Abstract
Background
The associations of the 5,10-methylenetetrahydrofolate reductase gene (MTHFR) rs1801133 polymorphism with coronary artery disease (CAD) and plasma lipid levels have been widely investigated, but the results were inconsistent and inconclusive. This meta-analysis aimed to clarify the relationships of the rs1801133 polymorphism with CAD and plasma lipid levels.
Methods
By searching in PubMed, Google Scholar, Web of Science, Cochrane Library, Wanfang, VIP and CNKI databases, 123 studies (87,020 subjects) and 65 studies (85,554 subjects) were identified for the CAD association analysis and the lipid association analysis, respectively. Odds ratio (OR) and standardized mean difference (SMD) were used to determine the effects of the rs1801133 polymorphism on CAD risk and lipid levels, respectively.
Results
The variant T allele of the rs1801133 polymorphism was associated with increased risk of CAD under allelic model [OR = 1.11, 95% confidence interval (CI) = 1.06–1.17, P < 0.01], additive model (OR = 1.25, 95% CI = 1.14–1.37, P < 0.01), dominant model (OR = 1.11, 95% CI = 1.04–1.17, P < 0.01), and recessive model (OR = 1.22, 95% CI = 1.12–1.32, P < 0.01). The T carriers had higher levels of total cholesterol (TC) (SMD = 0.04, 95% CI = 0.01–0.07, P = 0.02) and low-density lipoprotein cholesterol (LDL-C) (SMD = 0.07, 95% CI = 0.01–0.12, P = 0.01) than the non-carriers.
Conclusions
The meta-analysis suggested that the T allele of the rs1801133 polymorphism is a risk factor for CAD, which is possibly and partly mediated by abnormal lipid levels.
Electronic supplementary material
The online version of this article (10.1186/s12944-018-0837-y) contains supplementary material, which is available to authorized users.
Keywords: 5,10-Methylenetetrahydrofolate reductase; Polymorphism; rs1801133; Coronary artery disease; Lipid
Background
Coronary artery disease (CAD) is currently the leading cause of death in developed countries, and in some developing countries like China [1]. CAD is a multifactorial disease and a number of risk factors have been identified in the past few decades. Genetic polymorphism and dyslipidemia are the two most important risk factors for CAD [2, 3]. Genetic polymorphism is a term used to describe multiple forms of a single gene. Dyslipidemia is a state of abnormal amounts of lipids (e.g. triglycerides, cholesterol and/or phospholipids) in the blood, and is characterized by increased levels of triglycerides (TG), total cholesterol (TC) and low-density lipoprotein cholesterol (LDL-C), and/or decreased level of high-density lipoprotein cholesterol (HDL-C). Intensive efforts have been made in the scientific community to investigate the associations of the genetic polymorphisms in some specific genes with CAD risk and plasma lipid levels, but the results were inconsistent and inconclusive. It is difficult to identify the CAD- or dyslipidemia-related genetic polymorphisms successfully due to various reasons such as small sample sizes and ethnic differences.
5,10-methylenetetrahydrofolate reductase (MTHFR) is a rate-limiting enzyme in the one-carbon metabolism pathway and plays a key role in one-carbon metabolism by irreversibly catalyzing the conversion of 5,10-Methylenetetrahydrofolate (5,10-MTHF) to 5-Methyltetrahydrofolate (5-MTHF). 5-MTHF is a direct one-carbon donor (methyl group) for many substrates such as DNA [4], RNA [5] and proteins [6]. More importantly, 5-MTHF is the only one-carbon donor for the remethylation of homocysteine which is produced in methionine cycle. In methionine cycle, methionine first reacts with adenosine triphosphate to form S-adenosylmethionine (SAM) under the catalysis of methionine adenosyltransferase. The methyl group in SAM is activated, and SAM is called activated methionine. The activated methyl group of SAM can be transferred to target substrates such as DNA under the catalysis of methyltransferase, and SAM itself is converted into S-adenosine homocysteine after demethylation. Homocysteine is produced after the removal of adenosine from S-adenosine homocysteine under the catalysis of S-adenosylhomocysteine hydrolase. In the last step, homocysteine accepts the methyl group from 5-MTHF and methionine is formed again.
Homocysteine a potential risk factor for CAD and MTHFR has been reported to be associated with CAD risk and abnormal lipid levels [7–9]. Mikael et al. [8] reported that MTHFR(+/−) mice had significantly higher levels of plasma TG and more lipid deposition in aortic sinus compared with MTHFR (+/+) mice. In another study, Christensen et al. [9] found that high folic acid consumption led to pseudo-MTHFR deficiency in mice, and this deficiency resulted in altered lipid metabolism and liver injury.
The rs1801133 polymorphism (also known as the 677C > T polymorphism) is located in exon 4 of the MTHFR gene and formed by a transition from cytosine (C) to thymine (T). The 222nd genetic code of the MTHFR gene is changed accordingly from GCC to GTC, resulting in the replacement of alanine (Ala) by valine (Val) in the MTHFR polypeptide. A large number of studies have investigated the associations of the rs1801133 polymorphism with CAD and lipid levels. In some of these studies, the T allele of the rs1801133 polymorphism was reported to be associated with an increased risk of CAD [10–12] and elevated levels of TG [13, 14], TC [13–15] and LDL-C [13–16], and reduced levels of HDL-C [13, 17]. However, the results obtained from other studies did not support these findings [18–22]. Hence, a meta-analysis is required to clarify the relationships of the rs1801133 polymorphism with CAD and lipid levels.
Although two meta-analyses [23, 24] have addressed the issue of the association between the rs1801133 polymorphism and CAD in 2002 and 2005, respectively, their sample sizes were relatively small, and blood lipid variables were not considered in the analyses. In this study, a systematic review and updated meta-analysis was performed based on previous publications to investigate the associations of the rs1801133 polymorphism with CAD and lipid levels. The results of this meta-analysis can provide an opportunity to unveil the interrelationships among the rs1801133 polymorphism, dyslipidemia and susceptibility to CAD.
Result
Characteristics of the included studies
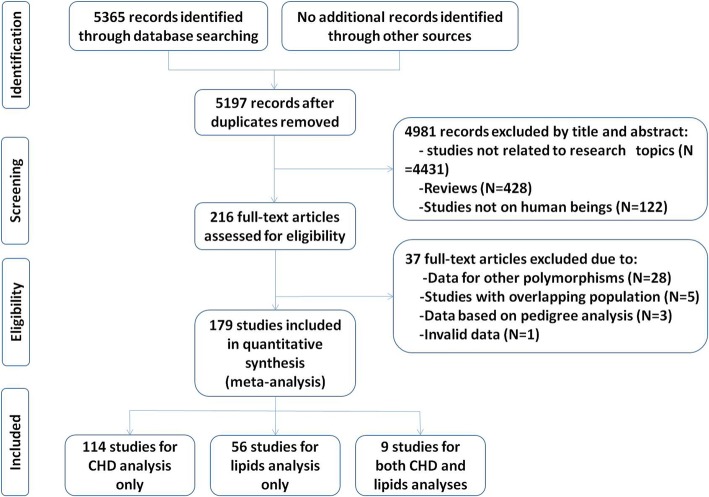
Initial search of the databases yielded 5197 articles. Four thousand nine hundred and eighty-one studies were excluded according to the titles and abstracts. Then full-text articles were retrieved and assessed on the basis of inclusion criteria. Thirty-seven studies were ineligible for the following reasons: 28 studies presented data for other polymorphisms; 5 studies had subjects overlapping with other publications; 3 studies were based on pedigree analysis; 1 study presented invalid data. In the end, 179 studies were selected for this meta-analysis (Fig. 1). One hundred twenty-three studies (87,020 subjects) of them were included in the CAD association analysis, and 65 studies (85,554 subjects) were included in the lipid association analysis. The references for the studies included in the present meta-analysis are listed in Additional file 1.
Fig. 1.
Flow diagram of the study selection process
The characteristics of the studies included in the CAD association analysis are summarized in Additional file 2: Table S1. Seventy-two studies, 35 studies, 5 studies and 11 studies involved in Caucasians, Asians, Africans and the subjects of other ethnic origins, respectively. The characteristics of the studies included in the lipid association analysis are summarized in Additional file 2: Table S2. The plasma lipid levels according to the genotypes of the rs1801133 polymorphism are presented in Additional file 2: Table S3. Twenty-four studies, 25 studies, 3 studies and 13 studies involved in Caucasians, Asians, Africans and the subjects of other ethnic origins, respectively. Eleven studies, 5 studies, 6 studies and 33 studies involved in CAD, diabetes, hypertension and healthy subjects, respectively. Fifty-three studies, 61 studies, 49 studies and 58 studies presented the data for TG, TC, LDL-C and HDL-C, respectively.
Association of the r1801133 polymorphism with CAD
The variant T allele of the rs1801133 polymorphism was associated with increased risk of CAD under allelic model [odds ratio (OR) = 1.11, 95% confidence interval (CI) = 1.06–1.17, P < 0.01], additive model (OR = 1.25, 95% CI = 1.14–1.37, P < 0.01), dominant model (OR = 1.11, 95% CI = 1.04–1.17, P < 0.01) and recessive model (OR = 1.22, 95% CI = 1.12–1.32, P < 0.01) (Table 1). When the analyses were limited to the studies in Hardy-Weinberg equilibrium (HWE), the association between the rs1801133 polymorphism and CAD under allelic model (OR = 1.11, 95% CI = 1.06–1.16, P < 0.01), additive model (OR = 1.23, 95% CI = 1.12–1.35, P < 0.01), dominant model (OR = 1.11, 95% CI = 1.05–1.17, P < 0.01) and recessive model (OR = 1.20, 95% CI = 1.11–1.30, P < 0.01) was also significant (Table 1). Then the subgroup analyses stratified by the ethnicity of the subjects were performed, and the results showed that the rs1801133 polymorphism was strongly associated with CAD in Asians, but not in Africans or the subjects of other ethnic origins. In Caucasians, the association between the rs1801133 polymorphism and CAD was ambiguous, i.e. the rs1801133 polymorphism was marginally significantly associated with CAD under additive model (OR = 1.08, 95% CI = 1.00–1.17, P = 0.05) and recessive model (OR = 1.09, 95% CI = 1.01–1.17, P = 0.02), but not under allelic model and dominant model (Table 1).
Table 1.
Meta-analysis between the MTHFR rs1801133 polymorphism and CAD risk
| Model | Group or subgroup | OR | 95% CI | P heterogeneity | P OR |
|---|---|---|---|---|---|
| Allelic model (T vs C) |
Total | 1.11 | 1.06–1.17 | < 0.01 | < 0.01 |
| Studies in HWE | 1.11 | 1.06–1.16 | < 0.01 | < 0.01 | |
| Male | 1.04 | 0.90–1.20 | < 0.01 | 0.59 | |
| Female | 0.98 | 0.88–1.08 | 0.40 | 0.63 | |
| Caucasian | 1.03 | 0.99–1.07 | < 0.01 | 0.10 | |
| Asian | 1.24 | 1.11–1.39 | < 0.01 | < 0.01 | |
| African | 1.30 | 0.84–2.00 | < 0.01 | 0.24 | |
| Other ethnicity | 1.30 | 0.96–1.76 | < 0.01 | 0.09 | |
| Additive model (TT vs CC) |
Total | 1.25 | 1.14–1.37 | < 0.01 | < 0.01 |
| Studies in HWE | 1.23 | 1.12–1.35 | < 0.01 | < 0.01 | |
| Male | 1.22 | 0.96–1.55 | < 0.01 | 0.10 | |
| Female | 0.93 | 0.75–1.16 | 0.65 | 0.53 | |
| Caucasian | 1.08 | 1.00–1.17 | 0.03 | 0.05 | |
| Asian | 1.46 | 1.20–1.76 | < 0.01 | < 0.01 | |
| African | 1.96 | 0.84–4.60 | < 0.01 | 0.12 | |
| Other ethnicity | 1.83 | 0.93–3.59 | < 0.01 | 0.08 | |
| Dominant model (TT + CT vs CC) |
Total | 1.11 | 1.04–1.17 | < 0.01 | < 0.01 |
| Studies in HWE | 1.11 | 1.05–1.17 | < 0.01 | < 0.01 | |
| Male | 1.00 | 0.80–1.25 | < 0.01 | 0.98 | |
| Female | 0.98 | 0.83–1.16 | 0.20 | 0.84 | |
| Caucasian | 1.02 | 0.97–1.06 | 0.05 | 0.45 | |
| Asian | 1.27 | 1.08–1.50 | < 0.01 | < 0.01 | |
| African | 1.22 | 0.78–1.92 | < 0.01 | 0.39 | |
| Other ethnicity | 1.30 | 0.95–1.77 | < 0.01 | 0.10 | |
| Recessive model (TT vs CT + CC) |
Total | 1.22 | 1.12–1.32 | < 0.01 | < 0.01 |
| Studies in HWE | 1.20 | 1.11–1.30 | < 0.01 | < 0.01 | |
| Female | 0.96 | 0.78–1.17 | 0.76 | 0.67 | |
| Male | 1.21 | 0.96–1.51 | < 0.01 | 0.10 | |
| Caucasian | 1.09 | 1.01–1.17 | 0.06 | 0.02 | |
| Asian | 1.36 | 1.14–1.62 | < 0.01 | < 0.01 | |
| African | 1.90 | 0.84–4.30 | < 0.01 | 0.13 | |
| Other ethnicity | 1.71 | 0.94–3.12 | < 0.01 | 0.08 |
MTHFR 5,10-methylenetetrahydrofolate reductase gene, CAD coronary artery disease, OR odds ratio, 95% CI 95% confidence interval, HWE Hardy-Weinberg equilibrium
Associations of the r1801133 polymorphism with plasma lipid levels
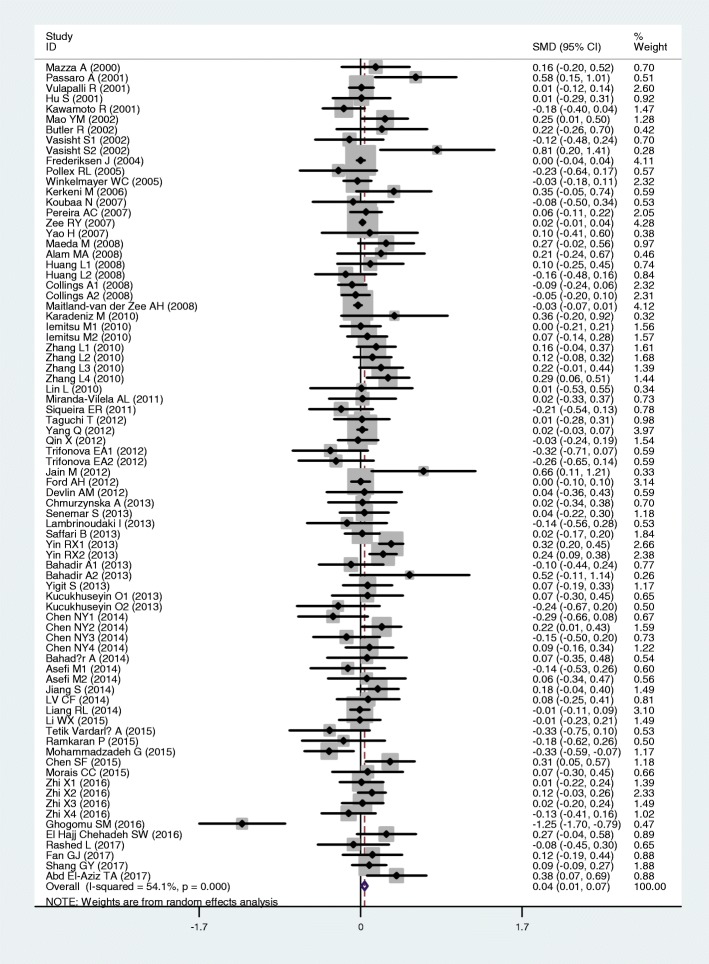
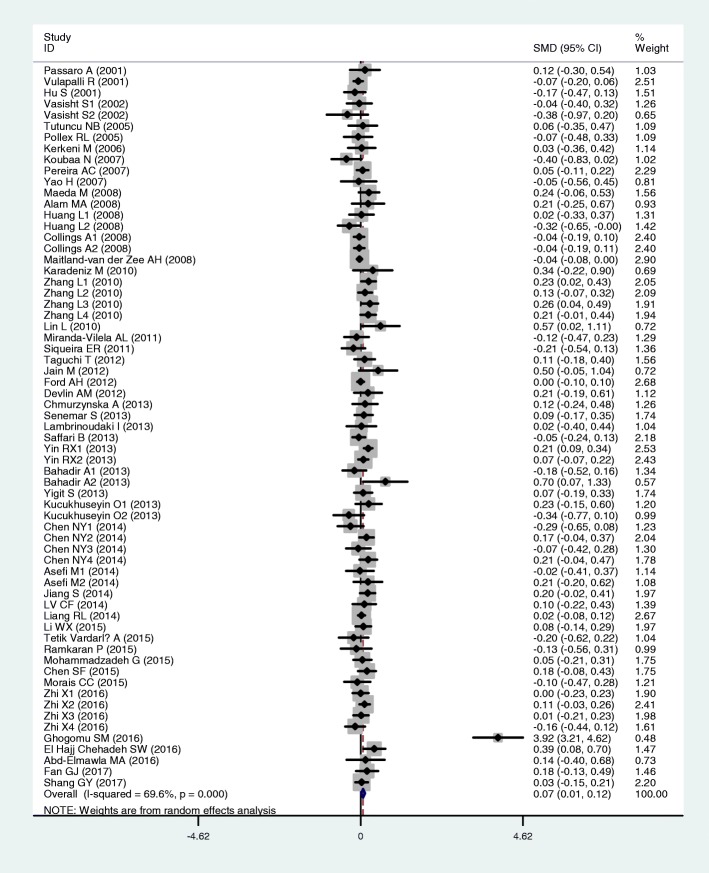
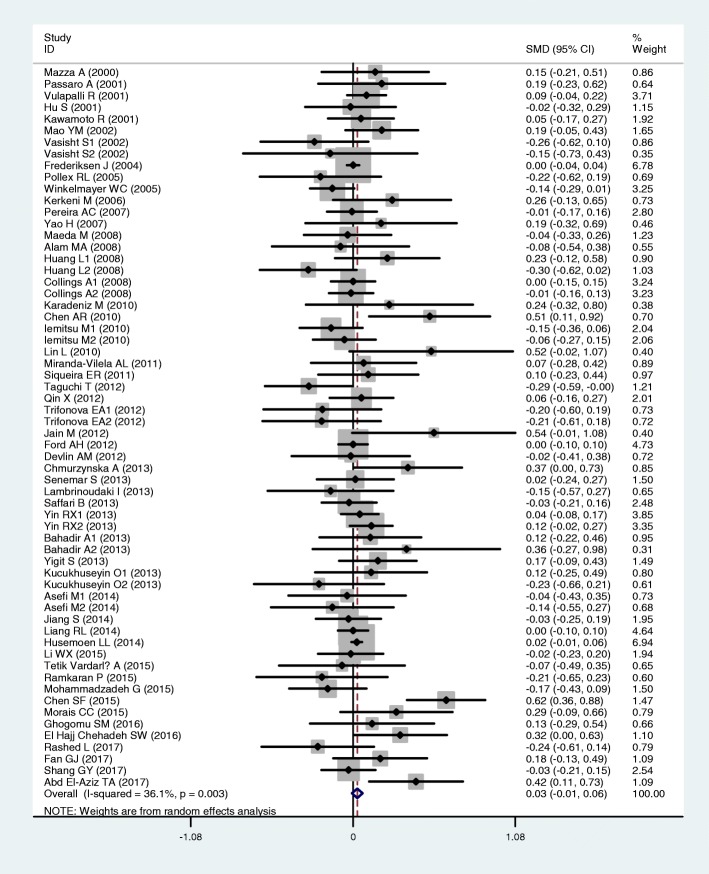
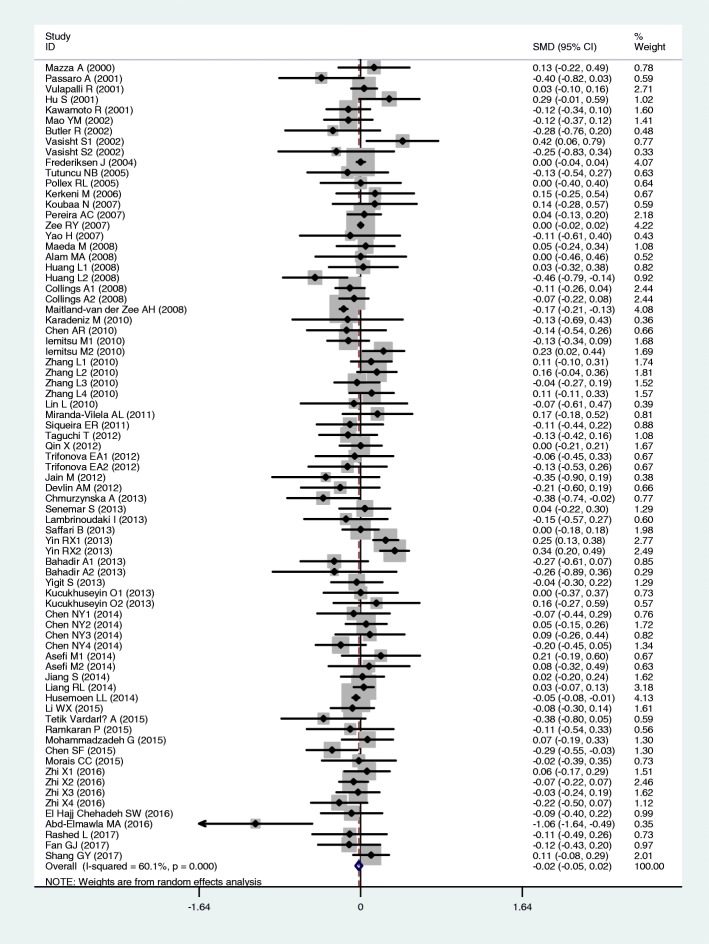
The outcomes of the analyses on all comparisons showed that the T allele carriers had higher levels of TC [standardized mean difference (SMD) = 0.04, 95% CI = 0.01–0.07, P = 0.02] and LDL-C (SMD = 0.07, 95% CI = 0.01–0.12, P = 0.01) than the non-carriers (Table 2, Figs. 2 and 3), and that there were no associations detected between the rs1801133 polymorphism and plasma levels of TG (SMD = 0.03, 95% CI = − 0.01-0.06, P = 0.11) and HDL-C (SMD = − 0.02, 95% CI = − 0.05-0.02, P = 0.30) (Table 2, Figs. 4 and 5). When the analyses were limited to the studies in HWE, the T allele carriers had higher levels of LDL-C (SMD = 0.04, 95% CI = 0.01–0.08, P = 0.03) and lower levels of HDL-C (SMD = − 0.02, 95% CI = − 0.03--0.00, P = 0.04) than the non-carriers (Table 2).
Table 2.
Meta-analysis between the MTHFR rs1801133 polymorphism and plasma lipid levels
| Group or subgroup | Comparison (Subjects) | SMD (95% CI) | P Heterogeneity | P SMD |
|---|---|---|---|---|
| TG | ||||
| All | 62 (39,760) | 0.03 (−0.01–0.06) | < 0.01 | 0.11 |
| Studies in HWE | 46 (34,889) | 0.01 (− 0.01–0.03) | 0.11 | 0.44 |
| Male | 4 (2518) | 0.00 (− 0.08–0.08) | 0.71 | 0.96 |
| Female | 8 (1589) | 0.13 (− 0.07–0.33) | 0.01 | 0.21 |
| Caucasian | 25 (29,136) | 0.00 (−0.04–0.04) | 0.11 | 0.93 |
| Asian | 24 (8639) | 0.05 (−0.02–0.13) | < 0.01 | 0.16 |
| African | 2 (191) | 0.20 (−0.09–0.48) | 0.65 | 0.18 |
| Other ethnicity | 11 (1794) | 0.07 (−0.03–0.17) | 0.79 | 0.15 |
| CAD | 10 (2907) | 0.01 (−0.06–0.09) | 0.54 | 0.74 |
| T2DM | 6 (812) | 0.07 (−0.14–0.28) | 0.09 | 0.51 |
| Hypertension | 4 (1522) | 0.03 (−0.09–0.15) | 0.70 | 0.63 |
| Healthy or control | 17 (26,612) | 0.01 (−0.02–0.04) | 0.33 | 0.50 |
| TC | ||||
| All | 79 (72,848) | 0.04 (0.01–0.07) | < 0.01 | 0.02 |
| Studies in HWE | 57 (51,283) | 0.01 (−0.00–0.03) | 0.13 | 0.12 |
| Male | 11 (5067) | 0.03 (−0.05–0.11) | 0.18 | 0.52 |
| Female | 15 (28,790) | 0.11 (0.02–0.19) | 0.01 | 0.02 |
| Caucasian | 26 (41,744) | 0.00 (−0.05–0.04) | 0.02 | 0.95 |
| Asian | 36 (13,385) | 0.10 (0.04–0.15) | 0.01 | < 0.01 |
| African | 3 (277) | −0.32 (−1.24–0.60) | < 0.01 | 0.49 |
| Other ethnicity | 14 (17,442) | −0.01 (− 0.04–0.02) | 0.48 | 0.55 |
| CAD | 9 (2676) | 0.02 (−0.06–0.10) | 0.45 | 0.66 |
| T2DM | 5 (705) | 0.03 (−0.20–0.27) | 0.09 | 0.77 |
| Hypertension | 6 (11,288) | −0.02 (− 0.06–0.02) | 0.47 | 0.34 |
| Healthy or control | 31 (49,859) | 0.07 (0.02–0.12) | < 0.01 | < 0.01 |
| LDL-C | ||||
| All | 65 (29,424) | 0.07 (0.01–0.12) | < 0.01 | 0.01 |
| Studies in HWE | 45 (13,946) | 0.04 (0.01–0.08) | 0.14 | 0.03 |
| Male | 10 (4999) | 0.04 (−0.04–0.12) | 0.13 | 0.36 |
| Female | 13 (3662) | 0.09 (0.01–0.16) | 0.43 | 0.02 |
| Caucasian | 18 (6226) | 0.00 (−0.05–0.05) | 0.71 | 0.95 |
| Asian | 31 (11,417) | 0.10 (0.05–0.15) | 0.10 | < 0.01 |
| African | 3 (277) | 1.16 (−0.94–3.25) | < 0.01 | 0.28 |
| Other ethnicity | 13 (11,504) | −0.02 (− 0.10–0.06) | 0.23 | 0.67 |
| CAD | 9 (2807) | 0.00 (−0.08–0.07) | 0.77 | 0.92 |
| T2DM | 6 (799) | 0.01 (−0.25–0.26) | 0.02 | 0.96 |
| Hypertension | 5 (10,843) | 0.06 (−0.05–0.18) | 0.12 | 0.30 |
| Healthy or control | 25 (8989) | 0.07 (0.01–0.13) | 0.02 | 0.02 |
| HDL-C | ||||
| All | 76 (76,811) | −0.02 (− 0.05–0.02) | < 0.01 | 0.30 |
| Studies in HWE | 57 (61,438) | −0.02 (− 0.03--0.00) | 0.72 | 0.04 |
| Male | 10 (3467) | −0.04 (− 0.11–0.03) | 0.79 | 0.29 |
| Female | 14 (28,630) | −0.05 (− 0.12–0.02) | 0.10 | 0.19 |
| Caucasian | 25 (51,768) | −0.03 (− 0.07–0.01) | 0.02 | 0.16 |
| Asian | 25 (9553) | 0.00 (−0.06–0.06) | < 0.01 | 0.92 |
| African | 2 (186) | 0.15 (− 0.14–0.43) | 0.99 | 0.32 |
| Other ethnicity | 3 (282) | −0.10 (− 0.18--0.02) | 0.26 | 0.01 |
| CAD | 8 (2570) | 0.05 (− 0.04–0.13) | 0.65 | 0.27 |
| T2DM | 6 (799) | 0.01 (−0.14–0.16) | 0.88 | 0.89 |
| Hypertension | 4 (1522) | −0.03 (− 0.15–0.08) | 0.85 | 0.56 |
| Healthy or control | 32 (56,554) | 0.00 (− 0.05–0.04) | < 0.01 | 0.97 |
MTHFR 5,10-methylenetetrahydrofolate reductase gene, SMD standardized mean difference, 95% CI 95% confidence interval, HWE Hardy-Weinberg equilibrium, TG triglyceride, TC total cholesterol, LDL-C low-density lipoprotein cholesterol, HDL-C high-density lipoprotein cholesterol, CAD coronary artery disease, T2DM type 2 diabetes mellitus
Fig. 2.
Forest plot of the meta-analysis between the MTHFR rs1801133 polymorphism and plasma total cholesterol (TC) levels
Fig. 3.
Forest plot of the meta-analysis between the MTHFR rs1801133 polymorphism and plasma low-density lipoprotein cholesterol (LDL-C) levels
Fig. 4.
Forest plot of the meta-analysis between the MTHFR rs1801133 polymorphism and triglycerides (TG) levels
Fig. 5.
Forest plot of the meta-analysis between the MTHFR rs1801133 polymorphism and plasma high-density lipoprotein cholesterol (HDL-C) levels
Then the subgroup analyses stratified by the characteristics of the subjects were performed. The significant associations of the rs1801133 polymorphism with higher levels of TC (SMD = 0.11, 95% CI = 0.02–0.19, P = 0.02) and LDL-C (SMD = 0.09, 95% CI = 0.01–0.16, P = 0.02) were detected in females, but not in males. The significant associations of the rs1801133 polymorphism with higher levels of TC (SMD = 0.10, 95% CI = 0.04–0.15, P < 0.01) and LDL-C (SMD = 0.10, 95% CI = 0.05–0.15, P < 0.01) were detected in Asians, but not in the subjects of other ethnic origins. However, the rs1801133 polymorphism was significantly associated with lower level of HDL-C (SMD = − 0.10, 95% CI = − 0.18--0.02, P = 0.01) only in the subjects of other ethnic origins. When health status was taken into account, the significant associations of the rs1801133 polymorphism with higher levels of TC (SMD = 0.07, 95% CI = 0.02–0.12, P < 0.01) and LDL-C (SMD = 0.07, 95% CI = 0.01–0.13, P = 0.02) were detected in healthy or control subjects, but not in the patients with CAD, type 2 diabetes mellitus (T2DM) and hypertension (Table 2).
Heterogeneity analysis
In the CAD association analysis, there was significant heterogeneity in allelic model (I2 = 68.2%, Pheterogeneity < 0.01), additive model (I2 = 55.6%, Pheterogeneity < 0.01), dominant model (I2 = 64.0%, Pheterogeneity < 0.01) and recessive model (I2 = 51.5%, Pheterogeneity < 0.01). Twenty-seven studies, 22 studies, 19 studies and 17 studies were identified as the main contributors to the heterogeneity for allelic model, additive model, dominant model and recessive model, respectively, by using Galbraith plots. The heterogeneity was effectively removed or decreased after exclusion of these outlier studies, but OR values and 95% CIs did not change substantially (allelic model: OR = 1.04, 95% CI = 1.01–1.07, Pheterogeneity = 0.12, POR = 0.01; additive model: OR = 1.12, 95% CI = 1.05–1.19, Pheterogeneity = 0.09, POR < 0.01; dominant model: OR = 1.04, 95% CI = 1.00–1.07, Pheterogeneity = 0.14, POR = 0.05; recessive model: OR = 1.12, 95% CI = 1.06–1.18, Pheterogeneity = 0.13, POR < 0.01) (Table 3).
Table 3.
Meta-analysis between the MTHFR rs1801133 polymorphism and CAD risk after excluding the studies with heterogeneity
| Comparisons | Ethnicity | OR | 95% CI | P heterogeneity | P OR |
|---|---|---|---|---|---|
| Allelic model (T vs C) |
Total | 1.04 | 1.01–1.07 | 0.12 | 0.01 |
| Male | 1.04 | 0.94–1.14 | 0.13 | 0.48 | |
| Female | 1.04 | 0.93–1.16 | 0.80 | 0.53 | |
| Caucasian | 1.03 | 1.00–1.06 | 0.32 | 0.07 | |
| Asian | 1.11 | 1.04–1.18 | 0.32 | < 0.01 | |
| African | 0.94 | 0.78–1.13 | 0.11 | 0.50 | |
| Other ethnicity | 0.97 | 0.83–1.14 | 0.13 | 0.74 | |
| Additive model (TT vs CC) |
Total | 1.12 | 1.05–1.19 | 0.09 | < 0.01 |
| Male | 1.07 | 0.91–1.27 | 0.23 | 0.42 | |
| Female | 1.02 | 0.79–1.31 | 0.82 | 0.87 | |
| Caucasian | 1.08 | 1.01–1.16 | 0.57 | 0.03 | |
| Asian | 1.28 | 1.12–1.46 | 0.05 | < 0.01 | |
| African | 0.79 | 0.50–1.26 | 0.26 | 0.32 | |
| Other ethnicity | 1.23 | 0.91–1.66 | 0.10 | 0.17 | |
| Dominant model (TT + CT vs CC) |
Total | 1.04 | 1.00–1.07 | 0.14 | 0.05 |
| Male | 1.04 | 0.94–1.15 | 0.16 | 0.44 | |
| Female | 1.07 | 0.91–1.25 | 0.51 | 0.42 | |
| Caucasian | 1.01 | 0.97–1.05 | 0.61 | 0.56 | |
| Asian | 1.15 | 1.05–1.25 | 0.14 | < 0.01 | |
| African | 1.01 | 0.81–1.26 | 0.07 | 0.94 | |
| Other ethnicity | 1.04 | 0.86–1.26 | 0.10 | 0.67 | |
| Recessive model (TT vs CT + CC) |
Total | 1.12 | 1.06–1.18 | 0.13 | < 0.01 |
| Male | 1.15 | 0.99–1.33 | 0.05 | 0.06 | |
| Female | 0.95 | 0.77–1.17 | 0.76 | 0.62 | |
| Caucasian | 1.07 | 1.01–1.14 | 0.68 | 0.03 | |
| Asian | 1.28 | 1.14–1.43 | 0.06 | < 0.01 | |
| African | 0.78 | 0.52–1.18 | 0.29 | 0.24 | |
| Other ethnicity | 1.22 | 0.93–1.60 | 0.18 | 0.16 |
MTHFR 5,10-methylenetetrahydrofolate reductase gene, CAD coronary artery disease, OR odds ratio, 95% CI 95% confidence interval
In the lipid association analysis, there was significant heterogeneity in the total comparisons for TG (I2 = 36.1%, Pheterogeneity < 0.01), TC (I2 = 54.1%, Pheterogeneity < 0.01), LDL-C (I2 = 69.6%, Pheterogeneity < 0.01) and HDL-C (I2 = 60.1%, Pheterogeneity < 0.01). Three comparisons, 9 comparisons, 3 comparisons and 6 comparisons were identified as the main contributors to the heterogeneity for TG, TC, LDL-C and HDL-C, respectively, by using Galbraith plots. SMD values and 95% CIs of TG and LDL-C did not change substantially after excluding the outlier comparisons. However, SMD values and 95% CIs of TC (SMD = 0.01, 95% CI = − 0.00-0.03, Pheterogeneity = 0.38, PSMD = 0.15) and HDL-C (SMD = − 0.02, 95% CI = − 0.03--0.00, Pheterogeneity = 0.48, PSMD = 0.05) changed significantly after excluding these outlier comparisons (Table 4).
Table 4.
Meta-analysis between the MTHFR rs1801133 polymorphism and plasma lipid levels after excluding the comparisons with heterogeneity
| Group or subgroup | Comparison (Subjects) |
SMD (95% CI) | P Heterogeneity | P SMD |
|---|---|---|---|---|
| TG | ||||
| All | 59 (39,196) | 0.01 (−0.01–0.03) | 0.32 | 0.22 |
| Male | 4 (2518) | 0.00 (− 0.08–0.08) | 0.71 | 0.96 |
| Female | 7 (1429) | 0.02 (− 0.09–0.13) | 0.06 | 0.74 |
| Caucasian | 24 (28,976) | 0.01 (− 0.02–0.03) | 0.31 | 0.54 |
| Asian | 22 (8235) | 0.02 (−0.03–0.06) | 0.17 | 0.50 |
| African | 2 (191) | 0.20 (−0.09–0.48) | 0.65 | 0.18 |
| Other ethnic | 11 (1794) | 0.07 (−0.03–0.17) | 0.79 | 0.15 |
| CAD | 10 (2907) | 0.01 (−0.06–0.09) | 0.54 | 0.74 |
| T2DM | 6 (812) | 0.07 (−0.08–0.22) | 0.09 | 0.35 |
| Hypertension | 4 (1522) | 0.03 (−0.09–0.15) | 0.70 | 0.63 |
| Healthy or control | 17 (26,612) | 0.01 (−0.01–0.04) | 0.33 | 0.28 |
| TC | ||||
| All | 70 (70,024) | 0.01 (−0.00–0.03) | 0.38 | 0.15 |
| Male | 11 (5067) | 0.02 (−0.04–0.08) | 0.18 | 0.44 |
| Female | 11 (28, 074) | 0.02 (−0.00–0.04) | 0.54 | 0.09 |
| Caucasian | 22 (41,176) | 0.01 (−0.01–0.03) | 0.81 | 0.42 |
| Asian | 32 (11,220) | 0.06 (0.02–0.10) | 0.34 | < 0.01 |
| African | 2 (186) | 0.15 (−0.14–0.44) | 0.15 | 0.31 |
| Other ethnic | 14 (17,442) | −0.01 (− 0.04–0.02) | 0.48 | 0.55 |
| CAD | 10 (2907) | 0.02 (−0.06–0.10) | 0.45 | 0.66 |
| T2DM | 5 (705) | 0.07 (−0.09–0.23) | 0.09 | 0.38 |
| Hypertension | 6 (11,288) | −0.02 (− 0.06–0.02) | 0.47 | 0.34 |
| Healthy or control | 26 (47,611) | 0.02 (−0.00–0.04) | 0.36 | 0.08 |
| LDL-C | ||||
| All | 62 (18,739) | 0.04 (0.01–0.07) | 0.10 | 0.01 |
| Male | 10 (4999) | 0.04 (−0.02–0.10) | 0.13 | 0.24 |
| Female | 13 (3662) | 0.09 (0.01–0.16) | 0.43 | 0.02 |
| Caucasian | 18 (6226) | 0.00 (−0.05–0.05) | 0.71 | 0.95 |
| Asian | 30 (10,439) | 0.08 (0.04–0.12) | 0.16 | < 0.01 |
| African | 2 (186) | −0.17 (−0.46–0.12) | 0.14 | 0.25 |
| Other ethnic | 12 (1888) | 0.00 (−0.10–0.09) | 0.19 | 0.98 |
| CAD | 9 (2807) | 0.00 (−0.08–0.07) | 0.77 | 0.92 |
| T2DM | 6 (799) | 0.06 (−0.09–0.21) | 0.02 | 0.43 |
| Hypertension | 4 (1227) | 0.14 (0.01–0.27) | 0.87 | 0.03 |
| Healthy or control | 24 (8011) | 0.06 (0.01–0.11) | 0.06 | 0.01 |
| HDL-C | ||||
| All | 70 (65,091) | −0.02 (−0.03--0.00) | 0.48 | 0.05 |
| Male | 10 (3467) | −0.04 (− 0.11–0.03) | 0.79 | 0.29 |
| Female | 14 (28,630) | −0.01 (− 0.03–0.02) | 0.10 | 0.61 |
| Caucasian | 23 (51,561) | −0.02 (− 0.03–0.00) | 0.49 | 0.08 |
| Asian | 33 (11,456) | −0.02 (− 0.06–0.02) | 0.22 | 0.42 |
| African | 2 (186) | 0.15 (−0.14–0.43) | 0.99 | 0.32 |
| Other ethnic | 12 (1888) | −0.04 (− 0.13–0.05) | 0.70 | 0.41 |
| CAD | 9 (2766) | 0.03 (−0.06–0.11) | 0.99 | 0.54 |
| T2DM | 6 (799) | 0.01 (−0.14–0.16) | 0.88 | 0.89 |
| Hypertension | 4 (1522) | −0.03 (− 0.15–0.08) | 0.85 | 0.56 |
| Healthy or control | 30 (54,825) | −0.02 (− 0.03–0.00) | 0.05 | 0.06 |
MTHFR 5,10-methylenetetrahydrofolate reductase gene, SMD standardized mean difference, 95% CI 95% confidence interval, TG triglyceride, TC total cholesterol, LDL-C low-density lipoprotein cholesterol, HDL-C high-density lipoprotein cholesterol, CAD coronary artery disease, T2DM type 2 diabetes mellitus
Publication bias test
Begg’s test and Egger’s test were used to evaluate the publication bias of the included studies, and the results showed that there might be a publication bias in the analysis between the rs1801133 polymorphism and CAD (P < 0.05 for all genetic models). To clarify this problem, a trim-and-fill method was used to adjust the results, and no trimming was performed and the results were unchanged. It indicates that there is no publication bias in the literature. The significant P values of Begg’s test and Egger’s test were originated from other factors, e.g. mixed ethnicity in some studies. No publication bias was detected in the lipid association analysis.
Discussion
In the present meta-analysis, the variant T allele of the rs1801133 polymorphism was associated with increased risk of CAD, and elevated levels of TC and LDL-C in the total population. It indicates that the T allele of the rs1801133 polymorphism is a risk factor for CAD, which is at least partly mediated by abnormal lipid levels.
A large number of studies have investigated the association between the rs1801133 polymorphism and CAD risk, as well as the underlying mechanisms, but most of them just focused on homosysteine. It was widely reported that the rs1801133 polymorphism influences the plasma levels of homosysteine in various populations such as Americans [25], Africans [17, 26, 27], Asians [18, 28, 29], Turkish [30, 31] and Brazilians [32]. In methionine cycle, homocysteine is formed after the removal of adenosine from S-adenosine homocysteine. Under normal condition, homocysteine is remethylated to methionine by accepting a methyl group from 5-MTHF. 5-MTHF is formed from the reduction of 5,10-MTHF under the catalysis of MTHFR. In the T allele carriers of the rs1801133 polymorphism, the function of MTHFR may be affected since the normal alanine residue is replaced by a valine residue in the polypeptide, which in turn affects the production of 5-MTHF and the remethylation of homcysteine, resulting in elevated plasma homosysteine levels. Studies have shown that homocysteine is a risk factor for CAD, and oxidative stress [33], vascular inflammation [34] and endothelial injury [35] are involved in the underlying mechanisms in which homocysteine causes CAD. All these events are likely to trigger the development of atherosclerosis and arterial thrombosis.
However, the role of homocysteine in the pathogenesis of CAD is controversial. Several studies [36–38] demonstrated no association between homocysteine and CAD. It indicates that some other risk factors are involved in the association between the rs1801133 polymorphism and CAD. In this meta-analysis, the results showed that the variant T allele carriers of the rs1801133 polymorphism have higher levels of TC and LDL-C than the non-carriers, which indicates that the abnormal lipid levels caused by the T allele of the rs1801133 polymorphism might be one of the important reasons in the development of CAD since dyslipidemia is closely associated with the progression of coronary atherosclerosis, and it accounts for around 50% of the population-attributable risk for CAD [39]. According to the 2013 ACC/AHA blood cholesterol guidelines [40] and the Adult Treatment Panel III (ATP III) Guidelines [41] of the United States, LDL-C was considered as a major cause of CAD and used as the primary target for therapy, and other lipid parameters were used as the secondary or supplementary targets.
The mechanisms in which the rs1801133 polymorphism is associated with plasma lipid levels have not been clarified yet. Several possible reasons can be proposed to explain the association between the rs1801133 polymorphism and plasma lipid levels. Firstly, the rs1801133 polymorphism may indirectly affect plasma lipid levels through the mediation of homocysteine [42–44]. Baszczuk et al. [42] reported that a daily administration of 15 mg of folic acid led to a considerable decrease in homocysteine levels, and a substantive increase in HDL-C levels in the patients with primary hypertension. In yeast cells, homocysteine supplementation increased cellular fatty acid and TG contents, induced a shift in fatty acid composition, and decreased the condensing enzymes involved in very long-chain fatty acid synthesis [43]. Secondly, the rs1801133 polymorphism may modulate the lipid metabolism by affecting the methylation state of DNA or proteins. 5-MTHF is not only the methyl donor for homocysteine, but for many other target molecules, including DNA and proteins [45]. Conceivably, the functions of the genes or proteins involved in lipid metabolism will be affected if their methylation state changes.
In most of the studies included in the lipid association analysis, a dominant model was used, i.e. TT + CT vs. CC. Therefore, a dominant model was also adopted in this meta-analysis. In subgroup analyses, we found that the differences in TC and LDL-C levels between the genotypes were mainly from Asian populations, whose SMD values were larger than those calculated in Caucasians, Africans and the subjects of other ethnicities (Table 2). The associations of the rs1801133 polymorphism with TC and LDL-C in Asians were consistently larger, which shows that there is a stronger association between the rs1801133 polymorphism and CAD in Asians as compared with other ethnicities (Table 1). Studies will be needed to elucidate the mechanisms that the rs1801133 polymorphism has different effects on blood lipid levels and CAD risk in different ethnicities.
In the lipid association analysis, subgroup analyses by gender and health status was performed since there might be important factors affecting the associations between the rs1801133 polymorphism and lipid levels. For example, the present meta-analysis indicates that gender might modulate the associations of the rs1801133 polymorphism with TC and LDL-C levels since there were significant associations existing only in females but not in males (Table 2). Health status might also modulate the associations of the rs1801133 polymorphism with TC and LDL-C levels. The significant associations of the rs1801133 polymorphism with TC and LDL-C levels only existed in healthy subjects, but not in the patients with CAD, T2DM and hypertension. The reason might be that the patients with CAD, T2DM and hypertension had serious metabolic disorders, which masked the effects of the rs1801133 polymorphism on TC and LDL-C levels. As compared with the diseases such as CAD, T2DM and hypertension, genetic polymorphisms usually have less impact on plasma lipid levels. In line with the results from the present study, several studies also reported that the rs1801133 polymorphism is associated TC and LDL-C levels in healthy subjects [13, 15, 46], but not in the patients with CAD [11, 20], T2DM [47, 48] and hypertension [49, 50]. Of the 179 studies included, 141 studies used polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method; 27 studies used real-time PCR method; 7 studies used DNA sequencing method; 1 study used gene chip method; and 3 studies did not report the genotyping method(s). Subgroup analyses stratified by the genotyping methods were conducted, and the results showed that there were differences in OR or SMD values among the studies with different genotyping methods (data not shown). In most cases, the results from the studies by PCR-RFLP method were in line with the results from all studies. The reason might be that most of the studies used PCR-RFLP method. The results from the studies by real-time PCR method or DNA sequencing method could have been affected by the small number of studies and small sample sizes.
Significant heterogeneity was detected in the total and subgroup analyses between the rs1801133 polymorphism and CAD, and the outlier studies were identified by using the Galbraith plots. No significant changes in OR values and 95% CIs were found after excluding the outlier studies (Table 3), which indicates that the association between the rs1801133 polymorphism and CAD is very strong. Significant heterogeneity was also detected in the total and some of the subgroup analyses between the rs1801133 polymorphism and plasma lipid levels. The outlier studies were identified and excluded, but SMD values and 95% CIs of LDL-C were not significantly changed in the total population or in Asians, which indicates that there is a strong association between the rs1801133 polymorphism and LDL-C levels, especially in Asians.
The associations of the rs1801133 polymorphism with CAD and plasma lipid levels are not likely to be type I errors (false-positive results). Firstly, the results from this meta-analysis are based on four different models for CAD association analysis, and on random effects model for lipid association analysis if the heterogeneity among the studies is significant (I2 > 50%). As compared with fixed effects model, the random effects model is a more conservative method and less likely to produce false-positive results. Secondly, 87,020 subjects and 85,554 subjects were included in the analysis for the CAD association analysis and the lipid association analysis, respectively. Among the subjects, 44.0% (CAD association analysis) and 52.3% (lipid association analysis) of them are the carriers of the variant T allele. Since the incidence of the T allele carriers is very high, type I errors could have been prevented in both the CAD and lipid association analyses. Thirdly, this meta-analysis only included the studies published in English and Chinese as it was very difficult to get the full papers published in various languages.
Conclusions
The current meta-analysis demonstrates that the rs1801133 polymorphism is associated with increased risk of CAD and elevated levels of TC and LDL-C. Further studies will be needed to elucidate the underlying mechanisms in which the rs1801133 polymorphism affects plasma lipid levels.
Methods
Literature search
The articles published before September 2017 on the associations of the rs1801133 polymorphism with CAD and/or plasma lipid levels were identified. The languages of the articles were limited to English and Chinese. A comprehensive search was conducted and nine electronic databases were searched to identify all relevant articles. The databases are as follows: PubMed, Embase, Baidu Scholar, Google Scholar, Web of Science, Cochrane Library, Wanfang, CBM and CNKI. The following keywords were used: (“5,10-Methylenetetrahydrofolate reductase” or “Methylenetetrahydrofolate reductase” or “MTHFR”), (“polymorphism” or “mutation” or “variant” or “C677T” or “rs1801133” or “Ala222Val”), (“coronary artery disease” or “coronary heart disease” or “heart disease” or “coronary disease” or “cardiovascular disease” or “angina pectoris” or “acute coronary syndrome” or “myocardial infarction” or “CAD” or “CHD” or “HD” or “CD” or “AP” or “ACS” or “MI”), (“plasma lipid” or “blood lipid” or “serum lipid”).
Inclusion and exclusion criteria
The inclusion criteria for the association analysis between the rs1801133 polymorphism and CAD are as follows: 1) studies using a population-based case-control design; 2) CAD cases were angiographically defined; 3) number or frequency of cases according to the r1801133 genotypes was available. The inclusion criteria for the association analysis between the rs1801133 polymorphism and lipid levels are as follows: 1) studies in which mean lipids and standard deviations (SD) or standard errors (SE) by the rs1801133 genotypes were available; 2) studies which reported at least one of the four lipid variables, i.e. TG, TC, LDL-C and HDL-C; 3) baseline data were used for interventional studies. All references cited by the included articles were reviewed to check the published work which was not indexed by PubMed, Embase, Baidu Scholar, Google Scholar, Web of Science, Cochrane Library, Wanfang, CBM and CNKI. Reports with incomplete data, studies based on pedigree data, case reports, review articles, abstracts and animal studies were excluded from the meta-analysis.
Data extraction
Data were extracted from each study by using a structured data collection form and by two investigators independently according to the pre-specified selection criteria. Decisions were compared and disagreements about study selection were resolved by consensus or by involving a third investigator. For the overlapping articles, only the publications that presented the most detailed information were included. In this meta-analysis, the data extracted from each of the included studies are as follows: first author, year of publication, age, ethnicity, gender, health status, type of study, genotyping method, lipid assay method, sample size, and mean with SD or SE according to the r1801133 genotypes. If data in a study were unconvincing, we attempted to contact the corresponding or first author by e-mail and telephone.
Data analysis
Statistical analysis was performed by using STATA version 12.0 (Stata Corporation LP, College Station, TX, USA). All the tests were two-sided and a P-value of less than 0.05 for any test or model was considered to be statistically significant. OR with 95% CI was used to evaluate the strength of the association between the rs1801133 polymorphism and CAD. The pooled OR was performed for allelic model (T vs C), additive model (TT vs CC), dominant model (TT + CT vs CC) and recessive model (TT vs CT + CC). SMD with 95% CI was used to assess the strength of the associations between the rs1801133 polymorphism and plasma lipid levels. A fixed-effect model (Mantel-Haenszel method) was used to evaluate the results if heterogeneity among the included studies was not significant (I2 < 50%). Otherwise, the random-effect model (DerSimonian-Laird method) was used [51]. Heterogeneity was investigated by Cochran’s χ2-based Q-statistic, and Galbraith plots were used to detect the potential sources of heterogeneity. OR and SMD values were recalculated after excluding the outlier studies. Subgroup analyses were performed according to ethnicity for CAD association analysis, and according to ethnicity, gender and health status for lipid association analysis. Ethnic subgroups were defined as Caucasian, Asian, African, and the subjects of other ethnic origin. Health status was defined as CAD, T2DM and hypertension. HWE was assessed by Fisher’s exact test. OR and SMD values were recalculated after excluding the studies which were not in HWE. Publication bias was tested by Begg’s funnel plots and Egger’s test [52].
Additional files
The reference list for the studies included in the present meta-analysis. (DOC 106 kb)
Table S1. Characteristics of the individual studies included in the meta-analysis between the MTHFR rs1801133 polymorphism and CAD; Table S2. Characteristics of the individual studies included in the meta-analysis between the MTHFR rs1801133 polymorphism and plasma lipid levels; Table S3. Plasma lipid levels according to the genotypes of the MTHFR rs1801133 polymorphism. (DOC 544 kb)
Acknowledgements
This research was supported by the grants from the Key Project of Education Department of Sichuan Province, P. R. China (17ZA0172) and the Cooperative Project on Scientific Research between Nanchong city and North Sichuan Medical College, P. R. China (NSMC20170403).
Funding
This research was supported by the grants from the Key Project of Education Department of Sichuan Province, P. R. China (17ZA0172) and the Cooperative Project on Scientific Research between Nanchong city and North Sichuan Medical College, P. R. China (NSMC20170403).
Availability to data and materials
All data generated or analysed during this study are included in this published article [and its supplementary information files].
Abbreviations
- 95% CI
95% confidence interval
- CAD
coronary artery disease
- HDL-C
high-density lipoprotein cholesterol
- HWE
Hardy-Weinberg equilibrium
- LDL-C
low-density lipoprotein cholesterol
- SMD
standardized mean difference
- T2DM
type 2 diabetes mellitus
- TC
total cholesterol
- TG
triglycerides
Authors’ contributions
Yongyan Song, Zhi Luo and Zhan Lu conceived of the study, participated in the design, and drafted the manuscript. Zhi Luo, Zhan Lu, Irfan Muhammad, Yun Chen, Qiuhong Chen and Jiaojiao Zhang carried out the study searches and collected the data. Yongyan Song performed the statistical analyses. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, Das SR, de Ferranti S, Després JP, Fullerton HJ, Howard VJ, Huffman MD, Isasi CR, Jiménez MC, Judd SE, Kissela BM, Lichtman JH, Lisabeth LD, Liu S, Mackey RH, Magid DJ, DK MG, Mohler ER, 3rd, Moy CS, Muntner P, Mussolino ME, Nasir K, Neumar RW, Nichol G, Palaniappan L, Pandey DK, Reeves MJ, Rodriguez CJ, Rosamond W, Sorlie PD, Stein J, Towfighi A, Turan TN, Virani SS, Woo D, Yeh RW, Turner MB. Heart Disease and Stroke Statistics-2016 Update: A Report From the American Heart Association. Circulation. 2016;133:e38–360. doi: 10.1161/CIR.0000000000000350. [DOI] [PubMed] [Google Scholar]
- 2.Erdmann J, Kessler T, Munoz Venegas L, Schunkert H. A decade of genome-wide association studies for coronary artery disease: the challenges ahead. Cardiovasc Res. 2018; 10.1093/cvr/cvy084. [Epub ahead of print] [DOI] [PubMed]
- 3.O'Donnell FL, Stahlman S, Oetting AA. Incidence rates of diagnoses of cardiovascular diseases and associated risk factors, active component, U.S. Armed Forces, 2007–2016. MSMR. 2018;25:12–18. [PubMed] [Google Scholar]
- 4.Wang L, Shangguan S, Chang S, Yu X, Wang Z, Lu X, Wu L, Zhang T. Determining the association between methylenetetrahydrofolate reductase (MTHFR) genepolymorphisms and genomic DNA methylation level: a meta-analysis. Birth Defects Res A Clin Mol Teratol. 2016;106:667–674. doi: 10.1002/bdra.23511. [DOI] [PubMed] [Google Scholar]
- 5.Xhemalce B. From histones to RNA: role of methylation in cancer. Brief Funct Genomics. 2013;12:244–253. doi: 10.1093/bfgp/els064. [DOI] [PubMed] [Google Scholar]
- 6.Lanouette S, Mongeon V, Figeys D, Couture JF. The functional diversity of protein lysine methylation. Mol Syst Biol. 2014;10:724. doi: 10.1002/msb.134974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kukrele P, Sharma RS. High homocysteine level- an important risk factor for coronary artery disease in vegetarians. J Assoc Physicians India. 2016;64:75. [Google Scholar]
- 8.Mikael LG, Wang XL, Wu Q, Jiang H, Maclean KN, Rozen R. Hyperhomocysteinemia is associated with hypertriglyceridemia in mice with methylenetetrahydrofolate reductase deficiency. Mol Genet Metab. 2009;98:187–194. doi: 10.1016/j.ymgme.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 9.Christensen KE, Mikael LG, Leung KY, Lévesque N, Deng L, Wu Q, Malysheva OV, Best A, Caudill MA, Greene ND, Rozen R. High folic acid consumption leads to pseudo-MTHFR deficiency, altered lipid metabolism, and liver injury in mice. Am J Clin Nutr. 2015;101:646–658. doi: 10.3945/ajcn.114.086603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Husemoen LL, Skaaby T, Jørgensen T, Thuesen BH, Fenger M, Grarup N, Sandholt CH, Hansen T, Pedersen O, Linneberg A. MTHFR C677T genotype and cardiovascular risk in a general population without mandatory folic acid fortification. Eur J Nutr. 2014;53:1549–1559. doi: 10.1007/s00394-014-0659-2. [DOI] [PubMed] [Google Scholar]
- 11.Ramkaran P, Phulukdaree A, Khan S, Moodley D, Chuturgoon AA. Methylenetetrahydrofolate reductase C677T polymorphism is associated with increased risk of coronary artery disease in young south African Indians. Gene. 2015;571:28–32. doi: 10.1016/j.gene.2015.06.044. [DOI] [PubMed] [Google Scholar]
- 12.Chen W, Hua K, Gu H, Zhang J, Wang L. Methylenetetrahydrofolate reductase C667T polymorphism is associated with increased risk of coronary artery disease in a Chinese population. Scand J Immunol. 2014;80:346–353. doi: 10.1111/sji.12215. [DOI] [PubMed] [Google Scholar]
- 13.Zhi X, Yang B, Fan S, Wang Y, Wei J, Zheng Q, Sun G. Gender-specific interactions of MTHFR C677T and MTRR A66G polymorphisms with overweight/obesity on serum lipid levels in a Chinese Han population. Lipids Health Dis. 2016;15:185. doi: 10.1186/s12944-016-0354-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin RX, Wu DF, Miao L, Htet Aung LH, Cao XL, Yan TT, Long XJ, Liu WY, Zhang L, Li M. Interactions of several single nucleotide polymorphisms and high body mass index on serum lipid traits. Biofactors. 2013;39:315–325. doi: 10.1002/biof.1073. [DOI] [PubMed] [Google Scholar]
- 15.Chen NY, Liu CW, Du LL, Xiao LP, Ge L, Wang YY, Wei Z, Wu HY, Luo CY, Liang L, Peng JH, Luo XQ, Yin RX, Nguyen CP, Pan SL. Enrichment of MTHFR 677 T in a Chinese long-lived cohort and its association with lipid modulation. Lipids Health Dis. 2014;13:104. doi: 10.1186/1476-511X-13-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang S, Zhao R, Pan M, Venners SA, Zhong G, Hsu YH. Associations of MTHFR and MTRR polymorphisms with serum lipid levels in Chinese hypertensive patients. Clin Appl Thromb Hemost. 2014;20:400–410. doi: 10.1177/1076029612467226. [DOI] [PubMed] [Google Scholar]
- 17.Abd-Elmawla MA, Rizk SM, Youssry I, Shaheen AA. Impact of genetic polymorphism of methylenetetrahydrofolate reductase C677T on development of Hyperhomocysteinemia and related oxidative changes in Egyptian β-thalassemia major patients. PLoS One. 2016;11:e0155070. doi: 10.1371/journal.pone.0155070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saffari B, Senemar S, Karimi M, Bahari M, Jooyan N, Yavarian M. An MTHFR variant, plasma homocysteine levels and late-onset coronary artery disease in subjects from southern Iran. Pak J Biol Sci. 2013;16:788–795. doi: 10.3923/pjbs.2013.788.795. [DOI] [PubMed] [Google Scholar]
- 19.Senemar S, Saffari B, Sharifkazemi MB, Bahari M, Jooyan N, Dehaghani ED, Yavarian M. 5,10-methylene tetrahydrofolate reductase C677T gene polymorphism, homocysteine concentration and the extent of premature coronary artery disease in southern Iran. EXCLI J. 2013;12:437–448. [PMC free article] [PubMed] [Google Scholar]
- 20.Kucukhuseyin O, Kurnaz O, Akadam-Teker AB, Isbir T, Bugra Z, Ozturk O, Yilmaz-Aydogan H. The association of MTHFR C677T gene variants and lipid profiles or body mass index in patients with diabetic and nondiabetic coronary heart disease. J Clin Lab Anal. 2013;27:427–434. doi: 10.1002/jcla.21623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frederiksen J, Juul K, Grande P, Jensen GB, Schroeder TV, Tybjaerg-Hansen A, Nordestgaard BG. Methylenetetrahydrofolate reductase polymorphism (C677T), hyperhomocysteinemia, and risk of ischemic cardiovascular disease and venous thromboembolism: prospective and case-control studies from the Copenhagen City heart study. Blood. 2004;104:3046–3051. doi: 10.1182/blood-2004-03-0897. [DOI] [PubMed] [Google Scholar]
- 22.Vasisht S, Gulati R, Narang R, Srivastava N, Srivastava LM, Manchanda SC, Agarwal DP. Polymorphism (C677T) in the 5,10-methylenetetrahydrofolate reductase (MTHFR) gene: a preliminary study on north Indian men. Indian J Clin Biochem. 2002;17:99–107. doi: 10.1007/BF02867949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klerk M, Verhoef P, Clarke R, Blom HJ, Kok FJ, Schouten EG, MTHFR Studies Collaboration Group MTHFR 677C-->T polymorphism and risk of coronary heart disease: a meta-analysis. JAMA. 2002;288:2023–2031. doi: 10.1001/jama.288.16.2023. [DOI] [PubMed] [Google Scholar]
- 24.Lewis SJ, Ebrahim S, Davey SG. Meta-analysis of MTHFR 677C->T polymorphism and coronary heart disease: does totality of evidence support causal role for homocysteine and preventive potential of folate? BMJ. 2005;331:1053. doi: 10.1136/bmj.38611.658947.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brilakis ES, Berger PB, Ballman KV, Rozen R. Methylenetetrahydrofolate reductase (MTHFR) 677C>T and methionine synthase reductase (MTRR) 66A>G polymorphisms: association with serum homocysteine and angiographic coronary artery disease in the era of flour products fortified with folic acid. Atherosclerosis. 2003;168:315–322. doi: 10.1016/S0021-9150(03)00098-4. [DOI] [PubMed] [Google Scholar]
- 26.Bennouar N, Allami A, Azeddoug H, Bendris A, Laraqui A, El Jaffali A, El Kadiri N, Benzidia R, Benomar A, Fellat S, Benomar M. Thermolabile methylenetetrahydrofolate reductase C677T polymorphism and homocysteine are risk factors for coronary artery disease in Moroccan population. J Biomed Biotechnol. 2007;2007:80687. doi: 10.1155/2007/80687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghazouani L, Abboud N, Mtiraoui N, Zammiti W, Addad F, Amin H, Almawi WY, Mahjoub T. Homocysteine and methylenetetrahydrofolate reductase C677T and A1298C polymorphisms in Tunisian patients with severe coronary artery disease. J Thromb Thrombolysis. 2009;27:191–197. doi: 10.1007/s11239-008-0194-1. [DOI] [PubMed] [Google Scholar]
- 28.Yadav S, Hasan N, Marjot T, Khan MS, Prasad K, Bentley P, Sharma P. Detailed analysis of gene polymorphisms associated with ischemic stroke in south Asians. PLoS One. 2013;8:e57305. doi: 10.1371/journal.pone.0057305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biswas A, Ranjan R, Meena A, Akhter MS, Yadav BK, Munisamy M, Subbiah V, Behari M, Saxena R. Homocystine levels, polymorphisms and the risk of ischemic stroke in young Asian Indians. J Stroke Cerebrovasc Dis. 2009;18:103–110. doi: 10.1016/j.jstrokecerebrovasdis.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 30.Ilhan N, Kucuksu M, Kaman D, Ilhan N, Ozbay Y. The 677 C/T MTHFR polymorphism is associated with essential hypertension, coronary artery disease, and higher homocysteine levels. Arch Med Res. 2008;39:125–130. doi: 10.1016/j.arcmed.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 31.Ozbek Z, Kucukali CI, Ozkok E, Orhan N, Aydin M, Kilic G, Sazci A, Kara I. Effect of the methylenetetrahydrofolate reductase gene polymorphisms on homocysteine, folate and vitamin B12 in patients with bipolar disorder and relatives. Prog Neuro-Psychopharmacol Biol Psychiatry. 2008;32:1331–1337. doi: 10.1016/j.pnpbp.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 32.Pereira AC, Miyakawa AA, Lopes NH, Soares PR, de Oliveira SA, Cesar LA, Ramires JF, Hueb W, Krieger JE. Dynamic regulation of MTHFR mRNA expression and C677T genotype modulate mortality in coronary artery disease patients after revascularization. Thromb Res. 2007;121:25–32. doi: 10.1016/j.thromres.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 33.Ma SC, Hao YJ, Jiao Y, Wang YH, Xu LB, Mao CY, Yang XL, Yang AN, Tian J, Zhang MH, Jin SJ, Xu H, Jiang YD, Zhang HP. Homocysteine induced oxidative stress through TLR4/NF κB/DNMT1 mediated LOX 1 DNA methylation in endothelial cells. Mol Med Rep. 2017;16:9181–9188. doi: 10.3892/mmr.2017.7753. [DOI] [PubMed] [Google Scholar]
- 34.Leng YP, Ma YS, Li XG, Chen RF, Zeng PY, Li XH, Qiu CF, Li YP, Zhang Z, Chen AF. l-Homocysteine-induced cathepsin V mediates the vascular endothelial inflammation inhyperhomocysteinaemia. Br J Pharmacol. 2018;175:1157–1172. doi: 10.1111/bph.13920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lian Z, Lv FF, Yu J, Wang JW. The anti-inflammatory effect of microRNA-383-3p interacting with IL1R2 against homocysteine-induced endothelial injury in rat coronary arteries. J Cell Biochem. 2018;119:6684–94. doi: 10.1002/jcb.26854. [DOI] [PubMed] [Google Scholar]
- 36.Chen CJ, Yang TC, Chang C, Lu SC, Chang PY. Homocysteine is a bystander for ST-segment elevation myocardial infarction: a case-control study. BMC Cardiovasc Disord. 2018;18:33. doi: 10.1186/s12872-018-0774-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clarke R, Bennett DA, Parish S, Verhoef P, Dötsch-Klerk M, Lathrop M, Xu P, Nordestgaard BG, Holm H, Hopewell JC, Saleheen D, Tanaka T, Anand SS, Chambers JC, Kleber ME, Ouwehand WH, Yamada Y, Elbers C, Peters B, Stewart AF, Reilly MM, Thorand B, Yusuf S, Engert JC, Assimes TL, Kooner J, Danesh J, Watkins H, Samani NJ, Collins R, Peto R, MTHFR Studies Collaborative Group Homocysteine and coronary heart disease: meta-analysis of MTHFR case-control studies, avoiding publication bias. PLoS Med. 2012;9:e1001177. doi: 10.1371/journal.pmed.1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han L, Wu Q, Wang C, Hao Y, Zhao J, Zhang L, Fan R, Liu Y, Li R, Chen Z, Zhang T, Chen S, Ma J, Liu S, Peng X, Duan S. Homocysteine, ischemic stroke, and coronary heart disease in hypertensive patients: APopulation-based. Prospective Cohort Study Stroke. 2015;46:1777–1786. doi: 10.1161/STROKEAHA.115.009111. [DOI] [PubMed] [Google Scholar]
- 39.Yusuf S, Hawken S, Ounpuu S, Dans T, Avezum A, Lanas F, McQueen M, Budaj A, Pais P, Varigos J, Lisheng L, INTERHEART Study Investigators Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries(the INTERHEART study): case-control study. Lancet. 2004;364:937–952. doi: 10.1016/S0140-6736(04)17018-9. [DOI] [PubMed] [Google Scholar]
- 40.Stone NJ, Robinson JG, Lichtenstein AH, Bairey Merz CN, Blum CB, Eckel RH, Goldberg AC, Gordon D, Levy D, Lloyd-Jones DM, McBride P, Schwartz JS, Shero ST, Smith SC, Jr, Watson K, Wilson PW, Eddleman KM, Jarrett NM, LaBresh K, Nevo L, Wnek J, Anderson JL, Halperin JL, Albert NM, Bozkurt B, Brindis RG, Curtis LH, DeMets D, Hochman JS, Kovacs RJ, Ohman EM, Pressler SJ, Sellke FW, Shen WK, Smith SC, Jr, Tomaselli GF, American College ofCardiology/American Heart Association Task Force on Practice Guidelines 2013 ACC/AHA guideline on the treatment of blood cholesterol to reduce atherosclerotic cardiovascular risk in adults: a report of the American College of Cardiology/American Heart Association task force on practice guidelines. Circulation. 2014;129:S1–45. doi: 10.1161/01.cir.0000437738.63853.7a. [DOI] [PubMed] [Google Scholar]
- 41.National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III) Third Report of the National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III) final report. Circulation. 2002;106:3143–3421. [PubMed] [Google Scholar]
- 42.Baszczuk A, Thielemann A, Musialik K, Kopczynski J, Bielawska L, Dzumak A, Kopczynski Z, Wysocka E. The Impact of Supplementation with Folic Acid on Homocysteine Concentration and Selected Lipoprotein Parameters in Patients with Primary Hypertension. J Nutr Sci Vitaminol (Tokyo) 2017;63:96–103. doi: 10.3177/jnsv.63.96. [DOI] [PubMed] [Google Scholar]
- 43.Visram M, Radulovic M, Steiner S, Malanovic N, Eichmann TO, Wolinski H, Rechberger GN, Tehlivets O. Homocysteine regulates fatty acid and lipid metabolism in yeast. J Biol Chem. 2018;293:5544–5555. doi: 10.1074/jbc.M117.809236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vijayakumar A, Kim EK, Kim H, Choi YJ, Huh KB, Chang N. Effects of folic acid supplementation on serum homocysteine levels, lipid profiles, and vascular parameters in post-menopausal Korean women with type 2 diabetes mellitus. Nutr Res Pract. 2017;11:327–333. doi: 10.4162/nrp.2017.11.4.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Llanos AA, Marian C, Brasky TM, Dumitrescu RG, Liu Z, Mason JB, Makambi KH, Spear SL, Kallakury BV, Freudenheim JL, Shields PG. Associations between genetic variation in one-carbon metabolism and LINE-1 DNA methylation inhistologically normal breast tissues. Epigenetics. 2015;10:727–735. doi: 10.1080/15592294.2015.1062205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang L, Yin RX, Liu WY, Miao L, Wu DF, Aung LH, Hu XJ, Cao XL, Wu JZ, Pan SL. Association of methylenetetrahydrofolate reductase C677T polymorphism and serum lipid levels in the Guangxi Bai Ku Yao and Han populations. Lipids Health Dis. 2010;9:123. doi: 10.1186/1476-511X-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pollex RL, Mamakeesick M, Zinman B, Harris SB, Hanley AJ, Hegele RA. Methylenetetrahydrofolate reductase polymorphism 677C>T is associated with peripheral arterial disease in type 2 diabetes. Cardiovasc Diabetol. 2005;4:17. doi: 10.1186/1475-2840-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maeda M, Yamamoto I, Fukuda M, Motomura T, Nishida M, Nonen S, Fujio Y, Kasayama S, Azuma J. MTHFR gene polymorphism is susceptible to diabetic retinopathy but not to diabetic nephropathyin Japanese type 2 diabetic patients. J Diabetes Complicat. 2008;22:119–125. doi: 10.1016/j.jdiacomp.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 49.Lv CF, Zhang T, Li LL, Wang CY, Liu K. Correlation of metabolism-related indicators with MTHFR gene polymorphism in H type hypertension patients. China Trop Med. 2014;14:86–88. [Google Scholar]
- 50.Fan GJ, Xu R, Zhang Q, Yun L, Cao YL, Zhang CM. Correlation between methylenetetrahydrofolate reductase gene C677T polymorphism and blood lipid abnormality among hypertensives. Chin J Arterioscler. 2017;25:153–158. [Google Scholar]
- 51.DerSimonian R, Kacker R. Random-effects model for meta-analysis of clinical trials: an update. Contemp Clin Trials. 2007;28:105–114. doi: 10.1016/j.cct.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 52.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. doi: 10.2307/2533446. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The reference list for the studies included in the present meta-analysis. (DOC 106 kb)
Table S1. Characteristics of the individual studies included in the meta-analysis between the MTHFR rs1801133 polymorphism and CAD; Table S2. Characteristics of the individual studies included in the meta-analysis between the MTHFR rs1801133 polymorphism and plasma lipid levels; Table S3. Plasma lipid levels according to the genotypes of the MTHFR rs1801133 polymorphism. (DOC 544 kb)