Abstract
Background
The clinical implication of plasma ESR1 mutations in the estrogen receptor (ER)-positive metastatic breast cancer (MBC) patients who had progressed after prior aromatase inhibitor (AI)-based therapy remains controversial. We conducted the first meta-analysis to investigate the prognostic significance and predictive role of plasma ESR1 mutations in MBC patients with prior exposure to AI therapy.
Materials and methods
We searched PubMed, Embase, and Cochrane Library databases for eligible studies. Meta-analysis was conducted to calculate combined hazard ratios (HRs) with 95% CIs for progression-free survival (PFS) and overall survival (OS). Subgroup and sensitivity analyses were also performed.
Results
This study enrolled a total of 1,530 patients with ER-positive MBC cases from six articles, including 429 ESR1 mutation carriers (28.04%). Meta-analysis demonstrated that plasma ESR1 mutation carriers had significantly worse PFS (HR: 1.40, 95% CI: 1.17–1.66; P<0.0001) and OS (HR: 1.65, 95% CI: 1.36–2.01; P<0.0001) compared to wild-type ESR1. Subgroup analysis showed that plasma ESR1 mutations were associated with shorter PFS after AI-based treatment, but were not significantly predictive of outcome on fulvestrant-containing therapy (HR: 1.26, 95% CI: 0.98–1.62; P=0.077). As for different ESR1 mutations, D538G mutation implied significantly worse PFS (HR: 1.50, 95% CI: 1.18–1.91; P=0.01), while Y537S mutation was not correlated with PFS (HR: 1.65, 95% CI: 0.87–1.73; P=0.134).
Conclusion
The meta-analysis indicated that plasma ESR1 mutation assessment may have prognostic significance and clinical value in guiding further endocrine therapy choice in ER+ MBC patients who received prior AI therapy.
Keywords: ESR1 mutations, breast carcinoma, cfDNA, ctDNA, meta analysis
Introduction
Approximately 75% of breast cancers are estrogen receptor (ER) positive, and the mainstay of treatment for this population is endocrine therapies such as selective ER modulators/downregulators, or estrogen deprivation by aromatase inhibitors (AIs).1,2 For patients with metastatic breast cancer (MBC), endocrine therapy is the preferred initial treatment, but unfortunately, almost all patients in this setting will develop endocrine resistance during treatment.3–5
Although several mechanisms have been linked to endocrine resistance, no biomarker has reached wide clinical use.6,7 Recent studies have identified a set of mutations in the ESR1 gene, which encodes ERα, from patients with endocrine-refractory MBC.8 Compared to primary breast cancers, ESR1 mutations are more prevalent in MBCs, particularly in those previously treated with AIs. According to studies using digital polymerase chain reaction techniques, ESR1 mutations were detected in 20%–55% of biopsies of ER-positive MBC patients.9–12 Most of these mutations are observed in the ligand-binding domain of the ESR1 gene, with D538G and Y537S being the most frequent ones.10,13,14 Recently, research has focused on detecting ESR1 mutations in “liquid biopsies” such as circulating cell-free DNA (cfDNA) or circulating tumor DNA (ctDNA) as an alternative to metastatic tumor tissue, which enables easier sampling.15–18 Several studies that investigated the association of plasma ESR1 mutations with the outcomes of endocrine therapies have shown that detection of ESR1 mutations at baseline blood draw predicts a shorter progression-free survival (PFS) after treatment with AIs.19–23
However, consensus has not been reached regarding the reliability of ESR1 mutations in cfDNA/ctDNA as predictive biomarkers due to some limitations with the current evidence.12 First, most of these reports included a limited number of patients, and thus, inconsistent findings exist. Moreover, these studies also differ in many aspects, such as mutations assessed, drugs administered, methods to process plasma, and techniques to detect mutations. These drawbacks make it underpowered to analyze the differential effects of different ESR1 mutations and their predictive value for distinct therapeutic agents such as AIs or fulvestrant.
Considering the weakness of the individual studies, we carried out a meta-analysis to evaluate the impact of ESR1 mutation status in cfDNA or ctDNA on disease-free survival and overall survival (OS) in patients with ER-positive MBC. Subgroup analyses were performed to assess the clinical relevance of the two most frequent ESR1 mutations (Y537S and D538G) and to elucidate the predictive significance of ESR1 mutations on AI-based and fulvestrant-containing therapies.
Materials and methods
Publication search
The electronic databases of PubMed, Embase, and Cochrane Library were comprehensively searched for relevant studies between 1990 and 2017 using the following keywords and their combinations: “breast cancer” OR “breast neoplasm” OR“ breast tumor”, “ctDNA” OR “cfDNA” OR “cell free DNA” OR “circulating” OR “plasma”, “ESR1” OR “Estrogen receptor α” OR “ERα”, and “therapy” OR “therapies”. Additionally, a manual search was performed to further retrieve eligible studies using reference lists of the publications identified from the original search.
Criteria for inclusion and exclusion
Records identified from databases and reference lists were firstly screened by titles and abstracts and then further reviewed by full-text articles. Eligibility criteria were as follows: 1) studies enrolled patients with ER-positive MBC; 2) ESR1 mutation status should be detected by cfDNA or ctDNA at baseline of certain endocrine therapy; and 3) the association between ESR1 mutations and survival status was investigated.
Studies were excluded if they were non-English articles, reviews, commentaries, or case reports. Other exclusion criteria included: 1) articles not available of hazard ratio (HR) with 95% CI for PFS and/or OS; 2) lacking treatment information after baseline ESR1 mutation analysis; and 3) duplicate reports from one study.
Quality assessment of studies
The quality of all relevant articles was evaluated independently by two authors (KZ and RH) using the Newcastle–Ottawa Scale (NOS) Quality Assessment Scale.24 The NOS consists of three parts: selection, comparability, and outcome (cohort studies). We used total scores to assess the quality of eligible studies. Study with a score of 7 or higher was regarded as high quality.
Data extraction
Two investigators (KZ and RH) independently extracted the following data from the original studies: first author, publication year, ESR1 mutations assessed, techniques used for ESR1 mutation detection in ctDNA/cfDNA, number of patients enrolled, number of patients with plasma ESR1 mutations, subsequent therapeutic regimens, and outcome data (HRs and 95% CIs for PFS/OS). If not reported by the articles, survival data were extracted from the Kaplan–Meier curves using the methods designed by Tierney et al.25 Disagreements between two authors were resolved through common sense with the third investigator (SW).
Statistical analysis
Statistical analysis was carried out according to the guidelines of the Meta-Analysis of Observational Studies in Epidemiology group.26 HRs and 95% CIs were performed for PFS and OS. I2 and Q tests were performed to evaluate heterogeneity. P<0.1 and I2≥50% indicated substantial heterogeneity according to the Cochrane Handbook.27 Fixed-effects or random-effects models were applied to estimate the HRs, depending on whether heterogeneity existed or not. Fixed-effects model was used if there was no evidence of heterogeneity between studies; otherwise, random-effects model was used. For all these analyses, P-value <0.05 was considered statistically significant. Funnel test, Begg’s test, and Egger’s test were carried out to assess publication bias. All statistical analyses were performed using STATA 12.0 software (StataCorp, College Station, TX, USA).
Results
Literature selection
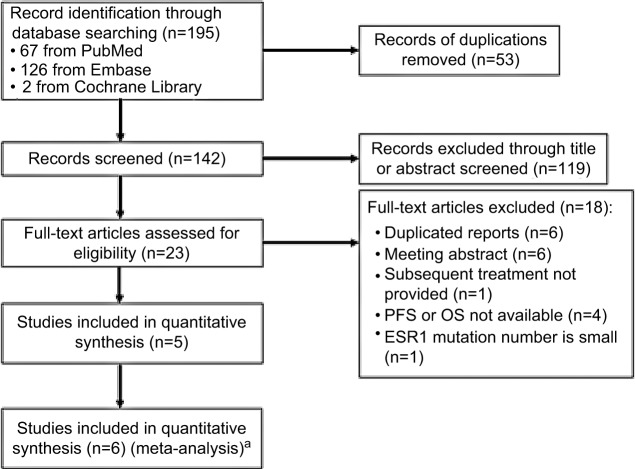
The process of literature screening and study selection is shown in Figure 1. After preliminary online search, 195 citations were retrieved out of Embase, MEDLINE, and the Cochrane Library databases. After the deletion of duplicate results, 142 records remained. By reviewing the titles and abstracts, 119 records were excluded. Full texts of the remaining 23 articles were carefully reviewed and assessed, and 17 articles were further eliminated due to several reasons, leaving 5 eligible articles. In the report presented by Fribbens et al,22 data from two randomized studies (SoFEA and PALOMA3) were included, which were analyzed independently. Therefore, six eligible studies were finally enrolled in the meta-analysis.28
Figure 1.
Flow diagram of literature review and study selection.
Notes: aThe report presented by Fribbens et al22 included data from two randomized studies (SoFEA and PALOMA3), which were analyzed independently. Therefore, six eligible studies were enrolled in the meta-analysis.
Abbreviations: OS, overall survival; PFS, progression-free survival.
Characteristics of included studies and patients
Characteristics of eligible studies are summarized in Table 1. The selected studies, published between 2015 and 2017, assessed the impact of ESR1 mutations on the outcome to therapies in a total of 1,546 ER+ MBC patients. All the studies were retrospective, including four prospective-retrospective analyses using archived baseline plasma from randomized clinical trials. Sample types were cfDNA or ctDNA. There were 429 ESR1 mutation carriers (28.04%). All patients received prior AIs. Subsequent therapeutic drugs were listed. The NOS scores of five included studies varied from 8 to 9 and the average NOS score was 8.5. An agreement between the two reviewers was 90% for study selection and 100% for quality assessment of these studies.
Table 1.
Characteristics of included studies
| Author (trial) | Year | ESR1 mutations panel | No. of patients analyzed for ESR1 | No. of ESR1 mutations-positive patients | Subsequent treatment | Sample type | Detection methods | Sampling time | Outcome | NOS |
|---|---|---|---|---|---|---|---|---|---|---|
| Schiavon et al19 | 2016 | D538G Y537S L536R Y537N Y537C | 171 | 18 | AI-based therapy | ctDNA | ddPCR | Baseline | PFS | 9 |
| Clatot et al21 | 2016 | D538G Y537S Y537N Y537C | 144 | 44 | AI based, chemotherapy, others | cfDNA | ddPCR | Baseline | PFS, OS | 8 |
| Fribbens et al22 (SoFEA) | 2016 | D538G Y537S Y537N E380Q | 161 | 63 | Fulvestrant + anastrozole/placebo, exemestane | ctDNA | Multiple xddPCR | Baseline | PFS, OS | 9 |
| Fribbens et al22 (PALOMA3) | 2016 | D538G Y537S Y537N E380Q | 360 | 91 | Fulvestrant + palbociclib/placebo | ctDNA | Multiple xddPCR | Baseline | PFS | 9 |
| Spoerke et al23 (FERGI) | 2016 | D538G Y537S E380Q Y537N Y537C P535H L536H L536P L536Q L536R | 153 | 57 | Fulvestrant + pictilisib/placebo | ctDNA | BEAMing | Baseline | PFS | 9 |
| Chandarlapaty et al20 (BOLERO-2) | 2016 | D538G Y537S | 541 | 156 | Exemestane + placebo/everolimus | cfDNA | ddPCR | Baseline | PFS, OS | 9 |
Abbreviations: AI, aromatase inhibitor; ct, circulating tumor; cf, cell free; dd, droplet digital; NOS, Newcastle–Ottawa Scale; OS, overall survival; PCR, polymerase chain reaction; PFS, progression-free survival.
ESR1 mutations and PFS
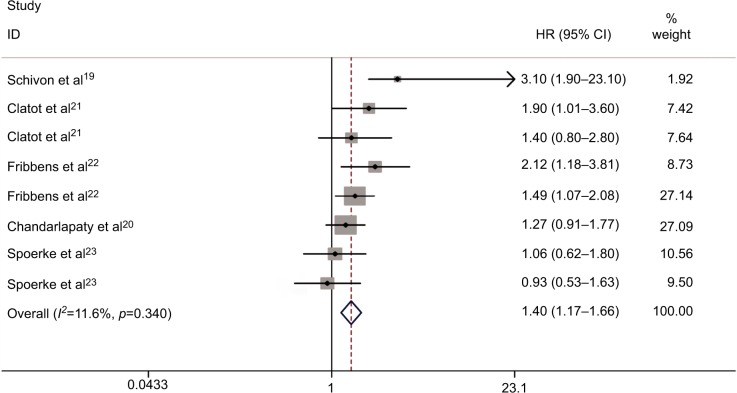
Figure 2 shows the forest plot of the analysis about the relationship between circulating ESR1 mutations and PFS. A total of six studies were subjected to this analysis. There was no obvious heterogeneity in the HRs for PFS among these studies (P=0.400, I2=4.2%). So, we used the fixed-effects model to calculate the pooled effect size. Our pooled analysis showed that plasma ESR1 mutations were significantly associated with worse PFS compared with wild-type (WT) ESR1 (HR: 1.40, 95% CI: 1.17–1.66; P<0.0001) in ER(+) MBC patients.
Figure 2.
Forest plot for PFS between plasma ESR1 mutation and wild-type ESR1.
Abbreviations: HR, hazard ratio; PFS, progression-free survival.
ESR1 mutations and OS
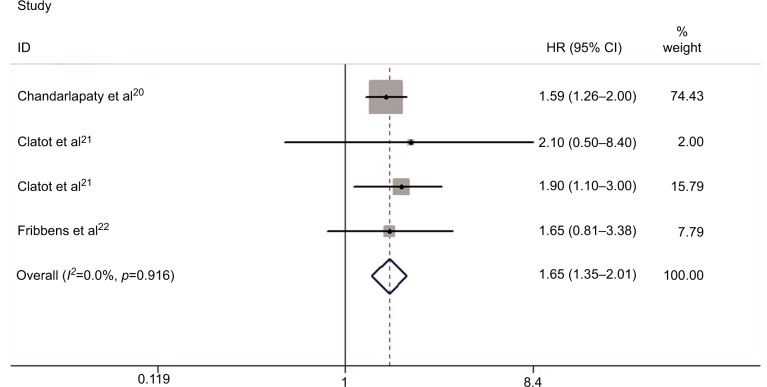
Figure 3 displays the forest plot of the analysis about OS. Five studies were subjected to the analysis. There was no evidence of heterogeneity in the HRs for OS among the individual trials (P=0.971, I2=0%). The combined HR with fixed-effects model for OS was 1.65 (95% CI: 1.36–2.01; P<0.001), indicating a prognostic association of ESR1 mutations in patients with ER+ MBC.
Figure 3.
Forest plot for OS between plasma ESR1 mutation and wild-type ESR1.
Abbreviations: HR, hazard ratio; OS, overall survival.
Subgroup analyses
We conducted subgroup analyses according to the subsequent treatment for MBC patients after baseline sample collection (Table 2). The pooled HR was 1.51 (95% CI: 1.15–2.00; P<0.001) for PFS and 1.61 (95% CI: 1.29–2.00; P<0.001) for OS, suggesting that plasma ESR1 mutation status was predictive of worse outcome on subsequent AI-based therapy. We then analyzed data from the three studies in which fulvestrant-containing therapies were administered. It turned out that the ESR1 mutations were not significantly correlated with PFS, with the pooled HR for PFS being 1.26 (95% CI: 0.98–1.62; P=0.077).
Table 2.
Results of subgroup analyses for effects of subsequent lines of treatment on PFS or OS
| Subsequent lines of treatment | PFS
|
OS
|
||||
|---|---|---|---|---|---|---|
| df | HR (95% CI) | Test for heterogeneity (I2) | df | HR (95% CI) | Test for heterogeneity (I2) | |
| AI-based therapy | 3 | 1.51 (1.15–2.00) | 26.7% | 2 | 1.61 (1.29–2.00) | 0.0% |
| Fulvestrant-containing therapy | 2 | 1.26 (0.98–1.62) | 20.1% | ------------ | ||
Abbreviations: AI, aromatase inhibitor; df, degrees of freedom; HR, hazard ratio; OS, overall survival; PFS, progression-free survival.
Subgroup analysis was conducted to further elucidate the impact of each ESR1 amino acid mutation on treatment outcome, which is demonstrated in Table 3. Only two articles provided data on association between individual mutations and PFS, which were included in the subgroup analysis.20–22 The fixed-effects model was adopted to calculate the pooled HRs about D538G mutation due to absence of heterogeneity (I2=0.0%, P=0.605). D538G mutation suggested significantly worse PFS (HR: 1.50, 95% CI: 1.18–1.91; P=0.01) compared with WT ESR1 in MBC patients receiving endocrine therapy. However, for Y537S mutation analysis, obvious heterogeneity was detected (I2=83.7%, P=0.002). Our result indicated that Y537S mutation was not correlated with PFS (HR: 1.65, 95% CI: 0.87–1.73; P=0.134).
Table 3.
Results of subgroup analyses for effects of ESR1 mutation on PFS
| ESR1 mutation | ESR1 mutation numbers | df | Model | HR (95% CI) | Test for heterogeneity (I2) |
|---|---|---|---|---|---|
| D538G vs WT ESR1 | 121 | 2 | Fixed | 1.50 (1.18–1.91) | 0.0% |
| Y537S vs WT ESR1 | 61 | 2 | Random | 1.65 (0.87–1.73) | 83.7% |
Abbreviations: df, degrees of freedom; HR, hazard ratio; PFS, progression-free survival; WT, wild type.
Publication bias and sensitivity analysis
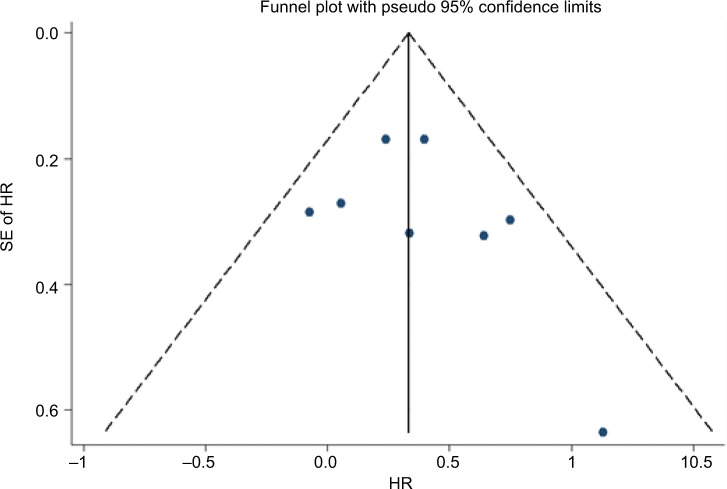
Funnel plot, Begg’s test, and Egger’s test were performed to assess the publication bias of the included studies. There was no evidence of publication bias according to funnel plot (Figure 4), Egger’s test (P=0.357), and Begg’s test (0.386) for PFS. Similarly, publication bias was not significant for OS studies according to Egger’s test (P=0.255) and Begg’s test (P=0.734). Each study was sequentially omitted to evaluate whether any single study had a significant impact on the pooled OS or PFS. Our results showed that the pooled HRs were not influenced by any individual study.
Figure 4.
Funnel plot to detect publication bias for PFS in the plasma ESR1 mutations vs wild-type ESR1.
Abbreviations: PFS, progression-free survival; SE, standard error; HR, hazard ratio.
Discussion
This meta-analysis evaluated the association between plasma ESR1 mutations and survival of MBC patients. Our results showed that plasma ESR1 mutations represent a worse prognostic factor in patients with MBC who received subsequent AI therapy. The combined HRs were 1.40 (95% CI: 1.18–1.67; P<0.0001) for disease-free survival and 1.66 (95% CI: 1.36–2.01; P<0.0001) for OS between mutated ESR1 and WT ESR1 in advanced ER-positive MBC after treatment containing endocrine agents, suggesting that breast cancer with and without ESR1 mutations are distinct subtypes that respond differently to therapies.
For patients who failed prior AI therapy, PFS after AI rechallenge was significantly decreased in those harboring ESR1 mutations (HR: 1.51, 95% CI: 1.15–2.00; P<0.001). But in the fulvestrant treatment subgroup, the pooled HR (1.26, 95% CI: 0.98–1.62; P=0.077) did not significantly favor those with a WT ESR1, suggesting that ESR1 mutations may not be correlated with innate or acquired fulves-trant resistance. This was consistent with previous in vitro studies that reported growth inhibition with fulvestrant in ESR1 mutant cell lines.28 These findings support the choice of fulvestrant over AI in treating MBC patients who had progressed after AI therapy and highlight the clinical implication of using plasma ESR1 mutation testing to guide further endocrine-based therapy.
Detecting ESR1 mutations in cfDNA/ctDNA has several obvious advantages over metastatic biopsies. Several studies have suggested that cfDNA or ctDNA sometimes reveal additional mutations compared with matching metastatic tumor biopsies due to tumor heterogeneity.15–18 This indicates that cfDNA/ctDNA may be more representative of the whole tumor landscape. In addition, serum samples can be easily obtained in a noninvasive way for multiple times, enabling assessment of mutations not only at baseline, but also sequentially during therapy. A rise in ESR1 mutations during treatment may indicate disease progression as a result of evolution through selective AI treatment. Upon treatment with fulvestrant, if resistant ESR1 mutant clones decay, this could possibly allow rechallenges with AIs in this subset of patients.
Another major question is whether the various ESR1 mutations lead to distinctive phenotypes; the D538G mutation implied shorter PFS compared to WT ESR1 (HR: 1.50, 95% CI: 1.18–1.91; P=0.01) for breast cancer patients, but the Y537S mutation did not relate to shorter PFS (HR: 1.65, 95% CI: 0.87–1.73; P=0.134). This subgroup analysis may provide clinicians a precise guideline for those patients who have different ESR1 mutations. However, several basic studies have shown that Y537S mutation represents the strongest ER activity under an estrogen-deprived condition, which is inconsistent with these findings.11,24 As for the Y537S mutation analysis, obvious statistical heterogeneity was detected (I2=83.7%), and in these two studies for Y537S mutations, there might be obvious heterogeneity between patients in the two different clinical studies.20,22 Moreover, owing to the limited number of articles, the relationship between different ESR1 mutations and the prognosis of breast cancer cannot be very clearly elucidated. Functions of other alterations, such as E380Q, Y537N, Y537C, and L536R in plasma DNA, have not been addressed in the current studies. Thus, future studies evaluating all ligand-binding domain mutations in ESR1 and their potential distinctive effects are of interest.
As we all know, ERa is an important gene for ER+ breast cancer, and gene signatures-derived cancer hallmarks are good prognostic biomarkers. Wang et al33 created an algorithm to identify prognostic gene signature sets for ER-negative and -positive breast cancers. Also, eTumorType could be set aside for tumor diagnosis based upon the copy number variations of captured CTCs or cfDNAs in blood samples. The molecular mechanism of PIK3CA mutations in luminal A patients can participate in a positive regulatory loop (PDGF-D/FLT1/SHC1), and moreover, the regulatory loop has a potential power to predict the survival of luminal A patients with PIK3CA mutated. The next step may use these algorithms to identify prognostic gene signature for ER+ subtype. A cancer hallmark network framework for modeling genome sequencing data forecast clinical phenotypes and cancer progression to better design patient treatment.29–33
Our results are promising to a certain extent, but several limitations of this meta-analysis still need to be discussed. First, the number of enrolled studies was limited and the sample size was relatively small, which might not have provided sufficient power of test to evaluate the associations between the plasma ESR1 mutations and clinical outcome in ER+ MBC patients. Second, our meta-analysis is based on data from published articles, so publication bias and selection bias may exist. Third, these studies were retrospectively conducted, which used different patient selection criteria and involved various treatments. Finally, some data were extracted from the survival curves and, thus, might not be accurate.
In conclusion, our results suggest that plasma ESR1 mutation analysis may have prognostic value and clinical implication in directing further endocrine therapy choice in ER+ MBC patients who failed prior treatment with AIs. Patients with ESR1 mutations and WT ESR1 seem to be different subtypes of advanced ER-positive breast cancer. Future clinical trials may use plasma DNA analysis to guide treatment selection according to ESR1 mutation status.
Acknowledgments
We thank Dr WG Deng and Dr M Chen (Sun Yat-Sen University Cancer Center, The State Key Laboratory of Oncology in South China) for professional editing of this manuscript.
This research received no specific grant from any funding agency in the public, commercial, or from not-for-profit sectors.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Burstein HJ, Prestrud AA, Seidenfeld J, et al. American Society of Clinical Oncology Clinical Practice Guideline: Update on Adjuvant Endocrine Therapy for Women With Hormone Receptor–Positive Breast Cancer. Journal of Clinical Oncology. 2010;28(23):3784–3796. doi: 10.1200/JCO.2009.26.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith IE, Dowsett M. Aromatase inhibitors in breast cancer. N Engl J Med. 2003;348(24):2431–2442. doi: 10.1056/NEJMra023246. [DOI] [PubMed] [Google Scholar]
- 4.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG): Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365(9472):1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 5.Burstein HJ, Temin S, Anderson H, et al. Adjuvant Endocrine Therapy for Women With Hormone Receptor–Positive Breast Cancer: American Society of Clinical Oncology Clinical Practice Guideline Focused Update. Journal of Clinical Oncology. 2014;32(21):2255–2269. doi: 10.1200/JCO.2013.54.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Laurentiis M, Arpino G, Massarelli E, et al. A meta-analysis on the interaction between HER-2 expression and response to endocrine treatment in advanced breast cancer. Clin Cancer Res. 2005;11(13):4741–4748. doi: 10.1158/1078-0432.CCR-04-2569. [DOI] [PubMed] [Google Scholar]
- 7.Amir E, Miller N, Geddie W, et al. Prospective study evaluating the impact of tissue confirmation of metastatic disease in patients with breast cancer. J Clin Oncol. 2012;30(6):587–592. doi: 10.1200/JCO.2010.33.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koboldt DC, Fulton RS, Mclellan MD, Schmidt MD, et al. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li S, Shen D, Shao J, et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep. 2013;4(6):1116–1130. doi: 10.1016/j.celrep.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merenbakh-Lamin K, Ben-Baruch N, Yeheskel A, et al. D538G mutation in estrogen receptor-α: A novel mechanism for acquired endocrine resistance in breast cancer. Cancer Res. 2013;73(23):6856–6864. doi: 10.1158/0008-5472.CAN-13-1197. [DOI] [PubMed] [Google Scholar]
- 11.Toy W, Shen Y, Won H, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet. 2013;45(12):1439–1445. doi: 10.1038/ng.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Angus L, Beije N, Jager A, Martens JW, Sleijfer S. ESR1 mutations: Moving towards guiding treatment decision-making in metastatic breast cancer patients. Cancer Treat Rev. 2017;52:33–40. doi: 10.1016/j.ctrv.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Forshew T, Murtaza M, Parkinson C, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4(136):ra68. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- 14.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haber DA, Velculescu VE. Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov. 2014;4(6):650–661. doi: 10.1158/2159-8290.CD-13-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sundaresan TK, Sequist LV, Heymach JV, et al. Detection of T790M, the Acquired Resistance EGFR Mutation, by Tumor Biopsy versus Noninvasive Blood-Based Analyses. Clin Cancer Res. 2016;22(5):1103–1110. doi: 10.1158/1078-0432.CCR-15-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guttery DS, Page K, Hills A, et al. Noninvasive detection of activating estrogen receptor 1 (ESR1) mutations in estrogen receptor-positive metastatic breast cancer. Clin Chem. 2015;61(7):974–982. doi: 10.1373/clinchem.2015.238717. [DOI] [PubMed] [Google Scholar]
- 18.Jeselsohn R, Yelensky R, Buchwalter G, et al. Emergence of constitutively active estrogen receptor-α mutations in pretreated advanced estrogen receptor-positive breast cancer. Clin Cancer Res. 2014;20(7):1757–1767. doi: 10.1158/1078-0432.CCR-13-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schiavon G, Hrebien S, Garcia-Murillas I, et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci Transl Med. 2015;7(313):313ra182. doi: 10.1126/scitranslmed.aac7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandarlapaty S, Chen D, He W, et al. Prevalence of ESR1 Mutations in Cell-Free DNA and Outcomes in Metastatic Breast Cancer: A Secondary Analysis of the BOLERO-2 Clinical Trial. JAMA Oncol. 2016;2(10):1310–1315. doi: 10.1001/jamaoncol.2016.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clatot F, Perdrix A, Augusto L, et al. Kinetics, prognostic and predictive values of ESR1 circulating mutations in metastatic breast cancer patients progressing on aromatase inhibitor. Oncotarget. 2016;7(46):74448–74459. doi: 10.18632/oncotarget.12950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fribbens C, O’Leary B, Kilburn L, et al. Plasma ESR1 Mutations and the Treatment of Estrogen Receptor-Positive Advanced Breast Cancer. J Clin Oncol. 2016;34(25):2961–2968. doi: 10.1200/JCO.2016.67.3061. [DOI] [PubMed] [Google Scholar]
- 23.Spoerke JM, Gendreau S, Walter K, et al. Heterogeneity and clinical significance of ESR1 mutations in ER-positive metastatic breast cancer patients receiving fulvestrant. Nat Commun. 2016;7:11579. doi: 10.1038/ncomms11579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wells GA, Shea B, O’Connell D, et al. The Newcastle–Ottawa Scale (NOS) for assessing the quality if nonrandomized studies in meta-analyses. 2011. [Accessed July 23, 2018]. Available from: http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp.
- 25.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stroup DF, Berlin JA, Morton SC, et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA. 2000;283(15):2008–2012. doi: 10.1001/jama.283.15.2008. [DOI] [PubMed] [Google Scholar]
- 27.Higgins JPT, Green S, editors. Cochrane Handbook for Systematic Reviews of Interventions. Version 5.1.0 [updated March 2011] The Cochrane Collaboration; 2011. [Accessed July 23, 2018]. Available from: http://handbook-5-1.cochrane.org/ [Google Scholar]
- 28.Yates LR, Gerstung M, Knappskog S, et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med. 2015;21(7):751–759. doi: 10.1038/nm.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J, Lenferink AEG, Deng Y, et al. Erratum: Corrigendum: Identification of high-quality cancer prognostic markers and metastasis network modules. Nat Commun. 2012;3(1) doi: 10.1038/ncomms1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zou J, Wang E. eTumorType, An Algorithm of Discriminating Cancer Types for Circulating Tumor Cells or Cell-free DNAs in Blood. Genomics Proteomics Bioinformatics. 2017;15(2):130–140. doi: 10.1016/j.gpb.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao S, Tibiche C, Zou J, et al. Identification and Construction of Combinatory Cancer Hallmark-Based Gene Signature Sets to Predict Recurrence and Chemotherapy Benefit in Stage II Colorectal Cancer. JAMA Oncol. 2016;2(1):37–45. doi: 10.1001/jamaoncol.2015.3413. [DOI] [PubMed] [Google Scholar]
- 32.Mcgee SR, Tibiche C, Trifiro M, Wang E. Network Analysis Reveals A Signaling Regulatory Loop in the PIK3CA-mutated Breast Cancer Predicting Survival Outcome. Gen Prot Bioinform. 2017;15(2):121–129. doi: 10.1016/j.gpb.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang E, Zaman N, Mcgee S, Milanese JS, Masoudi-Nejad A, O’Connor-McCourt M. Predictive genomics: a cancer hallmark network framework for predicting tumor clinical phenotypes using genome sequencing data. Semin Cancer Biol. 2015;30:4–12. doi: 10.1016/j.semcancer.2014.04.002. [DOI] [PubMed] [Google Scholar]