Abstract
Peritoneal metastasis is one of the major patterns of unresectability in colorectal cancer (CRC) and a cause of death in advanced CRC. Identification of distinct gene expressions between primary CRC and peritoneal seeding metastasis is to predict the metastatic potential of primary human CRC. Three pairs of primary CRC (SNU-2335A, SNU-2404A, and SNU-2414A) and corresponding peritoneal seeding (SNU-2335D, SNU-2404B, and SNU-2414B) cell lines were established to determine the different gene expressions and resulting aberrated signaling pathways in peritoneal metastasis tumor using whole exome sequencing and microarray. Whole exome sequencing detected that mutation in CYP2A7 was exclusively shared in peritoneal seeding cell lines. Microarray identified that there were five upregulated genes (CNN3, SORBS1, BST2, EPSTI1, and KLHL5) and two downregulated genes (TRY6 and STYL5) in the peritoneal metastatic cell lines. CNN3 expression was highly augmented in both mRNA and protein levels in peritoneal metastasis cells. Knockdown of Calponin 3 resulted in augmented level of E-cadherin in peritoneal metastasis cells, and migration and invasiveness decreased accordingly. We suggest that CNN3 takes part in cell projection and movement, and the detection and distribution of CNN3 may render prognostic information for predicting peritoneal seeding metastasis from primary colorectal cancer.
Introduction
Colorectal cancer (CRC) is the second most common cause of cancer-related deaths in developed countries. In Korea, CRC incidents are increasing annually as the third most common cancer [1]. Peritoneal metastasis is one of the major patterns of unresectability in CRC and a cause of death in advanced CRC. In spite of improvements in chemotherapy and surgical techniques, prognoses for peritoneal metastasis remain unfavorable [2]. Peritoneal carcinomatosis is considered as a sequence of events that together form a peritoneal metastatic cascade. Initial exfoliation of malignant cells is reported to involve several adhesion molecules, including E-cadherin, CD44, Selectins, and various leukocyte-associated antigens [3], [4]. Intraperitoneal tumor dissemination is partly promoted by tumor-induced mesothelial apoptosis mediated via Fas/FasL mechanism [5]. Attachment to the submesothelial tissue is mainly mediated by adhesion molecules such as ICAM-1, PECAM-1, and VCAM-1, which are expressed by mesothelial cells. Integrin also plays important role in adhesion to the basement membrane [6], [7]. Invasion into the subperitoneal space is induced by HGF/SF produced by mesothelial cells. Subsequent infiltration of the peritoneal-blood barrier occurs via degradation by proteases [8]. Recent genomic profiling studies have identified distinct gene expression patterns, determining CRC spreading to the liver, the peritoneum, or both. For instance, Diep et al. profiled that peritoneal carcinomatoses and liver metastases tend to have more DNA copy number changes than original lesions [9]. Kleivi et al. reported that chromosome arm 5p gains are common in peritoneal carcinomatoses, and 20 genes (including PTGER4, SKP2, and ZNF622) mapping in this region were overexpressed in the tumors [10]. Nevertheless, the complexity of peritoneal metastatic cascade and molecular cross talk between tumor cells and host elements demands for the development of new therapeutic targets for peritoneal seeding [11]. Thus, establishing peritoneal metastatic cell lines and identifying distinct gene expression patterns in the peritoneal seeding compared to the matched primary CRC will improve our understanding of the mechanisms responsible for peritoneal seeding metastases [9], [10].
In this paper, three pairs of primary CRC and corresponding peritoneal seeding cell lines were established and analyzed by the whole exome sequencing and microarray to identify distinct mutational statuses and gene expressions between primary CRC and peritoneal seeding metastasis.
Methods
Cell Line Establishment
Cell lines were established from pathologically proven colorectal carcinomas. Detailed procedure was described previously [12]. The locations and stages of original tumors were listed in Table 1.
Table 1.
Clinicopathologic Characteristics of Three Paired Colon Cancer Cell Lines
|
In Vitro |
In Vivo |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Growth |
Doubling |
Cell |
Sex | Age | Original Site of Cell Line | Pathology | TNM |
MSI |
|
| Pattern | Time (h) | Morphology | Stage | Status | |||||
| SNU-2335A | Adherent | 45 | Stellate | F | 55 | Sigmoid colon mass | M/D Adenocarcinoma | T3N2M1 | MSS |
| SNU-2335D | Adherent | 48 | Polygonal | F | 55 | Omental seeding nodule | T3N2M1 | MSS | |
| SNU-2335E | Adherent | 40 | Polygonal | F | 55 | Krukenberg tumor mass | T3N2M1 | MSS | |
| SNU-2404A | Adherent | 55 | Polygonal | F | 62 | Ascending colon mass | M/D Adenocarcinoma | T4bN2M1 | MSS |
| SNU-2404B | Adherent/ | 67 | Round/Polygonal | F | 62 | Omental seeding nodule | T4bN2M1 | MSS | |
| floating | |||||||||
| SNU-2414A | Adherent | 45 | Polygonal | M | 76 | Sigmoid colon mass | P/D Adenocarcinoma | T4bN2M1 | MSS |
| SNU-2414B | Adherent | 29 | Polygonal | M | 76 | Omental seeding nodule | T4bN2M1 | MSS | |
M/D, moderately differentiated; P/D, poorly differentiated; MSS, microsatellite stable.
DNA Fingerprinting Analysis
The genomic DNA from each cell line was amplified using the AmpFlSTR identifiler PCR amplification kit (Applied Biosystems, Foster City, CA). Detailed procedure was described previously [12]. The STR profiles of established peritoneal metastasis cancer cell lines and their parental colorectal cancer cell lines are listed in supplementary Table 23.
Cell Growth Properties
Suspensions of 5×104 cells were added to 35-mm tissue culture dishes containing culture medium. The number of cells was counted in triplicate at 24-hour intervals for at least 10 days. The doubling time of the cells was calculated from the growth phase. Cell viability was determined with 0.4% trypan blue dye, and the number of viable cells was counted using countess automated cell counter (Invitrogen, Carlsbad,CA). The morphology of cells grown in T-75-cm2 culture flasks was observed daily by phase-contrast microscopy. In addition, mycoplasma contamination was tested by the 16S-rRNA gene-based PCR amplification method using the e-Myco Mycoplasma PCR detection kit (Intron Biotechnology, Gyeonggi, Korea).
Cell Cultures
Forty-eight human CRC cell lines (SNU-61, SNU-70, SNU-81, SNU-175, SNU-254, SNU-283, SNU-407, SNU-479, SNU-503, SNU-769A, SNU-769B, SNU-796, SNU-977, SNU-1033, SNU-1040, SNU-1047, SNU-1181, SNU-1197A, SNU-1235, SNU-1411, SNU-1406, SNU-1544, SNU-1684, SNU-1746, SNU-C1, SNU-C2A, SNU-C4, SNU-C5, Caco-2, COLO201, COLO205, COLO320, DLD1, HCT-8, HCT-15, HCT-116, HT-29, LoVo, LS174T, NCI-H716, SW-403, SW-480, SW-620, SW-1116, WiDr, KM12C, KM12SM, and KM12L4) were obtained from the Korean Cell Line Bank. All cell lines were maintained in RPMI1640 media with 10% fetal bovine serum at 37°C and 5% CO2. All cell lines were tested and authenticated by Korean Cell Line Bank using DNA finger printing analysis.
Nucleic Acid Isolation and Complementary DNA Synthesis
Genomic DNA was extracted from the cell lines using QIAamp DNA Mini Kit (Qiagen, Hilden, Germany), and RNA was extracted using the TRIzol (Life Technologies, Carlsbad, CA) and RNeasy Plus Mini Kit (Qiagen) according to manufacturer’s protocol. For cDNA synthesis, QuantiTect Reverse Transcription Kit (Qiagen) was used. One microgram of total RNA, 2 μl of gDNA Wipeout Buffer, and DEPC water up to 14 μl were mixed together and incubated at 42°C for 2 minutes. The mixture was mixed with Quantiscript RT Buffer, RT Primer Mix, and Quantiscript Reverse Transcriptase and incubated at 42°C for 45 minutes. Finally, the mixture was incubated at 95°C for 2 minutes and cooled down to room temperature.
Microarray Analyses
Total RNA was extracted using TRIzol (Life Technologies, Carlsbad, CA) and purified with the RNeasy Mini Kit (Qiagen, Hilden, Germany). RNA integrity was assessed with an Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA). High-quality RNA (RNA integrity number > 9.0) was used for the gene expression microarray analysis (Affymetrix, Inc., Santa Clara, CA). After 16 hours of hybridization at 45°C, the arrays were washed and stained on a GeneChip Fluidics Station (Affymetrix, Inc.) and scanned using the Gene Chip Scanner 3000 (Affymetrix, Inc.). The CEL intensity data extracted by GCOS (Gene Chip Operating Software) and Affymetrix Expression Console software with the default RMA parameters were used for data analysis.
Whole Exome Sequencing
SureSelect sequencing libraries were prepared according to manufacturer’s instructions (Agilent sureselect all Exon kit 50 Mb) using the Bravo automated liquid handler. Three micrograms of genomic DNA was fragmented to a median size of 150 bp using the Covaris-S2 instrument (Covaris, Woburn, MA). The adapter ligated DNA was amplified by PCR, and the PCR product quality was assessed by capillary electrophoresis (Bioanalyzer, Agilent). The hybridization buffer and DNA blocker mix were incubated for 5 minute at 95°C and then for 10 minutes at 65°C in a thermal cycler. The hybridization mixture was added to the bead suspension and incubated for 30 minutes at RT while mixing. The beads were washed, and DNA was eluted with 50 ml SureSelect elution buffer (Agilent). The flow cell loaded on HISEQ 2500 sequencing system (Illumina).
qRT-PCR
Detailed procedure was described previously [12]. The primers that were used in this study are listed in supplementary table 21.
Protein Isolation and Western Blotting
Cells were harvested with a cell scraper after washing with cold PBS. Whole protein was extracted with EzRIPA buffer (ATTO Co., Tokyo, JAPAN) supplied with 1% protease inhibitor and 1% of phosphatase inhibitor. The protein concentration was calculated by SMART micro BCA protein assay kit (Intron Biotechnology, Gyeonggi, Korea). Equal amounts of protein were loaded on 4%-15% Mini-PROTEAN TGX Precast Gels (BIO-RAD, Hercules, CA) and blotted at 50 V for 2 hours. Proteins were transferred to a Trans-Blot Turbo Transfer Pack (BIO-RAD) using the Trans-Blot Turbo Transfer System V1.02 machine (BIO-RAD) at 2.5 A and 25 V. The membrane was incubated in 2.5% skim milk containing 0.5 % Tween 20 for an hour at room temperature. Primary antibodies against Calponin 3 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) (1:2000), Sorbin and SH3 Domain Containing 1 (Life Technologies, Carlsbad, CA) (1:1000), E-cadherin (Abcam, Cambridge, United Kingdom) (1:1000), and β-actin (Applied Biological Materials Inc., Richmond, BC, Canada) (1:5000) were introduced to the membrane and incubated at room temperature for 1 hour. Peroxidase-conjugated mouse or rabbit IgG antibody (Jackson Immunoresearch, West Grove, PA) (1:5000) was added as a secondary antibody and incubated at room temperature for 1 hour. Chemiluminescent working solution, WESTZOL (Intron Biotechnology), was decanted to the membrane. The membrane was exposed to Fuji RX film for 1-5 minutes.
Knockdown of CNN3 by Short Hairpin RNA Transduction
Forty thousand 293FT cells with 10 ml of DMEM supplied with 10% of FBS and 1% of penicillin streptomycin were seeded on a 100 pi tissue culture dish. After incubating for 24 hours at 37°C and 5% CO2, the culture medium was changed to 10 ml of Opti-MEM, and short hairpin RNA targeting CNN3 and control vector were treated using ViraSafe Lentiviral Packaging System, Pantropic (CELL BIOLABS, INC., San Diego, CA), and Lipofectamine 3000 (Invitrogen) in accordance with manufacturer’s protocol. After 48 hours, the viral soup was harvested and filtered through a 0.45-μm pored filter (Sartorius Stedim Biotech SA, Göttingen, Germany). The harvested viral soup was aliquoted into a 1.5-ml tube and kept at −70°C. Fifty thousand peritoneal metastatic cells were seeded on 24-well tissue culture plate with 0.5 ml of RPMI1460 medium and incubated at 37°C in an atmosphere of 5% CO2 and 95% air for 24 hours. Viral transduction was performed using ViraDuctin (CELL BIOLABS, INC., San Diego, CA) according to manufacturer’s protocol.
Confocal Assay
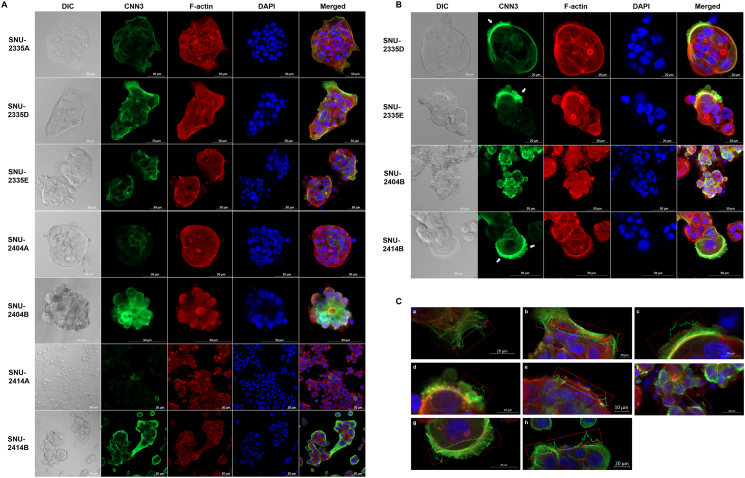
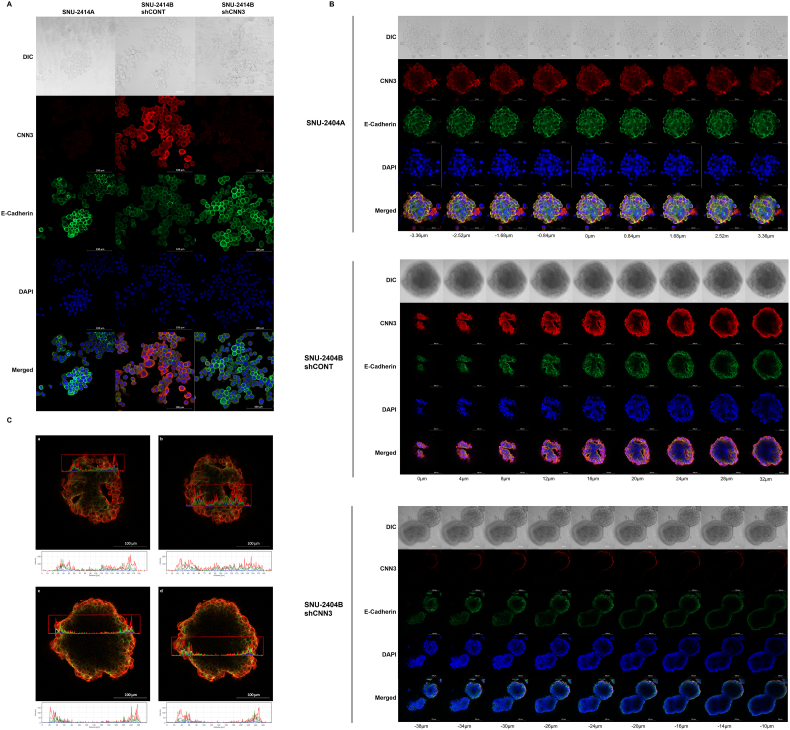
Four thousand cells were seeded on chambered coverglass (Thermo Fisher Scientific, Waltham, MA). The chambered coverglass was designed to be hydrophilic, and no ECM component was treated before seeding. Once 70% confluency had been reached, cells were washed with cold DPBS three times. Then, cells were fixed and permeabilized with BD Cytofix/Cytoperm (BD Science, San Jose, CA). After cells were washed with washing solution (BD Science), DPBS containing 2% FBS (GE Healthcare Life Sciences, Buckinghamshire, UK) was applied for an hour. After cells were washed with cold DPBS, Calponin 3 (Santa Cruz Biotechnology, Santa Cruz, CA) (1:500) and E-cadherin antibody (Abcam, Cambridge, United Kingdom) (1:400) diluted in 0.05% of PBST was applied for 1.5 hours in room temperature. Thereafter, cells were washed with 0.05% of PBST, and Alexa 488 and Alexa 568 secondary antibodies (Thermo Fisher Scientific, Waltham, MA) (1:500) diluted in 0.05% of PBS.T were applied for an hour in room temperature. DAPI and Rhodamine-conjugated Phalloidin (Sigma-Aldrich, MO, USA) were diluted in distilled water and applied for 30 minutes in room temperature. Cells were washed with DPBS three times and applied under confocal microscope. LSM800 Confocal Laser Scanning Microscope and ZEN software (Carl Zeiss, Oberkochen, Germany) were used to analyze images. Digital resolution, scan speed, and the number of pictures averaged were set to 1024 × 1024, 40 seconds per one channel, and 8 pictures, respectively. Diverse magnifications were used in accordance with growth patterns and sizes of cells. Scale bar is inserted into each image processed. The intensity of each channel was fixed for comparing target protein expression between samples. The pictures were focused on the bottom of the fixed cells for investigating protruding region of cell colonies and the location of Calponin 3 and E-cadherin. Z-stacking was applied for SNU-2404 cell lines due to their aggregating nature. Distances from the initial focal point are indicated separately in Figure 3.
Figure 3.
The expression of Calponin 3 is enhanced at the edge of the colonized cell population. (A) Confocal images represent relative expression and localization of Calponin 3 (green) in vitro. Red and blue indicate F-actin and nucleus, respectively. Calponin 3 was distinct at the edge of the colonized cell population. Scale bar is inserted into each image. (B) Confocal microscopy of metastatic peritoneal seeding cell lines. Metastatic cancer cell lines showed recruitment of Calponin 3 at the protruding regions of the colonized cell population (indicated by arrow). (C) Enlarged images from A and B indicated that the intensity of Calponin 3 (green) signal is prominent compared to F-actin (red) signal. (a) SNU-2335A. (b) (c) SNU-2335D. (d) SNU-2335E. (e) SNU-2404A. (f) SNU-2404B. (g, h) SNU-2414B. (SNU-2414A was omitted since the intensity of Calponin 3 was too low).
Gap Closure Migration Assay
Gap closure migration assay was performed using CytoSelect 24-Well Wound Healing Assay kit (CELL BIOLABS, INC., San Diego, CA) according to manufacturer’s protocol. Briefly, SNU-2414B naïve, SNU-2414B shControl, and SNU-2414B shCNN3 cells were diluted to 2 × 105 cells/ml in RPMI1640 supplied with 10% of FBS and 1% of penicillin streptomycin. Once the inserts provided in the assay kit were put into each well of 24-well tissue culture plate, 500 μl of previously diluted cell solution was dispensed to each side of the insert, making a total of 1 ml of cell solution each well. After 24 hours of incubation at 37°C in an atmosphere of 5% CO2 and 95% air, inserts were removed. Immediately, cells were washed with complete media twice. With 24-hour interval for 5 days, the wound closing process was recorded under phase contrast microscope. The closed wound area was calculated with ImageJ version 1.8.0. The whole procedures were independently triplicated, and average closed wound area was analyzed using GraphPad Prism version 7.00 for Windows (GraphPad Software, La Jolla, CA).
3D Spheroid Cell Invasion Assay
3D spheroid cell invasion assay was performed using Cultrex 96-Well 3D Spheroid BME Cell Invasion Assay kit (Trevigen, Gaithersburg, MD) according to manufacturer’s protocol. Dilute 1 × 106 cells per ml in cell culture medium. 10× Spheroid Formation ECM is thawed on ice at 4°C and diluted with RPMI1640 supplied with 10% of FBS and 1% of penicillin streptomycin chilled to 4°C. Spheroid Formation ECM solution is composed of proprietary mixture of extracellular matrix proteins derived from murine EHS sarcoma cells. Cells are resuspended in 1× Spheroid Formation ECM, and 50 μl of cell suspension is added to each well of the 3D Culture Qualified 96 Well Spheroid Formation Plate. Centrifuge at 200×g for 3 minutes at room temperature in a swinging bucket rotor, and incubate at 37°C in a tissue culture incubator for 72 hours to promote spheroid formation. After 72 hours, the 3D Culture Qualified 96 Well Spheroid Formation Plate is placed on ice for 15 minutes to cool wells. Working on ice, 50 μl of Invasion Matrix is added per well of the 3D Culture Qualified 96 Well Spheroid Formation Plate. The plate is centrifuged at 300×g at 4°C for 5 minutes in a swinging bucket rotor to eliminate bubbles and position spheroids within the Invasion Matrix towards the middle of the well. The plate is transferred to a tissue culture incubator set at 37°C for 1 hour to promote gel formation of the Invasion Matrix. After 1 hour, 100 μl of warm (37°C) cell culture medium is added containing 10% FBS as the chemoattractant. The spheroid in each well was photographed every 24 hours for 5 days under phase contrast microscope using the 4× objective. Images were analyzed using ImageJ version 1.8.0 (http://rsb.info.nih.gov/ij/). The area of the spheroid structure (μm2) was analyzed using GraphPad Prism version 7.00 for Windows, (GraphPad Software, La Jolla, CA). The entire procedures were independently triplicated.
Statistical Analysis
Independent replicates refer to independent cell samples seeded, treated, and imaged on different days. Differences between effects of shCONT and shCNN3 were assessed with two-way ANOVA test. P values < .05 were considered statistically significant and indicated with asterisks using GraphPad Prism version 7.00 for Windows (GraphPad Software, La Jolla, CA). Microarray visualization and analysis were performed using R program version 3.3.1 (R Foundation for Statistical Computing, Vienna, Austria) with various packages including iplot, gplot, lattice, and RCircos. The microarray data were evaluated using Kruskal-Wallis test and Mann-Whitney U test with a Bonferroni correction for pathway analysis. The analysis for different mRNA species was performed using a two-tailed Spearman’s rank correlation test, and differences were considered statistically significant at *P < .05.
Results
General Characteristics of Cell Lines
The clinicopathologic characteristics of the cell lines are summarized in Table 1. The morphologies of each cell lines are described in figure legends (Figure 1A). All cell lines were free from bacteria or Mycoplasma contamination, and not cross-contaminated (Supplementary Table 23).
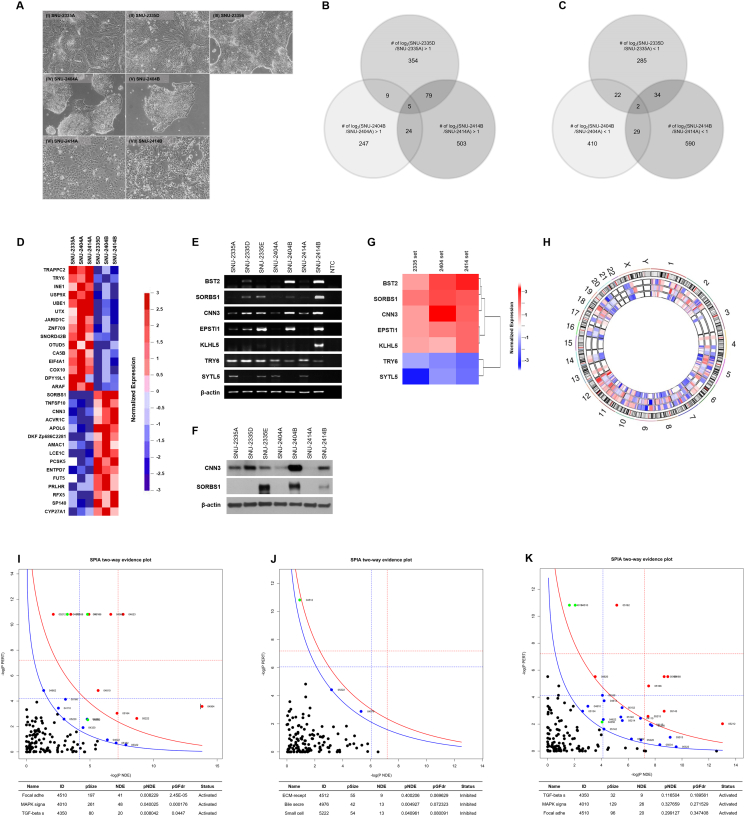
Figure 1.
Selection and verification of distinct gene expressions and aberrated signaling pathways in peritoneal seeding cell lines. (A) Phase-contrast microscopy of paired colorectal cancer cell lines before being applied to whole exome sequencing and microarray. The majority of tumor cells displayed a polygonal shape and had round-to-oval nuclei with prominent single-to-double nucleoli. (AI) SNU-2335A, (AII) SNU-2335D, and (AIII) SNU-2335E grew as monolayers composed of epithelial cell islands. (AIV) SNU-2404A grew as tightly packed floating cell aggregates. (AV) SNU-2404B, which was derived from ascending colon mass, grew as irregular floating clumps. (AVI) SNU-2414A and (AVII) SNU-2414B proliferated as islands of tightly packed epithelial cells and eventually formed dense monolayers. (B) Venn diagram shows the overlap of upregulated genes and (C) downregulated genes using two-fold change threshold. (D) Heatmap indicating classified primary and metastatic cell lines was drawn with 156 probe sets having *P < .05 by using paired t test. Among them, 15 genes with the highest score in primary and metastatic classes were identified. (E) The mRNA expressions of classified seven genes in established primary CRC and peritoneal cancer cell lines. (F) The protein levels of CNN3 and SORBS1 in the primary CRC and peritoneal cancer cell lines. (G) Heatmap represents the selected seven genes that were differently expressed in all three metastatic CRC cell lines. Expression level was converted to log2 scale. Each selected gene was clustered in accordance with similarity of expression levels. (H) Circos plot shows chromosomal area where certain sets of genes were up- or downregulated in all three metastatic cell lines. The outermost circle represents the entire human chromosome (hg19). The second, third, and fourth outermost circles represent chromosomal regions of SNU-2335, SNU-2404, and SNU-2414 sets, respectively. (I) SPIA evidence plot for the microarray result of SNU-2335 set. Each pathway is represented by one dot. The pathways at the right of the red curve are significant after Bonferroni correction. The pathways at the right of the blue curve line are significant after a FDR correction of the global P values. (J) SPIA evidence plot for the microarray result of SNU-2404 set. Each pathway is represented by one dot. The pathways at the right of the red curve are significant after Bonferroni correction. The pathways at the right of the blue curve line are significant after a FDR correction of the global P values. (K) SPIA evidence plot for the microarray result of SNU-2414 set. Each pathway is represented by one dot. The pathways at the right of the red curve are significant after Bonferroni correction. The pathways at the right of the blue curve line are significant after a FDR correction of the global P values.
Whole Exome Sequencing Data Analysis
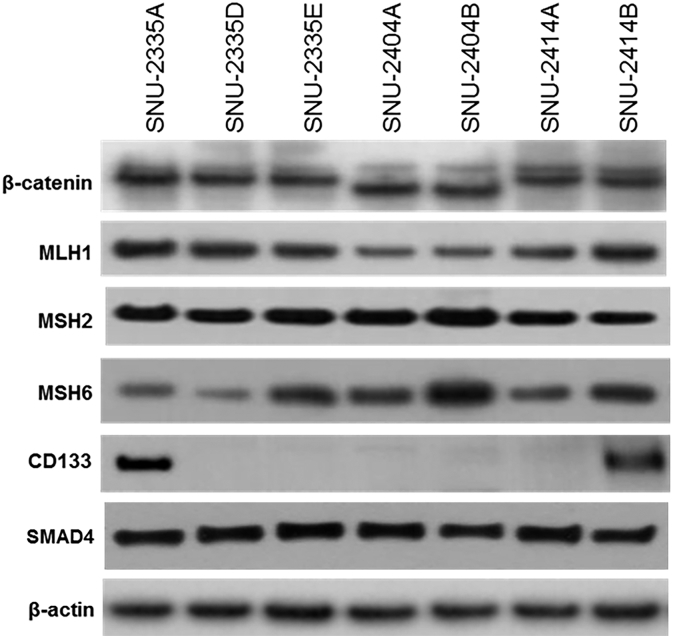
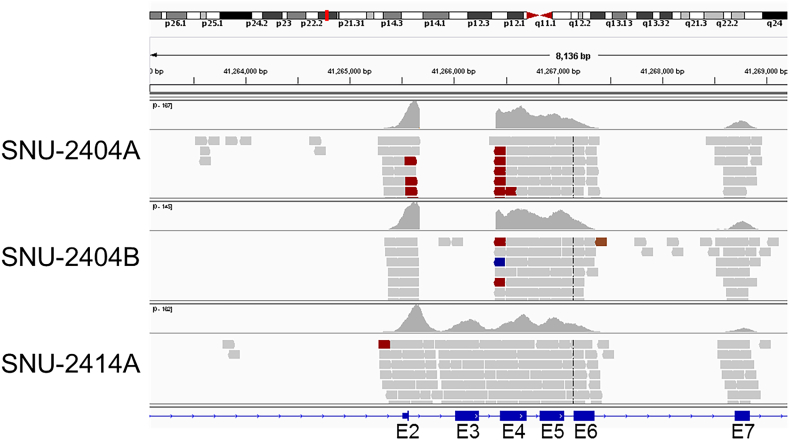
Thirty-one fundamental genes in developing CRC are selected from referring to previous research [13] (reference genes are listed in supplementary Table 23). SNU-2404 pair had the whole deletion of exon 3 in CTNNB1 as confirmed by Western blotting and IGV program (Supplementary Figure 1, Supplementary Figure 2). None of the TP53 mutations was located in hot spots containing CpG dinucleotides, and all mutations were homozygous. Detailed results of the whole exome sequencing are summarized in Supplementary Tables 1-5. Moreover, shared mutations in peritoneal metastasis cell lines were screened (Supplementary Tables 6-8). Only one missense mutation of CYP2A7 (c.181T>A/p.Phe61Ile) was exclusively shared in all three peritoneal metastatic cancer cell lines.
Supplementary Figure 1.
Western blotting images of primary CRC and peritoneal metastasis cancer cell lines. Established cell lines were examined to estimate the expression of mismatch repair proteins (MLH1, MSH2 and MSH6), SMAD4, β-catenin, and CD133.
Supplementary Figure 2.
IGV view of SNU-2404A, SNU-2404B, and SNU-2414A cell lines. IGV program was used to discover exon 3 deletion of CTNNB1 in SNU-2404A and SNU-2404B cell lines. SNU-2414A cell line was used as positive control.
Microarray Data Analysis
Two-folds change classification was used to select distinctly expressed genes in peritoneal metastasis cell lines. The number of up- and downregulated genes with their intersection between two different peritoneal metastasis cell lines is summarized in Figure 1, B and C. The number of upregulated genes in the intersection between SNU-2404 set and other two pairs was much lower than that of shared genes between SNU-2335 set and SNU-2414 set (Figure 1B). It was reencapsulated in the circos-plot presenting distinct mRNA expressions of classified genes (Figure 1H). There were multiple genomic regions such as 4q24-4q26, 5p15.33-5p14.1, 6p22.3-6p22.1, and 7p21.3 in which genes identified from SNU-2404 pair were reversely regulated in SNU-2335 and SNU-2414 sets. Given that the adherent growth pattern of SNU-2404A is changed to floating aggregates in SNU-2404B, it could be assumed that SNU-2404A had different kinds of alternation in extracellular matrix binding molecules or/and epithelial mesenchymal transition factors. This prospect leads to pathway analysis to investigate which signaling pathways are aberrated in peritoneal metastasis (Figure 1, I, J, K and Supplementary Figure 5, Supplementary Figure 6, Supplementary Figure 7, Supplementary Figure 8, Supplementary Figure 9, Supplementary Figure 10, Supplementary Figure 11). The result from the pathway analysis certified the assumption that the SNU-2404 set has distinct deregulated pathways from the other two pairs. Both SNU-2335 and SNU-2414 sets had activated focal adhesion, MAPK, and TGF-beta signaling (Figure 1, I and K). On the contrary, SNU-2404 set had inhibited ECM receptor binding, bile secretion, and small cell signaling (Figure 1J). There were five upregulated and two downregulated genes in all three peritoneal metastasis cell lines (Table 2, Figure 1, B and C), and these genes may play a role in connecting diverse deregulated pathways. For those seven genes, a heatmap was drawn to visualize fold changes (Figure 1G). Their expressions in mRNA level were reconfirmed by RT-PCR (Figure 1E). CNN3 became gene of interest since its mRNA level was remarkably augmented in SNU-2404B cells (10.6-folds) compared to that of SNU-2335D (2.5-folds) and SNU-2414B cells (4.2-folds) and was likely to explain the morphologic change of the SNU-2404 pair. To further classify primary cell lines and metastatic cell lines for target gene selection, 156 probe sets with *P<.05 using paired t test were selected. Among them, 15 genes with the highest scores in primary and metastatic classes were identified (Figure 1D). CNN3, SORBS1, and TRY6 were identified in both fold change selection and classification using a probe set, and their protein levels in isolated peritoneal pairs were confirmed with Western blotting. Protein expression of TRY6 was not detectable, and the only results from CNN3 and SORBS1 were shown (Figure 1F). Interestingly, SORBS1 band was absent in SNU-2335D, while it was highly increased in SNU-2335E, which is an ovarian metastasis cancer cell line. On the other hand, CNN3 band was clearly amplified in SNU-2335D compared to SNU-2335A, and SNU-2335E had similar expression level with SNU-2335A. Therefore, CNN3 was pursued for specific marker for peritoneal metastasis, and functional study was continued.
Supplementary Figure 5.
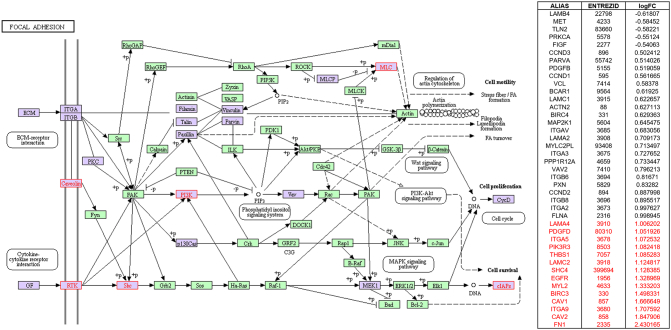
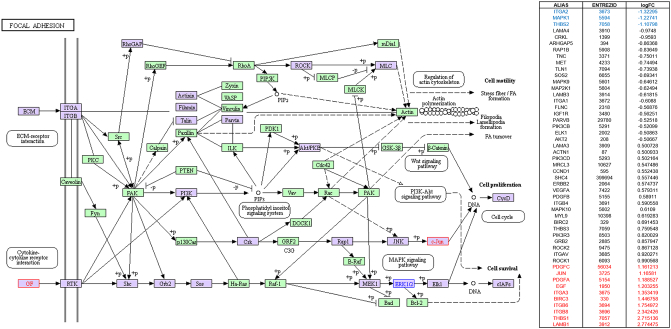
Focal adhesion signaling highlighted in KEGG pathway analysis in parallel with SPIA pathway analysis by using microarray result of SNU-2335 set. Genes that were upregulated more than two-fold were highlighted in red in KEGG diagram and the list from SPIA pathway. Genes that were downregulated more than two-fold were highlighted in blue in KEGG diagram and the list from SPIA pathway. Genes that were up- or downregulated more than 1.5-fold were highlighted in purple in KEGG diagram.
Supplementary Figure 6.
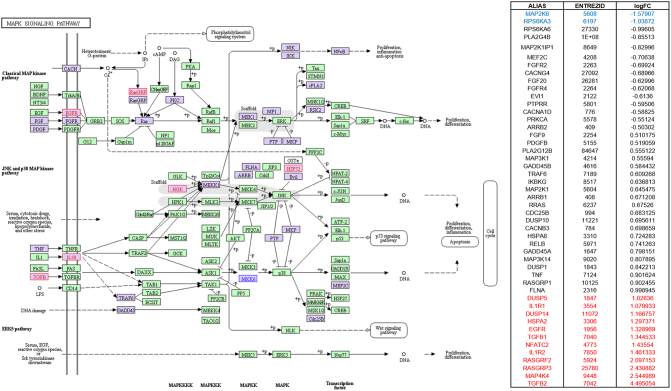
MAPK signaling highlighted in KEGG pathway analysis in parallel with SPIA pathway analysis by using microarray result of SNU-2335 set. Genes that were upregulated more than two-fold were highlighted in red in KEGG diagram and the list from SPIA pathway. Genes that were downregulated more than two-fold were highlighted in blue in KEGG diagram and the list from SPIA pathway. Genes that were up- or downregulated more than 1.5-fold were highlighted in purple in KEGG diagram.
Supplementary Figure 7.
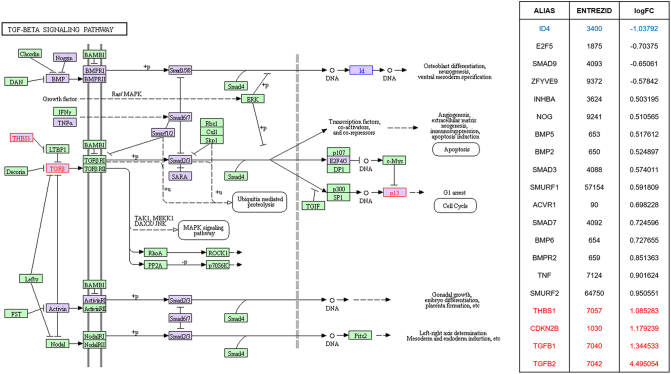
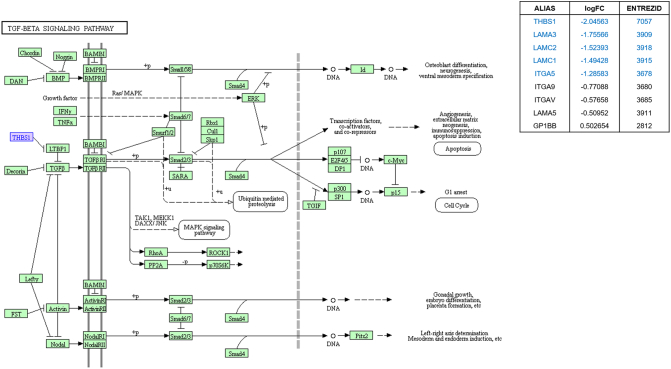
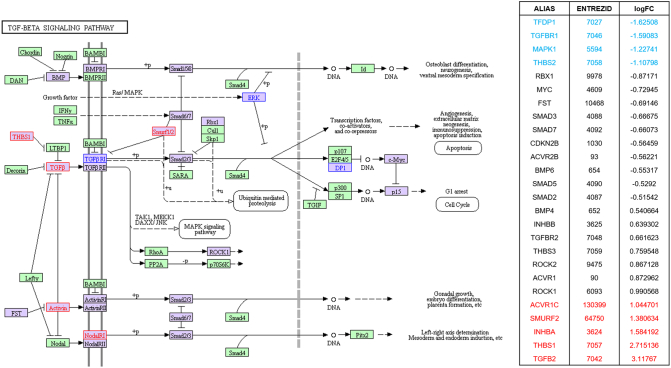
TGF-beta signaling highlighted in KEGG pathway analysis in parallel with SPIA pathway analysis by using microarray result of SNU-2335 set. Genes that were upregulated more than two-fold were highlighted in red in KEGG diagram and the list from SPIA pathway. Genes that were downregulated more than two-fold were highlighted in blue in KEGG diagram and the list from SPIA pathway. Genes that were up- or downregulated more than 1.5-fold were highlighted in purple in KEGG diagram.
Supplementary Figure 8.
TGF-beta adhesion signaling highlighted in KEGG pathway analysis in parallel with SPIA pathway analysis by using microarray result of SNU-2404 set. Genes that were upregulated more than two-fold were highlighted in red in KEGG diagram and the list from SPIA pathway. Genes that were downregulated more than two-fold were highlighted in blue in KEGG diagram and the list from SPIA pathway. Genes that were up- or downregulated more than 1.5-fold were highlighted in purple in KEGG diagram.
Supplementary Figure 9.
Focal adhesion signaling highlighted in KEGG pathway analysis in parallel with SPIA pathway analysis by using microarray result of SNU-2414 set. Genes that were upregulated more than two-fold were highlighted in red in KEGG diagram and the list from SPIA pathway. Genes that were downregulated more than two-fold were highlighted in blue in KEGG diagram and the list from SPIA pathway. Genes that were up- or downregulated more than 1.5-fold were highlighted in purple in KEGG diagram.
Supplementary Figure 10.
MAPK signaling highlighted in KEGG pathway analysis in parallel with SPIA pathway analysis by using microarray result of SNU-2414 set. Genes that were upregulated more than two-fold were highlighted in red in KEGG diagram and the list from SPIA pathway. Genes that were downregulated more than two-fold were highlighted in blue in KEGG diagram and the list from SPIA pathway. Genes that were up- or downregulated more than 1.5-fold were highlighted in purple in KEGG diagram.
Supplementary Figure 11.
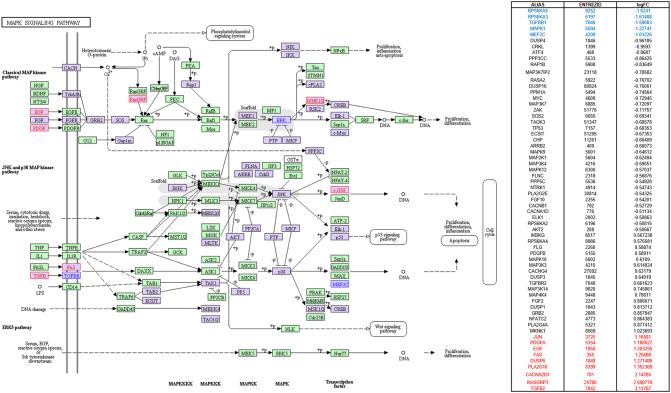
TGF-beta signaling highlighted in KEGG pathway analysis in parallel with SPIA pathway analysis by using microarray result of SNU-2414 set. Genes that were upregulated more than two-fold were highlighted in red in KEGG diagram and the list from SPIA pathway. Genes that were downregulated more than two-fold were highlighted in blue in KEGG diagram and the list from SPIA pathway. Genes that were up- or downregulated more than 1.5-fold were highlighted in purple in KEGG diagram.
Table 2.
Genes Selected from Microarray Data Analysis
| Probe Set ID | Symbol | Gene Description | GO Term Function | Fold Change |
||
|---|---|---|---|---|---|---|
| SNU-2335D | SNU-2404B | SNU-2414B | ||||
| Upregulated genes in all three cell lines | ||||||
| 7917885 | CNN3 | calponin 3, acidic | actin binding | 2.5 | 10.6 | 4.2 |
| 7935188 | SORBS1 | sorbin and SH3 domain containing 1 | actin binding | 3.5 | 5.3 | 4.6 |
| 7971296 | EPSTI1 | epithelial stromal interaction (breast) | 2.5 | 3.1 | 3.9 | |
| 8035304 | BST2 | bone marrow stromal cell antigen 2 | signal transducer activity | 2.5 | 5.5 | 8.2 |
| 8094625 | KLHL5 | kelch-like 5 (Drosophila) | actin binding | 2.3 | 2.1 | 4 |
| Probe Set ID | Symbol | Gene Description | Function | Fold Change |
||
| SNU-2335D | SNU-2404B | SNU-2414B | ||||
| Downregulated genes in all three cell lines | ||||||
| 8136790 | TRY6 | trypsinogen C | -2.2 | -2.6 | -4 | |
| 8166747 | SYTL5 | synaptotagmin-like 5 | protein binding | -8.5 | -2.7 | -3.6 |
Log2-fold change was computed based on the mRNA expressions of metastatic CRC cell lines (SNU-2335D, SNU-2404B, and SNU-2414B) compared to primary CRC cell lines (SNU-2335A, SNU-2404A, and SNU-2414A).
CNN3 Expression Analysis in Colorectal Cancer Cell Lines by RT-PCR and Western Blotting
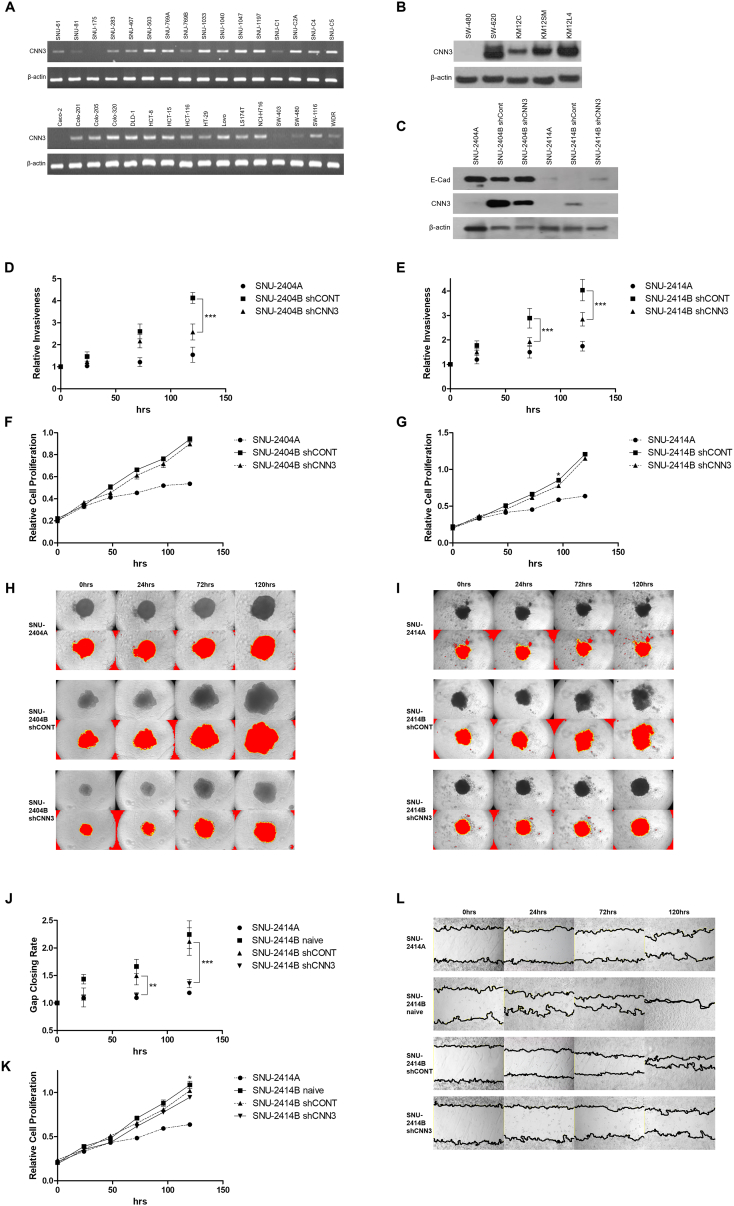
The expression of CNN3 in mRNA level was examined in 32 CRC cell lines (Figure 2A). Of the used 32 CRC cell lines, molecular profiles of 14 CRC cell lines were deposited in the canSAR database (https://cansar.icr.ac.uk). Mutational features of 14 cell lines were analyzed in accordance with their mRNA levels and described in the discussion section. The mRNA level of CNN3 was distinctly higher in SNU-769A (lymph node metastasis) than SNU-769B (primary CRC tissue). In order to verify if the Calponin 3 is amplified in other metastasis pair of CRC cell lines, SW-480, SW-620, KM12C, KM12SM, and KM12L4 cell lines were obtained from Korean Cell Line Bank. Consistently, Calponin 3 was increased in metastatic cancer cell lines (Figure 2B).
Figure 2.
The potential involvement of Calponin 3 in cell invasion and migration. (A) The mRNA level of CNN3 in 32 previously reported colorectal cancer cell lines. Amplified CNN3 fragments were absent in 3 of the 32 CRC cell lines (i.e., in SNU-175, Caco-2, and SW-403). Of the remaining 29 cell lines, 10 (SNU-61, SNU-81, SNU-283, SNU-407, SNU-769B, SNU-C1, Colo-201, HT-29, SW-480, and WiDr) had relatively low expressions, whereas 19 of the CRC cell lines (SNU-503, SNU-769A, SNU-1033, SNU-1040, SNU-1047, SNU-1197, SNU-C2A, SNU-C4, SNU-C5, Colo-205, Colo-320, DLD1, HCT-8, HCT-15, HCT-116, LoVo, LS174T, NCI-H716, and SW-1116) exhibited strong expressions. (B) The protein level of CNN3 in two pairs of metastasis colorectal cancer cell lines. Calponin 3 was increased in metastatic CRC cell lines (SW-620, KM12SM, and KM12L4) than primary CRC cell lines (SW-480 and KM12C). (C) The efficacy of short hairpin RNA to knockdown Calponin 3 was accessed with Western blotting. The E-cadherin level was confirmed in comparison to Calponin 3. E-cadherin and Calponin 3 were inversely related. The result of 3D spheroid cell invasion assay indicated that knockdown of Calponin 3 resulted in decreased invasiveness in both (D) SNU-2404 and (E) SNU-2414 set. Cell proliferation was measured in parallel with 3D spheroid cell invasion assay to distinguish invasion ability from the growth rate in both (F) SNU-2404 and (G) SNU-2414 set. Error bars and P values are calculated with independently repeated experiments (N = 3, *P<.05, ***P<.001) Besides, peritoneal metastasis cells with downregulated Calponin 3 formed spheroid in different shape from original peritoneal metastasis cells. Cells were aggregated more tightly, and the spheroid maintained intact round shape compared to original peritoneal metastasis cells in both (H) SNU-2404 and (I) SNU-2414 set. (J, L) Downregulating Calponin 3 in SNU-2414B hindered the wound closing rate compared to SNU-2414B naive and SNU-2414B with control vector. The experiment was triplicated and averaged. (K) Cell proliferation was measured in parallel with wound healing assay to distinguish migration ability from growth rate. The experiment was triplicated and averaged. Error bars and P values are calculated with independently repeated experiments (N = 3, *P<.05, **P<.01, ***P<.001)
3D Spheroid Cell Invasion Assay and Gap Closure Migration Assay
In order to further investigate the role of Calponin 3 in peritoneal metastasis cancer cells, short hairpin RNA targeting CNN3 (shCNN3) was transducted to the peritoneal metastasis cell lines. SNU-2335 pair was excluded since knockdown of Calponin 3 caused cell death. 3D spheroid cell invasion assay signified that knockdown of Calponin 3 resulted in decreased invasiveness in both SNU-2404 (Figure 2D) and SNU-2414 pairs (Figure 2E). Cell proliferation was measured in parallel with 3D spheroid cell invasion assay to distinguish invasion ability from the growth rate in both SNU-2404 (Figure 2F) and SNU-2414 set (Figure 2G). Besides, peritoneal metastasis cells with downregulated Calponin 3 aggregated more tightly, and the spheroid maintained intact round shape compared to parental peritoneal metastasis cells (Figure 2, H and I). SNU-2404 set was ruled out for gap closure migration assay for their floating aggregate nature. In parallel with 3D spheroid cell invasion assay, downregulating Calponin 3 in SNU-2414B hindered the wound closing rate compared to SNU-2414B naive and SNU-2414B with control vector (Figure 2, J and L). Entire procedure was independently triplicated and averaged. Cell proliferation was measured in parallel with wound healing assay to distinguish migration ability from growth rate (Figure 2K).
Calponin 3 Analysis in Established CRC Cell Lines by Immunofluorescence
The location and relative expression of Calponin 3 were examined by immunofluorescence. Consistent with prior findings, cell lines derived from peritoneal metastasis CRC had augmented Calponin 3 compared to the primary CRC cell lines (Figure 3A). In SNU-2335 and SNU-2414 sets, colocalization of Calponin 3 and F-actin was mostly restricted to intercellular junction, and Calponin 3 was most prominent at the edge of the colonized cell population (Figure 3, A, B and C).
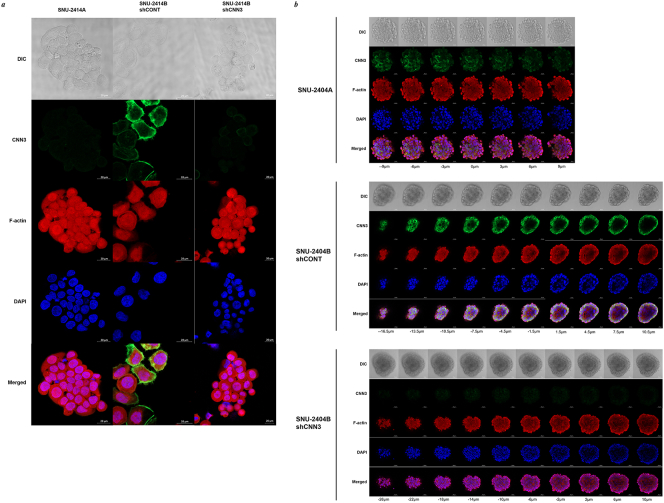
Referring to 3D invasion assay, knockdown of Calponin 3 resulted in tightly packed spheroid in both SNU-2404B and SNU-2414B, which led to the potential relationship of Calponin 3 and E-cadherin. Remarkably, knockdown of Calponin 3 in SNU-2414B cell line increased E-cadherin level in both Western blotting (Figure 2C) and immunofluorescence assay (Figure 4A). This was observed in SNU-2404B cell line in Western blotting as well (Figure 2C). To obtain entirety of floating SNU-2404B cell colonies, Z-stacking was applied. Within a single spheroid population, Calponin 3 was expressed in outer region of the SNU-2404B shCONT aggregate compared to E-cadherin (Figure 4, B and C). SPIA pathway analysis indicated SNU-2404 set had inhibited ECM binding signaling (Figure 1J), and this result may imply that Calponin 3 plays a role in impeding ECM binding of SNU-2404B cells. Downregulating Calponin 3 barely affected F-actin structure in both SNU-2404 and SNU-2414 sets (Supplementary Figure 4, A and B). Nevertheless, it could be observed that SNU-2414B shCNN3 cells had more circular shape than SNU-2414B shCONT cells (Supplementary Figure 4A).
Figure 4.
Immunofluorescence was performed to examine the relationship between Calponin 3 and E-cadherin. (A) Knockdown of Calponin 3 in SNU-2414B cell line increased E-cadherin expression. (B) To obtain entirety of floating SNU-2404B cell colonies, Z-stacking was applied. Within a single spheroid population, Calponin 3 was mostly located in the outer shell of the spheroid, and E-cadherin remained in the inner shell of the aggregate. (C) The intensity of Calponin 3 (Red) and E-Cadherin (Green) signals was measured and graphed through one axis. (a, b) Two random points were selected on the mid-transverse section of SNU-2404B shCONT spheroid image (16 μm). Peaks on the graph indicated that Calponin 3 signal is located in the outer region compared to E-cadherin signal. (c, d) Two random points were selected on the bottom-transverse section of SNU-2404B shCONT spheroid image (32 μm). Peaks on the graph indicated that Calponin 3 signal is located outer region compared to E-cadherin signal.
Supplementary Figure 4.
In terms of cell structure, downregulating Calponin 3 barely affected F-actin in both (A) SNU-2414 and (B) SNU-2404 sets. Nevertheless, it could be observed that SNU-2414B shCNN3 cells remained round in shape, and the protruding region at the edge of the cells was significantly diminished compared to SNU-2414B shCONT.
Discussion
Recently, there have been many reports regarding the use of novel gene markers for diagnostic tools, chemotherapeutic targets, and predicting prognosis [14], [15], [16], [17], [18]. Although genetic studies of colorectal carcinogenesis have focused on the differences between normal colonic mucosa and primary carcinomas [14], there are few studies on the genetic alteration of primary carcinomas to peritoneal metastases. Diep et al. [9] profiled that peritoneal carcinomatoses and liver metastases usually have more DNA copy number changes than original lesions. Kleivi et al. reported that chromosome arm 5p gains are common in peritoneal carcinomatoses, and 20 genes (including PTGER4, SKP2, and ZNF622) mapping in this region were overexpressed in the tumors [10]. The purpose of this study was to find novel markers for identifying peritoneal metastases in CRC. We identified 117 upregulated genes (Supplementary Table 19) and 87 downregulated genes (Supplementary Table 20) in more than two metastatic cell lines by microarray. Five upregulated (CNN3, SORBS1, BST2, EPSTI1, and KLHL5) and two downregulated genes (TRY6 and STYL5) were differentially expressed in all three metastatic cell lines (Table 2).
Microarray Data Analysis
According to Gene Ontology term analysis, three upregulated genes (CNN3, SORBS1 and KLHL5) in the three peritoneal metastasis cell lines were associated with actin binding (Table 2). KLHL5 was only present in SNU-2414B, whereas CNN3 and SORBS1 were detected in all peritoneal metastasis cell lines (Figure 1E). KLHL5 gene was reported to be related with various cellular processes, such as actin cytoskeleton interaction, cytoplasmic sequestration of transcription factors, and cell morphology [19]. Given that the growth pattern of SNU-2414B (monolayer) was different from that of SNU-2335D (multilayer) and SNU-2404B (floating), KLHL5 might be related to the contact inhibition. The exclusive expression pattern of BST2 and STYL5 in SNU-2335 triple was noteworthy. BST2 was present only in SNU-2335D, while STYL5 was expressed in SNU-2335A and SNU-2335E (Figure 1E). It was reported that the expression of STYL5 in mRNA level was increased in invasive breast carcinoma in response to estradiol, and it affected diverse aspects of vesicle trafficking including exocytosis [20]. Given that SNU-2335E was derived from a Krukenberg tumor mass, estradiol may be responsible for the SYTL5 expression in SNU-2335E. Furthermore, SYTL5 is known as an effector molecule that specifically binds and regulates the GTP-bound form of the GTPase RAB27A/B [21]. Recently, Dong W et al. reported that decreased expression of RAB27A/B correlates with metastasis in colorectal cancer [22]. SYTL5 was exclusively absent in all peritoneal metastasis cell lines in mRNA level (Figure 1E). Our findings may indicate that SYTL5 deregulates RAB27A/B in primary CRC and promotes proceeding to peritoneal metastasis. BST2 has been reported to mediate metastasis in several types of cancer [23], [24] Walter-Yohrling et al. revealed that BST2 expression was highly augmented in metastatic ovarian cancer [25]. Nevertheless, in SNU-2335 set, BST2 expression was selectively increased in peritoneal metastasis, and SNU-2335E was analogous to SNU-2335A in both microarray and RT-PCR. The absence of BST2 in Krukenberg tumor mass is to be elucidated.
Whole Exome Sequencing Data Analysis
Mutations that were previously reported in developing metastatic colorectal cancer were screened. The proposed functions of mutations and references are listed in Supplementary Table 9 [26], [27], [28], [29], [30], [31], [32]. CTNNB1 codes β-catenin, which plays a major role in Wnt signaling, and its status is closely related to distant metastasis. SNU-2335A harbored c.1974C>A/p.Asp658Glu missense mutation in CTNNB1, which was not detected in its paired metastatic cell lines. The c.1974C>A/p.Asp658Glu missense mutation was reported previously, but its functional role has not been discovered. Although all cell lines had MSH3 missense mutations, cells were identified as microsatellite stable. According to dbSNP database, clinical significance of one mutation (c.235A>G/p.Ile79Val) was known to be benign, and that of other two mutations (c.2846A>G/p.Gln949Arg and c.3133G>A/p.Ala1045Thr) was unknown. Even though c.3133G>A/p.Ala1045Thr mutation was reported to increase cancer risk in multiple organs such as colorectal cancer, breast cancer, prostate cancer, ovarian cancer, and esophageal cancer [33], none was reported to be related with inactivation of MSH3. One missense mutation in the CYP2A7 was shared in all three peritoneal metastatic cell lines. CYP2A7 is expressed mainly in liver tissue, and its role in colorectal cancer remains unclear. We took advantage of publically available cancer cell line database (canSAR) and drew comparison between mutational status of CYP2A7 gene and CNN3 expression level. Although the genomic positions of mutation were various, missense mutations of CYP2A7 were found in SNU-1040 (c.306C>T/p.Gly102Gly), SNU-C5 (c.164A>T/p.Tyr55Phe), and HCT-15 (c.230G>A/p.Arg77Gln) cell lines among 14 available CRC cell lines from the canSAR database. These cell lines had relatively high CNN3 level compared to the rest of 11 cell lines analyzed (Figure 2A). This may imply that mutational status of CYP2A7 gene is connected to augmented CNN3 expression.
We also compared our WES data to findings by Nicholas E. Navin’s lab which reported genomic alterations in distant metastases in a single cell level [34]. They detected 13 mutations that were exclusively found in primary CRC and 37 mutations that were selectively harbored in metastatic cells by deep-exome sequencing. Although genomic positions of each mutation varied, 5 genes were found to have missense mutations in our primary CRC cell lines, and 9 genes were found to have missense mutations in our peritoneal metastasis cell lines (Supplementary Table 10-15). They also reported genes that mediated clonal evolution in the metastatic site by ultra-deep targeted sequencing. We screened the variant allele frequency of those genes in our primary CRC and peritoneal metastasis cell lines (Supplementary Tables 16-18). In SNU-2414 set, PIK3CG and LINGO2 mutations were detected in primary CRC, and SPEN mutation was found in metastasis cell line, which was concordant with their findings. Moreover, variant allele frequencies of three missense mutations in EYS gene (c.5705A>T, c.4256T>C, c.1712A>G) were changed from ~0.5 in SNU-2414A to 1 in SNU-2414B, which may indicate clonal evolution at metastatic site. The potential role of EYS gene in metastasis should be elucidated.
CNN3 as a Potential Metastasis Marker
As one of three isoforms (basic/h1-calponin, neutral/h2-calponin, and acidic/h3-calponin) in the calponin family that functions as calmodulin and F-actin binding partners, Calponin 3 is expressed in smooth muscle and nonmuscle cells [35]. While the role of the calponin family in regulating smooth muscle contractility has been extensively investigated, the function and regulation in nonmuscle cells are much less understood. Recent studies have demonstrated that calponin family plays a regulatory role in nonmuscle cell motility, which makes calponin family as an attractive target for controlling cell proliferation, metastases, and cancer treatment [36].
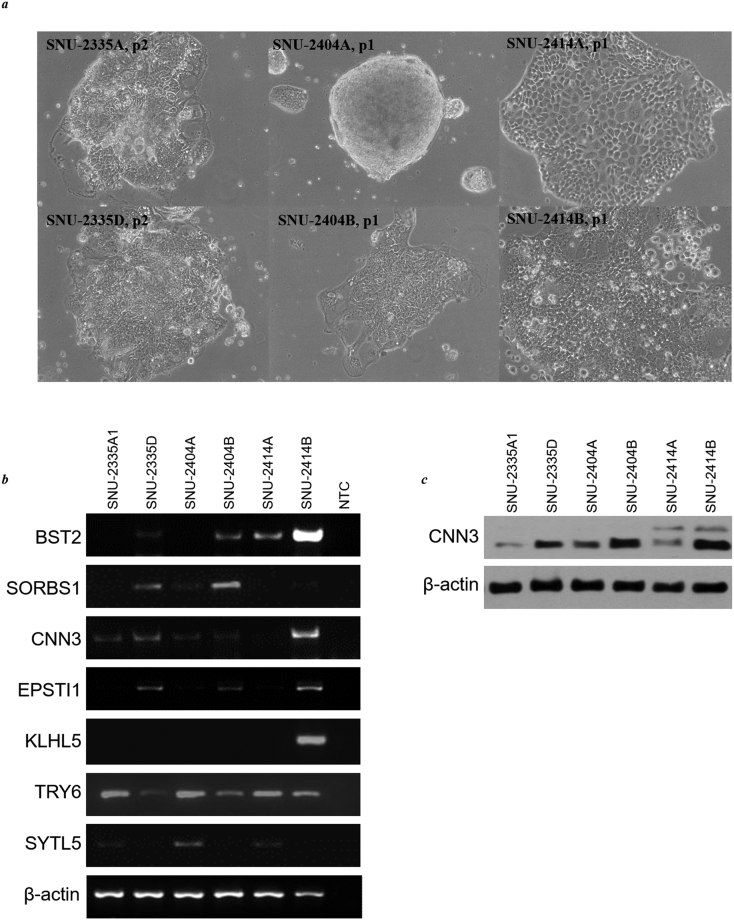
In order to confirm the expression profile in the original histological patient samples, we obtained very initial passages of each cell line (passage 1 or 2) and checked the differential expression of candidates in mRNA level (Supplementary Figure 3B). Expression profiles of all seven identified candidates in the initial passages of each cell line were analogous to passaged samples. Calponin 3 was the main target of our research and was confirmed in protein level as well (Supplementary Figure 3C). In all three pairs, Calponin 3 was augmented in the peritoneal metastasis cell lines.
Supplementary Figure 3.
(A) Phase-contrast microscopy of initially passaged peritoneal colorectal cancer cell lines. (B) The mRNA expressions of classified seven genes in initially passaged peritoneal colorectal cancer cell lines. (C) The protein levels of CNN3 in initially passaged peritoneal colorectal cancer cell lines.
3D spheroid cell invasion assay and Western blotting indicated that the E-cadherin level is inversely related to Calponin 3 expression (Figure 2C). The peritoneal metastatic cascade begins with the detachment of singular or clumps of tumor cells from the primary tumor [37]. Spontaneous exfoliation of malignant cells can be promoted by the downregulation of intracellular adhesion molecules on the tumor cell surfaces, more specifically E-cadherin [4]. Calponin 3 may participate in the initial dissemination of tumor cell from the primary tumor by downregulating the level of E-cadherin. Moreover, confocal images of SNU-2404 and SNU-2414 sets indicated that Calponin 3 was mostly located in outer boundaries, whereas E-cadherin remained at the cellular surface of inner cells within a colonized cell population (Figure 4, A and C). It was previously reported that the Calponin 3 was upregulated in response to mechanical stress such as high fluid pressure around tumor mass [38], [39]. Cells at the outmost region of the cell colonies experience sheer force, which may account for the selective expression of Calponin 3. In terms of the role of Calponin 3 in peritoneal metastasis cascade, the level of Calponin 3 within cells at the outer shell of tumor clumps can be increased by mechanical stress around the tumor mass, and augmented Calponin 3 deregulates the expression of E-cadherin to promote spontaneous tumor cell shedding from the primary tumor. The mechanism of Calponin 3 to decrease E-cadherin level should be further investigated. Moreover, the formation of actin-rich membrane protrusions such as lammelipodia and filipodia is important to peritoneal transport of detached tumor cells [40]. The confocal images of SNU-2414 set indicated that the protruding region at the edge of SNU-2414B shCNN3 was significantly diminished compared to SNU-2414B shCONT (Supplementary Figure 4A). This may imply that CNN3 took part in lamellipodia formation.
Taken together, we propose that the higher Calponin 3 expression may play a role in increasing CRC cell invasion partially due to its association with lower E-cadherin level, and suggest that Calponin 3 can be linked to a positive function in the peritoneal metastasis of CRC.
The following are the supplementary data related to this article.
Supplementary Table 1. Abnormalities of APC and TP53. The c.215C>G/p.Pro72Arg Transversion Mutation Was Previously Reported in the p53 Mutation Database (http://www-p53.iarc.fr) and Catalogue of Somatic Mutations in Cancer (http://cancer.sanger.ac.uk/cosmic) To Be Found in Five Cell Lines (SNU-2335A, SNU-2335D, SNU-2335E, SNU-2414A, and SNU-2414B).
Supplementary Table 2. Abnormalities of MSH3 and MSH6
Supplementary Table 3. Abnormalities of CASP8 and GPC6
Supplementary Table 4. Abnormalities of KRAS and NRAS (W, Wild Type)
Supplementary Table 5. Abnormalities of SMAD4 and β-Catenin (CTNNB1) (W, Wild Type)
Supplementary Table 6. Common Mutations in SNU-2335D and SNU-2404B
Supplementary Table 7. Common Mutations in SNU-2335D and SNU-2414B
Supplementary Table 8. Common Mutations in SNU-2404B and SNU-2414B
Supplementary Table 9. The Proposed Functions of Mutations That Are Previously Reported to Increase CRC Risk and Mediate Metastasis
Supplementary Table 10. WES Result of SNU-2335A Was Analyzed in Reference to the Genes That Had Missense Mutations Specifically in Primary CRC
Supplementary Table 11. WES Result of SNU-2335D Was Analyzed in Reference to the Genes That Had Missense Mutations Specifically in Metastatic Cancer
Supplementary Table 12. WES Result of SNU-2404A Was Analyzed in Reference to the Genes That Had Missense Mutations Specifically in Primary CRC
Supplementary Table 13. WES Result of SNU-2404B Was Analyzed in Reference to the Genes That Had Missense Mutations Specifically in Metastatic Cancer
Supplementary Table 14. WES Result of SNU-2414A Was Analyzed in Reference to the Genes That Had Missense Mutations Specifically in Primary CRC
Supplementary Table 15. WES Result of SNU-2414B Was Analyzed in Reference to the Genes That Had Missense Mutations Specifically in Metastatic Cancer
Supplementary Table 16. Variant Allele Frequency of Genes That Potentially Mediate Clonal Evolution in Metastasis Site in SNU-2335 Set
Supplementary Table 17. Variant Allele Frequency of Genes That Potentially Mediate Clonal Evolution in Metastasis Site in SNU-2404 Set
Supplementary Table 18. Variant Allele Frequency of Genes That Potentially Mediate Clonal Evolution in Metastasis Site in SNU-2414 Set
Supplementary Table 19. Upregulated Genes in More Than Two Sets of Cell Lines
Supplementary Table 20. Downregulated Genes in More Than Two Sets of Cell Lines
Supplementary Table 21. Primer Sequences That Were Used for RT-PCR
Supplementary Table 22. Genes That Were Used for Mutation Analysis
Supplementary Table 23. STR Profile of Peritoneal Omental Seeding Cancer Cell Line
Supplementary Table 24. Aberrated Signaling Pathways in SNU-2335 Set
Supplementary Table 25. Aberrated Signaling Pathways in SNU-2404 Set.
Supplementary Table 26. Aberrated Signaling Pathways in SNU-2414 Set
Ethics Approval and Consent to Participate
The study protocol was approved by the Institutional Review Board of Seoul National University Hospital (IRB No. H-1102-098-357). The study was performed in accordance with the Declaration of Helsinki.
Consent for Publication
Not applicable.
Availability of Data and Materials
Data supporting the results reported in the article can be accessed by the authors upon demand.
Conflict of Interest
The authors declare no conflict of interest.
Authorship
S. C. designed and performed the experiment and write the manuscript. C. W. performed surgical removal of primary and peritoneal metastasis tumor and helped designed the experiment. S. G. helped analyze microarray data. Y. A. helped knock down of Calponin 3. B. C. helped design the invasion assay. Y. K. provided CRC cell lines. S. Y. helped design the experiment. J. L. designed the experiment. J. G. designed the experiment.
Acknowledgements
Not applicable.
Footnotes
Funding: This study was funded in part by the Korean Cell Line Research Foundation and the Priority Research Center Program (2009-0093820) through a National Research Foundation of Korea grant funded by the MSIP. The first author received a scholarship from the BK21-plus education program provided by the National Research Foundation of Korea.
Contributor Information
Ja-Lok Ku, Email: kujalok@snu.ac.kr.
Jae-Gahb Park, Email: jgpark88@gmail.com.
References
- 1.Jung KW, Won YJ, Oh CM, Kong HJ, Lee DH, Lee KH. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2014. Cancer Res Treat. 2017;49:292–305. doi: 10.4143/crt.2017.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shih W, Chetty R, Tsao MS. Expression profiling by microarrays in colorectal cancer (Review) Oncol Rep. 2005;13:517–524. [PubMed] [Google Scholar]
- 3.Brabletz T, Hlubek F, Spaderna S, Schmalhofer O, Hiendlmeyer E, Jung A, Kirchner T. Invasion and metastasis in colorectal cancer: epithelial-mesenchymal transition, mesenchymal-epithelial transition, stem cells and beta-catenin. Cells Tissues Organs. 2005;179:56–65. doi: 10.1159/000084509. [DOI] [PubMed] [Google Scholar]
- 4.Hirohashi S. Inactivation of the E-cadherin-mediated cell adhesion system in human cancers. Am J Pathol. 1998;153:333–339. doi: 10.1016/S0002-9440(10)65575-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heath RM, Jayne DG, O'Leary R, Morrison EE, Guillou PJ. Tumour-induced apoptosis in human mesothelial cells: a mechanism of peritoneal invasion by Fas Ligand/Fas interaction. Br J Cancer. 2004;90:1437–1442. doi: 10.1038/sj.bjc.6601635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bracke ME. Role of adhesion molecules in locoregional cancer spread. Cancer Treat Res. 2007;134:35–49. doi: 10.1007/978-0-387-48993-3_3. [DOI] [PubMed] [Google Scholar]
- 7.Muller J, Yoshida T. Interaction of murine peritoneal leukocytes and mesothelial cells: in vitro model system to survey cellular events on serosal membranes during inflammation. Clin Immunol Immunopathol. 1995;75:231–238. doi: 10.1006/clin.1995.1076. [DOI] [PubMed] [Google Scholar]
- 8.Kataoka H, Tanaka H, Nagaike K, Uchiyama S, Itoh H. Role of cancer cell-stroma interaction in invasive growth of cancer cells. Hum Cell. 2003;16:1–14. doi: 10.1111/j.1749-0774.2003.tb00123.x. [DOI] [PubMed] [Google Scholar]
- 9.Diep CB, Teixeira MR, Thorstensen L, Wiig JN, Eknaes M, Nesland JM, Giercksky KE, Johansson B, Lothe RA. Genome characteristics of primary carcinomas, local recurrences, carcinomatoses, and liver metastases from colorectal cancer patients. Mol Cancer. 2004;3:6. doi: 10.1186/1476-4598-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kleivi K, Lind GE, Diep CB, Meling GI, Brandal LT, Nesland JM, Myklebost O, Rognum TO, Giercksky KE, Skotheim RI. Gene expression profiles of primary colorectal carcinomas, liver metastases, and carcinomatoses. Mol Cancer. 2007;6:2. doi: 10.1186/1476-4598-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lemoine L, Sugarbaker P, Van der Speeten K. Pathophysiology of colorectal peritoneal carcinomatosis: role of the peritoneum. World J Gastroenterol. 2016;22:7692–7707. doi: 10.3748/wjg.v22.i34.7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ku JL, Shin YK, Kim DW, Kim KH, Choi JS, Hong SH, Jeon YK, Kim SH, Kim HS, Park JH. Establishment and characterization of 13 human colorectal carcinoma cell lines: mutations of genes and expressions of drug-sensitivity genes and cancer stem cell markers. Carcinogenesis. 2010;31:1003–1009. doi: 10.1093/carcin/bgq043. [DOI] [PubMed] [Google Scholar]
- 13.Comprehensive molecular characterization of human colon and rectal cancerNature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buckhaults P, Rago C, St Croix B, Romans KE, Saha S, Zhang L, Vogelstein B, Kinzler KW. Secreted and cell surface genes expressed in benign and malignant colorectal tumors. Cancer Res. 2001;61:6996–7001. [PubMed] [Google Scholar]
- 15.Kitahara O, Furukawa Y, Tanaka T, Kihara C, Ono K, Yanagawa R, Nita ME, Takagi T, Nakamura Y, Tsunoda T. Alterations of gene expression during colorectal carcinogenesis revealed by cDNA microarrays after laser-capture microdissection of tumor tissues and normal epithelia. Cancer Res. 2001;61:3544–3549. [PubMed] [Google Scholar]
- 16.Koehler A, Bataille F, Schmid C, Ruemmele P, Waldeck A, Blaszyk H, Hartmann A, Hofstaedter F, Dietmaier W. Gene expression profiling of colorectal cancer and metastases divides tumours according to their clinicopathological stage. J Pathol. 2004;204:65–74. doi: 10.1002/path.1606. [DOI] [PubMed] [Google Scholar]
- 17.Williams NS, Gaynor RB, Scoggin S, Verma U, Gokaslan T, Simmang C, Fleming J, Tavana D, Frenkel E, Becerra C. Identification and validation of genes involved in the pathogenesis of colorectal cancer using cDNA microarrays and RNA interference. Clin Cancer Res. 2003;9:931–946. [PubMed] [Google Scholar]
- 18.Ichikawa Y, Ishikawa T, Takahashi S, Hamaguchi Y, Morita T, Nishizuka I, Yamaguchi S, Endo I, Ike H, Togo S. Identification of genes regulating colorectal carcinogenesis by using the algorithm for diagnosing malignant state method. Biochem Biophys Res Commun. 2002;296:497–506. doi: 10.1016/s0006-291x(02)00732-5. [DOI] [PubMed] [Google Scholar]
- 19.Xu J, Gu S, Wang S, Dai J, Ji C, Jin Y, Qian J, Wang L, Ye X, Xie Y. Characterization of a novel splicing variant of KLHL5, a member of the kelch protein family. Mol Biol Rep. 2003;30:239–242. doi: 10.1023/a:1026372901766. [DOI] [PubMed] [Google Scholar]
- 20.Wright PK, May FE, Darby S, Saif R, Lennard TW, Westley BR. Estrogen regulates vesicle trafficking gene expression in EFF-3, EFM-19 and MCF-7 breast cancer cells. Int J Clin Exp Pathol. 2009;2:463–475. [PMC free article] [PubMed] [Google Scholar]
- 21.Kuroda TS, Fukuda M, Ariga H, Mikoshiba K. Synaptotagmin-like protein 5: a novel Rab27A effector with C-terminal tandem C2 domains. Biochem Biophys Res Commun. 2002;293:899–906. doi: 10.1016/S0006-291X(02)00320-0. [DOI] [PubMed] [Google Scholar]
- 22.Dong W, Cui J, Yang J, Li W, Wang S, Wang X, Li X, Lu Y, Xiao W. Decreased expression of Rab27A and Rab27B correlates with metastasis and poor prognosis in colorectal cancer. Discov Med. 2015;20:357–367. [PubMed] [Google Scholar]
- 23.Hundemer M, Schmidt S, Condomines M, Lupu A, Hose D, Moos M, Cremer F, Kleist C, Terness P, Belle S. Identification of a new HLA-A2-restricted T-cell epitope within HM1.24 as immunotherapy target for multiple myeloma. Exp Hematol. 2006;34:486–496. doi: 10.1016/j.exphem.2006.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai D, Cao J, Li Z, Zheng X, Yao Y, Li W, Yuan Z. Up-regulation of bone marrow stromal protein 2 (BST2) in breast cancer with bone metastasis. BMC Cancer. 2009;9:102. doi: 10.1186/1471-2407-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walter-Yohrling J, Cao X, Callahan M, Weber W, Morgenbesser S, Madden SL, Wang C, Teicher BA. Identification of genes expressed in malignant cells that promote invasion. Cancer Res. 2003;63:8939–8947. [PubMed] [Google Scholar]
- 26.Jang EK, Song DE, Sim SY, Kwon H, Choi YM, Jeon MJ, Han JM, Kim WG, Kim TY, Shong YK. NRAS codon 61 mutation is associated with distant metastasis in patients with follicular thyroid carcinoma. Thyroid. 2014;24:1275–1281. doi: 10.1089/thy.2014.0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghidini M, Personeni N, Bozzarelli S, Baretti M, Basso G, Bianchi P, Tronconi MC, Pressiani T, Grizzi F, Giordano L. KRAS mutation in lung metastases from colorectal cancer: prognostic implications. Cancer Med. 2016;5:256–264. doi: 10.1002/cam4.592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ormanns S, Neumann J, Horst D, Kirchner T, Jung A. WNT signaling and distant metastasis in colon cancer through transcriptional activity of nuclear beta-Catenin depend on active PI3K signaling. Oncotarget. 2014;5:2999–3011. doi: 10.18632/oncotarget.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 30.Grimm WA, Messer JS, Murphy SF, Nero T, Lodolce JP, Weber CR, Logsdon MF, Bartulis S, Sylvester BE, Springer A. The Thr300Ala variant in ATG16L1 is associated with improved survival in human colorectal cancer and enhanced production of type I interferon. Gut. 2016;65:456–464. doi: 10.1136/gutjnl-2014-308735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Rolle AF, Klempner SJ, Garrett CR, Seery T, Sanford EM, Balasubramanian S, Ross JS, Stephens PJ, Miller VA, Ali SM. Identification and characterization of RET fusions in advanced colorectal cancer. Oncotarget. 2015;6:28929–28937. doi: 10.18632/oncotarget.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li FF, Yan P, Zhao ZX, Liu Z, Song DW, Zhao XW, Wang XS, Wang GY, Liu SL. Polymorphisms in the CHIT1 gene: associations with colorectal cancer. Oncotarget. 2016;7:39572–39581. doi: 10.18632/oncotarget.9138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miao HK, Chen LP, Cai DP, Kong WJ, Xiao L, Lin J. MSH3 rs26279 polymorphism increases cancer risk: a meta-analysis. Int J Clin Exp Pathol. 2015;8:11060–11067. [PMC free article] [PubMed] [Google Scholar]
- 34.Leung ML, Davis A, Gao R, Casasent A, Wang Y, Sei E, Vilar E, Maru D, Kopetz S, Navin NE. Single-cell DNA sequencing reveals a late-dissemination model in metastatic colorectal cancer. Genome Res. 2017;27:1287–1299. doi: 10.1101/gr.209973.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin JP, Zhang Z, Bautista JA. Isoform diversity, regulation, and functional adaptation of troponin and calponin. Crit Rev Eukaryot Gene Expr. 2008;18:93–124. doi: 10.1615/critreveukargeneexpr.v18.i2.10. [DOI] [PubMed] [Google Scholar]
- 36.Liu R, Jin JP. Calponin isoforms CNN1, CNN2 and CNN3: regulators for actin cytoskeleton functions in smooth muscle and non-muscle cells. Gene. 2016;585:143–153. doi: 10.1016/j.gene.2016.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jayne D. Molecular biology of peritoneal carcinomatosis. Cancer Treat Res. 2007;134:21–33. doi: 10.1007/978-0-387-48993-3_2. [DOI] [PubMed] [Google Scholar]
- 38.Daimon E, Shibukawa Y, Wada Y. Calponin 3 regulates stress fiber formation in dermal fibroblasts during wound healing. Archives Dermatol Res. 2013;305:571–584. doi: 10.1007/s00403-013-1343-8. [DOI] [PubMed] [Google Scholar]
- 39.Khawar IA, Kim JH, Kuh HJ. Improving drug delivery to solid tumors: priming the tumor microenvironment. J Control Release. 2015;201:78–89. doi: 10.1016/j.jconrel.2014.12.018. [DOI] [PubMed] [Google Scholar]
- 40.Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009;28:15–33. doi: 10.1007/s10555-008-9169-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Abnormalities of APC and TP53. The c.215C>G/p.Pro72Arg Transversion Mutation Was Previously Reported in the p53 Mutation Database (http://www-p53.iarc.fr) and Catalogue of Somatic Mutations in Cancer (http://cancer.sanger.ac.uk/cosmic) To Be Found in Five Cell Lines (SNU-2335A, SNU-2335D, SNU-2335E, SNU-2414A, and SNU-2414B).
Supplementary Table 2. Abnormalities of MSH3 and MSH6
Supplementary Table 3. Abnormalities of CASP8 and GPC6
Supplementary Table 4. Abnormalities of KRAS and NRAS (W, Wild Type)
Supplementary Table 5. Abnormalities of SMAD4 and β-Catenin (CTNNB1) (W, Wild Type)
Supplementary Table 6. Common Mutations in SNU-2335D and SNU-2404B
Supplementary Table 7. Common Mutations in SNU-2335D and SNU-2414B
Supplementary Table 8. Common Mutations in SNU-2404B and SNU-2414B
Supplementary Table 9. The Proposed Functions of Mutations That Are Previously Reported to Increase CRC Risk and Mediate Metastasis
Supplementary Table 10. WES Result of SNU-2335A Was Analyzed in Reference to the Genes That Had Missense Mutations Specifically in Primary CRC
Supplementary Table 11. WES Result of SNU-2335D Was Analyzed in Reference to the Genes That Had Missense Mutations Specifically in Metastatic Cancer
Supplementary Table 12. WES Result of SNU-2404A Was Analyzed in Reference to the Genes That Had Missense Mutations Specifically in Primary CRC
Supplementary Table 13. WES Result of SNU-2404B Was Analyzed in Reference to the Genes That Had Missense Mutations Specifically in Metastatic Cancer
Supplementary Table 14. WES Result of SNU-2414A Was Analyzed in Reference to the Genes That Had Missense Mutations Specifically in Primary CRC
Supplementary Table 15. WES Result of SNU-2414B Was Analyzed in Reference to the Genes That Had Missense Mutations Specifically in Metastatic Cancer
Supplementary Table 16. Variant Allele Frequency of Genes That Potentially Mediate Clonal Evolution in Metastasis Site in SNU-2335 Set
Supplementary Table 17. Variant Allele Frequency of Genes That Potentially Mediate Clonal Evolution in Metastasis Site in SNU-2404 Set
Supplementary Table 18. Variant Allele Frequency of Genes That Potentially Mediate Clonal Evolution in Metastasis Site in SNU-2414 Set
Supplementary Table 19. Upregulated Genes in More Than Two Sets of Cell Lines
Supplementary Table 20. Downregulated Genes in More Than Two Sets of Cell Lines
Supplementary Table 21. Primer Sequences That Were Used for RT-PCR
Supplementary Table 22. Genes That Were Used for Mutation Analysis
Supplementary Table 23. STR Profile of Peritoneal Omental Seeding Cancer Cell Line
Supplementary Table 24. Aberrated Signaling Pathways in SNU-2335 Set
Supplementary Table 25. Aberrated Signaling Pathways in SNU-2404 Set.
Supplementary Table 26. Aberrated Signaling Pathways in SNU-2414 Set
Data Availability Statement
Data supporting the results reported in the article can be accessed by the authors upon demand.