JH, Hu LY, Tsai SJ. A nationwide population-based retrospective cohort study of the risk of uterine, ovarian and breast cancer in women with polycystic ovary syndrome. Oncologist. 2015;20:45–9. doi: 10.1634/theoncologist.2014-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng Y, Xu H, Zeng X. Induced abortion and breast cancer: An updated meta-analysis. Medicine (Baltimore) 2018;97:e9613. doi: 10.1097/MD.0000000000009613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang MD, Lin KC, Lu MC, Jeng LB, Hsiao CL, Yueh TC. et al. Contribution of matrix metalloproteinases-1 genotypes to gastric cancer susceptibility in Taiwan. BioMedicine. 2017;7:10. doi: 10.1051/bmdcn/2017070203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen CM, Lai CH, Wu HJ, Wu LT. Genetic characteristic of class 1 integrons in proteus mirabilis isolates from urine samples. BioMedicine. 2017;7:9. doi: 10.1051/bmdcn/2017070202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang WS, Liu LC, Hsiao CL, Su CH, Wang HC, Ji HX. et al. The contributions of the tissue inhibitor of metalloproteinase-1 genotypes to triple negative breast cancer risk. BioMedicine. 2016;6:4. doi: 10.7603/s40681-016-0004-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagy R, Sweet K, Eng C. Highly penetrant hereditary cancer syndromes. Oncogene. 2004;23:6445–70. doi: 10.1038/sj.onc.1207714. [DOI] [PubMed] [Google Scholar]
- 9.Antoniou AC, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL. et al. Breast and ovarian cancer risks to carriers of the BRCA1 5382insC and 185delAG and BRCA2 6174delT mutations: a combined analysis of 22 population based studies. J Med Genet. 2005;42:602–3. doi: 10.1136/jmg.2004.024133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang CQ, Tang CH, Wang Y, Jin L, Wang Q, Li X. et al. FSCN1 gene polymorphisms: biomarkers for the development and progression of breast cancer. Scientific reports. 2017;7:15887. doi: 10.1038/s41598-017-16196-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang B, Chou Y-E, Lien M-Y, Su C-M, Yang S-F, Tang C-H. Impacts of CCL4 gene polymorphisms on hepatocellular carcinoma susceptibility and development. International journal of medical sciences. 2017;14:880. doi: 10.7150/ijms.19620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lien M-Y, Lin C-W, Tsai H-C, Chen Y-T, Tsai M-H, Hua C-H. et al. Impact of CCL4 gene polymorphisms and environmental factors on oral cancer development and clinical characteristics. Oncotarget. 2017;8:31424. doi: 10.18632/oncotarget.15615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pedrosa E, Carretero-Iglesia L, Boada A, Colobran R, Faner R, Pujol-Autonell I. et al. CCL4L polymorphisms and CCL4/CCL4L serum levels are associated with psoriasis severity. Journal of Investigative Dermatology. 2011;131:1830–7. doi: 10.1038/jid.2011.127. [DOI] [PubMed] [Google Scholar]
- 14.Lien MY, Tsai HC, Chang AC, Tsai MH, Hua CH, Wang SW. et al. Chemokine CCL4 Induces Vascular Endothelial Growth Factor C Expression and Lymphangiogenesis by miR-195-3p in Oral Squamous Cell Carcinoma. Frontiers in immunology. 2018;9:412. doi: 10.3389/fimmu.2018.00412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menten P, Wuyts A, Van Damme J. Macrophage inflammatory protein-1. Cytokine & Growth Factor Reviews. 2002;13:455–81. doi: 10.1016/s1359-6101(02)00045-x. [DOI] [PubMed] [Google Scholar]
- 16.Maurer M, Von Stebut E. Macrophage inflammatory protein-1. Int J Biochem Cell Biol. 2004;36:1882–6. doi: 10.1016/j.biocel.2003.10.019. [DOI] [PubMed] [Google Scholar]
- 17.Borzì RM, Mazzetti I, Macor S, Silvestri T, Bassi A, Cattini L. et al. Flow cytometric analysis of intracellular chemokines in chondrocytes in vivo: constitutive expression and enhancement in osteoarthritis and rheumatoid arthritis. FEBS Letters. 1999;455:238–42. doi: 10.1016/s0014-5793(99)00886-8. [DOI] [PubMed] [Google Scholar]
- 18.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19:403–10. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 19.Wang CQ, Tang CH, Wang Y, Jin L, Wang Q, Li X. et al. FSCN1 gene polymorphisms: biomarkers for the development and progression of breast cancer. Sci Rep. 2017;7:15887. doi: 10.1038/s41598-017-16196-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang CQ, Li Y, Huang BF, Zhao YM, Yuan H, Guo D. et al. EGFR conjunct FSCN1 as a Novel Therapeutic Strategy in Triple-Negative Breast Cancer. Scientific reports. 2017;7:15654. doi: 10.1038/s41598-017-15939-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang CQ, Tang CH, Chang HT, Li XN, Zhao YM, Su CM. et al. Fascin-1 as a novel diagnostic marker of triple-negative breast cancer. Cancer Med. 2016;5:1983–8. doi: 10.1002/cam4.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu W, Liu PY, Yang YC, Chen PC, Su CM, Chao CC. et al. Association of HMGB1 Gene Polymorphisms with Lung Cancer Susceptibility and Clinical Aspects. Int J Med Sci. 2017;14:1197–202. doi: 10.7150/ijms.20933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang LH, Wu MH, Chen PC, Su CM, Xu G, Huang CC. et al. Prognostic significance of high-mobility group box protein 1 genetic polymorphisms in rheumatoid arthritis disease outcome. International journal of medical sciences. 2017;14:1382–8. doi: 10.7150/ijms.21773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wood DE. National Comprehensive Cancer Network (NCCN) Clinical Practice Guidelines for Lung Cancer Screening. Thorac Surg Clin. 2015;25:185–97. doi: 10.1016/j.thorsurg.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 25.Moyer VA. Medications to decrease the risk for breast cancer in women: recommendations from the U.S. Preventive Services Task Force recommendation statement. Ann Intern Med. 2013;159:698–708. doi: 10.7326/0003-4819-159-10-201311190-00717. [DOI] [PubMed] [Google Scholar]
- 26.Shifman S, Bronstein M, Sternfeld M, Pisante-Shalom A, Lev-Lehman E, Weizman A. et al. A highly significant association between a COMT haplotype and schizophrenia. Am J Hum Genet. 2002;71:1296–302. doi: 10.1086/344514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang L, Zhang Y, Tang CH, Su CM. RAD52 gene polymorphisms are associated with risk of colorectal cancer in a Chinese Han population. Medicine (Baltimore) 2017;96:e8994. doi: 10.1097/MD.0000000000008994. [DOI] [PMC free article] [PubMed] [Google Scholar]