We present a comprehensive analysis of variation in genes and gene expression of allergens and immunoresponsive proteins in wheat.
Abstract
Wheat is an important staple grain for humankind globally because of its end-use quality and nutritional properties and its adaptability to diverse climates. For a small proportion of the population, specific wheat proteins can trigger adverse immune responses and clinical manifestations such as celiac disease, wheat allergy, baker’s asthma, and wheat-dependent exercise-induced anaphylaxis (WDEIA). Establishing the content and distribution of the immunostimulatory regions in wheat has been hampered by the complexity of the wheat genome and the lack of complete genome sequence information. We provide novel insights into the wheat grain proteins based on a comprehensive analysis and annotation of the wheat prolamin Pfam clan grain proteins and other non-prolamin allergens implicated in these disorders using the new International Wheat Genome Sequencing Consortium bread wheat reference genome sequence, RefSeq v1.0. Celiac disease and WDEIA genes are primarily expressed in the starchy endosperm and show wide variation in protein- and transcript-level expression in response to temperature stress. Nonspecific lipid transfer proteins and α-amylase trypsin inhibitor gene families, implicated in baker’s asthma, are primarily expressed in the aleurone layer and transfer cells of grains and are more sensitive to cold temperature. The study establishes a new reference map for immunostimulatory wheat proteins and provides a fresh basis for selecting wheat lines and developing diagnostics for products with more favorable consumer attributes.
INTRODUCTION
Wheat is a major staple cereal grain consumed worldwide that provides a major source of high-quality nutrition to humankind. However, for a small subset of the population, a range of wheat components, principally proteins, are associated with a number of important medical illnesses that can affect patient health and quality of life and, in some cases, can be life-threatening. There has been a large expenditure of research effort into understanding and characterizing these proteins associated with human disease. However, the complexity of the wheat genome and the lack of complete genome sequence information have meant that a detailed description of these proteins and their content and distribution within wheat remains poorly described. With the availability of the high-quality International Wheat Genome Sequencing Consortium (IWGSC) RefSeq (reference sequence) v1.0 reference genome, we have (1) used a comprehensive analysis workflow to identify and precisely characterize the allergens and antigens in wheat proteins associated with or implicated in human disease. Understanding the complete complement of proteins provides immense value for linking them to specific clinical effects and to understand disease pathogenesis. This knowledge also helps to underpin strategies that aim to modify or reduce the potential harmful effects of these proteins through approaches such as selective breeding or improved targeted genetic modification.
The most common human diseases associated with wheat are celiac disease and wheat allergy, where the latter encompasses immunoglobulin E (IgE)–mediated wheat allergy, baker’s asthma, and wheat-dependent exercise-induced anaphylaxis (WDEIA) (Fig. 1A). In recent years, the major allergenic and antigenic components of wheat that drive these illnesses have been well defined and are primarily found within the proline-rich wheat storage prolamin proteins, gliadin and glutenin, although other non-gluten proteins have been implicated in some allergic responses as well (Fig. 1B) (2). Celiac disease is a chronic inflammatory disorder with autoimmune-like features characterized by villous atrophy of the small intestine (3). It results from a CD4+ T cell–mediated reaction to specific gluten peptides from wheat, barley, and rye (4). Celiac disease is a global disease with a prevalence that varies with sex, age, and geographic location. The frequency of predisposing human leukocyte antigen human leukocyte antigen (HLA) haplotypes in the general population and per-capita wheat consumption are the two main determinants of prevalence based on reports of celiac disease in Western and Eastern Europe, North America, South America, Asia, Oceania, and Africa. The condition appears to be uncommon in Southeast Asia and sub-Saharan Africa. In a systematic review and meta-analysis, the global seroprevalence and more definitive biopsy-confirmed prevalence were estimated to be 1.4 and 0.7%, respectively (5). The seroprevalence of celiac disease in the United States from National Health and Nutrition Examination Surveys was 0.7% and showed that most cases remain undiagnosed in the community (6). Current treatment involves a lifelong and strict gluten-free diet to minimize the harmful effects of chronic inflammation caused by gluten peptides in affected patients.
Fig. 1. The prolamin superfamily and its relation to clinical diseases and allergen protein families.
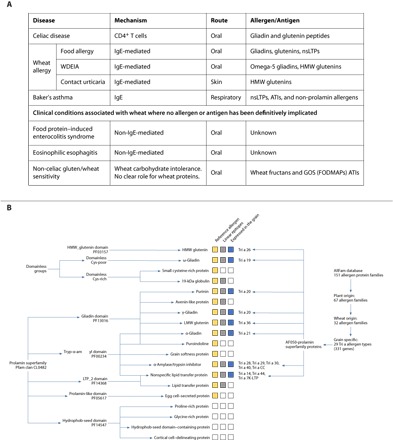
(A) Clinical syndromes associated with wheat ingestion or exposure. Mechanisms of wheat-related clinical syndromes, route of exposure, and major allergens and antigens are presented. (B) Protein groups primarily expressed in the seed are highlighted in yellow. Protein types with immunoreactive peptides in their sequence are highlighted in gray, and reference allergen homologs identified based on the AllFam database are highlighted in blue. “Tri a” labeling of the individual groups follows the nomenclature system of the World Health Organization/International Union of Immunological Societies (WHO/IUIS) Allergen Nomenclature Database.
Distinct from celiac disease is the clinical entity referred to as non-celiac gluten sensitivity or non-celiac wheat sensitivity (NCGS or NCWS, respectively) (7). This condition is defined by self-reported symptoms following ingestion of wheat, rye, or barley, typically gastrointestinal upset or fatigue and improvement with dietary removal of gluten-containing cereals. As there is no formal method of securing the diagnosis, its true prevalence remains unknown. However, gluten and/or wheat avoidance is common in Western communities; for example, in Australia, 11% of adults avoid wheat (half of whom are gluten-free), mainly in an attempt to alleviate gastrointestinal symptoms and fatigue (8). The evidence to implicate gluten as a cause of this syndrome is weak, and high-quality randomized feeding trials indicate that it is more likely to be driven by the poorly absorbed carbohydrate component of wheat, fructans, and galacto-oligosaccharides (GOS) in wheat flour (9). Additional wheat components may also drive gluten or wheat sensitivity. For example, α-amylase trypsin inhibitors (ATIs) can activate the innate immune system in vitro and potentially promote intestinal inflammation (10). However, human clinical trials are needed to confirm these in vitro reports.
Allergic responses to wheat can manifest in a variety of ways, depending on the route of exposure, that is, ingested (food allergy, either IgE-mediated or non–IgE-mediated), respiratory (baker’s asthma), and skin (contact urticaria) (Fig. 1A). The offending allergens often encompass the gliadins and glutenins but can involve other non-gluten proteins. Food wheat allergy is classically IgE-mediated and affects approximately 0.5% of children and mostly resolves by adulthood (11). Symptoms are typically acute (within minutes of ingestion of wheat) and carry the risk of a life-threatening anaphylactic response. Several uncommon non–IgE-mediated gastrointestinal conditions associated with ingested wheat hypersensitivity have also been reported and include eosinophilic esophagitis and food protein–induced enterocolitis syndrome; however, the wheat proteins associated with these disorders have not been clearly defined (Fig. 1A) (12). Treatment often involves dietary exclusion of wheat and often other food proteins. Baker’s asthma is among the most common occupational allergies manifesting with respiratory symptoms following wheat inhalation and primarily triggered by members of lipid transfer and nonspecific lipid transfer proteins (LTPs and nsLTPs, respectively) and ATIs (13). WDEIA is a rare type of food allergy occurring when wheat ingestion is accompanied by physical activity. Wheat allergens associated with WDEIA have been mainly linked to ω-5 gliadins or high–molecular weight glutenins (HMW glutenins). It is more common in adults, but data on prevalence are scarce (14).
Various approaches have been used to identify immunogenic peptide content and distribution in bread wheat and related species (1, 14–17). The known T cell, IgA/IgG, and IgE immunogenic peptides are deposited in curated databases such as the Immune Epitope Database and Analysis Resource (www.iedb.org), AllergenOnline (www.allergenonline.org), and the prolamin peptide epitope database ProPepper [https://propepper.net; (18)]. Tye-Din et al. (15) used an in vivo epitope mapping approach to establish a hierarchy of peptides derived from the known wheat, rye, and barley protein genes immunogenic in patients with the common genetic version (HLA-DQ2.5) of celiac disease. These immune-response “road maps” provide the basis for development of novel diagnostics, therapeutics, or genetically modified grains that have lower immunotoxicity in patients with celiac disease.
Bread wheat is a highly complex allohexaploid species that evolved by hybridization of three species, each contributing one of its subgenomes (A, B, and D) (19). Generating a full overview of the proteins and their genes associated with adverse allergic and immune responses in the subset of people with wheat-associated disorders has been significantly hampered by the inability to discriminate between the A, B, and D homologs of each gene and their encoded proteins. Here, we overcome this problem by using the recently published high-resolution IWGSC RefSeq v1.0 genome sequence of the cultivar Chinese Spring (1). Using this sequence in combination with public databases for wheat proteins/peptides, implicated in human disorders, we present a comprehensive analysis of wheat genes encoding these proteins and their chromosomal locations. Moreover, we highlight the crucial role of genotype and environment on their expression and provide new insight into the effect of biotic and climatic stress factors such as heat or drought (20) on grain protein composition. The study establishes a basis for new diagnostic tools to characterize or fingerprint wheat varieties for the food industry.
RESULTS
Annotation and chromosomal mapping of bread wheat genes encoding proteins implicated in human allergies and immune responses
We used the newly published high-quality bread wheat IWGSC genome RefSeq v1.0 (1) to expand the identification of the genes coding for proteins implicated in wheat-related food disorders based on information in the database of Allergen Families (AllFam; www.meduniwien.ac.at/allfam), supplemented by the data in the AllergenOnline FARRP database (www.allergenonline.org) defining 67 plant food allergen families, including 29 families with a Tri a assignment using the nomenclature of the WHO/IUIS Allergen Nomenclature Database (www.allergen.org). We refer to these latter families as the “reference allergens and antigens.” The reference allergens and antigens include proteins with Pfam domains (pfam.xfam.org) of the prolamin gene superfamily (Pfam clan CL0482), HMW glutenins (PF03157), and other protein families with various enzyme and metabolic functions (Fig. 1B) (20). Because of the repetitive sequence composition of many of these gene families, automatic annotation has been problematic. We therefore used the domain signatures in manual curation to identify the bread wheat genes encoding these proteins in the IWGSC RefSeq v1.0. In total, 356 genes encoding reference food and food-pollen cross allergens were identified and mapped to their chromosomal loci (Fig. 2 and data files S1 and S2). This is the “IWGSC v1.0 reference allergen map.” We refer to this map as the IWGSC v1.0 reference allergen map for bread wheat. Genes encoding reference allergens map to all 21 chromosomes of the A, B, and D subgenomes (Fig. 2).
Fig. 2. Reference allergen map of bread wheat.
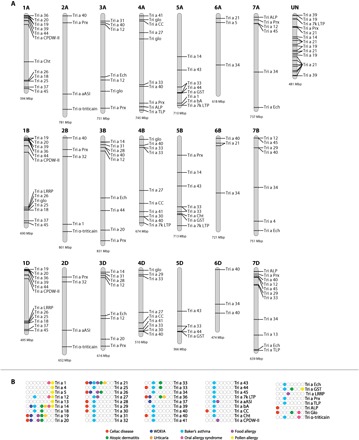
(A) Genome distribution of food disease–related reference allergens in the wheat genome. Only genes with presence of multiple disease-associated epitopes and over 70% sequence homology to reference allergens are presented. (B) Disease association of reference allergens.
As many as 226 of the 356 allergen genes belong to the prolamin gene superfamily (Figs. 1 and 2). Of these, the IWGSC v1.0 reference allergen map adds 127 previously unannotated genes and corrects 222 genes to IWGSC RefSeq v1.0 (19, 21). The 356 reference immunoresponsive gene homologs related to celiac disease, WDEIA, baker’s asthma, and food allergy are positioned in highly conserved gene clusters. As a general feature, genes implicated in food-related immune responses are located in linkage blocks that are enriched toward the telomere regions (Fig. 2). Major immunostimulatory proteins (for example, Tri a 19, Tri a 20, Tri a 21, and Tri a 14) representing various gliadin types and a few nsLTPs are implicated in most of the immune responses. Seventy-five genes with the PF13016 domain and 67 genes with the PF00234 domain from Pfam clan CL0482 were identified as encoding reference allergens and antigens mainly related to celiac disease, WDEIA, and baker’s asthma (fig. S1 and data files S1 and S2). Thirty of 263 genes that encode proteins with an LTP_2 domain represent reference allergens including Tri a 7k-LTP or Tri a 14 that provoke strong immune response in baker’s asthma (data file S2). Among the 35 ATI genes in the IWGSC v1.0 reference allergen map, 15 show high sequence identity to ATIs with positive IgA response in celiac disease. Proteins representing these celiac-related sequences belong to ATI 0.19 (labeled as Tri a 28), ATI CM2 (Tri a 29), and ATI CM3 (Tri a 30) subtypes (Fig. 2A). In addition to members of the prolamin superfamily, genes encoding non-prolamin allergen protein family members [for example, serine-protease inhibitors (serpins, Tri a 33)] have been implicated in baker’s asthma (Fig. 2 and data file S2). Furthermore, Glo-3 globulins (Tri a Glo in Fig. 2A) have been associated with celiac disease (22).
To identify all potential allergens and antigen proteins within the prolamin superfamily, we identified known and previously unknown proteins using Pfam domains PF13016, PF00234, PF14368, PF14547, and PF05617. In total, we detected 828 genes, of which 244, 321, and 229 were distributed across the A, B, and D subgenomes, respectively. These genes encode major prolamins; α-, γ-, and ω-gliadins; HMW and low–molecular weight (LMW) glutenins; the minor prolamin classes purinins and avenin-like proteins (ALPs); the prolamin superfamily members of ATIs, LTPs, and nsLTPs; proline-rich proteins; hydrophob-seed domain–containing proteins; egg cell–secreted proteins; and cortical cell–delineating proteins. For distribution of chromosomal loci, see the Supplementary Materials, fig S1, data file S2, and (1).
Wheat genes associated with specific human disorders
Here, we provide a detailed look at grain allergens and antigens present in the wheat genome belonging to the prolamin superfamily and other non-prolamin families that have been confirmed to elicit adverse immune reactions in a subset of people. Accurate and diagnostic fingerprints from these proteins are expected to support claims concerning health attributes associated with wheat flour samples.
Celiac disease proteins
To identify the celiac antigen domain proteins, linear epitope mapping was performed using reported T cell and B cell epitopes and peptides with known level of immune responses (section S1, figs. S2 and S3, and data file S3) (15). Epitopes were mapped using 100% sequence identity threshold. Major prolamin groups such as gliadins and glutenins are known to carry epitopes causing celiac disease (T cell epitopes) and epitopes involved in the IgA or IgG anti-gliadin responses (18). The ability to isolate disease-relevant T cells from the blood of celiac disease patients after oral gluten ingestion enabled a comprehensive assessment and ranking of the immunostimulatory peptides in gluten (15). The peptide’s capacity to stimulate T cells was measured in an interferon-γ (IFN-γ) enzyme-linked immunospot (ELISPOT) assay that quantifies individual T cells responding to antigen, expressed as “spot-forming units” (SFU). On the basis of the extensive annotation of the food immune response–related protein families in wheat, it was possible to extend the toxicity map concept to all proteins with a known relationship to celiac disease. Using the database of scored peptides for toxicity (15) in our analysis (fig. S2 and data file S2; deposited in the ProPepper database), we detect peptides with known immunoreactivity primarily mapping to the repetitive regions of α-, γ-, and ω-gliadins and LMW glutenins (see also section S1). In contrast, peptides with weaker immunoreactive signals are detected close to the C-terminal ends of most of the major prolamin protein sequences (fig. S2). Proteins containing peptides with strong immunoreactivity are abundant in the D subgenome and less frequent on B subgenome sequences. Peptides with high immune response are detected in 12 proteins, all representing α- and ω-gliadins from the D subgenome. The peptide YLQLQPFPQPQLPYPQPQLP that induces the highest IFN-γ ELISPOT response partially overlaps with a highly immunogenic 33-mer sequence from α-gliadin and its component immunodominant epitopes (DQ2.5-glia-α1a, DQ2.5-glia-α2, and DQ2.5-glia-α1b). Three ω-gliadin sequences from chromosome 1D contain multiple peptide regions that induce a high IFN-γ response (fig. S2). These peptides are enriched in repeats containing the QQPFP sequence that can be quantified by commercial enzyme-linked immunosorbent assay (ELISA) kits using the R5 monoclonal antibody (mAb) (23). Celiac disease–associated B cell epitopes are present in all the major prolamin types and were also present in ALPs and Glo-3 seed storage globulins. Immunostimulatory proteins without known epitopes were found among ATIs and serpins (Fig. 2 and data files S2 and S3).
Baker’s asthma proteins
A diverse range of grain proteins are associated with baker’s asthma, among which only a small number of proteins contain identified linear epitopes. Altogether, 63 linear epitopes related to baker’s asthma were mapped to the translated gene models presented here. Hits are mainly found in ATIs that belong to allergen groups of Tri a 28, Tri a 29, Tri a 40, and Tri a CC. Major baker’s asthma–associated epitopes are present in chromosome group 3 and chromosome group 6 genes encoding ATIs (data file S2).
We identified 30 genes encoding an LTP_2 domain that are homologs to known reference allergens (Tri a 7k LTP or Tri a 14 LTPs) that are also related to baker’s asthma. Immunogenic peptides, including LKCGVNLPYT and VKNLHNQARS, are present in a chromosome 5B–encoded nsLTP (Tri a 14). Tri a 44 allergens represent weak allergens encoded by PR60 nsLTPs located on the short arms of chromosome group 1. Serpins were reported as minor allergens in baker’s asthma (24), and epitopes are detected in serpin genes encoded on chromosome group 5 (data file S3). The chromosomal location and protein family information of the additional non-prolamin allergen families without known epitopes are collected in data file S2.
Food allergy proteins
Food allergy–related linear B cell epitopes are prominent in all the main prolamin types, with the highest number present in ω-gliadins, followed by γ- and α-gliadins (data file S3). Peptides including PQQPFP, QPQQPFP, and QQFPQQQ represent the most frequent food allergy–related epitopes in the wheat genome (data file S3). Epitopes such as QQQPP are also present in some nsLTP sequences encoded on chromosomes 4 and 5 groups and in b-type ALPs. Among non-prolamin allergen groups, we mapped epitopes to serpins encoded on the chromosome 5 group, and some serpins were also associated with celiac disease (see data file S3 for more annotation and mapping details of non-prolamin–type food allergy–related proteins).
WDEIA proteins
One hundred twenty-five linear epitopes related to WDEIA were retrieved from the ProPepper database and mapped to the translated wheat proteins using 100% peptide sequence matching. ω-5 gliadins (Tri a 19) are known as major contributors to WDEIA (25). WDEIA epitopes (for example, QQFPQQQ) were detected in large numbers in chromosome 1B–specific ω-5 gliadins, but they are not identified in ω-1,2 gliadins (Fig. 3 and data file S3). In contrast, we have found WDEIA epitopes that were originally identified in HMW glutenins, and γ-gliadins in ω-gliadin sequences that are known immunostimulatory proteins in celiac disease. The frequency of ω-5 WDEIA epitopes is extremely high (maximum of 101 epitopes per sequence) in chromosome 1B–specific ω-gliadins (data file S3). Protein fragments containing peptides like QQPGQ and QQSGQ related to WDEIA are present in significantly larger numbers in the x-type HMW glutenins (75 to 143 epitopes per sequence) than in y-type HMW glutenins (48 to 53 epitopes per sequence; data file S3).
Fig. 3. Epitope mapping and phylogenetic analysis in Prolamin clan (CL0482) protein families, HMW glutenins, and ω-gliadins.
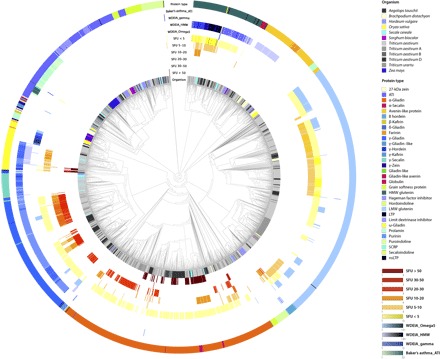
Protein sequences with gliadin (PF13016), protease inhibitor, seed storage and lipid transfer (PF00234), HMW glutenin (PF03157) domains, and ω-gliadins were used to analyze the expansion of the epitope content and composition. Protein sequences were retrieved from UniProt and used along with the reference genome sequence data of bread wheat, T. urartu, A. tauschii, barley, rye, and other grasses such as rice, Brachypodium, maize, and sorghum for phylogenetic analysis. Peptides that induce IFN-γ responses were grouped into six immune response groups (based on median SFU) and colored separately. Linear epitopes related to WDEIA and baker’s asthma are also labeled. The number of peptides per sequence is highlighted by color intensity changes. Linear epitopes related to WDEIA and baker’s asthma are also labeled. SCRP, small cysteine-rich protein.
NCWS proteins
α-Amylase/trypsin inhibitor subclasses ATI 0.19 and ATI CM3 have been related recently to NCWS (10). Their genes are encoded on chromosome groups 3, 4, and 7.
The allergenic and immunogenic peptides of bread wheat are characteristic of the Triticeae species
Phylogenetic analysis and epitope mapping using peptides related to celiac disease, WDEIA, and baker’s asthma identified three broad monophyletic clades within the Triticeae species and other grasses, including rice, Brachypodium, maize, and sorghum (Fig. 3, fig S3, data file S3, and the Supplementary Materials). Among these protein families, only proteins with Gliadin, Tryp_alpha_amyl, and LTP_2 domains have known linear epitopes for celiac disease, baker’s asthma, WDEIA, or food allergy in their sequences. On the basis of immune-response differences in peptides with known immunoreactivity measured as IFNγ-ELISPOT response in individual patients (15), the proteins can be further classified into six toxicity strength groups (Fig. 3). Proteins containing peptides that induce the strongest immune responses (arbitrarily defined by an SFU above 30 based on the IFNγ-ELISPOT assay) were only found in α- and ω-gliadins of bread wheat, the donors of A and D subgenomes (that is, Triticum urartu and Aegilops tauschii, respectively), and rye, but not in barley. Proteins with a medium level (median SFU between 10 and 30) of immune response were identified in all three gliadin types of chromosome groups 1 and 6, in some of the HMW glutenins, and in a few barley B hordeins. Weak antigen-producing peptides were mapped to HMW and LMW glutenins of wheat, rye, Brachypodium, and barley, and to all the gliadin types.
Linear WDEIA-γ epitopes are present mainly in γ-gliadins of bread wheat, T. urartu, and A. tauschii (Fig. 3). A few occurrences are detected in some α-gliadin, barley B hordein, and wheat LMW glutenin sequences. A large number of HMW glutenin–specific WDEIA epitopes are detected in HMW glutenins in all Triticeae. However, they are also characteristic of ALPs, ω-gliadins, and some γ-gliadins of the same taxa. WDEIA-related ω-5 epitopes are present in wheat, barley, and Brachypodium sequences. Baker’s asthma epitopes were found in all Triticeae ATIs. The clinical significance of these epitopes in non-wheat cereals for patient management remains unclear, but the findings suggest that immune reactions could be triggered by these other cereal proteins.
The presence of the α-gliadin–specific 33-mer peptide was investigated along with its five overlapping immunodominant T cell epitopes (fig. S4B). We identified α-gliadin–like prolamin sequences only in bread wheat, its genome donors, and rye. Altogether, 21 of the 534 investigated α-gliadin sequences contain this peptide, all of them with D subgenome origin (data file S3). Using the ω-gliadin sequences identified in the reference genome along with ω-gliadin sequences of A. tauschii, T. urartu, and ω-secalins in rye, we clustered the proteins into two major groups (fig. S4A). The branch in blue labels highly allergenic ω-5 gliadins, the major contributors of WDEIA. ω-1,2 gliadins are grouped into two subgroups from which the subcluster labeled in red represents an ω-gliadin group highly immunogenic in celiac disease. Notably, peptides found in these proteins have a similar level of immunogenicity in celiac disease to the alpha 33-mer peptides (15), whereas ω-secalins contain relatively weak immunostimulatory peptides.
Genetic variation in the gliadin and glutenin families among wheat cultivars
To investigate the effect of genetic variation on allergen/antigen potential in bread wheat cultivars, we compared the sequences of 133 gliadins and glutenins among the reference genotype Chinese Spring and the Norwegian bread wheat cultivars Bjarne and Berserk. In total, 395 single-nucleotide polymorphisms (SNPs) were detected (see Materials and Methods). In Bjarne, 70 gliadin and glutenin genes show allelic variations compared to the reference genome of Chinese Spring. In Berserk, 353 SNPs covering 73 genes were identified compared to the reference genome. SNPs were most frequently enriched in pseudo genes for α- and ω-gliadins. Seventeen α-gliadin, 6 γ-gliadin, and 16 ω-gliadin sequences were identical in Berserk and Bjarne but differed from those in Chinese Spring. In addition, 12 α-gliadins, 2 ω-gliadin, and 4 γ-gliadin sequences carried unique SNPs in Berserk. In Bjarne, unique changes in orthologous genes were detected in 12 α-gliadin, 1 γ-gliadin, and 2 ω-gliadin sequences. The major immunoreactive regions including the α-gliadin–specific 33-mer peptide and the highly toxic ω-gliadin regions were not different among the genotypes. In the γ-gliadins, some of the genetic variations affected the composition of the epitopes with low immunoreactivity. Some SNPs cause a modified number of cysteine residues that are predicted to have a direct effect on the functional properties.
Influence of growth temperature on grain allergen and antigen-response proteins
The reference allergen and antigen map described above provides an opportunity to explore the influence of temperature regime on these proteins in the two Norwegian bread wheat cultivars, Bjarne and Berserk, and Chinese Spring. For this, we used matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry (MALDI-TOF-MS) profile analysis of fractions of ω- and α-gliadins with strong immunoreactivity collected by reverse-phase high-performance liquid chromatography (RP-HPLC). The protein content and composition data in the three cultivars under the different temperature regimes are provided in section S1 and table S1.
The immunoreactive ω-gliadins were retrieved between retention times of 25.3 and 25.4 min and in the mass range of 41 to 44 kDa in all three cultivars (Fig. 4A and table S1). Using peak analysis of the MALDI profiles, we also identified proteins with small molecular weight from the same fraction that may represent fast ω-gliadins and other small sulfur-rich proteins without highly immunoreactive peptides (Fig. 4A). Under normal conditions (20°C/16°C day/night), the portion of immunoreactive ω-gliadins in Berserk, Bjarne, and Chinese Spring comprise 3.7, 6.6, and 5.4% of total protein, respectively. Low temperature had greatly decreased the levels of toxic ω-gliadins by 23.3% (from 0.6 to 0.46%), 41.3% (from 1.09 to 0.64%), and 17.8% (from 0.9 to 0.74%) in Berserk, Bjarne, and Chinese Spring, respectively. The effect of high temperature was more pronounced in Chinese Spring and Berserk, with an increase of 25.6 and 13.3%, respectively. Only a small increase of 3.7% was observed in Bjarne. These data show that strong antigen ω-gliadins are expressed in high amounts in grains and that their expression level is significantly affected by temperature.
Fig. 4. Quantification and protein profiling of major immunoreactive protein types in Chinese Spring, Bjarne, and Berserk.
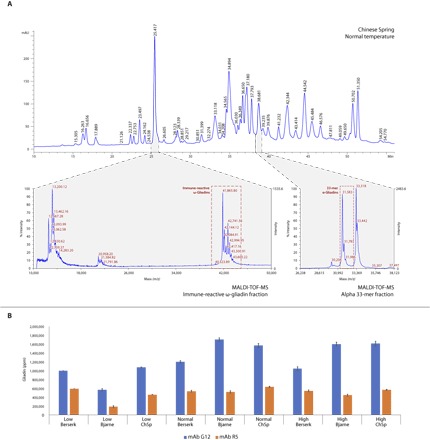
(A) MALDI-TOF analysis of major immunoreactive protein fractions using fractions collected in the RP-HPLC analysis. (B) Peptides measured by R5 and G12 mAbs are characteristic of main immunoreactive proteins related to celiac disease and WDEIA. Expression changes of these proteins were measured in three temperature regimes. m/z, mass/charge ratio.
Immunotoxic 33-mer–containing α-gliadins with monoisotopic mass values of 31.6 and 31.8 kDa were identified with retention times of 38 to 38.8 min in all three cultivars (Fig. 4A). This RP-HPLC peak represents the major α-gliadin fraction and is composed of six individual α-gliadin proteins within a molecular mass range of 30.2 to 33.4 kDa. Under normal temperature conditions, this fraction comprises 2.7 to 3.1% of the total protein content in all three cultivars. The level of decrease in response to low temperature for all three cultivars was similar and in the range of 28 to 30%.
Overall expression levels of allergen and antigen-response epitopes are routinely measured by the R5 or G12 mAbs (23). Of these, R5 mAb primarily detects QQPFP peptides that are present in 67% of α-gliadins in Chinese Spring. This peptide is found in as many as 90% of the γ-gliadin sequences and 28% of the complete and functional ω-gliadin sequences, but is absent from HMW glutenins (data file S3). Our peptide mapping results detect quantitative variation in the underlying proteins between three cultivars. Under normal conditions, Bjarne produced a stronger G12 mAb response, while Berserk showed a lower G12 mAb response (Fig. 4B). Low temperature led to a significant decrease in the G12 peptide level in all three cultivars, while high temperature resulted in a moderate decrease. Thus, we measured 30% less R5 peptide content in Bjarne and 16% less R5 peptide content in Berserk compared to Chinese Spring under normal conditions. This level was decreased by 30% under low temperature in Bjarne but did not change significantly in Berserk. High-temperature conditions had a slightly negative impact on R5 mAb response (Fig. 4B).
Transcript abundance for wheat allergens and antigens varies between genotypes, grain cell types, and growth temperature
The endosperm is the source of flour for baking and consists of three major cell types: (i) starchy endosperm, which stores gluten protein and starch; (ii) aleurone cells, a lipid storage tissue that secrets enzymes to recruit sugar and amino acids upon grain germination; and (iii) transfer cells facilitating uptake of sucrose from the photosynthetic tissues (25). To assess the influence of temperature and genotype on transcript abundance of 356 reference wheat allergens, we carried out an RNA sequencing (RNA-seq) analysis using the Bjarne, Berserk, and Chinese Spring genotypes grown under three different temperature regimes (see above). For each genotype and temperature, we extracted RNA from three cell types: starchy endosperm, aleurone, and transfer cells (Fig. 5). The transcripts for the majority of these reference allergen homologs were expressed in the starchy endosperm and in transfer cells. The nsLTP transcripts fall into two patterns based on their cell specificity of expression, with the first group showing highest expression in transfer cells (A in Fig. 5). The second group, also including transcripts encoding chitinases, globulins, and transcription elongation factors, is mainly expressed in aleurone cells (B in Fig. 5). In addition, the expression level of α-gliadins significantly differs between the three wheat genotypes, with a group of 21 α-gliadins being down-regulated in Bjarne compared to Chinese Spring and Berserk (C in Fig. 5). Also, a clear difference in expression levels for transcripts encoding α-, γ-, and ω-gliadins; LMW and HMW glutenins; ALPs; and ATIs exists in the starchy endosperm between Chinese Spring on the one hand and Bjarne and Berserk on the other (D in Fig. 5). For a majority of transcripts in aleurone cells, the expression level is lower in Bjarne and Berserk compared to Chinese Spring (Fig. 5). Gene expression in Chinese Spring is most severely reduced by low temperature in the starchy endosperm, with over 70% of the endosperm transcript down-regulated compared to normal temperature. Transcripts encoding nsLTPs, ALPs, and chitinases are the most severely reduced by low temperature. In starchy endosperm cells, high temperature significantly decreased the expression level of transcripts encoding chitinases, glutathione S-transferases, ALPs, and ATIs and increased the level of serine carboxypeptidases, peroxidases, and some ATIs transcripts (Fig. 5). ATIs associated with celiac disease and baker’s asthma are mainly expressed in transfer cells and starchy endosperm. In transfer cells, most of these transcripts show increased expression under low temperature and reduced expression under high temperature (E in Fig. 5).
Fig. 5. Effect of cell type, genotype, and temperature on transcript levels of genes encoding grain allergens.
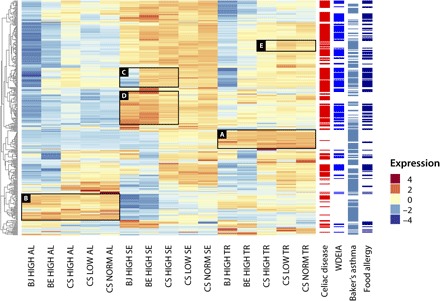
Heat map showing relative transcript levels of genes encoding reference allergens across cell types, genotypes (BJ, Bjarne; BE, Berserk; and CS, Chinese Spring), and temperatures (CS only). Association of reference allergen transcripts with celiac disease, WDEIA, Baker’s asthma, and food allergy.
The cumulative level of expression of the 54 transcripts encoding 63 peptides with known immunoreactivity strength (IFNγ-ELISPOT response in median SFU value) was calculated for the Chinese Spring, Bjarne, and Berserk (Fig. 6A and data file S4). The highest levels of expression of transcripts encoding these immunoreactive peptides are found in starchy endosperm cells, while their lowest levels are found in aleurone cells. In starchy endosperm, the influence of genotype is most marked for Berserk, with a higher expression level for most of the transcripts encoding the immunoreactive peptides (Fig. 6A). Five peptide-encoding transcripts with increased expression in Berserk compared to Bjarne are present in α-gliadins and one ω-gliadin with low to medium immunoreactivity, except for the peptide QPFPQPQQPFPWQPQQPFPQ, which represents a highly immunoreactive peptide. For transcripts with higher expression in Bjarne compared to Berserk, four mapped to γ-gliadins with low to medium immunoreactivity and one mapped to peptide YLQLQPFPQPQLPYSQPQP representing an α-gliadin. The relative levels of transcript described here reveal substantial differences between genotypes as well as between cell types in developing grains.
Fig. 6. Expression profile of the 54 genes encoding the 63 identified immunoreactive gliadin and glutenin peptides in the cells of the endosperm of Bjarne and Berserk at high temperature and Chinese Spring at high, low, and normal temperatures.
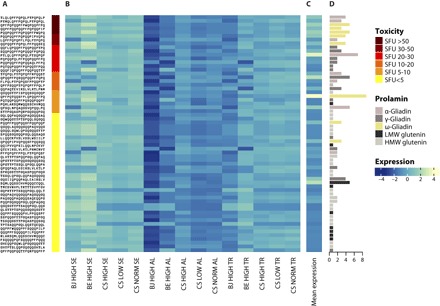
(A) Peptide identity and IFNγ-ELISPOT responses in median SFU values representing the immunoreactivity of peptides against patients’ blood sera according to Tye-Din et al. (15). Dark red represents strong immunoreactivity values, and yellow represents weak values. (B) Heat map showing the relative cumulative expression of the genes encoding each peptide across cell types, genotypes (BJ, Bjarne; BE, Berserk; and CS, Chinese Spring), and temperatures. (C) Heat map showing the scaled average expression level of the immunoreactive peptides across all endosperm cell types. (D) Number and identity of proteins containing the individual immunoreactive peptides.
One approach to evaluating commercial wheat cultivars for their potential to stimulate an allergic or adverse immune response is to measure the cumulative level of transcripts encoding peptides causing these reactions. By calculating the mean level of gene expression for the transcripts encoding each peptide, we detected relatively high levels of the transcripts encoding the most allergenic peptides (Fig. 6C). Among the top five selected peptides, transcripts encoding proteins with the strong immunoreactivity value peptides QPFPQPQQPFPWQPQQPFPQ and PQQPQQPFPQPQQPFPWQPQ are present in five ω-gliadins from the D subgenome (data file S4). For all 63 peptides investigated, expression levels decreased in the starchy endosperm in the low-temperature growth regime. In high temperature, no clear pattern could be detected. No significant difference was observed in starchy endosperm between genes encoding high-immunoreactivity peptides and those encoding low-immunoreactivity peptides in gene expression changes due to temperature and genotype.
To see if there is correspondence between the transcript abundance and protein expression, we have compared the transcriptional and translational expression profiles for immunoresponsive gliadins and glutenins. Both analyses (Supplementary Materials and table S1) show significantly reduced expression levels under a low-temperature regime and increased expression levels in response to high temperature. The transcriptome profile of gliadins shows reduced expression in Bjarne compared to Berserk and Chinese Spring, which is also reflected by proteomics. For glutenins, protein levels are much higher in Chinese Spring compared to Bjarne and Berserk, while the transcript abundances show slightly higher expression in Bjarne. The correspondence between transcript and protein levels suggests that transcript steady-state levels may be used to represent genotype allergen/antigen levels.
DISCUSSION
Wheat remains a major food crop around the world because of its favorable nutritional properties and adaptability to a range of climates. Its high gluten content imparts excellent rheological properties favorable for baking and enhances a range of food textures and palatability. While it is the primary cereal consumed in Europe, North America, and the Middle East, it is also becoming increasingly popular in Asia. Although only a small proportion of the global population cannot consume or be directly exposed to wheat due to specific medical illnesses, characterizing the genome regions that contribute to both disease and its favorable nutritional aspects is of importance given the prominence and importance of this grain in the human diet. To accomplish this analysis, we use the newly developed high-quality reference wheat genome data set to map the genomic regions associated with or implicated in human wheat-associated disease and examine the factors that affect gene expression.
Using the high-quality IWGSC RefSeq v1.0 reference genome (1) combined with a comprehensive analysis workflow, we have identified known and previously unknown members of the prolamin superfamily, the major contributors of food- and inhalation-related diseases. Homology-based analysis against known references and precise manual mapping were performed to compile a reference allergen/antigen map of wheat. The allocation of Pfam domains and linear epitopes to the complete wheat genome has helped us identify a reference allergy/immunostimulatory gene set and thus facilitate the identification of major chromosomal regions as potential targets for breeding programs. Genes associated with the same illness often cluster together at the telomere region of the chromosomes. The genome-scale identification and mapping of proteins related to food intolerance in wheat have enabled the detailed identification of so far unknown or less-characterized syntenic genes in related cereal species, such as durum, barley, or rye. Because of their common ancestry and over 70% sequence similarity, this reference allergen/antigen map can also facilitate the identification of the immunostimulatory regions in barley, rye, and other wheat-related species. When peptide sets with known IFN-γ responses (15) were used for the mapping, strong antigen proteins were mainly found in the A and D subgenomes of bread wheat, its genome donors, and rye. Peptides with medium and weak responses were also found in barley. These findings highlight the importance of cross-reactive peptides and proteins in wheat species and related cereals.
Chromosome 1 group glutenins and gliadins play a significant role in dough functionality but are also primary contributors to food intolerances in wheat. Our precise manual mapping of γ-gliadins, ω-gliadins, purinins, and LMW glutenins highlighted a more complex organization of the Glu-3 and Gli-1 loci. Compared to previous studies using sequencing of bacterial artificial chromosome (26, 27), the reference sequence assembly provided evidence for LMW glutenin genes and ω-gliadins not forming separate clusters, as is the case of γ-gliadins. While the genomic regions coding the LMW glutenin and ω-gliadin genes are strongly enriched in NLR genes related to pathogen stress (1), no NLR protein-coding genes were found within the γ-gliadin cluster. Our exact characterization of α-gliadin gene families on chromosomes 6A and 6B enabled the identification and precise mapping of the homeolog gene cluster on chromosome 6D.
We have found that the 33–amino acid–long peptide considered to be one of the most immunogenic gluten peptides (LQLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF) is in fact not commonly found in the α-gliadin sequences examined in our study, although it is often present in the bread wheat cultivars (17). Therefore, this provides an important candidate target for deletion from the genome (28). In the Chinese Spring reference genome, only 2 of the 59 α-gliadin sequences contained the peptide, both encoded by chromosome 6D, confirming previous results. The SNP variant analysis proved the presence of the 33-mer–containing α-gliadins in both Norwegian cultivars, Berserk and Bjarne. Our phylogenomic analyses indicated that the epitopes composing the alpha 33-mer peptide are present in different numbers and combinations in the genome donors of bread wheat. However, the complete peptide is characteristic only of the D genome that evolved 0.5 million years ago (29–31). The early origin of these epitopes in the history of the hexaploid wheat genome explains the high frequency of bread wheat cultivars with this highly immunogenic region (17). A stable isotope dilution assay in modern and old wheat cultivars and spelt samples (17) indicates approximately 0.42 g of the 33-mer containing protein in 100 g of flour. In our analysis, we collected a relatively small protein fraction with a retention time frame of 38 to 38.8 min containing six α-gliadin proteins. The molecular mass range was from 31.5 to 33.7 kDa and consisted of 33-mers amounting to 0.51 g per 100 g of flour. Using quantification of this fraction along with the highly immunoreactive ω-gliadin fraction by RP-HPLC, we therefore provide the basis for a relatively affordable diagnostic assay that can also be used in breeding programs. Additional prolamin gene clusters, such as the gliadin-like proteins on chromosome 3 and ALPs located on chromosomes 4A, 7A, and 7D, can also contribute to immune responses and represent cysteine-rich, grain-specific, prolamin protein families requiring further investigation.
Transcriptome data established the spatial and temporal expression patterns of reference allergens and antigens in three cell types of the wheat grain. Although transcripts of the major prolamin classes (gliadins and glutenins) were primarily enriched in the starchy endosperm, they were also detected in aleurone and transfer cells in the developing grain. Other members of the prolamin superfamily, including LTPs, ATIs, or the ALPs, were enriched in the aleurone and transfer cell layers compared to starchy endosperm cells. For the latter proteins, removal or reduction by milling techniques may be feasible. Significant changes in their spatial and temporal expression pattern under temperature stress conditions indicate their possible stress-related function during seed development or in seed germination.
Climate change and the increase in global mean temperatures accompanied by increased severity and frequency of extreme temperatures can result in two major forms of temperature stress on crops: more frequent heat stress and less frequent cold temperature stress. Temperature stress applied before flowering primarily affects the formation and number of spikelets, while temperature stress at flowering mainly affects floral development and grain number. Post-anthesis temperature stress, however, has a significant effect on starch and protein accumulation, as well as on protein composition. Timing and length of the stress directly affect the final protein content and composition, showing a more elevated effect when stress occurs during mid to late grain-filling period. Although these changes often result in decreased starch and increased protein contents, the protein composition shows a fine-tuned response in the end-product quality. Previous reports demonstrated the significant effect of high-temperature stress on grain development (20, 32), generally resulting in shorter maturation times, increased storage protein accumulation combined with a loss of metabolism-related proteins, and increased level of stress defense–related proteins. In most of these studies, increased α-gliadin and HMW glutenin accumulation was coupled with lower levels of LMW glutenin expression. In contrast, low-temperature stress decreases the seed N accumulation rate per day, resulting in a prolonged duration of grain filling and protein accumulation. The gene and cellular level responses identified in our study confirm the effect on prolamin superfamily gene expression. Knowledge of the effect of growth conditions on allergen content may provide selection criteria for grains to particularly sensitive consumers.
The results of this study demonstrate significant temperature effects on gene transcript steady-state levels and protein content. The effect of low- and high-temperature environment stress on seed protein composition of Bjarne and Berserk was previously studied by Uhlen et al. (33). Differences in grain weight, protein content, and dough quality parameters between the two environments studied were related to temperature. Here, we demonstrate the effect of temperature on food allergen and antigen content. Under high-temperature stress conditions, the changes in seed storage protein accumulation resulted in slightly increased expression of ω- and α-gliadins, the primary triggers of celiac disease and important contributors to occupational asthma and food allergies. In high temperature, the amount of 33-mer containing protein fraction increased by 25 to 33%, and the toxic ω-gliadin content increased by 3 to 26%. Bjarne, a high-protein Norwegian cultivar showed significantly different expression patterns in these major allergens. Although the effect of high temperature was not significant, the effect of low-temperature conditions resulted in 43% less toxic ω-gliadin content. Low-temperature conditions during seed development decreased the level of protein fractions primarily associated with celiac disease but increased the content of protein families related to WDEIA or baker’s asthma, like nsLTPs, ATIs, hydrolases, and peroxidases. The precise chromosomal mapping and functional annotation of these immunostimulatory gene families can be used in selection programs targeting traits, such as producing low-gliadin lines. Although RNA interference or genome editing eliminates the complete gliadin gene loci by using null lines, it may have an overall negative impact on storage protein accumulation; it does not necessarily result in significant loss of mixing properties (34, 35). However, the compensatory effect, resulting in modified expression levels of other prolamin classes, can have a negative impact on the total immunoreactive peptide content and composition. Mapping peptides with known strength in allergen/antigen response can now be applied to develop wheat lines with a significantly lowered allergen/immune response and gluten content and can directly enhance the method development for the food industry to precisely characterize and quantify the disease-associated protein content (28). The combined use of genome sequence and epitope databases, in silico prediction methodology, and cereal chemistry as presented in this manuscript results in a better understanding of the level of proteins that have the potential to induce significant immune responses present in the end products from wheat flour. The genetic variability and quantitation of these protein classes and understanding their environmental stability will enhance the production of food that can be used as a safe and healthy alternative to a currently highly restrictive approach that relies on absolute wheat and gluten avoidance.
MATERIALS AND METHODS
Identification of prolamin superfamily member genes
Domain alignments of Pfam Prolamin clan (CL0482) members characterized by a conserved disulfide-bonding patterning—including Pfam domains of Gliadin (PF13016), LTP_2 (PF14368), Hydrophob_seed (PF14547), Tryp_alpha_amyl (PF00234), and Prolamin_like (PF05617)—were used to identify gene members of the prolamin superfamily. HMW glutenin genes were identified using the domain alignments of Glutenin_HMW (PF03157). The presence of signal peptide regions and conserved cysteine-rich pattern was used to confirm features characteristic of prolamin superfamily members. Pattern (-C-C-Xn-C-Xn-C-) was used to identify small cysteine-rich sequences without a proper Pfam domain structure. Hits were manually checked; protein family and subfamily classes were aligned with the prolamin protein collection of the ProPepper database using ClustalW algorithm (18, 36).
Identification of potential and reference allergen and antigene genes using reference allergen Pfam signals
AllFam database and Pfam protein family domains related to known allergens were used to identify reference allergens and antigens (37). Pfam domain search in the reference genome was performed using profile hidden Markov models (HMMER3), as described in IWGSC 2018 (1).
Reference allergen homologs were identified using a 70% sequence similarity threshold. Similarly, proteins with known immunoreactivity in celiac disease patients with or without known linear epitope regions were also used to identify homologs with over 70% sequence identity. Similarities in protein size, position of cysteine residues, presence of a signal peptide region, and conserved secondary structural elements were also considered. In addition, known linear allergen and antigen epitopes were mapped. Position of genes encoding identified reference allergens were represented in a genome-based allergen map visualization in Adobe Illustrator.
Epitope mapping
Annotated linear epitope collection was retrieved from the ProPepper database (18) and used to map known immunoreactive protein sections. Celiac disease–associated HLA-DQ2–specific, HLA-DQ8–specific, and anti-gliadin epitopes were mapped to the protein sequences. Similarly, known peptide sequences related to WDEIA, baker’s asthma, or food allergy were used to map epitopes to the translated gene sequences. Only hits with 100% sequence identity were used for further analysis. Peptide hits covering unsequenced regions or with inner stop codons were excluded from the analyses. Epitope hits were visualized using the Motif search algorithm of CLC Genomics Workbench (v10.1). Similarly, peptides detected by the mAbs of the commercial kits R5 (R-Biopharm) and G12 (Romerlabs) were also mapped to the translated gene sequences using 100% sequence identity threshold. Epitope prevalence and epitope frequency were evaluated for each protein individually. Epitope count by protein sequence values are summarized in data file S3.
Epitope toxicity analyses
Peptide sequences with known individual IFN-γ responses measured on the ELISPOT assay in median SFU per million peripheral blood mononuclear cells values were obtained from (15). Peptide sets of wheat, secalin, and hordein secondary response peptides and the top 50 immunoreactive gliadin and glutenin peptides were used in the analysis. Median values considering all the patient responses were calculated. Individual peptide strength values were binned in six immune strength groups: SFU > 50, SFU 30 to 50, SFU 20 to 30, SFU 10 to 20, SFU 5 to 10, and SFU < 5 (data file S3). Peptide collections were mapped to α-, γ-, and ω-gliadin, and HMW and LMW glutenin sequences obtained from the translated prolamin gene sequences. Epitope expression values were calculated as a sum of gene expression values in which the encoded peptide sequence is present.
Phylogenetic analysis of the prolamin superfamily genes
The predicted protein sequences from each of the bread wheat subgenomes v1.0 (1) were combined with the predicted protein sequences from nine other Poaceae species (A. tauschii, T. urartu, Secale cereale, Hordeum vulgare subsp. vulgare cv. Morex, H. vulgare var. nudum, Brachypodium distachyon, Oryza sativa, Sorghum bicolor, and Zea mays) for phylogenomic analysis. The bread wheat subgenomes were handled as independent taxa. The protein sequences were screened and filtered as described in IWGSC 2018 (1). From the orthologous groups, all those with proteins containing one or more of the domains defining Prolamin clan (CL0482) were extracted. In addition, groups with a Glutenin_hmw (PF03157) domain were also considered. From this data set, the bread wheat v1.0 protein sequences were replaced by the manually curated v1.1 translated gene models from this study. Sequence alignment and tree constructions were performed as described elsewhere (1). Visualization and annotation of the trees were then carried out using CLC Genomics Workbench v10.1.
Protein sequences from the abovementioned analysis plus public sequences originating from the same taxa deposited in the UniProt database with PF13016, PF00234, and PF03157 domains were used along with domain-less ω-gliadins to perform a second phylogenetic analysis using the same methods as described above to explore epitope prevalence and expansion in bread wheat and related species. Epitope mapping described above was used for the annotation; specific epitope types and their frequency were counted.
Plant material growth conditions, dissection, and RNA extraction
Chinese Spring, the genotype used for the IWGSC sequencing, was grown along with two Norwegian cultivars, Bjarne (SvB87293/Bastian, released in 2002) and Berserk (Bastian/KN6//SW35128, released in 2007). Plants were grown in the phytotron at the University of Oslo. Four replicates were planted, each replicate with nine pots and four seedlings per pot organized on separate trolleys in the phytotron chamber. The plants were grown at 15°C (8 hours, night)/20°C (16 hours, day) until the heading of the first ear, and the temperature regime was then shifted to 20°C (8 hours, night)/26°C (16 hours, day). Single ears were tagged at anthesis to be harvested at the precise development stage during grain development. Developmental stages were determined on the basis of the heat sum received by the plants from anthesis. Ear samples were harvested immature, corresponding to 20 days after pollination (DAP) at a temperature regime of 20°C/15°C (16 hours, day/8 hours, night), yielding 367 day degrees. For the temperature regime reported here (26°C/20°C day/night), the ears were harvested at 15 DAP (corresponding to 367 day degrees). For Chinese Spring, a total of nine ears were harvested from each replicate. For Bjarne and Berserk, five ears were harvested from each replicate. Individual cell type and RNA isolation were performed as described in Pfeifer et al. (38). A total of 27 samples (3 genotypes × 3 replicates × 3 cell types) were submitted for RNA-seq analysis using paired-end libraries with 200–base pair inserts and an Illumina HiSeq2000 sequencer (Illumina). In addition, Chinese Spring was grown under two different temperature regimes, 15°C/10°C day/night and 20°C/15°C day/night (normal temperature). The plants were grown in normal temperature until the heading of the first ear and then temperature was changed. For the normal condition regime, ears were harvested at 20 DAP, and for the 15°C/10°C day/night regime, ears were harvested at 29 DAP (corresponding to 367 day degrees in the 26°C/20°C temperature regime). Ears were collected and dissected as described above. In total, 18 samples (2 temperatures × 3 replicates × 3 cell types) were submitted for RNA-seq analysis using paired-end libraries with 200–base pair inserts and an Illumina HiSeq2000 sequencer (Illumina).
Read mapping
RNA-seq short reads were trimmed using Trimmomatic with the parameters (ILLUMINACLIP:TrueSeq3-PE-2.fa:2:30:10:8: true LEADING:3 SLIDINGWINDOW:20:20 MINLEN:40). RNA-seq reads were mapped with kallisto 0.43.1 (default parameters) to the RefSeq v1.0 transcriptome including corrected transcript models for 984 genes to obtain transcript abundances. These counts were used as input DESeq2 to obtain log-transformed normalized expression counts.
Gene variant detection and SNP analysis
Gene variant analysis of α- and γ-gliadin and LMW glutenin classes was performed using Basic Variant Detection followed by the Low Frequency Variant Detection tool (CLC Genomics Workbench v10.1). Sequencing errors were excluded using an error model estimation with a statistical significance value of 1%. RNA-seq reads from biological replicates were mapped individually, and variants present in all three replicates were considered for further analyses. Single-nucleotide variants, multiple-nucleotide variants, and short- and medium-sized insertions/deletions were annotated as the cultivar-specific variants. Translated CDS sequences were used to check the changes in amino acid order secondary and protein structure. Nonspecific matches with below-threshold similarity (0.8) and length fractions (0.9) were ignored, and read maps obtained from the tree tissue libraries were merged for each replicate. Single-nucleotide variations and insertions/deletions resulting in nonsynonymous codon changes were only considered if variants were present in all three replicates.
Protein content measurements and protein extraction
Mature grains were milled to whole meal in a Laboratory Mill 3100 (Perten Instruments AB) with a 0.8-mm sieve. Total N content of flour was measured by the Dumas method according to Bremner and Mulvaney (39) and expressed as percent dry matter. Protein content of whole meal (% PC) was calculated by multiplying the N content by 5.7.
Size-exclusion high-performance liquid chromatography
Proteins were extracted from 100 mg of whole meal using 1 ml of 0.05 M phosphate-buffered saline buffer (pH 6.9) with 0.05% SDS. The separation and quantification of protein extracts were performed by HPLCq using an Agilent 1200 LC system (Agilent Technologies; http://www.agilent.com) following the method described by Larroque and Békés (40). Extracts (10 μl) were injected into a Bio SEC-5 (4.6 × 300 mm, 300 Å; Agilent Technologies) column maintained at room temperature. The analysis was performed in a buffer system using 0.5% SDS-phosphate buffer (pH 6.9). Supernatants were filtered through 0.45-μm polyvinylidene difluoride filters before HPLC analysis. The eluents used were ultrapure water (solvent A) and acetonitrile (ACN) (solvent B), each containing 0.1% trifluoroacetic acid (TFA) (HPLC-grade, Sigma-Aldrich). The flow rate was adjusted to 0.350 μl/min. Protein was separated by using a constant gradient with 50% of solvent A and 50% of solvent B in 15 min and detected by ultraviolet (UV) absorbance at 210 nm.
Reverse-phase high-performance liquid chromatography
Flour (60 mg) was extracted using 70% ethanol and vortex for 30 min in a horizontal vortex (MO BIO Laboratories Inc., Vortex-Genie 2). Samples were centrifuged for 15 min at 13,000 rpm using an Eppendorf Centrifuge 5424. Supernatant was filtered using a 0.45-μl filter into an HPLC glass vial. The protein extracts were separated using an Agilent 1200 LC system (Agilent Technologies) and the method of Larroque et al. (41). Extract (10 μl) was injected into a C18 reverse-phase ZORBAX 300SB-C18 column (4.6 × 150 mm, 5 μm, 300 Å; Agilent Technologies) maintained at 60°C. The eluents used were ultrapure water (solvent A) and ACN (solvent B), each containing 0.1% TFA (HPLC-grade, Sigma-Aldrich). The flow rate was adjusted to 1 ml/min. Protein was separated using a linear gradient from 21 to 47% of solvent B in 55 min and detected by UV absorbance at 210 nm. Every sample was sequentially injected twice for technical replication. RP-HPLC peak areas (expressed in arbitrary units) under the chromatograms were used to calculate gliadin amounts. ω-Gliadins were considered between 15 and 30 min; γ-gliadins, between 40 and 55 min; and α-gliadins, between 30 and 40 min. Individual peaks were collected as separate RP-HPLC fractions, freeze-dried, and resuspended in 100 μl of 70% ethanol.
Protein profiling using MALDI-TOF-MS analysis
Gliadin extraction was performed using 70% EtOH. MALDI-TOF-MS was used to obtain the mass spectra of the gliadin extracts and also those of their RP-HPLC fractions collected at different retention times. Sinapinic acid (SA) in 50% ACN and 0.05% TFA (10 mg/ml) was used as matrix. Samples were mixed in 1:10 proportion with SA dissolved in 50% ACN containing 0.1% TFA. Supernatant (1 μl) was spotted onto a 100-spot plate and dried at room temperature. An additional 1 μl of sample layer was applied. Biosystems Voyager DE Pro MALDI-TOF-MS was operated in linear high mass positive mode using 2050 V laser intensity, an acceleration voltage of 25 kV, grid at 93%, and guide wire at 0.2 settings, with a delay time of 700 ns. One thousand spectra were captured per profile. Each sample was replicated three times to avoid experimental errors. The detection mass range was set between 10,000 and 60,000 Da (m/z).
Enzyme-linked immunosorbent assay
The R5 Ridascreen Gliadin (R-Biopharm) sandwich enzyme immunoassay and the AgraQuant Gluten G12 (Romer Labs) sandwich enzyme assay were used to measure differences in the toxic epitope content in the various treatments and cultivars. Prolamin extracts were prepared in four replicates using the methodology provided by the kit suppliers. To measure the gliadin content of wheat flour samples, extracts were diluted by 1:5000 in the case of R5 and 1:10,000 in the case of G12 because of the different detection limits. ELISAs were performed as outlined in the manuals of the assays provided by the manufacturers. Results were interpreted by interpolating optical density (OD) values from the standard values, corrected by the dilution factor used for the flour samples. Calculated gliadin contents determined by the ELISAs were normalized by the protein content and gliadin/gluten ratio of each sample.
Supplementary Material
Acknowledgments
We thank A.-K. Uhlen for planning the experiments and performing grain protein analysis and C. Borjigin for dissecting the endosperm material. A.J. thanks the “Improving wheat quality for processing and health” Expert Working Group of the Wheat Initiative for the valuable discussions. Funding: We thank Graminor AS and the Norwegian Research Council (NFR) for financial support for NFR project 199387; Australian Government Department of Industry, Innovation, Science, Research and Tertiary Education funding agreement ACSRF00542; and Grain Research Development Corporation agreement UMU00037. G.G. was supported by the Hungarian National Research Fund (OTKA PD 115641). A.J., G.G., and Z.B. also thank the European Union together with the European Social Fund (grant nos. TÁMOP-4.2.4.A/2-11/1-2012-0001 and GINOP-2.3.2-15-2016-00028). Author contributions: A.J. and T.B. contributed equally to experimental design, data analysis, and interpretation/writing of the manuscript. A.M.: plant growth and tissue dissection; C.G.F., J.O., G.G., and Z.B.: protein and epitope analyses; C.M.: data analysis and chromosome map construction; I.F., D.L., K.F.X.M., and M.S.: phylogenomics analyses; G.K.-G: IWGSC genome assembly analyses. J.J.: AK58 genome analysis. P.G. and J.A.T.-D.: medical interpretation of data; W.M., R.A., and O.-A.O: planning of experiments and writing of the manuscript. Competing interests: J.A.T.-D. has served as a consultant and scientific advisory board member for ImmusanT Inc. and owns shares in Nexpep Pty Ltd. He is a co-inventor of patents pertaining to the use of gluten peptides in celiac disease therapeutics, diagnostics, and nontoxic gluten. Nexpep Pty. Ltd. and ImmusanT Inc. were formed to develop novel diagnostics and treatments for celiac disease. All other authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The RNA-seq data are submitted to the IWGSC Data Repository hosted at URGI: https://wheat-urgi.versailles.inra.fr/Seq-Repository with ID SRP150569. Translated gene sequences of reference allergens and antigens along with peptide sequences with known IFNγ-ELISPOT responses will be available for epitope and peptide analyses at https://genome.propepper.net. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/8/eaar8602/DC1
Section S1. Manual annotation and curation of the Prolamin superfamily genes in the reference genome
Section S2. Organization of major prolamin gene clusters of short arms of chromosomes 1 and 6
Section S3. Phylogenetic analysis of Prolamin superfamily genes
Section S4. Phylogenetic analysis of highly immunogenic α-gliadin sequences
Section S5. Impact of temperature stress on protein composition
Fig. S1. Chromosomal location of major food disease–related protein families on chromosome groups 1 and 6.
Fig. S2. Mapping of peptides with known IFNγ-ELISPOT response on the gliadin and glutenin sequences of Chinese Spring.
Fig. S3. Phylogenetic analysis of prolamin superfamily gene models.
Fig. S4. Phylogenetic analysis and epitope distribution of highly immunostimulatory gliadin and gliadin proteins in wheat and related species.
Table S1. Effect of temperature stress on protein content and composition.
Data file S1. CDS sequences of prolamin superfamily gene models and identified reference allergens in fasta file format.
Data file S2. Annotation of prolamin superfamily genes and reference allergens used in this study.
Data file S3. Epitope annotation table of sequences used for the phylogenetic analyses.
Data file S4. Expression of peptides with known IFNγ-ELISPOT responses.
Science article authors that are IWGSC members
REFERENCES AND NOTES
- 1.International Wheat Genome Sequencing Consortium , Shifting the limits in wheat research and breeding through a fully annotated and anchored reference genome sequence. Science (2018) 10.1126/science.aar7191.
- 2.Larré C., Lupi R., Gombaud G., Brossard C., Branlard G., Moneret-Vautrin D. A., Rogniaux H., Denery-Papini S., Assessment of allergenicity of diploid and hexaploid wheat genotypes: Identification of allergens in the albumin/globulin fraction. J. Proteomics 74, 1279–1289 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Ludvigsson J. F., Leffler D. A., Bai J., Biagi F., Fasano A., Green P. H., Hadjivassilou M., Kaukinen K., Kelly C. P., Leonard J. N., Lundin K. E. A., Murray J. A., Sanders D. S., Walker M. M., Zingone F., Ciacci C., The Oslo definitions for coeliac disease and related terms. Gut 62, 43–52 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hardy M. Y., Tye-Din J. A., Coeliac disease: A unique model for investigating broken tolerance in autoimmunity. Clin. Transl. Immunology 5, e112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh P., Arora A., Strand T. A., Leffler D. A., Catassi C., Green P. H., Kelly C. P., Ahuja V., Makharia G. K., Global prevalence of celiac disease: Systematic review and meta-analysis. Clin. Gastroenterol. Hepatol. 16, 832–836.e2 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Rubio-Tapia A., Ludvigsson J. F., Brantner T. L., Murray J. A., Everhart J. E., The prevalence of celiac disease in the United States. Am. J. Gastroenterol. 107, 1538–1544 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Gibson P. R., Skodje G. I., Lundin K. E. A., Non-coeliac gluten sensitivity. J. Gastroenterol Hepatol. 32 (suppl. 1), 86–89 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Golley S., Corsini N., Topping D., Morell M., Mohr P., Motivations for avoiding wheat consumption in Australia: Results from a population survey. Public Health Nutr. 18, 490–499 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skodje G. I., Sarna V. K., Minelle I. H., Rolfsen K. L., Muir J. G., Gibson P. R., Veierød M. B., Henriksen C., Lundin K. E. A., Fructan, rather than gluten, induces symptoms in patients with self-reported non-celiac gluten sensitivity. Gastroenterology 154, 529–539.e2 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Zevallos V. F., Raker V., Tenzer S., Jimenz-Calvente C., Ashfaq-Khan M., Rüssel N., Pickert G., Schild H., Steinbrink K., Schuppan D., Nutritional wheat amylase-trypsin inhibitors promote intestinal inflammation via activation of myeloid cells. Gastroenterology 152, 1100–1113.e12 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Longo G., Berti I., Burks A. W., Krauss B., Barbi E., IgE- mediated food allergy in children. Lancet 382, 1656–1664 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Sicherer S. H., Leung D. Y. M., Advances in allergic skin disease, anaphylaxis, and hypersensitivity reactions to foods, drugs, and insects in 2013. J. Allergy Clin. Immunol. 133, 324–334 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Armentia, D. de Luis, J. Crespo, L. Inglada, J. Castrodeza, S. Martin-Armentia, Wheat allergy, in Bioactive Food as Dietary Interventions for Arthritis and Related Inflammatory Diseases, R. R. Watson, V. R. Preedy, Eds. (Elsevier Science, 2012), pp. 189–202. [Google Scholar]
- 14.Scherf K. A., Brockow K., Biedermann T., Koehler P., Wieser H., Wheat-dependent exercise-induced anaphylaxis. Clin. Exp. Allergy 46, 10–20 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Tye-Din J. A., Stewart J. A., Dromey J. A., Beissbarth T., van Heel D. A., Tatham A., Henderson K., Mannering S. I., Gianfrani C., Jewell D. P., Hill A. V. S., McCluskey J., Rossjohn J., Anderson R. P., Comprehensive, quantitative mapping of T cell epitopes in gluten in celiac disease. Sci. Transl. Med. 2, 41ra51 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Constantin C., Quirce S., Poorafshar M., Touraev A., Niggemann B., Mari A., Ebner C., Akerström H., Heberle-Bors E., Nystrand M., Valenta R., Micro-arrayed wheat seed and grass pollen allergens for component-resolved diagnosis. Allergy 64, 1030–1037 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Schalk K., Lang C., Wieser H., Koehler P., Scherf K. A., Quantitation of the immunodominant 33-mer peptide from α-gliadin in wheat flours by liquid chromatography tandem mass spectrometry. Sci. Rep. 7, 45092 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Juhász A., Haraszi R., Maulis C., ProPepper: A curated database for identification and analysis of peptide and immune-responsive epitope composition of cereal grain protein families. Database 2015, bav100 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dubcovsky J., Dvorak J., Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 316, 1862–1866 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altenbach S. B., New insights into the effects of high temperature, drought and post-anthesis fertilizer on wheat grain development. J. Cereal Sci. 56, 39–50 (2012). [Google Scholar]
- 21.Zimin A. V., Puiu D., Hall R., Kingan S., Clavijo B. J., Salzberg S. L., The first near-complete assembly of the hexaploid bread wheat genome, Triticum aestivum. Gigascience 6, 1–7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huebener S., Tanaka C. K., Uhde M., Zone J. J., Vensel W. H., Kasarda D. D., Beams L., Briani C., Green P. H. R., Altenbach S. B., Alaedini A., Specific nongluten proteins of wheat are novel target antigens in celiac disease humoral response. J. Proteome Res. 14, 503–511 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osman A., Uhlig H. H., Valdes I., Amin M., Méndez E., Mothes T., A monoclonal antibody that recognizes a potential coeliac-toxic repetitive pentapeptide epitope in gliadins. Eur. J. Gastroenterol. Hepatol. 13, 1189–1193 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Sander I., Rozynek P., Rhis H. P., van Kampen V., Chew F. T., Lee W. S., Kotschy-Lang N., Merget R., Brüning T., Raulf-Heimsoth M., Multiple wheat flour allergens and cross-reactive carbohydrate determinants bind IgE in baker’s asthma. Allergy 66, 1208–1215 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Matsuo H., Kohno K., Morita E., Molecular cloning, recombinant expression and IgE-binding epitope of ω-5 gliadin, a major allergen in wheat-dependent exercise-induced anaphylaxis. FEBS J. 272, 4431–4438 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Liu C. Y., Shepherd K. W., Inheritance of B subunits of glutenin and omega- and gamma-gliadins in tetraploid wheat. Theor. Appl. Genet. 90, 1149–01157 (1995). [DOI] [PubMed] [Google Scholar]
- 27.Gao S., Gu Y. Q., Wu J., Coleman-Derr D., Huo N., Crossman C., Jia J., Zuo Q., Ren Z., Anderson O. D., Kong X., Rapid evolution and complex structural organization in genomic regions harboring multiple prolamin genes in the polyploid wheat genome. Plant Mol. Biol. 65, 189–203 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Sánchez-León S., Gil-Humanes J., Ozuna C. V., Giménez M. J., Sousa C., Voytas D. F., Barro F., Low-gluten, non-transgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol. J. 16, 902–910 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marcussen T., Sandve S. R., Heier L., Spannagl M., Pfeifer M. International Wheat Genome Sequencing Consortium, Jakobsen K. S., Wulff B. B. H., Steuernagel B., Mayer K. F. X., Olsen O.-A., Ancient hybridizations among the ancestral genomes of bread wheat. Science 345, 1250092 (2014). [DOI] [PubMed] [Google Scholar]
- 30.van Herpen T. W. J. M., Goryunova S. V., van der Schoot J., Mitreva M., Salentijn E., Vorst O., Schenk M. F., van Veelen P. A., Koning F., van Soest L. J. M, Vosman B., Bosch D., Hamer R. J., Gilissen L. J. W. J, Smulders M. J. M., Alpha-gliadin genes from the A, B, and D genomes of wheat contain different sets of celiac disease epitopes. BMC Genomics 7, 1 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molberg Ø., Uhlen A. K., Jensen T., Flæte N. S., Fleckenstein B. , Arentz-Hansen H., Raki M., Lundin K. E. A., Sollid L. M., Mapping of gluten T-cell epitopes in the bread wheat ancestors: Implications for celiac disease. Gastroenterology 128, 393–401 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y., Pan J., Huang X., Guo D., Lou H., Hou Z., Su M., Liang R., Xie C., You M., Li B., Differential effects of a post-anthesis heat stress on wheat (Triticum aestivum L.) grain proteome determined by iTRAQ. Sci. Rep. 7, 3468 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uhlen A. K., Dieseth J. A., Koga S., Böcker U., Hoel B., Anderson J. A., Moldestad A., Variation in gluten quality parameters of spring wheat varieties of different origin grown in contrasting environments. J. Cereal Sci. 62, 110–116 (2015). [Google Scholar]
- 34.Altenbach S. B., Tanaka C. K., Seabourn B. W., Silencing of omega-5 gliadins in transgenic wheat eliminates a major source of environmental variability and improves dough mixing properties of flour. BMC Plant Biol. 14, 393 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gil-Humanes J., Pistón F., Giménez M. J., Martín A., Barro F., The introgression of RNAi silencing of γ-gliadins into commercial lines of bread wheat changes the mixing and technological properties of the dough. PLOS ONE 7, e45937 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G., Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Radauer C., Bublin M., Wagner S., Mari A., Breiteneder H., Allergens are distributed into few protein families and possess a restricted number of biochemical functions. J. Allergy Clin. Immunol. 121, 847–852.e7 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Pfeifer M., Kugler K. G., Sandve S. R., Zhan B., Rudi H., Hvidsten T. R. International Wheat Genome Sequencing Consortium, Mayer K. F. X., Olsen O.-A., Genome interplay in the grain transcriptome of hexaploid bread wheat. Science 345, 1250091 (2014). [DOI] [PubMed] [Google Scholar]
- 39.J. M. Bremner, C. S. Mulvaney, Nitrogen—Total, in Methods of Soil Analysis, A. L. Page, R. H. Miller, D. R. Keeney, Eds. (American Society of Agronomy, Soil Science Society of America, ed. 2, 1982), pp. 595–624. [Google Scholar]
- 40.Larroque O. R., Békés F., Rapid size-exclusion chromatography analysis of molecular size distribution for wheat endosperm protein. Cereal Chem. 77, 451–453 (2000). [Google Scholar]
- 41.O. R. Larroque, F. Bekes, C. W. Wrigley, W. G. Rathmell, Analysis of gluten proteins in grain and flour blends by RP-HPLC, in Wheat Gluten Proceedings 7th International Workshop, P. R. Shewry, A. S. Tatham, Eds. (Royal Society of Chemistry, 2000), pp. 136–139. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/8/eaar8602/DC1
Section S1. Manual annotation and curation of the Prolamin superfamily genes in the reference genome
Section S2. Organization of major prolamin gene clusters of short arms of chromosomes 1 and 6
Section S3. Phylogenetic analysis of Prolamin superfamily genes
Section S4. Phylogenetic analysis of highly immunogenic α-gliadin sequences
Section S5. Impact of temperature stress on protein composition
Fig. S1. Chromosomal location of major food disease–related protein families on chromosome groups 1 and 6.
Fig. S2. Mapping of peptides with known IFNγ-ELISPOT response on the gliadin and glutenin sequences of Chinese Spring.
Fig. S3. Phylogenetic analysis of prolamin superfamily gene models.
Fig. S4. Phylogenetic analysis and epitope distribution of highly immunostimulatory gliadin and gliadin proteins in wheat and related species.
Table S1. Effect of temperature stress on protein content and composition.
Data file S1. CDS sequences of prolamin superfamily gene models and identified reference allergens in fasta file format.
Data file S2. Annotation of prolamin superfamily genes and reference allergens used in this study.
Data file S3. Epitope annotation table of sequences used for the phylogenetic analyses.
Data file S4. Expression of peptides with known IFNγ-ELISPOT responses.
Science article authors that are IWGSC members


