Abstract
We have developed ascatNgs to aid researchers in carrying out Allele-Specific Copy number Analysis of Tumours (ASCAT). ASCAT is capable of detecting DNA copy number changes affecting a tumor genome when comparing to a matched normal sample. Additionally, the algorithm estimates the amount of tumor DNA in the sample, known as Aberrant Cell Fraction (ACF). ASCAT itself is an R-package which requires the generation of many file types. Here, we present a suite of tools to help handle this for the user. Our code is available on our GitHub site (https://github.com/cancerit). This unit describes both ‘one-shot’ execution and approaches more suitable for large-scale compute farms.
Keywords: somatic, sequencing, cancer, copy-number
Introduction
Allele-Specific Copy number Analysis of Tumours (ASCAT) uses the sequencing read depth at Single Nucleotide Polymorphisms (SNPs) to calculate allele-specific copy number changes (Van Loo et al., 2010). The acsatNgs package provides an optimized workflow suitable for use with BAM (Li et al., 2009) or CRAM (Fritz et al., 2011) inputs containing whole-genome sequence (WGS). Additionally ascatNgs automates conversion of output to Variant Call Format, VCF (Danecek et al., 2011), which is not handled by ASCAT itself.
There are three main steps to the standard workflow: allele counting, the ASCAT core algorithm, and file conversion/clean up.
Allele counting (see Basic Protocol) is carried out using the alleleCount package (http://cancerit.github.io/alleleCount/). The code takes a list of known SNP positions and records the number of reads and genotype at each location. As this is a compute-intensive step, helper code in the ascatNgs package enables parallel processing of this step rather than processing over one million loci in one batch.
The ascatNgs pipeline uses the allele count files to generate the following:
Normalized log transform of read depth (LogR)—Tumor/Normal.
Allele frequencies (BAF)—Tumor/Normal.
This data is then processed with functions from ASCAT algorithm as follows:
GCcorrection
Plot LogR/BAF values (labeled red dot plots in Figs. 15.9.2 to 15.9.4)
Segmentation using allele-specific piecewise constant fitting (ASPCF). This is described in the ASCAT paper (Van Loo et al., 2010).
Plot the segmented LogR/BAF (red dot plots with green segments overlaid, Figs. 15.9.2 to 15.9.4).
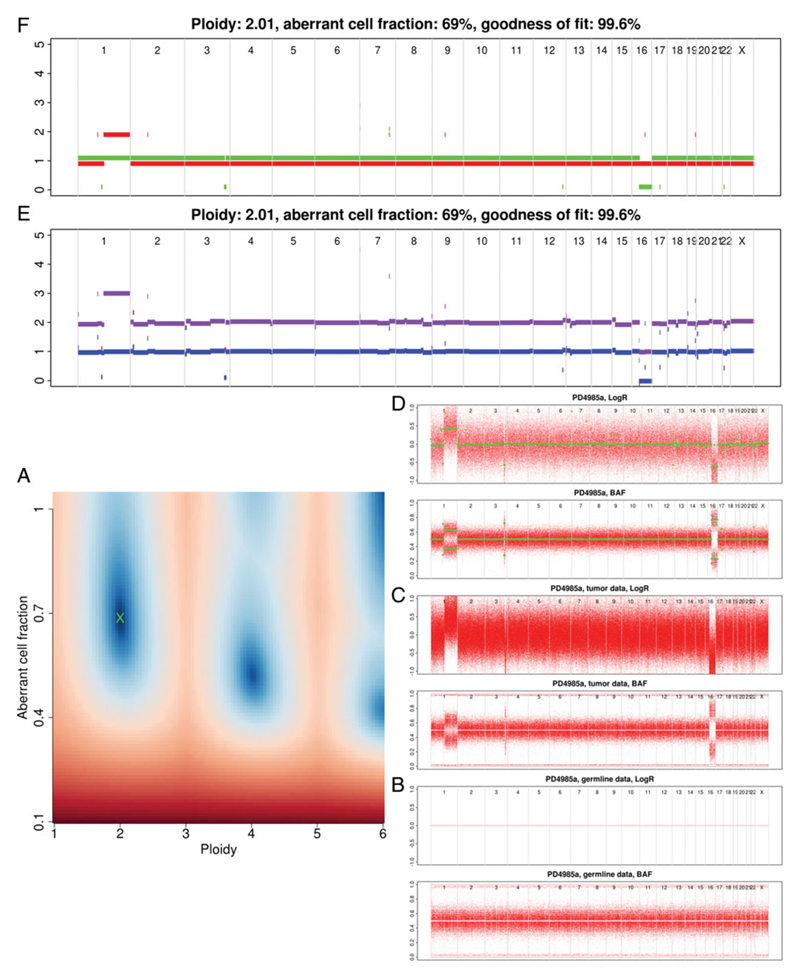
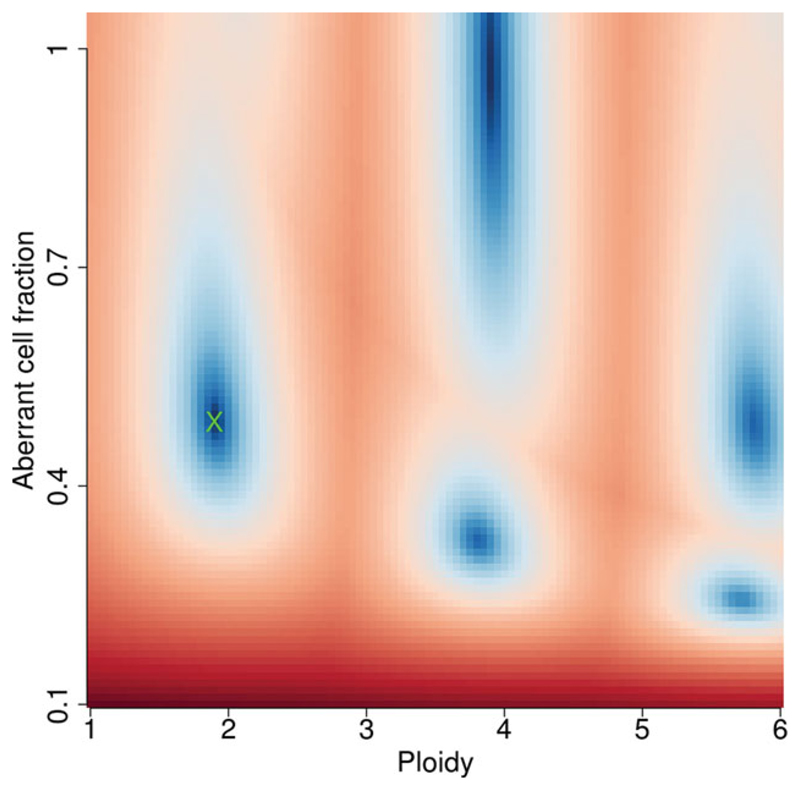
Figure 15.9.2.
A cancer sample pair with very few copy number aberrations and an overall ploidy of close to 2. Sunrise plots (A) tend to have banding of the decreasing intensity blue ‘good’ solution regions around multiples of the correct ploidy.
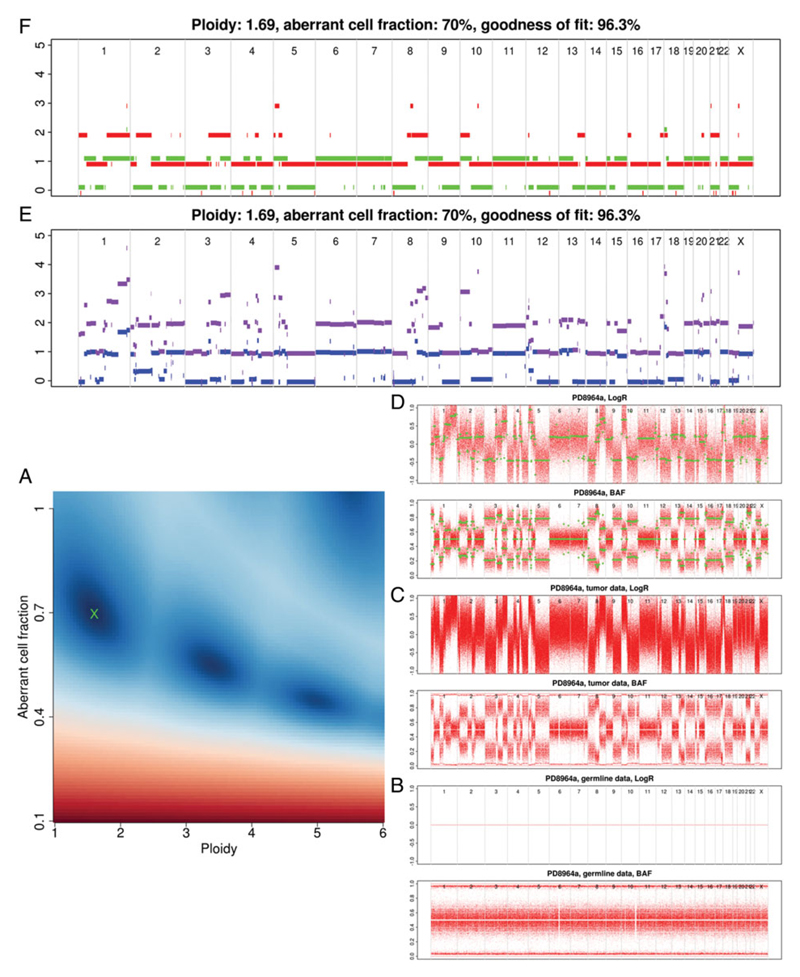
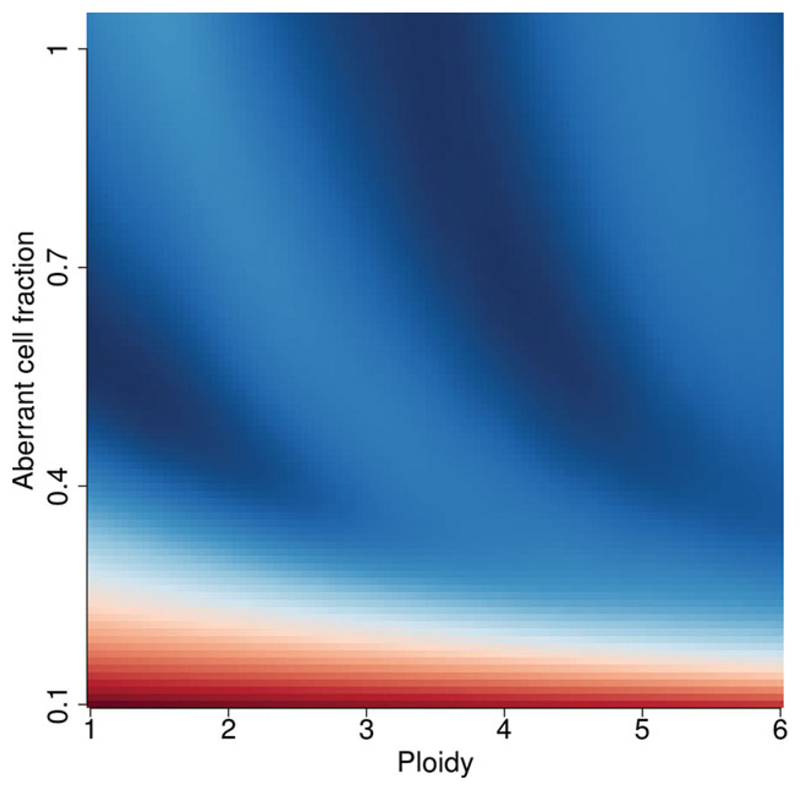
Figure 15.9.4.
A cancer sample pair with an overall copy loss. Note the banding of multiples of the ploidy in the sunrise plot (A).
Using this data, the ASCAT algorithm generates a copy number estimate (ploidy) for the whole sample, e.g., haploid, diploid, triploid, etc., and an estimate of purity (also known as Aberrant Cell Fraction, ACF). These are generated by creating a grid of possible values and evaluating the goodness-of-fit for both parameters. Occasionally, ASCAT needs assistance determining the correct aberrant cell fraction and ploidy, resulting in either a poor result or a failure to find a solution. ASCAT produces multiple plots to aid with this. Failure to obtain the correct values for these parameters can result in incorrect copy number states in the final results.
The final output file set is described in Table 15.9.1.
Table 15.9.1. Result Files.
| File extension | Type | Description |
|---|---|---|
| ASCATprofile.png | Image | Final copy number profile with integer (clonal) copy number states |
| ASPCF.png | Image | Plots of LogR and BAF values overlaid with segmented LogR and BAF |
| germline.png | Image | Plot of LogR and BAF values for normal sample |
| rawprofile.png | Image | Copy number profile without rounding to whole numbers. This, in our opinion, is the most valuable plot. |
| sunrise.png | Image | Goodness-of-fit plot of purity vs. ploidy. Blue indicates good fit, red bad. |
| tumour.png | Image | Plot of LogR and BAF values for tumor sample |
| copynumber.caveman.csv | Comma-separated values | Simple form of copy number segments in format:
|
| copynumber.caveman.vcf.gz | Bgzip | Bgzip’ed VCF file of copy number segments based on copynumber.caveman.csv |
| copynumber.caveman.vcf.gz.tbi | Tabix index | Tabix index for vcf.gz file. |
| copynumber.txt | Tab separated | Detailed output of LogR and BAF data correlated with segment information |
| samplestatistics.txt | Summary of key statistics | See Table 15.9.3 |
Interpretation of results and evaluation of ploidy and ACF values will be covered under Guidelines for Understanding Results.
The ascatNgs package performs a full normalization, segmentation, and copy number alteration analysis, used successfully within the Cancer Genome Project (CGP) and the International Cancer Genome Consortium (ICGC) PanCancer project. All components are wrapped to reduce the number of commands to one, for basic usage.
See Support Protocol 1 for installation instructions.
Once installed, running the following will list available options:
ascat.pl -hBasic Protocol: Calling Copy Number Segments with a Single Command for a Tumor/normal Sample Pair
ascatNgs is primarily used to provide copy number segments along with a prediction of tumor purity/ACF for a matched tumor/normal sample pair. This section describes how to achieve this using a single command.
Necessary Resources
Hardware
Estimates of hardware resources are based on a pair of tumor and normal WGS sequencing BAM/CRAM at 30- to 40-fold coverage for Human Genome Reference GRCh37d5
Minimum requirements:
A Linux computer with at least 5 GB of RAM
1 core
Processing storage of 2 GB
Run-time, 10 hr
Recommended:
A Linux computer with at least 20 GB of RAM
4 core
Processing storage of 2 GB
Run-time, 3.5 hr
Software
PCAP-core (v2+): https://github.com/ICGC-TCGA-PanCancer/PCAP-core/releases (specifically used to leverage the generic thread, log, and command management support common to many of the CancerIT tools)
cgpVcf (v2+): https://github.com/cancerit/cgpVcf/releases (contains VCF utilities to ensure consistent header information between all CancerIT tools)
alleleCount (v3+): https://github.com/cancerit/alleleCount/releases (provides the C allele counting program used to generate the counts used by ASCAT)
ascatNgs (v2+): https://github.com/cancerit/ascatNgs/releases (the tool being discussed here)
Each of these tools installs its own dependencies including:
biobambam2: https://github.com/gt1/biobambam2 (not used here)
bwa: https://github.com/lh3/bwa (not used here)
samtools v1.2+: https://github.com/samtools/samtools (provides the API for accessing BAM/CRAM files)
kentUtils: https://github.com/ENCODE-DCC/kentUtils (not used here)
VCFtools: http://vcftools.sourceforge.net/downloads.html (provides VCF validate tool)
Various perl libraries
Files
Static Reference files, see Support Protocol 2:
genome.fa: reference genome (with associated *.fai index). This must be the reference used during mapping of the input data
gender.loci: a small number of Y-specific loci to be used in automatic determination of gender when unknown. A default file is included in the distribution
SnpGcCorrections.tsv: GC correction windows for each SNP position You can find an example set at ftp://ftp.sanger.ac.uk/pub/cancer/support-files/CPIB/ascatNgs/Human/GRCh37/reference.tar.gz
The total size of these files will depend on the genome being analyzed. For Human GRCh37, the total space is ~3.1 GB
Sample data
BAM/CRAM files must have read-group entries including the sample name field ‘SM’:
<Tumour>.[b|cr]am: aligned whole-genome sequencing for tumor sample
<Normal>.[b|cr]am: aligned whole-genome sequencing for normal sample
BWA-mem (Li, 2013) and BWA-backtrack (Li and Durbin, 2009) have been tested; any other aligner using MAPQ and per-base-quality values appropriately should be suitable
Example data of COLO-829/COLO-829-BL (Pleasance et al., 2010) BAM files aligned with BWA-mem can be found at ftp://ftp.sanger.ac.uk/pub/cancer/support-files/CPIB/ascatNgs/ascatNgs_CPBI_exampleData.tar (be aware that ASCAT was not designed for use with cell-line data and this has been provided as a working example only due to access restrictions placed on human non-cell-line data)
An example result for the provided sample and reference is available at ftp://ftp.sanger.ac.uk/pub/cancer/support-files/CPIB/ascatNgs/ascatNgs_CPBI_exampleResult.tar.gz
See Table 15.9.1 and Guidelines for Understanding Results for a description of this data.
NOTE: Other than system commands, the user only interacts directly with ascatNgs via ascat.pl in this protocol.
NOTE: Steps 1 to 3 should be modified as appropriate for your download and output locations.
- Set an environment variable pointing to the reference files (downloaded or otherwise), e.g.:
export REF=/refarea - Set an environment variable for the base of the output area, e.g.:
export POUT=/workspace - Set an environment variable for the example data, e.g.:
export ASCEX=/exampleData - Create the output folder:
mkdir $POUT - Build the ascat.pl command (this example uses 4 cores):
ascat.pl \ -outdir $POUT/result \ -tumour $ASCEX/tumour/COLO-829.bam \ -normal $ASCEX/normal/COLO-829-BL.bam \ -reference $REF/genome.fa \ -snp_gc $REF/SnpGcCorrections.tsv \ -gender XX \ -genderChr Y \ -protocol WGS \ -platform ILLUMINA \ -species Human \ -assembly GRCh37d5 \ -cpus 4
When running your own data, please refer to the command line help ascat.pl -h and modify options appropriately. Alternate Protocol 1 gives more detail for the gender options.
Some arguments are populated from the BAM file headers where possible. If the information is not available in the header, the code will request that they be provided on the command line. The example above provides these explicitly, as the BAM files provided are what should be considered a minimum state with respect to header information. The optional items are described in Table 15.9.2.
Table 15.9.2. Parameters for Fields that are Optional in BAM Headers.
| Parameter | Detail | Values |
|---|---|---|
| -species | Species of source data. Normally in @SQ line of BAM header. | Free text, ensure strings are quoted if multiple words such as Homo sapiens |
| -assembly | The reference assembly used in mapping. Normally in @SQ line of BAM header. | E.g., GRCh37d5 |
| -platform | The sequencing platform. Normally in @RG line of BAM header | E.g., ILLUMINA, refer to the BAM/SAM specification for full value list |
All failures result in a non-zero exit code. A successfully completed run will have no $POUT/result/tmpAscat folder (unless the special option -noclean is in operation). See Troubleshooting for further details.
Interpretation of results is described in Guidelines for Understanding Results.
Alternate Protocol 1: Automatic Gender Determination
ASCAT needs to know the gender of the data being analyzed to give reliable results. The Basic Protocol specifies the gender as ‘XX’ for female (use ‘XY’ for male) but the accessory code can determine this with a high degree of accuracy by interrogating Y-specific loci in the normal BAM file.
Necessary Resources
A small set of Y-specific loci needs to be provided. These are required to be determined on a species/assembly basis. In the case of Human GRCh37, these are included in the ascatNgs distribution under:
~/perl/share/gender/GRCh37d5_Y.loci.
The selected loci should reliably have no reads mapped when data is from a female Once determined a simple tab delimited file is created:
<chr><tab><1-based-pos>
The file does not need to be sorted
Follow steps 1 to 4 of Basic Protocol 1, then modify the command in step 5 as follows:
Set -gender to L (meaning determine from loci).
Specify -locus as the path to the file described above.
Remove -genderChr as now determined from -locus file.
Alternate Protocol 2: Using ascatNgs with Compute Farm Infrastructure
Executing the complete workflow in a single command can be inefficient due to the latter step only utilizing a single CPU. For this reason, it is possible for more advanced users to break down the work into subcomponents to allow more efficient use of resources under a compute farm infrastructure.
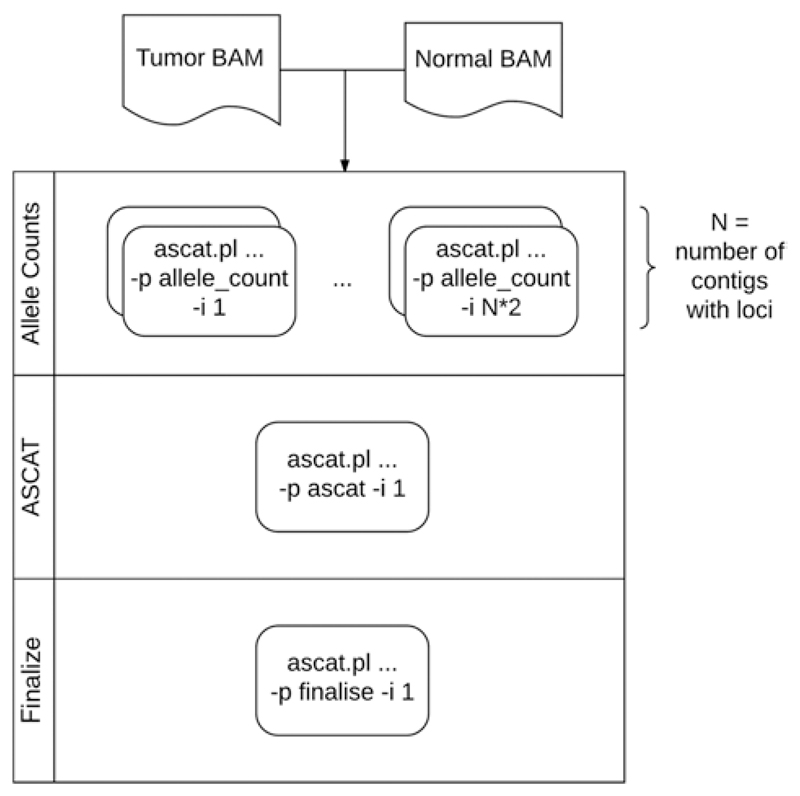
Figure 15.9.1 illustrates the different elements of the workflow.
Figure 15.9.1.
ascatNgs processing workflow. Individual components are executed automatically when run without the -p/-i options. The workflow automatically recovers to the last successful point on restart if killed for any reason. Please see Alternate Protocol 2 for further detail.
Necessary Resources
See Basic Protocol and Alternate Protocol 1; however, individual steps have different requirements that need modification on a per species/build basis
Follow Basic Protocol 1, steps 1 to 4 (or Alternate Protocol 1).
- Determine the number of chromosomes to be analyzed based on the reference files:
$ export CHRCNT='cut -f 2 $REF/SnpGcCorrections.tsv | uniq | wc -l' Remove -cpus 4 from the command in step 5 of the Basic Protocol.
-
Run the allele_count steps specifying:
-p allele_count -i Nwhere N = 1..(2*$CHRCNT)
- Once complete, execute ascat:
-p ascat -i 1 -
Finalize the dataset (moves data and builds relevant archives):
-p finalise -i 1Step 4 can be executed using a round-robin approach by setting a wrap limit. To do this, additionally specify -l and ensure that -i does not exceed this value, e.g.:-p allele_count -l 5 -i 1 -p allele_count -l 5 -i 5ascatNgs.pl will internally stack the allele_count jobs, for example, index 1 will process chr1, chr6, chr11 …
Support Protocol 1: Installation of acatNgs and Dependencies
ascatNgs has been packaged to minimize the complexity of installation. The examples below use the versions available at the time of publication. Please see the repositories for current versions.
In the following examples, please modify /your/scratcharea and ~/install-Base to appropriate locations. ~/installBase should be the location you would like to install to and should be the same for all of these steps.
Necessary Resources
Linux-based system with Web access
- Install PCAP-core (contains the thread framework for ascatNgs):
$ cd /your/scratcharea $ wget https://github.com/ICGC-TCGA-PanCancer/PCAP-core/archive/v3.0.1.tar.gz $ tar -zxf v3.0.1.tar.gz $ rm v3.0.1.tar.gz $ cd PCAP-core-3.0.1 $./setup.sh ~/installBase - Install cgpVcf (reusable VCF manipulation tools common to many CGP projects):
$ cd /your/scratcharea $ wget https://github.com/cancerit/cgpVcf/archive/v2.1.1.tar.gz $ tar -zxf v2.1.1.tar.gz $ rm v2.1.1.tar.gz $ cd cgpVcf-2.1.1 $./setup.sh ~/installBase - Install alleleCount (C allele counting of specified loci):
$ cd /your/scratcharea $ wget https://github.com/cancerit/alleleCount/archive/v3.1.1.tar.gz $ tar -zxf v3.1.1.tar.gz $ rm v3.1.1.tar.gz $ cd alleleCount-3.1.1 $./setup.sh ~/installBase Install R and the R-library RColorBrewer. Please discuss this with your local systems administrator if you are unsure how to proceed.
- Install ascatNgs (simplified use of ascat.R):
$ cd /your/scratcharea $ wget https://github.com/cancerit/ascatNgs/archive/v3.0.3.tar.gz $ tar -zxf v3.0.3.tar.gz $ rm v3.0.3.tar.gz $ cd ascatNgs-3.0.3 $./setup.sh ~/installBase
Support Protocol 2: Static Reference Files
The genome reference file is an essential requirement to run the algorithm. The following are recommended for WGS analysis.
Please note the chromosome names in files provided on the ftp site indicated in the Basic Protocol do not have a chr prefix.
genome.fa
This is the reference assembly as used for the mapping of the whole-genome sequencing data. The fasta index (fai) is also required. This can be generated by executing:
samtools faidx genome.fa
samtools is included in the install detailed in Support Protocol 1.
SnpGcCorrections.tsv
As the SNPs contained in this file may change over time, see the documentation on the ascatNgs wiki (https://github.com/cancerit/ascatNgs/wiki). This includes:
Generation from public SNP resources
Generation from BAM/CRAM normal data.
Guidelines for Understanding Results
ascatNgs generates multiple plots and data files on completion (see Table 15.9.1). Here we describe the format of the plots and files as well as providing some guidance for problematic samples.
Interpreting Plots
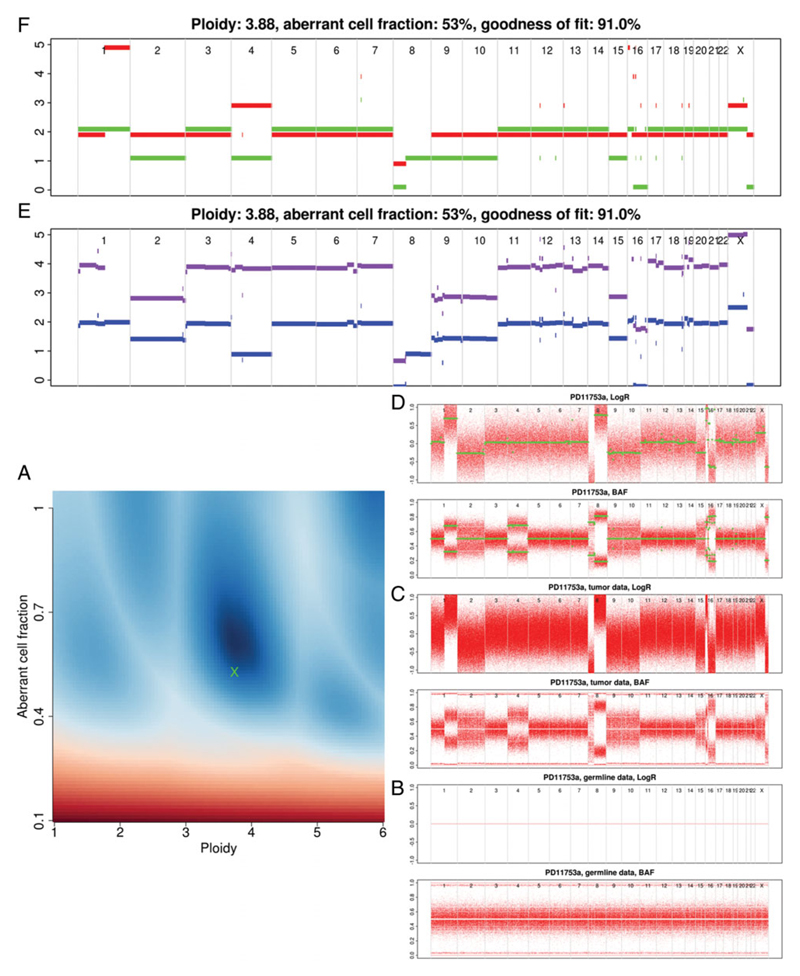
Figures 15.9.2 to 15.9.4 show several valid results for varying complexity of copy number aberrations in published cancer genomes (Nik-Zainal et al., 2016). Here, using Figure 15.9.2 as a reference, each of the plots is discussed in detail.
The sunrise plot (Fig. 15.9.2A) is discussed under ‘Checking Solution’ (see below). Each of the remaining plots present genomic position along the x axis in an ordered but un-scaled fashion. As you move up the figure the data becomes progressively more processed.
At the bottom right (Fig. 15.9.2B) is the germline LogR and BAF pair of plots (SAMPLE.germline.png). A LogR plot presents the normalized read counts. Due to the germline being used as the reference sample, we expect a line crossing the y axis at 0. The BAF plot describes the B-allele fraction for each of the SNP positions. For germline the plot should mostly consist of 3 horizontal bands:
~1 = Homozygous for B-allele
~0 = Homozygous for A-allele
0.5 ± 0.1 = heterozygous (always low density around 0.5).
If the germline BAF plot does not have this profile it is unlikely that ASCAT will give a valid solution. Reasons for this include:
Poor coverage in the normal (even coverage, >10× is recommended)
In-sufficient heterozygous SNPs in the sample (cell-lines, highly inbred strains)
Sample swaps (tumor swapped for normal, DNA or BAMs)
Normal contaminated with tumor (or other donor entirely).
Moving on to the tumor LogR plot (Fig. 15.9.2C, SAMPLE.tumour.png), we see that there is a large spread of read depth but areas of increase (1q) and decrease (16q) are clearly visible. How these regions correspond between plots will become clear as we progress. In the tumor, BAF plot regions with a shift in BAF correlate with changes in the read depth highlighted by the LogR plot.
The tumor BAF plot is more variable than that of the germline. This is due to the fraction of reads carrying a SNP being impacted by copy number aberrations (Van Loo et al., 2010).
The third pair of plots to consider are the segmented LogR and BAF (Fig. 15.9.2D, SAMPLE.ASPCF.png). In these, all points that are not heterozygous are removed before segmentation. This can result in some chromosomes having very few positions remaining, which presents as an uneven sizing of the chromosome blocks in the plot. This is often seen in highly inbred strains and cell lines. Due to the removal of homozygous positions, the regions of change are more clear even before segmentation has been applied (green points).
All of the plots described so far are part of pre-processing and are useful for diagnosing why ASCAT may fail to generate a solution (along with the sunrise plot).
The final two plots are very similar. First, the raw profile (Fig. 15.9.2E, SAMPLE.rawprofile.png) shows the total and minor copy number (purple/blue respectively). The ASCAT profile (Fig. 15.9.2F, SAMPLE.ASCATprofile.png) reports major and minor copy number (red/green respectively), after rounding to whole-number copy number states. Total copy number is the total number of copies of a genomic region in your sample. Major copy number differs in that it considers how many copies of the most prevalent allele are present in the sample. This is illustrated in Figure 15.9.2:
1q
Major/Minor = 2/1
Total/Minor = 3/1.
16q
Major/Minor = 1/0
Total/Minor = 1/0.
Data Files
The data used to generate the ASCAT profile is written to SAMPLE.caveman.copynumber.csv file with the column order of:
Segment #
Chr
Start (1-based)
End (1-based)
Germline Major
Germline Minor
Tumour Major
Tumour Minor.
The same information is also written to VCF following the specification (Danecek et al., 2011).
SAMPLE.copynumber.txt contains the following data (file header uses slightly different nomenclature):
Snp identifier
Chromosome
Position (1-based)
LogR*
Segmented LogR
BAF*
Segmented BAF*
Copy number
Minor allele
Raw copynumber.
Columns marked ‘*’ may contain ‘NA’ due to insufficient data for that calculation.
SAMPLE.samplestatistics.txt contains values shown on the plots so that they can be accessed by relevant downstream tools. These are described in Table 15.9.3.
Table 15.9.3. Sample Statistics Values, Written in this Form to Allow Automatic Parsing by Down-stream Tools.
| Label | Value | Detail |
|---|---|---|
| NormalContamination | Fraction | Estimate of normal cells contaminating sample |
| Ploidy | Decimal | Tumor ploidy (average copy number state across the genome) |
| rho | Fraction | Aberrant cell fraction |
| psi | Decimal | Internal ASCAT ploidy parameter |
| goodnessOfFit | Percentage | Confidence metric |
| GenderChr | Text | Name of the gender chromosome, which is never diploid, e.g., chrY/Y in Human, chrW/W in Chicken Note: the core ascat.R code does not support non-XX/XY genomes at present |
| GenderChrFound | Y/N | Was the GenderChr found or specified. |
Successful completion of ascatNgs and the underlying ASCAT algorithm does not guarantee an appropriate result. There are often several possible solutions, which are guided by setting appropriate purity and ploidy values (as discussed in the introduction). The following section describes how the sunrise plot is interpreted in these cases.
Checking Solution
When ASCAT completes, you should examine the ‘sunrise’ plot (SAMPLE.sunrise.png) to confirm that the appropriate ploidy and purity value has been chosen. If the solution is incorrect, the code can be re-run, manually specifying the more likely ploidy/purity values.
Panel A in Figures 15.9.2 to 15.9.4 all show common profiles for a sunrise plot. Generally, the upper section is predominately blue with a sloped delineation at the horizon (red/blue interface), with a single well-defined dark blue region. The blue indicates a good solution in this area; red indicates a bad solution using a goodness-of-fit model (Van Loo et al., 2010). Figure 15.9.2 is slightly unusual, as it is an exceptionally clean result without any bleed through between possible ploidies.
In this case, there would be no need to re-run.
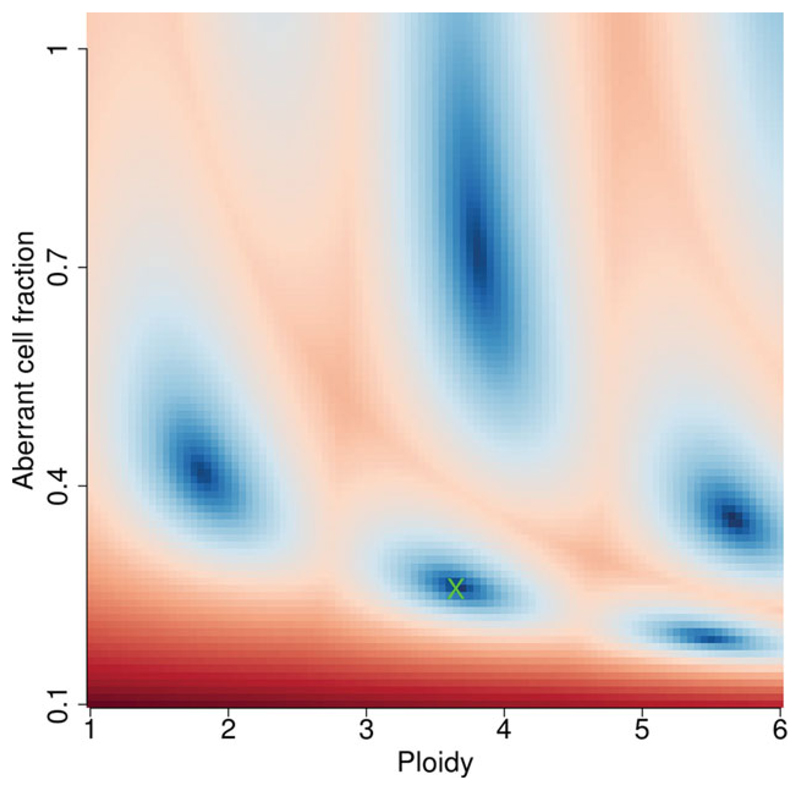
In some cases, additional runs with modified ploidy/purity values guided by the sunrise plot may be helpful. Please note that, ideally, you should have some estimation of the tumor cellularity of the original tissue samples to work from, based on histological data. For instance, Figure 15.9.5 has alterative regions that could be selected. If you have access to the histological information for the original tumor tissue sample that indicates the aberrant cell fraction or ‘purity’ is approximately 50%, then selecting the closest ‘good’ blue regions to that value is appropriate, e.g.
Figure 15.9.5.
A poorly resolved ‘sunrise’ plot with incorrect selection of ACF/Ploidy. Note multiple dark blue regions of similar color depth. In this situation, the correct solution is generally the region with the lower ploidy.
-ploidy 2
-purity 0.4
Figure 15.9.6 shows that after refitting with these values, the selected best solution is in this region and purity is lifted to ~50%.
Figure 15.9.6.
Repeat of data presented in Fig. 15.9.5 after refitting with purity = 0.4 and ploidy = 2.
In other data, particularly low-purity, low-sequencing-depth, or poor-quality samples, the algorithm cannot identify a solution (Fig. 15.9.7). Other solutions can be selected, but this should be done only if the user is able to identify a more suitable solution from the plot without ignoring anticipated values for purity from other sources.
Figure 15.9.7.
A ‘sunrise’ plot where it has not been possible to select a best solution. Note the absence of the green cross.
It should be noted that underlying data and sample-purity issues cannot be addressed by manual refitting, and caution is required in using this option. There is no way to handle poor-quality input data, and knowledge of your sequencing quality and tissue sampling data is required when determining if refitting is appropriate.
Much of this information has been distilled from Van Loo et al. (2012).
The complete set of files generated are described in Table 15.9.1.
Commentary
Background Information
Originally, the core ASCAT R script was embedded in an analysis pipeline developed by the group which was tightly linked to internal infrastructure. In early 2014, development began to make ASCAT suitable for use in the ICGC/TCGA PanCancer project, a systematic analysis of 2,500 WGS Tumor/Normal sample pairs (http://icgc.org).
ascatNgs was the result of this effort, and has been extended to allow ‘hands-off’ processing when a valid solution is not automatically produced. In these events, a default profile is generated allowing dependent analysis algorithms such as CaVEMan (see unit 15.10) to continue.
Critical Parameters
ascatNgs only works with whole-genome sequencing data, and has only been tested with data generated using the Illumina paired-end protocol.
Troubleshooting
ascat.pl gave a non-zero exit code
See the base process stdout/stderr and also the internal processing log files found here:
$POUT/result/tmpAscat/logs/
Be aware that every file contains the executed commands, so that the source of messages and errors are clear. There are *.out and *.err files for each stage. Identify the logs of interest by searching for a non-zero exit in these files:
grep -lF 'Command exited
with non-zero status' $POUT/
result/tmpAscat/logs/*
ascat.pl indicates failure during ‘finalize’ step
If BAM/CRAM files do not have complete header information, you may be required to define additional parameters during the processing of the ‘finalize’ step. The error message will indicate the relevant parameter that needs setting in these instances.
SSL connect error during install steps
This is an uncommon issue normally resolved by retry.
Figure 15.9.3.
A cancer sample pair with an overall ploidy of 4. Here we see that the sunrise plot (A) only offers one solution in the expected window of purity/ploidy.
Acknowledgement
We thank Kerstin Haase (The Francis Crick Institute, London), the current maintainer of ascat.R, the core algorithm.
This work was supported by the Wellcome Trust grant [098051].
Literature Cited
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz MH-Y, Leinonen R, Cochrane G, Birney E. Efficient storage of high throughput DNA sequencing data using reference-based compression. Genome Res. 2011;21:734–740. doi: 10.1101/gr.114819.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997. 2013 [q-bio]. Available at: http://arxiv.org/abs/1303.3997. [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with BurrowsWheeler transform. Bioinformatics (Oxford, England) 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, Martincorena I, Alexandrov LB, Martin S, Wedge DC, Van Loo P, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin M-L, Ordóñez GR, Bignell GR, Ye K, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loo P, Nilsen G, Nordgard S, Vollan H, Børresen-Dale A-L, Kristensen V, Lingjærde O. Analyzing cancer samples with SNP arrays. In: Wang J, Tan AC, Tian T, editors. Next Generation Microarray Bioinformatics Methods in Molecular Biology. Humana Press; Totowa, N.J.: 2012. pp. 57–72. [DOI] [Google Scholar]
- Van Loo P, Nordgard SH, Lingjærde OC, Russnes HG, Rye IH, Sun W, Weigman VJ, Marynen P, Zetterberg A, Naume B, Perou CM, et al. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci. 2010;107:16910–16915.. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Internet Resources
- https://github.com/canceritRepository for Wellcome Trust Sanger Institute Cancer Genome Project public projects.
- http://cancerit.github.io/ascatNgs/ascatNgs Web site, linking to repository.
- https://www.crick.ac.uk/research/a-z-researchers/researchers-v-y/peter-van-loo/software/ASCAT Web site.
- https://github.com/Crick-CancerGenomics/ascatRepository for the core ASCAT algorithm.