Abstract
Aim: Long non-coding RNAs (LncRNAs) have been identified to play a crucial role in tumorigenesis and the progression of many types of tumors. However, the clinical significance and biological function of lncRNA nuclear-enriched abundant transcript 1(NEAT1) in human osteosarcoma remains unknown. Here, we investigated the role of NEAT1 in human osteosarcoma cell lines and clinical tumor samples.
Methods: In this study, expression of NEAT1 was analyzed in 19 osteosarcoma tissues and paired adjacent non-tumor tissues by using quantitative real-time PCR. Additionally, knockdown of NEAT1 expression using Lentivirus-mediated siRNA was performed in order to explore the biological function of NEAT1 on osteosarcoma cell proliferation and metastasis through MTT, colony formation assay and transwell assay.
Results: NEAT1 was over-expressed in osteosarcoma tissues compared with adjacent non-tumor tissues. In addition, knockdown of NEAT1 expression could suppress cell proliferation, migration and invasion in vitro.
Conclusion: LncRNA NEAT1 was up-regulated in osteosarcoma tissue, promoting proliferation and metastasis of osteosarcoma cells. These findings indicate the role of this substance, as a growth regulator in osteosarcoma, and thus it may serve as a novel biomarker, and drug target for developing osteosarcoma therapies.
Keywords: NEAT1, Long non-coding RNA, Osteosarcoma
Introduction
Osteosarcoma is the most common malignant bone tumor of mesenchymal origin, usually occurring in children and adolescents with early occurrence commonly leading to pulmonary metastasis and a poor prognosis overall 1. Combination of limb salvage and neoadjuvant chemotherapy as a treatment strategy for this condition has increased five-year disease-free survival rates to 70% 2-4. Unfortunately, the toxic effects of chemotherapy, resistance to treatment, and the spread of metastasis remain some of the main obstacles to treatment of the disease. Therefore, it is necessary to explore the potential molecular mechanisms of this condition to discover novel biomarkers which may lead to the development of diagnosis and treatment of osteosarcoma.
LncRNAs are more than 200 nucleotides in length, containing non-protein-coding capacity transcripts. Recent studies have demonstrated that lncRNAs are emerging as new regulators within cellular processes such as cell proliferation, differentiation, apoptosis, and disease pathogenesis. Further evidence has shown that aberrantly expressed lncRNAs are associated with tumorigenesis in numerous types of cancers such as gastric cancer, cervical cancer 5, lung cancer and hepatocellular carcinoma 6, suggesting that they may perform an important regulatory function in tumorigenesis and cancer progression.
LncRNA nuclear-enriched abundant transcript 1 (NEAT1), which has two isoforms, 3.7 kb NEAT1_1 and 23 kb NEAT1_2, identified from the multiple endocrine neoplasia located at chromosome 11q13.1 7, is an crucial element of nuclear paraspeckles 8. Some studies have showed that aberrant expression of NEAT1 is correlated to a range of cancers diagnoses. For example, Fu et al. found that lncRNA NEAT1 was overexpressed in gastric cancer tissues and cell lines as well as clinical stage and distant metastases 9. Li et al. also showed that NEAT1 was overexpressed in, and correlated with poor prognosis in, endometrial cancer. Further studies suggested that NEAT1 promoted cell proliferation, invasion, migration and inhibited cell apoptosis in endometrial cancer 10. Additionally, Guo found lncRNA NEAT1 expression was increased in hepatocellular carcinoma tissues and related to tumor size and TNM stage 11. These findings suggest that NEAT1 may participate in the progression of various human cancers. To date however, the emerging potential role and mechanism of NEAT1 in osteosarcoma is yet unclear.
In this study, the biological functions of lncRNA NEAT1 in osteosarcoma development have been explored, by examining the expression pattern of NEAT1 in osteosarcoma tissues, followed by correlation analysis with respect to clinicopathological features of the disease. Moreover, in vitro assays were performed to demonstrate the biological functions of NEAT1 in osteosarcoma cell lines.
Materials and methods
Patients and samples
Osteosarcoma tissues and their adjacent non-tumor tissues were obtained from 19 pathologically diagnosed osteosarcoma patients who underwent resection of osteosarcoma within the First Hospital of China Medical University, (Shenyang, China) between July 2010 and July 2014. None of these patients had received preoperative therapy before surgery. Patients at stage IIB/III were included, and patients with any other primary disease were excluded. All tissues were immediately stored at -80°C. Related clinical data was also retrieved from each patient's medical records. This study was approved by the ethics committee of First Hospital of China Medical University, and informed consent was obtained from all of the patients.
Cell culture
The human osteosarcoma cell line U-2 OS was purchased from Shanghai Gefan Biotechnology. All cell lines were cultured in RPMI-1640 (GIBCO) medium supplemented with 10% fetal bovine serum (VAN Biotech) at 37°C in CO2 (5%).
Cell transfection
Osteosarcoma cells U-2 OS were transfected with shRNAs which were synthesized and inserted into the lentivirus core vector (hU6-MCS-CMV-RFP), purchased from GeneChem (Shanghai, China). The sequence of shRNA for NEAT1 was “TGGCTAGCTCAGGGCTTCAG”. Osteosarcoma cells U-2 OS were transfected at a multiplicity of infection of 20, and then the transfected U-2 OS cells were normal cultured 72 h to perform the following assays.
Cell proliferation analysis
Cell viability was measured using an “MTT kit” (Keygen Biotech, Jiangsu China) according to the manufacturer's instruction. A total of 3,000 transfected cells/well were seeded in 96‑well plates and incubated at 37˚C in 5% CO2 for 1 day. Then, 50 ml MTT solution was added into the medium for 4 h incubation at 37˚C in 5% CO2, and then each well was replaced with 150 ml dimethylsulfoxide. Cell proliferation was then monitored at 24, 48, 72 and 96 h. The absorbance value (OD) of each well was then measured at 490 nm.
Cell formation assay
Transfected cells (0.5x103) were placed into each well of 6-well plates and cultured for two weeks, with nutrient media being replaced every 4 days. Colonies were then fixed with 10% formaldehyde for 20mins, followed by staining with 0.1% crystal violet in phosphate buffered saline (PBS) for 5 minutes. The total number of stained colonies was then counted.
Cell migration and invasion assays
Osteosarcoma cell lines were harvested and collected 48 h after transfection with si-NEAT1 or si-NC. In the migration assay, 1x105 cells were seeded on a fibronectin-coated polycarbonate membrane insert within a transwell apparatus (Corning). In the invasion assay, the upper chamber of the apparatus was pre-coated with 24mg/ml Matrigel (Corning), followed by 1x105 transfected cells being placed into the upper chamber. Inserts were then placed into the bottom chamber wells of the apparatus containing 1640-medium with 10% FBS. After the cells were incubated for 24 h, the cells remaining on the upper membrane were removed by scrubbing with a cotton swab. Giemsa-stained cells adhering to the lower surface were counted under a microscope in five predetermined fields.
Quantitative real-time PCR analyses (qRT-PCR)
Total RNA was extracted from tissues or cells using TRIzol reagent (Invitrogen). cDNA synthesis was performed using a PrimeScript RT Reagent Kit with gDNA Eraser (Takara). Real-time PCR analyses were performed with SYBR Premix Ex Taq (Takara). Relative expression was calculated via the comparative cycle threshold method, and results were normalized to the expression of GAPDH. The sequences of specific RNA primers for NEAT1 or GAPDH were as follows: NEAT1 sense, 5′-TGGCTAGCTCAGGGCTTCAG-3′ and reverse, 5′-TCTCCTTGCCAAGCTTCCTTC-3′; GAPDH sense, 5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse, 5′-GAAGATGGTGATGGGATTTC-3′. The qRT-PCR and data collection were performed on Applied Biosystems 7900 Fast Real-Time PCR System (Applied Biosystems). Relative expression was calculated by using the 2-ΔΔCt method.
Statistical analysis
All experiments were performed in triplicate. SPSS 19.0 software and GraphPad Prism 6 were used to analyze data for statistical significance. The two-tailed Student's t test was used for comparisons of two independent groups. A p value of less than 0.05 denoted significance.
Results
LncRNA NEAT1 overexpression in osteosarcoma tissues
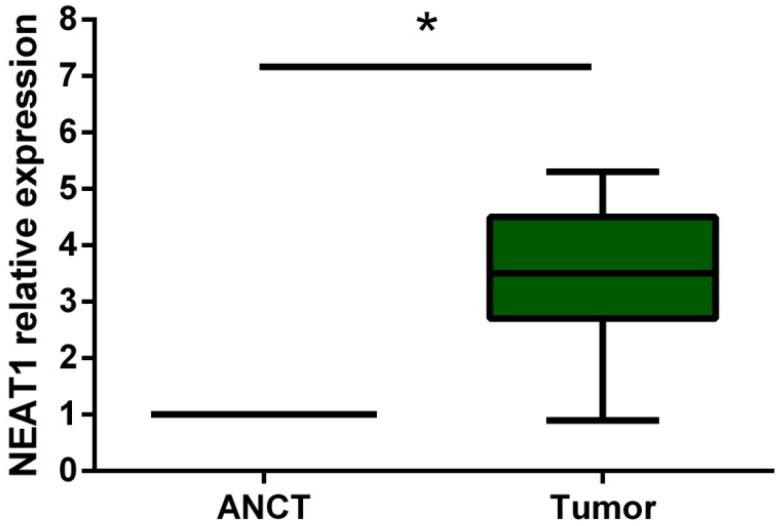
Real-Time PCR was performed to detect the expression level of NEAT1 in 19 osteosarcoma tissues and their adjacent non-tumor tissues (ANCT). Results indicated that the lncRNA NEAT1 expression level was significantly increased compared to that in ANCT (p<0.05) (Figure 1).
Figure 1.
Relative expression of lncRNA NEAT1 in osteosarcoma tissues in comparison with ANCT. LncRNA NEAT1 expression was examined by qRT-PCR and normalized to GAPDH expression. * p<0.05.
Silencing of lncRNA NEAT1 and inhibition of proliferation of osteosarcoma cells
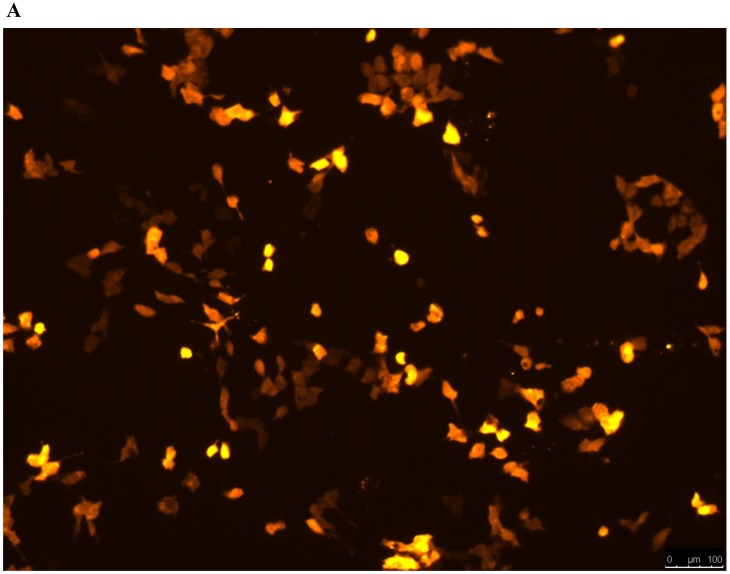
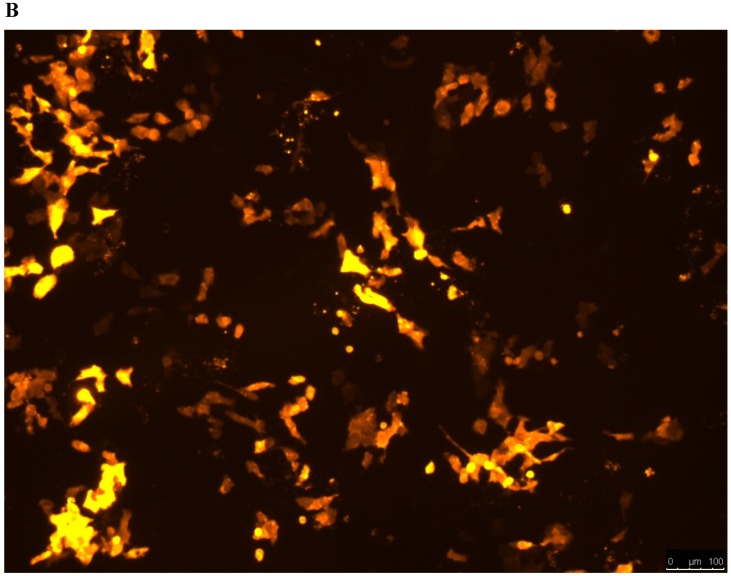
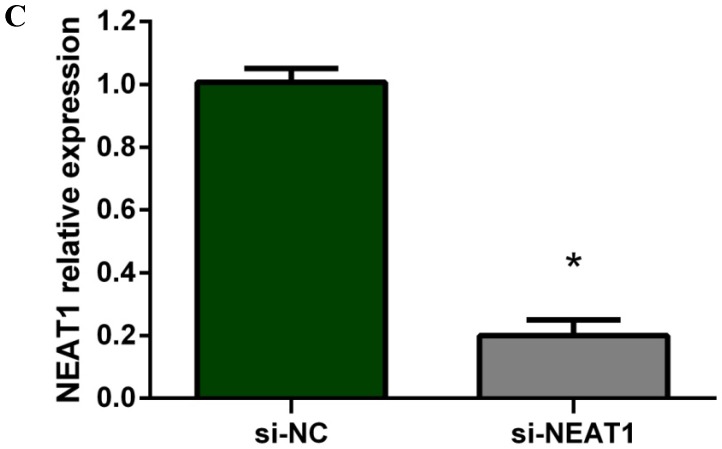
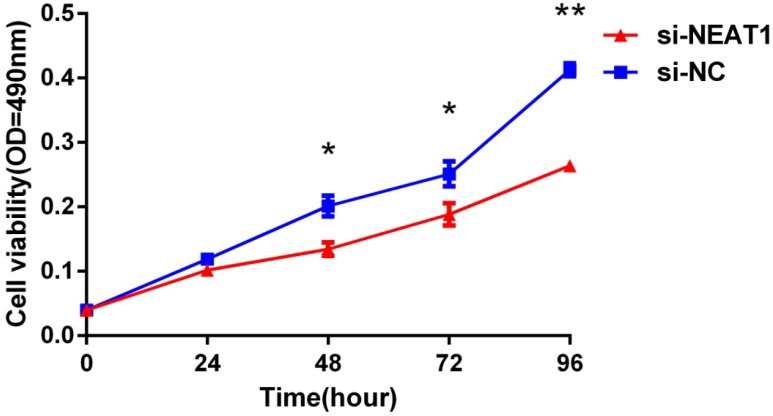
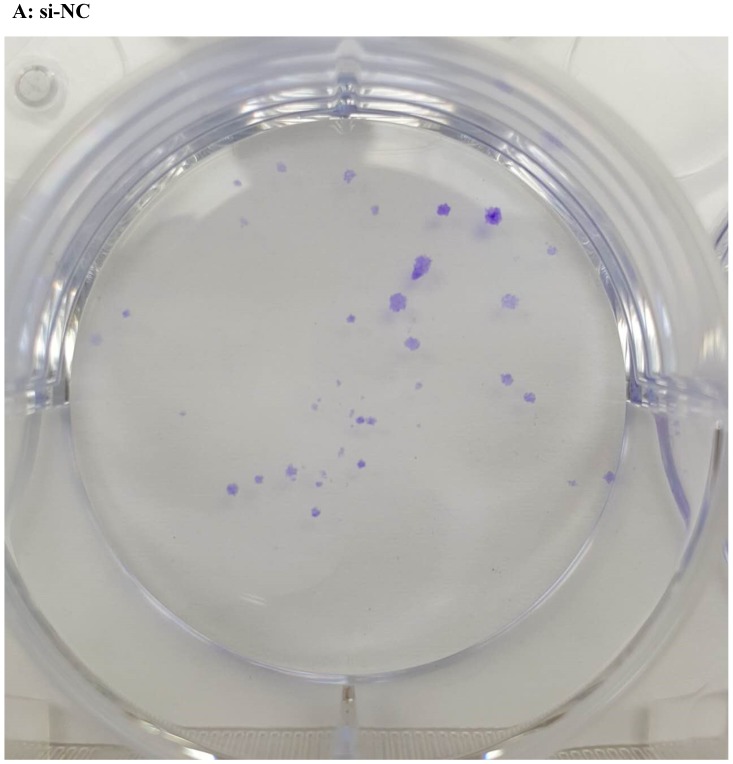
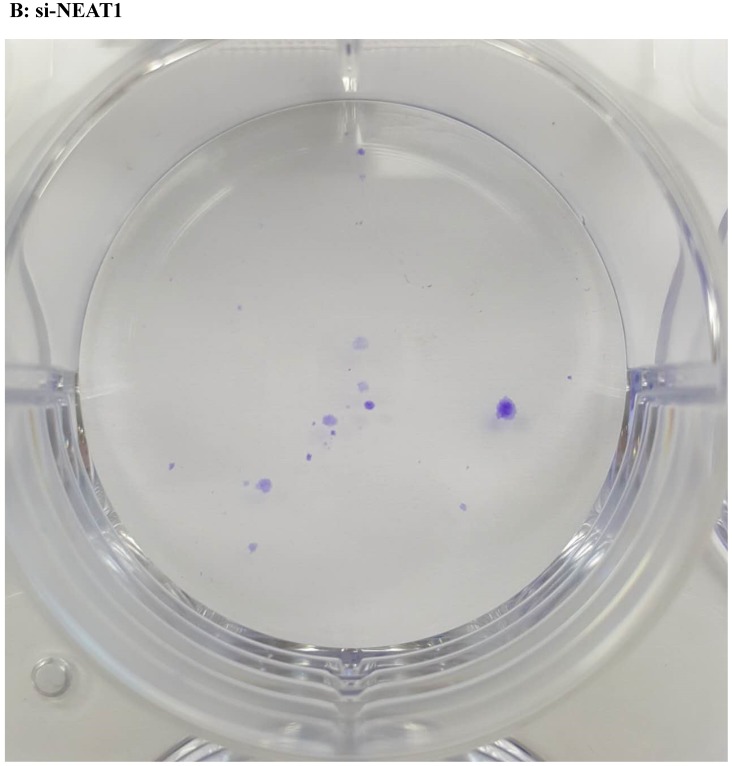
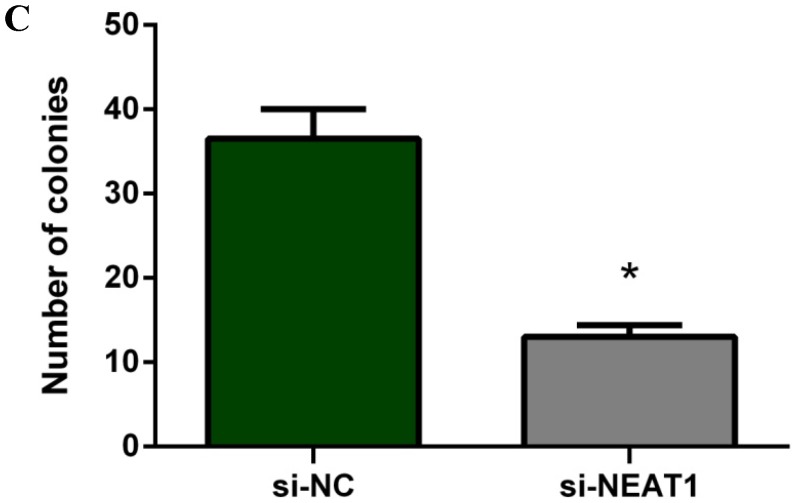
Lentivirus-mediated silencing of NEAT1 expression in osteosarcoma cell lines (U-2 OS) was used to investigate the function of NEAT1 on osteosarcoma cell proliferation. Real-Time PCR results indicated that expression level of NEAT1 in U-2 OS transfected with si-NEAT1 was significantly reduced (p<0.05) (Figure 2A, Figure 2B, Figure 2C). MTT assay results also indicated that silencing of NEAT1 expression significantly suppressed cell proliferation in U-2 OS compared with the negative control group (p<0.05) (Figure 3). Further, colony formation assays suggested that silencing of NEAT1 significantly decreased the number of colonies formed by U-2 OS (p<0.05) (Figure 4A, Figure 4B, Figure 4C).
Figure 2.
A, B: Lentivirus-mediated siRNA decreased the expression of NEAT1 in U-2 OS and relative NEAT1 expression of U-2 OS cell after transfection. The transfection efficiency was determined 72 h after incubation with lentivirus at an MOI of 20. C: Total RNA was extracted 72 h after transfection, the relative NEAT1 expression was determined using quantitative real-time PCR, and NEAT1 expression levels were normalized against GAPDH. * p<0.05.
Figure 3.
Downregulation of NEAT1 inhibited osteosarcoma cell U-2 OS proliferation. MTT assay was performed to determine the proliferation of transfected U-2OS cells. * p<0.05, ** p<0.01.
Figure 4.
A, B, C: Downregulation of NEAT1 inhibited osteosarcoma cell U-2 OS proliferation. Colony formation assay was performed to detect the proliferation of transfected U-2OS cells. * p<0.05.
Reduced lncRNA NEAT1 expression and inhibition of invasion and migration of osteosarcoma cells
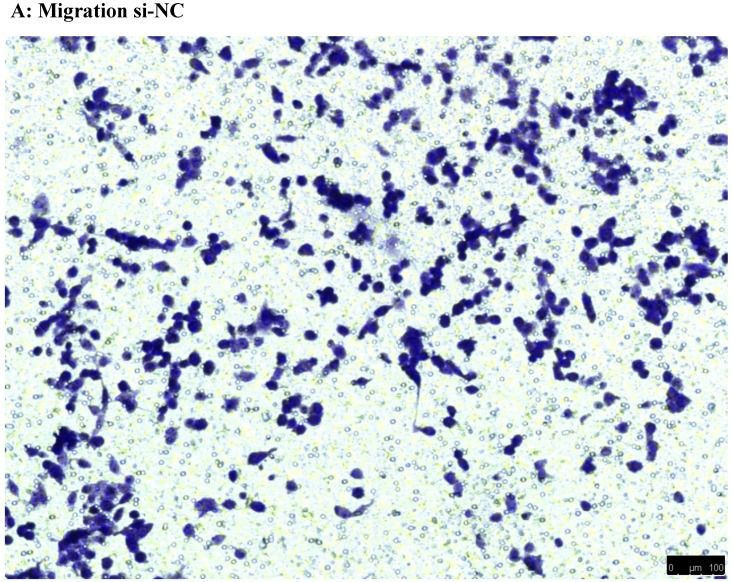
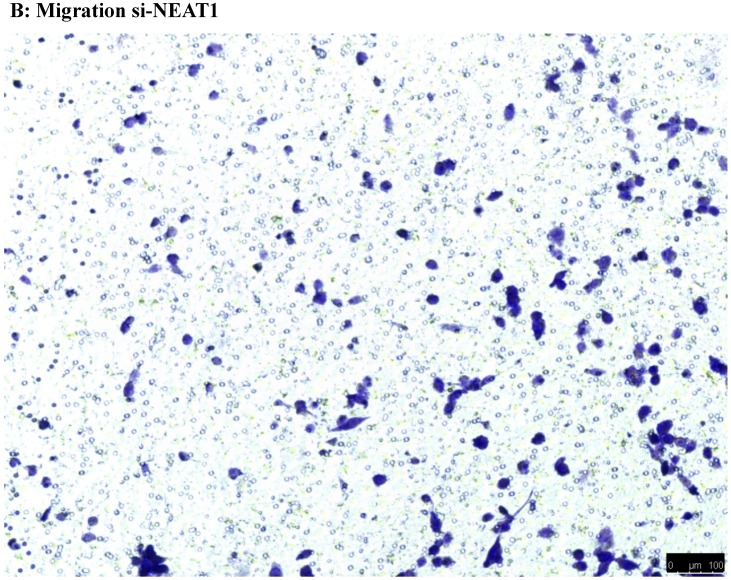
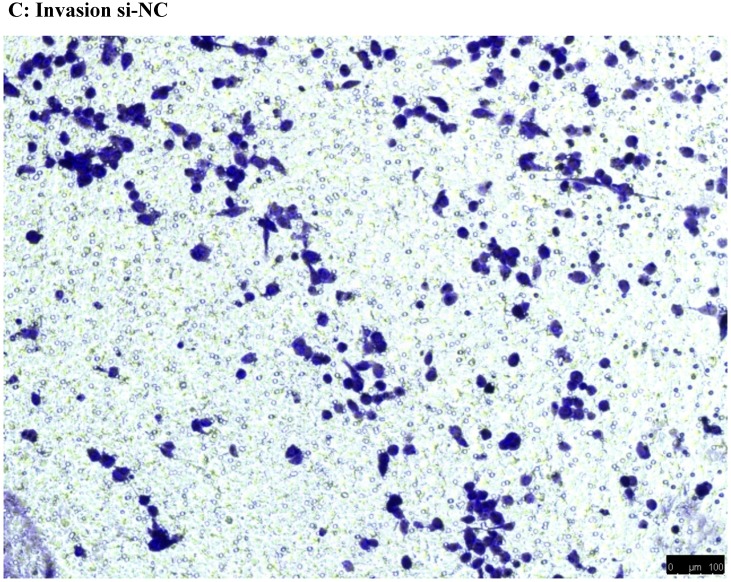
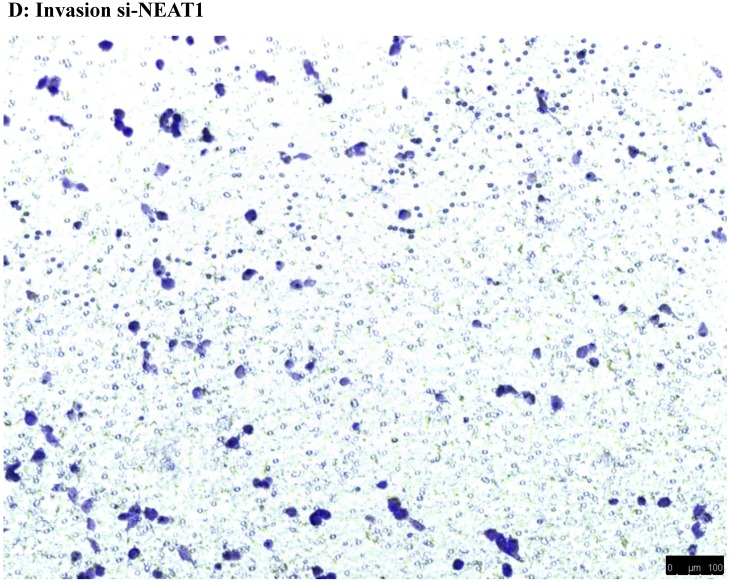
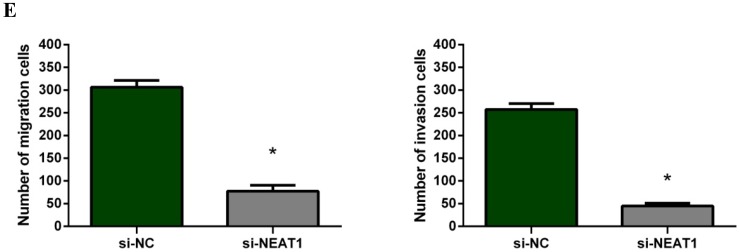
To investigate the effect of NEAT1 silencing on osteosarcoma cell invasion and migration, transwell invasion and migration assays were performed. Results indicated that the migration ability of osteosarcoma cell U-2 OS transfected with si-NEAT1 (Figure 5B) was significantly reduced compared with si-NC group (Figure 5A). Similar results were also seen in invasion assays, with U-2OS cells transfected with si-NEAT1 (Figure 5D) exhibiting a lower ability than that those in the si-NC group (Figure 5C). These results suggest that NEAT1 has a role in the promotion of metastasis of osteosarcoma.
Figure 5.
Downregulation of NEAT1 inhibited osteosarcoma cell U-2 OS migration and invasion. A, B: Transwell migration assays were used to investigate the migration of transfected U-2 OS cells, C, D: Transwell invasion assays were used to investigate the invasion of transfected U-2 OS cells. E: The number of cells passed through Transwell chamber * p<0.05.
Discussion
LncRNAs are more than 200 nucleotides in length, possessing a non-protein-coding capacity, and lacking an open reading frame 12. In recent years, more and more evidence has suggested that long non-coding RNAs play important roles in gene imprinting, cell differentiation, immune response, the occurrence of human disease, tumors, and other biological functions 13-14. Further work has also indicated that an absence of regulation of lncRNAs is involved in tumorigenesis and cancer progression 15. For example, Qiu G et al showed that the lncRNA MATAL1 was overexpressed in colorectal carcinoma, with high levels of MATAL1 expression being associated with the TNM stage of the disease 16. Moreover, research conducted by J Qiu et al suggested that the levels of expression of the lncRNA HOTRIL were significantly high within epithelial ovarian cancer tissues in comparison to normal ovarian tissues, with high expression of HOTAIR being closely associated with the occurrence, development, invasion, metastasis, and prognosis in epithelial ovarian cancer 17. In addition, Matouk IJ et al suggested that the lncRNA HULC was upregulated in hepatocellular carcinoma and hepatic colorectal metastasis, indicating that HULC could likely to become a new marker of hepatic colorectal metastasis 18. Several studies have also indicated that some transcription factors could be involved in the regulation of expression of lncRNAs, such as c-myc, which has been suggested to play a role in the transcription of HOTAIR, and further, that p53 could have a function in the regulation of PVT-1 expression 19-20. All of the studies identified here suggest that aberrant expression of lncRNA is correlated with tumors progression, and thus, they can be used as prognostic indicators. However, the function of NEAT1 in osteosarcoma has, to this point, remained unknown.
LncRNA NEAT1, locus on chromosome 11, is transcribed from the familial tumor syndrome. As a new class of sub-nuclear bodies, paraspeckles contain RNA-binding proteins in mammalian cells. Paraspeckles can regulate gene expression via maintaining mRNAs for rewriting in the nucleus. NEAT1 serves as a crucial component of paraspeckles 21-23. Indeed, aberrant expression of NEAT1 has been reported play an important function within various human cancers 24. With regards to existing literature, Chakravarty D et al found that NEAT1 was upregulated in prostate cancer; and further studies have shown that NEAT1 has induced oncogenic growth by altering the epigenetic landscape of target gene promoters to favour transcription 25. An additional study postulated that the expression of NEAT1 is cooperatively controlled by HuR and miR-124-3p, and has responsibilities within the regulation of ovarian carcinogenesis 26. In lung cancer, a strong correlation of high NEAT1 expression in tumors with poor survival was confirmed in NSCLC tissues, indicating that NEAT1 expression levels could be a helpful prognostic biomarker for high risk of NSCLC patients 27. Unfortunately, the effects of NEAT1 in osteosarcoma were not tracked in the same study.
NEAT1 has two isoforms, 3.7 kb NEAT1_1 and 23 kb NEAT1_2. These two isoforms of NEAT1 share the same transcriptional start site and the same RNA polymerase II promoter. They just differ in the location of their 3' ends. The NEAT1_2 has a tRNA-like structure at its 3' end that is processed by RNaseP cleavage rather than a poly-A tail 28. The primers for NEAT1 and shRNA we designed had included these two isoforms and mRNA measurements had covered both isoforms as well.
In this work, we have identified that the expression level of NEAT1 was frequently overexpressed within osteosarcoma tissues, compared with adjacent non-tumor tissues, suggesting that NEAT1 may also play a significant role in osteosarcoma development and progression. To further explore the underlying mechanism of lncRNA NEAT1 in osteosarcoma progression, we silenced NEAT1 expression in the osteosarcoma cell U-2 OS by way of Lentivirus-mediated RNA interference, followed by real-time PCR in order to detect the expression of NEAT1. We observed the fluorescence carefully under the inverted microscope and using RT-PCR to ensure the infection efficiency. MTT assays, colony formation assays and transwell assays were all employed to detect proliferation, migration and invasion capacity of osteosarcoma U-2 OS cells, with these assays indicating that silencing of NEAT1 significantly suppressed the proliferation, migration and invasion of osteosarcoma cells in vitro. These data further support the importance of NEAT1 in cellular biology and oncogenesis of osteosarcoma cells. However, further study is required in order to identify the exact pathway through which NEAT1 influences osteosarcoma and how this pathway works.
In summary, we have demonstrated that NEAT1 is commonly upregulated in human osteosarcoma, allowing this to be potentially harnessed as a prognostic factor for this condition and further, the silencing of NEAT1 may act as a potentially significant target for the inhibition of osteosarcoma cell proliferation and metastasis in vivo. These results underscore the importance of NEAT1 in osteosarcoma development.
Acknowledgments
This paper was supported by the natural science foundation of science and technology department of Liaoning province.
References
- 1.Rasalkar DD, Chu WC, Lee V, Paunipagar BK, Cheng FW, Li CK. Pulmonary metastases in children with osteosarcoma: characteristics and impact on patient survival. Pediatr Radiol. 2011;41:227–36. doi: 10.1007/s00247-010-1809-1. [DOI] [PubMed] [Google Scholar]
- 2.Choeyprasert W, Natesirinilkul R, Charoenkwan P, Sittipreechacharn S. Carboplatin and doxorubicin in treatment of pediatric osteosarcoma: a 9-year single institute experience in the Northern Region of Thailand. Asian Pac J Cancer Prev. 2013;14:1101–6. doi: 10.7314/apjcp.2013.14.2.1101. [DOI] [PubMed] [Google Scholar]
- 3.Bacci G, Balladelli A, Palmerini E, Alberghini M, Pollastri P, Galletti S. et al. Neoadjuvant chemotherapy for osteosarcoma of the extremities in preadolescent patients: the Rizzoli Institute experience. J Pediatr Hematol Oncol. 2008;30:908–12. doi: 10.1097/MPH.0b013e31817e4aee. [DOI] [PubMed] [Google Scholar]
- 4.Bacci G, Ferrari S, Mercuri M, Longhi A, Fabbri N, Galletti S. et al. Neoadjuvant chemotherapy for osteosarcoma of the extremities in patients aged 41-60 years: outcome in 34 cases treated with adriamycin, cisplatinum and ifosfamide between 1984 and 1999. Acta Orthop. 2007;78:377–84. doi: 10.1080/17453670710013960. [DOI] [PubMed] [Google Scholar]
- 5.Lee M, Kim HJ, Kim SW, Park SA, Chun KH, Cho NH. et al. The long non-coding RNA HOTAIR increases tumour growth and invasion in cervical cancer by targeting the Notch pathway. Oncotarget. 2016;7:44558–71. doi: 10.18632/oncotarget.10065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang M, Wang W, Li T, Yu X, Zhu Y, Ding F. et al. Long noncoding RNA SNHG1 predicts a poor prognosis and promotes hepatocellular carcinoma tumorigenesis. Biomed Pharmacother. 2016;80:73–9. doi: 10.1016/j.biopha.2016.02.036. [DOI] [PubMed] [Google Scholar]
- 7.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A. et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–26. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci U S A. 2009;106:2525–30. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu JW, Kong Y, Sun X. Long noncoding RNA NEAT1 is an unfavorable prognostic factor and regulates migration and invasion in gastric cancer. J Cancer Res Clin Oncol. 2016;142:1571–9. doi: 10.1007/s00432-016-2152-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Z, Wei D, Yang C, Sun H, Lu T, Kuang D. Overexpression of long noncoding RNA, NEAT1 promotes cell proliferation, invasion and migration in endometrial endometrioid adenocarcinoma. Biomed Pharmacother. 2016;84:244–51. doi: 10.1016/j.biopha.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 11.Guo S, Chen W, Luo Y, Ren F, Zhong T, Rong M. et al. Clinical implication of long non-coding RNA NEAT1 expression in hepatocellular carcinoma patients. Int J Clin Exp Pathol. 2015;8:5395–402. [PMC free article] [PubMed] [Google Scholar]
- 12.Zaratiegui M, Irvine DV, Martienssen RA. Noncoding RNAs and gene silencing. Cell. 2007;128:763–76. doi: 10.1016/j.cell.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 13.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–46. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–74. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 15.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiu G, Zhang XB, Zhang SQ, Liu PL, Wu W, Zhang JY. et al. Dysregulation of MALAT1 and miR-619-5p as a prognostic indicator in advanced colorectal carcinoma. Oncol Lett. 2016;12:5036–42. doi: 10.3892/ol.2016.5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu JJ, Wang Y, Ding JX, Jin HY, Yang G, Hua KQ. The long non-coding RNA HOTAIR promotes the proliferation of serous ovarian cancer cells through the regulation of cell cycle arrest and apoptosis. Exp Cell Res. 2015;333:238–48. doi: 10.1016/j.yexcr.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 18.Matouk IJ, Abbasi I, Hochberg A, Galun E, Dweik H, Akkawi M. Highly upregulated in liver cancer noncoding RNA is overexpressed in hepatic colorectal metastasis. Eur J Gastroenterol Hepatol. 2009;21:688–92. doi: 10.1097/meg.0b013e328306a3a2. [DOI] [PubMed] [Google Scholar]
- 19.Ma MZ, Li CX, Zhang Y, Weng MZ, Zhang MD, Qin YY. et al. Long non-coding RNA HOTAIR, a c-Myc activated driver of malignancy, negatively regulates miRNA-130a in gallbladder cancer. Mol Cancer. 2014;13:156. doi: 10.1186/1476-4598-13-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barsotti AM, Beckerman R, Laptenko O, Huppi K, Caplen NJ, Prives C. p53-Dependent induction of PVT1 and miR-1204. J Biol Chem. 2012;287:2509–19. doi: 10.1074/jbc.M111.322875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choudhry H, Albukhari A, Morotti M, Haider S, Moralli D, Smythies J. et al. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2alpha dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene. 2015;34:4546. doi: 10.1038/onc.2014.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35:467–78. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeng C, Xu Y, Xu L, Yu X, Cheng J, Yang L. et al. Inhibition of long non-coding RNA NEAT1 impairs myeloid differentiation in acute promyelocytic leukemia cells. BMC Cancer. 2014;14:693. doi: 10.1186/1471-2407-14-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chakravarty D, Sboner A, Nair SS, Giannopoulou E, Li R, Hennig S. et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun. 2014;5:5383. doi: 10.1038/ncomms6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chai Y, Liu J, Zhang Z, Liu L. HuR-regulated lncRNA NEAT1 stability in tumorigenesis and progression of ovarian cancer. Cancer Med. 2016;5:1588–98. doi: 10.1002/cam4.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun C, Li S, Zhang F, Xi Y, Wang L, Bi Y. et al. Long non-coding RNA NEAT1 promotes non-small cell lung cancer progression through regulation of miR-377-3p-E2F3 pathway. Oncotarget. 2016;7:51784–814. doi: 10.18632/oncotarget.10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–59. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]