Abstract
Dust and sandstorm events inject substantial quantities of foreign microorganisms into global ecosystems, with the ability to impact distant environments. The majority of these microorganisms originate from deserts and drylands where the soil is laden with highly stress-resistant microbes capable of thriving under extreme environmental conditions, and a substantial portion of them survive long journeys through the atmosphere. This large-scale transmission of highly resilient alien microbial contaminants raises concerns with regards to the invasion of sensitive and/or pristine sink environments, and to human health—concerns exacerbated by increases in the rate of desertification. Further increases in the transport of dust-associated microbiota could extend the spread of foreign microbes to new ecosystems, increase their load in present sink environments, disrupt ecosystem balance, and potentially introduce new pathogens. Our present understanding of these microorganisms, their phylogenic affiliations and functional significance, is insufficient to determine their impact. The purpose of this review is to provide an overview of available data regarding dust and sandstorm microbiota and their potential ramifications on human and ecosystem health. We conclude by discussing current gaps in dust and sandstorm microbiota research, and the need for collaborative studies involving high-resolution meta-omic approaches in conjunction with extensive ecological time-series studies to advance the field towards an improved and sufficient understanding of these invisible atmospheric travelers and their global ramifications.
Keywords: dust and sandstorms microbiota, global spread, stress resistant and pathogenic microorganisms, human and ecosystem health, metagenomics and multiomics, monitoring
Introduction
Dust and sandstorm events inject substantial quantities of foreign microorganisms to downwind atmosphere, terrestrial, and aquatic environments, and are known as one of the most far-reaching vehicles for transport of highly stress resistant and potentially invasive/pathogenic microorganisms across the globe (Kellogg and Griffin 2006; Favet et al. 2013; Smith et al. 2013; Weil et al. 2017). Significant increases in the concentration of bacteria and fungi are commonly detected in dust clouds during sandstorm events (Kellogg and Griffin 2006; Griffin 2007; Vijayakumar et al. 2017; Tang et al. 2018). Since microorganisms are fundamental players in ecosystem processes (Schulz et al. 2013; Bernhard and Kelly 2016; Graham et al. 2016), large-scale transport of highly stress resistant, foreign, and potentially invasive/pathogenic microorganisms could have wide ranging impacts on downwind environments and our health (Griffin and Kellogg 2004; Gonzalez-Martin et al. 2014). African dusts were linked to the meningitis outbreaks in sub-Saharan Africa (Agier et al. 2013; Jusot et al. 2017) and the coral reef decline in the Caribbean (Shinn et al. 2000). Dust storms are known to correlate with increased hospital emergency visits due to asthma exacerbations and other respiratory and cardiovascular complications (Gyan et al. 2005; Mallone et al. 2011; Tam et al. 2012; Lee et al. 2013; Meo et al. 2013). However, the few studies investigating the roles of dust storm microbiota in disease outbreaks found correlation, but did not explore clinical evidence for causal effects (Chen et al. 2010; Rodo et al. 2014; Wang et al. 2016). Although these studies provide a link between sandstorms and disease outbreaks, they were constrained in terms of number and scope.
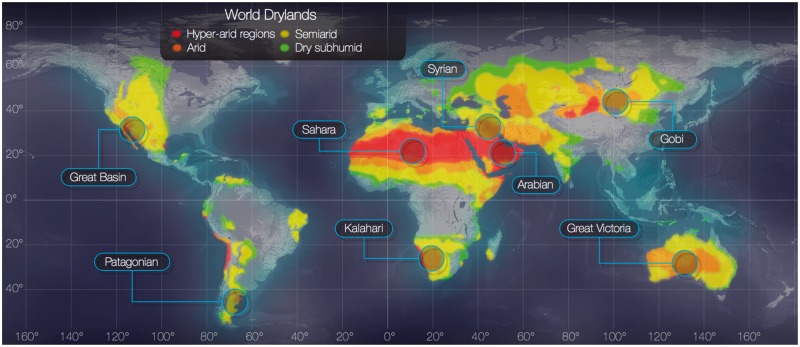
Expanded study is needed since dust activity is predicted to rise substantially in the future due to global climate changes and anthropogenic causes. The world’s drylands have expanded over the past 60 years, and their expansion is projected to continue well into the 21st century (Huang et al. 2017). Dryland regions cover approximately 40% of earth’s total landmass (White and Nackoney 2003), and are spread throughout the globe, with the largest percentage localized in Africa and Asia, as shown in figure 1. Increased aridity, frequent draughts, low precipitation, low soil fertility, and excessive anthropogenic exploitations have made dryland soils exceptionally vulnerable to desertification, degradation, and erosion by wind (Fu and An 2002; Maestre et al. 2014; Tao 2014; Zhou et al. 2016). Approximately 12x106 hectares of arable land is lost annually to desertification, which is 30–35 times greater than expected based on historic rates (UNCCD). Desertification generates new grounds for dust activities (Moulin and Chiapello 2006; McConnell et al. 2007; Johnson et al. 2011; Issanova et al. 2015; Sharma et al. 2015), and could increase microbial load in the atmosphere. The overall escalation in dust activities could alter the equilibrium in sink environments by increasing their share of foreign microbes, extremophiles, and potentially invasive/pathogenic species. It could also spread dust masses beyond present sink environments affecting new ecosystems.
Fig. 1.
—World map of drylands. Colors highlight regions with varying degrees of aridity. Major deserts (excluding Antarctica and the Arctic) are annotated. http://www.naturalearthdata.com/, last accessed July 21, 2018.
The ramifications of dust and sandstorm microbiota are potentially significant. In order to understand impact, we need to have detailed information on the agents of impact. The research in the field of dust and sandstorm aero-microbiology and its impact on downwind ecology is still in its infancy. Our current knowledge of these microorganisms and their functional attributes is insufficient to determine a cause and effect relationship between their spread and threats to downwind ecosystems. Understanding the breadth of microbial diversity in dust clouds, their viability, invasiveness/pathogenicity, and their long-term impacts on global ecosystems will require comprehensive, collaborative studies and novel approaches. In recent years, with the advent of culture-independent metagenomics, we are beginning to realize the vast extent of microbial diversity in sandstorm dust clouds and the extent of their reach (Smith et al. 2013; Rosselli et al. 2015; Cha et al. 2017; Maki et al. 2017b; Weil et al. 2017). These novel findings opened our eyes to a frontier of previously unknown microbial diversity, and the research in the field is gaining momentum due to pertinent discoveries. We aim to draw attention to this novel frontier of microbial ecology by examining: 1) The current understanding of dust and sandstorm derived microbiota (DSM), and their global spread; 2) Potential ramifications of DSM on human and ecosystem health; and 3) Perspectives to guide future research towards comprehensive understanding of DSM role at a global level.
Dust and Sandstorm Derived Microbiota (DSM) and Their Global Spread
Dust and Sandstorm Derived Microbiota
The majority of DSM originates from large deserts (Griffin 2007). Deserts are laden with highly stress resistant microorganisms capable of thriving in harsh environmental conditions with restricted water and nutrients availability, extremes of temperatures, and UV irradiation (Kuske et al. 1997; Dose et al. 2001; Varin et al. 2012; Makhalanyane et al. 2015; Etemadifar et al. 2016). Desert microbiota have increased abundance of genes involved in osmoregulation and dormancy, which likely contribute to their survival in hostile desert soil (Fierer et al. 2012). Microorganisms play significant roles in soil ecosystems, from their vital symbiotic interactions with plants, to their major roles in maintenance of biological soil crust, biodegradation of organic matters, and biogeochemical cycling of nutrients (Ortiz-Castro et al. 2009; Abed et al. 2010; Schulz et al. 2013). Microbial diversity in soil is dependent on its pH, nutrient, and moisture contents (Fierer and Jackson 2006; Clark et al. 2009; Lauber et al. 2009; Angel et al. 2010; Rousk et al. 2010). Some variations exist in the diversity of taxa that dwell in different deserts, with cold arctic deserts bearing lower microbial diversity compared with hot deserts (Pointing et al. 2009; Fierer et al. 2012; Lee et al. 2012). Some of the major bacterial taxa that frequently dwell in desert soil with high relative abundance include Actinobacteria, Bacteroidetes, Proteobacteria, Firmicutes, and cyanobacteria (Fierer et al. 2012; An et al. 2013). Archaea also appear to be equally abundant, the majority of which are chemolithoautotrophic ammonia oxidizers potentially involved in biogeochemical cycling of nitrogen and carbon (Makhalanyane et al. 2015). Desert soil also harbors diverse communities of fungi that can withstand adverse environmental conditions (Chan et al. 2013); some of the most common fungal genera in deserts include Alternaria, Aspergillus, Cladosporium, and Penicillium (Conley et al. 2006; Sterflinger et al. 2012; Makhalanyane et al. 2015). Taxonomically diverse selections of viruses were also found, although viral diversity and function in desert ecosystems is poorly understood (Fierer et al. 2007; Zablocki et al. 2016).
In addition to desert microbiome, sandstorms carry large quantities of the airborne microbiota encountered along their intermediate path. By some estimate, a cubic meter of air contains hundreds of thousands of microorganisms (Burrows et al. 2009; Prussin et al. 2015), with a diversity of taxa similar to that found in soil (Franzetti et al. 2011). The majority of these microbes originate from local sources, that is, soil, aquatic environments, plants, and anthropogenic pollution (Maron et al. 2005; Brodie et al. 2007). Some of the more dominant bacterial taxa in air include Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria, and Cyanobacteria (Bowers et al. 2009; Smith et al. 2013; Lee et al. 2017). Air also harbors diverse communities of fungi including Cladosporium, Aspergilllus, and Penicillium, some of which are potential allergens (Oh et al. 2014). Archaea including Thaumarchaeota and Euryarchaeota were also found in the air (Frohlich-Nowoisky et al. 2014). DNA sequences from a diversity of viruses, including those related to plant-associated geminiviruses and animal-infecting circoviruses, were also found in air (Whon et al. 2012). Airborne microbiota are known to resist adverse atmospheric conditions (Rothschild and Mancinelli 2001), and the atmosphere is believed to be a source of both beneficial and pathogenic microorganisms (Polymenakou 2012; Nicolaisen et al. 2017).
There is limited data on the extent of microbial diversity and taxonomic details of DSM. These details are required to study the functional and ecological significance of these microbes in their native environments and larger ecosystems.
Global Spread of DSM and Their Wide-Ranging Reach
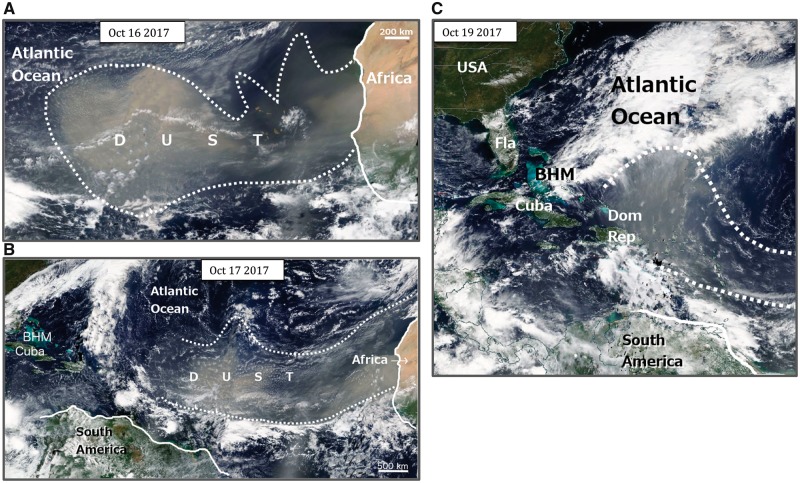
According to different studies, each year between 0.5 billion and 5 billion tons of dust is injected into the atmosphere (Perkins 2001), the majority of which originate from the Sahara Deserts and Sahel regions in Africa, and the Gobi and Taklamakan Deserts in Asia (Griffin 2007). Sandstorms from the Sahara Deserts transmit roughly a billion tons of dust across the atmosphere, and are considered as one of the major sources of the intercontinental dust transport (Moulin et al. 1997; Griffin 2007). Figure 2 shows NASA satellite images of a strong sandstorm that blew off the west coast of Africa on October 13, 2017, carrying massive amounts of African dusts across the Atlantic Ocean. The dust plume generated from this particular storm stretched for a few thousand kilometers over the Atlantic, and reached the Caribbean within 5–6 days. Sandstorms such as this are common occurrences in the region, and can transport substantial quantities of African dust thousands of kilometers from the source; depending on the strength and direction of the wind, these dust can move westward across the Atlantic Ocean to the Americas and the Caribbean (Prospero and Carlson 1981; Swap et al. 1992; Perry et al. 1997; Prenni et al. 2009), or north and eastward over the Mediterranean Sea to Europe and the Middle East, respectively (Ganor et al. 1991; Franzen et al. 1995; LoyePilot and Martin 1996; Kubilay et al. 2000; Rodrı´guez et al. 2001). Gobi and Taklamakan Deserts in Asia, the second largest source of long-range dust transport, inject approximately 100–460 Mt/year (Laurent et al. 2006) to 800 Mt/year (Zhang et al. 1997) of dust into the atmosphere (Bishop et al. 2002; Griffin 2007). The dust plumes generated from Asia sandstorms frequently disturb the air quality in East Asia (Iwasaka et al. 1983; Choi et al. 2001; Trochkine et al. 2003), and can reach as far as the Americas (Husar et al. 2001; VanCuren and Cahill 2002), the Alps (Grousset et al. 2003), the Arctic (Huang et al. 2015), and even circle around the globe (Uno et al. 2009). Strong sandstorms originating from African and Asian deserts are known to carry a large enough quantity of dust to affect air quality in destinations thousands of kilometers away from the source (Prospero 1999; Griffin 2007). Considering that one gram of soil can harbour 108–109 prokaryotes (Whitman et al. 1998), this could translate into transport of trillions upon trillions of microbes into the air and downwind destinations along their intermediate path. Increases in the concentration of microbes in sandstorm dust clouds have been documented extensively by both culture dependent and independent approaches (Kellogg and Griffin 2006; Griffin 2007; Nishimura et al. 2010; Yamaguchi et al. 2014).
Fig. 2.
—A massive sandstorm blows off the west coast of Africa and over the Atlantic Ocean towards the Caribbean, on October 13, 2017. (A) A dust plume greater than 2,000 km in length could be seen over the Atlantic Ocean, 3 days later on October 16. (B) The dust plume was stretched over 4,000 km by day 4, October 17. The dust clouds reached the Caribbean by day 5 on October 18, covering Dominican Republic (Dom Rep) by October 19 (C), and moving towards the Bahamas (BHM) and Florida (Fla.) in the United States (USA). The natural color images were captured with the Moderate Resolution Imaging Spectroradiometer (MODIS) abroad NASA’s Satellites. Images courtesy of NASA Earthdata: https://worldview.earthdata.nasa.gov/, last accessed July 21, 2018.
A few studies examined the impact of increased transport of DSM on ambient airborne microbiota in leeward environments. A recent metagenomic analysis of air samples collected during an Asian dust event in South Korea showed a significant increase in the number of bacterial Operational Taxonomic Units (OTU) with significantly different microbial composition including potential human pathogens, compared with control nondust samples (Cha et al. 2017). A similar analysis of air samples in Northern China demonstrated substantial increases in airborne bacterial diversity and concentration in dust versus nondust air samples (Tang et al. 2018). Asian dust was shown to carry substantial increases in the proportion of endospore forming Bacillus as well as other bacterial genera including Modestobacter (Cha et al. 2016), which are resistant to desiccation and UV irradiation (Gtari et al. 2012). Examination of two transpacific Asian air plumes at Mt. Bachelor Observatory (2.8 km above sea level) in North America revealed a richness of microbiota tantamount to those found in surface environments; the plumes carried increased level of Gram-positive bacteria that included many spore forming species adapted to survive in extreme conditions (Smith et al. 2013). Similarly, analysis of airborne bacterial communities in the free troposphere at high altitude (500–3,000 m above ground level) over the Noto Peninsula in Japan during Asian dust days showed a high diversity of bacteria, dominated by natural-sand/terrestrial-associated taxa including endospore forming Bacillus members (Maki et al. 2017a). Hara et al. (2015) isolated a number of UV tolerant endospore forming culturable bacteria in air samples collected over the East China Sea during Asian dust events, demonstrating that dust storms carry viable bacteria.
At present, we do not know exactly what percentage of DSM survives the long journey through the atmosphere. It is also uncertain whether these alien microbial contaminants can adapt to disparate new environmental conditions upon arrival, and if they could significantly alter their new host ecosystems. Findings from a recent culture independent metagenomic study by Weil et al. revealed that a massive sandstorm originating from the Sahara Deserts deposited a large fraction of the entire microbial communities from Saharan soil to snow covered Alpine mountains in Italy (Weil et al. 2017). Within these deposits, the authors found a number of putative pathogens and some of the most highly stress resistant microorganisms ever detected on the planet. Surprisingly, some of these microorganisms might have survived in the frozen Alpine soil and still remained viable upon melting of the snow (Weil et al. 2017). In a similar study, the snow samples collected at Mont Blanc glaciers (French Alps) during Sahara dust storms showed high indices of bacterial diversity with a dominance of phylotypes commonly found in desert and arid soils (Chuvochina et al. 2011a, 2011b), some of which belonged to genera known to carry resistance to adverse environmental conditions including extreme cold, UV irradiation, and desiccation; these traits could enable them to remain viable during atmospheric transport and subsequently adapt to the snow covered environment despite the physical and physiological stress associated with these events. Comparison of microbial data with their closest relatives in the GenBank identified 15 phylotypes as potential snow colonizers (Chuvochina et al. 2011b). In another study, sandstorm originating from the Sahara Desert was shown to change the bacterial assemblage and composition of a pristine fresh water lake in Austrian Alps, and increased viable bacterial concentration in these waters (Peter et al. 2014). These findings raise significant concerns over invasion and alteration of pristine and sensitive sink environments, and further suggest the spread of human diseases by alien microbial contaminants. However, the prevailing data on sandstorm microbiota is exceedingly sparse and insufficient to estimate the potential global effect exerted by these microorganisms.
Potential Ramifications of DSM on Human and Ecosystem Health
The Impact of DSM on Human Health
A primary concern with the dispersal of microorganisms via sandstorms is the potential impact on human health. A few studies have found correlations between DSM and disease outbreaks, but no major attempts were made to establish cause and affect relationships. The annual meningitis outbreaks in Sahel region of Africa were linked to the occurrence of seasonal changes including high temperatures, dry air, and increased dust activities from African deserts (Sultan et al. 2005; Griffin 2007; Agier et al. 2013; Jusot et al. 2017). Inhalation of hot and dry dusty air is thought to impair host immune response, leading to an increase in the nasopharyngeal carriage of precolonized or airborne bacterial pathogens, their direct aspiration into the lung, and dissemination into blood and brain (Jusot et al. 2017). The potential role of dust borne microbiota in the spread of meningitis in the region, however, is not clear and requires detailed investigations. Interestingly, several OTUs potentially belonging to meningitis pathogens were detected in the Sahara dust deposits within the snow packs in Swiss Alps (Meola et al. 2015). The outbreaks of Kawasaki disease (KD), a serious heart complication acquired in childhood, in Japanese children was linked to Candida species found in tropospheric winds originating from China (Rodo et al. 2014). The seasonal occurrence of KD in US children similarly showed a significant association with the strong wind currents that originated from central Asia (Rodo et al. 2011; Rodo et al. 2014). Another infectious disease presumably caused by wind dissemination of microorganisms is Valley Fever, whose fungal causative agents Coccidioides immitis and Coccidioides posadasii are primarily found in hot and arid desert soil (Kirkland and Fierer 1996). In particular, positive correlations were found between the intensification of sandstorms and the epidemics of Valley Fever in Southern United States in the last decade (Tong et al. 2017). The incidence of measles in Western China was positively correlated with dust events in the region (Ma et al. 2017). The epidemics of pulmonary tuberculosis, a chronic infectious disease of the lung caused by Mycobacterium tuberculosis, was similarly linked to Asian dust storms (ADS) in China (Wang et al. 2016). A significant increase in the concentration of ambient influenza type A virus was detected in air samples collected in Taiwan during ADS (Chen et al. 2010); and the highly pathogenic H5N1 avian influenza virus outbreaks in 2003–2005 in Japan and South Korea, positioned downwind of ADS, were potentially caused by increases in the influenza virus load during ADS (Chen et al. 2010). A strong association between increased particulate matter (PM2.5) levels and the risk of acquiring influenza-like illness was reported in Beijing, China across all the age categories studied (Feng et al. 2016). Outbreak of the foot-and-mouth disease (FMD) in Miyazaki, Japan in July–March 2010 which led to the slaughter of 289,000 domestic animals was linked to transport of the FMD virus via yellow sandstorms that originated from China (Maki et al. 2012). Dust events typically lead to increased hospital emergency visits due to asthma exacerbations and other respiratory and cardiovascular complications (Yang et al. 2005; Kanatani et al. 2010; Tam et al. 2012; Meo et al. 2013), but the potential link between sandstorm microbiota and these human health concerns have rarely been investigated even though mounting evidence suggest that microorganisms contribute to the establishment and/or exacerbations of asthma (Glezen et al. 2000; Denning et al. 2006; Hilty et al. 2010; Huang et al. 2011). Airborne dissemination of microorganisms could also greatly increase the range and influence of horizontal gene transfer. This would be particularly problematic when antibiotic resistant genes (ARGs) are acquired by pathogenic microorganisms rendering them resistant to antibiotic treatments (Martinez 2008).
Current data on the potential link between DSM and disease outbreaks show correlation, but are insufficient for establishing causation. Establishing a cause-and-effect relationship between DSM and human health concerns is difficult since the majority of these microorganisms are not yet characterized, and their viability, pathogenic potential, and geospatial distributions remain mostly unknown. A comprehensive database of DSM, environmental metadata, and implementation of disease surveillance strategies in downwind environments are all prerequisites for establishing a clear link between DSM and human health concerns. Surveillance strategies based on emerging biosensor technologies with high sensitivity and specificity for detection of windborne pathogens could provide advanced warning system for at-risk populations (Yoo et al. 2017).
The Impact of DSM on Aquatic Environments
Over two-third of our planet is covered with aquatic ecosystems including oceans, marines, lakes, rivers, and estuaries. Microorganisms are essential players in the natural functioning of these ecosystems from their major roles in biogeochemical cycling of nutrients, to their involvement in food chains through primary productivity, and the sequestration and export of atmospheric CO2. Dust storms could have significant impacts on these processes through: 1) replenishing of essential nutrients (i.e., P and N) and trace metals (i.e., Fe) needed for growth and propagation of indigenous microorganisms; and 2) deposit of a large supply of foreign microbes, including potentially pathogenic microorganisms.
The majority of previous research on dust impact in aquatic environments primarily examined the influence of dust nutrients on indigenous microorganisms with little to no focus on the potential impact of DSM. These studies suggest that dust nutrients could promote or inhibit propagation of certain species over others depending on the trophic status of marine environments (oligotrophic vs. eutrophic) and the richness/toxicity/anthropogenic content of the dust source (Herut et al. 2005; Paytan et al. 2009; Lekunberri et al. 2010; Romero et al. 2011; Gallisai et al. 2014; Guieu et al. 2014; Westrich et al. 2016). Concomitant with the deposit of nutrients, dust storms can deliver copious supplies of highly stress resistant foreign microorganisms to aquatic environments. In a microcosm bioassay experiment, when dust samples from different deserts with distinct microbial communities were added to presterilized Mediterranean seawater, a significant increase in bacterial production and nitrogen fixation was detected within a short period following dust addition suggesting that dust borne microorganisms were alive and metabolically active (Rahav et al. 2016). Peter et al. (2014) compared the viability, composition, and abundance of bacteria in a fresh water lake in Austrian Alps during rain events influenced by Saharan versus non-Saharan dust days, and showed that the bacterial assemblage and composition in the lake differed significantly between the two events, with Gamma Proteobacteria dominating the Saharan dust intrusion events and Beta Proteobacteria dominating the non-Saharan events. The authors found a rapid increase in bacterial cell count in sterile lake water samples upon addition of the rain from Saharan dust days demonstrating that the bacteria in the wet deposits were alive and able to readily propagate. These studies, although limited in their number, focus, and temporal scope, suggest that sandstorms could significantly impact the makeup of microbial community structures in downwind aquatic environments in part through deposit of large supplies of foreign microorganisms. Intrusion of large quantities of potentially stress resistant foreign microorganisms could affect microbial community interactions and equilibrium in these environments; but the long-term impact or threats to the ecology of pristine settings such as the alpine lakes is not known.
In addition to their predicted impact on ecosystem maintenance and sustainability, dust borne microorganisms could have pathogenic ramifications on aquatic organisms. One of the best studied examples is the potential link between increased dust activities and the rapid decline in the coral reef status in the Caribbean, which receives hundreds of millions of tons of dust from Sahara Deserts annually (Shinn et al. 2000). Aspergillus sydowii, the fungal causative agent of Aspergillosis or sea fan disease, was isolated from both the Sahara-derived dust samples in the Caribbean and from the diseased sea fan corals in the region (Weir et al. 2000; Garrison et al. 2003). Later, it was shown that inoculation of healthy sea fans with Aspergillus spp. isolated from dust samples in the Caribbean produced Aspergillosis-like disease morphology in the infected corals and the fungi was successfully reisolated from the diseased animals (Weir-Brush et al. 2004). In a tissue culture bioassay, A. sydowii was shown to impede the motility of dinoflagellate symbiodinium, the coral symbiont that plays essential roles in coral fitness and energy production (Hallegraeff et al. 2014). Considering that coral reefs house some of the most vibrant marine ecosystems, these studies support the notion that escalation in dust activities could have a negative impact on marine health through increased deposit of dust borne pathogens.
Long term monitoring of sandstorm spread using advanced satellite technologies combined with fieldwork analysis of DSM in downwind environments over the Ocean and terrestrial regions, and close observations of infectious outbreaks could reveal more concrete links between pathogenic DSM and major disease outbreaks in aquatic ecosystems.
The Impact of DSM on Agriculture
Long-range transport of atmospheric dust could have both positive and negative implication on plants and agriculture in sink environments. A clear example of the positive impact of intercontinental dust transport is the continuous fertilization of nutrient depleted soils in the Amazon. By transport of nutrients, in particular phosphorus, and topsoil to the Amazon, Sahara sandstorms are believed to be major contributors to the fertility and productivity of the Amazon rainforest (Yu et al. 2015). However, dust storms can also have negative impacts on downwind agriculture through transport of substantial quantities of foreign, stress resistant and/or pathogenic microorganisms, which could potentially change the make-up of soil microbial communities or introduce pathogenic microorganisms to crops and plants. Since modern crops lack genetic diversity and are susceptible to similar set of pathogens, aerial dispersion of plant pathogens and invasive microorganisms by sandstorms could have major implications in agricultural productivity and the global economy. Understanding the mechanisms involved in regional and long-range aerial transport of crop pathogens, and monitoring their spread could help instigate remediation strategies to combat the global spread of plant infectious diseases.
The majority of the studies on aerial dispersal of plant pathogens (phytopathogens) have primarily focused on fungi, since over a thousand species of fungi are responsible for 70% of all known plant diseases (Shi et al. 2016). Fungi form spores that can withstand adverse environmental conditions (Wainwright et al. 2003; Griffin 2004; Sterflinger et al. 2012), enabling them to survive long journeys through the atmosphere (Griffin et al. 2001; Prospero et al. 2005). Several species of fungi known to cause common plant diseases, such as leaf spot, rust, rot, wilt, inhibition of growth, black mold, and early blights, were identified in Asian and African dusts collected thousands of kilometers in sink environments (Yeo and Kim 2002; Kellogg et al. 2004; Wu et al. 2004; Ho et al. 2005; Griffin 2007; Kakikawa et al. 2008; Palmero et al. 2011; Grishkan et al. 2012; Smith et al. 2012). Brown and Hovmøller surveyed the global outbreaks of plant diseases caused by intercontinental dispersion of fungal spores and uncovered a number of fungal diseases that affected plantations of tobacco, coffee, banana, sugarcane, potato, wheat and cereal worldwide (Brown and Hovmoller 2002). Many species of the genus Puccinia with well-known long-range wind dispersion patterns are the causative agents of major crop disease outbreaks worldwide. Long distance dissemination of airborne Puccinia spores by winds, for example, were responsible for the crop stem rust in the North American continent (Eversmeyer and Kramer 2000), the wheat leaf rust in the Indian subcontinent (Nagarajan and Singh 1990), the sugarcane rust, stem rust, and stripe rust across different continents (Watson and De Sousa 1983; Purdy et al. 1985; Wellings and McIntosh 1990), and the loss of the cereal crops worldwide (Narayanasamy 2011). Other important fungal crop pathogens such as Hemileia vastatrix causing the coffee leaf rust (Bowden et al. 1971) and Phakopsora pachyrhizi causing the soybean rust (Pan et al. 2006) were introduced to North America by windborne dissemination from Africa and Asia, respectively. Since the long-range spread of crop diseases by pathogenic fungi could cause significant economic downturn and food insecurities globally, effective surveillance and management strategies are needed to halt the spread of infections (Mahaffee and Stoll 2016). Periodic monitoring of air with molecular approaches, such as quantitative PCR (qPCR) to identify specific pathogens and/or culture independent metagenomics to screen the entire airborne microbiota, are powerful tools that could be more commonly implemented to monitor and identify potential threats as early as possible (Nicolaisen et al. 2017). Early pathogen detection would enable implementation of well-informed disease management strategies to combat infections.
Studies on potential long distant aerial spread of plant pathogenic bacteria have not been as extensive as those of fungi. Although a large number of putative bacterial plant pathogens were found in regional and intercontinental dust samples (Griffin et al. 2001; Kellogg et al. 2004; Kakikawa et al. 2008; Chuvochina et al. 2011b; Munday et al. 2013), no evidence of a direct link between the long-range aerial transport of bacteria and the spread of plant infectious diseases has been found. The majority of well-known phytopathogenic bacteria, including Agrobacterium, Xanthomonas, Pseudomonas, Erwinia, Clavibacter, and Rhodococcus species (Francis et al. 2010; Mansfield et al. 2012), do not form endospores, which make them unlikely candidates for long distant transport, unless they possess other mechanisms for resisting adverse atmospheric conditions. Microorganisms can improve their viability in the atmosphere by acting as cloud condensation nuclei (CCN), which can help increase the relative humidity of their surrounding environment, and protect them against desiccation and harmful UV irradiation. The ability to form CCN potentially played a role in the long distant aerial spread of Erwinia carotovora, the causal agent of potato blackleg disease, from the source waters off the west coast of the United States (US) to inland destinations in Colorado (US), where the disease was prevalent (Franc 1994). Pseudomonas syringae, another known plant pathogen with the ability to act as CCN (Lindow et al. 1982), could use a similar strategy for long-range aerial transport and disease spread. The mechanisms involved in long distance aerial spread of phytopathogenic bacteria warrant closer examination in light of their potential ramifications on disease spread.
Unlike fungi and bacteria, plant viruses usually require vectors for transmission of infection. The main route for transmission of plant viruses is by means of insect vectors including aphids, whiteflies, leafhoppers, beetles, and many other insects (Maramorosch 1980; Agrios 2005; Ng and Perry 2004). Aphids are known to carry and spread a large diversity of viruses including the barley yellow dwarf viruses (BYDV) that affects wide varieties of cereal crops worldwide (Miller and Rasochova 1997; Hall et al. 2010). Plant viruses can also transmit infections by means of infected seeds and pollens (Mink 1993). Little is known about the potential long-range atmospheric transport of plant pathogenic viruses and disease transmission in sink environment, although large number of viral DNA sequences, in particular those belonging to plant associated viruses, have been found in the air (Whon et al. 2012). The identification of airborne plant viral DNA sequences does not necessarily indicate the presence of viable viruses that could transmit infection. Viral particles are particularly sensitive to lengthy exposure to desiccation, extreme temperatures, and UV irradiation (Prussin et al. 2014), and are extremely unlikely to remain viable during long-range atmospheric transport unless they are carried inside host vectors such as plant debris, pollen, bacteria, fungi, or insects. Before any decision can be made regarding the potential ramifications of aerial spread of viral plant pathogens on global agriculture, we need to acquire a more extensive knowledge of the airborne viral community structures, their role in the atmosphere, and the potential mechanisms involved in their viability, virulence, and spread throughout the atmosphere.
Perspectives
Our present knowledge of sandstorm microbiota is limited with regard to their diversity, function, and impact. The majority of our knowledge comes from culture-based studies. By many accounts, over 90% of environmental microbiota cannot be cultured in laboratories; also since microorganisms are grown in isolation in growth culture media, culture-based approaches cannot provide information on microbial community structures in their natural environments. This information is essential for understanding their potential impact on host ecosystems. Regardless of their potential limitations, these studies confirmed that a large number of viable microorganisms, including known and opportunistic human and plant pathogens, are transported via sandstorms (Kellogg and Griffin 2006; Vijayakumar et al. 2017). Culture-based approaches in their current state can only provide a small glimpse into sandstorm-associated microbial diversity and function, unless significant improvements are made to make it possible to grow the majority of environmental microbiota (Vester et al. 2015).
In recent years, with the advent and significant progress in culture-independent metagenomic approaches, we are beginning to realize the vast extent of microbial diversity spread via sandstorms. Table 1 provides a list of some of the most recent publications on dust and sandstorm microbiology, showing that culture independent metagenomics approaches are gaining momentum. As can be seen from the table, the majority of recent studies were conducted on Asian and African dust events, with a few focusing on Arabian and Australian dust storms. Endospore forming, stress resistant microorganisms were commonly found in dust samples from different studies. These studies, although promising, are limited in terms of number and scope, and in particular studies of viral metagenomics is noticeably lacking. Metagenomics could provide the ability to study the entire microbiota and their community composition through direct sequencing and characterization of environmental DNA without the need for prior cultivation. In this paper, we use the term metagenomics to denote both the targeted amplicons sequencing (TAS) and the whole-genome shotgun sequencing (WGS) approaches. Although metagenomics is frequently used in studies of microbiome from different environments, few studies have adopted it for microbial analysis of air and sandstorm samples (Behzad et al. 2015). Culture-independent phylogenetic studies using TAS revealed that surprisingly diverse microbial communities are transported by sandstorm; in particular those by Weil et al. (2017) who showed that a strong sandstorm originating from the Sahara Deserts in Africa deposited a large fraction of the entire desert microbiome to a pristine remote mountain in Italy. In order to understand “impact”, we need to more fully characterize microbial diversity and function. Few of the published sandstorm metagenomic studies thus far have provided detailed phylogenetic analysis of sandstorm microbiota at species and strain levels, which is needed for determining the ecological significance and invasiveness/pathogenicity of these microorganisms. This could be due to the sole use of TAS, which is known for its low-resolution power amongst closely related species, sequencing errors, amplification biases, and inability to detect viruses (Hong et al. 2009; Quince et al. 2009; Quince et al. 2011; Wylie et al. 2012; Logares et al. 2014; Sharpton 2014). A more insightful approach, which is not commonly in practice in sandstorm research, is analysis of the entire microbial genomes in the environmental samples using WGS. This approach has significantly improved our understanding of microbial community structure, genes, and function in many environments including marine and soil (Simon and Daniel 2011) and would benefit sandstorm research. Although considerable advances in DNA sequencing platforms and bioinformatics analysis have been made in recent years, the approach still suffers from an inability to completely characterize the entire microbiota in environmental samples mainly due to: 1) inefficient assembly of the entire sample metagenome; and 2) inability to annotate a substantial fraction of these metagenomes since current databases lack representative sequences for many environmental microorganisms. Further improvements in air/dust sample collection (Behzad et al. 2015), DNA extraction methodologies, sequencing approaches and platforms, bioinformatics tools and software, and the availability of larger database of annotated environmental microbiota could help further advance sandstorm metagenomic research. A combined metagenomic and single cell genomic approach could also overcome the current shortcomings of metagenomics by improving the single cell genome and metagenome assembly and annotations (Lasken and McLean 2014). Metatranscriptomics, meta-proteomics, and metabolomics are other omic approaches rarely used in studies of sandstorm aeromicrobiology. The multi-omic approaches could help unravel not only the entire community and their genome makeup but also the functional and ecological significance of these microorganisms in their native environments and global biosphere.
| Viruses | Dust Source | Collection Site | Methods of identification | Potential Impact | Reference |
|---|---|---|---|---|---|
| Wide diversity of DNA and RNA viruses shed from a range of hosts, including animals, arthropods, bacteria, fungi, humans, plants, and protists | Indoor dusts | Indoor |
|
|
(Rosario et al. 2018) |
| ssDNA geminivirus-related viruses; circovirus-related sequences; nanoviruses; microphages-related genomes | Ambient air and rainwater samples | Seoul, Korea |
|
Plant | (Whon et al. 2012) |
| Influenza A | Asian dust | Taiwan | Real time qPCR | Human | (Chen et al. 2010) |
N/D: Not Determined.
Table 1.
Culture Independent Metagenomic Approaches Are Increasingly Used in More Recent Studies to Detect DSM
| Bacteria or Archaea | Fungi | Dust Source | Collection Site | Methods of Identification | Potential Impact | References |
|---|---|---|---|---|---|---|
| Actinobacteria; Bacteroidetes; Proteobacteria; Acidobacteria; Chloroflexi | Asian dust | Northern China |
|
|
(Tang et al. 2018) | |
| Deinococcus-Thermus; Chloroflexi; Cyanobacteria; Gemmatimonadetes; Acidobacteria; |
|
Israel |
|
Human | (Gat et al. 2017) | |
| Geodermatophilus; Bacillus; Deinococcus; Gemmatimonas; Arthrobacter; Nocardiodes; Rubrobacter; Solirubrobacter | Aurebasidium; Periconia; Pleosporaceae; Montagnulaceae; Embellisia; Davidiella | Sahara dust | Dolomite Alps, Italy |
|
|
(Weil et al. 2017) |
| Dominated by Bacillus members | Asian dust |
|
|
N/D | (Maki et al. 2017a) | |
|
Arabian dust | Saudi Arabia | Fungal culture | Human | (Vijayakumar et al. 2017) | |
| Bacillus subtilis | Asian dust | Carbon nanotubes (SWCNTs)-based electrochemical biosensor | Human | (Yoo et al. 2017) | ||
|
Asian dust | Beijing, China |
|
N/D | (Zhen et al. 2017) | |
| Bacillus; Bacillus circulans; Modestobacter; Methylobacterium iners; Sphingomonas starnbergensis; Micrococcus terreus; Rubellimicrobium roseum; Rubellimicrobium aerolatum | Asian dust | Seoul, Korea |
|
Human | (Cha et al. 2017) | |
|
Asian dust | Mongolia |
|
N/D | (Maki et al. 2017b) | |
| Gemmatimonadetes; Adhaeribacter; Arthrobacter; Bacillus; Balneimonas; Cellulomonas; Geodermatophilus; Pontibacter; Rubellimicrobium; Rubrobacter; Sporosarcina | Sahara dust | Eastern Mediterranean |
|
Human | (Mazar et al. 2016) | |
|
Asian dust | Beijing, China |
|
|
(Yamaguchi et al. 2016) | |
|
Asian dust | S. Korea |
|
Human | (Cha et al. 2016) | |
|
Asian dust | Osaka, Japan |
|
N/D | (Park et al. 2016) | |
| Alternaria; Ulocladium; Aspergillus; Penicillium; Fusarium; Acremonium; Phoma | North Westerly dust | Kuwait |
|
Agriculture | (Al-Bader et al. 2016) | |
| Bacillus spp. | Mycosporium spp | Arabian dust | Iran | Culture dependent | Human | (Nourmoradi et al. 2015) |
|
Aspergillus spp. | African dust | Sardinia, Italy |
|
Human | (Rosselli et al. 2015) |
|
Asian dust | Over East China Sea |
|
N/D | (Hara et al. 2015) | |
|
Asian dust |
|
|
Human | (An et al. 2014) | |
|
Sahara dust | Swiss Alps |
|
Human | (Meola et al. 2015) | |
|
Asian dust (Kosa) | Japan |
|
|
(Maki et al. 2014) | |
| Aspergillus sydowii | Australian dust | Australian coastal waters | Culture dependent: Molecular sequencing | Marine | (Hallegraeff et al. 2014) | |
|
|
|
Israel |
|
|
(Katra et al. 2014) |
|
Sahara dust |
|
|
Aqueous ecosystems | (Peter et al. 2014) | |
|
Sahara dust | Pyrenees, Spain |
|
Aqueous ecosystems | (Vila-Costa et al. 2013) | |
| Proteobacteria; Firmicutes (Bacillus, Sporosarcina) | Sahara dust | South of Spain |
|
N/D | (Sánchez de la Campa et al. 2013) | |
| Proteobacteria (Comamonadaceae; Pseudomonadaceae, Enterobacteriaceae); Actinobacteria (Corynebacteriaceae, Streptomycetaceae); Firmicutes (Bacillaceae, Lachnospiraceae, Staphylococcaceae, Streptococcaceae) | Transpacific air plumes from Asia | Central Oregon, USA |
|
N/D | (Smith et al. 2013) | |
| Firmicutes-Bacillaceae; Actinobacteria-Geodermatophilaceae, Nocardiodaceae, Solirubrobacteraceae; Proteobacteria-Oxalobacteraceae, Rhizobiales, Sphingomonadaceae; Bacteroidetes-Cytophagaceae | Ascomycota; Basidiomycota; Chytridiomycota; Microsporidia; Glomeromycota | African dust |
|
|
|
(Favet et al. 2013) |
| Actinobacteria; Firmicutes (B. Subtillus); Proteobacteria; | Asian dust (Kosa) | Mt. Tateyama, Japan |
|
|
(Maki et al. 2011) | |
|
Sahara dust |
|
|
Ecosystem | (Chuvochina et al. 2011a) | |
|
Asian dust | Seoul, Korea |
|
N/D | (Jeon et al. 2011) | |
| Bacillus species; Pseudomonas species | Australian dust | Canberra and Melbourne (Australia) |
|
|
(Lim et al. 2011) |
In order to exert impact, DSM needs to survive the long journey across the atmosphere, and continue to remain viable and propagate in their new host ecosystems. On the subject of viability, current culture dependent and independent approaches do not provide accurate representation of the percent-viability of DSM before and upon arrival in destination. Knowledge of microbial viability is essential for determining the spread and propagation of foreign, pathogenic or invasive microorganisms. The presence of DNA sequences belonging to pathogenic microorganisms in environmental metagenome does not necessarily indicate that the pathogen is alive and able to propagate. Culture dependent approaches could provide better metrics for assessment of viability but currently greater than 90% of environmental microbes cannot be cultured using standard culturing media. Dust samples contain diverse groups of microorganisms that propagate in different environmental conditions; whereas some microbes prefer standard conditioned media for growth, others have specific growth requirements that need to be met. A combination of culture dependent and independent methodologies could be employed to circumvent the shortcomings associated with either of the two approaches (Vester et al. 2015). Culturing requirements could be improved by using culture independent multi-omic approaches to obtain information on the complexity of the microbiota in dust samples and their metabolic needs; this information could then be used to design specific culture media formulations to improve percent cultivability of environmental microbiota (Gutleben et al. 2018). This would help resolve questions pertaining to viability, and also advance studies of microbial diversity and function in culture settings.
Since sandstorms are recurring events and their impacts potentially additive, time-series studies are needed to determine potential long-term ramifications of increased DSM transmission on the ecology of downwind ecosystems (Chuvochina et al. 2011b; Weil et al. 2017). These studies could establish microbial fingerprints of various environments and enable tracking variations in microbial patterns in response to different environmental changes, including sandstorms (Faust et al. 2015). Comparative analysis of microbiota and environmental metadata from source and destination environments could enable identification of influential variables, including invasive/pathogenic microorganisms, involved in ecosystem alterations. The ideal settings for such studies would be environments with minimal external influence, and reduced seasonal variability (Hervas et al. 2009; Mladenov et al. 2011).
Time series studies place particular demands on consistency and reproducibility of data since they typical involve collaboration among multiple laboratories. Such collaborations require standardized approaches to enable comparison of data collected across different time and space, by multiple laboratories. Metagenomic approaches are particularly sensitive to variability in methodology; metagenomic studies that use different approaches and protocols generate different outcomes concerning microbial quantity, diversity, and composition (Clooney et al. 2016). Currently there is no consensus within the field on best methodologies, or standardization between multiple laboratories. Standardized Operating Protocols (SOP) would help unify the methodologies used by all the collaborators involved in the field. These SOPs require considerable investments of time and resources to develop and perfect; however, their use by wider research community could provide long-term benefits that could substantially advance DSM research. Collaborative work, in particular time-series studies, should address potential relationships between global climate changes, expansion of desertification, increased and widespread injection of DSM into downwind ecosystems, and their human health and ecological ramifications.
Conclusions
Dust activities are expected to rise globally due to climate changes and the rise in desertification caused by natural and anthropogenic events. This could lead to a substantial increase in transport of foreign, invasive, and potentially new pathogenic microorganisms that could alter equilibrium balance in downwind ecosystems, remodel pristine environments, and/or affect human and ecosystem health. Previous attempts to make concrete connections between DSM and human and environmental health concerns have been largely unsuccessful due to lack of substantive data. Inadequate knowledge of sandstorm microbiota limits our understanding of their broader significance. In recent years, TAS metagenomic studies have uncovered a large diversity of microbiota in dust clouds but the significance of these findings remains unknown due to the low-resolution power of TAS-based approaches. To gain a better insight into the phylogenetic diversity and functional and ecological significance of DSM, we recommend a combination of high-resolution strategies including WGS metagenomics, single cell genomics, and multiomic approaches in conjunction with improved culture-based methodologies. Given the increased prevalence of these highly stress resistant, diverse, and potentially pathogenic microorganisms in global atmosphere and downwind ecosystems, there is now a clear need for large-scale collaborative efforts to address potential relationships between the widespread injection of DSM in global atmosphere, ecosystem alterations, and global climate changes, through extensive ecological time-series studies.
Acknowledgments
The research reported in this publication was supported by fundings from King Abdullah University of Science and Technology (KAUST), under award numbers BAS/1/1059/01/01 and URF/1/1976/03/01. We thank Martin Ibarra for his assistance with literature search. Figure 1 was produced by Ivan Gromicho, scientific illustrator at King Abdullah University of Science and Technology (KAUST).
Literature Cited
- Abed RMM, Al Kharusi S, Schramm A, Robinson MD.. 2010. Bacterial diversity, pigments and nitrogen fixation of biological desert crusts from the sultanate of Oman. Fems Microbiol Ecol. 72(3):418–428. [DOI] [PubMed] [Google Scholar]
- Agier L, et al. 2013. Seasonality of meningitis in Africa and climate forcing: aerosols stand out. J R Soc Interface 10(79):20120814.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrios G. 2005. Plant pathology. Boston, MA: Elsevier Academic Press. [Google Scholar]
- Al-Bader D, Alqodaiby A, Suleman P.. 2016. Characterization of fungi transferred by dust storms in Kuwait and their plant pathogenicity. Aerobiologia 32(2):335–345. [Google Scholar]
- An S, Couteau C, Luo F, Neveu J, DuBow MS.. 2013. Bacterial diversity of surface sand samples from the Gobi and Taklamaken deserts. Microb Ecol. 66(4):850–860. [DOI] [PubMed] [Google Scholar]
- An S, Sin HH, DuBow MS.. 2014. Modification of atmospheric sand-associated bacterial communities during Asian sandstorms in China and South Korea. Heredity 114(5):460–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel R, Soares MIM, Ungar ED, Gillor O.. 2010. Biogeography of soil archaea and bacteria along a steep precipitation gradient. ISME J. 4(4):553–563. [DOI] [PubMed] [Google Scholar]
- Behzad H, Gojobori T, Mineta K.. 2015. Challenges and opportunities of airborne metagenomics. Genome Biol Evol. doi: 10.1093/gbe/evv064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhard AE, Kelly JJ.. 2016. Editorial: linking ecosystem function to microbial diversity (vol 7, 1041, 2016). Front Microbiol. 7:1041.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop JKB, Davis RE, Sherman JT.. 2002. Robotic observations of dust storm enhancement of carbon biomass in the north pacific. Science (New York, NY) 298(5594):817–821. [DOI] [PubMed] [Google Scholar]
- Bowden J, Gregory PH, Johnson CG.. 1971. Possible wind transport of coffee leaf rust across the Atlantic Ocean. Nature 229(5285):500–501. [DOI] [PubMed] [Google Scholar]
- Bowers RM, et al. 2009. Characterization of airborne microbial communities at a high-elevation site and their potential to act as atmospheric ice nuclei. Appl Environ Microbiol. 75(15):5121–5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodie EL, et al. 2007. Urban aerosols harbor diverse and dynamic bacterial populations. Proc Natl Acad Sci USA. 104(1):299–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JKM, Hovmoller MS.. 2002. Epidemiology: aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science (New York, NY) 297(5581):537–541. [DOI] [PubMed] [Google Scholar]
- Burrows SM, Elbert W, Lawrence MG, Pöschl U.. 2009. Bacteria in the global atmosphere—part 1: review and synthesis of literature data for different ecosystems. Atmos Chem Phys. 9(23):9263–9280. [Google Scholar]
- Cha S, et al. 2016. Alterations in the airborne bacterial community during Asian dust events occurring between February and March 2015 in South Korea. Sci Rep. 6(1): 37271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha S, et al. 2017. Metagenomic analysis of airborne bacterial community and diversity in Seoul, Korea, during December 2014, Asian dust event. PLoS One 12(1):e0170693.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y, Van Nostrand JD, Zhou J, Pointing SB, Farrell RL.. 2013. Functional ecology of an antarctic dry valley. Proc Natl Acad Sci USA. 110(22):8990–8995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PS, et al. 2010. Ambient influenza and Avian influenza virus during dust storm days and background days. Environ Health Perspect. 118(9):1211–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JC, Lee M, Chun Y, Kim J, Oh S.. 2001. Chemical composition and source signature of spring aerosol in Seoul, Korea. J Geophys Res. 106(D16):18067–18074. [Google Scholar]
- Chuvochina MS, Alekhina IA, Normand P, Petit JR, Bulat SA.. 2011. Three events of Saharan dust deposition on the Mont Blanc glacier associated with different snow-colonizing bacterial phylotypes. Microbiology 80(1):125–131. [Google Scholar]
- Chuvochina MS, et al. 2011. Community variability of bacteria in Alpine snow (Mont Blanc) containing Saharan dust deposition and their snow colonisation potential. Microbes Environ. 26(3):237–247. [DOI] [PubMed] [Google Scholar]
- Clark JS, Campbell JH, Grizzle H, Acosta-Martìnez V, Zak JC.. 2009. Soil microbial community response to drought and precipitation variability in the Chihuahuan desert. Microb Ecol. 57(2):248–260. [DOI] [PubMed] [Google Scholar]
- Clooney AG, et al. 2016. Comparing apples and oranges?: next generation sequencing and its impact on microbiome analysis. PLoS One 11(2):e0148028.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley CA, Ishkhanova G, McKay CP, Cullings K.. 2006. A preliminary survey of non-lichenized fungi cultured from the hyperarid Atacama Desert of Chile. Astrobiology 6(4):521–526. [DOI] [PubMed] [Google Scholar]
- Denning DW, O'Driscoll BR, Hogaboam CM, Bowyer P, Niven RM.. 2006. The link between fungi and severe asthma: a summary of the evidence. Eur Respir J. 27(3):615–626. [DOI] [PubMed] [Google Scholar]
- Dose K, et al. 2001. Survival of microorganisms under the extreme conditions of the Atacama Desert. Origins Life Evol B. 31(3):287–303. [DOI] [PubMed] [Google Scholar]
- Etemadifar Z, Gholami M, Derikvand P.. 2016. UV-resistant bacteria with multiple-stress tolerance isolated from desert areas in Iran. Geomicrobiol J. 33(7):1–598. [Google Scholar]
- Eversmeyer MG, Kramer CL.. 2000. Epidemiology of wheat leaf and stem rust in the Central Great Plains of the USA. Annu Rev Phytopathol. 38:491–513. [DOI] [PubMed] [Google Scholar]
- Faust K, Lahti L, Gonze D, de Vos WM, Raes J.. 2015. Metagenomics meets time series analysis: unraveling microbial community dynamics. Curr Opin Microbiol. 25:56–66. [DOI] [PubMed] [Google Scholar]
- Favet J, et al. 2013. Microbial hitchhikers on intercontinental dust: catching a lift in chad. ISME J. 7(4):850–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C, Li J, Sun WJ, Zhang Y, Wang QY.. 2016. Impact of ambient fine particulate matter (pm2.5) exposure on the risk of influenza-like-illness: a time-series analysis in Beijing, China. Environ Health Glob. 15(1): [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, et al. 2007. Metagenomic and small-subunit rRNA analyses reveal the genetic diversity of bacteria, archaea, fungi, and viruses in soil. Appl Environ Microbiol. 73(21):7059–7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Jackson RB.. 2006. The diversity and biogeography of soil bacterial communities. ProcNatl Acad Sci USA. 103(3):626–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, et al. 2012. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc Natl Acad Sci USA. 109(52):21390–21395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franc GD. 1988. Long distance transport of Erwinia carotovora in the atmosphere and surface water. Ph.D. dissertation, Colorado State University, Fort Collins, CO, 131 pp. [Available from Dept. of Bioagricultural Sciences and Pest Management, Fort Collins, CO 80523–1177.]
- Franc GD. 1994. Atmospheric transport of Erwinia carotovora. Proceedings 21st Conference on Agricultural and Forest Meteorology and 11th Conference on Biometeorology and Aerobiology, San Diego, CA, Amer. Meteor. Soc., 435–437.
- Francis I, Holsters M, Vereecke D.. 2010. The Gram-positive side of plant-microbe interactions. Environ Microbiol. 12(1):1–12. [DOI] [PubMed] [Google Scholar]
- Franzen LG, et al. 1995. The Saharan dust episode of south and central Europe, and northern Scandinavia, March 1991. Weather 50(9):313–318. [Google Scholar]
- Franzetti A, Gandolfi I, Gaspari E, Ambrosini R, Bestetti G.. 2011. Seasonal variability of bacteria in fine and coarse urban air particulate matter. Appl Microbiol Biotechnol. 90(2):745–753. [DOI] [PubMed] [Google Scholar]
- Frohlich-Nowoisky J, et al. 2014. Diversity and seasonal dynamics of airborne archaea. Biogeosciences 11(21):6067–6079. [Google Scholar]
- Fu C, An Z.. 2002. Study of aridization in northern china—a global change issue facing directly the demand of nation. Earth Sci Front. 9(2):271–275. [Google Scholar]
- Gallisai R, Peters F, Volpe G, Basart S, Baldasano JM.. 2014. Saharan dust deposition may affect phytoplankton growth in the Mediterranean Sea at ecological time scales. PLoS One 9(10):e110762.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganor E, Foner HA, Brenner S, Neeman E, Lavi N.. 1991. The chemical-composition of aerosols settling in Israel following dust storms. Atmos Environ. 25(12):2665–2670. [Google Scholar]
- Garrison VH, et al. 2003. African and Asian dust: from desert soils to coral reefs. Bioscience 53(5):469–480. [Google Scholar]
- Gat D, Mazar Y, Cytryn E, Rudich Y.. 2017. Origin-dependent variations in the atmospheric microbiome community in Eastern Mediterranean dust storms. Environ Sci Technol. 51(12):6709–6718. [DOI] [PubMed] [Google Scholar]
- Glezen WP, Greenberg SB, Atmar RL, Piedra PA, Couch RB.. 2000. Impact of respiratory virus infections on persons with chronic underlying conditions. JAMA 283(4):499–505. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Martin C, Teigell-Perez N, Valladares B, Griffin DW.. 2014. The global dispersion of pathogenic microorganisms by dust storms and its relevance to agriculture. Adv Agron. 127:1–41. [Google Scholar]
- Graham EB, et al. 2016. Microbes as engines of ecosystem function: when does community structure enhance predictions of ecosystem processes?. Front Microbiol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin DW. 2004. Terrestrial microorganisms at an altitude of 20,000 m in earth's atmosphere. Aerobiologia 20(2):135–140. [Google Scholar]
- Griffin DW. 2007. Atmospheric movement of microorganisms in clouds of desert dust and implications for human health. Clin Microbiol Rev. 20(3):459–477. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin DW, Kellogg CA.. 2004. Dust storms and their impact on ocean and human health: dust in earth’s atmosphere. EcoHealth 1(3):284–295. [Google Scholar]
- Griffin DW, Garrison VH, Herman JR, Shinn EA.. 2001. African desert dust in the Caribbean atmosphere: microbiology and public health. Aerobiologia 17:203–213. [Google Scholar]
- Grishkan I, Schlesinger P, Mamane Y.. 2012. Influence of dust storms on concentration and content of fungi in the atmosphere of Haifa, Israel. Aerobiologia 28(4):557–564. [Google Scholar]
- Grousset FE, Ginoux P, Bory A, Biscaye PE.. 2003. Case study of a Chinese dust plume reaching the french Alps. Geophys Res Lett. 30(6):1277. [Google Scholar]
- Gtari M, et al. 2012. Contrasted resistance of stone-dwelling geodermatophilaceae species to stresses known to give rise to reactive oxygen species. Fems Microbiol Ecol. 80(3):566–577. [DOI] [PubMed] [Google Scholar]
- Guieu C, et al. 2014. The significance of the episodic nature of atmospheric deposition to low nutrient low chlorophyll regions. Global Biogeochem Cycles 28:11. [Google Scholar]
- Gutleben J, et al. 2018. The multi-omics promise in context: from sequence to microbial isolate. Crit Rev Microbiol. 44(2):212–229. [DOI] [PubMed] [Google Scholar]
- Gyan K, et al. 2005. African dust clouds are associated with increased paediatric asthma accident and emergency admissions on the Caribbean island of Trinidad. Int J Biometeorol. 49(6):371–376. [DOI] [PubMed] [Google Scholar]
- Hall GS, Peters JS, Little DP, Power AG.. 2010. Plant community diversity influences vector behaviour and barley yellow dwarf virus population structure. Plant Pathol. 59(6):1152–1158. [Google Scholar]
- Hallegraeff G, et al. 2014. Australian dust storm associated with extensive aspergillus sydowii fungal “bloom” in coastal waters. Appl Environ Microbiol. 80(11):3315e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, et al. 2015. UV-tolerant culturable bacteria in an Asian dust plume transported over the East China Sea. Aerosol Air Qual Res. 15(2):591–599. [Google Scholar]
- Herut B, et al. 2005. Response of east mediterranean surface water to Saharan dust: on-board microcosm experiment and field observations. Deep Sea Res II 52(22–23):3024–3040. [Google Scholar]
- Hervas A, Camarero L, Reche I, Casamayor EO.. 2009. Viability and potential for immigration of airborne bacteria from Africa that reach high mountain lakes in Europe. Environ Microbiol. 11(6):1612–1623. [DOI] [PubMed] [Google Scholar]
- Hilty M, et al. 2010. Disordered microbial communities in asthmatic airways. PLoS One 5(1):e8578.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho HM, et al. 2005. Characteristics and determinants of ambient fungal spores in Hualien, Taiwan. Atmos Environ. 39(32):5839–5850. [Google Scholar]
- Hong SH, Bunge J, Leslin C, Jeon S, Epstein SS.. 2009. Polymerase chain reaction primers miss half of rRNA microbial diversity. ISME J. 3(12):1365–1373. [DOI] [PubMed] [Google Scholar]
- Huang J, et al. 2017. Dryland climate change: recent progress and challenges. Rev Geophys. 55(3):719–778. [Google Scholar]
- Huang YJ, et al. 2011. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immun. 127(2):372–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZW, et al. 2015. Short-cut transport path for Asian dust directly to the arctic: a case study. Environ Res Lett. 10(11):114018. [Google Scholar]
- Husar RB, et al. 2001. Asian dust events of April 1998. J Geophys Res. 106(D16):18317–18330. [Google Scholar]
- Issanova G, Abuduwaili J, Galayeva O, Semenov O, Bazarbayeva T.. 2015. Aeolian transportation of sand and dust in the Aral Sea region. Int J Environ Sci Technol. 12(10):3213–3224. [Google Scholar]
- Iwasaka Y, Minoura H, Nagaya K.. 1983. The transport and spacial scale of Asian dust-storm clouds: a case-study of the dust-storm event of April 1979. Tellus B 35(3):189–196. [Google Scholar]
- Jeon EM, et al. 2011. Impact of Asian dust events on airborne bacterial community assessed by molecular analyses. Atmos Environ. 45(25):4313–4321. [Google Scholar]
- Johnson MS, Meskhidze N, Kiliyanpilakkil VP, Gasso S.. 2011. Understanding the transport of patagonian dust and its influence on marine biological activity in the South Atlantic Ocean. Atmos Chem Phys. 11(6):2487–2502. [Google Scholar]
- Jusot JF, et al. 2017. Airborne dust and high temperatures are risk factors for invasive bacterial disease. J Allergy Clin Immun. 139(3):977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakikawa M, et al. 2008. Dustborne microorganisms in the atmosphere over an Asian dust source region, Dunhuang. Air Qual Atmos Health 1(4):195–202. [Google Scholar]
- Kanatani KT, et al. 2010. Desert dust exposure is associated with increased risk of asthma hospitalization in children. Am J Resp Crit Care 182(12):1475–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katra I, et al. 2014. Richness and diversity in dust stormborne biomes at the southeast mediterranean. Sci Rep. 4(1):5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg C, et al. 2004. Characterization of aerosolized bacteria and fungi from desert dust events in Mali, West Africa. Aerobiologica 20(2):99–110. [Google Scholar]
- Kellogg CA, Griffin DW.. 2006. Aerobiology and the global transport of desert dust. Trends Ecol Evol. 21(11):638–644. [DOI] [PubMed] [Google Scholar]
- Kirkland TN, Fierer J.. 1996. Coccidioidomycosis: a reemerging infectious disease. Emerg Infect Dis. 2(3):192–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubilay N, Nickovic S, Moulin C, Dulac F.. 2000. An illustration of the transport and deposition of mineral dust onto the eastern mediterranean. Atmos Environ. 34(8):1293–1303. [Google Scholar]
- Kuske CR, Barns SM, Busch JD.. 1997. Diverse uncultivated bacterial groups from soils of the arid southwestern united states that are present in many geographic regions. Appl Environ Microbiol. 63(9):3614–3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasken RS, McLean JS.. 2014. Recent advances in genomic DNA sequencing of microbial species from single cells. Nat Rev Genet. 15(9):577–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber CL, Hamady M, Knight R, Fierer N.. 2009. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol. 75(15):5111–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent B, Marticorena B, Bergametti G, Mei F.. 2006. Modeling mineral dust emissions from Chinese and mongolian deserts. Global Planet Change 52(1-4):121–141. [Google Scholar]
- Lee CK, Barbier BA, Bottos EM, McDonald IR, Cary SC.. 2012. The inter-valley soil comparative survey: the ecology of dry valley edaphic microbial communities. ISME J. 6(5):1046–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Kim H, Honda Y, Lim YH, Yi S.. 2013. Effect of Asian dust storms on daily mortality in seven metropolitan cities of Korea. Atmos Environ. 79:510–517. [Google Scholar]
- Lee JY, et al. 2017. Airborne bacterial communities in three East Asian cities of China, South Korea, and Japan. Sci Rep. 7(1). doi: 10.1038/s41598-017-05862-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekunberri I, et al. 2010. Effects of a dust deposition event on coastal marine microbial abundance and activity, bacterial community structure and ecosystem function. J Plankton Res. 32(4):381–396. [Google Scholar]
- Lim N, et al. 2011. Microbiological and meteorological analysis of two Australian dust storms in April 2009. Sci Total Environ. 412–413:223–231. [DOI] [PubMed] [Google Scholar]
- Lindow SE, Arny DC, Upper CD.. 1982. Bacterial ice nucleation—a factor in frost injury to plants. Plant Physiol. 70(4):1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logares R, et al. 2014. Metagenomic 16s rDNA illumina tags are a powerful alternative to amplicon sequencing to explore diversity and structure of microbial communities. Environ Microbiol. 16(9):2659–2671. [DOI] [PubMed] [Google Scholar]
- LoyePilot MD, Martin JM.. 1996. Saharan dust input to the western mediterranean: an eleven years record in Corsica. Environ Sci Tech Lib. 11:191–199. [Google Scholar]
- Ma YX, Zhou JD, Yang SX, Zhao YX, Zheng XD.. 2017. Assessment for the impact of dust events on measles incidence in western china. Atmos Environ. 157:1–9. [Google Scholar]
- Maestre FT, et al. 2014. Changes in biocrust cover drive carbon cycle responses to climate change in drylands (vol. 19, pg 3835, 2013). Global Change Biol. 20(8):2697–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahaffee WF, Stoll R.. 2016. The Ebb and flow of airborne pathogens: monitoring and use in disease management decisions. Phytopathology 106(5):420–431. [DOI] [PubMed] [Google Scholar]
- Makhalanyane TP, et al. 2015. Microbial ecology of hot desert edaphic systems. FEMS Microbiol Rev. 39(2):203–221. [DOI] [PubMed] [Google Scholar]
- Maki T, et al. 2011. Characterization of halotolerant and oligotrophic bacterial communities in Asian desert dust (kosa) bioaerosol accumulated in layers of snow on mount Tateyama, central Japan. Aerobiologia 27(4):277–290. [Google Scholar]
- Maki T, et al. 2017a. Variations in airborne bacterial communities at high altitudes over the noto peninsula (Japan) in response to Asian dust events. Atmos Chem Phys. 17(19):11877–11897. [Google Scholar]
- Maki T, et al. 2012. Outbreak of foot-and-mouth disease in Miyazaki from March to July 2010—effect of yellow sand and local surface wind. J Arid Land Stud. 22(1):167–170. [Google Scholar]
- Maki T, et al. 2017. Variations in the structure of airborne bacterial communities in tsogt-ovoo of Gobi desert area during dust events. Air Qual Atmos Health 10(3):249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki T, et al. 2014. Variations in the structure of airborne bacterial communities in a downwind area during an Asian dust (Kosa) event. Sci Total Environ. 488-489:75–84. [DOI] [PubMed] [Google Scholar]
- Mallone S, et al. 2011. Saharan dust and associations between particulate matter and daily mortality in Rome, Italy. Environ Health Perspect. 119(10):1409–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield J, et al. 2012. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol. 13(6):614–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maramorosch K. 1980. Spread of plant viruses and spiroplasmas through airborne vectors. Ann N Y Acad Sci. 353(1):179–185. [Google Scholar]
- Maron PA, et al. 2005. Assessing genetic structure and diversity of airborne bacterial communities by DNA fingerprinting and 16s rDNA clone library. Atmos Environ. 39(20):3687–3695. [Google Scholar]
- Martinez JL. 2008. Antibiotics and antibiotic resistance genes in natural environments. Science 321(5887):365–367. [DOI] [PubMed] [Google Scholar]
- Mazar Y, Cytryn E, Ere Y, Rudich Y.. 2016. Effect of dust storms on the atmospheric microbiome in the eastern mediterranean. Environ Sci Technol. 50(8):4194–4202. [DOI] [PubMed] [Google Scholar]
- McConnell JR, Aristarain AJ, Banta JR, Edwards PR, Simoes JC.. 2007. 20th-century doubling in dust archived in an Antarctic Peninsula ice core parallels climate change and desertification in South America. Proc Natl Acad Sci USA. 104(14):5743–5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meo SA, Al-Kheraiji MF, Alfaraj ZF, Alwehaibi NA, Aldereihim AA.. 2013. Respiratory and general health complaints in subjects exposed to sandstorm at Riyadh, Saudi Arabia. Pakistan J Med Sci. 29(2):642–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meola M, Lazzaro A, Zeyer J.. 2015. Bacterial composition and survival on Sahara dust particles transported to the European Alps. Front Microbiol. 6:1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W, Rasochova L.. 1997. Barley yellow Dwarf viruses. Ann Rev Phytopathol. 35:167–190. [DOI] [PubMed] [Google Scholar]
- Mink GI. 1993. Pollen-transmitted and seed-transmitted viruses and viroids. Annu Rev Phytopathol. 31:375–402. [DOI] [PubMed] [Google Scholar]
- Mladenov N, et al. 2011. Dust inputs and bacteria influence dissolved organic matter in clear Alpine lakes. Nat Commun. 2(1):405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulin C, Chiapello I.. 2006. Impact of human-induced desertification on the intensification of Sahel dust emission and export over the last decades. Geophys Res Lett. 33(18):L18808. [Google Scholar]
- Moulin C, Lambert CE, Dulac F, Dayan U.. 1997. Control of atmospheric export of dust from North Africa by the north Atlantic oscillation. Nature 387(6634):691–694. [Google Scholar]
- Munday CI, O'Loingsigh T, Tapper NJ, De Deckker P, Allison GE.. 2013. Utilisation of rep-pcr to track microbes in aerosols collected adjacent to their source, a saline lake in victoria, Australia. Sci Total Environ. 450–451:317–325. [DOI] [PubMed] [Google Scholar]
- Nagarajan S, Singh DV.. 1990. Long-distance dispersion of rust pathogens. Annu Rev Phytopathol. 28:139–153. [DOI] [PubMed] [Google Scholar]
- Narayanasamy P. 2011. Microbial Plant Pathogens-Detection and Disease Diagnosis: Fungal Pathogens, Vol. 1, Springer Science+Business Media B. Netherlands: Springer.
- Ng JCK, Perry KL.. 2004. Transmission of plant viruses by aphid vectors. Mol Plant Pathol. 5(5):505–511. [DOI] [PubMed] [Google Scholar]
- Nicolaisen M, et al. 2017. Fungal communities including plant pathogens in near surface air are similar across northwestern Europe. Front Microbiol. 8:1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y, et al. 2010. Similarity of bacterial community structure between Asian dust and its sources determined by rRNA gene-targeted approaches. Microbes Environ. 25(1):22–27. [DOI] [PubMed] [Google Scholar]
- Nourmoradi H, et al. 2015. The effect of dust storm on the microbial quality of ambient air in Sanandaj: a city located in the west of Iran. Glob J Health Sci. 7:114–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SY, Fong JJ, Park MS, Chang L, Lim YW.. 2014. Identifying airborne fungi in Seoul, Korea using metagenomics. J Microbiol. 52(6):465–472. [DOI] [PubMed] [Google Scholar]
- Ortiz-Castro R, Contreras-Cornejo HA, Macias-Rodriguez L, Lopez-Bucio J.. 2009. The role of microbial signals in plant growth and development. Plant Signal Behav. 4(8):701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmero D, et al. 2011. Fungal microbiota from rain water and pathogenicity of Fusarium species isolated from atmospheric dust and rainfall dust. J Ind Microbiol Biotechnol. 38(1):13–20. [DOI] [PubMed] [Google Scholar]
- Pan Z, et al. 2006. Long-term prediction of soybean rust entry into the continental United States. Plant Dis. 90(7):840–846. [DOI] [PubMed] [Google Scholar]
- Park J, Ichijo T, Nasu M, Yamaguchi N.. 2016. Investigation of bacterial effects of Asian dust events through comparison with seasonal variability in outdoor airborne bacterial community. Sci Rep. 6(1): [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paytan A, et al. 2009. Toxicity of atmospheric aerosols on marine phytoplankton. Proc Natl Acad Sci USA. 106(12):4601–4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins S. 2001. Dust, the thermostat. Sci News 160(13):200–201. [Google Scholar]
- Perry KD, Cahill TA, Eldred RA, Dutcher DD, Gill TE.. 1997. Long-range transport of North African dust to the eastern United States. J Geophys Res. 102(D10):11225–11238. [Google Scholar]
- Peter H, Hortnagl P, Reche I, Sommaruga R.. 2014. Bacterial diversity and composition during rain events with and without Saharan dust influence reaching a high mountain lake in the Alps. Env Microbiol Rep. 6(6):618–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pointing SB, et al. 2009. Highly specialized microbial diversity in hyper-arid polar desert. Proc Natl Acad Sci USA. 106(47):19964–19969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenakou PN. 2012. Atmosphere: a source of pathogenic or beneficial microbes?. Atmosphere 3(1):87–102. [Google Scholar]
- Prenni AJ, et al. 2009. Relative roles of biogenic emissions and Saharan dust as ice nuclei in the amazon basin. Nat Geosci. 2(6):402–404. [Google Scholar]
- Prospero JM. 1999. Long-range transport of mineral dust in the global atmosphere: impact of African dust on the environment of the southeastern united states. Proc Natl Acad Sci USA. 96(7):3396–3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prospero JM, Blades E, Mathison G, Naidu R.. 2005. Interhemispheric transport of viable fungi and bacteria from Africa to the Caribbean with soil dust. Aerobiologia 21(1):1–19. [Google Scholar]
- Prospero JM, Carlson TN.. 1981. Saharan air outbreaks over the tropical north-atlantic. Pure Appl Geophys. 119(3):677–691. [Google Scholar]
- Prussin AJ, Garcia EB, Marr LC.. 2015. Total concentrations of virus and bacteria in indoor and outdoor air. Environ Sci Tech Lett. 2(4):84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prussin AJ, Marr LC, Bibby KJ.. 2014. Challenges of studying viral aerosol metagenomics and communities in comparison with bacterial and fungal aerosols. Fems Microbiol Lett. 357(1):1–9. [DOI] [PubMed] [Google Scholar]
- Purdy LH, Krupa SV, Dean JL.. 1985. Introduction of sugarcane rust into the Americas and its spread to Florida. Plant Dis. 69(8):689–693. [Google Scholar]
- Quince C, et al. 2009. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods 6(9):639.. [DOI] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ.. 2011. Removing noise from pyrosequenced amplicons. BMC Bioinformatics 12(1):38.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahav E, Ovadia G, Paytan A, Herut B.. 2016. Contribution of airborne microbes to bacterial production and n2 fixation in seawater upon aerosol deposition. Geophys Res Lett. 43(2):719–727. [Google Scholar]
- Rodo X, et al. 2011. Association of Kawasaki disease with tropospheric wind patterns. Sci Rep. 1(1):152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodo X, et al. 2014. Tropospheric winds from northeastern china carry the etiologic agent of Kawasaki disease from its source to japan. Proc Natl Acad Sci USA. 111(22):7952–7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrı´guez S, Querol X, Alastuey A, Kallos G, Kakaliagou O.. 2001. Saharan dust contributions to PM10 and TSP levels is southern and eastern Spain. Atmos Environ. 35(14):2433–2447. [Google Scholar]
- Romero E, et al. 2011. Coastal mediterranean plankton stimulation dynamics through a dust storm event: an experimental simulation. Estuar Coast Shelf Sci. 93(1):27–39. [Google Scholar]
- Rosario K, Fierer N, Miller S, Luongo J, Breitbart M.. 2018. Diversity of DNA and RNA viruses in indoor air as assessed via metagenomic sequencing. Environ Sci Technol. 52(3):1014–1027. [DOI] [PubMed] [Google Scholar]
- Rosselli R, et al. 2015. Microbial immigration across the mediterranean via airborne dust. Sci Rep. 5(1):16306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothschild LJ, Mancinelli RL.. 2001. Life in extreme environments. Nature 409(6823):1092–1101. [DOI] [PubMed] [Google Scholar]
- Rousk J, et al. 2010. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 4(10):1340–1351. [DOI] [PubMed] [Google Scholar]
- Sánchez de la Campa A, García-Salamanca A, Solano J, de la Rosa J, Ramos J-L.. 2013. Chemical and microbiological characterization of atmospheric particulate matter during an intense African dust event in southern spain. Environ Sci Technol. 47(8):3630–3638. [DOI] [PubMed] [Google Scholar]
- Schulz S, et al. 2013. The role of microorganisms at different stages of ecosystem development for soil formation. Biogeosciences 10(6):3983–3996. [Google Scholar]
- Sharma H, Burark SS, Meena GL.. 2015. Land degradation and sustainable agriculture in Rajasthan, India. J Indus Pollut Control 31(1):7–11. [Google Scholar]
- Sharpton TJ. 2014. An introduction to the analysis of shotgun metagenomic data. Front Plant Sci. 5:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, et al. 2016. Autophagy in Plant Pathogenic Fungi, Autophagy in Current Trends in Cellular Physiology and Pathology Nikolai Gorbunov, IntechOpen, DOI: 10.5772/64634.
- Shinn EA, et al. 2000. African dust and the demise of Caribbean coral reefs. Geophys Res Lett. 27(19):3029–3032. [Google Scholar]
- Simon C, Daniel R.. 2011. Metagenomic analyses: past and future trends. Appl Environ Microbiol. 77(4):1153–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, et al. 2012. Free tropospheric transport of microorganisms from Asia to North America. Microb Ecol. 64(4):973–985. [DOI] [PubMed] [Google Scholar]
- Smith DJ, et al. 2013. Intercontinental dispersal of bacteria and archaea by transpacific winds. Appl Environ Microbiol. 79(4):1134–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterflinger K, Tesei D, Zakharova K.. 2012. Fungi in hot and cold deserts with particular reference to microcolonial fungi. Fungal Ecol. 5(4):453–462. [Google Scholar]
- Sultan B, Labadi K, Guegan JF, Janicot S.. 2005. Climate drives the meningitis epidemics onset in West Africa. PLoS Med. 2(1):e6.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swap R, Garstang M, Greco S, Talbot R, Kållberg P.. 1992. Saharan dust in the amazon basin. Tellus B 44(2):133–149. [Google Scholar]
- Tam WWS, Wong TW, Wong AHS, Hui DSC.. 2012. Effect of dust storm events on daily emergency admissions for respiratory diseases. Respirology 17(1):143–148. [DOI] [PubMed] [Google Scholar]
- Tang K, et al. 2018. Characterization of atmospheric bioaerosols along the transport pathway of Asian dust during the dust-bioaerosol 2016 campaign. Atmos Chem Phys. 18:7131–7148. doi: 10.5194/acp-18-7131. [Google Scholar]
- Tao W. 2014. Aeolian desertification and its control in northern China. Int Soil Water Conserv Res. 2(4):34–41. [Google Scholar]
- Tong DQ, Wang JXL, Gill TE, Lei H, Wang BY.. 2017. Intensified dust storm activity and valley fever infection in the southwestern United States. Geophys Res Lett. 44(9):4304–4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trochkine D, et al. 2003. Mineral aerosol particles collected in Dunhuang, China, and their comparison with chemically modified particles collected over Japan. J Geophys Res. 108(D23):8642. [Google Scholar]
- UNCCD. The United Nations Decade for Deserts (2010–2020) and the fight against Desertification. Available from: https://www.unccd.int/actions/united-nations-decade-deserts-2010-2020-and-fight-against-desertification, last accessed July 21, 2018.
- Uno I, et al. 2009. Asian dust transported one full circuit around the globe. Nat Geosci. 2(8):557–560. [Google Scholar]
- VanCuren RA, Cahill TA.. 2002. Asian aerosols in North America: frequency and concentration of fine dust. J Geophys Res. 107(D24):AAC-19-1–AAC-19–16. [Google Scholar]
- Varin T, Lovejoy C, Jungblut AD, Vincent WF, Corbeil J.. 2012. Metagenomic analysis of stress genes in microbial mat communities from Antarctica and the high arctic. Appl Environ Microbiol. 78(2):549–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vester JK, Glaring MA, Stougaard P.. 2015. Improved cultivation and metagenomics as new tools for bioprospecting in cold environments. Extremophiles 19(1):17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayakumar R, Al-Aboody M, Alturaiki W, Alsagaby S, Sandle T.. 2017. A study of airborne fungal allergens in sandstorm dust in Al-Zulfi, central region of Saudi Arabia. J Environ Occup Sci. 6(1):27–33. [Google Scholar]
- Vila-Costa M, et al. 2013. Bacterial and archaeal community structure in the surface microlayer of high mountain lakes examined under two atmospheric aerosol loading scenarios. FEMS Microbiol Ecol. 84(2):387–397. [DOI] [PubMed] [Google Scholar]
- Wainwright M, Wickramasinghe NC, Narlikar JV, Rajaratnam P.. 2003. Microorganisms cultured from stratospheric air samples obtained at 41 km. FEMS Microbiol Lett. 218(1):161–165. [DOI] [PubMed] [Google Scholar]
- Wang Y, et al. 2016. Effects of dust storm events on weekly clinic visits related to pulmonary tuberculosis disease in Minqin, China. Atmos Environ. 127:205–212. [Google Scholar]
- Watson I, De Sousa CNA.. 1983. Long distance transport of spores of Puccinia graminis tritici in the southern hemisphere. Proc Linn Soc NSW 106:10. [Google Scholar]
- Weil T, et al. 2017. Legal immigrants: invasion of alien microbial communities during winter occurring desert dust storms. Microbiome 5(1):32.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir J, Garrison V, Shinn E, Smith G.. 2000. The relationship between gorgonian coral (cnidaria: Gorgonacea) diseases and African dust events. In: Abstracts of the ninth international coral reef symposium, Bali, Indonesia. p. 78.
- Weir-Brush JR, Garrison VH, Smith GW, Shinn EA.. 2004. The relationship between gorgonian coral (Cnidaria: Gorgonacea) diseases and African dust storms. Aerobiologia 20(2):119–126. [Google Scholar]
- Wellings CR, McIntosh RA.. 1990. Puccinia striiformis f. Sp. Tritici in Australasia: pathogenic changes during the first 10 years. Plant Pathol. 39(2):316. [Google Scholar]
- Westrich JR, et al. 2016. Saharan dust nutrients promote vibrio bloom formation in marine surface waters. Proc Natl Acad Sci USA. 113(21):5964–5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RP, Nackoney J.. 2003. Drylands, people, and ecosystem goods and services: a web-based geospatial analysis. Technical report, Washington DC, USA.
- Whitman WB, Coleman DC, Wiebe WJ.. 1998. Prokaryotes: the unseen majority. Proc Natl Acad Sci USA. 95(12):6578–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whon TW, et al. 2012. Metagenomic characterization of airborne viral DNA diversity in the near-surface atmosphere. J Virol. 86(15):8221–8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu PC, Tsai JC, Li FC, Lung SC, Su HJ.. 2004. Increased levels of ambient fungal spores in Taiwan are associated with dust events from China. Atmos Environ. 38(29):4879–4886. [Google Scholar]
- Wylie KM, et al. 2012. Novel bacterial taxa in the human microbiome. PLoS One 7(6):e35294.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N, et al. 2016. Abundance and community structure of bacteria on Asian dust particles collected in Beijing, China, during the Asian dust season. Biol Pharm Bull. 39(1):68–77. [DOI] [PubMed] [Google Scholar]
- Yamaguchi N, et al. 2014. Changes in the airborne bacterial community in outdoor environments following Asian dust events. Microbes Environ. 29(1):82–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CY, Chen YS, Chiu HF, Goggins WB.. 2005. Effects of Asian dust storm events on daily stroke admissions in Taipei, Taiwan. Environ Res. 99(1):79–84. [DOI] [PubMed] [Google Scholar]
- Yeo HG, Kim JH.. 2002. SPM and fungal spores in the ambient air of West Korea during the Asian dust (yellow sand) period. Atmos Environ. 36(35):5437–5442. [Google Scholar]
- Yoo MS, et al. 2017. Development of electrochemical biosensor for detection of pathogenic microorganism in Asian dust events. Chemosphere 175:269–274. [DOI] [PubMed] [Google Scholar]
- Yu HB, et al. 2015. The fertilizing role of African dust in the amazon rainforest: a first multiyear assessment based on data from cloud-aerosol lidar and infrared pathfinder satellite observations. Geophys Res Lett. 42(6):1984–1991. [Google Scholar]
- Zablocki O, Adriaenssens EM, Cowan D.. 2016. Diversity and ecology of viruses in hyperarid desert soils. Appl Environ Microbiol. 82(3):770–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XY, Arimoto R, An ZS.. 1997. Dust emission from Chinese desert sources linked to variations in atmospheric circulation. J Geophys Res. 102(D23):28041–28047. [Google Scholar]
- Zhen Q, et al. 2017. Meteorological factors had more impact on airborne bacterial communities than air pollutants. Sci Total Environ. 601–602:703–712. [DOI] [PubMed] [Google Scholar]
- Zhou LM, Chen HS, Hua WJ, Dai YJ, Wei N.. 2016. Mechanisms for stronger warming over drier ecoregions observed since 1979. Clim Dynam. 47(9–10):2955–2974. [Google Scholar]